Abstract
Anopheles gambiae devotes over 2% of its protein coding genes to its 298 structural cuticular proteins (CPs). This paper provides new LC-MS/MS data on two adult structures, proboscises and palps, as well as three larval samples – 4th instar larvae, just their terminal segment, and a preparation enriched in their tracheae. These data were combined with our previously published results of proteins from five other adult structures, whole adults, and two preparations chosen for their relatively clean cuticle, the larval head capsules left behind after ecdysis and the pupal cuticles left behind after adult eclosion. Peptides from 28 CPs were recovered in all adult structures; 24 CPs were identified for the first time, 6 of these were members of the TWDL family. Most newly identified proteins came from the larval sources. Based solely on peptide recovery, from our data and from other investigators, most available on VectorBase, there were only 4 CPs that were restricted to a single adult structure. More were restricted to a single metamorphic stage, 14 in larvae, 0 in pupa and 32 in adults. Expression data from our earlier RT-qPCR studies reduces these numbers. Charting restriction of CPs to stage or structure is a step forward in establishing their specific roles.
Keywords: proboscis, palp, tracheae, larvae, LC-MS/MS
Graphical Abstract
1. Introduction
The cuticle of arthropods is an essential structure because it shapes and covers the morphological features that enable the breadth of arthropod habitats and lifestyles. In addition to its role as exoskeleton, it serves as the barrier between the internal organs and the environment, not only in terms of physical factors but also pathogens, parasites and predators. There is increasing evidence from analyses of cuticular protein (CP) transcripts and measurement of cuticle thickness, that alterations of the cuticle play a role in insecticide resistance (reviewed in Balabanidou et al., 2018). It is the composition of the structural CPs that interact with chitin that underlies the physical characteristics of individual regions of the exoskeleton and are so numerous as to constitute a significant fraction of the protein coding genes, over 2% in Anopheles gambiae (Willis, 2010; Willis et al., 2012). One possible explanation for the multiplicity of CP genes would be that individual proteins serve specific functions in a particular structure. An earlier study (Zhou et al., 2016) employed LC-MS/MS to examine the CPs of five structures from 5–6 day old adults (Johnston’s organ, the rest of the antenna, eye lens, legs and wings) and identified 132 CPs. Of these, 11 CPs restricted to only one of these structures and 43 were present in all. We also examined the total proteome of 5–6 day old adult An. gambiae in a study designed to probe the relative solubility of different CPs (Zhou et al., 2017). We have now examined two more adult structures, proboscises and palps, separating males and females, and have added a preparation enriched in larval tracheae as well as intact 4th instar larvae and justthe terminal segment of these larvae bearing spiracles, dorsal tufts, ventral brushes and tracheal gills.
An early LC-MS/MS study from our laboratory on An. gambiae allowed us to define a group of authentic (in contrast to “putative”) cuticular proteins because they were recovered from relatively clean cuticle preparations, cast larval head capsules and pupal cuticles left behind following eclosion to the adult (He et al., 2007). These preparations also contained muscle proteins and some enzymes associated with cuticle, but the majority of the peptides belonged to proteins that had characteristics of previously identified CP families (Andersen et al., 1995; Togawa et al., 2007) or a group of new families characterized by their low complexity (Cornman and Willis, 2009).
One complication of working with the CPs, especially those of mosquitoes, is that there are many proteins that have almost identical sequences yet are clearly coded for by distinct genes. We have named sets of such genes “sequence clusters” (Cornman et al., 2008; Cornman and Willis, 2008, 2009). There are 8 such clusters for the CPR family, two for the CPLCG family, one cluster, interspersed among the CPLCGs, encompassing all 9 of the CPLCWs, another encompassing 6/7 of the CPFLs, and one with 14 members in the CPLCP family. Their presence means that for many proteins we were unable to identify any unique peptide and had to settle for using peptides shared among them.
The data from all of these studies are shown in Table 1 and are summarized in Table 2. The current study increased the number of CPs to 298 with the addition of 16 proteins not previously classified as CPs. A combination of our data with data on VectorBase (https://www.vectorbase.org/), in Mastrobuoni et al. (2013), and in Masson et al. (2018, Supplementary data 3), reduced the number of CPs that appear to be restricted to a single adult structure to 4. The study of Masson et al. was especially useful as they carried out proteomics analyses on antennae (minus Johnston’s organ), legs and wings of An. gambiae to verify the efficacy of a new proteomics method that uses very small sample sizes and simplified preparatory steps. They published a detailed comparison of their results to those from Zhou et al. (2016) and were careful to distinguish between recovery of unique and shared peptides.
TABLE 1.
Cuticular proteins identified in different structures
| Updated data from references below. | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Source of data | This paper | He et al. 2007 | Zhou et al. 2016 | 2017 | ||||||||||
| Protein name | seq cluster | Prob | Palp | Larva - terminal seg | Larva - whole 4th | Larval tracheae | Larval head capsules | Cast pupal cuticles | Antenna with JO* | Eye lens | Leg | Wing | whole adults | Up-to-date summary |
| CPR1 | 2RA | US | US | US | S | US | US | US | US | |||||
| CPR2/4 | S | S | U | US | S | S | S | S | US | |||||
| CPR3 | US | US | US | S | US | US | S | S | US | |||||
| CPR5 | S | S | S | S | S | S | S | S | S | |||||
| CPR6 | US | US | US | US | S | S | S | S | US | |||||
| CPR7 | no | |||||||||||||
| CPR8 | [fUmU1] | U | U | U | ||||||||||
| CPR9-PA-PB | bU | mU | U | U | U | U | U | U | U | U | ||||
| CPR10 | U | U | U | U | U | |||||||||
| CPR11 | U | U | U1 | U | U | |||||||||
| CPR12/13 | U | U | U | US | S | US | U | US | ||||||
| CPR14 | U1 | U | U | |||||||||||
| CPR15 | bU | mU1 | U | U | US | S | US | US | U | U | U | US | ||
| CPR16 | bU | bU | U | U | U1 | U | U | U | U | U | U | U | ||
| CPR17 | 2LA | no | ||||||||||||
| CPR18 | no | |||||||||||||
| CPR19 | no | |||||||||||||
| CPR20 | no | |||||||||||||
| CPR21 | fU1 | U | U | U1 | U | U | U | |||||||
| CPR22 | U | U | U1 | V&W | U | |||||||||
| CPR23 | fU1 | U | U | U1 | U | U | U | U | U | |||||
| CPR24 | [U] | U | U | |||||||||||
| CPR25 | [U] | U1 | U | |||||||||||
| CPR26 | U | U | U | U | U | |||||||||
| CPR27 | no | |||||||||||||
| CPR28 | no | |||||||||||||
| CPR29 | no | |||||||||||||
| CPR30 | bU | U | U | {U} | U | U | ||||||||
| CPR31 | U | U | ||||||||||||
| CPR32 | no | |||||||||||||
| CPR33 | no | |||||||||||||
| CPR34 | 2LC | [S] | {S} | S | ||||||||||
| CPR35 | S | S | S | |||||||||||
| CPR36 | [S] | {S} | S | |||||||||||
| CPR37 | {S} | no | ||||||||||||
| CPR38 | {U} | no | ||||||||||||
| CPR39 | [S] | S | ||||||||||||
| CPR40/43/45/46 | S | S | S | |||||||||||
| CPR41 | S | S | S | |||||||||||
| CPR42 | S | S | S | |||||||||||
| CPR44 | S | S | S | |||||||||||
| CPR47 | 2LB | {U} | no | |||||||||||
| CPR48 | S | S | S | |||||||||||
| CPR49 | S | S | S | |||||||||||
| CPR50 | S | S | S | |||||||||||
| CPR51 | S | S | S | |||||||||||
| CPR52 | S | S | S | |||||||||||
| CPR53 | S | S | S | |||||||||||
| CPR54 | S | S | S | |||||||||||
| CPR55 | no | |||||||||||||
| CPR56 | no | |||||||||||||
| CPR57 | no | |||||||||||||
| CPR58 | fU | fUmU1 | US | U1S | US | S | US | US | US | US | U | US | ||
| CPR59 | bUS | bUS | US | US | US | US | US | US | US | US | US | U | US | |
| CPR60 | bS | bS | S | S | U1S | S | U1S | US | US | S | S | US | ||
| CPR61 | U | U | V&W | U | ||||||||||
| CPR62 | fU | bU | U | U | U | U | U | {U} | U | U | ||||
| CPR63 | no | |||||||||||||
| CPR64 | U1 | U | ||||||||||||
| CPR65 | 2LC | [S] | {S} | S | ||||||||||
| CPR66 | S | S | S | |||||||||||
| CPR67 | {U} | no | ||||||||||||
| CPR68 | no | |||||||||||||
| CPR69 | fU1mU | bU | U | U | U | U | U | U | ||||||
| CPR70 | bUS | fU1mUbS | US | US | US | US | US | US | US | US | U | US | ||
| CPR71 | no | |||||||||||||
| CPR72 | 2LC | [S] | S | |||||||||||
| CPR73 | U | U | U | |||||||||||
| CPR74 | [U] | [U] | U | |||||||||||
| CPR75 | [U] | U | U | |||||||||||
| CPR76 | U | U | ||||||||||||
| CPR77 | U | U | U | |||||||||||
| CPR78 | [fU] | U | U | |||||||||||
| CPR79 | U1 | U | U | {U} | {U} | U | ||||||||
| CPR80 | [fU] | {U} | U | |||||||||||
| CPR81 | fU | fU | {U} | U | U | |||||||||
| CPR82 | 3RA | S | US | US | U | US | ||||||||
| CPR83 | mS1 | S | S | S | US | U | U | US | ||||||
| CPR84/108 | mU1 | US | US | US | U | US | ||||||||
| CPR85 | no | |||||||||||||
| CPR86 | 3RB | S | S | S | ||||||||||
| CPR87 | S | S | S | |||||||||||
| CPR88 | S | S | S | |||||||||||
| CPR89 | S | S | S | |||||||||||
| CPR90 | S | S | S | |||||||||||
| CPR91 | S | S | S | |||||||||||
| CPR92 | 3RC | S | S | V&W | V&W | S | ||||||||
| CPR93 | S | S | S | |||||||||||
| CPR94 | S | S | S | |||||||||||
| CPR95 | S | S | S | |||||||||||
| CPR96 | S | S | S | |||||||||||
| CPR97 | S | S | S | |||||||||||
| CPR98 | S | US | US | {S} | S | |||||||||
| CPR99 | 3RC | S | S | V&W | V&W | S | ||||||||
| CPR100 | S | S | S | |||||||||||
| CPR101 | no | |||||||||||||
| CPR102 | no | |||||||||||||
| CPR103 | no | |||||||||||||
| CPR104 | no | |||||||||||||
| CPR105 | U1 | U | ||||||||||||
| CPR106 | U | U | U1 | U | U | U | ||||||||
| CPR107 | mS1 | US | S1 | S | S | US | S | US | ||||||
| CPR109 | S | S | S | |||||||||||
| CPR110 | bU | fUmU1 | U | U1 | U | U | U | U | U | U | U | U | ||
| CPR111 | U | U | {U} | U1 | U | |||||||||
| CPR112 | no | |||||||||||||
| CPR113 | bU | fU | U | U | U | {U} | {U} | U | U | |||||
| CPR114 | U | U | U | U | U | U | ||||||||
| CPR115 | 2RB | bS | bS | S | S | {S} | S | S | US | S | US | |||
| CPR116 | bU | U | U | U | U | U | U | U | U | |||||
| CPR117/154 | 2RB | bS | bS | S | S | S | S | S | S | S | S | |||
| CPR118/119/121/158 | bS | bS | S | S | S | S | S | S | S | S | ||||
| CPR120 | fUbS | bS | US | US | S | US | US | US | US | US | ||||
| CPR122 | fUmU1 | bS1 | S | S | US | US | US | US | US | US | ||||
| CPR123 | fU1bS | bS | US | US | US | US | US | US | US | US | ||||
| CPR124 | bU | bU | U | U | U | U | U | U | {U} | U | U | |||
| CPR125 | bU | bU | U | U | U | U | U | U | U | U | U | U | ||
| CPR126 | bU | bU | U | U | U | U | U | U | U | U | U | U | ||
| CPR127 | bU | bU | U | U | U | U | U | U | U | U | U | |||
| CPR128 | [mU] | {U} | {U} | U | U | |||||||||
| CPR129 | U | U | ||||||||||||
| CPR130 | bU | bU | U | U | U | U | U | U | U | U | U | U | ||
| CPR131 | U | U | U | U | U | U | U | |||||||
| CPR132 | U | U | U | U | ||||||||||
| CPR133/153 | [U] | [U] | U | V&W | U | U | ||||||||
| CPR134 | [U] | [U] | [U] | U | ||||||||||
| CPR135 | bU | bU | U | U | U | U | U | U | U | U | U | U | U | |
| CPR136 | 2LB | S | S | S | ||||||||||
| CPR137 | no | |||||||||||||
| CPR138 | U | {U} | U | |||||||||||
| CPR139 | U | U | ||||||||||||
| CPR140 | bU | bU | U | U | U | U | U | U | U | U | U | U | ||
| CPR141 | no | |||||||||||||
| CPR142 | 3RC | S | S | V&W | V&W | S | ||||||||
| CPR143 | [U] | U | U | |||||||||||
| CPR144 | no | |||||||||||||
| CPR145 | 2LC | no | ||||||||||||
| CPR146 | fU1 | U1 | U | U | U | U | U | U | U1 | U | ||||
| CPR147 | bU | U | {U} | {U} | U | U | ||||||||
| CPR148 | 3RB | S | S | S | ||||||||||
| CPR149 | S | S | S | |||||||||||
| CPR150 | S | S | S | {S} | S | |||||||||
| CPR151 | U | U | U | U | U | |||||||||
| CPR152 | U | U | ||||||||||||
| CPR155 | no | |||||||||||||
| CPR156 | 3RB | mU1 | U | |||||||||||
| CPR157 | fU | U | U | {U} | {U} | U | ||||||||
| CPR159 | no | |||||||||||||
| CPR160 | bU | bU | U | U | U | U | U | U | ||||||
| CPR161 | [fUmU1] | {U} | U1 | U | ||||||||||
| CPR162 | bU | bU | U | U | U | U | U | U | U | U | ||||
| CPR163 | U | U | U | U | U | U | ||||||||
| CPR164 | fU | fU | U1 | U | ||||||||||
| CPAP1-A | U | U | [U1] | U | ||||||||||
| CPAP1-B1 | no | |||||||||||||
| CPAP1-B2 | [U] | U1 | U | |||||||||||
| CPAP1-C | no | |||||||||||||
| CPAP1-F | U | U | {U} | U | ||||||||||
| CPAP1-G | U | U | U1 | U | U | U | U | |||||||
| CPAP1-H | no | |||||||||||||
| CPAP1-J | no | |||||||||||||
| CPAP1-K-PA | [US] | US | ||||||||||||
| CPAP1-K-PB | [S] | S | ||||||||||||
| CPAP1-M | [mU] | U | U | U | U | |||||||||
| CPAP1-N | no | |||||||||||||
| CPAP1-O | mU | U | {U} | U | ||||||||||
| CPAP3-A1a | U | U1 | U | U | U | U | U | U | ||||||
| CPAP3-A1b | S | US | {S} | US | ||||||||||
| CPAP3-A1c | S | US | U | {U} | {U} | US | ||||||||
| CPAP3-B | U | U | U | U | ||||||||||
| CPAP3-C-PA-PC | U | U | U1 | U | U | U | U | U | U | |||||
| CPAP3-D1 | U | U | U | U | U | U | U | U | ||||||
| CPAP3-D2-PA-PB | no | |||||||||||||
| CPAP3-E | U | U | U | [U] | {U} | U | ||||||||
| CPCFC1 | fU | U | U | U | U | U | U | |||||||
| CPF1 | US | US | US | US | S | S | S | US | ||||||
| CPF2 | fU | mU | US | US | US | US | US | US | U | US | ||||
| CPF3 | fU1 | bU | U | U | U | U | U | U | U | U | ||||
| CPF4 | bU | bU | U | U | U | U | U | U | U | U | ||||
| CPFL1 | bU | mU | U | U | U | U | U | U | {U} | U | U | |||
| CPFL2 | a | [fS] | U1S | S | U | U | US | |||||||
| CPFL3 | b | [fS] | S | S | S | S | S | |||||||
| CPFL4/6 | b | [fS] | S | S | S | S | S | |||||||
| CPFL5 | b | [fS] | S | S | S | S | S | |||||||
| CPFL7 | a | [fS] | S | S | S | S | S | |||||||
| CPLCA1 | bS | mUbS | US | US | US | US | US | S | US | {U} | US | US | ||
| CPLCA2 | bS | bUS | US | US | US | US | S | US | S | {U} | S | US | ||
| CPLCA3 | bU | U | U | U | U | U | U | U | ||||||
| CPLCG1 | bU | U | U | U | U | U | U | |||||||
| CPLCG2 | [U] | U | U | |||||||||||
| CPLCG3 | [U] | {S} | {S} | {S} | U | |||||||||
| CPLCG4 | fU1 | fU1mU | U | U | U | U | U | U | U | U | ||||
| CPLCG5 | bU | bU | U | U | U | U | U | U | U | U | ||||
| CPLCG6 | A | S | S | S | S | S | S | |||||||
| CPLCG7 | B | S | S | S | S | S | ||||||||
| CPLCG8 | A | S | S | S | S | S | S | |||||||
| CPLCG9 | A | S | S | S | S | S | S | |||||||
| CPLCG10 | B | S | S | S | S | S | ||||||||
| CPLCG11 | A | S | S | S | S | S | S | |||||||
| CPLCG12 | B | US | US | S | S | US | ||||||||
| CPLCG13 | A | S | S | S | S | S | S | |||||||
| CPLCG14 | U | U | US | US | U | {U} | U | U | U | |||||
| CPLCG15 | mU1 | fU1mU | U | U | US | U | U | U | U | US | ||||
| CPLCG16 | U | U | US | US | US | |||||||||
| CPLCG17 | A | U1S | S | S | S | S | US | |||||||
| CPLCG18 | A | S | S | S | S | S | S | |||||||
| CPLCG19 | A | S | S | S | S | S | S | |||||||
| CPLCG20 | B | S | S | S | S | S | ||||||||
| CPLCG21 | A | US | US | S | S | S | US | |||||||
| CPLCG22 | U | U | U | U | U1 | U | ||||||||
| CPLCG23 | no | |||||||||||||
| CPLCG24 | no | |||||||||||||
| CPLCG25 | A | S | S | S | S | S | S | |||||||
| CPLCG26 | A | US | US | S | S | {S} | US | |||||||
| CPLCG27 | A | US | S | S | S | US | ||||||||
| CPLCG28 | S | S | [U1] | U | ||||||||||
| CPLCG29 | {U} | no | ||||||||||||
| CPLCP1 | S | S | S | S | {U} | S | ||||||||
| CPLCP2 | S | US | S1 | US | ||||||||||
| CPLCP3 | US | US | US | US | {U} | S1 | US | |||||||
| CPLCP4 | no | |||||||||||||
| CPLCP5 | no | |||||||||||||
| CPLCP6 | S | S | S | S | S | S | S | S | S | |||||
| CPLCP7 | {U1S} | {U1S} | S | {S} | {S} | US | ||||||||
| CPLCP8 | US | S | US | US | U | US | US | U | US | |||||
| CPLCP9 | US | S | US | US | S1 | US | ||||||||
| CPLCP10 | bS | US | US | S | US | US | S | S | US | US | ||||
| CPLCP11 | bUS | US | S | U1S | U | US | US | US | US | US | US | |||
| CPLCP12 | fUS | bUS | US | US | US | U | US | US | US | US | US | US | ||
| CPLCP13 | C | S | S | S | S | S | ||||||||
| CPLCP14 | C | S | S | S | S | S | ||||||||
| CPLCP15 | C | S | S | S | S | S | ||||||||
| CPLCP16 | C | S | S | S | S | S | ||||||||
| CPLCP17 | C | S | S | S | S | S | ||||||||
| CPLCP18/19/23 | C | S | S | S | S | S | ||||||||
| CPLCP20 | C | S | S | S | S | S | ||||||||
| CPLCP21 | C | S | S | S | S | S | ||||||||
| CPLCP22 | C | S | S | S | S | S | ||||||||
| CPLCP24 | C | S | S1 | S | S | |||||||||
| CPLCP25 | C | US | US | S | S | US | ||||||||
| CPLCP26 | C | U1S | U1S | S | S | S | S | S | S1 | US | ||||
| CPLCP27 | US | US | US | |||||||||||
| CPLCP28 | no | |||||||||||||
| CPLCW1 | {US} | {US} | S | {U} | {S} | US | ||||||||
| CPLCW2/3 | {S} | {S} | {S} | {S} | S | |||||||||
| CPLCW4 | {S} | {S} | {S} | {S} | S | |||||||||
| CPLCW5 | {S} | {S} | {S} | {U} | S | |||||||||
| CPLCW6//8 | S | S | S | S | ||||||||||
| CPLCW7 | {US} | {US} | {U} | {S} | US | |||||||||
| CPLCW9 | {S} | {S} | {S} | {S} | S | |||||||||
| CPLCX1 | {U1} | U | {U} | U | ||||||||||
| CPLCX2 | bU | bU | U | U | U | U | U | U | {U} | U | U | U | ||
| CPLCX3 | bU | bU | U | U | U | U | U | U | U | U | U | U | U | |
| CPLCX4 | bU | U | U | U | U | |||||||||
| CPLCX5 | fU | U | U | U | U | U | U | U | ||||||
| CPLCX6 | [no] | |||||||||||||
| CPLCX7 | [U] | U | ||||||||||||
| CPLCX8 | [no] | |||||||||||||
| CPLCX9 | [U] | {U} | U | |||||||||||
| CPLCX10 | bU | bU | U1 | U | U | U | U | U | U | {U} | U | U | ||
| CPLCX11 | U | U | ||||||||||||
| CPLCX12 | [no] | |||||||||||||
| CPLCX13 | [U] | [U] | U | |||||||||||
| CPLCX14 | no | |||||||||||||
| CPLCX15 | no | |||||||||||||
| CPLCX16 | no | |||||||||||||
| CPLCX17 | no | |||||||||||||
| CPLCX18 | no | |||||||||||||
| CPLCX19 | U | U1 | U1 | U | U | U | U | U | ||||||
| CPLCX20 | U | U | U | U | U | |||||||||
| CPLCX21 | no | |||||||||||||
| CPLCX22 | [bU] | U | ||||||||||||
| TWDL1 | fU | U | U | U | U | U | U | U | U | U | U | U | ||
| TWDL2 | [fU] | US | US | US | US | |||||||||
| TWDL3 | fS1 | fS | US | US | US | US | ||||||||
| TWDL4 | fS | fS | S | S | S | S | ||||||||
| TWDL5 | US | US | U1S | US | ||||||||||
| TWDL6 | S | S | S | S | ||||||||||
| TWDL7 | US | US | US | US | ||||||||||
| TWDL8 | {S} | {S} | {S} | no | ||||||||||
| TWDL9 | bU | fU1 | U | U | U | U | U | U | {U} | U | U | U | ||
| TWDL10 | {S} | S1 | S | |||||||||||
| TWDL11 | bU | U | U | U | U | U | US | US | US | US | US | |||
| TWDL12 | fU1 | U | U | U | U | U | U | U | {S} | U | U | |||
| CPTC1 | U | U | U | |||||||||||
| CPTC2 | U | U | U | |||||||||||
| CPTC3 | U | U | U | |||||||||||
| CPTC4 | U | U | U | |||||||||||
| CPRC1 | fU1 | fU1 | U1 | [U] | U | |||||||||
| CPRC2 | U1 | U | ||||||||||||
| CPRC3 | U | U | U | |||||||||||
| CPRC4 | mU | U | U | U | U | U | ||||||||
| CPX1 | fU | U | U | U | U | U | U | U | U | U1 | U | |||
| CPX2 | U | U | U | U | U | U | ||||||||
| yellow | fU | bU1 | U | U | U | U | U | U | U | U | U | U | ||
| yellow-e | fUmU1 | U | U | U | U | U | U | U | U | U | U | |||
| yellow-f | [fU] | U | U | U | U | U | ||||||||
| yellow-h | U | U | U | U | U | U | U | U | ||||||
| SUMMARY | 19 no | |||||||||||||
| 35 no | ||||||||||||||
| 24 NEW | ||||||||||||||
U - unique peptide found in protein; U1 - only one peptide detected in only one sample.
S - peptide shared among proteins. S (bolded) - at least one peptide not found in a protein that had a unique peptide.
“m”, “f’ and “b” indicate source (male, female, both) of peptides found in proboscises or palps.
CPR protein groups are designated with color type in names: RR-1, green; RR-2, brown; unclassified, blue. Those scoring below cut-off score in CutProt-FamPred are in italics. Gray highlighting in “Protein Name” column indicates that peptides from that protein were identified in all six adult structures.
Colored highlighting indicates that the protein had not been identified in any other structure or was restricted to larvae. Square brackets [ ] in colored box indicate that VectorBase or Mastrobuoni et al., 2013 had peptides for this protein in other stages/structures.
Curly brackets { } surrounding U or S in boxes for antennae, legs or wings indicate that Masson et al., 2018 found peptide(s) for this structure when we had not. These brackets in larval columns indicate that the protein was not restricted to larvae based on Masson et al.
Antenna includes Johnston’s organ (JO). They were analyzed separately in Zhou et al. 2016 and JO not analyzed in Masson et al., 2018.
Data for whole adults from Zhou et al., 2017 S6_Table.
Gray highlighting in the” Up-to-date Summary” column indicates the 24 proteins identified by us for the first time in this study.
Gray highlighting in cells in the “whole adults” column indicates that the protein was identified only in adults in this and other studies.
Bolded “no” in Up-to-date summary column indicates that protein had no peptides in Mastrobuoni et al., 2013 or Masson et al., 2018 and/or on VectorBase as of5/16/18.
V&W indicates that even though no peptides were detected, antibodies raised against a peptide in that protein or sequence cluster were found in that structure by EM immunolocalization (Vannini and Willis, 2016).
A no in square brackets, means that trypsin peptides were unlikely. See Supplementary File 3.
AGAP numbers and complete amino acid sequences are in Supplementary File 3.
TABLE 2.
Summary of distribution of CPs, organized by families, in different preparations
| CPFAMILY | #UNIQUE PROTEINS (GENES) |
PROB | PALP | LTERM SEG |
4TII INSTAR LARVA |
L TRACHEA enriched |
L HEAD CAP |
CAST PUPAL CUT |
WHOLE ANTENNA |
EYE LENS |
LEG | WING | INTACT ADULTS |
PROTEINS NOT VERIFIED BYus |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CPR | ||||||||||||||
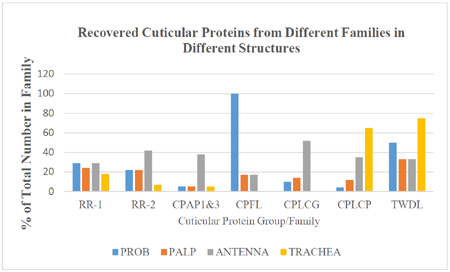
| RR-1 | 45(47) | 13 | 11 | 20 | 19 | 8 | 12 | 11 | 13 | 17 | 8 | 6 | 20 | 10 22.2% |
| RR-2 | 104 (113) | 23 | 23 | 24 | 19 | 7 | 66 | 48 | 44 | 30 | 23 | 22 | 35 | 22 21.2% |
| RR-UNCL | 4 (4) | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 2 | 3 | 2 | 2 | 2 | 0 |
| CPAP1 | 13(12) | 1 | 1 | 2 | 2 | 0 | 2 | 1 | 2 | 4 | 1 | 2 | 3 | 5 38.5% |
| CPAP3 | 8 (8) | 0 | 0 | 2 | 4 | 1 | 4 | 2 | 6 | 6 | 3 | 4 | 2 | 1 12.5% |
| CPCFC | 1 (1) | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 |
| CPF | 4 (4) | 3 | 3 | 2 | 2 | 0 | 4 | 4 | 4 | 2 | 2 | 4 | 4 | 0 |
| CPFL | 6 (7) | 6 | 1 | 6 | 6 | 0 | 6 | 6 | 1 | 1 | 0 | 0 | 1 | 0 |
| CPLCA | 3 (3) | 2 | 3 | 3 | 2 | 0 | 2 | 2 | 3 | 3 | 3 | 1 | 3 | 0 |
| CPLCG | 29 (29) | 3 | 4 | 22 | 22 | 0 | 21 | 21 | 15 | 5 | 4 | 5 | 7 | 3 10.3% |
| CPLCP | 26 (28) | 1 | 3 | 22 | 22 | 17 | 16 | 3 | 9 | 8 | 6 | 5 | 8 | 3 11.5% |
| CPLCW | 7 (9) | 0 | 0 | 7 | 7 | 7? | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| CPLCX | 22 (22) | 6 | 3 | 8 | 7 | 8 | 3 | 4 | 4 | 6 | 3 | 4 | 6 | 9 40.9% |
| TWDL | 12(12) | 6 | 4 | 10 | 10 | 9 | 4 | 3 | 4 | 3 | 3 | 3 | 5 | 1 8.3% |
| CPTC | 4 (4) | 0 | 0 | 0 | 0 | 0 | 4 | 4 | 0 | 0 | 0 | 0 | 0 | 0 |
| CPRC | 4 (4) | 1 | 2 | 1 | 3 | 0 | 0 | 0 | 1 | 0 | 1 | 2 | 1 | 0 |
| CPX | 2 (2) | 1 | 0 | 1 | 2 | 2 | 0 | 1 | 1 | 1 | 2 | 2 | 2 | 0 |
| YELLOW | 4 (4) | 1 | 3 | 3 | 3 | 1 | 4 | 4 | 3 | 3 | 2 | 3 | 3 | 0 |
| Totals | 298 (313) | 68 | 63 | 134 | 131 | 53 or up to 60 | 156 | 114 | 113 | 93 | 64 | 66 | 103 | 54 18.1% |
| Data Source | This paper | He et al., 2007 | Zhou et al., 2016 | Zhou et al., 2017 | ||||||||||
See Table 1 for complete data for individual proteins. Data for CPs enriched in a limited number of preparations are in bold
For our recent studies, we analyzed the data with a program ProteoIQ® (http://www.premierbiosoft.com/protein_quantification_software/index.html), which allowed us to obtain relative abundance of the recovered proteins quantitatively.
2. Materials and methods
2.1. Mosquito rearing, tissue collection, CP nomenclature
An. gambiae (G3 strain) were maintained in the insectary at the University of Georgia Entomology Department. Newly hatched first instar larvae were kept in an incubator at 27 °C at 12 h L:D and fed with ground Koi food Staple Diet (Foster and Smith Aquatics, Rhinelander, WI USA). Adults had access to water and an 8% (wt/vol) fructose solution. Fourth-instar larvae and adults, 5–6 days after eclosion, were collected and frozen at −80 °C until use.
To collect tracheal tissues from the larvae, we first cut off and retained the terminal abdominal segment, then used #5 fine forceps to remove the trachea from inside of the body. Proboscis and maxillary palp were cut from the heads of female and male adult mosquitoes using micro dissecting spring scissors (Roboz). Dissected structures were placed in PBS in 1.5 mL Kontes centrifuge tubes immediately after removal and frozen until enough were produced for further processing. For each of the two biological replicates, trachea and the terminal abdominal segment were collected from about 1050 and 550 larvae, respectively; 390 females and 400 males were used to collect both proboscises and maxillary palps. Each replicate was dissected by a different individual. For the two biological replicates, 210 and 240 frozen larvae were used as the starting material for intact 4th instar larvae.
We used the Web Site CutProtFam-Pred (http://aias.biol.uoa.gr/CutProtFam-Pred/home.php; Ioannidou et al., 2014) to aid in classifying CPs.
2.2. Sample preparation
PBS was removed from the tube containing the dissected structures following centrifugation. Proboscises, maxillary palps and tracheae were resuspended in 100–200 μL of 1% SDS containing 50 mM dithiothreitol (DTT) and homogenized with Kontes pestles. Each larval sample (whole and terminal segments) was homogenized in 450 μL of the same solution. The tubes were placed in a boiling water bath for 20 min for protein extraction. The tubes were centrifuged at the maximum speed for 5 min, and the supernatants were transferred to new tubes. The larvae, partial and whole, were extracted the same way 3 more times in 300 μL of the same solution. Final pellets were washed 3 times in 200–400 μL of ddH2O to remove the SDS. The extracts (second extract for the larval samples) and the final pellets were used for LC-MS/MS analysis.
An approximation of the protein concentration in the extracts was obtained with a NanoDrop N-1000 (Thermo Scientific, USA) using the protein A280 method. SDS extracts were resolved in NuPAGE 4–12% Bis-Tris Gels (NP0323BOX, Invitrogen). Photographs of gels showing distribution of proteins after a 45 min run are in Supplementary File 1. For LC-MS/MS analysis, the same sample was loaded onto 3 adjacent lanes. The gels were run at a constant voltage of 100 volts for about 20 min and stained with Coomassie (Quick Coomassie Stain, Generon, Ltd, UK). The entire area containing the proteins was cut into small pieces (~ 1 mm2). Further processing was the same as the procedure we used previously (Zhou et al., 2016). In brief, this involved destaining with water, rinsing with 50% acetonitrile in water, followed by 25 mM NH4HCO3 and then acetonitrile (ACN). Each time the volume used was sufficient to cover the pieces. Alkylation and reduction were carried out 10 mM DTT/25 mM NH4HCO3 at 65 °C for 1 h, followed by 55 mM IDA (iodoacetamide)/25 mM NH4HCO3 in the dark for 1 h. This was followed by washing with 25 mM NH4HCO3, dehydration with acetonitrile, and incubation with sequencing grade trypsin (Promega) (50:1 w/w protein/trypsin) overnight at 37°C. Extraction of digested peptides from the gel pieces was done with 50% acetonitrile/0.1% formic acid. Pooled supernatants were dried in a Savant SpeedVac (Thermo Scientific) and re-dissolved in 15 μL of 0.1% formic acid/10mM ammonium formate plus 1 μL 80% ACN/0.1%formic acid/10mM ammonium formate solution before LC-MS/MS analysis. The final pellets were washed 5 times with 25 mM ammonium bicarbonate, reduced with 10 mM DTT, alkylated with 55 mM IDA, digested with trypsin (50:1 w/w protein/trypsin) and then filtered through a 30 kDA spin filter (EMB Millipore). Supernatants were dried and re-suspended as above.
2.3. LC-MS/MS analysis
After resuspension, samples were loaded into an Ultimate 3000 LC System paired with an Orbitrap Fusion Tribrid Mass Spectrometer (Thermo Scientific) for LC-MS/MS analysis. Analysis was performed utilizing a nanospray ionization source. Six μL of each sample were injected and separated using a 75-μm × 150-mm Acclaim PepMap RSLC column packed with 2-μm diameter superficially porous particles (Thermo Scientific). The gradient used was 0–55% 80% acetonitrile/0.1% formic acid over 90 min at a flow rate of approximately 300 nL/min. The top 20 most intense ions were fragmented with CID (collision-induced dissociation) and the resulting MS/MS spectra were recorded. Dynamic exclusion was use to exclude precursor ions from the selection process.
2.4. Database searching
The Trans-Proteomic Pipeline (Seattle Proteome Center) was used to convert raw tandem mass spectra. We used Mascot (Matrix Scientific, Boston, MA, USA) to search MS/MS spectra using target and decoy protein databases. Searches were against two databases, An. gambiae peptides (AgamP4.3) from VectorBase and a locally constructed database of 304 putative and previously verified structural cuticular proteins used previously (Zhou et al., 2016, 2017). Decoy databases were created by reversing the protein sequences of corresponding target databases.
The following parameters were utilized in Mascot searching: a fragment tolerance of 0.6 Da;a peptide tolerance of 100 ppm; monoisotopic mass search; a maximum of two missed cleavages by trypsin; a fixed modification of carbamidomethylation of cysteine; and variable modifications of oxidation of methionine and deamidation of asparagine or glutamine. Resulting Mascot files were analyzed using ProteolQ (Premiere Biosoft; http://www.premierbiosoft.com/protein_quantification_software/index.html), where a 5% protein false discovery rate (FDR) was employed.
Numbers of CPs in different structures or stages was based on peptide recovery, although other types of data are discussed.
The permanent AGAP#s for CPs are in Supplementary File 3 along with the complete protein sequence. In some cases, the sequences are not identical to those currently on VectorBase.
All proteomics data are available with the following accession numbers: MasslVE accession #: MSV000082587, ProteomeXchange accession #: PXD010346
3. Results and Discussion
3.1. Overview
Our main purpose in this study was to find whether specific proteins could be identified as restricted to specific structures with particular structural requirements or constraints of size, complexity or shape (e.g., tubular) or to a specific stage of development. Beyond that we wished to supplement and organize the totality of the An. gambiae CP data acquired so far, both by us and other investigators.
3.2. Are any cuticular proteins used exclusively in one anatomical structure or metamorphic stage?
One explanation for the large number of cuticular proteins would be that their properties allow them to be used in a very specialized way. Table 1 reveals that such a relationship is at best rare. Of the 298 distinct CPs, only 4 are possibly restricted to a single adult structure (CPR156 from the palps, CPR139 from the legs and CPR64 and CPR152 from the antennae). Another protein (CPLCX11) was found only in the tracheal preparation. These are shown with plain colored boxes in Table 1.
Data on VectorBase from three other studies (Chaerkady et al., 2011, Champion et al., 2015 and Rund et al., 2013) as well as data from Mastrobuoni et al. (2013) and Masson et al. (2018), eliminated several that we had thought were unique for a single structure (colored boxes with U or S in brackets in Table 1).
One protein, CPR152, does appear to be unique in that by in situ hybridization it was restricted to Johnston’s organ and EM immunolocalization revealed it to be localized in several, but not all, discrete components of that structure (Vannini and Willis, 2016). We also recovered peptides for CPR152 from the rest of the antennae but nowhere else.
Our EM immunolocalization analysis showed how precise the deployment of CPs can be, not only in Johnston’s organ but also in the corneal lens. But, except for CPR152, the CPs we visualized were also found in other structures. Thus it appears that, in the main, different structures are built with different and specific combinations of different CPs rather than structure-restricted CPs, but in a tiny number of cases a specific protein may be required for specific components of a structure.
Stage-specificity is summarized in Table 3. We recovered peptides from 33 CPs (including 7 CPLCWs) that appeared to be restricted to larvae (highlighted in green in Table1). Data from other studies eliminated 19 of these. Those eliminated are shown with brackets around the U or S in the green colored cells on Table 1, leaving only 14 as larval-restricted. Furthermore, our EM immunolocalization studies revealed that antibodies raised against CPR22, CPR61, and CPR133/CPR153 reacted with proteins in adult structures (Vannini and Willis, 2016, 2017), revealing that they are not restricted to larvae, although this information was not used in our numerical summaries. We failed to detect any CPs that might be restricted to pupae.
TABLE 3.
CPs FOUND IN SINGLE AND MULTIPLE STAGES
| LARVAL | PUPAL | ADULT | ALL STAGES | |
|---|---|---|---|---|
| # by stage | 14 | 0 | 32 | 28 |
| Total # recovered | 182 | 114 | 152 | 244 |
Complete data are in Table 1
Recovery of peptides allowed our identification of 32 CPs only in adults. These are indicated with gray shading of their cells in the “whole adults” column in Table 1. Twelve of these had not been recovered in our analysis of whole adults. Their absence is not surprising since 8 had only been recovered by us from a single adult structure, and 3 of the others from only two structures.
This very limited number of possibly stage-specific CPs is further evidence for the employment of CPs irrespective of metamorphic stage, as forecast from earlier studies of electrophoretic banding patterns of CPs from the Cecropia silkmoth, Hyalophora cecropia (Willis 1986).
3.3. Can we be certain that a particular protein is present when the only evidence is based on peptides shared among proteins?
The presence of sequence clusters of CPs with almost identical sequences, and thus few or no unique peptides, complicates the assignment of proteins to a particular structure. This is best illustrated with the CPLCP family for which 28 genes have been annotated coding for 26 distinct proteins. In our previous analysis of adult structures (Zhou et al., 2016), we had found only a single peptide (KVPVYVEK) for 14 CPLCPs. Since that peptide was also present in two CPLCPs along with unique peptides, we did not report the other 12 proteins as present. In the current study, the presence of CPLCPs was more robust. We identified unique peptides for 10 CPLCPs. Multiple shared peptides were identified that belonged to all but 4 of the others (Supplementary Files 2 and 3). Yet of these 12 CPLCPs identified by only shared peptides, all but 4 had peptides shared with proteins that also had unique peptides, limiting our ability to declare them present. But, at least one of the four proteins with only shared peptides (CPLCP13, 14, 17, 20) must have also been present in the three preparations from larvae to account for the one or two peptides shared only among these four proteins. (These peptides are shown with purple type in Supplementary Files 2 and 3).
3.4. Are any cuticular proteins used in all adult structures?
There were 22 proteins plus the 6 in the 2RB sequence cluster from which we recovered peptides in all 6 adult structures (Table 1, highlighted in the column with the Protein name.) All of these proteins, except CPR160, had also been recovered in our current larval or original larval and/or pupal preparations. Thus they represent fundamental building blocks of cuticle. In our previous study of 5 adult structures, we had recovered peptides from 41 CPs in all structures. The absence of about half of these in proboscises or palps accounts for the reduction we now report. Thus this use of common and a subset of shared CPs to build different adult structures is further evidence for cuticular proteins combining in diverse proportions to provide properties required for a particular structure.
3.5. What is special about proboscises and palps?
The addition of two different adult structures to the 5 we had analyzed with LC-MS/MS (Zhou et al., 2016) augmented the data that showed how specific the deposition of CPs can be. Most striking was the recovery of peptides for CPFL2–7 in the female proboscis, proteins that had not been recovered in any of the other adult structures. These proteins are so similar (98–100% in their aligned regions) that we could only identify a single peptide that was unique for one of them. These proteins could be organized in two groups based on recovered peptides: i) CPFL2 and/or CPFL7 and ii) CPFL3 and/or CPFL4 and/or CPFL5 and/or CPFL6. The protein sequences and recovered peptides are shown in Supplementary File 3. Their major contribution to the female proboscis is documented in the quantitative data (discussed in detail in Section 3.8). Here the rank-ordered ProteoIQ data show that CPFL2–7 were the most abundant proteins in the final pellet (Table 4; Supplementary Files 4 and 5) and close to the top from the SDS-soluble gel preparations (Supplementary File 4). While these CPFL proteins were retrieved from only one adult structure, proboscises, peptides from them had also been recovered from the larval samples. With the exception of one of the larval samples, they were not among the top five larval constituents. In addition, they had previously been recovered from larval head capsules and cast pupal cuticles (Table 1).
Table 4.
The five most abundant CPs in each preparation
| Rank order of CPs in final pellets | Rank order of CPs in SDS gels | ||||||||||
| From CP database | |||||||||||
| Rank order | 1 | 2 | 3 | 4 | 5 | 1 | 2 | 3 | 4 | 5 | |
| ♀ proboscis 1 | CPFL2–7 | CPFL1 | CPR113 | CPR59 | CPR 70–43% | CPR16 | CPR70 | CPLCG5 | CPR124 | CPFL1–38% | |
| ♀ proboscis 2 | CPFL2–7 | CPFL1 | CPR113 | CPR59 | CPLCG5–39% | CPR16 | CPLCG5 | CPR124 | CPR70 | CPFL1–40% | |
| ♂ proboscis 1 | CPR113 | TWDL11 | 2RB | CPR59 | CPR16–26% | CPR70 | CPR59 | CPLCG5 | CPR16 | CPFL1–50% | |
| ♂ proboscis 2 | CPR113 | 2RB | CPLCG5 | TWDL9 | CPR15–32% | CPR70 | CPR16 | CPR59 | CPLCX3 | CPR60–40% | |
| ♀ palp 1 | CPR59 | 2RB | CPLCA3 | CPLCG5 | CPR110–67% | CPLCG5 | CPR81 | CPR59 | CPR130 | CPLCA2–40% | |
| ♀ palp 2 | CPLCG5 | CPR59 | CPLCA3 | CPR110 | CPLCX3–43% | CPLCG5 | CPR130 | CPR81 | CPR59 | CPLCA3–46% | |
| ♂ palp 1 | CPLCG5 | CPLCA3 | CPR59 | 2RB | CPLCX3–17% | 2RB | CPLCG5 | CPR59 | CPLCA3 | CPR130–14% | |
| ♂ palp 2 | CPLCG5 | CPLCA3 | CPR135 | CPLCX3 | CPR125–14% | CPR130 | CPLCG5 | CPLCA3 | CPR59 | CPR62–44% | |
| Rank order | 1 | 2 | 3 | 4 | 5 | 1 | 2 | 3 | 4 | 5 | |
| Tracheae 1 | CPR124 | TWDL12 | CPLCP8 | CPLCX5 | TWDL9–10% | CPR124 | CPLCX5 | TWDL3 | CPR116 | CPLCX3–9% | |
| Tracheae 2 | CPR124 | TWDL12 | CPLCP8 | CPLCX5 | CPLCP6–9% | CPR124 | CPLCX5 | TWDL12 | TWDL3 | TWDL4–12% | |
| Terminal segment 1 | CPLCW | CPLCA2 | 2RA | CPLCA1 | CPR59–59% | CPLCW | CPR21 | TWDL3 | 2RA | TWDL4–66% | |
| Terminal segment 2 | CPR124 | 2RA | CPR59 | CPLCW | CPLCA1–64% | TWDL3 | TWDL4 | TWDL2 | TWDL7 | TWDL5–42% | |
| Larva 1 | TWDL2 | 2RA | CPLCW | TWDL3 | TWDL4–68% | CPR23 | CPR21 | CPLCA1 | CPLCA2 | TWDL3–30% | |
| Larva 2 | 2RA | CPLCW | CPR59 | CPFL2–7 | CPF1–36% | CPR23 | 2RA | CPR124 | CPR21 | CPLCA1–31% | |
| Rank order of CPs in final pellets | Rank order of CPs in SDS gels | ||||||||||
| From P4.3 database | |||||||||||
| Rank order | 1 | 2 | 3 | 4 | 5 | 1 | 2 | 3 | 4 | 5 | |
| ♀ proboscis 1 | CPFL2–7 | CPR113 | CPR59 | CPFL1 | CPR70 | CPR16 | CPLCG5 | CPR124 | CPR70 | CPFL1 | |
| ♀ proboscis 2 | CPFL2–7 | CPR113 | CPFL1 | CPR59 | CPLCG5 | CPR16 | CPLCG5 | CPR124 | CPFL1 | CPR70 | |
| ♂ proboscis 1 | CPR113 | TWDL11 | CPR59 | CPR16 | CPLCX10 | CPR70 | CPR59 | CPR16 | CPLCG5 | CPR60 | |
| ♂ proboscis 2 | CPR113 | CPR59 | CPR15 | CPR124 | 2RB | CPR70 | CPR16 | CPR59 | CPR60 | CPR113 | |
| ♀ palp 1 | CPR59 | CPLCG5 | 2RB | CPLCA3 | CPR110 | CPLCG5 | CPR81 | CPR59 | CPR130 | CPLCA2 | |
| ♀ palp 2 | CPLCG5 | CPR59 | CPR110 | CPLCX3 | CPR140 | CPLCG5 | CPR130 | CPR81 | CPLCA3 | CPR59 | |
| ♂ palp 1 | CPLCG5 | CPLCA3 | CPR59 | 2RB | CPLCX3 | 2RB | CPLCG5 | CPR59 | CPLCA3 | CPR130 | |
| ♂ palp 2 | CPLCG5 | CPLCA3 | CPR135 | CPLCX3 | CPR59 | CPR130 | CPLCG5 | CPLCA3 | CPR59 | CPR62 | |
| Rank order | 1 | 2 | 3 | 4 | 5 | 1 | 2 | 3 | 4 | 5 | |
| Tracheae 1 | CPR124 | CPLCX20* | TWDL12 | CPLCP8 | CPX1* | CPR124 | CPLCX20* | CPLCX5 | CPX1* | CPLCX3 | |
| Tracheae 2 | CPR124 | CPLCX20* | TWDL12 | CPLCP8 | CPX1* | CPR124 | CPLCX20* | CPLCX5 | TWDL12 | TWDL3 | |
| Terminal segment 1 | CPLCW | 2RA | CPLCA2 | CPLCA1 | CPR59 | CPLCW | CPR21 | TWDL2 | TWDL3 | 2RA | |
| Terminal segment 2 | CPR124 | 2RA | CPR59 | CPLCW | CPLCA1 | TWDL3 | TWDL4 | CPLCW | CPR21 | CPLCA1 | |
| Larva 1 | TWDL2 | CPLCW | 2RA | TWDL3 | TWDL4 | CPR23 | CPR21 | CPLCA1 | CPLCA2 | TWDL3 | |
| Larva 2 | 2RA | CPLCW | CPR59 | CPFL2–7 | CPLCA1 | CPR23 | 2RA | CPR124 | CPR21 | CPLCA1 | |
Proteins highlighted in gray were found in all 6 adult structures examined. CPR protein names in green are RR-1, those in brown are RR-2. Proteins with name followed by a * are not in the CP database. Percentages are normalized spectral counts relative to the most abundant protein. 1and 2 after specimen name are replicate number.
CPFL1 was also a major constituent in the proboscis and was recovered from two of the other adult preparations. But, it is clearly distinct from the other 5 CPFLs, with only 25–30% amino acid identity, and at a different location on chromosome 3L.
There was another protein, AGAP002399, found abundantly in the proboscis that we had not found in the other preparations. Based on sequence, it appears to be a CP, and we have named it CPLCX22 due to its high coverage by low complexity regions. As presently annotated it lacks a signal peptide. According to VectorBase, numerous peptides were detected in larvae and several adult structures by Chaerkady et al. (2011). Thus it cannot be regarded as restricted to the proboscis.
A comparison of the 80 proteins recovered from proboscises and palps revealed that we had found 17 solely in the proboscises (in addition to CPFL2–7) and 17 solely in palps, but only shared peptides or a single peptide in a single replicate had been recovered from a few of these. In addition to the 5 proteins from CPFL2–7, there were 15 proteins that we had not recovered in the other 4 adult structures (Table 1). Five of these were only found in the proboscises and 5 solely in the palps, and 5 were recovered from both structures. Proteomics data from other studies revealed the presence of all but TWDL3 and TWDL4 in other adult structures. These two TWDLs had been recovered from larvae. Conversely, there were 20 proteins that we had identified with unique peptide(s) in only one of the five structures previously examined (Zhou et al., 2016) and 2 of these were detected in the proboscises, one in the palps, and 2 in both, removing them from the one-structure category.
Although we analyzed proboscises and palps from males and females separately, apart from CPFL2–7 found only in the female proboscis, there was insufficient recovery of other peptides to provide a meaningful comparison between the two sexes (Table 1, Supplementary File 2).
There is a recent paper on the proboscis of another dipteran, (Awuoche et al., 2017). Not surprisingly, since they studied mature adults, transcripts for only two CPs were identified. Both had orthologs present in the proboscises of An. gambiae (GM0Y002920 – CPR130 and CM0Y004676 – CPR157).
3.6. What is special about the CPs of larvae?
We analyzed three different larval samples. The first was the region lying posterior to larval abdominal segment 7 that we have abbreviated TERM SEG (terminal segment). It included the saddle, anal papillae, grids and dorsal and ventral brushes (Figure 65 in Harbach and Knight, 1980). The second consisted of whole larvae from the final (L4) instar. The third preparation was a sample enriched in tracheae, to be discussed in Section 3.7. We were interested in the two larval samples because our earlier work had only included the head capsule left behind after larvae molted, and we anticipated, and indeed found, that there would be many more proteins when more larval structures were examined.
At the time our first analysis was carried out (He et al., 2007), annotation of the An. gambiae genome was at an early stage. We had identified 17 proteins that we called CPLC. Subsequently (Cornman and Willis, 2009), these were separated into 5 distinct low complexity families, the first, TWDL, used a name established in the Drosophila literature, the others were given names that began CPLC, namely CPLCA, CPLCG, CPLCP, and CPLCW. There was also a group of CPs with low complexity that did not have characteristics that we could group into families so we just called them CPLCX. We now recognize 103 low complexity CP genes and 99 distinct proteins (See Table 1). We also had named only 149 CPRs, and now have 164 genes and 153 distinct proteins. Fortunately, peptides from proteins that we had not recognized as CPs in our initial proteomics analysis are displayed on VectorBase with the peptides we had identified, and we have included this information in Table 1.
Many proteins found in the head capsule preparations were present in one or both of the new larval samples and vice versa. Two large sequence clusters 2LC (14 distinct proteins) and 2LB (9 distinct proteins) were identified in the head capsules. There was only a single peptide identified in 8/9 of the 2LB group. Three peptides were identified, but only one per protein in 11/14 of the 2LC group. No peptides from these two sequence clusters were detected in the new larval samples. Furthermore, our data on peptides from adult antennae and data from pupae on VectorBase from Chaerkady et al. (2011) revealed that none of these proteins were restricted to larvae.
Conversely, we found unique peptides for 45 proteins in one or both of the larval samples that we had not detected in the head capsules (Table 1). It could be that some of these proteins had been so heavily cross-linked in the hard head capsule that they were not released with trypsin, or that, as in studies in Tribolium castaneum, there are distinct differences between proteins found in hard and soft cuticles (Dittmer et al., 2012). It is equally likely that some or most of this increase in identified proteins is that the mass spectrometers used in the current study were more powerful.
In the present study, we were interested in learning whether the presence of several distinct anatomical structures on the terminal segment (saddle, anal papillae, grids and dorsal and ventral brushes) could have influenced the peptides recovered in the terminal segment versus the entire 4th instar larva. These new results (Table 1) show that peptides for 13 CPs were recovered in the terminal segments and not the whole larvae, and 10 found in the whole larvae were not found in the terminal segments. All but two (CPR31 and CPR129) had also been identified in adult structures. Those two were found only in the whole larval preparation, suggesting the presence of a specific but unidentified structure.
There were two groups of proteins that we had not found in adult structures that were present in the larval samples. There is a group of CPLCPs (13–26) classified as group C by Cornman and Willis (2009). We recovered shared peptides in both larval preparations and the larval tracheae. The CPLCWs, discussed above, had not been identified in any adult structure nor the cast pupal cuticles, but they were found in the antennae by Masson et al. (2018) with a unique peptide for CPLCW5 and shared peptides for all but CPLCW6/CPLCW8. The only members of the 12-member TWDL family from which we had previously obtained peptides from the adult structures were TWDL1, 9, 11, and 12. These were recovered in the larval samples, along with TWDLs 2–7. Unique peptides were recovered for 8 TWDLs, shared peptides for an additional 2.
3.7. Peptides recovered from larval tracheae
Our first attempt to isolate larval tracheae resulted in recovering many peptides from the CPLCW family. Since we knew from in situ hybridization that transcripts from this family are restricted to the lateral bristles and the grid structures at the end of the abdomen, we assumed that our early samples had included epidermis underlying these structures. We thus repeated the dissections by first removing the posterior region and saving it for separate analysis. Then the remaining abdomen was opened and the mass of tracheae removed. The new procedure yielded just two CPLCW peptides in but one of the two “tracheal” samples (Supplementary File 2). Their abundance was just 1.5% of the top tracheal protein, but they are among the top five in both larvae and their terminal segments (Table 4; Supplementary Files 4, 5). Thus, it is unlikely that CPLCWs are actual components of tracheae.
The number of proteins identified in the tracheae-enriched sample (53 or up to 60 if the CPLCWs are included.) was smaller than from several of the other structures we have studied (Table 2) and 11 proteins were identified by only a single unique peptide. There were four previously named CPs that we found exclusively in tracheae (CPR143, CPLCX7, CPLCX9 and CPLCX11), but they were not particularly abundant (Supplementary Files 4 and 5) and peptides for the first three had been recovered previously in adults by Chaerkady et al. (2011). It is interesting that although all of the structures and larvae we analyzed have tracheae, peptides for these 4 proteins were restricted to tracheae in our analyses.
The CP content of the tracheae was dominated by a small number of proteins. In the final pellet samples, there were only 5 proteins in the top 90% of abundance based on spectral counts. In Table 4 we give the abundance of the fifth ranked CP relative to the first. Thus for the final pellet of the first tracheal sample the spectral counts were 28/282 for TWDL9/CPR124 (Supplementary File 4). We did not do a comparable analysis with the results from the P4.3 database because we were only interested in relative abundance of the CPs, and that database encompassed all proteins, many of which were far more abundant than the CPs (Supplementary File 5). The top CP in both final pellet and SDS-soluble samples was CPR124 that had also been found in many adult structures (Table 1). The second most abundant protein from the tracheae, in both the final pellet and the gel fractions was AGAP005786 (recognized only with the P4.3 database). This protein has long stretches of low complexity, and thus will now be named CPLCX20. It was also found in the terminal segments and whole larvae, but as a far less abundant protein. A Blast search reveals only one match in the entire NCBI database, to a protein from An. sinensis (KFB48567.1). VectorBase, however, displays one2one orthologs in 14 Anopheles species. If it is truly restricted to the genus Anopheles, it has the potential for exploitation in control strategies.
The fifth most abundant protein in tracheae was AGAP008253, a tiny protein that minus the signal peptide has only 78 amino acids. We had found it in the cast pupal cuticles (as ENSANGP00000006757), making it an authentic CP, although we had not recognized it as such (He et al., 2007). It will now be named CPX1. In contrast to CPLCX20, An. gambiae had numerous peptides for this protein displayed on VectorBase, primarily from total head appendages (VB lists them as olfactory). The top 100 results from a Blast search at NCBI brought up sequences with >90% coverage and >55% identity. After a few top hits to mosquitoes, the hits were to a diversity of insects including Hymenoptera, Coleoptera and Lepidoptera as well as other Diptera. The match for D. melanogaster was to CG17127, a mature protein almost the same length, but with no information other than peptides had been recovered from the head. If the Blast search excludes Holometabola, there are 11 hits as good as the first 100 in terms of coverage and identity. But there are no significant hits if Insecta are excluded, a finding that may perhaps be a reflection of its location in the tracheal system. The conservation of this protein and its broad taxonomic distribution indicate that it would be worthy of further study.
In addition to the abundant CPR124, we also recovered peptides from CPR116, a protein that by in situ hybridization is localized in the tracheoles that are associated with the sarcosomes (giant mitochondria) of the adult flight muscles (Supplementary File 7). CPR116 is the closest relative of CPR124 in a neighbor-joining tree of the An. gambiae RR-2 CPs. (Fig. 8 in Cornman et al., 2008).
Larval tracheae have also been isolated from Bombyx mori and their proteins analyzed with LC-MS/MS by Fu et al. (2011). They identified 20 CPs in the tracheae, of these An. gambiae only has good orthologs for 3, CPR162, TWDL1, and CPLCX19. The latter two were recovered from the larval tracheae. Cinege et al. (2017) identified a subset of 4 Nimrod proteins expressed in the developing tracheal system of D. melanogaster as well as the surface cuticle. They classified three proteins from An. gambiae as “orthologs.” We found peptides from two of these (CPLCP11 and 12) in our tracheal samples, but not the third (CPLCP3). An earlier study examined the genes affected in the embryos of the trachealess mutant of D. melanogaster. Five CPs were among them, An. gambiae’s orthologous groups were the 2RA and 2RB sequence clusters, CPAP3-A1a, b, c and CPAP3C. None of these had peptides from our larval tracheal analysis. Hence the cuticular properties that contribute to tracheae appears to be rather species specific, and it will be a challenge to learn how their properties contribute to this essential structure.
3.8. Do the quantitative data on peptide recovery help us to understand the deployment of CPs?
We have made use of the normalized Spectral Counts generated by ProteoIQ to assess the relative contribution of different CPs. Complete Rank Order data are in Supplementary Files 4 and 5. We summarized these data in Table 4 to show the top five in Rank Order for each of the 14 preparations (7 different samples done in duplicate). For this summary, in order to facilitate recognition of different proteins, we treated all members of the 2RA and 2RB sequence clusters, the CPLCW family and CPFL2–7 as single entries, and rank ordered them by the appearance of the first member of each of those groups. Data from the CP and P4.3 databases are summarized separately. In addition to presenting their rank orders, we also noted (gray highlighting) whether the protein had been found in all of the six adult structures we had analyzed. Over half of the most abundant proteins recognized in proboscises and palps were among those found in all 6 adult structures.
The situation was quite different for the larval tracheae and the two larval samples. Of those proteins found in all 6 adult structures, only CPR59 and CPLCX3 were among the most abundant in the samples from larvae. In these larval samples, members of protein families other than CPRs predominated, TWDLs were abundant, as were the conglomerate group of CPLCX. CPFL2–7 that were unique among adult structures for the female proboscis were among the most abundant in one preparation of 4th instar larvae.
The terminal segment has several distinct structures, the saddle, anal papillae, grids and dorsal and ventral brushes. We knew from in situ analyses (illustrated in Neafsey et al., 2015, Figure S19) that transcripts from CPLCW and CPLCG (Group A) were present in these structures. We anticipated that we might find higher ranking peptides for these two groups in the terminal segment than in the whole fourth instar larvae. But CPLCWs were at or near the top in both preparations (Table 4), and group A of the CPLCGs had about the same rankings farther down. Perhaps the fact that another transcript location for these two groups is the lateral bristles on thorax and abdomen compensated for the abundance of specialized structures in the terminal segment built in part from these proteins.
3.9. Are “yellow”proteins structural cuticularproteins?
Our analyses of the six adult structures turned up peptides from four proteins belonging to the “yellow” family. Based on the normalized spectral counts (Zhou et al., 2017), yellow-h (AGAP007549) was the most abundant protein in the antennal preparations and was present in all of the adult structures we first analyzed, but no peptides were detected from proboscises or palps, nor in any of the larval samples. We had previously detected peptides belonging to these four “yellow proteins” in the larval head capsules and cast pupal cuticles (He et al., 2007), and these data were sufficient for us to now call them CPs. The role of yellow family members other than as a component of royal jelly or in pigmentation is complex and unresolved (Ferguson et al., 2011). But data increasingly appear to be pointing to a role in cuticle structure (Xia et al., 2006; Arakane et al., 2010; Noh et al., 2015; Hinaux et al., 2018).
3.10. What did the use of the P4.3 database reveal about the abundance of CPs?
Mapping of recovered peptides to proteins was carried out with two different databases. The first had been constructed with CPs known at the time the study began, the second was the VectorBase P4.3 version of the entire proteome of An. gambiae. Only the P4.3 would reveal proteins that had not previously been recognized as CPs. Indeed, we added 18 proteins to the CP repertoire based on this database.
We naively anticipated that we might find odorant receptors (ORs) or odorant binding proteins (OBPs) or gustatory receptors (GRs) among the recovered proteins in proboscises and palps. VectorBase lists 60 GRs and 76 ORs, and 63 OBPs. Of these, only two OBPs (OBP9 and OBP 26) were recovered, both in the female palp. Data on Vector Base revealed that both had been detected in olfactory tissue by Rund et al. (2013), but peptides for OBP9 were widely distributed in several adult structures as well as pupae and larvae. Further examination of data on VectorBase revealed that even studies originally directed towards these receptors had not found them (Rund et al., 2013; Champion et al., 2015). Mastrobuoni et al. (2013) had more success in identifying these proteins in antennae, and OBP9 was by far the most abundant OBP they recovered. This paucity of identifications merely indicates that these receptors are not abundant and their presence is masked by the abundant proteins, muscle, ribosomal, and histones in addition to CPs. One thing that was evident was that our protocol of first isolating proteins in a boiling SDS buffer enriched the final pellet for CPs. This can be visualized in Supplementary File 5 where the CPs are highlighted in blue, while the others are in orange, green and gold respectively.
3.11. Do transcript expression data correlate well with MS data?
We have published RT-qPCR data for many of the CPs (Cornman and Willis, 2009; Togawa et al., 2007, 2008; Vannini et al., 2015). In addition we have RNA-Seq data for 5–6 da old adult females (Vannini et al., 2014). Supplementary File 6 compiles this expression data. First, we examined the stage-specificity of CPs. All 14 of the proteins we had designated as restricted to the larval stage had by far their highest expression in larval stages. Peptides from proteins that had been recovered in all developmental stages also had good transcript levels at all stages, with the possible exception of CPF3 and 4.
The situation with adult restriction was not as clear. We identified 32 proteins only in adults, using all available proteomics data, and have expression data for 20. Three of these had their highest expression in larvae. Peptides for these had been identified in limited structures, CPR80 in the palps, CPR139 in the leg, CPR147 in the proboscis and antenna. Masson et al. (2018), however, had found peptides from other adult structures (Table 1). It is likely that these genes are contributing to other structures in young larvae, for their transcripts with good expression were in larvae younger than the 4th instar that we had used for proteomics. CPCFC1 that we had detected only in adults, had high transcript levels at the start of L4 and P0. In situ hybridization had also shown abundant transcript in larval structures (Vannini et al., 2015). Some of the other adult-restricted proteins, based on peptide recovery, had high expression at P0, but several adult structures in Anopheles are visible at that time, with differentiation having started prior to pupation.
Finally we asked whether our expression data might help to explain the 35 CPs for which no peptides had been identified in our work or the work of others. These are indicated with bolded NO in Table 1. We have expression data for 24 of these. All but seven had substantial levels of transcripts (>50 and as high as 2400). These seven are indicated with red highlighting in Supplementary File 6. Four genes (blue highlighting) had their highest transcript levels in larvae younger than we examined. But, for the others with good expression, especially the 4 in the 2LA sequence cluster (CPR17–20, highlighted in green), it may be that these proteins become so heavily sclerotized or cross-linked in other ways that no peptides were accessible, although each protein had many possible trypsin peptides (Supplementary File 3). Alternatively, but less likely and less interestingly, we may just be seeing a limitation of MS/MS studies
3.12. What is the current status of the number of CPs in An. gambiae?
These new data allowed us to add 16 new CPs to our list, going from 282 to 298 distinct proteins designated as CPs, with two distinct proteins from each of two genes (CPAP1-K and CPAP3-D2). We now include four members of the “yellow” family, all yielded peptides that had previously been detected in our two, relatively clean cuticle samples, larval head capsules and cast pupal cuticles. We added two additional proteins so prominent in the tracheal fraction that they are deserving of elevation to the status of CPs, now named CPX1 and CPX2. CPX1 had been found in the cast pupal cuticles. Finally, and more problematically we have increased the number of CPLCX proteins from 10 to 22, most based solely on amino acid sequence. All CPs now recognized in An. gambiae with their AGAP designation are in Supplementary File 3.
Table 1 and Table 2 reveal that by combining the four LC-MS/MS studies we have carried out, we had recovered peptides, either unique or shared, for all but 54 of these annotated CPs, 82% of all annotated CPs are thus represented by peptides. As summarized in Table 2, we failed to detect peptides for about 40% of the members of two CP groups, the family CPAP1 and the assorted proteins named CPLCX. In this study, we recovered peptides from 24 CPs that we had not found before. Three of these were only found in proboscises and/or palps, 18 were from larval samples (3 only in tracheae) and 3 in larval samples plus one or both of the two new adult structures. These “new” CPs are indicated with gray highlighting in the “Up-to-date summary” column in Table 1.
Data on VectorBase from three other studies (Chaerkady et al., 2011; Champion et al., 2015; Rund et al., 2013) as well as data from Mastrobuoni et al. (2013) on proteins recovered from the antenna and data from Masson et al. (2018) on legs, wings and antennae reduced the number of CPs without recovered peptides from any structure/region to 35 (bolded NO in Table 1). RT-qPCR information on these unrecovered proteins was discussed in Section 3.11.
3.13. Do other studies support the finding of so many CPs in individual structures?
While the data and papers underlying our conclusions took over a decade to produce, a recent paper on the CPs of the brown planthopper (Nilaparvata lugens) incorporated de novo annotation, verification with PCR, RT-qPCR of transcripts from several stages, proteomics, RNAi to identify essential proteins and EM visualization of cuticle from defective animals. The 140 CPs identified could be assigned to 8 families, 32 of these CP genes were essential for development or egg production (Pan et al., 2018).
More relevant is a recent paper with extensive data relative to CP expression (Sobala and Adler, 2016) that used RNA-Seq coupled with transmission electron microscopy to chart the contribution of individual proteins to specific regions of the developing wings of Drosophila melanogaster. Their paper provides further evidence for the large number of CPs that contribute to a single structure. They found expression of 83 of the 146 genes that had been annotated as CPs in the developing adult wing. By proteomics, we identified 66 CPs in the mature wing of An gambiae, but out of a far larger number of CPs (Table 2).
4. Conclusions
LC-MS/MS has proven to be an useful method for charting the distribution of CPs in different adult structures and different metamorphic stages. It allowed us to find peptides for 82% of all proteins annotated as CPs. It revealed that very few CPs are restricted to a single stage and far fewer to a single adult structure. Most adult structures shared a group of abundant CPs. Further interpretation of these data employed RT-qPCR from published studies. Such data reinforced the classification of some CPs restricted to larvae and others found at all stages. They do not provide an explanation for the proteins that were not recovered. Of special interest are the CPs present in all developmental stages and all adult structures we examined. Many were the most abundant proteins and thus must be making important contributions to the structure of cuticle. Of equal interest are the far rarer cases of proteins restricted to specific structures, developmental stage or (in one case) taxon, for what further studies may reveal about their specialized contributions.
Supplementary Material
Supplementary File 1 – Photographs of gels
Supplementary File 2 – Recovered peptides organized by protein from all sources
Supplementary File 3 – CP sequences showing recovered peptides and AGAP#s
Supplementary File 4 – Rank order CP database for all samples
Supplementary File 5 – Rank order P4.3 database for all samples
Supplementary File 6 – RT-qPCR data from previous work– larval restricted, adult restricted, all stages, no peptides detected
Supplementary File 7 – CPR116 figure
HIGHLIGHTS.
Peptides have been identified from 244 of the 298 cuticular proteins (CPs) in Anopheles gambiae.
Most CPs are shared among structures and metamorphic stages; only 4 CPs were restricted to a single adult structure.
Twenty-eight CPs were in all adult structures; only in a single stage were 32 CPs in adults, 14 in larvae, none in pupae.
Five CPFL family members were abundant in the female proboscis, absent in other adult structures, but abundant in larvae.
ACKNOWLEDGEMENTS
We thank John S. Willis for suggesting that the proboscis might be a special case worth analyzing and for comments on the MS. We are grateful to Laura Vannini for help with the dissections and to Mark R. Brown and Anne Elliot for maintaining the mosquito facility from which the mosquitoes were obtained. We are also grateful to Parastoo Azadi who provided access to the Orbitrap Fusion in the UGA Analytical Services and Training Laboratory and to Stephanie Archer for her assistance in running the samples. This work was supported by a grant from the U.S. National Institutes of Health R01AI055624.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Andersen SO, Hojrup P, Roepstorff P, 1995. Insect cuticular proteins. Insect Biochem. Mol. Biol 25, 153–76. [DOI] [PubMed] [Google Scholar]
- Arakane Y, Dittmer NT, Tomoyasu Y, Kramer KJ, Muthukrishnan S, Beeman R, Kanost MR, 2010. Identification, mRNA expression and functional analysis of several yellow family genes in Tribolium castaneum. Insect Biochem. Mol. Biol 40, 259–266. [DOI] [PubMed] [Google Scholar]
- Awuoche EO, Weiss BL, Vigneron A, Aksoy E, Nyambega B, Attardo GM, Wu Y, O’Neill M, Murilla G, Aksoy S, 2017. Molecular characterization of tsetse’s proboscis and its response to Trypanosoma congolense infection. PLoS Negl. Trop. Dis 11, e0006057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balabanidou V, Grigoraki L, Vontas J, 2018. Insect cuticle: a critical determinant of insecticide resistance. Curr. Opin. Insect Sci 27, 68–74 [DOI] [PubMed] [Google Scholar]
- Chaerkady R, Kelkar DS, Muthusamy B, Kandasamy K, Dwivedi SB, Sahasrabuddhe NA, Kim MS, Renuse S, Pinto SM, Sharma R, Pawar H, Sekhar NR, Mohanty AK, Getnet D, Yang Y, Zhong J, Dash AP, MacCallum RM, Delanghe B, Mlambo G, Kumar A, Keshava Prasad TS, Okulate M, Kumar N, Pandey A, 2011. A proteogenomic analysis of Anopheles gambiae using high-resolution Fourier transform mass spectrometry. Genome Res 21, 1872–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champion MM, Sheppard AD, Rund SSC, Freed SA, O’Tousa JE, Duffield GE, 2015. Qualitative and quantitative proteomics Methods for the Analysis of the Anopheles gambiae mosquito proteome, in: Raman C, Goldsmith MR, Agunbiade TA (Eds.), Short Views on Insect Genomics and Proteomics Insect Proteomics Springer, pp. 37–62. [Google Scholar]
- Cinege G, Zsamboki J, Vidal-Quadras M, Uv A, Csordas G, Honti V, Gabor E, Hegedus Z, Varga GIB, Kovacs AL, Juhasz G, Williams MJ, Ando I, Kurucz E, 2017. Genes encoding cuticular proteins are components of the Nimrod gene cluster in Drosophila. Insect Biochem. Mol. Biol 87, 45–54. [DOI] [PubMed] [Google Scholar]
- Cornman RS, Togawa T, Dunn WA, He N, Emmons AC, Willis JH, 2008. Annotation and analysis of a large cuticular protein family with the R&R Consensus in Anopheles gambiae. BMC Genomics 9, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornman RS, Willis JH, 2008. Extensive gene amplification and concerted evolution within the CPR family of cuticular proteins in mosquitoes. Insect Biochem. Mol. Biol 38, 661–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornman RS, Willis JH, 2009. Annotation and analysis of low-complexity protein families of Anopheles gambiae that are associated with cuticle. Insect Mol. Biol 18, 607–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmer NT, Hiromasa Y, Tomich JM, Lu N, Beeman RW, Kramer KJ, Kanost MR, 2012. Proteomic and transcriptomic analyses of rigid and membranous cuticles and epidermis from the elytra and hindwings of the red flour beetle, Tribolium castaneum. J. Proteome Res 11, 269–278. [DOI] [PubMed] [Google Scholar]
- Ferguson LC, Green J, Surridge A, Jiggins CD, 2011. Evolution of the insect yellow gene family. Mol. Biol. Evol 28, 257–272. [DOI] [PubMed] [Google Scholar]
- Fu Q, Li P, Xu Y, Zhang S, Jia L, Zha X, Xiang Z, He N, 2011. Proteomic analysis of larval integument, trachea and adult scale from the silkworm, Bombyx mori. Proteomics 11, 3761–3967. [DOI] [PubMed] [Google Scholar]
- Harbach RE, Knight KL, 1980. Taxonomist’s glossary of mosquito anatomy, Plexus Publishing, Inc; Marlton, New Jersey. [Google Scholar]
- He N, Botelho JM, McNall RJ, Belozerov V, Dunn WA, Mize T, Orlando R, Willis JH, 2007. Proteomic analysis of cast cuticles from Anopheles gambiae by tandem mass spectrometry. Insect Biochem. Mol. Biol 37, 135–146. [DOI] [PubMed] [Google Scholar]
- Hinaux H, Bachem K, Battistara M, Rossi M, Xin Y, Jaenichen R, Le Poul Y, Arnoult L, Kobler JM, Grunwald Kadow IC, Rodermund L, Prud’homme B, Gompel N, 2018. Revisiting the developmental and cellular role of the pigmentation gene yellow in Drosophila using a tagged allele. Dev. Biol 438, 111–123. [DOI] [PubMed] [Google Scholar]
- Ioannidou ZS, Theodoropoulou MC, Papandreou NC, Willis JH, Hamodrakas SJ, 2014. CutProtFam-Pred: Detection and classification of putative structural cuticular proteins from sequence alone, based on profile Hidden Markov Models. Insect Biochem. Mol. Biol 52, 51–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson V, Arafah K, Voisin S, Bulet P, 2018. Comparative proteomics studies of insect cuticle by tandem mass spectrometry: Application of a novel proteomics approach to the pea aphid cuticular proteins. Proteomics 18(3–4). [DOI] [PubMed] [Google Scholar]
- Mastrobuoni G, Qiao H, Iovinella I, Sagona S, Niccolini A, Boscaro F, Caputo B, Orejuela MR, Della Torre A, Kempa S, Felicioli A, Pelosi P, Moneti G, Dani FR, 2013. A proteomic investigation of soluble olfactory proteins in Anopheles gambiae. PLoS One 8, e75162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neafsey DE, Waterhouse RM, Abai MR, Aganezov SS, Alekseyev MA, et al. , 2015. Mosquito genomics. Highly evolvable malaria vectors: the genomes of 16 Anopheles mosquitoes. Science 347, 1258522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh MY, Kramer KJ, Muthukrishnan S, Beeman RW, Kanost MR, Arakane Y, 2015. Loss of function of the yellow-e gene causes dehydration-induced mortality of adult Tribolium castaneum. Dev Biol 399, 315–324. [DOI] [PubMed] [Google Scholar]
- Pan PL, Ye YX, Lou YH, Lu JB, Cheng C, Shen Y, Moussian B, Zhang CX, 2018. A comprehensive omics analysis and functional survey of cuticular proteins in the brown planthopper. Proc. Natl. Acad. Sci. USA 115, 5175–5180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rund SS, Bonar NA, Champion MM, Ghazi JP, Houk CM, Leming MT, Syed Z, Duffield GE, 2013. Daily rhythms in antennal protein and olfactory sensitivity in the malaria mosquito Anopheles gambiae. Sci. Rep 3, 2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobala LF, Adler PN, 2016. The Gene Expression Program for the Formation of Wing Cuticle in Drosophila. PLoS Genet 12 (5) e 1006100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Togawa T, Dunn WA, Emmons AC, Nagao J, Willis JH, 2008. Developmental expression patterns of cuticular protein genes with the R&R Consensus from Anopheles gambiae. Insect Biochem. Mol. Biol 38, 508–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Togawa T, Dunn WA, Emmons AC, Willis JH, 2007, CPF and CPFL, two related gene families encoding cuticular proteins of Anopheles gambiae and other insects. Insect Biochem. Mol. Biol 37, 675–88. [DOI] [PubMed] [Google Scholar]
- Vannini L, Bowen JH, Reed TW, Willis JH, 2015. The CPCFC cuticular protein family: Anatomical and cuticular locations in Anopheles gambiae and distribution throughout Pancrustacea. Insect Biochem. Mol. Biol 65, 57–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannini L, Dunn WA, Reed TW, Willis JH, 2014. Changes in transcript abundance for cuticular proteins and other genes three hours after a blood meal in Anopheles gambiae. Insect Biochem. Mol. Biol 44, 33–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannini L and Willis JH, 2016. Immunolocalization of cuticular proteins in Johnston’s organ and the corneal lens of Anopheles gambiae. Arthropod Struct. Dev 45, 519–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannini L and Willis JH, 2017. Localization of RR-1 and RR-2 cuticular proteins within the cuticle of Anopheles gambiae. Arthropod Struct. Dev 46, 13–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis JH, 1986. The paradigm of stage-specific gene sets in insect metamorphosis time for revision. Arch. Insect Biochem. Physiol (Suppl 1), 47–58. [Google Scholar]
- Willis JH, 2010. Structural cuticular proteins from arthropods: annotation, nomenclature, and sequence characteristics in the genomics era. Insect Biochem. Mol. Biol 40, 189–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis JH, Papandreou NC, Iconomidou VA, Hamodrakas SJ, 2012. Cuticular Proteins, in: Gilbert LI (Ed.), Insect Molecular Biology and Biochemistry Academic Press, London, Waltham & San Diego, pp. 134–166. [Google Scholar]
- Xia AH, Zhou QX, Yu LL, Li WG, Yi YZ, Zhang YZ, Zhang ZF, 2006. Identification and analysis of YELLOW protein family genes in the silkworm, Bombyx mori. BMC Genomics 7:195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Badgett MJ, Bowen JH, Vannini L, Orlando R, Willis JH, 2016. Distribution of cuticular proteins in different structures of adult Anopheles gambiae. Insect Biochem. Mol. Biol 75, 45–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Badgett MJ Billard L, Bowen JH, Orlando R Willis JH, 2017. Properties of the cuticular proteins of Anopheles gambiae as revealed by serial extraction of adults. PlosOne 12(4):e0175423. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary File 1 – Photographs of gels
Supplementary File 2 – Recovered peptides organized by protein from all sources
Supplementary File 3 – CP sequences showing recovered peptides and AGAP#s
Supplementary File 4 – Rank order CP database for all samples
Supplementary File 5 – Rank order P4.3 database for all samples
Supplementary File 6 – RT-qPCR data from previous work– larval restricted, adult restricted, all stages, no peptides detected
Supplementary File 7 – CPR116 figure


