Abstract
Innate immune response is triggered by pathogen components, like lipopolysaccharides (LPS) of gram-negative bacteria. LPS initiates Toll-like receptor 4 (TLR4) signaling, which involves mitogen activated protein kinases (MAPK) and nuclear factor kappa B (NFκB) in different pathway branches and ultimately induces inflammatory cytokine and chemokine expression, macrophage migration and phagocytosis. Timely gene transcription and post-transcriptional control of gene expression confer the adequate synthesis of signaling molecules. As trans-acting factors RNA binding proteins (RBPs) contribute significantly to the surveillance of gene expression. RBPs are involved in the regulation of mRNA processing, localization, stability and translation. Thereby they enable rapid cellular responses to inflammatory mediators and facilitate a coordinated systemic immune response. Specific RBP binding to conserved sequence motifs in their target mRNAs is mediated by RNA binding domains, like Zink-finger domains, RNA recognition motifs (RRM), and hnRNP K homology domains (KH), often arranged in modular arrays. In this review, we focus on RBPs Tristetraprolin (TTP), human antigen R (HUR), T-cell intracellular antigen 1 related protein (TIAR), and heterogeneous ribonuclear protein K (hnRNP K) in LPS induced macrophages as primary responding immune cells. We discuss recent experiments employing RNA immunoprecipitation and microarray analysis (RIP-Chip) and newly developed individual-nucleotide resolution crosslinking and immunoprecipitation (iCLIP), photoactivatable ribonucleoside-enhanced crosslinking (PAR-iCLIP) and RNA sequencing techniques (RNA-Seq). The global mRNA interaction profile analysis of TTP, HUR, TIAR, and hnRNP K exhibited valuable information about the post-transcriptional control of inflammation related gene expression with a broad impact on intracellular signaling and temporal cytokine expression.
Keywords: RNA-binding proteins, post-transcriptional regulation, inflammation, bacterial lipopolysaccharides, macrophage activation
Introduction
The immune responses against bacteria, viruses and parasites require tight regulation, because uncontrolled, excessive or persisting immune reactions provoke inflammatory diseases (Zanotti et al., 2002). As a central component of the innate immune response, macrophages sense pathogen components such as lipopolysaccharides (LPS), an essential constituent of the outer membrane of gram-negative bacteria. Recognition of LPS by TLR4 on the macrophage surface results in the activation of MAPK and NFκB dependent signaling pathways, which activate inflammation related genes encoding pro- and anti-inflammatory cytokines and chemokines (Medzhitov and Horng, 2009; Takeuchi and Akira, 2010; Smale, 2012; Vaure and Liu, 2014). The underlying genome-wide changes in macrophage gene expression (Reynier et al., 2012; Rutledge et al., 2012) require downstream post-transcriptional checkpoints, which are critical for the appropriate modulation of immune reactions (Carpenter et al., 2014; Kafasla et al., 2014). Emerging experimental evidence highlights the impact of RNA binding proteins (RBPs) on the post-transcriptional control of the immune response (Fu and Blackshear, 2017; Garcia-Maurino et al., 2017; Diaz-Munoz and Turner, 2018; Mino and Takeuchi, 2018; Turner and Diaz-Munoz, 2018).
By analyzing RNA-protein interaction profiling and RNA sequencing experiments with TTP, HUR, TIAR, and hnRNP K we provide an overview on their target mRNAs, which are regulated at the level of mRNA stability and translation in LPS activated macrophages.
Zink-Finger Protein TTP Controls Target mRNA Decay in Inflammation
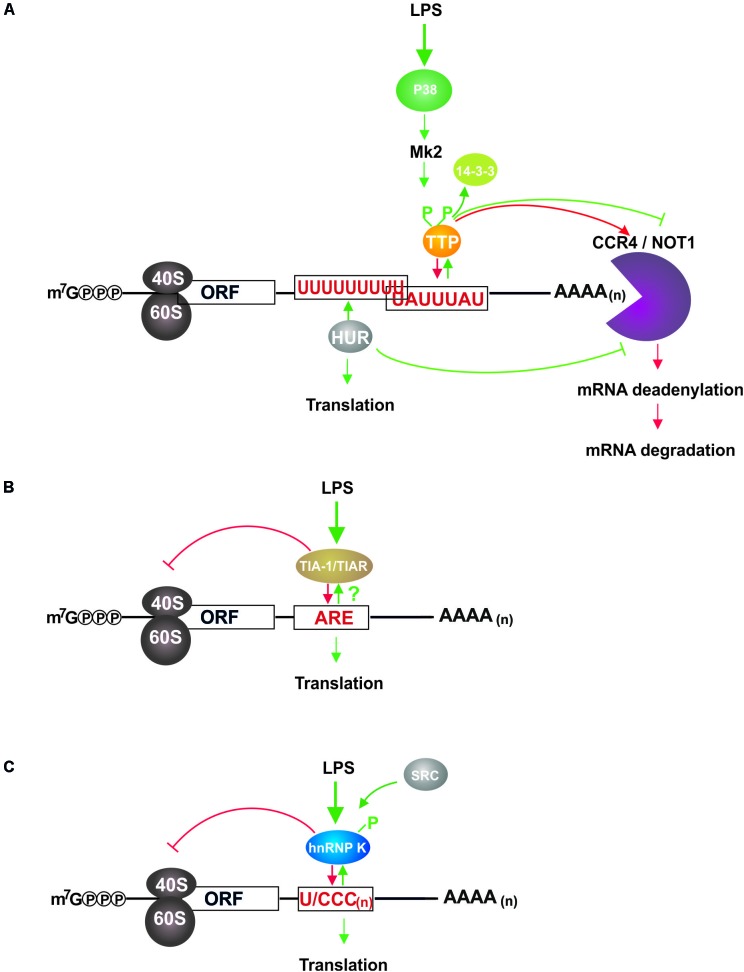
Tristetraprolin, encoded by the gene Zfp36, has been characterized as critical mRNA destabilizing protein in immune cells (Blackshear, 2002; Brooks and Blackshear, 2013). To initiate target mRNA decay, TTP mediates the recruitment of deadenylation and decapping complexes to the mRNA 3′ untranslated region (3′UTR) and 5′UTR, respectively (Fenger-Gron et al., 2005; Fabian et al., 2013). Tandem CCCH-type zinc-finger domains of TTP interact with AU-rich elements (ARE) that are mainly located in mRNA 3′UTRs (Lai et al., 1999; Worthington et al., 2002). In macrophages, target mRNAs primarily encode proteins related to inflammation response, among them cytokines and chemokines (Carballo et al., 1998; Lai et al., 1999; Stoecklin et al., 2008; Kratochvill et al., 2011; Sedlyarov et al., 2016; Tiedje et al., 2016). Under steady state conditions TTP is ubiquitously expressed at a basal level. Inflammatory stimuli like LPS and cytokines mediate transcriptional and post-transcriptional induction of TTP expression (Mahtani et al., 2001; Schaljo et al., 2009; Sedlyarov et al., 2016). AREs in TTP mRNA 3′UTR represent bona fide functional TTP binding sites. An auto-inhibitory feedback regulation established through the interaction of TTP with these AREs secures a decrease in TTP expression when inflammatory stimuli decline. Thereby TTP contributes to regulatory circuits, which prevent the development of chronic inflammation (Tchen et al., 2004; Schott et al., 2014). TTP deficiency in mice causes a systemic inflammatory syndrome, which is in part attributable to the absence of TTP-controlled tumor necrosis factor (TNFα) mRNA destabilization (Taylor et al., 1996; Carballo et al., 1997). In macrophages, LPS triggered TLR4 signaling leads to the stabilization of TTP target mRNAs and their enhanced translation (Tiedje et al., 2012, 2016). TTP serine phosphorylation catalyzed by TLR4 downstream kinase MK2 induces its sequestration by 14-3-3 proteins and target mRNA release (Chrestensen et al., 2004; Stoecklin et al., 2004). Hence CCR4-Not1 deadenylation complex recruitment is abrogated (Marchese et al., 2010; Clement et al., 2011; Sandler et al., 2011), target mRNAs are stabilized (Brook et al., 2006; Hitti et al., 2006) and translation-promoting factors replace TTP (Tiedje et al., 2012; Figure 1A).
FIGURE 1.
LPS induced impact of TTP, HUR, TIAR, and hnRNP K on mRNAs in macrophages. (A) TTP and HUR interact with U-rich elements in the 3′UTR of target mRNAs and regulate their stability and translation (Stoecklin et al., 2008; Tiedje et al., 2012, 2016; Sedlyarov et al., 2016). In non-induced cells bound TTP recruits the CCR4/NOT1 complex, which initiates 3′UTR deadenylation and 3′ to 5′ mRNA decay. LPS induced TLR4 signaling activates MK2, which phosphorylates TTP leading to its sequestration by 14-3-3 proteins and subsequent abolition of deadenylation complex association. Following T118 phosphorylation by MK2 HUR accumulates in the cytoplasm where it mediates mRNA stabilization, disables decay complex formation and enhances translation. By controlling mRNA decay, TTP curtails the synthesis of inflammation related proteins in non-induced macrophages, and regulates their balanced expression in response to LPS conjointly with HUR. (B) TIAR binding to AREs causes translational repression of target mRNAs, which is diminished by a so far unknown mechanism in LPS activated macrophages, resulting in elevated synthesis of cytokines (Piecyk et al., 2000; Kharraz et al., 2016). TIAR dampens the expression of immune response associated proteins, which can be activated immediately in response to LPS. (C) HnRNP K bound to U/CCC(n) elements in the mRNA 3′UTR inhibits target mRNA translation, but is released from the binding site following c-Src catalyzed tyrosine phosphorylation that is initiated in response to LPS dependent macrophage activation (Liepelt et al., 2014). Thereby a rapid LPS response facilitated by straight signaling molecule synthesis can be established.
Four studies examined the impact of TTP on inflammation-related pathways: (I) (Stoecklin et al., 2008) identified TTP bound mRNAs in untreated and LPS induced murine RAW264.7 macrophages by TTP co-immunoprecipitation and RIP-Chip analysis (Table 1). (II) To investigate TTP-driven mRNA decay (Kratochvill et al., 2011) treated bone marrow derived macrophages (BMDM) with actinomycin D for different times prior to RIP-chip (Table 1). (III) Employing iCLIP (Tiedje et al., 2016), identified TTP bound mRNAs in LPS treated BMDM from mice expressing GFP-TTP or the non-MK2 substrate variant (Table 1). The impact of TTP phosphorylation on global mRNA stability and mRNA translation was examined integrating iCLIP, RNASeq and Ribosome profiling (RiboSeq) (Tiedje et al., 2016). (IV) To map mRNA binding sites of endogenous TTP precisely and to unveil its role in inflammation resolution (Sedlyarov et al., 2016) applied PAR-iCLIP and RNASeq in BMDM of TTP(wt) and TTP(-/-) mice exposed to LPS for different times (Table 1).
Table 1.
Experimental strategies, tools and conditions employed in the studies discussed.
| RBP | Study | Mouse genotype | Cell type | LPS | Analytical strategy | Antibody | Data deposition |
|---|---|---|---|---|---|---|---|
| TTP | Stoecklin et al., 2008 | RAW264.7 mouse macrophages | 2 h,10 ng/ml LPS, E. coli 026:B6 (Sigma) | RIP-Chip Affymetrix Mouse Genome 430 2.0 array covering 39.000 transcripts | Rabbit anti-TTP | doi: 10.1074/jbc.M709657200 | |
| Kratochvill et al., 2011 | C57BL/6 mice TTP(wt) and TTP(-/-) | BMDM | 3 h, 10 ng/ml LPS, E. coli 055:B5 (Sigma) | RIP-Chip Affymetrix Mouse Gene ST 1.0 array covering 28 853 genes | Rabbit anti-TTP (Schaljo et al., 2009) | http://www.ncbi.nlm.nih.gov/geo/acc. GSE28880. | |
| Tiedje et al., 2016 | C57BL/6 TTP(-/-) mice expr. GFP-TTP(wt) and GFP-TTP(S52,178A) | BMDM | 1 h, 1 μg/μl LPS | iCLIP | GFP-nanobodies | http://www.ncbi.nlm.nih.gov/geo/acc. GSE81250 TTP-atlas: http://ttp-atlas.univie.ac.at | |
| Sedlyarov et al., 2016 | C57BL/6 mice TTP(wt) and TTP(ΔM) | BMDM | 3 h, 6 h, 10 ng/ml LPS (Sigma) | PAR-iCLIP | Rabbit anti-TTP (Kratochvill et al., 2011) | http://www.ncbi.nlm.nih.gov/geo/acc. GSE63468 TTP-atlas: http://ttp-atlas.univie.ac.at | |
| HUR | Sedlyarov et al., 2016 | C57BL/6 mice TTP(wt) and TTP(ΔM) | BMDM | 3 h, 6 h, 10 ng/ml LPS (Sigma) | PAR-iCLIP | HUR, clone 3A2 (Thermo Fisher Sci.) | http://www.ncbi.nlm.nih.gov/geo/acc. GSE63468 TTP-atlas: http://ttp-atlas.univie.ac.at |
| TIAR | Kharraz et al., 2016 | RAW264.7, expr. TIAR- or TIAR-ΔRRM-FLAG | 2 h, 100 ng/ml LPS, E. coli 0127:B8 (Sigma) | RIP-Chip Mouse Exonic Evidence Based Oligonucleotide (MEEBO) array covering 38.784 70mer probes (Stanford University, United States) | Anti-FLAG M2 agarose (Sigma) | http://www.ncbi.nlm.nih.gov/geo/acc.GSE77577 | |
| hnRNP K | Liepelt et al., 2014 | RAW264.7 mouse macrophages | 6 h, 10 ng/ml LPS, E. coli 0111:B4 (Sigma) | RIP-Chip Affymetrix Mouse Genome 430 2.0 array covering 39.000 transcripts | monoclonal hnRNP K (Naarmann et al., 2008) | http://www.ncbi.nlm.nih.gov/geo/acc.GSE48463 | |
Information related to experimental tools and conditions as provided in the original papers.
Detailed inspection of sequence motifs in TTP bound mRNAs, which were detected in the different studies (Table 1) revealed AU-rich TTP binding sites represented by AUUUA pentamers and UUAUUUAUU nonamers (Stoecklin et al., 2008). In target mRNA 3′UTRs UAUUUAU heptamers (Sedlyarov et al., 2016) are highly enriched as well. MRNAs encoding checkpoint regulators of LPS induced inflammation response, e.g., TNFα, IL-10, IL-15, CXCL2, and CCL2 were identified with all applied experimental and data validation strategies. Interestingly Kratochvill et al. (2011), reported that 25% of LPS induced transcripts were unstable. Among those displaying a TTP dependent decay were TNFα, IL-6, IL-10, TTP, CXCL1, CXCL2, CSF2, IER3 encoding transcripts (Kratochvill et al., 2011), which were also identified in the study of Tiedje et al. (2016). These data support the hypothesis that TTP functions in the elimination of inflammation related mRNAs, the maintenance of a balanced LPS response and the resolution of inflammation. Related pathways corresponding to enriched mRNAs include TNFα-, NFκB-, Wnt- and chemokine signaling, the formation of focal adhesions, apoptosis and mRNA processing. They were as well covered by mRNAs detected in RiboSeq experiments (Tiedje et al., 2016). The top 25 mRNAs, which were differentially bound by GFP-TTP and GFP-TTP(S52,178A) upon LPS stimulation include not only TNFα and NFκB-related signaling molecules like TNF, CXCL2, CXCL3, but also IER3 and DUSP1, which encode feedback inhibitors of the inflammatory response (Tiedje et al., 2016). These findings emphasize the importance of the MK2 dependent TTP release from target mRNAs to safeguard accurate feedback regulation of the inflammatory response. Remarkably, in the study of Sedlyarov et al. (2016) 343 TTP target mRNAs were identified through intron sequences bound by TTP. Of this group only 1% exhibited TTP dependent destabilization, suggesting that TTP binding to intron sequences does not affect intron processing. To characterize TTP impact on early and late LPS response (Sedlyarov et al., 2016) applied LPS treatment for 3 and 6 h. In the early phase only a few drivers of inflammation, such as TNFα, which activates central transcription inducers, e.g., NFκB are strongly controlled by TTP. In early and late response, the GO term coverage of target mRNA mostly overlapped. However, GO terms taxis and chemotaxis, which characterize the perpetuation of inflammation were only represented at the late response time point (Sedlyarov et al., 2016). From their analysis the authors conclude that TTP supports a switch to inflammation resolution by destabilizing mRNAs that encode migration-associated proteins, thereby impeding chronic inflammation.
HUR a Versatile RRM Domain Protein Modulates mRNA Stability
The ubiquitously expressed protein HUR, which is encoded by Elavl1, consists of two consecutive N-terminal RNA recognition motifs (RRM), a central less conserved basic hinge region and a third C-terminal RRM (Ma et al., 1996). Whereas RRM1 and RRM2 function in RNA binding, RRM3 contributes to RNA-protein complex stabilization and protein-protein interactions, including HUR dimerization (Pabis et al., 2018). The basic hinge region includes a shuttling domain (Fan and Steitz, 1998), which in response to stress and mitogen signaling, facilitates nuclear-cytoplasmic shuttling of the predominantly nuclear HUR (Keene, 1999). Cytoplasmic HUR accumulation, which is induced by p38 and MK2 dependent T118 phosphorylation in response to γ-irradiation and oxidative stress, augments its binding to p21, urokinase and urokinase receptor mRNAs and their stabilization (Tran et al., 2003; Lafarga et al., 2009). HUR binds AREs, mostly located in the target mRNA 3′UTR (Fan and Steitz, 1998; Lopez de Silanes et al., 2004), but also in intron sequences. HUR binding can contribute to alternative pre-mRNA splicing for specific genes like ZNF207, GANAB, DST and PTBP2 (Lebedeva et al., 2011) and the differential stabilization of 3′UTR ARE containing c-Fos and c-Jun mRNAs (Peng et al., 1998). Systematic mapping and functional evaluation of HUR-RNA interactions by PAR-Clip and RIP-Chip experiments employing HEK293 cells confirmed that HUR mediates the modulation of nuclear pre-mRNA processing and stabilizes cytoplasmic mRNAs, which bear both intronic and 3′UTR binding sites (Mukherjee et al., 2011). Interestingly, as shown for miR-7 that is encoded in the last HNRNPK exon, HUR binding to specific intronic miRNA precursors is implicated in their processing. HUR depletion from HeLa cells results in upregulation of miR-7, whereas hnRNP K expression remains unaffected, suggesting that HUR controls miR-7 precursor processing (Lebedeva et al., 2011). Implementing a refined digestion optimized RIP-seq protocol (DO-RIP-seq) (Nicholson et al., 2017a,b) were able to quantify HUR binding sites transcriptome-wide. Since HUR target mRNAs encode proteins implicated in cell cycle control, cell death and differentiation, post-translational HUR modifications and dysregulated functions are associated with a broad range of pathologic conditions (Srikantan and Gorospe, 2012; Grammatikakis et al., 2017). Disease-linked HUR phosphorylation, methylation and proteolytic cleavage not only regulate the subcellular localization of HUR, but affect as well its RNA-binding (reviewed in Grammatikakis et al., 2017). Notably, in HeLa cells, fibroblasts and carcinoma tissues HUR controls the stability and interactions of lncRNAs, such as HOTAIR (Yoon et al., 2013), LincRNA-p21 (Yoon et al., 2012), and 7SL (Abdelmohsen et al., 2014) and adjusts their function in gene expression control. Furthermore, HUR and specific miRNAs cooperate or compete in mRNA regulation (Srikantan et al., 2012). Modulation of miRNA binding by HUR has been reported in human MCF-7 epithelial and Huh7 liver cells (Poria et al., 2016), as well as in murine BMDM (Lu et al., 2014). Remarkably, in BMDM LPS induced MK2 catalyzed TTP phosphorylation causes a shift of the competitive binding equilibrium between HUR and TTP toward HUR, which stabilizes TNFα mRNA and stimulates its translation (Tiedje et al., 2012; Figure 1A). This finding corroborates a functional relevance of a regulated crosstalk between HUR and TTP in the LPS induced macrophage immune response. In their comprehensive PAR-iCLIP and RNASeq analysis of the BMDM response to LPS (Sedlyarov et al., 2016) mapped HUR and TTP mRNA binding sites comparatively. The study revealed that a UUUUUUUUU nonamer is the most overrepresented HUR binding motif. With 78% the majority of HUR binding sites was located in 3′UTRs, which exceeds two times the number of TTP 3′UTR sites, whereas in intron sequences only 17% of the HUR sites were identified. Binding sites for both, TTP and HUR were determined in 59 target mRNAs. 552 and 120 binding sites for HUR and TTP, respectively, were not overlapping and 118 sites did overlap by at least 1 nt (Sedlyarov et al., 2016). This overlap applied to 40 targets, including TNFα and CXCL2 mRNA, for which simultaneous TTP and HUR binding were confirmed experimentally. Stability and expression of mRNAs bearing solely TTP binding sites did not significantly differ from mRNAs with overlapping motifs, suggesting no co-regulation of mRNA stability (Sedlyarov et al., 2016) in macrophage inflammatory response, but possibly at the level of mRNA translation as shown for TNFα mRNA (Tiedje et al., 2012).
TIAR, a RRM Domain Protein Contributes to mRNA Translation Control
The two closely related DNA/RNA-binding proteins, T-cell intracellular antigen 1 (TIA-1) (Anderson et al., 1990) and TIA-1 related protein (TIAR), contain three N-terminal RRMs, which mediate oligonucleotide binding and a C-terminal Q-rich prion-related domain that enables participation in stress granule formation (Waris et al., 2014). TIAR RRM1 preferentially interacts with T-rich ssDNA and functions in transcription activation (Suswam et al., 2005). RRM2 displays affinity for U- and RRM3 for C-rich motifs (Dember et al., 1996; Cruz-Gallardo et al., 2014), whereas the RRM23-tandem domain binds mainly UC-rich sequences (Waris et al., 2017).
RRM2 and RRM3 contribute to nuclear accumulation of TIA proteins and nuclear export, respectively (Zhang et al., 2005). Both interact with U-rich stretches near mRNA 5′-splice sites (Del Gatto-Konczak et al., 2000) and modulate alternative splicing of mRNAs encoding FAS in murine fibroblasts (Forch et al., 2000), NF1 in rat neuronal cells (Zhu et al., 2008), human chondrocyte COL2A1 (McAlinden et al., 2007), liver CFTR (Zuccato et al., 2004), and CGRP in HeLa cells (Zhu et al., 2003). Furthermore, TIA proteins control TIAR and TIA-1 isoform expression tissue- and cell type specific (Izquierdo and Valcarcel, 2007).
In the cytoplasm, TIA proteins interact with 3′UTR AREs of mRNAs encoding inflammation related proteins. TIAR was shown to bind to the TNFα mRNA 3′UTR in RAW264.7 cells (Lewis et al., 1998; Gueydan et al., 1999) and peritoneal macrophages (Piecyk et al., 2000) in an LPS dependent manner. Enhanced TNFα synthesis in macrophages of TIAR(-/-) mice (Piecyk et al., 2000) suggests that in unstimulated cells TIAR impedes TNFα mRNA translation, which can be activated to drive inflammatory cytokine expression upon TLR4 mediated recognition of bacterial LPS (Figure 1B). TIA protein mediated control of TNFα expression is demonstrated by impaired TNFα mRNA regulation in TNFα(ΔARE) mice, where it is implicated in chronic inflammation (Kontoyiannis et al., 1999). Furthermore, mRNAs encoding inflammation related COX-2 (Cok et al., 2003; Dixon et al., 2003) and HMMP-13 mRNA (Yu et al., 2003) are TIAR targets in primary murine fibroblasts and human mesangial cells, respectively.
Interestingly, TIA proteins have also been shown to contribute to global translation regulation under amino acid starvation in HEK293S cells. TIA proteins bind to the 5′-oligopyrimidine tract of 5′-TOP mRNAs, which encode critical components of the translational apparatus, like ribosomal proteins and PABP-C1 and induce the release of these target mRNAs from actively translating polysomes (Damgaard and Lykke-Andersen, 2011). Besides that, TIA proteins are involved in the formation of stress granules, which sequester mRNAs that are translationally stalled by specific mRNPs under starvation-induced stress (Damgaard and Lykke-Andersen, 2011), heat shock and arsenide stress in fibroblasts (Kedersha et al., 1999), in LPS activated B-cells (Diaz-Munoz et al., 2017) and other adverse conditions including hypoxia and viral infection (Anderson and Kedersha, 2002; Waris et al., 2014).
RIP-Chip experiments were performed by Kharraz et al. (2016) to identify mRNAs specifically bound by TIAR in unstimulated and LPS induced murine RAW264.7 macrophages stably expressing TIAR(wt)-FLAG and TIAR(ΔRRM2)-FLAG. RRM2 of TIAR, which is required for high affinity mRNA binding (Dember et al., 1996; Kim et al., 2013) was deleted to discard all mRNAs that bind with low affinity. The analysis revealed that 351 mRNAs were bound by TIAR in unstimulated macrophages and 779 in LPS induced cells, with 8 transcripts exclusively bound in unstimulated and 436 in LPS induced cells, respectively. Binding of TNFα mRNA could be validated, also the binding of the mRNA that encodes MAPK phosphatase 1 (MKP-1 also termed CL100, VHV1, 3CH134, and DUSP1), for which an interaction with TIAR has been shown before in HeLa cells (Kuwano et al., 2008). The mRNAs encoding TLR4 and the serine/threonine phosphatase 2A catalytic subunit 2β could be identified as new targets (Kharraz et al., 2016). GO term analysis of TIAR target mRNAs shows that under both condition the GO terms catabolic process, cell cycle and regulation of apoptosis were enriched, which cover proteins involved in inflammatory response, cell proliferation, cell death and metabolism. However, exclusively in LPS induced cells mRNAs bound by TIAR encoded proteins within the GO term category positive regulation of IkB kinase/NFκB cascade (Kharraz et al., 2016).
T-cell intracellular antigen 1 related protein ARE binding specificity was lower than that of TTP (Stoecklin et al., 2008), but not affected by LPS treatment (Kharraz et al., 2016). The high number of mRNAs bound in response to LPS suggests that LPS directly modulates TIAR mRNA binding and that TIAR interacts, mediated by RRM1 and RRM3, with additional ARE-independent sequence motifs (Kharraz et al., 2016). TIAR mediated regulation of alternative mRNA splicing and inhibition of mRNA translation, which were shown for inflammation related proteins (Gueydan et al., 1999; Piecyk et al., 2000; Cok et al., 2003; Suswam et al., 2005), indicate that TIAR modulates the inflammatory response and contributes to its rapid decline when the stimulus disappears.
KH Domain Protein hnRNP K Modulates mRNA Translation
Heterogeneous ribonuclear protein K was first described as a structural component of nuclear ribonucleoprotein complexes associated with heterogeneous nuclear RNA (Pinol-Roma et al., 1988; Matunis et al., 1992). The protein contains three KH domains consisting of 65–70 amino acids (Gibson et al., 1993; Siomi et al., 1993; Dejgaard and Leffers, 1996), which occur with two distinct folding topologies (Baber et al., 1999; Grishin, 2001). In SELEX experiments UC3-4 RNA motifs were determined as optimal ligands for KH3 (Thisted et al., 2001). Binding of hnRNP K KH domains to ssDNA (Braddock et al., 2002; Backe et al., 2005) and RNA (Messias et al., 2006; Moritz et al., 2014) has been analyzed systematically. Quantitative evaluation indicated that the KH domains of hnRNP K contribute differentially to RNA binding, with KH1-KH2 acting as a tandem domain and KH3 as an individual binding domain (Moritz et al., 2014). The affinity of full-length hnRNP K is in the nanomolar range, while KD values for the isolated domain KH3 are micromolar (Backe et al., 2005; Moritz et al., 2014). Two U/CCC motifs within 19 nts confer hnRNP K binding, whereas four U/CCC motifs within 38 nts are necessary and sufficient for translational regulation (Ostareck et al., 1997). Bidirectional nuclear-cytoplasmic transport of hnRNP K is mediated by an N-terminal nuclear localization motif and a hnRNP K-specific shuttling domain (Michael et al., 1995, 1997).
As multifunctional protein hnRNP K is associated with transcription activation (Moumen et al., 2005), pre-mRNA splicing (Expert-Bezancon et al., 2002), mRNA stability control (Skalweit et al., 2003) and regulation of mRNA translation (Ostareck et al., 1997, 2001; Collier et al., 1998; Naarmann et al., 2008, 2010; Naarmann-de Vries et al., 2016). HnRNP K functions are modulated by mRNA specific associated mRNP components (Ostareck-Lederer and Ostareck, 2012) and by post-translational modifications. ERK dependent phosphorylation of S284,353 drives its cytoplasmic accumulation as prerequisite for hnRNP K-mediated mRNA translation regulation (Habelhah et al., 2001). Phosphorylation of KH3 Y458 by c-Src (Ostareck-Lederer et al., 2002; Messias et al., 2006; Adolph et al., 2007) and caspase-3 catalyzed cleavage (Naarmann-de Vries et al., 2013) control hnRNP K release from translational regulated target mRNAs and site-specific arginine methylation by PRMT1 regulates hnRNP K protein-protein interactions (Ostareck-Lederer et al., 2006).
In human Thp-1 monocytes, hnRNP K was shown to be associated with the COX-2 promoter and to control cytoplasmic COX-2 mRNA stability by modulating miRNA binding to the 3′UTR (Shanmugam et al., 2008).
RIP-Chip analysis of differential mRNA binding in untreated and LPS induced RAW264.7 macrophages demonstrated that 1901 mRNAs were differential bound by hnRNP K in response to LPS. GO term annotation allocated them to biological processes related to metabolism, cell communication, transport, cell cycle, development and immune response (Liepelt et al., 2014). Strikingly, whereas cytokines and chemokines were underrepresented among the 163 mRNAs related to immune response, 21 mRNAs encoded kinases and modulators in TLR4 signaling, of which 7 equally expressed mRNAs encoding IRAK4, IRAK1BP1, ERC1, CARM1/PRMT4, PI3KCA, AKT3, and TAK1 were specifically enriched in non-induced cells (Liepelt et al., 2014). A detailed analysis of differential hnRNP K association with the mRNA of transforming growth factor-ß-activated kinase 1 (TAK1), a central kinase in TLR4 signaling, revealed that KH domain 3 interacts with U/CCC elements in the TAK1 mRNA 3′UTR. Silencing of hnRNP K expression in macrophages and BMDM had no impact on the level of TAK1 mRNA, but endogenous TAK1 mRNA accumulated in actively translating polysomal fractions, resulting in an increased TAK1 expression. Through the regulation of TAK1 mRNA translation hnRNP K affects the phosphorylation of the TAK1 downstream target p38 and finally inflammatory cytokine gene transcription, i.e., TNFα, IL-1ß, and IL-10 (Liepelt et al., 2014), thereby hnRNP K modulates the LPS response of macrophages (Figure 1C).
Conclusion and Perspectives
As primary responding cells of the innate immune system, macrophages recognize pathogens and become activated to initiate and coordinate the organism-wide systemic immune response by cytokine and chemokine secretion, migration and phagocytosis. These processes require highly coordinated gene expression, which is achieved at the post-transcriptional level by regulated functional RBP-RNA interactions. Specific RBPs, TTP, HUR, TIAR, and hnRNP K, regulate the fate of their cellular target RNAs from synthesis to turnover and translation, thereby contributing to the coordination of the rapid and purposeful immune cell responses. LPS molecules of gram-negative bacteria are abundant and specific ligands that activate macrophages through TLR4 receptor signaling. Systematic analyses of RBP-RNA interaction in untreated and LPS-induced macrophages employing RIP-Chip, iCLIP, PAR-iCLIP, RNAseq, and RiboSeq studies revealed a first insight in the complex protein-mRNA networks that are established by RBPs, which bind mRNAs with different specificities for AREs and U-rich elements, like TTP, HUR, and TIAR and pyrimidine-rich sequence motifs, like hnRNP K based on their individual domain composition. These specific interactions ensure the simultaneous modulation of various target mRNAs, which encode proteins that function in defined biological processes related to the induction and resolution of immune response. Whereas TIAR and hnRNP K mainly modulate mRNA translation, which enables the direct regulation of signaling protein synthesis to initiate immune response, TTP and HUR are primarily involved in the control of mRNA decay and stability that is required to balance and resolve immune reactions.
These processes have been studied so far only for a limited number of mRNAs differentially bound by RBPs in response to LPS. It will be essential and informative to investigate the regulation of further target mRNAs of TTP, HUR, TIAR, and hnRNP K discovered in these studies to get more insight in regulatory feedback mechanisms that coordinate the balanced immune response and its dysregulation in chronic inflammation.
To this end it is interesting to note that a number of unconventional RBPs, have been identified recently, for which RNA related functions that will expand our understanding of post-transcriptional gene regulation still need to be elucidated (Castello et al., 2015; Albihlal and Gerber, 2018; Hentze et al., 2018).
In macrophages, 19 new putative RBPs, which lack well characterized RNA binding domains were identified by RNA interactome capture (Liepelt et al., 2016). Panther protein class annotation revealed that they are involved in signaling, enzymatic functions and cytoskeletal remodeling. It will be interesting to identify their target mRNAs and to study their potential functions in LPS induced macrophage response.
In addition, post-transcriptional RNA modifications (Nachtergaele and He, 2018) might add a further layer of regulation in LPS-induced macrophages, affecting RBP binding and thereby the fate of their target mRNAs.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We apologize to colleagues whose work was not cited due to length constrictions.
Footnotes
Funding. Financial support was provided by a grant from the Deutsche Forschungsgemeinschaft (DFG) SPP 1935, OS 290/6-1.
References
- Abdelmohsen K., Panda A. C., Kang M. J., Guo R., Kim J., Grammatikakis I., et al. (2014). 7SL RNA represses p53 translation by competing with HuR. Nucleic Acids Res. 42 10099–10111. 10.1093/nar/gku686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adolph D., Flach N., Mueller K., Ostareck D. H., Ostareck-Lederer A. (2007). Deciphering the cross talk between hnRNP K and c-Src: the c-Src activation domain in hnRNP K is distinct from a second interaction site. Mol. Cell. Biol. 27 1758–1770. 10.1128/MCB.02014-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albihlal W. S., Gerber A. P. (2018). Unconventional RNA-binding proteins: an uncharted zone in RNA biology. FEBS Lett. 592 2917–2931. 10.1002/1873-3468.13161 [DOI] [PubMed] [Google Scholar]
- Anderson P., Kedersha N. (2002). Stressful initiations. J. Cell Sci. 115(Pt 16), 3227–3234. [DOI] [PubMed] [Google Scholar]
- Anderson P., Nagler-Anderson C., O’Brien C., Levine H., Watkins S., Slayter H. S., et al. (1990). A monoclonal antibody reactive with a 15-kDa cytoplasmic granule-associated protein defines a subpopulation of CD8+ T lymphocytes. J. Immunol. 144 574–582. [PubMed] [Google Scholar]
- Baber J. L., Libutti D., Levens D., Tjandra N. (1999). High precision solution structure of the C-terminal KH domain of heterogeneous nuclear ribonucleoprotein K, a c-myc transcription factor. J. Mol. Biol. 289 949–962. 10.1006/jmbi.1999.2818 [DOI] [PubMed] [Google Scholar]
- Backe P. H., Messias A. C., Ravelli R. B., Sattler M., Cusack S. (2005). X-ray crystallographic and NMR studies of the third KH domain of hnRNP K in complex with single-stranded nucleic acids. Structure 13 1055–1067. 10.1016/j.str.2005.04.008 [DOI] [PubMed] [Google Scholar]
- Blackshear P. J. (2002). Tristetraprolin and other CCCH tandem zinc-finger proteins in the regulation of mRNA turnover. Biochem. Soc. Trans. 30(Pt 6), 945–952. [DOI] [PubMed] [Google Scholar]
- Braddock D. T., Baber J. L., Levens D., Clore G. M. (2002). Molecular basis of sequence-specific single-stranded DNA recognition by KH domains: solution structure of a complex between hnRNP K KH3 and single-stranded DNA. EMBO J. 21 3476–3485. 10.1093/emboj/cdf352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brook M., Tchen C. R., Santalucia T., McIlrath J., Arthur J. S., Saklatvala J., et al. (2006). Posttranslational regulation of tristetraprolin subcellular localization and protein stability by p38 mitogen-activated protein kinase and extracellular signal-regulated kinase pathways. Mol. Cell. Biol. 26 2408–2418. 10.1128/MCB.26.6.2408-2418.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks S. A., Blackshear P. J. (2013). Tristetraprolin (TTP): interactions with mRNA and proteins, and current thoughts on mechanisms of action. Biochim. Biophys. Acta 1829 666–679. 10.1016/j.bbagrm.2013.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballo E., Gilkeson G. S., Blackshear P. J. (1997). Bone marrow transplantation reproduces the tristetraprolin-deficiency syndrome in recombination activating gene-2 (-/-) mice. Evidence that monocyte/macrophage progenitors may be responsible for TNFalpha overproduction. J. Clin. Invest. 100 986–995. 10.1172/JCI119649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballo E., Lai W. S., Blackshear P. J. (1998). Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science 281 1001–1005. 10.1126/science.281.5379.1001 [DOI] [PubMed] [Google Scholar]
- Carpenter S., Ricci E. P., Mercier B. C., Moore M. J., Fitzgerald K. A. (2014). Post-transcriptional regulation of gene expression in innate immunity. Nat. Rev. Immunol. 14 361–376. 10.1038/nri3682 [DOI] [PubMed] [Google Scholar]
- Castello A., Hentze M. W., Preiss T. (2015). Metabolic enzymes enjoying new partnerships as RNA-binding proteins. Trends Endocrinol. Metab. 26 746–757. 10.1016/j.tem.2015.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrestensen C. A., Schroeder M. J., Shabanowitz J., Hunt D. F., Pelo J. W., Worthington M. T., et al. (2004). MAPKAP kinase 2 phosphorylates tristetraprolin on in vivo sites including Ser178, a site required for 14-3-3 binding. J. Biol. Chem. 279 10176–10184. 10.1074/jbc.M310486200 [DOI] [PubMed] [Google Scholar]
- Clement S. L., Scheckel C., Stoecklin G., Lykke-Andersen J. (2011). Phosphorylation of tristetraprolin by MK2 impairs AU-rich element mRNA decay by preventing deadenylase recruitment. Mol. Cell. Biol. 31 256–266. 10.1128/MCB.00717-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cok S. J., Acton S. J., Morrison A. R. (2003). The proximal region of the 3′-untranslated region of cyclooxygenase-2 is recognized by a multimeric protein complex containing HuR, TIA-1, TIAR, and the heterogeneous nuclear ribonucleoprotein U. J. Biol. Chem. 278 36157–36162. 10.1074/jbc.M302547200 [DOI] [PubMed] [Google Scholar]
- Collier B., Goobar-Larsson L., Sokolowski M., Schwartz S. (1998). Translational inhibition in vitro of human papillomavirus type 16 L2 mRNA mediated through interaction with heterogenous ribonucleoprotein K and poly(rC)-binding proteins 1 and 2. J. Biol. Chem. 273 22648–22656. 10.1074/jbc.273.35.22648 [DOI] [PubMed] [Google Scholar]
- Cruz-Gallardo I., Aroca A., Gunzburg M. J., Sivakumaran A., Yoon J. H., Angulo J., et al. (2014). The binding of TIA-1 to RNA C-rich sequences is driven by its C-terminal RRM domain. RNA Biol. 11 766–776. 10.4161/rna.28801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damgaard C. K., Lykke-Andersen J. (2011). Translational coregulation of 5′TOP mRNAs by TIA-1 and TIAR. Genes Dev. 25 2057–2068. 10.1101/gad.17355911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejgaard K., Leffers H. (1996). Characterisation of the nucleic-acid-binding activity of KH domains. Different properties of different domains. Eur. J. Biochem. 241 425–431. 10.1111/j.1432-1033.1996.00425.x [DOI] [PubMed] [Google Scholar]
- Del Gatto-Konczak F., Bourgeois C. F., Le Guiner C., Kister L., Gesnel M. C., Stevenin J., et al. (2000). The RNA-binding protein TIA-1 is a novel mammalian splicing regulator acting through intron sequences adjacent to a 5′ splice site. Mol. Cell. Biol. 20 6287–6299. 10.1128/MCB.20.17.6287-6299.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dember L. M., Kim N. D., Liu K. Q., Anderson P. (1996). Individual RNA recognition motifs of TIA-1 and TIAR have different RNA binding specificities. J. Biol. Chem. 271 2783–2788. 10.1074/jbc.271.5.2783 [DOI] [PubMed] [Google Scholar]
- Diaz-Munoz M. D., Kiselev V. Y., Le Novere N., Curk T., Ule J., Turner M. (2017). Tia1 dependent regulation of mRNA subcellular location and translation controls p53 expression in B cells. Nat. Commun. 8:530. 10.1038/s41467-017-00454-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Munoz M. D., Turner M. (2018). Uncovering the role of rna-binding proteins in gene expression in the immune system. Front. Immunol. 9:1094. 10.3389/fimmu.2018.01094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon D. A., Balch G. C., Kedersha N., Anderson P., Zimmerman G. A., Beauchamp R. D., et al. (2003). Regulation of cyclooxygenase-2 expression by the translational silencer TIA-1. J. Exp. Med. 198 475–481. 10.1084/jem.20030616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Expert-Bezancon A., Le J., Caer P., Marie J. (2002). Heterogeneous nuclear ribonucleoprotein (hnRNP) K is a component of an intronic splicing enhancer complex that activates the splicing of the alternative exon 6A from chicken beta-tropomyosin pre-mRNA. J. Biol. Chem. 277 16614–16623. 10.1074/jbc.M201083200 [DOI] [PubMed] [Google Scholar]
- Fabian M. R., Frank F., Rouya C., Siddiqui N., Lai W. S., Karetnikov A., et al. (2013). Structural basis for the recruitment of the human CCR4-NOT deadenylase complex by tristetraprolin. Nat. Struct. Mol. Biol. 20 735–739. 10.1038/nsmb.2572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X. C., Steitz J. A. (1998). Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 17 3448–3460. 10.1093/emboj/17.12.3448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenger-Gron M., Fillman C., Norrild B., Lykke-Andersen J. (2005). Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol. Cell 20 905–915. 10.1016/j.molcel.2005.10.031 [DOI] [PubMed] [Google Scholar]
- Forch P., Puig O., Kedersha N., Martinez C., Granneman S., Seraphin B., et al. (2000). The apoptosis-promoting factor TIA-1 is a regulator of alternative pre-mRNA splicing. Mol. Cell 6 1089–1098. 10.1016/S1097-2765(00)00107-6 [DOI] [PubMed] [Google Scholar]
- Fu M., Blackshear P. J. (2017). RNA-binding proteins in immune regulation: a focus on CCCH zinc finger proteins. Nat. Rev. Immunol. 17 130–143. 10.1038/nri.2016.129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Maurino S. M., Rivero-Rodriguez F., Velazquez-Cruz A., Hernandez-Vellisca M., Diaz-Quintana A., De la Rosa M. A., et al. (2017). RNA binding protein regulation and cross-talk in the control of AU-rich mRNA fate. Front. Mol. Biosci. 4:71 10.3389/fmolb.2017.00071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson T. J., Thompson J. D., Heringa J. (1993). The KH domain occurs in a diverse set of RNA-binding proteins that include the antiterminator NusA and is probably involved in binding to nucleic acid. FEBS Lett. 324 361–366. 10.1016/0014-5793(93)80152-K [DOI] [PubMed] [Google Scholar]
- Grammatikakis I., Abdelmohsen K., Gorospe M. (2017). Posttranslational control of HuR function. Wiley Interdiscip. Rev. RNA 8 1–11. 10.1002/wrna.1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishin N. V. (2001). KH domain: one motif, two folds. Nucleic Acids Res. 29 638–643. 10.1093/nar/29.3.638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueydan C., Droogmans L., Chalon P., Huez G., Caput D., Kruys V. (1999). Identification of TIAR as a protein binding to the translational regulatory AU-rich element of tumor necrosis factor alpha mRNA. J. Biol. Chem. 274 2322–2326. 10.1074/jbc.274.4.2322 [DOI] [PubMed] [Google Scholar]
- Habelhah H., Shah K., Huang L., Ostareck-Lederer A., Burlingame A. L., Shokat K. M., et al. (2001). ERK phosphorylation drives cytoplasmic accumulation of hnRNP-K and inhibition of mRNA translation. Nat. Cell Biol. 3 325–330. 10.1038/35060131 [DOI] [PubMed] [Google Scholar]
- Hentze M. W., Castello A., Schwarzl T., Preiss T. (2018). A brave new world of RNA-binding proteins. Nat. Rev. Mol. Cell. Biol. 19 327–341. 10.1038/nrm.2017.130 [DOI] [PubMed] [Google Scholar]
- Hitti E., Iakovleva T., Brook M., Deppenmeier S., Gruber A. D., Radzioch D., et al. (2006). Mitogen-activated protein kinase-activated protein kinase 2 regulates tumor necrosis factor mRNA stability and translation mainly by altering tristetraprolin expression, stability, and binding to adenine/uridine-rich element. Mol. Cell. Biol. 26 2399–2407. 10.1128/MCB.26.6.2399-2407.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izquierdo J. M., Valcarcel J. (2007). Two isoforms of the T-cell intracellular antigen 1 (TIA-1) splicing factor display distinct splicing regulation activities. Control of TIA-1 isoform ratio by TIA-1-related protein. J. Biol. Chem. 282 19410–19417. 10.1074/jbc.M700688200 [DOI] [PubMed] [Google Scholar]
- Kafasla P., Skliris A., Kontoyiannis D. L. (2014). Post-transcriptional coordination of immunological responses by RNA-binding proteins. Nat. Immunol. 15 492–502. 10.1038/ni.2884 [DOI] [PubMed] [Google Scholar]
- Kedersha N. L., Gupta M., Li W., Miller I., Anderson P. (1999). RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J. Cell Biol. 147 1431–1442. 10.1083/jcb.147.7.1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene J. D. (1999). Why is Hu where? Shuttling of early-response-gene messenger RNA subsets. Proc. Natl. Acad. Sci. U.S.A. 96 5–7. 10.1073/pnas.96.1.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharraz Y., Lefort A., Libert F., Mann C. J., Gueydan C., Kruys V. (2016). Genome-wide analysis of TIAR RNA ligands in mouse macrophages before and after LPS stimulation. Genom. Data 7 297–300. 10.1016/j.gdata.2016.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. S., Headey S. J., Yoga Y. M., Scanlon M. J., Gorospe M., Wilce M. C., et al. (2013). Distinct binding properties of TIAR RRMs and linker region. RNA Biol. 10 579–589. 10.4161/rna.24341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontoyiannis D., Pasparakis M., Pizarro T. T., Cominelli F., Kollias G. (1999). Impaired on/off regulation of TNF biosynthesis in mice lacking TNF AU-rich elements: implications for joint and gut-associated immunopathologies. Immunity 10 387–398. 10.1016/S1074-7613(00)80038-2 [DOI] [PubMed] [Google Scholar]
- Kratochvill F., Machacek C., Vogl C., Ebner F., Sedlyarov V., Gruber A. R., et al. (2011). Tristetraprolin-driven regulatory circuit controls quality and timing of mRNA decay in inflammation. Mol. Syst. Biol. 7:560. 10.1038/msb.2011.93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwano Y., Kim H. H., Abdelmohsen K., Pullmann R., Jr., Martindale J. L., Yang X., et al. (2008). MKP-1 mRNA stabilization and translational control by RNA-binding proteins HuR and NF90. Mol. Cell. Biol. 28 4562–4575. 10.1128/MCB.00165-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafarga V., Cuadrado A., Lopez de Silanes I., Bengoechea R., Fernandez-Capetillo O., Nebreda A. R. (2009). p38 Mitogen-activated protein kinase- and HuR-dependent stabilization of p21(Cip1) mRNA mediates the G(1)/S checkpoint. Mol. Cell. Biol. 29 4341–4351. 10.1128/MCB.00210-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai W. S., Carballo E., Strum J. R., Kennington E. A., Phillips R. S., Blackshear P. J. (1999). Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor alpha mRNA. Mol. Cell. Biol. 19 4311–4323. 10.1128/MCB.19.6.4311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebedeva S., Jens M., Theil K., Schwanhausser B., Selbach M., Landthaler M., et al. (2011). Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein HuR. Mol. Cell 43 340–352. 10.1016/j.molcel.2011.06.008 [DOI] [PubMed] [Google Scholar]
- Lewis T., Gueydan C., Huez G., Toulme J. J., Kruys V. (1998). Mapping of a minimal AU-rich sequence required for lipopolysaccharide-induced binding of a 55-kDa protein on tumor necrosis factor-alpha mRNA. J. Biol. Chem. 273 13781–13786. 10.1074/jbc.273.22.13781 [DOI] [PubMed] [Google Scholar]
- Liepelt A., Mossanen J. C., Denecke B., Heymann F., De Santis R., Tacke F., et al. (2014). Translation control of TAK1 mRNA by hnRNP K modulates LPS-induced macrophage activation. RNA 20 899–911. 10.1261/rna.042788.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liepelt A., Naarmann-de Vries I. S., Simons N., Eichelbaum K., Fohr S., Archer S. K., et al. (2016). Identification of RNA-binding proteins in macrophages by interactome capture. Mol. Cell. Proteomics 15 2699–2714. 10.1074/mcp.M115.056564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez de Silanes I., Zhan M., Lal A., Yang X., Gorospe M. (2004). Identification of a target RNA motif for RNA-binding protein HuR. Proc. Natl. Acad. Sci. U.S.A. 101 2987–2992. 10.1073/pnas.0306453101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y. C., Chang S. H., Hafner M., Li X., Tuschl T., Elemento O., et al. (2014). ELAVL1 modulates transcriptome-wide miRNA binding in murine macrophages. Cell. Rep. 9 2330–2343. 10.1016/j.celrep.2014.11.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W. J., Cheng S., Campbell C., Wright A., Furneaux H. (1996). Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein. J. Biol. Chem. 271 8144–8151. 10.1074/jbc.271.14.8144 [DOI] [PubMed] [Google Scholar]
- Mahtani K. R., Brook M., Dean J. L., Sully G., Saklatvala J., Clark A. R. (2001). Mitogen-activated protein kinase p38 controls the expression and posttranslational modification of tristetraprolin, a regulator of tumor necrosis factor alpha mRNA stability. Mol. Cell. Biol. 21 6461–6469. 10.1128/MCB.21.9.6461-6469.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchese F. P., Aubareda A., Tudor C., Saklatvala J., Clark A. R., Dean J. L. (2010). MAPKAP kinase 2 blocks tristetraprolin-directed mRNA decay by inhibiting CAF1 deadenylase recruitment. J. Biol. Chem. 285 27590–27600. 10.1074/jbc.M110.136473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matunis M. J., Michael W. M., Dreyfuss G. (1992). Characterization and primary structure of the poly(C)-binding heterogeneous nuclear ribonucleoprotein complex K protein. Mol. Cell. Biol. 12 164–171. 10.1101/SQB.1995.060.01.071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAlinden A., Liang L., Mukudai Y., Imamura T., Sandell L. J. (2007). Nuclear protein TIA-1 regulates COL2A1 alternative splicing and interacts with precursor mRNA and genomic DNA. J. Biol. Chem. 282 24444–24454. 10.1074/jbc.M702717200 [DOI] [PubMed] [Google Scholar]
- Medzhitov R., Horng T. (2009). Transcriptional control of the inflammatory response. Nat. Rev. Immunol. 9 692–703. 10.1038/nri2634 [DOI] [PubMed] [Google Scholar]
- Messias A. C., Harnisch C., Ostareck-Lederer A., Sattler M., Ostareck D. H. (2006). The DICE-binding activity of KH domain 3 of hnRNP K is affected by c-Src-mediated tyrosine phosphorylation. J. Mol. Biol. 361 470–481. 10.1016/j.jmb.2006.06.025 [DOI] [PubMed] [Google Scholar]
- Michael W. M., Eder P. S., Dreyfuss G. (1997). The K nuclear shuttling domain: a novel signal for nuclear import and nuclear export in the hnRNP K protein. EMBO J. 16 3587–3598. 10.1093/emboj/16.12.3587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael W. M., Siomi H., Choi M., Pinol-Roma S., Nakielny S., Liu Q., et al. (1995). Signal sequences that target nuclear import and nuclear export of pre-mRNA-binding proteins. Cold Spring Harb. Symp. Quant. Biol. 60 663–668. 10.1074/jbc.M710328200 [DOI] [PubMed] [Google Scholar]
- Mino T., Takeuchi O. (2018). Post-transcriptional regulation of immune responses by RNA binding proteins. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 94 248–258. 10.2183/pjab.94.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz B., Lilie H., Naarmann-de Vries I. S., Urlaub H., Wahle E., Ostareck-Lederer A., et al. (2014). Biophysical and biochemical analysis of hnRNP K: arginine methylation, reversible aggregation and combinatorial binding to nucleic acids. Biol. Chem. 395 837–853. 10.1515/hsz-2014-0146 [DOI] [PubMed] [Google Scholar]
- Moumen A., Masterson P., O’Connor M. J., Jackson S. P. (2005). hnRNP K: an HDM2 target and transcriptional coactivator of p53 in response to DNA damage. Cell 123 1065–1078. 10.1016/j.cell.2005.09.032 [DOI] [PubMed] [Google Scholar]
- Mukherjee N., Corcoran D. L., Nusbaum J. D., Reid D. W., Georgiev S., Hafner M., et al. (2011). Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol. Cell 43 327–339. 10.1016/j.molcel.2011.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naarmann I. S., Harnisch C., Flach N., Kremmer E., Kuhn H., Ostareck D. H., et al. (2008). mRNA silencing in human erythroid cell maturation: heterogeneous nuclear ribonucleoprotein K controls the expression of its regulator c-Src. J. Biol. Chem. 283 18461–18472. 10.1074/jbc.M710328200 [DOI] [PubMed] [Google Scholar]
- Naarmann I. S., Harnisch C., Muller-Newen G., Urlaub H., Ostareck-Lederer A., Ostareck D. H. (2010). DDX6 recruits translational silenced human reticulocyte 15-lipoxygenase mRNA to RNP granules. RNA 16 2189–2204. 10.1261/rna.2211110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naarmann-de Vries I. S., Brendle A., Bahr-Ivacevic T., Benes V., Ostareck D. H., Ostareck-Lederer A. (2016). Translational control mediated by hnRNP K links NMHC IIA to erythroid enucleation. J. Cell Sci. 129 1141–1154. 10.1242/jcs.174995 [DOI] [PubMed] [Google Scholar]
- Naarmann-de Vries I. S., Urlaub H., Ostareck D. H., Ostareck-Lederer A. (2013). Caspase-3 cleaves hnRNP K in erythroid differentiation. Cell Death Dis. 4:e548. 10.1038/cddis.2013.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachtergaele S., He C. (2018). Chemical modifications in the life of an mRNA transcript. Annu. Rev. Genet. 52 349–372. 10.1146/annurev-genet-120417-031522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson C. O., Friedersdorf M. B., Bisogno L. S., Keene J. D. (2017a). DO-RIP-seq to quantify RNA binding sites transcriptome-wide. Methods 118–119, 16–23. 10.1016/j.ymeth.2016.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson C. O., Friedersdorf M., Keene J. D. (2017b). Quantifying RNA binding sites transcriptome-wide using DO-RIP-seq. RNA 23 32–46. 10.1261/rna.058115.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostareck D. H., Ostareck-Lederer A., Shatsky I. N., Hentze M. W. (2001). Lipoxygenase mRNA silencing in erythroid differentiation: the 3′UTR regulatory complex controls 60S ribosomal subunit joining. Cell 104 281–290. 10.1093/nar/gky1138 [DOI] [PubMed] [Google Scholar]
- Ostareck D. H., Ostareck-Lederer A., Wilm M., Thiele B. J., Mann M., Hentze M. W. (1997). mRNA silencing in erythroid differentiation: hnRNP K and hnRNP E1 regulate 15-lipoxygenase translation from the 3′ end. Cell 89 597–606. 10.1093/emboj/17.12.3461 [DOI] [PubMed] [Google Scholar]
- Ostareck-Lederer A., Ostareck D. H. (2012). Precision mechanics with multifunctional tools: how hnRNP K and hnRNPs E1/E2 contribute to post-transcriptional control of gene expression in hematopoiesis. Curr. Protein Pept. Sci. 13 391–400. 10.1093/emboj/19.15.4154 [DOI] [PubMed] [Google Scholar]
- Ostareck-Lederer A., Ostareck D. H., Cans C., Neubauer G., Bomsztyk K., Superti-Furga G., et al. (2002). c-Src-mediated phosphorylation of hnRNP K drives translational activation of specifically silenced mRNAs. Mol. Cell. Biol. 22 4535–4543. 10.1101/gad.2.2.215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostareck-Lederer A., Ostareck D. H., Rucknagel K. P., Schierhorn A., Moritz B., Huttelmaier S., et al. (2006). Asymmetric arginine dimethylation of heterogeneous nuclear ribonucleoprotein K by protein-arginine methyltransferase 1 inhibits its interaction with c-Src. J. Biol. Chem. 281 11115–11125. 10.1074/jbc.M513053200 [DOI] [PubMed] [Google Scholar]
- Pabis M., Popowicz G. M., Stehle R., Fernandez-Ramos D., Asami S., Warner L., et al. (2018). HuR biological function involves RRM3-mediated dimerization and RNA binding by all three RRMs. Nucleic Acids Res. 10.1093/nar/gky1138 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng S. S., Chen C. Y., Xu N., Shyu A. B. (1998). RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. EMBO J. 17 3461–3470. 10.1093/emboj/17.12.3461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piecyk M., Wax S., Beck A. R., Kedersha N., Gupta M., Maritim B., et al. (2000). TIA-1 is a translational silencer that selectively regulates the expression of TNF-alpha. EMBO J. 19 4154–4163. 10.1093/emboj/19.15.4154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinol-Roma S., Choi Y. D., Matunis M. J., Dreyfuss G. (1988). Immunopurification of heterogeneous nuclear ribonucleoprotein particles reveals an assortment of RNA-binding proteins. Genes Dev. 2 215–227. 10.4049/jimmunol.0803883 [DOI] [PubMed] [Google Scholar]
- Poria D. K., Guha A., Nandi I., Ray P. S. (2016). RNA-binding protein HuR sequesters microRNA-21 to prevent translation repression of proinflammatory tumor suppressor gene programmed cell death 4. Oncogene 35 1703–1715. 10.1038/onc.2015.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynier F., de Vos A. F., Hoogerwerf J. J., Bresser P., van der Zee J. S., Paye M., et al. (2012). Gene expression profiles in alveolar macrophages induced by lipopolysaccharide in humans. Mol. Med. 18 1303–1311. 10.2119/molmed.2012.00230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutledge H. R., Jiang W., Yang J., Warg L. A., Schwartz D. A., Pisetsky D. S., et al. (2012). Gene expression profiles of RAW264.7 macrophages stimulated with preparations of LPS differing in isolation and purity. Innate Immun. 18 80–88. 10.1177/1753425910393540 [DOI] [PubMed] [Google Scholar]
- Sandler H., Kreth J., Timmers H. T., Stoecklin G. (2011). Not1 mediates recruitment of the deadenylase Caf1 to mRNAs targeted for degradation by tristetraprolin. Nucleic Acids Res. 39 4373–4386. 10.1093/nar/gkr011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaljo B., Kratochvill F., Gratz N., Sadzak I., Sauer I., Hammer M., et al. (2009). Tristetraprolin is required for full anti-inflammatory response of murine macrophages to IL-10. J. Immunol. 183 1197–1206. 10.4049/jimmunol.0803883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schott J., Reitter S., Philipp J., Haneke K., Schafer H., Stoecklin G. (2014). Translational regulation of specific mRNAs controls feedback inhibition and survival during macrophage activation. PLoS Genet. 10:e1004368. 10.1371/journal.pgen.1004368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedlyarov V., Fallmann J., Ebner F., Huemer J., Sneezum L., Ivin M., et al. (2016). Tristetraprolin binding site atlas in the macrophage transcriptome reveals a switch for inflammation resolution. Mol. Syst. Biol. 12:868. 10.15252/msb.20156628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanmugam N., Reddy M. A., Natarajan R. (2008). Distinct roles of heterogeneous nuclear ribonuclear protein K and microRNA-16 in cyclooxygenase-2 RNA stability induced by S100b, a ligand of the receptor for advanced glycation end products. J. Biol. Chem. 283 36221–36233. 10.1074/jbc.M806322200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi H., Matunis M. J., Michael W. M., Dreyfuss G. (1993). The pre-mRNA binding K protein contains a novel evolutionarily conserved motif. Nucleic Acids Res. 21 1193–1198. 10.1038/sj.emboj.7600163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalweit A., Doller A., Huth A., Kahne T., Persson P. B., Thiele B. J. (2003). Posttranscriptional control of renin synthesis: identification of proteins interacting with renin mRNA 3′-untranslated region. Circ. Res. 92 419–427. 10.1161/01.RES.0000059300.67152.4E [DOI] [PubMed] [Google Scholar]
- Smale S. T. (2012). Transcriptional regulation in the innate immune system. Curr. Opin. Immunol. 24 51–57. 10.1016/j.coi.2011.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikantan S., Gorospe M. (2012). HuR function in disease. Front. Biosci. 17:189–205. 10.1016/j.cell.2010.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikantan S., Tominaga K., Gorospe M. (2012). Functional interplay between RNA-binding protein HuR and microRNAs. Curr. Protein Pept. Sci. 13 372–379. 10.1016/S1074-7613(00)80411-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoecklin G., Stubbs T., Kedersha N., Wax S., Rigby W. F., Blackwell T. K., et al. (2004). MK2-induced tristetraprolin:14-3-3 complexes prevent stress granule association and ARE-mRNA decay. EMBO J. 23 1313–1324. 10.1038/sj.emboj.7600163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoecklin G., Tenenbaum S. A., Mayo T., Chittur S. V., George A. D., Baroni T. E., et al. (2008). Genome-wide analysis identifies interleukin-10 mRNA as target of tristetraprolin. J. Biol. Chem. 283 11689–11699. 10.1074/jbc.M709657200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suswam E. A., Li Y. Y., Mahtani H., King P. H. (2005). Novel DNA-binding properties of the RNA-binding protein TIAR. Nucleic Acids Res. 33 4507–4518. 10.1093/nar/gki763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O., Akira S. (2010). Pattern recognition receptors and inflammation. Cell 140 805–820. 10.1016/j.cell.2010.01.022 [DOI] [PubMed] [Google Scholar]
- Taylor G. A., Carballo E., Lee D. M., Lai W. S., Thompson M. J., Patel D. D., et al. (1996). A pathogenetic role for TNF alpha in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity 4 445–454. 10.1128/MCB.23.20.7177-7188.2003 [DOI] [PubMed] [Google Scholar]
- Tchen C. R., Brook M., Saklatvala J., Clark A. R. (2004). The stability of tristetraprolin mRNA is regulated by mitogen-activated protein kinase p38 and by tristetraprolin itself. J. Biol. Chem. 279 32393–32400. 10.1074/jbc.M402059200 [DOI] [PubMed] [Google Scholar]
- Thisted T., Lyakhov D. L., Liebhaber S. A. (2001). Optimized RNA targets of two closely related triple KH domain proteins, heterogeneous nuclear ribonucleoprotein K and alphaCP-2KL, suggest Distinct modes of RNA recognition. J. Biol. Chem. 276 17484–17496. 10.1074/jbc.M010594200 [DOI] [PubMed] [Google Scholar]
- Tiedje C., Diaz-Munoz M. D., Trulley P., Ahlfors H., Laass K., Blackshear P. J., et al. (2016). The RNA-binding protein TTP is a global post-transcriptional regulator of feedback control in inflammation. Nucleic Acids Res. 44 7418–7440. 10.1093/nar/gkw474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiedje C., Ronkina N., Tehrani M., Dhamija S., Laass K., Holtmann H., et al. (2012). The p38/MK2-driven exchange between tristetraprolin and HuR regulates AU-rich element-dependent translation. PLoS Genet. 8:e1002977. 10.1371/journal.pgen.1002977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran H., Maurer F., Nagamine Y. (2003). Stabilization of urokinase and urokinase receptor mRNAs by HuR is linked to its cytoplasmic accumulation induced by activated mitogen-activated protein kinase-activated protein kinase 2. Mol. Cell. Biol. 23 7177–7188. 10.1074/jbc.M206505200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner M., Diaz-Munoz M. D. (2018). RNA-binding proteins control gene expression and cell fate in the immune system. Nat. Immunol. 19 120–129. 10.1038/s41590-017-0028-4 [DOI] [PubMed] [Google Scholar]
- Vaure C., Liu Y. (2014). A comparative review of toll-like receptor 4 expression and functionality in different animal species. Front. Immunol. 5:316. 10.3389/fimmu.2014.00316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waris S., Garcia-Maurino S. M., Sivakumaran A., Beckham S. A., Loughlin F. E., Gorospe M., et al. (2017). TIA-1 RRM23 binding and recognition of target oligonucleotides. Nucleic Acids Res. 45 4944–4957. 10.1093/nar/gkx102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waris S., Wilce M. C., Wilce J. A. (2014). RNA recognition and stress granule formation by TIA proteins. Int. J. Mol. Sci. 15 23377–23388. 10.3390/ijms151223377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worthington M. T., Pelo J. W., Sachedina M. A., Applegate J. L., Arseneau K. O., Pizarro T. T. (2002). RNA binding properties of the AU-rich element-binding recombinant Nup475/TIS11/tristetraprolin protein. J. Biol. Chem. 277 48558–48564. 10.1074/jbc.M206505200 [DOI] [PubMed] [Google Scholar]
- Yoon J. H., Abdelmohsen K., Kim J., Yang X., Martindale J. L., Tominaga-Yamanaka K., et al. (2013). Scaffold function of long non-coding RNA HOTAIR in protein ubiquitination. Nat. Commun. 4:2939. 10.1038/ncomms3939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon J. H., Abdelmohsen K., Srikantan S., Yang X., Martindale J. L., De S., et al. (2012). LincRNA-p21 suppresses target mRNA translation. Mol. Cell 47 648–655. 10.1016/j.molcel.2012.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q., Cok S. J., Zeng C., Morrison A. R. (2003). Translational repression of human matrix metalloproteinases-13 by an alternatively spliced form of T-cell-restricted intracellular antigen-related protein (TIAR). J. Biol. Chem. 278 1579–1584. 10.1074/jbc.M203526200 [DOI] [PubMed] [Google Scholar]
- Zanotti S., Kumar A., Kumar A. (2002). Cytokine modulation in sepsis and septic shock. Expert Opin. Investig. Drugs 11 1061–1075. 10.1517/13543784.11.8.1061 [DOI] [PubMed] [Google Scholar]
- Zhang T., Delestienne N., Huez G., Kruys V., Gueydan C. (2005). Identification of the sequence determinants mediating the nucleo-cytoplasmic shuttling of TIAR and TIA-1 RNA-binding proteins. J. Cell Sci. 118(Pt 23), 5453–5463. 10.1242/jcs.02669 [DOI] [PubMed] [Google Scholar]
- Zhu H., Hasman R. A., Young K. M., Kedersha N. L., Lou H. (2003). U1 snRNP-dependent function of TIAR in the regulation of alternative RNA processing of the human calcitonin/CGRP pre-mRNA. Mol. Cell. Biol. 23 5959–5971. 10.1128/MCB.23.17.5959-5971.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Hinman M. N., Hasman R. A., Mehta P., Lou H. (2008). Regulation of neuron-specific alternative splicing of neurofibromatosis type 1 pre-mRNA. Mol. Cell. Biol. 28 1240–1251. 10.1128/MCB.01509-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccato E., Buratti E., Stuani C., Baralle F. E., Pagani F. (2004). An intronic polypyrimidine-rich element downstream of the donor site modulates cystic fibrosis transmembrane conductance regulator exon 9 alternative splicing. J. Biol. Chem. 279 16980–16988. 10.1074/jbc.M313439200 [DOI] [PubMed] [Google Scholar]