Abstract
One of the exciting movements in microbial sciences has been a refocusing and revitalization of efforts to mine the fungal secondary metabolome. The magnitude of biosynthetic gene clusters (BGCs) in a single filamentous fungal genome combined with the historic number of sequenced genomes suggests that the secondary metabolite wealth of filamentous fungi is largely untapped. Mining algorithms and scalable expression platforms have greatly expanded access to the chemical repertoire of fungal-derived secondary metabolites. In this Review, I discuss new insights into the transcriptional and epigenetic regulation of BGCs and the ecological roles of fungal secondary metabolites in warfare, defence and development. I also explore avenues for the identification of new fungal metabolites and the challenges in harvesting fungal-derived secondary metabolites.
Fungi have a long and intimate connection with humankind, particularly at the chemical level. The realization that fungi were the source of both harmful and beneficial compounds was brought to light by the aflatoxin poisoning event Turkey X disease in the 1960s1 and the discovery of the first broad-spectrum antibiotic, penicillin, considered the ‘wonder drug’ of World War II2. These bioactive molecules, termed secondary metabolites (also known as natural products), are produced by specific fungal taxa, predominately by filamentous fungi that belong to the Pezizomycotina Ascomycete class, and several Basidiomycete classes (for example, Agaricomycetes and Exobasidiomycetes), as well as by unexpected taxa such as Kluyveromyces lactis, in which the pulcherrimin gene cluster was recently discovered3.
Secondary metabolites are derived from central metabolic pathways and primary metabolite pools, with acyl-CoAs being the critical initial building blocks that feed into the synthesis of polyketide (for example, aflatoxin) and terpene (for example, carotene) secondary metabolites and amino acids being used for the synthesis of non-ribosomal peptide secondary metabolites (for example, penicillin) (FIG. 1a). In contrast to genes that are required for the synthesis of a primary metabolite that are dispersed throughout the fungal genome, the genes encoding the enzymatic activities to produce any secondary metabolite are arranged in a contiguous fashion as a biosynthetic gene cluster (BGC), such as the aflatoxin BGC4 (FIG. 1b). Secondary metabolites are crucial players in fungal development and actively shape interactions with other organisms. Indeed, genes within a BGC are often co-regulated in accordance with the ecological function of their encoded secondary metabolite. For example, the BGCs that encode pigments in Aspergillus fumigatus are induced during spore synthesis in this fungus5, the BGC that encodes the Fusarium graminearum virulence factor trichothecene is upregulated during colonization of plants6 and the Fusarium spp. BGC that encodes the antibacterial compound bikaverin is expressed during confrontations with the bacterium Ralstonia solanacearum7.
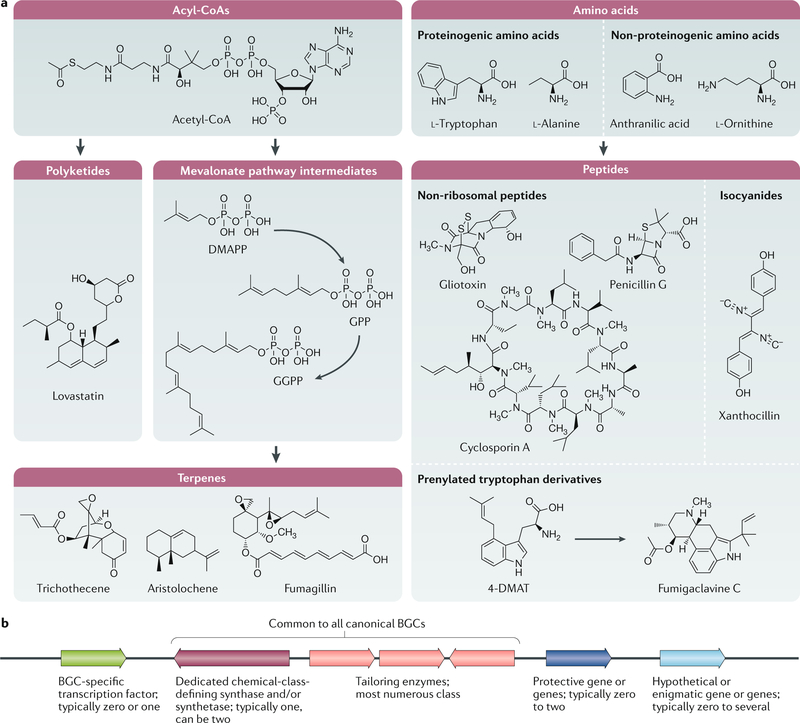
Fig. 1 |. The typical building blocks of secondary metabolites and a schematic overview of a biosynthetic gene cluster.
a | Most secondary metabolites can be grouped into three chemical categories: polyketides derived from acylCoAs, terpenes derived from acyl-CoAs and small peptides derived from amino acids. Hybrid molecules (polyketide– terpene, non-ribosomal peptide–polyketides and polyketide–fatty acid) are not shown. Fatty acid synthases (not shown) can occasionally contribute to the biosynthesis of secondary metabolites (for example, aflatoxin and sterigmatocystin are polyketide–fatty acid hybrids). b | Biosynthetic gene clusters (BGCs) are minimally composed of a chemically defining synthase and/or synthetase (polyketide synthase, terpene synthase and/or cyclase, non-ribosomal synthetase and isocyanide synthase) that use primary metabolites to form carbon backbones that are further modified by tailoring enzymes (for example, methyltransferases, p450 monooxygenases, hydroxylases and epimerases). Some BGCs contain cluster-specific transcription factors that typically positively regulate the other genes within the BGC; genes that encode proteins that mitigate the toxic property of the BGC secondary metabolite; and incongruous genes with hypothetical functions that are not obviously involved in the production of secondary metabolites or protection from the encoded metabolite. DMAPP, dimethylallyl diphosphate; DMAT, dimethylallyl tryptophan; GGPP, geranylgeranyl diphosphate; GPP, geranyl diphosphate. Part a adapted with permission from reF.30, Springer Nature Limited.
An often cited literature survey8 showed that of the 1,500 compounds that have been isolated from fungi between 1993 and 2001, more than half displayed antibacterial, antifungal or antitumour activity. A newer review9 covering fungal natural products that were discovered between 2009 and 2013 confirms the enormous potential of the fungal secondary metabolome. Although the underlying interest in fungal secondary metabolites is multivariate, these reviews highlight the predominate interest in fungal secondary metabolites — drug discovery. This interest has escalated in the past 10 years as a consequence of advances in genome sequencing, bioinformatic algorithms, phylogenetic sleuthing and increasing ease in fungal genome manipulations10.
In this Review, I present a universal view of key discoveries in the field of fungal secondary metabolites that have led to the prevailing worldwide focus of extracting the fungal secondary metabolome for profit and understanding of microbial communication. I describe the classification and genetics of secondary metabolites and BGCs, the transcriptional and epigenetic regulation of clusters and the ecological roles of secondary metabolites in defence, warfare and development. Finally, I explore avenues for the identification of new fungal metabolites with potential pharmaceutical application and the challenges in characterizing BGCs.
Chemistry and genetics
The synthesis of secondary metabolites primarily involves the polymerization of primary metabolites by dedicated enzymes (often referred to as backbone or core enzymes). The metabolites generated by the backbone enzymes are further ‘decorated’ by additional enzymes that can vastly alter the bioactivities of the metabolites. The backbone enzyme defines the chemical class of the generated secondary metabolite. For example, polyketide synthases (PKSs) produce polyketides from acyl-CoAs, non-ribosomal peptide synthetases (NRPSs) generate non-ribosomal peptides from amino acids and terpene synthases and terpene cyclases (TSs and TCs, respectively) generate terpenes from activated isoprene units. Some secondary metabolites are hybrids that are synthesized from two synthases and/or synthetases such as fumagillin (PKS–TC hybrid) or echinocandin (PKS–NRPS hybrid). The PKS–NRPS-derived hybrid secondary metabolites are often referred to as lipopeptides, but care is needed in assuming the origins of lipopeptides as the term also refers to molecules formed from ribosomally derived peptides attached to fatty acids or isoprenoids (such as canonical fungal mating pheromones)11. The two enzymes may be separate (for example, NRPS and PKS, generating valactamide)12 or fused (NRPS–PKS, generating tenuazonic acid)13. These backbone enzymes define the ‘typical’ classes of secondary metabolites, and the reader is referred to reviews focusing on the interesting chemistry involved in the synthesis of these metabolites14–17. Fungal secondary metabolites that are not generated by the synthases or synthetases discussed above include the ribosomally derived peptide ustiloxin18, fatty-acid-derived oxylipins19 and the recently identified isocyanide xanthocillin, which requires an isocyanide synthase20.
The genes involved in the biosynthesis of fungal secondary metabolites are typically arranged in BGCs, a chromosomal architecture that has facilitated the development of algorithms to predict BGCs that encode conserved synthases and/or synthetases in fungal genomes. Of note, algorithms are merely predictive and may underpredict or overpredict inclusion of genes within a BGC. True association with a cluster can be confirmed only through gene deletion. BGCs can range from two genes (for example, the valactamide BGC)12 to over 20 genes (for example, the aflatoxin BGC)21. The smaller clusters (two to four genes) contain genes encoding synthases or synthetases and tailoring enzymes that decorate initial synthase or synthetase products of the secondary metabolite. Larger BGCs contain not only synthases and/or synthetases and many tailoring genes but also frequently a gene that encodes a cluster-specific transcription factor as well as several genes inconsistent with chemical structure formation and/or hypothetical in nature. For example, although several genes in the sterigmatocystin BGC are co-regulated during the production of sterigmatocystin, there is no defining phenotype following gene deletion (for example, stcC, stcM and stcR)22. Many of these uncharacterized genes encode hypothetical, fungal and even species-specific proteins such as Cpur_05425 and Cpur_05426 in the ergochrome BGC23 and AFLA_022990, AFLA_023060 and AFLA_023090 in the aspergillic acid BGC24. By contrast, some of the proteins encoded by these other incongruous genes that were once overlooked are now known to provide protection from — or localization and/or destination of — toxic BGC natural products25 (see below).
Although most natural products are produced from a series of contiguous genes, there are notable exceptions to this arrangement. A. fumigatus possesses a ‘supercluster’ in which the genes encoding the secondary metabolites fumagillin and pseurotin are intertwined in one genomic locus26. Moreover, in Aspergillus nidulans, the synthesis of nidulanin A requires the enzymatic activity encoded on two chromosomes27 and in various Fusarium species, trichothecene genes are located in a primary tri5 BGC, but at least three genes are located outside of this cluster28. Dothiostromin, a phytotoxin structurally related to aflatoxin, is encoded by genes that are fragmented into three mini-c lusters on a single chromosome of the pine pathogen Dothistroma septosporum29.
Cluster regulation
Environmental signals.
The transcriptional and epigenetic activation of BGCs is a consequence of environmental stimuli and is dependent on the developmental stage of the producing fungus30,31. Nutritional input has long been realized to be important as reflected in the one strain-many compounds (OSMAC) approach to metabolite mining32. Temperature and light have been observed to induce or repress the synthesis of natural products since studies of aflatoxin production in Aspergillus flavus decades ago33. The regulatory pathway in response to changes in temperature has been shown to be dependent on the Velvet complex (see below) in A. fumigatus34 and may have an impact on virulence (for example, the production of at least two A. fumigatus spore secondary metabolites, the toxin trypacidin and the immunomodulator endocrocin is temperaturedependent)35,36 or mycotoxin contamination (exemplified by the temperature-dependent production of the terpene T-2 toxin in Fusarium species)37.
A considerable number of research groups have identified roles for red and blue light photoreceptors and/or their respective signal transduction pathways during the synthesis of fungal secondary metabolites38. The aflatoxin and related sterigmatocystin mycotoxin BGCs are among the well-known clusters that are repressed by white light39 (FIG. 2), whereas the Alternaria alternata mycotoxins alternariol and altertoxin are stimulated by white light (specifically blue light)40. The photoresponse pathways can intersect with known transcription factors such as CreA and/or Cre1 (the carbon catabolite regulator in fungi, called CreA in some species and Cre1 in other species) to regulate the synthesis of natural products such as the polyketide dihydrotrichotetro-nine41. The finding that the A. nidulans phytochrome FphA forms a complex with both blue light receptors and VeA, which is a member of the Velvet complex, provides a mechanistic model for how light sensing and the synthesis of secondary metabolites are conjoined42,43. The transcriptional responses of BGCs to changing environmental stress pathways, particularly oxidative stress, support the notion that secondary metabolites (for example, aflatoxin) function as protective agents from reactive oxygen species44,45. Taken together, published data clearly demonstrate that environmental signals that affect the synthesis of secondary metabolites are interdependent46.
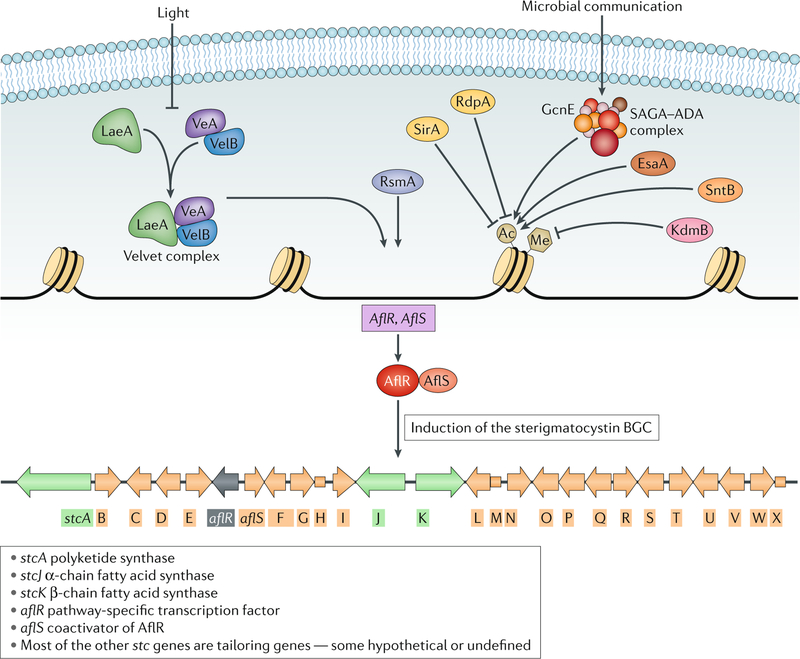
Fig. 2 |. Regulation of the sterigmatocystin biosynthetic gene cluster.
The Aspergillus nidulans sterigmatocystin biosynthetic gene cluster (BGC) is one of the most thoroughly studied BGCs at the regulatory level. The pathway-specific regulatory transcription factor, AflR, and its partner, AflS, are induced by specific proteins (for example, RsmA, a basic leucine zipper transcription factor146) and are epigenetically regulated by the Velvet complex66 and chromatin modifiers, including the histone 3 demethylase KdmB63, the histone 4 acetylase EsaA147, the histone deacetylases RdpA65 and SirA148 and the histone reader SntB149. Environmental factors such as light and interactions with other microorganisms or insects also affect the induction of the sterigmatocystin BGC. For example, fungus–bacteria interactions induce the cluster through the histone acetyltransferase GcnE, a member of the histone acetyltransferase SAGA–ADA (Spt–Ada–Gcn5–acetyltransferase–ADA) complex69, whereas white light can repress expression of some BGC-encoded genes39. A schematic of the sterigmatocystin BGC details the structure and encoded genes. Adapted with permission from reF.150, Springer Nature Limited.
Transcriptional regulation.
Our understanding of the regulation of BGCs47 continues to expand at a fast pace. Insights that have been gained from several studies present a hierarchical genetic circuitry from cluster-specific regulators to global transcriptional complexes (FIG. 2). Approximately, up to 50% of fungal BGCs contain a cluster-specific transcription factor, most commonly a C6-zinc cluster protein that recognizes palindromic motifs in cluster gene promoters48 (FIG. 1b). Occasionally a BGC contains more than one transcription factor49,50. Although originally thought to solely (and positively) regulate the genes within a given BGC, it is clear that some C6 transcription factors regulate genes within other BGCs and several metabolic pathways51,52. These studies suggest that a deeper analysis of the circuitry regulated by cluster-specific transcription factors could reveal unexpected insights into metabolic programmes that link BGC product formation with primary metabolism. For example, RNA sequencing (RNA-seq) and chemical analysis of overexpression of HasA, the C6 transcription factor that positively regulates the ironbinding secondary metabolite hexadehydroastechrome encoded within the has BGC, showed that HasA is a key component of a metabolic feedback circuitry that balances iron pools in A. fumigatus. This study exposed the concept that metal availability, in this case iron, could be a trigger for the production of secondary metabolites51. Metal starvation, specifically copper insufficiency, induces the synthesis of xanthocillin, presumably through the regulation of the xan BGC C6 transcription factor XanC20.
A number of ‘broad-domain’ transcription factors contribute to both positive and negative regulation of several BGCs, including the pH regulator PacC, CreA, the nitrogen regulator AreA and the CAAX basic leucine zipper protein HapX47. However, by far the most influential transcriptional complex that affects global regulation of secondary metabolites across every fungal genera studied thus far is the Velvet complex. This complex was first described in A. nidulans39 and is composed of LaeA (in Aspergillus spp., also known as Lae1 in most other fungi), VeA (or Vel1) and VelB (or Vel2). Detailed studies have primarily focused on LaeA (Lae1)53 and VeA (Vel1), which is postulated to be the DNA-binding partner of the complex. In general, these proteins are associated with global positive regulation of BGCs with some exceptions54. In contrast to the Velvet complex, McrA seems to be a global negative regulator of BGCs in Aspergillus and Penicillium species55.
Follow-up studies of microarray and RNA-seq cascades of regulators involved in the production of secondary metabolite have yielded unpredictable insights into BGC regulation. BrlA is a C2H2 transcription factor required for conidiophore development in Aspergillus and Penicillium species. Although several studies over the past 10 years have shown a contribution of BrlA to the production of secondary metabolites, particularly the regulation of metabolites involved in spore formation56,57, the magnitude of this regulation was only recently revealed in a study linking LaeA-mediated regulation of BrlA to the genome-wide control and biosynthesis of BGCs in A. fumigatus5. BrlA-dependent regulation of BGCs showed remarkable concordance with LaeA-dependent regulation of BGCs active during spore formation and during the non-developmental vegetative state.
Epigenetic regulation.
Efforts to remodel the fungal chromosome landscape by altering transcriptional accessibility to BGCs have been particularly fruitful in identifying metabolites from cryptic clusters58. The idea that heterochromatin and euchromatin could be important features in BGC accessibility arose from the gene cluster architecture, which suggested a co-regulation mechanism of expression associated with chromosome structure. This hypothesis was supported by a study showing a correlation between histone H4 acetylation of aflatoxin BGC promoters and the transcriptional order of their expression59. In 2007, the first study to address the potential of epigenetic manipulation in fungal secondary metabolite discovery60 reported that deletion of hdaA, which encodes a histone deacetylase, resulted in the transcriptional activation and consequent increase in the expression of multiple BGCs and their products in A. nidulans. Since then, a plethora of studies have expanded the concept of epigenetic regulation of BGCs, either by characterizing histonemodifying enzymes involved in activation or silencing of chromatin tracks or by treatment of fungal cultures with chemical inhibitors of these enzymes58,61–63. Most of these studies identified cryptic metabolites that were synthesized by deleting or overexpressing genes encoding histone acetylases, deacetylases, methylases and demethylases61,64. However, it is likely that an equal number of compounds are dampened through epigenetic regulation. For example, downregulation of the A. nidulans histone deacetylase RpdA resulted in equal numbers of metabolites being upregulated and downregulated by 100-fold65.
Several studies have coupled secondary metabolite regulatory networks with the epigenome. In 2010, a study66 showed that during the early-phase growth, the A. nidulans sterigmatocystin BGC is silent with methylation marks on histone H3 on residue K9 (indicative of heterochromatin), and those chromatin marks are reversed (indicative of euchromatin) as the fungus switches to the stationary, secondary metabolitepromoting growth phase; this reversal requires the global secondary metabolite regulator LaeA. Lae1, the functional homologue of LaeA, is also involved in regulating chromatin marks in Trichoderma reesei67, and in Fusarium fujikuroi, overexpression of the histone acetyltransferase HAT1 histone acetyltransferase could restore secondary metabolism in a Δlae1 background68. A pioneering study aimed at uncovering cryptic BGCs69 showed that the bacterium Streptomyces rapamycinicus induced the expression of the silent orsellinic acid BGC in A. nidulans through the activation of the histone acetyltransferase GcnE, a member of the histone acetyltransferase SAGA–ADA (Spt–Ada–Gcn5–acetyltransferase–ADA) complex. The SAGA–ADA complex also induced the expression of the sterigmatocystin BGC (FIG. 2). This work thus tied interspecies communication to the epigenetic machinery70. The LaeA partner VeA has also been connected to GcnE activity in suppression of the silent orsellinic acid BGC71.
Functions of secondary metabolites
There are several lines of evidence supporting ecological fitness roles for fungal secondary metabolites, including the assessment of regulatory cascades, genetic studies of BGC mutants and interaction studies with other organisms25,30,47. These studies showed that many genes that encode secondary metabolites are regulated in a manner congruent with fungal development or in response to stressors (both abiotic and biotic) and that loss or overproduction of specific secondary metabolites can alter fungal development, survival or interkingdom and intrakingdom encounters.
Protection from UV damage.
Melanin is a natural pigment typically found in spores or hyphae, and it is derived from polyketide or l-3,4-dihydroxyphenylalanine (l-DOPA) pathways. Melanin biosynthesis genes are frequently arranged in BGCs72,73. One of the first ecologically minded studies to show a protective role of spore melanin reported that an albino mutant of the maize pathogen Cochliobolus heterostrophus (formerly Bipolaris maydis) was unable to survive in the field74. Since this initial discovery, many studies have demonstrated the role of melanins and other aromatic natural products in photoprotection and/or protection from antioxidant chemicals75,76 (FIG. 3a). The same properties that protect from oxidizing radiation also have been shown to provide protection from host defence molecules in pathogenic fungi77 and have provided the foundation for the application of melanin as ultraviolet protectants for human skin and in food colouring, bioelectronics and cosmetics78.
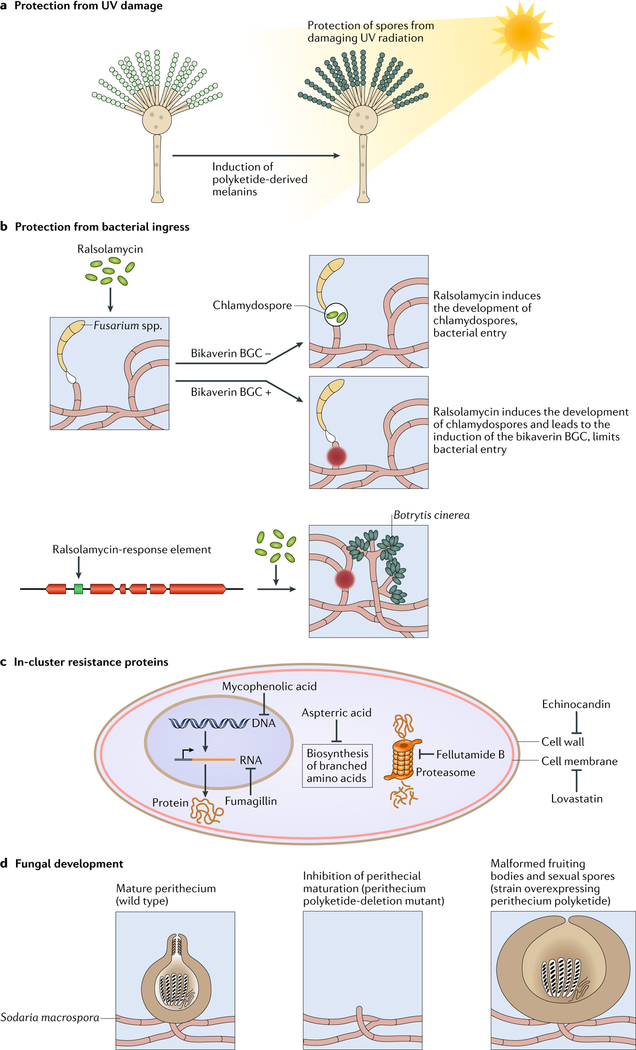
Fig. 3 |. The ecological roles of secondary metabolites.
a | Many fungi produce polyketide-derived melanins, a natural pigment that protects spores from damaging ultraviolet (UV) radiation. b | The bacterium Ralstonia solanacearum secretes the lipopeptide ralsolamycin that induces chlamydospore formation in fungi and expression of the bikaverin gene cluster in Fusarium spp.7,85. Bikaverin reduces bacterial entry and growth. Both the bikaverin biosynthetic gene cluster (BGC) and ralsolamycin response (that is, protection from bacterial ingress) have been transferred to some Botrytis species. c | For fungi to protect themselves from their own BGC-encoded antifungal secondary metabolites, they have evolved various self-protection strategies, including duplicated, resistant copies of target proteins within a BGC. The diagram of a fungal cell shows the cellular processes that are targeted by six secondary metabolites containing in-cluster resistant genes. Lovastatin interferes with ergosterol biosynthesis and thus cell membrane integrity by inhibiting 3-hydroxy-3-methylglutaryl-CoA (HMG-CoA) reductase98 (not shown), echinocandin targets cell wall synthesis by inhibiting β−1,3-d-glucan synthase100 (not shown), fellutamide is a proteasome inhibitor targeting the proteasome subunit C5 (REF.99), aspterric acid interferes with protein synthesis by targeting the branched chain amino acid synthesis enzyme dihydroxy-acid dehydratase101 (not shown), fumagillin inhibits RNA synthesis by targeting methionine aminopeptidase26 (not shown) and mycophenolic acid interferes with purine synthesis by inhibiting inosine-5ʹ-monophosphate dehydrogenase102 (not shown). d | Secondary metabolites can affect developmental processes in fungi. Deletion of a polyketide synthase in the fungus Sordaria macrospora inhibits perithecial formation, whereas its overexpression results in malformed fruiting bodies that lack the usual perithecial neck106.
Defence and weapons.
Protective and weaponized natural products help fungi obtain land rights in highly competitive ecological niches. Although hundreds of studies note the antibiotic properties of fungal metabolites, most studies are correlative and consist of testing extracts against a multitude of bacteria and fungi. Additionally, tests of extracts are biased in the form of concentrations relative to ecological concentrations, and it may well be that physiologically relevant concentrations of a metabolite could function as a signal rather than as a toxin. This possibility is illustrated by the gradient-dependent effects of Pseudomonas aeruginosa phenazine on Aspergillus, whereby a high concentration of phenazine is antifungal but a moderate concentration induces prolific sporulation in the fungus79. Similarly, the literature has dozens of examples of induction of fungal metabolites when fungi are confronted by other microorganisms and insects. Genetic, ecological and mechanistic studies now provide indisputable evidence that fungal chemicals compose the core element of microbial ‘language’ (fungi and microorganisms80; fungi and plants81; and fungi and insects82). Informative insect–fungal and bacterial–fungal chemical encounters illustrate the sophistication of this language as detailed below.
The production of either fungal or bacterial natural products has ecological outcomes on each microorganism. A series of elegant studies have addressed the curious symbiosis of the fungus Rhizopus microsporus with the endosymbiotic bacterium Burkholderia rhizoxinica.R. microsporus causes rice seedling blight, which is exacerbated by the microbial toxin rhizoxin. Although originally thought to be synthesized by the fungus, it is the endosymbiont that actually produces the polyketide macrolide83. To add further complexity to the system, rhizoxin is further chemically altered by some host fungi to enhance phytotoxicity in a mutually beneficial outcome for the two microorganisms80,84.
In what seems to be a more confrontational interaction, a succession of studies have unveiled a critical chemical role in the fraught interaction of the soil phytobacterium R. solanacearum with agricultural fungi. The lipopeptide ralsolamycin produced by R. solanacearum induces fungal development of a bacterial ‘housing’ chamber (chlamydospore)85 and is also the signal for either suppression (for example, imizoquin synthesis in A. flavus)86 or induction (for example, bikaverin in Fusarium spp.)7 of antibacterial fungal secondary metabolites. Intriguingly, induction of bikaverin by ralsolamycin is conserved in the fungus Botrytis cinerea, which horizontally acquired the bikaverin BGC from Fusarium spp.87,88. This study raises the concept that horizontally acquired BGCs may also transfer conserved regulatory responses, possibly as fitness responses (in this case production of an antibacterial compound), which promotes success in polymicrobial conflicts (FIG. 3b).
Another example of bacterial–fungal interaction is the unique multi-kingdom symbiosis of the Attine (fungus-farming) ant system. The ants are colonized by antibiotic-producing actinobacteria that protect the fungal garden from colonization by mycoparasites, including the fungus Escovopsis sp. The symbiotic bacterium Pseudonocardia sp. produces various antifungals (for example, dentigerumycin) that specifically target Escovopsis but not the garden fungus Leucoagaricus gongylophorus89. Escovopsis in turn also produces natural products that target both the actinobacteria and L. gongylophorus90.
Fungal–bacterial interactions may also be more subtle. The above interactions illustrate responses of Ascomycete fungi to various bacteria. Basidiomycete fungi are also known to alter their metabolic profile in bacterial encounters. Pigment synthesis is induced in the brown rot fungus Serpula lacrymans and one of the induced pigments, variegatic acid, inhibited swarming and biofilm spreading of Bacillus subtilis91,92.
The entomopathogenic pathogen Beauveria bassiana not only kills its insect prey — with toxic secondary metabolites being part of the arsenal — but also then poisons the cadaver with the antibacterial polyketide oosporein to limit microbial competition for its food supply93. Aflatoxin produced by A. flavus is another secondary metabolite with toxic properties towards insects, and a recent study provided evidence that aflatoxin provides a fitness advantage to A. flavus when the fungus encounters insects82,94. Together, these studies found that fungal fitness increased 26-fold in competition tests with the insect in an aflatoxin-rich environment and that aflatoxigenic strains of the fungus caused higher mortality in Drosophila melanogaster and held a measurable fitness advantage over non-toxigenic strains of the fungus during competition.
Protection from toxic natural products.
If so many of the BGC products are antifungal, how do fungi protect themselves from their own weapons? Many BGCs carry a mechanism to ensure self-protection against toxic BGC products (FIG. 3c). The three primary in-cluster self-protection strategies include efflux transporters (for example, GliA, an efflux pump important in resistance to gliotoxin95, and Tri12, which provides protection to trichothecenes28) or cellular BGC intermediate transporters (for example, CefM, which translocates the intermediate penicillin N from a microbody to the cytosol, where it is converted into the end product cephalosporin C)96; detoxifying enzymes that alter the chemical structure of the secondary metabolite to reduce critical target-binding properties (for example, GliT, an oxidoreductase that modifies the gliotoxin structure)97; and duplicated copies of the target protein (for example, incluster 3-hydroxy-3-methylglutarylCoA (HMG-CoA) reductase in the lovastatin BGC)98. And if the word clever can be applied to fungi, certainly the finding of duplicated, resistant target genes within a BGC fits this definition99. This genetic arrangement is particularly propitious for drug discovery as the duplicated function reveals an immediate natural-product eukaryotic target. Fungal BGCs containing known or postulated target proteins, in addition to the extra copy of HMG-CoA reductase in the statin BGCs, include the fumagillin BGC (target methionine aminopeptidases)26, fetullamide (the experimental validation of the protective value was shown by deleting inpE, the in-cluster copy of the proteasome subunit targeted by fetullamide)99, echinocandin (target β−1,3-d-glucan synthase)100, aspterric acid (target dihydroxy-acid dehydratase)101 and mycophenolic acid (target inosine-5ʹ-monophosphate dehydrogenase)102. All of these metabolites are antifungal; indeed, echinocandin derivatives are widely used as one of the few effective treatments of invasive human pathogens and pathogenic biofilms103.
Development.
Fungi produce many natural products that are either incorporated into developmental structures or function as signals to initiate developmental programmes. In addition to the 1,8-dihydroxynaphthalene (DHN) melanin spore metabolites, several BGCs encode pigments or toxic metabolites that provide protection for fungal sexual structures from environmental extremes or insect predation. Fusarubins are required for pigmentation of the perithecia in Fusarium species104, the perithecial toxin furocoumarin (neurosporin A) deters fungivore feeding on Neurospora crassa sexual stage105 and deletion of a PKS in the fungus Sordaria macrospora inhibits perithecial maturation, whereas its overexpression resulted in malformed fruiting bodies106 (FIG. 3d). Similarly, deletion of several BGC backbone genes results in aberrant sclerotial development in A. flavus43. Natural products are also important in spore germination, although most commonly in an offensive move whereby a fungus or bacterium will secrete a secondary metabolite that inhibits the spore germination of another fungus such as the polyketide GKK1032A from Penicillium that inhibits germination of the rice blast pathogen Magnaporthe oryzae107. By contrast, imizoquins are endogenous metabolites that are required for normal germination in A. flavus, in which loss delays and overproduction accelerates germination. Interestingly, the imizoquin BGC is repressed by the R. solanacearum lipopeptide ralsolamycin, and subsequently germination is delayed during A. flavus co-culture with the wild-type bacterium compared with co-culture of the fungus with a ralsolamycin-deficient mutant86.
Drug discovery
Since ancient times, health practices have incorporated the medicinal properties of fungi. Several potent fungal secondary metabolites (for example, penicillin, statins, cyclosporin and mycophenolic acid) were knowingly used in large-scale efforts to extend human life starting in the 20th century. However, progress is slow in identifying new fungal metabolites that can be advanced to the clinical stage, in part owing to an inability to identify the BGCs and/or express the desired secondary metabolites to high enough levels. However, the recent advances in genetic tools that enable the exploitation of the fungal metabolome (TABLE 1) coupled with the astounding numbers of BGCs revealed by fungal genome sequences108 (BOX 1) has created great optimism towards our ability to lucratively harvest fungal-derived pharmaceuticals. This ability is reflected not only in an avalanche of publications but also in the establishment of companies that focus solely on capturing these fungal treasures.
Table 1 |.
Tools and techniques for mining the fungal secondary metabolome
| Genetic and Bioinformatic tool | Feature | Refs |
|---|---|---|
| Endogenous expression systems | ||
| Targeted promoter exchange | Synthetic promoter | 99 |
| Transcription factor OE | Synthetic promoter | 50,118,149 |
| Epigenetic remodelling | Chromatin in repressed state | 58,60,62–66,147,148,151–153 |
| Global regulator OE | Synthetic promoter | 55,119 |
| Expression methods for heterologous hosts | ||
| Yeast stitching | Synthetic BGC, universal host | 122 |
| FAC | Scalable expression in Aspergillus | 12 |
| CoIN | Co-induction of sterigmatocystin promoters | 154 |
| Hex | Scalable expression in Saccharomyces | 125 |
| BGC mining algorithms | ||
| SMURF | Synthases (NRPS, PKS, DMATS and NRPS–PKS) and coordinates of genes | 112 |
| AntiSMASH | Synthases and substrate predictors | 155 |
| PRISM | NRPS, PKS and NRPS–PKS dereplication | 156 |
| SMIPS and CASSIS | DNA regulatory site | 157 |
| MIDDAS | Gene annotation, proteins and transcriptome data | 18 |
| FunGeneClusterS | NRPS, PKS, DMATS and co-expression data | 158 |
| SeMPI | Modular PKS | 159 |
| DEREPLICATOR+ | Dereplication strategies | 117 |
| Databases with fungal and bacterial BGCs | ||
| ClusterMine360 | 297 BGCs | 160 |
| IMG-ABC | 2,489 BGCs | 161 |
| MIBiG | 1,393 BGCs | 109 |
| Substrate predictors | ||
| AntiSMASH | Incorporates NRPS predictor | 161 |
| NP.searcher | Bacteria, output chemical structure | 100 |
| Pep2Path | MassSpec guided peptidic natural products | 162 |
| PRISM | NRPS, PKS and NRPS–PKS dereplication | 156 |
| GRAPE | Works with PRISM, retrobiosynthesis PKs and NRPs | 163 |
| GARLIC | Compares PRISM and GRAPE outputs for likelihood of backbone prediction | 163 |
| SeMPI | Modular PKS | 159 |
| NRPS predictor | A domain specificity | 164 |
BGC, biosynthetic gene cluster; CASSIS, cluster assignment by islands of sites; CoIN, co-inducible nitrate; DMATS, dimethylallyl tryptophan synthase; FAC, fungal artificial chromosome; NRP, non-ribosomal peptide; NRPS, non-ribosomal peptide synthetase; OE, overexpression; PK; polyketide; PKS, polyketide synthase; SMIPS, secondary metabolites by InterProScan.
Box 1 |. How many biosynthetic gene clusters do fungi contain?
In 1990, two independent research groups identified the first secondary metabolite cluster, the penicillin biosynthetic gene cluster (BGC), from the fungus Penicillium chrysogenum136,137. the clustering motif was considered uncommon at the time. By the mid-1990s, a half dozen additional secondary metabolite clusters had been identified, including the trichothecene, aflatoxin and sterigmatocystin mycotoxin clusters and several melanin BGCs138. However, at that time, the research community was not aware of the vast number of BGCs waiting to be discovered. indeed, the first sequenced fungal genome published in 1996 (Saccharomyces cerevisiae) was devoid of natural-product BGCs139. in 2005, an awakening to the incredible wealth of the fungal secondary metabolome arose from the genome sequences of three Aspergillus species114–116. Now algorithms suggest between 30 and 70 BGCs per species of fungi that are rich in secondary metabolites140. there are no studies of precisely how many species contain BGCs, but just considering Aspergillus and Penicillium alone, these genera are estimated to contain 339 and 354 species, respectively141,142. averaging the number of clusters to a conservative 50 BGCs per species, that would be 34,650 BGCs. even assuming that up to 25% of the clusters are duplicates, this brings the number to approximately 25,000 in these 2 genera. adding secondary-metabolite-rich classes such as Dothideomycetes (for example, Alternaria, Cochliobolus, Cercospora and Cladosporium) and sordariomycetes (for example, Fusarium, Tolypocladium, Magnaporthe and Pestalotiopsis) with conservatively 20,000 species143,144 and estimating that half of this number of species contain substantial chemical wealth adds another 500,000 BGCs. Lichenized fungi (which include 20,000 species) such as those in the class Lecanoromycetes (for example, Cyanodermella) are predicted to hold the equivalent if not more secondary metabolome space145. Considering the additional BGC-rich ascomycete taxa and Basidiomycetes, there is no doubt that the numbers of fungal BGCs lie over several million, a potentially several-fold underestimation.
Tools and techniques to access the fungal secondary metabolome.
The ability to capture, produce and characterize fungal secondary metabolites requires use of algorithms to identify BGCs, further bioinformatic tools to assist in structure prediction, genetic expression and induction techniques, host fungi for expression and instrumentation to detect and elucidate structure (FIG. 1; TABLE 1). Several recent reviews provide extensive details in the technological development of mining capabilities109–111.
The initial algorithms to identify BGCs were published in 2010 (REF.112) (SMURF) and 2011 (REF.113) (AntiSMASH), coming well after the 2005 publications of three Aspergillus spp. genomes, which alerted the research community to the vast number of (cryptic) BGCs in fungi114–116. Both algorithms are based on identifying conserved synthase and/or synthetaseencoding genes that are aligned next to genes that are likely to encode decorating enzymes within specified locational parameters. As these programmes do not capture all BGCs nor address dereplication concerns, additional programmes have been developed such as the new programme DEREPLICATOR + 117 (TABLE 1).
Once a BGC of interest is identified, the genes must be activated, which can be achieved in either the producing fungus (endogenous activation) or heterologous hosts. Endogenous activation is largely limited to fungi with accessible genetic systems (such as Aspergillus, Fusarium, Trichoderma, Penicillium and Pestalotiopsis spp.) by using knowledge of the regulatory systems (see above). Success in ‘turning on’ cryptic clusters in these species has been mostly attributed to the overexpression of genes encoding transcription factors86,118, promoter exchange99, manipulation of global regulatory complexes55,119, epigenetic manipulation61,64, chemical induction (for example, quorum molecule)120 and co-culture (see above and REF.121).
However, many fungi are recalcitrant to genetic manipulation, and the best method is to place and express their BGCs in heterologous systems, most commonly Saccharomyces cerevisiae or Aspergillus species110,122,123 (FIG. 4). Various synthetic approaches were successful, often taking advantage of the yeast recombination system to ‘stitch’ together BGCs using appropriate promoters for the host model124. These methods lead to the expression of a single BGC. A pioneering and scalable approach used a variation of the bacterial artificial chromosome system to design the FAC-MS (fungal artificial chromosome-metabolic scoring) system, which enabled the placement of 56 BGCs yielding 15 new compounds in an A. nidulans host12. Another scalable system, termed HEx, has been described using the host S. cerevisiae and led to the detection of 22 compounds from 41 BGCs125 (TABLE 1).
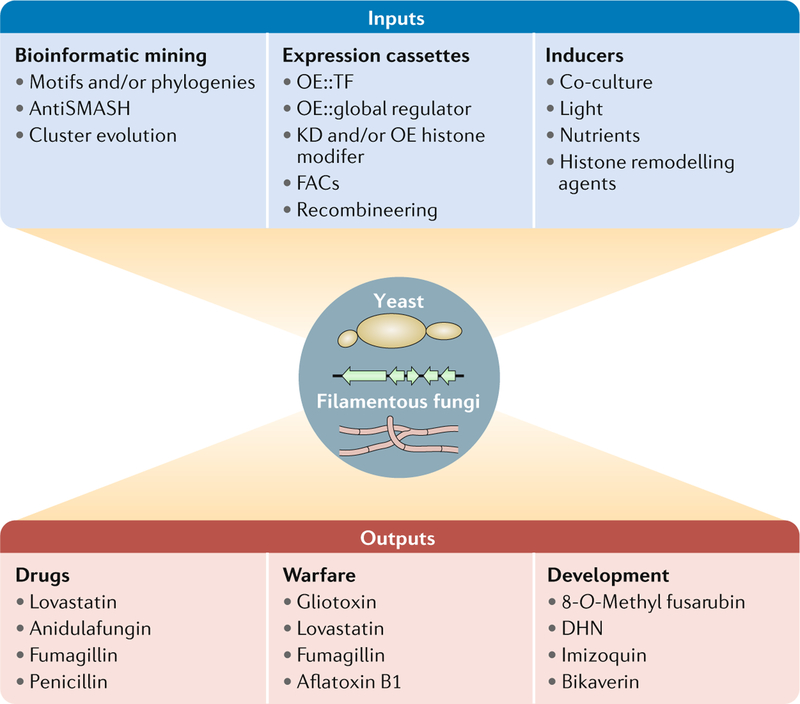
Fig. 4 |. Integration of genome mining with fungal biology yields valuable secondary metabolites.
Biosynthetic gene clusters (BGCs) can be expressed in either heterologous hosts (typically yeast and Aspergillus spp.) or endogenous filamentous hosts (middle circle). Key input features to select BGCs of interest start with bioinformatic mining of sequenced genomes to eliminate replication and identify uniquegenes. Expression cassettes can be used for gene overexpression, gene deletion, yeast recombineering and fungal artificial chromosome (FAC) construction. Inducers that can activate cryptic BGCs include abiotic stress, epigenetic chemicals, nutrients and co-cultures. Outputs include new drugs that may overlap with endogenous function of the fungus, including warfare and developmental signals. DHN, 1,8-dihydroxynaphthalene; KD, knockdown; OE, overexpression; TF, transcription factor.
Limitations to characterizing fungal BGCs.
One challenge of ‘picking’ the right BGC is that it may already have been characterized. The wealth of sequenced fungal genomes has enabled advanced bioinformatic studies that have defined evolutionary trends in BGC maintenance, formation and decay126. Many BGCs and their variants are conserved in disparate fungi, often as a result of horizontal transfer events88,127,128, thus a bioinformatic search for a cluster homologue is a critical factor in BGC selection. What is not yet achievable is knowing whether a cryptic cluster will actually produce a metabolite given the availability of all techniques at hand. What is rarely addressed — at least in public — are the numerous failures despite successful application of these genetic tools. In some cases, all genes in one BGC are heterologously expressed but no product is formed. Certainly post-transcriptional mechanisms, including intron splicing, misfolding of peptide, lack of precursors and cellular trafficking, could contribute to difficulties in execution, perhaps more so in S. cerevisiae, which has not evolved to synthesize complex natural products. But can there be other reasons? Are some BGCs on an evolutionary dead end or, alternatively, on a path to synthesis?
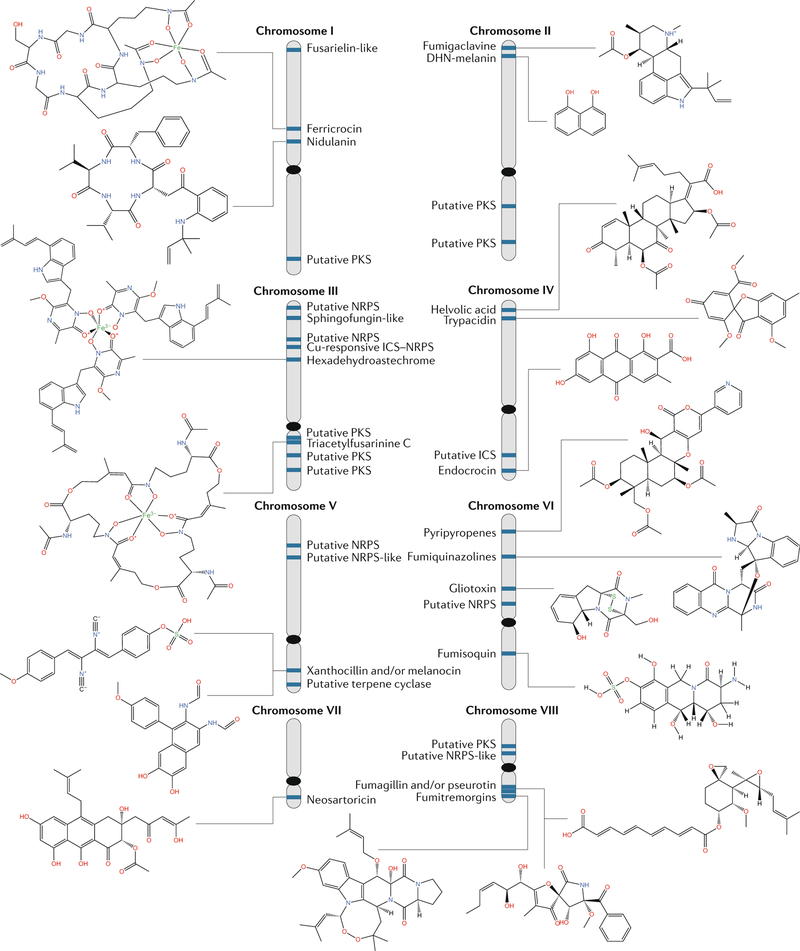
The BGCs of A. fumigatus provide a framework for speculation on this matter. This fungus is an important human pathogen, and genome sequences of over 100 isolates have been completed and substantial efforts have been made to identify its (potentially toxic) BGC products. Currently, over 50% of the secondary metabolites encoded in the known BGCs are assigned20,129,130 (FIG. 5). Detailed examination of BGC sequences of 66 isolates of A. fumigatus revealed seven types of BGC variation, including SNP, gene loss, gene gain, cluster loss, cluster mobility (transfer to another chromosome), fusion of clusters and idiomorphic (multi-allelic) clusters131. Of the 36 BGCs, only 20 showed no or little variation and consist primarily of characterized clusters, including the two siderophore clusters (ferricrocin and fusarinine C), fumigaclavine, hexadehydroastechrome, endocrocin, fumisoquin, DHN melanin, gliotoxin, fumiquinazoline, trypacidin, pyripyropene A, neosartoricin and the intertwined fumagillin–pseurotin supercluster. For the remaining 16 clusters, variation ranged from loss to multiple polymorphisms among the 66 strains of A. fumigatus, and only 2 of these BGCs are characterized (fumitremorgin and helvolic acid). It is instructive to look at the remaining 14 BGCs. For example, one of uncharacterized BGCs, called the fusarielin-like BGC, has five genes with homology to the six-gene fusarielin cluster in F. graminearum132. The cluster is absent in 4 of the 66 strains and has SNPs in 43 other strains, most of which, upon inspection, would lead to dysfunction of one or more gene products. Perhaps this cluster represents a death event caught in evolutionary time and no efforts to genetically manipulate the cluster would result in product formation in at least 43 of the strains. The fusarielin-like BGC may be reflective of an ancient BGC horizontal transfer from Fusarium to B. cinerea, where the bikaverin BGC is repeatedly decayed in numerous isolates of B. cinerea88,133.
Fig. 5 |. Chromosomal position of Aspergillus fumigatus biosynthetic gene clusters and their known or predicted products.
The predicted product nidulanin is based on a near-identical copy of the Aspergillus nidulans nidulanin A biosynthetic gene cluster (BGC)27 in Aspergillus fumigatus. Details of other clusters can be found in REFS20,129. DHN, 1,8-dihydroxynaphthalene; ICS, isocyanide synthase; NRPS, non-ribosomal synthetase; PKS, polyketide synthase. Adapted with permission from reF.47, Annual Reviews.
Outlook
Nearly 30 years have passed since the discovery of the penicillin BGC. What was then considered a rare event — the contiguous arrangement of genes dedicated to a single metabolite — is now known to be a common motif in the filamentous fungal genome, providing a bounty of wealth for the expansion of the pharmaceutical repertoire. The excitement and consequent investment of funds in fungal drug discovery opportunities are expected to continue unabated owing to the advanced and fairly inexpensive genomic tools available for fungal research that have successfully led to many elegant studies and the discovery of fascinating molecules12,125.
Existing challenges include avoiding replication (re-discovery) of known metabolites, expression of cryptic BGCs and the identification of BGCs composed of unexplored gene composition. Whereas mass spectrometry can attend in part to the dereplication issue117,134 and use of dereplication host strains can boost detection of novel compounds135, it is wise to address this potential concern before extensive genetic efforts. Phylogenetic analyses and bioinformatic programmes (TABLE 1) offer assistance in replication issues and insight into birth and death events in BGC evolution. Avoidance of characterizing synthases and/or synthetases of high identity to known enzymes could help in reducing re-discovery of known compounds, although consideration of the tailoring enzymes within a BGC is important as desired bioactivities can be attributable to modifying steps of the initial polyketide, non-ribosomal or terpene backbone. Additionally, metabolites derived from less common and, as yet, unknown chemistry most assuredly sidestep the replication issue but will require additional programming and machine learning efforts to identify this potential chemical space.
Metabolome.
The total number of small molecules in a biological sample.
Primary metabolites.
Metabolites that are produced by many unrelated taxa and are required for normal growth, development and reproduction.
Tailoring enzymes.
Enzymes that modify non-ribosomal peptides, polyketide backbones and/or terpenoid backbones after chain elongation from respective synthetases, synthases or cyclases.
Velvet complex.
A conserved transcriptional complex in filamentous fungi that is critical for the regulation of fungal secondary metabolism and reproduction in response to light and other environmental signals.
Phytochrome.
A red-light photoreceptor found in fungi, bacteria and plants.
Conidiophore.
The asexual spore (called conidium) bearing structure that is produced by many filamentous fungi. Specific secondary metabolites are associated with asexual spore formation.
Heterochromatin.
Highly condensed chromatin tightly wound around histones and less available to the transcriptional machinery. The heterochromatin state is dependent on specific post-translational histone modifications, such as deacetylation.
Euchromatin.
Lightly packed chromatin with looser arrangement around histones and accessible to the transcriptional machinery. The euchromatin state is dependent on specific post-translational histone modifications, such as acetylation and methylation.
Perithecia.
Sexual fruiting bodies containing sexual spores of some Ascomycete fungi.
Dereplication.
A screen in secondary metabolite analysis to eliminate already-known compounds from the discovery process.
Acknowledgements
The author thanks F. Y. Lim for generating the original figure 5 and J. Winans and C. D. Nwagwu for help with formatting the text. N.P.K. is funded by US National Institutes of Health (NIH) grants R01GM112739-01 and R01 AI065728-01.
Footnotes
Competing interests
There is no competing interest.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Reviewer information
Nature Reviews Microbiology thanks M. Andersen, J. Cary, D. Hoffmeister and the other anonymous reviewer(s) for their contribution to the peer review of this work.
References
- 1.Nesbitt BF, O’Kelly J, Sargeant K & Sheridan A Aspergillus flavus and turkey X disease: toxic metabolites of Aspergillus flavus. Nature 195, 1062–1063 (1962). [DOI] [PubMed] [Google Scholar]
- 2.Quinn R Rethinking antibiotic research and development: World War II and the penicillin collaborative. Am. J. Public Health 103, 426–434 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krause DJ et al. Functional and evolutionary characterization of a secondary metabolite gene cluster in budding yeasts. Proc. Natl Acad. Sci. USA 115, 11030–11035 (2018).This study characterizes the pulcherrimin cluster in K. lactis, a yeast that belongs to a taxon not associated with secondary metabolism.
- 4.Trail F et al. Physical and transcriptional map of an aflatoxin gene cluster in Aspergillus parasiticus and functional disruption of a gene involved early in the aflatoxin pathway. Appl. Environ. Microbiol 61, 2665–2673 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lind AL, Lim FY, Soukup AA, Keller NP & Rokas A An LaeA- and BrlA-dependent cellular network governs tissue-specific secondary metabolism in the human pathogen Aspergillus fumigatus. mSphere 3, e00050–18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lysøe E, Seong K-Y & Kistler HC The transcriptome of Fusarium graminearum during the infection of wheat. Mol. Plant Microbe Interact 24, 995–1000 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Spraker JE et al. Conserved responses in a war of small molecules between fungi and a bacterium. mBio 9, e00820–18 (2018).The paper reports the conserved induction of an antibacterial secondary metabolite cluster across disparate fungal genera in response to a lipopeptide that is secreted by the invading bacterium.
- 8.Pelaez F in Handbook of Industrial Mycology (ed. Zhiqiang A) (Marcel Dekker, New York, NY, 2005). [Google Scholar]
- 9.Schueffler A & Anke T Fungal natural products in research and development. Nat. Prod. Rep 31, 1425–1448 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Kück U, Bloemendal S & Teichert I Putting fungi to work: harvesting a cornucopia of drugs, toxins, and antibiotics. PLOS Pathog 10, e1003950 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caldwell GA, Naider F & Becker JM Fungal lipopeptide mating pheromones: a model system for the study of protein prenylation. Microbiol. Rev 59, 406–422 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clevenger KD et al. A scalable platform to identify fungal secondary metabolites and their gene clusters. Nat. Chem. Biol 13, 895–901 (2017).This paper presents a method to capture the entire secondary metabolome of a single species using FAC-MS technology.
- 13.Yun C-S, Motoyama T & Osada H Biosynthesis of the mycotoxin tenuazonic acid by a fungal NRPS-PKS hybrid enzyme. Nat. Commun 6, 8758 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hur GH, Vickery CR & Burkart MD Explorations of catalytic domains in non-ribosomal peptide synthetase enzymology. Nat. Prod. Rep 29, 1074 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmidt-Dannert C Biosynthesis of terpenoid natural products in fungi. Adv. Biochem. Eng. Biotechnol 148, 19–61 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Li X-W, Ear A & Nay B Hirsutellones and beyond: figuring out the biological and synthetic logics toward chemical complexity in fungal PKS-NRPS compounds. Nat. Prod. Rep 30, 765 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Chiang Y-M, Oakley BR, Keller NP & Wang CCC Unraveling polyketide synthesis in members of the genus Aspergillus. Appl. Microbiol. Biotechnol 86, 1719–1736 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Umemura M et al. Characterization of the biosynthetic gene cluster for the ribosomally synthesized cyclic peptide ustiloxin B in Aspergillus flavus. Fungal Genet. Biol 68, 23–30 (2014).This study identifies the first BGC that produces a ribosomally encoded cyclic peptide.
- 19.Pettit RK Small-molecule elicitation of microbial secondary metabolites. Microb. Biotechnol 4, 471–478 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lim FY et al. Fungal isocyanide synthases and xanthocillin biosynthesis in Aspergillus fumigatus. mBio 9, e00785–18 (2018).This study identifies novel BGCs that contain isocyanide synthases.
- 21.Yu J et al. Clustered pathway genes in aflatoxin biosynthesis. Appl. Environ. Microbiol 70, 1253–1262 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amaike S, Affeldt KJ & Keller NP in The Mycota: Agricultural Applications 2nd edn Vol. 11 (ed. Kempken F) 59–74 (Springer, Berlin, 2013). [Google Scholar]
- 23.Neubauer L, Dopstadt J, Humpf H-U & Tudzynski P Identification and characterization of the ergochrome gene cluster in the plant pathogenic fungus Claviceps purpurea. Fungal Biol. Biotechnol 3, 2 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lebar MD et al. Identification and functional analysis of the aspergillic acid gene cluster in Aspergillus flavus. Fungal Genet. Biol 116, 14–23 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Keller NP Translating biosynthetic gene clusters into fungal armor and weaponry. Nat. Chem. Biol 11, 671–677 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wiemann P et al. Prototype of an intertwined secondary-metabolite supercluster. Proc. Natl Acad. Sci. USA 110, 17065–17070 (2013).This report describes a supercluster in which the genes encoding the secondary metabolites fumagillin and pseurotin are intertwined.
- 27.Andersen MR et al. Accurate prediction of secondary metabolite gene clusters in filamentous fungi. Proc. Natl Acad. Sci. USA 110, E99–E107 (2013).This study identifies non-contiguous members within a BGC through expression data.
- 28.Ohsato S et al. Transgenic rice plants expressing trichothecene 3-O-acetyltransferase show resistance to the Fusarium phytotoxin deoxynivalenol. Plant Cell Rep 26, 531–538 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Bradshaw RE et al. Fragmentation of an aflatoxin-like gene cluster in a forest pathogen. New Phytol 198, 525–535 (2013).This study reports the fragmentation of a gene cluster dedicated to the production of a secondary metabolite.
- 30.Lim FY & Keller NP Spatial and temporal control of fungal natural product synthesis. Nat. Prod. Rep 31, 1277–1286 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kalinina SA, Jagels A, Cramer B, Geisen R & Humpf H-U Influence of environmental factors on the production of penitrems A–F by Penicillium crustosum. Toxins 9, 210 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hewage RT, Aree T, Mahidol C, Ruchirawat S & Kittakoop P One strain-many compounds (OSMAC) method for production of polyketides, azaphilones, and an isochromanone using the endophytic fungus Dothideomycete sp. Phytochemistry 108, 87–94 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Joffe AZ & Lisker N Effects of light, temperature, and pH value on aflatoxin production in vitro. Appl. Microbiol 18, 517–518 (1969). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lind AL, Smith TD, Saterlee T, Calvo AM & Rokas A Regulation of secondary metabolism by the Velvet complex is temperature-responsive in Aspergillus. G3 6, 4023–4033 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hagiwara D et al. Temperature during conidiation affects stress tolerance, pigmentation, and trypacidin accumulation in the conidia of the airborne pathogen Aspergillus fumigatus. PLOS ONE 12, e0177050 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berthier E et al. Low-volume toolbox for the discovery of immunosuppressive fungal secondary metabolites. PLOS Pathog 9, e1003289 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nazari L, Manstretta V & Rossi V A non-linear model for temperature-dependent sporulation and T-2 and HT-2 production of Fusarium langsethiae and Fusarium sporotrichioides. Fungal Biol 120, 562–571 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Bazafkan H et al. SUB1 has photoreceptor dependent and independent functions in sexual development and secondary metabolism in Trichoderma reesei. Mol. Microbiol 106, 742–759 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Bayram O et al. VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science 320, 1504–1506 (2008).This paper describes the identification of a conserved transcriptional complex that coordinates global regulation of secondary metabolism.
- 40.Pruss S et al. Role of the Alternaria alternata blue-light receptor LreA (white-collar 1) in spore formation and secondary metabolism. Appl. Environ. Microbiol 80, 2582–2591 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Monroy AA, Stappler E, Schuster A, Sulyok M & Schmoll MA CRE1-regulated cluster is responsible for light dependent production of dihydrotrichotetronin in Trichoderma reesei. PLOS ONE 12, e0182530 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Purschwitz J et al. Functional and physical interaction of blue- and red-light sensors in Aspergillus nidulans. Curr. Biol 18, 255–259 (2008). [DOI] [PubMed] [Google Scholar]
- 43.Calvo AM & Cary JW Association of fungal secondary metabolism and sclerotial biology. Front. Microbiol 6, 62 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kenne G et al. Activation of aflatoxin biosynthesis alleviates total ROS in Aspergillus parasiticus. Toxins 10, 57 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Montibus M, Pinson-Gadais L, Richard-Forget F, Barreau C & Ponts N Coupling of transcriptional response to oxidative stress and secondary metabolism regulation in filamentous fungi. Crit. Rev. Microbiol 41, 295–308 (2015). [DOI] [PubMed] [Google Scholar]
- 46.Fountain JC et al. Oxidative stress and carbon metabolism influence Aspergillus flavus transcriptome composition and secondary metabolite production. Sci. Rep 6, 38747 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Macheleidt J et al. Regulation and role of fungal secondary metabolites. Annu. Rev. Genet 50, 371–392 (2016). [DOI] [PubMed] [Google Scholar]
- 48.Fernandes M, Keller NP & Adams TH Sequence-specific binding by Aspergillus nidulans AflR, a C6 zinc cluster protein regulating mycotoxin biosynthesis. Mol. Microbiol 28, 1355–1365 (1998). [DOI] [PubMed] [Google Scholar]
- 49.Brown DW et al. Identification of a 12-gene fusaric acid biosynthetic gene cluster in Fusarium species through comparative and functional genomics. Mol. Plant Microbe Interact 28, 319–332 (2015). [DOI] [PubMed] [Google Scholar]
- 50.Yin W-B et al. A nonribosomal peptide synthetase-derived iron(III) complex from the pathogenic fungus. Aspergillus fumigatus. J. Am. Chem. Soc 135, 2064–2067 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wiemann P et al. Perturbations in small molecule synthesis uncovers an iron-responsive secondary metabolite network in Aspergillus fumigatus. Front. Microbiol 5, 530 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bergmann S et al. Activation of a silent fungal polyketide biosynthesis pathway through regulatory cross talk with a cryptic nonribosomal peptide synthetase gene cluster. Appl. Environ. Microbiol 76, 8143–8149 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bok JW & Keller NP in The Mycota: Biochemistry and Molecular Biology 3rd edn Vol. 3 (ed. Hoffmeister D) 21–29 (Springer International, Switzerland, 2016). [Google Scholar]
- 54.Chettri P & Bradshaw RE LaeA negatively regulates dothistromin production in the pine needle pathogen Dothistroma septosporum. Fungal Genet. Biol 97, 24–32 (2016). [DOI] [PubMed] [Google Scholar]
- 55.Oakley CE et al. Discovery of McrA, a master regulator of Aspergillus secondary metabolism. Mol. Microbiol 103, 347–365 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lim FY, Ames B, Walsh CT & Keller NP Co-ordination between BrlA regulation and secretion of the oxidoreductase FmqD directs selective accumulation of fumiquinazoline C to conidial tissues in Aspergillus fumigatus. Cell. Microbiol 16, 1267–1283 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mulinti P et al. Accumulation of ergot alkaloids during conidiophore development in Aspergillus fumigatus. Curr. Microbiol 68, 1–5 (2014). [DOI] [PubMed] [Google Scholar]
- 58.Cichewicz RH Epigenome manipulation as a pathway to new natural product scaffolds and their congeners. Nat. Prod. Rep 27, 11–22 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Roze LV, Arthur AE, Hong S-Y, Chanda A & Linz JE The initiation and pattern of spread of histone H4 acetylation parallel the order of transcriptional activation of genes in the aflatoxin cluster. Mol. Microbiol 66, 713–726 (2007). [DOI] [PubMed] [Google Scholar]
- 60.Shwab EK et al. Histone deacetylase activity regulates chemical diversity in Aspergillus. Eukaryot. Cell 6, 1656–1664 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gacek A & Strauss J The chromatin code of fungal secondary metabolite gene clusters. Appl. Microbiol. Biotechnol 95, 1389–1404 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fan A et al. Deletion of a histone acetyltransferase leads to the pleiotropic activation of natural products in Metarhizium robertsii. Org. Lett 19, 1686–1689 (2017). [DOI] [PubMed] [Google Scholar]
- 63.Gacek-Matthews A et al. KdmB, a Jumonji histone H3 demethylase, regulates genome-wide H3K4 trimethylation and is required for normal induction of secondary metabolism in Aspergillus nidulans. PLOS Genet 12, e1006222 (2016).Using genome-wide chromatin immunoprecipitation coupled with RNA-seq and liquid chromatography with tandem mass spectrometry (LC-MS/MS), this study presents unprecedented insight into the global epigenetic regulation of cryptic BGCs in one species.
- 64.Williams RB, Henrikson JC, Hoover AR, Lee AE & Cichewicz RH Epigenetic remodeling of the fungal secondary metabolome. Org. Biomol. Chem 6, 1895 (2008). [DOI] [PubMed] [Google Scholar]
- 65.Albright JC et al. Large-scale metabolomics reveals a complex response of Aspergillus nidulans to epigenetic perturbation. ACS Chem. Biol 10, 1535–1541 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reyes-Dominguez Y et al. Heterochromatic marks are associated with the repression of secondary metabolism clusters in Aspergillus nidulans. Mol. Microbiol 76, 1376–1386 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Karimi-Aghcheh R et al. Functional analyses of Trichoderma reesei LAE1 reveal conserved and contrasting roles of this regulator. G3 3, 369–378 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Niehaus E-M et al. Analysis of the global regulator Lae1 uncovers a connection between Lae1 and the histone acetyltransferase HAT1 in Fusarium fujikuroi. Appl. Microbiol. Biotechnol 102, 279–295 (2018). [DOI] [PubMed] [Google Scholar]
- 69.Nützmann H-W et al. Bacteria-induced natural product formation in the fungus Aspergillus nidulans requires Saga/Ada-mediated histone acetylation. Proc. Natl Acad. Sci. USA 108, 14282–14287 (2011).This paper reports the bacterial induction of a cryptic BGC via a chromatin remodelling enzyme complex.
- 70.Netzker T et al. Microbial communication leading to the activation of silent fungal secondary metabolite gene clusters. Front. Microbiol 6, 299 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bok JW et al. VeA and MvlA repression of the cryptic orsellinic acid gene cluster in Aspergillus nidulans involves histone 3 acetylation. Mol. Microbiol 89, 963–974 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tsai HF, Wheeler MH, Chang YC & Kwon-Chung KJ A developmentally regulated gene cluster involved in conidial pigment biosynthesis in Aspergillus fumigatus. J. Bacteriol 181, 6469–6477 (1999).This article presents the first identification of a BGC required for fungal development.
- 73.Zhang P et al. A cryptic pigment biosynthetic pathway uncovered by heterologous expression is essential for conidial development in Pestalotiopsis fici. Mol. Microbiol 105, 469–483 (2017). [DOI] [PubMed] [Google Scholar]
- 74.Leonard KJ Virulence, temperature optima, and competitive abilities of isolines of races T and 0 of Bipolaris maydis. Phytopathology 67, 1273–1279 (1977). [Google Scholar]
- 75.Shukla S et al. Total phenolic content, antioxidant, tyrosinase and α-glucosidase inhibitory activities of water soluble extracts of noble starter culture Doenjang, a Korean fermented soybean sauce variety. Food Control 59, 854–861 (2016). [Google Scholar]
- 76.Eisenman HC & Casadevall A Synthesis and assembly of fungal melanin. Appl. Microbiol. Biotechnol 93, 931–940 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jacobson ES Pathogenic roles for fungal melanins. Clin. Microbiol. Rev 13, 708–717 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhao L, Kim J-C, Paik M-J, Lee W & Hur J-SA Multifunctional and possible skin UV protectant, (3R)-5-hydroxymellein, produced by an endolichenic fungus isolated from Parmotrema austrosinense. Molecules 22, 26 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zheng H et al. Redox metabolites signal polymicrobial biofilm development via the NapA oxidative stress cascade in Aspergillus. Curr. Biol 25, 29–37 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Scherlach K & Hertweck C Mediators of mutualistic microbe-microbe interactions. Nat. Prod. Rep 35, 303–308 (2018). [DOI] [PubMed] [Google Scholar]
- 81.Zeilinger S et al. Friends or foes? Emerging insights from fungal interactions with plants. FEMS Microbiol. Rev 40, 182–207 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rohlfs M Fungal secondary metabolite dynamics in fungus-grazer interactions: novel insights and unanswered questions. Front. Microbiol 5, 788 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Partida-Martinez LP & Hertweck C Pathogenic fungus harbours endosymbiotic bacteria for toxin production. Nature 437, 884–888 (2005). [DOI] [PubMed] [Google Scholar]
- 84.Scherlach K, Busch B, Lackner G, Paszkowski U & Hertweck C Symbiotic cooperation in the biosynthesis of a phytotoxin. Angew. Chem. Int. Ed 51, 9615–9618 (2012). [DOI] [PubMed] [Google Scholar]
- 85.Spraker JE, Sanchez LM, Lowe TM, Dorrestein PC & Keller NP Ralstonia solanacearum lipopeptide induces chlamydospore development in fungi and facilitates bacterial entry into fungal tissues. ISME J 10, 2317–2330 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Khalid S et al. NRPS-derived isoquinolines and lipopetides mediate antagonism between plant pathogenic fungi and bacteria. ACS Chem. Biol 13, 171–179 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schumacher J et al. A functional bikaverin biosynthesis gene cluster in rare strains of Botrytis cinerea is positively controlled by VELVET. PLOS ONE 8, e53729 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Campbell MA, Rokas A & Slot JC Horizontal transfer and death of a fungal secondary metabolic gene cluster. Genome Biol. Evol 4, 289–293 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Oh D-C, Poulsen M, Currie CR & Clardy J Dentigerumycin: a bacterial mediator of an ant-fungus symbiosis. Nat. Chem. Biol 5, 391–393 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dhodary B, Schilg M, Wirth R & Spiteller D Secondary metabolites from Escovopsis weberi and their role in attacking the garden fungus of leaf-cutting ants. Chemistry 24, 4445–4452 (2018). [DOI] [PubMed] [Google Scholar]
- 91.Tauber JP, Gallegos-Monterrosa R, Kovács ÁT, Shelest E & Hoffmeister D Dissimilar pigment regulation in Serpula lacrymans and Paxillus involutus during inter-kingdom interactions. Microbiology 164, 65–77 (2018). [DOI] [PubMed] [Google Scholar]
- 92.Tauber JP, Schroeckh V, Shelest E, Brakhage AA & Hoffmeister D Bacteria induce pigment formation in the basidiomycete Serpula lacrymans. Environ. Microbiol 18, 5218–5227 (2016). [DOI] [PubMed] [Google Scholar]
- 93.Fan Y et al. Regulatory cascade and biological activity of Beauveria bassiana oosporein that limits bacterial growth after host death. Proc. Natl Acad. Sci. USA 114, E1578–E1586 (2017).This paper reports the finding that a fungus-derived antibacterial compound poisons the food supply to limit microbial competition.
- 94.Drott MT, Lazzaro BP, Brown DL, Carbone I & Milgroom MG Balancing selection for aflatoxin in Aspergillus flavus is maintained through interference competition with, and fungivory by insects. Proc. Biol. Sci 284, 20172408 (2017).This article provides evidence that a toxic secondary metabolite provides a fitness advantage to the fungus during confrontations with insects.
- 95.Dolan SK, O’Keeffe G, Jones GW & Doyle S Resistance is not futile: gliotoxin biosynthesis, functionality and utility. Trends Microbiol 23, 419–428 (2015). [DOI] [PubMed] [Google Scholar]
- 96.Teijeira F et al. The transporter CefM involved in translocation of biosynthetic intermediates is essential for cephalosporin production. Biochem. J 418, 113–124 (2009). [DOI] [PubMed] [Google Scholar]
- 97.Scharf DH et al. Transannular disulfide formation in gliotoxin biosynthesis and its role in self-resistance of the human pathogen Aspergillus fumigatus. J. Am. Chem. Soc 132, 10136–10141 (2010). [DOI] [PubMed] [Google Scholar]
- 98.Abe Y et al. Effect of increased dosage of the ML-236B (compactin) biosynthetic gene cluster on ML-236B production in Penicillium citrinum. Mol. Genet. Genomics 268, 130–137 (2002). [DOI] [PubMed] [Google Scholar]
- 99.Yeh H-H et al. Resistance gene-guided genome mining: serial promoter exchanges in Aspergillus nidulans reveal the biosynthetic pathway for fellutamide B, a proteasome inhibitor. ACS Chem. Biol 11, 2275–2284 (2016).This paper provides the first evidence that duplicated, resistant target genes within a BGC provide self-protection.
- 100.Yue Q et al. Genomics-driven discovery of a novel self-resistance mechanism in the echinocandin-producing fungus Pezicula radicicola. Environ. Microbiol 20, 3154–3167 (2018). [DOI] [PubMed] [Google Scholar]
- 101.Yan Y et al. Resistance-gene-directed discovery of a natural-product herbicide with a new mode of action. Nature 559, 415–418 (2018).This study describes a genomics approach to identify duplicated resistance genes and the discovery of a bioactive natural-product herbicide.
- 102.Hansen BG et al. A new class of IMP dehydrogenase with a role in self-resistance of mycophenolic acid producing fungi. BMC Microbiol 11, 202 (2011).This study reports on the initial demonstration that a duplicated target gene within a BGC can provide resistance to the BGC product using a heterologous host.
- 103.Larkin EL, Dharmaiah S & Ghannoum MA Biofilms and beyond: expanding echinocandin utility. J. Antimicrob. Chemother 73, i73–i81 (2018). [DOI] [PubMed] [Google Scholar]
- 104.Studt L, Wiemann P, Kleigrewe K, Humpf H-U & Tudzynski B Biosynthesis of fusarubins accounts for pigmentation of Fusarium fujikuroi perithecia. Appl. Environ. Microbiol 78, 4468–4480 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhao Y et al. Production of a fungal furocoumarin by a polyketide synthase gene cluster confers the chemo-resistance of Neurospora crassa to the predation by fungivorous arthropods. Environ. Microbiol 19, 3920–3929 (2017). [DOI] [PubMed] [Google Scholar]
- 106.Schindler D & Nowrousian M The polyketide synthase gene pks4 is essential for sexual development and regulates fruiting body morphology in Sordaria macrospora. Fungal Genet. Biol 68, 48–59 (2014). [DOI] [PubMed] [Google Scholar]
- 107.Becker J, Liermann JC, Opatz T, Anke H & Thines E GKK1032A2, a secondary metabolite from Penicillium sp. IBWF-029–96, inhibits conidial germination in the rice blast fungus Magnaporthe oryzae. J. Antibiot 65, 99–102 (2012). [DOI] [PubMed] [Google Scholar]
- 108.Nielsen JC et al. Global analysis of biosynthetic gene clusters reveals vast potential of secondary metabolite production in Penicillium species. Nat. Microbiol 2, 17044 (2017). [DOI] [PubMed] [Google Scholar]
- 109.Medema MH et al. Minimum information about a biosynthetic gene cluster. Nat. Chem. Biol 11, 625–631 (2015).This article presents a community effort to standardize annotations and metadata on BGCs and their products.
- 110.Alberti F, Foster GD & Bailey AM Natural products from filamentous fungi and production by heterologous expression. Appl. Microbiol. Biotechnol 101, 493–500 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chavali AK & Rhee SY Bioinformatics tools for the identification of gene clusters that biosynthesize specialized metabolites. Brief. Bioinform 19, 1022–1034 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Khaldi N et al. SMURF: genomic mapping of fungal secondary metabolite clusters. Fungal Genet. Biol 47, 736–741 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Medema MH et al. antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res 39, W339–W346 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Galagan JE et al. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 438, 1105–1115 (2005). [DOI] [PubMed] [Google Scholar]
- 115.Machida M et al. Genome sequencing and analysis of Aspergillus oryzae. Nature 438, 1157–1161 (2005). [DOI] [PubMed] [Google Scholar]
- 116.Nierman WC et al. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 438, 1151–1156 (2005). [DOI] [PubMed] [Google Scholar]
- 117.Mohimani H et al. Dereplication of microbial metabolites through database search of mass spectra. Nat. Commun 9, 4035 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Janevska S et al. Establishment of the inducible Tet-On system for the activation of the silent trichosetin gene cluster in Fusarium fujikuroi. Toxins 9, 126 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Jiang T et al. Overexpression of the global regulator LaeA in Chaetomium globosum leads to the biosynthesis of chaetoglobosin Z. J. Nat. Prod 79, 2487–2494 (2016). [DOI] [PubMed] [Google Scholar]
- 120.Palonen EK et al. Transcriptomic complexity of Aspergillus terreus Velvet gene family under the influence of butyrolactone I. Microorganisms 5, 12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Adnani N, Rajski SR & Bugni TS Symbiosis-inspired approaches to antibiotic discovery. Nat. Prod. Rep 34, 784–814 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Billingsley JM, DeNicola AB & Tang Y Technology development for natural product biosynthesis in Saccharomyces cerevisiae. Curr. Opin. Biotechnol 42, 74–83 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.He Y et al. Recent advances in reconstructing microbial secondary metabolites biosynthesis in Aspergillus spp. Biotechnol. Adv 36, 739–783 (2018). [DOI] [PubMed] [Google Scholar]
- 124.Yin W-B et al. Discovery of cryptic polyketide metabolites from dermatophytes using heterologous expression in Aspergillus nidulans. ACS Synth. Biol 2, 629–634 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Harvey CJB et al. HEx: a heterologous expression platform for the discovery of fungal natural products. Sci. Adv 4, eaar5459 (2018).This paper describes the tool and protocol development that led to the expression of 41 BGCs and 22 compounds in a yeast heterologous expression system.
- 126.Stepien Ł The use of Fusarium secondary metabolite biosynthetic genes in chemotypic and phylogenetic studies. Crit. Rev. Microbiol 40, 176–185 (2014). [DOI] [PubMed] [Google Scholar]
- 127.Khaldi N, Collemare J, Lebrun M-H & Wolfe KH Evidence for horizontal transfer of a secondary metabolite gene cluster between fungi. Genome Biol 9, R18 (2008).This early phylogenetic study provides evidence for horizontal transfer of natural-product BGCs in fungi.
- 128.Reynolds HT et al. Differential retention of gene functions in a secondary metabolite cluster. Mol. Biol. Evol 34, 2002–2015 (2017). [DOI] [PubMed] [Google Scholar]
- 129.Bignell E, Cairns TC, Throckmorton K, Nierman WC & Keller NP Secondary metabolite arsenal of an opportunistic pathogenic fungus. Phil. Trans. R. Soc. B 371, 20160023 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Perrin RM et al. Transcriptional regulation of chemical diversity in Aspergillus fumigatus by LaeA. PLOS Pathog 3, e50 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lind AL et al. Drivers of genetic diversity in secondary metabolic gene clusters within a fungal species. PLOS Biol 15, e2003583 (2017).This study compares DNA sequences of the BGCs of 66 A. fumigatus isolates and establishes 5 drivers of genetic diversity that explain BGC macroevolutionary patterns.
- 132.Droce A et al. Functional analysis of the fusarielin biosynthetic gene cluster. Molecules 21, 1710 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Campbell MA, Staats M, van Kan JAL, Rokas A & Slot JC Repeated loss of an anciently horizontally transferred gene cluster in Botrytis. Mycologia 105, 1126–1134 (2013). [DOI] [PubMed] [Google Scholar]
- 134.Nielsen KF & Larsen TO The importance of mass spectrometric dereplication in fungal secondary metabolite analysis. Front. Microbiol 6, 71 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chiang Y-M et al. Development of genetic dereplication strains in Aspergillus nidulans results in the discovery of aspercryptin. Angew. Chem. Int. Ed 55, 1662–1665 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Díez B et al. The cluster of penicillin biosynthetic genes: identification and characterization of the pcbAB gene encoding the alpha-aminoadipyl-cysteinyl-valine synthetase and linkage to the pcbC and penDE genes. J. Biol. Chem 265, 16358–16365 (1990). [PubMed] [Google Scholar]
- 137.Smith DJ, Burnham MK, Edwards J, Earl AJ & Turner G Cloning and heterologous expression of the penicillin biosynthetic gene cluster from Penicillum chrysogenum. Biotechnology 8, 39–41 (1990). [DOI] [PubMed] [Google Scholar]
- 138.Keller NP & Hohn TM Metabolic pathway gene clusters in filamentous fungi. Fungal Genet. Biol 21, 17–29 (1997). [PubMed] [Google Scholar]
- 139.Goffeau A et al. Life with 6000 genes. Science 274, 546–567 (1996). [DOI] [PubMed] [Google Scholar]
- 140.Inglis DO et al. Comprehensive annotation of secondary metabolite biosynthetic genes and gene clusters of Aspergillus nidulans, A. fumigatus, A. niger and A. oryzae. BMC Microbiol 13, 91 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Samson RA et al. Phylogeny, identification and nomenclature of the genus Aspergillus. Stud. Mycol 78, 141–173 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Visagie CM et al. Identification and nomenclature of the genus Penicillium. Stud. Mycol 78, 343–371 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kirk PM, Cannon PF, David JC & Stalpers JA (eds) Ainsworth & Bisby’s Dictionary of the Fungi 9th edn (CABI, 2001). [Google Scholar]
- 144.Schoch CL et al. A class-wide phylogenetic assessment of Dothideomycetes. Stud. Mycol 64, 1–15 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Jahn L et al. Linking secondary metabolites to biosynthesis genes in the fungal endophyte Cyanodermella asteris: the anti-cancer bisanthraquinone skyrin. J. Biotechnol 257, 233–239 (2017). [DOI] [PubMed] [Google Scholar]
- 146.Yin W-B et al. An Aspergillus nidulans bZIP response pathway hardwired for defensive secondary metabolism operates through aflR. Mol. Microbiol 83, 1024–1034 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Soukup AA et al. Overexpression of the Aspergillus nidulans histone 4 acetyltransferase EsaA increases activation of secondary metabolite production. Mol. Microbiol 86, 314–330 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Itoh E et al. Sirtuin A regulates secondary metabolite production by Aspergillus nidulans. J. Gen. Appl. Microbiol 63, 228–235 (2017). [DOI] [PubMed] [Google Scholar]
- 149.Ahuja M et al. Illuminating the diversity of aromatic polyketide synthases in Aspergillus nidulans. J. Am. Chem. Soc 134, 8212–8221 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Brakhage AA Regulation of fungal secondary metabolism. Nat. Rev. Microbiol 11, 21–32 (2013). [DOI] [PubMed] [Google Scholar]
- 151.Pfannenstiel BT et al. Revitalization of a forward genetic screen identifies three new regulators of fungal secondary metabolism in the genus Aspergillus. mBio 8, e01246–17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Gacek-Matthews A et al. KdmA, a histone H3 demethylase with bipartite function, differentially regulates primary and secondary metabolism in Aspergillus nidulans. Mol. Microbiol 96, 839–860 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Strauss J & Reyes-Dominguez Y Regulation of secondary metabolism by chromatin structure and epigenetic codes. Fungal Genet. Biol 48, 62–69 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wiemann P et al. CoIN: co-inducible nitrate expression system for secondary metabolites in Aspergillus nidulans. Fungal Biol. Biotechnol 5, 6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Blin K et al. antiSMASH 4.0 — improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res 45, W36–W41 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Skinnider MA et al. Genomes to natural products prediction informatics for secondary metabolomes (PRISM). Nucleic Acids Res 43, 9645–9662 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Wolf T, Shelest V, Nath N & Shelest E CASSIS and SMIPS: promoter-based prediction of secondary metabolite gene clusters in eukaryotic genomes. Bioinformatics 32, 1138–1143 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Vesth TC, Brandl J & Andersen MR FunGeneClusterS: predicting fungal gene clusters from genome and transcriptome data. Synth. Syst. Biotechnol 1, 122–129 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Zierep PF et al. SeMPI: a genome-based secondary metabolite prediction and identification web server. Nucleic Acids Res 45, W64–W71 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Conway KR & Boddy CN ClusterMine360: a database of microbial PKS/NRPS biosynthesis. Nucleic Acids Res 41, D402–D407 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Hadjithomas M et al. IMG-ABC: a knowledge base to fuel discovery of biosynthetic gene clusters and novel secondary metabolites. mBio 6, e00932 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Medema MH et al. Pep2Path: automated mass spectrometry-guided genome mining of peptidic natural products. PLOS Comput. Biol 10, e1003822 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Dejong CA et al. Polyketide and nonribosomal peptide retro-biosynthesis and global gene cluster matching. Nat. Chem. Biol 12, 1007–1014 (2016). [DOI] [PubMed] [Google Scholar]
- 164.Röttig M et al. NRPSpredictor2 — a web server for predicting NRPS adenylation domain specificity. Nucleic Acids Res 39, W362–W367 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]