Summary
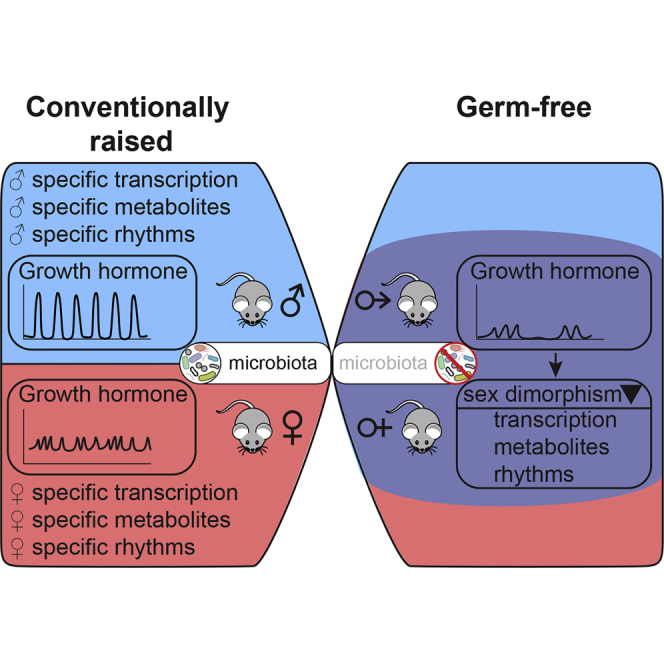
The circadian clock and associated feeding rhythms have a profound impact on metabolism and the gut microbiome. To what extent microbiota reciprocally affect daily rhythms of physiology in the host remains elusive. Here, we analyzed transcriptome and metabolome profiles of male and female germ-free mice. While mRNA expression of circadian clock genes revealed subtle changes in liver, intestine, and white adipose tissue, germ-free mice showed considerably altered expression of genes associated with rhythmic physiology. Strikingly, the absence of the microbiome attenuated liver sexual dimorphism and sex-specific rhythmicity. The resulting feminization of male and masculinization of female germ-free animals is likely caused by altered sexual development and growth hormone secretion, associated with differential activation of xenobiotic receptors. This defines a novel mechanism by which the microbiome regulates host metabolism.
Keywords: circadian clock, microbiota, liver, sexual dimorphism, growth hormone, ghrelin, sexual maturation, symbiosis, Bcl6
Graphical Abstract
Highlights
-
•
The microbiome is required for sexual dimorphism in gene expression and metabolism
-
•
Most already-described changes in GF mice are hallmarks of a feminized metabolism
-
•
Altered sexual maturation and GH secretion cause the damping of sexual dimorphism
-
•
Microbiota-derived metabolites and ghrelin likely drive these alterations
Physiology is dynamic over the day and different between sexes. Weger et al. show that the microbiome play a key role in sustaining these sex differences in gene expression and metabolism by ensuring proper sexual maturation and growth hormone secretion. Microbiota-derived metabolites and ghrelin likely drive these sexually dimorphic dynamics.
Introduction
Obesity-related metabolic diseases cause major public health problems. Dysregulation of the microbiome (Winer et al., 2016) and the circadian clock have been implicated in the progression of these diseases (Laermans and Depoortere, 2016). Many physiological processes, including fatty acid (FA) and bile acid (BA) metabolism, as well as cytokine and corticosteroid secretion, are regulated by both circadian rhythms and microbiota. Remarkably, the composition and metabolic activity of commensal bacteria show diurnal variations that depend on the circadian clock (Liang et al., 2015, Thaiss et al., 2014, Voigt et al., 2016), feeding rhythms (Thaiss et al., 2014, Thaiss et al., 2016, Voigt et al., 2014, Zarrinpar et al., 2014), and the nutritional composition of the diet (Leone et al., 2015, Voigt et al., 2014, Zarrinpar et al., 2014). Vice versa, microbiota can feed back on clock gene expression in the host. Indeed, germ-free (GF) or antibiotic-treated mice show disturbed circadian clock gene expression in liver and intestine (Björkholm et al., 2009, Joyce et al., 2014, Leone et al., 2015, Montagner et al., 2016, Mukherji et al., 2013). Moreover, conditions altering the composition of the microbiome, such as high-fat diet (HFD) (Hildebrandt et al., 2009), obesity (Ley et al., 2005), and Roux-en-Y gastric bypass surgery (Tremaroli et al., 2015) also affect host circadian clock gene expression (Ando et al., 2011, Kim et al., 2015, Kohsaka et al., 2007). However, effects of microbiome on the host circadian clock in peripheral tissues show striking inconsistencies.
To further study the relationship between the host-microbiota relationship and the circadian clock, we used RNA sequencing (RNA-seq) of conventionally raised (ConvR) and GF mice to compare temporal gene expression in liver, duodenum, ileum, and perigonadal white adipose tissue (WAT). We found that the molecular clock was globally unaffected, but genes involved in key metabolic processes exhibited an altered rhythmic expression in GF mice. Strikingly, we observed a feminization of gene expression (i.e., downregulation of male-biased and upregulation of female-biased genes) in the liver and WAT of GF male mice. In females, our analysis confirmed an attenuation of sexually dimorphic rhythmic gene expression and metabolic activities in GF mice. This was a consequence of altered sex-hormone and growth hormone (GH) signaling, likely due to the defective sexual maturation of GF mice. These results highlight the key role of the microbiota on the establishment of sexually dimorphic liver metabolism and tentatively elucidate several unexplained phenotypes of GF mice that are also known to be sexually dimorphic, such as resistance to liver cancer (Grant and Roe, 1969), xenobiotic detoxification (Carmody and Turnbaugh, 2014), metabolic differences (Nieuwdorp et al., 2014), and poor reproduction (Shimizu et al., 1998).
Results
Global Alteration of Gene Expression in the Digestive Tract of GF Mice
To study the impact of microbiota on host gene expression, we compared temporal gene expression in the liver, duodenum, ileum, and WAT of GF and ConvR male mice (Figure S1A). The transcriptomes of GF and ConvR could be distinguished in the liver, duodenum, and ileum but less so in WAT (Figure S1B). We first analyzed constitutive changes between GF and ConvR mice in the different tissues. In the duodenum and ileum of GF mice, most affected genes displayed a significant decrease in expression (Figure S1C and Table S1). Specifically, genes associated with immune response and immune cell mobility were downregulated in the intestine of GF mice (Figure S1D). In the liver, we detected a significant alteration for genes involved in lipid and cholesterol metabolism, confirming the role of microbiota in these pathways (Joyce et al., 2014). In WAT, genes related to immune response were downregulated, consistent with previous reports (Caesar et al., 2015).
Microbiota Depletion Alters Rhythmic Gene Expression
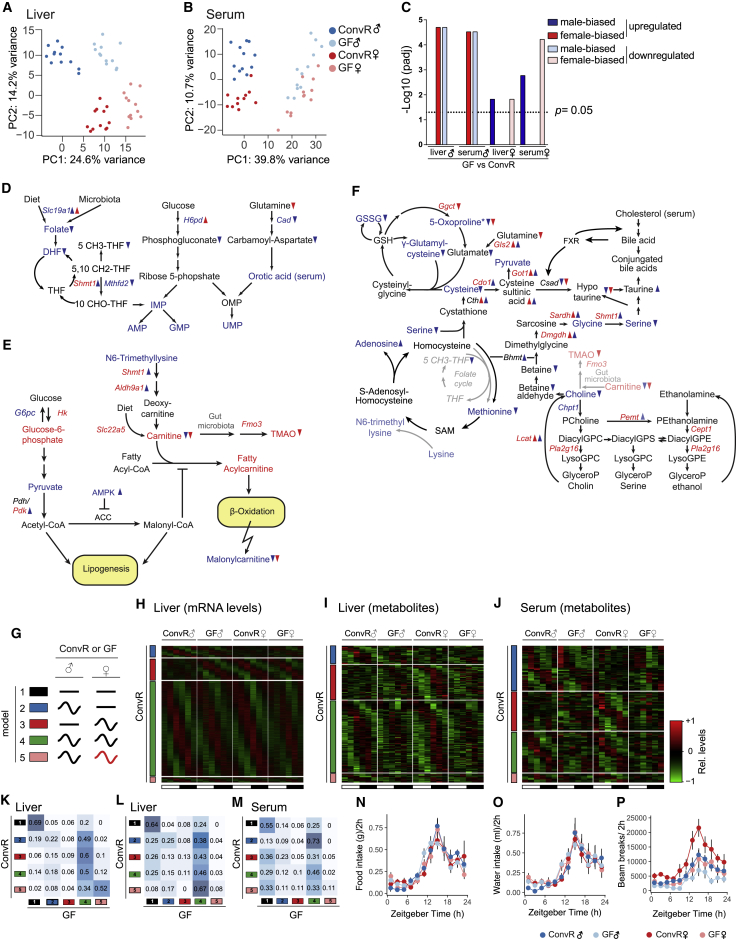
Next, we quantified rhythmic gene expression in ConvR and GF mice (see STAR Methods and (Atger et al., 2015)) (Figure 1A and Table S1). The analysis revealed that most rhythmic transcripts are unaffected (model 4). Interestingly, a higher proportion of genes gained rather than lost rhythmicity in the duodenum and ileum of GF mice (model 2). In the liver and WAT, this ratio was reversed (Figures 1B and 1C). Circadian clock and clock output genes showed only subtle changes in phase and amplitude of mRNA abundance across tissues (Figures 1D and S1E–S1H). Most of the genes that lost rhythmicity in the duodenum of GF mice were associated with ribosome biogenesis, a rhythmically orchestrated process in the liver (Sinturel et al., 2017, Wang et al., 2017), and glutamine metabolism, reported to correlate with colonization of the intestine (El Aidy et al., 2013) (Table S2). In the ileum, the genes that lost rhythmicity were involved in the organization of the extracellular matrix, potentially reflecting its rhythmic regeneration after its degradation by contacts with bacteria (Thaiss et al., 2016). Moreover, genes involved in carbohydrate metabolism gained rhythmicity in the ileum of GF mice, suggesting that microbiota, known to regulate glucose transport and metabolism (Donohoe et al., 2012), dampen rhythmic glucose metabolism in ConvR mice. In WAT of GF mice, the loss of rhythmicity in genes involved in cytokine signaling might reflect microbiota-dependent rhythmic inflammatory processes (Caesar et al., 2015). Meanwhile, genes associated with deoxyribonucleotide metabolism, a process showing increased activity during gut colonization (El Aidy et al., 2013), gained rhythmicity in GF mice.
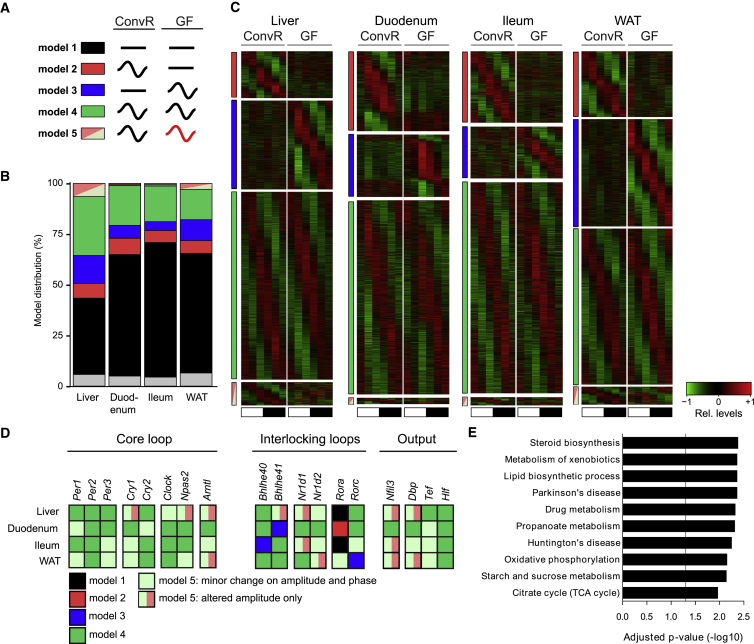
Figure 1.
Gut Microbiota Alters Rhythmic Gene Expression in Peripheral Organs but Mildly Affects Clock Gene Expression
(A) Alteration of rhythmic gene expression in male GF (GF♂) mice is assessed by model selection (model 1–5): black line, stable transcription; black wave, rhythmic transcription; red wave, rhythmic profiles with different rhythmic parameters (i.e., phase and/or amplitude).
(B) Model distribution of all expressed genes in the examined organs. Coloring indicates the corresponding models as shown in (A); gray, genes not assigned to any specific model.
(C) Heatmaps of normalized mRNA levels in male ConvR (ConvR♂) and GF♂ mice (red, high expression; green, low expression). Colored bars indicate the corresponding models. Black and white bars represent light conditions.
(D) The assignment of circadian clock genes to their corresponding models shows that their temporal profiles are only mildly affected.
(E) Liver genes with altered rhythmicity (models 2, 3, and 5) in GF mice are associated with metabolic and steroidogenic pathways and neurological disorders.
GF livers showed a loss of rhythmicity in genes involved in chromatin assembly and nucleosome assembly, including several histone and MCM proteins. These genes are induced during the S phase of the cell cycle (Costa et al., 2013) and during liver regeneration after partial hepatectomy (Huang et al., 2015), potentially explaining defective liver regeneration in GF mice (Cornell et al., 1990). Interestingly, several liver transcription factors (TFs) gained rhythmic expression in GF mice, including ChREBP (encoded by the Mlxipl gene), which could contribute to an increased number of rhythmic transcripts in these animals. Genes involved in polysaccharide, amino acid (AA), lipid, and steroid metabolism, in addition to drug metabolism, lost rhythmicity in GF animals (Figure 1E). Several of these biological functions are modulated in livers of GF mice (Joyce et al., 2014, Kübeck et al., 2016, Selwyn et al., 2015, Thaiss et al., 2016). Strikingly, the same functions are sexually dimorphic in mouse liver (Lichanska and Waters, 2008, Yang et al., 2006), suggesting the involvement of bacteria colonization in maintenance of sexual dimorphism in liver.
Sex-Specific Gene Expression Is Damped in GF Mice
To analyze altered sexually dimorphic gene expression in GF mice, we first defined hepatic female- and male-biased genes (STAR Methods and Table S3) and then studied their differential expression in ConvR and GF mice (Figure 2A). Remarkably, the genes upregulated in the liver of GF male mice were significantly enriched in female-biased genes, while those downregulated were enriched in male-biased genes (Figures 2B and 2D). The inverse observation in females confirmed an attenuation of sex-biased gene expression in livers of GF mice (Figures 2C and 2D). To substantiate this finding, we reanalyzed published data using GF and ConvR males under conventional chow diet (Leone et al., 2015) or HFD (Rabot et al., 2010) (Figures S2A, S2B, and S2F). Both datasets confirmed that male livers were feminized in the absence of microbiota. We also compared these sexually dimorphic gene expression signatures with additional conditions known to alter gut microbiota. When examining mice that grew up on an HFD (Cox et al., 2014) and were then subjected to vertical sleeve gastrectomy (VSG) (Myronovych et al., 2014), or fed with resistant starch (Kieffer et al., 2016), we found that HFD (Ley et al., 2005) led to a feminization of hepatic gene expression (Figures S2C and S2F). Both VSG and resistant starch feeding change gut microbiota composition (Kieffer et al., 2016, Tremaroli et al., 2015) and reverse the HFD-induced feminization of obese animals (Figures S2D–S2F). The observed feminization was not specific to the liver, as the sexually dimorphic WAT (Yang et al., 2006) in male GF animals also showed a feminized expression pattern (Figures S2G and S2H).
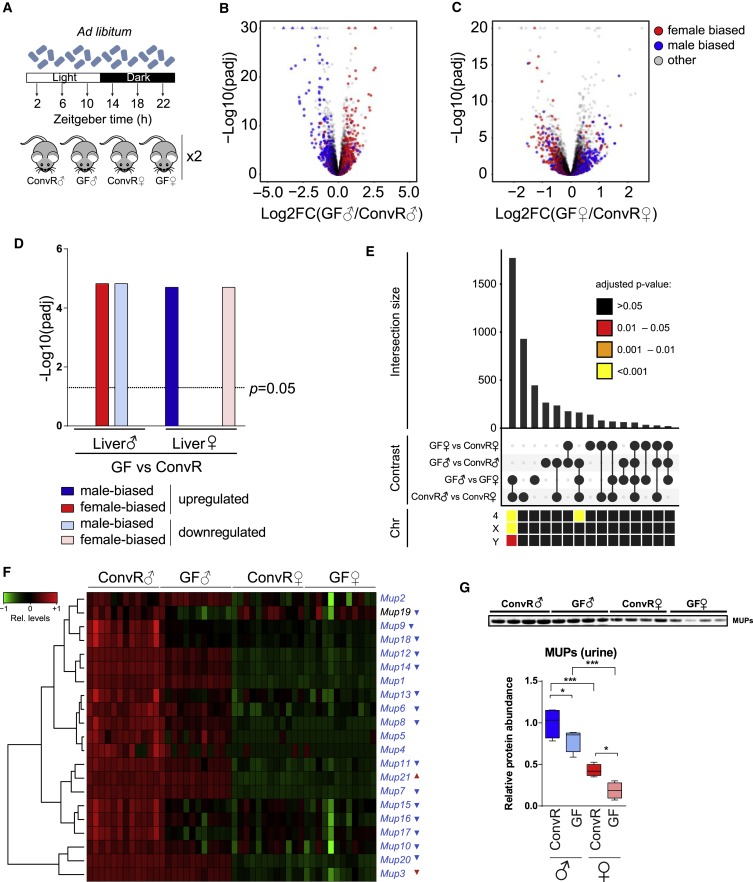
Figure 2.
A Lack of Gut Microbiota Decreases Sex-Specific Gene Expression by Feminizing Male and Masculinizing Female Hepatic Gene Expression
(A) Experimental design to detect sex-specific changes.
(B) Volcano plot of differences in mRNA expression between ConvR♂ and GF♂ categorized by sex-biased expression in ConvR mice (ConvR♂ versus ConvR female [ConvR♀]). Colors indicate male-biased (blue), female-biased (red), and unbiased (gray) expressed genes.
(C) Volcano plot of expression difference in mRNA expression between female GF (GF♀) and ConvR♀ mice. Color code is identical to (B).
(D) Statistical analysis of male- and female-biased genes showing a significant inversion of sex-biased genes toward the opposite sex in GF mice.
(E) Number of the shared differentially expressed genes (intersections size) between the four indicated contrasts. Lower heatmap indicates the enrichment for chromosomal location (Chr) for the corresponding intersection.
(F) Expression changes of differentially expressed Mup genes in GF♂ mice as assessed by RNA-seq. Most Mup genes are downregulated in GF♂ mice. The highest expressed Mup in female liver (Mup3) is downregulated in GF♀ mice. Colored gene name indicates unbiased (black) and male-biased (blue) expression. Triangle indicates the direction of significant change in GF♂ (blue) and GF♀ (red) mice in comparison with ConvR.
(G) Protein levels of MUP in the urine of GF♂ and GF♀ mice are significantly downregulated in male and female mice compared with their ConvR counterparts (each Tukey boxplot represents four independent biological replicates per condition). ∗p ≤ 0.05, ∗∗∗p ≤ 0.001.
To identify mechanisms involved in the establishment of sexual dimorphism by gut microbiota, we wondered whether sex-specific microbiota could explain sexually dimorphic gene expression. We profiled fecal microbiota of male and female mice during sex-specific cohousing and after a 2-week period when beddings across all cages were daily mixed. While beta diversity during sex separated housing conditions showed some statistical differences between male and female, individual variation was the main driver of this diversity (Figures S2I and S2J). Indeed, no statistical difference in the individual microbiota composition was detectable after housing the animals with the same bedding material, confirming that cohousing of the animals has a major impact on gut microbiota composition and diversity. Therefore, it is likely that the complete lack of gut microbiota, rather than sex difference in microbiota, alters sex-dimorphic gene expression.
Genes on sex chromosomes were not likely involved in the attenuation of sexual dimorphism since the sex-biased genes localized on gonosomes (about 40%) were unchanged between ConvR and GF conditions (Figure 2E). Sex-specific genes were also enriched on chromosome 4, where the male-specific genes encoding mouse urinary proteins (MUPs) are located in two clusters (Bishop et al., 1982). Concordantly, the vast majority of Mup genes (Figure 2F) and MUP proteins in urine displayed reduced levels in both GF males and females (Figure 2G). Among the biological functions associated with sexually dimorphic genes altered in GF mice, lipid, cholesterol, and steroid metabolism, as well as xenobiotic detoxification, were the most prevalent (Table S4).
Characterization of Signaling Pathways Involved in Damping of Liver Sexual Dimorphism in GF Mice
To identify TFs regulating the observed attenuation of sexual dimorphism in GF mice, we performed TF-binding site analysis near (±500 bp) transcription start sites (TSSs) of sexually dimorphic genes (STAR Methods). Among the predicted TF activities showing differences between ConvR males and females, we focused on those with reduced sex-differential activity in GF animals (Table S5). This identified the known sexually dimorphic GH-STAT5/BCL6 pathway and the associated TFs CUX2 and FOXA2 (Sugathan and Waxman, 2013), nuclear respiratory factor 1 (NRF1) and 2 (NRF2 also known as GABP, a member of the ETS family of TFs), aryl hydrocarbon receptor (AHR), and androgen receptor (AR) (Figure S3A). Testosterone activates AR and thereby induces sex-specific postnatal DNA demethylation in mouse liver, which plays an important role in epigenetic imprinting of sexual dimorphism in the liver (Reizel et al., 2015). Interestingly, DNA binding of NRF1 and GABP strongly depends on DNA methylation in mouse embryonic stem cells (Domcke et al., 2015), which potentially explains sex- and methylation-dependent transcriptional activity of NRF1 and GABP (Yokomori et al., 1995). Likewise, AHR transcriptional activation is sex-biased (Lee et al., 2015), inhibits GH signaling (Nukaya et al., 2004), and initiates crosstalk with sex-hormone signaling through direct interaction with estrogen receptor (ER) (Ohtake et al., 2011). While levels of 17β-estradiol (E2) have not been previously measured in GF animals, sexually dimorphic circulating testosterone is strongly blunted in GF animals (Markle et al., 2013, Yurkovetskiy et al., 2013), potentially explaining the differential activation of these TFs. To confirm such deregulation of sex hormones, we measured testosterone and E2 in serum from ConvR and GF males and females. We found that, in addition to testosterone, sexual dimorphism in E2 levels was also blunted, suggesting a potential role of sex hormones in the loss of sexually dimorphic activation of signaling pathways in GF mice (Figures 3A and 3B).
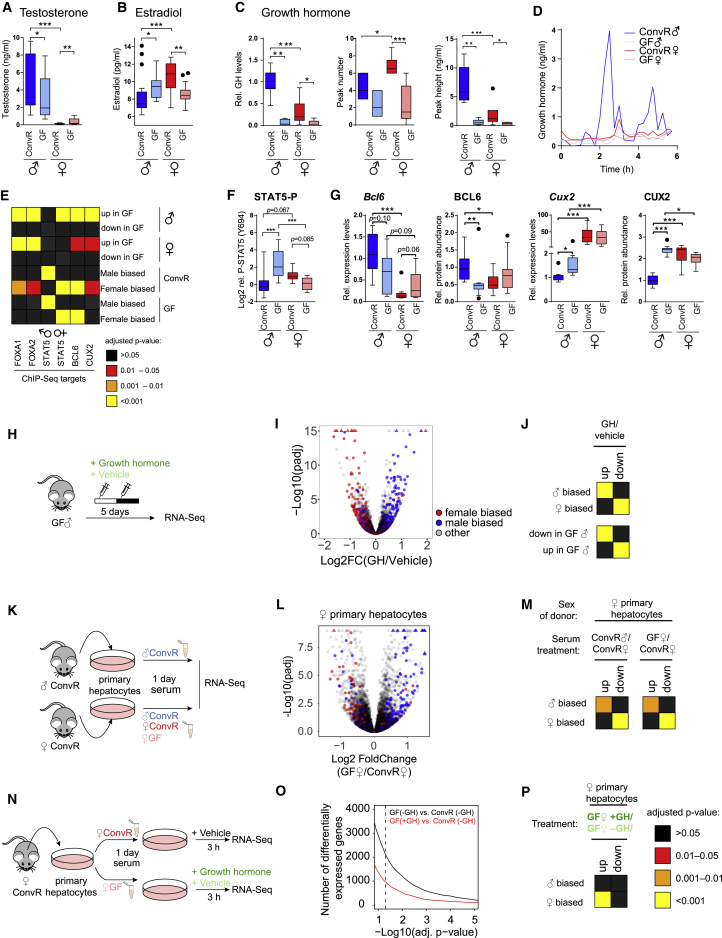
Figure 3.
Sex Hormones and the GH Signaling Network Are Perturbed in GF Mice
(A and B) Serum levels of testosterone (A) and E2 (B) in male and female ConvR and GF mice.
(C) Circulating GH total secretion, peak number, and levels in male and female ConvR and GF mice.
(D) Representative examples of temporal profiles of GH secretion in ConvR♂, GF♂, ConvR♀, and GF♀ animals.
(E) Enrichment analysis of FOXA1, FOXA2, BCL6, CUX2, and STAT5 (male and female specific) target genes for differentially expressed genes of indicated contrasts (♂ = GF♂ versus ConvR♂; ♀ = GF♀ versus ConvR♀; ConvR = ConvR♂ versus ConvR♀; GF = GF♂ versus GF♀). Target genes have been identified by published chromatin immunoprecipitation sequencing (ChIP-seq) data.
(F) Phosphorylation of STAT5 (Tyr694) in male and female ConvR and GF mice.
(G) Hepatic mRNA and protein levels of the other GH-dependent TFs BCL6 and CUX2 that are involved in sex-dimorphic gene expression for the indicated conditions.
(H) Experimental design to mimic male-specific GH pulses in GF mice.
(I) Volcano plot of expression difference in mRNA expression between GH-injected and vehicle-injected GF male mice show a rescue in sex-biased gene expression.
(J) Enrichment analysis of male- and female-specific genes (♂ biased, ♀ biased) or repressed or induced genes in GF mice (down or up in GF♂) for differentially expressed genes of the contrast GH- versus vehicle-injected GF♂ mice.
(K) Experimental design of an in vitro system of primary hepatocytes treated with sera of different sources (ConvR♂, ConvR♀, and GF♀).
(L) Volcano plot of expression difference in mRNA expression of female hepatocytes treated with GF♀ serum versus ConvR♀ serum. Color code corresponds to (B).
(M) Enrichment analysis of male- and female-specific genes (♂ biased, ♀ biased) for differentially expressed genes of the indicated contrasts (serum treatment: ConvR♂ versus ConvR♀ or GF♀ versus ConvR♀) in female hepatocytes.
(N) Cartoon depicting the experimental design to evaluate the effect of a tonic GH treatment.
(O) Number of differentially expressed genes in function of an adjusted p value threshold for the indicated contrast. GH treatment diminished the number of differentially expressed genes between female hepatocytes treated with GF♀ serum and ConvR♀ serum.
(P) Enrichment analysis of male- and female-specific genes (♂ biased, ♀ biased) for differentially expressed genes between GH-treated versus untreated female hepatocytes.
All boxplots represent at least 12 independent biological replicates per condition, except for (G), for which at least six biological replicates were used. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001. All boxplots are Tukey boxplots.
Liver sexual dimorphism is mainly regulated through the secretory pattern of GH by the pituitary male-specific pulsatile secretion versus more continuous secretion in females. This difference leads to sex-specific activation of GH signaling, as reflected in the activation and phosphorylation of the STAT5 TF, namely high and transient phosphorylation in males versus low and constant phosphorylation in females (Lichanska and Waters, 2008, Zhang et al., 2012). We envisioned that differential GH-STAT5 activation might underlie the decreased liver sexual dimorphism of GF mice. Thus, we first measured GH receptor (GHR) expression, which showed no significant differences in ConvR versus GF animals at protein levels and only a slight reduction in expression in females at the mRNA level (Figures S3B and S3C). We therefore speculated that gut microbiota could affect sexually dimorphic GH secretion. Surprisingly, both GF males and females secreted low levels of GH, and GF females displayed a male-like temporal secretion pattern, which was characterized by a more pulsatile secretion with a lower number of peaks (Figures 3C and 3D). This disturbed GF secretion therefore likely contributes to the damped sexual dimorphism of the liver.
We next studied downstream signaling pathways and measured the enrichment of male- and female-specific STAT5 target genes (Zhang et al., 2012) among up- and downregulated genes in GF mice. We found a significant enrichment of STAT5-regulated genes among upregulated genes in GF males, which coincided mainly with genes usually expressed in female liver and enriched for female STAT5 sites (Figure 3E). We further observed a characteristic bursty activation pattern of STAT5 in male ConvR mice, presumably reflecting male-specific GH secretion. Moreover, while GF males displayed a female-like baseline phosphorylation, GF females exhibited constant low phosphorylated STAT5, confirming the overall altered sexually dimorphic STAT5 activity (Figures 3F and S3D).
To establish causality between the observed altered GH secretion and the attenuated sexual dimorphism in GF mice, we mimicked male-specific GH pulses in GF males with two daily GH injections (Jarukamjorn et al., 2006) (Figure 3H). This treatment globally restored sexually dimorphic liver gene expression (Figures 3I, 3J, and S3G). We evaluated the effect of GH in females in vitro by incubating primary hepatocytes from female mice with serum from female (ConvR and GF) and male (ConvR) mice in the presence or absence of constant level of GH. This in vitro approach recapitulated the in vivo observation that sexual dimorphism is altered in GF mice, confirming that circulating factors play a key role (Figures 3K–3M and S3H). In addition, continuous treatment with GH mitigated the masculinized gene expression profile of female hepatocytes treated with GF female serum, establishing causality between altered GH secretion and loss of sexual dimorphism in GF female mice (Figures 3N–3P and S3I).
Regulation of Bcl6 Transcription through GH-Regulated Antisense Transcription
The male-biased repressor BCL6 and the female-biased activators FOXA1 (HNF3α), FOXA2 (HNF3β), ONECUT2 (HNF6β), and repressor CUX2 also play a key role in sexually dimorphic gene expression (Conforto et al., 2015, Melia et al., 2016, Sugathan and Waxman, 2013, Zhang et al., 2012). Among these TFs, Bcl6 and Cux2 show the most pronounced sexually dimorphic expression and are altered in GF mice at both mRNA and protein levels (Figures 3G, S3E, and S3F). Binding sites for these TFs were enriched in sex-biased genes that are upregulated in GF mice (Figure 3E). Among them, BCL6, in association with STAT5, was the best predictor for the decreased sexual dimorphism of GF mice. Bcl6 expression is regulated by GH at the transcriptional elongation level (Meyer et al., 2009). Quantification of the transcripts at the Bcl6 locus revealed, surprisingly, two non-annotated antisense (AS) transcripts, one starting at the TSS (as-2) and one inside the gene body (as-1) (Figure 4A). These two transcripts overlapped with the previously described highly transcribed region of the gene in female, regulated by GH (Meyer et al., 2009). In fact, all sense intronic and exonic signals were correlated across the different conditions. However, while as-2 was not correlated with the sense signals, as-1 was anti-correlated with Bcl6 exons and introns (Figures 4B and 4C), suggesting that the as-1 transcript interferes with the elongation of the sense transcript, potentially by modulating the sense transcript dynamics by AS transcription (Brown et al., 2018). Analysis of chromatin states near the as-1 TSS revealed the presence of a cryptic promoter (Figure S3J). The 3′ end of the as-1 transcript coincided with a male-specific DNase I hypersensitive site and STAT5 binding site, which disappeared under chronic GH treatment mimicking female GH secretion (Figure S3J). Interestingly, STAT5 binding to the Bcl6 gene represses its transcription (Lin et al., 2014), potentially explaining the repression of as-1 in males and the inhibition of Bcl6 elongation by GH (Meyer et al., 2009).
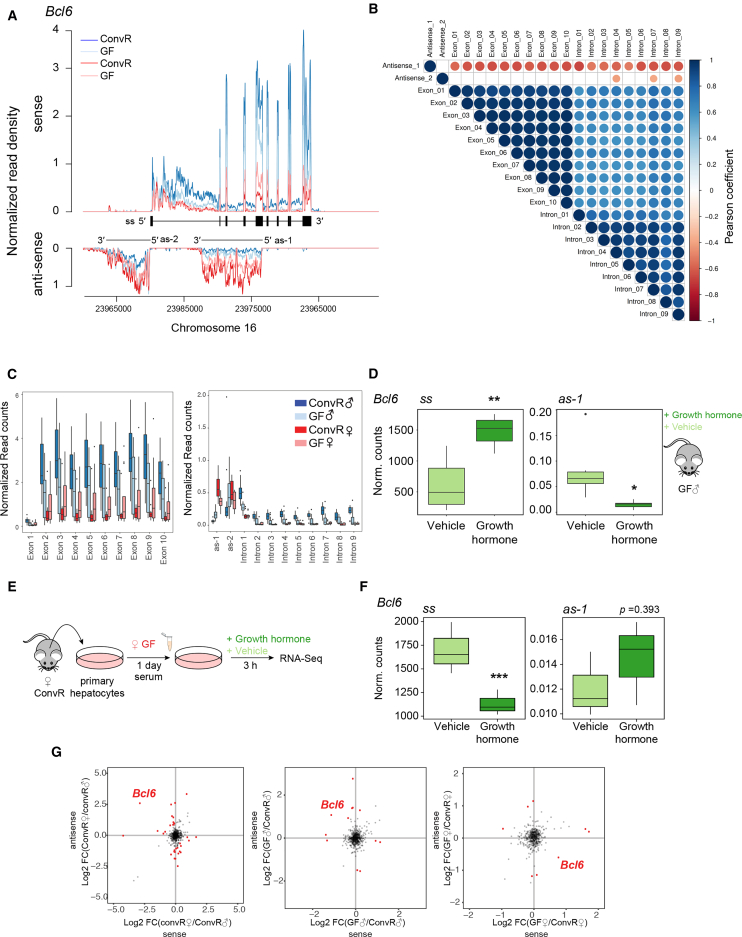
Figure 4.
Expression of the Male-Specific Repressor Bcl6 Anti-correlates with a GH-Responsive AS Transcript
(A) RNA-seq read mapping to the Bcl6 gene locus. The tracks are color coded for the indicated conditions. The log2 normalized read count versus chromosomal position is plotted across the y axis for sense (upper part) and AS (lower part) transcription. The schematic represents exons and introns of Bcl6 (ss) and the two AS transcripts (as-1, as-2).
(B) Correlation matrix based on Pearson's rank correlation coefficient test between the expression levels of the indicated exon, intron, and AS transcript. Colors indicate positive (blue) and negative (red) correlation. Size of the circles correspond to more significant p values, only correlation with an adjusted p value lower than 0.05 are depicted with a circle.
(C) Normalized read counts per exon (upper graph), intron, and AS transcript (lower graph) of the Bcl6 gene locus in liver of ConvR♂ (dark blue), ConvR♀ (dark red), GF♂ (light blue), and GF♀ mice (light red).
(D) Normalized read counts of GH-treated and vehicle-injected GF♂ mice for sense (left) and AS-1 transcript (right).
(E) Experimental design.
(F) Normalized read counts of GH- and vehicle-treated female hepatocytes under GF♀ serum incubation for sense (left) and antisense-1 transcript (right).
(G) Changes in sense and AS transcript levels between ConvR♂ versus ConvR (left), GF♂ versus ConvR♂ (middle), and GF♀ versus ConvR♀ (right). Significantly upregulated and downregulated genes on the level of Δsense–Δanti-sense are shown in red (false discovery rate<0.01 and logFCinteraction > 1). Gene names are depicted for genes with opposite behavior in the x and y axis (logFCx > 0.5 and logFCy < −0.5) or (logFCx < −0.5 and logFCy > 0.5). Globally, only Bcl6 showed robust differences between sense and AS transcript levels across all conditions indicating that a sex-specific AS transcript that potentially regulates sex-biased expression of a gene is unique.
All boxplots represent at least 12 independent biological replicates per condition, except for (F) for which three biological replicates were used. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001. All boxplots are Tukey boxplots.
GH injection in GF male mice decreased as-1 Bcl6 transcript, whereas the sense transcript increased its expression (Figure 4D). Conversely, female hepatocytes treated with GF female serum decreased sense transcript levels upon GH treatment, while low as-1 expression was unaltered. These results demonstrate the regulation of Bcl6 by GH and indicate an involvement of as-1 transcript in the regulation of Bcl6 sense transcript (Figures 4E and 4F). The sex-specific regulation of the as-1 transcript thus potentially provides a novel mechanism regulating sexually dimorphic gene expression. As such internal AS transcripts are quite common in mouse, we sought for additional similarly regulated sexually dimorphic genes in the liver. However, no additional transcripts showing similar sexually dimorphic sense/AS transcript levels were found, making Bcl6 unique among sexually dimorphic genes (Figure 4G).
Impact of Microbiota on the Sexually Dimorphic Metabolome
We next analyzed the impact of microbiota on sex-dimorphic metabolomes in liver and serum from ConvR and GF male and female mice. The observed sexual dimorphism in metabolite levels was markedly decreased in GF conditions (Figures 5A and 5B); notably, female-biased metabolites were upregulated in GF males and male-biased metabolites downregulated in both liver and serum (Figures 5C and S4A). Sexual dimorphism was also diminished in GF females, albeit to a lower extent (Figures 5C and S4B). To identify metabolic pathways with a microbiota-dependent sex-biased profile, we calculated a proxy for metabolic pathway activity and tested for statistical differences across all conditions (Table S6). This identified three clusters of interconnected metabolic pathways. The first cluster involves folate (vitamin B9), purine, and pyrimidine metabolism (Figures 5D and S4D). Folate and its metabolite DHF showed higher levels in ConvR male than in female mice, which was attenuated in GF condition. The male-biased concentration correlated with male-biased mRNA expression of the folate transporter SLC19A1 (RFC1) and the key enzyme for purine metabolism MTHFD2 (methylenetetrahydrofolate dehydrogenase and cyclohydrolase) (Ben-Sahra et al., 2016). Testosterone might be the driving force in establishing sexual dimorphism for folate metabolism, as it regulates MTHFD2 (Rovinetti et al., 1972). Microbiota is an important supplier of de novo synthesized folate to the host (LeBlanc et al., 2013), which likely led to the lower levels of folate and DHF in GF male mice. Increased concentrations in purine (inosine monophosphate, AMP, and guanosine monophosphate [GMP]) and pyrimidine (orotic acid and uridine monophosphate [UMP]) derivatives in ConvR male mice were slightly affected in absence of gut microbiota. The trifunctional key enzyme CAD (carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase) for pyrimidine synthesis (Ng et al., 2015) was expressed in a male-biased manner, potentially leading to the increased UMP concentration in males. Moreover, the known activation of the pentose phosphate pathway by testosterone (Kelly and Jones, 2013) likely increases the synthesis of ribose and boosts AMP, GMP, and UMP synthesis.
Figure 5.
Gut Microbiota Modulates the Sex-Dimorphic Metabolome
(A and B) Principal-component analysis score plots of mass spectroscopy-based metabolic profiling of male (♂) and female (♀) ConvR and GF mice in liver (A) and serum (B).
(C) Enrichment analysis of sex-biased metabolites in the upregulated (dark blue and red) and downregulated (light blue and red) fractions of metabolites in GF animals (GF versus ConvR) in the indicated tissue (liver or serum).
(D–F) Schematic depicting regulation by sex and microbiota for folate, purine, and pyrimidine metabolism (D), glycolysis and FA oxidation (E), and the metabolic network of glutathione (GSH, methionine, PL, and BA) (F). Metabolites and genes involved in the steps where genes or metabolites showing sex-biased or microbiota-dependent changes. All gene names and metabolites are color coded for male-biased (blue), female-biased (red), and sex-unbiased (black) levels in the liver. The triangles that follow a gene or metabolite name indicate the direction of change in GF versus ConvR male (blue) and female (red) mice.
(G) Sex-biased rhythmicity in gene expression was assessed by model selection (models 1–5) in both ConvR and GF mice: black line, stable transcription; black wave, rhythmic transcription; red wave, rhythmic profiles with different rhythmic parameters.
(H) Heatmap of normalized mRNA levels in ConvR♂ and GF♂, and ConvR♀ and GF♀, across a day. Colored bars indicate the corresponding sex-specific models (2–5) in ConvR mice. Black and white bars on the bottom represent light conditions.
(I and J) Heatmaps of relative metabolite levels in liver (I) and serum (J) sorted by the sex-specific models 2–5.
(K) Fraction of genes corresponding to models 1–5 in ConvR and their assigned model in GF mice. The blue color in each box represents the fraction size of genes between ConvR and GF mice, with darkest blue corresponding to highest fraction.
(L and M) Fraction of metabolites that are assigned to specific models for ConvR and GF mice in liver (E) and serum (F).
(N–P) Feeding (N) and drinking (O) rhythms are not different between ConvR and GF mice (p > 0.05). Locomotor activity is higher activity in ConvR♀ mice (p < 0.001) but not different from males under GF conditions (P). Error bars represent SEM.
A second pathway cluster involves glycolysis and FA oxidation (Figures 5E and S4E). Females exhibited increased Hexokinases (Hks), but decreased Glucose-6-phosphatase (G6pc) mRNA levels in comparison with male animals. This opposing sexually dimorphic expression pattern correlates with increased G6P levels. However, synthesis of acetyl-coenzyme A (CoA) is inhibited by an increased expression of Pyruvate Dehydrogenase Kinases (Pdk1, Pdk2) in females due to sexually dimorphic GH signaling (Kim et al., 2012). Furthermore, AMPK inhibits such GH regulation of Pdk expression. We observed a lower phosphorylation of AMPK in females independently of microbiota (Figure S4C), probably due to significantly decreased AMP levels (Figures 5D and S4D) that might further reinforce the inhibition of acetyl-CoA production and thus de novo lipogenesis in females (Harrison and Sinnett-Smith, 1990). Decreased lipogenesis in females would be in line with the reported female-biased increase in mitochondrial activity and lipid β-oxidation (Justo et al., 2005), and potentially reflects the positive effect of GH on these activities (Katkocin et al., 1979). We used carnitine derivatives as a readout of mitochondrial activity (Koves et al., 2008). Females showed lower levels of malonylcarnitine and a global increase of acylcarnitines in comparison with males, indicating increased mitochondrial activity and β-oxidation, respectively. What might be the cause of the observed sex bias in carnitine levels? Carnitine can originate from food or de novo synthesis (Reuter and Evans, 2012). The high level of carnitine in females is likely caused by E2 regulation of the main carnitine transporter SLC22A5 (OCTN2) (Wang et al., 2012). We found key enzymes of the de novo synthesis of carnitine to be expressed in a female-biased manner, thereby reinforcing mitochondrial activity. In addition, microbiota is able to metabolize and synthesize carnitine (Reuter and Evans, 2012) and plays a role in its absorption and availability. Consistent with these reports, we detected low levels of carnitine in GF mice. Interestingly, ConvR females showed high levels of the atherosclerosis-associated metabolite trimethylamine N-oxide (TMAO), likely produced by the female-specific Flavin-containing Monooxygenase 3 (FMO3) from carnitine. Altogether, our data corroborated the reported sexual dimorphism in energy homeostasis (Mauvais-Jarvis, 2015) and highlighted the role of microbiota in this pathway.
Finally, a cluster containing glutathione (GSH), AA, phospholipid (PL), and BA metabolism was profoundly sexually dimorphic and modulated by microbiota (Figures 5F and S4F–S4I). GH affects methionine and GSH metabolism (Brown-Borg et al., 2014), explaining the sex bias in this pathway. In addition, E2 increases GSH peroxidase activity and peroxide production (Pinto and Bartley, 1969), potentially leading to the observed higher GSH oxidation in males. Interestingly, colonization of the gut was shown to influence host intestinal GSH metabolism (Mardinoglu et al., 2015), explaining the generally lower levels of metabolites in this pathway in GF mice. Sexually dimorphic AA metabolism affects taurine and BA metabolism; these pathways are subject to regulation by GH (Rudling et al., 1997) and microbiota (Sayin et al., 2013). Consequently, taurine-conjugated BAs that inhibit farnesoid X receptor (FXR) activity (Sayin et al., 2013) were increased in GF mice (Figures S4G and S4H), probably repressing genes involved in BA synthesis and cysteine sulfinic acid decarboxylase, the rate-limiting enzyme of taurine metabolism. We also observed that PLs were globally higher in males (Figure S4I). High PLs are correlated with male-biased expression of Choline Phosphotransferase 1 (CHPT1), whereas the phospholipase PLA2G16 involved in the hydrolysis of these PL (Uyama et al., 2009) showed a female-biased expression. Through the metabolism of choline (Romano et al., 2015), gut microbiota affected choline absorption and transport, globally modulating PL in GF mice. All three metabolic clusters discussed here exemplify the regulation of sexually dimorphic liver metabolism by the microbiota through direct impact on nutrient metabolism and transport, but also indirectly through the modulation of GH and sex-hormone signaling.
GF Mice Showed an Attenuated Sexual Dimorphism in Rhythmic Gene Expression and Metabolome
Although sexual dimorphism of liver gene expression was extensively studied in animal models (Lichanska and Waters, 2008), little is known about the influence of sex on rhythmic gene expression. We thus performed a genome-wide comparative analysis of sexually dimorphic liver gene expression under ConvR and GF conditions (Figure 5G). While 71% of all rhythmic genes were rhythmic in both sexes in ConvR mice (model 4), 9% of the genes were rhythmically expressed only in males (model 2, red) and 16% only in females (model 3). Furthermore, 4% were diurnally expressed in both sexes with different phases or amplitudes (model 5) (Figure 5H). These sex-specific rhythms were mainly involved in steroid hormone and drug metabolism in males (model 2) and protein transport and cell signaling in females (model 3) (Table S4). Interestingly, genes exhibiting differentially rhythmic parameters between male and female (model 5) were enriched for circadian core clock genes. Some of these genes, namely Cry1, Npas2, Nr1d2, and Nfil3, showed a higher amplitude in females (Figures S5A–S5C). Surprisingly, when we studied these sex-specific rhythmic genes in GF mice, we found that a majority lost their sex-biased rhythmic expression and became similar in males and females (Figures 5K, S5D, and S5E). These observations indicate that sex-specific rhythmic mRNA expression was also influenced by the microbiome.
Since such rhythmically expressed genes likely influence metabolism, we studied the rhythmicity of serum and liver metabolomes under different conditions (Figures 5I and 5J and Table S6). Around 34% of metabolites were found to oscillate in abundance in the liver and 20% in the serum. Interestingly, while 55% of all rhythmic liver metabolites were rhythmic in both sexes in ConvR mice (model 4), only 29% were rhythmic in the serum. These differences may reflect the influence of sexual dimorphism in the production of serum metabolites by additional tissues, including sexual organs. While 11% of the metabolites were rhythmic in males only in the liver (model 2), 32% were rhythmic in the serum. Meanwhile, an equal proportion of 31% and 32% of measured metabolites were rhythmic exclusively in female liver and serum (model 3), respectively. Similar to what we observed for mRNA expression, this sexual dimorphism was strongly damped in GF mice, with most of these metabolites becoming rhythmic in both sexes (Figures 5L and 5M). Altogether, these data further corroborate that microbiota control sexually dimorphic rhythmic gene expression and metabolism.
As feeding rhythms strongly affect rhythmic gene expression, we performed a behavioral phenotyping. All groups of mice showed similar feeding and drinking rhythms (Figures 5N and 5O). We also reproduced the described sexual dimorphism on locomotor activity (Broida and Svare, 1984). Strikingly, the increased activity of ConvR females was lost in GF females, the latter thus showing a similar activity pattern as males (Figure 5P).
To characterize mechanisms underlying these changes, we performed TF-binding site analysis near TSSs of the sexually dimorphic rhythmic genes. Among the enriched TF-binding sites showing a predicted sexually dimorphic rhythmic activity in ConvR mice that is attenuated in GF mice, we noted the presence of consensus sequences for numerous receptors of hormones or metabolites (Table S5). In males, motifs for AR, AHR, and the nuclear receptor NR6A1, which shares its core DNA-binding motif with the xenobiotic receptors PXR and CAR (Figure S5F), showed a male-specific rhythmic activity in ConvR but not in GF mice (Figure S5G). Conversely, in females, the Estrogen-Related Receptors α (ESRRA), sharing its DNA-binding motif with the Estrogen Receptor α (ERα) (Vanacker et al., 1999), HNF1A and KLF4 displayed a rhythmic activity only in ConvR females, but in both sexes in GF conditions (Figure S5H). Rhythmic AR activation was likely a consequence of rhythmic testosterone secretion, and testosterone levels lose their sexual dimorphism in GF mice (Figure 3A). This is also likely true for ERα and E2 (Figure 3B). While rhythmic KLF4 expression has never been reported, KLF3, which shares the same DNA-binding motif and is more highly expressed, has been found rhythmic (Wang et al., 2017). Both of these TFs preferentially bind methylated DNA (Spruijt et al., 2013) and are therefore likely more active in females due to the absence of testosterone-induced DNA demethylation (Reizel et al., 2015). HNF1A is crucial for GH signaling (Lee et al., 1998) and likely participates in female-specific gene expression, as shown for the Cyp2c12 gene in rat (Waxman et al., 1996). Male mice are more sensitive to AHR ligands (Lee et al., 2015, Pohjanvirta et al., 2012) and AHR activation is time and circadian clock dependent (Qu et al., 2010, Tanimura et al., 2011). The same holds for PXR and CAR, which are also more efficiently activated in males (Anakk et al., 2007) and clock dependent (Gachon et al., 2006, Kriebs et al., 2017). Moreover, both pathways are stimulated by microbiota-derived molecules and metabolites (Moura-Alves et al., 2014, Venkatesh et al., 2014, Zelante et al., 2013). By looking at sex-biased and rhythmic metabolites in ConvR and GF mice, we identified AHR modulators such as picolinic acid, kynurenine, and daidzein, as well as PXR modulators such as indole propionate (IPA), β/γ tocopherol, and ferulate (Figure S5I). To test the involvement of these receptors, we incubated female primary hepatocytes with different types of sera. Sera from ConvR females induced a higher activation of both the AHR and PXR-related DNA motifs compared with ConvR male and GF female sera (Figure S5J). It is therefore likely that both the hormone disturbance and differential metabolism of food-derived molecules contribute to the alteration of sexually dimorphic rhythmic gene expression in GF mice.
Microbiota-Derived Metabolites Affect Sexual Maturation
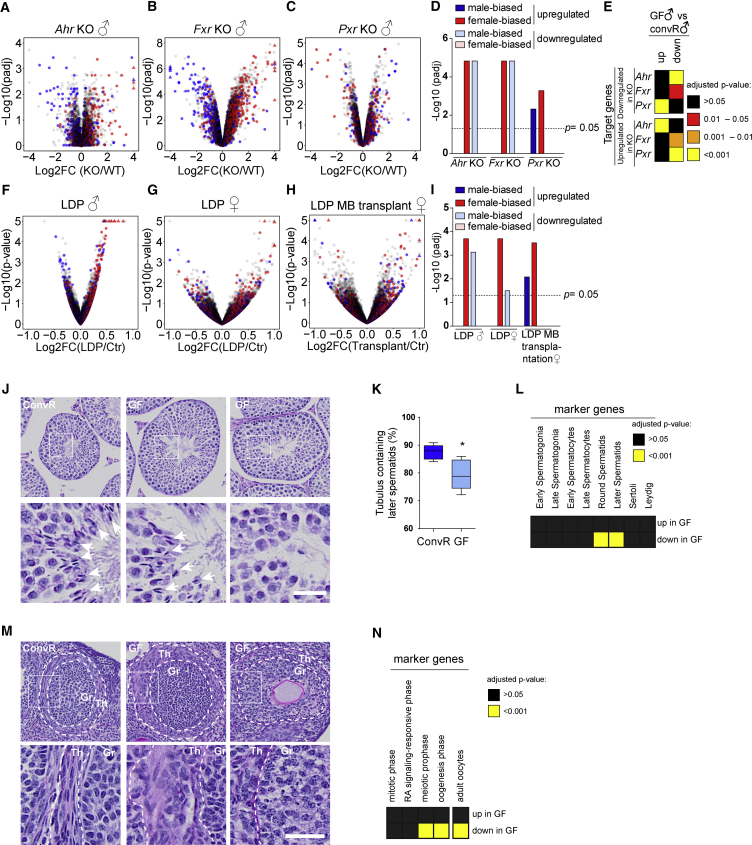
What causes attenuated sexual dimorphism and altered action of testosterone, E2, and GH in GF animals? Sexual dimorphism in the liver appears during puberty (Conforto and Waxman, 2012) when sex hormones imprint sex-specific and pulsatile GH secretion patterns (Jansson et al., 1985). Given the significant impact of the microbiome on sexual dimorphism of liver gene expression, microbiota-derived metabolites appear attractive agents to interfere with sexual development and the imprinting of GH secretion. Several signaling pathways regulated by microbiota-derived metabolites are particularly interesting. These include the activation of AHR and PXR by bacteria-derived molecules (Moura-Alves et al., 2014, Venkatesh et al., 2014, Zelante et al., 2013) and the activation of FXR or TGR5 (Sayin et al., 2013) and PXR (Staudinger et al., 2001) by BA. Indeed, in addition to their impact on liver metabolism, AHR (Karman et al., 2012), its activator (e.g., dioxin) (Petersen et al., 2006), PXR, and its agonist PCN (Zhang et al., 2010) also play a role in the maturation of sexual organs. The same holds for BA (Baptissart et al., 2014, Martinot et al., 2017). Therefore, we speculated that the differential activation of these pathways in GF mice during the prepubertal period could participate in establishing sexual dimorphism in the liver. Indeed, our analysis of the liver transcriptome in male Ahr (Tijet et al., 2006) and Fxr (Ijssennagger et al., 2016) knockout (KO) mice revealed a feminization of gene expression (Figures 6A, 6B, and 6D), while Pxr KO mice partly showed a masculinization (Figures 6C and 6D). Analysis of the expression of up- and downregulated genes in Ahr, Fxr, and Pxr KO mice in GF animals revealed that AHR-regulated genes showed an expression in GF mice similar to that observed in Ahr KO mice, whereas the opposite is observed in Pxr KO animals (Figures 6E and S6A). A less significant directional change in GF mice was observed for genes deregulated in Fxr KO mice (Figures 6E and S6A). These analyses suggest that the activation of AHR and PXR signaling pathways is increased and decreased in GF mice, respectively, corroborating that Ahr deletion is associated with loss of sexual dimorphism (Huang et al., 2016) and PXR activation decreases testosterone secretion (Zhang et al., 2010). It therefore appears likely that the modified activation of AHR and PXR during the prepubertal period is responsible for the attenuated sexual dimorphism in GF mice. Concordantly, male GF mice conventionalized at 5 weeks of age showed increased expression of female-biased genes compared with mice conventionalized just after birth (Figures S6B and S6E). Moreover, treatment with low-dose penicillin (LDP), which alters microbiota during early ages (Cox et al., 2014), induces a feminization of both male and female liver gene expression (Figures 6F, 6G, and 6I). We observed a similar increase of female gene expression by treatments with other antibiotics such as amoxicillin and tylosin in females (Figures S6C–S6E). This effect persists in female mice transplanted with fecal microbiota from LDP animals (Figures 6H and 6I). These observations suggest that changes in microbiota composition cause the hepatic feminization rather than the antibiotic treatment itself. Interestingly, LDP-treated males exhibited increased and decreased activation, respectively, of the AHR and PXR pathways, exactly as in GF mice (Figures S6F and S6G).
Figure 6.
Microbiota-Depleted Mice Show Differential Activation of Xenobiotic Receptors and Alteration in Sexual Development
(A–C) Changes in hepatic gene expression (log2FC) and adjusted p values of Ahr KO (A), Fxr KO (B), and Pxr KO (C) mouse models presented as a volcano plot.
(D) Adjusted p value for the enrichment of male- and female-biased genes in the indicated receptor KO model.
(E) Heatmap illustrating the statistical overrepresentation of target genes from different xenobiotic sensing nuclear receptors in differentially expressed genes of GF♂ mice liver (GF♂ versus ConvR♂). Target genes have been defined as significantly up- or downregulated in the respective KO model.
(F–H) Volcano plots depicting sex-biased hepatic gene expression across different conditions; for plot details see (A)–(C). (F and G) LDP-treated male (F) and female (G) animals versus their untreated counterparts. (H) Comparison between transfaunation of cecal microbiota into GF♀ mice from LDP-treated and -untreated animals.
(I) Adjusted p value for the enrichment of male- and female-biased genes under the indicated antibiotics treatment.
(J–L) GF mice show an alteration in spermatid differentiation. (J) Periodic acid–Schiff (PAS)-stained sections of testis from ConvR (left) and GF mice (middle and right) show that more seminiferous tubules of GF animals lack spermatids (white arrows) (right) than in their ConvR counterparts (left). (K) Tukey boxplots of at least five animals per group quantifying this difference (p < 0.05, Mann-Whitney U test). (L) Heatmap illustrating the statistical overrepresentation of marker genes of different stages of spermatogenesis in differentially up- or downregulated genes of testis in GF animals.
(M and N) GF mice present defects in follicle organization. (M) PAS-stained sections of follicles from ConvR (left) and GF mice (middle and right) show severe effects on the theca (Th) organization (left and right) and disorganization of some follicles in their granulosa (Gr) in GF animals. (N) Statistical overrepresentation of gene markers for fetal germ cells to oocyte development indicate defects in oocytes of GF mice.
Scale bars, 100 μm (lower magnification) and 320 μm (higher magnification).
Therefore, we hypothesized that activation of specific cell signaling pathways by microbiota-derived molecules during the prepubertal period is crucial for sexual maturation and downstream metabolism. We thus studied gene expression and morphology of the gonads of GF mice. Testis seminiferous tubules from GF male mice showed a significantly lower proportion of fully differentiated spermatids compared with ConvR males (Figures 6J and 6K). This was confirmed by the lower expression of genes specific to round and later spermatids, corroborating a deficient sexual maturation of GF males (Figure 6L, Table S7). In females, the organization of growing follicles was compromised, with disorganized granulosa cells and the presence of round-nuclei theca cells (Figure 6M). This disorganization was associated with the downregulation of genes specific to the late development stage and mature oocytes, confirming the disorganized follicular development in GF females (Figure 6N). Interestingly, both GH (Keene et al., 2002, Zaczek et al., 2002) and Ahr (Baba et al., 2008, Barnett et al., 2007) deficiencies lead to comparable deficient sexual maturation, suggesting that both pathways could be involved in the appearance of this phenotype.
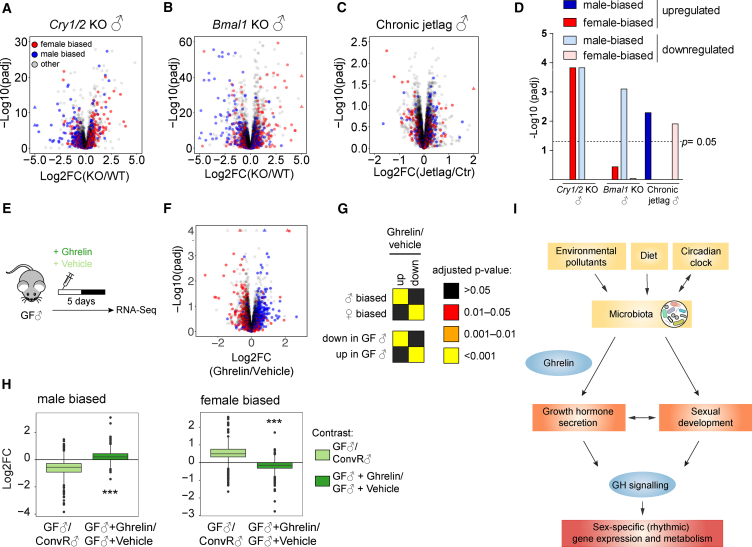
Hepatic expression of Ahr and Pxr is sexually dimorphic and rhythmic (Lu et al., 2013). AHR expression and activity (Qu et al., 2010, Tanimura et al., 2011) and Pxr expression are regulated by the circadian clock (Figure S7A). In addition, the circadian clock regulated BA homeostasis (Le Martelot et al., 2009) and several microbiota-derived metabolites known to activate AHR and PXR displayed a sexually dimorphic diurnal accumulation (Figure 5I). Therefore, we evaluated if disruption of the circadian clock and its downstream regulation of xenobiotic receptors also lead to change in liver sexual dimorphism. Cry1/Cry2 KO male mice exhibited a feminized liver transcriptome (Figures 7A and 7D). This feminization was also present in male Bmal1 KO mice (Figure 7B). Interestingly, Bmal1 KO mice show reduced GH levels as already shown for Cry1/Cry2 KO (Bur et al., 2009) (Figure S7B). Furthermore, a mouse model of chronic jet lag exhibited a masculinized liver transcriptome (Figures 7C and 7D). Underlying activities of the AHR and PXR pathways in Cry1/Cry2 KO mice were similar to the ones observed in GF mice (Figures S7C and S7G), while Bmal1 KO mice showed a decrease in the activities of both AHR- and PXR-dependent pathways (Figures S7D and S7G), and chronic jet lag mice exhibited no clear regulations of these pathways (Figures S7E and S7G). These data suggest that the dysregulation of AHR in clock mutants is likely involved in their feminized liver transcriptomes.
Figure 7.
Sexual Dimorphism Is Attenuated in Clock-Deprived Mice, and Ghrelin Plays an Important Role in Sustaining Sexually Dimorphic Gene Expression
(A–C) Changes in hepatic gene expression (log2FC) and adjusted p values of the clock-deprived models Cry1/Cry2 KO (A) and Bmal1 KO (B) compared with wild-type and chronically jet-lagged versus non-jet-lagged male animals (C) presented as a volcano plot. WT, wild-type.
(D) Graph depicting the adjusted p value of the analysis assessing the enrichment of male- and female-biased genes in the different conditions shown in (A)–(C).
(E–H) Ghrelin blunts the feminized expression signature of GF♂ mice. (E) Experimental design to evaluate the effect of ghrelin on hepatic sexually dimorphic gene expression. (F) Changes in hepatic gene expression (log2FC) and adjusted p values of ghrelin-injected GF♂ mice presented as a volcano plot. (G) Enrichment analysis of male- and female-specific genes (♂ biased, ♀ biased) or repressed or induced genes in GF mice (down or up in GF♂) for differentially expressed genes of the contrast ghrelin- versus vehicle-injected males. (H) The differences of differentially expressed sex-biased genes between GF and ConvR male mice are strongly attenuated by ghrelin injections (∗∗∗p < 0.001, paired t test).
(I) Working model for the control of sex-specific rhythmic gene expression and metabolism by microbiota.
To link these phenomena, we searched for a common pathway that might explain the observed phenotype in all described conditions and acting upstream of GH secretion. A likely candidate was ghrelin. GF mice have low ghrelin levels (Perry et al., 2016), a characteristic shared in obese individuals (Tschöp et al., 2001). Moreover, ghrelin secretion is stimulated by AHR activation (Lindén et al., 2014), whereas shift work (Crispim et al., 2011, Daniela et al., 2013) and Bmal1 deletion (Laermans et al., 2015) are associated with low and non-rhythmic ghrelin levels. Ghrelin has been shown to be an activator of GH secretion (Kojima et al., 1999), contributing to the amplitude of the secretion pattern (Farhy and Veldhuis, 2005, Xie et al., 2015). In addition, ghrelin secretion is stimulated by testosterone (Pagotto et al., 2003), whereas E2 inhibits ghrelin action in males (Clegg et al., 2007). Given the detected sex-hormone levels in GF mice, we hypothesized that ghrelin signaling alters GH secretion pattern in GF animals. We injected acetylated ghrelin in GF male mice and studied the impact on global hepatic sexual dimorphism. Ghrelin injection rescued the observed feminization of GF male mice, suggesting that ghrelin plays a key role in sustaining hepatic sexual dimorphism (Figures 7E–7H).
Discussion
We have shown that the absence of microbiota leads to alterations in sex-dimorphic gene expression and metabolism. Moreover, we gained novel insights into female- and male-specific rhythms of gene expression and metabolites, which also depend on the presence of microbiota. Indeed, the absence of microbiota altered GH secretion and sexual maturation and, consequently, sexual dimorphism in gene expression. Interestingly, such attenuated sexual dimorphism was also found in obese or circadian clock-deprived mice. Altered microbiota conditions also showed a perturbed secretion of the GH-releasing factor ghrelin, and we showed that ghrelin injections could restore hepatic sexual dimorphism in GF mice, indicating an important role of ghrelin in the maintenance of sexual dimorphism.
We found that the absence of microbiota perturbed hepatic gene expression in rhythmic and sexually dimorphic pathways such as lipid and xenobiotic metabolism. Although the signals responsible for this phenomenon are not well characterized, a role of microbiome-derived signals has been suggested (Leone et al., 2015, Montagner et al., 2016, Sayin et al., 2013, Thaiss et al., 2016, Venkatesh et al., 2014, Zelante et al., 2013). Here, we showed that sexual dimorphic gene expression and metabolism are altered in GF mice. Indeed, several metabolic characteristics of GF mice, such as reduced growth (Yan et al., 2016), reduced body fat, liver lipogenesis, increased mitochondrial activity, insulin sensitivity (Bäckhed et al., 2004), increased brown adipose tissue (Suarez-Zamorano et al., 2015), and resistance to HFD-induced obesity (Bäckhed et al., 2007, Rabot et al., 2010) are all hallmarks of feminized metabolism (Mauvais-Jarvis, 2015). Also, sex bias in sexually dimorphic xenobiotic detoxification (Blanck et al., 1986, Selwyn et al., 2015) and male-prone liver cancer (Grant and Roe, 1969, Vesselinovitch, 1987) is lost in GF and young age-castrated animals. All of these sex-specific diseases showed altered sex-hormone regulation and GH signaling. Therefore, we hypothesized that activation of cell signaling pathways by microbiota-derived molecules during the prepubertal period is crucial for sexual maturation and downstream metabolism. Microbiota-derived metabolites such as butyrate and other short-chain FAs (SCFAs) potentiate GH secretion in the pituitary (Miletta et al., 2014) and its stimulating factor ghrelin (Perry et al., 2016). Such regulation of secretion is potentially responsible for the low GH secretion in GF animals (Figures 3C and 3D). Consequently, GF mice displayed poor growth, paralleled by differences in the presence of SCFAs (Yan et al., 2016), potentially through the impact of SCFAs on GH secretion.
We also identified other microbiota-activated pathways, besides GH signaling, which play crucial roles in sexual development. For example, AhR KO (Karman et al., 2012) and Fxr KO (Martinot et al., 2017) mice suffer from defective sexual maturation. In addition, prenatal exposure to environmental pollutants that activate AHR or xenobiotic receptors leads to impaired sexual development in both mice and humans (Fishman and Yanai, 1983, La Rocca et al., 2014, La Rocca et al., 2015, Petersen et al., 2006, Winneke et al., 2014, Zhang et al., 2010). Interestingly, these so-called endocrine disruptors also affect GH secretion (Agrawal et al., 1995, Takeda et al., 2014) and the composition of the microbiome (Li et al., 2011, Murray et al., 2016, Zhang et al., 2015). Altered circadian and sleep-wake rhythms are similarly associated with impaired sex-hormone secretion and gonadal development (Alvarez et al., 2008, Miller et al., 2004), GH secretion (Bur et al., 2009, Weibel et al., 1997), alteration of microbiota (Liang et al., 2015, Poroyko et al., 2016, Thaiss et al., 2014, Voigt et al., 2016), and obesity and metabolic syndrome (Laermans and Depoortere, 2016). Similarly, genetic (Finkelstein et al., 1986) and HFD-induced obesity (De Schepper et al., 1998) are associated with reduced pulsatile GH secretion, reduced sexual dimorphism of sex-hormone production, gonadal deficiency (Escobar-Morreale et al., 2014), and altered gut microbiota (Winer et al., 2016). We showed that GF mice (Figures 6E and S6A), animals treated with antibiotics (Figures S6F and S6G), circadian clock-deficient animals (Figures S7C, S7D, and S7G), and mice fed with HFD (Figures S7F and S7G) all show a blunted response to AHR and PXR activators. In addition, both AHR (Zhou, 2016) and PXR (Venkatesh et al., 2014) are involved in immune responses, which lose sex bias in GF mice (Markle et al., 2013, Yurkovetskiy et al., 2013). The interplay between microbiota and sex-biased signaling pathways is schematized in a working model (Figure 7I).
Is this link between microbiome bacteria and sexual development conserved among animals throughout their evolution? Bacteria are known to influence sexual maturation in arthropods and nematodes. For example, Wolbachia and other non-pathologic parasites function as regulators of sex determination in insects (Kageyama et al., 2012). Interestingly, pathways involved in nematode innate immunity, such as the DAF-2/DAF-16 pathway (Garsin et al., 2003, Rae et al., 2012), also regulate genes involved in xenobiotic detoxification, lipid metabolism, and stress resistance (Murphy et al., 2003) and can affect fertility (Tissenbaum and Ruvkun, 1998). Of note, these pathways are also associated with GH signaling. Moreover, the NHR-25 nuclear receptor, a sensor of microbial metabolites (Lin and Wang, 2017), is involved in the sexual development of nematodes (Gissendanner and Sluder, 2000). SF-1 and LRH-1, the mammalian homologs of NHR-25, PL receptors (Krylova et al., 2005) controlled by BA (Lu et al., 2000), regulated intestinal immune response (Coste et al., 2007) and sexual development (Fayard et al., 2004). Both PL and BA are modified by sex and microbiota (Figures 5F, S4G–S4I, and S6). Thus, NHR-25 homologs could also play a role in microbiota-associated sexual dimorphism. Since specific bacteria species can rescue the sexual performance of sterile flies (Ben Ami et al., 2009) and gonad development and reproduction in zebrafish (Carnevali et al., 2013), it is likely that a role of the microbiota in sexual maturation is evolutionarily conserved.
Limitations of Study
We found that the mouse microbiome plays a critical role in sexual dimorphism through the regulation of GH secretion and sexual maturation. While we provide evidence that microbiota-derived metabolites are likely involved in sustaining sexual dimorphism through the activation of the xenobiotic receptors AHR and PXR, the exact mechanism of their action remains elusive. Moreover, the implication of deficient ghrelin secretion in attenuated sexually dimorphic expression patterns that we describe for several conditions will be a topic for future studies. Despite the described limitations of our study, the importance of microbiota in sustaining sexual dimorphism is unexpected and might bear relevance for human perturbation of microbiota diversity at an early age.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| STAT5 | Cell Signaling Technology | Cat# 9363S; RRID:AB_10693321 |
| Phospho-STAT5 (Tyr694) | Cell Signaling Technology | Cat# 9359S; RRID:AB_823649 |
| BCL6 | Cell Signaling Technology | Cat# 5650S; RRID:AB_10949970 |
| FOXA2 | Cell Signaling Technology | Cat# 8186S; RRID:AB_10892612 |
| AMPKα | Cell Signaling Technology | Cat# 2532; RRID:AB_330331 |
| Phospho-AMPKα (Thr172) | Cell Signaling Technology | Cat# 2535; RRID:AB_331250 |
| CUX2 | Thermo Fisher Scientific (Proteintech) | #24902-1-AP |
| anti-rGH (monkey) | National Hormone & Peptide Program (NHPP) | #AFP-411S; RRID:AB_2665564 |
| rabbit antiserum to rGH | National Hormone & Peptide Program (NHPP) | #AFP-5641801 |
| Anti-rabbit IgG, HRP-linked Antibody | Cell Signaling Technology | Cat# 7074; RRID:AB_2099233 |
| Chemicals, Peptides and Recombinant Proteins | ||
| rat acetylated Ghrelin | Tocris | #1465 |
| recombinant human Growth Hormone | BioVision | #4769 |
| mouse Growth Hormone | National Hormone & Peptide Program (NHPP) | #AFP-10783B |
| Critical Commercial Assays | ||
| Testosterone ELISA kit | Enzo | #ADI-900-065 |
| Mouse/Rat Estradiol ELISA | Calbiotech | #ES180S-100 |
| MiSeq Reagent kit V2 (500 cycles) | Illumina | #MS-102-2003 |
| QIAamp DNA Stool Mini Kit | Qiagen | #51504 |
| miRNeasy Mini Kit | Qiagen | #74106 |
| PE Cluster Kit v3 cBot-HS | Illumina | #FC-401-3001 |
| TruSeq SBS Kit v3-HS | Illumina | #PE-401-3001 |
| Ribo-Zero Gold | Illumina | #RS-122-2303 |
| Deposited Data | ||
| Raw and processed data (RNA-seq) of this paper | GEO (http://www.ncbi.nlm.nih.gov/geo/) | GEO: GSE77221 |
| Raw data (16S rRNA sequencing) of this paper | https://www.ncbi.nlm.nih.gov/sra | SRP151275 |
| Raw and processed data (Microarray); sexual dimorphism in white adipose tissue (male and female WAT) | GEO (http://www.ncbi.nlm.nih.gov/geo/) | GEO: GSE3086 |
| Raw and processed data (Microarray); germ-free mice vs conventionally raised (male liver) | GEO (http://www.ncbi.nlm.nih.gov/geo/) | GEO: GSE53590 |
| Raw and processed data (Microarray); germ-free mice on high-fat diet (male liver) | GEO (http://www.ncbi.nlm.nih.gov/geo/) | GEO: GSE19038 |
| Raw and processed data (RNA-seq); vertical sleeve gastrectomy on high-fat diet (male liver) | GEO (http://www.ncbi.nlm.nih.gov/geo/) | GEO: GSE53782 |
| Raw and processed data (RNA-seq); high-fat diet (male liver) | GEO (http://www.ncbi.nlm.nih.gov/geo/) | GEO: GSE87565 |
| Processed data (Microarray); resistant starch at high-fat diet (male liver) | Kieffer et al., 2016 | Available upon request from the authors |
| Raw and processed data (RNA-seq); Bmal1 KO (male liver) | GEO (http://www.ncbi.nlm.nih.gov/geo/) | GEO: GSE73554 |
| Raw and processed data (Microarray); Ahr KO mice (male liver) | GEO (http://www.ncbi.nlm.nih.gov/geo/) | GEO: GSE10082 |
| Raw and processed data (Microarray); Fxr KO mice (male liver) | GEO (http://www.ncbi.nlm.nih.gov/geo/) | GEO: GSE76162 |
| Raw and processed data (Microarray); Pxr KO mice (male liver) | GEO (http://www.ncbi.nlm.nih.gov/geo/) | GEO: GSE55746 |
| Raw and processed data (RNA-seq); low-dose penicillin treatment (LDP) (male and female liver) and microbiota transfaunation after LDP treatment (female liver) | GEO (http://www.ncbi.nlm.nih.gov/geo/) | GEO: GSE58089 |
| Raw and processed data (RNA-seq); early-life pulsed antibiotic (amoxicillin/tylosin) treatment in (male liver) | GEO (http://www.ncbi.nlm.nih.gov/geo/) | GEO: GSE68603 |
| Raw and processed data (Microarray); germ-free mice with bacterial reconstitution at 5 weeks of age | CIBEX (http://cibex.nig.ac.jp) | CIBEX: CBX256 |
| Raw and processed data (Microarray); chronic jet lag | GEO (http://www.ncbi.nlm.nih.gov/geo/) | GEO: GSE75475 |
| Broad single-cell transcriptome view of the male mouse germline | GEO (http://www.ncbi.nlm.nih.gov/geo/) | GEO: GSE104556 |
| Raw and processed data; ChIP-seq FOXA1/FOXA2 | ArrayExpress (https://www.ebi.ac.uk/arrayexpress/) | AE: E-MTAB-805 |
| Raw and processed data; ChIP-seq STAT5 and Bcl6 (male and female liver) | GEO (http://www.ncbi.nlm.nih.gov/geo/) | GEO: GSE31578 |
| Processed data; ChIP-seq CUX2 (female liver) | Conforto et al., 2012 | Provided as Supplemental Information |
| Raw and processed data; ChIP-seq H3K4m1, H3K4m3, H3K9a, H3K27a, H3K27m3, H3K36m3, H3K79m2, CTCF, POLII (male liver) | GEO (http://www.ncbi.nlm.nih.gov/geo/); ENCODE Project Consortium, 2012, Shen et al., 2012 | GEO: GSE29218; GEO: GSE31039 |
| Raw and processed data; DNase I Seq (male and female liver) | GEO (http://www.ncbi.nlm.nih.gov/geo/) | GEO: GSE21777 |
| Raw and processed data; POLII ChIP-seq (male and female liver) | GEO (http://www.ncbi.nlm.nih.gov/geo/) | GEO: GSE47899 |
| Positional weight matrices of transcription factors (SwissRegulon) | Pachkov et al., 2013; http://swissregulon.unibas.ch/fcgi/sr/downloads | N/A |
| Processed data; marker genes of germ cell differentiation in mouse testis | Lukassen et al., 2018 | N/A |
| Raw data; mouse transcriptome of different tissues including oocytes | GEO (http://www.ncbi.nlm.nih.gov/geo/) | GEO: GSE1133 |
| Processed data; fetal germline cells stages to oocytes | Li et al., 2017 | N/A |
| Experimental Models: Cell Lines | ||
| Mouse (CD-1) Cryopreserved Hepatocytes, Plateable Female | Thermo Fisher Scientific | #MSCP20 |
| Mouse (CD-1) Cryopreserved Hepatocytes, plateable male | Thermo Fisher Scientific | #MSCP10 |
| Experimental Models: Organisms/Strains | ||
| Conventionally raised mice (male and female) (C57BL/6 background) | Charles River Laboratories | C57BL/6J |
| Germ free (male and female) (C57BL/6 background) | University of Bern, CNRS TAAM, Orleans | N/A |
| Cry1/Cry2 KO mice (C57BL/6 background) | Prof. G. van der Horst (Erasmus University Medical Center, Rotterdam, the Netherlands) | N/A |
| Sterile chow diet | SAFE | #U8213G10R |
| Oligonucleotides | ||
| Forward_16S_rRNA: AATGATACGGCGACCACCGAGATCTACAC [8 base barcode]TATGGTAATTTGCCTACG GGNGGCWGCAG |
Kozich et al., 2013 | N /A |
| Reverse_16S_rRNA: CAAGCAGAAGACGGCATACGAGAT [8 base barcode] AGTCAGTCAGGCGACTACH VGGGTATCTAATCC |
Kozich et al., 2013 | N/A |
| Seq_p_Read1_V3 TATGGTAATTTGCCTACGG GNGGCWGCAG |
Caporaso et al., 2012 | N/A |
| Seq_p_Read2_V4 AGTCAGTCAGGCGACTACHVGGGTATCTAATCC |
Caporaso et al., 2012 | N/A |
| Seq_p_Index2_V4 GGATTAGATACCCBDGTAGTCGCCTGACTGACT |
Caporaso et al., 2012 | N/A |
| Software and Algorithms | ||
| Mothur (v1.33.0) | Schloss et al., 2009 | https://www.mothur.org/ |
| STAR (v2.5.3.a) | Dobin et al., 2013 | https://code.google.com/archive/p/rna-star/ |
| QIIME (v1.8) | Caporaso et al., 2010 | http://qiime.org/ |
| DESeq2 | Love et al., 2014 | https://doi.org/10.18129/B9.bioc.DESeq2 |
| Limma | Ritchie et al., 2015 | https://doi.org/10.18129/B9.bioc.limma |
| MotEvo (v1.03) | Arnold et al., 2012 | N/A |
| MARA (custom implementation) | Balwierz et al., 2014 | Custom scripts |
| Fiji | Schindelin et al., 2012 | https://imagej.net/Fiji |
| Ingenuity Pathway Analysis (IPA) | Qiagen | https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis |
| Enrichr | Kuleshov et al., 2016 | http://amp.pharm.mssm.edu/Enrichr/ |
| RDAVIDWebService | Jiao et al., 2012 | https://doi.org/10.18129/B9.bioc.RDAVIDWebService |
| R | R Core Team, 2018 | https://www.r-project.org |
| SAMtools | Li et al., 2009 | http://www.htslib.org/ |
| edgR | Robinson et al., 2010 | https://doi.org/10.18129/B9.bioc.edgeR |
| biomaRt | Durinck et al., 2009 | https://doi.org/10.18129/B9.bioc.biomaRt |
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to the Lead Contact, Frederic Gachon (f.gachon@uq.edu.au).
Experimental Model and Subject Details
The studies were conducted in accordance with the regulations of the Nestlé ethical committee and the veterinary office of the Canton of Vaud.
Mouse Strains
C57BL/6J male and female GF and ConvR mice were kept in gnotobiotic isolators under diurnal lighting conditions (12-hr light 12-hr dark) at a temperature of 21°C ± 2°C. The mice had free access to a sterile chow diet (irridated with >40 kiloGrays) and water. Sterility of GF animals was weekly assessed. All mice used in the experiments were between 9 and 16 weeks old. Liver, duodenum, jejenum, testis, ovaries, and perigonadal WAT were snap-frozen in liquid nitrogen and stored at −80° until further processing for RNA and protein extraction or fixed in formalin for histology.
Primary Hepatocytes
Cryopreserved hepatocytes from a female or male CD-1 donor mouse were purchased from Thermo Scientific (#MSCP10/20) and stored in liquid nitrogen before usage.
Method Details
Growth Hormone Serum Kinetics
Pulsatile secretion of GH was assessed with 9 to 11 weeks old C57BL/6J mice. Twenty-five sequential blood samples (4 μL) were taken at 15 min intervals starting at ZT 0.5 for each individual mouse. GH was measured using an ELISA assay described below.
Growth Hormone and Ghrelin Injections
Male GF animals received daily a subcutaneous injection with rat acetylated Ghrelin (10 μg/mouse) or PBS (at ZT 2) over a period of 5 days. Injections with recombinant human GH (50 μg/mouse) or PBS were applied twice a day (ZT 2 and ZT 14) over a period of 5 days.
Phenotyping Studies
Activity and food intake was measured with an automated measurement system (Phenomaster, TSE) equipped with feeding and drinking sensors, infrared beam based activity monitoring system and ventilated isopositive HEPA cages systems. Prior to use, cages were disassembled and autoclaved. Electronic sensors for phenotypic measurements were sterilized with hydrogen peroxide, acetic acid, and peracetic acid vapors sterilization technique. Mice were transferred into sterile isopositive phenomaster cages and measurements were started after an acclimation period of at least 18 hr.
Feces Collection for Microbiota Analysis
To assess gut microbiota diversity (16S rRNA Sequencing) we collected and froze fresh feces from male and female ConvR mice at the arrival (time point 1) of the animals from the provider and subsequently separated the animals into single cages. During the two following weeks, the bedding of the animals was mixed daily and distributed between all cages. Then, a second collection of feces was performed (time point 2).
Serum and GH Treatment in Primary Hepatocytes
Cryopreserved hepatocytes were seeded into Collagen I coated 24-well plates (Thermo Scientific #CM1024) (200,000 cells/well) and incubated in plating medium (Williams’ E Medium supplemented with 5% Fetal Bovine Serum, 1 μM Dexamethasone, 1% Penicillin/Streptomycin, 4 μg/mL Human Recombinant Insulin, 2 mM GlutaMAX and 15 mM HEPES (pH7.4)) without further passaging. After a 6 hr incubation period to allow the cells to attach to the plate, medium was changed to a defined incubation medium (Williams E Medium supplemented with 0.1 μM Dexamethasone, 0.5% Penicillin/Streptomycin, 6.25 μg/mL human recombinant insulin, 6.25 μg/mL human transferrin, 6.25 ng/mL selenous acid, 1.25 mg/mL bovine serum albumin, 5.35 μg/mL linoleic acid, 2 mM GlutaMAX and 15 mM HEPES (pH7.4)) and a Geltrex matrix overlay (Thermo Scientific #A1413201) was applied to the plated cells. The next day, the maintenance medium supplemented with 25% sterile-filtered serum from ConvR male, ConvR female or GF female mice was applied to the cells. After 24 hr cells were then either treated with 8 ng/mL mouse growth hormone or vehicle (PBS) for 2 hr before RNA extraction. Cells were kept at 37°C in a humidified atmosphere of 95% air and 5% CO2.
Total Protein Extraction and Analysis
Frozen pieces of liver were homogenized in a 20 mM HEPES buffer (pH 7.6) supplemented with 100 mM KCl, 0.1 mM EDTA, 1 mM NaF, 1 mM sodium orthovanadate, 1% Triton X-100, 0.5% Nonidet P-40, 0.15 mM spermin, 0.5 mM spermidine, 1 mM DTT, and a protease inhibitor cocktail. Homogenates were incubated on ice for 30 min and then cleared by a 10 min centrifugation at 21,000 g. After fractionation by SDS-PAGE, 65 μg of the cleared protein extracts were transferred to a PVDF membrane for western blot analysis. Primary antibodies were used at the following dilutions: 1:500 (STAT5), 1:500 (Phospho-STAT5 (Tyr694)), 1:500 (BCL6), 1:1000 (FOXA2), 1:1000 (AMPKα), 1:1000 (Phospho-AMPKα (Thr172)), and 1:500 (CUX2).
Measurement of MUPs Level in Urine
Urine was frozen after collection and stored at −80°C. Before usage, urine were cleared by a brief centrifugation (5 min at 9,000 g). 1 μL of the supernatant was fractionated by SDS-PAGE. Proteins were then visualized by Coomassie Blue staining. The level of MUPs was analyzed by quantifying the bands at a size of approximately 20 kDa.
Hormonal Assays
Circulation free testosterone and E2 were measured with commercially available ELISA kits (Key Resources Table) following the manufacturers’ protocols. Serum levels of GH were determined with a sandwich ELISA as described (Steyn et al., 2011). Specifically, a 96-well plate was coated with anti-rGH (monkey) as a capture antibody at a dilution of 1:40,000 and incubated overnight at 4°C. The next day, wells were blocked with blocking buffer (5% skim milk powder diluted in 0.05% PBS with Tween-20). After blocking, samples and standards (mouse Growth Hormone) were applied to each well. The bound standards and samples were then incubated with the detection antibody (rabbit antiserum to rGH (1:40,000)). The bound sandwich complex was incubated was anti-rabbit HRP conjugated antibody at a final dilution of 1:2,000. O-phenylenediamine served as a substrate of the enzymatic colorimetric rection which was stopped by adding HCl. Absorbance was measured at 490 nm. Secretion peaks were defined by a minimal two-fold increase in GH between two consecutive time points.
Microbiota Analysis by 16S rRNA Genes Sequencing
16S rRNA genes analysis was performed as described elsewhere (Kieser et al., 2018). Specifically, mouse feces were mechanically disrupted before DNA extraction. The V3 to V4 region of the 16S rRNA gene was amplified with region specific primers that included the Illumina flowcell adapter sequences and barcode sequences for dual indexing with 8 nt (Key Resources Table) following the suggestions of Kozich et al., 2013. After equimolar pooling, the PCR products were purified using magnetic beads Agencourt AMPure XP (Beckman Coulter). Library was quantified with a QuBit Fluorometer (ThermoScientific). The purified pool (10 nM) was sequenced in paired end (2∗250 cycles) on the MiSeq Illumina sequencer using the MiSeq Reagent kit V2 500 cycles as suggested in Caporaso et al., 2012. Paired-end sequences were joined and chimera sequences were removed using Mothur V.1.33.0 (Schloss et al., 2009). We used QIIME V.1.8 (Caporaso et al., 2010) to generate and annotate Operational Taxonomic Units (OUT) at 97% identity. A phylogenetic tree was built on the multiple alignment of the OTU representative sequences. Phylogenetic distances between samples were computed as weighted UniFrac distances (Lozupone and Knight, 2005) to calculate the principal coordinates using QIIME. Then, we defined the variance explained by individuality and sex using Adonis tests on the weighted UniFrac distances in QIIME.
Histology
Mouse testes and ovaries were fixed 24 hr with 10% formalin (Sigma-Aldrich), rinsed twice with PBS and then embedded in paraffin and cut into 4 mm thick sections. Tissue sections were deparaffinized, rehydrated in ethanol series and then washed with phosphate buffered saline PBS. Cell nuclei and polysaccharide staining was done with Mayer’s hematoxylin and Periodic Acid Schiff solution, respectively, using a Tissue-Tek Prisma autostainer from Sakura linked to a G2-coverslipper. The following reagents were used: 1% Periodic acid in water, Schiff reagent (Bio-Optica) and Hématoxyline solution (Harris Gill II PAP1Biosystems). The program is summarized below. Imaging was performed with an Olympus slide scanner at 20x magnification and pictures were analyzed using Fiji.
| Step | Station | Solution | Time | Delay | Mix |
|---|---|---|---|---|---|
| 1 | S∗ | Start Station | --:--:-- | ||
| 2 | D∗ | Drying Station | 00:10:00 | ∗∗ | OFF |
| 3 | 44 | Xylene | 00:03:00 | ∗∗ | OFF |
| 4 | 42 | Xylene | 00:03:00 | ∗∗ | OFF |
| 5 | 40 | Xylene | 00:03:00 | ∗∗ | OFF |
| 6 | 38 | Ethanol 100% | 00:01:00 | ∗∗ | OFF |
| 7 | 36 | Ethanol 100% | 00:01:00 | ∗∗ | OFF |
| 8 | 34 | Ethanol 96% | 00:01:00 | ∗∗ | OFF |
| 9 | 33 | Ethanol 70% | 00:01:00 | ∗∗ | OFF |
| 10 | W∗ | Wash Station | 00:01:00 | ∗∗ | EW |
| 11 | 35 | A. dest. | 00:00:30 | ∗∗ | OFF |
| 12 | 39 | Periodic Acid | 00:05:00 | == | ON |
| 13 | W∗ | Wash Station | 00:04:00 | 50% | EW |
| 14 | 37 | A. dest. | 00:01:00 | ∗∗ | EW |
| 15 | 21 | Schiff's | 00:15:00 | == | ON |
| 16 | W∗ | Wash Station | 00:10:00 | 50% | EW |
| 17 | 18 | A. dest. | 00:00:30 | ∗∗ | OFF |
| 18 | 4 | Hematoxylin | 00:05:00 | == | ON |
| 19 | W∗ | Wash Station | 00:00:30 | ∗∗ | EW |
| 20 | 6 | Acid Alcohol | 00:00:03 | == | ON |
| 21 | W∗ | Wash Station | 00:10:00 | ∗∗ | EW |
| 22 | 1 | Ethanol 70% | 00:01:00 | ∗∗ | OFF |
| 23 | 2 | Ethanol 96% | 00:01:00 | ∗∗ | OFF |
| 24 | 15 | Ethanol 100% | 00:01:00 | ∗∗ | OFF |
| 25 | 16 | Ethanol 100% | 00:02:00 | ∗∗ | OFF |
| 26 | 31 | Xylene | 00:01:00 | ∗∗ | OFF |
| 27 | 32 | Xylene | 00:01:00 | ∗∗ | OFF |
| 28 | E∗ | End Station | --:--:-- |
RNA Extraction and Total RNA Sequencing
Liver RNA was extracted as previously described (Gachon et al., 2006). Approximately 100 mg of frozen liver was grinded in extraction buffer (3.9 M Guanidium thiocyanate, 0.03 M Sodium citrate, 0.2 M Sodium acetate, 1% (v/v) 2-Mercaptoethanol) using a Polytron PT 2500 E homogenizer. An equal volume of phenol (saturated in H2O) and 0.5 volume of chloroform/isoamylalcohol (49:1 (v/v)) were added to the homogenate. The mixture was vigorously vortexed subsequently. Phase separation was accomplished by a centrifugation step at 4°C 12,000 g for 20 min. RNA of the aqueous phase was precipitated at −20°C during at least 20 min using an equal volume of isopropanol. The precipitate was pelleted by centrifugation at 12,000 g for 15 min at 4°C. Subsequently, the pellet was resuspended in 4M LiCl and subsequently re-pelleted during a centrifugation step of 12,000 g for 15 min at 4°C. The pellet was washed with 75% ethanol with a subsequent centrifugation at 12,000 g for 15 min at 4°C. The washed pellet was then dried at room temperature and dissolved in RNAse/DNAse free water. Frozen duodenum, ileum, testis, ovary, and perigonadal WAT were homogenized in Qiazol with the help of a tissue lyser II (Qiagen). Primary hepatocytes were directly lysed in Qiazol as recommended by the manufacturer. Subsequently, RNA was extracted using the miRNeasy Mini Kit following the manufacturer's protocol. Total RNA was quantified with Ribogreen (Life Technologies) and quality was assessed on a Fragment Analyzer (Advances Analytical). Sequencing libraries were prepared from 250 ng total RNA using the TruSeq Stranded Total LT Sample Prep Kit with the Ribo-Zero Gold depletion set. The procedure was automated on a Sciclone NGS Workstation (Perkin Elmer). The manufacturer’s protocol was followed, except for the PCR amplification step. The latter was run for 12 cycles with the KAPA HiFi HotStart ReadyMix (Kapa BioSystems). This optimal PCR cycle number was evaluated using the Cycler Correction Factor method as previously described (Atger et al., 2015). Purified libraries were quantified with Picogreen (Life Technologies) and the size pattern was checked with the DNA High Sensitivity Reagent kit on a LabChip GX (Perkin Elmer). After being pooled, 24 libraries were clustered at a concentration of 7 pmol on 8 lanes of a v3 paired-end sequencing flow cell (Illumina). Sequencing was performed for 2 x 100 or 2 x 125 cycles on a HiSeq 2500, following Illumina’s recommendations.
Metabolic Profiling
Frozen serum aliquots and liver pieces were analyzed by Metabolon as described previously (Evans et al., 2009). Specifically, samples were prepared using an automated MicroLab STAR system (Hamiliton Company). Methanol was added to the samples for protein precipitation. After centrifugation, the supernatant split into sufficient number of aliquots in order to measure them using four different Ultrahigh Performance Liquid Chromatography-Tandem Mass Spectroscopy (UPLC-MS/MS) methods. Organic solvent was subsequently removed using a TurboVap (Zymark) from the subsamples. The sample extracts were stored overnight under nitrogen before preparation for analysis. Sample extract were reconstituted in solvents compatible to each of the four methods. All methods were run on a Waters ACQUITY UPLC and a Thermo Scientific Q-Exactive high resolution/accurate mass spectrometer interfaced with a heated electrospray ionization (HESI-II) source and Orbitrap mass analyzer operated at 35,000 mass resolution.
(1)The first method was optimized for hydrophilic compounds using acidic positive ion conditions. Here, the extract was gradient eluted from a C18 column (Waters UPLC BEH C18-2.1x100 mm, 1.7 μm) using water and methanol, containing 0.05% perfluoropentanoic acid (PFPA) and 0.1% formic acid (FA).
(2)Hydrophobic compounds were the focus of the second method. Using the same C18 column (see 1) the extract was gradient eluted using methanol, acetonitrile, water, 0.05% PFPA and 0.01% FA at an overall higher organic content compared to method 1.
(3)Method 3 used basic negative ion with optimized conditions using a dedicated separate C18 column. Using methanol and water with 6.5mM Ammonium Bicarbonate at pH 8, basic extracts were gradient eluted from the column.
(4)The fourth method used a negative ionization with a subsequent elution from a HILIC column (Waters UPLC BEH Amide 2.1x150 mm, 1.7 μm). To this end, a gradient consisting of water and acetonitrile was used with 10 mM Ammonium Formate, pH 10.8. For all methods the scan range was between 70 and 1000 m/z.
Metabolomics Data Processing and Analysis
We restricted the downstream analysis to metabolites that were detected in at least 6 samples of an experimental group. Data were median normalized for each metabolite and missing values were imputed by k-nearest neighbors yielding a relative quantification of each metabolite. A two-factor linear model was fitted with the factors sex and hygiene status and applied to log2-transformed metabolite levels. In order to compare the means of specific contrasts (i.e. GF male vs. ConvR male, GF female vs. GF female, ConvR male vs. ConvR female and GF male vs. GF female) were used to make specific comparisons of treatments within a linear model. The retrieved p values were corrected for multiple testing (Benjamini and Hochberg, 1995). In order to assess changes on a pathway level, we employed a simple statistical approach based on aggregated z-scores as described (Krumsiek et al., 2015). Specifically, relative metabolite levels across all samples were transformed into z-scores. Z-scores of all metabolites sharing a specific sub- or super pathway were averaged. These values were used as a proxy for average activity in that specific pathway for each sample. The averaged z-scores were subjected to the same linear regression analysis used for single metabolites.
RNA-Seq Data Processing
Paired-end reads were mapped onto the Ensembl Mus musculus genome annotation (GRCm38/mm10) using STAR 2.3.8 (Dobin et al., 2013). Uniquely mapped reads were counted for each gene locus as described (Atger et al., 2015). Specifically, using a custom Perl script we applied SAMtools view (Li et al., 2009) to assigned unique mapped paired-end reads to exons considering read orientation. The count data were normalized by size factor and a variance stabilizing transformation was applied for downstream analysis. Differential gene expression was assessed by DESEq2 (Love et al., 2014). Data from other RNA-seq based studies that were used for subsequent gene set enrichment analysis were retrieved from indicated sources in Key Ressources Table and analyzed as described above.
Microarray Data Processing
Liver expression microarray data from other studies were retrieved from the indicated sources (Key Resources Table) and reanalyzed using the statistical package limma (Ritchie et al., 2015). Annotation was retrieved from biomaRt (Durinck et al., 2009) or, if not available, we used the annotation provided by the indicated repository. In case of probes hitting the same gene, the most significant probe was selected for further analysis. We applied a basic differential gene expression analysis comparing two conditions (e.g. antibiotica treated vs untreated, HFD vs chow diet). Statistical analysis and differential expression were assessed using an empirical Bayes method (Ritchie et al., 2015).
Functional and Gene Set Enrichment Analysis
Analysis for functional enrichment were performed with Ingenuity Pathway Analysis (Qiagen), Enrichr (Kuleshov et al., 2016), and RDAVIDWebService (Jiao et al., 2012). For the latter, we calculated enrichment for GO terms for biological processes and KEGG pathway annotations. p values were adjusted for multiple testing (Benjamini and Hochberg, 1995). To perform gene set enrichment analysis for sex-biased genes and TFs target genes, we employed the geneSetTest function of the limma package on a pre-ranked gene lists defined according to the DESeq2 or limma results for RNA-seq-derived or microarray-derived datasets, respectively. Gene sets were derived from several sources: gene sets for Ahr, Fxr, and Pxr KO regulated genes are derived from the list of differentially expressed genes from the indicated datasets (Key Resources Table). The gene set list of sexually dimorphic liver genes is based on the comparison between ConvR males and ConvR females. Sexually dimorphic genes for WAT were derived from the indicated reanalyzed microarray dataset (Key Resources Table). Genes that show significantly different expression patterns (adjusted p value ≤ 0.05 and log2 fold change > 0.5) between males and females were considered as male and female-biased, respectively. Public available ChIP-seq data were used to define downstream target gene sets of BCL6, STAT5, FOXA1/2, and CUX2 (Key Resources Table). We tested whether these gene sets are enriched in the up- or downregulated genes of the indicated comparisons. If neither of the alternatives showed a significant tendency, we tested whether the genes in the set tended to be differentially expressed without regard for direction (mixed).
Bcl6 Locus Analysis
Density Profile
Density of reads along the Bcl6 locus was computed for each sample. Briefly, paired-end reads mapped uniquely in the Bcl6 region with no mismatch were used. Samtools with "view" parameter was called on the bam files to retrieve position of the reads and mapping length. For each pair of reads, the nucleotides corresponding to the respective mapped sequence (position + mapping length) were incremented of one count in a strand specific manner. Total counts were normalized by total library size, size correction factor and an arbitrary constant (106) to improve readability using edgeR (Robinson et al., 2010). Median of the signal for each condition is reported after smoothing with a moving average of 50 nucleotides.
Correlation
Correlation matrix between the different features of the Bcl6 locus was computed and depicted using the corrplot R package (Wei and Simko, 2016) across the liver samples. Correlation (Pearson coefficient) was considered as significant if its respective p value was below 0.01 (Fisher’s Z transform).
Genome-Wide Sense-AS Analysis
In order to detect genes with similar response pattern as Bcl6 in the sense-antisense signal, we performed a genome-wide differential analysis of the sense-AS signal in liver. Uniquely mapped reads were counted for introns and exons features using SAMtools view option on the bam file as described above (density profile). The respective sense and AS reads counts were reported separately. Sum of exonic regions was used for the respective sense read counts while the sum of exonic and intronic reads count was used for the respective AS read counts. Reads counts were normalized using library size factor and differential expression between the different conditions was assessed using the general linear model framework of EdgeR (Robinson et al., 2010). In order to avoid false positive candidates, a strict genes selection based on reads count were applied. Genes with less average reads count on the respective sense signal than antisense signal were discarded. Genes with an average of at least 100 read counts in GF or ConvR mice for sense and AS were selected. Finally, genes with ratio sense/AS larger than 10 were discarded. Likelihood ratio test was applied by the glmLRT() function in EdgeR to test for the interaction term between strand (sense, AS) and conditions (e.g Male, Female) as described (Gaidatzis et al., 2015). FDR was computed using the topTags() function with Benjamini and Hochberg correction (Benjamini and Hochberg, 1995).
Motif Activity Analysis
For predictions of TF binding sites near promoters, we used MotEvo version 1.03 (Arnold et al., 2012) to scan ± 500 bp around the TSS. We used promoters (Balwierz et al., 2009) and weight matrices of TFs defined by SwissRegulon (Pachkov et al., 2013) (http://swissregulon.unibas.ch/fcgi/sr/downloads). We applied a penalized regression model as previously described (Balwierz et al., 2014) which uses an L2 penalty, allowing an estimate of the standard deviations.
Rhythmicity Assessment in Different Experimental Groups
The rhythmicity of gene mRNA levels and metabolites in male and female GF or ConvR mice were assessed as described (Atger et al., 2015). This method is based on multiple linear regression and a subsequent models selction that is based on the Bayesian information criterion. The full model is y(t) = μ + αcos((2π/24 h)t) + βsin((2π/24 h)t) + noise (y is the normalized and variance stabilized data, μ is mean, t is Zeitgeber time, α and β are the coefficients of the cosine and sine functions, respectively). Rhythmicity was compared across the different conditions (e.g. hygiene status and sex), we defined different model for two (GF and ConvR) or four conditions (GF female, ConvR female, GF male and ConvR male). Models were defined to have either zero (non-rhythmic pattern) or non-zero (rhythmic pattern) α and β coefficients for each analyzed condition. Moreover, for some models the values of α and β can be also shared within any combination of the two or four conditions. We used α and β to determine phase and log2 peak-to-trough amplitude as of gene expression. Linear regression was used to solve each of the defined models. We used a BIC based model selection to consider model complexity (Kass and Raftery, 1995) (RSS is the residual sum of squares of the linear regression, n is the number of time points and k is the number of parameters). To assess the confidence of the selected model we calculated the Schwarz weight using the formula , with . Genes that did not reach a BIC weight (BICW) above 0.4 were categorized as “ambiguous” (model 0). Genes that were identified as having an alteration in amplitude or phase between GF and ConvR mice were subdivided into groups of strongly altered, mildly altered, only altered in phase and only altered in amplitude. Modification was considered mild if the change was less than 4 hr for the period or less than 2 fold in amplitude between the two conditions.
Data and Software Availability
The accession number for the entire RNA-seq dataset reported in this manuscript is GSE77221. To ease accessibility, we divided the data into the following subseries: (1) Temporal profile of GF and ConvR male and female mice (duodenum and ileum – GSE114399, liver – GSE114400, WAT – GSE114401). (2) Temporal profile of Liver of Cry1/Cry2 KO mice (GSE114402). (3) GF male mice liver injected with GH (GSE114610). (4) GF male mice liver injected with Ghrelin (GSE114611). (5) Testes and ovaries of ConvR and GF female mice (GSE114612). (6) GH and mouse serum treatment on primary hepatocytes (GSE114613). 16s rRNA data have been deposited on the SRA database under the reference SRP151275.
Quantification and Statistical Analysis
If not otherwise stated, statistical analysis comparing the means between experimental groups (ConvR male, GF male, ConvR female and GF female) was performed with one-way analysis of variance (ANOVA) followed by the Holm multiple comparison test comparing selected comparisons (ConvR male vs. GF male, ConvR male vs. ConvR female, ConvR female vs. GF female and GF male vs. GF female) using R (R Core Team, 2018). Intersecting gene sets were visualized using the R-package UpSetR (Lex et al., 2014). All data are expressed as mean ± standard error of the mean (SEM) if not otherwise stated.
Acknowledgments
We thank Mathieu Membrez, Florian Atger, and Meltem Weger for scientific discussions; Julie Deuquet for project management; Alexandra Pinto and Nuno Silvano for taking care of GF animals; and Michèle Delley for technical assistance. We also thank Kathleen McCoy for her expertise in GF animals, Xavier Bonnefont for advice, and Sylvie Rabot and Ueli Schibler for advice and critical reading of the manuscript. Histology was performed at the EPFL Histology Core Facility. This research was supported by the European Research Council through individual Starting Grants ERC-2010-StG-260988 to F.G. F.N.’s laboratory was supported by the Swiss National Science Foundation grant 310030_173079. J.Y. benefits from the Natural Sciences and Engineering Research Council of Canada Postgraduate Studies Doctoral Scholarship.
Author Contributions
B.D.W., C.J.C., and F.G. designed the research. B.D.W., E.M., S.J., B. Betrisey, F.F., A.B., A.F., and A.C. performed the research. B.D.W., C.G., J.Y., B. Berger, B.B.-B., F.N., and F.G. analyzed the data. B.D.W. and F.G. wrote the paper. All authors reviewed and edited the manuscript.
Declaration of Interests
B.D.W., C.G., E.M., S.J., B. Betrisey, F.F., B. Berger, A.C., C.J.C., and F.G. are employees of Nestec, a subsidiary company of the Nestlé group.
Published: October 18, 2018
Footnotes
Supplemental Information includes seven figures and seven tables and can be found with this article online at https://doi.org/10.1016/j.cmet.2018.09.023.
Supplemental Information
Sheet 1. Detailed table legend.
Sheet 2. Results of the differential expression analysis using DESeq2 of GF versus ConvR male mice in liver.
Sheet 3. Results of the differential expression analysis using DESeq2 of GF versus ConvR male mice in duodenum.
Sheet 4. Results of the differential expression analysis using DESeq2 of GF versus ConvR male mice in ileum.
Sheet 5. Results of the differential expression analysis using DESeq2 of GF versus ConvR male mice in WAT.
Sheet 6. Results of the rhythmicity analysis using multiple linear regression and model selection in liver of male GF and ConvR mice.
Sheet 7. Results of the rhythmicity analysis using multiple linear regression and model selection in duodenum of male GF and ConvR mice.
Sheet 8. Results of the rhythmicity analysis using multiple linear regression and model selection in ileum of male GF and ConvR mice.
Sheet 9. Results of the rhythmicity analysis using multiple linear regression and model selection in WAT of male GF and ConvR mice.
Sheet 1. Detailed table legend.
Sheet 2. Gene Ontology (GO) classification related to the comparison of ConvR and GF male mice in liver.
Sheet 3. GO classification related to the comparison of ConvR and GF male mice in duodenum.
Sheet 4. GO classification related to the comparison of ConvR and GF male mice ileum.
Sheet 5. GO classification related to the comparison of ConvR and GF male mice in WAT.
Sheet 1. Detailed table legend.
Sheet 2. Results of the differential expression analysis using DESeq2 contrasting male ConvR versus female ConvR mice in liver.
Sheet 3. Results of the differential expression analysis using DESeq2 contrasting male GF versus female GF mice in liver.
Sheet 4. Results of the differential expression analysis using DESeq2 contrasting female GF versus female ConvR mice in liver.
Sheet 5. Results of the differential expression analysis using DESeq2 to test for interaction between sex and hygienic status.
Sheet 6. Results of the rhythmicity analysis using multiple linear regression and model selection in liver of male ConvR, male GF, female ConvR, and female GF mice.
Sheet 1. Detailed table legend.
Sheet 2. GO classification related to hepatic differentially expressed genes that show a significant interaction between sex and hygienic status.
Sheet 3. GO classification related to the comparison of ConvR and GF male mice in liver.
Sheet 1. Detailed table legend.
Sheet 2. Results of the differential expression analysis using a linear model (∼genotype∗status) to contrast: male ConvR and female ConvR, male GF and female GF, male GF versus male ConvR, female GF versus female ConvR.
Sheet 3. Results of the rhythmic activity analysis TF motifs using complex-valued singular value decomposition in male-specific rhythmic genes.
Sheet 4. Results of the rhythmic activity analysis of TF motifs using complex-valued singular value decomposition in female-specific rhythmic genes.
Sheet 1. Detailed table legend.
Sheet 2. Relative abundance of liver metabolites and statistics in ConvR male, female, GF male and female mice.
Sheet 3. Relative abundance of serum metabolites and statistics in ConvR male, female, GF male and female mice.
Sheet 4. Results of the rhythmicity analysis using multiple linear regression and model selection for metabolites in liver of ConvR male, female, GF male and female mice.
Sheet 5. Results of the rhythmicity analysis using multiple linear regression and model selection for metabolites in serum of ConvR male, female, GF male and female mice.
Sheet 6. Results of liver metabolite subpathway analysis using a linear model (∼genotype∗status) to contrast: male ConvR and female ConvR, male GF and female GF, male GF versus male ConvR, female GF versus female ConvR.
Sheet 7. Results of serum metabolite subpathway analysis using a linear model (∼genotype∗status) to contrast: male ConvR and female ConvR, male GF and female GF, male GF versus male ConvR, female GF versus female ConvR.
Sheet 1. Detailed table legend.
Sheet 2. Results of the differential expression analysis using DESeq2 of GH-injected male GF mice in liver.
Sheet 3. Results of the differential expression analysis using DESeq2 of ConvR male serum versus ConvR female serum-treated female primary hepatocytes.
Sheet 4. Results of the differential expression analysis using DESeq2 of GF female serum versus ConvR female serum-treated female primary hepatocytes.
Sheet 5. Results of the differential expression analysis using DESeq2 of GF female serum-treated female primary hepatocytes incubated with or in the absence of GH.
Sheet 6. Results of the differential expression analysis using DESeq2 of testis in GF versus ConvR mice.
Sheet 7. Results of the differential expression analysis using DESeq2 of ovaries in GF versus ConvR mice.
Sheet 8. Results of the differential expression analysis using DESeq2 of ghrelin-injected male GF mice in liver.
References
- Agrawal A.K., Pampori N.A., Shapiro B.H. Neonatal phenobarbital-induced defects in age- and sex-specific growth hormone profiles regulating monooxygenases. Am. J. Physiol. 1995;268:E439–E445. doi: 10.1152/ajpendo.1995.268.3.E439. [DOI] [PubMed] [Google Scholar]
- El Aidy S., Derrien M., Merrifield C.A., Levenez F., Dore J., Boekschoten M.V., Dekker J., Holmes E., Zoetendal E.G., van Baarlen P. Gut bacteria-host metabolic interplay during conventionalisation of the mouse germfree colon. ISME J. 2013;7:743–755. doi: 10.1038/ismej.2012.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez J.D., Hansen A., Ord T., Bebas P., Chappell P.E., Giebultowicz J.M., Williams C., Moss S., Sehgal A. The circadian clock protein BMAL1 is necessary for fertility and proper testosterone production in mice. J. Biol. Rhythms. 2008;23:26–36. doi: 10.1177/0748730407311254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anakk S., Huang W., Staudinger J.L., Tan K., Cole T.J., Moore D.D., Strobel H.W. Gender dictates the nuclear receptor-mediated regulation of CYP3A44. Drug Metab. Dispos. 2007;35:36–42. doi: 10.1124/dmd.106.011270. [DOI] [PubMed] [Google Scholar]
- Ando H., Kumazaki M., Motosugi Y., Ushijima K., Maekawa T., Ishikawa E., Fujimura A. Impairment of peripheral circadian clocks precedes metabolic abnormalities in ob/ob mice. Endocrinology. 2011;152:1347–1354. doi: 10.1210/en.2010-1068. [DOI] [PubMed] [Google Scholar]
- Arnold P., Erb I., Pachkov M., Molina N., van Nimwegen E. MotEvo: integrated Bayesian probabilistic methods for inferring regulatory sites and motifs on multiple alignments of DNA sequences. Bioinformatics. 2012;28:487–494. doi: 10.1093/bioinformatics/btr695. [DOI] [PubMed] [Google Scholar]
- Atger F., Gobet C., Marquis J., Martin E., Wang J., Weger B., Lefebvre G., Descombes P., Naef F., Gachon F. Circadian and feeding rhythms differentially affect rhythmic mRNA transcription and translation in mouse liver. Proc. Natl. Acad. Sci. U S A. 2015;112:E6579–E6588. doi: 10.1073/pnas.1515308112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T., Shima Y., Owaki A., Mimura J., Oshima M., Fujii-Kuriyama Y., Morohashi K.I. Disruption of aryl hydrocarbon receptor (AhR) induces regression of the seminal vesicle in aged male mice. Sex Dev. 2008;2:1–11. doi: 10.1159/000117714. [DOI] [PubMed] [Google Scholar]
- Bäckhed F., Ding H., Wang T., Hooper L.V., Koh G.Y., Nagy A., Semenkovich C.F., Gordon J.I. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. U S A. 2004;101:15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäckhed F., Manchester J.K., Semenkovich C.F., Gordon J.I. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc. Natl. Acad. Sci. U S A. 2007;104:979–984. doi: 10.1073/pnas.0605374104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balwierz P.J., Carninci P., Daub C.O., Kawai J., Hayashizaki Y., Van Belle W., Beisel C., van Nimwegen E. Methods for analyzing deep sequencing expression data: constructing the human and mouse promoterome with deepCAGE data. Genome Biol. 2009;10:R79. doi: 10.1186/gb-2009-10-7-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balwierz P.J., Pachkov M., Arnold P., Gruber A.J., Zavolan M., van Nimwegen E. ISMARA: automated modeling of genomic signals as a democracy of regulatory motifs. Genome Res. 2014;24:869–884. doi: 10.1101/gr.169508.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baptissart M., Vega A., Martinot E., Pommier A.J., Houten S.M., Marceau G., Haze A.d., Baron S., Schoonjans K., Lobaccaro J.-M.A. Bile acids alter male fertility through G-protein-coupled bile acid receptor 1 signaling pathways in mice. Hepatology. 2014;60:1054–1065. doi: 10.1002/hep.27204. [DOI] [PubMed] [Google Scholar]
- Barnett K.R., Tomic D., Gupta R.K., Miller K.P., Meachum S., Paulose T., Flaws J.A. The aryl hydrocarbon receptor affects mouse ovarian follicle growth via mechanisms involving estradiol regulation and responsiveness. Biol. Reprod. 2007;76:1062–1070. doi: 10.1095/biolreprod.106.057687. [DOI] [PubMed] [Google Scholar]
- Ben Ami E., Yuval B., Jurkevitch E. Manipulation of the microbiota of mass-reared Mediterranean fruit flies Ceratitis capitata (Diptera: Tephritidae) improves sterile male sexual performance. ISME J. 2009;4:28–37. doi: 10.1038/ismej.2009.82. [DOI] [PubMed] [Google Scholar]
- Ben-Sahra I., Hoxhaj G., Ricoult S.J.H., Asara J.M., Manning B.D. mTORC1 induces purine synthesis through control of the mitochondrial tetrahydrofolate cycle. Science. 2016;351:728–733. doi: 10.1126/science.aad0489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B. 1995;57:289–300. [Google Scholar]
- Bishop J.O., Clark A.J., Clissold P.M., Hainey S., Francke U. Two main groups of mouse major urinary protein genes, both largely located on chromosome 4. EMBO J. 1982;1:615–620. doi: 10.1002/j.1460-2075.1982.tb01217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björkholm B., Bok C.M., Lundin A., Rafter J., Hibberd M.L., Pettersson S. Intestinal microbiota regulate xenobiotic metabolism in the liver. PLoS One. 2009;4:e6958. doi: 10.1371/journal.pone.0006958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanck A., Åström A., Hansson T. Effects of neonatal and adult castration on the in vitro metabolism of steroids and xenobiotics in rat liver. Cancer Res. 1986;46:5072–5076. [PubMed] [Google Scholar]
- Broida J., Svare B. Sex differences in the activity of mice: modulation by postnatal gonadal hormones. Horm. Behav. 1984;18:65–78. doi: 10.1016/0018-506x(84)90051-5. [DOI] [PubMed] [Google Scholar]
- Brown T., Howe F.S., Murray S.C., Wouters M., Lorenz P., Seward E., Rata S., Angel A., Mellor J. Antisense transcription-dependent chromatin signature modulates sense transcript dynamics. Mol. Syst. Biol. 2018;14:e8007. doi: 10.15252/msb.20178007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown-Borg H.M., Rakoczy S., Wonderlich J.A., Armstrong V., Rojanathammanee L. Altered dietary methionine differentially impacts glutathione and methionine metabolism in long-living growth hormone-deficient Ames dwarf and wild-type mice. Longev. Healthspan. 2014;3:10. doi: 10.1186/2046-2395-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bur I.M., Cohen-Solal A.M., Carmignac D., Abecassis P.-Y., Chauvet N., Martin A.O., van der Horst G.T.J., Robinson I.C.A.F., Maurel P., Mollard P. The circadian clock components CRY1 and CRY2 are necessary to sustain sex dimorphism in mouse liver metabolism. J. Biol. Chem. 2009;284:9066–9073. doi: 10.1074/jbc.M808360200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caesar R., Tremaroli V., Kovatcheva-Datchary P., Cani P.D., Bäckhed F. Crosstalk between gut microbiota and dietary lipids aggravates WAT inflammation through TLR signaling. Cell Metab. 2015;22:658–668. doi: 10.1016/j.cmet.2015.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., Fierer N., Peña A.G., Goodrich J.K., Gordon J.I. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J.G., Lauber C.L., Walters W.A., Berg-Lyons D., Huntley J., Fierer N., Owens S.M., Betley J., Fraser L., Bauer M. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmody R.N., Turnbaugh P.J. Host-microbial interactions in the metabolism of therapeutic and diet-derived xenobiotics. J. Clin. Invest. 2014;124:4173–4181. doi: 10.1172/JCI72335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnevali O., Avella M.A., Gioacchini G. Effects of probiotic administration on zebrafish development and reproduction. Gen. Comp. Endocrinol. 2013;188:297–302. doi: 10.1016/j.ygcen.2013.02.022. [DOI] [PubMed] [Google Scholar]
- Clegg D.J., Brown L.M., Zigman J.M., Kemp C.J., Strader A.D., Benoit S.C., Woods S.C., Mangiaracina M., Geary N. Estradiol-dependent decrease in the orexigenic potency of ghrelin in female rats. Diabetes. 2007;56:1051–1058. doi: 10.2337/db06-0015. [DOI] [PubMed] [Google Scholar]
- Conforto T.L., Waxman D.J. Sex-specific mouse liver gene expression: genome-wide analysis of developmental changes from pre-pubertal period to young adulthood. Biol. Sex Differ. 2012;3:9. doi: 10.1186/2042-6410-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conforto T.L., Zhang Y., Sherman J., Waxman D.J. Impact of CUX2 on the female mouse liver transcriptome: activation of female-biased genes and repression of male-biased genes. Mol. Cell. Biol. 2012;32:4611–4627. doi: 10.1128/MCB.00886-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conforto T.L., Steinhardt I.V.G.F., Waxman D.J. Cross talk between GH-regulated transcription factors HNF6 and CUX2 in adult mouse liver. Mol. Endocrinol. 2015;29:1286–1302. doi: 10.1210/me.2015-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornell R.P., Liljequist B.L., Bartizal K.F. Depressed liver regeneration after partial hepatectomy of germ-free, athymic and lipopolysaccharide-resistant mice. Hepatology. 1990;11:916–922. doi: 10.1002/hep.1840110603. [DOI] [PubMed] [Google Scholar]
- Costa A., Hood I.V., Berger J.M. Mechanisms for initiating cellular DNA replication. Annu. Rev. Biochem. 2013;82:25–54. doi: 10.1146/annurev-biochem-052610-094414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coste A., Dubuquoy L., Barnouin R., Annicotte J.-S., Magnier B., Notti M., Corazza N., Antal M.C., Metzger D., Desreumaux P. LRH-1-mediated glucocorticoid synthesis in enterocytes protects against inflammatory bowel disease. Proc. Natl. Acad. Sci. U S A. 2007;104:13098–13103. doi: 10.1073/pnas.0702440104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox L.M., Yamanishi S., Sohn J., Alekseyenko A.V., Leung J.M., Cho I., Kim S.G., Li H., Gao Z., Mahana D. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell. 2014;158:705–721. doi: 10.1016/j.cell.2014.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crispim C.A., Waterhouse J., Dâmaso A.R., Zimberg I.Z., Padilha H.G., Oyama L.M., Tufik S., de Mello M.T. Hormonal appetite control is altered by shift work: a preliminary study. Metabolism. 2011;60:1726–1735. doi: 10.1016/j.metabol.2011.04.014. [DOI] [PubMed] [Google Scholar]
- Daniela S.C., Lima M.M., Carlos P.J., Bruno G. Appetite-regulating hormones from the upper gut: disrupted control of xenin and ghrelin in night workers. Clin. Endocrinol. (Oxf.) 2013;79:807–811. doi: 10.1111/cen.12114. [DOI] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domcke S., Bardet A.F., Adrian Ginno P., Hartl D., Burger L., Schübeler D. Competition between DNA methylation and transcription factors determines binding of NRF1. Nature. 2015;528:575–579. doi: 10.1038/nature16462. [DOI] [PubMed] [Google Scholar]
- Donohoe D.R., Wali A., Brylawski B.P., Bultman S.J. Microbial regulation of glucose metabolism and cell-cycle progression in mammalian colonocytes. PLoS One. 2012;7:e46589. doi: 10.1371/journal.pone.0046589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durinck S., Spellman P.T., Birney E., Huber W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 2009;4:1184. doi: 10.1038/nprot.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar-Morreale H.F., Álvarez-Blasco F., Botella-Carretero J.I., Luque-Ramírez M. The striking similarities in the metabolic associations of female androgen excess and male androgen deficiency. Hum. Reprod. 2014;29:2083–2091. doi: 10.1093/humrep/deu198. [DOI] [PubMed] [Google Scholar]
- Evans A.M., DeHaven C.D., Barrett T., Mitchell M., Milgram E. Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Anal. Chem. 2009;81:6656–6667. doi: 10.1021/ac901536h. [DOI] [PubMed] [Google Scholar]
- Farhy L.S., Veldhuis J.D. Deterministic construct of amplifying actions of ghrelin on pulsatile growth hormone secretion. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2005;288:R1649–R1663. doi: 10.1152/ajpregu.00451.2004. [DOI] [PubMed] [Google Scholar]
- Fayard E., Auwerx J., Schoonjans K. LRH-1: an orphan nuclear receptor involved in development, metabolism and steroidogenesis. Trends Cell Biol. 2004;14:250–260. doi: 10.1016/j.tcb.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Finkelstein J.A., Jervois P., Menadue M., Willoughby J.O. Growth hormone and prolactin secretion in genetically obese Zucker rats. Endocrinology. 1986;118:1233–1236. doi: 10.1210/endo-118-3-1233. [DOI] [PubMed] [Google Scholar]
- Fishman R.H., Yanai J. Long-lasting effects of early barbiturates on central nervous system and behavior. Neurosci. Biobehav. Rev. 1983;7:19–28. doi: 10.1016/0149-7634(83)90004-0. [DOI] [PubMed] [Google Scholar]
- Gachon F., Fleury Olela F., Schaad O., Descombes P., Schibler U. The circadian PAR-domain basic leucine zipper transcription factors DBP, TEF, and HLF modulate basal and inducible xenobiotic detoxification. Cell Metab. 2006;4:25–36. doi: 10.1016/j.cmet.2006.04.015. [DOI] [PubMed] [Google Scholar]
- Gaidatzis D., Burger L., Florescu M., Stadler M.B. Analysis of intronic and exonic reads in RNA-seq data characterizes transcriptional and post-transcriptional regulation. Nat. Biotechnol. 2015;33:722–729. doi: 10.1038/nbt.3269. [DOI] [PubMed] [Google Scholar]
- Garsin D.A., Villanueva J.M., Begun J., Kim D.H., Sifri C.D., Calderwood S.B., Ruvkun G., Ausubel F.M. Long-lived C. elegans daf-2 mutants are resistant to bacterial pathogens. Science. 2003;300:1921. doi: 10.1126/science.1080147. [DOI] [PubMed] [Google Scholar]
- Gissendanner C.R., Sluder A.E. nhr-25, the Caenorhabditis elegans ortholog of ftz-f1, is required for epidermal and somatic gonad development. Dev. Biol. 2000;221:259–272. doi: 10.1006/dbio.2000.9679. [DOI] [PubMed] [Google Scholar]
- Grant G.A., Roe F.J. Influence of germ-free status on hepatoma induction by 7,12-dimethylbenz(a)anthracene in C3H mice. Nature. 1969;222:1282–1283. doi: 10.1038/2221282a0. [DOI] [PubMed] [Google Scholar]
- Harrison L.M., Sinnett-Smith P.A. Rates of lipogenesis and plasma insulin concentrations in inbred lines of mice differing in fatness. Biochim. Biophys. Acta. 1990;1046:202–206. doi: 10.1016/0005-2760(90)90190-9. [DOI] [PubMed] [Google Scholar]
- Hildebrandt M.A., Hoffmann C., Sherrill–Mix S.A., Keilbaugh S.A., Hamady M., Chen Y.Y., Knight R., Ahima R.S., Bushman F., Wu G.D. High-fat diet determines the composition of the murine gut microbiome independently of obesity. Gastroenterology. 2009;137:1716–1724.e12. doi: 10.1053/j.gastro.2009.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L., Damle S.S., Booten S., Singh P., Sabripour M., Hsu J., Jo M., Katz M., Watt A., Hart C.E. Partial hepatectomy induced long noncoding RNA inhibits hepatocyte proliferation during liver regeneration. PLoS One. 2015;10:e0132798. doi: 10.1371/journal.pone.0132798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B., Butler R., Miao Y., Dai Y., Wu W., Su W., Fujii-Kuriyama Y., Warner M., Gustafsson J.-Å. Dysregulation of notch and ERα signaling in AhR−/− male mice. Proc. Natl. Acad. Sci. U S A. 2016;113:11883–11888. doi: 10.1073/pnas.1613269113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ijssennagger N., Janssen A.W.F., Milona A., Ramos Pittol J.M., Hollman D.A.A., Mokry M., Betzel B., Berends F.J., Janssen I.M., van Mil S.W.C. Gene expression profiling in human precision cut liver slices in response to the FXR agonist obeticholic acid. J. Hepatol. 2016;64:1158–1166. doi: 10.1016/j.jhep.2016.01.016. [DOI] [PubMed] [Google Scholar]
- Jansson J.-O., Edén S., Isaksson O. Sexual dimorphism in the control of growth hormone secretion. Endocr. Rev. 1985;6:128–150. doi: 10.1210/edrv-6-2-128. [DOI] [PubMed] [Google Scholar]
- Jarukamjorn K., Sakuma T., Jaruchotikamol A., Ishino Y., Oguro M., Nemoto N. Modified expression of cytochrome P450 mRNAs by growth hormone in mouse liver. Toxicology. 2006;219:97–105. doi: 10.1016/j.tox.2005.11.014. [DOI] [PubMed] [Google Scholar]
- Jiao X., Sherman B.T., Huang D.W., Stephens R., Baseler M.W., Lane H.C., Lempicki R.A. DAVID-WS: a stateful web service to facilitate gene/protein list analysis. Bioinformatics. 2012;28:1805–1806. doi: 10.1093/bioinformatics/bts251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce S.A., MacSharry J., Casey P.G., Kinsella M., Murphy E.F., Shanahan F., Hill C., Gahan C.G.M. Regulation of host weight gain and lipid metabolism by bacterial bile acid modification in the gut. Proc. Natl. Acad. Sci. U S A. 2014;111:7421–7426. doi: 10.1073/pnas.1323599111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justo R., Boada J., Frontera M., Oliver J., Bermúdez J., Gianotti M. Gender dimorphism in rat liver mitochondrial oxidative metabolism and biogenesis. Am. J. Physiol. Cell Physiol. 2005;289:C372–C378. doi: 10.1152/ajpcell.00035.2005. [DOI] [PubMed] [Google Scholar]
- Kageyama D., Narita S., Watanabe M. Insect sex determination manipulated by their endosymbionts: incidences, mechanisms and implications. Insects. 2012;3:161–199. doi: 10.3390/insects3010161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karman B.N., Hernández-Ochoa I., Ziv-Gal A., Flaws J.A. Involvement of the AHR in development and functioning of the female and male reproductive systems. In: Pohjanvirta R., editor. The AH Receptor in Biology and Toxicology. John Wiley; 2012. pp. 437–466. [Google Scholar]
- Kass R.E., Raftery A.E. Bayes factors. J. Am. Stat. Assoc. 1995;90:773–795. [Google Scholar]
- Katkocin D.M., Gupta K.M., Collipp P.J., Maddaiah V.T. Effects of growth hormone on respiration and ATPase activity of rat liver and heart mitochondria. Biochem. Med. 1979;22:134–144. doi: 10.1016/0006-2944(79)90046-2. [DOI] [PubMed] [Google Scholar]
- Keene D.E., Suescun M.O., Bostwick M.G., Chandrashekar V., Bartke A., Kopchick J.J. Puberty is delayed in male growth hormone receptor gene-disrupted mice. J. Androl. 2002;23:661–668. [PubMed] [Google Scholar]
- Kelly D.M., Jones T.H. Testosterone: a metabolic hormone in health and disease. J. Endocrinol. 2013;217:R25–R45. doi: 10.1530/JOE-12-0455. [DOI] [PubMed] [Google Scholar]
- Kieffer D.A., Piccolo B.D., Marco M.L., Kim E.B., Goodson M.L., Keenan M.J., Dunn T.N., Knudsen K.E.B., Martin R.J., Adams S.H. Mice fed a high-fat diet supplemented with resistant starch display marked shifts in the liver metabolome concurrent with altered gut bacteria. J. Nutr. 2016;146:2476–2490. doi: 10.3945/jn.116.238931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieser S., Sarker S.A., Sakwinska O., Foata F., Sultana S., Khan Z., Islam S., Porta N., Combremont S., Betrisey B. Bangladeshi children with acute diarrhea show fecal microbiomes with increased Streptococcus abundance, irrespective of diarrhea etiology. Environ. Microbiol. 2018 doi: 10.1111/1462-2920.14274. [DOI] [PubMed] [Google Scholar]
- Kim Y.D., Kim Y.-H., Tadi S., Yu J.H., Yim Y.-H., Jeoung N.H., Shong M., Hennighausen L., Harris R.A., Lee I.-K. Metformin inhibits growth hormone–mediated hepatic PDK4 gene expression through induction of orphan nuclear receptor small heterodimer partner. Diabetes. 2012;61:2484–2494. doi: 10.2337/db11-1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M., Son Y.G., Kang Y.N., Ha T.K., Ha E. Changes in glucose transporters, gluconeogenesis, and circadian clock after duodenal–jejunal bypass surgery. Obes. Surg. 2015;25:635–641. doi: 10.1007/s11695-014-1434-4. [DOI] [PubMed] [Google Scholar]
- Kohsaka A., Laposky A.D., Ramsey K.M., Estrada C., Joshu C., Kobayashi Y., Turek F.W., Bass J. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab. 2007;6:414–421. doi: 10.1016/j.cmet.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Kojima M., Hosoda H., Date Y., Nakazato M., Matsuo H., Kangawa K. Ghrelin is a growth-hormone-releasing acylated peptide from stomach. Nature. 1999;402:656–660. doi: 10.1038/45230. [DOI] [PubMed] [Google Scholar]
- Koves T.R., Ussher J.R., Noland R.C., Slentz D., Mosedale M., Ilkayeva O., Bain J., Stevens R., Dyck J.R.B., Newgard C.B. Mitochondrial overload and incomplete fatty acid oxidation contribute to skeletal muscle insulin resistance. Cell Metab. 2008;7:45–56. doi: 10.1016/j.cmet.2007.10.013. [DOI] [PubMed] [Google Scholar]
- Kozich J.J., Westcott S.L., Baxter N.T., Highlander S.K., Schloss P.D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 2013;79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriebs A., Jordan S.D., Soto E., Henriksson E., Sandate C.R., Vaughan M.E., Chan A.B., Duglan D., Papp S.J., Huber A.-L. Circadian repressors CRY1 and CRY2 broadly interact with nuclear receptors and modulate transcriptional activity. Proc. Natl. Acad. Sci. U S A. 2017;114:8776–8781. doi: 10.1073/pnas.1704955114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumsiek J., Mittelstrass K., Do K.T., Stückler F., Ried J., Adamski J., Peters A., Illig T., Kronenberg F., Friedrich N. Gender-specific pathway differences in the human serum metabolome. Metabolomics. 2015;11:1815–1833. doi: 10.1007/s11306-015-0829-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krylova I.N., Sablin E.P., Moore J., Xu R.X., Waitt G.M., MacKay J.A., Juzumiene D., Bynum J.M., Madauss K., Montana V. Structural analyses reveal phosphatidyl inositols as ligands for the NR5 orphan receptors SF-1 and LRH-1. Cell. 2005;120:343–355. doi: 10.1016/j.cell.2005.01.024. [DOI] [PubMed] [Google Scholar]
- Kübeck R., Bonet-Ripoll C., Hoffmann C., Walker A., Müller V.M., Schüppel V.L., Lagkouvardos I., Scholz B., Engel K.-H., Daniel H. Dietary fat and gut microbiota interactions determine diet-induced obesity in mice. Mol. Metab. 2016;5:1162–1174. doi: 10.1016/j.molmet.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laermans J., Depoortere I. Chronobesity: role of the circadian system in the obesity epidemic. Obes. Rev. 2016;17:108–125. doi: 10.1111/obr.12351. [DOI] [PubMed] [Google Scholar]
- Laermans J., Vancleef L., Tack J., Depoortere I. Role of the clock gene Bmal1 and the gastric ghrelin-secreting cell in the circadian regulation of the ghrelin-GOAT system. Sci. Rep. 2015;5:16748. doi: 10.1038/srep16748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc J.G., Milani C., de Giori G.S., Sesma F., van Sinderen D., Ventura M. Bacteria as vitamin suppliers to their host: a gut microbiota perspective. Curr. Opin. Biotechnol. 2013;24:160–168. doi: 10.1016/j.copbio.2012.08.005. [DOI] [PubMed] [Google Scholar]
- Lee Y.-H., Sauer B., Gonzalez F.J. Laron dwarfism and non-insulin-dependent diabetes mellitus in the Hnf-1α knockout mouse. Mol. Cell. Biol. 1998;18:3059–3068. doi: 10.1128/mcb.18.5.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Prokopec S.D., Watson J.D., Sun R.X., Pohjanvirta R., Boutros P.C. Male and female mice show significant differences in hepatic transcriptomic response to 2,3,7,8-tetrachlorodibenzo-p-dioxin. BMC Genomics. 2015;16:625. doi: 10.1186/s12864-015-1840-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone V., Gibbons S.M., Martinez K., Hutchison A.L., Huang E.Y., Cham C.M., Pierre J.F., Heneghan A.F., Nadimpalli A., Hubert N. Effects of diurnal variation of gut microbes and high-fat feeding on host circadian clock function and metabolism. Cell Host Microbe. 2015;17:681–689. doi: 10.1016/j.chom.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lex A., Gehlenborg N., Strobelt H., Vuillemot R., Pfister H. UpSet: visualization of intersecting sets. IEEE Trans. Vis. Comput. Graph. 2014;20:1983–1992. doi: 10.1109/TVCG.2014.2346248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley R.E., Bäckhed F., Turnbaugh P., Lozupone C.A., Knight R.D., Gordon J.I. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. U S A. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., Subgroup G.P.D.P. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Innocentin S., Withers D.R., Roberts N.A., Gallagher A.R., Grigorieva E.F., Wilhelm C., Veldhoen M. Exogenous stimuli maintain intraepithelial lymphocytes via aryl hydrocarbon receptor activation. Cell. 2011;147:629–640. doi: 10.1016/j.cell.2011.09.025. [DOI] [PubMed] [Google Scholar]
- Li L., Dong J., Yan L., Yong J., Liu X., Hu Y., Fan X., Wu X., Guo H., Wang X. Single-cell RNA-seq analysis maps development of human germline cells and gonadal niche interactions. Cell Stem Cell. 2017;20:858–873.e4. doi: 10.1016/j.stem.2017.03.007. [DOI] [PubMed] [Google Scholar]
- Liang X., Bushman F.D., FitzGerald G.A. Rhythmicity of the intestinal microbiota is regulated by gender and the host circadian clock. Proc. Natl. Acad. Sci. U S A. 2015;112:10479–10484. doi: 10.1073/pnas.1501305112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichanska A.M., Waters M.J. How growth hormone controls growth, obesity and sexual dimorphism. Trends Genet. 2008;24:41–47. doi: 10.1016/j.tig.2007.10.006. [DOI] [PubMed] [Google Scholar]
- Lin C.-C.J., Wang M.C. Microbial metabolites regulate host lipid metabolism through NR5A-Hedgehog signalling. Nat. Cell Biol. 2017;19:550–557. doi: 10.1038/ncb3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin G., LaPensee C.R., Qin Z.S., Schwartz J. Reciprocal occupancy of BCL6 and STAT5 on growth hormone target genes: contrasting transcriptional outcomes and promoter-specific roles of p300 and HDAC3. Mol. Cell Endocrinol. 2014;395:19–31. doi: 10.1016/j.mce.2014.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindén J., Lensu S., Pohjanvirta R. Effect of 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) on hormones of energy balance in a TCDD-sensitive and a TCDD-resistant rat strain. Int. J. Mol. Sci. 2014;15:13938–13966. doi: 10.3390/ijms150813938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C., Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T.T., Makishima M., Repa J.J., Schoonjans K., Kerr T.A., Auwerx J., Mangelsdorf D.J. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol. Cell. 2000;6:507–515. doi: 10.1016/s1097-2765(00)00050-2. [DOI] [PubMed] [Google Scholar]
- Lu Y.-F., Jin T., Xu Y., Zhang D., Wu Q., Zhang Y.-K.J., Liu J. Sex differences in the circadian variation of cytochrome P450 genes and corresponding nuclear receptors in mouse liver. Chronobiol. Int. 2013;30:1135–1143. doi: 10.3109/07420528.2013.805762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukassen S., Bosch E., Ekici A.B., Winterpacht A. Characterization of germ cell differentiation in the male mouse through single-cell RNA sequencing. Sci. Rep. 2018;8:6521. doi: 10.1038/s41598-018-24725-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardinoglu A., Shoaie S., Bergentall M., Ghaffari P., Zhang C., Larsson E., Bäckhed F., Nielsen J. The gut microbiota modulates host amino acid and glutathione metabolism in mice. Mol. Syst. Biol. 2015;11:834. doi: 10.15252/msb.20156487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markle J.G.M., Frank D.N., Mortin-Toth S., Robertson C.E., Feazel L.M., Rolle-Kampczyk U., von Bergen M., McCoy K.D., Macpherson A.J., Danska J.S. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science. 2013;339:1084–1088. doi: 10.1126/science.1233521. [DOI] [PubMed] [Google Scholar]
- Le Martelot G., Claudel T., Gatfield D., Schaad O., Kornmann B., Sasso G.L., Moschetta A., Schibler U. REV-ERBα participates in circadian SREBP signaling and bile acid homeostasis. PLoS Biol. 2009;7:e1000181. doi: 10.1371/journal.pbio.1000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinot E., Baptissart M., Véga A., Sèdes L., Rouaisnel B., Vaz F., Saru J.-P., de Haze A., Baron S., Caira F. Bile acid homeostasis controls CAR signaling pathways in mouse testis through FXRalpha. Sci. Rep. 2017;7:42182. doi: 10.1038/srep42182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauvais-Jarvis F. Sex differences in metabolic homeostasis, diabetes, and obesity. Biol. Sex Differ. 2015;6:14. doi: 10.1186/s13293-015-0033-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melia T., Hao P., Yilmaz F., Waxman D.J. Hepatic long intergenic noncoding RNAs: high promoter conservation and dynamic, sex-dependent transcriptional regulation by growth hormone. Mol. Cell. Biol. 2016;36:50–69. doi: 10.1128/MCB.00861-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer R.D., Laz E.V., Su T., Waxman D.J. Male-specific hepatic Bcl6: growth hormone-induced block of transcription elongation in females and binding to target genes inversely coordinated with STAT5. Mol. Endocrinol. 2009;23:1914–1926. doi: 10.1210/me.2009-0242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miletta M.C., Petkovic V., Eblé A., Ammann R.A., Flück C.E., Mullis P.-E. Butyrate increases intracellular calcium levels and enhances growth hormone release from rat anterior pituitary cells via the G-protein-coupled receptors GPR41 and 43. PLoS One. 2014;9:e107388. doi: 10.1371/journal.pone.0107388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller B.H., Olson S.L., Turek F.W., Levine J.E., Horton T.H., Takahashi J.S. Circadian clock mutation disrupts estrous cyclicity and maintenance of pregnancy. Curr. Biol. 2004;14:1367–1373. doi: 10.1016/j.cub.2004.07.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagner A., Korecka A., Polizzi A., Lippi Y., Blum Y., Canlet C., Tremblay-Franco M., Gautier-Stein A., Burcelin R., Yen Y.-C. Hepatic circadian clock oscillators and nuclear receptors integrate microbiome-derived signals. Sci. Rep. 2016;6:20127. doi: 10.1038/srep20127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moura-Alves P., Fae K., Houthuys E., Dorhoi A., Kreuchwig A., Furkert J., Barison N., Diehl A., Munder A., Constant P. AhR sensing of bacterial pigments regulates antibacterial defence. Nature. 2014;512:387–392. doi: 10.1038/nature13684. [DOI] [PubMed] [Google Scholar]
- Mukherji A., Kobiita A., Ye T., Chambon P. Homeostasis in intestinal epithelium is orchestrated by the circadian clock and microbiota cues transduced by TLRs. Cell. 2013;153:812–827. doi: 10.1016/j.cell.2013.04.020. [DOI] [PubMed] [Google Scholar]
- Murphy C.T., McCarroll S.A., Bargmann C.I., Fraser A., Kamath R.S., Ahringer J., Li H., Kenyon C. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–283. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- Murray I.A., Nichols R.G., Zhang L., Patterson A.D., Perdew G.H. Expression of the aryl hydrocarbon receptor contributes to the establishment of intestinal microbial community structure in mice. Sci. Rep. 2016;6:33969. doi: 10.1038/srep33969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myronovych A., Kirby M., Ryan K.K., Zhang W., Jha P., Setchell K.D.R., Dexheimer P.J., Aronow B., Seeley R.J., Kohli R. Vertical sleeve gastrectomy reduces hepatic steatosis while increasing serum bile acids in a weight-loss-independent manner. Obesity. 2014;22:390–400. doi: 10.1002/oby.20548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng B.G., Wolfe L.A., Ichikawa M., Markello T., He M., Tifft C.J., Gahl W.A., Freeze H.H. Biallelic mutations in CAD, impair de novo pyrimidine biosynthesis and decrease glycosylation precursors. Hum. Mol. Genet. 2015;24:3050–3057. doi: 10.1093/hmg/ddv057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwdorp M., Gilijamse P.W., Pai N., Kaplan L.M. Role of the microbiome in energy regulation and metabolism. Gastroenterology. 2014;146:1525–1533. doi: 10.1053/j.gastro.2014.02.008. [DOI] [PubMed] [Google Scholar]
- Nukaya M., Takahashi Y., Gonzalez F.J., Kamataki T. Aryl hydrocarbon receptor-mediated suppression of GH receptor and Janus kinase 2 expression in mice. FEBS Lett. 2004;558:96–100. doi: 10.1016/S0014-5793(03)01528-X. [DOI] [PubMed] [Google Scholar]
- Ohtake F., Fujii-Kuriyama Y., Kawajiri K., Kato S. Cross-talk of dioxin and estrogen receptor signals through the ubiquitin system. J. Steroid Biochem. Mol. Biol. 2011;127:102–107. doi: 10.1016/j.jsbmb.2011.03.007. [DOI] [PubMed] [Google Scholar]
- Pachkov M., Balwierz P.J., Arnold P., Ozonov E., van Nimwegen E. SwissRegulon, a database of genome-wide annotations of regulatory sites: recent updates. Nucleic Acids Res. 2013;41:D214–D220. doi: 10.1093/nar/gks1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagotto U., Gambineri A., Pelusi C., Genghini S., Cacciari M., Otto B., Castañeda T., Tschöp M., Pasquali R. Testosterone replacement therapy restores normal ghrelin in hypogonadal men. J. Clin. Endocrinol. Metab. 2003;88:4139–4143. doi: 10.1210/jc.2003-030554. [DOI] [PubMed] [Google Scholar]
- Perry R.J., Peng L., Barry N.A., Cline G.W., Zhang D., Cardone R.L., Petersen K.F., Kibbey R.G., Goodman A.L., Shulman G.I. Acetate mediates a microbiome-brain-β-cell axis to promote metabolic syndrome. Nature. 2016;534:213–217. doi: 10.1038/nature18309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen S.L., Krishnan S., Hudgens E.D. The aryl hydrocarbon receptor pathway and sexual differentiation of neuroendocrine functions. Endocrinology. 2006;147:s33–s42. doi: 10.1210/en.2005-1157. [DOI] [PubMed] [Google Scholar]
- Pinto R.E., Bartley W. The nature of the sex-linked differences in glutathione peroxidase activity and aerobic oxidation of glutathione in male and female rat liver. Biochem. J. 1969;115:449–456. doi: 10.1042/bj1150449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohjanvirta R., Miettinen H., Sankari S., Hegde N., Lindén J. Unexpected gender difference in sensitivity to the acute toxicity of dioxin in mice. Toxicol. Appl. Pharmacol. 2012;262:167–176. doi: 10.1016/j.taap.2012.04.032. [DOI] [PubMed] [Google Scholar]
- Poroyko V.A., Carreras A., Khalyfa A., Khalyfa A.A., Leone V., Peris E., Almendros I., Gileles-Hillel A., Qiao Z., Hubert N. Chronic sleep disruption alters gut microbiota, induces systemic and adipose tissue inflammation and insulin resistance in mice. Sci. Rep. 2016;6:35405. doi: 10.1038/srep35405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X., Metz R.P., Porter W.W., Neuendorff N., Earnest B.J., Earnest D.J. The clock genes period 1 and period 2 mediate diurnal rhythms in dioxin-induced Cyp1A1 expression in the mouse mammary gland and liver. Toxicol. Lett. 2010;196:28–32. doi: 10.1016/j.toxlet.2010.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabot S., Membrez M., Bruneau A., Gérard P., Harach T., Moser M., Raymond F., Mansourian R., Chou C.J. Germ-free C57BL/6J mice are resistant to high-fat-diet-induced insulin resistance and have altered cholesterol metabolism. FASEB J. 2010;24:4948–4959. doi: 10.1096/fj.10-164921. [DOI] [PubMed] [Google Scholar]
- Rae R., Sinha A., Sommer R.J. Genome-wide analysis of germline signaling genes regulating longevity and innate immunity in the nematode Pristionchus pacificus. PLoS Pathog. 2012;8:e1002864. doi: 10.1371/journal.ppat.1002864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . R Foundation for Statistical Computing; 2018. R: A Language and Environment for Statistical Computing. [Google Scholar]
- Reizel Y., Spiro A., Sabag O., Skversky Y., Hecht M., Keshet I., Berman B.P., Cedar H. Gender-specific postnatal demethylation and establishment of epigenetic memory. Genes Dev. 2015;29:923–933. doi: 10.1101/gad.259309.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter S.E., Evans A.M. Carnitine and acylcarnitines: pharmacokinetic, pharmacological and clinical aspects. Clin. Pharmacokinet. 2012;51:553–572. doi: 10.1007/BF03261931. [DOI] [PubMed] [Google Scholar]
- Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rocca C., Tait S., Guerranti C., Busani L., Ciardo F., Bergamasco B., Stecca L., Perra G., Mancini F., Marci R. Exposure to endocrine disrupters and nuclear receptor gene expression in infertile and fertile women from different Italian areas. Int. J. Environ. Res. Public Health. 2014;11:10146–10164. doi: 10.3390/ijerph111010146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rocca C., Tait S., Guerranti C., Busani L., Ciardo F., Bergamasco B., Perra G., Mancini F., Marci R., Bordi G. Exposure to endocrine disruptors and nuclear receptors gene expression in infertile and fertile men from Italian areas with different environmental features. Int. J. Environ. Res. Public Health. 2015;12:12426–12445. doi: 10.3390/ijerph121012426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano K.A., Vivas E.I., Amador-Noguez D., Rey F.E. Intestinal microbiota composition modulates choline bioavailability from diet and accumulation of the proatherogenic metabolite trimethylamine-N-oxide. mBio. 2015;6 doi: 10.1128/mBio.02481-14. e02481–e02414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovinetti C., Bovina C., Tolomelli B., Marchetti M. Effects of testosterone on the metabolism of folate coenzymes in the rat. Biochem. J. 1972;126:291–294. doi: 10.1042/bj1260291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudling M., Parini P., Angelin B. Growth hormone and bile acid synthesis. Key role for the activity of hepatic microsomal cholesterol 7α-hydroxylase in the rat. J. Clin. Invest. 1997;99:2239–2245. doi: 10.1172/JCI119398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayin S.I., Wahlström A., Felin J., Jäntti S., Marschall H.-U., Bamberg K., Angelin B., Hyötyläinen T., Orešič M., Bäckhed F. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013;17:225–235. doi: 10.1016/j.cmet.2013.01.003. [DOI] [PubMed] [Google Scholar]
- De Schepper J.A., Smitz J.P., Zhou X.L., Louis O., Velkeniers B.E., Vanhaelst L. Cafeteria diet-induced obesity is associated with a low spontaneous growth hormone secretion and normal plasma insulin-like growth factor-I concentrations. Growth Horm. IGF Res. 1998;8:397–401. doi: 10.1016/s1096-6374(98)80310-9. [DOI] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss P.D., Westcott S.L., Ryabin T., Hall J.R., Hartmann M., Hollister E.B., Lesniewski R.A., Oakley B.B., Parks D.H., Robinson C.J. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selwyn F.P., Cheng S.L., Bammler T.K., Prasad B., Vrana M., Klaassen C., Cui J.Y. Developmental regulation of drug-processing genes in livers of germ-free mice. Toxicol. Sci. 2015;147:84–103. doi: 10.1093/toxsci/kfv110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y., Yue F., McCleary D.F., Ye Z., Edsall L., Kuan S., Wagner U., Dixon J., Lee L., Lobanenkov V.V. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu K., Muranaka Y., Fujimura R., Ishida H., Tazume S., Shimamura T. Normalization of reproductive function in germfree mice following bacterial contamination. Exp. Anim. 1998;47:151–158. doi: 10.1538/expanim.47.151. [DOI] [PubMed] [Google Scholar]
- Sinturel F., Gerber A., Mauvoisin D., Wang J., Gatfield D., Stubblefield J.J., Green C.B., Gachon F., Schibler U. Diurnal oscillations in liver mass and cell size accompany ribosome assembly cycles. Cell. 2017;169:651–663.e14. doi: 10.1016/j.cell.2017.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spruijt C.G., Gnerlich F., Smits A.H., Pfaffeneder T., Jansen P.W.T.C., Bauer C., Münzel M., Wagner M., Müller M., Khan F. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell. 2013;152:1146–1159. doi: 10.1016/j.cell.2013.02.004. [DOI] [PubMed] [Google Scholar]
- Staudinger J.L., Goodwin B., Jones S.A., Hawkins-Brown D., MacKenzie K.I., LaTour A., Liu Y., Klaassen C.D., Brown K.K., Reinhard J. The nuclear receptor PXR is a lithocholic acid sensor that protects against liver toxicity. Proc. Natl. Acad. Sci. U S A. 2001;98:3369–3374. doi: 10.1073/pnas.051551698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steyn F.J., Huang L., Ngo S.T., Leong J.W., Tan H.Y., Xie T.Y., Parlow A.F., Veldhuis J.D., Waters M.J., Chen C. Development of a method for the determination of pulsatile growth hormone secretion in mice. Endocrinology. 2011;152:3165–3171. doi: 10.1210/en.2011-0253. [DOI] [PubMed] [Google Scholar]
- Suarez-Zamorano N., Fabbiano S., Chevalier C., Stojanovic O., Colin D.J., Stevanovic A., Veyrat-Durebex C., Tarallo V., Rigo D., Germain S. Microbiota depletion promotes browning of white adipose tissue and reduces obesity. Nat. Med. 2015;21:1497–1501. doi: 10.1038/nm.3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugathan A., Waxman D.J. Genome-wide analysis of chromatin states reveals distinct mechanisms of sex-dependent gene regulation in male and female mouse liver. Mol. Cell. Biol. 2013;33:3594–3610. doi: 10.1128/MCB.00280-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda T., Taura J., Hattori Y., Ishii Y., Yamada H. Dioxin-induced retardation of development through a reduction in the expression of pituitary hormones and possible involvement of an aryl hydrocarbon receptor in this defect: a comparative study using two strains of mice with different sensitivities to dioxin. Toxicol. Appl. Pharmacol. 2014;278:220–229. doi: 10.1016/j.taap.2014.04.022. [DOI] [PubMed] [Google Scholar]
- Tanimura N., Kusunose N., Matsunaga N., Koyanagi S., Ohdo S. Aryl hydrocarbon receptor-mediated Cyp1a1 expression is modulated in a CLOCK-dependent circadian manner. Toxicology. 2011;290:203–207. doi: 10.1016/j.tox.2011.09.007. [DOI] [PubMed] [Google Scholar]
- Thaiss C.A., Zeevi D., Levy M., Zilberman-Schapira G., Suez J., Tengeler A.C., Abramson L., Katz M.N., Korem T., Zmora N. Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell. 2014;159:514–529. doi: 10.1016/j.cell.2014.09.048. [DOI] [PubMed] [Google Scholar]
- Thaiss C.A., Levy M., Korem T., Dohnalová L., Shapiro H., Jaitin D.A., David E., Winter D.R., Gury-BenAri M., Tatirovsky E. Microbiota diurnal rhythmicity programs host transcriptome oscillations. Cell. 2016;167:1495–1510.e12. doi: 10.1016/j.cell.2016.11.003. [DOI] [PubMed] [Google Scholar]
- Tijet N., Boutros P.C., Moffat I.D., Okey A.B., Tuomisto J., Pohjanvirta R. Aryl hydrocarbon receptor regulates distinct dioxin-dependent and dioxin-independent gene batteries. Mol. Pharmacol. 2006;69:140–153. doi: 10.1124/mol.105.018705. [DOI] [PubMed] [Google Scholar]
- Tissenbaum H.A., Ruvkun G. An insulin-like signaling pathway affects both longevity and reproduction in Caenorhabditis elegans. Genetics. 1998;148:703–717. doi: 10.1093/genetics/148.2.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremaroli V., Karlsson F., Werling M., Ståhlman M., Kovatcheva-Datchary P., Olbers T., Fändriks L., le Roux Carel W., Nielsen J., Bäckhed F. Roux-en-Y gastric bypass and vertical banded gastroplasty induce long-term changes on the human gut microbiome contributing to fat mass regulation. Cell Metab. 2015;22:228–238. doi: 10.1016/j.cmet.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschöp M., Weyer C., Tataranni P.A., Devanarayan V., Ravussin E., Heiman M.L. Circulating ghrelin levels are decreased in human obesity. Diabetes. 2001;50:707–709. doi: 10.2337/diabetes.50.4.707. [DOI] [PubMed] [Google Scholar]
- Uyama T., Jin X.-H., Tsuboi K., Tonai T., Ueda N. Characterization of the human tumor suppressors TIG3 and HRASLS2 as phospholipid-metabolizing enzymes. Biochim. Biophys. Acta. 2009;1791:1114–1124. doi: 10.1016/j.bbalip.2009.07.001. [DOI] [PubMed] [Google Scholar]
- Vanacker J.M., Pettersson K., Gustafsson J.A., Laudet V. Transcriptional targets shared by estrogen receptor- related receptors (ERRs) and estrogen receptor (ER) α, but not by ERβ. EMBO J. 1999;18:4270–4279. doi: 10.1093/emboj/18.15.4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh M., Mukherjee S., Wang H., Li H., Sun K., Benechet A.P., Qiu Z., Maher L., Redinbo M.R., Phillips R.S. Symbiotic bacterial metabolites regulate gastrointestinal barrier function via the xenobiotic sensor PXR and toll-like receptor 4. Immunity. 2014;41:296–310. doi: 10.1016/j.immuni.2014.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vesselinovitch S.D. Certain aspects of hepatocarcinogenesis in the infant mouse model. Toxicol. Pathol. 1987;15:221–228. doi: 10.1177/019262338701500216. [DOI] [PubMed] [Google Scholar]
- Voigt R.M., Forsyth C.B., Green S.J., Mutlu E., Engen P., Vitaterna M.H., Turek F.W., Keshavarzian A. Circadian disorganization alters intestinal microbiota. PLoS One. 2014;9:e97500. doi: 10.1371/journal.pone.0097500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt R.M., Summa K.C., Forsyth C.B., Green S.J., Engen P., Naqib A., Vitaterna M.H., Turek F.W., Keshavarzian A. The circadian Clock mutation promotes intestinal dysbiosis. Alcohol Clin. Exp. Res. 2016;40:335–347. doi: 10.1111/acer.12943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Uray I.P., Mazumdar A., Mayer J.A., Brown P.H. SLC22A5/OCTN2 expression in breast cancer is induced by estrogen via a novel intronic estrogen-response element (ERE) Breast Cancer Res. Treat. 2012;134:101–115. doi: 10.1007/s10549-011-1925-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Mauvoisin D., Martin E., Atger F., Galindo A.N., Dayon L., Sizzano F., Palini A., Kussmann M., Waridel P. Nuclear proteomics uncovers diurnal regulatory landscapes in mouse liver. Cell Metab. 2017;25:102–117. doi: 10.1016/j.cmet.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waxman D.J., Zhao S., Choi H.K. Interaction of a novel sex-dependent, growth hormone-regulated liver nuclear factor with CYP2C12 promoter. J. Biol. Chem. 1996;271:29978–29987. doi: 10.1074/jbc.271.47.29978. [DOI] [PubMed] [Google Scholar]
- Wei, T., and Simko, V. (2016). Corrplot: visualization of a correlation matrix. R Package version 0.77. https://CRAN.R-project.org/package=corrplot.
- Weibel L., Follenius M., Spiegel K., Gronfier C., Brandenberger G. Growth hormone secretion in night workers. Chronobiol. Int. 1997;14:49–60. doi: 10.3109/07420529709040541. [DOI] [PubMed] [Google Scholar]
- Winer D.A., Luck H., Tsai S., Winer S. The intestinal immune system in obesity and insulin resistance. Cell Metab. 2016;23:413–426. doi: 10.1016/j.cmet.2016.01.003. [DOI] [PubMed] [Google Scholar]
- Winneke G., Ranft U., Wittsiepe J., Kasper-Sonnenberg M., Furst P., Kramer U., Seitner G., Wilhelm M. Behavioral sexual dimorphism in school-age children and early developmental exposure to dioxins and PCBs: a follow-up study of the Duisburg Cohort. Environ. Health Perspect. 2014;122:292–298. doi: 10.1289/ehp.1306533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie T.Y., Ngo S.T., Veldhuis J.D., Jeffery P.L., Chopin L.K., Tschöp M., Waters M.J., Tolle V., Epelbaum J., Chen C. Effect of deletion of ghrelin-O-acyltransferase on the pulsatile release of growth hormone in mice. J. Neuroendocrinol. 2015;27:872–886. doi: 10.1111/jne.12327. [DOI] [PubMed] [Google Scholar]
- Yan J., Herzog J.W., Tsang K., Brennan C.A., Bower M.A., Garrett W.S., Sartor B.R., Aliprantis A.O., Charles J.F. Gut microbiota induce IGF-1 and promote bone formation and growth. Proc. Natl. Acad. Sci. U S A. 2016;113:E7554–E7563. doi: 10.1073/pnas.1607235113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Schadt E.E., Wang S., Wang H., Arnold A.P., Ingram-Drake L., Drake T.A., Lusis A.J. Tissue-specific expression and regulation of sexually dimorphic genes in mice. Genome Res. 2006;16:995–1004. doi: 10.1101/gr.5217506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokomori N., Kobayashi R., Moore R., Sueyoshi T., Negishi M. A DNA methylation site in the male-specific P450 (Cyp 2d-9) promoter and binding of the heteromeric transcription factor GABP. Mol. Cell. Biol. 1995;15:5355–5362. doi: 10.1128/mcb.15.10.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurkovetskiy L., Burrows M., Khan A.A., Graham L., Volchkov P., Becker L., Antonopoulos D., Umesaki Y., Chervonsky A.V. Gender bias in autoimmunity is influenced by microbiota. Immunity. 2013;39:400–412. doi: 10.1016/j.immuni.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaczek D., Hammond J., Suen L., Wandji S., Service D., Bartke A., Chandrashekar V., Coschigano K., Kopchick J. Impact of growth hormone resistance on female reproductive function: new insights from growth hormone receptor knockout mice. Biol. Reprod. 2002;67:1115–1124. doi: 10.1095/biolreprod67.4.1115. [DOI] [PubMed] [Google Scholar]
- Zarrinpar A., Chaix A., Yooseph S., Panda S. Diet and feeding pattern affect the diurnal dynamics of the gut microbiome. Cell Metab. 2014;20:1006–1017. doi: 10.1016/j.cmet.2014.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelante T., Iannitti R.G., Cunha C., De Luca A., Giovannini G., Pieraccini G., Zecchi R., D’Angelo C., Massi-Benedetti C., Fallarino F. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. 2013;39:372–385. doi: 10.1016/j.immuni.2013.08.003. [DOI] [PubMed] [Google Scholar]
- Zhang B., Cheng Q., Ou Z., Lee J.H., Xu M., Kochhar U., Ren S., Huang M., Pflug B.R., Xie W. Pregnane X receptor as a therapeutic target to inhibit androgen activity. Endocrinology. 2010;151:5721–5729. doi: 10.1210/en.2010-0708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Laz E.V., Waxman D.J. Dynamic, sex-differential STAT5 and BCL6 binding to sex-biased, growth hormone-regulated genes in adult mouse liver. Mol. Cell. Biol. 2012;32:880–896. doi: 10.1128/MCB.06312-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Nichols R.G., Correll J., Murray I.A., Tanaka N., Smith P.B., Hubbard T.D., Sebastian A., Albert I., Hatzakis E. Persistent organic pollutants modify gut microbiota–host metabolic homeostasis in mice through aryl hydrocarbon receptor activation. Environ. Health Perspect. 2015;123:679–688. doi: 10.1289/ehp.1409055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L. AHR function in lymphocytes: emerging concepts. Trends Immunol. 2016;37:17–31. doi: 10.1016/j.it.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sheet 1. Detailed table legend.
Sheet 2. Results of the differential expression analysis using DESeq2 of GF versus ConvR male mice in liver.
Sheet 3. Results of the differential expression analysis using DESeq2 of GF versus ConvR male mice in duodenum.
Sheet 4. Results of the differential expression analysis using DESeq2 of GF versus ConvR male mice in ileum.
Sheet 5. Results of the differential expression analysis using DESeq2 of GF versus ConvR male mice in WAT.
Sheet 6. Results of the rhythmicity analysis using multiple linear regression and model selection in liver of male GF and ConvR mice.
Sheet 7. Results of the rhythmicity analysis using multiple linear regression and model selection in duodenum of male GF and ConvR mice.
Sheet 8. Results of the rhythmicity analysis using multiple linear regression and model selection in ileum of male GF and ConvR mice.
Sheet 9. Results of the rhythmicity analysis using multiple linear regression and model selection in WAT of male GF and ConvR mice.
Sheet 1. Detailed table legend.
Sheet 2. Gene Ontology (GO) classification related to the comparison of ConvR and GF male mice in liver.
Sheet 3. GO classification related to the comparison of ConvR and GF male mice in duodenum.
Sheet 4. GO classification related to the comparison of ConvR and GF male mice ileum.
Sheet 5. GO classification related to the comparison of ConvR and GF male mice in WAT.
Sheet 1. Detailed table legend.
Sheet 2. Results of the differential expression analysis using DESeq2 contrasting male ConvR versus female ConvR mice in liver.
Sheet 3. Results of the differential expression analysis using DESeq2 contrasting male GF versus female GF mice in liver.
Sheet 4. Results of the differential expression analysis using DESeq2 contrasting female GF versus female ConvR mice in liver.
Sheet 5. Results of the differential expression analysis using DESeq2 to test for interaction between sex and hygienic status.
Sheet 6. Results of the rhythmicity analysis using multiple linear regression and model selection in liver of male ConvR, male GF, female ConvR, and female GF mice.
Sheet 1. Detailed table legend.
Sheet 2. GO classification related to hepatic differentially expressed genes that show a significant interaction between sex and hygienic status.
Sheet 3. GO classification related to the comparison of ConvR and GF male mice in liver.
Sheet 1. Detailed table legend.
Sheet 2. Results of the differential expression analysis using a linear model (∼genotype∗status) to contrast: male ConvR and female ConvR, male GF and female GF, male GF versus male ConvR, female GF versus female ConvR.
Sheet 3. Results of the rhythmic activity analysis TF motifs using complex-valued singular value decomposition in male-specific rhythmic genes.
Sheet 4. Results of the rhythmic activity analysis of TF motifs using complex-valued singular value decomposition in female-specific rhythmic genes.
Sheet 1. Detailed table legend.
Sheet 2. Relative abundance of liver metabolites and statistics in ConvR male, female, GF male and female mice.
Sheet 3. Relative abundance of serum metabolites and statistics in ConvR male, female, GF male and female mice.
Sheet 4. Results of the rhythmicity analysis using multiple linear regression and model selection for metabolites in liver of ConvR male, female, GF male and female mice.
Sheet 5. Results of the rhythmicity analysis using multiple linear regression and model selection for metabolites in serum of ConvR male, female, GF male and female mice.
Sheet 6. Results of liver metabolite subpathway analysis using a linear model (∼genotype∗status) to contrast: male ConvR and female ConvR, male GF and female GF, male GF versus male ConvR, female GF versus female ConvR.
Sheet 7. Results of serum metabolite subpathway analysis using a linear model (∼genotype∗status) to contrast: male ConvR and female ConvR, male GF and female GF, male GF versus male ConvR, female GF versus female ConvR.
Sheet 1. Detailed table legend.
Sheet 2. Results of the differential expression analysis using DESeq2 of GH-injected male GF mice in liver.
Sheet 3. Results of the differential expression analysis using DESeq2 of ConvR male serum versus ConvR female serum-treated female primary hepatocytes.
Sheet 4. Results of the differential expression analysis using DESeq2 of GF female serum versus ConvR female serum-treated female primary hepatocytes.
Sheet 5. Results of the differential expression analysis using DESeq2 of GF female serum-treated female primary hepatocytes incubated with or in the absence of GH.
Sheet 6. Results of the differential expression analysis using DESeq2 of testis in GF versus ConvR mice.
Sheet 7. Results of the differential expression analysis using DESeq2 of ovaries in GF versus ConvR mice.
Sheet 8. Results of the differential expression analysis using DESeq2 of ghrelin-injected male GF mice in liver.
Data Availability Statement
The accession number for the entire RNA-seq dataset reported in this manuscript is GSE77221. To ease accessibility, we divided the data into the following subseries: (1) Temporal profile of GF and ConvR male and female mice (duodenum and ileum – GSE114399, liver – GSE114400, WAT – GSE114401). (2) Temporal profile of Liver of Cry1/Cry2 KO mice (GSE114402). (3) GF male mice liver injected with GH (GSE114610). (4) GF male mice liver injected with Ghrelin (GSE114611). (5) Testes and ovaries of ConvR and GF female mice (GSE114612). (6) GH and mouse serum treatment on primary hepatocytes (GSE114613). 16s rRNA data have been deposited on the SRA database under the reference SRP151275.