Abstract
Molecular tools are commonly directed at refining taxonomies and the species that constitute their fundamental units. This has been especially insightful for groups for which species hypotheses are ambiguous and have largely been based on morphological differences between certain life stages or sexes, and has added importance when taxa are a focus of conservation efforts. Here, we examine the taxonomic status of Arsapnia arapahoe, a winter stonefly in the family Capniidae that is a species of conservation concern because of its limited abundance and restricted range in northern Colorado, USA. Phylogenetic analyses of sequences of mitochondrial and nuclear genes of this and other capniid stoneflies from this region and elsewhere in western North America indicated extensive haplotype sharing, limited genetic differences, and a lack of reciprocal monophyly between A. arapahoe and the sympatric A. decepta, despite distinctive and consistent morphological differences in the sexual apparatus of males of both species. Analyses of autosomal and sex‐linked single nucleotide polymorphisms detected using genotyping by sequencing indicated that all individuals of A. arapahoe consisted of F1 hybrids between female A. decepta and males of another sympatric stonefly, Capnia gracilaria. Rather than constitute a self‐sustaining evolutionary lineage, A. arapahoe appears to represent the product of nonintrogressive hybridization in the limited area of syntopy between two widely distributed taxa. This offers a cautionary tale for taxonomists and conservation biologists working on the less‐studied components of the global fauna.
Keywords: cryptic taxa, ESA, GBS, hybrid zone, nonintrogressive hybridization, stonefly
1. INTRODUCTION
Within biology, arguably the most fundamental undertaking is that of defining a species. Proposals on what constitutes a species are legion; although the particulars vary depending on which of the many forms of evidence are used to delineate them—morphological, genetic, ecological, reproductive, geographical, or some combination—there is consensus that species constitute independently evolving lineages to which are assigned names (De Queiroz, 2007). Because all species are hypotheses, a species name is no more than the label for a particular hypothesis. But given the central role of species to biology, these names profoundly influence how we think about the elements and conservation of biodiversity.
Although most taxonomies of organisms are based on morphological characters, genetic tools are essential to refining these taxonomies by parsing genotypic variation across demographic, geographic, and taxonomic scales, discerning recent and ancient introgression, revealing cryptic taxa, and synonymizing dubious ones (Kjer, Simon, Yavorskaya, & Beutel, 2016). Perhaps most straightforward has been the genetic assignment of specimens to known lineages by linking their genotypes to those of named and phenotypically identified voucher specimens. This is the province of DNA barcoding—assigning individuals to species based on their genetic sequences, generally of the cytochrome c oxidase subunit 1 (COI) mitochondrial region—and it has been applied to taxa across all of life, with an emphasis on animals (Hebert, Cywinska, & Ball, 2003). One of the first demonstrations of its efficacy involved insects (Hebert, Penton, Burns, Janzen, & Hallwachs, 2004), and they have been the focus of initiatives to associate DNA barcodes with individual species across higher taxonomic categories and throughout particular geographies (Smith et al., 2008; Zhou et al., 2016). These efforts have been broadly successful (Blagoev et al., 2016; Webb et al., 2012), in part because they permit linking morphologically cryptic larvae or females with their more easily recognized adult male counterparts (Gamboa & Monaghan, 2014). For some groups of insects, however, these broad assessments are in their infancy and concordance between morphological and genetic identifications of forms has been uneven (Geiger et al., 2016). In part, this reflects taxonomies erected on morphological grounds that have not yet been evaluated from a molecular perspective (Schlick‐Steiner et al., 2010). This pattern also derives, however, from discord in the phylogenetic signal among different genes, especially for taxa of recent origin or exposed to introgressive hybridization (Funk & Omland, 2003). Taxonomic instability is expected given that all names are hypotheses about evolutionary lineages, and their revision is straightforward, albeit nontrivial, from a scientific perspective (Valdecasas, Williams, & Wheeler, 2008). But getting the names right is more than an academic exercise; society makes outsize investments in some of these hypotheses, particularly when they are associated with at‐risk taxa that are the focus of conservation (Mace, 2004; Schwartz & Boness, 2017).
An exemplar of many these issues is the Arapahoe snowfly, Arsapnia arapahoe Nelson & Kondratieff (Plecoptera: Capniidae), one of eight species of stoneflies in the western North American genus Arsapnia that were formerly assigned to the Capnia decepta Banks group (Murányi, Gamboa, & Orci, 2014; Nelson & Baumann, 1989). This species was originally described from single male specimens collected in two streams in 1986 and 1987 in the Cache la Poudre River basin in north‐central Colorado (Nelson & Kondratieff, 1988), and not observed again in these streams until March 2009, when the first putative females were also collected (Heinold & Kondratieff, 2010). More recent surveys for adults extended this distribution to 19 additional sites as far south as the South Platte River basin in the Colorado Front Range (Fairchild, Belcher, Zuellig, Vieira, & Kondratieff, 2017); larval forms remain unknown. Where present, this species was outnumbered by orders of magnitude by two sympatric capniids, A. decepta and Capnia gracilaria Claassen (Fairchild et al., 2017; Heinold, Gill, & Belcher, 2014). Its restricted range and apparent rarity led to a petition and subsequent candidacy for its listing under the U.S. Endangered Species Act (U.S. Fish and Wildlife Service, 2012).
The life history, morphology, and systematics of capniid stoneflies, however, make assessing the conservation status of many species, including A. arapahoe, a challenge. Members of this family tend to emerge as adults in late winter and early spring and can be locally abundant (Baumann, Gaufin, & Surdick, 1977), but the mating period is brief and synchronized at any particular location. Larvae apparently occupy hyporheic habitats that make them difficult to capture in benthic surveys, and this life stage can rarely be identified to species (Stewart & Stark, 1988). This is also true of adult females, for example, differences between A. arapahoe and A. decepta are thought to be subtle at best (Heinold & Kondratieff, 2010), and females of different species in this genus may be easily mistaken for one another and for females in confamilial taxa (Nelson & Baumann, 1989). Even male identification can be problematic. The shape, size, and ornamentation of the male reproductive organ often constitutes the basis for describing and identifying species, yet this structure can exhibit substantial local and range‐wide variation within taxa (Baumann & Stark, 2017). Male A. arapahoe, however, are readily identifiable because the epiproct lacks the mesal bulbous projections typical of all other members of this genus (Nelson & Kondratieff, 1988). This characteristic is so distinctive that it led Nelson and Kondratieff (1988) to speculate that A. arapahoe was the sister taxon to all other members of Arsapnia, and perhaps most closely related to A. sequoia Nelson & Baumann and A. utahensis Gaufin & Jewett, not the sympatric A. decepta. The position of these species within Capniidae is also ambiguous; systematists have long regarded Capniidae as a synthetic, paraphyletic assemblage in need of revision (Murányi et al., 2014; Nelson & Baumann, 1989).
Confusion about the taxonomic position of A. arapahoe grew when an attempt to use genetic tools to identify it (Heinold, Gill, Belcher, & Verdone, 2014) produced unexpected results: The interspecific distance between sympatric male A. arapahoe and A. decepta (0.10%) was typical of variation found within species of stoneflies (0.35%–0.53%; Gill et al., 2013; Morinière et al., 2017; Zhou, Jacobus, DeWalt, Adamowicz, & Hebert, 2010, but see Gill, Sandberg, & Kondratieff, 2015 for much higher estimates), not between species (generally ≥2%; Zhou, Adamowicz, Jacobus, DeWalt, & Hebert, 2009). Paradoxically, a putative female allotype of A. arapahoe shared the haplotype of a distantly related species in a different genus, Capnura wanica. The incidence of interspecific haplotype sharing and low interspecific divergence is rare in many arthropods (e.g., <2% among Canadian spiders (Blagoev et al., 2016) and virtually nonexistent in North American mayflies (Webb et al., 2012)), making assignment of different sexes to different species highly unusual and difficult to ascribe to a single mechanism. More likely is that a combination of factors is responsible, among them misidentification of voucher specimens, incomplete lineage sorting between recently diverged species, interspecific haplotype sharing caused by infection by the bacterium Wolbachia, or hybridization (Funk & Omland, 2003; Smith et al., 2012). Heinold et al. (2014) favored incomplete lineage sorting, arguing that the dramatic morphological differences between males were reliable and obvious evidence of speciation, haplotypes were not those of the markedly divergent Wolbachia sequences (and selective sweeps driven by Wolbachia infection are extraordinarily rare; Smith et al., 2012), and hybridization was not likely because of the lack of morphological intermediates between male A. arapahoe and A. decepta. But this interpretation did not account for the differences among male and female genotypes and failed to satisfy the requirement that an integrative taxonomy provides an evolutionary explanation for all aspects of morphological and molecular discord (Schlick‐Steiner et al., 2010).
A simple explanation for haplotype sharing among these taxa would be hybridization, but this phenomenon has been regarded as rare among stoneflies and other aquatic arthropods (Dijkstra, Monaghan, & Pauls, 2014; Hughes, Finn, Monaghan, Schultheis, & Sweeney, 2014), and until recently was thought to be confined to a few species pairs of eastern North American Allocapnia (Ross & Ricker, 1971). That hybridization might be more prevalent is discouraged by the biological species concept with its predisposition to view hybridization as a rare accident (Mallet, 2005) and by notions that behavioral or anatomical differences constitute intrinsic reproductive barriers. For example, conspecific drumming signals used by male and female stoneflies for mate recognition are regarded as species‐specific and likely to ensure pre‐zygotic reproductive isolation (Boumans & Johnsen, 2014; Stewart, 2001). Nonetheless, there are many examples of arthropod taxa with elaborate pre‐mating and putatively species‐specific displays that nonetheless result in attempted mating between species (Masly, 2012), including between stoneflies in different genera (Zeigler, 1990). Even if hybridization does not ensue, assuming species identity based on temporary male–female association could lead to misidentification of the less morphologically distinct females. There may be a stronger argument for the rarity of hybridization based on the “lock and key” hypothesis (Sota & Kubota, 1998), which posits that anatomical complementarity of male and female terminalia is required for successful reproduction, but again there are a host of examples demonstrating hybridization between anatomically disparate taxa (Shapiro & Porter, 1989). Nonetheless, the success of heterospecific crosses may be asymmetric because of pre‐ or post‐zygotic reproductive incompatibilities, that is, crosses between a male of one species and a female of the other may exhibit lower female survival, likelihood of insemination, or fitness of offspring than does the opposite pairing (Masly, 2012).
Our goal was to resolve the taxonomic ambiguity surrounding A. arapahoe via an iterative approach to integrative taxonomy (Yeates et al., 2011). We treated the morphological identifications as authoritative hypotheses to be evaluated in light of molecular data from A. arapahoe and related capniid stoneflies. To that end, we analyzed sequences of two mitochondrial regions and one nuclear gene. Because these results were inconclusive, we used genotyping by sequencing to more thoroughly explore the evolutionary origin and taxonomic validity of A. arapahoe.
2. MATERIALS AND METHODS
2.1. Sample collection and sequence selection
Specimens for sequencing were collected for this study or drawn from the collections at the C.P. Gillette Museum of Arthropod Diversity, Colorado State University, Fort Collins, Colorado (Figure 1; Supporting Information Table S1). All individuals were identified to species by taxonomic experts at this facility. Furthermore, we re‐examined every specimen under a dissecting microscope to confirm the sex of the individuals being genetically sequenced, and of all A. arapahoe specimens to ensure they were of this species. We examined specimens of: (a) A. arapahoe from across its northern Colorado range; (b) A. decepta from northern Colorado and Arizona; (c) additional members of the genus Arsapnia; and (d) other capniids from the range of A. arapahoe (e.g., Capnia gracilaria, Capnura wanica Frison) and elsewhere in western North America. These samples were supplemented with sequences of other specimens from these groups that were available in public databases (Supporting Information Table S2). There are two caveats. First, not all members of all groups were used in every analysis because of cost and because genotyping was not universally successful. Second, because our phylogenetic analyses (see below) revealed that polytomies were evident throughout the broad suite of species in different genera of Capniidae, we were uncertain as to which species might be relevant to understanding the phylogenetic identity of A. arapahoe. To that end, our phylogenetic analyses using two mitochondrial genes and one nuclear gene (Supporting Information Figures S2 and S3) of western North American capniid stoneflies identified a monophyletic clade that consisted of all species of the genus Arsapnia (A. arapahoe, A. coyote Nelson & Baumann, A. decepta, A. pileata Jewett, A. sequoia, A. teresa Claassen, A. tumida Claassen, and A. utahensis), two species of Sierracapnia (S. barberi Claassen and S. palomar Nelson & Baumann 1987), and three species of Capnia (C. elongata Claassen, C. gracilaria, and C. promota Frison), and we restrict presentation of our results to these taxa (hereafter, Arsapnia group), except where references to additional taxa are pertinent.
Figure 1.
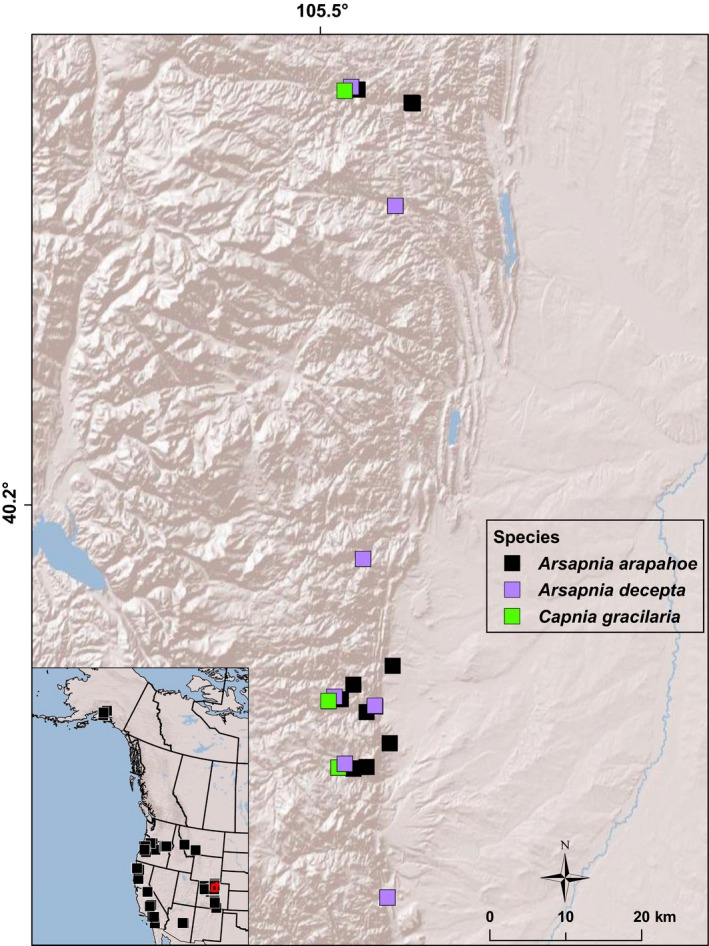
Locations of all specimens of capniid stoneflies examined in this study (lower left) and along the northern Colorado (USA) Front Range (inset area in red, expanded)
2.2. DNA sequencing
We used the QIAGEN DNeasy Blood and Tissue kit (QIAGEN Inc.) to extract genomic DNA from whole hind legs or the thorax of individual specimens, following the manufacturer's instructions for tissue. We sequenced COI and cytochrome b (cyt b) to facilitate comparisons with sequences in existing databases (Ratnasingham & Hebert, 2007) and to increase the taxonomic resolution of the mitochondrial data (Hillis, Pollock, McGuire, & Zwickl, 2003). We sequenced the ribosomal internal transcribed spacer (ITS1) to permit comparison of a nuclear phylogeny to those from the mitochondrial genes and to assess whether hybridization or sharing of mitochondrial genes associated with Wolbachia infection were evident. We amplified COI using the standard primers (LCO1490/HCO2198 (Folmer, Black, Hoeh, Lutz, & Vrijenhoek, 1994) or LepF1/LepR1 (Hebert et al., 2004)), cyt b using primers developed by Jordan et al. (2016), and ITS1 using primers developed by Pilgrim, Roush, and Krane (2002). We used reaction volumes of 50 μl containing 50–100 ng DNA, 1 × reaction buffer (Life Technologies), 2.5 mm MgCl2, 200 µm each dNTP, 1 µm each primer, and 1 U Taq polymerase (Life Technologies). The PCR program was 94°C/5 min, [94°C/1 min, 55°C/1 min, 72°C/1 min 30 s] × 34 cycles, and 72°C/5 min. The quality and quantity of template DNA were determined by 1.6% agarose gel electrophoresis. Products from PCR were purified with Exo‐Sap‐IT (Affymetrix‐USB Corporation) according to the manufacturer's instructions.
These PCR products were sequenced at Eurofins Genomics (Louisville, KY). Sequences for COI and cyt b were initially aligned with Sequencher (Gene Codes Corp.), with minor manual adjustments. Mitochondrial sequences were also translated to inspect them for ambiguous amino acids or stop codons; none were observed. The ITS1 sequences were aligned in the online version of MAFFT 7 (Katoh, Rozewicki, & Yamada, 2017) under the Q‐INS‐i algorithm with the default settings. Gaps were coded as characters following the simple method of Simmons and Ochoterena (2000) and appended to the nucleotide sequences using FastGap 1.2 (Borchsenius, 2009).
2.3. Genotyping by sequencing
To produce a much larger dataset from across the nuclear genome to refine our understanding of the identity of A. arapahoe, we used tunable genotyping by sequencing (GBS; Ott et al., 2017) on 180 specimens of capniid stoneflies (Data2Bio, Ames, Iowa). We focused on A. arapahoe and A. decepta, because of extensive haplotype overlap between these taxa, and Capnia gracilaria and Capnura wanica, because these were the most abundant confamilial stoneflies in this area (Fairchild et al., 2017) and the most likely contributors to heterospecific crosses. We also included representatives of all other members of the genus Arsapnia with unique haplotypes in the COI analysis and of most other capniid stoneflies present in Colorado (Supporting Information Table S1).
Initial sequencing produced ~501.7 M raw reads. After trimming reads with bases having PHRED quality scores of <15, ~440.6 M reads (mean length, 115 bp) remained. Consensus sequences were generated for A. arapahoe because of the lack of a suitable reference genome for alignment and SNP calling. Trimmed sequence reads from all samples were combined and normalized to a maximum of 50x coverage, using diginorm (Brown, Howe, Zhang, Pyrkosz, & Brom, 2012). The sequencing errors in the reads were then corrected using Fiona (Schulz et al., 2014). The coverage‐normalized and error‐corrected reads were condensed using CD‐HIT‐454 (Fu, Niu, Zhu, Wu, & Li, 2012) with ≥96% identity to form consensus clusters. Clusters with fewer than 10 component reads and shorter than 50 bp were discarded. Trimmed reads were aligned to the consensus reference sequence using GSNAP (Wu & Nacu, 2010). Confidently mapped reads were filtered if each mapped uniquely (≤2 mismatches every 36 bp and <5 bases for every 75 bp as tails) and were used for subsequent analyses.
Any site that was polymorphic (homozygous or heterozygous) relative to the reference genome sequence in at least one sample was considered a SNP. Putative homozygous and heterozygous SNPs were retained if the most common allele (or two alleles in heterozygotes) was supported by at least 80% of all the aligned reads covering that position, at least five unique reads supported the most common allele (or two most common alleles), and each polymorphic base had a PHRED base quality value ≥20. Polymorphisms in the first and last 3 bp of each read were ignored. Polymorphic sites were filtered further based on a minimum allele frequency of 1%, a constrained heterozygosity rate (ranging from zero to twice the product of the frequency of the two most common alleles), and a minimum call rate of 20%, that is, each locus was genotyped in 20% of all specimens.
2.4. Phylogenetic analyses
Our initial assessment of the validity and evolutionary position of A. arapahoe was based on four sets of analyses: (a) confinement to a statistical parsimony network independent of all other species and lack of haplotype sharing with other taxa; (b) reciprocal monophyly from all other stonefly species in the maximum‐likelihood phylogenetic trees of ITS1 and concatenated mtDNA sequences; (c) genetic differences commensurate with species‐level differences from all other taxa in the mitochondrial and ITS1 sequences; and (d) the presence of nucleotides diagnostic for this species in these sequences. First, we used TCS 1.21 (Clement, Posada, & Crandall, 2000) to construct 95% maximum parsimony networks based on COI sequences from field samples and public sequence libraries. Independent networks using this threshold have been regarded as representing single species, although this approach can underestimate species diversity because of the greater tendency to combine distinct taxa than to split a single taxon (Chen et al., 2010; Hart & Sunday, 2007). Sequences with ambiguous nucleotides were excluded from maximum parsimony networks to avoid spurious networks (Joly, Stevens, & van Vuuren, 2007). Second, we developed maximum‐likelihood phylogenetic trees for the ITS1 sequences and for concatenated sequences of COI and cyt b. Analyses were restricted to unique haplotypes which we identified using DAMBE version 6 (Xia, 2017). We used PartitionFinder 2.0 (Lanfear, Frandsen, Wright, Senfeld, & Calcott, 2016) to select the best‐fitting partitioning scheme as measured by AICc, constrained to the suite of evolutionary models considered by RAxML (Stamatakis, 2014) and excluding all outgroups, that is, nonmembers of the Arsapnia group. There were six data subsets for the concatenated mitochondrial sequences based on codon position and gene, and two subsets for ITS1 based on nucleotides and recoded gap characters. Because RAxML will only consider a single evolutionary model for the entire suite of partitions, we compared AICc scores among maximum‐likelihood models using the GTR, GTRGAMMA, and GTRGAMMAI evolutionary models to choose a best model. We then ran RAxML version 8.1.21 implemented through RAxMLGUI (Silvestro & Michalak, 2012) and set for rapid bootstrapping (1,000 bootstraps) and a thorough ML search. Third, we calculated pairwise genetic distances between haplotypes of specimens in terminal clades and among species identified based on morphology using MEGA 7.0 (Kumar, Stecher, & Tamura, 2016) independently for the three gene regions. We based these calculations on uncorrected p‐distances because these have been shown to perform as well or better for detection of barcode gaps indicative of species‐level divergence than the more broadly used Kimura‐2‐parameter model (Collins, Boykin, Cruickshank, & Armstrong, 2012). We focused on distances within and between A. arapahoe, A. decepta, Capnia gracilaria, and Capnura wanica. For A. decepta and Capnia gracilaria, we considered samples from outside Colorado separately, to avoid inflating distance estimates by including cryptic, divergent lineages. Finally, we searched for pure diagnostic nucleotide characters (DeSalle, Egan, & Siddall, 2005) to identify A. arapahoe, that is, those nucleotide–position combinations found in all haplotypes of this species and in no other member of this family (Wong, Shivji, & Hanner, 2009).
2.5. SNP analyses
Results of these analyses were not consistent with the prevailing hypothesis for A. arapahoe as a distinct species despite pronounced morphological differences among males. Hence, we pursued GBS to evaluate an alternative hypothesis consistent with the analyses of the mitochondrial data (see below): A. arapahoe is the result of a heterospecific cross in which the female parent was A. decepta. Using the GBS data, we undertook three approaches to evaluate other taxa that might be involved and to determine whether hybridization was introgressive. First, using SNPs with a minimum call rate of 20% across the 180 specimens (n = 123,726 SNPs genotyped in 20% of all specimens and averaging >56 reads/SNP), we examined the number of shared SNPs between A. arapahoe and the three common syntopic species, with the prediction that potentially parental taxa would share the most SNPs with A. arapahoe (Huang & Knowles, 2014). Next, we used principle coordinate analysis in genalex 6 (Peakall & Smouse, 2006) and inferred potential parental taxa from their position relative to specimens of A. arapahoe in two‐dimensional coordinate space (Payseur & Rieseberg, 2016). We restricted these analyses to a single SNP at each locus with a minimum call rate of 90% (n = 1,788) to avoid issues with linkage and to minimize the influence of missing data on potential patterns in hybridization. For the likely parental taxa identified in these analyses, we examined SNPs present in every specimen (minimum call rate = 100%) that were fixed for alternate alleles in each taxon and thus potentially diagnostic to permit precise estimates of the levels of introgression and heterozygosity within individuals (Hohenlohe, Amish, Catchen, Allendorf, & Luikart, 2011). We examined the distribution of alleles at these loci only in male A. arapahoe because the two putative A. arapahoe females were assigned by mitochondrial sequences to members of other, morphologically similar species (Nelson & Baumann, 1989). Another female phenotypically identified as A. decepta, however, had a mitochondrial and nuclear genotype matching that of male A. arapahoe and was considered a female representative of this taxon (see below). This led to identification of sex chromosome‐linked SNPs, the allelic patterns of which differed from those in autosomal loci (Carmichael et al., 2012) but were consistent with the interpretation of the origin of A. arapahoe.
3. RESULTS
The 563‐nucleotide COI dataset consisted of 556 sequences constituting 135 haplotypes, the 918‐nucleotide cyt b dataset included 99 sequences representing 96 haplotypes, and the 428‐nucleotide ITS1 dataset contained 88 sequences and 67 haplotypes. The ITS1 sequences were bracketed by 27 nucleotides of r18S and 36 nucleotides of r5.8S; these regions were invariant in all taxa. In contrast, the ITS1 region was of variable length (273–308 nucleotides) because of the extensive insertions and deletions typical of noncoding regions.
Analyses of 95% maximum parsimony networks of the Arsapnia group did not support recognizing A. arapahoe as a distinct taxon. Although 13 phenotypically identified species were included, the analysis produced only three separate networks (Figure 2, Supporting Information Figure S1). In the network with A. arapahoe, all specimens of that species shared haplotypes with A. decepta, and that network included four other species of Arsapnia, one species of Sierracapnia, and two species of Capnia. This network also included two specimens of A. decepta from Arizona, which were closely related to but distinct from those in Colorado. Interspecific patterns of relationships, however, were largely concordant with those in other sequence‐based analyses.
Figure 2.
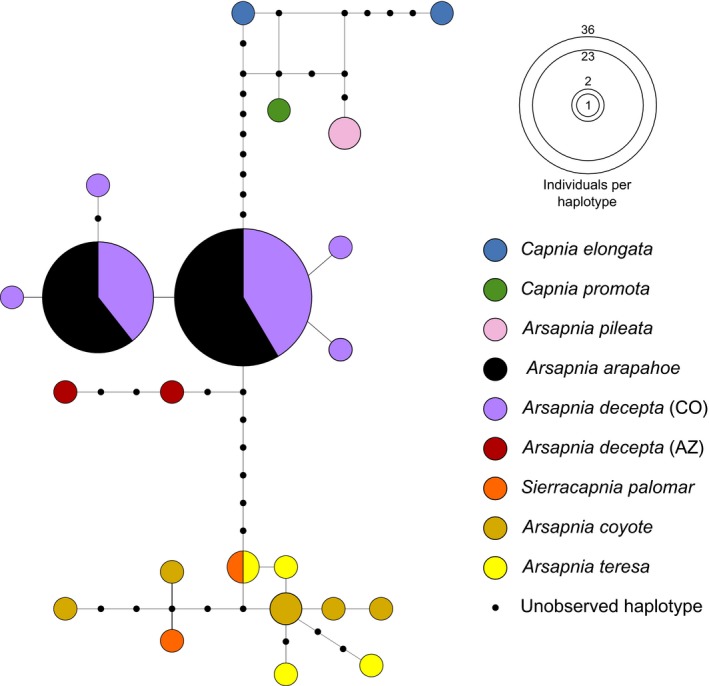
About 95% maximum parsimony network of cytochrome c oxidase subunit 1 sequences for haplotypes of capniid stoneflies from the clade representing the Arsapnia group. Only those specimens linked to the network containing A. arapahoe (n = 22) are shown. Each circle represents a haplotype, sizes are proportional to the number of individuals with that haplotype, and phenotypes associated with each haplotype are identified by color. Haplotype labels are in Supporting Information Tables S1 and S2. Each line segment represents a single mutation, and small black dots represent unobserved haplotypes. The remaining networks representing the Arsapnia group are in Supporting Information Figure S1
Both the mitochondrial and ITS1 phylogenies strongly supported the Arsapnia group (Figure 3). The mitochondrial phylogenies provided greater resolution at lower taxonomic levels, as would be expected because mitochondrial genes have smaller effective population sizes and thus diverge more rapidly relative to nuclear genes. In both analyses, Capnia gracilaria was supported as a member of (ITS1) or sister to (COI + cyt b) the remainder of the Arsapnia group. The terminal clades in the mitochondrial trees were not always concordant with the phenotypic identifications of specimens, especially of females (also see Supporting Information Table S1). This included two females phenotypically assigned to A. arapahoe that shared COI mitochondrial haplotypes with species in other genera: specimen 222 with Capnia gracilaria and specimen 225 with Capnura wanica. These sequences were unlikely to represent paralogous nuclear sequences because of their exact match to haplotypes of each of these taxa; thus, we considered them misidentified. Even ignoring these specimens, in neither phylogeny was A. arapahoe reciprocally monophyletic; it always (mitochondrial phylogeny) or usually (ITS1 phylogeny) occupied the same terminal clade as A. decepta.
Figure 3.
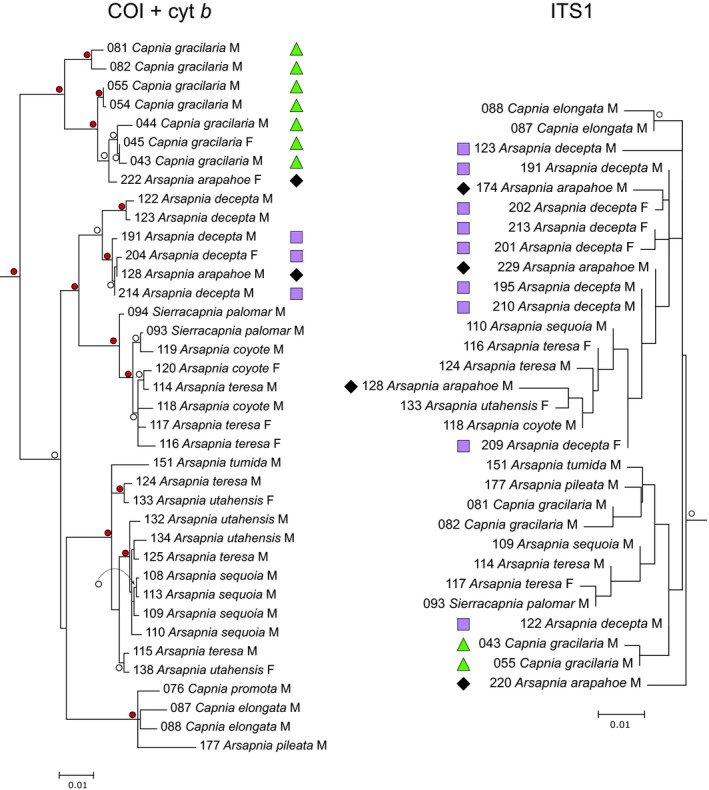
A portion of the best‐scoring phylogenetic tree inferred from a data‐partitioned maximum‐likelihood analysis (with 1,000 bootstrap replicates) of 91 sequences (1,419 nucleotides) of the concatenated mitochondrial genes cytochrome c oxidase subunit 1 and cytochrome b (COI + cyt b) and of 64 sequences (442 nucleotides and 87 gap‐coded positions) of the nuclear first internal transcribed spacer and adjacent portions of the r18S and r5.8S regions (ITS1). Only that portion of each tree representing the Arsapnia group is displayed. Sex of a specimen is indicated by M (male) or F (female). Symbols highlight specimens phenotypically identified as A. arapahoe (black diamonds), A. decepta (purple squares), or Capnia gracilaria (green triangles) in Colorado. Branches with bootstrap support >70% are labeled (○, bootstrap support 70%–90%;  , bootstrap support >90%). Branch lengths are proportional to the number of substitutions per site. For sequence numbers, see Supporting Information Table S1.
, bootstrap support >90%). Branch lengths are proportional to the number of substitutions per site. For sequence numbers, see Supporting Information Table S1.
Pairwise genetic distances supported a lack of divergence between A. arapahoe and A. decepta. Intraspecific differences in mitochondrial sequences between A. arapahoe and A. decepta from Colorado were minor (0.19%–0.31%), substantially smaller than those between A. decepta in Colorado and Arizona (1.11%–1.56%) or between Capnia gracilaria in Colorado and Oregon (1.74%–2.60%; Table 1). Mitochondrial differences between A. arapahoe and Colorado forms of Capnia gracilaria (2.96%–5.14%) and Capnura wanica (10.71%–13.79%) were also substantial. The ITS1 sequences revealed a slightly different pattern, in that the intraspecific variation in A. arapahoe (0.70%) was larger than its interspecific distance with respect to Colorado specimens of A. decepta (0.26%) and Capnia gracilaria (0.42%), yet still markedly smaller than the distance to Capnura wanica (7.18%).
Table 1.
Mean pairwise genetic differences (%) among haplotypes within (on the diagonal) and between (below the diagonal) Arsapnia arapahoe and related or sympatric capniid stoneflies for two mitochondrial genes and one nuclear gene
| Gene | Species | Arsapnia arapahoe | Arsapnia decepta (CO) | Arsapnia decepta (AZ) | Capnia gracilaria (CO) | Capnia gracilaria (OR) | Capnura wanica |
|---|---|---|---|---|---|---|---|
| COI | Arsapnia arapahoe | 0.18 | |||||
| Arsapnia decepta (CO) | 0.31 | 0.56 | |||||
| Arsapnia decepta (AZ) | 0.80 | 1.11 | 0.53 | ||||
| Capnia gracilaria (CO) | 2.96 | 3.18 | 3.13 | 0.73 | |||
| Capnia gracilaria (OR) | 2.84 | 3.06 | 3.02 | 1.74 | 0.18 | ||
| Capnura wanica | 10.71 | 10.80 | 10.87 | 10.77 | 10.53 | 0.36 | |
| cyt b | Arsapnia arapahoe | 0 | |||||
| Arsapnia decepta (CO) | 0.19 | 0.23 | |||||
| Arsapnia decepta (AZ) | 1.52 | 1.56 | 0.23 | ||||
| Capnia gracilaria (CO) | 5.14 | 5.18 | 5.43 | 0.70 | |||
| Capnia gracilaria (OR) | 4.67 | 4.71 | 5.14 | 2.60 | 1.05 | ||
| Capnura wanica | 13.79 | 13.67 | 14.14 | 15.60 | 15.25 | 1.17 | |
| ITS1 | Arsapnia arapahoe | 0.70 | |||||
| Arsapnia decepta (CO) | 0.26 | 0.09 | |||||
| Arsapnia decepta (AZ) | 0.53 | 0.40 | 0.53 | ||||
| Capnia gracilaria (CO) | 0.42 | 0.31 | 0.53 | 0 | |||
| Capnia gracilaria (OR) | 0.79 | 0.70 | 0.79 | 0.79 | 0 | ||
| Capnura wanica | 7.18 | 7.12 | 6.73 | 6.86 | 6.86 | 0 |
There were no single nucleotides in the COI or ITS1 analyses that were diagnostic for A. arapahoe or for the combination of A. arapahoe and A. decepta. A single nucleotide in the cyt b sequences was diagnostic for this combination (position 213, third codon, C; all other taxa, A or T). The public cyt b database, however, was represented by relatively few sequences and taxa, and this diagnostic position may have been represented in other taxa had larger numbers been available to examine.
Analyses of genome‐wide SNPs clarified the origin of A. arapahoe. Of the 123,726 SNPs across the entire sample of specimens with a minimum call rate of 20%, 54,293 were present in A. arapahoe. Large numbers of these were shared by A. decepta (20,965 SNPs) and C. gracilaria (18,925 SNPs), but not by Capnura wanica (62 SNPs), indicating that the latter species did not contribute to the A. arapahoe lineage. The principle coordinate analyses based on SNPs with a 90% minimum call rate positioned A. arapahoe midway between A. decepta and Capnia gracilaria, with all three taxa distantly related to most other members of the Arsapnia group (Figure 4). Specimen 279, originally identified as a female A. decepta, grouped with A. arapahoe and shared a mitochondrial haplotype with other A. arapahoe (Supporting Information Table S1). Thus, we considered this specimen to be a female A. arapahoe.
Figure 4.
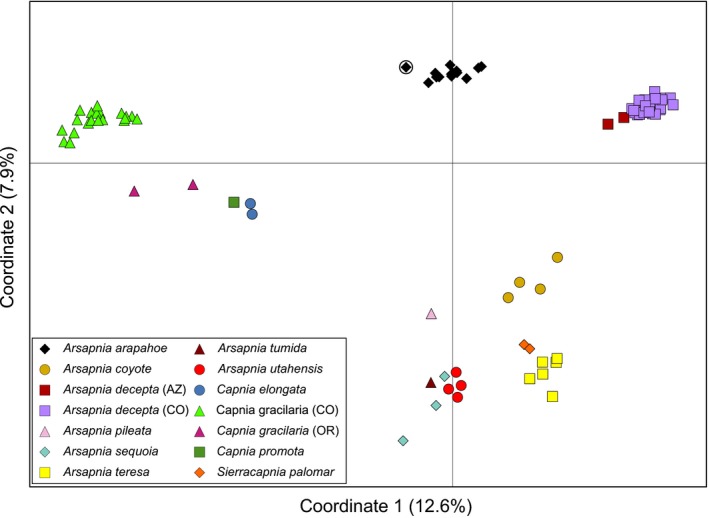
Scatterplot of specimens of the Arsapnia group on the first two principle coordinates derived from genetic distances based on 1,788 SNPs from 98 individuals. Samples are coded by phenotypically identified species, except a female (specimen 279) phenotypically identified as A. decepta with a mitochondrial and SNP genotype of A. arapahoe (circled)
Candidate diagnostic SNPs demonstrated that A. arapahoe was of hybrid origin. When considering all 145 SNPs fixed for different alleles in A. decepta and Capnia gracilaria, 60 were diagnostic when using A. arapahoe as a reference, that is, every specimen of A. arapahoe had half of its alleles from each of these species, and each SNP position was heterozygous (Figure 5). For an additional 51 SNPs, every male A. arapahoe was homozygous for the alleles from A. decepta, whereas the single female of this species was heterozygous for alleles from both parental species, indicating that these were sex‐linked SNPs and consistent with a form of X0 sex determination (Blackmon, Ross, & Bachtrog, 2016). The remaining SNPs were nondiagnostic or may have exhibited occasional genotyping errors, yet their allelic distributions were demonstrative of a first‐generation hybrid origin of A. arapahoe. For the nondiagnostic SNPs, about half of the alleles in A. arapahoe were from A. decepta (mean 53.6%, range, 48.5%–59.4%) and specimens were heterozygous at the majority of these SNPs (mean 85.8%, range 73.5%–96.8%). No individual of A. arapahoe for which we had SNP data had a genotype indicative of any level of introgression resulting from backcrosses to either parental species (in which case an individual would have ≥75% of the diagnostic alleles of one parent) or mating between hybrid individuals (in which case levels of heterozygosity would be ≤25%; Figure 5). Finally, all SNPs fixed for single allele in A. arapahoe (n = 1,275) were shared with A. decepta and Capnia gracilaria, that is, no SNPs were diagnostic for A. arapahoe.
Figure 5.
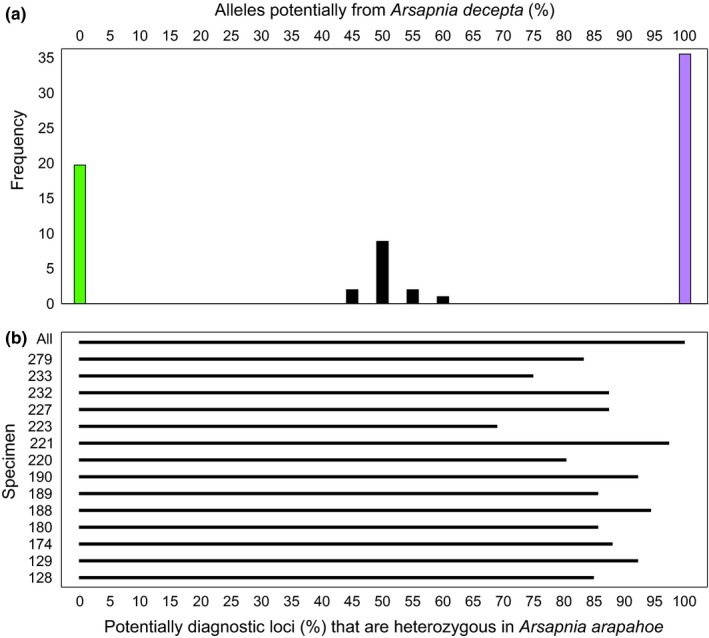
Patterns of genome‐wide SNPs (n = 94) in Arsapnia arapahoe (n = 14; black bars) that were fixed for alternate (and potentially diagnostic) alleles in Capnia gracilaria (n = 20; green bars) and A. decepta (n = 36; purple bars) in samples of these species from Colorado. (a) The percentage of alleles potentially diagnostic for A. decepta. That value subtracted from 100 equals the percentage of loci potentially derived from Capnia gracilaria. The central peak represents A. arapahoe with equal contributions of alleles from both potential parental species. (b) The percentage of loci at which A. arapahoe specimens are heterozygous. For 60 presumably autosomal loci, all A. arapahoe specimens (all) were heterozygous at all loci, indicating that alleles at these loci are diagnostic. For an additional 34 nondiagnostic autosomal SNPs, percentages of loci that were heterozygous are given for each specimen. Not shown are results for 51 sex‐linked SNPs for which all male A. arapahoe (specimens 128–233) are fixed for the alleles from A. decepta. The single female A. arapahoe (specimen 279) was heterozygous for each of these loci
4. DISCUSSION
Collectively, the genetic analyses of specimens from nine separate locations demonstrated that A. arapahoe consisted of individuals that were the first‐generation progeny of female A. decepta and male Capnia gracilaria. The phylogenetic analyses of two mitochondrial genes and one nuclear gene did not delineate A. arapahoe as a taxon distinct from its more common, widespread, and sympatric congener, A. decepta. Specimens of A. arapahoe often shared haplotypes with A. decepta and always occupied the same terminal clades. Interspecific differences between A. arapahoe and A. decepta were typical of differences found within, not between, other species of stoneflies, and were comparable to intraspecific mitochondrial variation throughout much of the animal kingdom (Goldstein & DeSalle, 2010), providing no evidence of any degree of lineage sorting. Although analyses of sequences of ITS1 were less informative, they did reveal monophyly among the Arsapnia group, which included Capnia gracilaria. In contrast, analyses of genome‐wide, autosomal and sex‐linked SNPs identified ongoing hybridization as the source of A. arapahoe, delineated the two parental taxa and (in conjunction with the mitochondrial data) the directionality of matings, and revealed that hybridization was nonintrogressive. This understanding offers a cautionary tale for practitioners of taxonomy and conservation of this and similar groups of little‐studied organisms.
We note that neither DNA barcoding nor morphological diagnosis of Arsapnia arapahoe failed wholly; rather, neither could fully elucidate the process that generated this species hypothesis. The mitochondrial contribution of female A. decepta to A. arapahoe was indicated by the absence of divergence between them, but was moot on whether this was from hybridization, recent divergence, or a faulty taxonomy. In retrospect, the combination in male A. arapahoe of many Arsapnia‐like anatomical characters (Murányi et al., 2014) and a relatively long epiproct reminiscent of Capnia gracilaria (and other species of Capnia; Nelson & Baumann, 1989) might have been interpreted as evidence of the hybrid origin of A. arapahoe, albeit one difficult to recognize amidst a background of morphological variability (Baumann & Stark, 2017). Regardless, the rise of genetic and genomic tools has made the detection of hybridization relatively straightforward, which in turn has revealed that hybridization and even hybrid speciation are a regular occurrence among many insect taxa (The Heliconius Research Consortium, 2012). Ours and other recent studies (Boumans & Tierno de Figueroa, 2016; Dussex, Chuah, & Waters, 2016) extend observations of these phenomena to Plecoptera across the globe and require their consideration in taxonomic and ecological studies, especially in instances of morphological and molecular (or cytonuclear) disagreement. Although we focused on the origins of A. arapahoe, other taxa included in our analyses have patterns that strongly parallel those that were indicative of hybridization (Figures 2 and 3). For example, male Sierracapnia palomar have a long, narrow epiproct markedly different from other members of Sierracapnia (Bottorff & Baumann, 2015), the species is a local endemic that is geographically disjunct from other congeners, and its haplotypes cluster with species of Arsapnia. Evaluating whether hybridization influences this species hypothesis, however, will require focused sampling of additional capniid stoneflies in and around its small range in central California.
Zoogeographic patterns suggest that the hybrids constituting A. arapahoe may be relatively restricted in their distribution. Arsapnia decepta is primarily a species of small and sometimes intermittent streams of the southwestern United States and adjacent Mexico that makes its most northerly advance along the Colorado Front Range. In contrast, Capnia gracilaria appears to occur in larger streams and the majority of its range is from South Dakota to Oregon and north through the Rocky Mountains to Alaska, with scattered observations in Arizona, New Mexico, and the Great Basin (Baumann, Sheldon, & Bottorff, 2017; DeWalt, Maehr, Neu‐Becker, & Stueber, 2018; Kondratieff & Baumann, 2002; Nelson & Baumann, 1989). This limits potential sympatry to basins in the central and southern U.S. Rocky Mountains, and even their areas of contact may be limited. The hybrid zones in northern Colorado streams are highly local (Fairchild et al., 2017), hinting at strong abiotic controls on the distribution of each species that restrict hybridization to short reaches of small streams where adult emergence of each species is synchronized by elevation or stream temperature. If so, it should be possible to predict the locations of hybrid zones in areas that have not yet been sampled or to forecast environmentally driven hybrid zone dynamics (cf. Young et al., 2016).
Confirmation of the presence of these hybrid zones will rely almost entirely on finding rare male A. arapahoe. In sampling targeted at A. arapahoe, only 41 of 26,170 specimens were morphologically identified as that taxon, and all were male (Fairchild et al., 2017). Females may have been equally represented, but the lack of distinguishing characteristics among many female capniid stoneflies may result in fewer being identified or being identified correctly. Genotyping of putative female A. arapahoe in the present study (and previously; Heinold et al., 2014) revealed that they possessed mitochondrial haplotypes identical to those of other capniid stoneflies. Similarly, a female identified as A. decepta based on morphological characters represented the only molecularly supported example of a female A. arapahoe. A third misidentification involved one of the 14 females phenotyped as A. decepta, which phylogenetically clustered with Capnia coloradensis Claassen. This particular error has precedent; Nelson and Baumann (1989) remarked that two observations of female A. decepta in locations geographically disjunct from their core range were likely attributable to misidentifications of Capnia coloradensis. Even for taxonomic experts, reliable identification of female capniid stoneflies may not be possible without resorting to molecular methods. This suggests some caution in accepting identifications of morphologically ambiguous individuals in museum collections unless supported by genetic data, and in assuming that reference sequences in public databases are correctly identified (Kvist, Oceguera‐Figueroa, Siddall, & Erséus, 2010), especially if the sex of those specimens is not recorded.
Nevertheless, such collections are the foundation of ecological, genetic, and taxonomic exploration for many taxa. Our interrogation of the status of A. arapahoe was motivated by the conflict between morphological and molecular interpretations of this species and made possible by a comprehensive museum collection. Despite recent calls to bolster the ranks of traditional taxonomists and the collections, they steward (Morrison, Sillett, Funk, Ghalambor, & Rick, 2017), the taxonomic impediment remains. There are too few taxonomists, and they are confronted by waves of genetic data simultaneously suggesting candidate species and challenging long‐standing species hypotheses. Technological advances that facilitate recovering genome‐wide data for many species, including from environmental samples for which detected taxa are never observed (Deiner et al., 2017), and the increasingly sophisticated algorithms for genetically driven species delimitation (Luo, Ling, Ho, & Zhu, 2018), make it likely that species discovery and revision will increasingly be crowd‐sourced to nontaxonomists such as ecologists and geneticists. The lack of consensus among species concepts and the variation in how genetic data are interpreted (Sukumaran & Knowles, 2017), however, suggest that an integrative taxonomy using multiple data sources (Padial, Miralles, De la Riva, & Vences, 2010) should be the standard, and emphasizes that expertise in morphological assessment remains indispensable (Zhou et al., 2016). Robust taxonomies also rely on the taxonomist's exploration of novel habitats and the ecologist's systematic inventory of representative ones (Sheldon, 2016) to produce the comprehensive zoogeographical sampling that is paramount to delimiting and describing biodiversity (Young, McKelvey, Pilgrim, & Schwartz, 2013).
Such an intensive approach may only be practical for taxa, or the hypotheses they represent, that are of intense conservation interest. This level of attention is being directed at many groups of freshwater species, including stoneflies, because they are disproportionately represented in lists of imperiled taxa (Strayer & Dudgeon, 2010; Williams et al., 2011). In the U.S., federal listing under the Endangered Species Act is typically reserved for those rare and declining taxa, or the distinct populations segments thereof, that appear most at risk. The highly restricted range and limited abundance of A. arapahoe, in light of the existing and proposed developments in this region, met those requirements and elevated this species to candidacy for listing (U.S. Fish and Wildlife Service, 2012). This status, however, rests on the notion that it represents a valid taxon to which a name may be applied. With respect to animal taxa of hybrid origin, the International Commission on Zoological Nomenclature has concluded that the zoological code does not provide for the naming of hybrids (http://iczn.org/content/article-301), except perhaps in instances of hybrid speciation leading to self‐sustaining lineages. We have no evidence, however, that A. arapahoe constitutes such a lineage; instead, the lack of later‐generation hybrids or backcrosses is indicative of nonintrogressive hybridization. Consequently, the individuals formerly recognized at A. arapahoe instead should be referred to as first‐generation hybrids between female A. decepta and male Capnia gracilaria (Frank‐Thorston Krell, International Commission on Zoological Nomenclature, personal communication). More broadly, these individuals are further evidence of the ubiquity of hybridization even between species thought to be reproductively isolated by morphology and behavior, and a reminder to consider this phenomenon as a potential source of variation in taxonomic and phylogenetic studies. We have little doubt that further instances of unrecognized hybridization, and its taxonomic consequences, will become apparent as genomic exploration of understudied groups continues.
CONFLICT OF INTEREST
None declared.
AUTHOR CONTRIBUTIONS
Designed research: MKY, KLP, MF, MKS; Performed research: MKY, RJS, KLP, MF; Analyzed data: MKY; Wrote the paper: MKY, RJS, KLP, MF, MKS.
Supporting information
ACKNOWLEDGMENTS
We thank Boris Kondratieff, Chris Verdone, and staff at the C.P. Gillette Museum of Arthropod Diversity, Colorado State University, Fort Collins, Colorado, for providing specimens, morphological identification, and specimen metadata, and David Wright for assistance with microscopy. Thomas Franklin helped prepare the figures. We also thank Taylor Wilcox and Leslie Ellwood for helpful comments on an earlier version of this manuscript, and Frank‐Thorston Krell for guidance on taxonomic decisions. This project was funded by the U.S. Fish and Wildlife Service and the U.S. Forest Service's Arapaho‐Roosevelt National Forest and Rocky Mountain Research Station.
Young MK, Smith RJ, Pilgrim KL, Fairchild MP, Schwartz MK. Integrative taxonomy refutes a species hypothesis: The asymmetric hybrid origin of Arsapnia arapahoe (Plecoptera, Capniidae). Ecol Evol. 2019;9:1364–1377. 10.1002/ece3.4852
DATA ACCESSIBILITY
Sequences of COI, cyt b, and ITS1 were deposited as GenBank accessions MK275688–276068. The ITS1 alignment is available in Dryad (https://doi.org/10.5061/dryad.c317v31).
REFERENCES
- Baumann, R. W. , Gaufin, A. R. , & Surdick, R. F. (1977). The stoneflies (Plecoptera) of the Rocky Mountains. Memoirs of the American Entomological Society, 31, 1–208. [Google Scholar]
- Baumann, R. W. , Sheldon, A. L. , & Bottorff, R. L. (2017). Stoneflies (Plecoptera) of Nevada. Monographs of the Western North American Naturalist, 10, 1–38. 10.3398/042.010.0101 [DOI] [Google Scholar]
- Baumann, R. W. , & Stark, B. P. (2017). Variation in the epiproct of Arsapnia decepta Banks, 1897 (Plecoptera: Capniidae), with comments on Arsapnia coyote (Nelson & Baumann 1987). Illiesia, 13, 1–21. [Google Scholar]
- Blackmon, H. , Ross, L. , & Bachtrog, D. (2016). Sex determination, sex chromosomes, and karyotype evolution in insects. Journal of Heredity, 108, 78–93. 10.1093/jhered/esw047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagoev, G. A. , deWaard, J. R. , Ratnasingham, S. , deWaard, S. L. , Lu, L. , Robertson, J. , … Hebert, P. D. N. (2016). Untangling taxonomy: A DNA barcode reference library for Canadian spiders. Molecular Ecology Resources, 16, 325–341. 10.1111/1755-0998.12444 [DOI] [PubMed] [Google Scholar]
- Borchsenius, F. (2009). FastGap 1.2 . Updated 7 February 2012. Retrieved from http://www.aubot.dk/FastGap_home.htm
- Bottorff, R. L. , & Baumann, R. W. (2015). Sierracapnia, a new genus of Capniidae (Plecoptera) from western North America. Illiesia, 11, 104–125. [Google Scholar]
- Boumans, L. , & Johnsen, A. (2014). Species‐specific communication bars interspecific mating between syntopic species of Zwicknia stoneflies (Plecoptera: Capniidae). Biological Journal of the Linnean Society, 113, 969–980. [Google Scholar]
- Boumans, L. , & Tierno de Figueroa, J. M. (2016). Introgression and species demarcation in western European Leuctra fusca (Linnaeus, 1758) and L. digitata Kempny, 1899 (Plecoptera: Leuctridae). Aquatic Insects, 37, 115–126. [Google Scholar]
- Brown, C. T. , Howe, A. , Zhang, Q. , Pyrkosz, A. B. , & Brom, T. H. (2012). A reference‐free Algorithm for Computational Normalization of Shotgun Sequencing Data.arXiv, 1203.4802v2.
- Carmichael, S. N. , Bekaert, M. , Taggart, J. B. , Christie, H. R. , Bassett, D. I. , Bron, J. E. , … Sturm, A. (2012). Identification of a sex‐linked SNP marker in the salmon louse (Lepeophtheirus salmonis) using RAD sequencing. PLoS ONE, 8, e77832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H. , Strand, M. , Norenburg, J. L. , Sun, S. , Kajihara, H. , Chernyshev, A. V. , … Sundberg, P. (2010). Statistical parsimony networks and species assemblages in cephalotrichid nemerteans (Nemertea). PLoS ONE, 5, e12885 10.1371/journal.pone.0012885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement, M. , Posada, D. , & Crandall, K. A. (2000). TCS: A computer program to estimate gene genealogies. Molecular Ecology, 9, 1657–1659. 10.1046/j.1365-294x.2000.01020.x [DOI] [PubMed] [Google Scholar]
- Collins, R. A. , Boykin, L. M. , Cruickshank, R. H. , & Armstrong, K. F. (2012). Barcoding’s next top model: An evaluation of nucleotide substitution models for specimen identification. Methods in Ecology and Evolution, 3, 457–465. 10.1111/j.2041-210X.2011.00176.x [DOI] [Google Scholar]
- De Queiroz, K. (2007). Species concepts and species delimitation. Systematic Biology, 56, 879–886. 10.1080/10635150701701083 [DOI] [PubMed] [Google Scholar]
- Deiner, K. , Renshaw, M. A. , Li, Y. , Olds, B. P. , Lodge, D. M. , & Pfrender, M. E. (2017). Long‐range PCR allows sequencing of mitochondrial genomes from environmental DNA. Methods in Ecology and Evolution, 8, 1888–1898. 10.1111/2041-210X.12836 [DOI] [Google Scholar]
- DeSalle, R. , Egan, M. G. , & Siddall, M. (2005). The unholy trinity: Taxonomy, species delimitation and DNA barcoding. Philosophical Transactions of the Royal Society of London, Series B: Biological Sciences, 360, 1905–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWalt, R. E. , Maehr, M. D. , Neu‐Becker, U. , & Stueber, G. (2018). Plecoptera species file online . Version 5.0/5.0, Retrieved from http://Plecoptera.SpeciesFile.org
- Dijkstra, K. D. , Monaghan, M. T. , & Pauls, S. U. (2014). Freshwater biodiversity and aquatic insect diversification. Annual Review of Entomology, 59, 143–163. 10.1146/annurev-ento-011613-161958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dussex, N. , Chuah, A. , & Waters, J. M. (2016). Genome‐wide SNPs reveal fine‐scale differentiation among wingless alpine stonefly populations and introgression between winged and wingless forms. Evolution, 70, 38–47. 10.1111/evo.12826 [DOI] [PubMed] [Google Scholar]
- Fairchild, M. P. , Belcher, T. P. , Zuellig, R. E. , Vieira, N. M. K. , & Kondratieff, B. C. (2017). A rare and cryptic endemic of the central Rocky Mountains, U.S.A.: The distribution of the Arapahoe snowfly, Arsapnia arapahoe (Nelson & Kondratieff, 1988) (Plecoptera: Capniidae). Illiesia, 13, 50–58. [Google Scholar]
- Folmer, O. , Black, M. , Hoeh, W. , Lutz, R. , & Vrijenhoek, R. (1994). DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Molecular Marine Biology and Biotechnology, 3, 294–299. [PubMed] [Google Scholar]
- Fu, L. , Niu, B. , Zhu, Z. , Wu, S. , & Li, W. (2012). CD‐HIT: Accelerated for clustering the next‐generation sequencing data. Bioinformatics, 28, 3150–3152. 10.1093/bioinformatics/bts565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk, D. J. , & Omland, D. E. (2003). Species‐level paraphyly and polyphyly: Frequency, causes, and consequences, with insights from animal mitochondrial DNA. Annual Review in Ecology, Evolution, and Systematics, 34, 397–423. 10.1146/annurev.ecolsys.34.011802.132421 [DOI] [Google Scholar]
- Gamboa, M. , & Monaghan, M. T. (2014). Association of adult female and male stoneflies (Plecoptera) from an alpine river using wing morphometrics and mitochondrial DNA. Aquatic Insects, 36, 1–8. 10.1080/01650424.2015.1013038 [DOI] [Google Scholar]
- Geiger, M. F. , Morinière, J. , Hausmann, A. , Haszprunar, G. , Wägele, W. , Hebert, P. D. , & Rulik, B. (2016). Testing the Global Malaise Trap Program‐How well does the current barcode reference library identify flying insects in Germany? Biodiversity Data Journal, 4, e10671 10.3897/BDJ.4.e10671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill, B. A. , Harrington, R. A. , Kondratieff, B. C. , Zamudio, K. R. , Poff, L. N. , & Funk, C. W. (2013). Morphological taxonomy, DNA barcoding, and species diversity in southern Rocky Mountain headwater streams. Freshwater Science, 33, 288–301. 10.1086/674526 [DOI] [Google Scholar]
- Gill, B. A. , Sandberg, J. B. , & Kondratieff, B. C. (2015). Evaluation of the morphological species concepts of 16 western Nearctic Isoperla species (Plecoptera: Perlodidae) and their respective species groups using DNA barcoding. Illesia, 11, 130–146. [Google Scholar]
- Goldstein, P. Z. , & DeSalle, R. (2010). Integrating DNA barcode data and taxonomic practice: Determination, discovery, and description. BioEssays, 33, 135–147. 10.1002/bies.201000036 [DOI] [PubMed] [Google Scholar]
- Hart, M. W. , & Sunday, J. (2007). Things fall apart: Biological species form unconnected parsimony networks. Biology Letters, 3, 509–512. 10.1098/rsbl.2007.0307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert, P. D. , Cywinska, A. , & Ball, S. L. (2003). Biological identifications through DNA barcodes. Proceedings of the Royal Society of London B: Biological Sciences, 270, 313–321. 10.1098/rspb.2002.2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert, P. D. N. , Penton, E. H. , Burns, J. M. , Janzen, D. H. , & Hallwachs, W. (2004). Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator . Proceedings of the National Academy of Sciences of the United States of America, 101, 14812–14817. 10.1073/pnas.0406166101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinold, B. D. , Gill, B. A. , Belcher, T. P. , & Verdone, C. J. (2014). Discovery of new populations and DNA barcoding of the Arapahoe snowfly Arsapnia arapahoe (Plecoptera: Capniidae). Zootaxa, 3866, 131–137. [DOI] [PubMed] [Google Scholar]
- Heinold, B. D. , & Kondratieff, B. C. (2010). Description of the female of Capnia arapahoe (Plecoptera: Capniidae). Entomological News, 121, 281–283. [Google Scholar]
- Hillis, D. M. , Pollock, D. D. , McGuire, J. A. , & Zwickl, D. J. (2003). Is sparse taxon sampling a problem for phylogenetic inference? Systematic Biology, 52, 124–126. 10.1080/10635150390132911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohenlohe, P. A. , Amish, S. J. , Catchen, J. M. , Allendorf, F. W. , & Luikart, G. (2011). Next‐generation RAD sequencing identifies thousands of SNPs for assessing hybridization between rainbow and westslope cutthroat trout. Molecular Ecology Resources, 11(s1), 117–122. 10.1111/j.1755-0998.2010.02967.x [DOI] [PubMed] [Google Scholar]
- Huang, H. , & Knowles, L. L. (2014). Unforeseen consequences of excluding missing data from next‐generation sequences: Simulation study of RAD sequences. Systematic Biology, 65, 357–365. 10.1093/sysbio/syu046 [DOI] [PubMed] [Google Scholar]
- Hughes, J. M. , Finn, D. S. , Monaghan, M. T. , Schultheis, A. , & Sweeney, B. W. (2014). Basic and applied uses of molecular approaches in freshwater ecology. Freshwater Science, 33, 168–171. 10.1086/675254 [DOI] [Google Scholar]
- Joly, S. , Stevens, M. I. , & van Vuuren, B. J. (2007). Haplotype networks can be misleading in the presence of missing data. Systematic Biology, 56, 857–862. 10.1080/10635150701633153 [DOI] [PubMed] [Google Scholar]
- Jordan, S. , Giersch, J. J. , Muhlfeld, C. C. , Hotaling, S. , Fanning, L. , Tappenbeck, T. H. , & Luikart, G. (2016). Loss of genetic diversity and increased subdivision in an endemic alpine stonefly threatened by climate change. PLoS ONE, 11, e0157386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh, K. , Rozewicki, J. , & Yamada, K. D. (2017). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Briefings in Bioinformatics, bbx108. 10.1093/bib/bbx108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjer, K. M. , Simon, C. , Yavorskaya, M. , & Beutel, R. G. (2016). Progress, pitfalls and parallel universes: A history of insect phylogenetics. Journal of the Royal Society Interface, 13, 20160363 10.1098/rsif.2016.0363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondratieff, B. C. , & Baumann, R. W. (2002). A review of the stoneflies of Colorado with description of a new species of Capnia (Plecoptera: Capniidae). Transactions of the American Entomological Society, 128, 385–401. [Google Scholar]
- Kumar, S. , Stecher, G. , & Tamura, K. (2016). MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Molecular Biology and Evolution, 33, 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvist, S. , Oceguera‐Figueroa, A. , Siddall, M. E. , & Erséus, C. (2010). Barcoding, types and the Hirudo files: Using information content to critically evaluate the identity of DNA barcodes. Mitochondrial DNA, 21, 1998–2005. [DOI] [PubMed] [Google Scholar]
- Lanfear, R. , Frandsen, P. B. , Wright, A. M. , Senfeld, T. , & Calcott, B. (2016). PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Molecular Biology and Evolution, 34, 772–773. 10.1093/molbev/msw260 [DOI] [PubMed] [Google Scholar]
- Luo, A. , Ling, C. , Ho, S. Y. , & Zhu, C. (2018). Comparison of methods for molecular species delimitation across a range of speciation scenarios. Systematic Biology, 67, 830–846. 10.1093/sysbio/syy011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mace, G. M. (2004). The role of taxonomy in species conservation. Philosophical Transactions of the Royal Society B: Biological Sciences, 359, 711–719. 10.1098/rstb.2003.1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallet, J. (2005). Hybridization as an invasion of the genome. Trends in Ecology & Evolution, 20, 229–237. 10.1016/j.tree.2005.02.010 [DOI] [PubMed] [Google Scholar]
- Masly, J. P. (2012). 170 years of “lock‐and‐key”: Genital morphology and reproductive isolation. International Journal of Evolutionary Biology, 2012, 247352 10.1155/2012/247352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morinière, J. , Hendrich, L. , Balke, M. , Beermann, A. J. , König, T. , Hess, M. , … Hausmann, A. (2017). A DNA barcode library for Germany′ s mayflies, stoneflies and caddisflies (Ephemeroptera, Plecoptera & Trichoptera). Molecular Ecology Resources, 17, 1293–1307. [DOI] [PubMed] [Google Scholar]
- Morrison, S. A. , Sillett, T. S. , Funk, W. C. , Ghalambor, C. K. , & Rick, T. C. (2017). Equipping the 22nd‐century historical ecologist. Trends in Ecology & Evolution, 32, 578–588. 10.1016/j.tree.2017.05.006 [DOI] [PubMed] [Google Scholar]
- Murányi, D. , Gamboa, M. , & Orci, K. M. (2014). Zwicknia gen. n., a new genus for the Capnia bifrons species group, with descriptions of three new species based on morphology, drumming signals and molecular genetics, and a synopsis of the West Palaearctic and Nearctic genera of Capniidae (Plecoptera). Zootaxa, 3812, 1–82. [DOI] [PubMed] [Google Scholar]
- Nelson, C. R. , & Baumann, R. W. (1989). Systematics and distribution of the winter stonefly genus Capnia (Plecoptera: Capniidae) in North America. Western North American Naturalist, 49, 289–363. [Google Scholar]
- Nelson, C. R. , & Kondratieff, B. C. (1988). A new species of Capnia (Plecoptera: Capniidae) from the Rocky Mountains of Colorado. Entomological News, 99, 77–80. [Google Scholar]
- Ott, A. , Liu, S. , Schnable, J. C. , Yeh, C.‐T. , Wang, K.‐S. , & Schnable, P. S. (2017). tGBS® genotyping‐by‐sequencing () enables reliable genotyping of heterozygous loci. Nucleic Acids Research, 45, e178 10.1093/nar/gkx853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padial, J. M. , Miralles, A. , De la Riva, I. , & Vences, M. (2010). The integrative future of taxonomy. Frontiers in Zoology, 7, 16 10.1186/1742-9994-7-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payseur, B. A. , & Rieseberg, L. H. (2016). A genomic perspective on hybridization and speciation. Molecular Ecology, 25, 2337–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peakall, R. , & Smouse, P. E. (2006). genalex 6: Genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes, 6, 288–295. 10.1111/j.1471-8286.2005.01155.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilgrim, E. M. , Roush, S. A. , & Krane, D. E. (2002). Combining DNA sequences and morphology in systematics: Testing the validity of the dragonfly species Cordulegaster bilineata . Heredity, 89, 184–190. 10.1038/sj.hdy.6800112 [DOI] [PubMed] [Google Scholar]
- Ratnasingham, S. , & Hebert, P. D. N. (2007). BOLD: The barcoding of life data system (www.barcodinglife.org). Molecular Ecology Notes, 7, 355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, H. H. , & Ricker, W. E. (1971). The classification, evolution, and dispersal of the winter stonefly genus Allocapnia. Illinois Biological Monograph 45. University of Illinois Press.
- Schlick‐Steiner, B. C. , Steiner, F. M. , Seifert, B. , Stauffer, C. , Christian, E. , & Crozier, R. H. (2010). Integrative taxonomy: A multisource approach to exploring biodiversity. Annual Review of Entomology, 55, 421–438. 10.1146/annurev-ento-112408-085432 [DOI] [PubMed] [Google Scholar]
- Schulz, M. H. , Weese, D. , Holtgrewe, M. , Dimitrova, V. , Nie, S. , Reinert, K. , & Richard, H. (2014). Fiona: A parallel and automatic strategy for read error correction. Bioinformatics, 30, i356–i363. 10.1093/bioinformatics/btu440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, M. K. , & Boness, D. J. (2017). Marine mammal subspecies in the age of genetics: Introductory remarks from the Associate Editor and Editor‐in‐Chief of Marine Mammal Science. Marine Mammal Science, 33(S1), 7–11. [Google Scholar]
- Shapiro, A. M. , & Porter, A. H. (1989). The lock‐and‐key hypothesis: Evolutionary and biosystematic interpretation of insect genitalia. Annual Review of Entomology, 34, 231–245. 10.1146/annurev.en.34.010189.001311 [DOI] [Google Scholar]
- Sheldon, A. L. (2016). Mutualism (carpooling) of ecologists and taxonomists. Biodiversity and Conservation, 25, 187–191. 10.1007/s10531-015-1032-3 [DOI] [Google Scholar]
- Silvestro, D. , & Michalak, I. (2012). raxmlGUI: A graphical front‐end for RAxML. Organisms Diversity & Evolution, 12, 335–337. 10.1007/s13127-011-0056-0 [DOI] [Google Scholar]
- Simmons, M. P. , & Ochoterena, H. (2000). Gaps as characters in sequence‐based phylogenetic analyses. Systematic Biology, 49, 369–381. 10.1093/sysbio/49.2.369 [DOI] [PubMed] [Google Scholar]
- Smith, M. A. , Rodriguez, J. J. , Whitfield, J. B. , Deans, A. R. , Janzen, D. H. , Hallwachs, W. , & Hebert, P. D. (2008). Extreme diversity of tropical parasitoid wasps exposed by iterative integration of natural history, DNA barcoding, morphology, and collections. Proceedings of the National Academy of Sciences of the United States of America, 105, 12359–12364. 10.1073/pnas.0805319105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, M. A. , Bertrand, C. , Crosby, K. , Eveleigh, E. S. , Fernandez‐Triana, J. , Fisher, B. L. , … Hrcek, J. (2012). Wolbachia and DNA barcoding insects: Patterns, potential, and problems. PLoS ONE, 7, e36514 10.1371/journal.pone.0036514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sota, T. , & Kubota, K. (1998). Genital lock‐and‐key as a selective agent against hybridization. Evolution, 52, 1507–1513. 10.1111/j.1558-5646.1998.tb02033.x [DOI] [PubMed] [Google Scholar]
- Stamatakis, A. (2014). RAxML version 8: A tool for phylogenetic analysis and post‐analysis of large phylogenies. Bioinformatics, 30, 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart, K. W. (2001). Vibrational communication (drumming) and mate‐searching behavior of stoneflies (Plecoptera); evolutionary considerations In Dominguez E. (Ed.), Trends in research in Ephemeroptera and Plecoptera (pp. 217–225). Boston, MA: Springer. [Google Scholar]
- Stewart, K. W. , & Stark, B. P. (1988). Nymphs of North American stonefly genera (Plecoptera) . Entomological Society of America.
- Strayer, D. L. , & Dudgeon, D. (2010). Freshwater biodiversity conservation: Recent progress and future challenges. Journal of the North American Benthological Society, 29, 344–358. 10.1899/08-171.1 [DOI] [Google Scholar]
- Sukumaran, J. , & Knowles, L. L. (2017). Multispecies coalescent delimits structure, not species. Proceedings of the National Academy of Sciences of the United States of America, 114, 1607–1612. 10.1073/pnas.1607921114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Heliconius Research Consortium (2012). Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature, 487, 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. Fish and Wildlife Service (2012). Endangered and threatened wildlife and plants; 12‐month finding on a petition to list the Arapahoe snowfly as threatened or endangered. Federal Register, 77, 27386–27403. [Google Scholar]
- Valdecasas, A. G. , Williams, D. , & Wheeler, Q. D. (2008). ‘Integrative taxonomy’ then and now: A response to Dayrat (2005). Biological Journal of the Linnean Society, 93, 211–216. 10.1111/j.1095-8312.2007.00919.x [DOI] [Google Scholar]
- Webb, J. M. , Jacobus, L. M. , Funk, D. H. , Zhou, X. , Kondratieff, B. , Geraci, C. J. , … Hebert, P. D. (2012). A DNA barcode library for North American Ephemeroptera: Progress and prospects. PLoS ONE, 7, e38063 10.1371/journal.pone.0038063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, J. E. , Williams, R. N. , Thurow, R. F. , Elwell, L. , Philipp, D. P. , Harris, F. A. , … Frissell, C. A. (2011). Native fish conservation areas: A vision for large‐scale conservation of native fish communities. Fisheries, 36, 267–277. 10.1080/03632415.2011.582398 [DOI] [Google Scholar]
- Wong, E. H. K. , Shivji, M. S. , & Hanner, R. H. (2009). Identifying sharks with DNA barcodes: Assessing the utility of a nucleotide diagnostic approach. Molecular Ecology Resources, 9, 243–256. 10.1111/j.1755-0998.2009.02653.x [DOI] [PubMed] [Google Scholar]
- Wu, T. D. , & Nacu, S. (2010). Fast and SNP‐tolerant detection of complex variants and splicing in short reads. Bioinformatics, 26, 873–881. 10.1093/bioinformatics/btq057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, X. (2017). DAMBE6: New tools for microbial genomics, phylogenetics, and molecular evolution. Journal of Heredity, 108, 431–437. 10.1093/jhered/esx033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeates, D. K. , Seago, A. , Nelson, L. , Cameron, S. L. , Joseph, L. E. , & Trueman, J. W. (2011). Integrative taxonomy, or iterative taxonomy? Systematic Entomology, 36, 209–217. 10.1111/j.1365-3113.2010.00558.x [DOI] [Google Scholar]
- Young, M. K. , McKelvey, K. S. , Pilgrim, K. L. , & Schwartz, M. K. (2013). DNA barcoding at riverscape scales: Assessing biodiversity among fishes of the genus Cottus (Teleostei) in northern Rocky Mountain streams. Molecular Ecology Resources, 13, 583–595. [DOI] [PubMed] [Google Scholar]
- Young, M. K. , Isaak, D. J. , McKelvey, K. S. , Wilcox, T. M. , Pilgrim, K. L. , Carim, K. J. , … Schwartz, M. K. (2016). Climate, demography, and zoogeography predict introgression thresholds in salmonid hybrid zones in Rocky Mountain streams. PLoS ONE, 11, e0163563 10.1371/journal.pone.0163563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeigler, D. D. (1990). Observations pertinent to the role of sexual selection in the stonefly Pteronarcella badia (Plecoptera: Pteronarcyidae). Entomological News, 101, 283–287. [Google Scholar]
- Zhou, X. , Adamowicz, S. J. , Jacobus, L. M. , DeWalt, R. E. , & Hebert, P. D. (2009). Towards a comprehensive barcode library for Arctic life‐Ephemeroptera, Plecoptera, and Trichoptera of Churchill, Manitoba, Canada. Frontiers in Zoology, 6, 30 10.1186/1742-9994-6-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, X. , Jacobus, L. M. , DeWalt, R. E. , Adamowicz, S. J. , & Hebert, P. D. (2010). Ephemeroptera, Plecoptera, and Trichoptera fauna of Churchill (Manitoba, Canada): Insights into biodiversity patterns from DNA barcoding. Journal of the North American Benthological Society, 29, 814–837. 10.1899/09-121.1 [DOI] [Google Scholar]
- Zhou, X. , Frandsen, P. B. , Holzenthal, R. W. , Beet, C. R. , Bennett, K. R. , Blahnik, R. J. , … Collins, G. E. (2016). The Trichoptera barcode initiative: A strategy for generating a species‐level Tree of Life. Philosophical Transactions of the Royal Society B: Biological Sciences, 371, 20160025 10.1098/rstb.2016.0025 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequences of COI, cyt b, and ITS1 were deposited as GenBank accessions MK275688–276068. The ITS1 alignment is available in Dryad (https://doi.org/10.5061/dryad.c317v31).


