Abstract
Sarcocystis spp. are protozoan parasites with an obligatory two-host life cycle, with herbivores as intermediate hosts and carnivores as definitive hosts. Cattle are intermediate hosts for several species of Sarcocystis: indeed, in addition to S. cruzi, S. hirsuta and S. hominis, at least four new species were recently identified in bovine muscle: S. bovifelis, S. rommeli, S. bovini and S.heydorni. Since is not possible to unambiguously discriminate between S. hominis and the new species either morphologically or by the analysis of the 18S ribosomial (rRNA) gene, the aim of the present study was to use molecular techniques to discriminate cattle Sarcocystis species, taking advantage of the higher discriminative power of the Cytochrome C Oxidase subunit I mitochondrial (mtDNA COI) gene. Therefore, 119 bovine muscle samples were tested to identify S. hominis-like sarcocystis using a multiplex PCR of the 18S rRNA gene; later, positive samples were tested using a newly designed primer set for the PCR amplification of COI gene. Species identification was achieved by sequencing the amplified products: 16 sequences were confirmed to belong to S. bovifelis, while 12 sequences didn’t constitute the best BLAST match of any of the published sequences, allowing to speculate the possible presence of S. hominis. This study confirms the higher discriminatory power of COI mitochondrial gene; besides, our work provides the first report of S. bovifelis in Italy.
Key words: Sarcocystis, COI gene, PCR, Cattle
Introduction
Sarcocystosis is a parasitic disease distributed worldwide, whose causative agents are protozoan parasites belonging to the phylum Apicomplexa. Sarcocystis spp. have an obligatory two-host life cycle: in the intermediate host occurs the asexual cycle with cyst formation, while the sexual cycle occurs in the definitive host (Dubey, 2015).
Definitive hosts, comprising carnivores and omnivores as well as humans, usually become infected by ingesting the sarcocysts in muscles, while intermediate hosts become infected through the ingestion of oocysts and sporocysts in feed or water contaminated by infected feces (Dubey, 2015).
Cattle are intermediate hosts for different species of Sarcocystis, among which S. hominis and S. heydorni are considered zoonotic; besides, Sarcocystis species have been suggested to be associated with bovine eosinophilic myositis, an inflammatory myopathy characterized by multifocal greygreen lesions in striated muscle which result in considerable economic losses (Vangeel et al., 2012).
Among cattle Sarcocystis, full life cycles are known only for S. cruzi, S. hirsuta and S.hominis. Indeed, during the last three years, five new Sarcocystis species have been named in cattle: S. bovifelis, S. sinensis (actually considered a water buffaloes species), S. rommeli, S. bovini and S.heydorni, leading to considerable confusion about the identification of cattle Sarcocystis because of incomplete descriptions and lack of morphological and molecular information. The timeline in Figure 1 reports the first evidence in scientific literature of cattle species known to date.
Figure 1.
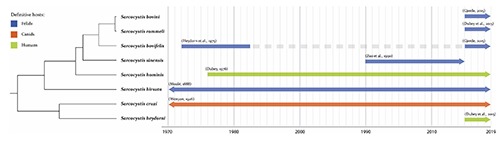
First evidence in scientific literature of cattle species, definitive host of the species and evolutionary history inferred by unweighted pair group method with arithmetic mean (UPGMA), based on COI partial sequences (850 bp).
As reported above, morphological diagnosis of cattle Sarcocystis is difficult as some of them share sarcocyst morphologies (Moré et al., 2014; Gjerde, 2016); for example, sarcocysts of S. bovifelis are morphologically indiscriminable from S. sinensis in water buffaloes and from the other S. sinensis- like species reported in cattle in recent years (S. bovini and S. rommeli), and they can’t either be discriminated by S. hominis sarcocysts using light microscopy, while just some differences can be identified by transmission electron microscopy (TEM) (Moré et al., 2014; Gjerde, 2016).
Molecular methods, particularly sequencing of 18S ribosomial RNA (rRNA), have been increasingly used to complement morphological data in the identification of Sarcocystis species. However, different methods and DNA regions used to compare the same taxonomic groups can lead to conflicting results.
For example, 18S rRNA gene is well suited for discrimination between more distant species but less for closely related species, where very few molecular differences are observed; on the other hand, in the last decade Cytochrome C Oxidase subunit I mitochondrial gene (mtDNA COI) has been chosen as the standard barcoding marker due to its high interspecific variation, low intraspecific variation (Hebert et al., 2005). The COI gene has already proved to be a useful genetic marker for the Sarcocystidae in ruminants (Gjerde, 2013), and it is better suited than the nuclear 18S rRNA gene to differentiate between closely related Sarcocystis spp., such as the mentioned S. sinensis-like species of cattle and S. hominis (Gjerde, 2016).
As they can’t be unambiguously discriminated by the analysis of the 18S rRNA gene, referring to S. hominis, S. bovifelis, S. bovini and S. rommeli we will use the term “Sarcocystis hominis-like” from now on, not including S. sinensis as it is now no longer considered a cattle species (Dubey et al., 2014).
The aim of the present study was to use molecular techniques to discriminate cattle Sarcocystis species, taking advantage of the higher discriminative power of the COI gene to discriminate S. hominis-like sarcocysts, considering the recent taxonomic revision of cattle Sarcocystis.
Materials and Methods
Samples collection and DNA extraction
A convenience sampling of 119 bovine carcasses was carried out over a 7 years’ time span (2011-2018); 25 gr sample of striated muscle were collected directly from different slaughterhouses in Piedmont region (North-West Italy) and stored at - 20°C. DNA extraction was performed using DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany), according to manufacturer’s tissue protocol; the lysis step was carried out at 56°C overnight with Proteinase K. DNA samples were kept frozen at -20°C.
PCR amplification of 18S sequences
The DNA samples obtained were initially tested to discriminate S. cruzi, S. hirsuta and S. hominis-like using the multiplex PCR protocol described by Chiesa et al. (2013). The samples producing amplicons of 180 bp, corresponding to the S. hominislike species, were subsequently tested using the primers developed for the amplification of the mitochondrial COI gene (see below).
PCR amplification of COI sequences
Primer design
As previously reported, S. hominis, S. bovifelis, S. rommeli and S. bovini can only be distinguished using the mitochondrial COI gene; therefore, our initial aim was to design a set of primer that would amplify a COI gene fragment from cattle Sarcocystis. To achieve this, we aligned the barcode region of COI from Sarcocystis partial COI sequences available from Genbank to identify conserved regions suitable for primer designing (Table 1).
Table 1.
COI sequences accession numbers in Genbank used for primers design.
| Species | Genbank accession number |
|---|---|
| S. cruzi | KT901095.1, KC209600.1, LC171861.1 |
| S. hirsuta | KT901077.1, KC209634.1, KT901023.1 |
| S. rommeli | KY120286.1, KY120291.1 |
| S. bovini | KT901022.1, KT901288.1, KT900999.1 |
| S. bovifelis | KT901287.1, KC209696.1, KT900997.1 |
| S. heydorni | KX057995.1, KX057994.1 |
| Toxoplasma gondii | JX473253.1, AF023246.1 |
Sequences of S. cruzi, S. hirsuta, S. rommeli, S. bovini, S. bovifelis, and S. heydorni were aligned using MEGA Software. Besides, the sequences of another member of the phylum Apicomplexa (Toxoplasma gondii, GenBank accession no. KT363924.1) were included in the alignment to evaluate possible cross reactions.
Based on these sequences, we designed different sets of primers with Primer3Plus software (http://www.bioinformatics.nl/cgibin/primer3plus/primer3plus.cgi); specificity of the primers for the Sarcocystis genus was examined in-silico using Primer- BLAST (http://www.ncbi.nlm.nih.gov/tools/primer-blast). The sequences of the primers pair, selected for specificity and amplicon parameters, are as follows:
i) forward COI primer: 5′-TGTACATACTTACGGCAGGT- 3’ corresponding to positions 11-30 of S. bovini sequence (GenBank accession no. KT901022.1); ii) reverse COI primer: 5′-CCGTAGGTATGGCGATCAT- 3′, matching positions 895-913 of S. bovini sequence (GenBank accession no. KT901022.1).
The primers were synthesized by Sigma Aldrich (St. Louis, MO).
Optimization of the PCR amplification conditions
The new PCR protocol was therefore optimized in order to obtain the expected amplicon. The final PCR mixture contained 2.5 μl of template DNA (5-20 ng/μL), 0,5 mM of each primer, 2.5 mM MgCl2, 0.2 mM of each dNTP, 1 U recombinant Taq DNA polymerase, 10 × PCR Buffer and distilled water to a total volume of 25 μL.
The final thermal cycle conditions used were: initial denaturation of 94° for 5 min; 35 cycles of 95° for 1 min, 60° for 45 s and 72° for 30 s, and final extension of 72° for 5 min.
PCR amplification
PCR amplification of COI sequences was performed on the samples previously resulted positive for S. hominis-like species after the amplification of the 18S rRNA gene (Chiesa et al. 2013).
PCR products were observed in 2% agarose gel stained with SYBR safe stain (Invitrogen; Groningen, The Netherlands) and observed in a blue light transilluminator (Invitrogen; Groningen, The Netherlands).
Sanger sequencing
Species identification was achieved by sequencing the amplified products: they were purified with Exo-Sap (USB Europe, Staufen, Germany) treatment according to the manufacturer’s recommendations. Forward and reverse sequencing reactions were performed using ABI Prism BigDye Terminator Cycle Sequencing Ready Reaction Kit, version 1.1 (Applied Biosystems, Foster City, CA). Sequenced fragments were purified by DyeEX (Qiagen, Hilden, Germany) and resolved by capillary electrophoresis using an ABI 310 Genetic Analyser (Applied Biosystems, Foster City, CA). The nucleotide sequences were analyzed using the BLASTN sequence similarity search at the NCBI database (Altschul et al., 1990).
Average evolutionary divergence
Finally, the Average Evolutionary Divergence over all Sequence Pairs of cattle Sarcocystis, based on the 18S rRNA gene and on the COI gene was estimated by MEGA 7.0 software (Kumar et al., 2016).
Results
The amplification of the 18S rRNA gene resulted in bands of ~180 bp in 30 samples, corresponding to S. hominis-like. After amplification with the new protocol, COI amplicons of the expected size (~850bp) were produced in all the samples (Figure 2).
Figure 2.
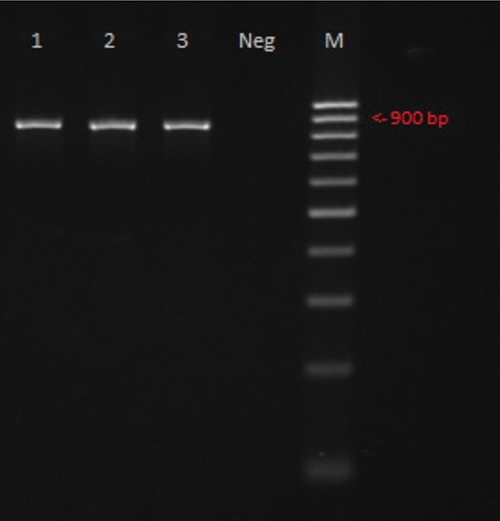
Amplification result of the Sarcocystis COI gene. M = 100 bp ladder; 1,2,3= positive samples; Neg = negative control.
The sequencing of the amplified products resulted in 28 readable sequences; among those, 16 showed 100% identity with published sequences from S. bovifelis (accession number KT900992.1) while 12 showed a homology of 89% to S. bovifelis (accession number KT900992.1) (Figure 3) and of 88% to S. bovini (accession number KT901021.1). Two samples did not result in readable sequences.
Figure 3.
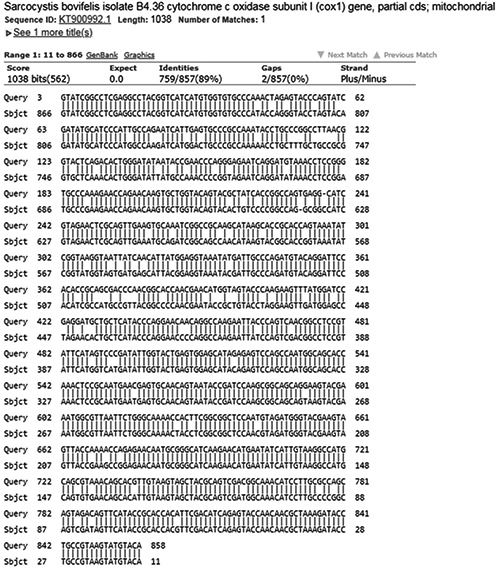
The comparison of one of the twelve nucleotide sequences of the Sarcocystis COI gene with NCBI information (89% homology with sequences of S. bovifelis).
The Average Evolutionary Divergence over all Sequence Pairs of the 7 cattle species, based on the 18S rRNA gene, was 0.039 (Table 2), while the Average Evolutionary Divergence based on the COI gene was 0.226 (Table 3).
Table 2.
The number of base substitutions per site from averaging over all sequence pairs are shown, based on the 18S rRNA gene.
| Genbank ID | Species | 1 | 2 | 2 | 4 | 5 | 6 | 7 | |
|---|---|---|---|---|---|---|---|---|---|
| KT901021.1 | S. bovini | 1 | |||||||
| KY120285.1 | S. rommeli | 2 | 0,01 | ||||||
| KT900992.1 | S. bovifelis | 3 | 0,007 | 0,014 | |||||
| KF954731.1 | S. hominis | 4 | 0,025 | 0,028 | 0,025 | ||||
| KT901158.1 | S. hirsuta | 5 | 0,039 | 0,043 | 0,041 | 0,041 | |||
| KX057997.1 | S. heydorni | 6 | 0,052 | 0,049 | 0,052 | 0,055 | 0,062 | ||
| KT901167.1 | S. cruzi | 7 | 0,048 | 0,051 | 0,046 | 0,05 | 0,059 | 0,024 |
Table 3.
The number of base substitutions per site from averaging over all sequence pairs are shown, based on the COI gene.
| Genbank ID | Species | 1 | 2 | 2 | 4 | 5 | 6 | 7 | |
|---|---|---|---|---|---|---|---|---|---|
| KT901021.1 | S. bovini | 1 | |||||||
| KY120292.1 | S. rommeli | 2 | 0,004 | ||||||
| KT900992.1 | S. bovifelis | 3 | 0,059 | 0,057 | |||||
| This study | Presuntive S. hominis | 4 | 0,134 | 0,132 | 0,129 | ||||
| KT901077.1 | S. hirsuta | 5 | 0,238 | 0,237 | 0,24 | 0,228 | |||
| KT901095.1 | S. cruzi | 6 | 0,313 | 0,315 | 0,337 | 0,333 | 0,35 | ||
| KX057995.1 | S. heydorni | 7 | 0,306 | 0,304 | 0,325 | 0,339 | 0,368 | 0,19 |
Discussion and Conclusions
DNA sequencing have been successfully applied in the classification of foodborne pathogens, improving diagnostic methods and phylogenetic analyses. Sarcocystis spp. is not a front-page parasite, but can lead to serious economic outcomes in the beef sector (Chiesa et al., 2013). The recent rise of new species in cattle posed a new challenge for parasitologists and food safety scholars, as applying different methods and analyzing different genes to compare the same taxonomic groups can often lead to conflicting results (Gjerde, 2016).
Until recently, molecular species identification of various Sarcocystis spp. have been based almost exclusively upon sequences of the nuclear rDNA unit, particularly the 18S rRNA. However, the mitochondrial COI gene appears to have a better phylogenetic signal than the other more used genes: as previously described by some authors, the evolution of the COI gene looks like to be sufficiently rapid to discriminate between closely related species and investigate intraspecific diversity (Hebert et al., 2003).
The use of COI gene for identifying protozoa is still in its initial stage, but has already proven to be a useful genetic marker for the Sarcocystidae family (Gjerde, 2013, 2016), leading to the discrimination of closely-related species previously impossible to differentiate, such as S. bovifelis, S. bovini, S. rommeli and S. sinensis, and to the definition of new Sarcocystis species (Gjerde, 2016; Hu et al., 2016, 2017).
Moreover, the discrimination of Sarcocystis species, and, of particular interest for this study, cattle Sarcocystis species, assumes a much more important role because humans can act as definitive host for two cattle sarcosporidian species, S. hominis and S. heydorni, with a resulting zoonotic risk for consumers of raw or undercooked meat (Bucca et al., 2011).
The high presence of S. hominis in North-West Italian cattle, tested with molecular methods based on the analysis of 18S rRNA gene, has previously brought into question the zoonotic characteristics of this species (Chiesa et al., 2013); besides, further doubts are due to the absence of reports, by the network of surveillance for foodborne diseases, of cases referable to sarcocystosis in Piedmont, a North-West Italian region characterized by a high consumption of raw beef. This has been explained by the low pathogenicity of S. hominis and the possibility that the infection goes unnoticed, as the symptoms reported in literature are common to most foodborne pathogens (Chiesa et al., 2013). On the top of that it can now be numbered the overestimation of the prevalence of S. hominis, likely misidentified in consequence of the low discriminative power of 18S rRNA gene, which was, until recently, the standard target gene for molecular diagnosis of cattle sarcocystosis.
As clarified by the different estimates of Average Evolutionary Divergence based on the 18S rRNA gene and on the COI gene, this study confirm the higher discriminatory power of COI mitochondrial gene, because of the greater number of bases substitutions per site between sequences of different cattle Sarcocystis species: considering the recent taxonomic confusion, the identification of a gene which can be used as the reference gene for the unambiguous identification of Sarcocystis species which are phylogenetically very close is of great importance.
Besides, this study demonstrates the presence of S. bovifelis in italian cattle: this is, to our knowledge, the first report of S. bovifelis in Italy and thus the role of domestic felids in the cattle sarcosporidiosis will need to be reconsidered. We couldn’t achieve an unambiguous identification of 12 sequences: these were all identical and did not constitute the best BLAST match of any of the published sequences, with the best hit being the sequences of S. bovifelis with 89% of homology. In this regard, an explanation could be found in the current absence of mtDNA COI gene sequences for S. hominis in Genbank : our hypothesis is that the 12 sequences correspond to the partial COI gene of S. hominis. The estimated phylogenetic distance between our sequences and the other cattle Sarcocystis sequences is consistent with this hypothesis; further studies are ongoing to confirm it. Finally, it wasn’t possible to obtain readable sequences from two samples, probably because of the presence of coinfections in bovine muscles (Chiesa et al., 2013), which could result in the overlapping between two or more sequences.
The molecular characterization of the COI gene of S. hominis should be considered of primary importance for the taxonomy clarification of cattle Sarcocystis species, allowing an appropriate risk assessment of public health issues arising from consumption of contaminated beef and providing new insights on the role of Sarcocystis species in the etiology of the eosinophilic myosistis in cattle.
Funding Statement
Funding: none.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ, 1990. Basic local alignment search tool. J Mol Biol 215:403-10. [DOI] [PubMed] [Google Scholar]
- Bucca M, Brianti E, Giuffrida A, Ziino G, Cicciari S, Panebianco A, 2011. Prevalence and distribution of Sarcocystis spp. cysts in several muscles of cattle slaughtered in Sicily, Southern Italy. Food Control 22:105-8. [Google Scholar]
- Chiesa F, Muratore E, Dalmasso A, Civera T, 2013. A new molecular approach to assess the occurrence of Sarcocystis spp. in cattle and products thereof: preliminary data. Ital J Food Saf 2:41. [Google Scholar]
- Dubey JP, 2015. Foodborne and waterborne zoonotic sarcocystosis. Food Water Parasitol 1:2-11. [Google Scholar]
- Dubey JP, Fayer R, Rosenthal BM, Calero-Bernal R, Uggla A, 2014. Identity of Sarcocystis species of the water buffalo (Bubalus bubalis) and cattle (Bos taurus) and the suppression of Sarcocystis sinensis as a nomen nudum. Vet Parasitol 205:1-6. [DOI] [PubMed] [Google Scholar]
- Gjerde B, 2013. Phylogenetic relationships among Sarcocystis species in cervids, cattle and sheep inferred from the mitochondrial cytochrome c oxidase subunit I gene. Int J Parasitol 43:579-91. [DOI] [PubMed] [Google Scholar]
- Gjerde B, 2016. Molecular characterisation of Sarcocystis bovifelis, Sarcocystis bovini n. sp., Sarcocystis hirsuta and Sarcocystis cruzi from cattle (Bos taurus) and Sarcocystis sinensis from water buffaloes (Bubalus bubalis). Parasitol Res 115:1473-92. [DOI] [PubMed] [Google Scholar]
- Hebert PDN, Cywinska A, Ball SL, deWaard JR, 2003. Biological identifications through DNA barcodes. P Roy Soc Lond B Bio 270:313-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert PDN, Gregory TR, Savolainen V, 2005. The Promise of DNA Barcoding for Taxonomy. Syst Biol 54:852-9. [DOI] [PubMed] [Google Scholar]
- Hu JJ, Huang S, Wen T, Esch GW, Liang Y, Li HL, 2017. Morphology, Molecular Characteristics, and Demonstration of a Definitive Host for Sarcocystis rommeli from Cattle (Bos taurus) in China. J Parasitol 103:471-6. [DOI] [PubMed] [Google Scholar]
- Hu JJ, Wen T, Chen XW, Liu TT, Esch GW, Huang S, 2016. Prevalance, Morphology, and Molecular Characterization of Sarcocystis heydorni Sarcocysts from Cattle (Bos Taurus) in China. J Parasitol 102:545-8. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K, 2016. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol 33:1870-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moré G, Pantchev A, Skuballa J, Langenmayer MC, Maksimov P, Conraths FJ, Venturini MC, Schares G, 2014. Sarcocystis sinensis is the most prevalent thick-walled Sarcocystis species in beef on sale for consumers in Germany. Parasitol Res 113:2223-30. [DOI] [PubMed] [Google Scholar]
- Vangeel L, Houf K, Geldhof P, Nollet H, Vercruysse J, Ducatelle R, Chiers K, 2012. Intramuscular inoculation of cattle with Sarcocystis antigen results in focal eosinophilic myositis. Vet Parasitol [DOI] [PubMed] [Google Scholar]


