Abstract
Chronic benzene exposure is associated with hematotoxicity and the development of aplastic anemia and leukemia. However, the signaling pathways underlying benzene-induced hematotoxicity remain to be defined. Here, we investigated the role of protein phosphatase 2A (PP2A) in the regulation of benzene-induced hematotoxicity in a murine model. Male mice with a hepatocyte-specific homozygous deletion of the Ppp2r1a gene (encoding PP2A Aα subunit) (HO) and matched wildtype (WT) mice were exposed to benzene via inhalation at doses of 1, 10, and 100 ppm for 28 days. Peripheral white blood cell counts and activation of bone marrow progenitors were attenuated in the HO mice, indicating that Ppp2r1a deletion protects against benzene-induced hematotoxicity. Moreover, elevation of urinary S-phenyl mercapturic acid, a benzene metabolite, was much greater in WT mice than in HO mice. Real-time exhalation analysis revealed more exhaled benzene but fewer benzene metabolites in HO mice than in WT mice, possibly because of the down-regulation of Cyp2e1, encoding cytochrome P4502E1, in hepatocytes of the HO mice. Loss-of-function screening disclosed that PP2A complexes containing the B56α subunit participate in regulating Cyp2e1 expression. Notably, PP2A–B56α suppression in HepG2 cells resulted in persistent β-catenin phosphorylation at Ser33-Ser37-Thr41 in response to CYP2E1 agonists. In parallel, nuclear translocation of β-catenin was inhibited, concomitant with a remarkable decrease of Cyp2e1 expression. These findings support the notion that a regulatory cascade comprising PP2A–B56α, β-catenin, and Cyp2e1 is involved in benzene-induced hematotoxicity, providing critical insight into the role of PP2A in responses to the environmental chemicals.
Keywords: beta-catenin (β-catenin), hematopoiesis, metabolism, phosphorylation, protein phosphatase 2 (PP2A), gene regulation, β-catenin phosphorylation, benzene, cyp2e1, hematotoxicity, protein phosphatase 2A
Introduction
Benzene is an important industrial material and environmental pollutant, which is released into the environment from cigarette smoke, industrial waste, crude oil, architectural ornaments, and automobile emissions (1). The population is exposed to benzene mostly through ambient vapors by inhalation and polluted foods or water ingestion. Benzene has been classified as a human hematological carcinogen by the International Agency for Research on Cancer (IARC) (2012). Epidemiological studies support a strong association between benzene exposure and aplastic anemia, myelodysplastic syndromes, leukemia, and other blood disorders (2–4). Mechanistic studies of benzene-induced toxicity have demonstrated that exposure to benzene and its metabolites resulted in chromosomal aberrations and gene mutations (5–7). Alterations in gene expression, oxidative stress, immune suppression, or perturbation of the fatty acid β-oxidation pathway (8–10) have also been implicated in benzene-induced hematotoxicity. Previously, we reported that epigenetic dysregulation via hypermethylation of O6-methylguanine-DNA methyltransferase and altered H3K4me3 modification were identified in the peripheral lymphocytes of low-dose benzene-exposed workers, resulting in perturbation of DNA damage repair-related signaling pathways, ultimately contributing to dysfunction of hematopoietic stem/progenitor cells (11, 12). Nevertheless, the critical targets and signaling pathways underlying benzene-induced hematotoxicity remain to be defined.
Reversible protein phosphorylation mediated by protein kinase and phosphatase plays a critical role in the regulation of many cellular processes in eukaryotes. Dysregulation of protein phosphorylation can be triggered by environmental pollutants, thus mediating toxic effects (13, 14). Prior studies have shown that benzene and its metabolites may activate several key signaling pathways such as PI3K-AKT, p38 MAPK, or JNK, subsequently triggering apoptosis of marrow cells or malignant progression of human leukemia cells (15, 16), suggesting the involvement of protein kinase in benzene-induced toxicity.
Protein phosphatase 2A (PP2A)3 is a ubiquitously expressed holoenzyme that is responsible for the majority of Ser/Thr phosphatase activity in eukaryotic cells and has been implicated in the regulation of diverse signaling pathways (17). PP2A holoenzymes contain a catalytic C subunit, a structural scaffolding A subunit, and a variable B regulatory subunit. The Ppp2r1a gene encodes the scaffolding subunit Aα, which is responsible for 90% of core/holoenzyme PP2A assemblies (18). The interactions between a variety of B subunits with the core AC dimer dynamically determine the target specificity and subcellular localization of individual PP2A holoenzymes. Perturbation of signaling pathways controlled by specific PP2A complexes has been linked to the development and progression of human diseases (19, 20). For example, the suppression of specific PP2A B subunits or the inhibition of PP2A phosphatase activity has been implicated in the development of acute myeloid leukemia (AML) (21–23). Recent studies have revealed that activation of PP2A could be a potential target for development of therapeutic drugs against AML (21, 24). Gene expression studies demonstrate that benzene exposure alters Ppp2r1a mRNA expression (25). Taken together, these observations suggest that PP2A may be a key modulator of benzene-induced hematotoxicity.
In this study, we constructed a mouse model with the hepatocyte-specific deletion of Ppp2r1a (encoding PP2A Aα), generating a useful tool to define the role of PP2A in the regulation of benzene-induced toxicity. We report that knockout of the PP2A Aα in murine hepatocytes leads to a decrease in benzene-induced hematotoxicity. Particularly, specific PP2A complexes containing the B56α subunit suppressed Cyp2e1 expression via perturbation of the dephosphorylation of β-catenin at Ser33-Ser37-Thr41. These data support a novel signaling pathway regulated by PP2A B56α complexes that plays an important role in benzene metabolic activation.
Results
Comparison between inhalation and oral gavage of benzene-exposure routes
To establish the benzene-exposure mouse model, we selected two independent cohorts of male FVB background mice and exposed them to benzene by inhalation or oral gavage administration. Male mice were exposed to benzene vapor at concentrations of 0, 1, 10, and 100 ppm, whereas the oral gavage group were treated with benzene at 0, 1, 10, and 100 mg/kg body weight/day, respectively, for 28 days. There was no significant change in body weight between the control and treatment groups of mice over the exposure time period. Peripheral blood cytometry was employed to assess the hematotoxicity induced by benzene. As shown in Table 1, mice in the inhalation benzene-exposed group displayed a significant decrease in the number of white blood cells (WBC) and lymphocytes (LYM) in a dose-dependent manner (Ptrend < 0.001), indicating that WBC and LYM counts were sensitive to benzene exposure. By contrast, we did not observe a parallel decrease in the number of red blood cells (RBC) except for the 100 ppm group compared with the control group via inhalation. Similar results were obtained in mice administered benzene by gavage (Table 1; Table S1). In addition to blood cell counts, we also measured urine SPMA levels, an internal exposure marker of benzene. The concentration of SPMA in the benzene-exposed group via inhalation was increased by 5-, 105-, and 265-fold, respectively, compared with that in control mice (Ptrend < 0.001) (Table 2). Similar results were found in benzene-exposed mice via oral gavage, with SPMA levels elevated by 3.5-, 108-, or 230.8-fold (Ptrend = 0.044), respectively. Thus, we successfully established benzene-induced hematotoxicity models through both oral gavage and inhalation routes. Importantly, we clarified the dose relevance between the two exposure approaches based on the content of metabolite and the extent of hematotoxicity.
Table 1.
Periphery blood cytometry in mice exposed to benzene by inhalation and oral gavage
| Exposure route | Exposure level (mean ± S.D., ppm or mg/kg) |
|||
|---|---|---|---|---|
| 0 | 1 | 10 | 100 | |
| Dynamic inhalation (n = 10) | ||||
| WBC (109/liter) | 4.69 ± 0.79 | 4.01 ± 0.69a | 3.67 ± 0.62a | 1.68 ± 0.43a |
| RBC (1012/liter) | 7.43 ± 0.58 | 7.04 ± 1.16 | 6.97 ± 0.78 | 5.08 ± 0.84a |
| Platelets (109/liter) | 1249.07 ± 124.90 | 1271.52 ± 188.36 | 1256.22 ± 83.59 | 1022.59 ± 134.67a |
| LYM (109/liter) | 2.62 ± 0.39 | 2.00 ± 0.38a | 1.71 ± 0.45a | 0.81 ± 0.28a |
| MCV (f l) | 51.42 ± 2.22 | 52.89 ± 2.74 | 56.88 ± 3.6a | 57.45 ± 3.30a |
| Oral gavage (n = 6) | ||||
| WBC (109/liter) | 4.83 ± 0.97 | 4.10 ± 1.13 | 3.47 ± 0.85a | 1.73 ± 0.31a |
| RBC (1012/liter) | 7.15 ± 0.23 | 6.77 ± 0.47 | 6.72 ± 0.26 | 5.15 ± 0.69a |
| Platelets (109/liter) | 1234.20 ± 81.71 | 1200.50 ± 70.44 | 1219.10 ± 86.55 | 1219.87 ± 108.16 |
| LYM (109/liter) | 2.57 ± 0.63 | 1.76 ± 0.65a | 1.70 ± 0.70a | 0.77 ± 0.22a |
| MCV (f l) | 54.87 ± 1.68 | 52.66 ± 2.36 | 57.68 ± 1.51a | 58.18 ± 3.11a |
a p < 0.05, compared with the corresponding control group, as determined by independent-sample t test or assessed with one-way ANOVA followed by Bonferroni post-test.
Table 2.
The amount of urinary SPMA in mice exposed to benzene (μg/g creatinine, mean ± S.D., n = 6)
| Exposure route | Dose (ppm/mg/kg) |
Ptrend | |||
|---|---|---|---|---|---|
| 0 | 1 | 10 | 100 | ||
| Inhalation | 0.32 ± 0.11 | 1.77 ± 1.17 | 31.66 ± 9.12a | 79.74 ± 9.65a | <0.0001 |
| Oral gavage | 0.40 ± 0.10 | 1.80 ± 1.40 | 43.63 ± 12.06a | 92.51 ± 5.53a | 0.044 |
a p < 0.05, compared with the corresponding control group, as determined by independent-sample t test or assessed with one-way ANOVA followed by Bonferroni post-test.
The effects of PP2A Aα deletion on regulation of benzene-induced hematotoxicity
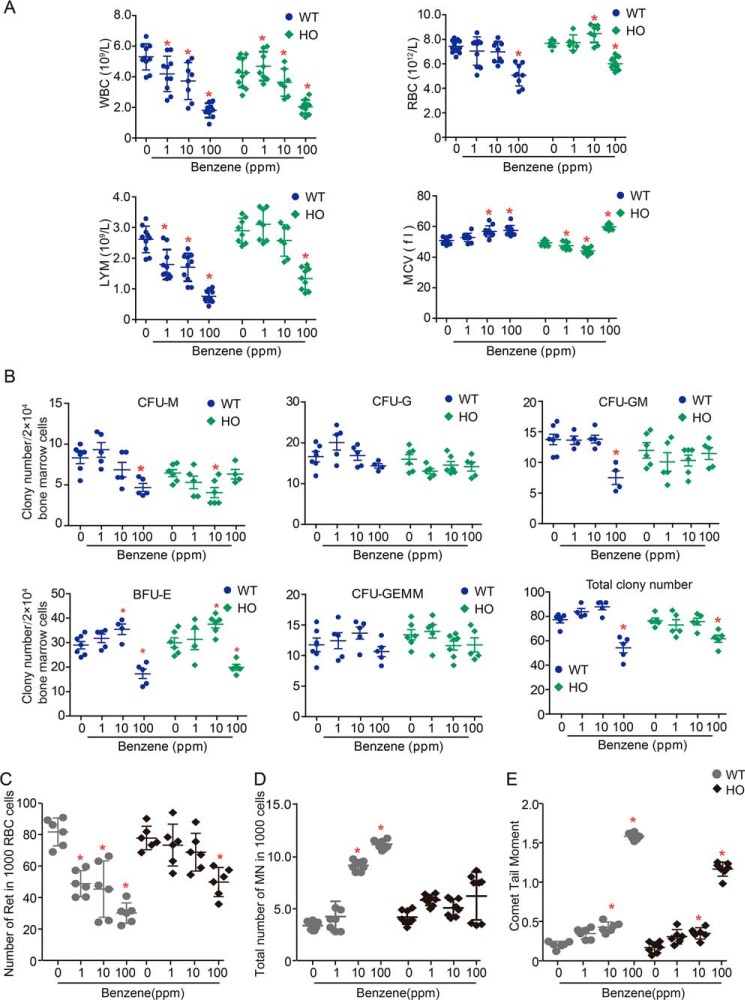
The mouse model with hepatocyte-specific deletion of the Ppp2r1a (coding PP2A Aα) gene was generated as described under “Experimental procedures.” Ppp2r1a deletion in the hepatocyte had no significant impact on mouse phenotype, behavior, growth rate, or fertility within 6 months after birth. To explore the role of PP2A in regulation of benzene-induced hematotoxicity, male mice with hepatocyte-specific deletion of Ppp2r1a (HO) and matched wildtype (WT) mice were exposed to benzene via inhalation at doses of 0, 1, 10, or 100 ppm for 28 days. As shown in Fig. 1A, WBC counts in WT mice decreased by 23.8, 30.8, and 61.5%, respectively, in WT mice exposed to 1, 10, or 100 ppm benzene compared with the vehicle control mice (Ptrend < 0.001). In HO mice, WBC counts increased by 9.6% in the 1 ppm group but declined by 16.3 and 48.8% in the 10 and 100 ppm group, suggesting that only high dose benzene treatment resulted in hematotoxicity. We noted that the decrease of WBC counts in WT mice was more profound than that in HO mice. Similarly, LYM counts in benzene-treated WT mice declined in a dose-dependent manner (Ptrend < 0.001), whereas there was no significant decrease in the LYM counts in HO mice except for the 100 ppm treatment group (Fig. 1A). Moreover, the decline of RBC counts in WT and HO mice only appeared in the 100-ppm treatment group and HO mice seemed to be more sensitive to benzene treatment. Taken together, these results indicate that suppression of PP2A Aα seems to confer resistance to benzene-induced hematotoxicity in these mice.
Figure 1.
PP2A is involved in regulation of benzene-induced hematotoxicity. WT and HO mice were exposed to benzene via inhalation at doses of 0, 1, 10, and 100 ppm, respectively, for 28 days. A, peripheral blood counts including WBC, RBC, MCV, and LYM (n = 8). B, bone marrow cells were plated in methylcellulose and the colony numbers of CFU-E, CFU-GM, CFU-G, and CFU-GEMM (n = 8) were counted following a 14-day culturing. C, the number of reticulocytes in 1000 red blood cells (n = 8) was counted and shown as mean ± S.D. D, the frequency of MN in 1000 bone marrow cells. *, p < 0.05, compared with the respective control group. E, the degree of DNA damage in benzene-exposed mice (n = 6) was determined by Comet assay, and comet tail moments of 75 peripheral blood lymphocytes are shown as mean ± S.D.
Both hematopoietic stem or progenitor cells (HS/PCs) and stromal cells in bone marrow (BM) are critical targets of benzene metabolites. We next assessed the proliferation capacities of BM progenitors upon benzene treatment using the colony-forming assay. By this method, a slight increase in colony number of BM progenitors in WT mice was noted in the 1- and 10-ppm groups, whereas a significant reduction was evident in the 100-ppm treatment group (Fig. 1B). In particular, benzene exposure decreased the number of CFU-M, CFU-GM, and CFU-E progenitors, as well as the total colony number by 48.0, 40.6, 44.6, or 26.3% only in the 100-ppm treatment group of WT mice (p < 0.05) compared with the control group. There were no significant changes with respect to CFU-G and CFU-GEMM upon benzene treatment. Notably, the colony number of CFU-E and total colony counts in HO mice exhibited a 33.5 and 13.5% decline at 100 ppm (p < 0.05), whereas the colony number of CFU-M, CFU-GM, CFU-G, and CFU-GEMM displayed no obvious changes. These observations indicate that PP2A Aα deficiency suppresses the toxicity of BM progenitor cells induced by benzene exposure. Consistently, we demonstrated that reticulocyte counts, a marker for hematopoietic potency of bone marrow erythroid, dramatically declined in WT mice exposed to benzene at 1, 10, and 100 ppm compared with the respective control group (p < 0.05) (Fig. 1C). In contrast, we only observed a 35.8% decrease of reticulocyte counts in the 100-ppm benzene-treated HO mice. These results were in agreement with examination of peripheral blood indicating that deletion of the PP2A Aα subunit led to the attenuation of benzene-induced hematopoietic toxicity.
In addition, we also examined the degree of genetic damage and genomic instability by determining the frequencies of micronuclei formation and performing Comet assays using bone marrow cells and peripheral blood lymphocytes of mice exposed to benzene. As a result, we found that the frequencies of micronuclei (MN) in bone marrow cells were 4.26, 9.15, and 11.21%, respectively, in WT mice exposed to benzene via inhalation at doses of 1, 10, and 100 ppm (Ptrend < 0.001) (Fig. 1D). In contrast, we did not observe significant changes in the frequencies of MN in HO mice exposed to benzene at any dose (Fig. 1D), indicating that PP2A Aα deficiency protected bone marrow cells from DNA damage induced by benzene. These results were further confirmed by a Comet assay, the Tail Moment values of peripheral blood cells were significantly higher in WT mice compared with that in HO mice (Fig. 1e). Taken together, these observations indicate an involvement of PP2A in the regulation of benzene-induced toxicity.
PP2A Aα deletion leads to the suppression of the metabolic activation of benzene
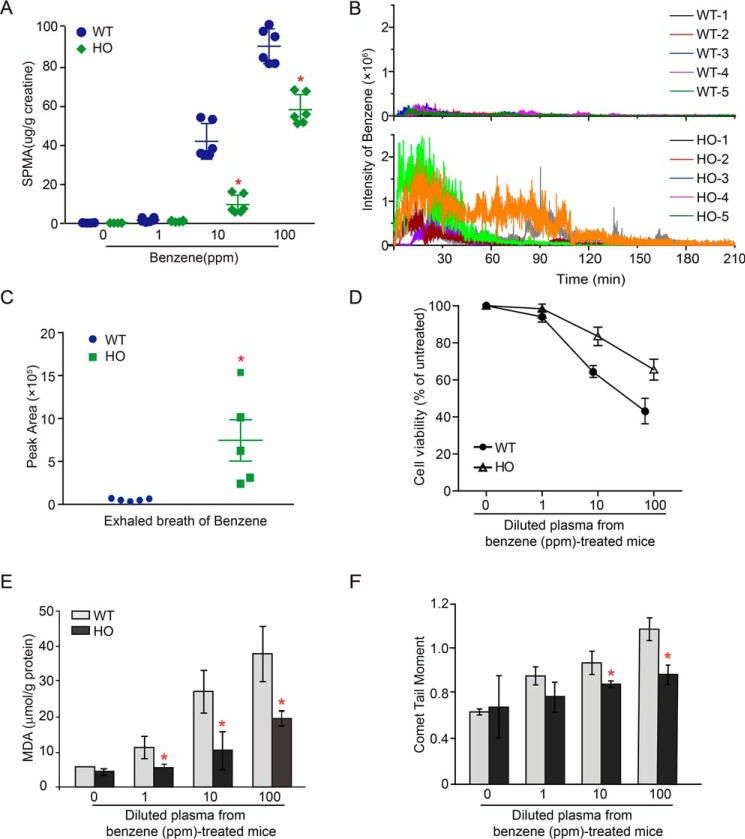
Metabolism of benzene to its toxic metabolites is generally thought to be a critical event in benzene-induced toxicity (26). To address whether PP2A Aα deletion has impact on benzene metabolism, we compared the amount of urinary SPMA in WT and HO mice upon benzene treatment. As shown in Fig. 2A, the urinary SPMA concentration increased in a dose-response manner in both WT and HO mice (Ptrend < 0.05). However, the increment of increase in urinary SPMA was enhanced in WT mice compared with HO mice. Remarkably, the urinary SPMA concentration in HO mice was decreased by 42.6% compared with that in WT mice in the 100-ppm treatment group (Fig. 2A). These results implicate the involvement of the PP2A Aα subunit in the regulation of benzene metabolism. To further assess the effects of PP2A Aα on metabolic activation of benzene, we employed a novel system to detect the components of exhalation dynamically by using secondary nanoelectrospray ionization coupled with an ultra-high resolution MS (Sec-nanoESI-UHRMS). The molecular ions of benzene and its metabolites, including phenol, trans, transmuconic acid (tt-MA), and hydroquinone (HQ) were identified through accurate mass measurement (Table S2). As shown in Fig. 2B, the dynamic changes of benzene in mouse exhalation upon benzene exposure showed that the amount of benzene in exhalation achieved the highest level 15–20 min after benzene administration via oral gavage and required 210 min to return to the basal level. Strikingly, the total amount of exhaled benzene was 7-fold higher in HO mice compared with the WT mice within 210 min (Fig. 2C). In parallel, we also detected the presence of three benzene metabolites in mouse exhalation, including phenol, tt-MA, and HQ (Fig. S2, A–C). The amount of exhaled phenol, tt-MA, and HQ in HO mice were decreased by 70, 33.3, or 54.5%, respectively, compared with those in WT mice (p < 0.01) (Fig. S2D), which was inversely proportional to the total amount of benzene over 210 min of observation. These findings support the idea that deficiency of the PP2A Aα subunit in hepatocytes leads to an inhibition of the metabolic activation of benzene. Thus, we speculate that a decline in the cytotoxicity of BM and peripheral blood cells might be present in HO mice due to lower levels of activated metabolites of benzene. To test this hypothesis, we examined the viability in HL60 cells that were cultured in medium supplemented with 2% plasma collected from WT or HO mice exposed to benzene at doses of 1, 10, and 100 ppm. As a result, we found that the relative cell viability was decreased by 3.2, 18.6, and 38.7%, respectively, in HL60 cells treated with diluted plasma derived from WT mice exposed to benzene at 1, 10, and 100 ppm (Fig. 2D). In contrast, HL60 cells displayed a reduced relative cell viability (5.4, 36.0, and 58.9%, respectively) following treatment with diluted plasma isolated from 1, 10, and 100 ppm benzene-treated HO mice (Fig. 2D). In addition, we examined the level of malondialdehyde (MDA) to compare the degree of oxidative stress induced by benzene-treated mouse plasma from WT and HO mice. As shown in Fig. 2E, the levels of MDA significantly increased in a dose-dependent manner in WT mice (Ptrend < 0.05). However, there was little effect on the increment of MDA in HO mice (p < 0.05) (Fig. 2E). Moreover, we demonstrated that genotoxicity, indicated by Tail Moment values, were enhanced in HL60 cells treated with plasma from benzene-treated WT mice compared with HO mice (Fig. 2F). Taken together, these results indicate that the plasma from benzene-treated HO mice contained fewer toxic metabolites than plasma from WT mice. The attenuation of the effects, such as oxidative stress, DNA damage, and cytotoxicity in HO mice might be attributable to the inhibition of metabolic activation of benzene. Collectively, these observations demonstrate that PP2A mediates benzene-induced hematotoxicity by regulating the metabolic activation of benzene.
Figure 2.
PP2A Aα deletion leads to suppression of the metabolic activation of benzene. A, the levels of urinary SPMA (n = 6) were normalized by urinary creatinine and presented as micrograms/g of creatinine. B, WT and HO mice (n = 5) were administrated 100 mg/kg of benzene. Exhaled benzene from mouse breath was measured in real-time by SE-SI-HRMS. Mass spectrum data were present as two-dimensional data with standardized m/z intensity and time. C, the level of exhaled benzene was quantified by peak area. *, p < 0.05 compared with WT mice, as determined by Mann-Whitney U test. HL60 cells were incubated with a medium containing 2% plasma isolated from the benzene-treated mice (n = 5) for 24 h, and subjected to analyses including cytotoxicity, MDA level, and DNA damage. D, relative cell viability was measured by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. E, MDA content was calculated as micromole/g of protein. F, Comet tail moments (TM) from 150 HL60 cells counted. Data were shown as mean ± S.D. from three independent experiments. *, p < 0.05, compared with the control cells.
PP2A regulates benzene-induced hematotoxicity by suppression of Cyp2e1 expression
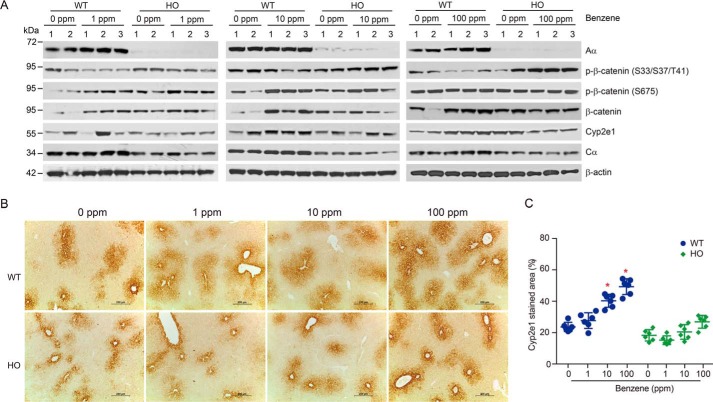
Because PP2A Aα subunit deficiency in the hepatocyte perturbs the metabolism of benzene, we speculated that PP2A may regulate the expression of key enzymes involved in benzene metabolism. Thus, we examined gene expression levels of 17 metabolic enzymes (listed in Table S2) in liver tissues of mice treated with different doses of benzene via inhalation. As shown in Fig. S3, the mRNA levels of Cyp2e1, gsta, and gstm4 in WT mouse liver were increased in a dose-response manner (p < 0.05). However, PP2A Aα deletion led to an inhibition of the induction of Cyp2e1 in benzene-treated HO mice (Fig. S3), indicating that PP2A might be involved in modulating Cyp2e1 expression. Next, we performed immunoblotting analysis to examine putative changes in Cyp2e1 protein levels in mouse liver tissues resulting from benzene inhalation. We found no induction of Cyp2e1 when WT or HO mice were exposed to 1 ppm benzene. However, Cyp2e1 induction was profound in WT mice exposed to 10 or 100 ppm benzene. Conversely, Cyp2e1 induction was abolished in HO mice exposed to benzene at any dose (Fig. 3A). Parallel immunohistochemical examination revealed that Cyp2e1-positive staining was decreased by 53.8 and 92.3%, respectively, in HO mice exposed to 10 and 100 ppm benzene compared with WT mice (Fig. 3B). Taken together, these findings demonstrate that PP2A Aα participates in the regulation of transcriptional activation of Cyp2e1 and, in turn, plays a critical role in mediating benzene-induced hematotoxicity.
Figure 3.
PP2A regulated benzene-induced hematotoxicity by suppression of Cyp2e1 expression. A, immunoblotting analysis of Aα, β-catenin, phosphorylated β-catenin, and Cyp2e1 in WT and HO mice liver treated with benzene at the indicated doses. B, the representative images are shown for liver sections stained with Cyp2e1 antibody (×100 magnification, scale bar, 200 μm). C, quantification of the Cyp2e1-stained area expressed as percentage of liver fields occupied (mean ± S.D., n = 6). *, p < 0.05, compared with the corresponding control group.
Previous studies have demonstrated that the transcriptional activation of Cyp2e1 is regulated by β-catenin signaling (27, 28). We examined the levels of β-catenin in liver tissues isolated from the benzene-treated WT and HO mice. As shown in Fig. 3A, benzene exposure led to a remarkable increase in β-catenin expression, which correlated with the suppression of Cyp2e1. These results suggest that β-catenin signaling participates in the activation of cyp2e1 transcription. In addition, we found that Wnt/β-catenin-interacted components including dickkopf-related protein 1 (DKK1), Axin, phosphorylated GSK3β at Ser9, and GSK3β were accordingly altered and might contribute to activation of the Wnt/β-catenin pathway in WT mice upon benzene exposure (Fig. S4), implicating a regulatory role of the Wnt/β-catenin pathway in benzene-induced hematotoxicity. In contrast, this effect was greatly attenuated in HO mice. To further demonstrate the role of β-catenin in benzene-induced hematotoxicity, we treated WT mice with 100 mg/kg of benzene and 5 mg/kg of XAV-939, a known inhibitor of β-catenin inhibitor for 7 days. As shown in Fig. S5, XAV-939 treatment reversed the decline of WBC and LYM counts in WT mice exposed to benzene. Furthermore, we conducted in vitro studies in HL60 cells and revealed that XAV-939 treatment attenuated HQ-induced cytotoxicity (Fig. S6). Taken together, these results support the notion that PP2A was involved in regulation of benzene-induced hematotoxicity by regulating the Wnt/β-catenin pathway.
Prior studies have reported that the phosphorylation of β-catenin at Ser33-Ser37-Thr41 is essential for ubiquitination-mediated degradation of β-catenin (29, 30) and the phosphorylation of β-catenin at Ser675 is responsible for β-catenin transcriptional activation (31). In our study, we found that the phosphorylated β-catenin at Ser33-Ser37-Thr41 decreased by 20.1, 42.6, or 65.6%, respectively, in the livers of WT mice treated with 1, 10, or 100 ppm benzene via inhalation. However, there was no difference between WT and HO mice with respect to phosphorylated β-catenin at Ser675 (Fig. 3A). Importantly, we found that the phosphorylated β-catenin at Ser33-Ser37-Thr41 in benzene-treated HO mouse liver was significantly enhanced, suggesting that PP2A might be responsible for dephosphorylation of β-catenin at Ser33-Ser37-Thr41 involved in Cyp2e1 induction. These observations suggest that PP2A regulates benzene-induced hematotoxicity through the β-catenin pathway and, in turn, represses the transactivation of cyp2e1 expression.
Specific PP2A complexes containing B56α participate in regulation of cyp2e1 expression
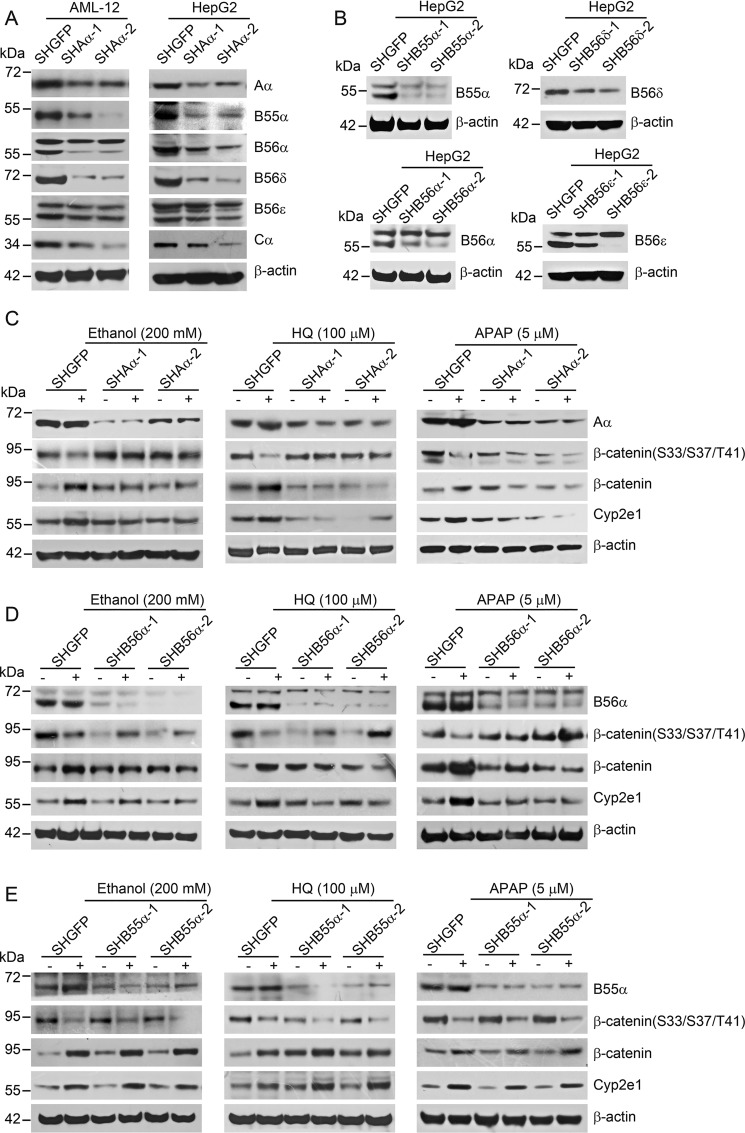
To identify specific PP2A complexes participating in the regulation of Cyp2e1 expression, we performed a loss-of-function screen in AML-12 (mouse liver cell line) and HepG2 cells (human liver tumor cell line) expressing shRNAs against a subset of PP2A regulatory subunits. Our preliminary results demonstrated that PP2A Aα deletion in mouse hepatocytes led to a remarkable decrease in protein levels of several B subunits, including B55α, B56α, B56ϵ, and B56δ. Next, we created stable AML-12 and HepG2 cell lines expressing shAα through retroviral infection and assessed the effect of Aα suppression on the expression of various B subunits. Two independent shRNAs that targeted different sequences of the PP2A Aα subunit were chosen to eliminate the off-target effects. As a result, the levels of Aα in AML-12–shAα-1 and AML-12–shAα-2 cells decreased by 54 and 90%, respectively, compared with AML-12 cells expressing a control vector (AML-12–shGFP) (Fig. 4A). Consistent with the results from mouse liver tissues, expression of B55α, B56α, B56ϵ, and B56δ subunits were dramatically down-regulated in AML-12–shAα-1 and AML-12–shAα-1 cells (Fig. 4A). Similar results were obtained in HepG2 cells expressing PP2A Aα shRNAs (Fig. 4B). In addition, we generated stable HepG2 cells expressing shB55α, shB56α, shB56ϵ, and shB56δ. The effects of gene suppression by introduction of shRNAs were confirmed by immunoblotting analysis (Fig. 4B). As shown in Fig. S7, Cyp2e1 expression was profoundly declined in HepG2 cells expressing shB56α, suggesting a role of PP2A B56α holoenzyme in the regulation of transcriptional activation of cyp2e1.
Figure 4.
Identification of specific PP2A complexes involved in the regulation of Cyp2e1 expression. A, AML-12 cells or HepG2 cells were transfected with vectors encoding shRNAs targeting GFP, or two independent shAα to generate stable cell lines as indicated. The protein level was determined by immunoblotting analysis with the indicated antibodies. B, HepG2 cells stably expressing shB55α, shB56α, shB56δ, and shB56ϵ. Immunoblotting analysis was performed to detect the protein levels in the indicated cell lines. HepG2-SHGFP cells, and (C) HepG2–SHAα-1 and HepG2–SHAα-2 cells, (D) HepG2–SHB56α-1 and HepG2–SHB56α-2 cells, (E) HepG2–SHB55α-1 and HepG2–SHB55α-2 cells were treated with 200 mm ethanol, 50 μm HQ, and 5 μm APAP for 24 h, respectively. Immunoblotting analysis was performed with the indicated antibodies.
Next, we assessed whether the PP2A B56α subunit mediated transcriptional activation of cyp2e1 upon treatment with known Cyp2e1 agonists. As expected, we observed a dose-dependent induction of Cyp2e1 expression in HepG2 cells upon treatment with ethanol, HQ, and acetaminophen (APAP) (Fig. S8A). In particular, PP2A Aα or B56α suppression led to a 62.5 or 78.6% decline in Cyp2e1 induction in HepG2 cells treated with 50 μm HQ (Fig. 4, B and D). Similar results were obtained when HepG2 cells were treated with 200 mm ethanol or 5 μm APAP (Fig. 4, C and D). As a control subunit, B55α suppression had no effect on the induction of Cyp2e1 protein expression (Fig. 4E). These results support the hypothesis that PP2A B56α holoenzyme might be responsible for the transcriptional activation of Cyp2e1.
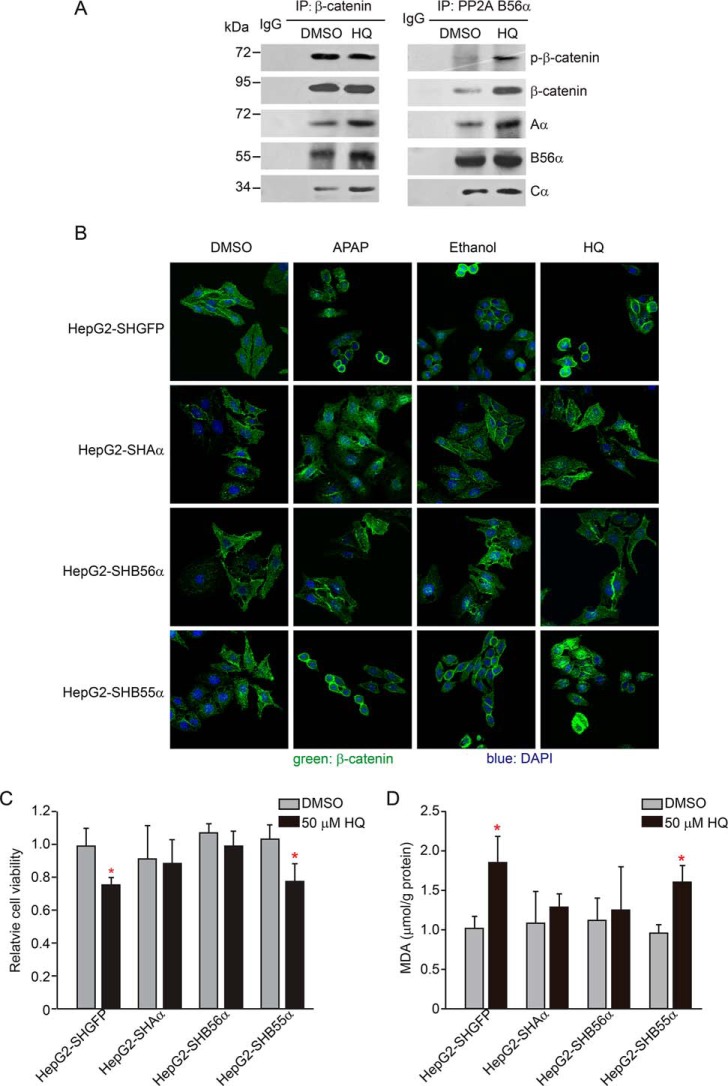
Concurrent with Cyp2e1 induction, we found that β-catenin phosphorylation at Ser33-Ser37-Thr41 significantly decreased in HepG2 cells treated with ethanol, HQ, or APAP (Fig. S8A). In contrast, up-regulation of the β-catenin protein was evident in HepG2 cells treated with three agonists, indicating that β-catenin phosphorylation at Ser33-Ser37-Thr41 negatively regulated Cyp2e1 expression. Moreover, we found that β-catenin mRNA remained unchanged upon HQ treatment, suggesting that the metabolite of benzene might up-regulate β-catenin through suppression of β-catenin degradation (Fig. S8, B–D). Notably, enhanced phosphorylated β-catenin at Ser33-Ser37-Thr41, coupled with decreased β-catenin, was also present in HepG2–SHAα and HepG2–SHB56α cells following ethanol, HQ, or APAP treatment (Fig. 4C). These findings demonstrate that the increased β-catenin and Cyp2e1 induction were abolished by suppression of PP2A Aα or B56α in HepG2 cells. In control experiments, we did not detect induction of Cyp2e1 in HepG2 cells expressing shB55α. Moreover, a co-immunoprecipitation assay confirmed the interaction between PP2A and β-catenin (Fig. 5A), showing that the B56α subunit was in complex with β-catenin at Ser33-Ser37-Thr41 upon APAP treatment (Fig. 5A). Consistent with these results, we visualized the β-catenin translocation from the cytoplasmic membrane to the nucleus when HepG2 cells were treated with 5 μm APAP, 200 mm ethanol, or 50 μm HQ under a laser scan confocal microscopy. Both the suppression of PP2A Aα or B56α subunit perturbed β-catenin translocation (Fig. 5B). In parallel, B56α subunit suppression inhibited the transcriptional activation of cyp2e1 upon these agonists' treatments (Fig. S8, B–D). In contrast, suppression of B55α had no effect on the translocation of β-catenin in response to APAP treatment (Fig. 5B). In parallel, we revealed the cell viability was decreased by 25.3 or 27.6%, respectively, in HepG2–SHGFP or HepG2–SHB55α cells treated with 50 μm HQ compared with the control cells. However, it displayed on obvious effect in cells expressing shAα and shB56α (Fig. 5C). Similar results were obtained when we examined the MDA levels in HepG2 cells (Fig. 5D), indicating that suppression of PP2A B56α sensitized the cells to HQ-induced cytotoxicity and oxidative damage. Taken together, these observations demonstrate that PP2A B56α complexes were indispensable for regulating the transcriptional activation of cyp2e1. In summary, we identified specific PP2A complexes containing the B56α subunit that directly contributes to regulating Cype21 expression through dephosphorylation of β-catenin at Ser33-Ser37-Thr41.
Figure 5.
PP2A B56α complexes interact with β-catenin and regulate β-catenin translocation. A, HepG2 cells were treated with 50 μm HQ for 24 h. Co-immunoprecipitation (IP) assays were performed with antibodies against β-catenin and PP2A B56α and followed by immunoblotting with antibodies targeting the indicated proteins. B, indicated cell lines were treated with 200 mm ethanol, 50 μm HQ, and 5 μm APAP for 24 h, respectively. Immunofluorescence analysis was performed using an antibody against β-catenin (green) and the images were visualized under the laser-scanning confocal microscopy. The nuclei (blue) were stained with 4′,6-diamidino-2-phenylindole (DAPI). HepG2–SHGFP and HepG2–SHB56α cells were treated with ethanol, HQ, and APAP at the indicated concentrations for 24 h, respectively, and followed by the examination, including (C) relative cell viability and (D) MDA content. Data were shown as mean ± S.D. from three independent experiments. *, p < 0.05, compared with the control cells.
Discussion
Dysregulation of protein phosphorylation in signaling pathways has been linked to the development of many human diseases (32, 33). Although several types of protein kinases have been shown to be directly involved in benzene-induced hematotoxicity, the key events with respect to protein phosphatase remain largely unknown. In the present study, we demonstrated that the perturbation of this regulatory pathway in a murine model with hepatocyte-specific homozygous deletion of Ppp2r1a gene confers resistance to benzene-induced hematotoxicity. We also identified specific PP2A complexes containing the B56α subunit that participates in regulation of Cyp2e1 expression through direct dephosphorylation of β-catenin at Ser31-Ser37-Thr41. These findings reveal a novel pathway mediated by protein phosphatases 2A in regulating benzene-induced hematotoxicity.
PP2A represent a major fraction of cellular Ser/Thr phosphatase activity in rodent and human tissues and is involved in the regulation of diverse biological and physiological processes (34). However, the roles of PP2A in the regulation of dephosphorylation of a given substrate in a given cell or tissue remain largely unknown. Although many PP2A transgenic, knockout, or knock-in mice have been generated (35), few models are available for investigating the tissue-specific functions of particular PP2A subunits. In this study, we generated a mouse model with a hepatocyte-specific deletion of the Ppp2r1a gene (coding PP2A Aα) to clarify the molecular pathways involved in regulation of benzene-induced hemototoxicity. Although the HO mice exhibit normal phenotype, behavior, growth rate, liver functions, and fertility within 3 months after birth, we found that these mice developed liver fibrosis at 12 months of age.4 Here, we demonstrate that depletion of PP2A Aα in the hepatocyte attenuated benzene-induced hematotoxicity, demonstrating a critical role of PP2A in mediating the chemical-induced hematotoxicity. Because the mouse hepatocytes are responsible for metabolic activation or suppression of drugs and environmental chemicals, we focused on the effects of PP2A Aα depletion on benzene metabolism.
Benzene toxicity is strongly dependent on biotransformation. Benzene metabolism produces a variety of intermediate compounds, including benzene oxide, phenol, catechol, HQ, and benzoquinone (36). Cytochrome P4502E1 (CYP2E1) plays a critical role in metabolic activation of benzene in rodents and humans (37). Low dose benzene tends to be metabolized rapidly and excreted primarily as conjugated metabolites. In contrast, metabolic capacity becomes saturated or perturbed at higher doses of benzene exposure, and exhalation of unmetabolized benzene was thought to be the primary route of excretion (36). With the aid of new technology, we were able to examine exhaled benzene and diverse metabolites dynamically. Notably, we demonstrated that greater amounts of benzene exhaled from HO mice was due to the suppression of benzene metabolic activation in hepatocytes resulting from the depletion of the PP2A Aα subunit. In accordance with these observations, several benzene metabolites, including phenol, tt-MA, and HQ in the exhalation declined dramatically. This approach provides a powerful tool for studying chemical biotransformation. Our findings also suggest that exhalable benzene might be a feasible and noninvasive biomarker for monitoring occupational exposure.
Previous studies reported that PP2A could modulate fetal liver erythropoiesis and maintain the survival of committed erythroid cells through the STAT5 pathway (38). This line of evidence indicates that inactivation of PP2A or dysregulation of specific pathways mediated by PP2A plays a key role in the recurrence of acute myeloid leukemia (21). PP2A-activating drugs markedly reduced survival and self-renewal of quiescent HSCs by targeting the JAK2/PP2A/β-catenin pathways (39). These findings indicate that PP2A-mediated pathways might function as critical regulators for determining blood or bone marrow cells fate. Here, we demonstrated that PP2A was involved in the regulation of benzene-induced hematotoxicity by modulating the transcriptional activation of cyp2e1, the key enzyme of benzene metabolism. Other studies have shown that PP2A is involved in the transcriptional activation of metabolic enzymes. For example, PP2A-mediated dephosphorylation of Sp1 at Ser59 was critical in tetrachlorodibenzo-p-dioxin-induced CYP1A1 transcription (40). In addition, the cytoplasmic CAR–Hsp90 complex recruits PP2A upon phenobarbital treatment and dephosphorylates CAR at Thr38 or Ser202, leading to transactivation of metabolic enzymes (41). Our findings demonstrate that PP2A contributed to chemical-induced toxicity by mediating the pathways involved in regulation of metabolic enzymes.
Previous studies reported that the transcriptional activation of cyp2e1 is regulated mainly by the Wnt/β-catenin pathway (27). Consistent with these findings, we demonstrate that activation of the Wnt/β-catenin pathway contributed to benzene-induced hematotoxicity. It has been reported that regulation of β-catenin is tightly associated with stress-induced posttranslational modifications, including phosphorylation (42). β-Catenin was sequentially phosphorylated at Ser33-Ser37-Thr41 by CKIα and GSK3, leading to degradation via the ubiquitination/proteasome machinery (43). In contrast, phosphorylation of β-catenin at Ser675 promoted the nuclear accumulation of β-catenin and activation of target gene transcription (31). In the current study, we found that the dephosphorylation of β-catenin at Ser33/Ser37/Thr41 in benzene-treated mouse liver led to up-regulation of β-catenin and, in turn, activation of cyp2e1 transcription. PP2A Aα or B56α subunits lead to abolishment of β-catenin degradation. Given that suppression of PP2A Aα or B56α subunits lead to abolishment of β-catenin degradation, we conclude that β-catenin dephosphorylation at Ser33-Ser37-Thr41 is a prerequisite for the transcriptional activation of cyp2e1 induced by HQ or other agonists. These observations indicate that dysregulation of the β-catenin pathway mediated by PP2A might contribute to benzene-induced hematotoxicity.
We previously found that free PP2A C and B subunits were unstable in cells, and assembling of PP2A holoenzyme led to the stabilization of C and B subunits. PP2A Aα deficiency led to the down-regulation of B subunits including B55α, B56α, B56ϵ, and B56δ, and consequently the perturbation of a specific signaling pathway (44). In this study, we identified a novel role of PP2A B56α holoenzyme in benzene-induced hematotoxicity by regulation of Cyp2e1 expression. It has been reported that the PP2A B56α subunit may function as a putative tumor suppressor by negatively regulating several key oncoproteins. For instance, PP2A-B56α was shown to be one component of a degradation complex for β-catenin mediated by the scaffold protein Axin1 (45). Here, we demonstrated that PP2A B56α directly interacted with β-catenin and controlled the degradation of β-catenin. Suppression of B56α markedly attenuated β-catenin dephosphorylation and its translocation in cells treated with Cyp2e1 inducers. These observations suggest that the PP2A B56α holoenzyme is the key factor in regulating benzene-induced hematotoxicity. Previous studies support a role for the PP2A B55α complex in Drosophila Wg signaling by directly interacting with and dephosphorylating β-catenin (46) and maternal PP2A:B56ϵ was required for Wnt-dependent accumulation of β-catenin protein and Xenopus dorsal development (47). A recent study reported that PP2A holoenzyme containing the B55β subunit regulated β-catenin phosphorylation in adenoid cystic carcinoma (48). However, in this study, we did not address whether B55α, B55β, or B56ϵ subunits participated in β-catenin dephosphorylation and transcriptional activation of cyp2e1. These findings indicate that regulation of the Wnt/β-catenin pathway by specific PP2A B subunits varies depending on different cell types or different species. Distinct PP2A complexes might be involved in β-catenin dephosphorylation at different residues.
In summary, we identified a novel pathway and key targets involved in benzene-induced hematotoxicity. Deletion of the Ppp2r1a gene in murine hepatocytes attenuated benzene-induced hematotoxicity by suppressing cyp2e1 transcription. Specific PP2A complexes containing the B56α subunit regulated Cyp2e1 expression through dephosphorylation of β-catenin at Ser33-Ser37-Thr41. These findings shed light on our understanding of the signaling pathway by which specific protein phosphatase regulates gene expression and cellular functions in response to environmental stress.
Experimental procedures
Animals
In this study, male mice with homozygous hepatocyte-specific deletion of the Ppp2r1a (encoding PP2A Aα) gene were generated by mating Ppp2r1aflox/flox mice (purchased from the Jackson Laboratory) with Alb-Cre mice. Ppp2r1aflox/flox mice have a mixed FVB/NJ and 129S4/SvJae genomic background, whereas Alb-Cre mice have a C57BL/6J genomic background. Ppp2r1aflox/flox; Alb+/− mice are hereafter termed HO mice. Ppp2r1aflox/flox; Alb −/− or Ppp2r1a+/+; Alb+/− mice obtained from the same breeding were used as controls and termed wildtype (WT) mice. All animal protocols were approved by the Animal Care and Use Committee of the Model Animal Research Center of Sun Yat-sen University.
Animal treatments
Benzene (Sigma) was diluted in corn oil (Sigma). Male WT and HO mice (8 weeks old) were randomly divided into four groups and treated with benzene by dynamic inhalation or oral gavage. For inhalation exposure, the dose of benzene employed was 1, 10, and 100 ppm. The duration of exposure was 28 days, at 6 h/day and 6 days/week. The benzene aerosol was generated by a Permease Permeater device (PD-1B, GASTEC CORP, Ayase, Japan) connected with filter-dried air as carrier gas in a 300-liter compressed gas cylinder. The generated aerosol was imported into the whole-body inhalation chambers (Guangzhou JIUFANG Electronics Co., Ltd., Guangzhou, China). The benzene concentrations in exposure chambers were monitored every 2 h and analyzed by GC-MS (GC-MS) (Agilent Technologies, Santa Clara, CA). The daily mean concentration of benzene is shown in Fig. S1. In addition, WT and HO mice were administered benzene by oral gavage in a volume of 10 ml/kg/body weight at doses of 1, 10, and 100 mg/kg, which was equivalent to 0, 0.108, 1.08, 10.8, and 108 ppm at an estimated inhalation exposure level (8-h total weight average) (U.S. EPA 1999), 6 days/week for 28 days. More than 8 mice per group were used for each experiment. The body weight of mice was recorded once a week during the period of benzene exposure. To clarify the role of β-catenin in benzene-induced hematotoxicity, 24 male WT mice were divided into 4 groups and treated with: 1) corn oil, 2) 100 mg/kg of benzene, 3) 5 mg/kg of XAV-939, and 4) 100 mg/kg of benzene, and 5 mg/kg of XAV-939, respectively, for 7 days.
Peripheral blood analysis
Mice were injected with 10% pentobarbital sodium before being sacrificed. Peripheral blood was collected from the inferior vena cava and analyzed in a Sysmex XE-2100 automatic hematology analyzer (Sysmex, Kobe, Japan). The peripheral blood counts (RBC, hemoglobin; hematocrit; mean corpuscular volume (MCV); average concentration of hemoglobin (MCH); mean corpuscular hemoglobin concentration (MCHC); platelets; reticulocytes; WBC; LYM; neutrophils; monocytes; and eosinophils) were determined. After peripheral blood analysis, plasma was isolated from whole blood by centrifugation at 450 × g for 10 min at room temperature and stored at −80 °C before use.
Determination of urinary SPMA
Urinary SPMA was measured according to a previously described protocol, with minor modifications (49). Briefly, the urine samples were thawed, vortexed, and centrifuged at 800 × g for 5 min and the supernatant was collected. Then, 0.5 ml of 10 mm sodium acetate buffer (pH 6.3) was mixed with 0.5 ml of supernatant, followed by the addition of 50 μl of 10 μg/liter of SPMA-d5 working solution. After solid phase extraction (Waters Oasis® MAX, Milford, MA), samples were analyzed by LC/electrospray tandem MS (LC-MS/MS) with a 0.01 μg/liter limit of detection. The urinary SPMA was normalized to urinary creatinine and expressed as μg/g of creatinine.
Hematopoietic colony assays
The colony forming cell assay was performed to assess the ability of hematopoietic progenitors to proliferate and differentiate into colonies in response to benzene. Briefly, BM cells from mice femur and tibia were flushed with mPBS buffer (2% fetal bovine serum in PBS) and filtered with a 70-μm cell strainer (BD Pharmingen, San Jose, CA). RBC-depleted bone marrow cells were plated at a final concentration of 5 × 105 cells/ml in triplicate and cultured in methylcellulose-based semisolid culture medium (R&D Systems) in an atmosphere of 37 °C with 5% CO2. 14 days after incubation, the colonies formed were viewed and scored under an inverted microscope. Hematopoietic colonies were classified as CFU-E (erythroid colonies), CFU-GM (mixed colonies of granulocytic and macrophage), CFU-G (pure granulocytic colonies), and CFU-GEMM (mixed type of colonies containing both the erythroid and myeloid cells).
Stable cell line establishment and cell treatment
To create stable AML-12 and HepG2 cells expressing shRNAs against PP2A Aα or B subunit, pLKO-shRNAs (14) were introduced into AML-12 or HepG2 cells by lentiviral infection and selected with puromycin (1 μg/ml). For the induction of Cyp2e1 expression, HepG2 cells were seeded in 6-cm plates with a density of 8 × 105 and treated with three types of Cyp2e1 agonists for 24 h. The chemicals and their concentrations were: ethanol (0, 100, 200, and 400 mm), acetaminophen (0, 1.25, 2.5, and 5 μm), and hydroquinone (0, 6.25, 12.5, and 25 μm).
Immunoblotting analysis and co-immunoprecipitation
Upon sacrifice, liver tissue was excised and homogenized in ice-cold RIPA lysis buffer (150 mmol/liter of NaCl, 1% Triton X-100, 0.5% deoxycholate, 0.1% SDS, and 50 mm Tris (pH 7.4)) containing protease inhibitors. The lysates were centrifuged at 12,000 × g for 20 min at 4 °C. Soluble proteins (80 μg) were subjected to 4–12% gradient acrylamide gel for SDS-PAGE before immunoblotting. The following antibodies were used: mouse anti-PP2A Cα (1D6) (Upstate Biotechnology), rabbit anti-Aα, and rabbit anti-β-catenin, β-catenin (Ser33/Ser37/Thr41), and β-catenin (Ser675) (Cell Signaling Technology), and rabbit anti-Cyp2e1 (Proteintech Group, Chicago, IL).
For immunoprecipitation analysis, 3 mg of proteins were incubated with specific antibodies overnight at 4 °C. 100 μl of prewashed 1:1 slurry of protein G-Sepharose was added to the mixture and incubated for an additional 2 h. The protein G beads–protein complexes were washed three times and eluted in 2× SDS loading buffer, followed by SDS-PAGE and immunoblotting.
Laser scanning confocal microscopy analysis
HepG2 cells (1 × 105) were seeded and grown overnight on coverslips and fixed in 3.7% formaldehyde for 15 min and permeabilized in 0.2% Triton X-100. The cells were then incubated in a blocking solution (3% FBS in PBS) for 30 min at 37 °C, followed by incubation with specific anti-β-catenin antibody (1:500) overnight. Alexa Fluor 488-conjugated IgG second antibody (1:1000) was incubated for 1 h. The slides were counterstained with 4′,6-diamidino-2-phenylindole (1 μg/ml) and observed under a LSM510 META laser scanning confocal microscope (Leica TCS SP5).
Immunohistochemistry analysis
Mouse livers were fixed in 4% buffered paraformaldehyde phosphate for 24 h, followed by decalcification in 10% EDTA for 2 weeks, then dehydrated, embedded in paraffin, and sectioned at 5-μm thick slides. The slides were incubated with primary antibodies against Cyp2e1 (1:150, Proteintech Group). Quantification of fluorescence was performed using ImageJ software (National Institute of Mental Health, Bethesda, MD) as observed and depicted as percent of relative expression.
Micronucleus assay
After mice were sacrificed, bone marrow smears were immediately made and stained with Giemsa. All slides were scored blindly. 1000 cells for each subject were examined for MN according to the criteria in the HUMN Project (50) under a microscope.
Neutral Comet assay
5 μl of whole blood was mixed with 200 μl of 0.8% low-melting point agarose and spread onto a CometAssay® HT 20-well slide (Trevigen). Slides were subsequently immersed in a pre-cooled lysis buffer (2 m NaCl, 30 mm Na2EDTA, 10 mm Tris-HCl, 1% Triton X-100, and 10% DMSO, pH 8.0) at 4 °C for 1 h. Next, the slides were subjected to electrophoresis, neutralization, dehydration, and staining, and viewed under a fluorescence microscope (Nikon Eclipse Ti-E). 150 lymphocytes randomly selected per slide were scored by Comet Assay Software Project 1.2.2 (University of Wroclaw, Poland) for each sample. Olive tail moment was selected as the parameter for assessing the degree of DNA damage.
Quantitative real-time PCR analysis
Total RNA was isolated using TRIzol Reagent (Invitrogen), and reverse transcription was carried out using the PrimeScriptTM II First Strand cDNA synthesis kit (Takara Bio, Inc., Tokyo, Japan) according to the manufacturers' instructions. The mRNA expression levels were examined using Toyobo Real-time PCR Master Mix (TOYOBO, Osaka, Japan) and analyzed using the Applied Biosystems 7500 Real-time PCR System. Actin was used as an internal control to determine relative mRNA expression. The primers for amplifying metabolic enzyme genes are detailed in Table S2.
Ex vivo assay for measurement of MDA and cytotoxicity in HL60 or HepG2 cells
3 × 104 HL60 cells/well were seeded onto a 96-well plate in quadruplicate for 2 h and incubated with medium containing 2% plasma isolated from the benzene-treated mice or HQ for 24 h. Cell Proliferation kit (WST-1) was used for the cell viability assay according to the manufacturer's protocol. MDA concentration was measured using the Lipid Peroxidation (MDA) Assay Kit (Beyotime, Nantong, China). After harvesting, the cell pellet was resuspended with ice-cold RIPA lysis buffer, followed by the addition of 200 μl of thiobarbituric acid reaction solution. The mixture was incubated in a water bath at 100 °C for 15 min and the supernatant was collected and subjected to absorbance detection at 532 nm using a Microplate Spectrophotometer (BIOTEC, USA). MDA content was expressed as micromole/g of protein.
Real-time breath analysis of exhaled benzene and its metabolites
Exhaled benzene in mouse breath was measured according to a protocol previously described (51). In brief, 8-week-old male mice obtained from the same breeding with the same approximate body weight were treated with benzene at a dose of 100 mg/kg via oral administration and immediately placed in a 50-ml Falcon conical centrifuge tube (∼25 °C, ∼40% RH). The mouse exhalation was collected through PTFE tubing (4 mm inner diameter) by carrier gas (indoor air) at 1 liter/min into a device equipped with a membrane inlet single photon ionization TOF-MS (MI-SPI-TOF-MS). Mass spectrum data were processed with SPIMS_V3.05 software and two-dimensional data with standardized m/z intensity and time were obtained. Exhaled benzene and its metabolites were analyzed and determined by charge-to-mass ratio (m/z) and reported in Table S3.
Statistical analysis
Data are shown as the mean ± S.D. All statistical analysis was performed using SPSS 20.0 statistical software (IBM, New York) and GraphPad Prism Software (La Jolla, CA). Differences of continuous variables (peripheral blood count, colony forming cell count, and gene expression level) between the groups were analyzed by independent-sample t test or assessed with one-way ANOVA followed by Bonferroni post-test. Kruskal-Wallis H test was used for statistical analysis of SPMA, MDA, Comet assay, and the Mann-Whitney U test was used for pairwise comparisons. Differences were considered statistically significant at p < 0.05.
Author contributions
L. C., X. Li, Z. He, and W. C. funding acquisition; L. C. writing-original draft; L. C., X. Li, and W. C. project administration; P. G., J. F., F. W., and S. W. data curation; P. G., H. Z., W. L., C. G., J. F., Y. Z., X. Liu, F. W., S. W., Q. Li, Z. He, H. L., S. C., X. W., L. Y., and Q.Li investigation; P. G., W. L., C. G., Z. Huang, Y. Z., and X. Li methodology; J. F. formal analysis; H. T., Q. W., G. D., and D. L. conceptualization; Y. X., W. C., and D. L. supervision; W. C. writing-review and editing.
Supplementary Material
This work was supported by Key Program of the National Natural Science Foundation of China Grant 81430079, Guangdong Provincial Natural Science Foundation Team Project Grant 2018B030312005, National Natural Science Foundation of China (NSFC) Grants 31401213, 81602877, 81372962, 81402715, and 81602882, the Fundamental Research Funds for the Central Universities Grant 17ykpy13, and the Natural Science Foundation of Guangdong Province Doctor Staring Program Grant 2016A030310163. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S8 and Tables S1–S3
L. Chen, P. Guo, H. Zhang, W. Li, C. Gao, Z. Huang, J. Fan, Y. Zhang, S. Li, X. Liu, F. Wang, S. Wang, Q. Li, Z. He, H. Li, S. Chen, X. Wu, L. Ye, Q. Li, H. Tang, Q. Wang, G. Dong, Y. Xiao, W. Chen, and D. Li, unpublished data.
- PP2A
- protein phosphatase 2A
- AML
- acute myeloid leukemia
- WBC
- white blood cells
- LYM
- lymphocytes
- RBC
- red blood cell
- SPMA
- S-phenyl mercapturic acid
- BM
- bone marrow
- MN
- micronuclei
- tt-MA
- trans-, transmuconic acid
- HQ
- hydroquinone
- MDA
- malondialdehyde
- GSK3β
- glycogen synthase kinase 3β
- shRNA
- short hairpin RNA
- APAP
- acetaminophen
- CYP2E1
- cytochrome P4502E1
- MCV
- mean corpuscular volume
- ANOVA
- analysis of variance.
References
- 1. Snyder R. (2012) Leukemia and benzene. Int. J. Environ. Res. Public Health 9, 2875–2893 10.3390/ijerph9082875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Vlaanderen J., Lan Q., Kromhout H., Rothman N., and Vermeulen R. (2012) Occupational benzene exposure and the risk of chronic myeloid leukemia: a meta-analysis of cohort studies incorporating study quality dimensions. Am. J. Indust. Med. 55, 779–785 10.1002/ajim.22087 [DOI] [PubMed] [Google Scholar]
- 3. Schnatter A. R., Glass D. C., Tang G., Irons R. D., and Rushton L. (2012) Myelodysplastic syndrome and benzene exposure among petroleum workers: an international pooled analysis. J. Natl. Cancer Inst. 104, 1724–1737 10.1093/jnci/djs411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bahadar H., Mostafalou S., and Abdollahi M. (2014) Current understandings and perspectives on non-cancer health effects of benzene: a global concern. Toxicol. Appl. Pharmacol. 276, 83–94 10.1016/j.taap.2014.02.012 [DOI] [PubMed] [Google Scholar]
- 5. Billet S., Paget V., Garçon G., Heutte N., André V., Shirali P., and Sichel F. (2010) Benzene-induced mutational pattern in the tumour suppressor gene TP53 analysed by use of a functional assay, the functional analysis of separated alleles in yeast, in human lung cells. Arch. Toxicol. 84, 99–107 10.1007/s00204-009-0478-z [DOI] [PubMed] [Google Scholar]
- 6. Zhang L., Eastmond D. A., and Smith M. T. (2002) The nature of chromosomal aberrations detected in humans exposed to benzene. Crit. Rev. Toxicol. 32, 1–42 10.1080/20024091064165 [DOI] [PubMed] [Google Scholar]
- 7. Bahadar H., Maqbool F., Mostafalou S., Baeeri M., Rahimifard M., Navaei-Nigjeh M., and Abdollahi M. (2015) Assessment of benzene induced oxidative impairment in rat isolated pancreatic islets and effect on insulin secretion. Environ. Toxicol. Pharmacol. 39, 1161–1169 10.1016/j.etap.2015.04.010 [DOI] [PubMed] [Google Scholar]
- 8. Sun R., Cao M., Zhang J., Yang W., Wei H., Meng X., Yin L., and Pu Y. (2016) Benzene exposure alters expression of enzymes involved in fatty acid oxidation in male C3H/He mice. Int. J. Environ. Res. Public Health 13, e1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McHale C. M., Zhang L., Lan Q., Vermeulen R., Li G., Hubbard A. E., Porter K. E., Thomas R., Portier C. J., Shen M., Rappaport S. M., Yin S., Smith M. T., and Rothman N. (2011) Global gene expression profiling of a population exposed to a range of benzene levels. Environ. Health Perspect. 119, 628–634 10.1289/ehp.1002546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Badham H. J., Renaud S. J., Wan J., and Winn L. M. (2010) Benzene-initiated oxidative stress: effects on embryonic signaling pathways. Chem-Biol. Interact. 184, 218–221 10.1016/j.cbi.2009.11.005 [DOI] [PubMed] [Google Scholar]
- 11. Li J., Zhang X., He Z., Sun Q., Qin F., Huang Z., Zhang X., Sun X., Liu L., Chen L., Gao C., Wang S., Wang F., Li D., Zeng X., Deng Q., et al. (2017) MGMT hypomethylation is associated with DNA damage in workers exposed to low-dose benzene. Biomarkers 22, 470–475 10.1080/1354750X.2016.1274335 [DOI] [PubMed] [Google Scholar]
- 12. Li J., Xing X., Zhang X., Liang B., He Z., Gao C., Wang S., Wang F., Zhang H., Zeng S., Fan J., Chen L., Zhang Z., Zhang B., Liu C., et al. (2018) Enhanced H3K4me3 modifications are involved in the transactivation of DNA damage responsive genes in workers exposed to low-level benzene. Environ. Pollut. 234, 127–135 10.1016/j.envpol.2017.11.042 [DOI] [PubMed] [Google Scholar]
- 13. Zhu X. N., Chen L. P., Bai Q., Ma L., Li D. C., Zhang J. M., Gao C., Lei Z. N., Zhang Z. B., Xing X. M., Liu C. X., He Z. N., Li J., Xiao Y. M., Zhang A. H., Zeng X. W., and Chen W. (2014) PP2A-AMPKα-HSF1 axis regulates the metal-inducible expression of HSPs and ROS clearance. Cell. Signal. 26, 825–832 10.1016/j.cellsig.2014.01.002 [DOI] [PubMed] [Google Scholar]
- 14. Chen L., Ma L., Bai Q., Zhu X., Zhang J., Wei Q., Li D., Gao C., Li J., Zhang Z., Liu C., He Z., Zeng X., Zhang A., Qu W., Zhuang Z., Chen W., and Xiao Y. (2014) Heavy metal-induced metallothionein expression is regulated by specific protein phosphatase 2A complexes. J. Biol. Chem. 289, 22413–22426 10.1074/jbc.M114.548677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li J., Jiang S., Chen Y., Ma R., Chen J., Qian S., Shi Y., Han Y., Zhang S., and Yu K. (2018) Benzene metabolite hydroquinone induces apoptosis of bone marrow mononuclear cells through inhibition of β-catenin signaling. Toxicol. In Vitro 46, 361–369 10.1016/j.tiv.2017.08.018 [DOI] [PubMed] [Google Scholar]
- 16. Liu W. H., Chou W. M., and Chang L. S. (2013) p38 MAPK/PP2Acα/TTP pathway on the connection of TNF-α and caspases activation on hydroquinone-induced apoptosis. Carcinogenesis 34, 818–827 10.1093/carcin/bgs409 [DOI] [PubMed] [Google Scholar]
- 17. Shi Y. (2009) Assembly and structure of protein phosphatase 2A. Sci. China Ser. C Life Sci. 52, 135–146 10.1007/s11427-009-0018-3 [DOI] [PubMed] [Google Scholar]
- 18. Zhou J., Pham H. T., Ruediger R., and Walter G. (2003) Characterization of the Aα and Aβ subunit isoforms of protein phosphatase 2A: differences in expression, subunit interaction, and evolution. Biochem. J. 369, 387–398 10.1042/bj20021244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sablina A. A., Hector M., Colpaert N., and Hahn W. C. (2010) Identification of PP2A complexes and pathways involved in cell transformation. Cancer Res. 70, 10474–10484 10.1158/0008-5472.CAN-10-2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu C., and Götz J. (2013) How it all started: tau and protein phosphatase 2A. J. Alzheimer's Dis. 37, 483–494 [DOI] [PubMed] [Google Scholar]
- 21. Arriazu E., Pippa R., and Odero M. D. (2016) Protein phosphatase 2A as a therapeutic target in acute myeloid leukemia. Front. Oncol. 6, 78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Toop H. D., Dun M. D., Ross B. K., Flanagan H. M., Verrills N. M., and Morris J. C. (2016) Development of novel PP2A activators for use in the treatment of acute myeloid leukaemia. Org. Biomol. Chem. 14, 4605–4616 10.1039/C6OB00556J [DOI] [PubMed] [Google Scholar]
- 23. Ruvolo P. P., Ruvolo V. R., Jacamo R., Burks J. K., Zeng Z., Duvvuri S. R., Zhou L., Qiu Y., Coombes K. R., Zhang N., Yoo S. Y., Pan R., Hail N. Jr., Konopleva M., Calin G., Kornblau S. M., and Andreeff M. (2014) The protein phosphatase 2A regulatory subunit B55alpha is a modulator of signaling and microRNA expression in acute myeloid leukemia cells. Biochim. Biophys. Acta 1843, 1969–1977 10.1016/j.bbamcr.2014.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ramaswamy K., Spitzer B., and Kentsis A. (2015) Therapeutic re-activation of protein phosphatase 2A in acute myeloid leukemia. Front. Oncol. 5, 16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Heijne W. H., Jonker D., Stierum R. H., van Ommen B., and Groten J. P. (2005) Toxicogenomic analysis of gene expression changes in rat liver after a 28-day oral benzene exposure. Mutat. Res. 575, 85–101 10.1016/j.mrfmmm.2005.02.003 [DOI] [PubMed] [Google Scholar]
- 26. Ross D. (2000) The role of metabolism and specific metabolites in benzene-induced toxicity: evidence and issues. J. Toxicol. Environ. Health A 61, 357–372 10.1080/00984100050166361 [DOI] [PubMed] [Google Scholar]
- 27. Ganzenberg K., Singh Y., and Braeuning A. (2013) The time point of β-catenin knockout in hepatocytes determines their response to xenobiotic activation of the constitutive androstane receptor. Toxicology 308, 113–121 10.1016/j.tox.2013.03.019 [DOI] [PubMed] [Google Scholar]
- 28. Groll N., Petrikat T., Vetter S., Colnot S., Weiss F., Poetz O., Joos T. O., Rothbauer U., Schwarz M., and Braeuning A. (2016) Coordinate regulation of Cyp2e1 by β-catenin- and hepatocyte nuclear factor 1α-dependent signaling. Toxicology 350–352, 40–48 [DOI] [PubMed] [Google Scholar]
- 29. Orford K., Crockett C., Jensen J. P., Weissman A. M., and Byers S. W. (1997) Serine phosphorylation-regulated ubiquitination and degradation of β-catenin. J. Biol. Chem. 272, 24735–24738 10.1074/jbc.272.40.24735 [DOI] [PubMed] [Google Scholar]
- 30. Stamos J. L., and Weis W. I. (2013) The β-catenin destruction complex. Cold Spring Harbor Perspect. Biol. 5, a007898 10.1101/cshperspect.a007898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zheng Y., Zhou C., Yu X. X., Wu C., Jia H. L., Gao X. M., Yang J. M., Wang C. Q., Luo Q., Zhu Y., Zhang Y., Wei J. W., Sheng Y. Y., Dong Q. Z., and Qin L. X. (2018) Osteopontin promotes metastasis of intrahepatic cholangiocarcinoma through recruiting MAPK1 and mediating Ser675 phosphorylation of β-catenin. Cell Death Dis. 9, 179 10.1038/s41419-017-0226-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Engin A. (2017) Human protein kinases and obesity. Adv. Exp. Med. Biol. 960, 111–134 10.1007/978-3-319-48382-5_5 [DOI] [PubMed] [Google Scholar]
- 33. Cordeiro M. H., Smith R. J., and Saurin A. T. (2018) A fine balancing act: a delicate kinase-phosphatase equilibrium that protects against chromosomal instability and cancer. Int. J. Biochem. Cell Biol. 96, 148–156 10.1016/j.biocel.2017.10.017 [DOI] [PubMed] [Google Scholar]
- 34. Janssens V., and Goris J. (2001) Protein phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem. J. 353, 417–439 10.1042/bj3530417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Reynhout S., and Janssens V. (2019) Physiologic functions of PP2A: lessons from genetically modified mice. Biochim. Biophys. Acta Mol. Cell Res. 1866, 31–50 [DOI] [PubMed] [Google Scholar]
- 36. Environment Protection Agency (2002) Toxicological review of benzene (non-cancer effects). United State Environment Protection Agency, Washington, D. C. [Google Scholar]
- 37. Gut I., Nedelcheva V., Soucek P., Stopka P., Vodicka P., Gelboin H. V., and Ingelman-Sundberg M. (1996) The role of CYP2E1 and 2B1 in metabolic activation of benzene derivatives. Arch. Toxicol. 71, 45–56 10.1007/s002040050357 [DOI] [PubMed] [Google Scholar]
- 38. Chen W., Gu P., Jiang X., Ruan H. B., Li C., and Gao X. (2011) Protein phosphatase 2A catalytic subunit α (PP2Acα) maintains survival of committed erythroid cells in fetal liver erythropoiesis through the STAT5 pathway. Am. J. Pathol. 178, 2333–2343 10.1016/j.ajpath.2011.01.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Neviani P., Harb J. G., Oaks J. J., Santhanam R., Walker C. J., Ellis J. J., Ferenchak G., Dorrance A. M., Paisie C. A., Eiring A. M., Ma Y., Mao H. C., Zhang B., Wunderlich M., May P. C., et al. (2013) PP2A-activating drugs selectively eradicate TKI-resistant chronic myeloid leukemic stem cells. J. Clin. Investig. 123, 4144–4157 10.1172/JCI68951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shimoyama S., Kasai S., Kahn-Perlès B., and Kikuchi H. (2014) Dephosphorylation of Sp1 at Ser-59 by protein phosphatase 2A (PP2A) is required for induction of CYP1A1 transcription after treatment with 2,3,7,8-tetrachlorodibenzo-p-dioxin or omeprazole. Biochim. Biophys. Acta 1839, 107–115 10.1016/j.bbagrm.2013.12.004 [DOI] [PubMed] [Google Scholar]
- 41. Mutoh S., Sobhany M., Moore R., Perera L., Pedersen L., Sueyoshi T., and Negishi M. (2013) Phenobarbital indirectly activates the constitutive active androstane receptor (CAR) by inhibition of epidermal growth factor receptor signaling. Sci. Signal. 6, ra31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Huang H., and He X. (2008) Wnt/β-catenin signaling: new (and old) players and new insights. Curr. Opin. Cell Biol. 20, 119–125 10.1016/j.ceb.2008.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Liu C., Li Y., Semenov M., Han C., Baeg G. H., Tan Y., Zhang Z., Lin X., and He X. (2002) Control of β-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell 108, 837–847 10.1016/S0092-8674(02)00685-2 [DOI] [PubMed] [Google Scholar]
- 44. Chen W., Possemato R., Campbell K. T., Plattner C. A., Pallas D. C., and Hahn W. C. (2004) Identification of specific PP2A complexes involved in human cell transformation. Cancer Cell 5, 127–136 10.1016/S1535-6108(04)00026-1 [DOI] [PubMed] [Google Scholar]
- 45. Seeling J. M., Miller J. R., Gil R., Moon R. T., White R., and Virshup D. M. (1999) Regulation of β-catenin signaling by the B56 subunit of protein phosphatase 2A. Science 283, 2089–2091 10.1126/science.283.5410.2089 [DOI] [PubMed] [Google Scholar]
- 46. Zhang W., Yang J., Liu Y., Chen X., Yu T., Jia J., and Liu C. (2009) PR55α, a regulatory subunit of PP2A, specifically regulates PP2A-mediated β-catenin dephosphorylation. J. Biol. Chem. 284, 22649–22656 10.1074/jbc.M109.013698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yang J., Wu J., Tan C., and Klein P. S. (2003) PP2A:B56ϵ is required for Wnt/β-catenin signaling during embryonic development. Development 130, 5569–5578 10.1242/dev.00762 [DOI] [PubMed] [Google Scholar]
- 48. Ishibashi K., Ishii K., Sugiyama G., Kamata Y. U., Suzuki A., Kumamaru W., Ohyama Y., Nakano H., Kiyoshima T., Sumida T., Yamada T., and Mori Y. (2018) Regulation of β-catenin phosphorylation by PR55β in adenoid cystic carcinoma. Cancer Genomics Proteomics 15, 53–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Schettgen T., Musiol A., Alt A., and Kraus T. (2008) Fast determination of urinary S-phenylmercapturic acid (S-PMA) and S-benzylmercapturic acid (S-BMA) by column-switching liquid chromatography-tandem mass spectrometry. J. Chromatogr. B Anal. Technolog. Biomed. Life Sci. 863, 283–292 10.1016/j.jchromb.2008.01.024 [DOI] [PubMed] [Google Scholar]
- 50. Kanaar R., and Hoeijmakers J. H. (1998) Genetic recombination. From competition to collaboration. Nature 391, 337–338 10.1038/34794 [DOI] [PubMed] [Google Scholar]
- 51. Zhang Y., Sun W., Shang D., Gao H., Zhou Z., and Li X. (2017) Investigation on the influence of time-of-day on benzene metabolic pharmacokinetics by direct breath analysis in mice. Chemosphere 184, 93–98 10.1016/j.chemosphere.2017.05.135 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.