Abstract
Airway epithelium structure/function can be altered by local inflammatory/immune signals, and this process is called epithelial remodeling. The mechanism by which this innate response is regulated, which causes mucin/mucus overproduction, is largely unknown. Exosomes are nanovesicles that can be secreted and internalized by cells to transport cellular cargo, such as proteins, lipids, and miRNA. The objective of this study was to understand the role exosomes play in airway remodeling through cell–cell communication. We used two different human airway cell cultures: primary human tracheobronchial (HTBE) cells, and a cultured airway epithelial cell line (Calu-3). After intercellular exosomal transfer, comprehensive proteomic and genomic characterization of cell secretions and exosomes was performed. Quantitative proteomics and exosomal miRNA analysis profiles indicated that the two cell types are fundamentally distinct. HTBE cell secretions were typically dominated by fundamental innate/protective proteins, including mucin MUC5B, and Calu-3 cell secretions were dominated by pathology-associated proteins, including mucin MUC5AC. After exosomal transfer/intake, approximately 20% of proteins, including MUC5AC and MUC5B, were significantly altered in HTBE secretions. After exosome transfer, approximately 90 miRNAs (∼4%) were upregulated in HTBE exosomes, whereas Calu-3 exosomes exhibited a preserved miRNA profile. Together, our data suggest that the transfer of exosomal cargo between airway epithelial cells significantly alters the qualitative and quantitative profiles of airway secretions, including mucin hypersecretion, and the miRNA cargo of exosomes in target cells. This finding indicates that cellular information can be carried between airway epithelial cells via exosomes, which may play an important role in airway biology and epithelial remodeling.
Keywords: exosomes, airway remodeling, mucin hypersecretion, asthma, chronic obstructive pulmonary disease
The human airway mucosa is lined by diverse epithelial cells, including ciliated and secretory goblet cells on the surface and serous and mucous cells in the submucosal glands (1). This organization of cells and secreted products contributes to mucociliary clearance and mediates an effective innate immune response to environmental exposure that helps maintain pulmonary homeostasis in both health and disease states (2). Respiratory epithelial cells modulate innate immune responses by releasing protective molecules, such as mucins, proteins (3), chemokines, cytokines, and growth factors, that regulate the migration and activation of diverse immune cells to sites of insult and injury (1, 4). Traditionally, cells were thought to interact through small, secreted molecules, such as hormones or growth factors, or through membrane junctions (5). However, the mechanism through which airway cells maintain this complex environment/interaction is poorly understood.
Exosomes, microvesicles, and apoptotic bodies are all secreted membrane-bound extracellular vesicles by definition, but their size, cellular origin, synthesis, cargo and function are distinct. Exosomes have been shown to be 40- to 120-nm, cup-shaped, endosomal, membranous vesicles that are secreted by nearly all cells, and to be present (6) in many biological fluids, including plasma, urine, saliva (7), BAL fluid (8), semen, cerebral spinal fluid (9), breast milk (10), and airway secretions (11). The function of extracellular vesicles, exosomes in particular, as novel mediators of intercellular communication has become a recent focus of extensive scientific research (12, 13).
Exosomes appear to be capable of interacting with and being internalized by nearly all cell types to transport cellular cargo, such as proteins, lipids, and nucleic acids (miRNA and mRNA) (13). Depending on their origin, exosomes contain specific sets of known proteins, including tetraspanins, heat shock proteins (Hsps), annexins and membrane-bound mucins; however, the role of the individual components of exosomes is still unknown. The functional role of exosomes is well studied in various diseases, such as cancer (14), diabetes (15), renal disorders (16), and inflammation (17). However, the role of exosomes in intercellular communication in the airway epithelium and their contributions to the innate defense against and/or pathogenesis of lung diseases are largely unknown.
We previously reported that cultured primary airway epithelial cells (human tracheobronchial [HTBE] cells) release exosome-like vesicles with innate immune properties (11). Here, we hypothesize that airway epithelium–derived exosomes play a major role in cell-to-cell communication in the lung, and that these exosomes are involved in regulating airway homeostasis and remodeling by transferring cellular cargo (miRNA or proteins). To test this hypothesis, we used two phenotypically distinct airway epithelial cell types: primary HTBE cells and a cultured human airway epithelial cell line (Calu-3). After controlled in vitro intercellular exosomal transfer, comprehensive proteomic and genomic characterization of cell secretions and exosomes was performed to understand alterations in the cell microenvironment mediated by cellular cross-talk through exosomes.
Methods
Additional details are provided in the data supplemental materials.
Cell Culture
Two different airway cell culture systems that secrete mucus were used in this study: primary HTBE cells and the Calu-3 cell line. HTBE cells were isolated and cultured as previously described (11, 18). Calu-3 cells, derived from human lung adenocarcinoma, were maintained at the air–liquid interface for at least 3 weeks, as previously described (19). Apical secretions were obtained by performing two sequential 1-ml PBS washes on the surface of the cultures. Culture washings obtained from HTBE cells from five individuals (each with a unique code identifier) were used.
Isolation and Characterization of Exosomes
Exosomes were isolated from HTBE and Calu-3 secretions using differential centrifugation (11). Nanoparticle tracking analysis was used for size and concentration analysis of the isolated exosomes using a NanoSight NS300 system (Malvern Instruments), as described previously (6). Each experiment was performed in triplicate. Electron microscopy (EM) analysis of exosomes was essentially performed as previously described (6).
Exosome Transfer between Cells
Cells were washed three times with PBS, and 1 × 108 exosomes in 100 μl of PBS were added to each well (n = 5) for 3 days; 100 μl of PBS was added to the control wells (n = 5). The cells were thoroughly washed with PBS on the fourth day, and the apical cell washings were collected after 30 minutes of incubation with 1 ml of PBS. Cell washings were collected for 3 days. The washings from each well from all collection times were pooled and aliquoted for analysis.
Exosome Labeling and Uptake
To observe the exosomal uptake by the recipient cells, Calu-3 exosomes were labeled with SYTO RNASelect green fluorescent stain and BODIPY TR ceramide red fluorescent stain. The exosomes were incubated at 37°C for 15 minutes and then purified from the excess dye using a Sepharose CL-2B 10/30 gel filtration column. As a control, PBS was mixed with dyes and purified similarly. HTBE cells were washed with PBS and treated with 1 × 108 labeled Calu-3 exosomes or the same volume of SYTO RNASelect-BODIPY TR-PBS control for 1.5 hours. Cells were washed twice with PBS, fixed with 100% ice-cold methanol, permeabilized with 0.1% Triton X-100, and blocked with 3% BSA. Cells were incubated with primary mouse anti–β-tubulin IV antibody (BioGenex) overnight at 4°C and then stained with secondary donkey anti-mouse Alexa 647 antibody and Hoechst. The z-stack confocal images were acquired using an Olympus FluoView FV1000 microscope with a 60× objective. Volocity software was used to construct three-dimensional images and XZ projections.
Mass Spectrometry
The samples were reduced, alkylated, and trypsinized, and label-free quantitative mass spectrometry (MS) was performed as described previously (20). The raw file was analyzed with Proteome Discoverer 1.3 software using the UniProt-human complete FASTA databank, and trypsin was set as the enzyme, with two max missed cleavage sites; methionine oxidation (+15.995 Da) was set as the dynamic modification, and carbamidomethylation (+57.021 Da) was set as the static modification.
Exosomal miRNA Library
A miRNA library was constructed according to the HTG EdgeSeq miRNA WTA ILM kit instructions. Briefly, 15 μl of lysis buffer was added to the 15-μl exosome sample. Tubes were then heated to 95°C for 15 minutes. Next, 1.5 μl of proteinase K was added, and the sample was mixed well by pipetting and incubated for 30 minutes at 50°C in an orbital shaker. A total of 25 μl of working lysate was transferred to each well of the HTG EdgeSeq scanning plate. Appropriate kit components for preparing miRNA libraries were loaded into the system platform, and the HTG EdgeSeq program was started. The sequencing adaptors and barcodes were added to the sequencing libraries upon completion of the HTG EdgeSeq run, and samples were amplified using the PCR method. After the PCR step, sequencing libraries were concentrated, pooled, and then sequenced on an MiSeq or HiSeq2500 RR Illumina sequencing system using the Single End 50 cycles setting.
Bioinformatics and Statistical Analysis
The miRNA raw count data were used as input for differential expression analysis with the Bioconductor R package, edgeR (21), which models the count data based on the negative binomial distribution. Differential expression between comparison groups was analyzed with the sample subsets using exact tests (22), and multiple tests were corrected using the Benjamini-Hochberg false discovery rate. Statistical analyses comparing two groups of proteomic data sets were performed using a t test.
Results
Our experimental strategy is summarized in Figure E1 in the data supplement. Briefly, HTBE primary cell cultures and the Calu-3 cell line were grown on an air–liquid interface. Label-free quantitative proteomics analysis of the secretions from both cultures was performed before and after exosomal exchange experiments. Exosomes secreted from both HTBE (HTBE-exo) and Calu-3 (Calu-3-exo) cells were isolated and characterized, and their protein and miRNA cargoes were studied.
Isolation and Initial Characterization of Exosomes/Vesicles
Nanoparticle tracking analysis indicated that the average size of HTBE-exo was 325 nm, and the average size of Calu-3-exo was 135 nm (Figures E2A and E2B). Electron microscopy images of negatively stained exosomal preparations showed typical, cup-shaped nanovesicular structures with a diameter in the range of 40–100 nm in both exosome populations. A proportion of HTBE-exo had membrane-tethered mucins that increased their overall radius of hydration in light scattering measurements (Figure E2C). HTBE cell secretions contained significantly more exosomes than Calu-3 cell secretions (Figure E2D).
Proteomics Analysis of HTBE-exo and Calu-3-exo
Proteomics analysis of the exosome preparations identified approximately 57 proteins in HTBE-exo and 63 proteins in Calu-3-exo, with 49 common proteins (Figure E3). MS analysis showed the presence of exosome-specific markers, such as CD59, annexins, Hsps (Hsp70 and Hsp90), cytoskeletal proteins, and PLUNC, in exosomes from both cell types. The proteomic profiles of both exosome types were largely similar (Table E1). HTBE-exo contained lysozyme C, Hsp70, and prostate stem cell antigen at higher levels than found in Calu-3-exo. In addition, HTBE-exo was highly enriched in membrane-tethered mucins (MUC1, MUC4, and MUC16). HTBE-exo contained unique proteins related to transmembrane ion transport and ion channel activity, such as sodium and chloride amino acid transporter protein, sodium-dependent phosphate transport protein 2B, erythrocyte band 7 integral membrane protein, the regulatory protein Na(+)/H(+) exchanger regulatory cofactor (NHERF1), and certain immune-related proteins, such as complement C3, polymeric Ig receptor (PIGR), and carcinoembryonic antigen–related cell adhesion molecule (CEACAM6) (Table E2). Proteins present exclusively in Calu-3-exo include MUC13, cell metabolic proteins (γ-glutamyltranspeptidase-1, glyceraldehyde-3-phosphate dehydrogenase), the cell surface peptidase, Neprilysin (CD10), transmembrane glycoprotein prominin-1 (CD133 antigen), the molecular chaperone endoplasmin, and immune-regulating proteins (protein S100-A9, dipeptidyl peptidase 4, and short PLUNC [SPLUNC1/BPIFA1]) (Table E3).
Proteomics Analysis of Apical Secretions from HTBE and Calu-3 Cells
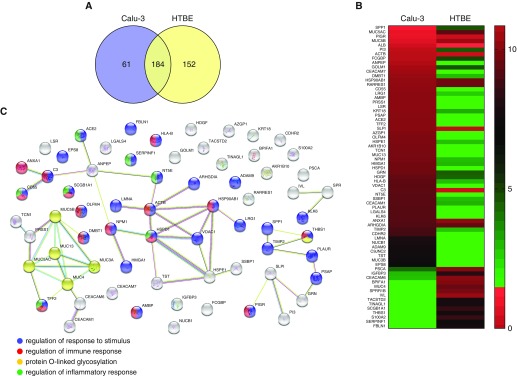
In total, 245 and 336 proteins were identified in secretions from HTBE cells and Calu-3 cells, respectively. Of these, 152 proteins were unique to Calu-3 cells, and 61 were unique to HTBE secretions. Proteomics analysis of the secretions indicated that the two-cell cultures had secretomes that were approximately 50% distinct (Figure 1A). Figure 1B shows unique and differentially expressed proteins in the HTBE and Calu-3 secretomes. Although proteins, such as SPLUNC1, mucin 4, involucrin, protein S-100-A2, uteroglobin, thrombospondin-1, fibulin-1, pigment epithelium-derived factor, and tumor-associated calcium signal transducer-2, were uniquely expressed in HTBE cells, other proteins, such as trefoil factor-2, olfactomedin-4, MUC13, transcobalamin-1, MUC3B, voltage-dependent anion-selective channel protein-1, CEACAM7, aminopeptidase N, protein AMBP, kallikrein-6, complement decay–accelerating factor (CD55), nucleobindin-1, and galectin-4, were present only in Calu-3 cells.
Figure 1.
Proteomics analysis of human tracheobronchial (HTBE) and Calu-3 cell secretions. (A) Venn diagram showing the total number of secreted proteins identified in the secretions of HTBE and Calu-3 cells. (B) Heat map of proteins displaying significantly (t test P ≤ 0.05) altered levels in HTBE and Calu-3 secretions. (C) Interaction network of altered proteins demonstrating biological processes.
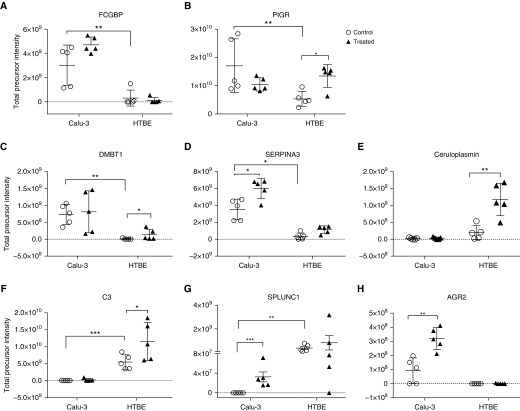
Label-free quantitative MS analysis indicated that 30 proteins were differentially expressed in both secretions (Figures 1B and 1C) at the baseline. The expression of the main gel-forming mucins of the airways, MUC5AC and MUC5B, was significantly higher in Calu-3 secretions than in HTBE secretions (Figures 2A and 2B). The MUC5AC:MUC5B ratio was also approximately 15-fold higher in Calu-3 secretions than in HTBE secretions (Figure 2C). In addition, innate immune response proteins, such as IgGFc-binding protein (FCGBP), PIGR, and deleted in malignant brain tumors (DMBT) 1, along with the antiprotease alpha-1 antichymotrypsin (SERPINA3), were significantly higher in Calu-3 secretions (Figures 3A–3D). In contrast, complement C3 and SPLUNC1 expression levels were higher in HTBE secretions (Figures 3E and 3F).
Figure 2.
Label-free quantitation of major gel-forming mucins in HTBE and Calu-3 cell secretions at baseline and after controlled exosome transfer. (A) MUC5AC and (B) MUC5B expression levels were significantly higher in Calu-3 cells than in HTBE cells. After the exosome transfer experiment, the expression levels of (D) MUC5AC and (E) MUC5B were significantly increased in HTBE secretions. (C) The ratio of MUC5AC and MUC5B were approximately 15-times higher in Calu-3 cell secretions compared to HTBE secretions. (F) The ratio in the Calu-3 cells further increased after the cells were treated with HTBE-exo. Solid circles and solid squares denote Calu-3 and HTBE control secretions, respectively. Open circles and solid triangles denote control and treated cultures, respectively. Statistical significance was determined by t tests; *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.005. NS = not significant.
Figure 3.
Relative abundance of certain innate defense proteins at baseline and after exosomal exchange. (A) FCGBP, (B) poly IgG receptor, (C) DMBT1, (D) alpha-1 antitrypsin, (E) ceruloplasmin, (F) complement C3, (G) SPLUNC1, and (H) AGR2. The mean and SEM values are indicated by major and minor horizontal bars, respectively. Open circles and solid triangles denote control and treated cultures, respectively. Statistical significance was determined by one-way ANOVA; *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.005.
Exosome Uptake by Recipient Cells
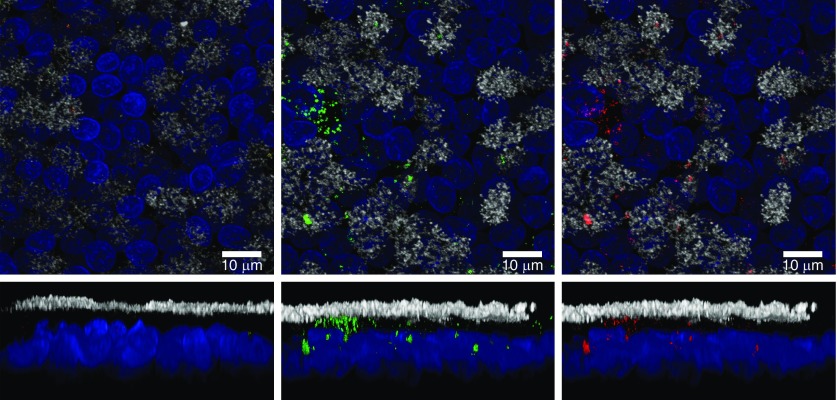
To investigate whether the exosomes were taken up by recipient cells, we labeled both exosomal RNA and membranes using SYTO RNASelect stain (green) and BODIPY TR ceramide stain (red), respectively. Confocal microscopy showed uptake of Calu-3 exosomes by HTBE cells, shown by strong green and red fluorescent signals in the whole-mount staining compared with the dye control (Figure 4). The RNA and membrane staining were generally colocalized in the cells. Cilia (tubulin) and nuclei (Hoechst) staining was used as a cell marker to determine whether exosomes were inside the cell or on the surface.
Figure 4.
Uptake of labeled exogenous Calu-3 exosomes by well-differentiated HTBE cells. Calu-3 exosomes (1 × 108) were labeled with SYTO RNASelect for RNA (green) and with BODIPY TR ceramide for membranes (red) before incubation with HTBE cells. Labeled exosomes or an equal volume of dye control were added to each well and incubated with cells for 1.5 hours at 37°C; the uptake of the labeled exosomes by HTBE cells was detected by confocal microcopy. Image of HTBE cells after incubation with a dye control (left panel) and labeled exosomes (middle and right panels). Top panels are extended focus XY projections, whereas the bottom panels are XZ projections. Ciliary tubulin (anti–β-tubulin) is stained in white, and nuclei (DAPI) are shown in blue. Scale bars: 10 μm.
Figure 4 indicates that the staining is mostly localized in the nonciliated areas, but there was also apparent staining in the ciliated cells.
The Secretomes of Both Cultures Change after Exosomal Exchange: Exosome-mediated Information Exchange and Altered Phenotype
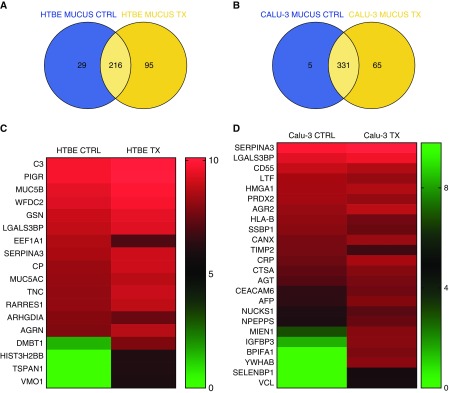
To elucidate the mechanism of intercellular communication between different airway cell types, we transferred HTBE-exo and Calu-3-exo to Calu-3 and HTBE cells, respectively. Treatment with the exosomes resulted in differential expression/regulation of certain proteins in the target cells. Label-free quantitative MS analysis of secretions after treatment of the HTBE cells with Calu-3-exo indicated that 95 new and unique proteins were secreted by the HTBE cells, and 66 of 311 proteins were significantly increased in HTBE secretions. A similar trend was observed in Calu-3 cells after treatment with HTBE-exo; specifically, 65 new and unique proteins were detected in the treated cells (Figures 5A and 5B).
Figure 5.
Label-free quantitation analysis of innate defense proteins in HTBE and Calu-3 cell secretions before and after exosomal exchange. (A and B) Venn diagram showing the total number of proteins expressed in the mucus secretions of HTBE and Calu-3 cells before and after exosome transfer, respectively. (C and D) Heat map showing the differentially expressed proteins in Calu-3 and HTBE cell secretions before and after treatment, respectively; P < 0.05. CTRL = control; TX = treatment.
The proteins that were significantly upregulated in HTBE cells after Calu-3-exo treatment were PIGR, WAP four-disulfide core domain protein 2, retinoic acid receptor responder protein 1, ceruloplasmin, DMBT1, complement C3, the mucins, MUC5B and MUC5AC, galectin-3–binding protein, vitelline membrane outer layer protein 1, histone H2B type 3-B, and tetraspanin-1 (Figures 2D, 2E, 3, and 5C).
SERPINA3, insulin-like growth factor–binding protein 3 (IGFBP3), CEACAM6, 14-3-3 proteins, BPI fold–containing family A member 1 (BPIFA1, SPLUNC1), anterior gradient protein-2 (AGR2), vinculin, migration and invasion enhancer 1, and selenium-binding protein 1 were increased in Calu-3 cell secretions after exosome exchange, whereas lactotransferrin, CD55, and peroxiredoxin-2 were decreased (Figures 3 and 5D).
miRNA Analysis of HTBE-exo and Calu-3-exo
A comprehensive miRNA comparison of the two types of exosome-like vesicles was performed to obtain an overview of differences in miRNA expression patterns that may have roles in intercellular communication between different airway epithelial cells and in disease pathogenesis. These experiments were performed using HTG EdgeSeq technology, and differential expression was evaluated using EdgeR software analysis.
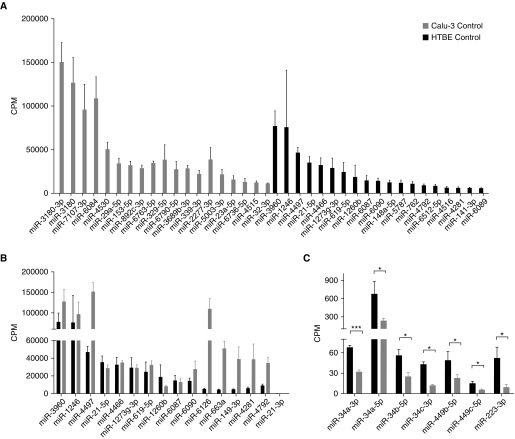
Similarities and significant differences were observed in miRNA expression between Calu-3-exo and HTBE-exo (Figure 6). More than 2,000 miRNAs were detected in each exosome population. miR-3960, miR-1246, miR-4497, miR-21, and miR-4466 were among the most abundant miRNAs in both exosomes (Figure 7A). miR-6126, miR-663a, miR-149, miR-4281, and miR-4792 were abundant in Calu-3-exo, but not in HTBE-exo (Figure 7B). Approximately 240 miRNAs were differentially expressed in HTBE and Calu-3 exosomes at baseline. Further analysis indicated that miRNAs related to inflammation and mucin production, such as miR-18a-5p, miR-19a-3p, miR-141-3p, miR-200a-3p, miR-200c-3p, miR-29a-3p, and miR-29b-1-5p, which are involved in cancer (23, 24) and respiratory disease–related pathways, were more abundantly expressed in Calu-3 cells. The miRNA families, miR-34a, -b, and -c and miR-449b and -c, which are known to be involved in ciliogenesis (25), were significantly more abundant in HTBE-exo than in Calu-3-exo (Figure 7C). The list of the top 50 miRNAs that were differentially expressed in HTBE versus Calu-3 control exosomes is shown in Table 1. Pathway analysis using Diana miRPath showed that these miRNAs were involved in a number of pathways, including pathways related to cancer and respiratory disease (Figure E4).
Figure 6.
Exosomal miRNA analysis. (A) Venn diagram showing changes in the number of miRNAs in the exosomes of HTBE and Calu-3 cells before and after exosome transfer. (B) Log ratio and mean average of the values (MA) plot showing differential miRNA expression in Calu-3-exo and HTBE-exo controls, (C) differential miRNA expression in HTBE-exo after exosome exchange, and (D) differential miRNA expression in Calu-3-exo after exosome exchange. Horizontal lines denote fold change of 2, while red/blue highlighted dots represent significantly increased/decreased miRNAs at false discovery rate <0.01. Approximately 240 miRNAs were differentially expressed in HTBE and Calu-3-exo. After exosome transfer, approximately 93 miRNAs were upregulated in HTBE-exo. CPM = counts per million; FC = fold change.
Figure 7.
Expression levels of the abundant miRNAs in HTBE and Calu-3 exosomes. (A) The expression levels of the most abundant miRNAs unique to each group. (B) A total of 16 of the top 50 most abundant miRNAs were common in both HTBE and Calu-3 exosomes, though most of these were orders of magnitude higher in Calu-3 exosomes. (C) The expression levels of the miR-34/449 group (associated with ciliogenesis) and miR223 (associated with acute lung injury) were upregulated in HTBE exosomes. Statistical significance was determined by t tests; *P ≤ 0.05, ***P ≤ 0.005.
Table 1.
List of top 50 miRs differentially expressed in Calu-3 versus HTBE exosomes
| Probe | logFC | P Value | FDR |
|---|---|---|---|
| miR-192-5p | 7.983572 | 7.02E−44 | 1.58E−40 |
| miR-194-5p | 8.065975 | 1.83E−43 | 2.06E−40 |
| miR-205-5p | −8.65767 | 1.13E−32 | 8.50E−30 |
| miR-375 | 5.741463 | 2.83E−30 | 1.60E−27 |
| miR-215-5p | 5.463103 | 5.80E−30 | 2.62E−27 |
| miR-6727-5p | 5.157095 | 8.50E−29 | 3.20E−26 |
| miR-6780b-5p | 5.43189 | 6.90E−24 | 2.22E−21 |
| miR-6126 | 5.028427 | 1.09E−20 | 3.07E−18 |
| miR-130b-3p | 3.93613 | 5.64E−18 | 1.41E−15 |
| miR-663a | 4.146252 | 1.31E−17 | 2.97E−15 |
| miR-6752-5p | 5.546187 | 5.11E−16 | 1.05E−13 |
| miR-10b-5p | 3.140973 | 5.24E−15 | 9.85E−13 |
| miR-15b-5p | 3.282913 | 1.73E−13 | 3.01E−11 |
| miR-6799-5p | 4.257163 | 2.45E−13 | 3.95E−11 |
| miR-192-3p | 2.847477 | 3.00E−13 | 4.51E−11 |
| miR-92a-3p | 2.968258 | 9.69E−13 | 1.37E−10 |
| miR-654-5p | 4.461773 | 1.22E−12 | 1.62E−10 |
| miR-101-3p | 2.92049 | 1.61E−12 | 2.02E−10 |
| miR-4634 | 3.120529 | 2.82E−12 | 3.35E−10 |
| miR-149-3p | 3.657877 | 8.64E−12 | 9.74E−10 |
| miR-10a-5p | 4.732037 | 1.77E−11 | 1.91E−09 |
| miR-30b-5p | 2.815723 | 2.29E−11 | 2.35E−09 |
| miR-320c | 2.932861 | 2.85E−11 | 2.80E−09 |
| miR-320b | 2.735203 | 3.09E−11 | 2.91E−09 |
| miR-28-5p | 2.661624 | 1.72E−10 | 1.56E−08 |
| miR-1237-5p | 2.899386 | 3.82E−10 | 3.31E−08 |
| miR-20a-5p | 2.6251 | 4.13E−10 | 3.45E−08 |
| miR-564 | 2.852591 | 4.57E−10 | 3.68E−08 |
| miR-424-5p | 2.642337 | 5.55E−10 | 4.32E−08 |
| miR-19a-3p | 2.488099 | 1.06E−09 | 7.94E−08 |
| miR-3197 | 2.773846 | 1.12E−09 | 8.18E−08 |
| miR-186-5p | 2.534553 | 1.49E−09 | 1.05E−07 |
| miR-19b-3p | 2.59162 | 1.81E−09 | 1.24E−07 |
| miR-4463 | 2.833971 | 2.80E−09 | 1.86E−07 |
| miR-138-5p | −2.97975 | 3.21E−09 | 2.07E−07 |
| miR-193b-3p | 2.5112 | 3.37E−09 | 2.11E−07 |
| miR-320e | 2.421153 | 5.36E−09 | 3.27E−07 |
| miR-4306 | 2.354884 | 6.13E−09 | 3.64E−07 |
| let-7i-5p | 2.684296 | 6.51E−09 | 3.76E−07 |
| miR-638 | 2.963232 | 8.70E−09 | 4.91E−07 |
| miR-362-3p | 2.325304 | 9.16E−09 | 5.04E−07 |
| miR-320d | 2.545148 | 1.00E−08 | 5.39E−07 |
| miR-17-5p | 2.391188 | 1.09E−08 | 5.72E−07 |
| miR-7-5p | 2.337118 | 1.28E−08 | 6.59E−07 |
| miR-148b-3p | 2.28763 | 1.41E−08 | 7.07E−07 |
| miR-20b-5p | 2.273357 | 2.03E−08 | 9.72E−07 |
| miR-29a-3p | 2.437546 | 2.02E−08 | 9.72E−07 |
| miR-148a-3p | 2.253075 | 2.20E−08 | 1.03E−06 |
| miR-4461 | 2.535988 | 2.68E−08 | 1.23E−06 |
| miR-4281 | 3.299971 | 3.25E−08 | 1.47E−06 |
Definition of abbreviations: FC = fold change; FDR = false discovery rate; HTBE = human tracheobronchial; miR = microRNA.
After exosomal transfer, few significant differences were found between Calu-3–treated exosomes; however, interestingly, approximately 90 miRNAs were upregulated in HTBE-exo compared with Calu-3-exo (Figures 6C and 6D) after exosomal transfer. Bioinformatic analysis of the miRNA library showed that the expression levels of miR-31-5p, miR-27b-3b, miR-21-5p, miR-21-3p, miR-100-5p, miR-34, and miR-449 were increased, and the expression levels of miR-375, miR-215-5p, and miR-192-5p were decreased significantly in HTBE-exo after the transfer. miR-3180 and miR-3180–3p increased in Calu-3 treated- exosomes. miR-18a-5p, 19a-3p, miR-141-3p, miR-200a-3p, miR-200c-3p, miR29a-3p, and miR-29b-1-5p were overexpressed in Calu-3-exo and were found to be upregulated in HTBE-exo after treatment with Calu-3-exo.
Discussion
Airway epithelial cells form an essential barrier for effective innate protection of the lung. In response to chronic infection and inflammation, the epithelial layer is remodeled. Airway cells may contribute to the pathogenesis of major chronic lung disorders, including chronic obstructive pulmonary disease, cystic fibrosis, asthma, and bronchogenic carcinoma. In this compromised state, the airway epithelium becomes activated by various unknown paracrine signaling molecules, leading to various structural and functional changes at the gene and protein level. Identification of these signaling molecules is critical to understanding the pathogenesis of these diseases and to developing strategies to treat diseases characterized by goblet cell remodeling and mucus hypersecretion. The focus of this study was to understand the physiological relevance of exosomes in intercellular communication and to explore new mechanisms involved in lung homeostasis, disease pathogenesis, and airway remodeling.
The airway epithelial layer does not function as an independent entity, but rather as an interdependent functional unit with differentiated epithelial cells (ciliated and goblet cells), mesenchymal cells, and endothelial cells, and with the extracellular matrix, that form the bronchial walls. How these cells coordinate to effect innate protection is not known. Therefore, in this study, to elucidate the mechanism of intercellular communication between different airway cells, we transferred HTBE-exo and Calu-3-exo to Calu-3 and HTBE cells, respectively. Multiple novel findings were evident from these experiments. First, we showed that exosomes can deliver their cargo to recipient cells. Second, we demonstrated that the two airway cell lines are distinct and treatment with exosomes derived from different cell types resulted in differential expression/regulation of certain proteins and miRNAs in the target cells, especially the ones related to innate defense, injury, and ciliogenesis. Last, we observed that transferring the information carried by exosomes/vesicles from one cell type to another affects the secretory phenotype of the recipient cells.
The MS-based proteome analysis indicated that Calu-3 and HTBE cells secreted qualitatively and quantitatively distinct sets of proteins that were broadly related to innate defense, stress/injury, and regulatory pathways. Approximately 213 proteins were unique to both secretions, which accounted for a distinct secretome of approximately 50% (Figures 1A and 1B). The mucin, MUC4, is known to be associated with cilia (26) and was unique to HTBE culture secretions. However, the mucins, MUC13 and MUC3B, were unique to Calu-3 secretions, which may suggest their role in cancer pathogenesis. The proteins trefoil factor-2, kallikrein-6, CD55, nucleobindin-1, and galectin-4 were present only in Calu-3. Quantitative MS data indicated that approximately 30 proteins were differentially expressed in the secretions, including the gel-forming mucins, MUC5B and MUC5AC. MUC5AC and MUC5B contribute to the viscoelastic properties of mucus and are found at elevated levels in the airways of individuals with inflammation, chronic respiratory diseases (27), and cancer (28). MUC5B is essential for the formation of a flowing mucus gel that is vital for epithelial protection and mucociliary clearance (29). The expression of both mucins was significantly higher in Calu-3 secretions compared with HTBE secretions. In the lung, as well as in HTBE cultures, MUC5B is the main gel-forming mucin. Increased MUC5AC (which dominates Calu-3 secretions) and increased MUC5AC:MUC5B ratio (15-fold higher in Calu3 secretions compared with HTBE secretions) in the lung generally associated with pathologies (27). Other glycoproteins, such as AMBP, zinc-alpha-2 glycoprotein, saposin D, and leucine-rich alpha-2 glycoprotein, were also significantly higher in Calu-3 cultures, and these glycoproteins are found to be upregulated in cancer cells (30). Furthermore, innate response- and injury-related proteins, such as PIGR, DMBT1, and FCGBP, were significantly higher in Calu-3 secretions, whereas complement C3, SPLUNC1, and prostate stem cell antigens were higher in HTBE secretions. Overall, the differential expression of mucins and related innate defense proteins suggests that HTBE and Calu-3 cells secrete fundamentally distinct secretions with unique protective/pathogenic profiles.
After exchanging exosomes, both HTBE and Calu-3 secretions were significantly altered. Approximately 20% of the proteins were significantly increased, whereas 10% were significantly decreased. In HTBE secretions, 95 unique proteins were apparently elevated after exosome transfer from Calu-3 cells. Importantly, MUC5AC and MUC5B were significantly elevated in HTBE secretions after the transfer of Calu-3-exo. Given that mucin hypersecretion is an aspect of airway remodeling in chronic hypersecretory conditions, including lung diseases induced by cigarette smoking, we can surmise that exosomes may play role in this pathological process and contribute to the pathogenesis of lung diseases characterized by airway remodeling and mucin hypersecretion. Similarly, Calu-3 cell secretions were significantly altered after treatment with HTBE-exo. Innate defense proteins and proteins related to the inflammatory response were increased in Calu-3 cells after treatment with HTBE-exo, whereas some stress/injury–related proteins were decreased upon treatment. BPIFA1 (SPLUNC1), an important innate defense protein in the airways (31, 32), was not detected in Calu-3 cells at baseline, but was markedly and significantly increased after HTBE exosome transfer, providing further insight into exosome-mediated changes in protein expression. Although the dominant Calu-3 mucin, MUC5AC, is slightly, but not significantly, increased after the exosome treatment, AGR2, a protein associated with MUC5AC (over) production (33), was significantly increased after the transfer. The observed changes in the proteome profiles and mucin secretions after exosomal transfer suggests that exosomal cargo may directly impact the protein expression of recipient cells. Therefore, the qualitative and quantitative alterations in all these proteins mediated by exosomes may be associated with airway epithelial remodeling. Because these alterations are correlated with the miRNA profile, the response after the transfer could be a result of reprogramming of the target cells rather than physical stimulus. However, the latter cannot be ruled out by the current data, and needs to be investigated further. In other words, the surface proteins on exosomes may interact with recipient cells, thereby potentially stimulating a certain cell response.
miRNAs play fundamental roles in several pulmonary diseases, including interstitial lung disease, chronic obstructive pulmonary disease, and asthma (34). Exosomal miRNAs delivered to target cells can significantly affect biological pathways within target/recipient cells, resulting in altered cellular function and the development of a pathological state. miRNAs are small, noncoding, endogenous, single-stranded, conserved sequences that mainly function by directly downregulating (34) or indirectly upregulating gene expression at the post-transcriptional level through binding to mRNAs and preventing their translation into proteins. A single miRNA can regulate several genes, and multiple miRNAs can regulate the same gene with additive or synergistic effects. To understand the role of miRNAs in the unique secretory phenotype of these two distinct cell types and to understand the role of secretome changes after exosomal exchange, we used exosomal miRNA profiling. Two observations were evident from the miRNA profiles: 1) the two cell types released distinct sets of miRNAs through their exosomes; and 2) after exosomal exchange, approximately 5% of the miRNA profile was altered in the HTBE cultures compared with the less than a 1% alteration observed in Calu-3 cells. We previously showed that airway epithelial exosomes carry small RNAs (11). Here, we demonstrated that these exosomes also carry miRNAs. Microarray analysis followed by next-generation sequencing detected more than 2,000 miRNAs in these cultures. Similar to their proteome content, these two distinct cell cultures exhibited distinct miRNA profiles. When we compared the differentially expressed miRNAs and proteins between Calu-3 and HTBE cells through pathway analyses, we observed that miRNAs, which are upstream regulators of protein expression, affected the cell state and function. Proteins present at lower levels in Calu-3 cells, such as IGFBP3 and SLP1, were found to be regulated by a number of miRNAs, including miRNA-4497, miR-4792, miR-6752-5p, miR-143-3p, miR-125b-5p (35), miR-34a-5p (25), and miR-664-3p, using ingenuity pathway analysis software. mir-196a, mir-101-3p, miR-31-5p, and certain miRNAs from the 17–92 cluster (24) were observed at higher levels in Calu-3-exo. These miRNAs are known to be overexpressed in human lung cancer and to promote cell proliferation. miR-3180 has been found to be involved in the proliferation of smooth muscle cells in the bladder through the Cdk2 signaling pathway (36). miR-19a and miR-19b-3p (37), along with miR-141 and miR-200c, are known to be involved in cancer metastasis and to play a role in epithelial–mesenchymal transition. miR-21, miR-146a, miR-146b, and miR-32-5p have been observed to be higher in patients with acute lung injury. Therefore, the miRNAs that were upregulated in HTBE-exo suggest that HTBE cells sustain some form of injury after treatment with exosomes from the cancerous Calu-3 cell line. In addition, we investigated the molecular network that can be activated by miRNAs that are differentially expressed in HTBE and Calu-3-exo. MYC and the NF-κβ complex seemed to be central regulators in the molecular network.
It is apparent from the quantitative proteomic analysis that Calu-3 cells secrete significantly more gel-forming mucins, MUC5B (∼4-fold) and MUC5AC (∼90-fold). This could be due to the genotype/phenotype of Calu-3 cells, which may tend to promote secretion of more mucins/mucus, and because Calu-3 cells exhibit a secretory goblet cell–rich phenotype compared with HTBE cells, 60–80% of which are ciliated. After the exosomal exchange, MUC5AC and MUC5B levels increased in the HTBE secretions. Therefore, it is critical to know the relationship between these mucins and exosomes. To further understand this finding, we attempted to correlate this tendency with the miRNA profiles of the two cell lines. Unfortunately, limited information is available on the relationship between miRNA and mucin secretion. Using structural and functional pathway analyses, miR-149-3p was predicted to regulate the expression of MUC5B (Figure E5), and no correlation was found for the regulation of MUC5AC. Although limited information is available in the literature associating miRNAs with mucin expression and secretion, these differentially expressed miRNAs in Calu-3 cells may represent a future target for screening studies. In addition, an interaction between the abundant gel-forming mucins and the exosomes is possible. The presence of MUC5AC and MUC5B in the exosomal preparation (Tables E1–E3) suggests a copurification, likely via an interaction between them. Validation of such interactions and consequences requires further studies. Ciliated cells are terminal cells that are affected by insult and injury as a part of airway remodeling in addition to goblet cell hyperplasia. Recently, miR-34/449 was shown to be important in ciliogenesis by altering the expression of a centriolar protein known to suppress cilia assembly (38, 39). In addition, vesicular miR-223 was recently shown to protect against lung injuries in mice (40). These miRNAs were significantly more abundant in HTBE-exo, and interestingly, we observed that the baseline level of miR-34/449 was upregulated in HTBE-exo, suggesting upregulated cilia biogenesis in these cells after treatment. Downregulation of miR-29 is associated with pulmonary fibrosis (41). miR-29 was detected in both Calu3 and HTBE cells, and was increased in HTBE cell–derived exosomes after exosomal exchange.
Exosome-mediated information transfer and cellular cross-talk have previously been shown in other systems, such as human and mouse mast cells (13), tumor microenvironments, tumor malignancies (42), and cancer metastasis (43). To the best of our knowledge, this is the first report of these phenomena in airway epithelial cells. The lung is a unique organ, considering the broad range of cells that are found within the parenchyma and airway structures. Cell–cell communication is essential for optimal functioning of the lung, and exosomes are therefore expected to be important players in lung biology and function (44). We previously showed that exosomes in the airway can play a role in innate defenses. Our present study highlights a new role for the cargo of airway epithelial–derived exosomes as modulating/regulating factors in the airway remodeling process. This study focused on cultured epithelial cell exosomes. However, other cells, such as macrophages and neutrophils, are also important sources of exosomes in the lung. Therefore, further studies combining all these factors are necessary to clarify the contribution of airway exosomes derived from resident epithelia and blood cells in lung homeostasis and in the innate response to infection and inflammation.
Exosome and miRNA research is an emerging field, despite its technical and biological challenges. Currently, more than 2,000 miRNAs have been characterized, but the identification of their target genes and functions is still in its infancy. Future studies focusing on the role of exosomal cargo–, miRNA-, and protein-mediated intercellular communication between different airway epithelial cells and between macrophages/neutrophils and airway epithelial cells are desperately needed to better understand the coordinatedinnate immune response of the lung. In conclusion, our data suggest that transfer of exosomal cargo between cells significantly alters protein expression, and reorganizes the miRNA cargo of exosomes in target cells, indicating that cellular information can be carried between airway epithelial cells via exosomes. Therefore, exosomes may play an important role in airway biology and epithelial remodeling.
Acknowledgments
Acknowledgment
The authors thank the University of North Carolina Marsico Lung Institute Tissue Procurement and cell culture core for cells and technical expertise.
Footnotes
This work was partially supported by American Lung Association grant RG-167538-N and National Institutes of Health grants R01HL103940 and P30DK065988.
Author Contributions: M.K., R.G., and S.H.R. designed the research studies; S.H.R. provided the cells; M.K. and R.G. wrote the manuscript; R.G., G.R., S.A., and J.T.S. performed the cell culture experiments and exosome isolation and characterization; R.G., G.R., and M.K. analyzed the proteomics data; R.G., J.C., and M.C. performed confocal microscopy experiments; P.A.M. performed the miRNA identification; and H.D. analyzed the miRNA data.
This article has a data supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1165/rcmb.2018-0156OC on September 19, 2018
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Whitsett JA, Alenghat T. Respiratory epithelial cells orchestrate pulmonary innate immunity. Nat Immunol. 2015;16:27–35. doi: 10.1038/ni.3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rajavelu P, Chen G, Xu Y, Kitzmiller JA, Korfhagen TR, Whitsett JA. Airway epithelial SPDEF integrates goblet cell differentiation and pulmonary Th2 inflammation. J Clin Invest. 2015;125:2021–2031. doi: 10.1172/JCI79422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Radicioni G, Cao R, Carpenter J, Ford AA, Wang T, Li L, et al. The innate immune properties of airway mucosal surfaces are regulated by dynamic interactions between mucins and interacting proteins: the mucin interactome. Mucosal Immunol. 2016;9:1442–1454. doi: 10.1038/mi.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kato A, Schleimer RP. Beyond inflammation: airway epithelial cells are at the interface of innate and adaptive immunity. Curr Opin Immunol. 2007;19:711–720. doi: 10.1016/j.coi.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rajasekaran SA, Beyenbach KW, Rajasekaran AK. Interactions of tight junctions with membrane channels and transporters. Biochim Biophys Acta. 2008;1778:757–769. doi: 10.1016/j.bbamem.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 6.Kesimer M, Gupta R. Physical characterization and profiling of airway epithelial derived exosomes using light scattering. Methods. 2015;87:59–63. doi: 10.1016/j.ymeth.2015.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ogawa Y, Taketomi Y, Murakami M, Tsujimoto M, Yanoshita R. Small RNA transcriptomes of two types of exosomes in human whole saliva determined by next generation sequencing. Biol Pharm Bull. 2013;36:66–75. doi: 10.1248/bpb.b12-00607. [DOI] [PubMed] [Google Scholar]
- 8.Rodríguez M, Silva J, López-Alfonso A, López-Muñiz MB, Peña C, Domínguez G, et al. Different exosome cargo from plasma/bronchoalveolar lavage in non–small-cell lung cancer. Genes Chromosomes Cancer. 2014;53:713–724. doi: 10.1002/gcc.22181. [DOI] [PubMed] [Google Scholar]
- 9.Gui Y, Liu H, Zhang L, Lv W, Hu X. Altered microRNA profiles in cerebrospinal fluid exosome in Parkinson disease and Alzheimer disease. Oncotarget. 2015;6:37043–37053. doi: 10.18632/oncotarget.6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Torregrosa Paredes P, Gutzeit C, Johansson S, Admyre C, Stenius F, Alm J, et al. Differences in exosome populations in human breast milk in relation to allergic sensitization and lifestyle. Allergy. 2014;69:463–471. doi: 10.1111/all.12357. [DOI] [PubMed] [Google Scholar]
- 11.Kesimer M, Scull M, Brighton B, DeMaria G, Burns K, O’Neal W, et al. Characterization of exosome-like vesicles released from human tracheobronchial ciliated epithelium: a possible role in innate defense. FASEB J. 2009;23:1858–1868. doi: 10.1096/fj.08-119131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pitt JM, Kroemer G, Zitvogel L. Extracellular vesicles: masters of intercellular communication and potential clinical interventions. J Clin Invest. 2016;126:1139–1143. doi: 10.1172/JCI87316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 14.Yang C, Robbins PD. The roles of tumor-derived exosomes in cancer pathogenesis. Clin Dev Immunol. 2011;2011:842849. doi: 10.1155/2011/842849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delić D, Eisele C, Schmid R, Baum P, Wiech F, Gerl M, et al. Urinary exosomal miRNA signature in type II diabetic nephropathy patients. PLoS One. 2016;11:e0150154. doi: 10.1371/journal.pone.0150154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Santucci L, Bruschi M, Candiano G, Lugani F, Petretto A, Bonanni A, et al. Urine proteome biomarkers in kidney diseases. I. Limits, perspectives, and first focus on normal urine. Biomark Insights. 2016;11:41–48. doi: 10.4137/BMI.S26229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Szul T, Bratcher PE, Fraser KB, Kong M, Tirouvanziam R, Ingersoll S, et al. Toll-like receptor 4 engagement mediates prolyl endopeptidase release from airway epithelia via exosomes. Am J Respir Cell Mol Biol. 2016;54:359–369. doi: 10.1165/rcmb.2015-0108OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fulcher ML, Gabriel S, Burns KA, Yankaskas JR, Randell SH. Well-differentiated human airway epithelial cell cultures. Methods Mol Med. 2005;107:183–206. doi: 10.1385/1-59259-861-7:183. [DOI] [PubMed] [Google Scholar]
- 19.Kreda SM, Okada SF, van Heusden CA, O’Neal W, Gabriel S, Abdullah L, et al. Coordinated release of nucleotides and mucin from human airway epithelial Calu-3 cells. J Physiol. 2007;584:245–259. doi: 10.1113/jphysiol.2007.139840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kesimer M, Cullen J, Cao R, Radicioni G, Mathews KG, Seiler G, et al. Excess secretion of gel-forming mucins and associated innate defense proteins with defective mucin un-packaging underpin gallbladder mucocele formation in dogs. PLoS One. 2015;10:e0138988. doi: 10.1371/journal.pone.0138988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40:4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robinson MD, Smyth GK. Small-sample estimation of negative binomial dispersion, with applications to SAGE data. Biostatistics. 2008;9:321–332. doi: 10.1093/biostatistics/kxm030. [DOI] [PubMed] [Google Scholar]
- 23.Tejero R, Navarro A, Campayo M, Viñolas N, Marrades RM, Cordeiro A, et al. miR-141 and miR-200c as markers of overall survival in early stage non–small cell lung cancer adenocarcinoma. PLoS One. 2014;9:e101899. doi: 10.1371/journal.pone.0101899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fuziwara CS, Kimura ET. Insights into regulation of the miR-17-92 cluster of miRNAs in cancer. Front Med (Lausanne) 2015;2:64. doi: 10.3389/fmed.2015.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yin H, Zhang S, Sun Y, Li S, Ning Y, Dong Y, et al. MicroRNA-34/449 targets IGFBP-3 and attenuates airway remodeling by suppressing Nur77-mediated autophagy. Cell Death Dis. 2017;8:e2998. doi: 10.1038/cddis.2017.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kesimer M, Ehre C, Burns KA, Davis CW, Sheehan JK, Pickles RJ. Molecular organization of the mucins and glycocalyx underlying mucus transport over mucosal surfaces of the airways. Mucosal Immunol. 2013;6:379–392. doi: 10.1038/mi.2012.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kesimer M, Ford AA, Ceppe A, Radicioni G, Cao R, Davis CW, et al. Airway mucin concentration as a marker of chronic bronchitis. N Engl J Med. 2017;377:911–922. doi: 10.1056/NEJMoa1701632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walsh MD, Clendenning M, Williamson E, Pearson SA, Walters RJ, Nagler B, et al. Expression of MUC2, MUC5AC, MUC5B, and MUC6 mucins in colorectal cancers and their association with the CpG island methylator phenotype. Mod Pathol. 2013;26:1642–1656. doi: 10.1038/modpathol.2013.101. [DOI] [PubMed] [Google Scholar]
- 29.Roy MG, Livraghi-Butrico A, Fletcher AA, McElwee MM, Evans SE, Boerner RM, et al. Muc5b is required for airway defence. Nature. 2014;505:412–416. doi: 10.1038/nature12807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ayyub A, Saleem M, Fatima I, Tariq A, Hashmi N, Musharraf SG. Glycosylated alpha-1-acid glycoprotein 1 as a potential lung cancer serum biomarker. Int J Biochem Cell Biol. 2016;70:68–75. doi: 10.1016/j.biocel.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 31.Kesimer M, Kirkham S, Pickles RJ, Henderson AG, Alexis NE, Demaria G, et al. Tracheobronchial air–liquid interface cell culture: a model for innate mucosal defense of the upper airways? Am J Physiol Lung Cell Mol Physiol. 2009;296:L92–L100. doi: 10.1152/ajplung.90388.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu T, Huang J, Moore PJ, Little MS, Walton WG, Fellner RC, et al. Identification of BPIFA1/SPLUNC1 as an epithelium-derived smooth muscle relaxing factor. Nat Commun. 2017;8:14118. doi: 10.1038/ncomms14118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schroeder BW, Verhaeghe C, Park SW, Nguyenvu LT, Huang X, Zhen G, et al. AGR2 is induced in asthma and promotes allergen-induced mucin overproduction. Am J Respir Cell Mol Biol. 2012;47:178–185. doi: 10.1165/rcmb.2011-0421OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sessa R, Hata A. Role of microRNAs in lung development and pulmonary diseases. Pulm Circ. 2013;3:315–328. doi: 10.4103/2045-8932.114758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hermeking H. MicroRNAs in the p53 network: micromanagement of tumour suppression. Nat Rev Cancer. 2012;12:613–626. doi: 10.1038/nrc3318. [DOI] [PubMed] [Google Scholar]
- 36.Sun Y, Luo DY, Zhu YC, Zhou L, Yang TX, Tang C, et al. MiR 3180-5p promotes proliferation in human bladder smooth muscle cell by targeting PODN under hydrodynamic pressure. Sci Rep. 2016;6:33042. doi: 10.1038/srep33042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li J, Yang S, Yan W, Yang J, Qin YJ, Lin XL, et al. MicroRNA-19 triggers epithelial-mesenchymal transition of lung cancer cells accompanied by growth inhibition. Lab Invest. 2015;95:1056–1070. doi: 10.1038/labinvest.2015.76. [DOI] [PubMed] [Google Scholar]
- 38.Wu J, Bao J, Kim M, Yuan S, Tang C, Zheng H, et al. Two miRNA clusters, miR-34b/c and miR-449, are essential for normal brain development, motile ciliogenesis, and spermatogenesis. Proc Natl Acad Sci USA. 2014;111:E2851–E2857. doi: 10.1073/pnas.1407777111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Song R, Walentek P, Sponer N, Klimke A, Lee JS, Dixon G, et al. miR-34/449 miRNAs are required for motile ciliogenesis by repressing cp110. Nature. 2014;510:115–120. doi: 10.1038/nature13413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Neudecker V, Brodsky KS, Clambey ET, Schmidt EP, Packard TA, Davenport B, et al. Neutrophil transfer of miR-223 to lung epithelial cells dampens acute lung injury in mice. Sci Transl Med. 2017;9:pii: eaah5360. doi: 10.1126/scitranslmed.aah5360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cushing L, Kuang PP, Qian J, Shao F, Wu J, Little F, et al. miR-29 is a major regulator of genes associated with pulmonary fibrosis. Am J Respir Cell Mol Biol. 2011;45:287–294. doi: 10.1165/rcmb.2010-0323OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Felicetti F, De Feo A, Coscia C, Puglisi R, Pedini F, Pasquini L, et al. Exosome-mediated transfer of miR-222 is sufficient to increase tumor malignancy in melanoma. J Transl Med. 2016;14:56. doi: 10.1186/s12967-016-0811-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hoshino A, Costa-Silva B, Shen TL, Rodrigues G, Hashimoto A, Tesic Mark M, et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527:329–335. doi: 10.1038/nature15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eissa NT. The exosome in lung diseases: message in a bottle. J Allergy Clin Immunol. 2013;131:904–905. doi: 10.1016/j.jaci.2013.01.021. [DOI] [PubMed] [Google Scholar]