Abstract
Ricin is a fast-acting protein toxin classified by the Centers for Disease Control and Prevention as a biothreat agent. In this report, we describe five new mouse mAbs directed against an immunodominant region, so-called epitope cluster II, on the surface of ricin’s ribosome-inactivating enzymatic subunit A (RTA). The five mAbs were tested alongside four previously described cluster II—specific mAbs for their capacity to passively protect mice against 10× LD50 ricin challenge by injection. Only three of the mAbs (LE4, PH12, and TB12) afforded protection over the 7-d study period. Neither binding affinity nor in vitro toxin-neutralizing activity could fully account for LE4, PH12, and TB12’s potent in vivo activity relative to the other six mAbs. However, epitope mapping studies by hydrogen exchange-mass spectrometry revealed that LE4, PH12, and TB12 shared common contact points on RTA corresponding to RTA α-helices D and E and β-strands d and e located on the back side of RTA relative to the active site. The other six mAbs recognized overlapping epitopes on RTA, but none shared the same hydrogen exchange-mass spectrometry profile as LE4, PH12, and TB12. A high-density competition ELISA with a panel of ricin-specific, single-domain camelid Abs indicated that even though LE4, PH12, and TB12 make contact with similar secondary motifs, they likely approach RTA from different angles. These results underscore how subtle differences in epitope specificity can significantly impact Ab functionality in vivo. ImmunoHorizons, 2018, 2: 262–273
INTRODUCTION
Ricin is at the top of the list of potential biothreat agents, according to a North Atlantic Treaty Organization Biomedical Advisory council (1). Ricin toxin is a product of the castor bean plant (Ricinus communis), which is cultivated worldwide for its oils used in industrial and cosmetic applications. The toxin itself is a ~65 kDa glycoprotein consisting of two subunits, ribosome-inactivating enzymatic subunit A (RTA) and ricin toxin B (RTB), joined by a single disulfide bond (2). RTA is an extraordinarily efficient RNA N-glycosidase (enzyme classification 3.2.2.22) that cleaves the sarcin-ricin loop of 28S rRNA, resulting in ribosome inactivation (3, 4). RTB is a galactose/N-acetyl galactosamine-specific lectin that facilitates RTA endocytosis and retrograde transport from the plasma membrane to the endoplasmic reticulum (ER) of mammalian cells. In the ER, RTA is liberated from RTB, partially unfolded, and then retrotranslocated across the ER membrane into the cell cytoplasm, presumably via the Sec61p translocon (5). In rodents and nonhuman primates, the LD50 of ricin ranges from 1 to 10 μg/kg by injection or inhalation (6).
RTA is the focus of current efforts to develop a countermeasure for ricin, including a subunit vaccine for use by first responders and military personnel (7–9). RTA, 267 aa residues in length, is a globular protein with a total of 10 β-strands (a-j) and seven α-helices (A-G) (2, 10). The active site constitutes a shallow cleft on one side of the molecule. There are four distinct immunodominant regions or epitope clusters on the surface of RTA, originally identified through competition ELISAs with four different toxin-neutralizing mAbs (11–13). Cluster I is focused around RTA’s α-helix B (residues 94–107), a protruding element previously known to be a target of potent toxin-neutralizing Abs (14, 15). Cluster II isdefinedby themAb SyH7 and is located on the back side of RTA, relative to the active site pocket. Cluster III involves α-helices C and G on the front side of RTA, whereas cluster IV forms a diagonal sash from the front to back of the A subunit.
Prior to this report, cluster II consisted of overlapping epitopes defined by four mouse mAbs: SyH7, PA1, TB12, and PH12 (13). All four mAbs have in vitro toxin-neutralizing activities (TNA) and have been shown to passively protect mice from 5× LD50 ricin challenge by injection over a 72-h period (11, 12). However, recent epitope mapping studies using hydrogen exchange-mass spectrometry (HX-MS) have indicated that cluster II actually sectors into at least two distinct subclusters (13). SyH7 engages RTA residues 14–24 (corresponding to α-helix A) and residues 184–207 (corresponding to a loop between α-helices F and G). PA1 also engages residues 184–207, whereas PH12 and TB12 contact RTA residues 62–69 (corresponding to a loop between β-strands d and e) and residues 154–164 (corresponding to a loop between α-helices D and E). Furthermore, we identified a collection of RTA-specific, single-domain camelid Abs (VHHs) that compete with SyH7, PA1, TB12, and PH12 to various degrees for bindingto ricin (16). The majority of the cluster II VHHs are devoid of TNA. Overall, these results indicate that cluster II is much more complex than originally anticipated, encompassing a large amount of surface area on RTA with numerous binding sites for neutralizing and nonneutralizing Abs. Therefore, the goal of the current study was to interrogate cluster II with additional mAbs in an effort to better define which specific structural elements are most associated with in vivo protection.
MATERIALS AND METHODS
Chemicals, reagents, and cell lines
Ricin (R. communis agglutinin II) was purchased from Vector Laboratories (Burlingame, CA) and dialyzed against PBS using a Slide-A-Lyzer Dialysis Cassette (Pierce, Rockford, IL) prior to use in animal experiments and cytotoxicity assays. Goat serum (New Zealand origin) was purchased from Life Technologies (Carlsbad, CA). Cell culture media was prepared by the Wadsworth Center Media Services Facility. MAbs were affinity purified from hybridoma supernatants by endotoxin-free protein G chromatography at the Dana Farber Cancer Institute’s mAb Core Facility (Boston, MA). African green monkey kidney (Vero) cells were obtained from the American Type Culture Collection (Manassas, VA). All other chemicals were purchased from MilliporeSigma (St. Louis, MO), unless otherwise specified.
Animal care and B cell hybridoma production
All mouse experiments were conducted in accordance with the Wadsworth Center’s Institutional Animal Care and Use Committee guidelines. Mice were housed under conventional, specific pathogen-free conditions. Six-week-old female BALB/c or Swiss Webster (SW) mice (Taconic Biosciences, Albany, NY) were administered sublethal amounts of ricin by i.p. injection as follows: 0.1 μg on days 0, 10, and 20; 0.2 μg for BALB/c; and 0.3 μg for SW on day 35. Mice were retro-orbitally bled on day 45; serum was tested by ELISA and toxin-neutralization assay to confirm seroconversion. As a final boost, mice were injected i.p. with the equivalent of ~10× LD50 ricin (2 μg) and then euthanized 4 d later by CO2 asphyxiation. Splenocytes were fused with mouse myeloma cells using Hybri-Max polyethylene glycol. Fusion products were seeded into 96-well, tissue culture-treated plates and cultured/selected in RPMI 1640 (Life Technologies) media supplemented with UltraCruz Hybridoma Cloning Supplement (Santa Cruz Biotechnology, Dallas, TX) containing FCS, oxaloacetate, sodium pyruvate, bovine insulin, hypoxanthine/ aminopterin/thymidine, and penicillin/streptomycin. Hypoxanthine/ aminopterin/thymidine was gradually replaced with hypoxanthine-thymidine; after which, surviving hybridomas secreting Abs of interest were cloned by limiting dilution and expanded in RPMI 1640 media without hypoxanthine-thymidine. Bulk hybridoma line expansions were cultured in serum-,protein-,and antibioticfree CD media (Life Technologies) and resulting supernatants were cleared by filtration before being submitted for affinity purification.
Direct and competitive ELISAs
ELISAs were performed as described (17). For direct ELISAs, Nunc MaxiSorp F96 microtiter plates (Thermo Fisher Scientific, Pittsburgh, PA) were coated with 1 μg/ml mAb or ricin diluted in PBS (pH 7.4). Plates were blocked with 2% goat serum (Life Technologies) in PBS/Tween (0.1%). Medium containing mAb or biotinylated ricin (biotin-R) was then applied to wells neat or diluted into block solution and incubated at room temperature (RT). HRP-labeled, goat anti-mouse, IgG-specific polyclonal Abs (SouthernBiotech, Birmingham, AL) or avidin/ HRP (Thermo Fisher Scientific) were used as secondary reagents along with 3,3’,5,5’-tetramethylbenzidine (Kirkegaard & Perry Laboratories, Gaithersburg, MD) as colorimetric detection substrate; a 1-M phosphoric acid solution was added to each well to stop the reaction. Plates were read on a VersaMax spectrophotometer and analyzed using SoftMax Pro 5.4.5 Software (Molecular Devices, Sunnyvale, CA). Half maximal effective concentration (EC50) values were determined by nonlinear regression of soluble ricin binding curves using least-squares method within the ECanything function of GraphPad Prism 7.01.
Epitope profiling immune-competition capture was performed as follows: Immulon 4 HBX 96-well microtiter plates (Thermo Fisher Scientific) were coated with capture mAb (1 μg/ml) diluted in PBS (pH 7.4) and incubated at RT. Wells were then blocked with 2% goat serum/PBS/Tween (0.1%) solution overnight at 4°C. The biotin-R-limiting concentration used for this capture assay was equal to the EC90 concentration for each coated mAb (range, 30–200 ng/ml); this concentration was kept constant across all wells, and each biotin-R solution was diluted in blocking solution containing 2% goat serum. Ten-fold excess of competitor mAb solutions were made in separate tubes; mAbs were diluted to 10 μg/ml in their respective EC90 values in solution, incubated 15 min, and then applied to wells in duplicate. A series of at least four wells per coated mAb were overlaid with biotin-R EC90-only solution as 100% binding controls for the purpose of calculating binding inhibition. Plates were incubated at RT for 1 h. Wells were then washed three times with PBS with 0.1% Tween-20 and overlaid with HRP/avidin (1 μg/ml) followed by 3,3’,5,5’- tetramethylbenzidine. Plates were analyzed with a VersaMax spectrophotometer using SoftMax Pro 5.2.5 Software. Ricin binding inhibition was calculated as a percentage of biotin-R binding to the capture mAb, in which [100-(OD450C/OD450B) × 100] = ricin binding inhibited by competitor (%); C, competed; B, biotin-R EC90 control.
Vero cell toxin-neutralization assay
Ricin TNA were performed as described (17). Opaque tissue culture-treated 96-well plates (Corning) containing confluent layers of Vero cells were treated with ricin (10 ng/ml), ricin mAb mixtures (in duplicate), or medium alone (as negative control), then incubated for 2 h at 37°C. Initial treatments were then aspirated; wells were overlaid with DMEM supplemented with 10% FBS and penicillin/streptomycin and incubated for 48 h at 37°C. Cell viability was assessed using CellTiter-Glo Reagent (Promega, Madison, WI); plates were read on a SpectraMax L Luminometer (Molecular Devices) and analyzed using SoftMax Pro 5.2.5 Software. 0ne hundred percent viability was defined as the average value of all wells treated with medium only. IC50 values were determined by nonlinear regression of cell viability curves using least squares method within the ECanything junction of GraphPad Prism 7.01.
Surface plasmon resonance
mAb association and dissociation rates for ricin toxin were determined by surface plasmon resonance using the Prote0n XPR36 (Bio-Rad Laboratories, Hercules, CA) as described (18). For ricin immobilization, general layer compact chips were equilibrated in running buffer PBS/0.005% Tween (pH 7.4) at a flow rate of 30 μl/min. Following EDAC (200 mM) sulfo-NHS (50 mM) activation (3 min), ricin was diluted in 10 mM sodium acetate (pH 5) at two different concentrations (4 and 2 μg/ml) and immobilized (2 min). A third vertical channel received only acetate buffer and served as a reference channel. The surfaces were deactivated using 1 M ethanolamine (5 min). The ProteOn multichannel module was then rotated to the horizontal orientation for Ab experiments. Each mAb was serially diluted in running buffer and injected at 50 μl/min for 180 s, followed by 1–3 h of dissociation. After each experiment, the chip surface was regenerated with 10 mM glycine (pH 1.5), each at 100 μl/min for 18 s, until the resonance unit values returned to baseline. All kinetic experiments were performed at 25°C. Kinetic constants for the Ab/ricin interactions were obtained with the ProteON Manager software 3.1.0 (Bio-Rad Laboratories).
Passive protection studies
mAbs (10 or 25 μg) were diluted in endotoxin-free PBS and administered by i.p. injection in a final volume of 0.4 ml to 8-wk-old female BALB/c mice (Taconic Biosciences). Six hours later, mice received the equivalent of ~10× LD50 of ricin (2 μg permouse)by i.p. injection. Following ricin challenge, mice were weighed once and scored for morbidity twice daily for 7 d; mice were euthanized when they became overtly moribund and/or weight loss was >20% prechallenge weight, as mandated by the Wadsworth Center’s Institutional Animal Care and Use Committee.
HX-MS
HX-MS experiments were conducted using a LEAP H/D-X PAL system (Carrboro, NC) and a quadrupole time of flight mass spectrometer (Agilent Technologies, Santa Clara, CA). HX-MS workflow and data processing for mapping epitopes of recombinant RTA by mAbs were carried out as described previously (13). For reasons of safety, HX-MS was conducted on a recombinant version of RTA carrying two attenuating point mutations (V76M and Y80A) (19). In brief, regions of RTA that exhibited significantly slower (protection) or faster (deprotection) HX in the presence of mAbs were identified using a combination of k-means clustering and significance testing based on time-averaged HX measurements, , quantifying the difference between the mAb/RiVax complex and unbound recombinant RTA. Unlike in the previous work, in this study, the results are filtered for solvent accessibility of the RTA residues (R Toth IV, S.K. Angalakurthi, N.J. Mantis, and D. Weis, manuscript in preparation). The k-means clustering was used to classify the effect of the mAb on the HX of RiVax from strongly protected to deprotected. Protected regions were used to define the epitopes. All epitopes identified in this study will be submitted to the Immune Epitope Database (IEDB) and Analysis Resource (http://www.iedb.org) (20).
Statistical analysis
Differences in survival between groups were determined with Kaplan-Meier analysis and log-rank testing. Pair-wise comparisons between groups were performed with log-rank tests, and the resulting p values were adjusted for multiple comparisons with the Benjamini-Hochberg procedure to control the false discovery rate. For all analyses, p values <0.05 were considered significant. All statistical analysis was carried out in GraphPad Prism 7 (GraphPad Software, San Diego, CA) or R version 3.4.2 (21).
RESULTS
To generate additional cluster II mAbs, we screened B cell hybridomas derived from BALB/c and SW mice that had been hyperimmunized with sublethal amounts of ricin toxin. Ricin-specific mAbs were identified by direct ELISA, whereas cluster Il-specific mAbs were classified initially by competition ELISA with SyH7 (data not shown). In total, we identified four new cluster II mAbs: LE4, CH1, SWB1, and 6C4 (Table I). A filth mAb, WECH1, was resurrected from a previous hybridoma screen (11). In addition to competition with SyH7, we performed cross-competition analysis among the five new mAbs themselves (Fig. 1), which confirmed that WECH1, LE4, CH1, SWB1, and 6C4 recognize overlapping epitopes on RTA.
TABLE I.
Relationship between epitope specificity and TNA
| RTA Contact Pointse | ||||||||
|---|---|---|---|---|---|---|---|---|
| mAba | Isotype | b | IC50c | In Vivod | a and A | d and e | D and E | F and G |
| PH12 | IgGl | 9 | 0.08 | 9/10 | – | + | + | – |
| TB12 | IgGl | 42 | 0.11 | 9/10 | – | + | + | – |
| LE4 | IgGl | 37 | 0.3 | 9/11 | – | + | + | – |
| CH1 | IgGl | 95 | >10 | 2/11 | – | – | + | – |
| SWB1 | IgGl | 200 | >10 | 1/5 | – | – | + | – |
| PA1 | IgGl | 9 | 0.07 | 1/11 | – | – | – | + |
| 6C4 | IgGl | 470 | >10 | 2/11 | – | + | – | + |
| WECH1 | IgGl | 370 | 1.7 | 1/11 | + | – | – | + |
| SyH7 | IgGl | 20 | 0.7 | 0/10 | + | – | – | + |
Underlines indicate the five new mAbs described in this study, whereas bold indicates passive protection up to day 7.
Apparent binding affinities (×10–12 M).
Micrograms per milliliter, as determined in Vero cell assay
Indicates number of survivors/total number in group.
Secondary structures on RTA associated with each mAb’s epitope. As per convention, capital letters refer to α-helices, and lowercase letters refer to β-strands.
FIGURE 1. Binning by cross-competition ELISAs of cluster II mAbs.
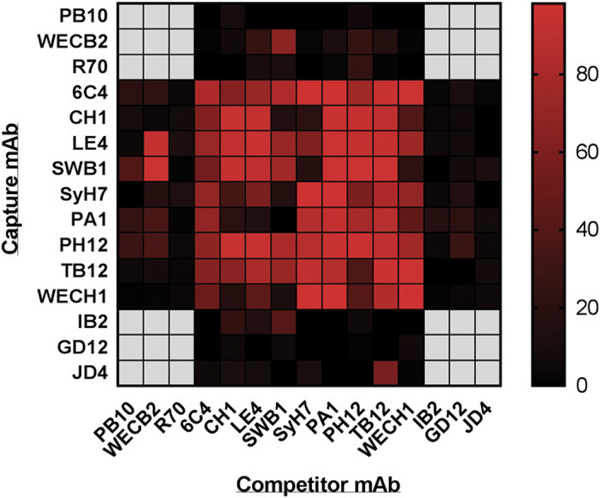
A heat map representation of a cross-competition ELISA with panel of RTA-specific mAbs, as described in the Materials and Methods. The mAbs listed on the vertical axis were coated onto microtiter plates and then assessed for the ability to capture soluble biotin-R in the presence of the indicated competitor mAb (horizontal axis). The percentage (%) of inhibition of biotin-R capture was calculated from the OD values as described in the Materials and Methods section. The values were plotted as a heat map using Prism 7 (GraphPad). The scale bar on the right indicates percentage of inhibition from no competition (black) to complete competition (bright red). The heat map is presented as a means of visualizing the relative competition groups or clusters (I — IV) referred to in the body of the article. PB10, WECB2, and R70 are in cluster I, IB2 in cluster III, and GD12 and JD4 in cluster IV. The remaining mAbs are in cluster II. Actual competition results are shown in Supplemental Fig. 1.
To formally assign the five new mAbs to cluster II, they were subject to a cross-competition capture ELISA with a panel of Abs representing clusters I (PB10, WECB2, and R70), II (SyH7, PA1, PH12, and TB12), III (IB2), and IV (GD12 and JD4). The epitopes recognized by these 10 RTA-specific mAbs were recently resolved using HX-MS, and the results were deposited in the IEDB (13,20). For the most part, the competition results were entirely consistent with the five new mAbs grouping exclusively within cluster II (Fig. 1, Supplemental Fig. 1). For example, the ability of 6C4 to capture soluble ricin was inhibited by the cluster II mAbs (SyH7, PA1, PH12, and TB12) as well as LE4, CH1, WECH1, and SWB1, but not by the representative cluster I, III, or IV mAbs. LE4’s competition profile was also unambiguous except for one instance of nonreciprocal competition with WECB2, one of the three cluster I mAbs. Specifically, soluble WECB2 prevented ricin capture by plate-bound LE4, although soluble LE4 did not prevent ricin capture by WECB2 (13). Another anomaly was the nonreciprocal competition between soluble SyH7/SWB1 and plate-bound CH1. Nonetheless, the overall competition profiles for WECH1, LE4, CH1, SWB1, and 6C4 are consistent with their grouping within cluster II.
The five new cluster II-specific mAbs were next examined for relative binding affinities (KD) and TNA. As expected, all five mAbs bound ricin toxin by direct ELISA (Fig. 2). EC50 values, determined by capture ELISA using biotin-labeled ricin, ranged from ~4 to 200 ng/ml (data not shown), whereas apparent binding affinities, as determined by surface plasmon resonance, ranged from 37 to 470 pM (Supplemental Fig. 2, Table I). TNA, as determined in a Vero cell cytotoxicity assay, ranged from strong for LE4 and WECH1 (IC50, 0.3–1.7 μg/ml) to weak (IC50, >10 μg/ml) for CH1, SWB1, and 6C4 (Fig. 2B, Table I). The four previously described cluster II “legacy” mAbs (PH12, TB12, PA1, and SyH7) each had strong TNA (Fig. 2C).
FIGURE 2. Relative binding profiles and TNA of cluster II-specific mAbs.
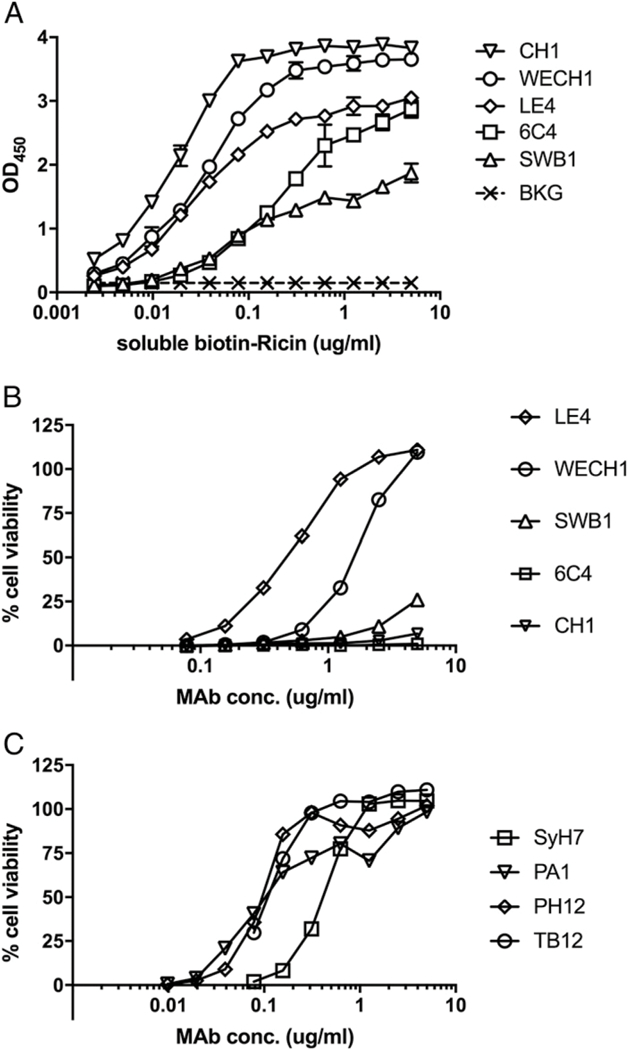
(A) Microtiter plates were coated with indicated mAbs (1 μg/ml) and then assessed for the ability to capture biotin-ricin at concentrations indicated on the x-ordinate. Captured biotin-ricin was detected with saturating amounts of avidin/HRP. BKG, background. (B and C) Indicated mAbs at concentrations shown on the x-axis were mixed with ricin toxin (10 ng/ml) and applied to Vero cells for 2 h. The cells were washed and incubated for ~48 h before being assessed for viability. Shown are representative cytotoxicity assays. Actual IC50 values are presented in Table I.
To assess the in vivo TNA of the five new cluster II mAbs, we performed passive protection studies in which groups of mice received individual mAbs by i.p. injection ~ 6h prior toa10 × LD50 ricin challenge by the same route. Mice were monitored for a period of 7 d for mortality and weight loss (22, 23). For the sake of comparison, the four cluster II legacy mAbs SyH7, PA1, TB12, and PH12 were also included in the study. It is important to note that previous passive protection studies with the cluster II legacy mAbs were terminated alter 3–5 d, not the 7 d used in this study (11,12).
The results of the passive immunization studies indicated that eight of the nine cluster II mAbs conferred some benefit against ricin intoxication (the exception being 6C4), as compared with control mice that received ricin only (Fig. 3A-I, Tables I, II). However, using survival on day 7 as the singular metric, the mAbs stratified into two categories: 1) mice treated with PH12, TB12, or LE4 that were nearly completely protected (90% survival) from ricin-induced death and were by all accounts normal (e.g., feeding behavior, weight gain, and grooming) during a several-week observation period after the formal completion of the study; and 2) mice treated with one of the other six mAbs (CH1, SWB1, PA1,6C4, WECH1, SyH7, or PA1) that succumbed to ricin intoxication (0–20% survival) by day 7 and exhibited significant morbidity as demonstrated by cumulative weight loss during the experimental observation period (Fig. 3J).
FIGURE 3. Potential of cluster II mAbs to passively protect mice against ricin challenge.
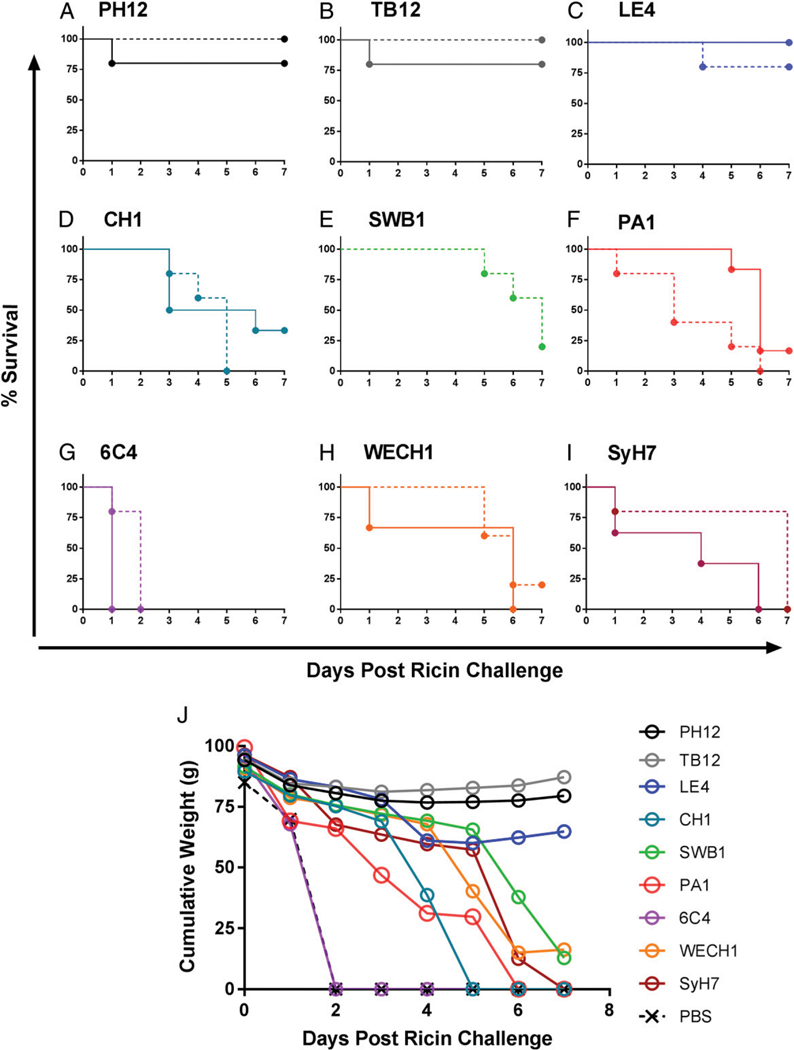
(A-I) Groups of mice were treated with 10 μg (solid) or 25 μg (dashed) of indicated mAbs ~6 h before ricin challenge (day 0). Following ricin challenge, mice were weighed daily for 7 d; mice were euthanized when they became overtly moribund and/or weight loss was >20% prechallenge weight. Control animals that received vehicle only (saline) and then were challenged with ricin succumbed to ricin intoxication within 48 h (data not shown). (J) Cumulative weight loss (i.e., sum of daily weights of mice within a given treatment group) of mice that received 25 μg mAb.
TABLE II.
Pairwise analysis of mAbs in passive protection studies
| PH12 | TB12 | LE4 | CH1 | SWB1 | PA1 | 6C4 | WECH1 | SyH7 | |
|---|---|---|---|---|---|---|---|---|---|
| TB12 | 1.00000a | — | — | — | — | — | — | — | — |
| LE4 | 0.93744a | 0.93744a | — | — | — | — | — | — | — |
| CH1 | 0.01468 | 0.01468 | 0.00528 | — | — | — | — | — | — |
| SWB1 | 0.01891 | 0.01891 | 0.04341 | 0.23219 | — | — | — | — | — |
| PA1 | 0.00195 | 0.00195 | 0.00094 | 0.93744 | 0.10706 | — | — | — | — |
| 6C4 | 0.00020 | 0.00020 | 1.8 × 10–5 | 1.8 × 10–5 | 0.00087 | 0.00016 | — | — | — |
| WECH1 | 0.00228 | 0.00228 | 0.00149 | 0.46408 | 0.29947 | 0.65094 | 1.80 × 10–5 | — | — |
| SyH7 | 0.00067 | 0.00067 | 0.00149 | 0.36137 | 0.32140 | 0.23258 | 0.00020 | 0.71816 | — |
| —b | 0.00020 | 0.00020 | 1.8 × 10–5 | 1.8 × 10–5 | 0.00067 | 0.00015 | (0.0339)c | 1.8 × 10–5 | 0.00020 |
Survival analysis between groups using pairwise comparisons with the log-rank test. The p values were adjusted for multiple testing with the Benjamini-Hochberg procedure.
The three MAbs (TB12, PH12, and LE4) were not significantly different from each other in the protection assay but were more effective in vivo than the other six mAbs tested, as noted by the boldface text.
Dash (—) indicates control mice that received ricin only.
Parentheses indicate that mice treated with 6C4 died significantly earlier than control mice.
The poor outcome of mice treated with CH1, SWB1, and 6C4 was not unexpected considering these mAbs’ relatively modest (or lack of) in vitro TNA and suboptimal apparent binding affinities. However, the failure of SyH7, PA1, and WECH1 to passively protect mice through day 7 postchallenge was surprising considering that their in vitro profiles were similar to PH12 and TB12. For example, all four mAbs have similar ricin toxin apparent binding affinities (KD) and roughly equivalent TNA. Thus, it was not immediately apparent what distinguished PH12, TB12, and LE4 from PA1, SyH7, and WECH1.
We had originally assumed, based on competition ELISAs, that SyH7, PA1, PH12, and TB12 recognize the same or nearly the same epitopes (11). This turned out not to be the case, as revealed through recent high-resolution epitope-mapping studies using HX-MS. By HX-MS, SyH7 protected RTA’s α-helix A (residues 14–24) and α-helices F and G (residues 184–207). PA1 interacted only with α-helices F and G (residues 184–207), whereas PH12 and TB12 protected a loop between α-helices D and E (154–164) and a loop between RTA’s β-strands d and e (residues 62–69) (13). The fact that SyH7/ PA1 and PH12/TB12 recognize spatially distinct epitopes within cluster II prompted us to define the actual binding sites of the five new cluster II mAbs. Therefore, LE4, CH1, SWB1, 6C4, and WECH1 were subjected to epitope mapping by HX-MS (13). A depiction of each RTA peptide map generated in the absence or presence of mAbs LE4, CH1, SWB1, 6C4, and WECH1 is shown in Fig. 4 and summarized in Table III, with the spatial location of the protected solvent-accessible residues mapped on the surface of RTA shown in Fig. 5.
FIGURE 4. HX-MS analysis of cluster II–specific mAbs (A) LE4, (B) CH1, (C) SWB1, (D) 6C4, and (E) WECH1.
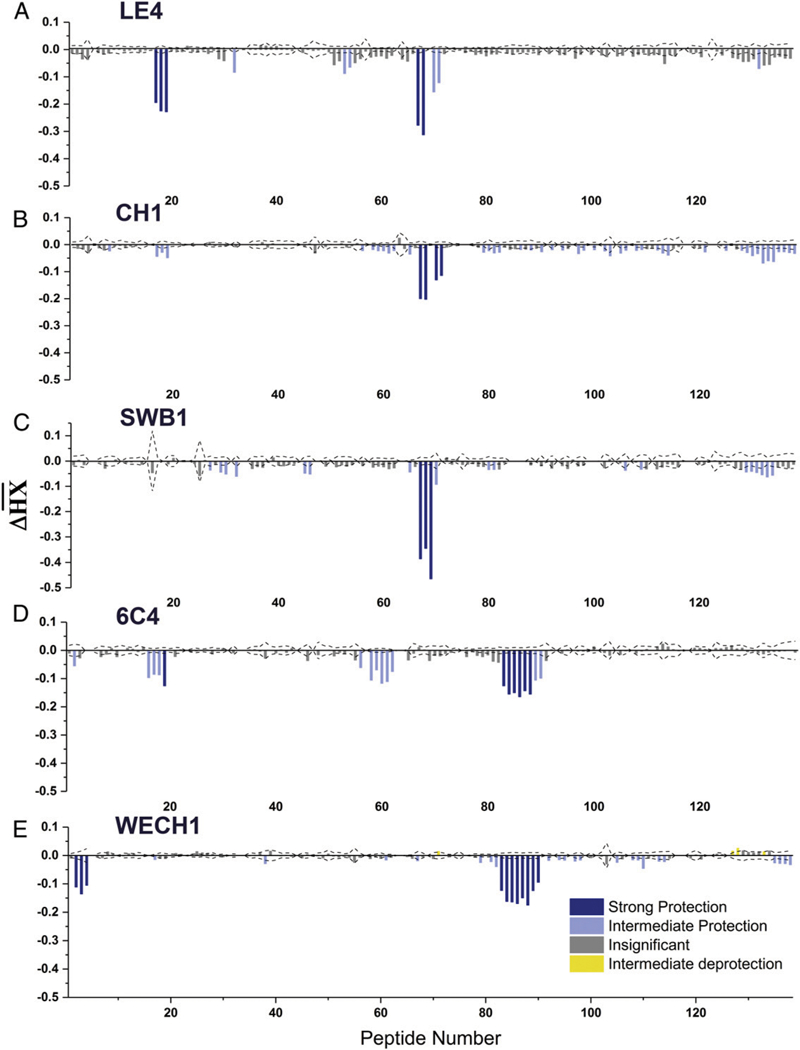
Relative levels of protection of RTA peptides by mAbs as defined by HX-MS. The values are clustered using k-means clustering into four categories: strong protection (deep blue), intermediate protection (light blue), insignificant protection (gray), and intermediate deprotection (yellow). The dotted lines represent the threshold for statistically significant changes in HX. The RTA peptides are indexed sequentially from the N terminus to C terminus as shown in Supplemental Table I.
TABLE III.
HX-MS analysis of cluster II–specific mAbs LE4, CH1, SWB1, 6C4, and WECH1
| Strongly Protected Elementsa | ||||
|---|---|---|---|---|
| mAb | Cluster | Peptides | Residues | Proximity |
| LE4 | II | 17–19 | 63–69 | β-Strand d |
| 67, 68 | 154–161, 163 | α-Helices D and E | ||
| CH1 | II | 67, 68, 70, 71 | 154–161, 163, 166, 167 | α-Helices D and E |
| SWB1 | II | 67–69 | 154–161, 163 | α-Helices D and E |
| 6C4 | II | 19 | 64–68 | β-Strand d |
| 83–88 | 184–187, 189–199, 210, 203, 205–206 | α-Helices F and G | ||
| WECH1 | II | 2–4 | 14–20, 22–24 | α-Helix A |
| 83–90 | 185–187, 189–199, 201, 203, 205–207 | α-Helices F and G | ||
| . | ||||
Actual exchange differences and entire RTA peptide list and corresponding residues are provided in Fig. 4 and Supplemental Table I
FIGURE 5. Epitope positioning of cluster II–specific mAbs on RTA.
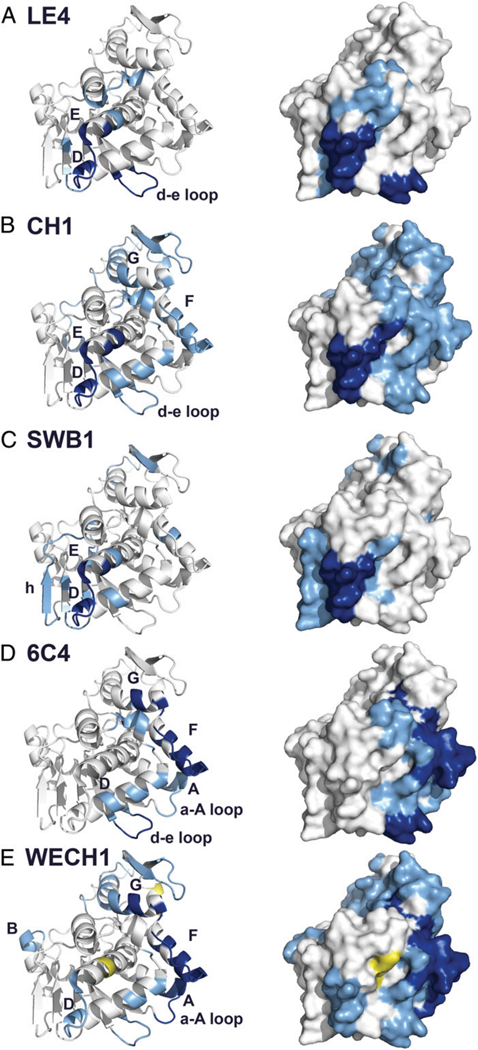
Epitopes on RTA as determined by HX-MS shown using ribbon (left) and surface (right) representations for the five new cluster II mAbs: (A) LE4, (B) CH1, (C) SWB1, (D) 6C4, and (E) WECH1. Degrees of protection are color coded: deep blue, strong protection; light blue, intermediate protection; and yellow, intermediate deprotection. Gray indicates insignificant protection. RTA was modeled with PyMOL using Protein DataBank identifier 3SRP.
HX-MS analysis revealed four distinct protection profiles within the cluster II mAbs examined. LE4 protected RTA peptides encompassing amino acid residues 63–69, which corresponds to a bend between β-strands d and e and residues 154–161 and 163, which spans the terminus of α-helix D to the proximal residues of α-helix E. In this respect, LE4 is nearly identical to PH12 and TB12. CH1 and SWB1 were similar to each other in that they protected RTA residues 154–167, corresponding to the loop between α-helix D and α-helix E. 6C4 strongly protected peptides corresponding to RTA residues 64–68 and 184–206, corresponding to the loop between β-strands d and e (residues 64–68) and the region between α-helices F and G (residues 184–206). Finally, HX-MS analysis indicated that WECH1 contacted RTA residues 14–24, corresponding to a loop between β-strand a and α-helix A and residues 184–207, corresponding to the region between α-helices F and G, a profile identical to SyH7.
A clear pattern emerged when the HX-MS epitope profiles of the five new cluster II mAbs were aligned with the epitopes of the four previously described cluster II mAbs (PH12, TB12, PA1, and SyH7), as shown in Table I. Specifically, PH12, TB12, and LE4, the three mAbs with the most potent in vivo TNA, had identical HX-MS epitope profiles involving contact with RTA residues 63–69 and 154–163. The other six mAbs either did not engage with residues 63–69 or 154–163 or engaged with just one (but not both) of those particular secondary elements. For example, CH1 and SWB1 protected residues 154–163, but not residues 63–69, whereas 6C4 engaged with residues 63–69, but not 154–163. SyH7, PA1, and WECH1 do not interact with either residues 154–163 or 63–69. It is tempting to speculate that simultaneous binding of an Ab to residues 63–69 and 154–163 (or at least in close proximity to these residues) is a determining factor in Ab potency in vivo. We will touch on this topic in the Discussion.
At the level of resolution afforded by HX-MS, PH12, TB12, and LE4 appear to have the same epitope. However, we reported that PH12 and TB12 have different profiles in competition ELISAs with a panel of ~60 VHHs, suggesting that the two mAbs do have distinct binding sites on RTA or different angles of approach (16). To better resolve the LE4 epitope vis-à-vis PH12 and TB12, we subjected LE4 to similar competition ELISAs with a subset of VHHs. As shown in Fig. 6, LE4, PH12, and TB12 each had unique competition profiles. For example, PH12 and TB12 did not compete with VHH JNM-E4, whereas LE4 did. TB12 did not compete with VHH JIY-D9, whereas PH12 and LE4 did. Finally, LE4 and PH12 did not compete with VHHs JIZ-B7 and V5E1, but TB12 did. Thus, PH12, TB12, and LE4 are in fact distinct mAbs with subtle differences in epitope specificity and/or different angles of approach on ricin toxin.
FIGURE 6. Refinement of cluster II epitopes by VHH competition ELISA.
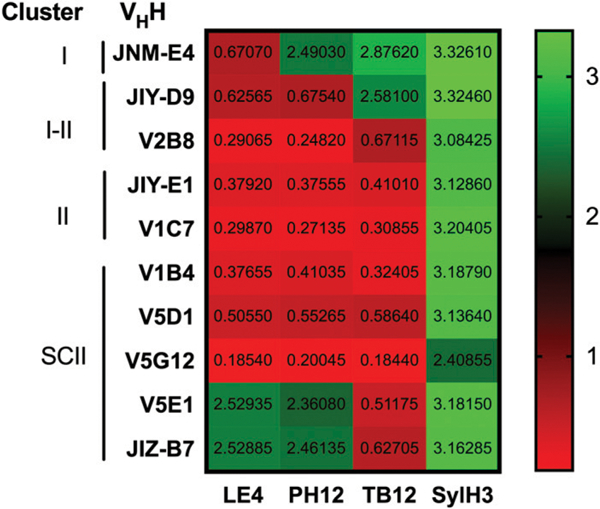
Ricin was captured onto microtiter wells coated with LE4, PH12, or TB12 and then probed with VHHs against epitope clusters I, I/II, II, or SCII (16). In the far-right column, ricin was captured with SylH3, an RTBspecific mAb that does not interfere with the panel of VHHs shown in the figure. The heat map is color coded from green (no competition) to red (competition). The actual OD obtained from the ELISA is presented in each box. The results shown are from one experiment of three biological replicates with essentially identical results.
DISCUSSION
Establishing comprehensive, high-resolution B cell epitope maps ofpathogen-associated Ags and toxins provides a basis for rational vaccine design and reveals possible targets for drug candidates (24, 25). Ricin toxin’s enzymatic subunit, RTA, is of particular interest in this regard because it is the foundation for two recombinant subunit vaccines under development (i.e., RiVax and RVEc), as well as a prime target for candidate immunotherapeutics (8). In previous reports, we defined four neutralizing “hotspots” on the surface of ricin toxin’s enzymatic subunit, RTA, that we have referred to as epitope clusters I-IV. Evidence suggests that epitope clusters I and II are the most immunodominant regions of ricin (13–16,18,26–29). Although cluster I epitopes have been defined at high resolution through a combination of peptide mapping, X-ray crystallography, and HX-MS, cluster II epitopes, until recently, were loosely defined based on limited peptide mapping studies and competition ELISAs (11, 30).
In this report, we characterized five new cluster II mAbs (LE4, CH1, SWB1, 6C4, and WECH1) and tested them side by side with the four previously described cluster II mAbs in a mouse model of ricin toxin challenge. The challenge studies indicated that the nine cluster II mAbs segregated into two groups (Table I): those that were able to passively protect mice for the duration of the 7-d study (LE4, TB12, and PH12), and those that were not (CH1, SWB1, PA1, 6C4, WECH1, and SyH7). The two groups varied in their relative binding affinities and in their capacity to neutralize ricin in vitro. However, the most notable difference between the groups was their contact sites on RTA. The protective mAbs, LE4, TB12, and PH12 each make strong contact with RTA residues 63–69 and 154–163, whereas the other six mAbs did not. Therefore, this report is significant in that it assigns for the first time, to our knowledge, specific Ab contacts on RTA secondary elements within cluster II that are associated with potent in vivo ricin TNA and passive protection in a mouse model.
We conclude that cluster II as a whole constitutes a relatively large patch situated on the back side of RTA relative to the active site (Fig. 7A). When ricin holotoxin is positioned with RTB at its base, the TB12, PH12, and LE4 epitopes are located toward the toxin’s apex along with CH1 and SWB1 (Fig. 7B, 7C), whereas the other epitopes within cluster II are situated in closer proximity to RTB (Fig. 7D-F). Unfortunately, there are no “landmarks” within or in proximity to cluster II that might explain why Ab occupancy in this region differentially attenuates ricin’s toxicity. In other words, why do TB12, PH12, and LE4 have potent in vivo TNA, whereas PA1 and SyH7 do not? TB12, PH12, and LE4 make two primary contact points with RTA: residues 63–69 and residues 154–163. Residues 63–69 correspond to abend between β-strands d and e, which are part of a six-stranded β-sheet (strands a and d-h) that dominate the first of RTA’s three folding domains (10). No particular junction has been ascribed to this six-stranded β-sheet, even though it clearly is integral to the overall tertiary structure of RTA. Elimination of the bend between β-strands d and e by site-directed mutagenesis (Δ62–66) did not impact RTA’s ability to depurinate ribosomes in a cell-free assay (31). Residues 154–163 encompass the C terminus of α-helix D, a short intervening loop (residues 157–161), and the proximal residues of α-helix E. Removal of residues 152–156 or 157–161 did not impact RTA activity in vitro, although perturbing α-helix E renders the subunit inactive (31). We do postulate that the surface area delineated by cluster II is important for ricin cytotoxicity, possibly playing a role in intracellular transport. We have demonstrated, for example, that SyH7, when bound to ricin, affects the efficiency of toxin transport from the plasma membrane to the TGN (32). SyH7 also interferes with in vitro protein disulfide isomerase-mediated reduction of the disulfide bond that links RTA to RTB, an event that normally occurs in the ER (33). Unfortunately, neither TB12, PH12, nor LE4 have been tested yet in these types of assays.
FIGURE 7. Surface depiction of cluster II epitopes on the surface of ricin toxin.
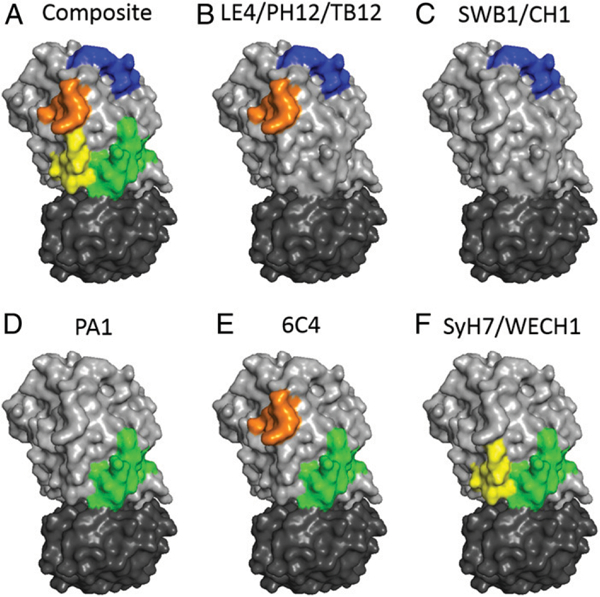
(A) Composite image of the four cluster II structural contact points listed in Table I mapped onto the structure of ricin (Protein DataBank identifier 2AAI) using PyMOL. (B–F) Individual epitopes for indicated mAbs. Colors: RTA, light gray; RTB, dark gray; β-strand a/α-helix A, yellow; d and e, orange; D and E, blue; and F and G, green.
The resolution afforded by HX-MS analysis is such that we were unable to distinguish differences in epitope specificity between LE4, PH12, and TB12. However, competition ELISAs with a collection of VHHs demonstrate that LE4, PH12, and TB12 are indeed different from each other. Based on available high-resolution VHH epitope maps, generated in some cases by X-ray crystallography, we can speculate as to how LE4, PH12, and TB12 may differentially engage RTA at residues 63–69 and 154–163. All three mAbs compete with VHHs V1C7 and JIY-E1, which are known from X-ray crystallography to contact RTA’s d and e and D and E loops. By contrast, JNM-E4 competes with LE4, but not PH12 or TB12. JNM-E4 is grouped within epitope cluster I and targets the “top” of RTA. Thus, LE4 likely approaches ricin from the top-down, relative to PH12 and TB12. TB12 competes V5E1 and JIZ-B7, whereas LE4 and PH12 do not. V5E1 and JIZ-B7 recognize epitopes at the RTA/RTB interface (34, 35), suggesting TB12 approaches RTA from the side or even underside. Finally, we postulate that PH12 attacks ricin at an angle somewhere between LE4 and TB12 based on competition with JIY-D9. Ultimately, assigning exact epitopes to LE4, PH12, and TB12 will require X-ray crystal structures of the mAbs in complex with RTA or ricin holotoxin.
Based on HX-MS analysis, the other six mAbs in cluster II (CH1, SWB1, PA1, 6C4, WECH1, and SyH7) recognize at least four different epitopes on RTA. Three of the mAbs, PA1, WECH1, and SyH7, have strong in vitro TNA, whereas the other three are devoid of activity, which is explained in large part by differences in relative binding affinities. In previous studies, we concluded that SyH7 and PA1 were, in fact, protective in our mouse model. However, it is now apparent that those conclusions were incorrect, because the experiments were terminated prematurely. Moreover, we failed to use body weight as a marker of morbidity, which other investigators have used successfully (23). From the current study, it is clear that mice treated with even relatively high doses of SyH7, CH1, SWB1, PA1, 6C4, or WECH1 begin to lose weight almost immediately after ricin challenge, whereas mice treated with TB12, PH12, or LE4 maintained normal body weights. Although the actual basis for ricin-induced death following systemic toxin exposure remains unknown, it most likely pertains to liver or kidney failure (36). Therefore, it is interesting that SyH7 is unable to neutralize ricin when mouse liver sinusoidal endothelial cells or Kupffer cells are the target cells, at least ex vivo (B. Mooney and N. Mantis, manuscript in preparation). We have just started examining the other cluster II mAbs like LE4, PH12, and TB12 for the ability to protect liver sinusoidal endothelial cells and Kupffer cells from ricin.
It should be noted that cluster II is actually more complicated than has been presented up to this point in the Discussion. Recent comprehensive epitope mapping studies along with X-ray crystallography have revealed that certain cluster II Abs recognize quaternary epitopes involving residues on RTA and RTB, an aggregate of epitopes we refer to as supercluster II (SCII) (16, 34, 35). A prime example is V5E1, a camelid VhH whose CDR1 and CDR2 elements contact RTA along α-helix A (residues 18–32), α-helix F (182–194), and the F/G loop, which explains competitive interference with SyH7 (35). At the same time, V5E1’s CDR3 straddles the RTA/RTB interface and docks in close proximity to RTB’s high affinity galactose/N-acetyl galactosamine lectin element. Conversely, JIZ-B7 is an example of a VHH whose primary target is RTB, but whose binding to ricin holotoxin is inhibited by SyH7 (which defines a cluster II Ab) (16, 34). The X-ray crystal structure of JIZ-B7 bound to ricin holotoxin is not available, although the structures of six other SCII VHHs bound to ricin holotoxin have been solved (M. Rudolph, D. Vance, and N. Mantis, manuscript in preparation).
Awaiting further analysis is the potential of the cluster II mAbs like LE4, PH12, and TB12 to protect against ricin administered by aerosol, the route of exposure that is most relevant to the military and civilian biodefense community (9,37). Inhalation of ricin elicits the clinical equivalent of acute respiratory distress syndrome, characterized by widespread apoptosis of alveolar macrophages, intra-alveolar edema, neutrophilic infiltration, accumulation of proinflammatory cytokines, and fibrinous exudate (38, 39). There are several examples in the literature in which passive administration of anti-RTA mAbs by injection have been shown to rescue mice from aerosolized ricin challenge, as long as the mAbs are delivered within a relatively short time (e.g., 4–6 h) following ricin exposure (40–43). In our hands, SyH7 was only moderately effective as a therapeutic in a pulmonary ricin-intoxication model (42). In fact, a cluster I mAb, called PB10, proved to be several-fold more effective than SyH7 when the two mAbs were tested side by side. Moreover, a fully humanized version of PB10 has recently been shown to rescue nonhuman primates from an aerosolized lethal-dose ricin challenge (C. Roy, N. Bohorova, O. Bohorov, D.Kim, M. Pauly, K. Whaley, D.J. Ehrbar, Y. Rong, P.J. Didier, L. Doyle, L. Zeitlin, and N. Mantis, submitted for publication). Thus, it will be imperative that PB10 serve as the “gold standard” when evaluating LE4, PH12, and TB12 as therapeutic mAbs.
According to the IEDB (http://www.iedb.org), more than 60 B cell epitopes have been localized on RTA and RTB subunits. Our current study now adds an additional five epitopes to that list. In total, we estimate that B cell epitopes have been assigned to roughly half the surface area of RTA (11,12,15,16,30,34,44–49). At least a subset of these epitopes is conserved across species (e.g., mouse, nonhuman primates, and humans), including several of the cluster I and II epitopes (50). Within the context of vaccine development, it still remains to be determined which specific epitopes on RTA are most important in mediating protective immunity and whether epitope usage differs between systemic and mucosal compartments.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Renjie Song in the Wadsworth Center’s Immunology Core Facility for assisting with surface plasmon resonance studies. We thank Dr. Jenny Tang of the Wadsworth Center’s Cell Culture Facility and Dr. Ed Greenfield of the Dana–Farber Cancer Institute mAb core facility for assistance in purification of mAbs. We also thank the Wadsworth Center animal care staff for pre–and poststudy animal maintenance.
This work was supported by Contract HHSN272201400021C and Grant Award 5R01AI125190 from the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (NIH). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Abbreviations used in this article:
- biotin-R
biotinylated ricin
- EC
effective concentration
- ER
endoplasmic reticulum
- HX
hydrogen exchange
- HX-MS
hydrogen exchange-mass spectrometry
- IEDB
Immune Epitope Database
- RT
room temperature
- RTA
ribosome-inactivating enzymatic subunit A
- RTB
ricin toxin B
- SCII
supercluster II
- SW
Swiss Webster
- TNA
toxin-neutralizing activity
Footnotes
DISCLOSURES
The authors have no financial conflicts of interest.
The epitope data presented in this article have been submitted to the Immune Epitope Database and Analysis Resource (http://www.iedb.org).
REFERENCES
- 1.Cieslak TJ, Kortepeter MG, Wojtyk RJ, Jansen HJ, Reyes RA, and Smith JO; And the NATO Biological Medical Advisory Panel. 2018. Beyond the dirty dozen: a proposed methodology for assessing future bioweapon threats. Mil. Med. 183: e59–e65. [DOI] [PubMed] [Google Scholar]
- 2.Rutenber E, Katzin BJ, Ernst S, Collins EJ, Mlsna D, Ready MP, and Robertus JD. 1991. Crystallographic refinement of ricin to 2.5 A. Proteins 10: 240–250. [DOI] [PubMed] [Google Scholar]
- 3.Endo Y, Mitsui K, Motizuki M, and Tsurugi K. 1987. The mechanism of action of ricin and related toxic lectins on eukaryotic ribosomes. The site and the characteristics of the modification in 28 S ribosomal RNA caused by the toxins. J. Biol. Chem. 262: 5908–5912. [PubMed] [Google Scholar]
- 4.Endo Y, and Tsurugi K. 1987. RNA N-glycosidase activity of ricin A-chain. Mechanism of action of the toxic lectin ricin on eukaryotic ribosomes. J. Biol. Chem. 262: 8128–8130. [PubMed] [Google Scholar]
- 5.Spooner RA, and Lord JM. 2012. How ricin and Shiga toxin reach the cytosol of target cells: retrotranslocation from the endoplasmic reticulum. Curr. Top. Microbiol. Immunol. 357: 19–40. [DOI] [PubMed] [Google Scholar]
- 6.Audi J, Belson M, Patel M, Schier J, and Osterloh J. 2005. Ricin poisoning: a comprehensive review. JAMA 294: 2342–2351. [DOI] [PubMed] [Google Scholar]
- 7.Brey III RN, Mantis NJ, Pincus SH, Vitetta ES, Smith LA, and Roy CJ. 2016. Recent advances in the development of vaccines against ricin. Hum. Vaccin. Immunother. 12: 1196–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vance DJ, and Mantis NJ. 2016. Progress and challenges associated with the development of ricin toxin subunit vaccines. Expert Rev. Vaccines 15: 1213–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wolfe DN, Florence W, and Bryant P. 2013. Current biodefense vaccine programs and challenges. Hum. Vaccin. Immunother. 9: 1591–1597. [DOI] [PubMed] [Google Scholar]
- 10.Montfort W, Villafranca JE, Monzingo AF, Ernst SR, Katzin B, Rutenber E, Xuong NH, Hamlin R, and Robertus JD. 1987. The three-dimensional structure of ricin at 2.8 A. J. Biol. Chem. 262: 5398–5403. [PubMed] [Google Scholar]
- 11.O’Hara JM, Kasten-Jolly JC, Reynolds CE, and Mantis NJ. 2014. Localization of non-linear neutralizing B cell epitopes on ricin toxin’s enzymatic subunit (RTA). Immunol. Lett. 158: 7–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Hara JM, Neal LM, McCarthy EA, Kasten-Jolly JA, Brey III RN, and Mantis NJ. 2010. Folding domains within the ricin toxin A subunit as targets of protective antibodies. Vaccine 28: 7035–7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Toth IV RT, Angalakurthi SK, Van Slyke G, Vance DJ, Hickey JM, Joshi SB, Middaugh CR, Volkin DB, Weis DD, and Mantis NJ. 2017. High-definition mapping of four spatially distinct neutralizing epitope clusters on RiVax, a candidate Ricin toxin subunit vaccine. Clin. Vaccine Immunol. 24: e00237–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lemley PV, Amanatides P, and Wright DC. 1994. Identification and characterization of a monoclonal antibody that neutralizes ricin toxicity in vitro and in vivo. Hybridoma 13: 417–421 [DOI] [PubMed] [Google Scholar]
- 15.Vance DJ, and Mantis NJ. 2012. Resolution of two overlapping neutralizing B cell epitopes within a solvent exposed, immunodominant α-helix in ricin toxin’s enzymatic subunit. Toxicon 60: 874–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vance DJ, Tremblay JM, Rong Y, Angalakurthi SK, Volkin DB, Middaugh CR, Weis DD, Shoemaker CB, and Mantis NJ. 2017. High-resolution epitope positioning of a large collection of neutralizing and nonneutralizing single-domain antibodies on the enzymatic and binding subunits of Ricin toxin. Clin. Vaccine Immunol. 24: e00236–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neal LM, O’Hara J, Brey III RN, and Mantis NJ. 2010. A monoclonal immunoglobulin G antibody directed against an immune-dominant linear epitope on the ricin A chain confers systemic and mucosal immunity to ricin. Infect. Immun. 78: 552–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rudolph MJ, Vance DJ, Cheung J, Franklin MC, Burshteyn F, Cassidy MS, Gary EN, Herrera C, Shoemaker CB, and Mantis NJ. 2014. Crystal structures of ricin toxin’s enzymatic subunit (RTA) in complex with neutralizing and non-neutralizing single-chain anti-bodies. J. Mol Biol. 426: 3057–3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smallshaw JE, Firan A, Fulmer JR, Ruback SL, Ghetie V, and Vitetta ES. 2002. A novel recombinant vaccine which protects mice against ricin intoxication. Vaccine 20: 3422–3427. [DOI] [PubMed] [Google Scholar]
- 20.Vita R, Zarebski L, Greenbaum JA, Emami H, Hoof I, Salimi N, Damle R, Sette A, and Peters B. 2010. The immune epitope database 2.0. Nucleic Acids Res. 38(Suppl. 1): D854–D862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.R Core Team. 2014. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 22.Smallshaw JE, Richardson JA, Pincus S, Schindler J, and Vitetta ES. 2005. Preclinical toxicity and efficacy testing of RiVax, a recombinant protein vaccine against ricin. Vaccine 23: 4775–4784. [DOI] [PubMed] [Google Scholar]
- 23.Smallshaw JE, Richardson JA, and Vitetta ES. 2007. RiVax, a recombinant ricin subunit vaccine, protects mice against ricin delivered by gavage or aerosol. Vaccine 25: 7459–7469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sesterhenn F, Bonet J, and Correia BE. 2018. Structure-based immunogen design-leading the way to the new age of precision vaccines. Curr. Opin. Struct. Biol. 51: 163–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zuverink M, and Barbieri JT. 2017. Protein structure facilitates high-resolution immunological mapping. Clin. Vaccine Immunol. 24: e00275–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dai J, Zhao L, Yang H, Guo H, Fan K, Wang H, Qian W, Zhang D, B Li, Wang H, and Guo Y. 2011. Identification of a novel functional domain of ricin responsible for its potent toxicity. J. Biol. Chem. 286: 12166–12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lebeda FJ, and Olson MA. 1999. Prediction of a conserved, neutralizing epitope in ribosome-inactivating proteins. Int. J. Biol. Macromol. 24: 19–26. [DOI] [PubMed] [Google Scholar]
- 28.Rudolph MJ, Vance DJ, Cassidy MS, Rong Y, Shoemaker CB, and Mantis NJ. 2016. Structural analysis of nested neutralizing and non-neutralizing B cell epitopes on ricin toxin’s enzymatic subunit. Proteins 84: 1162–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu Y, Dai J, Zhang T, Li X, Fang P, Wang H, Jiang Y, Yu X, Xia T, Niu L, et al. 2013. Structural insights into the neutralization mechanism of monoclonal antibody 6C2 against ricin. J. Biol. Chem. 288: 25165–25172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maddaloni M, Cooke C, Wilkinson R, Stout AV, Eng L, and Pincus SH. 2004. Immunological characteristics associated with the protective efficacy of antibodies to ricin. J. Immunol. 172: 6221–6228. [DOI] [PubMed] [Google Scholar]
- 31.Morris KN, and Wool IG. 1992. Determination by systematic deletion of the amino acids essential for catalysis by ricin A chain. Proc. Natl. Acad. Sci. USA 89: 4869–4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yermakova A, Klokk TI, O’Hara JM, Cole R, Sandvig K, and Mantis NJ. 2016. Neutralizing monoclonal antibodies against disparate epitopes on Ricin toxin’s enzymatic subunit interfere with intracellular toxin transport. Sci. Rep. 6: 22721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O’Hara JM, and Mantis NJ. 2013. Neutralizing monoclonal anti-bodies against ricin’s enzymatic subunit interfere with protein disulfide isomerase-mediated reduction of ricin holotoxin in vitro. J. Immunol. Methods 395: 71–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poon AY, Vance DJ, Rong Y, Ehrbar D, and Mantis NJ. 2017. A supercluster of neutralizing epitopes at the interface of Ricin’s enzymatic (RTA) and binding (RTB) subunits. Toxins (Basel) 9: E378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rudolph MJ, Vance DJ, Cassidy MS, Rong Y, and Mantis NJ. 2017. Structural analysis of single domain antibodies bound to a second neutralizing hot spot on Ricin toxin’s enzymatic subunit. J. Biol. Chem. 292: 872–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skilleter DN, and Foxwell BM. 1986. Selective uptake of ricin A-chain by hepatic non-parenchymal cells in vitro. Importance of mannose oligosaccharides in the toxin. FEBS Lett. 196: 344–348. [DOI] [PubMed] [Google Scholar]
- 37.Reisler RB, and Smith LA. 2012. The need for continued development of ricin countermeasures. Adv. Prev. Med. 2012: 149737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gal Y, Mazor O, Falach R, Sapoznikov A, Kronman C, and Sabo T. 2017. Treatments for pulmonary Ricin intoxication: current aspects and future prospects. Toxins (Basel) 9: E311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pincus SH, Bhaskaran M, Brey III RN, Didier PJ, Doyle-Meyers LA, and Roy CJ. 2015. Clinical and pathological findings associated with aerosol exposure of macaques to Ricin toxin. Toxins (Basel) 7: 2121–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gal Y, Mazor O, Alcalay R, Seliger N, Aftalion M, Sapoznikov A, Falach R, Kronman C, and Sabo T. 2014. Antibody/doxycycline combined therapy for pulmonary ricinosis: Attenuation of inflammation improves survival of ricin-intoxicated mice. Toxicol. Rep. 1: 496–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pratt TS, Pincus SH, Hale ML, Moreira AL, Roy CJ, and Tchou-Wong KM. 2007. Oropharyngeal aspiration of ricin as a lung challenge model for evaluation of the therapeutic index of antibodies against ricin A-chain for post-exposure treatment. Exp. Lung Res. 33: 459–481. [DOI] [PubMed] [Google Scholar]
- 42.Sully EK, Whaley KJ, Bohorova N, Bohorov O, Goodman C, Kim DH, Pauly MH, Velasco J, Hiatt E, Morton J, et al. 2014. Chimeric plantibody passively protects mice against aerosolized ricin challenge. Clin. Vaccine Immunol. 21: 777–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van Slyke G, Sully EK, Bohorova N, Bohorov O, Kim D, Pauly MH, Whaley KJ, Zeitlin L, and Mantis NJ. 2016. Humanized monoclonal antibody that passively protects mice against systemic and intranasal Ricin toxin challenge. Clin. Vaccine Immunol. 23: 795–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cohen O, Mechaly A, Sabo T, Alcalay R, Aloni-Grinstein R, Seliger N, Kronman C, and Mazor O. 2014. Characterization and epitope mapping of the polyclonal antibody repertoire elicited by ricin holotoxin-based vaccination. Clin. Vaccine Immunol. 21: 1534–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hu WG, Yin J, Chau D, Hu CC, Lillico D, Yu J, Negrych LM, and Cherwonogrodzky JW. 2013. Conformation-dependent high-affinity potent ricin-neutralizing monoclonal antibodies. BioMed Res. Int. 2013: 471346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Noy-Porat T, Rosenfeld R, Ariel N, Epstein E, Alcalay R, Zvi A, C Kronman, Ordentlich A, and Mazor O. 2016. Isolation of anti-Ricin protective antibodies exhibiting high affinity from immunized non-human primates. Toxins (Basel) 8: E64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rong Y, Van Slyke G, Vance DJ, Westfall J, Ehrbar D, and Mantis NJ. 2017. Spatial location of neutralizing and non-neutralizing B cell epitopes on domain 1 of ricin toxin’s binding subunit. PLoS One 12: e0180999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yermakova A, and Mantis NJ. 2011. Protective immunity to ricin toxin conferred by antibodies against the toxin’s binding subunit (RTB). Vaccine 29: 7925–7935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yermakova A, Vance DJ, and Mantis NJ. 2012. Sub-domains of ricin’s B subunit as targets of toxin neutralizing and non-neutralizing monoclonal antibodies. PLoS One 7: e44317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roy CJ, Brey RN, Mantis NJ, Mapes K, Pop IV, Pop LM,Ruback S, Killeen SZ, Doyle-Meyers L, Vinet-Oliphant HS, et al. 2015. Thermostable ricin vaccine protects rhesus macaques against aerosolized ricin: Epitope-specific neutralizing antibodies correlate with protection. Proc. Natl. Acad. Sci. USA 112: 3782–3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


