Summary
Production of interleukin-17 (IL-17) and IL-22 by T helper 17 (Th17) cells and group 3 innate lymphoid cells (ILC3s) in response to the gut microbiota ensures maintenance of intestinal barrier function. Here, we examined the mechanisms whereby the immune system detects microbiota in the steady state. A Syk-kinase-coupled signaling pathway in dendritic cells (DCs) was critical for commensal-dependent production of IL-17 and IL-22 by CD4+ T cells. The Syk-coupled C-type lectin receptor Mincle detected mucosal-resident commensals in the Peyer’s patches (PPs), triggered IL-6 and IL-23p19 expression, and thereby regulated function of intestinal Th17- and IL-17-secreting ILCs. Mice deficient in Mincle or with selective depletion of Syk in CD11c+ cells had impaired production of intestinal RegIIIγ and IgA and increased systemic translocation of gut microbiota. Consequently, Mincle deficiency led to liver inflammation and deregulated lipid metabolism. Thus, sensing of commensals by Mincle and Syk signaling in CD11c+ cells reinforces intestinal immune barrier and promotes host-microbiota mutualism, preventing systemic inflammation.
Keywords: intestinal barrier, dendritic cell, T lymphocyte, innate lymphoid cells, Mincle, Syk kinase, IL-17, IL-22, antimicrobial defense, gut microbiota translocation, liver inflammation, lipid metabolism
Graphical Abstract
Highlights
-
•
Mincle detects mucosal commensals and triggers IL-6 and IL-23p19 in PPs
-
•
LysoDC and CD11b+ DC from PPs prime a microbiota- and Mincle-Syk-dependent IL-17
-
•
Gut Th17 and ILCs producing IL-17 and IL-22 require Mincle and Syk in CD11c+ cells
-
•
Mincle promotes the gut barrier, limiting microbial translocation and liver inflammation
Martínez-López et al. explore host signaling pathways linking recognition of commensal microbes and Th17 differentiation. They find that the Mincle-Syk axis in Peyer’s patch DCs detects mucosal-resident bacteria, inducing IL-6 and IL-23p19 and stimulating IL-17 and IL-22 production by intestinal T cells and ILCs, which control the intestinal immune barrier function.
Introduction
The intestinal microbiota contributes to host metabolism, resistance to pathogen colonization, and host immune system development and homeostasis (Bäumler and Sperandio, 2016, Honda and Littman, 2016). However, its containment is essential, and increased microbial translocation is associated with systemic inflammation (Sonnenberg et al., 2012). The intestinal epithelial cells (IECs), the mucus layer, the generation of antimicrobial peptides, and the synthesis of microbe-specific immunoglobulin A (IgA) in the Peyer’s patches (PPs) constitute the “mucosal firewall” (Belkaid and Hand, 2014). This barrier is regulated by RORγt-dependent cells that include group 3 innate lymphoid cells (ILC3s) and T helper 17 (Th17) cells, which produce IL-17 and IL-22 and modulate antimicrobial peptide secretion by IECs and IgA production in the gut (Hirota et al., 2013, Kruglov et al., 2013). Therefore, the absence of RORγt-dependent cells or their mediators can lead to a breach of the intestinal barrier, commensal translocation, and systemic inflammation (Lochner et al., 2011, Sonnenberg et al., 2012).
The intestinal microbiota critically contributes to the generation and function of RORγt-dependent cells (Ivanov et al., 2008, Sanos et al., 2009, Satoh-Takayama et al., 2008). For instance, segmented filamentous bacteria (SFB) induce the release of serum amyloid A (SAA) proteins that promote local IL-17 production, myeloid-mediated IL-22 secretion by ILC3, and IgA responses (Atarashi et al., 2015, Gaboriau-Routhiau et al., 2009, Ivanov et al., 2009, Sano et al., 2015). Myeloid cells are critical to integrating the intestinal microenvironment and adaptive immune responses. Macrophages or dendritic cells (DCs) can be located in the lamina propria (LP) and/or in gut-associated lymphoid tissues, including PPs (Bekiaris et al., 2014). On the basis of the expression of lysozyme and CD11b, PPs contain particular subsets of DCs (Bonnardel et al., 2015, Da Silva et al., 2017). Th17 cell differentiation requires antigen recognition in the context of major histocompatibility complex II (MHCII), as well as IL-6 and transforming growth factor-β (TGF-β) from DCs (Ivanov et al., 2006, Persson et al., 2013). In contrast, activation of ILC3s is antigen independent but, in turn, requires the production of IL-23 by myeloid cells to release IL-22 and/or IL-17 (Longman et al., 2014, Satpathy et al., 2013). However, the particular subset of myeloid cells and the host signaling pathways linking recognition of commensal microbes to the induction of IL-17 and IL-22 production by RORγt+ cells in the steady state are poorly understood.
Some Syk-kinase-coupled C-type lectin receptors (CLRs), such as Dectin-1 (Clec7a), Dectin-2 (Clec4n), and Mincle (Clec4e), preferentially induce myeloid IL-6, IL-1β, and IL-23 secretion upon recognition of different components of the fungal or bacterial pathogen cell wall, promoting Th17 polarization (Geijtenbeek and Gringhuis, 2016, LeibundGut-Landmann et al., 2007). Therefore, we investigated whether Syk-coupled CLRs could contribute to myeloid cell sensing of the intestinal microbiota to promote functional responses in RORγt+ cells under homeostatic conditions. We found that sensing of mucosa-associated commensals by the Mincle and Syk pathway in lysozyme-expressing DCs (LysoDCs) and CD11b+ dome DCs from PPs contributes to IL-6 and IL-23p19 production by these cells. These myeloid-cell-derived cytokines regulate IL-17 and IL-22 production by T cells and ILCs, promoting intestinal barrier function and limiting microbial translocation, which is associated with inflammation and deregulated metabolism in the liver.
Results
Mincle and Syk Signaling in DCs Controls Microbiota-Driven Th17 Differentiation
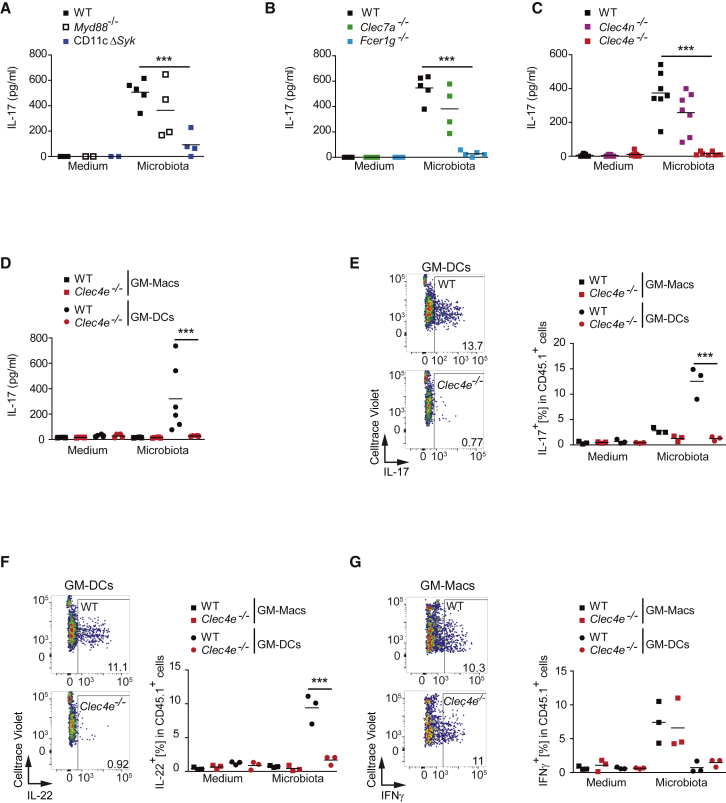
We assessed host signaling pathways with the potential to link recognition of commensal microbes by myeloid cells with Th17 differentiation. CD11c+ cells derived from bone marrow cultured with GM-CSF (GM-BMs) from mice lacking MyD88 (Myd88−/−) (Adachi et al., 1998) or Syk in the CD11c+ compartment (CD11cCre Sykflox/flox, named CD11cΔSyk mice) (Iborra et al., 2012, Whitney et al., 2014) or GM-BMs from wild-type (WT) littermates were loaded with chicken ovalbumin class-II peptide (OVA323–339) and co-cultured with naive OVA-specific (OT-II) CD4+ T cells in the presence or absence of specific-pathogen-free (SPF) gut microbiota. Although the proliferation of OT-II cells did not differ upon co-culture with different GM-BMs (Figure S1A), OT-II cell capacity to produce IL-17 after priming in the presence of gut microbiota was specifically blunted in the absence of Syk, but not MyD88, in GM-BMs (Figure 1A).
Figure 1.
Mincle and Syk Signaling in DCs Control Microbiota-Driven Th17 Differentiation
(A–C) Naive OT-II T cells were co-cultured with GM-BMs (1:2 ratio) from: (A) WT mice or mice lacking MyD88 (Myd88−/−) or Syk in the CD11c+ compartment (CD11cΔSyk); (B) WT mice or mice lacking Clec7a or mice lacking Fcer1g; (C) WT mice or mice lacking Clec4n or Clec4e, the indicated genotypes loaded with OVA peptide in the presence or absence of microbiota (10:1 GM-BM ratio), and IL-17 was measured by ELISA in the supernatant 3 days later.
(D–G) Naive OT-II T cells were co-cultured with GM-Macs or GM-DCs (1:1 ratio) loaded with OVA peptide in the presence or absence of microbiota (10:1 DC ratio) , and IL-17 was measured by ELISA in the supernatant 3 days later (D). IL-17 (E), IL-22 (F), and IFN-γ (G) production after re-stimulation was measured by intracellular staining and flow cytometry in OT-II T cells from the co-cultures. To the left is a representative plot, and to the right quantification is shown.
Individual data represent data generated from independent GM-BM cultures (biological replicates) in pools of at least two independent experiments. Individual data and arithmetic mean are shown. ∗∗∗p < 0.001 (one-way ANOVA and Bonferroni post hoc test). See also Figure S1.
Syk signaling following pattern recognition is characteristic of some CLRs. Syk binds to immunoreceptor tyrosine-based activation motif (ITAM) domains present in the FcRγ chain (Fcer1g) adaptor that couples to Dectin-2 (Clec4n) or Mincle (Clec4e) or to hemITAMs borne by CLRs, such as Dectin-1 (Clec7a) (Iborra and Sancho, 2015). Thus, we explored the ability of GM-BMs lacking Dectin-1 or the FcRγ chain to promote microbiota-driven Th17 differentiation from naive OT-II cells as above. Although OT-II cell proliferation following stimulation with GM-BMs was similar among the different GM-BM genotypes tested (Figure S1B), microbiota-induced Th17 differentiation was dependent on the expression of the FcRγ chain, but not Dectin-1, in GM-BMs (Figure 1B). We subsequently found that, upstream of the FcRγ chain, Mincle, but not Dectin-2, was needed in GM-BMs for microbiota-driven Th17 differentiation in vitro (Figures 1C and S1C). Notably, exposure to microbiota induced Syk phosphorylation in GM-BMs in a Mincle-dependent manner (Figure S1D).
GM-BMs comprise conventional DCs (GM-DCs) and monocyte-derived macrophages (GM-Macs) (Helft et al., 2015). GM-Macs expressed Mincle constitutively, whereas intestinal microbiota stimulation induced Mincle expression in GM-DCs (Figure S1E), as expected (Helft et al., 2015). In addition, we found that GM-DCs efficiently primed IL-17 and IL-22 production by OT-II cells in response to microbiota and in a Mincle-dependent fashion (Figures 1D–1F). In contrast, GM-Macs promoted IFN-γ-producing OT-II cells in a Mincle-independent manner (Figure 1G). These results suggest that the Mincle-FcRγ-chain-Syk axis in GM-DCs drives Th17 differentiation in response to intestinal commensals.
Mincle Senses Mucosa-Associated Commensals
We tested whether the intestinal microbiota contains a functional ligand for Mincle by analyzing the capacity of commensals to activate GM-BMs. Upregulation of MHCII, CCR7, and CD86 in GM-BMs by microbiota was significantly reduced in the absence of Mincle (Figure S2A). As expected in controls for the experiment, activation of GM-BMs by the Mincle ligand trehalose-6,6-dibehenate (TDB) was Mincle dependent, whereas activation mediated by lipopolysaccharide (LPS) was Mincle independent (Figure S2A). These results suggest that Mincle senses microbiota and thereby contributes to DC activation.
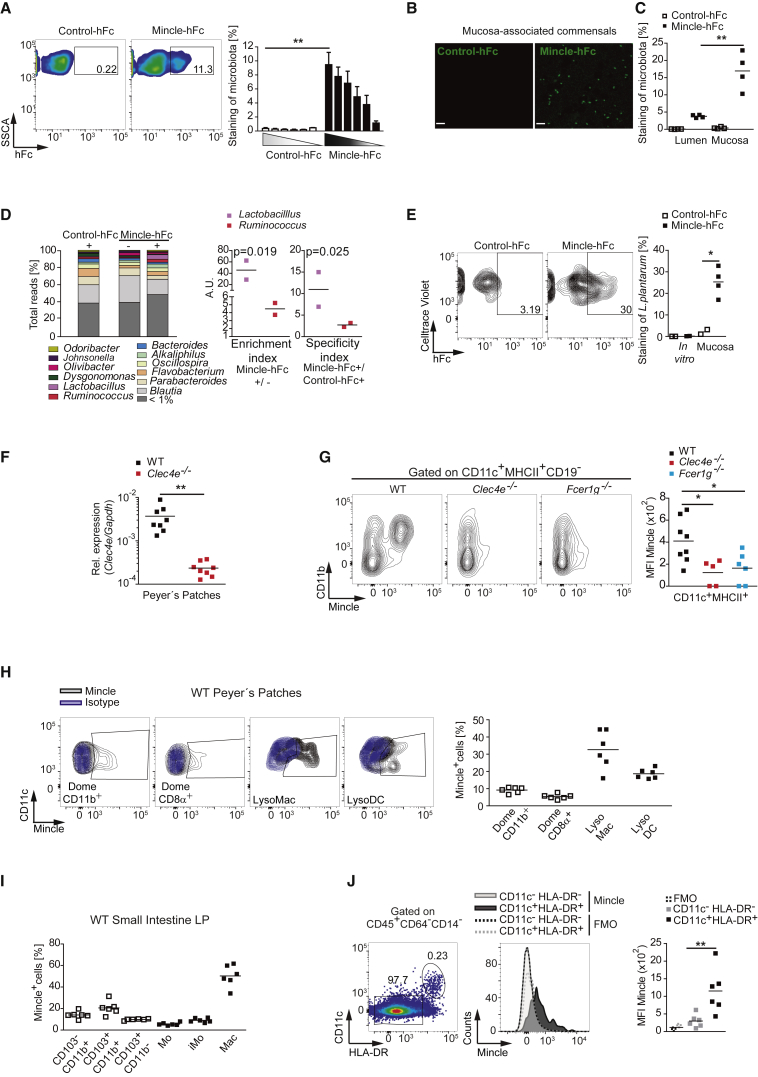
We next investigated whether Mincle could bind to the intestinal microbiota from our SPF mice. Mincle-ectodomain-human-Fc chimera (Mincle-hFc) recognized the microbiota in a dose-dependent manner (Figures 2A and S2B). Pre-incubation of Mincle-hFc with 2F2 anti-Mincle antibody or with the Mincle ligand TDB specifically prevented its binding to the microbiota (Figure S2C). In addition, Mincle-hFc did not bind to the gastrointestinal content from germ-free mice (Figure S2C). Notably, the analysis of small intestine mucosa from SPF mice revealed a more than 3-fold average enrichment in Mincle-hFc-labeled commensals compared with the luminal fraction (Figures 2B, 2C, and S2D). We additionally found that a fraction of luminal but not mucosa-associated microbiota was detected by hFc chimeras of the Syk-coupled CLRs Dectin-1 and Dectin-2 (Figure S2E).
Figure 2.
Mucosa-Associated Commensals Are Sensed by PP DCs Expressing Mincle
(A) Representative plots (left) and graph depicting the frequency of SPF microbiota stained with control-hFc or Mincle-hFc. Shown is the arithmetic mean + SEM of a pool of three replicates from two independent experiments.
(B) Analysis by stimulated emission depletion super-resolution microscopy of SPF-mouse mucosa-associated commensals labeled with control-hFc or Mincle-hFc. Scale bar, 2 μm.
(C) Frequency of SPF-mouse luminal and mucosal microbiota stained with control-hFc or Mincle-hFc by flow cytometry.
(D) Luminal microbiota was stained as in (A), sorted into Mincle-hFc-enriched (Mincle-hFc+), Mincle-hFc-depleted (Mincle-hFc−), and control-hFc-enriched (control-hFc+) fractions, and analyzed by 16S sequencing. Shown on the left is the relative abundance of each genus from two independent experiments. To the right are the enrichment index and specificity index, calculated as explained in the STAR Methods.
(E) Mucosa-associated commensals from WT SPF mice gavaged with Celltrace-violet-labeled Lactobacillus plantarum (L. plantarum) or in-vitro-grown L. plantarum were stained with control-hFc or Mincle-hFc and analyzed by flow cytometry. Shown on the left is representative staining. Shown on the right is the frequency of bacteria positive for the indicated staining and pre-gated on cell-violet-positive bacteria.
(F) Mincle expression in PPs from Mincle-deficient (Clec4e−/−) mice and WT littermates analyzed by qPCR and normalized to Gapdh.
(G) Mincle expression in the indicated genotypes by flow cytometry in PPs. On the left are representative plots. On the right, mean fluorescence intensity (MFI) is shown.
(H) Mincle expression by flow cytometry in the myeloid populations from WT mice PPs. On the left is a representative plot. On the right, the frequency of Mincle+ cells in the depicted subset is shown.
(I) Frequency of Mincle+ cells in the myeloid subsets from WT small intestine LP.
(J) Mincle expression in LP mononuclear cells from three colonic and three duodenal samples from healthy individuals by flow cytometry. On the left, the gating strategy is shown. In the middle, Mincle versus fluorescence minus one (FMO) expression in the CD11c−HLA-DR− and CD11c+HLA-DR+ populations is shown. Shown on the right is the MFI of Mincle expression or the FMO in six independent samples.
Data represent one representative experiment of two performed (B, C, E, F, and J) or were pooled from at least two independent experiments (G, H, and I). ∗p < 0.05, ∗∗p < 0.01 (unpaired two-tailed Student’s t test). See also Figure S2.
Comparative 16S sequencing analysis of Mincle-hFc-bound and -depleted microbial fractions, along with a control-hFc-bound fraction, revealed that Lactobacillus was one of the main genera enriched in Mincle-hFc-bound fractions (Figure 2D). Mincle-hFc-stained Lactobacillus was recovered from the small intestine epithelium of mice orally gavaged with Celltrace-Violet-labeled L. plantarum but, intriguingly, not the same bacteria growing in MRS Broth (Figures 2E and S2F). These results indicate that some phyla of mucosa-associated commensals, including Lactobacillus, might contain ligands for Mincle (Shah et al., 2016).
Mincle Is Expressed in DCs from PPs
Lactobacillus exhibits a preferential binding to the follicle-associated epithelium of the PPs (Plant and Conway, 2001). We hypothesized that Mincle-expressing cells that sense mucosa-resident commensals could be located in the gut-associated lymphoid tissue. Indeed, Mincle was expressed in PPs (Figure 2F), and FcRγ-chain-dependent Mincle expression was restricted to the CD11c+MHCII+CD19−CD11b+ subset in PPs (Figure 2G). Confocal microscopy of PP whole-mount preparations confirmed the selective Mincle expression in the CD11c+CD11b+ immune compartment (Figure S2G). Concurring with previously published results (Bonnardel et al., 2015), Mincle was mainly expressed by some lysozyme-expressing macrophages (LysoMacs), lysosyme-expressing DCs (LysoDCs), and dome CD11b+ DCs, but not dome CD8α+ DCs (Figures 2H and S2H). In contrast, Mincle expression in small intestine LP was mainly found on macrophages but not in CD64− DC subsets (Figures 2I and S2I). Mincle expression in human intestinal samples from healthy donors was revealed in a fraction of CD45+CD64−CD14−CD11c+HLA-DR+ cells by flow cytometry and immunohistochemistry (IHC) (Figures 2J and S2J). These results indicate that Mincle is expressed by macrophages in PPs and small intestine LP and by some DCs in the mouse PPs.
DCs from PPs Instruct Mincle- and Syk-Dependent Th17 Differentiation
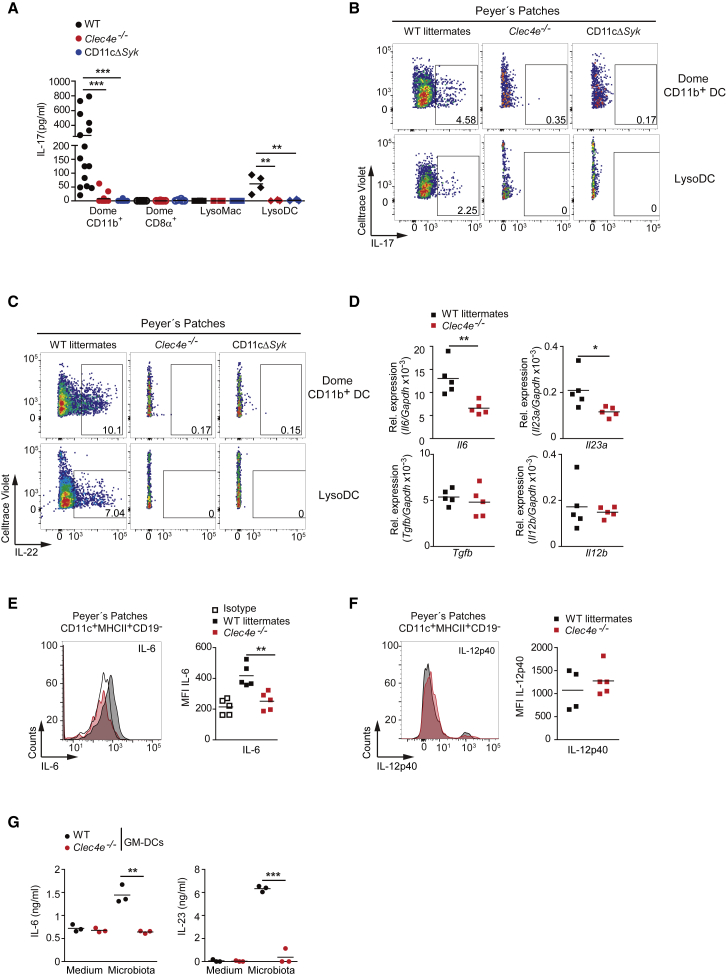
Next, we explored the contribution of freshly isolated dome CD11b+ DCs, dome CD8α+ DCs, LysoMacs, and LysoDCs from PPs to prime OT-II CD4+ T cells ex vivo. Dome CD11b+ DCs and LysoDCs had a higher capacity to induce IL-17 production by CD4+ T cells than did CD8α+ DCs and LysoMacs (Figure 3A). Moreover, the ability of CD11b+ DCs and LysoDCs to promote Th17 differentiation was dependent on Mincle and Syk (Figures 3A–3C, S3A, and S3B), whereas Th1 differentiation induced by these cells was Mincle and Syk independent (Figures S3C and S3D). These results suggest that dome CD11b+ DCs and LysoDCs located in PPs mediate Mincle- and Syk-dependent Th17 differentiation.
Figure 3.
DCs from PPs Instruct Mincle- and Syk-Dependent Th17 Differentiation
(A–C) Naive OT-II T cells were co-cultured for 3 days with dome CD11b+ DCs, CD8α+ DCs, LysoMacs, or LysoDCs from the indicated genotypes, sorted from PPs (1:1 ratio), and loaded with OVA323–339 peptide. (A) IL-17 secretion by ELISA. Each dot represents an independent co-culture where myeloid cells were from different mice from two independent experiments. (B and C) Representative FACS plots of IL-17 (B) and IL-22 (C) intracellular staining after OTII re-stimulation.
(D) Il6, Il23a, Tgfb, and Il12b transcripts in PPs of the indicated genotypes by qPCR; they were normalized to Gapdh.
(E and F) Analysis of IL-6 (E) and IL-12p40 (F) intracellular staining in CD11c+MHC-II+CD19− cells from PPs in the indicated genotypes. Shown on the left are representative histograms. On the right is MFI of staining.
(G) ELISA of IL-6 and IL-23 production by sorted GM-DCs from WT and Mincle-deficient (Clec4e−/−) mice untreated (medium) or stimulated with gut microbiota (10:1 DC ratio) for 12 h.
Data represent two independent pooled experiments (A) or one representative experiment of at least two performed (B–G). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 (A: one-way ANOVA and Bonferroni post hoc test; D, E, and G: unpaired two-tailed Student’s t test). See also Figure S3.
Mincle Fosters IL-6 and IL-23p19 Production by DCs in Response to Microbiota
The cytokine IL-6 plays a non-redundant role in Th17 differentiation in gut mucosa (Hu et al., 2011, Persson et al., 2013), and CD11b+ DCs from PPs are better at producing IL-6 than other DC subsets from PPs or splenic DCs (Sato et al., 2003). Conversely, activation of ILC3s is independent of IL-6 but dependent on microbiota-induced IL-23p19 (Klose and Artis, 2016). Therefore, we explored whether both instructing signals are produced in a Mincle-dependent manner in PPs. Mincle-deficient mice exhibited reduced IL-6 and IL-23p19, but their TGF-β and IL-12p40 transcripts in PPs were similar to those in WT littermates (Figure 3D). Consistent with this, IL-6 production by CD19−CD11c+MHCII+ DCs in PPs was diminished in Mincle-deficient mice compared with WT littermates, whereas IL-12p40 production was not affected (Figures 3E, 3F, and S3E). Accordingly, microbiota induced Mincle-dependent IL-6 and IL-23 production by GM-DCs (Figure 3G). These results indicate that the steady-state microbiota triggers Mincle-dependent IL-6 and IL-23p19 expression, which could instruct IL-17 and IL-22 production.
Mincle and Syk in DCs Are Needed for Intestinal IL-17 and IL-22 Production
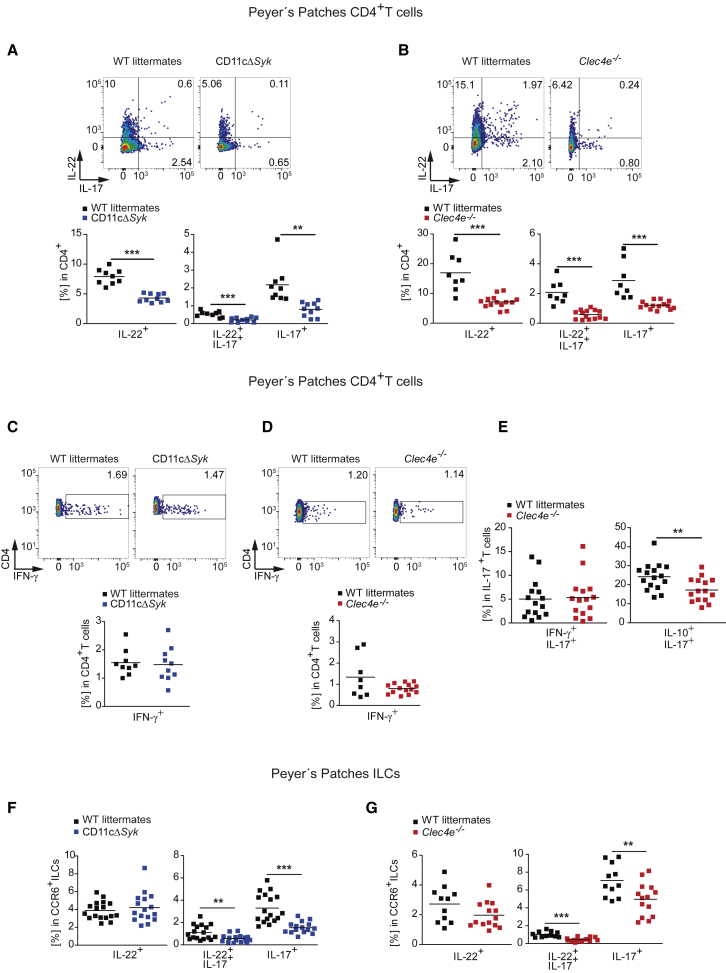
Because CD11cΔSyk mice selectively deplete Syk in spleen CD11c+ cells, but not in B or T cells (Whitney et al., 2014), we explored whether the absence of Mincle and Syk in the CD11c+ compartment could affect Th17 differentiation in the steady-state intestine in vivo. IL-17 and IL-22 production by both CD4+ T cells from PPs and sorted CD3+ T cells from the small intestine LP was reduced both in CD11cΔSyk mice (Figures 4A and S4A) and in Mincle-deficient mice (Figures 4B and S4B) compared with WT littermates, whereas IFN-γ production was not altered (Figures 4C, 4D, and S4C). IL-17-producing T cells generated in a Mincle-dependent manner in the steady state mainly co-produced IL-10, but not IFN-γ, indicating that these T cells were non-pathogenic (Figure 4E) (McGeachy et al., 2007).
Figure 4.
Mincle and Syk in DCs Are Needed for Intestinal IL-17 and IL-22 Steady-State Production
(A–E) Representative plots and summary graph of IL-17 and IL-22 (A and B), IFN-γ (C and D), and IL-17, IFN-γ, and IL-10 (E) production by intracellular staining after phorbol 12-myristate 13-acetate (PMA) and ionomycin stimulation in CD4+ T cells from PPs in the indicated genotypes.
(F and G) Frequency of CCR6+ ILCs from PPs producing IL-22 and IL-17 in the steady state by intracellular staining in the indicated genotypes.
At least two independent experiments were pooled. Each symbol represents an individual mouse. The arithmetic mean for each group is indicated. ∗∗p < 0.01, ∗∗∗p < 0.001 (unpaired two-tailed Student’s t test). See also Figure S4.
The frequencies of T cells in PPs and small intestine LP in the absence of Mincle and Syk were comparable to those of WT littermates (Figures S4D and S4E). Naive CD4+ T cells from both CD11cΔSyk and Mincle-deficient mice differentiated normally into IL-17- and IL-22-secreting effector cells in the presence of Th17-polarizing cytokines (Figure S4F). Furthermore, adoptively transferred naive CD4+ T cells from CD11cΔSyk mice or WT littermates (CD45.2+) similarly produced IL-17 and IL-22 in PPs of WT recipients (CD45.1) (Figure S4G). These results exclude an intrinsic defect of cytokine production in the T cells of these mice.
Additional sources of IL-17 and IL-22 in the gut are CCR6-expressing ILC3s that are mainly located in PPs (Klose and Artis, 2016). CD11cΔSyk and Mincle-deficient mice exhibited fewer IL-17- or IL-17- and IL-22-producing ILCs both in PPs and in small intestine LP than did WT littermates, whereas ILCs producing only IL-22 were comparable between genotypes (Figures 4F, 4G, S4H, and S4I). Frequencies of CCR6+ ILCs in PPs or CD3−CD90.2+ ILCs from the small intestine in the absence of the Mincle-Syk pathway were comparable to those in WT littermate controls (Figures S4J and S4K). These results reveal that the Mincle-Syk signaling pathway regulates IL-17 and IL-22 production by both innate and adaptive cellular sources in the steady-state small intestine and PPs.
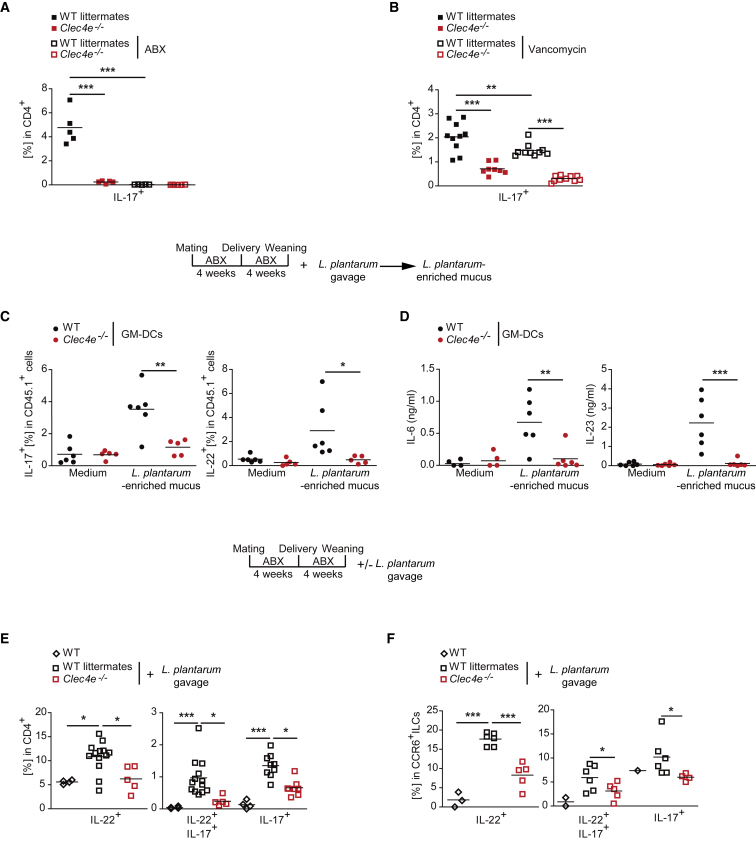
Mincle-Dependent IL-17-Producing Cells in PPs Require Commensal Bacteria
Treatment with a broad-spectrum antibiotic cocktail to disrupt the gut microbiota abolished IL-17 production by T cells from PPs in WT littermates to the levels found in Mincle-deficient and CD11cΔSyk mice (Figures 5A and S5A), suggesting that PP Th17 cell development requires intestinal bacteria. Given the prominent role of SFB in Th17 cell differentiation in PPs (Ivanov et al., 2009), we investigated the potential involvement of SFB in the Mincle-dependent Th17 differentiation. SFB were detected in our mouse colonies by PCR (IDEXX Bioresearch). Consistently, Saa1 ileal transcripts, which are linked to SFB presence (Ivanov et al., 2009), were found in our mouse colonies and even increased in the absence of the Mincle-Syk pathway (Figure S5B). SFB are sensitive to vancomycin treatment (Ivanov et al., 2008), as we demonstrated in our model by the reduction of the Saa1 ileal transcripts and the decrease in SFB content in feces after treatment (Figures S5C and S5D). Notably, vancomycin treatment slightly reduced IL-17 production by CD4+ T cells from PPs of WT littermates, but the remaining IL-17 production was dependent on Mincle (Figure 5B). Mincle-independent IFN-γ production by PP CD4+ T cells was not affected by vancomycin treatment (Figure S5E). These results show that vancomycin-resistant bacteria mediate Mincle-dependent IL-17 production by T cells from PPs.
Figure 5.
Commensal Bacteria Are Required for Mincle-Dependent Th17 Generation in PPs
(A and B) Frequency of IL-17 production by intracellular staining in re-stimulated CD4+ T cells from PPs of the indicated genotypes after treatment with an antibiotic cocktail (ABX) containing ampicillin, neomycin, metronidazole, and vancomycin (A) or vancomycin only (B) in the drinking water over 4 weeks.
(C) Frequency of IL-17 and IL-22 production by intracellular staining of re-stimulated OT-II T cells co-cultured with sorted WT and Mincle-deficient (Clec4e−/−) GM-DCs loaded with OVA323–339 peptide, as in Figures 1D and 1E; the cells were pulsed or not (medium) with mucosal-associated commensals from WT mice treated with ABX during gestation and lactation and gavaged with L. plantarum at weaning as indicated (L. plantarum-enriched mucus; 10:1 DC ratio).
(D) ELISA of IL-6 and IL-23 from the co-culture of GM-DC in (C).
(E and F) WT littermates and Mincle-deficient (Clec4e−/−) mice were treated with ABX during gestation and lactation and were gavaged with L. plantarum (1 × 106) at weaning (+L. plantarum gavage) or not (WT), as indicated in the figure. The frequency of IL-17- and IL-22-producing re-stimulated CD4+ T cells (E) or CCR6+ ILCs (F) from PPs by intracellular staining in the indicated genotypes and conditions is shown.
Data represent one representative experiment of two performed (A and F) or were pooled from at least two independent experiments (B–E). Each symbol represents an individual mouse. The arithmetic mean for each group is indicated. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 (A–E: one-way ANOVA and Bonferroni post hoc test; F: unpaired two-tailed Student’s t test). See also Figure S5.
Given the binding of Mincle-hFc to L. plantarum (Figure 2E), we tested the ability of mucosal-associated commensals from WT mice treated with ABX during gestation and lactation and gavaged with L. plantarum at weaning (L. plantarum-enriched mucus) to induce Th17 differentiation in vitro. For this, we stimulated naive OT-II T cells with sorted WT and Mincle-deficient (Clec4e−/−) GM-DCs loaded with OVA323–339 peptide and pulsed or not (medium) with L. plantarum-enriched mucus. We found that L. plantarum-enriched mucus induced IL-17 and IL-22 production by naive OT-II cells upon culture with WT but not Mincle-deficient GM-DCs (Figures 5C and S5F), whereas IFN-γ production was similar between both genotypes (Figure S5G). Induction of Th17 differentiation correlated with Mincle-dependent production of IL-6 and IL-23 by GM-DCs (Figure 5D). Moreover, compared with controls not gavaged with L. plantarum, mice treated with ABX and gavaged with L. plantarum at weaning as indicated above showed a Mincle-dependent induction of IL-17 and IL-22 production by CD4+ T cells and CCR6+ ILCs in PPs (Figures 5E and 5F) without affecting IFN-γ production (Figure S5H). These results suggest that mucosa-associated bacteria, including L. plantarum, drive Mincle-dependent IL-17- and IL-22-producing cells in PPs.
The Mincle-Syk Axis Contributes to Intestinal Barrier Function
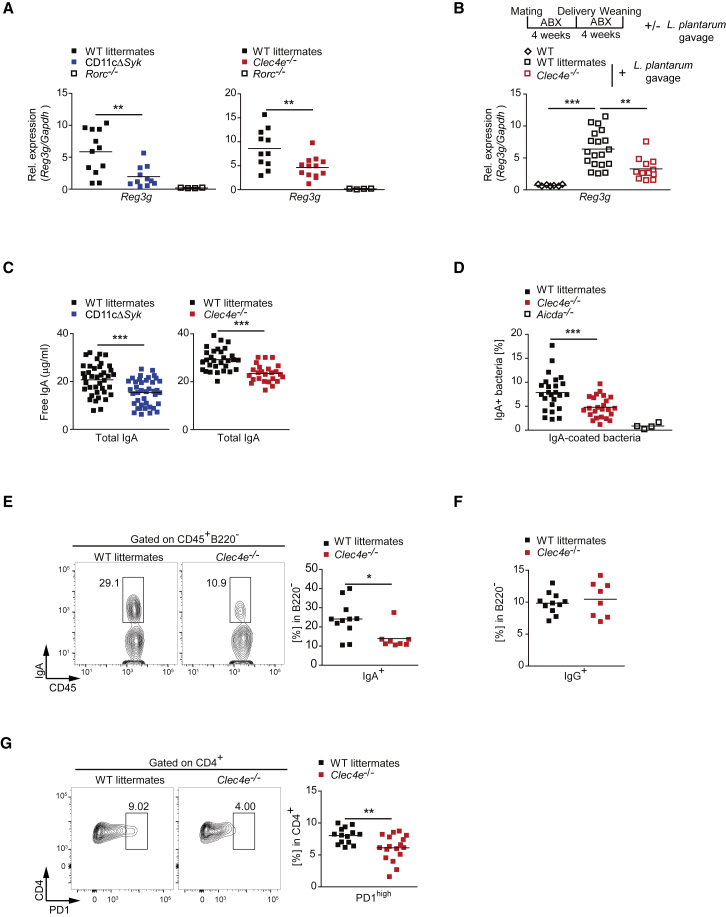
We next explored whether the defective IL-17 and IL-22 expression in Mincle-deficient and CD11cΔSyk mice affects intestinal barrier function. To assess the physical intestinal barrier integrity, we administered FITC-dextran by oral gavage and found that Mincle-deficient mice and WT littermates had similar serum FITC concentration at different time points (Figure S6A). To explore the immunological barrier, we examined the expression of intestinal antimicrobial peptides, specifically RegIIIγ, whose production relies on RORγt-dependent cells (Sanos et al., 2009). We found that steady-state Reg3g expression was reduced in the absence of Mincle or Syk in CD11c+ cells compared with WT cells (Figure 6A). Notably, oral administration of L. plantarum to ABX-treated weaned mice as above promoted Reg3g expression in a Mincle-dependent manner (Figure 6B).
Figure 6.
The Mincle-Syk Axis Contributes to Intestinal Barrier Function
(A and B) Reg3g transcripts were measured by qPCR and normalized to Gapdh in the jejunum of the indicated genotypes during the steady state (A) or after L. plantarum gavage, as described in the scheme (B).
(C) Quantification of total IgA measured by ELISA in the intestinal lumen of the indicated genotypes.
(D) Frequency of IgA+ bacteria by flow cytometry in the indicated genotypes.
(E and F) Representative plots and summary graphs of CD45+B220−IgA+ (E) or IgG+ (F) plasmatic cells from the small intestine LP of Mincle-deficient (Clec4e−/−) mice and WT littermate controls.
(G) Representative plots and summary graph of CD4+PD-1high T cells from PPs of the indicated genotypes.
Data were pooled from at least two independent experiments. Individual mice and the arithmetic mean of each group are indicated. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 (A and B: one-way ANOVA and Bonferroni post hoc test; C–H: unpaired two-tailed Student’s t test). See also Figure S6.
IgA secretion can be influenced directly by Th17 cells and ILC3s (Hirota et al., 2013, Kruglov et al., 2013). Total IgA was reduced in the intestinal lumen in Mincle-deficient and CD11cΔSyk mice compared with WT littermate controls (Figure 6C). Remarkably, Mincle deficiency decreased the generation of IgA against gut-microbiota-specific antigens, such as LPS and flagellin (Figure S6B). Flow-cytometry analysis of the frequency of IgA-coated intestinal bacteria, which was dependent on Aicda, revealed a specific reduction of IgA in the absence of Mincle (Figures 6D and S6C). Frequency of IgA+ plasma cells in the small intestine LP of Mincle-deficient and CD11cΔSyk mice was lower than in WT littermates (Figures 6E and S6D), and IgG+ plasma cells were similar (Figure 6F). This correlated with reduced percentages of PD-1highCD4+ T cells in PPs (Figures 6G and S6E), a marker of T follicular helper (Tfh) cells, which are a T cell subset that promotes IgA-producing germinal center B cells (Hirota et al., 2013). These differences in gut immunity did not result in significant differences in the luminal microbiota composition at the genus level between Mincle-deficient mice and their WT littermates (Figure S6F). These results indicate that the Mincle-Syk signaling pathway contributes to maintaining a functional intestinal immune barrier but does not play a major role in regulating intestinal microbiome composition.
The Mincle-Syk Pathway Promotes Commensal Bacteria Containment
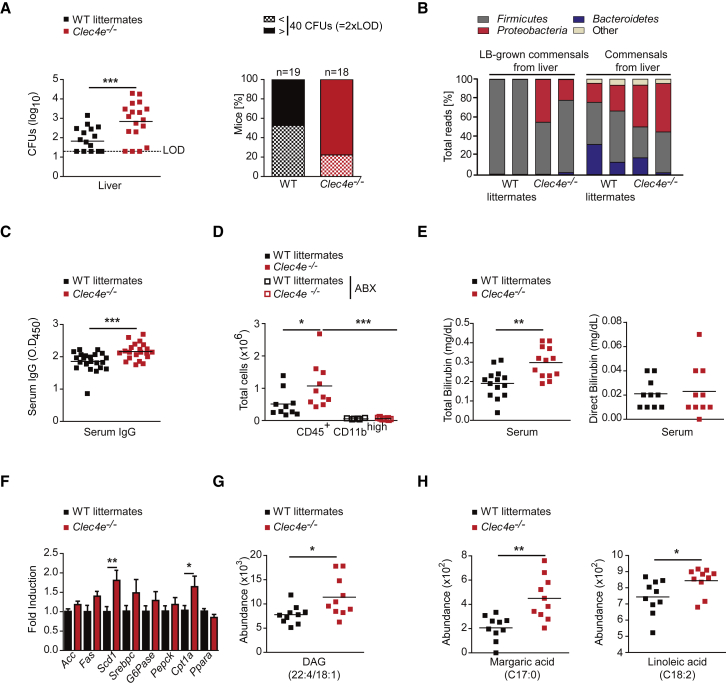
To explore whether the functional impairment of the intestinal barrier in Mincle-deficient and CD11cΔSyk mice affects microbial containment in the intestine, we first analyzed the presence of live facultative aerobe bacteria in extra-intestinal organs. We found increased microbe translocation to the liver in Mincle-deficient or CD11cΔSyk mice compared with littermate controls (Figures 7A and S7A). Deep 16S rRNA sequencing revealed that most of the surplus disseminated aerobe bacteria found in the liver of Mincle-deficient mice belonged to the phylum Proteobacteria either after growth in lysogeny broth (LB) or by direct sequencing of fresh liver homogenates (Figure 7B). As an additional readout for systemic dissemination of commensals (Zeng et al., 2016), we found increased intestinal-bacteria-specific IgG in the sera of Mincle-deficient and CD11cΔSyk mice compared with WT littermates (Figures 7C and S7B).
Figure 7.
Mincle-Syk Pathway Promotes Commensal Bacteria Containment
(A) Bacterial translocation into the liver of Mincle-deficient (Clec4e−/−) mice and WT littermates. On the left is the bacterial load as colony-forming units (CFUs) per organ, indicating the limit of detection (LOD). On the right, the frequencies of the mice of each genotype show more than 40 CFUs per organ (2× LOD).
(B) 16S rRNA sequencing analysis of LB-grown commensals from the liver (left) or bacterial DNA directly extracted from the liver (right) in the indicated genotypes. The graph shows the percentage of total reads corresponding to each phylum. Each bar represents four pooled LB plates per mouse and four mice per sample of each genotype (LB-grown commensals) or six pooled mice per genotype (commensals from the liver).
(C) ELISA of serum IgG against intestinal bacteria in the indicated genotypes.
(D) Total numbers of CD45+CD11bhigh cells infiltrated in the liver of the indicated genotypes in the steady state or after administration of an antibiotic cocktail (ABX).
(E) Total (left) or direct (right) bilirubin in the indicated genotypes.
(F) Acc, Fas, Scd1, Srebpc, G6pase, Pepck, Cpt1a, and Ppara transcripts from the liver of 15 mice of the indicated genotypes were analyzed by qPCR and normalized to Gapdh in three independent experiments; the graph shows fold induction compared with the WT.
(G and H) Individual lipid species of diacylglycerides (DAGs) (G) and the margaric (C17:0) and linoleic (C18:2) acid classes of free fatty acids (H) measured by a liquid chromatography-mass spectrometry (LC-MS) and gas chromatography-mass spectrometry (GC-MS) metabolomics-based profiling approach. Data are presented as the metabolites’ abundance.
Data were pooled from at least two independent experiments. Individual mice and the arithmetic mean of each group are shown (A, C–E, G, and H). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 (A–C and E–H [margaric acid]: unpaired two-tailed Student’s t test; H [linoleic acid]: Mann-Whitney U test; D: one-way ANOVA and Bonferroni post hoc test). See also Figure S7.
Next, we analyzed whether the presence of increased intestinal commensals in the liver of Mincle-deficient mice can lead to patho-physiological alterations. CD45+CD11bhigh myeloid cells, mainly neutrophils and inflammatory monocytes, were increased in the liver of Mincle-deficient mice compared with WT littermates in an ABX-dependent manner (Figures 7D and S7C). Liver inflammation in the absence of Mincle correlated with moderate liver malfunction reflected by an increased total, but not direct, serum bilirubin (Figure 7E) without affecting serum levels of alanine aminotransferase (ALT) or aspartate aminotransferase (AST) (Figure S7D). The expression of some lipid-metabolism-related genes, such as Scd1 (stearoyl-CoA desaturase-1) or Cpt1a (carnitine palmitoyltransferase 1), was significantly increased in the liver of Mincle-deficient mice (Figure 7F). This augmented expression of lipogenic genes correlated with an accumulation of diacylglycerides (DAGs) (Figure 7G) and some fatty acids, including margaric and linoleic acids, in Mincle-deficient mice (Figure 7H), as revealed by metabolomics assays (Figure S7E). These results indicate that the intestinal immune barrier fostered by the Mincle-Syk axis limits microbial translocation, preventing systemic inflammation and promoting host-microbiota mutualism.
Discussion
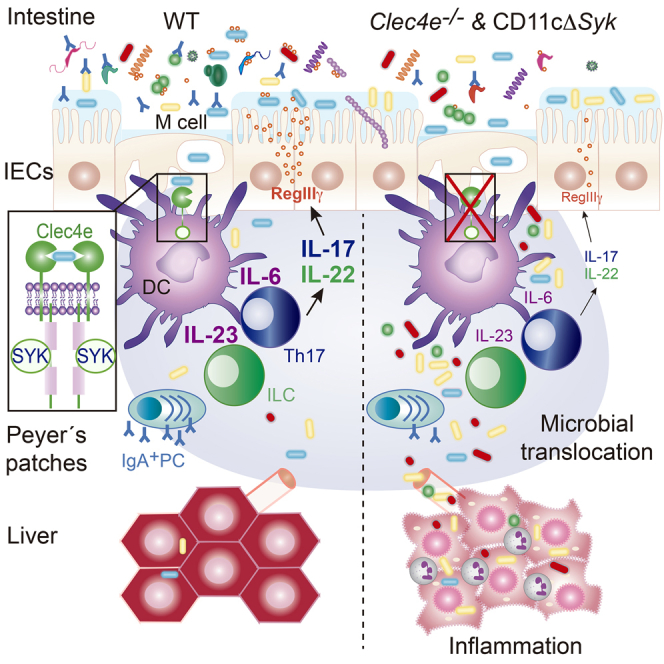
IL-17 and IL-22 contribute to intestinal barrier function in response to microbiota, regulating the expression of antimicrobial peptides and mucin genes and influencing IgA responses (Hirota et al., 2013, Kruglov et al., 2013). However, the host receptors and signaling pathways that link intestinal microbiota with regulation of IL-17 and IL-22 production to maintain a homeostatic host-microbiota relationship need to be further explored. Here, we provide evidence that the sensing of mucosal-associated bacteria by the Mincle-Syk signaling axis in LysoDCs and dome CD11b+ DCs in PPs induces IL-6 and IL-23, cytokines that stimulate intestinal T cells and ILCs to produce IL-17 and IL-22. Defective IL-17 and IL-22 production in the absence of a functional Mincle-Syk axis in DCs was associated with impaired intestinal immune barrier function, resulting in increased systemic translocation of microbiota, hepatic inflammation, and metabolic alterations. We found that Syk kinase, but not MyD88, behaved as a non-redundant innate signal for driving Th17 differentiation in response to gut microbial recognition by DCs. This is consistent with previous studies revealing a critical role for MyD88 signaling in T cells, but not in DCs, in Th17 commitment (Hu et al., 2011, Shaw et al., 2012). The Syk pathway is utilized by several CLRs, which play important roles in bridging innate and adaptive immunity after infection whereby they prominently modulate Th17 effector cell generation (Geijtenbeek and Gringhuis, 2016, LeibundGut-Landmann et al., 2007). Remarkably, Mincle-deficient mice phenocopied all defects observed in CD11cΔSyk mice, indicating that Mincle is most likely upstream of Syk in CD11c+ cells inducing IL-17 and IL-22 production in response to intestinal microbiota, although additional receptors sensing microbiota could trigger Syk.
The differential spatial distribution of commensal microbes in the intestine influences their capacity to shape host immune responses (Atarashi et al., 2015, Fung et al., 2014, Ivanov et al., 2008). Our results suggest a preferential location of commensal bacteria containing Mincle ligands close to the intestinal mucosa. The 16S rRNA sequencing of bacteria enriched by the extracellular domain of Mincle indicates that the small-intestine-adherent Lactobacillus (Donaldson et al., 2016) contains a ligand for Mincle. Intriguingly, exposition of the ligand seems to require transit through the intestine, resembling the described structural adaptations in the cell-surface composition of bacteria upon different environmental situations (Sengupta et al., 2013). Our results concur with an L. plantarum-derived glycolipid that signals through Mincle (Shah et al., 2016) and with previous reports indicating that Lactobacillus activates Syk in DCs (Weiss et al., 2012).
Some Lactobacillus species have preferential association with the follicle-associated epithelium of PPs (Plant and Conway, 2001). We showed Mincle expression in LysoDCs and LysoMacs and, to a lesser extent, in dome CD11b+ DCs. These myeloid cell subsets are efficient in the capture of antigens and essential to driving the generation of the mucosal immune response (Da Silva et al., 2017). We found that dome CD11b+ DCs and LysoDCs, but not LysoMacs, from PPs induced Th17 cell differentiation from naive T cells in a Mincle- and Syk-dependent fashion, consistent with their T cell priming capacity (Da Silva et al., 2017). Consistent with this, Mincle expression by DCs, but not macrophages, contributes to the generation of splenic immunity during Mycobacterium bovis infection in mice (Behler et al., 2015).
The intestinal microbiota is critical for IL-17 and IL-22 production by RORγt-dependent cells, and SFB are a key example (Atarashi et al., 2015, Gaboriau-Routhiau et al., 2009, Ivanov et al., 2008, Ivanov et al., 2009, Sano et al., 2015, Sawa et al., 2010). Vancomycin-resistant intestinal bacteria promoted Mincle-dependent Th17 generation in PPs, suggesting that it is at least partially independent of SFB. However, we cannot rule out that SFB launched a Th17 response that can be maintained by other vancomycin-resistant mucosa-associated bacteria after vancomycin administration, as suggested previously (Lécuyer et al., 2014).
In contrast, L. plantarum, which possesses a ligand for Mincle (Shah et al., 2016), was able to induce Mincle-dependent Th17 differentiation both in vitro and in ABX-treated mice. Mechanistically, IL-6 from CD103+CD11b+ DCs is key for Th17 differentiation in the intestine (Persson et al., 2013, Schlitzer et al., 2013). Compared with other DC subsets in PPs and spleen (Sato et al., 2003), LysoDCs and dome CD11b+ DCs from PPs are a major source of IL-6 (Da Silva et al., 2017). Our data indicate that Mincle contributes to IL-6 and IL-23p19 production by PPs DCs, which might explain the selective effect in the differentiation of Th17 but not Th1 cells. On the other hand, the regulation of IL-23p19 might affect ILC function, which requires IL-23R signaling for IL-22 and/or IL-17 secretion (Longman et al., 2014, Satpathy et al., 2013). The mucosal localization of Lactobacillus could underlie its role as a relevant conditioning factor for DCs in the PPs, but other bacteria genera at different locations might also condition the immune response locally or even systemically.
We found that reduced production of IL-17 and IL-22 in the absence of the Mincle-Syk axis in CD11c+ cells led to impaired Reg3g expression, which is regulated by IL-22 (Sanos et al., 2009). Impaired IgA generation correlated with a reduction in the frequency of IgA+ plasma cells in the small intestine LP and the proportion of CD4+PD-1high cells, which mark Tfh cells, in PPs in the absence of Mincle and Syk in CD11c+ cells. Tfh differentiation requires interaction with CD11b+ DCs and IL-6 for Bcl-6 expression (Krishnaswamy et al., 2017). Reduced IL-6 production by CD11b+ DCs, together with the contribution of the Th17 cell differentiation into Tfh in PPs (Hirota et al., 2013), could contribute to the defective IgA generation in the absence of Mincle. However, these differences in gut immunity did not result in significant differences in the luminal microbiota composition between Mincle-deficient mice and their WT littermates, although there could be regional changes in microbiota composition. For instance, Reg3g deficiency causes significant differences in mucosa-associated bacteria, but not in the composition of luminal bacterial communities. (Vaishnava et al., 2011).
We found that IL-17 and IL-22 deficiency correlated with increased peripheral dissemination of commensal bacteria and, in turn, with serum commensal-specific IgG antibodies, consistent with previous results (Lochner et al., 2011, Sonnenberg et al., 2012). Commensal translocation correlated with higher liver infiltration of neutrophils and monocytes in Mincle-deficient mice, concurring with a recent report indicating that Lactobacillus-dependent IL-22 secretion by ILCs can prevent liver inflammation by promoting the intestinal barrier (Nakamoto et al., 2017). Liver inflammation was associated with moderate liver malfunction reflected systemically by an increased total serum bilirubin and by the accumulation of DAG and some fatty acids in the liver of Mincle-deficient mice. These results suggest a putative link between commensal-driven inflammation and hepatic lipid metabolism, consistent with previous studies indicating that increased intestinal permeability, bacterial translocation, and subsequent inflammation can affect hepatic metabolism (Pierantonelli et al., 2017, Talukdar et al., 2012).
Our findings establish that Mincle and Syk signaling couples the sensing of mucosa-associated bacteria by DCs in PPs with the production of IL-6 and IL-23, cytokines that regulate IL-22 and IL-17 production through T cells and ILCs. This interaction promotes the intestinal immune barrier, limiting microbial translocation and preventing systemic inflammation and its metabolic consequences. The elucidation of this new host-commensal mechanism of interaction, which is essential to maintaining the homeostatic host-microbiota mutualism, has potential implications for diseases associated with increased bacterial translocation, such as metabolic disorders.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-mouse CD3, biotin, clone 17A2 | BioLegend | Cat# 100244; RRID: AB_2563947 |
| anti-mouse CD19, biotin, clone 6D5 | BioLegend | Cat# 115504; RRID: AB_312823 |
| anti-mouse Ly-6G and Ly-6C, biotin, clone RB6-8C5 | BD Biosciences | Cat# 553124; RRID: AB_394640 |
| anti-mouse CD16/CD32, biotin, clone 2.4G2 | BD Biosciences | Cat# 553143; RRID: AB_394658 |
| anti-mouse CD11b, biotin, clone M1/70 | BD Biosciences | Cat# 553309; RRID: AB_394773 |
| anti-mouse I-A/I-E(MHCII), biotin, clone 2G9 | BD Biosciences | Cat# 553143; RRID: AB_394957 |
| anti-mouse CD11c, biotin, clone HL3 | BD Biosciences | Cat# 553800; RRID: AB_395059 |
| anti-mouse CD45R/B220, biotin, clone RA3-6B2 | BD Biosciences | Cat# 553086; RRID: AB_394616 |
| anti-mouse CD8a, biotin, clone 53-6.7 | BD Biosciences | Cat# 553029; RRID: AB_394567 |
| anti-mouse CD3, clone 145-2C11 | BioXCell | Cat# BE0001-1; RRID: AB_1107634 |
| anti-mouse CD28, clone 37.51 | BioXCell | Cat# BE0015-1; RRID: AB_1107624 |
| anti-mouse IFN-γ, clone XMG1.2 | BioLegend | Cat# 505801; RRID: AB_315395 |
| anti-mouse IL-4, clone 11B11 | BioLegend | Cat# 504101; RRID: AB_315315 |
| anti-mouse IL-12/IL-23 p40, PE, clone C17.8 | Tonbo | Cat# 50-7123; RRID: AB_11218284 |
| anti-mouse IL-6, APC, clone MP5-20F3 | BioLegend | Cat# 504508; RRID: AB_10694868 |
| Rat IgG1, κ Isotype Control, APC, clone RTK2071 | BioLegend | Cat# 400412; RRID: AB_326518 |
| anti-mouse CD45.1, APC, clone A20 | eBiosciences | Cat# 17-0453-81; RRID: AB_469397 |
| anti-mouse CD45.1, PerCP-Cyanine5.5, clone A20 | eBiosciences | Cat# 45-0453-82; RRID: AB_1107003 |
| anti-mouse CD45.1, eFluor 450, clone A20 | eBiosciences | Cat# 48-0453-82; RRID: AB_1272189 |
| anti-mouse CD45, eFluor 450, clone 30-F11 | eBiosciences | Cat# 48-0451-82; RRID: AB_1518806 |
| anti-mouse CD45, APC, clone 30-F11 | BD Biosciences | Cat# 561018; RRID: AB_10584326 |
| anti-mouse CD16/CD32, clone 2.4G2 | Tonbo | Cat# 70-0161; RRID: AB_2621487 |
| Human Fc Block | BD Biosciences | Cat# 564219; RRID: AB_2728082 |
| anti-mouse CD44, FITC, clone IM7 | eBiosciences | Cat# 11-0441-81; RRID: AB_465044 |
| anti-mouse CD44, v450, clone IM7 | BD Biosciences | Cat# 560452; RRID: AB_1645274 |
| anti-mouse CD62L (L-Selectin), PE, clone MEL-14 | eBiosciences | Cat# 12-0621-83; RRID: AB_465722 |
| anti-mouse CD3e, FITC, clone 145-2C11 | Tonbo | Cat# 35-0031; RRID: AB_2621659 |
| anti-mouse CD8a, PE, clone 53-6.7 | eBiosciences | Cat# 12-0081-83; RRID: AB_465531 |
| anti-mouse CD11b, APC, clone M1/70 | eBiosciences | Cat# 17-0112-83; RRID: AB_469344 |
| anti-mouse CD197 (CCR7), biotin, 4B12 | eBiosciences | Cat# 13-1971-80; RRID: AB_466641 |
| anti-mouse IFN gamma, APC, clone XMG1.2 | eBiosciences | Cat# 17-7311-82; RRID: AB_469504 |
| anti-mouse CD172a (SIRP alpha), APC, clone P84 | eBiosciences | Cat# 17-1721-82; RRID: AB_10733158 |
| anti-mouse CD172a (SIRP alpha), biotin, clone P84 | eBiosciences | Cat# 13-1721-82; RRID: AB_1963572 |
| anti-mouse Ly6C, PerCP-Cyanine5.5, clone HK1.4 | eBiosciences | Cat# 45-5932-82; RRID: AB_2723343 |
| anti-mouse IgA, PE, clone mA-6E1 | eBiosciences | Cat# 12-4204-83; RRID: AB_465918 |
| anti-human IgG Fc, PE, polyclonal | eBiosciences | Cat# 12-4998-82; RRID: AB_465926 |
| anti-human IgG Fc, biotin, polyclonal | eBiosciences | Cat# 13-4998-83; RRID: AB_466660 |
| anti-mouse I-A/I-E (MHCII), FITC, clone 2G9 | BD Biosciences | Cat# 553623; RRID: AB_394958 |
| anti-mouse CD44, v450, clone IM7 | BD Biosciences | Cat# 560452; RRID: AB_1645274 |
| anti-mouse CD4, PerCP-Cy5.5, clone RM4-5 | BD Biosciences | Cat# 550954; RRID: AB_393977 |
| anti-mouse CD4, APC, clone RM4-5 | BD Biosciences | Cat# 553051; RRID: AB_398528 |
| anti-mouse CD19, PE, clone 1D3 | BD Biosciences | Cat# 561736; RRID: AB_10896141 |
| anti-mouse CD11c, APC, clone HL3 | BD Biosciences | Cat# 550261; RRID: AB_398460 |
| anti-mouse CD11c, PE, clone HL3 | BD Biosciences | Cat# 553802; RRID: AB_395061 |
| anti-mouse Siglec-F, BV421, clone E50-2440 | BD Biosciences | Cat# 562681; RRID: AB_2722581 |
| anti-mouse CD103, BUV395, clone M290 | BD Biosciences | Cat#740238; RRID: AB_2739985 |
| anti-mouse CD11c, BUV395, clone HL3 | BD Biosciences | Cat#564080; RRID: AB_2738580 |
| anti-mouse CD40, APC, clone 44986 | BD Biosciences | Cat# 558695; RRID: AB_164522 |
| anti-mouse IL-17A, PE, clone TC11-18H10 | BD Biosciences | Cat# 561020; RRID: AB_10584331 |
| anti-mouse IL-17A, APC-Cy7, clone TC11-18H10 | BD Biosciences | Cat# 560821; RRID: AB_2034016 |
| anti-mouse CD45R/B220, BV421, clone RA3-6B2 | BD Biosciences | Cat# 562922; RRID: AB_2737894 |
| anti-mouse CD11b, FITC, clone M1/70 | BD Biosciences | Cat# 11-0112-85; RRID: AB_464936 |
| anti-mouse Ly6G, PE, clone 1A8 | BD Biosciences | Cat# 551461; RRID: AB_394208 |
| anti-mouse F4/80, FITC, clone BM8 | BioLegend | Cat# 123107; RRID: AB_893500 |
| anti-mouse CD64 (FcγRI), PE/Cy7, clone X54-5/7.1 | BioLegend | Cat# 139313; RRID: AB_2563903 |
| anti-mouse I-A/I-E (MHCII), AF700, clone M5/114.15.2 | BioLegend | Cat# 107621; RRID: AB_493726 |
| anti-mouse Ly-6G, BV510, clone 1A8 | BioLegend | Cat# 127633; RRID: AB_2562937 |
| anti-mouse/human CD11b, BV605, clone M1/70 | BioLegend | Cat# 101237; RRID: AB_11126744 |
| anti-mouse CD172a (SIRPα), PerCP/Cy5.5, clone P84 | BioLegend | Cat# 144009; RRID: AB_2563547 |
| anti-mouse CD4, PE/Cy7, clone RM4-5 | BioLegend | Cat# 100528; RRID: AB_312729 |
| anti-mouse CD45, BV510, clone 30-F11 | BioLegend | Cat# 563891; RRID: AB_2734134 |
| anti-mouse CD90.2 (Thy-1.2), APC, clone 53-2.1 | BioLegend | Cat# 140312; RRID: AB_10640728 |
| anti-mouse CD196 (CCR6), PE, clone 29-2L17 | BioLegend | Cat# 129804; RRID: AB_1279137 |
| anti-mouse IL-17A, BV605, clone TC11-18H10 | BioLegend | Cat# 506927; RRID: AB_11126144 |
| anti-mouse IL-22, PE, clone Poly5164 | BioLegend | Cat# 516404; RRID: AB_2124255 |
| anti-mouse IL-22, PerCP/Cy5.5, clone Poly5164 | BioLegend | Cat# 516411; RRID: AB_2563373 |
| anti-mouse CD185 (CXCR5), PE, clone L138D7 | BioLegend | Cat# 145504; RRID: AB_2561968 |
| anti-mouse CD185 (CXCR5), APC, clone L138D7 | BioLegend | Cat# 145506; RRID: AB_2561970 |
| anti-mouse CD279 (PD-1), biotin, clone RMP1-30 | BioLegend | Cat# 109106; RRID: AB_313423 |
| anti-mouse CD279 (PD-1), BV421, clone 29F.1A12 | BioLegend | Cat# 135221; RRID: AB_2562568 |
| anti-mouse IgG1, PerCP/Cyanine5.5, clone RMG1-1 | BioLegend | Cat# 406612; RRID: AB_2562000 |
| anti-mouse/human CD11b, APC/Cy7, clone M1/70 | BioLegend | Cat# 101226; RRID: AB_830642 |
| anti-mouse MERTK (Mer), PE, clone 2B10C42 | BioLegend | Cat# 151506; RRID: AB_2617037 |
| anti-mouse CD317 (BST2, PDCA-1), APC, clone 927 | BioLegend | Cat# 127016; RRID: AB_1967127 |
| anti-mouse CD197 (CCR7), PE, clone 4B12 | BioLegend | Cat# 120106; RRID: AB_389358 |
| anti-human HLA-DR, BV570, clone L243 | BioLegend | Cat# 307638; RRID: AB_2650882 |
| anti-human CD11c, AF700, clone Bu15 | BioLegend | Cat# 337220; RRID: AB_2561503 |
| anti-human CD45, FITC, clone HI30 | BioLegend | Cat# 304006; RRID: AB_314394 |
| anti-mouse CD86, PE, clone GL-1 | Tonbo | Cat# 50-0862; RRID: AB_2621775 |
| anti-mouse CD4, PE, clone RM4-5 | Tonbo | Cat# 50-0042; RRID: AB_2621737 |
| anti-mouse CD115 (c-fms), biotin, clone AFS98 | ThermoFisher | Cat# 13-1152-81; RRID: AB_466563 |
| anti-mouse Mincle, clone 1B6 | MBL | Cat# D266-3B; RRID: AB_10950261 |
| Rat IgG1, κ Isotype Control Antibody, biotin, clone RTK2071 | BioLegend | Cat# 400403; RRID: AB_326509 |
| anti-human IgG Fc, PE, polyclonal | eBiosciences | Cat# 12-4998-82; RRID: AB_465926 |
| anti-human Mincle, clone 2F2 | Sigma-Aldrich | Cat# SAB1405010; RRID: AB_10738959 |
| Mouse IgM Negative Control, clone GC323 | Sigma-Aldrich | Cat# MABC008; RRID: AB_97858 |
| anti-phospho-Syk (Tyr525/526), clone C87C1 | Cell Signaling | Cat# 2710; RRID: AB_2197222 |
| anti- mouse CD11c, clone 3.9 | Abcam | Cat# ab11029; RRID: AB_297683 |
| anti-mouse CD11b, PE, clone M1/70 | BD Biosciences | Cat# 553311; RRID: AB_39477 |
| anti-mouse IgG (H+L), Alexa Fluor 647, polyclonal | Molecular Probes | Cat# A-21463; RRID: AB_2535869 |
| anti-human mincle, clone OTI2A8 | Abcam | Cat# ab131570; RRID: AB_11157581 |
| anti-mouse IL-23 p19, clone 5B2 | ThermoFisher | Cat# 14-7233-81; RRID: AB_842745 |
| anti-mouse IL-12/IL-23 p40, biotin, clone C17.8 | ThermoFisher | Cat# 13-7123-81; RRID: AB_466928 |
| anti-mouse IL-6, clone MP5-20F3 | BD Biosciences | Cat# 554400; RRID: AB_398549 |
| anti-mouse IL-6, biotin, clone MP5-32C11, | ThermoFisher | Cat# 13-7062-81; RRID: AB_466910 |
| anti-mouse IgA, polyclonal | Bethyl | Cat# A90-103A; RRID: AB_67136 |
| mouse IgA, κ Isotype Control, clone M18-254 | BD Biosciences | Cat# 553476; RRID: AB_479590 |
| anti-mouse IgA, HRP, polyclonal | SouthernBiotech | Cat# 1040-05; RRID: AB_2714213 |
| anti-mouse IgG-heavy and light chain, HR, polyclonal | Bethyl | Cat# A90-216P; RRID: AB_67184 |
| anti-mouse CD25, biotin, clone PC61 | BioLegend | Cat# 102004; RRID: AB_312853 |
| anti-mouse CD44, biotin, clone IM7 | BioLegend | Cat# 103004; RRID: AB_312955 |
| Bacterial and Virus Strains | ||
| Lactobacillus plantarum (Lp 39 [IAM 12477]) | ATCC | ATCC14917 |
| Biological Samples | ||
| Healthy adult intestine tissue | Hospital Universitario de La Princesa (Madrid, Spain) | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Liberase™ TM Research Grade | Sigma-Aldrich | Cat# 5401119001 |
| DNase I, Bovine Pancreas, > 2000U/MG | Biomatik | Cat# A4193 |
| Collagenase from Clostridium histolyticum, type 4 | Sigma-Aldrich | Cat# C5138 |
| Collagenase, Type 2 | Worthington | Cat# LS004176 |
| Collagenase D from Clostridium histolyticum | Sigma-Aldrich | Cat# 11088858001 |
| CellTrace™ Violet Cell Proliferation Kit | Invitrogen | Cat# C34557 |
| Recombinant murine GM-CSF | Miltenyi Biotec | Cat# 130-095-746 |
| Trehalose-6,6-dibehenate (TDB) Mincle Agonist | InvivoGen | Cat# tlrl-tdb |
| Phorbol 12-myristate 13-acetate (PMA) | Sigma-Aldrich | Cat# P8139 |
| Ionomycin calcium salt | Sigma-Aldrich | Cat# I0634 |
| Recombinant Mouse TGF-β | BioLegend | Cat# 763102 |
| Recombinant Mouse IL-6 | BioLegend | Cat# 575702 |
| Recombinant Mouse IL-23 | BioLegend | Cat# 589002 |
| Streptavidin APC | eBioscience | Cat# 17-4317-82 |
| Streptavidin PE | eBioscience | Cat# 12-4317-87 |
| Streptavidin PerCP-Cyanine5.5 | eBioscience | Cat# 45-4317-82; RRID: AB_10311495 |
| Streptavidin BV786 | BD Biosciences | Cat# 563858 |
| LIVE/DEAD™ Fixable Near-IR Dead Cell Stain Kit | Molecular Probes | Cat# 10154363 |
| Hoechst 33258 | ThermoFisher | Cat# H3569; RRID: AB_2651133 |
| Mincle (Clec4e)-human Fc Chimera protein | Iborra et al., 2016, this paper | N/A |
| Dectin-1(Clec7a)-human Fc Chimera protein | Iborra et al., 2016, this paper | N/A |
| Recombinant Human CLEC4E Fc Chimera Protein | R&D | Cat# 8995-CL-050 |
| Recombinant Human IgG1 Fc Protein | R&D | Cat# 110-HG-100; RRID: AB_276244 |
| Recombinant Human Dectin-2/CLEC6A Protein | R&D | Cat# 3114-DC-050 |
| SYTO™ 61 Red Fluorescent Nucleic Acid Stain | ThermoFisher | Cat# S11343 |
| Cimetidine hydrochloride | Sigma-Aldrich | Cat# 70059-30-2 |
| Sincalide | Tebu-bio | Cat# HOR-274 |
| Poly-L-lysine hydrobromide | Sigma-Aldrich | Cat# P1524 |
| Streptavidin, Alexa Fluor™ 488 | ThermoFisher | Cat# S11223; RRID: AB_2336881 |
| DAPI (4’,6-Diamidino-2-Phenylindole, Dihydrochloride) | ThermoFisher | Cat# D1306; RRID: AB_2629482 |
| SureBlue™ TMB 1- Peroxidase Substrate | SeraCare | Cat# 5120-0075 |
| TMB Stop Solution | SeraCare | Cat# 5150-0020 |
| Ampicilin (Gobemicin) | Normon | Cat# 882829.6 |
| Neomycin Sulfate | Sigma-Aldrich | Cat# 1405-10-3 |
| Metronidazole (Flagyl) | Sanofi | Cat# 667717.9 |
| Vancomycin | Normon | Cat# 850941.6 |
| Sucralose | Sigma-Aldrich | Cat# 56038-13-2 |
| Fluorescein isothiocyanate–dextran | Sigma-Aldrich | Cat# 46944 |
| Critical Commercial Assays | ||
| Biotinylation Kit | Abcam | Cat# ab201795 |
| Bacteria Counting Kit | ThermoFisher | Cat# B7277 |
| RNeasy Mini Kit | QIAGEN | Cat# 74104 |
| High-Capacity cDNA Reverse Transcription Kit | Applied Biosystems | Cat# 4368814 |
| GoTaq qPCR Master Mix | Promega | Cat# A6001 |
| Tyramide signal amplification (TSA) detection kit Alexa Fluor 568 | Molecular Probes | Cat# T20949 |
| Mouse IL-17A (homodimer) ELISA Ready-SET-Go | Affymetrix | Cat# 88-7371 |
| Mouse IFN-γ ELISA Set | BD Biosciences | Cat# 555138 |
| QIAamp PowerFecal Pro DNA Kit | QIAGEN | Cat# 51804 |
| Zero Blunt™ TOPO™ PCR Cloning Kit | ThemoFisher | Cat# K280002 |
| Ultra-Deep Microbiome Prep | Molzym | Cat# G-030-050 |
| QIAamp DNA Mini Kit | QIAGEN | Cat# 51304 |
| QIAamp DNA Stool Mini Kit | QIAGEN | Cat# 51504 |
| 2x Kapa HiFi Hot Start Ready Mix | Kapa Biosystems | Cat# KK5702 |
| Nextera XT DNA Library Preparation Kit | Illumina | Cat# FC-131-1024 |
| Experimental Models: Organisms/Strains | ||
| Mouse: Clec4e−/− (B6.Cg-Clec4etm1.1Cfg) | Wells et al., 2008 | N/A |
| Mouse: CD11cΔSyk (B6.Cg-Tg(Itgax-cre)1-1Reiz/J Syktm1.2Tara/J) | Iborra et al., 2012 | N/A |
| Mouse: OT-II (B6.Cg-Tg (TcraTcrb) 425Cbn/J) | Jackson Laboratory | Cat# 004194 |
| Mouse: Rorc−/− (B6.Cg -Rorctm1Litt/J) | Sun et al., 2000 | N/A |
| Mouse: Aicda−/− (C57BL/6 Aicdatm3.1Mnz/J) | Revy et al., 2000 | N/A |
| Oligonucleotides | ||
| Primers for qRT-PCR, see Table S1 | This paper | N/A |
| Software and Algorithms | ||
| FlowJo v10 | Tree Star | https://www.flowjo.com/solutions/flowjo/downloads |
| BaseSpace Application 16S Metagenomics v1.0 | Illumina | https://support.illumina.com/downloads/local-run-manager-16s-metagenomics-module.html |
| Quantitative Insights into Microbial Ecology v1.9.0 (QIIME) | Caporaso et al., 2010 | http://qiime.org/ |
| GraphPad Prism v5 | GraphPad Software | N/A |
| Other | ||
| Streptavidin MicroBeads | Miltenyi Biotec | Cat# 130-048-101 |
| Anti-mouse CD11c MicroBeads | Miltenyi Biotec | Cat# 130-097-059 |
| LPS-EB Ultrapure (Ultrapure LPS, E. coli 0111:B4) | InvivoGen | Cat# tlrl-3pelps |
| Zymosan | InvivoGen | Cat# tlrl-zyn |
| Brefeldin A | Sigma-Aldrich | Cat# B7651 |
| Anti-PE MicroBeads | Miltenyi Biotec | Cat# 130-048-801; RRID: AB_244373 |
| ProLong™ Gold Antifade Mountant | Life Technologies | Cat# P36930 |
| Standard flagellin from S. typhimurium | InvivoGen | Cat# tlrl-stfla |
| AMPure XP beads | Beckman Coulter | Cat# A63882 |
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, David Sancho (dsancho@cnic.es). The mouse lines obtained from other laboratories are described below and may require a Material Transfer Agreement (MTA) with the providing scientists.
Experimental Model and Subject Details
Experimental Animals
Mice were bred and maintained in groups of 2-5 animals per cage at the CNIC under specific pathogen-free conditions. Unless otherwise stated, males and females of 7-9 weeks old were used. Animal studies were approved by the local ethics committee. All animal procedures conformed to EU Directive 2010/63EU and Recommendation 2007/526/EC regarding the protection of animals used for experimental and other scientific purposes, enforced in Spanish law under Real Decreto 1201/2005.
Colonies included Clec4e−/− (B6.Cg-Clec4etm1.1Cfg) mice backcrossed more than 10 times to C57BL/6J-Crl were kindly provided by the Scripps Research Institute, through R. Ashman and C. Wells (Griffiths University, Australia) (Wells et al., 2008). CD11cΔSyk (Iborra et al., 2012, Whitney et al., 2014) and Clec4e−/− mice were generated along with WT littermates by heterozygous matings. OT-II CD4+ TCR transgenic mice in C57BL/6 background (B6.Cg-Tg (TcraTcrb) 425Cbn/J) were from The Jackson Laboratory. OT-II mice were mated with B6/SJL expressing the CD45.1 congenic marker (The Jackson Laboratory) to facilitate cell tracking. Rorc−/− mice (Sun et al., 2000) and Aicda−/− mice (Revy et al., 2000) were kindly provided by Dr. B. Becher (UZH, Switzerland) and Dr. A. Ramiro (CNIC, Spain), respectively. All the mice were backcrossed more than 10 generations to C57BL/6J-Crl.
Human samples
Human intestinal samples were obtained from healthy controls (3 male age 43, 26, 59; 3 female age 26, 48, 58) with no known autoimmune diseases or malignancies who had been referred to the endoscopy unit for screening of gastrointestinal diseases, although in all cases they had macroscopically and histologically normal (non-inflamed) intestines. Samples were obtained following informed consent after ethical approval (BER-CDEII-2015) from Hospital Universitario de La Princesa (Madrid, Spain).
Microbe strains
For the indicated experiments, Lactobacillus plantarum (ATCC14917) was grown overnight on MRS Broth medium (Oxoid) at 37°C in aerobic conditions to approximately OD600nm of 0.5
Methods Details
Mouse cell isolation and purification
Small intestine LP cells were isolated as previously described (Goodyear et al., 2014). Briefly, small intestine was opened longitudinally and washed with HBSS (Thermo Fisher Scientific), cut and placed into HBSS 5 mM DTT (Sigma Aldrich). Next, tissues were washed 3 times with HBSS 2% fetal bovine serum (FBS), 5 mM EDTA, minced into fine pieces and digested in HBSS supplemented with 0.2 Wünsch units/mL Liberase TM (Sigma-Aldrich) and 200 Kunitz/mL DNase I (Biomatik). Cells were passed through a 70 μm cell strainer (Falcon Products) and washed with RPMI complete medium supplemented with 20% FBS, 2 mM L-glutamine, 100 U/mL penicillin, 100 μg/mL streptomycin, 50 μM 2-mercaptoethanol.
In the indicated experiments, liver cell dissociation was performed as previously described (Allen et al., 2017). Briefly, liver was perfused through the heart with 20 mL of PBS. The liver was extracted, minced into fine pieces and digested in HBSS containing 0.5 mg/mL of type IV Collagenase from Clostridium histolyticum (Sigma Aldrich) and 40 μg/mL DNase I (Biomatik) during 20 min at room temperature. The digestion was stopped with FBS. The cells were filtered through a 70 μm cell strainer and centrifuged at 54 x g 2 min at 4°C. The supernatant was collected and centrifuged at the same speed twice. The final supernatant (non-parenchymal cells) was centrifuged at 300 x g 10 min at 4°C and used for flow cytometry analysis.
Spleen and skin-draining lymph nodes or Peyer’s Patches (PPs) (5 per mouse/ 3 mice per sample) collected in RPMI complete medium were mechanically dissociated using tweezers and a syringe plunger. Tissue homogenates were filtered through a 70 μm cell strainer. In indicated experiments, PPs were digested as described (Bonnardel et al., 2015) using 100 μg/mL of type 2 collagenase from Clostridium histolyticum (Worthington Biochemical Corporation) and 140 μg/mL of DNase I (Biomatik) for 40 min at room temperature and myeloid cells were enriched by negative selection using a cocktail of biotin-conjugated antibodies (anti-CD3, CD19, Biolegend) and Streptavidin-microbeads (Miltenyi Biotec).
Where further purification of naive CD4+ T cells from spleen and lymph nodes was required, cells were first enriched by negative selection using a cocktail of biotin-conjugated antibodies (anti-CD11c, CD11b, B220, MHC-II, CD8, GR1, CD16/32, BD Bioscience) and Streptavidin-microbeads (Miltenyi Biotec) and then sorted based on expression of CD44 and CD62L using a FACs Aria cell sorter. Where indicated, cells were labeled with CellTracer Violet (5 μM, Thermo Fisher Scientific). Dome CD11b+, CD8α+ DCs, LysoDCs and LysoMacs from PPs and CD3+ T cells or CD3- CD90.2+ ILCs from small intestine LP were purified using the FACs Aria cell sorter.
Human intestinal samples
Human biopsies were collected in ice-chilled complete medium Dutch-modified RPMI-1640 containing 100 μg/mL penicillin/streptomycin, 2 mM L-glutamine, 50 μg/mL gentamicine and 10% FBS (All reagents from Sigma-Aldrich) and processed immediately in the laboratory by incubating them twice in HBSS containing 1 mM DTT and 1 mM EDTA for 30 min at 37°C with 250 rpm rotation. Intestinal biopsies were then digested in RPMI supplemented with 1 mg/mL of collagenase D (Sigma-Aldrich) for a maximum of 90 min at 37°C with 250 rpm rotation. Intestinal LP cells were then passed through 100 μm cell strainers (Fisher) and washed in FACS buffer (ice-cold PBS supplemented with 2.5 mM EDTA, 5% FBS, 0.2% sodium azide) before proceeding with the antibody staining.
GM-CSF Bone Marrow-derived cells generation and stimulation
Cell suspensions from bone marrow of the indicated genotypes including WT, Myd88−/− (Adachi et al., 1998), CD11cΔSyk (Iborra et al., 2012), Fcer1g−/− (B6; 129P2-Fcer1gtm1Rav/J), Clec7a−/− (Marakalala et al., 2013), Clec4n−/− (Saijo et al., 2010) and Clec4e−/− (Wells et al., 2008) were cultured on tissue culture flasks (Falcon Products) in the presence of 20 ng/mL recombinant GM-CSF (Miltenyi Biotec). GM-CSF BM-derived cells were collected on day 6, purified by positive selection with anti-CD11c-microbeads (Miltenyi Biotec) and plated 16 h before stimulation.
In the indicated experiments, GM-CSF BM-derived CD11c+ cells (GM-BM, 2x106/mL) were stimulated with LPS EB (100 ng/mL, InvivoGen), plated TDB (1 μg/well, InvivoGen), zymosan (10 μg/mL, InvivoGen) or by co-culture with the indicated ratio of total microbiota from SPF mice, quantified by the optical density at 600 nm (OD600nm) together with quantification by flow cytometry, in the presence of 100 U/mL Penicillin and 100 μg/mL Streptomycin. Activation of GM-BMs was assessed by quantifying the upregulation of MHCII, CD40, CD86, CCR7 or Syk phosphorylation.
For some experiments, GM-BMs were FACS-sorted based on the expression of MERTK and CD115 into conventional DCs (GM-DCs) and monocyte-derived macrophages (GM-Macs) as previously described (Helft et al., 2015).
Cell stimulation and intracellular cytokine detection
For intracellular cytokine analysis, sorted T cells or ILCs from the small intestine LP were stimulated with PMA (Sigma Aldrich, 100 ng/mL) and ionomycin (Sigma Aldrich, 500 ng/mL) for 4 hours, in the presence of Brefeldin A in the last 3.5 hours (Sigma Aldrich, 5μg/mL). Cells were fixed with 4% paraformaldehyde (PFA) and incubated with anti-IL-17, anti-IL-22 or anti-IFN-γ during permeabilization with 0.1% saponin. For in vitro Th17 priming, splenic naive CD4+ cells were purified using MACS technology (Miltenyi Biotec) according to manufacturers’ instructions. 104 CD4+ cells were incubated with plate-bound anti-CD3 (Clone 145-2C11, BioxCell, 1μg/mL) as well as soluble anti-CD28 (Clone 37.51, BioxCell, 2 μg/mL), anti-IFN-γ (10 μg/mL), anti-IL-4 (10 μg/mL), TGF-β (2 ng/mL), IL-6 (5 ng/mL) and IL-23 (20 ng/mL). All recombinant cytokines used in this assay were from Biolegend. IL-17A and IL-22 production was assessed after 5 days by flow cytometry.
FACS-sorted Dome CD11b+, CD8α+ DCs, LysoDCs and LysoMacs from PPs (5 × 103), GM-DCs and GM-Macs (5 × 104) or MACS-purified GM-CSF BM-derived CD11c+ cells (1 x105) were co-cultured with FACS-sorted Celltrace Violet-labeled naive CD4+CD62L+ CD44- OT-II T (5x103 or 5 x104, respectively), and chicken ovalbumin peptide (OVA323-339, 100 μM) in 200 μL RPMI complete medium in 96-well round-bottom plates. GM-CSF BM-derived CD11c+ cells, GM-DCs and GM-Macs were loaded previously with mucosa-associated commensals at a 10:1 ratio, in the presence of 100 U/mL penicilin, 100 μg/mL streptomycin and 50 μg/mL gentamycin. After 72h, cell proliferation was analyzed by Celltrace Violet dilution (flow cytometry) and supernatants assessed for IL-17 or IFN-γ quantification by ELISA. For re-stimulation, cells harvested after 72 h of primary culture were stimulated with PMA and ionomycin for 6 h (intracellular cytokine detection) in the presence of Brefeldin A in the last 4 hours of culture. Cells were stained for CD4, fixed with 4% PFA, and incubated with anti-IL-17, anti-IL-22 or anti-IFN-γ during permeabilization with 0.1% saponin.
For intracellular IL-12p40 and IL-6 staining in vivo, mice were intraperitoneally inoculated with Brefeldin A (125 μg/mouse). PPs were recovered 12 h after brefeldin A injection. The cells obtained were fixed and permeabilized and were stained with PE conjugated anti-IL-12p40 antibody (Tonbo) or APC anti-mouse IL-6 antibody or its isotype control antibody APC Rat IgG1, κ (Biolegend).
Flow cytometry
Cells were pre-incubated for 10 min at 4°C with anti-mouse CD16/CD32 (clone 2.4G2, Tonbo Bioscience) or anti human Fc block (BD Biosciences) to block unspecific antibody binding toward murine or human cells, respectively; before staining with the appropriate antibodies. Samples for flow cytometry were stained in FACs buffer. Anti-mouse antibodies to CD45.1 (APC, PerCP-Cy5.5, efluor450), CD45 efluor450, CD44 FITC, CD62L PE, CD8α PE, CD11b APC, biotin CCR7, IFN-γ APC, CD172a (SIRPα)(biotin, APC), Ly6C PerCP-Cy5.5, IgA PE and anti-human IgG (Fc gamma-specific) PE or biotin, were obtained from eBioscience.
Anti-mouse I-A/I-E (MHCII) FITC, CD44 V450, CD4 (APC, PerCP-Cy5.5), CD19 PE, CD11c (APC, PE), Siglec-F BV421, CD103 BUV395, CD11c BUV395, CD40 APC, IL17A (PE, APC-Cy7), CD45R/B220 BV421, CD11b FITC, Ly6G PE, and Streptavidin BV786 were from BD Biosciences.
Anti-mouse F4/80 FITC, CD64 PECy7, I-A/I-E (MHCII) AF700, Ly6G BV510, CD11b BV605, CD172a (Sirpα) PerCPCy5.5, CD4 PECy7, CD45 BV510, CD90.2 (Thy-1.2) APC, CD196 (CCR6) PE, IL-17A BV605, IL-22 (PE, PerCP-Cy5.5), CD185 (CXCR5) (PE, APC), CD279 (PD-1) (biotin, BV421), IgG1 PerCP-Cy5.5, CD11b APC-Cy7, MERTK (Mer) PE, CD317 (BST2, PDCA-1) APC, CD197 (CCR7) PE and anti-human HLA-DR BV570, CD11c AF700, CD45 FITC were from Biolegend.
Anti-mouse CD86, CD4 PE, CD3 FITC was from Tonbo Biosciences. Anti-mouse CD115 biotin was from ThermoFisher. Mouse and human Mincle were stained with 1B6 anti-Mincle (MBL) biotinylated by using a biotinylation Kit (Abcam) or its isotype control antibody biotin rat IgG1, κ (Biolegend). Non-cell permeant Hoechst 33258 (0.1 μM, ThermoFisher) was used as a counterstain to detect murine necrotic cells, while human dead cells were excluded from the analysis using a live/dead fixable near-infrared dead cell stain kit (Molecular Probes). Data was acquired on a LSR Fortessa (BD Biosciences) and analyzed with FlowJo software version 10 (TreeStar).
Microbiota and mucosa-associated commensals preparation and binding studies
For total microbiota analysis, fresh content from the whole intestine, except caecum, of SPF or germ-free mice was collected from mice with ad libitum access to food and water. For mucosa-associated commensals isolation, SPF mice or mice treated with antibiotics in utero until the weaning and gavaged with Lactobacillus plantarum (L. plantarum) were starved for 18 hours prior to collecting the mucus by gentle scraping of the epithelium from the small intestine as previously described (Cohen and Laux, 1995). In both cases, the samples were vortexed, filtered by 70 μm filter, centrifuged at 600 g 1 min to remove large particles, the supernatant was centrifuged at 13000 g for 3 min and the pellet was diluted in to an OD600nm of 0.6 or count by flow cytometry by using Bacteria Counting Kit (Thermo Fisher Scientific) as described (Veal et al., 2000). The analysis by flow cytometry was done as previously described (Eriksson et al., 2013). Mincle-hFc, Dectin-1-hFc generated as described (Iborra et al., 2016), Dectin-2-hFc or Mincle-hFc (R&D Systems) and control-hFc (R&D Systems) were prepared in 10% skimmed milk in PBS were incubated overnight at 4°C on a rotating wheel, washed three times and labeled with PE-conjugated goat anti-hFc antibody (eBioscience). SYTO 61 red fluorescent nucleic acid stain (ThermoFisher) at 2.5 mM was then incorporated and washed three time in PBS. For IgA-coated bacteria analysis, anti-mouse IgA PE (eBioscience) was added together with SYTO 61 red fluorescent nucleic acid stain.
For Mincle-hFc bound bacteria purification, anti-hFc stained bacteria were incubated with anti-PE microbeads (Miltenyi), washed twice and then purified. After MACS separation, part of the negative and the positive fractions were collected for 16S sequencing analysis.
Human Mincle-Fc chimera was blocked with anti-human Mincle 2F2 clone (Sigma-Aldrich); isotype matched antibody (clone GC323, mouse IgM) was used as a control (Sigma Aldrich) or with Trehalose-6,6-dibehenate (TDB) (InvivoGen) prior microbiota staining assay.
CellTracer Violet Labeling of Lactobacillus plantarum
L. plantarum (ATCC14917) was grown overnight on MRS Broth medium (Oxoid) at 37°C in aerobic conditions to approximately OD600nm of 0.5. Bacteria were labeled with CellTracer Violet (100 μg/mL, Invitrogen). Bacteria were incubated 10 min at 30°C with constant agitation, washed twice and inoculated by gavage in SPF WT mice starved for 18 hours and previously intraperitoneally injected with 3 mg of cimetidine HCl (Sigma Aldrich) and 0.02 mg of sincalide (Tebu-bio). The mucosa-associated commensals were collected 6 hours later for flow cytometry analysis.
RNA isolation and quantitative PCR
Total RNA from the liver parenchymal fraction, small intestine, ileum or PPs was isolated using Trizol (Thermo Fisher Scientific) in combination with RNeasy Mini Kit (QIAGEN) accordingly to manufacturer’s instructions. RNA was reverse transcribed to cDNA using random hexamers and High Capacity cDNA Reverse Transcription Kit (Applied Byosystem). Quantitative PCR amplification was performed with the GoTaq qPCR Master Mix (Promega) in a 7900HT Fast Real-Time PCR System (Applied Byosystem). All reactions were done in triplicate, following the manufacturer’s instructions. Data were normalized to Gadph or bactin indicated in each case and displayed as relative values. Primer sequences can be found in Table S1.
Immunofluorescence, confocal microscopy, and histology
For immunofluorescence of phospho-Syk (P-Syk), purified CD11c+ GM-BMs were adhered to coverslip and fixed with Fixation/Permeabilization Solution Kit (BD biosciences) after stimulation. Anti-P-Syk (C87C1) antibody from Cell Signaling was added and incubated at 4°C overnight. Samples were washed and anti-P-Syk was detected using the tyramide signal amplification (TSA) detection kit Alexa Fluor 568 (Molecular Probes) as recommended by the manufacturer. For mucosa-associated commensals, mucus was adhered to coverslips coated with poly-L lysine (50 μg/mL, Sigma Aldrich) overnight at 4°C. Coverslips were washed in PBS and fixed in 2% PFA for 10 min at RT. Samples were then incubated with blocking solution (2% skimmed milk in PBS) for 60 min at room temperature and stained with Mincle-hFc or Control-hFc followed by anti-human IgG (Fc gamma-specific) biotin (eBioscience) and Streptavidin-Alexa 488 (ThermoFisher). Finally, samples were covered with ProLong Gold Antifade Reagent liquid mountant (Life Technologies) and visualized using a Leica SP8 STED super resolution microscope. For Mincle expression analysis, PPs whole mount preparations were done as described (Rios et al., 2016) and stained with anti-CD11c antibody (Clone 3.9, Abcam), biotin anti-mouse Mincle (Clone 1B6, MBL), PE labeled anti-CD11b (Clone M1/70, BD biosciences), Alexa Fluor 647 Chicken Anti Mouse IgG (H+L) (Molecular Probes), Alexa Fluor 488 Streptavidin (Thermo Fisher Scientific) and DAPI (Thermofisher). The samples were fixed, covered as described and visualized using a Zeiss LSM 780 confocal microscope. Finally, anti-human Mincle antibody (2A8, Abcam) was used for IHC on paraffin sections of healthy human colon samples as previously described (Iborra et al., 2016).
Enzyme-linked immunosorbent assays (ELISA)
IL-17 or IFN-γ levels were determined using the mouse IL-17A (homodimer) ELISA Ready-SET-Go (Affymetrix) or the mouse IFN-γ ELISA Set (BD OptEIA) according to the manufacturer’s instructions. IL-23 and IL-6 were quantified using matched antibody pair ELISA. Anti-mouse IL-23 p19 (5B2) together with biotinylated anti-mouse IL-12/IL-23 p40 (C17.8) both from Thermo Fisher Scientific. Anti-Mouse IL-6 (MP5-20F3) (BD Biosciences) with biotinylated anti-mouse IL-6 (MP5-32C11) (Thermo Fisher Scientific). Flagellin or LPS specific IgA were measured in the intestinal lavage as previously described (Cullender et al., 2013). Briefly, corning 96 well EIA/RIA plates (Cultek) were coated overnight at 4°C with 1 μg/mL of purified flagellin from Salmonella Typhimurium (InvivoGen) or 1 μg/well of LPS EB (InvivoGen). To measure total IgA, the plates were coated with goat anti-mouse IgA antibody (Bethyl) and the standard curve done by using purified mouse IgA, κ isotype control (BD PharMingen) as previously described (Bunker et al., 2015). To analyze serum commensal bacteria-specific IgG, intestinal bacteria were prepared to coat ELISA plates, after heat-killing at 85°C for 1 hour, following a modified protocol from the previously described (Zeng et al., 2016). The plates were washed 3 times with PBS containing 0.05% Tween 20 and blocked with PBS containing 10% FBS for 1 hour at room temperature. Intestinal lavage was prepared by vortexing the intestinal content collected in 3 mL of PBS during 3 min. Samples were centrifuged at 8,000 g for 2 min and removed the supernatant for assaying. Intestinal content supernatants or mice serum (to IgG analysis) were applied to the wells after the blocking step and incubated at 4°C o/n. The antibody used for detection was goat anti mouse IgA HRP (Southern Biotech) or goat anti mouse IgG-heavy and light chain HRP (Bethyl) together with Sure Blue TMB 1-Component Microwell Peroxidase Substrate and TMB Stop Solution (SeraCare). The absorbance was read at 450 nm.
Adoptive transfer experiments
Naive CD4+ T cells from B6/SJL mice expressing CD45.1 were negatively selected using a cocktail of biotin-conjugated antibodies (anti-CD11c, CD11b, B220, MHC-II, CD8, GR1, CD16/32 (BD Bioscience) and anti-CD25, CD44 (Biolegend) followed by separation with Streptavidin-microbeads (Miltenyi Biotec). 6x106 cells were adoptively transferred into recipient mice. IL-17 and IL-22 intracellular production by flow cytometry in PPs T cells was determined 14 days after transfer by PMA and Ionomycin re-stimulation and intracellular cytokine staining followed by flow cytometry.
Antibiotic Treatment, SFB quantification, and Lactobacillus plantarum colonization
For ablation of intestinal bacteria, an antibiotic cocktail of 1 g/L each of Ampicillin (Normon), Neomycin sulfate (Sigma Aldrich), Metronidazole (Sanofi); of 0.5 g/L Vancomycin (Normon) and of 4 mg/mL Sucralose (Sigma Aldrich) was used as previously described (Rakoff-Nahoum et al., 2004). For selective depletion, Vancomycin alone was also used as previously described (Ivanov et al., 2008). Antibiotics were added into the drinking water on a weekly basis. For treatment from birth, breeding pairs were kept with the antibiotic cocktail. Control cages were kept on regular water plus sucralose.
SFB in feces was detected by PCR (IDEXX Bioresearch). For SFB quantification in feces, microbial DNA was isolated from mouse fecal samples using QIAamp PowerFecal Pro DNA kit (QIAGEN), according to manufacturer’s instructions. SFB DNA was then detected by quantitative PCR with SFB specific primers (see Table S1) and SYBR-Green PCR master mix. SFB quantification was performed with reference to a standard curve generated with quantified plasmid DNA where SFB 16S rRNA gene has been cloned in PCR-blunt II vector (ThermoFisher) according to suppliers’ instructions.
Mice treated with antibiotics in utero until the weaning, were inoculated by gavage with L. plantarum. One hour prior to the bacterial gavage mice were injected intraperitoneally with 3 mg of cimetidine HCl (Sigma Aldrich) and 0.02 mg of sincalide (Tebu-bio) in 100 μl of PBS to inhibit stomach acid secretion and empty the gallbladder to improve the colonization as previously described (Iida et al., 2013).
For preparation of bacterial inocula, L. plantarum was grown overnight on MRS Broth medium (Oxoid) at 37°C in aerobic conditions. The bacteria were suspended in PBS. Mice were gavaged with 200 μL of inoculum (dose 1x106). Mice received doses every two days during the four weeks.
FITC-Dextran permeability assay
Mice were orally gavaged with 4-kDa fluorescein isothiocyanate (FITC)–dextran probe (Sigma-Aldrich) (60 mg/100 g body weight). Blood was collected 1 and 4 hours after and FITC-dextran measurements were performed in duplicate by fluorometry. Dilutions of FITC-dextran in PBS were used as a standard curve, and absorption of 100 μL of serum or standard was measured in a fluorimeter (Fluoroskan ascent, Thermo Scientific) at 488 nm.
Analysis of bacterial translocation
Liver was aseptically removed and homogenized in sterile-filtered 0.05% NP40 1% BSA PBS before plating serial dilutions on LB agar plates, which were then incubated at 37°C in aerobic conditions without light.
Bacterial DNA isolation and 16S gene sequencing
Ultra-Deep Microbiome Prep10 (Molzym) was used accordingly to manufacturer’s instructions to isolate enriched microbial DNA from fresh liver and to remove of animal host DNA. Microbial genomic DNA from bound Control-hFc, Mincle-hFc or not bound Mincle-hFc fractions or Mincle deficient mice and their WT littermates intestinal content was isolated by using QIAamp DNA Mini Kit or QIAamp DNA Stool Mini Kit (QIAGEN) respectively. PCR and sequencing were performed following the 16S Metagenomic Sequencing Library Preparation guide (Illumina Inc.) The V3-V4 region of 16S rDNA was amplified using the primers recommended in this guide (see Table S1). One PCR reaction was performed per sample and one single amplicon of approximately 460 bp was created. 2x Kapa HiFi Hot Start Ready Mix (Kapa Biosystems) was used for the first amplification step. AMPure XP beads (Beckman Coulter) were used to purify the amplicons and to remove free primers and dimer species. Each amplicon (5 μl) were used as template in a second Index PCR. This second PCR attached dual indices and Illumina sequencing adapters to each amplicon using the Nextera XT index Kit (Illumina). An additional clean-up step was performed with the AMPure XP beads. All the libraries were run in a Bioanalyzer DNA 1000 chip (Agilent Tecnologies) to verify the size of the product. Libraries were quantified by fluorometric method (Qubit dsDNA) and quality was check in the 2100 Agilent Bioanalyzer. All libraries were diluted to the same concentration, using Illumina Resuspension Buffer (RSB) and were combine an equal volume of all of them in the same pool to sequence. The sample pool (10 nM) was denatured with 0.2 N NaOH, then diluted to 6 pM and combined with 50% (v/v) denatured 6 pM PhiX, prepared following Illumina guidelines. Samples were sequenced on the MiSeq sequencing platform, using a 2 × 300 cycle V3 kit, following standard Illumina sequencing protocols.
Processing of 16S sequencing data
Samples were analyzed using BaseSpace Application 16S Metagenomics v1.0 (Illumina) to determine the bacterial taxonomical composition. This program use Illumina-curated version of the GreenGenes taxonomic database and RDP Classifier (Ribosomal Database Project). For some additional data analysis, the “Quantitative Insights into Microbial Ecology” software (QIIME version 1.9.0) was used (Caporaso et al., 2010). Processed reads were then clustered in Operational taxonomic units (OTUs) using UCLUST with a similarity threshold of 0.97 and were aligned using PyNast against 16S reference database GreenGenes version 13.8 using default parameters. Enrichment index was obtained by comparing the relative abundance of each genus in Mincle-hFc-enriched and depleted fractions. Specificity index was calculated by comparing the relative abundance of each genus in Mincle-hFc and Control-hFc enriched fractions.
Serum hepatic parameters and metabolomics profiling
Alanine aminotransferase (ALT), Aspartate aminotransferase (AST), total and indirect bilirubin were measured by using an automatic biochemical analyzer (Dimension RxL Max Integrated Chemistry System, Siemens).
For metabolomics profiling, liver was collected after perfusion through the heart with cold PBS. An aliquot of approx. 20 mg was immediately freeze-clamped in liquid nitrogen and stored at −80°C until lipid extraction and analysis. Liver tissue was lysed in cold MeOH:H2O (1:1, sample:solvent 1:10 ratio) with a TissueLyser LT homogenizer (QIAGEN). Afterward, liver homogenate was split in two aliquots for the metabolomic profiling of diacylglycerides (DAG) and free fatty acids (FFA) using HPLC-ESI-MS-QTOF and GC-EI-MS-QTOF (Agilent Technologies) as previously reported (González-Peña et al., 2017). For DAG profiling, the resulting data matrix is composed of all individual species of DAGs detected in the samples sorted by their characteristic retention time and neutral mass, and the abundance of each compound for each sample. DAGs were identified by the exact mass and by the elucidation of MS/MS spectra obtained using a LC-MS/MS analysis in production mode. In contrast, for FFA profiling, the resulting matrix is made of all individual species of FFAs detected in the samples sorted by their characteristic retention time and target ion, and the abundance of each compound for each sample. FFAs were identified by comparing their retention time, retention index and mass fragmentation patterns with those available in an in-house library as described (Mastrangelo et al., 2015)
Quantification and Statistical Analysis
Statistical analysis was performed using GraphPad Prism 5 (GraphPad Software). Statistical significance was determined by parametric Student’s t test, by Mann & Whitney’s U test or one-way ANOVA with Bonferroni post hoc test unless otherwise stated. Differences with p values ≤ 0.05 were considered significant. ∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001. n represents number of biological replicates.
Acknowledgments
We are grateful to members of the D.S. laboratory and Dr. E. Fernández-Malavé for discussions and critical reading of the manuscript. We appreciate the support of A. Tomás-Loba, G. Sabio, P. Martín, A. Tsilingiri, A.R. Ramiro, C.L. Abram, C.A. Lowell, J.M. García-Lobo, M. Molina, and M.C. Rodríguez for providing reagents and support. We thank the staff at the Fundación Centro Nacional de Investigaciones Cardiovasculares Carlos III (CNIC) facilities for technical support. M.M.-L. received a Formación de Personal Universitario (FPU) fellowship (AP2010-5935) from the Spanish Ministerio de Educación. S.I. is funded by grant SAF2015-74561-JIN from the Spanish Ministerio de Ciencia, Innovación, y Universidades (MCIU) and Fondos Europeos de Desarrollo Regional (FEDER). G.D.B and D.M.R. are supported by the Wellcome Trust and the MRC Centre for Medical Mycology at the University of Aberdeen. S.L.L. is supported by the Swiss National Science Foundation (PP00P3_150758). Work in the D.S. laboratory is funded by the CNIC and grant SAF2016-79040-R from MCIU, the Agencia Estatal de Investigación, and FEDER; B2017/BMD-3733 Immunothercan-CM from Comunidad de Madrid; RD16/0015/0018-REEM from FIS-Instituto de Salud Carlos III, MCIU, and FEDER; the Acteria Foundation; the Constantes y Vitales prize (Atresmedia); La Marató de TV3 Foundation (201723); the European Commission (635122-PROCROP H2020), and the European Research Council (ERC-2016-Consolidator Grant 725091). The CNIC is supported by the MCIU and the Pro-CNIC Foundation and is a Severo Ochoa Center of Excellence (SEV-2015-0505).
Author Contributions
Conceptualization, M.M.-L., S.I., S.L.-L., and D.S.; Methodology, all authors; Investigation, M.M.-L., S.I., R.C.-G., A.M., E.R.M., and V.G.-R.; Writing – Original Draft, M.M.-L., S.I., D.S.; Writing – Review & Editing, all authors; Main Funding Acquisition, D.S. and S.I.; Resources, D.M.R., G.D.B., D.B., M.C., and J.P.G.; Supervision S.I., S.L.-L., and D.S.
Declaration of Interests
B.K. and M.J.R. are employees of MedImmune and are AstraZeneca shareholders. L.M. and A.D. are employees of Miltenyi Biotec. The rest of the authors declare no conflict of interests.
Published: January 29, 2019
Footnotes
Supplemental Information includes seven figures and one table and can be found with the article online at https://doi.org/10.1016/j.immuni.2018.12.020.
Contributor Information
Salvador Iborra, Email: siborra@ucm.es.
David Sancho, Email: dsancho@cnic.es.
Supplemental Information
References
- Adachi O., Kawai T., Takeda K., Matsumoto M., Tsutsui H., Sakagami M., Nakanishi K., Akira S. Targeted disruption of the MyD88 gene results in loss of IL-1- and IL-18-mediated function. Immunity. 1998;9:143–150. doi: 10.1016/s1074-7613(00)80596-8. [DOI] [PubMed] [Google Scholar]
- Allen J.N., Dey A., Nissly R., Fraser J., Yu S., Balandaram G., Peters J.M., Hankey-Giblin P.A. Isolation, characterization, and purification of macrophages from tissues affected by obesity-related inflammation. J. Vis. Exp. 2017;(122) doi: 10.3791/55445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K., Tanoue T., Ando M., Kamada N., Nagano Y., Narushima S., Suda W., Imaoka A., Setoyama H., Nagamori T. Th17 cell induction by adhesion of microbes to intestinal epithelial cells. Cell. 2015;163:367–380. doi: 10.1016/j.cell.2015.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäumler A.J., Sperandio V. Interactions between the microbiota and pathogenic bacteria in the gut. Nature. 2016;535:85–93. doi: 10.1038/nature18849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behler F., Maus R., Bohling J., Knippenberg S., Kirchhof G., Nagata M., Jonigk D., Izykowski N., Mägel L., Welte T. Macrophage-inducible C-type lectin Mincle-expressing dendritic cells contribute to control of splenic Mycobacterium bovis BCG infection in mice. Infect. Immun. 2015;83:184–196. doi: 10.1128/IAI.02500-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekiaris V., Persson E.K., Agace W.W. Intestinal dendritic cells in the regulation of mucosal immunity. Immunol. Rev. 2014;260:86–101. doi: 10.1111/imr.12194. [DOI] [PubMed] [Google Scholar]
- Belkaid Y., Hand T.W. Role of the microbiota in immunity and inflammation. Cell. 2014;157:121–141. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnardel J., Da Silva C., Henri S., Tamoutounour S., Chasson L., Montañana-Sanchis F., Gorvel J.P., Lelouard H. Innate and adaptive immune functions of peyer’s patch monocyte-derived cells. Cell Rep. 2015;11:770–784. doi: 10.1016/j.celrep.2015.03.067. [DOI] [PubMed] [Google Scholar]
- Bunker J.J., Flynn T.M., Koval J.C., Shaw D.G., Meisel M., McDonald B.D., Ishizuka I.E., Dent A.L., Wilson P.C., Jabri B. Innate and adaptive humoral responses coat distinct commensal bacteria with immunoglobulin A. Immunity. 2015;43:541–553. doi: 10.1016/j.immuni.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., Fierer N., Peña A.G., Goodrich J.K., Gordon J.I. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen P.S., Laux D.C. Bacterial adhesion to and penetration of intestinal mucus in vitro. Methods Enzymol. 1995;253:309–314. doi: 10.1016/s0076-6879(95)53026-6. [DOI] [PubMed] [Google Scholar]
- Cullender T.C., Chassaing B., Janzon A., Kumar K., Muller C.E., Werner J.J., Angenent L.T., Bell M.E., Hay A.G., Peterson D.A. Innate and adaptive immunity interact to quench microbiome flagellar motility in the gut. Cell Host Microbe. 2013;14:571–581. doi: 10.1016/j.chom.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Silva C., Wagner C., Bonnardel J., Gorvel J.P., Lelouard H. The Peyer’s patch mononuclear phagocyte system at steady state and during infection. Front. Immunol. 2017;8:1254. doi: 10.3389/fimmu.2017.01254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson G.P., Lee S.M., Mazmanian S.K. Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol. 2016;14:20–32. doi: 10.1038/nrmicro3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson M., Johannssen T., von Smolinski D., Gruber A.D., Seeberger P.H., Lepenies B. The C-Type lectin receptor SIGNR3 binds to fungi present in commensal microbiota and influences immune regulation in experimental colitis. Front. Immunol. 2013;4:196. doi: 10.3389/fimmu.2013.00196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung T.C., Artis D., Sonnenberg G.F. Anatomical localization of commensal bacteria in immune cell homeostasis and disease. Immunol. Rev. 2014;260:35–49. doi: 10.1111/imr.12186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaboriau-Routhiau V., Rakotobe S., Lécuyer E., Mulder I., Lan A., Bridonneau C., Rochet V., Pisi A., De Paepe M., Brandi G. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677–689. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- Geijtenbeek T.B., Gringhuis S.I. C-type lectin receptors in the control of T helper cell differentiation. Nat. Rev. Immunol. 2016;16:433–448. doi: 10.1038/nri.2016.55. [DOI] [PubMed] [Google Scholar]
- González-Peña D., Dudzik D., García A., Ancos B., Barbas C., Sánchez-Moreno C. Metabolomic fingerprinting in the comprehensive study of liver changes associated with onion supplementation in hypercholesterolemic wistar rats. Int. J. Mol. Sci. 2017;18:18. doi: 10.3390/ijms18020267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodyear A.W., Kumar A., Dow S., Ryan E.P. Optimization of murine small intestine leukocyte isolation for global immune phenotype analysis. J. Immunol. Methods. 2014;405:97–108. doi: 10.1016/j.jim.2014.01.014. [DOI] [PubMed] [Google Scholar]
- Helft J., Böttcher J., Chakravarty P., Zelenay S., Huotari J., Schraml B.U., Goubau D., Reis e Sousa C. GM-CSF mouse bone marrow cultures comprise a heterogeneous population of CD11c(+)MHCII(+) macrophages and dendritic cells. Immunity. 2015;42:1197–1211. doi: 10.1016/j.immuni.2015.05.018. [DOI] [PubMed] [Google Scholar]
- Hirota K., Turner J.E., Villa M., Duarte J.H., Demengeot J., Steinmetz O.M., Stockinger B. Plasticity of Th17 cells in Peyer’s patches is responsible for the induction of T cell-dependent IgA responses. Nat. Immunol. 2013;14:372–379. doi: 10.1038/ni.2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda K., Littman D.R. The microbiota in adaptive immune homeostasis and disease. Nature. 2016;535:75–84. doi: 10.1038/nature18848. [DOI] [PubMed] [Google Scholar]
- Hu W., Troutman T.D., Edukulla R., Pasare C. Priming microenvironments dictate cytokine requirements for T helper 17 cell lineage commitment. Immunity. 2011;35:1010–1022. doi: 10.1016/j.immuni.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iborra S., Sancho D. Signalling versatility following self and non-self sensing by myeloid C-type lectin receptors. Immunobiology. 2015;220:175–184. doi: 10.1016/j.imbio.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iborra S., Izquierdo H.M., Martínez-López M., Blanco-Menéndez N., Reis e Sousa C., Sancho D. The DC receptor DNGR-1 mediates cross-priming of CTLs during vaccinia virus infection in mice. J. Clin. Invest. 2012;122:1628–1643. doi: 10.1172/JCI60660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iborra S., Martínez-López M., Cueto F.J., Conde-Garrosa R., Del Fresno C., Izquierdo H.M., Abram C.L., Mori D., Campos-Martín Y., Reguera R.M. Leishmania uses mincle to target an inhibitory ITAM signaling pathway in dendritic cells that dampens adaptive immunity to infection. Immunity. 2016;45:788–801. doi: 10.1016/j.immuni.2016.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida N., Dzutsev A., Stewart C.A., Smith L., Bouladoux N., Weingarten R.A., Molina D.A., Salcedo R., Back T., Cramer S. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science. 2013;342:967–970. doi: 10.1126/science.1240527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov I.I., McKenzie B.S., Zhou L., Tadokoro C.E., Lepelley A., Lafaille J.J., Cua D.J., Littman D.R. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- Ivanov I.I., Frutos Rde.L., Manel N., Yoshinaga K., Rifkin D.B., Sartor R.B., Finlay B.B., Littman D.R. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. 2008;4:337–349. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov I.I., Atarashi K., Manel N., Brodie E.L., Shima T., Karaoz U., Wei D., Goldfarb K.C., Santee C.A., Lynch S.V. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose C.S., Artis D. Innate lymphoid cells as regulators of immunity, inflammation and tissue homeostasis. Nat. Immunol. 2016;17:765–774. doi: 10.1038/ni.3489. [DOI] [PubMed] [Google Scholar]
- Krishnaswamy J.K., Gowthaman U., Zhang B., Mattsson J., Szeponik L., Liu D., Wu R., White T., Calabro S., Xu L. Migratory CD11b+ conventional dendritic cells induce T follicular helper cell-dependent antibody responses. Sci. Immunol. 2017;2:2. doi: 10.1126/sciimmunol.aam9169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruglov A.A., Grivennikov S.I., Kuprash D.V., Winsauer C., Prepens S., Seleznik G.M., Eberl G., Littman D.R., Heikenwalder M., Tumanov A.V., Nedospasov S.A. Nonredundant function of soluble LTα3 produced by innate lymphoid cells in intestinal homeostasis. Science. 2013;342:1243–1246. doi: 10.1126/science.1243364. [DOI] [PubMed] [Google Scholar]
- Lécuyer E., Rakotobe S., Lengliné-Garnier H., Lebreton C., Picard M., Juste C., Fritzen R., Eberl G., McCoy K.D., Macpherson A.J. Segmented filamentous bacterium uses secondary and tertiary lymphoid tissues to induce gut IgA and specific T helper 17 cell responses. Immunity. 2014;40:608–620. doi: 10.1016/j.immuni.2014.03.009. [DOI] [PubMed] [Google Scholar]
- LeibundGut-Landmann S., Gross O., Robinson M.J., Osorio F., Slack E.C., Tsoni S.V., Schweighoffer E., Tybulewicz V., Brown G.D., Ruland J., Reis e Sousa C. Syk- and CARD9-dependent coupling of innate immunity to the induction of T helper cells that produce interleukin 17. Nat. Immunol. 2007;8:630–638. doi: 10.1038/ni1460. [DOI] [PubMed] [Google Scholar]
- Lochner M., Ohnmacht C., Presley L., Bruhns P., Si-Tahar M., Sawa S., Eberl G. Microbiota-induced tertiary lymphoid tissues aggravate inflammatory disease in the absence of RORgamma t and LTi cells. J. Exp. Med. 2011;208:125–134. doi: 10.1084/jem.20100052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longman R.S., Diehl G.E., Victorio D.A., Huh J.R., Galan C., Miraldi E.R., Swaminath A., Bonneau R., Scherl E.J., Littman D.R. CX₃CR1⁺ mononuclear phagocytes support colitis-associated innate lymphoid cell production of IL-22. J. Exp. Med. 2014;211:1571–1583. doi: 10.1084/jem.20140678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marakalala M.J., Vautier S., Potrykus J., Walker L.A., Shepardson K.M., Hopke A., Mora-Montes H.M., Kerrigan A., Netea M.G., Murray G.I. Differential adaptation of Candida albicans in vivo modulates immune recognition by dectin-1. PLoS Pathog. 2013;9:e1003315. doi: 10.1371/journal.ppat.1003315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastrangelo A., Ferrarini A., Rey-Stolle F., García A., Barbas C. From sample treatment to biomarker discovery: A tutorial for untargeted metabolomics based on GC-(EI)-Q-MS. Anal. Chim. Acta. 2015;900:21–35. doi: 10.1016/j.aca.2015.10.001. [DOI] [PubMed] [Google Scholar]
- McGeachy M.J., Bak-Jensen K.S., Chen Y., Tato C.M., Blumenschein W., McClanahan T., Cua D.J. TGF-beta and IL-6 drive the production of IL-17 and IL-10 by T cells and restrain T(H)-17 cell-mediated pathology. Nat. Immunol. 2007;8:1390–1397. doi: 10.1038/ni1539. [DOI] [PubMed] [Google Scholar]
- Nakamoto N., Amiya T., Aoki R., Taniki N., Koda Y., Miyamoto K., Teratani T., Suzuki T., Chiba S., Chu P.S. Commensal lactobacillus controls immune tolerance during acute liver injury in mice. Cell Rep. 2017;21:1215–1226. doi: 10.1016/j.celrep.2017.10.022. [DOI] [PubMed] [Google Scholar]
- Persson E.K., Uronen-Hansson H., Semmrich M., Rivollier A., Hägerbrand K., Marsal J., Gudjonsson S., Håkansson U., Reizis B., Kotarsky K., Agace W.W. IRF4 transcription-factor-dependent CD103(+)CD11b(+) dendritic cells drive mucosal T helper 17 cell differentiation. Immunity. 2013;38:958–969. doi: 10.1016/j.immuni.2013.03.009. [DOI] [PubMed] [Google Scholar]
- Pierantonelli I., Rychlicki C., Agostinelli L., Giordano D.M., Gaggini M., Fraumene C., Saponaro C., Manghina V., Sartini L., Mingarelli E. Author Correction: Lack of NLRP3-inflammasome leads to gut-liver axis derangement, gut dysbiosis and a worsened phenotype in a mouse model of NAFLD. Sci. Rep. 2017;7:17568. doi: 10.1038/s41598-017-17187-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plant L., Conway P. Association of Lactobacillus spp. with Peyer’s patches in mice. Clin. Diagn. Lab. Immunol. 2001;8:320–324. doi: 10.1128/CDLI.8.2.320-324.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakoff-Nahoum S., Paglino J., Eslami-Varzaneh F., Edberg S., Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Revy P., Muto T., Levy Y., Geissmann F., Plebani A., Sanal O., Catalan N., Forveille M., Dufourcq-Labelouse R., Gennery A. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2) Cell. 2000;102:565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- Rios D., Wood M.B., Li J., Chassaing B., Gewirtz A.T., Williams I.R. Antigen sampling by intestinal M cells is the principal pathway initiating mucosal IgA production to commensal enteric bacteria. Mucosal Immunol. 2016;9:907–916. doi: 10.1038/mi.2015.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo S., Ikeda S., Yamabe K., Kakuta S., Ishigame H., Akitsu A., Fujikado N., Kusaka T., Kubo S., Chung S.H. Dectin-2 recognition of alpha-mannans and induction of Th17 cell differentiation is essential for host defense against Candida albicans. Immunity. 2010;32:681–691. doi: 10.1016/j.immuni.2010.05.001. [DOI] [PubMed] [Google Scholar]
- Sano T., Huang W., Hall J.A., Yang Y., Chen A., Gavzy S.J., Lee J.-Y., Ziel J.W., Miraldi E.R., Domingos A.I. An IL-23R/IL-22 circuit regulates epithelial serum amyloid a to promote local effector Th17 responses. Cell. 2015;163:381–393. doi: 10.1016/j.cell.2015.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanos S.L., Bui V.L., Mortha A., Oberle K., Heners C., Johner C., Diefenbach A. RORgammat and commensal microflora are required for the differentiation of mucosal interleukin 22-producing NKp46+ cells. Nat. Immunol. 2009;10:83–91. doi: 10.1038/ni.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato A., Hashiguchi M., Toda E., Iwasaki A., Hachimura S., Kaminogawa S. CD11b+ Peyer’s patch dendritic cells secrete IL-6 and induce IgA secretion from naive B cells. J. Immunol. 2003;171:3684–3690. doi: 10.4049/jimmunol.171.7.3684. [DOI] [PubMed] [Google Scholar]
- Satoh-Takayama N., Vosshenrich C.A., Lesjean-Pottier S., Sawa S., Lochner M., Rattis F., Mention J.J., Thiam K., Cerf-Bensussan N., Mandelboim O. Microbial flora drives interleukin 22 production in intestinal NKp46+ cells that provide innate mucosal immune defense. Immunity. 2008;29:958–970. doi: 10.1016/j.immuni.2008.11.001. [DOI] [PubMed] [Google Scholar]
- Satpathy A.T., Briseño C.G., Lee J.S., Ng D., Manieri N.A., Kc W., Wu X., Thomas S.R., Lee W.L., Turkoz M. Notch2-dependent classical dendritic cells orchestrate intestinal immunity to attaching-and-effacing bacterial pathogens. Nat. Immunol. 2013;14:937–948. doi: 10.1038/ni.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa S., Cherrier M., Lochner M., Satoh-Takayama N., Fehling H.J., Langa F., Di Santo J.P., Eberl G. Lineage relationship analysis of RORgammat+ innate lymphoid cells. Science. 2010;330:665–669. doi: 10.1126/science.1194597. [DOI] [PubMed] [Google Scholar]
- Schlitzer A., McGovern N., Teo P., Zelante T., Atarashi K., Low D., Ho A.W.S., See P., Shin A., Wasan P.S. IRF4 transcription factor-dependent CD11b+ dendritic cells in human and mouse control mucosal IL-17 cytokine responses. Immunity. 2013;38:970–983. doi: 10.1016/j.immuni.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta R., Altermann E., Anderson R.C., McNabb W.C., Moughan P.J., Roy N.C. The role of cell surface architecture of lactobacilli in host-microbe interactions in the gastrointestinal tract. Mediators Inflamm. 2013;2013:237921. doi: 10.1155/2013/237921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah S., Nagata M., Yamasaki S., Williams S.J. Total synthesis of a cyclopropane-fatty acid α-glucosyl diglyceride from Lactobacillus plantarum and identification of its ability to signal through Mincle. Chem. Commun. (Camb.) 2016;52:10902–10905. doi: 10.1039/c6cc05631h. [DOI] [PubMed] [Google Scholar]
- Shaw M.H., Kamada N., Kim Y.G., Núñez G. Microbiota-induced IL-1β, but not IL-6, is critical for the development of steady-state TH17 cells in the intestine. J. Exp. Med. 2012;209:251–258. doi: 10.1084/jem.20111703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenberg G.F., Monticelli L.A., Alenghat T., Fung T.C., Hutnick N.A., Kunisawa J., Shibata N., Grunberg S., Sinha R., Zahm A.M. Innate lymphoid cells promote anatomical containment of lymphoid-resident commensal bacteria. Science. 2012;336:1321–1325. doi: 10.1126/science.1222551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z., Unutmaz D., Zou Y.R., Sunshine M.J., Pierani A., Brenner-Morton S., Mebius R.E., Littman D.R. Requirement for RORgamma in thymocyte survival and lymphoid organ development. Science. 2000;288:2369–2373. doi: 10.1126/science.288.5475.2369. [DOI] [PubMed] [Google Scholar]
- Talukdar S., Oh D.Y., Bandyopadhyay G., Li D., Xu J., McNelis J., Lu M., Li P., Yan Q., Zhu Y. Neutrophils mediate insulin resistance in mice fed a high-fat diet through secreted elastase. Nat. Med. 2012;18:1407–1412. doi: 10.1038/nm.2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaishnava S., Yamamoto M., Severson K.M., Ruhn K.A., Yu X., Koren O., Ley R., Wakeland E.K., Hooper L.V. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334:255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veal D.A., Deere D., Ferrari B., Piper J., Attfield P.V. Fluorescence staining and flow cytometry for monitoring microbial cells. J. Immunol. Methods. 2000;243:191–210. doi: 10.1016/s0022-1759(00)00234-9. [DOI] [PubMed] [Google Scholar]
- Weiss G., Maaetoft-Udsen K., Stifter S.A., Hertzog P., Goriely S., Thomsen A.R., Paludan S.R., Frøkiær H. MyD88 drives the IFN-β response to Lactobacillus acidophilus in dendritic cells through a mechanism involving IRF1, IRF3, and IRF7. J. Immunol. 2012;189:2860–2868. doi: 10.4049/jimmunol.1103491. [DOI] [PubMed] [Google Scholar]
- Wells C.A., Salvage-Jones J.A., Li X., Hitchens K., Butcher S., Murray R.Z., Beckhouse A.G., Lo Y.-L.-S., Manzanero S., Cobbold C. The macrophage-inducible C-type lectin, mincle, is an essential component of the innate immune response to Candida albicans. J. Immunol. 2008;180:7404–7413. doi: 10.4049/jimmunol.180.11.7404. [DOI] [PubMed] [Google Scholar]
- Whitney P.G., Bär E., Osorio F., Rogers N.C., Schraml B.U., Deddouche S., LeibundGut-Landmann S., Reis e Sousa C. Syk signaling in dendritic cells orchestrates innate resistance to systemic fungal infection. PLoS Pathog. 2014;10:e1004276. doi: 10.1371/journal.ppat.1004276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng M.Y., Cisalpino D., Varadarajan S., Hellman J., Warren H.S., Cascalho M., Inohara N., Núñez G. Gut microbiota-induced immunoglobulin G controls systemic infection by symbiotic bacteria and pathogens. Immunity. 2016;44:647–658. doi: 10.1016/j.immuni.2016.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.