ABSTRACT
The Ran-binding protein 2 (Ranbp2/Nup358) is a cytoplasmic and peripheral nucleoporin comprised of 4 Ran-GTP-binding domains (RBDs) that are interspersed among diverse structural domains with multifunctional activities. Our prior studies found that the RBD2 and RBD3 of Ranbp2 control mitochondrial motility independently of Ran-GTP-binding in cultured cells, whereas loss of Ran-GTP-binding to RBD2 and RBD3 are essential to support cone photoreceptor development and the survival of mature retinal pigment epithelium (RPE) in mice. Here, we uncover that loss of Ran-GTP-binding to RBD3 alone promotes the robust age-dependent increase of ubiquitylated substrates and S1 subunit (Pmsd1) of the 19S cap of the proteasome in the retina and RPE and that such loss in RBD3 also compromises the structural integrity of the outer segment compartment of cone photoreceptors only and without affecting the viability of these neurons. We also found that the E2-ligase and partner of Ranbp2, ubc9, is localized prominently in the mitochondrial-rich ellipsoid compartment of photoreceptors, where Ranbp2 is also known to localize with and modulate the activity of mitochondrial proteins. However, the natures of Ranbp2 and ubc9 isoforms to the mitochondria are heretofore elusive. Subcellular fractionation, co-immunolocalization and immunoaffinity purification of Ranbp2 complexes show that novel isoforms of Ranbp2 and ubc9 with molecular masses distinct from the large Ranbp2 and unmodified ubc9 isoforms localize specifically to the mitochondrial fraction or associate with mitochondrial components, whereas unmodified and SUMOylated Ran GTPase are excluded from the mitochondrial fraction. Further, liposome-mediated intracellular delivery of an antibody against a domain shared by the mitochondrial and nuclear pore isoforms of Ranbp2 causes the profound fragmentation of mitochondria and their delocalization from Ranbp2 and without affecting Ranbp2 localization at the nuclear pores. Collectively, the data support that Ran GTPase-dependent and independent and moonlighting roles of Ranbp2 or domains thereof and ubc9 control selectively age-dependent, neural-type and mitochondrial functions.
KEYWORDS: mitochondria, nucleoporin, photoreceptor neuron, Ran-binding protein 2, Ran GTPase, transgenic mice, ubiquitin proteostasis, ubiquitin-conjugating enzyme E2I (ubc9)
The Ranbp2 (also called Nup358) is a multifunctional nuclear protein that is localized to cytoplasmic filaments emanating the nuclear pore complex (NPC),1 where it controls rate-limiting steps of nucleocytoplasmic trafficking.2-5 Ran GTPase is a central driver of transport of substrates between the nuclear and cytoplasmic compartments. Specifically, co-activation of Ran-GTP hydrolysis by the structurally and biochemically equivalent RBDs of Ranbp2 promotes the release of Ran GTPase from the nuclear import receptor, importin-β, and nuclear cargoes bound to the nuclear export receptor, exportin-1/CRM1.4,6 Although the control of these nuclear transport processes are seemingly of ubiquitous relevance to support vital cellular functions from yeast to man, multiples lines of evidence support that Ranbp2 is not evolutionary conserved and that Ranbp2 or domains thereof control selectively the expression cell-type, stress-elicited or disease traits in mice and humans. For example, mutations that uncouple RBD2 and RBD3 of Ranbp2 from Ran-GTP impair the development of cone photoreceptor neurons, while the structures and functions of mature cones appear unaffected in mice.7 By contrast, these mutations do not impair the maturation of the retinal pigment epithelium (RPE), but they cause the degeneration of the mature RPE in mice.7 Further, semi-dominant mutations in the leucine-rich domain of Ranbp2 predispose the development of necrotic encephalopathies in humans upon exposure to infectious stressors and coupling defects of oxidative phosphorylation have been reported in patients.8,9
Recent studies have shown that the RBDs and other domains of Ranbp2 harbor moonlighting functions. For example, we found recently that ectopic expression of Ranbp2 constructs comprising the kinesin-binding domain (KBD) together with its flanking domains, RBD2 and RBD3, in cultured cells control mitochondrial motility independently of Ran-GTP binding.10 This effect arises from the direct stimulation by RBD2 and RBD3 of the motor (ATPase) activity of the microtubule-based motor isoforms of kinesins, KIF5B and KIF5C that mediate mitochondrial transport.10-13 The underpinnings of RBDs' moonlighting are unclear, but a possibility is that moonlighting activities of domains of Ranbp2 arise from distinct subcellular localizations of Ranbp2. In this regard, our earlier work found that Ranbp2 is not exclusively localized to the NPC. For example, various antibodies against Ranbp2 co-localize with mitochondrial markers, such as the mitochondrial heat shock protein 70 (mHsp70) and mitochondrial-associated hexokinase I (HKI), to the mitochondrial-rich compartment (ellipsoid) of photoreceptor neurons and mitochondria in cultured cells.14,15 Further, the leucine-rich domain of Ranbp2 associates with the nuclear-encoded and mitochondrial metallochaperone, cox11, which promotes copper metalation of the subunit 1 of cytochrome c oxidase.14 Recently, we have found that Ranbp2 also localizes inside the nucleus of spinal motor neurons and that deletion of Ranbp2 from these neurons in mice depletes the short-lived Ranbp2 from NPCs and triggers the redistribution of a long-lived and nuclear isoform of Ranbp2 to the cytoplasmic compartment, where it becomes co-localized to the mitochondria.16
To dissect the biological and physiological functions arising from the multifunctional activities of Ranbp2 and domains thereof, such as the RBD3, we performed genetic complementation studies to examine transgenic mice, TgRBD3*-HA::−/−, which express in a null Ranbp2 background the Ranbp2 gene in a bacterial artificial chromosome (BAC) with the mutation, W2186R, in RBD3 (Fig. 1A). This mutation in RBD3 uncouples its binding to Ran-GTP (Fig. 1B). TgRBD3*-HA::−/− mice rescue the lethality caused by loss of native Ranbp2 and these mice lack overt behavioral and motor phenotypes compared with other Ranbp2 transgenic lines.7 Since the RBD3 flanks upstream the cyclophilin-like domain (CLD), which associates specifically with the S1 (also called Pmsd1) and other subunits of the 19S cap of the proteasome,17,18 we examined the longitudinal effect(s) of RBD3 in ubiquitin proteostasis (free ubiquitin and ubiquitylated substrates). As shown in Fig. 1C, we found a strong age-dependent rise of total ubiquitin levels in the retina and RPE tissues of TgRBD3*-HA::−/−. By 9-month of age, there was a robust increase of the total ubiquitin levels by 5-fold in the retina (p = 0.03) and by 2-fold in the RPE (p = 0.007) of TgRBD3*-HA::−/− compared with wild-type mice (+/+) (Fig. 1C). These effects were also accompanied by a significant increase of the levels of the S1 subunit of the 19S cap of the proteasome in both the retina (p = 0.04) and RPE (p = 0.03) of TgRBD3*-HA::−/− (Fig. 1D, E). Although we also found a significant decrease of the levels of 2 other partners of Ranbp2, SUMOylated-Ran GAP (p = 0.03) and Stat3 (p = 0.04), in the RPE of TgRBD3*-HA::−/−, the downregulations of these partners were not observed in the retina of the same TgRBD3*-HA::−/− mice (Fig. 1D, E). These data indicate that age-dependent regulation of ubiquitin proteostasis by RBD3 of Ranbp2 do not promote changes in SUMOylation of substrates of Ranbp2, such as RanGAP.
Figure 1.
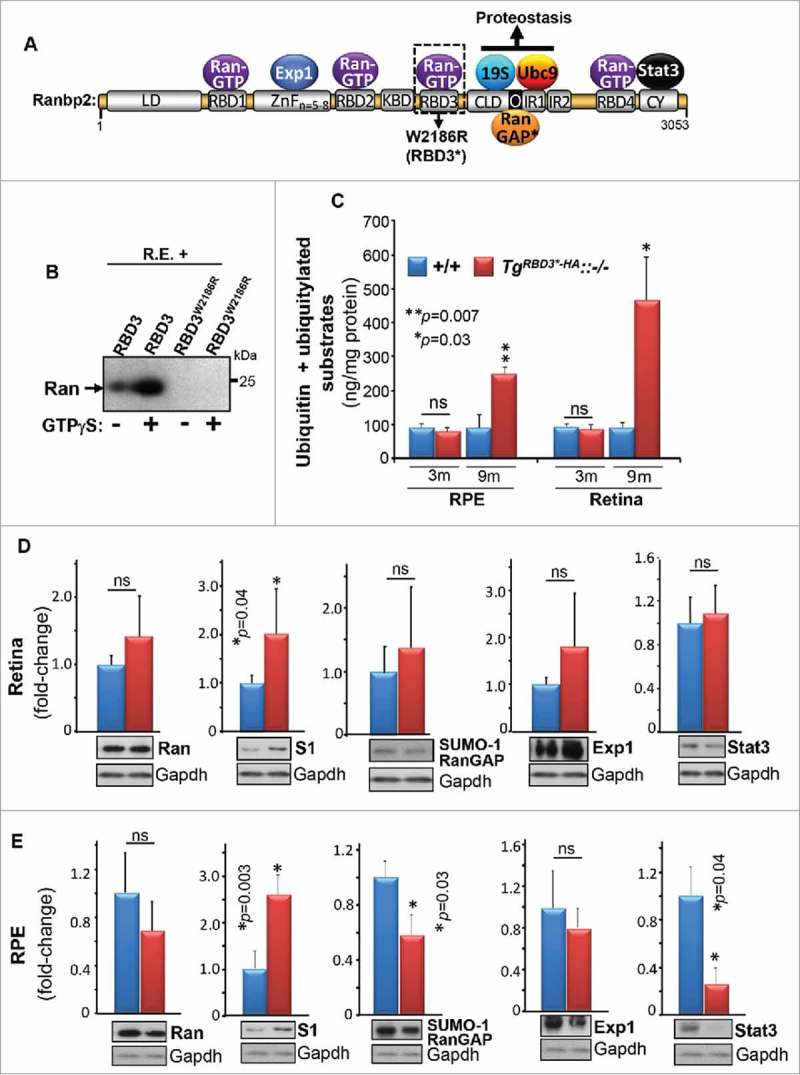
Age-dependent deregulation of the ubiquitin-proteasome system (UPS) by Ran-GTP-dependent loss of RBD3 function of Ranbp2. (A) Primary structure of Ranbp2 and partners of domains of Ranbp2. The W2186R mutation in RBD3 of Ranbp2 is shown. LD, leucine-rich domain; RBDn = 1–4, Ran GTPase-binding domains, n = 1–4; ZnFn = 5–8 zinc-finger-rich domain – the number of zinc-fingers vary from 5 to 8 depending on the species (n = 5–8) and they conform to the consensus motif W-XC-X(2,4)-C-X(3)-N-X(6)-C-X (2)-C; KBD, kinesin-1-binding domain; CLD, cyclophilin-like domain; IR, internal repeat; O, overlapping domain between CLD and IR1; CY, cyclophilin domain. (B) GST-fused RDB3 binds Ran GTPase in the presence of the non-hydrolyzable GTP analog, GTP-γ-S, whereas the W2186R mutation in RBD3 abolishes its association with Ran-GTP in GST pull-down assays with retinal extracts (R.E.). (C) Age-dependent accumulation of ubiquitin and ubiquitylated substrates in the RPE and retina. Results are given as means ± s.d.; n = 4. (D, E) The retina (D) and RPE (E) of TgRBD3*-HA::−/− mice share the upregulation of the S1 subunit (also called Psmd1) of the 19S cap of the proteasome. Representative immunoblots are shown below the graphs. Results are given as mean ± s.d.; n = 4. Legend: Exp1, exportin-1/CRM1; S1, S1 subunit of the 19S cap of the proteasome (also called Pmsd1); Ran, Ran GTPase; SUMO-1 RanGAP, SUMOylated Ran GTPase-activating protein (90 kDa); Stat3, signal transducer and activator of transcription 3; m, months, n.s., not significant.
Our prior studies found that Ranbp2 is essential to the survival of cone photoreceptors19 and that uncoupling the binding of Ran-GTP to both RBD2 and RBD3 of Ranbp2 impairs the development of M- and S-cone photoreceptor neurons without affecting apparently the physiologic functions of the surviving and mature M and S-cone photoreceptors.7 Hence, we examined the effects of loss of binding of Ran-GTP to RBD3 alone of Ranbp2 in the development and morphological structures of M- and S-cone photoreceptor neurons. We examined retinal sections by confocal microscopy of 8-week old wild-type mice (+/+), transgenic mice with loss of Ran-GTP-binding to RBD3 alone in a constitutive null Ranbp2 background (TgRBD3*-HA::−/−) and transgenic mice with loss of Ran-GTP-binding to both RBD2 and RBD3 in cone photoreceptors lacking Ranbp2 (TgRBD2/3*-HA::cone-cre::−/−). Retinal sections were co-immunostained with the pan-cone photoreceptor marker, arrestin (Arr4) (Fig. 2A, a-a”), and the E2-ligase and the Ranbp2 partner, ubc9 (Fig. 2A, b-b”). In contrast to +/+ mice, which showed uniform and intense Arr4+-labeling of cones (Fig. 2A, a), the cones of TgRBD3*-HA::−/− had uneven immunolabeling of Arr4, in particular at the level of the outer segments (Fig. 2A, a’), but this line did not show the degeneration of Arr4+-cones like in age-matched TgRBD2/3*-HA::cone-cre::−/− mice (Fig. 2A, a”). By contrast, there were no overt differences in ubc9 immunostaining of retinal neurons between genotypes (Fig. 2A, b-b”). Parenthetically, we observed that ubc9 immunostaining was prominent at the apical regions of nuclei and inner segments (ellipsoid) of cone photoreceptors (Fig. 2A, b; inset image). Then, we examined signs of degeneration of cone photoreceptors by carrying out morphometric analyses of M and S-cones and by determining their densities and length of the outer segments (OS) across the superior, central and ventral regions of the retina with antibodies against the abundant markers of OS of M- and S-cones, M and S-opsin, respectively (Fig. 2B). These analyses showed that there were no significant differences in the number of M and S-cones and length of their OS across the dorsal, ventral and central regions of the retina between +/+ and TgRBD3*-HA::−/− mice.
Figure 2.
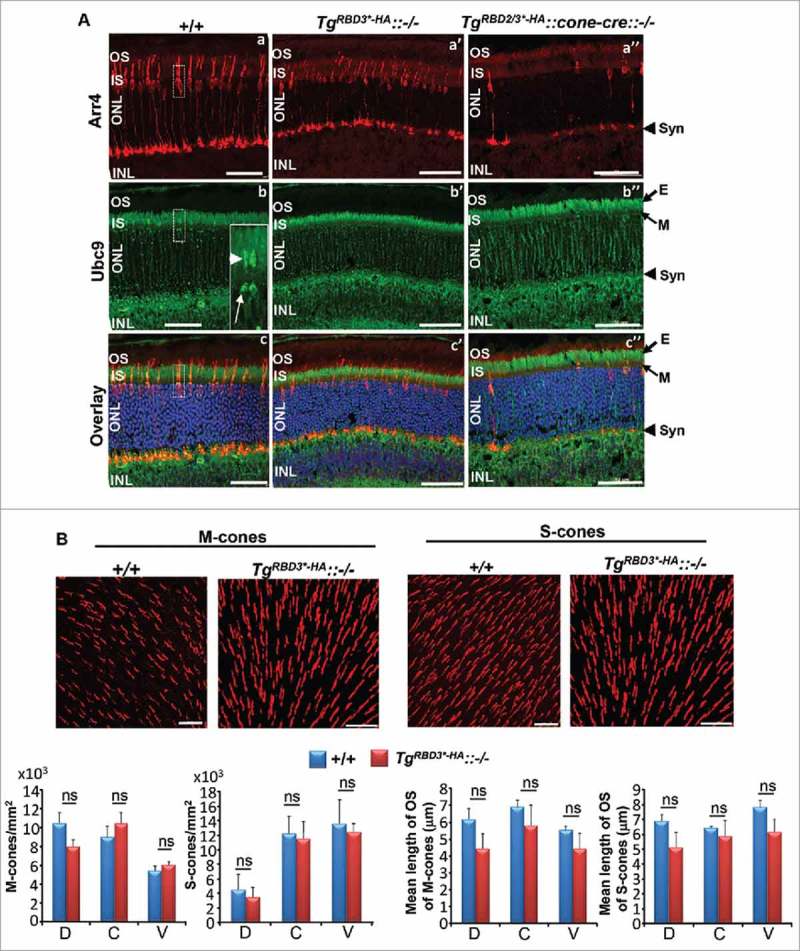
Confocal microscopy and morphometric analyses of M- and S-cone photoreceptor neurons of 8-week old TgRBD3*-HA::−/− mice. (A) Confocal images of radial sections of the central region of retinae of +/+, TgRBD3*-HA::−/− and TgRBD2/3*-HA::cone-cre::−/− mice immunostained with the pan-cone marker, arrestin (Arr4), and ubc9. +/+ mice present intense and uniform immunostaining of Arr4, whereas such staining is sparser in TgRBD3*-HA::−/− in which cones appear disordered compared with +/+ mice. TgRBD2/3*-HA::cone-cre::−/− mice present much fewer cones than TgRBD3*-HA::−/− mice. The overall distribution of ubc9 in retinal neurons is indistinguishable between genotypes. Note the enrichment of ubc9 in the mitochondrial-rich ellipsoid (E) compartment of rod and cone photoreceptors. Inset in (b) denotes the dash box region of a cone photoreceptor in a-c and the strong labeling of the apical nuclear region (arrow) and ellipsoid (arrowhead). (B) Topographic distribution and length of outer segments of M and S-cone photoreceptors in retinae of +/+ and TgRBD3*-HA::−/− mice. There were no significant differences in the distribution and length of outer segments of M and S-cones in the dorsal (D), central (C) and ventral (V) regions of retinae between genotypes. Images are representative confocal z-stacks images of M and S-cones in the central region of the retina (top panel), whereas quantitative analyses of cones are shown below image panels. Results are given as mean ± s.d.; n = 4. Legend: OS, outer segments; IS, inner segments; ONL, outer nuclear layer (nuclei of photoreceptors); INL, inner nuclear layer (nuclei of inner retinal neurons); E, ellipsoid compartment (of IS); M, myoid compartment (of IS); Syn, synaptiC-terminal; D, dorsal; C, central; V, ventral; ns, non-significant; TgRBD3*-HA, Tg-Ranbp2RBD3*-HA; TgRBD2/3*-HA, Tg-Ranbp2RBD2/3*-HA; −/−, Ranbp2−/−; cone-cre, HRGP-cre; +/+, wild-type; n.s., not significant. Scale bars: 50 μm (A), 20 μm (B).
To probe the apparent disparity in the distribution of immunolabeling between Arr4 and markers of the OS (e.g., M and S-opsin) of +/+ and TgRBD3*-HA::−/− mice, we performed histological and morphological analysis of fixed retinal sections by light and ultrastructural microscopy. As shown in Fig. 3A, the overall structure and organization of the retina and RPE in semi-thin sections by light microscopy under standard cardiac perfusion and post-fixation conditions (1:1 paraformaldehyde and glutaraldehyde at 2%) appeared normal between age-matched +/+ and TgRBD3*-HA::−/− mice, with the exception of OS of cone photoreceptors of TgRBD3*-HA::−/− that presented prominent lacunae. Then, to probe the strength of aldehyde cross-linking agents in the genotype-dependent development of lacunae in OS, we perfused mice and post-fixed their retinas only with the strong aldehyde fixative agent, glutaraldehyde, and at a higher concentration. To this effect, the lacunae were greatly attenuated in OS of cones of TgRBD3*-HA::−/− (Fig. 3B). Together these data indicate that compared with +/+ mice, the cone OS of TgRBD3*-HA::−/− mice are less susceptible to withstand osmotic stress of dehydration and infiltration during specimen processing. Finally, we examined cone photoreceptors and in particular, the highly membrane-rich OS of +/+ and TgRBD3*-HA::−/− mice at ultrastructural level by transmission electron microscopy. Retinal sections fixed with 4% glutaraldehyde underwent secondary fixation with osmium tetraoxide, which reacts with unsaturated fatty acids and thus acts as a membrane contrasting agent.20 As shown in Fig. 3C, the cone OS of +/+ mice were characterized by membrane-rich stacks of discs, whereas cones of TgRBD3*-HA::−/− mice had lacunae with highly electron-lucent lumina within regions of OS. Remnants of membrane debris were also seen in the lacunae of OS of TgRBD3*-HA::−/− (Fig. 3C, right panel).
Figure 3.
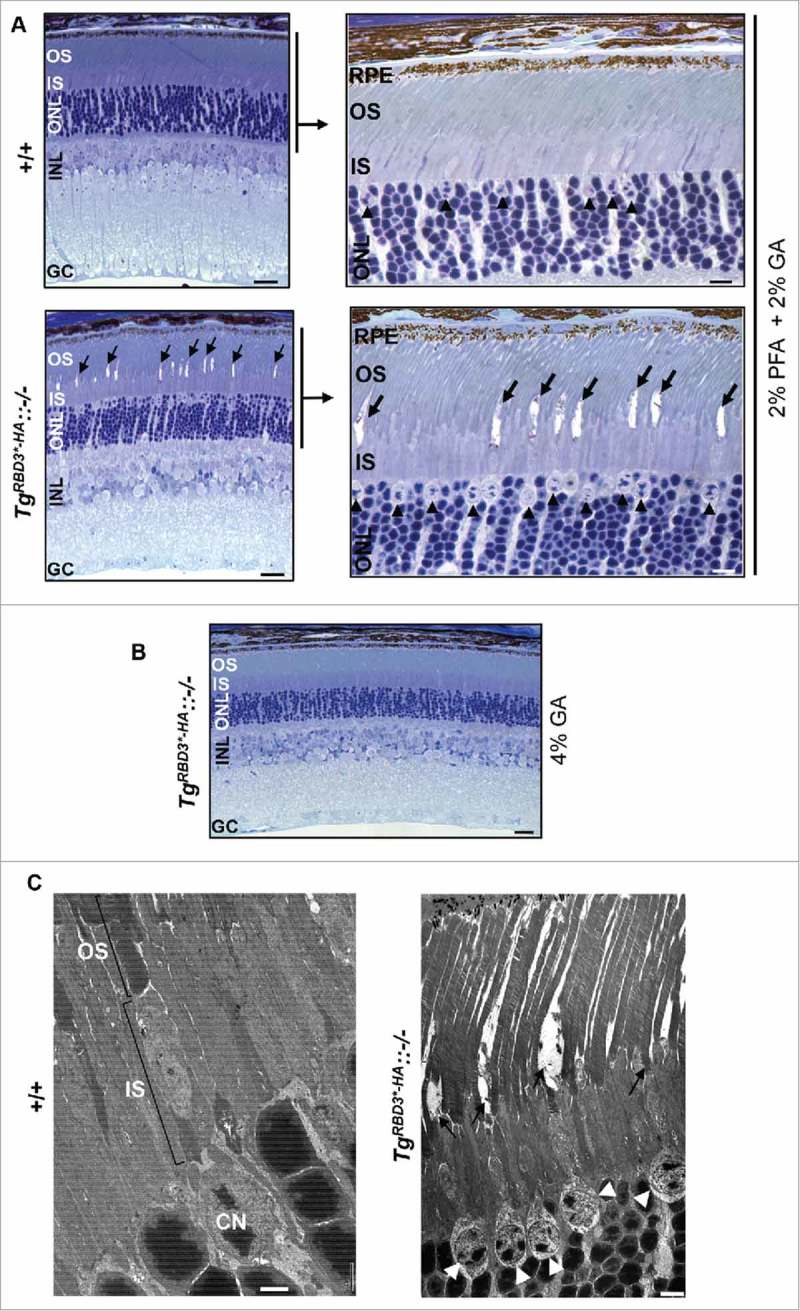
Light and ultrastructural microscopy of retinas and photoreceptors of +/+ and TgRBD3*-HA::−/− mice. (A, B) Low (A, left panels) and high magnifications (A, right panels) of light microscopy images of methylene blue-stained sections of retinae of 5-month old mice perfused and fixed in 2% paraformaldehyde and 2% glutaraldehyde (A) and 4% glutaraldehyde (B). Note that compared with +/+ mice and when fixed with 2% paraformaldehyde and 2% glutaraldehyde, cone photoreceptors of TgRBD3*-HA::−/− mice present prominent lacunae in OS (arrows) (A), whereas the lacunae in OS are greatly reduced in cones of TgRBD3*-HA::−/− by fixation with 4% glutaraldehyde (B). Black arrowheads point to euchromatic nuclei of cone photoreceptors typically localized in the distal margin of the ONL (A, right panels). (C) Ultrastructural images of photoreceptors of 5-month old mice acquired by transmission electron microscopy. Electrolucent lumina of prominent lacunae in the outer segments (OS) of cone photoreceptors are present only in TgRBD3*-HA::−/− mice. White arrowheads point to euchromatic nuclei of cone photoreceptors with electrolucent chromatin and they contrast with the heterochromatic and electrodense nuclei of rod photoreceptors. Legend: OS, outer segments; IS, inner segments; ONL, outer nuclear layer (nuclei of photoreceptors); INL, inner nuclear layer (nuclei of inner retinal neurons); GC, ganglion neurons; RPE, retinal pigment epithelium; CN, cone nucleus; GA, glutaraldehyde; PA, paraformaldehyde; TgRBD3*-HA, Tg-Ranbp2RBD3*-HA; −/−, Ranbp2−/−; +/+, wild-type. Scale bars: 50 μm (A, left panel; B), 10 μm (A, right panel), 2 μm (C, left panel); 4 μm (C, right panel).
As noted previously, our prior work showed that Ranbp2 interacts and localizes also with mitochondrial proteins14 and its tripartite domain, RBD2-KBD-RBD3, modulates directly the motor activity of kinesin-1 and mitochondrial motility in a Ran-GTP-independent manner.10-12 In this study, we also found that the subcellular localization of another partner of Ranbp2, ubc9, was enriched in the ellipsoid compartment of photoreceptors (Fig. 2A), where mitochondria are clustered. We (and others) have consistently observed at least 2 isoforms of ubc9 that resolve in SDS-PAGE with apparent molecular masses below and above 25 and 32 kDa, respectively (Fig. 4A).21,29 The 36 kDa specie is thought to represent an ubc9-SUMO thioester specie or ubc9 dimers that are sensitive to reducing agents, whereas the small molecular mass of ubc9 (∼17 kDa) is unmodified ubc9.21 To gain better understanding of the nature of the ubc9 specie(s) that localize to the mitochondrial-rich (ellipsoid) compartment of photoreceptors, we examined the distribution of ubc9 species between mitochondrial and cytosolic fractions of retinae. These subcellular fractionation assays showed that the ubc9 isoform of ∼36 kDa was exclusively localized to the mitochondrial fraction (Fig. 4B). Conversely, the ubc9 isoform of ∼17 kDa was excluded from mitochondrial fraction and it was found instead in the cytosolic fraction (Fig. 4B). These observations were also recapitulated with subcellular fractions of cultured NIH 3T3 fibroblasts (Fig. 4C, left panel). We extended also the subcellular partition analysis to Ran GTPase, because our prior work showed that the control of mitochondrial motility and kinesin-1 activation by Ranbp2 is independent of Ran-GTP-binding to Ranbp2.10 We found that the unmodified and SUMOylated isoforms of Ran GTPase of 24 and 34 kDa, respectively,22 were excluded from the mitochondrial fraction and were found instead in the cytosolic fraction (Fig. 4C, middle panel). These data cement the notion of Ran GTPase-independent and ubc9-dependent roles in mitochondrial function.
Figure 4.
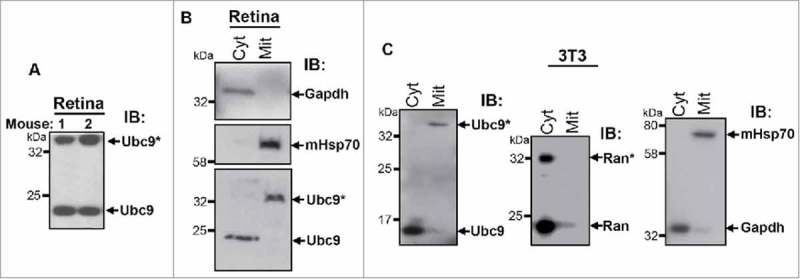
Identification of an ubc9 isoform, which selectively localizes to the mitochondrial fraction. (A) Immunoblot of ubc9 of mouse retinal homogenates shows that ubc9 resolves in SDS-PAGE as 2 species of molecular masses of < 25 and >35 kDa. (B) Immunoblot of Ubc9 of subcellular fractions of mouse retinal extracts shows that the low 17 kDa (ubc9) and high molecular ∼36 kDa isoforms (ubc9*) of ubc9 localize to the cytosolic (Cyt) and mitochondrial fractions (Mit), respectively. Gadph and mHsp70 are cytosolic and mitochondrial markers, respectively. (C) Immunoblot of Ubc9 of subcellular fractions of 3T3 fibroblast extracts shows that the ∼17 (ubc9) and ∼36 kDa isoforms (ubc9*) of ubc9 localize to the cytosolic (Cyt) and mitochondrial fractions (Mit), respectively (left panel). The 24 and 34 kDa isoforms of unmodified (Ran) and SUMOylated Ran GTPase (Ran*), respectively, are excluded from the mitochondrial fraction and both are found in the cytosolic fraction (middle panel). Gadph and mHsp70 are cytosolic and mitochondrial markers, respectively (right panel). Legend: Ran, Ran GTPase; Gadph, glyceraldehyde 3-phosphate dehydrogenase; mHsp70, mitochondrial heat shock protein 70.
To gain further understanding of the localization of Ranbp2 and some of its partners with the mitochondria, we performed co-immunoprecipitation assays with retinal extracts and purified mitochondria and multiple antibodies raised against expressed domains of Ranbp2 (Fig. 5A) and mitochondrial proteins. Some of these mitochondrial proteins were previously found to be partners or substrates of Ranbp2 and their activities were controlled by Ranbp2 (e.g., cox11 and HK1).14 Consistent with other studies,23 the monoclonal antibody, mAb414, recognizes 3 nucleoporins, Ranbp2/Nup358, Nup153 and Nup62 (Fig. 5B). Control immunoprecipitation reactions with mAb414 followed by immunoblot analyses with an antibody against the ZnF domain of Ranbp2, ZnF Ab09 (Fig. 5A), shows that as anticipated the ZnF Ab09 is specific toward Ranbp2 only (Fig. 5C, lane 2). However, the ZnF Ab09 recognizes 2 immunological isoforms of Ranbp2 with apparent molecular masses of ∼358 and 80 kDa (hereafter denoted as Ranbp2i358 and Ranbp2i80, respectively) in co-immunoprecipitation assays with ZnF Ab09 (Fig. 5C, lane 3) and with another antibody, KBD Ab41, against the KBD domain of Ranbp2 (Fig. 5C, lane 4). By contrast, co-immunoprecipitation reactions with antibodies against mHsp70, cox11 and HK1, followed by immunoblot analysis with the ZnF Ab09 showed that mHsp70 and cox11 co-precipitate Ranbp2i80 only (Fig. 5C, lanes 5 and 6), while HK1 did not co-precipitate Ranbp2i80 (Fig. 5C, lane 7). To further probe the immunological properties of Ranbp2i80, we examined co-precipitates of mHsp70 with KBD Ab41 (Fig. 5D) and ZnF Ab09, which was pre-blocked with its ZnF antigen (Fig. 5E). To this effect, KBD Ab41 detected also Ranbp2i80 (Fig. 5D), whereas the blocked ZnF Ab09 failed to detect Ranbp2i80 (Fig. 5E). Then, we purified mitochondria by differential ultracentrifugation and immunoprecipitated purified mitochondria with cox11 and mHsp70 antibodies followed by immunoblot analysis with another antibody, ZnF Ab08 (Fig. 5A), which was independently raised against ZnF of Ranbp2.24 These assays showed that Ranbp2i80 was present in the mitochondrial fraction and that Ranbp2i80 co-precipitated with cox11 and mHsp70 (Fig. 5F). Finally, we performed reciprocal co-immunoprecipitation assays with ZnF Ab08 and KBD Ab41 followed by immunoblot analysis with mHsp70 antibody. These assays showed also that mHsp70 co-precipitated with Ranbp2i80 (Fig. 5G).
Figure 5.
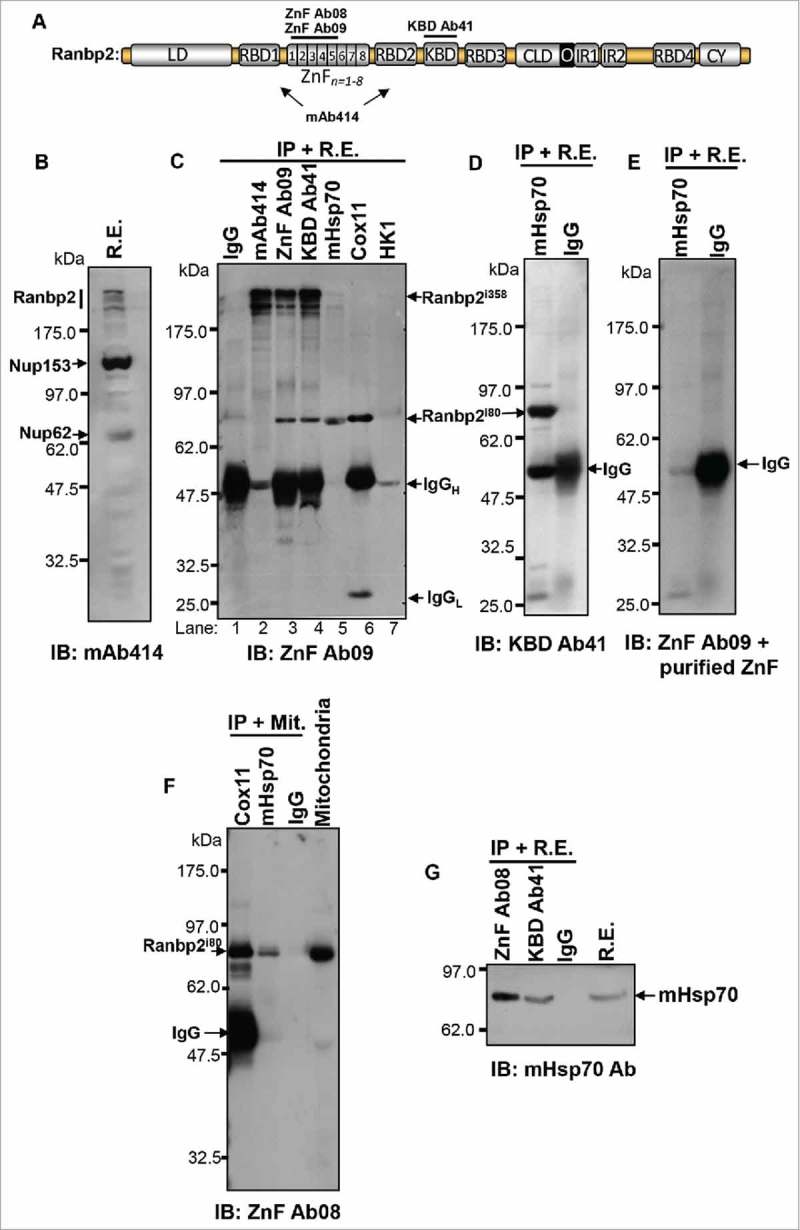
Identification of a Ranbp2 isoform, which selectively associates with mitochondrial markers and localizes to the mitochondrial fraction. (A) Antibodies against Ranbp2 and selective domains thereof. The antibodies, ZnF Ab08 and ZnF Ab09, were raised against the first 5 zinc-finger (ZnF) domains of Ranbp2, KBD Ab41 was raised against the KBD domain of Ranbp2 and mAb414 recognizes undefined epitopes shared by Ranbp2 (Nup358), Nup68 and Nup153. (B) The mAb414 recognizes Ranbp2 (Nup358), Nup68 and Nup153 in retinal homogenates. (C) The ZnF Ab09 recognizes a low molecular mass isoform of Ranbp2 of ∼80 kDa (Ranbp2i80) in co-immunoprecipitates of retinal extracts with the Ranbp2 antibodies, ZnF Ab09 and KBD Ab41, and antibodies against the mitochondrial proteins, mHsp70 and cox11. The large 358 kDa isoform of Ranbp2 (Ranbp2i358) is not co-immunoprecipitated from extracts by the antibodies against mHsp70, cox11and HK1 (lanes 5, 6 and 7). HK1 antibody does not co-immunoprecipitate Ranbp2i80 (lane 7). Ranbp2i358 is co-immunoprecipitated from retinal extracts with the antibodies, mAb414, ZnF Ab09 and KBD Ab41. (D) The Ranbp2 antibody, KBD Ab41, detects only the small Ranbp2i80 in co-immunoprecipitates of retinal extracts with the mHsp70 antibody. (E) Blocking of antibody, ZnF Ab09, with purified recombinant ZnF protein abolishes the detection of Ranbp2i80 co-immunoprecipitated from retinal extracts with mHsp70 antibody. (F) The Ranbp2 antibody, ZnF Ab09, detects only the Ranbp2i80 isoform in purified mitochondria (Mit) immunoprecipitated with antibodies against mHsp70 and cox11. (G) mHsp70 is co-immunoprecipitated from retinal extracts by the Ranbp2 antibodies, ZnF Ab09 and KBD Ab41. IgG (rabbit) is a control antibody in C-G. The murine origin of the monoclonal antibodies, mAb414, mHsp70 and HK1, leads to weak signals of IgGH in panels, (C)and F, owing to poor cross-reactivity with the goat anti-rabbit enzyme (HRP)-conjugated secondary antibody. Legends: R.E., retinal extracts; IgGH, IgG heavy-chain; IgGL, IgG light-chain; IB, immunoblot; IP, immunoprecipitation; HK1, hexokinase-1; mHsp70, mitochondrial heat shock protein 70.
Our prior work has also shown that ectopic expression of KBD of Ranbp2 in cell cultures specifically promotes the perinuclear clustering of mitochondria and stimulates mitochondrial motility by regulation of the coupling of KBD of Ranbp2 with the mitochondrial and microtubule-based motor protein, kinesin-1.10-12,24 Since co-immunoprecipitation experiments showed that the ZnF and KBD domains of Ranbp2 are shared by the Ranbp2i358 and Ranbp2i80 isoforms (Fig. 5C), we examined the colocalization of Ranbp2 to the mitochondria with the antibody, ZnF Ab09 (Fig. 6A), and the functional effect(s) of intracellular delivery of KBD Ab41 on distribution of mitochondria and Ranbp2 in the live cone photoreceptor cell line, 661W (Fig. 6B, C).25,26 As shown in Fig. 6A, there was extensive colocalization of Ranbp2 with archetypal worm-like mitochondria in 661W cells. Further, liposome-mediated delivery of KBD Ab41 to 661W cells promoted the prominent fragmentation and clustering of mitochondria (Fig. 6B, g; C, e) and the delocalization of Ranbp2 from the mitochondria (Fig. 6B, h; C, f), but without affecting the localization of Ranbp2 at the nuclear rim (Fig. 6B, f; C, d). By contrast, mock-liposome transfected cells showed widespread colocalization of Ranbp2 with worm-like mitochondria (Fig. 6B, b-d; C, b and c). The fragmentation of mitochondria and their delocalization from Ranbp2 also resulted in widespread atrophy of cells transfected with KBD Ab41 (Fig. 6B, e), but not of mock transfected cells (Fig. 6B, a). Hence, these results provide additional mechanistic evidence of an isoform of Ranbp2, which shares the ZnF and KBD domains, in the regulation of mitochondrial distribution and function.
Figure 6.
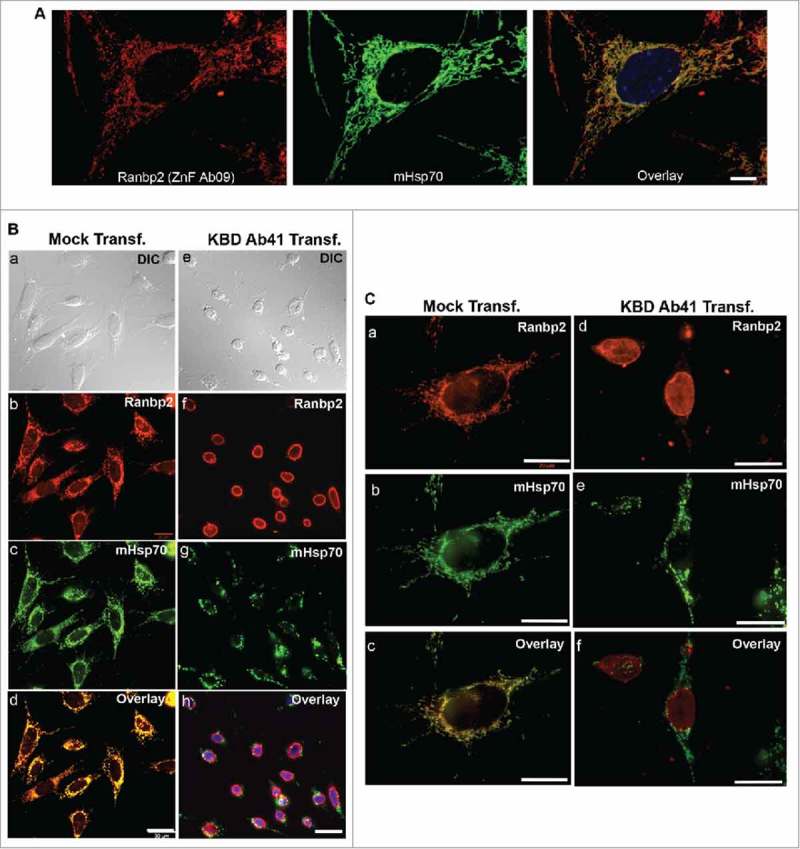
Liposome-mediated delivery of anti-KBD antibody of Ranbp2 into 661W neurons leads to mitochondrial fragmentation, delocalization of RanBP2 from mitochondria and neuronal atrophy. (A) Ranbp2 colocalizes extensively with mHsp70-immunostained worm-like mitochondria in 661W neurons. Ranbp2 was detected with ZnF Ab09. (B) Ranbp2 colocalizes extensively with mHsp70-immunostained worm-like mitochondria in liposome mock-treated cells and cells present healthy morphology (a-d). Liposome-mediated delivery of KBD Ab41 into 661W neurons (e-h) causes strong neural atrophy (e), mitochondrial fragmentation (g) and the delocalization of Ranbp2 from the mitochondria (h) without affecting the localization of Ranbp2 at the nuclear rim (f). Internalized KBD Ab41 was detected with Alexa-conjugated goat anti-rabbit antibody (f). (C) High-magnification images of liposome mock and KBD Ab41-treated 661W neurons immunostained for Ranbp2 and mHsp70 as shown in image panels of B. Legends: KBD Ab41 Transf., cells with liposome-mediated delivery of Ranbp2 antibody, KBD Ab41; Mock Transf., liposome mock-treated cells. DIC, differential interference contrast (Nomarski optics). Scale bars: 10 μm (A), 30 μm (B), 20 μm (C).
Here we show that loss of Ran-GTP-binding to RBD3 alone of Ranbp2 promotes the ordinary and age-dependent increase of ubiquitylated substrates and S1 subunit of the 19S cap of the proteasome without causing overt morphological manifestations in tissues of varied cellular heterogeneity, such as retina and RPE, while specifically rendering the OS of cone photoreceptors susceptible to structural disintegration before a rise of total ubiquitin levels ensued in the retina. Hence, the coupling of the co-activation of Ran GTPase by RBD3 of Ranbp2 controls primarily functions related to the structural maintenance of cone photoreceptors and secondarily broader and age-dependent functions in ubiquitin proteostasis across tissues. Our prior studies have shown that the age-dependent functions of Ranbp2 are the result of a confluence of activities of various domains of Ranbp2 and partners thereof and aging or environmental stressors.27-30 Future studies are needed to parse the molecular bases of the age-dependent activities of Ranbp2.
Our previous studies with cell cultures have shown that deletion of RBD3 from the neighboring domains of Ranbp2, CLD and IR1+2, that specifically bind to subunits of the 19S cap of the proteasome and ubc9 (Fig. 1A), respectively, cause the accumulation of ubiquitylated and properly folded reporter substrates in live cells;17 thus indicating that RBD3 controls functions of the 19S cap of the proteasome. Physiologically, this and other lines of works suggest that the selective loss of Ran GTPase co-activation by RBD3 of Ranbp2 together with the age-dependent increase in oxidative stress reduce proteasome activity owing likely to a decline in competency of nucleocytoplasmic shuttling and partition of substrates by Ran GTPase.31-34 This decline likely promotes the age-dependent accumulation of selective substrates and the compensatory upregulation of 19S cap subunits and ubiquitylated substrates.
In rod photoreceptors, the degeneration of OS is invariably followed by the death of these neurons in mouse models and humans with retinal degenerative diseases, such as retinitis pigmentosa.35-37 However, we have recently shown in mice that loss of the SUMO-binding motif of Ranbp2 can stave off rod photoreceptor death from OS degeneration evoked by light-stress.27 In cone photoreceptors, mounting evidence support that these photosensory neurons present distinct cell-death mechanisms from rods19 and that degeneration of the OS of cones do not trigger cone death.38,39 For example, loss of the Drosha/Dgcr8 miRNA-processing machinery in cone photoreceptors of adult mice results in the degeneration of the OS without loss of cone photoreceptors.38 Consistent with this observation, our study found that the structural disintegration of OS of TgRBD3*-HA::−/− mice did not promote changes in the number of M- and S-cones. The mechanisms by which RBD3 controls specifically the structural integrity of cone photoreceptors needs to be worked out. However, mounting evidence supports that Ranbp2 controls the levels of free fatty acids in retinal and spinal motor neurons,16,29,30 and the cell type-dependent nucleocytoplasmic partition and proteostasis of selective heterogeneous nuclear ribonucleic proteins, such as hnRNPA2B1 and hnRNPH3.16,28 Further, other recent findings support that SUMOylated hnRNPA2B1 binds and controls the loading of miRNAs to exosomes40 and that several SUMO-binding motifs of Ranbp2 bind to argonaute protein and thereby promoting the coupling of miRNA-induced silencing complex (miRISC) with target mRNAs.41 The biologic relevance of the promiscuous binding of argonaute to SUMO-binding motifs of Ranbp2 and other proteins is unknown41 and such binding contrasts with the physiologic and biologic control of SUMOylated RanGAP, HDAC4 and the ubiquitin-proteasome system by a specific SUMO-binding motif of Ranbp2.7,17,27 Regardless, we have parenthetically found that loss of Ranbp2 also causes the strong upregulation of pre-miRNA variants of miR-124a in cones19 and the strong downregulation of miRNAs, such as miR-224 and mi-R200b, in the RPE.7 Further, the nucleocytoplasmic partition of hnRNPA1 controls the Sterol Regulatory Element-Binding Protein 1a (SREBP-1a)-mediated expression of lipogenic genes implicated in the synthesis of fatty acids, triglycerides and cholesterol.42 Taken together, these data suggest an attractive model by which the RBD3 of Ranbp2 controls the maintenance of OS of cones by regulating the nucleocytoplasmic transport of cone-selective factors with roles in gene silencing of selective lipogenic genes that are important to OS maintenance. Importantly, this study also demonstrates that it is possible to parse genetically and physiologically Ran-GTP-dependent roles of RBD2 and RBD3 of Ranbp2 in developmental and mature functions of cones. Specifically, uncoupling Ran GTPase from RBD2 and RBD3 causes deficits in developing cones, while loss of Ran-GTP-dependent function of RBD3 alone affects the structural integrity of OS of mature cones without causing developmental deficits in the number of cones.
Finally, our study found the existence and association of an ubc9 isoform of ∼36 kDa and of a novel and smaller isoform of Ranbp2, Ranbp2i80, with the mitochondria and components thereof, whereas Ran GTPase was excluded from the mitochondrial fraction. Additional studies are needed to characterize exactly the Ranbp2i80 isoform in the mitochondrial fraction, the nature of the post-translation modification(s) of ubc9 associated to mitochondria and the interplay between the mitochondrial-associated Ranbp2i80 and ubc9 isoforms in mitochondrial motility and functions. It is possible that Ranbp2i80 represents a splice-variant of Ranbp2, since alternative splice variants of Ranbp2 have been isolated.43 Regardless, the association of Ranbp2i80 with mitochondrial substrates, such as cox11, poses an apparent topological conundrum, because cox11 is a nuclear-encoded protein resident in the intermembrane space of mitochondria.44 It is possible that the Ranbp2i358 and Ranbp2i80 isoforms perform complementary roles in co-chaperoning the transport or import of selective substrates to the mitochondria or that these interactions take place at closely apposed inner and outer mitochondrial membranes, where mitochondrial protein import occurs. Further, modification of ubc9 by SUMOylation or other factors guided by Ranbp2i80 may promote discrimination of ubc9 substrates that control mitochondrial fission or kinesin-1-mediated mitochondrial motility events by regulation of the interaction of the KBD of Ranbp2i80 with microtubule-based motor and kinesin-1 isoforms, KIF5B/KIF5C.10-12,24,45 The discovery of novel immunological-related isoforms of ubc9 and Ranbp2 to the mitochondria will open new avenues to unanticipated moonlighting roles of these proteins or domains thereof in mitochondrial biogenesis, motility or function(s).
Materials and methods
Transgenic mice
The generation of Ranbp2−/− and transgenic mice, TgRBD3*-HA::−/− and TgRBD2/3*-HA::cone-cre::−/−, by BAC recombineering were described previously.7,14,19 Mice were in a mixed genetic background. Animal protocols were approved by the Institutional Animal Care and Use Committees at Duke University in adherence to NIH guidelines for the care and use of laboratory animals and ARVO guidelines for the Use of Animals in Vision Research. Mice were reared in a pathogen-free barrier facility and cyclic light (12h:12h; <70 lx) and given ad libitum access to water and chow diet 5LJ5 (Purina, Saint Louis, MO). Mice of either sex were used by this study.
Antibodies
The following and previously characterized antibodies were used for immunofluorescence (IF), immunoprecipitation (IP) or immunoblots (IB): rabbit affinity-purified anti-Ranbp2 (ZnF) antibodies, ZnF Ab09 and ZnF Ab08 [generated by the Ferreira laboratory and previously called ZnF-20909 and ZnF-20908;24 5 μg (IP), 215 ng/ml (IB), 10 μg/ml (IF)]; rabbit affinity-purified anti-Ranbp2 antibody, KBD Ab41 [generated by the Ferreira laboratory and previously called JX41 Ab;24 5 μg (IP), 240 ng/ml (IB), 5 μg/ml (IF)]; rabbit affinity-purified anti-cox11 (no. 4) [generated by the Ferreira laboratory;14 5 μg (IP)]; rabbit anti-L/M opsin no. 21069 [generated by the Ferreira laboratory;15 1:500, (IF)]; mouse mAb414 against nuclear pore complex proteins Ranbp2/Nup358, Nup62 and Nup153 [400 ng/ml (IB), 5 μg (IP), Covance, Emeryville, CA; Cat: MMS-120P]; rabbit anti-CRM1 (1:1,000 (IB), Santa Cruz Biotechnology, Santa Cruz, CA; Cat: sc-5595); mouse anti-Ran GTPase (1:4,000 (IB), BD Biosciences; Cat: 610341); rabbit anti-Stat3 [1:100 (IF), 1:1000 (IB), Cell Signaling, Boston, MA; Cat: 4904]; rabbit anti-Gapdh [1:500 (IB), Santa Cruz Biotechnology, Inc., Santa Cruz, CA; Cat: sc-25778]; rabbit anti-19S proteasome S1 [1:1000 (IB), Thermo Scientific, Rockford, IL; Cat: PA1–973]; mouse anti-GMP-1 [1:1000 (IB), Thermo Scientific, Rockford, IL; Cat: 33–2400]; rabbit anti-S opsin [1:100 (IF) Millipore, Temecula, CA; Cat: 5407]; mouse anti-mtHsp70 [1:600 (IB), 5 μg (IP), 1:50 (IF), Thermo Scientific, Rockford, IL; Cat: MA3–028]; mouse monoclonal anti-HK1 (clone 2B) [gift from J. Wilson, 5 μg (IP)]; mouse anti-ubc9 [1:100 (IF), BD Transduction Laboratories, Cat: 610749]; rabbit anti-cone arrestin (Arr4) [1:100 (IF) Millipore, Temecula, CA; Cat: 15282]; Alexa Fluor-conjugated secondary antibodies (488 and 594) and DAPI were from Invitrogen (Carlsbad, CA); goat horseradish peroxidase-conjugated anti-rabbit IgG [25 ng/ml (IB)] and rabbit IgG [5 μg (IP)] were from Jackson ImmunoResearch Laboratories, Inc., West Grove, PA.
Ubiquitylation assays
Total ubiquitin levels (free ubiquitin and ubiquitylated protein conjugates) in the RPE and retinae were determined by the UbiQuant ELISA kit as directed by the manufacturer and described elsewhere (LifeSensors, Malvern, PA).16
Cell culture and subcellular fractions
NIH 3T3 fibroblasts (passage #133; ATCC, Manassas, VA) and 661W cells25,26 were cultured in Dulbeco's modified Eagle's medium (DMEM; Gibco/Thermo Scientific, Rockford, IL) supplemented with 10% fetal calf serum at 37°C in presence of 5% CO2 and 100% humidity. Confluent fibroblasts were harvested, washed with 1x PBS and cytosolic and mitochondrial subcellular fractions were prepared with the Cell Fractionation Kit (MitoSciences, Eugene, OR; Cat#MS861) as directed by the manufacturer and described elsewhere.19 Subcellular fractions were resolved in premade 5–15% gradient Criterion SDS-polyacrylamide gels (Bio-Rad, Hercules, CA).
GST-pull down assays
GST-RBD3 or GST-RBD3W2186R were expressed and purified from E. coli as described.46 GST–pull down assays were performed with fresh bovine retinae homogenized in CHAPS-homogenization buffer (20 mM Tris-HCl, pH 6.8; 0.25M NaCl, 2mM 2-mercaptoethanol, 0.75% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid; 0.02% sodium azide, 5% glycerol, 200 μM Pefabloc (Roche Applied Science) at 4oC as described.46 Briefly, CHAPS-solubilized retinal extracts (2mg; ∼80 μl) were incubated with 1.1 µM of GST-RBD3 or GST-RBD3W2186R in the presence or absence of guanosine 5′-3-O-(thio)triphosphate (GTPγS; 10 μM) at 4oC on a nutator. After for 20 min, 60 µl of 50% slurry of glutathione-S1-sepharose 4B beads (GE Healthcare) equilibrated in incubation buffer (100 mM NaCl, 50 mM Tris-HCl, pH 7.5, 2 mM MgCl2, 2 mM CaCl2 0.5% CHAPS) were added and incubated for another 20 min at 4oC on a nutator. Sepharose beads were centrifuged and washed thrice in washing buffer (100 mM NaCl, 50 mM Tris-HCl, pH 7.5, 2 mM MgCl2, 2 mM CaCl2 0.2% Triton-X100). Co-precipitates were eluted with Laemmli buffer with 100mM DTT and resolved by electrophoresis in 10% SDS-PAGE with a Hoefer SE600 electrophoresis apparatus (Holliston, MA). Protein concentrations were determined by the Bradford method using BSA as the standard.
Co-immunoprecipitation assays
Co-immunoprecipitation assays were performed with fresh bovine retinae homogenized in Nonidet P-40 homogenization buffer (50 mM Tris-HCl, pH 8.0; 150 mM NaCl, 1.0% Nonidet P-40) at 4oC as described24 or with whole mitochondria purified from bovine retinae and resuspended in 1xPBS. Briefly, Nonidet P-40-solubilized retinal extracts (3.0 mg) were pre-cleared with 2 µg of non-immunized IgG and 50 µl of a 50% sepharose-4B-bead slurry (GE Healthcare). Precleared retinal extracts (3.0 mg) or purified mitochondria (OD260 ∼0.017) were incubated with antibodies (5 µg per reaction) and 40 µl of a 50% sepharose-4B-bead slurry for 60 min at 4oC in a nutator. Co-immunoprecipitates from retinal extracts and bound to sepharose-4B-beads were loaded onto spin filters (CytoSignal, Irvine CA), centrifuged and washed thrice in Nonidet P-40 homogenization buffer. Co-precipitates of purified mitochondria were collected by gravity precipitation of incubation reactions and gentle washing in 1xPBS. Co-precipitates were eluted with Laemmli buffer with 100mM DTT. Immunoprecipitates were resolved by electrophoresis in 9% SDS-PAGE with a Hoefer SE600 electrophoresis apparatus (Holliston, MA). Protein concentrations of retinal extracts were determined by the Bradford method using BSA as the standard.
Immunoblotting and relative protein quantification
Protein complexes purified by GST–pull down and immunoprecipitation assays and cell and tissue extracts were blotted on PVDF membrane after separation by SDS-PAGE as described.46 Membranes were blocked with non-fat dry milk block solution (Bio-Rad, Hercules, CA) and when applicable, blots with pre-stained molecular mass markers were cut and different sections were incubated with different antibodies as described elsewhere. Blots were reprobed for Gapdh or other antibodies when applicable. For antibody blocking, the antibody, ZnF Ab09, was pre-incubated with purified recombinant ZnF of Ranbp2 (20 μg) for 30 min before probing the membrane. Blots were developed with a SuperSignal chemiluminescence substrate (Thermo Scientific, Rockford, IL). Unsaturated band intensities were quantified by densitometry with Metamorph v7.0 (Molecular Devices). Integrated density values (idv) of representative bands were normalized to idv of Gapdh and background as described.7,19
Immunohistochemistry, confocal microscopy and morphometry
Mice were anesthetized with ketamine (100 mg/kg) and xylazine (10 mg/kg) and perfused intra-cardiacally with 2% paraformaldehyde in 1xPBS. The mouse cornea was burned in the superior region with a Low Temperature Cautery (Bovie Medical Corporation, St. Peterburg, FL). Eyes were dissected, lens removed and incubated in 2% paraformaldehyde/1xPBS for 4 h at room temperature, followed by infiltration with 5% sucrose for 1 h and 30% sucrose for 12–16 hr at 4oC. Then, eyes were embedded and frozen in Tissue-Tek O.C.T. compound (Sakura,Torrance) and stored at -80oC. Cryosections (10 μm thick) along the vertical meridian of the eyecup were collected on lysine-coated glass slides with a cryotome (Microm HM550). Retinal flat mounts were prepared by removing retinas from fixed eyeballs and using the caruncle as an orientation landmark. The retinas were cut in a 4-quadrant cloverleaf pattern and fixed in a 24-well plate for 5 min. Cryosections and flatmounts were washed with 1xPBS, permeabilized and blocked in 1xPBS/0.1% Triton X-100/10% normal goat serum followed by incubation with primary antibodies, washes in 1xPBS (thrice), incubation with anti-rabbit or anti-mouse AlexaFluor-488 or AlexaFluor-594-conjugated secondary antibodies and final washes in 1xPBS (thrice) as described.19 Sections were counter-stained with DAPI (Invitrogen, CA). Specimens were mounted on glass slides with Fluoromount-G (Southern Biotech). Images were acquired with a Nikon C1+ laser-scanning confocal microscope coupled with a LU4A4 launching base of 4 solid state diode lasers (407 nm/100 milliwatts, 488 nm/50 milliwatts, 561 nm/50 milliwatts, and 640 nm/40 milliwatts) and controlled by Nikon EZC1.3.10 software (version 6.4). Morphometric analyses of outer segments of M- and S-cones that were immunostained for M or S-opsin in retinal flat mounts were performed from 3 image fields of ∼127 × 127 μm from each retinal region. The optical slices were 3-dimensionally reconstructed for the whole length of outer segments (∼19–24 μm, step size of 0.5 μm) with the post-acquisition Nikon Elements AR software (version 4.0).
Semi-thin sections and transmission electron microscopy
Mice were anesthetized like described previously, perfused intra-cardiacally with 2.0% glutaraldehyde and 2% paraformaldehyde (1:1) or 4.0% glutaraldehyde in 0.1 M sodium cacodylate buffer, pH 7.4 followed by fixation of the eyes in the same fixative buffer used for perfusion overnight at 4oC. Semi-thin sections (0.5 μm) were cut along the vertical meridian, mounted on glass slides and stained with 1% methylene blue. Images were captured with an Axiopan-2 light microscope controlled by Axovision Rel 4.6 and coupled to an AxioCam HRc digital camera (Carl Zeiss, Germany). For electron microscopy, specimens were post-fixed in 2% osmium tetraoxide in 0.1% cacodylate buffer and embedded in Spurr resin. Ultrathin sections (60 nm-thick) were cut with a Leica Ultracut S (Leica Microsystems, Waltzer, Germany), stained with 2% uranyl acetate and 4% lead citrate and examined on a Phillips BioTwin CM120 electron microscope equipped with Gatan Orius and Olympus Morada digital cameras. Images of high magnification were stitched with Photoshop (Adobe Corp)
Preparation of retinal and 3T3 fibroblast subcellular fractions
Cytosolic and mitochondrial subcellular fractions of mouse retinae and 3T3 fibroblasts were prepared with the Cell Fractionation Kit (MitoSciences, Eugene, OR; Cat#MS861) as directed by the manufacturer and described elsewhere.19 For analyses of Ranbp2 complexes immunoprecipitated from mitochondria, crude mitochondrial fractions were purified from bovine retinae homogenized in TKM buffer (0.25M sucrose in 50mM Tris, pH 7.5 containing 25mM KCl, 5mM MgCl2) and ultracentrifuged in a discontinuous sucrose gradient as described elsewhere.47 The absorbance at OD260 of purified mitochondria was determined. Mitochondrial, retinal and cell culture homogenates were prepared in RIPA buffer [25mM Tris, pH 8.2, 50mM NaCl, 0.5% Nonidet P-40 (NP-40), 0.5% deoxycholate and 0.1% SDS] containing 10mM iodoacetamide and concentrations were determined by the BCA method using BSA as the standard.
Liposome-mediated delivery of antibodies into 661W cells and microscopy
For liposome-mediated delivery of the Ranbp2 antibody, KBD Ab41, the ProVectin kit (Imgenex, San Diego, CA) was used as per manufacturer's instructions. About 10 μg of antibody was used at ∼100 μg/ml. Mock lipofections were performed exactly the same way with the exception that KBD Ab41 was omitted from the Provectin reagent. Upon lipofection, cells were placed back in the incubator with fresh conditioned medium at 37°C in presence of 5% CO2 and 100% humidity. About 12 hours later, cells were fixed with 2% paraformaldehyde/1xPBS and processed for immunocytochemistry as described elsewhere.14 For the localization of the delivered KBD Ab41 into cells, only Alexa fluorescent-conjugated goat anti-rabbit secondary antibody (2.5 μg/ml; Molecular Probes) was used after fixation of cells. mHsp70 in KBD Ab41 and mock-treated cells was detected with mouse monoclonal anti-mtHsp70 (2.5 μg/ml). Ranbp2 in mock transfected cells was detected with KBD Ab41 (2.5 μg/ml). Cells and proteins were visualized by DIC/Nomarski and wide-field epifluorescence microscopy, respectively, on a TE2000U inverted research microscope, equipped with Nomarski optics, Plan Apochromat objectives with NA of 1.4 and appropriate excitation and emission filter wheels. Images were captured at non-saturating integration levels and similar exposure times for each type of staining, 12-bit mono black/white and then pseudocolored. Images were captured with a charge-coupled device camera (CoolSNAP HQ; Ropers Scientific). Images were acquired with METAMORPH Software v6.3 (Molecular Devices).
Statistical analyses
Student's t-test for 2 groups was used. Data are reported as average values ± SD. Differences among the groups were considered statistically significant when p-value ≤ 0.05.
Abbreviations
- Ranbp2
Ran-binding protein 2
- RBD3
Ran-binding domain 3
- KBD
kinesin-binding domain
- ZnF
zinc-finger-rich domain
- Ran
Ras-related nuclear protein
- Ran GAP
Ran GTPase-activating protein
- ubc9
ubiquitin-conjugating enzyme E2I (ubiquitin-conjugating protein 9)
- Arr4
cone arrestin
- RPE
retinal pigment epithelium
- NPC
nuclear pore complex
- HK1
hexokinase type 1
- mHsp70
mitochondrial heat shock protein 70
- Nup
nucleoporin
- TgRBD3*-HA::−/−
transgenic BAC of Ranbp2 with a Ran-GTP-binding mutation in RBD3 and expressed in a null Ranbp2 background (Ranbp2−/−)
- TgRBD2/3*-HA::cone-cre::−/−
transgenic BAC of Ranbp2 with Ran-GTP-binding mutations in RBD2 and RBD3 and expressed in cones with a null Ranbp2 background
- +/+
wild-type
- −/−
Ranbp2−/−
- BAC
bacterial artificial chromosome
- OS
outer segment of photoreceptor neurons.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Ying Hao for help with the processing of the specimens for transmission electron microscopy (Duke University, Durham, NC).
Funding
The study was funded by National Institutes of Health Grants EY019492 and GM083165 to P.A.F.
References
- [1].Delphin C, Guan T, Melchior F, Gerace L. RanGTP targets p97 to RanBP2, a filamentous protein localized at the cytoplasmic periphery of the nuclear pore complex. Mol Biol Cell. 1997;8(12):2379-90. doi: 10.1091/mbc.8.12.2379. PMID:9398662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Vetter IR, Nowak C, Nishimoto T, Kuhlmann J, Wittinghofer A. Structure of a Ran-binding domain complexed with Ran bound to a GTP analogue: implications for nuclear transport. Nature. 1999;398(6722):39-46. doi: 10.1038/17969. PMID:10078529 [DOI] [PubMed] [Google Scholar]
- [3].Hamada M, Haeger A, Jeganathan KB, van Ree JH, Malureanu L, Walde S, Joseph J, Kehlenbach RH, van Deursen JM. Ran-dependent docking of importin-beta to RanBP2/Nup358 filaments is essential for protein import and cell viability. J Cell Biol. 2011;194(4):597-612. doi: 10.1083/jcb.201102018. PMID:21859863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Ritterhoff T, Das H, Hofhaus G, Schroder RR, Flotho A, Melchior F. The RanBP2/RanGAP1*SUMO1/Ubc9 SUMO E3 ligase is a disassembly machine for Crm1-dependent nuclear export complexes. Nat Commun. 2016;7:11482. doi: 10.1038/ncomms11482. PMID:27160050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Singh BB, Patel HH, Roepman R, Schick D, Ferreira PA. The zinc finger cluster domain of RanBP2 is a specific docking site for the nuclear export factor, exportin-1. J Biol Chem. 1999;274(52):37370-8. doi: 10.1074/jbc.274.52.37370. PMID:10601307 [DOI] [PubMed] [Google Scholar]
- [6].Villa Braslavsky CI, Nowak C, Gorlich D, Wittinghofer A, Kuhlmann J. Different structural and kinetic requirements for the interaction of Ran with the Ran-binding domains from RanBP2 and importin-beta. Biochemistry (Mosc). 2000;39(38):11629-39. doi: 10.1021/bi001010f [DOI] [PubMed] [Google Scholar]
- [7].Patil H, Saha A, Senda E, Cho KI, Haque M, Yu M, Qiu S, Yoon D, Hao Y, Peachey NS, et al. Selective Impairment of a subset of Ran-GTP-binding domains of Ran-binding protein 2 (Ranbp2) suffices to recapitulate the degeneration of the Retinal Pigment Epithelium (RPE) triggered by Ranbp2 ablation. J Biol Chem. 2014;298:29767-89. doi: 10.1074/jbc.M114.586834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Neilson DE, Adams MD, Orr CM, Schelling DK, Eiben RM, Kerr DS, Anderson J, Bassuk AG, Bye AM, Childs AM, et al. Infection-triggered familial or recurrent cases of acute necrotizing encephalopathy caused by mutations in a component of the nuclear pore, RANBP2. Am J Hum Genet. 2009;84(1):44-51. doi: 10.1016/j.ajhg.2008.12.009. PMID:19118815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Neilson DE, Eiben RM, Waniewski S, Hoppel CL, Varnes ME, Bangert BA, Wiznitzer M, Warman ML, Kerr DS. Autosomal dominant acute necrotizing encephalopathy. Neurology. 2003;61(2):226-30. doi: 10.1212/01.WNL.0000073544.28775.1A. PMID:12874403 [DOI] [PubMed] [Google Scholar]
- [10].Patil H, Cho KI, Lee J, Yang Y, Orry A, Ferreira PA. Kinesin-1 and mitochondrial motility control by discrimination of structurally equivalent but distinct subdomains in Ran-GTP-binding domains of Ran-binding protein 2. Open Biol. 2013;3(3):120183. doi: 10.1098/rsob.120183. PMID:23536549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Cho KI, Yi H, Desai R, Hand AR, Haas AL, Ferreira PA. RANBP2 is an allosteric activator of the conventional kinesin-1 motor protein, KIF5B, in a minimal cell-free system. EMBO reports. 2009;10(5):480-6. doi: 10.1038/embor.2009.29. PMID:19305391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Cho KI, Cai Y, Yi H, Yeh A, Aslanukov A, Ferreira PA. Association of the kinesin-binding domain of RanBP2 to KIF5B and KIF5C determines mitochondria localization and function. Traffic. 2007;8:1722-35. doi: 10.1111/j.1600-0854.2007.00647.x. PMID:17887960 [DOI] [PubMed] [Google Scholar]
- [13].Tanaka Y, Kanai Y, Okada Y, Nonaka S, Takeda S, Harada A, Hirokawa N. Targeted disruption of mouse conventional kinesin heavy chain, kif5B, results in abnormal perinuclear clustering of mitochondria. Cell. 1998;93(7):1147-58. doi: 10.1016/S0092-8674(00)81459-2. PMID:9657148 [DOI] [PubMed] [Google Scholar]
- [14].Aslanukov A, Bhowmick R, Guruju M, Oswald J, Raz D, Bush RA, Sieving PA, Lu X, Bock CB, Ferreira PA. RanBP2 modulates Cox11 and hexokinase I activities and haploinsufficiency of RanBP2 causes deficits in glucose metabolism. PLoS Genet. 2006;2(10):e177. doi: 10.1371/journal.pgen.0020177. PMID:17069463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Mavlyutov TA, Cai Y, Ferreira PA. Identification of RanBP2- and kinesin-mediated transport pathways with restricted neuronal and subcellular localization. Traffic. 2002;3(9):630-40. doi: 10.1034/j.1600-0854.2002.30905.x. PMID:12191015 [DOI] [PubMed] [Google Scholar]
- [16].Cho KI, Yoon D, Qiu S, Danziger Z, Grill WM, Wetsel WC, Ferreira PA. Loss of Ranbp2 in motoneurons causes disruption of nucleocytoplasmic and chemokine signaling, proteostasis of hnRNPH3 and Mmp28, and development of amyotrophic lateral sclerosis-like syndromes. Dis Model Mech. 2017;10(5):559-79. doi: 10.1242/dmm.027730. PMID:28100513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Yi H, Friedman JL, Ferreira PA. The cyclophilin-like domain of Ran-binding protein-2 modulates selectively the activity of the ubiquitin-proteasome system and protein biogenesis. J Biol Chem. 2007;282:34770-8. doi: 10.1074/jbc.M706903200. PMID:17911097 [DOI] [PubMed] [Google Scholar]
- [18].Ferreira PA, Yunfei C, Schick D, Roepman R. The cyclophilin-like domain mediates the association of Ran-binding protein 2 with subunits of the 19 S regulatory complex of the proteasome. J Biol Chem. 1998;273(38):24676-82. doi: 10.1074/jbc.273.38.24676. PMID:9733766 [DOI] [PubMed] [Google Scholar]
- [19].Cho KI, Haque M, Wang J, Yu M, Hao Y, Qiu S, Pillai IC, Peachey NS, Ferreira PA. Distinct and atypical intrinsic and extrinsic cell death pathways between photoreceptor cell types upon specific ablation of Ranbp2 in cone photoreceptors. PLoS Genet. 2013;9(6):e1003555. doi: 10.1371/journal.pgen.1003555. PMID:23818861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Belazi D, Sole-Domenech S, Johansson B, Schalling M, Sjovall P. Chemical analysis of osmium tetroxide staining in adipose tissue using imaging ToF-SIMS. Histochem Cell Biol. 2009;132(1):105-15. doi: 10.1007/s00418-009-0587-z. PMID:19319557 [DOI] [PubMed] [Google Scholar]
- [21].Bossis G, Melchior F. Regulation of SUMOylation by reversible oxidation of SUMO conjugating enzymes. Mol Cell. 2006;21(3):349-57. doi: 10.1016/j.molcel.2005.12.019. PMID:16455490 [DOI] [PubMed] [Google Scholar]
- [22].Sakin V, Richter SM, Hsiao HH, Urlaub H, Melchior F. Sumoylation of the GTPase Ran by the RanBP2 SUMO E3 Ligase Complex. J Biol Chem. 2015;290(39):23589-602. doi: 10.1074/jbc.M115.660118. PMID:26251516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Davis LI, Blobel G. Nuclear pore complex contains a family of glycoproteins that includes p62: glycosylation through a previously unidentified cellular pathway. Proc Natl Acad Sci U S A. 1987;84(21):7552-6. doi: 10.1073/pnas.84.21.7552. PMID:3313397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Cai Y, Singh BB, Aslanukov A, Zhao H, Ferreira PA. The docking of kinesins, KIF5B and KIF5C, to Ran-binding protein 2 (RanBP2) is mediated via a novel RanBP2 domain. J Biol Chem. 2001;276(45):41594-602. doi: 10.1074/jbc.M104514200. PMID:11553612 [DOI] [PubMed] [Google Scholar]
- [25].Tan E, Ding XQ, Saadi A, Agarwal N, Naash MI, Al-Ubaidi MR. Expression of cone-photoreceptor-specific antigens in a cell line derived from retinal tumors in transgenic mice. Invest Ophthalmol Vis Sci. 2004;45(3):764-8. doi: 10.1167/iovs.03-1114. PMID:14985288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].al-Ubaidi MR, Font RL, Quiambao AB, Keener MJ, Liou GI, Overbeek PA, Baehr W. Bilateral retinal and brain tumors in transgenic mice expressing simian virus 40 large T antigen under control of the human interphotoreceptor retinoid-binding protein promoter. J Cell Biol. 1992;119(6):1681-7. doi: 10.1083/jcb.119.6.1681. PMID:1334963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Cho KI, Haney V, Yoon D, Hao Y, Ferreira PA. Uncoupling phototoxicity-elicited neural dysmorphology and death by insidious function and selective impairment of Ran-binding protein 2 (Ranbp2). FEBS Lett. 2015;589(24 Pt B):3959-68. doi: 10.1016/j.febslet.2015.11.037. PMID:26632511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Cho KI, Patil H, Senda E, Wang J, Yi H, Qiu S, Yoon D, Yu M, Orry A, Peachey NS, et al. Differential loss of prolyl isomerase or chaperone activity of Ran-binding protein 2 (Ranbp2) unveils distinct physiological roles of its cyclophilin domain in proteostasis. J Biol Chem. 2014;289(8):4600-25. doi: 10.1074/jbc.M113.538215. PMID:24403063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Cho KI, Yi H, Tserentsoodol N, Searle K, Ferreira PA. Neuroprotection resulting from insufficiency of RANBP2 is associated with the modulation of protein and lipid homeostasis of functionally diverse but linked pathways in response to oxidative stress. Dis Model Mech. 2010;3(9-10):595-604. doi: 10.1242/dmm.004648. PMID:20682751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Cho KI, Yi H, Yeh A, Tserentsoodol N, Cuadrado L, Searle K, Hao Y, Ferreira PA. Haploinsufficiency of RanBP2 is neuroprotective against light-elicited and age-dependent degeneration of photoreceptor neurons. Cell Death Differ. 2009;16(2):287-97. doi: 10.1038/cdd.2008.153. PMID:18949001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Datta S, Snow CJ, Paschal BM. A pathway linking oxidative stress and the Ran GTPase system in progeria. Mol Biol Cell. 2014;25(8):1202-15. doi: 10.1091/mbc.E13-07-0430. PMID:24523287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Nagai M, Yoneda Y. Downregulation of the small GTPase ras-related nuclear protein accelerates cellular ageing. Biochim Biophys Acta. 2013;1830(3):2813-9. doi: 10.1016/j.bbagen.2012.11.001. PMID:23160023 [DOI] [PubMed] [Google Scholar]
- [33].Fichtman B, Harel A. Stress and aging at the nuclear gateway. Mech Ageing Dev. 2014;135:24-32. doi: 10.1016/j.mad.2014.01.003. PMID:24447784 [DOI] [PubMed] [Google Scholar]
- [34].D'Angelo MA, Raices M, Panowski SH, Hetzer MW. Age-dependent deterioration of nuclear pore complexes causes a loss of nuclear integrity in postmitotic cells. Cell. 2009;136(2):284-95. doi: 10.1016/j.cell.2008.11.037. PMID:19167330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Wright AF, Chakarova CF, Abd El-Aziz MM, Bhattacharya SS. Photoreceptor degeneration: genetic and mechanistic dissection of a complex trait. Nat Rev Genet. 2010;11(4):273-84. doi: 10.1038/nrg2717. PMID:20212494 [DOI] [PubMed] [Google Scholar]
- [36].Bramall AN, Wright AF, Jacobson SG, McInnes RR. The genomic, biochemical, and cellular responses of the retina in inherited photoreceptor degenerations and prospects for the treatment of these disorders. Annu Rev Neurosci. 2010;33:441-72. doi: 10.1146/annurev-neuro-060909-153227. PMID:20572772 [DOI] [PubMed] [Google Scholar]
- [37].Hartong DT, Berson EL, Dryja TP. Retinitis pigmentosa. Lancet. 2006;368(9549):1795-809. doi: 10.1016/S0140-6736(06)69740-7. PMID:17113430 [DOI] [PubMed] [Google Scholar]
- [38].Busskamp V, Krol J, Nelidova D, Daum J, Szikra T, Tsuda B, Jüttner J, Farrow K, Scherf BG, Alvarez CP, et al. miRNAs 182 and 183 are necessary to maintain adult cone photoreceptor outer segments and visual function. Neuron. 2014;83(3):586-600. doi: 10.1016/j.neuron.2014.06.020. PMID:25002228 [DOI] [PubMed] [Google Scholar]
- [39].Moore KB, Vetter ML. MicroRNA maintenance of cone outer segments. Neuron. 2014;83(3):510-2. doi: 10.1016/j.neuron.2014.07.031. PMID:25102555 [DOI] [PubMed] [Google Scholar]
- [40].Villarroya-Beltri C, Gutierrez-Vazquez C, Sanchez-Cabo F, Perez-Hernandez D, Vazquez J, Martin-Cofreces N, et al. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun. 2013;4:2980. doi: 10.1038/ncomms3980. PMID:24356509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Sahoo MR, Gaikwad S, Khuperkar D, Ashok M, Helen M, Yadav SK, Singh A, Magre I, Deshmukh P, Dhanvijay S, et al. Nup358 binds to AGO proteins through its SUMO-interacting motifs and promotes the association of target mRNA with miRISC. EMBO Reports. 2017;18(2):241-63. doi: 10.15252/embr.201642386. PMID:28039207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Siculella L, Tocci R, Rochira A, Testini M, Gnoni A, Damiano F. Lipid accumulation stimulates the cap-independent translation of SREBP-1a mRNA by promoting hnRNP A1 binding to its 5'-UTR in a cellular model of hepatic steatosis. Biochim Biophys Acta. 2016;1861(5):471-81. doi: 10.1016/j.bbalip.2016.02.003. PMID:26869449 [DOI] [PubMed] [Google Scholar]
- [43].Ferreira PA, Hom JT, Pak WL. Retina-specifically expressed novel subtypes of bovine cyclophilin. J Biol Chem. 1995;270(39):23179-88. doi: 10.1074/jbc.270.39.23179. PMID:7559465 [DOI] [PubMed] [Google Scholar]
- [44].Bode M, Woellhaf MW, Bohnert M, van der Laan M, Sommer F, Jung M, Zimmermann R, Schroda M, Herrmann JM. Redox-regulated dynamic interplay between Cox19 and the copper-binding protein Cox11 in the intermembrane space of mitochondria facilitates biogenesis of cytochrome c oxidase. Mol Biol Cell. 2015;26(13):2385-401. doi: 10.1091/mbc.E14-11-1526. PMID:25926683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Knipscheer P, Flotho A, Klug H, Olsen JV, van Dijk WJ, Fish A, Johnson ES, Mann M, Sixma TK, Pichler A. Ubc9 sumoylation regulates SUMO target discrimination. Mol Cell. 2008;31(3):371-82. doi: 10.1016/j.molcel.2008.05.022. PMID:18691969 [DOI] [PubMed] [Google Scholar]
- [46].Ferreira PA. Characterization of RanBP2-associated molecular components in neuroretina. Methods Enzymol. 2000;315:455-68. doi: 10.1016/S0076-6879(00)15861-6. PMID:10736720 [DOI] [PubMed] [Google Scholar]
- [47].Castagnet P, Mavlyutov T, Cai Y, Zhong F, Ferreira P. RPGRIP1s with distinct neuronal localization and biochemical properties associate selectively with RanBP2 in amacrine neurons. Hum Mol Genet. 2003;12(15):1847-63. doi: 10.1093/hmg/ddg202. PMID:12874105 [DOI] [PubMed] [Google Scholar]


