Abstract
Genome-wide-association studies (GWASs), epigenetic, gene-expression and gene–gene interaction projects, nutritional genomics and investigations of the gut microbiota have increased our knowledge of the pathophysiology of eating disorders (EDs). However, compared with anorexia nervosa, genetic studies in patients with bulimia nervosa and binge-eating disorder are relatively scarce, with the exception of a few formal genetic and small-sized candidate–gene-association studies. In this article, we review important findings derived from formal and molecular genetics in order to outline a genetics-based pathophysiological model of EDs. This model takes into account environmental and nutritional factors, genetic factors related to the microbiome, the metabolic and endocrine system, the immune system, and the brain, in addition to phenotypical traits of EDs. Shortcomings and advantages of genetic research in EDs are discussed against the historical background, but also in light of potential future treatment options for patients with EDs.
Keywords: anorexia nervosa, binge-eating disorder, bulimia nervosa, genetics, GWAS, microbiome
Introduction
Historical reflection on psychiatric genetics
Ancestry plays a decisive role in early historic attempts to create a sense of purpose and identity. This approach is found in several religions, including prehistoric cults, Buddhism, Islam, Judaism and Christianity.1 In the book of Genesis, for example, which was reportedly written in the 5th and 4th centuries BC, the descendants of Abraham are recorded.2 One way to interpret such genealogies may be to value them as early evidence of the idea that ancestry comes with the inheritance of traits, characteristics and competences that indicate individuals for certain assignments.
However, the first significant step towards scientific genetics was made by Gregor Johann Mendel (1822–1884), who gained posthumous recognition as the founder of formal genetics for conducting and publishing the results of his ‘experiments in plant hybridization’, from which he demonstrated dominant and recessive inheritance.3 In psychiatry, formal genetics became an important research focus in the early 20th century4,5 leading to compulsory sterilization of the mentally ill to prevent the occurrence of mental disorders.6 In this context, it is important to mention Ernst Rüdin (1874–1952), a member of the German Nazi Party, who has been credited as the originator of modern psychiatric genetics.7 He promoted the prevention of assumed hereditary mental illnesses by the prohibition of marriage or sterilization,8 despite his knowledge that major mental disorders, including affective disorders, do not follow a simple pattern of Mendelian inheritance.9
The next major step in scientific genetics following the discovery of statistical patterns of inheritance was to elucidate the molecular basis of heredity. Deoxyribonucleic acid (DNA), the molecule that carries genetic information, was first isolated in 1869 by the Swiss physician Friedrich Miescher (1844–1895) and termed ‘nuclein’.10 In 1953, Watson and Crick suggested the double-helix structure of this molecule.11 Further work by Crick and coworkers showed that the genetic code was based on nonoverlapping triplets of bases, allowing Khorana and colleagues to decipher this genetic code.12 These developments laid the foundations for genetic association studies, which have been used to identify candidate genes or genome regions that were hypothesized as contributing to a specific disease.13 In the 1980s, early molecular genetic studies in psychiatry focused on candidate genes that were considered to be involved in neurotransmitter metabolism, neurotransmitter receptors and neuropeptides.14 However, the candidate–gene approach is currently regarded as obsolete and problematic in the field of psychiatry due to its insufficient statistical power and difficulties with replication.
Around the turn of the millennium, researchers began to investigate the entire genome in a noncandidate-driven approach by performing genome-wide association studies (GWASs). The first successful GWAS was published by Klein and colleagues in 2005 on age-related macular degeneration.15 Since then, ~4000 human GWASs have examined ~2000 diseases and traits, and thousands of single-nucleotide polymorphism (SNP) associations have been identified.16 As GWASs require a large number of study participants, study centres need to forge consortia in order to achieve those numbers. As a result, the Psychiatric Genomics Consortium (PGC) has become the largest consortium in the history of psychiatry. The PGC is dedicated to elucidating the fundamental biology of psychiatric disorders, informing clinical practice, and delivering novel therapeutic targets on the basis of genomic research16 with GWASs currently being the gold standard and the workhorse of psychiatric genetics.
In 1986, Hegstrand and Hine published a groundbreaking study wherein they demonstrated significant differences in hypothalamic histamine levels between germ-free and conventionally raised animals.17 Thus, they proved that microbes influence brain chemistry18 and specifically, histaminergic neurotransmission, which is particularly important for appetite and weight regulation.17,19 From the 1980s until now, multiple important synergistic activities between humans and the microbes living in and on them have been identified;20–23 as a result, following completion of the human genome sequence, researchers felt that the achievement was incomplete, as it neglected the genetic material of the microbiota. Therefore, they called for the Human Microbiome Project as a ‘second human genome project’ that ‘would entail a comprehensive inventory of microbial genes and genomes at the four major sites of microbial colonization in the human body: mouth, gut, vagina, and skin’.21 As there is substantial evidence on the significance of the ecological community of microorganisms that share our body space,20–23 this review on genetic risk factors for eating disorders (EDs) will also take into account the microbiome, which comprises the genetic material within our microbiota.
The latest genetic research in the field of EDs does not stop at examining the triplets of bases of DNA sequences in human cells and other organisms within the human body and contributing to its function, it also takes considers gene function, gene expression, gene–gene interaction and the interaction between genes and food.
Eating disorders
EDs are characterized by a persistent disrupted eating behaviour, which leads to changes in dietary intake, impaired physical health and psychosocial problems. The Diagnostic and Statistical Manual of Mental Disorders (DSM-5) currently recognises three primary EDs: anorexia nervosa (AN), bulimia nervosa (BN), and binge-eating disorder (BED).24 Additionally, the DSM-5 mentions avoidant/restrictive food-intake disorder (ARFID; an eating disturbance manifested by a restrictive eating pattern with persistent failure to meet appropriate nutritional needs), pica (the consumption of nonfood) and rumination disorder (regurgitation and rechewing food).24,25 However, as we are not aware of any genetic research on ARFID, pica or rumination disorder, the current review will not focus on these conditions.
The main criteria for the diagnosis of AN are a significantly low body weight in the context of age, sex, and physical health, intense fear of weight gain and disturbed body perception.24,25 The dietary deficit is often accompanied by significant physical health issues, including growth retardation, osteopenia, amenorrhoea and renal insufficiency, in addition to changes in laboratory parameters, cardiac arrhythmia and disturbances of thyroid function.26 The main criteria for the diagnosis of BN are recurrent binges (the consumption of an unusually large amount of food within a short time interval associated with loss of control) associated with compensatory behaviours, including vomiting, excessive physical activity or fasting at least once a week for three months, and excessive preoccupation with shape and weight.24,25 BED is characterized by the intermittent consumption of large quantities of food, with a sense of loss of control (binge eating), at least once a week for three months without the use of the extreme compensatory strategies, and the experience of marked distress regarding the binge eating; three or more of the following symptoms have to be present: eating more rapidly than normal, eating until feeling uncomfortably full, eating large amounts of food when not feeling physically hungry, eating alone because of being embarrassed by how much one is eating, and feeling disgusted with oneself, depressed, or very guilty after overeating.24
Given that transitions between diagnostic categories are common, some theorists have proposed dimensional models of EDs or, in other words, a spectrum of disordered eating. This view is supported by the fact that there are many overlapping risk factors and symptoms25,27,28 and by estimates of the genetic correlation between AN and BN, based on data derived from the Swedish Twin Study of Adults, which show that common comorbidity of AN and BN can in part be accounted for by shared genetic and environmental influences.29 Therefore, this review will not only include specific EDs, but will also include the main pathophysiological factors that may apply for the spectrum of EDs.
Aim of this review
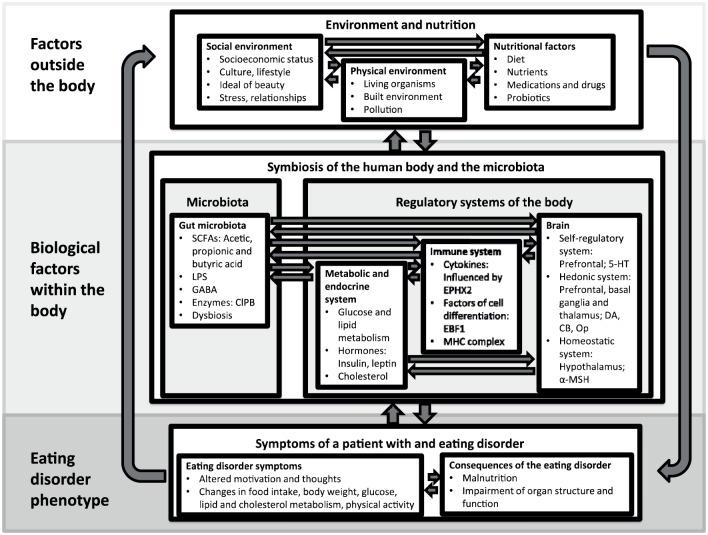
This review summarises the results of genetic research in order to develop a hypothetical pathophysiological model of EDs (Figure 1). This hypothetical model will outline the wider picture of biological factors contributing to the development of EDs. As explained above, we have included microbiome studies in this review, as the importance of the microbiome for psychiatric disorders, and specifically for EDs, is increasingly acknowledged by researchers,30–34 and the majority of microbiota research uses genetic methods. We do not aim to provide a detailed and comprehensive summary of genetic and microbiome research and their methods, as detailed summaries have already been written in previous years on genetic35–40 and gut microbiota34 research in EDs.
Figure 1.
Schematic and simplified depiction of a pathophysiological model of eating disorders based on genetic findings.
SCFAs, short chain fatty acids; LPS, lipopolysaccharides; GABA, gamma-aminobutyric acid; B ClpB, caseinolytic protease; EBF1, early B-cell factor 1; EPHX2, epoxide hydrolase 2; MHC, major histocompatibility complex; 5-HT, serotonin; DA, dopamine; CB, cannabinoid; Op, opioids; α-MSH, melanocyte-stimulating hormone.
For further details see section ‘A pathophysiological model of eating disorders based on genetic findings’.
Formal genetic studies
Family studies
The familial nature of AN has been well established; the first-degree relatives of individuals with AN have an approximately 10-fold greater lifetime risk of falling ill with an ED than relatives of unaffected individuals.41–43 However, family studies are unable to determine the extent to which the observed familial aggregation is due to genetic or environmental factors.
Twin studies
To determine the genetic risk, twin studies have been performed, which have yielded a high heritability of 50–60%; for example, a large twin study performed by Bulik and colleagues led to a heritability estimation for AN of 56%, with the remaining variance attributable to both shared and unique environment factors.44 Holland and colleagues showed concordance rates for AN of 0.71 for monozygotic twins and 0.1 for dizygotic twins.45
Twin studies also revealed that genetic effects and environmental factors contribute to the liability to BN.36 BN aggregates in families of affected patients. The relative risks for BN have been reported as 4.2 and 4.4 for first-degree female relatives of patients with AN and BN, respectively.42 Twin studies of BED have reported heritability estimates between 41% and 57%, depending on the criteria for BED.46–48
Adoption studies
To the best of our knowledge, adoption studies are lacking in EDs. However, in studies investigating disordered eating symptoms, an adoption study with 123 adopted and 56 biological female sibling pairs was published by Klump and colleagues.49 Eating pathology, body dissatisfaction, weight preoccupation and binge eating were assessed with the Minnesota Eating Behaviors Survey.50 Using biometric model fitting, the authors calculated significant genetic influences between 59% and 82% on all forms of disordered eating. Nonshared environmental factors accounted for the remaining variance. These data demonstrate the significant genetic effect on disordered eating.49
Molecular genetic studies
Linkage studies
Linkage studies aim to identify genomic regions that have an increased likelihood of containing genes that are associated with a disorder or trait. Genetic linkage means the tendency of DNA sequences to be inherited together, if they are located close together in the same region on a chromosome. A linkage analysis is conducted on samples of related individuals, including parent–offspring trios, affected sibling pairs or dense pedigrees. Linkage analyses in EDs have detected signals on chromosomes 1, 2, 3, 4 and 13 for AN and on chromosome 10 for BN.39,51–54 Fine mapping of one of these regions on chromosome 1 identified the serotonin 1D (HTR1D) and the opioid delta 1 receptor (OPRD1) genes.52 However, without subsequent fine mapping, a key limitation of linkage analysis is its inability to narrow down regions to an experimentally feasible number of genes.40
Candidate–gene association studies
Candidate–gene association studies are hypothesis-driven evaluations of specific variants in a particular gene, most often selected on the basis of a biological hypothesis, as a candidate gene is a prespecified gene of interest that may be associated with a phenotype or disease state. Candidate–gene association studies have been most successful for diseases in which the relevance of the selected gene to the biology of the disorder was already well established.55 For example, candidate–gene studies have identified common and rare alleles in genes already known to influence serum lipid phenotypes, including levels of low-density lipoprotein and triglycerides.56 For psychiatric disorders in which the underlying biology remains fully elucidated, candidate–gene studies involve more speculation regarding the relevance of the selected gene and thus have a smaller prior probability of detecting a true association.55
Candidate–gene studies in anorexia nervosa
A systematic search for human genetic candidate–gene associations studies in AN found associations of AN with (a) genes related to neurotransmitters, including serotonin, norepinephrine and glutamate, (b) genes related to hunger regulatory systems, including leptin, agouti-related peptide (AGRP), melanocyte-stimulating hormone (α-MSH), the melanocortin-4 receptor (MC4R), neuropeptide Y (NPY) and ghrelin, (c) genes related to feeding motivation- and reward-related systems, including opioids, cannabinoids and dopamine, (d) genes related to systems regulating energy metabolism, (e) to neuroendocrine systems, and (f) to the immune system and inflammatory response.57 Overall, the studies summarized in this review examine only one or a few markers and have small samples. The majority of these results have not been replicated in larger samples, and these associations do not emerge in GWASs. Therefore, no far-reaching conclusions should be drawn on the basis of these results.
Candidate–gene studies in bulimia nervosa
In BN, candidate–gene association studies revealed nonsignificant or conflicting results for the serotonin transporter, dopamine receptor D4, catechol-O-methyltransferase, leptin receptor, MC4R, AGRP, proopiomelanocortin (POMC), oestrogen receptor 1, and the brain-derived neurotrophic factor (BDNF) (for review see40). Positive results were obtained in genetic studies investigating polymorphisms in ghrelin,58 oestrogen receptor ER2,59 cannabinoid receptor CR1,60 and the fat mass and obesity-associated gene.61,62 However, none of these positive results have been independently replicated in larger samples.
Candidate–gene studies in binge-eating disorder
In BED, candidate–gene association studies revealed nonsignificant or conflicting results for ankyrin repeat and kinase domain containing 1, ghrelin, MC4R, and BDNF genes (for review, see the work by Yilmaz et al.40). Positive results were obtained in genetic studies investigating polymorphisms in the serotonin transporter,60 dopamine receptor D263 and μ1-opioid receptor.64 Similar to the positive results derived from candidate–gene studies in BN, none of these results have been replicated.
Genome-wide association studies
GWASs have evolved over the last 10 years into a useful tool for investigating the genetic architecture of human disease and can be described as the current workhorse of psychiatric genetics.65 A GWAS is an observational study of a genome-wide set of genetic variants in different individuals to determine whether any variant is associated with a trait. Like candidate–gene associations studies, GWASs compare cases and controls, but they investigate up to 1,000,000 genetic markers in hundreds of thousands of study participants using a hypothesis-free approach. As with the majority of psychiatric disorders, GWASs suggest that AN is polygenic in nature, with several genetic variants of small effect being involved.66–68
In a GWAS comprising 12 cohorts (3495 AN cases and 10,982 controls), only one genome-wide significant locus on chromosome 12 in a region harbouring a previously reported type 1 diabetes and autoimmune disorder locus was identified.68 Multiple genes in the region around this top SNP rs4622308 reached gene-based significance. The top locus in this region and the fourth top locus in the SNP-based analyses of this study was FAM19A2, a putative chemokine or cytokine.68 Candidate genes from previous studies did not reach gene-based significance in this study (for further details see the work by Duncan et al.68).
Li and colleagues published the results of a GWAS of 692 female AN cases and 3570 female matched controls. They conducted an analysis of phenotypic variability which led to the identification of a specific genetic risk factor that approached genome-wide significance. This SNP, rs929626, is located in the sixth intron of the early b-cell factor 1 (EBF1) gene which influences leptin signalling. Thus, the authors concluded that their finding suggests that a variant-mediated dysregulation of leptin signalling may be involved in the pathophysiology of AN.69 However, EBF1 is also important in the immune system for B-cell maturation.70,71
The latest GWAS in AN based on 2158 cases from nine populations of European origin and 15,485 ancestrally matched controls was published by Huckins and colleagues and focused on common, low-frequency and rare genome-wide variation in AN.72 Although no results reached genome-wide significance, two notable common variants were identified. The first was an intronic variant associated with the OPCML gene that codes for an opioid-binding protein, which is localized in the plasma membrane and may have an accessory role in opioid receptor function. The second was an intergenic variant associated with the ANKRD50 gene, which is a protein-coding gene involved in the endosome-to-plasma membrane trafficking and recycling of cargo proteins, including glucose transporter 1 (GLUT1).72,73
GWASs in EDs have been used to calculate genetic correlations between different disorders, or between disorders and physical traits. Significant positive genetic correlations were observed between AN and schizophrenia, obsessive–compulsive disorder (OCD), neuroticism, educational attainment, and high-density lipoprotein cholesterol. Significant negative genetic correlations were obtained between AN and body mass index (BMI), insulin sensitivity, glucose metabolism, and lipid phenotypes.68,74,75
Genetic studies using state-of-the-art technologies in BN or BED are limited and, to the best of our knowledge, no large GWASs have been performed in BN and BED. However, Wade and colleagues conducted a GWAS in 2564 female twins.76 They did not establish a diagnosis for BN or BED, although participants completed a health and lifestyle self-report questionnaire which included symptoms of disordered eating of the AN spectrum, BN spectrum and substance-induced purging. However, no association between symptoms of disordered eating and genetic variants reached genome-wide significance in this study. The authors concluded that larger samples and meta-analyses are required to identify genes and pathways contributing to predisposition to EDs.76
Although GWASs are considered an appropriate tool to identify genetic variants of genome-wide significance for the investigated disorder, the effect of certain genetic variants contributing to the overall heritability are too small to be detected by GWASs. This phenomenon is referred to as missing heritability, which should be taken into account while interpreting GWAS data.77,78 Statisticians are currently developing solutions on how to combine such weak but possibly important signals to increase the power of GWASs.77–81
Epigenetics
Epigenetics examines changes in gene function that do not involve changes in the DNA sequence.82 Epigenetic mechanisms include methylation of the DNA, histone modification, and the regulation of gene expression at the transcriptional and post-transcriptional level by noncoding ribonucleic acids (RNAs), including micro-RNAs (miRNAs), short-interfering RNAs (siRNAs), and piwi-interacting RNAs (piRNAs).83
Global methylation studies
The majority of epigenetic studies in EDs have measured the global DNA methylation level. Methylation can change the activity of a DNA segment without changing the sequence. When located in a gene promoter, DNA methylation typically acts to repress gene transcription. However, to measure global methylation means quantifying the 5-methylcytosine content across the whole DNA. This approach does not look at specific DNA segments or genes. Frieling and colleagues identified global DNA hypomethylation in patients with AN compared with controls.84 In this study, patients with AN showed a trend towards a lower global methylation compared with patients with BN, and no difference was found between patients with BN and controls. A reduction of global DNA methylation was also found in a trial performed by Tremolizzo and colleagues in adolescents with AN.85 However, no evidence for altered global DNA methylation in association with AN was found by Saffrey and colleagues,86 whereas increased global methylation in AN was reported by Booij and colleagues.87
Epigenome-wide association studies
Epigenome-wide association studies allow for the determination of global DNA methylation and also to regional changes in DNA methylation throughout the genome. These studies can lead to epigenetic results relating to a specific gene. For example, the abovementioned study by Booij and colleagues87 measured not only global methylation, but also methylation of specific genes. In women with AN, they identified hypermethylated regions representing genes associated with histone acetylation, RNA modification, cholesterol storage and lipid transport, and dopamine and glutamate signalling. Of note, they specifically detected hypermethylated sites at the Tenascin XB (TNXB) gene.87 This result was confirmed by Kesselmeier and colleagues in a high-throughput DNA methylation analysis of blood from 47 females with AN, 47 lean females without AN and 100 controls.88 TNXB encodes an extracellular matrix glycoprotein with antiadhesive effects.89 Absence of this protein in humans has been associated with the Ehlers-Danlos syndrome, a connective tissue disorder.90 The gene is located on chromosome 6 within the major histocompatibility complex (MHC) class III region.91
Although the results of global methylation studies and epigenome-wide association studies are promising, it is important to take into account that the sample sizes of the reported studies are small and that epigenetic changes are tissue specific. Additionally, epigenetic changes are associated with diurnal and seasonal factors, medication, exercise, diseases, diet, psychological and socioeconomic factors.92 It may be the case that these factors account for the contradictory results obtained in the abovementioned global methylation studies.84–87
Gene expression studies
Gene expression is the process by which genetic instructions, as they are coded on the DNA, are used to synthesize gene products such as messenger RNA (mRNA), functional RNA or proteins. To our knowledge, most of the gene expression studies in EDs measured mRNA expression.
Messenger ribonucleic acid expression studies
Changes in the mRNA expression have been reported in atrial natriuretic peptide (ANP),93 cannabinoid 1 receptor (CB1),94 POMC,95 and dopamine transporter (DAT) and D2 receptor (DRD2)96 genes in people with AN. Additionally, the expression of leptin receptor-coding gene was shown to differ significantly between binge–purge AN subtype and restricting subtype patients, and between AN restricting subtype patients and control participants.97 Kahl and colleagues observed an increase in mRNA expression of tumour necrosis factor (TNF)-α and interleukin (IL)-6 in patients with AN at hospital admission compared with controls. The mRNA expression of TNF-α remained significantly higher, whereas that of IL-6 decreased in weight-restored patients.38,98
Gene–gene interaction studies
Genes do not function alone but exert effects on disease traits together with other genes (gene–gene interactions) and through interactions with environmental factors. Mercander and colleagues performed a family-based association study with 151 TagSNPs covering 10 neurotrophin signalling genes in 371 EDs trios of Spanish, French and German origin. They found that the nerve growth factor (NGFB) gene modified the AN risk conferred by certain neurotrophic receptor tyrosine kinase 3 (NTRK3)-risk genotypes by a synergistic gene–gene interaction.99
A study by Urwin and Nunn provided evidence for an important interaction between the monoamine oxidase A (MAOA) and serotonin transporter (SERT) genes.100 They assessed 114 patients with AN and their biological parents, and found that a SERT gene genotype with no apparent individual effect was preferentially transmitted to children with AN when the more active MAOA gene variant was also transmitted. In this study, the increased risk of developing the disorder was up to eight times greater than the risk imposed by the MAOA gene variant alone.100
Cui and colleagues combined linkage analyses and methods to examine gene–gene interactions.101 DNA was collected from two large families affected by EDs, and mutations segregating with illness were identified using a combination of microarray linkage and whole-genome sequencing. In the first family, linkage analysis of 20 members across three generations identified a rare missense mutation in the oestrogen-related receptor-α (ESRRA) gene that segregated with illness. In the second family, the analysis of eight members across four generations identified a missense mutation in the histone deacetylase 4 (HDAC4) gene that segregated with illness. The authors followed up on these findings with a transcriptional analysis. The HDAC4 transcript was found to repress the expression of ESRRA in vitro and in in vivo animal experiments.101
Nutritional genomics
The science of nutritional genomics includes nutrigenetics and nutrigenomics. Nutrigenetic studies examine the effect of genetic variation on diet, and the response to certain nutrients. Nutrigenomic studies aim to assess the influence of nutrients on the expression of genes.38,102,103
Nutrigenetic studies
Nutrigenetic research in EDs remains in its infancy. However, the first studies have revealed promising results. For example, Scott-Van Zeeland and colleagues reported gene variants within the epoxide hydrolase 2 (EPHX2) gene were associated with susceptibility to AN,104 and data from the same study group showed that, on eating a meal, an increase of pro-inflammatory molecules was observed in AN patients, but not in healthy controls, depending on EPHX2 enzyme activity and the consumed polyunsaturated fatty acids (PUFAs).105 An EPHX2-dependent inflammatory response following a meal in those with AN, but not in healthy controls, suggests that the genetically determined way our body reacts to certain nutrients may contribute to the development of an ED.
Nutrigenomic studies
The general aim of nutrigenomics is to identify the effects of nutrients, including macronutrients and micronutrients, on the genome.103 However, nutrigenomic studies, including studies in other genetic subdisciplines, require large sample sizes to identify those interactions and to reveal fundamental biological insights.38,16 Although nutrigenomic studies may offer a promising approach to elucidate the effects of diet on health, to the best our knowledge, no such nutrigenomic studies have been performed in the field of EDs in order to determine the influence of dietary ingredients on the genome.
Genetics of the microbiome
In microbiome studies, stool samples are usually collected from study participants, and the bacterial DNA is extracted and determined using commercially available kits.
Gut microbiota has been demonstrated as involved in different metabolic functions, including the regulation of weight gain, energy harvest from the diet and insulin secretion, and is heavily influenced by diet and lifestyle.33,106
The microbiota are reported to produce an array of bioactive metabolic products capable of entering the systemic circulation. These metabolic products can have profound effects on host metabolism, immune function, and gene expression in several organ systems, including the central nervous system (CNS).107 Short chain fatty acids (SCFAs) are volatile fatty acids produced by bacteria in the bowel. Acetic acid, propionic acid, and butyric acid are the most abundant.108 Butyrate is rapidly used as an energy source for colonocytes, whereas the majority of acetate and propionate enter the portal circulation.109 Butyrate is an HDAC inhibitor with potential effects on gene expression in human cells.110 Propionate crosses the blood–brain barrier, enters the CNS and affects various physiological processes, including cell signalling, neurotransmitter synthesis and release, free-radical production and mitochondrial function. Propionate is a precursor for cholesterol synthesis regulation and gluconeogenesis in the liver.111 Acetate is the main SCFA in the blood and has a key metabolic role in peripheral tissues where it acts as a substrate for lipogenesis.31,112 Therefore, microbiota are capable of modulating the brain and the metabolic system of the body.
Enteroendocrine cells express specific receptors for bacterial products, and these cells modify the secretion of hormones that regulate hunger and satiety according to the obtained receptor signals.113 Additionally, bacterial products, including lipopolysaccharides (LPS) modulate the function of the blood–brain barrier and increase its permeability, thus increasing the effect of circulating cytokines to appetite regulation.114 Furthermore, the host produces antibodies against microbial peptides, which can act as autoantibodies against appetite-regulating hormones including α-MSH.115–118 Therefore, it can be concluded that the microbiota can influence major processes within the immune system.
However, literature in this area remains limited, with only a small number of studies measuring gut bacterial profiles in patients with EDs.112,118–121 These studies identified novel bacterial species in patients with AN admitted to hospital,112 profound microbial perturbations in patients with AN compared with controls, and disturbed SCFA profiles in patients with AN.30–33
In addition to SCFAs, there are other molecules produced by human microbiota that may influence the brain, in addition to appetite and weight regulation. For example, caseinolytic protease B (ClpB) produced by enterobacteria has been identified as a mimetic of α-MSH. In a study by Breton and colleagues, plasma concentrations of ClpB were measured in female patients with AN, BN, and BED, and were compared with healthy participants. ClpB concentrations were elevated in patients with ED, without significant differences in patient subgroups. Plasma ClpB concentrations correlated with ED psychopathology and α-MSH. These results suggest a link between bacterial ClpB and the ED pathophysiology.121
In this context, it is important to refer to a study that examined the use of antimicrobial medication in a large patient cohort treated for BED, BN, and AN over the 5-year period preceding ED treatment. A total of 1592 patients with an ED were compared with 6368 matched controls from the general population. The study population was linked to the prescription data of antibacterial, antifungal and antiviral medication. Individuals with BN and BED had received antimicrobial medication prescriptions more often than their matched controls, whereas no significant difference was found in AN. Regarding the defined daily doses per individual, patients with BN, BED, and males with AN had significantly higher total antimicrobial medication use. It was concluded that changes in intestinal microbiota due to infections, inflammation, or antibacterial medications may contribute to EDs.122
A pathophysiological model of eating disorders based on genetic and gut microbiome findings
Family studies, twin studies and GWASs all suggest that genetics plays a major role in the development of EDs. In order to create a pathophysiological model based on the results of genetic studies, we have rearranged the findings to include factors outside the human body, the biology of the body which is significantly influenced by genes, and the resulting ED phenotype (Figure 1). Significant nongenetic risk factors contributing to the aetiopathogenesis of EDs will be mentioned, but not explained in detail, as they are not the focus of this review. The reader may refer to the cited literature.
Factors outside the body
Relevant factors in the development of an ED from outside the body include the social environment, the biological environment and nutritional factors. Within the social environment, the socioeconomic status, culture and lifestyle, a certain ideal of beauty, relationships, and psychosocial stresses including academic challenges, which can have an impact on the emergence of an ED.123,124 Additional relevant factors outside the human body mutually influencing each other include the biological environment, diet, and the use of medication and probiotics.125–128
The above environmental factors are able to directly precede ED symptoms, for example, when a teenage girl does not fit the beauty ideal of her peers, is bullied for having ‘fat thighs’ and starts a diet, they can develop symptoms of AN, as it has been shown that bullying can lead to dieting, which is a major risk factor for the development of an ED.129,130 These factors may also indirectly lead to an ED by influencing the biology of the body. For example, several cases have been reported in the literature, where streptococcus infection triggered an autoimmune response in the body that lead to AN.131
Biological factors within the body
As described in the previous sections of this article, the most important elements of the biology of the human body for the development of an ED from a genetic perspective include the microbiome, the metabolic system, the immune system and the brain (middle of Figure 1).
Microbiota
It has been hypothesized that microbes in the gastrointestinal tract manipulate their host’s eating behaviour to increase microbe fitness. Microbes may do this through generating cravings for foods that they specialize on or foods that suppress their competitors, or inducing dysphoria until individuals eat foods for the benefit of the microbes in their body.115 Such nutritional and environmental factors can lead to dysbiosis of the microbiome, which, as a consequence, alters the production of SCFAs, including acetic, propionic and butyric acid (an HDAC inhibitor),110 and of certain enzymes, including ClpB (a mimetic of α-MSH).121 As explained previously, the bacteria and the molecules they produce influence the metabolic system, the immune system, and the brain.30–32,106–121
Metabolic and endocrine system
The metabolic system comprises a number of metabolic and endocrine organs, including the gastrointestinal tract, the liver, endocrine glands, including the pancreas and the thyroid, and the fatty tissue, in addition to certain cell functions, including the transport of glucose into the cell. This system is influenced by food intake, diet, medication, physical activity, the homeostatic system of the brain and the immune system. It can alter the metabolism of cholesterol and lipids, and can send various hormonal signals, including ghrelin, insulin and leptin, to the brain and the immune system.58,97,132 The findings of associations between a variant of the ANKRD50 gene with AN and polymorphisms in the ghrelin58 and the fat mass and obesity-associated genes61,62 with BN indicate the importance of the metabolic and endocrine system in the development of AN and BN from a genetic perspective.72,73
Immune system
The immune system reacts to stress, food intake, components of bacteria (LPS) or molecules produced by microbiota by altering its production of cytokines, including TNF-α and IL-6, chemokines and factors of cell differentiation, which have an influence on the brain.105,98,133–136 EPHX2 enzyme activity has been demonstrated having a diet-dependent modulating influence on cytokine production.104 The fact that epigenetic research has identified a hypermethylated region at the TNXB gene, which is localized within the MHC class III region, is a further indication for the importance of the immune system.87,88,91 The MHC class III molecules include several proteins with immune functions, including cytokines. These proteins are involved in global and specific inflammatory responses.137 In this respect, it is interesting to mention that the strongest genetic association in schizophrenia at the population level involves variation in the MHC locus.138
In order to understand the possible significance of the immune system in the pathophysiology of EDs as indicated by genetic research, it is important to include the ways in which the immune system has been shown to influence other bodily systems, including the brain and the metabolic system. For example, in accordance with Kahl and colleagues, who observed an increase in the mRNA expression of TNF-α and IL-6 in patients with AN,98 meta-analyses have found elevated levels of TNF-α, IL-1 and IL-6 in patients with AN.139,140 These changes may appear as a consequence of acute, chronic or social stress.135,136,141 Additionally, bacteria from the gut microbiota can stimulate the production of cytokines, for example by the release of LPS, which binds to pattern recognition receptors including Toll-like receptor 4, on circulating monocytes and macrophages.142 In turn, cytokines can influence the signal transduction of afferent nerves, including the vagus nerve, which convey information to the brain; they are also capable of permeating the blood–brain barrier, and influencing brain metabolism and signal processing by activating monoamine reuptake, stimulating the hypothalamic–pituitary–adrenocortical (HPA) axis and decreasing the production of serotonin via the increased activity of indolamine-2,3-dioxygenase.133,134 The importance of the interplay between the immune system and the brain is further underlined by the fact that changes in the concentrations of pro-inflammatory cytokines have been found in the serum or plasma of patients with different psychiatric disorders, including depression,134,143 post-traumatic stress disorder and schizophrenia.144–147
The immune system is also closely linked to the metabolic and endocrine systems. Cytokines, including TNF-α, which are released during infection and inflammation, activate the HPA system at the hypothalamic, pituitary, and adrenal level, resulting in the release of cortisol as the most important negative feedback signal to prevent an overshoot of the ongoing host defence;148–151 glucocorticoids, in turn, suppress the production of pro-inflammatory cytokines, and a chronically activated HPA axis in chronic stress results in defective immune system responses to an inflammatory challenge.151–155 These interactions may partly explain the influence exerted by HPA axis hormones on natural killer cell (NK) activity in patients with AN.156 Similar to depression, a disturbed interplay between cytokines and the HPA axis has been found in AN.151,157
Brain
In order to sort and interpret genetic findings on the brain and neurotransmitter systems, we refer to an approach based on brain-imaging studies, which illustrates the importance of three specific brain systems for EDs: the self-regulatory, the hedonic and the homeostatic systems.25 In this approach, the self-regulatory system is located mainly in the prefrontal areas, which embeds eating into the social context, creates individual values and provides self-regulatory control in response to environmental, social and cultural factors. Important transmitters of this system are serotonin, noradrenalin, acetylcholine and glutamate, of which serotonin and glutamate have support from genetic studies. The hedonic system consists of regions in the prefrontal cortex, the basal ganglia and the thalamus. It is relevant for increasing the desire to eat and pleasure during food consumption on the basis of information from the sensory organs and the hippocampus using dopamine, opioids, and cannabinoids, which have been identified in genetic research of EDs, as neurotransmitters. The hypothalamic homeostatic system integrates peripheral signals of food consumption and energy storages, including leptin, in order to regulate appetite. α-MSH, of which POMC is a precursor protein and has been shown to be of importance in the development of AN in gene-expression studies, is an important signalling molecule of the homeostatic system.25
Genetic findings can be sorted according to these three brain systems. According to this classification, genetically predisposed alterations in brain function can be found in patients with an ED in the self-regulatory system, including modified serotonin signalling in patients with AN and BED,39,40,53,60,81,100 and altered norepinephrine and glutamate signalling in patients with AN.57 Regarding the hedonic system, positive significant associations in AN, BN and BED have been found for dopamine, cannabinoid and opioid signalling.60,63,64,72,94–96 The importance of the homeostatic system is demonstrated by associations of candidate genes in studies of patients with AN, in which genes associated with AGRP, α-MSH, MC4R, NPY appeared to be significantly associated;57 and in patients with BN, where positive results have been obtained for ghrelin,58 which unfolds its orexigenic effects in the hypothalamus.132 α-MSH signalling seems to be of specific importance, as this molecule is also relevant in the crosstalk between the microbiome and the brain.115–118
Eating disorder phenotype
The combination of the biological processes which are influenced by environmental and nutritional factors lead to the behavioural traits of EDs, including an altered response to food intake due to inflammatory processes, to changes in the regulation of weight and metabolism, or an altered motivation towards food intake and physical activity. In EDs, however, the phenotype is not only defined by the psychological and behavioural symptoms of the disorder, but also by its consequence, for example, in the case of AN, malnutrition and an impairment of organ structure and function26 and thus, again, influencing the biology of the human body. EDs also put a strain on the patients’ relationships and lead to social withdrawal.158
Taken together, Figure 1 shows the major factors of the aetiopathogenesis EDs. These factors are related to the environment, nutrition, microbiota, regulatory systems of the body and ED symptoms, and they influence each other. The depicted information might assist in interpreting genetic findings regarding the microbiota and the human genome against the background of environmental and biological factors and the different facets of the ED phenotype.
Discussion
The pathophysiological model
Even though the contemporary genetic knowledge regarding EDs has not led to specific diagnostic methods or therapies, it contributes to valuable novel insights into the pathophysiology of EDs and assists in interpreting findings from other research areas, including brain imaging, neuroimmunological and neuroendocrine research, psychopharmacology, and neuropsychology. On the basis of the reviewed literature, we propose a biological disease model based first and foremost on contemporary genetic findings and research on the microbiome. Although this model explains the connection between environmental and nutritional factors, microbiota and the metabolic system, the immune system, the CNS, and ED pathology, it has shortcomings due to uncertainties in the genetic knowledge and due to assumptions made regarding the nature of EDs.
Shortcomings
Shortcomings in the genetic knowledge of EDs arise from the lack of certain types of genetic studies. For example, in other psychiatric disciplines, brain-imaging studies have revealed genetic effects on brain structure and function, thus implicating neural pathways and causal mechanisms.159 Despite substantial progress in neuroimaging studies,160,161 published imaging genetic studies on EDs are limited. However, within the Enhancing Neuro Imaging Genetics by Meta-Analysis (ENIGMA) Consortium, there is an ENIGMA–AN imaging genetics working group dedicated to improving the understanding of structural brain changes in patients with AN.162
The same is unfortunately the case for pharmacogenetic studies, which may partly be due to a lack of consensus on effective pharmacological treatment strategies.25 Other areas of research lacking in EDs include the examination of common and rare variations, copy number variation, cross-disorder analysis, and gene-environment interplay, specifically the interplay between gene expression, genetic imprinting, and pre- and postnatal environmental factors.
The majority of genetic studies in EDs have been performed in AN, with only a limited number of studies in BN or BED. For example, no GWAS has been published for BN or BED, with the exception of symptoms of the BN spectrum.67 The reported GWASs in AN67–69,72 remain limited in their power to identify a meaningful set of genes contributing to AN, as one single significant locus in a study of thousands of patients with AN is a small outcome for such a large effort.
In addition, ED entities remain an area of discussion.27,28 For example, AN may not be a purely psychiatric disorder, but a disorder with significant psychological, brain–anatomical, metabolic or immunological aspects. It may be the case that these traits will assist in subclassifying AN in the future. However, as highlighted in our explanations relating to Figure 1, these systems are so closely linked together, that such subclassification may not work in practice.
Current efforts for genetic research in EDs
Currently, the PGC is performing GWASs in EDs. It is the largest consortium in the history of psychiatry and has recently commenced a programme of research designed to deliver ‘actionable’ findings, with genomic results that reveal fundamental biology, inform clinical practice, and deliver novel therapeutic targets. The central idea of the PGC is to convert genetic findings into biologically, clinically, and therapeutically meaningful insights.16
Examples of promising contemporary studies collecting genetic data include the Avon Longitudinal Study of Parents and Children (ALSPAC), the Norwegian Mother and Child Cohort Study (MoBa), the Anorexia Nervosa Genetics Initiative (ANGI), the Charlotte’s Helix Project, the Binge Eating Genetics INitiative (BEGIN), and the abovementioned ENIGMA–AN work group. The ALSPAC is a prospective pregnancy cohort (baseline 1991–1992) charting the health of 14,500 families in the Bristol area.163 MoBa is a study for which >90,000 pregnant women were recruited between 1998 and 2008, and in which >70,000 of fathers participated.164 ANGI is the largest and most rigorous genetic investigation of EDs conducted to date. This international collaboration collected blood samples and clinical information from 13,364 individuals with lifetime AN as well as ancestrally matched controls. ANGI has thus substantially augmented the PGC AN sample collection. ANGI recruited from the United States, Australia, New Zealand, Sweden and Denmark via national registers, treatment centres, social and traditional media.165 The Charlotte’s Helix Project was founded in the UK to honour the ED advocacy work of Charlotte Bevan and is part of a global effort to collect DNA samples from >25,000 individuals with AN.166 BEGIN will conduct a large GWAS in 2500 individuals with binge-type ED and matched controls, coupled with microbiota sampling, and deep phenotyping using an ED recovery application programmed on Apple watches.167
The sample sizes, phenotyping and statistical approaches of these initiatives are promising, and these projects are likely to elucidate further important pathophysiological factors leading to the development of EDs.
Historical and philosophical considerations
It can be argued that heritability has already been considered in biblical times and that we are still unable to transform this relatively old idea into an effective therapy for patients, however, there is legitimate hope that novel genetic methods, including GWASs, will reveal crucial pathophysiological pathways for EDs and lead to novel therapeutic strategies in the near future.
Publication bias is widespread in biomedical research, including genetics.168,169 The study by Rüdin9 was mentioned as a historic example of unpublished negative results in order not to challenge one’s political beliefs and risk one’s own carrier.9 Publication bias was not only a problem during the time of Nazi pervasion, it is an ongoing issue.170
The discovery of the unconscious and the development of antidepressants were significant breakthroughs in psychiatry, which had a marked impact not only on psychiatric treatment, but also on the way we see ourselves.171,172 The elucidation that the microbiome has a significant impact on us may have a similar implication, as it shows that we are not a single entity, but more symbiosis of billions of organisms.
Therapeutic implications
It may be the case that important discoveries have already been made, but we cannot realise them, in a manner similar to Gregor Johann Mendel, who gained recognition as the founder of formal genetics only posthumously. For example, the increase in the mRNA expression of TNF-α in patients with AN98 may have an impact on the development of novel drug targets for EDs as available anti-TNF-α drugs, for example etanercept, have been used in other psychiatric disorders and may, in principle, be assessed in EDs also.173
In addition to TNF-α and IL-6,98 we have discussed LPS,114 SCFAs,107–111 ClpB,121 EPHX2,104 EBF1,69,70 TNXB,87,88 which, in addition to other MHC proteins, are pathophysiological key molecules. These molecules may offer potential as future drug targets, if inhibitors or analogues of these molecules become available.
Zheng and colleagues performed experiments in germ-free mice. The transplantation of microbiota from patients suffering from a depressive episode resulted in depression-like behaviours, whereas colonization with microbiota derived from healthy controls did not.174 Therefore, faecal microbiota transplant may become a future therapeutic option for patients with EDs. In a more specific approach, it has been shown that Lactobacillus rhamnosus alleviates OCD-like and anxious behaviours in animals, possibly by altering gamma-aminobutyric acid (GABA) receptors or even synthesizing and releasing GABA.175 Therefore, L. rhamnosus may have beneficial psychological effects in patients suffering from psychiatric disorders with these symptoms, including those with OCD, anxiety or AN.
Taken together, genetic research in EDs has changed the way we think about AN, BN and BED, as it clearly demonstrates the importance of biological factors in the pathophysiology of EDs. This knowledge will hopefully lead to novel therapeutic ideas and eventually put an end to the management of life-threatening medical complications, nutritional rehabilitation, including tube feeding and other unpleasant coercive measures, and often unsuccessful psychosocial therapy being the only treatment options for EDs like AN.
Footnotes
Funding: This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Conflict of interest statement: Janet Treasure declares that she will be the Principal Investigator of a randomized controlled trial evaluating the effects of naloxone hydrochloride nasal spray on eating behaviours in bulimia nervosa sponsored by Opiant Pharmaceuticals. All other authors declare no conflict of interest.
Contributor Information
Hubertus Himmerich, Department of Psychological Medicine, King’s College London, London, UK.
Jessica Bentley, Department of Psychological Medicine, King’s College London, London, UK.
Carol Kan, Department of Psychological Medicine, King’s College London, London, UK.
References
- 1. Hardacre H. Ancestors: ancestor worship. In: Jones L. (ed.) Encyclopedia of religion. 2nd ed. Detroit: Thomson Gale, 2005, pp. 320–325. [Google Scholar]
- 2. Catholic Truth Society. The CTS new catholic bible. 1st ed London: Catholic Truth Society, 2007, pp. 12–13. [Google Scholar]
- 3. Mendel JG. Versuche über Pflanzenhybriden. Verh Naturforsch Ver Brünn 1866; 4: 3–47. [Google Scholar]
- 4. Mott FW. The inborn factors of nervous and mental disease. Brain 1911; 34: 73–101. [Google Scholar]
- 5. Schulze TG, Fangerau H, Propping P. From degeneration to genetic susceptibility, from eugenics to genethics, from Bezugsziffer to LOD score: the history of psychiatric genetics. Int Rev Psychiatry 2004; 16: 246–259. [DOI] [PubMed] [Google Scholar]
- 6. Schmuhl H. Rassenhygiene, Nationalsozialismus, Euthanasie: Von der Verhütung zur Vernichtung “lebensunwerten Lebens”. 2nd ed. Göttingen: Vandenhoeck and Ruprecht, 1992, p. 152. [Google Scholar]
- 7. Weber MM. Ernst Rüdin, 1874–1952: a German psychiatrist and geneticist. Am J Med Genet A 1996; 67: 323–331. [DOI] [PubMed] [Google Scholar]
- 8. Schulz B. Ernst Rüdin. Arch Psychiatr Neurol 1953; 190: 187–195. [DOI] [PubMed] [Google Scholar]
- 9. Kösters G, Steinberg H, Kirkby KC, et al. Ernst Rüdin’s unpublished 1922–1925 study ‘Inheritance of manic-depressive insanity’: genetic research findings subordinated to eugenic ideology. PLoS Genet 2015; 11: e1005524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dahm R. Discovering DNA: Friedrich Miescher and the early years of nucleic acid research. Hum Genet 2008; 122: 565–581. [DOI] [PubMed] [Google Scholar]
- 11. Watson JD, Crick FH. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature 1953; 171: 737–738. [DOI] [PubMed] [Google Scholar]
- 12. Raju TN. The Nobel Chronicles. 1968: Har Khorana (b 1922); Robert Holley (1922-93); Marshall Nirenberg (b 1927). Lancet 1999; 354: 690. [DOI] [PubMed] [Google Scholar]
- 13. Lewis CM, Knight J. Introduction to genetic association studies. Cold Spring Harb Protoc 2012; 2012: 297–306. [DOI] [PubMed] [Google Scholar]
- 14. Gurling H. Candidate genes and favoured loci: strategies for molecular genetic research into schizophrenia, manic depression, autism, alcoholism and Alzheimer’s disease. Psychiatr Dev 1986; 4: 289–309. [PubMed] [Google Scholar]
- 15. Klein RJ, Zeiss C, Chew EY, et al. Complement factor H polymorphism in age-related macular degeneration. Science 2005; 308: 385–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sullivan PF, Agrawal A, Bulik CM, et al. Psychiatric genomics: an update and an agenda. Am J Psychiatry 2018; 175: 15–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hegstrand LR, Hine RJ. Variations of brain histamine levels in germ-free and nephrectomized rats. Neurochem Res 1986; 11:185–191. [DOI] [PubMed] [Google Scholar]
- 18. Prescott SL. History of medicine: origin of the term microbiome and why it matters. Hum Microbiome J 2017; 4: 24–25. [Google Scholar]
- 19. Himmerich H, Minkwitz J, Kirkby KC. Weight gain and metabolic changes during treatment with antipsychotics and antidepressants. Endocr Metab Immune Disord Drug Targets 2015; 15: 252–260. [DOI] [PubMed] [Google Scholar]
- 20. Savage DC. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol 1977; 31: 107–133. [DOI] [PubMed] [Google Scholar]
- 21. NIH HMP Working Group, Peterson J, Garges S, et al. The NIH human microbiome project. Genome Res 2009; 19: 2317–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schmidt TSB, Raes J, Bork P. The human gut microbiome: from association to modulation. Cell 2018; 172: 1198–1215. [DOI] [PubMed] [Google Scholar]
- 23. Relman DA, Falkow S. The meaning and impact of the human genome sequence for microbiology. Trends Microbiol 2001; 9: 206–208. [DOI] [PubMed] [Google Scholar]
- 24. American Psychiatric Association. Diagnostic and statistical manual of mental disorders. 5th ed Arlington: APA Publishing, 2013. [Google Scholar]
- 25. Himmerich H, Treasure J. Psychopharmacological advances in eating disorders. Expert Rev Clin Pharmacol 2018; 11: 95–108. [DOI] [PubMed] [Google Scholar]
- 26. Himmerich H, Schönknecht P, Heitmann S, et al. Laboratory parameters and appetite regulators in patients with anorexia nervosa. J Psychiatr Pract 2010; 16: 82–92. [DOI] [PubMed] [Google Scholar]
- 27. Tozzi F, Thornton L, Klump K, et al. Symptom fluctuation in eating disorders: correlates of diagnostic crossover. Am J Psychiatry 2005; 162: 732–740. [DOI] [PubMed] [Google Scholar]
- 28. Hay P, Fairburn C. The validity of the DSM-IV scheme for classifying bulimic eating disorders. Int J Eat Disord 1998; 23: 7–15. [DOI] [PubMed] [Google Scholar]
- 29. Bulik CM, Thornton LM, Root TL, et al. Understanding the relation between anorexia nervosa and bulimia nervosa in a Swedish national twin sample. Biol Psychiatry 2010; 67: 71–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Glenny EM, Bulik-Sullivan EC, Tang Q, et al. Eating disorders and the intestinal microbiota: mechanisms of energy homeostasis and behavioral influence. Curr Psychiatry Rep 2017; 19: 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Borgo F, Riva A, Benetti A, et al. Microbiota in anorexia nervosa: the triangle between bacterial species, metabolites and psychological tests. PLoS One 2017; 12: e0179739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kleiman SC, Glenny EM, Bulik-Sullivan EC, et al. Daily changes in composition and diversity of the intestinal microbiota in patients with anorexia nervosa: a series of three cases. Eur Eat Disord Rev 2017; 25: 423–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tremaroli V, Bäckhed F. Functional interactions between the gut microbiota and host metabolism. Nature 2012; 489: 242–249. [DOI] [PubMed] [Google Scholar]
- 34. Lam YY, Maguire S, Palacios T, et al. Are the gut bacteria telling us to eat or not to eat? Reviewing the role of gut microbiota in the etiology, disease progression and treatment of eating disorders. Nutrients 2017; 14; 9: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Trace SE, Baker JH, Peñas-Lledó E, et al. The genetics of eating disorders. Annu Rev Clin Psychol 2013; 9: 589–620. [DOI] [PubMed] [Google Scholar]
- 36. Bulik CM, Sullivan PF, Wade TD, et al. Twin studies of eating disorders: a review. Int J Eat Disord 2000; 27: 1–20. [DOI] [PubMed] [Google Scholar]
- 37. Bulik CM, Kleiman SC, Yilmaz Z. Genetic epidemiology of eating disorders. Curr Opin Psychiatry 2016; 29: 383–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shih PA, Woodside DB. Contemporary views on the genetics of anorexia nervosa. Eur Neuropsychopharmacol 2016; 26: 663–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hinney A, Friedel S, Remschmidt H, et al. Genetic risk factors in eating disorders. Am J Pharmacogenomics 2004; 4: 209–223. [DOI] [PubMed] [Google Scholar]
- 40. Yilmaz Z, Hardaway JA, Bulik CM. Genetics and epigenetics of eating disorders. Adv Genomics Genet 2015; 5: 131–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lilenfeld L, Kaye W, Greeno C, et al. A controlled family study of restricting anorexia and bulimia nervosa: comorbidity in probands and disorders in first-degree relatives. Arch Gen Psychiatry 1998; 55: 603–610. [DOI] [PubMed] [Google Scholar]
- 42. Strober M, Freeman R, Lampert C, et al. Controlled family study of anorexia nervosa and bulimia nervosa: evidence of shared liability and transmission of partial syndromes. Am J Psychiatry 2000; 157: 393–401. [DOI] [PubMed] [Google Scholar]
- 43. Strober M, Freeman R, Lampert C, et al. Males with anorexia nervosa: a controlled study of eating disorders in first-degree relatives. Int J Eat Disord 2001; 29: 263–269. [DOI] [PubMed] [Google Scholar]
- 44. Bulik CM, Sullivan PF, Tozzi F, et al. Prevalence, heritability, and prospective risk factors for anorexia nervosa. Arch Gen Psychiatry 2006; 63: 305–312. [DOI] [PubMed] [Google Scholar]
- 45. Holland AJ, Hall A, Murray R, et al. Crisp Anorexia nervosa: a study of 34 twin pairs and one set of triplets. Br J Psychiatry 1984; 145: 414–419. [DOI] [PubMed] [Google Scholar]
- 46. Javaras KN, Laird NM, Reichborn-Kjennerud T, et al. Familiality and heritability of binge eating disorder: results of a case-control family study and a twin study. Int J Eat Disord 2008; 41: 174–179. [DOI] [PubMed] [Google Scholar]
- 47. Reichborn-Kjennerud T, Bulik C, Tambs K, et al. Genetic and environmental influences on binge eating in the absence of compensatory behaviours: a population-based twin study. Int J Eat Disord 2004; 36: 307–314. [DOI] [PubMed] [Google Scholar]
- 48. Thornton LM, Mazzeo SE, Bulik CM. The heritability of eating disorders: methods and current findings. Curr Top Behav Neurosci 2011; 6: 141–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Klump KL, Suisman JL, Burt SA. Genetic and environmental influences on disordered eating: an adoption study. J Abnorm Psychol 2009; 118: 797–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Von Ranson KM, Klump KL, Iacono WG, et al. The Minnesota eating behavior survey: a brief measure of disordered eating attitudes and behaviors. Eat Behav 2005; 6: 373–392. [DOI] [PubMed] [Google Scholar]
- 51. Grice DE, Halmi KA, Fichter MM, et al. Evidence for a susceptibility gene for anorexia nervosa on chromosome 1. Am J Hum Genet 2002; 70: 787–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bergen AW, Van den Bree MB, Yeager M, et al. Candidate genes for anorexia nervosa in the 1p33–36 linkage region: serotonin 1D and delta opioid receptor loci exhibit significant association to anorexia nervosa. Mol Psychiatry 2003; 8: 397–406. [DOI] [PubMed] [Google Scholar]
- 53. Devlin B, Bacanu SA, Klump KL, et al. Linkage analysis of anorexia nervosa incorporating behavioral covariates. Hum Mol Genet 2002; 11: 689–696. [DOI] [PubMed] [Google Scholar]
- 54. Bulik CM, Devlin B, Bacanu SA, et al. Significant linkage on chromosome 10p in families with bulimia nervosa. Am J Hum Genet 2003; 72: 200–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. McCarroll SA, Feng G, Hyman SE. Genome-scale neurogenetics: methodology and meaning. Nat Neurosci 2014; 17: 756–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cohen J, Pertsemlidis A, Kotowski IK, et al. Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat Genet 2005; 37: 161–165. [DOI] [PubMed] [Google Scholar]
- 57. Rask-Andersen M, Olszewski PK, Levine AS, et al. Molecular mechanisms underlying anorexia nervosa: focus on human gene association studies and systems controlling food intake. Brain Res Rev 2010; 62: 147–164. [DOI] [PubMed] [Google Scholar]
- 58. Ando T, Komaki G, Naruo T, et al. Possible role of preproghrelin gene polymorphisms in susceptibility to bulimia nervosa. Am J Med Genet B Neuropsychiatr Genet 2006; 141B: 929–934. [DOI] [PubMed] [Google Scholar]
- 59. Nilsson M, Naessen S, Dahlman I, et al. Association of estrogen receptor beta gene polymorphisms with bulimic disease in women. Mol Psychiatry 2004; 9: 28–34. [DOI] [PubMed] [Google Scholar]
- 60. Monteleone P, Tortorella A, Castaldo E, et al. Association of a functional serotonin transporter gene polymorphism with binge eating disorder. Am J Med Genet B Neuropsychiatr Genet 2006; 141B: 7–9. [DOI] [PubMed] [Google Scholar]
- 61. Muller TD, Greene BH, Bellodi L, et al. Fat mass and obesity-associated gene (FTO) in eating disorders: evidence for association of the rs9939609 obesity risk allele with bulimia nervosa and anorexia nervosa. Obes Facts 2012; 5: 408–419. [DOI] [PubMed] [Google Scholar]
- 62. Jonassaint CR, Szatkiewicz JP, Bulik CM, et al. Absence of association between specific common variants of the obesity-related FTO gene and psychological and behavioral eating disorder phenotypes. Am J Med Genet B Neuropsychiatr Genet 2011; 156B: 454–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Davis C, Levitan RD, Yilmaz Z, et al. Binge eating disorder and the dopamine D2 receptor: genotypes and sub-phenotypes. Prog Neuropsychopharmacol Biol Psychiatry 2012; 38: 328–335. [DOI] [PubMed] [Google Scholar]
- 64. Davis CA, Levitan RD, Reid C, et al. Dopamine for ‘wanting’ and opioids for ‘liking’: a comparison of obese adults with and without binge eating. Obesity 2009; 17: 1220–1225. [DOI] [PubMed] [Google Scholar]
- 65. Bush WS, Moore JH. Chapter 11: Genome-wide association studies. PLoS Comput Biol 2012; 8: e1002822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wang K, Zhang H, Bloss CS, et al. A genome-wide association study on common SNPs and rare CNVs in anorexia nervosa. Mol Psychiatry 2011; 16: 949–959. [DOI] [PubMed] [Google Scholar]
- 67. Boraska V, Franklin CS, Floyd JA, et al. A genome-wide association study of anorexia nervosa. Mol Psychiatry 2014; 19: 1085–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Duncan L, Yilmaz Z, Gaspar H, et al. Significant locus and metabolic genetic correlations revealed in genome-wide association study of anorexia nervosa. Am J Psychiatry 2017; 174: 850–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Li D, Chang X, Connolly JJ, et al. A genome-wide association study of anorexia nervosa suggests a risk locus implicated in dysregulated leptin signaling. Sci Rep 2017; 7: 3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Hagman J, Belanger C, Travis A, et al. Cloning and functional characterization of early B-cell factor, a regulator of lymphocyte-specific gene expression. Genes Dev 1993; 7: 760–773. [DOI] [PubMed] [Google Scholar]
- 71. Kikuchi H, Nakayama M, Takami Y, et al. EBF1 acts as a powerful repressor of Blimp-1 gene expression in immature B cells. Biochem Biophys Res Commun 2012; 422: 780–785. [DOI] [PubMed] [Google Scholar]
- 72. Huckins LM, Hatzikotoulas K, Southam L, et al. Investigation of common, low-frequency and rare genome-wide variation in anorexia nervosa. Mol Psychiatry 2018; 23: 1169–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. McGough IJ, Steinberg F, Gallon M, et al. Identification of molecular heterogeneity in SNX27-retromer-mediated endosome-to-plasma-membrane recycling. J Cell Sci 2014; 127: 4940–4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Bulik-Sullivan B, Finucane HK, Anttila V, et al. An atlas of genetic correlations across human diseases and traits. Nat Genet 2015; 47: 1236–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Nordsletten AE, Larsson H, Crowley JJ, et al. Patterns of nonrandom mating within and across 11 major psychiatric disorders. JAMA Psychiatry 2016; 73: 354–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Wade TD, Gordon S, Medland S, et al. Genetic variants associated with disordered eating. Int J Eat Disord 2013; 46: 594–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Kao PY, Leung KH, Chan LW, et al. Pathway analysis of complex diseases for GWAS, extending to consider rare variants, multi-omics and interactions. Biochim Biophys Acta 2017; 1861: 335–353. [DOI] [PubMed] [Google Scholar]
- 78. Manolio TA, Collins FS, Cox NJ, et al. Finding the missing heritability of complex diseases. Nature 2009; 461: 747–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Zeng P, Zhao Y, Qian C, et al. Statistical analysis for genome-wide association study. J Biomed Res 2015; 29: 2285–2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Wang K, Li M, Hakonarson H. Analysing biological pathways in genome-wide association studies. Nat Rev Genet 2010; 11: 843–854. [DOI] [PubMed] [Google Scholar]
- 81. Pinheiro AP, Bulik CM, Thornton LM, et al. Association study of 182 candidate genes in anorexia nervosa. Am J Med Genet B Neuropsychiatr Genet 2010; 153: 1070–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Dupont C, Armant DR, Brenner CA. Epigenetics: definition, mechanisms and clinical perspective. Semin Reprod Med 2009; 27: 351–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Gomes AQ, Nolasco S, Soares H. Non-coding RNAs: multi-tasking molecules in the cell. Int J Mol Sci 2013; 14: 16010–16039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Frieling H, Gozner A, Römer KD, et al. Global DNA hypomethylation and DNA hypermethylation of the alpha synuclein promoter in females with anorexia nervosa. Mol Psychiatry 2007; 12: 229–230. [DOI] [PubMed] [Google Scholar]
- 85. Tremolizzo L, Conti E, Bomba M, et al. Decreased whole-blood global DNA methylation is related to serum hormones in anorexia nervosa adolescents. World J Biol Psychiatry 2014; 15: 327–333. [DOI] [PubMed] [Google Scholar]
- 86. Saffrey R, Novakovic B, Wade TD. Assessing global and gene specific DNA methylation in anorexia nervosa: a pilot study. Int J Eat Disord 2014; 47: 206–210. [DOI] [PubMed] [Google Scholar]
- 87. Booij L, Casey KF, Antunes JM, et al. DNA methylation in individuals with anorexia nervosa and in matched normal-eater controls: a genome-wide study. Int J Eat Disord 2015; 48: 874–882. [DOI] [PubMed] [Google Scholar]
- 88. Kesselmeier M, Pütter C, Volckmar AL, et al. High-throughput DNA methylation analysis in anorexia nervosa confirms TNXB hypermethylation. World J Biol Psychiatry 2018; 19: 187–199. [DOI] [PubMed] [Google Scholar]
- 89. Bristow J, Tee MK, Gitelman SE, et al. Tenascin-X - a novel extracellular-matrix protein encoded by the human Xb gene overlapping P450c21b. J Cell Biol 1993; 122: 265–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Chen WY, Kim MS, Shanbhag S, et al. The phenotypic spectrum of contiguous deletion of CYP21A2 and tenascin XB: quadricuspid aortic valve and other midline defects. Am J Med Genet A 2009; 149: 2803–2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Weissensteiner T, Lanchbury JS. Allelic polymorphism of two multifunctional regions in the central human MHC: tenascin X, XB-S and YB, and their duplicated fragments XA and YA. Eur J Immunogenet 1997; 24: 201–209. [DOI] [PubMed] [Google Scholar]
- 92. Quach A, Levine ME, Tanaka T, et al. Epigenetic clock analysis of diet, exercise, education, and lifestyle factors. Aging 2017; 9: 419–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Frieling H, Bleich S, Otten J, et al. Epigenetic downregulation of atrial natriuretic peptide but not vasopressin mRNA expression in females with eating disorders is related to impulsivity. Neuropsychopharmacology 2008; 33: 2605–2609. [DOI] [PubMed] [Google Scholar]
- 94. Frieling H, Albrecht H, Jedtberg S, et al. Elevated cannabinoid 1 receptor mRNA is linked to eating disorder related behavior and attitudes in females with eating disorders. Psychoneuroendocrinology 2009; 34: 620–624. [DOI] [PubMed] [Google Scholar]
- 95. Ehrlich S, Weiss D, Burghardt R, et al. Promoter specific DNA methylation and gene expression of POMC in acutely underweight and recovered patients with anorexia nervosa. J Psychiatr Res 2010; 44: 827–833. [DOI] [PubMed] [Google Scholar]
- 96. Frieling H, Romer KD, Scholz S, et al. Epigenetic dysregulation of dopaminergic genes in eating disorders. Int J Eat Disord 2010; 43: 577–583. [DOI] [PubMed] [Google Scholar]
- 97. Janas-Kozik M, Stachowicz M, Mazurek U, et al. Preliminary study of the expression of genes connected with the orexigenic and anorexigenic system using microarray technique in anorexia nervosa. Neuropsychobiology 2008; 57: 116–120. [DOI] [PubMed] [Google Scholar]
- 98. Kahl KG, Kruse N, Rieckmann P, et al. Cytokine mRNA expression patterns in the disease course of female adolescents with anorexia nervosa. Psychoneuroendocrinology 2004; 29: 13–20. [DOI] [PubMed] [Google Scholar]
- 99. Mercader JM, Saus E, Agüera Z, et al. Association of NTRK3 and its interaction with NGF suggest an altered cross-regulation of the neurotrophin signaling pathway in eating disorders. Hum Mol Genet 2008; 17: 1234–1244. [DOI] [PubMed] [Google Scholar]
- 100. Urwin RE, Nunn KP. Epistatic interaction between the monoamine oxidase A and serotonin transporter genes in anorexia nervosa. Eur J Hum Genet 2005; 13: 370–375. [DOI] [PubMed] [Google Scholar]
- 101. Cui H, Moore J, Ashimi SS, et al. Eating disorder predisposition is associated with ESRRA and HDAC4 mutations. J Clin Invest 2013; 123: 4706–4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Fenech M, El-Sohemy A, Cahill L, et al. Nutrigenetics and nutrigenomics: viewpoints on the current status and applications in nutrition research and practice. J Nutrigenet Nutrigenomics 2011; 4: 69–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Mutch DM, Wahli W, Williamson G. Nutrigenomics and nutrigenetics: the emerging faces of nutrition. FASEB J 2005; 19: 1602–1616. [DOI] [PubMed] [Google Scholar]
- 104. Scott-Van Zeeland AA, Bloss CS, Tewhey R, et al. Evidence for the role of EPHX2 gene variants in anorexia nervosa. Mol Psychiatry 2014; 19: 724–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Yang J, Hammock BD, Halmi KA, et al. Substrate-dependent postprandial oxylipin responses reveal the potential role of nutrient-gene interaction in Anorexia Nervosa. Paper presented at the American College of Neuropsychopharmacology Annual Meeting, 2015, Hollywood, US. [Google Scholar]
- 106. Lozupone CA, Stombaugh JI, Gordon JI, et al. Diversity, stability and resilience of the human gut microbiota. Nature 2012; 489: 220–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. MacFabe DF, Cain NE, Boon F, et al. Effects of the enteric bacterial metabolic product propionic acid on object-directed behavior, social behavior, cognition, and neuroinflammation in adolescent rats: relevance to autism spectrum disorder. Behav Brain Res 2011; 217: 47–54. [DOI] [PubMed] [Google Scholar]
- 108. Ríos-Covián D, Ruas-Madiedo P, Margolles A, et al. Intestinal short chain fatty acids and their link with diet and human health. Front Microbiol 2016; 7: 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Topping DL, Clifton PM. Short-chain fatty acids and human colonic function: roles of resistant starch and nonstarch polysaccharides. Physiol Rev 2001; 81: 1031–1064. [DOI] [PubMed] [Google Scholar]
- 110. Waldecker M, Kautenburger T, Daumann H, et al. Histone-deacetylase inhibition and butyrate formation: fecal slurry incubations with apple pectin and apple juice extracts. Nutrition 2008; 24: 366–374. [DOI] [PubMed] [Google Scholar]
- 111. Puertollano E, Kolida S, Yaqoob P. Biological significance of short-chain fatty acid metabolism by the intestinal microbiome. Curr Opin Clin Nutr Metab Care 2014; 17: 139–144. [DOI] [PubMed] [Google Scholar]
- 112. Pfleiderer A, Lagier JC, Armougom F, et al. Culturomics identified 11 new bacterial species from a single anorexia nervosa stool sample. Eur J Clin Microbiol Infect Dis 2013; 32: 1471–1481. [DOI] [PubMed] [Google Scholar]
- 113. Raybould HE. Gut chemosensing: interactions between gut endocrine cells and visceral afferents. Auton Neurosci 2010; 153: 41–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Banks WA, Gray AM, Erickson MA, et al. Lipopolysaccharide-induced blood-brain barrier disruption: roles of cyclooxygenase, oxidative stress, neuroinflammation, and elements of the neurovascular unit. J Neuroinflammation 2015; 12: 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Alcock J, Maley CC, Aktipis CA. Is eating behavior manipulated by the gastrointestinal microbiota? Evolutionary pressures and potential mechanisms. Bioessays 2014; 36: 940–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Fetissov SO, Hallman J, Oreland L, et al. Autoantibodies against alpha-MSH, ACTH, and LHRH in anorexia and bulimia nervosa patients. Proc Natl Acad Sci USA 2002; 99: 17155–17160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Fetissov SO, Harro J, Jaanisk M, et al. Autoantibodies against neuropeptides are associated with psychological traits in eating disorders. Proc Natl Acad Sci USA 2005; 102: 14865–14870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Armougom F, Henry M, Vialettes B, et al. Monitoring bacterial community of human gut microbiota reveals an increase in Lactobacillus in obese patients and Methanogens in anorexic patients. PLoS One 2009; 4: e7125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Mack I, Cuntz U, Grämer C, et al. Weight gain in anorexia nervosa does not ameliorate the faecal microbiota, branched chain fatty acid profiles, and gastrointestinal complaints. Sci Rep 2016; 6: 26752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Morita C, Tsuji H, Hata T, et al. Gut dysbiosis in patients with anorexia nervosa. PLoS One 2015; 10: e0145274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Breton J, Legrand R, Akkermann K, et al. Elevated plasma concentrations of bacterial ClpB protein in patients with eating disorders. Int J Eat Disord 2016; 49: 805–808. [DOI] [PubMed] [Google Scholar]
- 122. Raevuori A, Lukkariniemi L, Suokas JT, et al. Increased use of antimicrobial medication in bulimia nervosa and binge-eating disorder prior to the eating disorder treatment. Int J Eat Disord 2016; 49: 542–552. [DOI] [PubMed] [Google Scholar]
- 123. Levine MP, Smolak L, Moodey AF, et al. Normative developmental challenges and dieting and eating disturbances in middle school girls. Int J Eat Disord 1994; 15: 11–20. [DOI] [PubMed] [Google Scholar]
- 124. Miller MN, Pumariega AJ. Culture and eating disorders: a historical and cross-cultural review. Psychiatry 2001; 64: 93–110. [DOI] [PubMed] [Google Scholar]
- 125. Yacoub R, Kaji D, Patel SN, et al. Association between probiotic and yogurt consumption and kidney disease: insights from NHANES. Nutr J 2016; 15: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Faith MS, Kral TVE. Social environmental and genetic influences on obesity and obesity-promoting behaviors: fostering research integration. In: Genes, behavior, and the social environment: moving beyond the nature/nurture debate. National Academy of Sciences, 2006. [Google Scholar]
- 127. Nagasawa M, Smith MC, Barnes JH, Jr, et al. Meta-analysis of correlates of diabetes patients’ compliance with prescribed medications. Diabetes Educ 1990; 16: 192–200. [DOI] [PubMed] [Google Scholar]
- 128. Rosi A, Mena P, Pellegrini N, et al. Environmental impact of omnivorous, ovo-lacto-vegetarian, and vegan diet. Sci Rep 2017; 7: 6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Copeland WE, Bulik CM, Zucker N, et al. Does childhood bullying predict eating disorder symptoms? A prospective, longitudinal analysis. Int J Eat Disord 2015; 48: 1141–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Lloyd EC, Frampton I, Verplanken B, et al. How extreme dieting becomes compulsive: a novel hypothesis for the role of anxiety in the development and maintenance of anorexia nervosa. Med Hypotheses 2017; 108: 144–150. [DOI] [PubMed] [Google Scholar]
- 131. Sokol MS. Infection-triggered anorexia nervosa in children: clinical description of four cases. J Child Adolesc Psychopharmacol 2000; 10: 133–145. [DOI] [PubMed] [Google Scholar]
- 132. Schwartz MW, Woods SC, Porte D, Jr, et al. Central nervous system control of food intake. Nature 2000; 404: 661–671. [DOI] [PubMed] [Google Scholar]
- 133. Capuron L, Miller AH. Immune system to brain signaling: neuropsychopharmacological implications. Pharmacol Ther 2011; 130: 226–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Lichtblau N, Schmidt FM, Schumann R, et al. Cytokines as biomarkers in depressive disorder: current standing and prospects. Int Rev Psychiatry 2013; 25: 592–603. [DOI] [PubMed] [Google Scholar]
- 135. Himmerich H, Fischer J, Bauer K, et al. Stress-induced cytokine changes in rats. Eur Cytokine Netw 2013; 24: 97–103. [DOI] [PubMed] [Google Scholar]
- 136. Krügel U, Fischer J, Bauer K, et al. The impact of social isolation on immunological parameters in rats. Arch Toxicol 2014; 88: 853–855. [DOI] [PubMed] [Google Scholar]
- 137. Gruen JR, Weissman SM. Human MHC class III and IV genes and disease associations. Front Biosci 2001; 6: 960–972. [DOI] [PubMed] [Google Scholar]
- 138. Sekar A, Bialas AR, de Rivera H, et al. Schizophrenia risk from complex variation of complement component 4. Nature 2016; 530: 177–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Solmi M, Veronese N, Favaro A, et al. Inflammatory cytokines and anorexia nervosa: a meta-analysis of cross-sectional and longitudinal studies. Psychoneuroendocrinology 2015; 51: 237–252. [DOI] [PubMed] [Google Scholar]
- 140. Dalton B, Bartholdy S, Robinson L, et al. A meta-analysis of cytokine concentrations in eating disorders. J Psychiatr Res 2018; 103: 252–264. [DOI] [PubMed] [Google Scholar]
- 141. Krügel U, Fischer J, Radicke S, et al. Antidepressant effects of TNF-alpha blockade in an animal model of depression. J Psychiatr Res 2013; 47: 611–616. [DOI] [PubMed] [Google Scholar]
- 142. Sherwin E, Rea K, Dinan TG, et al. A gut (microbiome) feeling about the brain. Curr Opin Gastroenterol 2016; 32: 96–102. [DOI] [PubMed] [Google Scholar]
- 143. Schmidt FM, Lichtblau N, Minkwitz J, et al. Cytokine levels in depressed and non-depressed subjects, and masking effects of obesity. J Psychiatr Res 2014; 55: 29–34. [DOI] [PubMed] [Google Scholar]
- 144. Waheed A, Dalton B, Wesemann U, et al. A systematic review of interleukin-1β in post-traumatic stress disorder: evidence from human and animal studies. J Interferon Cytokine Res 2018; 38: 1–11. [DOI] [PubMed] [Google Scholar]
- 145. Hussein S, Dalton B, Willmund GD, et al. A systematic review of tumor necrosis factor-α in post-traumatic stress disorder: evidence from human and animal studies. Psychiatr Danub 2017; 29: 407–420. [DOI] [PubMed] [Google Scholar]
- 146. Müller N, Weidinger E, Leitner B, et al. The role of inflammation in schizophrenia. Front Neurosci 2015; 9: 372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Borovcanin M, Jovanovic I, Dejanovic SD, et al. Increase systemic levels of IL-23 as a possible constitutive marker in schizophrenia. Psychoneuroendocrinology 2015; 56: 143–147. [DOI] [PubMed] [Google Scholar]
- 148. Kapcala LP, Chautard T, Eskay RL. The protective role of the hypothalamic-pituitary-adrenal axis against lethality produced by immune, infectious, and inflammatory stress. Ann N Y Acad Sci 1995; 771: 419 –437. [DOI] [PubMed] [Google Scholar]
- 149. Tilders FJ, DeRijk RH, Van Dam AM, et al. Activation of the hypothalamus-pituitary-adrenal axis by bacterial endotoxins: routes and intermediate signals. Psychoneuroendocrinology 1994; 9: 209 –232. [DOI] [PubMed] [Google Scholar]
- 150. Silverman MN, Pearce BD, Biron CA, et al. Immune modulation of the hypothalamic-pituitary-adrenal (HPA) axis during viral infection. Viral Immunol 2005; 18: 41–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Himmerich H, Binder EB, Künzel HE, et al. Successful antidepressant therapy restores the disturbed interplay between TNF-alpha system and HPA axis. Biol Psychiatry 2006; 60: 882–888. [DOI] [PubMed] [Google Scholar]
- 152. Arzt E, Kovalovsky D, IgazL M, et al. Functional cross-talk among cytokines, T-cell receptor, and glucocorticoid receptor transcriptional activity and action. Ann N Y Acad Sci 2000; 917: 672–677. [DOI] [PubMed] [Google Scholar]
- 153. Besedovsky HO, Del Rey A. The cytokine-HPA axis feed-back circuit. Z Rheumatol 2000; 59(Suppl. 2): 26 –30. [DOI] [PubMed] [Google Scholar]
- 154. Reul JM, Labeur MS, Wiegers GJ, et al. Altered neuroimmunoendocrine communication during a condition of chronically increased brain corticotropin-releasing hormone drive. Ann N Y Acad Sci 1998; 840: 444–455. [DOI] [PubMed] [Google Scholar]
- 155. Schöbitz B, Reul JM, Holsboer F. The role of the hypothalamic-pituitary-adrenocortical system during inflammatory conditions. Crit Rev Neurobiol 1994; 8: 263–291. [PubMed] [Google Scholar]
- 156. Staurenghi AH, Masera RG, Prolo P, et al. Hypothalamic-pituitary-adrenal axis function, psychopathological traits, and natural killer (NK) cell activity in anorexia nervosa. Psychoneuroendocrinology 1997; 22: 575–590. [DOI] [PubMed] [Google Scholar]
- 157. Limone P, Biglino A, Bottino F, et al. Evidence for a positive correlation between serum cortisol levels and IL-1beta production by peripheral mononuclear cells in anorexia nervosa. J Endocrinol Invest 2000; 23: 422–427. [DOI] [PubMed] [Google Scholar]
- 158. Corcos M, Flament MF, Giraud MJ, et al. Early psychopathological signs in bulimia nervosa. A retrospective comparison of the period of puberty in bulimic and control girls. Eur Child Adolesc Psychiatry 2000; 9: 115–121. [DOI] [PubMed] [Google Scholar]
- 159. Medland SE, Jahanshad N, Neale BM, et al. Whole-genome analyses of whole-brain data: working within an expanded search space. Nat Neurosci 2014; 17: 791–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Frank GK. Advances from neuroimaging studies in eating disorders. CNS Spectr 2015; 20: 391–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. King JA, Frank GKW, Thompson PM, et al. Structural neuroimaging of anorexia nervosa: future directions in the quest for mechanisms underlying dynamic alterations. Biol Psychiatry 2018; 83: 224–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. The ENIGMA Consortium. Enhancing Neuro Imaging Genetics by MetaAnalysis, http://enigma.ini.usc.edu/ (accessed 5 September 2018).
- 163. University of Bristol. Avon Longitudinal Study of Parents and Children, http://www.bristol.ac.uk/alspac/ (accessed 5 September 2018).
- 164. Norwegian Institute of Public Health. Norwegian Mother and Child Cohort Study, https://www.fhi.no/en/studies/moba/ (accessed 5 September 2018).
- 165. Thornton LM, Munn-Chernoff MA, Baker JH, et al. The Anorexia Nervosa Genetics Initiative (ANGI): Overview and methods. Contemp Clin Trials 2018; 74: 61–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Charlotte’s Helix Team. Charlotte’s Helix Project, https://www.charlotteshelix.net/ (accessed 5 September 2018).
- 167. Bulik C. The Binge Eating Genetics INitiative, https://walkforhope.com/begin-binge-eating-genetics-initiative/ (accessed 5 September 2018).
- 168. Thornton A, Lee P. Publication bias in meta-analysis: its causes and consequences. J Clin Epidemiol 2000; 53: 207–216. [DOI] [PubMed] [Google Scholar]
- 169. Duncan LE, Keller MC. A critical review of the first 10 years of candidate gene-by-environment interaction research in psychiatry. Am J Psychiatry 2011; 168: 1041–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Coburn KM, Vevea JL. Publication bias as a function of study characteristics. Psychol Methods 2015; 20: 310–330. [DOI] [PubMed] [Google Scholar]
- 171. Luborsky L, Barrett MS. The history and empirical status of key psychoanalytic concepts. Annu Rev Clin Psychol 2006; 2: 1–19. [DOI] [PubMed] [Google Scholar]
- 172. Steinberg H, Himmerich H. Roland Kuhn-100th birthday of an innovator of clinical psychopharmacology. Psychopharmacol Bull 2012; 45: 48–50. [PMC free article] [PubMed] [Google Scholar]
- 173. Schmidt FM, Kirkby KC, Himmerich H. The TNF-alpha inhibitor etanercept as monotherapy in treatment-resistant depression - report of two cases. Psychiatr Danub 2014; 26: 288–290. [PubMed] [Google Scholar]
- 174. Zheng P, Zeng B, Zhou C, et al. Gut microbiome remodeling induces depressive-like behaviors through a pathway mediated by the host’s metabolism. Mol Psychiatry 2016; 21: 786–796. [DOI] [PubMed] [Google Scholar]
- 175. Kantak PA, Bobrow DN, Nyby JG. Obsessive-compulsive-like behaviors in house mice are attenuated by a probiotic (Lactobacillus rhamnosus GG).Behav Pharmacol 2014; 25: 71–79. [DOI] [PubMed] [Google Scholar]