Abstract
The saliva of hematophagous arthropods injected during blood feeding contains potent pharmacologically active components to counteract the host hemostatic and inflammatory systems. In the present study, dominant salivary gland transcripts of Panstrongylus chinai, a vector of Chagas disease, were analyzed by sequencing randomly selected clones of the salivary gland cDNA library. This analysis showed that 56.5% of the isolated transcripts coded for putative secreted proteins, of which 73.7% coded for proteins belonging to the lipocalin family. The most abundant transcript of lipocalin family proteins was a homologue of pallidipin 2, an inhibitor of collagen-induced platelet aggregation of Triatoma pallidipennis. In addition, homologues of triafestin, an inhibitor of the kallikrein-kinin system of T. infestans, were identified as the dominant transcript. Other salivary transcripts encoding lipocalin family proteins had homology to triplatin (an inhibitor of platelet aggregation) and others with unknown function. Other than lipocalin family proteins, homologues of a Kazal-type serine protease inhibitor (putative anticoagulant), a hemolysin-like protein (unknown function), inositol polyphosphate 5-related protein (a regulator of membrane phosphoinositide), antigen 5-related protein (unknown function) and apyrase (platelet aggregation inhibitor) were identified.
Keywords: Panstrongylus chinai, Saliva, Transcriptome, Bioinformatics, cDNA library
1. Introduction
Hematophagous arthropods have evolved a wide set of pharmacologically active molecules to counteract host hemostatic processes (Ribeiro, 1995). When probing in the host skin prior to blood sucking, they inject saliva containing various physiologically active agents such as vasodilators, anticoagulants and platelet aggregation inhibitors (Ribeiro, 1995; Ribeiro and Francischetti, 2003; Valenzuela, 2004, 2005; Champagne, 2005). In addition, other bioactive agents, such as inhibitors of biogenic amine, complement system inhibitors and immunomodulators, have been identified in their saliva (Ribeiro, 1995; Ribeiro and Francischetti, 2003; Valenzuela, 2004, 2005; Champagne, 2005). Since hematophagous arthropods have evolved their feeding strategy independently, each species has developed distinctive salivary components to overcome host defense mechanisms (Ribeiro, 1995; Ribeiro and Francischetti, 2003; Valenzuela, 2004, 2005; Champagne, 2005). Therefore, salivary molecules of blood feeders have been extensively studied in various species to understand their universal and unique biological activities and to discover novel candidates for natural pharmacological agents (Ribeiro, 1995; Ribeiro and Francischetti, 2003; Valenzuela, 2004, 2005; Champagne, 2005).
Triatomine bugs, commonly known as kissing bugs, are blood sucking insects including the three major genera of the Chagas disease vector in Central and South America; genus Rhodnius in tribe Rhodniini and genera Triatoma and Panstrongylus in tribe Triatomini (Beard, 2005; Patterson et al., 2009; Schofield and Galvão, 2009). Of these, the genus Panstrongylus is composed of 13 species, and Panstrongylus chinai is known as a vector species of Chagas disease in Ecuador, Peru, and Venezuela (Abad-Franch et al., 2001; Grijalva et al., 2005, 2015; Patterson et al., 2009). Different from some other blood feeders such as mosquitoes, kissing bugs including P. chinai take 20–30 min for blood-sucking (Beard, 2005); however, they take blood efficiently regardless of host hemostatic and immune responses, suggesting their unique strategy to counteract host responses.
Salivary proteins, which are closely involved in their blood sucking strategy, have been explored in kissing bugs, especially in Rhodnius prolixus, which has characteristic cherry red-colored salivary glands (Champagne et al., 1995; Montfort et al., 2000; Ribeiro et al., 2004a; Andersen et al., 2005). Their functional analyses resulted in discovery of unique pharmacologically active components; for example, nitrophorin 2 (prolixin-S), one of the nitrophorins that are nitric oxide (NO)-binding heme proteins abundant in R. prolixus, was characterized to function both as an anticoagulant by the protein itself and a vasodilator by NO released from the protein (Champagne et al., 1995; Kaneko et al., 1999; Isawa et al., 2000; Ribeiro et al., 2004a). Different from Rhodnius species, salivary proteins of Triatoma and Panstrongylus species in the tribe Triatomini are colorless (Champagne et al., 1995). To identify the variations in blood sucking strategies in kissing bugs, salivary gland transcripts were recently extensively explored in Triatoma species, T. brasiliensis (Santos et al., 2007), T. infestans (Ribeiro et al., 2004a; Assumpção et al., 2008; Schwarz et al., 2014), T. dimidiata (Kato et al., 2010), T. matogrossensis (Assumpção et al., 2012) and T. rubida (Ribeiro et al., 2012). In addition, sialotranscriptome analysis was performed, more recently in a Panstrongylus species, P. megistus (Ribeiro et al., 2015). In the present study, to obtain further insight into the salivary biochemical and pharmacological complexity of triatomine bugs, the salivary gland transcripts were analyzed in a Panstrongylus species, P. chinai, which is a vector of Chagas disease mainly distributed west of the Andean Cordillera in South America (Schofield and Galvão, 2009).
2. Materials and methods
2.1. Panstrongylus chinai salivary glands
Panstrongylus chinai were reared in an insectary room at Hokkaido University in Japan and maintained at 26 ± 2 °C with uncontrolled humidity. They were fed at two-week intervals on rats. Salivary glands were dissected from fourth and fifth-instar nymphs after 2 weeks of feeding, transferred to an RNAlater reagent (Ambion, Austin, TX) and stored at −20° C until mRNA extraction.
2.2. Construction of a salivary gland cDNA library
Panstrongylus chinai salivary gland mRNA was isolated from 22 sets of the salivary glands using a Micro FastTrack mRNA isolation kit (Invitrogen, San Diego, CA). The PCR-based cDNA library was prepared following the instructions for the SMART cDNA library construction kit (BD-Clontech, Palo Alto, CA) with some modifications (Valenzuela et al., 2004). The quality of the cDNA was checked by agarose gel electrophoresis and the absence of smaller fragments derived from degraded mRNA was confirmed. The obtained cDNA library was fractionated using a Chromaspin 1000 column (BD-Clontech) into small (approximately 400–800 bp), medium (approximately 800–1200 bp) and large (> 1200 bp) transcripts based on their electrophoresis pro-files on a 1.1% agarose gel. Pooled fractions were ligated into Lambda TriplEx2 vector (BD-Clontech) and packaged into lambda phage (Stratagene, La Jolla, CA).
2.3. Sequence analysis of the cDNA library
Single, isolated phage plaques were picked from the plate using sterile wooden sticks and placed into 50 μl of water. Amplification of cDNA was performed in a volume of 20 μl using a pair of primers, PT2F1 (5′-AAG TAC TCT AGC AAT TGT GAG C-3′) and PT2R1 (5′-CTC TTC GCT ATT ACG CCA GCT G-3′), Premix Taq (Takara Bio, Shiga, Japan) and 4 μl of phage suspension as templates. After an initial denaturation at 75 °C for 3 min and following 95 °C for 4 min, PCR amplification was performed with 33 cycles of denaturation (95 °C, 1 min), annealing (55 °C, 1 min) and polymerization (72 °C, 1 min 30 s). PCR products were cleaned using Multiscreen PCR cleaning plates (Millipore Corporation, Bedford, MA), and the cleaned PCR product was suspended in 50 μl of water. The products were submitted to Macrogen Inc. (Seoul, Korea) for sequence analyses with a PT2F3 primer (5′-TCT CGG GAA GCG CGC CAT TGT-3′).
2.4. Bioinformatics
Expressed sequence tags (ESTs) were trimmed of primer and vector sequences and clustered. The ESTs were grouped based on nucleotide homology of 95% identity over 100 residues using the BLASTn algorithm (Altschul et al., 1997). The assembly of the ESTs into transcript contigs was done using the CAP3 algorithm, generating a consensus sequence (Huang, 1992). Contigs and singletons (contig containing only one sequence) were compared using BLASTx or BLASTn (Altschul et al., 1997) of the non-redundant (NR) protein database of the National Center of Biological Information (NCBI), the gene ontology database (GO) (Ashburner et al., 2000), and the Conserved Domains Database (CDD) that include all Pfam (Bateman and Birney, 2000), SMART (Schultz et al., 1998) and COG protein domains in the NCBI (Marchler-Bauer et al., 2002). Additionally, contigs were compared using BLASTn (Altschul et al., 1997) to custom databases of mitochondrial (mit-pla) and rRNA (rrna) nucleotide sequences. Identification of putative secreted proteins was conducted using SignalP server (Bendtsen et al., 2004).
2.5. Phylogenetic analysis
The sequences that had homologies with secreted proteins by BLASTx analyses were aligned with CLUSTAL W software (Thompson et al., 1994) and examined using Molecular Evolutionary Genetics Analysis (MEGA) version 5.2 (Tamura et al., 2011). Phylogenetic trees by the neighbor-joining method were constructed with the distance algorithms available in the MEGA package. Bootstrap values were determined on 1000 replicates of the data sets.
3. Results and discussion
3.1. Sequence analysis of the P. chinai salivary gland cDNA library
A Panstrongylus chinai salivary gland cDNA library was constructed and sequence analysis was performed on 1152 randomly selected clones. As a result, 773 high-quality sequences were obtained and classified into 302 contigs. Three categories of expressed genes were derived from the manual annotation of the contigs: secreted, housekeeping and unknown. The putative secreted category comprised 437 clones (56.5%) in 64 contigs. High ratios of transcripts encoding secreted proteins were reported in other triatomine bugs (Ribeiro et al., 2004a, 2012, 2015; Santos et al., 2007; Assumpção et al., 2008, 2011, 2012; Kato et al., 2010; Santiago et al., 2016). The housekeeping category had 155 clones (20.1%) in 111 contigs, and the category of “unknowns” comprised 181 clones (23.4%) in 127 contigs.
3.2. Housekeeping genes
The contigs of housekeeping genes (111 gene contigs with 155 sequences in total) were divided into 17 subgroups on the basis of their possible function (Table 1). The largest subgroup was associated with “translation, ribosomal structure and biogenesis” (50 sequences in 33 contigs), and followed by “energy production and conversion” (24 sequences in 11 contigs), “posttranslational modification, protein turnover, chaperones” (11 sequences in 8 contigs), and “signal transduction mechanisms” (11 sequences in 8 contigs). Twenty-three sequences in 17 contigs, which represent conserved proteins with unknown function were classified as “unknown conserved”. Other sequences identified with homology to housekeeping genes were associated with cell structure, transport, metabolism, and signal transduction.
Table 1.
Functional classification of housekeeping genes expressed in Panstrongylus chinai salivary glands.
| Type of transcripts | Clusters | Sequences | % Sequences |
|---|---|---|---|
| Translation, ribosomal structure and biogenesis | 33 | 50 | 32.3 |
| Energy production and conversion | 11 | 24 | 15.5 |
| Posttranslational modification, protein turnover, chaperones | 8 | 11 | 7.1 |
| Signal transduction mechanisms | 8 | 11 | 7.1 |
| Cytoskeleton | 6 | 6 | 3.9 |
| Intracellular trafficking, secretion, and vesicular transport | 4 | 6 | 3.9 |
| Inorganic ion transport and metabolism | 4 | 4 | 2.6 |
| RNA processing and modification | 4 | 4 | 2.6 |
| Amino acid transport and metabolism | 3 | 3 | 1.9 |
| Carbohydrate transport and metabolism | 3 | 3 | 1.9 |
| Transcription | 3 | 3 | 1.9 |
| Cell cycle control, cell division, chromosome partitioning | 2 | 2 | 1.3 |
| Cell wall/membrane/envelope biogenesis | 2 | 2 | 1.3 |
| Chromatin structure and dynamics | 1 | 1 | 0.6 |
| Lipid transport and metabolism | 1 | 1 | 0.6 |
| Nuclear structure, Intracellular trafficking, secretion, and vesicular transport | 1 | 1 | 0.6 |
| Unknown conserved | 17 | 23 | 14.9 |
| Total | 111 | 155 | 100.0 |
3.3. Putative secreted proteins
The transcripts coding for secreted proteins were further analyzed using the BLASTx algorithm for comparison to the NCBI non-redundant protein database. Table 2 shows the classification of transcripts coding for putative secreted proteins in P. chinai salivary glands. Remarkably, out of 437 transcripts associated with putative secreted proteins, 322 transcripts (73.7%) coded for the lipocalin family of proteins. Other transcripts encoding for secreted proteins were homologous to kazal-domain peptides, hemolysin, inositol polyphosphate 5-phosphatase, antigen 5-related protein, apyrase, trypsin, heme-binding protein, triplatin, and others (Table 2). Lipocalins, the most abundant transcripts in this cDNA library were also reported to be present in the salivary glands of ticks (Mans, 2005; Mans and Ribeiro, 2008; Mans et al., 2008), but not in the saliva of mosquitoes and sand flies (Valenzuela et al., 2003, 2004; Ribeiro et al., 2004b, 2007; Arcà et al., 2005, 2007; Anderson et al., 2006; Kato et al., 2006, 2013; Calvo et al., 2009; Hostomská et al., 2009; Rohoušová et al., 2012; Chagas et al., 2013; de Moura et al., 2013). A high percentage of secreted lipocalins were also reported in other Triatoma and Panstrongylus species: 55.0% in T. infestans (Assumpção et al., 2008), 93.8% in T. brasiliensis (Santos et al., 2007), 89.9% in T. dimidiata (Kato et al., 2010), 53.0% in T. matogrossensis (Assumpção et al., 2012), 65.3% in T. rubida (Ribeiro et al., 2012) and 87.2% in P. megistus (Ribeiro et al., 2015).
Table 2.
Classification of transcripts coding for putative secreted proteins in Panstrongylus chinai salivary glands.
| Type of transcripts | Clusters | Sequences | % Sequences |
|---|---|---|---|
| Lipocalin family | 41 | 322 | 73.7 |
| Kazal peptide | 3 | 34 | 7.8 |
| Hemolysin | 3 | 34 | 7.8 |
| Inositol polyphosphate 5-phosphatase | 5 | 20 | 4.6 |
| Antigen 5-related | 2 | 13 | 3.0 |
| Apyrase | 3 | 5 | 1.1 |
| Trypsin | 1 | 2 | 0.5 |
| Heme-binding protein | 1 | 1 | 0.2 |
| Triplatin | 1 | 1 | 0.2 |
| Others | 4 | 5 | 1.1 |
| Total | 64 | 437 | 100.0 |
3.3.1. Lipocalins
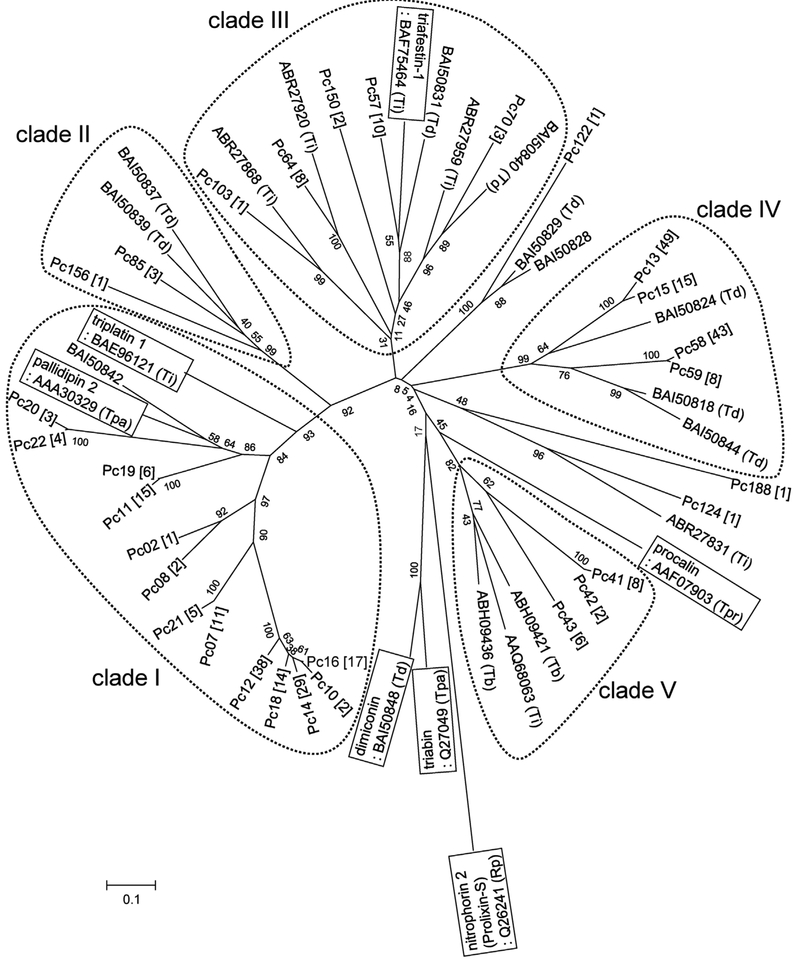
Of the transcripts coding for lipocalin-like molecules from this cDNA library, homologues of pallidipin 2, an inhibitor of collagen-induced platelet aggregation in T. pallidipennis saliva (Noeske-Jungblut et al., 1994; Haendler et al., 1995), were the most abundant transcripts accounting for 47.2% from the transcripts coding for this family of proteins (Table 3, and Table 1 in Data in Brief). The second most abundant transcript (36.3%) was similar to triatin-like salivary lipocalin identi-fied in the saliva of T. dimidiata (Kato et al., 2010). Other transcripts had homologies to molecules similar to salivary lipocalin from T. dimidiata (7.7%), lipocalin-like TiLipo37 from T. infestans (5.0%), and molecules similar to pallidipin-like salivary lipocalin from T. dimidiata(1.9%) (Table 1 in Data in Brief). Phylogenetic analysis of P. chinai salivary lipocalins together with other representative triatomine lipocalins resulted in the formation of five major clades (Fig. 1, Table 1 in Data in Brief). Further characteristics of P. chinai lipocalin-like clades are described below.
Table 3.
Most abundant salivary gland transcripts from Panstrongylus chinai.
| Cluster | No. of seq in cluster | pI | Mature Mw (kDa) | Cleavage position | Best match to NR protein database: Accession No. | E value | % identity | Accession No. |
|---|---|---|---|---|---|---|---|---|
| Pc13 | 49 | 4.83 | 18.7 | 18–19 | Td18, similar to triatin-like salivary lipocalin (Triatoma dimidiata): BAI50824 | 8e-71 | 61 | AB999669 |
| Pc58 | 43 | 7.10 | 19.8 | 18–19 | Td11, similar to triatin-like salivary lipocalin (Triatoma dimidiata): BAI50818 | 7e-83 | 66 | AB999688 |
| Pc12 | 38 | 8.44 | 19.3 | 18–19 | pallidipin 2 (Triatoma pallidipennis): AAA30329 | 2e-49 | 46 | AB999668 |
| Pc77 | 32 | 8.30 | 5.7 | 27–28 | salivary kazal-type proteinase inhibitor (Triatoma infestans): ABR27896 | 3e-18 | 56 | AB999695 |
| Pc14 | 29 | 7.72 | 19.3 | 18–19 | pallidipin 2 (Triatoma pallidipennis): AAA30329 | 2e-49 | 45 | AB999670 |
| Pc16 | 17 | 8.44 | 20.2 | 18–19 | pallidipin 2 (Triatoma pallidipennis): AAA30329 | 5e-49 | 46 | AB999672 |
| Pc60 | 17 | 5.95 | 22.9 | 18–19 | hemolysin-like secreted salivary protein 1 (Triatoma infestansy): ABR27902 | 1e-75 | 56 | AB999690 |
| Pc61 | 16 | 5.07 | 18.8 | 20–21 | salivary secreted protein (Triatoma infestansy): ABR27926 | 4e-46 | 59 | AB999691 |
| Pc11 | 15 | 5.29 | 18.7 | 16–17 | pallidipin 2 (Triatoma pallidipennis): AAA30329 | 5e-80 | 64 | AB999667 |
| Pc15 | 15 | 5.31 | 20.1 | 18–19 | Td18, similar to triatin-like salivary lipocalin (Triatoma dimidiata): BAI50824 | 3e-78 | 61 | AB999671 |
| Pc32 | 15 | 9.10 | 34.6 | 16–17 | salivary inositol polyphosphate 5-phosphatase (Triatoma infestans): ABR27973 | 5e-146 | 68 | AB999682 |
| Pc18 | 14 | 5.68 | 19.4 | 18–19 | pallidipin 2 (Triatoma pallidipennis): AAA30329 | 6e-50 | 45 | AB999674 |
| Pc80 | 12 | 9.13 | 24.1 | 24–25 | Td16, similar to antigen 5-like protein (Triatoma dimidiata): BAI50822 | 1e-79 | 55 | AB999696 |
| Pc07 | 11 | 4.94 | 19.2 | 18–19 | pallidipin 2 (Triatoma pallidipennis): AAA30329 | 2e-46 | 44 | AB999663 |
| Pc57 | 10 | 8.95 | 22.0 | 18–19 | Td26, similar to salivary lipocalin (Triatoma dimidiata): BAI50831 | 1e-75 | 57 | AB999687 |
| Pc41 | 8 | 8.66 | 18.6 | 18–19 | lipocalin-like TiLipo37 (Triatoma infestans): AAQ68063 | 7e-57 | 51 | AB999683 |
| Pc59 | 8 | 7.10 | 19.9 | 18–19 | Td45, similar to pallidipin-like salivary lipocalin (Triatoma dimidiata): BAI50844 | 1e-81 | 64 | AB999689 |
| Pc64 | 8 | 9.19 | 19.2 | 15–16 | salivary lipocalin (Triatoma infestans): ABR27920 | 3e-73 | 63 | AB999692 |
| Pc19 | 6 | 5.33 | 18.7 | 16–17 | pallidipin 2 (Triatoma pallidipennis): AAA30329 | 1e-79 | 64 | AB999675 |
| Pc43 | 6 | 8.65 | 18.5 | 18–19 | salivary lipocalin (Triatoma brasiliensis): ABH09436 | 4e-52 | 50 | AB999685 |
| Pc21 | 5 | 4.95 | 19.2 | 18–19 | pallidipin 2 (Triatoma pallidipennis): AAA30329 | 4e-44 | 43 | AB999677 |
| Pc22 | 4 | 7.05 | 19.3 | 18–19 | pallidipin 2 (Triatoma pallidipennis): AAA30329 | 2e-60 | 55 | AB999678 |
| Pc20 | 3 | 7.72 | 19.2 | 18–19 | pallidipin 2 (Triatoma pallidipennis): AAA30329 | 8e-60 | 54 | AB999676 |
| Pc70 | 3 | 8.65 | 21.5 | 18–19 | Td40, similar to triabin-like lipocalin 4a precursor (Triatoma dimidiata): BAI50840 | 3e-98 | 68 | AB999693 |
| Pc85 | 3 | 5.14 | 22.4 | 16–17 | Td38, similar to pallidipin-like salivary lipocalin (Triatoma dimidiata): BAI50839 | 1e-53 | 50 | AB999697 |
| Pcl59a | 3 | 79 kDa salivary apyrase precursor (Triatoma infestans): CAE46445 | 2e-25 | 57 | AB999709 | |||
| Pc08 | 2 | 8.67 | 19.8 | 18–19 | pallidipin 2 (Triatoma pallidipennis): AAA30329 | 2e-62 | 53 | AB999664 |
| Pc09 | 2 | 8.90 | 6.7 | 20–21 | No matches found | AB999665 | ||
| Pc10 | 2 | 6.49 | 19.4 | 18–19 | pallidipin 2 (Triatoma pallidipennis): AAA30329 | 7e-49 | 46 | AB999666 |
| Pc31 | 2 | 9.23 | 34.2 | 16–17 | salivary inositol polyphosphate 5-phosphatase (Triatoma infestans): ABR27973 | 1e-144 | 68 | AB999681 |
| Pc42 | 2 | 8.20 | 18.7 | 18–19 | lipocalin-like TiLipo37 (Triatoma infestans): AAQ68063 | 2e-57 | 52 | AB999684 |
| Pc97 | 2 | 5.74 | 34.3 | 18–19 | serine protease (Creona’ades dilutus): AAL15154 | 1e-66 | 45 | AB999698 |
| Pc150 | 2 | 8.44 | 21.9 | 16–17 | triabin-like lipocalin 4a precursor (Triatoma infestans): ABR27959 | 4e-38 | 44 | AB999706 |
| Pc17a | 2 | pallidipin 2 (Triatoma pallidipennis): AAA30329 | 1e-37 | 61 | AB999673 | |||
| Pc173a | 2 | Td38, similar to pallidipin-like salivary lipocalin (Triatoma dimidiata): BAI50839 | 1e-04 | 47 | AB999710 | |||
| Pc02 | 1 | 8.97 | 19.1 | 18–19 | pallidipin 2 (Triatoma pallidipennis): AAA30329 | 7e-62 | 55 | AB999658 |
| Pc03 | 1 | 8.68 | 16.6 | 19–20 | pallidipin 2 (Triatoma pallidipennis): AAA30329 | 3e-35 | 58 | AB999659 |
| Pc24 | 1 | 6.11 | 35.3 | 17–18 | salivary inositol polyphosphate 5-phosphatase (Rhodnius prolixus): AAB08434 | 2e-78 | 47 | AB999679 |
| Pc29 | 1 | 8.93 | 34.5 | 16–17 | salivary inositol polyphosphate 5-phosphatase (Triatoma infestans): ABR27973 | 2e-145 | 68 | AB999680 |
| Pc75 | 1 | 3.88 | 14.7 | 21–22 | salivary protein MYS3 precursor (Rhodnius prolixus): AAQ20837 | 6e-11 | 31 | AB999694 |
| Pc103 | 1 | 9.42 | 21.7 | 18–19 | salivary lipocalin (Triatoma infestans): ABR27868 | 5e-64 | 55 | AB999699 |
| Pc122 | 1 | 9.24 | 18.4 | 18–19 | Td24, similar to salivary lipocalin (Triatoma dimidiata): BAI50829 | 2e-68 | 74 | AB999702 |
| Pc124 | 1 | 9.07 | 20.1 | 19–20 | salivary lipocalin (Triatoma infestans): ABR27831 | 1e-54 | 49 | AB999704 |
| Pe134 | 1 | 6.12 | 16.5 | 21–22 | salivary protein MYS2 precursor (Triatoma brasiliensis): ABH09417 | 6e-95 | 87 | AB999705 |
| Pc156 | 1 | 4.72 | 21.4 | 28–19 | Td33, similar to pallidipin-like salivary lipocalin (Triatoma dimidiata): BAI50837 | 9e-48 | 49 | AB999707 |
| Pc237 | 1 | 6.17 | 13.1 | 19–20 | Td76, similar to heme-binding protein (Triatoma dimidiata): BAI53116 | 3e-49 | 71 | AB999713 |
| Pc266 | 1 | 4.86 | 3.5 | 28–29 | salivary kazal-type proteinase inhibitor (Triatoma infestans): ABR27896 | 3e-05 | 38 | AB999714 |
| Pc294 | 1 | 10.51 | 8.6 | 24–25 | antigen-5-like protein precursor (Triatoma infestans): ABR27897 | 1e-15 | 48 | AB999718 |
| Pc188 | 1 | 6.18 | 17.4 | 20–21 | salivary lipocalin 1 (Triatoma brasiliensis): ABH09421 | 4e-26 | 35 | AB999711 |
Truncated in the 5′ region.
Fig. 1.
Phylogenetic tree of the Panstrongylus chinai salivary lipocalins with those from other triatomine bugs. The sequences of P. chinai lipocalin obtained in the present study were aligned with representative triatomine lipocalin sequences obtained from the non-redundant protein database, and a phylogenetic tree was constructed. The number of sequences in each contig was shown in square brackets. The sequences from the database are represented by “GenBank accession number (abbreviation of the species name)”. Td, Triatoma dimidiata; Ti, T. infestans; Tb, T. brasiliensis; Tpa, T. pallidipennis; Tpr, T. protracta; Rp, Rhodnius prolixus. The scale bar represents 0.1% divergence. Bootstrap values are shown around branches.
3.3.1.1. Pc lipocalin clade I.
This group is the most abundant transcript in P. chinai salivary glands, and is composed of 152 clones in 17 contigs. The molecules showed a similarity to pallidipin 2, an inhibitor of collagen-induced platelet aggregation, identified in T. pallidipennis saliva (Noeske-Jungblut et al., 1994; Haendler et al., 1995). Sixteen contigs (Pc02, Pc05-Pc08, Pc10-Pc12, Pc14 and Pc16-Pc22) showed the highest homology to pallidipin 2 while Pc03 was homologous with Td42, a molecule similar to pallidipin 2, in T. dimidiata saliva with unknown function. The sequences of Pc05, Pc06, and Pc17 were truncated in the 5′ region. In the alignment of the encoding proteins with pallidipin 2, all six cysteine residues were conserved, suggesting a similar protein structure based on the disulfide bonds (Fig. 1 in Data in Brief). Phylogenetic analysis resulted in the formation of four distinct subclades; the first containing Pc11, Pc19, Pc20 and Pc22 that were closer to pallidipin 2, the second containing Pc02 and Pc08, the third containing Pc07 and Pc21, and the fourth containing Pc10, Pc12, Pc14, Pc16 and Pc18 (Fig. 1, and Fig. 1 in Data in Brief).
3.3.1.2. Pc lipocalin clade II.
This clade consisted of 6 clones in 3 contigs (Pc85, Pc156 and Pc173), and had homology with a lipocalin Td38 in T. dimidiata saliva with unknown function. The sequence of Pc173 was truncated in the 5′ region. Alignment of the encoding proteins with Td38 showed that all six cysteine residues were conserved (Fig. 2 in Data in Brief).
3.3.1.3. Pc lipocalin clade III.
This clade consisted of 25 clones in 6 contigs (Pc56, Pc57, Pc64, Pc70, Pc103 and Pc150), and had a homology with triafestin-1, an inhibitor of kallikrein–kinin system activation in T. infestans saliva (Isawa et al., 2007). Two contigs (Pc56 and Pc57) and Pc70 showed the highest homology to Td26 and Td40 from T. dimidiata saliva, respectively, the function of which are unknown. On the other hand, two contigs (Pc64 and Pc103) and Pc150 were homologous with salivary lipocalin and triabin-like lipocalin 4a precursor from T. infestans, respectively, neither of which have identified functions. The sequence of Pc56 was truncated in the 5′ region. Alignment of the encoding proteins with triafestin-1 showed that the GXW motif, which is often observed in lipocalins, was conserved in 4 contigs (Pc57, Pc64, Pc70 and Pc150), and all six cysteine residues were located at the corresponding positions in all molecules (Fig. 3 in Data in Brief). Phylogenetic analysis resulted in the formation of close (Pc56, Pc57, Pc70 and Pc150) and relatively distinct (Pc64 and Pc103) subclades to triafestin-1 (Fig. 1, and Fig. 3 in Data in Brief).
3.3.1.4. Pc lipocalin clade IV.
This clade consisted of 117 clones in 6 contigs (Pc04, Pc13, Pc15, Pc58, Pc59 and Pc226), and had homology with Td18, a salivary protein from T. dimidiata with unknown function (Kato et al., 2010). The sequences of Pc04 and Pc226 were truncated in the 5′ region. In the alignment of the encoding proteins with Td18, the GXW motif was conserved, and all six cysteine residues were located at the corresponding positions in all molecules (Fig. 4 in Data in Brief). Phylogenetic analysis resulted in the formation of close (Pc13 and Pc15) and distinct (Pc58 and Pc59) subclades to Td18 (Fig. 1, and Fig. 4 in Data in Brief).
3.3.1.5. Pc lipocalin clade V.
This clade consisted of 16 clones in 3 contigs (Pc41, Pc42 and Pc43), and had homology with lipocalin-like TiLipo37 from T. infestans saliva with unknown function. Pc41 and Pc42 showed the highest homology to lipocalin-like TiLipo37, while Pc43 was homologous with salivary lipocalin from T. brasiliensis with unknown function. The GXW motif was conserved, and all six cysteine residues were located at the corresponding positions in all molecules (Fig. 5 in Data in Brief). Phylogenetic analysis resulted in the formation of distinct subclades (Pc41, Pc42 and Pc43) to lipocalin-like TiLipo37 (Fig. 1, and Fig. 5 in Data in Brief).
3.3.1.6. Other lipocalins.
Two contigs (Pc122 and Pc301) and Pc123 had homologies with Td24 and Td23 from T. dimidiata saliva with unknown function, respectively (Table 3). On the other hand, Pc124 and Pc276 were homologous with salivary lipocalin and triabin-like salivary lipocalin from T. infestans, respectively, neither of which have known functions, and Pc188 showed homology with salivary lipocalin 1 from T. brasiliensis with unknown function (Table 3).
3.3.2. Kazal-type serine protease inhibitor
Thirty four clones in 3 contigs (Pc76, Pc77 and Pc266) contained a Kazal domain in the sequences. Kazal-type serine protease inhibitors are small proteins, typically 40–60 amino acids in length, with characteristic sequences CX1–6CX6–9PVCGX8–15CX3–6CX7–16C, in which Xn are integral numbers of amino acid residues. Each set of cysteine residues, 1st and 5th, 2nd and 4th, and 3rd and 6th, forms a disulfide bond, resulting in a characteristic three-dimensional structure (Rimphanitchayakit and Tassanakajon, 2010). Kazal-type serine pro-tease inhibitors identified in mosquito and leech saliva and in the midgut of kissing bugs have been characterized to work as anticoagulants during blood feeding (Friedrich et al., 1993; Campos et al., 2002; Nowak and Schrör, 2007; Santos et al., 2007; Rimphanitchayakit and Tassanakajon, 2010; Watanabe et al., 2010, 2011). Although homologous proteins to Kazal-type serine protease inhibitors have been identified in the saliva of kissing bugs (Santos et al., 2007; Assumpção et al., 2008, 2012; Schwarz et al., 2014; Ribeiro et al., 2015; Santiago et al., 2016), their bioactivities have not yet been characterized. In the P. chinai salivary gland transcripts coding for secretory proteins, a higher ratio of this molecule (7.8%) was identified when compared to other triatomine bugs (0.5–2.8%) (Santos et al., 2007; Assumpção et al., 2008, 2012; Schwarz et al., 2014; Ribeiro et al., 2015; Santiago et al., 2016), suggesting its important role in blood feeding in this species. The sequence of Pc76 was truncated in the 5′ region. Alignment of the encoding proteins with Kazal-type serine protease inhibitor-like protein from T. infestans showed that all six cysteine residues and PVCG sequences were conserved in Pc77 and Pc266 (Fig. S1).
3.3.3. Hemolysin-like protein
Thirty-four clones in 3 contigs (Pc60, Pc61 and Pc118) shared homology with a hemolysin-like protein identified in T. infestans saliva, of which the function is not known (Tables 2 and 3). The sequence of Pc76 was truncated in the 5′ region. Hemolysin is a pore-forming toxin identified in microorganisms (Gouaux, 1998; Melton et al., 2004; Wassenaar, 2005). The role of this protein in the saliva of triatomine bugs is not known; however, it is speculated that the protein may act as a cytolytic protein, causing erythrocyte lysis and aiding in the early steps of the digestion process (Assumpção et al., 2008). Alternatively, it may work as an antimicrobial peptide in the saliva. The putative amino acid sequence of Pc60 showed a high homology with T. infestans hemolysin-like protein with similar molecular weight, whereas Pc61 showed lower similarity and lacked its C-terminal region (Fig. S2).
3.3.4. Inositol polyphosphate 5-phosphatase-like protein
Twenty clones in 5 contigs (Pc24, Pc29, Pc31, Pc32 and Pc292) shared homology with an inositol polyphosphate 5-phosphatase (IP5P)-like protein identified in T. infestans saliva (Tables 2 and 3). The sequence of Pc292 was truncated in the 5′ region. Although the role of salivary IP5Ps remains unclear, the enzyme is well-conserved in kissing bugs, suggesting that it plays an important role in the blood sucking process of this insect (Ribeiro et al., 2004a, 2012, 2015; Andersen and Ribeiro, 2006; Santos et al., 2007; Assumpção et al., 2008, 2011, 2012; Kato et al., 2010; Schwarz et al., 2014; Santiago et al., 2016). In the alignment of the encoding proteins with IP5P-like protein identified in T. infestans saliva, two cysteine residues were located at the corresponding positions in all molecules (Fig. S3). Phylogenetic analysis resulted in two subclades; three contigs (Pcs29, Pc31 and Pc32) showed a closer relationship to IP5P-like protein from T. infestans saliva whereas a singleton (Pc24) composed a separate subclade (Fig. S3).
3.3.5. Antigen 5-related protein
Thirteen clones in 2 contigs (Pc80 and Pc294) shared homology with an antigen 5-related protein identified in T. infestans saliva (Tables 2 and 3). This family of proteins belongs to the cysteine-rich secretory proteins (CRISPs) that are related to venom allergens in social wasps and ants (Lu et al., 1993; Hoffman, 1993; King and Spangfort, 2000). Transcripts coding for this family of protein have been identified in the salivary glands of blood sucking insects including kissing bugs (Ribeiro et al., 2004a, 2012, 2015; Santos et al., 2007; Assumpção et al., 2008, 2011, 2012; Kato et al., 2010; Schwarz et al., 2014; Santiago et al., 2016). Salivary antigen 5 family proteins form kissing bugs were shown to inhibit collagen-induced platelet aggregation and neutrophil oxidative burst (Assumpção et al., 2013). Alignment and phylogenetic analysis of antigen 5-related proteins from various species indicated that P. chinai salivary antigen 5-related proteins had closer relationships with those from kissing bugs than those from saliva of other blood sucking arthropods such as mosquitoes and sand flies (Fig. S4).
3.3.6. Apyrase-like protein
Five clones in 3 contigs (Pc158, Pc159 and Pc270) shared homology with a 79 kDa salivary apyrase in T. infestans (Tables 2 and 3). Apyrase is a member of the nucleoside triphosphate-diphosphohydrolase family present in a variety of organisms. Salivary apyrase has been identified in a variety of blood-sucking arthropods and functions to hydrolyze ADP resulting in inhibition of ADP-induced platelet aggregation when injected into the host (Ribeiro and Francischetti, 2003; Faudry et al., 2004).
3.3.7. Trypsin-like protein
Two clones in a contig Pc97 showed homology with a serine pro-tease in the saliva of Creontiades dilutus, phytophagous Hemiptera, and a trypsin-like protein in T. infestans saliva (Tables 2 and 3). In phytophagous insects, salivary trypsin-like proteins injected into plant tissue are shown to play important roles in extra-intestinal digestion to facilitate efficient nutrition intake in the gut (Zhu et al., 2003). Although the biological function of this enzyme in blood sucking arthropods has yet to be determined, it is speculated that this protein is related to digestion and absorption of nutrients from host blood. Alignment of these three proteins showed that all nine cysteine residues were conserved among them (Fig. S5).
3.3.8. Heme-binding protein
A singleton Pc237 shared homology with Td76 from T. dimidiate saliva with unknown function (Tables 2 and 3, Fig. S6). The homo-logues were reported in the salivary glands of T. brasiliensis and T. dimidiata (Santos et al., 2007; Kato et al., 2010) and in the hemolymph and oocytes of R. prolixus (Paiva-Silva et al., 2002). The roles of these salivary proteins for blood-feeding remains to be clarified, but it may function as a vasodilator by the effect of nitric oxide potentially associated with heme binding on the salivary protein as reported in nitrophorin 2 (prolixin-S), a heme-binding protein of R. prolixus saliva (Champagne et al., 1995; Yuda et al., 1997; Kaneko et al., 1999).
3.3.9. Triplatin-binding protein
A singleton Pc01 had homology with triplatin from T. infestans saliva although the 5′ region was truncated. Triplatin was identified as a collagen-induced platelet aggregation inhibitor (Morita et al., 2006).
3.3.10. Other putative secreted proteins
Singletons Pc75 and Pc134 shared homology with salivary proteins MYS3 from R. prolixus and MYS2 from T. brasiliensis, respectively (Table 3). A singleton Pc115 was homologous to the glutathione peroxidase 6-like protein of Cimex lectularius. A contig, Pc09, did not have marked similarity to known proteins.
4. Conclusions
The present study identified dominant transcripts containing a variety of lipocalin in the salivary glands of P. chinai, a vector of Chagas disease in Ecuador and Peru (Grijalva et al., 2005, 2015). Most P. chinai salivary components were similar to those of Triatoma species when compared to those of Rhodnius species (Ribeiro et al., 2004a, 2012, 2015; Santos et al., 2007; Assumpção et al., 2008, 2011, 2012; Kato et al., 2010; Schwarz et al., 2014; Santiago et al., 2016), corresponding with morphology and phylogenetic relationships of mitochondrial DNAs (García et al., 2001; Schofield and Galvão, 2009; Justi et al., 2014). In addition, salivary proteins of Panstrongylus species are colorless as observed in Triatoma species, whereas those of Rhodnius species are cherry red due to dominant components of heme-binding proteins in the saliva, which is indicative of common feeding strategies in Panstrongylus and Triatoma species. Salivary proteins homologous to procalin, which was identified as the major allergen of T. protracta saliva, were abundant in Triatoma species; e.g., approximately 60% in T. dimidiata (Kato et al., 2010; Assumpção et al., 2012). On the other hand, the most abundant salivary protein in P. chinai was a homologue of pallidipin 2, an inhibitor of collagen-induced platelet aggregation of T. pallidipennis, accounting for about 50% of transcripts encoding secreted proteins, suggesting diverse feeding strategies among kissing bugs.
In the P. chinai salivary gland transcripts, a higher ratio of transcripts encoding a Kazal domain-containing protein was identified when compared to other kissing bugs (Santos et al., 2007; Assumpção et al., 2008, 2012; Schwarz et al., 2014; Ribeiro et al., 2015; Santiago et al., 2016). Several Kazal-type serine protease inhibitors containing 2–8 Kazal domains were identified from midgut of kissing bugs; rhodniin from R. prolixus (Friedrich et al., 1993), infestin from T. infestans (Campos et al., 2002), brasiliensin from T. brasiliensis (Araujo et al., 2007) and dipetalogastin from Dipetalogaster maximus (Mende et al., 1999), and were characterized to inhibit several enzymes such as thrombin, trypsin and chymotrypsin (Rimphanitchayakit and Tassanakajon, 2010). On the other hand, Kazal-type serine protease inhibitors identified from saliva of kissing bugs contain only one Kazal domain and have not been characterized (Santos et al., 2007; Assumpção et al., 2008, 2012; Schwarz et al., 2014; Ribeiro et al., 2015; Santiago et al., 2016). Functional analysis will reveal enzymatic properties and contribute to further study on the target specificity of these enzymes.
In conclusion, the most abundant proteins of P. chinai saliva were identified in this study. The result will provide further insights into the evolution of salivary components in blood sucking arthropods. In addition, cDNAs and recombinant proteins prepared from these transcripts will result in the discovery of novel pharmacologically active compounds, as well as the development of biomarkers following exposure to P. chinai.
Supplementary Material
Acknowledgements
We are grateful to Dr. José M. C. Ribeiro (Vector Biology Section, Laboratory of Malaria and Vector Research, NIAID, NIH, USA) for the development and training of all custom bioinformatics programs used in this research. We also thank Dr. Cesar Suarez and Mrs. Erika Sanchez (Laboratory of Medical Entomology, National Institute of Health and Tropical Medicine, Ecuador) for providing Panstrongylus chinai. This study was supported by the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan (Grant Nos. 25257501 and 15H04588).
Footnotes
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.actatropica.2017.06.022.
References
- Abad-Franch F, Paucar A, Carpio C, Cuba CA, Aguilar HM, Miles MA, 2001. Biogeography of Triatominae (Hemiptera: Reduviidae) in Ecuador: implications for the design of control strategies. Mem. Inst. Oswaldo Cruz 96, 611–620. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ, 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen JF, Ribeiro JMC, 2006. A secreted salivary inositol polyphosphate 5-phosphatase from a blood-feeding insect: allosteric activation by soluble phosphoinositides and phosphatidylserine. Biochemistry 45, 5450–5457. [DOI] [PubMed] [Google Scholar]
- Andersen JF, Gudderra NP, Francischetti IMB, Ribeiro JMC, 2005. The role of salivary lipocalins in blood feeding by Rhodnius prolixus. Arch. Insect Biochem. Physiol 58, 97–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JM, Oliveira F, Kamhawi S, Mans BJ, Reynoso D, Seitz AE, Lawyer P, Garfield M, Pham M, Valenzuela JG, 2006. Comparative salivary gland transcriptomics of sandfly vectors of visceral leishmaniasis. BMC Genomics 7, 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araujo RN, Campos IT, Tanaka AS, Santos A, Gontijo NF, Lehane MJ, Pereira MH, 2007. Brasiliensin: a novel intestinal thrombin inhibitor from Triatoma brasiliensis (Hemiptera: Reduviidae) with an important role in blood intake. Int. J. Parasitol 37, 1351–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcà B, Lombardo F, Valenzuela JG, Francischetti IMB, Marinotti O, Coluzzi M, Ribeiro JMC, 2005. An updated catalogue of salivary gland transcripts in the adult female mosquito, Anopheles gambiae. J. Exp. Biol 208, 3971–3986. [DOI] [PubMed] [Google Scholar]
- Arcà B, Lombardo F, Francischetti IMB, Pham VM, Mestres-Simon M, Andersen JF, Ribeiro JMC, 2007. An insight into the sialome of the adult female mosquito Aedes albopictus. Insect Biochem. Mol. Biol 37, 107–127. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G, 2000. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assumpção TCF, Francischetti IMB, Andersen JF, Schwarz A, Santana JM,Ribeiro JMC, 2008. An insight into the sialome of the blood-sucking bug Triatoma infestans, a vector of Chagas’ disease. Insect Biochem. Mol. Biol 38, 213–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assumpção TCF, Charneau S, Santiago PB, Francischetti IMB, Meng Z, Araújo CN, Pham VM, Queiroz RM, de Castro CN, Ricart CA, Santana JM, Ribeiro JMC, 2011. Insight into the salivary transcriptome and proteome of Dipetalogaster maxima. J. Proteome Res. 10, 669–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assumpção TCF, Eaton DP, Pham VM, Francischetti IMB, Aoki V, Hans-Filho G, Rivitti EA, Valenzuela JG, Diaz LA, Ribeiro JMC, 2012. An insight into the sialotranscriptome of Triatoma matogrossensis, a kissing bug associated with fogo selvagem in South America. Am. J. Trop. Med. Hyg 86, 1005–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assumpção TCF, Ma D, Schwarz A, Reiter K, Santana JM, Andersen JF, Ribeiro JMC, Nardone G, Yu LL, Francischetti IMB, 2013. Salivary antigen-5/CAP family members are Cu2+-dependent antioxidant enzymes that scavenge O2 ⨪ and inhibit collagen-induced platelet aggregation and neutrophil oxidative burst. J. Biol. Chem 288, 14341–14361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A, Birney E, 2000. Searching databases to find protein domain organization.Adv. Protein Chem. 54, 137–157. [DOI] [PubMed] [Google Scholar]
- Beard CB, 2005. Kissing bugs and bedbugs, Heteroptera In: Marquardt WC, Black WC, Freier JE, Hagedorn HH, Hemingway J, Higgs S, James AA, Kondratieff B, Moore CG (Eds.), Biology of Disease Vectors, second edition. Elsevier, San Diego, CA, pp. 57–65. [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S, 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol 340, 783–795. [DOI] [PubMed] [Google Scholar]
- Calvo E, Pham VM, Marinotti O, Andersen JF, Ribeiro JMC, 2009. The salivary gland transcriptome of the neotropical malaria vector Anopheles darlingi reveals accelerated evolution of genes relevant to hematophagy. BMC Genomics 10, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos IT, Amino R, Sampaio CA, Auerswald EA, Friedrich T, Lemaire HG, Schenkman S, Tanaka AS, 2002. Infestin, a thrombin inhibitor presents in Triatoma infestans midgut, a Chagas’ disease vector: gene cloning, expression and characterization of the inhibitor. Insect Biochem. Mol. Biol 32, 991–997. [DOI] [PubMed] [Google Scholar]
- Chagas AC, Calvo E, Rios-Velásquez CM, Pessoa FA, Medeiros JF, Ribeiro JMC, 2013. A deep insight into the sialotranscriptome of the mosquito, Psorophora albipes. BMC Genomics 14, 875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champagne DE, Nussenzveig RH, Ribeiro JMC, 1995. Purification, partial characterization, and cloning of nitric oxide-carrying heme proteins (nitrophorins) from salivary glands of the blood-sucking insect Rhodnius prolixus. J. Biol. Chem 270, 8691–8695. [DOI] [PubMed] [Google Scholar]
- Champagne DE, 2005. Antihemostatic molecules from saliva of blood-feeding arthropods. Pathophysiol. Haemost. Thromb 34, 221–227. [DOI] [PubMed] [Google Scholar]
- de Moura TR, Oliveira F, Carneiro MW, Miranda JC, Clarêncio J, Barral-Netto M, Brodskyn C, Barral A, Ribeiro JMC, Valenzuela JG, de Oliveira CI, 2013. Functional transcriptomics of wild-caught Lutzomyia intermedia salivary glands: identification of a protective salivary protein against Leishmania braziliensis infection. PLoS Negl. Trop. Dis 7, e2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faudry E, Lozzi SP, Santana JM, D’Souza-Ault M, Kieffer S, Felix CR, Ricart CAO, Sousa MV, Vernet T, Teixeira ARL, 2004. Triatoma infestans apyrases belong to the 5′-nucleotidase family. J. Biol. Chem 279, 19607–19613. [DOI] [PubMed] [Google Scholar]
- Friedrich T, Kröger B, Bialojan S, Lemaire HG, Höffken HW, Reuschenbach P, Otte M, Dodt J, 1993. A Kazal-type inhibitor with thrombin specificity from Rhodnius prolixus. J. Biol. Chem 268, 16216–16222. [PubMed] [Google Scholar]
- García BA, Moriyama EN, Powell JR, 2001. Mitochondrial DNA sequences of triatomines (Hemiptera: Reduviidae): phylogenetic relationships. J. Med. Entomol 38, 675–683. [DOI] [PubMed] [Google Scholar]
- Gouaux E, 1998. α-Hemolysin from Staphylococcus aureus: an archetype of beta-barrel, channel-forming toxins. J. Struct. Biol 121, 110–122. [DOI] [PubMed] [Google Scholar]
- Grijalva MJ, Palomeque-Rodríguez FS, Costales JA, Davila S, Arcos-Teran L,2005. High household infestation rates by synanthropic vectors of Chagas disease in southern Ecuador. J. Med. Entomol 42, 68–74. [DOI] [PubMed] [Google Scholar]
- Grijalva MJ, Villacis AG, Ocaña-Mayorga S, Yumiseva CA, Moncayo AL, Baus EG, 2015. Comprehensive survey of domiciliary triatomine species capable of transmitting Chagas disease in southern Ecuador. PLoS Negl. Trop. Dis 9, e0004142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haendler B, Becker A, Noeske-Jungblut C, Krätzschmar J, Donner P, Schleuning WD, 1995. Expression of active recombinant pallidipin, a novel platelet aggregation inhibitor, in the periplasm of Escherichia coli. Biochem. J 307, 465–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman DR, 1993. Allergens in Hymenoptera venom. XXV: the amino acid sequences of antigen 5 molecules and the structural basis of antigenic cross-reactivity. J. Allergy Clin. Immunol 92, 707–716. [DOI] [PubMed] [Google Scholar]
- Hostomská J, Volfová V, Mu J, Garfield M, Rohousová I, Volf P, Valenzuela JG, Jochim RC, 2009. Analysis of salivary transcripts and antigens of the sand fly Phlebotomus arabicus. BMC Genomics 10, 282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, 1992. A contig assembly program based on sensitive detection of fragment overlaps. Genomics 14, 18–25. [DOI] [PubMed] [Google Scholar]
- Isawa H, Yuda M, Yoneda K, Chinzei Y, 2000. The insect salivary protein, prolixin-S, inhibits factor IXa generation and Xase complex formation in the blood coagulation pathway. J. Biol. Chem 275, 6636–6641. [DOI] [PubMed] [Google Scholar]
- Isawa H, Orito Y, Jingushi N, Iwanaga S, Morita A, Chinzei Y, Yuda M, 2007. Identification and characterization of plasma kallikrein-kinin system inhibitors from salivary glands of the blood-sucking insect Triatoma infestans. FEBS J. 274, 4271–4286. [DOI] [PubMed] [Google Scholar]
- Justi SA, Russo CA, Mallet JR, Obara MT, Galvão C, 2014. Molecular phylogeny of Triatomini (Hemiptera: Reduviidae: Triatominae). Parasites Vectors 7, 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko Y, Shojo H, Yuda M, Chinzei Y, 1999. Effects of recombinant nitrophorin-2 nitric oxide complex on vascular smooth muscle. Biosci. Biotechnol. Biochem 63, 1488–1490. [DOI] [PubMed] [Google Scholar]
- Kato H, Anderson JM, Kamhawi S, Oliveira F, Lawyer PG, Pham VM, Sangare CS, Samake S, Sissoko I, Garfield M, Sigutova L, Volf P, Doumbia S, Valenzuela JG, 2006. High degree of conservancy among secreted salivary gland proteins from two geographically distant Phlebotomus duboscqi sandflies populations (Mali and Kenya). BMC Genomics 7, 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Jochim RC, Gomez EA, Sakoda R, Iwata H, Valenzuela JG, Hashiguchi Y, 2010. A repertoire of the dominant transcripts from the salivary glands of the blood-sucking bug, Triatoma dimidiata, a vector of Chagas disease. Infect. Genet. Evol 10, 184–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Jochim RC, Gomez EA, Uezato H, Mimori T, Korenaga M, Sakurai T, Katakura K, Valenzuela JG, Hashiguchi Y, 2013. Analysis of salivary gland transcripts of the sand fly Lutzomyia ayacuchensis, a vector of Andean-type cutaneous leishmaniasis. Infect. Genet. Evol 13, 56–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Jochim RC, Gomez EA, Tsunekawa S, Valenzuela JG, Hashiguchi Y, Transcripts coding for lipocalin family proteins in salivary glands of Panstrongylus chinai, a vector of Chagas disease. Data Brief. Submitted 10.1016/j.actatropica.2017.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King TP, Spangfort MD, 2000. Structure and biology of stinging insect venom allergens. Int. Arch. Allergy Immunol. 123, 99–106. [DOI] [PubMed] [Google Scholar]
- Lu G, Villalba M, Coscia MR, Hoffman DR, King TP, 1993. Sequence analysis and antigenic cross-reactivity of a venom allergen, antigen 5, from hornets, wasps, and yellow jackets. J. Immunol 150, 2823–2830. [PubMed] [Google Scholar]
- Mans BJ, Ribeiro JMC, 2008. Function, mechanism and evolution of the moubatinclade of soft tick lipocalins. Insect Biochem. Mol. Biol 38, 841–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mans BJ, Andersen JF, Francischetti IMB, Valenzuela JG, Schwan TG, Pham VM, Garfield MK, Hammer CH, Ribeiro JMC, 2008. Comparative sialomics between hard and soft ticks: implications for the evolution of blood-feeding behavior. Insect Biochem. Mol. Biol 38, 42–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mans BJ, 2005. Tick histamine-binding proteins and related lipocalins: potential as therapeutic agents. Curr. Opin. Invest. Drugs 6, 1131–1135. [PubMed] [Google Scholar]
- Marchler-Bauer A, Panchenko AR, Ariel N, Bryant SH, 2002. Comparison of sequence and structure alignments for protein domains. Proteins 48, 439–446. [DOI] [PubMed] [Google Scholar]
- Melton JA, Parker MW, Rossjohn J, Buckley JT, Tweten RK, 2004. The identi-fication and structure of the membrane-spanning domain of the Clostridium septicum α toxin. J. Biol. Chem 279, 14315–14322. [DOI] [PubMed] [Google Scholar]
- Mende K, Petoukhova O, Koulitchkova V, Schaub GA, Lange U, Kaufmann R, Nowak G, 1999. Dipetalogastin, a potent thrombin inhibitor from the blood-sucking insect. Dipetalogaster maximus cDNA cloning, expression and characterization. Eur. J. Biochem 266, 583–590. [DOI] [PubMed] [Google Scholar]
- Montfort WR, Weichsel A, Andersen JF, 2000. Nitrophorins and related antihemo-static lipocalins from Rhodnius prolixus and other blood-sucking arthropods. Biochim. Biophys. Acta 1482, 110–118. [DOI] [PubMed] [Google Scholar]
- Morita A, Isawa H, Orito Y, Iwanaga S, Chinzei Y, Yuda M, 2006. Identification and characterization of a collagen-induced platelet aggregation inhibitor triplatin, from salivary glands of the assassin bug, Triatoma infestans. FEBS J. 273, 2955–2962. [DOI] [PubMed] [Google Scholar]
- Noeske-Jungblut C, Krätzschmar J, Haendler B, Alagon A, Possani L, Verhallen P, Donner P, Schleuning WD, 1994. An inhibitor of collagen-induced platelet aggregation from the saliva of Triatoma pallidipennis. J. Biol. Chem 269, 5050–5053. [PubMed] [Google Scholar]
- Nowak G, Schrör K, 2007. Hirudin—the long and stony way from an anticoagulant peptide in the saliva of medicinal leech to a recombinant drug and beyond. A historical piece. Thromb. Haemost 98, 116–119. [PubMed] [Google Scholar]
- Paiva-Silva GO, Sorgine MHF, Benedetti CE, Meneghini R, Almeida IC,Machado EA, Dansa-Petretski M, Yepiz-Plascencia G, Law JH, Oliveira PL, Masuda H, 2002. On the biosynthesis of Rhodnius prolixus heme-binding protein. Insect Biochem. Mol. Biol 32, 1533–1541. [DOI] [PubMed] [Google Scholar]
- Patterson JS, Barbosa SE, Feliciangeli MD, 2009. On the genus Panstrongylus Berg 1879: evolution, ecology and epidemiological significance. Acta Trop. 110, 187–199. [DOI] [PubMed] [Google Scholar]
- Ribeiro JMC, Francischetti IMB, 2003. Role of arthropod saliva in blood feeding: sialome and post-sialome perspectives. Annu. Rev. Entomol 48, 73–88. [DOI] [PubMed] [Google Scholar]
- Ribeiro JMC, Andersen J, Silva-Neto MAC, Pham VM, Garfield MK, Valenzuela JG, 2004a. Exploring the sialome of the blood-sucking bug Rhodnius prolixus. Insect Biochem. Mol. Biol 34, 61–79. [DOI] [PubMed] [Google Scholar]
- Ribeiro JMC, Charlab R, Pham VM, Garfield M, Valenzuela JG, 2004b. An insight into the salivary transcriptome and proteome of the adult female mosquito Culex pipiens quinquefasciatus. Insect Biochem. Mol. Biol 34, 543–563. [DOI] [PubMed] [Google Scholar]
- Ribeiro JMC, Arcà B, Lombardo F, Calvo E, Pham VM, Chandra PK, Wikel SK,2007. An annotated catalogue of salivary gland transcripts in the adult female mosquito, Aedes aegypti. BMC Genomics 8, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro JMC, Assumpção TC, Pham VM, Francischetti IMB, Reisenman CE,2012. An insight into the sialotranscriptome of Triatoma rubida (Hemiptera: Heteroptera). J. Med. Entomol 49, 563–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro JMC, Schwarz A, Francischetti IMB, 2015. A deep insight into the sialotranscriptome of the Chagas disease vector, Panstrongylus megistus (Hemiptera: Heteroptera). J. Med. Entomol 52, 351–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro JMC, 1995. Blood-feeding arthropods: live syringes or invertebrate pharmacologists? Infect. Agents Dis. 4, 143–152. [PubMed] [Google Scholar]
- Rimphanitchayakit V, Tassanakajon A, 2010. Structure and function of invertebrate Kazal-type serine proteinase inhibitors. Dev. Comp. Immunol 34, 377–386. [DOI] [PubMed] [Google Scholar]
- Rohoušová I, Subrahmanyam S, Volfová V, Mu J, Volf P, Valenzuela JG, Jochim RC, 2012. Salivary gland transcriptomes and proteomes of Phlebotomus tobbi and Phlebotomus sergenti, vectors of leishmaniasis. PLoS Negl. Trop. Dis 6, e1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago PB, Assumpção TC, de Araújo CN, Bastos IM, Neves D, da Silva IG, Charneau S, Queiroz RM, Raiol T, Oliveira JV, de Sousa MV, Calvo E, Ribeiro JMC, Santana JM, 2016. A deep insight into the sialome of Rhodnius neglectus, a vector of Chagas disease. PLoS Negl. Trop. Dis 10, e0004581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos A, Ribeiro JMC, Lehane MJ, Gontijo NF, Veloso AB, Sant’ Anna MRV, Nascimento Araujo R, Grisard EC, Pereira MH, 2007. The sialotranscriptome of the blood-sucking bug Triatoma brasiliensis (Hemiptera, Triatominae). Insect Biochem. Mol. Biol 37, 702–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield CJ, Galvão C, 2009. Classification, evolution, and species groups within theTriatominae. Acta Trop. 110, 88–100. [DOI] [PubMed] [Google Scholar]
- Schultz J, Milpetz F, Bork P, Ponting CP, 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. U. S. A 95, 5857–5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz A, Medrano-Mercado N, Schaub GA, Struchiner CJ, Bargues MD, Levy MZ, Ribeiro JMC, 2014. An updated insight into the sialotranscriptome of Triatoma infestans: developmental stage and geographic variations. PLoS Negl. Trop. Dis 8, e3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S, 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol 28, 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ, 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela JG, Francischetti IMB, Pham VM, Garfield MK, Ribeiro JMC, 2003. Exploring the salivary gland transcriptome and proteome of the Anopheles stephensi mosquito. Insect Biochem. Mol. Biol 33, 717–732. [DOI] [PubMed] [Google Scholar]
- Valenzuela JG, Garfield M, Rowton ED, Pham VM, 2004. Identification of the most abundant secreted proteins from the salivary glands of the sand fly Lutzomyia long-ipalpis, vector of Leishmania chagasi. J. Exp. Biol 207, 3717–3729. [DOI] [PubMed] [Google Scholar]
- Valenzuela JG, 2004. Exploring tick saliva: from biochemistry to ‘sialomes’ and functional genomics. Parasitology 129, S83–94. [DOI] [PubMed] [Google Scholar]
- Valenzuela JG, 2005. Blood-feeding arthropod salivary glands and saliva In:Marquardt WC, Black WC, Freier JE, Hagedorn HH, Hemingway J, Higgs S, James AA, Kondratieff B, Moore CG (Eds.), Biology of Disease Vectors, second edition. Elsevier, San Diego, CA, pp. 377–386. [Google Scholar]
- Wassenaar TM, 2005. Use of antimicrobial agents in veterinary medicine and implications for human health. Crit. Rev. Microbiol 31, 155–169. [DOI] [PubMed] [Google Scholar]
- Watanabe RM, Soares TS, Morais-Zani K, Tanaka-Azevedo AM, Maciel C,Capurro ML, Torquato RJ, Tanaka AS, 2010. A novel trypsin Kazal-type inhibitor from Aedes aegypti with thrombin coagulant inhibitory activity. Biochimie 92, 933–939. [DOI] [PubMed] [Google Scholar]
- Watanabe RM, Tanaka-Azevedo AM, Araujo MS, Juliano MA, Tanaka AS, 2011. Characterization of thrombin inhibitory mechanism of rAaTI, a Kazal-type inhibitor from Aedes aegypti with anticoagulant activity. Biochimie 93, 618–623. [DOI] [PubMed] [Google Scholar]
- Yuda M, Higuchi K, Sun J, Kureishi Y, Ito M, Chinzei Y, 1997. Expression, reconstitution and characterization of prolixin-S as a vasodilator—a salivary gland nitric-oxide-binding hemoprotein of Rhodnius prolixus. Eur. J. Biochem 249, 337–342. [DOI] [PubMed] [Google Scholar]
- Zhu YC, Zeng F, Oppert B, 2003. Molecular cloning of trypsin-like cDNAs and comparison of proteinase activities in the salivary glands and gut of the tarnished plant bug Lygus lineolaris (Heteroptera: Miridae). Insect Biochem. Mol. Biol 33, 889–899. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.