Summary
Live cell imaging has been widely used to generate data for quantitative understanding of cellular dynamics. Various applications have been developed to perform automated imaging data analysis, which often requires tedious manual correction. It remains a challenge to develop an efficient curation method that can analyze massive imaging datasets with high accuracy. Here, we present eDetect, a fast error detection and correction tool that provides a powerful and convenient solution for the curation of live cell imaging analysis results. In eDetect, we propose a gating strategy to distinguish correct and incorrect image analysis results by visualizing image features based on principal component analysis. We demonstrate that this approach can substantially accelerate the data correction process and improve the accuracy of imaging data analysis. eDetect is well documented and designed to be user friendly for non-expert users. It is freely available at https://sites.google.com/view/edetect/ and https://github.com/Zi-Lab/eDetect.
Subject Areas: Automation in Bioinformatics, Bioinformatics, Cell Biology
Graphical Abstract
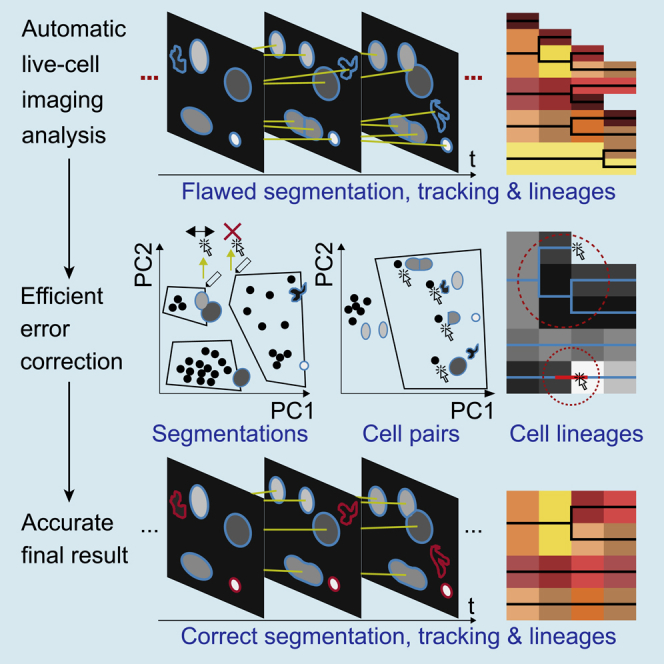
Highlights
-
•
eDetect is a user-friendly software for live cell imaging data analysis
-
•
It provides an easy solution to detect and correct errors in imaging data analysis
-
•
It enables precise cell lineage reconstruction from long-term imaging datasets
-
•
It is compatible with CellProfiler and performs well in different imaging datasets
Automation in Bioinformatics; Bioinformatics; Cell Biology
Introduction
Live cell imaging has been widely used in monitoring cellular processes and dynamics at the single-cell level (Ni et al., 2018, Specht et al., 2017, White et al., 2018). During the course of these studies, large volumes of time-lapse images are often generated by automatic microscopes, which poses a challenge in quantifying cellular dynamics from these datasets with high precision (Skylaki et al., 2016). In the past, computational tools have been developed to enable automated analysis of large image datasets (Blanchoud et al., 2015, Carpenter et al., 2006, Cooper et al., 2017, de Chaumont et al., 2012, Held et al., 2010, Hilsenbeck et al., 2017, Liepe et al., 2016, Winter et al., 2016). However, the errors in the automated analyses impaired the accuracy in quantifying long-term single-cell dynamics. In most cases, fully automatic algorithms cannot guarantee 100% accuracy (Ulman et al., 2017). Even a small portion of errors can result in a much larger amount of false cell lineages (Skylaki et al., 2016). Therefore manual inspection and correction are needed (Hilsenbeck et al., 2016).
Several tools have provided functions to manually correct image data analysis results. For example, CellProfiler and qTfy allow the users to manually edit the contours of segmented objects one by one (Carpenter et al., 2006, Hilsenbeck et al., 2016). In addition, CellTracker, NucliTrack, LEVER, and tTt support manual track editing (Cooper et al., 2017, Hilsenbeck et al., 2016, Piccinini et al., 2016, Winter et al., 2016). These tools are useful, but an approach is still needed to improve the efficiency of data curation.
Here, we report an error detection and correction tool for live cell imaging data analysis (eDetect). In eDetect, we proposed a new gating approach to classify flawed segmentation and tracking results by visualizing image features in scatterplots based on principal-component analysis (PCA). As cell objects with similar appearance often aggregate in the scatterplot, we can easily detect a group of errors from voluminous time-lapse images. Similar errors can be efficiently corrected with batch operations in eDetect. In addition, eDetect uses multiple quality control steps to achieve high accuracy in the final live cell imaging analysis results. In summary, eDetect is a comprehensive software tool that integrates cell segmentation, feature extraction, cell tracking, and efficient data curation.
Results
eDetect Provides a Fast Way to Detect Segmentation Errors
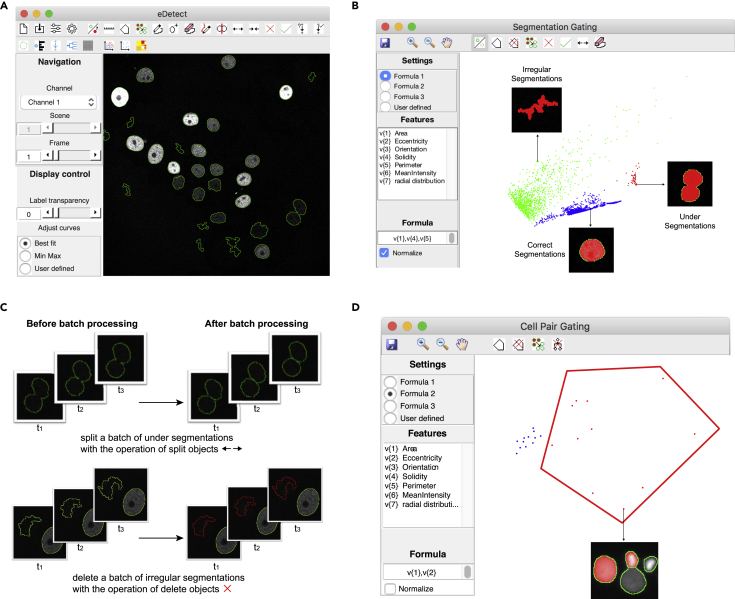
We first used eDetect to perform cell segmentation on the live cell imaging data of mouse stem cells, which is a benchmark dataset (Fluo-N2DH-GOWT1) from the Cell Tracking Challenge (Maška et al., 2014, Ulman et al., 2017). The results of automatic segmentations are shown in the image display window (Figure 1A). We found that the application of automatic segmentation algorithms can result in different types of errors such as under-segmentations (multiple adjacent nuclei are labeled as one object), over-segmentations (one nucleus is divided into multiple objects), and irregular shapes. To efficiently detect these errors, eDetect provides a function of segmentation gating, which uses a scatterplot to visualize image features based on PCA. Each point in this plot represents a segmented object. As shown in Figure 1B, the correct and wrong segmentation objects are well separated and displayed as different groups. By clicking on the points in the scatterplot, the corresponding segmentation objects are highlighted in the image display window. In this way, the users can easily inspect and identify different groups of segmentation errors.
Figure 1.
Main Features of eDetect
(A) Main interface of eDetect.
(B) Segmentation gating helps the users to separate correct and incorrect segmentation objects into different groups. Representative correct and incorrect segmentation objects are shown for different groups.
(C) Batch correction of segmentation errors. Segmentation gating enables splitting a batch of under-segmentations (red points in panel B) or deleting irregular segmentations (green points in panel B) in all frames with one batch operation.
(D) Cell pair gating function in eDetect. The red polygon gate indicates the likely misassigned cell divisions in the cell tracking. An example of misassigned cell division is shown.
eDetect Can Correct a Group of Errors with Batch Operations
To further improve the efficiency of error correction, eDetect provides both manual and batch operations for splitting and deleting cell segmentations. When the same type of segmentation errors are grouped together in the PCA plot, eDetect can correct these types of errors with a batch operation after selecting them with a polygon. For example, the cluster of under-segmentations can be corrected by one click of splitting objects in eDetect (Figure 1C). Similarly, the irregular non-cell segmentations can be easily discarded with one “Delete objects” operation.
Multiple Quality Control Steps Improve the Accuracy of Live Cell Imaging Data Analysis
Although segmentation gating can assist the users to correct segmentation errors efficiently, some mistakes may still exist. Therefore eDetect provides additional steps to improve the accuracy of data analysis. To illustrate this feature, we applied eDetect to analyze a live cell imaging dataset (HaCaT-FUCCI) from human HaCaT cells that stably express the CFP-H2B nuclear marker and mCherry-Geminin FUCCI cell cycle indicator proteins (Sakaue-Sawano et al., 2008).
To improve the precision of cell tracking, eDetect uses cell pair gating to visualize identified cell divisions in another PCA-based scatterplot. eDetect automatically locates and marks the corresponding pair of cells in the original image when the user points at a cell division pair (Figure 1D). By inspecting the highlighted pairs of cells, the users can find and correct misassigned cell divisions.
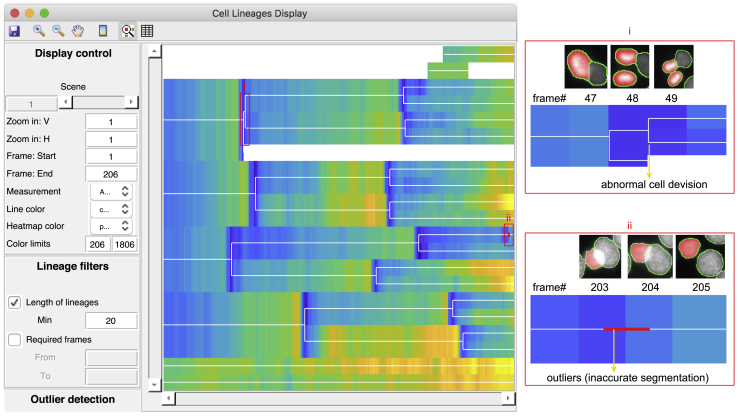
As the last quality control module in eDetect, cell lineages display window assists the users to identify missed errors from previous steps. This is implemented by visualizing different measurements (e.g., nuclear area) of the objects in cell lineages with a heatmap. In general, nuclear area changes gradually under normal cell growth and only shows abrupt changes during cell divisions. The users can easily find abnormalities such as abnormal cell divisions, abrupt changes of nuclear sizes, and incomplete cell lineages (Figure 2). eDetect can assist us to check and correct these abnormalities in convenient ways. For example, eDetect has an automatic outlier detection function to mark abrupt changes within cell lineage branches. In addition, it can generate a synchrogram (a sequence of images of the selected cell lineage), which facilitates the check on the correctness of cell lineages (Sigal et al., 2006). After multiple error correction steps, we can accurately quantify the dynamics of the cell cycle sensor protein mCherry-Geminin in individual HaCaT cells (Figure S1).
Figure 2.
Cell Lineages Display in eDetect Improves the Accuracy of Live Cell Imaging Analysis Results
eDetect enables the detection of other overlooked errors with heatmap visualization. Two examples of errors in cell lineages results are zoomed in and shown on the right: (i) abnormal cell divisions (the cell division between frame #48 and #49 is misassigned), and (ii) outliers (the highlighted nucleus in frame #203 has an inaccurate segmentation).
Compatibility of eDetect with CellProfiler
eDetect can be used together with CellProfiler to maximize its interoperability. The users can first perform image preprocessing and segmentation with CellProfiler and then import the results into eDetect for error detection and correction. As CellProfiler provides advanced algorithms and pipelines for image analysis, the compatibility of eDetect with CellProfiler will improve the broad application of eDetect for live cell imaging analysis.
Evaluation of eDetect Performance to Detect and Correct Errors in Live Cell Imaging Data Analysis
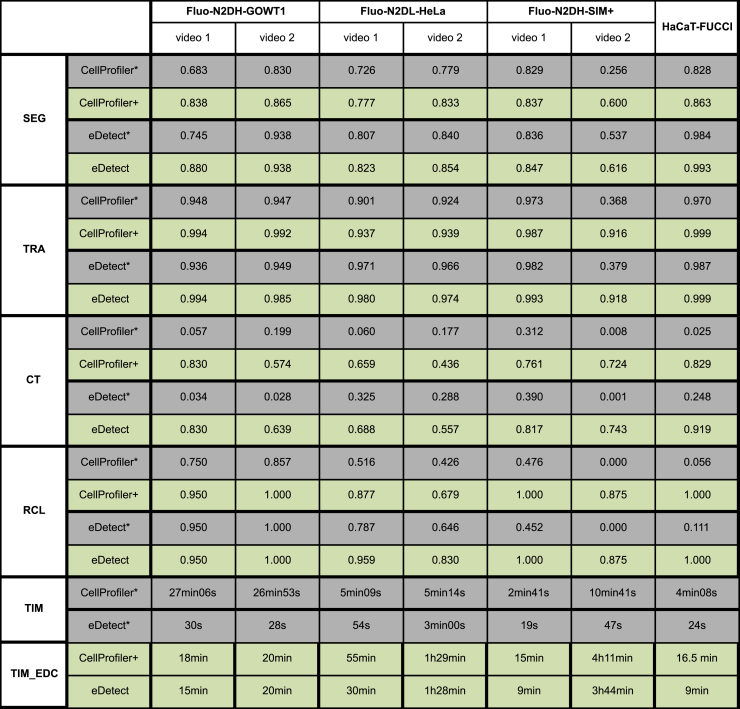
To evaluate the effectiveness and time cost of eDetect in error detection and correction, we first performed automatic data analysis in CellProfiler (CellProfiler*) and eDetect (eDetect*) without any corrections. In parallel, we imported the automatic analysis results from CellProfiler and then performed additional error correction with eDetect (CellProfiler+). In addition, we implemented automatic image analysis and error correction with eDetect (eDetect). Performance was quantified based on segmentation accuracy (SEG), tracking accuracy (TRA), complete tracks (CT) and recall of complete lineages (RCL). The RCL score measures the fraction of cells (including their descendants) that are correctly tracked from the first frame to the last frame of the movie. A detailed description of the performance metrics is provided in the Transparent Methods. The performance scores were calculated for four datasets, consisting of an imaging dataset HaCaT-FUCCI and three training datasets from the Cell Tracking Challenge.
As shown in Figure 3, eDetect can improve the performance of data analyses on different datasets by implementing error correction (comparing eDetect* and eDetect). The average values of SEG and TRA for the tested four datasets are increased by 0.04 and 0.10, respectively. More importantly, the average values of CT and RCL are increased to a larger extent, by 0.55 and 0.38, respectively. Similar performance augmentation is observed by comparing CellProfiler* and CellProfiler+. The improvement of CT and RCL values is more apparent when a long-term live cell imaging dataset is analyzed. For the 2-day imaging dataset HaCaT-FUCCI, although automatic algorithms in CellProfiler* and eDetect* reached very good SEG and TRA scores, the corresponding CT and RCL values are very low. This result indicates that a small portion of segmentation and tracking errors occurring in the automatic analysis can lead to a lot of errors in CT, which need to be corrected to reach valid conclusions. By performing error detection and correction in eDetect, the RCL value was increased from 0.111 to 1 with about 9 min of error correction on HaCaT-FUCCI dataset. The RCL value is especially important to draw biological conclusions when a reconstruction of complete cell lineages is needed. Therefore, eDetect is very useful especially for long-term imaging data analysis.
Figure 3.
The Performance of Error Detection and Correction Function in eDetect
The performance values were obtained with or without additional error correction steps in eDetect. CellProfiler*: automatic pipeline analysis with CellProfiler. CellProfiler+: automatic pipeline analysis was performed with CellProfiler first and then the results were imported to eDetect and additional error detection and correction was performed. eDetect*: automatic analysis with eDetect. eDetect: automatic analysis plus error detection and correction in eDetect.
In general, eDetect is efficient and has a low time cost (Figure 3). The execution time (TIM) for the automatic analysis step is less than 1 min for six tested imaging videos running on windows with eight CPU cores (Intel Core i7-6,700 Processor, 3.4 GHz). The time used in the error detection and correction steps (TIM_EDC) depends on the performance of automatic segmentation algorithms and the size of the image dataset. The error correction steps took about 10–90 min for six of the analyzed videos with total number of frames ranging between 65 and 206. For the video #2 of Fluo-N2DH-SIM+ dataset, the performance measures are relative poor when automatic analyses were performed in both CellProfiler* and eDetect*. This might be caused by the artificial noise introduced in this simulated dataset. In this case, several hours of manual correction of segmentations are needed to substantially improve the accuracy of data analysis.
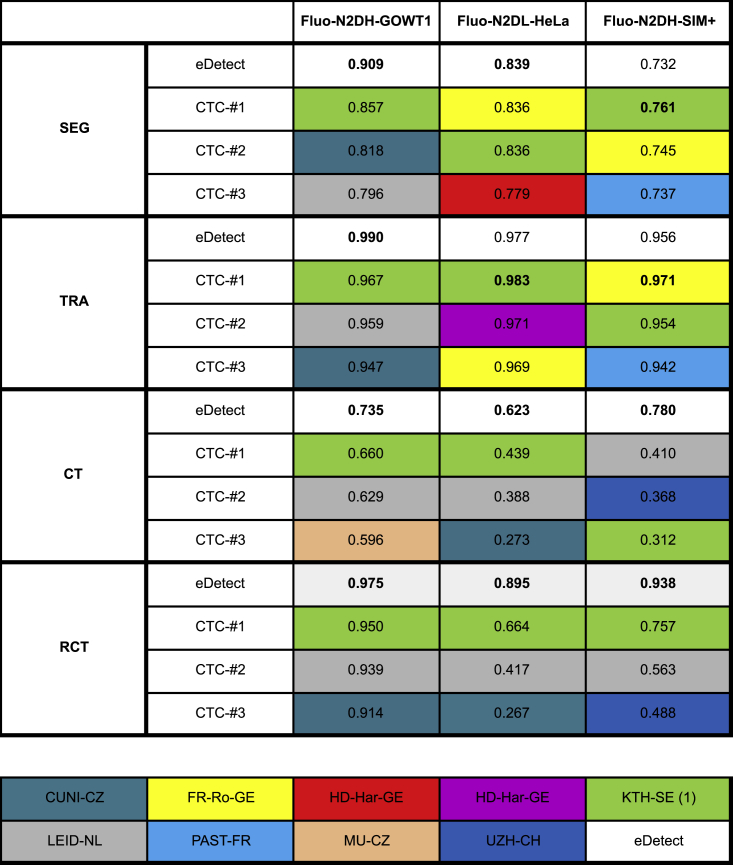
We next compared eDetect with the top-three performing methods from Cell Tracking Challenge using three benchmarking datasets (Figure 4). In terms of the SEG and TRA scores, eDetect has comparable performance to those of top-performing methods. It is important to note that, with the function of error detection and correction, eDetect could achieve the best performance in the measurements of complete tracks and recall of complete lineages. These results highlight the importance of error correction in long-term live cell imaging data analysis.
Figure 4.
Comparison of eDetect with Top Three Performing Methods from the Cell Tracking Challenge
For each dataset, the performance scores were computed by the average of scores from two training videos. The color code below indicates the top three performing methods submitted by the corresponding participants in Cell Tracking Challenge.
Discussion
The quantification of long-term single-cell dynamics is crucial for understanding cell signaling dynamics and cell fate in development. Although improving the accuracy of automatic algorithms is very important for live cell imaging data analysis (Maška et al., 2014, Ulman et al., 2017), an efficient curation tool is also urgently needed because a small fraction of errors, especially occurring earlier in the videos, could lead to a large fraction of errors in the complete cell lineages. In eDetect, we proposed a PCA-based approach to detect and correct errors, which substantially reduces the time for manually pinpointing errors from massive image datasets. In addition, eDetect can help us to correct or remove errors with one-click batch operations. Last, but not least, multiple quality control steps ensure a high precision in the final imaging analysis results.
Despite its effectiveness, segmentation gating in eDetect is inherently limited: it requires the users to customize PCA input variables. In practice, it is difficult to completely separate correct and incorrect segmentations at once. The users can achieve accurate segmentations by applying multiple gating steps with different PCA inputs even if each round of gating is not ideal. Moreover, overlooked errors can still be found in subsequent quality control steps. On the other hand, supervised learning methods have been previously used for phenotypic analysis of high-content imaging data (Piccinini et al., 2017, Ramo et al., 2009, Smith et al., 2018). In the future, these methods could be potentially used in eDetect to detect segmentation errors.
Here, we demonstrated eDetect's capability to detect and correct errors in live cell imaging data analysis. Making sense of large imaging datasets is a demanding challenge that requires powerful and efficient computational tools. The development of eDetect will be valuable for improving the precision of live cell imaging data analysis.
Limitations of This Study
Current release of eDetect does not provide multiple algorithms for segmentation and tracking. The performance of automatic analysis in eDetect may not be suitable for certain datasets. The compatibility of eDetect with CellProfiler offers the choice of various segmentation and tracking methods from CellProfiler, but it requires additional pipeline setup work. In the future, eDetect could be improved by integrating more established methods for segmentation and tracking. In addition, there is also a strong demand for the analysis of high-throughput time-lapse datasets. Although eDetect can in principle handle these datasets, it would require a large amount of manual labor. To address this challenge, it would be helpful to apply machine learning methods for error detection and correction.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
The authors want to thank Dr. E. Bártová for providing a dataset from the Cell Tracking Challenge and Dr. Xuedong Liu and Adrian Ramirez for proofreading of the manuscript. We are grateful to anonymous reviewers for their constructive suggestions, which have helped us to improve eDetect. This work was supported by grants to Z.Z. from the Federal Ministry of Education and Research (BMBF, Germany)-funded e:Bio SyBioT project (031A309) and the German Research Foundation (DFG, GRK 1772, Computational Systems Biology). H.H. was supported by a fellowship from International Max Planck Research School for Computational Biology and Scientific Computing.
Author Contributions
H.H. developed the software and implemented performance evaluation on benchmark datasets. G.W. generated some live cell imaging data for testing this software. Y.L. tested the tool. Z.Z. conceived and supervised the project, provided input on the design and features of the application, and tested the tool. Z.Z. wrote the paper with input from H.H.
Declaration of Interests
The authors declare no competing interests.
Published: March 29, 2019
Footnotes
Supplemental Information includes Transparent Methods, one figure, and one table and can be found with this article online at https://doi.org/10.1016/j.isci.2019.02.004.
Supplemental Information
References
- Blanchoud S., Nicolas D., Zoller B., Tidin O., Naef F. CAST: an automated segmentation and tracking tool for the analysis of transcriptional kinetics from single-cell time-lapse recordings. Methods. 2015;85:3–11. doi: 10.1016/j.ymeth.2015.04.023. [DOI] [PubMed] [Google Scholar]
- Carpenter A.E., Jones T.R., Lamprecht M.R., Clarke C., Kang I.H., Friman O., Guertin D.A., Chang J.H., Lindquist R.A., Moffat J. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper S., Barr A.R., Glen R., Bakal C. NucliTrack: an integrated nuclei tracking application. Bioinformatics. 2017;33:3320–3322. doi: 10.1093/bioinformatics/btx404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Chaumont F., Dallongeville S., Chenouard N., Herve N., Pop S., Provoost T., Meas-Yedid V., Pankajakshan P., Lecomte T., Le Montagner Y. Icy: an open bioimage informatics platform for extended reproducible research. Nat. Methods. 2012;9:690–696. doi: 10.1038/nmeth.2075. [DOI] [PubMed] [Google Scholar]
- Held M., Schmitz M.H., Fischer B., Walter T., Neumann B., Olma M.H., Peter M., Ellenberg J., Gerlich D.W. CellCognition: time-resolved phenotype annotation in high-throughput live cell imaging. Nat. Methods. 2010;7:747–754. doi: 10.1038/nmeth.1486. [DOI] [PubMed] [Google Scholar]
- Hilsenbeck O., Schwarzfischer M., Loeffler D., Dimopoulos S., Hastreiter S., Marr C., Theis F.J., Schroeder T. fastER: a user-friendly tool for ultrafast and robust cell segmentation in large-scale microscopy. Bioinformatics. 2017;33:2020–2028. doi: 10.1093/bioinformatics/btx107. [DOI] [PubMed] [Google Scholar]
- Hilsenbeck O., Schwarzfischer M., Skylaki S., Schauberger B., Hoppe P.S., Loeffler D., Kokkaliaris K.D., Hastreiter S., Skylaki E., Filipczyk A. Software tools for single-cell tracking and quantification of cellular and molecular properties. Nat. Biotechnol. 2016;34:703–706. doi: 10.1038/nbt.3626. [DOI] [PubMed] [Google Scholar]
- Liepe J., Sim A., Weavers H., Ward L., Martin P., Stumpf M.P. Accurate reconstruction of cell and particle tracks from 3D live imaging data. Cell Syst. 2016;3:102–107. doi: 10.1016/j.cels.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maška M., Ulman V., Svoboda D., Matula P., Matula P., Ederra C., Urbiola A., España T., Venkatesan S., Balak D.M. A benchmark for comparison of cell tracking algorithms. Bioinformatics. 2014;30:1609–1617. doi: 10.1093/bioinformatics/btu080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Q., Mehta S., Zhang J. Live-cell imaging of cell signaling using genetically encoded fluorescent reporters. FEBS J. 2018;285:203–219. doi: 10.1111/febs.14134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccinini F., Balassa T., Szkalisity A., Molnar C., Paavolainen L., Kujala K., Buzas K., Sarazova M., Pietiainen V., Kutay U. Advanced cell classifier: user-friendly machine-learning-based software for discovering phenotypes in high-content imaging data. Cell Syst. 2017;4:651–655.e5. doi: 10.1016/j.cels.2017.05.012. [DOI] [PubMed] [Google Scholar]
- Piccinini F., Kiss A., Horvath P. CellTracker (not only) for dummies. Bioinformatics. 2016;32:955–957. doi: 10.1093/bioinformatics/btv686. [DOI] [PubMed] [Google Scholar]
- Ramo P., Sacher R., Snijder B., Begemann B., Pelkmans L. CellClassifier: supervised learning of cellular phenotypes. Bioinformatics. 2009;25:3028–3030. doi: 10.1093/bioinformatics/btp524. [DOI] [PubMed] [Google Scholar]
- Sakaue-Sawano A., Kurokawa H., Morimura T., Hanyu A., Hama H., Osawa H., Kashiwagi S., Fukami K., Miyata T., Miyoshi H. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell. 2008;132:487–498. doi: 10.1016/j.cell.2007.12.033. [DOI] [PubMed] [Google Scholar]
- Sigal A., Milo R., Cohen A., Geva-Zatorsky N., Klein Y., Alaluf I., Swerdlin N., Perzov N., Danon T., Liron Y. Dynamic proteomics in individual human cells uncovers widespread cell-cycle dependence of nuclear proteins. Nat. Methods. 2006;3:525–531. doi: 10.1038/nmeth892. [DOI] [PubMed] [Google Scholar]
- Skylaki S., Hilsenbeck O., Schroeder T. Challenges in long-term imaging and quantification of single-cell dynamics. Nat. Biotechnol. 2016;34:1137–1144. doi: 10.1038/nbt.3713. [DOI] [PubMed] [Google Scholar]
- Smith K., Piccinini F., Balassa T., Koos K., Danka T., Azizpour H., Horvath P. Phenotypic image analysis software tools for exploring and understanding big image data from cell-based assays. Cell Syst. 2018;6:636–653. doi: 10.1016/j.cels.2018.06.001. [DOI] [PubMed] [Google Scholar]
- Specht E.A., Braselmann E., Palmer A.E. A Critical and comparative review of fluorescent tools for live-cell imaging. Annu. Rev. Physiol. 2017;79:93–117. doi: 10.1146/annurev-physiol-022516-034055. [DOI] [PubMed] [Google Scholar]
- Ulman V., Maška M., Magnusson K.E., Ronneberger O., Haubold C., Harder N., Matula P., Matula P., Svoboda D., Radojevic M. An objective comparison of cell-tracking algorithms. Nat. Methods. 2017;14:1141–1152. doi: 10.1038/nmeth.4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White M.D., Zhao Z.W., Plachta N. In vivo imaging of single mammalian cells in development and disease. Trends Mol. Med. 2018;24:278–293. doi: 10.1016/j.molmed.2018.01.003. [DOI] [PubMed] [Google Scholar]
- Winter M., Mankowski W., Wait E., Temple S., Cohen A.R. LEVER: software tools for segmentation, tracking and lineaging of proliferating cells. Bioinformatics. 2016;32:3530–3531. doi: 10.1093/bioinformatics/btw406. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.