Abstract
Background
G protein-coupled receptor 35 (GPR35) is an orphan receptor and is vastly expressed in immune cells and gastrointestinal cells, suggesting the potential physiological importance of GPR35 in these cells. Here, we tested the hypothesis that the lack of GPR35 expression in the colon mucosa exacerbates the severity of dextran sulfate sodium (DSS)-induced experimental colitis in mice.
Methods
Colitis was induced in GPR35 wild-type (GPR35+/+) and GPR35 knockout (GPR35−/−) mice through the administration of DSS in drinking water for 5 days followed by regular facility water for 1 day. Induction of colitis was evaluated by measuring relative body weight loss, clinical illness scores, and morphological changes in the colon. Abolition of Gpr35 gene expression in the colon mucosa of GPR35−/− mice was confirmed by quantitative real-time PCR (qPCR). Gene expressions of inflammatory and tissue remodeling cytokines were detected by qPCR. Human colorectal epithelial Caco cells were transfected with siRNA against GPR35 before treated with 1% DSS in vitro. Protein expressions were measured using Western blot.
Results
GPR35−/− mice receiving DSS showed a significantly worsened colitis disease with profound loss of body weight and a considerable amount of severe clinical illness compared to GPR35+/+ mice that received DSS. The histology of colon sections from GPR35−/− mice showed extensive pathological changes including submucosal edema, diffuse ulcerations, and evidence of complete loss of crypts compared to wild-type mice. The mean histopathological score was significantly higher in GPR35−/− mice as compared to GPR35+/+ mice. The qPCR data revealed significant expression of pro-inflammatory and tissue remodeling cytokines in GPR35−/− colon mucosa, including IL-1β, CXCL1, CXCL2, CCL2, HMGB1, TGFβ1, TGFβ3, MMP1/9/12. The protein expressions of Zonula occludens-1, E-cadherin, Claudin1 were decreased upon knocking down GPR35 with or without 1% DSS treatment.
Conclusions
Our experimental data suggest that lack of GPR35 resulted in worsened disease outcome in DSS-induced experimental colitis, indicating that GPR35 could play a crucial role in protecting from colonic inflammation and serve as a therapeutic target.
Keywords: G protein-coupled receptor 35, Inflammatory bowel disease, Colitis, Dextran sulfate sodium, Colon inflammation
Introduction
Inflammatory bowel disease (IBD) is an intricate multifactorial, chronic, and relapsing disease that involves inflammation of the gastrointestinal tract [1, 2]. It comprises ulcerative colitis (UC) and Crohn’s disease (CD) triggered through various genetic and environmental elements [3–5]. According to Crohn’s and Colitis Foundation of America (CCFA) report, about 1.6 million Americans are afflicted with IBD. Furthermore, IBD is commonly recognized as one of the leading causative risk factors for the progression of colorectal cancer mediated through chronic intestinal inflammation [6, 7]. Despite significant advances in patient care, clinical data showed low remission rates at best of 40%. There is a compelling need to understand molecular mechanisms of IBD in order to identify novel therapeutic targets and effective treatment strategies.
G protein-coupled receptor 35 (GPR35) is an “orphan” receptor identified in 1998 [8] and has gained significant attention as a vital therapeutic task largely due to its close pathophysiological association with a plethora of diseases [9]. Nonetheless, the limitation in pharmacological approach and the lack of known endogenous ligands have led to a poor understanding of the therapeutic potential targeting this receptor. Recently, several endogenous ligands have been discovered stimulating GPR35, but have not been recognized as pivotal molecules largely due to deficiency of biological specificity [10, 11]. The functionality of GPR35 remains largely unknown. Several studies have shown that GPR35 is highly expressed in colon, spleen, urinary system, liver, thymus, and immune cells [11–13]. By in situ hybridization, GPR35-specific signaling has been revealed in the epithelial cells located in the crypts and villi, but not in lamina propria [11]. This group has also found that GPR35 is expressed in peripheral leukocytes, monocytes, T cells, neutrophils, dendritic cells, and eosinophils. Its ligands such as kynurenic acid and CXCL17 can also be detected in the gastrointestinal tract. Because significantly higher expression levels of GPR35 have been found in the gastrointestinal (GI) tract [14], it is speculated that GPR35 may play a substantial role in gut homeostasis. Interestingly, GPR35 has been recognized as a crucial element facilitating chronic inflammatory conditions in the intestinal region, which in turn leads to IBD. Two earlier investigations used genome-wide association study (GWAS) to identify a Gpr35 single nucleotide polymorphism (SNP) related to the early onset of ulcerative colitis [15, 16]. Further GWAS studies have shown that GPR35 is also associated with other diseases such as type 2 diabetes [17] and coronary artery disease [18]. However, the exact role of GPR35 playing in the onset of ulcerative colitis is not clear.
In our current study, we investigated whether GPR35 expression in the colon mucosa plays any key role in protecting the colon from inflammation. We tested the hypothesis that deletion of GPR35 deteriorates the severity of dextran sulfate sodium (DSS)-induced colitis disease. To our knowledge, this is the first in vivo investigation in animals to observe the impact of GPR35 on the onset of experimental colitis using GPR35 knockout (KO) mice. Histological analyses have been used to evaluate the severity of colon injuries. Several inflammatory and tissue remodeling cytokines have been detected as the first attempt to explore the potentially important role of GPR35 under inflammatory conditions.
Materials and Methods
Mice and Colitis Induction
Specific pathogen-free male C57BL6 mice (6–8 weeks of age) were procured from The Jackson Laboratory (Bar Harbor, ME). GPR35 knockout mice (GPR35−/−, on the C57BL6 background) were purchased from Wellcome Trust Sanger Institute (Cambridge, England) [19, 20]. GPR35−/− mice were bred with C57BL/6 to generate GPR35−/− mice and GPR35+/+ mice. All mice were bred and maintained in the Eugene Applebaum College of Pharmacy and Health Science Animal Care Facility. Colitis was induced by the administration of 4% (w/v) dextran sulfate sodium (DSS, 36,000–50,000 mol.wt; MP Biomedicals, Irvine, CA) in drinking water for 5 consecutive days in GPR35−/− mice and GPR35+/+ mice, followed by the administration of regular facility water for 1 day [21, 22]. On day 7, all experimental mice were euthanized, followed by excision of their colons, measurement, and preparation for histology. All animal procedures were performed according to Wayne State University Institutional Animal Care and Use Committee (IACUC) guidelines.
Assessment of Disease Severity
Daily clinical illness scoring was evaluated based on a distinctive combination of body weight loss, stool consistency, and presence or absence of blood in stool [21–23]. The scoring details are shown in Table 1 as previously described [23]. Animals exhibiting clinical illness noticeable by weight loss of 20% combined with diarrhea and gross bleeding were given a maximal score of 10 (Weight loss > 20%; stool consistency, diarrhea; and presence of blood, occult+/Gross) and were promptly euthanized.
Table 1.
Clinical illness scores
| Score | Weight loss (%) | Stool consistency | Blood in stool |
|---|---|---|---|
| 0 | None | Normal | None |
| 1 | None | Normal | Occult+ |
| 2 | 1–5 | Normal | None |
| 3 | 1–5 | Soft | None |
| 4 | 1–5 | Soft | Occult+ |
| 5 | 1–5 | Diarrhea | Occult+/Gross |
| 6 | 5–10 | Soft | Occult+/Gross |
| 7 | 5–10 | Diarrhea | Occult+/Gross |
| 8 | 10–20 | Soft | Occult+/Gross |
| 9 | 10–20 | Diarrhea | Occult+/Gross |
| 10 | > 20 | Diarrhea | Occult+/Gross |
Histopathology and Scoring
The “Swiss rolls” method was used to prepare the sections of the mouse colons for histology (exclusive of cecum) [21, 22]. Briefly, a longitudinal excision was made to cut open the whole colon along the mesenteric axis into two full-length colon halves. The colon halves were prepared into a Swiss roll and fixed with 10% buffered formalin. The fixed colon samples were further processed to prepare paraffin-embedded slides (5-μm-thick sections), and hematoxylin and eosin (H&E) were used to stain sections. The inflammatory features were evaluated by four simply detectable pathological measures such as cellular infiltration (0–5), deterioration of crypt architecture (crypt damage, 0–4), extent of mucosal ulceration (0–3), and absence or presence of submucosal edema (0 and 1) as described previously [21, 22]. The evaluation was performed by two blinded examiners.
Congo Red Staining to Differentiate Eosinophils and Neutrophils
Differentiation between neutrophils and eosinophil granulocytes was performed on paraffin-embedded colon slides (5-μm-thick sections) using Congo red staining kit (IHC World, MD, USA). Neutrophils and eosinophils were enumerated in the submucosa and mucosa at the different stages of deteriorating crypt architecture (crypt damage, 0–4), with cells in at least 5 different field areas of each crypt damage score counted within high-power fields (400x) [22]. The damage scores were determined by two pathologists in a blinded way.
Quantitative Reverse Transcriptase PCR (qPCR) Analyses
Total RNA was isolated from mouse colon mucosa using RNeasy Mini kit (Qiagen, MD, USA). Total RNA was reverse-transcribed to a single-stranded cDNA using the high-capacity cDNA Reverse Transcription kit (Applied Biosystems, Life Technologies, CA, USA). The qPCR was carried out by PrimeTime qPCR Assay kit (Integrated DNA Technologies, Iowa, USA). Mouse GPR35 primer and probe sequences are 5’-ATC ACA GGT AAA CTC TCA GAC ACC AAC T-3′ (forward), 5′-CTT GAA CGC TTC CTG GAA CTC T-3′ (reverse) and 5′-TGG ATG CCA TCT GTT ACT ACT ACA TGG CCA-3′ (probe) [11]. The β-actin primer and probe sequences were 5′-CAT CTT GGC CTC ACT GTC CAC-3′ (forward), 5′-GGG CCG GAC TCA TCG TAC T-3′(reverse), 5′-TGC TTG CTG ATC CAC ATC TGC TGG A-3′ (probe). PCR products were separated in agarose gel to confirm the absence of GPR35 DNA in the knockout mice. For gene expressions of cytokines, inflammatory and remodeling factors, qPCR was carried out by using specific real-time PCR primers synthesized by Integrated DNA Technologies. The primer sequences are shown in Table 2. Amplification and detection of specific products were performed with the ABI PRISM 7500 Sequence Detection System using Fast SYBR™ Green Master Mix (Thermo Fisher Scientific). Fluorescent signals were normalized to an internal reference, and the threshold cycle (Ct) was set within the exponential phase of the PCR. The target gene PCR Ct value was normalized by subtracting the β-actin Ct value, which gave the ΔCt value. The relative expression levels of each target between groups were then calculated using the following equation: relative gene expression = 2−(ΔCt_treatments-ΔCt_controls).
Table 2.
Primer sequences for qPCRs
| Gene name | Forward primer | Reverse primer |
|---|---|---|
| Tnf-α | 5′-CCAACGCCCTCCTGGCCAAC-3′ | 5′-GAGCACGTAGTCGGGGCAGC-3′ |
| Il-1β | 5′-GGAGAACCAAGCAACGACAAAATA-3′ | 5′-TGGGGAACTCTGCAGACTCAAAC-3′ |
| Il-6 | 5′-CCCAATTTCCAATGCTCTCCT-3′ | 5′-TGAATTGGATGGTCTTGGTCC-3′ |
| Cxcl1 | 5′-AGACCATGGCTGGGATTCAC-3′ | 5′-CGCGACCATTCTTGAGTGTG-3′ |
| Cxcl2 | 5′-CTGCATCTGCCCTAAGGTCT-3′ | 5′-AGTGCTTGAGGTGGTTGTGG-3′ |
| Ccl2 | 5′-CTGCATCTGCCCTAAGGTCT-3′ | 5′-AGTGCTTGAGGTGGTTGTGG-3′ |
| Tgfβ1 | 5′-GATGGATCACACGTCCTTCCC-3′ | GAAGTGCGTCTGAACTCTTGT-3′ |
| Tgfβ2 | 5′-GTGGCCGAGCAGCGGATTGA-3′ | TGCACAGCGTCTGTCACGTCG-3′ |
| Tgfβ3 | 5′-CAACCCCAGCTCCAAGCGCA-3′ | GCCATTCAGCGGTGCCCCTT-3′ |
| Mmp1 | 5′-TGATGAGACGTGGACCAACAG-3′ | 5′-GAATGAGTGAGCCCAAGGGA-3′ |
| Mmp2 | 5′-GCTGTATTCCCGACCGTTGA-3′ | TGGTCCGCGTAAAGTATGGG-3′ |
| Mmp9 | 5′-TGAAAACCTCCAACCTCACGG-3′ | 5′-AGAGACTGCTTCTCTCCCATCA-3′ |
| Mmp12 | 5′-TTGGATTATTGGAATGCTGC-3′ | 5′-GCACATTTTGATGAGGCAGA-3′ |
| Hmgb1 | 5′-CCGGATGCTTCTGTCAACTT-3′ | 5′-GGGCGGTACTCAGAACAGAA-3′ |
| Egf | 5′-TCCTGACCCCGAACTTCTCA-3′ | 5′-TTAACGTGTAGCCCTCAGCG-3′ |
| Pdgf | 5′-TCAAGGTGGCCAAAGTGGAG-3′ | 5′-CTGGGTTCAGGTTGGAGGTC-3′ |
| Fgf1 | 5′-GGGAGATCACAACCTTCGCA-3′ | 5′-GGATCCTCAAGAAGTGGCCC-3′ |
| Fgf2 | 5′-GGCTGCTGGCTTCTAAGTGT-3′ | 5′-AGTGCCACATACCAACTGGAG-3′ |
Human Colorectal Epithelial Cell Culture In Vitro and Treatments
Human colorectal epithelial cells Caco2 cells were obtained from American Type Culture Collection (ATCC) (Manassas, VA) and maintained in Minimum Essential Medium (MEM) (Corning) supplemented with 10% heat-inactivated fetal bovine serum (FBS, Sigma) and penicillin/streptomycin (Corning) at 37 °C with 5% CO2. When Caco2 cells reached 80% confluence under the microscope, 0.25% trypsin was used for cell digestion and passage. Caco2 cells were transfected with scrambled siRNA (small interfering RNA) or GPR35 siRNA for 48 h, followed by 1% DSS treatment for 4 h.
Small Interfering RNA Transfection
A scrambled siRNA and GPR35 siRNA used in the experiment were predesigned and synthesized by MilliporeSigma. One day before transfection, Caco2 cells were seeded into a 6-well plate till the cell confluence reached 80%. Transfection was performed using Mission siRNA transfection reagent (Sigma) according to the manufacturer’s instructions.
Western Blot Analyses
Forty-eight hours after transfection, Caco2 cells were treated with 1% DSS for 4 h and were lysed for western blot. Total cell lysate was heated at 42 °C for 15 min for denaturation. Electrophoresis was performed with 7.5% polyacrylamide gel, and proteins were transferred onto the polyvinylidene difluoride (PVDF) membrane. The membrane was blocked for 1 h in 5% goat serum before the addition of primary antibody for overnight incubation at 4 °C. The following day, the membrane was incubating with secondary antibodies for 1 h. The ECL chemiluminescent kit was used for development, and the images were taken in Amersham imager 600 system (GE health care). Antibodies against Zonula occludens-1 (ZO-1, #8193), β-catenin (#8480), Caludin1 (#13255), and E-cadherin (#3195) were from Cell Signaling Technology. Antibody against GPR35 was from Novus (NBP2–24640), and antibody against GAPDH was from Thermo Fisher scientific (AM4300). At least three biological repeats were obtained.
Statistical Analyses
Data were expressed as Mean ± SEM. The unpaired t test and one-way ANOVA were used to evaluate comparisons between all animal groups using GraphPad Prism software (version 7.0). P values below 0.05 were considered statistically significant (*p < 0.05; #p < 0.05).
Results
GPR35−/− Mice Demonstrated Weight Loss and Exhibited Increased Clinical Illness Scores in DSS-Induced Colitis
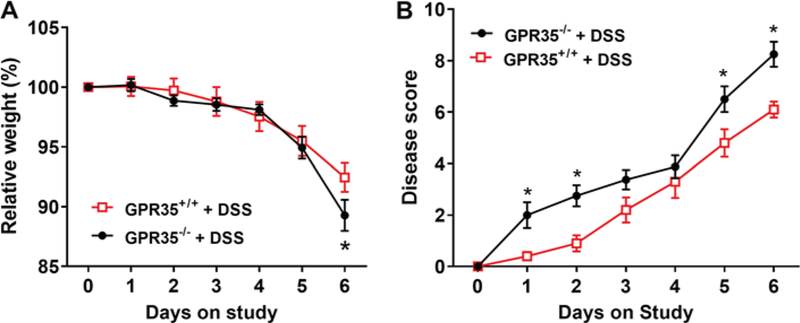
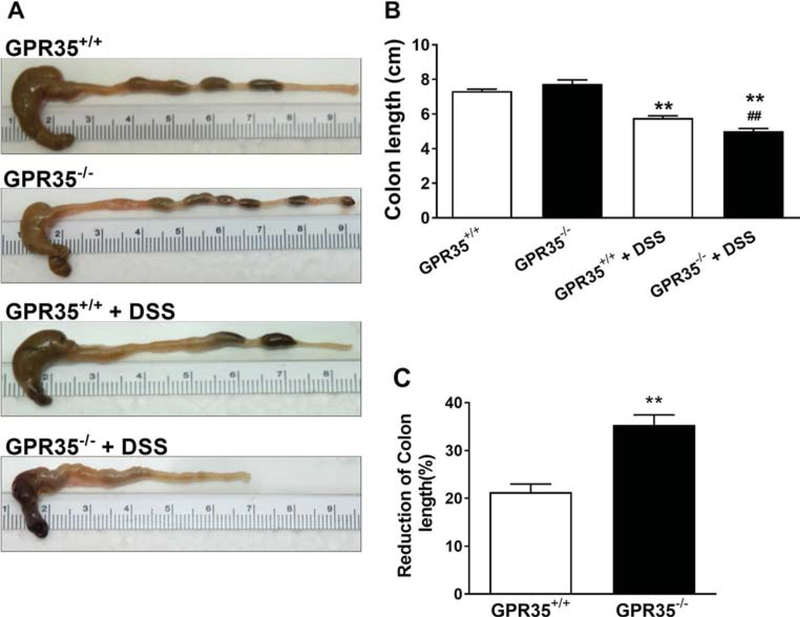
Real-time PCR and DNA electrophoresis were performed in the colon tissues to confirm the deletion of GPR35 gene in GPR35−/− mice using GPR35+/+ mice as controls (Supplemental Figure 1). The expression of GPR35 in the mucosal part of the colon was not significantly different between GPR35+/+ mice administered DSS and GPR35+/+ with control treatment. Administration of DSS through drinking water resulted in a decrease in body weight in both strains of mice (Fig. 1a). A significant difference in body weight loss across the strains was observed only on day 6 (*p < 0.05). On day 6, the mean weight loss between GPR35+/+ and GPR35−/− mice was 7 and 11%, respectively. Interestingly, GPR35−/− mice showed signs of soft stool and occult blood at an early stage (on day 1 or 2), and 75% of these mice developed bloody diarrhea after day 4. However, the GPR35+/+ mice showed soft stools with occult blood, but only 20% of these mice exhibited signs of diarrhea after day 4. During the experimental period, the severity of daily clinical changes was scored in each mouse based on weight loss, stool consistency, and the presence of occult or gross blood. The daily clinical illness scores of the GPR35−/− mice were found to be augmented significantly on days 1, 2, 5, and 6 as compared with the GPR35+/+ mice (Fig. 1b). The colons of GPR35−/− mice showed significant shortening compared to GPR35+/+ mice upon DSS administration (Fig. 2a, b). The average percentage of colon length reduction in DSS-administered GPR35−/− and GPR35+/+ mice was 35 and 21%, respectively. The increased clinical illness scores and reduction of colon length in GPR35−/− mice support the hypothesis that the deletion of GPR35 impairs colon functions in this model of colitis. There was no significant change in body weight (data not shown) or colon length (Fig. 2a, b) in control treatment groups of either strain (GPR35−/− and GPR35+/+ with control treatment).
Fig. 1.
Pattern of daily body weight changes and daily clinical illness scores of GPR35+/+ and GPR35−/− mice receiving DSS. Colitis was induced by administration of 4% (w/v) DSS in drinking water for 5 consecutive days in GPR35−/− mice and GPR35+/+ mice, followed by administration of regular facility water for 1 day as previously described [21, 22]. a The percentage of daily animal body weight changes during the experimental period. A significant difference in body weight loss across the strains was observed only on day 6 (*p < 0.05 vs. GPR35+/+ + DSS). b Summary of daily clinical illness scores during the experimental period. The severity of daily clinical changes was scored in each mouse based on weight loss, stool consistency, and presence of occult or gross blood. After DSS administration, the daily clinical illness scores of the GPR35−/− mice were augmented significantly on days 1, 2, 5, and 6 as compared with the GPR35+/+ mice. N = 10 (GPR35+/+ + DSS), n = 8 (GPR35−/− + DSS); *p < 0.05 versus GPR35+/+ + DSS
Fig. 2.
Macroscopic observations of colon length and reduction of colon length in GPR35+/+ and GPR35−/− mice administered DSS. a Representative macroscopic observations of colon length. b Bar graph showing colon length of GPR35+/+ alone, GPR35−/− alone, GPR35+/+ + DSS, and GPR35−/− + DSS groups. After administration with 5 days of DSS and 1 day of water, the colons of GPR35−/− mice showed significant shortening compared to those of GPR35+/+ mice. **p < 0.01 versus GPR35+/+ alone; ##p < 0.01 versus GPR35+/+ + DSS. c Percentage of colon length reduction in GPR35+/+ and GPR35−/− mice after DSS administration. A significant reduction of colon length was observed in GPR35−/− compared to GPR35+/+ mice upon DSS administration. N = 10 (GPR35+/+ alone), n = 6 (GPR35−/− alone), n = 10 (GPR35+/+ + DSS), n = 8 (GPR35−/− + DSS). *p < 0.05 versus GPR35+/+ alone; #p < 0.05 versus GPR35+/+ + DSS
GPR35−/− Mice Exhibited Extensive Ulcers and Colonic Inflammation Compared with GPR35+/+ Mice upon DSS Administration
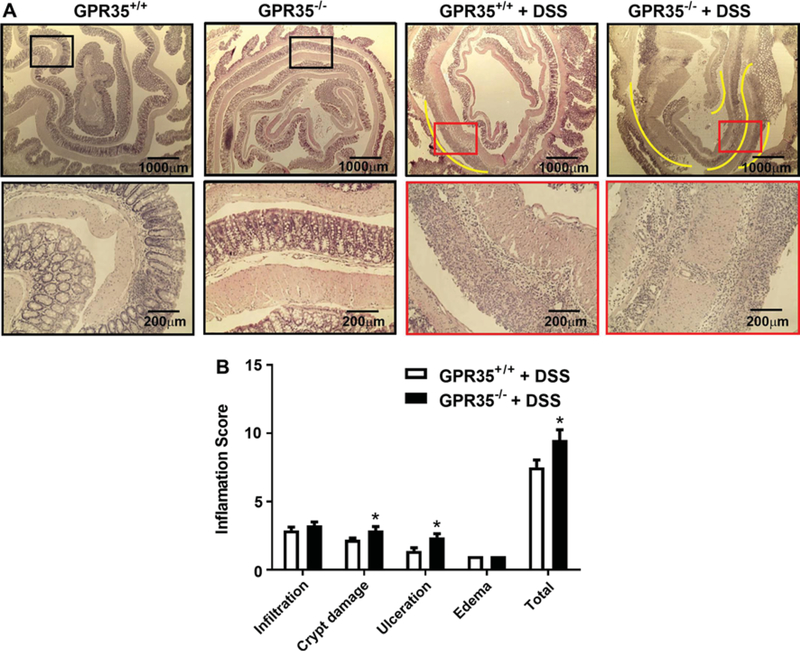
Next, we investigated if the GPR35−/− mice had exacerbated colon tissue injury in DSS-induced colitis. The colonic histology of GPR35−/− and GPR35+/+ (without DSS treatment) groups showed normal architecture of the crypt with no signs of inflammation and/or ulceration (Fig. 3a). However, the histology sections of DSS-administered GPR35−/− mice colons showed extensive pathological changes (Fig. 3a) including submucosal edema, diffuse ulcerations (indicated by yellow lines), with most of the ulcerated areas showing signs of extensive inflammatory cell infiltration (up to mucosal part of the colon). Further, numerous places in the mid-colon area showed evidence of complete loss of crypts (damage score up to 4). With DSS administration, the mean histopathological score was significantly higher in GPR35−/− mice as compared to wild-type mice (Fig. 3b). In GPR35+/+ mice with DSS administration, the colons showed small ulcers (yellow line), with inflammatory cell infiltration only seen in the ulcerated parts. Most of the areas showed an intact structure of crypt in GPR35+/+ colons. Compared to GPR35−/− mice, the average histopathology score was significantly lower in GPR35+/+ mice (Fig. 3b).
Fig. 3.
Colon histology and inflammation score of GPR35+/+ and GPR35−/− mice receiving DSS. a The colon histology sections of GPR35+/+ alone, GPR35−/− alone, GPR35+/+ + DSS, and GPR35−/− + DSS groups. After DSS or vehicle administration, colon sections were stained with hematoxylin and eosin. The colon histology sections of GPR35−/− mice revealed severe pathology with observation of submucosal edema and huge ulcerations (yellow lines) with extensive inflammatory cell infiltration. Further, numerous places in the mid-colon area in GPR35−/− mice showed evidence of complete loss of crypts (scored 4). Black frames indicate normal mucosal structure, and red frames indicate an ulceration area in the mucosal structure. The frame indicates the region magnified in the bottom panel. b Inflammation scores of GPR35+/+ + DSS and GPR35−/− + DSS. The total histopathological score was significantly higher in GPR35−/− + DSS as compared to GPR35+/+ + DSS. N (GPR35+/+ + DSS) = 10, n (GPR35−/− + DSS) = 8, *p < 0.05 versus GPR35+/+ + DSS
Deletion of GPR35 Did Not Alter Neutrophil or Eosinophil Infiltration into Colon Tissues
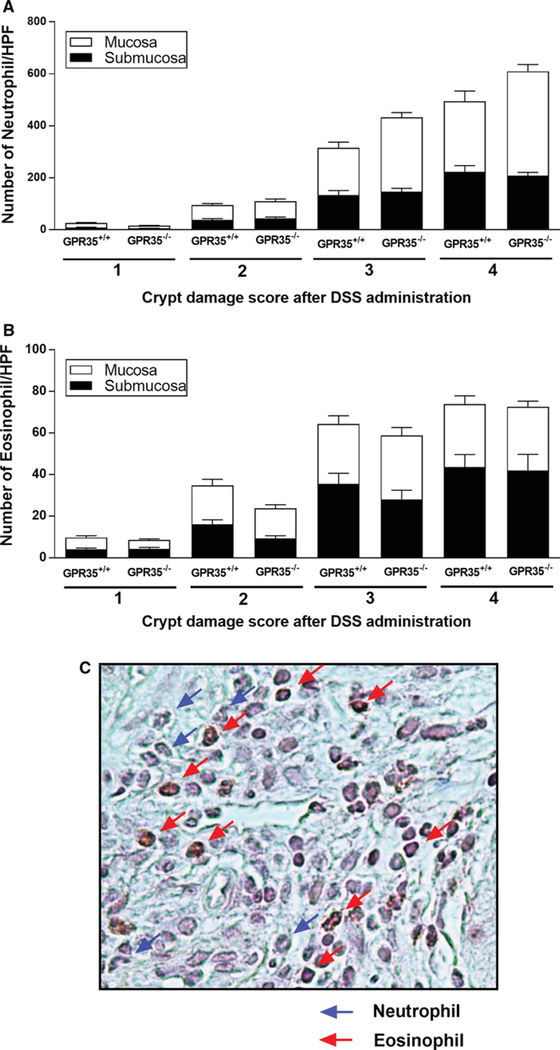
Congo red staining was performed in colon sections to differentiate between neutrophils and eosinophil granulocytes. (Representative picture of colon mucosa from DSS-administered GPR35+/+ mice is shown in Fig. 4c.) As shown in Fig. 4a, neutrophil infiltration into mucosa and submucosa areas was closely associated with the degree of crypt damage in the DSS-induced colitis in both GPR35−/− and GPR35+/+ mice, but there was no significant difference between GPR35−/− and GPR35+/+ mice with the same crypt damage score. However, the GPR35−/− mice did show a trend of increased neutrophil infiltration into the mucosa at crypt damage degree of 3 and 4, as compared with GPR35+/+ mice (Fig. 4a). Similarly, the eosinophil infiltration into mucosa and submucosa areas was increased at crypt damage degrees of 2 and above but did not differ significantly between GPR35−/− and GPR35+/+ mice (Fig. 4b).
Fig. 4.
Neutrophil and eosinophil numbers in the colonic submucosa and mucosa in the colons of GPR35+/+ and GPR35−/− mice administered DSS. Multiple high-power fields (HPF, 400 ×) of regions representing the different scores of crypt damage in the mucosa and submucosa were pooled from each mouse into a single score, and the scores of different mice were pooled to derive a mean ± SEM. The numbers of neutrophils or eosinophils were counted in each area of the different scores of crypt damage. a The numbers of neutrophils in the submucosa and mucosa in each score category of crypt damage. b The numbers of eosinophils in the submucosa and mucosa in each score category of crypt damage. In both graphs, no statistical significance has been found between GPR35+/+ and GPR35−/− mice administered with DSS in each score category of crypt damage. c Representative high-power photomicrograph of Congo redstained section in DSS-inflamed GPR35+/+ mouse colon mucosa. Red staining distinguishes eosinophils (red arrows) from unstained neutrophils (blue arrows)
Upregulation of Pro-inflammatory Factors by GPR35 Deletion in DSS-Induced Colitis
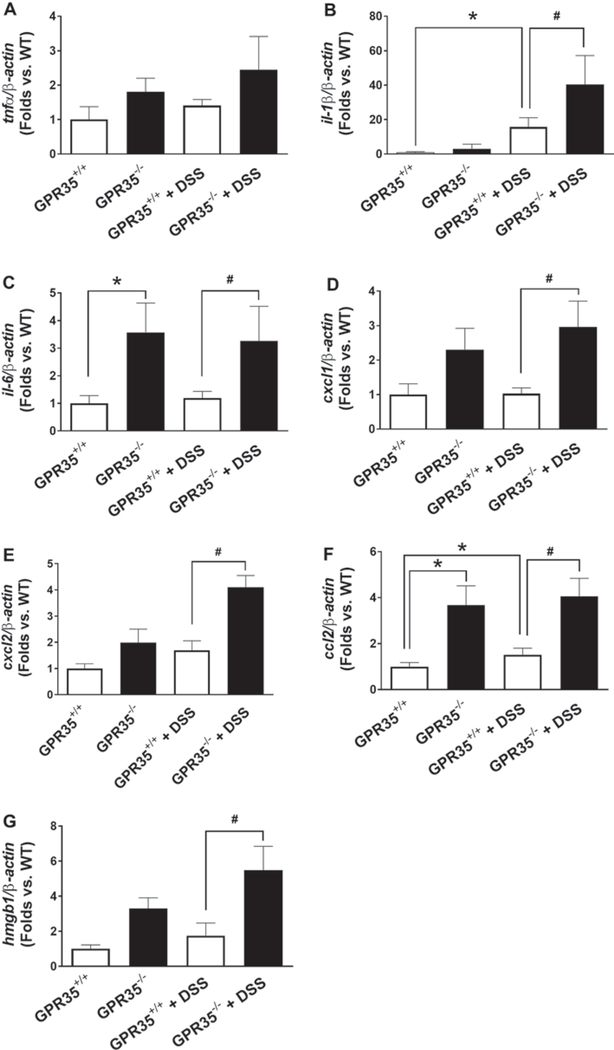
In order to compare the secretion of the inflammatory cytokines in GPR35−/− mice, seven different cytokines in the mouse colon mucosa of GPR35−/− mice with or without DSS administration were examined by qPCR using colon mucosa from GPR35+/+ mice as controls. These cytokines include tumor necrosis factor (TNF-α), interleukin-1β (IL-1β), interleukin-6 (IL-6), C-X-C motif chemokine ligand 1 (CXCL1), C-X-C motif chemokine ligand 2 (CXCL2), C–C motif chemokine ligand 2 (CCL2), and high-mobility group box 1 (HMGB1) (Fig. 5). Upon DSS administration, GPR35−/− mice demonstrated significantly higher expression of IL-1β, CXCL1, CXCL2, CCL2, and HMGB1 at mRNA levels compared to GPR35+/+ mice (Fig. 5c–g). These data suggest an inhibitory role of GPR35 on inflammation under the setting of DSS-induced colitis.
Fig. 5.
Gene expressions of inflammatory factors by GPR35 deletion in DSS-induced colitis. Total RNA was extracted from colonic mucosa in GPR35−/− mice with or without DSS treatment, GPR35+/+ mice with or without DSS treatment. Expression levels of cytokines including tumor necrosis factor (TNF-α), interleukin-1β (IL-1β), interleukin-6 (IL-6), C-X-C motif chemokine ligand 1 (CXCL1), C-X-C motif chemokine ligand 2 (CXCL2), C–C motif chemokine ligand 2 (CCL2), and high-mobility group box 1 (HMGB1) were detected by qPCR, using β-actin as internal control. qPCR analyses of mRNA encoding TNF-α (a), IL-1β (b), IL-6 (c), CXCL1 (d), CXCL2 (e), CCL-2 (f), and HMGB1 (g) are shown in bar graph. N = 5 (GPR35+/+ alone), n = 3 (GPR35−/− alone), n = 5 (GPR35+/+ + DSS), n = 4 (GPR35−/− + DSS). *p < 0.05 versus GPR35+/+; #p < 0.05 versus GPR35+/+ + DSS
Upregulation of Tissue Remodeling Factors by GPR35 Deletion in DSS-Induced Colitis
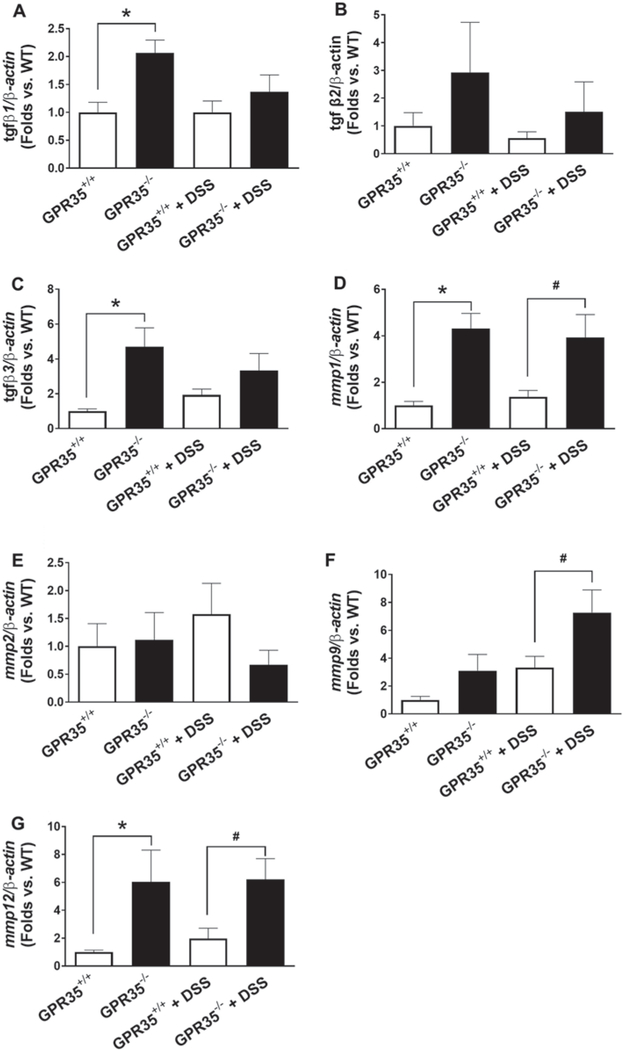
To further dissect the role of GPR35 in the regulation of tissue damage during DSS-induced colitis, seven tissue remodeling factors were examined by qPCR, including transforming growth factor β (TGFβ) 1, 2, and 3, and matrix metalloproteinase (MMP) 1, 2, 9, and 12. As shown in Fig. 6, DSS administration resulted in higher expression of MMP1, MMP9, and MMP12 in GRP35−/− mice as compared with GPR35+/+ mice. Meanwhile, most of these cytokines were also increased significantly in GPR35−/− mice compared to GPR35+/+ mice with control treatment. Among them, TGFβ1 (Fig. 6a), TGFβ3 (Fig. 6c), MMP1 (Fig. 6d), and MMP12 (Fig. 6g) are the most dramatically upregulated cytokines. TGFβ2 also demonstrated an increase in levels in GRP35−/− mice as compared with GPR35+/+ mice at both control and DSS treatment, although with no statistical significance (Fig. 6b).
Fig. 6.
Gene expressions of tissue remodeling factors by GPR35 deletion in DSS-induced colitis. Total RNA was extracted from colonic mucosa in GPR35−/− mice with or without DSS treatment, GPR35+/+ mice with or without DSS treatment. qPCR was performed using β-actin as internal control. Seven tissue remodeling factors in colon mucosa in GPR35−/− mice with or without DSS, comprising transforming growth factor β (TGFβ) 1, 2, and 3; matrix metalloproteinase (MMP) 1, 2, 9, and 12. The levels of TGFβ1 (a), TGFβ2 (b), TGFβ3 (c), MMP1 (d), MMP2 (e), MMP9 (f), and MMP12 (g) in colons from GPR35+/+ and GPR35−/− with DSS administration or control treatment are shown in bar graphs. N = 5 (GPR35+/+ alone), n = 3 (GPR35−/− alone), n = 5 (GPR35+/+ + DSS), n = 4 (GPR35−/− + DSS). *p < 0.05 versus GPR35+/+; #p < 0.05 versus GPR35+/+ + DSS
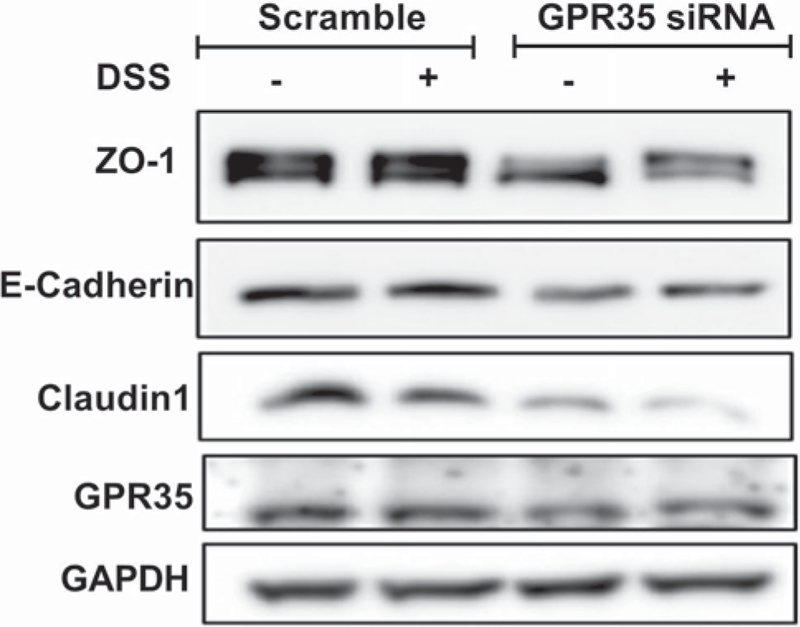
GPR35 Downregulation Decreases the Expression of ZO-1, E-cadherin, and Claudin1 in Human Colorectal Epithelial Cells In Vitro
In human colorectal epithelial Caco2 cells cultured in vitro, we knocked down GRP35 and examined the expression of cell–cell adhesion molecules such as ZO-1, E-cadherin, β-catenin, and Claudin1 using Western blot. We found that those molecules were downregulated in GPR35 knockdown colorectal epithelial cells (Fig. 7). When the cells were exposed to DSS (1%, 4 h), greater downregulation of ZO-1 and Claudin1 was observed in GPR35 knockdown cells than that in control cells.
Fig. 7.
GPR35 deficiency affects the expression of cell junction molecules. Colorectal epithelial cells (Caco2) were transfected with siRNA targeting GPR35 (GPR35 KD) or non-targeting siRNA which was used as control (control). Then the cells were treated with or without 1% DSS for 4 h (n = 3 biological repeats). The expression of ZO-1, E-cadherin, and Claudin1 was decreased in GPR35 deficiency cells. DSS treatment resulted in decrease in ZO-1, E-cadherin, and Claudin1, but the decreases in ZO-1 and Claudin1 were more severe in GPR35 knockdown cells
Discussion
The IBD has affected millions of people worldwide. Currently, the remission rate is still low considering all the advances in patient care. GPR35 has been implicated in inflammatory, neurological, and cardiovascular diseases, but the signaling and functions of GPR35 have remained unclear for decades. We tested whether GPR35 mutant mice are more prone to develop a DSS-induced experimental colitis. Our results suggested a lower body weight, diarrhea, and blood stool in GPR35−/− mice with DSS administration. The colon length was significantly reduced upon DSS administration. Detailed histological analyses in the colons of the GPR35−/− mice unveiled significantly destructed colon mucosa structure and a diffused ulcerative area. Further gene analyses suggested upregulation of a subset of proinflammatory cytokines and tissue remodeling factors. To our knowledge, this is the first study to examine the in vivo role of GPR35 in the regulation of tissue damage and inflammation in experimental IBD.
Recent findings from clinical studies have indicated GPR35 as a risk factor for chronic inflammatory disorders such as IBD and ulcerative colitis [15, 16], providing a strong rationale to look at the role of GPR35 in the pathogenesis and progression of IBD. Most recently, Tsukahara et al. has reported that administration of GPR35 agonist could attenuate the DSS-induced colitis in mice. Moreover, they have shown that GPR35 agonists can promote intestinal epithelial cell migration in vitro which probably contribute to the injury repair of colitis [24]. In the present study, we examined the onset and progression of DSS-induced experimental colitis in the genetically modified GPR35−/− mice. We found, without DSS administration, the GPR35−/− mice showed no apparent colonic alterations from the GPR35+/+ mice. There was no apparent difference in crypt cells and no signs of stool hydration or diarrhea (Fig. 2). However, following 5 days of DSS treatment and 1 day of regular water administration, the GPR35−/− mice became severely ill as demonstrated by more weight loss, higher disease scores (Fig. 1), and significantly shortened colon length compared with GPR35+/+ mice with DSS administration (Fig. 2). Histological analysis revealed that there was a major loss of the colon epithelial layer and diffused and large ulcerative areas in GPR35−/− mice, while the GPR35+/+ mice had comparatively less epithelial layer damage and scattered ulcerative areas (Fig. 3). These data clearly support the notion that the deletion of GPR35 exacerbates colon epithelial damage and ulcerations in DSS-induced colitis.
One interesting finding from our study is the disassociation between the severe tissue damage, elevation of pro-inflammatory and remodeling cytokine, and moderate inflammatory cell infiltration. The inflammation in mucosa and submucosa areas was increased by DSS administration in both GPR35+/+ mice and GPR35−/− mice, but there was no significant difference between the two mouse strains (Fig. 4). Current findings were observed in DSS-induced colitis animal model, in which the adaptive immune system is not required for development of colitis in this model, as colonic inflammation can occur in the absence of inflammatory cells [25]. Furthermore, Congo red staining suggested that neutrophil and eosinophil infiltration into mucosa and submucosa areas was also comparable between GPR35+/+ and GPR35−/− mice (Fig. 4), suggesting that the numbers of inflammatory cells recruited to the colon were not altered by GPR35 deletion. On the other hand, the qPCR data showed that in untreated GPR35−/− mice, the pro-inflammatory cytokines such as IL-6, CXCL1, CCL2, HMGB1 (Fig. 5), as well as tissue remodeling factors such as TGFβ1, TGFβ3, MMP1/12 (Fig. 6), were much higher compared with untreated GPR35+/+ mice. The inhibitory effects of GPR35 on inflammatory responses in our studies are in line with previous reports [11, 26, 27]. IL-6 and CCL2 are two pro-inflammatory factors that are most dramatically elevated in GPR35−/− colons. It has been reported that these two cytokines induce each other and collaborate to promote the polarization of monocytes into M2 macrophages [28]. MMP1, MMP9, and MMP12 were found elevated in GPR35−/− mice, especially after DSS administration. MMPs are potent factors that promote inflammation by cleavage of protein substrates [29]. The cleaved products can often result in gain of function, such as TNF-α and IL-1β, but they can also lead to degradation or conversion of the active chemokine to an antagonist, such as CXCL12 and CCL2 (also known as MCP-1) [30, 31]. With high levels of pro-inflammatory cytokines and remodeling cytokines in the GPR35−/− colons, there was no significant difference in inflammatory cell infiltration into colon tissues between GPR35+/+ and GPR35−/− mice. It is possible that GPR35 also facilitates inflammatory cell trafficking in mucosa area, but the underlying mechanisms are not clear and may require more investigations in future. On the other hand, GPR35 ablation may also attenuate the recovery of epithelial injuries caused by DSS administration. The overall effect of global GPR35−/− is a combination of the inhibited inflammatory response and attenuated epithelial cell recovery from DSS-induced injuries. Further mechanistic studies are needed to explain why deletion of GPR35 can affect epithelial cell recovery and mucosal barrier repair in addition to its regulatory role in inflammatory response.
In this animal model, we established that GPR35 is a potentially important target for intervention in IBD. However, DSS administration per se did not impair GPR35 mRNA levels (Supplemental Figure 1A), suggesting that DSS-induced changes in GPR35 actions either occur at post-translational levels or through modulating its ligands. Recent discoveries have characterized several endogenous ligands such as kynurenic acid (KYNA), zaprinast, and lysophosphatidic acid (LPA) [10, 11, 13]. Using a high throughput approach, Wang et al. have revealed that KYNA can serve as a putative ligand for human and murine GPR35 [11]. However, the physiological roles of KYNA in activating human GPR35 are not clear since human GPR35 requires a much higher concentration of KYNA to achieve modest activation, compared with rat GPR35 [32]. Oka et al. reported that LPA acted as a ligand for GPR35 but it seems that LPA acts unselectively toward GPR35 and in a manner distinct from other GPR35 ligands [10]. Therefore, there is a need to clarify the relevance of these reported effects of ligands at GPR35 and their significant species selectivity in future studies. It has been reported that GPR35 is expressed in CXCL17-responsive monocytes and in the THP-1 monocytoid cell line and may function as a novel chemokine receptor of the mucosal chemokine CXCL17 [33]. Thus, GPR35 function in macrophages/immune cells is pro-inflammatory, and there-fore, its loss in macrophages would be expected to reduce inflammation and promote intestinal regeneration. How-ever, the loss of pro-growth functions of GPR35 in intestinal epithelial cells (IECs) could contribute to attenuated GI injury recovery and aggravated inflammatory response. Therefore, the overall IBD response in global GPR35-deficient mice may be caused by the combinatorial protective and destructive effects of multiple cell types including IECs, macrophages, and other cell types. This is very likely the reason for the observed modest effect of GPR35 deficiency in our present studies. In the future, analysis of cell-type-specific KO of the Gpr35 gene will be required to clearly define the cell autonomous functions of Gpr35 in intestinal lesion development and IBD progression.
In summary, our study provides the first in vivo evidence for an essential role of GPR35 in the protection against DSS-induced experimental colitis. Deletion of GPR35 exacerbates the disease progression, which is associated with an outburst of a subset of inflammatory and tissue remodeling cytokines. We believe that GPR35 is a promising candidate for therapeutic intervention given its involvement in a variety of physiological and pathological processes. As more pharmacological tools are being developed and made available to the scientific community, we will be able to further probe for GPR35 actions in physiological and disease models. Eventually, our future studies will aim to translate these findings from a mouse model of experimental colitis into clinical settings, in order to truly assess the therapeutic value of GPR35.
Supplementary Material
Acknowledgments
We thank the Wellcome Trust Sanger Institute Mouse Genetics Project (Sanger MGP) and its funders for providing the mutant mouse line (Allele: GPR35) and INFRAFRONTIER/EMMA (www.infrafrontier.eu). Funding information may be found at www. sanger.ac.uk/mouseportal and associated primary phenotypic information at www.mousephenotype.org.
Funding This work was supported in part by American Heart Association Scientist Development Grant 13SDG16930098 (to J.M.W.), NIH/ NIDDK R01 DK109036 (To J.M.W.), Wayne State University Faculty Start-up Fund (To J.M.W.), and NIH/NHLBI R01 HL128647 (To C.L.).
Abbreviations
- cDNA
Complementary deoxyribonucleic acid
- CXCL17
C-X-C motif chemokine 17
- DSS
Dextran sulfate sodium
- GPR35
G protein-coupled receptor 35
- HEK 293 cells
Human embryonic kidney cells 293
- KO
Knockout
- KYNA
Kynurenic acid
- LPA
Lysophosphatidic acid
- PCR
Polymerase chain reaction
- RNA
Ribonucleic acid
- WT
Wild type
Footnotes
Electronic supplementary material The online version of this article (https://doi.org/10.1007/s10620-018-5216-z) contains supplementary material, which is available to authorized users.
Compliance with ethical standards
Conflict of interest No conflicts of interest, financial or otherwise, are declared by the authors.
References
- 1.Novak EA, Mollen KP. Mitochondrial dysfunction in inflammatory bowel disease. Front Cell Dev Biol 2015;3:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Danese S, Grisham M, Hodge J, Telliez JB. JAK inhibition using tofacitinib for inflammatory bowel disease treatment : a hub for multiple inflammatory cytokines. Am J Physiol Gastrointest Liver Physiol 2016;310:G155–G162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doecke JD, Simms LA, Zhao ZZ, et al. Genetic susceptibility in IBD: overlap between ulcerative colitis and Crohn’s disease. Inflamm Bowel Dis 2013;19:240–245. [DOI] [PubMed] [Google Scholar]
- 4.Rogler G, Zeitz J, Biedermann L. The search for causative environmental factors in inflammatory bowel disease. Dig Dis Sci 2016;34:48–55. [DOI] [PubMed] [Google Scholar]
- 5.Gearry RB. IBD and environment: are there differences between east and west. Dig Dis Sci 2016;34:84–89. [DOI] [PubMed] [Google Scholar]
- 6.Kim ER, Chang DK. Colorectal cancer in inflammatory bowel disease: the risk, pathogenesis, prevention and diagnosis. World J Gastroenterol 2014;20:9872–9881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yashiro M Ulcerative colitis-associated colorectal cancer. World J Gastroenterol 2014;20:16389–16397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Divorty N, Mackenzie AE, Nicklin SA, Milligan G. G protein-coupled receptor 35: an emerging target in inflammatory and cardiovascular disease. Front Pharmacol 2015;6:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Milligan G Orthologue selectivity and ligand bias: translating the pharmacology of GPR35. Trends Pharmacol Sci 2011;32:317–325. [DOI] [PubMed] [Google Scholar]
- 10.Oka S, Ota R, Shima M, Yamashita A, Sugiura T. GPR35 is a novel lysophosphatidic acid receptor. Biochem Biophys Res Commun 2010;395:232–237. [DOI] [PubMed] [Google Scholar]
- 11.Wang J, Simonavicius N, Wu X, et al. Kynurenic acid as a ligand for orphan G protein-coupled receptor GPR35. J Biol Chem 2006;281:22021–22028. [DOI] [PubMed] [Google Scholar]
- 12.Barth MC, Ahluwalia N, Anderson TJ, et al. Kynurenic acid triggers firm arrest of leukocytes to vascular endothelium under flow conditions. J Biol Chem 2009;284:19189–19195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taniguchi Y, Tonai-Kachi H, Shinjo K. Zaprinast, a well-known cyclic guanosine monophosphate-specific phospho-diesterase inhibitor, is an agonist for GPR35. FEBS Lett 2006;580:5003–5008. [DOI] [PubMed] [Google Scholar]
- 14.O’Dowd BF, Nguyen T, Marchese A, et al. Discovery of three novel G-protein-coupled receptor genes. Genomics 1998;47:310–313. [DOI] [PubMed] [Google Scholar]
- 15.Imielinski M, Baldassano RN, Griffiths A, et al. Common variants at five new loci associated with early-onset inflammatory bowel disease. Nat Genet 2009;41:1335–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ellinghaus D, Folseraas T, Holm K, et al. Genome-wide association analysis in primary sclerosing cholangitis and ulcerative colitis identifies risk loci at GPR35 and TCF4. Hepatology 2013;58:1074–1083. [DOI] [PubMed] [Google Scholar]
- 17.Horikawa Y, Oda N, Cox NJ, et al. Genetic variation in the gene encoding calpain-10 is associated with type 2 diabetes mellitus. Nat Genet 2000;26:163–175. [DOI] [PubMed] [Google Scholar]
- 18.Sun YV, Bielak LF, Peyser PA, et al. Application of machine learning algorithms to predict coronary artery calcification with a sibship-based design. Genet Epidemiol 2008;32:350–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Skarnes WC, Rosen B, West AP, et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature 2011;474:337–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.White JK, Gerdin AK, Karp NA, et al. Genome-wide generation and systematic phenotyping of knockout mice reveals new roles for many genes. Cell 2013;154:452–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farooq SM, Stadnyk AW. Neutrophil infiltration of the colon is independent of the FPR1 yet FPR1 deficient mice show differential susceptibilities to acute versus chronic induced colitis. Dig Dis Sci 2012;57:1802–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farooq SM, Stillie R, Svensson M, Svanborg C, Strieter RM, Stadnyk AW. Therapeutic effect of blocking CXCR2 on neutrophil recruitment and dextran sodium sulfate-induced colitis. J Pharmacol Exp Ther 2009;329:123–129. [DOI] [PubMed] [Google Scholar]
- 23.Stillie R, Stadnyk AW. Role of TNF receptors, TNFR1 and TNFR2, in dextran sodium sulfate-induced colitis. Inflamm Bowel Dis 2009;15:1515–1525. [DOI] [PubMed] [Google Scholar]
- 24.Tsukahara T, Hamouda N, Utsumi D, Matsumoto K, Amagase K, Kato S. G protein-coupled receptor 35 contributes to mucosal repair in mice via migration of colonic epithelial cells. Pharmacol Res 2017;123:27–39. [DOI] [PubMed] [Google Scholar]
- 25.Dieleman LA, Pena AS, Meuwissen SG, van Rees EP. Role of animal models for the pathogenesis and treatment of inflammatory bowel disease. Scand J Gastroenterol Suppl 1997;223:99–104. [PubMed] [Google Scholar]
- 26.Thorburn AN, Macia L, Mackay CR. Diet, metabolites, and “western-lifestyle” inflammatory diseases. Immunity 2014;40:833–842. [DOI] [PubMed] [Google Scholar]
- 27.Kuc D, Zgrajka W, Parada-Turska J, Urbanik-Sypniewska T, Turski WA. Micromolar concentration of kynurenic acid in rat small intestine. Amino Acids 2008;35:503–505. [DOI] [PubMed] [Google Scholar]
- 28.Roca H, Varsos ZS, Sud S, Craig MJ, Ying C, Pienta KJ. CCL2 and interleukin-6 promote survival of human CD11b+ peripheral blood mononuclear cells and induce M2-type macrophage polarization. J Biol Chem 2009;284:34342–34354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parks WC, Wilson CL, Lopez-Boado YS. Matrix metalloproteinases as modulators of inflammation and innate immunity. Nat Rev Immunol 2004;4:617–629. [DOI] [PubMed] [Google Scholar]
- 30.McQuibban GA, Butler GS, Gong JH, et al. Matrix metalloproteinase activity inactivates the CXC chemokine stromal cell-derived factor-1. J Biol Chem 2001;276:43503–43508. [DOI] [PubMed] [Google Scholar]
- 31.McQuibban GA, Gong JH, Wong JP, Wallace JL, Clark-Lewis I, Overall CM. Matrix metalloproteinase processing of monocyte chemoattractant proteins generates CC chemokine receptor antagonists with anti-inflammatory properties in vivo. Blood 2002;100:1160–1167. [PubMed] [Google Scholar]
- 32.Turski MP, Turska M, Paluszkiewicz P, Parada-Turska J, Oxenkrug GF. Kynurenic Acid in the digestive system-new facts, new challenges. Int J Tryptophan Res 2013;6:47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maravillas-Montero JL, Burkhardt AM, Hevezi PA, Carnevale CD, Smit MJ, Zlotnik A. Cutting edge: GPR35/CXCR8 is the receptor of the mucosal chemokine CXCL17. J Immunol 2015;194:29–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.