Abstract
Anaerobic and aerobic bacteria were quantitated in respiratory samples across three cystic fibrosis (CF) centres using extended culture methods. Subjects, ages 1–69 years, who were clinically stable provided sputum (n=200) or bronchoalveolar lavage (n=55). Eighteen anaerobic and 39 aerobic genera were cultured from 59% and 95% of samples, respectively; 16/57 genera had a ≥5% prevalence across centres. Analyses of microbial communities using co-occurrence networks in sputum samples showed groupings of oral, including anaerobic, bacteria whereas typical CF pathogens formed distinct entities. Pseudomonas was associated with worse nutrition and F508del genotype, whereas anaerobe prevalence was positively associated with pancreatic sufficiency, better nutrition and better lung function. A higher ratio of total anaerobe/total aerobe colony forming units was associated with pancreatic sufficiency and better nutrition. Subjects grouped by factor analysis who had relative dominance of anaerobes over aerobes had milder disease compared to a Pseudomonas-dominated group with similar proportions of subjects being homozygous for F508del. In summary, anaerobic bacteria occurred at an early age. In sputum producing subjects anaerobic bacteria were associated with milder disease suggesting that targeted eradication of anaerobes may not be warranted in sputum producing CF subjects.
Keywords: Cultured microbiota, multi-centre, Prevotella, bronchoalveolar lavage, child, disease outcomes
Introduction
Antibiotic therapies for cystic fibrosis (CF) respiratory infections are targeted at specific bacteria, typically Staphylococcus aureus, Pseudomonas aeruginosa, and other gram-negative species. However, both culture-dependent and culture-independent molecular methods have redefined CF respiratory infections as polymicrobial, with a diverse lower airway bacterial community [1]. Anaerobic conditions have been described in CF lower airways [2, 3]. Importantly, obligate and facultative anaerobic bacteria, typically associated with the oral cavity, have been cultured in abundance from CF respiratory samples [4, 5]. Indeed, extended anaerobic culture methods identified anaerobic bacteria in high numbers in >60% of CF sputum samples from adults [6].
The role of anaerobic bacteria in CF lower airways disease is not clearly understood. Studies have identified anaerobic bacteria from direct lung samples, suggesting their presence does not simply reflect salivary contamination during sampling [7]. In vitro studies have demonstrated that anaerobes release virulence factors, including proteases and pro-inflammatory short chain fatty-acids, [8, 9] suggesting that anaerobes may be pathogenic in the CF lung. In contrast, in vivo molecular studies indicate that greater anaerobe diversity is associated with milder disease. [10, 11]. Molecular methods highlight the breadth of bacteria and novel preparation methods and metagenomics address a short-coming of conventional next-generation sequencing, i.e. the amplification of sequences of non-viable bacteria [12–14]. Bacterial abundance can be measured by qPCR; however, 16S rRNA copy numbers per genome vary between genera and quantitation of different genera in multi-species infections remains imprecise [15, 16]. Culture allows enumeration of the different bacterial genera in a poly-microbial infection, demonstrates viability and growth in lower airways and enables subsequent in vitro studies [17, 18]. Culturing anaerobic and bacteria not typically considered as pathogenic is also relevant to clinical care; i.e. would knowledge about abundance of these bacteria be useful for treatment?
Here we assessed the association of strict anaerobes with disease severity in CF across ages at three CF centers with different genetic and geographic backgrounds and medications. Although “CF pathogens” can grow under anaerobic conditions, this likely reflects adaptation to the micro-environment and not distinct genera/infections. We focused on strict anaerobic bacteria to match genera reported in molecular studies and to ascertain the load and viability of these anaerobes in lower airways in relation to typical CF pathogens. We used extended culture methods attempting to investigate the role of anaerobes in CF lung disease. The key questions investigated were: 1) does the prevalence of anaerobes vs. aerobes in subjects with CF change as a function of age and site/climate; 2) are there associations between prevalence or quantity of anaerobes and genotype, disease severity, and medications; and 3) are community structures established by anaerobes and do they differ across ages, disease severity, or associations with aerobes?
Methods
Subjects and sample collection
People diagnosed with CF attending CF Centres in Belfast (Adult and Paediatric CF Clinics, Belfast Health and Social Care Trust), University of North Carolina (UNC) at Chapel Hill (UNC Hospital Pediatric and Adult Clinics) and Dublin (Beaumont Hospital and Our Lady’s Children’s Hospital, Crumlin) were prospectively enrolled into this study between July 2010 and November 2013. Subjects who had undergone transplant, were taking cystic fibrosis transmembrane conductance regulator potentiators/modulators, or enrolled in interventional therapeutic trials were excluded. Ethical approval for the study was obtained at each institution and informed consent/assent obtained from all adults, parents, and paediatric subjects, respectively. Either bronchoalveolar lavage (BAL) or sputum (if able to expectorate), and concomitant clinical information were collected when subjects were clinically stable, defined as not having received antibiotics beyond chronic maintenance therapy and ≥6 weeks post-completion of IV antibiotic therapy. Bronchoscopy was performed at UNC if a child had a procedure requiring anaesthesia, and for routine surveillance at Dublin. At both sites, bronchoscopy was performed utilizing a laryngeal mask airway (LMA) or endotracheal tube (ETT).
Clinical data
Demographics, anthropometrics, lung function, medication history, and disease complications were entered into a centralised database. Genotype was categorised by number of F508del alleles. Disease severity was assessed by nutritional status, pancreatic status, and lung function (forced expiratory volume in 1 second; FEV1) if subjects were able to reliably perform spirometry per ATS/ERS criteria [19]. For comparisons of body mass index (BMI) across paediatric and adult age ranges, BMI was stratified into poor, acceptable, and well-nourished categories based on percentiles in children (≤5 %ile, 6–50 %ile, >50 %ile) and absolute BMI in adults [20].
Bacterial isolation and identification
Standardised culture protocols were used across the three sites as previously described [6] with further details provided in the Supplement. Briefly, samples were treated for 15 minutes with dithiothreitol prior to serial dilution and plating on both, selective and non-selective media (Table S1). Plates were incubated under aerobic- microaerophilic and anaerobic conditions for between 2 and 7 days. All bacteria detected were quantified (colony forming units/gram sputum; CFU/g or CFU/mL in BAL) by total viable count (TVC). Each distinct colonial morphotype was identified by full length16S rRNA gene sequencing. Strict anaerobes were defined as bacteria known not to survive under atmospheric oxygen tension, whereas facultative anaerobic and aerobic bacteria that grew under anaerobic conditions were included as aerobes in all analyses.
Statistical analysis
Demographics, markers of disease severity, and bacterial prevalence (presence of bacteria at CFU >0) were compared across clinical sites using chi-square or Fisher’s exact tests for categorical variables and Mantel-Haenszel mean score chi-square for ordinal variables. Associations between presence of bacteria and markers of disease severity were assessed by a Mantel-Haenszel mean score chi-square test. Analyses were stratified by site as described.
Multivariable logistic regression was used to test for factors that predict prevalence of specific bacterial genera. Predictors were identified via a stepwise model selection (with significance level for entry or staying in the model of p=0.10 and p=0.05, respectively).
Analyses that included bacterial quantities/density were log transformed to account for non-normal distributions using log10(CFU/g+1) to incorporate samples with no detectable bacteria. Bacterial diversity measures included richness (number of counted taxa), evenness and diversity (Shannon-Wiener index). Analyses of bacterial communities (network analyses and factor analysis) were conducted on sputum bacterial quantity for the most common genera. Co-occurrence (network analysis) between taxonomic groups was calculated as previously described [21]. Factor analysis as an unsupervised method to group genera was conducted using varimax rotations to achieve independent factor groupings. These factors were used to assign each subject to a group defined by their dominating factor based on similarity of bacterial quantities (see details in Supplement).
Results
Subjects and samples
Two-hundred and fifty-five subjects were enrolled. Sputum samples (n=200) were obtained at all three sites whereas BAL (n=55) samples were collected only at UNC (n=24) and Dublin (n=31). All samples from children <6 years were BAL (n=39). Either sputum (n=34) or BAL (n=12) were collected from subjects in the 6–<18 years group; for subjects ≥18 years, 166 sputum and 4 BAL samples were collected.
Subject characteristics by site are shown in Table 1. Site-specific differences in the study subjects included older age and fewer subjects positive for F508del mutations in Belfast compared with UNC or Dublin. Fewer subjects in Belfast were undernourished, and, despite being an older cohort, FEV1 values in Belfast were similar to those in UNC and Dublin, consistent with milder disease in Belfast. Use of chronic medications was lower in those contributing BAL versus sputum (Table S2A, B).
Table 1:
Patient Demographics by site
| Clinical Site | |||||
|---|---|---|---|---|---|
| Overall N=255 N (%) |
B N=75 N (%) |
C N=65 N (%) |
D N=115 N (%) |
p values1 | |
| Age(years) | |||||
| Mean | 21.9 | 29.1 | 16.1 | 20.5 | |
| Std Dev | 12.7 | 12.2 | 10.3 | 12.1 | |
| Median | 21.6 | 26.2 | 14.5 | 22.0 | |
| Minimum | 1.0 | 8.3 | 1.0 | 1.0 | |
| Maximum | 68.2 | 68.2 | 50.0 | 61.2 | |
| Age(years) | 255 | 75 | 65 | 115 | <.0001 |
| 0–<6 | 39 (15%) | 0 (0%) | 14 (22%) | 25 (22%) | |
| 6–<13 | 21 (8%) | 4 (5%) | 13 (20%) | 4 (3%) | |
| 13–<18 | 25 (10%) | 6 (8%) | 9 (14%) | 10 (9%) | |
| 18–<25 | 77 (30%) | 22 (29%) | 18 (28%) | 37 (32%) | |
| 25–<30 | 44 (17%) | 19 (25%) | 5 (8%) | 20 (17%) | |
| 30+ | 49 (19%) | 24 (32%) | 6 (9%) | 19 (17%) | |
| Gender | 255 | 75 | 65 | 115 | 0.13 |
| Female | 115 (45%) | 29 (39%) | 36 (55%) | 50 (43%) | |
| Male | 140 (55%) | 46 (61%) | 29 (45%) | 65 (57%) | |
| F508del mutation | 251 | 74 | 63 | 114 | 0.001 |
| Homozygote | 131 (52%) | 29 (39%) | 45 (71%) | 57 (50%) | |
| Heterozygote | 99 (39%) | 34 (46%) | 15 (24%) | 50 (44%) | |
| None | 21 (8%) | 11 (15%) | 3 (5%) | 7 (6%) | |
| Pancreatic status | 255 | 75 | 65 | 115 | 0.61 |
| PI | 223 (87%) | 64 (85%) | 59 (91%) | 100 (87%) | |
| PS | 32 (13%) | 11 (15%) | 6 (9%) | 15 (13%) | |
| BMI | 235 | 74 | 65 | 96 | 0.01 |
| undernourished | 26 (11%) | 3 (4%) | 9 (14%) | 14 (15%) | |
| acceptable | 118 (50%) | 34 (46%) | 37 (57%) | 47 (49%) | |
| well nourished | 91 (39%) | 37 (50%) | 19 (29%) | 35 (36%) | |
| FEV1 % predicted GLI | 212 | 74 | 51 | 87 | 0.02 |
| <41% | 42 (20%) | 10 (14%) | 5 (10%) | 27 (31%) | |
| 41–80% | 115 (54%) | 48 (65%) | 28 (55%) | 39 (45%) | |
| >80% | 55 (26%) | 16 (22%) | 18 (35%) | 21 (24%) | |
| Sample Type | 255 | 75 | 65 | 115 | <0.0001 |
| BAL | 55 (22%) | 0 (0%) | 24 (37%) | 31 (27%) | |
| Sputum | 200 (78%) | 75 (100%) | 41 (63%) | 84 (73%) | |
| Chronic antibiotics | 173 (68%) | 58 (77%) | 43 (66%) | 72 (63%) | 0.10 |
| Flucloxacillin | 18 (7%) | 8 (11%) | 1 (2%) | 9 (8%) | 0.09 |
| Azithromycin | 127 (50%) | 44 (59%) | 37 (57%) | 46 (40%) | 0.02 |
| Inhaled antibiotics2 | 130 (52%) | 42 (56%) | 30 (47%) | 58 (52%) | 0.58 |
| Any mucolytic | 205 (80%) | 59 (79%) | 54 (83%) | 92 (80%) | 0.79 |
| DNAse | 164 (64%) | 54 (72%) | 43 (66%) | 67 (58%) | 0.15 |
| Hypertonic saline | 109 (43%) | 15 (20%) | 37 (57%) | 57 (50%) | <0.0001 |
|
Inhaled corticosteroids |
105 (41%) | 31 (41%) | 36 (55%) | 38 (33%) | 0.004 |
| Antacid3 | 129 (51%) | 32 (43%) | 41 (63%) | 56 (49%) | 0.05 |
| Insulin | 34 (13%) | 7 (9%) | 8 (12%) | 19 (17%) | 0.35 |
B=Belfast, Northern Ireland; C=Chapel Hill, USA; D=Dublin, Ireland. PS=pancreatic sufficiency. PI=pancreatic insufficiency. BMI=body mass index. FEV1-forced expiratory volume in 1 second expressed as % predicted based on GLI=global lung initiative reference values[45]. BAL=bronchoalveolar lavage AZM=azithromycin. Patients were continuously enrolled at each site, not necessarily reflecting the study site population.
: Comparisons across sites using chi- square, Mantel-Haenszel mean score chi-square, or Fisher’s exact test.
: tobramycin, colistin and aztreonam.
: antacids, H2-blockers and proton pump inhibitors.
Prevalence and abundance of genera with age
Eighteen anaerobic and 39 aerobic genera were cultured (Table S3) representing 167 species (Table S4). The prevalence of one or more strict anaerobic genus at any CFU in a sample was 59% and was higher in sputum (67%) than BAL samples (31%), p<0.001. Aerobic bacteria were present at any CFU in 197/200 (99%) of sputum and in 51/55 (93%) BAL samples. Mean±SEM TVC were higher for aerobic than anaerobic bacteria in sputum (2.27×108±3.80×107 vs. 2.86×107±2.07×107 CFU/g; p<0.001), and BAL (3.97×107±3.71×107 vs. 2.27×104±1.61×104 CFU/ml; p<0.001).
Ratio of anaerobic to aerobic viable counts for BAL and sputum showed no differences across age groups (Fig S1A). Bacterial richness was lower in BAL (3.03±0.31) compared with sputum (5.18±0.16), p<0.001 but similar within each sample type over age sextiles that approximate clinical progression (Figure 1A). Shannon diversities showed similar trends (Figure S1B). Sixteen genera, which had a ≥5% prevalence across BAL and sputum samples, are shown by age sextiles in Table S5. The most prevalent anaerobic and aerobic genera across all ages were Prevotella (51%) and Streptococcus (82%), respectively. More prevalent bacteria were more abundant with the exception of Burkholderia, with a 5% prevalence but being most abundant when present, and Prevotella with high prevalence but low abundance. Changes in prevalence of the most frequently cultured anaerobes and aerobes with age showed decreasing prevalence from childhood to mid-adulthood with subsequent increases for anaerobic genera and Streptococcus. These trends were inverse for Pseudomonas (Figure 1B).
Figure 1A: Richness by age groups across all sites and sample types.
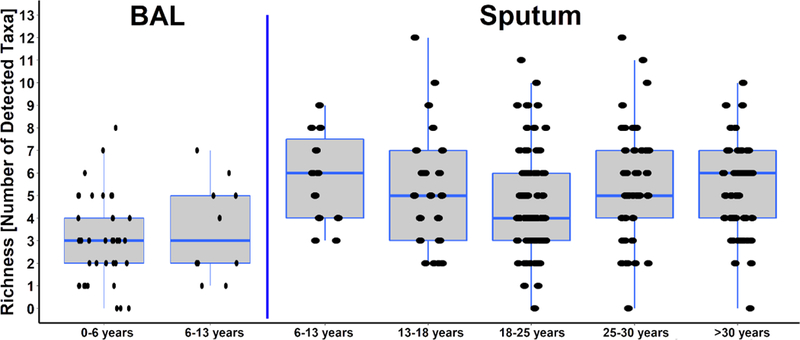
Richness defined as number of anaerobic and aerobic genera detected at any CFU per sample and did not differ between the age groups within sample types.
Figure 1B: Prevalence of most frequently cultured bacteria by age groups across all sites and sample types.
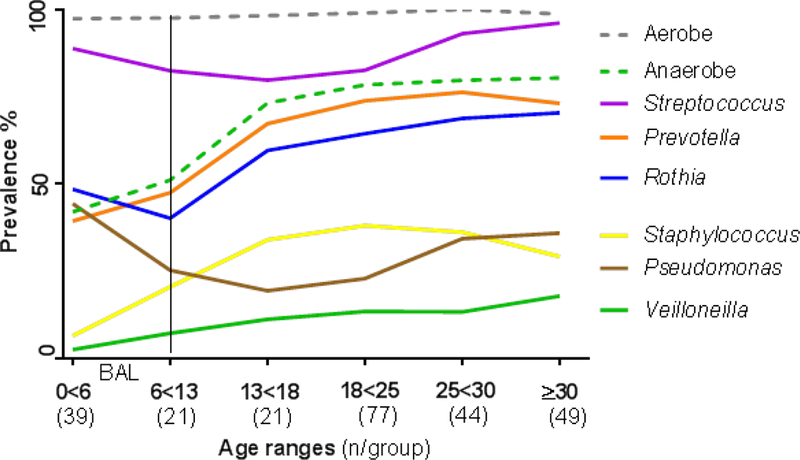
Prevalence for most prevalent bacteria are shown by age groups/range in years. The number of samples for each group is provided in parenthesis. Only BAL samples are included in subjects <6 years, as indicated by the vertical line.
Analyses of samples by sites revealed differences in bacterial richness across sites but similar diversity in sputum samples (Figure S2A). There were no differences in richness nor diversity in BAL samples from Dublin versus UNC (Figure S2B). The prevalence of Pseudomonas was highest at UNC with Burkholderia, Haemophilus, Actinomyces, Rothia and Gemella being more prevalent in Belfast. Neisseria prevalence was lowest in Belfast. Dublin had the lowest prevalence of Prevotella and Veillonella. Further details are shown by site and sample type in Table S3 and Figure S3.
Relationship between bacterial prevalence, abundance and disease severity
In subjects who produced sputum (n=200) anaerobes were associated with phenotypically milder disease, e.g. better lung function and BMI and with pancreatic sufficiency, absence of insulin and chronic antibiotic use, but not genotype (Figure 2A). H. influenzae, an aerobe observed in early CF disease followed a similar pattern (Figure 2B). Staphylococcus exhibited an intermediate pattern with negative associations, i.e. was less prevalent, with use of chronic antibiotics (Figure 2C). The pattern for Pseudomonas; however, was distinctly different from anaerobes. Pseudomonas prevalence was positively associated with worse BMI, number of F508del alleles, and chronic antibiotics (Figure 2D).
Figure 2: Prevalence as percent of subjects with any CFU/g sputum for a) Anaerobes, b) Haemophilus, c) Staphylococcus, and d) Pseudomonas in sputum and their association with clinical characteristics.
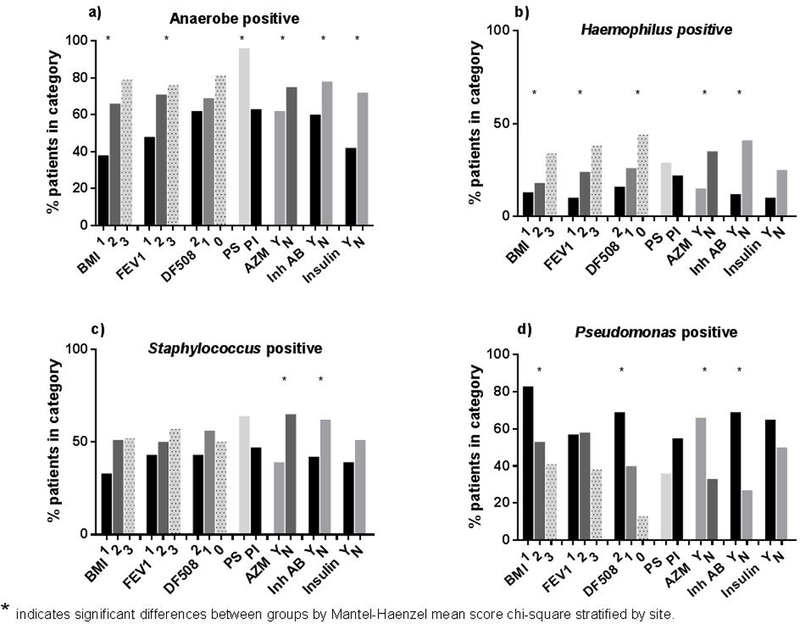
BMI are 1 - poor nutritional status; 2 - adequate; 3 - well-nourished. Categories for FEV1 are 1: <40% predicted; 2: 40-80%; 3: >80%. Numbers per category are given as percentage of subjects positive for this characteristic. Comparisons are by Mantel-Haenzel mean score chi-square test stratified by site.
Logistic regression adjusting for covariates with p <0.05 identified through stepwise modeling revealed a lower prevalence of anaerobes in pancreatic insufficient subjects (Table 2). With respect to aerobes, a lower prevalence of Haemophilus and Staphylococcus was associated with use of azithromycin. Pseudomonas prevalence again was associated with F508del alleles and azithromycin and inhaled antibiotic use. For BAL, clinical or site variables were not associated with bacterial prevalence.
Table 2:
Odds ratios from logistic regression models of bacterial prevalence in sputum
|
Anaerobes N=197 |
Haemophilus N=197 |
Staphylococcus N=200 |
Pseudomonas N=195 |
|
|---|---|---|---|---|
| F508del (1 vs 0) | -- | -- | -- | 8.24 (1.00, 68.20) |
| F508del (2 vs 0) | -- | -- | -- | 17.63 (2.13,146.04) |
| Pancreatic insufficiency | 0.12 (0.03, 0.53) | -- | -- | -- |
| Azithromycin | -- | 0.41 (0.18, 0.92) | 0.35 (0.19, 0.62) | 2.26 (1.10, 4.67) |
| Inhaled antibiotics | -- | 0.26 (0.12, 0.57) | -- | 3.97 (1.96, 8.05) |
| Center C vs. B | 0.58 (0.22, 1.52)1 | 0.41 (0.15, 1.08)1 | -- | -- |
| Centre D vs. B | 0.17 (0.08, 0.38) | 0.17 (0.07, 0.41) | -- | -- |
Variables evaluated for model inclusion were: gender, age, centre (B=Belfast; C=UNC; D=Dublin), FEV1 and BMI as categories (as in Figure 2), F508del, pancreatic status, chronic use of flucloxacillin, azithromycin, inhaled antibiotics, DNase, hypertonic saline, corticosteroids and antacids. “ – “ indicates these variables were not selected for the final model because they were not significant predictors as determined by stepwise model selection with p=0.10 for entry and p=0.05 for staying in the model.
Difference is not significant but comparison included in model for comparison to D vs. B.
No associations were observed for absolute anaerobic or aerobic viable counts and clinical variables or study site in sputum or BAL. However, the calculation of ratio of anaerobe/aerobe CFU showed that subjects with relatively higher load of anaerobes had milder disease (pancreatic sufficiency and higher BMI) than aerobe-dominated subjects (Figure S4).
Community structure in sputum and association with subject characteristics
Bacterial communities reflecting the 16 genera with prevalence of ≥5% were explored in sputum samples (n=200) using microbial co-occurrence network analysis. Bacteria typical of the oropharynx e.g. Streptococcus, Rothia, and Gemella associated with each other. In contrast, Staphylococcus, Pseudomonas and Burkholderia each formed distinct entities (Figure 3).
Figure 3: Bacterial co-occurrence network between genera detected by extended microbial culture.
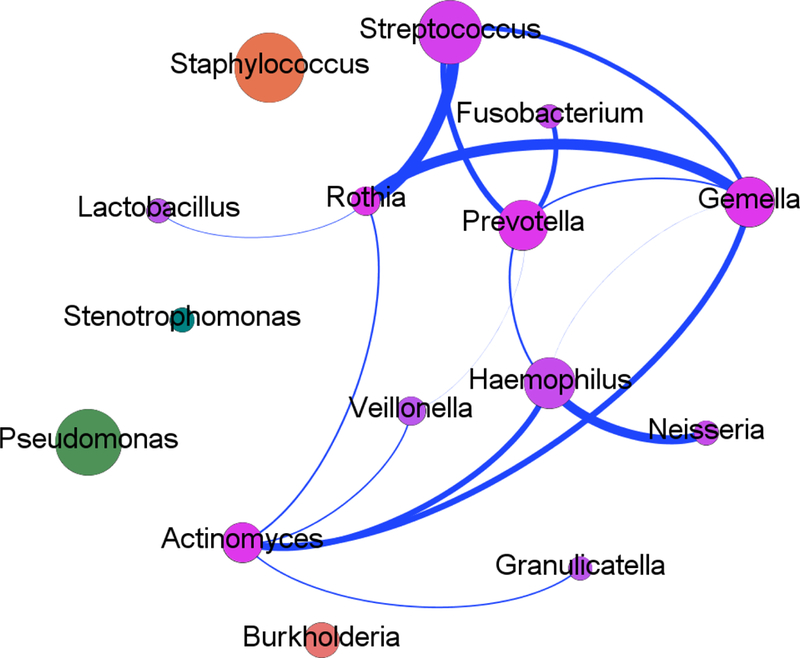
The sixteen genera detected in ≥5% of sputum samples are contained within the network. Co-occurring microbial taxa are shown with nodes (circles) denoting a particular taxon within the network and each line (edge) a significant co-occurrence relationship (Spearman's rank correlation coefficient for positive correlations (blue lines); adjusted p-value <0.05 [FDR correction]). The size of the corresponding nodes demonstrates the relative proportion of each genera within the current study and the thickness of edges represents the strength of the corresponding associations.
Next, factor analysis was used to reduce the complexity of the microbiota per sample into a set of variables that best defined sample community composition. The bacterial community compositions were generally consistent with the co-occurrence networks. Five components/factors explained 49% of the variation (Table 3). Factors 1 and 2 were dominated by oral-associated bacteria, including Streptococcus, Gemella, and Rothia. Factor 3 included Haemophilus and Neisseria. Factor 4 was dominated by obligate anaerobes. Pseudomonas defined factor 5, which was also negatively associated with Staphylococcus and Burkholderia.
Table 3:
Loadings from Factor Analysis showing groupings of bacteria with each other based on quantity in sputum (n=200).
| Factor 1 | Factor 2 | Factor 3 | Factor 4 | Factor 5 | |
|---|---|---|---|---|---|
| Streptococcus | 0.672 | −0.056 | 0.166 | −0.032 | −0.076 |
| Gemella | 0.574 | 0.274 | 0.087 | 0.109 | −0.098 |
| Rothia | 0.550 | 0.434 | −0.162 | 0.121 | 0.004 |
| Stenotrophomonas | 0.496 | −0.329 | −0.123 | −0.138 | 0.300 |
| Granulicatella | 0.035 | 0.608 | 0.219 | −0.095 | 0.150 |
| Lactobacillus | −0.085 | 0.597 | −0.282 | 0.122 | −0.081 |
| Actinomyces | 0.320 | 0.562 | 0.135 | −0.094 | −0.235 |
| Neisseria | 0.028 | −0.075 | 0.736 | −0.057 | 0.106 |
| Haemophilus | 0.162 | 0.263 | 0.65 | 0.123 | −0.166 |
| Fusobacterium | −0.182 | 0.101 | 0.049 | 0.654 | 0.209 |
| Porphyromonas | 0.014 | −0.201 | −0.162 | 0.630 | 0.022 |
| Prevotella | 0.378 | −0.027 | 0.352 | 0.557 | −0.092 |
| Veillonella | 0.165 | 0.275 | 0.108 | 0.454 | −0.236 |
| Pseudomonas | −0.034 | −0.097 | −0.051 | 0.051 | 0.735 |
| Staphylococcus | −0.245 | −0.265 | 0.325 | 0.160 | −0.491 |
| Burkholderia | 0.179 | 0.039 | −0.318 | −0.097 | −0.533 |
| Eigen-Value | 2.42 | 1.62 | 1.37 | 1.31 | 1.16 |
| Variance explained | 11.41% | 9.94% | 9.80% | 9.31% | 8.90% |
Five factors explain 49% of the total variation. Blue indicating positive associations and red negative associations with darker colour indicating the robustness of associations.
To define the clinical relevance of these microbial communities, patient sputum samples were defined by their dominating factor (Table 4). Thus, each sample was assigned to groups 1 through 5. There were no differences in pancreatic status, use of azithromycin, mucolytics, and anaerobic or aerobic bacterial load/density between groups. Groups 4 and 5 had the highest percentage of F508del homozygous subjects yet differed significantly with regard to both anaerobes and Pseudomonas. The group with higher anaerobic density/diversity (Group 4) tended to have higher FEV1 and a lower incidence of poor nutritional status compared with the Pseudomonas dominated Group 5. Notably, Group 3 included the fewest subjects with poor nutritional status and most with high FEV1, compared with other groups, but had the highest proportion of subjects <20 yrs.
Table 4:
Patient characteristics within the groups characterised by bacterial communities identified by Factor Analysis
| Group 1 | Group 2 | Group 3 | Group 4 | Group 5 | p-value a | |
|---|---|---|---|---|---|---|
| N per group | 48 | 35 | 46 | 27 | 44 | |
| Age | 27.56±1.49 | 28.62±1.75 | 22.99±1.53 | 24.34±1.99 | 26.82±1.56 | 0.089 |
| <20 years (% ) | 27% | 17% | 43% | 37% | 20% | 0.048 |
| % homozygote F508del | 40 | 54 | 37 | 64 | 61 | 0.046 |
| % null F508del | 15 | 11 | 6 | 4 | 2 | 0.2 |
| FEV1 % (GLI) | 59.47±3.17e | 55.79±3.78c | 75.64±3.28c, d,e | 61.58±4.26 | 57.08±3.35d | 0.0002 |
| BMI (m/kg2) | 21.9±0.52 | 22.9±0.7 | 21.8±0.5 | 20.8±0.7 | 20.8±0.5 | 0.1 |
| BMI category (% poor nutritional status) |
10 | 6 | 2 | 15 | 29 | 0.0033 |
| inh. antibiotics (% use) | 67 | 66 | 35 | 56 | 78 | 0.0007 |
| Pseudomonas pos. (%) | 38 | 37 | 39 | 59 | 91 | <0.0001 |
| Staphylococcus pos. (%) | 31 | 49 | 78 | 70 | 27 | <0.0001 |
| Any anaerobe (%) | 81 | 60 | 72 | 100 | 32 | <0.0001 |
| Aerobe richness | 6.83±0.43c | 6.69±0.50d | 6.07±0.44 | 5.7±0.57b | 4.57±0.45c,d | 0.0038 |
| Anaerobe richness | 1.75±1.19c | 1.20±0.23 | 1.59±0.20d | 3.22±0.26b | 0.68±0.21c,d | <0.0001 |
Group 1, 2 dominated by oral bacteria; Group 3 Haemophilus-Neisseria; Group 4 anaerobes; Group 5 Pseudomonas-dominated.
– p-values for comparison of categorical (Pearson) or continuous variables (ANOVA) across groups. Adjusted difference (Tukey’s):
– differs from all other groups;
Indicate differences to corresponding letter by row,
Indicate differences to corresponding letter by row,
– Indicate differences to corresponding letter by row.
BMI=body mass index. FEV1-forced expiratory volume in 1 second expressed as % predicted based on GLI=global lung initiative reference values [45]. Inh. Antibiotics=inhaled antibiotics including colistin, tobramycin, aztrenoam,
Discussion:
Anaerobic and aerobic bacteria were cultured from respiratory samples across all age ranges to assess bacterial prevalence and quantify density in CF respiratory secretions. The prevalence and quantities of both anaerobic and aerobic bacteria were lower in BAL compared to sputum. Interestingly, ratio of anaerobes/aerobes in BAL i.e. incorporating quantity were similar to sputum. Facultative anaerobes and micro-aerophilic bacteria were also cultured in both sample types (Table S3) and were included with aerobes, potentially underestimating the true anaerobic burden in the CF lung. Pseudomonas and Staphylococcus also grow anaerobically, a growth-mode associated with persistence in chronic infection but the focus in this study was on strict anaerobes as described in molecular studies. The prevalence of anaerobes and Streptococcus declined from early childhood to early adult-hood, i.e. a time when many patients experience worsening disease. In older ages, anaerobes were more prevalent again, possibly related to less severe disease (Fig 1B) as reflected by the higher anaerobe/aerobe ratio in subjects with better nutritional status (Fig. S4). Anaerobic and streptococcal species are highly abundant in oral secretions and are detected in the oral cavity in the first months of life [22]. Although oral secretions are a source of contamination of sputum and BAL, the high abundance of anaerobic genera makes sample contamination an unlikely explanation for our findings. Although BAL samples were obtained via ETT or LMA, which decreases the risk of contamination, the lower anaerobe prevalence in BAL compared with sputum likely reflects the younger BAL subject age with lower bacterial load and sample dilution. Aerobe prevalence and abundance was also lower in BAL than sputum. Typically, BAL is assumed to represent a 30–80 fold dilution of epithelial lining fluid [23, 24].
The source of CF lower airway oral bacteria likely reflects micro-aspiration as reported in healthy subjects, in whom oral bacteria are found in proximal airways but continuous muco-ciliary clearance prevents growth to high densities [25, 26]. We suggest that starting in infancy, abnormal muco-ciliary clearance in CF enhances retention of aspirated flora in an anaerobic airway environment, allowing anaerobic bacteria to proliferate. Such a scenario is consistent with recent CF infant BAL microbiome analyses [27–29].
The contribution of anaerobic bacteria to lung disease is not clearly understood. This large study, which analyzed samples from 255 CF subjects across 3 sites and a wide age range, characterised the anaerobic and aerobic microbial community composition in the CF airways using extended culture methods. A prior molecular study had shown geographical differences in the CF microbiome [30]. In our study, differences occurred by study sites but not by continent, making climate a less relevant factor than genotype (e.g. F508 del) and associated disease severity. We observed that anaerobic bacteria were more prevalent in sputum of subjects with milder disease. Higher lung function, increased BMI, pancreatic sufficiency and absence of requirement for insulin were associated with higher prevalence of anaerobes in univariate analyses (Figure 2). Lower prevalence of anaerobes and Haemophilus in subjects on azithromycin could be due to the antibacterial activity of azithromycin. Azithromycin exhibits good activity against those organisms in non-CF disease; however, high rates of resistance are seen in CF that may mitigate its activity [31, 32]. Notably, associations with pancreatic sufficiency remained significant when adjusting for multiple medications and clinical parameters (Table 2). A sensitivity analyses with p=0.1 for staying in the model, included FEV1 for Pseudomonas, azithromycin and insulin for anaerobes, and hypertonic saline for Haemophilus (Table S6).
Our culture based findings are consistent with observations from molecular microbiome studies describing higher diversity with less severe CF lung disease [5, 33], and decreasing diversity in patients with more rapid lung function decline [5, 11]. Our quantitative cultures allowed these relationships to be extended to anaerobic and aerobic bacterial density, anaerobe/aerobe ratio and diversity. A higher anaerobe/aerobe ratio was associated with pancreatic sufficiency and better nutrition, with trends for higher lung function in those with higher anaerobes/aerobe ratio (Figure S4). However, associations between absolute aerobic or anaerobic bacterial concentrations/density and FEV1 were not detected. We hypothesise that the absence of such an association partly reflects the spatial heterogeneity of CF lung disease, with bacterial density reflecting disease severity in focal bronchiectatic areas, whereas FEV1 reflects the overall extent of diseased lung. The hypothesis that FEV1 is not sufficiently sensitive to detect regional, bronchiectatic disease is supported by a study that reported associations between anaerobic density in CF patients and lung clearance index but not FEV1 [34]. Moreover, it should be noted that age-based CF-specific lung function parameters developed for children [35] are not available for adults, and effects of genetic mutations on lung function were not included. Thus, although we used GLI references for comparison across body size and centre, analyses of relationships of bacterial density with FEV1 may have been more sensitive if CF- and age-specific references were available.
Analyses of anaerobe pathogenicity in vitro may also yield insight into the relationships between anaerobes and CF disease severity. Prevotella was the most abundant and prevalent anaerobic genus in this study and has been reported consistently in other studies [5, 6, 10, 36–38]. In vitro studies of laboratory strains of Prevotella demonstrated that they produce short-chain fatty acids and proteases that may be pro-inflammatory [9]. However, Prevotella elicits less pro-inflammatory responses in respiratory cell lines and murine models than classic aerobic pathogens [39]. Thus, conceivably the relatively lower virulence of anaerobes contributes to their association with a milder CF phenotype.
Co-occurrence networks to visualise potential bacterial interactions showed groupings of orally-derived anaerobic bacteria, which occupied niches distinct from the known pathogens (e.g. Pseudomonas, Staphylococcus) that were themselves highly dissimilar (Figure 3). Both, Burkholderia and Pseudomonas release products that are toxic to other bacteria, which may contribute to their distinct niches as well as their dominance [40]. CF Registry data also demonstrate decreasing prevalence of Staphylococcus in older patients as Pseudomonas prevalence increases, suggesting a similar community competition for these two bacteria [41, 42].
Factor analyses (Table 3) revealed bacterial groupings similar to the co-occurrence networks. These factors enabled testing of associations with clinical parameters based on samples defined by the highest factor loadings (Table 4). Group 4 with highest prevalence and diversity of anaerobes had milder disease than the Pseudomonas-dominated group but not the mildest disease, possibly reflecting that group 4 exhibited the highest proportion of F508del homozygote subjects and moderate/high rates of Pseudomonas.
A challenge to the present study is inclusion of paediatric to adult ages necessitating comparisons between BAL and sputum. For age wide comparisons prevalence was used as surrogate in addition to ratio of anaerobic/aerobic bacterial load. Similar to prior studies, this study is limited by its observational design and thus cannot determine association versus causality with respect to anaerobe-induced disease severity. This difference impacts consideration of targeted anaerobe antibiotic therapy in subjects with CF. A recent study in infants with CF demonstrated more airway inflammation in children whose airway microbiome was dominated by anaerobes compared with “sterile” airways [29]. Anaerobes also may condition the lower CF airway to promote infection with more classic pathogens [43]. Such data may support treatment of anaerobes in early CF disease. In contrast, bacterial culture and molecular studies in older, sputum-producing subjects suggest that a diverse lung microbiota with relatively abundant anaerobes is associated with milder disease. Conceivably, higher anaerobic diversity in adults with milder disease reflects lower antibiotic use in such subjects [44] and/or lower intrinsic virulence of anaerobes [39]. Therefore targeted “eradication” of anaerobes in older patients when clinically at baseline might result in little clinical benefit and more resistant organisms. Anaerobes may increase at onset of exacerbations; therefore their role may fluctuate with short term changes in disease state [6, 14]. Anaerobes are not routinely cultured for in the clinical setting. This approach may be justified in older subjects if antimicrobial therapy is not being considered.
In summary, anaerobic bacteria were detected in CF respiratory secretions at all ages. In the age groups investigated in this study, aerobes dominated over anaerobes with respect to bacterial numbers. The prevalence of anaerobic genera was associated with milder disease and less use of antibiotics, suggesting that a diverse anaerobic microbiota may be a marker of better health in older subjects with CF. Sputum anaerobic/aerobic bacterial genera clustered into communities, which were associated with clinical outcomes. Cross-sectional studies do not permit distinction of anaerobes as markers versus causative factors in the severity of lung disease. Collectively, our data suggest that targeted eradication of anaerobes in the context of aerobic pathogenic bacteria may not be warranted until further data are available.
Supplementary Material
Acknowledgements
We would like to thank the patients and families who participated in this study and the clinical teams at each site who facilitated completion of the study. We acknowledge all staff involved in laboratory culture of bacteria and patient recruitment.
Funding: Work at UNC was supported by the National Institutes of Health (HL084934, HL100809, P30DK065988, and P0HL108808). Research at Queen’s University Belfast was funded by the Health and Social Care Research and Development Division, Public Health Agency, Northern Ireland and the Medical Research Council through a US-Ireland Partnership Grant. MMT was supported by a Health and Social Care Research and Development, Public Health Agency, Northern Ireland-funded UK National Institute for Health Research Career Scientist Award. Research at RCSI, Dublin, was supported by Science Foundation Ireland and the Health Research Board under grant SFI/08/US/B1676 and by the National Institutes of Health under grant 5R01 HL092964–04.
Abbreviations
- CF
Cystic fibrosis
- FEV1
forced expiratory volume in 1 second
- BAL
bronchoalveolar lavage
- GLI
Global Lung Initiative
- CFU
colony forming units/gram sputum
- TVC
total viable count
Footnotes
Take home message: Anaerobic bacteria are cultured across all ages, occur as communities and correlate with milder disease in adults.
REFERENCES:
- 1.Sherrard LJ, Tunney MM, Elborn JS. Antimicrobial resistance in the respiratory microbiota of people with cystic fibrosis. Lancet 2014: 384(9944): 703–713. [DOI] [PubMed] [Google Scholar]
- 2.Worlitzsch D, Tarran R, Ulrich M, et al. Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. The Journal of clinical investigation 2002: 109(3): 317–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cowley ES, Kopf SH, LaRiviere A, et al. Pediatric Cystic Fibrosis Sputum Can Be Chemically Dynamic, Anoxic, and Extremely Reduced Due to Hydrogen Sulfide Formation. MBio 2015: 6(4): e00767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Worlitzsch D, Rintelen C, Bohm K, et al. Antibiotic-resistant obligate anaerobes during exacerbations of cystic fibrosis patients. Clinical microbiology and infection : the official publication of the European Society of Clinical Microbiology and Infectious Diseases 2009: 15(5): 454–460. [DOI] [PubMed] [Google Scholar]
- 5.Coburn B, Wang PW, Diaz Caballero J, et al. Lung microbiota across age and disease stage in cystic fibrosis. Sci Rep 2015: 5: 10241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tunney MM, Field TR, Moriarty TF, et al. Detection of anaerobic bacteria in high numbers in sputum from patients with cystic fibrosis. American journal of respiratory and critical care medicine 2008: 177(9): 995–1001. [DOI] [PubMed] [Google Scholar]
- 7.Brown PS, Pope CE, Marsh RL, et al. Directly sampling the lung of a young child with cystic fibrosis reveals diverse microbiota. Annals of the American Thoracic Society 2014: 11(7): 1049–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghorbani P, Santhakumar P, Hu Q, et al. Short-chain fatty acids affect cystic fibrosis airway inflammation and bacterial growth. Eur Respir J 2015: 46(4): 1033–1045. [DOI] [PubMed] [Google Scholar]
- 9.Mirkovic B, Murray MA, Lavelle GM, et al. The Role of Short-Chain Fatty Acids, Produced by Anaerobic Bacteria, in the Cystic Fibrosis Airway. American journal of respiratory and critical care medicine 2015: 192(11): 1314–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cox MJ, Allgaier M, Taylor B, et al. Airway microbiota and pathogen abundance in age-stratified cystic fibrosis patients. PloS one 2010: 5(6): e11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao J, Schloss PD, Kalikin LM, et al. Decade-long bacterial community dynamics in cystic fibrosis airways. Proceedings of the National Academy of Sciences of the United States of America 2012: 109(15): 5809–5814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Young GR, Smith DL, Embleton ND, et al. Reducing Viability Bias in Analysis of Gut Microbiota in Preterm Infants at Risk of NEC and Sepsis. Frontiers in cellular and infection microbiology 2017: 7: 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nguyen LD, Deschaght P, Merlin S, et al. Effects of Propidium Monoazide (PMA) Treatment on Mycobiome and Bacteriome Analysis of Cystic Fibrosis Airways during Exacerbation. PloS one 2016: 11(12): e0168860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lim YW, Evangelista JS 3rd, Schmieder R, et al. Clinical insights from metagenomic analysis of sputum samples from patients with cystic fibrosis. Journal of clinical microbiology 2014: 52(2): 425–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klappenbach JA, Saxman PR, Cole JR, et al. rrndb: the Ribosomal RNA Operon Copy Number Database. Nucleic Acids Res 2001: 29(1): 181–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vetrovsky T, Baldrian P. The variability of the 16S rRNA gene in bacterial genomes and its consequences for bacterial community analyses. PloS one 2013: 8(2): e57923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sherrard LJ, Schaible B, Graham KA, et al. Mechanisms of reduced susceptibility and genotypic prediction of antibiotic resistance in Prevotella isolated from cystic fibrosis (CF) and non-CF patients. The Journal of antimicrobial chemotherapy 2014: 69(10): 2690–2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sherrard LJ, McGrath SJ, McIlreavey L, et al. Production of extended-spectrum beta-lactamases and the potential indirect pathogenic role of Prevotella isolates from the cystic fibrosis respiratory microbiota. Int J Antimicrob Agents 2016: 47(2): 140–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laszlo G Standardisation of lung function testing: helpful guidance from the ATS/ERS Task Force. Thorax 2006: 61(9): 744–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Turck D, Braegger CP, Colombo C, et al. ESPEN-ESPGHAN-ECFS guidelines on nutrition care for infants, children, and adults with cystic fibrosis. Clin Nutr 2016: 35(3): 557–577. [DOI] [PubMed] [Google Scholar]
- 21.Einarsson GG, Comer DM, McIlreavey L, et al. Community dynamics and the lower airway microbiota in stable chronic obstructive pulmonary disease, smokers and healthy non-smokers. Thorax 2016: 71(9): 795–803. [DOI] [PubMed] [Google Scholar]
- 22.Kononen E Development of oral bacterial flora in young children. Ann Med 2000: 32(2): 107–112. [DOI] [PubMed] [Google Scholar]
- 23.Singh S, Grover V, Christie L, et al. A comparative study of bronchoscopic microsample probe versus bronchoalveolar lavage in patients with burns-related inhalational injury, acute lung injury and chronic stable lung disease. Respiration; international review of thoracic diseases 2015: 89(1): 19–26. [DOI] [PubMed] [Google Scholar]
- 24.Braun J, Mehnert A, Dalhoff K, et al. Different BALF protein composition in normal children and adults. Respiration; international review of thoracic diseases 1997: 64(5): 350–357. [DOI] [PubMed] [Google Scholar]
- 25.Dickson RP, Erb-Downward JR, Freeman CM, et al. Spatial Variation in the Healthy Human Lung Microbiome and the Adapted Island Model of Lung Biogeography. Annals of the American Thoracic Society 2015: 12(6): 821–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bassis CM, Erb-Downward JR, Dickson RP, et al. Analysis of the upper respiratory tract microbiotas as the source of the lung and gastric microbiotas in healthy individuals. MBio 2015: 6(2): e00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Subbarao P, Milla C, Aurora P, et al. Multiple-Breath Washout as a Lung Function Test in Cystic Fibrosis. A Cystic Fibrosis Foundation Workshop Report. Annals of the American Thoracic Society 2015: 12(6): 932–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frayman KB, Armstrong DS, Carzino R, et al. The lower airway microbiota in early cystic fibrosis lung disease: a longitudinal analysis. Thorax 2017: 72(12): 1104–1112. [DOI] [PubMed] [Google Scholar]
- 29.Muhlebach MS, Zorn BT, Esther CR, et al. Initial acquisition and succession of the cystic fibrosis lung microbiome is associated with disease progression in infants and preschool children. PLoS pathogens 2018: 14(1): e1006798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stressmann FA, Rogers GB, Klem ER, et al. Analysis of the bacterial communities present in lungs of patients with cystic fibrosis from American and British centers. Journal of clinical microbiology 2011: 49(1): 281–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Farrell DJ, Flamm RK, Sader HS, et al. Results from the Solithromycin International Surveillance Program (2014). Antimicrobial agents and chemotherapy 2016: 60(6): 3662–3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sherrard LJ, Graham KA, McGrath SJ, et al. Antibiotic resistance in Prevotella species isolated from patients with cystic fibrosis. The Journal of antimicrobial chemotherapy 2013: 68(10): 2369–2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fodor AA, Klem ER, Gilpin DF, et al. The adult cystic fibrosis airway microbiota is stable over time and infection type, and highly resilient to antibiotic treatment of exacerbations. PloS one 2012: 7(9): e45001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O’Neill K, Bradley JM, Johnston E, et al. Reduced bacterial colony count of anaerobic bacteria is associated with a worsening in lung clearance index and inflammation in cystic fibrosis. PloS one 2015: 10(5): e0126980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kulich M, Rosenfeld M, Campbell J, et al. Disease-specific reference equations for lung function in patients with cystic fibrosis. American journal of respiratory and critical care medicine 2005: 172(7): 885–891. [DOI] [PubMed] [Google Scholar]
- 36.Tunney MM, Klem ER, Fodor AA, et al. Use of culture and molecular analysis to determine the effect of antibiotic treatment on microbial community diversity and abundance during exacerbation in patients with cystic fibrosis. Thorax 2011: 66(7): 579–584. [DOI] [PubMed] [Google Scholar]
- 37.Zemanick ET, Wagner BD, Robertson CE, et al. Airway microbiota across age and disease spectrum in cystic fibrosis. Eur Respir J 2017: 50(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harris JK, De Groote MA, Sagel SD, et al. Molecular identification of bacteria in bronchoalveolar lavage fluid from children with cystic fibrosis. Proceedings of the National Academy of Sciences of the United States of America 2007: 104(51): 20529–20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Larsen JM, Musavian HS, Butt TM, et al. Chronic obstructive pulmonary disease and asthma-associated Proteobacteria, but not commensal Prevotella spp., promote Toll-like receptor 2-independent lung inflammation and pathology. Immunology 2015: 144(2): 333–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de la Fuente-Nunez C, Reffuveille F, Fernandez L, et al. Bacterial biofilm development as a multicellular adaptation: antibiotic resistance and new therapeutic strategies. Current opinion in microbiology 2013: 16(5): 580–589. [DOI] [PubMed] [Google Scholar]
- 41.CF-Foundation. Patient Registry Annual Report 2013. https://wwwcfforg/2013_CFF_Patient_Registry_Annual_Data_Reportpdf 2013.
- 42.UK. Cystic Fibrosis Registry 2015 Annual Report https://wwwcysticfibrosisorguk/the-work-we-do/uk-cf-registry 2015: Accessed Jan 2018. [Google Scholar]
- 43.Flynn JM, Niccum D, Dunitz JM, et al. Evidence and Role for Bacterial Mucin Degradation in Cystic Fibrosis Airway Disease. PLoS pathogens 2016: 12(8): e1005846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao J, Murray S, Lipuma JJ. Modeling the impact of antibiotic exposure on human microbiota. Sci Rep 2014: 4: 4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quanjer PH, Hall GL, Stanojevic S, et al. Age- and height-based prediction bias in spirometry reference equations. Eur Respir J 2012: 40(1): 190–197. [DOI] [PubMed] [Google Scholar]
- 46.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Statist Soc Ser B Statist Methodol 1995: 57: 289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


