Abstract
Histidine phosphorylation, the so-called hidden phosphoproteome, is a poorly characterized post-translational modification of proteins1,2. Here we describe a role of histidine phosphorylation in tumorigenesis. Proteomic analysis of 12 tumours from an mTOR-driven hepatocellular carcinoma mouse model revealed that NME1 and NME2, the only known mammalian histidine kinases, were upregulated. Conversely, expression of the putative histidine phosphatase LHPP was downregulated specifically in the tumours. We demonstrate that LHPP is indeed a protein histidine phosphatase. Consistent with these observations, global histidine phosphorylation was significantly upregulated in the liver tumours. Sustained, hepatic expression of LHPP in the hepatocellular carcinoma mouse model reduced tumour burden and prevented the loss of liver function. Finally, in patients with hepatocellular carcinoma, low expression of LHPP correlated with increased tumour severity and reduced overall survival. Thus, LHPP is a protein histidine phosphatase and tumour suppressor, suggesting that deregulated histidine phosphorylation is oncogenic.
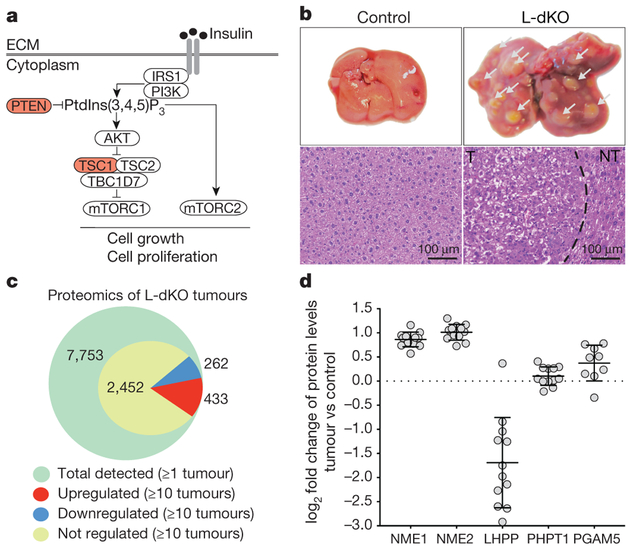
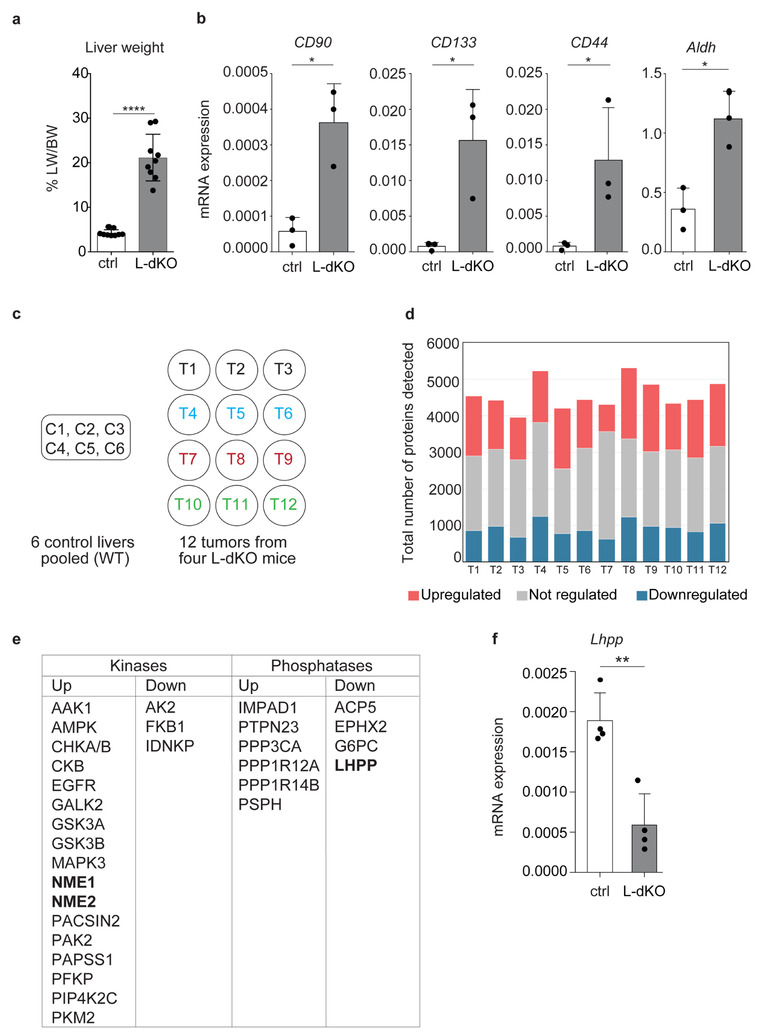
Liver cancer is the second leading cause of cancer-related deaths globally. Hepatocellular carcinoma (HCC) represents approximately 90% of primary liver cancer cases3. Recent studies indicate that almost 50% of HCC cases display aberrant PI3K–AKT–mTOR signalling4, including loss of the tumour suppressors PTEN, TSC1, and TSC23 (Fig. 1a). We generated an HCC mouse model by liver-specific deletion of PTEN and TSC1, using the neonatally expressed albumin promoter as a cre driver (see Methods). We hereafter refer to this model as liver-specific double-knockout (L-dKO) mice. L-dKO mice invariably exhibited hepatomegaly and advanced liver tumours at 6 and 20 weeks of age, respectively (Fig. 1b and Extended Data Fig. 1a). Histopathological analysis (Fig. 1b) and molecular classification based on mRNA expression of the liver cancer markers CD133 (also known as Prom1), cD90 (Thy1), Cd44 and Aldh1 (Aldh1a1) revealed that the L-dKO liver tumours indeed mimicked poorly differentiated human HCC (Extended Data Fig. 1b), consistent with other studies5,6.
Figure 1 |. Proteomics on L-dKO tumours.
a, Diagram of mTOR signalling showing TSC and PTEN tumour suppressors. b, Top, representative images of whole livers from 20-week-old L-dKO (tumours indicated with arrowheads) and control mice. Bottom, representative photomicrographs of haematoxylin and eosin (H&E)-stained liver from control mice, tumour (T) and adjacent non-tumour (NT) tissue from L-dKO mice (20 weeks). Similar results were obtained in tumours from nine mice. c, Number of tumour proteins detected and quantified by mass spectrometry. A total of 7,753 proteins were quantified in all tumours (n = 12). 3,147 proteins were quantified in a minimum of 10 tumours, of which 433 proteins were upregulated (red), 262 were downregulated (blue), and 2,452 were unchanged (yellow). ANOVA-based two-sample t-test with a false discovery rate (FDR) of 2% was used to determine regulation. d, Mass spectrometry-determined fold change in independent L-dKO tumours (n = 12 for NME1, NME2, LHPP and PHPT1; n = 9 for PGAM5) compared to control livers (n = 6);
data are mean ± s.d. PHPT1 and PGAM5 are two defined mammalian histidine phosphatases.
We performed quantitative proteomic analysis on three tumours each from four 20-week-old, L-dKO mice (12 tumours in total; see Methods). Liver protein extracts from six cre-negative age- and sex-matched littermates were pooled in equal amounts and used as a control (Extended Data Fig. 1c). We identified approximately 4,500 proteins per tumour (Extended Data Fig. 1d). Of the 3,147 proteins that were detected in a minimum of 10 tumours, 433 proteins were upregulated and 262 proteins were downregulated compared to the control (Fig. 1c). We manually curated a list of known or putative phosphatases and kinases significantly deregulated in a minimum of ten tumours (Extended Data Fig. 1e). Particularly intriguing in this list were the upregulated histidine kinases NME1 and NME2 (also known as NDPKA and NDPKB, respectively) (Fig. 1d), and the downregulated putative histidine phosphatase LHPP (phospholysine phosphohistidine inorganic pyrophosphate phosphatase).
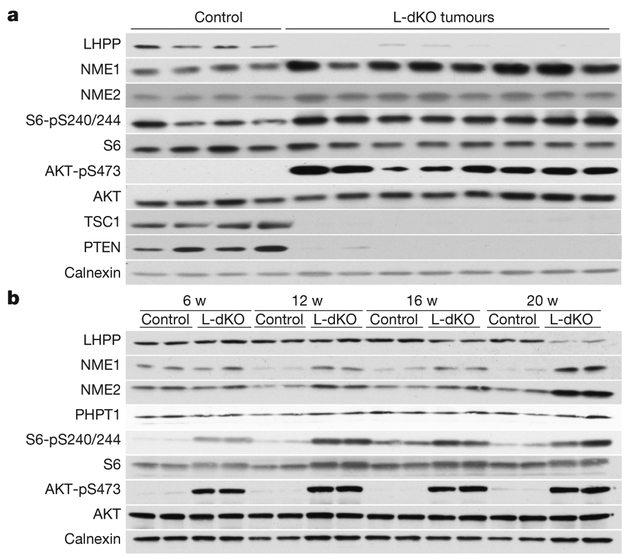
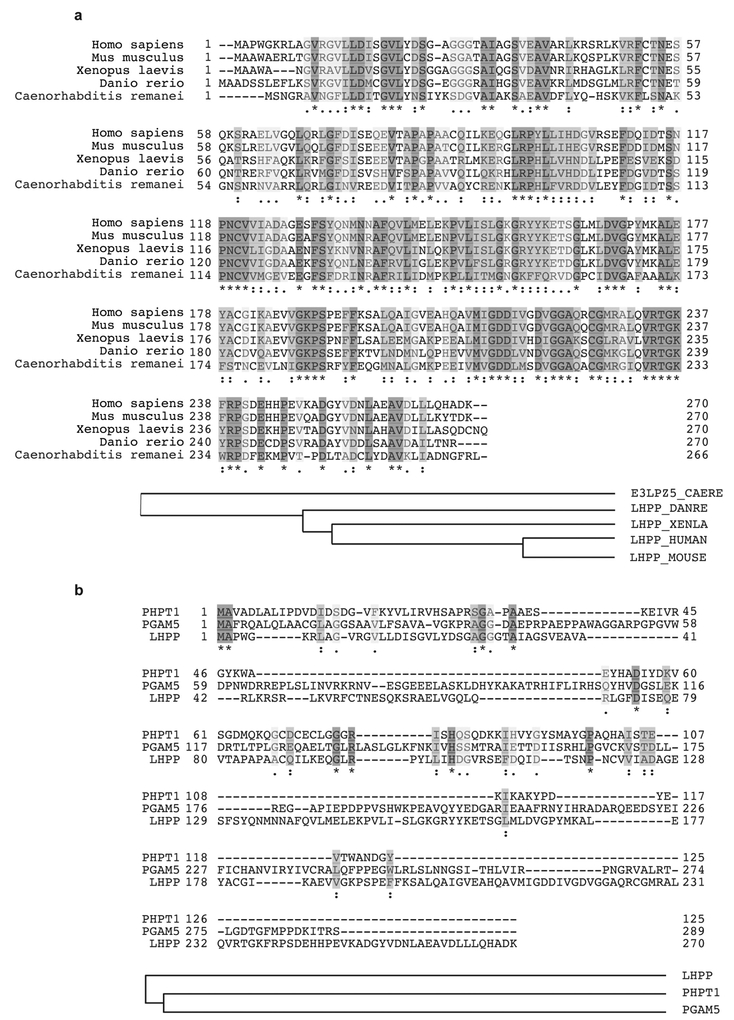
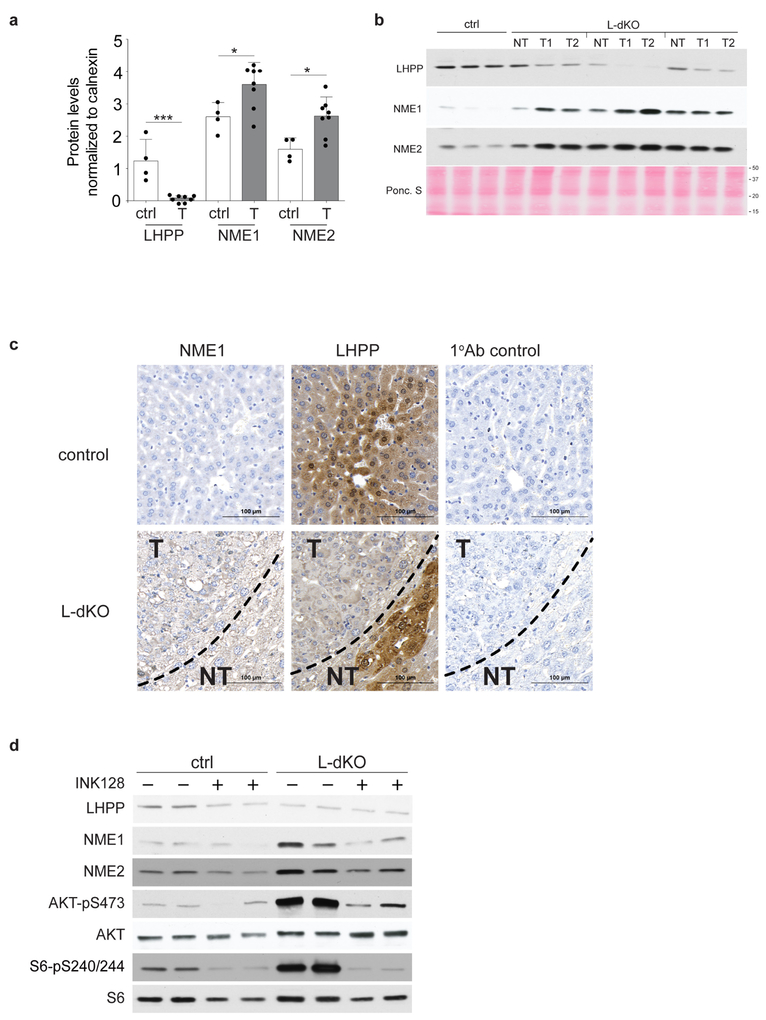
LHPP has been purified from bovine liver and shown to hydrolyse P–N bonds in synthetic substrates in vitro7–9, but is otherwise poorly characterized. LHPP is evolutionarily conserved from worm to human (overall 28% identity) (Extended Data Fig. 2a), but only weakly homologous to the two defined mammalian histidine phosphatases PHPT1 (8.1% identity) and PGAM5 (6.4% identity)10–12 (Extended Data Fig. 2b). We note that PHPT1 and PGAM5 levels were unchanged in tumours from L-dKO mice (Fig. 1d). NME1 and NME2 are homologous proteins (88% identical) that function as histidine kinases as either homo- or hetero-oligomers. NME1 and NME2 are the only mammalian protein histidine kinases reported so far13–15. Immunoblot analysis confirmed that LHPP was downregulated and NME1 and NME2 were upregulated (Fig. 2a and Extended Data Fig. 3a) in L-dKO tumours, as initially observed by proteomic analysis. Immunoblot analysis of S6 protein phosphorylated at Ser240 and Ser244 (S6-pS240/244) and AKT phosphorylated at Ser473 (AKT-pS473) confirmed high mTORC1 and mTORC2 activity, respectively, in L-dKO liver tissue and tumours (Fig. 2a, b). To determine whether the loss of LHPP expression coincided with tumorigenesis, we examined whole liver tissue (6, 12 and16 weeks) and isolated tumours (20 weeks) from L-dKO mice at different ages and hence at different stages of tumour development (Fig. 2b). Age-matched cre-negative littermates served as controls. LHPP expression was mildly reduced in the livers of L-dKO mice at 16 weeks, at which age macroscopic tumours were first visible. As observed previously, LHPP was strongly decreased in tumours of 20-week-old mice. As expected, mTOR signalling was high at all ages. Thus, the reduction in LHPP expression coincided with tumour development, but did not correlate with mTOR activity. Hepatic NME1 and NME2 expression decreased in control mice in an age-dependent manner, but remained high in L-dKO liver tissue and tumours. Thus, increased NME1 and NME2 expression in L-dKO mice correlated with increased mTOR activity rather than with tumour development (Fig. 2b), suggesting that mTOR may control NME1 and NME2 expression. To determine whether loss of LHPP expression was specific to tumours, we performed immunoblot analysis on dissected tumour and non-tumour liver tissue from 20-week L-dKO mice. LHPP expression was reduced (or absent) in the tumour, compared to the non-tumour liver tissue (Extended Data Fig. 3b). Immunohistochemistry on liver from 20-week L-dKO mice confirmed that LHPP was strongly downregulated (undetectable) in tumours compared to non-tumour tissue (Extended Data Fig. 3c). Like mTOR activity, NME1 and NME2 expression was similarly upregulated in both tumour and non-tumour L-dKO liver tissue compared to control liver tissue (Extended Data Fig. 3b, c), again indicating that the expression of NME1 and NME2 correlated with mTOR activity rather than tumorigenicity. These findings suggest that LHPP inactivation, in a background of constitutively high NME1 and NME2 activity in L-dKO liver, is a key event in tumorigenesis.
Figure 2 |. Loss of LHPP expression is confined to L-dKO tumours.
a, Immunoblot analysis indicates reduced LHPP and increased NME1and NME2 expression in tumours compared to age-matched littermate control (liver samples from control mice (n = 4) and two tumour samples each from four 20-week-old L-dKO mice (n = 8)). For full scans, see Supplementary Fig. 1. b, Immunoblot analysis indicates reduced LHPP expression (16 and 20 weeks (w)) and increased NME1 and NME2 expression (12, 16 and 20 weeks) in L-dKO liver (6, 12 and 16 weeks) and tumours (20 weeks) compared to age-matched control mice (6, 12 and 16 weeks, n = 4, liver tissues from two mice are pooled per lane). For full scans, see Supplementary Fig. 2.
The above findings suggest that the expression of NME1 and NME2, but not LHPP, is controlled by mTOR. To investigate this further, we acutely treated (24 h) L-dKO mice with the mTORC1 inhibitor rapamycin or the pan-mTOR inhibitor INK128. INK128 treatment reduced NME1 and NME2 expression in L-dKO tumours, but had no effect on LHPP (Extended Data Fig. 3d). Rapamycin had no effect on the level of NME1, NME2 or LHPP (data not shown). Thus, the expression of NME1 and NME2 seems to be mTORC2 dependent, whereas LHPP expression is mTOR independent.
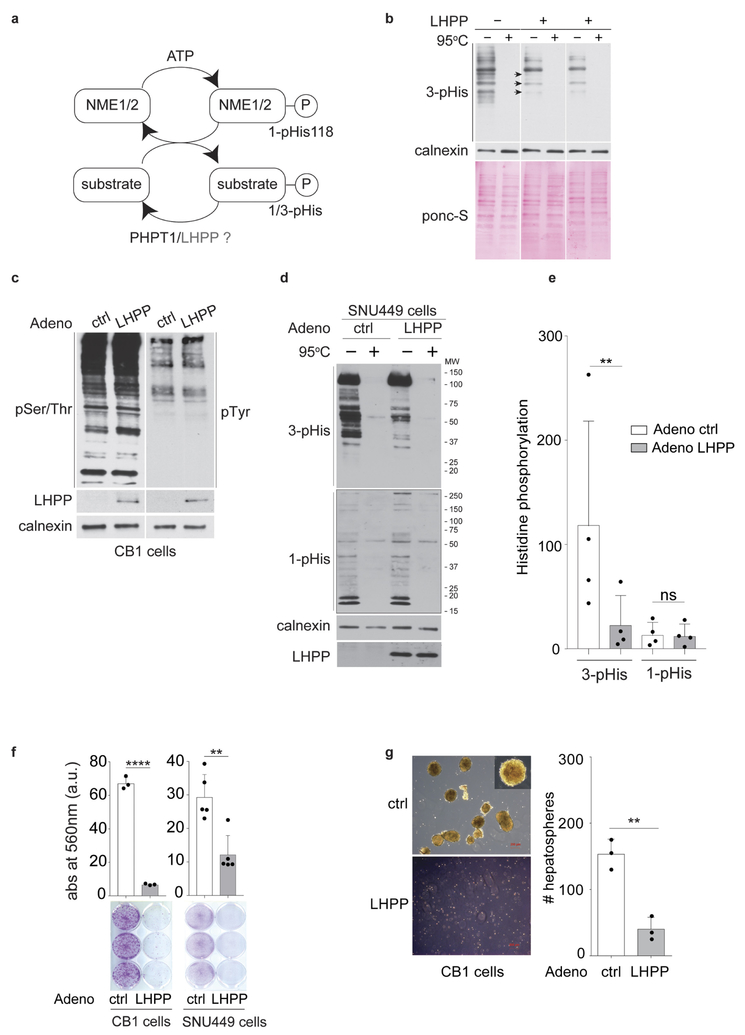
The paired upregulation of NME1 and NME2 (histidine kinases) and downregulation of LHPP (putative histidine phosphatase) prompted us to investigate protein histidine phosphorylation in L-dKO tumours1,16. Phosphohistidine (pHis) exists in two isomers, 1-pHis and 3-pHis, depending on the position of the phospho-acceptor nitrogen in the histidine imidazole ring (N1 and N3 positions, respectively)2,17. Unlike the phosphoester (P–O) bond in other phosphoamino acids, the phosphoramidate (P–N) bond in phosphohistidine is heat- and acid-labile, thus making pHis difficult to detect in biological samples18 This difficulty has been largely circumvented by the recent development of monoclonal antibodies that specifically recognize 1-pHis or 3-pHis2. The new antibodies were used to assess histidine phosphorylation (1-pHis and 3-pHis) in L-dKO tumours and control liver (Fig. 3a) (see Methods). The immunoblot signals of both 1-pHis and 3-pHis were high in tumours compared to control tissue (Fig. 3a, b). Heating sample-buffer-solubilized lysates at 95 °C for 10 min before SDS–PAGE caused dephosphorylation, indicating that the signals detected in the unheated samples are indeed histidine-phosphorylated proteins (Fig. 3a). As reported previously with cultured cells2, multiple 3-pHis-positive proteins but only one or a few 1-pHis proteins were detected in tumours and liver tissue. This could be due to the lower thermodynamic stability of 1-pHis than of 3-pHis19,20, or the existence of fewer proteins phosphorylated at the N1 position. The major band observed with the 1-pHis antibody is likely to be co-migrating NME1-pHis118 and NME2-pHis118 (Fig. 3a), because it displays an appropriate molecular mass (18 kDa) for NME1 and NME2 and was lost upon NME1 and NME2 knockdown (data not shown). NME1 and NME2 normally autophosphorylate position N1 of His118, and then transfer this phosphate to histidine (N1 or N3) in target proteins (Extended Data Fig. 4a). Thus, consistent with increased levels of NME1-pHis118 and NME2-pHis118 and decreased LHPP levels, histidine phosphorylation was significantly increased in L-dKO tumours.
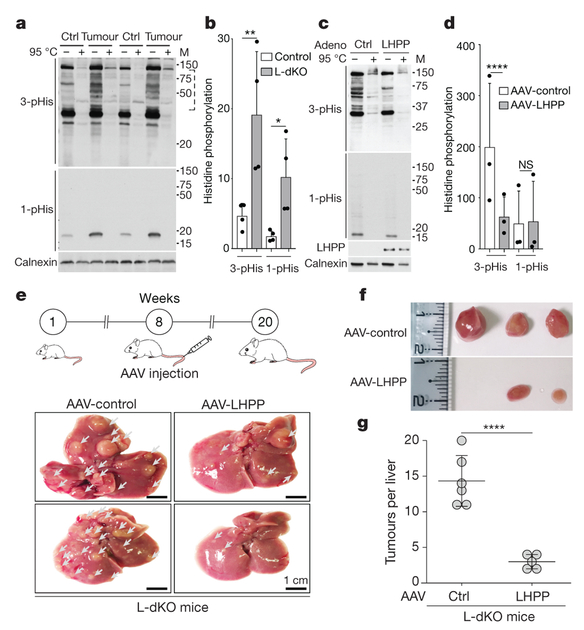
Figure 3 |. LHPP is a protein histidine phosphatase and a tumour suppressor.
a, b, Immunoblot analysis (a) and quantification (b), shows increased 3-pHis and 1-pHis signals in L-dKO tumours (20 weeks) compared to control (ctrl) littermates. Similar results were observed in tumours from L-dKO mice (n = 4). Tissue lysates in 2× sample buffer (pH 8.8) heated at 95 °C for 10 min were dephosphorylated and served as a control. For full scans, see Supplementary Fig. 5. In the 3-pHis blot, the area enclosed in the dashed line was used for quantification (n = 4). Adjusted *P = 0.0460, **P = 0.0051, two-sided, Holm-Sidak’smultiple comparisons test. Data are mean ± s.d. M, molecular mass. c, Immunoblotting shows a reduction in 3-pHis levels in CB1 cells infected with adenovirus (adeno) containing LHPP, compared to cells infected with a control adenovirus. Similar results were observed in three biological experiments. For full scans, see Supplementary Fig. 7. d, Quantification of immunoblots indicate that 3-pHis signals are significantly reduced upon LHPP re-expression compared to control-infected cells (n = 3 biological repeats). ****P < 0.0001, two-sided ratio paired t-test. NS, not significant. Data are mean ± s.d. e, Top, experimental timeline. AAV-control or AAV-LHPP was injected into the tail vein of 8-week-old L-dKO and littermate control mice. Bottom, representative images of whole livers showing reduction in L-dKO liver tumour burden after infection with AAV-LHPP compared to infection with AAV-control (arrowheads indicate tumours). Similar results were observed in five different L-dKO mice infected with AAV-LHPP. f, Photographs of tumours isolated from L-dKO mice infected with AAV-LHPP and AAV-control. g, Quantification of tumours in L-dKO mice showing a significant reduction in tumour burden after infection with AAV-LHPP (n = 5 mice), compared to AAV-control (n = 6 mice). ****P < 0.0001, two-sided unpaired t-test. Data are mean ± s.d.
To identify proteins in which histidine phosphorylation is increased in tumours, we used a combination of anti-1-pHis and anti-3-pHis monoclonal antibodies to immunoprecipitate pHis proteins from four L-dKO tumours and control tissue. Subsequent mass spectrometry identified 1,173 and 823 proteins from tumour and control liver tissue lysates, respectively (Supplementary Tables 1 and 2). In total, 260 proteins were more abundant (at least twofold) in tumours compared to control tissue (Supplementary Table 3). Only 18 proteins were more abundant in control tissue (see below for more discussion).
We next investigated whether LHPP is indeed a protein phosphohistidine phosphatase. First, we assayed LHPP phosphohistidine phosphatase activity in viíro (see Methods). Recombinant LHPP treatment reduced 3-pHis levels in an electrophoretically fractionated CB1 cell lysate transferred to a membrane (Extended Data Fig. 4b). CB1 is a cell line derived from L-dKO tumours (see Methods) and thus displays high levels of histidine phosphorylation. Second, we assessed LHPP phosphatase activity in vivo. Restoration of Lhpp expression in CB1 cells, using an adenovirus delivery system, reduced 3-pHis 3.1-fold with negligible effect on 1-pHis, pSer/Thr or pTyr (Fig. 3c, d, Extended Data Fig. 4c). Similar results were obtained with regard to pHis (3.9-fold drop in 3-pHis, no effect on 1-pHis) after introducing mouse Lhpp into human SNU449 cells, a liver cancer cell line with low intrinsic LHPP levels (Extended Data Fig. 4d, e). Immunoblot analysis confirmed the expression of recombinant Lhpp in both cell lines (Fig. 3c and Extended Data Fig. 4d). Collectively, these results indicate that LHPP is a histidine phosphatase broadly acting on N3-phosphorylated proteins. We note that PGAM5 was previously demonstrated12 to be a 1-pHis phosphatase acting specifically on NME2-pHis118.
To identify potential LHPP targets, we immunoprecipitated N3-phosphorylated proteins from CB1 cells lacking and re-expressing LHPP Mass spectrometry revealed nine proteins that were more abundant (at least 1.7-fold) in CB1 cells that lack LHPP (Supplementary Table 4). These included ACLY (ATP citrate lyase), known to be N3-phosphorylated at His760 (see below).
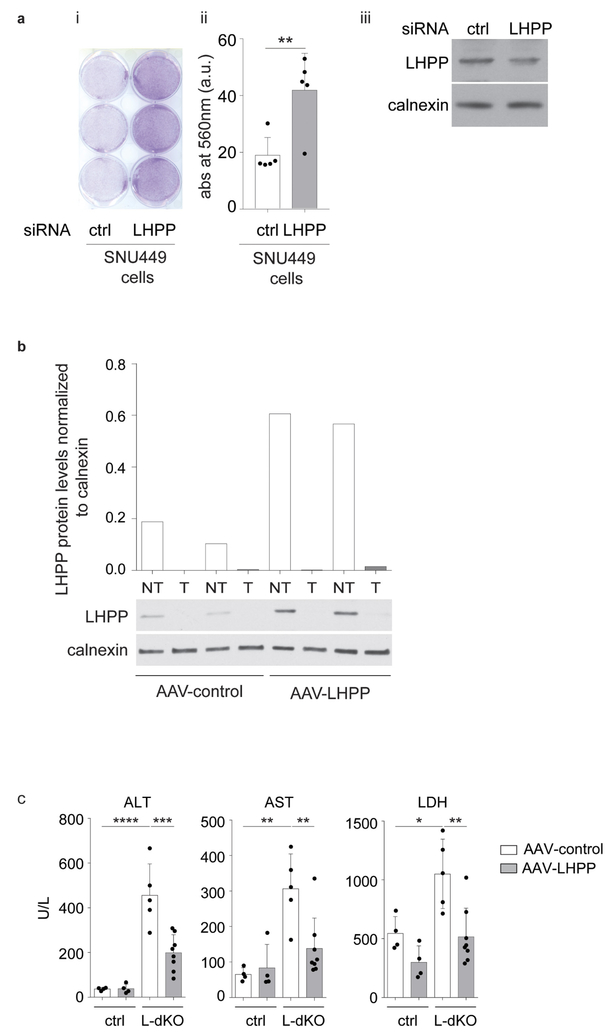
We next examined further the role of the phosphatase LHPP in proliferation and tumorigenesis. Increased expression of LHPP in CB1 and SNU449 cells significantly impaired cell proliferation and, in CB1 cells, reduced hepatosphere formation (Extended Data Fig. 4f, g). Conversely, knocking down the already low level of LHPP expression in SNU449 cells enhanced cell proliferation (Extended Data Fig. 5a). Next, we investigated the effect of LHPP expression on tumour formation in L-dKO mice. Hepatocyte-specific, adeno-associated virus (AAV) expressing either red fluorescent protein (RFP) (AAV/DJ-ALBp-RFP, hereafter referred to as AAV-control) or RFP and murine LHPP (AAV/DJ-ALBp-RFP-2A-mLhpp, hereafter referred to as AAV-LHPP) was injected into the tail vein of 8-week-old L-dKO and littermate control mice (Fig. 3e). Tumour burden was assessed at 20 weeks of age. Hepatic overexpression of LHPP in AAV-LHPP-infected L-dKO mice (20 weeks) was confirmed by immunoblotting (Extended Data Fig. 5b). Macroscopic and histological examination of livers revealed few and small (micronodular) tumours in AAV-LHPP-infected L-dKO mice (Fig. 3e–g). The few ‘escaper’ tumours that arose in AAV-LHPP-infected L-dKO mice displayed reduced LHPP expression, again suggesting that LHPP downregulation is essential for tumorigenesis. AAV-control-infected L-dKO livers exhibited severe HCC with multiple large neoplastic (macronodular) tumours. Serum levels of alanine amino transferase (ALT), aspartate amino transferase (AST) and lactate dehydrogenase (LDH), well characterized indicators of liver damage, were also reduced in AAV-LHPP-infected L-dKO mice compared to AAV-control-infected mice (Extended Data Fig. 5c). No detrimental effect was observed in control mice injected with either AAV-control or AAV-LHPP (Extended Data Fig. 5c). Thus, the expression of LHPP prevents tumour formation and maintains liver function in L-dKO mice, suggesting that the protein histidine phosphatase LHPP is a tumour suppressor.
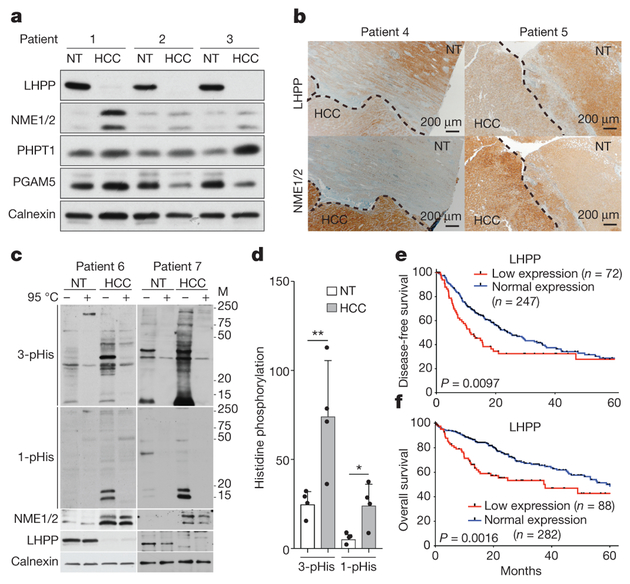
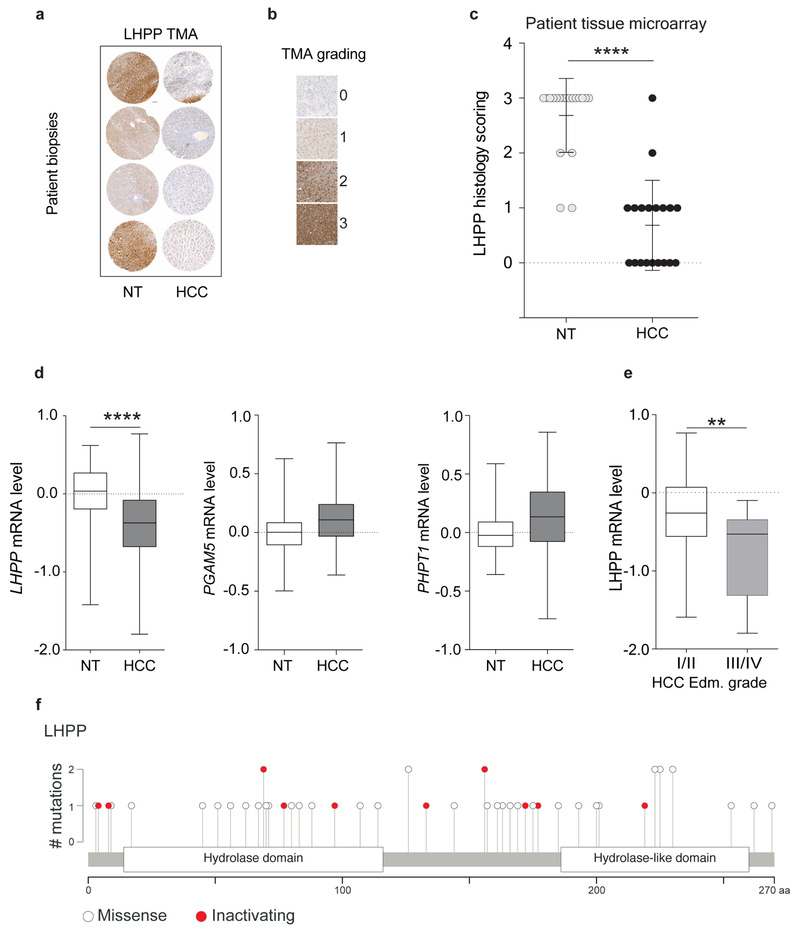
We next examined NME1, NME2 and LHPP expression in human liver tumours, from patients with HCC. Resected HCC tissue and adjacent non-tumour tissue were assessed for LHPP, NME1 and NME2 levels by immunoblot analysis. LHPP was low in the tumour (HCC) compared to non-tumour tissue in several patients (Fig. 4a), as observed in L-dKO mice. NME1 and NME2 expression was high in the tumours. We note that human NME1 and NME2, unlike their mouse counterparts, do not co-migrate (Fig. 4a). In agreement with the immunoblot analysis, immunohistochemistry also showed that LHPP expression was low and NME1 and NME2 expression was high in HCC compared to adjacent non-tumour tissue from two additional patients (Fig. 4b). Furthermore, using a tissue microarray assay, we evaluated LHPP expression in tumour and matched non-tumour liver samples from 20 patients with HCC. Tumour sections that had greater than 70% tumour cells and non-tumour sections that contained greater than 70% normal cells were chosen for the array (see Methods). LHPP expression was significantly downregulated in HCC compared to matched non-tumour tissue (Extended Data Fig. 6a–c). Similarly, on the basis of a published mRNA expression dataset from 37 patients with HCC21, LHPP mRNA levels (but not PHPT1 or PGAM5 mRNA levels) were significantly reduced in tumour tissue compared to matched non-tumour tissue (Extended Data Fig. 6d). We note that, in L-dKO mice, reduced LHPP protein expression also correlated with reduced Lhpp mRNA levels (Extended Data Fig. 1f). Finally, consistent with increased levels of NME1 and NME2 and decreased levels of LHPP (Fig. 4c), we observed significantly higher 1-pHis and 3-pHis signals in human HCC compared to matched non-tumour tissue (Fig. 4c, d). Collectively, the above findings suggest that histidine phosphorylation is upregulatedin both mouse and human tumours.
Figure 4 |. Loss of LHPP expression in human HCC correlates with reduced patient survival.
a, b, Immunoblot (n = 3 patients) (a) and immunohistochemistry (b) (n = 2 patients) analyses showing reduced LHPP and increased NME1 and NME2 expression in liver tissue from patients with HCC compared to adjacent non-tumour liver tissue in a total of n = 7 HCC patients (n = 2 other patients in panel c). For full scans, see Supplementary Fig. 12. c, Immunoblot analysis showing increased 3-pHis and 1-pHis signals in human HCC compared to adjacent non-tumour tissue. Similar results were observed in tumours from two additional HCC patients (total n = 4 patients). For full scans, see Supplementary Fig. 13. Tissue lysates in 2 × sample buffer (pH 8.8) heated at 95 °C for 10 min to dephosphorylate pHis served as control. The monoclonal antibodiesused to detect 3-pHis and 1-pHis are SC44–1 and SC1–1, respectively. d, Quantification of immunoblot (from c and two other western blots, n = 4 in total) indicates that 3-pHis and 1-pHis signals are significantly higher in human HCC compared to adjacent non-tumoral tissue (band intensities in each lane are normalized to the intensity of corresponding calnexin protein levels after subtracting the intensities from corresponding heated lane). The y axis represents band intensities. *P = 0.0368, **P = 0.0064, two-sided ratio paired t-test. Data are mean ± s.d. e, f, Kaplan-Meier curves, log-rank test, showing the correlation between LHPP expression and clinical outcome, as analysed for disease-free survival (e) and overall survival (f).
We next investigated clinical correlates of LHPP expression. HCC is scored based on Edmondson grades I to IV in which grade IV is the most severe or aggressive. Examination of a previously published mRNA dataset21 revealed that low LHPP expression is associated with more severe tumours (Edmondson grades III/IV versus I/II) (Extended Data Fig. 6e). These results, combined with our earlier observation that LHPP expression in L-dKO mice reduced tumorigenicity (Fig. 3e–g), suggest that low LHPP expression could correlate with reduced patient survival. We referred to The Cancer Genome Atlas (TCGA) to determine whether there is a correlation between LHPP expression and overall survival or disease-free survival (see Methods). LHPP downregulation was associated with faster disease progression and worse overall survival in 319 and 370 patients with HCC, respectively (Fig. 4e, f). Median disease-free survival and overall survival of patients with HCC decreased from 25 to 12 months and from 61 to 38 months, respectively, after the loss of LHPP expression. Thus, decreased LHPP expression in human HCC is associated with increased histidine phosphorylation and reduced survival.
We identified 49 LHPP mutations in datasets from TCGA and the International Cancer Genome Consortium (ICGC). Twelve (24.5%) of the mutations, found in oesophagus (2), head and neck (2), stomach (2), bladder (1), breast (1), skin (1), liver (1), lung (1), and pancreas (1), are predicted to be inactivating mutations (Extended Data Fig. 6f), that is, nonsense, frameshift or splice site mutations (Supplementary Table 5). The >20% prevalence of inactivating mutations suggests that LHPP is a tumour suppressor gene22. This suggestion is strengthened by two genome-wide association studies (GWAS) indicating that the LHPP locus is associated with oral cancer, pharyngeal cancer and acute lymphoblastic leukaemia23,24. Whole-genome sequencing previously identified the LHPP locus as a risk factor for major depressive disorder25,26. These observations suggest that loss of LHPP and thus increased histidine phosphorylation is oncogenic and possibly also involved in other disorders. Importantly, the results collectively suggest that LHPP is a histidine phosphatase and tumour suppressor.
How might histidine phosphorylation be related to cancer? The histidine-phosphorylated proteins we detected preferentially in tumours include the crucial DNA replication factors MCM3, MCM6 and MCM7, the p53 inhibitor AROS, the cytidine deaminase ABEC3 that converts cytosine to uracil and may induce mutations, the receptor-mediated endocytosis and NOTCH signalling protein AAK1, the mTORC1 effector RSK227, and CHTOP. The potential LHPP targets we detected that might be important for tumorigenesis include the essential fatty acid biosynthetic enzyme ACLY6, the proto-oncoprotein and RNA polymerase-associated protein LEO128, a variant histone H2A commonly expressed in HCC29, and the inflammation-related and HCC-promoting HMGB1 protein30 (Supplementary Table 4).
METHODS
Animals.
Liver-specific Tsc1 and Pten double-knockout mice were obtained by crossing Tsc1lox/lox mice (exons 17 and 18)31 with Ptenloxllox mice (exons 4 and 5)32 to transgenic mice expressing Cre recombinase under the control of the hepatocyte-specific albumin promoter (Alb-CreTg/0)33, to generate liver-specific double-knockout (Tsc1lox/loxPten lox/lox Alb-CreTg/0, in short L-dKO) mice. Age- and sex-matched littermate Tsc1Iox/loxPtenlox/lox mice without the cre genes were used as controls in all animal experiments. No other randomization protocol or blinding was applied beyond this criterion. The mice produced were on mixed genetic background (C57BL/6J, 129/SvJae, BALB/cJ). Mice were housed under temperature and humidity-controlled conditions, in a 12-h light/dark cycle with lights switched on between 06:00 to 18:00. All experiments were conducted on male mice (age between 6 and 20 weeks). In all experiments, mice were fasted overnight before euthanasia by CO2 inhalation. All relevant ethical regulations were followed in this study. The tumour harvest time limit was set at 20 weeks. In addition, the clinical conditions of mice were monitored according to the score sheet as stipulated by the Institutional Animal Care and Use Committee (IACUC), Kantonales Veterinaeramt of Kanton Basel-Stadt. None of the guidelines was exceeded in any experiment performed. For all animal experiments, respecting the animal ethical rules, a minimum number of animals required to get a statistically meaningful result were used. All animal experiments were performed in accordance with the federal ethical guidelines and were approved by the Kantonales Veterinaeramt of Kanton Basel-Stadt.
Mass spectrometry.
Sample preparation for proteome analysis of tumours.
Tumours from 20-week-old L-dKO mice and whole liver from littermate control mice were carefully dissected under magnifying lens and snap-frozen in liquid nitrogen. Frozen tissue was pulverized in a metal plate cooled on dry ice and was transferred into a cooled microcentrifuge tube. The powdered tissue was resuspended in 8 M urea (Applichem, A1086) (containing 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 1 mM PMSF, 1 × Complete Mini Protease Inhibitors (Roche),1 × PhosSTOP (Roche)) and homogenized using a Polytron (PT 10–35 GT) at 500g for 2 min. The sample was placed on ice during this process. Lysates were then subjected to rotation at 4 °C with regular vortexing. Lysates were then centrifuged at 10,000g for 10 min at 4 °C to remove cell debris. Protein concentration in the supernatant was determined using Bradford assay. Equal amounts (5 mg each) of ‘pooled control’ and tumour tissue were reduced with 2.5 mM dithiothreitol (DTT) (Sigma) for 45 min at 56 °C, cooled for 5–10 min at room temperature and alkylated with 7.5 mM iodoacetamide (IAA) (Sigma) for 45 min at room temperature (reaction protected from light). The urea concentration was reduced to 4 M by diluting with 25 mM Tris-HCl, pH 8.0. Proteins were treated with endoproteinase LysC (1:100 (w/w), Wako) at 37 °C for 2 h. The urea concentration of the LysC digest was reduced to 1 M. Later, trypsin (Worthington) digestion (1:100; w/w) was performed for 2 h at 37 °C, followed by a second round of trypsin digestion (1:100; w/w) overnight at 37 °C. Digestion was stopped by adding 0.4% trifluoroacetic acid (TFA) (Applied Biosystems) (v/v) that lowered the pH of the solution to below pH 2.0. Later, the digest was centrifuged at 10,000g for 10 min at room temperature and the supernatant was desalted on a C18 reverse-phase SepPak-200mg column (Waters). First, the cartridge was primed with 2 ml 100% acetonitrile (ACN) (Biosolve), followed by 4 ml of 50% acetonitrile containing 0.5% acetic acid (Merck) and equilibrated with 4 ml 0.1% TFA. Peptides were loaded twice in 0.4% TFA and the cartridge was washed with 10 ml of 0.1% TFA. Later, the cartridge was washed with 0.5% acetic acid and the peptides were eluted with 4 ml of 80% ACN, 0.5% acetic acid. The peptides were dried in a SpeedVac and subjected to strong cation exchange chromatography (SCX). SCX fractionation was performed according to34 with a few modifications detailed below. Storage buffer was removed from a HiTrap SP cartridge (GE Healthcare, 17115101) and washed twice with 2 ml of SCX buffer A (5 mM KH2PO4 (pH 2.65), 30% can). Later, the cartridge was equilibrated twice with 2 ml of SCX buffer B (5 mM KH2PO4 (pH 2.65), 30% ACN containing 500 mM KCl), and the cartridge was re-equilibrated with 2 ml of SCX buffer A. The peptides were resuspended in 1 ml of SCX buffer A, centrifuged at 10,000g for 2 min to remove any unwanted debris. The peptides were then loaded with a syringe onto the HiTrap SP cartridge. The flow-through was then collected. The bound peptides were then desorbed in a stepwise manner: with 1 ml each of SCX buffer A containing 50 mM, 100 mM, 150 mM, 250 mM and 350 mM KCl. Each fraction was collected and the peptide concentration was estimated at 280 nm. The fractions were dried in a SpeedVac and peptides were then desalted on Microspin cartridges (Vydac). 10 μg of each fraction was removed for liquid chromatography–tandem mass spectrometry (LC-MS/MS proteome analysis).
LC–MS/MS analysis.
Dried peptides were dissolved in 20 μl of 0.1% acetic acid, 0.005% TFA. The peptides were analysed by capillary LC–MS/MS using a homemade separating column (0.075 mm × 18 cm) packed with Reprosil C18 reverse-phase material (2.4 μm particle size). The column was connected on line to an Orbitrap FT hybrid instrument (Thermo Scientific). Solvent A (0.1% acetic acid in water, 0.005% TFA) and solvent B (0.1% acetic acid, 0.005% TFA and 80% acetonitrile in water) were used for peptide separation. 2 μl of peptide digest were injected with a Proxeon Easy-LC capillary pump (Thermo Scientific) set to 0.3 μl min−1. A gradient from 0 to 40% solvent B in solvent A in 190 min was delivered with the nano pump at a flow rate of 300 nl min−1. The percentage of solvent B was increased to 75% in 10 min at the end of the gradient. Later, the eluting peptides were ionized at 2.5 kV and mass spectrometer was operated in data-dependent mode. The precursor scan was done in the Orbitrap set to a 60,000 resolution, while the fragment ions were mass analysed in the LTQ instrument. A top twenty method was used for fragmentation.
Protein identification.
The LC–MS/MS data were searched with Proteome Discoverer 1.4 (Thermo Scientific) set to Mascot and Sequest HT search engines with 10 ppm precursor ion tolerance. The fragment ions were set to 0.6 Da tolerance. Medium confidence (5% FDR) was used for peptide search. Swiss-Prot KB database set to M. musculus was used for Mascot. For Sequest HT, an inhouse databank was constructed by extracting M. musculus entries from UniProtKB.
Quantification.
The raw files from the LC–MS/MS were first passed through Progenesis QI software (Nonlinear Dynamics, Waters) and the matched SCX fractions between ‘pooled control’ and tumours were aligned. A minimum of 75% alignment was set for analysis. During alignment, the search identifications from Proteome Discoverer (version 1.4) were imported into Progenesis for matching the aligned features with the identified peptides.
Statistical analysis.
R-based Perseus program, version 1.4.0.2, was used for statistical data analysis. An ANOVA-based two-sample t-test was performed, adjusting S0 to 1, the number of randomizations to 250 (default), and the FDR to 5% for the proteome35,36. *P < 0.05, **P < 0.01, ***P < 0.001, ****P< 0.0001.
p-His antibody immunoprecipitation of proteins for mass spectrometry.
Tissue and cell lysis for pHis immunoprecipitation.
Tumours from 20-week-old L-dKO mice and whole liver from littermate control mice were carefully dissected under magnifying lens and snap-frozen in liquid nitrogen. Frozen tissue was pulverized in a metal plate cooled on dry ice and was transferred into a cooled microcentrifuge tube. The powdered tissue was resuspended in 25 mM Tris-HCl (pH 8.5); 100 mM NaCl, pH: 8.5; 1 mM PMSF, 1 × Complete Mini Protease Inhibitors (Roche), 1 × PhosSTOP (Roche) and homogenized using a Polytron (PT 10–35 GT) at 500g for 2 min. Precautions were taken to preserve pHis (pH 8.5 and constant 4 °C) and reagents (such as detergents) that can interfere with mass spectrometry. The sample was placed on ice during this process. Lysates were then incubated at 4 °C on an end-over-end rotator for 2 h with regular vortexing. Lysates were then centrifuged at 10,000g for 10 min at 4 °C to remove cell debris. Protein concentration in the supernatant was determined using BCA assay. Tissue lysate from four different littermate control animals was pooled to obtain a pooled control. For immunoprecipitation of pHis proteins from CB1 cells with and without LHPP, 2 mg for each condition was used.
Bead preparation and preclearing of the lysate.
One millilitre of protein A Sepharose beads (Thermo, 101042) was mixed with 1 ml of lysis buffer (25 mM Tris-HCl (pH 8.5); 100 mM NaCl, pH: 8.5; 1 mM PMSF, 1 × Complete Mini Protease Inhibitors (Roche), 1× PhosSTOP (Roche)) and washed twice (by spinning at 2,000g, 4 min, 4 °C) and resuspended in 2 ml of lysis buffer. For pre-clearing, the protein lysate (10 mg for tissues, 2 mg for cells) was incubated with 200 μl of prepared beads and incubated at 4 °C on an end-over-end rotator for 2 h. Later, the lysate was spun (2,000g, 4 min, 4 °C) and the precleared lysate (supernatant) was divided into two different tubes (5 mg each for tissues, 1 mg each for cells).
Antibody-lysate incubation.
For tissue lysates, both 1-pHis and 3-pHis (clones SC1–1 and SC44–1) antibodies were used. For cell lysates, only 3-pHis antibody was used. Lysates were incubated with pHis antibody (30 μg of 3-pHis plus 10 μg of 1-pHis for tissue lysate; 30 μg of 3-pHis for cell lysate) or IgG antibody (40 μg for tissue lysate and 30 μg for cell lysate). Lysates were incubated at 4 °C on an end-over-end rotator for 2 h. Later, 200 μl of beads in lysis buffer were added and incubation continued at 4 °C on an end-over-end rotator for 12 h.
Washing the beads and elution of the proteins.
The column was washed four times with 2 ml of lysis buffer (pre-cooled) at 4 °C. After the third wash, the fourth wash was performed in a new microcentrifuge tube to avoid any contamination from proteins attached to the wall of the microcentrifuge tube. Proteins were eluted in two fractions (E1 and E2) with 2 × 600 μl 100 mM triethylamine (TEA), pH 11. The fractions were dried in a SpeedVac and resuspended in 50 mM Tris-HCl (pH 7.5).
Sample preparation.
The eluate in 50 mM Tris-HCl was treated with endoproteinase LysC (1:100 (w/w), Wako) at 37 °C for 2 h. Later, trypsin (Sigma) digestion (1:100; w/w) was performed for 2 h at 37 °C (pH 8.0), followed by a second round of trypsin digestion (1:100; w/w) overnight at 37 °C. Digestion was stopped by adding 0.4% TFA (Applied Biosystems) (v/v) that lowered the pH of the solution to below pH 2.0. Later, the digest was centrifuged at 10,000g for 10 min at room temperature and the supernatant was desalted on Microspin cartridges (Vydac).
First, the cartridge was activated with 100 μl methanol, followed by 100 μl 80% ACN, 0.5% acetic acid and the column was equilibrated with 2 × 100 μl 0.1% TFA, 1% ACN. Peptides were loaded twice in 0.4% TFA and the cartridge was washed five times with 100 μl 0.1% TFA, 1% ACN. Later, the peptides were eluted with 2 × 100 μl 80% ACN, 0.5% acetic acid. The peptides were dried in a SpeedVac and subjected to LC-MS/MS proteome analysis. For further details, please see the ‘LC–MS/MS analysis’ section above.
Protein identification.
The LC–MS/MS data were searched with MaxQuant37, version 1.5.3.8 and searched against the mouse Swiss-Prot database 2017-05-09. A mass tolerance of 20 p.p.m. was allowed for the first search, and a mass tolerance of 10 p.p.m. was allowed for the main search. Two missed cleavages were allowed. Peptide FDR, protein FDR, and site decoy fraction were set to 2%. For protein quantification, a ratio count of 1 was applied. The fragment ions were set to 0.6 Da tolerance. Medium confidence (5% FDR) was used for peptide search.
Quantification.
The protein groups datasets were exported into a FileMaker Pro-12 databank.
Identification of proteins with histidine phosphorylation in tumours.
Samples were run in triplicates. Label-free quantification (LFQ) intensities were obtained for each protein from MaxQuant. A mean of LFQ intensities was calculated and used for all further calculations. Proteins without any ratio owing to no detection in either of the samples were also included in the analysis. Mass spectrometry analysis identified 3,265 proteins in the tumour immunoprecipitate from a total of 4 different L-dKO tumours. A total of 1,508 proteins were at least twofold-enriched in the pHis immunoprecipitate compared to the control IgG immunoprecipitate in more than 1 tumour. Out of these, 1,173 proteins were detected and enriched (minimum twofold) in 2 out of 2 tumours (100%), or 3 out of 3 tumours (100%), or 3 out of 4 tumours (75%, 100%) (Supplementary Table 2).
Identification of proteins with histidine phosphorylation in control tissue.
Mass spectrometry analysis identified 3,011 proteins in the control tissue immunoprecipitate from a total of 4 different control animals. A total of 1,141 proteins were at least twofold-enriched in the pHis immunoprecipitate compared to the control IgG immunoprecipitate in more than 1 tumour. Out of these, 823 proteins were detected and enriched (minimum twofold) in 2 out of 2 control tissues (100%), or 3 out of 3 control tissues (100%), or 3 out of 4 control tissues (75%, 100%) (Supplementary Table 1).
Identification of proteins with increased histidine phosphorylation in tumours.
Out of the proteins detected in tumour (1,173) and control (823) tissue, 236 proteins were detected and enriched (minimum twofold) in 2 out of 2 experiments (100%), or 3 out of 3 experiments (100%), or 3 out of 4 experiments (75%, 100%) (Supplementary Table 3). A total of 24 proteins that are exclusively detected in tumour tissue and hence had no ratio are indicated in the list shown in Supplementary Table 3.
Identification of proteins with decreased histidine phosphorylation in CB1 cells upon LHPP re-expression.
Mass spectrometry analysis identified 295 proteins in the CB1 immunoprecipitate from a total of 5 different experiments. Nine proteins were at least 1.7-fold enriched in the CB1 cells lacking LHPP compared to the control IgG immunoprecipitate and LHPP re-expressing cells in minimum 50% of the experiments (Supplementary Table 4).
Antibodies.
PTEN (CST, 9188), TSC1 (Bethyl, A300–316A), phospho-S6 (CST, 4856), total S6 (CST, 2217), pAKT S473 (CST, 9271), total AKT (CST, 9272), Calnexin (Enzo, ADI-SPA-860), NME1 (CST, 3345), NME2 (Abcam, ab60602), and LHPP (Novus, NBP1–83273) were obtained commercially. Phospho-histidine monoclonal antibodies against N1 and N3 positions were as described previously2. The monoclonal antibodies used to detect 3-pHis and 1-pHis were SC44–1 and SC1–1, respectively. Horseradish peroxidase (HRP)-coupled anti-mouse (115–035774) and anti-rabbit (211-032-171) secondary antibodies were purchased from Jackson laboratories.
Immunoblot analysis.
For regular immunoblot analysis, the liver tissue was homogenized in T-PER (ThermoFisher scientific, 78510) supplemented with 1 mM PMSF, 1× Complete Mini Protease Inhibitors (Roche), 1× PhosSTOP (Roche) using a Polytron (PT 10–35 GT) at 500g for 2 min. Equal amounts of homogenate were SDS-PAGE fractionated and transferred onto a nitrocellulose membrane that was incubated, after blocking (5% BSA in TBST), with appropriate antibodies. For phosphohistidine analysis, tissues (equal wet weights) and equal number of cells were lysed directly in 2 × pHis sample buffer (5 × pHis sample buffer: 250 mM Tris-HCl pH 8.8, 0.02% bromophenol blue, 50% glycerol, 50 mM EDTA, 500 mM DTT and 10% SDS), incubated on ice and sonicated (15 s × 3 times, Hielscher VialTweeter) to disrupt cells and shear DNA. Lysates were clarified by centrifugation (10,000g for 15 min at 4 °C). Fresh lysates were resolved immediately using polyacrylamide gels at 4 °C with a modified stacking gel, pH 8.8 and 10% resolving gel, pH 8.8. All electrophoresis and protein transfer was strictly carried out at 4 °C. In addition, the running and transfer buffers were precooled before use. The nitrocellulose membranes were blocked in casein blocking buffer (0.1% casein, 10% PBS, pH 8.8). Primary antibodies were resuspended in casein blocking buffer with 0.1% Tween-20, incubated overnight at 4°C. The monoclonal antibodies used to detect 3-pHis and 1-pHis were SC44–1 and SC1–1, respectively at a dilution of 0.5 μg ml−1. The membranes were washed three times (15 min per wash at 4 °C) with TBST (modification, pH 8.8) and incubated with anti-rabbit HRP conjugated secondary antibody (1:10,000) at 4 °C. Lysates in sample buffer (pH 8.8) heated at 95 °C for 10 min to dephosphorylate pHis served as a control. The bands in the heated lane are either heat-resistant pHis bands or a non-specific signal. For quantification of the pHis immunoblots, band intensities in each lane are normalized to intensity of corresponding calnexin protein levels after subtracting the intensities from corresponding heated lane; the y-axis represents band intensities.
Immunoblot analysis on Lhpp adenovirus-infected cells.
A 50–60% confluent dish was first infected, 24 h after infection, the cells (CB1 or SNU449) were trypsinized and 2 × 105 cells were seeded into a 6-well adherent tissue culture dish and 24 h later lysed in pHis compatible buffer (2 × pHis sample buffer, for buffer composition, see the ‘Immunoblot analysis’ section above).
Crystal violet staining.
On Lhpp-adenovirus infected cells.
Twelve hours after infection, CB1 (2 × 104 per well) and SNU449 (4 × 104 per well) cells were seeded into a 6-well plate and stained with crystal violet (2% crystal violet in 20% methanol) on day 7 (n = 4). The plate was left to dry at room temperature. Absorbance was measured at 560 nm on a Tecan Infinite M1000 machine. The software used to control was Tecan i-control, version 1.11.1.0, according to the manufacturer’s instructions. Reference (or blank) was set with an empty 6-well plate. Multiple reads were read per well (15 × 15 reads) and a border of 3,000 μm was left in each well. Number of flashes was set to 25 with 10 ms of settle time.
On LHPP-siRNA-transfected cells.
LHPP siRNA (human LHPP ON-TARGET plus set of 4, pooled, catalogue number LQ-018950-02-0005; siRNA 1 CAACCCAAACUGUGUGGUA, siRNA 2 CAUGAAGGCGCUUGAGUAU, siRNA 3 GCAGCACGCCGACAAGUGA, siRNA 4 CUGAAGCGUUCCCG GCUGA) and negative control siRNA (ON-TARGET plus non-targeting pool) were purchased from Dharmacon. SNU449 cells were transfected in the presence of the transfection reagent jetPRIME (PolyPlus transfection) as per the manufacturer’s instructions. Cells were then re-suspended in complete DMEM plus 10% FBS medium. Then 36 h after transfection, SNU449 cells were re-transfected with LHPP siRNA and were seeded in a 6-well plate (4 × 104 per well). The cells were stained with crystal violet (2% crystal violet in 20% methanol) on day 3.
Cell culture.
CB1 cells were established from 20-week-old L-dKO tumours. Multiple L-dKO tumours were chopped finely into small pieces and washed in warm 20 ml PBS. The processed tumour tissues were seeded in a 6-well tissue culture dish in DMEM with 10% FBS. Six different clones, CB1-CB6 were selected by limited trypsinization and amplified in DMEM with 10% FBS. Clone CB1 (transformed in vivo) was used for all future assays. SNU449 cells were obtained from ATCC and grown in DMEM with 10% FBS. We did not authenticate the cell line in our laboratory. The CB1 cell line was authenticated as hepatocytes by probing for the markers TSC1 and PTEN that were knocked out only in hepatocytes. CB1 and SNU449 cell lines were tested and found to be free of mycoplasma contamination.
In vitro phosphatase assay.
CB1 cells were lysed in 2 × sample buffer (pH 8.8) and electrophoresed in two sets using a 10% modified SDS gel (see the ‘Immunoblot analysis’ section above). The proteins were transferred onto a nitrocellulose membrane. After transfer, the membrane was cut into two halves. One half of the membrane was treated with 0.5 μg of recombinant LHPP (Abcam, ab116175) in 500 μl Tris-MgCl2 buffer (20 mM Tris-HCl, pH 8.0, and 5 mM MgCl2). The other half of the membrane was treated with 1 ml Tris-MgCl2 buffer. The assay was carried out at room temperature. After 4 h of treatment with LHPP, the blots were washed in casein blocking buffer (pH 8.8) and immunoblotted with 3-pHis antibody. The monoclonal antibody used to detect 3-pHis was SC44–1.
INK128 treatment and AAV administration.
For acute INK128 administration, 20-week-old L-dKO (n = 8) and control animals (n = 8) were injected with INK128 (1 mg kg−1 of body weight) or with the vehicle (intraperitonial injection). INK128 was dissolved in 5% 1-methyl-2-pyrrolidionone, 15% polyvinylpyrrolidone K30, and 80% water. INK128 injection was performed twice (06:00 and 18:00) over 24 h. For AAV administration, 8-week-old control and L-dKO mice were infected with AAV/DJ-ALBp-RFP (AAV-control) or AAV/DJ-ALBp-RFP-2A-mLhpp (AAV–LHPP) (5 × 1011 genome copies per mouse) via tail vein injection.
Hepatosphere assays.
CB1 cells (2 × 105 cells per well) were seeded in 6-well ultra-low attachment plates (Corning, 3471, Costar 6 Well Plate with lid) in serum-free DMEM/F12 with recombinant murine epidermal growth factor (20 ng ml−1; R&D Systems), murine hepatocyte growth factor (Sigma-Aldrich), and 1 × B27 serum-free supplement (Invitrogen). Cells were cultured at 37 °C, in 5% CO2. On the fifth day, hepatospheres measuring greater than 100 μm were counted for the assay.
Histopathology and immunohistochemistry.
After euthanization of mice, the livers were fixed in 4% paraformaldehyde, dehydrated and embedded into paraffin wax blocks. Embedded-tissues were cut into 4-μm-thick sections placed on SuperFrost slides (Thermo Scientific) and stained with H&E (Merck). Imunohistochemistry was performed using the antibodies NME1 (CST, 3345, dilution 1:100), NME2 (Abcam, ab60602, 1:100), and LHPP (Novus, NBP1–83273, 1:200).
HCC tissue specimens and tissue microarrays.
Formalin-fixed and paraffin-embedded tumour specimens, as well as fresh frozen tissue were obtained from the Institute of Pathology, University of Basel, Switzerland. For tissue microarray construction, a representative tumour area was selected on an H&E-stained slide of the donor block. Afterwards, a core punch with a diameter of 0.6 mm was taken from the tumour, and in selected cases from the non-tumoral liver tissue. Core punches were transferred to a new paraffin recipient block by using a programmed tissue arrayer (Beecher Instruments). The tissue microarray contained 30 HCC samples and 30 non-tumour liver control samples. Microarrays were cut in sections of 4 μm thickness and stained with a polyclonal LHPP antibody diluted at 1:200 using an automatic Benchmark XT staining machine (Ventana Medical Systems Inc.). LHPP staining intensity was evaluated by a clinical pathologist (M.S.M,) and graded semiquantitatively into: 0 for negative staining, 1 for weak positive staining, 2 for moderate positive staining and 3 for strong positive staining (Extended Data Fig. 6b). Ten pairs of samples in which one of the tissue sections was washed away during staining were ignored during evaluation. While scoring for staining intensity, the pathologist was double blinded to the antigen that was probed and the sample identity.
Re-analysis of transcriptomic profiling data.
LHPP expression was evaluated in 59 HCC needle biopsies, 37 of which were matched with their corresponding non-neoplastic liver parenchyma (cirrhotic tissues), using published transcriptomic data (GSE64041)21. CEL files were normalized using the Qlucore Omics Explorer software (Qlucore AB). LHPP expression was extracted for each sample. RNA sequencing gene expression data including outcomes from 370 hepatocellular carcinomas were retrieved from The Cancer Genome Atlas dataset (TCGA, Provisional) using the cbioportal (http://www.cbioportal.org, accessed 4 November 2016) website. Downregulation of LHPP was defined as z-score ≤ −1.
LHPP mutation analysis.
We interrogated the cBioPortal38,39 and ICGC data portal (https://dcc.icgc.org) for deletions at the LHPP locus in samples across 33 cancer types. We identified 32 cases harbouring deletion at the LHPP locus. Furthermore, we identified 49 mutations across the datasets. We removed duplicate TCGA samples that appear in both data portals and variants with >1% population frequencies in the Exome Aggregation Consortium data set, ExAC40. 12 out of 49 (24.5%) of the mutations are inactivating mutations (Supplementary Table 5).
Serum analysis.
Liver damage enzymes ALT, AST and LDH were measured using a biochemical analyser (Cobas c 111 analyser, Roche).
Patient material and ethics.
All relevant ethical regulations were followed in this study. All the experiments with human tissue samples reported in this study were approved by the ethics commission of Northwestern Switzerland (EKNZ, approval No. 361/12). An informed consent was obtained from the human subjects.
Data availability.
All data generated during this study are included in this paper and its figures, Extended Data and Supplementary Information. For gel source data, see Supplementary Figs 1–13. In addition, publicly available data were analysed in this study and are cited wherever relevant.
Extended Data
Extended Data Figure 1 |. Proteomics on L-dKO tumours.
a, Liver weight (LW) to body weight (BW) ratio (percentage) of 20-week-old L-dKO mice (n = 9) and age-matched control mice (n = 9). ****P < 0.0001, two-sided unpaired t-test. b, mRNA expression analysis in 20-week-old L-dKO tumours (n = 3) compared to livers from age-matched control mice (n = 3). CD90 (also known as Thy1; *P = 0.0104), CD133 (also known as Prom1; *P = 0.0227), Cd44 (*P = 0.0473) and Aldh1 (also known as Aldh1a1; *P = 0.0107), two-sided unpaired t-test. Data are mean ± s.d. c, Diagram showing the scheme used for mass spectrometry-based proteomic analysis. Hepatic proteomes from control mice (n = 6) were combined and used as a pooled control against tumours (n = 12) from four different L-dKO mice. d, Graph showing number of proteins upregulated (red), downregulated (blue) and not regulated (grey) in 12 tumours from c. ANOVA-based two-sample t-test with a FDR of 2%, S0 = 1 was used to determine regulation (n = 12). e, List of kinases and phosphatases (including putative phosphatases) regulated in a minimum of 10 out of 12 tumours. f, Lhpp mRNA expression analysis in 20-week-old L-dKO tumours compared to livers from age-matched control mice (n = 4). P = 0.0022, two-sided unpaired t-test. Data are mean ± s.d.
Extended Data Figure 2 |. LHPP protein is conserved in eukaryotes.
a, LHPP protein is highly conserved in eukaryotes, from worm to human. b, Human LHPP protein shows weak homology to known human histidine phosphatases PHPT1 and PGAM5.
Extended Data Figure 3 |. Loss of LHPP expression is confined to L-dKO tumours.
a, Quantification of immunoblot (from Fig. 2a) indicates reduced LHPP (***P = 0.0004) and increased NME1 (*P = 0.0254) and NME2 (*P = 0.0105) expression in tumours compared to age-matched control littermates (band intensities in each lane are normalized to intensity of corresponding calnexin protein levels), control mice (n = 4), L-dKO tumours (n = 8). P values are from a two-sided unpaired t-test. Data are mean ± s.d. b, Immunoblot analysis ofliver tumour (n = 6)and non-tumour (n = 3) tissue from three independent 20-week-old L-dKO mice shows tumour-specific loss of LHPP expression. Ponceau S image of the whole blot is also included. For full scan, see Supplementary Fig. 3. c, Immunohistochemistry analyses showing increased NME1 expression in L-dKO liver compared to liver from control mice. Although L-dKO tumours displayed reduced expression of LHPP, hepatic LHPP expression in the non-tumour region was comparable to control mice. Parallel tissue sections that were processed similarly (without primary antibody) served as antibody control. Similar results were obtained in livers from 4 independent control mice and L-dKO mice. d, Immunoblot analysis of liver from control mice and L-dKO liver tumours treated with INK128 (1 mg kg−1 body weight; two intraperitonial injections in 24 h; n = 4 mice per condition, lysates from two mice are pooled into one lane). LHPP expression remains unchanged, but NME1 and NME2 expressionis reduced considerably in L-dKO mice after INK128 treatment. For full scans, see Supplementary Fig. 4.
Extended Data Figure 4 |. LHPP is a protein histidine phosphatase.
a, Diagram of NME1 and NME2 autophosphorylation on His118. The high-energy phosphate is later transferred from the histidine kinase dimer pair to the histidine on the substrate. b, CB1 cell lysate was electrophoresed and transferred onto a nitrocellulose membrane. Two sets of the membrane were treated with recombinant full-length LHPP (for 4 h) in TMD buffer (see Methods) and the other set of the membrane was treated with plain TMD buffer (pH 8.5). Immunoblotting analysis of both the membranes with 3-pHis antibody (clone SC44–1) indicates that LHPP is a protein pHis phosphatase. For full scans, see Supplementary Fig. 6.c, Immunoblotting shows no apparent change pSer/Thr or pTyr levels in CB1 cells infected with adenovirus containing LHPP compared to cells infected with RFP-only control. Similar result was observed in two independent experiments. For full scans, see Supplementary Fig. 9. d, e, Immunoblotting (d) and quantification (e) show a reduction in 3-pHis levels in SNU449 cells infected with adenovirus containing LHPP compared to control-infected cells. Similar results were observed in four independent biological experiments. For full scans, see Supplementary Fig. 8. A 50–60% confluent dish was first infected, 24 h after infection, the cells were trypsinized and 2 × 105 cells were seeded into a 6-well adherent tissue culture dish and 24 h later lysed in pHis compatible buffer (see Methods). The monoclonal antibodies used to detect 3-pHis and 1-pHis were SC44–1 and SC1–1, respectively (n = 4). **P = 0.0019, two-sided ratio paired t-test. Data are mean ± s.d. f, Colony-forming assay in CB1 cells (n = 3 biologically independent experiments) and SNU449 (n = 5 biologically independent experiments) cells shows reduced proliferation upon LHPP over expression. Twelve hours after infection, CB1 (2 × 104 per well) and SNU449 (4 × 104 per well) cells were seeded into a 6-well plate tissue culture plate and stained with crystal violet (2% crystal violet in 20% methanol) on day 7. **P = 0.0026, ****P < 0.0001, two-sided unpaired t-test. Data are mean ± s.d. See Methods for further details. g, Photomicrographs of LHPP-overexpressing CB1 cells (left) displayed reduced ability to form hepatospheres in ultralow attachment plates; n = 3 biologically independent experiments.**P = 0.0024, two-sided unpaired t-test. Data are mean ± s.d. (see Methods).
Extended Data Figure 5 |. LHPP is a tumour suppressor.
a, Left and middle, colony-forming assay in SNU449 cells shows increased proliferation upon LHPP knockdown using pooled short interfering RNA (siRNA). Then 36 h after infection, SNU449 cells were re-transfected with LHPP siRNA and 4 × 104 cells per well were seeded into a 6-well tissue culture plate and allowed to proliferate for another 3 days. At the end of the third day, cells were stained with crystal violet (2% crystal violet in 20% methanol), and similar results were obtained from five biologically independent experiments. **P = 0.0075, two-sided unpaired t-test. Data are mean ± s.d. Right, immunoblot analysis (n = 1) revealed a reduction in LHPP protein levels after siRNA transfection. Longer exposure ofthe LHPP blot relative to the blot in Extended Data Fig. 4d. In addition to immunobloting, a reduction in LHPP transcript levels after siRNA transfection was also confirmed in two independent experiments (data not shown). For full scans, see Supplementary Fig. 10. b, Immunoblot analysis (bottom) and quantification (top) of non-tumour and tumour liver lysates confirmed LHPP overexpression in 20-week-old AAV-LHPP-infected mice (as in Fig. 3e). For full scans, see Supplementary Fig. 11.c, Serum extracted from mice in (Fig. 3e) and analysed for ALT, AST and LDH levels shows a reduction in liver damage upon AAV-LHPP infection, unit per litre (U l−1) (control mice infected with AAV-control and AAV-LHPP virus (n = 4), L-dKO mice infected with AAV-control (n = 5) and AAV-LHPP virus (n = 8)). Data are mean ±s.d. P values are by one-way ANOVA with Tukey’s multiple-comparison test. ALT (adjusted ***P = 0.004, ****P< 0.0001), AST (adjusted **P = 0.013 for control mice versus L-dKO mice transfected with AAV-control; adjusted **P = 0.0075 for L-dKO mice transfected with AAV-control versus L-dKO mice transfected with AAV-LHPP), LDH (adjusted *P = 0.0196, **P = 0.0037).
Extended Data Figure 6 |. LHPP is a tumour suppressor in human HCC.
a, Representative immunohistochemistry images from the LHPP-stained tissue microarray. Scale bar, 100 μm. b, Representative images and staining intensities of LHPP staining in the tissue microarray. c, Tissue microarray (TMA) indicates that LHPP is significantly downregulated in human HCC compared to adjacent non-tumour tissue (n = 20 HCC patients, ****P < 0.0001, two-sided paired t-test). d, Box plots showing a significant reduction in LHPP expression in HCC tissue compared to adjacent non-tumour tissue (n = 59). ****P < 0.0001, two-sided Mann-Whitney U test. Non-tumour (minimum, −1.422, median, 0.03329, maximum, 0.6179, lower 95% confidence interval (CI) of mean −0.0931, upper 95% CI of mean 0.0931) and HCC (minimum, −1.797, median, −0.3729, maximum, 0.7683, lower 95% CI of mean −0.5328, upper 95% CI of mean −0.2506). Expression of PHPT1 and PGAM5 was not reduced in HCC. e, Box plots showing a strong reduction in LHPP expression in Edmondson grade III/IV tumours (aggressive tumours, n = 17) compared to Edmondson grade I/II tumours (less aggressive tumours, n = 42).**P = 0.0036, two-sided Mann-Whitney U test. Edmondson grade I/II (minimum, −1.592, median, −0.2594, maximum 0.7683, lower 95% CI of mean −0.3987, upper 95% CI of mean −0.0986) and III/IV (minimum, −1.797, median, −0.5281, maximum, −0.09671, lower 95% CI of mean −1.018, upper 95% CI of mean −0.4717). f, Lollipop plot showing the distribution of mutations in LHPP across pan-cancer datasets in the TCGA and ICGC portal. Solid white circles indicate missense mutations, red circles indicate inactivating mutations (including nonsense mutations, splice site mutations and frameshift deletions; detailed list of mutations is provided in Supplementary Table 5).
Supplementary Material
Acknowledgements
M.N.H. acknowledges the Louis Jeantet Foundation, the Swiss National Science Foundation, SystemsX.ch, and the European Research Council (MERiC). T.H. acknowledges USPHS grants CA080100, CA082683 and CA194584 from the NCI. T.H. is an American Cancer Society Professor, and holds the Renato Dulbecco Chair in Cancer Research. S.P acknowledges the Swiss National Science Foundation (Ambizione grant number PZ00P3_168165).
Footnotes
Supplementary Information is available in the online version of the paper.
Online Content Methods, along with any additional Extended Data display items and Source Data, are available in the online version of the paper; references unique to these sections appear only in the online paper.
The authors declare no competing financial interests. Readers are welcome to comment on the online version of the paper.
Publisher's Disclaimer: Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kee J-M & Muir TW Chasing phosphohistidine, an elusive sibling in the phosphoamino acid family. ACS Chem. Biol 7, 44–51 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fuhs SR et al. Monoclonal 1- and 3-phosphohistidine antibodies: new tools to study histidine phosphorylation. Cell 162, 198–210 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Llovet JM et al. Hepatocellular carcinoma. Nat. Rev. Dis. Primers 2, 16018–16023 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Schulze K et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat. Genet 47, 505–511 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kenerson HL et al. Akt and mTORC1 have different roles during liver tumorigenesis in mice. Gastroenterology 144, 1055–1065 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guri Y et al. mTORC2 promotes tumorigenesis via lipid synthesis. Cancer Cell 32, 807–823 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Yokoi F, Hiraishi H & Izuhara K Molecular cloning of a cDNA for the human phospholysine phosphohistidine inorganic pyrophosphate phosphatase. J. Biochem 133, 607–614 (2003). [DOI] [PubMed] [Google Scholar]
- 8.Hiraishi H, Yokoi F & Kumon A 3-phosphohistidine and 6-phospholysine are substrates of a 56-kDa inorganic pyrophosphatase from bovine liver. Arch. Biochem. Biophys 349, 381–387 (1998). [DOI] [PubMed] [Google Scholar]
- 9.Hiraishi H, Ohmagari T, Otsuka Y, Yokoi F & Kumon A Purification and characterization of hepatic inorganic pyrophosphatase hydrolyzing imidodiphosphate. Arch. Biochem. Biophys 341, 153–159 (1997). [DOI] [PubMed] [Google Scholar]
- 10.Ek P et al. Identification and characterization of a mammalian 14-kDa phosphohistidine phosphatase. Eur. J. Biochem 269, 5016–5023 (2002). [DOI] [PubMed] [Google Scholar]
- 11.Klumpp S et al. Protein histidine phosphatase: a novel enzyme with potency for neuronal signaling. J. Cereb. Blood Flow Metab 22, 1420–1424 (2002). [DOI] [PubMed] [Google Scholar]
- 12.Panda S et al. Identification of PGAM5 as a mammalian protein histidine phosphatase that plays a central role to negatively regulate CD4+ T Cells. Mol. Cell 63, 457–469 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cai X, Srivastava S, Surindran S, Li Z & Skolnik EY Regulation of the epithelial Ca2+ channel TRPV5 by reversible histidine phosphorylation mediated by NDPK-B and PHPT1. Mol. Biol. Cell 25, 1244–1250 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hartsough MT et al. Nm23-H1 metastasis suppressor phosphorylation of kinase suppressor of Ras via a histidine protein kinase pathway. J. Biol. Chem 277, 32389–32399 (2002). [DOI] [PubMed] [Google Scholar]
- 15.Wagner PD & Vu ND Phosphorylation of ATP-citrate lyase by nucleoside diphosphate kinase. J. Biol. Chem 270, 21758–21764 (1995). [DOI] [PubMed] [Google Scholar]
- 16.Fuhs SR & Hunter T pHisphorylation: the emergence of histidine phosphorylation as a reversible regulatory modification. Curr. Opin. Cell Biol 45, 8–16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kee J-M, Villani B, Carpenter LR & Muir TW Development of stable phosphohistidine analogues. J. Am. Chem. Soc 132, 14327–14329 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Riley NM & Coon JJ Phosphoproteomics in the age of rapid and deep proteome profiling. Anal. Chem 88, 74–94 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Attwood PV, Piggott MJ, Zu XL & Besant PG Focus on phosphohistidine. Amino Acids 32, 145–156 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Besant PG & Attwood PV Detection and analysis of protein histidine phosphorylation. Mol. Cell. Biochem 329, 93–106 (2009). [DOI] [PubMed] [Google Scholar]
- 21.Makowska Z et al. Gene expression analysis of biopsy samples reveals critical limitations of transcriptome-based molecular classifications of hepatocellular carcinoma. J. Pathol. Clin. Res 2, 80–92 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vogelstein B et al. Cancer genome landscapes. Science 339, 1546–1558 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vijayakrishnan J et al. A genome-wide association study identifies risk loci for childhood acute lymphoblastic leukemia at 10q26.13 and 12q23.1. Leukemia 31, 573–579 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lesseur C et al. Genome-wide association analyses identify new susceptibility loci for oral cavity and pharyngeal cancer. Nat. Genet 48, 1544–1550 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neff CD et al. Evidence for HTR1A and LHPP as interacting genetic risk factors in major depression. Mol. Psychiatry 14, 621–630 (2009). [DOI] [PubMed] [Google Scholar]
- 26.CONVERGE consortium. Sparse whole-genome sequencing identifies two loci for major depressive disorder. Nature 523, 588–591 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park I-H, Bachmann R, Shirazi H & Chen J Regulation of ribosomal S6 kinase 2 by mammalian target of rapamycin. J. Biol. Chem 277, 31423–31429 (2002). [DOI] [PubMed] [Google Scholar]
- 28.Chong PSY et al. LEO1 is regulated by PRL-3 and mediates its oncogenic properties in acute myelogenous leukemia. Cancer Res. 74, 3043–3053 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Lo Re, O. et al. Induction of cancer cell stemness by depletion of macrohistone H2A1 in hepatocellular carcinoma. Hepatology 67, 10.1002/hep.29519 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Chen M et al. High-Mobility Group Box 1 promotes hepatocellular carcinoma progression through miR-21-mediated matrix metalloproteinase activity. Cancer Res. 75, 1645–1656 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kwiatkowski DJ et al. A mouse model of TSC1 reveals sex-dependent lethality from liver hemangiomas, and up-regulation of p70S6 kinase activity in Tsc1 null cells. Hum. Mol. Genet 11, 525–534 (2002). [DOI] [PubMed] [Google Scholar]
- 32.Horie Y et al. Hepatocyte-specific Pten deficiency results in steatohepatitis and hepatocellular carcinomas. J. Clin. Invest 113, 1774–1783 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Postic C & Magnuson MA DNA excision in liver by an albumin-Cre transgene occurs progressively with age. Genesis 26, 149–150 (2000). [DOI] [PubMed] [Google Scholar]
- 34.Dephoure N & Gygi SP A solid phase extraction-based platform for rapid phosphoproteomic analysis. Methods 54, 379–386 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tyanova S et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods 13, 731–740 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Tusher VG, Tibshirani R & Chu G Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl Acad. Sci. USA 98, 5116–5121 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cox J & Mann M MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 26, 1367–1372 (2008). [DOI] [PubMed] [Google Scholar]
- 38.Gao J et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sc. Signal 6, pl1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cerami E et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lek M et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated during this study are included in this paper and its figures, Extended Data and Supplementary Information. For gel source data, see Supplementary Figs 1–13. In addition, publicly available data were analysed in this study and are cited wherever relevant.