Abstract
Background & Aims:
Crohn’s disease (CD) presents as chronic and often progressive intestinal inflammation, but the contributing pathogenic mechanisms are unclear. We aimed to identify alterations in intestinal cells that could contribute to the chronic and progressive course of CD.
Methods:
We took an unbiased, system-wide approach, performing sequence analysis of RNA extracted from formalin-fixed, paraffin-embedded ileal tissue sections from patients with CD (n=36) and without CD (controls, n=32). We selected relatively uninflamed samples, based on histology, before gene expression profiling; validation studies were performed using adjacent serial tissue sections. A separate set of samples (3 controls and 4 CD) was analyzed by transmission electron microscopy. We developed methods to visualize an overlapping modular network of genes dysregulated in the CD samples. We validated our findings using biopsy samples (110 CD samples for gene expression analysis and 54 for histologic analysis) from the UNITI-2 phase 3 trial of ustekinumab for patients with CD and healthy individuals (26 samples used in gene expression analysis).
Results:
We identified gene clusters that were altered in nearly all CD samples. One cluster encoded genes associated with the enterocyte brush border, leading us to investigate microvilli. In ileal tissues from patients with CD, the microvilli were of reduced length and had ultrastructural defects, compared to tissues from controls. Microvilli length correlated with expression of genes that regulate microvilli structure and function. Network analysis linked the microvilli cluster to several other down-regulated clusters associated with altered intra-cellular trafficking and cellular metabolism. Enrichment of a core microvilli gene set was also lower in the UNITI-2 trial CD samples compared with controls; expression of microvilli genes correlated with microvilli length and endoscopy score, and associated with response to treatment.
Conclusions:
In a transcriptome analysis of formalin-fixed, paraffin-embedded ileal tissues from patients with CD and controls, we associated transcriptional alterations with histologic alterations, such as differences in microvilli length. Decreased microvilli length and reduced expression of the microvilli gene set might contribute to epithelial malfunction and the chronic, progressive disease course in patients with CD.
Keywords: Inflammatory bowel diseases, FFPE, next-generation sequencing, RNA-seq
INTRODUCTION
Crohn’s disease (CD) is a form of inflammatory bowel disease (IBD) primarily characterized by clinical symptoms and pathological signs, such as transmural influx of inflammatory cells 1. Even with treatment, CD subjects experience periods of remission and periods of active inflammation. Factors driving the chronic, relapsing nature of CD may include dysbiosis of the intestinal microbiome, which is a persistent pathological feature of CD intestine 2; but it is less clear if analogous persistent defects occur in host cells. We propose that uninflamed intestinal tissue regions will be a rich source for uncovering such defects that could contribute to the chronic nature of CD.
We adopted a molecular profiling approach to predict alterations in small intestinal host cells because this platform provides an unbiased, system-wide view of disease-associated processes and has been successfully applied to personalized medicine approaches for other diseases 3. However, the interpretation of transcriptional data from subjects with complex, inflammatory diseases is challenging, as the gene expression levels obtained from a heterogeneous sample are influenced by disease state as well as proportional abundances of constituent cells and relative transcript abundances within these cells 4. Immune cell infiltration as well as tissue damage or remodeling that can occur downstream of active inflammation influence bulk gene expression data. This confounding effect has been clearly demonstrated in molecular profiling studies of several complex inflammatory diseases, including scleroderma 5, rheumatoid arthritis 6, and CD 7-9. To overcome this challenge, we adopted a molecular profiling approach that allows researchers to match intact tissue morphology to gene expression data.
Formalin-fixed, paraffin-embedded (FFPE) tissue is the standard preservation method used for routine surgical pathology. Recent technical developments have enabled extraction of DNA, RNA and protein from FFPE tissues 10, thereby providing an abundant tissue source for genomic analyses of clinical samples. Several studies have compared transcriptional profiles generated from fresh frozen and FFPE samples from the same individual and found that the two preservation methods produced concordant data, despite the higher degree of RNA degradation in the FFPE samples 11-14. A key advantage of FFPE molecular profiling for complex, inflammatory disease samples is that it allows for definitive selection using histological criteria of the exact samples to be profiled. Traditionally, selection of relatively uninflamed intestinal tissues for CD molecular profiling studies relies on gross endoscopic evaluation and/or subsequent histological assessment of a tissue site near to the one used for RNA isolation. Furthermore, whereas the entire tissue sample is lysed during traditional RNA extraction methods, only a small portion of the FFPE tissue sample is used for RNA extraction. Thus, serial FFPE tissue sections are available to correlate gene expression findings with histological features predicted to be altered.
In this study, we generated RNA-seq transcriptional profiles using standard, archival FFPE pathology tissue specimens from CD and control subjects who underwent ileo-colic resection surgery. We chose resection samples because these contain whole-thickness tissue, including the muscularis propria, and thus all intestinal host cells could be examined (in contrast to biopsy samples). Histological assessment prior to molecular profiling was performed to select samples only from tissue regions uninvolved in active inflammatory disease based on the current clinical guidelines used by pathologists. We surmised that this pre-screening process would minimize molecular signatures driven by altered proportional abundances of cell subsets and potentially unmask those driven by underlying defects in host cells, especially if these consisted of small magnitude gene expression changes. Such defects might contribute to the chronic, progressive nature of CD. We identified several common gene signatures altered in CD intestine compared to controls, one of which led us to identify previously unrecognized morphometric features of CD, reduced length and altered ultrastructure of epithelial microvilli. We validated the reduced enrichment of this gene signature and its correlation with microvilli length in an independent cohort of CD patients with biopsy samples. Together, these data supported the hypothesis that there are persistent alterations of host cells in relatively uninflamed CD intestinal tissue.
METHODS
Additional details are provided in Supplementary Methods.
Study subjects and histological sample selection
We retrospectively obtained de-identified FFPE tissue samples and demographic data from subjects who underwent their first ileo-colic resection surgery at Barnes-Jewish Hospital, St. Louis between 2006 and 2010. The set of H&E-stained tissue sections from each resection were examined by two pathologists to identify the terminal ileum, which was subjected to additional histological criteria to select samples with intact mucosa and minimal to absent active or chronic inflammatory disease. For transmission electron microscopy (TEM), ileal tissues were obtained from resection or biopsy specimen regions that were visually and endoscopically uninvolved with active disease. For the UNITI-2 cohort 15, ileal biopsies from patients with ileal or ileo-colonic CD were obtained from representative regions as determined by endoscopy. One set of biopsies was prepared for microarray and the other for FFPE H&E-stained tissue sections. Ileal biopsies from 26 healthy subjects were used as a control group for the gene expression analysis. Written informed consent was obtained from all study participants prior to inclusion in these studies, which were approved by the IRB.
Molecular profiling
For the resection specimens, two unstained 5-μm FFPE tissue sections were used for RNA extraction and DNase treatment with the RNeasy FFPE kit (Qiagen) according to the manufacturer’s instructions. The Genome Technology Access Center (GTAC) at Washington University performed RNA-seq library preparation, sequencing and read alignment. Sequencing was performed on an Illumina HiSeq2000 SR42 using single reads extending 42 bases. Microarrays were performed as previously described 16. For the biopsy specimens, ileal biopsies were stored in RNAlater at −80°C until RNA extracti on with the RNeasy Fibrous Tissue Mini Kit (Qiagen) and hybridization to Affymetrix GeneChip Human Genome U133 PM arrays.
Partitional clustering and functional enrichment analyses
The mapped gene-level transcript matrix file was filtered to exclude transcripts with a mean of 0 cpm and those that were “non-protein coding and non-annotated”. An unpaired twotailed Mann-Whitney t test was used to identify annotated, protein-coding transcripts with an unadjusted P <.05 between CD and Non-IBD to use as the input for principal components analysis (PCA) and fuzzy c-means partitional clustering, which were performed with Partek® Genomics Suite® software v6.6 Copyright © 2017 (St. Louis, MO, USA). Enrichr (http://amp.pharm.mssm.edu/Enrichr/) 17, the Human Protein Atlas (HPA) v16.1 18 and published brush border proteomics studies 19, 20 were used to assess functional enrichment.
Imaging
Image acquisition and quantification was performed in a blinded fashion for all histological metrics. Brightfield images were acquired with an Olympus BX51 microscope equipped with UPlanFL 10X/0.30, 20X/0.50, 40X/0.75, and 100X/1.30 Oil Iris objective lenses, an Olympus DP70 or DP22 camera and DP Controller or cellSens Standard v1.17 software. Confocal images were obtained with a Zeiss LSM880 laser scanning confocal microscope equipped with a 63X/1.4 Zeiss Plan Apochromat oil objective lens. TEM images were acquired with a JEOL model 1400EX electron microscope and AMT Advantage HR (Advanced Microscopy Technology) high definition CCD, 11 megapixel TEM camera. Adobe Photoshop CS6 was used to adjust brightness and contrast, sharpen and crop images.
Co-expression module network
A co-expression network was constructed using a novel approach termed correlation graphlet analysis (CGA). A full description of CGA as well as the computer code and sample data are provided as supplementary materials. The correlation gene network contained 1,326 nodes and 39,567 edges prior to compression into modules. Module construction resulted in 84 modules (29 of which contained at least 10 genes) connected by 87 edges. The networks were visualized with Cytoscape 21 using a force-directed layout.
Statistics
Graphpad Prism v7 was used to perform statistical analysis with P <.05 considered to be significant, unless otherwise indicated. Parametric or nonparametric statistical tests were applied as appropriate after testing for normal distribution of data, as described in the figure legends. Transcriptional enrichment scores for the UST phase 3 microarray dataset were computed with Gene Set Variation Analysis (GSVA) 22 and correlated to the Simple Endoscopic Score for Crohn’s Disease (SES-CD) with Spearman correlation. Differences in enrichment scores between phenotypes were analyzed using parametric tests in ArrayStudio v10 (OmicSoft Corp).
RESULTS
Selection of tissue samples for transcriptional profiling
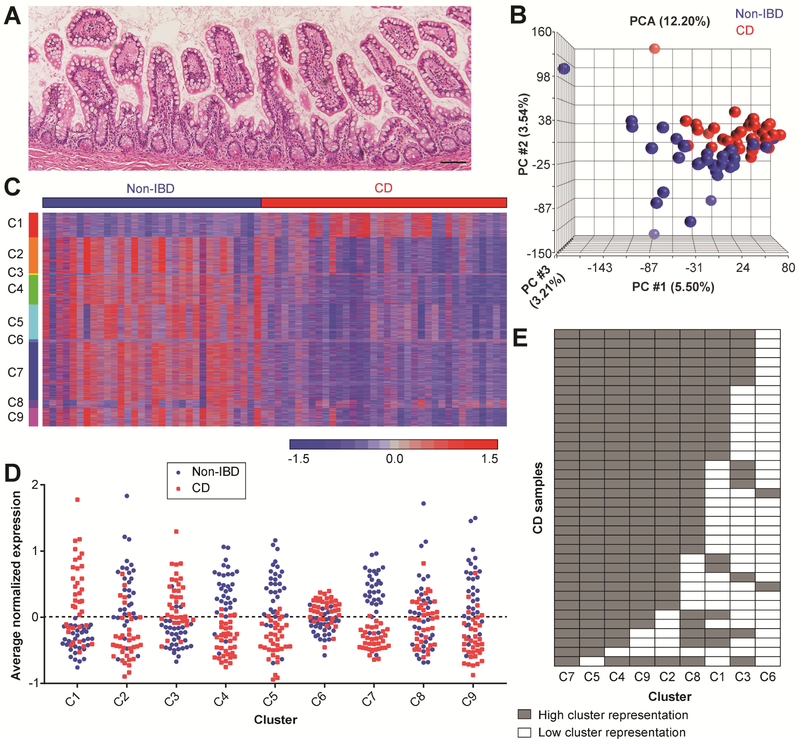
We retrospectively procured FFPE intestinal tissue from CD subjects and control subjects without IBD (Non-IBD) who underwent surgical resection at Washington University. Histological screening of H&E-stained tissue sections was performed to identify ileal tissue regions with relatively uninflamed mucosa, which we surmised would be most likely to harbor early molecular changes contributing to disease chronicity. We identified 68 samples (one per subject) with absent to minimal active or chronic inflammatory disease as defined by well-accepted clinical pathology parameters (Figure 1A; Figure S1A) and passed RNA quality control metrics (Figure S1B-E). There was no difference in the proportion of Non-IBD (n=32) and CD (n=36) samples that passed quality control (P =.4747 by Fisher’s exact test). The demographic data for these CD subjects were comparable to the spectrum of data previously reported 23, 24 (Table S1). The average age at the time of surgery was significantly different between the Non-IBD and CD groups (Table S1); however, no significant differences in transcript abundances were identified within either diagnosis group based on this variable. Principal components analysis (PCA) of the gene expression data showed distinct but overlapping grouping of the samples based on diagnosis (Figure 1B). No further subgroups were apparent.
Figure 1. Identification of common gene clusters in non-inflamed ileal resection tissue samples from CD subjects.
(A) Representative image of H&E-stained ileal resection margin tissue that met the histological inclusion criteria for this study. Bar, 100 μm. (B) PCA of the CD (n=36; red) and Non-IBD (n=32; blue) samples included in the final gene expression analysis. (C) Intensity plot of normalized mRNA expression levels for 6,493 genes grouped by the 9 clusters (labeled C1-C9) generated with partitional clustering. (D) Graph of the average normalized gene expression for each sample for Clusters 1-9. The cluster centroid is at 0; values greater than 0 are relatively up-regulated and values less than 0 are relatively down-regulated in CD compared to Non-IBD. (E) A binary representation of the data in (D). Cells are colored gray if the cluster has high representation in a sample (≥∣0.2∣ average normalized gene expression in ≥40% of the cluster genes).
Identification of common molecular features in Crohn’s disease
To gain biological insight into the molecular features that distinguished the CD from the Non-IBD samples, we used a broad set of differentially expressed genes as input for fuzzy cmeans partitional clustering. Of the 9 gene clusters generated, 4 were relatively up-regulated (1, 3, 6, 8) and 5 were relatively down-regulated (2, 4, 5, 7, and 9) in CD vs. Non-IBD (Figure 1C, Table S2). Although this algorithm allows genes to belong to multiple clusters, we observed very little overlap (Table S3). Human transcriptional profiling data reflects inter-individual variation due to genetics, environmental exposures, disease state, etc. 4, 25. Thus, as expected, the cluster genes were variably expressed among the samples, resulting in varying cluster representation. We were most interested in identifying common host cell alterations, so we assessed cluster representation first by calculating the average normalized expression of the genes forming each cluster (Figure 1D). Clusters 4, 5 and 7 were most commonly altered in CD samples (i.e. had the greatest degree of separation between Non-IBD and CD). We next determined whether the average cluster gene expression resulted from many genes with potentially small but consistent expression changes or from a small number of genes with large expression changes. For this, we generated an arbitrary, binary cut-off for the cluster representation of each sample: a highly represented cluster had at least 40% of its genes with >∣0.2∣ normalized gene expression difference (Figure 1E). This analysis again showed that Clusters 4, 5 and 7 were most commonly altered in CD, with 97%-100% of the samples meeting or exceeding these criteria, indicating many genes having potentially small, but consistent, changes in expression. Functional enrichment analysis (selected results in Table 1; full results with statistics in Table S4) showed that Clusters 4 and 7 were enriched for gene ontology (GO) cellular component terms related to mitochondria and pathways related to the tricarboxylic acid (TCA) cycle. Cluster 7 was also enriched for terms related to cellular trafficking. Cluster 5 was enriched for several metabolism pathways, cell-cell junction organization, and the GO cellular component terms plasma membrane, brush border and cell-cell junction.
Table 1.
Selected functional enrichment of partitional cluster genes.
| Cluster | # of Genes |
Pathway | GO cellular component |
Tissue |
|---|---|---|---|---|
| 1 | 743 | Complement cascade, innate immune system, response to hypoxia, TNF signaling, IFN signaling | Extracellular vesicle, nucleoplasm, cytosol | Small intestine, whole blood |
| 2 | 1103 | Chromatin organization, cell cycle, gene expression, mRNA processing | Nucleolus, cytosol, ribosome | CD8+ T cells, CD4+ T cells |
| 3 | 43 | None | Mitochrondrion, intracellular organelle | None |
| 4 | 902 | Respiratory electron transport, glycolysis, TCA cycle | Mitochrondrion, intracellular organelle | None |
| 5 | 1056 | Metabolic pathways, drug metabolism, bile secretion, fatty acid degradation, cell junction organization, PPAR signaling | Plasma membrane, brush border, cell-cell junction | Small intestine |
| 6 | 89 | Netrin-1 signaling, complement cascade | None | None |
| 7 | 1758 | TCA cycle, VEGF signaling, membrane trafficking | Mitochrondrion, cell-substrate junction, vesicle membrane | None |
| 8 | 243 | Focal adhesion, extracellular matrix organization, integrin-mediated adhesion, TGFb signaling | Extracellular matrix, basement membrane | Smooth muscle, adipose |
| 9 | 556 | mRNA processing, translation, ribosome, spliceosome | Nucleolus, cytosol, ribosome | None |
Microvilli gene expression and length are reduced in uninflamed Crohn’s ileum
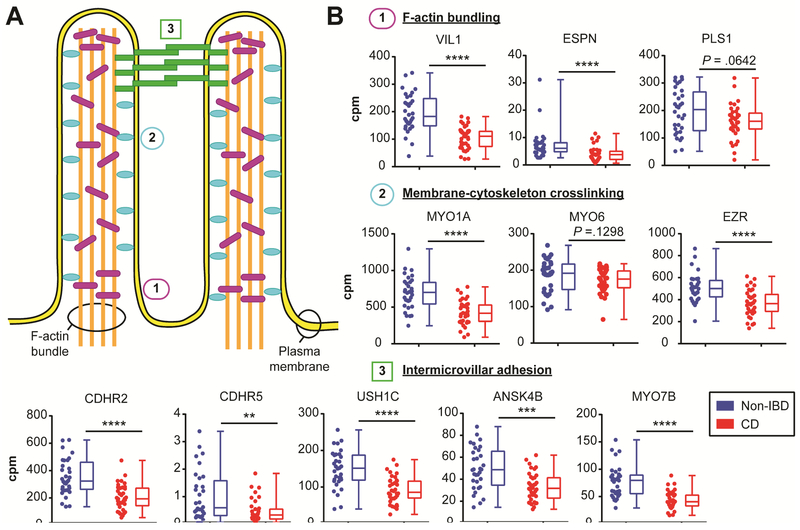
We performed a more focused analysis of Cluster 5 because it was commonly altered in CD and associated with small intestine tissue. Many of the Cluster 5 genes are well-known to be specifically or highly expressed in enterocytes, including alkaline phosphatase (ALPI) 26, epithelial cell adhesion molecule (EPCAM) 27, fatty acid binding protein 2 (FABP2) 28, keratin 20 (KRT20) 29 and villin 1 (VIL1) 30. To determine if there was indeed an enrichment for enterocyte genes in this gene set, we utilized the HPA 18 and published literature to examine the products of a subset of Cluster 5 genes with reliable protein localization data in the small intestine (see Supplementary Methods; Table S5). Of these 172 proteins, all were expressed in epithelium, with 171 expressed in enterocytes and 1 expressed in goblet cells. The majority (79%; 136/172) were enriched in epithelial compared to non-epithelial cells, and 44% (75/172) were enriched specifically at the enterocyte brush border. The brush border membrane is organized into apical F-actin-supported protrusions called microvilli that increase the surface area and functional capacity of enterocytes lining the intestinal tract 31. This analyses strongly indicated microvilli as a feature for further exploration. We further validated the brush border enrichment by comparing Cluster 5 to a gene set identified from 2 proteomic studies of mouse brush border 19, 20. We observed significant overlap between these gene sets (P <1.66 × 10−45 by hypergeometric distribution test) (Figure S2A). The formation, organization and stabilization of microvilli is achieved through several molecular mechanisms, including F-actin bundling, membranecytoskeleton crosslinking, and intermicrovillar adhesion 31 (Figure 2A). Nearly all of the genes known to participate in these molecular functions had decreased expression in CD compared to Non-IBD samples (Figure 2B), which we confirmed by microarray (Figure S2B), and belonged to Cluster 5.
Figure 2. Decreased expression of genes related to microvilli formation in CD intestine.
(A) Schematic of intestinal microvilli structure highlighting the F-actin bundling components (1; purple rods), membrane-cytoskeleton crosslinking (2; blue ovals), and intermicrovillar adhesion complex (3; green rectangles). (B) Graphs of RNA-seq gene expression levels displayed as counts per million (cpm) for the indicated microvilli component genes. The same data is displayed side-by-side as scatterplots and box-and-whiskers plots for CD (n=36; red) and Non-IBD (n=32; blue). **P <.01, ***P <.001, ****p <.0001 by unpaired twotailed Mann-Whitney t-test. The exact P-value is indicated for non-significant comparisons.
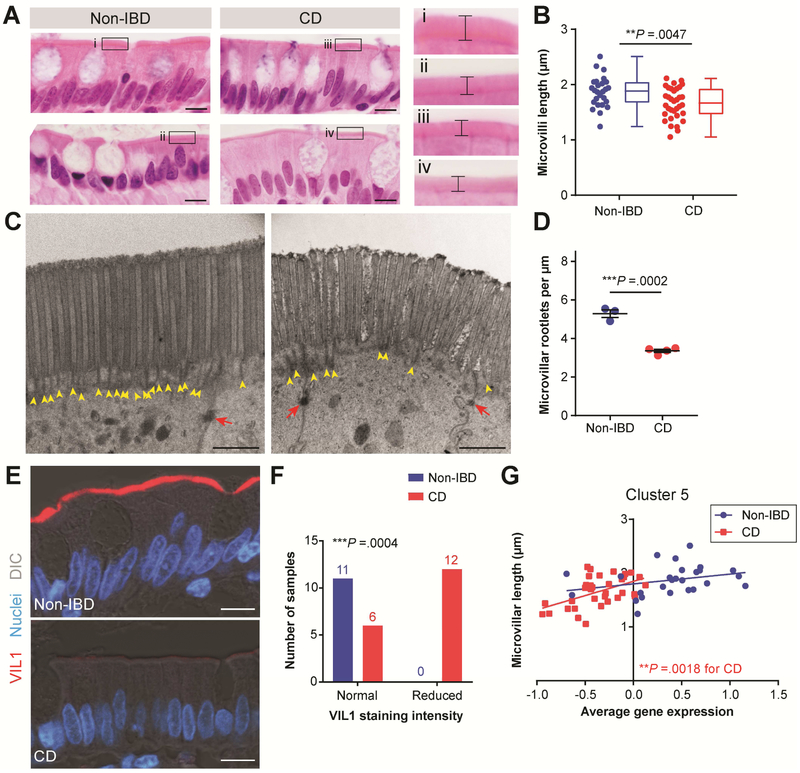
To determine if the transcriptional alterations in microvilli-associated genes corresponded to histological alteration of microvilli, we performed a quantitative analysis of microvilli length using serial tissue sections adjacent to those used for transcriptional analysis. CD enterocytes had a ~11% decrease in average microvilli length compared to Non-IBD (Figure 4A, B). To support our ability to precisely determine average microvilli length, we performed rigorous statistical validation of our sampling method (Figure S3 and Figure S4) and demonstrated high inter-observer reproducibility (Figure S5). We next performed transmission electron microscopy (TEM) of enterocytes from a separate set of Non-IBD and CD uninvolved ileal samples to determine if there might be an underlying ultrastructural alteration of microvilli in CD. In CD samples, we observed a significant reduction in the density of the microvilli rootlets, which are bundles of actin filaments that extend into the apical cytoplasm (Figure 3C, D and Figure S6A, C), whereas other ultrastructural features were similar compared to Non-IBD (Figure S6B). Microvilli rootlets were easily identifiable, electron dense structures in Non-IBD enterocytes; however, the electron dense nature of the rootlets appeared qualitatively reduced in the CD samples (Figure 4C, Figure S6A), potentially reflective of reduced actin bundling. As a staining control, desmosomes appeared similar between CD and Non-IBD. Accordingly, we next assessed protein localization of the actin bundling protein VIL1 with confocal microscopy using serial tissue sections to those used for FFPE RNA extraction (Figure 3E, F). VIL1 protein staining was intensely and consistently localized to the epithelial brush border in Non-IBD samples. In contrast, VIL1 staining was strongly reduced in many CD samples, despite these samples being stained at the same time and imaged with the same settings as the Non-IBD samples. This observation was consistent with the reduced VIL1 mRNA expression (Figure 2B) and potential alteration in microvilli rootlet actin bundling in CD samples. Overall, these data suggest that the microvilli gene expression and length phenotypes might be indicators of a potentially multi-factorial defect occurring at the enterocyte apical surface in CD.
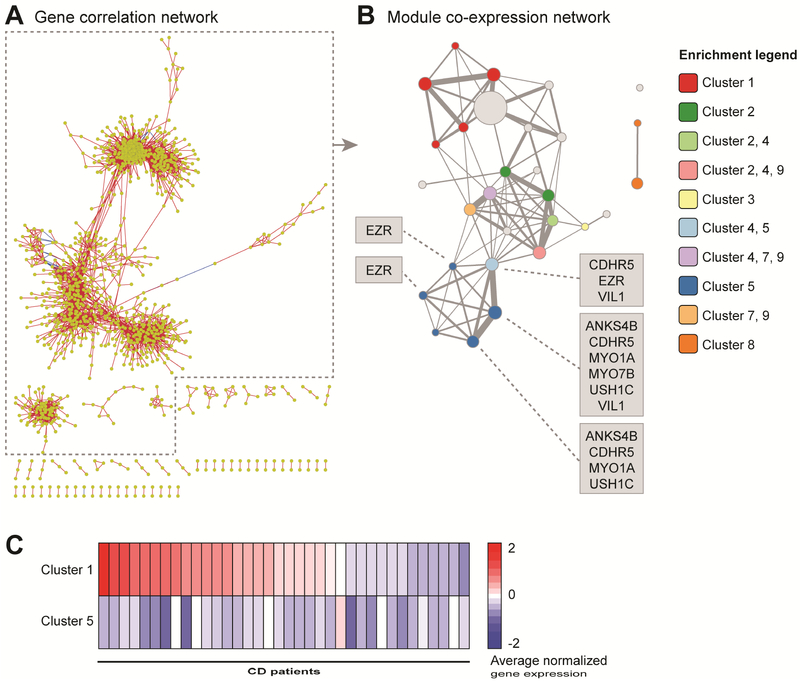
Figure 4. The common CD gene clusters are connected in a modular network.
(A) Visual organization of the gene correlation network with nodes representing genes and edges representing an absolute Spearman correlation of ≥0.75 between genes. (B) Visual organization of the co-expression module network with nodes representing modules and edges representing shared genes between modules. Node size and edge thickness are relative to the number of genes represented by each. The node color indicates significant enrichment by one-sided hypergeometric test for partitional cluster genes, as described in the legend. (C) Intensity plot of the average normalized gene expression values for partitional Cluster 1 and Cluster 5 in CD samples. The samples are arranged in the same order for each gene cluster according to their Cluster 1 rank.
Figure 3. Decreased microvilli length in CD correlates with Cluster 5 average gene expression.
(A) Representative images of H&E-stained ileal tissue regions used to measure microvilli length. Bars, 10 μm. A magnified view of the boxed region in each image is shown to the right; brackets indicate microvilli length. (B) Graph of average microvilli length (μm) with the same data displayed side-by-side as scatterplots and as box-and-whisker plots for Non-IBD (n=26; blue) and CD (n=34; red) samples. **P =.0047 by unpaired two-tailed Mann-Whitney t-test. (C, D) TEM images of enterocytes captured from the upper portion of villi from Non-IBD (n=3) and CD (n=4) samples. (C) Representative images of the epithelial apical surface. Yellow arrowheads designate rootlet ends and red arrows designate desmosomes. Bars, 1 μm. (D) Graph of average microvilli rootlet densities quantified from TEM images and displayed as mean ± s.e.m. ***P =.0002 by unpaired two-tailed parametric t test. (E) Representative confocal images of Non-IBD and CD intestinal epithelial cells co-stained for VIL1 (red). Nuclei are visualized with bisbenzimide (blue). Tissue structure is visualized with a DIC image overlay (gray). Bars, 10 μm. (F) Contingency graph of Non-IBD and CD samples with normal or reduced VIL1 staining intensity. ***P =.0004 by Fisher’s exact test comparing Non-IBD and CD (sample size is indicated above bars). (G) X-Y graph of average microvilli length and Cluster 5 average normalized gene expression for the samples in (B). CD: **P =.0018 and r2=.2666 and Non-IBD: P =.1444 (not significant) and r2=.0866 by linear regression.
Correlation of microvillar gene signature expression and length
A deeper probe into the relationship between our gene expression and histological findings showed that microvilli length was directly correlated to the average Cluster 5 gene expression in CD samples (Figure 4F) and inversely correlated to the average gene expression of Clusters 2 and 9 (Figure S7A). In contrast, average enterocyte cell height was similar between CD and Non-IBD samples and did not correlate with Cluster 5 gene expression (Figure S7B, C). Because altered gene expression patterns associated with a particular cell type can arise if the proportion of constituent cells are altered within heterogeneous tissues, we investigated whether a proportional change in enterocytes or other cell type could account for Cluster 5. Our histological inclusion criteria selected for samples with generally intact epithelium, and, supporting this, we identified several enterocyte marker genes in our RNA-seq data set which were not changed in CD compared to Non-IBD (Figure S8). We also examined the proportions of goblet cells and Paneth cells, which have been implicated in CD pathogenesis 32 and are the next most common epithelial subsets in the small intestine. Neither the number of these cells nor the gene expression of their markers correlated with Cluster 5 (Figure S9, Figure S10). Taken together, these data highlight the specificity of the correlation between Cluster 5 and microvilli length.
Network analysis shows common molecular features of Crohn’s disease are linked
To investigate potential co-regulation of diminished and elevated gene expression in CD relative to Non-IBD samples, we performed network analysis. We first constructed a gene correlation network with a more stringent cutoff than used for the partitional clustering. However, interactions were still difficult to visualize or interpret due to the complexity of the network (Figure 4A). Therefore, we developed a novel network analysis approach termed correlation graphlet analysis (CGA). With this approach, simpler networks are produced by empirical selection of a correlation threshold followed by collapsing correlated genes into modules and constructing the modules into a network where edges represent overlapping genes between modules (see Supplementary Methods and Figures S11 and S12). The resulting module network consisted of a large, primary component composed of three highly connected regions as well as some smaller, isolated components (Figure 4B and Table S6).
We used the module network to perform an independent validation of the partitional clustering results and to investigate relationships between the gene clusters by determining which network modules significantly overlapped with the cluster genes (Figure 4B, Figure S13). Several interconnected modules were highly enriched for Cluster 5 genes, including the microvilli structural component genes. The Cluster 5-enriched region connected to a second network region containing modules with overlapping enrichment for the genes of the commonly down-regulated Clusters 2, 4, 7 and 9, which were functionally enriched for cellular trafficking, cellular metabolism, TCA cycle and mitochondria-related genes. The interconnectedness of these regions suggests these gene expression alterations and cellular functions might be coupled to each other and potentially to microvilli length. The third region of the large, connected component of the module network was enriched for Cluster 1, a cluster relatively up-regulated in CD and enriched for immune-related terms, including innate immune response as well as TNF and IFN signaling. This region was indirectly connected to the Cluster 5-enriched region through the Cluster 2-4-7-9-enriched region. As Cluster 1 was represented only in ~53% of the CD samples in contrast to the down-regulated clusters being represented in nearly all CD samples, it is unclear whether these up-regulated genes were a cause or consequence of the down-regulated gene modules. Supporting a lack of direct linkage between Clusters 1 and 5, only a few subjects demonstrated an inverse correlation between partitional Cluster 1 and Cluster 5 average gene expression, whereas the majority of the samples did not exhibit this relationship (Figure 4C). Therefore, the evidence does not support the gene expression differences in Cluster 1 being causal of those in Cluster 5.
The microvilli signature associates with microvilli length and CD clinical parameters in an independent cohort with biopsy samples
Our findings with the resection cohort raised several questions: can these findings be corroborated in an independent cohort; can biopsy material be utilized; and what is the potential clinical relevance of this transcriptional signature and microvilli defect in CD? To address these questions, we performed a post hoc analysis of gene expression and microvilli length data from biopsy specimens obtained from the UST phase 3 studies in CD patients who had failed conventional therapies (UNITI-2 cohort). UST phase 3 studies established that UST, a monoclonal antibody to the p40 subunit of interleukin-12 and interleukin-23, is an effective treatment for moderate-to-severe CD 15. Here, we analyzed two subsets of patients from the full UNITI-2 cohort that had ileal or ileal-colonic disease and had endoscopic biopsy samples that had been processed for gene expression and/or histological analyses. These patient subsets had similar demographics and disease characteristics to the full UNITI-2 cohort (Table S7), with the exception that fecal calprotectin levels were significantly lower in the two subsets. Additionally, the histological subset had significantly lower endoscopic disease activity in the terminal ileum compared to the gene expression subset, likely a result of the inclusion criteria used to select these subsets.
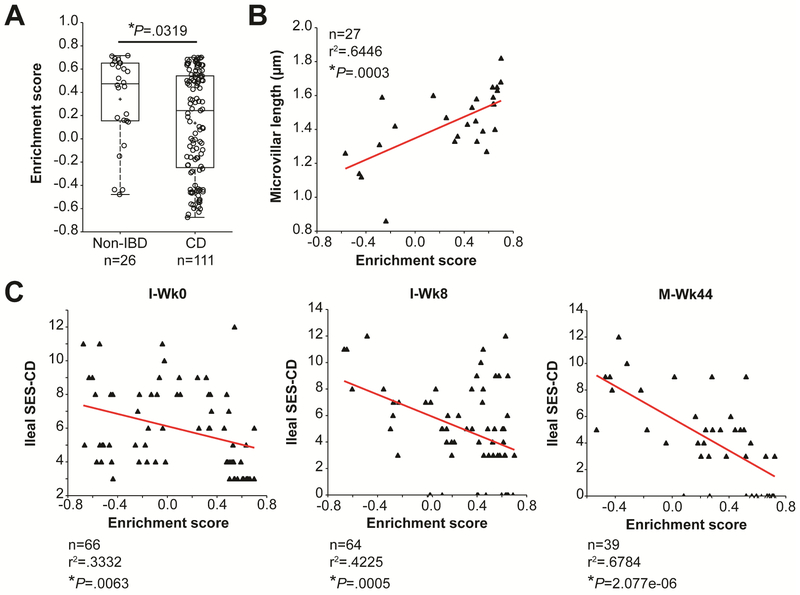
Gene set variation analysis (GSVA) is a method to assess the enrichment of a gene set across a study population in an unsupervised manner 22. We performed GSVA using the gene set generated by the overlap of Cluster 5 with the co-expression module network (herein referred to as the core Cluster 5 gene set; Table S8) because the latter had been generated with more stringent criteria than the partitional clustering. The enrichment score for the core Cluster 5 gene set was significantly lower in CD versus healthy control biopsies (Figure 5A) and was also significantly correlated with microvilli length (Figure 5B). These results using biopsy samples corroborated the findings in the resection specimens.
Figure 5. Core Cluster 5 expression and microvilli length correlate in an independent, phase 3 clinical trial CD cohort with ileal biopsy samples.
(A) Graph of enrichment scores from GSVA of the core Cluster 5 gene set in the UNITI-2 gene expression subset and healthy controls. (B) X-Y graph of GSVA enrichment scores and average microvilli lengths for the UNITI-2 histology subset. (C) X-Y graphs of GSVA enrichment scores and SES-CD scores at the indicated time points. An unpaired two-tailed parametric t test (A) and Spearman’s correlation (B, C) were used to determine statistical significance.
An additional advantage of the UNITI-2 cohort is that longitudinal samples and robust clinical information are available. For example, disease severity was determined endoscopically by the SES-CD score 33 at three time points: induction baseline (I-Wk0), induction week 8 (I-Wk8), and maintenance week 44 (M-Wk44). The SES-CD score negatively correlated with the core Cluster 5 gene set enrichment score at all 3 time points (Figure 5C), demonstrating that worse disease (i.e. high SES-CD score) was associated with de-enrichment of the core Cluster 5 gene set. Conversely, minimal disease activity (i.e. lower SES-CD score) was associated with gene set enrichment scores more similar to the healthy control samples. We observed a stronger correlation at M-Wk44 compared to I-Wk8 or I-Wk0, likely due to the larger dynamic range of the data following UST treatment.
Impact of UST on the microvilli gene signature and microvilli length
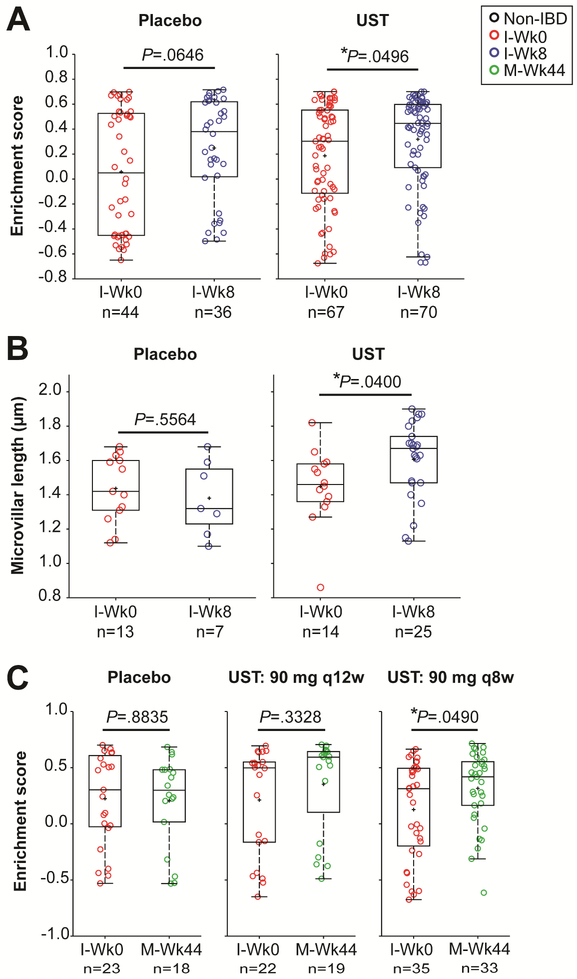
We next tested if core Cluster 5 gene set enrichment differed between placebo and UST-treated subjects. In subjects who received UST induction therapy, the enrichment score was higher (i.e. more similar to Non-IBD controls) at I-Wk8 compared to I-Wk0 (Figure 6A). A significant increase in the average microvilli length was also observed in the UST-treated subjects at I-Wk8 compared to baseline (Figure 6B). Patients who received placebo did not show significant alterations in core Cluster 5 gene set enrichment or microvilli length (Figure 6B, C). Similar effects were observed with gene set enrichment in patients at M-Wk44 who received UST maintenance therapy at 8-week intervals (Figure 6C), but the sample size (n=5) was insufficient to assess microvilli length at the M-Wk44 time point.
Figure 6. Ustekinumab (UST) induction and maintenance therapies impact core Cluster 5 gene expression and microvilli length.
(A, B) Graphs of the enrichment scores for the core Cluster 5 gene set in Placebo and UST-treated samples at I-Wk0 vs. I-Wk8 (A) and at I-Wk0 vs. M-Wk44, with two different dosing regimens (90 mg q12w or 90 mg q8w) used during weeks 8 to 44 (B). (C) Graphs of the average microvilli lengths in Placebo and UST-treated samples at I-Wk0 vs. I-Wk8. (A-C) An unpaired two-tailed parametric t test was used to determine statistical significance.
Because the gene set enrichment changed in patients who received UST, we next evaluated whether this was associated with clinical response to UST therapy. In the UST phase 3 study, patients were defined as responders if they experienced a decrease of 100 points from their baseline value or a value <150 by the CDAI. We observed that CDAI-defined responders had gene set enrichment scores similar to Non-IBD controls at I-Wk0 and I-Wk8, and thus, these enrichment scores did not significantly change between the I-Wk0 and I-Wk8 time points (Figure S14A). Gene set enrichment scores also did not significantly change in CDAI-defined responders or non-responders who received placebo (Figure S14A). In contrast, gene set enrichment scores improved in the non-responders between these time points, despite the lack of clinical improvement. A difference in core Cluster 5 gene set enrichment was observed between CDAI-defined responders and non-responders at I-Wk0 (Figure S14B), prior to UST therapy, whereas CDAI and SES-CD scores were similar between these groups at baseline (Figure S14C, D). Sample size was insufficient to perform a similar analysis of the NR and R groups at the M-Wk44 time point. These data suggest that core Cluster 5 gene enrichment has potential to discriminate between CDAI-defined UST responders and non-responders.
DISCUSSION
We performed RNA-seq profiling of ileal tissue from CD and control subjects to determine if persistent defects in host cells were present in subjects who underwent resection surgery. Such defects may underlie the chronic, relapsing or progressive nature of CD. One advance of this study was the use of FFPE molecular profiling technology, which allowed us to histologically select samples with relatively uninflamed mucosa and minimize potential confounding effects due to the altered cellular proportions. Because only a small portion of the tissue sample was required for RNA extraction, serial tissue sections from the same set of FFPE tissue blocks could be used for direct histological validation of identified transcriptional signatures. The power of such definitive, histological validation is exemplified by our discovery of reduced microvilli length in CD subjects, a histologic feature that directly correlated to the enrichment for an enterocyte-associated gene signature. A similar approach combining histological screening and FFPE transcriptomics could aid future studies examining gene expression in heterogeneous tissue samples from subjects with complex, inflammatory diseases.
We identified several common molecular signatures in the CD resection tissue samples. These signatures were comprised of many genes with small, but consistent alterations in expression, a pattern reminiscent of the adult CD genetic landscape, where a large number of genetic polymorphisms are thought to have small, but cumulative effects 34. We focused our validation efforts on Cluster 5 because it was a common molecular signature in the CD subjects that localized to a unique region in the module network and enrichment analysis pointed towards a particular cell type and subcellular location, the enterocyte brush border. This gene set was relatively de-enriched in CD compared to controls in our surgical resection cohort, and we validated this result in an independent cohort with biopsy samples. The extension of our findings to biopsy samples establishes that Cluster 5 gene expression de-enrichment is not limited to “late-stage disease” complicated by stenosis or fistula and requiring surgical intervention. Furthermore, this observation demonstrates that the gene signatures identified by the FFPE transcriptional profiling approach (i.e. purposeful sampling normalization via histological selection) can subsequently be assessed in data sets generated from samples that did not undergo the same histological selection process. The additional analysis with the UNITI-2 UST CD phase 3 trial specimens also suggested that core Cluster 5 gene set enrichment and microvilli length may have utility in discriminating responses to therapy; however, additional analysis in other drug trial cohorts is required. These CD-associated phenotypes should be further explored as potential objective endpoints.
Our analysis of the Cluster 5 gene set led to an informed pathological analysis of enterocyte microvilli length, which we discovered to be reduced in CD compared to controls. TEM analysis further demonstrated a deficit in the rootlets of CD microvilli. A similar, although more extreme, phenotype was observed in a mouse model deficient for expression of Plastin 1 (Pls1), which had a 20% reduction in microvilli length and lacked microvilli rootlets 35. Despite this relatively subtle microvillar phenotype, the Pls1-deficient mouse displayed increased damage in the DSS intestinal injury model. The biological consequence of reduced microvilli length in CD subjects could not be inferred from the samples in this study; however, the literature supports that reduced microvilli length and stability are also detrimental to human intestinal homeostasis. For example, microvillous inclusion disease (caused by deleterious gene mutations in Myosin 5b (MYO5B) and in Syntaxin 3 (STX3)) presents as patchy loss of microvilli and is associated with persistent diarrhea and failure to thrive 36, 37. USH1C, which has recently been reported to be a critical organizer of the intermicrovillar adhesion complex that stabilizes the brush border 38, 39, is one of the causative genes for Usher syndrome. Usher syndrome is primarily associated with deafness, but a subset of patients exhibit enteropathy 40, 41. In this study, PLS1, MYO5B, STX3, and USH1C all had diminished expression in CD relative to Non-IBD and belonged to Cluster 5. Together, these studies indicate that epithelial microvilli should be explored further to determine how defects in these structures might contribute to the chronic, relapsing nature CD.
Module network analysis enabled us to intuit relationships between the gene clusters identified in this study. We found that the microvilli cluster (Cluster 5) directly connected to a network region enriched for the other commonly down-regulated gene clusters in CD, suggesting these gene expression alterations occurred in the same cell type or were caused by a similar biological driver. As many of the cellular functions associated with these modules cannot be measured histologically, the measurement of microvillar length could represent a surrogate histological feature that synthesizes the effects of a multitude of gene regulatory changes. We were not able to definitively distinguish the biological driver of the down-regulated gene expression clusters; however, one possibility is that they are driven by inflammatory mediators originating from regions of active disease elsewhere in the gut in a “field effect”. Supporting this notion, the up-regulated genes associated with immune activation in Cluster 1 were linked to the down-regulated gene regions in the network. Cluster 1 was only strongly represented in about half of the study samples, but a low level of inflammatory activity could potentially drive the microvillar alterations. Alternatively, as microvilli are plastic and remodel in response to the cellular environment 42, non-host factors also represent potential biological drivers of the observed gene expression alterations. Nutritional signals, such as the fasting and refeeding cycle in reptiles 43, total parenteral nutrition 44, and small bowel resection 45 as well as altered microbiota composition 46 have all been reported to affect microvilli length. This study emphasizes the need to develop additional experimental model systems to study the biological drivers and consequences of microvillar alterations in intestinal disease.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank the members of the Washington University core facilities, in particular, the Digestive Disease Research Core Center for subject recruitment and provision of study samples, the Genome Technology Access Center in the Department of Genetics for assistance with genomic analysis, and Karen Green in the Electron Microscopy Facility in the Department of Pathology and Immunology for assistance with TEM.
Grant support:
This work was supported by a grant from the Crohn’s and Colitis Foundation to T. S. Stappenbeck. The GTAC is partially supported by the NCI (P30 CA91842 to the Siteman Cancer Center) and the NIH National Center for Research Resources (UL1 TR000448 to the Washington University ICST/CTSA) and the NIH Roadmap for Medical Research. The Washington University Digestive Disease Research Core Center (DDRCC) is supported by grant P30DK052574 from the NIDDK. K. L. VanDussen was supported by a Crohn’s and Colitis Foundation Research Fellowship Award (# 290895) and a K01 from the NIH (DK109081). M.A. Ciorba is supported by the NIDDK (DK109384), the Daniel H. Present Senior Research Award from the Crohn’s and Colitis Foundation, and the philanthropic Givin’ it all for Guts Foundation.
Abbreviations:
- CD
Crohn’s disease
- CGA
correlation graphlet analysis
- FFPE
formalin-fixed, paraffin-embedded
- GO
gene ontology
- GSVA
gene set variation analysis
- HPA
Human Protein Atlas
- H&E
hematoxylin and eosin
- IBD
inflammatory bowel disease
- MVID
microvillous inclusion disease
- Non-IBD
control subjects without inflammatory bowel disease
- PCA
principal components analysis
- TCA
tricarboxylic acid
- TEM
transmission electron microscopy
- UST
ustekinumab
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures:
K.L.V, T.C.L., R.D.H. and T.S.S. have filed a provisional patent related to this work.
A.S., K.L., J.G.P., J.R.F and J.E.T. are employees of Janssen Research and Development, LLC.
P.K.K., B.D.M, K.F.S., and M.A.C. have nothing to disclose.
Transcript profiling: Sequencing files and microarray data from the resection specimens are deposited at ArrayExpress with the accession numbers E-MTAB-5783 and E-MTAB-5790, respectively. Microarray data from the biopsy specimens are deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus with the accession number GSE112366.
Reviewers can access these data sets with the following log-in credentials.
RNA-sequencing data from the resection specimens
Username: Reviewer_E-MTAB-5783
Password: 31ppX9ER
Microarray data from the resection specimens
Username: Reviewer_E-MTAB-5790
Password: ovxyJmzz
Microarray data from the biopsy specimens
Link: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE112366
Password: yzgtymusnjcbvyh
Writing assistance: Not applicable.
REFERENCES
- 1.Abraham C, Cho JH. Inflammatory bowel disease. N Engl J Med 2009;361:2066–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sartor RB, Wu GD. Roles for Intestinal Bacteria, Viruses, and Fungi in Pathogenesis of Inflammatory Bowel Diseases and Therapeutic Approaches. Gastroenterology 2017;152:327–339 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cardoso F, van’t Veer LJ, Bogaerts J, et al. 70-Gene Signature as an Aid to Treatment Decisions in Early-Stage Breast Cancer. New England Journal of Medicine 2016;375:717–729. [DOI] [PubMed] [Google Scholar]
- 4.Shen-Orr SS, Gaujoux R. Computational deconvolution: extracting cell type-specific information from heterogeneous samples. Curr Opin Immunol 2013;25:571–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Milano A, Pendergrass SA, Sargent JL, et al. Molecular subsets in the gene expression signatures of scleroderma skin. PLoS One 2008;3:e2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lindberg J, af Klint E, Ulfgren AK, et al. Variability in synovial inflammation in rheumatoid arthritis investigated by microarray technology. Arthritis Res Ther 2006;8:R47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simms LA, Doecke JD, Walsh MD, et al. Reduced alpha-defensin expression is associated with inflammation and not NOD2 mutation status in ileal Crohn’s disease. Gut 2008;57:903–10. [DOI] [PubMed] [Google Scholar]
- 8.Arijs I, De Hertogh G, Machiels K, et al. Mucosal gene expression of cell adhesion molecules, chemokines, and chemokine receptors in patients with inflammatory bowel disease before and after infliximab treatment. Am J Gastroenterol 2011;106:748–61. [DOI] [PubMed] [Google Scholar]
- 9.Arijs I, Quintens R, Van Lommel L, et al. Predictive value of epithelial gene expression profiles for response to infliximab in Crohn’s disease. Inflamm Bowel Dis 2010;16:2090–8. [DOI] [PubMed] [Google Scholar]
- 10.Hewitt SM, Lewis FA, Cao Y, et al. Tissue handling and specimen preparation in surgical pathology: issues concerning the recovery of nucleic acids from formalin-fixed, paraffin-embedded tissue. Arch Pathol Lab Med 2008;132:1929–35. [DOI] [PubMed] [Google Scholar]
- 11.Liu Y, Noon AP, Aguiar Cabeza E, et al. Next-generation RNA sequencing of archival formalin-fixed paraffin-embedded urothelial bladder cancer. Eur Urol 2014;66:982–6. [DOI] [PubMed] [Google Scholar]
- 12.Zhao W, He X, Hoadley KA, et al. Comparison of RNA-Seq by poly (A) capture, ribosomal RNA depletion, and DNA microarray for expression profiling. BMC Genomics 2014;15:419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li P, Conley A, Zhang H, et al. Whole-Transcriptome profiling of formalin-fixed, paraffin-embedded renal cell carcinoma by RNA-seq. BMC Genomics 2014;15:1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Williams PM, Li R, Johnson NA, et al. A novel method of amplification of FFPET-derived RNA enables accurate disease classification with microarrays. J Mol Diagn 2010;12:680–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feagan BG, Sandborn WJ, Gasink C, et al. Ustekinumab as Induction and Maintenance Therapy for Crohn’s Disease. N Engl J Med 2016;375:1946–1960. [DOI] [PubMed] [Google Scholar]
- 16.VanDussen KL, Liu TC, Li D, et al. Genetic variants synthesize to produce paneth cell phenotypes that define subtypes of Crohn’s disease. Gastroenterology 2014;146:200–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen EY, Tan CM, Kou Y, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 2013;14:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uhlen M, Oksvold P, Fagerberg L, et al. Towards a knowledge-based Human Protein Atlas. Nat Biotechnol 2010;28:1248–50. [DOI] [PubMed] [Google Scholar]
- 19.McConnell RE, Benesh AE, Mao S, et al. Proteomic analysis of the enterocyte brush border. Am J Physiol Gastrointest Liver Physiol 2011;300:G914–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoshida S, Fukutomi T, Kimura T, et al. Comprehensive proteome analysis of brush border membrane fraction of ileum of ezrin knockdown mice. Biomed Res 2016;37:127–39. [DOI] [PubMed] [Google Scholar]
- 21.Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 2013;14:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Freeman HJ. Application of the Montreal classification for Crohn’s disease to a single clinician database of 1015 patients. Can J Gastroenterol 2007;21:363–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manser CN, Frei P, Grandinetti T, et al. Risk factors for repetitive ileocolic resection in patients with Crohn’s disease: results of an observational cohort study. Inflamm Bowel Dis 2014;20:1548–54. [DOI] [PubMed] [Google Scholar]
- 25.Whitney AR, Diehn M, Popper SJ, et al. Individuality and variation in gene expression patterns in human blood. Proc Natl Acad Sci U S A 2003;100:1896–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Revenu C, Ubelmann F, Hurbain I, et al. A new role for the architecture of microvillar actin bundles in apical retention of membrane proteins. Mol Biol Cell 2012;23:324–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schnell U, Cirulli V, Giepmans BN. EpCAM: structure and function in health and disease. Biochim Biophys Acta 2013;1828:1989–2001. [DOI] [PubMed] [Google Scholar]
- 28.Levy E, Menard D, Delvin E, et al. Localization, function and regulation of the two intestinal fatty acid-binding protein types. Histochem Cell Biol 2009;132:351–67. [DOI] [PubMed] [Google Scholar]
- 29.Zhou Q, Toivola DM, Feng N, et al. Keratin 20 helps maintain intermediate filament organization in intestinal epithelia. Mol Biol Cell 2003;14:2959–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Madison BB, Dunbar L, Qiao XT, et al. Cis elements of the villin gene control expression in restricted domains of the vertical (crypt) and horizontal (duodenum, cecum) axes of the intestine. J Biol Chem 2002;277:33275–83. [DOI] [PubMed] [Google Scholar]
- 31.Crawley SW, Mooseker MS, Tyska MJ. Shaping the intestinal brush border. J Cell Biol 2014;207:441–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maloy KJ, Powrie F. Intestinal homeostasis and its breakdown in inflammatory bowel disease. Nature 2011;474:298–306. [DOI] [PubMed] [Google Scholar]
- 33.Daperno M, D’Haens G, Van Assche G, et al. Development and validation of a new, simplified endoscopic activity score for Crohn’s disease: the SES-CD. Gastrointest Endosc 2004;60:505–12. [DOI] [PubMed] [Google Scholar]
- 34.McGovern DP, Kugathasan S, Cho JH. Genetics of Inflammatory Bowel Diseases. Gastroenterology 2015;149:1163–1176 e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grimm-Gunter EMS, Revenu C, Ramos S, et al. Plastin 1 Binds to Keratin and Is Required for Terminal Web Assembly in the Intestinal Epithelium. Molecular Biology of the Cell 2009;20:2549–2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muller T, Hess MW, Schiefermeier N, et al. MYO5B mutations cause microvillus inclusion disease and disrupt epithelial cell polarity. Nat Genet 2008;40:1163–5. [DOI] [PubMed] [Google Scholar]
- 37.Wiegerinck CL, Janecke AR, Schneeberger K, et al. Loss of syntaxin 3 causes variant microvillus inclusion disease. Gastroenterology 2014;147:65–68 e10. [DOI] [PubMed] [Google Scholar]
- 38.Crawley SW, Shifrin DA Jr., Grega-Larson NE, et al. Intestinal brush border assembly driven by protocadherin-based intermicrovillar adhesion. Cell 2014;157:433–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li J, He Y, Lu Q, et al. Mechanistic Basis of Organization of the Harmonin/USH1C-Mediated Brush Border Microvilli Tip-Link Complex. Dev Cell 2016;36:179–89. [DOI] [PubMed] [Google Scholar]
- 40.Bitner-Glindzicz M, Lindley KJ, Rutland P, et al. A recessive contiguous gene deletion causing infantile hyperinsulinism, enteropathy and deafness identifies the Usher type 1C gene. Nat Genet 2000;26:56–60. [DOI] [PubMed] [Google Scholar]
- 41.Mathur P, Yang J. Usher syndrome: Hearing loss, retinal degeneration and associated abnormalities. Biochim Biophys Acta 2015;1852:406–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Delacour D, Salomon J, Robine S, et al. Plasticity of the brush border - the yin and yang of intestinal homeostasis. Nat Rev Gastroenterol Hepatol 2016;13:161–74. [DOI] [PubMed] [Google Scholar]
- 43.Secor SM. Evolutionary and cellular mechanisms regulating intestinal performance of amphibians and reptiles. Integr Comp Biol 2005;45:282–94. [DOI] [PubMed] [Google Scholar]
- 44.Guedon C, Schmitz J, Lerebours E, et al. Decreased brush border hydrolase activities without gross morphologic changes in human intestinal mucosa after prolonged total parenteral nutrition of adults. Gastroenterology 1986;90:373–8. [DOI] [PubMed] [Google Scholar]
- 45.Hardin JA, Chung B, O’Loughlin E V, et al. The effect of epidermal growth factor on brush border surface area and function in the distal remnant following resection in the rabbit. Gut 1999;44:26–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chevalier C, Stojanovic O, Colin DJ, et al. Gut Microbiota Orchestrates Energy Homeostasis during Cold. Cell 2015;163:1360–74. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.