Abstract
The role of long non-coding RNAs (lncRNAs) in regulating endothelial function through the DNA damage response (DDR) remains poorly understood. In this study, we demonstrate that lncRNA maternally expressed gene 3 (Meg3) interacts with the RNA binding protein polypyrimidine tract binding protein 3 (PTBP3) to regulate gene expression and endothelial function through p53 signaling ─ a major coordinator of apoptosis and cell proliferation triggered by the DDR. Meg3 expression is induced in endothelial cells (ECs) upon p53 activation. Meg3 silencing induces DNA damage, activates p53 signaling, increases the expression of p53 target genes, promotes EC apoptosis, and inhibits EC proliferation. Mechanistically, Meg3 silencing reduces the interaction of p53 with Mdm2, induces p53 expression, and promotes the association of p53 with the promoters of a subset of p53 target genes. PTBP3 silencing recapitulates the effects of Meg3 deficiency on the expression of p53 target genes, EC apoptosis and proliferation. The Meg3-dependent association of PTBP3 with the promoters of p53 target genes suggests that Meg3 and PTBP3 restrain p53 activation. Our studies reveal a novel role of Meg3 and PTBP3 in regulating p53 signaling and endothelial function, which may serve as novel targets for therapies to restore endothelial homeostasis.
INTRODUCTION
The vascular endothelium plays an important role in maintaining the proper functions of different organs. Endothelial dysfunction contributes to the pathogenesis of major chronic diseases such as obesity, diabetes and atherosclerosis (1–4). Different insults, such as chronic inflammatory disease states, oxidative stress, metabolic dysfunction and aging, promote endothelial dysfunction through transcriptional reprogramming (5–8). In response to these insults, DNA damage may ensue and adversely contribute to cardiovascular and metabolic disease (9–12). However, the molecular mechanisms and signaling events by which DNA damage regulates endothelial function are not completely understood.
The cellular response to DNA damage involves the activation of multiple signaling cascades that orchestrate the appropriate repair of DNA damage with signaling events involved in apoptosis and proliferation (13,14). The DNA damage response (DDR) resulting from genotoxic, oxidative, and metabolic stress is controlled by three phosphoinositide 3-kinase (PI3K)-related kinases: Ataxia-telangiectasia mutated (ATM), ATM- and Rad3-related (ATR), and DNA-dependent protein kinase (DNA-PK) (12,13). Formation of DNA double-strand breaks (DSB) is a severe type of DNA damage. ATM is vigorously activated following DSB and interacts with many downstream effectors including p53 (14). p53 protein is a stress- and DNA damage-responsive transcriptional factor that activates cell cycle arrest, DNA repair, and apoptosis (15). High glucose or palmitic acid induces p53 expression in human endothelial cells (ECs) (16). Expression of p53 is also induced in vascular endothelium in mice fed a high-calorie diet (16). Inhibition of endothelial p53 attenuated metabolic abnormalities associated with dietary obesity (16). Deletion of p53 in the vascular endothelium reduced the number of apoptotic ECs in mice, and protects mice from cardiac dysfunction after pressure overload (17). Therefore, discovering new mechanisms by which p53 signaling regulates endothelial function may provide new targets for therapeutic intervention.
Long non-coding RNAs (lncRNAs) are an important class of RNA transcripts that regulate gene expression and signal transduction. Changes in their expression and function contribute to the pathogenesis of a range of disease states (18–21). LncRNAs participate in several aspects of the DDR and regulate the expression of key components of related pathways (22,23). LncRNA maternally expressed gene 3 (Meg3) is an imprinted gene located at chromosome 12 in mice and chromosome 14 in humans (24). The role of Meg3 has been examined in different cell types including cancer cells, neurons, hepatocytes, cardiac fibroblasts, and ECs (25–30). Meg3 regulates p53 signaling in a cell-specific manner. Meg3 interacts with p53 in cancer cells and neurons to activate p53-mediated inhibition of cell proliferation and induction of apoptosis (25,27,28). In contrast, Meg3 interacts with p53 in cardiac fibroblasts exerting no effects on a p53 response, cell apoptosis, or proliferation (26). These studies also highlight that Meg3 may regulate p53 signaling in a cell-specific manner. However, it remains unknown how Meg3 determines the transcriptional output of p53 signaling in the regulation of EC proliferation and apoptosis in response to DNA damage. Moreover, the reciprocal regulation of Meg3 and DNA damage remains poorly understood. In this study, we find that Meg3 cooperates with polypyrimidine tract binding protein 3 (PTBP3) to control the DDR, thereby protecting endothelial function.
MATERIALS AND METHODS
Reagents
Nutlin-3 and doxorubicin were purchased from Sigma (St Louis, MO, USA). Palmitic acids were purchased from Nu-Chek Prep, Inc. (Waterville, MN, USA). Albumin (bovine serum, fraction v, fatty acid-poor, endotoxin-free) was purchased from EMD Millipore (Burlington, MA, USA). GapmeR Meg3 (5′-GTAAGACAAGCAAGAG-3′) and GapmeR negative control A (5′-AACACGTCTATACGC-3′) were purchased from Exiqon (Vedbaek, Denmark). Silencer™ Select Negative Control No. 1 siRNA, PTBP1 siRNA (Assay ID: s11435), and PTBP3 siRNA (Assay ID: s19414) were purchased from Thermo Fisher Scientific Life Sciences (Waltham, MA, USA). Recombinant human TNF-α was purchased from R&D Systems (210-TA/CF; Minneapolis, MN, USA). Lipofectamine 2000 and TRIzol reagents were from Thermo Fisher Scientific Life Sciences. Protease inhibitor cocktail tablets were from Roche (Basel, Switzerland). Phosphatase inhibitor was from Active Motif (Carlsbad, CA, USA). ATM inhibitor KU-55933 (Cat# 118500) were from MilliporeSigma (Burlington, MA, USA).
Cell culture and transfection
Human Umbilical Vein Cells (HUVECs) (cc-2159) were obtained from Lonza (Morristown, NJ, USA) and cultured in EC growth medium EGM-2 (cc-3162). Cells used for all experiments were subcultured <8 times. HUVECs (90 000/well) were plated into 12-well plates, transfected with 10 nM GapmeR, or 30 nM small interfering RNAs (siRNAs) when cells reached 80–90% confluency. Lipofectamine 2000 was used following the manufacturer instructions. Cells were grown for 36 h before treatment with TNF-α (10 ng/ml), nutlin-3 (10 μM), doxorubicin (0.2, 0.6 or 2.0 μM), or palmitic acid (100 μM) for different time points. Palmitic acid/BSA complexes were prepared as previously described (31). In one experiment, cells were treated with ATM inhibitor at 10 μM for 3 h.
Lentivirus production and cell transduction
pLKO.1-TRC cloning vector was a gift from Dr David Root (Addgene plasmid #10878) (32). pMD2.G was a gift from Dr Didier Trono (Addgene plasmid #12259). psPAX2 was a gift from Dr Didier Trono (Addgene plasmid #12260). shp53 pLKO.1 puro was a gift from Dr Bob Weinberg (Addgene plasmid #19119) (33). Meg3 shRNA oligos or negative control oligos were cloned into pLKO.1-TRC cloning vector between AgeI and EcoRI sites. Lentivirus for pLKO.1-ctl shRNA or pLKO.1-Meg3 shRNA was generated by cotransfection of 293T cells (ATCC CRL-3216) using pMD2.G and psPAX2 in a 3:2:1 ratio, respectively. Plasmids were transfected using Lipofectamine 2000. Transfection mix was added dropwise to the dish and the medium was changed ∼16 h later. Supernatant was collected 2 days later by filtering through a 0.45 μm filter and stored at −80°C. Transduction of HUVECs was carried out in 6-well plates adding 1 ml lentiviral supernatant to 1 ml medium in combination with 8 μg/ml PolybreneTM (AB01643; American Bio, Natick, MA, USA). The medium was changed 16 h later.
Neutral comet assay
HUVECs transfected with GapmeRs or siRNA were further treated with or without TNF-α (10 ng/ml) for 16 h. Neutral comet assay was performed according to the manufacturer's protocol using the Single Cell Gel Electrophoresis kit (Enzo Life Sciences Inc. Farmingdale NY, USA). Briefly, the cells were trypsinized, washed and resuspended in ice-cold phosphate-buffered saline. Further, the cells were combined with low melting point agarose and applied on pre-coated comet slides to solidify. After cell lysis and alkaline denaturation for 40 min, electrophoresis was performed at 15 volts (1 volt per cm) for 20 min in 1× TBE, pH 10.0 buffer. Subsequently, the slides were dehydrated by immersing in 70% ethanol, air-dried and stained with CYGREEN® green dye. DNA strand break was assessed by measuring tail moment and tail length using fluorescence microscope for at least 150 comets per condition for each experiment. Comets were analyzed using OpenComet software.
Transcriptomic analysis by microarray
Arraystar Human LncRNA Microarray v4.0, performed by Arraystar Inc. (Rockville, MD, USA), is designed for the global expression profiling of 40 173 human lncRNA and 20,730 protein-coding mRNA transcripts. HUVECs were transfected with 10 nM control GapmeRs or Meg3 GapmeRs and treated with 10 ng/ml TNF-α for 3 h. Cells were lysed with TRIzol reagent and total RNA was isolated according to the manufacturer instructions. Sample-labeling and array hybridization were performed according to the Agilent One-Color Microarray-Based Gene Expression Analysis protocol (Agilent Technology, Santa Clara, CA, USA) with minor modifications. Briefly, mRNA was purified from total RNA after removal of rRNA (mRNA-ONLY™ Eukaryotic mRNA Isolation Kit, Epicentre, Madison WI, USA). Then, each sample was amplified and transcribed into fluorescent cRNA along the entire length of the transcripts without 3′ bias utilizing a random priming method (Arraystar Flash RNA Labeling Kit, Arraystar, Rockville, MD, USA). The labeled cRNAs were purified by RNeasy Mini Kit (Qiagen, Hilden, Germany). The concentration and specific activity of the labeled cRNAs (pmol Cy3/μg cRNA) were measured using a NanoDrop™ ND-1000. 1 μg of each labeled cRNA was fragmented by adding 5 μl 10× Blocking Agent and 1 μl of 25× Fragmentation Buffer, then heated at 60°C for 30 min. Finally, 25 μl 2× GE Hybridization buffer was added to dilute the labeled cRNA. 50 μl of hybridization solution was dispensed into the gasket slide and assembled into the lncRNA expression microarray slide. The slides were incubated for 17 h at 65°C in an Agilent Hybridization Oven. The hybridized arrays were washed, fixed and scanned using the Agilent DNA Microarray Scanner (part number G2505C). Agilent Feature Extraction software (version 11.0.1.1) was used to analyze acquired array images. Quantile normalization and subsequent data processing were performed using the GeneSpring GX v12.1 software package (Agilent Technologies).
The predicted target genes above were input into the Database for Annotation, Visualization and Integrated Discovery (DAVID v6.8; https://david.ncifcrf.gov/) for pathway analysis. We used the KEGG (Kyoto Encyclopedia of Genes and Genomes) database to analyze the potential functions of these target genes in the pathways (34,35). Values of P < 0.05 were considered to be statistically significant; with the lower P values indicating greater significance of the pathway.
Proteomic analysis using TMT10-plex labeling and liquid chromatography-tandem mass spectrometry (LC-MS/MS)
HUVECs were transfected with 10 nM control GapmeRs or Meg3 GapmeRs and treated with or without 10 ng/ml TNF-α for 4 h. Cells were washed 6 times with ice-cold 1× PBS. Then cells were scraped into ice-cold 1× PBS and spun at 300 × g for 5 min. Cell pellets were extracted for 20 min on ice using RIPA buffer (ThermoFisher) containing 1× protease inhibitor (EDTA-free Protease Inhibitor Cocktail, Sigma). Protein was precipitated and washed to remove detergent using the ProteoExtract kit (EMD-MilliPore). Pellets were then redissolved, the protein content assayed and 50 μg from each sample were reduced with 10 mM DTT and alkylated with 20 mM iodoacetamide. The proteins were digested with 1 μg of trypsin for 16 h at 37°C, before addition of a further 0.5 μg of trypsin for a further 6 h. The peptides were labeled using the TMT10-plex reagent (ThermoFisher) according to the manufacturer's instructions. The three biological replicates for control samples treated with TNF-α were labeled with the 129N, 130N and 131 labels. The 3 biological replicates for the Meg3 knockdown treated with TNF-α were labeled with the 126, 127N and 128N labels. Finally, two biological replicates of the control and Meg3 knockdown cells without TNF-α treatment, were labeled with the 128C, 129C and 127C, 130C labels, respectively. All labeled samples were combined equally and the mixture was fractionated into 60 fractions using high-pH reverse phase chromatography and recombined to give a total of 12 final fractions according to the strategy of Yang et al. (36). Mass spectrometric analysis of each fraction was carried out using a Dionex U3000 nanoRSLC running a 2 h gradient through a 0.075 mm × 250 mm C18 Waters CSH130 peptide column feeding into a Q-Exactive HF mass spectrometer (ThermoScientific).
The protein identification and quantification were processed using Mascot Server software (Matrix Science, London, UK; version 2.5.1) and Proteome Discoverer (ThermoScientific, version 2.1), respectively. The mass spectra were searched against the common contaminants cRAP_20150130 database (www.theGPM.org) and the SwissProt database (selected for Human, April 2017) using trypsin, a fragment ion mass tolerance of 0.02 Da and a parent ion tolerance of 10 ppm. The TMT label was specified as a fixed modification for both the N-terminus and the lysine sidechain, with oxidation of methionine and carbamidomethyl of cysteine specified as variable modifications. The data were searched using a decoy database to set the false discovery rate to 1% (high confidence) and 5% (medium confidence). The co-isolation threshold was set to 50%, and the average S/N to 10. Protein abundances were normalized using the total abundance from all the channels. Differentially expressed proteins were selected if the P-value was <0.05 and the change >20%.
RNA immunoprecipitation
HUVECs were cross-linked with 0.3% formaldehyde at room temperature for 10 min and neutralized with 0.125 M of glycine at room temperature for 5 min. Cross-linked cells were washed with 1× PBS, collected by scraping and lysed in polysome lysis buffer containing 100 mM KCl, 5 mM MgCl2, 10 mM HEPES at pH 7.0, 0.5% Nonidet P-40, 1 mM dithiothreitol (DTT), 200 units/ml RNase OUT (Invitrogen Cat. No. 10777-019) and Complete Mini, EDTA-free Protease Inhibitor Tablet (11836170001, Roche) for 30 min on ice. Lysate was diluted to 1 ml in NT-2 buffer containing 50 mM Tris–HCl, pH 7.4, 150 mM NaCl, 1 mM MgCl2, 0.5% NP-40, 20 mM EDTA at pH 8.0, 1 mM DTT, 200 units/ml RNase OUT and sonicated for 20 s at 20% power using Branson 250 Ultrasonic Sonifier. Lysates were cleared by centrifugation at 10 000 × g at 4°C for 10 min. Cleared lysates were split equally, incubated with control Isotype IgG or PTBP3 antibody pre-coupled to Dynabeads™ Protein G and tumbled at 4°C for 12 h. The immuno-complex was washed twice with NT-2 buffer and subsequently with NT-2 buffer supplemented with 500 mM NaCl. Washed complex was resuspended in 150 μl of 1× NT-2 buffer supplemented with 1% sodium dodecyl sulfate (SDS) and 1.2 mg/ml proteinase K and incubated at 55°C for 30 min. RNA was extracted using the TRIzol reagent. RNA was reverse-transcribed using Superscript II reverse transcriptase. Quantitative PCR was performed to measure the amount of lncRNA Meg3 and the results represent the percentage of Meg3 in the PTBP3 interacting RNA fraction over that of input.
Caspase3/7 activity assay
HUVECs transfected with GapmeRs or siRNAs were further treated with or without TNF-α (10 ng/ml) for 16 h. Caspase 3/7 activity was measured using caspase-Glo® 3/7 assay kit (Promega, Madison, WI, USA) according to the manufacturer's instructions. Relative luminescence intensity was recorded using a fluorometric plate reader (BioTek Instruments, Winooski, VT, USA).
Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL)
HUVECs transfected with GapmeRs or siRNAs were further treated with or without TNF-α (10 ng/ml) for 16 h. TUNEL staining was performed according to the manufacturer's protocol using In Situ Cell Death Detection Kit, TMR red (Sigma). Briefly, the cells were fixed with 4% paraformaldehyde for 30 min at room temperature and subsequently permeabilized with 0.25% Triton X-100 in 0.1% sodium citrate for 5 min. Subsequently, the cells were stained with TUNEL Enzyme-TMR label mixture and counter stained with DAPI. Cells were analyzed by using a fluorescent microscope (EVOS) with appropriate filters. The numbers of TUNEL positive cells in eight random fields were counted, the results were represented as the average number of TUNEL positive cells per High Power Field (HPF).
EdU incorporation
HUVECs were seeded into 12-well plates (1 × 105 cells/well) 24 h prior to transfection with either control or Meg3 GapmeR. After 24 h transfection, cells were re-plated (8 × 104/ml) and cultured for 12 h in an incubator with 5% CO2 at 37°C. Then cells were treated with or without TNF-α (10 ng/ml) for 12 h, and EdU labeling was performed using Click-iT™ EdU Alexa Fluor™ 555 Imaging Kit (Thermo Fisher Scientific, Waltham, MA, USA). The media was replaced with medium containing 10 μM EdU and incubated for 1.5 h in an incubator with 5% CO2 at 37°C, followed by fixation, permeabilization, EdU detection and DNA staining according to the manufacturer's instructions. Stained cells were photographed using a fluorescent microscope (EVOS) with 10× magnification.
Real-time qPCR
HUVECs were lysed with TRIzol reagent and total RNA was isolated according to manufacturer instructions. High-Capacity cDNA Reverse Transcription Kit Cat# 4368814 from Thermo Fisher Scientific was used to generate cDNAs. GoTaq qPCR Master Mix (Cat# A6001) from Promega was used for real-time quantitative PCR (qPCR) with Bio-Rad CFX96 Touch Real-time PCR Detection System. To detect Meg3, GoTaq® Probe qPCR Master Mix (Promega) and TaqMan® Gene Expression Assays (ID# Hs00292028_m1) were used. See Supplementary Table S7 for primer sequences.
Cell cycle analysis
HUVECs were seeded into 6-well plates (3 × 105 cells/well) 24 h prior to transfection with either control or Meg3 GapmeR. The medium was replaced 24 h after transfection and the cells were cultured for another 24 h. EdU incorporation and DNA staining were performed according to the manufacturer's instructions of Click-iT EdU Alexa Fluor 488 flow cytometry assay Kit. The cells were incubated in medium with 10 μM EdU for 1.5 h with 5% CO2 at 37°C. Before fixation for 15 min at RT, the cells were trypsinized, harvested by putting 1 × 106 cells per tube and washed once with PBS containing 1% BSA. Then the cells were permeabilized and stained with Click-iT reaction cocktail, DNA was stained with FxCycle violet. Cell cycle was analyzed on a BD Cytek FACSort.
LncRNA pull-down assay and mass spectrometry
Meg3 and its deletion mutants (primers are listed in Supplementary Table S7) were cloned into pGEM-3Zf(+) (Promega). Linearized pGEM-3Zf(+) expressing Meg3 or Meg3 deletion mutants was used as a template to synthesize biotinylated Meg3 or Meg3 antisense (AS) control RNA using Riboprobe® In Vitro Transcription Systems (Promega). A lncRNA pull-down assay was performed as previously described (37). Twenty-million cells were harvested and lysed in 5 ml cell swelling buffer (20 mM Tris–Cl, pH 7.4, 1.5 mM MgCl2, 10 mM KCl, 0.5% NP-40, 1 mM DTT) on ice for 30 min followed by dounce homogenizing 20 times to release nuclei. The nuclei pellet was lysed in 2 ml nuclear lysis buffer – NLB buffer (50 mM Tris–Cl, pH 8.0, 140 mM NaCl, 1 mM EDTA, 1% Triton-X100, 0.1% sodium deoxycholate, 0.05% SDS, 1 mM DTT, protease inhibitor) on ice for 15 min and centrifuged at 14 000 × g for 10 min to clarify nuclear lysates. One milliliter lysates were pre-cleared with T1-beads for 1 h at 4°C and incubated with 2.5 μg of folded biotinylated RNA (sense Meg3 or antisense Meg3 RNA) for 2 h at 4°C. Then the RNA–protein complexes were captured by yeast tRNA (0.2 μg/μl) blocked T1-beads by rotating at 4°C for 30 min. The beads were washed with 0.5 ml NLB buffer once and 0.5 ml RIP buffer (20 mM Tris–Cl, pH 7.4, 140 mM NaCl, 1 mM EDTA, 0.5% NP-40, 1 mM DTT) for 3 times, then re-suspended in 2× Laemmli buffer and heated at 95°C for 5 min. The retrieved proteins were subjected to silver stain or identified by mass spectrometry. Gel bands were digested using 1.5 μg trypsin, dried completely in a speedvac, and resuspended with 21 μl of 0.1% formic acid pH 3.0. 5 μl was injected per run using an Easy-nLC 1200 UPLC system. Samples were loaded directly onto a 45 cm long 75 μm inner diameter nano capillary column packed with 1.9 μm C18-AQ (Dr. Maisch, Germany) mated to metal emitter in-line with an Orbitrap Fusion Lumos (Thermo Scientific, USA). The mass spectrometer was operated in data dependent mode with the 120 000 resolution MS1 scan (AGC 4e5, Max IT 50 ms, 400–1500 m/z) in the Orbitrap followed by up to 20 MS/MS scans with CID fragmentation in the ion trap. Dynamic exclusion list was invoked to exclude previously sequenced peptides for 60 s if sequenced within the last 30 s and maximum cycle time of 3 s was used. Peptide were isolated for fragmentation using the quadrupole (1.6 Da window). Ion-trap was operated in Rapid mode with AGC 2e3, maximum IT of 300ms and minimum of 5000 ions. Raw files were searched using Byonic and Sequest HT algorithms within the Proteome Discoverer 2.1 suite (Thermo Scientific, USA). 10 ppm MS1 and 0.4 Da MS2 mass tolerances were specified. Caramidomethylation of cysteine was used as fixed modification, oxidation of methionine, acetylation of protein N-termini, conversion of glutamine to pyro-glutamate and deamidation of asparagine were specified as dynamic modifications. Trypsin digestion with maximum of two missed cleavages were allowed. Files searched against the Uniprot homo sapiens database downloaded Feb 23rd, 2017 and supplemented with common contaminants. Scaffold (version Scaffold_4.8.4, Proteome Software Inc., Portland, OR) was used to validate MS/MS based peptide and protein identifications. Peptide identifications were accepted if they could be established at >90% probability by the Scaffold Local FDR algorithm. Protein identifications were accepted if they could be established at >99% probability and contained at least two identified peptides. Protein probabilities were assigned by the Protein Prophet algorithm. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony.
Chromatin immunoprecipitation (ChIP)
HUVECs were treated with TNF-α (10 ng/ml) for 3 h. Formaldehyde was added directly to cell culture media to a final concentration of 1% and the cells were cross-linked for 10 min at room temperature. Cross-linked cells were neutralized by addition of glycine to a final concentration of 0.125 M and incubated at room temperature for 5 min. Fixed cells were rinsed twice with cold 1× PBS and scraped into 1× PBS containing Protease Inhibitor Cocktail (Roche). Cells were collected by centrifugation at 800 × g and resuspended in cell lysis buffer containing 10 mM HEPES pH 7.9, 0.5% NP-40, 1.5 mM MgCl2, 10 mM KCl and incubated on ice for 15 min. Nuclei were collected by centrifugation at 800 × g and resuspended in nuclear lysis buffer containing 1% SDS, 10 mM EDTA, 50 mM Tris–HCl (pH 8.1), with protease inhibitor cocktail and the samples were sonicated using a Branson 250 Sonifier for 7 cycles with each cycle comprising 20 s sonication at 20% power and 1 min rest to yield an average DNA length of 500–1000 bp. Samples were then cleared by centrifuging at 14 000 × g for 15 min. 10 μg of chromatin was diluted to 1 ml in ChIP dilution buffer containing 0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris–HCl, pH 8.1, 167 mM NaCl and incubated with 4 μg Isotype control IgG or PTBP3 antibody pre-coupled to 20 μl of Dynabeads™ Protein G (10003D, Invitrogen) and tumbled at 4°C overnight. Prior to antibody addition 1% of diluted chromatin was saved as input. The immuno-precipitates were washed once with low salt immune complex wash buffer containing 0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris–HCl, pH 8.1, 150 mM NaCl; once with high salt immune complex wash buffer containing 0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris–HCl, pH 8.1, 500 mM NaCl; twice with LiCl wash buffer containing 0.25 M LiCl, 1% IGEPAL-CA630, 1% sodium deoxycholic acid, 1 mM EDTA, 10 mM Tris–HCl, pH 8.1; and twice with TE buffer containing 10 mM Tris–HCl, 1 mM EDTA, pH 8.0.
Washed samples were resuspended in elution buffer containing 1% SDS and 0.1 M NaHCO3 with 20 μl of RNase and incubated at 37°C for 30 min to remove RNA. Further, proteinase K was added to a final concentration of 0.2 mg/ml and reverse cross-linked by incubating at 62°C for 2 h followed by incubating at 95°C for 10 min. DNA was purified using Nucleospin columns using the manufacturer's protocol. The amount of DNA targets in the eluted sample was measured by quantitative PCR using primers specifically targeting p53 binding regions for the respective genes or negative control primers amplifying actin promoter or chromosome 22 intergenic sequence (Supplementary Table S7).
In situ RNA detection
HUVECs were cultured in Nunc® Lab-Tek® II Chamber Slide™ 8-well system. In situ RNA detection was performed using RNAscope® 2.5 HD Reagent Kit-RED (Advanced Cell Diagnostics) and RNAscope® Probe - Hs-MEG3 following the manufacturer's instructions. RNAscope® Negative Control Probe DapB (E. Coli RNA) was used a negative control and RNAscope® Positive Control Probe Hs-PPIB as a positive control. For co-staining of Meg3 and PTBP3, permeabilized cells were not treated with RNAscope® protease III. After Meg3 staining, cells were blocked with RNase-free 5% goat serum in 1× PBS followed by the primary antibodies against PTBP3 in blocking buffer at 4°C for overnight. Then cells were washed and incubated with the secondary antibodies Alexa Fluor 488-AffiniPure Donkey Anti-Mouse IgG and DAPI at room temperature for 1 h. Stained samples were mounted with Fluoromount-G for confocal imaging. Images were acquired using a Nikon A1R-Ti2 live cell imaging confocal system. This system has an integrated six-solid-state laser package (with 405, 440, 488, 514, 561 and 640 nm laser lines) and the ultra-sensitive GaAsp hybrid 4+1 channel detector system.
Chromatin isolation by RNA purification (ChIRP)
ChIRP was done as previously described (38). Meg3 antisense oligos with a 3′-Biotin-TEG modification were designed and synthesized as described (39) (Supplementary Table S7).
Co-immunoprecipitation
Cells were washed once with ice cold 1× PBS and collected in 1× PBS containing protease and phosphatase inhibitor cocktail (Roche). After centrifugation at 200 × g, the cells were resuspended in RIPA lysis buffer (Thermo Scientific). Subsequently, cell lysates were collected after spinning at 13 000 × g for 10 min. Protein concentrations were measured by using the Pierce BCA Protein Assay Kit. After saving 1% of the immunoprecipitation volume as input, lysates were split into two parts containing equal amount of total protein. Protein A/G Dynabeads™ (Invitrogen) were incubated with IgG or specific antibody at room temperature for 30 min. Beads coupled with antibodies were cross-linked by incubating with 25 mM DMP in 0.2 M triethanolamine, pH 8.2 for 45 min. Further, the antibody coupled beads were incubated in 0.1 M ethanolamine, pH 8.2 for 30 min. The cross-linked antibody-bead complexes were washed with 1× PBS containing 0.02% Tween-20 and combined with the protein lysates and rotated overnight at 4°C. Subsequently, the immunoprecipitation complexes were washed three times with ice cold RIPA buffer. Immuno-precipitated proteins were eluted in elution buffer containing 125 mM Tris–HCl, 5% SDS, 20% glycerol and 0.01% Bromophenol Blue for 10 min at room temperature with gentle agitation. 40 mM DTT was added to the eluted protein and the lysates were incubated at 95°C for 5 min for western blot analysis. Anti-light chain-specific horseradish peroxidase-conjugated antibodies (Jackson ImmunoResearch) were used for detection.
Histone H3 and PTBP3 association assay
The method used for crosslinking, nuclear lysate preparation, immunoprecipitation and washing were the same as described in the ChIP assay with the following modifications. The collected nuclear lysates were sonicated using a Branson 250 Sonifier for 3 cycles with each cycle comprising sonication for 20 s at 20% power. The immunoprecipitated samples using anti-histone H3 were heated in protein gel loading sample buffer containing DTT at 95°C for 5 min and analyzed by SDS-PAGE. Anti-light chain-specific horseradish peroxidase-conjugated antibodies (Jackson ImmunoResearch) were used for detection.
Western blot analysis and antibodies
HUVECs were scraped and lysed in RIPA buffer that was supplemented with protease and phosphatase inhibitors. Cell lysates were collected after spinning at 13 000 × g for 10 min. Protein concentrations were measured by using the Pierce BCA Protein Assay Kit. Lysates were analyzed by SDS-PAGE using 8% or 10% gels. Proteins were electroblotted onto PVDF membranes (Bio-Rad, Hercules, CA, USA) and then incubated with corresponding antibodies. ECL Plus Western blotting detection reagents (RPN2132; GE Healthcare, Pittsburg, PA, USA) were used to visualize proteins. ImageJ software (National Institutes of Health, Bethesda, MD, USA) was used to analyze protein abundance. Antibodies used in this study include anti-phospho-ATM, anti-ATM, anti-phospho-ATR, anti-ATR, anti-phospho-DNA-PK, anti-DNA-PK, anti-phospho-H2AX, anti-H2AX, anti-GAPDH, anti-p53, anti-MDM2, anti-p21, anti-PTBP3 and others. See Supplementary Table S7 for more information about antibodies.
Statistical analysis
Paired or unpaired Student's t tests for single comparisons and ANOVA followed by Bonferroni post hoc test for multiple comparisons were used to determine significant differences (P < 0.05) for data with a normal distribution. Normality was checked using the Kolmogorov–Smirnov test. Data are reported as mean ± S.E.M.
RESULTS
Meg3 expression is induced by p53-mediated transcription
The human Meg3 gene produces 16 different transcript variants (Supplemental Figure S1A). We found that the transcript variants 1 and 6 of the Meg3 gene are dominantly expressed in HUVECs using rapid amplification of complementary DNA ends (Supplemental Figure S1B). This was confirmed with a pair of primers that can amplify all the transcript variants followed by DNA sequencing (Supplementary Figure S1C). A subcellular fractionation assay detected Meg3 predominantly in the nuclear fraction, while GAPDH mRNA was found predominantly in the cytoplasmic fraction (Supplementary Figure S1D). Consistently, Meg3 in situ hybridization verified that the majority of Meg3 is expressed in the nucleus in HUVECs which was dramatically reduced by RNase treatment or Meg3 GapmeR-mediated knockdown (Supplementary Figure S1E).
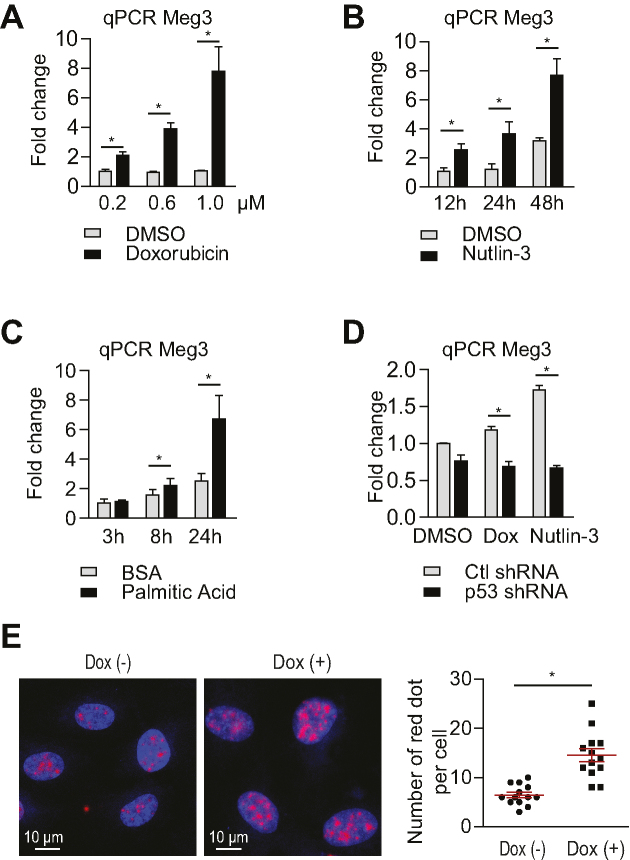
Meg3 expression is elevated in senescent HUVECs compared with early passage cells (30), and in HUVECs exposed to oxidative stress (40) and hypoxia (41). In this study, we further examined Meg3 expression in response to different stimuli and the signaling pathways that lead to Meg3 induction in ECs. Doxorubicin is a chemotherapy drug that can induce DNA damage response including p53 activation. Doxorubicin treatment led to the induction of Meg3 in HUVECs (Figure 1A). Meg3 expression was 2.1-, 3.8- and 6.3-fold higher in HUVECs that were treated with doxorubicin at 0.2, 0.6 and 1 μM, respectively, for 12 h compared to control cells. Nutlin-3, a pharmacological inhibitor of MDM2 that is a negative regulator of p53, causes p53 accumulation without inducing DNA damage. Meg3 expression was induced by 2.4-, 3.0- and 2.7-fold after 12, 24 and 48 h, respectively, of 10 μM nutlin-3 treatment (Figure 1B). 100 μM palmitic acid induced Meg3 expression by 1.4-fold after 8 h and 2.7-fold after 24 h treatment (Figure 1C). To determine whether Meg3 is induced in a p53-dependent manner, we examined Meg3 expression in HUVECs after shRNA-mediated silencing of p53. Indeed, p53 knockdown blocked the induction of Meg3 by nutlin-3 (10 μM for 12 h) and doxorubicin (40 nM for 12 h) (Figure 1D). We also examined Meg3 expression by in situ hybridization in HUVECs in response to doxorubicin treatment at 0.2 μM for 12 h. Meg3 expression was induced by 2.3-fold in the nucleus (Figure 1E). To determine whether the Meg3 promoter mediates the induction of Meg3, the Meg3 promoter was cloned for luciferase reporter assay. As shown in the Supplemental Figure S2A, nutlin-3 treatment had no effect on Meg3 promoter activity. We performed chromatin immunoprecipitation (ChIP) followed by qPCR to examine the binding of p53 at the Meg3 promoter. However, no binding of p53 at the Meg3 promoter was observed under both basal and doxorubicin conditions (Supplemental Figure S2B). The data suggest that this ∼3.2 kb region (–2997 to +264) of the Meg3 promoter is not likely involved in the induction of Meg3 in response to DNA damage or p53 accumulation. In summary, Meg3 expression is induced by DNA damage-inducing stimuli in a p53 dependent manner through a distal p53 cis-element. These data suggest a potential role of Meg3 in regulating the DDR and p53 signaling.
Figure 1.
Meg3 expression is induced by the DNA damage response in a p53 dependent manner. (A) qPCR analysis of Meg3 expression in HUVECs treated with 200 nM, 600 nM and 1 μM of doxorubicin for 12 h. (B) qPCR analysis of Meg3 expression in HUVECs treated with 10 μM nutlin-3 for 12, 24 and 48 h. (C) qPCR analysis of Meg3 expression in HUVECs treated with 100 μM palmitic acid for 12, 24 and 48 h. (D) qPCR analysis of Meg3 expression in HUVECs treated with doxorubicin (40 nM for 12 h) or nutlin-3 (10 μM for 12 h) with or without lentiviral knockdown of p53. (E) In situ hybridization of Meg3 in HUVECs treated with or without 0.2 μM doxorubicin for 12 h. The number of red dots per nucleus from 13 cells was counted for each condition. Data show mean ± S.E.M., n = 3 (A–D), n = 13 (E); *P < 0.05.
Meg3 knockdown induces DNA damage
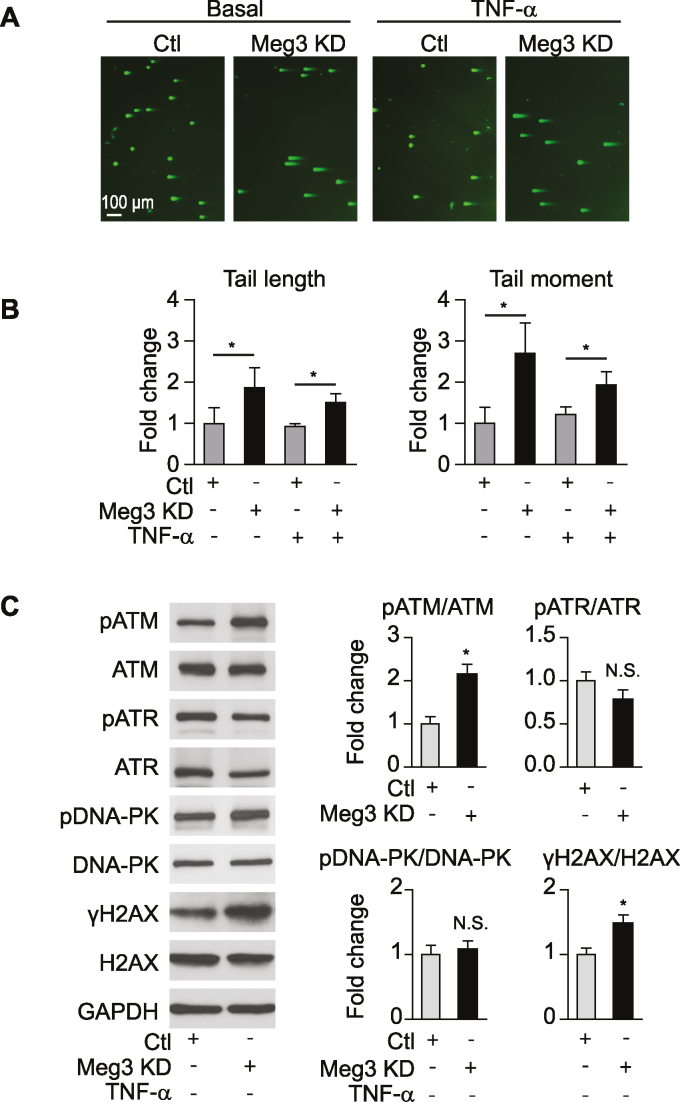
Since Meg3 expression is induced by activators of the DDR, we next examined the role of Meg3 in DNA damage. DNA double-strand breaks (DSB) are a severe form of DNA damage, which can be detected by the neutral comet assay (42,43). An increase in DNA tail length and tail moment represents a higher burden of DSBs. Meg3 knockdown by 10 nM GapmeRs (chemically modified antisense oligonucleotides) led to 1.9- and 2.7-fold increases in tail length and tail moment, respectively, under basal conditions (Figure 2A, B). In the presence of TNF-α, both the tail length and tail moment were increased by 1.6-fold (Figure 2A, B). Phosphorylation of histone H2AX at serine 19 (known as γH2AX), a marker of DSB damage and repair (44–47), is also increased by Meg3 silencing in ECs (Figure 2C). DDR is controlled by three kinases ATM, ATR, and DNA-PK (13), and serine 139 of H2AX can be phosphorylated by all three kinases (48). ATM, but not DNA-PK and ATR, is activated upon Meg3 silencing (Figure 2C). We also examined the effects of Meg3 knockdown on DNA damage using GapmeRs at 2 nM concentration. Meg3 GapmeRs reduced Meg3 expression by 88% at 2 nM and 94% at 10 nM compared to control GapmeRs (Supplementary Figure S3A). Meg3 reduction by 2 nM GapmeRs resulted in a 2.4-fold increase in tail length, 2.9-fold increase in tail moment, 1.9-fold increase in ATM phosphorylation, and 1.7-fold increase in γH2AX under basal conditions (Supplementary Figure S3B, C). These data demonstrate that Meg3 protects DNA from damage.
Figure 2.
Meg3 knockdown induces DNA damage. HUVECs were transfected with control or Meg3 GapmeRs for neutral comet assay (A and B) and western blot analysis (C). (A) Representative images of DNA tail length and moment under basal condition and in ECs treated with 10 ng/ml TNF-α for 16 h. (B) Quantification of DNA tail length and moment under basal conditions and in ECs treated with 10 ng/ml TNF-α for 16 h. (C) Western blot analysis of key proteins involved in the DNA damage response under basal conditions. Data show mean ± S.E.M., n ≥ 3; *P < 0.05.
Meg3 knockdown activates p53 signaling in ECs under inflammatory conditions
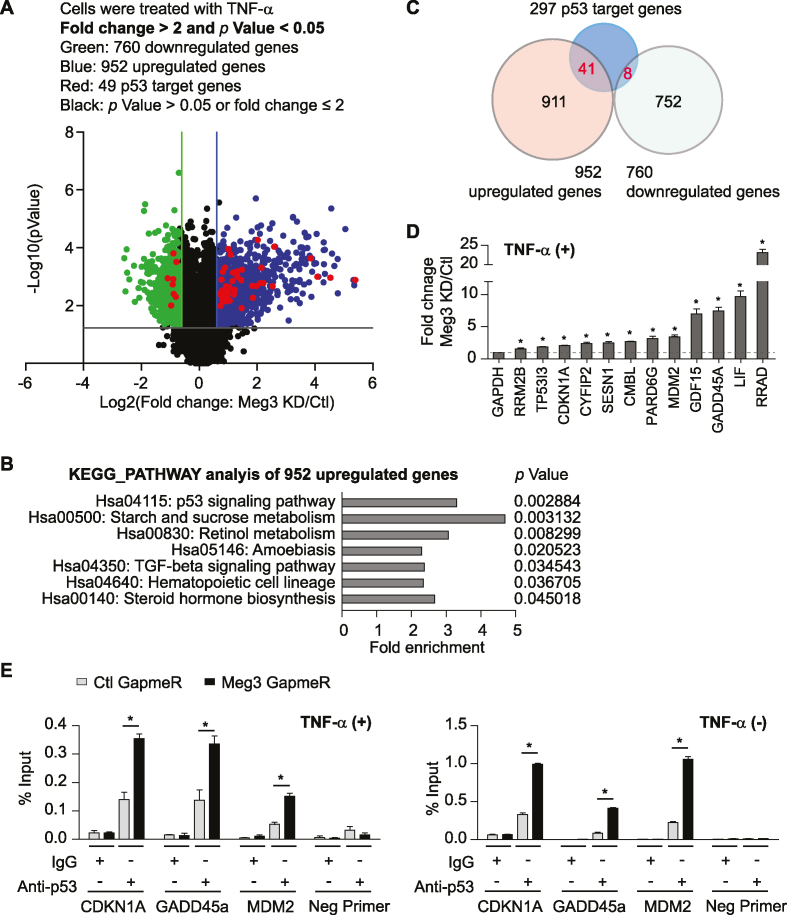
To determine the transcriptome that is regulated by Meg3, we performed transcriptome microarray profiling in HUVECs transfected with negative control or Meg3 GapmeRs and stimulated with TNF-α for 3 h. Meg3 GapmeRs used in this study have been validated by a previous study to examine the role of Meg3 in angiogenesis (30). Meg3 expression was reduced by 94% (Supplementary Figure S3A), which is associated with the changes of expression of 1712 genes (P-value < 0.05, fold change > 2) (Figure 3A and Supplementary Table S1). Among these genes, 952 genes were upregulated, and 760 genes were down-regulated. KEGG pathway analysis of up-regulated genes identified the p53 signaling pathway as the most significant among seven signaling pathways in HUVECs after Meg3 silencing (Figure 3B and Supplementary Table S2). The genome-wide p53 binding sites and p53 target genes in ECs have not been examined. However, there are 297 p53 target genes identified in other cell types by previous studies (49,50), and many of them were induced by Meg3 knockdown in ECs. From 952 upregulated genes, 41 were identified as p53 target genes, and eight were identified from 760 reduced genes (Figure 3C and Supplementary Table S3). Real-time qPCR analysis confirmed that the expression of many p53 target genes were significantly induced in ECs when Meg3 expression was reduced (Figure 3D). Meg3 knockdown also led to the induction of p53 target genes under basal conditions (Supplementary Figure S4A). Interestingly, Meg3 knockdown also induced the expression of TNF-α at the mRNA level (Supplementary Figure S4B). We next explored the possibility that Meg3 controls the expression of p53 target genes by regulating p53-mediated transcription. We performed chromatin immunoprecipitation (ChIP) followed by qPCR to examine p53 binding at the promoter regions of several p53 target genes in the presence or absence of Meg3 silencing. In response to Meg3 silencing, the amount of p53 bound to the promoters of CDKN1A, GADD45A, and MDM2 was significantly increased in ECs under basal and TNF-α-treated conditions (Figure 3E). Our data suggest that Meg3 restrains the expression of a subset of p53 target genes by inhibiting the binding of p53 to the promoters of these genes.
Figure 3.
Transcriptome analysis identifies p53 signaling as the most significantly upregulated pathway upon Meg3 knockdown. HUVECs were transfected with control or Meg3 GapmeRs. 24 h after transfection, cells were treated with 10 ng/ml TNF-α for 3 h and collected for microarray gene chip analysis. Chromatin immunoprecipitation was performed using transfected cells with or without 10 ng/ml TNF-α treatment for 1 h. (A) Volcano plot shows differentially expressed mRNAs in ECs upon Meg3 knockdown. (B) KEGG signaling pathways analysis identified significantly regulated pathways among upregulated genes upon Meg3 knockdown. (C) Venn diagram shows p53 target genes that are regulated by Meg3. (D) qPCR analysis of a group of p53 target genes that were induced upon Meg3 knockdown. (E) Enrichment of p53 at the promoters of indicated genes was identified by chromatin immunoprecipitation using anti-p53 antibodies followed by qPCR analysis. Data show mean ± S.E.M., n = 3; *P < 0.05.
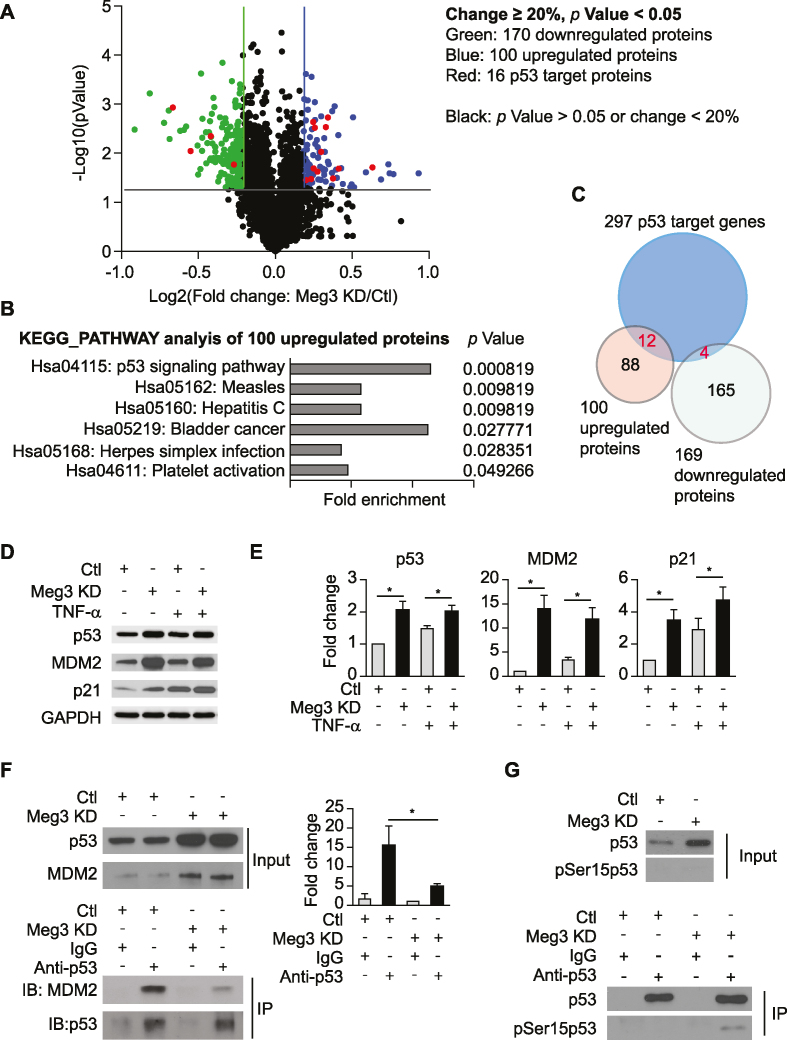
To examine the effects of Meg3 knockdown on the expression of proteins, quantitative proteomic analysis was conducted using TMT10-plex labelling followed by mass spectrometry. TNF-α treatment led to 166 differentially expressed proteins (Supplementary Table S4 sheet 1). Common NF-kB target genes including VCAM1, ICAM1, SELE, and CCL2 were induce by 6.6-, 5.3-, 3.7- and 3.2-fold, respectively, at the protein level. KEGG pathway analysis identified three TNF-α responsive pathways including ‘TNF signaling pathway’, ‘NF-kappaB signaling pathway’, and ‘Cell adhesion molecules’ as significantly enriched signaling pathways among others, suggesting that our quantitative proteomics analysis can successfully detect differentially expressed proteins (Supplementary Table S4 sheet 2). Under basal conditions (without TNF-α treatment), Meg3 knockdown resulted in 209 differentially expressed proteins (Supplementary Table S4 sheet 3). KEGG pathway analysis revealed four significantly enriched signaling pathways including: ribosome biogenesis in eukaryotes, endocytosis, glucagon signaling pathway, biosynthesis of antibiotics, and platelet activation (Supplementary Table S4 sheet 4). In response to TNF-α, Meg3 knockdown resulted in 269 differentially expressed proteins (Figure 4A and Supplementary Table S4 sheet 5). Among them, 100 proteins were upregulated, and 169 proteins were downregulated. Gene ontology and pathway analysis also identified the p53 signaling pathway as the most significantly regulated pathway in HUVECs after Meg3 knockdown (Figure 4B and Supplementary Table S4 sheet 6). Among 100 upregulated proteins, 12 were p53 targets; while only 4 out of 169 downregulated proteins were p53 targets (Figure 4C and Supplementary Table S5). Indeed, Western blot analysis verified that Meg3 silencing leads to an increase in expression of p53, MDM2, and p21 at the protein level under both basal and TNF-α-treated conditions (Figure 4D, E). The amount of p53 protein in cells is determined mainly by the rate at which it is degraded. MDM2 binds to p53 and stimulates its ubiquitination and degradation(51). We reasoned that the interaction between MDM2 and p53 is reduced upon Meg3 silencing. Indeed, we found that after immunoprecipitation (IP) for p53, the level of p53-associated MDM2 was significantly reduced in HUVECs transfected with Meg3 GapmeRs (Figure 4F). The phosphorylation of p53 at serine 15 resulting from DNA damage attenuates the interaction between p53 and MDM2, which leads to the accumulation of p53 (52). We examined the phosphorylation of p53 at serine 15 in HUVECs and found that Meg3 depletion led to an increase of the phosphorylated p53 at serine 15 (Figure 4G). We also generated lentiviral shRNA to knockdown Meg3 expression. Consistent with Meg3 silencing by GapmeRs, lentiviral Meg3 shRNA induced the expression of p53 target genes, which is associated with a 56% reduction of Meg3 in HUVECs (Supplementary Figure S4C, D). Both our transcriptomic and proteomic analysis revealed that p53 signaling is one of the dominant signaling pathways upregulated by Meg3 knockdown, suggesting that endogenous Meg3 restrains p53 activation in HUVECs.
Figure 4.
Quantitative proteomics analysis identifies p53 signaling as the most significantly regulated pathway upon Meg3 silencing. HUVECs were transfected with control or Meg3 GapmeRs. 24 h after transfection, cells were treated with or without 10 ng/ml TNF-α for 4 h. After treatment, cells were collected for TMT10-plex labeling and mass spectrometry analysis, western blot analysis, or immunoprecipitation. (A) Volcano plot shows differentially expressed proteins in ECs upon Meg3 silencing under TNF-α-treated condition. (B) KEGG signaling pathways were identified with significant enrichment among upregulated proteins upon Meg3 silencing under TNF-α-treated condition. (C) Venn diagram shows proteins encoded by p53 target genes that are regulated by Meg3 under TNF-α-treated condition. (D) Western blot analysis of p53, MDM2, and p21. (E) Quantifications for (D). (F) Co-immunoprecipitation was performed to examine the interaction between p53 and MDM2 in TNF-α-treated HUVECs. (G) The level of phosphorylated p53 at serine 15 was examined in immunoprecipitated total p53 in TNF-α-treated HUVECs transfected with control or Meg3 GapmeRs. Data show mean ± S.E.M., n = 3; *P < 0.05.
In response to DNA damage, ATM is activated to phosphorylate a range of downstream protein substrates to control and fine-tune a complex signaling network (14). We examined the effects of ATM inhibition or depletion on p53 signaling and its target gene expression that are regulated by Meg3. Consistent with our finding that p53 signaling is the dominant signaling pathway regulated by Meg3, p53 depletion completely blocked the induction of p53 target genes upon Meg3 knockdown (Supplementary Figure S5A). In contrast, ATM depletion did not have the blocking effects (Supplementary Figure S5B). Consistent with the result, ATM inhibition by ATM inhibitor KU-55933 did not attenuate the increase in p53 expression and the level of γH2AX resulted from Meg3 knockdown (Supplementary Figure S5C). These data suggest that the effects of Meg3 knockdown on p53 and γH2AX may function independently from ATM signaling (13,14).
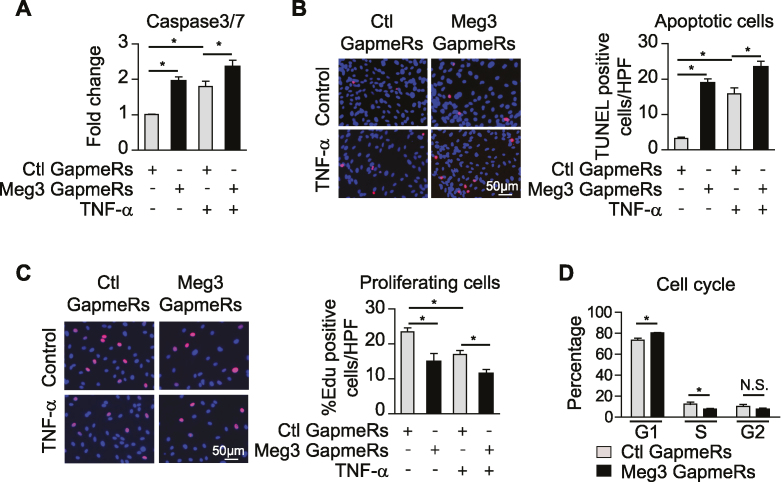
Meg3 knockdown promotes EC apoptosis and inhibits EC proliferation
Transcription factor p53 is a key player in cellular responses to DNA damage and pathological stress, and its activation leads to cell-cycle arrest and apoptosis (15,53–55). We examined the effects of Meg3 knockdown on EC apoptosis and proliferation. Meg3 knockdown increased the activities of caspase-3/7 by 94% and 30% in ECs under basal and TNF-α-treated conditions, respectively, compared to control cells, indicating increased apoptosis resulted from Meg3 knockdown (Figure 5A). Cell apoptosis was also examined by TdT-mediated dUTP nick end labelling assay (TUNEL). The numbers of TUNEL-positive cells increased by 6.6- and 1.4-fold in HUVECs with Meg3 knockdown (Figure 5B). Since we found Meg3 deficiency induced the expression of p21, which is an important mediator of cell-cycle arrest induced by p53 (56,57), we reasoned that the Meg3 knockdown may inhibit cell proliferation. Indeed, the number of proliferating cells were reduced by 37% (basal) and 33% (TNF-α) in ECs by Meg3 knockdown revealed by EdU incorporation (Figure 5C). To assess whether Meg3 knockdown has any effect on the phases of cell cycle, we performed flow cytometric analysis of cell cycle. In accordance with the inductive effect on p53 expression, HUVECs transfected with Meg3 GapmeRs had an increased population of cells in G1 phase (Meg3 knockdown versus control: 80.3 ± 0.3% versus 73.4 ± 1.8%), a decreased population of cells in S phage (Meg3 knockdown versus control: 7.5 ± 1.0% versus 12.3 ± 1.9%), while the percentage of cells in G2/M phase was not significantly changed (Meg3 knockdown versus control: 7.8 ± 1.0% versus 10.1 ± 2.1%) (Figure 5D). One important function of ECs is to form capillary-like structures in response to angiogenic signals during angiogenesis which can be determined in vitro by the Matrigel network tube formation assay. We examined the effects of Meg3 on EC tube formation. As shown in the Supplementary Figure S6, Meg3 knockdown impaired EC tuber formation under both basal and TNF-α-treated conditions indicating a protective role of endogenous Meg3 in endothelial function. These data demonstrate that Meg3 plays important roles in EC apoptosis and proliferation by regulating p53 signaling.
Figure 5.
Meg3 knockdown promotes apoptosis and inhibits proliferation in ECs. HUVECs were transfected with control or Meg3 GapmeRs, treated with or without 10 ng/ml TNF-α for 12 or 16 h. (A) Caspase 3/7 activities reflected by luminescence levels were measured, and fold changes were calculated relative to that in ECs transfected with control GapmeRs without TNF-α treatment. (B) The number of TUNEL positive cells per high power view were shown. TUNEL positive cells are in red color due to presence of DNA breaks, and DAPI-stained nuclei appear in blue. (C) The percentages of EdU positive cells (red) among Hoechst 33342 (blue) stained cells were calculated in transfected cells with or without 12 h TNF-α treatment. (D) Cell cycle distribution revealed by flow cytometry. Data show mean ± S.E.M., n = 3; *P < 0.05.
Meg3 binds to PTBP3
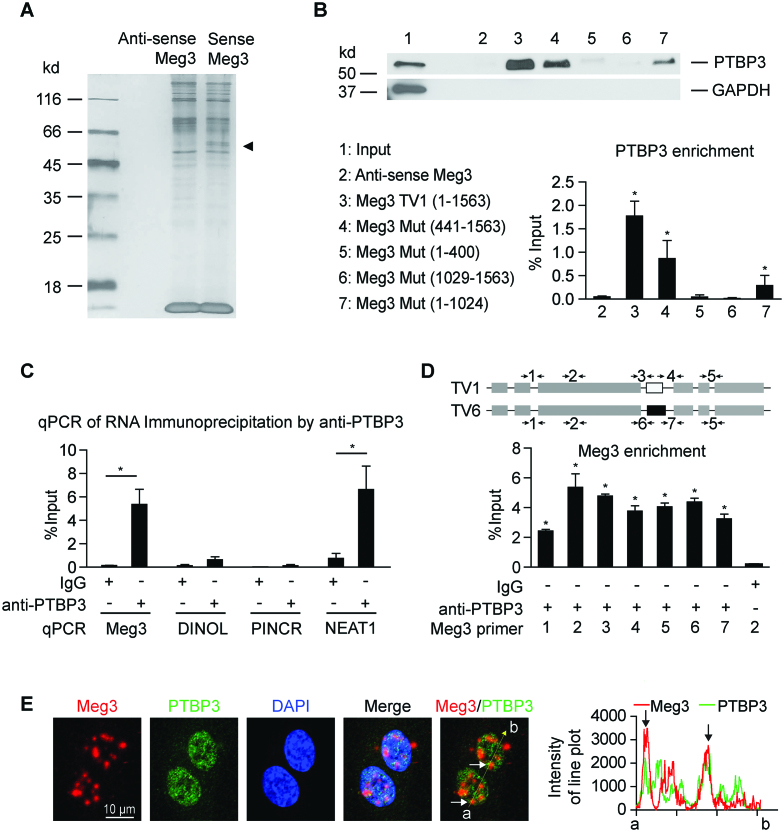
LncRNAs regulate gene expression through different mechanisms (20,23,58,59). LncRNAs can regulate local chromatin structure and/or the expression of neighboring genes in cis, or regulate cellular functions in trans after leaving the site of transcription through the interaction with other RNA or protein molecules (60). Meg3 silencing did not affect the expression of Dlk1, a nearest-neighbor protein-coding gene of Meg3 (data not shown), suggesting Meg3 likely regulates the expression of p53 target genes in trans. First, an lncRNA pull-down assay and mass spectrometry were conducted to identify Meg3 binding proteins. The RNA–protein complexes were subjected to silver staining after separation on a gel (Figure 6A). In parallel, the RNA–protein complexes were subjected to mass spectrometry to identify proteins binding exclusively to sense Meg3 RNAs compared with antisense Meg3 RNAs (Supplementary Table S6). Polypyrimidine Tract Binding protein 3 (PTBP3) was identified as a new Meg3 binding partner. Western blot analysis confirmed the enrichment of PTBP3 in the RNA–protein complexes pulled-down by sense Meg3 RNAs (Figure 6B). To explore the interaction between PTBP3 and Meg3, we conducted a lncRNA pull-down assay using Meg3 deletion mutants followed by western blot analysis of PTBP3. This analysis revealed that the region encompassing Meg3 441–1024 nucleotides is critical for the interaction between Meg3 and PTBP3 (Figure 6B). To confirm the interaction of endogenous Meg3 and PTBP3, we performed RNA immunoprecipitation, and found robust enrichment of Meg3 in the PTBP3-interacting RNA fraction compared to IgG control (Figure 6C). A well-documented PTBP3 interacting lncRNA, NEAT1 (61), was included as a positive control and it was enriched by PTBP3 RNA immunoprecipitation (Figure 6C). We identified that Meg3 transcript variants 1 and 6 are highly expressed in HUVECs (Supplemental Figure S1B, C). The sequence in Meg3 transcript variant 1 (936–1049 nucleotide region) encoded by exon 4 is different from that in Meg3 transcript variant 6. The interaction of PTBP3 with two Meg3 transcript variants were examined using seven different primer pairs by RNA immunoprecipitation. Both transcript variants were enriched in RNA immunoprecipitation using PTBP3 antibodies (Figure 6D). Partial colocalization of Meg3 and PTBP3 was observed in HUVECs by dual-staining (Figure 6E). Meg3 knockdown did not change PTBP3 expression (Supplemental Figure S7A). PTBP3 was highly expressed in the nucleus, and its expression and distribution were not changed by Meg3 depletion and in response to different stimuli (Supplementary Figure S7B, C). PTBP3 knockdown had no effect on the levels of total p53, phosphorylated p53 at serine 15, and acetylated p53 at lysine 382 in whole lysates (Supplementary Figure S7D). Interestingly, we observed that PTBP3 was immunoprecipitated by anti-histone H3 antibodies, suggesting an interaction between PTBP3 and chromatin (Supplementary Figure S7E). However, the interaction was not affected by Meg3 depletion or doxorubicin treatment at the whole chromatin level (Supplementary Figure S7E). A few proteins have been identified as Meg3 binding partners previously, including p53 and PTBP1 (28,29). PTBP1 was indeed one of the top-ranked proteins identified by our lncRNA pull-down and mass spectrometry (Supplementary Table S6). However, p53 was neither identified by proteomics (Supplementary Table S6) nor western blot analysis following lncRNA pull-down in HUVECs (Supplementary Figure S8A). In contrast, a smaller fraction of Meg3 was detected by p53 RNA immunoprecipitation (Supplementary Figure S8B). LncRNA DINOL was used as a positive control which has been identified as a p53 binding partner (62). Taken together, the in vitro and in vivo binding experiments demonstrate that PTBP3 is a protein binding partner of Meg3 in ECs.
Figure 6.
LncRNA pull-down assay identifies PTBP3 as a binding partner of Meg3. (A) LncRNA pull-down assay was used to identify the proteins associated with Meg3 transcript variant 1 (TV1) by incubating the cell lysates with biotinylated sense or antisense Meg3 RNA. The RNA–protein complexes captured by T-1 beads were subjected to silver stain after separation on SDS-PAGE gel. (B) The RNA–protein complexes from lncRNA pulldown using antisense Meg3 (negative control RNA), sense Meg3 (TV1), and Meg3 deletion mutants were subjected to western blot analysis of PTBP3 and GAPDH after separation on SDS-PAGE gel. GAPDH was examined as a negative control protein. (C) The interaction of endogenous PTBP3 with Meg3 was detected by RNA immunoprecipitation. EC lysates were immunoprecipitated with anti-PTBP3 antibody or Isotype matched control IgG. Meg3 was examined by qPCR in the immuno-precipitates using primer set 2 as shown in (D). LncRNA Neat1 was used as a positive control RNA that interacts with PTBP3. (D) Different sets of Meg3 primers were used to detect Meg3 by qPCR following RNA immunoprecipitation using anti-PTBP3 antibodies. Primer sets 1, 2, 5 detect Meg3 transcript variants 1 and 6 (TV1 and TV6); primer sets 3 and 4 detect Meg3 TV1; and primer sets 6 and 7 detect Meg3 TV6. (E) Dual-staining of Meg3 and PTBP3 in HUVECs. Linear trajectories (yellow line) crossing the cells with the intensities of Meg3 and PTBP3 signals were presented at the right side of images. White and black arrows indicate partial colocalization of Meg3 and PTBP3 in the nucleus of HUVECs. Data show mean ± S.E.M., n = 3; *P < 0.05.
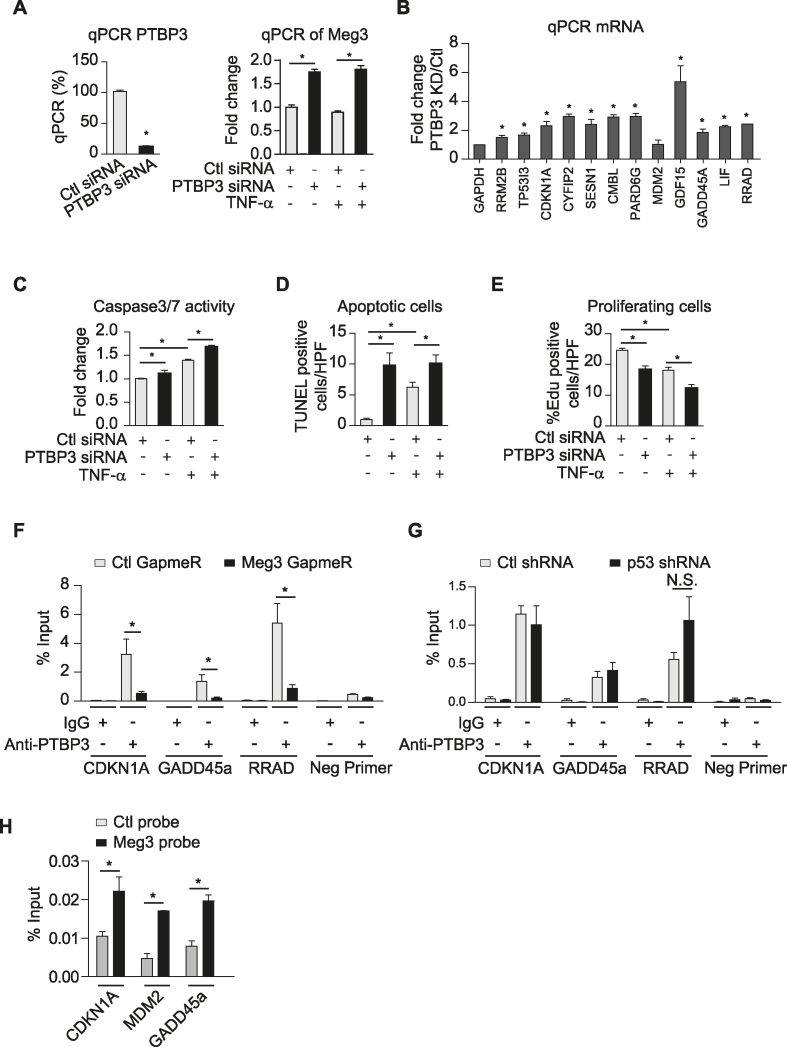
PTBP3 silencing activates p53 signaling, promotes EC apoptosis, and inhibits EC proliferation
PTBP3 is a RNA binding protein, and its function is poorly understood (63). To examine if PTBP3 mediates the effects of Meg3 in ECs, we first examined the effects of siRNA-mediated PTBP3 silencing on the expression of p53 target genes. PTBP3 expression was reduced by 87.7% at the mRNA level (Figure 7A), which is associated with increased expression of p53 target genes as indicated in Figure 7B. Interestingly, we found that Meg3 expression was increased by PTBP3 knockdown (Figure 7A). Caspase3/7 activities were significantly increased upon PTBP3 knockdown under basal and TNF-α-treated conditions (Figure 7C). The TUNEL assay revealed that PTBP3 silencing led to a 9.8-fold (basal) and 1.6-fold (TNF-α) increase of TUNEL-positive cells (Figure 7D). The role of PTBP3 in EC proliferation was examined using the EdU incorporation assay. The numbers of EdU-positive cells were decreased by 25.5% and 31.9% under basal and TNF-α-treated conditions, respectively, after PTBP3 silencing (Figure 7E). To determine the mechanism by which PTBP3 knockdown increases the expression of p53 target genes, we performed a ChIP assay to examine the association of PTBP3 with the promoters of p53 target genes. PTBP3 can bind to the promoters of p53 target genes including CDKN1A, GADD45a, and RRAD in a Meg3-dependent manner (Figure 7F), suggesting that PTBP3 could function as a regulator that restrains p53 activation. Interestingly, we found that p53 knockdown has no effect on the binding of PTBP3 at the promoters of p53 target genes including CDKN1A, GADD45a, and RRAD (Figure 7G). Chromatin isolation by RNA purification revealed the interaction of Meg3 with the promoters of p53 target genes, including CDKN1A, GADD45a, and MDM2 (Figure 7H). Since PTBP1 was identified as a Meg3 binding partner (29), we also examined whether PTBP1 is involved in mediating Meg3′s effects. PTBP1 expression was reduced by 83.4% and 82.8% under basal and TNF-α-treated conditions, respectively (Supplementary Figure S9A). PTBP1 knockdown did not induce the expression of p53 target genes except CMBL which was induced by 37% (Supplementary Figure S9B) and did not affect EC proliferation in EdU incorporation assay (Supplementary Figure S9C). PTBP1 knockdown reduced caspase3/7 activities suggesting that PTBP1 reduction inhibits EC apoptosis (Supplementary Figure S9D). Our data demonstrate that PTBP3 rather than PTBP1 knockdown recapitulates the effects of Meg3 knockdown on EC p53 signaling, apoptosis, and proliferation, suggesting that Meg3 and PTBP3 play an important role in regulating p53 signaling and endothelial function (Figure 8).
Figure 7.
PTBP3 knockdown phenocopies Meg3′s effects on EC apoptosis and proliferation. HUVECs were transfected with control or PTBP3 siRNAs (A–E), treated with or without TNF-α for 12 or 16 h. HUVECs were transfected with control GapmeRs or Meg3 GapmeRs (F). (A) PTBP3 knockdown by siRNA significantly reduces PTBP3 mRNA. (B) qPCR analysis of a group of p53 target genes in TNF-α-treated HUVECs. (C) Caspase 3/7 activities reflected by luminescence levels were measured, and fold changes were calculated relative to that in ECs transfected with control siRNAs without TNF-α treatment. (D) The number of TUNEL positive cells per high power view were shown. TUNEL positive cells are in red color due to presence of DNA breaks, and DAPI-stained nuclei appear in blue. (E) The percentages of EdU positive cells (red) among Hoechst 33342 (blue) stained cells were calculated in transfected cells with or without 12 h TNF-α treatment. (F) Enrichment of PTBP3 at the promoters of indicated genes was identified by chromatin immunoprecipitation using anti-PTBP3 antibodies followed by qPCR analysis in TNF-α-treated HUVECs transfected with Ctl GapmeRs or Meg3 GapmeRs. (G) Enrichment of PTBP3 at the promoters of indicated genes was identified by chromatin immunoprecipitation using anti-PTBP3 antibodies followed by qPCR analysis in TNF-α-treated HUVECs transduced with lentivirus expressing Ctl shRNA or p53 shRNA. N.S., non-significant. (H) qPCR analysis of CDKN1A, MDM2, and GADD45a promoters after chromatin isolation by RNA purification using control probes or Meg3 probes. Data show mean ± S.E.M., n = 3 or 4 (A–G) or = 2 (H); *P < 0.05.
Figure 8.
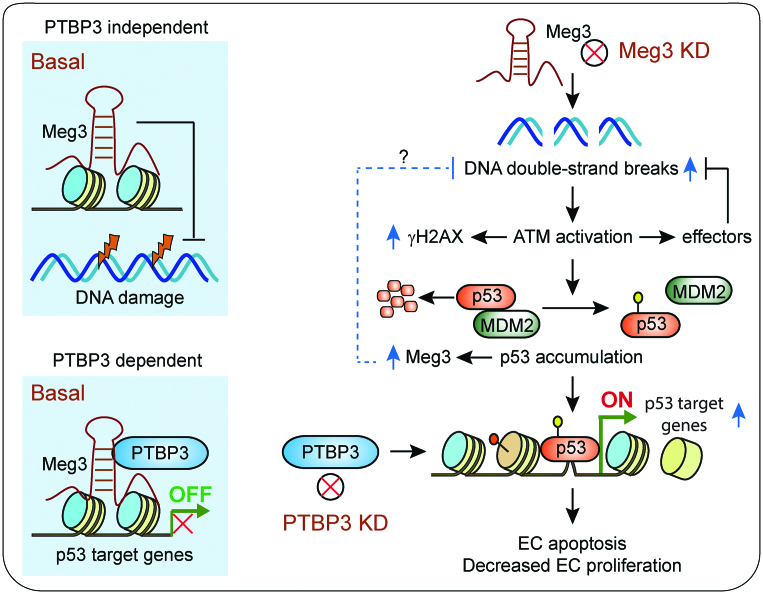
Schematic summary of this study. Under basal conditions, Meg3 protects DNA from damage independent of PTBP3, while it restrains the expression of p53 target genes in cooperation with PTBP3 likely through interactions with the promoters of p53 target genes. DNA damage induces Meg3 expression via a p53 dependent mechanism. Induced Meg3 is likely involved in restoring the homeostasis of DNA damage response through an unknown mechanism, as indicated by a question mark. Meg3 knockdown leads to DNA damage and ATM activation, which in turn stabilizes p53 by disrupting p53 and MDM2 interaction. Activation of p53 signaling results in the expression of p53 target genes, EC apoptosis, and decreased EC proliferation. PTBP3 knockdown phenocopies Meg3′s effects on the activation of p53 signaling, the expression of p53 target genes, EC apoptosis, and decreased EC proliferation.
DISCUSSION
In this study, we examined the role of Meg3 in regulating endothelial function through p53 signaling ─ one of the major coordinators of the DDR (14,64). Previous studies using other cell types indicate that Meg3 displays positive, negative, or no effects on p53 signaling. For example, Meg3 overexpression induces the expression of p53 and its target genes in cancer cells leading to the inhibition of cell proliferation (25). In mouse neurons, Meg3 interacts with p53, and its overexpression induces p53 and enhances its transcription activity resulting in an increase in cell death (28). However, the role of endogenous Meg3 was not examined in both these studies. In a recent study (26), Meg3 was found to interact with p53 in mouse cardiac fibroblasts. While inhibition of endogenous Meg3 reduced the expression of matrix metalloproteinase-2 through the inhibition of p53 binding at its promoter, there was no effect on cell apoptosis or proliferation (26). In osteosarcoma tumor cells, Meg3 silencing suppressed cell growth and promoted apoptosis (65). Because of the divergent effects of Meg3 on p53 in specific cell types, it is important to identify the molecular basis by which Meg3 controls p53 signaling in ECs. Our global transcriptomic and proteomic analysis revealed that p53 signaling is the most significantly upregulated signaling pathway upon Meg3 silencing (Figures 3 and 4). This is not due to off-target effects of Meg3 GapmeRs because Meg3 knockdown mediated by lentiviral Meg3 shRNA also activates p53 signaling (Supplementary Figure S4C, D). Our data suggest that endogenous Meg3 restrains p53 activation in response to inflammatory stimuli in ECs.
Meg3 is a DNA damage-responsive gene (Figure 1), and its expression is induced in a p53-dependent manner (Figure 1D). Allen et al. demonstrated that p53 binding sites can be within 50 kb downstream or upstream of the transcription start sites of p53 direct target genes (49). Although we have not identified the cis-element that mediates the induction of Meg3 by p53 (Supplementary Figure S2), our data suggest that Meg3 induction is mediated by the binding of p53 to the distal cis-elements.
We found that Meg3 silencing induces p53 expression at the protein level (Figure 4D). This is very likely due to the activation of ATM (14) (Figure 2C). There are several potential mechanisms for p53 induction in the presence of Meg3 deficiency: (i) post-translational modification of either Mdm2 or p53. Mdm2 is a negative regulator of p53 expression by binding to p53 and stimulating its ubiquitination and degradation (51), an effect that can be blocked by the post-translational modification of either p53 or MDM2 (66–68); (ii) sequestration of Mdm2 by other proteins including ribosomal protein L-5 or L-11 (RPL-5/11) (69) or CDKN2A/ARF (70) resulting in stabilization of p53; or (iii) an increased rate in p53 synthesis. For example, the association of p53 mRNA with ribosome or ribosome proteins such as ribosomal protein L-26 (RPL-26) can increase the translation of p53 mRNA (71). Among these three potential mechanisms, we found that the interaction between p53 and MDM2 is markedly reduced while the phosphorylation of p53 at serine 15 is increased by Meg3 silencing (Figure 4F, G), thereby highlighting that post-translational modifications is likely having a dominant role.
PTBP3 is an RNA-binding protein, and its function remains largely unknown (63). A recent study reported that inhibition of PTBP3 induced apoptosis and cell cycle arrest in tumor cells (72). We found that PTBP3 knockdown induced the expression of p53 target genes as indicated in Figure 7B. PTBP3 silencing also led to increased apoptosis and decreased cell proliferation (Figure 7C–E). PUMA and NOXA are effectors of p53 signaling that contribute to p53 induced-apoptosis (73). However, deletion of these p53 effectors cannot fully recapitulate the effects of loss of p53 (73), suggesting that other effectors are involved. It remains unknown what effectors contribute to the effects of Meg3 or PTBP3 silencing on cell proliferation and apoptosis.
We found that Meg3 controls p53 signaling under inflammatory states by regulating p53 binding at the promoters of several p53 target genes (Figure 3E). There are two possibilities for how the interaction of Meg3 and PTBP3 regulate the association of p53 with its binding sites. First, Meg3 is potentially recruited to the regulatory cis-element(s) of these genes in cooperation with PTBP3 that maintains a repressed chromatin state (Figure 8). Indeed, it was reported that Meg3 can directly interact with regulatory sequences of many genes (39). Second, the Meg3 and PTBP3 interaction potentially sequesters p53 from binding to the promoters of a subset of genes. Since Meg3 can interact with p53 directly (25,26,28), it is possible that the interaction between Meg3 and p53 could competitively prevent the binding of p53 to the promoters of these p53-regulated genes, although this mechanism itself cannot explain the selective induction of p53-regulated genes upon Meg3 or PTBP3 silencing. Furthermore, we found that Meg3 has a very week interaction with p53 in ECs (Supplementary Figure S8). Because Meg3 cooperates with PTBP3 to restrain p53 activation, PTBP3 could restrain p53 activation through an unknown mechanism. Indeed, we found that PTBP3 binds to the promoters of p53 target genes in a Meg3-dependent manner (Figure 7F).
It has been reported that Meg3 binds to EZH2 and JARID2 (39,74,75). The histone methyltransferase EZH2 is a core subunit of the polycomb repressive complex 2 (PRC2) that can trimethylate histone H3 lysine 27 (H3K27) and lead to a repressive chromatin state coinciding with gene silencing (76). JARID2 is an accessory component of PRC2. JARID2 can recruit PRC2 complexes to a heterologous promoter, thereby increasing H3K27 trimethylation levels on target genes, which in turn decreases gene expression (77). Indeed, we can detect the interactions of Meg3 with JARID2 (not EZH2) in lncRNA pull-down assays (data not shown). This raises the possibility that Meg3 could recruit PRC2 via JARID2 to specific genomic loci to restrain gene expression in ECs by introducing repressive chromatin modification such as H3K27 trimethylation. Future investigation will be required to explore how Meg3 regulates endothelial cell responses through these multiple non-p53 dependent pathways.
In summary, we demonstrated that Meg3 cooperates with PTBP3 to control the DNA damage response thereby protecting endothelial function. We found that Meg3 restrains p53 activation in response to inflammatory stimuli, which is not observed in other cell types or contrary to its role in cancer cells and neurons. Our data highlight the possibility that a change in endothelial Meg3 expression may contribute to endothelial dysfunction related to cardiovascular disease and obesity. These data suggest that therapeutic interventions that maintain Meg3 expression in the vascular endothelium may provide opportunities for preventing endothelial DNA damage and vascular dysfunction without compromising the beneficial effects of p53 signaling in other cell types such as in cancer (78).
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Dr Dirk Anderson for cell cycle analysis, Dr Reinier Boon and Dr Stefanie Dimmeler for providing the target sequence of Meg3 GapmeRs. Proteomics experiments in Figure 6 were carried out by the Proteomics and Mass Spectrometry Core facility at Princeton University. We also thank the Morrison Microscopy Core, Center for Biotechnology, UNL for confocal imaging.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health Funded COBRE [1P20GM104320 to the Nebraska Center for the Prevention of Obesity-related Diseases through Dietary Molecules]; American Heart Association [SDG#15SDG25400012 to X.S.]; Layman Fund from the University of Nebraska Foundation [to X.S.]; National Institutes of Health [HL115141, HL117994, HL134849, GM115605 to M.W.F.]; Arthur K. Watson Charitable Trust [to M.W.F.]; Dr Ralph & Marian Falk Medical Research Trust, Bank of America, N.A. Trustee [to M.W.F.]. Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1. Apovian C.M., Bigornia S., Mott M., Meyers M.R., Ulloor J., Gagua M., McDonnell M., Hess D., Joseph L., Gokce N.. Adipose macrophage infiltration is associated with insulin resistance and vascular endothelial dysfunction in obese subjects. Arterioscler. Thromb. Vasc. Biol. 2008; 28:1654–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tabit C.E., Chung W.B., Hamburg N.M., Vita J.A.. Endothelial dysfunction in diabetes mellitus: Molecular mechanisms and clinical implications. Rev. Endocr. Metab. Disord. 2010; 11:61–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sun X., Belkin N., Feinberg M.W.. Endothelial microRNAs and atherosclerosis. Curr. Atheroscler. Rep. 2013; 15:372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gimbrone M.A. Jr, Garcia-Cardena G.. Endothelial cell dysfunction and the pathobiology of atherosclerosis. Circ. Res. 2016; 118:620–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Engin A. Endothelial dysfunction in obesity. Adv. Exp. Med. Biol. 2017; 960:345–379. [DOI] [PubMed] [Google Scholar]
- 6. Ungvari Z., Tarantini S., Kiss T., Wren J.D., Giles C.B., Griffin C.T., Murfee W.L., Pacher P., Csiszar A.. Endothelial dysfunction and angiogenesis impairment in the ageing vasculature. Nat. Rev. Cardiol. 2018; 15:555–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Amodio G., Moltedo O., Faraonio R., Remondelli P.. Targeting the endoplasmic reticulum unfolded protein response to counteract the oxidative Stress-Induced endothelial dysfunction. Oxid. Med. Cell Longev. 2018; 2018:4946289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Castellon X., Bogdanova V.. Chronic inflammatory diseases and endothelial dysfunction. Aging Dis. 2016; 7:81–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shah A., Gray K., Figg N., Finigan A., Starks L., Bennett M.. Defective base excision repair of oxidative DNA damage in vascular smooth muscle cells promotes atherosclerosis. Circulation. 2018; 138:1446–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shah A.V., Bennett M.R.. DNA damage-dependent mechanisms of ageing and disease in the macro- and microvasculature. Eur. J. Pharmacol. 2017; 816:116–128. [DOI] [PubMed] [Google Scholar]
- 11. Uryga A., Gray K., Bennett M.. DNA Damage and repair in vascular disease. Annu. Rev. Physiol. 2016; 78:45–66. [DOI] [PubMed] [Google Scholar]
- 12. Shimizu I., Yoshida Y., Suda M., Minamino T.. DNA damage response and metabolic disease. Cell Metab. 2014; 20:967–977. [DOI] [PubMed] [Google Scholar]
- 13. Blackford A.N., Jackson S.P.. ATM, ATR, and DNA-PK: The trinity at the heart of the DNA damage response. Mol. Cell. 2017; 66:801–817. [DOI] [PubMed] [Google Scholar]
- 14. Shiloh Y., Ziv Y.. The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat. Rev. Mol. Cell Biol. 2013; 14:197–210. [PubMed] [Google Scholar]
- 15. Kruiswijk F., Labuschagne C.F., Vousden K.H.. Nat. Rev. Mol. Cell Biol. 16:393–405. [DOI] [PubMed] [Google Scholar]
- 16. Yokoyama M., Okada S., Nakagomi A., Moriya J., Shimizu I., Nojima A., Yoshida Y., Ichimiya H., Kamimura N., Kobayashi Y. et al. Inhibition of endothelial p53 improves metabolic abnormalities related to dietary obesity. Cell Rep. 2014; 7:1691–1703. [DOI] [PubMed] [Google Scholar]
- 17. Gogiraju R., Xu X., Bochenek M.L., Steinbrecher J.H., Lehnart S.E., Wenzel P., Kessel M., Zeisberg E.M., Dobbelstein M., Schafer K.. Endothelial p53 deletion improves angiogenesis and prevents cardiac fibrosis and heart failure induced by pressure overload in mice. J. Am. Heart Assoc. 2015; 4:e001770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lorenzen J.M., Thum T.. Long noncoding RNAs in kidney and cardiovascular diseases. Nat. Rev. Nephrol. 2016; 12:360–373. [DOI] [PubMed] [Google Scholar]
- 19. Simion V., Haemmig S., Feinberg M.W.. LncRNAs in vascular biology and disease. Vascul. Pharmacol. 2018; doi:10.1016/j.vph.2018.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sallam T., Sandhu J., Tontonoz P.. Long noncoding RNA discovery in cardiovascular disease: Decoding form to function. Circ. Res. 2018; 122:155–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Boon R.A., Jae N., Holdt L., Dimmeler S.. Long noncoding RNAs: From clinical genetics to therapeutic targets. J. Am. Coll. Cardiol. 2016; 67:1214–1226. [DOI] [PubMed] [Google Scholar]
- 22. Dianatpour A., Ghafouri-Fard S.. The role of long non coding RNAs in the repair of DNA double strand breaks. Int. J. Mol. Cell Med. 2017; 6:1–12. [PMC free article] [PubMed] [Google Scholar]
- 23. Sun X., Haider Ali M.S.S., Moran M.. The role of interactions of long non-coding RNAs and heterogeneous nuclear ribonucleoproteins in regulating cellular functions. Biochem. J. 2017; 474:2925–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kobayashi S., Wagatsuma H., Ono R., Ichikawa H., Yamazaki M., Tashiro H., Aisaka K., Miyoshi N., Kohda T., Ogura A. et al. Mouse Peg9/Dlk1 and human PEG9/DLK1 are paternally expressed imprinted genes closely located to the maternally expressed imprinted genes: mouse Meg3/Gtl2 and human MEG3. Genes Cells. 2000; 5:1029–1037. [DOI] [PubMed] [Google Scholar]
- 25. Zhou Y., Zhong Y., Wang Y., Zhang X., Batista D.L., Gejman R., Ansell P.J., Zhao J., Weng C., Klibanski A.. Activation of p53 by MEG3 non-coding RNA. J. Biol. Chem. 2007; 282:24731–24742. [DOI] [PubMed] [Google Scholar]
- 26. Piccoli M.T., Gupta S.K., Viereck J., Foinquinos A., Samolovac S., Kramer F.L., Garg A., Remke J., Zimmer K., Batkai S. et al. Inhibition of the cardiac Fibroblast-Enriched lncRNA Meg3 prevents cardiac fibrosis and diastolic dysfunction. Circ. Res. 2017; 121:575–583. [DOI] [PubMed] [Google Scholar]
- 27. Zhu J., Liu S., Ye F., Shen Y., Tie Y., Wei L., Jin Y., Fu H., Wu Y., Zheng X.. Long noncoding RNA MEG3 interacts with p53 protein and regulates partial p53 target genes in hepatoma cells. PLoS One. 2015; 10:e0139790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yan H., Yuan J., Gao L., Rao J., Hu J.. Long noncoding RNA MEG3 activation of p53 mediates ischemic neuronal death in stroke. Neuroscience. 2016; 337:191–199. [DOI] [PubMed] [Google Scholar]
- 29. Zhang L., Yang Z., Trottier J., Barbier O., Wang L.. Long noncoding RNA MEG3 induces cholestatic liver injury by interaction with PTBP1 to facilitate shp mRNA decay. Hepatology. 2017; 65:604–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Boon R.A., Hofmann P., Michalik K.M., Lozano-Vidal N., Berghauser D., Fischer A., Knau A., Jae N., Schurmann C., Dimmeler S.. Long noncoding RNA Meg3 controls endothelial cell aging and function: Implications for regenerative angiogenesis. J. Am. Coll. Cardiol. 2016; 68:2589–2591. [DOI] [PubMed] [Google Scholar]
- 31. Kim F., Tysseling K.A., Rice J., Pham M., Haji L., Gallis B.M., Baas A.S., Paramsothy P., Giachelli C.M., Corson M.A. et al. Free fatty acid impairment of nitric oxide production in endothelial cells is mediated by IKKbeta. Arterioscler. Thromb. Vasc. Biol. 2005; 25:989–994. [DOI] [PubMed] [Google Scholar]
- 32. Moffat J., Grueneberg D.A., Yang X., Kim S.Y., Kloepfer A.M., Hinkle G., Piqani B., Eisenhaure T.M., Luo B., Grenier J.K. et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006; 124:1283–1298. [DOI] [PubMed] [Google Scholar]
- 33. Godar S., Ince T.A., Bell G.W., Feldser D., Donaher J.L., Bergh J., Liu A., Miu K., Watnick R.S., Reinhardt F. et al. Growth-inhibitory and tumor- suppressive functions of p53 depend on its repression of CD44 expression. Cell. 2008; 134:62–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Huang da W., Sherman B.T., Lempicki R.A.. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009; 4:44–57. [DOI] [PubMed] [Google Scholar]
- 35. Huang da W., Sherman B.T., Lempicki R.A.. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009; 37:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yang F., Shen Y., Camp D.G. 2nd, Smith R.D.. High-pH reversed-phase chromatography with fraction concatenation for 2D proteomic analysis. Expert Rev. Proteomics. 2012; 9:129–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Atianand M.K., Hu W., Satpathy A.T., Shen Y., Ricci E.P., Alvarez-Dominguez J.R., Bhatta A., Schattgen S.A., McGowan J.D., Blin J. et al. A long noncoding RNA lincRNA-EPS acts as a transcriptional brake to restrain inflammation. Cell. 2016; 165:1672–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chu C., Quinn J., Chang H.Y.. Chromatin isolation by RNA purification (ChIRP). J. Vis. Exp. 2012; 61:e3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mondal T., Subhash S., Vaid R., Enroth S., Uday S., Reinius B., Mitra S., Mohammed A., James A.R., Hoberg E. et al. MEG3 long noncoding RNA regulates the TGF-beta pathway genes through formation of RNA-DNA triplex structures. Nat. Commun. 2015; 6:7743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fuschi P., Carrara M., Voellenkle C., Garcia-Manteiga J.M., Righini P., Maimone B., Sangalli E., Villa F., Specchia C., Picozza M. et al. Central role of the p53 pathway in the noncoding-RNA response to oxidative stress. Aging. 2017; 9:2559–2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Neumann P., Jae N., Knau A., Glaser S.F., Fouani Y., Rossbach O., Kruger M., John D., Bindereif A., Grote P. et al. The lncRNA GATA6-AS epigenetically regulates endothelial gene expression via interaction with LOXL2. Nat. Commun. 2018; 9:237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ribas-Maynou J., Garcia-Peiro A., Abad C., Amengual M.J., Navarro J., Benet J.. Alkaline and neutral Comet assay profiles of sperm DNA damage in clinical groups. Hum. Reprod. 2012; 27:652–658. [DOI] [PubMed] [Google Scholar]
- 43. Olive P.L., Banath J.P.. The comet assay: A method to measure DNA damage in individual cells. Nat. Protoc. 2006; 1:23–29. [DOI] [PubMed] [Google Scholar]
- 44. Sharma A., Singh K., Almasan A.. Histone H2AX phosphorylation: a marker for DNA damage. Methods Mol. Biol. 2012; 920:613–626. [DOI] [PubMed] [Google Scholar]
- 45. Mah L.J., El-Osta A., Karagiannis T.C.. gammaH2AX: a sensitive molecular marker of DNA damage and repair. Leukemia. 2010; 24:679–686. [DOI] [PubMed] [Google Scholar]
- 46. Turinetto V., Giachino C.. Multiple facets of histone variant H2AX: A DNA double-strand-break marker with several biological functions. Nucleic Acids Res. 2015; 43:2489–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Valdiglesias V., Giunta S., Fenech M., Neri M., Bonassi S.. gammaH2AX as a marker of DNA double strand breaks and genomic instability in human population studies. Mutat. Res. 2013; 753:24–40. [DOI] [PubMed] [Google Scholar]
- 48. Georgoulis A., Vorgias C.E., Chrousos G.P., Rogakou E.P.. Genome Instability and gammaH2AX. Int. J. Mol. Sci. 2017; 18:1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Allen M.A., Andrysik Z., Dengler V.L., Mellert H.S., Guarnieri A., Freeman J.A., Sullivan K.D., Galbraith M.D., Luo X., Kraus W.L. et al. Global analysis of p53-regulated transcription identifies its direct targets and unexpected regulatory mechanisms. eLife. 2014; 3:e02200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Riley T., Sontag E., Chen P., Levine A.. Transcriptional control of human p53-regulated genes. Nat. Rev. Mol. Cell Biol. 2008; 9:402–412. [DOI] [PubMed] [Google Scholar]
- 51. Momand J., Wu H.H., Dasgupta G.. MDM2–master regulator of the p53 tumor suppressor protein. Gene. 2000; 242:15–29. [DOI] [PubMed] [Google Scholar]
- 52. Shieh S.Y., Ikeda M., Taya Y., Prives C.. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell. 1997; 91:325–334. [DOI] [PubMed] [Google Scholar]
- 53. Lin K., Hsu P.P., Chen B.P., Yuan S., Usami S., Shyy J.Y., Li Y.S., Chien S.. Molecular mechanism of endothelial growth arrest by laminar shear stress. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:9385–9389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Damico R., Simms T., Kim B.S., Tekeste Z., Amankwan H., Damarla M., Hassoun P.M.. p53 mediates cigarette smoke-induced apoptosis of pulmonary endothelial cells: inhibitory effects of macrophage migration inhibitor factor. Am. J. Respir. Cell Mol. Biol. 2011; 44:323–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lorenzo E., Ruiz-Ruiz C., Quesada A.J., Hernandez G., Rodriguez A., Lopez-Rivas A., Redondo J.M.. Doxorubicin induces apoptosis and CD95 gene expression in human primary endothelial cells through a p53-dependent mechanism. J. Biol. Chem. 2002; 277:10883–10892. [DOI] [PubMed] [Google Scholar]
- 56. Deng C., Zhang P., Harper J.W., Elledge S.J., Leder P.. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell. 1995; 82:675–684. [DOI] [PubMed] [Google Scholar]
- 57. Gartel A.L., Radhakrishnan S.K.. Lost in transcription: p21 repression, mechanisms, and consequences. Cancer Res. 2005; 65:3980–3985. [DOI] [PubMed] [Google Scholar]
- 58. Sun Q., Hao Q., Prasanth K.V.. Nuclear long noncoding RNAs: Key regulators of gene expression. Trends Genet. 2017; 34:142–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Haemmig S., Simion V., Yang D., Deng Y., Feinberg M.W.. Long noncoding RNAs in cardiovascular disease, diagnosis, and therapy. Curr. Opin. Cardiol. 2017; 32:776–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kopp F., Mendell J.T.. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018; 172:393–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yang X., Qu S., Wang L., Zhang H., Yang Z., Wang J., Dai B., Tao K., Shang R., Liu Z. et al. PTBP3 splicing factor promotes hepatocellular carcinoma by destroying the splicing balance of NEAT1 and pre-miR-612. Oncogene. 2018; doi:10.1038/s41388-018-0416-8. [DOI] [PubMed] [Google Scholar]
- 62. Schmitt A.M., Garcia J.T., Hung T., Flynn R.A., Shen Y., Qu K., Payumo A.Y., Peres-da-Silva A., Broz D.K., Baum R. et al. An inducible long noncoding RNA amplifies DNA damage signaling. Nat. Genet. 2016; 48:1370–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tan L.Y., Whitfield P., Llorian M., Monzon-Casanova E., Diaz-Munoz M.D., Turner M., Smith C.W.. Generation of functionally distinct isoforms of PTBP3 by alternative splicing and translation initiation. Nucleic Acids Res. 2015; 43:5586–5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Meek D.W. Tumour suppression by p53: A role for the DNA damage response. Nat. Rev. Cancer. 2009; 9:714–723. [DOI] [PubMed] [Google Scholar]
- 65. Wang Y., Kong D.. Knockdown of lncRNA MEG3 inhibits viability, migration, and invasion and promotes apoptosis by sponging miR-127 in osteosarcoma cell. J. Cell. Biochem. 2018; 119:669–679. [DOI] [PubMed] [Google Scholar]
- 66. Meek D.W., Anderson C.W.. Posttranslational modification of p53: cooperative integrators of function. Cold Spring Harb. Perspect. Biol. 2009; 1:a000950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Cheng Q., Cross B., Li B., Chen L., Li Z., Chen J.. Regulation of MDM2 E3 ligase activity by phosphorylation after DNA damage. Mol. Cell. Biol. 2011; 31:4951–4963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Cheng Q., Chen L., Li Z., Lane W.S., Chen J.. ATM activates p53 by regulating MDM2 oligomerization and E3 processivity. EMBO J. 2009; 28:3857–3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Horn H.F., Vousden K.H.. Cooperation between the ribosomal proteins L5 and L11 in the p53 pathway. Oncogene. 2008; 27:5774–5784. [DOI] [PubMed] [Google Scholar]
- 70. Honda R., Yasuda H.. Association of p19(ARF) with Mdm2 inhibits ubiquitin ligase activity of Mdm2 for tumor suppressor p53. EMBO J. 1999; 18:22–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ofir-Rosenfeld Y., Boggs K., Michael D., Kastan M.B., Oren M.. Mdm2 regulates p53 mRNA translation through inhibitory interactions with ribosomal protein L26. Mol. Cell. 2008; 32:180–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Liang X., Shi H., Yang L., Qiu C., Lin S., Qi Y., Li J., Zhao A., Liu J.. Inhibition of polypyrimidine tract-binding protein 3 induces apoptosis and cell cycle arrest, and enhances the cytotoxicity of 5- fluorouracil in gastric cancer cells. Br. J. Cancer. 2017; 116:903–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Aubrey B.J., Kelly G.L., Janic A., Herold M.J., Strasser A.. How does p53 induce apoptosis and how does this relate to p53-mediated tumour suppression. Cell Death Differ. 2018; 25:104–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kaneko S., Bonasio R., Saldana-Meyer R., Yoshida T., Son J., Nishino K., Umezawa A., Reinberg D.. Interactions between JARID2 and noncoding RNAs regulate PRC2 recruitment to chromatin. Mol. Cell. 2014; 53:290–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zhao J., Ohsumi T.K., Kung J.T., Ogawa Y., Grau D.J., Sarma K., Song J.J., Kingston R.E., Borowsky M., Lee J.T.. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol. Cell. 2010; 40:939–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Margueron R., Reinberg D.. The Polycomb complex PRC2 and its mark in life. Nature. 2011; 469:343–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Pasini D., Cloos P.A., Walfridsson J., Olsson L., Bukowski J.P., Johansen J.V., Bak M., Tommerup N., Rappsilber J., Helin K.. JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature. 2010; 464:306–310. [DOI] [PubMed] [Google Scholar]
- 78. Khoo K.H., Verma C.S., Lane D.P.. Drugging the p53 pathway: understanding the route to clinical efficacy. Nat. Rev. Drug Discov. 2014; 13:217–236. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.