Abstract
During homeostasis, the colonic epithelium is replenished every 3–5 days by rapidly cycling Lgr5 + stem cells. However, various insults can lead to depletion of Lgr5 + stem cells, and colonic epithelium can be regenerated from Lgr5‐negative cells. While studies in the small intestine have addressed the lineage identity of the Lgr5‐negative regenerative cell population, in the colon this question has remained unanswered. Here, we set out to identify which cell(s) contribute to colonic regeneration by performing genetic fate‐mapping studies of progenitor populations in mice. First, using keratin‐19 (Krt19) to mark a heterogeneous population of cells, we found that Lgr5‐negative cells can regenerate colonic crypts and give rise to Lgr5 + stem cells. Notch1 + absorptive progenitor cells did not contribute to epithelial repair after injury, whereas Atoh1 + secretory progenitors did contribute to this process. Additionally, while colonic Atoh1 + cells contributed minimally to other lineages during homeostasis, they displayed plasticity and contributed to epithelial repair during injury, independent of Lgr5 + cells. Our findings suggest that promotion of secretory progenitor plasticity could enable gut healing in colitis.
Keywords: Atoh1, colitis, Krt19, Notch1, stem cells
Subject Categories: Development & Differentiation, Molecular Biology of Disease, Stem Cells
Introduction
The colonic epithelium turns over every 5–7 days, driven by dividing stem cells located at the crypt base. Genetic lineage tracing studies in mice have demonstrated that leucine‐rich repeat‐containing G‐protein‐coupled receptor (Lgr5)‐expressing crypt‐based columnar (CBC) cells are the main cells that replenish both the small intestinal and colonic epithelium in normal homeostasis (Barker & Clevers, 2010). Additionally, other genes, such as mTert, Lrig1, or Krt19, have been reported to mark stem/progenitor cells that contribute to epithelial turnover in the colon (Montgomery et al, 2011; Powell et al, 2012; Asfaha et al, 2015). However, the inability to simultaneously use more than one Cre‐based mouse lineage tracing model has led to challenges in distinguishing among the role of different progenitor or stem cell pools (Barker et al, 2007; Sangiorgi & Capecchi, 2008; Buczacki et al, 2013; Barriga et al, 2017).
We have previously demonstrated that when Lgr5 + cells are ablated, cells marked by genes including Bmi1 or Krt19 can regenerate Lgr5 + cells and their lineages in the small intestine or colon, respectively (Tian et al, 2011; Asfaha et al, 2015). In the context of injury, secretory (Dll1 or Prox1) and absorptive (Alpi) cell lineages, as well as Lgr5‐expressing Paneth cells, have been proposed to renew all the cells along the crypt–villus axis in the small intestine (van Es et al, 2012; Buczacki et al, 2013; Metcalfe et al, 2014; Tetteh et al, 2016; Yan et al, 2017). These data point to cellular plasticity in the intestinal epithelium, and, in particular, to the ability of more differentiated cells to revert to a stem cell fate within the crypt (Fre et al, 2011; Furuyama et al, 2011; Montgomery et al, 2011; Takeda et al, 2011; Powell et al, 2012). Notably, we and others have also reported that Lgr5 + stem cells are highly sensitive to epithelial injury induced by radiation or colitis (Yan et al, 2012; Asfaha et al, 2015; Ishibashi et al, 2018), suggesting that an Lgr5‐negative cell population is responsible, at least in part, for colonic regeneration. More recently, work by two groups suggested that secretory cells marked by the transcription factor Atoh1 contribute to colonic epithelial repair (Ishibashi et al, 2018 and Tomic et al, 2018). However, neither study directly addressed whether Atoh1 + cells are dependent on Lgr5 + stem cells for this epithelial repair. It also remains unknown whether Atoh1 + progenitors are required for colonic epithelial repair and whether the cellular plasticity of these cells extends to epithelial cells of the absorptive cell lineage.
Identifying Lgr5‐negative progenitors in the colon, as opposed to small intestine, has proven challenging due to the paucity of distinct stem cell markers. Here, we confirm that Lgr5 + stem cells are dispensable for colonic regeneration in colitis and determine the contribution of various Lgr5‐negative colonic progenitors to epithelial regeneration following colonic injury. Using genetic fate‐mapping models for labeling of keratin‐19 (Krt19) expressing cells or absorptive (Notch1) and secretory (Atoh1) progenitors, we carried out lineage tracing and cell‐specific ablation studies in the context of DSS colitis to test the functional contribution of each progenitor or stem cell population to epithelial regeneration.
Results
Lrg5 + stem cells are sensitive to colonic injury and dispensable for epithelial regeneration post‐colitis
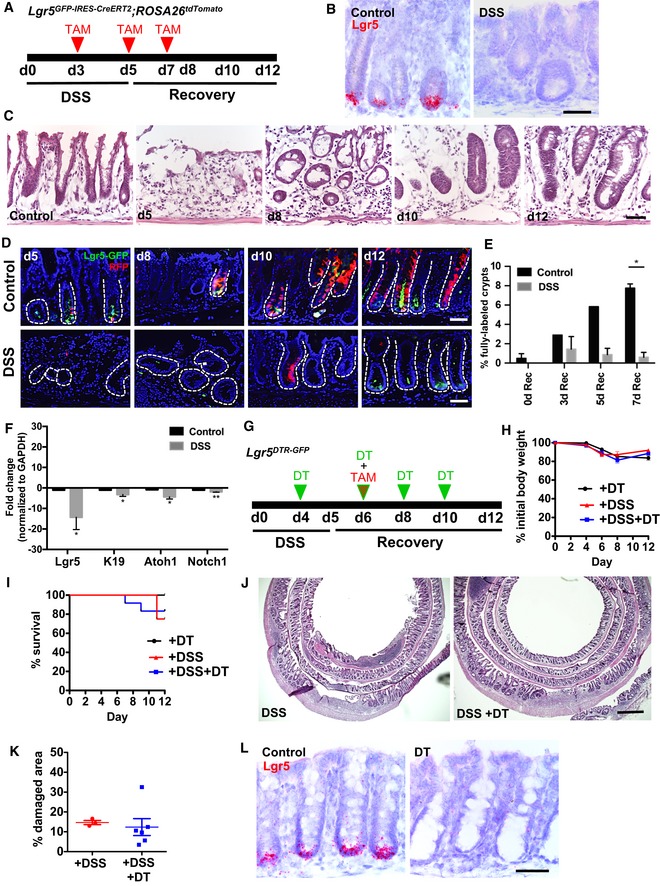
To determine whether Lgr5 + cells contribute to colonic epithelial regeneration post‐colitis, we performed genetic fate‐mapping studies using Lgr5 GFP‐IRES‐CreERT2 ;ROSA26 tdTomato (Barker et al, 2007; Madisen et al, 2010) mice that were administered 5 days of 2.5–3% dextran sodium sulfate (DSS) in the drinking water (Fig 1A). Compared to control mice that were administered regular drinking water, DSS‐treated mice showed complete crypt destruction and epithelial damage with a marked infiltration of inflammatory cells within the lamina propria at days 5–10, and regeneration of crypts occurred by days 10–12 (Fig 1C). Consistent with previous reports (Okayasu et al, 1990; Cooper et al, 1993; Laroui et al, 2012), we found that the distal colon was the most severely injured colonic segment after DSS. Therefore, we focused our analysis on the distal colon. After 5 days of DSS colitis, we observed complete loss of Lgr5 mRNA expression predominantly in the distal colon, as assessed by RNAscope single molecule in situ hybridization (Fig 1B). Consistent with our RNAscope data, we observed that Lgr5‐GFP+ cells were absent from the crypt base immediately following and up until 5 days post‐DSS (day 10; Fig 1D and E). Control Lgr5 GFP‐IRES‐CreERT2 ;ROSA26 tdTomato mice showed fully labeled tdTomato+ lineage‐traced crypts as early as 4 days after initial tamoxifen induction of Cre recombinase (day 8; Fig 1D and E), whereas in DSS‐treated mice, tdTomato+ lineage‐traced crypts were not detectable within the colonic epithelium at any of the time points examined after treatment was begun (Fig 1D and E), consistent with the absence of Lgr5 + cells (Davidson et al, 2012). In line with recent work on the regeneration of intestinal Lgr5 + cells following injury or ablation (Tian et al, 2011; Metcalfe et al, 2014; Asfaha et al, 2015) by 7 days post‐DSS (day 12), we observed a return of Lgr5‐GFP+ cells at the crypt base (Fig 1D). Indeed, the loss of Lgr5 + cells post‐DSS colitis was consistent with our RT–PCR analysis of stem and progenitor cell markers, which revealed that Lgr5 mRNA expression was the most reduced transcript post‐DSS, with nearly a 15‐fold reduction compared to control mice (Fig 1F).
Figure 1. Lgr5 + stem cells are sensitive to colonic injury and dispensable for epithelial regeneration in colitis.
-
AIllustration of experimental protocol outlining DSS‐induced colitis in Lgr5 GFP‐CreERT2 ;ROSA26 tdTomato mice.
-
BRNAscope analysis of Lgr5 mRNA in DSS colitis demonstrating loss of Lgr5 transcript (N = 3 per condition).
-
CHematoxylin and eosin staining of the colonic epithelium following DSS‐induced damage. Crypts are dramatically injured by d5. At d8 through d12, regenerating crypts are observed.
-
D, ECompared to untreated controls, DSS treatment ablates Lgr5‐GFP+ stem cells and results in functional loss of tdTomato+ lineage tracing as early as d5. Lgr5‐GFP+ stem cells reappear by d12 (N = 3–4 per condition).
-
FEffects of DSS colitis injury on RNA expression levels of stem (Lgr5 and Krt19) and progenitor (Atoh1 and Notch1) cell markers. Lgr5 mRNA expression is significantly decreased after exposure to DSS (N = 6–7 per condition).
-
GDSS and DT treatment of Lgr5 DTR‐GFP mice.
-
H, IMice body weight (H) and survival (I) are comparable between three treatments: DSS + DT (N = 12), only DSS (N = 8), or only DT (N = 3).
-
JHematoxylin and eosin staining of the colonic epithelium shows no morphological difference between DSS alone and DSS + DT‐treated mice.
-
KThe extent of epithelial damage is similar in mice treated with DSS alone versus DSS + DT (to ablate Lgr5 + stem cells; N = 3 DSS; N = 6 DSS + DT).
-
LRNAscope for Lgr5 in Lgr5 DTR‐GFP mice showing loss of Lgr5 transcripts after DT treatment (N = 3–4 per condition).
Given that Lgr5 + stem cells were sensitive to colitis injury, we tested whether these cells were functionally required for colonic epithelial regeneration. We administered diphtheria toxin (DT) to ablate Lgr5 + cells in Lgr5 DTR‐GFP transgenic mice during and for 5 days following DSS, and we examined the effects on body weight, colonic histology, and overall survival (Fig 1G–K). We observed no difference in body weight or overall survival up to 12 days following initiation of DSS after ablating Lgr5 + cells (Fig 1H and I). Confirming previously reported findings, we did not detect any difference in the degree of colonic epithelial injury or inflammation between saline‐ and DT‐treated groups as assessed by histology (Fig 1J and K; Metcalfe et al, 2014). To conclusively assess the absence of Lgr5 + stem cells post‐DT treatment, we carried out RNAscope to examine Lgr5 mRNA expression along the antero‐posterior colonic epithelium. In contrast to DSS injury, DT treatment resulted in complete loss of Lgr5 expression throughout the colon (Fig 1L).
Lgr5‐negative/Krt19 + cells contribute to colonic epithelial regeneration in colitis
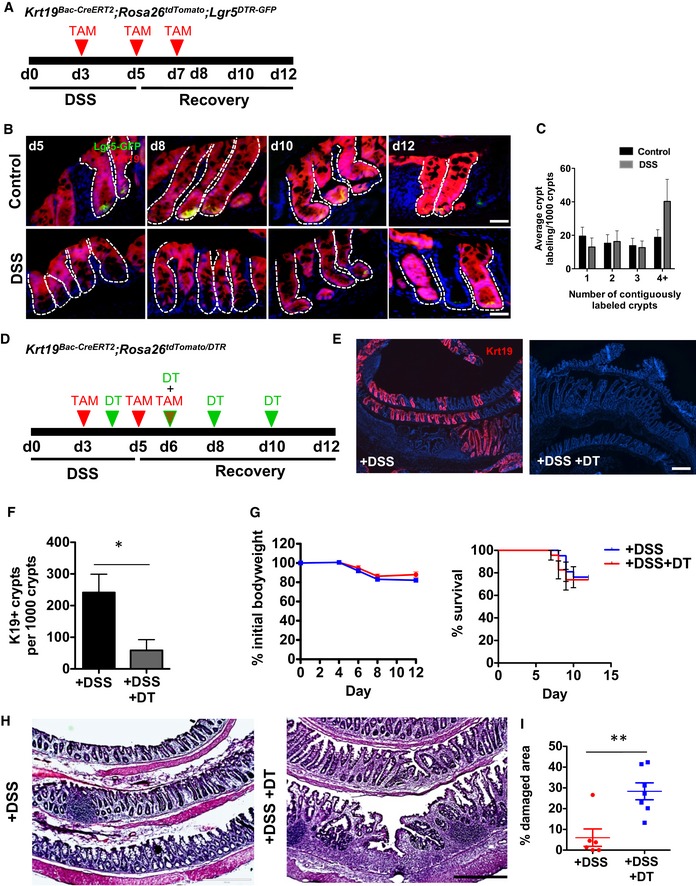
Since Lgr5 + cells were dispensable for colonic epithelial regeneration in colitis, we next asked which Lgr5‐negative cell population was responsible for colonic epithelial regeneration post‐colitis. We first examined the role of Krt19 + cells, as we previously showed that this marker labels intestinal and colonic crypt cells, but excludes rapidly cycling Lgr5 + stem cells (Asfaha et al, 2015; Figs 2B and EV1A–C). To examine the role of Krt19 + cells in colonic regeneration, we crossed Krt19 BAC‐CreERT2 mice (Asfaha et al, 2015) to ROSA26 tdTomato reporter mice and performed genetic lineage tracing studies in the setting of DSS colitis (Fig 2A). Consistent with our previously reported findings, during homeostasis the Krt19 + cells showed the capacity to lineage trace entire colonic crypts, including Lgr5‐GFP+ cells, at 8–10 days following tamoxifen induction (Fig 2B). Krt19 + cells continued to lineage trace entire colonic crypts throughout the regenerative period following DSS colitis as well as give rise to new Lgr5‐GFP+ cells by 12 days after initiation of DSS (Fig 2B and C). Quantification of contiguously labeled Krt19 + crypts, indicative of crypt fission as previously described (Okayasu et al, 1990; Park et al, 1995; Hirata et al, 2013), revealed an increase in lineage‐traced crypts, consistent with regeneration over the 12‐day time interval examined (Fig 2C).
Figure 2. Lgr5‐negative/Krt19 + cells contribute to epithelial renewal during homeostasis and following colitis.
-
AIllustration of experimental protocol outlining DSS‐induced colitis in Krt19 BAC‐CreERT2 ;ROSA26 tdTomato mice.
-
B, CKrt19 + cells lineage trace entire colonic crypts during both normal homeostasis and injury (N = 3–4 per condition).
-
DDSS and DT treatment of Krt19 CreERT2 ;ROSA26 tdTomato/DTR mice.
-
E, FDT ablation of Krt19 + cells significantly reduced the number of lineage‐labeled crypts (N ≥ 3 per condition).
-
GMice body weight and survival remain unchanged between DSS and DSS + DT‐treated groups (N = 6 control; N = 7 DT).
-
H, IHematoxylin and eosin staining of the colonic epithelium after DSS versus DSS + DT (H) and quantification of damaged epithelial area (I), showing more damage is seen in mice treated with both DSS + DT treatment (N = 6 control; N = 7 DT).
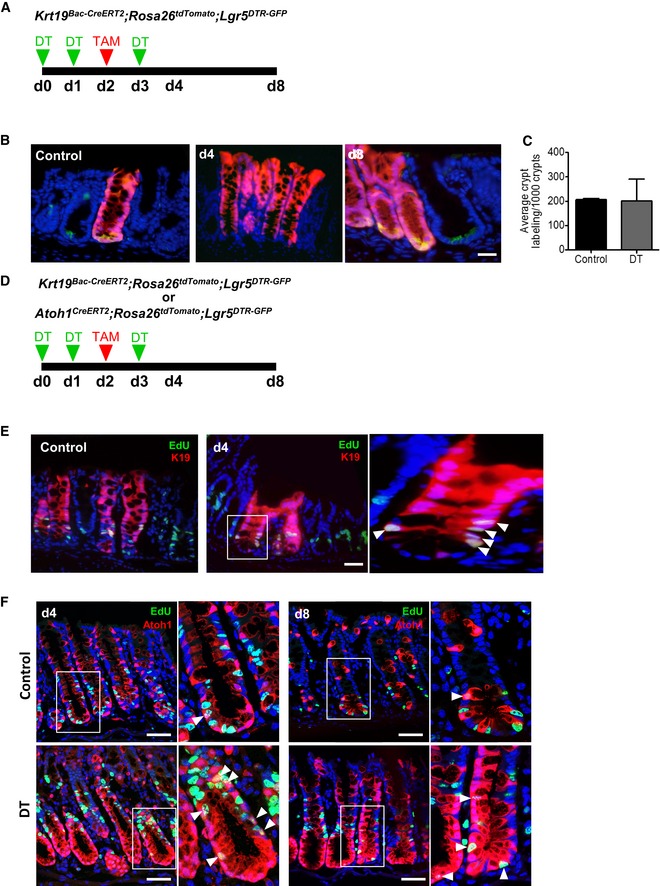
Figure EV1. Lgr5‐negative/Krt19 + and Atoh1 + cells show colonic epithelium renewal capacity in homeostasis and injury.
-
AIllustration of experimental protocol outlining DT treatment of Krt19 BAC‐CreERT2 ;ROSA tdTomato ;Lgr5 DTR‐GFP mice.
-
B, CIn control conditions and after Lgr5 + stem cell ablation, Krt19 + cells form crypt ribbons and give rise to new Lgr5 + stem cells (N = 2 per time point).
-
DIllustration of experimental protocol outlining DT treatment of Krt19 BAC‐CreERT2 or Atoh1 CreERT2 mice.
-
E, FRare proliferating Krt19 + or Atoh1 + cells (insets, arrowheads) are found in the transit‐amplifying zone of control crypts, and none are present at the bottom of crypts. At d4 of DT treatment, more proliferating Krt19 + or Atoh1 + cells (insets, arrowheads) are found in the transit‐amplifying zone and the crypts are devoid of proliferating cells. By d8 of DT treatment, proliferating Atoh1 + cells (insets, arrowheads) are found at the bottom of crypts.
Given that Krt19 + cells renewed the epithelium post‐DSS colitis, we next examined the effects of conditional Krt19 + cell ablation in the setting of DSS‐induced colitis. We crossed Krt19 BAC‐CreERT2 ;ROSA26 tdTomato mice and ROSA26 DTR mice in order to allow us to selectively ablate DTR‐expressing cells following DT administration. We administered tamoxifen to induce Cre‐mediated expression of DTR, followed by DT administration (Fig 2D), which ablated nearly all Krt19‐tdTomato+ cells in Krt19 BAC‐CreERT2 ;ROSA26 tdTomato/DTR mice (Fig 2E and F). Surprisingly, DT ablation of Krt19 + cells did not alter the survival or colonic histology of mice with colitis (Fig 2G and H), suggesting that Krt19 + cells may also be dispensable for renewal. Concurrent DSS colitis and Krt19 + cells ablation did not affect body weight and overall animal survival (Fig 2G); however, it did cause a significant increase in colonic histologic damage when compared to saline‐treated controls (Fig 2H and I). Taken together, these data indicate that Krt19‐expressing cells contribute to epithelial regeneration within the colon.
We previously showed that Krt19 labels a heterogeneous population of cells within the crypt (Asfaha et al, 2015), including both absorptive and secretory cells. Thus, we further assessed whether Krt19 is expressed in all secretory cells or only a subset of cells within the colon. We examined colonic tissue sections from Krt19 BAC‐CreERT2 ;ROSA26 tdTomato mice 24 h post‐tamoxifen and looked for co‐localization of various secretory cell markers with TdTomato+ cells. By immunofluorescence staining, we detected overlap of TdTomato+ with ChgA, Dclk1, and Muc2 expression, indicating that Krt19 indeed marks a variety of secretory cells including enteroendocrine, tuft, and goblet cells, respectively (Fig EV2A and B).
Figure EV2. Krt19‐expressing cells include multiple secretory cell types in the colon and small intestine.
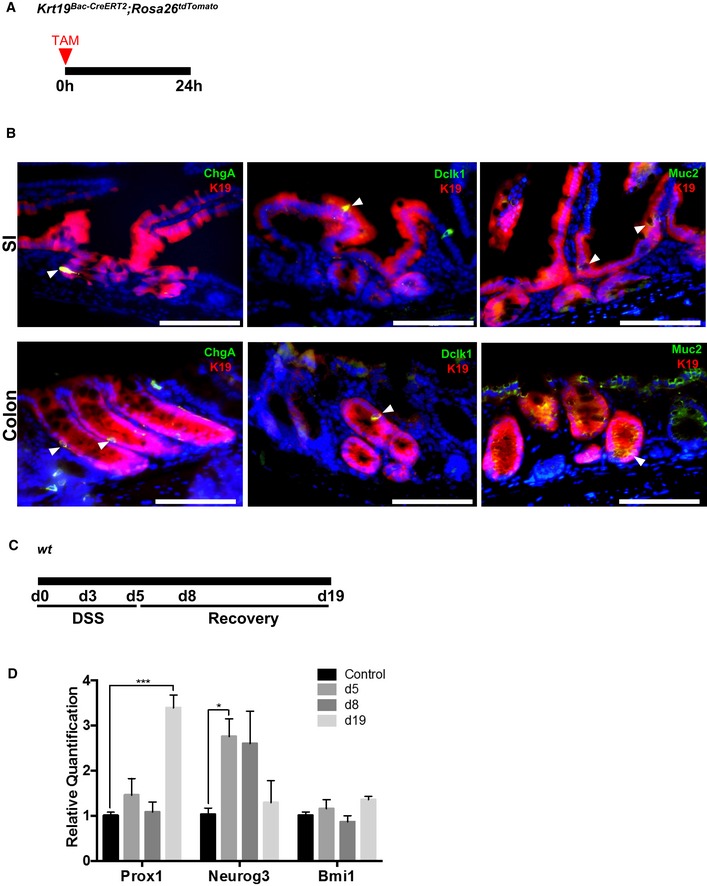
- Illustration of experimental protocol outlining tamoxifen induction of Krt19 BAC‐CreERT2 ;ROSA26 tdTomato mice.
- Immunofluorescence staining showing co‐localization of Krt19 + cells with ChgA +, Dclk1 +, and Muc2 + secretory cells in the small intestine and colon (arrowheads).
- Illustration of experimental protocol outlining DSS‐induced colitis in wild‐type mice.
- Effects of DSS colitis on RNA expression levels of various secretory (Prox1, Neurog3, Bmi1) cell markers. Neurog3 expression is significantly increased acutely post‐DSS (d5), whereas Prox1 expression is significantly increased during late recovery (d19). Bmi1 expression is not affected by DSS colitis.
Colonic Notch1 + absorptive progenitors do not contribute to epithelial repair in homeostasis or injury
We next sought to precisely define the Lgr5‐negative/Krt19 + population that contributes to epithelial regeneration in colitis and assessed whether an absorptive or progenitor cell plays a major role in epithelial repair. We assayed the contribution of absorptive progenitors, defined by expression of Notch1 (van Es et al, 2005; Riccio et al, 2008; Pellegrinet et al, 2011), in colonic regeneration, by using Notch1 CreERT2 mice (Fre et al, 2011) crossed to ROSA26 tdTomato mice. In control mice, at day 5 (2 days following the initial tamoxifen dose), Notch1 + cells were found dispersed throughout the colonic crypt, consistent with the labeling of absorptive progenitors and their progeny (Fig 3B). In rare instances (~1–2% of crypts), we observed entire crypts lineage traced by day 12, consistent with previous reports of Notch1 expression within Lgr5 + stem cells (Fig 3B; Itzkovitz et al, 2012), and we detected rare Notch1‐expressing tdTomato + cells that co‐localized with Lgr5‐GFP+ expression (Fig 3B). To further assess whether Notch1 + absorptive progenitors and/or their progeny contribute to colonic regeneration following colitis injury, we administered DSS to Notch1 CreERT2 ;ROSA26 tdTomato mice and examined tdTomato+ lineage tracing (Fig 3B). In the setting of DSS colitis, Notch1 + cells appeared sensitive to injury, with very rare tdTomato+ cells being detectable by day 10 (5 days post‐DSS) and no fully lineage‐traced colonic crypts being detected at days 10–12 (Fig 3B and C). Next, we carried out Notch1 + lineage tracing studies in the presence or absence of Lgr5 + cell ablation with DT in Notch1 CreERT2 ;ROSA26 tdTomato crossed to Lgr5 DTR‐GFP mice (Fig 3D). Indeed, Notch1 + cell lineage tracing was no longer detectable upon DT ablation of Lgr5 + cells (Fig 3E and F), suggesting that some Notch1‐expressing cells also express Lgr5. Taken together, these findings indicate that Notch1 + absorptive cells do not play a major role in colonic epithelial regeneration following DSS‐induced colitis.
Figure 3. Colonic Notch1 + absorptive progenitors aid in homeostatic renewal but do not show renewal capacity in injury.
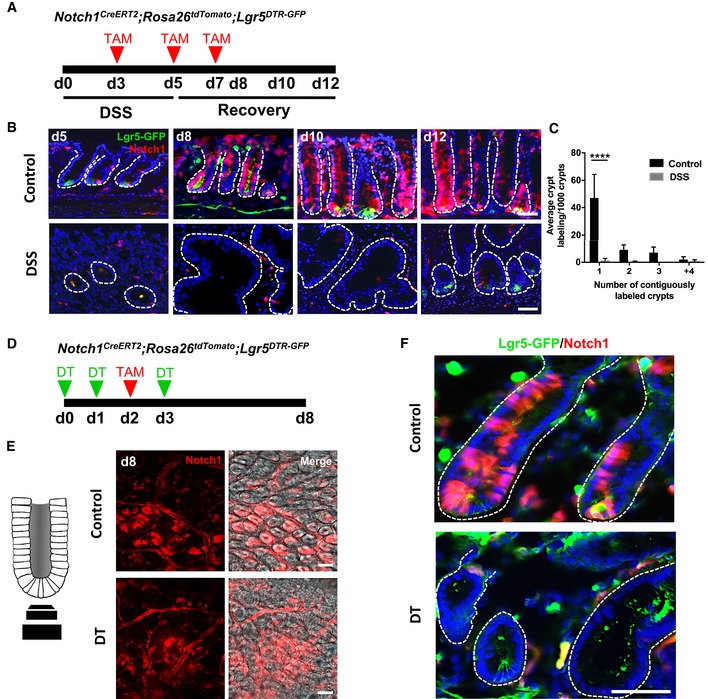
-
AIllustration of experimental protocol outlining DSS‐induced colitis in Notch1 CreERT2 ;ROSA26 tdTomato ;Lgr5 DTR‐GFP mice.
-
B, CNotch1 + progenitors are able to generate Lgr5 + stem cells and contribute to epithelial renewal in homeostasis, however, do not contribute to regeneration after colitis (N = 4 per condition).
-
DIllustration of experimental protocol outlining DT ablation of Lgr5 + cells in the context of Notch1 + cell lineage tracing using Notch1 CreERT2 ;ROSA26 tdTomato ;Lgr5 DTR‐GFP mice.
-
E, FImages of whole mount (E) and tissue sections (F) of Notch1 CreERT2 ;ROSA26 tdTomato ;Lgr5 DTR‐GFP mice showing Notch1 lineage‐labeled crypts. No fully labeled crypts were found after Lgr5 + stem cell ablation.
Atoh1 + secretory progenitors display plasticity during both homeostasis and injury in colonic regeneration
Next, to determine the role of secretory progenitors and their progeny in colonic epithelial homeostasis and injury, we carried out genetic lineage tracing studies using Atoh1 CreERT2 mice (Fujiyama et al, 2009) crossed to ROSA26 tdTomato mice. We examined tdTomato+ lineage tracing in Atoh1 CreERT2 ;ROSA26 tdTomato mice administered DSS following the protocol illustrated in Figs 2A and 3A. In control mice, at day 5 (2 days after the initial tamoxifen dose), numerous Atoh1 + cells were found dispersed throughout the colonic crypt, consistent with the labeling of secretory progenitors and their progeny (Figs 4A and EV3A and B). Interestingly, while only rare fully lineage‐traced crypts (< 5%) were observed at 7 days post‐tamoxifen, by 30 days post‐tamoxifen this increased to ~20% of total colonic crypts (Fig EV3A and B). This finding suggests that the Atoh1 + cell population may overlap with the stem cell population during homeostasis, as previously shown in the small intestine (Kim et al, 2016; Ishibashi et al, 2018). In the setting of DSS colitis, day 12 lineage tracing of entire colonic crypts from Atoh1 + cells remained unchanged from control mice without colitis (Fig 4A and B). Atoh1 + cells also gave rise to newly regenerated Lgr5 + cells 1 week post‐DSS (day 12), demonstrating that Atoh1 + progenitors are resistant to DSS colitis and contribute to the colonic epithelial regeneration (Fig 4A and B). Notably, by 30 days post‐DSS, quantification of tdTomato+ crypts, as a proxy of progenitor cells giving rise to multiple lineages, revealed that Atoh1 + lineage tracing significantly increased post‐colitis, consistent with the expansion of Atoh1 + progenitors during colonic regeneration (Fig 4C and D). Taken together, our data show that Atoh1 + progenitors can contribute to renewal during both homeostasis and injury.
Figure 4. Atoh1 + secretory progenitors display plasticity and contribute to colonic regeneration in homeostasis and injury.
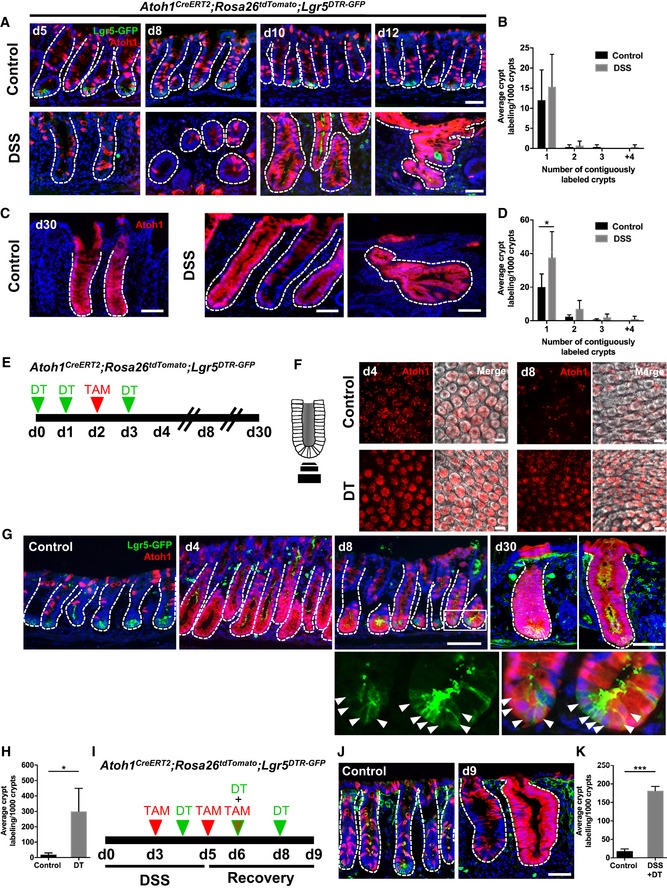
-
A, BAtoh1 + progenitors show stem cell capacity during normal homeostasis and regeneration. At early stages of recovery, d10–12, Atoh1 + cells lineage trace the colonic crypt (N = 4 per condition).
-
C, DBy d30 after DSS, Atoh1‐derived stem cells continue to lineage trace complete colonic crypts (N = 4 per condition).
-
EIllustration of experimental protocol outlining DT treatment of Atoh1 CreERT2 ;ROSA26 tdTomato ;Lgr5 DTR‐GFP mice.
-
F, GWhole mount (F) and section (G) images of Atoh1 lineage‐labeled crypts. Majority of crypts are labeled after Lgr5 + stem cell ablation and remain label with the appearance of Lgr5 + stem cells (inset, arrowheads). Atoh1 lineage‐labeled crypts remain by d30.
-
HSignificantly increased labeling of crypt base was observed at d8 in DT‐treated mice (N = 4 per condition).
-
IDSS and DT treatment of Atoh1 CreERT2 ;ROSA26 tdTomato ;Lgr5 DTR‐GFP mice.
-
J, KScattered labeling of Atoh1 + cells in control conditions, while DSS + DT injury lead to Atoh1 lineage‐labeled crypts lacking Lgr5 + stem cells.
Figure EV3. Atoh1 + cells show rare renewal capacity in homeostasis, yet post‐injury Atoh1 + cells acquire stemness.
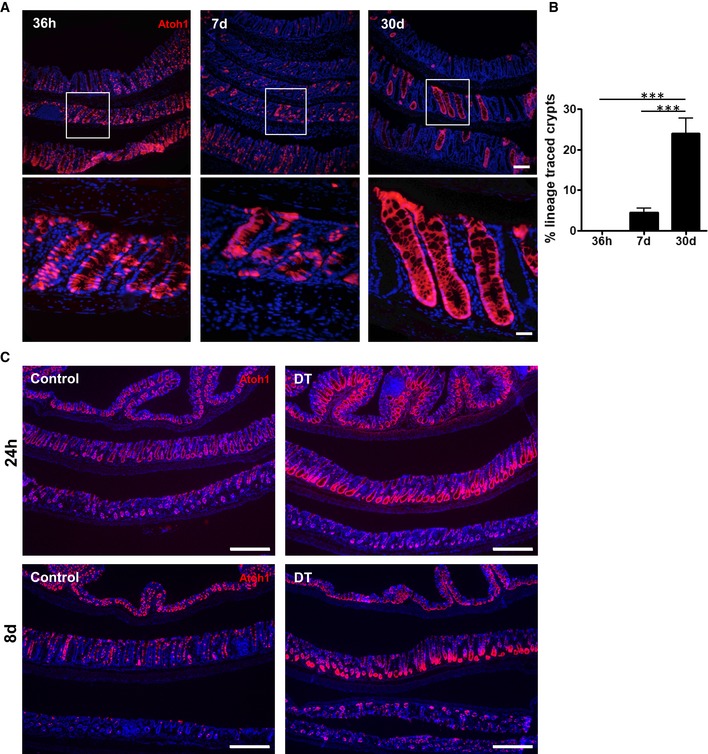
-
A, BOccasionally, Atoh1 + cells are able to form crypt ribbons that persist up to 30 days after lineage labeling in homeostasis (N = 4 per condition).
-
CFollowing DT ablation of Lgr5 + cells, the majority of crypts are renewed and maintained by Atoh1 + lineage.
To further investigate which secretory cell type is involved in Lgr5 + cell‐independent regeneration following DSS colitis, we analyzed the expression of the secretory cell markers Prox1, Neurog3, and Bmi1 (Yan et al, 2017) in colonic tissue following DSS recovery by qRT–PCR. We observed increased expression of Neurog3 and Prox1 post‐DSS at day 5 and day 19, respectively, while Bmi1 expression remained unchanged (Fig EV2C and D). These data suggest that Prox1 + and/or Neurog3 + secretory cell populations may overlap with cells expressing Atoh1, and cells expressing either or both of these markers could be important for epithelial regeneration after injury.
To definitively distinguish between lineage tracing from Atoh1 + progenitors and Lgr5 + cells in the colon, we assessed Atoh1 + lineage tracing studies in the context of Lgr5 + cell ablation. Using Atoh1 CreERT2 ;ROSA26 tdTomato ;Lgr5 DTR‐GFP mice, we administered DT to ablate Lgr5 + cells while simultaneously lineage tracing from Atoh1 + cells (Fig 4E). Our experimental paradigm effectively ablated all Lgr5‐expressing cells from the colon, with no Lgr5‐GFP+ cells being seen in the colon 24 h following the last dose of DT (Fig 4F and G). DT ablation of Lgr5 + cells resulted in extensive Atoh1 + lineage labeling of crypts at day 4, followed by Atoh1 + progenitor cell lineage tracing the colonic crypt base of nearly all crypts examined (Figs 4F–H and EV3C). This was in contrast to saline‐treated controls, in which only rare tdTomato + crypts were detectable at day 8 post‐tamoxifen, and consistent with previous reports of dedifferentiation in small intestine (Fig 4G and H; van Es et al, 2012; Tetteh et al, 2016). Indeed, tdTomato+ lineage tracing significantly increased by nearly 15‐fold following Lgr5 + cell ablation compared to controls (Fig 4H). Interestingly, Atoh1 + lineage localization at the crypt base at day 8 was correlated with the reemergence of Lgr5 + cells and coincided with our observations in DSS colitis, where new Lgr5‐GFP+ cells derive from Atoh1 + lineage (Fig 4G, arrowheads within inset). Further, homeostasis is maintained by Atoh1‐derived Lgr5 + stem cells, demonstrated by 30‐day labeled crypts (Fig 4G). Remarkably, this self‐renewal capacity of Atoh1 + cells post‐colitis was unaffected by simultaneous Lgr5 + cell ablation (Fig 4I–K).
EdU labeling revealed that, at baseline, many tdTomato+ Krt19‐expressing cells within the crypt were proliferating. This was in contrast to the rare tdTomato+ Atoh1‐expressing cells which were EdU+ at baseline, but more easily detectable above the crypt base by day 4 after Lgr5 + cell ablation (Fig EV1D–F). Notably, at day 8 of Lgr5 + cell ablation, EdU+ Atoh1‐expressing cells were seen at the crypt base (Fig EV1F). These data suggest that, unlike crypt base Lgr5 + stem cells that are susceptible to injury, Atoh1 + progenitor cells are located higher up within the crypt and are critical to colonic epithelial regeneration in the setting of injury.
Atoh1 + colonic progenitor cells show cellular plasticity in response to injury in vitro
Our in vivo observations indicated that colonic Atoh1 + cells have limited renewal capacity during homeostasis but show significantly increased contribution in the setting of colonic epithelial injury. Therefore, to determine whether the response of Atoh1 + cells was intrinsic to these cells and independent of the stromal niche, we examined the behavior of Atoh1 + cells after injury in vitro. First, we isolated colonic epithelial cells from Atoh1 CreERT2 ;ROSA26 tdTomato mice to generate colonic organoids. Then, using 4‐hydroxytamoxifen (4‐OHT), we induced recombination in organoids and compared the contribution of Atoh1‐tdTomato+ cells under control conditions or following 4 Gy of irradiation (Fig 5A). We could not detect any tdTomato fully labeled colonic organoids up to 12 days after initial culture (pre‐passage). However, following 4 Gy of irradiation, we observed a significant expansion of Atoh1‐derived tdTomato+ cells by day 12 (Fig 5B and C), indicating that in vitro, Atoh1 + progenitors display cellular plasticity and contribute to epithelial regeneration in the setting of injury. The expansion of Atoh1 + cells was similarly evident upon irradiation of small intestinal organoids, confirming that Atoh1 + secretory progenitors in small intestinal organoids display plasticity comparable to colonic organoids (Fig EV4A–C). We observed a similar phenomenon following mechanical stress induced by passaging of organoids (Sambuy et al, 2005; Dupont et al, 2011; Zachos et al, 2016), such that by days 14–15 Atoh1‐derived tdTomato+ cells in small intestinal (Fig EV4D–F) and colonic (Fig EV5A–C) organoids had undergone a significant expansion. This was in striking contrast to initial culture of small intestinal and colonic organoids, in which there was a gradual decrease in the number of tdTomato+ cells over time, consistent with the labeling of short‐lived progenitors and/or secretory cells (Figs EV4E and EV5B). Indeed, tdTomato+ cells gradually disappeared prior to organoid passage.
Figure 5. Atoh1 + colonic progenitor cells show renewal capacity in vitro upon injury.
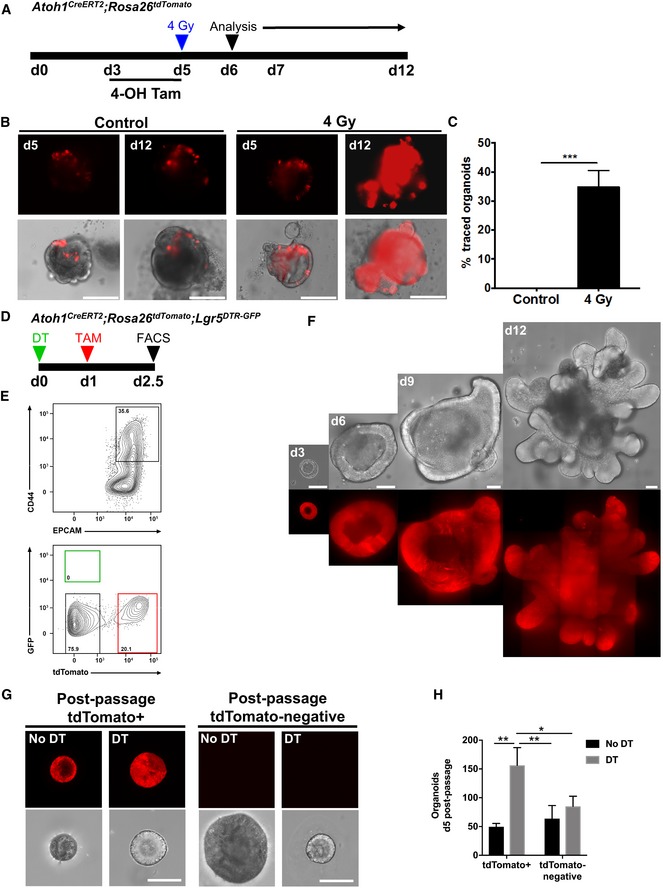
-
AIllustration of experimental protocol outlining 4‐OHT induction and irradiation of Atoh1 CreERT2 ;ROSA26 tdTomato colonic organoids.
-
B, CSporadic labeling of Atoh1 + cells after 4‐OHT induction. Following irradiation, entire colonic organoids exhibit full lineage labeling of Atoh1 + cells, characteristic of multipotent stem cells (N = 3 per condition).
-
D, EFlow cytometry for tdTomato+ cells in crypt epithelium from DT‐treated Atoh1 CreERT2 ;Lgr5 DTR‐GFP ;ROSA26 tdTomato mice.
-
FtdTomato+ colonic organoids from single Atoh1‐tdTomato+/Lgr5‐negative cells (N = 1; n = 3 technical replicates per condition).
-
GPost‐passage colonic organoids established from single tdTomato+cells of control or DT‐treated mice continue growing. tdTomato‐negative cells of control or DT‐treated mice also form colonic organoids after passaging.
-
HtdTomato+ colonic organoids established from single cells from mice in which Lgr5 + stem cells have been ablated show higher growth efficiency compared to their colonic organoid counterparts (N = 1; n = 3 technical replicates per condition).
Figure EV4. Small intestinal Atoh1 + cells show renewal activity following injury in vitro .
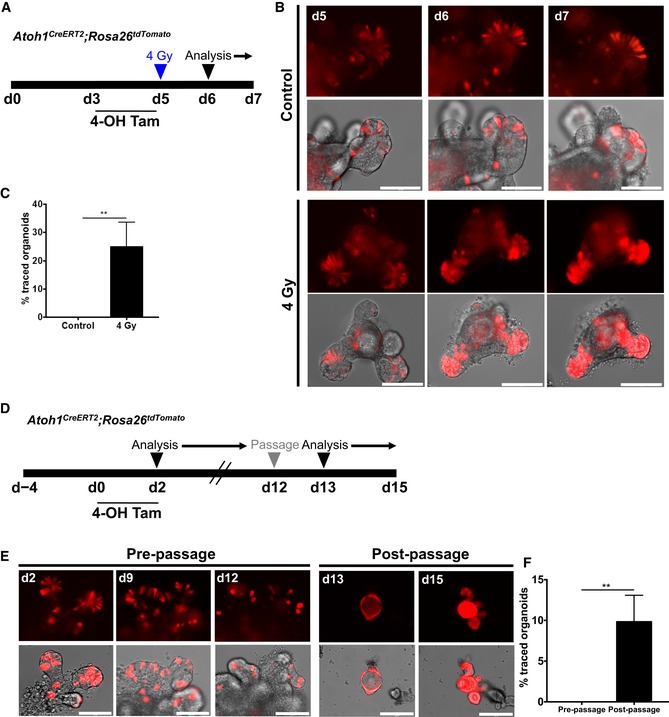
-
AIllustration of experimental protocol outlining 4‐OHT induction and irradiation of Atoh1 CreERT2 ;ROSA26 tdTomato small intestinal organoids.
-
BAtoh1‐tdTomato+ cells within crypts and villus regions of unirradiated organoids are scattered among tdTomato‐negative cells. After irradiation‐induced damaged, Atoh1‐tdTomato+ cells are able to acquire stemness and give rise to fully labeled crypts.
-
CAfter radiation‐induced damage, 25% of organoids show robust lineage labeling (N = 3 control; N = 4 irradiated; n = 3 technical replicates per condition).
-
DIllustration of experimental protocol outlining 4‐OHT induction of Atoh1 CreERT2 ;ROSA26 tdTomato small intestinal organoids.
-
E, FPre‐passage organoids contain sporadic Atoh1‐tdTomato+ cells interspersed between non‐labeled cells. Post‐passaging, fully labeled spheroids develop into fully labeled organoids (N = 3; n = 3 technical replicates per condition).
Figure EV5. Atoh1 + colonic progenitor cells show renewal capacity post‐stress induced passaging in vitro .
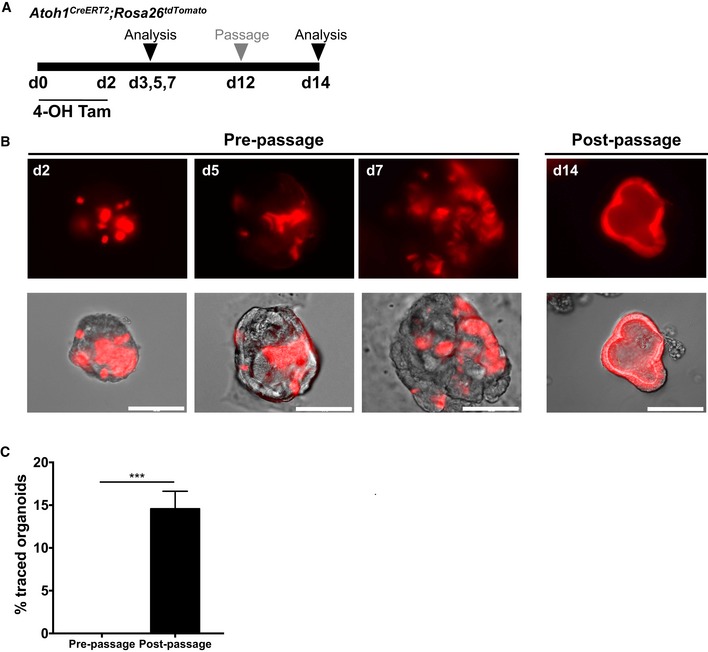
-
AIllustration of experimental protocol outlining 4‐OHT induction of Atoh1 CreERT2 ;ROSA tdTomato colonic organoids.
-
B, CPre‐passage colonic organoids exhibit lineage labeling of Atoh1 + cells that is limited to single cells. Following passage, Atoh1 + progenitors give rise to fully labeled colonic organoids (N = 3; n = 3 technical replicates per condition).
Next, to test whether Atoh1 + cells in the colon have renewal capacity in vitro, we carried out single cell organoid cultures, as previously described (Sato et al, 2009). Using Atoh1 CreERT2 ;ROSA26 tdTomato ;Lgr5 DTR‐GFP mice treated with tamoxifen, we dissociated colonic epithelia into single cells and isolated tdTomato+ cells by flow cytometry (FACS; Fig 5D and E). We then seeded 104 single Atoh1‐expressing tdTomato+ cells per well and found that Atoh1 + cells displayed the capacity to form colonic organoids (Fig 5F), as did occasional tdTomato‐negative cells. We then compared colonic organoid formation as a measure of stem cell capacity of Atoh1 + or Atoh1‐negative crypt cells from DT‐treated and control mice and found significantly increased colonic organoid formation of Atoh1 + cells after Lgr5 + stem cell ablation (Fig 5G and H). Taken together, these data demonstrate that, whereas Atoh1 + secretory cells show infrequent renewal capacity during normal homeostasis, they have a significantly increased capacity to renew the epithelium and dedifferentiate to stem cells upon injury or ablation of the Lgr5‐expressing stem cells.
Atoh1 + colonic progenitor cells are essential for epithelial regeneration post‐injury
Given our observation that Atoh1 + secretory progenitors display renewal capacity in the setting of colonic epithelial injury, we next set out to determine whether Atoh1 + cells are dispensable for colonic regeneration in colitis. To examine this, we crossed Atoh1 CreERT2 ;ROSA26 tdTomato with ROSA26 DTR mice, which allowed us to conditionally and selectively ablate Atoh1‐expressing cells upon treatment with DT. Following the protocol outlined in Fig 6A, we examined the effects of Atoh1 + cell ablation in the setting of DSS colitis in Atoh1 CreERT2 ;ROSA26 tdTomato/DTR mice. Using tdTomato+ cells as a surrogate marker of Cre‐induced DTR expression in Atoh1‐expressing cells, we observed that following DT treatment, Atoh1‐tdTomato+ cell lineage tracing was only marginally decreased by day 12 (Fig 6B), which may be the result of higher sensitivity of ROSA26 tdTomato to recombination than the ROSA26 DTR allele. Nevertheless, concurrent Atoh1 + cell ablation with DSS colitis resulted in exacerbation of colitis as seen by significantly increased weight loss and decreased mouse survival by day 9 (Fig 6C). Moreover, Atoh1 + cell ablation resulted in significantly increased histologic damage (Fig 6D, arrow and E), indicating that Atoh1 + cells play an important functional role in epithelial regeneration after colitis.
Figure 6. Atoh1 + colonic progenitor cells are essential for epithelial regeneration post‐injury.
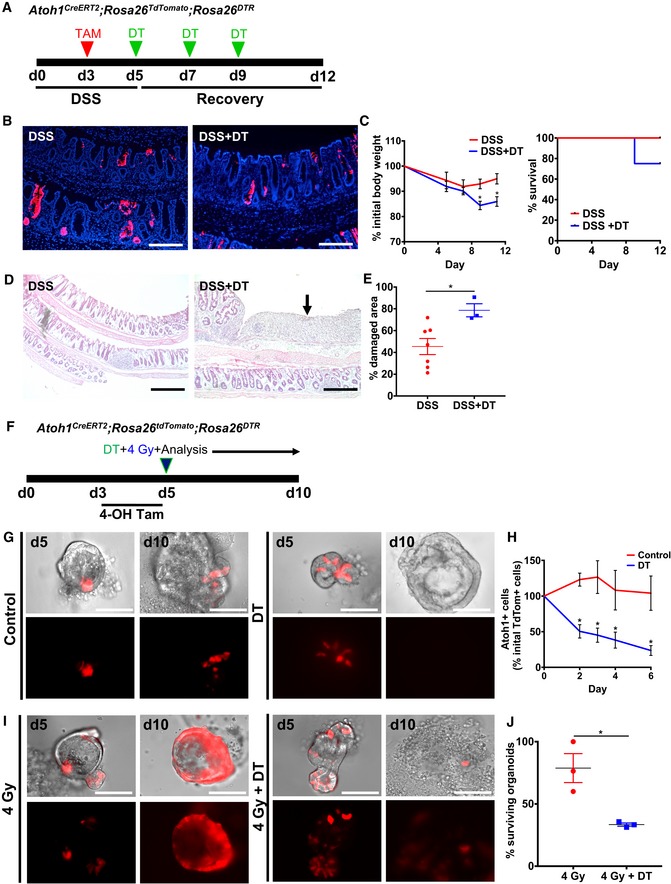
-
AIllustration of experimental protocol outlining DSS‐induced colitis in Atoh1 CreERT2 ;ROSA26 tdTomat/DTR mice.
-
BAtoh1 + lineage labeling post‐DSS and DSS + DT.
-
CDSS + DT‐treated mice exhibit significant weight loss and lower survival compared to DSS alone (N = 4–7 control; N = 3–4 DSS).
-
D, EHematoxylin and eosin staining of colonic epithelium after DSS versus DSS + DT (D). Extensive epithelial damage was observed (D, arrow) and quantified in DSS + DT‐treated mice (N = 7 control; N = 4 DSS).
-
FIllustration of experimental protocol outlining 4‐OHT induction and irradiation in Atoh1 CreERT2 ;ROSA26 tdTomato/DTR colonic organoids.
-
G, HDT treatment effectively ablates Atoh1 + cells shown by reduced number of tdTomato+ cells per organoid.
-
I, JConcurrent irradiation injury and Atoh1 + cell ablation result in significantly reduced organoid survival (N = 3; n = 3 technical replicates per condition).
To further test whether Atoh1 + cells are required for epithelial regeneration post‐injury, we isolated colonic epithelial cells from Atoh1 CreERT2 ;ROSA26 tdTomato/DTR mice and generated colonic organoids. Following 4‐OHT induction of organoids to lineage label Atoh1 + cells (Fig 6F), we determined that DT effectively ablated Atoh1 + cells in vitro, as seen by a significant reduction in tdTomato+ cells at day 2 post‐DT (Fig 6G and H). To then examine the effect of Atoh1 + cell ablation in the setting of injury, we subjected the organoids to 4 Gy irradiation (Fig 6F). As we previously demonstrated, Atoh1 + cells exhibit self‐renewal capacity and expand to give rise to entire organoids in the setting of radiation injury. However, the combination of irradiation and DT‐induced Atoh1 + cell ablation resulted in impaired regenerative capacity and ultimately colonic organoid death, in contrast to saline‐treated control colonic organoids (Fig 6I and J). These data demonstrate that Atoh1 + cell renewal capacity is required for the regenerative capacity of the colonic epithelium after injury affecting Lgr5 + stem cells.
Discussion
It is broadly accepted that Lgr5 + stem cells at the crypt base maintain epithelial turnover during homeostasis. Upon epithelial insult, however, Lgr5 + cells are susceptible to injury, and they must be regenerated before homeostasis can resume. In the small intestine, this renewal can occur through dedifferentiation of committed cells and perhaps from a population of reserve stem cells that have been suggested to reside higher up within the intestinal crypt (Li & Clevers, 2010; Yan et al, 2012; Barker, 2014; Asfaha et al, 2015). Furthermore, in the small intestine, newly formed Lgr5 + cells are indispensable for regeneration following radiation injury (Metcalfe et al, 2014). In contrast, following DSS‐induced colonic injury, DT ablation of colonic Lgr5 + stem cells in Lgr5 DTR mice has no apparent effect on regeneration (Metcalfe et al, 2014). Additionally, colonic epithelial injury resulting in Lgr5 + stem cell disruption promotes crypt hyperplasia and hyperproliferation. Together, these data suggest that colonic Lgr5 + stem cells are dispensable for epithelial regeneration and that epithelial repair can be fueled by an Lgr5‐negative cell population. In order to further delineate which cell population is the principal contributor to colonic epithelial regeneration, we used genetic fate‐mapping mouse models to examine the role of secretory and absorptive progenitor cells following injury.
Using an allele that labels Krt19‐expressing cells, including progenitors and stem cells that are Lgr5‐negative (Asfaha et al, 2015), we determined that a cell population distinct from Lgr5 + stem cells contributes to colonic epithelial regeneration post‐colitis. Notably, Lgr5‐negative, Krt19‐expressing cells are required for proper colonic regeneration to occur, as ablation of Krt19 + cells results in impaired colonic regeneration. These findings are consistent with previous reports in the small intestine showing that plasticity of secretory and absorptive lineages contributes to epithelial regeneration when Lgr5 + stem cells are injured (van Es et al, 2012; Tetteh et al, 2016; Yan et al, 2017). These data confirm that an Lgr5‐negative cell within the colonic crypt has the capacity to revert to a stem cell state to regenerate all epithelial cells, including Lgr5 + cells.
To more precisely define the subset of Lgr5‐negative cells that contribute to colonic regeneration, we examined the fate of Notch1 + absorptive and Atoh1 + secretory progenitors following colitis. Our findings demonstrate that Notch1 + absorptive progenitors do not play a role in colonic epithelial repair, whereas Atoh1 + secretory cells proliferate and expand to contribute to epithelial regeneration during colitis. Interestingly, in the colon, Atoh1 + secretory cells also show infrequent renewal capacity during normal homeostasis, in contrast to the small intestine, in which Atoh1 + secretory cells do not lineage trace at baseline. Upon injury, however, both colonic and intestinal Atoh1 + cells expand to renew the epithelium, consistent with plasticity required for crypt regeneration. Our observations are consistent with the recent findings reported by two other groups demonstrating that Atoh1 + secretory cells are able to contribute to colonic regeneration post‐colitis (Ishibashi et al, 2018; Tomic et al, 2018). Thus, a subset of Atoh1 + cells in the colon can renew the epithelium during normal homeostasis, whereas in the small intestine, Atoh1 + cells behave similarly to intestinal Dll1 + progenitors cells that are short‐lived and only lineage trace upon epithelial injury (van Es et al, 2012).
Complementing the recent findings showing that phosphorylation of ATOH1 regulates the plasticity of secretory progenitors (Tomic et al, 2018), we found that renewal by Atoh1 + cells is independent of Lgr5 + stem cells and that Atoh1 + cells are multipotent in terms of their renewal capacity. This is demonstrated by their ability to give rise to colonic organoids in vitro, as well as to lineage trace in vivo in the absence of Lgr5 + cells by DT ablation during regeneration. Overall, our results support the idea that during colonic regeneration, Atoh1 + progenitors undergo a proliferative phase 4 days post‐injury, which takes place in complete absence of the bona fide Lgr5 + stem cells. Subsequently, Atoh1 + cells enter a normalization phase ~8 days post‐injury, characterized by reemergence of Lgr5 + stem cells and resumption of steady state. A similar phenomenon is also seen in intestinal regeneration after irradiation (Kim et al, 2017).
Notably, Atoh1‐negative cells formed colonic organoids in the absence of Lgr5 + cells, albeit to a smaller degree than Atoh1 + cells. This suggests that, in the context of injury, the presence of additional cells within the crypt may also aid in colonic regeneration, as we and others have recently proposed (Yui et al, 2018). Importantly, we demonstrate for the first time that Atoh1 + cells are essential for colonic regeneration, as their ablation during colitis significantly impairs colonic healing. Moreover, our data also shows that the combination of radiation or DSS colitis plus Atoh1 + cell ablation significantly impairs colonic healing, thus suggesting that simultaneous elimination of both Lgr5 + and Atoh1 + cell populations is detrimental to colonic healing.
In summary, we have found that the renewal capacity of Atoh1 + secretory progenitors, which is modest during normal homeostasis, is critical for colonic epithelial regeneration during colitis. Importantly, colonic regeneration in this context is independent of Lgr5 + stem cells, and Notch1 + absorptive progenitors do not significantly contribute to this process. These findings suggest that future therapy aimed at promoting colonic regeneration in colitis may be enhanced by promoting the survival and function of Atoh1 + secretory progenitors.
Materials and Methods
Animals
Mouse lines used include combinations of the following alleles or transgenes: Lgr5 GFP‐IRES‐CreERT2 (Jax 008875; Barker et al, 2007), Lgr5 DTR‐GFP (Tian et al, 2011), Krt19 BAC‐CreERT2 (Asfaha et al, 2015), Notch1 CreERT2 (Fre et al, 2011), Atoh1 CreERT2 (Fujiyama et al, 2009), ROSA26 DTR (Jax 007900; Buch et al, 2005), and ROSA26 tdTomato (Jax 007905; Madisen et al, 2010). Mice were 8–16 weeks of age at the start of each experiment. Rodent work was done in accordance with approved protocols by the Institutional Animal Care and Use Committees at University of California San Francisco and University of Western Ontario.
DSS‐induced colitis and tamoxifen induction
Mice were exposed to 2.5–3% (w/v) DSS (DS1004, Gojira Fine Chemicals or 02160110, MP Biomedicals) in their drinking water for 5 days (D0–D5). The mice were closely monitored during the treatment and recovery periods using body weight and other established criteria. Mice were oral gavaged with three doses of 1–6 mg tamoxifen (0215673894, MP Biomedicals or T5648, Sigma) dissolved in corn oil, and colonic tissue was harvested at the indicated time points during recovery.
Diphtheria toxin cell ablation
Using the Lgr5 DTR‐GFP allele, the diphtheria toxin receptor is constitutively expressed in all Lgr5‐expressing cells. For ablation studies, mice were treated with 200–1,000 ng of diphtheria toxin (322326, Sigma‐Aldrich) at regular intervals and monitored as described above. Using Atoh1 CreERT2 ;ROSA26 DTR mice, we are able to conditionally express the diphtheria toxin receptor in Atoh1‐expressing cells upon tamoxifen treatment. For ablation studies in vivo, mice were treated with 1,000 ng of diphtheria toxin at regular intervals and monitored as described above. For treatment of organoids in vitro, diphtheria toxin was added to the culture medium at a final concentration of 200 ng/ml every other day.
Tissue preparation for immunofluorescence and histology
Harvested intestinal and colonic tissues were perfusion‐fixed or fresh‐frozen. For perfusion fixation, animals were anesthetized by intraperitoneal (i.p.) injection of 250 mg/kg of body weight avertin (2,2,2‐tribromoethanol) and transcardially perfused with 4% paraformaldehyde (PFA) in 0.1 M phosphate‐buffered saline (PBS). Dissected tissues were post‐fixed in 4% PFA for 3 h at 4°C and cryoprotected in 30% sucrose in 1× PBS overnight at 4°C. Tissue was embedded in OCT compound (4583, Sakura or 23‐730‐571, Thermo Fisher Scientific), frozen, and stored at −80°C. Fresh tissues were rinsed in sterile ice‐cold 1× phosphate‐buffered saline (PBS), embedded in OCT compound, frozen, and stored at −80°C. Fresh tissue sections were fixed with 4% paraformaldehyde. For paraffin‐embedded tissues, we fixed tissues with 4% paraformaldehyde for 24–48 h, followed by paraffin processing, using a standard protocol.
Immunofluorescence and histology
Immunofluorescence was performed on 6‐ to 7‐μm cryosections. Primary antibodies and dilutions used are as follows: chicken anti‐GFP (1:1,000; GFP‐1020, Aves Labs), rabbit anti‐DCLK1 (1:200; ab31704, Abcam), rabbit anti‐chromogranin A (1:100; 20085, ImmunoStar), and rabbit anti‐Muc2 (1:100; ab76774, Abcam). Appropriate secondary antibodies from Thermo Fisher Scientific were used at 1:1,000. Sections were counterstained with Dapi (1:10,000; D9542, Sigma) and coverslipped with Vectashield (H‐1000, Vector Labs) or ProLong Gold Antifade (P36930, Thermo Fisher Scientific). For histology assessment, 6‐ to 7‐μm paraffin or frozen sections were prepared and stained with hematoxylin and eosin using standard methods. Slides were coverslipped using Permount (SP15‐100, Fisher Scientific).
Tissue preparation for RNAscope
Intestinal and colonic tissues were immersed in 4% PFA for 24 h at room temperature, followed by standard dehydration and paraffin embedding.
RNAscope and quantification of Lgr5 transcripts
RNA in situ hybridization for Lgr5 expression was performed on 7‐μm paraffin sections using RNAscope® 2.5 High Definition (HD)—Red Assay (322350, Advanced Cell Diagnostics). Manufacturer's protocol was followed with 15 min of target retrieval and 30 min of protease digestion, using the following probe: Mm Lgr5 (312171, Advanced Cell Diagnostics). Quantification of Lgr5 mRNA transcripts was performed using the open‐source platform Fiji (Schindelin et al, 2012) and the analysis guidelines from Advanced Cell Diagnostics. Area of single probes was measured and used to determine total probe count within probe clusters. Total probe clusters containing at least 10 probes were quantified and normalized to number of crypts analyzed in proximal, mid, and distal colon.
Intestinal and colonic organoid cultures
Intestinal or colonic crypts were isolated from mouse as previously described by Bjerknes and Cheng with some modifications, and cultured in the presence of EGF 50 ng/ml (PMG8043, Invitrogen), mNoggin 100 ng/ml (250‐38, PeproTech), R‐spondin 1 μg/ml, and Wnt3a 100 ng/ml (315‐20, PeproTech) for colonic cultures, as previously described (Sato et al, 2009). Organoids were treated with 4‐hydroxytamoxifen (4‐OHT) 1 μM (H6278, Sigma) to induce Cre‐recombinase activity. Organoids were irradiated using a Cobalt‐60 Irradiator (Gammacell) at 80.3 cGy/min for a total dose of 4 Gy. Traced organoids were quantified as the percentage of organoids displaying entire tdTomato+ crypt tracing over the total number of organoids with detectable Atoh1‐tdTomato+ cells per well.
Single cell colonic organoid cultures
Flow cytometry
Colon from Atoh1 CreERT2 ;ROSA26 tdTomato ;Lgr5 DTR‐GFP mice was dissected, flushed with cold 1× PBS, opened lengthwise, and subjected to 10 mM DTT in complete HBSS (14185‐020, Gibco; HBSS, 10 mM HEPES pH 8, 2% FBS) at 37°C for 20 min followed by 10‐min wash in complete HBSS. Next, colonic tissue was cut into 0.5‐mm pieces and subjected to 10 mM EDTA in complete HBSS. After EDTA incubation, the solution was subjected to vigorous mechanical dissociation. The suspension was filtered through a 40‐μm cell strainer, the volume was adjusted to 45 ml with cold RPMI (10‐543Q, Lonza), and cells were pelleted by centrifugation at 350 × g for 5 min. The resulting cell pellet was resuspended in complete HBSS containing 5 mM EDTA and stained for flow cytometry using CD45‐BV605 (30‐F11, 103139, BioLegend), CD326/EpCAM‐APC (G8.8, 118213, BioLegend), and CD44‐PeCy7 (IM7, 103029, BioLegend) for 30 min on ice. After washing, Dapi (1:10,000; D9542, Sigma) was added. Cells were subsequently sorted on a FACSAria II (BD Bioscience) cell sorter into 500 μl advanced DMEM/F12 medium (12634‐010, Thermo Fisher Scientific) supplemented with 1% GlutaMAX (35050061, Thermo Fisher Scientific) and 1% penicillin/streptomycin (15140148, Thermo Fisher Scientific).
Culture
Cells were spun down and resuspended in 50 μl Phenol‐free Matrigel (356231, Cornig) supplemented with 10 μM Jagged‐1 (188‐204, Anaspec,), 500 ng/ml human EGF (AF‐100‐15, PeproTech), 1 μg/ml murine Noggin (250‐38, PeproTech), 10% mouse R‐spondin1 conditioned medium (gift of Noah Shroyer, Baylor College of Medicine), and 10% murine Wnt3a conditioned medium (CRL‐2647, ATCC). Matrigel drops of 50 μl were plated in a 12‐well cell culture plate and left to set at 37°C for 15 min. Organoids were cultured in advanced DMEM/F12 in the presence of 50 ng/ml human EGF, 100 ng/ml murine Noggin, 5% mouse R‐spondin1 conditioned medium, 50% murine Wnt3a conditioned medium, 10 mM nicotinamide (N3376, Sigma), 1% N‐2 supplement (17502048, Thermo Fisher Scientific), and 2% B‐27 supplement (17504044, Thermo Fisher Scientific). During first 3 days of culture, 2.5 μM Y‐27632 dihydrochloride (Y0503, Sigma), 2.5 μM Chir99021 (SML1046, Sigma), and 1 μM Jagged‐1 were used, and medium was changed every 3 days.
Image acquisition and analysis
Fluorescence and bright‐field images were acquired using a Leica DM5000 B and Leica DFC 500 with LAS V4.9 software or EVOS‐FL Auto microscope. Confocal images were obtained as a z‐stack of 0.76‐μm optical sections acquired sequentially using a Leica TCS SP5 II confocal microscope with LAS X software. Ventral images of whole mount crypts were acquired with a Zeiss Observer Z1 with ZEN blue software.
To determine the severity of colitis, mouse body weight and survival were assessed during the experimental time course. Furthermore, colonic tissue was examined for percentage of damaged area. Assessment of damage throughout the entire length of the colon was made possible using cross section through the colonic Swiss roll preparation. Total colonic area and total damaged area were obtained using EVOS FL Auto area detection software (Life Technologies), and total damaged area was expressed as a percentage of total colonic area. The number of contiguously labeled crypts was used as a determinant of tissue regeneration and was obtained by manually counting the number of contiguously lineage‐traced crypts and expressing these values per 1,000 crypts.
Statistical analysis
Normally distributed data were analyzed using parametric tests including two‐tailed Student's t‐test with Welch's correction, one‐way ANOVA with Bonferroni correction, or two‐way ANOVA with Sidak's multiple comparisons test. The non‐parametric Mann–Whitney U‐test was used if the data did not fit a normal distribution. Significance was taken as P < 0.05 with a confidence interval of 95%. Data are presented as mean ± SD for parametric data or as mean ± SEM for non‐parametric data.
Author contributions
Conceptualization: DC‐A, ENF, RN, HJG, ODK, SA; Methodology and data acquisition: DC‐A, ENF, RN, HJG, TW, MAP, ODK, SA; Analysis: DC‐A, ENF, RN, HJG, ODK, SA; Investigation: DC‐A, ENF, RN, HJG, FJdS, ODK, SA; Writing: DC‐A, ENF, HJG, ODK, SA.
Conflict of interest
F.J.d.S. is an employee of Genentech and owns shares in Roche.
Supporting information
Expanded View Figures PDF
Review Process File
Acknowledgements
We thank Nicholas Wang, Asoka Rathnayake, and Liyue Zhang for technical assistance and Drs. Ysbrand M. Nusse, Jimmy Kuang‐Hsien Hu, Amnon Sharir and Kara L. McKinley for helpful discussions. This work was supported by operating grants from CIHR and Cancer Research Society awarded to S.A. and by the National Institutes of Health (R35‐DE026602, U01‐DK103147) and the California Institute for Regenerative Medicine (RN3‐06525) to O.D.K. Research reported in this publication was supported by National Institute of Diabetes and Digestive and Kidney Disorders (NIDDK) and National Institute of Allergy and Infectious Diseases (NIAID) of the National Institutes of Health under grant number U01DK103147. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. E.F. was supported by a Cancer Research and Technology Transfer (CaRTT) Strategic Training Program Stipend, and H.G. was supported in part by a studentship from the Lawson Internal Research Fund.
The EMBO Journal (2019) 38: e99984
Contributor Information
Ophir D Klein, Email: ophir.klein@ucsf.edu.
Samuel Asfaha, Email: sasfaha2@uwo.ca.
References
- Asfaha S, Hayakawa Y, Muley A, Stokes S, Graham TA, Ericksen RE, Westphalen CB, von Burstin J, Mastracci TL, Worthley DL, Guha C, Quante M, Rustgi AK, Wang TC (2015) Krt19(+)/Lgr5(‐) cells are radioresistant cancer‐initiating stem cells in the colon and intestine. Cell Stem Cell 16: 627–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, Clevers H (2007) Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449: 1003–1007 [DOI] [PubMed] [Google Scholar]
- Barker N, Clevers H (2010) Leucine‐rich repeat‐containing G‐protein‐coupled receptors as markers of adult stem cells. Gastroenterology 138: 1681–1696 [DOI] [PubMed] [Google Scholar]
- Barker N (2014) Adult intestinal stem cells: critical drivers of epithelial homeostasis and regeneration. Nat Rev Mol Cell Biol 15: 19–33 [DOI] [PubMed] [Google Scholar]
- Barriga FM, Montagni E, Mana M, Mendez‐Lago M, Hernando‐Momblona X, Sevillano M, Guillaumet‐Adkins A, Rodriguez‐Esteban G, Buczacki SJA, Gut M, Heyn H, Winton DJ, Yilmaz OH, Attolini CS, Gut I, Batlle E (2017) Mex3a marks a slowly dividing subpopulation of Lgr5+ intestinal stem cells. Cell Stem Cell 20: 801–816 e807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buch T, Heppner FL, Tertilt C, Heinen TJAJ, Kremer M, Wunderlich FT, Jung S, Waisman A (2005) A Cre‐inducible diphtheria toxin receptor mediates cell lineage ablation after toxin administration. Nature Methods 2: 419–426 [DOI] [PubMed] [Google Scholar]
- Buczacki SJ, Zecchini HI, Nicholson AM, Russell R, Vermeulen L, Kemp R, Winton DJ (2013) Intestinal label‐retaining cells are secretory precursors expressing Lgr5. Nature 495: 65–69 [DOI] [PubMed] [Google Scholar]
- Cooper HS, Murthy SN, Shah RS, Sedergran DJ (1993) Clinicopathologic study of dextran sulfate sodium experimental murine colitis. Lab Invest 69: 238–249 [PubMed] [Google Scholar]
- Davidson LA, Goldsby JS, Callaway ES, Shah MS, Barker N, Chapkin RS (2012) Alteration of colonic stem cell gene signatures during the regenerative response to injury. Biochem Biophys Acta 1822: 1600–1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, Elvassore N, Piccolo S (2011) Role of YAP/TAZ in mechanotransduction. Nature 474: 179–183 [DOI] [PubMed] [Google Scholar]
- van Es JH, van Gijn ME, Riccio O, van den Born M, Vooijs M, Begthel H, Cozijnsen M, Robine S, Winton DJ, Radtke F, Clevers H (2005) Notch/gamma‐secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature 435: 959–963 [DOI] [PubMed] [Google Scholar]
- van Es JH, Sato T, van de Wetering M, Lyubimova A, Nee AN, Gregorieff A, Sasaki N, Zeinstra L, van den Born M, Korving J, Martens AC, Barker N, van Oudenaarden A, Clevers H (2012) Dll1+ secretory progenitor cells revert to stem cells upon crypt damage. Nat Cell Biol 14: 1099–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fre S, Hannezo E, Sale S, Huyghe M, Lafkas D, Kissel H, Louvi A, Greve J, Louvard D, Artavanis‐Tsakonas S (2011) Notch lineages and activity in intestinal stem cells determined by a new set of knock‐in mice. PLoS One 6: e25785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiyama T, Yamada M, Terao M, Terashima T, Hioki H, Inoue YU, Inoue T, Masuyama N, Obata K, Yanagawa Y, Kawaguchi Y, Nabeshima Y, Hoshino M (2009) Inhibitory and excitatory subtypes of cochlear nucleus neurons are defined by distinct bHLH transcription factors, Ptf1a and Atoh1. Development 136: 2049–2058 [DOI] [PubMed] [Google Scholar]
- Furuyama K, Kawaguchi Y, Akiyama H, Horiguchi M, Kodama S, Kuhara T, Hosokawa S, Elbahrawy A, Soeda T, Koizumi M, Masui T, Kawaguchi M, Takaori K, Doi R, Nishi E, Kakinoki R, Deng JM, Behringer RR, Nakamura T, Uemoto S (2011) Continuous cell supply from a Sox9‐expressing progenitor zone in adult liver, exocrine pancreas and intestine. Nat Genet 43: 34–41 [DOI] [PubMed] [Google Scholar]
- Hirata A, Utikal J, Yamashita S, Aoki H, Watanabe A, Yamamoto T, Okano H, Bardeesy N, Kunisada T, Ushijima T, Hara A, Jaenisch R, Hochedlinger K, Yamada Y (2013) Dose‐dependent roles for canonical Wnt signalling in de novo crypt formation and cell cycle properties of the colonic epithelium. Development 140: 66–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi F, Shimizu H, Nakata T, Fujii S, Suzuki K, Kawamoto A, Anzai S, Kuno R, Nagata S, Ito G, Murano T, Mizutani T, Oshima S, Tsuchiya K, Nakamura T, Watanabe M, Okamoto R (2018) Contribution of ATOH1(+) cells to the homeostasis, repair, and tumorigenesis of the colonic epithelium. Stem Cell Reports 10: 27–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itzkovitz S, Lyubimova A, Blat IC, Maynard M, van Es J, Lees J, Jacks T, Clevers H, van Oudenaarden A (2012) Single‐molecule transcript counting of stem‐cell markers in the mouse intestine. Nat Cell Biol 14: 106–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Saadatpour A, Guo G, Saxena M, Cavazza A, Desai N, Jadhav U, Jiang L, Rivera MN, Orkin SH, Yuan GC, Shivdasani RA (2016) Single‐cell transcript profiles reveal multilineage priming in early progenitors derived from Lgr5(+) intestinal stem cells. Cell Rep 16: 2053–2060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CK, Yang VW, Bialkowska AB (2017) The role of intestinal stem cells in epithelial regeneration following radiation‐induced gut injury. Curr Stem Cell Rep 3: 320–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laroui H, Ingersoll SA, Liu HC, Baker MT, Ayyadurai S, Charania MA, Laroui F , Yan Y, Sitaraman SV, Merlin D (2012) Dextran sodium sulfate (DSS) induces colitis in mice by forming nano‐lipocomplexes with medium‐chain‐length fatty acids in the colon. PLoS One 7: e32084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Clevers H (2010) Coexistence of quiescent and active adult stem cells in mammals. Science 327: 542–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES, Zeng H (2010) A robust and high‐throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 13: 133–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalfe C, Kljavin NM, Ybarra R, de Sauvage FJ (2014) Lgr5+ stem cells are indispensable for radiation‐induced intestinal regeneration. Cell Stem Cell 14: 149–159 [DOI] [PubMed] [Google Scholar]
- Montgomery RK, Carlone DL, Richmond CA, Farilla L, Kranendonk ME, Henderson DE, Baffour‐Awuah NY, Ambruzs DM, Fogli LK, Algra S, Breault DT (2011) Mouse telomerase reverse transcriptase (mTert) expression marks slowly cycling intestinal stem cells. Proc Natl Acad Sci USA 108: 179–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okayasu I, Hatakeyama S, Yamada M, Ohkusa T, Inagaki Y, Nakaya R (1990) A novel method in the induction of reliable experimental acute and chronic ulcerative colitis in mice. Gastroenterology 98: 694–702 [DOI] [PubMed] [Google Scholar]
- Park HS, Goodlad RA, Wright NA (1995) Crypt fission in the small intestine and colon. A mechanism for the emergence of G6PD locus‐mutated crypts after treatment with mutagens. Am J Pathol 147: 1416–1427 [PMC free article] [PubMed] [Google Scholar]
- Pellegrinet L, Rodilla V, Liu Z, Chen S, Koch U, Espinosa L, Kaestner KH, Kopan R, Lewis J, Radtke F (2011) Dll1‐ and dll4‐mediated notch signaling are required for homeostasis of intestinal stem cells. Gastroenterology 140: 1230–1240 e1231–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell AE, Wang Y, Li Y, Poulin EJ, Means AL, Washington MK, Higginbotham JN, Juchheim A, Prasad N, Levy SE, Guo Y, Shyr Y, Aronow BJ, Haigis KM, Franklin JL, Coffey RJ (2012) The pan‐ErbB negative regulator Lrig1 is an intestinal stem cell marker that functions as a tumor suppressor. Cell 149: 146–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riccio O, van Gijn ME, Bezdek AC, Pellegrinet L, van Es JH, Zimber‐Strobl U, Strobl LJ, Honjo T, Clevers H, Radtke F (2008) Loss of intestinal crypt progenitor cells owing to inactivation of both Notch1 and Notch2 is accompanied by derepression of CDK inhibitors p27Kip1 and p57Kip2. EMBO Rep 9: 377–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambuy Y, De Angelis I, Ranaldi G, Scarino ML, Stammati A, Zucco F (2005) The Caco‐2 cell line as a model of the intestinal barrier: influence of cell and culture‐related factors on Caco‐2 cell functional characteristics. Cell Biol Toxicol 21: 1–26 [DOI] [PubMed] [Google Scholar]
- Sangiorgi E, Capecchi MR (2008) Bmi1 is expressed in vivo in intestinal stem cells. Nat Genet 40: 915–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, Clevers H (2009) Single Lgr5 stem cells build crypt‐villus structures in vitro without a mesenchymal niche. Nature 459: 262–265 [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda‐Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A (2012) Fiji: an open‐source platform for biological‐image analysis. Nat Methods 9: 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda N, Jain R, LeBoeuf MR, Wang Q, Lu MM, Epstein JA (2011) Interconversion between intestinal stem cell populations in distinct niches. Science 334: 1420–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetteh PW, Basak O, Farin HF, Wiebrands K, Kretzschmar K, Begthel H, van den Born M, Korving J, de Sauvage F, van Es JH, van Oudenaarden A, Clevers H (2016) Replacement of lost Lgr5‐positive stem cells through plasticity of their enterocyte‐lineage daughters. Cell Stem Cell 18: 203–213 [DOI] [PubMed] [Google Scholar]
- Tian H, Biehs B, Warming S, Leong KG, Rangell L, Klein OD, de Sauvage FJ (2011) A reserve stem cell population in small intestine renders Lgr5‐positive cells dispensable. Nature 478: 255–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomic G, Morrissey E, Kozar S, Ben‐Moshe S, Hoyle A, Azzarelli R, Kemp R, Chilamakuri CSR, Itzkovitz S, Philpott A, Winton DJ (2018) Phospho‐regulation of ATOH1 is required for plasticity of secretory progenitors and tissue regeneration. Cell Stem Cell 23: 436–443 e437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan KS, Chia LA, Li X, Ootani A, Su J, Lee JY, Su N, Luo Y, Heilshorn SC, Amieva MR, Sangiorgi E, Capecchi MR, Kuo CJ (2012) The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations. Proc Natl Acad Sci USA 109: 466–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan KS, Gevaert O, Zheng GXY, Anchang B, Probert CS, Larkin KA, Davies PS, Cheng ZF, Kaddis JS, Han A, Roelf K, Calderon RI, Cynn E, Hu X, Mandleywala K, Wilhelmy J, Grimes SM, Corney DC, Boutet SC, Terry JM et al (2017) Intestinal enteroendocrine lineage cells possess homeostatic and injury‐inducible stem cell activity. Cell Stem Cell 21: 78–90 e76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yui S, Azzolin L, Maimets M, Pedersen MT, Fordham RP, Hansen SL, Larsen HL, Guiu J, Alves MRP, Rundsten CF, Johansen JV, Li Y, Madsen CD, Nakamura T, Watanabe M, Nielsen OH, Schweiger PJ, Piccolo S, Jensen KB (2018) YAP/TAZ‐dependent reprogramming of colonic epithelium links ECM remodeling to tissue regeneration. Cell Stem Cell 22: 35–49 e37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachos NC, Kovbasnjuk O, Foulke‐Abel J, In J, Blutt SE, de Jonge HR, Estes MK, Donowitz M (2016) Human enteroids/colonoids and intestinal organoids functionally recapitulate normal intestinal physiology and pathophysiology. J Biol Chem 291: 3759–3766 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expanded View Figures PDF
Review Process File


