Nitrate is of central importance in bacterial physiology. Previous studies indicated that movements toward nitrate are due to energy taxis, which is based on the cytosolic sensing of consequences of nitrate metabolism. Here we present the first report on nitrate chemotaxis. This process is initiated by specific nitrate binding to the periplasmic ligand binding domain (LBD) of McpN. Nitrate chemotaxis is highly regulated and occurred only under nitrate starvation conditions, which is helpful information to explore nitrate chemotaxis in other bacteria. We present the three-dimensional structure of the McpN-LBD in complex with nitrate, which is the first structure of a chemoreceptor PilJ-type domain. This structure reveals striking similarities to that of the abundant 4-helix bundle domain but employs a different sensing mechanism. Since McpN homologues show a wide phylogenetic distribution, nitrate chemotaxis is likely a widespread phenomenon with importance for the life cycle of ecologically diverse bacteria.
KEYWORDS: Pseudomonas aeruginosa, chemoreceptor, chemotaxis, nitrate
ABSTRACT
Chemotaxis and energy taxis permit directed bacterial movements in gradients of environmental cues. Nitrate is a final electron acceptor for anaerobic respiration and can also serve as a nitrogen source for aerobic growth. Previous studies indicated that bacterial nitrate taxis is mediated by energy taxis mechanisms, which are based on the cytosolic detection of consequences of nitrate metabolism. Here we show that Pseudomonas aeruginosa PAO1 mediates nitrate chemotaxis on the basis of specific nitrate sensing by the periplasmic PilJ domain of the PA2788/McpN chemoreceptor. The presence of nitrate reduced mcpN transcript levels, and McpN-mediated taxis occurred only under nitrate starvation conditions. In contrast to the NarX and NarQ sensor kinases, McpN bound nitrate specifically and showed no affinity for other ligands such as nitrite. We report the three-dimensional structure of the McpN ligand binding domain (LBD) at 1.3-Å resolution in complex with nitrate. Although structurally similar to 4-helix bundle domains, the ligand binding mode differs since a single nitrate molecule is bound to a site on the dimer symmetry axis. As for 4-helix bundle domains, ligand binding stabilized the McpN-LBD dimer. McpN homologues showed a wide phylogenetic distribution, indicating that nitrate chemotaxis is a widespread phenotype. These homologues were particularly abundant in bacteria that couple sulfide/sulfur oxidation with nitrate reduction. This work expands the range of known chemotaxis effectors and forms the basis for the exploration of nitrate chemotaxis in other bacteria and for the study of its physiological role.
INTRODUCTION
Many bacteria are capable of flagellum-driven tactic movements in stimulus gradients. Genome analyses have revealed that more than half of the bacterial genomes contain genes necessary for taxis (1). The canonical form of taxis is based on stimulus reception by the chemoreceptor that leads to changes in the activity of the CheA autokinase, which subsequently modulates transphosphorylation to the CheY response regulator. The phosphorylated form of CheY binds to the flagellar motor, ultimately causing taxis toward or away from the stimulus (2).
Many bacteria possess the capacity to perform chemotaxis and energy taxis. The typical chemotaxis mechanism is initiated by the recognition of specific chemoeffectors at periplasmic ligand binding domains (LBDs), leading to receptor activation. Chemoreceptor function is largely determined by the nature of the chemoeffectors recognized, and the chemoreceptors specific for different compound classes such as amino acids, tricarboxylic acid (TCA) cycle intermediates, polyamines, purines, or inorganic phosphate have been identified (3). Alternatively, energy taxis is a metabolism-dependent form of taxis and represent directed movement in gradients of physicochemical parameters that affect metabolism (4). In contrast to chemotaxis, it is not the chemoeffector that is sensed per se but the consequences of its metabolism. Energy taxis occurs in response to a very wide range of stimuli, including metabolizable substrates such as sugars, organic acids, and amino acids; electron acceptors such as oxygen, nitrate, fumarate, and dimethyl sulfoxide; and compounds that affect metabolism otherwise, such as light or metabolic inhibitors (5–7).
Escherichia coli, the model organism traditionally used to study chemotaxis, has 4 chemoreceptors with a periplasmic LBD that mediate chemotaxis primarily with respect to amino acids, sugars, or dipeptides (8). In addition, it has an Aer chemoreceptor that mediates energy taxis by sensing redox changes via a flavin adenine dinucleotide (FAD)-containing cytosolic PAS domain (8). However, genome analyses have indicated that many other bacteria have significantly more chemoreceptors (up to 80) than E. coli (5). In addition, these chemoreceptors are characterized by diversity in the LBD type since more than 80 different LBD types were found to form part of chemoreceptors (9).
The elevated number of chemoreceptors and their diversity in the LBDs suggest that the chemosensory capacity of many bacteria is very extensive but remains in general largely unexplored. The scientific community is now only at the beginning of the process of identifying the chemotactic spectra of many bacteria, establishing links between chemoeffectors and LBD types, and identifying the physiological relevance of chemotaxis to newly identified chemoeffectors (9).
Here, we have addressed this issue using the opportunistic human pathogen Pseudomonas aeruginosa PAO1 as a model (9, 10). This bacterium has 26 chemoreceptors that feed into four chemosensory pathways (11). Two pathways, corresponding to Che and Che2, were shown to play a role in chemotaxis. Whereas the Che pathway appears to be essential for chemotaxis (12, 13), the Che2 pathway was found to be required for optimal chemotactic responses (14). Alternatively, the Chp pathway was associated with twitching motility (15–18), whereas the Wsp pathway modulates cyclic diguanylate monophosphate (c-di-GMP) levels (19). P. aeruginosa chemoreceptors employ together 11 different LBD types for signal sensing (9), with the 4-helix bundle (4HB) (20), CACHE (21), and helical bimodular (HBM) (22) domains being most abundant (9). Several of these receptors have been functionally annotated, and all were found to mediate chemoattraction. The 4HB domain containing receptor CtpH responded specifically to inorganic phosphate (Pi) (23, 24), the HBM domains containing McpK and CtpL were identified as α-ketoglutarate (25)- and Pi (23, 24)-specific receptors, and the paralogous dCACHE domains containing receptors PctA, PctB, and PctC were shown to mediate chemotaxis to amino acids (26–29), whereas the sCACHE domain containing receptor PA2652 was found to mediate taxis to different C4-carboxylic acids (30, 31).
P. aeruginosa has two chemoreceptors, PA2788 and PA0411/PilJ, that possess a PilJ-type LBD (Pfam signature PF13675) (9). This domain is annotated in the Pfam database (32) as “Type IV pili methyl-accepting chemotaxis transducer N-ter.” The PilJ receptor feeds into the Chp pathway and is not related to chemotaxis (18), whereas PA2788 was predicted to feed into the Che pathway (11). However, the ligands recognized by the PilJ and PA2788 are unknown. Bioinformatic analyses have shown that PilJ domains represent 2% of all chemoreceptor LBDs (9). In addition, a search in the Pfam database revealed that PilJ domains are also employed by other bacterial sensor proteins such as sensor kinases, diguanylate cyclases, and transcriptional regulators. In this study, we aimed at identifying the function of the PilJ domain containing chemoreceptors in P. aeruginosa.
RESULTS
Nitrate is a specific ligand for PA2788-LBD.
To identify ligands that bind to PA2788, we cloned the DNA sequence encoding the LBD of PA2788 into an expression vector. Protein was expressed in E. coli and purified by affinity chromatography. High-throughput ligand screening assays were then conducted using the thermal shift method (33). In this method, changes in the melting temperature (Tm) of a protein, representing the midpoint of thermal protein unfolding, are recorded. The binding of ligands typically increases the Tm, and shifts of greater than 2°C are considered significant. We screened 480 ligands from Biolog compound arrays PM1, PM2A, PM3B, PM4A, and PM5 that served as bacterial carbon, nitrogen, phosphorus, and sulfur sources.
In the absence of ligand, the Tm of PA2788-LBD was 43.5°C. Fig. 1A shows the changes in Tm caused by each of the 95 compounds of array PM3B comprising nitrogen sources. In the presence of NaNO3, the Tm was increased by 3.5°C, whereas NaNO2 caused only a minor increase of 0.5°C. No significant Tm shifts were obtained for the compounds in arrays PM1, PM2A, PM4A, and PM5.
FIG 1.
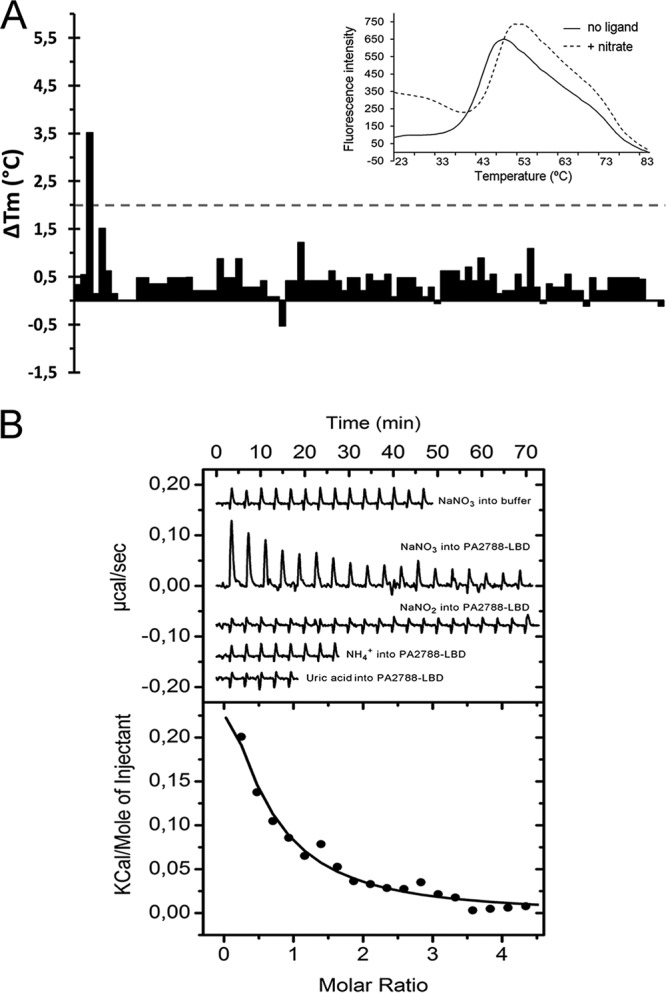
Identification of nitrate as a PA2788-LBD ligand. (A) Thermal shift assays using compounds of Biolog compound array PM3B. Shown are the Tm changes with respect to the ligand-free protein. The insert shows the thermal unfolding curves of ligand-free PA2788-LBD and in the presence of nitrate. (B) Microcalorimetric binding studies of PA2788-LBD. The upper panel shows the heat changes caused by the injection of 2 mM (12.8-µl aliquots) NaNO3 into buffer and 36 µM PA2788-LBD as well as the titration of PA2788-LBD with 2 mM NaNO2, 2 mM ammonia, and 1 mM uric acid. The lower panel depicts the concentration-normalized and dilution heat-corrected integrated peak areas of the PA2788-LBD titration data with NaNO3. The line corresponds to the best fit using the “One binding site model” of the MicroCal version of ORIGIN.
To verify ligand binding, isothermal titration calorimetry (ITC) experiments were conducted. In an initial control experiment, the heat changes derived from the injection of 2 mM NaNO3 into buffer were recorded (Fig. 1B), with results showing that the levels of dilution heat were low and uniform. Titration of PA2788-LBD with the same ligand caused endothermic heat changes (ΔH = 0.38 ± 0.1 kcal/mol) indicative of an entropy-driven binding process (TΔS = 6.4 kcal/mol) characterized by a dissociation constant of 47 ± 8 µM. The same protein was also titrated with NaNO2, ammonia and uric acid (Fig. 1B), but an absence of binding was noted in all cases, confirming the thermal shift assay results. We were intrigued by the failure of PA2788-LBD to sense nitrite since other sensor proteins were found to sense nitrate as well as nitrite (34–36). ITC experiments performed with ligands provide information only on higher-affinity binding events. To confirm that PA2788-LBD does not bind nitrite with low affinity, we performed titrations of PA2788-LBD with nitrate in the absence and presence of 20 mM nitrite. In cases of nitrite binding, this would alter nitrate recognition. However, this was not the case (see Fig. S1A in the supplemental material), confirming that PA2788-LBD is a nitrate-specific receptor. Following the demonstration that PA2788 specifically binds nitrate, this chemoreceptor was named McpN.
Specificity of nitrate recognition at McpN-LBD. (A) Microcalorimetric titration of McpN-LBD with NaNO3 in the absence and presence of 20 mM NaNO2. (Upper panel) In black, raw data for the titration of 36 µM McpN-LBD with 12.8-μl aliquots of 2 mM NaNO3; in red, repetition of the experiment with both ligands containing in addition 20 mM NaNO2. (Lower panel) Concentration data representing normalized and dilution heat-corrected integrated peak areas of raw data. The continuous lines represent the curve fits obtained using the “One binding site model” of ORIGIN. (B) Superimposition of the nitrate molecule in the structures of McpN-LBD (in blue) and NarX-LBD (in orange; PDB ID 3EZH). Shown are the amino acid residues in the vicinity of bound nitrate. Hydrogen bonds are indicated by dotted lines. Download FIG S1, TIF file, 2.4 MB (2.4MB, tif) .
Copyright © 2019 Martín-Mora et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Nitrate is not recognized by the LBDs of the PilJ and PA4520 chemoreceptors.
Inspection of the chemoreceptor repertoire of P. aeruginosa (9) suggests that two other chemoreceptors may also bind nitrate. First, the LBD of the PilJ (PA0411) receptor is composed of two consecutive PilJ domains (9). Second, chemoreceptor PA4520 was predicted to contain a NIT LBD (Pfam PF08376). This domain, representing approximately 3% of all extracellular prokaryotic LBDs (21), was predicted to recognize nitrate and nitrite (37).
To verify whether these receptors also bind nitrate, we generated the purified individual LBDs of both receptors for thermal shift assay ligand screening using the PM3B array. In the absence of ligand, PilJ-LBD and PA4520-LBD unfolded with Tm values of 41.8 and 34.4°C, respectively. In both cases, however, no significant ligand-induced Tm shifts were noted (Fig. S2A and B) and microcalorimetric titrations confirmed the absence of NaNO3 and NaNO2 binding (Fig. S2C). To exclude the possibility that endothermic and exothermic contributions to binding canceled out each other at a given analysis temperature, the experiments were repeated at 15°C; however, the same result was produced, confirming the absence of nitrate/nitrite binding to PilJ-LBD and PA4520-LBD.
The absence of nitrite and nitrate binding to PilJ-LBD and PA4520-LBD. (A and B) Results from thermal shift assays. Shown are changes in the Tm values of PilJ-LBD (A) and PA4520-LBD (B) in the presence of compounds from Biolog array PM3B. (C) Microcalorimetric titration of PilJ-LBD and PA4520-LBD with NaNO3 and NaNO2. As a control, the titration of buffer with NaNO3 is shown. The injection volume was 19.2 µl. Download FIG S2, TIF file, 2.6 MB (2.6MB, tif) .
Copyright © 2019 Martín-Mora et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
McpN mediates nitrate chemotaxis under nitrate starvation conditions.
To assess the function of McpN, we conducted quantitative capillary chemotaxis assays for NaNO3 using the wild-type (wt) strain as well as a mutant deficient in the mcpN gene. These assays were conducted using the standard conditions that we routinely employ to study P. aeruginosa chemotaxis. This assay involves cell culture in MS minimal medium (4.2 g/liter Na2HPO4, 2.8 g/liter KH2PO4, 2.0 g/liter NH4NO3, 0.2 g/liter MgSO4 7H2O, 17.0 mg/liter FeCl3 6H2O, 0.8 mg/CoCl2 6H2O, 0.6 mg/liter CaCl2 2H2O, 0.3 mg/liter Na2MoO4 2H2O, 0.1 mg/liter H3BO3, 0.2 mg/liter ZnSO4 7H2O, 0.2 mg/liter CuSO4 7H2O, 0.2 mg/liter MnSO4 7H2O) supplemented with glucose (note that this medium also contains 25 mM NH4NO3 as a nitrogen source). However, only very minor, nonsignificant responses were detected over the entire nitrate concentration range (Fig. S3).
Quantitative capillary chemotaxis assays of P. aeruginosa PAO1 and the mcpN mutant under conditions of abundant nitrate. Cells were grown in M9 minimal medium supplemented with glucose as a carbon source. This medium contains 25 mM ammonium nitrate as a nitrogen source. Data were corrected using the number of cells that swam into buffer containing capillaries (1,992 ± 298). Download FIG S3, TIF file, 0.07 MB (70.2KB, tif) .
Copyright © 2019 Martín-Mora et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Previous studies have shown that chemotaxis to Pi in PAO1 was not observed in rich medium containing significant amounts of Pi but was induced by Pi starvation (23). We hypothesized that this might also be the case for nitrate chemotaxis and followed an approach similar to that described previously by Wu et al. (23). Thus, cells were precultured in rich 2× YT medium (10 g yeast extract liter−1, 16 g Bacto tryptone liter−1, 10 g NaCl liter−1) and then diluted 133-fold into N0 medium (which lacks nitrogen sources) and continued to grow for another 3 h. Under these conditions, strong chemotactic responses to NaNO3 were obtained. Initial significant responses were obtained at a NaNO3 concentration of 5 µM, whereas maximal responses were observed at 500 µM (Fig. 2). No nitrate chemotaxis was observed for the mcpN mutant, suggesting that it is the sole nitrate chemotaxis receptor. The complementation of this mutant with a plasmid harboring the mcpN gene restored nitrate chemotaxis (Fig. 2).
FIG 2.
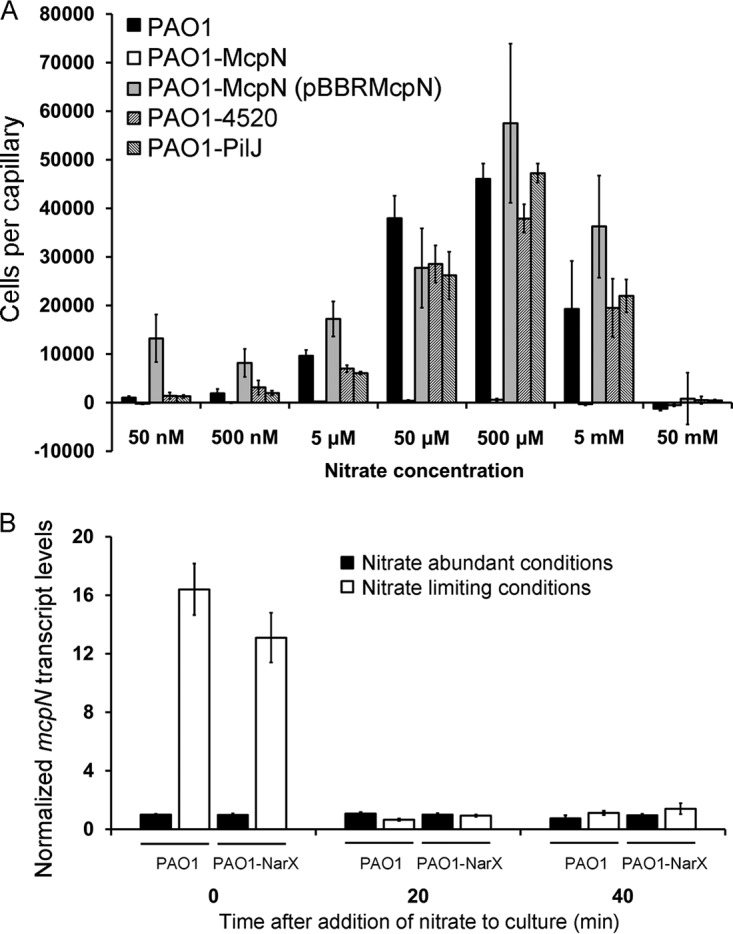
The McpN chemoreceptor of P. aeruginosa mediates nitrate chemotaxis. (A) Quantitative capillary chemotaxis assays of different P. aeruginosa PAO1 strains at different NaNO3 concentrations. Cells were grown in rich 2× YT medium and then diluted 133-fold into N0 medium (deficient in nitrogen sources). Data represent means of results from three biological replicates conducted in triplicate. (B) RT-qPCR analysis of the mcpN transcript in the wild-type strain and in a mutant defective in the NarX sensor kinase. Cells were gown in MS medium supplemented with glucose (containing 25 mM NH4NO3) or in nitrate-deficient N0 medium (inoculated using a culture grown in 2× YT medium) until an OD600 of 0.15 was reached (time zero), at which point NaNO3 was added to reach a final concentration of 1 mM. Further samples were taken after 20 and 40 min. Shown are mcpN transcript levels normalized with respect to the transcript levels of the rpoD reference gene at time zero under conditions of nitrate abundance. Data represent means and standard deviations of results from three biological replicates conducted in triplicate.
To identify the possible roles of the PilJ receptor and the NIT domain containing PA4520 chemoreceptor in nitrate chemotaxis, single mutants with mutations of the corresponding genes were also analyzed. As shown in Fig. 2A, their responses to nitrate were similar to those seen with the wt strain, confirming that the observed nitrate chemotaxis was mediated solely by McpN.
Nitrate reduces mcpN transcript levels.
To explain the absence of taxis under conditions of nitrate abundance, we hypothesized that nitrate might repress expression of the mcpN gene. To verify this hypothesis, we quantified mcpN transcript levels by reverse transcription-quantitative PCR (RT-qPCR). These assays were carried out using RNA from cells grown using the same protocol used for the chemotaxis assays under conditions of nitrate abundance and limitation. As shown in Fig. 2B, mcpN transcript levels were approximately 16 times higher under nitrate-limiting conditions than under nitrate-abundant conditions. To verify that the absence or presence of nitrate was the cause for these differences, NaNO3 was added to these cultures to reach a final concentration of 1 mM and samples were taken for RT-qPCR experiments after additional periods of growth of 20 and 40 min. The results showed that the addition of nitrate to cells grown under nitrate-limiting conditions reduced mcpN transcript levels to those seen under nitrate-abundant conditions, indicating that nitrate reduces mcpN expression (Fig. 2B).
The NarX/NarL two-component system (TCS) senses nitrate and regulates genes involved in nitrate metabolism (38). To identify a potential role of this TCS in mcpN expression, we quantified mcpN transcript levels in a mutant defective in the gene encoding the NarX sensor kinase. However, RT-qPCR data revealed no statistical differences in the transcript levels of mcpN (Fig. 2B).
McpN signals through the Che pathway.
The 26 PAO1 chemoreceptors signal through four different chemosensory pathways, and McpN was predicted to signal through the Che pathway (11). To verify this prediction, we conducted chemotaxis assays for NaNO3 using mutants defective in the genes encoding the CheA paralogues of the Che1 (CheA1) and Che2 (CheA2) pathways. As shown in Fig. S4, no nitrate chemotaxis was observed in the cheA1 mutant, whereas the responses of the cheA2 mutant were comparable to wt levels. These results thus confirm that McpN signals through the Che pathway (11).
Quantitative capillary chemotaxis assays of P. aeruginosa PAO1 and mutants deficient in cheA1 (PA1458) and cheA2 (PA0178) in the presence of 500 µM NaNO3. Data represent means and standard deviations of results from three biological replicates conducted in triplicate. Download FIG S4, TIF file, 1.4 MB (1.4MB, tif) .
Copyright © 2019 Martín-Mora et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The three-dimensional (3D) structure of McpN-LBD.
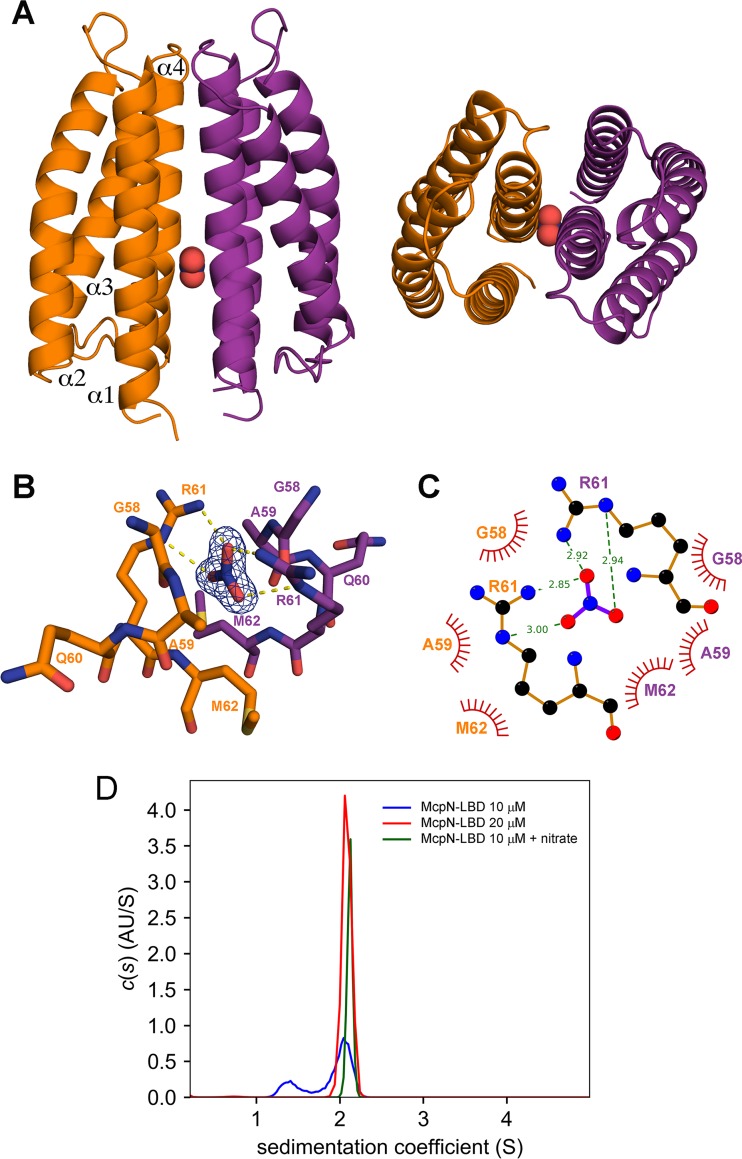
McpN-LBD in complex with nitrate was crystallized in a buffer at pH 7.5, and its structure was resolved by X-ray crystallography to a resolution of 1.3 Å. According to the Matthews coefficient, the unit cell accommodates three chains. A structural alignment of these three chains resulted in root mean square deviation (RMSD) values below 0.5, indicating that these chains can be considered identical. Chains A and B of the unit cell form a dimer (Fig. 3A), whereas chain C forms another dimer with a symmetry-related chain. The McpN-LBD monomer is composed of 4 α-helices that pack into a 4-helix bundle. Dimerization is achieved through the interaction of 22 residues of chains A and B that establish 16 hydrogen bonds and occlude a surface of approximately 1,100 Å2 in each monomer.
FIG 3.
The three-dimensional structure of McpN-LBD in complex with nitrate. (A) Side (left) and top (right) views of the overall structure. Nitrate is shown in red. (B) The nitrate binding pocket. Shown are amino acids from both chains that interact with nitrate. The |2Fo-Fc| electron density of nitrate is contoured at 1.5 sigma. (C) Schematic representation of amino acids involved in hydrogen bonding to nitrate, shown as dotted lines, while the spoked arcs represent residues that make nonbonded contacts. (D) Analysis of the oligomeric state of McpN-LBD using sedimentation velocity analytical ultracentrifugation (AU). Shown are the sedimentation velocity c(s) profiles of ligand-free McpN-LBD at different concentrations and protein in complex with nitrate.
A single molecule of nitrate is bound to a site with a positive surface charge at the dimer interface. The binding site is situated on the dimer symmetry axis; consequently, the same amino acids from the two monomers establish interactions with bound nitrate. Whereas G58, A59, and M62 establish nonbonded contacts, R61 played a key role in recognition since it forms two hydrogen bonds with nitrate (Fig. 3B). To verify the role of R61, we generated a McpN-LBD R61A mutant. The intrinsic tryptophan fluorescence emission spectrum (Fig. S5A) and the thermal unfolding properties of the mutant protein (Fig. 1; see also Fig. S5B) were comparable to those seen with the native protein, indicating that this amino acid replacement did not cause major changes to the overall protein structure. Analysis of this protein by the thermal shift assay and ITC showed that this protein was unable to recognize nitrate (Fig. S5B and C).
Analysis of the McpN-LBD R61A mutant. (A) Intrinsic tryptophan fluorescence emission spectra of McpN-LBD and McpN-LBD R61A. (B) Thermal shift assay of McpN-LBD R61A in the absence and presence of NaNO3. (C) Microcalorimetric titration of McpN-LBD R61A with NaNO3. Download FIG S5, TIF file, 2.9 MB (2.9MB, tif) .
Copyright © 2019 Martín-Mora et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
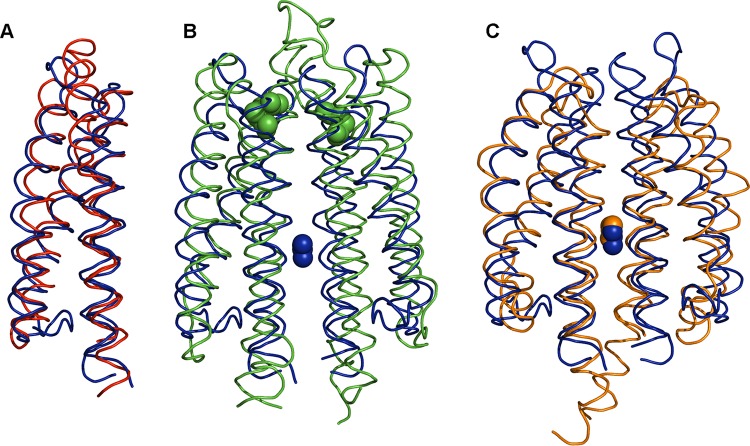
The McpN-LBD structure was aligned to all structures currently deposited in the protein data bank using the DALI algorithm, and the closest structural homologues are listed in Table 1. Surprisingly, the closest structure was a LBD of a histidine kinase that belonged to a different family, namely, CHASE3. The high level of structural similarity between this domain and McpN-LBD is illustrated in Fig. 4A. The only chemoreceptor LBD with significant structural similarity was the HBM domain of the McpS chemoreceptor, which is composed of two 4-helix bundles (39).
TABLE 1.
Structural alignment of McpN-LBD with structures deposited in the Protein Data Banka
| PDB ID |
Protein type | Species | Ligand | Pfam/InterPro ID | Z- score |
No. of aligned residues |
Sequence identity (%) |
Reference |
|---|---|---|---|---|---|---|---|---|
| 3VA9 | LBD of HK9 SK | Rhodopseudomonas palustris | CHASE3 (PF05227) | 13.6 | 114 | 13 | Unpublished | |
| 5XSJ | LBD of LytS SK | Clostridium beijerinckii | XylFII ligand binding protein |
Unannotated | 13.3 | 112 | 6 | 82 |
| 4K0D | LBD of Adeh_2942 SK |
Anaeromyxobacter dehalogenans | Unannotated | 12.3 | 117 | 17 | 83 | |
| 2YFB | LBD of McpS CR1 | Pseudomonas putida | TCA cycle intermediates |
HBM (PF16591) | 12.0 | 113 | 12 | 39 |
| 3EZH | LBD of NarX SK | Escherichia coli | Nitrate/nitrite | PilJ (PF13675) | 11.9 | 104 | 21 | 72 |
| 3O1J | LBD of TorS SK | Vibrio parahaemolyticus | TorT periplasmic binding protein |
TorS-like (IPR038188) | 11.2 | 111 | 14 | 84 |
| 4IGG | α-Catenin | Homo sapiens | β-Catenin | Vinculin (PF01044) | 10.7 | 114 | 5 | 85 |
| 5JEQ | LBD of NarQ SK | Escherichia coli | Nitrate/nitrite | PilJ (PF13675) | 10.6 | 103 | 15 | 73 |
| 5XA5 | α-Catenin | Caenorhabditis elegans | β-Catenin | Vinculin (PF01044) | 10.4 | 110 | 7 | 85 |
| 5XFL | α-Catenin | Mus musculus | β-Catenin | Vinculin (PF01044) | 10.3 | 114 | 5 | 85 |
Shown are the structures with a Z-score above 10. The listed structures share less than 90% sequence similarity. SK, sensor kinase; CR, chemoreceptor.
FIG 4.
Structural alignment of the McpN-LBD C∝ chain with structural homologues. In all cases, McpN-LBD is shown in blue. (A) Alignment with a CHASE3 domain of an uncharacterized histidine kinase of Rhodopseudomonas palustris (PDB ID 3VA9), the closest structural homologue found in a DALI search (Table 1). (B) Alignment with Tar-LBD (PDB ID 1VLT). Bound aspartate (Tar) is shown in green, whereas bound nitrate (McpN) is shown in blue. (C) Alignment with the sensor domain of the NarX histidine kinase (PDB ID 3EZH). Bound nitrates overlap and are shown in blue (McpN) and orange (NarX).
Nitrate binding promotes McpN-LBD dimerization.
Analytical ultracentrifugation (AUC) experiments were conducted to assess the oligomeric state of McpN-LBD. Initially, we used the three-dimensional structure of McpN-LBD to calculate the expected sedimentation (S) coefficients using HYDROPRO (40) software. This analysis resulted in s20,w values of 1.7 S and 2.8 S (at 20 degrees and using water) for the monomeric and dimeric species, respectively.
Sedimentation velocity experiments were performed on 5 µM to 40 µM McpN-LBD in the absence of ligand, and data obtained with 10 and 20 µM protein are shown in Fig. 3D. At 10 µM, two peaks could be identified with s20,w values of 1.85 S and 2.69 S, which fit well with the values determined for the monomer and dimer, respectively. At 20 µM and 40 µM, only protein dimers were observed, indicating that the equilibrium had shifted completely to this oligomeric state. To assess the effect of nitrate binding on the oligomeric state, the experiments described above were repeated in the presence of saturating nitrate concentrations. No changes in oligomeric state occurred at the 20 and 40 µM concentrations, whereas a single peak with s20,w = 2.79 S (Fig. 3D) was obtained at 10 µM protein, indicating that nitrate binding had shifted the equilibrium entirely to the dimeric state. Taken together, the data indicate that McpN-LBD was present in a monomer-dimer equilibrium and that nitrate binding stabilized the dimeric state.
Definition of the N-box.
To identify potential McpN homologues in other species, we conducted a BLAST-P search of McpN-LBD in the NCBI database of nonredundant protein sequences, excluding members of the Pseudomonas genus. An alignment of the top 87 sequences is shown in Fig. S6. All sequences belonged to the PilJ family and formed part of chemoreceptors. Most of the corresponding species were marine bacteria, and a significant proportion of them are able to oxidize elemental sulfur or sulfite (see Table S1 in the supplemental material). Furthermore, a number of human pathogens such as Enterobacter cloacae, Streptococcus pneumoniae, and Eggerthia catenaformis were among the species that harbor McpN homologues (Table S1).
Alignment of the McpN-LBD sequences with homologues from other species. The alignment is based on a BLAST-P search of McpN-LBD in the NCBI database of nonredundant sequences, excluding species of the genus Pseudomonas. The alignment was done using the CLUSTALW algorithm of npsa software (C. Combet, C. Blanchet, C. Geourjon, and G. Deléage, Trends Biochem Sci 25:147–150, 2000, https://www.cell.com/trends/biochemical-sciences/fulltext/S0968-0004(99)01540-6?_returnURL=https%3A%2F%2Flinkinghub.elsevier.com%2Fretrieve%2Fpii%2FS0968000499015406%3Fshowall%3Dtrue). The BLOSUM62 matrix was used, and the settings used for analysis of gap costs were defined as follows: existence, 11; extension, 1. The McpU-LBD sequence is shaded in yellow. Download FIG S6, PDF file, 0.02 MB (18KB, pdf) .
Copyright © 2019 Martín-Mora et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Characteristics of species that contain McpN homologues. A BLAST-P search was conducted in the NCBI database of nonredundant protein sequences (excluding species of the genus Pseudomonas), and the top 87 sequences are listed in Fig. S6. This table shows the characteristics of the corresponding bacterial species. Human pathogens are highlighted in green, and bacteria that oxidize sulfurous compounds are highlighted in yellow. Download Table S1, DOCX file, 0.09 MB (91.5KB, docx) .
Copyright © 2019 Martín-Mora et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The sequence alignment of McpN-LBD homologues revealed only a very modest level of overall sequence identity of approximately 5%. However, the zone around the nitrate binding site, which we have termed the N-box, was highly conserved and the corresponding sequence logo is shown in Fig. 5A. We have shown above that not all PilJ domains bind nitrate, since no binding was observed for the PilJ LBD, which is composed of two PilJ domains (Fig. S2). As shown in Fig. S7, the N-box was not conserved in either of the PilJ domains of the PilJ chemoreceptor. We then scanned the TrEMBL database using PROSITE (41) and the following consensus pattern for the N-box: [IVL]-[ND]-x-A-G-x-Q-R-M-L-[ST]-Q. The random statistical probability of a match was well below 1 sequence.
FIG 5.
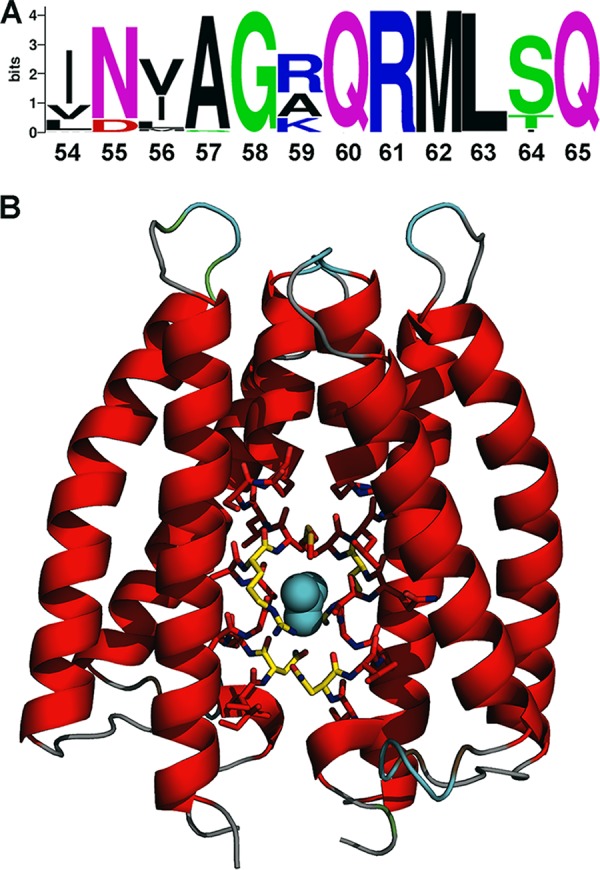
Definition of the N-box of PilJ domains. (A) The sequence logo of the N-box as derived from the alignment shown in Fig. S6. (B) Structure of McpN-LBD in which the 12 amino acids of the N-box are shown as sticks together with bound nitrate.
Sequence alignment of the PilJ domains of the McpN and PilJ chemoreceptors of P. aeruginosa PAO1. PilJ-pilJ1 corresponds to the N-terminal PilJ domain (amino acids 36 to 163), which is followed in sequence by the PilJ-pilJ2 domain (amino acids 164 to 314). The N-box in McpN is shaded in yellow. Red, identical residues; green, highly similar residues; blue, weakly similar residues. The alignment was done using the CLUSTALW algorithm of npsa software (Combet et al., 2000). The BLOSUM62 matrix was used, and the settings used for analysis of gap costs were defined as follows: existence, 11; extension, 1. Download FIG S7, TIF file, 0.6 MB (591.2KB, tif) .
Copyright © 2019 Martín-Mora et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
However, 941 sequences containing PilJ domains which are likely to be nitrate binding domains were retrieved. The retrieved sequences formed part of all major families of signal transduction systems, namely, transcriptional regulators, sensor kinases, chemoreceptors, and diguanylate cyclases. There were 1,135 protein sequences with at least one PilJ domain in Pfam at the time of the search, and the N-box may be usable as a means to identify PilJ domains that are able to bind nitrate.
Nitrate chemotaxis in other bacterial species.
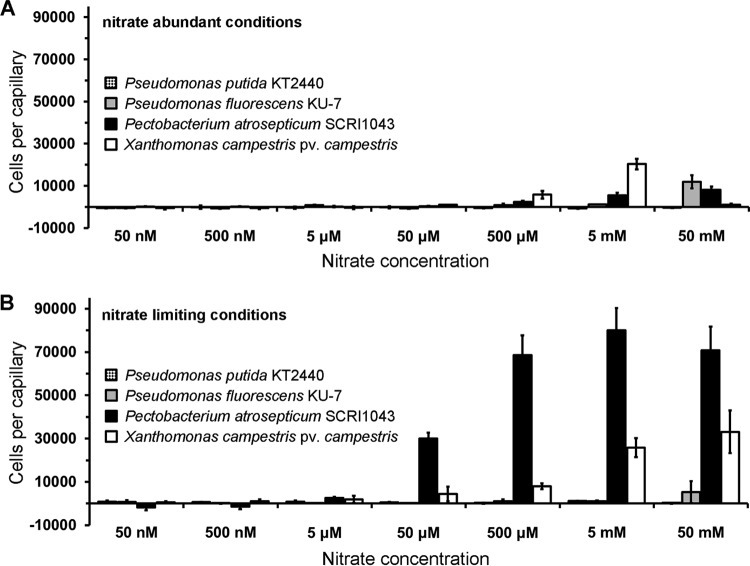
Subsequent work was aimed at assessing nitrate chemotaxis in other species. To that end, we conducted quantitative capillary chemotaxis assays using different strains grown under conditions of nitrate abundance and limitation. Pseudomonas putida KT2440 and Pseudomonas fluorescens KU-7 do not have an McpN homologue but contain, as in the case of P. aeruginosa, a NIT domain containing chemoreceptor. Our experiments showed that P. putida KT2440 was devoid of nitrate chemotaxis whereas P. fluorescens showed only minor responses to 50 mM nitrate (Fig. 6).
FIG 6.
Nitrate chemotaxis in other species. Quantitative capillary chemotaxis assays of different strains to NaNO3. (A) Cells grown under conditions of nitrate abundance. (B) Cells grown under conditions of nitrate limitation. Data represent means and standard deviations of results from three biological replicates conducted in triplicate.
We then studied two bacterial species that are among the top 10 plant-pathogenic bacteria (42), namely, Pectobacterium atrosepticum and Xanthomonas campestris pv. campestris. Interestingly, the responses of P. atrosepticum were very similar to those of PAO1, since only very minor responses were observed under nitrate abundance conditions but strong responses were observed under nitrate starvation conditions (Fig. 6). There is no McpN homologue among the 36 chemoreceptors of this strain, but there is a single receptor with a NIT domain. X. campestris pv. campestris also showed significant chemotaxis under nitrate-limiting conditions and only minor responses under nitrate abundance conditions (Fig. 6). Altogether, these data suggest that the induction of nitrate chemotaxis by nitrate limitation is common to other bacteria.
DISCUSSION
Nitrate is a final electron acceptor for anaerobic respiration and also serves as a nitrogen source for aerobic growth. Taxis to nitrate has been observed for a significant number of bacteria such as E. coli, Salmonella enterica serovar Typhimurium (43, 44), Pseudomonas spp. (45–48), Shewanella spp. (49, 50), Azospirillum brasilense (51), Rhodobacter sphaeroides, Agrobacterium tumefaciens (52), Thioploca spp. (53), and Synechococcus spp. (54). The three major bacterial pathways for nitrate metabolism include respiratory, assimilatory, and dissimilatory nitrate reduction (55), and any type of metabolism can lead to energy taxis. In some of the cases, it has been demonstrated that bacterial nitrate taxis is based on energy taxis (44, 49, 51, 52). For example, deletion or inhibition of enzymes that participate in nitrate metabolism abolished nitrate taxis (49). In other reports, the molecular mechanism of nitrate taxis, such as in the case of, for example, lake water bacteria (46) or different denitrifying strains (45, 47), is unclear. It is possible that those observations were based on chemotaxis. Here we identify the molecular mechanism of nitrate-specific chemotaxis that is initiated by the specific recognition of nitrate at a periplasmic chemoreceptor LBD. McpN homologues show a broad phylogenetic distribution, including those of archaea and bacteria belonging to the Firmicutes and Proteobacteria phyla (see Table S1 in the supplemental material), which indicates that nitrate chemotaxis may be a widespread mechanism.
Interestingly, among the species that harbor McpN homologues were a significant number of bacteria isolated from marine sediments that are able to oxidize sulfide or elemental sulfur (Table S1). There is evidence that the oxidation of reduced sulfur compounds in these bacteria is coupled to the reduction of electron acceptors such as nitrate (56). As a consequence, some sulfide oxidizers were found to store nitrate in vacuoles (57) at concentrations of up to 370 mM (58, 59). This intracellular nitrate is used to oxidize sulfide in deeper anoxic zones of sediments. This process has been particularly well studied in Beggiatoa spp. (60), which are also among the species that contain an McpN homologue (Table S1). On the basis of experiments performed with nitrate-reducing/sulfide-oxidizing shelf sediment bacteria belonging to the Thioploca genus, a functional model was proposed (53). The authors showed that the nitrate concentration in the sediment was lower than that in the flume water and that nitrate chemotaxis directed bacteria to the sediment surface, where they filled their vacuoles with nitrate. They then migrated back into deeper sediment layers, where they oxidized sulfide to sulfate until the nitrate was depleted, which induced the upward movement. Taken together, the data thus suggest the particular importance of nitrate chemotaxis in marine sulfide/sulfur-oxidizing bacteria.
Nitrate serves PAO1 as the sole nitrogen source for growth, and the anaerobic growth of this strain is accomplished through the denitrification enzyme pathway that catalyzes the sequential reduction of nitrate to nitrogen gas (61). Nitrate chemotaxis was observed in pathogenic P. aeruginosa bacteria but not in the nonpathogenic P. putida and P. fluorescens, suggesting that it may be related to virulence. Previous studies have shown a link between virulence and nitrate metabolism for anaerobically grown PAO1, since a mutant with a mutation in the nitrate reductase gene was avirulent in Caenorhabditis elegans (61). PAO1 causes airway infections in cystic fibrosis patients, and the sputum nitrate/nitrite concentration was 774 µM in cystic fibrosis patients, well above the concentration seen with the healthy control group (421 µM) (62). Importantly, these concentrations are in the range of the optimal chemotaxis responses measured here (Fig. 2), indicating that nitrate chemotaxis may be related to pathogenicity, as in the case of S. Typhimurium, where taxis to host-derived nitrate is required for efficient host infection (44).
PAO1 nitrate chemotaxis was observed only under nitrate starvation conditions (Fig. 2A), whereas no taxis was observed in under nitrate abundance conditions (see Fig. S3 in the supplemental material), and similar results were seen for P. atrosepticum (Fig. 6). This is unusual, since chemotactic behaviors are typically either constitutive or inducible by the chemoeffector (63, 64). However, striking similarities exist between Pi and nitrate chemotaxis in PAO1. Pi taxis was observed only under Pi starvation conditions and not under Pi abundance conditions (23, 24). As in the case of mcpN, the presence of Pi was shown to decrease the transcript levels of both Pi chemoreceptor genes, i.e., ctpL and ctpH (65). Pi was identified as a key signal molecule that controls the expression of many virulence genes and features in PAO1 (65, 66). Pi and nitrate are both inorganic anions, and it is tempting to speculate that chemotaxis repression mediated by chemoeffector abundance is a feature of this compound family.
Almost one-third of all chemoreceptor LBDs are recognized by the Pfam 4HB domain signature (9). Signaling of chemoreceptors with this domain has been extensively studied, and the 3D structure reveals a 4-helix antiparallel bundle (67, 68). Although the McpN-LBD sequence is not recognized by the Pfam 4HB signature, its structure superimposes very well on that of the 4HB Tar-LBD (Fig. 4B). This, together with the fact that the closest structural McpN-LBD homologue is a CHASE3 domain (Table 1), demonstrates that the 4-helix bundle is a conserved structural motif for ligand sensing formed by members of different LBD families. Although conserved in structure, the modes of ligand binding for McpN-LBD and Tar-LBD are different. The Tar-LBD dimer recognizes two signal molecules with high negative cooperativity that bind to the dimer interface at two sites that are not on the dimer symmetry axis (68, 69). In contrast, a single molecule of nitrate binds to a single site located at the dimer symmetry axis of McpN-LBD (Fig. 3A). However, 4HB domains and McpN-LBD (Fig. 3D) have in common that the individual domains are present in a monomer-dimer equilibrium and that ligand binding shifts this equilibrium to the dimeric state (69, 70).
The NarX/NarL and NarQ/NarP two-component systems control transcriptional responses to nitrate and nitrite, which are the preferred anaerobic electron acceptors in E. coli (71). The LBDs of the NarX and NarQ sensor kinases are structural homologs of McpN-LBD (Table 1), and their 3D structures in complex with nitrate have been solved (72, 73). Although McpN-LBD and NarX-LBD share only 21% sequence identity, their structures align very well and the nitrate binding site is conserved (Fig. 4C). McpN-LBD differs from NarX-LBD in several aspects. Our AUC studies showed that McpN-LBD has an intrinsic propensity to dimerize which is enhanced in the presence of nitrate. In contrast, NarX-LBD is monomeric even at a concentration of 10 mM and in the presence of nitrate (72). NarX and NarQ are characterized by a certain plasticity in ligand recognition, since they bind to nitrate, nitrite, and sulfite (34–36). In contrast, McpN-LBD recognizes nitrate exclusively and has no physiologically relevant affinity for nitrite (Fig. 1; see also Fig. S1A). The superimposition of the ligand binding pockets of NarX-LBD and McpN-LBD (Fig. S1B) did not provide any obvious reason for this difference in ligand specificity data.
The NIT domain is present in different signal transduction protein families and was previously proposed to be a sensor domain for nitrate and nitrite (37). However, the recombinant NIT domain of PA4520 did not bind nitrate or nitrite (Fig. S2) and a mutant defective in this receptor was not affected in nitrate chemotaxis (Fig. 2). In addition, P. putida and P. fluorescens both possess a NIT domain containing a chemoreceptor which, however, did not mediate nitrate chemotaxis under the experimental conditions tested (Fig. 6). The NIT domain may thus represent a superfamily that contains subfamilies with different ligand binding properties and biological functions.
The demonstration of specific nitrate chemotaxis as reported here widens the range of known chemoeffectors and provides the basis for an assessment of this phenotype in other bacteria and for the elucidation of its physiological relevance.
MATERIALS AND METHODS
Bacterial strains, culture media, and growth conditions.
Bacterial strains used are listed in Table 2. Bacteria were grown aerobically at 30°C or 37°C, unless otherwise specified, in lysogeny broth (LB), 2× YT medium (10 g yeast extract liter−1, 16 g Bacto tryptone liter−1, 10 g NaCl liter−1), or MS medium (4.2 g/liter Na2HPO4, 2.8 g/liter KH2PO4, 2.0 g/liter NH4NO3, 0.2 g/liter MgSO4 7H2O, 17.0 mg/liter FeCl3 6H2O, 0.8 mg/CoCl2 6H2O, 0.6 mg/liter CaCl2 2H2O, 0.3 mg/liter Na2MoO4 2H2O, 0.1 mg/liter H3BO3, 0.2 mg/liter ZnSO4 7H2O, 0.2 mg/liter CuSO4 7H2O, 0.2 mg/liter MnSO4 7H2O) supplemented with 20 mM d-glucose as a carbon source. Alternatively, Xanthomonas campestris was grown in M9 minimal medium supplemented with 20 mM d-glucose, 5 mM NaNO3 and 5% (vol/vol) LB medium. E. coli DH5α was used as a host for gene cloning. When necessary, antibiotics were used at the following final concentrations (in micrograms per milliliter): ampicillin, 100; kanamycin, 50, tetracycline, 40.
TABLE 2.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or relevant characteristic(s)a | Reference or source |
|---|---|---|
| Strains | ||
| Escherichia coli BL21(DE3) | F–
ompT gal dcm lon hsdSB(rB– mB–) λ(DE3 [lacI lacUV5-T7p07 ind1 sam7 nin5]) [malB+]K-12(λS) |
86 |
| Escherichia coli DH5α | F–
endA1 glnV44 thi-1 recA1 relA1 gyrA96 deoR nupG purB20 φ80dlacZΔM15 Δ(lacZYA-argF)U169, hsdR17(rK– mK+), λ– |
87 |
| Pseudomonas putida KT2440 | Wild-type strain | 88 |
| Pseudomonas aeruginosa PAO1 | Wild-type strain | 89 |
| Pseudomonas aeruginosa PAO1-McpN | PAO1 transposon mutant pa2788::ISlacZ/hah; Tcr | 90 |
| Pseudomonas aeruginosa PAO1-PilJ | PAO1 transposon mutant pa0411::ISphoA/hah; Tcr | 90 |
| Pseudomonas aeruginosa PAO1-PA4520 | PAO1 transposon mutant pa4520::ISphoA/hah; Tcr | 90 |
| Pseudomonas aeruginosa PAO1-NarX | PAO1 transposon mutant pa3878::ISphoA/hah; Tcr | 90 |
| Pseudomonas aeruginosa PAO1-CheA1 | PAO1 transposon mutant pa1458::ISphoA/hah; Tcr | 90 |
| Pseudomonas aeruginosa PAO1-CheA2 | PAO1 transposon mutant pa0178::ISlacZ/hah; Tcr | 90 |
| Pseudomonas fluorescens KU-7 | Wild-type strain | 91 |
| Pectobacterium atrosepticum SCRI1043 | Wild-type strain | 92 |
| Xanthomonas campestris pv. campestris | Wild-type strain | M. Milagros- Lopez (IVIA, Spain) |
| Plasmids | ||
| pET28b(+) | Protein expression plasmid; Kmr | Novagen |
| pMcpN-LBD | pET28b(+) derivative containing a DNA fragment encoding McpN-LBD cloned into the NdeI/XhoI sites; N-terminal His6 tag; Kmr |
This study |
| pPilJ-LBD | pET28b(+) derivative containing a DNA fragment encoding PilJ-LBD cloned into the NdeI/EcoRI sites; N-terminal His6 tag; Kmr |
This study |
| pET4520-LBD | pET28b(+) derivative containing a DNA fragment encoding PA4520-LBD cloned into the NdeI/SalI sites; N-terminal His6 tag; Kmr |
This study |
| pBBR1MCS2_START | oriRK2 mobRK2; Kmr | 93 |
| pBBRMcpN | pBBR1MCS2_START derivative containing mcpN gene; Kmr | This study |
| pCR2.1-TOPO | PCR cloning vector; ori pUC ori f1 lacZα; Apr, Kmr | Invitrogen |
| pCR-McpN-LBD | pTOPO derivative containing a DNA fragment encoding McpN-LBD; Apr, Kmr | This study |
| pCR-McpN-R61A | pTOPO derivative containing a DNA fragment encoding McpN-LBD (R61A); Apr, Kmr |
This study |
| pMcpN-R61A | pET28b derivative containing a DNA fragment encoding His-tagged McpN-LBD (R61A); Kmr |
This study |
Ap, ampicillin; Km, kanamycin; Tc, tetracycline.
Plasmid construction.
The plasmids and oligonucleotides used are listed in Table 2 and in Table S2 in the supplemental material, respectively. Protein expression plasmids were constructed by amplification from genomic DNA of P. aeruginosa PAO1 for the DNA fragments encoding the LBDs of PilJ (amino acids 36 to 315), McpN (amino acids 44 to 179), and PA4520 (amino acids 38 to 321). The resulting PCR products were cloned into pET28(+) to generate plasmids pPilJ-LBD, pMcpN-LBD, and pET4520-LBD. In all cases, plasmids were verified by sequencing. For the construction of the complementing plasmid pBBRMcpN, the mcpN gene was amplified using primers listed in Table S2. The resulting PCR fragment was cloned into the NdeI and BamHI sites of pBBR1MCS2_START, and the plasmid was transformed into P. aeruginosa PAO1-McpN by electroporation.
Oligonucleotides used in this study. Download Table S2, DOCX file, 0.02 MB (20.3KB, docx) .
Copyright © 2019 Martín-Mora et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Protein overexpression and purification.
E. coli BL21(DE3) was transformed with the expression plasmids, and the resulting strains were grown in 2-liter Erlenmeyer flasks containing 400 ml LB medium supplemented with kanamycin. Cultures were grown under conditions of continuous stirring (200 rpm) at 30°C. The growth temperature was lowered to 16°C when an optical density at 600 nm (OD600) of 0.5 was reached, and protein expression was induced after 30 min by the addition of 0.1 mM isopropyl β-d-1-thiogalactopyranoside. Cultures were grown for another 14 h prior to harvesting of cells by centrifugation at 10,000 × g and 4°C for 30 min. Cell pellets were resuspended in buffer A (20 mM Tris-HCl, 0.1 mM EDTA, 300 mM NaCl, 10 mM imidazole, 5% [vol/vol] glycerol, pH 7.6) and broken by French press treatment at a gauge pressure of 62.5 lb/in2. After centrifugation at 20,000 × g for 1 h, the supernatant was loaded onto a 5-ml HisTrap column (Amersham Bioscience) previously equilibrated with buffer A. After washing with buffer A containing 35 mM imidazole was performed, protein was eluted by the use of a 35 to 500 mM imidazole gradient in buffer A. Proteins were dialyzed into the following buffers for analysis: for PA2788-LBD, 20 mM Tris-HCl (pH 7.4); for PA4520-LBD, 5 mM Tris-HCl, 5 mM MES (morpholineethanesulfonic acid), and 5 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)] (pH 7.5); for PA0411-LBD, 50 mM HEPES (pH 7.5).
Differential scanning fluorimetry (DSF).
DSF assays were performed on a MyIQ2 Real-Time PCR instrument (Bio-Rad). Compounds from different arrays (Biolog, Hayward, CA, USA) were dissolved in 50 μl water, which, according to the manufacturer, corresponds to a concentration of 10 to 20 mM. The composition of these arrays is provided in http://208.106.130.253/pdf/pm_lit/PM1-PM10.pdf. Screening was performed using 96-well plates. Each well contained 2.5 μl of the dissolved compound, 20.5 μl protein, and 2 μl SYPRO Orange (Life Technologies). The control well contained protein without ligand. Samples were heated from 23°C to 85°C at a scan rate of 1°C/min, and fluorescence changes were monitored. Tm values correspond to the minima of the first derivatives of the raw data.
Isothermal titration calorimetry (ITC).
Experiments were performed on a VP microcalorimeter (Microcal, Amherst, MA, USA) at 25°C. Proteins were placed into the sample cell (36 to 65 µM). Compound solutions (1 to 5 mM) were prepared in dialysis buffer and placed into the injector syringe. Titrations involved the injection of 9.6-µl to 19.2-µl aliquots of compound solution into the protein. In cases in which no binding was observed, the experiment was repeated at an analysis temperature of 15°C. The mean enthalpy values from the titration of buffer with compounds were subtracted from raw titration data prior to data analysis performed with the “One binding site model” of the MicroCal version of ORIGIN.
Analytical ultracentrifugation (AUC).
Experiments were performed on a Beckman Coulter Optima XL-I analytical ultracentrifuge (Beckman-Coulter, Palo Alto, CA, USA) equipped with UV-visible light absorbance and interference optics detection systems, using an An50Ti 8-hole rotor and 12-mm-path-length charcoal-filled epon double-sector centerpieces. The experiments were carried out at 10°C using 5 μM to 40 μM McpN-LBD in the absence and presence of 0.6 mM NaNO3.
Sedimentation velocity (SV) runs were carried out at a rotor speed of 48,000 rpm using 400-µl samples with the dialysis buffer as the reference. A laser was used at a wavelength of 235 nm in the absorbance optics mode. Least-squares boundary modeling of the SV data was used to calculate sedimentation coefficient distributions with the size-distribution c(s) method (74) implemented in SEDFIT v14.1 software. Buffer density (ρ = 1.003 g/ml [0.99989 g/ml in the presence of NaNO3]) and viscosity [η = 0.013137 poise [0.01313 poise in the presence of NaNO3]) at 10°C were estimated using SEDNTERP software (75) for the buffer components. The partial specific volume used was 0.7192 ml/g as calculated from the amino acid sequence using SEDNTERP software.
Intrinsic tryptophan fluorescence spectroscopy.
McpN-LBD and McpN-LBD R61A mutants were dialyzed into 20 mM Tris-HCl (pH 7.4), and the reaction mixtures were adjusted to a concentration of 5 μM. Proteins were placed into a PTI QM-2003 fluorimeter (Photon Technology International, Lawrenceville, NJ), and emission spectra were recorded at wavelengths of 305 to 400 nm following excitation at 295 nm. Spectra were recorded at 20°C using a slit width of 4 nm with a scan speed of 1 nm/s. Spectra were corrected with the buffer emission spectrum.
Quantitative capillary chemotaxis assays.
Assays were conducted using two different protocols that differed under the cell culture conditions. Under conditions of nitrate abundance, overnight cultures in MS minimal medium supplemented with 20 mM glucose as a carbon source (note that this medium contains 25 mM NH4NO3) were used to inoculate fresh medium to reach an OD600 of 0.05. Cells were cultured at 30°C or 37°C until an OD600 of 0.4 to 0.5 was reached. Under conditions of nitrate limitation, 150 µl of an overnight culture in rich 2× YT medium was used to inoculate 20 ml of N0 medium (MS lacking a nitrogen source). Growth was continued for 3 h (pseudomonads) or 4.5 h (P. atrosepticum), at which point the cells had reached an OD600 of 0.15 to 0.2. For X. campestris, M9 minimal medium supplemented with 20 mM d-glucose, 5 mM NaNO3, and 5% (vol/vol) LB was used for the conditions of nitrate abundance, whereas M8 minimal medium (M9 without nitrogen source) supplemented with 20 mM d-glucose and 5% (vol/vol) LB was used for the conditions of nitrate limitation. Cells were grown for 6 h until the OD600 reached 0.25 to 0.3.
Under both conditions, cells were washed twice by centrifugation (1,667 × g and 6 min at 4°C) and resuspension in chemotaxis buffer (50 mM potassium phosphate, 20 µM EDTA, 0.05% [vol/vol] glycerol, pH 7.0) and then resuspended in the same buffer to reach an OD600 of 0.1. Aliquots (230 µl) of the resulting cell suspension were placed into the wells of a 96-well microtiter plate. Capillaries (Microcaps; Drummond Scientific [reference P1424]) (1 μl) were heat-sealed at one end and filled with buffer (control) or chemoeffector solution prepared in chemotaxis buffer. The capillaries were immersed into the bacterial suspensions at its open end. After 30 min, capillaries were removed from the wells, rinsed with sterile water, and emptied into 1 ml of chemotaxis buffer. Serial dilutions were plated onto M9 minimal medium plates supplemented with 20 mM glucose and incubated overnight at 30 or 37°C. CFU counts were determined and corrected with the buffer control.
RT-qPCR gene expression analysis.
Total RNA was extracted using a High Pure RNA isolation kit (Roche Diagnostics) and treated with Turbo DNase (Invitrogen). RNA quality was verified by agarose gel electrophoresis and quantified spectrophotometrically. Subsequently, cDNA was synthesized from 500 ng RNA using SuperScript II reverse transcriptase (Invitrogen) and 200 ng of random hexamer primers (Roche) following the instructions of the manufacturers. Quantitative PCR was performed using iQ SYBR green supermix (Bio-Rad) in a MyiQ2 thermal cycler (Bio-Rad). The following protocol was used: 95°C (5 min), 35 cycles of 95°C (10 s) and 61°C (30 s), and melting curve analysis from 55 to 95°C, with an increment of 0.5°C/10 s. Gene expression data were normalized to expression of the rpoD reference gene. The primers used are listed in Table S2.
McpN-LBD crystallization and structure resolution.
Crystallization conditions were screened using the capillary counter-diffusion technique and commercially available crystallization kits GCB-CSK, PEG448-49, and AS-49 (Triana Science & Technology, Granada, Spain). The protein, maintained at 1.5 mg/ml in 20 mM Tris-HCl–200 mM NaCl (pH 7.5), was incubated at 4°C with 1.7 mM NaNO3, and the excess of NaNO3 was removed by centrifugation using Amicon concentrators (3-kDa cutoff). The protein-ligand complex was loaded into 0.2-mm-inner-diameter capillaries, and crystals of sufficient size appeared in 0.82 M K/phosphate–0.82 M Na/phosphate (0.1 M Na/HEPES, pH 7.5). Crystals were extracted from the capillary, flash-cooled in liquid nitrogen, and stored until data collection. Crystals were diffracted at beam line ID23-1 of the European Synchrotron Radiation Facility (ESRF). Data were indexed and integrated with XDS (76) and scaled with SCALA (77). Attempts at molecular replacement using homology models generated using the NarX sensor domain (PDB identifier [ID] 3EZI) and the NarQ sensor domain (PDB ID 5IJI) were unsuccessful. Phases were obtained using Arcimboldo (78) and searching for two helices that were 30 amino acids in length. Refinement was initiated with Refmac (79) and finalized with phenix.refine (80), tracking the quality with MolProbity (81). Refinement statistics and quality indicators are summarized in Table S3.
Data collection and refinement statistics of the 3D structure of the McpN-LBD-nitrate complex. Values in parentheses represent the highest-resolution shell. Download Table S3, DOCX file, 0.02 MB (21KB, docx) .
Copyright © 2019 Martín-Mora et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Site-directed mutagenesis.
An overlapping PCR mutagenesis approach was used to construct the alanine substitution mutant McpN-LBD R61A. First, a NdeI/XhoI DNA fragment of pMcpN-LBD was cloned into the same sites of pCR2.1-TOPO and transformed into E. coli DH5α (dam positive [dam+]). Next, the resulting pCR-McpN-LBD plasmid was fully amplified by PCR using a complementary primer pair carrying the mutation. The parental plasmid was cleaved using DpnI, and plasmids with the desired mutation were recirculated with T4 DNA ligase (Roche). The presence of the mutation in the resulting plasmid, pCR-McpN-R61A, was confirmed by sequencing prior to cloning into the NdeI/XhoI sites of pET28(+) to generate pMcpN-R61A (Table 2).
Data availability.
Coordinates and structure factors of McpN-LBD were deposited at the PDB with accession code 6GCV.
ACKNOWLEDGMENTS
We acknowledge the European Synchrotron Radiation Facility (ID23-1 and ID30A-1,3) and thank the beam line staff members for their invaluable support.
This work was supported by FEDER funds and Fondo Social Europeo through grants from the Junta de Andalucía (grant CVI-7335) and the Spanish Ministry for Economy and Competitiveness (grants BIO2013-42297, BIO2016-76779-P, and BIO2016-74875-P). We furthermore acknowledge NIH grant P30 DK089507, which financed the generation of bacterial mutants.
Footnotes
Citation Martín-Mora D, Ortega Á, Matilla MA, Martínez-Rodríguez S, Gavira JA, Krell T. 2019. The molecular mechanism of nitrate chemotaxis via direct ligand binding to the PilJ domain of McpN. mBio 10:e02334-18. https://doi.org/10.1128/mBio.02334-18.
REFERENCES
- 1.Wuichet K, Zhulin IB. 2010. Origins and diversification of a complex signal transduction system in prokaryotes. Sci Signal 3:ra50. doi: 10.1126/scisignal.2000724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bi S, Sourjik V. 2018. Stimulus sensing and signal processing in bacterial chemotaxis. Curr Opin Microbiol 45:22–29. doi: 10.1016/j.mib.2018.02.002. [DOI] [PubMed] [Google Scholar]
- 3.Matilla MA, Krell T. 2017. Chemoreceptor-based signal sensing. Curr Opin Biotechnol 45:8–14. doi: 10.1016/j.copbio.2016.11.021. [DOI] [PubMed] [Google Scholar]
- 4.Alexandre G. 2010. Coupling metabolism and chemotaxis-dependent behaviours by energy taxis receptors. Microbiology 156:2283–2293. doi: 10.1099/mic.0.039214-0. [DOI] [PubMed] [Google Scholar]
- 5.Alexandre G, Greer-Phillips S, Zhulin IB. 2004. Ecological role of energy taxis in microorganisms. FEMS Microbiol Rev 28:113–126. doi: 10.1016/j.femsre.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 6.Taylor BL, Zhulin IB, Johnson MS. 1999. Aerotaxis and other energy-sensing behavior in bacteria. Annu Rev Microbiol 53:103–128. doi: 10.1146/annurev.micro.53.1.103. [DOI] [PubMed] [Google Scholar]
- 7.Schweinitzer T, Josenhans C. 2010. Bacterial energy taxis: a global strategy? Arch Microbiol 192:507–520. doi: 10.1007/s00203-010-0575-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parkinson JS, Hazelbauer GL, Falke JJ. 2015. Signaling and sensory adaptation in Escherichia coli chemoreceptors: 2015 update. Trends Microbiol 23:257–266. doi: 10.1016/j.tim.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ortega A, Zhulin IB, Krell T. 2017. Sensory repertoire of bacterial chemoreceptors. Microbiol Mol Biol Rev 81:e00033-17. doi: 10.1128/MMBR.00033-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kato J, Kim HE, Takiguchi N, Kuroda A, Ohtake H. 2008. Pseudomonas aeruginosa as a model microorganism for investigation of chemotactic behaviors in ecosystem. J Biosci Bioeng 106:1–7. doi: 10.1263/jbb.106.1. [DOI] [PubMed] [Google Scholar]
- 11.Ortega DR, Fleetwood AD, Krell T, Harwood CS, Jensen GJ, Zhulin IB. 2017. Assigning chemoreceptors to chemosensory pathways in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 114:12809–12814. doi: 10.1073/pnas.1708842114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kato J, Nakamura T, Kuroda A, Ohtake H. 1999. Cloning and characterization of chemotaxis genes in Pseudomonas aeruginosa. Biosci Biotechnol Biochem 63:155–161. doi: 10.1271/bbb.63.155. [DOI] [PubMed] [Google Scholar]
- 13.Masduki A, Nakamura J, Ohga T, Umezaki R, Kato J, Ohtake H. 1995. Isolation and characterization of chemotaxis mutants and genes of Pseudomonas aeruginosa. J Bacteriol 177:948–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferrandez A, Hawkins AC, Summerfield DT, Harwood CS. 2002. Cluster II che genes from Pseudomonas aeruginosa are required for an optimal chemotactic response. J Bacteriol 184:4374–4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whitchurch CB, Leech AJ, Young MD, Kennedy D, Sargent JL, Bertrand JJ, Semmler AB, Mellick AS, Martin PR, Alm RA, Hobbs M, Beatson SA, Huang B, Nguyen L, Commolli JC, Engel JN, Darzins A, Mattick JS. 2004. Characterization of a complex chemosensory signal transduction system which controls twitching motility in Pseudomonas aeruginosa. Mol Microbiol 52:873–893. doi: 10.1111/j.1365-2958.2004.04026.x. [DOI] [PubMed] [Google Scholar]
- 16.Fulcher NB, Holliday PM, Klem E, Cann MJ, Wolfgang MC. 2010. The Pseudomonas aeruginosa Chp chemosensory system regulates intracellular cAMP levels by modulating adenylate cyclase activity. Mol Microbiol 76:889–904. doi: 10.1111/j.1365-2958.2010.07135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jansari VH, Potharla VY, Riddell GT, Bardy SL. 2016. Twitching motility and cAMP levels: signal transduction through a single methyl-accepting chemotaxis protein. FEMS Microbiol Lett 363:fnw119. doi: 10.1093/femsle/fnw119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Persat A, Inclan YF, Engel JN, Stone HA, Gitai Z. 2015. Type IV pili mechanochemically regulate virulence factors in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 112:7563–7568. doi: 10.1073/pnas.1502025112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hickman JW, Tifrea DF, Harwood CS. 2005. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc Natl Acad Sci U S A 102:14422–14427. doi: 10.1073/pnas.0507170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ulrich LE, Zhulin IB. 2005. Four-helix bundle: a ubiquitous sensory module in prokaryotic signal transduction. Bioinformatics 21:iii45. doi: 10.1093/bioinformatics/bti1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Upadhyay AA, Fleetwood AD, Adebali O, Finn RD, Zhulin IB. 2016. Cache domains that are homologous to, but different from PAS domains comprise the largest superfamily of extracellular sensors in prokaryotes. PLoS Comput Biol 12:e1004862. doi: 10.1371/journal.pcbi.1004862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ortega A, Krell T. 2014. The HBM domain: introducing bimodularity to bacterial sensing. Protein Sci 23:332–336. doi: 10.1002/pro.2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu H, Kato J, Kuroda A, Ikeda T, Takiguchi N, Ohtake H. 2000. Identification and characterization of two chemotactic transducers for inorganic phosphate in Pseudomonas aeruginosa. J Bacteriol 182:3400–3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rico-Jiménez M, Reyes-Darias JA, Ortega Á, Díez Peña AI, Morel B, Krell T. 2016. Two different mechanisms mediate chemotaxis to inorganic phosphate in Pseudomonas aeruginosa. Sci Rep 6:28967. doi: 10.1038/srep28967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin-Mora D, Ortega A, Reyes-Darias JA, García V, López-Farfán D, Matilla MA, Krell T. 2016. Identification of a chemoreceptor in Pseudomonas aeruginosa that specifically mediates chemotaxis towards alpha-ketoglutarate. Front Microbiol 7:1937. doi: 10.3389/fmicb.2016.01937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taguchi K, Fukutomi H, Kuroda A, Kato J, Ohtake H. 1997. Genetic identification of chemotactic transducers for amino acids in Pseudomonas aeruginosa. Microbiology 143:3223–3229. doi: 10.1099/00221287-143-10-3223. [DOI] [PubMed] [Google Scholar]
- 27.Rico-Jimenez M, Munoz-Martinez F, Garcia-Fontana C, Fernandez M, Morel B, Ortega A, Ramos JL, Krell T. 2013. Paralogous chemoreceptors mediate chemotaxis towards protein amino acids and the non-protein amino acid gamma-aminobutyrate (GABA). Mol Microbiol 88:1230–1243. doi: 10.1111/mmi.12255. [DOI] [PubMed] [Google Scholar]
- 28.Reyes-Darias JA, Yang Y, Sourjik V, Krell T. 2015. Correlation between signal input and output in PctA and PctB amino acid chemoreceptor of Pseudomonas aeruginosa. Mol Microbiol 96:513–525. doi: 10.1111/mmi.12953. [DOI] [PubMed] [Google Scholar]
- 29.Reyes-Darias JA, García V, Rico-Jiménez M, Corral-Lugo A, Lesouhaitier O, Juárez-Hernández D, Yang Y, Bi S, Feuilloley M, Muñoz-Rojas J, Sourjik V, Krell T. 2015. Specific gamma-aminobutyrate chemotaxis in pseudomonads with different lifestyle. Mol Microbiol 97:488–501. doi: 10.1111/mmi.13045. [DOI] [PubMed] [Google Scholar]
- 30.Alvarez-Ortega C, Harwood CS. 2007. Identification of a malate chemoreceptor in Pseudomonas aeruginosa by screening for chemotaxis defects in an energy taxis-deficient mutant. Appl Environ Microbiol 73:7793–7795. doi: 10.1128/AEM.01898-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin-Mora D, Ortega A, Perez-Maldonado FJ, Krell T, Matilla MA. 2018. The activity of the C4-dicarboxylic acid chemoreceptor of Pseudomonas aeruginosa is controlled by chemoattractants and antagonists. Sci Rep 8:2102. doi: 10.1038/s41598-018-20283-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, Salazar GA, Tate J, Bateman A. 2016. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res 44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krell T. 2015. Tackling the bottleneck in bacterial signal transduction research: high-throughput identification of signal molecules. Mol Microbiol 96:685–688. doi: 10.1111/mmi.12975. [DOI] [PubMed] [Google Scholar]
- 34.Lee AI, Delgado A, Gunsalus RP. 1999. Signal-dependent phosphorylation of the membrane-bound NarX two-component sensor-transmitter protein of Escherichia coli: nitrate elicits a superior anion ligand response compared to nitrite. J Bacteriol 181:5309–5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stewart V. 2003. Biochemical Society Special Lecture. Nitrate- and nitrite-responsive sensors NarX and NarQ of proteobacteria. Biochem Soc Trans 31:1–10. doi: 10.1042/bst0310001. [DOI] [PubMed] [Google Scholar]
- 36.Williams SB, Stewart V. 1997. Discrimination between structurally related ligands nitrate and nitrite controls autokinase activity of the NarX transmembrane signal transducer of Escherichia coli K-12. Mol Microbiol 26:911–925. [DOI] [PubMed] [Google Scholar]
- 37.Shu CJ, Ulrich LE, Zhulin IB. 2003. The NIT domain: a predicted nitrate-responsive module in bacterial sensory receptors. Trends Biochem Sci 28:121–124. doi: 10.1016/S0968-0004(03)00032-X. [DOI] [PubMed] [Google Scholar]
- 38.Van Alst NE, Sherrill LA, Iglewski BH, Haidaris CG. 2009. Compensatory periplasmic nitrate reductase activity supports anaerobic growth of Pseudomonas aeruginosa PAO1 in the absence of membrane nitrate reductase. Can J Microbiol 55:1133–1144. doi: 10.1139/w09-065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pineda-Molina E, Reyes-Darias J-A, Lacal J, Ramos JL, García-Ruiz JM, Gavira JA, Krell T. 2012. Evidence for chemoreceptors with bimodular ligand-binding regions harboring two signal-binding sites. Proc Natl Acad Sci U S A 109:18926–18931. doi: 10.1073/pnas.1201400109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ortega A, Amoros D, Garcia de la Torre J. 2011. Prediction of hydrodynamic and other solution properties of rigid proteins from atomic- and residue-level models. Biophys J 101:892–898. doi: 10.1016/j.bpj.2011.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sigrist CJ, de Castro E, Cerutti L, Cuche BA, Hulo N, Bridge A, Bougueleret L, Xenarios I. 2013. New and continuing developments at PROSITE. Nucleic Acids Res 41:D344–D347. doi: 10.1093/nar/gks1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mansfield J, Genin S, Magori S, Citovsky V, Sriariyanum M, Ronald P, Dow M, Verdier V, Beer SV, Machado MA, Toth I, Salmond G, Foster GD. 2012. Top 10 plant pathogenic bacteria in molecular plant pathology. Mol Plant Pathol 13:614–629. doi: 10.1111/j.1364-3703.2012.00804.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taylor BL, Miller JB, Warrick HM, Koshland DE Jr.. 1979. Electron acceptor taxis and blue light effect on bacterial chemotaxis. J Bacteriol 140:567–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rivera-Chavez F, Lopez CA, Zhang LF, Garcia-Pastor L, Chavez-Arroyo A, Lokken KL, Tsolis RM, Winter SE, Baumler AJ. 2016. Energy taxis toward host-derived nitrate supports a Salmonella pathogenicity island 1-independent mechanism of invasion. mBio 7:e00960-16. doi: 10.1128/mBio.00960-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roush CJ, Lastoskie CM, Worden RM. 2006. Denitrification and chemotaxis of Pseudomonas stutzeri KC in porous media. J Environ Sci Health A Tox Hazard Subst Environ Eng 41:967–983. doi: 10.1080/10934520600689258. [DOI] [PubMed] [Google Scholar]
- 46.Dennis PG, Seymour J, Kumbun K, Tyson GW. 2013. Diverse populations of lake water bacteria exhibit chemotaxis towards inorganic nutrients. ISME J 7:1661–1664. doi: 10.1038/ismej.2013.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kennedy MJ, Lawless JG. 1985. Role of chemotaxis in the ecology of denitrifiers. Appl Environ Microbiol 49:109–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Emerson D. 1999. Complex pattern formation by Pseudomonas strain KC in response to nitrate and nitrite. Microbiology 145:633–641. doi: 10.1099/13500872-145-3-633. [DOI] [PubMed] [Google Scholar]
- 49.Baraquet C, Théraulaz L, Iobbi-Nivol C, Méjean V, Jourlin-Castelli C. 2009. Unexpected chemoreceptors mediate energy taxis towards electron acceptors in Shewanella oneidensis. Mol Microbiol 73:278–290. doi: 10.1111/j.1365-2958.2009.06770.x. [DOI] [PubMed] [Google Scholar]
- 50.Nealson KH, Moser DP, Saffarini DA. 1995. Anaerobic electron acceptor chemotaxis in Shewanella putrefaciens. Appl Environ Microbiol 61:1551–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Greer-Phillips SE, Stephens BB, Alexandre G. 2004. An energy taxis transducer promotes root colonization by Azospirillum brasilense. J Bacteriol 186:6595–6604. doi: 10.1128/JB.186.19.6595-6604.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee DY, Ramos A, Macomber L, Shapleigh JP. 2002. Taxis response of various denitrifying bacteria to nitrate and nitrite. Appl Environ Microbiol 68:2140–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zopfi J, Kjaer T, Nielsen LP, Jorgensen BB. 2001. Ecology of Thioploca spp.: nitrate and sulfur storage in relation to chemical microgradients and influence of Thioploca spp. on the sedimentary nitrogen cycle. Appl Environ Microbiol 67:5530–5537. doi: 10.1128/AEM.67.12.5530-5537.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Willey JM, Waterbury JB. 1989. Chemotaxis toward nitrogenous compounds by swimming strains of marine Synechococcus spp. Appl Environ Microbiol 55:1888–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sparacino-Watkins C, Stolz JF, Basu P. 2014. Nitrate and periplasmic nitrate reductases. Chem Soc Rev 43:676–706. doi: 10.1039/c3cs60249d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xie W, Wang F, Guo L, Chen Z, Sievert SM, Meng J, Huang G, Li Y, Yan Q, Wu S, Wang X, Chen S, He G, Xiao X, Xu A. 2011. Comparative metagenomics of microbial communities inhabiting deep-sea hydrothermal vent chimneys with contrasting chemistries. ISME J 5:414–426. doi: 10.1038/ismej.2010.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schulz HN, Brinkhoff T, Ferdelman TG, Marine MH, Teske A, Jorgensen BB. 1999. Dense populations of a giant sulfur bacterium in Namibian shelf sediments. Science 284:493–495. [DOI] [PubMed] [Google Scholar]
- 58.McHatton SC, Barry JP, Jannasch HW, Nelson DC. 1996. High nitrate concentrations in vacuolate, autotrophic marine Beggiatoa spp. Appl Environ Microbiol 62:954–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ahmad A, Barry JP, Nelson DC. 1999. Phylogenetic affinity of a wide, vacuolate, nitrate-accumulating Beggiatoa sp. from Monterey Canyon, California, with Thioploca spp. Appl Environ Microbiol 65:270–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Preisler A, de Beer D, Lichtschlag A, Lavik G, Boetius A, Jørgensen BB. 2007. Biological and chemical sulfide oxidation in a Beggiatoa inhabited marine sediment. ISME J 1:341–353. doi: 10.1038/ismej.2007.50. [DOI] [PubMed] [Google Scholar]
- 61.Van Alst NE, Picardo KF, Iglewski BH, Haidaris CG. 2007. Nitrate sensing and metabolism modulate motility, biofilm formation, and virulence in Pseudomonas aeruginosa. Infect Immun 75:3780–3790. doi: 10.1128/IAI.00201-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Linnane SJ, Keatings VM, Costello CM, Moynihan JB, O'Connor CM, Fitzgerald MX, McLoughlin P. 1998. Total sputum nitrate plus nitrite is raised during acute pulmonary infection in cystic fibrosis. Am J Respir Crit Care Med 158:207–212. doi: 10.1164/ajrccm.158.1.9707096. [DOI] [PubMed] [Google Scholar]
- 63.Parales RE. 2004. Nitrobenzoates and aminobenzoates are chemoattractants for Pseudomonas strains. Appl Environ Microbiol 70:285–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shi K, Fan X, Qiao Z, Han Y, McDermott TR, Wang Q, Wang G. 2017. Arsenite oxidation regulator AioR regulates bacterial chemotaxis towards arsenite in Agrobacterium tumefaciens GW4. Sci Rep 7:43252. doi: 10.1038/srep43252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bains M, Fernandez L, Hancock RE. 2012. Phosphate starvation promotes swarming motility and cytotoxicity of Pseudomonas aeruginosa. Appl Environ Microbiol 78:6762–6768. doi: 10.1128/AEM.01015-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zaborin A, Romanowski K, Gerdes S, Holbrook C, Lepine F, Long J, Poroyko V, Diggle SP, Wilke A, Righetti K, Morozova I, Babrowski T, Liu DC, Zaborina O, Alverdy JC. 2009. Red death in Caenorhabditis elegans caused by Pseudomonas aeruginosa PAO1. Proc Natl Acad Sci U S A 106:6327–6332. doi: 10.1073/pnas.0813199106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ottemann KM, Xiao W, Shin YK, Koshland DE Jr.. 1999. A piston model for transmembrane signaling of the aspartate receptor. Science 285:1751–1754. doi: 10.1126/science.285.5434.1751. [DOI] [PubMed] [Google Scholar]
- 68.Milburn MV, Prive GG, Milligan DL, Scott WG, Yeh J, Jancarik J, Koshland DE Jr, Kim SH. 1991. Three-dimensional structures of the ligand-binding domain of the bacterial aspartate receptor with and without a ligand. Science 254:1342–1347. doi: 10.1126/science.1660187. [DOI] [PubMed] [Google Scholar]
- 69.Milligan DL, Koshland DE Jr.. 1993. Purification and characterization of the periplasmic domain of the aspartate chemoreceptor. J Biol Chem 268:19991–19997. [PubMed] [Google Scholar]
- 70.Fernandez M, Matilla MA, Ortega A, Krell T. 2017. Metabolic value chemoattractants are preferentially recognized at broad ligand range chemoreceptor of Pseudomonas putida KT2440. Front Microbiol 8:990. doi: 10.3389/fmicb.2017.00990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Noriega CE, Lin HY, Chen LL, Williams SB, Stewart V. 2010. Asymmetric cross-regulation between the nitrate-responsive NarX-NarL and NarQ-NarP two-component regulatory systems from Escherichia coli K-12. Mol Microbiol 75:394–412. doi: 10.1111/j.1365-2958.2009.06987.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cheung J, Hendrickson WA. 2009. Structural analysis of ligand stimulation of the histidine kinase NarX. Structure 17:190–201. doi: 10.1016/j.str.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gushchin I, Melnikov I, Polovinkin V, Ishchenko A, Yuzhakova A, Buslaev P, Bourenkov G, Grudinin S, Round E, Balandin T, Borshchevskiy V, Willbold D, Leonard G, Buldt G, Popov A, Gordeliy V. 2017. Mechanism of transmembrane signaling by sensor histidine kinases. Science 356:eaah6345. doi: 10.1126/science.aah6345. [DOI] [PubMed] [Google Scholar]
- 74.Schuck P. 2000. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys J 78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Laue TM, Shah BD, Ridgeway TM, Pelletier SL. 1992. Computer-aided interpretation of analytical sedimentation data for proteins, p 90–125. In Harding SE, Rowe AJ, Horton JC (ed), Analytical ultracentrifugation in biochemistry and polymer science. Royal Society of Chemistry, Cambridge, United Kingdom. [Google Scholar]
- 76.Kabsch W. 2010. XDS. Acta Crystallogr D Biol Crystallogr 66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Evans P. 2006. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr 62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 78.Sammito M, Millán C, Frieske D, Rodríguez-Freire E, Borges RJ, Usón I. 2015. ARCIMBOLDO_LITE: single-workstation implementation and use. Acta Crystallogr D Biol Crystallogr 71:1921–1930. doi: 10.1107/S1399004715010846. [DOI] [PubMed] [Google Scholar]
- 79.Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. 2011. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr 67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD. 2012. Towards automated crystallographic structure refinement withphenix.refine. Acta Crystallogr D Biol Crystallogr 68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen VB, Arendall WB III, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. 2010. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li J, Wang C, Yang G, Sun Z, Guo H, Shao K, Gu Y, Jiang W, Zhang P. 2017. Molecular mechanism of environmental d-xylose perception by a XylFII-LytS complex in bacteria. Proc Natl Acad Sci U S A 114:8235–8240. doi: 10.1073/pnas.1620183114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pokkuluri PR, Dwulit-Smith J, Duke NE, Wilton R, Mack JC, Bearden J, Rakowski E, Babnigg G, Szurmant H, Joachimiak A, Schiffer M. 2013. Analysis of periplasmic sensor domains from Anaeromyxobacter dehalogenans 2CP-C: structure of one sensor domain from a histidine kinase and another from a chemotaxis protein. Microbiologyopen 2:766–777. doi: 10.1002/mbo3.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Moore JO, Hendrickson WA. 2012. An asymmetry-to-symmetry switch in signal transmission by the histidine kinase receptor for TMAO. Structure 20:729–741. doi: 10.1016/j.str.2012.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rangarajan ES, Izard T. 2013. Dimer asymmetry defines alpha-catenin interactions. Nat Struct Mol Biol 20:188–193. doi: 10.1038/nsmb.2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jeong H, Barbe V, Lee CH, Vallenet D, Yu DS, Choi SH, Couloux A, Lee SW, Yoon SH, Cattolico L, Hur CG, Park HS, Segurens B, Kim SC, Oh TK, Lenski RE, Studier FW, Daegelen P, Kim JF. 2009. Genome sequences of Escherichia coli B strains REL606 and BL21(DE3). J Mol Biol 394:644–652. doi: 10.1016/j.jmb.2009.09.052. [DOI] [PubMed] [Google Scholar]
- 87.Woodcock DM, Crowther PJ, Doherty J, Jefferson S, DeCruz E, Noyer-Weidner M, Smith SS, Michael MZ, Graham MW. 1989. Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res 17:3469–3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Belda E, van Heck RGA, José Lopez-Sanchez M, Cruveiller S, Barbe V, Fraser C, Klenk H-P, Petersen J, Morgat A, Nikel PI, Vallenet D, Rouy Z, Sekowska A, Martins dos Santos VAP, de Lorenzo V, Danchin A, Médigue C. 2016. The revisited genome of Pseudomonas putida KT2440 enlightens its value as a robust metabolic chassis. Environ Microbiol 18:3403–3424. doi: 10.1111/1462-2920.13230. [DOI] [PubMed] [Google Scholar]
- 89.Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, Brinkman FS, Hufnagle WO, Kowalik DJ, Lagrou M, Garber RL, Goltry L, Tolentino E, Westbrock-Wadman S, Yuan Y, Brody LL, Coulter SN, Folger KR, Kas A, Larbig K, Lim R, Smith K, Spencer D, Wong GK, Wu Z, Paulsen IT, Reizer J, Saier MH, Hancock RE, Lory S, Olson MV. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 90.Jacobs MA, Alwood A, Thaipisuttikul I, Spencer D, Haugen E, Ernst S, Will O, Kaul R, Raymond C, Levy R, Chun-Rong L, Guenthner D, Bovee D, Olson MV, Manoil C. 2003. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 100:14339–14344. doi: 10.1073/pnas.2036282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hasegawa Y, Muraki T, Tokuyama T, Iwaki H, Tatsuno M, Lau PC. 2000. A novel degradative pathway of 2-nitrobenzoate via 3-hydroxyanthranilate in Pseudomonas fluorescens strain KU-7. FEMS Microbiol Lett 190:185–190. doi: 10.1111/j.1574-6968.2000.tb09284.x. [DOI] [PubMed] [Google Scholar]
- 92.Bell KS, Sebaihia M, Pritchard L, Holden MT, Hyman LJ, Holeva MC, Thomson NR, Bentley SD, Churcher LJ, Mungall K, Atkin R, Bason N, Brooks K, Chillingworth T, Clark K, Doggett J, Fraser A, Hance Z, Hauser H, Jagels K, Moule S, Norbertczak H, Ormond D, Price C, Quail MA, Sanders M, Walker D, Whitehead S, Salmond GP, Birch PR, Parkhill J, Toth IK. 2004. Genome sequence of the enterobacterial phytopathogen Erwinia carotovora subsp. atroseptica and characterization of virulence factors. Proc Natl Acad Sci U S A 101:11105–11110. doi: 10.1073/pnas.0402424101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Obranic S, Babic F, Maravic-Vlahovicek G. 2013. Improvement of pBBR1MCS plasmids, a very useful series of broad-host-range cloning vectors. Plasmid 70:263–267. doi: 10.1016/j.plasmid.2013.04.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Specificity of nitrate recognition at McpN-LBD. (A) Microcalorimetric titration of McpN-LBD with NaNO3 in the absence and presence of 20 mM NaNO2. (Upper panel) In black, raw data for the titration of 36 µM McpN-LBD with 12.8-μl aliquots of 2 mM NaNO3; in red, repetition of the experiment with both ligands containing in addition 20 mM NaNO2. (Lower panel) Concentration data representing normalized and dilution heat-corrected integrated peak areas of raw data. The continuous lines represent the curve fits obtained using the “One binding site model” of ORIGIN. (B) Superimposition of the nitrate molecule in the structures of McpN-LBD (in blue) and NarX-LBD (in orange; PDB ID 3EZH). Shown are the amino acid residues in the vicinity of bound nitrate. Hydrogen bonds are indicated by dotted lines. Download FIG S1, TIF file, 2.4 MB (2.4MB, tif) .
Copyright © 2019 Martín-Mora et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The absence of nitrite and nitrate binding to PilJ-LBD and PA4520-LBD. (A and B) Results from thermal shift assays. Shown are changes in the Tm values of PilJ-LBD (A) and PA4520-LBD (B) in the presence of compounds from Biolog array PM3B. (C) Microcalorimetric titration of PilJ-LBD and PA4520-LBD with NaNO3 and NaNO2. As a control, the titration of buffer with NaNO3 is shown. The injection volume was 19.2 µl. Download FIG S2, TIF file, 2.6 MB (2.6MB, tif) .
Copyright © 2019 Martín-Mora et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Quantitative capillary chemotaxis assays of P. aeruginosa PAO1 and the mcpN mutant under conditions of abundant nitrate. Cells were grown in M9 minimal medium supplemented with glucose as a carbon source. This medium contains 25 mM ammonium nitrate as a nitrogen source. Data were corrected using the number of cells that swam into buffer containing capillaries (1,992 ± 298). Download FIG S3, TIF file, 0.07 MB (70.2KB, tif) .
Copyright © 2019 Martín-Mora et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Quantitative capillary chemotaxis assays of P. aeruginosa PAO1 and mutants deficient in cheA1 (PA1458) and cheA2 (PA0178) in the presence of 500 µM NaNO3. Data represent means and standard deviations of results from three biological replicates conducted in triplicate. Download FIG S4, TIF file, 1.4 MB (1.4MB, tif) .
Copyright © 2019 Martín-Mora et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Analysis of the McpN-LBD R61A mutant. (A) Intrinsic tryptophan fluorescence emission spectra of McpN-LBD and McpN-LBD R61A. (B) Thermal shift assay of McpN-LBD R61A in the absence and presence of NaNO3. (C) Microcalorimetric titration of McpN-LBD R61A with NaNO3. Download FIG S5, TIF file, 2.9 MB (2.9MB, tif) .
Copyright © 2019 Martín-Mora et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Alignment of the McpN-LBD sequences with homologues from other species. The alignment is based on a BLAST-P search of McpN-LBD in the NCBI database of nonredundant sequences, excluding species of the genus Pseudomonas. The alignment was done using the CLUSTALW algorithm of npsa software (C. Combet, C. Blanchet, C. Geourjon, and G. Deléage, Trends Biochem Sci 25:147–150, 2000, https://www.cell.com/trends/biochemical-sciences/fulltext/S0968-0004(99)01540-6?_returnURL=https%3A%2F%2Flinkinghub.elsevier.com%2Fretrieve%2Fpii%2FS0968000499015406%3Fshowall%3Dtrue). The BLOSUM62 matrix was used, and the settings used for analysis of gap costs were defined as follows: existence, 11; extension, 1. The McpU-LBD sequence is shaded in yellow. Download FIG S6, PDF file, 0.02 MB (18KB, pdf) .
Copyright © 2019 Martín-Mora et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Characteristics of species that contain McpN homologues. A BLAST-P search was conducted in the NCBI database of nonredundant protein sequences (excluding species of the genus Pseudomonas), and the top 87 sequences are listed in Fig. S6. This table shows the characteristics of the corresponding bacterial species. Human pathogens are highlighted in green, and bacteria that oxidize sulfurous compounds are highlighted in yellow. Download Table S1, DOCX file, 0.09 MB (91.5KB, docx) .
Copyright © 2019 Martín-Mora et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Sequence alignment of the PilJ domains of the McpN and PilJ chemoreceptors of P. aeruginosa PAO1. PilJ-pilJ1 corresponds to the N-terminal PilJ domain (amino acids 36 to 163), which is followed in sequence by the PilJ-pilJ2 domain (amino acids 164 to 314). The N-box in McpN is shaded in yellow. Red, identical residues; green, highly similar residues; blue, weakly similar residues. The alignment was done using the CLUSTALW algorithm of npsa software (Combet et al., 2000). The BLOSUM62 matrix was used, and the settings used for analysis of gap costs were defined as follows: existence, 11; extension, 1. Download FIG S7, TIF file, 0.6 MB (591.2KB, tif) .
Copyright © 2019 Martín-Mora et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Oligonucleotides used in this study. Download Table S2, DOCX file, 0.02 MB (20.3KB, docx) .
Copyright © 2019 Martín-Mora et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data collection and refinement statistics of the 3D structure of the McpN-LBD-nitrate complex. Values in parentheses represent the highest-resolution shell. Download Table S3, DOCX file, 0.02 MB (21KB, docx) .
Copyright © 2019 Martín-Mora et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
Coordinates and structure factors of McpN-LBD were deposited at the PDB with accession code 6GCV.