SUMMARY
The means by which the physicochemical properties of different cellular components together determine bacterial cell shape remain poorly understood. Here, we investigate a programmed cell-shape change during Bacillus subtilis sporulation, when a rod-shaped vegetative cell is transformed to an ovoid spore. Asymmetric cell division generates a bigger mother cell and a smaller, hemispherical forespore. The septum traps the forespore chromo-some, which is translocated to the forespore by SpoIIIE. Simultaneously, forespore size increases as it is reshaped into an ovoid. Using genetics, timelapse microscopy, cryo-electron tomography, and mathematical modeling, we demonstrate that forespore growth relies on membrane synthesis and SpoIIIE-mediated chromosome translocation, but not on peptidoglycan or protein synthesis. Our data suggest that the hydrated nucleoid swells and inflates the forespore, displacing ribosomes to the cell periphery, stretching septal peptidoglycan, and reshaping the forespore. Our results illustrate how simple biophysical interactions between core cellular components contribute to cellular morphology.
In Brief
DNA generates the turgor pressure that inflates the forespore in B. subtilis spore development.
Graphical Abstract
INTRODUCTION
Bacterial cells display an amazing variety of cellular morphologies, which are often the defining signatures of different species (Holt et al., 1994; Young, 2006). It is generally accepted that cell shape is determined by the peptidoglycan (PG) cell wall. The molecular mechanisms involved in cell wall homeostasis are starting to be deciphered, leading to the emergence of models for the maintenance of basic shapes (Amir and Nelson, 2012; Bartlett et al., 2017; Cabeen et al., 2009; Nguyen et al., 2015; Pinho et al., 2013; Ursell et al., 2014). However, it is unclear how the cell wall and other cellular components interact to generate the shape of bacterial cells.
Some bacteria modify their shape during specific developmental processes. A paradigmatic example is sporulation in Bacillus subtilis (Errington, 2003; Higgins and Dworkin, 2012; Tan and Ramamurthi, 2014) during which rod shaped cells transform into ovoid spores. The study of cell shape transformations during this process can provide insights into the mechanism of cell shape generation. Sporulation starts with an asymmetrically positioned cell division event (polar septation) that generates two cells of different size and fate (Figure 1A): the smaller forespore becomes a resilient spore, whereas the larger mother cell lyses after contributing to forespore development. After polar septation, the membrane of the mother cell migrates around the fore-spore in a phagocytosis-like process called engulfment until the leading edges of the engulfing membrane meet and fuse, releasing the forespore into the mother cell cytoplasm.
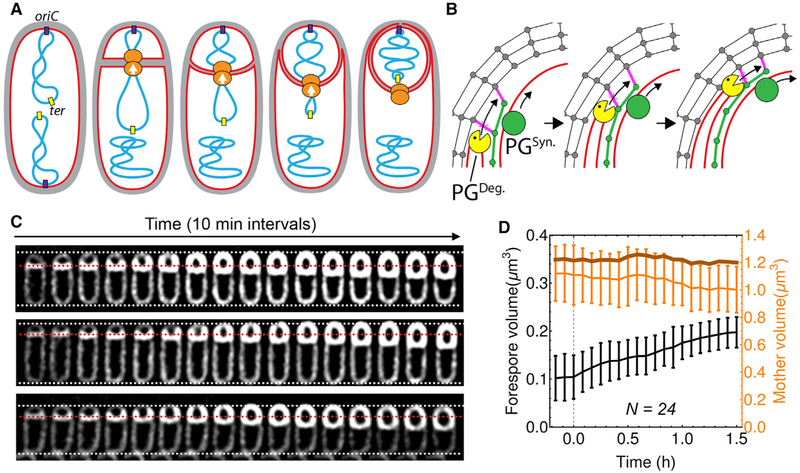
Figure 1. Forespore Growth during Sporulation.
(A) Diagram of polar septation, chromosome translocation, and engulfment showing membranes (red), PG (gray), chromosomes (blue), origin of replication (purple), terminus (yellow), and SpoIIIE (orange), with translocation polarity indicated by arrows.
(B) Model for engulfment membrane migration. New PG (green) is synthesized ahead of the engulfing membrane by forespore PG-biosynthetic machineries (PGSyn., green circle), crosslinked (magenta) to the lateral cell wall (gray), and degraded by SpoIIDMP (yellow Pacman), extending the septal PG and moving the septal junction around the forespore.
(C) Timelapse fluorescence microscopy of three wild-type sporangia throughout engulfment showing FM4–64 stained membranes (white). Dotted lines are added as references, with white indicating the total length of the sporangium and red the original position of the septum.
(D) Average forespore volume (black line, left y axis), mother cell volume (light orange line, right y axis) and forespore plus mother cell volume (dark orange line, right y axis) of wild-type sporangia over time. Time traces were aligned so that 0 hr was the onset of septum curving. Error bars represent standard deviation.
See also Figure S1 and S2.
Immediately after polar septation, the forespore is hemispherical (Figure 1A [second cell]). However, during and shortly after engulfment, it is reshaped into an ovoid (Pogliano et al., 1999). We have previously observed that a thin layer of PG persists between the mother cell and the forespore membranes throughout engulfment (Tocheva et al., 2013); this layer likely plays an important role in reshaping the forespore. Our recent data indicate that this layer originates as an extension of the septal PG by coordinated PG synthesis and degradation at the leading edge of the engulfing membrane (Figure 1B; Ojkic et al., 2016). During this process, synthesis is carried out by forespore biosynthetic complexes that move at a position that coincides with the leading edge of the engulfing mother cell membrane and PG degradation by a mother cell protein complex (SpoIIDMP) that localizes to the leading edge of the engulfing membrane. We propose that coordinated PG synthesis and degradation moves the junction between the septal PG and the lateral cell wall around the forespore, mediating membrane migration and extending the septal PG to fully enclose the fore-spore (Figure 1B), from now on referred to as the “extended septal PG.“
Before polar septation, the chromosomes are remodeled into an elongated axial filament (Kay and Warren, 1968; Pogliano et al., 2002; Ryter, 1965), with the origins of replication at the cell poles and the termini at midcell (Figure 1A [leftmost cell]; Webb et al., 1997). As a consequence, when the septum is formed, the origin-proximal part of the forespore chromosome is inside the forespore, and the origin distal ~2/3 of the chromo-some (Wu and Errington, 1994, 1998) remains in the mother cell (Figure 1A). Endospore-forming bacteria have evolved a system to deal with this topological problem: the SpoIIIE membrane protein is recruited to septal midpoint and moves the chromosome from the mother cell to the forespore (Wu and Errington, 1997; Bath et al., 2000; Massey et al., 2006). SpoIIIE assembles two subcomplexes, one in each cell (Yen Shin et al., 2015). Normally, only the mother cell subcomplex is active, transporting the chromosome into the forespore. However, in the absence of the mother cell subcomplex, the forespore subcomplex can transport the chromosome out of the forespore and into the mother cell, indicating that SpoIIIE can function as a reversible motor (Sharp and Pogliano, 2002; Yen Shin et al., 2015). DNA translocation is an energy intensive process that consumes one molecule of ATP per every two base pairs transported (or ~1.5 million ATP molecules total; Liu et al., 2015). The process commences immediately after polar septation, when the forespore is just ~0.1 μm3, packing the chromosome into a cell that is, at the onset of this process, just 1/10 the volume of the mother cell (Figure 1). It is unclear how this tiny cell accommodates the chromosome and its associated water and counterions.
Here, we study how different core cellular components contribute to the final forespore size and morphology. We propose a model for forespore remodeling in which chromosome translocation increases forespore turgor pressure, stretching the extended septal PG so that it obtains the shape dictated by the forespore biosynthetic complexes and accommodating new membrane that is synthesized during engulfment. In the absence of DNA translocation, the membranes surrounding the forespore appear wrinkled and floppy, which might hinder the completion of engulfment by providing a more complicated membrane morphology for the engulfing membranes to traverse. Our results demonstrate that DNA contributes to forespore size and morphology, not only through the information it carries, but also through the physicochemical properties of the DNA molecule itself.
RESULTS
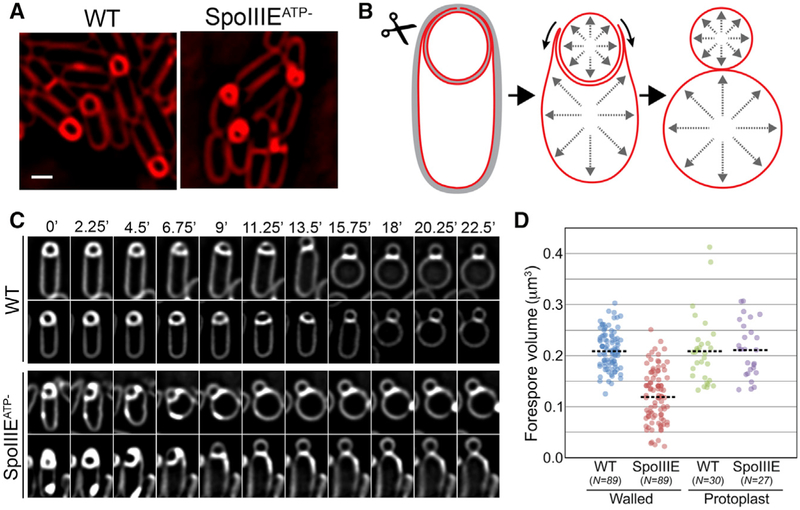
The Forespore Volume Increases in Detriment of the Mother Cell Volume
To visualize the transition in forespore shape from hemispherical to ovoid, we performed timelapse fluorescence microscopy using fluorescent membrane stains compatible with spore formation (Figures 1C, S1, and S2; Pogliano et al., 1999). Immediately after polar septation, the forespore volume is ~0.1 μm3, constituting just ~10% of the total volume of the sporangium (Figures 1C and 1D). However, the forespore starts elongating toward the mother cell, increasing its volume for ~3 hr. Forespore volume doubles by engulfment completion (Figures 1C and 1D) and continues increasing until it plateaus at ~0.3 μm3, roughly triple its initial volume (Figure S1). The mother cell volume decreases by the same amount as the forespore volume increases, therefore maintaining a constant total volume for the sporangium (Figures 1C and 1D). The reduction in mother cell volume contrasts with the ~25% increase in mother cell surface area during engulfment (Ojkic et al., 2016), which is required to allow the engulfing membrane to completely enclose the forespore.
Forespore Growth Requires Chromosome Translocation and Membrane Synthesis, but Not PG Synthesis
In previous work, we have observed that mutants impaired in chromosome translocation typically have small forespores (Becker and Pogliano, 2007; Sharp and Pogliano, 1999, 2002). To test if chromosome translocation was necessary for fore-spore growth, we performed timelapse microscopy of a strain unable to translocate the chromosome into the forespore due to a point mutation in SpoIIIE that abolishes ATPase activity (SpoIIIEATP−; Sharp and Pogliano, 1999). In this mutant, although chromosome translocation is blocked, SpoIIIE still assembles a translocation channel at the septum, maintaining separation of the forespore and mother cell membranes and cytoplasms (Fleming et al., 2010). Timelapse microscopy showed that SpoIIIEATP− forespores showed negligible volume increase compared to the wild-type (Figures 2A–2C, and S2). After membrane migration, an excess of membrane seemed to accumulate around the forespore, and a membrane blob was frequently observed at the mother cell distal tip of the sporangium (Figures 2B and S2). These results indicate that chromosome translocation is critical for forespore growth.
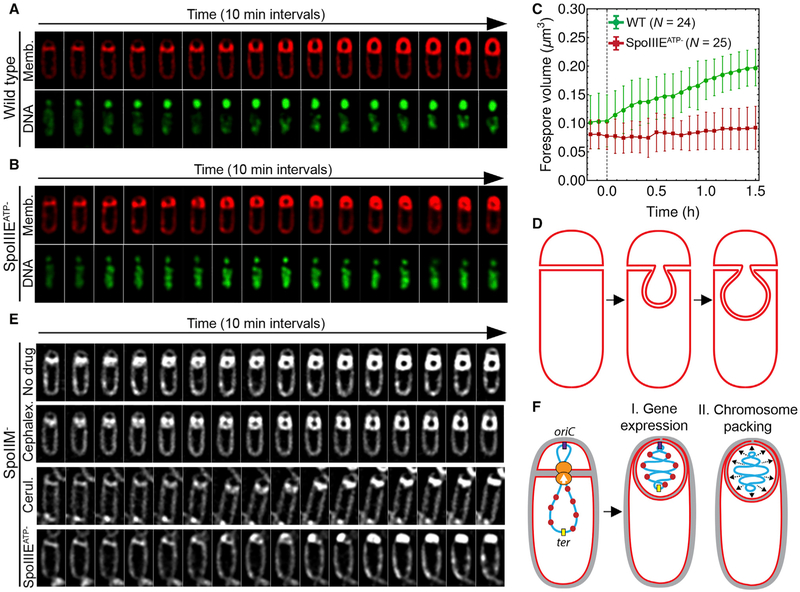
Figure 2. Processes Required for Forespore Growth.
(A and B) Timelapse fluorescence microscopy of wild-type (A) and SpoIIIEATP− (B) sporangia showing FM4–64-stained membranes (red) and SYTOX-green-stained DNA (green).
(C) Change in forespore volume for wild-type (WT, green) and SpoIIIEATP− (red). Time 0 hr was set as in Figure 1. Error bars represent standard deviation.
(D) Diagram of bulge formation in spoIID, spoIIM, and spoIIP mutants.
(E) Timelapse microscopy of spoIIM sporangia without drug, or in the presence of cephalexin (50 μg/mL) or cerulenin (30 μg/mL). The last row is spoIIM spoIIEATP− sporangia. Membranes were stained with FM4–64. See Figure S2 for additional antibiotics.
(F) Alternative models to explain the dependence of forespore growth on chromosome translocation. For simplicity, only the forespore chromosome is shown. Additional details are provided in the main text.
See also Figure S2.
We next tested if forespore growth, like vegetative growth, relied on PG and membrane synthesis. However, these processes are essential for engulfment (Ojkic et al., 2016), complicating the analysis. In order to uncouple forespore growth from engulfment, we used engulfment-defective strains that lacked one protein in the SpoIIDMP complex (Figure 1B). In the absence of any one of these three proteins, membrane migration is blocked, but the forespore continues to grow, forming a bulge into the mother cell cytoplasm (Frandsen and Stragier, 1995; Lopez-Diaz et al., 1986; Smith et al., 1993; Figure 2D). We monitored bulge formation by timelapse microscopy in strains lacking SpoIIM (Figures 2E and S2). Bulges started to form shortly after polar septation and continued growing for ~3 hr. Importantly, chromosome translocation was also required for bulge growth, as indicated by the failure to form bulges in SpoIIIEATP− SpoIIM−mutant (Figures 2E [bottom row] and S2). We then monitored bulge formation in the presence of antibiotics that block PG (cephalexin, fosfomycin, penicillin V, or bacitracin) and membrane (cerulenin) biosynthesis (Figures 2E [second and third rows] and S2) using antibiotic concentrations that affect the formation of polar septa (Ojkic et al., 2016). Bulges formed after treatment with all the PG synthesis inhibitors tested, but they failed to grow when membrane synthesis was inhibited (Figures 2E [second row] and S2). After inhibiting PG synthesis, bulges appeared unstable, collapsing or disintegrating at later time points (Figure S2). This instability would reduce the number of bulges observed in batch cultures, perhaps explaining why we previously concluded that PG synthesis was required for bulge formation (Meyer et al., 2010). Together, these results suggest that forespore growth depends on membrane synthesis and chromosome translocation, but not on PG synthesis.
Hypotheses to Explain the Requirement of Chromosome Translocation for Forespore Growth
While the requirement of membrane synthesis for forespore growth has a straightforward explanation, the role of chromo-some translocation is not clear. One possibility is that growth requires the forespore expression of genes that are not initially trapped in the forespore (Figure 2F [red dots in left and middle cells]). By this hypothesis (hereafter called the gene expression model), the inhibition of forespore growth in the absence of DNA translocation would be due to the failure of genes located in the origin-distal part of the chromosome to enter and be expressed in the forespores of SpoIIIEATP−sporangia. An alternative model—the chromosome packing model (Figure 2F)—posits that forespore growth is a physical consequence of packing the ~4-megabase B. subtilis chromosome into the forespore, which initially comprises just ~10% (Figure 1) of the total volume of the sporangium. Packing this long, negatively charged DNA polymer into the small forespore might generate a significant turgor pressure due to the requirement for additional water molecules and positively charged counterions to neutralize the charge. These models are not necessarily mutually exclusive.
Reverse Chromosome Translocation Results in Forespore Shrinking and Collapse
We next designed experiments to tease apart the relative contribution of forespore gene expression and chromosome packing to forespore growth. First, we employed a genetic strategy to reverse SpoIIIE-mediated chromosome translocation so that DNA is first moved into and then out of the forespore (Figure 3A; Yen Shin et al., 2015). To do so, we tagged SpoIIIE with a modified ssrA* sequence that is recognized by the E. coli adaptor protein SspBEc and delivered to the B. subtilis ClpXP protease for degradation (Griffith and Grossman, 2008). We then degraded SpoIIIE in the mother cell by expressing sspBEc from a mother cell specific promoter (PspoIID). As a consequence, degradation commences slightly after the initiation of chromosome translocation, which gives time for most sporangia to initially translocate the chromosome into the forespore, supporting forespore growth (Figure 3A). However, after mother cell SpoIIIE-ssrA* is degraded, the forespore SpoIIIE subcomplex transports the chromosome out of the forespore. If chromosome packing drives forespore growth, we expect that pumping the chromo-some out of this cell would relieve the turgor pressure, causing it to shrink or collapse (Figure 3A [chromosome packing model]). However, if growth is the consequence of de novo synthesis of specific gene products in the forespore that mediate the cell shape change, then reverse chromosome translocation should have no consequence on forespore shape, since the products would already be present in the forespore as evidenced by the initial growth of this cell (Figure 3A [gene expression model]).
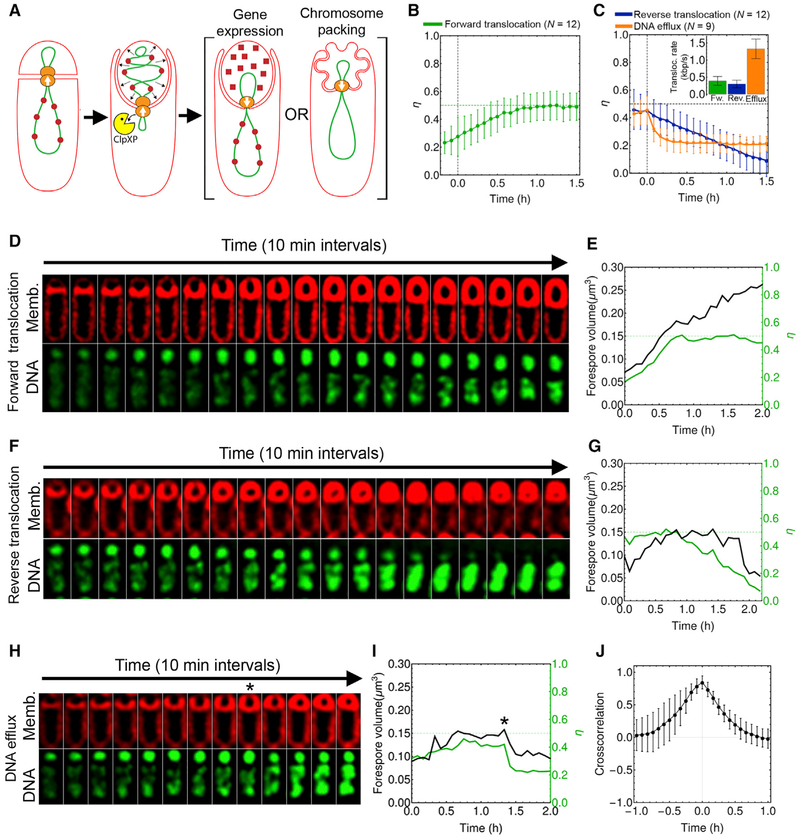
Figure 3. Reverse Chromosome Translocation Produces Forespore Shrinking and Collapse.
(A) Degradation of mother cell SpoIIIE reverses chromosome translocation (Yen Shin et al., 2015), with differing impacts on forespore volume predicted by the gene expression and the chromosome packing hypotheses.
(B) Mean forespore DNA fraction (η) over time during forward chromosome translocation. The green dotted line indicates DNA fraction after chromosome translocation (η= 0.5). Error bars represent standard deviation.
(C) Mean forespore DNA fraction over time during reverse chromosome translocation and DNA efflux. The inset shows translocation rates (absolute value, kb/s per chromosome arm) during forward translocation (Fw.), reverse translocation (Rev.), and DNA efflux. Error bars represent standard deviation.
(D) Timelapse microscopy of a sporangium stained with FM4–64 (red) and SYTOX green (green) without SpoIIIE degradation.
(E) Graphs showing forespore volume (black line, left y axis) and fraction of DNA in the forespore (η, green line, right y axis) of the sporangium shown in (D).
(F–I) Examples of outcomes after SpoIIIE degradation in the mother cell. (F) Slow chromosome translocation out of the forespore. (G) Graph showing forespore volume and forespore DNA fraction for the sporangium in (F). (H) Example of abrupt DNA efflux. The asterisk indicates the onset of reverse chromosome translocation. (I) Graph showing forespore volume and forespore DNA fraction of the sporangia shown in (H).
(J) Cross-correlation between η and forespore volume during abrupt DNA efflux. Error bars represent standard deviation.
See Figure S3 for additional examples.
We monitored chromosome translocation and forespore volume by timelapse microscopy, staining the DNA with SYTOX green and membranes with FM4–64 (Becker and Pogliano, 2007). In the absence of SpoIIIE degradation, the chromosome was translocated from the mother cell to the forespore in every sporangium with a forward translocation rate of 0.4 kb/s per chromosome arm (Figures 3B and 3C [inset]), and translocation was typically completed in less than 1 hr after polar septation (Figures 3B–3E and S3A). As expected, the forespore volume increased continuously as the chromosome was translocated to the forespore, continuing after translocation in a second phase of growth (Figures 3D, 3E, and S3A). When SpoIIIE was degraded in the mother cell, most sporangia initially showed forward chromosome translocation, which supported forespore growth (Figures 3F–3I, S3B, and S3C). After the initial forward translocation, we observed chromosome movement out of the forespore in a significant fraction of the sporangia, suggesting that the mother cell SpoIIIE subcomplex was degraded (Figures 3F–3I). We observed two distinct types of reverse translocation (Figures 3C and 3F–3I). In some sporangia, the chromosome was slowly and completely translocated out of the forespore, with a reverse translocation rate of −0.3 kb/s (Figure 3C [inset])—similar to the rate at which it had initially been translocated into the forespore—as expected for SpoIIIE-mediated reverse chromosome translocation (Figures 3C, 3F, 3G, and S3B). In these cases, the forespores gradually lost volume, becoming misshapen and ultimately collapsing (Figures 3F, 3G, and S3B). In other sporangia, parts of the chromosomes quickly moved out of the forespores (−1.3 kb/s; Figure 3C), with coincident and abrupt losses of forespore volume and no subsequent movements of the remainder of the chromosomes out of the forespores (Figures 3C, 3H, 3I, and S3C). Cross-correlation analysis showed that, during DNA efflux, there was no time delay between chromosome movement out of the forespore and the loss of forespore volume, suggesting a strong coupling between both processes (Figure 3J). This abrupt DNA efflux has not been previously observed. Our prior results indicate that SpoIIIE complexes in both cells are required to assemble a stable paired channel and to maintain daughter cell separation (Yen Shin et al., 2015). It is therefore possible that degradation of mother cell SpoIIIE causes the forespore SpoIIIE channel to disassemble in some sporangia, opening a pore in the septum that allows the immediate redistribution of the chromosome between the two cells in order to minimize the repulsive and hydration forces associated with packing the chromosome into the small forespore.
Thus, the loss of the forespore chromosome correlates with decreased fore-spore volume over two distinct time-scales, consistent with the hypothesis that DNA generates pressure within the forespore that is required to maintain its size and shape.
Sustained Translation in the Forespore Is Not Required for Growth
The above results show that chromosome packing in the fore-spore is critical for growth and suggest that expression of genes encoded in the region of the chromosome initially trapped in the mother cell might be dispensable for forespore growth. To further evaluate the gene expression model, we tested if sustained protein synthesis was required in the forespore to support growth. We used the cell-specific protein degradation system (Yen Shin et al., 2015) to specifically block translation in the forespore after polar septation (Figure 4A). To this end, we constructed a strain in which the sole isoleucine-tRNA synthetase, IleS, was tagged with ssrA*. The IleS-ssrA* fusion supports growth with a doubling time and cellular morphology identical to the parent strain, and production of SspBEc from a xylose-inducible promoter causes a rapid cessation of growth (Figures S4 and S4B; Lamsa et al., 2016). To block translation in the forespore, we expressed sspBEc from a forespore-specific promoter (PsspE(2G)). Degradation of IleS-ssrA* in the forespore dramatically reduced spore production, suggesting that the protein is efficiently degraded (Figure S4C).
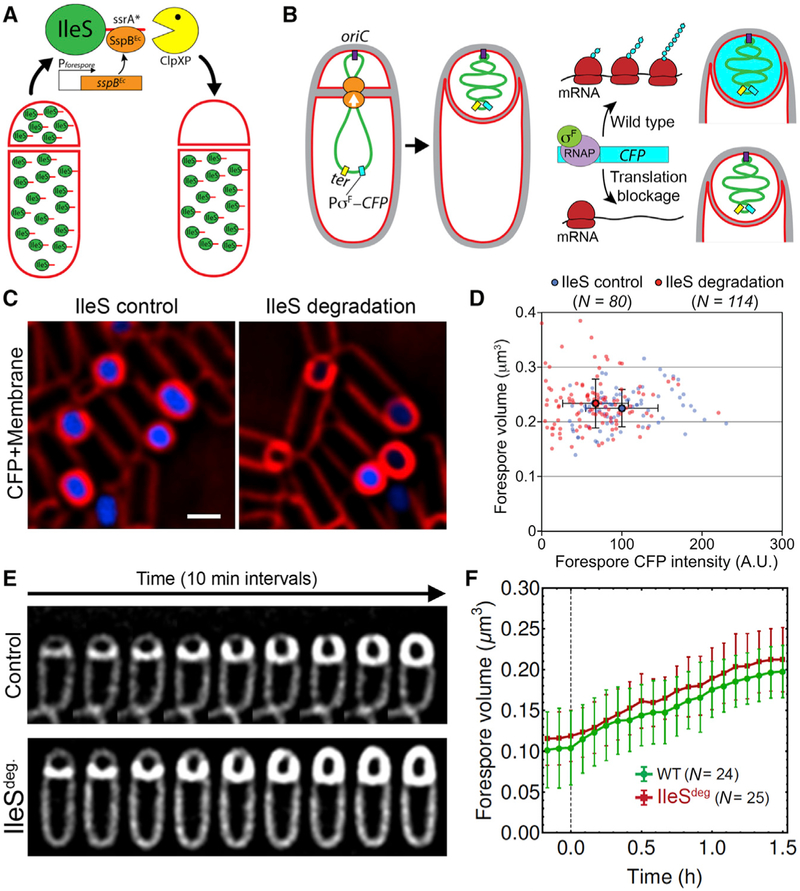
Figure 4. Sustained Protein Synthesis in the Forespore Is Not Required for Growth.
(A and B) Strategies to block translation in the forespore (A) and to determine the degree to which IleS-ssrA* degradation reduces forespore translation (B).
(C) Fluorescence microscopy of sporangia expressing PspoIIQ–CFP without IleS degradation (left) or with IleS degradation in the forespore (right). Membranes were stained with FM4–64 (red). CFP images (blue) used identical exposures and adjustments to directly compare fluorescence intensity. Scale bar, 1 μm.
(D) Total CFP fluorescence in the forespore (x axis) versus forespore volume (y axis) of sporangia without (blue) or with (red) IleS degradation in the forespore. Each dot represents an individual forespore. Solid dots represent the average CFP intensity and volume of wild-type (blue) and IleS-depleted forespores (red). Error bars represent standard deviations. IleS degradation significantly reduced CFP intensity (p < 0.0001) but has no effect on forespore size (p = 4625) compared to the control.
(E) Timelapse fluorescence microscopy of wild-type (control) and after IleS degradation in the forespore (IleSdeg.).
(F) Change in forespore volume during engulfment in wild-type (WT, green) and after forespore degradation of IleS (IleSdeg., red). Error bars represent standard deviation.
See also Figure S4.
To monitor the degree to which translation was impaired before chromosome translocation was completed, we placed a gene encoding the cyan fluorescent protein (CFP) under the control of a forespore-specific, σF-dependent promoter and integrated the construct into a site close to the chromosome terminus. Since the terminus is in the mother cell immediately after polar septation, CFP is not produced until the cfp gene reaches the forespore (Figure 4B). After forespore IleS degradation, we would expect a reduced CFP signal if translation was impaired by the time chromosome translocation finishes (Figure 4B). CFP intensity would therefore be indicative of the level of translation in individual forespores, allowing us to determine if there is a correlation between forespore size and the degree to which translation is inhibited. We plotted forespore CFP fluorescence versus forespore volume for individual sporangia about to complete engulfment with and without IleS degradation (Figures 4C and 4D). As expected, the average CFP intensity was shifted toward lower values after degradation (Figure 4D), and ~20% of the forespores showed negligible CFP signal, suggesting that in those fore-spores, translation was almost completely blocked before chromosome translocation finished. Importantly, there was no correlation between CFP intensity and forespore size, indicating that continuous protein synthesis in the forespore is not required for growth. Production of SspBEc in the forespore without ssrA* had no impact on CFP intensity or forespore size (Figure S4D).
We next performed timelapse microscopy to monitor fore-spore growth after IleS degradation, following only those 20% of sporangia that lacked detectable CFP expression at the end of the experiment (Figure 4E). Loss of forespore protein synthesis had no impact on forespore growth (Figures 4E and 4F). These results indicate that forespore growth does not require the continuous production of proteins in the forespore, suggesting that the chromosome packing is the main driver of forespore growth, at least during the first 1.5 hr of growth studied here.
Chromosome Packing Increases Forespore Volume by Extending the Forespore Membrane
Next, we explored if there was a connection between chromo-some packing and membrane synthesis during forespore growth. It is conceivable that increased turgor pressure in the forespore triggers the synthesis of the extra membrane required for growth. Alternatively, it is possible that the excess membrane required for forespore growth and engulfment is produced independently of chromosome translocation, in which case chromo-some packing might simply extend the forespore membrane to yield an effective volume increase. This second possibility seems to be supported by the accumulation of excess membrane around the forespores in the absence of chromosome translocation (Figures 2B, 5A, and S2). However, it is possible that the membrane excess is solely part of the mother cell engulfing membrane, which might wrinkle as it migrates around a smaller than normal forespore.
Figure 5. Chromosome Packing Increases Forespore Volume by Extending the Fore-spore Membrane.
(A) Fluorescence microscopy of wild-type and SpoIIIEATP− sporangia. Scale bar, 1 μm.
(B) Cartoon representing the transition between a walled (left) and protoplasted (right) sporangium upon enzymatic cell wall removal with lysozyme (scissors) at low osmolarity.
(C) Timelapse microscopy showing the transition to protoplasts for wild-type (WT, upper two rows) and SpoIIIEATP− sporangia (bottom two rows).
(D) Volumes of WT and SpoIIIEATP- forespores after engulfment membrane migration for intact (walled) and protoplasted (protoplast) sporangia. Each dot represents one sporangium. Dotted line, average of each set.
To distinguish between these two possibilities, we artificially extended the forespore membranes by enzymatically removing the cell wall under low-osmolarity conditions (Figures 5B and 5C). We have previously shown that sporulating cells do not lyse when the cell wall is removed in the absence of osmoprotectants (Ojkic et al., 2014). However, under those conditions, the engulfing membrane retracts (Figures 5B and 5C), suggesting that the turgor pressures of the mother cell and forespore increase (Ojkic et al., 2014), which would also allow both cells to fully extend their membranes during the transition to protoplasts. We measured the volume of wild-type and SpoIIIEATP− fore-spores from sporangia with an intact cell wall (Figure 5A) after completion of engulfment membrane migration, but not engulfment membrane fission (Figure 5D [walled]). As expected, the average volume of SpoIIIEATP− forespores was significantly reduced compared to wild-type forespores (Figure 5D). We then determined the volume of wild-type and SpoIIIEATP− fore-spores after digesting the cell wall under low-osmolarity conditions (Figure 5D [protoplast]). We monitored protoplast formation by timelapse microscopy, focusing on sporangia that had completed engulfment membrane migration by the time lysozyme was added (Figure 5C). As shown in Figure 5D, the average volumes of protoplasted wild-type and SpoIIIEATP− forespores are indistinguishable and equivalent to those of walled wild-type forespores. These results suggest that forespores synthesize the excess membrane required for growth independently of chromosome translocation and that chromosome packing might simply increase forespore turgor pressure to extend the forespore membrane, thereby increasing forespore volume.
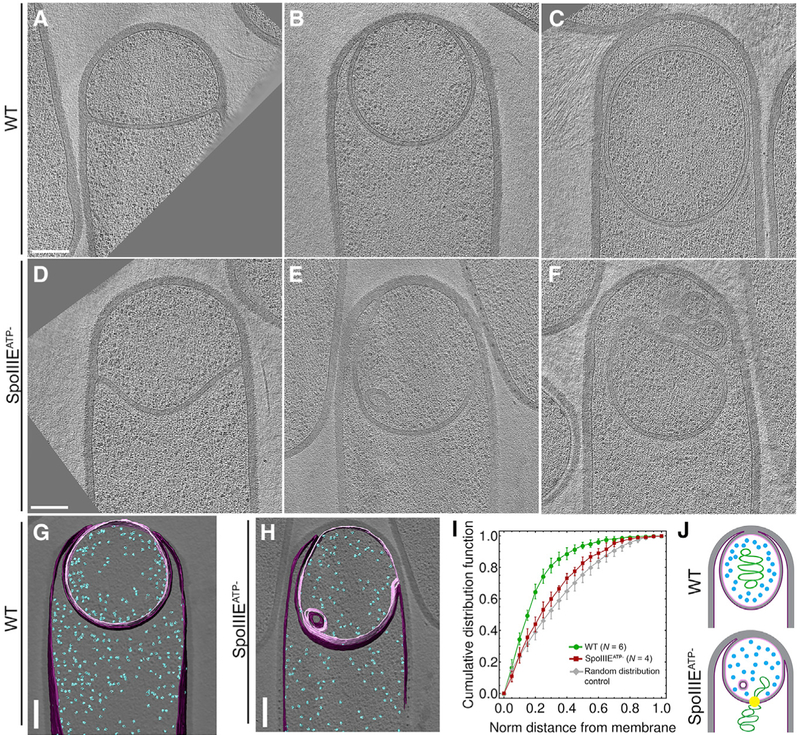
The Impact of DNA Translocation on Forespore Morphology Visualized by Cryo-electron Tomography
In order to better understand the impact of chromosome translocation on forespore morphology, we visualized wild-type and SpoIIIEATP− sporangia by cryo-electron tomography (CET). B. subtilis cells are too thick to obtain high-resolution tomograms using CET. To circumvent this limitation, we used cryo-focused ion beam (FIB) milling to thin the samples to a thickness compatible with CET, which allows visualization of cellular structures in a near-native state, at molecular resolution (Mahamid et al., 2016; Villa et al., 2013). The tomograms of wild-type sporangia show that, during engulfment, the septum curves smoothly toward the mother cell as the mother cell membrane engulfs the forespore (Figures 6A and 6B). After engulfment, forespores are roughly ovoid, and the membranes are completely extended (Figure 6C), with a thin layer of extended septal PG between the forespore and the mother cell membranes, as we previously observed in slender ponA mutant sporangia (Tocheva et al., 2013).
Figure 6. DNA Translocation Affects Ribo-some Distribution and Membrane Topography.
(A–F) Slices of cryo-electron tomograms depicting different stages of engulfment in wild-type sporangia (A–C) and corresponding states in spoIIIEATP− sporangia (D–F). Scale bar, 200 nm.
(G and H) Segmented tomogram of WT (G) and spoIIIEATP− (H) sporangia, showing ribosomes (blue), the forespore membrane (pink), and the mother cell membrane (purple).
(I) Cumulative distribution of ribosomes as a function of the distance from the forespore membrane in WT (green) and SpoIIIEATP− forespores. The gray line represents the cumulative distribution of randomly distributed sets of ribosomes generated in silico. Error bars represent standard deviation.
(J) Cartoons illustrating the exclusion of ribosomes by the forespore nucleoid.
See also Figure S5.
In spoIIIEATP− sporangia, we noted that the septum appeared wavy (Figures 6D and S5G) and less extended than wild-type. At later stages of engulfment, we observed invaginations of the double membrane that encloses the fore-spore (Figures 6E and 6F), and after engulfment, an excess of membrane accumulated, often at the sporangial cell pole (Figures 6F and S5H) or, sometimes, in folds of the two membranes into the forespore (Figures 6E and 6H). These observations are consistent with fluorescence microscopy results, where we typically observed membrane blobs at the mother cell distal tip of SpoIIIEATP− sporangia or elsewhere in the fore-spore (Figure S2). These results support the idea that, in the absence of chromosome translocation, newly synthesized fore-spore membrane cannot be fully extended, causing the fore-spore and engulfing mother cell membrane to wrinkle.
Ribosome Mapping Shows that the Hydrated Chromosome Occupies a Significant Volume of the Forespore Core
The bacterial nucleoid does not typically display a high-order organization and thus cannot be directly observed in our tomograms. However, previous studies have shown that the nucleoid excludes B. subtilis ribosomes (Lewis et al., 2000). The spatial organization of ribosomes therefore provides insight into the position and space occupied by the hydrated nucleoid. Ribosomes can be readily identified in our tomograms, using template matching and subtomogram averaging (Briggs, 2013). We therefore compared the spatial distribution of ribosomes in wild-type and SpoIIIEATP− sporangia (Figures 6G and 6H) and used this information to infer the space occupied by the forespore nucleoid. We focused on sporangia at late stages of engulfment to ensure that chromosome translocation was complete in wild-type and quantified ribosome distribution relative to the membrane. In SpoIIIEATP− sporangia, ribosomes were distributed uniformly across the forespore cytoplasm, indicating that the portion of the chromosome in the forespore occupies a small volume of the cell cytoplasm (Figures 6H–6J). In wild-type sporangia, forespore ribosomes were confined to the cell periphery, suggesting the full nucleoid occupies a significant fraction of the forespore core (Figures 6G, 6I, and 6J). Equivalent results were obtained by imaging GFP-labeled ribosomes with fluorescence microscopy (Figure S5).
Modeling the Impact of Chromosome-Generated Turgor Pressure on Septal PG and Forespore Shape
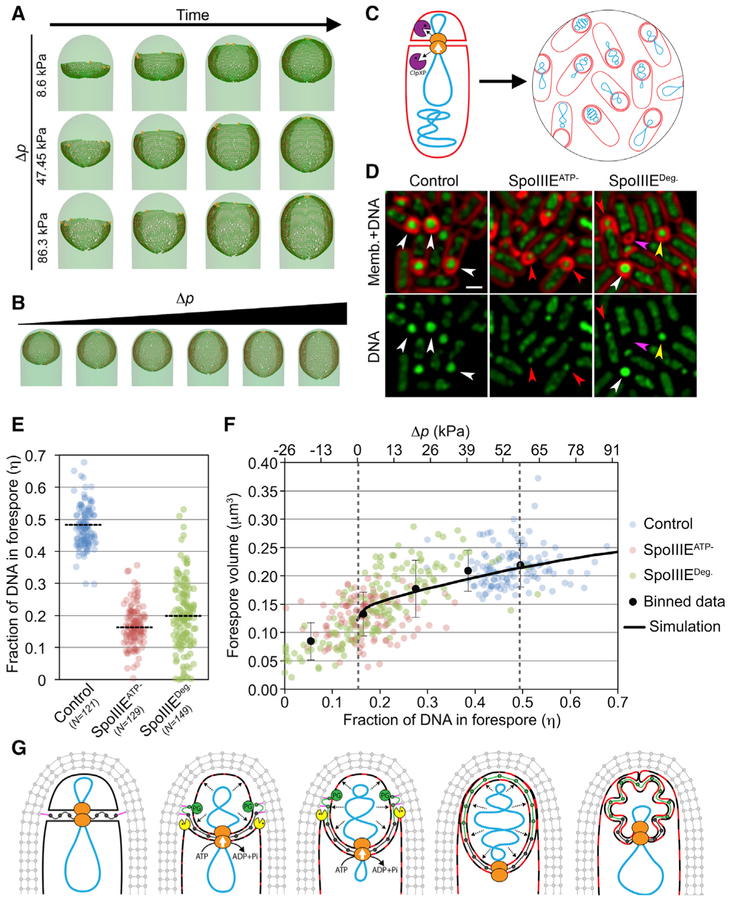
Taken together, our results suggest that the forespore chromosome effectively inflates the forespore, generating an increased turgor pressure that allows the cell to accommodate the additional membrane synthesized during engulfment and subsequent steps of spore formation. We estimated potential contributions of chromosome packing to the forespore turgor pressure (see STAR Methods). Our calculations indicate that packing a whole chromosome in the forespore can generate a pressure difference (Δp) between the forespore and mother cell between ~20 and ~80 kPa, mainly due to the osmotic effect of the counterions required to neutralize the negative charges of the DNA (Figure S6) and, to a lesser extent, due to the decreased entropy resulting from packing the chromosome into the small forespore. However, is this increase in Δp sufficient to explain observed engulfment dynamics and forespore morphology? To explore this, we modified our in silico mathematical model for engulfment (Ojkic et al., 2016). Briefly, using a simple model that accounts for cooperation between cell wall synthesis and degradation (Figure 1B), we proposed a mechanism for the progression of the leading edge of the engulfing membrane, in which the junction between the septal PG and the lateral cell wall moves around the forespore (Ojkic et al., 2016). Increased turgor pressure in the forespore pushes the septum and stretches the septal PG, leading to forespore expansion. It is important to note that, since the peptide-bond spring constant (kpep = 50 pN/nm) is much smaller than the glycan spring constant (kgly = 5570 pN/nm), the main volume change is due to elastic stretching of the peptide bond (Nguyen et al., 2015; Ojkic et al., 2016). In our model, newly synthesized peptide bonds are assumed to be relaxed and are subsequently stretched due to increasing forespore turgor pressure.
We simulated engulfment with different values of Δp between the forespore and the mother cell (Figures 7A and 7B). Our simulations assume that the same amount of extended septal PG is present in all cells and thus that any size difference is exclusively due to PG stretching. We observed drastic changes in forespore volume depending on Δp, indicating that the pressure differential between the two cells is a critical parameter in forespore size determination (Figure S7A). Our simulations with Δp around 60 kPa, which is within the Δp range that can be caused by chromosome translocation, produced forespore sizes in accordance to those found in vivo. Similarly, our simulations with low Δp yielded forespore sizes compatible with those found in SpoIIIEATP− sporangia.
Figure 7. Modeling the Impact of Chromo-some-Generated Turgor Pressure on Septal PG and Forespore Size.
(A) Simulation snapshots of differing stages of engulfment at increasing pressure difference (Δp) between the forespore and mother cell. Time between frames is 0.28 hr.
(B) Simulation snapshots for Δp in the range from 8.63–86.3 kPa. All forespores have the same amount of extended septal PG. For these pressures, peptide deformations are in the linear elastic regime, with negligible nonlinear peptide deformation (see Figure S7A).
(C) Simultaneous degradation of SpoIIIE in the mother cell and forespore produces sporangia with varying amounts of DNA in the forespore.
(D) Fluorescence microscopy showing a wild-type strain (wild-type), SpoIIIEATP−, and a strain in which SpoIIIE is simultaneously degraded in the mother cell and the forespore (SpoIIIEDeg.). Scale bar, 1 μm. Arrowheads represent forespores with completely translocated (white), non-translocated (red), partially translocated (yellow), or no (magenta) chromosome.
(E) Fraction of forespore DNA in wild-type sporangia (blue), SpoIIIEATP− sporangia (red), and SpoIIIEDeg. sporangia (green). Each dot represents a different sporangium. Dotted lines, average of each set.
(F) Correlation between the fraction of DNA in the forespore (primary × axis) and forespore volume of the sporangia analyzed in (E). Binned data for all strains are shown with black dots (error bars represent standard deviation) and compared with the simulation (black line) for different Δp (secondary × axis). Vertical dotted lines indicate the average fraction of DNA in SpoIIIEATP− (left line) and wild-type (right line) forespores.
(G) Model showing engulfment and reshaping of the forespore. After polar septation, the forespore is hemispherical and is separated from the mother cell by relaxed septal PG (dark gray). Chromosome translocation increases forespore turgor pressure, stretching the septal PG and allowing the accommodation of newly synthesized membrane (red patches on the membrane). Simultaneously, coordinated PG synthesis by forespore biosynthetic proteins (PG, green) and degradation by SpoIIDMP (yellow Pacman) moves the junction between the septum and the lateral cell wall (pink peptide crosslink) around the forespore, extending the septal PG (with new PG represented in green). In the absence of chromosome translocation, forespore turgor pressure remains low, preventing stretching of the extended septal PG and the accommodation of the excess membrane produced during engulfment.
See also Figure S6 and S7.
To see how well this model predicted forespore volume at varying chromosome-mediated pressure differences, we took advantage of B. subtilis strains that have different amounts of DNA in the forespore: a translocation proficient strain in which the chromosome is fully translocated to the forespore, a SpoIIIEATP− strain in which only 1/3 of the chromosome is in the forespore, and a strain in which SpoIIIE is tagged with ssrA* and simultaneously degraded in the mother cell and in the forespore by producing SspBEc in both cells (Yen Shin et al., 2015). Due to asynchronies between the onset of chromo-some translocation and SpoIIIE degradation, this last strain produces a plethora of chromosome translocation phenotypes, ranging from non-translocated to almost completely translocated (Figures 7C–7E) (Yen Shin et al., 2015). To assess the fraction of the chromosome translocated in each sporangium, we stained the DNA with SYTOX green and quantified the amount of DNA in the forespore relative to the total DNA in the sporangium (fraction of DNA in the forespore, η). We specifically selected sporangia that had completed engulfment membrane migration to compare sporangia at the same stage. By plotting h against forespore volume, we observed a clear positive correlation (correlation coefficient r = 0.74; Figure 7F). We searched for the best fit of our simulations to the data with Δp = 0 kPa corresponding to non-translocated DNA (η ~ 0.16), finding that, when the chromosome is completely translocated (η ~ 0.5), Δp is ~58 kPa. This pressure agrees well with our theoretical estimates of DNA-mediated osmotic pressure (see STAR Methods) and in vitro osmotic measurements (Dobrynin and Rubinstein, 2005; Hansen et al., 2001; Raspaud et al., 2000). Finally, we found that the model was able to accurately predict forespore growth in other Bacillus species with different cell volumes and chromosome sizes (Figure S7). Together, our simulations with different pressures and Bacillus species closely match experimental data, supporting the notion that increased forespore turgor pressure due to chromosome translocation deforms and extends the septal PG, allowing the forespore to adopt its final size and shape.
DISCUSSION
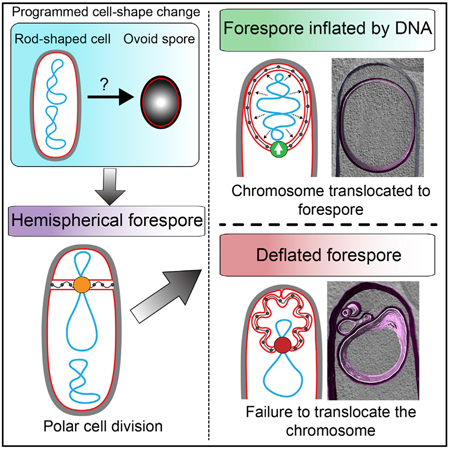
To understand the mechanisms controlling cell size and shape in bacteria, it is necessary to consider both biological regulatory mechanisms and the underlying biophysical principles of the system. Here, we studied the transition in forespore shape during B. subtilis sporulation, from hemispherical to ovoid, using a combination of cell biology, genetics, CET, and biophysical modeling. We propose a mechanism for forespore shape generation in which the final shape of the forespore is purely achieved through physiochemical interactions between three core cellular constituents: SpoIIIE-mediated DNA translocation, which effectively inflates the forespore like air in a balloon, stimulating forespore growth by extending the fore-spore membranes and stretching septal PG (Figure 7G). Our results reveal an unexpected role of DNA in increasing fore-spore pressure, allowing it to grow within the confines of the extended septal PG to adopt its final size and shape. First, we demonstrated that the chromosome needs to be present in the forespore for this cell to adopt its extended shape (Figure 3). Second, we showed that sustained translation in the forespore is not required for growth (Figure 4). Altogether, our observations indicate that the main contribution of chromosome translocation to forespore size and shape is the increase in forespore turgor pressure rather than the expression of genes located on the region of the chromosome that is initially excluded from the forespore.
According to our theoretical calculations, the main contribution of the chromosome to forespore pressure is osmotic (see STAR Methods). This can be conceptualized according to the Gibbs-Donnan effect (Figure S6; Castelnovo and Evilevitch, 2006): the translocation of a negatively charged polyelectrolyte, such as DNA, to the forespore would generate an electrostatic imbalance between the forespore and the mother cell, with the forespore negatively charged with respect to the mother cell. Such imbalance is compensated by a redistribution of the cytoplasmic cations between both cells, generating an effective osmotic Δp. Our CET images indicate that, upon chromosome translocation, the forespore nucleoid displaces the ribosomes to the cell periphery (Figure 6). This observation is compatible with the idea that the chromosome might in fact act as a sponge that swells by sequestering cations and water, causing the fore-spore to inflate.
Our calculations indicate that packing a full chromosome in the forespore could generate a difference of pressure between mother cell and forespore ranging from ~20 to ~80 kPa. At first sight, this Δp seems relatively minor in the context of a cell with a turgor pressure of ~2 MPa during vegetative growth (What-more and Reed, 1990). However, it is similar to the turgor pressure of E. coli cells (~30 kPa) (Deng et al., 2011), which is sufficient to deform the thin layer of septal PG from flat during division to curved after cell separation. Our CET images (Figure 6; Tocheva et al., 2013) show that the extended septal PG has a thickness similar to that of E. coli cell wall, suggesting that the Δp between forespore and mother cell should be sufficient to stretch the extended septal PG, making it curve into the mother cell. In agreement with this idea, our simulations indicate that PG stretching due to chromosome-generated turgor pressure is enough to explain the size difference between wild-type and SpoIIIEATP− forespores during engulfment. This is further supported by the rapid loss of forespore volume upon abrupt reverse chromosome translocation (Figures 3H–3J and S3C), which suggests that the chromosome produces an elastic deformation of the forespore. It is therefore possible that increased forespore turgor pressure does not trigger synthesis of extra PG but rather promotes the stretching of the septal PG—a hypothesis that is consistent with the independence of forespore growth from PG synthesis. A stretching-based model has also been proposed for the elongation of the E. coli PG sacculus (Rojas et al., 2014).
Chromosome translocation proceeds through the hydrolysis of vast amounts of ATP by the mother cell SpoIIIE subcomplex. Forespore growth could therefore be envisioned as an energy transduction process, from chemical energy stored in ATP to mechanical energy in the form of the expansion of the DNA with its associated counterions, similar to the hydration of a glass-like gel that in turn expands the membranes and stretches the septal PG (Parry et al., 2014; Taylor, 1923). In several bacteriophages, the ATP-driven packaging motor generates a force resulting in a continuous increase in internal pressure as DNA is packaged inside the phages (Evilevitch et al., 2003; Smith et al., 2001), and energy generated in this process is converted to work when the DNA is injected into the host cell. Analogously, the ATP-driven motor of SpoIIIE might also force the DNA inside the small volume of the forespore. In support of this idea, we have observed that the DNA is rapidly ejected from the forespore in some sporangia after degradation of SpoIIIE in the mother cell, presumably through an opening in the septum generated due to the disassembly of the translocation complex. The mechanical energy generated in the process of packaging the chromosome in the forespore may be converted into work done in stretching the septal PG and extending the forespore membrane to proceed with engulfment.
Overall, our results lead to a mechanism of forespore size and morphology determination in which the septal PG is stretched as a consequence of the DNA-mediated increase in forespore turgor pressure, allowing the membranes that surround the forespore to adopt an optimal smooth conformation to proceed with engulfment. Then, coordinated PG synthesis and degradation at the leading edge of the engulfing membrane allows the extension of the septal PG, moving the junction between the septal PG and lateral cell wall around the forespore and generating space for the movement of the mother cell engulfing membrane (Figure 7G). The results presented here illustrate how relatively simple biochemical processes and biophysical principles governing the interaction between core cellular components can together mediate complex processes, such as programmed changes in cellular morphology and engulfment.
STAR⋆METHODS
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides, and Recombinant Proteins | ||
| FM4–64 | Thermo Fisher Scientific | Cat#T13320 |
| SYTOX Green | Thermo Fisher Scientific | Cat#S7020 |
| Fosfomycin | MilliporeSigma | Cat#P5396 |
| Bacitracin | MilliporeSigma | Cat#B0125 |
| Penicillin V | MilliporeSigma | Cat#1504489 |
| Cephalexin | MilliporeSigma | Cat#C4895 |
| Cerulenin | MilliporeSigma | Cat#C2389 |
| Rifampicin | MilliporeSigma | Cat#R3501 |
| Experimental Models: Organisms/Strains | ||
| Bacillus subtilis PY79 | Youngman et al., 1984 | Tax. ID:1415167 |
| Bacillus megaterium QM B1551 | Dr. Peter Setlow at UConn Health | Tax. ID:545693 |
| Bacillus thuringiensis subsp. israelenis | Bacillus Genetic Stock Center | 4Q2; Tax. ID:339854 |
| Bacillus pumilus BL8 | Dr. Louise Temple at James Madison University | Tax. ID:1189615 |
| See Table S1 for a complete list of strains used in this paper, including all the Bacillus subtilis PY79 variants | N/A | N/A |
| Oligonucleotides | ||
| See Table S2 for oligonucleotides | N/A | N/A |
| Recombinant DNA | ||
| Plasmid: pJLG38: GFPΩkan | This paper | N/A |
| Plasmid: pJLG82: amyE::PsspE(2G)-sspBΩcat | This paper | N/A |
| Plasmid: pJLG113: ileS-ssrAΩkan | This paper | N/A |
| Plasmid: pER226: rpsB-GFPΩkan | This paper | N/A |
| Software and Algorithms | ||
| JFilament | Smith et al., 2010 | http://athena.physics.lehigh.edu/jfilament/ |
| volumeestimator | Ojkic et al., 2016 | https://elifesciences.org/articles/18657/figures#SD2-data |
| intensityestimator | This paper | Data S1 |
| IMOD | Mastronarde, 1997 | http://bio3d.colorado.edu/imod/; RRID: SCR_003297 |
| TomoSegMemTV | Martinez-Sanchez et al., 2014 | https://sites.google.com/site/3demimageprocessing/tomosegmemtv |
| Amira | Commercial software by Thermo Scientific (formerly FEI) | https://www.fei.com/software/amira-3d-for-life-sciences/; RRID: SCR_014305 |
| EMAN2 | Tang et al., 2007 | http://blake.bcm.edu/emanwiki/EMAN2 |
| Dynamo | Castaño-Díez et al., 2012 | https://wiki.dynamo.biozentrum.unibas.ch/w/index.php/Main_Page |
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Kit Pogliano (kpogliano@ucsd.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
We used four different Bacillus species as experimental models: Bacillus subtilis PY79, Bacillus megaterium QM B1551, Bacillus thuringiensis serotype israelensis, and Bacillus pumilus BL8. All of them were routinely grown in LB plates at 30°C. Sporulation was induced in two different ways: (i) For B. subtilis PY79, sporulation was induced by resuspension in A+B medium, after growing the bacteria in 1/4 diluted LB to O.D.600 ~ 0.5. For strains carrying an integration in thrC, the resuspension medium was supplemented with 50 μg/ml of threonine. Sporulation cultures were grown at 37°C for batch culture and protoplast experiments, and at 30°C for timelapse experiments. (ii) For comparing forespore sizes of all the model organisms (Figure S7), sporulation was induced by exhaustion in Difco Sporulation Medium (DSM) at 30°C. Relevant details about the different experimental models are provided below:
Bacillus subtilis PY79
Most of the work was done using this model, since it is highly genetically tractable. A list of the different Bacillus subtilis PY79 strains used in his study is provided in Table S1. The plasmids and oligonucleotides used to construct the different strains are provided in Tables S2 and S3, respectively. Table S4 includes detailed descriptions of plasmid construction.
Chromosome size: 4.03 megabases.
No plasmids.
Bacillus megaterium QM B1551
Chromosome size: 5.10 megabases.
Seven plasmids, with a combined size of 0.43 megabases.
Bacillus thuringiensis serotype israelensis
Chromosome size: 5.50 megabases.
Six plasmids, with a combined size of 0.84 megabases.
Bacillus pumilus BL8
Chromosome size: ~3.7–3.8 megabases.
METHOD DETAILS
Experimental procedures
Fluorescence microscopy from batch cultures
Cells were visualized on an Applied Precision DV Elite optical sectioning microscope equipped with a Photometrics CoolSNAP-HQ2 camera and deconvolved using SoftWoRx v5.5.1 (Applied Precision). For imaging, cells (12 μl) were transferred to 1.2% agarose pads prepared using sporulation resuspension medium. Cells were typically imaged ~3 hours after sporulation induction at 37°C. The median focal plane is shown. When appropriate, membranes were stained with 0.5 μg/ml of FM4–64, added directly to the agarose pad. DNA was stained with 0.5 μM of SYTOX green, added to the sporulating cultures ~1 hour after resuspension and to the agarose pad.
To observe ribosome distribution using a RPS2-GFP after transcription blockage with rifampicin (Figure S5), sporulating cultures of a strain expressing rpsB-GFP were split 2 hours after sporulation induction and incubated with and without 0.25 μg/ml of rifampicin for one extra hour. Cells were then imaged as described above.
Timelapse fluorescence microscopy
Sporulation was induced at 30°C. To visualize DNA, the culture was supplemented with 0.5 μM SYTOX green one hour after sporulation induction. SYTOX green is typically used as an indicator of dead cells, as it has a limited ability to cross the membrane of living cells. However, when added to sporulating cultures several hours before imaging, SYTOX green stains the chromosomes yielding a readily detectable fluorescence signal, without affecting the sporulation process. To visualize the membranes, 0.5 μg/ml FM4–64 was added to the culture ~2 hours after sporulation induction and incubation continued for another hour. Seven ml samples were taken 3 hours after resuspension and transferred to agarose pads prepared as follows: 2/3 volume of supernatant from the sporulation culture; 1/3 volume 3.6% agarose in fresh A+B sporulation medium; 0.17 μg/ml FM4–64; 0.17 μM SYTOX green. When appropriate, antibiotics were added to the pads to the final concentrations indicated in the figure legends. Pads were partially dried, covered with a glass slide and sealed with petroleum jelly to avoid dehydration during timelapse imaging. Petroleum jelly is not toxic and cannot be metabolized by B. subtilis, which poses an advantage over other commonly used sealing compounds, such as glycerol, which can be used as a carbon source and inhibit the initiation of sporulation. Pictures were taken in an environmental chamber at 30°C every 5 min for at least 5 hours. Excitation/emission filters were TRITC/CY5 for membrane imaging, and FITC/FITC for DNA imaging. Excitation light transmission was set to 5% to minimize phototoxicity. Exposure time was 0.1 s. For presentation purposes, sporangia were aligned vertically (with forespore on top) by rotating them using Photoshop.
Protoplast timelapse fluorescence microscopy
Sporulation was induced at 37°C. Samples were taken 3–4 hours after resuspension, spun at 7000 × g for 10 s, and resuspended in 25 μl of a buffer containing 20 mM maleic acid and 20 mM MgCl2, pH 6.5. 10 μl of the resuspended culture were placed on a poly-L-lysine-treated coverslip and mixed with lysozyme and FM 4–64 (final concentrations 1 mg/ml and 5 μg/ml, respectively). Images were taken at room temperature, every 45 s for one hour. Excitation light transmission was set to 32%. Exposure time was 0.1 s.
Cryo-Electron Tomography
Holey carbon coated QUANTIFOIL R 2/1 copper grids were glow discharged using Pelco easiGlow glow discharge cleaning system and sporulating B. subtilis PY79 cells were deposited on these grids. Samples were taken approximately 2–3 hours after resuspension for wild-type sporulating cells and at 2.5 hours after resuspension for spoIIIEATP- strain. Excess resuspension media was removed by manual blotting using Whatman No. 1 filter paper from the reverse side so that cells form a monolayer on the grids. Grids were then plunge-frozen using a custom-built vitrification device (Max Planck Institute for Biochemistry, Munich) by rapid immersion in ethane/propane mixture cooled by liquid nitrogen.
Grids were mounted into modified FEI Autogrids after vitrifcation. This prevents any mechanical damage to the delicate grids during subsequent transfer steps. Then, these clipped grids were transferred into a dual-beam (cryo-FIB/SEM) microscope (Scios, Thermo Fisher Scientific, formerly FEI) equipped with a cryogenic stage. Thin sections of 100–250 nm, or lamellae, were prepared as previously described in Rigort et al., 2012 (see protocol for lamella preparation, not wedges). Each lamella contains ~10–12 cells.
Tilt-series were collected from typically −66°to +66°with a tilt increment of 1.5°or 2°using SerialEM in a 300-keV Tecnai G2 Polara (Thermo Fisher Scientific, formerly FEI) equipped with post-column Quantum Energy Filter (Gatan) and a K2 Summit 4k × 4k direct detector camera (Gatan). Images were recorded at a nominal magnification of 34,000 with a pixel size of 0.61 nm or 22,500 with a pixel size of 0.92 nm. The dose rate was set to 10–12 e−/physical pixel at the camera level. Frame exposure was set to 0.1 s, with a total exposure in a frame set to be determined by an algorithm targeting an average count number. The total dose in a tomogram was typically ~100–150 e−/Ao2 with a defocus ranging from −5 to −6 mm. A total of 24 tomograms from 14 FIB-milled lamellae were collected for wild-type sporulating cells and 16 tomograms from 6 FIB-milled lamellae for spoIIIEATP- cells.
Tomograms were reconstructed in IMOD (Mastronarde, 1997) using patch tracking.
Image analysis
Determination of cell volume
To determine the volumes of the forespore and the mother cell, we extracted the membrane contours of the forespore and the mother cell using microscopy images in which membranes were stained with FM4–64. For this purpose, we used the semi-automated active contour software JFilament, available as ImageJ plugin (Smith et al., 2010). In the case of timelapse movies, the membrane position was extracted for every time frame. We then used a custom-built Mathematica software (volumeestimator) to calculate 3D volumes of forespore, by assuming rotational symmetry around the axis connecting the center of masses of forespore and mother cell. The code is available in Ojkic et al., 2016. In the case of the forespore volumes reported in Figures 4D, 5D, and S4D, forespore volume was extracted without considering the center of masses to determine the rotational axis. Instead, since we only focused on the fully engulfed forespores that have typical ovoid shape, the rotational axis was estimated by finding axis of rotation as minimum moment of inertia axis. This method was validated by comparing forespore volumes using the two different methods giving R2 > 0.99 (graph not shown).
Estimation of chromosome translocation
The forespore and mother cell contours were determined using JFilament, as above. We extended the Mathematica software to extract the total fluorescence intensity of the pixels enclosed by the contours (intensityestimator). In the case of timelapse movies, we used one optical section per time frame, and the total forespore and mother cell SYTOX green intensities were tracked over time. For measurements from batch culture microscopy experiments, we added the SYTOX green intensities of six consecutive optical sections, covering a total thickness of 0.75 μm. The background intensity was calculated using a rolling radius of 3.2 μm (ImageJ). From the total SYTOX green intensity within the forespore and the mother cell, we subtracted the total background intensity to obtain the net SYTOX green intensity in each cell. We defined the fraction of DNA in forespore (η) as [total net SYTOX green intensity in FS / (total net SYTOX green intensity in FS + total net SYTOX green intensity in MC)]. When chromosome translocation is completed, η is ~0.5 (Figures 3B and 7E).
Translocation rate measurements
From the measurement of fraction of DNA in the forespore (η) over time (see previous section) we calculated translocation speed per chromosome arm as v = N dη/dt, where N is the number of bp per DNA, and t is time. During forward and slow reverse translocation, chromosomes are translocated with constant rate (Figure 3B and3C). For each cell, we determined dη/dt using linear fit from h versus time graph, and we calculated the average ± SD of the different cells. The fast reverse translocation rate was calculated similarly as above, but for each cell the slopes were determined from three consecutive time points separated by 5 min intervals, after the onset of the reverse translocation.
Forespore CFP fluorescence intensity
To determine the CFP intensity in the forespore, the intensity of five optical sections was added, covering a total thickness of 0.6 μm. The forespore contour and the CFP intensity enclosed by it were calculated using intensityestimator, as explained in “Estimation of chromosome translocation“ subsection.
Fluorescently labeled ribosome distribution
Forespore and mother cell contours were extracted using the ImageJ plugin JFilament, as above. The long axis of every sporangium was rotated to be parallel with y-axis, with the forespore pointing up. Then, we aligned the centers of mass of every forespore, and normalized RPS2-GFP fluorescence at the medial focal plane. To calculate average fluorescence distribution of many forespores, we created a square grid with a meshwork size of 16 nm. For each meshwork point we binned pixel intensities that were within a radius of 64 nm. Subsequently, we calculated the average fluorescence for each meshwork point and plotted as a density plot (Figure S5J). Normalized intensity through the center of the forespore and perpendicular to the long axis of the cell (along y = 0 line) is plotted in Figure S5K.
Ribosome distribution in CET
Semi-automatic segmentation of membranes was done using TomosegmemTV (Martinez-Sanchez et al., 2014) and then manual refinement in Amira (FEI Visualization Sciences Group). Subtomograms containing ribosomes were manually picked using EMAN2 (Tang et al., 2007) and averaged, classified and placed in their original positions in the tomogram using Dynamo (Castaño-Díez et al., 2012) and Amira. We analyzed the distribution of the ribosomes in tomograms of six wild-type and four SpoIIIEATP− forespores. We determined the shortest distance between each ribosome and the segmented surface of the forespore in 3D. For each case, the distance between the ribosome and the forespore membrane was normalized to the largest possible distance from the membrane determined numerically for each forespore in 3D. As a control, we generated random distributions of ribosomes within the SpoIIIEATP- forespores wherein the same number of ribosomes (as found in the respective tomogram) were initiated randomly within the membrane region. The number of random sets for each SpoIIIEATP− forespores was ~300. A cumulative distribution of ribosomes (normalized to the total number of ribosomes) was then plotted against distance from the membrane for each forespore.
DNA mesh pore size calculation
Estimates for DNA mesh pore size in the forespore were made based on calculations in SI appendix, section S4 in Castellana et al., 2016. L denotes the total length of forespore DNA (~4.03 megabases or ~1.37 mm) and V is the volume of the forespore (0.1–0.2 μm3) wherein DNA is confined. As an approximation, DNA can be thought of as occupying a cubic lattice of volume V that consists of N cubic pores each with edge length a (pore size) so that V = Na3 and DNA length, L is given by L = 3aN for close packing. This provides an estimate of pore size as a
Based on these calculations, the forespore nucleoid can be thought of as a DNA mesh with an average pore diameter of ~15–20 nm.
Modeling, simulations & pressure calculations
Engulfment model and simulations are described in detail in Ojkic et al., 2016. Different Bacillus species were simulated using different initial forespore sizes, by varying the radius of the forespore to be 0.26, 0.32, 0.55 and 0.6 μm corresponding to B. pumilus, B. subilis, B. thuringiensis, and B. megaterium, respectively. The cell sizes were derived from fluorescence microscopy images of sporangia of the different species (Figures S7B and S7C). Below there is a detailed description of how chromosome-generated turgor pressure was calculated for B. subtilis. The same calculations where used for different species, taking into account their respective cell and genome sizes.
In B. subtilis, packing a long ~4-megabase DNA polymer into a forespore that is initially ~0.1 μm3 (Figure 1) creates a DNA concentration (CDNA) of ~45 g/l. Each base pair (bp) of the chromosome accompanies two negative phosphate charges, which makes the DNA molecule highly negatively charged. Based on those premises we explored entropic, elastic, electrostatic, and osmotic contributions to the total forespore pressure.
Entropic contribution
By packing the chromosome into the forespore the number of potential DNA conformations is reduced, leading to the increased entropic pressure. When fully translocated, DNA volume fraction (DNA volume divided by forespore volume) is ~6.2%. This produces an entropic pressure of ~2 kPa, estimated using Langevin simulations (Pereira et al., 2017). This estimate is in a good agreement with the experimental measurements in E. coli, which suggest that the entropic pressure generated by a 4.6-megabase chromosome is ~4 kPa (Pelletier et al., 2012). However, this estimated entropic pressure is not large enough to significantly deform peptidoglycan meshwork (Figure S7).
Elastic contribution
The DNA elastic bending stiffness tends to restore bent DNA molecules to their straight configuration. The DNA persistence length, lp ~50 nm, is much smaller than the typical linear size of the forespore, 460 nm. Additionally, the DNA persistence length decreases in ionic solutions (Manning, 2006). Since the forespore size is about an order of magnitude larger than the DNA persistence length, the DNA elastic bending contribution to the forespore pressure is negligible.
Electrostatic contribution
DNA is negatively charged and the electrostatic repulsion depends on the spacing between neighboring DNA strands. To estimate average DNA radial separation d, we approximate the DNA molecule by a cylinder of the same length as a whole molecule: d2 LDNA = Vfspore, where LDNA is the total DNA length (Purohit et al., 2003). Since bp linear size is 0.34 nm (Kempes et al., 2016; Raspaud et al., 2000), we obtain that the typical distance between two neighboring DNA strands is ~8.4 nm. DNA electrostatic interactions are screened in the ionic solutions due to counterions and the typical Debye’s electrostatic screening length of about 0.8 nm (Podgornik et al., 2016; Raspaud et al., 2000). Since the calculated distance d between neighboring DNA strands is much larger than the Debye’s screening length, the electrostatic contribution to the pressure is also negligible.
Osmotic contribution
DNA is negatively charged and attracts positively charged counterions that contribute to osmotic pressure. The DNA volume fraction in the forespore is ~6.2% corresponding to a semi-dilute polymer solution. In this regime, the osmotic pressure (p) has polymeric (pp) and an ionic contribution of the DNA counterions (pi) (Dobrynin and Rubinstein, 2005; Dobrynin et al., 1995). For DNA forespore concentrations, the polymeric contribution is negligible (Raspaud et al., 2000) therefore p ≈ pi. The expression for the osmotic pressure of DNA counterions is given by:
where R is the ideal gas constant, T is the temperature, ϕ is the osmotic coefficient, Cc is the counterion concentration, and Cs is the salt concentration (Dobrynin et al., 1995). The counterion concentration is equal to the DNA phosphate concentration (Raspaud et al., 2000). The above expression fits well with experimentally measured counterion osmotic pressure with ϕ = (0.245 ± 0.020) (Raspaud et al., 2000). Note that the dependence of the osmotic pressure due to DNA counterions on Cs accounts for the classical Gibbs-Donnan effect. Using experimentally measured salt concentration in E. coli (Lo et al., 2006) in the range of 2–20 mM and applying above expression we obtain that the forespore osmotic pressure is in the range 26–70 kPa. This estimate agrees with in vitro measurement of osmotic pressure of 63 kPa for the Cs = 10 mM and DNA forespore concentration (Hansen et al., 2001; Raspaud et al., 2000). In the lower range of salt concentration, Debye’s electrostatic screening length remains smaller than the typical distance between neighboring DNA strands.
QUANTIFICATION AND STATISTICAL ANALYSIS
For each experiment we had at least two biological replicas, and each one contained at least three technical replicas. Averages of individual cells, but not the averages of different replicas are reported. The number of cells analyzed (N) is indicated in each figure. Data represent the mean of N cells. The standard deviation is shown as dispersion measurement.
Supplementary Material
Highlights.
Chromosome translocation reversibly inflates the forespore
The forespore volume increases at the expense of the mother cell volume
Forespore growth relies on membrane, but not on peptidoglycan synthesis
DNA-generated turgor pressure reshapes the forespore
ACKNOWLEDGMENTS
We thank Davide Marenduzzo, Rudi Podgornik, and Rosalind Allen for discussion. This work was supported by ERC starting grant 280492-PPHPI (N.O. and R.G.E.), NIH grant R01-GM57045 (K.P.), NIH Director’s New Innovator Award 1DP2GM123494–01 (E.V.), and EMBO Long Term Fellowship ALTF 1274–2011 (J.L.-G). We used the UCSD Cryo-EM Facility (supported by NIH grant R01-GM33050 to Dr. Timothy S. Baker and the Agouron Institute) and the San Diego Nanotechnology Infrastructure of UCSD (supported by the NSF grant ECCS-1542148).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes seven figures, four tables, and one data set and can be found with this article online at https://doi.org/10.1016/j.cell.2018.01.027.
DECLARATION OF INTERESTS
K.P. is a co-founder, a shareholder, and a member of the advisory board of Linnaeus Bioscience (La Jolla, CA). The terms of this arrangement have been reviewed and are managed by the University of California, San Diego in accordance with its conflict of interest policies.
SUPPORTING CITATIONS
The following references appear in the Supplemental Information: Guérout-Fleury et al. (1996); Sandman et al. (1987).
REFERENCES
- Amir A, and Nelson DR (2012). Dislocation-mediated growth of bacterial cell walls. Proc. Natl. Acad. Sci. USA 109, 9833–9838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett TM, Bratton BP, Duvshani A, Miguel A, Sheng Y, Martin NR, Nguyen JP, Persat A, Desmarais SM, VanNieuwenhze MS, et al. (2017). A periplasmic polymer curves Vibrio cholerae and promotes pathogenesis. Cell 168, 172–185.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bath J, Wu LJ, Errington J, and Wang JC (2000). Role of Bacillus subtilis SpoIIIE in DNA transport across the mother cell-prespore division septum. Science 290, 995–997. [DOI] [PubMed] [Google Scholar]
- Becker EC, and Pogliano K (2007). Cell-specific SpoIIIE assembly and DNA translocation polarity are dictated by chromosome orientation. Mol. Microbiol 66, 1066–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs JAG (2013). Structural biology in situ–the potential of subtomogram averaging. Curr. Opin. Struct. Biol 23, 261–267. [DOI] [PubMed] [Google Scholar]
- Cabeen MT, Charbon G, Vollmer W, Born P, Ausmees N, Weibel DB, and Jacobs-Wagner C (2009). Bacterial cell curvature through mechanical control of cell growth. EMBO J. 28, 1208–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaño-Díez D, Kudryashev M, Arheit M, and Stahlberg H (2012). Dynamo: a flexible, user-friendly development tool for subtomogram averaging of cryo-EM data in high-performance computing environments. J. Struct. Biol 178, 139–151. [DOI] [PubMed] [Google Scholar]
- Castellana M, Hsin-Jung Li S, and Wingreen NS (2016). Spatial organization of bacterial transcription and translation. Proc. Natl. Acad. Sci. USA 113, 9286–9291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelnovo M, and Evilevitch A (2006). Binding effects in multivalent Gibbs-Donnan equilibrium. Europhys. Lett 73, 635–641. [Google Scholar]
- Deng Y, Sun M, and Shaevitz JW (2011). Direct measurement of cell wall stress stiffening and turgor pressure in live bacterial cells. Phys. Rev. Lett 107, 158101. [DOI] [PubMed] [Google Scholar]
- Dobrynin AV, and Rubinstein M (2005). Theory of polyelectrolytes in solutions and at surfaces. Prog. Polym. Sci 30, 1049–1118. [Google Scholar]
- Dobrynin AV, Colby RH, and Rubinstein M (1995). Scaling Theory of Poly-electrolyte Solutions. Macromolecules 28, 1859–1871. [Google Scholar]
- Errington J (2003). Regulation of endospore formation in Bacillus subtilis. Nat. Rev. Microbiol 1, 117–126. [DOI] [PubMed] [Google Scholar]
- Evilevitch A, Lavelle L, Knobler CM, Raspaud E, and Gelbart WM (2003). Osmotic pressure inhibition of DNA ejection from phage. Proc. Natl. Acad. Sci. USA 100, 9292–9295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming TC, Shin JY, Lee SH, Becker E, Huang KC, Bustamante C, and Pogliano K (2010). Dynamic SpoIIIE assembly mediates septal membrane fission during Bacillus subtilis sporulation. Genes Dev. 24, 1160–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frandsen N, and Stragier P (1995). Identification and characterization of the Bacillus subtilis spoIIP locus. J. Bacteriol 177, 716–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith KL, and Grossman AD (2008). Inducible protein degradation in Bacillus subtilis using heterologous peptide tags and adaptor proteins to target substrates to the protease ClpXP. Mol. Microbiol 70, 1012–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guérout-Fleury AM, Frandsen N, and Stragier P (1996). Plasmids for ectopic integration in Bacillus subtilis. Gene 180, 57–61. [DOI] [PubMed] [Google Scholar]
- Hansen PL, Podgornik R, and Parsegian VA (2001). Osmotic properties of DNA: critical evaluation of counterion condensation theory. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 64, 021907. [DOI] [PubMed] [Google Scholar]
- Higgins D, and Dworkin J (2012). Recent progress in Bacillus subtilis sporulation. FEMS Microbiol. Rev 36, 131–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt JH, Krieg NR, Sneath PHA, Staley JT, and Williams ST (1994). Bergey’s manual of determinative bacteriology, Ninth Edition (Lippincott Williams & Wilkins; ). [Google Scholar]
- Kay D, and Warren SC (1968). Sporulation in Bacillus subtilis. Morphological changes. Biochem. J 109, 819–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempes CP, Wang L, Amend JP, Doyle J, and Hoehler T (2016). Evolutionary tradeoffs in cellular composition across diverse bacteria. ISME J. 10, 2145–2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamsa A, Lopez-Garrido J, Quach D, Riley EP, Pogliano J, and Pogliano K (2016). Rapid Inhibition Profiling in Bacillus subtilis to identify the mechanism of action of new antimicrobials. ACS Chem. Biol 11, 2222–2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis PJ, Thaker SD, and Errington J (2000). Compartmentalization of transcription and translation in Bacillus subtilis. EMBO J. 19, 710–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Chistol G, and Bustamante C (2015). Two-subunit DNA escort mechanism and inactive subunit bypass in an ultra-fast ring ATPase. eLife 4, 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo CJ, Leake MC, and Berry RM (2006). Fluorescence measurement of intracellular sodium concentration in single Escherichia coli cells. Biophys. J 90, 357–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Diaz I, Clarke S, and Mandelstam J (1986). spoIID operon of Bacillus subtilis: cloning and sequence. J. Gen. Microbiol 132, 341–354. [DOI] [PubMed] [Google Scholar]
- Mahamid J, Pfeffer S, Schaffer M, Villa E, Danev R, Cuellar LK, Förster F, Hyman AA, Plitzko JM, and Baumeister W (2016). Visualizing the molecular sociology at the HeLa cell nuclear periphery. Science 351, 969–972. [DOI] [PubMed] [Google Scholar]
- Manning GS (2006). The persistence length of DNA is reached from the persistence length of its null isomer through an internal electrostatic stretching force. Biophys. J 91, 3607–3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Sanchez A, Garcia I, Asano S, Lucic V, and Fernandez JJ (2014). Robust membrane detection based on tensor voting for electron tomography. J. Struct. Biol 186, 49–61. [DOI] [PubMed] [Google Scholar]
- Massey TH, Mercogliano CP, Yates J, Sherratt DJ, and Löwe J (2006). Double-stranded DNA translocation: structure and mechanism of hexameric FtsK. Mol. Cell 23, 457–469. [DOI] [PubMed] [Google Scholar]
- Mastronarde DN (1997). Dual-axis tomography: an approach with alignment methods that preserve resolution. J. Struct. Biol 120, 343–352. [DOI] [PubMed] [Google Scholar]
- Meyer P, Gutierrez J, Pogliano K, and Dworkin J (2010). Cell wall synthesis is necessary for membrane dynamics during sporulation of Bacillus subtilis. Mol. Microbiol 76, 956–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LT, Gumbart JC, Beeby M, and Jensen GJ (2015). Coarse-grained simulations of bacterial cell wall growth reveal that local coordination alone can be sufficient to maintain rod shape. Proc. Natl. Acad. Sci. USA 112, E3689–E3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojkic N, López-Garrido J, Pogliano K, and Endres RG (2014). Bistable forespore engulfment in Bacillus subtilis by a zipper mechanism in absence of the cell wall. PLoS Comput. Biol 10, e1003912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojkic N, López-Garrido J, Pogliano K, and Endres RG (2016). Cell-wall remodeling drives engulfment during Bacillus subtilis sporulation. eLife 5, e18657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry BR, Surovtsev IV, Cabeen MT, O’Hern CS, Dufresne ER, and Jacobs-Wagner C (2014). The bacterial cytoplasm has glass-like properties and is fluidized by metabolic activity. Cell 156, 183–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier J, Halvorsen K, Ha B-Y, Paparcone R, Sandler SJ, Woldringh CL, Wong WP, and Jun S (2012). Physical manipulation of the Escherichia coli chromosome reveals its soft nature. Proc. Natl. Acad. Sci. USA 109, E2649–E2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira MCF, Brackley CA, Lintuvuori JS, Marenduzzo D, and Orlandini E (2017). Entropic elasticity and dynamics of the bacterial chromosome: A simulation study. J. Chem. Phys 147. [DOI] [PubMed] [Google Scholar]
- Pinho MG, Kjos M, and Veening J-W (2013). How to get (a)round: mechanisms controlling growth and division of coccoid bacteria. Nat. Rev. Micro-biol. 11, 601–614. [DOI] [PubMed] [Google Scholar]
- Podgornik R, Aksoyoglu MA, Yasar S, Svenšek D, and Parsegian VA (2016). DNA equation of state: In vitro vs in Viro. J. Phys. Chem. B 120, 6051–6060. [DOI] [PubMed] [Google Scholar]
- Pogliano J, Osborne N, Sharp MD, Abanes-De Mello A, Perez A, Sun YL, and Pogliano K (1999). A vital stain for studying membrane dynamics in bacteria: a novel mechanism controlling septation during Bacillus subtilis sporulation. Mol. Microbiol 31, 1149–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogliano J, Sharp MD, and Pogliano K (2002). Partitioning of chromosomal DNA during establishment of cellular asymmetry in Bacillus subtilis. J. Bacteriol 184, 1743–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purohit PK, Kondev J, and Phillips R (2003). Mechanics of DNA packaging in viruses. Proc. Natl. Acad. Sci. USA 100, 3173–3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raspaud E, da Conceiçao M, and Livolant F (2000). Do free DNA counterions control the osmotic pressure? Phys. Rev. Lett 84, 2533–2536. [DOI] [PubMed] [Google Scholar]
- Rigort A, Bäuerlein FJ, Villa E, Eibauer M, Laugks T, Baumeister W, and Plitzko JM (2012). Focused ion beam micromachining of eukaryotic cells for cryoelectron tomography. Proc. Natl. Acad. Sci. USA 109, 4449–4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas E, Theriot JA, and Huang KC (2014). Response of Escherichia coli growth rate to osmotic shock. Proc. Natl. Acad. Sci. USA 111, 7807–7812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryter A (1965). MORPHOLOGIC STUDY OF THE SPORULATION OF BACILLUS SUBTILIS. Ann. Inst. Pasteur (Paris) 108, 40–60. [PubMed] [Google Scholar]
- Sandman K, Losick R, and Youngman P (1987). Genetic analysis of Bacillus subtilis spo mutations generated by Tn917-mediated insertional mutagenesis. Genetics 117, 603–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp MD, and Pogliano K (1999). An in vivo membrane fusion assay implicates SpoIIIE in the final stages of engulfment during Bacillus subtilis sporulation. Proc. Natl. Acad. Sci. USA 96, 14553–14558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp MD, and Pogliano K (2002). Role of cell-specific SpoIIIE assembly in polarity of DNA transfer. Science 295, 137–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K, Bayer ME, and Youngman P (1993). Physical and functional characterization of the Bacillus subtilis spoIIM gene. J. Bacteriol 175, 3607–3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DE, Tans SJ, Smith SB, Grimes S, Anderson DL, and Bustamante C (2001). The bacteriophage straight phi29 portal motor can package DNA against a large internal force. Nature 413, 748–752. [DOI] [PubMed] [Google Scholar]
- Smith MB, Li H, Shen T, Huang X, Yusuf E, and Vavylonis D (2010). Segmentation and tracking of cytoskeletal filaments using open active contours. Cytoskeleton 67, 693–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan IS, and Ramamurthi KS (2014). Spore formation in Bacillus subtilis. Environ. Microbiol. Rep 6, 212–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, and Ludtke SJ (2007). EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol 157, 38–46. [DOI] [PubMed] [Google Scholar]
- Taylor CV (1923). The contractile vacuole in Euplotes: An example of the solgel reversibility of cytoplasm. J. Exp. Zool 37, 259–289. [Google Scholar]
- Tocheva EI, López-Garrido J, Hughes HV, Fredlund J, Kuru E, Vannieu-wenhze MS, Brun YV, Pogliano K, and Jensen GJ (2013). Peptidoglycan transformations during Bacillus subtilis sporulation. Mol. Microbiol 88, 673–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursell TS, Nguyen J, Monds RD, Colavin A, Billings G, Ouzounov N, Gitai Z, Shaevitz JW, and Huang KC (2014). Rod-like bacterial shape is maintained by feedback between cell curvature and cytoskeletal localization. Proc. Natl. Acad. Sci. USA 111, E1025–E1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa E, Schaffer M, Plitzko JM, and Baumeister W (2013). Opening windows into the cell: focused-ion-beam milling for cryo-electron tomography. Curr. Opin. Struct. Biol 23, 771–777. [DOI] [PubMed] [Google Scholar]
- Webb CD, Teleman A, Gordon S, Straight A, Belmont A, Lin DC, Grossman AD, Wright A, and Losick R (1997). Bipolar localization of the replication origin regions of chromosomes in vegetative and sporulating cells of B. subtilis. Cell 88, 667–674. [DOI] [PubMed] [Google Scholar]
- Whatmore AM, and Reed RH (1990). Determination of turgor pressure in Bacillus subtilis: a possible role for K+ in turgor regulation. J. Gen. Microbiol 136, 2521–2526. [DOI] [PubMed] [Google Scholar]
- Wu LJ, and Errington J (1994). Bacillus subtilis SpoIIIE protein required for DNA segregation during asymmetric cell division. Science 264, 572–575. [DOI] [PubMed] [Google Scholar]
- Wu LJ, and Errington J (1997). Septal localization of the SpoIIIE chromo-some partitioning protein in Bacillus subtilis. EMBO J. 16, 2161–2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu LJ, and Errington J (1998). Use of asymmetric cell division and spoIIIE mutants to probe chromosome orientation and organization in Bacillus subtilis. Mol. Microbiol 27, 777–786. [DOI] [PubMed] [Google Scholar]
- Yen Shin J, Lopez-Garrido J, Lee S-H, Diaz-Celis C, Fleming T, Bustamante C, and Pogliano K (2015). Visualization and functional dissection of coaxial paired SpoIIIE channels across the sporulation septum. eLife 4, e06474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young KD (2006). The selective value of bacterial shape. Microbiol. Mol. Biol. Rev 70, 660–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngman P, Perkins JB, and Losick R (1984). A novel method for the rapid cloning in Escherichia coli of Bacillus subtilis chromosomal DNA adjacent to Tn917 insertions. Mol. Gen. Genet 195, 424–433. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.