Summary
Due to their different lifestyles, effective defence against biotrophic pathogens normally leads to increased susceptibility to necrotrophs, and vice versa. Solving this trade‐off is a major challenge for obtaining broad‐spectrum resistance in crops and requires uncoupling the antagonism between the jasmonate (JA) and salicylate (SA) defence pathways. Pseudomonas syringae pv. tomato (Pto) DC3000, the causal agent of tomato bacterial speck disease, produces coronatine (COR) that stimulates stomata opening and facilitates bacterial leaf colonization. In Arabidopsis, stomata response to COR requires the COR co‐receptor AtJAZ2, and dominant AtJAZ2Δjas repressors resistant to proteasomal degradation prevent stomatal opening by COR. Here, we report the generation of a tomato variety resistant to the bacterial speck disease caused by Pto DC3000 without compromising resistance to necrotrophs. We identified the functional ortholog of AtJAZ2 in tomato, found that preferentially accumulates in stomata and proved that SlJAZ2 is a major co‐receptor of COR in stomatal guard cells. SlJAZ2 was edited using CRISPR/Cas9 to generate dominant JAZ2 repressors lacking the C‐terminal Jas domain (SlJAZ2Δjas). SlJAZ2Δjas prevented stomatal reopening by COR and provided resistance to Pto DC3000. Water transpiration rate and resistance to the necrotrophic fungal pathogen Botrytis cinerea, causal agent of the tomato gray mold, remained unaltered in Sljaz2Δjas plants. Our results solve the defence trade‐off in a crop, by spatially uncoupling the SA‐JA hormonal antagonism at the stomata, entry gates of specific microbes such as Pto DC3000. Moreover, our results also constitute a novel CRISPR/Cas‐based strategy for crop protection that could be readily implemented in the field.
Keywords: coronatine, CRISPR/Cas9, JAZ2, pseudomonas, stomata, tomato
Introduction
Pseudomonas syringae is a widespread bacterial pathogen that causes disease on a broad range of economically important plant species. Among them, P. syringae pv. tomato DC3000 (Pto DC3000) is the causative agent of the bacterial speck disease of tomato (Solanum lycopersicum) (Blancard, 2012). Susceptible tomato cultivars include the variety Moneymaker, which is agronomically and economically important. Outbreaks of bacterial speck on tomatoes occur in moderate temperatures (15–25°) and wet conditions (Jones et al., 1991). Under favourable environmental conditions, disease symptoms appear as small brown necrotic spots (specks) in leaf and fruits (Bender et al., 1987). This disease affects negatively the productivity and marketability of the tomatoes (Jones et al.,1991) and causes economic losses all over the world (Gitaitis et al., 1985; Schneider, 1977).
The initial infective process of P. syringae relies on natural openings and accidental wounds on the plant surface to colonize internal tissues (Melotto et al., 2008). Stomata are an example of such openings, providing one of the main routes through which the foliar pathogen P. syringae can penetrate the leaf epidermis and start multiplying aggressively in the apoplast (Melotto et al., 2017). Stomata are pores present on the surface of the leaves that are involved in gas exchange, regulating photosynthesis and water loss through transpiration stream (Kim et al., 2010). Besides this, stomata also play an active and dynamic role in defence against pathogens being an integral part of the plant innate immunity system (Liu et al., 2009; Melotto et al., 2006, 2008; Zhang et al., 2008). Upon microbial perception, achieved by the specific recognition of conserved microbial associated molecular patterns (MAMPS) such as bacterial flagellin, via surface‐localized receptors, plants rapidly close stomata (Melotto et al., 2006). This closure requires the salicylic acid (SA) and abscisic acid (ABA) plant hormones, and inhibits the entry of P. syringae restricting host tissue colonization (Du et al., 2014; Gimenez‐Ibanez et al., 2017; Melotto et al., 2006; Zeng and He, 2010; Zhang et al., 2008). Once in the apoplast, P. syringae encounters apoplastic plant immunity, which also relies on a plethora of plant hormones including SA and jasmonic acid (JA). In general terms, SA defences positively regulate resistance to biotrophic and hemi‐biotrophic microbes such as P. syringae, whereas a combination of JA and ethylene (ET) pathways activates resistance against necrotrophic pathogens such as the fungus Botrytis cinerea (Robert‐Seilaniantz et al., 2011). SA and JA/ET defence pathways generally antagonize each other, and consequently, elevated resistance to biotrophs is often correlated with increased susceptibility to necrotrophs, and vice versa (Glazebrook, 2005).
Pseudomonas syringae strains such as Pto DC3000 have evolved a refined strategy for manipulating hormonal crosstalk by producing coronatine (COR), a mimic of the bioactive JA hormone, JA‐isoleucine (JA‐Ile) (Fonseca et al., 2009a). By activating the JA pathway, COR inhibits SA‐dependent defences and thus stimulates the reopening of stomata to facilitate bacterial invasion and growth in the apoplast (Brooks et al., 2005; Gimenez‐Ibanez et al., 2017; Laurie‐Berry et al., 2006; Melotto et al., 2006, 2008; Zheng et al., 2012). The signalling cascade triggered by COR and JA‐Ile is well established. COR is perceived by a co‐receptor formed by the F‐box protein COI1 (CORONATINE‐INSENSITIVE 1) and JAZ (JASMONATE ZIM DOMAIN) repressor proteins (Chini et al., 2007; Katsir et al., 2008; Sheard et al., 2010; Thines et al., 2007; Xie et al., 1998). JAZ co‐receptors are COI1 substrates that negatively regulate the JA‐signalling pathway by directly interacting with and repressing transcription factors (TFs) such as MYC2/3/4 that control JA‐regulated genes (Chini et al., 2007; Fernández‐Calvo et al., 2011; Lorenzo et al., 2004; Sheard et al., 2010; Thines et al., 2007). Repression of TFs by JAZ is mediated by recruitment of the TOPLESS (TPL) co‐repressor complex, through the adaptor protein NINJA. JAZ also prevent the interaction of the TFs with the MEDIATOR complex through MED25 (Cevik et al., 2012; Chen et al., 2012; Pauwels et al., 2010; Zhang et al., 2015). Under stress conditions, COR or JA‐Ile promote the formation of JAZ‐COI1 complexes, triggering JAZ degradation via the 26S proteasome (Chini et al., 2007; Sheard et al., 2010; Thines et al., 2007). This leads to de‐repression of the TFs that initiate the transcription of JA‐dependent genes, and repression of SA‐dependent defences against the bacteria (Fonseca et al., 2009b; Gimenez‐Ibanez and Solano, 2013). Thus, COR acts as a potent virulence factor in plants by triggering the degradation of JAZs. Acquisition of COR by bacterial pathogens has been of tremendous adaptive importance during host‐pathogen evolution because it has allowed bacteria to manipulate the host hormonal network to promote susceptibility.
Among JAZ repressors, we recently showed that AtJAZ2 is a major COR/JA‐Ile co‐receptor in Arabidopsis controlling stomata dynamics during bacterial invasion. JAZ2 is constitutively expressed at the stomata and hijacked by bacterially produced COR to suppress SA‐dependent stomatal closure and promote bacterial penetration (Gimenez‐Ibanez et al., 2017). Arabidopsis jaz2 loss‐of‐function mutants are partially impaired in pathogen‐induced stomatal closing and more susceptible to Pseudomonas. In contrast, truncated JAZ2 forms lacking the C‐terminal Jas domain (JAZ2Δjas) act as gain‐of‐function mutations that prevent stomatal reopening by COR and are highly resistant to bacterial penetration. The C‐terminal Jas domain is responsible for the interaction with COI1 in the presence of hormone and thus, JAZ forms lacking this conserved domain are resistant to COR‐induced degradation, overcoming the action of the phytotoxin during the entry process of P. syringae (Gimenez‐Ibanez et al., 2017). The fact that JAZ2 function is mostly restricted to the stomata makes that alterations in this gene do not affect the SA‐JA crosstalk in other tissues and, therefore, jaz2Δjas mutants still retain unaltered resistance against necrotrophs. Our work suggested that gain‐of‐function mutations in JAZ2 could be used as a general strategy to spatially uncouple SA‐JA hormonal antagonism and to block the entry of P. syringae strains that produce COR without compromising resistance to necrotrophs, which is mostly apoplastic.
Genome editing technologies such as CRISPR/Cas9 (Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR‐associated endonuclease 9) enable precise and directed modifications of DNA sequences in vivo (Schiml and Puchta, 2016). CRISPR/Cas is emerging as an accurate method to improve crops because of its specificity, simplicity and versatility. The Cas9 protein functions as a nuclease and is directed to a target site by a specific guide RNA (gRNA), inducing site‐specific double‐strand breaks. This damage can be then repaired by non‐homologous end‐joining (NHEJ) or homologous recombination (HR), but in the case of NHEJ, the process of repair is error prone, which results in disruptive insertions or deletions at targeted loci that can result in translational frame shifts, amino acid replacements or deletions (Mahfouz et al., 2014).
Genetic manipulation of defence pathways has succeeded in enhancing resistance to specific kinds of pathogens. However, due to the antagonistic interactions between the SA and JA defence pathways, efforts to develop plants with broad‐spectrum resistance by manipulation of hormonal signalling genes has had limited success so far because enhancement of the SA‐dependent defences leads to a reduction in JA‐based resistance and vice versa (Robert‐Seilaniantz et al., 2011; Pieterse et al., 2012). Solving this trade‐off is a major challenge in agriculture and requires uncoupling the antagonism between hormonal pathways in crops.
In this study, we report on a solution to this trade‐off by spatially uncoupling the SA‐JA antagonism at the stomata and generating a tomato (Moneymaker) resistant to the bacterial speck disease caused by the pathogen Pto DC3000, without compromising resistance to necrotrophic pathogens. We identified the functional ortholog of the COR stomatal co‐receptor AtJAZ2 in tomato (SlJAZ2) and edited this gene with the CRISPR/Cas9 system to generate truncated JAZ2 forms lacking the C‐terminal Jas domain (SlJAZ2Δjas). This edited gain‐of‐function SlJAZ2Δjas mutant fully prevented stomatal reopening by COR and reduced bacterial entry through the stomata, increasing resistance to Pto DC3000 infection. Stomatal aperture in Sljaz2‐edited plants remained unaltered during transpiration. Moreover, since our strategy does not affect JA‐signalling outside the stomata, Sljaz2Δjas plants showed unaltered levels of resistance to the necrotrophic fungal pathogen B. cinerea, causal agent of the tomato gray mold. In addition to uncouple the SA‐JA defence antagonism in a crop, our results constitute a novel strategy for crop protection using CRISPR/Cas that could be readily implemented in the field.
Results
SlJAZ2 is the ortholog of AtJAZ2
The AtJAZ2 protein belongs to the TIFY super‐family that includes 12 canonical members in Arabidopsis. Similarly, 12 JAZ proteins have also been identified in tomato (Chini et al., 2017; Ishiga et al., 2013; Sun et al., 2011). However, correlation between specific versions of JAZ genes among plant species is unclear. In order to identify the ortholog of AtJAZ2 in tomato we performed phylogenetic analysis using protein sequences of all JAZs from both plant species. AtJAZ2 protein grouped into a clade that included AtJAZ1, and the SlJAZ1 and SlJAZ2 proteins as the closest tomato orthologs (Figure 1a and Figure S1). SlJAZ3 and SlJAZ4 were also closer to AtJAZ2 than to any other tomato or Arabidopsis protein. Among proteins in this clade, AtJAZ1, AtJAZ2 and SlJAZ2 showed a remarkable similar length (Figure 1a). This phylogenetic analysis indicated that the ortholog of AtJAZ2 in tomato was likely SlJAZ2 or SlJAZ1, although SlJAZ3 and SlJAZ4 cannot be discarded.
Figure 1.
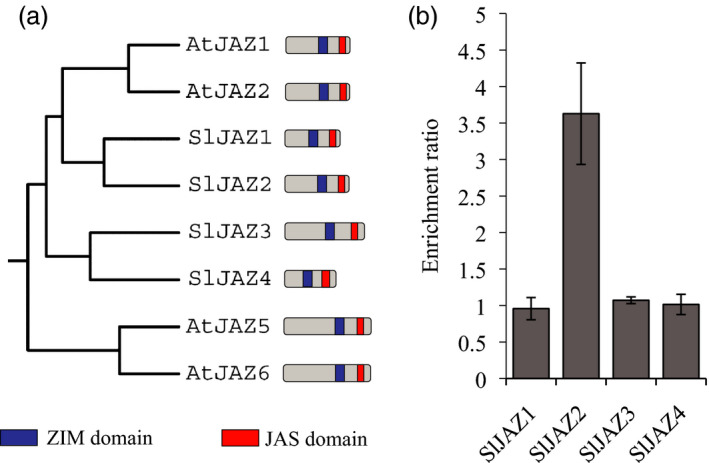
SlJAZ2 is the ortholog of AtJAZ2. (a) Phylogenetic tree of JAZ1, JAZ2, JAZ3 and JAZ4 from Arabidopsis and tomato. SlJAZ1 and SlJAZ2 are the closest proteins to AtJAZ2. The schematic representation indicates length and domains of each JAZ protein. (b) Enrichment ratio of RT‐PCR analyses comparing expression levels of SlJAZ1, SlJAZ2, SlJAZ3 and SlJAZ4 in tomato whole leaf tissue and epidermal peels, which are enriched in guard cells. This experiment was repeated twice with similar results.
JAZ2 is strongly expressed at stomatal guard cells compared to other JAZs in Arabidopsis (Gimenez‐Ibanez et al., 2017). Thus, we next analysed if any of the closest tomato candidate orthologs of AtJAZ2 were expressed at the stomata. To do this, we performed quantitative RT‐PCR analyses that compared the expression levels of these genes between tomato whole leaf tissue and epidermal peels, which are enriched in guard cells. SlJAZ2 was the only gene enriched in the stomata‐abundant epidermal peels fraction whereas SlJAZ1, SlJAZ3 and SlJAZ4 relative levels were similar among them (Figure 1b). This pinpointed SlJAZ2 as the functional tomato ortholog of AtJAZ2.
SLJAZ2 editing through CRISPR/Cas9
In Arabidopsis, a truncated form of AtJAZ2 lacking the Jas domain (AtJAZ2ΔJas) is resistant to AtCOI1‐dependent degradation after COR treatment. Arabidopsis plants overexpressing this degradation resistant JAZ2 dominant form are insensitive to the phytotoxin COR and more resistant to DC3000 (Gimenez‐Ibanez et al., 2017). We used CRISPR/Cas9 genome‐editing technology to generate sequence‐specific mutations that would disrupt the Jas domain of SlJAZ2, leading to a JAZ2ΔJas tomato variant in the commercial variety Moneymaker. To do this, we targeted the start site of the Jas domain in the SlJAZ2 locus using a dual gRNA strategy that facilitates the generation of homozygous deletions (Brooks et al., 2014). The two selected gRNA targets within SlJAZ2 were positioned on opposite DNA strands, and overlapped 7 nucleotides (Figure 2a). DNA sequence analysis of primary transformants identified two homozygous lines for SlJAZ2 containing deletions of seven and four nucleotides in the designated area (Figure 2b). Both deletions were predicted to disrupt the Jas domain of SlJAZ2 (Figure 2b). We further explored if these genome edited Sljaz2Δjas mutations were specifically being expressed in our tomato lines. To do this, we performed quantitative RT‐PCR experiments amplifying wild‐type (WT) SlJAZ2 or each one of the two specific Sljaz2Δjas mutations generated in both tomato lines. WT, Line 1 and Line 2 amplified exclusively their expected transcripts (Figure 2b), indicating both lines carry homozygous mutations at the SlJAZ2 locus generating forms of SlJAZ2Δjas that are effectively being transcribed. Further analysis of T1 and T2 Sljaz2Δjas lines by DNA sequencing of SlJAZ2 showed that the homozygous mutations were stably transmitted to the offspring.
Figure 2.
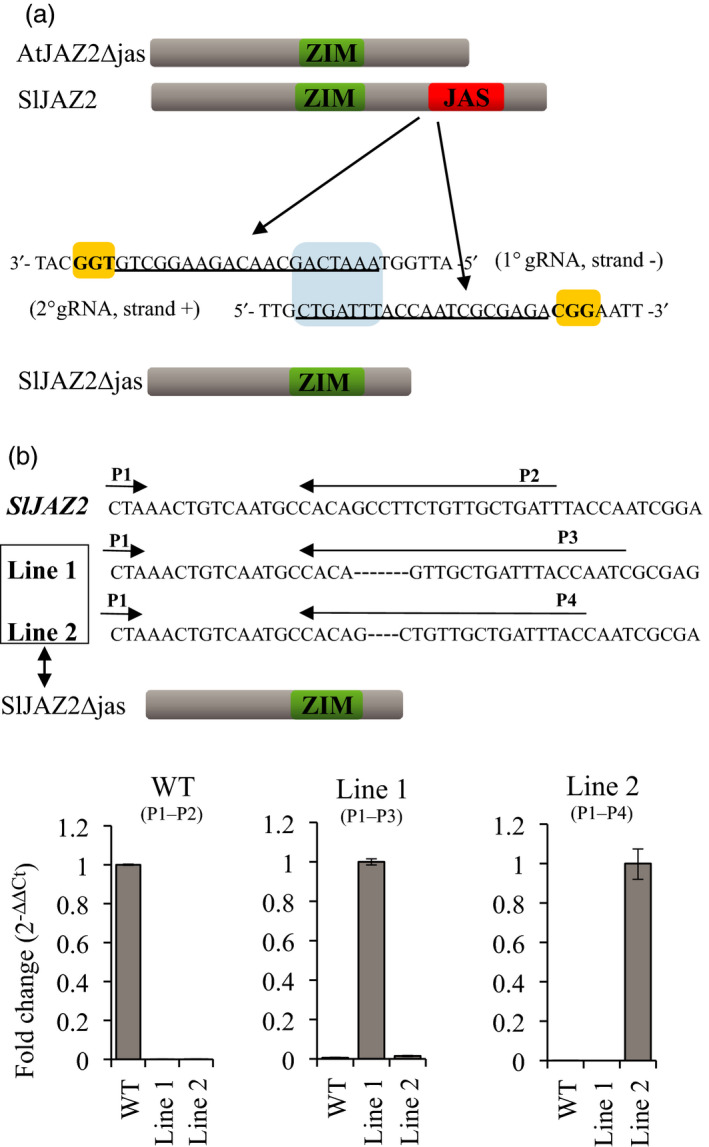
CRISPR/Cas9‐mediated editing of SlJAZ2. (a) Schematic representation of AtJAZ2Δjas, SlJAZ2, SlJAZ2Δjas, and position of both gRNAs designed to target the Jas domain in the SlJAZ2 locus. Nucleotide sequence indicates double strand DNA of SlJAZ2. Yellow boxes indicate the PAM sequence. Underlined sequence indicates the two gRNAs used. The 1° gRNA is targeted before the start of the Jas domain (strand −) while the 2°gRNA is targeted to the beginning of the Jas domain (strand +). The target area of both gRNAs overlaps seven nucleotides (blue box). (b) Transcriptional expression by RT‐PCR of SlJAZ2 and SlJAZ2Δjas forms in WT and SlJaz2Δjas plants (Line 1 and Line 2) using different combinations of primers as described in the figure.
Gain‐of‐function Sljaz2Δjas prevent stomatal reopening by COR
Pathogen perception induces stomatal closure to limit pathogen penetration. Some P. syringae strains produce COR as a critical virulence strategy to re‐open stomata and cause disease. Thus, we next evaluated the ability of both Sljaz2Δjas tomato lines to close stomata upon perception of the highly conserved flg22 peptide of bacterial flagellum (Chinchilla et al., 2007), and to reopen them in the presence of COR. We incubated leaves of WT plants and Sljaz2Δjas mutants with flg22, flg22 plus COR or a mock solution as a control. In mock conditions, the aperture range of the stomata was wide and similar between WT and both Sljaz2Δjas tomato lines. As expected, flg22 induced stomatal closure in all tomatoes, whereas COR induced stomatal re‐opening in WT plants (Figure 3a). In contrast to WT, both Sljaz2Δjas tomato lines were fully impaired in COR‐mediated stomatal reopening (Figure 3a). This indicates that, similar to Arabidopsis, SlJAZ2 has a key role regulating stomata dynamics in response to bacteria and that COR‐induced stomata reopening requires inhibition of SlJAZ2, which cannot be achieved in the dominant Sljaz2Δjas mutants.
Figure 3.
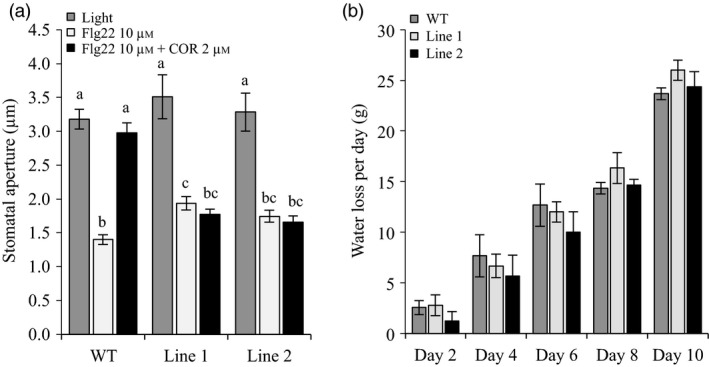
Sljaz2Δjas mutants prevent stomatal reopening by COR. (a) Stomatal aperture in tomato WT and Sljaz2Δjas mutants (Line 1 and Line 2) after 2 h of incubation with flg22, flg22 plus COR or a mock control. Error bars indicate standard error of the mean (SEM; n = 60). Letters above bars represent statistically distinct groups (P < 0.01, one‐way Anova with Tukey‐HSD). Similar results were obtained in three independent biological replicates (b) Water transpiration rate (water loss) in WT and Sljaz2Δjas (Line 1 and Line 2) tomato plants during a time course experiment of 10 days. Error bars indicate standard deviation (SD; n = 3). Not statistical difference was found between WT and mutant lines (one‐way Anova with Tukey‐HSD). Results are representative of two independent experiments.
Stomata are the major regulators of water transpiration in plants (Brodribb and McAdam, 2017). We next evaluated whether the rate of transpiration was affected in Sljaz2Δjas lines compared to WT tomato plants. Time‐course measurements showed that the rate of water loss in all plants was similar (Figure 3b). We also analysed leaf temperature, which is an indirect measure of stomata opening and water transpiration, using infrared thermography and found no differences related to stomatal apertures (Figure S2). Overall, this indicates that Sljaz2Δjas tomato lines are not affected in the aperture of the stomata during the process of transpiration, but are rather specifically affected in the response to bacteria.
Dominant Sljaz2Δjas mutants are resistant to P. syringae infection
In order to evaluate the resistance of adult Sljaz2Δjas plants towards bacterial pathogens such as P. syringae, we next compared bacterial replication of Pto DC3000 on WT and both Sljaz2Δjas lines infected by surface inoculation (dipping) or syringe infiltration. Surface infection mimics natural infection conditions and it is a very common technique to assess plant resistance to pathogens (Zipfel et al., 2004). In contrast, the infiltration technique overcomes the early stomatal level of regulation measuring mainly apoplastic cell‐based defences. WT plants infected by surface inoculation with Pto DC3000 showed extensive chlorosis and specks, typical symptoms of the bacterial speck disease in tomatoes (Figure 4a). In contrast, Sljaz2Δjas plants did not display these typical symptoms of disease suggesting their enhanced resistance (Figure 4a). Disease symptoms correlated well with bacterial titres. Sljaz2Δjas plants surface inoculated showed significantly lower bacterial titres than those in its respective WT control plants (Figure 4b). In the case of plants infected with Pto DC3000 by infiltration, bacteria grew to similar levels on both WT and Sljaz2Δjas lines (Figure 4c). Consistently, inoculation with the coronatine‐deficient Pto DC3000 strain AK87 (Pto DC3000 COR−) showed similar bacterial titles in WT and Sljaz2Δjas plants (Figure S3). These results supports the idea that SlJAZ2 is a key regulator of stomata dynamics during the penetration process of Pto DC3000 and indicates that Sljaz2Δjas plants promote bacterial resistance by blocking the action of the phytotoxin COR at the stomata.
Figure 4.
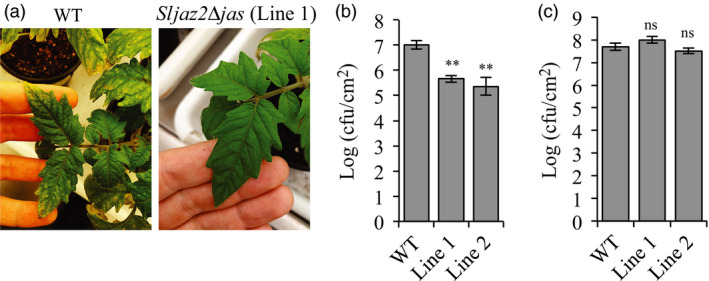
Sljaz2Δjas mutants are more resistant to Pto DC3000 when surface inoculated. (a) Pto DC3000 disease symptoms on WT and Sljaz2Δjas (Line 1) tomato plants after surface inoculation with Pto DC3000 bacteria at 108 colony‐forming units/mL (cfu/mL). Pictures were taken 6 days postinoculation and show typical specks caused by Pto DC3000. (b) Growth of Pto DC3000 on WT and Sljaz2Δjas (Line 1 and Line 2) tomato plants 6 days after surface inoculation by dipping with bacteria at 108 cfu/mL. (c) Growth of Pto DC3000 on WT and Sljaz2Δjas (Line 1 and Line 2) tomato plants 2 days after syringe infiltration with bacteria at 5 × 105 cfu/mL. Error bars indicate SEM (n = 7). Asterisks represent significant differences **P < 0.01 (Student's t‐test). Similar results were obtained in three independent experiments and representative results are shown.
Dominant Sljaz2Δjas tomato plants show unaltered levels of resistance against the necrotrophic pathogen Botrytis cinerea
The antagonism between JA and SA pathways is a barrier to improve plant immunity and generate broad‐spectrum resistance. To evaluate if the insensitivity to COR generated in the edited Sljaz2Δjas tomato lines affects resistance against necrotrophic fungal pathogens, we selected B. cinerea, causal agent of the gray mold of tomatoes. Thus, we measured the susceptibility of WT and Sljaz2Δjas lines to B. cinerea by quantifying leaf infected area. Lesions caused by the fungus indicated similar progression of disease symptoms (Figure 5a) and area affected by the pathogen in WT and Sljaz2Δjas lines (Figure 5b). These results indicate that SlJAZ2 function seems restricted to stomata guard cells and it plays a very limited or inexistent role in apoplastic defence responses against necrotrophic fungi.
Figure 5.
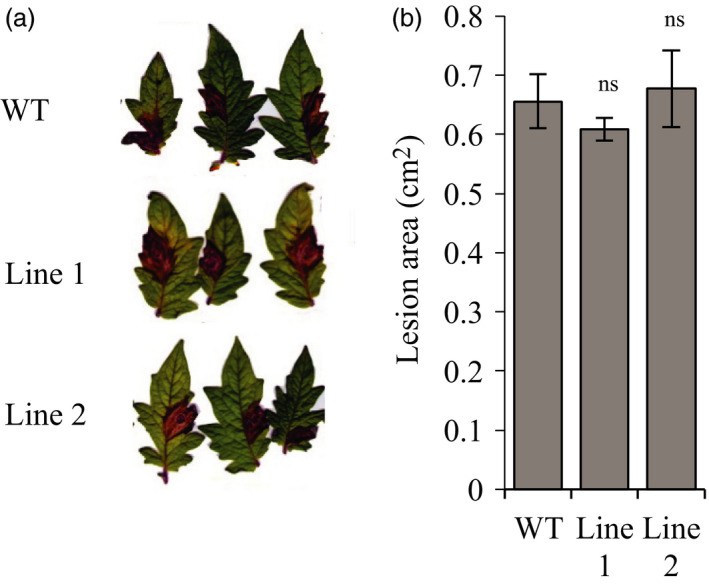
Sljaz2Δjas mutants retain WT resistance to necrotrophic fungi. (a) Botrytis cinerea disease symptoms on WT and Sljaz2Δjas mutants 4 days postinoculation (dpi) with 5 × 106 spores m/L. (b) Lesion Area produced by Botrytis cinerea on WT and Sljaz2Δjas mutants 4 days postinoculation (dpi) with 5 × 106 spores m/L. Error bars indicate standard error SEM (n = 8). (Student's t‐test, P < 0.05; ns, not significant). Results are representative of two independent experiments.
Discussion
One of the great challenges for food security in the 21st century is to improve yield through the development of disease‐resistant crops. However, novel strategies to improve plant immunity are particularly challenging. In vascular plants, hormonal crosstalk between JA and SA, the key two hormonal pathways controlling resistant to vastly all type of pathogens, often antagonize each other, and thus, enhanced resistance against biotrophic pathogens normally leads to increased susceptibility to necrotrophs, and vice versa (Glazebrook, 2005). Solving this trade‐off is a major challenge for obtaining broad‐spectrum resistance and requires uncoupling the antagonism between these hormonal pathways in crops.
Pto DC3000 is the natural causal agent of the bacterial speck disease of tomato. Bacterial spots on tomato fruits have been reported to cause up to 52% loss of fruit weight (Jones et al., 1986), whereas in the field, yield losses due to bacterial speck vary from 75% in plants infected at an early stage of growth to 5% in plants infected later in the season (Yunis et al., 1980). Therefore, it becomes necessary to uncover novel sources of resistance against this pathogen. Pto DC3000 uses COR as a virulent factor to induce stomata opening and facilitate entrance into the leaf apoplast. We have recently identified the specific JAZ co‐receptor of COR at the stomata in Arabidopsis (Gimenez‐Ibanez et al., 2017). A dominant mutation of this stomata‐specific AtJAZ2 lacking a C‐terminal Jas regulatory domain (Atjaz2Δjas) is insensitive to COR and cannot re‐open stomata after bacterial infection. This mutation confers resistance to biotrophic Pto DC3000 without affecting susceptibility to necrotrophs, due to its specific action at guard cells (Gimenez‐Ibanez et al., 2017).
In this study, we demonstrate that similar Δjas mutations in stomata‐accumulating JAZ proteins can be used as a general strategy to increase resistance to COR‐producing bacteria in crops. Thus, as a proof‐of‐concept, we identified the tomato ortholog of AtJAZ2 and successfully generated a bacterial speck resistant tomato in the commercial variety Moneymaker by CRISPR/Cas9‐mediated SlJAZ2 editing. Sljaz2Δjas plants were significantly more resistant to Pto DC3000 when plants where surface inoculated, but equally susceptible by infiltration. This indicates that this SlJAZ2 variant is mainly affecting the stomatal defence layer by counteracting the effect of COR. Consistently, Sljaz2Δjas tomato lines were fully impaired in COR‐mediated stomatal reopening. Remarkably, since the Sljaz2Δjas mutation only manipulates the JA‐dependent defences at the stomata, the JA‐SA antagonism is not affected in the mesophyll and, therefore, apoplastic resistance to the necrotrophic pathogen B. cinerea remains unaffected. These results suggest that similar to Arabidopsis, SlJAZ2 plays a major role in regulating stomatal aperture during biotic stresses, and that COR produced by Pto DC3000 hijacks SlCOI1‐SlJAZ2 co‐receptor to promote the entry of the bacteria to the internal tissues of tomato leaf. Moreover, these results prove that is possible to uncouple SA‐JA antagonism spatially to design novel strategies for protection against COR‐producing P. syringae strains in crops, without affecting resistance to necrotrophs.
Alteration of the regulation of plant stomatal aperture can lead to deleterious effects. A number of environment factors, including CO2 level, light, water and other abiotic stresses also regulate stomatal dynamics. In all these cases, ABA functions as a chemical messenger that induces stomata closure (Lim et al., 2015; Vishwakarma et al., 2017). However, several lines of evidence suggest that Sljaz2Δjas tomato lines are not affected in these ABA‐mediated physiological processes. First, in Arabidopsis, gain‐of‐function mutations in JAZ2 prevent stomatal reopening by COR by inhibiting SA‐dependent stomatal closure triggered upon specific microbial perception without affecting ABA signalling at the stomata (Gimenez‐Ibanez et al., 2017). Second, the rate of stomatal aperture in normal light conditions was not affected in Sljaz2Δjas tomato mutants. Third, neither water transpiration rate or leaf temperature was affected in Sljaz2Δjas compared to WT. Finally, Sljaz2Δjas plants displayed a complete phenotypically normal development compared to WT tomatoes (Figure S4). Taken together, these results suggest that the Sljaz2Δjas mutation impact on the regulation of stomatal aperture exclusively during biotic conditions, when SA is produced to execute resistance against biotrophic pathogens trying to invade the plant.
Current methods to control bacterial speck disease of tomato are based on removal of plant debris and weeds, crop rotation, and the use of non‐infected seeds and transplants. Chemical sprays based on copper are also commonly used, but these may not be effective in environmental conditions favourable for infection (Somodi et al., 1996). Moreover, copper can also kill plant cells if absorbed in sufficient quantities and can produce problematic toxicity in soil. In contrast, tomato resistant varieties offer the most effective means of management. P. syringae strains vary in their ability to establish and maintain epiphytic populations on the surface of the leave before infection of internal plant tissues. In this sense, it is becoming clear that transition from epiphytic to endophytic lifestyles is a critical process to establish a successful infection cycle. This transitional process is enhanced by the production of COR. In the case of Sljaz2Δjas, these plants would impede the entry on the bacteria, elongating its epiphytic phase where P. syringae encounters a harsh environment with limited nutrients. This elongated epiphytic phase on Sljaz2Δjas tomatoes should reduce survival of the bacteria further enhancing resistance.
Current resistant varieties are based on the introduction of specific disease‐resistant genes that recognized pathogenic molecules intracellularly. However, pathogens frequently adapt to and overcome genetic resistance especially when it is determined by major resistant genes (Brown, 2015). The ability of Sljaz2Δjas tomato plants to interfere with the penetration of the pathogen represents a new strategy for providing resistance that may be achieved by a low energetic cost for the plant and in a clean way for the environment. Ultimately, a hierarchical design containing multiple layers of resistance in a crop may be the most feasible way to achieve durable resistant in the field (Fuchs, 2017; Pilet‐Nayel et al., 2017).
Recent results demonstrate that novel techniques for genome editing could be successfully applied in crops to introduce directed resistance. For example, CRISPR/Cas has been recently used to generate powdery mildew resistant tomatoes (Nekrasov et al., 2017) and engineer resistance to geminiviruses in tobacco (Ali et al., 2015). Public policies are forcing researchers to develop transgene‐free improved crops. In this context DNA editing technologies (also called New Breeding Techniques) have emerged as a novel strategy to overcome this challenge. The status of genetically edited crop varieties is still debated by regulatory authorities, for example countries such as US consider transgene‐free genetically edited crops as non‐GMO. We hope that plant varieties such as the one presented here could be adopted worldwide to enhance plant productivity and resistance against pests. These superior varieties have the potential to fight against agricultural losses in the field due to pests, reducing the chemical inputs towards a more sustainable agriculture for the environment.
Altogether, we demonstrate the feasibility to create bacterial speck resistant tomatoes through CRISPR/Cas9‐mediated editing of SlJAZ2. Moreover, we also define a novel strategy to overcome the penetration of COR‐producing P. syringae strains through the stomata by spatially uncoupling SA‐JA antagonism.
Experimental procedures
Phylogenetic analysis
JAZ proteins sequences from Arabidopsis thaliana and Solanum lycopersicum (tomato) were aligned with Dialing software (http://www.genomatix.de/cgi-bin/dialign/dialign.pl). Phylogenetic tree was represented using the online tool ‘Phylodendron’, (http://iubio.bio.indiana.edu/treeapp/treeprint-form.html).
RT‐qPCR analysis
Leaf tissue or epidermal peels were harvested from 4‐ to 6‐week‐old plants and frozen in liquid nitrogen. RNA was extracted using a Plant total RNA purification mini kit (Favorgen®). One microgram of RNA was used for cDNA synthesis using a MultiScribe Reverse Transcriptase (Applied Biosystems). PCR was performed on a 7500 thermocycler using SYBR‐green (both from Applied Biosystems). SlActin was used as endogenous control. Oligonucleotides used in this study are described in Table S1. Data analysis shown was done using three technical replicates from one biological sample; similar results were obtained with at least two additional independent biological replicates.
Plasmid construction
Designed gRNAs (Table S2) targeting the Jas domain of SlJAZ2 (Solyc12 g009220) were cloned into the vector Bsa_Bbs_tandem gRNA_pBS. This vector confers resistance to ampicillin and gRNAs are expressed under different versions of the ubiquitin promoter. Then, a fragment containing both gRNAs and their promoters were cloned into the vector Pk7_CAS9‐TPC_polylinker pBKS. This vector confers resistance to spectinomycin in bacteria and kanamycin in plants. Cas9 is expressed under an ubiquitin promoter. Annealing of gRNAs and cloning was performed by ordinary methods using BsaI and BbsI restriction enzymes. Fragment with the two gRNAs together with their promoters were clone using the KpnI restriction enzyme. gRNAs were designed with the online tool ‘Breaking Cas’ (Oliveros et al., 2016) (http://bioinfogp.cnb.csic.es/tools/breakingcas/). gRNAs were unique and specific for the target region (Jas domain sequence of SlJAZ2). 1° gRNA had a score of 96.3% with the best possible off‐targets having a 0.2%. 2° gRNA had a score of 98.4%, with the best possible off‐targets having a 0.5%.
Plant growth and transformation
A tomato cultivar Moneymaker was transformed with the pK7_CAS9‐TPC_polylinker pBKS, containing both gRNAs for SlJAZ2, as previously described (Wittmann et al., 2016). Plants were grown under a 16‐h‐light/8‐h‐dark cycle in a growth chamber or greenhouse. Around 120 cotyledons (60 tomato plants) were initially used for transformation with Agrobacterium. Many cotyledons generated a callus and produced shots, which were transferred for root generation. In total, 23 regenerated plants were transferred to the greenhouse. PCR amplification of SlJAZ2 and DNA sequencing identified mutations in these plants and indicated that 8/23 were wild‐type, 12/23 were heterozygous or chimeras and 3/23 contained an homozygous mutation. T1 and T2 progeny of selected Sljaz2Δjas Lines (1 and 2) was further obtained and analysed by DNA sequencing of SlJAZ2 to confirm that homozygous mutations were stably transmitted to the offspring.
Measurements of stomatal aperture
After 2 days of stratification of seeds at 4°C, tomato WT and Sljaz2Δjas Lines 1 and 2 were grown on soil in a chamber with an 8 h light period at 23°C and a 16 h dark period at 19°C, relative humidity of 75%. The abaxial side of leave from 4‐ to 6‐week‐old tomato plants were collected and stuck on cover slips (peeled). Samples were incubated in a solution of 10 mM MES/Tris pH 6.0 and 30 mM KCl (working buffer). For promotion of closure assays, compounds (10μm flg22, 10μm flg22 + 2μm COR or a mock solution) were directly diluted in the working buffer in contact with epidermal peels during 2 h. Aperture of stomata and hydathode pores were determined under an optical microscope fitted with a camera lucida and a digitizing pad linked to a computer as described (Leonhardt et al., 1997). Values reported are the means of at least three independent experiments, for which 60 aperture widths were measured each time. Error bars represent standard deviations of the means. One‐way Anova with Tukey‐HSD was used for statistical analysis. Calculations were performed using a statistical calculator (http://astatsa.com/OneWay_Anova_with_TukeyHSD/).
Pseudomonas bacterial strains
Pseudomonas strains used in this study were Pseudomonas syringae pv. tomato (Pto) DC3000 and the coronatine‐deficient Pto DC3000 strain (Pto DC3000 COR−) which is a Pto DC3000 AK87 mutant that carries mutations in cmaA (coronamic acid A) and cfa6 (coronafacic acid 6) (Brooks et al., 2004).
Pseudomonas syringae DC3000 infection assays
Tomato plants for infection assays were grown in a cycle of 14 hours light and 10 hours darkness with 45‐60% humidity for 6‐7 weeks. Bacterial growth assays in tomato were performed as described previously with some modifications (Balmuth and Rathjen, 2007). For surface infection assays, plants were dipped in a bacterial suspension containing 108 colony‐forming units m/L (cfu/mL) bacteria (OD600 = 0.2) with 0.02% Silwet L‐77. For bacterial growth assays by syringe infiltration, leaves were syringe infiltrated with a bacterial suspension containing 5x105 cfu/mL bacteria (OD600 = 0.001). Leaf disks were collected 2 days (infiltration) and 6/7 days (surface infection) postinfection and bacterial growth was quantified as described previously (Gimenez‐Ibanez et al., 2009). Error bars indicate SEM (n = 7). These experiments were repeated at least three times with similar results, and representative results are shown.
Botrytis cinerea infection assays
Botrytis cinerea infection assays were performed as described previously (Monte et al., 2014) with some modifications. Five‐ to six‐week‐old tomato leaves were inoculated with 20 μL of a suspension of 5 × 106 spores m/L PDB (Difco, Le Pont de Claix, France). At least eight leaves were inoculated per treatment. Disease symptoms were scored 4 days postinoculations. Area of the lesion was measured using jimage software (https://imagej.nih.gov/ij/). This experiment was repeated twice with similar results.
Leaf temperature and water transpiration ratio
For leaf temperature assays, 3–4 weeks tomato plants were examined under Thermacam infrared camera (FLIR A655sc 25°, 50°Hz) during different days. Images were saved and analysed on a personal computer using the ResearchIR4 Max Software provided by FLIR. For the calculation of water transpiration rate, plants were stratified 2 days at 4°C in dark and grown in soils from 1 to 2 weeks. Then plants were transferred to pots with soil at 20% humidity (all pots weight the same and humidity content was calculated to be 20%) and keep some days for acclimatization. During at least 10 days plants weight was measured every day, after measuring water was added to restore 20% humidity in pots, a control (without plant) was used to measure the amount of water lost by exchange of the soil. The difference of the water lost compare to the negative control (without plant) is a measure of the water transpiration rate of the plants through the stomata. This experiment was repeated twice with similar results.
Statistical methods
Statistical significance based on Students's t test analysis was calculated using Excel software (*P < 0.05; **P < 0.01). Statistical significance based on one‐way Anova with Tukey‐HSD (*P < 0.05; **P < 0.01) was calculated using a statistical calculator (http://astatsa.com/OneWay_Anova_with_TukeyHSD/).
Conflict of interest
A.O, S.G‐I. and R.S. are inventors in a patent regarding the technology published in this paper.
Author contributions
R.S. designed the research. A.O, S.G‐I. and N.L. performed the experiments. All authors analyse the data. R.S. supervised the work. A.O., S.G‐I. and R.S. wrote the article. All authors read and edited the article.
Supporting information
Figure S1 Phylogenetic tree of all AtJAZs and SlJAZs. The schematic representation indicates length and domains of each JAZ protein.
Figure S2 False colour infrared image shows no differences between WT and Sljaz2Δjas plants. On the left, image of WT and Sljaz2Δjas tomato plants (Line 1 and Line 2), as depicted in the figure, taken with a reflex camera. On the right, the same plants analysed under an infrared camera to measure leaf temperature.
Figure S3 Sljaz2Δjas mutant is equally resistant to Pto DC3000 COR‐ when surface inoculated. Growth of Pto DC3000 COR‐ on WT and Sljaz2Δjas (Line 1) tomato plants 7 days after surface inoculation by dipping with bacteria at 108 cfu/mL. Error bars indicate SEM (n = 7).
Figure S4 Sljaz2Δjaz plants display a normal phenotype.
Table S1 RT‐PCR oligonucleotides used in this study.
Table S2 Oligonucleotides used as gRNAs in this study.
Acknowledgements
We thank Dr. Michael Nicolas and Dr. Pilar Cubas (CNB‐CSIC), for developing and sharing CRISPR/Cas vectors. We also thank Caterina Brancato and Dr. Kenneth Berendzen (Center for Plant Molecular Biology (ZMBP), University of Tübingen) for advice on tomato transformation. A.O. was supported by a ‘Severo Ochoa’ CNB PhD fellowship. S.G‐I. was supported by the Spanish Ministry of Science and Innovation Grant BIO2014‐55884‐JIN. Work in N.L.' s laboratory is funded by ANR program. Work in R.S.' s laboratory is funded by the Spanish Ministry for Science and Innovation grant BIO2016‐77216‐R (AEI/FEDER, UE) and Fundación UAM Grant 2015007.
References
- Ali, Z. , Abulfaraj, A. , Idris, A. , Ali, S. , Tashkandi, M. and Mahfouz, M.M. (2015) CRISPR/Cas9‐mediated viral interference in plants. Genome Biol. 16, 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmuth, A. and Rathjen, J.P. (2007) Genetic and molecular requirements for function of the Pto/Prf effector recognition complex in tomato and Nicotiana benthamiana. Plant J. 51, 978–990. [DOI] [PubMed] [Google Scholar]
- Bender, C.L. , Stone, H.E. , Sims, J.J. and Cooksey, D.A. (1987) Reduced pathogen fitness of Pseudomonas syringae pv. tomato Tn5 mutants defective in coronatine production. Physiol. Mol. Plant Pathol. 30, 273–283. [Google Scholar]
- Blancard, D. (2012). Tomato Diseases (Elsevier).
- Brodribb, T.J. and McAdam, S.A.M. (2017) Evolution of the stomatal regulation of plant water content. Plant Physiol. 174, 639–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks, D.M. , Hernández‐Guzmán, G. , Kloek, A.P. , Alarcón‐Chaidez, F. , Sreedharan, A. , Rangaswamy, V. , Peñaloza‐Vázquez, A. , et al. (2004) Identification and Characterization of a Well‐Defined Series of Coronatine Biosynthetic Mutants of Pseudomonas syringae pv. tomato DC3000. Mol. Plant‐Microbe Interact. 17, 162–174. [DOI] [PubMed] [Google Scholar]
- Brooks, D.M. , Bender, C.L. and Kunkel, B.N. (2005) The Pseudomonas syringae phytotoxin coronatine promotes virulence by overcoming salicylic acid‐dependent defences in Arabidopsis thaliana. Mol. Plant Pathol. 6, 629–639. [DOI] [PubMed] [Google Scholar]
- Brooks, C. , Nekrasov, V. , Lippman, Z.B. and Van Eck, J. (2014) Efficient gene editing in tomato in the first generation using the clustered regularly interspaced short palindromic repeats/CRISPR‐associated9 system. Plant Physiol. 166, 1292–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, J.K.M. (2015) Durable resistance of crops to disease: a darwinian perspective. Annu. Rev. Phytopathol. 53, 513–539. [DOI] [PubMed] [Google Scholar]
- Cevik, V. , Kidd, B.N. , Zhang, P. , Hill, C. , Kiddle, S. , Denby, K.J. , Holub, E.B. et al. (2012) MEDIATOR25 acts as an integrative hub for the regulation of jasmonate‐responsive gene expression in arabidopsis. Plant Physiol. 160, 541–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, R. , Jiang, H. , Li, L. , Zhai, Q. , Qi, L. , Zhou, W. , Liu, X. et al. (2012) The arabidopsis mediator subunit MED25 differentially regulates jasmonate and abscisic acid signaling through interacting with the MYC2 and ABI5 transcription factors. Plant Cell 24, 2898–2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla, D. , Zipfel, C. , Robatzek, S. , Kemmerling, B. , Nürnberger, T. , Jones, J.D.G. , Felix, G. et al. (2007) A flagellin‐induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448, 497–500. [DOI] [PubMed] [Google Scholar]
- Chini, A. , Fonseca, S. , Fernández, G. , Adie, B. , Chico, J.M. , Lorenzo, O. , García‐Casado, G. et al. (2007) The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448, 666–671. [DOI] [PubMed] [Google Scholar]
- Chini, A. , Ben‐Romdhane, W. , Hassairi, A. and Aboul‐Soud, M.A.M. (2017) Identification of TIFY/JAZ family genes in Solanum lycopersicum and their regulation in response to abiotic stresses. PLoS ONE 12, e0177381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, M. , Zhai, Q. , Deng, L. , Li, S. , Li, H. , Yan, L. , Huang, Z. et al. (2014) Closely related NAC transcription factors of tomato differentially regulate stomatal closure and reopening during pathogen attack. Plant Cell 26, 3167–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández‐Calvo, P. , Chini, A. , Fernández‐Barbero, G. , Chico, J.M. , Gimenez‐Ibanez, S. , Geerinck, J. , Eeckhout, D. et al. (2011) The arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23, 701–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca, S. , Chico, J.M. and Solano, R. (2009a) The jasmonate pathway: the ligand, the receptor and the core signalling module. Curr. Opin. Plant Biol. 12, 539–547. [DOI] [PubMed] [Google Scholar]
- Fonseca, S. , Chini, A. , Hamberg, M. , Adie, B. , Porzel, A. , Kramell, R. , Miersch, O. et al. (2009b) (+)‐7‐iso‐Jasmonoyl‐L‐isoleucine is the endogenous bioactive jasmonate. Nat. Chem. Biol. 5, 344–350. [DOI] [PubMed] [Google Scholar]
- Fuchs, M. (2017) Pyramiding resistance‐conferring gene sequences in crops. Curr. Opin. Virol. 26, 36–42. [DOI] [PubMed] [Google Scholar]
- Gimenez‐Ibanez, S. and Solano, R. (2013) Nuclear jasmonate and salicylate signaling and crosstalk in defense against pathogens. Front. Plant Sci. 4, 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimenez‐Ibanez, S. , Hann, D.R. , Ntoukakis, V. , Petutschnig, E. , Lipka, V. and Rathjen, J.P. (2009) AvrPtoB targets the LysM receptor kinase CERK1 to promote bacterial virulence on plants. Curr. Biol. 19, 423–429. [DOI] [PubMed] [Google Scholar]
- Gimenez‐Ibanez, S. , Boter, M. , Ortigosa, A. , García‐Casado, G. , Chini, A. , Lewsey, M.G. , Ecker, J.R. et al. (2017) JAZ2 controls stomata dynamics during bacterial invasion. New Phytol. 213, 1378–1392. [DOI] [PubMed] [Google Scholar]
- Gitaitis, R.D. , Jones, J.B. , Jaworski, C.A. and Phatak, S.C. (1985) Incidence and development of Pseudomonas syringae pv. syringae on tomato transplants in Georgia. Plant Dis. 69, 32–35. [Google Scholar]
- Glazebrook, J. (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227. [DOI] [PubMed] [Google Scholar]
- Ishiga, Y. , Ishiga, T. , Uppalapati, S.R. and Mysore, K.S. (2013) Jasmonate ZIM‐domain (JAZ) protein regulates host and nonhost pathogen‐induced cell death in tomato and nicotiana benthamiana. PLoS ONE 8, e75728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.B. , Pohronezny, K.L. , Stall, R.E. and Jones, J.P. (1986) Survival of Xanthomonas campestris pv. vesicatoria in Florida on tomato crop residue, weeds, seeds, and volunteer tomato plants. Phytopathology 76, 430–434. [Google Scholar]
- Jones, J.B. , Jones, J.P. , Stall, R.E. and Zitter, T.A. (1991) Compendium of tomato diseases. Am. Phytopathol. Soc. 84, 133. [Google Scholar]
- Katsir, L. , Schilmiller, A.L. , Staswick, P.E. , He, S.Y. and Howe, G.A. (2008) COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc. Natl Acad. Sci. 105, 7100–7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, T.‐H. , Böhmer, M. , Hu, H. , Nishimura, N. and Schroeder, J.I. (2010) Guard cell signal transduction network: advances in understanding abscisic acid, CO2, and Ca2+ Signaling. Annu. Rev. Plant Biol. 61, 561–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie‐Berry, N. , Joardar, V. , Street, I.H. and Kunkel, B.N. (2006) The Arabidopsis thaliana JASMONATE INSENSITIVE 1 gene Is required for suppression of salicylic acid‐dependent defenses during infection by Pseudomonas syringae . Mol. Plant‐Microbe Interact. 19, 789–800. [DOI] [PubMed] [Google Scholar]
- Leonhardt, N. , Marin, E. , Vavasseur, A. , and Forestier, C. (1997) Evidence for the existence of a sulfonylurea‐receptor‐like protein in plants: modulation of stomatal movements and guard cell potassium channels by sulfonylureas and potassium channel openers. Proc. Natl. Acad. Sci. U. S. A. 94, 14156‐14161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, C. , Baek, W. , Jung, J. , Kim, J.‐H. and Lee, S. (2015) Function of ABA in stomatal defense against biotic and drought stresses. Int. J. Mol. Sci. 16, 15251–15270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J. , Elmore, J.M. , Fuglsang, A.T. , Palmgren, M.G. , Staskawicz, B.J. and Coaker, G. (2009) RIN4 functions with plasma membrane H + ‐ATPases to regulate stomatal apertures during pathogen attack. PLoS Biol. 7, e1000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo, O. , Chico, J.M. , Sánchez‐Serrano, J.J. and Solano, R. (2004) JASMONATE‐INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate‐regulated defense responses in Arabidopsis. Plant Cell 16, 1938–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahfouz, M.M. , Piatek, A. and Stewart, C.N. (2014) Genome engineering via TALENs and CRISPR/Cas9 systems: challenges and perspectives. Plant Biotechnol. J. 12, 1006–1014. [DOI] [PubMed] [Google Scholar]
- Melotto, M. , Underwood, W. , Koczan, J. , Nomura, K. and He, S.Y. (2006) Plant stomata function in innate immunity against bacterial invasion. Cell 126, 969–980. [DOI] [PubMed] [Google Scholar]
- Melotto, M. , Underwood, W. and He, S.Y. (2008) Role of stomata in plant innate immunity and foliar bacterial diseases. Annu. Rev. Phytopathol. 46, 101–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melotto, M. , Zhang, L. , Oblessuc, P.R. and He, S.Y. (2017) Stomatal defense a decade later. Plant Physiol. 174, 561–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monte, I. , Hamberg, M. , Chini, A. , Gimenez‐Ibanez, S. , García‐Casado, G. , Porzel, A. , Pazos, F. et al. (2014) Rational design of a ligand‐based antagonist of jasmonate perception. Nat. Chem. Biol. 10, 671–676. [DOI] [PubMed] [Google Scholar]
- Nekrasov, V. , Wang, C. , Win, J. , Lanz, C. , Weigel, D. and Kamoun, S. (2017) Rapid generation of a transgene‐free powdery mildew resistant tomato by genome deletion. Sci. Rep. 7, 482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveros, J.C. , Franch, M. , Tabas‐Madrid, D. , San‐León, D. , Montoliu, L. , Cubas, P. and Pazos, F. (2016) Breaking‐Cas—interactive design of guide RNAs for CRISPR‐Cas experiments for ENSEMBL genomes. Nucleic Acids Res. 44, W267–W271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels, L. , Barbero, G.F. , Geerinck, J. , Tilleman, S. , Grunewald, W. , Pérez, A.C. , Chico, J.M. et al. (2010) NINJA connects the co‐repressor TOPLESS to jasmonate signalling. Nature 464, 788–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse, C.M.J. , Van der Does, D. , Zamioudis, C. , Leon‐Reyes, A. and Van Wees, S.C.M. (2012) Hormonal Modulation of Plant Immunity. Annu. Rev. Cell Dev. Biol. 28, 489–521. [DOI] [PubMed] [Google Scholar]
- Pilet‐Nayel, M.‐L. , Moury, B. , Caffier, V. , Montarry, J. , Kerlan, M.‐C. , Fournet, S. , Durel, C.‐E. et al. (2017) Quantitative resistance to plant pathogens in pyramiding strategies for durable crop protection. Front. Plant Sci. 8, 1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert‐Seilaniantz, A. , Grant, M. and Jones, J.D.G. (2011) Hormone crosstalk in plant disease and defense: more than just JASMONATE‐SALICYLATE antagonism. Annu. Rev. Phytopathol. 49, 317–343. [DOI] [PubMed] [Google Scholar]
- Schiml, S. and Puchta, H. (2016) Revolutionizing plant biology: multiple ways of genome engineering by CRISPR/Cas. Plant Methods 12, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, R.W. (1977) Bacterial speck of tomato: sources of inoculum and establishment of a resident population. Phytopathology 67, 388–394. [Google Scholar]
- Sheard, L.B. , Tan, X. , Mao, H. , Withers, J. , Ben‐Nissan, G. , Hinds, T.R. , Kobayashi, Y. et al. (2010) Jasmonate perception by inositol‐phosphate‐potentiated COI1–JAZ co‐receptor. Nature 468, 400–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somodi, G.C. , Jones, J.B. , Scott, J.W. , Wang, J.F. and Stall, R.E. (1996) Relationship between the hypersensitive reaction and field resistance to tomato race 1 of Xanthomonas campestris pv. vesicatoria. Plant Dis. 80, 1151–1154. [Google Scholar]
- Sun, J.‐Q. , Jiang, H.‐L. and Li, C.‐Y. (2011) Systemin/jasmonate‐mediated systemic defense signaling in tomato. Mol. Plant 4, 607–615. [DOI] [PubMed] [Google Scholar]
- Thines, B. , Katsir, L. , Melotto, M. , Niu, Y. , Mandaokar, A. , Liu, G. , Nomura, K. et al. (2007) JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signalling. Nature 448, 661–665. [DOI] [PubMed] [Google Scholar]
- Vishwakarma, K. , Upadhyay, N. , Kumar, N. , Yadav, G. , Singh, J. , Mishra, R.K. , Kumar, V. et al. (2017) Abscisic acid signaling and abiotic stress tolerance in plants: a review on current knowledge and future prospects. Front. Plant Sci. 8, 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann, J. , Brancato, C. , Berendzen, K.W. and Dreiseikelmann, B. (2016) Development of a tomato plant resistant to Clavibacter michiganensis using the endolysin gene of bacteriophage CMP1 as a transgene. Plant. Pathol. 65, 496–502. [Google Scholar]
- Xie, D.X. , Feys, B.F. , James, S. , Nieto‐Rostro, M. and Turner, J.G. (1998) COI1: an Arabidopsis gene required for jasmonate‐regulated defense and fertility. Science 280, 1091–1094. [DOI] [PubMed] [Google Scholar]
- Yunis, H. , Bashan, Y. , Okon, Y. , Henis, Y. and others. (1980) Weather dependence, yield losses and control of bacterial speck of tomato caused by Pseudomonas tomato. Plant Dis. 64, 937–939. [Google Scholar]
- Zeng, W. and He, S.Y. (2010) A prominent role of the flagellin receptor FLAGELLIN‐SENSING2 in mediating stomatal response to pseudomonas syringae pv tomato dc3000 in arabidopsis. Plant Physiol. 153, 1188–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, W. , He, S.Y. and Assmann, S.M. (2008) The plant innate immunity response in stomatal guard cells invokes G‐protein‐dependent ion channel regulation. Plant J. 56, 984–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, F. , Yao, J. , Ke, J. , Zhang, L. , Lam, V.Q. , Xin, X.F. , Zhou, X.E. , et al. (2015) Structural basis of JAZ repression of MYC transcription factors in jasmonate signalling. Nature 525, 269–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, X.Y. , Spivey, N.W. , Zeng, W. , Liu, P.P. , Fu, Z.Q. , Klessig, D.F. , He, S.Y. et al. (2012) Coronatine promotes pseudomonas syringae virulence in plants by activating a signaling cascade that inhibits salicylic acid accumulation. Cell Host Microbe 11, 587–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel, C. , Robatzek, S. , Navarro, L. , Oakeley, E.J. , Jones, J.D.G. , Felix, G. and Boller, T. (2004) Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428, 764–767. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Phylogenetic tree of all AtJAZs and SlJAZs. The schematic representation indicates length and domains of each JAZ protein.
Figure S2 False colour infrared image shows no differences between WT and Sljaz2Δjas plants. On the left, image of WT and Sljaz2Δjas tomato plants (Line 1 and Line 2), as depicted in the figure, taken with a reflex camera. On the right, the same plants analysed under an infrared camera to measure leaf temperature.
Figure S3 Sljaz2Δjas mutant is equally resistant to Pto DC3000 COR‐ when surface inoculated. Growth of Pto DC3000 COR‐ on WT and Sljaz2Δjas (Line 1) tomato plants 7 days after surface inoculation by dipping with bacteria at 108 cfu/mL. Error bars indicate SEM (n = 7).
Figure S4 Sljaz2Δjaz plants display a normal phenotype.
Table S1 RT‐PCR oligonucleotides used in this study.
Table S2 Oligonucleotides used as gRNAs in this study.


