Abstract
SOX4, together with SOX11 and SOX12, forms group C of SRY-related (SOX) transcription factors. They play key roles, often in redundancy, in multiple developmental pathways, including neurogenesis and skeletogenesis. De novo SOX11 heterozygous mutations have been shown to cause intellectual disability, growth deficiency, and dysmorphic features compatible with mild Coffin-Siris syndrome. Using trio-based exome sequencing, we here identify de novo SOX4 heterozygous missense variants in four children who share developmental delay, intellectual disability, and mild facial and digital morphological abnormalities. SOX4 is highly expressed in areas of active neurogenesis in human fetuses, and sox4 knockdown in Xenopus embryos diminishes brain and whole-body size. The SOX4 variants cluster in the highly conserved, SOX family-specific HMG domain, but each alters a different residue. In silico tools predict that each variant affects a distinct structural feature of this DNA-binding domain, and functional assays demonstrate that these SOX4 proteins carrying these variants are unable to bind DNA in vitro and transactivate SOX reporter genes in cultured cells. These variants are not found in the gnomAD database of individuals with presumably normal development, but 12 other SOX4 HMG-domain missense variants are recorded and all demonstrate partial to full activity in the reporter assay. Taken together, these findings point to specific SOX4 HMG-domain missense variants as the cause of a characteristic human neurodevelopmental disorder associated with mild facial and digital dysmorphism.
Keywords: QA keywords
Introduction
Corticogenesis and skeletogenesis are complex, tightly regulated developmental processes. Proper corticogenesis requires adequate production of neural progenitor cells, followed by guidance of these cells toward neurogenesis and specialization into distinct, fully functional neuronal subtypes.1 In the human fetus, neural progenitors develop in the ventricular and subventricular zones of the nascent central nervous system.2 Starting on embryonic day 42,1 these progenitors initiate their transition into proliferating neurons, which migrate outward into the cortical mantle and commit to a post-mitotic differentiated state. This process is largely completed by birth. Similarly, proper skeletogenesis requires adequate production of multipotent progenitor cells, controlled migration and amplification of these cells, correct differentiation into chondrocytes, osteoblasts, and other skeletal cell types, and coordinated activity of these and associated cell types. Skeletal progenitor cells arise from the neural crest and the paraxial and lateral plate mesoderm, and migrate into the future sites of the craniofacial, axial, and appendicular skeleton by the fifth or sixth week of human embryo gestation.3 They subsequently amplify and commit to specific cell types to ensure proper skeleton patterning, size and function. Both neurogenesis and skeletogenesis are controlled at each step by unique genetic programs.1, 4, 5 The uniqueness of these programs is determined by the combinations of transcription factors and other regulatory factors as much as by the individual factors themselves, as many of these factors contribute to distinct processes. To date, various types of developmental syndromes have been linked to genetic variants affecting the expression or activity of such factors, but the genetic origin of many diseases remains unknown.5, 6
The family of SOX transcription factors is comprised of 20 members.7 Its first identified member, SRY, is encoded by the sex-determining region of the Y chromosome. It features a DNA-binding domain related to that present in a class of high-mobility-group (HMG) proteins. SOX proteins are defined as having at least 50% identity with SRY in this HMG (also called SOX) domain. Most SOX genes have been shown using in vitro assays and experimental animal models to have key roles in determining cell fate and differentiation in discrete lineages such that, altogether, the SOX family participates in the control of virtually all progenitor/stem and differentiated cell types. Mutations within and around several SOX genes have been associated with severe human syndromes. Among them, SRY (MIM: 480000) mutations cause XY sex reversal (MIM: 400044);8 SOX9 (MIM: 608160) mutations cause campomelic dysplasia (generalized chondrodysplasia [MIM: 114290]) and XY sex reversal;9 SOX10 (MIM: 602229) mutations cause Waardenburg-Shah syndrome (pigmentary abnormalities, hearing loss, and Hirschsprung disease [MIM: 277580]);10 and SOX5 (MIM: 604975) mutations cause Lamb-Shaffer syndrome (intellectual disability, behavior abnormalities, and dysmorphic features [MIM: 616803]).11 Beside SRY and SOX3 (MIM: 313430), all SOX genes are located on autosomal chromosomes, and disease-causing mutations were determined in virtually all cases to be inactivating, heterozygous, and de novo. Most SOX-related diseases have thus been proposed to be due to gene haploinsufficiency. They are not nearly as severe as the phenotypes of mice lacking both gene copies, but their major impact on affected individuals has demonstrated the importance of carrying two intact gene copies for normal development. To date, mutations in almost a dozen of the twenty SOX genes have not been associated with a human disease yet.
SOX4 (MIM: 184430), together with SOX11 (MIM: 600898) and SOX12 (MIM: 601947), forms the SOXC group, one of the eight groups that compose the SOX family.12, 13 The three SOXC proteins have almost identical DNA-binding domains and are also highly conserved in their other known functional region, a transactivation domain located at their C terminus. Their genes overlap in expression in many cell types and are most active in progenitor cells. SOX12 has a weak transactivation domain and is dispensable for mouse development and adult physiology.2, 13, 14, 15, 16, 17, 18 In contrast, knockdown of either sox4 or sox11 in Xenopus laevis embryos causes microphthalmia with or without coloboma.19 Homozygous inactivation of Sox4 in the mouse is lethal at embryonic day 14 (early fetal stage) due to heart malformation,20 and Sox11 inactivation is lethal at birth due to marked underdevelopment of such vital organs as the heart, spleen, and lungs.17 Combined inactivation of Sox4 and Sox11 is lethal at embryonic day 10.5 due to a block in early organogenesis.15 Conditional gene inactivation studies have revealed additive and redundant roles for Sox4 and Sox11 in many developmental processes. During cerebral cortex formation, Sox4 and Sox11 are most highly expressed in intermediate progenitor cells.2 Sox4 inactivation affects the maintenance of these cells, and Sox11 inactivation reduces their proliferation and differentiation, resulting in a small brain with a thin cerebral cortex at birth.2 Combined inactivation of the two genes drastically impairs neuronal progenitor cell survival15 and activation of key neuronal differentiation genes.21 Regarding skeletogenesis, single inactivation of Sox4 or Sox11 in progenitor cells has mild if any consequences, whereas simultaneous inactivation of both genes severely reduces cell survival and affects downstream lineage specification, leading to abnormal patterning, growth, and maturation of skeletal primordia.22, 23 In humans, de novo SOX11 missense variants abolishing the DNA-binding capability of SOX11 have been associated with a neurodevelopmental disorder whose features—microcephaly, global developmental delay, intellectual disability, and facial and digital abnormalities—are compatible with mild Coffin-Siris syndrome (CSS [MIM: 135900]).24, 25
Here we report four individuals who carry distinct heterozygous de novo missense variants in SOX4 and who share global development delay, mild to severe intellectual disability (ID), facial dysmorphism, and fifth finger clinodactyly. Along with data from SOX4 RNA profiling in humans, sox4 knockdown assays in Xenopus embryos, in silico predictions of protein structural damage, and functional assays for transcriptional activity in vitro, these findings concur that SOX4 is a critical gene for human global, intellectual, and skeletal development.
Material and Methods
Ascertainment of De Novo SOX4 Sequence Variants and Statistical Analyses
Subject 1 was identified through trio-based exome sequencing performed on subjects with syndromic ID at the University of Washington Center for Medical Mendelian Genomics (UW-CMG). Parents provided consent according to the IRB protocol 3206/2016 at Policlinico S. Orsola-Malpighi (Bologna, Italy). Three other subjects were discovered through trio-based exome sequencing performed as part of the Deciphering Developmental Disorders (DDD) study (data freeze of 4,296 children).6 The DDD study had UK Research Ethics Committee approval (10/H0305/83, granted by the Cambridge South REC, and GEN/284/12 granted by the Republic of Ireland REC). Consent for publication of photographs was obtained from the parents of subjects 1, 2, and 4. The occurrence of SOX4 missense variants in neuro-developmental disorders and the clustering of these variants were assessed using DenovolyzeR26 and CLUMP,27 respectively.
In Silico Assessment of SOX4 Variant Pathogenicity
Evolutionary conservation of the SOX4 HMG-domain sequence was assessed with MacVector16 software using human and vertebrate orthologous sequences retrieved from the NCBI protein database (Tables S1 and S2). The presence of variants in the SOX4 coding sequence in the human healthy population was queried using ExAC and gnomAD.28 The effects of missense variants on protein structure and function were predicted using PolyPhen-2,29 HOPE,30 and SWISS-MODEL.31
Assessment of SOX4 Expression in the Human Brain
Variations in SOX4 transcript levels in the human brain among different developmental stages and anatomical regions were investigated using RNA-seq and RNA microarray data from the BrainSpan Atlas of the Developing Human Brain.32
sox4 Knockdown in Xenopus Embryos
Xenopus laevis embryos were generated, staged, and cultured according to standard protocols.33 For loss-of-function experiments, 40 ng of sox4 morpholino oligonucleotide (MO) or control MO (GeneTools) was injected into both dorso-animal blastomeres to target anterior neural tissue.19, 33, 34 The specificity of the sox4 MO was previously demonstrated by showing that the morphant phenotype could be rescued by co-injection of a human SOX4 construct.19 GFP RNA (0.5 ng) was co-injected as a lineage tracer.19 Embryos were collected at stage 43 and then either fixed with formaldehyde for whole-mount analysis or euthanized with ethyl 3-aminobenzoate methanesulfonate (5–10 g/L; Sigma-Aldrich) for brain analysis. Embryos and brains were imaged using a Zeiss Axiophot microscope. All measurements were done using NIH ImageJ software and compared using the Mann-Whitney U test (GraphPad Prism).
Functional Assessment of SOX4 Variants In Vitro
SOX4 mutations were introduced into a mouse 3FLAG-SOX4 expression plasmid23 using QuikChange Site-Directed Mutagenesis (Stratagene) and appropriate DNA primers (Table S3). Capillary sequencing was used to verify the SOX4 wild-type and variant sequences. A mouse POU3F2 expression plasmid and a 6FXO-p89Luc reporter plasmid were used as described.12
To assess the expression level and intracellular localization of SOX4 variants, COS-1 cells were plated at 300,000/well (6-well plates) in 2 mL of DMEM medium supplemented with 10% FCS. Eight hours later, they were transfected with mixtures containing 1 μg of empty or SOX4 expression plasmid and 3 μL of FuGENE 6 (Promega). The next day, cytoplasmic and nuclear extracts were prepared using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Fisher Scientific). They were tested by western blotting under standard conditions. Briefly, 1 μL of extract was subjected to 8% SDS-PAGE and transferred to PVDF membranes using iBLOT 2 Gel Transfer Device (Thermo Fisher Scientific). Membranes were blocked in Tris-Buffered Saline with 0.1% (v/v) Tween 20 (TBST) and 5% (w/v) nonfat dry milk for 1 hr and then incubated overnight at 4°C in blocking solution containing a 1:25,000 dilution of peroxidase-conjugated anti-FLAG M2 antibody (Sigma-Aldrich, A8592). FLAG-SOX4 signals were visualized on X-ray films using SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific).
The same extracts were used to assess the ability of SOX4 variants to bind DNA. Electrophoretic mobility shift assays (EMSAs) were carried out essentially as described.12 Briefly, 10 fmoles of FXO probe labeled with [α-32P]-dCTP was incubated with 1 μL of cytoplasmic or nuclear extract in the presence of 1 μg of poly(dG-dC).poly(dG-dC) (Sigma-Aldrich). Reactions were incubated for 30 min and protein/DNA complexes were resolved by electrophoresis under native conditions. Gels were exposed overnight to X-ray films at −80°C.
The transactivation capability of SOX4 variants was tested essentially as described.12 Briefly, COS-1 cells were transiently transfected with mixtures containing 500 ng of 6FXO-p89Luc reporter, 150 ng of pSV-beta-galactosidase (Promega), 0 or 150 ng of 3FLAG-mSOX4 and POU3F2 expression plasmids, and empty expression plasmid for a total of 1 μg of DNA. Forty hours later, cell extracts were made in 150 μL of Tropix Lysis buffer supplemented with 0.5 mM DTT (Applied Biosystems) and protease inhibitor cocktail (Thermo Fisher Scientific). Extracts were tested in Dual-Light luciferase and beta-galactosidase assay (Thermo Fisher Scientific).
Results
Identification of Four Subjects with a Neurodevelopmental Syndrome and SOX4 Variants
A child presenting with severe developmental delay, ID, and other clinical features was found through trio-based exome sequencing to carry a de novo missense SOX4 variant (g.chr6:21594963C>A [hg19], c.198C>A [p.Phe66Leu]). Three other children with a similar disease and also carrying a de novo SOX4 missense variant were identified through the DDD study (g.chr6:21595099G>C, c.334G>C [p.Ala112Pro]; g.chr6:21594941T>G, c.176T>G [p.Ile59Ser]; g.chr6:21595080G>T, c.315G>T [p.Lys105Asn]; GenBank: NM_003107.2). These four subjects were from unrelated families and their SOX4 variants were distinct. Detailed case reports can be found in Supplemental Note and a summary of clinical findings in Table 1. In brief, all four children had global development delay and ID, but at varying degrees, case subject 2 being very severe; case subject 1, severe; case subject 4, mild; and case subject 3, very mild. All case subjects also had characteristic facial dysmorphism, with anteverted nares, wide mouth with a cupid bow, posteriorly rotated ears, and fifth-finger clinodactyly (Figure 1). Additionally, the most severely affected children had hypotonia and other clinical features, such as ventricular septal defect (case subject 1) and spastic quadriparesis (case subject 2).
Table 1.
Summary of Demographic, Genetic, and Clinical Characteristics of the Subject Cohort
| Subject 1 | Subject 2 | Subject 3 | Subject 4 | |
|---|---|---|---|---|
| Gender | male | male | female | female |
| Age | 4 years 8 months | 6 years 8 months | 6 years 0 month | 6 years 10 months |
| Ancestry | Italian | Scottish-Hungarian | French | Scottish |
| SOX4 variant | c.198C>A (p.Phe66Leu) | c.334G>C (p.Ala112Pro) | c.176T>G (p.Ile59Ser) | c.315G>T (p.Lys105Asn) |
| Growth Delay | ||||
| Height | 100.7 cm (10th) | N.D. | 98.6 cm (6th) | 109.7 cm (3rd) |
| Weight | 15.0 kg (8th) | 14.8 kg (1st) [−3.66 SD] | 14.6 kg (9th) | 17 kg (3rd) |
| OFC | 47.6 cm (1st) [−2.5 SD] | 48.3 cm (0.4th) [−2.8 SD] | 50 cm (12th) [−0.65 SD] | 52.3 cm (45th) [−0.1 SD] |
| Speech delay | first words at 4 years | no speech | delayed speech at 3 years | first words at 2 years |
| Walking delay | first steps at 27 months | not ambulant | no delay | first steps at 21 months |
| Intellectual and neurological delay | mild ID (IQ: 68); epilepsy; myelination delay | severe ID; spastic quadriparesis; cerebellar atrophy; patchy changes in cerebral white matter | very mild learning difficulties | learning difficulties requiring educational support (IQ: 52) |
| Facial dysmorphism | microbrachycephaly; epicanthus; stellate iris pattern; short nose; upturned nares; wide mouth with cupid bow; and posteriorly rotated ears | microcephaly; trigonocephaly; metopic ridge; epicanthic folds; infra-orbital folds; and wide mouth with cupid bow | deep-set eyes; infra-orbital grooves; upturned nares; wide mouth with cupid bow and full lips | deep-set eyes; infra-orbital creases; malar flattening, upturned nares; wide mouth with cupid bow, and full lips; and posteriorly rotated ears |
| Hand and foot malformation | bilateral 5th finger clinodactyly | congenital vertical talus; bilateral 5th finger clinodactyly | mild 5th finger clinodactyly; dysplastic 5th toenails | bilateral 5th finger clinodactyly; mild camptodactyly |
| Hypotonia | mild, generalized | truncal hypotonia | not detected | not detected |
| Other abnormal features | ventricular septal defect; feeding difficulties and constipation; strabismus and keratoconus | feeding difficulties; laryngomalacia; delayed secondary dentition | none detected | none detected |
Parentheses indicate percentiles. Brackets indicate standard deviations. N.D., not determined. OFC, occipitofrontal head circumference.
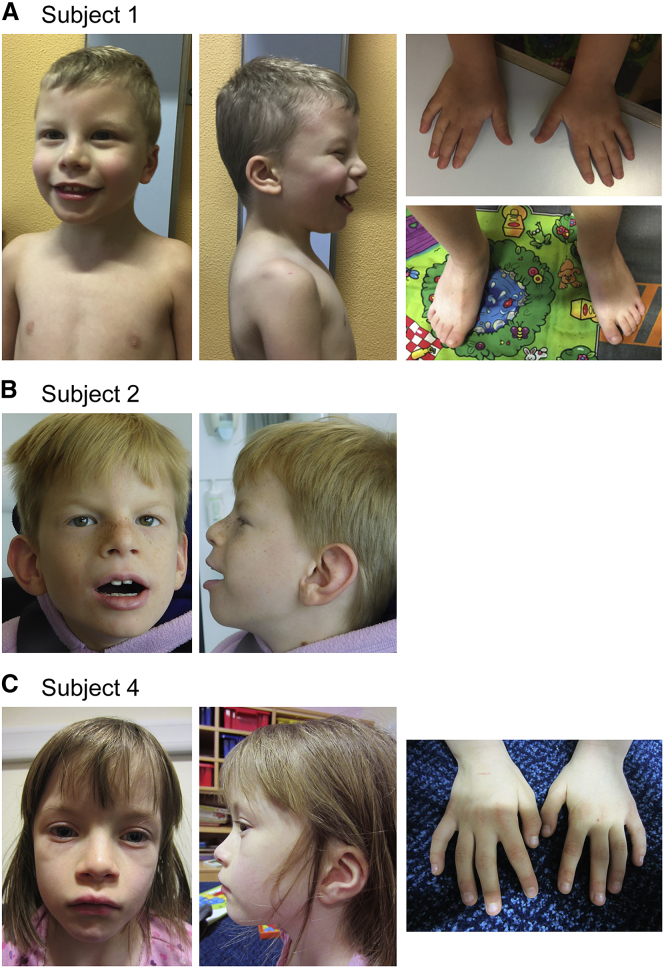
Figure1.
Pictures of Three Subjects
Subject 1 at 4 years and 8 months (A), subject 2 at 6 years and 8 months (B), and subject 4 at 6 years and 10 months (C). Frontal and profile views of the heads show mild facial dysmorphism,including anteverted nares, wide mouth with acupid bow, and posteriorly rotated ears. Pictures of the hands of subjects 1 and 4 show bilateral 5th finger clinodactyly. A picture of the feet of subject 1 shows normal morphology.
Based on these findings, we asked whether the occurrence of de novo SOX4 missense variants in individuals with a neuro-developmental disorder was significant. We applied denovolyzeR26 to all case subjects identified in the 4,293 DDD probands, as reported in denovo-db 1.6.1.35 We found after multiple-testing correction with the Bonferroni method for 19,618 genes that the occurrence of the de novo SOX4 missense variants was statistically significant (p = 0.012).
Together, these data strongly suggest that SOX4 missense variants underlie a specific form of human neurodevelopmental disorder associated with mild dysmorphism.
SOX4 Is under Marked Sequence Conservation Constraint in Humans
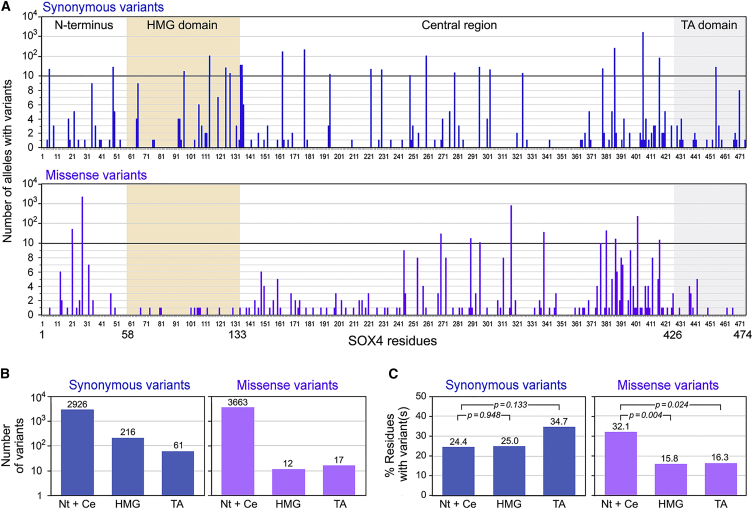
Genomic information recently collected from the general human population provides evidence that SOX4 is under tight sequence conservation constraint and is thus likely critical for normal development. ExAC, which contains exome sequences for 60,706 unrelated human individuals with no history of severe pediatric disease, indeed indicates that while 196.6 missense variants were expected for SOX4, only 90 were observed, for a constraint z-score of 3.72. Furthermore, while 4.6 loss-of-function (nonsense) variants were expected, only 1 was observed, resulting in a probability of loss-of-function intolerance of 0.38. We then used gnomAD, which provides genomic information for 138,632 individuals, including those from ExAC, to analyze the distribution of SOX4 variants in the presumably normal human population. Synonymous and missense variants were present throughout the SOX4 coding sequence (Figure 2A). The N-terminal and central regions of the protein, which are not known to be functionally involved, exhibited more missense variants than synonymous variants (3,663 versus 2,926; Figure 2B), whereas the DNA-binding and transactivation domains displayed fewer missense variants than synonymous variants (12 versus 216 and 17 versus 61, respectively). These differences, however, were not statistically significant in two-sample t tests, likely because the numbers of variants varied greatly per amino acid. Similar tests performed for the percentages of residues per domain featuring at least one variant also failed to detect statistically significant differences for synonymous variants between the HMG, transactivation, and other domains (Figure 2C). In contrast, the percentages of residues with at least one missense variant were significantly much lower in the HMG domain (p = 0.004) and lower in the transactivation domain (p = 0.024) than in the other domains. Together with the fact that each of the 12 missense variants located in the HMG domain was detected in a single individual and in the heterozygous state, these data suggest that genomic variants in the SOX4 HMG domain might seldom be compatible with normal development, even in the heterozygous state. Supporting this conclusion, no deletions of SOX4 were identified in a recently published copy-number-variant map of the human genome in normal individuals.36
Figure 2.
Analysis of SOX4 Missense Variants in the gnomAD Control Cohort
(A) Bar graphs showing the numbers of synonymous and missense SOX4 variants detected in gnomAD for each amino acid of the SOX4 protein sequence. The HMG domain is highlighted in yellow and the transactivation domain in gray.
(B) Comparison of the numbers of synonymous and missense gnomAD variants in the different SOX4 domains. Columns correspond to the N-terminal and central region (Nt + Ce), HMG domain (HMG), and transactivation domain (TA). Values are indicated at the top of each column.
(C) Percentages of residues per SOX region that feature at least one gnomAD variant. The p values obtained in two-sample t tests for the comparison of regions is shown.
SOX4 Is Strongly Expressed in Actively Developing Regions of the Human Brain
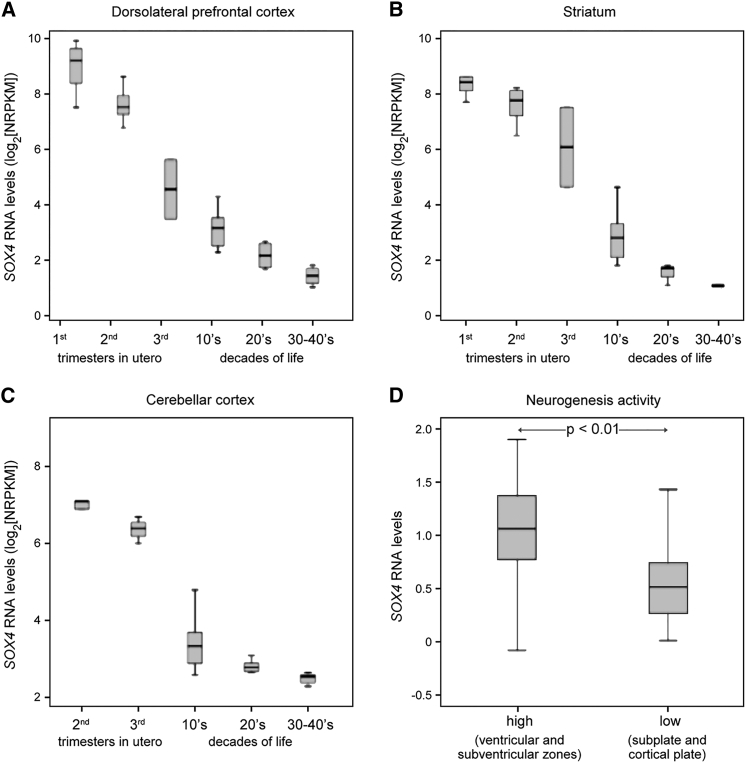
To add support to the proposition that SOX4 may be critical for human brain development, we examined its gene expression in specific regions of the human brain using RNA-seq and RNA-microarray data available in the BrainSpan Atlas of the Developing Human Brain.32 SOX4 expression was found to be high in all brain regions examined (dorsolateral prefrontal cortex, striatum, and cerebellar cortex) during the first two trimesters of embryonic gestation, and then to decrease progressively to reach a very low level by the 3rd and 4th decades of postnatal life (Figures 3A–3C). SOX4 expression was higher in areas of very active neurogenesis, including the ventricular and subventricular zones, than in less active areas, such as the cortical plate and subplate (Mann-Whitney U-test p < 0.01) (Figure 3D). While not proving that SOX4 has important functions in human brain development, these expression data nevertheless constitute a necessary argument to support the notion that SOX4 may directly control the development of several regions of the human brain.
Figure 3.
SOX4 Transcript Levels in the Developing and Adult Human Brain
(A–C) Changes in SOX4 expression levels during development and adult life in the dorsolateral prefrontal cortex, striatum, and cerebellar cortex, as assessed by RNA-seq. The first three samples were obtained during the three trimesters of fetal development, and the next three during the first four decades of life, as indicated on the x axis. No data were available for the cerebellar cortex in the first trimester of embryogenesis.
(D) RNA microarray data demonstrating that SOX4 expression is significantly higher in neuroanatomical regions with high-level neurogenesis (ventricular and subventricular zones) than in regions with low-level neurogenesis (subplate and cortical plate) at 21 weeks of gestation (Mann-Whitney U-test, p < 0.01).
Error bars in the standard box plots from PASW/SPSS represent the interquartile range.
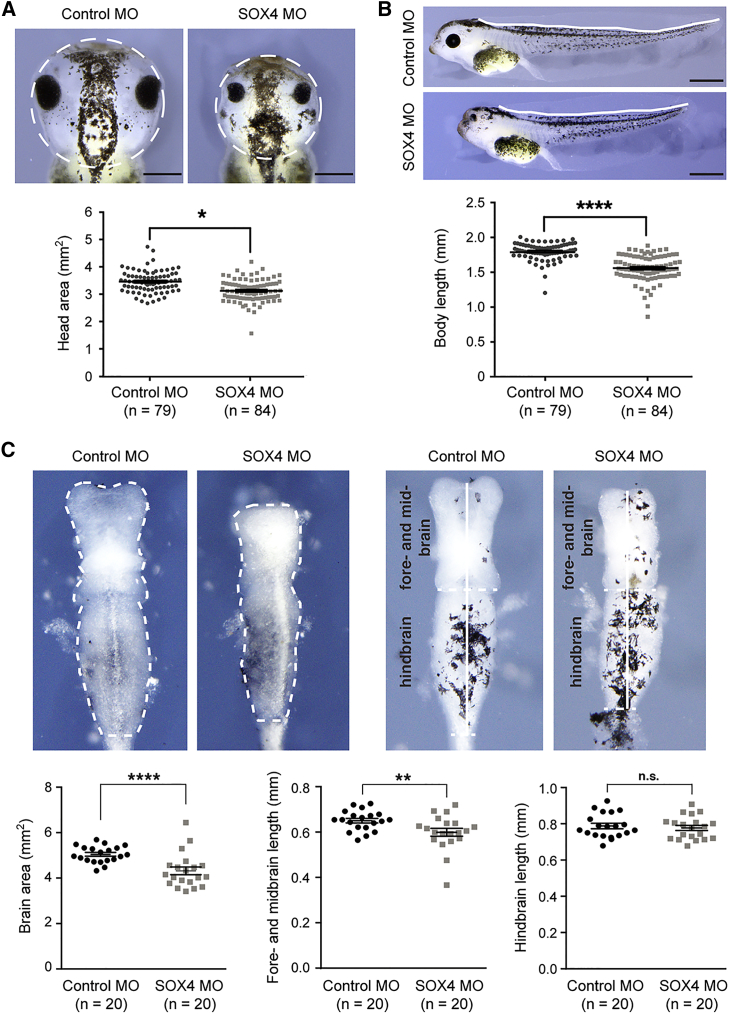
sox4 Knockdown in Xenopus Embryos Interferes with Brain and Whole-Body Development
To functionally test the importance of SOX4 in brain development in vivo, we knocked down its ortholog in Xenopus laevis embryos using a well-described morpholino oligonucleotide (MO).19 We injected either a control or a sox4 MO in the dorso-animal blastomeres of 8-cell-stage embryos to target anterior neural tissue.34 We previously reported that sox4 morphants had microphthalmia at stage 43.19 Further analysis revealed that they also had microcephaly and small bodies (Figures 4A and 4B). The reduced size of the brain in mutants compared to controls reflected underdevelopment of the forebrain and midbrain, but not hindbrain (Figure 4C). These data thus suggest that SOX4 may also significantly participate in neurogenesis and other aspects of embryonic development in humans.
Figure 4.
Effect of sox4 Knockdown on Xenopus laevis Embryo Development
(A) Top, representative pictures of stage-43 Xenopus embryos showing that bilateral injection of sox4 MO leads to a smaller head area (white dotted circles) and to microphthalmia compared to bilateral injection of control MO. Bottom, graph showing quantification of the head area for all tested embryos. n, number of embryos. The p value was calculated by a non-parametric Mann-Whitney rank sum test. ∗p ≤ 0.05. Scale bar, 1 mm.
(B) Sox4 depletion results in shorter body length. Data are presented as in (A). ∗∗∗∗p ≤ 0.0001. Scale bar, 1 mm.
(C) Sox4 deficiency impairs brain development. Top left, pictures showing that sox4 MO injections lead to a small brain area (dotted line). Top right, pictures showing that Sox4 depletion results in underdevelopment of the fore- and mid-brain, but not hindbrain (white vertical lines separated by a horizontal dotted line). Bottom panels, data quantification performed as in (A). n, number of embryos. ∗∗∗∗p ≤ 0.0001; ∗∗p ≤ 0.01; n.s., not significant.
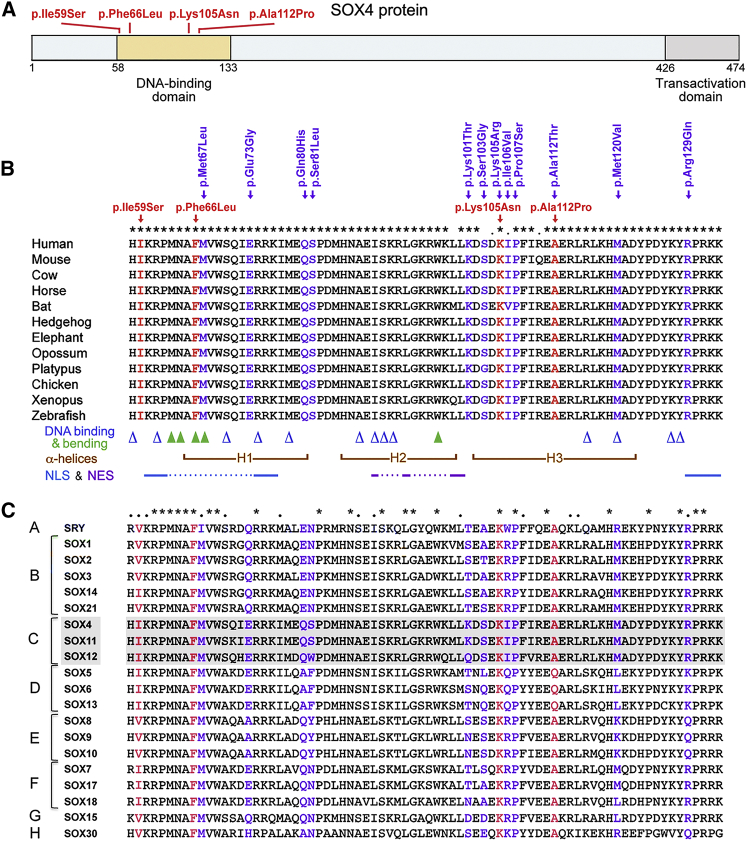
The Subjects’ SOX4 Missense Variants Cluster in the HMG Domain and Are Not Found in Control Individuals
The SOX4 missense variants identified in the four subjects were all located in the HMG domain (Figure 5A). This apparent clustering of variants was striking considering that missense variants in this domain were found to be significantly underrepresented in the control human population. We therefore asked whether it was also statistically significant. We compared the locations of the subjects’ missense variants to the location of gnomAD missense variants using CLUMP.27 Since our four case subjects’ variants were novel, we used only singleton variants as controls. Upon performing 10K permutations and correction for multiple testing with the Benjamini-Hochberg method, we obtained a statistically significant difference (p value = 0.017) between the average score of the four case subjects (1.04) and that of control subjects (3.73). This result thus supports the proposition that disease-causing SOX4 variants cluster in the HMG domain.
Figure 5.
Analysis of the Location of SOX4 Missense Variants Detected in Subjects and in gnomAD Individuals
(A) Schematic of the human SOX4 protein showing the location of the four subject missense variants in the HMG domain. Variants are shown in red and their positions are marked with bars. Numbers indicate the position of amino acids in the protein sequence.
(B) Alignment of the SOX4 HMG-domain sequences from various vertebrate species. Sequence accession numbers are listed in Table S1. SOX4 variants found in the study subjects and in the gnomAD control cohort are shown in red and purple, respectively, above the sequences, and amino acids matching the variants are similarly colored in the aligned sequences. Above the sequences, symbols denote fully conserved (asterisks) and semi-conserved (dots) amino acids. Below the sequences, residues important for DNA binding and bending are shown with blue open triangles and green closed triangles, respectively. Brown brackets demarcate the three α helices (H1, H2, and H3) that secondarily structure the DNA-binding domain. Key amino acids in the N-terminal and C-terminal nuclear localization signal sequences (NLS) and nuclear export signal sequence (NES) are shown with continued lines and are linked with dotted lines.
(C) Comparison of the HMG domain sequences of all human SOX proteins. Sequence accession numbers are listed in Table S2.
The four subjects’ variants were distinct from those reported in gnomAD and they affected residues fully conserved in SOX4 vertebrate orthologs (Figure 5B). Moreover, Phe66 and Lys105 are conserved in all human SOX proteins; Ala112 is replaced by Gln in the SOXD group, but is otherwise conserved in all human SOX proteins; and Ile59 is fully or semi-conserved in all human SOX proteins (Figure 5C). Two of the twelve gnomAD variants affected the same residues as those in subjects 2 and 4, but they were different (p.Lys105Arg instead of p.Lys105Asn and p.Ala112Thr instead of p.Ala112Pro); two others also affected highly conserved residues (p.Met67Leu and p.Pro107Ser); but the other eight affected residues are poorly conserved in the SOX family (Figures 5B and 5C). Further, three of the latter variants matched wild-type residues in other SOX proteins (p.Lys101Thr, p.Ser103Gly, and p.Arg129Gln). Thus, unlike the case subjects’ variants, only a subset of gnomAD variants affected highly conserved residues.
Since variants in other SOX HMG domains have been associated with disease, we asked whether some of them matched those detected in our subjects (Table 2). SRY variants in residues equivalent to those affected in our subjects were shown to cause disease, but only one (p.Phe112Leu) fully matched a SOX4 variant (p.Phe66Leu). Variants in residues equivalent to Phe66, Ala112, and Lys105 in SOX4 were also shown in SOX genes located on autosomal chromosomes, namely SOX9, SOX10, or SOX11, to cause disease at the heterozygous state. Again, only one (p.Lys150Asn in SOX10) resulted in the same substitution as in SOX4 (p.Lys105Asn). No disease-causing mutation in the residue equivalent to Ile59 in SOX4 has been reported for SOX genes located on autosomal chromosomes. Since p.Ile59Ser was detected in the least affected child (subject 3), it is conceivable that a heterozygous missense mutation of this residue might also be on the benign or mild-disease side for other SOX genes. We performed the same analysis for the 12 SOX4 HMG-domain gnomAD variants (Table S4). In brief, five SOX4 gnomAD variants affected residues that had no reported variant or only gnomAD variants in other SOX genes. The seven others had at least one disease-causing variant in another SOX gene, but only one was a full match: p.Ala112Thr in SOX4, p.Ala113Thr in SRY, and p.Ala158Thr in SOX9. Collectively, these findings support the notion that the four subjects’ SOX4 variants are likely pathogenic and that at least a subset of SOX4 gnomAD variants might also be pathogenic. This conclusion for the latter is plausible since the gnomAD database was established by excluding individuals with severe pediatric disease, leaving open the possibility that it includes individuals with mild, unreported disease.
Table 2.
Pathological Missense Variants in SRY and Other SOX Protein Residues Matching the SOX4 Variants Identified in Four Individuals with ID in This Study
| Protein | Variant | Phenotype | Reference |
|---|---|---|---|
| SOX4 | p.Phe66Leu | neurodevelopmental syndrome | this study |
| SRY | p.Phe67Val | gonadal dysgenesis, XY sex reversal | Scherer et al.48 |
| SOX9 | p.Phe112Leu | campomelic dysplasia | Kwok et al.9 |
| SOX10 | p.Phe111Val | Kallmann syndrome | Pingault et al.49 |
| SOX4 | p.Ala112Pro | neurodevelopmental syndrome | this study |
| SRY | p.Ala113Thr | gonadal dysgenesis | Zeng et al.50 |
| SOX9 | p.Ala158Thr | campomelic dysplasia, XY sex reversal | Preiss et al.51 |
| SOX9 | p.Ala158Val | campomelic dysplasia | Karaer et al.52 |
| SOX10 | p.Ala157Val | Waardenburg syndrome type IV | Morín et al.53 |
| SOX11 | p.Ala102Val | Coffin-Siris syndrome | Okamoto et al.54 |
| SOX4 | p.Ile59Ser | neurodevelopmental syndrome | this study |
| SRY | p.Val60Leu | XY sex reversal | Harley et al.55 |
| SOX4 | p.Lys105Asn | neurodevelopmental syndrome | this study |
| SRY | p.Lys106Ile | gonadal dysgenesis | Pontiggia et al.56 |
| SOX10 | p.Lys150Asn | Hirschsprung’s disease | Chaoui et al.57 |
In Silico Analyses Predict Damaging Structural Consequences for the Four Subjects’ SOX4 variants
The crystal structure of the SOX4 HMG domain bound to DNA was previously solved and shown to be very similar to that of other SOX proteins.37 Complementary assays in vitro for several SOX proteins have validated and extended the acquired knowledge by demonstrating which residues are critical for DNA binding, DNA bending, and protein shuttling between the cytoplasm and nucleus.7 Valuable analyses can therefore be performed in silico to predict the consequences of missense variants on the structure and hence the function of the SOX4 HMG domain. The phenylalanine changed into leucine in subject 1 (p.Phe66Leu) belongs to the so-called FM wedge, a protein motif intercalating within the minor groove of DNA and conferring on the SOX domain one of its characteristic properties, which is to bend the DNA helix (Figures S1A and S1B).37 Both phenylalanine and leucine are hydrophobic, but leucine has a short aliphatic chain whereas phenylalanine has a large aromatic chain (Figure S1C). HOPE30 predicted that this chain difference could disrupt protein function. The alanine residue changed into proline in subject 2 (p.Ala112Pro) is located within the third of three α helices that confer on the SOX domain an L-shape essential for DNA binding (Figures S1A and S1B). The replacement of a small hydrophobic amino acid by a larger, more rigid residue was predicted to remove a hydrogen bond and thereby to destabilize the helix (Figure S1D). The isoleucine residue changed into serine in subject 3 (p.Ile59Ser) occupies the second position in the N terminus of the SOX domain, close to two residues critical for DNA binding (His58 and Arg61). The p.Ile59Ser variant introduces a residue that is smaller and less hydrophobic than the wild-type residue and that might therefore destabilize the local β strand configuration and disrupt hydrophobic interactions in the core of the protein or on its surface (Figure S1E). The lysine residue changed into asparagine in subject 4 (p.Lys105Asn) is located in the middle of the third α helix of the SOX domain (Figures S1A and S1B). The replacement of a large, positively charged residue by a smaller, neutral residue was predicted to disrupt a salt bridge and hence the wild-type α-helical structure (Figure S1F).
In line with the HOPE predictions, PolyPhen-229 foresaw that three variants would be highly damaging (p.Ile59Ser: score of 0.997; p.Ala112Pro: score of 1; p.Lys105Asn: score of 0.99) and that the fourth one could be damaging (p.Phe66Leu: score of 0.499).
We applied the same prediction tools to the 12 SOX4 gnomAD variants. In brief, considering the location of the residues in the HMG domain with respect to the functional motifs (Figure S2A), the conservation of the residues in SOX proteins and the change in side-chain type caused by the variants (Figures S2B and S2C), three variants stood out as being most likely to be pathogenic: p.Met67Leu, p.Pro107Ser, and p.Ala112Thr. It is worth noting that all three residues cause disease when mutated in other SOX proteins but, as mentioned earlier, only p.Ala112Thr has known disease-causing matches (Table S4).
In conclusion, in silico tools predict that all SOX4 subject variants, but only a subset of gnomAD variants, could damage the structure and hence the activity of the SOX4 HMG domain.
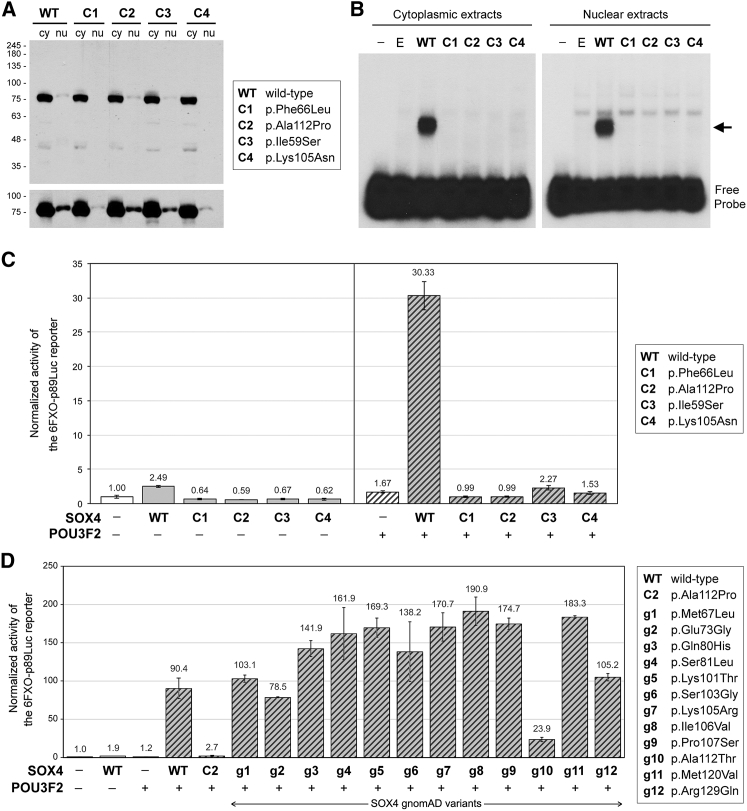
The Four Subjects’ SOX4 Variants Are Unable to Bind DNA and Transactivate
To test the functional impact of our subjects’ missense mutations, we generated a mammalian expression plasmid for each variant. When transiently transfected into COS-1 cells, the plasmids for the wild-type and four variants led to similar amounts of SOX4 in the cytoplasmic and nuclear compartments (Figure 6A), indicating that the missense mutations did not affect SOX4 synthesis, stability, and nuclear translocation. We then performed electrophoretic mobility shift assays to test the ability of the variants to bind DNA. We used an FXO DNA probe previously shown to bind SOX4 efficiently.12 This probe corresponds to a minimal FGF4 enhancer sequence (F) and features a SOX-binding site (X) adjacent to a POU-domain-binding site (O).38 Wild-type SOX4 formed a stable complex with the probe, as expected, but none of the variants did (Figure 6B). This result implies that the mutations sufficiently alter the structure of the HMG domain to prevent the variant proteins from binding to DNA in vitro.
Figure 6.
Consequences of Missense Mutations on SOX4 Protein Level and Activity
(A) Western blot of cytoplasmic (cy) and nuclear (nu) extracts from COS-1 cells transiently transfected with expression plasmids encoding wild-type (WT) or variant SOX4 proteins (C1 to C4) fused at the N terminus with a 3FLAG epitope. The proteins were identified using anti-FLAG antibody. The Mr of protein standards is indicated in k units. Note that each SOX4 protein exhibited the expected Mr of approximately 75 k. The bottom panel is a longer exposure of the same blot as in the top panel. It is limited to the SOX4 protein region and shows that SOX4 protein was present in all nuclear extracts, but at a lower level than in cytoplasmic extracts. Variant SOX4 proteins did not show significant differences in nuclear localization compared to wild-type SOX4 when several independent experiments were considered.
(B) EMSA comparing the binding efficiency of SOX4 wild-type and variants. The cell extracts were the same as in (A), but also included negative controls without SOX4 (−, no plasmid; E, empty plasmid). The arrow indicates the complex formed between wild-type SOX4 and the DNA probe.
(C) Assay of the ability of the four case subjects’ SOX4 variants to activate transcription. COS-1 cells were transiently transfected with a 6FXO-p89-Luc reporter, a pSV-beta-galactosidase plasmid, and expression plasmids for wild-type (WT) or variant SOX4 (C1 to C4), and POU3F2. Reporter activities are presented as the mean ± standard deviation obtained from triplicates for each condition. They were normalized for transfection efficiency and are reported as fold increase relative to the activity of the reporter in the absence of SOX4 and POU3F2. The presence (+) or absence (−) of SOX4 and POU3F2 plasmid is indicated beneath the bars. These data were reproduced in more than three independent experiments.
(D) Assay of the ability of the 12 gnomAD SOX4 HMG-domain missense variants (g1 to g12) to activate transcription. COS-1 cells were transfected as described in (C) and using SOX4 and POU3F2 expression plasmids as shown in the figure. Data were calculated and are presented as in (C). They were reproduced in four independent experiments. A western blot showing that similar amounts of SOX4 protein were made in all conditions is presented in Figure S3.
We next asked whether the variants could activate transcription in intact cells. We transiently transfected COS-1 cells with a reporter plasmid containing six tandem copies of the FXO sequence12 along with plasmids encoding no protein (empty), SOX4 wild-type or variants, and POU3F2 (also known as BRN-2). As shown previously,12 wild-type SOX4 and POU3F2 activated the reporter weakly when expressed individually and synergized to reach a high level of activation when co-expressed (Figure 6C). In contrast, none of the SOX4 variants could transactivate the reporter by itself or in synergy with POU3F2.
We then tested the SOX4 gnomAD variants in our reporter assay. Like wild-type SOX4, all 12 variants were well expressed in COS-1 cells (Figure S3). Interestingly, 11 of them were as competent or up to twice as competent as wild-type SOX4 in activating the reporter in synergy with POU3F2, whereas p.Ala112Thr was 4-fold less efficient than wild-type SOX4 but was still nine times as active as the p.Ala112Pro variant found in subject 2 (Figure 6D).
These functional assays in vitro thus suggest that our four subjects developed a disease due at least in part to the inability of their SOX4 variant to function as transcription factors. They also suggest that the individuals carrying the gnomAD variants had normal development, except perhaps the p.Ala112Thr carrier.
Discussion
The present study links SOX4 variants to a human developmental disease. We reported four unrelated children who presented with global developmental delay and intellectual disability associated with distinctive dysmorphic features. Each child was heterozygous for a different de novo SOX4 missense variant and none of these variants was detected in a large cohort of presumably healthy individuals. They significantly clustered in the HMG domain and were predicted in silico to drastically alter the structure of this domain. Functional assays in vitro revealed their inability to bind DNA and activate transcription. Based on these data and evidence that SOX4 is highly expressed in the developing human brain and is necessary in animal models for brain development, we propose that reduced expression of SOX4 target genes at critical points in embryonic and early postnatal development of the subjects led to the neurodevelopmental syndrome and associated dysmorphic features.
The HMG domain is the common feature and primary functional region of all SOX proteins.7, 39 Almost 50% of the 76 residues that compose this domain are conserved in all 20 human SOX proteins.40, 41, 42 These residues confer on the proteins their abilities to bind and bend DNA, to shuttle between the cytoplasm and nucleus, and to interact with other proteins. Numerous missense variants in SRY and other SOX genes have been associated with severe disease in humans, and in most cases, the variants were located in the HMG domain. Thus, a first and solid hint of possible pathogenicity was the fact that our subjects’ SOX4 variants were located in the HMG domain and affected critical residues, whereas SOX4 variants in this domain are significantly underrepresented in the general population. The most solid piece of evidence that we obtained in favor of pathogenicity was that none of our four subjects’ variants was able to transactivate a reporter gene in vitro, whereas 11 of the 12 variants detected in the HMG domain in gnomAD exhibited similar activity as wild-type SOX4, and the 12th retained significant activity compared to the subjects’ variants. In silico analyses of structural consequences of the variants had predicted damaging consequences for all subjects’ variants, but also for several gnomAD variants. Our reporter assay thus appears to be a more reliable predictor of pathogenicity for SOX4 variants in the HMG domain than current in silico tools. As genetic testing becomes more and more customary in the future, we surmise that more SOX4 variants will be identified in individuals with neurodevelopmental disease and possibly other disorders and that our reporter assay will be instrumental in helping discern pathogenic from nonpathogenic mutations.
The clinical features of our subjects suggest that the spectrum of severity of the neurodevelopmental disorder due to SOX4 variants is wide. No other disruptive variants that could explain the wide range of disease severity were identified in these children. Genotype-phenotype correlations are not evident at the moment and, as in many other genetic disorders, the causes of the wide range of clinical severity have yet to be elucidated. We envision two possible scenarios. The first is that the SOX4 variants may have different impacts in vivo, despite having similar damaging consequences in our functional assays in vitro. The second is that variants in other genes, which could have been present but were not flagged as possibly pathogenic, or variants possibly present in gene regulatory regions outside the exome may aggravate or lessen the impact of SOX4 variants on development. The analysis of multiple children carrying the same and additional SOX4 variants should help answer this question. Also, the sophistication of in silico prediction tools and the refinement of functional assays in animal models and in vitro, which would test SOX4 variants in a more physiological context, should also be helpful.
The neurodevelopmental defects of our four case subjects were detected from early infancy and were consistent with impaired development of the cerebral cortex, a region of the brain considerably more developed in humans than in other mammals and involved in higher-order functions such as thinking, cognition, memory, attention, and language. Neurogenesis is largely complete by mid-gestation, and a lifetime maximum number of neuronal connections are established by 3 years of age.43 In keeping with the proposition that SOX4 is likely to have important roles in development of the human cerebral cortex, we showed that SOX4 transcript levels were highest in the brain during early fetal development, steadily declined during postnatal life, and were lowest past the age of 20 years. We also showed that the expression of SOX4 in the brain of 21-week-old fetuses was highest in neurogenic niches.
All four subjects were small for their age and exhibited mild but distinct facial dysmorphism and fifth finger clinodactyly. Their small stature could have several origins since SOX4 was shown in animal models to have important roles in multiple processes. These processes include skeletogenesis and, strikingly, the morphological parameters of our children bear resemblance with those of mice lacking the SOXC genes specifically in skeletal cells. Inactivation of Sox4, Sox11, or both genes in differentiated chondrocytes impaired skeletal growth.44 Inactivation of either Sox4 or Sox11 in skeletal progenitor cells affected growth.23 Facial and digital defects matching those of our subjects were not noted but might have been overlooked. Notably, inactivation of both Sox4 and Sox11 in skeletal progenitor cells resulted in complete failure of skeletal primordia growth, articulation, and ossification. Clinodactyly, craniofacial dysmorphism, and short stature of our subjects could thus be due to a reduction in the global activity of SOX4 in skeletal cells.
The first process discovered to be impacted by Sox4 inactivation in the mouse is outflow track formation.20 Sox4-null mouse embryos indeed die in utero from heart septation defects known as common arterial trunk in humans. One can thus postulate that a reduction in SOX4 activity due to haploinsufficiency or another mechanism contributed to the peri-membranous ventricular septal defect detected in our subject 1. This abnormality was not reported in the other individuals, nor was it reported in Sox4+/− mice, suggesting that its penetrance is influenced by genetic background. With this in mind, as well as evidence from animal studies that SOX4 impacts many processes, close follow-up of individuals with SOX4 variants predicted to be damaging should be recommended to determine whether the variants predispose to cardiac and other problems besides neurological and skeletal issues in development and beyond.
The target genes of SOX4 in the development of the brain, skeleton, and other organs have not yet been fully defined. In mouse intermediate cortex progenitor cells, SOX4 was shown to transactivate Tbr2, a gene required in the mouse to produce adequate numbers of neurons in each cortical layer.2, 45 In cultured mouse neuronal stem cells, SOX4 was able to activate a DCX (MIM: 300121) reporter gene.14 DCX mutations are associated in humans with cortical malformations.46 Acting largely in redundancy, mouse SOX4 and SOX11 were found to activate a transcriptional program critical to specify the identity and connectivity of corticospinal neurons.16 They were also shown to control skeletal cell fate and differentiation by promoting signaling pathways of major importance in many processes, including canonical and non-canonical WNT signaling.23, 44 A genome-wide analysis of SOX4 targets in prostate cancer cells identified multiple genes of potential relevance to neuronal, skeletal, and many other processes.47 For example, SOX4 was shown to regulate WDR45 (MIM: 300526), a gene associated with neurodegeneration and iron accumulation in the brain; multiple components of the RNA-induced silencing (RISC) complex; and several genes involved in the transforming growth factor-β, Hedgehog, Notch, and WNT pathways. SOX4 variants thus have a clear potential to affect the expression of important genes in neurodevelopment, skeletogenesis, and other processes.
The close relationships existing between SOX4 and SOX11 make it relevant to compare the current work with earlier reports that associated SOX11 mutations with a form of mild Coffin-Siris syndrome.24, 25 For both SOX4 and SOX11, missense variants found in subjects were located in the HMG domain25 and functional studies indicated that the variant proteins lacked transcriptional activity. In both cases, individuals exhibited mild to severe intellectual disability, growth deficiency, specific dysmorphic facial features, and fifth-finger clinodactyly. Several SOX11 subjects were also described to have hypoplastic fifth-toe nails, which was seen in one of our SOX4 individuals, and syndactyly of the 2nd and 3rd toes, which was not seen in any of our case subjects. Thus, SOX4 and SOX11 mutations result in a series of similar clinical characteristics, but not all defects are alike in nature, penetrance, and severity. This finding is consistent with the overlapping expression patterns of the human SOX4 and SOX11 genes in development, and with the similar but not identical activities of the SOX4 and SOX11 proteins. A full description and comparison of the clinical phenotypes of larger cohorts of individuals will be helpful in the future to more precisely define the extent of neurodevelopmental, skeletal, and other clinical features caused by pathogenic variants in SOX4 and SOX11 and the degree of similarities and dissimilarities between the two types of diseases. Based on all data currently available, we propose that non-functional variants in SOX4 and SOX11 may cause a novel class of syndromes that have overlapping clinical features, notably in neurodevelopment and skeletogenesis.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
We thank the children and their families who contributed to this study. We also thank Okan Elci for help with statistical analyses. Sequencing was provided by the University of Washington Center for Mendelian Genomics (UW-CMG) and was funded by National Institutes of Health NHGRI and NHLBI (UM1 HG006493 and U24 HG008956 grants to the UW-CMG and to the CMG coordinating center). The DDD study presents independent research commissioned by the Health Innovation Challenge Fund (grant number HICF-1009-003), a parallel funding partnership between Wellcome and the Department of Health, and the Wellcome Sanger Institute (grant number WT098051). The research team acknowledges the support of the National Institute for Health Research, through the Comprehensive Clinical Research Network. This study makes use of DECIPHER, which is funded by the Wellcome. Part of this study was conducted in the former Lefebvre laboratory at and funded by the Cleveland Clinic Lerner Research Institute (Chair’s Innovative Research Award to V.L.). Funding was also received from the National Institutes of Health NIAMS (AR68308 grant to V.L.). The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies.
Published: January 17, 2019; corrected online: January 29, 2019
Footnotes
Supplemental Data include three figures, four tables, and Supplemental Note and can be found with this article online at https://doi.org/10.1016/j.ajhg.2018.12.014.
Contributor Information
Claudio Graziano, Email: claudio.graziano@unibo.it.
Véronique Lefebvre, Email: lefebvrev1@email.chop.edu.
Web Resources
CLUMP, https://omictools.com/clump-tool https://omictools.com/clump-tool
DECIPHER, https://decipher.sanger.ac.uk/
DenovolyzeR, http://denovolyzer.org
ExAC Browser, http://exac.broadinstitute.org/
gnomAD Browser, http://gnomad.broadinstitute.org/
OMIM, http://www.omim.org/
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
SWISS-MODEL, https://swissmodel.expasy.org
Supplemental Data
References
- 1.Urbán N., Guillemot F. Neurogenesis in the embryonic and adult brain: same regulators, different roles. Front. Cell. Neurosci. 2014;8:396. doi: 10.3389/fncel.2014.00396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen C., Lee G.A., Pourmorady A., Sock E., Donoghue M.J. Orchestration of neuronal differentiation and progenitor pool expansion in the developing cortex by SoxC genes. J. Neurosci. 2015;35:10629–10642. doi: 10.1523/JNEUROSCI.1663-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lefebvre V., Bhattaram P. Vertebrate skeletogenesis. Curr. Top. Dev. Biol. 2010;90:291–317. doi: 10.1016/S0070-2153(10)90008-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kozhemyakina E., Lassar A.B., Zelzer E. A pathway to bone: signaling molecules and transcription factors involved in chondrocyte development and maturation. Development. 2015;142:817–831. doi: 10.1242/dev.105536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Warman M.L., Cormier-Daire V., Hall C., Krakow D., Lachman R., LeMerrer M., Mortier G., Mundlos S., Nishimura G., Rimoin D.L. Nosology and classification of genetic skeletal disorders: 2010 revision. Am. J. Med. Genet. A. 2011;155A:943–968. doi: 10.1002/ajmg.a.33909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wright C.F., Fitzgerald T.W., Jones W.D., Clayton S., McRae J.F., van Kogelenberg M., King D.A., Ambridge K., Barrett D.M., Bayzetinova T., DDD study Genetic diagnosis of developmental disorders in the DDD study: a scalable analysis of genome-wide research data. Lancet. 2015;385:1305–1314. doi: 10.1016/S0140-6736(14)61705-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kamachi Y., Kondoh H. Sox proteins: regulators of cell fate specification and differentiation. Development. 2013;140:4129–4144. doi: 10.1242/dev.091793. [DOI] [PubMed] [Google Scholar]
- 8.Nikolova G., Vilain E. Mechanisms of disease: Transcription factors in sex determination--relevance to human disorders of sex development. Nat. Clin. Pract. Endocrinol. Metab. 2006;2:231–238. doi: 10.1038/ncpendmet0143. [DOI] [PubMed] [Google Scholar]
- 9.Kwok C., Weller P.A., Guioli S., Foster J.W., Mansour S., Zuffardi O., Punnett H.H., Dominguez-Steglich M.A., Brook J.D., Young I.D. Mutations in SOX9, the gene responsible for Campomelic dysplasia and autosomal sex reversal. Am. J. Hum. Genet. 1995;57:1028–1036. [PMC free article] [PubMed] [Google Scholar]
- 10.Pingault V., Bondurand N., Kuhlbrodt K., Goerich D.E., Préhu M.O., Puliti A., Herbarth B., Hermans-Borgmeyer I., Legius E., Matthijs G. SOX10 mutations in patients with Waardenburg-Hirschsprung disease. Nat. Genet. 1998;18:171–173. doi: 10.1038/ng0298-171. [DOI] [PubMed] [Google Scholar]
- 11.Lamb A.N., Rosenfeld J.A., Neill N.J., Talkowski M.E., Blumenthal I., Girirajan S., Keelean-Fuller D., Fan Z., Pouncey J., Stevens C. Haploinsufficiency of SOX5 at 12p12.1 is associated with developmental delays with prominent language delay, behavior problems, and mild dysmorphic features. Hum. Mutat. 2012;33:728–740. doi: 10.1002/humu.22037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dy P., Penzo-Méndez A., Wang H., Pedraza C.E., Macklin W.B., Lefebvre V. The three SoxC proteins--Sox4, Sox11 and Sox12--exhibit overlapping expression patterns and molecular properties. Nucleic Acids Res. 2008;36:3101–3117. doi: 10.1093/nar/gkn162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoser M., Potzner M.R., Koch J.M., Bösl M.R., Wegner M., Sock E. Sox12 deletion in the mouse reveals nonreciprocal redundancy with the related Sox4 and Sox11 transcription factors. Mol. Cell. Biol. 2008;28:4675–4687. doi: 10.1128/MCB.00338-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mu L., Berti L., Masserdotti G., Covic M., Michaelidis T.M., Doberauer K., Merz K., Rehfeld F., Haslinger A., Wegner M. SoxC transcription factors are required for neuronal differentiation in adult hippocampal neurogenesis. J. Neurosci. 2012;32:3067–3080. doi: 10.1523/JNEUROSCI.4679-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bhattaram P., Penzo-Méndez A., Sock E., Colmenares C., Kaneko K.J., Vassilev A., Depamphilis M.L., Wegner M., Lefebvre V. Organogenesis relies on SoxC transcription factors for the survival of neural and mesenchymal progenitors. Nat. Commun. 2010;1:9. doi: 10.1038/ncomms1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shim S., Kwan K.Y., Li M., Lefebvre V., Sestan N. Cis-regulatory control of corticospinal system development and evolution. Nature. 2012;486:74–79. doi: 10.1038/nature11094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sock E., Rettig S.D., Enderich J., Bösl M.R., Tamm E.R., Wegner M. Gene targeting reveals a widespread role for the high-mobility-group transcription factor Sox11 in tissue remodeling. Mol. Cell. Biol. 2004;24:6635–6644. doi: 10.1128/MCB.24.15.6635-6644.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen J., Ju H.L., Yuan X.Y., Wang T.J., Lai B.Q. SOX4 is a potential prognostic factor in human cancers: a systematic review and meta-analysis. Clin. Transl. Oncol. 2016;18:65–72. doi: 10.1007/s12094-015-1337-4. [DOI] [PubMed] [Google Scholar]
- 19.Cizelsky W., Hempel A., Metzig M., Tao S., Hollemann T., Kühl M., Kühl S.J. sox4 and sox11 function during Xenopus laevis eye development. PLoS ONE. 2013;8:e69372. doi: 10.1371/journal.pone.0069372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schilham M.W., Oosterwegel M.A., Moerer P., Ya J., de Boer P.A., van de Wetering M., Verbeek S., Lamers W.H., Kruisbeek A.M., Cumano A., Clevers H. Defects in cardiac outflow tract formation and pro-B-lymphocyte expansion in mice lacking Sox-4. Nature. 1996;380:711–714. doi: 10.1038/380711a0. [DOI] [PubMed] [Google Scholar]
- 21.Bergsland M., Werme M., Malewicz M., Perlmann T., Muhr J. The establishment of neuronal properties is controlled by Sox4 and Sox11. Genes Dev. 2006;20:3475–3486. doi: 10.1101/gad.403406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lefebvre V., Bhattaram P. SOXC genes and the control of skeletogenesis. Curr. Osteoporos. Rep. 2016;14:32–38. doi: 10.1007/s11914-016-0296-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhattaram P., Penzo-Méndez A., Kato K., Bandyopadhyay K., Gadi A., Taketo M.M., Lefebvre V. SOXC proteins amplify canonical WNT signaling to secure nonchondrocytic fates in skeletogenesis. J. Cell Biol. 2014;207:657–671. doi: 10.1083/jcb.201405098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsurusaki Y., Koshimizu E., Ohashi H., Phadke S., Kou I., Shiina M., Suzuki T., Okamoto N., Imamura S., Yamashita M. De novo SOX11 mutations cause Coffin-Siris syndrome. Nat. Commun. 2014;5:4011. doi: 10.1038/ncomms5011. [DOI] [PubMed] [Google Scholar]
- 25.Hempel A., Pagnamenta A.T., Blyth M., Mansour S., McConnell V., Kou I., Ikegawa S., Tsurusaki Y., Matsumoto N., Lo-Castro A., DDD Collaboration Deletions and de novo mutations of SOX11 are associated with a neurodevelopmental disorder with features of Coffin-Siris syndrome. J. Med. Genet. 2016;53:152–162. doi: 10.1136/jmedgenet-2015-103393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ware J.S., Samocha K.E., Homsy J., Daly M.J. Interpreting de novo variation in human disease using denovolyzeR. Curr. Protoc. Hum. Genet. 2015;87:1–15. doi: 10.1002/0471142905.hg0725s87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turner T.N., Douville C., Kim D., Stenson P.D., Cooper D.N., Chakravarti A., Karchin R. Proteins linked to autosomal dominant and autosomal recessive disorders harbor characteristic rare missense mutation distribution patterns. Hum. Mol. Genet. 2015;24:5995–6002. doi: 10.1093/hmg/ddv309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Venselaar H., Te Beek T.A., Kuipers R.K., Hekkelman M.L., Vriend G. Protein structure analysis of mutations causing inheritable diseases. An e-Science approach with life scientist friendly interfaces. BMC Bioinformatics. 2010;11:548. doi: 10.1186/1471-2105-11-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bienert S., Waterhouse A., de Beer T.A., Tauriello G., Studer G., Bordoli L., Schwede T. The SWISS-MODEL Repository-new features and functionality. Nucleic Acids Res. 2017;45(D1):D313–D319. doi: 10.1093/nar/gkw1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller J.A., Ding S.L., Sunkin S.M., Smith K.A., Ng L., Szafer A., Ebbert A., Riley Z.L., Royall J.J., Aiona K. Transcriptional landscape of the prenatal human brain. Nature. 2014;508:199–206. doi: 10.1038/nature13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoffmeister M., Krieg J., Ehrke A., Seigfried F.A., Wischmann L., Dietmann P., Kühl S.J., Oess S. Developmental neurogenesis in mouse and Xenopus is impaired in the absence of Nosip. Dev. Biol. 2017;429:200–212. doi: 10.1016/j.ydbio.2017.06.026. [DOI] [PubMed] [Google Scholar]
- 34.Moody S.A., Kline M.J. Segregation of fate during cleavage of frog (Xenopus laevis) blastomeres. Anat. Embryol. (Berl.) 1990;182:347–362. doi: 10.1007/BF02433495. [DOI] [PubMed] [Google Scholar]
- 35.Turner T.N., Yi Q., Krumm N., Huddleston J., Hoekzema K., F Stessman H.A., Doebley A.L., Bernier R.A., Nickerson D.A., Eichler E.E. denovo-db: a compendium of human de novo variants. Nucleic Acids Res. 2017;45(D1):D804–D811. doi: 10.1093/nar/gkw865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zarrei M., MacDonald J.R., Merico D., Scherer S.W. A copy number variation map of the human genome. Nat. Rev. Genet. 2015;16:172–183. doi: 10.1038/nrg3871. [DOI] [PubMed] [Google Scholar]
- 37.Jauch R., Ng C.K., Narasimhan K., Kolatkar P.R. The crystal structure of the Sox4 HMG domain-DNA complex suggests a mechanism for positional interdependence in DNA recognition. Biochem. J. 2012;443:39–47. doi: 10.1042/BJ20111768. [DOI] [PubMed] [Google Scholar]
- 38.Yuan H., Corbi N., Basilico C., Dailey L. Developmental-specific activity of the FGF-4 enhancer requires the synergistic action of Sox2 and Oct-3. Genes Dev. 1995;9:2635–2645. doi: 10.1101/gad.9.21.2635. [DOI] [PubMed] [Google Scholar]
- 39.Hou L., Srivastava Y., Jauch R. Molecular basis for the genome engagement by Sox proteins. Semin. Cell Dev. Biol. 2017;63:2–12. doi: 10.1016/j.semcdb.2016.08.005. [DOI] [PubMed] [Google Scholar]
- 40.Sock E., Pagon R.A., Keymolen K., Lissens W., Wegner M., Scherer G. Loss of DNA-dependent dimerization of the transcription factor SOX9 as a cause for campomelic dysplasia. Hum. Mol. Genet. 2003;12:1439–1447. doi: 10.1093/hmg/ddg158. [DOI] [PubMed] [Google Scholar]
- 41.Katoh-Fukui Y., Igarashi M., Nagasaki K., Horikawa R., Nagai T., Tsuchiya T., Suzuki E., Miyado M., Hata K., Nakabayashi K. Testicular dysgenesis/regression without campomelic dysplasia in patients carrying missense mutations and upstream deletion of SOX9. Mol. Genet. Genomic Med. 2015;3:550–557. doi: 10.1002/mgg3.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang X., Xue M., Zhao M., He F., Li C., Li X. Identification of a novel mutation (Ala66Thr) of SRY gene causes XY pure gonadal dysgenesis by affecting DNA binding activity and nuclear import. Gene. 2018;651:143–151. doi: 10.1016/j.gene.2018.01.076. [DOI] [PubMed] [Google Scholar]
- 43.Ortega J.A., Memi F., Radonjic N., Filipovic R., Bagasrawala I., Zecevic N., Jakovcevski I. The subventricular zone: A key player in human neocortical development. Neuroscientist. 2018;24:156–170. doi: 10.1177/1073858417691009. [DOI] [PubMed] [Google Scholar]
- 44.Kato K., Bhattaram P., Penzo-Méndez A., Gadi A., Lefebvre V. SOXC transcription factors induce cartilage growth plate formation in mouse embryos by promoting noncanonical WNT signaling. J. Bone Miner. Res. 2015;30:1560–1571. doi: 10.1002/jbmr.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arnold S.J., Huang G.J., Cheung A.F., Era T., Nishikawa S., Bikoff E.K., Molnár Z., Robertson E.J., Groszer M. The T-box transcription factor Eomes/Tbr2 regulates neurogenesis in the cortical subventricular zone. Genes Dev. 2008;22:2479–2484. doi: 10.1101/gad.475408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gleeson J.G., Allen K.M., Fox J.W., Lamperti E.D., Berkovic S., Scheffer I., Cooper E.C., Dobyns W.B., Minnerath S.R., Ross M.E., Walsh C.A. Doublecortin, a brain-specific gene mutated in human X-linked lissencephaly and double cortex syndrome, encodes a putative signaling protein. Cell. 1998;92:63–72. doi: 10.1016/s0092-8674(00)80899-5. [DOI] [PubMed] [Google Scholar]
- 47.Scharer C.D., McCabe C.D., Ali-Seyed M., Berger M.F., Bulyk M.L., Moreno C.S. Genome-wide promoter analysis of the SOX4 transcriptional network in prostate cancer cells. Cancer Res. 2009;69:709–717. doi: 10.1158/0008-5472.CAN-08-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scherer G., Held M., Erdel M., Meschede D., Horst J., Lesniewicz R., Midro A.T. Three novel SRY mutations in XY gonadal dysgenesis and the enigma of XY gonadal dysgenesis cases without SRY mutations. Cytogenet. Cell Genet. 1998;80:188–192. doi: 10.1159/000014978. [DOI] [PubMed] [Google Scholar]
- 49.Pingault V., Bodereau V., Baral V., Marcos S., Watanabe Y., Chaoui A., Fouveaut C., Leroy C., Vérier-Mine O., Francannet C. Loss-of-function mutations in SOX10 cause Kallmann syndrome with deafness. Am. J. Hum. Genet. 2013;92:707–724. doi: 10.1016/j.ajhg.2013.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zeng Y.T., Ren Z.R., Zhang M.L., Huang Y., Zeng F.Y., Huang S.Z. A new de novo mutation (A113T) in HMG box of the SRY gene leads to XY gonadal dysgenesis. J. Med. Genet. 1993;30:655–657. doi: 10.1136/jmg.30.8.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Preiss S., Argentaro A., Clayton A., John A., Jans D.A., Ogata T., Nagai T., Barroso I., Schafer A.J., Harley V.R. Compound effects of point mutations causing campomelic dysplasia/autosomal sex reversal upon SOX9 structure, nuclear transport, DNA binding, and transcriptional activation. J. Biol. Chem. 2001;276:27864–27872. doi: 10.1074/jbc.M101278200. [DOI] [PubMed] [Google Scholar]
- 52.Karaer K., Yüksel Z., Yalınbaş E., Scherer G. A case of campomelic dysplasia in whom a new mutation was found in the SOX9 gene. Turk Pediatri Ars. 2014;49:154–156. doi: 10.5152/tpa.2014.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morín M., Viñuela A., Rivera T., Villamar M., Moreno-Pelayo M.A., Moreno F., del Castillo I. A de novo missense mutation in the gene encoding the SOX10 transcription factor in a Spanish sporadic case of Waardenburg syndrome type IV. Am. J. Med. Genet. A. 2008;146A:1032–1037. doi: 10.1002/ajmg.a.32181. [DOI] [PubMed] [Google Scholar]
- 54.Okamoto N., Ehara E., Tsurusaki Y., Miyake N., Matsumoto N. Coffin-Siris syndrome and cardiac anomaly with a novel SOX11 mutation. Congenit. Anom. (Kyoto) 2018;58:105–107. doi: 10.1111/cga.12242. [DOI] [PubMed] [Google Scholar]
- 55.Harley V.R., Jackson D.I., Hextall P.J., Hawkins J.R., Berkovitz G.D., Sockanathan S., Lovell-Badge R., Goodfellow P.N. DNA binding activity of recombinant SRY from normal males and XY females. Science. 1992;255:453–456. doi: 10.1126/science.1734522. [DOI] [PubMed] [Google Scholar]
- 56.Pontiggia A., Rimini R., Harley V.R., Goodfellow P.N., Lovell-Badge R., Bianchi M.E. Sex-reversing mutations affect the architecture of SRY-DNA complexes. EMBO J. 1994;13:6115–6124. doi: 10.1002/j.1460-2075.1994.tb06958.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chaoui A., Watanabe Y., Touraine R., Baral V., Goossens M., Pingault V., Bondurand N. Identification and functional analysis of SOX10 missense mutations in different subtypes of Waardenburg syndrome. Hum. Mutat. 2011;32:1436–1449. doi: 10.1002/humu.21583. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.