Significance
Legionella pneumophila is a bacterial pathogen causing outbreaks of a lethal pneumonia. The genus Legionella comprises 65 species for which aquatic amoebae are the natural reservoirs. Using functional and comparative genomics to deconstruct the entire bacterial genus, we reveal the surprising parallel evolutionary trajectories that have led to the emergence of human pathogenic Legionella. An unexpectedly large and unique repository of secreted proteins (>18,000) containing eukaryotic-like proteins acquired from all domains of life (plant, animal, fungal, archaea) contrasts with a highly conserved type IV secretion system. This study reveals an unprecedented environmental reservoir of bacterial virulence factors and provides an understanding of how reshuffling and gene acquisition from environmental eukaryotic hosts may allow for the emergence of human pathogens.
Keywords: Legionella, protozoa, coevolution, horizontal gene transfer, human pathogen
Abstract
The genus Legionella comprises 65 species, among which Legionella pneumophila is a human pathogen causing severe pneumonia. To understand the evolution of an environmental to an accidental human pathogen, we have functionally analyzed 80 Legionella genomes spanning 58 species. Uniquely, an immense repository of 18,000 secreted proteins encoding 137 different eukaryotic-like domains and over 200 eukaryotic-like proteins is paired with a highly conserved type IV secretion system (T4SS). Specifically, we show that eukaryotic Rho- and Rab-GTPase domains are found nearly exclusively in eukaryotes and Legionella. Translocation assays for selected Rab-GTPase proteins revealed that they are indeed T4SS secreted substrates. Furthermore, F-box, U-box, and SET domains were present in >70% of all species, suggesting that manipulation of host signal transduction, protein turnover, and chromatin modification pathways are fundamental intracellular replication strategies for legionellae. In contrast, the Sec-7 domain was restricted to L. pneumophila and seven other species, indicating effector repertoire tailoring within different amoebae. Functional screening of 47 species revealed 60% were competent for intracellular replication in THP-1 cells, but interestingly, this phenotype was associated with diverse effector assemblages. These data, combined with evolutionary analysis, indicate that the capacity to infect eukaryotic cells has been acquired independently many times within the genus and that a highly conserved yet versatile T4SS secretes an exceptional number of different proteins shaped by interdomain gene transfer. Furthermore, we revealed the surprising extent to which legionellae have coopted genes and thus cellular functions from their eukaryotic hosts, providing an understanding of how dynamic reshuffling and gene acquisition have led to the emergence of major human pathogens.
Legionnaires’ disease or legionellosis is an atypical pneumonia caused by bacteria of the genus Legionella. Shortly after the discovery of Legionella pneumophila (1), it was reported that this bacterium is pathogenic for freshwater and soil amoebae of the genera Acanthamoeba and Naegleria (2). This finding led to a new perception in microbiology, whereby bacteria that parasitize protozoa can utilize similar processes to infect human cells. Sequencing and analyses of the L. pneumophila genome substantiated this idea when it revealed the presence of a large number and variety of eukaryotic-like domains within the predicted proteome (3). Many of these proteins, termed effector proteins, were shown to be secreted into the host cell where they facilitate Legionella intracellular replication within a specialized compartment termed the Legionella-containing vacuole (LCV) (3, 4). Overall, the type IV secretion system (T4SS) Dot/Icm secretes more than 300 different effector proteins into the host cell and is indispensable for virulence of L. pneumophila (5–8). The presence of the Dot/Icm T4SS in other L. pneumophila strains and in selected Legionella species has also been reported (9–12), but recent genome-scale studies of Legionella (13–15) have indicated that the T4SS is present in every Legionella strain analyzed.
Despite high conservation of the Dot/Icm system among different Legionella species, effector repertoires appear to vary greatly. An analysis of putative T4SS effectors of Legionella longbeachae, the second most frequent cause of Legionnaires’ disease, revealed that only about 50% of the virulence factors described in L. pneumophila were also present in the genome of L. longbeachae (16). Recently, Burstein et al. (14) analyzed 38 Legionella species using a machine learning approach to predict T4SS effectors, and Joseph et al. (15) examined Legionella genome dynamics; both groups concluded that DNA interchange between different species is rare. However, still little is known about the potential of the different species to cause human disease and about the impact and the specific characteristics of the T4SS effectors on the evolution of new human pathogens within this environmental bacterial genus.
Here, we present a comprehensive analysis of the Legionella genus genome, covering 80 Legionella strains belonging to 58 Legionella species and subspecies. We establish a pan-genus pool of putative T4SS effectors and show that this comprises over 18,000 proteins and we identify more than 200 eukaryotic-like proteins and 137 eukaryotic domains, including a unique class of putative bacterial Rab GTPases. We confirmed experimentally that a subset of these proteins translocate into the host cell upon infection. We conclude that the T4SS is highly conserved at the sequence level, but that the effector proteins secreted are highly diverse.
Results and Discussion
The Legionella Genus Genome Is Dynamic and Characterized by Frequent Genetic Exchange.
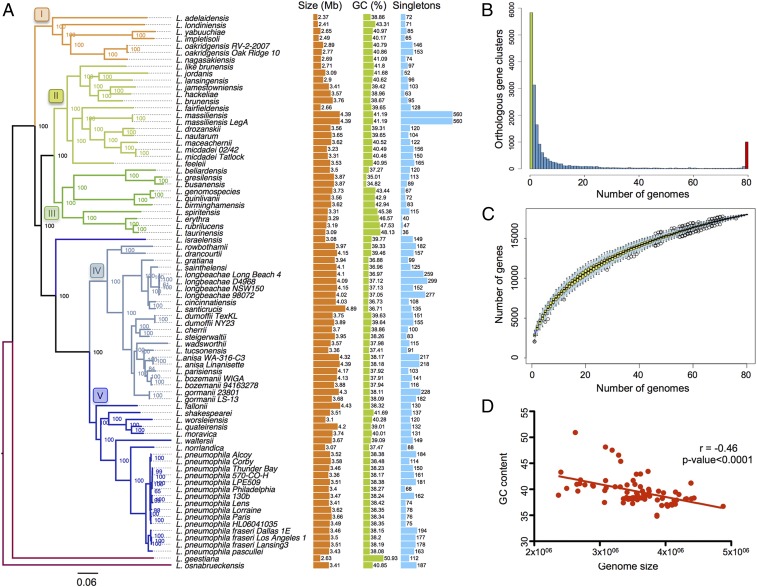
We sequenced 58 Legionella species and analyzed them in combination with all publicly available genomes (80 genomes in total) (SI Appendix, Table S1). The Legionella genomes were extremely diverse: The genome size varied from 2.37 Mb (Legionella adelaidensis) to 4.88 Mb (Legionella santicrucis), the GC content varied from 34.82% (Legionella busanensis) to 50.93% (Legionella geestiana), and the number of clusters of orthologous genes as defined with OrthoMCL was 17,992, of which 5,832 (32%) were strain specific (singletons) (Fig. 1A). Only 1,008 genes (6%) constituted the core genome (Fig. 1B) compared with an earlier analysis of 38 Legionella species that found 16,416 clusters of orthologs and 1,054 core genes (14). The addition of 40 genomes comprising 16 Legionella species sequenced in our study increased the number of orthologous gene clusters by over 1,576 and decreased the core genome by 46 genes, underlining the high diversity of the Legionella genus. This difference suggests that the Legionella genus pan-genome is far from fully described and that sequencing of additional Legionella species will increase the genus gene repertoire significantly. This is supported by the rarefaction curve that does not reach a plateau (Fig. 1C).
Fig. 1.
The Legionella genomes are diverse in size and gene content. (A) Phylogeny of the genus based on the core genome, genome size, GC content, and number of singletons of each species are depicted. Numbers represent bootstrap values. Branches are colored according to the clade they belong to. Genome size and GC content include plasmids if present in the corresponding species. The number of singletons is based on the results of OrthoMCL (takes into account orthologs and paralogs). Each species has been compared with the others without taking into account strains from the same species to avoid bias due to the number of strains sequenced within a species. (B) Occurrence of genes within the 80 analyzed Legionella genomes. Left end of the x axis (green bar), genes present in a single genome (strain-specific genes; 5,832, ∼32% of the pangenome); right end of the x axis (red bar), genes present in all 80 genomes (core genome; 1,008 genes, ∼6% of the pan-genome). (C) Gene accumulation curve for the total number of proteins of the 80 genomes. (D) Negative correlation between genome size and GC content, indicating high acquisition of foreign genes (Pearson’s correlation coefficient equal to −0.46 with P < 0.0001).
The highly dynamic nature of these genomes is also seen in the analysis of the strain-specific genes and the accessory genome because it highlights the presence of several mobile genetic elements, which are often associated with genes encoding for transfer regions/conjugative elements such as the type IVA secretion systems (T4ASSs). These T4ASSs [classified as T4SSF, -G, -I, and -T (17)] are present in each strain to varying degrees, indicating that they circulate among the different Legionella strains (SI Appendix, Table S2) and therefore drive genome dynamics and diversification. It has been suggested that the incorporation of foreign DNA via horizontal gene transfer (HGT) is responsible for an increase in the AT content and the increase in genome size (18). Indeed, we found a negative correlation between the genome size and the GC content for the Legionella genomes, which also suggests frequent HGT (Fig. 1D) (19). Despite the importance of flagella for transmission to new hosts, as shown for L. pneumophila, flagella encoding genes were not conserved in all species but showed a patchy distribution, as 23 of the 80 strains analyzed lacked flagella genes (SI Appendix, Fig. S1). The analyses showed that the Legionella genus genome is highly diverse, dynamic, and shaped by HGT.
The Genus Legionella Encodes Proteins with 137 Different Eukaryotic Domains.
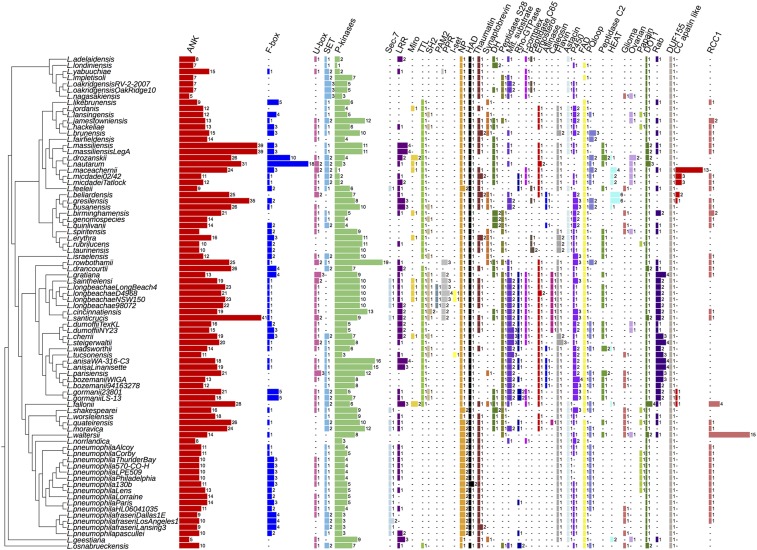
InterProScan analysis of all 58 Legionella species revealed the presence of 137 different eukaryotic motifs/domains in the genus Legionella (SI Appendix, Table S3) according to the definition that a eukaryotic domain is one that is found in >75% of eukaryotic genomes and in <25% of prokaryotic genomes. The most abundant eukaryotic domains identified were ankyrin repeats. Interestingly, Legionella santicrucis and Legionella massiliensis encoded 41 and 39 ankyrin domains, respectively (Fig. 2). Ankyrin motifs were found frequently associated with other eukaryotic motifs and thus constituted modular proteins associated with eukaryotic F-box, U-box, Rab, or SET domains. Notably, F-box and U-box domains were present in more than two-thirds of the species analyzed (Fig. 2), suggesting that manipulation of the host ubiquitin system is a fundamental virulence strategy of Legionella species. Generally, the genomes contained one to three F-box–containing proteins, with the exception of Legionella nautarum and Legionella drozanskii, which contained 18 and 10, respectively. The SET domain containing protein RomA of L. pneumophila that induces a unique host chromatin modification (20) is present in 46 of the 58 Legionella species, suggesting the ability of many Legionella species to manipulate host chromatin (Fig. 2). Interestingly, the Sec-7 domain present in the effector RalF, a bacterial exchange factor for the ADP-ribosylation factor family of guanosine triphosphatases and the first described Dot/Icm effector of L. pneumophila (21), was present in only eight (L. pneumophila, L. longbeachae, Legionella feelei, Legionella sainthelensi, L. santicrucis, Legionella shakespearei, Legionella quateirensis, and Legionella moravica) of the 58 Legionella species analyzed, suggesting that different effectors may compensate for RalF activity or that LCV biogenesis varies among different species (Fig. 2).
Fig. 2.
Eukaryotic domains have a diverse distribution within the genus Legionella, suggesting multiple acquisition events. The number and distribution of the 41 most frequently identified eukaryotic motifs within the genus Legionella are shown. Numbers represent the number of proteins containing this eukaryotic motif. ANK, ankyrin; Astacin, peptidase M12A astacin; C/C, clathrin/coatomer adaptor adaptin-like; DH, Dbl homology domain; DOT1, histone methylation DOT1; Ergosterol, ergosterol biosynthesis; FAD, cytokinin dehydrogenase 1 FAD/cytokinin binding domain; F-box, F-box domain; Flavin, flavin monooxygenase-like; Glioma, leucine-rich glioma-inactivated EPTP repeat; HAD, HAD-superfamily hydrolase; I-set, Ig I-set; LLR, leucine rich repeats; Miro, mitochondrial Rho domain; Mit. substrate, mitochondrial substrate/solute carrier; NP, nucleoside phosphatase gda1/cd39; Ovarian, ovarian tumor otubain; P450, cytochrome_P450; PAM2, ataxin-2 C-terminal; Papain, peptidase C1A papain C-terminal; Peptidase C2, calpain catalytic domain; Peptidase C65, peptidase C65 otubain; P-kinases (protein kinases); PPR, pentatricopeptide repeat; PQloop, PQ loop repeat; Rab, Rab small GTPases; RCC1, regulator of chromosome condensation; Rho-GTPase, Rho-GTPase domain; Sec-7, Sec-7 domain; SET, SET domain; SH2, Src homology 2; T-complex, T-complex 10/11; TTL, tubulin-tyrosine ligase; U-box, U-box domain.
One identified motif in Legionella was the ergosterol reductase ERG4/ERG24 (IPR001171) domain. Ergosterol is the primary sterol in the cell membranes of filamentous fungi, present in membranes of yeast and mitochondria (22). Importantly, it is also the major sterol of amoebae such as Acanthamoeba castellanii and Acanthamoeba polyphaga, the natural hosts of Legionella (23, 24). We found that 31 Legionella species encoded one or two proteins with the ERG4/ERG24 domain (Fig. 2). The L. longbeachae protein (Llo1320) containing this domain showed 56% amino acid identity to that encoded by the amoeba Naegleria gruberi and 30% amino acid identity to that encoded by A. castellanii strain Neff. This domain was also present in other amoebae-related bacteria such as Parachlamydia acanthamoebae and Protochlamydia naegleriophila, as well as Coxiella burnetii. Phylogenetic analyses suggest that L. longbeachae acquired this domain from amoeba (SI Appendix, Fig. S2A).
Phylogenetic analyses of the here-identified C-terminal alliinase or caleosin domains present in Legionella beliardensis and Legionella anisa or the L. longbeachae clade (Fig. 2), respectively, further supported acquisition of these domains from plants, amoeba, or fungi (SI Appendix, Fig. S2 B and C). They probably help Legionella to fight competitor bacteria or fungi in amoebae or in the environment. Together, our analyses highlight key domains preferentially present in protozoa, fungi, plants, or animals that have been acquired by different Legionella species.
A Unique Case in the Prokaryotic World: Legionella Encode Small GTPase-Like Domains.
The Ras-related small GTPase superfamily comprises more than 150 members in humans, which function as key regulators of signal transduction in almost all cellular processes (25). These enzymes bind and hydrolyze GTP to GDP and activate downstream effectors when bound to GTP. The first identified member was the p21-Ras protein, an evolutionary conserved small GTPase that controls cell proliferation, survival, and migration through its effector binding at RAF/MAPK and PI3K (26). The Ras protein superfamily is subdivided into at least five distinct branches: Ras, Rho, Rab, Arf, and Ran (27). Evolutionarily conserved orthologs are found in Drosophila, Caenorhabditis elegans, Saccharomyces cerevisiae, Schizosaccharomyces pombe, Dictyostelium, and plants (28).
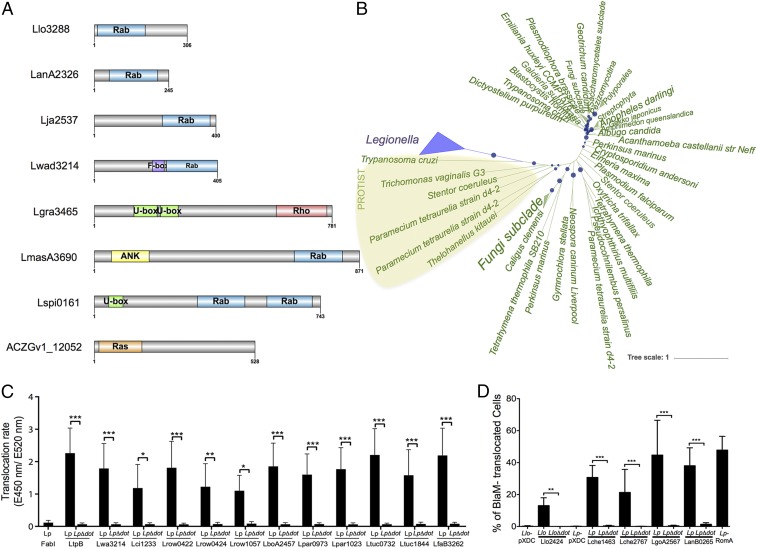
The only Rab-like protein in a prokaryotic genome was reported in the L. longbeachae genome sequence (16). However, upon analysis of our 80 Legionella strains, we identified 184 small GTPases, of which 104 could be classified with a very high confidence as Rho-, Ras-, or Rab-like proteins (1 Rho, 34 Ras, and 71 Rab domains) (SI Appendix, Fig. S3 and Table S4). Blastp analysis of these proteins in the National Center for Biotechnology Information database revealed that 149 of the 184 small GTPases of Legionella were exclusively present in Legionella and eukaryotic organisms (Table 1). The Rab domain was localized to different parts of the effector proteins, and a subset of Rab proteins carried additional domains such as U-box domains, ankyrin motifs, or F-box domains (Fig. 3A). Alignment of the different Rab domains identified in the Legionella genomes revealed that the structural features of eukaryotic Rab domains were conserved among the Legionella proteins (SI Appendix, Fig. S4).
Table 1.
Homology of Legionella Rab domain-containing proteins against protozoan Rab proteins
| Domain | Protein | First blast hit | Identity, % | Coverage, % | E value |
| Rab | Lade0491 | Entamoeba histolytica | 35 | 52 | 4.E-17 |
| Rab | LgoA0634 | Paramecium tetraurelia | 33 | 51 | 2.E-19 |
| Rab | Llo3288 | Ichthyophthirius multifiliis | 42 | 53 | 4.E-31 |
| Rab | Lstei0814 | Tetrahymena thermophila | 34 | 86 | 3.E-26 |
| Rab | Lstei2185 | Stentor coeruleus | 38 | 55 | 6.E-29 |
| Rab | Lbir2252 | Entamoeba invadens | 32 | 55 | 5.E-15 |
| Rab | Lges1860 | Entamoeba histolytica | 34 | 55 | 7.E-25 |
| Rab+ Fbox | Lwad3214 | Paramecium tetraurelia | 34 | 35 | 7.E-14 |
| Rab | Lgra2891 | Guillardia theta | 36 | 56 | 2.E-19 |
| Rab | Lgra3435 | Entamoeba histolytica | 35 | 59 | 2.E-27 |
| Rab | Lma1540 | Paramecium tetraurelia | 34 | 55 | 1.E-17 |
| Rab + ank | LmasA3690 | Oxytricha trifallax | 34 | 19 | 2.E-19 |
| Rab | Lqua0234 | Dictyostelium fasciculatum | 38 | 34 | 1.E-25 |
| Rab | Lquin3026 | Tetrahymena thermophila | 34 | 57 | 1.E-19 |
| Rab | Lspi0161 | Naegleria gruberi | 34 | 24 | 7.E-24 |
| Rab | Lwal3261 | Paramecium tetraurelia | 33 | 85 | 7.E-18 |
Each Rab protein listed in the table represents a different orthologous group. Results are based on blastp searches using the nonredundant National Center for Biotechnology Information database.
Fig. 3.
Domain organization of small GTPases in Legionella and phylogenetic analyses of the Llo3288 Rab proteins suggest eukaryotic origin. (A) Domain organization of the different small GTPase proteins identified. (B) Unrooted tree of Llo3288 and homologs recruited by blastp constructed using likelihood. Local support values are represented with circles on the corresponding branches, and size of circles is proportional to the values (only local support values of at least 0.7 are shown). (C) Translocation of selected proteins using the beta-lactamase translocation assay and infection of Raw264.7 cells for 1 h with Lp wild type or LpΔdotA expressing BlaM-effector fusions analyzed with a microplate reader. Three independent experiments (n = 9) were done. Statistical significance was determined by two-way ANOVA with multiple comparisons test (*P < 0.05; **P < 0.01; ***P < 0.001). (D) Translocation of selected proteins using the beta-lactamase translocation assay and infection of THP-1 cells at a multiplicity of infection of 50 during 1 h 30 min with Lp and Llo strains before addition of CCF4-AM and analyses by flow cytometry. Histograms show the frequency of BlaM- translocated, blue fluorescence-emitting cells as means ± SD of three independent experiments (n = 12). Statistical significance was determined by Wilcoxon matched pairs test (**P < 0.01; ***P < 0.001). Llo, L. longbeachae wild type; LloΔdot, L. longbeachae ΔdotA; Lp, L. pneumophila wild type; LpΔdot, L. pneumophila ΔdotA.
To analyze further the evolutionary history of the Ras-related domains in Legionella, we undertook phylogenetic analyses of these proteins. For example, the two L. longbeachae Rab proteins, Llo1716 and Llo3288, were present in all strains closely related to L. longbeachae, suggesting that they and their orthologs share a common origin and evolved from a gene acquired by the ancestor of all these species (SI Appendix, Fig. S5). Further phylogenetic analysis of 16 Rab proteins present in eight different Legionella species showed that these Rab domains were acquired by HGT, mainly from protozoa (Fig. 3B and SI Appendix, Fig. S6). Recently, a novel isoform of Rab5D was identified in the Acanthamoeba polyphaga mimivirus (APMV) and all group I members of the family Mimiviridae (29). Phylogenetic analyses suggested that the Rab GTPase was acquired by an ancestor of the Mimiviridae family, and Rabs from Mimiviridae, Plasmodium, and few lower eukaryotes form a separate clade (29). Thus, Legionella and APMV that both infect the protozoa Acanthamoeba encode Rab proteins, most likely to mimic and subvert host cell function. To substantiate that these proteins act in the host cell, we determined whether the Rab-containing proteins were bona fide substrates of the Dot/Icm T4SS by creating fusion proteins between the 16 different Rab proteins and the catalytic domain of the TEM-1 beta-lactamase (indicated by asterisks in SI Appendix, Fig. S5). Translocation assays were performed using wild-type L. pneumophila as a surrogate host and compared with an isogenic Dot/Icm mutant (∆dotA). All 16 Rab motif-containing proteins were translocated by L. pneumophila, but not by the ∆dotA mutant (Fig. 3 C and D).
More than 250 Different Eukaryotic-Like Proteins Are Encoded in Legionella Genomes.
In addition to modular effectors with eukaryotic domains, the Legionella genome encodes proteins that are similar to eukaryotic proteins, many of which are proven effectors of the Dot/Icm T4SS. A wider search for eukaryotic-like proteins in the Legionella genus identified 2,196 eukaryotic-like proteins representing more than 400 different orthologous groups that matched better to eukaryotes than to prokaryotes from a total of 6,809 different orthologous proteins that matched with eukaryotic proteins. Among these, we identified 156 proteins with a eukaryotic domain, and 210 eukaryotic-like proteins (SI Appendix, Table S5). Furthermore, 152 eukaryotic-like proteins detected possess a higher GC content (40 to 62%) than the rest of the genome, indicating recent HGT. Phylogenetic analysis of selected, identified proteins suggest that these were acquired from eukaryotes. As an example, SI Appendix, Fig. S7 shows the protein LanA0735 from L. anisa, a species frequently found in artificial water systems. This protein belongs to the pyridine nucleotide-disulfide oxidoreductase family, a subfamily of the FAD-dependent oxidoreductase family. LanA0735 showed some similarity to thioredoxin reductase, which exists as two major ubiquitous isoenzymes in higher eukaryotic cells—one cytosolic and the other mitochondrial. The cytosolic form has been implicated in interference with the acidification of the lysosomal compartment in C. elegans (30) and, thus, LanA0735 may help Legionella avoid vacuole acidification during infection.
Among the proteins defined as eukaryotic-like, two previously described phospholipases of L. pneumophila, PlcB (Lpp1411/Lpg1455) and PlcA (Lpp0565/Lpg0502), were identified in our analysis as eukaryotic proteins. The only other bacteria encoding these two enzymes are Pseudomonas and amoebae-associated bacteria. The two enzymes have phospholipase activity (31), but their role in infection is unknown. Here, they were predicted as phosphatidylcholine-hydrolyzing phospholipase C. Phosphatidylcholine is a eukaryotic membrane phospholipid that is present in only about 15% of prokaryotic species; in particular, bacteria interacting with eukaryotes (32). L. pneumophila belongs to the phosphatidylcholine-containing group of bacteria, which includes Francisella tularensis or Brucella abortus (33). These pathogens use the phosphatidylcholine synthase pathway exclusively for phosphatidylcholine formation and are thought to depend on choline supplied from the host cell (34). Indeed, it has been shown that phosphatidylcholine synthesis is required for L. pneumophila virulence (35). Thus, it is tempting to infer that the role of these enzymes may be to help acquire choline from the host cell.
Evolutionary History of Eukaryotic Domains and Eukaryotic Proteins.
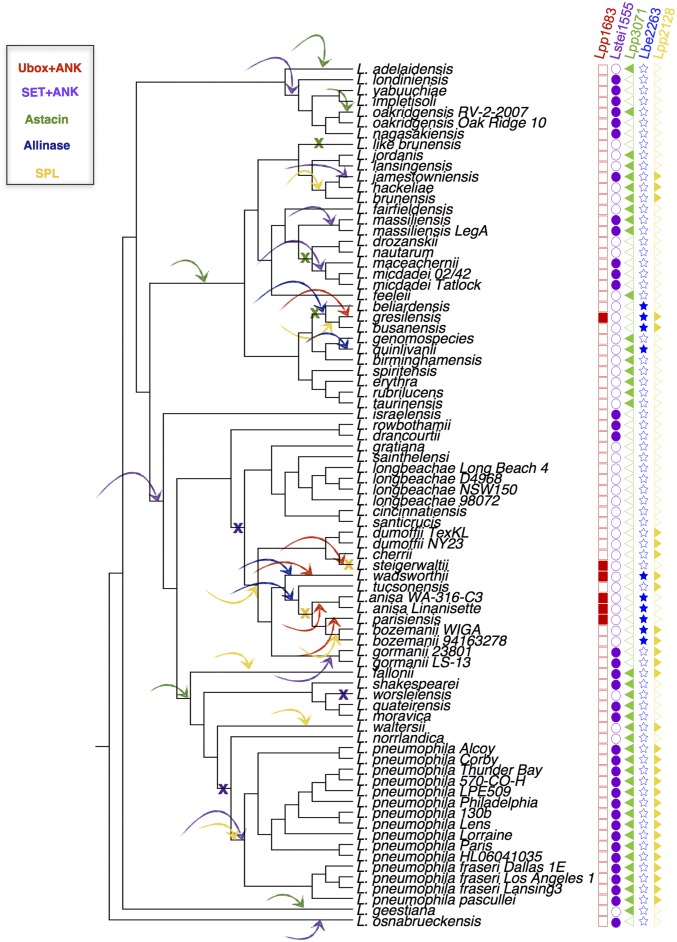
It is intriguing that Legionella species encode such a diverse repertoire of eukaryotic domains and eukaryotic-like proteins. To understand better this unique feature of the genus, we analyzed the evolutionary history of these proteins. After phylogenetic reconstruction of the genus Legionella based on the core genome (at least 50% identical) (Fig. 1A), we analyzed the distribution of the eukaryotic motifs and the eukaryotic proteins with respect to the evolution of the genus. For most, we found patchy distribution, as the repertoire of these proteins is variable among the different Legionella species (Fig. 2). Such a distribution is indicative of gain and loss events during the evolution of the genus. To analyze further how these proteins may have evolved in Legionella, we selected 25 eukaryotic motifs representing 2,837 different proteins in over 800 orthologous groups and used the program Gloome to analyze the gain and loss events for these proteins. We found that the number of gain events (1,197; 69%) considerably exceeded the number of loss events (549; 31%), a bias that was even stronger when using parsimony (1,628 gain events vs. 89 loss events) (SI Appendix, Fig. S8). These results were confirmed also when using a more conservative approach (by taking a probability cutoff for the stochastic model of 0.8 instead of 0.5) and when analyzing each motif separately.
An exemplary view of this result is shown in Fig. 4 for four proteins encoding different motifs (U-box and ankyrin repeat, SET domain and ankyrin repeat, astacin domain, and alliinase domain). In Fig. 4, loss events are indicated by a cross, and gain events by an arrow pointing to the branch. The number of gain events exceeds the number of loss events, indicating that in the Legionella genus, gene acquisition is dominant. Moreover, gene acquisition seems to be an ongoing and frequent process in the genus Legionella, given the high number of events we observed and the fact that most of them are localized in the terminal branches of the tree (SI Appendix, Fig. S8). To analyze whether eukaryotic-like proteins have the same evolutionary history, we took the sphingosine 1-phosphate lyse (LpSpl) (36, 37) as an example. Indeed, when running the same analyses, this gene also appeared to have been gained multiple times during the evolution of the genus (Fig. 4).
Fig. 4.
Gain–loss prediction for selected eukaryotic proteins and domain-containing proteins. Arrows pointing to the branches represent gain events, and crosses represent loss events. Filled squares, circles, triangles, or stars indicate the presence of the respective protein; empty squares, circles, triangles, or stars indicate that the protein is absent in this species.
Thus, in comparison with most prokaryotic species analyzed to date, more gene gain events are evident than loss events during evolution of the Legionella genus, which is also corroborated by the fact that the ancestral genomes were probably smaller (Fig. 1A, cluster I). Indeed, as seen in Fig. 1A, in each of the defined phylogenetic clusters, only few genomes have a larger size. For example, in cluster II, L. massiliensis is the only species with a big genome; thus, the most parsimonious explanation is that the ancestor of this clade had a small genome and, in the branch leading to L. massiliensis, gene gain occurred. This finding is similar to what was described for the adaptation of louse-borne intracellular pathogens and amoeba-associated bacteria. It is well known that the specialization of intracellular bacteria is associated with genome reduction, and extreme genome reduction can be seen in louse‐borne human specialists. In contrast, nonspecialized intraamoebal microorganisms exhibit a genome larger than their relatives due to gene conservation and acquisition (38).
The Dot/Icm Secretion System Is a Highly Conserved Machinery Secreting Thousands of Different Proteins.
The Dot/Icm T4SS is indispensable for intracellular replication of L. pneumophila in both amoeba and macrophages (39). In stark contrast to the high genetic diversity observed in the Legionella genomes, the Dot/Icm T4SS is part of the core genome because it is present in all species analyzed and the organization of the constituent proteins is highly conserved, even at the amino acid level. The proteins comprising the secretion machinery show an average amino acid identity of more than 50% and some even more than 90% compared with the L. pneumophila Dot/Icm components (SI Appendix, Fig. S9A and Table S6). The most conserved proteins are DotB, a secretion ATPase (86 to 100% amino acid identity), and IcmS, a small acidic cytoplasmic protein (74 to 98% amino acid identity). This high conservation is even seen with one of the few non-Legionella species that encodes a Dot/Icm system, C. burnetii.
The only gene of the Dot/Icm system that is not present in all Legionella species is icmR. IcmR interacts with IcmQ as a chaperone, preventing IcmQ self-dimerization (40). Although IcmQ is highly conserved, the gene encoding IcmR is frequently replaced by one or two nonhomologous genes encoding for proteins that are called FIR because they can functionally replace IcmR (41). When overlapping the occurrence of the different FIR genes with the phylogeny of the species, most phylogenetically closely related species share homologous FIR genes (SI Appendix, Fig. S10). Apart from two conserved regions (SI Appendix, Fig. S11), the absence of sequence homology among FIR proteins indicates that icmR is an extremely fast-evolving gene and, therefore, probably under positive selection. The reason why this gene is extremely divergent is still unknown but could be also linked to the high variety of Dot/Icm effectors described in this genus. Thus, except for the FIR genes, the Dot/Icm T4SS is highly conserved and encoded in a very dynamic genetic context.
It has been shown previously that the more than 300 substrates of the L. pneumophila Dot/Icm system are not universally present within the genus Legionella, because among 38 Legionella species, only seven core effectors were described (14). Surprisingly, when adding the 40 additional genomes and 16 Legionella species sequenced in this study, we identified eight core effectors instead of seven. A comparison of the two studies confirmed Lpg0103 (VipF), Lpg0107 (RavC), Lpg2300 (LegA3/AnkH/AnkW), and Lpg2815 (IroT/MavN) as core substrates (14) (SI Appendix, Fig. S9B and Table S7). Three of the previously defined core substrates (Lpg0140, Lpg2832, and Lpg3000) were present in two genomes as two consecutive genes instead of one; however, this fragmentation might be a sequencing error, and we thus considered these substrates also as core substrates (SI Appendix, Table S7). In our study, we identified one additional core effector gene, lpg1356/lpp1310. This protein was reported by Lifshitz et al. (42) as secreted protein, but had not been included in the Burstein et al. (14) effector search, which explains the different result (SI Appendix, Fig. S9B and Table S7). Similar to most of the other core substrates, their functions are not known, but Lpg1356 encodes eight eukaryotic Sel-1 motifs similar to LpnE, an L. pneumophila virulence determinant that influences vacuolar trafficking (43). Furthermore, seven other genes are present in all but one, two, or four genomes, thus they might have important functions in host–pathogen interactions (SI Appendix, Table S7). Interestingly, when the effector repertoire of several strains of one species is compared, the conservation of the effectors is very high (between 82% and 97%) (SI Appendix, Table S8). However, if more strains than two are available for a species, as is the case for L. pneumophila for which 11 strains could be compared, the conservation of the effector pool is only 65% (264 of the 408 different effectors identified in the 11 strains) (SI Appendix, Table S8). Thus, the L. pneumophila core effector set is also smaller than previously thought. Together, the genus Legionella has eight core substrates present in all genomes and seven additional ones that are present in nearly all genomes.
Interestingly, whereas the number of core Dot/Icm substrates is extremely small, the number and the diversity of predicted Dot/Icm substrates is extremely high. Indeed, through a machine learning approach, Burstein et al. (14) predicted that the Legionella genus would encode 5,885 effectors. Here, we extended these analyses and identified 4,767 proteins with eukaryotic motifs that have a high probability to be secreted effectors, as shown for the Rab-like proteins. If we consider that the orthologs of these proteins in each species are also effectors, then the number raises to 7,103 (representing 1,145 different orthologous proteins) (SI Appendix, Fig. S9C). Moreover, we identified 2,196 eukaryotic-like proteins representing 414 different orthologous genes, which, together with the above-mentioned eukaryotic motif carrying proteins, form 1,400 different putative orthologous substrates of the Dot/Icm T4SS. Finally, when adding to the effectors predicted in this study (based on their similarity to eukaryotic domains and proteins), the effectors previously described in L. pneumophila and their orthologs (more than 7,000 proteins representing about 300 different orthologs), as well as the effectors predicted by the machine learning approach and their orthologs (more than 10,000 proteins representing about 900 different orthologs) (14), the total number of different effectors rises to almost 18,000 proteins (more than 1,600 orthologous groups) (SI Appendix, Fig. S9C and Table S9). Therefore, the Legionella genus has by far the highest number and widest variety of effectors described for an intracellular bacterium. Furthermore, when calculating the growth-accumulation curve for Dot/Icm-predicted effectors, this number should still increase with the sequencing of new Legionella genomes, as the plateau is not yet reached (SI Appendix, Fig. S9D).
The Ability to Infect Human Cells Has Been Acquired Independently Several Times During the Evolution of the Genus Legionella.
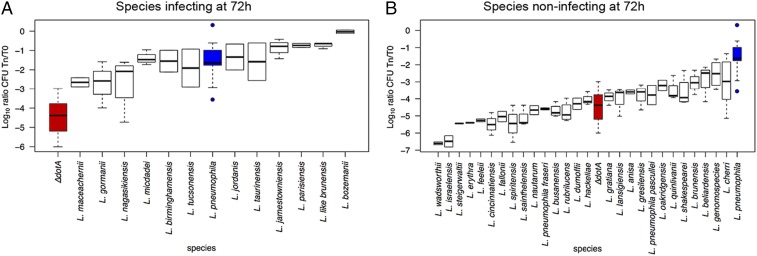
Among the 65 Legionella species known, L. pneumophila is responsible for over 90% of human disease, followed by L. longbeachae [2 to 7% of cases, except 30% in Australia and New Zealand (44)]. Certain Legionella species, such as Legionella micdadei, Legionella dumoffii, or Legionella bozemani, have once or sporadically been associated with human disease (44), and all other species seem to be environmental bacteria only. The reasons for these differences are not known. To explore whether all species are able to replicate in human cells, we chose the human macrophage-like cell line THP-1 as model and tested the replication capacity of 47 different Legionella species. Infections were carried out in duplicate or triplicate and colony-forming units were recorded at 24, 48, and 72 h postinfection. Levels of intracellular replication were compared with wild-type L. pneumophila strain Paris and an isogenic nonreplicating ∆dotA mutant as reference strains (Fig. 5 and SI Appendix, Figs. S12 and S13). Results were also compared with data previously reported for different Legionella species in THP-1, U937, A549, and Mono Mac 6 cells, mouse and guinea pig-derived macrophages, and in guinea pigs (SI Appendix, Table S10). When results at 72 h postinfection were analyzed, 28 of the 47 species tested were impaired for intracellular replication, whereas nine species replicated similarly to, or better than, L. pneumophila Paris (Fig. 5). These nine species were Legionella gormanii, Legionella jamestowniensis, Legionella jordanis, Legionella like brunensis, Legionella maceachernii, Legionella micdadei, Legionella nagasakiensis, Legionella parisiensis, and Legionella tucsonensis. Interestingly, L. jamestowniensis, for which one human case has been reported (45), replicated better than L. pneumophila Paris. Indeed, L. jamestowniensis productively infects human U937-derived phagocytes. The remaining eight species showed variable replication patterns, being significantly different from L. pneumophila Paris only in one or two of the three analyzed time points (SI Appendix, Fig. S12). Broadly, the species most frequently reported from human disease (L. pneumophila, L. longbeachae, L. micdadei, L. bozemanii, and L. dumoffii) are also those that replicated robustly in THP-1 cells. The only exception was the L. dumoffii strains that were impaired for replication in THP-1 cells but which have been shown to replicate in other cell types and guinea pigs. Together, there is a convincing correlation between the frequency of isolation from human disease and the ability to grow in macrophage-like cells.
Fig. 5.
The replicative capacity of the different Legionella species in THP-1 cells correlates with their epidemiological features. Replication of each strain at the time point 72 h after infection of THP-1 cells is shown (24 and 48 h postinfection shown in SI Appendix, Fig. S14). Intracellular replication was determined by recording the number of colony-forming units (CFU) after plating on buffered charcoal yeast extract agar. Blue boxes indicate L. pneumophila Paris, representative of a replicating strain; red boxes indicate L. pneumophila ∆dotA, representative of nonreplicating strain. The strains are ordered according to the mean replication values. (A) Legionella species replicating similar to, or significantly better than, L. pneumophila Paris. (B) Species with no replication capacity or significantly lower replication capacity compared to L. pneumophila Paris.
To analyze this further, we overlapped the replication results with the phylogeny of the genus. Apart from the small cluster containing L. beliardensis, Legionella gresilensis, and L. busanensis, which were all unable to grow in THP-1 cells, replicating and nonreplicating strains were mixed in the phylogeny (SI Appendix, Fig. S14). This suggests that the capacity to replicate in human cells has been acquired independently several times during evolution of the Legionella genus, possibly as a result of recruiting effectors that allow adaptation to particular niches. To understand whether a specific set of effectors is necessary to infect human cells, we further analyzed the combination of effectors present in the strains isolated from human disease and those present in strains capable of replicating in THP-1 cells. Surprisingly, no specific set of effectors could be attributed to strains capable of replicating in human cells or isolated from human disease, although among these strains, certain conserved motifs always present were identified, such as ankyrin motifs or F-box or SET domains, suggesting that common pathways need to be subverted to cause human infection. Thus, the capacity to infect human cells has been acquired independently, several times during the evolution of the genus Legionella.
In conclusion, the analysis of 80 Legionella strains representing 58 different Legionella species has revealed a contrasting picture of the Legionella genus. It encodes a highly conserved T4SS predicted to secrete more than 18,000 proteins, of which only eight are conserved throughout the genus. Together, the genomes portray an extremely diverse genus shaped by massive interdomain HGT, circulating mobile genetic elements and eukaryotic-like proteins. Our in-depth analyses of eukaryotic features of the Legionella genomes identified 137 different eukaryotic domains, of which Rab or Ras domain-containing proteins were quasi-unique to the genus Legionella. The secretion assays undertaken for 16 of these Rab or Ras domain-containing proteins confirmed that these were translocated Dot/Icm effectors. In addition to the eukaryotic domains, we identified 210 orthologous groups of eukaryotic-like proteins. If all these proteins in the different species and their orthologs are taken into account, we found more than 8,000 proteins that have been shaped by interdomain HGT in the genus Legionella. Thus, to our knowledge, the genus Legionella contains the widest variety and highest number of eukaryotic proteins and domains of any prokaryotic genus genome analyzed to date. Analyzing more strains per species will probably discover new unknown effectors, increasing our knowledge of the set of tools used by Legionella to infect eukaryotic cells. Although eukaryotic proteins and domains were a universal feature of the genus Legionella, the repertoire of these proteins for each species was different. Surprisingly, even when the same motif was present in different species, these were often present in different proteins with no orthology. In accordance with this finding, our evolutionary analysis of the presence/absence of these domains and proteins suggests that these proteins were mostly acquired through gene gain events.
When exploring the replication capacity of 47 different Legionella species in the human macrophage-like cell line THP-1, we found that the 23 species were capable of replicating in THP-1 cells. However, these did not cluster in the phylogeny, indicating that the capacity to replicate in macrophages can be achieved by different combinations of effectors and that this capacity has been acquired several times during the evolution of the Legionella genus. As humans are an accidental host for Legionella, the capacity to replicate in macrophages may also have been obtained by a coincidental acquisition of different virulence properties initially needed to adapt to a specific natural host, such as amoebae. Indeed, due to the high conservation of key signaling pathways in professional phagocytes such as amoebae and human macrophages, different combinations of effectors may allow Legionella species to infect higher eukaryotic cells by chance.
Here, we show that all Legionella species have acquired eukaryotic proteins that likely modulate specific host functions to allow intracellular survival and replication in eukaryotic host cells. At a certain point, the evolution of a combination of effector proteins that allow replication in human cells may inadvertently lead to the emergence of new human pathogens from environmental bacteria.
Materials and Methods
The materials and methods are described at length in SI Appendix, Materials and Methods. This includes sequencing and assembly, sequence processing and annotation, pan/core genome, ortholog and singleton definition, phylogenetic reconstruction and evolutionary analysis, phylogenetic analyses of Rab and eukaryotic-like proteins, infection assays, statistical analysis, and translocation assays. The raw sequence reads were deposited in the European Nucleotide Archive (46). The sequences and annotations can be accessed at https://github.com/bbi-ip/Legionella_genus_proteins.git (47).
Supplementary Material
Acknowledgments
We thank Tim P. Stinear for critical reading of the manuscript and helpful comments, and we acknowledge the receipt of 53 different Legionella strains from the Collection of the Institut Pasteur. Work in the C.B. laboratory is financed by the Institut Pasteur, the Agence Nationale de la Recherche Grant ANR-10-LABX-62-IBEID, and the Fondation pour la Recherche Médicale Grant DEQ20120323697.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The raw sequence reads have been deposited in the European Nucleotide Archive (accession no. PRJEB24896). The sequences and annotations can be accessed at https://github.com/bbi-ip/Legionella_genus_proteins.git.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1808016116/-/DCSupplemental.
References
- 1.Fraser DW, et al. Legionnaires’ disease: Description of an epidemic of pneumonia. N Engl J Med. 1977;297:1189–1197. doi: 10.1056/NEJM197712012972201. [DOI] [PubMed] [Google Scholar]
- 2.Rowbotham TJ. Preliminary report on the pathogenicity of Legionella pneumophila for freshwater and soil amoebae. J Clin Pathol. 1980;33:1179–1183. doi: 10.1136/jcp.33.12.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cazalet C, et al. Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat Genet. 2004;36:1165–1173. doi: 10.1038/ng1447. [DOI] [PubMed] [Google Scholar]
- 4.Brüggemann H, Cazalet C, Buchrieser C. Adaptation of Legionella pneumophila to the host environment: Role of protein secretion, effectors and eukaryotic-like proteins. Curr Opin Microbiol. 2006;9:86–94. doi: 10.1016/j.mib.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 5.Komano T, Yoshida T, Narahara K, Furuya N. The transfer region of IncI1 plasmid R64: Similarities between R64 tra and legionella icm/dot genes. Mol Microbiol. 2000;35:1348–1359. doi: 10.1046/j.1365-2958.2000.01769.x. [DOI] [PubMed] [Google Scholar]
- 6.Escoll P, Mondino S, Rolando M, Buchrieser C. Targeting of host organelles by pathogenic bacteria: A sophisticated subversion strategy. Nat Rev Microbiol. 2016;14:5–19. doi: 10.1038/nrmicro.2015.1. [DOI] [PubMed] [Google Scholar]
- 7.Finsel I, Hilbi H. Formation of a pathogen vacuole according to Legionella pneumophila: How to kill one bird with many stones. Cell Microbiol. 2015;17:935–950. doi: 10.1111/cmi.12450. [DOI] [PubMed] [Google Scholar]
- 8.Nora T, Lomma M, Gomez-Valero L, Buchrieser C. Molecular mimicry: An important virulence strategy employed by Legionella pneumophila to subvert host functions. Future Microbiol. 2009;4:691–701. doi: 10.2217/fmb.09.47. [DOI] [PubMed] [Google Scholar]
- 9.Burstein D, et al. Genome-scale identification of Legionella pneumophila effectors using a machine learning approach. PLoS Pathog. 2009;5:e1000508. doi: 10.1371/journal.ppat.1000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gomez-Valero L, et al. Extensive recombination events and horizontal gene transfer shaped the Legionella pneumophila genomes. BMC Genomics. 2011;12:536. doi: 10.1186/1471-2164-12-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gomez-Valero L, et al. Comparative analyses of Legionella species identifies genetic features of strains causing Legionnaires’ disease. Genome Biol. 2014;15:505. doi: 10.1186/s13059-014-0505-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morozova I, et al. Comparative sequence analysis of the icm/dot genes in Legionella. Plasmid. 2004;51:127–147. doi: 10.1016/j.plasmid.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 13.Sánchez-Busó L, Comas I, Jorques G, González-Candelas F. Recombination drives genome evolution in outbreak-related Legionella pneumophila isolates. Nat Genet. 2014;46:1205–1211. doi: 10.1038/ng.3114. [DOI] [PubMed] [Google Scholar]
- 14.Burstein D, et al. Genomic analysis of 38 Legionella species identifies large and diverse effector repertoires. Nat Genet. 2016;48:167–175. doi: 10.1038/ng.3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Joseph SJ, et al. Dynamics of genome change among Legionella species. Sci Rep. 2016;6:33442. doi: 10.1038/srep33442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cazalet C, et al. Analysis of the Legionella longbeachae genome and transcriptome uncovers unique strategies to cause Legionnaires’ disease. PLoS Genet. 2010;6:e1000851. doi: 10.1371/journal.pgen.1000851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guglielmini J, de la Cruz F, Rocha EP. Evolution of conjugation and type IV secretion systems. Mol Biol Evol. 2013;30:315–331. doi: 10.1093/molbev/mss221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bohlin J, Brynildsrud OB, Sekse C, Snipen L. An evolutionary analysis of genome expansion and pathogenicity in Escherichia coli. BMC Genomics. 2014;15:882. doi: 10.1186/1471-2164-15-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bohlin J, Sekse C, Skjerve E, Brynildsrud O. Positive correlations between genomic %AT and genome size within strains of bacterial species. Environ Microbiol Rep. 2014;6:278–286. doi: 10.1111/1758-2229.12145. [DOI] [PubMed] [Google Scholar]
- 20.Rolando M, et al. Legionella pneumophila effector RomA uniquely modifies host chromatin to repress gene expression and promote intracellular bacterial replication. Cell Host Microbe. 2013;13:395–405. doi: 10.1016/j.chom.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 21.Nagai H, Kagan JC, Zhu X, Kahn RA, Roy CR. A bacterial guanine nucleotide exchange factor activates ARF on Legionella phagosomes. Science. 2002;295:679–682. doi: 10.1126/science.1067025. [DOI] [PubMed] [Google Scholar]
- 22.Pasanen AL, Yli-Pietila K, Pasanen P, Kalliokoski P, Tarhanen J. Ergosterol content in various fungal species and biocontaminated building materials. Appl Environ Microbiol. 1999;65:138–142. doi: 10.1128/aem.65.1.138-142.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith FR, Korn ED. 7-Dehydrostigmasterol and ergosterol: The major sterols of an amoeba. J Lipid Res. 1968;9:405–408. [PubMed] [Google Scholar]
- 24.Thomson S, et al. Characterisation of sterol biosynthesis and validation of 14α-demethylase as a drug target in Acanthamoeba. Sci Rep. 2017;7:8247. doi: 10.1038/s41598-017-07495-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wennerberg K, Rossman KL, Der CJ. The Ras superfamily at a glance. J Cell Sci. 2005;118:843–846. doi: 10.1242/jcs.01660. [DOI] [PubMed] [Google Scholar]
- 26.Simanshu DK, Nissley DV, McCormick F. RAS proteins and their regulators in human disease. Cell. 2017;170:17–33. doi: 10.1016/j.cell.2017.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rojas AM, Fuentes G, Rausell A, Valencia A. The Ras protein superfamily: Evolutionary tree and role of conserved amino acids. J Cell Biol. 2012;196:189–201. doi: 10.1083/jcb.201103008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Colicelli J. Human RAS superfamily proteins and related GTPases. Sci STKE. 2004;2004:RE13. doi: 10.1126/stke.2502004re13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zade A, Sengupta M, Kondabagil K. Extensive in silico analysis of Mimivirus coded Rab GTPase homolog suggests a possible role in virion membrane biogenesis. Front Microbiol. 2015;6:929. doi: 10.3389/fmicb.2015.00929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li W, et al. Two thioredoxin reductases, trxr-1 and trxr-2, have differential physiological roles in Caenorhabditis elegans. Mol Cells. 2012;34:209–218. doi: 10.1007/s10059-012-0155-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hiller M, Lang C, Michel W, Flieger A. Secreted phospholipases of the lung pathogen Legionella pneumophila. Int J Med Microbiol. 2017:S1438-4221(17)30290-4. doi: 10.1016/j.ijmm.2017.10.002. [DOI] [PubMed] [Google Scholar]
- 32.Aktas M, et al. Phosphatidylcholine biosynthesis and its significance in bacteria interacting with eukaryotic cells. Eur J Cell Biol. 2010;89:888–894. doi: 10.1016/j.ejcb.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 33.Geiger O, López-Lara IM, Sohlenkamp C. Phosphatidylcholine biosynthesis and function in bacteria. Biochim Biophys Acta. 2013;1831:503–513. doi: 10.1016/j.bbalip.2012.08.009. [DOI] [PubMed] [Google Scholar]
- 34.Comerci DJ, Altabe S, de Mendoza D, Ugalde RA. Brucella abortus synthesizes phosphatidylcholine from choline provided by the host. J Bacteriol. 2006;188:1929–1934. doi: 10.1128/JB.188.5.1929-1934.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Conover GM, et al. Phosphatidylcholine synthesis is required for optimal function of Legionella pneumophila virulence determinants. Cell Microbiol. 2008;10:514–528. doi: 10.1111/j.1462-5822.2007.01066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Degtyar E, Zusman T, Ehrlich M, Segal G. A Legionella effector acquired from protozoa is involved in sphingolipids metabolism and is targeted to the host cell mitochondria. Cell Microbiol. 2009;11:1219–1235. doi: 10.1111/j.1462-5822.2009.01328.x. [DOI] [PubMed] [Google Scholar]
- 37.Rolando M, et al. Legionella pneumophila S1P-lyase targets host sphingolipid metabolism and restrains autophagy. Proc Natl Acad Sci USA. 2016;113:1901–1906. doi: 10.1073/pnas.1522067113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moliner C, Fournier PE, Raoult D. Genome analysis of microorganisms living in amoebae reveals a melting pot of evolution. FEMS Microbiol Rev. 2010;34:281–294. doi: 10.1111/j.1574-6976.2010.00209.x. [DOI] [PubMed] [Google Scholar]
- 39.Segal G, Shuman HA. Legionella pneumophila utilizes the same genes to multiply within Acanthamoeba castellanii and human macrophages. Infect Immun. 1999;67:2117–2124. doi: 10.1128/iai.67.5.2117-2124.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Duménil G, Isberg RR. The Legionella pneumophila IcmR protein exhibits chaperone activity for IcmQ by preventing its participation in high-molecular-weight complexes. Mol Microbiol. 2001;40:1113–1127. doi: 10.1046/j.1365-2958.2001.02454.x. [DOI] [PubMed] [Google Scholar]
- 41.Feldman M, Zusman T, Hagag S, Segal G. Coevolution between nonhomologous but functionally similar proteins and their conserved partners in the Legionella pathogenesis system. Proc Natl Acad Sci USA. 2005;102:12206–12211. doi: 10.1073/pnas.0501850102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lifshitz Z, et al. Computational modeling and experimental validation of the Legionella and Coxiella virulence-related type-IVB secretion signal. Proc Natl Acad Sci USA. 2013;110:E707–E715. doi: 10.1073/pnas.1215278110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Newton HJ, et al. Sel1 repeat protein LpnE is a Legionella pneumophila virulence determinant that influences vacuolar trafficking. Infect Immun. 2007;75:5575–5585. doi: 10.1128/IAI.00443-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu VL, et al. Distribution of Legionella species and serogroups isolated by culture in patients with sporadic community-acquired legionellosis: An international collaborative survey. J Infect Dis. 2002;186:127–128. doi: 10.1086/341087. [DOI] [PubMed] [Google Scholar]
- 45.Prochazka B, et al. Draft genome sequence of Legionella jamestowniensis isolated from a patient with chronic respiratory disease. Genome Announc. 2016;4:e01007-16. doi: 10.1128/genomeA.01007-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gomez-Valero L., Rusniok C, Buchrieser C. 2018 Comparative genomics of Legionella species. European Nucleotide Archive. Available at https://www.ebi.ac.uk/ena/data/view/PRJEB24896. Deposited December 21, 2018.
- 47.Gomez-Valero L, et al. 2018 Data from “Comparative genomics of Legionella species.” European Nucleotide Archive. Available at https://github.com/bbi-ip/Legionella_genus_proteins.git. Deposited October 22, 2018.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.