Abstract
How cell and tissue identity persist despite constant cell turnover is an important biologic question with cell therapy implications. Although many mechanisms exist, we investigated the controls for site-specific gene expression in skin, given its diverse structures and functions. For example, the transcriptome of in vivo pal-moplantar (i.e., volar) epidermis is globally unique, including Keratin 9 (KRT9). Although volar fibroblasts have the capacity to induce KRT9 in nonvolar keratinocytes, we show here that volar keratinocytes continue to express KRT9 in in vitro solo cultures. Despite this, KRT9 expression is lost with volar keratinocyte passaging, despite stable hypomethylation of its promoter. Coincident with KRT9 loss is a gain of the primitive keratin 7 and a signature of dsRNA sensing, including the double-stranded RNA (dsRNA) receptor DExD/H-Box Helicase 58 (DDX58/RIG-I). Exogenous dsRNA inhibits KRT9 expression in early passage volar keratinocytes or in vivo footpads of wild-type mice. Loss of DDX58 in passaged volar keratinocytes rescues KRT9 and inhibits KRT7 expression. Additionally, DDX58-null mice are resistant to the ability of dsRNA to inhibit KRT9 expression. These results show that the sensing of dsRNA is critical for loss of cell-specific gene expression;our results have important implications for how dsRNA sensing is important outside of immune pathways.
INTRODUCTION
How the epithelia and mesenchyme interact to control tissue identity is an important question in biology. The study into this control dates back to important studies in which investigators swapped epidermis and dermis in skin, for example, to try to identify which compartment is dominant in defining tissue architecture (Eames and Schneider, 2005). This question is timely now with the advent of cellular therapies, for example, to cure genetically based skin blistering diseases (Hirsch et al., 2017;Mavilio et al., 2006;Sebastiano et al., 2014;Umegaki-Arao et al., 2014). In these treatments, investigators remove cells from the affected individual, gene correct, and expand and graft them back onto the skin. The ultimate widespread use of these technologies, however, hinges on the understanding of how well these corrected cells will engraft and function. For example, if cells retain positional identity, can a single corrected clone be used across the body? Will epidermal-dermal interactions dictate the cell’s fate? The study of positional identity in tissues is important from many perspectives.
There are many suggestions that the mesenchyme is dominant in defining tissue function. For example, in quail and chicken swapping of epidermis and dermis, the dermis was found to control timing and pattern of feather formation (Eames and Schneider, 2005). Consistent with this, fibroblasts retain their positional identity in culture (Chang et al., 2002) and have been proposed to have powerful abilities to induce appendage formation even in adult animals (Higgins et al., 2013;Jahoda et al., 1984). However, compartment swapping experiments in several species showed a more mixed picture for partial control from both compartments or even epithelial dominance (Billingham and Silvers, 1963, 1967). This result is more consistent with current models high-lighting important sequential feedback events, for example, in appendage patterning (Millar, 2002). Nevertheless, despite all of this progress, the positional identity of keratinocytes has received scant attention.
In this article, we used volar (palmoplantar) skin identity as a model system to understand how tissue identity is controlled. Keratin (KRT) 9 has been proven to be specific for volar skin through its initial discovery (Knapp et al., 1986; Schweizer et al., 1989) and the study of patients with mutations in KRT9 who have symptoms mostly limited to the palms and soles (Codispoti et al., 2009). Similarly, Krt9-null mice have similar symptoms of compensatory thickened volar skin, highlighting the fact that KRT9 likely provides some of the unique structural support of volar skin (Fu et al., 2014). Yamaguchi et al. (1999) and Kim et al. (2016) previously showed that ectopic KRT9 can be induced by volar fibroblasts. However, whether volar keratinocytes have intrinsic positional memory and expression of KRT9 is unknown.
This study was motivated by our ongoing clinical trials using volar fibroblasts to induce a volar skin phenotype. We hope that inducing a volar phenotype at the stump site of amputees will enhance prosthetic use, given the pressure-and friction-adaptive properties of volar skin.
Although this effort builds on the work showing mesen-chymal contributions to epithelial gene expression, it will be important to explore the limitations of this approach given possible intrinsic epithelial positional identity.
Here, we find that volar keratinocytes do indeed retain positional identity and the ability to express KRT9. Although this is the case, volar fibroblasts still augment KRT9 expression, indicating that both epithelial and mesenchymal memory define positional tissue identity of volar skin. We also find that with prolonged culture, keratinocytes express the more primitive KRT7 and lose the site-specific KRT9. This is coincident with increased double-stranded RNA (dsRNA) signaling through its receptor DExD/H-Box Helicase 58 (DDX58/RIG-I), which is both sufficient and necessary for KRT9 expression loss. These results support a model in which dsRNA is capable for inducing stem cell behavior and is important early in regeneration (Nelson et al., 2015; Zhu et al., 2017)—but is also a hurdle for maintaining cell identity ex vivo.
RESULTS
KRT9 is among the most highly expressed transcripts in volar skin and persists in cultured volar keratinocytes in the absence of fibroblasts
A long-standing question in epithelial biology is the degree of autonomy or dependence on mesenchymal (fibroblast) signals for gene expression of the epithelia (Billingham and Silvers, 1967;Dhouailly, 1973). For example, although there are reports of fibroblast positional memory maintenance in vitro (Chang et al., 2002), there are no such reports for epithelial cultured cells. To begin testing this question, we sought to develop a model using human keratinocytes to test their positional memory. Given the unique features of pal-moplantar (volar) skin, we focused on volar keratinocytes.
KRT9 is a known cause of palmoplantar keratodermas and is characterized as a volar-specific protein, given the paucity of symptoms outside of the palms and soles in individuals with mutations in KRT9 (Codispoti et al., 2009;Reis et al.,1994). To verify this, we collected mRNA from whole epidermis in the volar skin of palms and soles and in the dorsum of hands and feet as a control (Kim et al., 2016). We performed gene array expression analyses of samples from three individuals and first plotted the data in a principal component analysis. We found that volar skin shares a common genetic signature where samples from the palm and sole cluster together and separate from the dorsal skin controls (Figure 1 a). Although KRT9 is the highest expressed gene by fold change (Kim et al., 2016), it is also among the highest expressed in volar skin as analyzed by z-score (Figure 1b). We confirmed the increase in KRT9 transcript by Western blotting (Figure 1c). These results verify that KRT9 is a robust marker of volar skin.
Figure 1. KRT9 is a robust marker of volar skin in vivo and is partially maintained in cultured volar keratinocytes in the absence of fibroblasts.
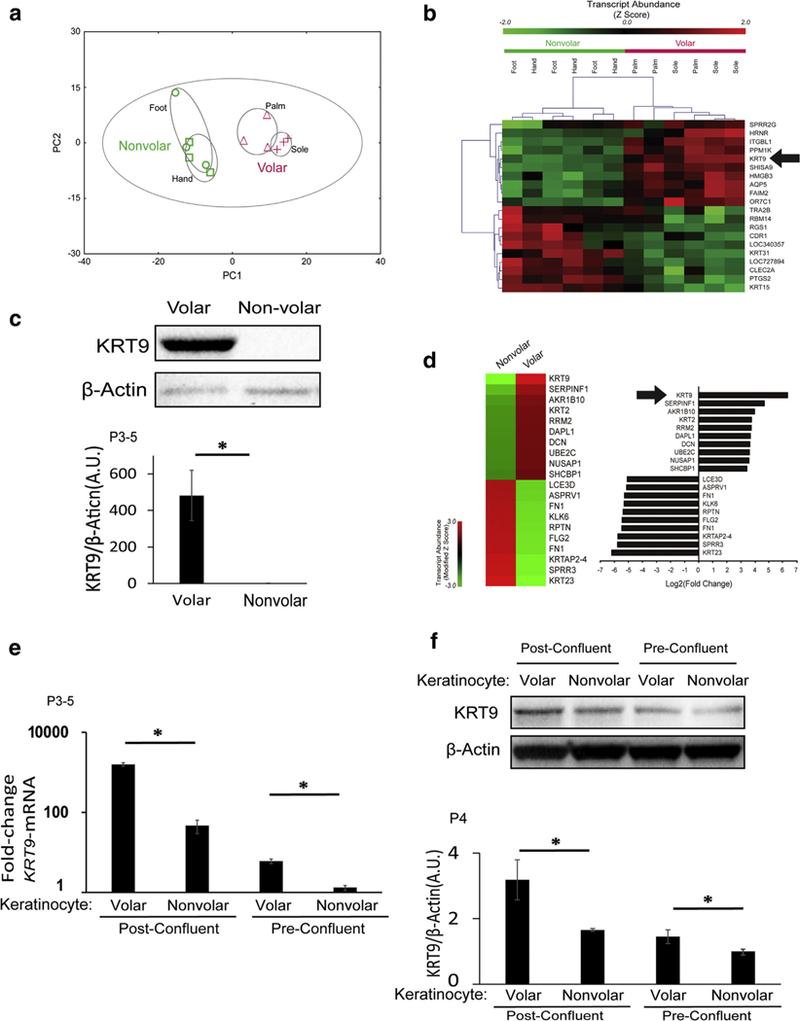
(a) Principle component analysis of microarray data from human volar and nonvolar epidermis show clear clustering of volar skin tissue. n = 3 samples from each site. (b) Unsupervised clustering of expression data from human volar and nonvolar tissue shows that KRT9 is one of the most unique genes in volar skin. n = 3 samples from each site). KRT9 had the highest fold change transcript in volar tissue (30.6-fold, P = 0.0002) (Kim et al., 2016 under GEO: GSE39267. (c) KRT9 protein expression is robust in volar compared with nonvolar human epidermis, as measured by Western blot (top) and normalized to b-Actin (bottom). n = 3, *P < 0.05. (d) In early passaged in vitro solo culture of keratinocytes, microarray analysis shows that KRT9 is maintained as the most unique transcript in volar keratinocytes compared with nonvolar keratinocytes, under GEO: GSE116055. (e) KRT9 mRNA expression of volar keratinocytes is higher than nonvolar keratinocytes both in preconfluent and postconfluent cultures, as measured by quantitative real-time reverse transcriptase–PCR and normalized to RPLP0. n = 3, *P < 0.05. (f) KRT9 protein expression is still elevated in volar compared with nonvolar keratinocytes in both postconfluent and preconfluent conditions, as measured by Western blot (top) and normalized to b-Actin (bottom). n = 3, *P < 0.05. A.U., arbitrary unit; GEO, Gene Expression Omnibus; KRT, Keratin; P, passage.
Because volar fibroblasts induce KRT9 in nonvolar kerati-nocytes (Kim et al., 2016;Yamaguchi et al., 1999), our first question was whether volar keratinocytes continue to express KRT9 in the absence of fibroblasts. We initiated keratinocyte cultures from biopsy samples of the sole to compare with nonvolar keratinocytes from foreskin. At passage 3, we purified mRNA for microarray gene expression analysis. We analyzed both fold change (Figure 1d, right) and modified z-score (Figure 1 d, left) and discovered that KRT9 expression was not only retained in volar keratinocytes but was actually still the singly most enriched gene in volar keratinocytes (Figure 1d). We next verified this in three separate individuals and noted that although confluence promotes KRT9 transcription, volar keratinocytes expressed more KRT9 compared with nonvolar keratinocytes in identical culture conditions (Figure 1 e). KRT9 protein abundance also matched the pattern of KRT9 transcription (Figure 1f). These results show that volar keratinocytes retain their positional memory for KRT9 expression in the absence of volar fibroblasts.
Volar fibroblasts stimulate KRT9 expression in both volar and nonvolar keratinocytes
Because volar keratinocytes have an intrinsic ability to induce KRT9, we next wondered if they were more responsive to KRT9 induction by volar fibroblasts. Using in vitro cocultures of keratinocytes and fibroblasts, we found that although volar keratinocytes express the highest amount of KRT9 irrespective of fibroblast identity, they express more KRT9 mRNA (Figure 2a) and protein (Figure 2b) in volar compared with nonvolar fibroblasts. These results show that site-specific gene expression of keratinocytes has both intrinsic and extrinsic controls in the case of KRT9.
Figure 2. Volar keratinocytes are still sensitive to KRT9 induction by volar fibroblasts and have stable demethylation of the KRT9 promoter in culture.
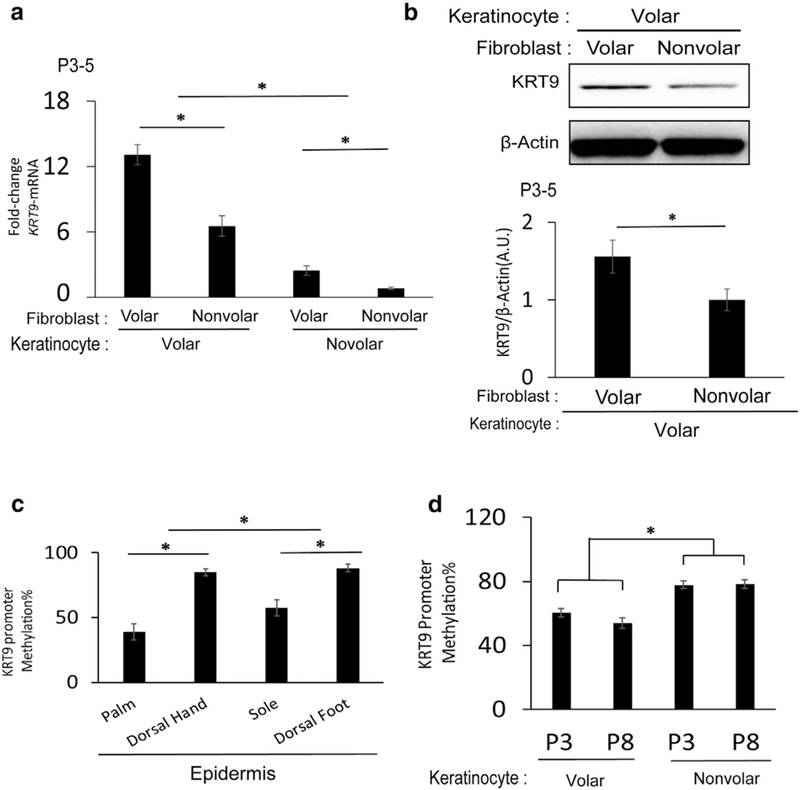
(a) KRT9 mRNA expression is elevated in volar keratinocytes without volar fibroblasts but is still induced more by volar fibroblasts in co-cultures. n = 3, *P < 0.05. (b) KRT9 protein expression of volar keratinocytes increases in co-culture with volar fibroblasts, as measured by Western blot (top) and normalized to β-Actin (bottom). n = 3, *P< 0.05. (c) KRT9 promoter is hypomethylated in volar human epidermis compared with nonvolar epidermis, as assessed by pyrosequencing. n = 3, *P < 0.05.(d) KRT9 promoter is also hypomethylated in cultured volar keratinocytes in both early and late passages, as assessed by pyrosequencing (n = 3, *P < 0.05). KRT, Keratin; P, passage.
VOLAR KERATINOCYTES HAVE STABLE DEMETHYLATION AT THE KRT9 PROMOTER
Epigenetic modifications are a likely candidate mechanism to explain how volar keratinocyte KRT9 expression is maintained in culture. In particular, we hypothesized that the KRT9 promoter might be hypomethylated in volar keratino-cytes. To test this, we identified CpG sites in the promoter that were candidate methylation sites and tested their methylation levels by pyrosequencing.
We first measured KRT9 promoter methylation in the epidermis of both palms and soles to compare versus dorsal hand and foot. We found statistically significant hypo-methylation of KRT9 in volar skin (Figure 2c). We next tested if this methylation pattern persists in vitro in cultured kerati-nocytes. Indeed, in both early (passage 3) and late (passage 8) passaged volar keratinocytes, the KRT9 promoter is significantly hypomethylated (Figure 2d). These results show that epigenetic promoter methylation is likely one mechanism where positional memory persists in volar keratinocytes in culture.
KRT9 expression decreases with keratinocytes passaging, coincident with a gene signature of dsRNA sensing
Despite the stable methylation of the KRT9 promoter, we found that KRT9 expression is lost during passaging of volar keratinocytes in vitro. This was true for both KRT9 mRNA (Figure 3a) and protein (Figure 3b). To define the mechanism, we performed gene array transcriptome analysis of early (passage 4) versus late (passage 8) passaged volar keratino-cytes to determine which gene changes were coincident with KRT9 loss (Figure 3c).
Figure 3. KRT9 expression in volar keratinocytes is lost with passaging coincident with a gene signature of dsRNA sensing; dsRNA inhibits KRT9 expression.
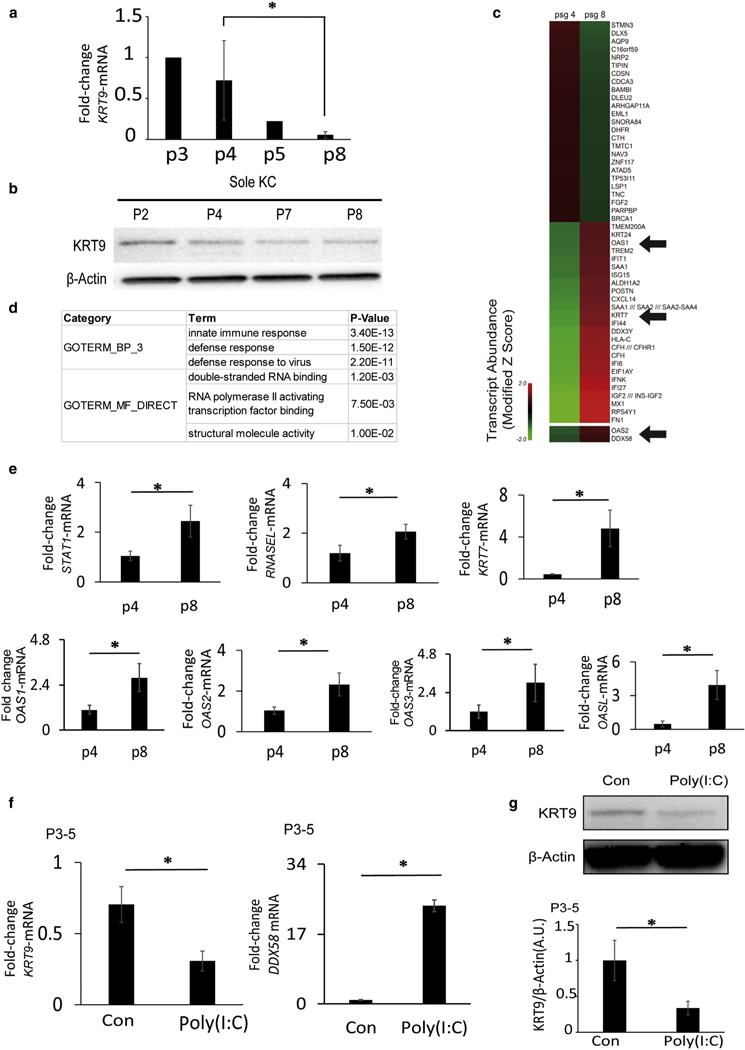
(a) KRT9 mRNA expression of volar keratinocytes decreases with increased cell passages, as detected by quantitative real-time reverse transcriptase–PCR. n = 3, *P < 0.05. (b) KRT9 protein expression in volar keratinocytes decreases with increases in cell passage. (c) Heat map of gene expression on early (p4) and late (p8) cultured volar keratinocytes shows increases in interferon and dsRNA response genes, under GEO: GSE116055. (d) In gene ontology analysis of the top 100 probe sets elevated in late passage, innate immune response and dsRNA binding are the most significantly represented in their categories. (e) Confirmation of microarray data with quantitative real-time reverse transcriptase–PCR gene expression shows elevation of STAT1, RNASEL, KRT7, OAS1, OAS2, OAS3, and OASL in passage 8 volar keratinocytes. n = 3, *P < 0.05. (f) dsRNA inhibits KRT9 and stimulates DDX58 transcription in early passage volar keratinocytes as measured by quantitative real-time reverse transcriptase–PCR. n = 3, *P < 0.05. (g) dsRNA inhibits KRT9 protein expression in early passage volar keratinocytes, as detected by Western blot (top) and normalized to b-Actin (bottom). n = 3, *P < 0.05. Con, control; dsRNA, double-stranded RNA; GEO, Gene Expression Omnibus; KC, Keratinocytes; KRT, Keratin; P, passage.
Although KRT9 loss in passage 8 keratinocytes was present on the array, one of the most significant genes lost was DLX5. Consistent with the roles of homeobox genes to define gene expression, we found that small interfering RNA (siRNA) depletion of DLX5 in early passage keratinocytes could also decrease KRT9 expression (see Supplementary Figure S1b online). DLX2 was also decreased on our arrays in late passaged keratinocytes, and this isoform was also necessary to maintain KRT9 expression (see Supplementary Figure S1a). These results confirm the importance of homeobox genes to define cell identity.
Among genes elevated in passage 8 keratinocytes, OAS1 was one of the most dramatic. The OAS family genes sense dsRNA and are important for viral defense and interferon activation. Indeed, other interferon response genes were noted as well (IFIT1 and ISG15 in Figure 3c). Thus, we performed unsupervised gene ontology analysis to identify broad categories of gene function that were modified in early versus late passage keratinocytes (Figure 3d). We found highly significant enrichment for innate immune activation (P = 3.4 × 10−13) but also dsRNA binding (P = 1.2 × 10−3). These results suggest that dsRNA activation of innate immune pathways might trigger a more primitive phenotype of keratinocytes in culture and loss of site-specific identity.
To test this we first verified these gene expression changes in volar keratinocytes from different donors after early or late passage. We found significant or suggestive increases in all four OAS family members (Figure 3e). Also, the OAS family is known to activate RNASEL, whose transcription was also elevated. Finally, KRT7 is a primitive keratin found in simple epithelia (Sandilands et al., 2013), and as evidence of loss of cellular identity, we found a concomitant increase of KRT7 expression (Figure 3e) as KRT9 expression decreased (Figure 3a and b). These results show a correlation between gain of primitive markers/loss of site-specific identity and dsRNA response pathways.
dsRNA is sufficient and its receptor DDX58 is necessary for KRT9 loss in passaged volar keratinocytes
For functional insight into the importance of dsRNA in the loss of keratinocyte identity, we first treated early passage keratinocytes with polyinosinic:polycytidylic acid (poly[I:C]) (a synthetic dsRNA) to see if this could recapitulate the loss of KRT9 expression. Indeed, dsRNA reduced KRT9 mRNA (Figure 3f) and protein (Figure 3g). In parallel, the dsRNA receptor DDX58—also up-regulated in late passage keratinocyte microarrays—also increased with dsRNA exposure (Figure 3f). This shows that dsRNA is sufficient to inhibit site-specific KRT9 expression.
Given the strong positive feedback for DDX58 expression with dsRNA exposure and the functional importance of DDX58 as an in vivo dsRNA receptor, we sought to investigate its role in inducing identity loss in volar keratinocytes. We first tested how DDX58 levels might change with passaging. We found that DDX58 mRNA abundance increases with passaging (Figure 4a) in a pattern inverse to that seen for KRT9 (Figure 3a and b). To test whether DDX58 is functionally required for loss of KRT9 expression, we treated late passage keratinocytes with DDX58 siRNA and increased KRT9 mRNA expression (Figure 4b). This also rescued KRT9 protein expression (Figure 4c). The effect on KRT7 was the opposite: KRT7 mRNA and protein decreased with DDX58 siRNA (Figure 4d). Finally, we tested if DDX58 siRNA could also rescue the OAS signature of late passaged keratinocytes. Indeed, OAS1, OAS2, and the downstream RNASEL transcript levels were decreased with siRNA treatment (Figure 4e). Although DDX58 appears functionally required, other dsRNA receptors such as TLR3 had no effect on KRT9 expression (see Supplementary Figure S2a online); similarly, although the OAS family genes appear to be good readout for dsRNA activity, they do not appear to regulate KRT9 expression (see Supplementary Figure S2b). These results show that DDX58 is uniquely required for loss of KRT9 and increase of OAS/KRT7 expression in volar keratinocytes with cell passaging.
Figure 4. The dsRNA receptor DDX58 increases with keratinocyte passaging; DDX58 loss rescues KRT9 expression.
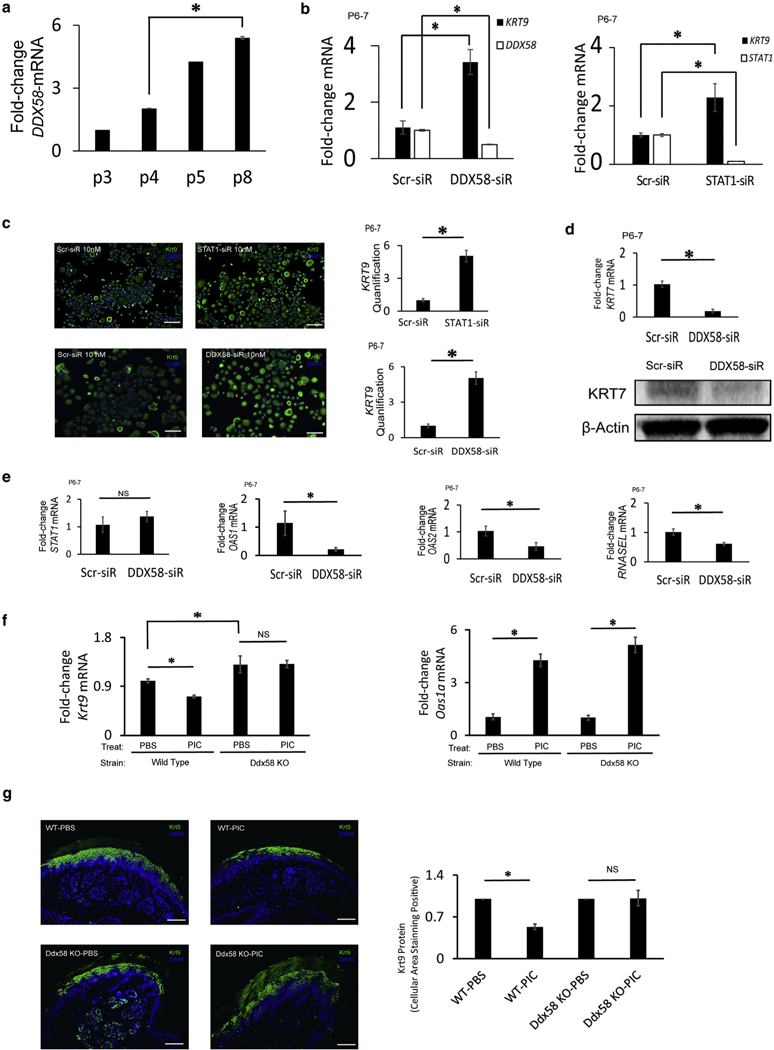
(a) DDX58 mRNA expression of volar keratinocytes increases with increased cell passages. n = 3, *P < 0.05. (b) DDX58 knockdown (DDX58-siR) in late passage volar keratinocytes increases KRT9 mRNA expression; downstream STAT1 knockdown (STAT1-siR) increases KRT9 mRNA expression also. n = 4, *P < 0.05. (c) KRT9 protein expression (green) increased when late passage volar keratinocytes were treated with DDX58-siR and STAT1-siR, as stained by immunofluorescence (left, blue nuclear DAPI stain) with signal quantification (right). n = 4, *P < 0.05. Scale bars = 200 μm. (d) DDX58 knockdown (DDX58-siR) in late passage volar keratinocytes inhibits KRT7 mRNA (top; n = 3, *P < 0.05) and protein expression (bottom). (e) DDX58 knockdown in late passage volar keratinocytes normalizes OAS and RNASEL expression, but not STAT1. n = 3, *P < 0.05. (f, g) In vivo, dsRNA inhibits (f) Krt9 mRNA and (g) protein (green) in wild-type but not Ddx58-null mice, despite equivalent inductions of Oas1a. n = 3, *P < 0.05. Scale bar = 100 μm. dsRNA, double-stranded RNA; KO, knockout; KRT, keratin; NS, not significant; p, passage; PBS, phosphate buffered saline; PIC, polyinosinic:polycytidylic acid; Scr, scramble; siR, small interfering RNA.
Although canonical signaling from DDX58 is through MAVS, siRNA treatment did not rescue KRT9 expression (see Supplementary Figure S2c). However, noncanonical interactions of DDX58 also occur with STAT1, although independently of the interferon receptor as an intermediate (Xu et al., 2018). We therefore tested STAT1 siRNA and discovered that KRT9 mRNA (Figure 4b) and protein (Figure 4c) are increased. IRF9, a STAT1/2 binding protein, did not, however, appear to be required (see Supplementary Figure S2d). These results indicate that the mechanism of KRT9 expression loss in passaged keratinocytes could be through a DDX58-STAT1 signaling pathway.
As a final experiment to test the in vivo relevance of these findings, we sought to test the ability of dsRNA to inhibit Krt9 expression in wild-type and DDX58-null strains. We injected dsRNA into mouse footpads and subsequently harvested the skin to test for gene and protein expression. We find that although Krt9 gene expression decreases in wild-type mice, it does not change in Ddx58-null mice, despite increases in Oasla expression (Figure 4f). Oas1a was induced by dsRNA in vivo in Ddx58-null mice but not in vitro in human kera-tinocytes (Figure 4e), possibly because of in vivo nonacute compensation. Regardless, dsRNA reduced Krt9 protein expression in wild-type mice but not in Ddx58-null mice (Figure 4g). These results corroborate our in vitro findings and show the physiologic relevance for how dsRNA in vivo inhibits site-specific gene expression of Krt9.
DISCUSSION
The question of whether the epidermal or mesenchymal layers contribute more to tissue identity is an important question with mixed previous findings. We show here that in the case of palmoplantar volar skin, both keratinocytes and fibroblasts contribute to tissue identity, as assayed by site-specific gene expression of KRT9. Despite the ability for keratinocytes to maintain positional memory, this is lost with passaging because of dsRNA sensing by its receptor DDX58. These results are significant in that they give information about the control of site-specific gene expression and spurious activation of dsRNA sensing with prolonged in vitro culture. Although cell passaging in vitro is an artificial condition, this study has important ramifications for the use of cellular therapy, and we argue that it provides an important context for comparison with in vivo dsRNA sensing pathways.
We have previously shown that dsRNA has the capacity to induce stem cell features in keratinocytes and regeneration/ morphogenesis during wound healing (Nelson et al., 2015; Zhu et al., 2017). In that work, for example, we noted that dsRNA had the ability to decrease markers of keratinocyte differentiation. We hypothesized that when released by damaged tissue, dsRNA maintained keratinocytes in an undifferentiated state and competent for morphogenesis at the time of re-epithelialization during wound closure. The present discovery of a strong dsRNA signature in an unbiased transcriptome analysis of identity loss in volar keratinocytes during cell passaging illustrates a second context in which dsRNA functions similarly and corroborates a physiologic role for dsRNA to maintain cells in a more undifferentiated state. This also dovetails with recent discoveries that antiviral pathways such as dsRNA sensing are a retained feature of a wide variety of stem cells (Wu et al., 2018). In sum, the present work adds to evidence that dsRNA controls differentiation pathways.
There are multiple implications of this work for current cellular therapy. First, efforts to modify skin identity such as at the stump site of amputees might benefit from dual cell therapy by including both keratinocytes and fibroblasts. There is no simple supremacy of dermal fibroblasts, at least in the case of volar skin, and reports of fibroblasts inducing dramatic morphogenesis might be compromised by “fibroblast only” fractions actually containing contaminating keratinocytes (Jahoda et al., 1984). Second, keratinocyte positional memory might complicate trials using gene-corrected keratinocyte grafts, for example, in blistering skin diseases. The use of a single highly expanded gene-corrected clone to correct broad swaths of skin might result in keratinocytes in which gene expression is not consistent with regional differences. This might also result in decreased long-term engraftment rates. Future work will need to assess the potential issues with anterior/posterior or dorsal/ventral axis mismatch cell therapy, for example.
Despite the presented findings, open questions persist. One is defining the exact role of homeobox genes such as the DLX family in maintaining volar identity. However, perhaps the most complicated is the identity of dsRNA sensed by DDX58. Although U1 spliceosomal RNA (U1) is proposed to be a ligand of multiple noninfectious contexts of dsRNA sensing (Bernard et al., 2012; Liu et al., 2016;Ranoa et al., 2016), we did not find elevations in U1 total levels or localization in late versus early passaged keratinocytes (see Supplementary Figure S3a and b online). Identifying the ligand for DDX58 in this context will require careful evaluation of not only potential ligand abundance but also cellular location. Another important question will be if dsRNA/ DDX58 loss of cell identity in culture is a broad mechanism in multiple cell types.
Another important question is defining the exact epigenetic changes and mechanisms that explain expression differences with passaging. An important candidate is the Polycomb complex (Botchkarev and Mardaryev, 2016;Dauber et al., 2016). We searched our microarrays of early versus late passage keratinocytes (Figure 3c) and found that multiple Polycomb members are lost with passaging (out of 49,325 transcripts, EZH2 (#204), CBX5 (#325), and PCGF5 (#725) most lost transcripts). Because skin Polycomb gene deletion in mice shows increases in nonepidermal gene expression, our demonstration of increases in primitive keratins such as KRT7 (and loss of skin-specific KRT9) with passaging could be in part related to decreased Polycomb activity. Hence, future work is important and necessary.
In conclusion, we have shown that keratinocytes retain positional memory but also receive important input from fibroblasts in controlling site-specific KRT9 expression (Knapp et al., 1986;Schweizer et al., 1989). We also show that dsRNA sensing by the DDX58 receptor inhibits positional memory with passaging. This work has implications for both future human cellular trials and basic studies of nonimmune roles of dsRNA sensing.
MATERIALS AND METHODS
Human samples
Human samples were taken under written informed consent and under a Johns Hopkins Medical Institution Review Board-approved protocol (NA_00033375) and followed Declaration of Helsinki principles. Skin biopsy specimens were obtained from foreskin and the dorsal and ventral positions of the foot and hand.
Microarray analysis
RNA was isolated from human volar and nonvolar epidermis (Fibrous Tissue RNA kit; Qiagen). Early and late passage volar keratinocyte RNA was submitted to the Deep Sequencing and Microarray core (Johns Hopkins Medical Institution) for Affymetrix Human Exon 1.0 ST microarray chips (Affymetrix, Santa Clara, CA). Gene expressions in the Affymetrix CEL files were extracted and normalized with Partek Genomics Suite software, version 6.6 (Partek, St. Louis, MO) using the robust multichip analysis algorithm. Analysis of variance was used to analyze the significance of gene expression. Microarray data is available at the National Center for Biotechnology Information Gene Expression Omnibus database under accession number GSE116055.
Primary cell isolation and culture
Skin tissues were separated using enzyme digestion methods as previously described (Kim et al., 2016). Human epidermal keratinocytes and dermal fibroblasts were isolated from newborn foreskin obtained after parents’ consent and from adult skin (scalp, hand, palm, foot and sole) with donors’ consent. Skin samples were incubated in 0.4% dispase II at 4 °C overnight. Next, epidermis was separated from dermis, and 0.025% trypsin-EDTA was used to isolate keratinocytes from epidermis. After removing epidermis, dermis was placed in 0.1% collagenase and incubated for 10 minutes at 37° C. Adult keratinocytes were cultured in KGM-GOLD (Lonza, Switzerland) media with 10 μmol/L of Rho kinase inhibitor (Y-27632, Cayman, Ann Arbor, MI). For all experiments, keratinocytes were used only after at least two passages to eliminate contaminating cells. Fibroblasts were cultured in DMEM with 10% fetal bovine serum and 1× antibiotic-antimycotic cocktail. Keratinocytes were plated by dropping 200,000 cells/20 μl of drops (three drops for a 12-well plate) using KBM (KGM media as described but without any supplements) media to induce differentiation (termed postconfluent). Unless otherwise specified, sole keratinocytes were used as volar keratinocytes. Foreskin keratinocytes were used as nonvolar keratinocytes. For co-culture experiments, the mixture of keratinocytes and fibroblasts (5:1 ratio) was plated by dropping as described and incubated for 48 hours in a 12-well plate. At the end of experiments, RNA and protein were isolated for further analysis. For volar keratinocytes, the sole was used. For nonvolar keratinocytes, foreskin was used and was found to be similar to dorsal hand skin.
RNA isolation and quantitative real-time reverse transcriptase–PCR
Total RNA was extracted from skin tissue or cells using RNeasy Mini Kit (Qiagen, Valencia, CA). The concentration of RNA was determined by Nano Drop 2000 (Thermo Scientific, West Palm Beach, FL). Next, RNA was reverse transcribed to cDNA using high-capacity cDNA Reverse Transcription Kit (Life Technologies, Waltham, MA). Quantitative real-time reverse transcriptase PCR analysis was performed using TaqMan probes and TaqMan Fast Reaction Master Mix (see Supplementary Table S1 online). Relative expressions of mRNA were analyzed by using target genes’ CT values, normalizing to RPLP0 for human and Actb for mouse as housekeeping genes and computed by ΔΔCT method.
Mouse experiments
All animal protocols are approved by the Johns Hopkins Medical Institution Animal Care and Use Committee. C57BL/6J mice (Jackson Laboratory, Bar Harbor, ME) were used as wild-type controls. Ddx58-nuII mice in 129Sv/C57BL/6 background were provided by the Adolfo García-Sastre laboratory (New York, NY). PoIy(I:C) or phosphate buffered saline was injected subcutaneously into paws; 20 μl of 1 μg/mI poIy(I:C) or phosphate buffered saline was injected into foreIimbs, and 40 μI of 1 μg/mI poIy(I:C) or phosphate buffered saIine was injected into hindIimbs of 4- to 5-week-oId maIe and femaIe mice. Mice were injected three times every other day. At 24 hours after the Iast injection, paw skin was used for quantitative reaI-time reverse transcriptase–PCR and immunostaining.
For pyrosequencing, siRNA, Western bIot, poIy(I:C) treatment, immunofluorescence in situ, Taqman probes, and siRNA probes, see the SuppIementary MateriaIs onIine.
Supplementary Material
ACKNOWLEDGMENTS
The laboratory of Adolfo García-Sastre kindly provided Ddx58-null mice. The authors also thank Conover Talbot, Jr. (JHMI Deep Sequencing and Microarray Core) for assistance with microarray analysis. Research reported in this publication was supported by the National Institute of Arthritis, MusculoskeIetal, and Skin Diseases, part of the National Institutes of Health, under R01AR064297 and AR068280 to LAG. This work was also supported by the Department of Defense, Armed Forces Institute of Regenerative Medicine, Extremities Regeneration (AFIRM2-ER11), CDMRMP W81XWH-16-C-0167, and Northrop Grumman Electronic Systems, as well as the Thomas Provost, MD Young Faculty Development Fund ofJohns Hopkins Dermatology to LAG. Dongwon Kim was supported by a postdoctoral fellowship from MaryIand Stem Cell Research Grant (2017-MSCRFF-3905).
Abbreviations:
- dsRNA
double-stranded RNA
- KRT
keratin
- poly(I:C)
poly-inosinic:polycytidylic acid
- siRNA
small interfering RNA
Footnotes
SUPPLEMENTARY MATERIAL
Supplementary material is linked to the online version of the paper at www.jidonline.org, and at https://doi.org/10.1016/j.jid.2018.07.021.
CONFLICT OF INTEREST
The authors state no conflict of interest
REFERENCES
- Bernard JJ, Cowing-Zitron C, Nakatsuji T, Muehleisen B, Muto J, Borkowski AW, et al. Ultraviolet radiation damages self noncoding RNA and is detected by TLR3. Nat Med 2012;18:1286–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billingham RE, Silvers WK. The origin and conservation of epidermal speci-ficities. N Engl J Med 1963;268:539–45. [DOI] [PubMed] [Google Scholar]
- Billingham RE, Silvers WK. Studies on the conservation of epidermal speci-ficities of skin and certain mucosas in adult mammals. J Exp Med 1967;125:429–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botchkarev VA, Mardaryev AN. Repressing the keratinocyte genome: how the Polycomb complex subunits operate in concert to control skin and hair follicle development. J Invest Dermatol 2016;136:1538–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HY, Chi JT, Dudoit S, Bondre C, van de Rijn M, Botstein D, et al. Diversity, topographic differentiation, and positional memory in human fibroblasts. Proc Natl Acad Sci USA 2002;99:12877–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codispoti A, Colombo E, Zocchi L, Serra V, Pertusi G, Leigheb G, et al. Knuckle pads, in an epidermal palmoplantar keratoderma patient with Keratin 9 R163W transgrediens expression. Eur J Dermatol 2009;19:114–8. [DOI] [PubMed] [Google Scholar]
- Dauber KL, Perdigoto CN, Valdes VJ, Santoriello FJ, Cohen I, Ezhkova E. Dissecting the roles of Polycomb repressive complex 2 subunits in the control of skin development. J Invest Dermatol 2016;136:1647–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhouailly D Dermo-epidermal interactions between birds and mammals: differentiation of cutaneous appendages. J Embryol Exp Morphol 1973;30: 587–603. [PubMed] [Google Scholar]
- Eames BF, Schneider RA. Quail-duck chimeras reveal spatiotemporal plasticity in molecular and histogenic programs of cranial feather development. Development 2005;132:1499–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu DJ, Thomson C, Lunny DP, Dopping-Hepenstal PJ, McGrath JA, Smith FJD, et al. Keratin 9 is required for the structural integrity and terminal differentiation of the palmoplantar epidermis. J Invest Dermatol 2014;134: 754–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins CA, Chen JC, Cerise JE, Jahoda CA, Christiano AM. Microenvironmental reprogramming by three-dimensional culture enables dermal papilla cells to induce de novo human hair-follicle growth. Proc Natl Acad Sci USA 2013;110:19679–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch T, Rothoeft T, Teig N, Bauer JW, Pellegrini G, De Rosa L, et al. Regeneration of the entire human epidermis using transgenic stem cells. Nature 2017;551:327–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahoda CA, Horne KA, Oliver RF. Induction of hair growth by implantation of cultured dermal papilla cells. Nature 1984;311:560–2. [DOI] [PubMed] [Google Scholar]
- Kim D, Hossain MZ, Nieves A, Gu L, Ratliff TS, Mi Oh S, et al. To control site-specific skin gene expression, autocrine mimics paracrine canonical Wnt signaling and is activated ectopically in skin disease. Am J Pathol 2016;186:1140–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp AC, Franke WW, Heid H, Hatzfeld M, Jorcano JL, Moll R. Cytokeratin no. 9, an epidermal type I keratin characteristic of a special program of keratinocyte differentiation displaying body site specificity. J Cell Biol 1986;103:657–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Gu Y, Han Y, Zhang Q, Jiang Z, Zhang X, et al. Tumor exosomal RNAs promote lung pre-metastatic niche formation by activating alveolar epithelial TLR3 to recruit neutrophils. Cancer Cell 2016;30:243–56. [DOI] [PubMed] [Google Scholar]
- Mavilio F, Pellegrini G, Ferrari S, Di Nunzio F, Di Iorio E, Recchia A, et al. Correction of junctional epidermolysis bullosa by transplantation of genetically modified epidermal stem cells. Nature Med 2006;12:1397. [DOI] [PubMed] [Google Scholar]
- Millar SE. Molecular mechanisms regulating hair follicle development. J Invest Dermatol 2002;118:216–25. [DOI] [PubMed] [Google Scholar]
- Nelson AM, Reddy SK, Ratliff TS, Hossain MZ, Katseff AS, Zhu AS, et al. dsRNA Released by Tissue Damage Activates TLR3 to Drive Skin Regeneration. Cell Stem Cell 2015;17:139–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranoa DR, Parekh AD, Pitroda SP, Huang X, Darga T, Wong AC, et al. Cancer therapies activate RIG-I-like receptor pathway through endogenous non-coding RNAs. Oncotarget 2016;7:26496–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis A, Hennies HC, Langbein L, Digweed M, Mischke D, Drechsler M, et al. Keratin 9 gene mutations in epidermolytic palmoplantar keratoderma (EPPK). Nat Genet 1994;6:174–9. [DOI] [PubMed] [Google Scholar]
- Sandilands A, Smith FJ, Lunny DP, Campbell LE, Davidson KM, MacCallum SF, et al. Generation and characterisation of keratin 7 (K7) knockout mice. PLoS One 2013;8:e64404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer J, Baust I, Winter H. Identification of murine type I keratin 9 (73 kDa) and its immunolocalization in neonatal and adult mouse foot sole epidermis. Exp Cell Res 1989;184:193–206. [DOI] [PubMed] [Google Scholar]
- Sebastiano V, Zhen HH, Haddad B, Bashkirova E, Melo SP, Wang P, et al. Human COL7A1-corrected induced pluripotent stem cells for the treatment of recessive dystrophic epidermolysis bullosa. Sci Transl Med 2014;6: 264ra163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umegaki-Arao N, Pasmooij AM, Itoh M, Cerise JE, Guo Z, Levy B, et al. Induced pluripotent stem cells from human revertant keratinocytes for the treatment of epidermolysis bullosa. Sci Transl Med 2014;6:264ra164. [DOI] [PubMed] [Google Scholar]
- Wu X, Dao Thi VL, Huang Y, Billerbeck E, Saha D, Hoffmann HH, et al. Intrinsic Immunity Shapes Viral Resistance of Stem Cells. Cell 2018;172: 423–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu XX, Wan H, Nie L, Shao T, Xiang LX, Shao JZ. RIG-I: a multifunctional protein beyond a pattern recognition receptor. Protein Cell 2018;9:246–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Itami S, Tarutani M, Hosokawa K, Miura H, Yoshikawa K. Regulation of keratin 9 in nonpalmoplantar keratinocytes by palmoplantar fibroblasts through epithelial-mesenchymal interactions. J Invest Dermatol 1999;112:483–8. [DOI] [PubMed] [Google Scholar]
- Zhu AS, Li A, Ratliff TS, Melsom M, Garza LA. After skin wounding, non-coding dsRNA coordinates prostaglandins and Wnts to promote regeneration. J Invest Dermatol 2017;137:1562–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


