Methicillin-resistant S. aureus continues to develop resistance to antimicrobials, including those in current clinical use as daptomycin (DAP). Resistance to DAP arises by mutations in cell membrane and cell wall genes and/or upregulation of the two-component VraSR system. However, less is known about the connection between the pathogen and virulence traits during DAP resistance development. We provide new insights into VraSR and its regulatory role for virulence factors during DAP resistance, highlighting coordinated interactions that favor the higher persistence of MRSA DAP-resistant strains in the infected host.
KEYWORDS: daptomycin, MRSA, VraSR, virulence
ABSTRACT
Methicillin-resistant Staphylococcus aureus (MRSA) threatens human health in hospital and community settings. The lipopeptide antibiotic daptomycin (DAP) is a frequently used treatment option for MRSA infection. DAP exposure can cause bacterial resistance because mutations are induced in genes implicated in cell membrane and cell wall metabolism. Adaptations aimed at surviving antimicrobial pressure can affect bacterial physiology and modify in vivo aptitude and pathogenesis. In this study, clinical DAP-susceptible (DAPs) and DAP-resistant (DAPr) MRSA isolates were used to investigate associations between DAP resistance and staphylococcal virulence. We previously found that VraSR is a critical sensor of cell membrane/wall homeostasis associated with DAP acquisition during MRSA infection. The present study found that DAPr CB1634 and CB5014 MRSA strains with vraSR upregulation were less virulent than their susceptible counterparts, CB1631 and CB5013. Differential gene-transcription profile analysis revealed that DAPr CB1634 had decreased agr two-component system expression, virulence factors, and highly suppressed hemolysis activity. Functional genetic analysis performed in DAPr CB1634 strains using vraSR inactivation followed by gene complementation found that vraSR acted as a transcriptional agrA regulator. These results indicated that VraSR has a broad range of regulatory functions. VraSR also appeared to affect DAPr adherence to epithelial cells, which would affect DAPr strain colonization and survival in the host. The correlation between DAP resistance and decreased virulence was also found in the CB5013 (DAPs) and CB5014 (DAPr) pair. Taken together, these findings are the first evidence that DAP resistance and MRSA virulence are tightly connected and involve compromised expression of regulatory and virulence determinants.
IMPORTANCE Methicillin-resistant S. aureus continues to develop resistance to antimicrobials, including those in current clinical use as daptomycin (DAP). Resistance to DAP arises by mutations in cell membrane and cell wall genes and/or upregulation of the two-component VraSR system. However, less is known about the connection between the pathogen and virulence traits during DAP resistance development. We provide new insights into VraSR and its regulatory role for virulence factors during DAP resistance, highlighting coordinated interactions that favor the higher persistence of MRSA DAP-resistant strains in the infected host.
INTRODUCTION
Staphylococcus aureus is a significant and ubiquitous opportunistic pathogen. The multidrug-resistant pathogen methicillin-resistant S. aureus (MRSA) is a major concern for public health in hospital and community settings and is associated with the development of numerous diseases (1). These diseases range from skin and soft tissue infections to severe life-threatening infections (e.g., pneumonia, endocarditis, and bacteremia) (2). Prevention of MRSA infection has improved, but infections caused by this pathogen remain challenging. The anti-MRSA antibiotics approved for different infections (e.g., complicated skin structure infections, bacteremia, and pneumonia) include vancomycin, linezolid, telavancin, ceftaroline, and daptomycin (DAP) (3).
DAP is a cyclic anionic lipopeptide that shares structural similarities with cationic antimicrobial peptides (CAMPs), a group of molecules produced by mammalian innate immune systems (4). DAP molecules first form micelles in the presence of physiological calcium concentrations. Next, phospholipid phosphatidylglycerol (PG) induces a structural transition in the DAP-calcium complex, allowing its binding to the cytoplasmic membrane (5), causing membrane depolarization, homeostasis imbalance, and cell death (4).
DAP-resistant S. aureus clinical isolates have been isolated from patients treated with DAP and other antibiotics (e.g., vancomycin) (6, 7). Although DAP resistance is rare, treatment failure occurs in more than 20% of the cases of resistance (8, 9) and still represents a challenge when encountered (10–12).
To resist DAP activity, the bacteria must impede the drug from reaching the cell membrane or penetrating it (5). The main factors described involving resistance to DAP, among other possible processes, include (i) production of a more positively charged cell surface to prevent DAP-Ca2+ insertion through electrostatic repulsion (13, 14), (ii) alteration of membrane fluidity by changing phospholipid content and asymmetry (13, 15, 16), (iii) decreased autolysis and increased thickening of the cell wall (17–19), and (iv) physiological and metabolic adaptations directed to increase the carbon flow to the synthesis of precursors needed for cell wall biosynthesis (18). Underlying these mechanisms are different nonsynonymous mutations in genes involved in the regulation of cell membrane structure and function, notably mprF, which is the most frequently described mutation in clinical DAP-resistant strains (14, 20–22). Other mechanistically relevant mutations can include those in cell wall-associated components (23, 24).
The success of a pathogen in overcoming a given antimicrobial therapy and continuing to spread during infections depends not only on the intrinsic and acquired resistance to the drug but also on additional factors, such as the resistance fitness costs, the pathogenicity of the strain, and the host conditions. The interplay between these mechanisms is poorly understood. Many studies have described a relationship between resistance mechanisms and virulence in several Gram-negative bacterial species, such as Pseudomonas aeruginosa (25–27), Acinetobacter baumannii (28–30), Escherichia coli (31), and Klebsiella pneumoniae (32, 33). For multidrug-resistant S. aureus, there is evidence demonstrating a tight connection between resistance to β-lactams, vancomycin, and glycopeptides and the pathogenicity of the MRSA strains (34–37). However, the impact of acquiring DAP resistance in clinical S. aureus and its correlation with pathogenicity and virulence have not been deeply explored.
We previously found that mprF mutation is not the only factor that determines DAP resistance. We provided functional evidence that upregulation of vraSR is a key factor associated with DAP and that inactivation results in increased DAP susceptibility. We also found that VraSR is a critical regulator of cell membrane homeostasis in response to alteration of membrane surface charges and reorganization of cell division proteins associated with cell wall synthesis (38). The accessory gene regulator (agr) is an important virulence regulator during S. aureus infection. RNA III is the effector of the system known to upregulate the expression of toxins and to downregulate genes encoding cell surface-associated proteins (39). The agr operon mutation has been commonly reported for VISA (vancomycin-intermediate S. aureus); agr dysfunction in the absence of mutation has been also described. The loss of agr that occurs frequently in clinical isolates enhances the survival of S. aureus during DAP treatment. This result compares with the rapid killing of wild-type S. aureus strains (40). In the present study, we used in vitro and in vivo experiments and found that acquisition of DAP resistance and virulence in MRSA is a tightly connected and regulated mechanism that includes a cross-talk regulatory pathway between vraSR and agr. This process may contribute to the persistence of DAP-resistant strains during infection.
RESULTS
Acquisition of DAP resistance impacts MRSA strain pathogenicity.
In previous studies, we explored the detailed mechanistic basis of DAP resistance in a set of clinical isogenic DAPs/DAPr strains (11, 38). However, the effect of acquiring DAP resistance on the virulence of the strain has not been examined. We used two of the previously characterized isogenic MRSA clinical strains, DAPs CB1631 and DAPr CB1634, which were isolated from a patient who had a DAP therapy failure (11) (Table 1). We evaluated the capacity of both strains to adhere to and invade the human epithelial cell line A529. While no significant differences in adhesion were observed between the two strains (Fig. 1A), when the strains where assessed for their ability to internalize into the A259 cells (Fig. 1B), DAPs CB1631 was more effective than its resistant CB1634 counterpart. This finding is in accordance with increased expression of fnbAB and isdABDE observed in CB1631 (Table 2), suggesting that the DAPs strain elicits more pronounced invasion traits than DAPr CB1634.
TABLE 1.
Bacterial strains used in this study and their MIC values obtained using the Etesta
| Strain or plasmid | Description | DAP MIC (µg/ml) |
Reference or source |
|---|---|---|---|
| Strains | |||
| S. aureus | |||
| N315 | Hospital-acquired methicillin-resistant SCCmec type II |
0.125 | 60 |
| ATCC 29213 | MSSA, standard strain for CLSI antimicrobial susceptibility testing |
0.125 | 61 |
| Newman | MSSA, isolated from a human infection |
0.25 | 62 |
| KVR | N315 ΔvraSR::cat | 60 | |
| CB1631 | DAPs, SCCmec type II | 0.25 | 41 |
| CB1634 | DAPr isogenic to CB1631 | 4 | 41 |
| CB1634+agr | CB1634 + expressing psk265 full-length agr |
0.094 | This study |
| CB5013 | DAPs, SCCmec type II | 0.25 | 41 |
| CB5014 | DAPr, isogenic to CB5013 | 4 | 41 |
| CB1634ΔvraSR | CB1634 ΔvraSR::cat | 0.25 | This study |
| CB1634ΔvraSR+ΔvraSR | CB1634 ΔvraSR::cat pVRASR-2 |
2 | This study |
| CB5014ΔvraSR | CB5014 ΔvraSR::cat | 0.25 | This study |
| CB5014ΔvraSR+ΔvraSR | CB5014 ΔvraSR::cat pVRASR-2 |
2 | This study |
| S. epidermidis Y1 | 0.25 | This study | |
| Plasmids | |||
| pCR-XL-2 TOPO | Cloning vector, Ampr Kanr |
ThermoFisher | |
| S. aureus RN4220(pVRASR-2) | Entire vraS/vraR cloned into pAW8-tet |
39 |
Abbreviations: cat, chloramphenicol resistant; Tetr, tetracycline resistant; Ampr, ampicillin resistant; Kanr, kanamycin resistant.
FIG 1.
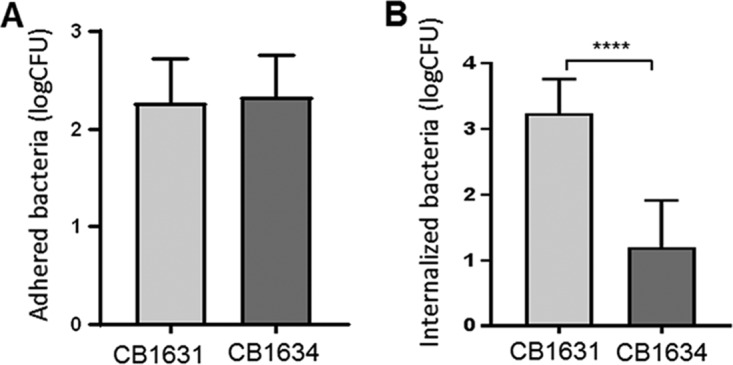
S. aureus susceptibility to DAP correlates with the virulence of MRSA strains. (A) Adhesion of S. aureus DAPs CB1631 and DAPr CB1634 to A459 human epithelial cells. (B) Internalization of S. aureus DAPs CB1631 and DAPr CB1634 into human A459 epithelial cells. Data represent the mean and standard deviation from three independent experiments. Statistically significant differences were determined using an unpaired Student t test (****, P < 0.0001).
TABLE 2.
Gene expression analysis of S. aureus DAPs CB1631 compared with DAPr CB1634 using RNA-seq
| ORF | Gene | Product or function | Fold change |
|---|---|---|---|
| SA1844 | agrA | Accessory gene regulator A | 4.501 |
| SA1842 | agrB | Accessory gene regulator B | 1.984 |
| SA1843 | agrC | Accessory gene regulator C | 2.218 |
| SAS066 | agrD | Accessory gene regulator R | 2.424 |
| SA2457 | capA | Capsular polysaccharide biosynthesis protein Cap5A | 1.359 |
| SA0145 | capB | Capsular polysaccharide biosynthesis protein Cap5B | 4.856 |
| SA0146 | capC | Capsular polysaccharide biosynthesis protein Cap5C | 1.444 |
| SA0147 | capD | Capsular polysaccharide biosynthesis protein Cap5D | 2.363 |
| SA0148 | capE | Capsular polysaccharide biosynthesis protein Cap5E | 2.672 |
| SA0149 | capF | Capsular polysaccharide biosynthesis protein Cap5F | 2.425 |
| SA0150 | capG | Capsular polysaccharide biosynthesis protein Cap5G | 1.798 |
| SA0151 | capH | Capsular polysaccharide biosynthesis protein Cap5H | 3.642 |
| SA0152 | capI | Capsular polysaccharide biosynthesis protein Cap5I | 1.641 |
| SA0153 | capJ | Capsular polysaccharide biosynthesis protein Cap5J | 1.288 |
| SA0154 | capK | Capsular polysaccharide biosynthesis protein Cap5K | 1.99 |
| SA0155 | capL | Capsular polysaccharide biosynthesis protein Cap5L | 1.213 |
| SA0156 | capM | Capsular polysaccharide biosynthesis protein Cap5M | 1.217 |
| SA0157 | capN | Capsular polysaccharide biosynthesis protein Cap5N | 2.879 |
| SA0159 | capP | Capsular polysaccharide biosynthesis protein Cap5P | 1.259 |
| SA0742 | clfA | Clumping factor A, fibrinogen binding protein | −1.977 |
| SA2423 | clfB | Clumping factor B, fibrinogen binding protein | 1.039 |
| SA0222 | coa | Staphylocoagulase precursor | 2.458 |
| SA2291 | fnbA | Fibronectin binding protein A | 1.742 |
| SA2290 | fnbB | Fibronectin binding protein B | 2.692 |
| SA0309 | geh | Lipase | 1.274 |
| SA1756 | hlb | Truncated β-hemolysin | 3.075 |
| SAS065 | hld | δ-Hemolysin | 8.235 |
| SA2207 | hlgA | γ-Hemolysin component A | 1.412 |
| SA2209 | hlgB | γ-Hemolysin component B | 2.195 |
| SA2208 | hlgC | γ-Hemolysin component C | 1.476 |
| SA2356 | isaA | Immunodominant antigen A | 2.629 |
| SA0977 | isdA | Iron-regulated surface determinant protein A | 2.582 |
| SA0976 | isdB | Iron-regulated surface determinant protein B | 2.841 |
| SA0979 | isdD | Iron-regulated surface determinant protein D | 3.817 |
| SA0980 | isdE | Iron-regulated surface determinant protein E | 2.702 |
| SA1637 | lukD | Leukotoxin | 3.537 |
| SA1638 | lukE | Leukotoxin | 3.515 |
| SA0661 | saeR | Response regulator SaeR | 4.824 |
| SA0660 | saeS | Sensor histidine kinase SaeS | 3.078 |
| SA2206 | sbi | Immunoglobulin G binding protein | 4.546 |
| SA0519 | sdrC | Serine-aspartate repeat-containing protein C, fibrinogen binding protein | 1.171 |
| SA0520 | sdrD | Serine-aspartate repeat-containing protein D, fibrinogen binding protein | 1.316 |
| SA0521 | sdrE | Serine-aspartate repeat-containing protein E, fibrinogen binding protein | 1.165 |
| SA1869 | sigB | RNA polymerase sigma factor | 1.066 |
| SA0107 | spa | Protein A | −1.264 |
| SA1631 | splA | Serine protease | 4.269 |
| SA1630 | splB | Serine protease | 2.244 |
| SA1629 | splC | Serine protease | 1.147 |
| SA1628 | splD | Serine protease | 3.059 |
| SA2093 | ssaA | Staphylococcal secretory antigen | 2.6 |
| SA1700 | vraR | Response regulator VraR | −3.021 |
| SA1701 | vraS | Sensor protein VraS | −1.816 |
| SA0018 | walK | Sensor protein kinase WalK (VicK, YycG) | −1.122 |
| SA0017 | walR | Response regulator WalR (VicR, YycF) | −1.289 |
DAPr MRSA strains exhibit decreased transcription of virulence genes.
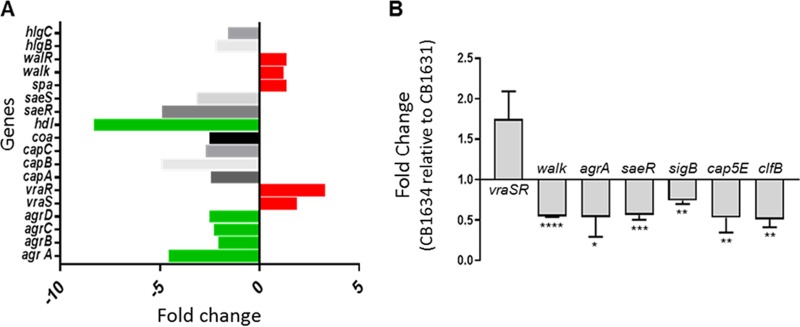
To interrogate whether differences in gene expression could reflect the potential factors linking virulence and DAP resistance, we analyzed the transcription levels of several major staphylococcal virulence factors in DAPs CB1631 and DAPr CB1634 strains using RNA-seq. Master gene regulators (agrA, saeR, and saeS), which control the global expression of multiple virulence factors, were upregulated in the CB1631 strain (Fig. 2A and Table 2). The genes with the highest upregulation were those coding for cytolytic proteins (hld, hlb, lukD, and lukE), serine proteases (splA, splC, and splD), coagulases (coa), surface adhesins important for host colonization (isdA, isdB, isdD, and isdE), capsular biosynthesis proteins (cap5), and innate immune response evasion factors (sbi). In contrast, the two-component system kinase sensor vraS and its response regulator vraR were downregulated, consistent with the observed DAPs phenotype of CB1631 (41). Notably, two important adhesins (spa and clfA) were downregulated in DAPs CB1631. To validate the RNA-seq results, we performed reverse transcription-quantitative PCR (qRT-PCR) for most of the virulence genes and master regulators. As shown in Fig. 2B, agrA, saeR, and sigB regulators showed significantly decreased levels of transcription in DAPr CB1634 compared with the DAPs strain while transcription levels of vraSR mRNA were increased.
FIG 2.
Gene expression analysis. (A) RNA-seq expression analysis comparing CB1634 with CB1631, expressed in fold changes. (B) Quantification of the mRNA expression of regulatory and virulence genes in the S. aureus CB1634 strain relative to its parental CB1631 strain using qRT-PCR. Statistically significant differences were determined using an unpaired Student t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001). Genes upregulated are denoted in red, and genes downregulated are shown in green.
Together, these results suggest that the acquisition of DAP resistance impacts the transcriptional profile and regulatory pathways of MRSA, which could influence the pathobiology of S. aureus.
Decreased hemolysis production and virulence in DAPr CB1634.
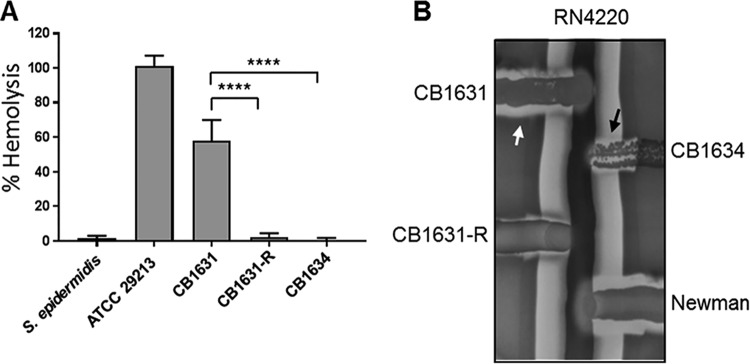
A feature of highly virulent strains is their ability to lyse red blood cells (RBCs) by secreting hemolysins. As mentioned previously, we found decreased transcription levels of agrA and saeR and a mutation in saeR in the DAPr S. aureus CB1634 strain. These two regulators are some of the primary activators of staphylococcal hemolysins. To further confirm the invasiveness of DAPs strains, we performed a hemolytic assay using rabbit RBCs (Fig. 3A). Staphylococcusepidermidis and S. aureus ATCC 29213 were used as nonhemolytic and hemolytic controls, respectively. In this experiment, we included the in vitro-generated CB1631-R strain. As shown in Fig. 3A, the S. aureus DAPs CB1631 strain elicited significantly more hemolysis than its in vivo CB1634 and in vitro-generated CB1631-R DAPr counterparts. These observations were followed by the measuring of hemolysis production in both DAPr and DAPs strains. As illustrated in Fig. 3B, the DAPs CB1631 strain produced α-hemolysin and δ-hemolysin, which is demonstrated by the clearing zone at the intersection of the β-hemolysis halo of the S. aureus RN4220 streak; similar effects were observed with the positive-control S. aureus Newman strain. For DAPr CB1634, there was only δ-hemolysis and absence of α-hemolysin. Similarly, in the in vitro-obtained DAPr mutant CB1631-R, its hemolysin production was markedly diminished compared with the parental strain (Fig. 3B). Thus, these results suggested a potential connection between levels of DAP resistance acquired either in vitro or in vivo and decreased pathogenicity in the host.
FIG 3.
Hemolysis of RBCs. (A) Rabbit erythrocytes were incubated with an equal volume of bacterial supernatants, and hemolysis was measured spectrophotometrically using absorbance at 540 nm. A standard curve was used to determine the percent hemolysis. S. epidermidis was used as a nonhemolytic control; S. aureus ATCC 29213 was used as a positive control. Data represent the means and standard deviations from at least three independent experiments. Statistically significant differences were determined using a one-way ANOVA; a Bonferroni a posteriori test was performed (****, P < 0.0001). (B) δ-hemolysin activity was present in CB1631, CB1634, and, to a lesser extent, in CB1631-R (black arrow). α-hemolysis was present in CB1631 (white arrow).
VraSR regulatory cross talk with agr determines increases in virulence and hemolysis.
Cameron et al. recently described an association between VraSR and VISA strain virulence (42). They demonstrated the capacity of VraSR to modulate S. aureus virulence by binding the P2-P3 intergenic region of the agr promoter, indicating that when S. aureus is subject to vancomycin induction, VraR binds and inhibits the function of the Agr quorum sensing system, causing reductions in the virulence of VISA/hVISA strains (42, 43). Given our previous findings revealing (i) that VraSR is a key factor during DAP resistance and (ii) the defective expression of agr found in CB1634, we postulated that VraSR may transcriptionally regulate agr expression in CB1634.
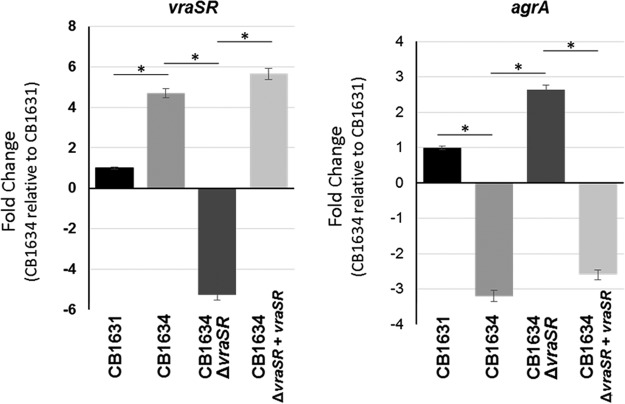
To test this hypothesis, we evaluated mRNA expression levels of both agrA and vraSR using real-time reverse transcription-PCR (RT-PCR) analysis (Fig. 4), showing consistency with results of RNA-seq analysis (Fig. 2A), i.e., decreased expression of agrA in CB1634 when vraSR was upregulated (Fig. 4). These effects were further analyzed using a vraSR mutant generated in the CB1634 strain, as described in Materials and Methods. We found that inactivation of vraSR (CB1634ΔvraSR) resulted in increased agr expression, while vraSR complementation (CB1634ΔvraSR+vraSR) reversed these effects and reduced agrA to levels comparable to those seen in the CB1634 strain. Similar results were obtained when additional DAPs/r strains (e.g., CB5013/CB5014) and corresponding mutant/transcomplemented strains were examined (data not shown).
FIG 4.
Quantitation of vraSR and agrA mRNA using real-time RT-PCR. RNA was prepared from cells of DAPs CB1631, DAPr CB1634, CB1634ΔvraSR, and complemented mutant CB1634ΔvraSR+vraSR strains collected during the exponential phase of growth. Relative fold change values of specific vraSR mRNA are shown on the vertical axis; 16S rRNA was used as an internal control (*, P < 0.001).
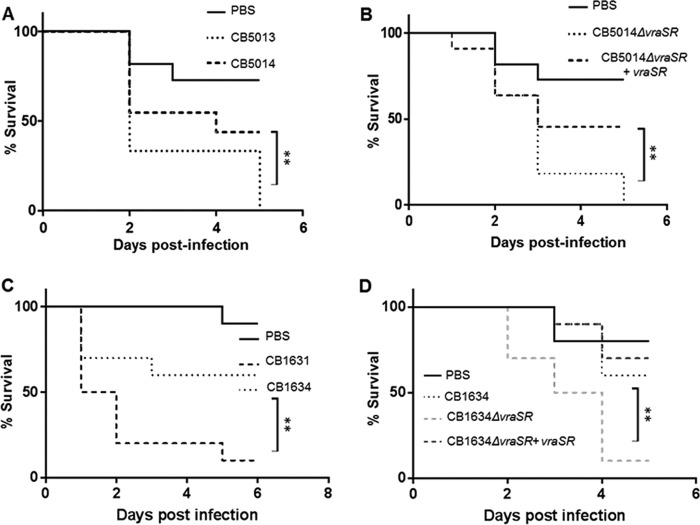
To evaluate the effects of vraSR/agrA on virulence traits, we tested the role of the aforementioned regulatory system in an in vivo model of Galleria mellonella infection. Groups of larvae were inoculated with a bacterial suspension containing the corresponding CB5013, CB5014, CB5014ΔvraSR, and CB5014ΔvraSR+vraSR strains (106 bacteria/worm), as previously described (11). An uninfected control group received a PBS treatment to control for multiple injections. Worms were monitored daily, and any deaths that occurred over the next 5 days were recorded. Worms injected with PBS showed 80% to 100% survival at day 5 (Fig. 5A and B). Groups injected with the parent CB5013 strain had low survival rates (0% to 30%, day 5; Fig. 5A), while in contrast, those worms infected with the CB5014 strain had survival rates of 40% to 70% at day 5. The CB5014ΔvraSR strain showed a similar trend as the one observed with the CB5013 parent strain (i.e., survival rate of 0% at day 5; Fig. 5B). Following CB5014ΔvraSR transcomplementation (CB5014ΔvraSR+vraSR), the survival rate was higher (50% to 90%; Fig. 5B), as observed with the worms injected with the CB5014 strains (Fig. 5A). Similar results were obtained with worms infected with CB1634, CB1634, CB1634ΔvraSR, and CB1634ΔvraSR+vraSR strains (106 bacteria/worm; Fig. 5C and D).
FIG 5.
G. mellonella infection with DAPs/r and derivative strains. Groups of larvae (10/group) were inoculated with 10 µl PBS (uninfected control group) or bacterial suspension containing 1.5 × 106 CFU/ml DAPs CB5013 and DAPr CB5014 (A) and its corresponding mutant and complemented strains, CB5014ΔvraSR and CB5014ΔvraSR+vraSR (B), into the last proleg and incubated at 37°C. Worms were checked daily, and any deaths were recorded, for a total of 10 days. A minimum of three independent experimental replicates were performed for each experiment. Similar analyses were performed with DAPs CB1631 and DAPr CB1634 (C) and its corresponding mutant and complemented strains, CB1634ΔvraSR and CB1634ΔvraSR+vraSR (D). Survival data were plotted using the Kaplan-Meier method and expressed as percentage of survival versus time. Statistically significant differences were determined using the log rank test (**, P < 0.01).
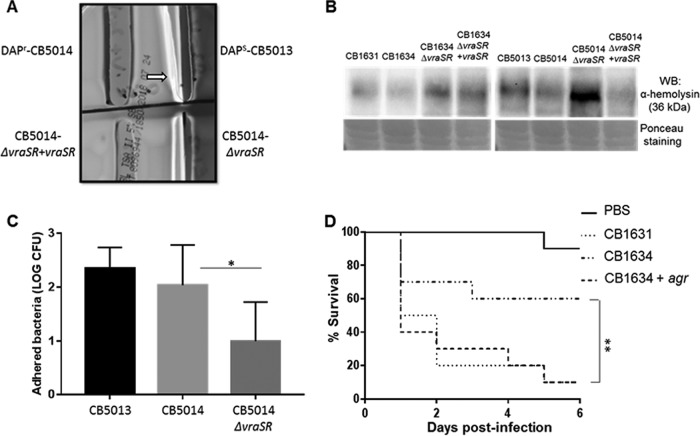
Taking into account the observations showing differences between DAPs/r strains in terms of hemolysis (Fig. 2), we further investigated whether vraSR may have affected α-hemolysin production. To test this hypothesis, Western blot analysis using a specific anti-α-hemolysin antibody was performed with lysates from both DAPr CB1634 and CB5014 strains and the latter’s corresponding vraSR mutant and transcomplemented strains, CB5014ΔvraSR and CB5014ΔvraSR+vraSR. As depicted in Fig. 6B, the DAPs CB1631 strain produced α-hemolysin to a greater extent than its DAPr counterpart CB1634 (Fig. 6B, lanes 1 and 2). Inactivation of vraSR in DAPr CB1634 (CB1634ΔvraSR) did correlate with increased α-hemolysin levels, similar to the DAPs CB1631 strain (lane 3), while vraSR transcomplementation (CB1634ΔvraSR+vraSR) showed similar levels as those seen in DAPr CB1634, i.e., decreased levels compared to CB1634ΔvraSR. Similar observations were made between strains CB5013, CB5014, and the corresponding vraSR mutant and transcomplemented strains (i.e., CB5014ΔvraSR and CB5014ΔvraSR+vraSR; lanes 5 to 8, Fig. 6B).
FIG 6.
Analysis of virulence factors of DAPr CB5014 and DAPr CB1634 and derivative strains. (A) Hemolysis in blood agar plates of CB5013, CB5014, mutant CB5014ΔvraSR, and transcomplemented CB5014ΔvraSR+vraSR. (B) Western blot analyses of α-hemolysin in supernatants collected and concentrated from CB1631, CB1634, CB1634ΔvraSR, CB1634ΔvraSR+vraSR, CB5013, CB5014, CB5014ΔvraSR, and CB5014ΔvraSR+vraSR derivative strains. Ponceau staining was used as a loading control. α-hemolysis was higher in CB1631, CB5013, CB1634ΔvraSR, and CB5014ΔvraSR; low levels of alpha-toxin are seen in CB1634, CB5014, and complemented strains CB1634ΔvraSR+vraSR and CB5014ΔvraSR+vraSR. (C) Adhesion of S. aureus DAPs CB5013 and DAPr CB5014 and its corresponding mutant CB5014ΔvraSR to A459 human epithelial lung cells. Reduced adhesion was found in CB5014ΔvraSR compared with its parent DAPr CB5014 counterpart (*, P < 0.01). (D) G. mellonella infection of groups of larvae (10/group) inoculated with PBS and bacterial suspension containing 1.5 × 106 CFU/ml DAPs CB1631, DAPr CB1634, and its overexpressed agr derivative CB1634+agr. Survival data were plotted using the Kaplan-Meier method and expressed as percentage of survival versus time. Statistically significant differences were determined using the log rank test (**, P < 0.01).
We then analyzed whether inactivation of VraSR affected the capacity of DAPr cells to adhere to A549 human lung epithelial cells, showing there was a statistically significant difference in adhesion levels between CB5014ΔvraSR and CB5014 (P < 0.01; Fig. 6C). CB5014ΔvraSR had low levels of adhesion to epithelial cells compared with its parent strain, CB5014. These results suggested that VraSR is associated with α-hemolysin production and that VraSR promotes the adhesion of the DAPr strain to epithelial cells.
To investigate the cross talk between vraSR and agr in relation to virulence during DAP resistance, extratemporal agr overexpression was performed in CB1634 using the pSK265 vector containing the wild-type copy of the agr operon. The results shown in Fig. 6D indicated that agr enforced expression in CB1634 (CB1634+agr) determined an increase in virulence with survival percentages similar to the parental CB1631 strain. However, α-hemolysis was not restored after agr complementation, suggesting that that although agr is associated with CB1634 defects in virulence, it was not able to restore α-hemolysis production, as there was no direct effect of agr on hemolysis during DAP resistance (data not shown). Moreover, we ruled out, by having performed saeR transcomplementation of saeR mutation in CB1634 (CB1634+psaeRSWT), that the δ-hemolysis phenotype was identical to its parental CB1634 strain and did not show restored capacity to produce α-hemolysis (data not shown). Taken together, these results suggested that virulence in DAPr strains is dependent on VraSR regulatory control of agrA.
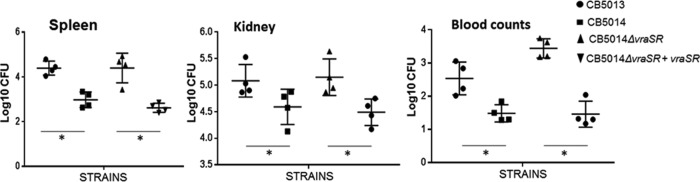
Finally, to determine the differences in response to infection between the DAPs and DAPr strains in mammalian tissues, we used an established murine septicemia model. Groups of 5 to 6 mice each were inoculated via tail injection with ∼1 × 10 to 2 × 107 CFU of DAPs CB5013, DAPr CB5014, and the corresponding vraSR mutant CB5014ΔvraSR and transcomplemented CB5014ΔvraSR+vraSR MRSA strains. After 72 h, mice were euthanized, and spleen, kidneys, and whole blood were collected aseptically, homogenized (spleen and kidneys), serially diluted in PBS, and plated onto tryptic soy agar (TSA) plates to determine the number of viable staphylococci. As depicted in Fig. 7, DAPs CB5013 cells proliferated in all cases to higher values during infection compared with the DAPr CB5014 isogenic strain; importantly, CB5014ΔvraSR displayed similar infection levels as those corresponding to CB5013, values that were significantly reduced when levels of vraSR were restored, i.e., CB5014ΔvraSR+vraSR. Similar results were obtained for CB1631, CB1634, and their corresponding mutant/complemented vraSR strains (data not shown). These results, together with the observations suggesting that MRSA DAPs strains (e.g., CB5013 and CB1631) are more prone to invading mammalian cells than their DAPr counterpart strains (e.g., CB1634; Fig. 1B), highlight first the attenuated in vivo virulence of the DAPr strain and second the mechanistic role played by vraSR in this process.
FIG 7.
In vivo sepsis mouse model showing the effect of DAP susceptibility on the colonization of kidney and spleen. Groups of six mice were used. Each group (n = 6) was inoculated via tail injection with ∼1 × 10 to 2 × 107 CFU of either the DAPs CB5013 or DAPr CB5014 MRSA strain grown in TSB at 37°C, 150 rpm. Mice were euthanized at 72 h postinfection. Kidneys, spleen, and whole blood were collected aseptically, homogenized (kidneys and spleen), serially diluted in PBS, and plated onto TSA plates to determine the number of viable staphylococci. Results are expressed as the logarithm of CFU per gram of organ (log CFU/g). Statistically significant differences were determined using the unpaired Student t test (*, P < 0.05).
DISCUSSION
When facing challenging environmental conditions, bacteria can adopt diverse adaptation strategies to survive. Particularly within a host, they can adjust expression of virulence factors at any time during infection (44). In addition, if they are exposed to antimicrobial agents, they are able to generate metabolic and/or genetic changes that promote survival when exposed to a certain drug while sustaining the infection within the host (18). The correlation between resistance to several antibiotics and virulence has been demonstrated previously in different bacterial species (34, 36, 45, 46). In the present study, we analyzed how DAP resistance impacted the pathogenicity of clinically derived MRSA strains obtained from cases of DAP treatment failure. Our results showed that the in vivo virulence of DAPr strains was notably attenuated compared with their DAPs counterparts, as shown in the in vivo G. mellonella invertebrate model and the murine septicemia model. This finding was consistent with the observation that the counterpart DAPs CB1631 strain was more hemolytic and had higher expression of different virulence determinants. This evidence was further supported by the results using an in vitro DAP-resistant mutant CB1631-R. A considerable rise in DAP MIC) (8 µg/ml) was associated with markedly attenuated virulence, lowered expression of virulence factors, and lowered hemolysis. These results were consistent with the results reported by Cameron et al. (45), in which clinical and in vitro DAPr strains were less virulent but more persistent in vivo than their DAPs progenitor strains. Similarly, DAPr Streptococcus mitis strains have been shown to display reduced in vitro and in vivo virulence in an endovascular infection model; the parental DAP- susceptible strain outcompeted the DAPr variant in all target organs (46). One explanation for the lowered virulence of DAPr staphylococcal strains could be that bacteria are more likely to prefer to sustain a chronic persistent infection instead of invasive acute infections once they acquire resistance. In the long term, this strategy might be more beneficial for DAPr strains from a fitness and survival perspective. The fitness costs of DAP resistance determine the aptitude of the strain in vivo and therefore influence the course and type of infection. In fact, the acquisition and maintenance of resistance is a costly process, and in order to survive the presence of antibiotics, bacteria sacrifice numerous proteins, thus losing certain abilities. In support of this hypothesis, it was shown that DAP-nonsusceptible strains isolated after DAP treatment failure had significant alterations in metabolic pathways needed to support resistance (18). In another report, in vitro DAPr mutants of strains isolated from hospitalized patients with bloodstream infections showed decreased fitness and pathogenicity (47). Regarding our strains, Roch et al. demonstrated that DAPs CB1631 and CB5013 are more fitness competent (48) and, as addressed in this work, more virulent than their isogenic DAPr strains. In the present study, we found that the pattern of reduced virulence seen in clinical DAP-resistant strains also occurred in the in vitro-generated DAP-resistant mutant. For the CB1631-R in vitro mutant, the fitness repercussion of DAP resistance was even more pronounced; the strain reached a high MIC value (8 µg/ml) by incorporating nonsynonymous mutations in three different genes (walK, rpoC, and mrpF) in its genome. These mutations compromised growth, the expression of numerous virulence factors, and the in vivo pathogenic competence (data not shown).
Another important new finding of our study is that VraSR appears to have an accessory role other than sensing membrane damage in DAP-resistant strains. To understand the molecular mechanism contributing to attenuated virulence in DAP- resistant strains, we centered our attention on vraSR and agr regulators. Expression of these regulators appeared to be uniformly altered in most of our DAPr strains, which ruled out an effect associated with strain background. Consistent with our findings, Pader et al. found that loss of agr quorum sensing, which occurs frequently in S. aureus isolates, enhances S. aureus survival during DAP treatment (40). They also demonstrated that as a mechanism of protection, defective agr mutants survive antibiotic exposure by releasing membrane phospholipids that bind and inactivate DAP (40).
In addition to maintaining the highly demanding process of DAP resistance, agr overexpression in the DAPr strain (CB1634+agr) not only increased its virulence but also negatively affected the in vivo persistence (Fig. 6D). These results suggested that through downregulation of agr, VraSR provides DAPr strains with advantageous survival traits. We found that the reduced virulence of DAPr strains was reversed in their counterpart vraSR mutants, highlighting the role of VraSR in virulence modulation. The findings of Chang et al.in Streptococcussuis support this hypothesis (49). They described that vraSR has an essential role in facilitating the resistance of S. suis to killing in human blood (49). Taken together, these findings and ours indicate that VraSR may initiate a regulatory response to counteract neutrophil defense by increasing the probability of DAPr strain survival in the host.
Overall, these results provide evidence that virulence and DAP resistance in MRSA are intrinsically related. It is likely that these two processes must be carefully regulated to be mutually associated. An understanding of how these mechanisms interconnect will contribute to the elucidation of the evolution of DAPr strains and potentially identify methods for the prevention and treatment of life-threatening MRSA infections.
MATERIALS AND METHODS
Bacterial strains, culture conditions, antibiotics, and plasmids.
All strains used in this study are listed in Table 1; some of these strains were reported previously (41). Strains were grown in tryptic soy broth (TSB) (BD, Sparks, MD), Mueller-Hinton broth (MH) (BD, Sparks, MD), tryptic soy agar (TSA) (BD, Sparks, MD), TSA with 5% sheep blood (BBL, Sparks, MD), and MH agar (MHA) (BD, Sparks, MD). DAP was provided by Merck (formerly Cubist Pharmaceuticals; Lexington, MA).
Overnight cultures grown in MH were used for inoculation to an initial optical density at 600 nm (OD600) of 0.05. Cultures were grown aerobically at 37°C in flasks with a 10:1 flask-to-volume ratio and with shaking at 250 rpm and supplemented when required with different concentrations of DAP and 50 μg/ml CaCl2. Cultures of mutants and transcomplemented VraSR strains were grown in tetracycline (5 μg/ml) and chloramphenicol (10 μg/ml), respectively. Bacterial growth was assessed by measuring OD600, and viability was measured by CFU/ml serial dilutions on MHA plates. Antibiotic MICs were determined using the Etest (bioMérieux, Marcy l’Etoile, France) and the broth microdilution method according to CLSI guidelines (50). ATCC 29213 was used as an internal control for MIC assays. Because CLSI has not yet established a resistance breakpoint for DAP, strains with MICs of ≥1 µg/ml were considered nonsusceptible (48). The term resistant is being used in the present study to simplify reading and understanding. Plasmid DNA was isolated from Escherichia coli strains using a QIAprep Spin miniprep kit (Qiagen, Valencia, CA) according to the manufacturer’s protocol. Plasmids were transformed into S. aureus RN4220 by electroporation using a previously described procedure (51). Plasmids were introduced in the final S. aureus strain using 80α-phage transduction (52). In vitro DAP-resistant mutant CB1631-R was obtained using progressive daily passages of DAPs CB1631 in subinhibitory concentrations of DAP (gradient concentrations were 0, 0.06, 0.125, 0.25, 0.5, 1, 2, 4, 8, 16, and 32 µg/ml) with 50 µg/ml CaCl2 in 24-well plates at 37°C for 15 days. Measurement of DAP MICs was performed according to CLSI guidelines to confirm the identity of the in vitro mutants and their corresponding parental strains.
Construction of DAP CB1634+agr complementation.
To generate the CB1634 ectopic agr-overexpressed construct CB1634+agr, the full length of agr was cloned into psk265 as previously described (53). Transaction of products required for complete agr activity, including an upstream region putative ribosomal binding site and the agr promoters (P2 and P3), was performed as previously described (53) (Table 1).
Construction of vraSR-null mutants and complementation.
A mutant (CB1634 ΔvraSR::cat) strain was obtained by transducing the deletion vraSR mutant (ΔvraSR::cat) by ϕ11 phage from strain KVR (54) into DAPr CB1634 (52), resulting in CB1634 vraSR. This mutant was transcomplemented using transduction and the pAW8 shuttle plasmid, which contained a 3.3-kb fragment corresponding to the entire vraSR operon (52); the CB1634ΔvraSR+vraSR strain was produced (Table 1). A similar procedure was performed into the CB5014 strain to produce a vraSR mutant (CB5014ΔvraSR) and a transcomplemented vraSR mutant, CB5014ΔvraSR+vraSR.
Galleria mellonella survival assay.
Galleria mellonella infections were performed as described previously (55). Briefly, groups of 10 Galleria mellonella larvae at their last instar stage (Knutson’s Live Bait, Brooklyn, MI) were inoculated with 10 µl of bacterial suspension (∼107 CFU/ml) during the last left proleg. Worms were incubated at 37°C and monitored every 24 h over a period of 5 days; those worms that did not move when touched and that were dark brown were considered dead. All trials included an uninfected control (injected with PBS). All experiments were performed in three independent replicates, and results were expressed as survival percentage versus time.
Epithelial adhesion and invasion assays.
A549 human lung epithelial cells (ATCC CCL 185) were used for cell culture assays. First, 24-well plates were seeded with 2 × 105 cells/well and incubated overnight at 37°C and 5% CO2. Once cells reached confluence (0.2 × 106), culture medium was removed, and cells were washed with DMEM and 10% fetal bovine serum. For adhesion experiments, overnight bacterial cultures in DMEM were added to the cellular monolayer at a multiplicity of infection (MOI) of 10. After a 1-h incubation at 37°C in 5% CO2, nonadherent cells were removed by washing three times with PBS. For invasion experiments, overnight bacterial cultures in DMEM were added to the cellular monolayer at an MOI of 40 and incubated for 2 h at 37°C in 5% CO2. Medium was aspirated and replaced with DMEM and 10% fetal bovine serum containing 100 µg/ml gentamicin and 5 µg/ml lysostaphin to remove noninternalized bacteria. Cells were incubated for 2 h at 37°C and 5% CO2. Eukaryotic cells were detached from the wells with 0.25% Trypsin in 1 mM EDTA and lysed with 0.1% Triton X-100. Extracts were vigorously vortexed, serially diluted in PBS, and plated onto TSA. Independent experiments were performed in triplicate.
Expression analysis using RNA-seq and qRT-PCR.
Overnight bacterial cultures grown in MH broth at 37°C and shaken at 150 rpm were diluted 1:100 in MH and incubated until they reached an OD600 of ∼0.5. RNAlater reagent (Sigma) was added to bacterial cell cultures to protect the cellular RNA. Total RNA extraction was performed using an RNeasy isolation kit (Qiagen), and the DNA was removed using a DNA-free DNA removal kit (Thermo Fisher Scientific). RNA concentrations were assessed by measuring absorbance at 260 and 280 nm using a NanoDrop 8000 (Thermo Fisher Scientific, Waltham, MA). For RNA-seq analysis, RNA was prepared from S. aureus CB1631 and CB1634 cells collected during the exponential phase of growth. The quality of the total RNA was determined using RNA Nano chips (Agilent Technologies, Santa Clara, CA) run with an Agilent 2100 Bioanalyzer and 2100 Expert software. The genome-wide transcript sequencing libraries were prepared according to the manufacturer’s recommendations (ScriptSeq; Epicentre) and sequenced on a MiSeq instrument (Illumina, San Diego, CA). Differential gene expression was determined using Lasergene (v14) software (DNAStar, Madison, WI) and the PATRIC web resource (56); differences of >1.5-fold and P < 0.05 after applying Bonferroni correction were considered significant. For qRT-PCR, real-time reverse transcription-PCR analysis was performed using a SensiFAST SYBR No-ROX one-step kit (Bioline, Taunton, MA). Probes and primers were synthetized by Eurofins Genomics (Thermo Fisher Scientific, Waltham, MA); the corresponding sequences are provided in Table 3. The level of gene expression for the studied strain compared with its parental strain (reference) was expressed as 2−ΔΔCT, where CT represents the threshold cycle value, ΔCT represents the difference in threshold cycle between the target gene and the control gene (16S), and ΔΔCT represents the difference in ΔCT between the studied strain and the parental strain. Values represent the means from three independent experiments.
TABLE 3.
Primers and probes used in this study
| Primer | Sequence (5′→3′) |
|---|---|
| agrA-F | CGCAACTGATAATATGAGGTGCTTGA |
| agrA-R | CAACTGGGTCATGCGAATTTCACTGC |
| clfB-F | GGTGGTGTAACTCTTGAATCGGAGTC |
| clfB-R | GGACTCAGACAGCGATTCAGATTCAG |
| cap5E-F | ATACGACAGAAGCGTAGAATCATTAG |
| cap5E-R | GTGTTGGCTTACACATATCGCCATC |
| hlb-F | AGCTACTCATCAACTGTTGCTG |
| hlb-R | GTTGCTATCATTATCGAATCCAC |
| Hld-F | GTTCACTGTGTCGATAATCCA |
| Hld-R | AGGAAGGAGTGATTTCAATGG |
| icaA-F | AAACTTGGTGCGGTTACAGG |
| icaA-R | GTAGCCAACGTCGACAACTG |
| psmB-F | TTATTTCAAAGGTGAGGGAGAGATTT |
| psmB-R | TTGTTGTGCAGCTTGCACAGT |
| saeR-F | AATACCATCATCAACCAGTT |
| saeR-R | CTCAAATTCCTTAATACGCATA |
| sigB-F | CTAAATCTTCGTGATGTGATTGTCG |
| sigB-R | AACCAATGGATTAAAGAACACCAAG |
| splA-F | AGGCGGAGGAAACTACGA |
| splA-R | ACTATCGCAAGGTCTTCT |
| vraSR-F | GGTGCAACGTTCCATATTGTATCATT |
| vraSR-R | GGCTTCAACTCATGGGCTTTGGCAA |
| walK-F | AAACAACTACAATCCCTTCATACTAA |
| walK-R | CTTGACGGTTGGCATACTCACTTAA |
| 16S-F | TCCGGAATTATTGGGCGTAA |
| 16S-R | CCACTTTCCTCTTCTGCACTCA |
| SaeR-Fw2 | TTGATATCATGGTACTTGATATCA |
| SaeR-Rv | CTCAAATTCCTTAATACGCATA |
Hemolysis assay.
Hemolytic activity was assayed as described previously (57). Briefly, bacteria were grown overnight in TSB at 37°C with shaking at 150 rpm. The OD600 was measured, and the OD of bacterial cultures was adjusted to the lowest value. Supernatants were filter sterilized (0.22 µm) and incubated with equal volumes of 2% RBC solution (Hardy Diagnostics, Santa Maria, CA) for 1 h at 37°C and 5% CO2. Cells were centrifuged at 13.000 rpm for 10 s. Released hemoglobin was measured by determining the absorbance at 540 nm. PBS and S. epidermidis Y1 were used as spontaneous hemolysis and nonhemolytic controls, respectively. Staphylococcusaureus subsp. aureus ATCC 29213 was used as the beta-hemolytic control. A standard curve was performed to determine the percentage of hemolysis. Independent experiments were performed in triplicate.
δ-Hemolysis assay.
Evaluation of δ-hemolysis production was used as an indirect test to determine agr functionality and was performed as described previously (58). Briefly, S. aureus RN4220 (beta-hemolytic) was streaked along the middle of a blood agar plate. Strains of interest were streaked perpendicular to RN4220 to determine presence or absence of δ-hemolysis.
Whole-genome sequencing.
Chromosomal DNA from staphylococcal strains grown in MH overnight at 37°C was prepared using the DNeasy Blood and Tissue kit (Qiagen). Library preparation and sequencing (MiSeq; Illumina) was performed by the Epigenetics and Genomics laboratory at Weill Cornell University, New York, NY. Genomes were assembled, annotated, and analyzed for nucleotide changes using Lasergene (v14) software (DNAStar, Madison, WI) and the PATRIC variation analysis service. The S. aureus N315 sequence (GenBank accession number BA000018; PATRIC ID 158879.11) was used as the reference sequence.
Secreted protein preparation and Western blot analysis.
Bacteria were grown in MH until reaching an OD600 of approximately 0.6. Then, the samples were centrifuged for 10 min at 4,000 rpm, and the supernatant was passed through 0.22-μm-pore-size membrane filters (Millex; Millipore Sigma, Burlington, MA). Samples were normalized by adjustment of the volume to equal the sample OD as previously reported (59). Samples were concentrated in Amicon 10,000-molecular-weight-cutoff centrifugal filters (Millipore Sigma) to a final volume of 40 μl. Ponceau staining was used as load control.
For Western blot analysis, 20 μg of proteins of each sample was loaded and separated using 4 to 12% SDS-PAGE electrophoresis gradient gels (ThermoFisher, Carlsbad, CA), after which they were blot transferred onto pure nitrocellulose blotting membranes (Fisher Scientific, Hampton, NH). The membranes were blocked using 5% low-fat milk in PBS. Alpha-toxin was probed with a polyclonal anti-alpha antibody (Millipore Sigma) at a 1/2,000 dilution, followed by incubation with a secondary goat anti-rabbit IgG(H+L) antibody at a 1/5,000 dilution. Protein bands were developed in autoradiography films (Denville Scientific Inc., South Plainfield, NJ).
Murine sepsis model.
A septicemia mouse model was used to determine the role of DAP susceptibility in S. aureus pathogenicity. Groups of six SCID Beige mice (Envigo, Houston, TX) were used. Each group (n = 6) was inoculated via tail injection with ∼1 × 107 to 2 × 107 CFU of either the DAPs CB5013 or DAPr CB5014 MRSA strain and mutant CB5014ΔvraSR and transcomplemented CB5014ΔvraSR+vraSR MRSA strains grown in TSB at 37°C, 150 rpm. Mice were euthanized at 72 h postinfection. Kidneys and spleen were collected aseptically, homogenized with a homogenizer (150 Homogenizer; Fisher Scientific), serially diluted in PBS, and plated onto TSA plates to determine the number of viable staphylococci. Results were expressed as the logarithm of CFU per gram of organ (log CFU/g).
Animal ethics statement.
All animal studies were approved by the Institutional Animal Care and Use Committee of the Houston Methodist Research Institute. To ensure protection and proper manipulation of animals, experiments were performed by trained personnel at the animal facility of the Houston Methodist Research Institute.
Statistical analysis.
The statistical analysis was performed using GraphPad Prism 7 software. The log rank test was used to assess significant differences (P < 0.05) for Kaplan-Meier survival curves. The unpaired Student t test was used to determine significant differences (P < 0.05) for adhered and internalized bacteria in cell culture assays, log CFU/g in the in vivo mouse sepsis model, and the mRNA expression of the CB1631 and CB1634 strains from the qRT-PCR analysis. One-way ANOVA (P < 0.05) was used to evaluate the significance of differences in hemolysis percentage and the mRNA expression of the CB5013 and CB5014 strains from the qRT-PCR analysis. To test the normality and homoscedasticity assumptions of the ANOVA, a Shapiro-Wilk test (P < 0.05) and a Brown-Forsythe test (P < 0.05), respectively, were performed. As an a posteriori analysis, the Bonferroni multiple-comparison test (P < 0.05) was performed.
ACKNOWLEDGMENTS
This study was funded in part by Merck (formerly Cubist Pharmaceuticals), Lexington, MA, and by an NIH grant (NIH-R56AI102503-01A1) to A. E. Rosato.
We thank the Epigenomic Core of Weill Cornell Medical College for their WGS service, Maria P. Martinez for her help with RNA-seq experiments, and Melanie Roch for her fruitful discussions.
REFERENCES
- 1.Horn J, Stelzner K, Rudel T, Fraunholz M. 2018. Inside job: Staphylococcus aureus host-pathogen interactions. Int J Med Microbiol 308:607–624. doi: 10.1016/j.ijmm.2017.11.009. [DOI] [PubMed] [Google Scholar]
- 2.Gordon RJ, Lowy FD. 2008. Pathogenesis of methicillin-resistant Staphylococcus aureus infection. Clin Infect Dis 46(Suppl 5):S350–S359. doi: 10.1086/533591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu C, Bayer A, Cosgrove SE, Daum RS, Fridkin SK, Gorwitz RJ, Kaplan SL, Karchmer AW, Levine DP, Murray BE, Rybak J, Talan DA, Chambers HF. 2011. Clinical practice guidelines by the Infectious Diseases Society of America for the treatment of methicillin-resistant Staphylococcus aureus infections in adults and children: executive summary. Clin Infect Dis 52:285–292. doi: 10.1093/cid/cir034. [DOI] [PubMed] [Google Scholar]
- 4.Miller WR, Bayer AS, Arias CA. 2016. Mechanism of action and resistance to daptomycin in Staphylococcus aureus and enterococci. Cold Spring Harb Perspect Med 6:a026997. doi: 10.1101/cshperspect.a026997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Foster TJ. 2017. Antibiotic resistance in Staphylococcus aureus. Current status and future prospects. FEMS Microbiol Rev 41:430–449. doi: 10.1093/femsre/fux007. [DOI] [PubMed] [Google Scholar]
- 6.Capone A, Cafiso V, Campanile F, Parisi G, Mariani B, Petrosillo N, Stefani S. 2016. In vivo development of daptomycin resistance in vancomycin-susceptible methicillin-resistant Staphylococcus aureus severe infections previously treated with glycopeptides. Eur J Clin Microbiol Infect Dis 35:625–631. doi: 10.1007/s10096-016-2581-4. [DOI] [PubMed] [Google Scholar]
- 7.Gómez Casanova N, Siller Ruiz M, Muñoz Bellido JL. 2017. Mechanisms of resistance to daptomycin in Staphylococcus aureus. Rev Esp Quimioter 30:391–396. [PubMed] [Google Scholar]
- 8.Stefani S, Campanile F, Santagati M, Mezzatesta ML, Cafiso V, Pacini G. 2015. Insights and clinical perspectives of daptomycin resistance in Staphylococcus aureus: a review of the available evidence. Int J Antimicrob Agents 46:278–289. doi: 10.1016/j.ijantimicag.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 9.Seaton RA, Menichetti F, Dalekos G, Beiras-Fernandez A, Nacinovich F, Pathan R, Hamed K. 2015. Evaluation of effectiveness and safety of high-dose daptomycin: results from patients included in the European Cubicin((R)) outcomes registry and experience. Adv Ther 32:1192–1205. doi: 10.1007/s12325-015-0267-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baltz RH. 2009. Daptomycin: mechanisms of action and resistance, and biosynthetic engineering. Curr Opin Chem Biol 13:144–151. doi: 10.1016/j.cbpa.2009.02.031. [DOI] [PubMed] [Google Scholar]
- 11.Mehta S, Singh C, Plata KB, Chanda PK, Paul A, Riosa S, Rosato RR, Rosato AE. 2012. Beta-lactams increase the antibacterial activity of daptomycin against clinical MRSA strains and prevent selection of DAP-resistant derivatives. Antimicrob Agents Chemother 56:6192–6200. doi: 10.1128/AAC.01525-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tran TT, Munita JM, Arias CA. 2015. Mechanisms of drug resistance: daptomycin resistance. Ann N Y Acad Sci 1354:32–53. doi: 10.1111/nyas.12948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones T, Yeaman MR, Sakoulas G, Yang SJ, Proctor RA, Sahl HG, Schrenzel J, Xiong YQ, Bayer AS. 2008. Failures in clinical treatment of Staphylococcus aureus infection with daptomycin are associated with alterations in surface charge, membrane phospholipid asymmetry, and drug binding. Antimicrob Agents Chemother 52:269–278. doi: 10.1128/AAC.00719-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patel D, Husain M, Vidaillac C, Steed ME, Rybak MJ, Seo SM, Kaatz GW. 2011. Mechanisms of in-vitro-selected daptomycin-non-susceptibility in Staphylococcus aureus. Int J Antimicrob Agents 38:442–446. doi: 10.1016/j.ijantimicag.2011.06.010. [DOI] [PubMed] [Google Scholar]
- 15.Ernst CM, Staubitz P, Mishra NN, Yang SJ, Hornig G, Kalbacher H, Bayer AS, Kraus D, Peschel A. 2009. The bacterial defensin resistance protein MprF consists of separable domains for lipid lysinylation and antimicrobial peptide repulsion. PLoS Pathog 5:e1000660. doi: 10.1371/journal.ppat.1000660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mishra NN, Bayer AS, Weidenmaier C, Grau T, Wanner S, Stefani S, Cafiso V, Bertuccio T, Yeaman MR, Nast CC, Yang SJ. 2014. Phenotypic and genotypic characterization of daptomycin-resistant methicillin-resistant Staphylococcus aureus strains: relative roles of mprF and dlt operons. PLoS One 9:e107426. doi: 10.1371/journal.pone.0107426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Camargo IL, Neoh HM, Cui L, Hiramatsu K. 2008. Serial daptomycin selection generates daptomycin-nonsusceptible Staphylococcus aureus strains with a heterogeneous vancomycin-intermediate phenotype. Antimicrob Agents Chemother 52:4289–4299. doi: 10.1128/AAC.00417-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaupp R, Lei S, Reed JM, Peisker H, Boyle-Vavra S, Bayer AS, Bischoff M, Herrmann M, Daum RS, Powers R, Somerville GA. 2015. Staphylococcus aureus metabolic adaptations during the transition from a daptomycin susceptibility phenotype to a daptomycin nonsusceptibility phenotype. Antimicrob Agents Chemother 59:4226–4238. doi: 10.1128/AAC.00160-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mishra NN, Yang SJ, Sawa A, Rubio A, Nast CC, Yeaman MR, Bayer AS. 2009. Analysis of cell membrane characteristics of in vitro-selected daptomycin-resistant strains of methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 53:2312–2318. doi: 10.1128/AAC.01682-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen CJ, Huang YC, Chiu CH. 2015. Multiple pathways of cross-resistance to glycopeptides and daptomycin in persistent MRSA bacteraemia. J Antimicrob Chemother 70:2965–2972. doi: 10.1093/jac/dkv225. [DOI] [PubMed] [Google Scholar]
- 21.Friedman L, Alder JD, Silverman JA. 2006. Genetic changes that correlate with reduced susceptibility to daptomycin in Staphylococcus aureus. Antimicrob Agents Chemother 50:2137–2145. doi: 10.1128/AAC.00039-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang SJ, Mishra NN, Rubio A, Bayer AS. 2013. Causal role of single nucleotide polymorphisms within the mprF gene of Staphylococcus aureus in daptomycin resistance. Antimicrob Agents Chemother 57:5658–5664. doi: 10.1128/AAC.01184-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berscheid A, Francois P, Strittmatter A, Gottschalk G, Schrenzel J, Sass P, Bierbaum G. 2014. Generation of a vancomycin-intermediate Staphylococcus aureus (VISA) strain by two amino acid exchanges in VraS. J Antimicrob Chemother 69:3190–3198. doi: 10.1093/jac/dku297. [DOI] [PubMed] [Google Scholar]
- 24.Su J, Iehara M, Yasukawa J, Matsumoto Y, Hamamoto H, Sekimizu K. 2015. A novel mutation in the vraS gene of Staphylococcus aureus contributes to reduce susceptibility against daptomycin. J Antibiot (Tokyo) 68:646–648. doi: 10.1038/ja.2015.42. [DOI] [PubMed] [Google Scholar]
- 25.Abdelraouf K, Kabbara S, Ledesma KR, Poole K, Tam VH. 2011. Effect of multidrug resistance-conferring mutations on the fitness and virulence of Pseudomonas aeruginosa. J Antimicrob Chemother 66:1311–1317. doi: 10.1093/jac/dkr105. [DOI] [PubMed] [Google Scholar]
- 26.Martínez-Ramos I, Mulet X, Moyá B, Barbier M, Oliver A, Albertí S. 2014. Overexpression of MexCD-OprJ reduces Pseudomonas aeruginosa virulence by increasing its susceptibility to complement-mediated killing. Antimicrob Agents Chemother 58:2426–2429. doi: 10.1128/AAC.02012-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sánchez P, Linares JF, Ruiz-Díez B, Campanario E, Navas A, Baquero F, Martínez JL. 2002. Fitness of in vitro selected Pseudomonas aeruginosa nalB and nfxB multidrug resistant mutants. J Antimicrob Chemother 50:657–664. doi: 10.1093/jac/dkf185. [DOI] [PubMed] [Google Scholar]
- 28.Hraiech S, Roch A, Lepidi H, Atieh T, Audoly G, Rolain JM, Raoult D, Brunel JM, Papazian L, Bregeon F. 2013. Impaired virulence and fitness of a colistin-resistant clinical isolate of Acinetobacter baumannii in a rat model of pneumonia. Antimicrob Agents Chemother 57:5120–5121. doi: 10.1128/AAC.00700-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.López-Rojas R, Domínguez-Herrera J, McConnell MJ, Docobo-Peréz F, Smani Y, Fernández-Reyes M, Rivas L, Pachón J. 2011. Impaired virulence and in vivo fitness of colistin-resistant Acinetobacter baumannii. J Infect Dis 203:545–548. doi: 10.1093/infdis/jiq086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lopez-Rojas R, McConnell MJ, Jimenez MME, Dominguez-Herrera J, Fernandez-Cuenca F, Pachon J. 2013. Colistin resistance in a clinical Acinetobacter baumannii strain appearing after colistin treatment: effect on virulence and bacterial fitness. Antimicrob Agents Chemother 57:4587–4589. doi: 10.1128/AAC.00543-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Da Silva GJ, Mendonça N. 2012. Association between antimicrobial resistance and virulence in Escherichia coli. Virulence 3:18–28. doi: 10.4161/viru.3.1.18382. [DOI] [PubMed] [Google Scholar]
- 32.Hennequin C, Robin F. 2016. Correlation between antimicrobial resistance and virulence in Klebsiella pneumoniae. Eur J Clin Microbiol Infect Dis 35:333–341. doi: 10.1007/s10096-015-2559-7. [DOI] [PubMed] [Google Scholar]
- 33.Kidd TJ, Mills G, Sa-Pessoa J, Dumigan A, Frank CG, Insua JL, Ingram R, Hobley L, Bengoechea JA. 2017. A Klebsiella pneumoniae antibiotic resistance mechanism that subdues host defences and promotes virulence. EMBO Mol Med 9:430–447. doi: 10.15252/emmm.201607336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Geisinger E, Isberg RR. 2017. Interplay between antibiotic resistance and virulence during disease promoted by multidrug-resistant bacteria. J Infect Dis 215:S9–S17. doi: 10.1093/infdis/jiw402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Majcherczyk PA, Barblan JL, Moreillon P, Entenza JM. 2008. Development of glycopeptide-intermediate resistance by Staphylococcus aureus leads to attenuated infectivity in a rat model of endocarditis. Microb Pathog 45:408–414. doi: 10.1016/j.micpath.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 36.Peleg AY, Jara S, Monga D, Eliopoulos GM, Moellering RC Jr, Mylonakis E. 2009. Galleria mellonella as a model system to study Acinetobacter baumannii pathogenesis and therapeutics. Antimicrob Agents Chemother 53:2605–2609. doi: 10.1128/AAC.01533-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qin L, McCausland JW, Cheung GY, Otto M. 2016. PSM-mec—a virulence determinant that connects transcriptional regulation, virulence, and antibiotic resistance in staphylococci. Front Microbiol 7:1293. doi: 10.3389/fmicb.2016.01293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Renzoni A, Kelley WL, Rosato RR, Martinez MP, Roch M, Fatouraei M, Haeusser DP, Margolin W, Fenn S, Turner RD, Foster SJ, Rosato AE. 2017. Molecular bases determining daptomycin resistance-mediated resensitization to beta-lactams (seesaw effect) in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 61:e01634-16. doi: 10.1128/AAC.01634-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Novick RP, Ross HF, Projan SJ, Kornblum J, Kreiswirth B, Moghazeh S. 1993. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J 12:3967–3975. doi: 10.1002/j.1460-2075.1993.tb06074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pader V, Hakim S, Painter KL, Wigneshweraraj S, Clarke TB, Edwards AM. 2016. Staphylococcus aureus inactivates daptomycin by releasing membrane phospholipids. Nat Microbiol 2:16194. doi: 10.1038/nmicrobiol.2016.194. [DOI] [PubMed] [Google Scholar]
- 41.Mehta S, Cuirolo AX, Plata KB, Riosa S, Silverman JA, Rubio A, Rosato RR, Rosato AE. 2012. VraSR two-component regulatory system contributes to mprF-mediated decreased susceptibility to daptomycin in-vivo-selected MRSA clinical strains. Antimicrob Agents Chemother 56:92–102. doi: 10.1128/AAC.00432-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cameron DR, Lin YH, Trouillet-Assant S, Tafani V, Kostoulias X, Mouhtouris E, Skinner N, Visvanathan K, Baines SL, Howden B, Monk IR, Laurent F, Stinear TP, Howden BP, Peleg AY. 2017. Vancomycin-intermediate Staphylococcus aureus isolates are attenuated for virulence when compared with susceptible progenitors. Clin Microbiol Infect 23:767–773. doi: 10.1016/j.cmi.2017.03.027. [DOI] [PubMed] [Google Scholar]
- 43.Dai Y, Chang W, Zhao C, Peng J, Xu L, Lu H, Zhou S, Ma X. 2017. VraR binding to the promoter region of agr inhibits its function in vancomycin-intermediate Staphylococcus aureus (VISA) and heterogeneous VISA. Antimicrob Agents Chemother 61:e02740-16. doi: 10.1128/AAC.02740-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tuchscherr L, Bischoff M, Lattar SM, Noto Llana M, Pförtner H, Niemann S, Geraci J, Van de Vyver H, Fraunholz MJ, Cheung AL, Herrmann M, Völker U, Sordelli DO, Peters G, Löffler B. 2015. Sigma factor SigB is crucial to mediate Staphylococcus aureus adaptation during chronic infections. PLoS Pathog 11:e1004870. doi: 10.1371/journal.ppat.1004870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cameron DR, Mortin LI, Rubio A, Mylonakis E, Moellering RC Jr, Eliopoulos GM, Peleg AY. 2015. Impact of daptomycin resistance on Staphylococcus aureus virulence. Virulence 6:127–131. doi: 10.1080/21505594.2015.1011532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Garcia-de-la-Maria C, Xiong YQ, Pericas JM, Armero Y, Moreno A, Mishra NN, Rybak MJ, Tran TT, Arias CA, Sullam PM, Bayer AS, Miro JM. 2017. Impact of high-level daptomycin resistance in the Streptococcus mitis group on virulence and survivability during daptomycin treatment in experimental infective endocarditis. Antimicrob Agents Chemother 61:e02418-16. doi: 10.1128/AAC.02418-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li S, Yin Y, Chen H, Wang Q, Wang X, Wang H. 2017. Fitness cost of daptomycin-resistant Staphylococcus aureus obtained from in vitro daptomycin selection pressure. Front Microbiol 8:2199. doi: 10.3389/fmicb.2017.02199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roch M, Gagetti P, Davis J, Ceriana P, Errecalde L, Corso A, Rosato AE. 2017. Daptomycin resistance in clinical MRSA strains is associated with a high biological fitness cost. Front Microbiol 8:2303. doi: 10.3389/fmicb.2017.02303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chang P, Li W, Shi G, Li H, Yang X, Xia Z, Ren Y, Li Z, Chen H, Bei W. 2018. The VraSR regulatory system contributes to virulence in Streptococcus suis via resistance to innate immune defenses. Virulence 9:771–782. doi: 10.1080/21505594.2018.1428519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.CLSI. 2015. M100-S25, performance standards for antimicrobial susceptibility testing, 25th informational supplement. CLSI, Wayne, PA. [Google Scholar]
- 51.Lee CY, Buranen SL, Ye ZH. 1991. Construction of single-copy integration vectors for Staphylococcus aureus. Gene 103:101–105. doi: 10.1016/0378-1119(91)90399-V. [DOI] [PubMed] [Google Scholar]
- 52.Novick R. 1967. Properties of a cryptic high-frequency transducing phage in Staphylococcus aureus. Virology 33:155–166. doi: 10.1016/0042-6822(67)90105-5. [DOI] [PubMed] [Google Scholar]
- 53.Plata KB, Rosato RR, Rosato AE. 2011. Fate of mutation rate depends on agr locus expression during oxacillin-mediated heterogeneous-homogeneous selection in methicillin-resistant Staphylococcus aureus clinical strains. Antimicrob Agents Chemother 55:3176–3186. doi: 10.1128/AAC.01119-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kuroda M, Kuroda H, Oshima T, Takeuchi F, Mori H, Hiramatsu K. 2003. Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. Mol Microbiol 49:807–821. [DOI] [PubMed] [Google Scholar]
- 55.Desbois AP, Coote PJ. 2011. Wax moth larva (Galleria mellonella): an in vivo model for assessing the efficacy of antistaphylococcal agents. J Antimicrob Chemother 66:1785–1790. doi: 10.1093/jac/dkr198. [DOI] [PubMed] [Google Scholar]
- 56.Wattam AR, Davis JJ, Assaf R, Boisvert S, Brettin T, Bun C, Conrad N, Dietrich EM, Disz T, Gabbard JL, Gerdes S, Henry CS, Kenyon RW, Machi D, Mao C, Nordberg EK, Olsen GJ, Murphy-Olson DE, Olson R, Overbeek R, Parrello B, Pusch GD, Shukla M, Vonstein V, Warren A, Xia F, Yoo H, Stevens RL. 2017. Improvements to PATRIC, the all-bacterial Bioinformatics Database and Analysis Resource Center. Nucleic Acids Res 45:D535–D542. doi: 10.1093/nar/gkw1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dean MA, Olsen RJ, Long SW, Rosato AE, Musser JM. 2014. Identification of point mutations in clinical Staphylococcus aureus strains that produce small-colony variants auxotrophic for menadione. Infect Immun 82:1600–1605. doi: 10.1128/IAI.01487-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Adhikari RP, Arvidson S, Novick RP. 2007. A nonsense mutation in agrA accounts for the defect in agr expression and the avirulence of Staphylococcus aureus 8325-4 traP::kan. Infect Immun 75:4534–4540. doi: 10.1128/IAI.00679-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jousselin A, Renzoni A, Andrey DO, Monod A, Lew DP, Kelley WL. 2012. The posttranslocational chaperone lipoprotein PrsA is involved in both glycopeptide and oxacillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother 56:3629–3640. doi: 10.1128/AAC.06264-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kuroda M, Ohta T, Uchiyama I, Baba T, Yuzawa H, Kobayashi I, Cui L, Oguchi A, Aoki K, Nagai Y, Lian J, Ito T, Kanamori M, Matsumaru H, Maruyama A, Murakami H, Hosoyama A, Mizutani-Ui Y, Takahashi NK, Sawano T, Inoue R, Kaito C, Sekimizu K, Hirakawa H, Kuhara S, Goto S, Yabuzaki J, Kanehisa M, Yamashita A, Oshima K, Furuya K, Yoshino C, Shiba T, Hattori M, Ogasawara N, Hayashi H, Hiramatsu K. 2001. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet 357:1225–1240. doi: 10.1016/S0140-6736(00)04403-2. [DOI] [PubMed] [Google Scholar]
- 61.Safdar N, Andes D, Craig WA. 2004. In vivo pharmacodynamic activity of daptomycin. Antimicrob Agents Chemother 48:63–68. doi: 10.1128/AAC.48.1.63-68.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Duthie ES, Lorenz LL. 1952. Staphylococcal coagulase; mode of action and antigenicity. J Gen Microbiol 6:95–107. doi: 10.1099/00221287-6-1-2-95. [DOI] [PubMed] [Google Scholar]