Summary
The HUB2 gene encoding histone H2B monoubiquitination E3 ligase is involved in seed dormancy, flowering timing, defence response and salt stress regulation in Arabidopsis thaliana. In this study, we used the cauliflower mosaic virus (CaMV) 35S promoter to drive AtHUB2 overexpression in cotton and found that it can significantly improve the agricultural traits of transgenic cotton plants under drought stress conditions, including increasing the fruit branch number, boll number, and boll‐setting rate and decreasing the boll abscission rate. In addition, survival and soluble sugar, proline and leaf relative water contents were increased in transgenic cotton plants after drought stress treatment. In contrast, RNAi knockdown of GhHUB2 genes reduced the drought resistance of transgenic cotton plants. AtHUB2 overexpression increased the global H2B monoubiquitination (H2Bub1) level through a direct interaction with GhH2B1 and up‐regulated the expression of drought‐related genes in transgenic cotton plants. Furthermore, we found a significant increase in H3K4me3 at the DREB locus in transgenic cotton, although no change in H3K4me3 was identified at the global level. These results demonstrated that AtHUB2 overexpression changed H2Bub1 and H3K4me3 levels at the GhDREB chromatin locus, leading the GhDREB gene to respond quickly to drought stress to improve transgenic cotton drought resistance, but had no influence on transgenic cotton development under normal growth conditions. Our findings also provide a useful route for breeding drought‐resistant transgenic plants.
Keywords: Gossypium hirsutum Linn., HISTONE MONOUBIQUITINATION 2 (HUB2), drought, transgenic plants, histone monoubiquitination, histone methylation
Introduction
As the global temperature has increased by approximately 0.13 °C per decade since 1950 and rainfall has decreased, drought is becoming one of the most serious threats to agricultural productivity worldwide (Lobell et al., 2011). Ten percent of the arable land globally has been affected by desertification, and plants growing in such unsuitable environments are unable to reach their full genetic potential, exhibiting reduced growth and a more than 50% decrease in production (Bartels and Sunkar, 2005; Zhu, 2002). Cotton (Gossypium hirsutum) is an important commercial crop, as its fibre is used in many textiles and its seeds are rich in protein and produce edible cottonseed oil. Cotton is mainly distributed in hot and dry regions such as Xinjiang Province in China, South Africa, and Central Asia, which makes cotton more tolerant to drought stress than other crops; however, drought is still one of the most restrictive factors for its production and fibre quality (Soth, 1999). Understanding the basic molecular mechanisms of drought stress and using genetic engineering to develop more productive stress‐tolerant crops will allow us to better address this tremendous challenge.
Traditional breeding has made some progress in developing plants that can cope with abiotic stress, but this ability is often transferred along with other undesirable traits, and reproductive barriers between species make it difficult to obtain desired traits quickly and effectively (Varshney et al., 2011). Studies of the molecular mechanisms by which plants respond to abiotic stress have already identified a series of genes involved in stress tolerance; these genes encode various kinds of proteins, including transcription factors (TFs), enzymes, molecular chaperones and other functional proteins. These findings have provided a solid foundation for genetic breeding and have helped accelerate breeding speed (Umezawa et al., 2006). Individual or combined expression of the Arabidopsis H+‐pyrophosphatase gene AtAVP1 and the vacuolar Na+/H+ antiporter gene AtNHX1 in cotton reduces the cell water potential and promotes the development of the root system to increase drought and salt tolerance (Pasapula et al., 2011; Shen et al., 2015). The expression of some genes involved in abscisic acid (ABA) biosynthesis (LOS5/ABA3) or signal transduction (ABI3 and ABI5) can enhance the activity of ABA‐dependent pathways to increase transgenic cotton drought tolerance and productivity (Mittal et al., 2014; Yue et al., 2012). Other genes encoding functional proteins can also be transformed into cotton to improve drought tolerance and agronomic traits (Divya et al., 2010; Parkhi et al., 2009; Yu et al., 2016; Zhang et al., 2014). Although genetic breeding has made some progress through the expression of several candidate abiotic stress response genes in cotton, the identification of new drought resistance genes would enrich our germplasm resources and deepen our understanding of drought‐resistant breeding.
The N‐termini of histones can be covalently modified by methylation, acetylation, phosphorylation, ADP ribosylation, biotinylation and monoubiquitination. Unlike polyubiquitination, which marks proteins for proteasome‐mediated degradation, monoubiquitination alters the subcellular localization or the biochemical or molecular functions of target proteins (Deshaies and Joazeiro, 2009). Histone monoubiquitination, especially H2B monoubiquitination (H2Bub1), is always associated with histone H3K4 or H3K36 methylation and cooperatively regulates gene expression. In the Arabidopsis genome, there are two C3HC4 RING‐type E3 ligases, HISTONE MONOUBIQUITINATION1 (HUB1) and HUB2, which can form heterotetramer catalyse histone H2B monoubiquitination in cooperation with the E2 ubiquitin‐conjugating enzyme (UBC1 and UBC2), exerting multiple effects on plant growth and development (Cao et al., 2008; Xu et al., 2009). Arabidopsis hub1 and hub2 mutants exhibit a global lack of H2Bub1, and this deficiency is associated with dysregulation of key regulators of the cell cycle G2‐to‐M transition and changes in the expression of seed dormancy‐related genes (Fleury et al., 2007; Liu et al., 2007). Deficiencies in H2Bub1 modification can also affect histone H3K4me3 and H3K36me3 levels, which modulate the expression of the floral repressor FLOWER LOCUS C (FLC) to regulate flowering time (Cao et al., 2008; Gu et al., 2009). Recent studies have shown that H2Bub1 affects photomorphogenesis, cutin and wax composition and the circadian clock through regulating the expression of related genes (Bourbousse et al., 2012; Himanen et al., 2012; Menard et al., 2014). In addition, HUB1 and HUB2 facilitate plant biotic stress responses through directly monoubiquitinating histone H2B at the R gene locus (Zou et al., 2014) and other defence‐related gene loci (Dhawan et al., 2009; Hu et al., 2014). In addition, its homologs in rice (Oryza sativa) can participate in late anther development through direct functions related to tapetum degradation‐related genes (Cao et al., 2015). AtHUB2 can also be induced by salt or mannitol and participates in salt stress regulation (Zhou et al., 2017).
As a range of cellular processes are regulated by HUB2, we ectopically expressed AtHUB2 in the cotton cultivar CCRI24 and found that AtHUB2 transgenic cotton plants exhibited significantly elevated drought tolerance under both greenhouse and field conditions. Using immunoblotting and Chromosome ImmunoPrecipitation (ChIP), we found that AtHUB2 transgenic cotton has a higher global level of H2Bub1, which led to an increase in H3K4me3 at the GhDREB gene, quickly triggering GhDREB expression under drought stress conditions. This work is the first report that AtHUB2 participates in the drought stress response and that heterologous expression of AtHUB2 improves drought resistance in transgenic cotton through influencing histone modifications.
Results
AtHUB2 plays a positive role in Arabidopsis under drought stress conditions
AtHUB2 is a pleiotropic gene that participates in many aspects of plant growth, developmental regulation and defence or immune response modulation. However, studies on the potential role of this gene in abiotic stress regulation remain limited, with the exception of one report showing that H2B monoubiquitination is involved in salt stress regulation (Zhou et al., 2017). To test whether AtHUB2 functions under drought stress in Arabidopsis, we analysed seedlings of the Arabidopsis Col‐0 (WT) line, the hub2 mutant line hub2‐2 and a complemented hub2‐2 mutant line (HUB2/hub2‐2) to determine their tolerance to osmotic and drought stress. Seeds of hub2‐2, HUB2/hub2‐2 and WT plants were grown in media containing different concentrations of polyethylene glycol 8000 (PEG‐8000). As the PEG‐8000 concentration increased from −0.25 to −0.7 MPa, the germination rates of hub2‐2, HUB2/hub2‐2 and WT seeds all decreased, but the germination rates of WT and HUB2/hub2‐2 seeds were significantly higher than that of hub2‐2 seeds. In addition, in medium without PEG‐8000, hub2‐2, HUB2/hub2‐2 and WT seeds all germinated (Figure 1a,b). Six‐day‐old normally growing WT, HUB2/hub2‐2 and hub2‐2 seedlings were transferred to media containing different concentrations of PEG‐8000. After 6 days of treatment, root growth was measured with ImageJ (NIH, USA) software, and the root growth of hub2‐2 mutants was found to be significantly inhibited compared with that of WT and HUB2/hub2‐2 plants. When osmotic stress reached −0.7 MPa, WT and HUB2/hub2‐2 seedlings had more green leaves than hub2 mutants. Under normal growth conditions, there were no differences in root length between WT and HUB2/hub2‐2, but their roots were slightly longer than those of hub2‐2 plants (Figure 1c,d).
Figure 1.
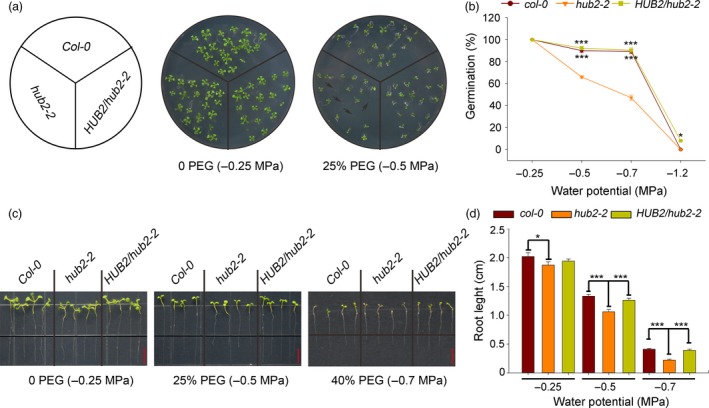
Response of Athub2 mutants to osmotic stress. (a) Seeds of the indicated genotypes were germinated on 1/2 MS medium or 1/2 MS previously infused with 25% PEG. Photographs were taken 7 days after germination. Black arrows indicate un‐germinated seeds. (b) Percentages of seeds that germinated in different concentrations of PEG. (c) Root growth of the indicated genotypes. Six‐day‐old seedlings were transferred from 1/2 MS to 1/2 MS medium previously infused with different concentrations of PEG. Photographs were taken 4 and 7 days after transfer, bar = 0.5 cm. (d) Quantification of relative root length in (c). Vertical bars represented standard deviation (SD). (n ≥ 30, *P < 0.05; ***P < 0.001).
Furthermore, soil‐grown hub2‐2 mutants were more sensitive to drought stress than WT and HUB2/hub2‐2 plants. Plants were allowed to germinate and grow normally for 14 days and were then subjected to drought treatment for 12 days and re‐watering for 3 days, and their survival rates were then recorded (Figure S1a). The survival rate of the WT plants was 91.94%, and that of the HUB2/hub2‐2 seedlings was 85.17%, while only 64.15% of the hub2‐2 mutants survived. The survival rates of the WT and HUB2/hub2‐2 plants were significantly higher than that of the hub2‐2 mutants (Figure S1b). We next examined the water loss rate of leaves by measuring the weight of the leaves at different drought time points. The hub2‐2 mutants lost water faster than the WT and HUB2/hub2‐2 plants, while there was no difference in the water loss rate between the WT and HUB2/hub2‐2 leaves (Figure S1c). These results demonstrated that AtHUB2 plays a positive role in the Arabidopsis drought stress response.
AtHUB2 overexpression improves the agricultural traits of transgenic cotton grown in a waterproof shed
Since AtHUB2 was found to be involved in drought stress responses, we generated transgenic cotton overexpressing AtHUB2 to test the effects on drought stress resistance. AtHUB2 has four alternative splicing isoforms in which key functional sites are conserved (Figure S2). To generate AtHUB2‐overexpressing transgenic cotton, the 1152 bp full‐length coding sequence (CDS) of one of AtHUB2 isoforms, At1g55250.2, with Myc tag added to its 5′ end was cloned into the pCAMBIA1305.1 vector under the control of the cauliflower mosaic virus (CaMV) 35S promoter. The pCAMBIA1305.1‐AtHUB2 construct was introduced into the cotton cultivar CCRI24 via Agrobacterium‐mediated transformation, and 12 independent transgenic lines (T0) were produced. We then used the T0 plants to raise T1 progeny via self‐fertilization, and the presence and integrity of the transgene were confirmed by PCR analysis of genomic DNA with primers specific for the CaMV 35S promoter and AtHUB2 (Figure S3a). Three independent homozygous T2 transgenic plants, line2, line4 and line6, were further identified by Southern blotting analysis with single‐ or two‐copy inserts (Figure S3b). Semi‐quantitative RT‐PCR and quantitative real‐time PCR (RT‐qPCR) showed that the three transgenic lines exhibited similar transcription levels (Figure S3c,d), and immunoblotting analyses with an anti‐Myc antibody showed that the AtHUB2 protein could be detected only in the transgenic lines (Figure S3e). These results showed that AtHUB2 was successfully transformed into the cotton cultivar CCRI24 and could be inherited in the offspring.
Phenotypic experiments were carried out at the Institute of Cotton Research of the Chinese Academy of Agricultural Science (CAAS), Anyang, Henan province. Three AtHUB2 transgenic lines (line2, line4 and line6), the control cultivar CCRI24 and the drought‐resistant control line ZR409 were grown on test land. Statistical analysis of agricultural characteristics indicated that under normal growth conditions, there were no obvious developmental differences between the transgenic and control plants, except that line6 had a lower plant height (Figure S4). However, when the plants were grown under drought conditions, as shown in Figure 2, the plant height, fruit branching and boll‐setting rate were significantly increased in all three transgenic lines. Moreover, compared with the control plants, line2 in particular exhibited remarkable increases and presented greener and less wilted leaves (Table S1). In addition, with the exception of line6, the abscission rate was significantly reduced in the transgenic lines compared with the control plants. These results suggest that AtHUB2 overexpression did not affect the agricultural traits of transgenic cotton under normal growth conditions but could improve these traits under drought stress conditions.
Figure 2.
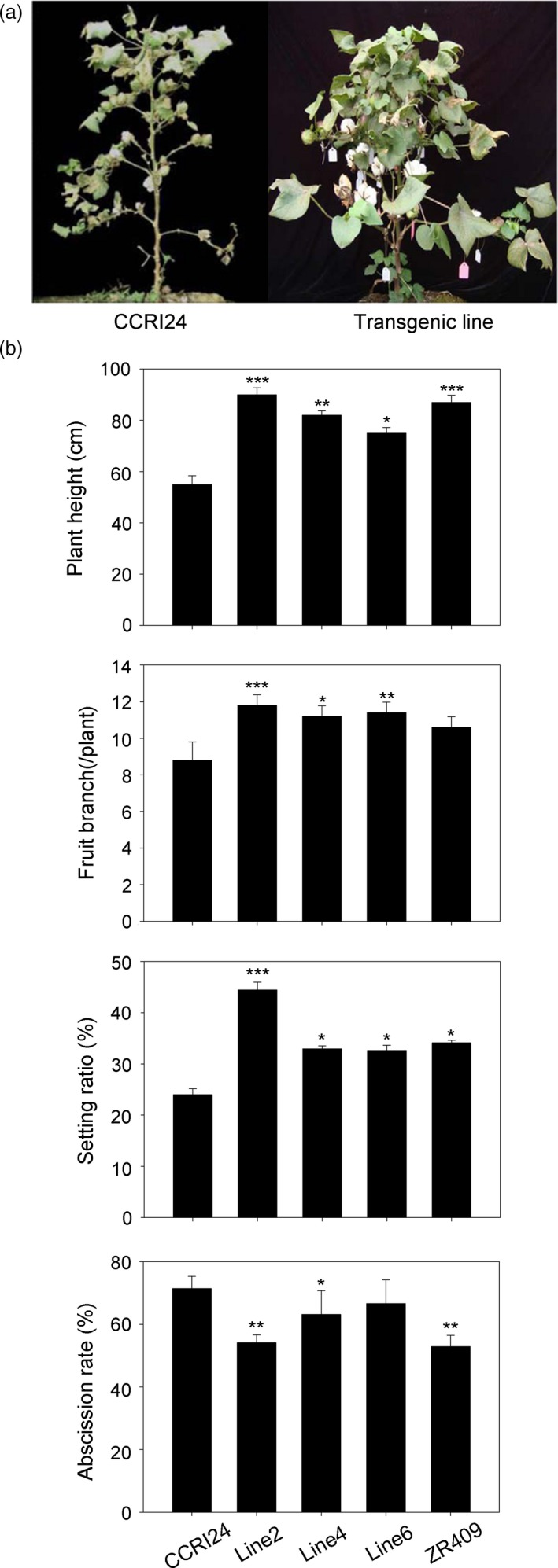
Agronomic traits of different lines grown in a waterproof shed. (a) Drought tolerance assay in the field under a waterproof shed in Anyang, Henan province, China. The photographs were taken 1 month after re‐watering. (b) Plant height, fruit branching, boll‐setting rates and abscission rates of controls (transgenic acceptor CCRI24 and drought‐resistant ZR409) and AtHUB2‐expressing cotton lines (line2, line4 and line6) in the field under drought stress conditions (n ≥ 30). Vertical bars represented SD. (*P < 0.05; **P < 0.01; ***P < 0.001).
AtHUB2 overexpression in cotton enhances drought tolerance and regulates drought stress‐related gene expression in the greenhouse
To further confirm that AtHUB2 overexpression can improve the drought resistance of transgenic cotton, we performed a drought tolerance assay in the greenhouse. The three transgenic lines described above as well as the controls were grown under controlled irrigation conditions and then subjected to drought stress 4 weeks after germination. As shown in Figure 3a, no obvious difference was observed between the transgenic and control plants under normal growth conditions, but after 20 and 28 days of drought stress, the control leaves were much more wilted than those of the transgenic lines. After 35 days of drought treatment, the leaves of CCRI24 plants were essentially withered, while those of the transgenic plants were still green and exhibited only slight wilting. After 1 week of recovery, most of the transgenic plants had recovered, showing survival rates from 80% to 97.78%, while the CCRI24 cultivar had a survival rate of only 47.74% (Figures 3c and S5).
Figure 3.
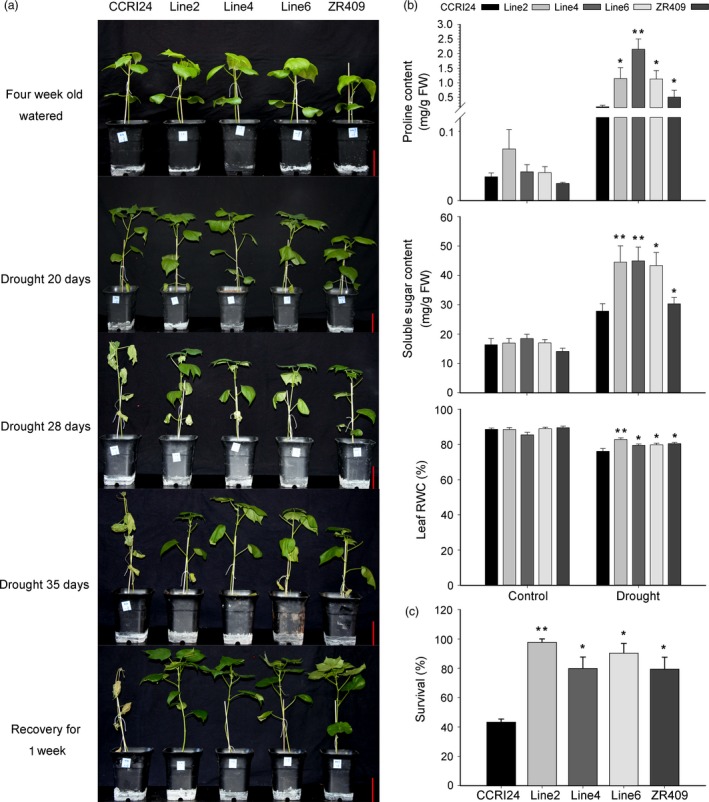
AtHUB2 significantly enhances the drought tolerance of transgenic cotton in the greenhouse. (a) Drought tolerance of control plants and AtHUB2‐expressing cotton plants. Photographs were taken at 4 weeks after germination, after 20, 28 and 35 days of drought stress, and after a 1‐week recovery period. Bar = 7.5 cm. (b) Leaf relative water content (RWC), proline content and soluble sugar content of transgenic cotton and control plants with or without drought treatment (n ≥ 30). (c) Survival rates of plants after 1 week of recovery (n ≥ 30). Vertical bars represent SD. (*P < 0.05; **P < 0.01).
We then measured and compared soluble sugar content, proline content and leaf relative water content (RWC) between the controls and transgenic cotton plants with or without drought treatment (Figure 3b). Without drought treatment, the soluble sugar and proline contents were low, and no differences were observed among CCRI24 and ZR409. During drought treatment, the proline and soluble sugar contents increased significantly in both transgenic and control plants, but the increases in transgenic plants were more significant than those in control plants. In addition, the leaf RWC decreased after drought treatment in both control and transgenic plants but was still significantly higher in transgenic lines than in control plants.
We next detected the expression of drought stress‐related genes. Without drought treatment, the expression levels of the ABA pathway‐dependent gene responsive to dehydration (GhRD22), the ABA pathway‐independent gene dehydration‐responsive element‐binding protein (GhDREB), the proline synthesis‐related gene δ1‐pyrroline‐5‐carboxylate synthetase (GhP5CS), the homologue of the rice negative drought stress regulation factor plasma membrane intrinsic protein 2;7 (OsPIP2;7) in cotton GhPIP2;7‐like (Ning et al., 2011) and mitogen‐activated protein kinase kinase 2 (GhMKK2) were not significantly different between the transgenic plants and control plants. After air‐drying for 4 h, the expression levels of GhRD22, GhDREB, GhP5CS and GhMKK2 were all up‐regulated in both the transgenic cotton and control plants, but the expression levels were significantly increased in the transgenic lines compared with the control line. In contrast, the expression of GhPIP2; 7‐like was significantly decreased in the transgenic lines compared with that in the control cotton (Figure 4a). Taken together, these results indicated that AtHUB2 overexpression can significantly improve the drought tolerance of transgenic cotton and influence drought‐related gene expression.
Figure 4.
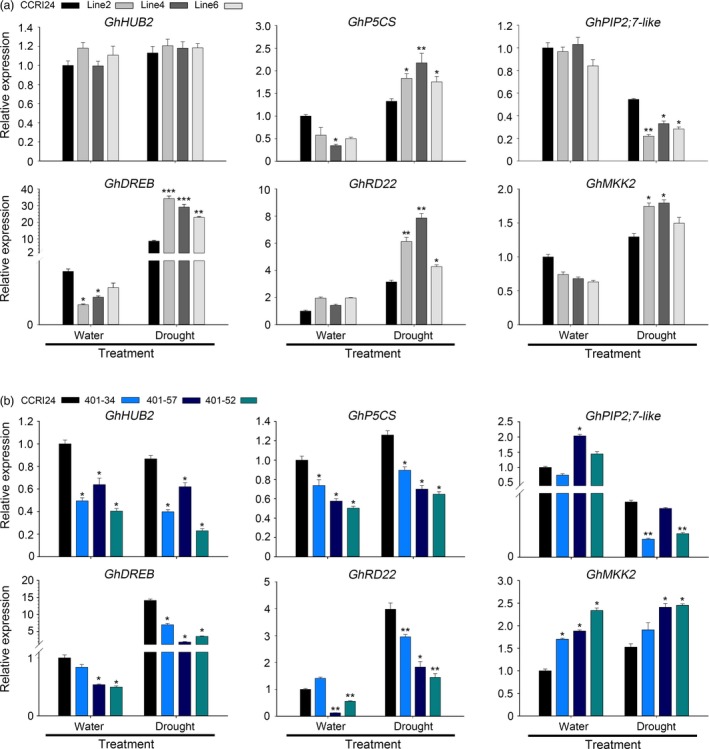
RT‐qPCR analyses of the expression of stress‐responsive genes in control, AtHUB2 overexpression lines (a) and GhHUB2‐knockdown lines (b). The RNA extracted from the leaves of the indicated genotypes (4 weeks old) before or after air‐drying for 4 h. Values determined via RT‐qPCR were normalized to the expression of GhUBI1. Vertical bars represent the SD. (*P < 0.05; **P < 0.01, ***P < 0.001).
GhHUB2‐specific RNAi reduces drought tolerance in transgenic cotton
To verify that the activity of AtHUB2 is the cause of the drought effect observed in transgenic cotton, we used GhHUB2‐specific RNA interference (RNAi) transgenic plants to perform drought tolerance assays. GhHUB2 RNAi transgenic cotton plants were generated previously in our lab. Two candidate AtHUB2 homologs, GhHUB2‐A and GhHUB2‐B, were identified in G. hirsutum through multiple sequence alignment against the TM‐1 genome using the AtHUB2 protein sequence. The multiple sequence alignment results showed that these two GhHUB2s share 98.6% nucleotide identity and 98.4% amino acid sequence identity and that both contain a conserved C3HC4 RING finger domain in the C‐terminal region. Therefore, the 272 bp region from the 3′ end, which included the RING domain shared by both GhHUB2s, was chosen for RNAi (Feng et al., 2018). Three T4 generation plants with significantly down‐regulated expression of GhHUB2 were used to perform drought tolerance assays (401‐34, 401‐57 and 401‐52). Under normal growth conditions, there were no developmental differences between WT and GhHUB2‐knockdown plants. However, after drought treatment, the GhHUB2‐knockdown plants exhibited more wilting than the WT plants (Figure 5a). While 50% of WT plants survived, only 11.11%–33.33% of GhHUB2‐knockdown plants survived after 25 days drought treatment and 3 days of recovery (Figure 5b). Moreover, the water loss rates of leaves detached from GhHUB2‐knockdown plants were higher than those of leaves from WT plants (Figure 5c). To confirm that the reduced drought resistance was due to down‐regulation of GhHUB2 expression, RT‐qPCR was performed to evaluate the transcription level of the GhHUB2 gene using template RNAs derived from the leaves of cotton plants exhibiting normal growth or exposed to air drought for 4 h. The results showed that a reduction in the expression of GhHUB2 to 40.35%–62.01% of WT expression was evident. The expression of GhHUB2 was not affected by drought in WT, GhHUB2‐knockdown plants or even AtHUB2‐overexpressing plants (Figure 4). Then, we examined the expression of drought‐related genes in GhHUB2‐knockdown and WT plants. As shown in Figure 4b, the expression levels of GhDREB, GhP5CS and GhRD22 were decreased compared with those in WT plants under normal growth conditions and were then up‐regulated by drought stress, whereas the expression levels of these genes were still significantly lower in GhHUB2‐knockdown plants than in WT plants. The expression levels of GhPIP2; 7‐like, which are negative regulatory factors of drought, were significantly down‐regulated in both GhHUB2‐knockdown plants and WT plants. The expression levels of GhMKK2 were higher in GhHUB2‐knockdown plants than in WT plants, whether with or without drought treatment. The RT‐qPCR results showed that the expression of GhDREB, GhRD22 and GhP5CS, but not GhPIP2; 7‐like and GhMKK2 was related to AtHUB2 overexpression. These results indicated that GhHUB2 was involved in the cotton drought stress response; however, the expression of GhHUB2 was not induced by drought stress, and overexpression of AtHUB2 was the reason for the improved drought resistance of transgenic cotton.
Figure 5.
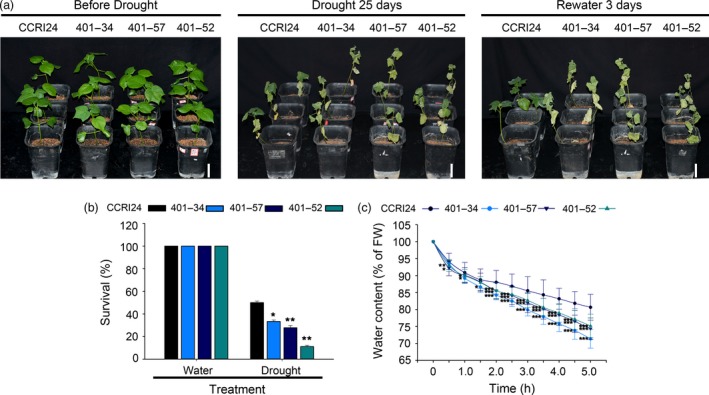
Response of GhHUB2‐knockdown lines to drought stress. (a) Drought tolerance of control CCRI24 and GhHUB2‐knockdown cotton plants. Photographs were taken at 4 weeks after germination, after 25 days of drought stress and re‐watering for 3 days. Bar = 7.5 cm. (b) Survival rates of plants after 25 days of drought stress and 3 days of recovery (n ≥ 30). (c) The volume of water lost from detached leaves of the indicated genotypes (4 weeks old) via air‐drying was measured as the percentage of change in the fresh weight (FW) of the leaves (n ≥ 30). Vertical bars represent SD. (*P < 0.05; **P < 0.01; ***P < 0.001).
AtHUB2 is a functional E3 ligase that increases transgenic cotton histone H2B monoubiquitination through interacting with GhH2B1
To understand how AtHUB2 overexpression improves transgenic cotton drought tolerance, we performed a yeast two‐hybrid assay to screen a cotton whole‐growth‐period cDNA library using AtHUB2‐pDEST32 (At1g55250.2) as bait, and a series of candidate genes were identified (Table S2). The cotton histone GhH2B1, which was previously identified as candidate GhHUB2 interact protein (Feng et al., 2018), was included in the group of 17 positive clones. Multiple sequence alignment and phylogenetic tree analysis showed that the GhH2B1 C‐terminus and key functional amino acid sites are conserved (Figure S6) (Hwang et al., 2003). And GhH2B1 is most closely related to AtHTB9 (AT3G45980), which was previously reported as an AtHUB2‐interacting protein (Cao et al., 2008). We then co‐transformed BD‐AtHUB2 (BD, pGBKT7, Clontech) with AD‐GhH2B1 (AD, pGADT7, Clontech) in AH109 yeast to confirm the interaction between AtHUB2 and GhH2B1 (Figure 6a). The results of in vitro pull‐down and split‐luciferase assays led to the same conclusion (Figure 6b,c).
Figure 6.
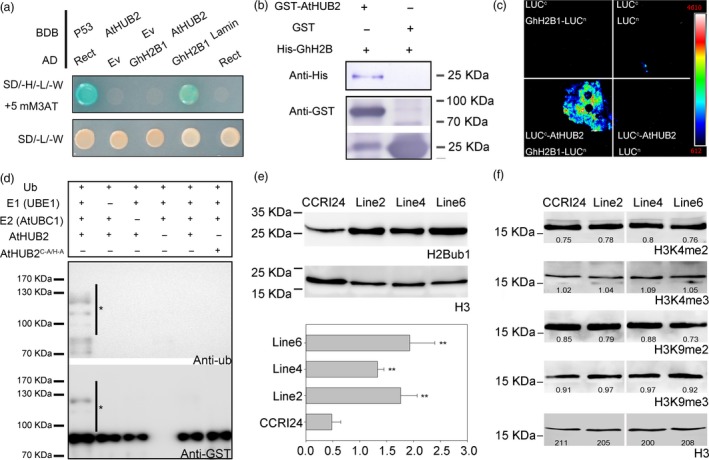
AtHUB2 is a functional E3 ligase. (a) Yeast two‐hybrid (Y2H) assays. GhH2B1‐pGADT7 was transformed into yeast cells with either the pGBKT7 vector alone (BDB) or AtHUB2‐pGBKT7, and growth was monitored on selective medium plates. (b) Pull‐down assay. GST‐HUB2 and GST alone were incubated with GhH2B1‐His. And detected with an anti‐GST antibody and anti‐His antibody. (c) Split‐luciferase assay. GhH2B1‐LUC n and LUC c‐AtHUB2 co‐injection tobacco. The combinations of LUC n and LUC c, LUC n and LUC c‐ AtHUB2, and GhH2B1‐LUC n and LUC c were used as negative controls. (d) AtHUB2 possesses E3 ubiquitin ligase activity in vitro. GST‐AtHUB2 and the point mutant GST‐HUB2 C346A/H348 were tested for E3 ubiquitin ligase activity. Anti‐Ub and anti‐GST antibodies were used to detect ubiquitinated proteins and GST‐AtHUB2 variants respectively. The asterisk indicates the polyubiquitinated proteins. (e) Global histone modification in the control (CCRI24) and transgenic lines. H2B monoubiquitination was detected with an anti‐H2Bub1 antibody, and H3 was used as a loading control. ImageJ software was used to analyse the greyscale value of each band. Vertical bars represent SD (**P < 0.01). (f) Detection of H3K4 methylation on a global scale in the control (CCRI24) and transgenic lines. And an anti‐H3K4me2 and anti‐H3K4me3 antibodies were used to detect target band, respectively. H3K9me2 and H3K9me3 were used as unchanged controls, and H3 was used as a loading control. The number below the lane indicates the greyscale value of each band.
AtHUB2 has a conserved C3HC4 RING‐type domain that controls histone H2B monoubiquitination. We tested the E3 ligase activity of AtHUB2 in vitro. A purified GST‐AtHUB2 fusion protein was mixed with ubiquitin (ub), rabbit E1 UBE1, and Arabidopsis E2 UBC1, and an AtHUB2 point mutant (C346A/H348A) was used as a negative control (Figure S2). Immunoblot analysis with anti‐ub and anti‐GST antibodies showed that ubiquitinated proteins were detectable only in the presence of all these components, while no ubiquitination was observed in the presence of HUB2C346A/H348A (Figure 6d). These results suggest that AtHUB2 could be autoubiquitinated in the presence of E1 and E2 enzymes and that the C3HC4 RING domain is essential for the protein ubiquitination ability of AtHUB2. We then examined the H2Bub1 modification pattern in the control cultivar CCRI24 and transgenic plants. An anti‐H2Bub1 antibody detected one clear band in all plants, but the level of H2Bub1 was higher in transgenic plants than in control plants. ImageJ software was used to analyse the greyscale value of each band and further confirmed that the H2Bub1 level was significantly improved in transgenic lines (Figure 6e). These findings indicated that AtHUB2 possesses E3 ubiquitin ligase activity and works directly on cotton histone H2B in AtHUB2‐overexpressing cotton.
The increase in H2Bub1 improves H3K4me3 modification at the GhDREB chromatin locus in AtHUB2 transgenic cotton
Previous studies have shown that H2Bub1 is associated with histone H3K4 methylation (Lee et al., 2007). To further explain how the global H2Bub1 level influences drought tolerance in transgenic cotton, we performed an immunoblotting assay to analyse H3K4 methylation levels. Anti‐H3K9me2 and anti‐H3K9me3 antibodies were used as negative controls. As shown in Figure 6f, there were no significant differences in the global H3K4 and H3K9 methylation levels between control and transgenic plants. This finding indicates that changes in the global H2Bub1 level had no effect on the global H3K4 methylation level.
The up‐regulated H2Bub1 had no influence on the global H3K4 methylation level, but the expression of drought stress‐related genes was significantly changed in transgenic cotton. Therefore, we examined whether histone trimethylation (H3K4me3) was altered at these drought‐related genes. A ChIP assay was performed with an anti‐H2Bub1 antibody and an anti‐H3K4me3 antibody against chromatin derived from four‐week‐old WT and AtHUB2‐overexpressing lines (line2 and line4) respectively. Then, we used RT‐qPCR to detect the enrichment of H2Bub1 and H3K4me3 in the chromatin of these drought‐related genes. The results indicated that H2Bub1 and H3K4me3 enrichment was only obviously detectable at the GhDREB chromatin locus, while no or only weak enrichment was detected at the other genes (date not shown). Moreover, a significant increase in the level of H2Bub1 was observed across the GhDREB P1‐P4 region in transgenic cotton (Figure 7b). In addition, the H3K4me3 level was increased at the promoter (P2) and transcription start site (TSS) (P3) and in the gene body (P4) of GhDREB (Figure 7c), whereas no difference was observed in the UBI1 chromatin region (Figure 7d). To determine the effect of H2Bub1 and H3K4me3 enrichment at the GhDREB chromatin locus, we detected the spatio‐temporal expression of GhDREB. As shown in Figure 7e, with a longer air‐drying time, the expression level of GhDREB was gradually increased in both WT and AtHUB2‐overexpressing plants, but the expression levels were higher in the latter. Additionally, the expression levels reached a maximum at 4 h in AtHUB2‐overexpressing plants compared to 5 h in WT. These results indicated that the H2Bub1 was associated with H3K4me3 at the GhDREB locus, and the higher H3K4me3 modification would trigger a quick response of GhDREB to drought stress in AtHUB2 transgenic cotton.
Figure 7.
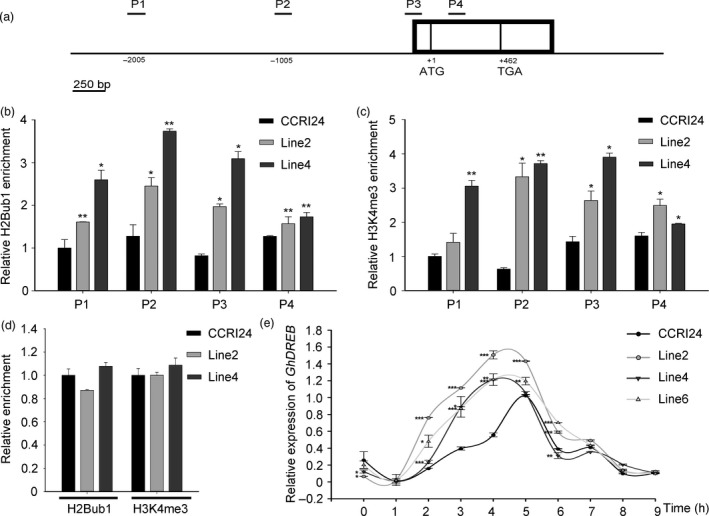
Detection of global histone H2Bub1 and H3K4me3 levels at the GhDREB chromatin locus. (a) Schematic diagram of the GhDREB gene. The exons are shown in boxes, and the indicated PCR fragments analysed in the ChIP assay are indicated with short black lines. P1: 2000 bp upstream of the ATG; P2: promoter region; P3: transcription start site (TSS) region; P4: gene body region. (b) H2Bub1 at the GhDREB locus in the CCRI24 and transgenic lines. Immunoprecipitated DNA was analysed via RT‐qPCR, and enrichment was determined as a percentage of the input. (c) H3K4me3 at the GhDREB locus in the CCRI24 and transgenic lines. (d) The GhUBI gene was selected as a housekeeping gene that was not differentially expressed in the transgenic lines. (e) Expression of GhDREB at different time points of air drought treatment in control and AtHUB2‐overexpressing lines. Real‐time RT‐qPCR quantification was normalized to the expression of GhUBI. Vertical bars represent SD. (*P < 0.05, **P < 0.01, ***P < 0.001).
Discussion
AtHUB1/2 are major enzymes responsible for histone H2B monoubiquitination in Arabidopsis, which affects many aspects of plant growth and development, including cellular proliferation, seed dormancy, and immune and salt stress responses. This study is the first report showing that AtHUB2 plays a role in drought stress and obtaining AtHUB2‐overexpressing drought‐resistant cotton plants with stable inheritance, as demonstrated by increased plant height, fruit branch numbers and boll numbers under drought conditions.
In plants, H2Bub1 is a marker of active chromatin and is broadly involved in the regulation of gene transcription. Our results showed that the TFs GhRD22 and GhDREB and the proline synthesis‐related gene GhP5CS were significantly up‐regulated in transgenic cotton, while these genes were lower in GhHUB2‐knockdown plants that encounter drought stress compared to the CCRI24 cultivar (Figure 4). The immunoblotting results showed that AtHUB2 overexpression increased global H2Bub1 levels, but we detected H2Bub1 enrichment only at the GhDREB gene locus. H2Bub1 enrichment at the other genes was almost undetectable. The drought stress response is a complex process involving multiple signalling and regulatory pathways. H2Bub1 may function at upstream genes or through an indirect pathway to influence the expression of these genes, and the rapid response of GhDREB after drought stress may provide a signal for other pathways to regulate the expression of the other genes.
The DREB belongs to a large group of plant‐specific TFs known as the APETALA 2/ethylene‐responsive element‐binding factor (AP2/ERF) family. In Arabidopsis, there are eight DREB2s, and DREB2A and DREB2B are highly induced by drought stress (Mizoi et al., 2012). However, their functions under drought stress remain unclear. There is a negative regulatory domain (NRD) in DREB2 that keeps the protein inactive under normal conditions. The activity of DREB2 can be modulated by posttranslational regulatory mechanisms, including ubiquitination, phosphorylation and alternative splicing (Agarwal et al., 2007; Matsukura et al., 2010; Qin et al., 2008). In Arabidopsis, the DREB2A‐interacting protein DBP3‐1 was found to form a transcriptional complex that contributes to the target gene selectivity of DREB2 under heat stress (Sato et al., 2014). In addition to posttranslational regulation, epigenetic modifications also participate in the transcriptional regulation of DREB family genes. Under osmotic stress, the levels of H3K9 and H4K5 acetylation in the ZmDREB2A promoter region increase quickly to activate ZmDREB2A expression (Zhao et al., 2014). Differential acetylation of histone H3 at the promoter can facilitate the activation of OsDREB1b transcription (Roy et al., 2014). According to ChIP‐Seq data published in 2010, 90% of annotated Arabidopsis genes carry H3K4 methylation marks; and H3K4me3 was broadly distribute on the nucleosomes of stress‐induced genes, including DREB2 (van Dijk et al., 2010). Our results further indicate that the epigenetic modifications participate in DREB gene transcriptional control in cotton. The GhDREB chromatin locus was enriched with H2Bub1 and H3K4me3 modifications. Increased H2Bub1 could elevate the H3K4me3 modification at the GhDREB chromatin locus in AtHUB2 transgenic cotton. And decreased H2Bub1 would reduce the H3K4me3 modification at the GhDREB chromatin locus in GhHUB2‐konckdown transgenic cotton (Figure S7). H3K4me3 always linked to the transcriptional activation of related genes. As show in Figures 7 and S7, elevated H3K4me3 promoted GhDREB expression, and when the H3K4me3 was impaired, the expression of GhDREB was lower than control. Hyper‐methylation of H3K4 is a mark of ‘stress memory’, as previous exposure to abiotic stress can make plants more resistant to future exposure, which is accompanied by high levels of H3K4me3 on nucleosomes (Ding et al., 2012; Liu et al., 2014). Our results demonstrated that the levels of H3K4me3 at the GhDREB chromatin locus were higher in AtHUB2 transgenic cotton. When the transgenic cotton encountered drought stress, the expression level of GhDREB was quickly up‐regulated because of ‘stress memory’ (Figure 7e).
The expression of a series of drought‐related genes is under the control of DREB, which makes it a useful tool for breeding stress‐resistant transgenic plants. However, the direct overexpression of DREBs in transgenic Arabidopsis leads to severe growth retardation. However, in our study, the overexpression of AtHUB2 in transgenic cotton changed the epigenetic modifications at the GhDREB chromatin locus, allowing a lower expression level under normal growth conditions without any other effects on plant development. When the transgenic cotton encounters drought stress, the increased H3K4me3 modification at the GhDREB chromatin locus allows it to quickly respond to drought stress to improve transgenic plant drought resistance. Our results provide useful tools for breeding drought‐resistant transgenic plants.
Experimental procedures
Plant materials
The full‐length AtHUB2 (NCBI accession number: BT029207) CDS was cloned into the pCambia1305.1 vector with a Myc tag added to its 5′ end, and the expression of the Myc‐AtHUB2 fusion protein was driven by the cauliflower mosaic virus (CaMV) 35S promoter. The AtHUB2 overexpression vector was transformed into the cotton cultivar CCRI24 via Agrobacterium‐mediated transformation, and 12 lines of T0 transgenic plants were obtained. The T0 plants were used to generate T1 progeny via self‐fertilization, and the presence and integrity of the transgene were confirmed by PCR analysis of genomic DNA with specific primers (Table S3). The AtHUB2 protein was detected with anti‐Myc antibody (Sigma).
Seeds of the wild‐type (WT) Arabidopsis line Columbia‐0 (Col‐0), the hub2‐2 (Salk_071289) mutant line and the hub2‐2 mutant line complemented with HUB2 (HUB2/hub2‐2) were used in the experiments (Col‐0 background). The T‐DNA insertion mutant of hub2‐2 (Salk_071289) was obtained from the ABRC (Ohio State University, Columbus, OH). The complemented line HUB2/hub2‐2 was produced by transforming hub2‐2 mutant plants with the pCAMBIA1200 vector expressing HUB2 genomic DNA (including both introns and exons) driven by the CaMV 35S promoter.
Plant growth conditions and drought treatment
Cotton seeds were placed on wet germination paper, incubated at 30 °C for 2 days for germination and then planted in 12*12*15 cm3 pots containing 1 L of soil mixture. These plants (≥30 for each line) were grown in a greenhouse at 28 ± 2 °C with a 16/8 h photoperiod for 28 days and then divided into two groups: a well‐watered group, as a control, and a drought treatment group. For drought treatment, water was withheld for 35 days, and photographs were taken on days 20, 28 and 35. The plants were then re‐watered for 1 week for recovery.
Field drought treatments were carried out in Anyang, Henan province, China. The control plants (cultivar CCRI24) and the homozygous transgenic cotton lines were divided into two groups. One group was well‐watered as a control, while the second group was grown in a waterproof shed. The waterproof shed was irrigated with 30 tons/acre water 1 week before sowing. When the soil water content was 16%–20%, the seeds were sown. The soil water content was then detected every 30 days, and re‐watering was performed after 3 months of drought treatment (35 tons/acre). The photographs were taken 1 month after re‐watering.
Seeds of Arabidopsis WT (Col‐0) plants, the hub2‐2 (Salk_071289) mutants and hub2 mutant complemented with HUB2 (HUB2/hub2‐2) were sterilized and incubated for 3 days at 4 °C in the dark. For seedling assays, sterile seeds were germinated on half‐strength MS medium for 3 days and then transferred to half‐strength MS agar medium infused with different concentrations of polyethylene glycol 8000 (PEG‐8000) (Verslues and Bray, 2004). The plates were incubated at 22 °C with a 16/8 h photoperiod. For soil dehydration assays, plants were germinated on half‐strength MS medium, grown in soil at 22 °C with a 16 h light photoperiod for 14 days and then grown under the same conditions with or without additional water for 12 days. Photographs were taken before drought treatment and on day 12 after drought treatment and re‐watered for 3 days.
Southern blotting
Genomic DNA was extracted from 4‐week‐old CCRI24 and AtHUB2 transgenic cotton plants according to Paterson's method (Paterson et al., 1993). Fifty micrograms of genomic DNA was digested with the BamHI restriction enzyme (NEB), separated on a 0.8% agarose gel, and transferred onto a Hybond‐N+ nylon membrane (Amersham) by capillary blotting. Hybridization was performed according to the DIG High Prime DNA Labeling and Detection Starter Kit II (Roche).
Quantitative real‐time PCR analysis
Total RNA was extracted from the leaves of cotton plants grown under normal conditions or plants subjected to drought treatment using an RNAprep Pure total RNA extraction kit for polysaccharide‐ and polyphenol‐rich plants (TIANGEN), and first‐strand cDNA synthesis was performed using 2.0 μg of total RNA, an Oligo‐dT (18) primer and M‐MLV reverse transcriptase (Promega). Quantitative real‐time RT‐PCR (RT‐qPCR) analysis was performed in a CFX‐96 real‐time system (Bio‐Rad) using SYBR Premix Ex Taq (TaKaRa). The expression levels of the genes mentioned above were examined using specific primers. GhUBI1 was used as an internal control (Wang et al., 2013). All the primers used in this study are shown in Table S3.
Measurement of proline, soluble sugars and relative water content (RWC)
Leaves from normal control or stress‐treated plants at similar developmental stages were used to measure proline content, soluble sugar content and RWC. Proline was measured according to the method of Bates (Bates et al., 1973). Soluble sugars were measured using the anthrone reagent (Dubois et al., 1956). The RWC of leaves was assayed as described by Parida (Parida et al., 2007).
Yeast two‐hybrid assays
A cotton whole‐growth‐period two‐hybrid library was constructed by Invitrogen (Shanghai, China). RNA was extracted from cotton at four stages of development. The pDEST22 vector system was then constructed. The bait gene was inserted into the pDEST32 vector via the Gateway® recombination reaction, and the two‐hybrid library was screened according to the protocol for the ProQuestTM two‐hybrid system provided by Invitrogen.
To test specific two‐hybrid interactions, AtHUB2 was inserted into the pGBKT7 vector as bait, and the interaction partners were inserted into the pGADT7 vector as prey. The bait and prey constructs were co‐transformed into yeast AH109 cells. Positive clones were identified by the ability to grow on SD medium lacking histidine/leucine/tryptophan (SD/‐His/‐Leu/‐Try) and containing 5 mM 3‐aminotriazole (3AT).
Split‐luciferase and pull‐down assays
AtHUB2 and GhH2B1 were fused to the C‐ and N‐termini of the firefly LUC enzyme to produce LUCc‐AtHUB2 and GhH2B1‐LUCn respectively. LUCc and LUCn were used as negative controls. LUCc‐AtHUB2, GhH2B1‐LUCn, LUCc and LUCn were transiently expressed in Nicotiana benthamiana. Leaves were collected 3 days after infiltration and infiltrated with 1 mm luciferin before being observed. Fluorescence was detected using a Tanon 5200 chemiluminescence imaging system (Tanon).
Recombinant (glutathione S‐transferase) GST‐AtHUB2 and GhH2B‐His (histidine) containing plasmids were transformed into Escherichia coli BL21 (DE3) cells, and expression was induced with 0.5 mm or 1 mm isopropyl β‐D‐1‐thiogalactopyranoside (IPTG). Recombinant fusion proteins were affinity purified from bacterial lysates according to the GE protocol (www.gelifesciences.com/protein-purification). Purified GST‐AtHUB2 and GhH2B‐His were used for in vitro binding experiments according to a previously described protocol (Xie et al., 1999).
In vitro ubiquitination and immunoblotting assays
Rabbit E1 (Boston Biochem, 200 ng), purified Arabidopsis E2 AtUBC1‐His (500 ng), purified GST‐AtHUB2 (1 mg) and recombinant plant ubiquitin (Boston Biochem, 80 μg) were used for the in vitro ubiquitination assay. An AtHUB2 protein carrying two point mutations (C345A & H347A) was used as the negative control for fully functional AtHUB2. Reactions were performed at 37 °C for 2 h and then stopped by the addition of an equal volume of loading buffer (40 mm Tris‐HCl, pH 6.8, 2% SDS, 10% glycerol, 0.1% Coomassie Brilliant Blue, 0.1% bromophenol blue and 1% β‐mercaptoethanol) and boiling for 10 min.
For total plant protein extraction, leaves were ground in liquid nitrogen and dissolved in denaturing buffer containing 50 mm Tris‐HCl, pH 7.5, 150 mm NaCl, 0.1% NP‐40, 4 m carbamide and 1 mm PMSF. Protein concentrations were determined using a Coomassie protein assay kit (Bradford). Each total protein sample was analysed by 10% SDS‐PAGE and immunoblotting using relevant antibodies. The specific antibodies employed in these assays were as follows: anti‐H3 (Cell Signaling Technology); anti‐H3K4me2 (Millipore); anti‐H3K4me3 (Millipore); anti‐H3K9me2 (Millipore); anti‐H3K9me3 (Millipore); anti‐ub (abcam); and anti‐GST (Cell Signaling Technology).
ChIP assay
ChIP experiments were performed as previously described (Saleh et al., 2008) with some modifications. A 3 g sample of leaves was fixed with cross‐linking buffer and 1% formaldehyde using vacuum infiltration two times for 15 min each, and the cross‐linking reaction was quenched with 0.125 m glycine. The leaves were ground in a mortar and pestle in liquid nitrogen, resuspended in nuclei isolation buffer and then filtered through Miracloth. After centrifugation, the pellets (nuclei) were resuspended in cold nuclei lysis buffer and sonicated (Bioruptor® Plus sonication device, Diagenode, Belgium). After centrifugation, the supernatant was pre‐cleared with Magna ChIP™ Protein A Magnetic Beads (EMD Millipore corporation, Temecula, CA), and specific antibodies were added and incubated overnight at 4 °C. The specific antibodies used were as follows: anti‐H2Bub1 (Cell Signaling Technology, Danvers, MA) and anti‐trimethyl‐H3K4 (Abcam). The enriched DNA fragments were detected by RT‐qPCR and compared with the input samples, the amount of immunoprecipitated chromatin as normalized to the total amount of chromatin used in GhDREB P1 region in wild‐type plants was given as 1. GhUBI1 was used as a negative control.
Phylogenetic analysis
The phylogenetic tree was constructed from the indicated amino acid sequences using the MEGA5 programme based on the neighbor‐joining method with 1000 bootstraps. The numbers beside the branches represent bootstrap values based on 1000 replicates. Sequences were obtained from the National Center for Biotechnology Information (NCBI) and The Arabidopsis Information Resource (TAIR) databases. The multiple sequence alignment of the indicated sequences was analysed with CLUSTAL W2 or CLC Sequence Viewer 7.0.
Statistical analysis
All experimental data are the means of at least three independent replicates, and statistical analyses were performed using Microsoft Office Excel 2007, Microsoft Corporation, Redmond, WA. Variations among transgenic and control plants subjected to different treatments were compared using one‐way analysis of variance (ANOVA). (*P < 0.05; **P < 0.01; ***P < 0.001).
Authors’ contributions
J‐L.D. and T.W.: led the study and revised the manuscript. H.C.: performed the main experiments and wrote the manuscript. H.F. participated in vector construction. X‐Y.Z.: participated in low‐generation detection and purification of transgenic materials. C‐J.Z.: participated in plant material preparation. All authors read and approved the final manuscript.
Conflict of interest
The authors declare no competing financial interests.
Supporting information
Figure S1 Response of Athub2 mutants to dehydration stress.
Figure S2 Characteristics of AtHUB2.
Figure S3 Generation and molecular characterization of transgenic cotton lines expressing AtHUB2.
Figure S4 Agronomic traits of different AtHUB2 transgenic lines in the field.
Figure S5 AtHUB2 significantly enhances the drought tolerance of transgenic cotton in the greenhouse.
Figure S6 Multiple sequence alignment and phylogenetic tree analysis of GhH2B1 with Arabidopsis histone H2Bs.
Figure S7 Detection GhDREB chromatin locus H2Bub1 and H3K4me3 levels in GhHUB2‐knockdown plants.
Table S1 Comparison of drought‐related indexes between transgenic cotton lines and controls.
Table S2 Positive clones from library screening.
Table S3 Primers used in this study.
Acknowledgements
We thank the Arabidopsis Biological Resource Center for providing us with the T‐DNA insertion mutants; Mrs Zhixia Wu from the Cotton Research Institute, Chinese Academy of Agriculture Sciences, for plant material examination and identification; Dr Changwang Deng and Dr Xi Chen from our laboratory for providing the AtHUB2 overexpression vector and HUB2/hub2‐2 seeds; and Dr. Shanwei Li from our laboratory for identifying part of the transgenic materials. This work was supported by the Ministry of Agriculture Transgenic Major Projects (2013ZX08005‐003 & 2016ZX08005‐003), the National Key Research and Development Program (2016YFD0101006) and the Project for Extramural Scientists of State Key Laboratory of Agrobiotechnology (2018SKLAB6‐17).
Contributor Information
Tao Wang, Email: wangt@cau.edu.cn.
Jiangli Dong, Email: dongjl@cau.edu.cn.
References
- Agarwal, P. , Agarwal, P.K. , Nair, S. , Sopory, S.K. and Reddy, M.K. (2007) Stress‐inducible DREB2A transcription factor from Pennisetum glaucum is a phosphoprotein and its phosphorylation negatively regulates its DNA‐binding activity. Mol. Genet. Genomics, 277, 189–198. [DOI] [PubMed] [Google Scholar]
- Bartels, D. and Sunkar, R. (2005) Drought and salt tolerance in plants. Crit. Rev. Plant Sci. 24, 23–58. [Google Scholar]
- Bates, L.S. , Waldren, R.P. and Teare, I.D. (1973) Rapid determination of free proline for water‐stress studies. Plant Soil, 39, 205–207. [Google Scholar]
- Bourbousse, C. , Ahmed, I. , Roudier, F. , Zabulon, G. , Blondet, E. , Balzergue, S. , Colot, V. et al (2012) Histone H2B monoubiquitination facilitates the rapid modulation of gene expression during Arabidopsis photomorphogenesis. PLoS Genet. 8, 1002825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, Y. , Dai, Y. , Cui, S.J. and Ma, L.G. (2008) Histone H2B monoubiquitination in the chromatin of FLOWERING LOCUS C regulates flowering time in Arabidopsis . Plant Cell, 20, 2586–2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, H. , Li, X. , Wang, Z. , Ding, M. , Sun, Y. , Dong, F. , Chen, F. et al (2015) Histone H2B monoubiquitination mediated by HISTONE MONOUBIQUITINATION1 and HISTONE MONOUBIQUITINATION2 Is involved in anther development by regulating tapetum degradation‐related genes in rice. Plant Physiol. 168, 1389–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies, R.J. and Joazeiro, C.A.P. (2009) RING Domain E3 Ubiquitin Ligases. Annu. Rev. Biochem. 78, 399–434. [DOI] [PubMed] [Google Scholar]
- Dhawan, R. , Luo, H.L. , Foerster, A.M. , AbuQamar, S. , Du, H.N. , Briggs, S.D. , Scheid, O.M. et al (2009) HISTONE MONOUBIQUITINATION1 interacts with a subunit of the mediator complex and regulates defense against necrotrophic fungal pathogens in Arabidopsis . Plant Cell, 21, 1000–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk, K. , Ding, Y. , Malkaram, S. , Riethoven, J.‐J.M. , Liu, R. , Yang, J. , Laczko, P. et al (2010) Dynamic changes in genome‐wide histone H3 lysine 4 methylation patterns in response to dehydration stress in Arabidopsis thaliana . BMC Plant Biol. 10, 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, Y. , Fromm, M. and Avramova, Z. (2012) Multiple exposures to drought ‘train’ transcriptional responses in Arabidopsis . Nat. Commun. 3, 740. [DOI] [PubMed] [Google Scholar]
- Divya, K. , Jami, S.K. and Kirti, P.B. (2010) Constitutive expression of mustard annexin, AnnBj1 enhances abiotic stress tolerance and fiber quality in cotton under stress. Plant Mol. Biol. 73, 293–308. [DOI] [PubMed] [Google Scholar]
- Dubois, M. , Gilles, K.A. , Hamilton, J.K. , Rebers, P.A. and Smith, F. (1956) Colorimetric method for determination of sugars and related substances. Anal. Chem. 28, 350–356. [Google Scholar]
- Feng, H. , Li, X. , Chen, H. , Deng, J. , Zhang, C. , Liu, J. , Wang, T. et al (2018) GhHUB2, a ubiquitin ligase, is involved in cotton fiber development via the ubiquitin–26S proteasome pathway. J. Exp. Bot. 10.1093/jxb/ery269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleury, D. , Himanen, K. , Cnops, G. , Nelissen, H. , Boccardi, T.M. , Maere, S. , Beemster, G.T.S. et al (2007) The Arabidopsis thaliana homolog of yeast BRE1 has a function in cell cycle regulation during early leaf and root growth. Plant Cell, 19, 417–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, S.Q. , Chen, M. , Xia, L.Q. , Xiu, H.J. , Xu, Z.S. , Li, L.C. , Zhao, C.P. et al (2009) A cotton (Gossypium hirsutum) DRE‐binding transcription factor gene, GhDREB, confers enhanced tolerance to drought, high salt, and freezing stresses in transgenic wheat. Plant Cell Rep., 28, 301–311. [DOI] [PubMed] [Google Scholar]
- Himanen, K. , Woloszynska, M. , Boccardi, T.M. , Groeve, S. , Nelissen, H. , Bruno, L. , Vuylsteke, M. et al (2012) Histone H2B monoubiquitination is required to reach maximal transcript levels of circadian clock genes in Arabidopsis . Plant J. 72, 249–260. [DOI] [PubMed] [Google Scholar]
- Hu, M. , Pei, B.L. , Zhang, L.F. and Li, Y.Z. (2014) Histone H2B monoubiquitination is involved in regulating the dynamics of microtubules during the defense response to Verticillium dahliae toxins in Arabidopsis . Plant Physiol. 164, 1857–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, W.W. , Venkatasubrahmanyam, S. , Ianculescu, A.G. , Tong, A. , Boone, C. and Madhani, H.D. (2003) A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol. Cell, 11, 261–266. [DOI] [PubMed] [Google Scholar]
- Lee, J.S. , Shukla, A. , Schneider, J. , Swanson, S.K. , Washburn, M.P. , Florens, L. , Bhaumik, S.R. et al (2007) Histone crosstalk between H2B monoubiquitination and H3 methylation mediated by COMPASS. Cell, 131, 1084–1096. [DOI] [PubMed] [Google Scholar]
- Liu, Y.X. , Koornneef, M. and Soppe, W.J.J. (2007) The absence of histone H2B monoubiquitination in the Arabidopsis hub1 (rdo4) mutant reveals a role for chromatin remodeling in seed dormancy. Plant Cell, 19, 433–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, N. , Fromm, M. and Avramova, Z. (2014) H3K27me3 and H3K4me3 chromatin environment at super‐induced dehydration stress memory genes of Arabidopsis thaliana . Mol. Plant, 7, 502–513. [DOI] [PubMed] [Google Scholar]
- Lobell, D.B. , Schlenker, W. and Costa‐Roberts, J. (2011) Climate trends and global crop production since 1980. Science, 333, 616–620. [DOI] [PubMed] [Google Scholar]
- Matsukura, S. , Mizoi, J. , Yoshida, T. , Todaka, D. , Ito, Y. , Maruyama, K. , Shinozaki, K. et al (2010) Comprehensive analysis of rice DREB2‐type genes that encode transcription factors involved in the expression of abiotic stress‐responsive genes. Mol. Genet. Genomics, 283, 185–196. [DOI] [PubMed] [Google Scholar]
- Menard, R. , Verdier, G. , Ors, M. , Erhardt, M. , Beisson, F. and Shen, W.H. (2014) Histone H2B Monoubiquitination is Involved in the Regulation of Cutin and Wax Composition in Arabidopsis thaliana . Plant Cell Physiol. 55, 455–466. [DOI] [PubMed] [Google Scholar]
- Mittal, A. , Gampala, S.S.L. , Ritchie, G.L. , Payton, P. , Burke, J.J. and Rock, C.D. (2014) Related to ABA‐Insensitive3 (ABI3)/Viviparous1 and AtABI5 transcription factor coexpression in cotton enhances drought stress adaptation. Plant Biotechnol. J. 12, 578–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoi, J. , Shinozaki, K. and Yamaguchi‐Shinozaki, K. (2012) AP2/ERF family transcription factors in plant abiotic stress responses. Biochem. Biophys. Acta. 1819, 86–96. [DOI] [PubMed] [Google Scholar]
- Ning, Y. , Jantasuriyarat, C. , Zhao, Q. , Zhang, H. , Chen, S. , Liu, J. , Liu, L. et al (2011) The SINA E3 Ligase OsDIS1 Negatively Regulates Drought Response in Rice. Plant Physiol. 157, 242–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parida, A.K. , Dagaonkar, V.S. , Phalak, M.S. , Umalkar, G.V. and Aurangabadkar, L.P. (2007) Alterations in photosynthetic pigments, protein and osmotic components in cotton genotypes subjected to short‐term drought stress followed by recovery. Plant Biotechnol. Rep. 1, 37–48. [Google Scholar]
- Parkhi, V. , Kumar, V. , Sunilkumar, G. , Campbell, L.M. , Singh, N.K. and Rathore, K.S. (2009) Expression of apoplastically secreted tobacco osmotin in cotton confers drought tolerance. Mol. Breeding, 23, 625–639. [Google Scholar]
- Pasapula, V. , Shen, G.X. , Kuppu, S. , Paez‐Valencia, J. , Mendoza, M. , Hou, P. , Chen, J.A. et al (2011) Expression of an Arabidopsis vacuolar H + ‐pyrophosphatase gene (AVP1) in cotton improves drought and salt tolerance and increases fibre yield in the field conditions. Plant Biotechnol. J. 9, 88–99. [DOI] [PubMed] [Google Scholar]
- Paterson, A.H. , Brubaker, C.L. and Wendel, J.F. (1993) A rapid method for extraction of cotton (Gossypium spp.) genomic DNA suitable for RFLP or PCR analysis. Plant Mol. Biol. Rep. 11, 122–127. [Google Scholar]
- Qin, F. , Sakuma, Y. , Tran, L.S.P. , Maruyama, K. , Kidokoro, S. , Fujita, Y. , Fujita, M. et al (2008) Arabidopsis DREB2A‐interacting proteins function as RING E3 ligases and negatively regulate plant drought stress‐responsive gene expression. Plant Cell, 20, 1693–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy, D. , Paul, A. , Roy, A. , Ghosh, R. , Ganguly, P. and Chaudhuri, S. (2014) Differential acetylation of histone H3 at the regulatory region of OsDREB1b promoter facilitates chromatin remodelling and transcription activation during cold stress. PLoS ONE, 9, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh, A. , Alvarez‐Venegas, R. and Avramova, Z. (2008) An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat. Protoc. 3, 1018–1025. [DOI] [PubMed] [Google Scholar]
- Sato, H. , Mizoi, J. , Tanaka, H. , Maruyama, K. , Qin, F. , Osakabe, Y. , Morimoto, K. et al (2014) Arabidopsis DPB3‐1, a DREB2A Interactor, specifically enhances heat stress‐induced gene expression by forming a heat stress‐specific transcriptional complex with NF‐Y subunits. Plant Cell, 26, 4954–4973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, G.X. , Wei, J. , Qiu, X.Y. , Hu, R.B. , Kuppu, S. , Auld, D. , Blumwald, E. et al (2015) Co‐overexpression of AVP1 and AtNHX1 in cotton further improves drought and salt tolerance in transgenic cotton plants. Plant Mol. Biol. Rep. 33, 167–177. [Google Scholar]
- Soth, G.C. (1999) The impact of cotton on freshwater resources and ecosystems: a preliminary synthesis.
- Umezawa, T. , Fujita, M. , Fujita, Y. , Yamaguchi‐Shinozaki, K. and Shinozaki, K. (2006) Engineering drought tolerance in plants: discovering and tailoring genes to unlock the future. Curr. Opin. Biotechnol. 17, 113–122. [DOI] [PubMed] [Google Scholar]
- Varshney, R.K. , Bansal, K.C. , Aggarwal, P.K. , Datta, S.K. and Craufurd, P.Q. (2011) Agricultural biotechnology for crop improvement in a variable climate: hope or hype? Trends Plant Sci. 16, 363–371. [DOI] [PubMed] [Google Scholar]
- Verslues, P.E. and Bray, E.A. (2004) LWR1 and LWR2 are required for osmoregulation and osmotic adjustment in Arabidopsis . Plant Physiol. 136, 2831–2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, M. , Wang, Q. and Zhang, B. (2013) Evaluation and selection of reliable reference genes for gene expression under abiotic stress in cotton (Gossypium hirsutum L.). Gene, 530, 44–50. [DOI] [PubMed] [Google Scholar]
- Xie, Q. , Sanz‐Burgos, A.P. , Guo, H.S. , Garcia, J.A. and Gutierrez, C. (1999) GRAB proteins, novel members of the NAC domain family, isolated by their interaction with a geminivirus protein. Plant Mol. Biol. 39, 647–656. [DOI] [PubMed] [Google Scholar]
- Xu, L. , Menard, R. , Berr, A. , Fuchs, J. , Cognat, V. , Meyer, D. and Shen, W.H. (2009) The E2 ubiquitin‐conjugating enzymes, AtUBC1 and AtUBC2, play redundant roles and are involved in activation of FLC expression and repression of flowering in Arabidopsis thaliana. Plant J. 57, 279–288. [DOI] [PubMed] [Google Scholar]
- Yu, L.H. , Wu, S.J. , Peng, Y.S. , Liu, R.N. , Chen, X. , Zhao, P. , Xu, P. et al (2016) Arabidopsis EDT1/HDG11 improves drought and salt tolerance in cotton and poplar and increases cotton yield in the field. Plant Biotechnol. J. 14, 72–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue, Y.S. , Zhang, M.C. , Zhang, J.C. , Tian, X.L. , Duan, L.S. and Li, Z.H. (2012) Overexpression of the AtLOS5 gene increased abscisic acid level and drought tolerance in transgenic cotton. J. Exp. Bot. 63, 3741–3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, D.Y. , Yang, H.L. , Li, X.S. , Li, H.Y. and Wang, Y.C. (2014) Overexpression of Tamarix albiflonum TaMnSOD increases drought tolerance in transgenic cotton. Mol. Breeding, 34, 1–11. [Google Scholar]
- Zhao, L. , Wang, P. , Yan, S.H. , Gao, F. , Li, H. , Hou, H.L. , Zhang, Q. et al (2014) Promoter‐associated histone acetylation is involved in the osmotic stress‐induced transcriptional regulation of the maize ZmDREB2A gene. Physiol. Plant. 151, 459–467. [DOI] [PubMed] [Google Scholar]
- Zhou, S. , Chen, Q. , Sun, Y. and Li, Y. (2017) Histone H2B monoubiquitination regulates salt stress‐induced microtubule depolymerization in Arabidopsis . Plant Cell Environ. 8, 1512–1530. [DOI] [PubMed] [Google Scholar]
- Zhu, J.K. (2002) Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53, 247–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, B.H. , Yang, D.L. , Shi, Z.Y. , Dong, H.S. and Hua, J. (2014) Monoubiquitination of histone 2B at the disease resistance gene locus regulates its expression and impacts immune responses in Arabidopsis . Plant Physiol. 165, 309–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Response of Athub2 mutants to dehydration stress.
Figure S2 Characteristics of AtHUB2.
Figure S3 Generation and molecular characterization of transgenic cotton lines expressing AtHUB2.
Figure S4 Agronomic traits of different AtHUB2 transgenic lines in the field.
Figure S5 AtHUB2 significantly enhances the drought tolerance of transgenic cotton in the greenhouse.
Figure S6 Multiple sequence alignment and phylogenetic tree analysis of GhH2B1 with Arabidopsis histone H2Bs.
Figure S7 Detection GhDREB chromatin locus H2Bub1 and H3K4me3 levels in GhHUB2‐knockdown plants.
Table S1 Comparison of drought‐related indexes between transgenic cotton lines and controls.
Table S2 Positive clones from library screening.
Table S3 Primers used in this study.


