 Characterization of a series of fluorinated compounds for competitive 19F NMR reveals the principles that can guide developing highly sensitive assays.
Characterization of a series of fluorinated compounds for competitive 19F NMR reveals the principles that can guide developing highly sensitive assays.
Abstract
Systematic characterization of a series of fluorinated VHL ligands, varying binding affinity and position of the trifluoromethyl group, qualifies a spy molecule for competitive 19F NMR screening and reveals guiding principles to develop highly sensitive assays with low material consumption.
Over the past few years, the application of nuclear magnetic resonance (NMR) to study small molecule interactions with biomolecular targets has increased. Both the large number of methods available and the ability to detect weak interactions imperceptible to other biophysical techniques make NMR a valuable resource for pharmaceutical research, from fragment-based drug discovery (FBDD) to lead optimization campaigns.1
A powerful approach to detect binders across a range of affinities consists in competition ligand-observed NMR experiments.2 Competitive ligands are detected by monitoring the signals of a known binder, also referred to as a reporter or spy molecule. When a competing ligand is present, the spy molecule will be displaced from its binding site. The increased relative population of free vs. bound state of the spy molecule can be detected by monitoring different NMR parameters and is dependent on the affinity of the competitor.3 If the binding affinity of the spy molecule is known, the affinity of the competitors can be estimated from the extent of the displacement.
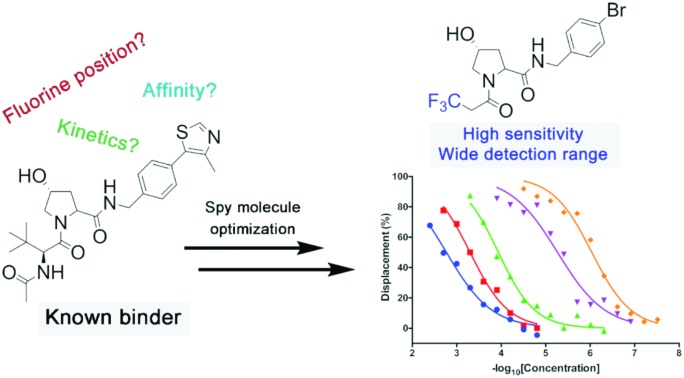
In this context, the usage of fluorinated compounds as spy molecules presents yet further advantages. Fluorine atoms are absent in most common solvents, buffer components and biomolecules, resulting in simpler datasets to analyze when compared to proton-based methods. Furthermore, the high chemical shift anisotropy (CSA) of fluorine yields very clear responses to changes in the chemical environment, making the binding detection extremely sensitive.4a Despite many successful applications of 19F NMR competition experiments,5 the properties required to achieve sensitive spy molecules remain understudied. To date, spy molecule selection has consisted in either picking hits from the screening of a fluorinated compound library, or preparing fluorinated analogues of a known ligand in an unguided manner.6 Although aspects of spy molecule design have been discussed, including use of CF or CF3 groups, residence time, and fluorine local environment,4,6 the majority of these features are evaluated solely from a theoretical point of view. It thus remains unclear how much optimization is needed to obtain the best spy molecule. Here, a series of fluorinated ligands of a well-characterized ligand–protein binding system were developed and their potential as spy molecules was evaluated.
The von Hippel Lindau (VHL) E3 ligase was chosen as the target, because ligand binding to its hydroxyproline (Hyp) recognition site is deeply characterized.7 Many Hyp containing ligands are available with crystallographically characterized binding modes and a wide range of dissociation constants (KD). Moreover, the development of a 19F NMR competition assay for probing the Hyp site could see many valuable applications in the development of novel VHL ligands and VHL-based chemical degraders,8 and distinguishing Hyp site binders from those interacting with other sites.7,9
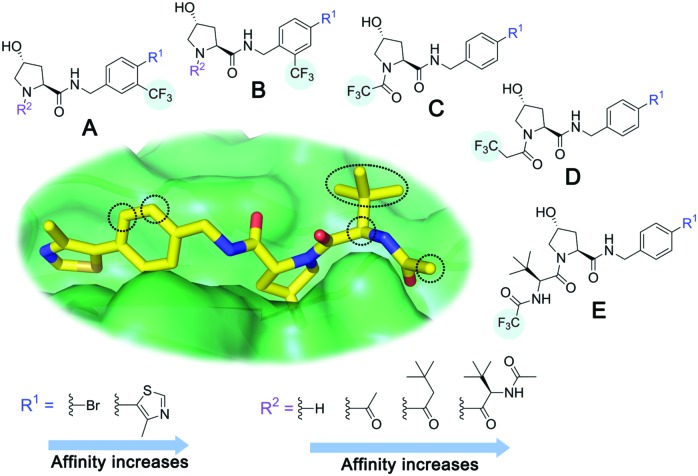
For spy molecule design, a trifluoromethyl (CF3) modification was preferred over, e.g., fluoromethyl to generate a larger set of analogues (high availability of CF3 containing starting materials) and to yield increased signal to background. The potent binder VH032 (PDB: ; 4W9H)7c was used as a template in the design, and five positions were chosen for placing the CF3 modification (Fig. 1). These positions were selected by considering potential clashes with the protein, truncating left- and right-hand side groups to modulate the binding affinity. In total twenty-two compounds were synthesized by adapting previously described synthetic routes to prepare VHL ligands.7
Fig. 1. Design of a fluorinated spy molecule series. The co-crystal structure of VHL binder VH032 (PDB: 4W9H) inspired the choice of five positions (dashed circles) to attach a CF3 group, either on aromatic (series A and B) or aliphatic (C–E) regions.
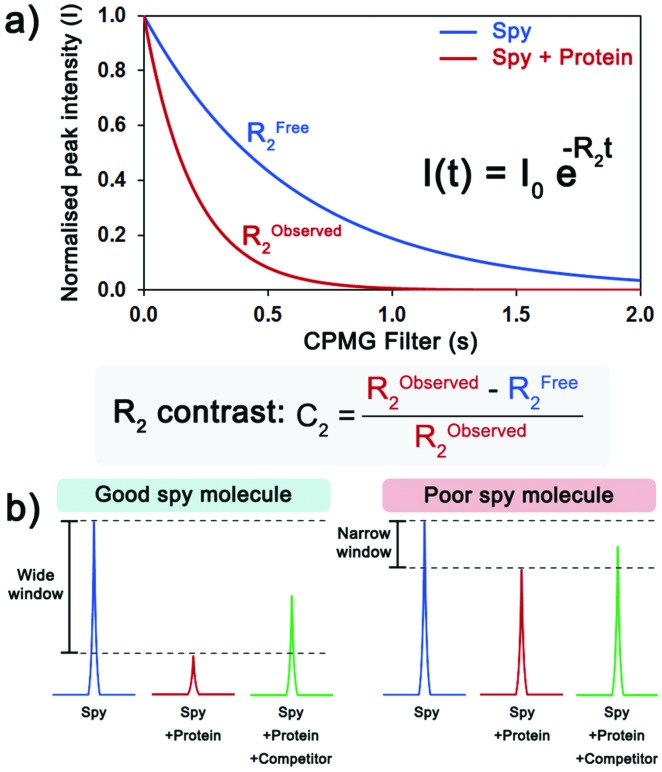
Due to the large CSA of fluorine and the significant differences in the fluorine isotropic chemical shift (δF) between the free and bound states, the transverse relaxation rate (R2) is a well-established sensitive parameter to detect ligand binding.4 Therefore, fluorine NMR experiments including a Carr–Purcell–Meiboom–Gill (CPMG) spin-echo filter10 before acquisition were employed to monitor ligand binding to VHL by measuring peak intensity, as changes in other NMR parameters, such as chemical shift and peak width, were not as significant. The R2 values of the CF3 peak of each compound in the absence and in the presence of protein were determined by performing multiple 19F CPMG experiments varying the CPMG filter (Fig. 2a). To quantify the shift in R2 upon binding, the R2 contrast (C2)11 was determined. Ideally, a good spy molecule should have a large increase in R2 (fast relaxation) when protein is present, resulting in a large assay window for the competition experiments (Fig. 2b).
Fig. 2. Evaluation of the sensitivity of a spy molecule. (a) Measurement of the transverse relaxation rate (R2) of the spy molecule in the absence (blue) and the presence of protein (red). The C2 quantifies the extent of the shift in R2 in the presence of protein. (b) Upon addition of protein, the 19F CPMG signal of a good spy molecule (left) is greatly reduced, resulting in a large window to detect and rank displacers, while for a poor spy molecule (right), the difference between the signals is too small.
The KD of each compound to VHL was determined by surface plasmon resonance (SPR) experiments. The KD of the majority of the compounds could be measured confidently (Table 1), with the exception of cases where the binding responses were too low (KD > 1.5 mM) or the responses were above the theoretical maximum response (promiscuous or unspecific binders).
Table 1. Characterisation of spy molecules by SPR and 19F NMR.
| Series | Compound | R1 | R2 | K D, SPR (μM) | C 2 a (%) |
| A | 1 |
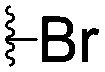
|
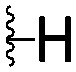
|
>1500 | 4.1 ± 3.1 |
| 2 |
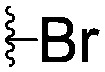
|
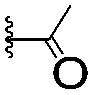
|
657 ± 49 b | 6.5 ± 2.2 | |
| 3 |
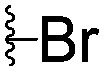
|
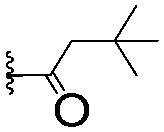
|
407 ± 5 | 28.2 ± 2.3 | |
| 4 |
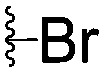
|
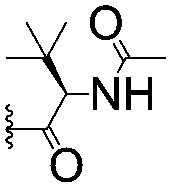
|
4.39 ± 0.12 | 38.9 ± 2.5 | |
| 5 |
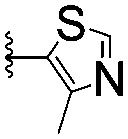
|
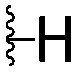
|
>1500 | 2.2 ± 1.8 | |
| 6 |
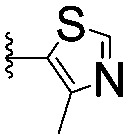
|
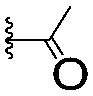
|
878 ± 98 b | 37.7 ± 2.6 | |
| 7 |
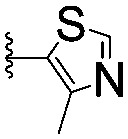
|
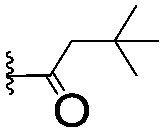
|
67.1 ± 14.0 | 62.1 ± 3.0 | |
| 8 |
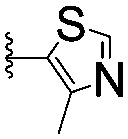
|
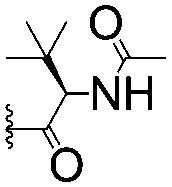
|
2.99 ± 0.75 | 17.7 ± 2.2 | |
| B | 9 |
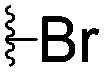
|
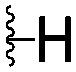
|
>1500 | 0.9 ± 2.7 |
| 10 |
|
|
1352 ± 94 b | 51.9 ± 2.1 | |
| 11 |
|
|
110 ± 9 | 47.9 ± 4.1 | |
| 12 |
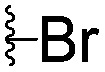
|
|
1.14 ± 0.12 | 7.6 ± 2.4 | |
| 13 |
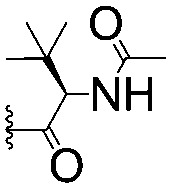
|
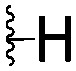
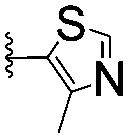
|
c | 3.2 ± 2.0 | |
| 14 |
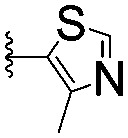
|
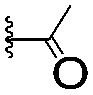
|
c | 53.9 ± 2.5 | |
| 15 |
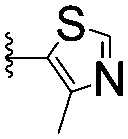
|
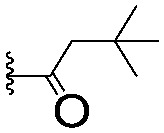
|
35.2 ± 7.0 | 41.3 ± 2.7 | |
| 16 |
|
|
0.268 ± 0.030 | 1.3 ± 2.4 | |
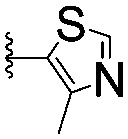
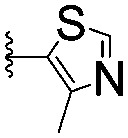
| C | 17 |
|
— | 645 ± 100 b | 62.0 ± 2.6 |
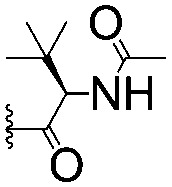
| 18 |
|
— | 24.8 ± 3.4 | 76.0 ± 3.2 | |
| D | 19 |
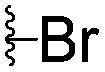
|
— | 145 ± 29 | 70.1 ± 4.5 |
| 20 |
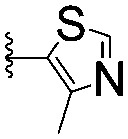
|
— | 12.4 ± 1.9 | 48.4 ± 5.6 | |
| E | 21 |
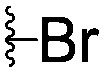
|
— | 0.447 ± 0.085 | 3.7 ± 8.8 |
| 22 |
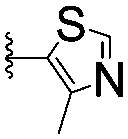
|
— | 0.0969 ± 0.0068 | 0.8 ± 8.6 |
aConditions: spy molecule at 100 μM in the absence or presence of protein at 1 μM.
bIntermediate-weak binders, KD obtained by fitting the data with a 1-to-1 binding model as the maximum response (RMAX) could not be obtained experimentally.
cHigh responses in the SPR experiments, promiscuous or unspecific binders.
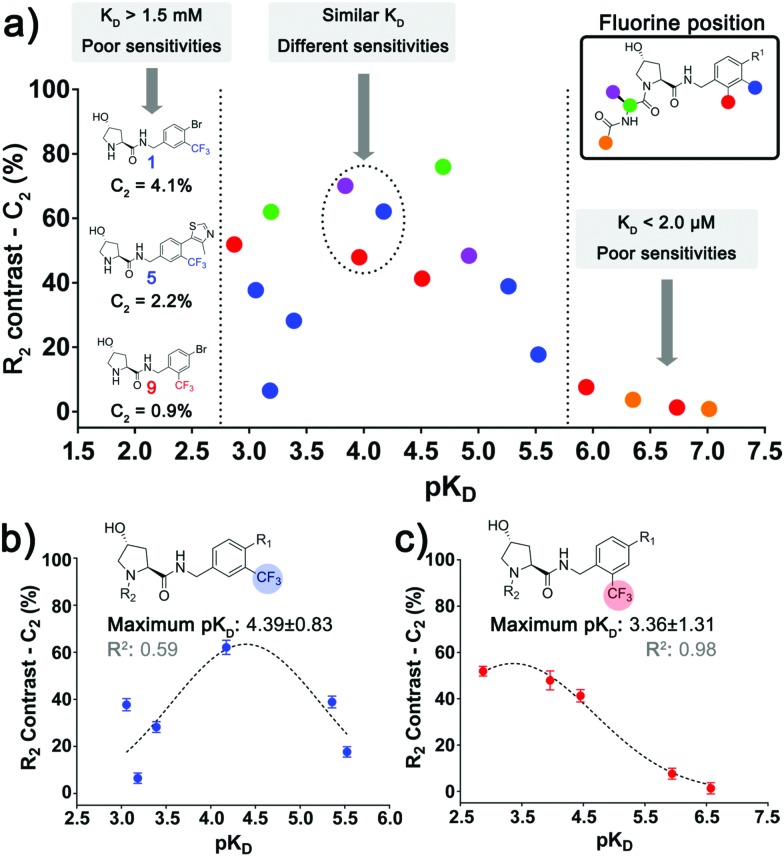
A number of trends can be observed. The weakest binders of the series (1, 5 and 9) were among the spy molecules with the lowest C2. In this case the concentration of the spy–protein complex was much lower than KD, so too small a percentage of the molecules contributed to the signal, resulting in a low C2 value. On the other hand, as expected the tightest binders of the series (12, 16, 21 and 22) also presented a very low C2. This is due to their higher residence time in the binding site, not allowing many molecules to interact with the protein during the time of the NMR experiment. Notably, despite the high affinities, the dissociation rates of these compounds were too high to be determined accurately by SPR, showing that only remarkably fast binding kinetics can yield a sensitive spy molecule at low protein concentrations.
By plotting C2versus pKD (Fig. 3a), two binding affinity limits where the spy molecule sensitivity decays could be observed, only molecules of pKD = 3.0–5.5 presented good values of C2. To understand the effect of the position of the CF3 group, we plotted C2versus pKD for ligands within the series A (Fig. 3b) and B (Fig. 3c). In both cases the data are distributed as a bell-shaped curve, whereby ideal spy molecules should be weak enough to possess very fast kinetics, but not too weak, so the amount of spy–protein complex is sufficiently populated to allow for detecting overall changes in relaxation. This observation agrees with the theoretical prediction by Dalvit et al. varying the residence time of the fluorinated ligand.4b
Fig. 3. Relationship between sensitivity, affinity and position of the fluorinated group. (a) Correlation between C2 and the respective pKD of all the spy molecules where the affinity could be estimated, highlighting the regions where the sensitivity of the spy molecules decays considerably. Dashed circles display molecules with similar pKD that possess different C2 due to the variation in the fluorine attachment. The same plot was also made for the aromatic series A (b) and B (c), where the trend of an ideal intermediate affinity can be observed.
Interestingly, the best affinity range varied across the two series, with optimal pKD ≈ 4.4 (KD ≈ 40 μM) for series A and pKD ≈ 3.4 (KD 400 μM) for series B. This result could be explained by differences in association rates (kon) or ΔδF (bound and free states) of equivalent compounds between the series, leading to different sensitivities at the same KD. This observation shows that not only the absolute affinity, but also the chemical environment surrounding the fluorine atom in the context of the bound ligand affected the sensitivity of the spy molecules. In further support of the importance of the fluorine position, 7, 11 and 19 (Fig. 3a, dashed circle) all had similar affinities (pKD ≈ 3.8–4.2) but different C2 due to varying CF3 attachments.
The most sensitive spy molecules (Table 1, highest C2) were 7, 17, 18 and 19. Although molecule 18 presented the overall highest sensitivity, molecule 19 was selected for setting the competition assay. As 19 is seven times weaker than 18, it was hypothesized that 19 would be more readily displaced by weak competitor fragments, allowing a wider range of KD detection. Molecule 19 was also preferred over 17 because its affinity could be determined more accurately by SPR (ESI,† Fig. S1).
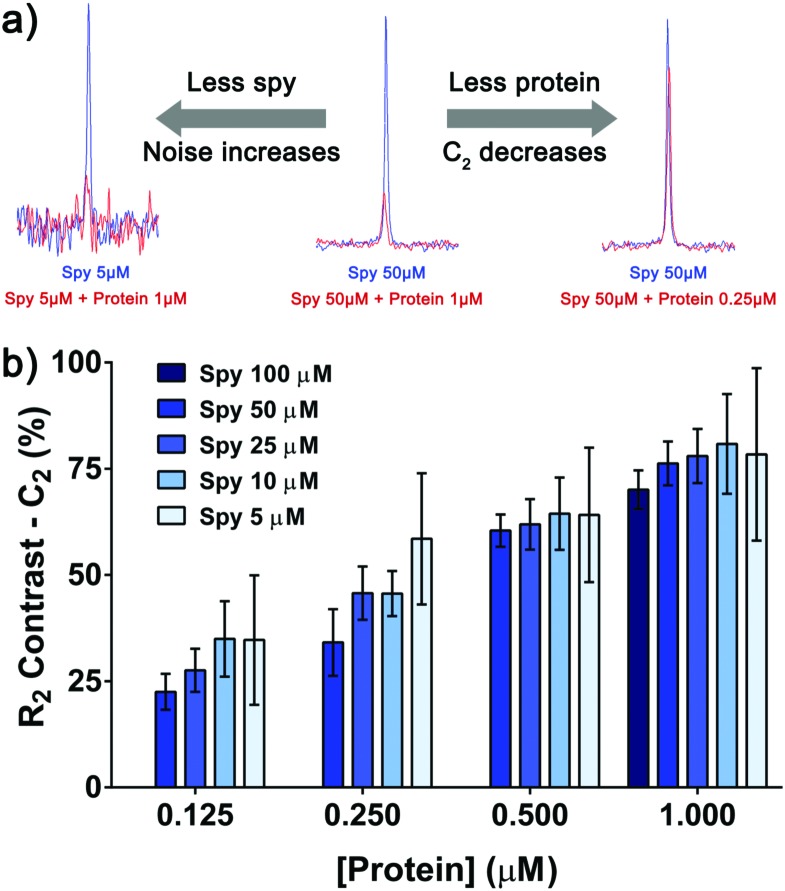
Spy molecule 19 consistently displayed good sensitivity under different conditions. When the concentrations of spy and protein were simultaneously lowered, a binding response could still be observed even at the lowest concentrations (19 at 5 μM, VHL at 125 nM), albeit with lower signal-to-noise ratio (S/N) and C2, requiring longer experiment times (Fig. 4a). Most conditions displayed a reasonable C2 (≥40%), allowing the detection of competitors even with low material consumption and number of scans (4 minutes per sample). In any case, conditions with C2 as high as possible should be aimed for whenever possible.
Fig. 4. Binding response can be observed at low concentrations of spy molecule 19 and protein. (a) Overlay of the 19F CPMG peak (200 ms filter) of spy molecule 19 in the absence (blue) and presence of protein (red). When lowering the concentrations of spy molecule and protein, the S/N and C2 values respectively decreased. (b) Measurement of the C2 of spy molecule 19 at different concentrations (in triplicates), in the presence of four concentrations of protein. The error progressively increases as the concentration of spy molecule goes down, due to the increase in noise of the NMR data.
Noticeably, the C2 values were affected to a great extent by the total amount of protein rather than the total amount of spy molecule (Fig. 4b). At 1.0 μM of protein the C2 values varied between 70 and 80% for all the concentrations of spy tested, even if they were around 1.5–15 times lower than the KD. In this way, for selecting the most sensitive spy molecule of a series, a single screening at fixed concentration of protein is adequate to determine the best compounds to set up the competition assay, even if affinities vary significantly and sub-KD concentrations are used.
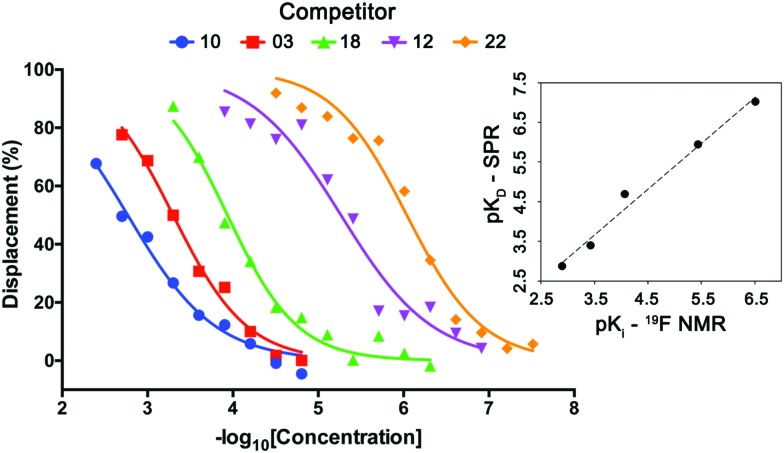
Lastly, the utility and scope of using spy molecule 19 to quantify binding of competitors across a wide range of affinities was evaluated (Fig. 5). Five compounds were titrated against 19 at 50 μM and VHL at 1.0 μM. In all cases the displacement of the spy molecule was concentration-dependent and the Ki (inhibition constant) values were derived from the respective IC50 using a competitive binding model.12 The measured binding affinities compared remarkably well with the respective values obtained by SPR across 4-log units. The same experiment was attempted with spy molecules 6 and 11 under the same conditions, and in these cases, the assay could not differentiate the binders as efficiently (ESI,† Fig. S2–S4), since this would require higher protein concentration. Binders of other pockets present in the VHL E3 ligase previously reported did not displace spy molecule 19 as expected, showing that the displacement is site-specific (ESI,† Fig. S5).
Fig. 5. Determination of the affinities of VHL binders using spy molecule 19. Displacement of spy molecule 19 in the presence of different concentrations of five VHL binders (molecules 3, 10, 12, 18 and 22) was concentration dependent. By knowing the concentrations of spy molecule (50 μM), and protein (1 μM) and the KD between spy molecule 19 and VHL, the Ki of each competitor was determined, having good correlation with the respective KD values obtained by SPR (top right inset graph).
In summary, we qualify a broad-scope high-quality spy molecule for the E3 ligase VHL able to differentiate a wide range of KD values with good correlation with orthogonal techniques. This advance will aid future screening efforts for novel VHL ligands and chemical degraders. More broadly, this work provides a blueprint for obtaining the most sensitive reporter for a given protein–ligand system, while minimizing protein and spy molecule consumption, and argues for exploring chemical space with fluorine at different positions of a binding ligand, especially in the early stages of projects where only weak binders might be available. This guidance will prove useful to many groups in academia and industry that develop NMR assays for FBDD or hit optimization campaigns.
This project has received funding from the European Research Council (ERC) under the European Union's Seventh Frame-work Programme (FP7/2007–2013) as a Starting Grant to A. C. (grant agreement no. ERC-2012-StG-311460 DrugE3CRLs) and the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES, PhD Studentship 7148-14-3 to G. V. C.). Biophysics and drug discovery activities were supported by Wellcome Trust strategic awards to Dundee (100476/Z/12/Z and 094090/Z/10/Z, respectively). We thank D. Fletcher and A. Bortoluzzi for support with NMR and helpful discussions.
Conflicts of interest
The Ciulli laboratory receives sponsored research support from Boehringer Ingelheim and Nurix, Inc. A. C. is a scientific founder, director and shareholder of Amphista Therapeutics, which is developing targeted protein degradation platforms.
Supplementary Material
Footnotes
†Electronic supplementary information (ESI) available: Compound synthesis and characterization, experimental procedures for protein expression and biophysical experiments (raw data and fitting). See DOI: 10.1039/c8cc09790a
References
- (a) Sanchez-Pedregal V. M., Reese M., Meiler J., Blommers M. J. J., Griesinger C., Carlomagno T. Angew. Chem., Int. Ed. 2005;44:4172. doi: 10.1002/anie.200500503. [DOI] [PubMed] [Google Scholar]; (b) Betz M., Saxena K., Schwalbe H. Curr. Opin. Chem. Biol. 2006;10:219. doi: 10.1016/j.cbpa.2006.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Dias D. M., Ciulli A. Prog. Biophys. Mol. Biol. 2014;116:101. doi: 10.1016/j.pbiomolbio.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Gossert A. D., Jahnke W. Prog. Nucl. Magn. Reson. Spectrosc. 2016;97:82. doi: 10.1016/j.pnmrs.2016.09.001. [DOI] [PubMed] [Google Scholar]
- (a) Mayer M., Meyer B. J. Am. Chem. Soc. 2001;123:6108. doi: 10.1021/ja0100120. [DOI] [PubMed] [Google Scholar]; (b) Dalvit C., Fasolini M., Flocco M., Knapp S., Pevarello P., Veronesi M. J. Med. Chem. 2002;45:2610. doi: 10.1021/jm011122k. [DOI] [PubMed] [Google Scholar]; (c) Siriwardena A. H., Tian F., Noble S., Prestegard J. H. Angew. Chem., Int. Ed. 2002;41:3454. doi: 10.1002/1521-3773(20020916)41:18<3454::AID-ANIE3454>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Dalvit C., Flocco M., Knapp S., Mostardini M., Perego R., Stockman B. J., Veronesi M., Varasi M. J. Am. Chem. Soc. 2002;124:7702. doi: 10.1021/ja020174b. [DOI] [PubMed] [Google Scholar]
- (a) Dalvit C. Prog. Nucl. Magn. Reson. Spectrosc. 2007;51:243. doi: 10.1016/j.pnmrs.2023.07.001. [DOI] [PubMed] [Google Scholar]; (b) Dalvit C., Vulpetti A. Chem. – Eur. J. 2016;22:7592. doi: 10.1002/chem.201600446. [DOI] [PubMed] [Google Scholar]; (c) Dalvit C., Fagerness P. E., Hadden D. T. A., Sarver R. W., Stockman B. J. J. Am. Chem. Soc. 2003;125:7696. doi: 10.1021/ja034646d. [DOI] [PubMed] [Google Scholar]
- (a) Jordan J. B., Poppe L., Xia X. Y., Cheng A. C., Sun Y., Michelsen K., Eastwood H., Schnier P. D., Nixey T., Zhong W. G. J. Med. Chem. 2012;55:678. doi: 10.1021/jm201441k. [DOI] [PubMed] [Google Scholar]; (b) Kim Y., Hilty C. Angew. Chem., Int. Ed. 2015;54:4941. doi: 10.1002/anie.201411424. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Buratto R., Mammoli D., Canet E., Bodenhausen G. J. Med. Chem. 2016;59:1960. doi: 10.1021/acs.jmedchem.5b01583. [DOI] [PubMed] [Google Scholar]; (d) Dalvit C., Vulpetti A. J. Med. Chem. 2018 doi: 10.1021/acs.jmedchem.8b01210. [DOI] [PubMed] [Google Scholar]
- Vulpetti A., Hommel U., Landrum G., Lewis R., Dalvit C. J. Am. Chem. Soc. 2009;131:12949. doi: 10.1021/ja905207t. [DOI] [PubMed] [Google Scholar]
- (a) Buckley D. L., Van Molle I., Gareiss P. C., Tae H. S., Michel J., Noblin D. J., Jorgensen W. L., Ciulli A., Crews C. M. J. Am. Chem. Soc. 2012;134:4465. doi: 10.1021/ja209924v. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Van Molle I., Thomann A., Buckley D. L., So E. C., Lang S., Crews C. M., Ciulli A. Chem. Biol. 2012;19:1300. doi: 10.1016/j.chembiol.2012.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Galdeano C., Gadd M. S., Soares P., Scaffidi S., Van Molle I., Birced I., Hewitt S., Dias D. M., Ciulli A. J. Med. Chem. 2014;57:8657. doi: 10.1021/jm5011258. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Dias D. M., Van Molle I., Baud M. G. J., Galdeano C., Geraldes C. F. G. C., Ciulli A. ACS Med. Chem. Lett. 2014;5:23. doi: 10.1021/ml400296c. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Soares P., Gadd M. S., Frost J., Galdeano C., Ellis L., Epemolu O., Rocha S., Read K. D., Ciulli A. J. Med. Chem. 2018;61:599. doi: 10.1021/acs.jmedchem.7b00675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (a) Toure M., Crews C. M. Angew. Chem., Int. Ed. 2016;55:1966. doi: 10.1002/anie.201507978. [DOI] [PubMed] [Google Scholar]; (b) Gadd M. S., Testa A., Lucas X., Chan K. H., Chen W., Lamont D. J., Zengerle M., Ciulli A. Nat. Chem. Biol. 2017;13:514. doi: 10.1038/nchembio.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Maniaci C., Hughes S. J., Testa A., Chen W., Lamont D. J., Rocha S., Alessi D. R., Romeo R., Ciulli A. Nat. Commun. 2017;8:830. doi: 10.1038/s41467-017-00954-1. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Testa A., Lucas X., Castro G. V., Chan K. H., Wright J. E., Runcie A. C., Gadd M. S., Harrison W. T. A., Ko E. J., Fletcher D., Ciulli A. J. Am. Chem. Soc. 2018;140:9299. doi: 10.1021/jacs.8b05807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (a) Lucas X., Van Molle I., Ciulli A. J. Med. Chem. 2018;61:7387–7393. doi: 10.1021/acs.jmedchem.8b00842. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Cardote T. A. F., Ciulli A. ChemMedChem. 2017;12:1491. doi: 10.1002/cmdc.201700359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (a) Carr H. Y., Purcell E. M. Phys. Rev. 1954;94:630–638. [Google Scholar]; (b) Meiboom S., Gill D. Rev. Sci. Instrum. 1958;29:688. [Google Scholar]
- Buratto R., Mammoli D., Chiarparin E., Williams G., Bodenhausen G. Angew. Chem., Int. Ed. 2014;53:11376. doi: 10.1002/anie.201404921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung I. K., Demetriades M., Hardy A. P., Lejeune C., Smart T. J., Szollossi A., Kawamura A., Schofield C. J., Claridge T. D. J. Med. Chem. 2013;56:547. doi: 10.1021/jm301583m. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.