Abstract
The transcription factor nuclear factor (NF)-κB promotes inflammatory and stress-responsive gene transcription across a range of cell types in response to the cytokine tumor necrosis factor (TNF). Although NF-κB signaling exhibits significant variability across single cells, some target genes supporting high levels of TNF-inducible transcription exhibit fold-change detection of NF-κB, which may buffer against stochastic variation in signaling molecules. It is unknown whether fold-change detection is maintained at NF-κB target genes with low levels of TNF-inducible transcription, for which stochastic promoter events may be more pronounced. Here, we used a microfluidic cell-trapping device to measure how TNF-induced activation of NF-κB controls transcription in single Jurkat T cells at the promoters of integrated HIV and the endogenous cytokine gene IL6, which produce only a few transcripts per cell. We tracked TNF-stimulated NF-κB RelA nuclear translocation by live-cell imaging and then quantified transcript number by RNA FISH in the same cell. We found that TNF-induced transcript abundance at 2 h for low- and high-abundance target genes correlates with similar strength with the fold change in nuclear NF-κB. A computational model of TNF-NF-κB signaling, which implements fold-change detection from competition for binding to κB motifs, could reproduce fold-change detection across the experimentally measured range of transcript outputs. However, multiple model parameters affecting transcription had to be simultaneously varied across promoters to maintain fold-change detection while also matching other trends in the single-cell data for low-abundance transcripts. Our results suggest that cells use multiple biological mechanisms to tune transcriptional output while maintaining robustness of NF-κB fold-change detection.
Introduction
Tumor necrosis factor (TNF) acts on a wide range of cell types and is a critical mediator of inflammatory and stress responses (1) and of the development of certain immune cells, including T lymphocytes (2, 3). TNF stimulation activates a signaling cascade that disrupts the interaction between the transcription factor nuclear factor-κB (NF-κB) and the inhibitor of NF-κB α (IκBα), thereby releasing NF-κB and allowing it to accumulate in the nucleus. NF-κB can then regulate the expression of hundreds of genes, including those encoding its own negative regulators IκBα (NFKBIA) and A20 (TNFAIP3) and inflammatory cytokines TNF (TNFA) and IL-6 (IL6) (4). Given its central role in development and immunity, understanding how cells decode responses to TNF signals is of fundamental interest.
Dynamic signaling and response measurements from single cells provide direct information about input-output signal-response functions (5, 6). By measuring dynamic nuclear translocation of the NF-κB RelA subunit and transcription in the same single cells, we previously demonstrated that the maximal fold change in nuclear RelA predicts transcript abundance of NFKBIA, TNFAIP3, and IL8 after TNF treatment in HeLa cells, a cell line of epithelial origin (7). This observation had significant implications for our understanding of RelA-mediated signaling in these cells because only two signaling circuits commonly observed in biological systems allow for fold-change detection (8). One is an incoherent type 1 feed-forward loop (I1-FFL), in which a molecular species increases with the input and also inhibits the output (9). By adapting a mathematical model of TNF-stimulated NF-κB RelA signaling (10), we showed that we could accurately reproduce fold-change detection for the measured RelA target genes by incorporating an I1-FFL mechanism via competition for binding to κB motifs (7). However, the generality of this mechanism across cell types and promoters has not been explored.
NF-κB is also a positive transcriptional regulator of human immunodeficiency virus-1 (HIV), which infects CD4+ T cells (11). We recently observed that TNF-activated NF-κB deterministically controls activation of an HIV reporter, even for latent viruses with very low TNF-inducible transcription (12). Interestingly, we observed significant differences in TNF-induced recruitment of RNA polymerase II at the promoters of these low-abundance HIV target genes in Jurkat cells as compared to the high-abundance target NFKBIA (12). Therefore, we were interested to see whether the transcription of low-abundance target genes also exhibited fold-change detection and whether the I1-FFL model of NF-κB-mediated transcription could accurately describe the HIV transcription patterns we observed in T cells.
These questions are challenging to address in T cells with a single-cell approach because T cells do not adhere to surfaces and are therefore difficult to track when perturbed. Consequently, we developed a workflow whereby we immobilized Jurkat T cells within a microfluidic cell trap array (13) and then tracked NF-κB RelA nuclear translocation by live-cell imaging and quantified transcript number by fluorescence in situ hybridization of single messenger RNAs (smFISH) in the same cell. Using this workflow, we measured how TNF-induced activation of NF-κB controls transcription in single Jurkat T cells across diverse NF-κB target promoters, including HIV and endogenous regulatory and cytokine genes, exhibiting a range of TNF-inducible transcript levels.
We found that all measured NF-κB target promoters responded to TNF-stimulated fold change in nuclear RelA, even for transcripts induced at an average messenger RNA (mRNA) level of six or fewer transcripts per cell after 2 h of TNF treatment. Our computational model of RelA signaling, with I1-FFL topology downstream of RelA, accurately recapitulated fold-change detection at low-abundance genes but only if multiple model parameters affecting transcription were varied simultaneously. Different trends in the single-cell data served to constrain the model parameter space. Our results suggest that cells use multiple biological mechanisms to tune transcriptional output while maintaining robustness of NF-κB fold-change detection.
Materials and Methods
Cell culture and pharmacological treatments
Jurkat T cell clones J65c 4.4 and 6.6 were created as previously described (12). Each cell line contains the same retroviral integration of a full-length human RelA fused at its N-terminus to mCherry (Ch-RelA) and a unique integration of a full-length, non-replication-competent reporter virus construct (psLTR-Tat-GFP) in which Nef has been replaced with green fluorescent protein (GFP) and the open reading frames of all other viral proteins except Tat are disrupted with premature stop codons. J65c cells were cultured in Roswell Park Memorial Institute 1640 (RPMI) medium (Thermo Fisher Scientific, Waltham, MA). All media were supplemented with 10% fetal bovine serum (Atlanta Biologicals, Flowery Branch, GA), 100 U/mL penicillin, and 100 μg/mL streptomycin (Thermo Fisher Scientific). Cells were maintained in 5% CO2 at 37°C and were never cultured beyond passage 12. J65c cells were grown to 5 × 105 cells/mL before treatment with recombinant human TNF (Peprotech, Rocky Hill, NJ). As in a previous study, we used 160 ng/mL cycloheximide (CHX; Sigma, St. Louis, MO) to block HIV Tat protein production and the resulting amplification of HIV gene expression (12) and to restrict our analysis to RelA-mediated transcription. This level of CHX is very low and does not change TNF-stimulated RelA translocation dynamics (Fig. S1 A), nor does it affect viability under our imaging conditions (no increased cell death was observed for CHX-treated cells versus untreated cells in our time-lapse images). For consistency, we used these same conditions throughout this study, irrespective of which transcript was assayed.
Microfluidic device fabrication
The silicon master was etched as previously described (13). After silanization (see (13) for conditions), a mixture of polydimethylsiloxane (PDMS) base and curing agent (Sylgard 184 Elastomer kit; Dow Corning, Midland, MI) was mixed 10:1 by weight and poured over the master. This was degassed and cured for 2 h at 75°C. The cured PDMS mold was peeled from the master and cut to fit a #1 glass coverslip. A 7 mm Harris Uni-Core punch (Ted Pella, Redding, CA) was used to punch holes for reservoirs at the inlet and outlet ports. A #1 25 × 75 mm microscope coverslip was rinsed with isopropanol and dried and cleared of dust with filtered air before being placed inside a PE-25 benchtop plasma cleaner (Plasma Etch) with the mold. The coverslip and PDMS mold were exposed to O2 plasma at 150 W, 200 mTorr for 2 min and adhered immediately upon removal from the plasma chamber. The channel and the inlet and outlet reservoirs were filled with deionized water to maintain hydrophilic surfaces.
Cell loading protocol
For each experiment, 5 × 106 cells were suspended in 1 mL of fresh RPMI medium before being loaded into the microfluidic device. Cell clumps were dispersed immediately before loading by pipetting the cell suspension vigorously and filtering cells through a 35 μm nylon mesh strainer cap tube (BD Falcon; BD Biosciences, Bedford, MA). 50 μL of cells was loaded into the inlet and allowed to flow through the channel for 10 min. Flow-through medium in the outlet was first removed to prevent backflow that can dislodge cells. The cell suspension was then removed from the inlet by gently scraping the bottom of the inlet reservoir with a pipette while withdrawing the medium. 100 μL of medium was added to the inlet to wash the cells for 3 min to remove any free-floating cells from the main channel and the inlet and outlet reservoirs; this wash step was repeated three times. 50 μL medium was added only to the inlet after final wash to ensure continuous flow and the device was moved onto the microscope stage for imaging and stimulation (see below). The continuous flow was maintained throughout imaging to prevent cells from dislodging from the traps.
Live-cell imaging
Cells were imaged on a Nikon Eclipse Ti spinning disk confocal microscope (Yokogawa CSU-W1 spinning disk) with a plan apochromatic 60× oil objective (numerical aperture [NA] 1.4; Nikon, Tokyo, Japan). Ch-RelA was excited with a 561 nm laser at 20–30% power, and images were collected with an Andor iXon Ultra888 EMCCD camera (Andor Technology, Belfast, Northern Ireland). Live-cell imaging was performed on an environmentally controlled stage at 37°C and 5% CO2 (Tokai Hit, Fujinomiya, Japan). Once the loaded device was secured to the stage insert, five regions of interest were identified, and their positions relative to fiducial markers on the outside of the channel were recorded. 12 μm Z-stacks with 1 μm slices were recorded for each position as a reference image. Fluid was removed from the outlet and then inlet, and 50 μL of RPMI medium containing TNF was added to the inlet and imaging immediately started. Based on our previous study, we estimated that TNF flows through the entire length of the channel within 30 s and that 50 μL of media was sufficient to maintain a nearly constant flow velocity of ∼300 μm/s for 2 h (13). 12 μm Z-stacks of each region of interest were recorded every 3 min for 110 min after stimulation; using Z-stacks ensured that we captured the nuclear signal near the cell equator even as cells moved in the z axis during imaging. Nikon Perfect Focus autofocus system was used to identify the same Z slice for every time point. After 110 min of imaging, cells were fixed by removing fluid from the outlet and then inlet and replacing with 50 μL of 4% formaldehyde in phosphate-buffered saline (PBS) for fixation before in situ hybridization.
smFISH
Stellaris RNA FISH probe sets (Biosearch Technologies, Novato, CA) targeting NFKBIA/IκBα, TNFAIP3/A20, env-GFP/HIV, and GAS6 were previously described (7, 12, 14). The IL6/IL-6 probe set was designed using online software (https://www.biosearchtech.com/) and previously described strategies (7, 15). Probes for env-GFP, NFKBIA, and IL6 were conjugated to carboxyfluorescein; probes for TNFAIP3 and GAS6 were conjugated to Quasar 670. Probe sets for the main targets are included in Table 1.
Table 1.
smFISH Probe Sets
| Probe (5′-3′) env-GFP | Probe (5′-3′) NFKBIA | Probe (5′-3′) TNFAIP3 | Probe (5′-3′) IL6 | ||||
|---|---|---|---|---|---|---|---|
| 1 | aggatccgttcactaatcga | 1 | agtagccgctccttcttcag | 1 | atacaaagcctgaggaagga | 1 | ggggttgagactctaatatt |
| 2 | cagatcgtcccagataagtg | 2 | tcgtctttcatggagtccag | 2 | acagctttccgcatattgct | 2 | atctccagtcctatatttat |
| 3 | tagctgaagaggcacaggct | 3 | cttgaccatctgctcgtact | 3 | gtgtatcggtgcatggtttt | 3 | gagggcagaatgagcctcag |
| 4 | agagtaagtctctcaagcgg | 4 | atgatggccaagtgcaggaa | 4 | ggcaagttctgaacatttcc | 4 | ttctctttcgttcccggtgg |
| 5 | tccacaatcctcgttacaat | 5 | ggtcagtgccttttcttcat | 5 | tcccgaaactgaggacaaaa | 5 | gctcctggaggggagataga |
| 6 | caccaatatttgagggcttc | 6 | ttcacctggcggatcacttc | 6 | tttctgtcgatgagggcttt | 6 | tggagaaggagttcatagct |
| 7 | tcctgactccaatactgtag | 7 | agttgaggaaggccaggtct | 7 | agtttcttctggctttccag | 7 | agaaggcaactggaccgaag |
| 8 | tatggctgtggcattgagca | 8 | agtctgctgcaggttgttct | 8 | tactgagaagtggcatgcat | 8 | aaggcagcaggcaacaccag |
| 9 | cttctataaccctatctgtc | 9 | tcagcaatttctggctggtt | 9 | taccaagtctgtgtcctgaa | 9 | cggctacatctttggaatct |
| 10 | agctctataagctgcttgta | 10 | atttcctcgaaagtctcgga | 10 | ttgcgtgtgtctgtttcctt | 10 | aagaggtgagtggctgtctg |
| 11 | tattcttctaggtatgtggc | 11 | ttggtagccttcaggatgga | 11 | cagttgccagcggaatttaa | 11 | atttgtttgtcaattcgttc |
| 12 | gcaccggtcttatagcaaaa | 12 | tagacacgtgtggccattgt | 12 | cccgtttcaacaaattcctg | 12 | agatgccgtcgaggatgtac |
| 13 | tcctcgcccttgctcaccat | 13 | tagccatggatagaggctaa | 13 | cagttccgagtatcatagca | 13 | atgtctcctttctcagggc |
| 14 | gggcaccaccccggtgaaca | 14 | attttgcaggtccactgcga | 14 | gtctgtggaagccattttga | 14 | tcacacatgttactcttgtt |
| 15 | cgccgtccagctcgaccagg | 15 | acacttcaacaggagtgaca | 15 | ttgtactgaagtccacttcg | 15 | ttctgccagtgcctctttgc |
| 16 | ctgaacttgtggccgtttac | 16 | taactctgttgacatcagcc | 16 | gaggatgttgcaaaggacaa | 16 | tctttggaaggttcaggttg |
| 17 | gccctcgccctcgccggaca | 17 | tagggagaatagccctggta | 17 | ggaacctgattccaaacttc | 17 | aagcatccatctttttcagc |
| 18 | tcagcttgccgtaggtggca | 18 | ttttctagtgtcagctggcc | 18 | attccacccactttcaaagg | 18 | ctcctcattgaatccagatt |
| 19 | gtggtgcagatgaacttcag | 19 | actctctggcagcatctgaa | 19 | atggggtatctgtagcattc | 19 | tgatgattttcaccaggcaa |
| 20 | ccagggcacgggcagcttgc | 20 | tgtgtcatagctctcctcat | 20 | aaatgatggctgtcatagcc | 20 | acctcaaactccaaaagacc |
| 21 | tcagggtggtcacgagggtg | 21 | tgaactccgtgaactctgac | 21 | aagtggaacagctcggattt | 21 | tgttctggaggtactctagg |
| 22 | ctgaagcactgcacgccgta | 22 | aacacacagtcatcataggg | 22 | aagtcttcaaatcttccccg | 22 | tgttcctcactactctcaaa |
| 23 | cttcatgtggtcggggtagc | 23 | ataacgtcagacgctggcct | 23 | cggggatttctatcaccatt | 23 | tactcatctgcacagctctg |
| 24 | cggacttgaagaagtcgtgc | 24 | atgttctttcagcccctttg | 24 | atgagatgagttgtgccatg | 24 | caggaactggatcaggactt |
| 25 | tggacgtagccttcgggcat | 25 | gtgtggatataagtacaccc | 25 | ttcatccaactttgcggcat | 25 | gggtggttattgcatctaga |
| 26 | cttgaagaagatggtgcgct | 26 | aataagacgttttgggccag | 26 | gccatttcttgtactcatgc | 26 | gctggcatttgtggttgggt |
| 27 | gggtcttgtagttgccgtcg | 27 | aaaatgagggctgatcctac | 27 | ttcacagacatgaagaaggg | 27 | tggttctgtgcctgcagctt |
| 28 | ccctcgaacttcacctcggc | 28 | gtcccctacaaaaagttcac | 28 | tgagcactcatggcataaag | 28 | tgagatgagttgtcatgtcc |
| 29 | gatgcggttcaccagggtgt | 29 | ggtgaggtttaaaagaagtt | 29 | tttgattcttttgccgcctc | 29 | aactccttaaagctgcgcag |
| 30 | ttgaagtcgatgcccttcag | 30 | aaccttctccaaaaacccca | 30 | gagttcagctttgggagttt | 30 | catgctacatttgccgaaga |
| 31 | ccccaggatgttgccgtcct | 31 | gtggtccttccatgaaattt | 31 | tcactgaacagaaaagggct | 31 | aacaacaacaatctgaggtg |
| 32 | tgtagttgtactccagcttg | 32 | cagtcactcgaagcacaata | 32 | tgctgcacattcagtgtgaa | 32 | acaggtttctgaccagaaga |
| 33 | catgatatagacgttgtggc | 33 | atgtcacaggataccactgg | 33 | gcaacgttcacaaaatccgt | 33 | aacataagttctgtgcccag |
| 34 | tgccgttcttctgcttgtcg | 34 | gcttaacactcctggctgtt | 34 | aatgtcctggtaacatcctg | 34 | cgctcatacttttagttctc |
| 35 | cggatcttgaagttcacctt | 35 | ttttcaccccacatcactga | 35 | tgaagcaagtactgcagatc | 35 | cttacataaattaactcagc |
| f36 | gctgccgtcctcgatgttgt | 36 | cacaaaccttgacaggtagt | 36 | tctggagtcccaaaatacac | 36 | caaactgcatagccactttc |
| 37 | tggtagtggtcggcgagctg | 37 | tacaccatttacaggagggt | 37 | aaacacagtgtgcaaaagcc | 37 | ccaagaaatgatctggctct |
| 38 | gtcgccgatgggggtgttct | 38 | gtaccaaaataattaccaac | 38 | tcaaacatggtgcttccaag | 38 | atttgaggtaagcctacact |
| 39 | ttgtcgggcagcagcacggg | – | – | 39 | gaaacacttctggcagtatc | 39 | aaaatatgtataagttagcc |
| 40 | ggactgggtgctcaggtagt | – | – | 40 | ggcctcatgaaatctctgat | – | – |
| 41 | cgttggggtctttgctcagg | – | – | 41 | tcagttgctcttctgtcctt | – | – |
| 42 | aggaccatgtgatcgcgctt | – | – | 42 | ttgaggtgctttgtgtggtt | – | – |
| 43 | ggcggcggtcacgaactcca | – | – | 43 | ttgccaaaatgatcacaggc | – | – |
| 44 | tcgtccatgccgagagtgat | – | – | 44 | aaagcattcgttgcagtagc | – | – |
After fixation in the passive-flow device, cells were washed three times with PBS, allowing the buffer to flow through the device for 3 min for each wash. Cells were permeabilized by adding 150 μL of 70% ethanol to the inlet and 50 μL of 70% ethanol to the outlet and placing the device for 16 h at 4°C. After permeabilization, cells were washed three times with PBS and then incubated with 10% formamide (Thermo Fisher Scientific) in 2× saline sodium citrate (SSC; Thermo Fisher Scientific) for 30 min at 37°C. Cells were hybridized by adding 50 μL of indicated probes (concentrations discussed below) in buffer containing 2× SSC, 10% formamide, and 100 mg/mL dextran sulfate (MW > 500,000; Millipore, Burlington, MA) at 37°C in the dark for 12 h. Cells were washed twice with 10% formamide in 2× SSC. Finally, cells were counterlabeled with 1 μg/mL Hoechst 33342 (Thermo Fisher Scientific) and imaged in either PBS or VectaShield mounting media (Vector Labs, Peterborough, UK).
We note here that a key practice to minimize the loss of flow and cells during the smFISH protocol was to implement a simple stepwise procedure that would minimize the loss of hydrostatic pressure in the microfluidic system during fluid replacement. At every buffer exchange step, the outlet was emptied first, and then the inlet was emptied and immediately refilled with the new buffer to reestablish flow. All solutions, including the viscous hybridization buffer, passed through the channel and could be displaced within 60 s by addition of a subsequent solution (Fig. S1 B). We confirmed that the multiple incubation and wash steps did not result in significant cell loss (Fig. S1 C).
Labeled cells were imaged on a Nikon Eclipse Ti spinning disk confocal microscope with a plan apochromatic 100× oil objective (NA 1.45; Nikon). Regions of interest imaged in the live-cell imaging step were reidentified using the fiducial markers. For each field, we acquired Z-stacks of 30–60 images with 0.3 μm intervals.
Implementing the smFISH protocol in the device, we found that moderate probe concentrations (100–200 nM) yielded labeling comparable to that achieved with cells on a dish; higher probe concentrations (500 nM) resulted in high background and the loss of distinct spots (Fig. S1 D). In addition, both in conventional culture and in the device, we found very low nonspecific probe binding when performing labeling for transcripts of GAS6, a gene not normally expressed in Jurkat cells (16) (200 nM probes; Fig. S1 E). Finally, validating the specificity of the env-GFP probe set, we found very few foci in parental Jurkat cells (averaging to <0.3 per cell; Fig. S1 F).
Image analysis
The Z-center for each cell was manually identified, and the mean fluorescence intensity (MFI) for nuclear Ch-RelA in this slice was quantified at every time point. Background was determined by measuring the MFI of an area equivalent to the cell nucleus but in an area of the channel with no objects in the same slice. The background MFI was then subtracted from the nuclear MFI. Cells that were displaced from the traps or divided during imaging were not analyzed. The features for Ch-RelA nuclear translocation were extracted from live-cell time courses as previously reported (7, 12). Fixed cells with labeled transcripts were registered to live cells based on fiducial markers. Cell outlines were traced manually, and transcripts were identified and counted using FISH-Quant software (17) as previously described (12).
Chromatin immunoprecipitation
EZ ChIP Kit (Upstate) reagents and protocols were used as described previously (12). 15 × 106 cells were fixed in 1% formaldehyde for 10 min. Unreacted formaldehyde was quenched with 125 mM glycine for 5 min. Cells were washed three times with PBS and lysed with 1% sodium dodecyl sulfate lysis buffer in the presence of cOmplete protease inhibitor cocktail (Roche, Basel, Switzerland). Lysed cells were sonicated with a Diagenode Bioruptor (settings: seven cycles of 5 min per cycle with 30 s pulses followed by 30 s of incubation at a high power). Samples were precleared with Protein G agarose (Millipore), and 1% of each sample was used as a percent input. Samples were incubated with antibody for 16 h at 4°C. Protein G agarose was then added to the samples and incubated for 1 h at 4°C. Beads were washed once each with low salt, high salt, and LiCl immune complex wash buffers included in the kit, and then washed twice with 10 mM Tris, 1 mM EDTA (pH 8.0). DNA-protein complexes were eluted from agarose beads with 1% sodium dodecyl sulfate and 100 nM NaHCO3. Crosslinks were reversed by incubating samples with 5 M NaCl overnight at 65°C. DNA was purified with DNA spin filters and eluted with 10 mM Tris, 1 mM EDTA (pH 8.0). DNA was quantified by quantitative PCR (iCycler, iQ5; BioRad, Hercules, CA) using SYBR Green Supermix (BioRad). qPCR was performed in triplicate, and melt curves were run to ensure product specificity. Antibodies and primers are listed in Table 2.
Table 2.
Antibody and Primers Used in ChIP Assays
| Antibody | Source | Catalog # |
|---|---|---|
| anti-NF-κB p65 rabbit monoclonal | Cell Signaling | 8242 |
| Primers | Sequence (5′-3′) |
|---|---|
| 5′ HIV LTR forward | TGCATCCGGAGTACTTCAAGAACTGC |
| 5′ HIV LTR reverse | CTAACCAGAGAGACCCAGTACAGGC |
| IL6 forward | CTAGCCTCAATGACGACCTAAGC |
| IL6 reverse | GAATGAGCCTCAGACATCTCCAG |
| TNFAIP3 forward | GTGACTTTGGAAAGTCCCGTGG |
| TNFAIP3 reverse | TTGGCCCGCCACGAAGACTG |
| NFKBIA forward | TGACCCTAGTGGCTCATCGC |
| NFKBIA reverse | GGACTGCTGTGGGCTCTG |
Computational modeling
To make predictions about fold-change detection, RelA promoter binding, and transcript variance across a range of target transcriptional outputs, we used a previously reported computational model of TNF-RelA signaling (7) (Fig. S2; Table S1). Ensemble simulations that included sampling to mimic cell-to-cell variability were run using the ode15s solver in MATLAB (The MathWorks, Natick, MA). Sensitivity analyses and the five-dimensional parameter scan were run using a Fortran encoding of the same model and the DLSODE solver, with outputs plotted in MATLAB.
Briefly, the transcription rate of the RelA inducible target promoter is given by the following ordinary differential equation:
| (1) |
in which the transcript synthesis rate c1t is modified by the fractional binding (FB) of nuclear RelA (nNFκB) to the target promoter and the target transcript (tIndTarget) is degraded at a rate c3t. Fractional binding of nNFκB depends on the affinity of RelA for the target promoter (k), the concentration of the competitor protein (Comp), the affinity of the competitor for the target promoter (k3), and the Hill coefficient (ht), which effectively varies the steepness of the change in transcriptional response over a range of nNFκB concentrations:
| (2) |
For the sensitivity analysis, the normalized local sensitivity index was calculated for features of both nuclear RelA (nNFκB; initial, Finit, maximal, Fmax, and fold change, Fmax/Finit) and the target transcript (tIndTarget; initial, mRNAinit, maximal, mRNAmax, at t = 2 h, mRNAt = 2h, and fold change, mRNAmax/mRNAinit) using the following:
| (3) |
Here, ki is the parameter whose relative influence on the selected Feature is calculated by varying the value of ki 10-fold around its nominal value while all other parameters are unchanged (using , as given in Table S1). The first term of the sensitivity index in Eq. 3 is the slope of the Feature-versus-ki curve at . The second term allows for normalization by the nominal value and the value of Feature at . The slope was calculated by polynomial approximation and differentiation routines in MATLAB; a second-order polynomial function yielded a satisfactory approximation of the Feature-versus-ki curve. We note that total NFκB (NF) and ka were kept constant and were chosen to represent an “average” cell with values of 0.24 μM and 10−4, respectively.
To predict behavior across a range of RelA target promoters with different transcriptional outputs, we first scanned across a range of relative affinities of competitor versus RelA for the target promoter (changing competitor affinity k3 while maintaining the same RelA affinity k) sufficient to produce the approximate absolute change in transcription output observed between NFKBIA and IL6 (scanning over two orders of magnitude centered over the nominal parameter value). For each value of k3, we simulated 450 “cells” by varying total NFκB (RelA) abundance and ka (a proxy for the strength of activation of pathways leading to IKK) but keeping other parameters at their default value, as described previously (7). After setting total NFκB abundance, the system is allowed to reach an unstimulated steady state with ka = 0, setting basal cell conditions, including basal (or “initial”) transcript numbers. Then, the stimulation is initiated by switching ka to its value for this “cell.” For each set of 450 “cells,” we estimated the predicted correlation of number of target transcripts at 120 min versus maximal fold change in nNFκB over this 120 min period. As a measure of the change in transcript distribution after stimulation, we also estimated a “relative variance” by dividing the variance of target transcripts at t = 120 min in the presence of TNF by the variance in transcripts in the basal state. Finally, we estimated the average fractional binding (FB) on the target promoter for each combination of scanned parameter values around the peak of nNFκB concentration using the time points 0, 30, and 60 min; FB was calculated based on Eq. 2.
For a two-dimensional parameter scan, we varied transcriptional output by scanning across both a range of competitor affinity to the inducible target promoter (k3), as well as a range of competitor affinity for the competitor promoter (k4, to modulate the relative abundance of RelA versus Comp, which is less well determined). As for the initial one-dimensional scan, we simulated ensembles of 450 “cells,” varying total NFκB and ka and scanned over a range of 10-fold below to 10-fold above the nominal parameter values.
Finally, we explored model behavior over a five-dimensional parameter space, varying k3 and k4 as well as c1t, c3t, and ht (see Eqs. 1 and 2). Each of the five parameters were varied in all possible combinations over a range of two orders of magnitude, centered near its default value (using multipliers 0.1000, 0.1359, 0.1848, 0.2511, 0.3414, 0.4640, 0.6307, 0.8572, 1.1652, 1.5838, 2.1528, 2.9262, 5.4063, and 9.9885, chosen among the 16 equally spaced points in log10 space). The other model parameters were maintained at their original value. For a selection of combinations at the extremes or “corners” of the parameter space, for boundary regions of the parameter spaces that yielded appropriate transcript numbers, and for regions that qualitatively fitted HIV LTR promoter behavior, we also ran ensemble simulations of 450 “cells” to obtain estimates of the correlation of induced target transcript number with fold change, average FB, and/or relative variance.
Statistical analysis
Differences between distributions of transcripts or time-course features were determined by Kolmogorov-Smirnov (K-S) test (p < 0.05). 95% confidence intervals (CIs) on descriptive statistics of RNA distributions were estimated from the 2.5 to 97.5% quantiles of copy numbers per cell bootstrapped by sampling 1000 times with replacement. Differences in the central tendency between pairs of distributions were determined using a Mann-Whitney U test (p < 0.05) to compare their median. To compare dynamics between Ch-RelA time courses, differences between distributions of tmax (defined as time to reach Fmax) and tduration (defined as the time between 50% Fmax rising and 50% Fmax falling) were determined by K-S test (p < 0.05). All statistical tests and regression and correlation analyses were performed in Prism (Graphpad) unless otherwise indicated.
Results
Implementation of smFISH for suspension T cells in a passive-flow microfluidic device
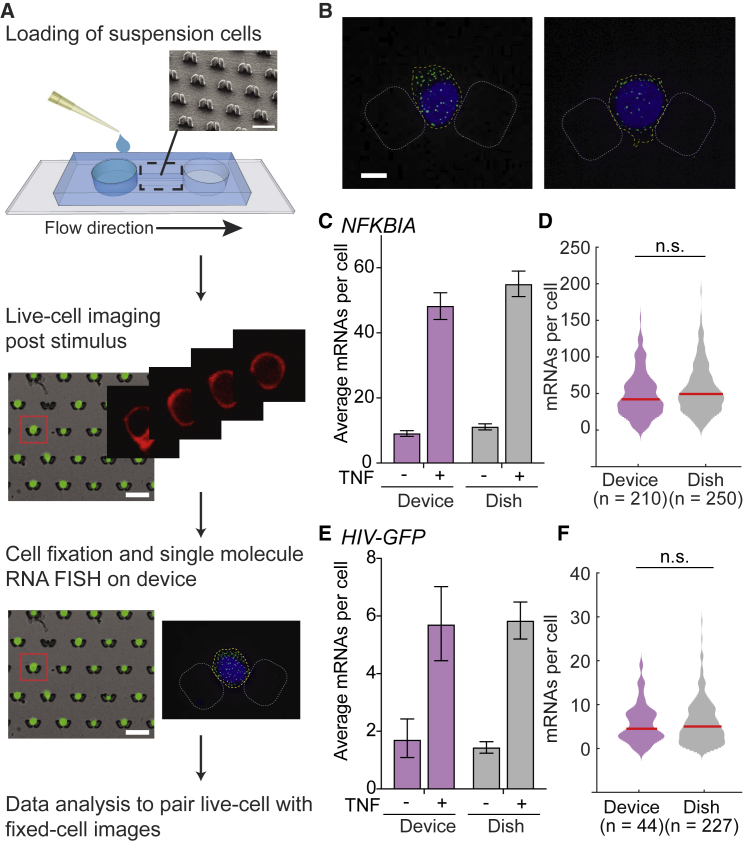
We previously developed a user-friendly microfluidic device that immobilizes suspension cells in an array of traps using passive gravity-driven flow between two reservoirs to implement long-term live-cell imaging of suspension cells (13). Our goal here was to collect paired data, tracking NF-κB RelA nuclear translocation dynamics by live-cell imaging and RelA-driven transcriptional output by smFISH in the same cells (Fig. 1 A), to gain insight into how NF-κB signaling dynamics are converted to transcriptional outputs in Jurkat T cells. A challenge posed by fixed-cell response assays such as smFISH is that they require multiple fluid exchanges within the device to allow for incubations with different reagents and multiple wash steps. After optimization of the workflow to implement smFISH in the device (see Materials and Methods), imaging Z-stacks on the device produced well-defined puncta, which were suitable for downstream analyses (Fig. 1 B). Previous studies have shown that these puncta represent individual mRNAs and thus allow accurate quantification of mRNA number in individual cells (15, 18, 19).
Figure 1.
Optimization of a microfluidic-device-enabled protocol to connect signaling dynamics to smFISH measurements in single suspension cells. (A) A schematic of the live-cell-to-fixed-cell protocol. A cell suspension is pipetted into the inlet of a microfluidic device and flows into the channel containing traps that catch cells via gravity. Upon stimulation with a medium exchange, live cells are imaged in the traps for a desired amount of time. Then, cells are fixed and assayed by single-molecule RNA fluorescence in situ hybridization (smFISH). Data are then processed, and the live-cell and fixed-cell images are analyzed together for the same single cells. (B) Representative images of Jurkat J65c cells treated with 20 ng/mL TNF for 1 h, fixed in traps, and labeled for single NFKBIA mRNAs with carboxyfluorescein-conjugated probes (green). Nuclei stained with Hoechst (blue); scale bars, 5 μm. (C) A bar graph of mean NFKBIA transcript counts, as measured by smFISH for unstimulated cells and cells stimulated with 20 ng/mL TNF for 1 h measured in the passive-flow device (purple) and in a tissue culture dish (gray). Data are presented as the mean ± 95% CIs obtained by bootstrapping. (D) Violin plots of TNF-stimulated NFKBIA transcript distributions measured by smFISH at 1 h in the passive-flow device (purple) or tissue culture dish (gray). The median is indicated by the red line. There are no significant differences between the distributions (p > 0.05 by Kolmogorov-Smirnov (K-S) test). (E and F) A bar graph of mean HIV-GFP transcript count (E) and violin plots of HIV-GFP transcript distributions (F) measured by smFISH after 2 h of TNF stimulation. All other details are as in (C) and (D).
To validate the smFISH implementation in the device, we quantified NFKBIA transcripts in J65c cells, a clonal population of Jurkat T lymphocytes stably expressing an NF-κB mCherry-RelA (Ch-RelA) reporter protein. We compared measurements collected in the device to our previously published measurements of NFKBIA transcripts collected in a dish (12). We found that mean NFKBIA transcript numbers and the distribution of NFKBIA transcripts before and 1 h after TNF stimulation were indistinguishable between the two formats (Fig. 1, C and D). A similar comparison for TNF-stimulated HIV transcripts from clonal J65c Jurkat cell populations containing a latent-but-inducible HIV integration with very low transcript abundance also showed no evidence of changes in transcriptional output between formats (12) (Fig. 1, E and F). Overall, our data show that smFISH for both high- and low-abundance inducible transcripts in T cells can be implemented in our passive-flow device, allowing for a live-cell-to-fixed-cell imaging workflow for nonadherent cells.
Jurkat cells exhibit fold-change detection of RelA at promoters with high-inducible transcription
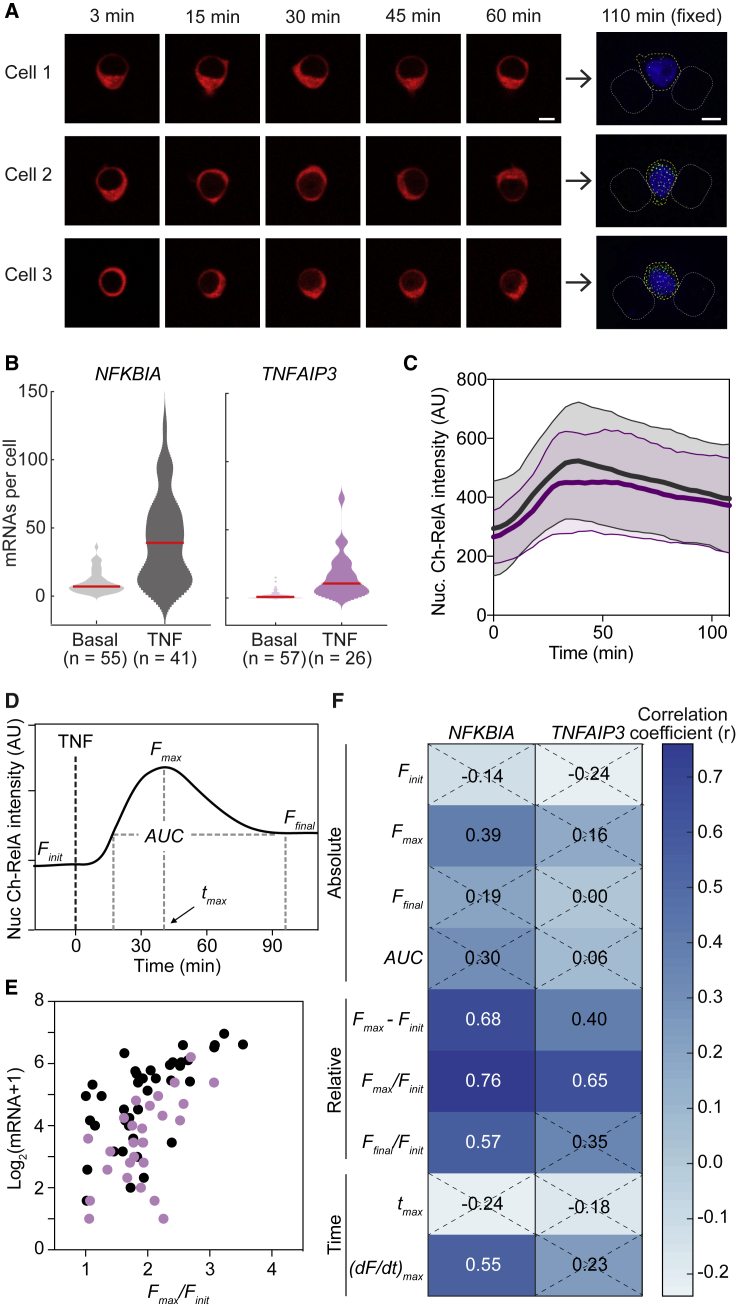
We proceeded to image TNF-induced NF-κB RelA translocation dynamics followed by smFISH in J65c cells to explore NF-κB RelA signal input-transcriptional output relationships. Ch-RelA translocation was imaged for 2 h after stimulation with 20 ng/mL TNF (Fig. 2 A, left). Cells were then fixed in the traps, and transcripts of the RelA target gene (here NFKBIA) were labeled by smFISH in the device to produce a set of live-cell signaling traces paired with transcriptional output (Fig. 2 A, right).
Figure 2.
Fold-change detection of nuclear RelA is observed in Jurkat T cells at promoters exhibiting high levels of TNF-inducible transcription. (A) Time-lapse images of Ch-RelA paired with smFISH images of NFKBIA transcripts at 2 h after 20 ng/mL TNF treatment from the same cells. Scale bars, 5 μm. (B) Violin plots of transcript number distributions for NFKBIA (gray) and TNFAIP3 (purple) measured by smFISH before and 2 h after TNF stimulation in the device. The median is indicated by the red line. (C) Time-course traces of nuclear Ch-RelA from J65c cells collected in the passive-flow device after 20 ng/mL TNF treatment for two independent experiments. Data are presented as the mean ± SD of individual cell traces (n = 41, traces linked to NFKBIA, gray; n = 26, traces linked to TNFAIP3, purple). Lack of statistical significance of differences in dynamics was determined by comparing distributions of tmax and tduration of nuclear RelA peak intensity between the data sets (p > 0.05 by K-S test). AU, arbitrary units. (D) A schematic indicating features calculated from individual nuclear Ch-RelA time courses including initial, maximal, and final fluorescence (Finit, Fmax, and Ffinal); time at which Fmax is reached (tmax); and area under the curve (AUC). (E) A scatter plot of smFISH-measured transcript number (2 h post-TNF addition) versus the maximal fold change in nuclear Ch-RelA (Fmax/Finit) for NFKBIA (gray) and TNFAIP3 (purple). (F) A heat map of Pearson correlation coefficients (r) with smFISH-measured transcript number for all extracted metrics describing same-cell nuclear Ch-RelA time courses. Nonsignificant correlations (p > 0.05) are indicated by a dashed X.
We first explored input-output relationships for the high-abundance targets NFKBIA and TNFAIP3. After 2 h of TNF stimulation, the average number of transcripts increased to 40 and 16 (observed maximum of 124 and 73) transcripts per cell, respectively, for NFKBIA and TNFAIP3 (Fig. 2 B). Ch-RelA nuclear translocation dynamics over that period were not significantly different between the two experiments (Fig. 2 C). We note that TNF-stimulated Ch-RelA translocation dynamics differed between the device and a tissue culture dish; nuclear Ch-RelA peaked 5–10 min later and was sustained over a longer period (monitored for 2 h) in the device versus in a dish (Fig. S3 A). This slower and more sustained pattern of nuclear Ch-RelA translocation was not due to a delay in TNF stimulation between the inlet and outlet because the average response of the cells near the inlet of the device was indistinguishable from the response of cells closer to the outlet (Fig. S3 B). Despite the slower and more sustained signaling dynamics observed in the device, transcript number distributions remained unchanged (Fig. 1, C–F), suggesting that T cells respond to a feature of the signaling dynamics that is conserved across formats.
We previously showed that maximal fold change in nuclear RelA is predictive of NFKBIA and TNFAIP3 early transcriptional output in HeLa cells (t = 60 min (7)) and of the fluorescent protein production from an HIV activation reporter construct in Jurkat T cells (12). To evaluate which aspects of RelA signaling are most predictive of NFKBIA and TNFAIP3 transcriptional output in Jurkat cells, we extracted features from individual TNF-stimulated Ch-RelA nuclear intensity time courses (Fig. 2 D) and matched them to the number of transcripts observed in the same cell. We included absolute nuclear RelA intensity (initial, maximal, and final), features describing relative changes in nuclear intensity (maximal and final intensity normalized to initial intensity), a feature that integrated nuclear intensity over time (area under the curve, AUC), and a feature that described pathway flux (the maximal rate of change). We verified that the distributions of feature values were not significantly different between NFKBIA and TNFAIP3 experiments (Fig. 2 C; Materials and Methods), confirming that Ch-RelA nuclear translocation was similar in both sets of sampled cells.
When we compared correlations of each Ch-RelA feature with transcript number, we found that the maximal fold change in RelA nuclear intensity (Fmax/Finit) was the strongest predictor for NFKBIA and TNFAIP3 early transcription levels across all features considered (for t = 2 h; Fig. 2, E and F), consistent with our observations in HeLa cells. Interestingly, we found that the distributions of Fmax/Finit for nuclear RelA were the same for translocation time courses collected in the tissue culture dish versus the device (Fig. S3 C), which may explain why the observed differences in translocation dynamics between the two formats did not significantly affect transcriptional output. Overall, we conclude that fold-change detection of nuclear RelA in response to TNF stimulation is maintained in Jurkat T cells for the high-abundance RelA target genes NFKBIA and TNFAIP3.
Fold-change detection is conserved at promoters exhibiting low inducible transcription
We recently studied RelA-mediated activation of latent HIV using J65c cells harboring unique integrations of an HIV lentiviral construct driving expression of GFP and the viral protein Tat. We specifically studied two clones, referred to as J65c 4.4 and 6.6 (12), which do not express virus in the basal state but are activated by TNF. 2 h of TNF treatment while blocking viral positive feedback induced similar, very low mean transcription at these two viral integration sites (Fig. 3 A; ∼6 transcripts per cell). We also observed a low TNF-induced transcript mean for the cytokine IL6 (Fig. 3 A), consistent with the expectation that IL6 is not strongly activated by TNF in immune cells (20); however, the increase in response to TNF was nonetheless significant (Fig. 3 B). Therefore, we wanted to explore whether fold-change detection between nuclear RelA and transcript number would be observed at these promoters, which support very low amounts of inducible transcription.
Figure 3.
Maximal fold change in nuclear RelA correlates with transcript output for gene promoters exhibiting low levels of TNF-inducible transcription. (A) A bar graph of mean smFISH-measured transcript counts of HIV 6.6 (blue), HIV 4.4 (yellow), and IL6 (green) before and 2 h after 20 ng/mL TNF stimulation. Data are presented as the mean ± 95% CIs obtained by bootstrapping. (B) Violin plots of transcript number distributions for HIV 6.6, HIV 4.4, and IL6 measured by smFISH, before and 2 h after TNF stimulation. The median is indicated by the red line. Statistical significance for differences in the medians of two distributions is indicated (∗P < 0.05; ∗∗∗∗P < 0.0001; Mann-Whitney U test). (C) Scatter plots of smFISH-measured transcript number (2 h post-TNF) for HIV 6.6 (blue), HIV 4.4 (yellow), and IL6 (green) versus maximal nuclear Ch-RelA abundance (Fmax; left), AUC (middle), and maximal fold change in nuclear Ch-RelA (Fmax/Finit; right). (D) A heat map of Pearson correlation coefficients (r) with smFISH-measured transcript numbers for all extracted metrics describing same-cell nuclear Ch-RelA time courses. Nonsignificant correlations (p > 0.05) indicated by a dashed X.
In addition to differences in TNF-induced average transcript numbers, the TNF-induced transcript number distributions also varied across promoters. J65c 6.6 exhibited a highly skewed distribution of HIV transcript numbers in response to TNF stimulation that was more similar to TNFAIP3, but J65c 4.4 HIV transcript number distribution and distributions for IL6 both exhibited much less skew (Fig. 3 B; Table 3). We previously observed such differences in transcript distributions and showed that they are associated with differences in the underlying mechanisms of RelA-mediated transcription (12), bringing into question whether the fold change in nuclear RelA will remain strongly correlated with transcript number at both promoters.
Table 3.
Statistical Moments for Experimentally Measured Transcript Distributions
| Target | Mean |
Variance |
Skewness |
|||
|---|---|---|---|---|---|---|
| Basal | +TNF | Basal | +TNF | Basal | +TNF | |
| NFKBIA | 10.1 (8.0, 12.3) | 40.6 (31.1, 50.4) | 64.5 (36.6, 93.8) | 984.9 (596, 1377) | 1.3 (0.7, 1.9) | 0.7 (0.2, 1.2) |
| TNFAIP3 | 2.0 (1.3, 2.8) | 16.2 (10.6, 22.7) | 8.6 (2.8, 16.0) | 267.9 (78.7, 515.3) | 2.5 (1.3, 3.4) | 1.8 (0.4, 2.7) |
| HIV 6.6 | 1.8 (1.1, 2.7) | 6.3 (3.2, 10.0) | 8.0 (1.8, 17.5) | 129.5 (17.0, 250.3) | 3.5 (0.8, 4.4) | 2.7 (1.3, 4.4) |
| HIV 4.4 | 1.7 (1.1, 2.4) | 5.7 (4.4, 7.0) | 6.8 (1.9, 14.1) | 19.1 (10.2, 28.4) | 3.0 (1.1, 4.1) | 1.0 (0.3, 1.6) |
| IL6 | 0.5 (0.3, 0.7) | 3.0 (2.3, 3.7) | 0.8 (0.4, 1.3) | 5.0 (3.0, 7.1) | 2.0 (1.1, 2.8) | 0.5 (−0.1, 1.0) |
Values for the upper and lower bounds of the 95% CIs obtained by bootstrapping are listed in parentheses.
When we compared correlations of each Ch-RelA feature with early transcript number for these low-abundance targets, we found that the maximal Ch-RelA nuclear intensity and AUC of Ch-RelA intensity were again only weak predictors of TNF-induced transcript number at 2 h (Fig. 3 C). In contrast, the maximal fold change in RelA nuclear intensity (Fmax/Finit) was a strong predictor for HIV and IL6 transcription (Fig. 3 C). For IL6, the correlation with maximal nuclear RelA fold change held despite the fact that transcript number ranged only from 0 to 10 transcripts per cell. As with high-abundance genes, we observed that only features describing relative changes in Ch-RelA nuclear intensity were strong predictors of output, with maximal fold change in nuclear RelA being the strongest predictor of all the features considered (Fig. 3 D). Surprisingly, this trend in the correlations was similar for all promoters despite the differences in their transcript distributions (Fig. 3 B; Table 3). We tested whether integrating the relative change in nuclear Ch-RelA over time (i.e., area under the curve of F/Finit or AUCFC) would improve the correlation over using only its maximum. However, consistent with the observation that there is a strong correlation between these two features (Fig. S3 D), we found no improvement in correlations. Overall, we conclude that RelA fold-change detection is conserved at genes supporting low TNF-inducible mean transcription and with different TNF-induced transcript distributions.
Mathematical model of I1-FFL RelA signaling reveals that varying relative RelA binding affinity alone is insufficient to recapitulate single-cell experimental data
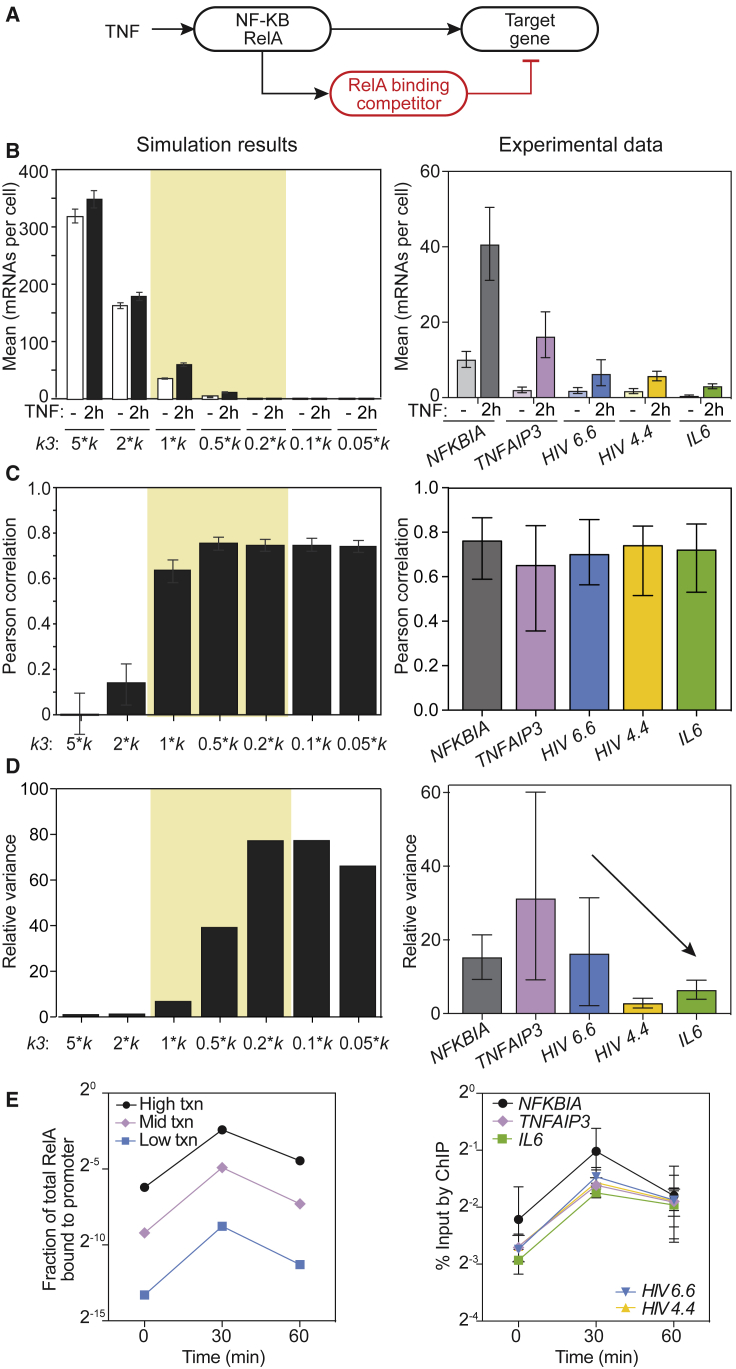
Fold-change detection of signals can be executed with an I1-FFL signaling motif (8). We previously implemented an I1-FFL in a mathematical model of TNF-stimulated NF-κB RelA activation and target gene transcription by incorporating a competitor protein that is induced by RelA and competes for binding to promoter κB sites, thereby inhibiting RelA-mediated transcription (7) (Fig. 4 A; Fig. S2). NF-κB p50, NF-κB p52, and BCL3 are potential candidates for this competitor protein because they are all induced by RelA (21), and they form complexes that suppress transcription of κB-site-containing genes, including IL6 and the HIV long terminal repeat (e.g., p52:p52 or p50:p50, which is stabilized by BCL3 (22, 23)). This model accurately described our observations in HeLa cells for RelA target genes with high early TNF-inducible transcription (7). We now sought to explore whether this model could account for our new experimental observations in Jurkat cells extending to RelA target genes with low early TNF-inducible transcription.
Figure 4.
Decreasing transcriptional output by decreasing relative RelA binding maintains fold-change detection but is inconsistent with experimental RelA binding data. (A) A schematic of how the I1-FFL motif was implemented in a computational model of NF-κB RelA signaling. (B) A bar graph of mean transcriptional output simulated by the model (left) and measured experimentally by smFISH (right) before and 2 h after TNF stimulation. Simulations were run for k4 = 1 × k and the indicated k3 values, where k4 is the affinity of competitor for the competitor promoter and k and k3 are the respective affinities of nNFκB and competitor for the induced target promoter; cell-to-cell variability was modeled by sampling for total RelA-containing NF-κB (NF) and TNF-induced IKK activity (ka). Values that reproduced the experimentally observed transcript range are shaded in yellow. Error bars represent the ±95% CIs obtained by bootstrapping. Experimental data are similarly presented as the mean ± 95% CIs obtained by bootstrapping. (C) A bar graph of Pearson correlations between Fmax/Finit and transcript abundance 2 h after TNF stimulation for simulated results (left) and experimental data (right). Simulations were run as described in (B). Error bars again represent 95% CIs. (D) A bar graph of relative variance in transcript distributions post- versus pre-TNF treatment (t = 2 h, 20 ng/mL) for simulation results (left) and experimental data (right). Relative variance is defined as variance at t = 2 h over the variance at t = 0; error bars represent 95% CIs. (E) Time courses of simulated RelA binding (left) and measured enrichment of RelA (% input measured by ChIP; right) after TNF addition. Simulations were run as described in (B) (error bars for 95% CIs are smaller than the markers). ChIP data are presented as mean ± standard error of independent biological duplicate (HIV 4.4 and HIV 6.6) or triplicate (endogenous promoters). There are no significant differences in measured time courses (p > 0.05, calculated by two-way ANOVA).
We previously showed that it is possible to tune transcriptional output of high-abundance RelA targets in the model while maintaining fold-change detection by simply varying the strength of competitor binding affinity relative to RelA binding affinity at its promoter (7). This is biologically plausible because the relative affinities of RelA and the candidate competitor proteins vary across promoter κB site sequences (24). To determine if this finding still held at low-abundance targets, we performed a model parameter scan (changing competitor affinity k3 while maintaining the same RelA affinity k) to find a range of competitor affinities for target promoters that reproduced the approximate absolute change in transcript abundance for NFKBIA, TNFAIP3, HIV, and IL6 observed in our experiments 2 h after addition of 20 ng/mL TNF (Fig. 4 B). At each competitor affinity value, we simulated cell-to-cell variability by sampling over a range of total RelA-containing NFκB abundance (NF) and ka (a proxy for the strength of activation of pathways leading to IKK), two parameters chosen based on the sensitivity of fold change of nuclear RelA (Fig. S5). All other parameters remained the same as previously reported (7) (Table S1).
We found that moderate to strong correlations between maximal fold change in nuclear RelA and transcript number were maintained as competitor affinity increased and TNF-induced transcript abundance decreased, consistent with experimental observations (Fig. 4 C). We also previously reported that the variance of TNF-mediated transcript number distributions relative to transcript variance in the basal state (referred to as relative transcript variance) increased with decreasing TNF-induced transcript abundance (7). Consistent with this, reducing average TNF-stimulated mRNA levels from 58 to 1 resulted in the model prediction that relative transcript variance would increase ∼8-fold (Fig. 4 D). However, our single-cell measurements show that for very low-abundance targets, relative transcript variance decreases, in contrast to model predictions (Fig. 4 D, right) and to our observations at high-abundance targets (7).
To explore this discrepancy, we noted that if we reduced transcript output solely by varying competitor affinity, the model predicted that the average fraction of RelA bound at 30 min after TNF treatment would need to be reduced nearly 80-fold (Fig. 4 E, left). We tested this prediction by using chromatin immunoprecipitation (ChIP) to measure average RelA binding at the promoters at 0, 30, and 60 min after TNF treatment. In contrast to the model prediction, we found no significant differences in RelA binding across all promoters after TNF treatment (Fig. 4 E, right). Because the relative abundances of RelA versus competitor proteins (determined by varying k4) is a less well-determined parameter and may differ between promoters, we also scanned across a range of k4 values to assess whether different trends in RelA binding and relative transcript variance could be found, but we found that all trends remained similar (Fig. S4). Thus, varying competitor affinity can reproduce the difference in transcript abundance at low- versus high-output targets and observations of fold-change detection, but it is inconsistent with experimental measurements of relative transcript variance and average RelA binding. We conclude that other mechanisms must be used to reduce transcript abundance to very low levels while maintaining fold-change detection.
Tuning multiple model parameters for individual target genes reproduces observed TNF-RelA-mediated behavior
To systematically determine other model parameters that strongly affect target transcript abundance, we performed a parameter sensitivity analysis (Fig. S5). We found that in addition to competitor binding affinity (k3) and relative abundance of RelA versus competitor (k4), target gene transcription in the model is most affected by target transcript synthesis rate (c1t), target transcript degradation rate (c3t), and a Hill coefficient determining the steepness of the transcription response with respect to the concentration of nuclear RelA (ht). We performed a five-way parameter scan across a range of values for the parameters identified above and examined the distributions of transcript abundance outputted by the model that fell within our experimentally observed ranges (Fig. 5 A; yellow-red heat map). Cell-to-cell heterogeneity in the model was again produced by varying the initial concentrations of total RelA (NF) and the TNF-induced IKK activity (ka). Within this broad parameter space, it was possible to simulate a range of TNF-mediated increases in transcript abundance that are associated with different patterns of fold-change detection, relative transcript variance, and fractional RelA binding (Fig. S6 A).
Figure 5.
Tuning multiple model parameters for each target promoter can reproduce experimentally observed behaviors. (A) A heat map of transcript output 2 h after TNF stimulation for a range of RelA target binding affinities (k3) and relative competitor abundance (k4) values (varied across small rectangles, subsampling our simulation results to multipliers 0.1000, 0.1848, 0.3414, 0.6307, 1.1652, 2.1528, and 5.4063) and target transcript synthesis (c1t) and degradation (c3t) rates (varied within small rectangles using the full set of simulations described in Materials and Methods). The heat map color scale is set to the experimentally observed range of transcript output for all transcripts combined. Areas in gray color scale are outside of the experimentally observed range. Simulations were done for a Hill coefficient of ht = 3.2. Small blue squares highlight areas further explored in Fig. S6. (B) Possible parameter sets for each target transcript are highlighted in color over the same k3-k4-c1t-c3t space as in (A) and for the indicated Hill coefficient (ht), if they reproduce experimentally observed baseline and TNF-induced transcript levels at 2 h and have a Pearson correlation coefficient of r > 0.6 for fold-change detection.
We next explored how the predicted transcript distributions compared to our experimental measurements across a range of transcriptional outputs. To do this, we identified parameter sets that reproduced the approximate absolute transcript abundance pre- and post-TNF addition for each target observed experimentally (with t = 2 h, 20 ng/mL; defined by the interquartile range of single-cell measurements; Fig. S6 B). We further refined these sets by requiring that the fold-change correlation be recapitulated, resulting in a reduced range of possible parameters for each promoter (Fig. 5 B). Fractional RelA binding, FB, was constant for all simulations with the same pair of k3 and k4 values and neighboring regions in the k3-k4 space had similar RelA binding values. By comparing across similar k3-k4 regions, it was possible to find regions within this space in which transcript abundance changed without significant changes in RelA binding (Fig. 5 B; Fig. S6 A). Based on these simulation results, we conclude that to maintain fold-change detection for all low-abundance targets, it becomes necessary to vary a combination of model parameters, rather than a single parameter as we previously reported (7).
Finally, we explored whether we could combine experimentally observed trends in relative transcript variance with prior biological knowledge to further constrain the parameter space. In particular, TNF-stimulated transcript number distributions and their relative variance trends are different for the two integrations of HIV, despite the fact that the promoter is identical and supports a similar average TNF-stimulated transcription. Therefore, we fixed RelA competitor affinity (k3) and competitor abundance (k4), as well as transcript degradation rate (c3t), for HIV, based on the assumption that these values would be the same for the same HIV promoter and transcript, respectively. We then varied transcript synthesis rate (c1t) and the Hill coefficient (ht) and searched for parameterizations that could reproduce the observed trends in relative transcript variance while still maintaining fold-change detection (Fig. 6, A–C). We found that by individually varying these two parameters for a specific set of k3, k4, and c3t, it was possible to obtain a similar increase in transcript abundance with different relative variances while maintaining fold-change detection (Fig. 6, D and E). Furthermore, it was possible to approximately reproduce all experimentally observed trends across the full range of transcript abundances with identical RelA binding (i.e., fixed values of k3 and k4). We note that these are not the only parameterizations that could reproduce the trends and that the assumption that k3, k4 and c3t are invariant is unlikely to hold for NFKBIA, TNFAIP3, and IL6, although the space is substantially constrained (Fig. 5 B). By using experimental measurements to fix additional model parameters (e.g., c3t for NFKBIA, TNFAIP3, and IL6), it may be possible to find, for each promoter, a well-constrained parameter set that could reproduce experimental trends.
Figure 6.
Parametrizations of an I1-FFL model of NF-κB-driven transcription can capture features of TNF-induced increases in HIV transcription as well as that of low-abundance endogenous genes. (A–D) Heat maps are shown of baseline (A) and post-TNF (t = 2 h; B) mean transcript number, relative variance (expressed as log10 (Vart = 2 h/Vart = 0 h) (C), and Pearson correlation of transcript number with fold change in nuclear RelA (r; D) for a range of target transcript transcription rates (c1t versus its default value c1td) and Hill coefficients (ht). Parametrizations with good fit for NFKBIA (gray), TNFAIP3 (lilac), HIV 6.6 (blue), HIV 4.4 (yellow), and IL6 (green) are outlined. All simulations were run using fixed values of k3 (0.3414 × k), k4 (1.1652 × k), and c3t (0.6307 × c3) chosen from a region that showed good fit to mean transcript numbers for HIV 6.6 and HIV 4.4 (Fig. 5B). Cell-to-cell variability was generated by sampling over ka and total RelA-containing NF-κB (parameter NF; Table S1) as described in Materials and Methods. (E) Two-dimensional scatter plots showing the relationship between target mRNA abundance and maximal fold change in nuclear NFκB (for t = 2 h) for model simulations with parametrizations chosen in (A)–(D) to recapitulate HIV 4.4 and HIV 6.6 data (top, yellow and blue, respectively) or simulations that show quantitatively and qualitatively different mRNAs versus maximal fold change distributions (bottom, gray). Selected parameter values are indicated (right); cell-to-cell variability was obtained by sampling total NFκB and ka values, and all other parameters were held at their default value in the model. (F) A schematic diagram showing some of the informative features of same-cell single-cell data measuring both an input and an output of a signaling pathway.
Overall, our quantitative single-cell data set of NF-κB signal-response relationships for low-abundance transcripts provided additional constraints for testing TNF-RelA model predictions. We conclude that to maintain fold-change detection across promoters with widely varying transcriptional outputs requires changes in multiple determinants of transcriptional outputs and that to accurately model NF-κB-driven gene transcription for a wide range of targets, measuring single-cell input-output relationship can help constrain model parameter values.
Discussion
Signal-response relationships measured in single cells provide insights into regulatory mechanisms. We previously discovered that RelA target promoters exhibit fold-change detection of TNF-stimulated signaling; here, we extended this finding to show that fold-change detection is conserved across cell types and even at promoters with low TNF-induced transcription. Acquiring signal-response relationships in single Jurkat T cells required development of a microfluidic-device-enabled imaging protocol to measure paired live-cell signaling dynamics and fixed-cell transcriptional output in the same suspension cells.
Our findings suggest that fold-change detection may be a general mechanism of TNF-induced NF-κB signal detection. Fold-change detection has also been observed in the extracellular regulated kinase pathway (25) and for cyclic adenosine 3′,5′-monophosphate in social amoebae (26), indicating that fold-change detection may confer broadly useful biological functionality. For example, fold-change detection has been proposed as a mechanism by which cells buffer against stochastic variation in signaling molecules by allowing transcriptional output to correlate with signal strength instead of absolute abundance of the signal-driving protein (7, 8). Our results show that this buffering is observed for low-abundance transcripts, even though gene expression noise has been observed to increase as mean expression decreases (27, 28).
Our results demonstrate some advantages of measuring transcript number for low-abundance transcripts rather than tracking a transcriptional reporter that requires protein expression. We previously quantified how TNF-stimulated RelA signaling features correlated to levels of HIV activation in J65c 4.4 and 6.6 using a GFP gene expression reporter that is subject to positive feedback amplification by the HIV-encoded protein Tat (12). A weakness of that approach was that only a minor fraction of the J65c 4.4 cells (25%) exhibited detectable reporter expression, even though most cells increased HIV transcription (12). Although we did find that the maximal fold change in RelA was the strongest predictor of the Tat-mediated GFP protein expression for both viral integrations, we observed that this correlation was significantly weaker for J65c 4.4 versus J65c 6.6 (r = 0.55 vs. r = 0.78). By directly measuring transcriptional output using smFISH, we now conclude that the RelA signal-to-transcript relationship is identical for both clones, and inefficient Tat-mediated positive feedback degrades the correlation for J65c 4.4. We note, however, that smFISH is a static, end-point measurement, whereas transcriptional reporters provide a way to track transcriptional dynamics, which may provide additional useful information in other biological scenarios.
To gain mechanistic insight, we compared our experimental measurements across a range of transcript abundances to simulations of a mathematical model of TNF-mediated NF-κB signaling (7). Tuning RelA binding affinity at the target promoters was sufficient to recapitulate our observation of fold-change detection across cells for low-abundance transcripts as previously reported (7) but was inconsistent with our experimental observation that average RelA binding did not change at the low-abundance targets. Instead, we were able to qualitatively reproduce experimental observations by varying multiple parameters independently for each target promoter.
Why might there be limited transcription at some promoters despite similar transcription factor occupancy? One potential simple explanation is that these different targets have different transcript stability, a parameter that has previously been shown to also strikingly affect the dynamics of transcript accumulation of NF-κB and p53 targets (29, 30, 31). However, a difference in transcript stability could not explain the differences in transcript count distributions between the two HIV LTR integrations that we studied because their outputs are the same transcripts. Here, an explanation might be differing local chromatin environments. Indeed, genome-wide studies of transcriptional reporters integrated at different genomic locations, including the HIV LTR, show that transcriptional activity is affected by features of the local chromatin environment (32, 33) and the stability of nucleosomes, which compact DNA and may inhibit access of transcriptional machinery (34). In our abstracted model of NF-κB-driven transcription, such effects can be reflected in differences in target transcription rates (c1t). Additionally, transcription factor (TF)-induced chromatin and nucleosome remodeling appears to be a rate-limiting step for transcriptional activation (35, 36). Because rates of chromatin and nucleosome remodeling are slower than TF binding (37), they may further impose an apparent threshold of TF binding required for transcription (38). In our model, such behavior can be captured by varying the Hill coefficient (ht) characterizing the transcriptional response at a target promoter.
There are several molecular mechanisms that underlie this chromatin remodeling that may limit the rate of RelA-mediated transcription. Although RelA cannot directly remodel chromatin, RelA has been shown to recruit the histone acetyltransferase p300, which acetylates histones to enhance transcriptional activity (23, 39). Moreover, the candidate competitor proteins in the incoherent feed-forward loop of NF-κB-driven transcription, p50, p52, and BCL3, have been reported to mediate their repressive transcriptional activity at the HIV LTR and other RelA target promoters by recruiting histone deacetylases (40, 41, 42). Thus, RelA has the potential to mediate chromatin remodeling and change transcriptional output on a timescale that is different from the lifetime of the RelA-bound promoter complex, effectively yielding target genes with different transcript abundances while under the control of the same RelA-DNA-binding activity.
Going forward, quantitative models of transcription activated by the NF-κB pathway or other gene regulatory pathways will need to incorporate many contributing factors (i.e., signal decoding, promoter architecture, and chromatin environment) to accurately predict input-output relationships for specific genes in individual cells of different types. This will require the integration of both cell-population “average” and same-cell single-cell experimental measurements as described here (Fig. 6 F). The recruitment of cofactor complexes and changes in chromatin environment at the promoter has been addressed mathematically by modeling promoters as switching between discrete states (43, 44, 45, 46, 47). Incorporating this multistate promoter structure into the TNF-RelA signaling model may more accurately capture how RelA activates its transcript targets as compared to varying Hill coefficients for a single-state promoter. A recent study demonstrated how a transcription factor that affects the frequency of transitions between promoter states can cause strong transcriptional responses even with weak binding (48). We recently showed that TNF likely affects both the frequency of transitions between promoter states and the size of the transcriptional bursts in the active promoter state, depending on the chromatin environment at the promoter (12). Accurately modeling how transcription factors influence switching between these states will require additional direct observations of signaling input-output relationships across cell types, signaling pathways, and other gene targets, as well as coupling with measurements and manipulations of additional regulatory factors such as chromatin environment and transcriptional cofactor binding.
Author Contributions
V.C.W., S.G., and K.M.-J. conceived of the study. V.C.W. performed experiments. V.C.W., S.M., S.G., and K.M.-J. performed data analysis. S.M. and S.G. performed computational modeling. R.R. contributed to microfluidic protocol design. V.C.W., S.M., S.G., and K.M.-J. prepared figures. S.G. and K.M.-J. wrote the manuscript with contributions from all authors. S.G. and K.M.-J. supervised the research.
Acknowledgments
This work was funded by the National Science Foundation (CBET-1454301 to K.M.-J.) and the National Institutes of Health (R01-GM104247 to S.G.). V.C.W. was supported by a National Institutes of Health Predoctoral Training Grant in Genetics (2T32GM007499-36, 5T32GM007499-34, 5T32GM007499-35).
Editor: Kevin Janes.
Footnotes
Victor C. Wong and Shibin Mathew contributed equally to this work.
Victor C. Wong’s present address is Janelia Research Campus, Howard Hughes Medical Institute, Ashburn, Virginia.
Ramesh Ramji’s present address is Illumina Inc., San Diego, California.
Six figures and one table are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(19)30027-X.
Contributor Information
Suzanne Gaudet, Email: suzanne_gaudet@dfci.harvard.edu.
Kathryn Miller-Jensen, Email: kathryn.miller-jensen@yale.edu.
Supporting Material
References
- 1.Aggarwal B.B. Signalling pathways of the TNF superfamily: a double-edged sword. Nat. Rev. Immunol. 2003;3:745–756. doi: 10.1038/nri1184. [DOI] [PubMed] [Google Scholar]
- 2.Webb L.V., Ley S.C., Seddon B. TNF activation of NF-κB is essential for development of single-positive thymocytes. J. Exp. Med. 2016;213:1399–1407. doi: 10.1084/jem.20151604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xing Y., Wang X., Hogquist K.A. Late stages of T cell maturation in the thymus involve NF-κB and tonic type I interferon signaling. Nat. Immunol. 2016;17:565–573. doi: 10.1038/ni.3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smale S.T. Hierarchies of NF-κB target-gene regulation. Nat. Immunol. 2011;12:689–694. doi: 10.1038/ni.2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gaudet S., Miller-Jensen K. Redefining signaling pathways with an expanding single-cell toolbox. Trends Biotechnol. 2016;34:458–469. doi: 10.1016/j.tibtech.2016.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lane K., Van Valen D., Covert M.W. Measuring signaling and RNA-seq in the same cell links gene expression to dynamic patterns of NF-κB activation. Cell Syst. 2017;4:458–469.e5. doi: 10.1016/j.cels.2017.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee R.E., Walker S.R., Gaudet S. Fold change of nuclear NF-κB determines TNF-induced transcription in single cells. Mol. Cell. 2014;53:867–879. doi: 10.1016/j.molcel.2014.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adler M., Szekely P., Alon U. Optimal regulatory circuit topologies for fold-change detection. Cell Syst. 2017;4:171–181.e8. doi: 10.1016/j.cels.2016.12.009. [DOI] [PubMed] [Google Scholar]
- 9.Goentoro L., Shoval O., Alon U. The incoherent feedforward loop can provide fold-change detection in gene regulation. Mol. Cell. 2009;36:894–899. doi: 10.1016/j.molcel.2009.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ashall L., Horton C.A., White M.R. Pulsatile stimulation determines timing and specificity of NF-kappaB-dependent transcription. Science. 2009;324:242–246. doi: 10.1126/science.1164860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan J.K., Greene W.C. NF-κB/Rel: agonist and antagonist roles in HIV-1 latency. Curr. Opin. HIV AIDS. 2011;6:12–18. doi: 10.1097/COH.0b013e32834124fd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wong V.C., Bass V.L., Miller-Jensen K. NF-κB-chromatin interactions drive diverse phenotypes by modulating transcriptional noise. Cell Rep. 2018;22:585–599. doi: 10.1016/j.celrep.2017.12.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramji R., Wong V.C., Miller-Jensen K. A passive-flow microfluidic device for imaging latent HIV activation dynamics in single T cells. Integr. Biol. 2015;7:998–1010. doi: 10.1039/c5ib00094g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Padovan-Merhar O., Nair G.P., Raj A. Single mammalian cells compensate for differences in cellular volume and DNA copy number through independent global transcriptional mechanisms. Mol. Cell. 2015;58:339–352. doi: 10.1016/j.molcel.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raj A., van den Bogaard P., Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Methods. 2008;5:877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dirks W., Rome D., Drexler H.G. Expression of the growth arrest-specific gene 6 (GAS6) in leukemia and lymphoma cell lines. Leuk. Res. 1999;23:643–651. doi: 10.1016/s0145-2126(99)00075-2. [DOI] [PubMed] [Google Scholar]
- 17.Mueller F., Senecal A., Zimmer C. FISH-quant: automatic counting of transcripts in 3D FISH images. Nat. Methods. 2013;10:277–278. doi: 10.1038/nmeth.2406. [DOI] [PubMed] [Google Scholar]
- 18.Zong C., So L.H., Golding I. Lysogen stability is determined by the frequency of activity bursts from the fate-determining gene. Mol. Syst. Biol. 2010;6:440. doi: 10.1038/msb.2010.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.So L.H., Ghosh A., Golding I. General properties of transcriptional time series in Escherichia coli. Nat. Genet. 2011;43:554–560. doi: 10.1038/ng.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramirez-Carrozzi V.R., Braas D., Smale S.T. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell. 2009;138:114–128. doi: 10.1016/j.cell.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ten R.M., Paya C.V., Israël A. The characterization of the promoter of the gene encoding the p50 subunit of NF-kappa B indicates that it participates in its own regulation. EMBO J. 1992;11:195–203. doi: 10.1002/j.1460-2075.1992.tb05042.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lewin S.R., Lambert P., Crowe S.M. Constitutive expression of p50 homodimer in freshly isolated human monocytes decreases with in vitro and in vivo differentiation: a possible mechanism influencing human immunodeficiency virus replication in monocytes and mature macrophages. J. Virol. 1997;71:2114–2119. doi: 10.1128/jvi.71.3.2114-2119.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhong H., May M.J., Ghosh S. The phosphorylation status of nuclear NF-kappa B determines its association with CBP/p300 or HDAC-1. Mol. Cell. 2002;9:625–636. doi: 10.1016/s1097-2765(02)00477-x. [DOI] [PubMed] [Google Scholar]
- 24.Siggers T., Chang A.B., Bulyk M.L. Principles of dimer-specific gene regulation revealed by a comprehensive characterization of NF-κB family DNA binding. Nat. Immunol. 2011;13:95–102. doi: 10.1038/ni.2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cohen-Saidon C., Cohen A.A., Alon U. Dynamics and variability of ERK2 response to EGF in individual living cells. Mol. Cell. 2009;36:885–893. doi: 10.1016/j.molcel.2009.11.025. [DOI] [PubMed] [Google Scholar]
- 26.Kamino K., Kondo Y., Sawai S. Fold-change detection and scale invariance of cell-cell signaling in social amoeba. Proc. Natl. Acad. Sci. USA. 2017;114:E4149–E4157. doi: 10.1073/pnas.1702181114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bar-Even A., Paulsson J., Barkai N. Noise in protein expression scales with natural protein abundance. Nat. Genet. 2006;38:636–643. doi: 10.1038/ng1807. [DOI] [PubMed] [Google Scholar]
- 28.Sanchez A., Golding I. Genetic determinants and cellular constraints in noisy gene expression. Science. 2013;342:1188–1193. doi: 10.1126/science.1242975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zambrano S., De Toma I., Agresti A. NF-κB oscillations translate into functionally related patterns of gene expression. eLife. 2016;5:e09100. doi: 10.7554/eLife.09100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Porter J.R., Fisher B.E., Batchelor E. p53 pulses diversify target gene expression dynamics in an mRNA half-life-dependent manner and delineate co-regulated target gene subnetworks. Cell Syst. 2016;2:272–282. doi: 10.1016/j.cels.2016.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hafner A., Stewart-Ornstein J., Lahav G. p53 pulses lead to distinct patterns of gene expression albeit similar DNA-binding dynamics. Nat. Struct. Mol. Biol. 2017;24:840–847. doi: 10.1038/nsmb.3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Akhtar W., de Jong J., van Steensel B. Chromatin position effects assayed by thousands of reporters integrated in parallel. Cell. 2013;154:914–927. doi: 10.1016/j.cell.2013.07.018. [DOI] [PubMed] [Google Scholar]
- 33.Lewinski M.K., Bisgrove D., Bushman F.D. Genome-wide analysis of chromosomal features repressing human immunodeficiency virus transcription. J. Virol. 2005;79:6610–6619. doi: 10.1128/JVI.79.11.6610-6619.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaplan N., Moore I.K., Segal E. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458:362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim H.D., O’Shea E.K. A quantitative model of transcription factor-activated gene expression. Nat. Struct. Mol. Biol. 2008;15:1192–1198. doi: 10.1038/nsmb.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boeger H., Griesenbeck J., Kornberg R.D. Nucleosome retention and the stochastic nature of promoter chromatin remodeling for transcription. Cell. 2008;133:716–726. doi: 10.1016/j.cell.2008.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Voss T.C., Schiltz R.L., Hager G.L. Dynamic exchange at regulatory elements during chromatin remodeling underlies assisted loading mechanism. Cell. 2011;146:544–554. doi: 10.1016/j.cell.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller-Jensen K., Dey S.S., Schaffer D.V. Chromatin accessibility at the HIV LTR promoter sets a threshold for NF-κB mediated viral gene expression. Integr. Biol. (Camb) 2012;4:661–671. doi: 10.1039/c2ib20009k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mukherjee S.P., Behar M., Ghosh G. Analysis of the RelA:CBP/p300 interaction reveals its involvement in NF-κB-driven transcription. PLoS Biol. 2013;11:e1001647. doi: 10.1371/journal.pbio.1001647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jamaluddin M., Choudhary S., Brasier A.R. Respiratory syncytial virus-inducible BCL-3 expression antagonizes the STAT/IRF and NF-kappaB signaling pathways by inducing histone deacetylase 1 recruitment to the interleukin-8 promoter. J. Virol. 2005;79:15302–15313. doi: 10.1128/JVI.79.24.15302-15313.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wessells J., Baer M., Johnson P.F. BCL-3 and NF-kappaB p50 attenuate lipopolysaccharide-induced inflammatory responses in macrophages. J. Biol. Chem. 2004;279:49995–50003. doi: 10.1074/jbc.M404246200. [DOI] [PubMed] [Google Scholar]
- 42.Williams S.A., Chen L.F., Greene W.C. NF-kappaB p50 promotes HIV latency through HDAC recruitment and repression of transcriptional initiation. EMBO J. 2006;25:139–149. doi: 10.1038/sj.emboj.7600900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peccoud J., Ycart B. Markovian modeling of gene-product synthesis. Theor. Popul. Biol. 1995;48:222–234. [Google Scholar]
- 44.Raj A., Peskin C.S., Tyagi S. Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 2006;4:e309. doi: 10.1371/journal.pbio.0040309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Singh A., Razooky B., Weinberger L.S. Transcriptional bursting from the HIV-1 promoter is a significant source of stochastic noise in HIV-1 gene expression. Biophys. J. 2010;98:L32–L34. doi: 10.1016/j.bpj.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Skupsky R., Burnett J.C., Arkin A.P. HIV promoter integration site primarily modulates transcriptional burst size rather than frequency. PLoS Comput. Biol. 2010;6:e1000952. doi: 10.1371/journal.pcbi.1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chavali A.K., Wong V.C., Miller-Jensen K. Distinct promoter activation mechanisms modulate noise-driven HIV gene expression. Sci. Rep. 2015;5:17661. doi: 10.1038/srep17661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li C., Cesbron F., Hofer T. Frequency modulation of transcriptional bursting enables sensitive and rapid gene regulation. Cell Syst. 2018;6:409–423.e11. doi: 10.1016/j.cels.2018.01.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.