Significance
Posttranscriptional RNA interference-mediated antiviral defense is well conserved, in which the key players, Dicer and Argonaute, act to digest viral RNAs. Utilizing a filamentous fungus model, we have previously shown the Spt–Ada–Gcn5 acetyltransferase (SAGA) transcriptional activator to regulate the upregulation of the two main antiviral RNA-silencing components, dicer-like 2 (dcl2) and argonaute-like 2 (agl2). Here, we show an additional distinct functional role for DCL2 in the virus-responsive, SAGA-mediated transcriptional upregulation of a subset of host genes. Strikingly, certain upregulated genes contribute to mitigate viral symptom induction. No involvement of Argonaute in this Dicer function is suggested by the agl2 disruption assay. Therefore, DCL2 is bifunctional in dual-layer antiviral defense to inhibit viral replication and alleviate symptom expression.
Keywords: RNA silencing, SAGA, antiviral defense, Cryphonectria parasitica, fungal virus
Abstract
In antiviral RNA interference (RNAi), Dicer plays a primary role in processing double-stranded RNA (dsRNA) molecules into small-interfering RNAs (siRNAs) that guide Argonaute effectors to posttranscriptional suppression of target viral genes. Here, we show a distinct role for Dicer in the siRNA-independent transcriptional induction of certain host genes upon viral infection in a filamentous fungus. Previous studies have shown that the two key players, dicer-like 2 (dcl2) and argonaute-like 2 (agl2), of antiviral RNAi in a phytopathogenic ascomycete, Cryphonectria parasitica, are highly transcriptionally induced upon infection with certain RNA mycoviruses, including the positive-stranded RNA hypovirus mutant lacking the RNAi suppressor (Cryphonectria hypovirus 1-Δp69, CHV1-Δp69). This induction is regulated by the Spt–Ada–Gcn5 acetyltransferase (SAGA) complex, a well-known transcriptional coactivator. The present study shows that diverse host genes, in addition to dcl2 and agl2, were up-regulated more than 10-fold by SAGA upon infection with CHV1-Δp69. Interestingly, DCL2, but not AGL2, was essential for SAGA-mediated global gene up-regulation. Moreover, deletion of certain virus-induced genes enhanced a CHV1-Δp69 symptom (growth rate) but not its accumulation. Constitutive, modest levels of dcl2 expression drastically reduced viral siRNA accumulation but were sufficient for full-scale up-regulation of host genes, suggesting that high induction of dcl2 and siRNA production are not essential for the transcriptional up-regulation function of DCL2. These data clearly demonstrate the dual functionality of DCL2: as a dsRNA-specific nuclease in posttranscriptional antiviral RNA silencing and as a key player in SAGA-mediated host gene induction, which independently represses viral replication and alleviates virus-induced symptom expression.
Dicer plays a pivotal role in the initiation of RNA silencing by recognizing double-stranded RNAs (dsRNAs) and cleaving them into small RNAs using its RNase III-like double-stranded RNA-specific nuclease activities. Small RNAs are largely classified into two groups: small-interfering RNAs (siRNAs) and microRNAs. These small RNAs are incorporated into the Argonaute (AGO) effector complex that subsequently either digests target mRNAs with its RNase H-like activities or suppresses their translation. This RNA silencing [also referred to as RNA interference (RNAi)] pathway is conserved across eukaryotic organisms (1–4), biologically functions to protect against molecular pathogens such as viruses and transposons, and is associated with many physiological regulatory pathways such as development and stress tolerance. Antiviral RNA silencing is the primary defense in plants, insects, and lower eukaryotes (5–7); thus, deficiencies in the key components of RNA silencing often result in enhanced susceptibility of hosts to viruses. This enhanced susceptibility generally involves increased viral replication, which is considered to cause more severe symptoms (8–11). RNA silencing appears to be regulated at various steps, for instance, transcriptional regulation of the key genes (7, 12) and their posttranscriptional regulation (13), although their mechanisms are not well studied. In this context, the Cryphonectria parasitica (a phytopathogenic ascomycete, chestnut blight fungus)/mycovirus provides a unique system for exploring the mechanisms underlying the regulation of RNA silencing (7, 14).
Similar to many other filamentous fungi, C. parasitica has several RNA silencing-related genes; two dicer-like (dcl), four argonaute-like 2 (agl), and four RNA-dependent RNA polymerase (rdr) genes (15). Unlike higher eukaryotes, the fungus can survive multiple simultaneous deletions of the key RNA-silencing genes without showing a phenotypic change, indicating the dispensability of these genes in vegetative growth (16–18). Among these, only dcl2 and agl2 are required for antiviral RNA silencing (15, 17), which are highly induced as a result of infection with certain RNA viruses (17, 19, 20). We have previously developed a genetic screening protocol for the identification of factors involved in the induction pathway from the sensing of viral infection to transcriptional up-regulation. This screen led to the identification of the universally conserved Spt–Ada–Gcn5 acetyltransferase (SAGA) complex as a regulator of this up-regulation (21), which interestingly requires DCL2 and AGL2 as positive feedback players. However, DCL2 and AGL2 requirement patterns are different among viruses; a dsRNA reovirus, mycoreovirus 1 (MyRV1, family Reoviridae) requires only DCL2 but not AGL2, while a positive-stranded RNA hypovirus lacking an RNA-silencing suppressor, p29 (Cryphonectria hypovirus 1-Δp69, CHV1-Δp69, family Hypoviridae) requires both DCL2 and AGL2 (21).
SAGA is a general transcriptional coactivator that forms a multiprotein complex comprising 18–20 subunits in a multimodular organization and is responsible for two histone-modifying activities: histone acetyltransferase (HAT) and histone deubiquitinase (DUB) (22–25). SAGA components are shared by transcription-related complexes such as general transcription factors and other coactivators (26). It has previously been shown that the SAGA complex up-regulates many genes in response to abiotic stress. In the case of the budding yeast, Saccharomyces cerevisiae, ∼10% of all genes have been reported to be under the control of SAGA (27, 28). Our previous study showed HAT activity, but not DUB activity, to be a major player in the SAGA-mediated induction of dcl2 upon viral infection in C. parasitica (21). HAT activity, as a major contributor, and DUB activity as a minor contributor, are involved in the SAGA-mediated up-regulation of agl2 (21).
In the present paper, we provide evidence that following virus infection, DCL2 plays an essential role, independently of AGL2, in the SAGA-mediated up-regulation of a subset of genes in C. parasitica, including the key RNA-silencing genes, some of which contribute to the alleviation of RNA viral symptom induction. These data clearly show dual functionality of DCL2 in the previously established posttranscriptional RNA silencing and in the currently revealed global SAGA-mediated transcriptional up-regulation, both of which contribute to antiviral defense.
Results
DCL2 Is Essential for SAGA-Mediated Gene Up-Regulation upon CHV1-Δp69 Infection in C. parasitica.
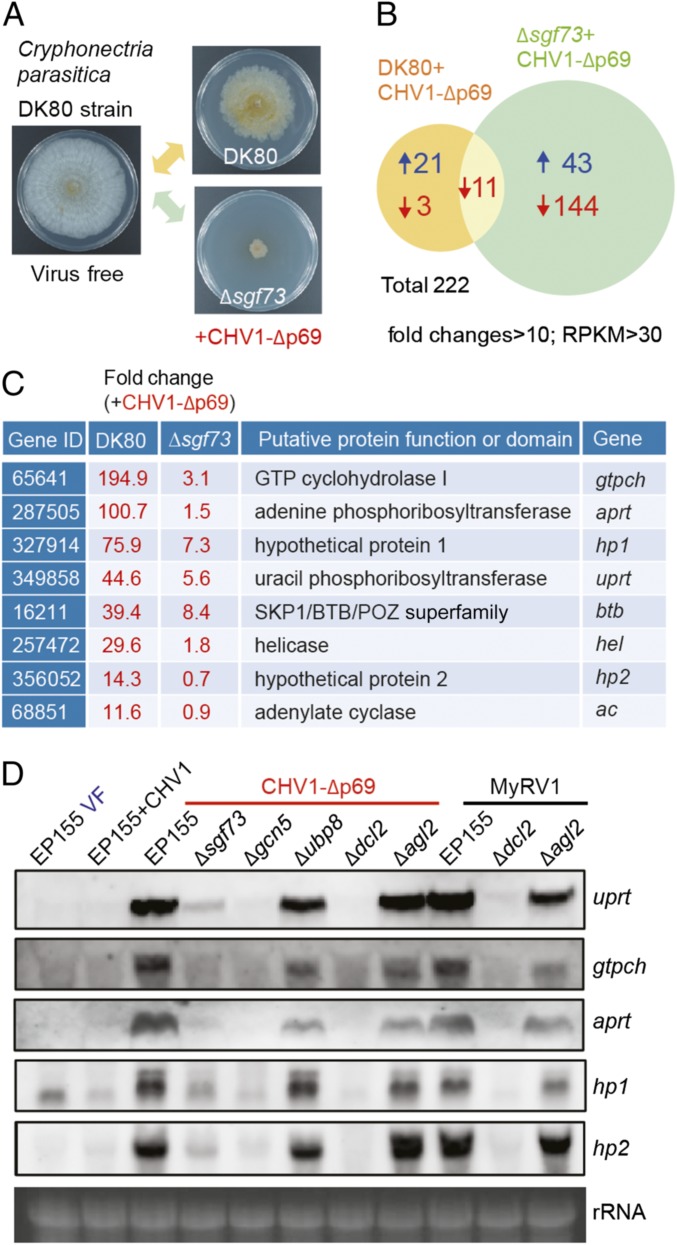
Using a genetic screening approach, we have previously shown that sgf73, a component of the SAGA, is essential for the high induction of antiviral RNA silencing (21). To investigate whether the SAGA complex is involved in transcriptional up-regulation of a wide range of genes following viral infection in C. parasitica, we analyzed the global transcriptome profiles of the CHV1-Δp69–infected Δsgf73 and DK80 (Δsgf73 parental strain) strains relative to that of the virus-free DK80 strain using high-throughput cDNA sequencing (RNA-seq) (Fig. 1A). There were 222 combined genes (or candidate genes), with more than 10-fold altered expression [P = 0 and reads per kilobase per million (RPKM) >30] in both virus-infected C. parasitica strains. Specifically, in the Δsgf73 strain, 144 and 43 genes showed reduced and elevated expression, respectively, while the transcription of these genes was not similarly altered in the virus-infected DK80 strain (Fig. 1B). This observation suggests that the SAGA complex is associated with the transcriptional regulation of these genes but that their expression is not implicated in CHV1-Δp69 infection. Only 21 genes showed exclusively elevated expression in the CHV1-Δp69–infected DK80 strain (but not in the Δp69-infected Δsgf73 strain) (Fig. 1B and SI Appendix, Table S1), implying that the SAGA complex is involved in up-regulation of these genes upon CHV1-Δp69 infection. From the results of semiquantitative RT-PCR analysis of these 21 genes, only 8 consistently showed the expected transcriptional expression pattern of high induction upon CHV1-Δp69 infection, and this induction was diminished by inactivation of the sgf73 gene (SI Appendix, Fig. S1 A–E). Such method-dependent differences are occasionally observed (29, 30). Based on a gene annotation and protein domain search, among these 8 genes, 6 are predicted to encode GTP cyclohydrolase I (gtpch), adenine phosphoribosyltransferase (aprt), uracil phosphoribosyltransferase (uprt), SKP1/BTB/POZ superfamily protein (btb), helicase (hel), and adenylate cyclase (ac), while the other two genes encode hypothetical proteins with unknown functions (hp1 and hp2) (Fig. 1C). Interestingly, the gtpch and aprt genes are located in very close genomic positions in a head-to-head direction (SI Appendix, Fig. S2).
Fig. 1.
The role of the SAGA complex and DCL2 in gene transcriptional up-regulation upon viral infection. (A) Colony morphology of the virus-free DK80 and the CHV1-Δp69–infected DK80 and Δsgf73 mutant C. parasitica strains. The fungal strains were grown on PDA plates for 7 d and photographed. (B) Transcriptomic analysis showing the number of genes with altered transcript expression in the DK80 and Δsgf73 strains upon CHV1-Δp69 infection. Transcript levels were compared between virus-free DK80 and each of CHV1-Δp69–infected DK80 and Δsgf73. (C) List of genes with highly induced expression in DK80 but not in Δsgf73 following CHV1-Δp69 infection. Gene IDs were obtained from the JGI Genome Portal (https://genome.jgi.doe.gov/Crypa2/Crypa2.home.html). (D) RNA blotting analysis of the uprt, gtpch, aprt, hp1, and hp2 gene transcripts in the wild-type strain (EP155) and the SAGA complex (Δsgf73, Δgcn5, and Δubp8)- and RNA silencing (Δdcl2 and Δagl2)-related fungal mutant strains infected with CHV1, CHV1-Δp69, and MyRV1, or virus-free (VF). The RNA gel was stained with ethidium bromide, and 28S rRNA is shown as a loading control (rRNA) in this and subsequent figures.
Next, the transcriptional induction of these CHV1-Δp69 up-regulated genes in the wild-type C. parasitica strain (EP155), SAGA complex (Δsgf73, Δgcn5, and Δubp8)-, and RNA silencing (Δdcl2 and Δagl2)-related fungal mutants was assayed by Northern blotting. From preliminary RNA blotting analysis, we detected the expression of uprt, gtpch, aprt, hp1, and hp2 transcripts but not btb, hel, or ac transcripts in CHV1-Δp69–infected DK80 and EP155 strains. The three undetectable genes were excluded from further RNA blotting analyses. Expression of the uprt, gtpch, aprt, hp1, and hp2 transcripts was highly induced in the CHV1-Δp69–infected EP155 strain but uninduced in the wild-type CHV1-infected EP155 strain (Fig. 1D), suggesting that CHV1-encoded p29 suppresses the transcriptional induction of these genes similarly to that previously observed for the suppression of dcl2 transcriptional induction by this protein (17, 20). Upon CHV1-Δp69 infection, the transcriptional induction of the uprt, gtpch, aprt, hp1, and hp2 genes was largely diminished in Δsgf73 and undetectable in Δgcn5 mutants, but was retained at a high level in Δubp8 mutants (Fig. 1D), indicating that transcriptional induction of these genes required the histone acetyltransferase (GCN5) but not histone deubiquitinase (UBP8) activity of the SAGA complex modules. Intriguingly, the gene transcripts were not induced in the Δdcl2 mutant but remained at a high level in the Δagl2 mutant (Fig. 1D). Similarly, upon dsRNA viral (MyRV1) infection, transcriptional induction was detected in wild-type EP155 and the Δagl2 mutant but not in the Δdcl2 mutant (Fig. 1D). RT-PCR analysis also confirmed that the transcriptional induction of the hel, ac, btb, and uprt genes was highly diminished in Δdcl2 (SI Appendix, Fig. S1F). Furthermore, this transcriptional up-regulation was observed in fungal transformants expressing inverted repeats (IR dsRNA) derived from an endogenous gene of C. parasitica, mitogen-activated protein kinase (CpMK1) gene (20) (SI Appendix, Fig. S3). Taken together, these observations reveal the roles of the SAGA complex and DCL2 in C. parasitica gene transcriptional induction following viral infection or dsRNA expression.
Deletion of Virus-Induced Genes Enhances CHV1-Δp69 Symptom Expression.
Since the transcriptional expression of these genes was up-regulated by viral infection, we investigated their roles in the host immune response. Disruption mutants of the uprt, gtpch, aprt, hp1, hp2, btb, hel, and ac genes (SI Appendix, Fig. S4) were generated from the parent C. parasitica DK80 strain that supports efficient homologous recombination-based targeted disruption (31). The disruption mutants were subsequently inoculated with CHV1-Δp69 through hyphal fusion. All virus-free disruptants showed similar growth and phenotype as the parental strain DK80 on potato dextrose agar (PDA) plates, with the exception of the Δhel mutant, which showed markedly slower growth and less mycelial pigmentation (Fig. 2A and SI Appendix, Fig. S5). Upon CHV1-Δp69 infection, most of the disruption mutants showed a slight-to-marked reduction in fungal growth compared with virus-infected DK80. In particular, Δaprt and Δhp2 exhibited the most prominent reduction in fungal growth following CHV1-Δp69 infection (Fig. 2 A and B). Northern blotting analysis showed that CHV1-Δp69 RNA accumulation levels in the disruption mutants were similar to those in the DK80 strain, and the dcl2 transcripts were also highly induced (Fig. 2C). These results suggest that the virus-induced genes have no direct antiviral role but instead are required for enhancement of the physiological tolerance of the host fungus against viral infection.
Fig. 2.
Symptom expression and RNA accumulation of CHV1-Δp69 in the C. parasitica mutants with deletions in the genes up-regulated by viral infection. (A) Phenotypic growth of DK80 and its disruption mutants of virus-induced genes (Δuprt, Δgtpch, Δaprt, Δhp1, Δhp2, Δbtb, Δhel, and Δac) infected with CHV1-Δp69. The fungal colonies were cultured on PDA plates for 5 d and photographed. See SI Appendix, Fig. S5 for colony morphologies of a subset of the virus-free mutant strains. (B) Colony size of the CHV1-Δp69–infected fungal mutants shown in A. Bars represent colony diameter (n = 3). Asterisk (*) indicates a significant difference (Student’s t test) at P < 0.1. (C) Northern blotting analysis of the CHV1-Δp69 RNA genome and dcl2 transcripts in the fungal mutants shown in A.
Previous studies in other organisms, including S. cerevisiae, have shown SAGA-regulated genes to be responsive to abiotic stresses (32, 33); thus, we investigated whether these virus-responsive genes are also induced by stress conditions and therefore implicated in abiotic stress tolerance in fungi. All of the disruptants (except Δhel) and the wild-type EP155 strain were grown under oxidative stress (0.05% H2O2 treatment) or osmotic stress (0.5 M KCl treatment). Under these conditions, the wild-type and all tested mutant C. parasitica strains showed similar growth, and the transcripts of dcl2 and these seven genes were uninduced (SI Appendix, Fig. S6), showing that they were not responsive to these stress conditions.
High-Level Induction of dcl2 Is Nonessential for Its Transcriptional Up-Regulation Function.
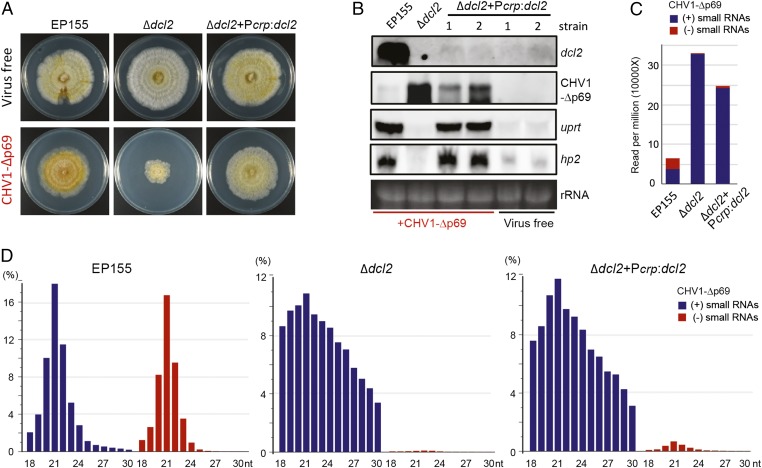
Our observations show that the transcriptional induction of the eight genes always occurs in accordance with high-level dcl2 transcriptional induction; thus, we questioned whether high induction of dcl2 is necessary for the transcriptional induction of these eight genes. To address this, a dcl2 expression construct (Pcrp:dcl2), in which a noninducible constitutive promoter of the cryparin (crp) gene from C. parasitica (34) was used to drive dcl2 cDNA expression, was used to transform the C. parasitica Δdcl2 mutant. The fungal transformants were subsequently inoculated with CHV1-Δp69 through hyphal fusion. Virus-free EP155 and Δdcl2 exhibited similar growth and phenotype; however, CHV1-Δp69 infection drastically hampered Δdcl2 but not EP155 growth (Fig. 3A). Notably, introduction of the Pcrp:dcl2 construct recovered Δdcl2 growth, following CHV1-Δp69 infection, to a similar level as that seen in virus-infected EP155 (Fig. 3A). Northern blotting detected high-level accumulation of the dcl2 transcript in CHV1-Δp69–infected EP155 but not in the virus-infected Δdcl2+Pcrp:dcl2 strain (Fig. 3B), which was expected from the cryparin promoter activity. CHV1-Δp69 RNA accumulated to a lower level in the Δdcl2+Pcrp:dcl2 than the Δdcl2 strain, but not as low as the CHV1-Δp69 RNA accumulation in EP155 (Fig. 3B). This observation indicates partial dcl2 complementation in Δdcl2+Pcrp:dcl2 and confirms the significance of DCL2 induction in strengthening antiviral defense. Strikingly, the uprt and hp2 transcripts were highly induced in the virus-infected Δdcl2+Pcrp:dcl2 strain, to a similar level as that seen in the virus-infected EP155 strain (Fig. 3B); thus, the high induction of the dcl2 transcript in C. parasitica is not required for the up-regulation of these genes.
Fig. 3.
Gene transcriptional up-regulation and viral small RNA production in the dcl2 complemented C. parasitica strain. (A) Colony morphology of the virus-free and CHV1-Δp69–infected EP155, Δdcl2, and Δdcl2+Pcrp:dcl2 (dcl2 complemented). The fungal strains were grown on PDA plates for 7 d. (B) Northern blotting analysis of the CHV1-Δp69 RNA genome and gene transcript accumulation in the fungal strains described in A. Two representative strains 1 and 2 of Δdcl2+Pcrp:dcl2 were analyzed. (C) Abundance of CHV1-Δp69–derived small RNAs. Viral small RNA reads (18–30 nucleotides) per million of the total small RNAs are shown for EP155, Δdcl2, and Δdcl2+Pcrp:dcl2. (D) Size distribution of CHV1-Δp69–derived small RNAs. “+” and “−” indicate small RNAs derived from the positive viral genomic (blue bars) or complementary (negative) strands (red bars), respectively.
To investigate the relationship between the dcl2 expression level and viral siRNA production, the total small RNAs in the CHV1-Δp69–infected EP155, Δdcl2, and Δdcl2+Pcrp:dcl2 strains were subjected to deep sequencing analysis. In EP155, there was a large number of CHV1-Δp69–derived small RNAs with typical characteristics of viral siRNA, such as a near equal proportion of both strands, predominantly 21 nt in length, and A/U bias at the 5′-terminal nucleotide (Fig. 3 C and D and SI Appendix, Fig. S7). Surprisingly, in the Δdcl2 samples, approximately fivefold more small RNAs were mapped to the CHV1-Δp69 genome, but they had unusual characteristics such that they had almost entirely positive-strand polarity with a wide-size distribution (18–30 nucleotides) and no preferential nucleotide at the 5′ terminus (Fig. 3 C and D and SI Appendix, Fig. S7). Moreover, the CHV1-Δp69–derived positive-strand small RNAs in the EP155 and Δdcl2 samples differed in distribution pattern across the viral genome (SI Appendix, Fig. S8A). A similar phenomenon was observed in a Dicer 2 mutant in the fly, Drosophila melanogaster (35). It is therefore likely that these virus-derived small RNAs in Δdcl2 are not bona fide viral siRNAs but rather the degradation products created by an unknown host endonuclease. The Δdcl2+Pcrp:dcl2 sample also contained a high number of virus-derived small RNAs with similar profiles to those in the Δdcl2 sample; however, there was a measurable increase in the proportion of negative-strand virus-derived small RNAs (Fig. 3 C and D and SI Appendix, Fig. S8B), indicating the production of a small amount of viral siRNAs by the dcl2 complementation. This observation suggests that low-level expression of dcl2 in C. parasitica is correlated with a lower production of viral siRNAs, also implying that high-level production of viral siRNAs is not linked to the role of DCL2 in the transcriptional up-regulation of host genes.
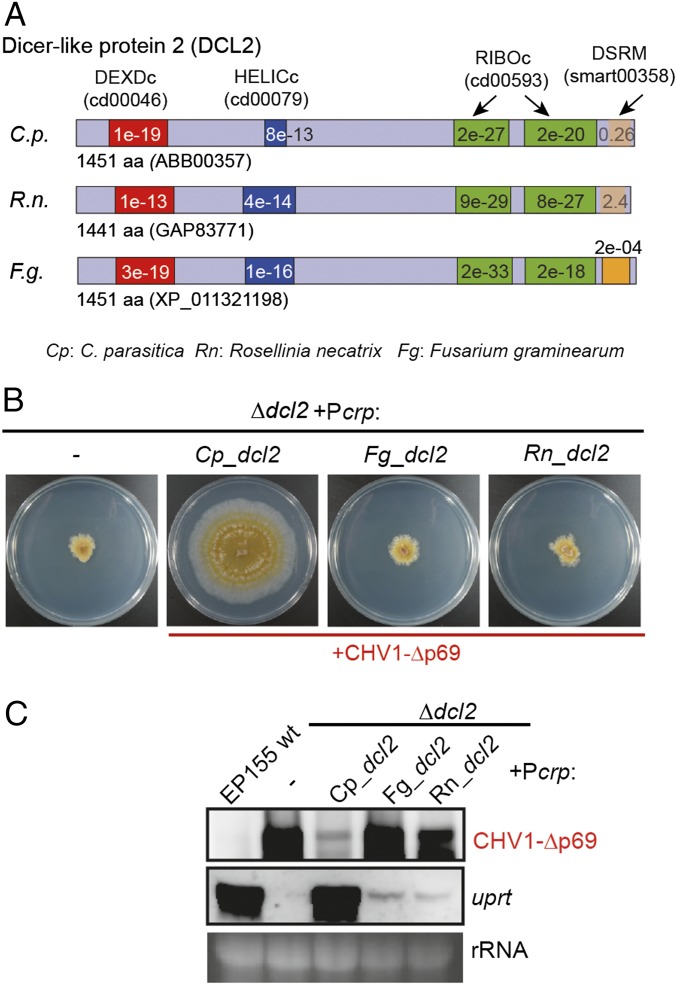
The protein sequence of DCL2 from C. parasitica contains two helicase (DEXDc and HELICc), two ribonuclease III (RNase III) (RIBOc), and one dsRNA binding motif (DSRM) (16) (Fig. 4A). To examine the importance of these motifs in the transcriptional up-regulation function of DCL2, five dcl2 mutant constructs were generated by introducing alanine substitutions (3–6 amino acids) into the conserved amino acid sequences of each of these motifs (SI Appendix, Figs. S9 and S10A). The dcl2 mutants were subsequently expressed in the Δdcl2 strain under the control of the cryparin promoter, as described above. Phenotypic observation and Northern blotting analysis show that none of these dcl2 mutants were able to restore Δdcl2 growth or transcriptional host gene induction upon CHV1-Δp69 infection (SI Appendix, Fig. S10 B–D), suggesting that these motifs are critical for such DCL2 functions.
Fig. 4.
Complementation using DCL2 homologs from other fungi. (A) Schematic diagrams showing helicase (DEXDc and HELICc), ribonuclease III (RIBOc), and DSRMs in DCL2 proteins from C. parasitica (ABB00357), R. necatrix (GAP83771), and F. graminearum (XP_011321198). (B) Phenotypic growth of C. parasitica Δdcl2 expressing homologous DCL2 from C. parasitica (+Pcrp:Cp_dcl2), and heterologous DCL2 from R. necatrix (+Pcrp:Rn_dcl2) or from F. graminearum (+Pcrp:Fg_dcl2) infected with CHV1-Δp69. The fungal strains were cultured on PDA plates for 7 d and photographed. (C) Northern blotting analysis of the CHV1-Δp69 RNA genome and uprt gene transcript accumulation in the fungal strains described in B.
dcl2 Homologs from Different Fungi Are Partially Functional in C. parasitica.
Next, we investigated whether dcl2 gene homologs from other fungi are functional in C. parasitica. Similar to C. parasitica, Rosellinia necatrix (white root rot fungus) and Fusarium graminearum (Fusarium head blight fungus) belong to the division Ascomycota but to the orders Xylariales and Hypocreales, respectively, whereas C. parasitica belongs to the order Diaporthales. The DCL2 proteins from R. necatrix and F. graminearum are similarly characterized by the presence of helicase, RNase III, and dsRNA binding motifs (Fig. 4A), which are distantly related to C. parasitica (SI Appendix, Fig. S11). Expression of dcl2 from R. necatrix or F. graminearum, under the control of the cryparin promoter (Pcrp:Rn_dcl2 and Pcrp:Fg_dcl2), in Δdcl2 was confirmed by RT-PCR (SI Appendix, Fig. S12). However, the heterologous dcl2 expression was unable to recover fungal growth upon CHV1-Δp69 infection (Fig. 4B) and led to virus accumulation comparable to noncomplemented Δdcl2 (Fig. 4C). Partial complementation by the homologs from different fungi was observed by slight increases in negative-strand virus-derived small RNA accumulation (SI Appendix, Fig. S13). Moreover, the expression of these heterologous dcl2 genes mediated only partial induction of the uprt gene compared with that in the EP155 strain (Fig. 4C). Thus, the combined results show that heterologous DCL2 proteins from R. necatrix and F. graminearum were able to partially complement C. parasitica DCL2 function.
Discussion
Dicer is known to down-regulate host and viral genes posttranscriptionally or transcriptionally with the aid of Argonaute via the biosynthesis of siRNAs or microRNAs. This primary function of Dicer therefore requires RNase III-like dsRNA-specific nuclease activity to digest dsRNA or structured single-stranded RNA molecules. In the present study, we show clearly that DCL2 is involved in the transcriptional up-regulation of a number of host C. parasitica genes, which is mediated by a general transcriptional SAGA complex. More interestingly, some of the up-regulated host genes contribute to the alleviation of symptom induction. An emerging picture of virus–host interactions from the present study is that a Dicer serves as a multifaceted guardian entity to protect hosts by repressing viral replication and mitigating symptom induction (Fig. 4).
The role for a Dicer revealed in the present study (hereafter termed “role A”) is distinct, with respect to a few points, from the well-established role (hereafter termed “role B”). First, role B requires Argonaute (AGL2), while role A appears not to require AGL2. This transcriptional induction role for Dicer is substantially different from the recently reported RNA-silencing components-mediated gene transcriptional up-regulation. In the flowering plant, Arabidopsis thaliana, AGO directly binds the chromatin of regulated genes with the guidance of small RNAs derived from them, thus requiring a small RNA-generating Dicer (36). In D. melanogaster, Dicer 2 appears to play an Argonaute 2-independent role, likely as a dsRNA sensor, to transcriptionally induce the Vago (a cytokine-like molecule) gene, which contributes to the inhibition of viral load in the body fat of host insects (35). Moreover, we have previously reported the virus-specific pathogen-associated molecular pattern (PAMP) requirement of AGL2 and DCL2, as positive feedback players, for the transcriptional induction of their genes in C. parasitica (21). Namely, full-scale induction of the agl2 and dcl2 genes by CHV1-Δp69 requires both DCL2 and AGL2, whereas that by MyRV1 requires only DCL2. Deletion of agl2 has been shown to lower the induction level of dcl2 upon CHV1-Δp69 infection. The present study clearly indicates the dispensability of AGL2 in role A of the SAGA-governed transcriptional induction of host genes upon infection with CHV1-Δp69 (Fig. 1D). Taken together, it can be concluded that relatively modest, steady-state levels of the dcl2 transcript are sufficient for C. parasitica host gene up-regulation (Fig. 3B), and that CHV1-Δp69 induces sufficient, albeit not full, levels of dcl2 in the absence of AGL2 (20).
Second, role B depends on small RNAs, while role A may be independent of small RNAs. We failed to uncouple the two DCL2 activities, i.e., roles A and B, by complementation analyses with constructs carrying mutations in the conserved domains such as RNase III (SI Appendix, Fig. S10); however, the nonessentiality of AGL2 strongly suggests no involvement of Dicer-generated small RNAs in transcriptional regulation. This hypothesis is supported by the observation that the considerable reduction in viral siRNA, resulting from the lack of dcl2 induction, has no effect on Dicer-associated transcriptional induction in C. parasitica (Fig. 3B). Further support comes from the observation that a number of host genes are under the regulation of Dicer–SAGA, suggesting global regulation rather than small RNA-mediated sequence-specific regulation. Third, role B is largely posttranscriptional down-regulation, likely in a dose-dependent manner at least in C. parasitica (20); when the expression of dcl2 is higher, C. parasitica becomes more resistant to susceptible viruses. Role A, on the other hand, is transcriptional up-regulation, and a relatively low level of Dicer expression is sufficient for this activity (Fig. 3B). Fourth, while role B is specifically involved in inhibition of viral replication, role A is involved in mitigation of a virus-induced symptom (Fig. 2A). Dicer deletion mutants often show enhanced host susceptibility to viruses as characterized by pronounced symptoms. It is generally accepted that this enhanced symptom induction results from increased virus replication; thus, the present study reveals an additional dimension to the Dicer-mediated alleviation of virus-induced symptoms.
Viral symptom induction is a result of complex interactions between a host and a virus and is primarily determined by their genotypes. We have previously identified a host C. parasitica gene encoding a possible Mg2+ transporter, nam1, involved in colony morphology that contributes to the alleviation of symptom induction by CHV1 strains (37). The present study also shows several genes to be regulated by SAGA in a Dicer-dependent manner and suggests involvement of some in the alleviation of symptom induction. Interestingly, many of these genes appear to be involved in the modification or degradation of nucleotides or RNA (Fig. 1C). Deletion of, for instance, the hp2 (JGI Genome Portal ID 356052) or aprt (JGI Genome Portal ID 287505) genes, resulted in a more severe phenotype as characterized by less growth and an irregular margin compared with the parental DK80 strain, when infected with CHV1-Δp69 (Fig. 2A). These phenotypic differences from the parental strain only appeared following infection with CHV1-Δp69 (Fig. 2A and SI Appendix, Fig. S5); therefore, these genes are concluded to be induced upon viral infection and to alleviate symptom induction (reduced growth rate). Importantly, no significant difference in CHV1-Δp69 accumulation was observed in these mutants, suggesting functional roles of these genes in fungal physiology rather than in the direct inhibition of viral replication. Although single deletion mutants of other genes show phenotypic alteration similar to parental DK80 following infection with CHV1-Δp69, there remains the possibility that these genes, in concert, mitigate viral symptom induction.
The SAGA complex is a universally conserved transcriptional coactivator involved in the up-regulation of many genes, and its role in abiotic stress responses has been relatively well studied (28, 38, 39). For instance, in S. cerevisiae, many genes are regulated by SAGA in response to osmotic, heat, and oxidative stress; and some of the responsive genes are important for the mitigation of such stress (32). However, our attempts to associate SAGA-mediated, virus-induced gene regulation with abiotic (osmotic and oxidative) stress responses failed (SI Appendix, Fig. S6). Noteworthy, there may be subsets of SAGA-regulated stress-responsive genes, whether Dicer independent or dependent, as reported for other organisms. Our current and previous studies show SAGA to play important roles in responses against positive single-stranded RNA (CHV1-Δp69, lacking the p29 RNA-silencing suppressor) and dsRNA (MyRV1) viruses, and to regulate the transcriptional induction of the two main antiviral RNA-silencing components, dcl2 and agl2, in C. parasitica. Another recent paper shows SAGA to be associated with plant gene regulation upon pathogen attack by an oomycete (Phytopthora sojae, a fungus-like eukaryotic microorganism) (21, 40), indicating that a pathogen-derived effector targets the SAGA pathway to benefit the infection process (40). In this regard, the activity of CHV1 p29, as an RNA-silencing suppressor to impair the up-regulation of dcl2 and possibly SAGA-mediated up-regulation of other host genes, is noteworthy. Therefore, CHV1 p29 is reminiscent of the oomycetous effector, although their mechanisms could be different. Inspection of Fig. 3 revealed that a constitutive level of dcl2 is sufficient for the up-regulation of SAGA-mediated host genes, restoration of fungal growth impaired by CHV1-Δp69, and suppression of CHV1-Δp69 replication. However, infection by the wild-type CHV1 failed to induce transcription up-regulation of a subset of host genes (Fig. 1D), despite the fact that CHV1 infection induced dcl2 by 10-fold compared with virus-free EP155 (17). These observations suggest that CHV p29 may directly block the up-regulation of SAGA-mediated host genes (Fig. 5).
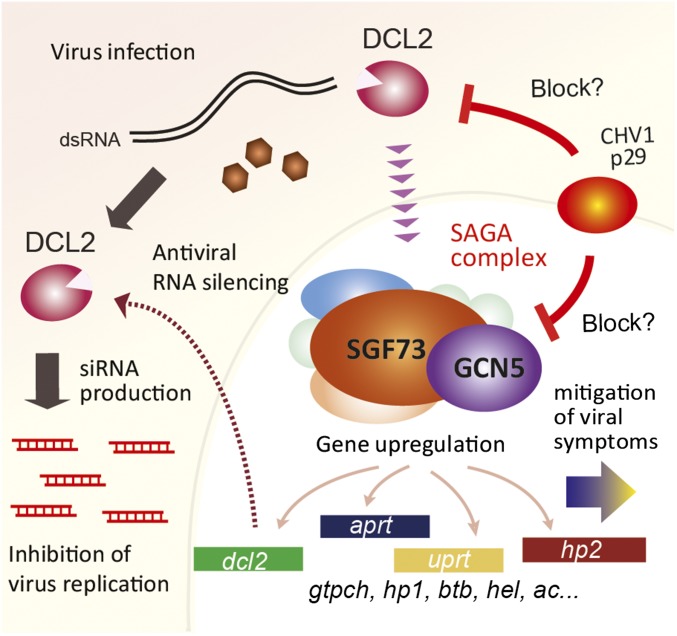
Fig. 5.
A model of the dual function of DCL2 in C. parasitica. Upon viral infection, DCL2 has a direct antiviral role in the processing of virus-derived dsRNA into siRNAs. DCL2 is also involved in gene transcriptional induction mediated by the SAGA complex. The dcl2 transcript is up-regulated to further strengthen the antiviral RNA-silencing response, while the transcript induction of a subset of genes enhances the physiological tolerance of the fungal host.
Given these observations and considerations, we propose a model to account for the regulatory pathway (Fig. 5) in which Dicer, in combination with SAGA, up-regulates a subset of virus-responsive host genes, including the key RNA-silencing genes. This up-regulation is cancelled out by the RNA-silencing suppressor, p29, encoded by CHV1 (Figs. 1D and 5). Collectively, the bilayer antiviral pathway is considered to be regulated by DCL2 in a small RNA-dependent (RNA silencing) and AGL2-independent manner (symptom mitigation). This may show a parallelism to multilayer antiviral immunity mechanisms in higher eukaryotes, for instance, RNA silencing and viral molecular pattern (dsRNA)-triggered immunity in plants and insects (41–43). Although it is possible that DCL2 serves as a dsRNA sensor, as is the case with two essential innate immune receptors, retinoic acid-inducible gene I (RIG-I) and melanoma differentiation-associated gene 5 (MDA5), in vertebrates (44) and possibly Dicer 2 in invertebrates (or insects) (35), the manner by which C. parasitica DCL2 participates in the transcriptional up-regulation pathway remains largely elusive. Little is known about whether the global SAGA-mediated transcriptional regulation upon viral infection is a universal phenomenon, or whether Dicer contributes to it. Recently, pattern requirement for antiviral RNA silencing, different even from that in C. parasitica, was revealed for a fungus belonging to a different family but to the same order or class (45, 46). Thus, although different functionality of Dicer can readily be predicted in different organisms, it will be of great interest to investigate whether Dicer is involved in transcriptional gene regulation in other organisms.
Materials and Methods
Viral and Fungal Strains.
Three viral strains were used in the present study: wild-type CHV1 (47); an ORF A mutant of CHV1 lacking the viral silencing suppressor, p29, (CHV1-Δp69b) (48); and a reovirus, MyRV1 (49, 50). The standard C. parasitica strain, EP155, and its RNA silencing-deficient derivatives, Δdcl2 and Δagl2 (16, 17, 31), were generous gifts from Donald L. Nuss, University of Maryland, College Park, MD. The C. parasitica strain, DK80, a mutant of EP155 disrupted in the cpk80 gene necessary for nonhomologous recombination (31), was a generous gift from Bao-shan Chen, Guangxi University, Nanning, China. CpMK1-IR transformants expressing inverted repeats (IR dsRNA) derived from an endogenous gene of C. parasitica, mitogen-activated protein kinase (CpMK1) gene were described previously (20). Fungal cultures were grown at 22–27 °C on PDA plates on the benchtop for maintenance and phenotypic observations and on PDA plates layered with cellophane for RNA preparation.
Plasmid Constructs and Fungal Transformation.
DNA fragments used for the generation of deletion (knockout) mutants through homologous recombination (neomycin selection) have been described previously (37). For the dcl2 complementation experiment, the DNA fragment corresponding to the C. parasitica dcl2 ORF was inserted into the HpaI site of pCPXNeo (14) using the in-fusion cloning system (Clontech). Site-directed mutagenesis of dcl2 was carried out using two PCR steps, as described previously (51).
Spheroplast isolation and transformation were performed as described previously (52).
RNA Analysis.
Total RNA was extracted as described previously (53). RNA blotting was performed, as described previously, using digoxigenin (DIG)-labeled PCR products (Roche) (54). RT-PCR was performed by the method of Andika et al. (21). All primers used in the present study are listed in SI Appendix, Table S2.
Transcriptome, Differential Expression, and Small RNA Analyses.
Total RNA was used for cDNA library construction using the TruSeq RNA Sample Preparation Kit (Illumina). In this method, cDNA is end repaired and adenylated before adaptor ligation, library construction, and amplification. The sequence-ready library was subjected to paired-end sequencing of 100 nucleotide reads using Illumina HiSEq. 2500 technology (Illumina). The cDNA library construction and deep sequencing analysis were carried out by Macrogen Japan, Ltd.
Transcriptome analysis was performed using the CLC Genomics Workbench 10 software (CLC bio). Following adaptor trimming, paired-end sequencing reads were mapped to the reference sequences from the annotated de novo transcriptome assembly of C. parasitica (https://genome.jgi.doe.gov/portal/Crypa2/Crypa2.download.ftp.html). Mapped read counts of each gene were converted to RPKM. RPKM values were used in two-group comparisons by Kal’s Z test to determine the significantly differentially expressed genes (P < 0.05 and fold change >10 with an RPKM >30).
Small RNA cDNA library construction and sequencing were performed by the Research Institute for Microbial Diseases of Osaka University using the Illumina TruSeq Small RNA Library Preparation Kit and the HiSEq. 2000 system (50-bp single-end reads). Treatment of raw data and subsequent viral-derived small RNA analysis were performed as described previously (21) using the CLC Genomics Workbench and the MISIS2 program (55).
Sequence and Phylogenetic Analyses.
Sequence data were analyzed using GENETYX-MAC (Genetyx Co) or Enzyme X v3.3.3 (nucleobytes.com/enzymex/index.html). The conserved protein domains were predicted by the National Center for Biotechnology Information (NCBI) conserved domain database (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Multiple sequence alignments were performed using the CLC Genomics Workbench or MAFFT v7 (56). Phylogenetic tree construction was performed as described previously (57).
Supplementary Material
Acknowledgments
We thank Drs. Donald L. Nuss and Satoko Kanematsu for the generous gift of the fungal/viral strains, Drs. Shota Nakamura, Daisuke Motooka, and Daisuke Okazaki for fruitful discussion, and the Next-Generation Sequencing core facility of the Genome Information Research Center at the Research Institute for Microbial Diseases of Osaka University for support in RNA sequencing.This study was supported in part by Yomogi, Inc. (N.S.) and Grants-in-Aid for Scientific Research (A) and Innovative Areas from the Japanese Ministry of Education, Culture, Sports, Science, and Technology (KAKENHI 25252011, 17H01463, 16H06436, 16H06429, and 16K21723 to N.S. and H.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1812407116/-/DCSupplemental.
References
- 1.Du P, et al. Viral infection induces expression of novel phased microRNAs from conserved cellular microRNA precursors. PLoS Pathog. 2011;7:e1002176. doi: 10.1371/journal.ppat.1002176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pumplin N, Voinnet O. RNA silencing suppression by plant pathogens: Defence, counter-defence and counter-counter-defence. Nat Rev Microbiol. 2013;11:745–760. doi: 10.1038/nrmicro3120. [DOI] [PubMed] [Google Scholar]
- 3.Aliyari R, et al. Mechanism of induction and suppression of antiviral immunity directed by virus-derived small RNAs in Drosophila. Cell Host Microbe. 2008;4:387–397. doi: 10.1016/j.chom.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deleris A, et al. Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science. 2006;313:68–71. doi: 10.1126/science.1128214. [DOI] [PubMed] [Google Scholar]
- 5.Ding SW. RNA-based antiviral immunity. Nat Rev Immunol. 2010;10:632–644. doi: 10.1038/nri2824. [DOI] [PubMed] [Google Scholar]
- 6.Baulcombe D. RNA silencing in plants. Nature. 2004;431:356–363. doi: 10.1038/nature02874. [DOI] [PubMed] [Google Scholar]
- 7.Nuss DL. Mycoviruses, RNA silencing, and viral RNA recombination. Adv Virus Res. 2011;80:25–48. doi: 10.1016/B978-0-12-385987-7.00002-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qu F, Ye X, Morris TJ. Arabidopsis DRB4, AGO1, AGO7, and RDR6 participate in a DCL4-initiated antiviral RNA silencing pathway negatively regulated by DCL1. Proc Natl Acad Sci USA. 2008;105:14732–14737. doi: 10.1073/pnas.0805760105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang XH, et al. RNA interference directs innate immunity against viruses in adult Drosophila. Science. 2006;312:452–454. doi: 10.1126/science.1125694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mourrain P, et al. Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell. 2000;101:533–542. doi: 10.1016/s0092-8674(00)80863-6. [DOI] [PubMed] [Google Scholar]
- 11.Dalmay T, Horsefield R, Braunstein TH, Baulcombe DC. SDE3 encodes an RNA helicase required for post-transcriptional gene silencing in Arabidopsis. EMBO J. 2001;20:2069–2078. doi: 10.1093/emboj/20.8.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spellberg MJ, Marr MT., 2nd FOXO regulates RNA interference in Drosophila and protects from RNA virus infection. Proc Natl Acad Sci USA. 2015;112:14587–14592. doi: 10.1073/pnas.1517124112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsuboyama K, Tadakuma H, Tomari Y. Conformational activation of Argonaute by distinct yet coordinated actions of the Hsp70 and Hsp90 chaperone systems. Mol Cell. 2018;70:722–729 e4. doi: 10.1016/j.molcel.2018.04.010. [DOI] [PubMed] [Google Scholar]
- 14.Eusebio-Cope A, et al. The chestnut blight fungus for studies on virus/host and virus/virus interactions: From a natural to a model host. Virology. 2015;477:164–175. doi: 10.1016/j.virol.2014.09.024. [DOI] [PubMed] [Google Scholar]
- 15.Nakayashiki H. RNA silencing in fungi: Mechanisms and applications. FEBS Lett. 2005;579:5950–5957. doi: 10.1016/j.febslet.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 16.Segers GC, Zhang X, Deng F, Sun Q, Nuss DL. Evidence that RNA silencing functions as an antiviral defense mechanism in fungi. Proc Natl Acad Sci USA. 2007;104:12902–12906. doi: 10.1073/pnas.0702500104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun Q, Choi GH, Nuss DL. A single Argonaute gene is required for induction of RNA silencing antiviral defense and promotes viral RNA recombination. Proc Natl Acad Sci USA. 2009;106:17927–17932. doi: 10.1073/pnas.0907552106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang DX, Spiering MJ, Nuss DL. Characterizing the roles of Cryphonectria parasitica RNA-dependent RNA polymerase-like genes in antiviral defense, viral recombination and transposon transcript accumulation. PLoS One. 2014;9:e108653. doi: 10.1371/journal.pone.0108653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang X, Shi D, Nuss DL. Variations in hypovirus interactions with the fungal-host RNA-silencing antiviral-defense response. J Virol. 2012;86:12933–12939. doi: 10.1128/JVI.00961-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chiba S, Suzuki N. Highly activated RNA silencing via strong induction of dicer by one virus can interfere with the replication of an unrelated virus. Proc Natl Acad Sci USA. 2015;112:E4911–E4918. doi: 10.1073/pnas.1509151112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andika IB, Jamal A, Kondo H, Suzuki N. SAGA complex mediates the transcriptional up-regulation of antiviral RNA silencing. Proc Natl Acad Sci USA. 2017;114:E3499–E3506. doi: 10.1073/pnas.1701196114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koutelou E, Hirsch CL, Dent SY. Multiple faces of the SAGA complex. Curr Opin Cell Biol. 2010;22:374–382. doi: 10.1016/j.ceb.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morgan MT, et al. Structural basis for histone H2B deubiquitination by the SAGA DUB module. Science. 2016;351:725–728. doi: 10.1126/science.aac5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McMahon SJ, Pray-Grant MG, Schieltz D, Yates JR, 3rd, Grant PA. Polyglutamine-expanded spinocerebellar ataxia-7 protein disrupts normal SAGA and SLIK histone acetyltransferase activity. Proc Natl Acad Sci USA. 2005;102:8478–8482. doi: 10.1073/pnas.0503493102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee KK, Swanson SK, Florens L, Washburn MP, Workman JL. Yeast Sgf73/Ataxin-7 serves to anchor the deubiquitination module into both SAGA and Slik(SALSA) HAT complexes. Epigenet Chromatin. 2009;2:2. doi: 10.1186/1756-8935-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Helmlinger D, Tora L. Sharing the SAGA. Trends Biochem Sci. 2017;42:850–861. doi: 10.1016/j.tibs.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee TI, et al. Redundant roles for the TFIID and SAGA complexes in global transcription. Nature. 2000;405:701–704. doi: 10.1038/35015104. [DOI] [PubMed] [Google Scholar]
- 28.Huisinga KL, Pugh BF. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol Cell. 2004;13:573–585. doi: 10.1016/s1097-2765(04)00087-5. [DOI] [PubMed] [Google Scholar]
- 29.Yates SA, et al. De novo assembly of red clover transcriptome based on RNA-Seq data provides insight into drought response, gene discovery and marker identification. BMC Genomics. 2014;15:453. doi: 10.1186/1471-2164-15-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zenoni S, et al. Characterization of transcriptional complexity during berry development in Vitis vinifera using RNA-Seq. Plant Physiol. 2010;152:1787–1795. doi: 10.1104/pp.109.149716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lan X, et al. Deletion of the cpku80 gene in the chestnut blight fungus, Cryphonectria parasitica, enhances gene disruption efficiency. Curr Genet. 2008;53:59–66. doi: 10.1007/s00294-007-0162-x. [DOI] [PubMed] [Google Scholar]
- 32.Rep M, et al. The Saccharomyces cerevisiae Sko1p transcription factor mediates HOG pathway-dependent osmotic regulation of a set of genes encoding enzymes implicated in protection from oxidative damage. Mol Microbiol. 2001;40:1067–1083. doi: 10.1046/j.1365-2958.2001.02384.x. [DOI] [PubMed] [Google Scholar]
- 33.Guha N, Desai P, Vancura A. Plc1p is required for SAGA recruitment and derepression of Sko1p-regulated genes. Mol Biol Cell. 2007;18:2419–2428. doi: 10.1091/mbc.E06-10-0946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kwon BR, et al. Assessment of the core cryparin promoter from Cryphonectria parasitica for heterologous expression in filamentous fungi. Appl Microbiol Biotechnol. 2009;83:339–348. doi: 10.1007/s00253-009-1906-y. [DOI] [PubMed] [Google Scholar]
- 35.Deddouche S, et al. The DExD/H-box helicase Dicer-2 mediates the induction of antiviral activity in drosophila. Nat Immunol. 2008;9:1425–1432. doi: 10.1038/ni.1664. [DOI] [PubMed] [Google Scholar]
- 36.Liu C, et al. Arabidopsis ARGONAUTE 1 binds chromatin to promote gene transcription in response to hormones and stresses. Dev Cell. 2018;44:348–361 e7. doi: 10.1016/j.devcel.2017.12.002. [DOI] [PubMed] [Google Scholar]
- 37.Faruk MI, Eusebio-Cope A, Suzuki N. A host factor involved in hypovirus symptom expression in the chestnut blight fungus, Cryphonectria parasitica. J Virol. 2008;82:740–754. doi: 10.1128/JVI.02015-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moraga F, Aquea F. Composition of the SAGA complex in plants and its role in controlling gene expression in response to abiotic stresses. Front Plant Sci. 2015;6:865. doi: 10.3389/fpls.2015.00865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gaupel AC, Begley TJ, Tenniswood M. Gcn5 modulates the cellular response to oxidative stress and histone deacetylase inhibition. J Cell Biochem. 2015;116:1982–1992. doi: 10.1002/jcb.25153. [DOI] [PubMed] [Google Scholar]
- 40.Kong L, et al. A Phytophthora effector manipulates host histone acetylation and reprograms defense gene expression to promote infection. Curr Biol. 2017;27:981–991. doi: 10.1016/j.cub.2017.02.044. [DOI] [PubMed] [Google Scholar]
- 41.Mussabekova A, Daeffler L, Imler JL. Innate and intrinsic antiviral immunity in Drosophila. Cell Mol Life Sci. 2017;74:2039–2054. doi: 10.1007/s00018-017-2453-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Niehl A, Wyrsch I, Boller T, Heinlein M. Double-stranded RNAs induce a pattern-triggered immune signaling pathway in plants. New Phytol. 2016;211:1008–1019. doi: 10.1111/nph.13944. [DOI] [PubMed] [Google Scholar]
- 43.Zambon RA, Nandakumar M, Vakharia VN, Wu LP. The Toll pathway is important for an antiviral response in Drosophila. Proc Natl Acad Sci USA. 2005;102:7257–7262. doi: 10.1073/pnas.0409181102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takeuchi O, Akira S. Innate immunity to virus infection. Immunol Rev. 2009;227:75–86. doi: 10.1111/j.1600-065X.2008.00737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu J, Lee KM, Cho WK, Park JY, Kim KH. Differential contribution of RNA interference components in response to distinct Fusarium graminearum virus infections. J Virol. 2018;92:e01756-17. doi: 10.1128/JVI.01756-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mochama P, Jadhav P, Neupane A, Marzano SYL. Mycoviruses as triggers and targets of RNA silencing in white mold fungus Sclerotinia sclerotiorum. Viruses. 2018;10 doi: 10.3390/v10040214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shapira R, Choi GH, Nuss DL. Virus-like genetic organization and expression strategy for a double-stranded RNA genetic element associated with biological control of chestnut blight. EMBO J. 1991;10:731–739. doi: 10.1002/j.1460-2075.1991.tb08004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suzuki N, Nuss DL. Contribution of protein p40 to hypovirus-mediated modulation of fungal host phenotype and viral RNA accumulation. J Virol. 2002;76:7747–7759. doi: 10.1128/JVI.76.15.7747-7759.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suzuki N, Supyani S, Maruyama K, Hillman BI. Complete genome sequence of Mycoreovirus-1/Cp9B21, a member of a novel genus within the family Reoviridae, isolated from the chestnut blight fungus Cryphonectria parasitica. J Gen Virol. 2004;85:3437–3448. doi: 10.1099/vir.0.80293-0. [DOI] [PubMed] [Google Scholar]
- 50.Hillman BI, Supyani S, Kondo H, Suzuki N. A reovirus of the fungus Cryphonectria parasitica that is infectious as particles and related to the Coltivirus genus of animal pathogens. J Virol. 2004;78:892–898. doi: 10.1128/JVI.78.2.892-898.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chiba S, Jamal A, Suzuki N. First Evidence for internal ribosomal entry sites in diverse fungal virus genomes. MBio. 2018;9:e02350-17. doi: 10.1128/mBio.02350-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eusebio-Cope A, Suzuki N. Mycoreovirus genome rearrangements associated with RNA silencing deficiency. Nucleic Acids Res. 2015;43:3802–3813. doi: 10.1093/nar/gkv239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eusebio-Cope A, Sun L, Hillman BI, Suzuki N. Mycoreovirus 1 S4-coded protein is dispensable for viral replication but necessary for efficient vertical transmission and normal symptom induction. Virology. 2010;397:399–408. doi: 10.1016/j.virol.2009.11.035. [DOI] [PubMed] [Google Scholar]
- 54.Salaipeth L, Chiba S, Eusebio-Cope A, Kanematsu S, Suzuki N. Biological properties and expression strategy of rosellinia necatrix megabirnavirus 1 analysed in an experimental host, Cryphonectria parasitica. J Gen Virol. 2014;95:740–750. doi: 10.1099/vir.0.058164-0. [DOI] [PubMed] [Google Scholar]
- 55.Seguin J, Otten P, Baerlocher L, Farinelli L, Pooggin MM. MISIS-2: A bioinformatics tool for in-depth analysis of small RNAs and representation of consensus master genome in viral quasispecies. J Virol Methods. 2016;233:37–40. doi: 10.1016/j.jviromet.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 56.Katoh K, Rozewicki J, Yamada KD. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. September 6, 2017 doi: 10.1093/bib/bbx1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kondo H, Chiba S, Suzuki N. Detection and analysis of non-retroviral RNA virus-like elements in plant, fungal, and insect genomes. Methods Mol Biol. 2015;1236:73–88. doi: 10.1007/978-1-4939-1743-3_7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.