Abstract
The overexpression of multidrug efflux pumps is an important mechanism of clinical resistance in Gram-negative bacteria. Recently, four small molecules were discovered that inhibit efflux in Escherichia coli and interact with the AcrAB-TolC efflux pump component AcrA. However, the binding site(s) for these molecules was not determined. Here, we combine ensemble docking and molecular dynamics simulations with tryptophan fluorescence spectroscopy, site-directed mutagenesis, and antibiotic susceptibility assays to probe binding sites and effects of binding of these molecules. We conclude that clorobiocin and SLU-258 likely bind at a site located between the lipoyl and β-barrel domains of AcrA.
Introduction
According to a report from the Centers for Disease Control and Prevention, approximately two million illnesses and 23,000 deaths are caused by antibiotic resistance each year in the United States alone (1). Some strains of Escherichia coli are now resistant to at least six different classes of antibiotics, and strains of Pseudomonas aeruginosa are now resistant to all classes of antibiotics (2).
One of the primary mechanisms for multidrug resistance in Gram-negative bacteria such as E. coli and P. aeruginosa is the overexpression of multidrug efflux pumps, which expel antibiotics taken up by the bacteria (2). In E. coli, the major multidrug efflux pump is AcrAB-TolC, and its counterpart in P. aeruginosa is MexAB-OprM (3). AcrAB-TolC consists of three proteins, AcrA, AcrB, and TolC, which upon assembly, span both membranes and the periplasm (4). A cryo-electron microscopy (cryo-EM) structure of the assembled pump in the apo state was resolved at 16 Å resolution (5). Subsequently, cryo-EM structures of both the apo state and inhibitor (MBX2931)-bound state were resolved at 6.5 Å (6). In agreement with biochemical and genetic studies, these structural analyses showed that the complex has a stoichiometry of 3:6:3 for AcrB, AcrA, and TolC, respectively. Each protomer of the AcrB homotrimer consists of both a transmembrane and a periplasmic domain, whereas each protomer of the TolC homotrimer consists of a β-barrel domain and a mostly α-helical periplasmic domain. The complex is assembled by the membrane fusion protein AcrA. Six AcrA protomers interact as a trimer of dimers in the assembled pump, connecting the inner membrane transporter, AcrB, to the outer membrane component, TolC (6).
AcrA consists of four domains connected by flexible linkers: the α-helical hairpin, lipoyl, β-barrel, and membrane-proximal (MP) domains (Fig. 1) (7). The α-helical hairpin domain interacts with the coiled-coil domains of TolC (5, 8). The lipoyl and β-barrel domains are the most conserved features of diverse membrane-fusion proteins. The MP domain is contained primarily within the C-terminal region of the AcrA sequence but also includes an N-terminal β-strand. This highly flexible domain binds directly to one monomer of the inner membrane transporter AcrB (9). The 6.5-Å cryo-EM structure of the apo complex revealed spacious gaps between each AcrA dimer through which antibiotics might be inadvertently expelled into the periplasm. However, in cryo-EM models with the inhibitor MBX2931 bound to AcrB, the gaps between AcrA dimers are absent and the channel in TolC is open, thus presumably allowing antibiotics and other efflux pump substrates to exit the cell (6).
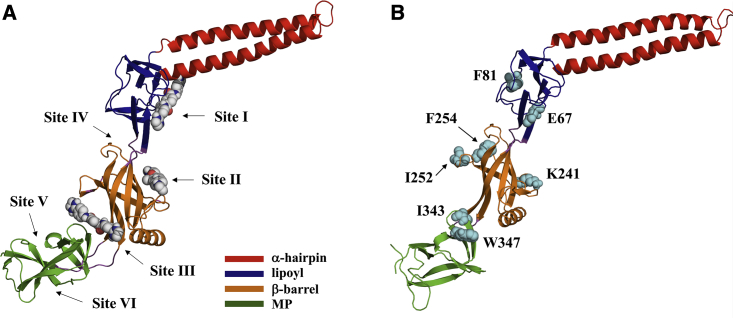
Figure 1.
(A) Inhibitor binding sites predicted from blind ensemble docking of SLU-258, clorobiocin, and novobiocin (sites I–III) and additional sites predicted by FTMap (sites IV–VI). Sites I, II, and III are located between the α-hairpin and lipoyl domains, in the β-barrel domain, and between the β-barrel and MP domain, respectively. Site IV is located between the β-barrel and lipoyl domain, and sites V and VI are located in the MP domain. For clarity, only SLU-258 is shown. (B) Residues mutated individually to Trp for fluorescence quenching experiments are labeled and shown as cyan spheres. The intrinsic Trp in wild-type AcrA, W347, was mutated to Phe. To see this figure in color, go online.
Whereas most efflux pump inhibitors target the transporter, AcrB, recently the assembly of the pump has been targeted by searching for molecules that bind to AcrA. A combination of virtual screening, antibiotic potentiation assays, and surface plasmon resonance led to the discovery of four molecules NSC227186, NSC60339, NSC33353, and NSC305798 that inhibit efflux in E. coli and interact with AcrA (10). Each molecule potentiated the activity (i.e., decreased the minimal inhibitory concentration (MIC)) of the antibiotics novobiocin and erythromycin and bound AcrA in vitro. NSC227186, also called clorobiocin, is an aminocoumarin antibiotic that differs from novobiocin by a chloro-substitution of the C-8′ methyl group and a pyrrole substitution of the 3-O″-carbamoyl group in novobiocin. Similar to novobiocin, NSC227186 is an excellent substrate for the AcrAB-TolC efflux pump and directly binds to AcrA, but only NSC227186 and not novobiocin inhibits efflux of erythromycin (10). Using an in vivo proteolysis approach, it was also found that the inhibitor NSC60339 (SLU-258) but not the other three compounds altered the structure of AcrA. In the absence of SLU-258, cleavage of AcrA by trypsin occurred at two sites, R296 and K396, whereas addition of this inhibitor also resulted in cleavage at K346, indicating that SLU-258 likely alters the structure of AcrA in a manner that increases the accessibility of K346. Subsequently, structure-activity relationships inferred from a docked pose of SLU-258 to AcrA helped guide the modification of the chemical scaffold of SLU-258 to develop second-generation efflux pump inhibitors targeting AcrA (11). Two additional synthesized analogs, SLU-417 and SLU-225, were identified that permeated the outer membrane, increased efflux inhibition compared to SLU-258, and potentiated novobiocin and erythromycin better than SLU-258.
Possible interactions between analogs and AcrA and the specific binding site(s) for these inhibitors remain to be definitively determined (10). Further development of improved efflux pump inhibitors that target AcrA would benefit from better binding site characterization and a knowledge of allosteric communication that might be inhibited by binding and disrupt pump assembly. Here, we combine structural modeling of AcrA and computational ensemble docking (12) with tryptophan (Trp) fluorescence spectroscopy, site-directed mutagenesis, and antibiotic susceptibility assays to examine potential binding sites and effects of binding of SLU-258 and clorobiocin.
Materials and Methods
Strains and plasmids
Wild-type and ΔAcrAB strains of E. coli and their hyperporinated (“-pore”) variants used in this study are derivatives of BW 25113. The E. coli BL21 (DE3) strain was used for overexpression and purification of AcrA variants.
Site-directed mutagenesis
All substitutions in the acrA gene were constructed by QuikChange Lightning Site-Directed Mutagenesis kit (Agilent, Santa Clara, CA) using either pET28AcrA (for in vitro analyses) or p151AcrAB (for in vivo studies) as templates (13). Introduced mutations and the lack of undesired mutations were verified by DNA sequencing (Oklahoma Medical Research Foundation, Oklahoma City, OK).
Antibacterial susceptibility assays
Susceptibilities of wild-type E. coli cells and all AcrA mutants were determined by a twofold broth dilution method (14). E. coli cells with native and hyperporinated outer membranes were used to analyze the activities of efflux pumps with low and high rates of influx across the outer membrane, respectively (15). Cells were grown in Luria-Bertani broth (tryptone, 10 g/L; yeast extract, 5 g/L; NaCl, 5 g/L) at 37°C with shaking at 200 rotations per minute. To induce expression of the pore, when the cells reached an optical density at 600 nm of 0.3–0.4, L-arabinose (final concentration of 0.1%) was added, and the cells were further incubated for 2–3 h until the optical density at 600 nm reached 1.0. The MICs were measured in 96-well microtiter plates. Exponentially growing cells were inoculated into wells containing Luria-Bertani medium in the presence of novobiocin, erythromycin, tetracycline, and sodium dodecyl sulfate (SDS) at a constant concentration of inducer. Cell growth after incubation at 37°C for 18 and 24 h was determined visually or using a Spark 10M microplate reader (Tecan, Männedorf, Switzerland).
Protein expression and purification
For protein expression and analyses, membrane fractions were isolated from E. coli cells and analyzed by SDS polyacrylamide gel electrophoresis followed by immunoblotting with primary monoclonal anti-AcrA (1:60,000 dilution) and anti-AcrB (1:4000 dilution) antibodies followed by a secondary alkaline phosphatase-conjugated anti-rabbit immunoglobulin antibody (Sigma, St. Louis, MO). 5-bromo-4-chloro-3-indolylphosphate and nitroblue tetrazolium were used to visualize the bands. All purified AcrA variants used in this study contained the previously reported S362C substitution and a C-terminal six-histidine tag to facilitate protein purification by metal-affinity chromatography (13). Proteins were purified as described previously (16).
Fluorescence spectroscopy
Steady-state fluorescence analyses were carried out using a RF-5301PC spectrofluorophotometer (Shimadzu, Japan). Proteins were analyzed at 0.2 and 0.5 μM concentrations in the presence of increasing concentrations of inhibitors from 0 to 50 μM. The fluorescence intensity was monitored at 340 nm with a 5-nm bandwidth (excitation at 280 nm with a 5-nm bandwidth) and corrected for sample dilution and inner filter effects (17). The equilibrium association constants for single titrations (Kas) were determined by fitting the theoretical dependence of the fluorescence intensity on the total concentration of the compound to the experimental data points using the following equation (18):
| (1) |
where F is measured fluorescence intensity, F0 is the initial fluorescence intensity, Δf is the difference between the fluorescence intensities of the apo-AcrA and the complex, and fL is the fluorescence intensity of the ligand.
The final KD values were calculated as weighted averages of three independent titrations, with the weights taken as the reciprocal SD squared. Numerical least-squares nonlinear regression analysis was performed using ORIGIN 9.0.
Time-resolved fluorescence measurements were done using a FluoTime 300 fluorometer (PicoQuant, Berlin, Germany). The excitation source was a pulsed 292-nm laser diode, whereas emission settings matched the maximum of the protein emission band: 330 nm for wild-type, F81, F254, and I343 and 350 nm for E67, K241, and I252, respectively. The lifetimes were acquired at magic angle conditions in 2- × 2-mm optical path cuvettes. The protein and ligand concentrations were 5 and 100 μM, respectively. Recorded fluorescence lifetime decays were analyzed and fitted with FluoFit software according to the following expression:
| (2) |
where I(t) is the intensity at time t, IRF is the instrument response function, Ai is the amplitude of component i, and τi is the lifetime component.
Fluorescent quenching by iodide was analyzed according to the Stern-Volmer relationship:
| (3) |
| (4) |
where F0 is the fluorescence intensity in the absence of the quencher, F is the fluorescence intensity with the quencher, kq is the quenching rate coefficient, τ0 is the fluorescence lifetime of the excited state, and KSV is the Stern-Volmer quenching constant (19).
Biolayer interferometry
Biolayer interferometry (BLI) experiments were carried out on an Octet Red96 system (ForteBio, Fremont, CA). AcrA was immobilized on the sensor tips (AR2G Amine) using the primary amine-coupling protocol recommended by the manufacturer. The assays were carried out in the binding buffer containing 20 mM 2-(N-morpholino)ethanesulfonic acid (MES-KOH; pH 6.0), 0.03% dodecyl maltoside, and 150 mM NaCl (MES buffer) at 24°C. The protein-loaded tips were sequentially placed in a buffer supplemented with 1% dimethyl sulfoxide for 1 min to reach a stable baseline, then in buffer supplemented with increasing concentrations of SLU-417 or clorobiocin and 1% dimethyl sulfoxide for 1 min, and then placed back into the baseline buffer to allow dissociation to occur for 2–3 min. Changes in refractive index were analyzed with ForteBio Data Analysis 9.0 by fitting the data to a simple 1:1 binding model. SLU-417 was used instead of SLU-258 because the latter demonstrated significant nonspecific binding to the sensor tip.
Homology modeling
A structural model of the AcrA monomer was generated from a partially complete x-ray crystal structure of AcrA (Protein Data Bank [PDB]: 2F1M (7)) and a full-length structure of MexA (PDB: 2V4D (20)) using Modeller 9.17 (21). The sequences of full-length wild-type AcrA and MexA were aligned with the SALIGN algorithm in Modeller (Fig. S1) (21). The sequence of wild-type AcrA was then threaded onto the 2F1M and 2V4D templates. One hundred models were generated, and the model with the lowest discrete optimized protein energy score (21) was used for molecular dynamics (MD) simulation.
MD simulation before docking
MD simulation was performed to explore the flexibility of the protein and to discover potential binding sites. A 50-ns MD simulation of the best-scoring model was carried out using the AMBER99SB force field (22) and TIP3P (transferable intermolecular potential with three points) water model (23) to describe the protein and solvent, respectively. Periodic boundary conditions were applied, and a cutoff of 1.4 nm was used for real-space nonbonded interactions. The particle mesh Ewald method with a grid spacing of 0.16 nm and the Verlet cutoff scheme (24) were used to describe long-range electrostatics. A long-range dispersion correction (25) was applied to the energy and pressure to account for truncation of the Lennard-Jones interactions. All bonds were constrained with the LINCS algorithm (26), which enabled the use of a 2-fs time step with the leapfrog integrator. The model was solvated in a triclinic box with a minimum of 3.0 nm between the protein and the nearest face of the box. The charge of the system was neutralized with three Cl− ions, and the energy was minimized with a tolerance of 1000 kJ mol−1 nm−1 and a step size of 0.01 nm. A two-step equilibration was carried out before data collection. In the first equilibration step, the temperature was adjusted over 1 ns to 298 K in an NVT (constant number of particles, temperature, and volume) ensemble using a velocity-rescale thermostat (27) with a time constant of 0.1 ps. In the second step, the pressure was adjusted over 1 ns to 1 bar using a Parrinello-Rahman barostat (28, 29) with a time constant of 2 ps. During both equilibration steps, harmonic restraints were applied to all heavy atoms of the protein with a force constant of 1000 kJ mol−1 nm−2. Production MD simulations for data collection were carried out in the NPT (constant number of particles, temperature, and pressure) ensemble without harmonic restraints. These MD simulations were performed with GROMACS 5.1.2 (30).
Ensemble docking
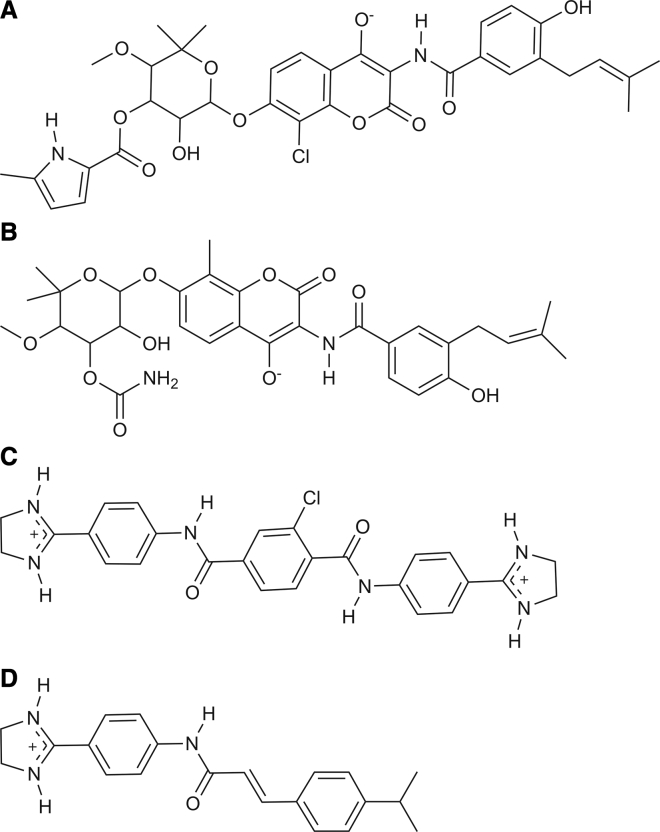
All-atom root mean-square deviation (RMSD) clustering of the MD trajectory was performed with gmx cluster (30) to select conformations for ensemble docking. Using an RMSD cutoff of 0.4 nm, 29 conformations were selected. The inhibitors SLU-258, clorobiocin, and novobiocin (Fig. 2) were docked to each conformation with VinaMPI, a massively parallel implementation of AutoDock Vina (31, 32). The initial structures for clorobiocin and novobiocin were obtained from the ZINC15 database (33) and SLU-258 was obtained from NCI Diversity Set V (33). The protonation state for each inhibitor was taken as the major predicted state at pH 7 from the ZINC15 database. Clorobiocin and novobiocin were assigned a charge of −1 and SLU-258 a charge of +2 (Fig. 2). Hydrogen atoms were added to SLU-258 with Molecular Operating Environment 2016 (34).
Figure 2.
Chemical structures of (A) clorobiocin, (B) novobiocin, (C) SLU-258, and (D) SLU-417.
The docking search space was centered at the center of mass of each conformation and included the entire protein except for the tip of the α-hairpin domain (Fig. S2), which was excluded because of limits on the size of the search space in AutoDock Tools (31). Each docking run was performed three times with an exhaustiveness of 1000 and a maximal number of poses for ligand and protein conformations of 25. The poses from all docking runs for each ligand were combined and clustered based on the RMSD of all ligand atoms with a cutoff of 0.25 nm. The centroid poses from two of the largest clusters were selected as the predicted binding sites. A third site referred to as site I was selected from ensemble docking based on previous work (10).
Binding site prediction with FTMap
A single, representative conformation obtained from RMSD clustering of the 50-ns MD simulation trajectory of the wild-type AcrA monomer was submitted to the FTMap (http://ftmap.bu.edu/home.php) web server (35) with default options.
Inhibitor docking
Clorobiocin and SLU-258 were docked to 29 conformations of AcrA near site IV, which is located between F81 and F254. The same conformations of AcrA and each inhibitor as the blind docking were used. The center of the docking search space was the center of mass of F81 and F254 for each conformation with grid dimensions 25 × 25 × 25 Å. The exhaustiveness was set to 10, and the maximal number of binding poses for each conformation was 25.
Extended MD simulations to explore AcrA conformations
For the starting structure of AcrA, we used one of the bound conformations from (36). The protein was then solvated with ∼157,000 TIP3P water molecules (23) in a cubic box of length 168 Å. For consistency with the earlier simulations (10), the CHARMM36m force field (37) was used to describe the protein. All systems were neutralized with 0.3 M NaCl. Hydrogen mass repartitioning (38) was used for all protein atoms to enable the use of a 4-fs timestep for all interactions. A 12-Å Lennard-Jones cutoff with switching beginning at 10 Å was used for van der Waals interactions. Simulations were performed with Amber 16 (39) on graphics processing units. The temperature was held constant at 300 K using a Langevin thermostat (40). Periodic boundary conditions were used along with the particle mesh Ewald method to describe long-range electrostatics (24). Bonds involving hydrogen atoms were constrained to their equilibrium length with the SETTLE algorithm (41) for water molecules and the SHAKE algorithm for all others (42). We first ran a pre-equilibration simulation of 1 ns of wild-type AcrA, after which AcrA was mutated accordingly. Finally, we ran 250-ns production runs of the wild-type and mutated systems.
Results and Discussion
Structural modeling of AcrA
We first generated a structural model of wild-type AcrA using the x-ray structures of truncated AcrA and full-length MexA as templates. Residues 1–37 at the N-terminus and 380–397 at the C-terminus were excluded from the model because neither template covered these regions (Fig. S1). The sequence identity between the MP domain of AcrA and MexA is 56%. AcrA was also modeled using x-ray structures of AcrA and MexA as templates in the 6.5-Å cryo-EM structure of apo AcrAB-TolC (6). Alignment and Cα superposition of snapshots from the 50-ns MD simulation of the wild-type model onto a single chain of AcrA from the assembled complex revealed an overall RMSD for the whole protein of around 6 Å (Fig. S3). However, the individual domain RMSDs were ∼3 Å for the β-barrel and MP domains and ∼2 Å or lower for the hairpin and lipoyl domains. Thus, the majority of the motion of the protein occurs at the interfaces between the domains, and the structures of the individual domains remained close to those in the starting model.
Calculation of potential binding sites
Using the structural model of AcrA, a combination of computational methods was used to locate potential binding sites of SLU-258 and clorobiocin. In addition, novobiocin was included as an antibiotic that binds to AcrA but does not inhibit the efflux pump activity (10). 29 conformations of wild-type AcrA were extracted from an MD simulation trajectory using RMSD clustering for use in blind ensemble docking of SLU-258, novobiocin, and clorobiocin (Fig. 2).
Three sites were selected by clustering of the docked poses by RMSD, which we refer to as sites I, II, and III (Fig. 1 A). Site I is located at the interface of the α-hairpin and lipoyl domains and has been found by site-directed mutagenesis to be functionally important (10). When residues at site I, such as E67, were mutated to Ala, the MIC of the antibiotics novobiocin and erythromycin decreased, indicating that the mutations impair efflux function and make the bacteria more susceptible to the antibiotics. We found previously that binding of SLU-258 at site I could result in a steric clash that would inhibit AcrA oligomerization (10), which would presumably disrupt the assembly of the full complex.
Sites II and III were the most highly populated sites in the ensemble docking of each inhibitor (Fig. S4). Site II is located in the β-barrel domain, and site III is located between the β-barrel and MP domains (Fig. 1 A; Fig. S5). Previously, it was found that in vivo proteolysis of the assembled AcrAB-TolC pump with trypsin resulted in a major fragment of AcrA (K46−R294/R296) (10). Upon addition of SLU-258, a fragment corresponding to residues K46−K346 of AcrA was also observed. These findings indicate that the inhibitor alters the structure of AcrA, presumably making K346 more accessible to trypsin. K346 is located in the MP domain near site III (Fig. S6). One possible explanation for these observations is that inhibitor binding at site III changes the structure of AcrA. In the assembled complex, the MP domain interacts with AcrB (20). If an inhibitor binds at site III, it could prevent the MP domain of AcrA from interacting properly with AcrB and thus prevent assembly of the pump. Alternatively, the change in structure may result from allosteric propagation of binding at another site to site III.
To search for additional binding sites that may have been overlooked by blind ensemble docking, we used the binding site identification program FTMap (35). FTMap indeed also identified sites I, II, and III but identified three additional sites located in the MP domain and between the β-barrel and lipoyl domains (Fig. 1 A). One site, site IV, is located between the β-barrel and lipoyl domain. The other two sites, V and VI, are located in the MP domain.
Truncated AcrA lacking the MP domain binds SLU-417 and clorobiocin
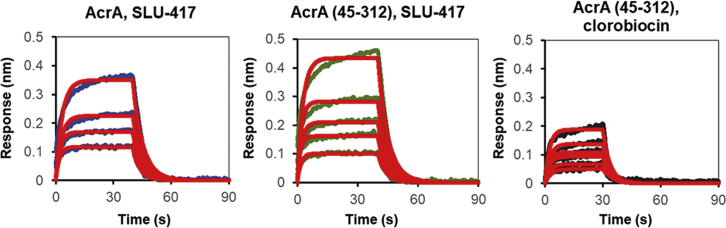
We assessed the binding of inhibitors (Fig. 2) to a truncated construct of AcrA lacking the MP domain (residues 45–312) (7) using BLI. In these experiments, SLU-258 was replaced with its truncated derivative SLU-417 because the former bound nonspecifically to sensors. Binding of both clorobiocin and SLU-417 to the truncated construct was observed with affinities similar to the full-length protein (Fig. 3), diminishing the likelihood that sites III, V, and VI are major inhibitor binding sites in AcrA.
Figure 3.
Biolayer interferometry (BLI) spectra for SLU-417, an analog of SLU-258, binding to full-length AcrA, SLU-417 binding to a truncated variant of AcrA lacking the MP domain (AcrA (45–312)), and clorobiocin binding to AcrA (45–312). Experimental data are shown in blue, green, and back lines. The data were fitted into a simple 1:1 binding model, and the fitted lines are shown in red. To see this figure in color, go online.
Mutations in site IV have the largest impacts on efflux pump function
We carried out site-directed mutagenesis of selected residues near sites I, II, III, and IV (Fig. 1 B) and performed Trp fluorescence spectroscopy on the mutants to detect inhibitor binding. Changes in Trp fluorescence can occur by three different mechanisms: static quenching, dynamic quenching, and changes in the local environment. Static quenching occurs when a fluorescent molecule (such as clorobiocin, novobiocin, or SLU-258) quenches fluorescence intensity through direct contact with the Trp residue (43). Dynamic quenching occurs when excitation of the fluorescent molecule transfers energy in its excited state from the Trp to the molecule without direct contact. Changes in the local environment of Trp that result in increased or decreased solvent exposure can also modify fluorescence properties (44).
Wild-type AcrA contains only one intrinsic Trp residue, W347, which is located near site III. Thus, W347 was mutated to Phe, and the following residues were mutated individually to Trp: E67W near site I, K241W near site II, I252W and I343W near site III, and F81W and F254W near site IV (Fig. 1). E67, K241, and I343 were chosen because they are on the surface of AcrA and are close enough for an inhibitor bound in a given site to interact directly with each of these residues through static quenching. F81, I252, and F254 were chosen because we expect that they could interact indirectly with inhibitors through an allosteric change in the local environment of the Trp or excitation energy transfer through dynamic quenching (Fig. 1 A).
All AcrA variants with single Trp substitutions were expressed at levels comparable to that of the wild-type (data not shown). To determine whether these Trp mutations in AcrA affect functionality of the efflux pump, we measured the MICs of selected antibiotics for strains carrying the AcrA variants and compared them to the wild-type. We measured the MICs for novobiocin, erythromycin, and tetracycline in cells with and without pores in their outer membrane. All three of these antibiotics are AcrAB-TolC substrates (45). The presence of pores diminishes the permeability barrier of the outer membrane, allowing antibiotic activity and the corresponding effects on efflux to be assessed directly (46). SDS, which is also an AcrAB-TolC substrate, was used as a nonantibiotic control. All selected mutants were expressed and retained at least partial efflux function compared to the wild-type (Table 1). However, the F81W mutant (site IV) exhibited the largest decrease in MIC for each antibiotic because cells carrying this AcrA variant were hypersusceptible to antibiotics even with the native outer membrane. In hyperporinated cells, which have increased influx of antibiotics into the periplasm, E67W (site I) provided weak protection against antibiotics. All other AcrA mutants containing Trp substitutions were functionally similar to the AcrA W347F variant and were two- to fourfold less active than wild-type AcrA (Table 1). These results show that the F81W and E67W substitutions decrease the activity of the AcrAB-TolC efflux pump. Thus, these two residues are functionally important for efflux.
Table 1.
Antibiotic Susceptibilities of Wild-Type AcrA and Selected Trp Mutants
| No Pore |
Pore |
|||||||
|---|---|---|---|---|---|---|---|---|
| NOV | ERY | TET | SDS | NOV | ERY | TET | SDS | |
| – | 1 | 0.5 | 0.25 | 19.5 | 0.5 | 0.25 | 0.06 | 9.7 |
| pUC18 | 1 | 1 | 0.25 | 39 | 0.12 | 0.12–0.2 | 0.06 | 9.7 |
| Wild-type | 256 | 64 | 0.5 | >10,000 | 32 | 8–16 | 0.5 | 312 |
| W347F | 128 | 16 | 0.5 | >10,000 | 8 | 2–4 | 0.25 | 312 |
| W347F K241W | 128 | 16 | 0.5 | >10,000 | 4 | 1 | 0.125 | 156 |
| W347F I252W | 128 | 32 | 1 | >10,000 | 4 | 1 | 0.25 | 156 |
| W347F F254W | 128 | 16 | 0.25–0.5 | >10,000 | 8 | 1 | 0.125 | 78 |
| I343W | 256 | 64 | 0.5 | >10,000 | 8 | 2–4 | 0.125 | 39–78 |
| W347F I343W | 128–256 | 16 | 0.25 | >10,000 | 8 | 1 | 0.25 | 156 |
| W347F E67W | 64–128 | 16–32 | 0.5 | >10,000 | 4 | 1 | 0.25 | 39 |
| W347F F81W | 8 | 2 | 0.25 | 39.1 | 1 | 0.125 | 0.25 | 15.6 |
NOV, novobiocin; ERY, erythromycin; TET, tetracycline.
Fluorescence of Trp residues in AcrA reports on binding of inhibitors
The fluorescence spectrum of apo wild-type AcrA presents a maximum at = 330 nm (Table 2), which indicates that the single W347 is not exposed to an aqueous environment but is buried in the protein. In agreement, substitution of this Trp in AcrA W347F abolished this maximum. The fluorescence emission spectra and maxima of the F81W and F254W variants were similar to that of the wild-type protein, suggesting that these Trp residues are also buried within the protein. Surprisingly, the I343W mutant exhibited peaks at 340 nm with a distinct bump at 310 nm, indicating two states in which this Trp residue has different exposure to the buffer (Fig. 4). Furthermore, these states differ from that of the wild-type protein. All the remaining mutants have values of 350 nm, indicating that their Trp residues are exposed on the surface of AcrA.
Table 2.
Fluorescence Properties of Single Trp Mutants of AcrA in the Absence and Presence of Inhibitors
| Wild-type | F81W | I252W | I343W | E67W | F254W | K241W | ||
|---|---|---|---|---|---|---|---|---|
| λmax (nm)a | apo | 330 | 330 | 350 | 310, 340 | 350 | 330 | 350 |
| Clo | 338 | 338 | 350 | 345 | 350 | 338 | 350 | |
| SLU | 338 | 341 | 350 | 348 | 353 | 341 | 353 | |
| Nov | 336 | 341 | 350 | 343 | 350 | 340 | 350 | |
| τ (ns)b | apo | 3.19; 2.60 | 2.36; 1.68 | 4.66; 3.91 | 2.99; 2.09 | 6.33 | 2.60; 2.12 | 4.59; 4.03 |
| Clo | 3.25; 2.61 | 1.88; 1.51 | 4.63; 4.0 | 2.83; 2.09 | 6.24 | 2.52; 2.11 | 4.52; 4.0 | |
| SLU | 3.18; 2.66 | 2.07; 1.53 | 4.49; 3.67 | 2.88; 2.13 | 6.22 | 2.53; 2.14 | 4.43; 3.87 | |
| Nov | 3.23; 2.62 | 2.0; 1.38 | 4.75; 4.11 | 2.95; 2.13 | 6.38 | 2.62; 2.12 | 4.54; 3.98 | |
| KD (μΜ)c | SLU | 67.2 ± 12.6 | 46.9 ± 11.3 | 49.2 ± 19.8 | 53.5 ± 16.8 | 40.6 ± 7.2 | 64.7 ± 27.8 | 45.9 ± 13.4 |
| Clo | 37.3 ± 23.3 | 13.5 ± 2.2 | 22.8 ± 7.7 | 20.9 ± 3.5 | 22.3 ± 9.0 | 23.6 ± 4.6 | 35.8 ± 2.5 | |
| Ksvd | apo | 1.19 ± 0.02 | 1.12 ± 0.05 | 4.8 ± 0.05 | 1.47 ± 0.06 | 12.97 ± 0.25 | 0.84 ± 0.01 | 2.44 ± 0.01 |
| Clo | 1.39 ± 0.05 | 0.65 ± 0.01 | 4.61 ± 0.03 | 1.21 ± 0.03 | 12.1 ± 0.26 | 0.72 ± 0.01 | 2.15 ± 0.03 | |
| SLU | 1.43 ± 0.03 | 1.28 ± 0.03 | 4.59 ± 0.05 | 1.29 ± 0.05 | 13.25 ± 0.34 | 0.95 ± 0.02 | 1.99 ± 0.04 |
Clo, clorobiocin; Nov, novobiocin; SLU, SLU-258.
Wavelength of fluorescence emission maximum.
Average intensity τ; average amplitude τ.
Dissociation constants calculated based on quenching of Trp fluorescence.
Stern-Volmer quenching constant.
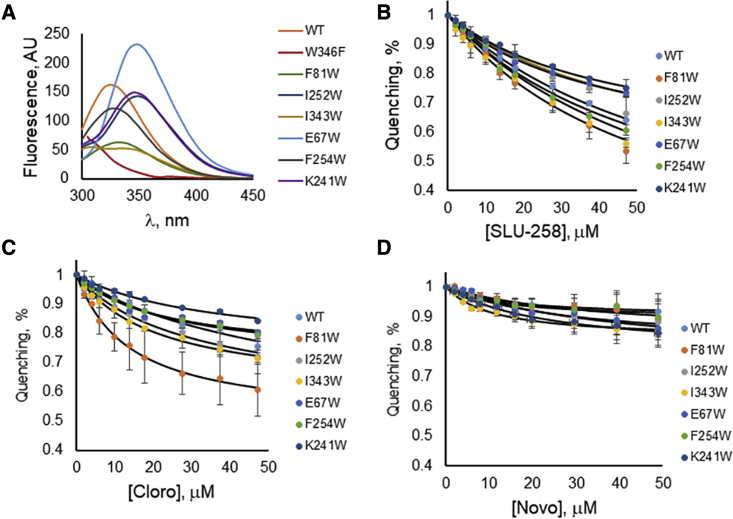
Figure 4.
Fluorescence of AcrA mutants carrying a single Trp substitution in the absence of inhibitors (A) and quenching in the presence of increasing concentrations of SLU-258 (B), clorobiocin (Cloro) (C), and novobiocin (Novo) (D). Error bars represent SD, n = 3. To see this figure in color, go online.
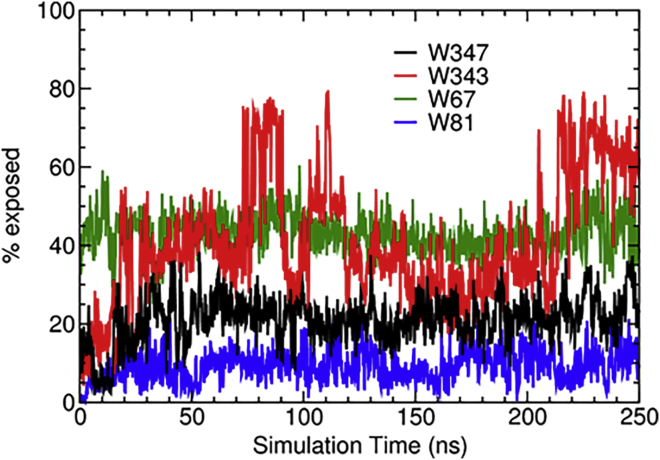
To investigate the effects of introducing Trp substitutions in the protein, we ran additional 250-ns simulations of wild-type AcrA as well as the I343W, E67W, and F81W mutants starting from an AcrB-bound state extracted from a cryo-EM structure of the tripartite complex (36) (see Materials and Methods). We tracked the solvent-accessible surface area of W347 and the respective mutated Trp side chains (Fig. 5). W347 and W81 remained less than ∼40% exposed throughout most of the simulation, whereas W67 was predictably higher, between 40 and 50% exposed. This behavior is in agreement with the red shift (λmax at 350 nm) in the fluorescence spectra observed for E67W, which indicates a more solvent-exposed side chain than that of W347 in the wild-type. On the other hand, W343 oscillated between low (∼40%) and high (∼70%) exposure, which is consistent with the two peaks observed in the fluorescence spectra for the I343W mutant (Table 2).
Figure 5.
Solvent-accessible surface area of Trp side chains for residues 347 (black; wild-type), 343 (red; I343W), 67 (green; E67W), and 81 (blue; F81W). To see this figure in color, go online.
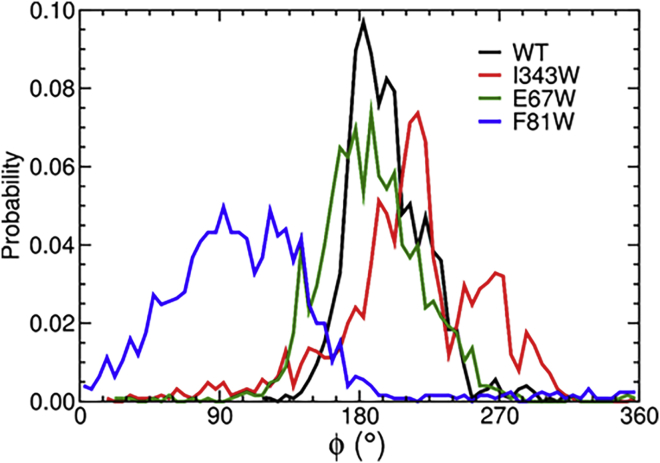
We also tracked global conformational changes in wild-type and mutant AcrA by monitoring the pseudodihedral angle defined by the center of mass of each domain in the simulations and found that, with the exception of F81W, all systems sampled roughly the same conformations (Fig. 6). Thus, the two peaks in the I343W fluorescence spectra are not due to changes in the conformation of AcrA but rather the two-state behavior of W343 itself.
Figure 6.
Dihedral angle (ϕ) between the membrane-proximal and α-hairpin domains about the axis defined by the lipoyl and β-barrel domains (47). To see this figure in color, go online.
With the addition of clorobiocin or SLU-258 to the wild-type, of W347 increased to 338 nm. Similarly, for the F81W and F254W mutants, increased to 338 nm for clorobiocin and 341 nm for SLU-258. Therefore, the binding of these inhibitors to wild-type, F81W, and F254W (site IV) changed their Trp fluorescence by exposing these residues to the solvent. At the same time, the fluorescence intensity of these AcrA variants decreased with increasing concentrations of clorobiocin and SLU-258 (Figs. 4 and S7–S10). The greatest Trp fluorescence quenching occurred for F81W (site IV) and I343W (site III) upon addition of clorobiocin and SLU-258, followed by F254W (site IV) (Fig. 4). F81W and I343W had quenching efficiencies of ∼40–45% for SLU-258, and F81W had an efficiency of ∼40% with clorobiocin. These results indicate that binding of SLU-258 and clorobiocin change the structure of AcrA. It is also possible that the inhibitor is in direct contact with each mutant residue concerned. No changes in the fluorescence maxima were detected for all other mutants, suggesting that these solvent-exposed Trp residues do not undergo large conformational changes in the presence of ligands. Quenching by novobiocin was marginal for all the mutants, suggesting that binding of this ligand does not significantly affect the structure of AcrA.
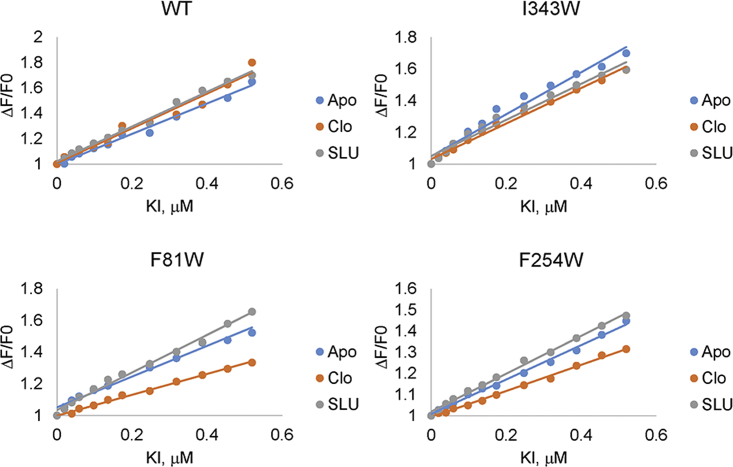
Trp fluorescence quenching was also used to estimate the affinities of clorobiocin and SLU-258 to different AcrA variants. We found that, independent of the Trp position, both ligands have comparable micromolar affinity to the protein, with clorobiocin binding to AcrA two to three times more tightly than SLU-258. A similar affinity of SLU-258 to AcrA (78 μM) was reported previously based on surface plasmon resonance measurements (10).
F81W, F254W, and I343W exhibited the lowest intensities, amplitudes, and lifetimes in the presence of clorobiocin and SLU-258 (Table 2). However, as described above, the W343 oscillates between low and high solvent exposure (Fig. 5). I343 is located in the MP domain, which is absent in the x-ray crystal structure of AcrA and is known to be conformationally flexible (7, 20). The MP domain is highly flexible in vitro and can change conformation regardless of whether a ligand is bound (20). In contrast, site IV, which is flanked by F81W and F254W, was resolved in the crystal structure and is therefore expected to be more rigid (7). Hence, fluorescence changes near site IV are more likely to be due to inhibitor binding.
F81W and F254W, both located in site IV, are protected from quenching when clorobiocin and SLU-258 are bound
To examine further whether clorobiocin and SLU-258 bind near F81W (site IV) or F254W (site IV), we analyzed quenching of the Trp fluorescence by increasing the concentration of potassium iodide with and without inhibitors. If an inhibitor binds near F81W or F254W, fluorescence of the Trp may be protected from iodide quenching by the inhibitor. Indeed, F81W was significantly protected (42% reduction in quenching) from iodide by clorobiocin (Fig. 7; Table 2). Weak protection by clorobiocin (15% reduction in quenching) was also observed for F254W. No significant changes in potassium iodide quenching were found for Trp residues at any other positions (Fig. S11).
Figure 7.
Trp fluorescence in the absence (Apo) and presence of clorobiocin (Clo) or SLU-258 (SLU) with increasing concentrations of potassium iodide (KI). To see this figure in color, go online.
In the presence of clorobiocin, Trp residues in F81W and F254W are more solvent-exposed than without this ligand, as indicated by red shifts in the respective fluorescence spectra (Table 2). Thus, it is unlikely that protection of iodide is a result of relocating these Trp residues deeper in the protein interior due to conformational changes or oligomerization. Clorobiocin is therefore likely to protect F81W directly from quenching by iodide. The effect of SLU-258 was different in that its presence resulted in somewhat enhanced quenching of F81W and F254W with iodide. Thus, clorobiocin and SLU-258 may have different binding poses from each other or interact with different residues in AcrA.
SLU-258 and clorobiocin likely bind near site IV.
The following observations all point to SLU-258 and clorobiocin binding at or near site IV:
-
1)
Both inhibitors bind to an AcrA construct lacking the MP domain.
-
2)
F81W impairs efflux.
-
3)
Fluorescence changes upon binding the inhibitors to F81W and F254W most resemble the corresponding changes in the wild-type.
-
4)
Both clorobiocin and SLU-258 interfere with the quenching of potassium iodide with F81W and F254W in site IV.
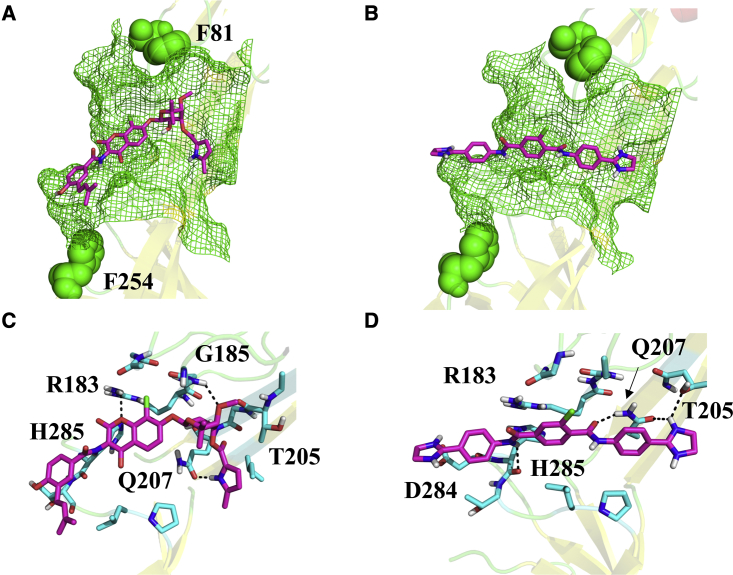
These results provide evidence that site IV is associated with clorobiocin and SLU-258 binding (Fig. 8). In the docked models, the inhibitors engage in largely different interactions with the binding site. Whereas the negatively charged clorobiocin forms hydrogen bonds and electrostatic interactions with R183, the R183 side chain is pointed away from the positively charged SLU-258 inhibitor (Fig. 8). In addition, the hydroxy group of T205 forms a hydrogen bonding interaction with clorobiocin but is oriented away from SLU-258. Nevertheless, clorobiocin and SLU-258 both form hydrogen bonding interactions with Q207, but clorobiocin also forms a hydrogen bond with the backbone of G185, whereas SLU-258 form a hydrogen bond with the backbone carbonyl of H285 (Fig. 8). Thus, site IV, which engages in different interactions for clorobiocin and SLU-258, provides the best explanation for the observed fluorescence changes, interference with potassium iodide quenching, and ultimately the binding site location of SLU-258 and clorobiocin.
Figure 8.
Docked models of clorobiocin and SLU-258 (magenta) bound at site IV between the lipoyl and β-barrel domains, flanked by F81 and F254 (green spheres). (A and B) Residues within 5 Å of each ligand are shown in green mesh. (C and D) Residues in contact with each ligand are shown in cyan sticks, and the interactions are indicated by black dashes. To see this figure in color, go online.
Conclusions
A combination of computational modeling and experimental Trp fluorescence spectroscopy has been used to identify likely binding sites of clorobiocin and SLU-258 to the membrane fusion protein AcrA of the AcrAB-TolC efflux pump in E. coli. Our findings point to site IV, which is located at the interface of the β-barrel and lipoyl domain, as the most likely binding site for the two inhibitors. The F81W mutation in site IV increased the susceptibility of antibiotics to E. coli, and this effect is correlated with the highest quenching of fluorescence intensity of all the Trp mutants. Both F81W and F254W mutants, which flank site IV, possessed values that are identical to the wild-type apo protein, with similar shifts relative to the wild-type in the presence of each inhibitor. The lifetimes for both residues were the shortest of all the mutants, and they were the only two mutants that interfered with quenching by potassium iodide.
The mechanism of inhibition for both inhibitors remains unknown. However, if the inhibitors indeed bind at site IV, a conformational change might occur upon binding that causes the protein to adopt a more closed state, which could interfere with assembly of the pump. In the model of the inhibitor-AcrA complex, SLU-258 forms a hydrogen bond with H285, and previous MD simulations have shown that H285 plays a role in the pH-dependent conformational changes of AcrA (47). Refinement of models of inhibitor binding to site IV may form the basis for identifying structure-activity relationships and designing improved inhibitors (11) with greater antibiotic potentiation activity.
Authors Contributions
Z.M.D., R.L.F., and J.K. designed AcrA mutants and performed fluorescence measurements. N.A. analyzed functionalities of AcrA variants. A.T.G. and A.H. performed molecular docking and simulations. Z.G., J.M.P., J.C.G., J.C.S., and H.I.Z. designed experiments and wrote the manuscript.
Acknowledgments
This work was supported by National Institutes of Health, United States grant AI052293 to H.I.Z. This work used resources of the Compute and Data Environment for Science at Oak Ridge National Laboratory, which is managed by UT-Battelle for the Department of Energy, United States under contract no. DE-AC05-00OR22725. A.H. and J.C.G. acknowledge support from the National Science Foundation, United States through grant MCB-1452464 as well as the Extreme Science and Engineering Discovery Environment, which is supported by National Science Foundation grant number OCI-1053575.
Editor: Alan Grossfield.
Footnotes
Zbigniew M. Darzynkiewicz and Adam T. Green contributed equally to this work.
Eleven figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(19)30026-8.
Supporting Material
References
- 1.Frieden, T. 2017. Antibiotic resistance threats in the United States. Report from the U.S. Department of Health and Human Services, Centers for Disease Control and Prevention, April 23, 2013. https://www.cdc.gov/drugresistance/threat-report-2013/pdf/ar-threats-2013-508.pdf.
- 2.Levy S.B., Marshall B. Antibacterial resistance worldwide: causes, challenges and responses. Nat. Med. 2004;10(Suppl):S122–S129. doi: 10.1038/nm1145. [DOI] [PubMed] [Google Scholar]
- 3.Kim J.S., Jeong H., Ha N.C. Structure of the tripartite multidrug efflux pump AcrAB-TolC suggests an alternative assembly mode. Mol. Cells. 2015;38:180–186. doi: 10.14348/molcells.2015.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Du D., van Veen H.W., Luisi B.F. Assembly and operation of bacterial tripartite multidrug efflux pumps. Trends Microbiol. 2015;23:311–319. doi: 10.1016/j.tim.2015.01.010. [DOI] [PubMed] [Google Scholar]
- 5.Du D., Wang Z., Luisi B.F. Structure of the AcrAB-TolC multidrug efflux pump. Nature. 2014;509:512–515. doi: 10.1038/nature13205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Z., Fan G., Du D. An allosteric transport mechanism for the AcrAB-TolC multidrug efflux pump. eLife. 2017;6 doi: 10.7554/eLife.24905. e24905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mikolosko J., Bobyk K., Ghosh P. Conformational flexibility in the multidrug efflux system protein AcrA. Structure. 2006;14:577–587. doi: 10.1016/j.str.2005.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lobedanz S., Bokma E., Koronakis V. A periplasmic coiled-coil interface underlying TolC recruitment and the assembly of bacterial drug efflux pumps. Proc. Natl. Acad. Sci. USA. 2007;104:4612–4617. doi: 10.1073/pnas.0610160104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ge Q., Yamada Y., Zgurskaya H. The C-terminal domain of AcrA is essential for the assembly and function of the multidrug efflux pump AcrAB-TolC. J. Bacteriol. 2009;191:4365–4371. doi: 10.1128/JB.00204-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abdali N., Parks J.M., Zgurskaya H.I. Reviving antibiotics: efflux pump inhibitors that interact with AcrA, a membrane fusion protein of the AcrAB-TolC multidrug efflux pump. ACS Infect. Dis. 2017;3:89–98. doi: 10.1021/acsinfecdis.6b00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haynes K.M., Abdali N., Walker J.K. Identification and structure-activity relationships of novel compounds that potentiate the activities of antibiotics in Escherichia coli. J. Med. Chem. 2017;60:6205–6219. doi: 10.1021/acs.jmedchem.7b00453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amaro R.E., Baudry J., Smith J.C. Ensemble docking in drug discovery. Biophys. J. 2018;114:2271–2278. doi: 10.1016/j.bpj.2018.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tikhonova E.B., Yamada Y., Zgurskaya H.I. Sequential mechanism of assembly of multidrug efflux pump AcrAB-TolC. Chem. Biol. 2011;18:454–463. doi: 10.1016/j.chembiol.2011.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tikhonova E.B., Wang Q., Zgurskaya H.I. Chimeric analysis of the multicomponent multidrug efflux transporters from gram-negative bacteria. J. Bacteriol. 2002;184:6499–6507. doi: 10.1128/JB.184.23.6499-6507.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Westfall D.A., Krishnamoorthy G., Rybenkov V.V. Bifurcation kinetics of drug uptake by Gram-negative bacteria. PLoS One. 2017;12:e0184671. doi: 10.1371/journal.pone.0184671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tikhonova E.B., Dastidar V., Zgurskaya H.I. Kinetic control of TolC recruitment by multidrug efflux complexes. Proc. Natl. Acad. Sci. USA. 2009;106:16416–16421. doi: 10.1073/pnas.0906601106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fonin A.V., Sulatskaya A.I., Turoverov K.K. Fluorescence of dyes in solutions with high absorbance. Inner filter effect correction. PLoS One. 2014;9:e103878. doi: 10.1371/journal.pone.0103878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Darzynkiewicz Z.M., Bojarska E., Darzynkiewicz E. Interaction of human decapping scavenger with 5′ mRNA cap analogues: structural requirements for catalytic activity. J. Phys. Condens. Matter. 2007;19:285217. [Google Scholar]
- 19.Keizer J. Nonlinear fluorescence quenching and the origin of positive curvature in Stern-Volmer plots. J. Am. Chem. Soc. 1983;105:1494–1498. [Google Scholar]
- 20.Symmons M.F., Bokma E., Koronakis V. The assembled structure of a complete tripartite bacterial multidrug efflux pump. Proc. Natl. Acad. Sci. USA. 2009;106:7173–7178. doi: 10.1073/pnas.0900693106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sali A., Blundell T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 22.Hornak V., Abel R., R., Simmerling C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jorgensen W.L., Chandrasekhar J., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 24.Darden T., York D., Pedersen L. Particle mesh Ewald: an N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 25.Abraham, M. J., D. van der Spoel, …, the GROMACS Development Team. 2018. GROMACS User Manual, Version 2018. www.gromacs.org.
- 26.Hess B. P-LINCS: a parallel linear constraint solver for molecular simulation. J. Chem. Theory Comput. 2008;4:116–122. doi: 10.1021/ct700200b. [DOI] [PubMed] [Google Scholar]
- 27.Bussi G., Donadio D., Parrinello M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007;126:014101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 28.Parrinello M., Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 1981;52:7182–7190. [Google Scholar]
- 29.Nosé S., Klein M.L. Constant pressure molecular dynamics for molecular systems. Mol. Phys. 1983;50:1055–1076. [Google Scholar]
- 30.Van Der Spoel D., Lindahl E., Berendsen H.J. GROMACS: fast, flexible, and free. J. Comput. Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 31.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ellingson S.R., Smith J.C., Baudry J. VinaMPI: facilitating multiple receptor high-throughput virtual docking on high-performance computers. J. Comput. Chem. 2013;34:2212–2221. doi: 10.1002/jcc.23367. [DOI] [PubMed] [Google Scholar]
- 33.Sterling T., Irwin J.J. ZINC 15--ligand discovery for everyone. J. Chem. Inf. Model. 2015;55:2324–2337. doi: 10.1021/acs.jcim.5b00559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Molecular Operating Environment (MOE). 2013. Chemical Computing Group Inc. (Montreal, QC, Canada).
- 35.Kozakov D., Grove L.E., Vajda S. The FTMap family of web servers for determining and characterizing ligand-binding hot spots of proteins. Nat. Protoc. 2015;10:733–755. doi: 10.1038/nprot.2015.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jeong H., Kim J.S., Ha N.C. Pseudoatomic structure of the tripartite multidrug efflux pump AcrAB-TolC reveals the intermeshing cogwheel-like interaction between AcrA and TolC. Structure. 2016;24:272–276. doi: 10.1016/j.str.2015.12.007. [DOI] [PubMed] [Google Scholar]
- 37.Huang J., Rauscher S., MacKerell A.D., Jr. CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat. Methods. 2017;14:71–73. doi: 10.1038/nmeth.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hopkins C.W., Le Grand S., Roitberg A.E. Long-time-step molecular dynamics through hydrogen mass repartitioning. J. Chem. Theory Comput. 2015;11:1864–1874. doi: 10.1021/ct5010406. [DOI] [PubMed] [Google Scholar]
- 39.Case D., Betz R., Kollman P. University of California; San Francisco, CA: 2016. AMBER 2016. [Google Scholar]
- 40.Feller S.E., Zhang Y.H., Brooks B.R. Constant pressure molecular dynamics simulation: the Langevin piston method. J. Chem. Phys. 1995;103:4613–4621. [Google Scholar]
- 41.Miyamoto S., Kollman P.A. Settle - an analytical version of the shake and rattle algorithm for rigid water models. J. Comput. Chem. 1992;13:952–962. [Google Scholar]
- 42.Ryckaert J.-P., Ciccotti G., Berendsen H.J.C. Numerical integration of the Cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 43.Vaiana A.C., Neuweiler H., Smith J.C. Fluorescence quenching of dyes by tryptophan: interactions at atomic detail from combination of experiment and computer simulation. J. Am. Chem. Soc. 2003;125:14564–14572. doi: 10.1021/ja036082j. [DOI] [PubMed] [Google Scholar]
- 44.van de Weert M., Stella L. Fluorescence quenching and ligand binding: a critical discussion of a popular methodology. J. Mol. Struct. 2011;998:144–150. [Google Scholar]
- 45.Takatsuka Y., Chen C., Nikaido H. Mechanism of recognition of compounds of diverse structures by the multidrug efflux pump AcrB of Escherichia coli. Proc. Natl. Acad. Sci. USA. 2010;107:6559–6565. doi: 10.1073/pnas.1001460107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krishnamoorthy G., Wolloscheck D., Zgurskaya H.I. Breaking the permeability barrier of Escherichia coli by controlled hyperporination of the outer membrane. Antimicrob. Agents Chemother. 2016;60:7372–7381. doi: 10.1128/AAC.01882-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang B., Weng J., Wang W. Interdomain flexibility and pH-induced conformational changes of AcrA revealed by molecular dynamics simulations. J. Phys. Chem. B. 2012;116:3411–3420. doi: 10.1021/jp212221v. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.