Abstract
DNA double-strand breaks (DSB) in bacteria can be repaired by non-homologous end-joining (NHEJ), a two-component system relying on Ku and LigD. While performing a genetic characterization of NHEJ in Sinorhizobium meliloti, a representative of bacterial species encoding several Ku and LigD orthologues, we found that at least two distinct functional NHEJ repair pathways co-exist: one is dependent on Ku2 and LigD2, while the other depends on Ku3, Ku4 and LigD4. Whereas Ku2 likely acts as canonical bacterial Ku homodimers, genetic evidences suggest that Ku3-Ku4 form eukaryotic-like heterodimers. Strikingly, we found that the efficiency of both NHEJ systems increases under stress conditions, including heat and nutrient starvation. We found that this stimulation results from the transcriptional up-regulation of the ku and/or ligD genes, and that some of these genes are controlled by the general stress response regulator RpoE2. Finally, we provided evidence that NHEJ not only repairs DSBs, but can also capture heterologous DNA fragments into genomic breaks. Our data therefore suggest that NHEJ could participate to horizontal gene transfer from distantly related species, bypassing the need of homology to integrate exogenous DNA. This supports the hypothesis that NHEJ contributes to evolution and adaptation of bacteria under adverse environmental conditions.
INTRODUCTION
DNA double-strand breaks (DSB) are potentially lethal DNA damages, and a source of genome instability (1). In bacteria, DSB repair mostly relies on homologous recombination (HR), a universal mechanism dependent on the conserved RecA protein, which involves exchange of genetic material between homologous DNA sequences (2). As HR requires the presence of a second intact genomic copy, it is effective at repairing DSB when multiple genome copies are present, i.e. essentially during exponential growth. In some bacterial species, a second pathway of DSB repair has been described, called non-homologous end joining (NHEJ; 3–5), which is a seemingly simplified version of the major eukaryotic DSB repair pathway. In eukaryotic cells, a heterodimer of the Ku70–Ku80 proteins binds DNA ends at DSBs, and recruits a number of additional proteins, including LigIV, to process and ligate the DNA ends (6). In contrast, in bacterial species in which NHEJ has been studied so far, a unique Ku protein binds DNA ends as homodimers, and recruits LigD, a multifunctional enzyme carrying nuclease, polymerase and ligase activities, which catalyzes processing and ligation of DNA ends (7,8). However, despite the apparent simplicity of this two-component repair system, 17% of sequenced bacterial genomes possessing ku genes actually encode several orthologues, and this proportion reaches 50 to >70% in some taxonomic classes such as α-Proteobacteria and Streptomycetes (9–11). Thus, Sinorhizobium meliloti encodes four putative Ku and LigD (Supplementary Figure S1A) while Streptomyces ambofaciens encodes three putative Ku and four putative LigD domains. This plurality of orthologues raises at least two questions, the first about their relative contributions to DSB repair and the second about the maintenance of multiple NHEJ systems in a single organism. Supporting the hypothesis that all Ku proteins might contribute to DSB repair, deletion analyses of ku genes showed that they all are involved in resistance against ionizing radiations (a particular cause of DSBs) of S. meliloti stationary phase bacteria and S. ambofaciens spores (9,12). However, the connections between the various Ku orthologues, their interplay with the LigD proteins and the specificities of the different systems they might define together remain completely unknown.
Here, we report a comprehensive genetic characterization of NHEJ in S. meliloti, and elucidate the respective contributions of the various ku and ligD genes. In particular, we show that several independent NHEJ pathways indeed co-exist, and we provide the first genetic experimental evidence that a Ku heterodimer may be formed in vivo, while bacterial NHEJ was thought to invariably involve a simple Ku homodimer. In addition, we also demonstrate that NHEJ is activated under stress conditions, including heat and nutrient starvation, and that part of this system is under the control of the general stress response regulator RpoE2. Finally, for the first time in bacteria, we show that NHEJ not only repairs DSBs, but can also catalyze the integration of heterologous DNA fragments into the breaks. Altogether, our data provide new insights into the mechanisms of DSB repair in bacteria, and suggest that NHEJ might contribute to the evolution of bacterial genomes under adverse environmental conditions by participating in the acquisition of foreign DNA from distantly related organisms during horizontal gene transfer events.
MATERIALS AND METHODS
Bacterial strains and growth conditions
The strains used in this study are detailed in Supplementary Table S1. Escherichia coli strains were grown in Luria-Bertani (LB) medium at 37°C. Sinorhizobium meliloti strains were grown at 28°C, either in LB medium supplemented with 2.5 mM CaCl2 and 2.5 mM MgCl2 (LBMC; used for strain constructions and precultures), in TY medium supplemented with 6 mM CaCl2 (TYC), or in Vincent minimal medium (VMM, 14). Antibiotics were added at the following final concentrations: 100 μg ml−1 streptomycin (Sm), 40 μg ml−1 gentamicin (Gm), 10 μg ml−1 tetracyclin (Tet), 40 μg ml−1 spectinomycin (Spec) or 50 μg ml−1 carbenicillin (Cb). For the plasmid-based NHEJ assay, X-gal (0.08 g l−1) and sucrose (50 g l−1) were added to the medium.
Strain and plasmid constructions
All plasmid constructions were performed in E. coli DH5α. Oligonucleotides used in this work are listed in Supplementary Table S2. The absence of mutations in all constructs was checked by DNA sequencing. Plasmids were introduced into S. meliloti by electrotransformation as described (13). Gene deletions were performed as described (14) using pJQ200mp19 derivatives. I-SceI site and complementing genes were introduced into the S. meliloti chromosome at the rhaS locus by a double recombination event of pJQ200mp19 derivatives containing the DNA sequences of interest flanked by ∼500 bp DNA fragments of the rhaS region. All plasmid and multiple mutant constructions of this work are detailed in Supplementary Information and Supplementary Table S3.
IR sensitivity
Bacteria were grown to stationary phase (24 h) in VMM medium supplemented with Sm. Cultures were washed and diluted at an OD600 of 0.3 in 0.85% NaCl, and aliquoted in 1.5 ml Eppendorf tubes. Bacteria were irradiated in a Biobeam 8000 gamma irradiator at a dose of 300 Gy. 100 μl of serial dilutions were plated on LBMC supplemented with Sm and incubated for 72 h at 28°C. The survival was calculated as the number of CFU obtained at 300 Gy divided by the total number of CFU obtained with bacteria that did not undergo IR treatment.
β-Galactosidase assays
WT and rpoE2 strains harboring promoter-lacZ reporter plasmids (described in supplementary material) were grown in TYC supplemented with Sm and Tet to OD600 = 0.6. For heat shock, bacteria were grown 75 min at 40°C. 100 μl samples of each culture were collected and β-galactosidase activity was assayed as described (15).
S. meliloti competent cells
For exponential phase S. meliloti competent cells preparation, bacteria were grown in TYC supplemented with Sm to OD600 = 0.6 and competent cells were prepared as described (13). For heat shock, bacteria were switched 75 min at 40°C before preparation of competent cells. For stationary phase S. meliloti competent cells, bacteria were maintained for 24 h in stationary phase (OD600 ∼ 3–4), diluted in fresh medium at OD600 = 0.3 and allowed to restart growth for one generation before preparation of competent cells. Competent cells directly prepared from stationary phase cultures were also used and found to display similar repair efficiencies as restarted bacteria, but with higher variability of transformation efficiency (not shown). For DNA integration assays, 10 μM of cumate was added to the cultures 30 min before preparation of competent cells and during all following steps. In order to increase transformation efficiencies in plasmid-based NHEJ assays, repair of I-SceI-mediated DSB, and DNA integration assays, competent cells were electro-transformed with DNA prepared from S. meliloti and gel-purified after agarose gel electrophoresis (13).
Plasmid-based NHEJ assays
pDP63, pDP64 and pDP65 are derivatives of pBBR1MCS-5 (GmR) containing a lacZ gene inactivated by insertion of the Bacillus subtilis sacB gene in the BamHI, SmaI or PstI site respectively. These plasmids were digested by BamHI (5′ protruding ends), SmaI (blunt ends) and PstI (3′ protruding ends), respectively, and linear plasmid DNA was gel-purified after agarose gel electrophoresis. Prior to electroporation, linear plasmid DNA was incubated for 10 min at 65°C to dissociate sticky ends. 10–25 ng of linear plasmid DNA were co-transformed with 1 ng of the circular plasmid pDP90 used as transformation control. 1/10 of the transformation mixture was plated on LBMC supplemented with Sm and Tet (circular plasmid transformant selection) and 9/10 on LBMC supplemented with Sm, Gm and sucrose (repaired linear plasmid transformant selection) ± X-gal and incubated for 72 h at 28°C. NHEJ efficiency was calculated as the ratio of the transformation efficiencies of linear versus circular plasmid. Repair fidelity was calculated as the percentage of Lac+ (blue) transformants. Unfaithful NHEJ repair junctions were amplified by PCR from white S. meliloti colonies using OCB1043 and OCB1178 primers, and the ∼600 bp PCR products were sequenced using M13rev as primer (∼100 nt upstream of the junction). We never detected any insertion or deletion of nucleotide triplet(s) in Lac− colonies, presumably because they are expected to restore the lacZ open reading frame. Sequencing of plasmid DNA from 51 randomly chosen blue (Lac+) clones obtained upon transformation of various strains with linear plasmids carrying all types of DNA ends revealed the WT sequence in every case, showing that the level of false positives resulting from addition or deletion of nucleotide triplet(s) is <2%.
I-SceI-mediated DSB repair assays
Competent cells of S. meliloti strains carrying a single I-SceI restriction site in their chromosome were electro-transformed with 25 ng of plasmid expressing the I-SceI gene (pLS273-25: details in supplementary material) or the empty vector (pLS257-1) as transformation control. Transformation mixtures of pLS273-25 (totality) or pLS257-1 (100 μl of a 10−2 dilution) were plated on LBMC supplemented with Sm and Tet. Plates were incubated for 72h at 28°C and the survival of each strain was calculated as the ratio of transformation efficiencies (in transformants μg−1) of pLS273-25 versus pLS257-1. I-SceI restriction site regions were amplified by PCR from 48 pLS273-25 transformants for each strain at 28 and 40°C (collected from 3 independent transformation experiments) using OCB1539 and OCB1547 as primers, and PCR products were digested with I-SceI to check the presence/absence of an intact restriction site. Out of 384 PCR reactions, only 6 did not give any reaction product (presumably because of a large deletion encompassing the PCR priming sequences), and 107 PCR products were resistant to I-SceI digestion (because of mutations in the restriction site). All the corresponding transformants derived from either the WT strain at 28°C and 40°C or the ku2 mutant at 40°C (see Figure 5B). Forty PCR products resistant to digestion were sequenced using OCB1539 as primer to determine the nature of the mutations (Supplementary Figure S3). Plasmid DNA was extracted from 29 transformants carrying an intact I-SceI site (from each strain at 28 and 40°C) and used to re-transform CBT2173. All plasmids transformed at a frequency equivalent to the control plasmid, suggesting that they all contained mutation(s) impairing the I-SceI meganuclease activity, which was confirmed by DNA sequencing (Supplementary Figure S4).
Figure 5.
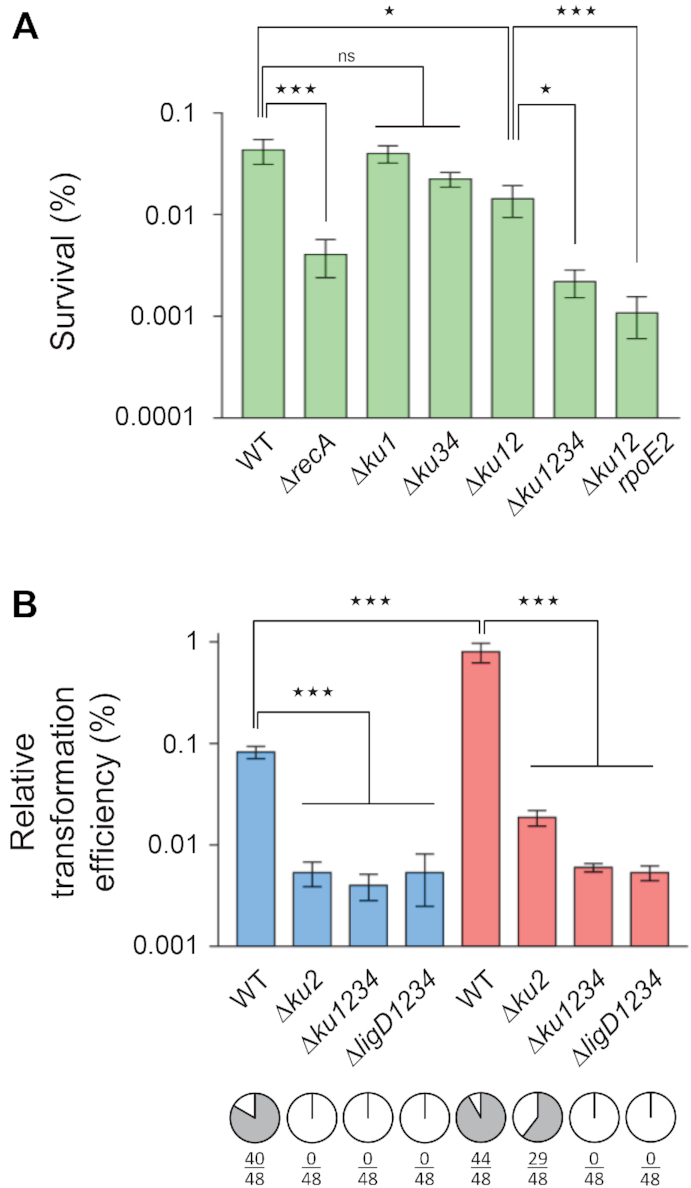
NHEJ on genomic double-strand breaks. (A) Stationary phase cultures of the indicated S. meliloti strains were exposed or not to ionizing radiations (IR) at a dose of 300 Gy, and bacteria were serial diluted and plated. Survival was calculated as the colony forming ability after exposure to 300 versus 0 Gy (expressed in %). Results shown are the means (± SEM) of the data obtained from independent experiments (n ≥ 3). One outlier value was excluded using Grubbs test (alpha = 0.05%). (B) S. meliloti strains CBT2173 (WT), CBT2175 (Δku2), CBT2177 (Δku1234) or CBT2496 (ΔligD1234), which all carry a single I-SceI site in the chromosome, were transformed either with pLS273-25, a plasmid encoding the I-SceI meganuclease, or with the empty vector pLS257-1 as a control. Competent cells were prepared from exponential phase cultures at either 28°C (blue) or 40°C (red). Transformation efficiencies with the control plasmid ranged from 106 to 107 transformants μg−1. Results are expressed as the relative transformation efficiencies of the two plasmids (in %, n = 3, mean ± SEM). 48 colonies in each instance were screened by PCR and I-SceI digestion for the presence (white) or absence (gray) of an intact I-SceI site (pie graphs; numbers indicate the number of fragments resistant to digestion, due to mutation of the I-SceI site, except in the 6 cases out of 384 where we did not get any PCR product, presumably because of a large deletion from the I-SceI site encompassing the PCR priming sequences). In each panel, statistical analyses were performed using a one-way analysis of variance (see Supplementary Table S4 for detailed statistical analysis results). The Bonferroni posttest was used to carry out the indicated comparisons (n.s., non significant; *P < 0.05; ***P < 0.001).
DNA integration assays
A 1.3 kb DNA fragment encoding a SpecR gene flanked by I-SceI compatible DNA ends was obtained by BstXI digestion of pLS278-9 (detailed in supplementary material). pLS273-25* carries the I-SceI* gene under the control of a cumate inducible promoter. I-SceI* encodes a truncated, presumably less active than WT form of the I-SceI meganuclease which makes possible to get transformants in the absence of cumate (described in supplementary material). Competent cells carrying pLS273-25* were co-transformed with the SpecR DNA fragment (25 ng) and pBBR1MCS-5 (0.1 ng) as a control. 1/10 of the transformation mixture was plated on LBMC supplemented with Sm and Gm (selection of pBBR1MCS-5 transformants) and 9/10 on LBMC supplemented with Sm and Spec (selection of integration events) and incubated for 72 h at 28°C. Integration events were checked by PCR amplification with OCB1539/1547 as primers, followed by BclI digestion to determine insert orientation. Junction sequences were determined on 29 PCR products using primers OCB1579/1580.
RESULTS
NHEJ is active in log phase S. meliloti bacteria
To characterize NHEJ in S. meliloti, we designed a plasmid-based transformation assay inspired from the one developed in mycobacteria (16,17). In this test, the transformation efficiency of a linear plasmid DNA molecule was taken as a measure of the bacterial ability to recircularize it by joining its DNA ends. Several plasmids (GmR) were constructed which contain a lacZ gene disrupted by the B. subtilis sacB gene at either the BamHI, SmaI or PstI site. Linear DNA was generated from these plasmids by digestion using the corresponding enzymes followed by gel-purification (to minimize contamination with uncut or partially digested plasmid) and used to electro-transform competent S. meliloti bacteria for GmR. The plates also contained sucrose to counter-select bacteria transformed by potential trace contaminations of residual circular plasmid DNA (as the sacB gene is toxic in the presence of sucrose), as well as X-gal in some of the experiments, to distinguish faithful from unfaithful repair events (blue Lac+ vs white/light blue Lac− colonies). A control circular plasmid (TetR) was included in transformation mixtures to normalize independent transformation data, and the repair rate was expressed as the ratio of linear to circular plasmid transformation efficiencies.
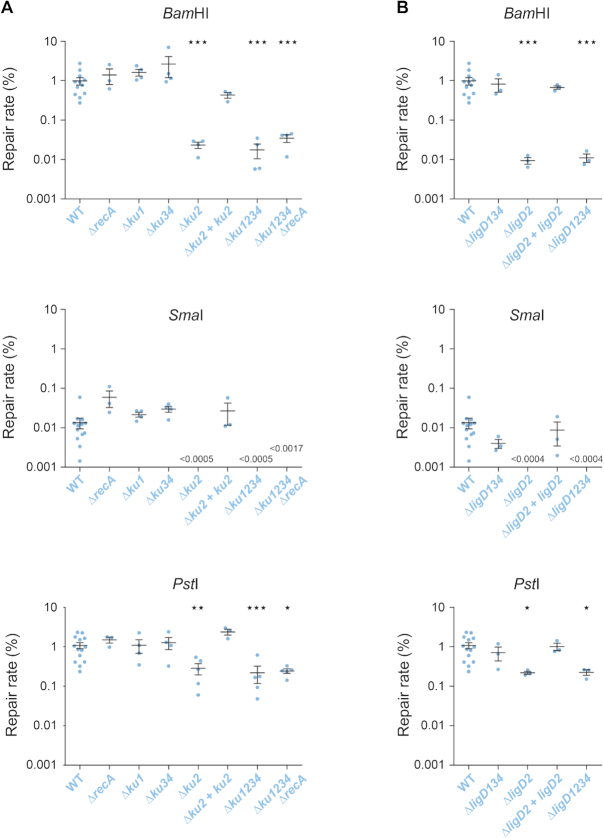
In wild type S. meliloti bacteria growing in log phase, compatible 5′ (BamHI) or 3′ (PstI) protruding ends were repaired at similar rates (∼1%), whereas blunt ends (SmaI) were repaired 100-fold less efficiently (Figure 1A). Repair was faithful, since 94, 78 and 98% of Lac+ transformants were obtained for 5′, blunt and 3′ ends, respectively (Supplementary Table S5). End repair was not due to homologous recombination, as it was not impaired in a recA mutant strain. In contrast, the efficiency of DNA repair decreased in a Δku1234 quadruple mutant, although to different extents depending on the DNA ends (Figure 1A).
Figure 1.
NHEJ in exponential phase. Linear plasmid DNA generated by restriction digest of either pDP63 by BamHI (5′ protruding ends) or pDP64 by SmaI (blunt ends) or pDP65 by PstI (3′ protruding ends) was used to transform (GmR) competent cells of the indicated S. meliloti strains, prepared from exponential phase cultures at 28°C. A control circular plasmid was included in the mixture, and served as an internal transformation control (TetR). The repair rate was calculated as the ratio of linear to circular plasmid transformation efficiencies (expressed in %). Transformation efficiencies with the control plasmid ranged from 105 to 107 transformants μg−1. Results shown are the means (±SEM) of the plotted values, obtained from independent experiments (n ≥ 3). The absence of value means that no colony was obtained (detection threshold indicated on the graph). Statistical analyses were performed using a one-way analysis of variance (see Supplementary Table S4 for detailed statistical analysis results). The Bonferroni posttest was used to compare each mutant strain to the WT (*P < 0.05; **P < 0.01; ***P < 0.001).
Together, these results show that logarithmically growing S. meliloti bacteria have the capacity to repair DNA ends by NHEJ.
Conventional NHEJ in log phase is mainly (if not only) dependent on Ku2 and LigD2
To determine the importance of the different Ku proteins in DNA repair of log phase bacteria, we tested single and multiple deletion mutants in our plasmid assay. End joining efficiency in the Δku1 and Δku3 Δku4 (referred to as ku34 hereafter) mutant strains did not differ significantly from that measured in the wild type (WT) strain, for all types of DNA ends. In contrast, in the strain carrying a ku2 deletion, DNA repair decreased to the level observed in the Δku1234 quadruple mutant (Figure 1A). Reintroduction of the ku2 gene at an ectopic locus in the ku2 mutant restored repair efficiency to near WT levels (Figure 1A). These results show that in log phase, plasmid DNA repair occurs mainly through conventional NHEJ, and that among the four Ku orthologues, only Ku2 significantly participates to NHEJ.
Sinorhizobium meliloti also encodes several LigD orthologues, whose contribution to DNA repair was not explored so far. To identify those involved in Ku2-dependent repair, we tested various ligD deletion mutants. For all types of DNA ends, the end joining efficiency of the ΔligD134 triple mutant appeared similar to that of the WT strain, whereas in the ΔligD2 and ΔligD1234 mutants, it was reduced to a level equivalent to that observed with the ku2 mutant (Figure 1B). Full complementation to WT repair levels upon reintroduction of an ectopic ligD2 gene confirmed that the phenotypes were not due to a polar effect of the ligD2 deletion on the downstream ku2 gene (Supplementary Figure S1A).
Since >99% of the repair events rely on the Ku2/LigD2-dependent NHEJ, the Lac− colonies likely resulted from unfaithful repair events of this system. Sequence analyses of plasmid repair junctions in these colonies revealed that they resulted from combinations of various classes of events including partial or complete filling-in of 5′ protruding ends, partial or complete resections of 5′ and 3′ protruding ends (with rare large deletions) and non-templated insertions of a few nucleotides (Supplementary Figure S2). These events are characteristic of NHEJ mistakes, as previously described in Mycobacterium (16,17).
Together, these data show that LigD2 and Ku2, encoded at the same chromosomal locus, work together during log phase in a main conventional NHEJ pathway in S. meliloti.
A high fidelity alternative end-joining pathway is active in S. meliloti
In the absence of Ku2 or LigD2, 5′ and 3′ protruding ends were still repaired at a detectable level, even in the quadruple ku1234 or ligD1234 mutants (Figure 1). This indicates that in addition to the Ku2/LigD2-dependent NHEJ, one or several Ku- and LigD-independent end-joining mechanisms exist, that we will thereafter refer to as Alternative End-Joining (A-EJ). Note however that we do not exclude the possibility that in the absence of LigD, Ku protects DNA ends that are then ligated by other cellular ligases, and conversely that in the absence of Ku, the LigD are still able to ligate DNA ends.
This alternative repair is not dependent on homologous recombination, as it was still active in a ku1234 recA mutant (Figure 1A). A-EJ was 20-fold more efficient with 3′ (∼0.2%) than 5′ protruding ends (∼0.01%). We could never observe any residual repair of blunt ends in all strains lacking ku2 or ligD2, which suggests that alternative repair of blunt ends, if any, occurs at a level below the detection threshold of our test (i.e. ∼0.0005%).
Interestingly, 100% of the transformants obtained with strains carrying a ku2 and/or a ligD2 mutation were Lac+, a value significantly higher than the 94–98% observed in the WT strain (Fisher's exact test, P < 0.0001; Supplementary Tables S5 and S6). This suggests that the A-EJ pathway is more faithful than the conventional NHEJ pathway.
Efficiency of conventional NHEJ (but not A-EJ) of S. meliloti increases in stress conditions
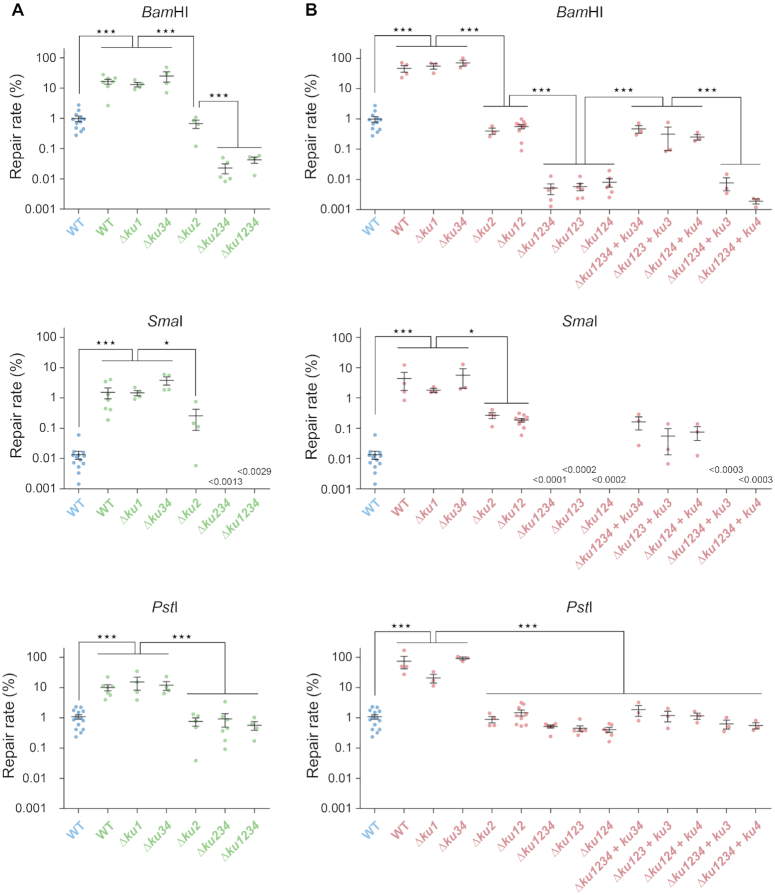
In an attempt to assess NHEJ efficiency under stress conditions, we repeated the plasmid assay with competent cells prepared from stationary phase cultures (nutrient stress), or from log phase cultures shifted from 28 to 40°C for 75 min (heat stress). Interestingly, linear plasmid DNA transformed more efficiently stressed than unstressed bacteria, irrespective of the type of DNA ends (Figure 2). 5′ or 3′ protruding ends were repaired 10–60 fold more efficiently by stressed versus unstressed bacteria, and blunt ends were even repaired >100-fold more efficiently by stressed bacteria. In both stationary phase and heat stressed bacteria, these high repair efficiencies dropped strongly in a Δku1234 or a ΔligD1234 mutant, showing that they were essentially due to conventional NHEJ (Figures 2 and 3). Accordingly, the frequency and type of repair infidelities in both conditions were similar to those observed in log phase at 28°C (Supplementary Table S5 and Supplementary Figure S2).
Figure 2.
NHEJ under stress conditions. Similar experiments as in Figure 1 were performed using competent cells prepared from stationary phase cultures (A; green) or exponential phase cultures after a 75min shift from 28 to 40°C (B; red). Values obtained with the WT strain in exponential phase at 28°C (blue) are the same as in Figure 1 and are reported as a reference. Results shown are the means (±SEM) of the plotted values, obtained from independent experiments (n ≥ 3). The absence of value means that no colony was obtained (detection threshold indicated on the graph). Statistical analyses were performed using a one-way analysis of variance (see Supplementary Table S4 for detailed statistical analysis results). The Bonferroni posttest was used to carry out the indicated comparisons (*P< 0.05; ***P < 0.001).
Figure 3.
Involvement of multiple LigD under stress conditions. Similar experiments as in Figures 1 and 2 were performed using competent cells prepared from heat stressed exponential phase cultures (red). (A) Ligases involved with Ku2: all ligD mutations are in a Ku+ background. Values obtained with the WT strain are the same as in Figure 2 and are indicated as references. (B) Ligases involved with Ku3–4: all ligD mutations are introduced in a ku1 ku2 mutant background. All results shown are the means (±SEM) of the plotted values, obtained from independent experiments (n ≥ 3). The absence of value means that no colony was obtained (detection threshold indicated on the graph). Statistical analyses were performed using a one-way analysis of variance (see Supplementary Table S4 for detailed statistical analysis results). The Bonferroni posttest was used to carry out the indicated comparisons (*P< 0.05; **P < 0.01; ***P < 0.001).
Whether in stationary phase or heat stressed bacteria, residual repair rates of ∼0.01% and ∼0.5% were still visible in the absence of conventional NHEJ (Δku1234 or ΔligD1234 mutants) for 5′ and 3′ protruding ends, respectively, while blunt end repair was undetectable (Figures 2 and 3). This residual repair was extremely faithful (significantly higher than the WT; Fisher's exact test, P < 0.0001; Supplementary Tables S5 and S6). These rate and fidelity of repair are comparable to those of the A-EJ measured in Δku1234 or ΔligD1234 mutant bacteria in log phase at 28°C, suggesting that in contrast to conventional NHEJ, A-EJ is not stimulated in stressed bacteria.
Formally, the increase of repair rates observed under stress conditions could result from either a stimulation of NHEJ activity, or an inhibition of cellular exonuclease activities resulting in a better chance to repair DNA. We think this second hypothesis unlikely as it would imply a higher basal level of A-EJ repair, and a less frequent occurrence of large deletions under stress conditions, that we did not observe. We therefore conclude that stress conditions lead to an increased capacity of S. meliloti to repair DNA ends by conventional NHEJ.
NHEJ repair in stressed bacteria involves mainly the Ku2 pathway and a second, Ku3-Ku4 dependent pathway
In unstressed growing bacteria, conventional NHEJ is dependent on Ku2. To test the Ku requirements in stressed bacteria, we repeated the plasmid transformation assays on various ku mutants. Whereas the repair rate was not significantly affected by Δku1 or Δku34 mutations, it was reduced by one-two orders of magnitude in the Δku2 mutant in comparison to WT cells (Figure 2). This shows that Ku2-dependent NHEJ is the major pathway of end repair in stressed bacteria.
However, 5′ and blunt ends were repaired more efficiently in the Δku2 than in the Δku1234 mutant, blunt ends being even repaired more efficiently than in unstressed WT bacteria (Figure 2). These results suggest that one or several additional Ku proteins are involved in these stress conditions. Deletion of the ku3 ku4 operon in the Δku2 mutant led to a reduction of the repair rate of 5′ protruding ends in stationary phase, which reached the level and fidelity of A-EJ repair measured in the Δku1234 mutant, whereas blunt end repair was no longer detected (Figure 2A and Supplementary Table S5). Likewise, reintroduction of the ku34 operon at an ectopic chromosomal locus in the ku1234 mutant restored repair to ku12 levels at 40°C (Figure 2B). Sequence analyses revealed typical NHEJ infidelity events in the ku12 background at 40°C, very similar to those observed in the WT strain, except at blunt ends where deletions were found instead of the 1 nucleotide insertions usually found in the WT (Supplementary Figure S2). Altogether, these results show that in addition to Ku2, the Ku3 and/or Ku4 proteins also contribute to conventional NHEJ in stressed bacteria.
To know whether ku3, ku4 or both are involved in the new stress-activated pathway(s), we tested additional single ku3 and ku4 in-frame deletions in the Δku12 background at 40°C, where only the Ku3-Ku4-dependent NHEJ is active. Interestingly, deletion of either ku3 or ku4 led to the same repair deficiencies of 5′ and blunt ends as the Δku34 deletion (Figure 2B). Bringing an ectopic copy of the deleted gene in these mutants restored their repair level to that of the Δku12 strain. In contrast, ectopic expression of either ku3 or ku4 alone in the Δku1234 mutant did not lead to complementation (Figure 2B). This shows that both ku3 and ku4 are required in the same pathway.
In contrast to 5′ and blunt ends, repair of 3′ protruding ends under stress conditions was not further reduced significantly by deletion of ku34 in a ku2 background (Figure 2). This could indicate that Ku3–Ku4 and associated ligases are not efficient at recognizing/repairing these DNA ends. However, we noticed that 3′ ends were repaired more faithfully in the absence than in the presence of ku34 in a ku2 background (Fisher's exact test, P ≤ 0.0014; see Supplementary Tables S5 and S6). This suggested to us that the unfaithful Ku3–Ku4-dependent NHEJ and the high-fidelity Ku-independent A-EJ were both active at similar levels in the residual repair of 3′ ends in a ku2 background.
Our observations therefore show that the stimulation of NHEJ observed in stressed bacteria mainly results from stimulation of the Ku2-dependent repair, and that an additional pathway dependent on both Ku3 and Ku4 (referred to as Ku3–4 hereafter) is activated in these conditions.
Multiple LigD proteins participate in NHEJ in stressed bacteria
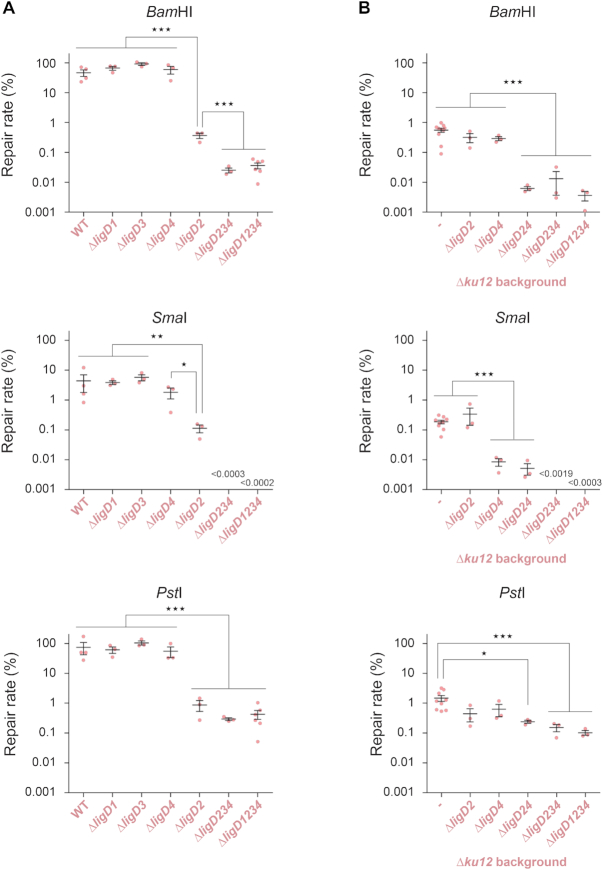
In unstressed growing bacteria, Ku2-dependent NHEJ only involves LigD2. We wondered whether other S. meliloti LigD orthologues are involved in NHEJ under stress conditions. In heat stressed bacteria, and for all types of DNA ends, the repair efficiency was not significantly altered in single ΔligD1, ΔligD3 and ΔligD4 mutants. In contrast, it was reduced in the ΔligD2 mutant to a level equivalent to that measured in the Δku2 mutant (Figure 3A). This indicates that, as in log phase bacteria at 28°C, LigD2 is the major enzyme involved in Ku2-dependent repair in stressed bacteria, irrespective of the DNA ends. However, for 5′ and blunt ends, the repair level was not reduced to that of the ku1234 mutant, which was reached only by the triple ligD234 and quadruple ligD1234 mutants. This suggested that one or more LigD proteins, different from LigD2, are involved in the Ku3–4-dependent pathway.
To analyze the LigD requirement of the Ku3–4 pathway, we tested the effect of deleting ligD genes in a Δku12 mutant at 40°C, in which the Ku3–4 pathway only is active. In this background, 5′ end repair was not significantly affected by ΔligD2 or ΔligD4 deletion, whereas it was reduced 100-fold in the double ΔligD24 mutant (Figure 3B). This shows that both LigD2 and LigD4 are active and redundant at repairing 5′ protruding ends through the Ku3–4 pathway. Additional deletion of ligD3 and ligD1 did not further decrease the repair rate, suggesting that they are not involved in 5′ end repair in these conditions. Similar trends were observed for 3′ ends, although differences were smaller (Figure 3B). Again, this could be because Ku3–4 and associated ligases are poorly efficient at recognizing/repairing 3′ ends, or because 3′ends are repaired at similar levels by conventional NHEJ and by A-EJ. In agreement with the second hypothesis, 3′ end repair was close to 100% faithful in the absence of both ligD2 and ligD4 in the ku12 background, a value significantly higher than that observed when either of the two genes is present (Fisher's exact test, P < 0.0001; Supplementary Tables S5 and S6), suggesting that both LigD are active with Ku3-Ku4 for repairing 3′ends.
In contrast, while blunt end repair was as efficient in the ΔligD2 as in the ligD2 proficient strain, it was reduced ∼30-fold in the ΔligD4 and ΔligD24 mutants (Figure 3B). This suggests that LigD2 is not able to repair blunt ends when associated to Ku3–4, and that LigD4 is the main enzyme active at repairing blunt ends through the Ku3–4 pathway. Additional deletion of ligD3 further reduced the repair of blunt ends to undetectable level, showing that LigD3 is also weakly active at working with Ku3–4 under these conditions.
Altogether, these results show that LigD2, LigD3 and LigD4 are active under stress conditions. LigD2 is the main (if not the only) enzyme active with Ku2, whereas all three enzymes can act with Ku3–4. In the latter case, both LigD2 and LigD4 can repair 5′ and probably 3′ ends, whereas both LigD4 and LigD3 are active on blunt ends. LigD4 is the only ligase active with Ku3–4 on both protruding and blunt ends.
Stress induction of ku2, ku3 ku4 and ligD4 expression, and role of the general stress response
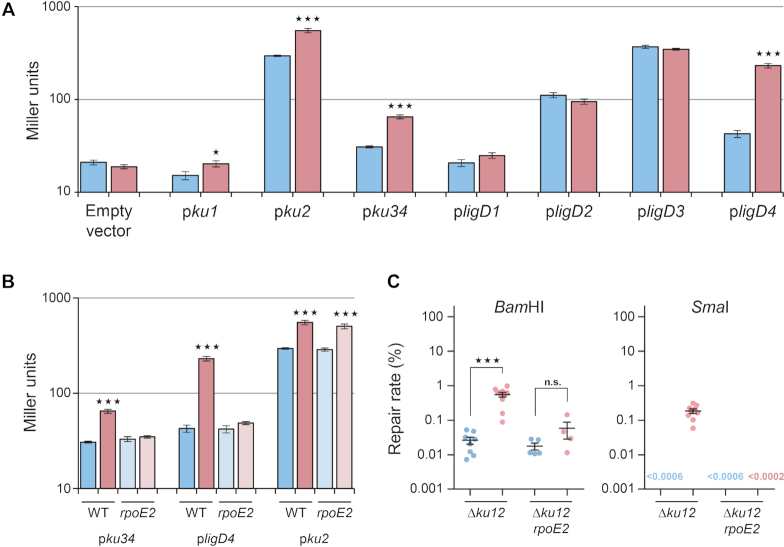
Among the S. meliloti ku and ligD orthologues, expression of the ku2 and ku34 genes was previously described to be induced when S. meliloti enters the stationary phase (9,15,18). Our observation that Ku2- and Ku3–4-dependent repair is activated in stationary phase suggests that it could result from an increase of ku gene expression. To experimentally test whether the expression level of ku and ligD genes is a major determinant of NHEJ activity, we constructed promoter-lacZ transcriptional fusions and used them to compare promoter activity in the absence or presence of stress.
We observed that: (i) ku2 is the most expressed ku gene in log phase at 28°C or 40°C, (ii) ku2 and ku34 are up-regulated at 40°C versus 28°C, (iii) ku1 expression is low (empty vector level) in both conditions (Figure 4A). This is consistent with Ku2-dependent repair being the major NHEJ pathway in all conditions, and with Ku3–4-dependent repair being detectable at 40°C only, while Ku1-dependent repair is undetectable. In contrast, ligD3 is the most expressed ligD gene at 28°C, followed by ligD2, ligD4 and ligD1. Consistently with its activity under stress conditions, ligD4 is strongly up-regulated at 40°C, at a level even higher than ligD2. There is therefore no strict correlation between the expression levels of ligD genes and their involvement in repair. This suggests that the NHEJ pathway active in a given condition is mostly determined by the Ku protein present in this condition, presumably because Ku binds DNA first (7).
Figure 4.
Transcriptional regulation of NHEJ. (A) Beta-galactosidase activities, expressed in Miller units, were measured in exponential phase cultures at either 28°C (blue) or after 75 min at 40°C (red) of WT bacteria carrying the indicated plasmid-borne promoter-lacZ fusions, or the promoter-less pCZ962 vector (n = 3, mean ± SEM). (B) Beta galactosidase activities were measured as in (A) in either WT or rpoE2 mutant bacteria carrying the pku34, pligD4 or pku2 lacZ fusions (n = 3, mean ± SEM). (C) Repair rates were measured as in Figure 1, using competent cells of Δku12 and Δku12 rpoE2 mutant strains prepared from exponential phase cultures either at 28°C (blue), or after a 75 min shift from 28 to 40°C (red). Results shown are the means (±SEM) of the plotted values, obtained from independent experiments (n ≥ 3). In each panel, statistical analyses were performed using a one-way analysis of variance (see Supplementary Table S4 for detailed statistical analysis results). The Bonferroni posttest was used to compare for each strain the results obtained in stress versus control conditions (n.s., non significant; *P< 0.05; ***P < 0.001).
The RpoE2 sigma factor is activated under many stress conditions, and responsible for regulating the general stress response in S. meliloti (15). Interestingly, putative promoter sequences recognized by RpoE2 are present upstream from the transcription start sites of the ku3–ku4 operon and the ligD4 gene (Supplementary Figure S1B) and ku3 as ligD4 were found up-regulated upon rpoE2 overexpression (15,19). This strongly suggested that the transcription of these genes could be under control of the general stress response regulator RpoE2. To validate this hypothesis, we measured the expression of the ku3 and ligD4 promoter-lacZ fusions in WT and rpoE2 mutant backgrounds. Whereas the ku3 and ligD4 promoters were activated upon heat stress in the WT strain, they were no longer inducible in the rpoE2 mutant (Figure 4B) while expression of the ku2 promoter, used as control, was similar in WT and rpoE2 mutant cells. Altogether, these results confirm that ku3 ku4 and ligD4 (but not ku2) are under the control of RpoE2.
To test whether stress-activation of the Ku3–4-dependent NHEJ pathway results from up-regulation of the corresponding genes we measured Ku3–4-dependent repair activity in rpoE2 deficient vs proficient strains. Experiments were performed in a ku12 background where the Ku3–4 pathway is the only one activated by stress. The repair efficiency of 5′ and blunt ends was increased at 40°C in the rpoE2 proficient strain, as shown above, but not in the rpoE2 mutant background (Figure 4C). This suggests that stress activation of the Ku3–4 pathway results from up-regulation of expression of its components.
Altogether, these observations show that the efficiency of NHEJ repair pathways is controlled, at least in part, at the level of transcription of the involved ku and ligD genes, and suggest that stress induction of NHEJ repair results from up-regulation of these genes. Whereas we do not know how ku2 is regulated, we showed that expression of ku3-ku4 and ligD4, and as a result the activity of the Ku3–4-dependent NHEJ pathway, are under the control of the general stress response regulator RpoE2.
The Ku2 and Ku3–4-dependent pathways are also active at repairing genomic DSB
To test whether the DNA repair measured using the plasmid transformation assay actually reflects what occurs at the genomic level, we first tested the sensitivity to ionizing radiations (IR) of various S. meliloti strains. We and others previously showed that in log phase, there was no difference in IR survival between the WT and the NHEJ mutant strains, whereas the recA mutant survival was strongly reduced, confirming that homologous recombination is the main pathway of DSB repair in growing bacteria (9,20). In contrast, we showed that in stationary phase, both NHEJ genes and recA contributed to IR resistance (20). Here we tested the involvement of the different NHEJ pathways in IR resistance of stationary phase bacteria. As expected, the recA mutant displayed a significant reduction of cell viability in comparison to the WT strain at 300 Gy, whereas the Δku1 mutant did not (Figure 5A). IR resistance decreased ∼30% in the Δku34 mutant, but this was found to be statistically non significantly different from the WT strain. However, the Δku12 mutant displayed a significant reduction of cell viability of 67%, and a further 30% reduction of survival was observed in the Δku1234 and Δku12 rpoE2 strains (Figure 5A). These observations therefore confirm that both the Ku2- and Ku3–4 NHEJ pathways contribute to IR resistance in stationary phase, with the Ku3–4 pathway playing a non-negligible role (∼30% of the events). As in the plasmid repair assay, we could not detect any influence of the ku1 deletion.
To more closely examine DNA repair of genomic DSB, we designed a S. meliloti strain carrying a single I-SceI restriction site at the chromosomal rhaS locus, and transformed this strain with a plasmid encoding the I-SceI meganuclease. This plasmid transformed the I-SceI-carrying strains ∼104-fold less efficiently than the empty vector (Figure 5B) whereas it transformed at a normal level the WT strain lacking the I-SceI restriction site (not shown). This shows that the DSB introduced at the chromosomal I-SceI site by the plasmid-encoded meganuclease is lethal for most transformed cells. Accordingly, analysis of viable transformants obtained in these conditions revealed that they contained either a mutation in the I-SceI site (rendering it resistant to the meganuclease) or a mutation in the meganuclease gene (impairing enzyme activity) (see Material and methods and Supplementary Figure S4). The plasmid transformation efficiency decreased ∼10-fold in the ku2 mutant, and the distribution of mutations differed from the WT, with no mutation of the I-SceI site. This result shows that mutations of the I-SceI site in the WT strain at 28°C resulted from unfaithful Ku2-dependent NHEJ repair. In agreement, the mutations observed were mainly deletions (Supplementary Figure S3) as expected for NHEJ mistakes on 3′ protruding ends. The transformation efficiency in the ku1234 mutant was similar as in the ku2 mutant, as expected if the Ku2 pathway is the only NHEJ repair pathway in log phase at 28°C. When similar experiments were performed with heat-stressed competent cells, a significantly 10-fold higher transformation efficiency was observed in WT cells, as expected if NHEJ is stimulated in this condition (Figure 5B and Supplementary Figure S3). Indeed, in the ku2 and ku1234 strains, the plasmid transformation frequency dropped back to background levels. However, the distribution of mutations was different from that observed at 28°C, with mutations of the I-SceI site still detected in the ku2 mutant, and no mutation isolated in the ku1234 strain. This indicates that the Ku3–4 NHEJ pathway participates in I-SceI site mutagenesis at 40°C. These data show that NHEJ is active in logarithmically growing bacteria for repairing DSB occurring in the genome, and that depending on the conditions, either only the Ku2- (28°C) or the Ku2- and to a lesser extent the Ku3–4-dependent pathways (40°C) are mobilized. Mutagenic NHEJ repair events were never detected with the ligD1234 strain, suggesting that the LigD enzymes are also involved in the NHEJ repair of genomic DSB, as described above for linear plasmid repair. Sequence analysis of mutations obtained in the ku2 mutant at 40°C showed a spectrum of mutations very similar to that observed in the WT strain at 28 or 40°C (Supplementary Figure S3). This confirms that the Ku3–4 and Ku2 pathways behave similarly on 3′ protruding ends, as already observed with plasmidic PstI ends (Supplementary Figure S2).
Altogether, these results show that the Ku2 and Ku3-Ku4-dependent pathways are both active at repairing genomic double-strand breaks in logarithmic and stationary phases of growth.
Exogenous DNA can be captured in genomic DNA breaks through NHEJ repair
Although NHEJ is well known to be involved in either programmed or accidental genome rearrangements in eukaryotic cells (21–26), its implication in prokaryotic cells has been suggested (27), but not demonstrated. To test the possibility that DNA fragments can be captured during NHEJ repair events of genomic DSB, we first generated S. meliloti strains carrying a single I-SceI site at the chromosomal rhaS locus and in which expression of the I-SceI meganuclease-encoding gene is inducible by addition of cumate (the meganuclease used here was a mutated form of I-SceI, presumably less active than the WT, see Supplementary Information). Whereas in the absence of cumate these strains grew at the same rate and displayed the same colony forming ability as strains carrying the empty vector, addition of cumate to the cultures led to a cessation of growth as soon as 6 hours after induction, and plating on cumate-containing plates led to a >103 reduction in cell viability (not shown). This indicates that genomic DSB are induced by cumate addition. Competent cells were prepared 30 minutes after cumate addition, from cultures grown at either 28 or 40°C, and were transformed with a linear DNA cassette carrying a Spec resistance gene flanked by I-SceI-compatible restriction sites generated with BstXI. All strains displayed similar transformation efficiencies with a control circular plasmid (Table 1). Four SpecR transformants were obtained with the WT strain at 28°C over six independent experiments, and the transformation efficiency increased 70-fold at 40°C (Table 1). In contrast, the ku2, ku1234 and ligD1234 mutants gave no transformants at 28°C, and only three transformants were obtained at 40°C with the ku2 mutant over six experiments. PCR analysis of the rhaS region in 285 SpecR transformants showed that they all resulted from insertion of the transforming DNA cassette in the chromosomal I-SceI site. Closer analysis of insertion events by restriction digest showed that most of them (257/285) had occurred in the expected orientation, and DNA sequencing of 23 of them revealed five deletion events at one insert extremity (Supplementary Figure S5). In 9% of the cases (26/285), the cassette was inserted in the opposite orientation, with mutations at both ends (mainly deletions) as expected for NHEJ repair of incompatible 3′ protruding ends (Supplementary Figure S5). Finally, two clones were found to have resulted from insertion of two SpecR cassettes in tandem. Altogether these data indicate that NHEJ, which is stimulated in stress conditions, can promote insertion of a heterologous DNA fragment in a chromosomal DSB.
Table 1.
NHEJ-promoted capture of heterologous DNA in a genomic DSB
| Transformation efficiencya | |||
|---|---|---|---|
| Condition | Genotype | Control plasmid pBBR1MCS-5 (GmR) | Linear DNA cassette (SpecR) |
| 28°C (log phase) | WT | 3.6 (±0.7) × 107 | 3.0 (±2.0) × 101 |
| Δku2 | 3.6 (±0.8) × 107 | <43 (±0.9) | |
| Δku1234 | 3.9 (±0.8) × 107 | <43 (±0.9) | |
| ΔligD1234 | 6.2 (±1.8) × 107 | <33 (±0.0) | |
| 40°C (log phase) | WT | 7.6 (±1.4) × 107 | 2.2 (±0.3) × 103 |
| Δku2 | 7.7 (±1.7) × 107 | 2.2 (±1.4) × 101 | |
| Δku1234 | 7.9 (±1.9) × 107 | <43 (±0.9) | |
| ΔligD1234 | 8.1 (±0.8) × 107 | <44 (±0.0) | |
aValues correspond to the means (± SEM) of the numbers of transformants μg−1 of DNA from three to six independent experiments.
DISCUSSION
Several end-joining mechanisms in S. meliloti
In this work, we provide a comprehensive genetic study of NHEJ in S. meliloti. We show that at least two conventional NHEJ repair systems co-exist in the same bacterial species, a major system dependent on Ku2 and LigD2, and a less efficient system dependent on Ku3 and Ku4, mainly associated to LigD4 (and specifically active in stress conditions). Apart from their different composition and efficiencies, we could not identify any distinctive qualitative repair properties of the two systems, except the higher propensity of the Ku3–4 pathway to generate deletions instead of 1-nucleotide insertions at blunt ended DSB (Supplementary Figure S2). This could reflect that a higher nuclease activity is associated to the Ku3–4 pathway. Although the Ku3–4 pathway appeared to play a minor role in linear plasmid DNA repair, it turned out to substantially contribute to IR resistance in stationary phase bacteria. This may reflect a better capacity to recognize, bind or repair the DNA ends of DSB generated by IR in comparison to those generated by enzymatic digestion. Alternatively, this may indicate that the Ku3-Ku4 pathway is involved in the repair of other IR-generated DNA damages. We did not detect any repair deficiency associated to ligD1 or ku1 deletion, in contrast to previous work (9), which may be explained by the different genetic backgrounds used (Rm1021 versus Rm2011). Although the Ku-LigD genetic interactions uncovered in this study likely reflect protein-protein interactions as described in other organisms, we cannot rule out the hypothesis that binding of Ku proteins on DNA ends simply protects them from exonucleolytic degradation, making possible their subsequent ligation by LigD proteins without physical Ku-LigD interaction. In vitro characterization of protein–protein–DNA interactions will be required to clarify the model.
We observed that in addition to conventional NHEJ, a high fidelity but poorly efficient Ku- and/or LigD-independent A-EJ mechanism is present in S. meliloti, which is reminiscent of the situation previously described in NHEJ mutants of M. smegmatis and in the NHEJ-free E. coli (16,28). It is not clear whether A-EJ repair represents a specific end-joining pathway, or simply the non-specific ligation of free DNA ends by cellular ligases. In E. coli and M. smegmatis, A-EJ involves the essential LigA ligase (16,28). The S. meliloti genome contains a ligA (SMc01878), but also a putative ligB (SMc03177), and three putative ligC (SMa0424, SMb20008 and SMb20912) genes. Further experiments are needed to identify the ligase(s) involved in A-EJ in S. meliloti. Nevertheless, this system contributes very little to DSB repair in S. meliloti.
We observed that Ku2-dependent NHEJ is active in exponential phase, not only for joining plasmid DNA ends, but also for repairing DSBs artificially introduced in the chromosome. This is consistent with previous observation that Ku2 binds DNA ends in exponentially growing bacteria exposed to ionizing radiations (9). NHEJ was also shown to be active in growing Mycobacterium cells (16,17). Nevertheless, as previously described in Mycobacterium (29), inactivation of NHEJ by deletion of the four ku genes did not sensitize S. meliloti to ionizing radiations in exponential phase, whereas inactivation of recA did (9,20). This suggests that when possible (i.e. when a second intact genomic copy is available) RecA-dependent HR is the preferred DSB repair pathway. How HR is preferred over NHEJ for DSB repair in exponential phase is a matter of debate. That NHEJ is inhibited to favor HR has been proposed (30), but we clearly observed that NHEJ is active in exponential phase in S. meliloti.
Stress-inducibility of conventional NHEJ
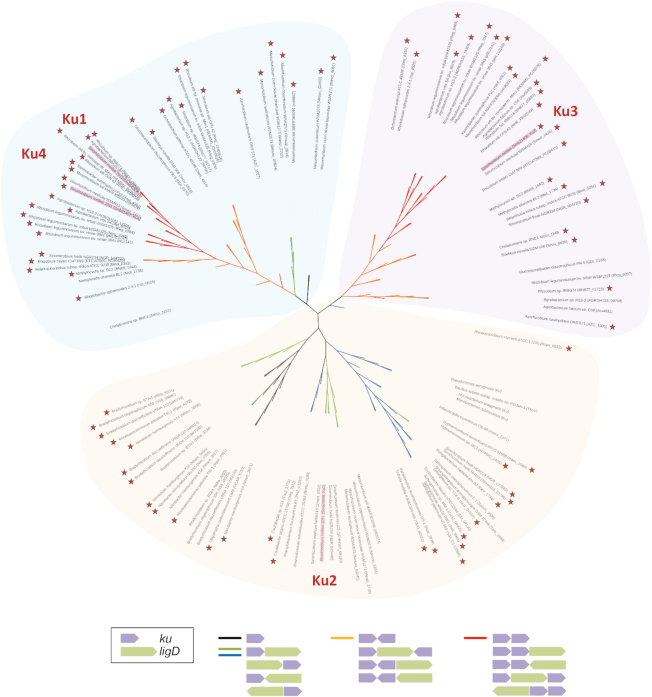
Remarkably, the activity of conventional NHEJ (in contrast to A-EJ) increases under stress conditions. Our study indicates that this activation could result from the up-regulation of ku and/or ligD gene expression. Little is known about the transcriptional regulation of NHEJ genes in bacteria, except in B. subtilis where ku is under the control of the sporulation sigma factor SigG (31). We show that the ku3 ku4 operon and ligD4 gene are part of the general stress regulon of S. meliloti, as their transcription is under control of the EcfG sigma factor RpoE2. EcfG-dependent regulation of ku genes can likely be generalized to many bacterial species, since in most (∼90%) alpha-Proteobacteria containing one or several putative ku genes in their genomes, at least one of them is preceded by putative EcfG binding sequences (Figure 6). In addition, some of these ku genes were found up-regulated in transcriptomic approaches under at least one stress condition (32,33). On the other hand, ku2 is not regulated by RpoE2 but its expression is increased under heat stress (15, this work), and in stationary phase (9,18) and whether ku2 could be induced under other environmental stress conditions remains to be tested. ku2 was not found among the genes under the control of the ‘heat-shock’ sigma factors RpoH1 or RpoH2 (34), or under the control of the RelA-dependent stringent response (35) and ku2 is probably not part of the SOS response, and not regulated by quorum-sensing (9). Finally, ku1 expression was very low and only weakly up-regulated under heat stress, but one cannot exclude that it would be strongly induced in so far untested conditions.
Figure 6.
Phylogenetic analysis of Ku proteins in alpha-Proteobacteria. Non-exhaustive unrooted phylogenic tree of Ku proteins from alpha-Proteobacteria based on the alignment of 111 putative Ku protein sequences retrieved from the NCBI data base (http://www.ncbi.nlm.nih.gov) using a blast analysis with the S. meliloti Ku3 protein. B. subtilis, M. smegmatis, M. tuberculosis and P. aeruginosa Ku proteins were added to the alpha-Proteobacterial Ku sequences. S. meliloti Ku proteins are highlighted in red. The branches of the tree are colored depending on ku and ligD gene organization (black: no other Ku or LigD is encoded in the vicinity of the ku gene; green: a LigD is encoded in the vicinity of the ku gene; Blue: a LigD is encoded in the vicinity of the ku gene, and the Ku protein is the only one encoded by the bacterial species; orange: another Ku is encoded in the vicinity but both ku genes are not organized in operon; red: the ku gene is organized in operon with another ku gene). The presence of putative RpoE2-like boxes in the promoter regions of ku or their putative operons is marked by red stars. Sequence alignment was completed with Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/) and the tree was constructed using the Neighbour Joining method and formatted with the iTOL online tool (http://itol.embl.de).
Stress conditions can lead to the inhibition of new cycles of DNA replication, bacterial growth arrest, and reduction of the genome copy number, as observed in stationary phase (36). Thus, DSB occurring either as a direct consequence of this stress, or later on because of another reason, will not be repaired by HR. Increasing NHEJ activity under stress conditions may therefore be necessary to achieve a sufficient repair efficiency of DSBs in the absence of HR.
Finally, although we clearly establish that NHEJ is transcriptionally regulated we cannot rule out the involvement of additional levels of regulation since M. smegmatis Ku activity has been reported to be regulated through acetylation/deacetylation of certain lysine residues (37,38) and eukaryotic Ku proteins are regulated by post-translational modifications through acetylation, ubiquitination and sumoylation (39–41).
Ku-Ku interactions: a possible bacterial Ku heterodimer?
In eukaryotic cells, Ku functions as a Ku70–Ku80 heterodimer (6). Most bacterial species studied so far (B. subtilis, Mycobacterium sp., Pseudomonas aeruginosa) encode only one Ku protein, which excludes the possibility of heterodimer formation. In S. meliloti, the presence of four putative Ku proteins opens the possibility of multiple interactions. Here, we show that Ku2 is enough to achieve the main NHEJ activity, which suggests that Ku2, if acting as a dimer as in other organisms, does so in a homodimeric form. A similar conclusion was also reached by Walker and coworkers (9) who observed the formation of Ku2-YFP foci in Δku134 bacteria exposed to IR, suggesting that Ku2 was active on its own for binding DNA ends. Strikingly, we also brought the genetic evidence that Ku3 and Ku4 work together in the same pathway. Remarkably, our phylogenetic analysis of Ku proteins in alpha-Proteobacteria reveals that Ku3 homologues always co-exist with a Ku4 homologue in these bacteria (Figure 6); moreover, these genes are most often organized in operons, or located in close proximity, and preceded by putative EcfG binding sites (Figure 6). Therefore, there seems to be a strong selection pressure for the co-maintenance and co-expression of these genes. We propose that in these bacteria, Ku3 and Ku4-like proteins act together as heterodimers, reminiscent of the situation in eukaryotic cells. In vitro biochemical characterization will be required to confirm this prediction.
The conservation of this putative bacterial Ku heterodimer in alpha-Proteobacteria, in spite of the low end-joining efficiency of the associated pathway, suggests that it brings a selective advantage to bacteria. Ku3 and Ku4 belong to different phylogenetic clades (Figure 6), reflecting significant differences between the proteins. In particular, the C-terminal end of Ku3, like that of Ku2, comprises a number of repeated motifs rich in lysine residues, previously described in M. smegmatis and B. subtilis to be important for DNA binding and interaction with LigD (10,42). These motifs are absent from Ku4, which could therefore display different DNA binding properties and/or LigD interactions. This could confer specific properties to the putative Ku3–Ku4 heterodimer. Interestingly, we noticed that the Ku3–4 pathway is comparatively more active on blunt ends than the Ku2 pathway (Figure 2). This could result from a difference in DNA ends recognition or binding. Moreover we showed that several LigD proteins are able to genetically interact with Ku3–4 (see below). This could also result from different interaction properties of Ku3 and Ku4. This may confer to the Ku2 and Ku3–4 systems different abilities to process or ligate DNA ends, the latter system having more flexibility for repairing complex DNA ends, such as those present at DSB generated by physical or chemical agents (43). In eukaryotic cells, Ku70 and Ku80 also present distinct characteristics, including different abilities to interact with various proteins. Finally, the putative Ku3–Ku4 heterodimer could be more stable than the Ku2 homodimer, which would offer a selective advantage during long periods of stress or starvation.
Multiple LigD-Ku interactions?
The main NHEJ pathway is governed by Ku2 and LigD2, which are encoded by neighboring genes. This is reminiscent of the situation previously described in other bacteria, where only one Ku-LigD pair is operational, encoded in the same operon. Note however that although in close proximity, ligD2 and ku2 in S. meliloti are differentially transcribed from independent promoters, as previously noticed (9). Accordingly, we showed here that ku2 transcription is up-regulated by stress, while ligD2 expression is not. Similarly to the Ku proteins, several LigD proteins are encoded by S. meliloti. This opens multiple possibilities of Ku-LigD associations. Here, we showed that only (or mainly) LigD2 works with Ku2. In contrast, our observations suggest that LigD4, LigD2 and LigD3 are able to function with Ku3–4. Nevertheless, we assume that LigD4 is the actual LigD acting with Ku3–4 since: (i) ligD4 expression is under the control of RpoE2, like that of ku3-ku4, (ii) LigD2 associates with Ku3–4 only to repair protruding ends, and this observation was made in a ku2 mutant background, which may not be physiological, iii) LigD3-dependent NHEJ activity is very low. Note that the two main S. meliloti LigD at work in NHEJ (LigD2 and LigD4) possess the canonic nuclease, ligase and polymerase domains found in the model Mycobacterium LigD (see Supplementary Figure S1) while the ‘accessory’ LigDs lack either the polymerase domain (LigD1; encoded by the neighbouring gene), or the nuclease domain (LigD3). This observation strengthens our proposal that LigD2 and LigD4 are the actual NHEJ ligases working with Ku2 and Ku3–4, respectively, and suggests that LigD1 and LigD3 may not be bona fide NHEJ ligases.
NHEJ, an underestimated tool of bacterial genome evolution
NHEJ is inherently unfaithful, as DNA ends of natural DSB often need to be processed before ligation (7,8). But this infidelity poorly contributes to mutagenesis under normal conditions of growth, since faithful HR is the major pathway of DSB repair in bacteria. However, it is well known from studies mainly performed in E. coli that under stress conditions, HR-dependent repair of DSBs can become mutagenic in a process dependent on the general stress response sigma factor RpoS (44). We showed here that in S. meliloti and probably other alpha-Proteobacteria, stress conditions also stimulate NHEJ, in a process partly dependent on the general stress response EcfG sigma factor RpoE2. We therefore assume that NHEJ significantly contributes to DSB-dependent stress-induced mutagenesis in these bacteria. Interestingly, NHEJ was recently suggested to participate in stationary phase mutagenesis in Pseudomonas putida (45).
In addition to the mutagenesis linked to NHEJ infidelity, evidence is provided for the first time here that NHEJ can promote integration of heterologous DNA into bacterial genomic DSBs. This may partially explain how horizontal gene transfer (HGT) occurs between genetically distant species. Indeed, because foreign DNA integration is mediated by HR, HGT is often limited by the level of genomes similarity between the donor and recipient organisms, restricting transfers to closely related species. Nevertheless, many examples of gene transfers between distantly related species have been documented. In a recent in silico analysis of several hundreds of prokaryotic genomes, Popa et al. found that gene transfer events from distant donors are more frequent when the recipient genome encodes NHEJ proteins. From this statement, they proposed that NHEJ could contribute to HGT (27). Our study provides the first molecular evidence that NHEJ can actually promote integration of heterologous DNA in a bacterial genome. Thus, we hypothesize that in addition to the inherent infidelity of NHEJ, its participation in the integration of heterologous DNA is another form of DSB-dependent stress-induced mutagenesis that can contribute to genome evolution and adaptation of bacteria, in particular to stressful environments. In addition, we cannot exclude that NHEJ also participates in the integration at DSB of mobile DNA fragments within species, in so far undescribed accidental or programmed genome rearrangements in bacteria, as described in ciliates (24). Finally, while NHEJ coupled to site directed DNA cleavage (using CRISPR-Cas systems for instance) has found applications in genome engineering by allowing deletion or point mutations at the cleavage site (46–48) our findings suggest that NHEJ could be also used for site-specific integration of DNA in bacterial genomes.
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of the CBR team for helpful discussions, and Ludovic Cottret for help with phylogenetic analyses. We thank Andreas Kaczmarczyk for providing plasmid pQF. We are grateful to Carine Pestourie and the Non-Invasive Exploration Service of the Genotoul-Anexplo Platform (US006/CREFRE INSERM/UPS) for the gamma irradiator.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Département Santé des Plantes et Environnement of INRA; French Laboratory of Excellence project TULIP [ANR-10-LABX-41]. Pierre Dupuy was recipient of a Contrat Jeune Scientifique from INRA. Funding for open access charge: Institut National de la Recherche Agronomique.
Conflict of interest statement. None declared.
REFERENCES
- 1. Bennett C.B., Lewis A.L., Baldwin K.K., Resnick M.A.. Lethality induced by a single site-specific double-strand break in a dispensable yeast plasmid. Proc. Natl. Acad. Sci. U.S.A. 1993; 90:5613–5617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Michel B., Leach D.. Homologous recombination-enzymes and pathways. EcoSal Plus. 2012; 5:doi:10.1128/ecosalplus.7.2.7. [DOI] [PubMed] [Google Scholar]
- 3. Della M., Palmbos P.L., Tseng H.-M., Tonkin L.M., Daley J.M., Topper L.M., Pitcher R.S., Tomkinson A.E., Wilson T.E., Doherty A.J.. Mycobacterial Ku and ligase proteins constitute a two-component NHEJ repair machine. Science. 2004; 306:683–685. [DOI] [PubMed] [Google Scholar]
- 4. Gong C., Martins A., Bongiorno P., Glickman M., Shuman S.. Biochemical and genetic analysis of the four DNA ligases of mycobacteria. J. Biol. Chem. 2004; 279:20594–20606. [DOI] [PubMed] [Google Scholar]
- 5. Weller G.R., Kysela B., Roy R., Tonkin L.M., Scanlan E., Della M., Devine S.K., Day J.P., Wilkinson A., d’Adda di Fagagna F. et al. Identification of a DNA nonhomologous end-joining complex in bacteria. Science. 2002; 297:1686–1689. [DOI] [PubMed] [Google Scholar]
- 6. Fell V.L., Schild-Poulter C.. The Ku heterodimer: function in DNA repair and beyond. Mutat. Res. Rev. Mutat. Res. 2015; 763:15–29. [DOI] [PubMed] [Google Scholar]
- 7. Pitcher R.S., Brissett N.C., Doherty A.J.. Nonhomologous end-joining in bacteria: a microbial perspective. Annu. Rev. Microbiol. 2007; 61:259–282. [DOI] [PubMed] [Google Scholar]
- 8. Shuman S., Glickman M.S.. Bacterial DNA repair by non-homologous end joining. Nat. Rev. Microbiol. 2007; 5:852–861. [DOI] [PubMed] [Google Scholar]
- 9. Kobayashi H., Simmons L.A., Yuan D.S., Broughton W.J., Walker G.C.. Multiple Ku orthologues mediate DNA non-homologous end-joining in the free-living form and during chronic infection of Sinorhizobiummeliloti. Mol. Microbiol. 2008; 67:350–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. McGovern S., Baconnais S., Roblin P., Nicolas P., Drevet P., Simonson H., Piétrement O., Charbonnier J.-B., Le Cam E., Noirot P. et al. C-terminal region of bacterial Ku controls DNA bridging, DNA threading and recruitment of DNA ligase D for double strand breaks repair. Nucleic Acids Res. 2016; 44:4785–4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhu H., Shuman S.. Characterization of Agrobacteriumtumefaciens DNA ligases C and D. Nucleic Acids Res. 2007; 35:3631–3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hoff G., Bertrand C., Zhang L., Piotrowski E., Chipot L., Bontemps C., Confalonieri F., McGovern S., Lecointe F., Thibessard A. et al. Multiple and variable NHEJ-like genes are involved in resistance to DNA damage in Streptomycesambofaciens. Front. Microbiol. 2016; 7:1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ferri L., Gori A., Biondi E.G., Mengoni A., Bazzicalupo M.. Plasmid electroporation of Sinorhizobium strains: The role of the restriction gene hsdR in type strain Rm1021. Plasmid. 2010; 63:128–135. [DOI] [PubMed] [Google Scholar]
- 14. Sauviac L., Bruand C.. A putative bifunctional histidine kinase/phosphatase of the HWE family exerts positive and negative control on the Sinorhizobiummeliloti general stress response. J. Bacteriol. 2014; 196:2526–2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sauviac L., Philippe H., Phok K., Bruand C.. An extracytoplasmic function sigma factor acts as a general stress response regulator in Sinorhizobiummeliloti. J. Bacteriol. 2007; 189:4204–4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Aniukwu J., Glickman M.S., Shuman S.. The pathways and outcomes of mycobacterial NHEJ depend on the structure of the broken DNA ends. Genes Dev. 2008; 22:512–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gong C., Bongiorno P., Martins A., Stephanou N.C., Zhu H., Shuman S., Glickman M.S.. Mechanism of nonhomologous end-joining in mycobacteria: a low-fidelity repair system driven by Ku, ligase D and ligase C. Nat. Struct. Mol. Biol. 2005; 12:304–312. [DOI] [PubMed] [Google Scholar]
- 18. Sallet E., Roux B., Sauviac L., Jardinaud M.-F., Carrère S., Faraut T., de Carvalho-Niebel F., Gouzy J., Gamas P., Capela D. et al. Next-generation annotation of prokaryotic genomes with EuGene-P: application to Sinorhizobiummeliloti 2011. DNA Res. 2013; 20:339–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schlüter J.-P., Reinkensmeier J., Barnett M.J., Lang C., Krol E., Giegerich R., Long S.R., Becker A.. Global mapping of transcription start sites and promoter motifs in the symbiotic α-proteobacterium Sinorhizobiummeliloti 1021. BMC Genomics. 2013; 14:156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dupuy P., Gourion B., Sauviac L., Bruand C.. DNA double-strand break repair is involved in desiccation resistance of Sinorhizobiummeliloti, but is not essential for its symbiotic interaction with Medicagotruncatula. Microbiology. 2017; 163:333–342. [DOI] [PubMed] [Google Scholar]
- 21. Hazkani-Covo E., Covo S.. Numt-mediated double-strand break repair mitigates deletions during primate genome evolution. PLoS Genet. 2008; 4:e1000237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lin Y., Waldman A.S.. Capture of DNA sequences at double-strand breaks in mammalian chromosomes. Genetics. 2001; 158:1665–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lin Y., Waldman A.S.. Promiscuous patching of broken chromosomes in mammalian cells with extrachromosomal DNA. Nucleic Acids Res. 2001; 29:3975–3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Marmignon A., Bischerour J., Silve A., Fojcik C., Dubois E., Arnaiz O., Kapusta A., Malinsky S., Bétermier M.. Ku-mediated coupling of DNA cleavage and repair during programmed genome rearrangements in the ciliate Parameciumtetraurelia. PLoS Genet. 2014; 10:e1004552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Moore J.K., Haber J.E.. Capture of retrotransposon DNA at the sites of chromosomal double-strand breaks. Nature. 1996; 383:644–646. [DOI] [PubMed] [Google Scholar]
- 26. Ricchetti M., Fairhead C., Dujon B.. Mitochondrial DNA repairs double-strand breaks in yeast chromosomes. Nature. 1999; 402:96–100. [DOI] [PubMed] [Google Scholar]
- 27. Popa O., Hazkani-Covo E., Landan G., Martin W., Dagan T.. Directed networks reveal genomic barriers and DNA repair bypasses to lateral gene transfer among prokaryotes. Genome Res. 2011; 21:599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chayot R., Montagne B., Mazel D., Ricchetti M.. An end-joining repair mechanism in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:2141–2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Stephanou N.C., Gao F., Bongiorno P., Ehrt S., Schnappinger D., Shuman S., Glickman M.S.. Mycobacterial nonhomologous end joining mediates mutagenic repair of chromosomal double-strand DNA breaks. J. Bacteriol. 2007; 189:5237–5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gupta R., Barkan D., Redelman-Sidi G., Shuman S., Glickman M.S.. Mycobacteria exploit three genetically distinct DNA double-strand break repair pathways. Mol. Microbiol. 2011; 79:316–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang S.T., Setlow B., Conlon E.M., Lyon J.L., Imamura D., Sato T., Setlow P., Losick R., Eichenberger P.. The forespore line of gene expression in Bacillussubtilis. J. Mol. Biol. 2006; 358:16–37. [DOI] [PubMed] [Google Scholar]
- 32. Alexandre A., Oliveira S.. Response to temperature stress in rhizobia. Crit. Rev. Microbiol. 2013; 39:219–228. [DOI] [PubMed] [Google Scholar]
- 33. Cytryn E.J., Sangurdekar D.P., Streeter J.G., Franck W.L., Chang W.-S., Stacey G., Emerich D.W., Joshi T., Xu D., Sadowsky M.J.. Transcriptional and physiological responses of Bradyrhizobiumjaponicum to desiccation-induced stress. J. Bacteriol. 2007; 189:6751–6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Barnett M.J., Bittner A.N., Toman C.J., Oke V., Long S.R.. Dual RpoH sigma factors and transcriptional plasticity in a symbiotic bacterium. J. Bacteriol. 2012; 194:4983–4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Krol E., Becker A.. ppGpp in Sinorhizobiummeliloti: biosynthesis in response to sudden nutritional downshifts and modulation of the transcriptome. Mol. Microbiol. 2011; 81:1233–1254. [DOI] [PubMed] [Google Scholar]
- 36. Hengge-Aronis R. Recent insights into the general stress response regulatory network in Escherichia coli. J. Mol. Microbiol. Biotechnol. 2002; 4:341–346. [PubMed] [Google Scholar]
- 37. Li Z., Wen J., Lin Y., Wang S., Xue P., Zhang Z., Zhou Y., Wang X., Sui L., Bi L.-J. et al. A Sir2-like protein participates in mycobacterial NHEJ. PLoS One. 2011; 6:e20045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhou Y., Chen T., Zhou L., Fleming J., Deng J., Wang X., Wang L., Wang Y., Zhang X., Wei W. et al. Discovery and characterization of Ku acetylation in Mycobacteriumsmegmatis. FEMS Microbiol. Lett. 2015; 362:fnu051. [DOI] [PubMed] [Google Scholar]
- 39. Hang L.E., Lopez C.R., Liu X., Williams J.M., Chung I., Wei L., Bertuch A.A., Zhao X.. Regulation of Ku-DNA association by Yku70 C-terminal tail and SUMO modification. J. Biol. Chem. 2014; 289:10308–10317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kim K.-B., Kim D.-W., Park J.W., Jeon Y.-J., Kim D., Rhee S., Chae J.-I., Seo S.-B.. Inhibition of Ku70 acetylation by INHAT subunit SET/TAF-Iβ regulates Ku70-mediated DNA damage response. Cell. Mol. Life Sci. CMLS. 2014; 71:2731–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Postow L., Ghenoiu C., Woo E.M., Krutchinsky A.N., Chait B.T., Funabiki H.. Ku80 removal from DNA through double strand break-induced ubiquitylation. J. Cell Biol. 2008; 182:467–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kushwaha A.K., Grove A.. C-terminal low-complexity sequence repeats of Mycobacteriumsmegmatis Ku modulate DNA binding. Biosci. Rep. 2013; 33:175–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schipler A., Iliakis G.. DNA double-strand-break complexity levels and their possible contributions to the probability for error-prone processing and repair pathway choice. Nucleic Acids Res. 2013; 41:7589–7605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shee C., Gibson J.L., Darrow M.C., Gonzalez C., Rosenberg S.M.. Impact of a stress-inducible switch to mutagenic repair of DNA breaks on mutation in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:13659–13664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Paris Ü., Mikkel K., Tavita K., Saumaa S., Teras R., Kivisaar M.. NHEJ enzymes LigD and Ku participate in stationary-phase mutagenesis in Pseudomonasputida. DNA Repair (Amst.). 2015; 31:11–18. [DOI] [PubMed] [Google Scholar]
- 46. Su T., Liu F., Gu P., Jin H., Chang Y., Wang Q., Liang Q., Qi Q.. A CRISPR-Cas9 assisted non-homologous end-joining strategy for one-step engineering of bacterial genome. Sci. Rep. 2016; 6:37895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tong Y., Charusanti P., Zhang L., Weber T., Lee S.Y.. CRISPR-Cas9 based engineering of actinomycetal genomes. ACS Synth. Biol. 2015; 4:1020–1029. [DOI] [PubMed] [Google Scholar]
- 48. Zheng X., Li S.-Y., Zhao G.-P., Wang J.. An efficient system for deletion of large DNA fragments in Escherichia coli via introduction of both Cas9 and the non-homologous end joining system from Mycobacteriumsmegmatis. Biochem. Biophys. Res. Commun. 2017; 485:768–774. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.