Abstract
A combination therapy of multiple drugs including isoniazid, rifampicin, ethambutol and pyrazinamide has been proven to be an effective option for the vast majority of tuberculosis (TB) patients. However, various adverse drug reactions (ADRs) limit its merit, with anti-TB drug-induced hepatotoxicity (ATDH) being a common and sometimes severe ADR. This study aimed to investigate the association between polymorphisms in two nuclear receptor genes, pregnane X receptor (PXR) and constitutive androstane receptor (CAR), and the risk of ATDH in a Chinese population. Subjects with or without hepatotoxicity during anti-TB treatment were recruited. DNA was extracted from peripheral blood and genotypes of the selected single nucleotide polymorphisms (SNPs) were determined by using the improved multiplex ligation detection reaction technique. Three genetic models (additive, dominant, and recessive) as well as haplotype, SNP-SNP interaction analyses were used to evaluate the genetic risk of ATDH. A total of 502 subjects (203 ATDH and 299 non-ATDH) were enrolled. The results showed that the minor allele of rs7643645 and the H0010001 haplotype in PXR were associated with decreased risk of ATDH, suggesting that drug-metabolizing enzymes regulated by PXR are involved in the pathogenesis of ATDH. More studies are required to verify this result.
Introduction
Tuberculosis (TB) remains one of the world’s biggest health problems, with 10 million new cases and approximately 1.6 million deaths in 2017 reported by WHO1. China had the second largest number of new TB cases in the world in 2017, with 889,000 estimated new cases and an incidence of 63/100,0001. Currently, 86% of newly diagnosed cases can be successfully treated, mainly due to the widespread use of standard chemotherapy (a combination of at least isoniazid, rifampicin, ethambutol and pyrazinamide for 2 months, followed by 4 months of at least isoniazid and rifampicin) in drug sensitive patients1. However, 15% of patients receiving treatment with these first line anti-TB drugs developed adverse drug reactions, resulting in suspension or discontinuation of one or more anti-TB drugs2, thereby increasing the risk of TB relapse, drug resistance, and even TB–related death3. Hepatotoxicity is a common and sometimes severe adverse drug reaction that occurs during anti-TB treatment, with an incidence of 2–28%, depending on the definition of hepatotoxicity and the specific population surveyed4.
There are four possible pathogenic mechanisms of drug-induced liver injury including the generation of reactive metabolites, oxidative stress, mitochondrial dysfunction and immunological response5. Unfortunately, the exact mechanisms of ATDH are not yet fully understood but multiple risk factors have been reported to associate with ATDH including genetic factors. Both environmental factors and genetic factors, as well as their interaction have been reported to be associated with ATDH6. Howerver, results from different study were inconsistent6. In addition, some studies indicated gene-gene and gene-environmental interaction may account for some variant susceptibility to ATDH in certain population7,8.
Genetic variants in drug metabolizing enzymes such as N-acetyltransferase 2 (NAT2), cytochrome P450 2E1 (CYP2E1) and glutathione S-transferases (GSTM1, GSTT1), have been the most widely studied in respect to ATDH susceptibility in the Chinese population9–11, as well as other populations12. However, these genetic biomarkers have shown poor reproducibility in some studies. Other genetic predictors of ATDH were implicated in certain populations, e.g. ATP binding cassette subfamily B member 1 in Brazilians or human leukocyte antigen in Indians13,14. In our previous study, we observed cytochrome P450 2B6 variants may associate with susceptibility to ATDH but only in males15. However, the findings to date may only represent a small minority of the genetic variants relevant to ATDH and a more reliable biomarker is needed for predicting ATDH. Genetic variants influencing the expression and/or activities of target proteins may result in a varying degree of accumulation of hepatotoxins in TB drug metabolism. However, ATDH remains unpredictable even when genetic and environmental factors are taken into account16. Therefore, we increased the sample size of our previous study to explore other candidate genes in addition to the phase I, II or III drug-metabolizing enzymes that were the focus of most previous studies.
The nuclear receptor subfamily 1 group I member 2 (NR1I2) gene, also known as pregnane X receptor (PXR), was first identified in 1988 as a member of the nuclear receptor superfamily, and is primarily expressed in the liver, intestine and kidney17. When activated by an agonist such as rifampicin, PXR up-regulates a large number of target drug-metabolizing enzyme genes including the cytochrome P450 family, carboxylesterases, glutathione S-transferases, UDP-glucuronosyltransferase, etc18, resulting in enhanced drug metabolism. Thus, during combination treatment with multiple anti-TB drugs, PXR may contribute to drug-drug interactions, and lead to variability in drug metabolism and disposition. Li et al. found that co-administration of rifampicin and isoniazid leads to higher levels of alanine aminotransferase (ALT), alkaline phosphatase, and bile plugs through a PXR-mediated minor metabolic pathway in a humanized PXR mouse model19. Moreover, rifampicin-mediated PXR activation can function as a negative regulator of inflammation and immunity, linking drug and xenobiotic metabolism to immune responses, through inhibition of the NF-κB signaling pathway20,21. In addition, a heightened sensitivity to oxidative toxicants was noted in both cell and mouse models upon PXR activation22. With improvement of genetic research methodology, a large number of single nucleotide polymorphisms (SNPs) in the PXR gene were proven to influence the function of PXR23, as well as an individual’s susceptibility to drug-associated liver injury24.
The nuclear receptor subfamily 1 group I member 3 (NR1I3), also known as the constitutive androstane receptor (CAR), is highly expressed in the liver and showed an overlapping set of target genes with PXR in response to potentially harmful chemicals25,26. It has been demonstrated that CAR gene polymorphisms may play a role in the inter-individual variability in the expression of many drug-metabolizing enzymes and in the susceptibility to drug-induced hepatotoxicity27. Therefore, the aim of present study was to investigate a possible correlation between PXR and CAR gene polymorphisms, together with the interaction of SNP-SNP, and risk of ATDH in Chinese TB patients.
Results
Patient characteristics
A total of 502 TB patients were enrolled in our study, including 203 ATDH and 299 non-ATDH. There were no statistically significant differences in the age, gender, ethnicity, height, weight, smoking, alcohol abuse and positive hepatitis B surface antigen (HbsAg) results between the patients with hepatotoxicity and those without hepatotoxicity (see Table 1 for details).
Table 1.
Clinical and demographic characteristics of the cases and controls.
| ATDH group (n = 203) | No-ATDH group (n = 299) | P value | |
|---|---|---|---|
| Age, years† | 38.58 ± 16.42 | 38.42 ± 16.81 | 0.913$ |
| Age, years# | 36, 25.5, 68 | 35, 26, 71 | |
| Female | 108 (53.2) | 146 (48.8) | 0.336& |
| Ethnicity‡ | 0.072*,§ | ||
| Han | 181 (89.2) | 280 (93.6) | |
| Tibetan | 13 (6.4) | 13 (4.3) | |
| Yi | 7 (3.4) | 6 (2.0) | |
| Others | 2 (1.0) | 0 (0) | |
| Smoker‡ | 57 (29.2) | 77 (26.4) | 0.489§ |
| Alcohol abuse‡ | 25 (12.8) | 33 (11.4) | 0.641§ |
| Height, centimeter† | 162.85 ± 7.42 | 164.98 ± 7.97 | 0.125$ |
| Weight, kilogram† | 54.78 ± 9.89 | 54.94 ± 10.30 | 0.866$ |
| Positive HBsAg‡ | 18 (9.4) | 15 (5.0) | 0.059§ |
†Data shown as mean ± standard deviation; #data shown as median, interquartile range, range; ‡data shown as number of cases (frequency) §analyzed by chi-square test; $analyzed by independent sample t test. *P value was calculated between the Han and the other populations.
ATDH: antituberculosis drug-induced hepatotoxicity.
The severity and timing of ATDH onset are presented in Table 2. Among the ATDH group, 25.6%, 38.4% and 17.2% presented grade 1, grade 2 and grade 3 hepatotoxicity, respectively and 18.7% developed severe liver injury with ALT >10 upper limit of normal (ULN). About half of the cases of ATDH occurred in the first month of anti-tuberculosis treatment (see Table 2 for details).
Table 2.
Frequency of hepatotoxicity according to severity and onset of antituberculosis drug-induced hepatotoxicity.
| N (total = 203) | Proportion | |
|---|---|---|
| Severity of hepatotoxicity† | ||
| Grade 1 (ALT 2–2.5 times ULN) | 52 | 25.6% |
| Grade 2 (ALT 2.5–5 times ULN) | 78 | 38.4% |
| Grade 3 (ALT 5–10 times ULN) | 35 | 17.2% |
| Grade 4 (ALT > 10 × ULN) | 38 | 18.7% |
| Onset of hepatotoxicity after antituberculosis drug usage | ||
| <30 days | 104 | 51.2% |
| 30–60 days | 60 | 29.6% |
| 60–90 days | 14 | 6.9% |
| >90 days | 25 | 12.3% |
†Severity of hepatotoxicity is classified according to the WHO Toxicity Classification Standards. The criterion for grade 1 was modulated to 2–2.5 times ULN in this study.
ULN: upper limit of normal.
Association of PXR and CAR genetic polymorphisms with the risk of ATDH
Relevant features of the selected SNPs of PXR and CAR are shown in Table S1. The concordance rate of genotype results for the blind repeated samples was 99.5%. Genotype distributions of the SNPs in the ATDH and non-ATDH groups are presented in Table 3, and all the SNPs conformed to Hardy-Weinberg equilibrium (HWE) (p > 0.05). As shown in Table 3, the rs7643645 SNP in PXR showed statistical significance (additive model: OR = 0.704, 95% CI 0.539–0.918, P = 0.010; Dominant model: OR = 0.609, 95% CI 0.405–0.917, P = 0.017). Compared with the major allele, carrying the minor (G) allele was significantly associated with decreased ATDH risk. We also observed that the proportion of rs7643645 allele G decreased as the severity of ATDH increased (trend P = 0.003): 50.5% in controls, 43.8% in grade 1 and 2 groups, and 38% in grade 3 and 4 groups. However, we did not detect any association between the other 12 SNPs and risk of ATDH (Table 3).
Table 3.
Association analysis between the PXR and CAR SNPs and antituberculosis-drug induced hepatotoxicity.
| SNP | Genotype†, n case/control | PHWE¶ | Additive* | Dominant* | Recessive* | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0/0 | 0/1 | 1/1 | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | |||
| PXR | |||||||||||
| rs7643645 | 70/73 | 97/150 | 36/76 | 0.785 | 0.704 (0.539–0.918) | 0.010 | 0.609 (0.405–0.917) | 0.017 | 0.645 (0.405–1.026) | 0.064 | |
| rs6785049 | 65/104 | 107/137 | 31/58 | 0.954 | 0.972 (0.743–1.271) | 0.833 | 1.208 (0.809–1.804) | 0.356 | 0.677 (0.408–1.123) | 0.131 | |
| rs3732357 | 88/131 | 97/139 | 18/29 | 0.143 | 1.012 (0.756–1.355) | 0.936 | 1.030 (0.706–1.503) | 0.878 | 0.973 (0.510–1.855) | 0.934 | |
| rs3814055 | 117/174 | 71/107 | 15/18 | 0.416 | 1.112 (0.823–1.502) | 0.489 | 1.068 (0.732–1.559) | 0.731 | 1.473 (0.707–3.066) | 0.301 | |
| rs2472682 | 71/100 | 102/149 | 30/50 | 0.447 | 0.950 (0.723–1.248) | 0.713 | 1.001 (0.674–1.487) | 0.995 | 0.835 (0.499–1.395) | 0.490 | |
| rs3814057 | 45/74 | 110/145 | 48/80 | 0.716 | 1.005 (0.770–1.312) | 0.970 | 1.299 (0.831–2.030) | 0.251 | 0.788 (0.509–1.221) | 0.286 | |
| rs2472677 | 82/133 | 88/142 | 33/44 | 0.230 | 0.968 (0.740–1.266) | 0.811 | 0.891 (0.607–1.306) | 0.552 | 1.097 (0.651–1.847) | 0.729 | |
| CAR | |||||||||||
| rs6686001 | 136/194 | 57/98 | 10/7 | 0.818 | 0.991 (0.704–1.395) | 0.958 | 0.912 (0.614–1.354) | 0.648 | 1.766 (0.619–5.034) | 0.288 | |
| rs2307424 | 59/81 | 94/150 | 50/68 | 0.560 | 0.937 (0.719–1.221) | 0.628 | 0.884 (0.584–1.337) | 0.559 | 0.957 (0.610–1.502) | 0.848 | |
| rs4073054 | 159/243 | 41/54 | 3/2 | 0.815 | 1.160 (0.752–1.789) | 0.502 | 1.149 (0.723–1.823) | 0.556 | 1.753 (0.243–12.64) | 0.578 | |
| rs3003596 | 59/90 | 96/147 | 48/62 | 0.560 | 1.032 (0.788–1.351) | 0.818 | 0.997 (0.661–1.504) | 0.989 | 1.104 (0.692–1.763) | 0.677 | |
| rs7530560 | 75/99 | 89/146 | 39/54 | 0.386 | 0.986 (0.754–1.289) | 0.919 | 0.917 (0.616–1.366) | 0.670 | 1.086 (0.670–1.760) | 0.739 | |
| rs2502805 | 94/129 | 82/127 | 27/43 | 0.066 | 0.975 (0.743–1.279) | 0.855 | 0.979 (0.668–1.434) | 0.913 | 0.943 (0.547–1.627) | 0.833 | |
†“0” represents the major allele, “1” represents the minor allele.
¶HWE: Hardy-Weinberg equilibrium. HWE was assessed by the χ2 goodness-of-fit test based on the genotype distributions in this study.
*Adjusted for ethnicity, age, gender, height, weight, smoking, drinking, and HbsAg status. Additive: minor allele homozygotes versus heterozygotes versus major allele homozygotes. Dominant: heterozygotes plus minor allele homozygotes versus major allele homozygotes. Recessive: minor allele homozygotes versus major allele homozygotes plus heterozygotes.
Association of the haplotypes of PXR and CAR with risk of ATDH
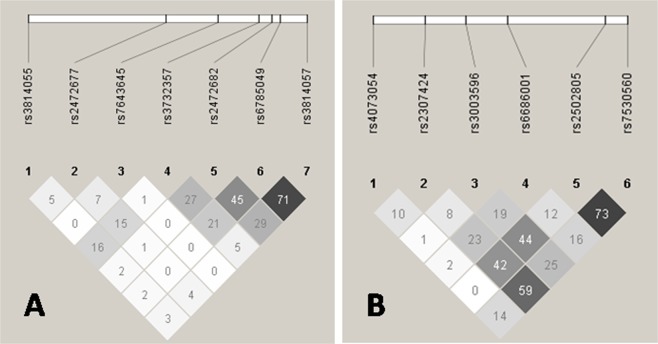
The linkage disequilibrium (LD) plots of selected PXR and CAR SNPs in the study population are shown in Fig. 1. Low LD among these SNPs was observed in both genes, except moderate LD between rs6785049 and rs3814057 in PXR, and between rs2502805 and rs7530560 in CAR. By haplotype analyses of these tagSNPs, we found 9 haplotypes in PXR and 6 haplotypes in CAR with a frequency more than 0.03 (Tables 4 and 5, respectively). Among these common haplotypes, one haplotype (H0010001) in the PXR gene had a significant association with decreased ATDH risk (OR = 0.591, 95% CI 0.377–0.927, P = 0.021).
Figure 1.
Linkage disequilibrium (LD) plots for PXR (A) and CAR (B). The LD plots were generated by Haploview 4.2. Polymorphisms are identified by their dbSNP rs numbers, and their relative positions are marked by vertical lines within the white horizontal bar. The numbers within squares indicate the r2 value, expressed as a percentile.
Table 4.
Analyses of derived haplotypes (frequency more than 3%) from seven polymorphisms of PXR with the risk of ATDH.
| Haplotypes† | ATDH group | Non-ATDH group | OR | 95% CI | P value | ||
|---|---|---|---|---|---|---|---|
| n | % | n | % | ||||
| H0000001 | 24 | 5.9 | 26 | 4.4 | 1.362 | 0.767–2.419 | 0.291 |
| H0010001 | 31 | 7.7 | 71 | 11.9 | 0.591 | 0.377–0.927 | 0.021 |
| H0010110 | 36 | 9.0 | 50 | 10.5 | 1.069 | 0.678–1.686 | 0.774 |
| H0100110 | 34 | 8.4 | 59 | 9.9 | 0.806 | 0.514–1.263 | 0.346 |
| H0101000 | 21 | 5.2 | 23 | 3.8 | 1.280 | 0.688–2.383 | 0.435 |
| H0101001 | 16 | 3.9 | 25 | 4.2 | 0.839 | 0.466–1.858 | 0.931 |
| H0110110 | 27 | 6.7 | 31 | 5.1 | 1.315 | 0.768–2.252 | 0.318 |
| H1101001 | 43 | 10.5 | 51 | 8.5 | 1.138 | 0.730–1.776 | 0.568 |
| H1111001 | 16 | 3.9 | 31 | 5.2 | 0.730 | 0.391–1.360 | 0.320 |
†“0” represents the major alleles, “1” represents the minor alleles.
Order of polymorphisms: rs3814055, rs2472677, rs7643645, rs3732357, rs2472682, rs6785049, rs3814057. ATDH: antituberculosis drug-induced hepatotoxicity.
Table 5.
Analyses of derived haplotypes (frequency more than 3%) from six examined polymorphisms of CAR with the risk of ATDH.
| Haplotypes† | ATDH group | Non-ATDH group | OR | 95% CI | P value | ||
|---|---|---|---|---|---|---|---|
| n | % | n | % | ||||
| H001000 | 114 | 28.0 | 169 | 28.4 | 0.946 | 0.714–1.254 | 0.699 |
| H011000 | 47 | 11.7 | 57 | 9.6 | 1.210 | 0.804–1.819 | 0.360 |
| H010011 | 115 | 28.2 | 182 | 30.5 | 0.863 | 0.653–1.141 | 0.300 |
| H111001 | 28 | 6.9 | 33 | 5.5 | 1.239 | 0.735–2.089 | 0.421 |
| H000100 | 76 | 19.0 | 107 | 17.9 | 1.040 | 0.751–1.440 | 0.815 |
| H110011 | 18 | 4.5 | 23 | 3.9 | 1.139 | 0.607–2.135 | 0.686 |
†“0” represents the major alleles, “1” represents the minor alleles.
Order of polymorphisms: rs4073054, rs2307424, rs3003596, rs6686001, rs2502805, rs7530560. ATDH: antituberculosis drug-induced hepatotoxicity.
SNP-SNP interactions with risk of ATDH
We carried out a multifactor dimensionality reduction (MDR) analysis with all the tested SNPs to investigate potential genetic interactions associated with ATDH. We limited the interaction models from two-way to five-way. However, we did not identify a multilocus model with receivable cross-validation consistency (from 2/10–5/10). Moreover, all these models did not reach the threshold value of statistical significance (Table S2).
Power analysis
We calculated the power of the sample size for the 13 selected SNPs under the allelic models (Figs S1 and S2). The results showed that our study has reasonable power (>90%) to draw conclusions with OR1.6 or above, except rs4073054 which has a low MAF of 0.105.
Discussion
ATDH remains a serious problem for TB prevention and treatment due to the limited understanding of its pathogenesis. Therefore, biomarkers which can detect patients with different susceptibility to ATDH are urgently needed to further understanding of the pathogenesis of this adverse event and for guiding clinical decision making28. In the present association study, we focused on the genetic polymorphisms of the nuclear receptor, which can influence a wide range of metabolizing enzymes in the liver that are involved in drug-drug interactions29. We found that rs7643645 and a haplotype of the PXR gene were associated with ATDH. The PXR rs7643645 G allele and individuals carrying the H0010001 haplotype showed significantly decreased risk for ATDH.
Genetic polymorphisms of NAT2, CYP2E1 and the GST family are the most studied in relation to ATDH, for they may affect the metabolism and elimination of isoniazid, or are involved in oxidant-antioxidant balance in liver cells after exposure to anti-TB drugs12. We previously demonstrated that CYP2E1, GSTP1 and CYP2B6 were associated with susceptibility to ATDH by a meta-analysis and a case-control study15,30,31. Moreover, NAT2 mediates isoniazid biotransformation to form acetylhydrazine, which then undergoes oxidization by CYP2E1 to form a hepatotoxic substance32, but there is little evidence to implicate these enzymes in the metabolism of other anti-TB drugs such as pyrazinamide, which account for approximately 60% of ATDH33. The incidence of liver injury was higher in patients with co-treatment of rifampicin and isoniazid than taking either drug alone34. A widely accepted explanation for this is based on the rifampicin-mediated up-regulation of metabolic enzymes, which may contribute to isoniazid bioactivation and toxicity35,36. PXR serves as the main acceptor of rifampicin, and may be involved in this drug-drug interaction and the pathogenesis of ATDH. A previous study suggested that multiple SNPs, including rs7643645, in the PXR gene influenced the expression of PXR and its target genes in donor livers with or without rifampicin pretreatment23. The G allele of rs7643645 showed loss of a potential hepatic nuclear factor 4α binding site and was associated with lower CYP3A, MDR1 and PXR mRNA expression, compared to the A allele37,38. The lower level of drug-drug interaction in individuals carrying the PXR rs7643645 GG genotype may be a potential protective mechanism, which is consistent with our result. Another potential pathogenesis of ATDH is the imbalance between production of reactive oxygen species and detoxification of reactive intermediates39. Gong et al. found that mice treated with a PXR agonist and cell lines with activated human PXR showed heightened sensitivity to oxidative toxicants, with an increased production of reactive oxygen species22. Therefore, further study is needed to determine whether the rs7643645 G allele confers lower PXR activity and expression, and reduced susceptibility to oxidative stress and ATDH.
Surprisingly, a previous study by Wang et al. concluded that the rs7643645 AA genotype was associated with the decreased risk of ATDH in females40. However, a large number of studies indicate that females have a higher risk of ATDH than males41,42. CYP3A activity was higher in females compared with males43. We reasoned that the rs7643645 GG genotype with lower CYP3A activity may lead to decreased risk of ATDH. This hypothesis should be tested in future studies. Moreover, we excluded the potential risk factors of ATDH such as cancer and renal failure, which made up nearly 30% of the subjects by Wang et al. study and this may make our results more reliable. However, some differences between two studies should also be taken into consideration, e.g. the study design (Wang et al. performed a prospectively conducted study40, while we utilized a case-control study) and the definition of ATDH (Wang et al. defined ATDH as ALT > 5 × ULN in asymptomatic patients and ALT > 3 × ULN in symptomatic patients40, while we used criteria recommended by an international consensus meeting which are described in the methods section). Another genetic association study by Zazuli et al. demonstrated that PXR rs3814055 was associated with risk of ATDH44, which is inconsistent with our result. We believe these conflicting findings may be due to differences in ethnicity and definition of ATDH between the studies. In the study by Zazuli et al., ATDH was designated as an increase in serum ALT and AST levels above the ULN. Moreover, if we raised the ALT threshold (ALT > 5 ULN) for ATDH, that recommended by the Chinese Society of Hepatology in 201545, to further explore the clinical utility of testing for rs7643645, we observed a stable positive result (cases: 73, controls 299; additive model: OR = 0.601, 95% CI = 0.416–0.817, P = 0.006; dominant model: OR = 0.400, 95% CI = 0.190–0.842, P = 0.013; recessive model: OR = 0.574, 95% CI = 0.335–0.986, P = 0.043).
We did not find any positive results for the SNPs in the CAR genetic region. There are several reasons to explain this lack of association. Firstly, although the sample size of our study gave an acceptable power (>90%) to detect a common risk allele with an OR of 1.6 in most SNPs, the analysis of rs4073054 was still underpowered (Figs S1 and S2). Secondly, antituberculosis drugs and their metabolites were metabolized by enzymes such as NAT2 or CYP2E128. CAR or its genetic variation may not play a pivotal role in this specific pathway. Given that combined analyses of SNPs may display a more complete picture of the candidate genes, we further conducted a haplotype analysis and, a SNP-SNP interaction analysis of the selected tagSNPs. We only found that a haplotype in PXR (H0010001) showed decreased risk of ATDH.
There are several strengths of our study. We limited the confounding factors by excluding the comorbidities of HIV infection, cancer, renal failure or cardiac dysfunction and estimated the association between genetic factors and risk of ATDH with adjustment for other confounders. Moreover, in order to avoid potential bias, those who were in charge of clinical data and those who were responsible for laboratory data worked independently in this study. In addition, SNPs in the Chinese Beijing Han population located between 3000 bp upstream and 300 bp downstream of each gene were systematically reviewed to maximize inclusion of functional SNPs. Although some non-Han subjects were included in this study (i.e. 26 Tibetan, 13 Yi, and 2 others), a previous genome-wide association study found that Chinese Han and Tibetans share a similar genetic background46. There is a paucity of similar studies for Yi and other Chinese populations. Therefore, we had no alternative but to choose the SNP information of the Beijing Han population. We believe that the small proportion of Yi and other groups (3%) had only a minor effect on our results. If the subgroup of Chinese Han only was analyzed, we found rs7643645 was still significantly associated with decreased risk of ATHD (cases: 181, control: 280; additive model: OR = 0.607, 95% CI = 0.461–0.800, P < 0.001; dominant model: OR = 0.504, 95% CI = 0.334–0.760, P = 0.001; recessive model: OR = 0.539, 95% CI = 0.332–0.875, P = 0.012).
Nevertheless, some potential limitations should be taken into consideration when interpreting our results. Firstly, the activity of enzymes including PXR, CAR and other related downstream metabolic enzymes were not evaluated in the individuals. Secondly, a large number of previous studies have shown that polymorphisms of multiple genes may be involved in ATDH, suggesting the occurrence of ATDH may result from the combination of variation in several susceptibility genes, such as NAT2 or CYP2E147. However, we did not genotype other candidate genes. Thirdly, in this retrospective case-control study, there were some inevitable biases in the characteristics of the patients. We limited these biases by using a combination of face-to-face questionnaires and screening the medical records, and excluded the subjects with incomplete or uncertain clinical/demographic data. In addition to HBV co-infection, alcohol consumption, and smoking (which we used as covariates in the analysis), hepatitis C virus (HCV) co-infection was also shown to be a risk factor for ATDH48. However, according to an epidemiologic study with a total of 236,920 individuals performed in China49, the incidence of HCV co-infection is just 3%. Therefore, we did not test for HCV co-infection in all the participants. Despite this, we searched the medical records in West China Hospital and found that 214 patients (96 in ATDH group and 118 in no-ATDH group) had been serologically tested for anti-HCV. The incidence of HCV co-infection was 3.1% (3/96) in ATDH group and 1.7% (2/118) in the no-ATDH group. Therefore, this risk factor may only have caused a minimal bias. We also analyzed the subjects without HBV/HCV co-infection, and found the result did not change.
In conclusion, PXR genetic polymorphism may be a valuable biomarker potentially involved in ATDH. More studies are required to verify our results.
Methods
Subjects
Ethical approval for this study was obtained from the Institutional Review Board of the West China Hospital of Sichuan University. Methods were carried out in accordance with the approved guidelines. Written informed consent was obtained from each subject. In this case-control study, we recruited TB patients who were diagnosed and treated in the West China Hospital, Chengdu, Sichuan Province, People’s Republic of China between October 2013 and October 2016. The inclusion criteria for the control group (non-ATDH group) were as follows: (a) Normal serum ALT, aspartate aminotransferase and bilirubin levels before anti-TB treatment. (b) Daily treatment with isoniazid, rifampicin, ethambutol and pyrazinamide for at least 2 months, followed by daily treatment with isoniazid, rifampicin and ethambutol, with the total course of treatment equal to or more than 6 months. The dosage of each drug was as follows: isoniazid 300 mg/d, rifampicin 450 mg/d ≤ 50 kg body weight and 600 mg/d > 50 kg body weight, ethambutol 750 mg/d, pyrazinamide 1500 mg/d. (c) At least monthly liver function tests and all with normal results. The inclusion criteria for the case group (ATDH group) were as follows: (a) Normal serum ALT, AST and bilirubin levels before anti-TB treatment. (b) During the period of first line anti-TB treatment (the dosage of each drug was the same as the no-ATDH group), patients meeting the criteria of ATDH reported by an international consensus meeting 199050. In detail, the definition of ATDH was at least one instance of ALT greater than 2 times the ULN and/or a combined increase in AST and total bilirubin, provided one of them was greater than 2 times the ULN, as previously defined. The cutoff of ULN was 50 IU/L for ALT and AST, 28 umol/L for total bilirubin. (c) No administration of other potentially hepatotoxic drugs in the two weeks before ATDH. (d) Liver function tests returned to normal after suspension of anti-TB drugs. Exclusion criteria were: a) Refusal to provide a blood sample or to sign an informed consent form. (b) Positivity for HIV/AIDS. (c) Pregnancy. (d) Cancer, renal failure or cardiac dysfunction that may cause liver dysfunction. (e) Incomplete clinical/demographic data. (f) age <14 year old. The definition of alcohol abuse was daily consumption of more than 60 g of ethanol per day which is based on the WHO criteria (Global Status Report on Alcohol and Health 2014, http://apps.who.int/iris/bitstream/10665/112736/1/9789240692763_eng.pdf?ua=1).
Sample collection and processing
After obtaining consent, we collected a 5 ml peripheral blood specimen from each patient in ethylene diamine tetraacetic acid coated tubes (BD Vacutainers, Franklin Lakes, NJ, USA). The plasma was isolated within 24 hours for determination of the HBsAg by commercial enzyme-linked immunosorbent assay kits (Abbott Park, Wiesbaden, Germany) and the rest of blood specimen was used for extraction of genomic DNA by commercial kit (Axygen Scientific Inc, Union City, CA, USA). The DNA samples were stored at −80 °C until further analysis.
Selection and genotyping of tagging SNPs
All eligible SNPs within 3000 bp upstream and 300 bp downstream of the PXR and CAR genes in the Chinese Han Beijing population were obtained from the HapMap database (Phase II + III Release 27; NCBIB 36; www.hapmap.org). All the SNPs in these regions were filtered using a pairwise tagging algorithm by the Haploview v4.2 software program51, with a minor allele frequency (MAF) ≥0.1 and a LD r2 measure ≥0.80. The LD plots of SNPs based on the data from the HapMap Project in the PXR and CAR genes are shown in Figs S3 and S4, respectively. After calculation by Haploview v4.2, we obtained 7 blocks and 6 blocks for PXR and CAR, respectively. The tagSNPs were selected from each block based on 3 principles. (a) Evidence from the literature that the SNP alters the expression or activity of PXR, CAR or its targets. (b) Association with drug-induced liver injury in a previous study. (c) Functional SNPs predicted by FastSNP52.
Genotyping of the selected SNPs was performed by laboratory technicians who were blind to the clinical data, using an improved multiplex ligation detection reaction (iMLDR) technique developed by Genesky Biotechnologies Inc. (Shanghai, China)53. We also used 10% blind repeated samples for quality control of genotyping. The primer and probe information is shown in Table S3. Raw data were analyzed using the GeneMapper 4.1 software program (Department of Computer Science, University of California at Berkeley, Berkeley, CA, 94720, USA).
Statistical analysis
Statistical analyses were performed using SPSS version 17.0 (Chicago, Illinois, USA). All tagSNPs were tested for agreement with HWE by a goodness-of-fit χ2 test. Quantitative variables were expressed as mean ± standard deviation (SD), and qualitative variables were presented as percentages. The demographic and clinical data of the ATDH group and the no-ATDH group were compared using the χ2 test and Student’s t-test. Three genetic models (additive, dominant, and recessive) were used for assessing the genotype risk of ATDH. Association between genotypes and ATDH risk was evaluated by P value and odds ratio (OR) as well as corresponding 95% confidence intervals (CI) by logistic regression. Covariates considered for inclusion in the regression analysis were ethnicity, age, gender, height, weight, smoking, drinking, and HbsAg result. LD among the selected SNPs was assessed by Haploview v4.2 (http://www.broad.mit.edu/mpg/haploview/)51. SHEsis online software was applied for the haplotype analysis of PXR and CAR gene polymorphisms (http://analysis.bio-x.cn/myAnalysis.php)54. Rare haplotypes with a frequency less than 0.03 were collapsed into one category in the final haplotype analyses. Two-sided values of p < 0.05 were considered statistically significance. Furthermore, we used the Multifactor Dimensionality Reduction Software (version 3.0.2) to analyze the SNP-SNP interactions associated with ATDH55. An overall best model was selected that had high cross-validation consistency and maximum testing balanced accuracy. P < 0.05 by comparing the observed average CVC of each chosen model with the distribution of average consistencies under the null hypothesis of no associations derived empirically from 1000 permutations was considered significant. We also calculated the power of our study design using Power and Simple Size Calculation Software (http://biostat.mc.vanderbilt.edu/PowerSampleSize).
Supplementary information
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant No. 81370121 and Grant No. 81170042), the Research Fund for the Doctoral Program of Higher Education of China (RFDP Grant No. 20130181110068) and the National Scientific and Technological Major Project of China (Grant No. 2012ZX10004-901).
Author Contributions
Y.W. carried out the molecular genetic studies and drafted the manuscript. X.X. carried out the molecular genetic studies and statistical analysis. W.H. carried out the molecular genetic studies. A.S. contributed to the study conception and interpretation of the data. J.Q.H. contributed to the study conception and helped revise the manuscript. M.Z. carried out the sample acquisition and genotyping assays. S.W. carried out the sample acquisition and genotyping assays. M.W. carried out the sample acquisition. G.C. carried out the sample acquisition.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yu Wang, Xi Xiang, and Wei-Wei Huang contributed equally.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-38452-z.
References
- 1.Global tuberculosis report 2018. Geneva: World Health Organization; 2018. Available from, http://www.who.int/tb/publications/global_report/en/ (Accessed on 7th, Dec 2018).
- 2.Lv X, et al. Adverse reactions due to directly observed treatment strategy therapy in Chinese tuberculosis patients: a prospective study. Plos one. 2013;8:e65037. doi: 10.1371/journal.pone.0065037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Okanurak K, Kitayaporn D, Akarasewi P. Factors contributing to treatment success among tuberculosis patients: a prospective cohort study in Bangkok. The international journal of tuberculosis and lung disease: the official journal of the International Union against Tuberculosis and Lung Disease. 2008;12:1160–1165. [PubMed] [Google Scholar]
- 4.Tostmann A, et al. Antituberculosis drug-induced hepatotoxicity: concise up-to-date review. Journal of gastroenterology and hepatology. 2008;23:192–202. doi: 10.1111/j.1440-1746.2007.05207.x. [DOI] [PubMed] [Google Scholar]
- 5.Yuan, L. & Kaplowitz, N. Mechanisms of drug-induced liver injury. Clinics in liver disease17, 507–518, vii, 10.1016/j.cld.2013.07.002 (2013). [DOI] [PMC free article] [PubMed]
- 6.Chen R, Wang J, Zhang Y, Tang S, Zhan S. Key factors of susceptibility to anti-tuberculosis drug-induced hepatotoxicity. Archives of toxicology. 2015;89:883–897. doi: 10.1007/s00204-015-1473-1. [DOI] [PubMed] [Google Scholar]
- 7.Chamorro JG, et al. Effect of gene-gene and gene-environment interactions associated with antituberculosis drug-induced hepatotoxicity. Pharmacogenetics and genomics. 2017;27:363–371. doi: 10.1097/FPC.0000000000000300. [DOI] [PubMed] [Google Scholar]
- 8.Costa GN, et al. Genetic interaction between NAT2, GSTM1, GSTT1, CYP2E1, and environmental factors is associated with adverse reactions to anti-tuberculosis drugs. Molecular diagnosis & therapy. 2012;16:241–250. doi: 10.2165/11634480-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 9.Du H, et al. Slow N-acetyltransferase 2 genotype contributes to anti-tuberculosis drug-induced hepatotoxicity: a meta-analysis. Molecular biology reports. 2013;40:3591–3596. doi: 10.1007/s11033-012-2433-y. [DOI] [PubMed] [Google Scholar]
- 10.Cai L, et al. Meta-Analysis-Based Preliminary Exploration of the Connection between ATDILI and Schizophrenia by GSTM1/T1 Gene Polymorphisms. Plos one. 2015;10:e0128643. doi: 10.1371/journal.pone.0128643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deng R, Yang T, Wang Y, Tang N. CYP2E1 RsaI/PstI polymorphism and risk of anti-tuberculosis drug-induced liver injury: a meta-analysis. The international journal of tuberculosis and lung disease: the official journal of the International Union against Tuberculosis and Lung Disease. 2012;16:1574–1581. doi: 10.5588/ijtld.12.0304. [DOI] [PubMed] [Google Scholar]
- 12.Richardson M, et al. Influence of genetic variants on toxicity to anti-tubercular agents: a systematic review and meta-analysis (protocol) Systematic reviews. 2017;6:142. doi: 10.1186/s13643-017-0533-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pontual Y, et al. ABCB1 gene polymorphism associated with clinical factors can predict drug-resistant tuberculosis. Clinical science. 2017;131:1831–1840. doi: 10.1042/CS20170277. [DOI] [PubMed] [Google Scholar]
- 14.Sharma SK, Balamurugan A, Saha PK, Pandey RM, Mehra NK. Evaluation of clinical and immunogenetic risk factors for the development of hepatotoxicity during antituberculosis treatment. American journal of respiratory and critical care medicine. 2002;166:916–919. doi: 10.1164/rccm.2108091. [DOI] [PubMed] [Google Scholar]
- 15.Wang Y, et al. Association of CYP2B6 gene polymorphisms and anti-tuberculosis drug-induced hepatotoxicity in a Chinese population. Infection, genetics and evolution: journal of molecular epidemiology and evolutionary genetics in infectious diseases. 2017;51:198–202. doi: 10.1016/j.meegid.2017.04.001. [DOI] [PubMed] [Google Scholar]
- 16.Lee WM. Drug-induced hepatotoxicity. The New England journal of medicine. 2003;349:474–485. doi: 10.1056/NEJMra021844. [DOI] [PubMed] [Google Scholar]
- 17.Kliewer SA, et al. An orphan nuclear receptor activated by pregnanes defines a novel steroid signaling pathway. Cell. 1998;92:73–82. doi: 10.1016/S0092-8674(00)80900-9. [DOI] [PubMed] [Google Scholar]
- 18.Zhang B, Xie W, Krasowski MD. PXR: a xenobiotic receptor of diverse function implicated in pharmacogenetics. Pharmacogenomics. 2008;9:1695–1709. doi: 10.2217/14622416.9.11.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li F, et al. Human PXR modulates hepatotoxicity associated with rifampicin and isoniazid co-therapy. Nature medicine. 2013;19:418–420. doi: 10.1038/nm.3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gupta S, Grieco MH, Siegel I. Suppression of T-lymphocyte rosettes by rifampin. Studies in normals and patients with tuberculosis. Annals of internal medicine. 1975;82:484–488. doi: 10.7326/0003-4819-82-4-484. [DOI] [PubMed] [Google Scholar]
- 21.Zhou C, et al. Mutual repression between steroid and xenobiotic receptor and NF-kappaB signaling pathways links xenobiotic metabolism and inflammation. The Journal of clinical investigation. 2006;116:2280–2289. doi: 10.1172/JCI26283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gong H, et al. Orphan nuclear receptor pregnane X receptor sensitizes oxidative stress responses in transgenic mice and cancerous cells. Molecular endocrinology. 2006;20:279–290. doi: 10.1210/me.2005-0205. [DOI] [PubMed] [Google Scholar]
- 23.Lamba J, Lamba V, Strom S, Venkataramanan R, Schuetz E. Novel single nucleotide polymorphisms in the promoter and intron 1 of human pregnane Xreceptor/NR1I2 and their association with CYP3A4 expression. Drug metabolism and disposition: the biological fate of chemicals. 2008;36:169–181. doi: 10.1124/dmd.107.016600. [DOI] [PubMed] [Google Scholar]
- 24.Andrews E, et al. A role for the pregnane X receptor in flucloxacillin-induced liver injury. Hepatology. 2010;51:1656–1664. doi: 10.1002/hep.23549. [DOI] [PubMed] [Google Scholar]
- 25.Swales K, Negishi M. CAR, driving into the future. Molecular endocrinology. 2004;18:1589–1598. doi: 10.1210/me.2003-0397. [DOI] [PubMed] [Google Scholar]
- 26.Timsit YE, Negishi M. CAR and PXR: the xenobiotic-sensing receptors. Steroids. 2007;72:231–246. doi: 10.1016/j.steroids.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ikeda S, et al. Functional analysis of four naturally occurring variants of human constitutive androstane receptor. Molecular genetics and metabolism. 2005;86:314–319. doi: 10.1016/j.ymgme.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 28.Ramappa V, Aithal GP. Hepatotoxicity Related to Anti-tuberculosisDrugs: Mechanisms and Management. Journal of clinical and experimental hepatology. 2013;3:37–49. doi: 10.1016/j.jceh.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.di Masi A, De Marinis E, Ascenzi P, Marino M. Nuclear receptors CAR and PXR: Molecular, functional, and biomedical aspects. Molecular aspects of medicine. 2009;30:297–343. doi: 10.1016/j.mam.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 30.Wang FJ, et al. Update meta-analysis of the CYP2E1 RsaI/PstI and DraI polymorphisms and risk of antituberculosis drug-induced hepatotoxicity: evidence from 26 studies. Journal of clinical pharmacy and therapeutics. 2016;41:334–340. doi: 10.1111/jcpt.12388. [DOI] [PubMed] [Google Scholar]
- 31.Wu S, et al. Genetic Polymorphisms of Glutathione S-Transferase P1 (GSTP1) and the Incidence of Anti-Tuberculosis Drug-Induced Hepatotoxicity. Plos one. 2016;11:e0157478. doi: 10.1371/journal.pone.0157478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blumberg HM, Leonard MK, Jr., Jasmer RM. Update on the treatment of tuberculosis and latent tuberculosis infection. Jama. 2005;293:2776–2784. doi: 10.1001/jama.293.22.2776. [DOI] [PubMed] [Google Scholar]
- 33.Lee SW, et al. NAT2 and CYP2E1 polymorphisms and susceptibility to first-line anti-tuberculosis drug-induced hepatitis. The international journal of tuberculosis and lung disease: the official journal of the International Union against Tuberculosis and Lung Disease. 2010;14:622–626. [PubMed] [Google Scholar]
- 34.Steele MA, Burk RF, DesPrez RM. Toxic hepatitis with isoniazid and rifampin. A meta-analysis. Chest. 1991;99:465–471. doi: 10.1378/chest.99.2.465. [DOI] [PubMed] [Google Scholar]
- 35.Miguet JP, Mavier P, Soussy CJ, Dhumeaux D. Induction of hepatic microsomal enzymes after brief administration of rifampicin in man. Gastroenterology. 1977;72:924–926. [PubMed] [Google Scholar]
- 36.Ono Y, Wu X, Noda A, Noda H, Yoshitani T. Participation of P450-dependent oxidation of isoniazid in isonicotinic acid formation in rat liver. Biological & pharmaceutical bulletin. 1998;21:421–425. doi: 10.1248/bpb.21.421. [DOI] [PubMed] [Google Scholar]
- 37.Tirona RG, et al. The orphan nuclear receptor HNF4alpha determines PXR- and CAR-mediated xenobiotic induction of CYP3A4. Nature medicine. 2003;9:220–224. doi: 10.1038/nm815. [DOI] [PubMed] [Google Scholar]
- 38.Kamiya A, Inoue Y, Gonzalez FJ. Role of the hepatocyte nuclear factor 4alpha in control of the pregnane X receptor during fetal liver development. Hepatology. 2003;37:1375–1384. doi: 10.1053/jhep.2003.50212. [DOI] [PubMed] [Google Scholar]
- 39.Chowdhury A, et al. Induction of oxidative stress in antitubercular drug-induced hepatotoxicity. Indian journal of gastroenterology: official journal of the Indian Society of Gastroenterology. 2001;20:97–100. [PubMed] [Google Scholar]
- 40.Wang JY, et al. Gender-Dimorphic Impact of PXR Genotype and Haplotype on Hepatotoxicity During Antituberculosis Treatment. Medicine. 2015;94:e982. doi: 10.1097/MD.0000000000000982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shakya R, Rao BS, Shrestha B. Incidence of hepatotoxicity due to antitubercular medicines and assessment of risk factors. The Annals of pharmacotherapy. 2004;38:1074–1079. doi: 10.1345/aph.1D525. [DOI] [PubMed] [Google Scholar]
- 42.Yee D, et al. Incidence of serious side effects from first-line antituberculosis drugs among patients treated for active tuberculosis. American journal of respiratory and critical care medicine. 2003;167:1472–1477. doi: 10.1164/rccm.200206-626OC. [DOI] [PubMed] [Google Scholar]
- 43.Hunt CM, Westerkam WR, Stave GM. Effect of age and gender on the activity of human hepatic CYP3A. Biochemical pharmacology. 1992;44:275–283. doi: 10.1016/0006-2952(92)90010-G. [DOI] [PubMed] [Google Scholar]
- 44.Zazuli Z, et al. Polymorphism of PXR gene associated with the increased risk of drug-induced liver injury in Indonesian pulmonary tuberculosis patients. Journal of clinical pharmacy and therapeutics. 2015;40:680–684. doi: 10.1111/jcpt.12325. [DOI] [PubMed] [Google Scholar]
- 45.Aithal GP, et al. Case definition and phenotype standardization in drug-induced liver injury. Clinical pharmacology and therapeutics. 2011;89:806–815. doi: 10.1038/clpt.2011.58. [DOI] [PubMed] [Google Scholar]
- 46.Xu S, et al. A genome-wide search for signals of high-altitude adaptation in Tibetans. Molecular biology and evolution. 2011;28:1003–1011. doi: 10.1093/molbev/msq277. [DOI] [PubMed] [Google Scholar]
- 47.Metushi I, Uetrecht J, Phillips E. Mechanism of isoniazid-induced hepatotoxicity: then and now. British journal of clinical pharmacology. 2016;81:1030–1036. doi: 10.1111/bcp.12885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim WS, et al. Hepatitis C and not Hepatitis B virus is a risk factor for anti-tuberculosis drug induced liver injury. BMC infectious diseases. 2016;16:50. doi: 10.1186/s12879-016-1344-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Q, et al. Epidemiology of Hepatitis B and Hepatitis C Infections and Benefits of Programs for Hepatitis Prevention in Northeastern China: A Cross-Sectional Study. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2016;62:305–312. doi: 10.1093/cid/civ859. [DOI] [PubMed] [Google Scholar]
- 50.Benichou C. Criteria of drug-induced liver disorders. Report of an international consensus meeting. Journal of hepatology. 1990;11:272–276. doi: 10.1016/0168-8278(90)90124-a. [DOI] [PubMed] [Google Scholar]
- 51.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 52.Yuan HY, et al. FASTSNP: an always up-to-date and extendable service for SNP function analysis and prioritization. Nucleic acids research. 2006;34:W635–641. doi: 10.1093/nar/gkl236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xie B, et al. Deltex1 Polymorphisms Are Associated with Hepatitis B Vaccination Non-Response in Southwest China. Plos one. 2016;11:e0149199. doi: 10.1371/journal.pone.0149199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shi YY, He L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell research. 2005;15:97–98. doi: 10.1038/sj.cr.7290272. [DOI] [PubMed] [Google Scholar]
- 55.Hahn LW, Ritchie MD, Moore JH. Multifactor dimensionality reduction software for detecting gene-gene and gene-environment interactions. Bioinformatics. 2003;19:376–382. doi: 10.1093/bioinformatics/btf869. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.