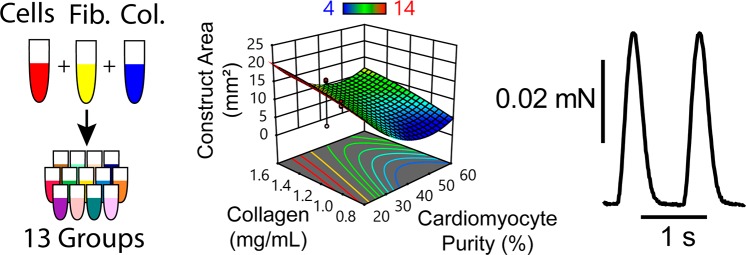
Abstract
Natural polymer hydrogels are used ubiquitously as scaffold materials for cardiac tissue engineering as well as for soft tissue engineering more broadly because of FDA approval, minimal immunogenicity, and well-defined physiological clearance pathways. However, the relationships between natural polymer hydrogels and resident cell populations in directing the development of engineered tissues are poorly defined. This interaction is of particular concern for tissues prepared with iPSC-derived cell populations, in which population purity and batch-to-batch variability become additional critical factors to consider. Herein, the design space for a blended fibrin and collagen scaffold is characterized for applications in creating engineered myocardium with human iPSC-derived cardiomyocytes. Stiffness values of the acellular hydrogel formulations approach those of native myocardium in compression, but deviate significantly in tension when compared to rat myocardium in both transverse and longitudinal fiber orientations. A response surface methodology approach to understanding the relationship between collagen concentration, fibrin concentration, seeding density, and cardiac purity found a statistically significant predictive model across three repeated studies that confirms that all of these factors contribute to tissue compaction. In these constructs, increased fibrin concentration and seeding density were each associated with increased compaction, while increased collagen concentration was associated with decreased compaction. Both the lowest (24.4% cTnT+) and highest (60.2% cTnT+) cardiomyocyte purities evaluated were associated with decreased compaction, whereas the greatest compaction was predicted to occur in constructs prepared with a 40–50% cTnT+ population. Constructs prepared with purified cardiomyocytes (≥75.5% cTnT+) compacted and formed syncytia well, although increased fibrin concentration in these groups was associated with decreased compaction, a reversal of the trend observed in unpurified cardiomyocytes. This study demonstrates an analytical approach to understanding cell–scaffold interactions in engineered tissues and provides a foundation for the development of more sophisticated and customized scaffold platforms for human cardiac tissue engineering.
Keywords: tissue engineering, hydrogel, collagen, fibrin, design of experiments, hiPSC-derived cardiomyocytes
1. Introduction
Over the past decade, the field of tissue engineering has witnessed increased focus on scaffold materials used to create engineered tissues, due to both improved understanding of the substantial role that substrates can play in cell development and function, as well as advances in material technologies in terms of customization, cost, and accessibility. Recent studies have shown that substrate material properties alone can significantly alter cell phenotype, growth rate, and differentiation pathway.1−3 As a result, substrate and scaffold properties such as stiffness, remodeling rate, and ligand density have expanded material selection considerations well beyond simple biologic inertness. Concurrently, fabrication techniques such as electrospinning, wet spinning, salt leeching, and 3D printing with biocompatible polymers have necessitated the development of scaffold materials with both complex and extremely diverse properties and geometries, allowing for increased control over these very same variables.4−7
However, these great strides in identifying, categorizing, and manipulating cell–material interactions have not been matched with a comparable understanding of the mechanisms of cell–dense engineered tissue formation. Attempts to define the role of substrate parameters in cell or tissue function have typically been singular in their approach, evaluating the influence of a single variable on tissue function, which oversimplifies the interconnected parameter space of engineered tissues.8−10 As a result, predictive tissue scaffold models have been limited in their scope.11−13 Many of these characterization studies focus on responses exclusively at the cellular level and employ analytical formats and materials unsuitable for direct translation to tissue scale application. Although more sophisticated statistical approaches are increasingly used to address other challenges related to tissue engineering, such as the optimization of stem cell differentiation,14,15 biomaterial fabrication,16 and imaging protocols,17 these same methods have not been applied to hydrogel-based cardiac tissues. This has led to the identification of tissue scaffold compositions for specific cell populations primarily through trial-and-error, and limited data has been published to justify the selected components.
Within the cardiac tissue engineering niche, there are diverse and evolving approaches to producing engineered myocardium. Once common to the field, poorly defined additives and supplements derived from basement membrane, such as Matrigel and Geltrex, are being eliminated because of an emphasis on reproducibility and translation. Although both natural and synthetic polymer scaffolds are often used, simple hydrogels are often favored because of their ease-of-use and the ability to seed a high number of cells in a small volume, relative to other scaffold modalities such as premade scaffolds seeded later with cells. Natural polymers have a high density of cell binding sites, relatively high rate of remodeling by resident cells with nontoxic byproducts, and off-the-shelf availability. However, they are typically lacking in mechanical stiffness and strength that can be achieved with synthetic materials. Both collagen hydrogels with concentrations ranging from 0.5 to 1.5 mg/mL18−23 and fibrin hydrogels with concentrations ranging from 2.0 to 4.0 mg/mL24−26 have been used frequently as cardiac tissue scaffolds, but there is no evidence in the literature of these two hydrogels used in blends.
Collagen I is the predominant structural protein in the human body, as well as in native myocardium. Characterized by the triple helical molecular structure typical of all collagens, collagen I self-assembles into fibrils composed of interleaved individual molecules, which are further organized into collagen fibers of varying diameters in native tissue. In tissues exposed to high mechanical stresses, such as tendons, ligaments, and muscle, these fibrils and fibers are oriented parallel to the stresses, maximizing their tensile mechanical contributions. In the myocardium, collagen fibers are the primary component of the endomysium, forming the long channels that surround the cardiac muscle bundles and contributing to the anisotropy of native myocardium. Although most collagen hydrogel scaffolds are initially unorganized, it has been demonstrated that organization can be imparted over time via stresses applied by resident cells.30
Fibrin is a critical component of the wound healing cascade, wherein it provides vital, temporary structural reinforcement and stability to blood clots for hemostasis before degrading via fibrinolysis as the vessel wall is repaired and new extracellular matrix (ECM) deposited. In vivo, fibrin can be completely degraded just hours after polymerization.31,32 Fibrin can also rapidly degrade in vitro due to plasmin and nonplasmin proteases present in cell media and produced by resident cells. When fibrin is used as a scaffold component it is typically used with a fibrinolysis inhibitor, such as aprotinin or aminocaproic acid, to increase fibrin persistence. Fibrin has great utility as a tissue scaffold because of its potential for rapid polymerization when exposed to thrombin (on the order of seconds to minutes) and due to its ability to be prepared at much greater concentrations than collagen (up to ∼100 mg/mL compared to ∼10 mg/mL), which facilitates its use in forming tissue constructs and allows it to contribute substantially to construct mechanical properties.
Beyond scaffold composition, engineered cardiac tissues also differ significantly from each other in their initial cell population and seeding density. It is well-accepted that there are functional differences between native cardiomyocytes and those derived from stem cells, but there can also be significant variability based on factors such as source animal species and age, passage number, purity, differentiation protocol, and even differentiation batch.33−35 Because of the low growth rate of mature cardiomyocytes (from ≤1% in adult native cardiomyocytes36 to 2.7% in 6 week old human embryonic stem cell derived cardiomyocytes37,38) the initial seeding density of cardiomyocytes in the fabrication of engineered myocardium is also a major factor in driving cell-scaffold interactions and tissue formation. Seeding density values from the literature range widely from 2.5 to 17 million cells/mL.21−23,25 For all of these reasons, there is unlikely to be a single engineered myocardium scaffold that yields desired results with all cell types and seeding densities, but rather a spectrum of scaffold parameter sets that can be tuned to compliment a given cell population.
Herein, we present an innovative approach to characterizing the impact of both cell population and scaffold composition on engineered myocardium formation and function. We believe that employing a multifactor design of experiments approach, entirely in a 3D format, will yield a more complete understanding of the ways in which cells and scaffolds interact to form functional engineered tissues. It is our aim that this study will serve as a foundation for the development of more sophisticated and highly functional engineered cardiac tissues in the future.
2. Materials and Methods
2.1. Fabrication of PDMS Tissue Molds
Custom polydimethylsiloxane (PDMS) molds with rectangular troughs 3 mm wide and either 9 mm or 17 mm long were used to prepare cellularized and acellular hydrogels, respectively, for tensile mechanics and compaction characterization. Circular molds 5 mm in diameter were fabricated for acellular compression mechanics. All molds were fabricated using a previously described technique utilizing laser cut acrylic master molds.39,40 Briefly, negative acrylic master molds were designed in Adobe Illustrator (Adobe System Inc., San Jose, CA) and then fabricated on a ULS 6.75 model laser cutter equipped with a 75 W laser. Sylgard 184 (Dow Corning, Midland, MI) PDMS was prepared according to the manufacturer’s specifications (10:1 base:curing agent) and cast over the etched surface of the master mold. Molds were cured overnight in a 60 °C oven, autoclaved, and then positioned at the bottom of the wells of an untreated polystyrene six well plate. No adhesive was necessary to hold the molds in place in the wells for the new PDMS molds, likely due to the hydrophobicity of both the PDMS and polystyrene surfaces.
2.2. Cardiac Differentiation and Lactate Purification
Cardiomyocytes were differentiated from human induced pluripotent stem cells (hiPSCs; Gibco human episomal iPSCs or GCaMP6 WTC hiPSCs from The Gladstone Institutes) using chemically defined conditions and small molecule modulation of the Wnt signaling pathway as previously described.23,39 Briefly, hiPSCs cultured on vitronectin (Gibco)-coated 10 cm polystyrene tissue culture plates were treated with 6 μM Chiron 99021 (Tocris), a glycogen synthase kinase 3 (GSK3) inhibitor at day 0, followed by 5 μM IWP2 (Tocris), a Wnt inhibitor at day 3. Cardiac phenotype, demonstrated by beating behavior, is typically first observed between day 10 and day 14. Cardiomyocytes differentiated from hiPSCs were used for the fabrication of cellularized constructs between differentiation day 14 and 18, unless designated for lactate purification. Cardiomyocytes designated for lactate purification were harvested on day 15 and replated to new culture vessels coated with Matrigel on day 15 in RPMI 1640 medium with B27 supplement (RPMI+B27; Gibco). These cells were deprived of media change from day 16 to 19 and were fed with lactate media (DMEM without glucose, l-glutamine, phenol red, sodium pyruvate and sodium bicarbonate (D-5030, Sigma) + 4 mM l-glutamine, 1X Non-Essential Amino Acids, and 4 mM lactate, pH 7.4) on day 20 and day 22. Lactate purified cells were fed with RPMI+B27 on day 24 and then harvested to seed tissues on day 26. Cardiac purity was measured via flow cytometry analysis on every batch of cells used for the preparation of cellularized constructs. Purity was defined by the percentage of cardiac troponin T (cTnT) positive cells.
2.3. Preparation of Collagen and Fibrin Hydrogels Seeded with hiPSC-Derived Cardiomyocytes
Detailed methods for preparing collagen and fibrin hydrogel constructs seeded with cardiomyocytes have been described previously.39,40 Briefly, prior to cellularized construct preparation, custom molds were prepared for tissue culture by placing autoclaved PDMS molds at the bottom of the wells of an untreated six-well plate and treating with oxygen plasma to reduce the hydrophobicity of the PDMS surface.
Blended collagen I and fibrin hydrogels seeded with cardiomyocytes were prepared by combining neutralized collagen, fibrinogen, thrombin, and cell precursor mixtures immediately prior to casting. The collagen precursor mix was prepared by neutralizing a 4.0 mg/mL rat collagen I commercial stock solution (Advanced Biomatrix, San Diego, CA) with 1 M sodium hydroxide to a pH of 7.4, supplementing with 10× RPMI 1640 medium, and further diluting with DI water to achieve 1X RPMI, as well as the targeted collagen concentration (ranging from 0.8 to 1.6 mg/mL) in the casting mix. The fibrinogen precursor mix was prepared by diluting a 100 mg/mL bovine fibrinogen stock solution in construct media (RPMI+B27, supplemented with bovine aprotinin to achieve 50 U/mL aprotinin) as well as the targeted fibrinogen concentration (ranging from 0 to 20 mg/mL) in the casting mix. Finally, the cell precursor mix was prepared by resuspending harvested cardiomyocytes in RPMI+B27 to achieve the desired seeding density (9, 12, or 15 million cells/mL) in the casting mix. The collagen precursor mix, cell precursor mix, and cast mix were all kept on ice prior to casting. In the case of the fractional factorial methodology studies, the precursor mixes were combined in the wells of a chilled, untreated, 96-well plate to accommodate the large number of unique formulations. Thrombin was added immediately prior to casting. Specific formulations for the fractional factorial and response surface methodology studies were preferentially chosen based on which would lend the greatest amount of statistical information to the respective models.41,42 Constructs were stimulated at 1 Hz (section 2.4) during culture at 37 °C and 5% CO2, fed with construct media every 48 h, and supplemented with 50 U/mL aprotinin every day between feeds to mitigate fibrinolysis.
2.4. Electrical Stimulation of Hydrogels Seeded with hiPSC-Derived Cardiomyocytes
All cellularized constructs were stimulated for the duration of the culture period using a C-Pace EP system with six-well C-dish electrode assembly lids (IonOptix). This system is composed of stimulator lids with pairs of carbon electrodes for each well of a six well plate. Carbon electrodes extend into the culture medium and are positioned on opposite sides of the engineered tissues. A ribbon cable connects the stimulator lid to the stimulator bank outside of the incubator. Constructs were field stimulated with a 1 Hz, 10.0 V, 4.0 ms duration bipolar pulse train for the full culture period.
2.5. Preparation of Acellular Collagen and Fibrin Hydrogels
Acellular hydrogels were prepared using the same methodology as the cellularized constructs with the cell precursor mix and construct media replaced by PBS. Additionally, rather than treating the PDMS surface with oxygen plasma prior to casting, the troughs of the PDMS molds were lightly coated with canola oil to facilitate hydrogel release.
2.6. Mechanical Characterization of Hydrogels and Constructs
2.6.1. Acellular Hydrogels
Acellular blended collagen and fibrin hydrogels were evaluated in both compression and tension. Compression testing was performed on 5 mm diameter hydrogel discs cast 1.88 ± 0.06 mm thick, prepared using custom PDMS molds. Hydrogels were allowed to polymerize for one hour at either 4 or 37 °C, and were then incubated for either 1 or 72 h at 37 °C in PBS in the dark. Sample thickness was measured with a micrometer immediately prior to testing. All compression testing was performed unconfined, between two flat platens, with a 1 kN load cell, at a constant strain rate of 1 mm/min (Instron E1000) until the limit of the load cell was reached. Compressive Young’s modulus was determined from a near-linear portion of the stress–strain curve in the physiological range, usually between 0 and 5% strain, and is referred to as compressive stiffness throughout.
A subset of the acellular hydrogel formulations tested in compression, chosen to span the range of compressive stiffness values observed, were selected for further evaluation under tension. Acellular tensile samples were cast in custom 3 mm × 17 mm PDMS troughs, 1.29 ± 0.05 mm thick, with 0.5 mm diameter posts at either end. Samples were allowed to polymerize for one hour at 37 °C and then incubated further for either 1 or 72 h of incubation at 37 °C. Prior to testing, samples were cut in half crosswise and thicknesses were measured with a micrometer while widths and lengths were measured with calipers. Each half of the construct was tested under tension on a micromechanical system (Aurora Scientific) equipped with a 5 mN load cell in a bath of PBS at 37 °C. All samples were tested at a constant strain rate of 1 mm/min until failure. Tensile Young’s modulus was determined from the linear region of the stress–strain curve, usually between 15 and 25% strain, and is referred to tensile stiffness throughout.
2.6.2. Cellularized Constructs
Both active and passive tensile mechanical properties were analyzed for beating cellularized cardiac tissue constructs. Cellularized constructs were prepared in custom PDMS molds with troughs 3 mm (W) × 9 mm (L), with 0.5 mm diameter posts at either end of the troughs. Constructs were mechanically evaluated after 6 days of culture at 37 °C with 1 Hz stimulation. Uniformity of beating was visually assessed and assigned a score of 0 for no beating, 1 for localized regional beating, or 2 for uniform beating throughout tissue. After mounting the tissue on the mechanical setup and immediately prior to testing, bright field optical microscopy images were collected (Olympus SZ40) for later image analysis to determine initial sample dimensions. Both active and passive mechanical testing was performed in a 37 °C bath of Tyrode’s solution (1.8 mM CaCl2, 1.0 mM MgCl2, 5.4 mM KCl, 140 mM NaCl, 0.33 mM NaH2PO4, 10 mM HEPES, 5 mM Glucose). Resting tension was set to 10% above slack length. Active mechanical testing was performed by measuring construct contraction force at strains of 5, 10, 15, 20, 25, and 30%, held for 120 s at each length to allow for stress relaxation and electrically stimulated at 1 Hz during the final 20 s of each step to capture twitch contractions. Peak active stress is calculated at 0.3 strain, normalized for cross-sectional area. The rate of force or calcium rise is reported as upstroke velocity, Vup (in units of stress/time or normalized calcium amplitude/time, respectively). Passive testing uses eight precondition cycles at a rate of 1 mm/min to 10% strain followed by pull-to-break at the same rate until sample failure. Young’s modulus was determined from the linear region of the stress–strain curve, usually between 15 and 25% strain.
2.7. Mechanical Analysis of Native Rat Myocardium
Passive mechanical properties were analyzed for samples of native rat myocardium. All animal procedures were conducted in accordance with the U.S. NIH Policy on Humane Care and Use of Laboratory Animals and the Brown University Institutional Animal Care and Use Committee (IACUC protocol No. 1310000025). Immediately after harvest, whole rat hearts were washed, ventricles opened to expose the endocardium, and stored in Tyrodes solution with a protease inhibitor cocktail at 4 °C. Tissue samples were dissected and analyzed within 6 h. Compression samples were collected as discs with sharp dissection via a 5 mm biopsy punch through the full thickness of the left ventricular myocardial layer. Compression testing was performed unconfined, between two flat platens, with a 1 kN load cell, at a constant strain rate of 1 mm/min until the limit of the load cell was reached. Young’s modulus was determined from the linear portion of the stress–strain curve, usually between 0 and 5% strain. Tensile samples were collected as strips approximately 2 mm × 10 mm via sharp dissection with microsurgical spring scissors from the epicardial layer (∼1/4 wall thickness). Special care was taken to collect tensile samples with a long axis either parallel to the circumferential epicardial muscle fibers (referred to as longitudinal samples) or perpendicular to these muscle fibers (referred to as transverse samples, refer to Figure S1 for further clarification). Tensile testing was performed in a 37 °C bath of Tyrode’s solution with eight preconditioning cycles at a rate of 1 mm/min to 10% strain followed by pull-to-break at the same rate until sample failure. Young’s modulus was determined from the linear region of the stress–strain curve, usually between 15 and 25% strain.
2.8. Construct Compaction Assay
Construct compaction was evaluated through a nondestructive, longitudinal imaging assay. Cellularized tissues cast in custom PDMS troughs 3 mm × 9 mm were prepared in six-well plates as described above. Constructs were imaged via bright field optical microscopy at 4, 24, 48, 72, 96, and 120 h time points. Images were then analyzed in ImageJ (Schneider, 2012) to determine visible two-dimensional construct areas.
2.9. Immunofluoresence Staining and Sarcomere Length Analysis
Immunostaining was performed as previously described.23 Briefly, cellular constructs were fixed immediately after mechanical testing in 4% paraformaldehyde (MilliporeSigma) for 10 min then processed into frozen blocks with OCT and sectioned into 5 μm slices. Sections underwent antigen retrieval with a proteinase K digest (10 μg/mL; Roche) for 5 min at 37 °C prior to staining for sarcomeric alpha-actinin (α-actinin), connexin 43 (Cx43), and cell nuclei (DAPI). Images were taken with an Olympus FV3000 Confocal Microscope and processed using ImageJ. Sarcomere length was analyzed by taking the average distance between the midpoint of continuous sarcomeres for regions with a minimum of 3 visible bands aligned with the major axis of the construct.
2.10. Statistical Analysis
All statistical analyses were performed in Prism 7 (GraphPad Inc., San Diego, CA). For comparison between two groups, Student’s t tests were used. For comparisons of more than two groups, one way analysis of variance (ANOVA) with multiple comparisons and Tukey’s posthoc test was used. Error bars represent standard error of the mean unless otherwise noted. Group differences were considered statistically significant for p-values <0.05.
3.0. Results
3.1. Collagen and Fibrin Modulate Acellular Construct Stiffness
Investigation of acellular blended hydrogel compressive mechanical properties was performed via a two-level fractional factorial approach to assess how these natural matrix proteins interact in the absence of cellular remodeling. A total of five hydrogel formulations and processing parameters were considered, namely collagen concentration, fibrin concentration, aprotinin concentration, polymerization temperature, and time in culture. Two levels were evaluated for each factor, referred to as “low” and “high” for convenience. Low and high factor levels were chosen based on the lower and upper bounds of tolerances necessary for cell viability, historical data from our lab,23,39 and values previously reported in the literature21,22,25−27,43 (Table 1). Higher levels of collagen and fibrin were chosen to emulate the density of ECM in compacted engineered tissues for analysis of acellular samples. The fractional factorial approach leverages linear regression statistics to reduce the number of trials that must be conducted to quantify the impact of each factor on a response, at the cost of the ability to isolate high order interactions from the system. According to the sparsity-of-effects principle, such interactions are rare and are unlikely to have a significant effect on a response of interest.41 Sixteen individual hydrogel formulations were defined as combinations of these high and low levels for each factor and tested in compression (Figure 1B).
Table 1. Composition of Engineered Cardiac Tissue Constructs Reported in the Literature.
| research group | hydrogel composition | protein concentration | cell type (s) | seeding density (cells/mL) |
|---|---|---|---|---|
| Black25 | fibrin | 3.3 mg/mL | neonatal rat CM | 4.9 × 106 |
| Coulombe23 | collagen I | 1.25–1.5 mg/mL | hiPSC-CM | 17 × 106 |
| Eschenhagen18,19 | collagen I and Matrigel | 0.56–0.89 mg/mL | neonatal rat CM | 1.1 × 106 to 11 × 106 |
| Murry20 | collagen I and Geltrex | 1.25 mg/mL | hESC/iPSC-CM | 20 × 106 |
| Radisic27,28 | Davol ultrafoam + Matrigel | 7.5 mg/mL | neonatal rat CM + C2C12 | 67 × 106 to 123 × 106 |
| Tranquillo24 | fibrin | 2–4 mg/mL | hiPSC-CM | 3 × 106 to 5 × 106 |
| Vunjak-Novakovic29 | porous PGS coated with laminin | N/A, solid | neonatal rat CM | 146 × 106 |
| Zimmerman21 | collagen I | 0.8–1.2 mg/mL | hiPSC-CM or neonatal rat/mouse | 2.5 × 106 to 3 × 106 |
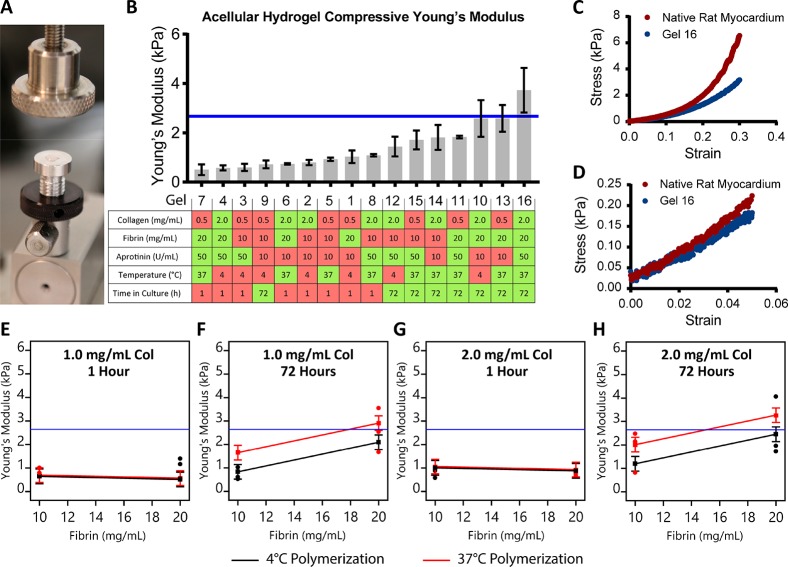
Figure 1.
Unconfined compression of acellular blended hydrogels and native rat myocardium. (A) Unconfined compression testing apparatus with hydrogel disc on platen. (B) Unconfined compressive Young’s modulus of hydrogel formulations shown from least to most stiff, with the formulation and processing parameters for each gel defined in the table below. Red boxes indicate low factor levels and green boxes indicate high factor levels. The blue line indicates the mean unconfined compressive Young’s modulus of adult rat myocardium tested under the same conditions. (C, D) Representative unconfined compressive stress–strain plots for rat myocardium and Gel 16 from 0 to 30% and 0 to 5% strain, respectively. (E–G) Interaction plots for the influence of polymerization temperature, time in culture, and fibrin concentration on hydrogel stiffness. Black symbols and lines represent the low polymerization temperature (4 °C), whereas red symbols and lines represent high polymerization temperature (37 °C). Blue lines represent mean compressive stiffness of adult rat myocardium.
Unconfined compressive Young’s modulus values ranged from 0.50 ± 0.22 to 3.73 ± 0.91 kPa across all formulations, spanning the mean value measured for native rat myocardium (2.67 ± 0.61 kPa, Figure 1B). Both hydrogels and native myocardium stress/strain curves demonstrated strain-stiffening phenomena typical of natural polymers and native tissue. High levels of both time in culture and polymerization temperature were associated with increased hydrogel compressive stiffness (green squares in table of Figure 1B), and were found to be significant factors (p < 0.05) in the fractional factorial analysis (see Table S4). Collagen concentration and aprotinin concentration had no significant impact on blended hydrogel compressive stiffness. The stiffest gel, number 16, showed good agreement with the nonlinear toe region of native rat myocardium (Figure 1C), particularly in the physiological range of strain (Figure 1D). Interestingly, high fibrin concentration also significantly increased hydrogel stiffness, but the increase was only apparent after 72 h of incubation in PBS in the dark (Figure 1E–H). Although the fibrin and thrombin concentrations were sufficient to induce polymerization within 5–10 min, well before the start of testing, it is possible that additional cross-links or molecular interactions (including fibrin–fibrin or fibrin-collagen interactions) developed during the extended incubation, which altered the compressive mechanical properties associated with initial fibrin concentration.
Native myocardium experiences both compressive and tensile forces in vivo,44 and for this reason, tensile testing was done on a subset of four formulations, spanning the range of measured compressive stiffness values and containing high (2 mg/mL) collagen. For comparison, rat cardiac tissue samples were characterized in both long and transverse fiber directions (see details in Methods, section 2.7) due to the anisotropic architecture of native myocardium.
Tensile stiffness ranged from 0.28 ± 0.04 to 3.48 ± 0.33 kPa, which was well-below the tensile stiffness values found for native rat myocardium (30.80 ± 2.71 kPa in the long fiber direction and 16.58 ± 1.85 kPa in the transverse fiber direction, Figure 2B-D). These stiffness values and the anisotropic ratio of approximately 2:1 are similar to values reported in the literature.45,46 Again, both the hydrogels and native myocardium stress/strain curves demonstrated the strain-stiffening phenomenon. Gels with low fibrin concentrations had lower stiffness values versus those with high fibrin concentrations at one hour (blue bars, Figure 2B). However, tensile stiffness of both of the low fibrin concentration gels increased after 72 h of culture, while there was no significant change in tensile stiffness of the other groups with culture time. This may indicate that collagen assembly enabled increased tensile stiffness at 72 h in gels with low fibrin (purple bars in gels 2 and 8, Figure 2B), and that either increased fibrin presence interfered with collagen fibril formation or there was no additive benefit of high fibrin to tensile stiffness at 72 h in gels with high fibrin (purple bars in gels 10 and 16, Figure 2B), limiting the tensile stiffness of the material.
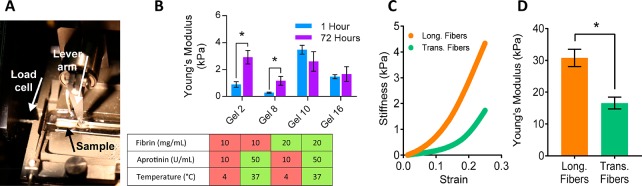
Figure 2.
Tensile analysis of acellular blended hydrogels and native rat myocardium. (A) Micromechanics setup consisting of a 37 °C PBS bath with platinum electrodes, lever arm of rapid stepper motor (right), and 5 mN load cell (left) used for mechanical analysis. (B) Young’s modulus for hydrogel formulations (n = 3 for all groups) for 1 h (blue) and 72 h (purple) samples (n = 3, *P < 0.05). (C) Representative stress/strain curves for longitudinal (Long., orange) and transverse (Trans., teal) fiber native rat myocardium samples. Note distinct toe regions under 20% strain. (D) Mean adult rat myocardial stiffness data collected from samples prepared with long and transverse fiber orientations collected from two adult rats (n = 5, *P < 0.05).
3.2. Tissue Formulation Influences Compaction
Analysis of the acellular blended hydrogels indicated that fibrin concentration was the most direct method of manipulating blended hydrogel tensile and compressive stiffnesses, with time in culture and collagen concentration playing lesser roles. Knowing that cellular adhesions to the matrix proteins would significantly remodel the tissue with time, we hypothesized that an optimum tissue formulation could be defined by testing a wider range of fibrin concentrations without changing other parameters using hiPSC-derived cardiomyocytes. However, when cellularized constructs were prepared with fibrin concentrations ranging from 5 to 20 mg/mL in the presence of 2 mg/mL collagen, constructs failed to compact and resume a beating phenotype two of the three times the experiment was repeated (Figure S2A). Flow cytometry analysis of cardiac purity based on cTnT protein expression revealed differences in construct diameter for cardiac purities ranging from 13 to 77% cTnT+, suggesting purity plays an important role in tissue development by modulating tissue compaction. Considering that tissue compaction, a process through which resident cells adhere to and remodel the surrounding matrix, is a necessary precursor to functional cardiac tissue formation, a pilot study was conducted to evaluate the role of collagen concentration and seeding density on tissue compaction (Figure S2B). This study had 49.7% cTnT+ cells and used a narrowed range of collagen, from 0.8 to 1.2 mg/mL, based on what is prevalent in the literature (see Table 1) and what has worked successfully for our lab in the past.23,39 This pilot study showed that higher cell density (pink lines) and lower collagen concentration (lighter colors) enabled the greatest compaction (i.e., smallest construct diameter; Figure S2). Together, these experiments with cellularized constructs suggest that collagen concentration, fibrin concentration, seeding density, and cell purity all impact construct compaction, and therefore merit inclusion as factors in a broader set of cell-based experiments.
To evaluate tissue formation, we chose a response surface methodology (RSM) approach to assess the impact of each factor and two-level factor interactions on tissue compaction rather than using an empirical approach that would require a higher number of test groups. Collagen concentration, fibrin concentration, and seeding density were each considered as factors, each with three evenly spaced levels, in each iteration of the compaction assay experiment. Low, medium, and high levels for each factor were selected based on values from the literature (Table 1) and informed by the acellular mechanics and pilot compaction studies (see Table 2). By traditional methods, this selection of factors and levels would have required the evaluation of 27 groups in triplicate, for a total of 81 constructs.
Table 2. Factors and Levels Considered in Each Run of the Compaction Assay.
| low | medium | high | |
|---|---|---|---|
| collagen (mg/mL) | 0.8 | 1.2 | 1.6 |
| fibrin (mg/mL) | 0 | 4 | 8 |
| seeding density (1 × 106/mL) | 9 | 12 | 15 |
Instead, specific formulations to test were selected based on a standard three factor, three level Box-Behnken design.41 This design format prescribed 13 construct formulations, each of which was prepared in triplicate in each run of the experiment. Additionally, one center point group was repeated across each of the two six-well plates used for each experiment as a control to assess and minimize variability between plates, for a total of 42 constructs in each run of the experiment with a single batch of hiPSC-derived cardiomyocytes (Figure 3).
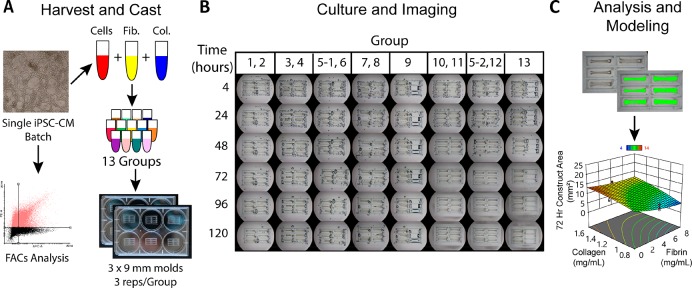
Figure 3.
Compaction assay schematic for a single hiPSC-CM batch. (A) One batch of hiPSC-CMs is harvested, a FACs sample is collected, and the remaining cells are incorporated into one of 13 sample groups dictated by the Box-Behnken design. Construct suspensions are each used to cast three 35 μL constructs in 3 × 9 mm PDMS wells (the center point Group 5 was repeated as 5–1 and 5–2 to account for variability between plates). (B) Throughout culture, samples were imaged at time points 4–120 h (left row labels) to monitor compaction. (C) Visible construct 2D area was measured via image analysis software as a metric of construct compaction. These results were used to build a response surface model (RSM) for compaction at each time point. Results from multiple experiments and hiPSC-CM batches were used to build a predictive model incorporating hiPSC-CM purity as a factor.
To incorporate cardiomyocyte purity as a fourth factor, the full experiment was repeated three times with discrete batches of hiPSC-derived cardiomyocytes. Flow cytometry samples collected during harvest were used to assign a cardiomyocyte purity to each run. Data collected across three of these runs with purities of 24.4, 30.2, and 60.2% was successfully incorporated into a single significant predictive model, with a nonsignificant lack-of-fit to the collected data (Figure 4).
Figure 4.
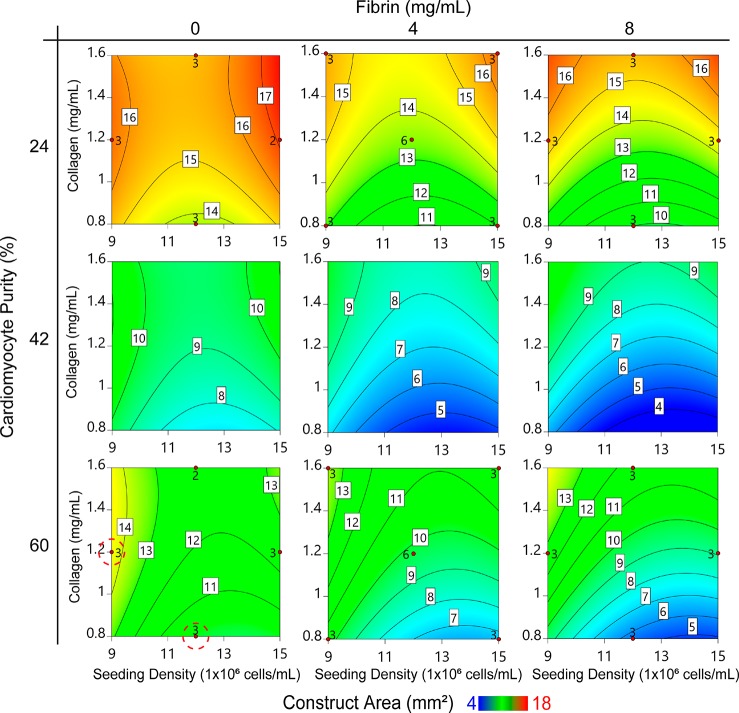
Contour plots describing predicted construct compaction after 72 h of culture. Data is organized by cardiac purity (major y-axis), fibrin concentration (major x-axis), collagen concentration (minor y-axes), and seeding density (minor x-axes) for unpurified cardiomyocytes. Contour labels and colors indicate the predicted construct two-dimensional area in mm2 (white boxed labels) after 72 h of culture. Red dots and adjacent numbers indicate the number of empirical data replicates for a corresponding factor set, in positions prescribed by the Box-Behnken design. Red dashed circles indicate contractile groups.
Figure 4 is a discrete representation of the continuous model generated from the data collected in this experiment. Each of the nine contour plots represents the model’s prediction of visible construct area in mm2 (a metric of compaction) for a given cardiac purity value (major y-axis) and fibrin concentration (major x-axis) after 72 h in culture, at which point compaction had stabilized. Within each contour plot, construct area values specific to a given collagen concentration (minor y-axis) and seeding density (minor x-axis) can be found. Blue areas represent small visible construct areas, associated with high degrees of compaction, whereas red areas represent high visible construct areas, associated with low degrees of compaction. A complete numeric representation of this and all other presented models can be found in the Supporting Information.
Increased fibrin concentration and initial seeding density were both associated with increased construct compaction, while increased collagen concentration was associated with decreased compaction. Cardiac purity was the predominant factor influencing construct compaction and our data shows nuances not yet reported in cardiac tissue engineering. The least compaction was observed with the lowest cardiomyocyte purity (24.4%), suggesting that cardiomyocyte-ECM adhesions are crucial for hydrogel remodeling and tissue compaction. Interestingly, the highest cardiomyocyte purity (60.2%) was associated with moderate compaction that increased with increasing fibrin concentration to a minimum area of about 5 mm2. The greatest compaction was predicted to occur with cardiomyocyte purities ranging from 40 to 50%, also demonstrating reduced area to a minimum of about 4 mm2 with increasing fibrin concentration (Figure S4).
While these experiments identified construct formulations that consistently yielded high degrees of compaction, only two formulations produced uniformly beating constructs across all three of the trials (Figure 4) and therefore predictive models for functional mechanics such as contractility were not generated from these experiments. We hypothesized that disruption of the cardiac electrical syncytium was occurring due to the noncardiomyocyte cell population in spite of high levels of compaction. Thus, we repeated this experiment a fourth time with lactate purified cardiomyocytes having 75.5% cTnT+ cells (Figure 5A).
Figure 5.
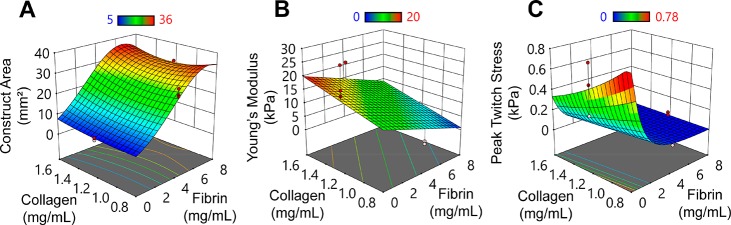
Response surface plots for cardiac tissue constructs prepared from purified hiPSC-derived cardiomyocytes (15 × 106 cells/mL) at day 6. (A) Visible construct area as measured via the medium-throughput compaction assay. See Figure S3 for an alternative representation of this data. (B) Tensile stiffness under a constant strain rate of 10% strain/min. (C) Force generation normalized by cross-sectional area at 1 Hz electrical stimulation. Red and pink dots represent data points above and below (respectively) those predicted by the model.
3.3. Robust Compaction and Beating is Associated with Increased Stiffness and Force Production
Passive and active mechanical characteristics were quantified in constructs containing lactate purified cardiomyocytes where functional cardiac tissues emerged in 9 of 13 groups (Figure S3). In addition to compaction, predictive models were generated for Young’s modulus and peak twitch force (Figure 5B,C).
Surprisingly, although compaction was dominated by fibrin concentration, high concentrations of fibrin reduced compaction with lactate purified cardiomyocytes, which is the opposite effect of fibrin to that observed in unpurified cardiomyocyte constructs. While again increased seeding density was associated with increased compaction and increased collagen concentration was associated with decreased compaction, their effects were small (Figure S3, Table S9). Further, data using the lactate purified cardiomyocytes could not be incorporated into a single predictive model with data from the unpurified cardiomyocyte experiments due to a statistically significant lack of fit (p < 0.0001). This result indicates that a quadratic or lower order model could not be found to describe the compaction response of both purified and unpurified cardiomyocytes, which we discuss below.
Predictive modeling for tensile Young’s modulus of cardiac tissue constructs associates increased stiffness with increasing collagen concentration and cell number but decreased fibrin concentration. Tensile stiffness of this hydrogel engineered tissue system evaluated empirically approached values of native rat myocardium (15–30 kPa; Figures 2D and Figure S4) in the 0 mg/mL fibrin, 1.2 mg/mL collagen, and 15 × 106 cells/mL group, where Young’s modulus was 21.79 ± 3.23 kPa (Group 11; Table S1, Figure S5D), approximated by the model as 16 kPa. Further, three other groups approached the lower limit of the physiological range of stiffness (Figure S5), suggesting an open landscape for mimicking the native myocardium stiffness.
Active twitch contractions at 1 Hz (equivalent to 60 beats per minute, a resting human heart rate) has a physiological positive force-length response 6 days after tissue formation (Figure S6). Predictive modeling for peak twitch stress (i.e., force normalized to cross-sectional area) associates high peak twitch forces with increased seeding densities and decreased fibrin concentrations (as with Young’s modulus) but also with decreased collagen concentration (which is the opposite of the Young’s modulus data and predictions; Figure 5B, C). The greatest peak twitch stress observed experimentally at 30% stretch beyond resting length is 0.54 ± 0.14 kPa in constructs produced with 0 mg/mL fibrin, 1.2 mg/mL collagen, and 15 × 106 cells/mL (Group 11), which is the same group as peak stiffness. However, peak twitch stress as high as 0.84 kPa is predicted for constructs with lower (0.8 mg/mL) collagen concentration. Peak twitch stress varies between groups as much as 5-fold (Table 3) demonstrating that matrix composition alone greatly influences not just tissue remodeling (compaction) but also cellular functional phenotype. The contraction upstroke velocity, Vup, is faster with higher peak stress but similar between groups when Vup is normalized by peak stress, suggesting that crossbridge binding and force generation kinetics are not altered in the different tissue groups. The twitch duration, measured by the time (in ms) from peak stress to 50 and 90% relaxation (T50 and T90, respectively) and controlled by calcium handling dynamics and thin filament deactivation, shows little change between groups, suggesting little to no dependence of twitch relaxation on force amplitude. This force data suggests that the ECM composition at tissue casting impacts the early (day 6) amplitude of force generation in engineered cardiac tissue with little impact on cardiomyocyte functional maturation.
Table 3. Lactate Purified Twitch Mechanics at 1 Hz for Select Groupsa.
| group | peak active stress (kPa) | Young’s modulus (kPa) | Vup (mN/mm2/s) | T50 (ms) | T90 (ms) |
|---|---|---|---|---|---|
| 1 | 0.49 ± 0.13 | 8.29 ± 0.025 | 2.29 ± 0.71 | 180 ± 11.1 | 318 ± 15.0 |
| 4 | 0.11 ± 0.05 | 13.03 ± 1.19 | 0.59 ± 0.30 | 124 ± 16.0 | 238 ± 11.4 |
| 7 | 0.18 ± 0.08 | 18.92 ± 1.02 | 1.18 ± 0.55 | 129 ± 24.7 | 237 ± 40.0 |
| 11 | 0.54 ± 0.14* | 21.79 ± 3.26 | 2.94 ± 0.68* | 156 ± 6.35 | 279 ± 3.48* |
Vup, upstroke velocity; T50 and T90, time to 50% and 90% relaxation, respectively. See Figure S6 for more information. n = 3 for all groups. * indicates p < 0.05 versus group 4.
3.4. Force Production Follows Calcium Kinetics
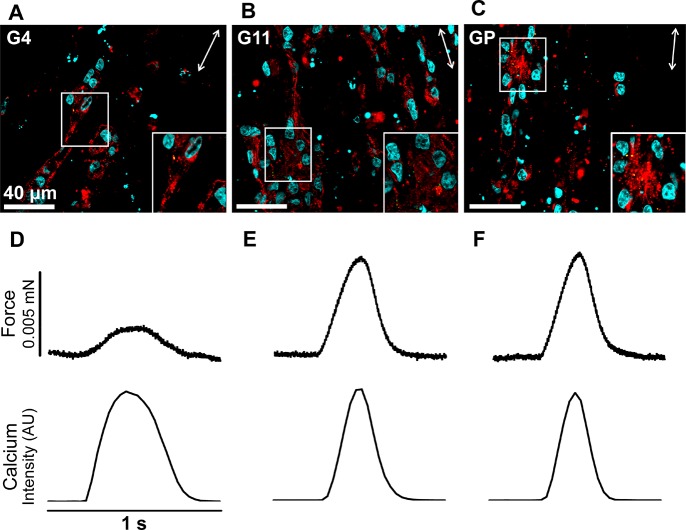
To directly evaluate calcium transient kinetics in engineered cardiac tissues prepared using the conditions described, tissues were prepared with cardiomyocytes differentiated from an hiPSC line genetically engineered to express a calcium indicator, GCaMP6 (GCaMP-CMs). The conditions associated with groups 11 and 4 were chosen for further evaluation based on the significant differences observed in force production (Tables 3, S3). Additionally, a new group was added (referred to as Group “Predicted”) based on the model’s prediction of conditions that would maximize force production (15e6 cells/mL in 0.8 mg/mL collagen only). All groups were cultured under the conditions previously described for 6 days and evaluated for cellular morphology (Figure 6A–C) and function (Figure 6D–F, Table S3). All groups formed syncytia and were visibly beating uniformly when evaluated on day 6. No differences are observed in alpha-actinin-positive cardiomyocyte content (cell number or size), alignment, and sarcomere length (data not shown). Connexin 43 shows no change in expression level or distribution of gap junctions (Figure 6A–C, insets), suggesting that force amplitude was not primarily altered by cardiomyocyte or myofilament content, alignment, or electrical connectivity at this relatively early time point after tissue formation.
Figure 6.
Histological analysis and simultaneous force/calcium transient analysis of cardiac tissue constructs prepared from GCaMP-CMs at day 6. (A–C) Frozen sections of tissue constructs prepared under group 4 conditions (G4, 9e6 cells/mL in 1.2 mg/mL collagen only), group 11 conditions (G11,15e6 cells/mL in 1.2 mg/mL collagen only), and the conditions predicted to lead to the highest force production by the model (GP, 15e6 cells/mL in 0.8 mg/mL collagen only). Immunofluorescence of tissue sections show α-actinin (red) labeled cardiomyocytes aligned with the tissue main axis (double-headed arrows), connexin 43 (green), and DAPI (cyan). Insets highlight striated cardiomyocytes and punctate connexin 43-positive gap junctions. (D–F) Force (top) and calcium transient (bottom) traces from representative contractions are generated from fluorescent video capture of GCaMP signal for each of the three groups (corresponding to A–C) at 15% strain and 1 Hz.
Active mechanical measurements are made with simultaneous fluorescent video collection to capture calcium transients from the GCaMP signal (Figure 6D–F). As expected, faster upstroke velocity, Vup, is required to reach higher peak stress in group 11 (Tables 3, Figure S3). However, significant differences in upstroke velocity of force disappear when Vup is normalized for stress amplitude (i.e., normalized by the number of crossbridges, since force is proportional to crossbridge number). These normalized force upstroke kinetics are similar to calcium kinetics (namely, calcium Vup is not significantly different between groups 4 and 11; Table S3). These data suggest no change in calcium release kinetics due to ECM conditions of engineered tissues; however, calcium amplitude was not measured and could explain force amplitude differences. No differences in twitch duration (T50, T90) suggest that calcium sequestration kinetics are not significantly impacted by ECM composition.
Within 72 h of casting, all but one of the six replicates prepared for the Predicted group (GP) failed due to necking and breaking, suggesting that the mechanical properties of the compacted hydrogel are insufficient to withstand the cell-generated tissue tension.47 The single surviving sample from the Predicted group produced the greatest peak contraction force, validating the model and supporting a critical role for collagen density in maintaining tissue integrity during early tissue formation. However, the model cannot account for an imbalance of cell tension and ECM strength during remodeling that causes breaking, so empirical evaluation must be completed.
4. Discussion
The study presented herein sought to understand the relationship between blended collagen and fibrin hydrogels and resident human iPSC-derived cardiomyocyte populations for the purpose of creating engineered cardiac tissues. Utilizing a response surface methodology to evaluate the impact and interactions of these factors, the results clearly demonstrate that scaffold protein composition and cell population both play critical roles in engineered cardiac tissue development, as measured by compaction, tissue stiffness, and active force production.
In the field of cardiac tissue engineering, the debate concerning what the optimal scaffold is ensues. How to define “optimum” will depend upon the desired outcome of the study, be it in vitro disease modeling or in vivo heart repair. Our study evaluated acellular and cellular constructs to assess how scaffold composition impacts stiffness (Young’s modulus), hydrogel compaction, and contractile force generation using the two most common natural polymer matrices, namely type 1 collagen and fibrin. Knowing that the stiffness of the environment impacts cellular phenotype,9 acellular compressive and tensile mechanics (Figures 1 and 2) demonstrated that the concentrations of collagen and fibrin dominated the time-dependent stiffness response. In cellular constructs, we found that (1) purity of the input hiPSC-derived cardiomyocyte population drives the success of tissue formation into a uniformly beating cardiac tissue and (2) trade-offs must be made between maximizing tensile stiffness (to match values from native adult myocardium) and maximizing contraction force. The range of acellular blended collagen and fibrin hydrogel formulations considered in this study approximated the compressive stiffness of native rat myocardium, but deviated significantly (P < 0.0001) from the tensile stiffness of native rat myocardial tissue in both longitudinal (30.80 ± 2.71 kPa) and transverse (16.58 ± 1.85 kPa) fiber orientations. However, tensile stiffness values of two groups of cellularized cardiac constructs (18.92 ± 1.02 kPa and 21.79 ± 3.26 kPa) approached the tensile stiffness values of native adult myocardium. Remodeling by resident cells and associated stresses resulting from isometric compaction (due to the custom-designed tissue molds39,40) increase scaffold protein density and provide alignment cues, which are both likely responsible for the increased tensile stiffness values in cellular constructs. It is important to note that the predictive model generated in this study associated increased initial collagen concentration with both increased tensile stiffness and decreased peak active twitch stress (Figure 5). Therefore, while high collagen concentrations yield constructs with passive mechanics that approximate those of native myocardium, lower collagen concentrations yield constructs with improved active contractile mechanics (Figures 2 and 5). A potential explanation for this trade-off of stiffness versus contractile force amplitude is that the hiPSC-derived cardiomyocytes are relatively immature at the 6-day evaluation time point used in this study, having structure and contractility akin to fetal tissue,38,48,49 and thus may function best in a stiffness environment that more closely matches that of the fetal heart (12 ± 4 kPa).50
The experiment design employed in this study aimed to identify a role for fibrin in a blended hydrogel system that was driven by the hypothesis that the stiffness provided by fibrin would improve cellular remodeling, tissue stiffness, and contractility upon tissue formation and prior to cellular maturation. An important consideration for scaffold composition is that prior experiments in our lab and others47 have demonstrated that sufficiently low scaffold protein concentrations and high seeding densities can lead to construct compaction to the point of extreme necking, breaking, and construct failure. Studies by other groups that use only fibrin as a scaffold material utilize an inhibitor of fibrinolysis (like aprotinin or aminocaproic acid) to inhibit fibrin degradation, presumably to enable tissue formation, prevent breaking, and allow sufficient time for cellular deposition of matrix.25,26 Thus, although this study did not assess fibrin-only constructs, we show that compaction of blended hydrogel constructs is aided by fibrin in these experimental conditions (Figure 4), potentially due to its ease of degradation and remodeling by the resident cell population, which is useful in many tissue engineering applications. Aprotinin was chosen as a fibrinolytic inhibitor for this study due to its widespread use in the field of cardiac tissue engineering.32,51,52 Aprotinin is known to inhibit several serine proteases including plasmin, trypsin, and chymotrypsin.53 Conversely, other fibrinolytic inhibitors such as aminocaproic acid54 and tranexamic acid55 are believed to influence a narrower set of proteases, which may be suited for mechanistic study of fibrin remodeling in engineered tissues.
The iterative DOE approach is a powerful tool for assessing the influence of scaffold composition on tissue formation and function, and our results clearly show modulation of the engineered tissue by both scaffold properties and cell population, specifically cell purity and seeding density, though likely also cell type, cell line, and species. Therefore, for the purpose of optimizing construct formulation for generating compact contractile human cardiac tissue with hiPSC-derived cardiomyocytes, our study demonstrates that a low collagen concentration (0.8 mg/mL) and high cardiomyocyte purity are required.
Using unpurified cardiomyocytes (25–60% cTnT-positive), increasing fibrin concentration and cell seeding density were found to increase tissue compaction, while increased collagen concentration was found to decrease compaction (Figure 4). This model predicts the greatest compaction occurs with a moderate cardiomyocyte purity (40–50%); however, this group did not correlate with uniform beating, which is visual evidence of a syncytium throughout the tissue. Only 60% pure cardiomyocytes with no fibrin demonstrated uniform beating in all three tissue samples (Figure 4, circled data points), suggesting a strong role for cardiac purity in predicting a functional cardiac tissue. This result may guide quality control parameters for cardiac tissue engineering. Importantly, the impact and influence of collagen and fibrin scaffold parameters on tissue formation, as measured by tissue compaction, changed as a result of higher cardiac purities, which was achieved through either efficient directed differentiation or with post-differentiation lactate-based metabolic purification. The inversion of fibrin’s effect on tissue compaction and the fact that constructs prepared with low versus high purity cardiomyocytes could not be described by a single response surface model suggest that hiPSC-derived cardiomyocytes alone are effective in binding collagen and compacting the hydrogel to form functional cardiac tissue. The lactate purification protocol used in this study requires 11 days of treatment beyond the 14–15 day protocol required for cardiomyocyte differentiation. As a result, the population of lactate purified cells was substantially older than the unpurified cell groups (26 days vs 14–18 days). However, these older cells performed well in collagen-only engineered tissues when purity was high, resulting in peak twitch forces of 0.54 ± 0.14 mN/mm2 (Figure 5). Constructs prepared with high-purity, unpurified cardiomyocytes (84.6% cTnT+) responded similarly to collagen hydrogel scaffolds with and without fibrin (Figure S7). How the age of cardiomyocytes entering tissues impacts tissue formation remains an open issue; a recent study by Vunjak-Novakovic and colleagues strongly supports using younger cardiomyocytes (10–12 days differentiation in 12 mg/mL fibrin hydrogel scaffold) to take advantage of their cellular plasticity as they adapt to the 3D environment and mature in the engineered tissue,51 but this approach compromises control over the input cell population and primarily it is purity. As we observed with high purity cardiomyocytes, contractility was greatly modulated by scaffold composition, emphasizing the importance of well-defined scaffold compositions in forming functional tissues with reproducibility.
Fully defined culture conditions (without serum or Matrigel) are used in this study to minimize unknown and variable protein constituents and are being adopted in a number of tissue engineering studies utilizing hiPSC-derived cells for future clinical applications. Indeed, the engineered cardiac tissues prepared in this study are cultured under defined conditions similar to those optimized by Zimmermann and colleagues.56 In agreement with their study, which used fibroblast doping to modulate cardiomyocyte content of engineered tissues to be 40–75% cardiomyocytes, we show that peak construct compaction occurs with constructs composed of 40–50% cTnT+ cardiomyocytes. We also show a consistently increased force amplitude with low collagen and no fibrin across batches of high purity cardiomyocytes (with and without lactate purification and from Gibco or GCaMP hiPSC lines), suggesting that low collagen is a critically important design parameter in the context of defined culture conditions that enables functional tissue formation despite variation in hiPSC-cardiomyocyte populations. Force generation kinetics followed the kinetics of the calcium transient, but were not different between groups when normalized by force amplitude, suggesting that differences in peak force were not due to alterations in calcium or force kinetics. Possible explanations for the differences in contractile force amplitude are the amplitude of the calcium release (not measured) or force transduction pathways through the cytoskeleton and ECM. Future work may elucidate these mechanisms and their relationship to “maturation” through longer culture periods, in addition to fibroblast supplementation and different biophysical culture conditions (e.g., Tiburcy et al. uses additional stretching of the tissue after compaction and stiff posts for maintenance or “maturation” culture). Here, we demonstrate that uniformly contracting tissues with good force generation are formed with an optimum, defined matrix composition by day 6, prior to maturation culture, while accounting for varying input cardiomyocyte populations.
The noncardiomyocyte hiPSC-derived cell population is largely dismissed as a “fibroblast-like” cell type, but heterogeneous cell and matrix interactions do impact engineered tissue formation. Construct compaction peaked at approximately 45% purity for all fibrin concentrations, suggesting that neither hiPSC-derived cardiomyocytes nor the noncardiac cells dominate the compaction response, but that cardiac tissue remodeling relies on a balance of heterogeneous cell interactions. Cardiac fibroblasts are primarily responsible for matrix remodeling in native myocardial tissue57,58 and show plasticity in their number and phenotype during aging and disease, such as postmyocardial infarction when they become “activated” to a more contractile and synthetic phenotype.59,60 Although the noncardiac population in these studies was 3.0–43.6% positive for alpha-smooth muscle actin (αSMA; a classic marker of activated, contractile fibroblasts61,62) they were not efficient at compacting matrix (as may be expected for fibroblasts) when they made up ∼75% of the cell population, suggesting that they may also have an immature or poorly defined phenotype. It is clear that under these in vitro conditions, cardiomyocytes play a prominent role in tissue remodeling and that heterogeneous interactions with fibroblasts and scaffold composition modulate cardiomyocyte-driven compaction and contractility. Although prior studies have suggested that human cardiomyocytes contribute to the process of matrix deposition and remodeling,63 to the best of our knowledge, this is the first study to demonstrate and quantify the network of interactions between cells and matrix components influencing the ability of hiPSC-derived cardiomyocytes to form and remodel engineered tissue.
5. Conclusions
Engineered cardiac tissue development is dictated by a complex relationship between scaffold composition, cell density, and cell purity. In collagen and fibrin hydrogel scaffolds seeded with unpurified hiPSC-derived cardiomyocytes, the greatest tissue compaction was predicted to occur with a relatively high concentration of fibrin (8 mg/mL), low concentration of collagen (0.8 mg/mL), high seeding density (15 × 106 cells/mL), and moderate cardiomyocyte purity (40–50%). However, beating tissues appeared only in pure collagen groups with higher cardiac purity (≥60%), and at 6 days after tissue formation peak contractile force was 0.54 ± 0.14 mN/mm2 with 15 × 106 cardiomyocytes/mL in 1.2 mg/mL collagen. These results provide robust methods for creating highly functional human iPSC-derived cardiac tissues for many translational applications, and emphasize the importance of matching scaffold composition to cell population to form tissues with optimal efficiency and function.
Acknowledgments
We gratefully acknowledge funding from NIH R01 HL135091 and the GCaMP6 hiPSC line from Bruce Conklin (The Gladstone Institutes).
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsbiomaterials.8b01112.
The Supporting Information contains figures and tables reporting data from pilot experiments, alternate visualizations and further details of data presented in the main manuscript, and detailed statistics describing the reported RSM models (PDF).
The authors declare no competing financial interest.
Supplementary Material
References
- Discher D. E.; Janmey P.; Wang Y. Tissue Cells Feel and Respond to the Stiffness of Their Substrate. Science 2005, 310 (5751), 1139–1143. 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- Gershlak J. R.; Resnikoff J. I.; Sullivan K. E.; Williams C.; Wang R. M.; Black L. D. III Mesenchymal Stem Cells Ability to Generate Traction Stress in Response to Substrate Stiffness Is Modulated by the Changing Extracellular Matrix Composition of the Heart during Development. Biochem. Biophys. Res. Commun. 2013, 439 (2), 161–166. 10.1016/j.bbrc.2013.08.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran J. M.; Chen R.; Hunt J. A. The Guidance of Human Mesenchymal Stem Cell Differentiation in Vitro by Controlled Modifications to the Cell Substrate. Biomaterials 2006, 27 (27), 4783–4793. 10.1016/j.biomaterials.2006.05.001. [DOI] [PubMed] [Google Scholar]
- Cheng J.; Jun Y.; Qin J.; Lee S.-H. Electrospinning versus Microfluidic Spinning of Functional Fibers for Biomedical Applications. Biomaterials 2017, 114, 121–143. 10.1016/j.biomaterials.2016.10.040. [DOI] [PubMed] [Google Scholar]
- Caves J. M.; Kumar V. A.; Martinez A. W.; Kim J.; Ripberger C. M.; Haller C. A.; Chaikof E. L. The Use of Microfiber Composites of Elastin-like Protein Matrix Reinforced with Synthetic Collagen in the Design of Vascular Grafts. Biomaterials 2010, 31 (27), 7175. 10.1016/j.biomaterials.2010.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears N. A.; Dhavalikar P. S.; Seshadri D.; Cosgriff-Hernandez E. A Review of 3D Printing of Tissue Engineering Constructs. Tissue Eng., Part B 2016, 22, 298. 10.1089/ten.teb.2015.0464. [DOI] [PubMed] [Google Scholar]
- Hwang P. T.; Murdock K.; Alexander G. C.; Salaam A. D.; Ng J. I.; Lim D.-J.; Dean D.; Jun H.-W. Poly(ε-Caprolactone)/Gelatin Composite Electrospun Scaffolds with Porous Crater-like Structures for Tissue Engineering. J. Biomed. Mater. Res., Part A 2016, 104, 1017. 10.1002/jbm.a.35614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhabra P.; Tyagi P.; Bhatnagar A.; Mittal G.; Kumar A. Optimization, Characterization, and Efficacy Evaluation of 2% Chitosan Scaffold for Tissue Engineering and Wound Healing. J. Pharm. BioAllied Sci. 2016, 8 (4), 300–308. 10.4103/0975-7406.199346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacot J. G.; McCulloch A. D.; Omens J. H. Substrate Stiffness Affects the Functional Maturation of Neonatal Rat Ventricular Myocytes. Biophys. J. 2008, 95 (7), 3479–3487. 10.1529/biophysj.107.124545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien S.-M.; Ko L.-Y.; Huang T.-J. Effect of Pore Size on ECM Secretion and Cell Growth in Gelatin Scaffold for Articular Cartilage Tissue Engineering. Acta Biomater. 2009, 5 (2), 670–679. 10.1016/j.actbio.2008.09.020. [DOI] [PubMed] [Google Scholar]
- Drury J. L.; Mooney D. J. Hydrogels for Tissue Engineering: Scaffold Design Variables and Applications. Biomaterials 2003, 24 (24), 4337–4351. 10.1016/S0142-9612(03)00340-5. [DOI] [PubMed] [Google Scholar]
- Guyot Y.; Papantoniou I.; Chai Y. C.; Van Bael S.; Schrooten J.; Geris L. A Computational Model for Cell/ECM Growth on 3D Surfaces Using the Level Set Method: A Bone Tissue Engineering Case Study. Biomech. Model. Mechanobiol. 2014, 13 (6), 1361–1371. 10.1007/s10237-014-0577-5. [DOI] [PubMed] [Google Scholar]
- Tartarini D.; Mele E.. Adult Stem Cell Therapies for Wound Healing: Biomaterials and Computational Models. Front. Bioeng. Biotechnol. 2016, 3, 10.3389/fbioe.2015.00206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J. P.; Hu D.; Domian I. J.; Ogle B. M. An Integrated Statistical Model for Enhanced Murine Cardiomyocyte Differentiation via Optimized Engagement of 3D Extracellular Matrices. Sci. Rep. 2016, 5, 18705. 10.1038/srep18705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J. P.; Bache-Wiig M. K.; Provenzano P. P.; Ogle B. M. Heterogeneous Differentiation of Human Mesenchymal Stem Cells in 3D Extracellular Matrix Composites. BioRes. Open Access 2016, 5 (1), 37–48. 10.1089/biores.2015.0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.; Ryu S.; Tokareva O.; Gronau G.; Jacobsen M. M.; Huang W.; Rizzo D. J.; Li D.; Staii C.; Pugno N. M.; et al. Predictive Modelling-Based Design and Experiments for Synthesis and Spinning of Bioinspired Silk Fibres. Nat. Commun. 2015, 6, 6892. 10.1038/ncomms7892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnaert M.; Kerckhofs G.; Papantoniou I.; Vlierberghe S. V.; Boterberg V.; Dubruel P.; Luyten F. P.; Schrooten J.; Geris L. Multifactorial Optimization of Contrast-Enhanced Nanofocus Computed Tomography for Quantitative Analysis of Neo-Tissue Formation in Tissue Engineering Constructs. PLoS One 2015, 10 (6), e0130227. 10.1371/journal.pone.0130227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eschenhagen T.; Didié M.; Münzel F.; Schubert P.; Schneiderbanger K.; Zimmermann W.-H. 3D Engineered Heart Tissue for Replacement Therapy. Basic Res. Cardiol. 2002, 97 (1), I146–I152. 10.1007/s003950200043. [DOI] [PubMed] [Google Scholar]
- Zimmermann W.-H.; Melnychenko I.; Wasmeier G.; Didié M.; Naito H.; Nixdorff U.; Hess A.; Budinsky L.; Brune K.; Michaelis B.; et al. Engineered Heart Tissue Grafts Improve Systolic and Diastolic Function in Infarcted Rat Hearts. Nat. Med. 2006, 12 (4), 452–458. 10.1038/nm1394. [DOI] [PubMed] [Google Scholar]
- Tulloch N. L.; Muskheli V.; Razumova M. V.; Korte F. S.; Regnier M.; Hauch K. D.; Pabon L.; Reinecke H.; Murry C. E. Growth of Engineered Human Myocardium with Mechanical Loading and Vascular Co-Culture. Circ. Res. 2011, 109 (1), 47–59. 10.1161/CIRCRESAHA.110.237206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiburcy M.; Meyer T.; Soong P. L.; Zimmermann W.-H. Collagen-Based Engineered Heart Muscle. Methods Mol. Biol. 2014, 1181, 167–176. 10.1007/978-1-4939-1047-2_15. [DOI] [PubMed] [Google Scholar]
- Naito H.; Melnychenko I.; Didié M.; Schneiderbanger K.; Schubert P.; Rosenkranz S.; Eschenhagen T.; Zimmermann W.-H. Optimizing Engineered Heart Tissue for Therapeutic Applications as Surrogate Heart Muscle. Circulation 2006, 114 (1 suppl), I-7–I-78. 10.1161/CIRCULATIONAHA.105.001560. [DOI] [PubMed] [Google Scholar]
- Rupert C. E.; Coulombe K. L. K. IGF1 and NRG1 Enhance Proliferation, Metabolic Maturity, and the Force-Frequency Response in hESC-Derived Engineered Cardiac Tissues. Stem Cells Int. 2017, 7648409 10.1155/2017/7648409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer J.; Tranquillo R. T. Tissue Contraction Force Microscopy for Optimization of Engineered Cardiac Tissue. Tissue Eng., Part C 2016, 22, 76. 10.1089/ten.tec.2015.0220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye K. Y.; Sullivan K. E.; Black L. D. Encapsulation of Cardiomyocytes in a Fibrin Hydrogel for Cardiac Tissue Engineering. J. Visualized Exp. 2011, 55, 3251. 10.3791/3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel J. S.; Ye L.; Zhang P.; Tranquillo R. T.; Zhang J. J. Functional Consequences of a Tissue-Engineered Myocardial Patch for Cardiac Repair in a Rat Infarct Model. Tissue Eng., Part A 2014, 20 (7–8), 1325–1335. 10.1089/ten.tea.2013.0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radisic M.; Park H.; Shing H.; Consi T.; Schoen F. J.; Langer R.; Freed L. E.; Vunjak-Novakovic G. Functional Assembly of Engineered Myocardium by Electrical Stimulation of Cardiac Myocytes Cultured on Scaffolds. Proc. Natl. Acad. Sci. U. S. A. 2004, 101 (52), 18129–18134. 10.1073/pnas.0407817101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radisic M.; Euloth M.; Yang L.; Langer R.; Freed L. E.; Vunjak-Novakovic G. High-Density Seeding of Myocyte Cells for Cardiac Tissue Engineering. Biotechnol. Bioeng. 2003, 82 (4), 403–414. 10.1002/bit.10594. [DOI] [PubMed] [Google Scholar]
- Marsano A.; Maidhof R.; Luo J.; Fujikara K.; Konofagou E. E.; Banfi A.; Vunjak-Novakovic G. The Effect of Controlled Expression of VEGF by Transduced Myoblasts in a Cardiac Patch on Vascularization in a Mouse Model of Myocardial Infarction. Biomaterials 2013, 34 (2), 393–401. 10.1016/j.biomaterials.2012.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Y.; Wang X.; Lee D.; Greisler H. P. Dynamic Quantitative Visualization of Single Cell Alignment and Migration and Matrix Remodeling in 3D Collagen Hydrogels under Mechanical Force. Biomaterials 2011, 32 (15), 3776–3783. 10.1016/j.biomaterials.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jockenhoevel S.; Zund G.; Hoerstrup S. P.; Chalabi K.; Sachweh J. S.; Demircan L.; Messmer B. J.; Turina M. Fibrin Gel – Advantages of a New Scaffold in Cardiovascular Tissue Engineering. Eur. J. Cardiothorac. Surg. 2001, 19 (4), 424–430. 10.1016/S1010-7940(01)00624-8. [DOI] [PubMed] [Google Scholar]
- Coffin S. T.; Gaudette G. R. Aprotinin Extends Mechanical Integrity Time of Cell Seeded Fibrin Sutures1. J. Biomed. Mater. Res., Part A 2016, 104, 2271. 10.1002/jbm.a.35754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D.; Wei H.; Lu J.; Ho S.; Zhang G.; Sun X.; Oh Y.; Tan S. H.; Ng M. L.; Shim W.; et al. Generation of Patient-Specific Induced Pluripotent Stem Cell-Derived Cardiomyocytes as a Cellular Model of Arrhythmogenic Right Ventricular Cardiomyopathy. Eur. Heart J. 2013, 34 (15), 1122–1133. 10.1093/eurheartj/ehs226. [DOI] [PubMed] [Google Scholar]
- Zuppinger C. 3D Culture for Cardiac Cells. Biochim. Biophys. Acta, Mol. Cell Res. 2016, 1863, 1873–1881. 10.1016/j.bbamcr.2015.11.036. [DOI] [PubMed] [Google Scholar]
- Feinberg A. W.; Ripplinger C. M.; van der Meer P.; Sheehy S. P.; Domian I.; Chien K. R.; Parker K. K. Functional Differences in Engineered Myocardium from Embryonic Stem Cell-Derived versus Neonatal Cardiomyocytes. Stem Cell Rep. 2013, 1 (5), 387–396. 10.1016/j.stemcr.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann O.; Bhardwaj R. D.; Bernard S.; Zdunek S.; Barnabe-Heider F.; Walsh S.; Zupicich J.; Alkass K.; Buchholz B. A.; Druid H.; et al. Evidence for Cardiomyocyte Renewal in Humans. Science 2009, 324 (5923), 98–102. 10.1126/science.1164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laflamme M. A.; Gold J.; Xu C.; Hassanipour M.; Rosler E.; Police S.; Muskheli V.; Murry C. E. Formation of Human Myocardium in the Rat Heart from Human Embryonic Stem Cells. Am. J. Pathol. 2005, 167 (3), 663–671. 10.1016/S0002-9440(10)62041-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson C.; Tran D. D.; George S. C. Concise Review: Maturation Phases of Human Pluripotent Stem Cell-Derived Cardiomyocytes. Stem Cells 2013, 31 (5), 829. 10.1002/stem.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munarin F.; Kaiser N. J.; Kim T. Y.; Choi B.-R.; Coulombe K. L. K. Laser-Etched Designs for Molding Hydrogel-Based Engineered Tissues. Tissue Eng., Part C 2017, 23 (5), 311–321. 10.1089/ten.tec.2017.0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser N. J.; Munarin F.; Coulombe K. L. K. Custom Engineered Tissue Culture Molds from Laser-Etched Masters. J. Visualized Exp. 2018, (135), e57239–e57239. 10.3791/57239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Box G. E. P.; Hunter J. S.; Hunter W. G.. Statistics for Experimenters: Design, Innovation, and Discovery; Wiley-Interscience, 2005. [Google Scholar]
- NIST/SEMATECH e-Handbook of Statistical Methods; http://www.itl.nist.gov/div898/handbook/index.htm (accessed Jan 22, 2018).
- Bursac N.; Loo Y.; Leong K.; Tung L. Novel Anisotropic Engineered Cardiac Tissues: Studies of Electrical Propagation. Biochem. Biophys. Res. Commun. 2007, 361 (4), 847–853. 10.1016/j.bbrc.2007.07.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce W. H. Body Forces and Pressures in Elastic Models of the Myocardium. Biophys. J. 1981, 34 (1), 35–59. 10.1016/S0006-3495(81)84836-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelmayr G. C.; Cheng M.; Bettinger C. J.; Borenstein J. T.; Langer R.; Freed L. E. Accordion-like Honeycombs for Tissue Engineering of Cardiac Anisotropy. Nat. Mater. 2008, 7 (12), 1003–1010. 10.1038/nmat2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhana B.; Iyer R. K.; Chen W. L. K.; Zhao R.; Sider K. L.; Likhitpanichkul M.; Simmons C. A.; Radisic M. Influence of substrate stiffness on the phenotype of heart cells. Biotechnol. Bioeng. 2010, 105 (6), 1148–1160. 10.1002/bit.22647. [DOI] [PubMed] [Google Scholar]
- Wang H.; Svoronos A. A.; Boudou T.; Sakar M. S.; Schell J. Y.; Morgan J. R.; Chen C. S.; Shenoy V. B. Necking and Failure of Constrained 3D Microtissues Induced by Cellular Tension. Proc. Natl. Acad. Sci. U. S. A. 2013, 110 (52), 20923–20928. 10.1073/pnas.1313662110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batalov I.; Feinberg A. W. Differentiation of Cardiomyocytes from Human Pluripotent Stem Cells Using Monolayer Culture. Biomarker Insights 2015, 10 (Suppl 1), 71–76. 10.4137/BMI.S20050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan J. M.; Lyon A. R.; Harding S. E. The Case for Induced Pluripotent Stem Cell-Derived Cardiomyocytes in Pharmacological Screening. Br. J. Pharmacol. 2013, 169 (2), 304–317. 10.1111/j.1476-5381.2012.02118.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacot J. G.; Martin J. C.; Hunt D. L. Mechanobiology of Cardiomyocyte Development. J. Biomech. 2010, 43 (1), 93. 10.1016/j.jbiomech.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronaldson-Bouchard K.; Ma S. P.; Yeager K.; Chen T.; Song L.; Sirabella D.; Morikawa K.; Teles D.; Yazawa M.; Vunjak-Novakovic G. Advanced Maturation of Human Cardiac Tissue Grown from Pluripotent Stem Cells. Nature 2018, 556 (7700), 239–243. 10.1038/s41586-018-0016-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.; Lee S. J.; Cheng H.-J.; Yoo J. J.; Atala A. 3D Bioprinted Functional and Contractile Cardiac Tissue Constructs. Acta Biomater. 2018, 70, 48–56. 10.1016/j.actbio.2018.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahdy A. M.; Webster N. R. Perioperative Systemic Haemostatic Agents. Br. J. Anaesth. 2004, 93 (6), 842–858. 10.1093/bja/aeh227. [DOI] [PubMed] [Google Scholar]
- Kupcsik L.; Alini M.; Stoddart M. J. Epsilon-Aminocaproic Acid Is a Useful Fibrin Degradation Inhibitor for Cartilage Tissue Engineering. Tissue Eng., Part A 2009, 15 (8), 2309–2313. 10.1089/ten.tea.2008.0400. [DOI] [PubMed] [Google Scholar]
- Cholewinski E.; Dietrich M.; Flanagan T. C.; Schmitz-Rode T.; Jockenhoevel S. Tranexamic Acid—An Alternative to Aprotinin in Fibrin-Based Cardiovascular Tissue Engineering. Tissue Eng., Part A 2009, 15 (11), 3645–3653. 10.1089/ten.tea.2009.0235. [DOI] [PubMed] [Google Scholar]
- Tiburcy M.; Hudson J. E.; Balfanz P.; Schlick S.; Meyer T.; Chang Liao M.-L.; Levent E.; Raad F.; Zeidler S.; Wingender E.; et al. Defined Engineered Human Myocardium With Advanced Maturation for Applications in Heart Failure Modeling and Repair. Circulation 2017, 135 (19), 1832–1847. 10.1161/CIRCULATIONAHA.116.024145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camelliti P.; Borg T. K.; Kohl P. Structural and Functional Characterisation of Cardiac Fibroblasts. Cardiovasc. Res. 2005, 65 (1), 40–51. 10.1016/j.cardiores.2004.08.020. [DOI] [PubMed] [Google Scholar]
- Fan D.; Takawale A.; Lee J.; Kassiri Z. Cardiac Fibroblasts, Fibrosis and Extracellular Matrix Remodeling in Heart Disease. Fibrog. Tissue Repair 2012, 5 (1), 15. 10.1186/1755-1536-5-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinde A. V.; Frangogiannis N. G. Fibroblasts in Myocardial Infarction: A Role in Inflammation and Repair. J. Mol. Cell. Cardiol. 2014, 70, 74–82. 10.1016/j.yjmcc.2013.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bujak M.; Frangogiannis N. G. The Role of TGF-β Signaling in Myocardial Infarction and Cardiac Remodeling. Cardiovasc. Res. 2007, 74 (2), 184–195. 10.1016/j.cardiores.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago J.-J.; Dangerfield A. L.; Rattan S. G.; Bathe K. L.; Cunnington R. H.; Raizman J. E.; Bedosky K. M.; Freed D. H.; Kardami E.; Dixon I. M. C. Cardiac Fibroblast to Myofibroblast Differentiation in Vivo and in Vitro: Expression of Focal Adhesion Components in Neonatal and Adult Rat Ventricular Myofibroblasts. Dev. Dyn. 2010, 239 (6), 1573–1584. 10.1002/dvdy.22280. [DOI] [PubMed] [Google Scholar]
- Willems I. E.; Havenith M. G.; De Mey J. G.; Daemen M. J. The Alpha-Smooth Muscle Actin-Positive Cells in Healing Human Myocardial Scars. Am. J. Pathol. 1994, 145 (4), 868–875. [PMC free article] [PubMed] [Google Scholar]
- Kreutziger K. L.; Muskheli V.; Johnson P.; Braun K.; Wight T. N.; Murry C. E. Developing Vasculature and Stroma in Engineered Human Myocardium. Tissue Eng., Part A 2011, 17 (9–10), 1219–1228. 10.1089/ten.tea.2010.0557. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.