Significance
Chlamydomonas reinhardtii is the premier reference organism for understanding unicellular green algae. Chlamydomonas is an important model for photosynthesis as well as fermentation and other anaerobic pathways under dark anoxic conditions. We have produced a diurnal transcriptome, validated by subproteomic analyses, and matched with measurements of pigments, select metabolites, and physiological parameters. We report that the majority of the algal genome is differentially expressed over the course of the day and the timing of specific genes is dictated by their biological function. We also discovered that fermentation rather than respiration is the preferred metabolic fate of starch-derived glycolytic pyruvate. We offer our rich dataset to the algal and plant communities.
Keywords: photobioreactor, systems biology, cell division, histone expression, chloroplast
Abstract
The unicellular green alga Chlamydomonas reinhardtii displays metabolic flexibility in response to a changing environment. We analyzed expression patterns of its three genomes in cells grown under light–dark cycles. Nearly 85% of transcribed genes show differential expression, with different sets of transcripts being up-regulated over the course of the day to coordinate cellular growth before undergoing cell division. Parallel measurements of select metabolites and pigments, physiological parameters, and a subset of proteins allow us to infer metabolic events and to evaluate the impact of the transcriptome on the proteome. Among the findings are the observations that Chlamydomonas exhibits lower respiratory activity at night compared with the day; multiple fermentation pathways, some oxygen-sensitive, are expressed at night in aerated cultures; we propose that the ferredoxin, FDX9, is potentially the electron donor to hydrogenases. The light stress-responsive genes PSBS, LHCSR1, and LHCSR3 show an acute response to lights-on at dawn under abrupt dark-to-light transitions, while LHCSR3 genes also exhibit a later, second burst in expression in the middle of the day dependent on light intensity. Each response to light (acute and sustained) can be selectively activated under specific conditions. Our expression dataset, complemented with coexpression networks and metabolite profiling, should constitute an excellent resource for the algal and plant communities.
Life evolved under the constant pressure of limited resources. The invention of oxygenic photosynthesis by early cyanobacteria partially relieved these limitations, placing biology under the dependence of the Sun. This had the unexpected benefit of bringing predictability to physiology and metabolic pathways. The rotation of the Earth around the Sun allowed the temporal separation of incompatible biochemical reactions over the course of a day. On a global scale, much of transcription was placed early under the control of molecular oscillators or circadian clocks to properly gate key physiological events to the right time of day (or night) and optimize resource allocation.
Over the course of evolution, the rhythmic fraction of the transcriptome has adjusted to the physiology and habitat of the organism. Recent technical advances, such as microarrays, transcriptomics, and untargeted proteomics have brought unprecedented detail to our understanding of circadian and diurnal rhythms in daily physiology and behavior. However, the intrinsic biology of more complex eukaryotic model systems comes with drawbacks: they are composed of several tissues whose individual signals are diluted at the whole-organism level. The strength of rhythms can also be affected by developmental stage.
In this regard, unicellular algae are well suited for studies of rhythmic transcription in response to a changing environment (1). Among them, Chlamydomonas reinhardtii is a workhorse in the fields of photosynthesis, chloroplast biology, ciliopathies, and metal homeostasis, and is a reference organism in biotechnology for production of high-value bio-products and biofuel (2, 3). Chlamydomonas is also metabolically flexible, with phototrophic, heterotrophic, and fermentation capabilities, the latter predominating under anoxic conditions that prevail in the dark (4). It can acclimate to changes in light intensity (5, 6). Located at the base of the green lineage, Chlamydomonas shares fundamental regulatory and metabolic pathways with other algae and land plants, and possesses the advantages of a unicellular microbial system that can be (i) grown in large volumes and (ii) easily synchronized with alternating light-dark cycles, both in terms of diurnal gene expression and cell division. Most of the algal primary metabolism, protein synthesis, DNA replication, and organelle biogenesis pathways are diurnally coordinated to sustain growth toward cell division (7, 8). Chlamydomonas cells can undergo several consecutive divisions at dusk, producing 2 to 32 daughter cells in 1 d, depending on growth conditions. Multiple divisions will cause groups of cells to fall out of synchrony with the population at night and decrease detection sensitivity of rhythmic behaviors (9, 10).
Here, we expose Chlamydomonas cultures to conditions that mimic a day in nature, with light–dark cycles superimposed with warm–cool cycles, and air levels of carbon dioxide. Our cells divide exactly once, increasing synchrony between cells and thus signal over noise. We find that most of the Chlamydomonas transcriptome, including chloroplast and mitochondria-encoded genes, undergoes diurnal changes according to biological function. We also provide molecular and physiological evidence for the integration of two light signals, revealed by the expression of light-harvesting protein genes. Finally, we propose that Chlamydomonas cells utilize much of their stored carbon for fermentation in the night rather than respiration to maintain the redox and energy balance they need during their resting phase. We invite our readers to look for their favorite genes in our dataset and discover coexpressed genes.
Results
How to Get Exactly Two Daughter Cells per Division.
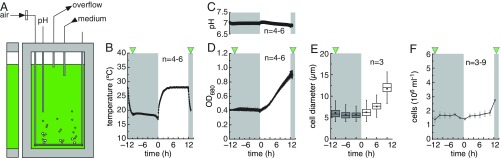
We exposed Chlamydomonas cultures grown in bioreactors to conditions they might experience in nature: warm days and cool nights, with 0.04% of carbon dioxide provided by bubbling with air (Fig. 1 A–C). Congruent light–dark and warm–cool cycles strengthen synchronization over the diurnal cycle. We did not provide a reduced carbon source to ensure that the number of daughter cells produced would be dictated by light intensity (photon flux density) and photoperiod, which modulate biomass production in the light (11, 12). We therefore optimized both parameters to achieve an exact doubling of cell number in 24 h. Cell growth is restricted to the light part of the diurnal cycle, as expected (Fig. 1 D and E). Cell density remains constant at 2–3 × 106 cells/mL, except shortly after dusk when it doubles, consistent with the birth of two daughter cells per mother cell (Fig. 1F). Reproducibility across independent experiments is excellent, even when conducted weeks or months apart (Fig. 1D). This provides us with the unique opportunity to collect physiological, metabolic, transcriptome, and proteome data across one full diurnal cycle according to the sampling scheme shown in SI Appendix, Fig. S1, and resolve functional relationships between genes, generate new hypotheses, and validate decades of algal research.
Fig. 1.
How to obtain exactly two daughter cells during the Chlamydomonas cell division cycle. (A) Schematic illustration of the flat-panel photobioreactor design. (B–F) Experimental parameters measured under our growth conditions. Real-time profiles of the growth medium temperature (B), pH (C), and optical density at 680 nm (D). (E) Distribution of cell size over the diurnal cycle, shown as a box plot. (F) Number of cells in the culture as a function of diurnal time. Data (except E) are shown as average ± SD (n = 3–9, specified in each panel). The green triangles indicate the timing of cell division.
Global Transcriptome Profiling over a Diurnal Cycle.
We measured transcript abundance of all mitochondrial, chloroplast, and nuclear genes in Chlamydomonas by deep-sequencing. We opted for a Ribo-depletion approach during library preparation to capture the full extent of transcriptional changes rather than the typical selection of poly-adenylylated transcripts, as several interesting transcripts lack a poly(A) tail. These include ribosomal RNAs generated by RNA polymerases I and III, replication-dependent histones (13), some long noncoding RNAs (14, 15), and organellar transcripts (16).
We collected samples in triplicate every 2 h over the course of a diurnal cycle, starting and ending 1 h after lights-off (CT−11 and CT+13, lights-on being referred to as CT0 by convention), with denser sampling around dawn to capture higher resolution at the dark to light transition (SI Appendix, Fig. S1). Reproducibility of expression estimates across replicates is excellent, with average correlation coefficients R2 ∼ 0.9 (SI Appendix, Fig. S2). Our dataset represents over 850,000 expression estimates across replicates and genomes.
Testing for Synchrony with DNA Replication.
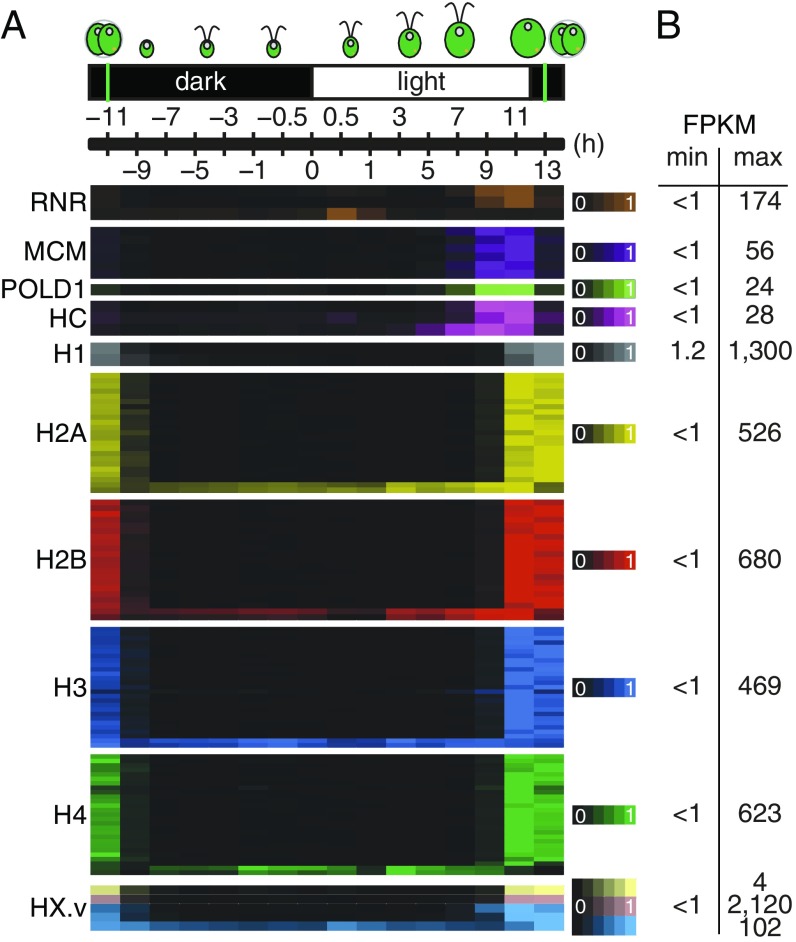
The exact doubling of cell number shortly after dusk suggested that our cell population was well-synchronized. To confirm these results at the molecular level, we extracted expression values for genes involved in cell division from our dataset. Before division can occur cells need to duplicate their genomes, which requires synthesis of deoxyribonucleotides via reduction of ribonucleotides, a step catalyzed by ribonucleotide reductases (RNR) (17, 18). Next, the minichromosome maintenance (MCM) complex, a conserved helicase, must unwind the DNA double-helix to allow access for the DNA-dependent DNA polymerase POLD1. Finally, newly synthesized DNA molecules become covered in core histones (H2A, H2B, H3, and H4) and linker histone H1 for nucleosome (re)assembly. This sequence of events is expected to take place within a 2-h window centered around CT+11 (19), and our results are remarkably consistent with this prediction: expression estimates for the large subunit and for one small subunit of Chlamydomonas RNR, all MCM2–MCM7 subunits, and POLD1 show peak expression between CT+9 and CT+11, followed by core and linker histones between CT+11 and CT+13 (Fig. 2). By untargeted proteomics, we detect peptides for RNR1 (the large subunit of RNR) and RIR2a (one of the two small subunits), and for all subunits of the MCM complex almost exclusively at CT+11 (SI Appendix, Fig. S3). Therefore, the Chlamydomonas MCM complex is controlled at the level of transcription and protein degradation, in contrast to budding yeast, where MCM proteins are present constitutively in the nucleus, and only a small fraction becomes associated with replicating DNA during each S phase (20).
Fig. 2.
High synchrony across cells illustrated by the expression of genes involved in DNA replication. (A) Heatmap showing transcript abundance for genes involved in DNA RNR (3 genes, brown), DNA helicase (MCM, 6 genes, purple), DNA polymerase (POLD1), histone chaperones (HC, three genes, magenta), histone H1 (2 genes, gray), histone H2A (22 genes, yellow), histone H2B (20 genes, red), histone H3 (26 genes, blue), and histone H4 (26 genes, green). Histone variants (HX.v) are shown in a lighter version of the color used to depict their respective histone groups. In all cases, expression was normalized to 1 for the highest value. (B) Dynamic expression range of replication genes. Minimum and maximum RNA abundance (in FPKM) from the average of three independent experiments is shown. The green vertical lines indicate the timing of cell division.
Histone deposition onto DNA is aided by a number of histone chaperones: chromatin assembly factor 1 (CAF-1), antisilencing factor 1 (ASF1), and histone regulator A (HIRA) (21). CAF-1 function is alone limited to DNA replication in higher plants and animal systems. In Chlamydomonas, the algal orthologs for CAF-1, ASF1, and HIRA (HIR1) are coexpressed with peak expression between CT+9 and CT+11, a pattern more consistent with replication-dependent histone deposition. However, ASF1 expression does rise transiently after dawn, and HIR1 expression starts to increase around CT+3, which could allow both proteins to participate in nucleosome deposition outside of DNA replication (Fig. 2).
Most, but not all genes involved in DNA replication share the same expression pattern. For example, the second Chlamydomonas RNR2 gene (RIR2b) is only expressed shortly after dawn (CT+0.5), which is matched by a secondary peak in RNR1 expression, suggesting a distinct role, possibly in DNA damage repair in response to photodamage. RNR1 protein is still present around CT+1, although at low levels, and can therefore associate with the morning-specific RIR2b subunit for RNR function (SI Appendix, Fig. S3B). Similarly, two genes per canonical core histone gene family and a single histone H3 variant are expressed constitutively outside the DNA replication window, and may provide an emergency pool of histones for epigenetic control in response to changes in the environment (Fig. 2A). Taken together, these results demonstrate the very high degree of synchronization of our cell population. These results also highlight the potential for gene discovery and for distinguishing functions of paralogs from our dataset, which we further explore in the next sections.
The Majority of Chlamydomonas Genes Exhibit a Rhythmic Expression Pattern.
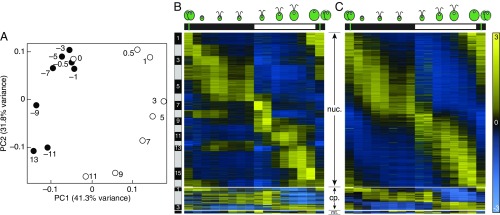
To parse out our rich dataset, we performed a principle component analysis (PCA) to reduce complexity, shown in Fig. 3A. A testament to the controlled experimental conditions is the grouping of time points CT−11 and CT+13, which are temporally identical over a diurnal cycle. In addition, we observed coclustering of samples collected later during the night, from CT−7 to CT−0.5, during which cells remain relatively more metabolically quiescent during the G0 phase of the cell cycle. The first two components capture over 73% of the total variance in expression: the first component separates samples between day and night, while the second component sorts them according to timing (Fig. 3A). A PCA of the Arabidopsis transcriptome displayed a very similar pattern when seedlings were grown under light–dark cycles (22), suggesting that our samples may cycle robustly, from synchronized cell division, diurnal or circadian rhythms, or both.
Fig. 3.
The Chlamydomonas transcriptome is highly dynamic and rhythmic over one diurnal cycle. (A) PCA of the Chlamydomonas transcriptome over the diurnal cycle: samples collected at night (●), samples collected during the day (○). The time of sample collection is indicated next to the corresponding circle. (B) Heatmap representation of gene expression for 10,465 differentially expressed genes over the diurnal cycle. Genes were included if at least 1,000 counts were detected in the experiment, and there was a Benjamini–Hochberg-adjusted P value < 0.01 for differential expression. Genes were grouped into 16 clusters of nuclear genes, 3 clusters of chloroplast genes, and 1 mitochondrial cluster. For easier visualization, the vertical scale of the organellar transcriptomes (chloroplast transcriptome, Middle; mitochondrial transcriptome, Bottom) is enlarged 50 times relative to the nuclear transcriptome. (C) Heatmap representation of gene expression for 6,916 genes based on peak time (phase) of expression, as determined by the algorithm JTK_CYCLE, with minimal expression of 1 FPKM for at least one time-point. For easier visualization, the vertical scale of the organellar transcriptomes (chloroplast transcriptome, Middle; mitochondrial transcriptome, Bottom) is enlarged 25 times relative to the nuclear transcriptome. The green vertical lines indicate the timing of cell division.
To increase the resolution of the analysis, we next turned to k-means clustering, yielding 11,377 differentially expressed genes, or 85% of all transcribed genes, that fall in 16 clusters for nucleus-encoded genes, 3 clusters for chloroplast genes, and a single cluster for mitochondrial genes. These numbers are comparable to those published in an earlier diurnal study where Chlamydomonas cells were grown at high CO2 (8). Visualizing the entire dataset as a heatmap emphasizes the degree to which the Chlamydomonas transcriptome is driven at a genome-wide level by diurnal oscillations (Fig. 3B). Because the timing of physiological and molecular events during the day is critical for cell growth and survival, we determined the time of peak expression, or phase, using the algorithm JTK_CYCLE (23). Updating the heatmap of the transcriptome sorted by phase rather than cluster essentially yields comparable results (Fig. 3C), and underscores the impact of diurnal rhythms on algal biology.
Functional Gene Clustering Around the Diurnal Cycle.
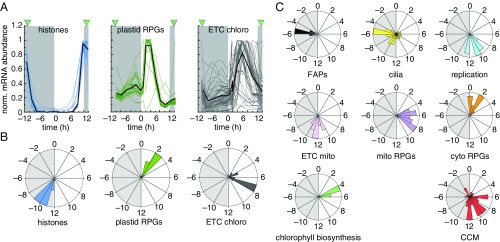
As illustrated with DNA replication genes (Fig. 2), temporal coexpression of genes across the diurnal cycle can point to similar function. We surveyed expression patterns and potential timing coincidence for a number of functional categories: cilia [from CiliaCut (24)] and other flagella-associated proteins (FAPs), nucleus-encoded ribosomal protein genes (RPGs), mitochondrial and chloroplast electron transfer chains (ETCs), and the carbon-concentrating mechanism (CCM). The expression of most FAPs peaks in the middle of the night between CT−5 and CT−6, following the completion of DNA replication (Fig. 4) and histone deposition on the newly synthesized DNA (Fig. 4A), as expected. Cells will have a fully functional pair of cilia by next dawn and will be ready for phototaxis and optimal photosynthesis. Nucleus-encoded cytosolic and plastid RPGs peak early during the day to sustain photosynthesis and incorporate newly fixed carbon into proteins critical for cell growth. Chlorophyll biosynthetic genes all peak during the first half of the day, and precede the observed rise in cellular chlorophyll content (SI Appendix, Fig. S4).
Fig. 4.
Genes with similar function are coexpressed. (A) Examples of synchronous gene-expression profiles for histone genes, nucleus-encoded plastid RPGs, and genes involved in the chloroplast ETC. All expression estimates are normalized to 1 for the highest value. The green triangles indicate the timing of cell division. (B and C) Phase distributions over the diurnal cycle for genes belonging to chosen gene categories. Phase values are based on the algorithm JTK_CYCLE, with a cut-off rate BH.Q of 1 × 10−5. Functional categories shown are: histones, DNA replication, FAPs, RPGs, mitochondrial and chloroplast ETC, chlorophyll biosynthesis, CCM, cilia (flagellar proteome, including all FAPs).
As the number of genes within a category increases, so does the spread of measured phases. For example, many genes involved in the CCM show peak expression during the day (0–9 h after lights-on), while others reach peak levels either during the night or shortly before dawn (Fig. 4C). Genes encoding proteins found in the cilia proteome (25) make up another example: although FAPs exhibit a clear phase preference for the middle of the night, the larger set of genes encoding ciliary proteins takes on a much broader phase distribution. Many support cilia regrowth following cell division, but a significant fraction shows peak expression during the day or shortly after dusk (Fig. 4C). This includes 1 of the 12 Chlamydomonas carbonic anhydrases, CAH6. CAHs catalyze the interconversion of CO2 into bicarbonate (HCO3−) and protons. The expression pattern of CAH6 is unique among CAHs: CAH6 is low during the day, when other CAHs (CAH1 to -5) reach their peak, and is much less induced by low CO2 than other CAHs, like the mitochrondria-localized CAH4 and CAH5 (SI Appendix, Fig. S5). CAH6 localizes to the cilia, where it interacts with FAP12 (26) and may contribute to chemotaxis toward HCO3−. Sensitivity of Chlamydomonas cells to HCO3− is under diurnal control with highest sensitivity at night, coincident with peak CAH6 expression (27).
Cells produce ATP via photosynthesis and respiration to fuel growth, for which they rely on their organelles. Most genes involved in photosynthetic electron transfer are expressed during the day (Fig. 4C). As cells prepare to shut down photosynthetic processes in anticipation of darkness, mitochondria-driven respiration appears to take over, as the expression of its electron transport chain constituents reaches maximum expression around dusk (Fig. 4C).
Our results demonstrate clustered expression of genes that participate in similar cellular functions. Furthermore, cellular events proceed in an orderly fashion during the day and night parts of the diurnal cycle, as hinted by PCA (Fig. 3A). Finally, our dataset enables association of putative functions with distinct family members or unknown genes (SI Appendix, Fig. S6) and allows the generation of testable hypotheses. For example, the expression pattern of mitochondrial electron transport chain genes suggests that respiration should be higher at night, during which time cells break down starch produced during the day as a carbon source, which we set out to test next.
Respiration Does Not Reach Maximum Capacity at Night.
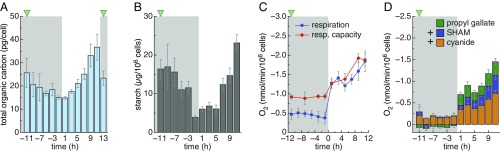
We first determined when cells accumulate starch. Total organic carbon per cell varies about twofold over the diurnal cycle, reaching its lowest levels at the end of the night and rising gradually during the day (Fig. 5A). Starch increases fivefold over the course of the day, particularly in the later part of the day, and at its peak accounts for about 25% of total organic carbon in the cell at the end of the day (Fig. 5B). Starch levels decrease continuously during the night, and are almost depleted by the beginning of the next day, as is also the case in Arabidopsis (28, 29). We therefore assumed that respiratory activity would follow starch degradation; the cultures were continuously aerated, so oxygen should not be limiting. Indeed, the truncated hemoglobin THB8, a marker for strict hypoxia (30), is not expressed in our cultures (Dataset S10). However, oxygen consumption, a quantitative measure of respiration, is in fact low in the night, despite cellular inability to produce ATP from photosynthesis (Fig. 5C). The potential for respiration (or respiratory capacity) is high, but the components are not engaged (Fig. 5C). Going back to our expression dataset, transcripts for the genes encoding all complexes of the mitochondrial respiratory chain, including cytochrome c oxidase (complex IV) and ATP synthase (complex V), display two distinct peaks, one early at night, and one in the middle of the day (SI Appendix, Fig. S7).
Fig. 5.
Chlamydomonas cells do not use respiration to full capacity in the dark. Total nonpurgeable organic carbon (A) and starch content (B) of Chlamydomonas cells over the diurnal cycle. Data shown on a per cell basis. The green triangles indicate the timing of cell division. (C) Oxygen consumption (blue line) and respiration (resp) capacity of cells treated with the mitochondrial uncoupling agent FCCP (red line). Oxygen consumption was measured on the same samples before and after addition of FCCP. Data shown on a per cell basis. (D) Relative contribution of cytochrome c oxidase and alternative oxidases in oxygen consumption. Potassium cyanide (cyanide in figure) inhibits cytochrome c oxidase, while SHAM and propyl gallate target mitochondrial and plastid terminal oxidases (AOX and PTOX, respectively). Data shown on a per cell basis.
We then analyzed the fraction of oxygen consumption directly attributable to cytochrome c oxidase or to alternative oxidases utilizing selective inhibitors, because oxygen consumption measurements reflect the combined activities of these enzymes. Each oxidase type accounts for half of total oxygen consumption during the day, while at night most of the oxygen consumption activity can be attributed to cytochrome c oxidase (Fig. 5D). However, because cytochrome c oxidase operates at capacity during the day, but not in the night (Fig. 5C), cytochrome c oxidase-dependent O2 consumption is still higher at each time point during the day compared with the night (Fig. 5D). Therefore, the increased oxygen consumption noted in the light period cannot be solely attributed to the alternative oxidases. We conclude that the high respiratory activity during the day reflects a greater cellular demand for ATP to facilitate maximal macromolecular metabolism and growth. During the dark portion of the diurnal cycle the demand for ATP is lower because the cells are in G0 (the metabolically more quiescent stage of the cell cycle), and even low levels of respiration may be sufficient to fulfill cellular ATP requirements. Nevertheless, this left open the question of how glycolytically generated NADH might be reoxidized and the fate of pyruvate, which we explored next.
Fermentation as the Path for Reoxidation of the NADH Generated During Glycolysis.
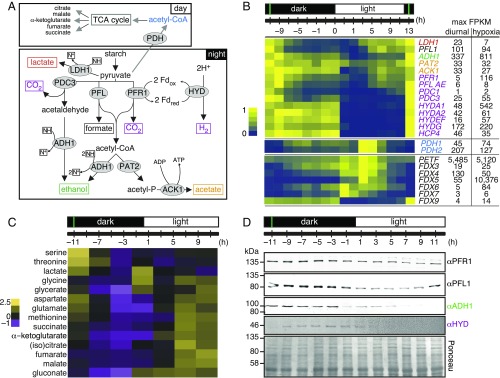
The oxidation of Glc to pyruvate generates NADH, which must be reoxidized to NAD+ to support continued glycolysis. In aerobic organisms, this occurs via respiration, which is coupled to ATP synthesis. Although the yield of ATP from aerobic oxidation of NADH is substantial, the process is slower than anaerobic pathways for NADH reoxidation (e.g., conversion of pyruvate to lactate or ethanol). These anaerobic pathways are well-studied in Chlamydomonas (Fig. 6A). We obtained validation of the fermentation hypothesis in the expression levels of Chlamydomonas fermentation genes, and discovered some unexpected additional complexity. All genes reach their peak accumulation after dusk and are predominantly more highly expressed at night, with the exception of LDH1 and PFL1 (which show some expression also in the day period). In addition, individual genes appear to be turned off sequentially (Fig. 6B). For many fermentation genes, high expression is restricted to the night, suggesting potential light-mediated mRNA degradation or transcriptional repression. We hypothesize that the proteins encoded by these genes offer multiple options over the course of the dark part of the diurnal cycle for the reoxidation of NADH to NAD+. Genes encoding enzymes involved in the conversion of pyruvate to lactate, ethanol, and formate, including PFL1, show peak expression right after dusk followed by a fast decline, while genes encoding enzymes generating acetate (PAT2 and ACK1) and CO2 (PDC3) reach maximal expression later during the night and over a longer time scale (Fig. 6B). One of the products of pyruvate metabolism, lactate, is the sole soluble metabolite that accumulates at night (Fig. 6C), which further substantiates the operation of anaerobic routes for pyruvate metabolism. RNAs encoding enzymes that are not sensitive to oxygen begin to rise already by day’s end, while RNAs encoding oxygen-sensitive enzymes like PFR1 do not accumulate until after dusk (Fig. 6B). Genes encoding the other oxygen-sensitive enzymes, PFL1 and PAT2, begin to be expressed during the day, but exhibit a large jump in their expression at dusk (421% for PFL1 and 360% for PAT2, between CT+11 and CT+13). These gene-expression patterns are roughly mirrored in protein accumulation, assessed immunologically for PFR1, PFL, ADH, and HYDA1/2 (Fig. 6D), and by proteomics analysis of soluble cell fractions for PFR1, HYDG, and HYDA2 (SI Appendix, Fig. S8). Because RNA half-lives are typically much shorter than that of proteins, unless there is a specific mechanism to degrade or destabilize the proteins, the proteins persist into the day period. Some proteins are lost more rapidly, perhaps because loss of their oxygen-labile cofactors renders them susceptible to proteolytic digestion. Expression of the genes encoding pyruvate dehydrogenase enzymes PDH1 and PDH2 (catalyzing the oxidative degradation of pyruvate to acetyl-CoA) is restricted to the middle of the day, preceding the accumulation of numerous TCA intermediates, as expected (Fig. 6 B and C).
Fig. 6.
Chlamydomonas cells use anaerobic routes for handling pyruvate. (A) Key pyruvate metabolism pathways according to refs. 36 and 46. Final fermentation products are shown in boxes and enzymes in gray ellipses. Abbreviations: ACK1, acetate kinase 1; ADH1, acetaldehyde/alcohol dehydrogenase; HYDA, [Fe–Fe]-hydrogenase; LDH1, d-lactate dehydrogenase; PAT2, phosphate acetyltransferase 2; PFL1, pyruvate formate lyase; PFR1, pyruvate ferredoxin oxidoreductase. N+, NAD+; NH, NADH. (B) Normalized expression of fermentation genes listed, shown as a heatmap. Numbers on the right side indicate maximum FPKM values in our samples (diurnal) and in cells grown under dark hypoxia for 6 h (34). (C) Changes in water-soluble metabolites over the diurnal cycle, analyzed by GC-MS. Results are shown as a heatmap of z-score normalized abundance for metabolites that changed significantly over the diurnal cycle. (D) Fermentation enzymes are more abundant in the dark. Total protein samples were separated by denaturing SDS/PAGE, followed by immune-detection with antibodies raised against PFR1, PFL, ADH, and HYDA1+2. Equal protein amounts were loaded and confirmed with Ponceau S stain. All data are shown as average ± SD (n = 3). The immune-detection was performed at least twice on independent samples. The green vertical lines indicate the timing of cell division.
Our cultures were not anaerobic, but genes encoding anaerobic routes for pyruvate metabolism are expressed at or near the same levels in these experiments as they are in anaerobic cells (Fig. 6B), with the exception of HYDA1 and PFR1, which are 10–20 times more highly expressed in strict hypoxia (31–34). We turned to other transcriptome datasets with samples collected in the dark to independently validate the hypothesis that anaerobic pyruvate catabolism represents the typical rather than exceptional metabolic program: we observed essentially the same pattern and abundance in an earlier diurnal time course, including the differential expression behavior of HYDA1, PFR1, and ADH1 (8). Other studies also detected significant expression of fermentation genes in nonhypoxic cultures (35, 36); all indicate that expression of fermentation genes is not restricted to strict anaerobic conditions, and that aerated Chlamydomonas cultures have the potential to metabolize pyruvate by fermentation rather than via the TCA cycle at night.
Ferredoxin is the central electron donor in chloroplast metabolism, and the assignment of a particular ferredoxin (of the 13 encoded in the genome) (37, 38) to the hydrogenase is still ambiguous, although FDX5 is the favored candidate because of its high expression in hypoxia and anaerobiosis. However, FDX5 shows peak expression during the day (like PETF) and is therefore more likely to play a role in photosynthesis (38). Of the seven isoforms predicted to localize to chloroplast (37), only the expression of FDX9 matches the expression pattern of HYDA1 and HYDA2 (Fig. 6B), and may be the more likely candidate.
Acute and Sustained Responses to Light Intensity.
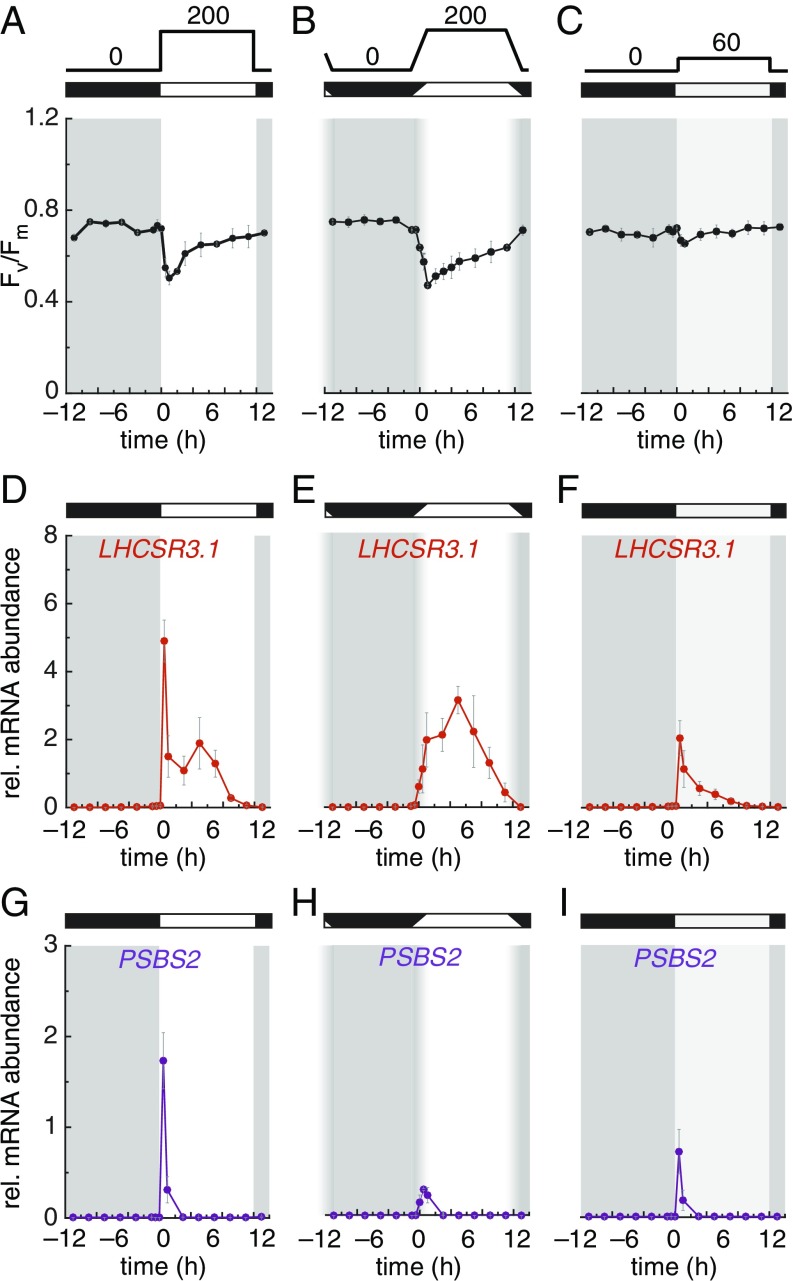
Light is the main source of cellular photooxidative damage, and excess light energy may be dissipated as chlorophyll fluorescence or as heat. We were surprised to find signs of light stress in our cultures, although light intensity was moderate and lacked a detectable UV component. Physiological indicators of light stress include a drop in photosystem II efficiency, as measured by Fv/Fm (Fig. 7A and SI Appendix, Fig. S9B) and a sharp rise in the plastoquinol pool (SI Appendix, Fig. S10 A and B), concomitant with the induction of the stress-responsive genes encoding light harvesting-like proteins, specifically LHCSR1, LHCSR3s, and PSBSs (Fig. 7 D and G and SI Appendix, Fig. S9) (39, 40). We only detected this transient induction because of the experimental design, involving CO2 limitation (air levels) and dense sampling of the culture at dawn, and find it is consistent with the function of PSBS in setting up photoprotection (41). PSBS protein follows PSBS expression, with a sharp, transient peak 1 h after lights-on; both messenger and protein are actively degraded within 3 h of light exposure. LHCSR3 proteins remain constant over the diurnal cycle but are clearly modified upon transfer to light, as expected (SI Appendix, Fig. S11) (42).
Fig. 7.
Chlamydomonas cells integrate two light inputs to cope with light stress. (A–C) Photosystem II capacity, as determined by Fv/Fm values over the diurnal cycle, in cultures exposed to 200 µmol photons/m2/s with abrupt (A) or gradual 2-h transition at dawn (B), or exposed to 60 µmol photons/m2/s with abrupt transition at dawn (C). Relative (rel) mRNA abundance for LHCSR3.1 (D–F) and PSBS2 (G–I) by quantitative RT-PCR, in samples collected from cells grown under diurnal conditions with 200 µmol photons/m2/s with abrupt transition at dawn (D and G), 200 µmol photons/m2/s with gradual transition at dawn (E and H), or 60 µmol photons/m2/s with abrupt transition at dawn (F and I). All data are shown as average ± SD (n = 3). rel, relative.
A build-up of a reduced plastoquinone pool is indicative of a saturation in the plastid ETC between PSII and PSI and overacidification of the plastid lumen, which will promote phosphorylation of LHCII antennae proteins, leading to their dissociation from PSII and association with PSI (43). Measuring chlorophyll fluorescence at 77 °K indeed supports this transition from state I to state II, as evidenced by a higher fluorescence of the PSI-LHC peak at 713 nm (SI Appendix, Fig. S10C). The photosynthetic apparatus therefore undergoes a major restructuring in the light to cope with and prevent photooxidative stress, which must occur even at a surface photon flux density of only 200 μmol photons/m2/s.
Accumulation of PSBS mRNA and PSBS protein is limited to dawn. Fourteen genes are coexpressed with PSBS and include two chlorophyll a/b-binding proteins, an E3 ubiquitin ligase and several genes encoding unknown functions (SI Appendix, Fig. S12 A and B). LHCSR3 genes show a more prolonged expression window that extends to the middle of the day, where they may function in long-term acclimation to light stress, as evidenced by the partial recovery of Fv/Fm parameters (Fig. 7 A–C). LHCSR3 genes share their expression patterns with 21 other genes, among them, a glutaredoxin (GRX4) and a fatty acid desaturase (FAD3) (SI Appendix, Fig. S12 C, D, and F). Other genes with strong induction at dawn participate in detoxification of reactive oxygen species, including a glutathione peroxidase (GPX5), superoxide dismutase (FSD1), and nucleoredoxins (NRX2 and NRX3) (SI Appendix, Fig. S12E).
Two Distinct Signaling Pathways Mediate Acclimation to Light Intensity.
We reasoned that the expression patterns of LHCSR3 and PSBS genes may reflect a response to (i) light intensity, (ii) the abrupt transition from darkness to light, or (iii) both. We therefore dissected the relative contribution of each signal by adjusting the experimental set-up. A gradual ramping from 0 to 200 µmol photons/m2/s over 2 h abrogates PSBS’s and LHCSR3’s acute induction, but still results in a drop in Fv/Fm that is comparable to that seen following an instantaneous transition (Fig. 7 D, E, G, and H and SI Appendix, Fig. S9). The second peak in LHCSR3 expression around CT+5 to CT+7 remains and likely represents a response to sustained light intensity (44). Indeed, dropping the fluence rate from 200 to 60 µmol photons/m2/s eliminates the second, later LHCSR3 peak, confirming our hypothesis (Fig. 7F). Under the lower fluence rate, Fv/Fm remains constant and high over the diurnal cycle (Fig. 7C), indicating that cells do not suffer from light stress.
The loss of the later LHCSR3 peak also uncovers the existence of an acute response at dawn (Fig. 7F), albeit at a lower amplitude than seen when cells are transferred to 200 µmol photons/m2/s (Fig. 7D). This acute response is echoed by a peak in PSBS expression, although again with a low amplitude (SI Appendix, Fig. S9). Cells can therefore measure photosynthetic photon flux over time, and adjust the amplitude of the acute response accordingly.
Discussion
Rhythmic gene expression in the green alga Chlamydomonas reflects their biological function and pathways (Fig. 4). Little attention has been given to nighttime physiology, as cells enter the metabolically quiescent phase (or G0) of the cell cycle. Our results demonstrate that a greater fraction of pyruvate generated from starch degradation in Chlamydomonas enters anaerobic rather than aerobic metabolism. Many fermentation enzymes are oxygen-sensitive, but their transcription is restricted to the night, which is reminiscent of the temporal separation of nitrogen fixation in cyanobacteria (45). For each molecule of pyruvate consumed, Chlamydomonas cells therefore produce much less ATP than they could from aerobic metabolism. We estimate that for each ATP produced via fermentation, five are produced via respiration (Fig. 5 and SI Appendix), based on the amounts of starch and oxygen consumed at night. Although respiration is still the main source of ATP production at night, we hypothesize that the main contribution of fermentation pathways, aside from ATP itself, is for the reoxidation of NADH. Is this a waste of fuel or is it an indication of unknown metabolic interactions in the natural environment? The products of pyruvate metabolism are typically excreted (46) and may support prokaryotes, which in turn may provide vitamins to the alga (47). Anaerobic pathways can oxidize NADH more rapidly than can oxidative phosphorylation, which may be a consideration at low oxygen tension, a situation that might prevail in the natural environment, and is likely compatible with the low ATP demand during the metabolically more quiescent part of the cell cycle (48).
Like in animals and the related unicellular alga Volvox, most Chlamydomonas histone transcripts are not poly-adenylylated (49, 50) and their expression has therefore been largely unexplored by most RNA-sequencing experiments. Because we used Ribo-depletion, we successfully documented their tight coexpression following DNA replication and cell division, and preceding cilia biogenesis in anticipation of the next dawn and photosynthesis-driven phototaxis and chemotaxis. Of considerable surprise was how greatly histone transcripts are induced: 100- to 1,000-fold, far greater than necessitated by the expected 2-fold increase in protein levels needed to complete mitosis. Their high abundance may, however, drive timely/immediate histone translation. This phenomenon, called RNA superinduction, is becoming recognized as a new regulatory mechanism that allows for competitive translation of particular mRNAs (51). We estimate that almost half of all of the cytosolic ribosomes are busy making more of themselves at dawn by translating mRNAs for ribosomal proteins (SI Appendix, Table S1). Regulating protein translation poses a conundrum to a growing cell: how many ribosomes to dedicate to the synthesis of new ribosomal proteins versus all other cellular proteins. Rhythmic expression may have been adopted as an early coping mechanism in unicellular algae: bursts of transcripts at the proper time of day along the diurnal cycle, to produce just enough proteins to make it through another day (52).
Materials and Methods
Strains and Culture Conditions.
Chlamydomonas strain CC-5390 [CC-4351 (cw15–325 mt+) rescued with the pCB412 cosmid carrying the ARG7 gene] was used for all experiments in this study. Cells were precultivated in 250-mL Erlenmeyer flasks containing 100 mL Tris-acetate-phosphate (TAP) medium with trace element solution, as described previously (53). These precultures were grown in TAP with constant agitation in an Innova incubator (160 rpm, New Brunswick Scientific) at 24 °C in continuous light (90 µmol photons/m2/s), provided by cool white fluorescent bulbs (4,100 K) and warm white fluorescent bulbs (3,000 K) in a 2:1 ratio until inoculation of photobioreactors.
Photobioreactors Operation and Monitoring.
All experiments were performed in presterilized flat-panel photobioreactors (Photobioreactor FMT 150 from Photon System Instruments). Each photobioreactor (working volume 0.4 L) was aerated and mixed with pressurized air at an airflow of 0.2 L/min with mass flow controllers (Smart TMF SLA5850; Brooks). Temperature was set to 28 °C during the day and 18 °C at night, while illumination was provided by a panel of red and blue LEDs (LED Light Source SL 3500; Photon System Instruments). Light fluence was set to 200 µmol photons/m2/s unless stated otherwise. For experiments with gradual adjustment of light intensity at dawn and dusk, we used the linear dimming option.
Cells were inoculated from TAP precultures at a starting optical density at 680 nm of 0.05 in high-salt medium (HSM) supplemented with a modified trace element solution (53, 54). Cells were allowed to grow in turbidostat mode for a minimum of 5 d under entraining conditions, until they reached an optical density of 0.4 (corresponding to 2–3 × 106 cells/mL). We determined cell number and size with a Beckman Coulter Multisizer 3 with a 50-µm orifice (Beckman Coulter). Samples were concentrated 10-fold in HSM medium before counting. Biological replicates refer to cultures from independent photobioreactors.
RNA Extraction and Library Preparation.
A total of 3 × 107 cells were collected at each time-point by centrifugation at 1,424 × g for 5 min at 4 °C. We extracted total RNA with the TRIzol reagent as previously described (55). RNA was DNase-treated with Turbo DNase (Ambion), followed by a cleaning and concentration step with the RNA Clean & Concentrator-5 kit (Zymo Research). RNA quality and concentration were determined on a Nanodrop 2000 (Thermo Fisher Scientific) and an RNA 6000 microfluidic chip on a Bioanalyzer 2100 (Agilent). The University of California, Los Angeles Neuroscience Genomics Core prepared stranded RNA-sequencing libraries using the RiboZero stranded TruSeq RNA Samples prep kit (Illumina). Library quality control was performed by Bioanalyzer with a DNA 1000 microfluidic chip. Libraries were quantified using a Qubit fluorimeter (Thermo Fisher Scientific), pooled and sequenced on a HiSeq 2000 sequencer as single-end 50 bp reads. Transcriptome data were deposited in the NCBI Gene Expression Omnibus (GEO) database under accession GSE112394 (56). Analysis of the resulting data were performed as described previously (16).
Transcriptome Data Analysis.
Multidimensional scaling.
Relative expression estimates were imported into the R package cummeRbund (56, 57), and subjected to multidimensional scaling with the method MDSplot with individual replicates and with mean expression per time point.
K-means clustering and heatmap.
Expression estimates for 10,394 nuclear genes (with a Benjamini–Hochberg adjusted P value < 0.01 and expression ≥1,000 total counts) were normalized and then subjected to k-means clustering with the Kmeans tool in the R package amap with 16 centers. The 16 resulting clusters were arranged manually in order of the peak of expression. Using the same inclusion criteria, 68 chloroplast genes were subjected to k-means clustering with three centers. Only three mitochondrial genes met the inclusion criteria. These were normalized and plotted but not subjected to k-means clustering. The resulting expression table of clustered genes was normalized by row and plotted as a heatmap with the heatmap.2 tool in the R package gplots.
Phase analysis.
The timing (or phase) of peak expression for each gene was determined with the R package MetaCycle, using the algorithm JTK_CYCLE (JTK). The three biological replicates were double-plotted to restrict phase-calling to within the diurnal range, set between 20 and 28 h. The JTK method does not allow for uneven sampling intervals or noninteger sampling times; we therefore (i) removed samples collected at CT−0.5 and CT+0.5 and (ii) added columns of missing values at every even-numbered hour, except at time 0, at which time a sample was collected (CT0 being the dark-to light transition). We applied a cut-off of BH.Q of 1 × 10−10 as described previously (8), followed by an expression estimate cut-off of ≥1 mean fragments per kilobase of transcript per million mapped reads (FPKM) for at least one sample over the whole time course. Initial estimated phase values were adjusted to the predicted period length of each gene, with the function adjustPhase = “predictedPer” within meta2d of MetaCycle. We followed the same strategy for chloroplast-encoded genes using mean expression across the three replicates, and looked for rhythmic genes with a period of about 24 h (range 20–28). We applied a cut-off of BH.Q of 0.001 (for 24-h rhythms) to account for the single expression estimates used for the analysis. We reanalyzed the Zones et al. dataset (8) with the same settings, keeping all hourly samples and excluding half-hour time points.
Quantitative real-time PCR.
Reverse transcription was initiated with an oligo dT18 primer with 2.5 µg total RNA as template and the reverse-transcriptase SuperScript III (Invitrogen), according to the manufacturer’s instructions. The resulting cDNAs were diluted 10-fold before use. Each reaction contained cDNAs corresponding to 100 ng total RNA, 6 pmol each of forward and reverse primers, 0.25 mM dNTPs, 1× Ex Taq buffer with Mg2+ (TaKaRa), 0.01% (wt/vol) SYBR Green I Nucleic Acid Stain (Cambrex Bio Science Rockland), 0.1% (vol/vol) Tween 20, 100 µg/mL BSA, 5% (vol/vol) DMSO, in a total volume of 20 µL. The following program was used: 95 °C for 5 min, followed by 40 cycles of 95 °C for 15 s, 65 °C for 60 s, with fluorescence measurement after each 65 °C step; a melting curve analysis was performed between 65 and 95 °C to confirm specific amplification. Relative abundances were calculated using LinReg, with EIF1A (Cre02.g103550) as reference transcript. Primer sequences are given in SI Appendix.
Proteomics.
Protein detection by LC-MS/MS.
We collected 4 × 107 cells by centrifugation at 1,450 × g at 4 °C for 4 min. The cell pellet was washed once with 1 mL 10 nM phosphate, pH 7.0, resuspended in 200 µL 10 nM phosphate, pH 7.0 and subjected to slow freeze–thaw cycles to quantitatively extract soluble proteins. Protein concentration of the soluble fraction was determined by BCA assay (Thermo Fisher Scientific). Urea and DTT were added to all samples at a final concentration of 8 M and 10 mM, respectively before incubation at 60 °C for 30 min with constant shaking (800 rpm). All samples were then diluted eightfold with 100 mM NH4HCO3 and 1 mM CaCl2, and digested with sequencing-grade modified porcine trypsin (Promega) provided at a 1:50 (wt/wt) trypsin-to-protein ratio for 3 h at 37 °C. Digested samples were desalted using a four-probe positive-pressure Gilson GX-274 ASPEC system (Gilson) with Discovery C18 100-mg/mL solid-phase extraction tubes (Supelco) as follows: columns were preconditioned with 3 mL methanol, followed by 2 mL 0.1% trifluoroacetic acid (TFA) in water. Samples were then loaded onto columns, followed by 4 mL 95:5 water:acetonitrile (ACN) 0.1% TFA. Samples were eluted with 1 mL 20:80 water:ACN 0.1% TFA, and concentrated to a final volume of ∼100 µL in a Speed Vac. After determination of peptide concentration by BCA assay, samples were diluted to 0.25 µg/µL with nanopore water for LC-MS/MS analysis [LC part: LC column of fused silica (360 µm × 70 cm) handpacked with Phenomenex Jupiter derivatized silica beads of 3-µm pore size (Phenomenex); HPLC part: HPLC NanoAcquity UPLC system (Waters); MS part: Q Exactive mass spectrometer (Thermo Fisher Scientific)]. Samples were loaded onto LC columns with 0.05% formic acid in water and eluted in 0.05% formic acid in ACN over 100 min. Twelve high-resolution (17.5 K nominal resolution) data-dependent MS/MS scans were recorded for each survey MS scan (35 K nominal resolution) using normalized collision energy of 30, isolation width of 2.0 m/z, and rolling exclusion window lasting 30 s before previously fragmented signals are eligible for reanalysis. Unassigned charge and singly charged precursor ions were ignored.
MS/MS spectra were interrogated against the Chlamydomonas proteome via a target-decoy approach; unique peptide sequences with their relative protein abundance as defined by “PeakMaxIntensity” by the in-house MASIC software are given in Dataset S2. The mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium (www.proteomexchange.org/) under accession no. PXD010794 (58). More details on peak matching and quantification are available in SI Appendix.
Total carbon content analysis.
Total nonpurgeable organic carbon content of cells was determined as described previously (59) with minor modifications. We collected 3 × 107 cells by centrifugation at 3,100 × g for 2 min at 4 °C, and washed the cell pellet once in 10 mM Phosphate buffer (pH 7.0). Each pellet was then overlaid with 0.9 mL 3 M HCl and digested at 65 °C for 16 h with constant agitation. Cell lysates were diluted 111-fold with MilliQ water, for a final hydrochloric acid concentration of 27 mM, and sparged to remove inorganic carbon. Some organic carbon may be purged from the sample by this method, but was not measured in our experimental set-up. We therefore report “nonpurgeable” organic carbon.
Starch measurements.
We collected 15-mL culture (∼5 × 107 cells) by centrifugation at 1,650 × g for 10 min at room temperature, and extracted starch by ethanolic extraction, as previously described (60). We digested starch into glucose with the addition of amyloglucosidase, and then measured glucose levels with the Glucose HK kit (Sigma) according to the manufacturer’s instructions.
Immunodetection of Chlamydomonas proteins.
We collected 15 mL of cultures at a cell density of 2 × 106 cells/mL by centrifugation at 1,650 × g at 4 °C. We extracted total proteins by resuspending cell pellets in 300 µL 50 mM Na-phosphate pH 7.0 with cOmplete EDTA-free protease inhibitor (Sigma), 2% (wt/vol) SDS, 10% (wt/vol) sucrose. Protein concentrations were determined with Pierce BCA assay against BSA as standard (Thermo Fisher Scientific). Proteins were separated by SDS/PAGE gels, loading 10 µg protein per lane, and transferred to nitrocellulose membranes by semidry electroblotting. Following blocking in 3% nonfat dried milk in 1× PBS with 0.1% (wt/vol) Tween 20 for 30 min at room temperature, membranes were incubated with primary and secondary antibodies in the same solution, with intervening washes in 1× PBS, 0.1% Tween 20. Primary antibodies were used at the following dilutions (with provenance laboratories): hydrogenase 1:2,000 (T. Happe, Ruhr-Universität Bochum, Bochum, Germany); PSBS 1:4,000 (P. Jahns, Heinrich-Heine-Universität in Düsseldorf, Düsseldorf, Germany); PFR 1:3,000 (A. Atteia, Marine Biodiversity, Exploitation and Conservation, Unité Mixte de Recherche, Sète, France); PFL 1:20,000 (A. Atteia); ADH 1:2,500 (A. Atteia); LHCSR3 1:4,000 (M. Hippler, University of Münster, Münster, Germany). A goat anti-rabbit secondary antibody, conjugated to alkaline phosphatase, was used at a dilution of 1:10,000 for detection according to the manufacturer’s instructions.
Photosynthetic parameters.
Imaging of maximum quantum efficiency of photosystem II was performed using a FluorCam 700 MF system (Photon Systems Instruments) using the Fv/Fm settings as described previously (61). Chlamydomonas strains were dark-adapted for 15 min before each experiment. Fluorescence parameters were calculated as follows: Fv/Fm = (Fm − Fo)/Fm, where Fv is the calculated variable fluorescence, Fm is the maximal fluorescence measured immediately after the saturating pulse, and Fo is the initial fluorescence of dark-adapted cells. Fluorescence emission spectra were collected at 77 °K, as described previously (62).
Oxygen consumption and evolution measurements.
Oxygen evolution rates were measured on a standard Clark-type electrode (Hansatech Oxygraph with a DW-1 chamber) and analyzed with Hansatech OxyLab software v1.15. All experiments were carried out on 2 mL of cells removed from the culture (density of ∼2–3 × 106 cells/mL) in the presence of 10 mM KHCO3 and under constant stirring. Respiration rates were measured as oxygen consumption over a period of at least 5 min in the dark (or until the observed rate stabilizes). We measured oxygen evolution on the same samples over 5 min by turning the built-in LED to provide 200 µmol photons/m2/s (as in the photobioreactors) and 500 µmol photons/m2/s (saturating light) after a 5-min dark acclimation period. The rate of photosynthetic oxygen evolution was calculated as the difference between oxygen evolution in the light and oxygen consumption in the dark for each sample.
For inhibitor studies, we prepared fresh stock solutions of all chemicals in 100% ethanol [100 mM n-propyl gallate, Sigma P3130; 400 mM salicylhydroxamic acid (SHAM), Sigma-Aldrich S7504; 10 mM carbonyl acyanide 4-(trifluoromethoxyl)phenylhydrazone (FCCP), Sigma-Aldrich C2920; or MilliQ water (200 mM potassium cyanide, Fisher P223-100)]. Final concentrations for inhibitors during the experiments were 1 mM n-Propyl gallate, 5 mM SHAM, 4 mM potassium cyanide and 5 µM FCCP (for respiratory capacity), and respiration rate was measured as oxygen consumption over 5 min in the dark, and again after an acclimation period in the presence of the inhibitors. Control samples received ethanol alone at a concentration of 1% to account for potential solvent effects.
Metabolites analysis by GC-MS.
Metabolites extraction was performed as previously described (63). Briefly, cells were collected from 10 mL of a culture that had reached a cell density of 2–4 × 106 cells/mL by vacuum filtration onto a PVDF filter (GVWP02500; Millipore). Filters were frozen immediately in liquid nitrogen. Extraction of metabolites was carried out by soaking filters in 600 µL of cold 70% methanol 30% chloroform (vol/vol) with vortexing at 4 °C for 70 min. Filters were removed, and samples collected by a short centrifugation before the addition of 300 µL cold LC-grade water, followed by two freeze–thaw cycles. After centrifugation, the aqueous polar phase was collected and dried in a Speed Vac. Derivatization and analysis of metabolites was performed as described (64) by gas chromatography-mass spectrometry (GC-MS) on a 7200 GC-QTOF instrument (Agilent). Data analysis was conducted with the help of the Mass Hunter Software (Agilent). For relative normalization, all metabolite peak areas were normalized to cell number.
Plastoquinone measurements.
We collected a total of 4–8 × 107 cells/mL Chlamydomonas cells by centrifugation at 1,424 × g for 5 min at 4 °C. Cell pellets were resuspended in 500 µL 95% (vol/vol) ethanol spiked with 50 µL of 121 µM ubiquinone-10 (final amount of 6.05 nmols) as internal standard, and homogenized in a 5-mL Pyrex tissue grinder. The grinder was rinsed with 500 µL 95% (vol/vol) ethanol, and combined with the initial homogenate. We separated cell debris by centrifugation at 18,000 × g for 5 min at 4 °C and immediately analyzed extracts by HPLC as described using 100 µL of each extract (65). We tested whether the centrifugation step may affect plastoquinol/plastoquinone ratios in the samples by measuring platoquinol and plastoquinone in matched sets with either centrifugation or direct quenching by the addition of ethanol (∼70% final concentration) to cell cultures. Plastoquinol/plastoquinone ratios were not significantly different between sets, indicating that the redox state of plastoquinone is not affected during sample collection under our conditions.
Data availability.
Transcriptome data were deposited at the National Center for Biotechnology Information’s Gene Expression Omnibus under accession GSE112394 (56).
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (66) partner repository with the dataset identifier PXD010794 and 10.6019/PXD010794 (58).
Supplementary Material
Acknowledgments
We thank Elisabeth Klemp, Katrin Weber, and Maria Graf for technical assistance during gas chromatography-mass spectrometry sample preparation and analysis. S.S.M., P.A.S., and S.D.G. were supported by a cooperative agreement with the US Department of Energy Office of Science, Office of Biological and Environmental Research program under Award DE-FC02-02ER63421. D.S. was supported by the European Molecular Biology Organization (ALTF 653-2013). G.J.B. and E.S. were supported by National Science Foundation Grants MCB-1608088 and MCB-1712608. Part of the research was performed using Environmental Molecular Sciences Laboratory (Ringgold ID 130367), a Department of Energy Office of Science User Facility at Pacific Northwest National Laboratory (PNNL) in Richland, WA, sponsored by the Office of Biological and Environmental Research.
Footnotes
The authors declare no conflict of interest.
Data deposition: Transcriptome data were deposited in the NCBI Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE112394). The mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium (www.proteomexchange.org/) via the PRIDE partner repository (accession no. PXD010794).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1815238116/-/DCSupplemental.
References
- 1.Noordally ZB, Millar AJ. Clocks in algae. Biochemistry. 2015;54:171–183. doi: 10.1021/bi501089x. [DOI] [PubMed] [Google Scholar]
- 2.Harris EC. The Chlamydomonas Sourcebook: Introduction into Chlamydomonas and Its Laboratory Use. Academic; New York: 2008. [Google Scholar]
- 3.Scranton MA, Ostrand JT, Fields FJ, Mayfield SP. Chlamydomonas as a model for biofuels and bio-products production. Plant J. 2015;82:523–531. doi: 10.1111/tpj.12780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atteia A, van Lis R, Tielens AG, Martin WF. Anaerobic energy metabolism in unicellular photosynthetic eukaryotes. Biochim Biophys Acta. 2013;1827:210–223. doi: 10.1016/j.bbabio.2012.08.002. [DOI] [PubMed] [Google Scholar]
- 5.Niyogi KK, Björkman O, Grossman AR. The roles of specific xanthophylls in photoprotection. Proc Natl Acad Sci USA. 1997;94:14162–14167. doi: 10.1073/pnas.94.25.14162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Erickson E, Wakao S, Niyogi KK. Light stress and photoprotection in Chlamydomonas reinhardtii. Plant J. 2015;82:449–465. doi: 10.1111/tpj.12825. [DOI] [PubMed] [Google Scholar]
- 7.Tulin F, Cross FR. A microbial avenue to cell cycle control in the plant superkingdom. Plant Cell. 2014;26:4019–4038. doi: 10.1105/tpc.114.129312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zones JM, Blaby IK, Merchant SS, Umen JG. High-resolution profiling of a synchronized diurnal transcriptome from Chlamydomonas reinhardtii reveals continuous cell and metabolic differentiation. Plant Cell. 2015;27:2743–2769. doi: 10.1105/tpc.15.00498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Howell SH, Walker LL. Transcription of the nuclear and chloroplast genomes during the vegetative cell cycle in Chlamydomonas reinhardi. Dev Biol. 1977;56:11–23. doi: 10.1016/0012-1606(77)90151-8. [DOI] [PubMed] [Google Scholar]
- 10.Hsu PY, Harmer SL. Circadian phase has profound effects on differential expression analysis. PLoS One. 2012;7:e49853. doi: 10.1371/journal.pone.0049853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donnan L, John PCL. Cell cycle control by timer and sizer in Chlamydomonas. Nature. 1983;304:630–633. doi: 10.1038/304630a0. [DOI] [PubMed] [Google Scholar]
- 12.Lemaire SD, et al. Analysis of light/dark synchronization of cell-wall-less Chlamydomonas reinhardtii (Chlorophyta) cells by flow cytometry. Eur J Phycol. 1999;34:279–286. [Google Scholar]
- 13.Marzluff WF, Wagner EJ, Duronio RJ. Metabolism and regulation of canonical histone mRNAs: Life without a poly(A) tail. Nat Rev Genet. 2008;9:843–854. doi: 10.1038/nrg2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sunwoo H, et al. MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009;19:347–359. doi: 10.1101/gr.087775.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilusz JE, Freier SM, Spector DL. 3′ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell. 2008;135:919–932. doi: 10.1016/j.cell.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gallaher SD, et al. High-throughput sequencing of the chloroplast and mitochondrion of Chlamydomonas reinhardtii to generate improved de novo assemblies, analyze expression patterns and transcript speciation, and evaluate diversity among laboratory strains and wild isolates. Plant J. 2018;93:545–565. doi: 10.1111/tpj.13788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reichard P. Interactions between deoxyribonucleotide and DNA synthesis. Annu Rev Biochem. 1988;57:349–374. doi: 10.1146/annurev.bi.57.070188.002025. [DOI] [PubMed] [Google Scholar]
- 18.Fontecave M. Ribonucleotide reductases and radical reactions. Cell Mol Life Sci. 1998;54:684–695. doi: 10.1007/s000180050195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bourguignon LYW, Palade GE. Incorporation of polypeptides into thylakoid membranes of Chlamydomonas reinhardtii. Cyclic variations. J Cell Biol. 1976;69:327–344. doi: 10.1083/jcb.69.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Young MR, Tye BK. Mcm2 and Mcm3 are constitutive nuclear proteins that exhibit distinct isoforms and bind chromatin during specific cell cycle stages of Saccharomyces cerevisiae. Mol Biol Cell. 1997;8:1587–1601. doi: 10.1091/mbc.8.8.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Otero S, Desvoyes B, Gutierrez C. Histone H3 dynamics in plant cell cycle and development. Cytogenet Genome Res. 2014;143:114–124. doi: 10.1159/000365264. [DOI] [PubMed] [Google Scholar]
- 22.Bläsing OE, et al. Sugars and circadian regulation make major contributions to the global regulation of diurnal gene expression in Arabidopsis. Plant Cell. 2005;17:3257–3281. doi: 10.1105/tpc.105.035261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hughes ME, Hogenesch JB, Kornacker K. JTK_CYCLE: An efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J Biol Rhythms. 2010;25:372–380. doi: 10.1177/0748730410379711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Merchant SS, et al. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007;318:245–250. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pazour GJ, Agrin N, Leszyk J, Witman GB. Proteomic analysis of a eukaryotic cilium. J Cell Biol. 2005;170:103–113. doi: 10.1083/jcb.200504008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mackinder LCM, et al. A spatial interactome reveals the protein organization of the algal CO2-concentrating mechanism. Cell. 2017;171:133–147.e14. doi: 10.1016/j.cell.2017.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choi HI, Kim JYH, Kwak HS, Sung YJ, Sim SJ. Quantitative analysis of the chemotaxis of a green alga, Chlamydomonas reinhardtii, to bicarbonate using diffusion-based microfluidic device. Biomicrofluidics. 2016;10:014121. doi: 10.1063/1.4942756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Graf A, Schlereth A, Stitt M, Smith AM. Circadian control of carbohydrate availability for growth in Arabidopsis plants at night. Proc Natl Acad Sci USA. 2010;107:9458–9463. doi: 10.1073/pnas.0914299107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sulpice R, et al. Arabidopsis coordinates the diurnal regulation of carbon allocation and growth across a wide range of photoperiods. Mol Plant. 2014;7:137–155. doi: 10.1093/mp/sst127. [DOI] [PubMed] [Google Scholar]
- 30.Hemschemeier A, et al. Hypoxic survival requires a 2-on-2 hemoglobin in a process involving nitric oxide. Proc Natl Acad Sci USA. 2013;110:10854–10859. doi: 10.1073/pnas.1302592110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Atteia A, et al. Pyruvate formate-lyase and a novel route of eukaryotic ATP synthesis in Chlamydomonas mitochondria. J Biol Chem. 2006;281:9909–9918. doi: 10.1074/jbc.M507862200. [DOI] [PubMed] [Google Scholar]
- 32.Hemschemeier A, Jacobs J, Happe T. Biochemical and physiological characterization of the pyruvate formate-lyase Pfl1 of Chlamydomonas reinhardtii, a typically bacterial enzyme in a eukaryotic alga. Eukaryot Cell. 2008;7:518–526. doi: 10.1128/EC.00368-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Lis R, et al. Concerted up-regulation of aldehyde/alcohol dehydrogenase (ADHE) and starch in Chlamydomonas reinhardtii increases survival under dark anoxia. J Biol Chem. 2017;292:2395–2410. doi: 10.1074/jbc.M116.766048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hemschemeier A, et al. Copper response regulator1-dependent and -independent responses of the Chlamydomonas reinhardtii transcriptome to dark anoxia. Plant Cell. 2013;25:3186–3211. doi: 10.1105/tpc.113.115741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Magneschi L, et al. A mutant in the ADH1 gene of Chlamydomonas reinhardtii elicits metabolic restructuring during anaerobiosis. Plant Physiol. 2012;158:1293–1305. doi: 10.1104/pp.111.191569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burgess SJ, et al. Identification of the elusive pyruvate reductase of Chlamydomonas reinhardtii chloroplasts. Plant Cell Physiol. 2016;57:82–94. doi: 10.1093/pcp/pcv167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang W, et al. Critical role of Chlamydomonas reinhardtii ferredoxin-5 in maintaining membrane structure and dark metabolism. Proc Natl Acad Sci USA. 2015;112:14978–14983. doi: 10.1073/pnas.1515240112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sawyer A, Winkler M. Evolution of Chlamydomonas reinhardtii ferredoxins and their interactions with [FeFe]-hydrogenases. Photosynth Res. 2017;134:307–316. doi: 10.1007/s11120-017-0409-4. [DOI] [PubMed] [Google Scholar]
- 39.Peers G, et al. An ancient light-harvesting protein is critical for the regulation of algal photosynthesis. Nature. 2009;462:518–521. doi: 10.1038/nature08587. [DOI] [PubMed] [Google Scholar]
- 40.Tibiletti T, Auroy P, Peltier G, Caffarri S. Chlamydomonas reinhardtii PsbS protein is functional and accumulates rapidly and transiently under high light. Plant Physiol. 2016;171:2717–2730. doi: 10.1104/pp.16.00572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Correa-Galvis V, et al. Photosystem II subunit PsbS is involved in the induction of LHCSR protein-dependent energy dissipation in Chlamydomonas reinhardtii. J Biol Chem. 2016;291:17478–17487. doi: 10.1074/jbc.M116.737312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bonente G, et al. Analysis of LhcSR3, a protein essential for feedback de-excitation in the green alga Chlamydomonas reinhardtii. PLoS Biol. 2011;9:e1000577. doi: 10.1371/journal.pbio.1000577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lemeille S, et al. Analysis of the chloroplast protein kinase Stt7 during state transitions. PLoS Biol. 2009;7:e45. doi: 10.1371/journal.pbio.1000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Allorent G, et al. A dual strategy to cope with high light in Chlamydomonas reinhardtii. Plant Cell. 2013;25:545–557. doi: 10.1105/tpc.112.108274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schneegurt MA, Sherman DM, Nayar S, Sherman LA. Oscillating behavior of carbohydrate granule formation and dinitrogen fixation in the cyanobacterium Cyanothece sp. strain ATCC 51142. J Bacteriol. 1994;176:1586–1597. doi: 10.1128/jb.176.6.1586-1597.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mus F, Dubini A, Seibert M, Posewitz MC, Grossman AR. Anaerobic acclimation in Chlamydomonas reinhardtii: Anoxic gene expression, hydrogenase induction, and metabolic pathways. J Biol Chem. 2007;282:25475–25486. doi: 10.1074/jbc.M701415200. [DOI] [PubMed] [Google Scholar]
- 47.Helliwell KE, Wheeler GL, Leptos KC, Goldstein RE, Smith AG. Insights into the evolution of vitamin B12 auxotrophy from sequenced algal genomes. Mol Biol Evol. 2011;28:2921–2933. doi: 10.1093/molbev/msr124. [DOI] [PubMed] [Google Scholar]
- 48.Aledo JC, del Valle AE. The ATP paradox is the expression of an economizing fuel mechanism. J Biol Chem. 2004;279:55372–55375. doi: 10.1074/jbc.M410479200. [DOI] [PubMed] [Google Scholar]
- 49.Müller K, Lindauer A, Brüderlein M, Schmitt R. Organization and transcription of Volvox histone-encoding genes: Similarities between algal and animal genes. Gene. 1990;93:167–175. doi: 10.1016/0378-1119(90)90221-c. [DOI] [PubMed] [Google Scholar]
- 50.Fabry S, et al. The organization structure and regulatory elements of Chlamydomonas histone genes reveal features linking plant and animal genes. Curr Genet. 1995;28:333–345. doi: 10.1007/BF00326431. [DOI] [PubMed] [Google Scholar]
- 51.Barry KC, Ingolia NT, Vance RE. Global analysis of gene expression reveals mRNA superinduction is required for the inducible immune response to a bacterial pathogen. eLife. 2017;6:e22707. doi: 10.7554/eLife.22707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Piques M, et al. Ribosome and transcript copy numbers, polysome occupancy and enzyme dynamics in Arabidopsis. Mol Syst Biol. 2009;5:314. doi: 10.1038/msb.2009.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kropat J, et al. A revised mineral nutrient supplement increases biomass and growth rate in Chlamydomonas reinhardtii. Plant J. 2011;66:770–780. doi: 10.1111/j.1365-313X.2011.04537.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sueoka N. Mitotic replication of deoxyribonucleic acid in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 1960;46:83–91. doi: 10.1073/pnas.46.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Strenkert D, Schmollinger S, Sommer F, Schulz-Raffelt M, Schroda M. Transcription factor-dependent chromatin remodeling at heat shock and copper-responsive promoters in Chlamydomonas reinhardtii. Plant Cell. 2011;23:2285–2301. doi: 10.1105/tpc.111.085266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gallaher S, Strenkert D. 2019 Transcriptomics analysis of the Chlamydomonas reinhardtii diurnal cycle. Gene Expression Omnibus (GEO). Available at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE112394. Deposited March 27, 2018.
- 57.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Monroe M, Strenkert S. 2018 Systems biology approach of synchronized Chlamydomonas cells. ProteomeXchange Consortium via the PRIDE. Available at https://www.ebi.ac.uk/pride/archive/projects/PXD010794. Deposited August 16, 2018.
- 59.Schmollinger S, et al. Nitrogen-sparing mechanisms in Chlamydomonas affect the transcriptome, the proteome, and photosynthetic metabolism. Plant Cell. 2014;26:1410–1435. doi: 10.1105/tpc.113.122523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kumar D, et al. A bioactive peptide amidating enzyme is required for ciliogenesis. eLife. 2017;6:e25728. doi: 10.7554/eLife.25728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kropat J, et al. A regulator of nutritional copper signaling in Chlamydomonas is an SBP domain protein that recognizes the GTAC core of copper response element. Proc Natl Acad Sci USA. 2005;102:18730–18735. doi: 10.1073/pnas.0507693102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Strenkert D, et al. Genetically programmed changes in photosynthetic cofactor metabolism in copper-deficient Chlamydomonas. J Biol Chem. 2016;291:19118–19131. doi: 10.1074/jbc.M116.717413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Veyel D, Erban A, Fehrle I, Kopka J, Schroda M. Rationales and approaches for studying metabolism in eukaryotic microalgae. Metabolites. 2014;4:184–217. doi: 10.3390/metabo4020184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fiehn O, et al. Metabolite profiling for plant functional genomics. Nat Biotechnol. 2000;18:1157–1161. doi: 10.1038/81137. [DOI] [PubMed] [Google Scholar]
- 65.Block A, et al. Functional modeling identifies paralogous solanesyl-diphosphate synthases that assemble the side chain of plastoquinone-9 in plastids. J Biol Chem. 2013;288:27594–27606. doi: 10.1074/jbc.M113.492769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vizcaíno JA, et al. 2016 update of the PRIDE database and related tools. Nucleic Acids Res. 2016;44:D447–D456. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Transcriptome data were deposited at the National Center for Biotechnology Information’s Gene Expression Omnibus under accession GSE112394 (56).
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (66) partner repository with the dataset identifier PXD010794 and 10.6019/PXD010794 (58).