ABSTRACT
Viroids are small infectious, non-protein-coding circular RNAs that replicate independently and, in some cases, incite diseases in plants. They are classified into two families: Pospiviroidae, composed of species that have a central conserved region (CCR) and replicate in the cell nucleus, and Avsunviroidae, containing species that lack a CCR and whose multimeric replicative intermediates of either polarity generated in plastids self-cleave through hammerhead ribozymes. The compact, rod-like or branched, secondary structures of viroid RNAs have been predicted by RNA folding algorithms and further examined using different in vitro and in vivo experimental techniques. However, direct data about their native tertiary structure remain scarce. Here we have applied atomic force microscopy (AFM) to image at single-molecule resolution different variant RNAs of three representative viroids: potato spindle tuber viroid (PSTVd, family Pospiviroidae), peach latent mosaic viroid and eggplant latent viroid (PLMVd and ELVd, family Avsunviroidae). Our results provide a direct visualization of their native, three-dimensional conformations at 0 and 4 mM Mg2+ and highlight the role that some elements of tertiary structure play in their stabilization. The AFM images show that addition of 4 mM Mg2+ to the folding buffer results in a size contraction in PSTVd and ELVd, as well as in PLMVd when the kissing-loop interaction that stabilizes its 3D structure is preserved.
KEYWORDS: Viroids, RNA structure, RNA structural/functional elements, kissing-loop interactions, ribozymes, atomic force microscopy, single-molecule approaches
Introduction
Viroids are the smallest known nucleic acid-based infectious agents, with their genomes consisting of a single-stranded (ss), circular, non-protein-coding RNA ranging in size from ~250 to 430 nucleotides (nt) [1–5]. Viroids replicate and invade systemically some host plants, often causing disease by altering pathways that mediate gene expression and development [6–10]. Viroid replication takes place in the nuclei (family Pospiviroidae) or in plastids, mostly chloroplasts (family Avsunviroidae), and proceeds through an entirely RNA-based rolling-circle mechanism [11–16]. In the best supported model [17] the infecting, most abundant monomeric circular (mc) strand, to which the (+) polarity is assigned by convention, is reiteratively transcribed by either the nuclear RNA polymerase II or a nuclear-encoded chloroplastic RNA polymerase (both redirected to accept RNA templates), thus producing multimeric (-) strands. In the family Pospiviroidae the latter serve directly as templates for generating multimeric (+) strands that are cleaved, within a central conserved region (CCR), into unit-length monomeric linear (ml) forms by an RNase of class III, and then circularized by DNA ligase 1 (redirected to accept RNA substrates). In the family Avsunviroidae the multimeric (-) strands are first cleaved by embedded hammerhead ribozymes, with a chloroplastic isoform of tRNA ligase catalyzing circularization of the resulting ml (-) into the mc (-) forms which, in turn, prime the second half of the replication cycle that is symmetric to the first one. Due to the presence of ribozymes, along with other properties (see below), viroids have been proposed to be relics of the RNA world [17,18] in the context of the origin and early evolution of life [19].
Data derived from different approaches support the notion that viroid RNA genomes are largely self-complementary, folding into highly-compact rod-shaped or branched secondary structures composed of double-stranded (ds) RNA stretches flanked by internal loops and bulges that are often stabilized by non-canonical interactions [20,21]. Potato spindle tuber viroid (PSTVd, 359 nt), the type member of the family Pospiviroidae [22,23], adopts a predicted rod-like secondary structure in silico, in vitro and in vivo [23–26]. Its terminal and internal loops, as well as the bulges, are critical for replication and systemic trafficking throughout the infected plant [27–30]. In contrast, members of the family Avsunviroidae, like peach latent mosaic viroid (PLMVd, 337 nt) [31] and eggplant latent viroid (ELVd, 335 nt) [32], adopt either multibranched or bifurcated secondary structures, respectively. Moreover, in PLMVd and the other member of the genus Pelamoviroid [33], the (+) strands are stabilized by a kissing-loop interaction required for in vitro folding and in vivo viability [34–36].
However, when it comes to single-molecule approaches, data on the three-dimensional (3D) native (or denatured) structures of viroid RNA genomes are very scarce. More specifically, transmission electron microscopy (TEM) has revealed that under non-denaturing conditions PSTVd adopts a ~ 50 nm long, rod-like secondary structure resulting from the high self-complementarity of the viroid RNA [37,38]. Under mild denaturing conditions, besides the ‘double-stranded’ rods, partially open molecules resembling ‘tennis rackets’ as well as completely denatured linear and circular single-stranded RNAs were observed [39,40].
Atomic force microscopy (AFM) is a type of scanning probe microscopy that allows structural and dynamic studies of single macromolecules at nanometer resolution [41,42]. One of the main advantages of AFM over electron microscopy-based techniques is that it offers a 3D surface profile of the imaged sample without requiring any staining or coating, thus minimizing structural disruption of the biological entity under study. AFM permits visualization and manipulation across length scales that range from biomolecules to cells. Therefore, this technology is increasingly used in different fields, including virology [43–45]. In particular, AFM has matured to provide nanometer spatial resolution of RNA molecules of different lengths and structures, as well as of RNA-RNA or RNA-protein complexes [46–49]. Within this framework, and based on our previous experience in imaging by AFM structured and functional RNA molecules of viral origin [50], here we have conducted a high resolution structural analysis of the three viroids mentioned above (PSTVd, PLMVd and ELVd), using different variants thereof and experimental conditions. This analysis has allowed the first AFM visualization of the native structures of single viroid RNA molecules, from which functional implications can be derived.
Results
AFM analysis of viroid structure
To investigate the native 3D structure at single-molecule resolution of representative viroid RNAs of the families Pospiviroidae (PSTVd) and Avsunviroidae (PLMVd and ELVd), a systematic and comparative AFM analysis was performed. RNA adsorption on mica surfaces was performed using 3-aminopropyltriethoxysilane (APTES), one of the currently available reagents that promotes a tight adhesion of RNA molecules via electrostatic interactions without damaging or disrupting their native structure [45,49,50]. Other reported routes for the surface binding of RNA, such as those comprising the addition of salts containing divalent cations (e.g. Mg2+, Zn2+ or Ni2+) [45,51,52], are not appropriate for this study given our interest in examining the effect exerted by a divalent cation (Mg2+) on viroid 2D and, particularly, 3D RNA structure.
After RNA thermal denaturation followed by renaturation in the folding buffer lacking Mg2+ or in some instances containing 4 mM Mg2+, at least three independent samples of each of the nine viroid RNA preparations (Table 1) were imaged by AFM. Besides the analysis of the shape and main structural features of the imaged viroid RNAs, the average length of 25 individual, full-length molecules from each viroid preparation was computed.
Table 1.
Main features of the viroid RNAs imaged by AFM in this work.
| Family | Viroid | Length (nt) | Variant RNA analyzed (GenBank acc no.) |
Features | [Mg2+] in the folding buffer* |
|---|---|---|---|---|---|
| Pospiviroidae | PSTVd | 359 |
PSTVd-ml(+) (U23058.1) |
ml (+) RNA, in vitro transcript (IVT) | 0 |
| 4 | |||||
|
PSTVd-mc(+) (AJ634596.1) |
mc (+) RNA, isolated from infected tissue | 4 | |||
| Avsunviroidae | PLMVd | 337 |
PLMVd-ml(+)wt (AJ005303.1) |
ml (+) RNA (wild type), IVT | 0 |
| 4 | |||||
| PLMVd-ml(+)mut | ml (+) RNA (mutant with kissing-loop disrupted), IVT | 4 | |||
| ELVd | 335 |
ELVd-ml(+) (AJ536613) |
ml (+) RNA, IVT | 0 | |
| 4 | |||||
|
ELVd-ml(-) (AJ536613) |
ml (-) RNA, IVT | 4 |
*The folding buffer is composed of 100 mM HEPES pH 7.4 and 100 mM NaCl, either lacking Mg2+ or supplemented with 4 mM MgCl2.
Potato spindle tuber viroid (family Pospiviroidae): a rod- or quasi-rod-shaped structure imaged as consecutive bumps
PSTVd is a 359 nt-long RNA with a theoretical length of approximately 49 nm (assuming a uniform A-dsRNA structure with a pitch of 3.0 nm and 11 bp/turn: 180 bp x 0.27 nm/bp = 48.6 nm) [48,53], resulting in a proposed rod-like secondary structure (Fig. 1A). Two RNAs of this viroid were imaged (Table 1): PSTVd-ml(+), the ml (+) RNA transcribed in vitro (linearized between positions C1 and G2), and PSTVd-mc(+), the mc (+) RNA isolated from infected plant leaves. The first RNA was renatured in the folding buffer lacking Mg2+ or containing 4 mM MgCl2, whereas the second one was only imaged in 4 mM Mg2+-containing buffer. Fig. 1B shows representative 2D and 3D AFM images in ambient conditions of PSTVd-ml(+) renatured in buffer without Mg2+, and Suppl. Fig. S1 depicts 25 single-molecule 2D images together with the measured length of each of them. Their topographic profiles reflect a majority of rod- or quasi-rod-like molecules composed of three consecutive, elongated bumps displaying different angular orientations (with one of the bumps generally forming an angle of 90° to 180° with respect to the other two). The bumps might correspond to dsRNA segments flanked by internal loops or bulges; although more than three bumps should be expected according with the number of internal loops or bulges predicted in vivo [26], some of them (in particular, the symmetrical or quasi-symmetrical internal loops) might not induce a pronounced bending detectable by AFM.
Figure 1.
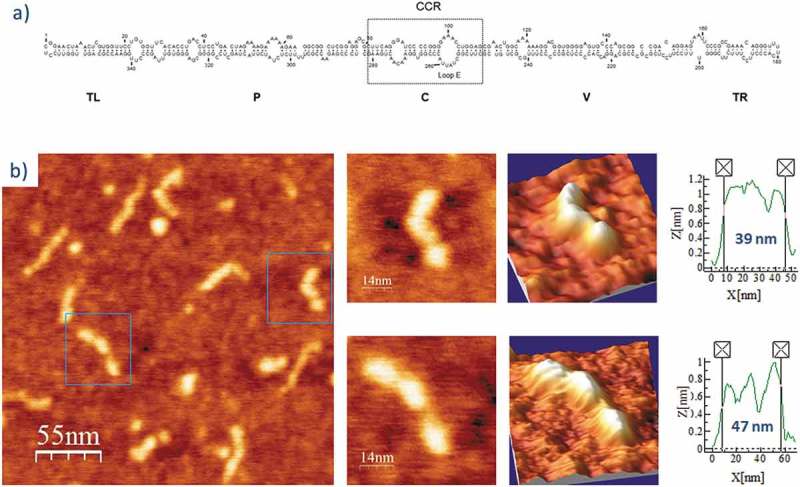
(A) Predicted secondary structure of PSTVd-ml(+). The 359 nt-long, rod-like secondary structure of PSTVd shows the five domains characteristic of members of the family Pospiviroidae: Terminal Left (TL), Pathogenic (P), Central (C), Variable (V), and Terminal Right (TR) [77]. The Central Conserved Region (CCR) is located within the C domain and contains an UV-sensitive loop E motif stabilized by non-canonical base-pairs. Adapted from [26]. (B) AFM images of PSTVd-ml(+), renatured in the absence of Mg2+. A field of 275 × 275 nm is shown on the left panel and two characteristic molecules are zoomed on the right one. 3D views of the imaged viroid RNAs, as well as profiles with their measured lengths (X [nm]) and heights [Z [nm]), are also displayed.
The average length of PSTVd-ml(+), calculated after measuring 25 unbiasedly chosen, full-length viroid molecules present in different preparations (Suppl. Fig. S1), was 43 nm (Table 2), being each individual bump 10 to 18 nm long and 0.6 to 1 nm high. In turn, PSTVd-ml(+) renatured in the buffer containing 4 mM Mg2+ showed a topologically analogous rod-like structure (Fig. 2 and Suppl. Fig. S2), but its average length was reduced to 35 nm (18.6% shorter than that of the same viroid RNA in the buffer lacking Mg2+, a statistically significant length reduction as revealed by an ANOVA test, see Table 2), and the internal bumps appeared less clear, thus indicating a Mg2+-induced molecular compaction.
Table 2.
Summary of the average molecular lengths of individual viroid RNAs (n = 25 in all cases) imaged by AFM (shown in Suppl. Figs S1 to S9).
| Viroid RNA variant |
[Mg2+] in the folding buffer | Measured length (nm) |
ANOVA test * |
||
|---|---|---|---|---|---|
| Mean ± SD | Median | F | F crit | ||
| PSTVd-ml(+) | 0 | 43 ± 6* | 42.0 | 15.85 | 3.12 |
| 4 | 35 ± 6 | 35.0 | |||
| PSTVd-mc(+) | 4 | 35 ± 6 | 34.7 | ||
| PLMVd-ml(+)wt | 0 | 34 ± 3 | 34.3 | 63.16 | 3.12 |
| 4 | 24 ± 4* | 24.1 | |||
| PLMVd-ml(+)mut | 4 | 33 ± 4 | 33.0 | ||
| ELVd-ml(+) | 0 | 30 ± 2* | 29.6 | 11.04 | 3.12 |
| 4 | 26 ± 5 | 26.2 | |||
| ELVd-ml(-) | 4 | 25 ± 4 | 25.0 | ||
*One-way ANOVA was used to test the null hypothesis that the mean length of the three variants within each viroid species is equal. The results show that F > F crit in all cases, thus evidencing that the mean length of one of the three variants (marked with an asterisk in the ‘Mean’ column) is significantly different than those of the other two.
Figure 2.
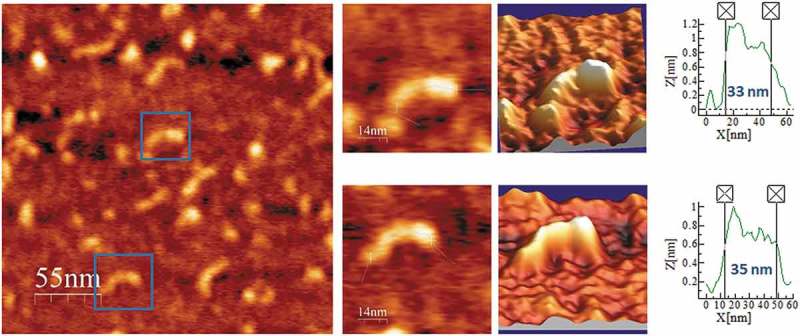
AFM images of PSTVd-ml(+) renatured in 4 mM Mg2+. A field of 275 × 275 nm is shown on the left panel and two characteristic molecules are zoomed on the right one. 3D views of the imaged viroid RNAs, as well as profiles with their measured lengths, are also displayed.
A different form of the same viroid, PSTVd-mc(+), the mc (+) RNA isolated from infected plant leaves, was purified and AFM imaged after renaturation in 4 mM Mg2+-containing buffer. The imaged rod-like features (Fig. 3 and Suppl. Fig. S3) were fairly indistinguishable from those of PSTVd-ml(+), thus suggesting that the covalent closing of the molecule occurring in vivo did not affect the overall topology of the viroid RNA. Its average molecular length was 35 nm, equivalent to that of PSTVd-ml(+) under the same Mg2+ concentration.
Figure 3.
AFM images of PSTVd-mc(+) renatured in 4 mM Mg2+. A field of 275 × 275 nm is shown on the left panel and two characteristic molecules are zoomed on the right. 3D views of the imaged viroid RNAs, as well as profiles with their measured lengths, are also displayed.
The size distribution of the imaged variants of PSTVd (depicted in Suppl. Figs. S1 to S3) is represented in Fig. 4A. This comparison of viroid lengths at single-molecule level reveals that the most frequently imaged PSTVd-ml(+) molecules are 36–40 nm long at 0 mM Mg2+, while the peak in the distribution is shifted to the 31–35 nm interval for both PSTVd-ml(+) and PSTVd-mc(+) at 4 mM Mg2+. This result reinforces the significant difference in mean lengths shown in Table 2.
Figure 4.
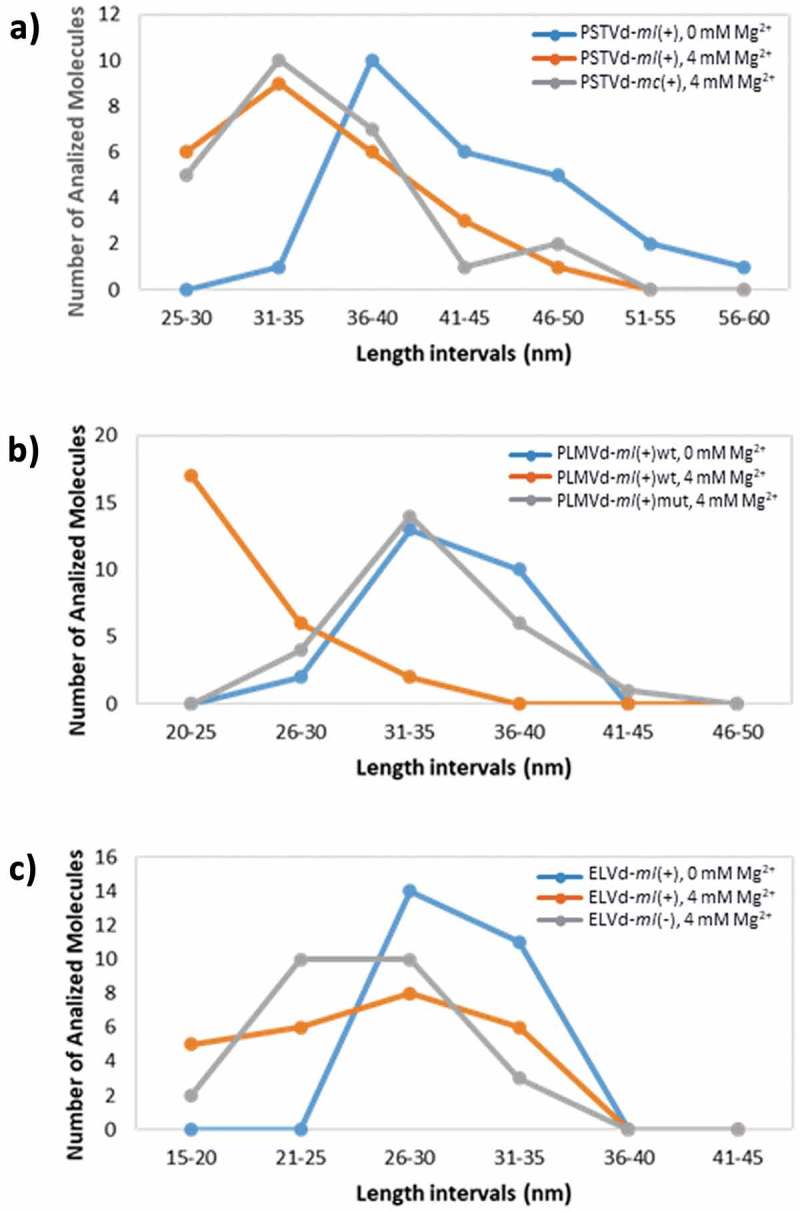
Length distribution of the imaged single molecules of PSTVd (A), PLMVd (B) and ELVd (C) at 0 and 4 mM Mg2+. Length intervals of 5 nm have been used for the comparative analysis of all the viroid variants.
Peach latent mosaic viroid (family Avsunviroidae): a kissing-loop interaction critical for stabilizing the 3D structure
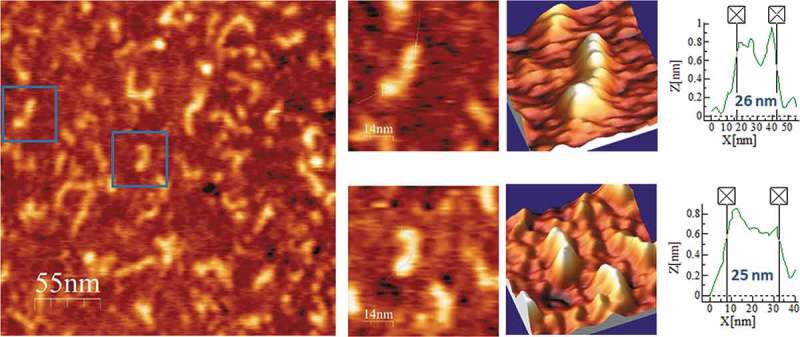
Following the same workflow, we performed the AFM imaging of PLMVd-ml(+)wt, the wild type ml (+) RNA resulting from self-cleavage during in vitro transcription of head-to-tail dimeric RNA. The predicted secondary structure of this viroid RNA (Fig. 5A) shows a ramified topology, with a rod-like domain (the so-called ‘hammerhead arm’, spanning positions 1–53 and 284–337) protruding from a multibranched domain spanning positions 54–283 [54]. This second domain is stabilized by a well-characterized kissing-loop interaction between nucleotides 176GCGG179 and 209CCGC212 in PLMVd-ml(+)wt [34], which is disrupted in the mutant PLMVd-ml(+)mut, wherein positions 209–212 are four consecutive As introduced by site-directed mutagenesis. The topographic image of PLMVd-ml(+)wt in the folding buffer without added Mg2+ (Fig. 5B and Suppl. Fig. S4) recalls the shape of a characteristic ‘spoon’ or ‘lollipop’, where the handle and the head would correspond to the rod-like and multibranched domains, respectively. The average measured length of PLMVd-ml(+)wt was 34 nm (Table 2), being the head imaged as a 15 × 20 nm ellipse (Suppl. Fig. S4) with a height exceeding 1.4 nm. Renaturation of PLMVd-ml(+)wt RNA in the folding buffer containing 4 mM Mg2+ rendered an overall topology where the ‘spoon’ shape was less evident (Fig. 6 and Suppl. Fig. S5), showing a marked reduction in the average length (24 nm). This Mg2+-induced compaction could result from the stabilization afforded to the whole RNA structure and, particularly, to the kissing-loop interaction.
Figure 5.
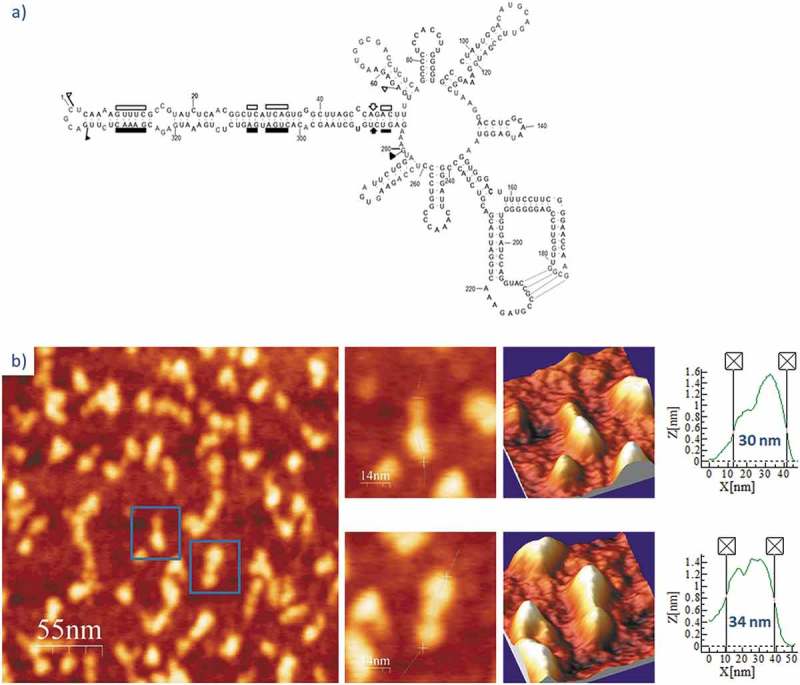
(A) Predicted secondary structure of PLMVd-ml(+)wt. The 337-nt, multibranched secondary structure of this ribozyme-containing member of the family Avsunviroidae is depicted. Boundaries of the (+) and (-) self-cleaving domains are indicated by flags, nucleotides conserved in most natural hammerhead structures are marked by bars, and the self-cleavage sites are identified by arrows. Filled and open symbols refer to (+) and (-) polarities, respectively. Nucleotides involved in a kissing-loop interaction supported by chemical probing [34] are identified by broken lines. Adapted from [78]. (B) AFM images of PLMVd-ml(+)wt renatured in the absence of Mg2+. A field of 275 × 275 nm is shown on the left panel and two characteristic molecules are zoomed on the right. 3D views of the imaged viroid RNAs, as well as profiles with their measured lengths are also displayed.
Figure 6.
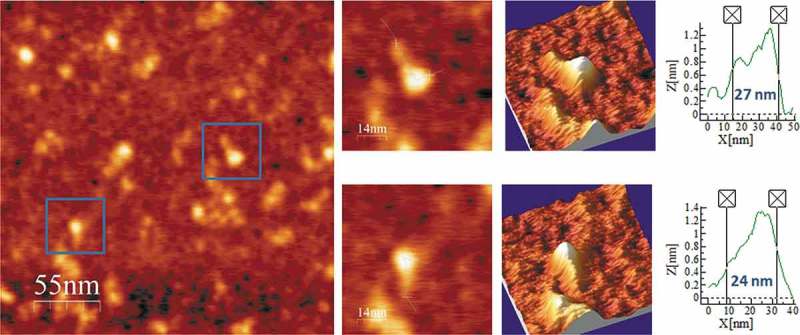
AFM images of PLMVd-ml(+)wt renatured in 4 mM Mg2+. A field of 275 × 275 nm is shown on the left panel and two characteristic molecules are zoomed on the right. 3D views of the imaged viroid RNAs, as well as profiles with their measured lengths are also displayed.
Interestingly, after renaturation in the folding buffer supplemented with 4 mM Mg2+ of PLMVd-ml(+)mut, in which the kissing-loop interaction was disrupted, the AFM analysis showed morphologies intermediate with respect to the two previous samples (Fig. 7 and Suppl. Fig. S6). Also, its computed average length was 33 nm, a 37.5% longer than that of PLMVd-ml(+)wt RNA renatured under the same Mg2+ concentration. Therefore, thanks to the detailed analysis performed and the high AFM resolution achieved, this study provides direct physical evidence on the stabilizing effects produced by a kissing-loop interaction in the overall 3D structure of PLMVd (+) RNA.
Figure 7.
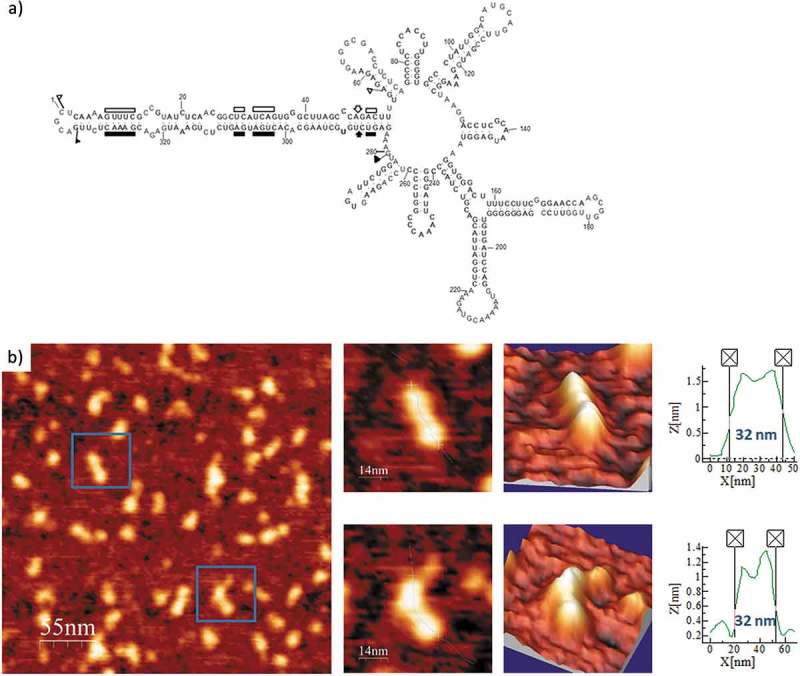
(A) Predicted secondary structure of PLMVd-ml(+)mut, with mutations that disrupt the kissing-loop interaction characteristic of PLMVd-ml(+)wt. The 337-nt, multibranched secondary structure of this mutant viroid is depicted (see details in legend of Figure 5A). (B) AFM images of PLMVd-ml(+)mut renatured in 4 mM Mg2+. A field of 275 × 275 nm is shown on the left panel and two characteristic molecules are zoomed on the right. 3D views of the imaged viroid RNAs, as well as profiles with their measured lengths are also displayed.
The size distribution of the imaged variants of PLMVd (Suppl. Figs. S4 to S6) depicted in Fig. 4B shows that the peak in the graph shifts to shorter lengths (20–25 nm) at 4 mM Mg2+ only in PLMVd-ml(+)wt, in which the kissing-loop interaction is allowed. In turn, the length distribution of variant PLMVd-ml(+)mut (with the kissing-loop interaction disrupted) at 4 mM Mg2+ shows a maximum in the 31–35 nm interval, as in PLMVd-ml(+)wt in a folding buffer lacking divalent ions. This observation reinforces the role played by the kissing-loop interaction in the stabilization of the 3D structure of PLMVd.
Eggplant latent viroid (family Avsunviroidae): similar conformations adopted by either polarity strand
Regarding ELVd, the predicted secondary structures of either polarity contain two terminal bifurcations that interrupt the overall elongated shape of the molecule. Representative AFM images of ELVd-ml(+), the ml (+) RNA (see secondary structure in Fig. 8A), renatured in the buffer without Mg2+, are shown in Fig. 8B and Suppl. Fig. S7. The topographic images reflect a majority of molecules with rod-like conformations, typically containing two or three aligned bumps, with an average length of 30 nm and a height lower than 1 nm (Table 2). ELVd-ml(+) was also imaged after being refolded in 4 mM Mg2+ and, as illustrated in Fig. 9 and Suppl. Fig. S8, this RNA maintained its main overall shape seen in the absence of divalent cations, though adopting a more compact conformation with one or two bumps and an average length of 26 nm, which correlated with the thickening (up to 10 nm wide and 3 nm high) observed in one of its terminal domains.
Figure 8.
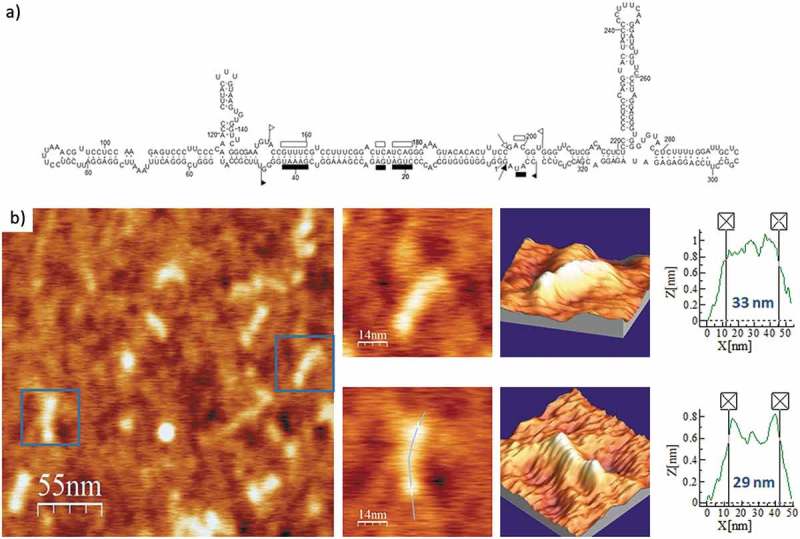
(A) Predicted secondary structure of ELVd-ml(+). In this ribozyme-containing member (335 nt) of the family Avsunviroidae the sequences involved in the hammerhead structures are delimited by flags, motifs conserved in natural hammerhead structures are denoted by bars, and self-cleavage sites are marked by arrows. Adapted from [32]. (B) AFM images of ELVd-ml(+) renatured in the absence of Mg2+. A field of 275 × 275 nm is shown on the left panel and two characteristic molecules are zoomed on the right. 3D views of the imaged viroid RNAs, as well as profiles with their measured lengths are also displayed.
Figure 9.
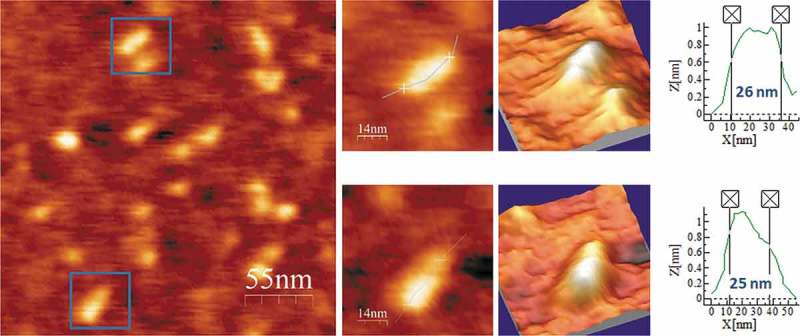
AFM images of ELVd-ml(+) renatured in 4 mM Mg2+. A field of 275 × 275 nm is shown on the left panel and two characteristic molecules are zoomed on the right. 3D views of the imaged viroid RNAs, as well as profiles with their measured lengths are also displayed.
Finally, ELVd-ml(-), the ml (-) RNA (see secondary structure in Fig. 10A), was imaged after refolding in the 4 mM Mg2+-containing buffer. As shown in Fig. 10B and Suppl. Fig. S9, the topology of the analyzed ELVd-ml(-) molecules was highly similar to that of ELVd-ml(+), with an average length of 25 nm (almost identical to that of ELVd-ml(+) in the same buffer), though slightly thicker in some of the imaged molecules. Therefore, AFM imaging did not reveal relevant differences in the conformation of ELVd RNAs of either polarity in 4 mM Mg2+.
Figure 10.
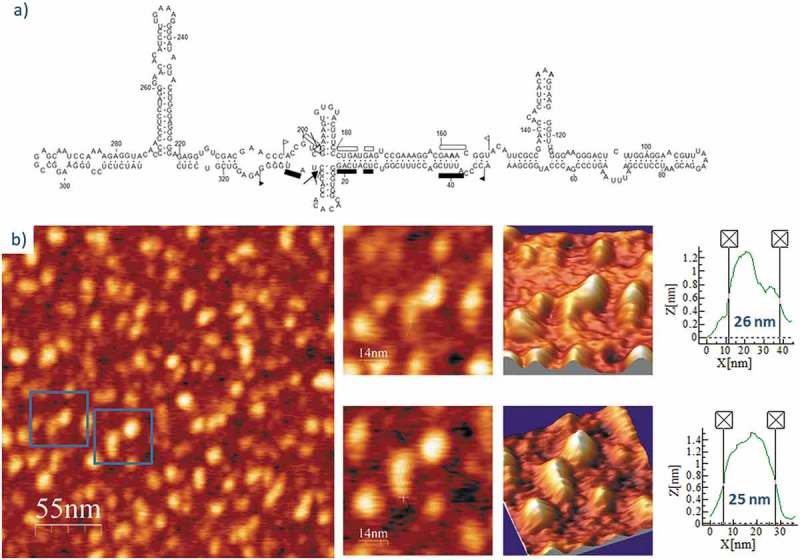
(A) Predicted secondary structure of ELVd-ml(-). For details, see legend of Fig. 7A. (B) AFM images of ELVd-ml(-) renatured in 4 mM Mg2+. A field of 275 × 275 nm is shown on the left panel and two characteristic molecules are zoomed on the right. 3D views of the imaged viroid RNAs, as well as profiles with their measured lengths are also displayed.
The size distribution of the imaged variants of ELVd (depicted in Suppl. Figs. S7 to S9) is summarized in Fig. 4C. For this viroid, the maximum number of individual molecules analyzed lies in the interval 26–30 nm, both in ELVd-ml(+) (at 0 mM Mg2+ and 4 mM Mg2+) and ELVd-ml(-) (at 4 mM Mg2+). However, in the presence of divalent cations, the overall distribution is slightly displaced towards shorter lengths in either viroid polarity.
Discussion
Because viroid RNAs do not code for any protein, the 3D conformation they adopt plays a critical role in the expression of their biological activity [20,21]. Dissection of viroid RNA structure has been previously addressed with three methodologies. First, in silico, by predicting the most stable secondary structure using RNA folding algorithms [55–57]. Second, in vitro, by biochemical examination in solution with bisulphite, RNases, dimethyl sulphate [23,24,34] and selective 2ʹ-hydroxyl acylation analyzed by primer extension (SHAPE) [25,58,59], as well as by biophysical approaches like nuclear magnetic resonance [60,61], thermal denaturation and temperature gradient gel electrophoresis [22,62–66] and Raman spectroscopy [67]. And third, in vivo, by searching for natural co-variations (and conversions of canonical into wobble base-pairs, or vice versa) that maintain the double-stranded stems [54,59,68–71], and for substitutions that preserve loop shapes in accordance with isostericity matrices predicting recurrent 3D motifs [27–29], as well as by SHAPE in situ with viroid-infected tissues [26,72]. However, all these techniques analyze samples composed of a very large ensemble of RNA molecules and generate average responses. In contrast, TEM (and more recently AFM, which provides 3D information) operate the other way around: they focus on isolated RNA molecules present in a given sample, and extract single-molecule information, from which individual shapes and sizes can be obtained to generate then average values. Both population-based and high-resolution single-molecule approaches are to a good extent complementary, and their combined data should strengthen the biological significance of the resulting inferences.
PSTVd, the first viroid identified and sequenced [22,23], has become the model for most studies, including its initial visualization by TEM. Under native conditions, TEM showed that the PSTVd mc (+) RNA (359 nt) folds into a rod-shaped conformation resembling that of a dsRNA of ~50 nm [37,38,40], while another viroid of the same family but slightly smaller (303 nt) adopts a similar conformation of ~35 nm in length [39]. These molecular sizes are consistent with each other, with that predicted theoretically for PSTVd assuming a uniform A-dsRNA conformation (49 nm), and with that first reported here by AFM for the PSTVd ml (+) RNA in the folding buffer lacking Mg2+ (43 nm). In the corresponding images, three bumps are clearly distinguishable (Fig. 1B and Suppl. Fig. S1), making it tempting to associate them with domains TL+P, C and V+ TR, respectively (see legend of Fig. 1), though more AFM-based data are needed to deepen into the PSTVd internal structure. Addition of 4 mM Mg2+ to the folding buffer resulted in a similar size contraction for the PSTVd-ml(+) and PSTVd-mc(+) RNAs, as expected for the shielding exerted by this cation on the repulsion between proximal negatively-charged phosphate groups of the RNA backbone. Indeed, thermal denaturation analysis has revealed that the melting temperature of viroid RNAs increases in the presence of Mg2+ [66].
Remarkably, in the absence of Mg2+, AFM provided for PLMVd-ml(+)wt a different conformation resembling a ‘spoon’ or ‘lollipop’ rather than a rod. This conformation is in excellent agreement with that initially predicted in silico [31,54] and then in vitro using distinct biochemical approaches, which additionally revealed the existence of a kissing-loop interaction stabilizing the multibranched domain of this RNA (Fig. 5A) [34,36,69,71,73]. A similar though more compact topology was observed by renaturing PLMVd-ml(+)wt in the presence of 4 mM Mg2+, showing a neat overall size reduction consistent with the stabilization of the kissing-loop exerted by the divalent cation. However, the resolution of this first AFM analysis of viroid structure is not sufficient to statistically correlate the shortening of the molecule with an increase in the diameter of the imaged ‘spoon head’, and/or with the reorientation of the stem-loops and hairpins forming the multibranched domain delimited by nucleotides 53–284 of PLMVd-ml(+)wt, a topic that deserves further investigation.
Interestingly, in the 4 mM Mg2+-containing buffer, the length of the mutant PLMVd-ml(+)mut (with the kissing-loop interaction disrupted, see secondary structure in Fig. 7A) was significantly longer (as revealed by an ANOVA test) than that of PLMVd-ml(+)wt under the same ionic conditions (see Table 2 and Fig. 4B), and the ‘spoon’ conformation was clearer than in the wild-type viroid RNA under the same ionic conditions. Also, a detailed analysis of the individual PLMVd-ml(+)mut molecules imaged (Suppl. Fig. S6 and data not shown), evidenced a short (10 to 15 nm long) and flat arm protruding from the head of some molecules, which might correspond to one of the hairpins of the multibranched domain in the absence of the kissing-loop interaction (Fig. 7A). Such feature was much less evident in the wild type variant PLMVd-ml(+)wt at 4 mM Mg2+ (Suppl. Fig. S5), as shown in the representative molecules imaged in Fig. 11. These observations highlight the relevance of elements of tertiary structure, like kissing-loop interactions that are characteristically strengthened by Mg2+, in enhancing the compactness of the 3D structure of some viroid RNAs. Moreover, the results shown here are consistent with previous observations by non-denaturing PAGE showing that disruption of the kissing-loop interaction in the other viroid of the same genus results in a relaxed conformation with slower electrophoretic mobility [35].
Figure 11.
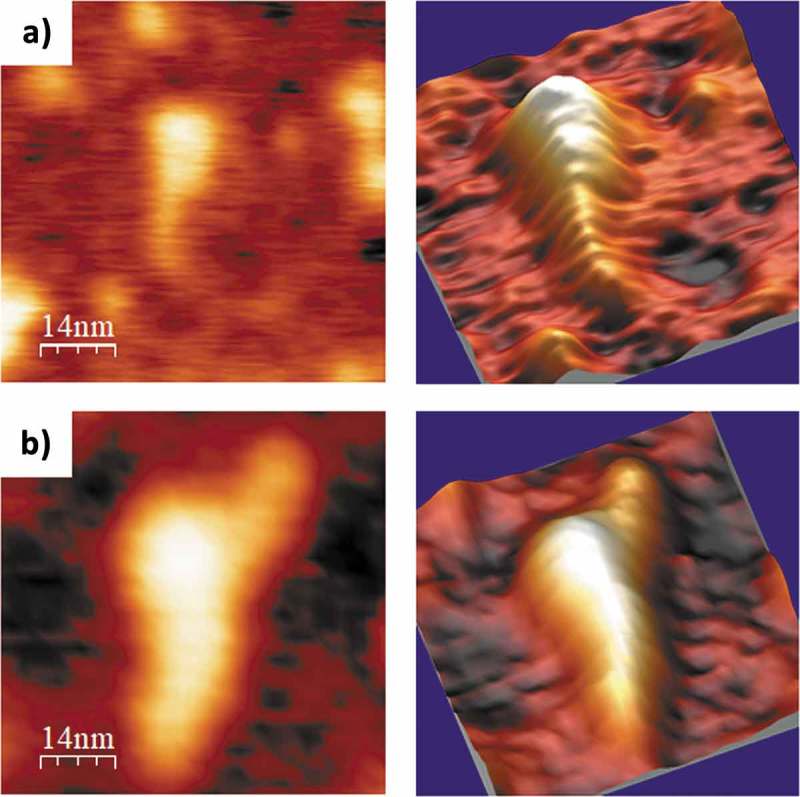
AFM images of selected PLMVd-ml(+)wt (A) and PLMVd-ml(+)mut (B) molecules, renatured in 4 mM Mg2+. 2D (left) and 3D (right) views are displayed, which clearly show the difference in compactness among the wild type (see secondary structure in Fig. 5A) and mutant (see secondary structure in Fig. 7A) viroid RNAs, as well as the presence of a short and flat arm protruding from the head of the latter.
Lastly, previous analyzes in silico, in vitro and in vivo predicted for ELVd ml (+) and (-) RNAs similar structures with terminal bifurcations of different size [32,58,59], somehow halfway between those reported for PSTVd-ml(+) (rod-like) and PLMVd-ml(+)wt (multibranched). Our AFM images of ELVd-ml(+) and ELVd-ml(-) are consistent with these predictions, showing comparable topology and size for either polarity strand of ELVd in Mg2+-containing buffer. Moreover, in the AFM images of some of the individual molecules, a sort of thickening in one of the terminal domains was observed that could be produced by one of the bifurcations of either polarity RNA, although even at the maximum AFM resolution no clearer distinction could be made.
In summary, the AFM data reported here support and complement, using a single-molecule approach, those previously obtained with other approaches in silico, in vitro and in vivo. Although for reasons stated before [59] the latter three approaches should not necessarily concord with each other when applied to any RNA in general, they do provide consistent results when dealing with three different viroids: PSTVd, ELVd, and avocado sunblotch viroid (ASBVd), the type member of the family Avsunviroidae [26,59,72]. Moreover, the main conclusion from two of these studies performed by in vivo SHAPE is that PSTVd and ASBVd RNAs accumulate in planta as free RNAs, adopting a rod-shaped secondary structure without tightly-bound host proteins. Thus, the potential distorting effects that the latter might exert on viroid RNA structure in vivo can be reasonably dismissed and, consequently, our AFM data obtained in vitro for the six viroid RNA variants analyzed can be fairly extrapolated to the in vivo habitat. To conclude, our results provide the first direct visualization at single-molecule resolution of viroid RNA 3D structure, and confirm the stabilizing role that elements of tertiary structure, like kissing-loop interactions, play in some of their functional conformations.
Materials and methods
Viroid samples
The viroid variants analyzed by AFM in this work are listed in Table 1. PLMVd (variant gds6, GenBank AJ005303.1) and ELVd (reference variant, GenBank AJ536613) ml RNAs of (+), or (+) and (-) polarities, respectively, were the unit-length self-cleavage products resulting from in vitro transcription (under the control of the T7 or T3 promoters [74]) of recombinant plasmids containing dimeric head-to-tail viroid-cDNA inserts. PSTVd (variant RG1, GenBank U23058.1) was made to contain flanking ribozymes [15] in order to generate during in vitro transcription ml (+) RNA opened between positions C1 and G2. A mutated version of the PLMVd-gds6 variant, with positions 209–212 (CCGC) replaced by four consecutive As, was obtained by PCR and abutted primers, one of which containing the changes to be introduced in its 5ʹ terminus. The unit-length strands were purified by denaturing polyacrylamide gel electrophoresis (PAGE) in 5% gels and subsequent elution. The mc (+) RNA of variant PSTVd-Nb (GenBank AJ634596.1) was isolated and purified from leaves of infected Nicotiana benthamiana by extraction with phenol-saturated buffer, fractionation with non-ionic cellulose, clarification with methoxyethanol, and double PAGE as reported before [26].
Sample preparation for AFM
Disks of muscovite mica Hi-Grade V2 (Monocomp Instrumentation) were attached to 13-mm steel pucks using Aron Alpha high-strength rapid bonding adhesive based on alpha cyanoacrylate (Agar Scientific Limited). Immediately prior to APTES functionalization, the top layer of the mica was cleaved using Scotch tape to reveal an atomically flat surface. Afterwards, the mica surface was treated with a 0.1% solution of APTES (SIGMA-Aldrich) for 15 min, washed with 2-propanol, rinsed with ultrapure, DEPC-treated milliQ water and dried at 37°C.
Purified viroid RNAs were diluted to 0.5 ng/μl in folding buffer (100 mM HEPES pH 7.4 and 100 mM NaCl), either magnesium-free or containing 4 mM MgCl2. They were then denatured by incubation at 95°C for 10 min, and renatured at 37°C for 10 min. Droplets (30 μl) of renatured viroid RNAs at 1–3 nM were deposited onto freshly cleaved, APTES-modified mica surfaces and incubated at 25°C for 20 min in a humidity chamber. Excess of RNA was rinsed off with DEPC-treated MilliQ water. The RNA-containing surfaces were then air-dried at a constant temperature of 25°C for 2 h [49,50,75].
AFM imaging
AFM analysis was performed in air, at room temperature, in the dynamic mode with a Nanoscope IIIA (Veeco) and an Agilent 5500 PicoPlus (Agilent Technologies) microscopes. Tapping mode AFM was carried out using silicon cantilevers with nominal curvature radius of 8 nm (Bruker), nominal force constant of 4 N/m and resonance frequency in the 50–80 kHz range. The set-points used were kept in the 0.3–0.6 V range, while the free amplitude values were in the 0.7–0.8 V range. The images (from 512 × 512 up to 2048 × 2048 pixels) were recorded at a scan rate of 1 line/s. A minimum of three independent samples of each viroid preparation at each buffer composition (nine in total) were used, thus rendering more than 27 samples analyzed by AFM, from which different AFM fields were imaged.
The influence of the tip radius is a relevant issue when imaging nanometer structures by AFM. In principle, with a nominal tip radius of 8 nm (which is considered the optimal one for imaging biological samples in air using tapping mode, and consequently selected for this work), distances below this value are difficult to resolve. Furthermore, for larger distances tip convolution is still present and leads to a widening of the imaged structure. This later aspect has indeed been taken into account here, since the length distances have been measured between points located at half of the height of the imaged structure (as an example, see graphs in Figs. 1–3 and 5–10). This procedure leaves out of the measurement approximately 8 nm (the tip nominal radius) of lateral extension at each of both extremes of the imaged molecule, thus minimizing the tip size effects.
Image analyses
The software package WSxM v5.0 (Nanotec) [76] was used to analyze all the AFM images, as well as to measure the length of 25 individual, full-length viroid RNA molecules that were randomly chosen among those present in different samples of each preparation. The selection of full-length molecules (using broad enough intervals of 25–60 nm in PSTVd, 20–50 nm in PLMVd and 15–40 nm in ELVd, see Fig. 4), which showed the viroid morphology characteristic of each species, excluded: i) shorter RNAs likely resulting from Mg2+-induced cleavage, ii) RNAs showing unusual molecular orientations on the modified mica surface, and iii) longer structures likely generated by bimolecular complexes or multimeric RNA aggregates.
Funding Statement
This work was supported by the Spanish Ministerio de Economía y Competitividad (MINECO) grants BIO2016-79618-R (funded by EU under the FEDER programme) to C.B. and BFU2104-56812-P to R.F., as well as by the Comunidad de Madrid grant S2018/NMT-4349 to L.V. CIBERehd is funded by the Instituto de Salud Carlos III (ISCIII).
Abbreviations
- AFM
atomic force microscopy
- APTES
3-aminopropyltriethoxysilane
- ASBVd
avocado sunblotch viroid
- CCR
central conserved region
- ELVd
eggplant latent viroid
- mc and ml
monomeric circular and linear viroid RNA, respectively
- PAGE
polyacrylamide gel electrophoresis
- PLMVd
peach latent mosaic viroid
- PSTVd
potato spindle tuber viroid
- SHAPE
selective 2ʹ-hydroxyl acylation analyzed by primer extension
- TEM
transmission electron microscopy
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplementary data
Supplemental data for this article can be accessed here.
References
- [1].Diener TO. Discovering viroids–a personal perspective. Nat Rev Microbiol. 2003. October;1(1):75–80.PubMed PMID: 15040183 [DOI] [PubMed] [Google Scholar]
- [2].Flores R, Hernandez C, Martinez de Alba AE, et al. Viroids and viroid-host interactions. Annu Rev Phytopathol. 2005;43:117–139. PubMed PMID: 16078879 [DOI] [PubMed] [Google Scholar]
- [3].Ding B. The biology of viroid-host interactions. Annu Rev Phytopathol. 2009;47:105–131. PubMed PMID: 19400635 [DOI] [PubMed] [Google Scholar]
- [4].Zhang Z, Qi S, Tang N, et al. Discovery of replicating circular RNAs by RNA-seq and computational algorithms. PLoS Pathog. 2014. December;10(12):e1004553 PubMed PMID: 25503469; PubMed Central PMCID: PMCPMC4263765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Serra P, Messmer A, Sanderson D, et al. Apple hammerhead viroid-like RNA is a bona fide viroid: autonomous replication and structural features support its inclusion as a new member in the genus Pelamoviroid. Virus Res. 2018. April2;249:8–15. PubMed PMID: 29510173. [DOI] [PubMed] [Google Scholar]
- [6].Hadidi A, Flores R, Randles JW, et al editors. Viroids and Satellites. Boston: Academic Press; 2017. [Google Scholar]
- [7].Flores R, Minoia S, Carbonell A, et al. Viroids, the simplest RNA replicons: how they manipulate their hosts for being propagated and how their hosts react for containing the infection. Virus Res. 2015. November2;209:136–145. PubMed PMID: 25738582. [DOI] [PubMed] [Google Scholar]
- [8].Hammann C, Steger G. Viroid-specific small RNA in plant disease. RNA Biol. 2012. June;9(6):809–819. PubMed PMID: 22617880 [DOI] [PubMed] [Google Scholar]
- [9].Kovalskaya N, Hammond RW. Molecular biology of viroid-host interactions and disease control strategies. Plant Sci. 2014. November;228:48–60. PubMed PMID: 25438785. [DOI] [PubMed] [Google Scholar]
- [10].Tsagris EM, Martinez de Alba AE, Gozmanova M, et al. Viroids. Cell Microbiol. 2008. November;10(11):2168–2179. PubMed PMID: 18764915. [DOI] [PubMed] [Google Scholar]
- [11].Grill LK, Semancik JS. RNA sequences complementary to citrus exocortis viroid in nucleic acid preparations from infected gynura aurantiaca. Proc Natl Acad Sci U S A. 1978. February;75(2):896–900. PubMed PMID: 16592500; PubMed Central PMCID: PMCPMC411364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Branch AD, Benenfeld BJ, Robertson HD. Evidence for a single rolling circle in the replication of potato spindle tuber viroid. Proc Natl Acad Sci U S A. 1988. December;85(23):9128–9132. PubMed PMID: 16594003; PubMed Central PMCID: PMCPMC282677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Branch AD, Robertson HD. A replication cycle for viroids and other small infectious RNA’s. Science. 1984. February 3;223(4635):450–455. PubMed PMID: 6197756 [DOI] [PubMed] [Google Scholar]
- [14].Daros JA, Marcos JF, Hernandez C, et al. Replication of avocado sunblotch viroid: evidence for a symmetric pathway with two rolling circles and hammerhead ribozyme processing. Proc Natl Acad Sci U S A. 1994. December 20;91(26):12813–12817. PubMed PMID: 7809126; PubMed Central PMCID: PMCPMC45530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Feldstein PA, Hu Y, Owens RA. Precisely full length, circularizable, complementary RNA: an infectious form of potato spindle tuber viroid. Proc Natl Acad Sci U S A. 1998. May 26;95(11):6560–6565. PubMed PMID: 9601006; PubMed Central PMCID: PMCPMC27879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Daros JA, Flores R. Arabidopsis thaliana has the enzymatic machinery for replicating representative viroid species of the family Pospiviroidae. Proc Natl Acad Sci U S A. 2004. April 27;101(17):6792–6797. PubMed PMID: 15096616; PubMed Central PMCID: PMCPMC404124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Flores R, Gago-Zachert S, Serra P, et al. Viroids: survivors from the RNA world? Annu Rev Microbiol. 2014;68:395–414. PubMed PMID: 25002087 [DOI] [PubMed] [Google Scholar]
- [18].Diener TO. Circular RNAs: relics of precellular evolution? Proc Natl Acad Sci U S A. 1989. December;86(23):9370–9374. PubMed PMID: 2480600; PubMed Central PMCID: PMCPMC298497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Ruiz-Mirazo K, Briones C, de la Escosura A. Prebiotic systems chemistry: new perspectives for the origins of life. Chem Rev. 2014. January 8;114(1):285–366. PubMed PMID: 24171674. [DOI] [PubMed] [Google Scholar]
- [20].Flores R, Serra P, Minoia S, et al. Viroids: from genotype to phenotype just relying on RNA sequence and structural motifs. Front Microbiol. 2012;3:217 PubMed PMID: 22719735; PubMed Central PMCID: PMCPMC3376415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Steger G, Perreault JP. Structure and associated biological functions of viroids. Adv Virus Res. 2016;94: 141–172. 2016/03/22 ed. [DOI] [PubMed] [Google Scholar]
- [22].Diener TO. Potato spindle tuber viroid. 8. Correlation of infectivity with a UV-absorbing component and thermal denaturation properties of the RNA. Virology. 1972. November;50(2):606–609. PubMed PMID: 4636118 [DOI] [PubMed] [Google Scholar]
- [23].Gross HJ, Domdey H, Lossow C, et al. Nucleotide sequence and secondary structure of potato spindle tuber viroid. Nature. 1978. May 18;273(5659):203–208. PubMed PMID: 643081 [DOI] [PubMed] [Google Scholar]
- [24].Gast FU, Kempe D, Spieker RL, et al. Secondary structure probing of potato spindle tuber viroid (PSTVd) and sequence comparison with other small pathogenic RNA replicons provides evidence for central non-canonical base-pairs, large A-rich loops, and a terminal branch. J Mol Biol. 1996. October 11;262(5):652–670. PubMed PMID: 8876645. [DOI] [PubMed] [Google Scholar]
- [25].Giguere T, Adkar-Purushothama CR, Perreault JP. Comprehensive secondary structure elucidation of four genera of the family Pospiviroidae. PLoS One. 2014;9(6):e98655 PubMed PMID: 24897295; PubMed Central PMCID: PMCPMC4045682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Lopez-Carrasco A, Flores R. Dissecting the secondary structure of the circular RNA of a nuclear viroid in vivo: A “naked” rod-like conformation similar but not identical to that observed in vitro. RNA Biol. 2017. August 3;14(8):1046–1054. PubMed PMID: 27574720; PubMed Central PMCID: PMCPMC5680722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Wang Y, Zirbel CL, Leontis NB, et al. RNA 3-dimensional structural motifs as a critical constraint of viroid RNA evolution. PLoS Pathog. 2018. February;14(2):e1006801 PubMed PMID: 29470541; PubMed Central PMCID: PMCPMC5823408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Zhong X, Leontis N, Qian S, et al. Tertiary structural and functional analyses of a viroid RNA motif by isostericity matrix and mutagenesis reveal its essential role in replication. J Virol. 2006. September;80(17):8566–8581. PubMed PMID: 16912306; PubMed Central PMCID: PMCPMC1563885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Zhong X, Tao X, Stombaugh J, et al. Tertiary structure and function of an RNA motif required for plant vascular entry to initiate systemic trafficking. Embo J. 2007. August 22;26(16):3836–3846. PubMed PMID: 17660743; PubMed Central PMCID: PMCPMC1952227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Zhong X, Archual AJ, Amin AA, et al. A genomic map of viroid RNA motifs critical for replication and systemic trafficking. Plant Cell. 2008;20(1):35–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Hernandez C, Flores R. Plus and minus RNAs of peach latent mosaic viroid self-cleave in vitro via hammerhead structures. Proc Natl Acad Sci U S A. 1992. May 1;89(9):3711–3715. PubMed PMID: 1373888; PubMed Central PMCID: PMCPMC525560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Fadda Z, Daros JA, Fagoaga C, et al. Eggplant latent viroid, the candidate type species for a new genus within the family Avsunviroidae (hammerhead viroids). J Virol. 2003. June;77(11):6528–6532. PubMed PMID: 12743309; PubMed Central PMCID: PMCPMC155007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Navarro B, Flores R. Chrysanthemum chlorotic mottle viroid: unusual structural properties of a subgroup of self-cleaving viroids with hammerhead ribozymes. Proc Natl Acad Sci U S A. 1997. October 14;94(21):11262–11267. PubMed PMID: 9326597; PubMed Central PMCID: PMCPMC23434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Bussiere F, Ouellet J, Cote F, et al. Mapping in solution shows the peach latent mosaic viroid to possess a new pseudoknot in a complex, branched secondary structure. J Virol. 2000. March;74(6):2647–2654. PubMed PMID: 10684279; PubMed Central PMCID: PMCPMC111753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Gago S, De la Pena M, Flores R. A kissing-loop interaction in a hammerhead viroid RNA critical for its in vitro folding and in vivo viability. Rna. 2005. July;11(7):1073–1083. PubMed PMID: 15928342; PubMed Central PMCID: PMCPMC1370792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Dubé A, Baumstark T, Bisaillon M, et al. The RNA strands of the plus and minus polarities of peach latent mosaic viroid fold into different structures. Rna. 2010. March;16(3):463–473. PubMed PMID: 20089682; PubMed Central PMCID: PMCPMC2822911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Sogo JM, Koller T, Diener TO. Potato spindle tuber viroid. X. Visualization and size determination by electron microscopy. Virology. 1973. September;55(1):70–80. PubMed PMID: 4728831 [DOI] [PubMed] [Google Scholar]
- [38].Goodman TC, Nagel L, Rappold W, et al. Viroid replication: equilibrium association constant and comparative activity measurements for the viroid-polymerase interaction. Nucleic Acids Res. 1984. August 10;12(15):6231–6246. PubMed PMID: 6473106; PubMed Central PMCID: PMCPMC320069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Sanger HL, Klotz G, Riesner D, et al. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A. 1976. November;73(11):3852–3856. PubMed PMID: 1069269; PubMed Central PMCID: PMCPMC431239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].McClements WL, Kaesberg P. Size and secondary structure of potato spindle tuber viroid. Virology. 1977. February;76(2):477–484. PubMed PMID: 841846 [DOI] [PubMed] [Google Scholar]
- [41].Bustamante C, Keller D. Scanning force microscopy in biology. Physics Today. 1995;48(12):32–38. [Google Scholar]
- [42].Hansma HG, Kasuya K, Oroudjev E. Atomic force microscopy imaging and pulling of nucleic acids. Curr Opin Struct Biol. 2004. June;14(3):380–385. PubMed PMID: 15193320 [DOI] [PubMed] [Google Scholar]
- [43].Kuznetsov YG, Daijogo S, Zhou J, et al. Atomic force microscopy analysis of icosahedral virus RNA. J Mol Biol. 2005;347(1):41–52. [DOI] [PubMed] [Google Scholar]
- [44].Alvarez DE, Lodeiro MF, Luduena SJ, et al. Long-range RNA-RNA interactions circularize the dengue virus genome. J Virol. 2005. June;79(11):6631–6643. PubMed PMID: 15890901; PubMed Central PMCID: PMCPMC1112138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Schon P. Atomic force microscopy of RNA: state of the art and recent advancements. Semin Cell Dev Biol. 2018. January;73:209–219. PubMed PMID: 28843977. [DOI] [PubMed] [Google Scholar]
- [46].Hansma HG, Oroudjev E, Baudrey S, et al. TectoRNA and ‘kissing-loop’ RNA: atomic force microscopy of self-assembling RNA structures. J Microsc. 2003. December;212(Pt 3):273–279. PubMed PMID: 14629553. [DOI] [PubMed] [Google Scholar]
- [47].Basame S, Wai-Lun Li P, Howard G, et al. Spatial assembly and RNA binding stoichiometry of a LINE-1 protein essential for retrotransposition. J Mol Biol. 2006. March 24;357(2):351–357. PubMed PMID: 16434051. [DOI] [PubMed] [Google Scholar]
- [48].Ares P, Fuentes-Perez ME, Herrero-Galan E, et al. High resolution atomic force microscopy of double-stranded RNA [10.1039/C5NR07445B]. Nanoscale. 2016;8(23):11818–11826. [DOI] [PubMed] [Google Scholar]
- [49].Gilmore JL, Yoshida A, Takahashi H, et al. Analyses of nuclear proteins and nucleic acid structures using atomic force microscopy In: Nakagawa S, Hirose Teditors. Nuclear bodies and noncoding RNAs: methods and protocols. New York, NY: Springer New York; 2015. p. 119–153. [DOI] [PubMed] [Google Scholar]
- [50].Garcia-Sacristan A, Moreno M, Ariza-Mateos A, et al. A magnesium-induced RNA conformational switch at the internal ribosome entry site of hepatitis C virus genome visualized by atomic force microscopy. Nucleic Acids Res. 2015. January;43(1):565–580. PubMed PMID: 25510496; PubMed Central PMCID: PMCPMC4288189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Cheng H, Zhang K, Libera JA, et al. Polynucleotide adsorption to negatively charged surfaces in divalent salt solutions. Biophys J. 2006. February 15;90(4):1164–1174. PubMed PMID: 16449197; PubMed Central PMCID: PMCPMC1367268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Lyubchenko YL, Shlyakhtenko LS, Ando T. Imaging of nucleic acids with atomic force microscopy. Methods. 2011. June;54(2):274–283. PubMed PMID: 21310240; PubMed Central PMCID: PMCPMC3114274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Lang D, Steely HT Jr., Kao CY, et al. Length, mass, and denaturation of double-stranded RNA molecules compared with DNA. Biochim Biophys Acta. 1987. December 8;910(3):271–281. PubMed PMID: 3118956 [DOI] [PubMed] [Google Scholar]
- [54].Ambros S, Hernandez C, Desvignes JC, et al. Genomic structure of three phenotypically different isolates of peach latent mosaic viroid: implications of the existence of constraints limiting the heterogeneity of viroid quasispecies. J Virol. 1998. September;72(9):7397–7406. PubMed PMID: 9696836; PubMed Central PMCID: PMCPMC109966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Lorenz R, Bernhart SH, Honer Zu Siederdissen C, et al. ViennaRNA package 2.0 [journal article]. Algorithms Mol Biol. 2011. November 24;6(1):26 PubMed PMID: 22115189; PubMed Central PMCID: PMCPMC3319429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Reuter JS, Mathews DH. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics. 2010. March 15;11:129 PubMed PMID: 20230624; PubMed Central PMCID: PMCPMC2984261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003. July 1;31(13):3406–3415. PubMed PMID: 12824337; PubMed Central PMCID: PMCPMC169194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Giguere T, Adkar-Purushothama CR, Bolduc F, et al. Elucidation of the structures of all members of the Avsunviroidae family. Mol Plant Pathol. 2014. October;15(8):767–779. PubMed PMID: 25346967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Lopez-Carrasco A, Gago-Zachert S, Mileti G, et al. The transcription initiation sites of eggplant latent viroid strands map within distinct motifs in their in vivo RNA conformations. RNA Biol. 2016;13(1):83–97. PubMed PMID: 26618399; PubMed Central PMCID: PMCPMC4829332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Dingley AJ, Steger G, Esters B, et al. Structural characterization of the 69 nucleotide potato spindle tuber viroid left-terminal domain by NMR and thermodynamic analysis. J Mol Biol. 2003. December 5;334(4):751–767. PubMed PMID: 14636600 [DOI] [PubMed] [Google Scholar]
- [61].Dufour D, de la Pena M, Gago S, et al. Structure-function analysis of the ribozymes of chrysanthemum chlorotic mottle viroid: a loop-loop interaction motif conserved in most natural hammerheads. Nucleic Acids Res. 2009. February;37(2):368–381. PubMed PMID: 19043070; PubMed Central PMCID: PMCPMC2632901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Baumstark T, Riesner D. Only one of four possible secondary structures of the central conserved region of potato spindle tuber viroid is a substrate for processing in a potato nuclear extract. Nucleic Acids Res. 1995. November 11;23(21):4246–4254. PubMed PMID: 7501442; PubMed Central PMCID: PMCPMC307376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Delan-Forino C, Deforges J, Benard L, et al. Structural analyses of avocado sunblotch viroid reveal differences in the folding of plus and minus RNA strands. Viruses. 2014. January 29;6(2):489–506. PubMed PMID: 24481250; PubMed Central PMCID: PMCPMC3939467. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Henco K, Sanger HL, Riesner D. Fine structure melting of viroids as studied by kinetic methods. Nucleic Acids Res. 1979. July 11;6(9):3041–3059. PubMed PMID: 493134; PubMed Central PMCID: PMCPMC327916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Riesner D, Henco K, Rokohl U, et al. Structure and structure formation of viroids. J Mol Biol. 1979. September 5;133(1):85–115. PubMed PMID: 529284 [DOI] [PubMed] [Google Scholar]
- [66].Semancik JS, Morris TJ, Weathers LG, et al. Physical properties of a minimal infectious RNA (viroid) associated with the exocortis disease. Virology. 1975. January;63(1):160–167. PubMed PMID: 1111210 [DOI] [PubMed] [Google Scholar]
- [67].Hui-Bon-Hoa G, Kaddour H, Vergne J, et al. Raman characterization of avocado sunblotchviroid and its response to external perturbations and self-cleavage [journal article]. BMC Biophys. 2014. March 21;7(1):2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].De la Pena M, Navarro B, Flores R. Mapping the molecular determinant of pathogenicity in a hammerhead viroid: A tetraloop within the in vivo branched RNA conformation. Proc Natl Acad Sci U S A. 1999;96(17):9960–9965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Fekih Hassen I, Massart S, Motard J, et al. Molecular features of new peach latent mosaic viroid variants suggest that recombination may have contributed to the evolution of this infectious RNA. Virology. 2007. March 30;360(1):50–57. PubMed PMID: 17113618. [DOI] [PubMed] [Google Scholar]
- [70].Pelchat M, Levesque D, Ouellet J, et al. Sequencing of peach latent mosaic viroid variants from nine North American peach cultivars shows that this RNA folds into a complex secondary structure. Virology. 2000. May 25;271(1):37–45. PubMed PMID: 10814568. [DOI] [PubMed] [Google Scholar]
- [71].Serra P, Bertolini E, Martinez MC, et al. Interference between variants of peach latent mosaic viroid reveals novel features of its fitness landscape: implications for detection. Sci Rep. 2017. February17;7:42825 PubMed PMID: 28211491; PubMed Central PMCID: PMCPMC5314366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Lopez-Carrasco A, Flores R. The predominant circular form of avocado sunblotch viroid accumulates in planta as a free RNA adopting a rod-shaped secondary structure unprotected by tightly bound host proteins. J Gen Virol. 2017. July;98(7):1913–1922. PubMed PMID: 28699864 [DOI] [PubMed] [Google Scholar]
- [73].Dube A, Bolduc F, Bisaillon M, et al. Mapping studies of the peach latent mosaic viroid reveal novel structural features. Mol Plant Pathol. 2011. September;12(7):688–701. PubMed PMID: 21726370; PubMed Central PMCID: PMCPMC3256235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Green MR, Sambrook J. Molecular cloning: A laboratory manual. Vol. 1 New York, NY: Cold Spring Harbor Laboratory Press; 2012. [Google Scholar]
- [75].Lyubchenko Y, Shlyakhtenko L, Harrington R, et al. Atomic force microscopy of long DNA: imaging in air and under water [article]. Proc Natl Acad Sci U S A. 1993. March 15;90(6):2137–2140. PubMed PMID: 8460119; PubMed Central PMCID: PMCPMC46040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Horcas I, Fernandez R, Gomez-Rodriguez JM, et al. WSXM: a software for scanning probe microscopy and a tool for nanotechnology. Rev Sci Instrum. 2007. January;78(1):013705 PubMed PMID: 17503926. [DOI] [PubMed] [Google Scholar]
- [77].Keese P, Symons RH. Domains in viroids: evidence of intermolecular RNA rearrangements and their contribution to viroid evolution. Proc Natl Acad Sci U S A. 1985. July;82(14):4582–4586. PubMed PMID: 3860809; PubMed Central PMCID: PMCPMC390429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Di Serio F, Gisel A, Navarro B, et al. Deep sequencing of the small RNAs derived from two symptomatic variants of a chloroplastic viroid: implications for their genesis and for pathogenesis. PLoS One. 2009. October 21;4(10):e7539 PubMed PMID: 19847296; PubMed Central PMCID: PMCPMC2760764. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


