Ribosyl 1,5-bisphosphate (PRibP) was discovered 65 years ago and was believed to be an important intermediate in ribonucleotide metabolism, a role immediately taken over by its “big brother” phosphoribosyldiphosphate. Only recently has PRibP come back into focus as an important player in the metabolism of ribonucleotides with the discovery of the pentose bisphosphate pathway that comprises, among others, the intermediates PRibP and ribulose 1,5-bisphosphate (cf.
KEYWORDS: Nudix hydrolase; PRPP; nucleoside catabolism; nucleotide metabolism; phosphonate catabolism; ribose 1,5-bisphosphate
SUMMARY
Ribosyl 1,5-bisphosphate (PRibP) was discovered 65 years ago and was believed to be an important intermediate in ribonucleotide metabolism, a role immediately taken over by its “big brother” phosphoribosyldiphosphate. Only recently has PRibP come back into focus as an important player in the metabolism of ribonucleotides with the discovery of the pentose bisphosphate pathway that comprises, among others, the intermediates PRibP and ribulose 1,5-bisphosphate (cf. ribose 5-phosphate and ribulose 5-phosphate of the pentose phosphate pathway). Enzymes of several pathways produce and utilize PRibP not only in ribonucleotide metabolism but also in the catabolism of phosphonates, i.e., compounds containing a carbon-phosphorus bond. Pathways for PRibP metabolism are found in all three domains of life, most prominently among organisms of the archaeal domain, where they have been identified either experimentally or by bioinformatic analysis within all of the four main taxonomic groups, Euryarchaeota, TACK, DPANN, and Asgard. Advances in molecular genetics of archaea have greatly improved the understanding of the physiology of PRibP metabolism, and reconciliation of molecular enzymology and three-dimensional structure analysis of enzymes producing or utilizing PRibP emphasize the versatility of the compound. Finally, PRibP is also an effector of several metabolic activities in many organisms, including higher organisms such as mammals. In the present review, we describe all aspects of PRibP metabolism, with emphasis on the biochemical, genetic, and physiological aspects of the enzymes that produce or utilize PRibP. The inclusion of high-resolution structures of relevant enzymes that bind PRibP provides evidence for the flexibility and importance of the compound in metabolism.
INTRODUCTION
The compound α-d-ribosyl 1,5-bisphosphate (5-phospho-α-d-ribosyl 1-phosphate [PRibP]) is formed in several metabolic pathways, by phosphorylation of ribosyl 1-phosphate, phosphorolysis of a variety of ribonucleoside 5′-monophosphates (NMPs), dephosphorylation of 5-phospho-α-d-ribosyl 1-diphosphate (PRPP), and catabolism of phosphonates by the carbon-phosphorus (C-P) lyase pathway (Table 1). PRibP was discovered in the 1950s by Hans Klenow while characterizing phosphoribomutase (Table 2) (1, 2). Leloir had suggested the existence of the compound while studying the metabolism of sugar phosphates (3). The formation of PRibP was shown to be catalyzed by phosphoglucomutase (Table 2) (4), and indeed, phosphoribomutase catalyzed the same reaction (5). In analogy with the mechanism of phosphoglucomutase (6), PRibP is a catalytic intermediate or coenzyme in the reversible phosphoribomutase catalytic cycle (ribose 5-phosphate → PRibP → ribosyl 1-phosphate) (5).
TABLE 1.
Enzymes producing or utilizing PRibP
| Enzyme (systematic name) | EC no. | PDB accession no. | Reference(s) |
|---|---|---|---|
| PRibP-producing enzymes | |||
| ADP-dependent ribosyl 1-phosphate 5-kinase (ADP:α-d-ribose 1-phosphate 5-phosphotransferase) | 2.7.1.212 | 49, 50 | |
| ATP-dependent ribosyl 1-phosphate 5-kinase (ATP:α-d-ribose 1-phosphate 5-phosphotransferase) | 53 | ||
| Ribose 5-phosphate 1-kinase | 30 | ||
| PRPP diphosphohydrolase (diphospho-myo-inositol-polyphosphate diphosphohydrolase) | 3.6.1.52 | 2Q9P, 2FVV, 1KT9 | 57–59 |
| AMP phosphorylase (AMP:phosphate α-d-ribosyl 5′-phosphate-transferase) | 2.4.2.57 | 4GA4, 4GA5, 4GA6 | 52, 72 |
| 5-Phosphoribosyl 1,2-cyclic phosphate phosphodiesterase (5-phospho-α-d-ribose 1,2-cyclic phosphate 2-phosphohydrolase [α-d-ribose 1,5-bisphosphate-forming]) | 3.1.4.55 | 3G1P | 44, 75, 76 |
| PRibP-utilizing enzymes | |||
| PRibP isomerase (α-d-ribose 1,5-bisphosphate aldose-ketose-isomerase) | 5.3.1.29 | 3VM6, 3A9C, 3A11 | 52, 93 |
| PRibP phosphokinase (ATP:ribose-1,5-bisphosphate phosphotransferase) | 2.7.4.23 | 66, 67 | |
| PRibP phosphohydrolase | 81 | ||
TABLE 2.
Relevant enzymes other than those producing or utilizing PRibP
| Enzyme (systematic name, EC no.) | Reaction | Reference(s) |
|---|---|---|
| Adenine phosphoribosyltransferase (AMP:diphosphate phospho-d-ribosyltransferase, EC 2.4.2.7) | AMP + PPi → adenine + PRPP | 180 |
| AMP nucleosidase (AMP phosphoribohydrolase, EC 3.2.2.4) | AMP → ribose 5-phosphate + adenine | 181 |
| Ribonucleoside kinases (ATP:ribonucleoside 5′-phosphotransferase, EC 2.7.1.N) | Ribonucleoside + ATP → ribonucleoside 5′-monophosphate + ADP | 51 |
| Bisphosphoglycerate mutase (3-phospho-d-glycerate 1,2-phosphomutase, EC 5.4.2.4) | Glycerate 1,3-bisphosphate → glycerate 2,3-bisphosphate | 133 |
| Glucose 1,6-bisphosphate synthase (3-phospho-d-glyceroyl-phosphate:α-d-glucose-1-phosphate 6-phosphotransferase, EC 2.7.1.106) | Glucose 1-phosphate + glycerate 1,3-bisphosphate → glucose 1,6-bisphosphate + glycerate 3-phosphate | 88–90 |
| 5-Methylthioribosyl 1-phosphate isomerase (S-methyl-5-thio-α-d-ribose-1-phosphate aldose-ketose-isomerase, EC 5.3.1.23) | 5-Methylthioribosyl 1-phosphate → 5-methylthioribulose 1-phosphate | 182 |
| Phosphoglucomutase (α-d-glucose 1,6-phosphomutase, EC 5.4.2.2) | Glucose 1-phosphate → glucose 6-phosphate | 4 |
| Phosphoglycerate mutase (glycolytic) (d-phosphoglycerate 2,3-phosphomutase [2,3-diphosphoglycerate independent], EC 5.4.2.12) | Glycerate 3-phosphate → glycerate 2-phosphate | 137 |
| Phosphoribomutase (α-d-ribose 1,5-phosphomutase, EC 5.4.2.7) | α-d-Ribose 1-phosphate → α-d-ribose 5-phosphate | 1, 2 |
| Phosphoribulokinase (ATP:d-ribulose-5-phosphate 1-phosphotransferase, EC 2.7.1.19) | Ribulose 5-phosphate + ATP → ribulose 1,5-bisphosphate | 183 |
| PRPP synthase (ATP:d-ribose-5-phosphate diphosphotransferase, EC 2.7.6.1) | Ribose 5-phosphate + ATP → PRPP + AMP | 71 |
| Purine nucleoside phosphorylase (purine-nucleoside:phosphate ribosyltransferase, EC 2.4.2.1) | Adenosine + Pi → adenine + ribosyl 1-phosphate | 51 |
| Rubisco (3-phospho-d-glycerate carboxy-lyase [dimerizing; d-ribulose-1,5-bisphosphate-forming], EC 4.1.1.39) | Ribulose 1,5-bisphosphate + CO2 + H2O → 2 glycerate 3-phosphate | |
| Form I | 184–187 | |
| Form II | 188 | |
| Form II/III | 150, 189 | |
| Form III | 111, 190, 191 |
As it became evident that ribonucleotides could be synthesized in the absence of ribonucleosides (7, 8), a search for the origin of the (phospho)ribose moiety of ribonucleotides was initiated, and Klenow suggested a reaction for the formation of ribonucleoside 5′-phosphates that involved PRibP as a component: nucleobase + PRibP → ribonucleoside 5′-monophosphate + inorganic phosphate (Pi) (4, 9). However, with the discovery of PRPP (10) and its roles in the conversion of purine and pyrimidine bases to ribonucleotides (10, 11) as well as in de novo ribonucleotide biosynthesis (12), research on the role of PRibP in ribonucleotide biosynthesis was abandoned (H. Klenow, personal communication) (13). Remarkably, 60 years after Klenow’s suggestion and surrender, microbial species, particularly among archaea, have been found to contain a phosphorylase that catalyzes the reaction that he suggested, adenine + PRibP → AMP + Pi, as described in detail below.
It is our intensions with this review to provide a comprehensive description of PRibP metabolism, with the inclusion of relevant biochemical reactions, properties, and structures of the enzymes catalyzing these reactions, and descriptions of PRibP as an effector molecule, the organization of relevant genes, and similarities and differences in PRibP-containing pathways among organisms and their evolution. In addition, we seek to understand the extent to which the two homologous compounds PRibP and PRPP may substitute for or complement one another. A number of bisphosphate compounds besides PRibP are prominent participants of cellular metabolism either as metabolites or as regulatory molecules (or both, as with PRibP). We therefore seek to briefly review common features of these molecules as well.
For the purpose of this review, nucleotide and amino acid sequence analyses were performed with BLAST (14), with the program packages of the Integrated Microbial Genomes (IMG) website (http://img.jgi.doe.gov/) (15, 16) and the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/) (17, 18), as well as with MultAlin (http://multalin.toulouse.inra.fr/multalin/) and DNA Strider (19, 20). Figures of three-dimensional structures were prepared with the PyMOL Molecular Viewer (http://www.pymol.org) (21). Multiple-sequence alignments were performed with the Muscle algorithm (22, 23), phylogenetic analysis was performed with MEGA 7 (24), and graphics were improved with the Interactive Tree of Life (iToL) program (25).
CHEMISTRY OF PRibP
PRibP can be synthesized chemically (26) or enzymatically from ribosyl 1-phosphate by the use of phosphoglucomutase or phosphoribomutase, as described below: ribosyl 1-phosphate + glucose 1,6-bisphosphate → PRibP + glucose 6-phosphate (2, 4). A procedure for the enzymatic preparation of 2-deoxyribosyl 1,5-bisphosphate has also been reported (27, 28). Chemical synthesis of PRibP can be achieved by treatment of an acetylated derivative of ribose 5-phosphate with crystalline phosphoric acid, which results in a derivative phosphorylated at the anomeric carbon. The procedure also works with several hexose 6-phosphate compounds (29).
Similar to other compounds that contain a C-1 phosphate ester, PRibP is labile and decomposes to ribose 5-phosphate and Pi, particularly under acidic conditions (4, 30). Ribosyl 1-phosphate decomposes to ribose and Pi (31), whereas PRPP decomposes to ribose 5-phosphate and inorganic diphosphate (PPi) (32, 33). In contrast, phosphate esters with C-5 of ribose are stable under acidic conditions (34). At alkaline pH, PRibP may be formed by decomposition of PRPP together with ribosyl 1-phosphate, ribose 5-phosphate, 5-phosphoribosyl 1,2-cyclic phosphate, and Pi (35). Under these conditions, PRibP is formed by hydrolysis of the decomposition intermediate 5-phosphoribosyl 1,2-cyclic phosphate: PRPP → 5-phosphoribosyl 1,2-cyclic phosphate → PRibP/ribosyl 1-phosphate/ribose 5-phosphate. Some researchers, however, were unable to detect PRibP formation upon PRPP decomposition, which they proposed to follow the pathway PRPP → 5-phosphoribosyl 1,2-cyclic phosphate → ribosyl 1-phosphate → ribose (36). As described below, several cases of physiological, nonenzymatic formation of PRibP have been described. Indeed, this process may well occur through the intermediate formation of 5-phosphoribosyl 1,2-cyclic phosphate, a process that is stimulated by divalent cations such as Mg2+ and nitrogen-containing compounds such as imidazole (35, 37, 38). Erythrocytes generate PRibP when incubated with inosine in the presence of Pi (39, 40), and PRibP formed under these conditions was postulated to originate from the decomposition of PRPP (41).
A few compounds that may be structural variants of PRibP have been identified: α-d-ribosyl 2,5-bisphosphate, α-d-ribosyl 1,5-cyclic phosphate, and α-d-ribosyl 1,2-cyclic phosphate. α-d-Ribosyl 2,5-bisphosphate is formed during chemical breakdown of PRPP (38) or as an intermediate during cleavage of the C-P lyase reaction product 5-phosphoribosyl 1,2-cyclic phosphate in the Gram-positive bacterial species Clostridium difficile and Eggerthella lenta, where the final product is ribose 5-phosphate (42). α-d-Ribosyl 1,5-cyclic phosphate has also been shown to be formed during spontaneous breakdown of PRPP (37). Finally, as described below, α-d-ribosyl 1,2-cyclic phosphate is the product of C-P lyase (EC 4.7.1.1; α-d-ribose-1-methylphosphonate-5-phosphate C-P-lyase [methane forming]) (43, 44).
As discussed below, PRibP may replace the role of PRPP in certain phosphoribosylation reactions. These PRibP-dependent reactions are particularly widespread among archaeal species, although they are not limited to these. It is possible that PRibP can be considered an “ancestor” of PRPP in early life and possibly also in prebiotic times. Contrary to the situation with PRPP, which can be formed under prebiotic conditions during one-pot synthesis of AMP (45), information on the formation of PRibP in prebiotic chemical simulations is scarce or nonexistent, as ribose appears to be an unstable compound and preferably is replaced by other compounds, such as glyceraldehyde (for a review, see reference 46). Altogether, it is plausible that PRibP may have served as an early phosphoribosyl donor and that modern organisms have acquired PRPP, resulting in more-efficient biochemical processes based on PPi as a leaving group rather than Pi.
BIOCHEMICAL PATHWAYS INVOLVING PRibP
Catabolism of Ribonucleoside 5′-Monophosphates and Ribonucleosides
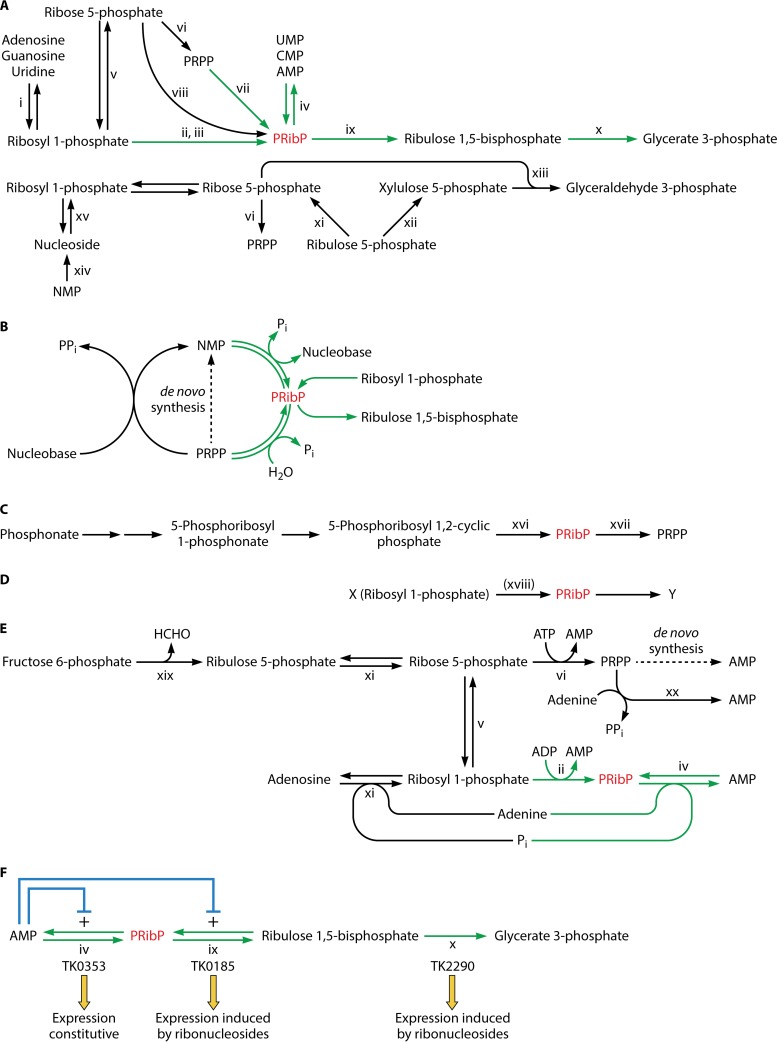
In several archaeal species, PRibP may be regarded as being located at the junction of a number of reactions involved in the conversion of pentose phosphates. Of these, the reactions of the hyperthermophilic archaeon Thermococcus kodakarensis KOD1 have been studied most thoroughly (47, 48). Figure 1A shows how PRibP, drawn in red, is produced and utilized in several biochemical pathways. The individual chemical reactions are described in greater detail in Fig. 2A (formation of PRibP) and Fig. 2B (utilization of PRibP).
FIG 1.
PRibP-containing pathways and their regulation. PRibP is shown in red. (A) Comparison of the pentose bisphosphate pathway and related feeder reactions with the pentose phosphate pathways. The pentose bisphosphate pathway leads from AMP to glycerate 3-phosphate and has PRibP as an intermediate. PRibP can also be formed by dephosphorylation of PRPP. These reactions as well as other reactions specific for or predominant in archaeal species are shown with green arrows. Enzyme activities are indicated by roman numerals (xi [lower part], purine nucleoside phosphorylases and uridine phosphorylase; ii, ADP-dependent ribosyl 1-phosphate 5-kinase; iii, ATP-dependent ribosyl 1-phosphate 5-kinase; iv, AMP phosphorylase; v, phosphoribomutase; vi, PRPP synthase; vii, PRPP diphosphohydrolase; viii, ribose 5-phosphate 1-kinase; ix, PRibP isomerase; x, form III Rubisco). The pentose phosphate pathway leads from ribulose 5-phosphate to glyceraldehyde 3-phosphate. The pathway has been extended with ribosyl 1-phosphate for comparison with the pentose bisphosphate pathway. Ribulose 5-phosphate is generated from glucose 6-phosphate through the oxidative branch of the pentose phosphate pathway or by phosphorolysis of ribonucleosides followed by isomerization of ribosyl phosphate. Ribonucleosides in turn may be generated by dephosphorylation of ribonucleotides. Enzyme activities are indicated by roman numerals (xi [upper part], ribose phosphate isomerase; xii, ribulose 5-phosphate 3-epimerase; xiii, transketolase [sedoheptulose 7-phosphate is also generated in this reaction and further converted by transaldolase]; xiv, ribonucleoside 5′-monophosphate phosphohydrolase; xv, nucleoside phosphorylase). (B) Nucleoside and nucleotide salvage pathways. A nucleobase may be phosphoribosylated by reaction with PRPP to a ribonucleoside 5′-monophosphate (NMP) in reactions catalyzed by phosphoribosyltransferases. Reactions specific for certain archaeal species (shown as green arrows) may reversibly convert NMP to PRibP (catalyzed by AMP phosphorylase). PRibP can be also formed by phosphorylation of ribosyl 1-phosphate (catalyzed by ribosyl 1-phosphate 5-kinase). (C) C-P lyase pathway for phosphonate catabolism with PRibP as an intermediate. Relevant enzyme activities are indicated with roman numerals (xvi, phosphoribosyl cyclic phosphodiesterase; xvii, ribosyl bisphosphate phosphokinase). (D) Alternative formation of PRibP in E. coli. PRibP maybe formed from an unknown compound, X, which could be ribosyl 1-phosphate in a reaction catalyzed by ribosyl 1-phosphate 5-kinase (reaction xviii). PRibP may be converted to a second unknown compound, Y. (E) De novo pathway and salvage of ribonucleotides. Enzyme activities are indicated by roman numerals (xi [lower part], purine nucleoside phosphorylase; ii, ADP-dependent ribosyl 1-phosphate 5-kinase; iv, AMP phosphorylase; v, phosphoribomutase; vi, PRPP synthase; xi [upper part], ribose phosphate isomerase; xix, bifunctional 6-phospho-3-hexuloisomerase,3-hexulose-6-phosphate synthase; xx, adenine phosphoribosyltransferase). The dotted arrow indicates the multistep de novo pathway leading from PRPP to AMP. (F) Regulation of the pentose bisphosphate pathway in T. kodakarensis, AMP phosphorylase (catalyzing reaction designated reaction iv [TK0353 gene]), PRibP isomerase (catalyzing reaction designated ix [TK0185 gene]), and Rubisco (catalyzing reaction designated x [TK2290 gene]). Angled blue arrows show the positive effects of AMP on the activities of AMP phosphorylase and PRibP isomerase. Below the gene designations, the mode of genetic regulation of each gene is indicated.
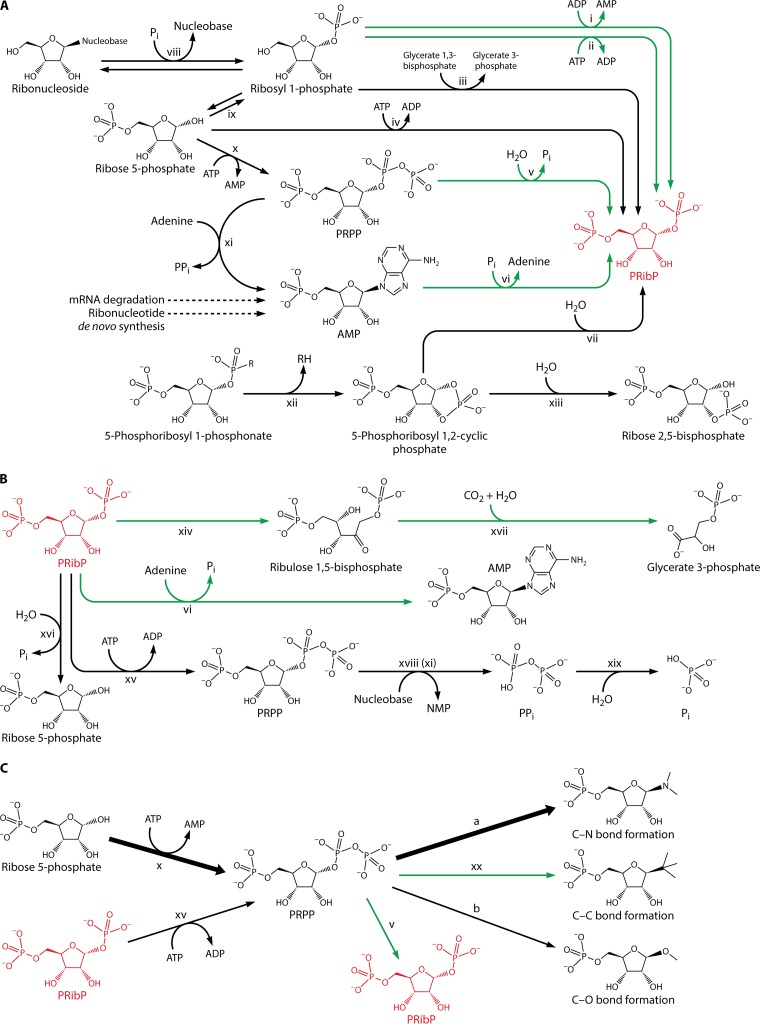
FIG 2.
Chemical compounds and enzymes of PRibP and PRPP metabolism. PRibP is shown in red. Green arrows indicate reactions that have been analyzed primarily in archaeal species. Dotted arrows indicate reaction pathways consisting of multiple enzymes. Enzymes are indicated by roman numerals. (A) Formation of PRibP. Reaction i, ADP-dependent ribosyl 1-phosphate 5-kinase; reaction ii, ATP-dependent ribosyl 1-phosphate 5-kinase (pyrimidine nucleoside kinase); reaction iii, glucose 1,6-bisphosphate synthase; reaction iv, ribose 5-phosphate 1-kinase; reaction v, PRPP diphosphohydrolase; reaction vi, AMP phosphorylase; reaction vii, 5-phosphoribosyl 1,2-cyclic phosphodiesterase; reaction viii, ribonucleoside phosphorylases; reaction ix, phosphoribomutase; reaction x, PRPP synthase; reaction xi, adenine phosphoribosyltransferase; reaction xii, C-P lyase; reaction xiii, 5-phosphoribosyl 1,2-cyclic phosphate hydrolase. (B) Utilization of PRibP. Reaction vi, AMP phosphorylase, in the reverse direction relative to that shown in panel A; reaction xiv, PRibP isomerase; reaction xv, PRibP phosphokinase; reaction xvi, PRibP phosphohydrolase; xvii, form III Rubisco; reaction xviii, phosphoribosyltransferase such as adenine phosphoribosyltransferase (panel A, reaction xi); reaction xix, diphosphatase. Reaction xiv (PRibP isomerase) is reversible but is not included in panel A, as the isomerization of ribulose 1,5-bisphosphate to PRibP has no apparent physiological function. (C) Summary of reactions involving PRPP. Formation of PRPP occurs by two enzymes, catalyzing reaction x, PRPP synthase, and reaction xv, PRibP phosphokinase. The reaction catalyzed by PRPP synthase is by far the most important of the two under normal physiological conditions, indicated by the heavy arrow. PRPP is a substrate for numerous phosphoribosyltransferases. Reactions resulting in the formation of N-glycosidic bonds by far outnumber those resulting in the formation of C- and O-glycosidic bonds, indicated by the heavy arrow labeled “a.” The activity of these phosphoribosyltransferases leads to purine and pyrimidine ribonucleotides, the amino acids histidine and tryptophan, and NAD. There are four types (types I to IV) of phosphoribosyltransferases. A number of reactions result in the formation O-glycosidic bonds, indicated by the arrow labeled “b.” Reaction v, PRPP diphosphohydrolase; reaction xx, 4-aminobenzoate phosphoribosyltransferase of the methanogenic archaea Methanosarcina thermophila, M. jannaschii, and Methanothermobacter thermoautotrophicus and the nonmethanogenic archaeon A. fulgidus. Readers interested in the metabolism of PRPP are encouraged to consult recent reviews (65, 121).
In catabolism, PRibP is converted to ribulose 1,5-bisphosphate in a reaction catalyzed by PRibP isomerase and then to glycerate 3-phosphate in a reaction catalyzed by ribulose 1,5-bisphosphate carboxylase/oxygenase (Rubisco) (Fig. 1A and 2B). Altogether, one pentose bisphosphate molecule is converted to two 3-carbon phosphates with an additional carbon originating from carbon dioxide in the carboxylation reaction catalyzed by Rubisco. Glycerate 3-phosphate in turn is an intermediate of glycolysis and gluconeogenesis, and thus, PRibP may be catabolized with the production of ATP (glycolysis), used for storage as a carbohydrate (gluconeogenesis), or used for biosynthetic purposes (glycolysis and gluconeogenesis) (49, 50). Additionally, the nucleobase moiety may be recycled by phosphoribosylation in reactions catalyzed by phosphoribosyltransferases (nucleobase + PRPP → NMP + PPi) (Fig. 1B) (51).
Several biochemical reactions are responsible for the formation of PRibP. First, AMP phosphorylase has affinity for a variety of ribonucleoside 5′-monophosphates in addition to AMP: NMP + Pi → nucleobase + PRibP. Phosphorolysis of deoxyribonucleoside 5′-monophosphates by AMP phosphorylase produces 2-deoxyribosyl 1,5-bisphosphate: dNMP + Pi → nucleobase + 2-deoxyribosyl 1,5-bisphosphate (49, 52). The fate of 2-deoxyribosyl 1,5-bisphosphate generated by the phosphorolysis of deoxyribonucleoside 5′-monophosphates remains unknown. Possibly, the compound may be cleaved by deoxyriboaldolase to yield glyceraldehyde 3-phosphate and acetyl phosphate.
Second, phosphorylation of ribosyl 1-phosphate leads to PRibP (Fig. 1A). This reaction is catalyzed by ADP-dependent ribosyl 1-phosphate 5-kinase (ribosyl 1-phosphate + ADP → PRibP + AMP [T. kodakarensis gene TK2029]) or by ATP-dependent ribosyl 1-phosphate 5-kinase of the hyperthermophilic archaeon Pyrobaculum calidifontis (ribosyl 1-phosphate + ATP → PRibP + ADP [gene Pcal_0041]) (53). Homologs of these two enzymes are restricted to archaeal species, as described further below.
Ribosyl 1-phosphate in turn may be formed by phosphorolysis of adenosine or guanosine (adenosine + Pi → adenine + ribosyl 1-phosphate [T. kodakarensis gene TK1895] and guanosine + Pi → guanine + ribosyl 1-phosphate [T. kodakarensis gene TK1482]). Uridine phosphorylase activity has also been identified (uridine + Pi → uracil + ribosyl 1-phosphate [T. kodakarensis gene TK1479]) (Fig. 1A and 2A), whereas cytidine phosphorylase activity appears to be absent (49). Ribonucleoside phosphorylase-catalyzed reactions are widespread among bacterial and eukaryotic species (51, 54).
Ribonucleoside kinases with affinities for adenosine, guanosine, and uridine appear to be absent from archaeal species. In contrast, cytidine kinase has been identified in T. kodakarensis (cytidine + ATP → CMP + ADP [gene TK1843]). The enzyme also phosphorylates deoxycytidine, although the Kmdeoxycytidine value is more than 2-fold higher than the Kmcytidine value. The TK1843 gene has been cloned and expressed in Escherichia coli, and the gene product has been characterized enzymatically. Despite this, the activity has not been demonstrated in cell extracts of T. kodakarensis, suggesting that the gene is expressed only under certain conditions. The presence of cytidine kinase and the absence of cytidine phosphorylase suggest that cytidine is catabolized differently from the nucleosides that are catabolized by phosphorolysis followed by phosphorylation by ADP-dependent ribosyl 1-phosphate 5-kinase (49). In addition, the fact that deoxycytidine may be phosphorylated, followed by phosphorolysis by AMP phosphorylase, suggests the possibility of the formation of 2-deoxyribosyl 1,5-bisphosphate. Further catabolism of this compound may involve deoxyriboaldolase, as described above.
The sequence of reactions leading from AMP to glycerate 3-phosphate has been designated the pentose bisphosphate pathway (49). This designation is reminiscent of the more familiar (nonoxidative) pentose phosphate pathway of bacterial and eukaryotic species, shown for comparison in Fig. 1A. ADP-dependent ribosyl 1-phosphate 5-kinase is important in this respect, as the activity of this enzyme provides PRibP to the pentose bisphosphate pathway. The presence of a complete pentose phosphate pathway appears to be missing in archaeal species (55). Although ribose 5-phosphate isomerase (ribulose 5-phosphate → ribose 5-phosphate) is commonly found in archaea, ribulose 5-phosphate 3-epimerase (ribulose 5-phosphate → xylulose 5-phosphate) as well as transaldolase (glyceraldehyde 3-phosphate + sedoheptulose 7-phosphate → fructose 6-phosphate + erythrose 4-phosphate) are absent, accounting for the inability of archaeal species to thrive on pentoses (56).
Cell extracts of T. kodakarensis indeed produce glycerate 3-phosphate when incubated with a ribonucleoside 5′-monophosphate (AMP, CMP, or UMP) or with PRibP or ribulose 1,5-bisphosphate (52). The formation of PRibP by the pentose bisphosphate pathway was furthermore demonstrated by the use of mutant strains of T. kodakarensis with knockout lesions of the genes encoding AMP phosphorylase (gene TK0353) or ADP-dependent ribosyl 1-phosphate 5-kinase (gene TK2029). In cell extracts, the ribose moiety of ribonucleosides is converted to PRibP via phosphorolysis followed by phosphorylation of ribosyl 1-phosphate to PRibP, rather than by phosphorylation of the ribonucleosides to the ribonucleotides and phosphorolysis (by AMP phosphorylase): PRibP formation from ribonucleosides was abolished in an ADP-dependent ribosyl 1-phosphate 5-kinase knockout mutant but not in an AMP phosphorylase knockout mutant (49). Furthermore, glycerate 3-phosphate formation from AMP was severely reduced by the addition of the Rubisco inhibitor 2-carboxyarabinitol 1,5-bisphosphate, demonstrating that glycerate 3-phosphate formation from nucleosides occurs by the four-enzyme pathway consisting of nucleoside phosphorylase, ADP-dependent ribosyl 1-phosphate 5-kinase, PRibP isomerase, and Rubisco, whereas glycerate 3-phosphate formation from ribonucleotides (AMP, CMP, or UMP) occurs by the three-enzyme pathway consisting of AMP phosphorylase, PRibP isomerase, and Rubisco, i.e., the pentose bisphosphate pathway (52).
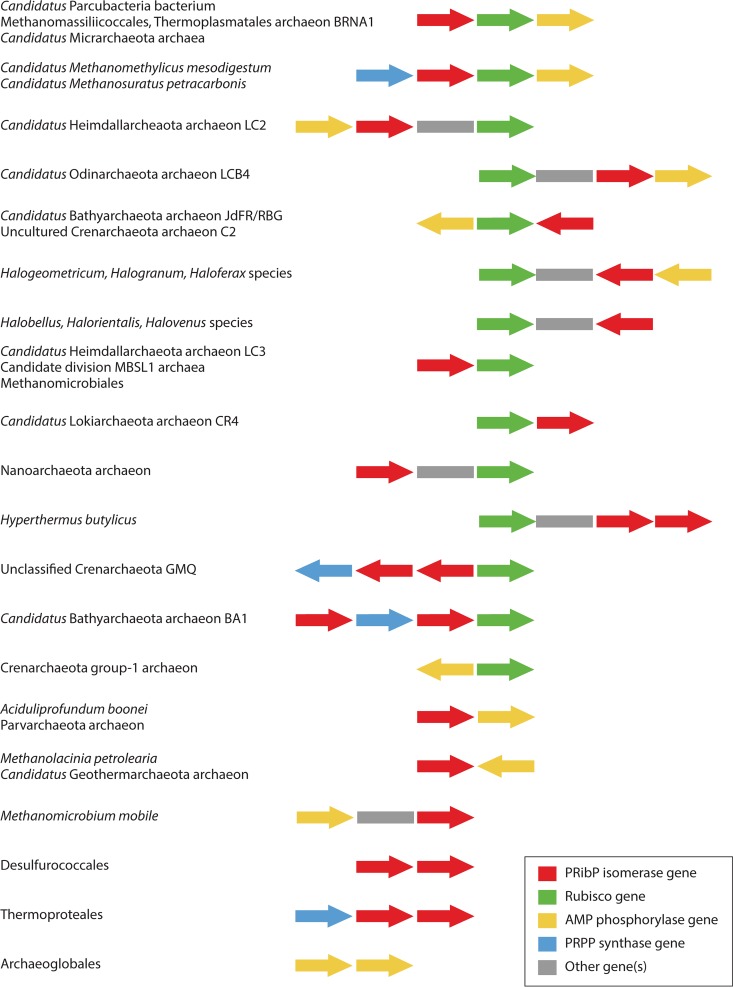
Although the pathway leading from ribonucleoside 5′-monophosphate or ribosyl 1-phosphate to glycerate 3-phosphate has been particularly studied in T. kodakarensis, a survey of the distribution of genes for AMP phosphorylase, PRibP isomerase, and Rubisco among archaeal species revealed that either all three genes were present or none were present at all, with only a few exceptions, and that the three genes are broadly distributed among the archaeal orders Archaeoglobales, Methanomicrobiales, Methanosarcinales, and Thermococcales as well as in several members of the orders Desulfurococcales, Halobacteriales, Methanococcales, and Thermoproteales (50). Indeed, as described below, the pentose bisphosphate pathway is found among species of all four of the major archaeal groups, Euryarchaeota, TACK (including Thaumarchaeota, Aigarchaeota, Crenarchaeota, and Korarchaeota, among others), DPANN (including Diapherotrites, Parvarchaeota, Aenigmarchaeota, Nanoarchaeota, and Nanohaloarchaeota, among others), and Asgard, as evaluated by bioinformatic analysis. ADP-dependent ribosyl 1-phosphate 5-kinase is present throughout the Thermococcus and Pyrococcus species. Some archaeal species contain PRibP isomerase and Rubisco but not ADP-dependent ribosyl 1-phosphate 5-kinase, suggesting the presence of an alternative enzyme, such as an ATP-dependent ribosyl 1-phosphate 5-kinase homologous to that of P. calidifontis, or a preference for using nucleoside kinases to phosphorylate the ribonucleosides, followed by the activity of AMP phosphorylase. On the other hand, nucleoside phosphorylases are present in essentially all archaeal species (49, 50). As revealed below, certain Eubacteria may contain the pentose bisphosphate pathway, as suggested from bioinformatic analysis.
Finally, hydrolysis of PRPP by a variety of enzymes produces PRibP (PRPP → PRibP + Pi, PRPP diphosphohydrolase, and Nudix hydrolases) (Fig. 1A and 2A) (57–60). The dephosphorylation of PRPP to PRibP may also occur nonenzymatically (61). A pathway of Methanocaldococcus jannaschii leading from PRPP to ribulose 1,5-bisphosphate has been shown to include the Mj0601 gene product (61), an enzyme catalyzing a rather complicated chemical reaction of thiazole synthesis and, thus, the thiamine biosynthetic pathway: NAD + glycine + S2− → ADP 5-(2-hydroxyethyl)-4-methylthiazole-2-carboxylate (adenylated thiazole) + nicotinamide (62, 63). It is difficult to reconcile the participation of the Mj0601 gene product and PRPP with ribulose 1,5-bisphosphate production, as the Mj0601 protein, when purified from E. coli, was unable to convert PRPP to ribulose 1,5-bisphosphate. In spite of these observations with the Mj0601 protein, the following pathway may lead to glycerate 3-phosphate: PRPP → PRibP → ribulose 1,5-bisphosphate → glycerate 3-phosphate. The inclusion of the adenine phosphoribosyltransferase reaction (Table 2) results in the following series of reactions: PRPP → AMP → PRibP → ribulose 1,5-bisphosphate → glycerate 3-phosphate (61). In support of this pathway is the finding that M. jannaschii harbors a gene specifying AMP phosphorylase (MJ0667) (64). Thus, PRibP may be generated from PRPP by both nonenzymatic and enzyme-catalyzed reactions.
To summarize, two reaction sequences may exist for the formation of ribulose 1,5-bisphosphate in archaeal species: AMP → PRibP → ribulose 1,5-bisphosphate and PRPP (→ AMP) → PRibP → ribulose 1,5-bisphosphate (Fig. 1B).
PRibP is a potent regulator of both glycolysis and pentose phosphate catabolism, as described in detail below (57). PRibP therefore may be the final product of a pathway, at least in some organisms (ribose 5-phosphate → PRPP → PRibP).
PRibP in Nucleobase Salvage Reactions
Ribonucleotides may be synthesized by de novo pathways or by salvage or auxiliary reactions in which preformed nucleobases or ribonucleosides are converted to ribonucleoside 5′-monophosphates. A large number of salvage or auxiliary reactions have been identified. For example, nucleosides may be phosphorolyzed to the nucleobase and ribosyl 1-phosphate by ribonucleoside phosphorylases, followed by phosphoribosylation of the nucleobase to the ribonucleoside 5′-monophosphate by specific phosphoribosyltransferases (Fig. 1B). Ribosyl 1-phosphate in turn may be converted to ribose 5-phosphate by phosphoribomutase, and ribose 5-phosphate may by diphosphorylated to PRPP by PRPP synthase (ATP:d-ribose-5-phosphate diphosphotransferase) (Table 2). Alternatively, ribonucleosides may be phosphorylated to ribonucleoside 5′-monophosphates by a number of ATP-dependent ribonucleoside kinases. These processes have been previously reviewed (51, 65).
The existence of PRibP adds a new type of salvage reaction, as shown in Fig. 1B. Thus, in addition to the phosphoribosyltransferase-catalyzed reactions (nucleobase + PRPP → NMP + PPi), ribonucleoside 5′-monophosphates may be synthesized by the activity of AMP phosphorylase acting on PRibP and nucleobases in the reverse direction relative to the degradation pathway described above: PRibP + nucleobase → NMP + Pi. As PRibP can be generated from ribosyl 1-phosphate, which in turn can be generated from ribose 5-phosphate, a new, versatile nucleobase salvage pathway exists in organisms containing the pentose bisphosphate pathway. As described below, AMP is an activator of PRibP isomerase activity, with the enzyme being virtually inactive in the absence of AMP. Therefore, the catabolic pathway (NMP → → → glycerate 3-phosphate) operates only with ample AMP present. Under all other conditions, AMP phosphorylase functions as a nucleobase salvage enzyme (49).
Catabolism of Phosphonates by the C-P Lyase Pathway
The catabolism of phosphonates by the C-P lyase pathway has been shown to involve PRibP as an intermediate (66). Phosphonates are compounds that contain a C-P bond. The C-containing part of the phosphonate may vary considerably, with the simplest phosphonate being methylphosphonate. In E. coli, the 14-cistron phnCDEFGHIJKLMNOP operon specifies enzymes and other proteins that are necessary for or involved in the internalization and catabolism of phosphonates (Fig. 1C) (67, 68). The phn-specified gene products of this pathway convert the phosphonate-phosphorus to the α-phosphate of the diphosphoryl group of C-1 of PRPP (Fig. 1C and 2A) (44, 66). The C-P lyase pathway may be divided into three stages: (i) activation of phosphonate in which the phosphonate becomes esterified to a phosphoribosyl moiety, resulting in the formation of 5-phosphoribosyl 1-phosphonate, the substrate of the C-P bond-cleaving enzyme; (ii) C-P bond cleavage by phnJ-specified C-P lyase and phosphodiester formation to yield 5-phosphoribosyl 1,2-cyclic phosphate and an alkane; and (iii) further processing to yield a useful phosphorus-containing product. Here 5-phosphoribosyl 1,2-cyclic phosphate is hydrolyzed to PRibP in a reaction catalyzed by phosphoribosyl cyclic phosphodiesterase, followed by phosphorylation of PRibP to PRPP in a reaction catalyzed by ribosyl bisphosphate phosphokinase (44, 66). For the sake of completeness, Pi originating from phosphonate can be generated from PRPP by a phosphoribosyltransferase-catalyzed reaction (PRPP + nucleobase → NMP + PPi), followed by the activity of diphosphatase (PPi → 2Pi) (67).
Other PRibP-Containing Pathways
Mutants of E. coli deleted for the phnN gene specifying ribosyl bisphosphate phosphokinase (PRibP + ATP → PRPP + ADP) appear to possess a leaky phenotype in the sense that they are able to grow with phosphonate as the sole phosphate source but grow at a reduced rate (68). This phenotype suggests that an alternative or redundant pathway for the utilization of PRibP exists. In addition, as described in detail below, PRPP-less strains of E. coli (Δprs) (69, 70) regain the ability to produce PRPP by the expression of the phnN gene, even in an otherwise Δphn genetic background. This observation suggests that the substrate for phnN-specified ribosyl bisphosphate phosphokinase is already present in cells, which are not exposed to phosphonate. A possible candidate for the catalysis of PRibP formation is ribosyl 1-phosphate 5-kinase. Presumably, PRibP can also be removed by an enzymatic activity in a reaction leading from PRibP to an unknown compound (Fig. 1D) (66).
ENZYMES CATALYZING THE SYNTHESIS AND UTILIZATION OF PRibP
ADP-Dependent Ribosyl 1-Phosphate 5-Kinase
ADP-dependent ribosyl 1-phosphate 5-kinase phosphorylates ribosyl 1-phosphate and, to a much lesser extent, 2-deoxyribosyl 1-phosphate with ADP as the preferred phosphoryl donor (ribosyl 1-phosphate + ADP → PRibP + AMP) (Fig. 2A, reaction i, and Table 1). The enzyme from T. kodakarensis contains 294 amino acid residues and has a molecular mass of 31.9 kDa. In addition to ADP, GDP may function as a phosphoryl donor, approximately half as effectively as ADP, as may UDP, which is approximately 1/10 as effective as ADP. Neither CDP nor PPi acts as a phosphoryl donor. The enzyme requires a divalent metal ion with Ca2+ > Mg2+ > Co2+. With Mg2+ present, the kinetic values were determined: a KMRib1P of 0.53 mM, a KMADP of 7.3 mM, and a Vmax of ∼1 mmol (min · mg protein)−1. An equilibrium constant was established at 85°C as 6 × 103, thus highly favoring phosphorylation of ribosyl 1-phosphate (49). A three-dimensional structure of ADP-dependent ribosyl 1-phosphate 5-kinase has not been reported.
The use of ADP rather than ATP as the phosphoryl donor by ribosyl 1-phosphate 5-kinase is a characteristic of kinases of archaea. Several of these enzymes are specific for ADP or other ribonucleoside diphosphates as phosphoryl donors, such as ADP-dependent glucokinase or ADP-dependent phosphofructokinase (50, 56). The use of ADP as a phosphoryl donor led to speculations about the pool size of AMP, which was suggested to be larger in archaeal organisms with high contents of ADP-dependent kinases (50).
ATP-Dependent Ribosyl 1-Phosphate 5-Kinase
The reaction catalyzed by ATP-dependent ribosyl 1-phosphate 5-kinase of P. calidifontis (gene Pcal_0041) resembles that described above, with the replacement of ADP for ATP: ribosyl 1-phosphate + ATP → PRibP + ADP (Fig. 2A, reaction ii, and Table 1). The enzyme also phosphorylates cytidine and uridine (Vmax values of 15, 12, and 11 μmol [min · mg protein]−1 for ribosyl 1-phosphate, cytidine, and uridine, respectively). The catalytic efficiencies (kcat/Km) were calculated to be 12, 3.1, and 5.1 mM−1 s−1 for ribosyl 1-phosphate, cytidine, and uridine, respectively. ATP is the most efficient phosphoryl donor, with GTP being slightly less efficient. Cell extracts of P. calidifontis were shown to be able to produce PRibP from ribosyl 1-phosphate in the presence of ATP, as coupling of the activities of PRibP isomerase and Rubisco resulted in the formation of glycerate 3-phosphate (ribosyl 1-phosphate → PRibP → ribulose 1,5-bisphosphate → glycerate 3-phosphate) (53). In the present context, the enzyme is named ATP-dependent ribosyl 1-phosphate 5-kinase. However, also considering the enzyme’s affinity for cytidine and uridine, a proper name may by ATP-dependent ribosyl 1-phosphate 5-kinase/pyrimidine nucleoside kinase.
The physiological function of ATP-dependent ribosyl 1-phosphate 5-kinase is presently only poorly understood. Thus, the pentose bisphosphate pathway appears to be absent in P. calidifontis, as evaluated by bioinformatic analysis. Those authors furthermore excluded PRibP as a final product and suggested that the compound might be a precursor of PRPP (53). However, P. calidifontis contains a bona fide PRPP synthase-encoding gene (Pcal_1127), as evaluated by a BLAST search with the M. jannaschii PRPP synthase amino acid sequence as the query (71). It therefore seems difficult to envision a function of PRibP in PRPP synthesis.
AMP Phosphorylase
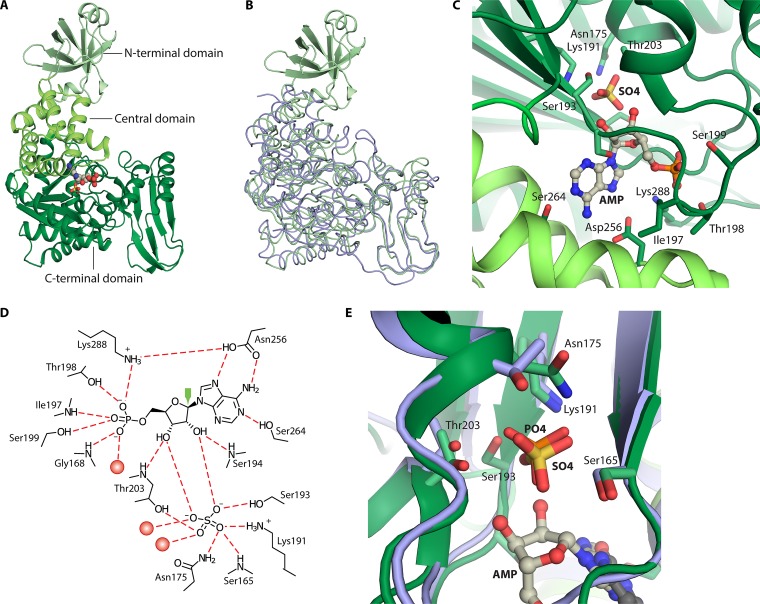
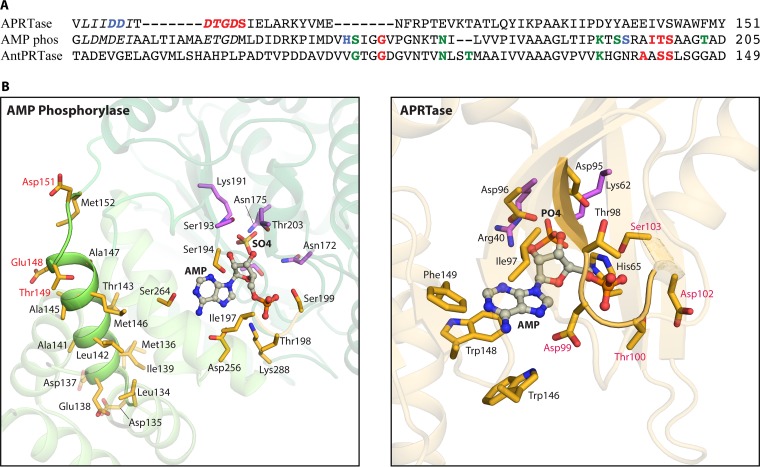
AMP phosphorylase (AMP + Pi → adenine + PRibP) (Fig. 2A, reaction vi, and Table 1), although widely distributed among archaea, as described below, has been studied only in T. kodakarensis. This enzyme contains 503 amino acid residues and has a calculated molecular mass of 53.6 kDa. AMP phosphorylase consists of three domains, an N-terminal domain of 84 amino acid residues, a central domain (residues 85 to 149 and 241 to 272), and a C-terminal domain (residues 161 to 233 and 276 to 503) (Fig. 3A). Amino acid sequence analysis has assigned AMP phosphorylase to nucleoside phosphorylase family ii, i.e., pyrimidine nucleoside phosphorylases. In this respect, the 84-amino-acid N-terminal domain appears to constitute an extension compared to the amino acid sequences of pyrimidine phosphorylases in general (50). Superimposition of T. kodakarensis AMP phosphorylase and Thermus thermophilus pyrimidine nucleoside phosphorylase revealed a root mean square deviation of 2.0 Å for 388 C-α atoms (total of 418 atoms) (Fig. 3B) (72).
FIG 3.
Structure of T. kodakarensis AMP phosphorylase and comparison with the nucleoside phosphorylase family ii member Geobacillus stearothermophilus pyrimidine nucleoside phosphorylase. The structure of AMP phosphorylase represents the reaction-inert variant Tk-AMP phosphorylase ΔC10. (A) Subunit structure with N-terminal, central, and C-terminal domains in various shades of green. AMP is shown as spheres of gray (carbon), red (oxygen), and blue (nitrogen). Sulfur is rendered in yellow. (B) Superimposition of the T. kodakarensis three-domain AMP phosphorylase (rendered in green) and two-domain nucleoside phosphorylase family ii member G. stearothermophilus pyrimidine nucleoside phosphorylase (rendered in blue) (118). (C) The active site of AMP phosphorylase. (D) Binding of AMP and Pi-mimicking sulfate to amino acid residues and water molecules (red spheres) at the active site of AMP phosphorylase. Dashed red lines represent hydrogen bonds. The green arrow points to C1′ of AMP, which is attacked by nucleophilic Pi represented by sulfate (72). (E) Superimposition of active sites of AMP phosphorylase and G. stearothermophilus pyrimidine nucleoside phosphorylase. PDB accession numbers are 4GA6 for AMP phosphorylase and 1BRW for pyrimidine nucleoside phosphorylase.
AMP phosphorylase displays specificity toward AMP, CMP, UMP, and dCMP. The specific activity obtained with CMP is twice that obtained with AMP. The enzyme also has very low activity with dAMP and GMP (52). Saturation kinetics of the enzyme with AMP or dCMP is sigmoidal, indicating cooperativity in the binding of these substrates and implying regulation of AMP phosphorylase activity by these two substrates (52).
In vitro, AMP phosphorylase forms large, soluble, enzymatically active, multimeric aggregates consisting of more than 40 subunits, which have hampered the crystallization of the enzyme. The three-dimensional structure was subsequently determined by combining the structure of AMP phosphorylase lacking the 84-amino-acid N-terminal domain with that lacking the 10 C-terminal amino acid residues. The deletion of either of these amino acid stretches resulted in the formation of proteins with lower-order quaternary structures, presumably hexamers (deletion of the N-terminal domain) or dimers (deletion of the 10 C-terminal amino acids). Removal of the N-terminal domain completely abolished enzymatic activity, whereas deletion of the 10 C-terminal amino acids had no effect on enzymatic activity. The formation of dimers is promoted by an intricate set of subunit-subunit interactions involving both the N-terminal and central domains (72).
The active site of AMP phosphorylase is located in a pocket between the central and C-terminal domains where substrate binding induces a closed conformation. This is achieved by movement of the N-terminal domain of a neighboring subunit similar to that seen for the pyrimidine nucleoside phosphorylases. The N-terminal domain is important not only for subunit multimerization but also for enzymatic activity and thermostability (72).
Both AMP and a sulfate ion were identified in the three-dimensional structure, with the sulfate ion presumably mimicking the substrate Pi. The binding of AMP in the active site is well described (Fig. 3C and D). The adenine moiety (N-1, N-7, and the amino group) makes hydrogen bonds to Ser264 and Asp256. Residues of AMP phosphorylase that form hydrogen bonds to the 5′-phosphate of AMP are Gly168, Ile197, Thr198, Ser199, and Lys288 (Fig. 3C). Thus, a 5′-phosphate binding motif may exist, 193-Ser-Ser-Arg-Ala-Ile-Thr-Ser-Ala-Ala-Gly-Thr-203, which is reminiscent of a subset of amino acid residues of the 5′-phosphate (or PRPP) loop of the PRPP binding site of M. jannaschii PRPP synthase, 217-Thr-Gly-Gly-Thr-220, i.e., a hydroxyl-carrying or small amino acid of a loop structure that wraps around the 5-phosphate moiety of PRPP (65).
The distance between the sulfate O-1 atom and the C-1′ atom of the ribose moiety is 3.3 Å, and thus, the sulfate ion is located within a reasonable distance for nucleophilic attack. The sulfate ion forms hydrogen bonds to Thr203 (located within the 5′-phosphate binding loop) and to Ser193, Lys191 (in or close to the 5′-phosphate binding loop), Asn175, and Ser165 as well as to the 2′- and 3′-hydroxyl groups of the ribose moiety of AMP. Finally, the two hydroxyl groups form hydrogen bonds to Ser193 and Thr203 (also within the 5′-phosphate binding loop) (Fig. 3C) (72). Altogether, the resemblance of the positions of Pi or sulfate and the adenine moiety of AMP in AMP phosphorylase and of uracil in uracil phosphorylase is evident from Fig. 3E.
An interesting aspect of the structure of AMP phosphorylase is that the large multimeric structures mentioned above occur as a result of a “domain swap” involving the C terminus. Thus, the last β-strand (β18) of one subunit protrudes into a neighboring subunit and forms a sheet with that subunit’s β7-β11 strands. Exactly this type of interaction is prevented by the deletion of the 10 C-terminal amino acid residues (72).
Ribose 5-Phosphate 1-Kinase
Studies of bisphosphate-containing metabolites of the Gram-negative soil bacterium Pelomonas saccharophila, formerly Pseudomonas saccharophila (73), revealed an enzyme capable of phosphorylating ribose 5-phosphate at the C-1 hydroxyl, hence ribose 5-phosphate 1-kinase (Table 1). Only ribose 5-phosphate and ATP are substrates for this kinase, and the product was identified as PRibP (ribose 5-phosphate + ATP → PRibP + ADP) (Fig. 2A, reaction iv, and Table 1). Although sharing substrates with PRPP synthase, the activity of ribose 5-phosphate 1-kinase is clearly distinguished from that of the former (ribose 5-phosphate + ATP → PRPP + AMP) by the following observations regarding the reaction product: stimulation of the activity of phosphoribomutase, electrophoretic mobility identical to that of authentic PRibP, acid lability with the formation of ribose 5-phosphate, the lack of activity with orotate phosphoribosyltransferase and orotate, and the lack of inhibition of orotate phosphoribosyltransferase with PRPP as the diphosphoryl donor. The enzyme displayed high apparent Km values for both the substrates ribose 5-phosphate and ATP, approximately 1 mM, and inhibition was observed at high substrate concentrations (30). A lack of an amino acid sequence of ribose 5-phosphate 1-kinase prevents a comparison of functional amino acid residues of this enzyme with those of PRPP synthase.
Little is known of the genome sequence of P. saccharophila, although a sequence representing a partial gene for the large subunit of form I Rubisco has been identified, consistent with the organism’s ability to grow autotrophically in the presence of carbon dioxide, dihydrogen, and oxygen (cbbL [GenBank accession number AM501463 {74}]). Presumably, this organism therefore may not be a member of the pentose bisphosphate pathway-containing organisms described above, and presumably, it contains a bona fide Calvin-Benson-Bassham cycle in which the Rubisco substrate ribulose 1,5-bisphosphate is recycled. Nevertheless, the existence of ribose 5-phosphate 1-kinase may suggest a role of PRibP in photosynthesis in P. saccharophila provided that the coding capacity of PRibP isomerase is also present in the genome. Alternatively, the formation of PRibP by ribose 5-phosphate 1-kinase may simply be a source of coenzyme for the phosphoribomutase-catalyzed reaction.
Phosphoribosyl 1,2-Cyclic Phosphate Phosphodiesterase
Phosphoribosyl 1,2-cyclic phosphate phosphodiesterase (phosphoribosyl 1,2-cyclic phosphate → PRibP) (Fig. 2A, reaction vii, and Table 1), encoded by phnP in E. coli, is an enzyme of the C-P lyase pathway. The enzyme hydrolyzes one of the products of the phnJ-specified C-P lyase, 5-phosphoribosyl 1,2-cyclic phosphate, to PRibP and as such functions in the above-mentioned stage (iii) of the C-P lyase pathway, i.e., processing to obtain a useful phosphorus-containing product.
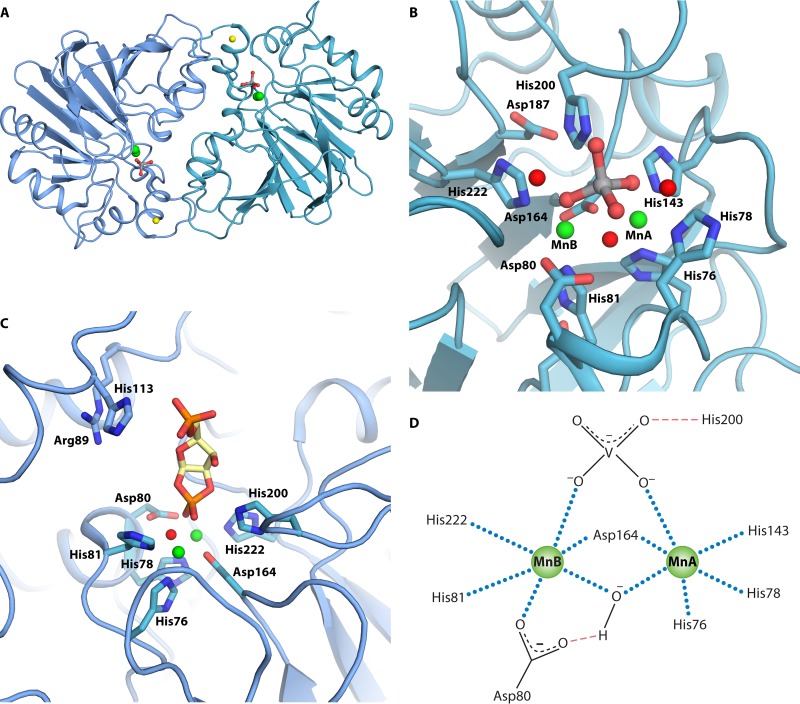
Phosphoribosyl 1,2-cyclic phosphate phosphodiesterase is a member of the β-lactamase family of metal-dependent hydrolases. The closest sequence identity is found within the tRNase Z phosphodiesterase family and the E. coli rbn gene product, RNase BN, which also is a phosphodiesterase. Typical for this family of metal-dependent hydrolases, E. coli phosphoribosyl 1,2-cyclic phosphate phosphodiesterase is a homodimer with a dinuclear active site containing two manganese ions. Ni2+ may replace Mn2+. A Zn2+, located approximately 17 Å away from the active site, is coordinated by three cysteine residues and one histidine residue close to the C-terminal end (Fig. 4A). This Zn2+ is believed to be important for the integrity of the dimeric structure of the enzyme. The active site is at the dimerization interface and, as mentioned above, contains two manganese ions. These two manganese ions are coordinated by five histidine residues (His76, His78, His81, His200, and His222) and two aspartate residues (Asp80 and Asp164) (75). The orthovanadate ion is an inhibitor of phosphoribosyl 1,2-cyclic phosphate phosphodiesterase activity, and crystals of the enzyme have been obtained in the presence of an orthovanadate ion. The active site of the enzyme with an orthovanadate ion surrounded by the histidine and aspartate residues is shown in Fig. 4B and D. The orthovanadate ion forms a bridge between the two manganese ions, which are also bridged by a water molecule. Additionally, the orthovanadate ion forms a hydrogen bond with His200. One Mn2+ (MnA) interacts with His143, His76, His78, Asp164, the orthovanadate ion, and a water molecule, whereas the second Mn2+ (MnB) interacts with His81, His222, Asp80, Asp164, the orthovanadate ion, and a water molecule. With this arrangement, the water molecule is well positioned for nucleophilic attack on the vanadium nucleus, mimicking the phosphorus nucleus of the natural substrate (Fig. 4B and D). Several amino acid residues of the active site of phosphoribosyl 1,2-cyclic phosphate phosphodiesterase have been altered. These amino residues are part of motif II (HxHxDH), where the His78Ala and Asp80Ala mutant variants were analyzed; of motif IV, where Asp164Ala was analyzed; and of motif V, where His222Ala was analyzed (Fig. 4B). In general, large reductions in the kcat/Km values were observed primarily due to changes in kcat. Additionally, the His200Ala variant revealed a 10-fold reduction in kcat/Km, which was quite modest given that this residue functions as a general acid catalyst in the reaction. Analysis of the variant active-site histidine and aspartate residues demonstrates the importance of these residues in catalysis. It is possible that the effects would be even more pronounced had the actual, physiological substrate been used to probe these active-site amino acid residues rather than artificial substrates (76).
FIG 4.
Structure and active site of phosphoribosyl 1,2-cyclic phosphate phosphodiesterase of E. coli. Structures are based on data reported under PDB accession number 3P2U (76). (A) Dimeric quaternary structure. Orthovanadate is shown as gray and red sticks, and manganese and zinc ions are shown as green and yellow spheres, respectively. (B) Orthovanadate bound to the active site. Water molecules are shown as red spheres, and the two manganese ions are shown as green spheres and indicated by MnA and MnB. (C) Binding of 5-phosphoribosyl 1,2-cyclic phosphate to the active site. The cyclic phosphate is modeled using orthovanadate coordinates (76). Carbons of 5-phosphoribosyl 1,2-cyclic phosphate are shown in yellow. (D) Interactions of active-site residues with Mn2+, orthovanadate, and water. Mn2+ coordinations are shown as blue dotted lines, and hydrogen bonds are shown by red dashed lines.
The physiological substrate for phosphoribosyl 1,2-cyclic phosphate phosphodiesterase was shown to be 5-phosphoribosyl 1,2-cyclic phosphate. To show this, the compound was synthesized in vitro and shown to by hydrolyzed by purified phosphoribosyl 1,2-cyclic phosphate phosphodiesterase. A series of one- and two-dimensional nuclear magnetic resonance (NMR) analyses assigned the product as PRibP (44). Besides 5-phosphoribosyl 1,2-cyclic phosphate, the enzyme has affinity for ribosyl 1,2-cyclic phosphate, i.e., a dephosphorylated derivative of the physiological substrate, as well as a number phosphodiesters, such as 2′,3′-cyclic CMP, 2′,3′-cAMP, 2′,3′-cyclic GMP, and a number of substituted methyl phenyl phosphodiesters (75, 76). Crystallization of phosphoribosyl 1,2-cyclic phosphate phosphodiesterase in the presence of 5-phosphoribosyl 1,2-cyclic phosphate failed, but an energy-minimized structure of the compound was manually docked into the enzyme structure with an alignment of the orthovanadate ion and the 1,2-cyclic phosphate and with an orientation appropriate for in-line attack by water (Fig. 4C) (76).
E. coli mutants defective in phnP have proven valuable for the isolation of small-molecule components of the C-P lyase pathway and the elucidation of their structures. Thus, phnP mutants accumulate not only substrates of phosphoribosyl 1,2-cyclic phosphate phosphodiesterase (44, 77) but also substrates of previous enzymes of the pathway, such as the substrates of C-P lyase. An example is 5′-phosphoribosyl 1′-(2-N-acetamidoethylphosphonate), which is formed in cells thriving on 2-aminoethylphosphonate as a phosphate source. In addition, the dephosphorylated derivatives ribosyl 1,2-cyclic phosphate, ribosyl 1′-(2-N-acetamidoethylphosphonate), and ribosyl 1′-ethylphosphonate have been identified in the growth medium of a phnP mutant strain. The latter compound accumulates in a phnP strain supplemented with ethylphosphonate (44). The dephosphorylated compounds very likely appear in the growth medium after intracellular buildup of the 5- or 5′-phosphorylated compounds and subsequent excretion with the simultaneous hydrolysis of the 5- or 5′-phosphate ester.
PRibP Phosphokinase
PRibP, the product of the activity of phosphoribosyl cyclic phosphodiesterase of the C-P lyase pathway (Fig. 1C), is a substrate for phnN-specified PRibP phosphokinase (PRibP + ATP → PRPP + ADP) (Fig. 2B, reaction xv, and Table 1) (66).
The biochemical reaction catalyzed by E. coli PRibP phosphokinase was established as follows. The pyridine nucleotide de novo pathway contains the reaction PRPP + quinolinate → nicotinate ribonucleotide + CO2 + PPi, a reaction catalyzed by quinolinate phosphoribosyltransferase. PRPP is provided by the reaction catalyzed by PRPP synthase (ribose 5-phosphate + ATP → PRPP + AMP) encoded by the prs gene (51, 65, 78). A PRPP-less strain (Δprs) therefore must be supplied with a pyridine source, such as NAD or nicotinamide mononucleotide, in addition to other compounds necessary to supply the PRPP-consuming pathways, i.e., purine and pyrimidine compounds, histidine, and tryptophan (69). Figure 2C summarizes the reactions involving PRPP. The expression of phnN in a Δprs strain relieved the requirement for pyridine. It was therefore postulated that with increased (i.e., constitutive) phnN gene expression, a small amount of PRPP was synthesized from some alternative source other than the “usual” prs-specified PRPP synthase-catalyzed reaction. Incubation in vitro of PRibP with ATP and purified PhnN resulted in the synthesis of a compound with thin-layer chromatographic properties identical to those of PRPP. Indeed, the compound formed was PRPP, as it was a substrate for purified xanthine phosphoribosyltransferase from Bacillus subtilis: xanthine + PRPP → XMP + PPi. The identification of the chemical reaction catalyzed by the phnN gene product was the first assignment of a biochemical function to any phnCDEFGHIJKLMNOP-specified gene product (66).
By far, the synthesis of cellular PRPP is catalyzed by PRPP synthase (51, 67). Very little PRPP is contributed by PRibP phosphokinase (66). PRPP synthase and PRibP phosphokinase catalyze the synthesis of PRPP from ribose 5-phosphate and PRibP, respectively. Alignment of the amino acid sequences of the two enzymes shows only low similarity, approximately 25%. Among the amino acid residues of B. subtilis PRPP synthase with a known function based on the crystal structure, very few, if any at all, can be recognized in the amino acid sequence of PRibP phosphokinase. Also, the similarity of PRibP phosphokinase to type I or type II phosphoribosyltransferases is very low, 16% with both adenine phosphoribosyltransferase (type I) (adenine + PRPP → AMP + PPi) and quinolinate phosphoribosyltransferase (type II). Both PRibP phosphokinase and PRibP isomerase must be able to bind PRibP prior to and during catalysis. However, alignment of the amino acid sequences of the two enzymes does not provide much information as to the function of specific residues of PRibP phosphokinase. The only hint appears to be the sequence 20-Arg-Gly-Ala-Gly-Lys-24 of the PRibP isomerase amino acid sequence. This sequence is involved in the binding of the 1-phosphate moiety of PRibP and resembles the PRibP phosphokinase sequence 12-Ser-Gly-Ser-Gly-Lys-16. Altogether, the evolutionary origin and mechanism of PRibP phosphokinase remain enigmatic, and a deeper understanding awaits the determination of the three-dimensional structure of the enzyme. The physiological importance of PRibP phosphokinase in the context of phosphonate catabolism and PRPP metabolism has been previously reviewed (65, 67, 79, 80).
PRPP Diphosphohydrolase
Some enzymes have been shown to hydrolyze the phosphoric anhydride bond of the diphosphoryl moiety of PRPP, which results in the formation of PRibP: PRPP → PRibP + Pi (Fig. 2A, reaction v, and Table 1). Most notably, a number of the Nudix hydrolases are capable of hydrolyzing PRPP to PRibP. Typically, these enzymes hydrolyze dinucleoside polyphosphate compounds of the type ApnA, as described by Fisher and colleagues (57), and include human diadenosine tetraphosphate (Ap4A) hydrolase; human diadenosine hexaphosphate hydrolase/diphosphoinositol polyphosphate (DIPP) hydrolase 1; DIPP-2α, DIPP-2β, DIPP-3α, and DIPP-3β hydrolases; Caenorhabditis elegans Ap4A hydrolase; Salmonella enterica serovar Typhimurium Ap4A hydrolase; Saccharomyces cerevisiae Ap6A hydrolase; and Deinococcus radiodurans ApnA hydrolase. The African swine fever virus g5R protein also has PRPP-phosphohydrolyzing activity. An enzyme with PRibP-forming activity from PRPP has also been identified in macrophages (81). The relation of this enzyme to the Nudix hydrolases remains undisclosed. The products of the activities of several of the enzymes were shown to be PRibP and Pi. Kinetic values suggest a physiological function of the PRibP-generating activity in addition to the ApnA- or diphosphoinositol polyphosphate-cleaving activity (57).
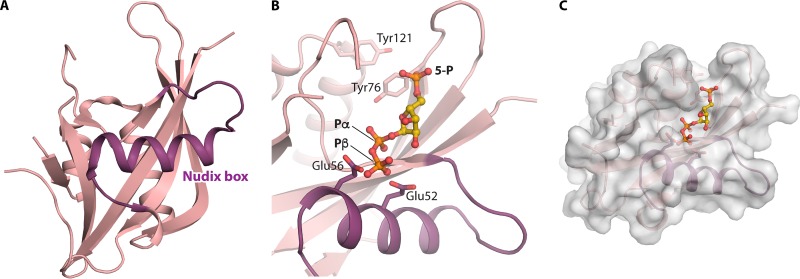
A conserved sequence of approximately 23 amino acid residues, the Nudix box, folds as a loop-helix-loop structure involved in the binding of divalent cations and the polyphosphate moiety of the substrate. The Nudix box has the consensus sequence GX5EX7REUXEEXGU (where U represents leucine, isoleucine, or valine and X may be any amino acid) and forms part of the catalytic site for diphosphate hydrolysis (82). The three-dimensional structures of numerous Nudix hydrolases have been determined (83), including the C. elegans Ap4A hydrolase (Fig. 5A) (60). Modeling of PRPP into the three-dimensional structure of this enzyme revealed a possibility of binding at the active site with the PRPP 5-phosphate located exactly at the position of the substrate Ap4A P1 phosphate and the PRPP α-1-phosphate located at the position of the substrate Ap4A P4 phosphate. In this Nudix hydrolase, P1 is the phosphate closest to the adenosyl residue whose adenine moiety is sandwiched between Tyr76 and Tyr121, while P4 is the phosphate farthest away and also the target for nucleophilic attack by a water molecule. Thus, the ribosyl moiety of PRPP occupies the positions of the Ap4A P2 and P3 phosphate groups, and the nucleophilic water is in position for attack at the α-phosphate of the C-1 diphosphoryl. The important residues of the enzyme, Glu52 and Glu56, are correctly located for catalysis (Fig. 5B and C) (57, 60).
FIG 5.
Structure of C. elegans Ap4A hydrolase and possible binding of PRPP. (A) Cartoon representation showing the overall fold of the enzyme, with the Nudix loop-helix-loop highlighted (PDB accession number 1KT9) (60). (B) PRPP modeled into the substrate site of the enzyme using the coordinates under PDB accession number 1FSG, as described previously (57). Tyr76 and Tyr121, which bind the adenine moiety of one of the nucleosides, and Glu56 and catalytic Glu52 are shown as sticks. 5-P, Pα, and Pβ indicate the phosphoryl groups of PRPP whose carbons are shown in yellow. (C) Surface representation of Ap4A hydrolase showing a possible cavity with binding of PRPP (57).
Nudix hydrolases are found in all three domains of life. However, relatively few of those of archaeal origin have been characterized. M. jannaschii and T. kodakarensis contain genes specifying proteins with a Nudix motif, ADP-ribose diphosphatase, encoded by the MJ1149 (M. jannaschii) and TK2284 (T. kodakarensis) genes, and the specified enzymes are very specific for ADP-ribose. Although the formation of PRibP has not been demonstrated, the enzymes apparently are unable to hydrolyze diadenosine polyphosphate compounds (84, 85). The results presented by Fisher and colleagues and those described above indicate that only Nudix hydrolases with activity toward dinucleoside polyphosphate have activity toward PRPP, and thus, it is unlikely that the M. jannaschii Nudix hydrolase is capable of hydrolyzing PRPP. Indeed, this is consistent with previously reported data for the conversion of PRPP to PRibP in M. jannaschii that indicated an abiotic process (61). Such a spontaneous dephosphorylation of PRPP to PRibP is chemically possible according to the results of an analysis of the spontaneous decomposition of PRPP described previously, which showed that divalent metal ions (e.g., Mg2+) and nitrogen-containing compounds (e.g., imidazole) stimulate the process (37). For reviews on the properties, catalysis, and evolution of Nudix hydrolases, see references 83, 86, and 87.
Glucose 1,6-Bisphosphate Synthase
Glucose 1,6-bisphosphate synthase activity (glucose 1-phosphate + glycerate 1,3-bisphosphate → glucose 1,6-bisphosphate + glycerate 3-phosphate) (Fig. 2A, reaction iii, and Table 2) of various vertebrate tissues was demonstrated quite some time ago (88–90). The mechanism of glucose 1,6-bisphosphate synthase resembles that of phosphoglucomutase, with a transient phosphorylation of a specific serine residue. The phosphoryl group is provided by glycerate 1,3-bisphosphate (91). Glucose 1,6-bisphosphate synthase has an amino acid sequence that is highly conserved among phosphohexomutases, 173-Thr-Ala-Ser-His-Asn-178, which contains the phosphorylatable serine residue. Glucose 1,6-bisphosphate synthase also catalyzes the synthesis of PRibP and 2-deoxyribosyl 1,5-bisphosphate: ribosyl 1-phosphate + glycerate 1,3-bisphosphate → PRibP + glycerate 3-phosphate and (2-deoxy)ribosyl 1-phosphate + glycerate 1,3-bisphosphate → (2-deoxy)PRibP + glycerate 3-phosphate. The kcat/Km values of the enzyme for glucose 1-phosphate, ribosyl 1-phosphate, and 2-deoxyribosyl 1-phosphate are 5.5, 5.4, and 5.6 s−1 μM−1, respectively. The enzyme also uses glucose 6-phosphate and ribose 5-phosphate as phosphoryl acceptors although less efficiently than the 1-phosphate compounds (92).
Human glucose 1,6-bisphosphate synthase is specified by the phosphoglucomutase 2-like no. 1 (PGM2L1) gene. The PGM2L1 gene is expressed in all tissues analyzed, most abundantly in brain tissue. Although phosphoglucomutase (glucose 1-phosphate → glucose 6-phosphate), specified by the human PGM2 gene, is also able to catalyze the synthesis of glucose 1,6-bisphosphate, the kinetic analysis favors the PGM2L1-specified synthase as the main source of glucose 1,6-phosphate and, therefore, presumably also of PRibP in human tissue (92).
Glucose 1,6-bisphosphate synthase appears to be limited to vertebrates (92). Indeed, protein BLAST analysis of prokaryotic genome sequences with human glucose 1,6-bisphosphate synthase as a query revealed no glucose 1,6-bisphosphate synthase sequences, whereas the most closely related phosphoglucomutase was abundant in the search results.
PRibP Isomerase
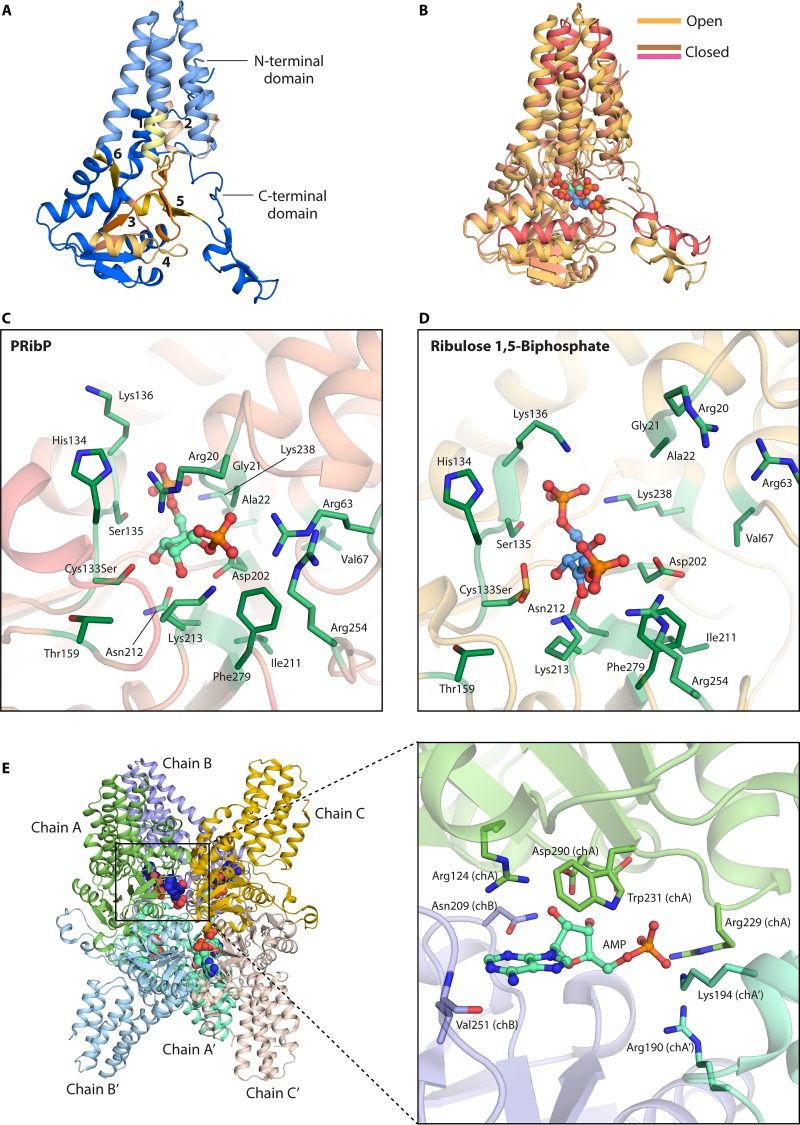
PRibP isomerase (PRibP → ribulose 1,5-bisphosphate) (Fig. 2B, reaction xiv, and Table 1) catalyzes the reversible interconversion of PRibP and ribulose 1,5-bisphosphate. T. kodakarensis PRibP isomerase contains 322 amino acid residues and has a calculated molecular mass of 36.3 kDa, while gel filtration chromatography revealed a molecular mass of 260 kDa. The asymmetric unit of the crystal of the apo form of the enzyme contained six subunit molecules, which, together with the gel filtration results, suggests that the enzyme exists as a homohexamer in solution (93). The activity of the enzyme is activated by AMP; the enzyme is hardly active in the absence of AMP. Although AMP is the most potent activator, other adenosyl-harboring compounds also stimulate the activity, with decreasing potency: dAMP > ADP > adenosine. GMP also activates the enzyme although to a lesser extent (52). Some kinetic constants were determined at 85°C in the presence of AMP, with a KmPRibP value of 0.6 mM and a kcat value of 29 s−1. The enzyme is specific for the α-anomer of PRibP (52, 93).
The three-dimensional structure of PRibP isomerase has been solved for the apoenzyme, the substrate (PRibP)-bound enzyme, and the product (ribulose 1,5-bisphosphate)-bound enzyme (93), as have the structures of the apoenzyme and the PRibP-bound and ribulose 1,5-bisphosphate-bound enzyme that also contained the activator AMP (94). To determine the closed PRibP-bound structure, the inactive Cys133Ser mutant variant was used rather than the wild-type enzyme, which converted PRibP to the product during crystallization. The two-domain structure of the apoenzyme is shown in Fig. 6A. The structures of the apo form and the product-bound form are essentially identical, showing an open conformation with respect to the active site, whereas the substrate-bound form adopts a closed conformation. The substrate- and product-bound structures are believed to represent pre- and postcatalytic enzyme forms, respectively (93). The binding of the substrate PRibP results in large conformational changes in each subunit (Fig. 6B).
FIG 6.
Three-dimensional structure of PRibP isomerase from T. kodakarensis. (A) Two-domain structure of the monomer in the absence of ligands with assignment of motifs 1 to 6 (93). (B) Dynamics of motion following binding of PRibP. Shown is a superimposition of monomers of wild-type PRibP isomerase with bound ribulose 1,5-bisphosphate (open conformation) and the Cys133Ser variant of PRibP isomerase with bound PRibP (closed conformation). Conformational differences are indicated in dark salmon. Carbons of PRibP and ribulose 1,5-bisphosphate are drawn in green and blue, respectively. PRibP binds to the closed conformation, whereas ribulose 1,5-bisphosphate binds to the open conformation. (C) Active site of PRibP isomerase with bound PRibP, shown in light green. The residues interacting with the phosphoryl and ribosyl groups of PRibP are represented in green sticks. (D) Amino acid residues interacting with ribulose 1,5-bisphosphate in open conformation, shown in green sticks. (E) Ribonucleotide binding to P. horikoshii PRibP isomerase. (Left) Three-dimensional hexameric structure of mutant PRibP isomerase Asp204Asn. Each trimer chain of the asymmetric unit (chains A, B, and C) is shown in bright colors; the hexamer is formed through crystallographic symmetry (shown in faded colors for chains A′, B′, and C′). AMP molecules are shown as spheres. (Right) Close-up of the AMP binding site. The residues interacting with AMP are shown as sticks, with the chain identifications given in parentheses. PDB accession numbers are 3A11 for the T. kodakarensis PRibP apo form, 3A9C for the open conformation, 3VM6 for the closed conformation (93, 95), and 5YFW for P. horikoshii (94).
The PRibP binding site is well defined, with the 5-phosphate coordinated by Ser135, Lys136, and Lys238 and the 1-phosphate coordinated by Arg20, Gly21, Arg63, Lys213, and Arg254. In addition, the ribose moiety interacts with Cys133, Asp202, and Asn212. Also, the binding of the product ribulose 1,5-bisphosphate in the “product” binding site is well defined. The structures of the active site with PRibP or ribulose 1,5-bisphosphate are shown in Fig. 6C.
Both Cys133 and Asp202 are important for catalytic activity since none of the mutant variants, Cys133Ser, Cys133Ala, or Asp202Asn, has detectable isomerization activity. Cys133 is close to Thr159 and His134, which promotes its deprotonation and which is necessary for catalysis. Asp202 is surrounded by Ala22, Val67, Ile211, and Phe279, the latter of which promotes the protonation of Asp202, which is also necessary for catalysis. The structure also shows that Asp202 is close to the ribosyl ring of PRibP and that Cys133 is close to C-1 of PRibP. The reaction is initiated by the transfer of a proton from the Asp202 carboxylate group to the ribosyl moiety, followed by abstraction of a proton from PRibP C-2 by the deprotonated Cys133 thiolate. This is followed by the formation of a PRibP C-1:C-2 double bond and the opening of the furanose ring, i.e., the formation of a cis-phosphoenolate intermediate that subsequently tautomerizes to the 2-keto isomer ribulose 1,5-bisphosphate. Thus, protonation and deprotonation of Asp202 and Cys133 are both important steps during catalysis by PRibP isomerase (Fig. 6C) (93, 95).
The alleged regulatory subunit of Pyrococcus horikoshii OT3 translation initiation factor 2B (aIF2B), encoded by the PH0208 gene and annotated after bioinformatic analysis (96), was shown to resemble PRibP isomerase, as in silico analysis demonstrated that all the amino acid residues necessary for substrate binding, catalysis, and subunit interactions known for T. kodakarensis PRibP isomerase were also present in the PH0208 gene product (95). The amino acid sequences specified by PH0208 and T. kodakarensis PRibP isomerase are 86% identical, and this identity is followed in the three-dimensional structure, as superimposition of the structures of the two enzymes revealed a root mean square deviation of 3.8 Å for the 316 C-α atoms. Binding of PRibP or ribulose 1,5-bisphosphate to the PH0208-specified protein was analyzed by isothermal titration calorimetry. The binding of the compounds to the wild-type enzyme was “delusional,” due to heat changes caused by the combined effects of binding of the substrate, catalysis, and release of the product. Rather, the reaction-inert Cys135Ser and Asp204Asn mutant variants revealed dissociation constants of 10 to 14 μM (binding of PRibP) or 7.2 to 7.5 μM (binding of ribulose 1,5-bisphosphate). Importantly, the structure of P. horikoshii PRibP isomerase has been determined with the potent activator AMP present. Figure 6E shows the hexameric quaternary structure of the enzyme with AMP molecules bound. Interestingly, AMP binding is mediated by amino acid residues contributed by three subunits, chain A, chain B, and chain A′. Chain A contributes Arg124 and Asp290, both of whose side chains undergo dramatic shifts when AMP binds to the enzyme. Arg124 and Asp290 form hydrogen bonds to O-2′ and O-3′ of the ribose moiety of AMP; Arg229 and Trp231 interact with the phosphoryl moiety of AMP. Chain B contributes Asn209 and Val251 that form hydrogen bonds to O-2′ and O-3′ of the ribose moiety and to N-1 and N-7 of the adenine of AMP, respectively. Chain A′ provides Lys194 and Arg190 that bind to the phosphoryl moiety of AMP. It was concluded that the binding of AMP provided structural stability to the enzyme (94).
PRibP isomerase and archaeal translation initiation factors.
PRibP isomerase was originally annotated as a member of the translation initiation factor 2 subunit protein family (PF01008), as described above for the PHO208 gene product, and named archaeal translation initiation factor 2B (50). This assignment was based on sequence alignment analysis rather than biochemical analysis of the reaction catalyzed by the enzyme. Indeed, the six amino acid sequence motifs characterizing the PF01008 protein family are well conserved in PRibP isomerase (97). The determination of the three-dimensional structure of PRibP isomerase allowed a detailed analysis of the structural context of the six sequence motifs (Fig. 6A). Motif 1 (residues 19 to 28) contains residues important for the closure of the active site following PRibP binding as well as two residues (Arg20 and Gly21) important for binding to the 1-phosphate of the substrate PRibP. Motif 2 (residues 61 to 73) contains Arg63 that is also important for binding to the 1-phosphate of the substrate PRibP. Motif 3 (residues 129 to 135) contains the catalytically important Cys133 residue as well as Ser135 that is involved in binding the 5-phosphate of the substrate. Motif 4 (residues 158 to 172) contains residues involved in dimerization, while motif 5 (residues 197 to 216) includes the catalytically important Asp202 residue as well as Asn212, which is involved substrate binding. Motifs 4, 5, and 6 (residues 278 to 296), all allocated in the C-terminal domain, furthermore contain a Rossman fold and are involved in subunit-subunit interactions. Altogether, at least motifs 1, 2, 3, and 5 come together and contribute to the formation of the active site (93).
The structure and function of translation initiation factors have been thoroughly reviewed, with descriptions of the functions of the six motifs (98, 99).
5-Methylthioribosyl 1-phosphate isomerase.
5-Methylthioribosyl 1-phosphate isomerase (Table 2) catalyzes the isomerization of 5-methylthioribosyl 1-phosphate and 5-methylthioribulose 1-phosphate. Thus, this reaction is similar to that catalyzed by PRibP isomerase, with the replacement of the 5-phosphate of PRibP with the methylthio moiety of 5-methylthioribosyl 1-phosphate. In metabolism, the latter compound originates from S-adenosylmethionine and is an intermediate of a salvage pathway for methionine synthesis (100). As with PRibP isomerase, 5-methylthioribosyl 1-phosphate isomerase has been previously annotated as an initiation factor 2 subunit after bioinformatic analysis, but experimental analysis revealed the catalytic capability of 5-methylthioribosyl 1-phosphate isomerase (101). The mechanisms of catalysis of the two isomerases are very likely similar (93, 101). 5-Methylthioribosyl 1-phosphate isomerase amino acid sequences may appear in searches for PRibP isomerase sequences in spite of their low overall amino acid sequence similarity. Typically, the amino acid sequences of the two enzymes are only 30% identical. Both enzymes contain the six motifs of the PF01008 protein family as well as the catalytically important cysteine and aspartate residues described above. The two enzymes, however, may be distinguished by careful analysis of sequence alignments. PRibP contains a 5-phosphate moiety, which binds to amino acid residues of motif 3. In 5-methylthioribosyl 1-phosphate, the hydrophilic phosphoryl group is replaced by the hydrophobic methylthio group. Consequently, an addition of a hydrophobic region of 8 amino acid residues immediately follows motif 3 in 5-methylthioribosyl 1-phosphate isomerase. Thus, motif 3 (shown in boldface type) and the addition of amino acid residues on the C-terminal side of motif 3 (shown in italic type) of the two T. kodakarensis enzymes are 128-VIMTHCHSK-137 (PRibP isomerase) and 162-NVLTHCNAGSLATVQLG-178 (5-methylthioribosyl 1-phosphate isomerase) (93). Finally, both PRibP isomerase and 5-methylthioribosyl 1-phosphate isomerase amino acid sequences can be distinguished from those of initiation factor 2 subunits by the lack of the catalytic cysteine and aspartate residues in the latter. Indeed, a remarkable three-dimensional structure similarity has been found between PRibP isomerase, 5-methylthioribosyl 1-phosphate isomerase, and initiation factor 2 subunits of the hyperthermophilic archaeon Archaeoglobus fulgidus and of humans (93, 95, 102, 103).
PRibP Phosphohydrolase
An enzymatic activity designated PRibP phosphohydrolase was discovered in macrophages exposed to hypoxia (PRibP → ribose 5-phosphate + Pi). As described below, the level of PRibP increases dramatically in macrophages immediately after the onset of hypoxia. The rise in the PRibP level is followed by an immediate decline. The authors of that study claimed that PRibP phosphohydrolase is responsible for this rapid decline in PRibP levels. There are no data reported on the characteristics of this enzyme (81).
RUBISCO
Although Rubisco does not use or produce PRibP, a description of the enzyme is included here, because it is an important component of the pathway for the conversion of the ribosyl moieties of ribonucleoside 5′-monophosphates to glycerate 3-phosphate, i.e., the pentose bisphosphate pathway.
Enzymatically, active Rubisco with carboxylating activity comes in three variants, forms I, II, and III, each catalyzing the carboxylation of one molecule of ribulose 1,5-bisphosphate with the formation of two molecules of glycerate 3-phosphate (ribulose 1,5-bisphosphate + CO2 + H2O → 2 glycerate 3-phosphate) (Fig. 2B, reaction xvii, and Table 2). Form I and II Rubiscos participate in primary carbon assimilation by the Calvin-Benson-Bassham cycle. Form III originally was found in archaea, but with the pervasive analysis of genomes of uncultivable organisms, genes encoding form III Rubisco have also been discovered among bacteria. This putative bacterial form III Rubisco, however, has not yet been characterized. Form III Rubisco does not participate in primary carbon assimilation. Rather, form III Rubiscos have a catabolic role in the removal of intermediates formed by ribonucleoside 5′-monophosphate catabolism and directing the ribose moiety carbons to central metabolism. The biochemical, structural, and phylogenetic relations of the Rubiscos have been reviewed in detail previously (104–109).
It came as a surprise that several archaeal species apparently contained genes encoding Rubiscos, particularly as bioinformatic analysis of the same species did not reveal genes for phosphoribulokinase (Table 2), the enzyme normally catalyzing the production of the Rubisco substrate ribulose 1,5-bisphosphate. After cloning the gene specifying T. kodakarensis Rubisco, purification and characterization of the enzyme (110), and determination of its three-dimensional structure (111), the form III Rubisco was established (112, 113). The archaea harboring this form III Rubisco are unable to carry out primary carbon assimilation by the Calvin-Benson-Bassham cycle, and the physiological importance of this form III Rubisco remained enigmatic until the discovery that the substrate for Rubisco, ribulose 1,5-bisphosphate, could be formed by dephosphorylation of PRPP (61). Subsequently, the pentose bisphosphate pathway was discovered and provided a role of form III Rubisco as the connecting enzyme in ribonucleoside 5′-monophosphate catabolism and central energy metabolism, as described above. It should be noted that some archaeal species, such as Methanospirillum hungatei, Methanoculleus marisnigri, and Methanosaeta thermophila, contain genes encoding phosphoribulokinase (114).
Altogether, the discovery that archaeal Rubisco is part of the pentose bisphosphate pathway has generated intense interest in the function of this Rubisco form in relation to forms I and II of primary carbon assimilation (114).
PHOSPHORYLASES, PYROPHOSPHORYLASES, AND HYDROLASES
Cleavage of N-glycosidic bonds may proceed by one of several mechanisms, phosphorolysis, pyrophosphorolysis, or hydrolysis, depending on the nature of the nucleophilic substrate. With AMP as the substrate, examples of these reactions are AMP + Pi → adenine + PRibP (AMP phosphorylase) (phosphorolysis), AMP + PPi → adenine + PRPP (adenine phosphoribosyltransferase) (pyrophosphorolysis), and AMP + H2O → adenine + ribose 5-phosphate (AMP nucleosidase) (hydrolysis). A variant of the phosphorolysis reaction is the purine nucleoside phosphorylase-catalyzed reaction adenosine + Pi → adenine + ribosyl 1-phosphate (Table 2).
AMP Phosphorylase and AMP Pyrophosphorylase
AMP phosphorylase and adenine phosphoribosyltransferase both use AMP as a substrate in one direction and PRibP or PRPP in the reverse reaction, respectively. It is therefore natural to assume that the two enzymes would show some structural similarity with respect to active-site architecture. As a type I phosphoribosyltransferase, Sulfolobus solfataricus adenine phosphoribosyltransferase contains a typical PRPP binding loop, 91-VLIIDDITDTGDS-103 (65, 115–117) (Fig. 7A). In this sequence, the 95-Asp-Asp-96 dipeptide that interacts with the 2- and 3-hydroxyls of PRPP is shown in italic type, while the 99-Asp-Thr-Gly-Asp-Ser-103 pentapeptide that wraps around the 5′-phosphate of AMP and the 5-phosphate of PRPP is shown in boldface type. Indeed, T. kodakarensis AMP phosphorylase contains a variant of the PRPP binding loop, 133-GLDMDEIAALTIAMAETGDM-152. In this sequence, the acidic dipeptide is 137-Asp-Glu-138, and the analogous 5-phosphate binding pentapeptide, 148-ETGDM-152, contains acidic glutamate and aspartate, glycine, and threonine residues. However, the two subsites are separated by seven residues, shown in subscript type (residues 141 to 147). Thus, the PRPP binding site is effectively corrupted, and indeed, the binding of the 5-phosphate of PRibP or the 5′-phosphate of AMP is mediated by a different amino acid sequence, including the motif 197-Ile-Thr-Ser-199, bearing some resemblance to the pentapeptide described above. These amino acid residues, as expected, are not at all conserved in phosphoribosyltransferases (Fig. 7A). The adenine moiety is partly held in place by stacking by Trp146 in adenine phosphoribosyltransferase, whereas in AMP phosphorylase, the binding of the adenine moiety does not involve aromatic amino acid residues (72, 115). A comparison of the binding of the substrate AMP in AMP phosphorylase and adenine phosphoribosyltransferase is shown in Fig. 7B. The helix shown in the left-hand panel contains the corrupted PRPP binding loop.
FIG 7.
Phosphorylase versus pyrophosphorylase. (A) Alignment of ribose phosphate binding sequences of AMP phosphorylase, anthranilate, and adenine phosphoribosyltransferases. Alignment was performed with MultAlin. APRTase, adenine phosphoribosyltransferase (type I) of S. solfataricus (115); AMP phos, AMP phosphorylase of T. kodakarensis (72); AntPRTase, anthranilate phosphoribosyltransferase (type IV) of Mycobacterium tuberculosis (159). Amino acid residues involved in the binding of the 5′-phosphate of AMP or the 5-phosphate of PRPP are shown in red, and residues involved in the binding of nucleophilic Pi (AMP phosphorylase) or PPi (anthranilate phosphoribosyltransferase) are shown in green, whereas residues involved in the binding of the ribose moiety of AMP or PRPP are shown in blue. Residues of the PRPP binding loop of adenine phosphoribosyltransferase are shown in italic type, and the homologous residues of AMP phosphorylase are also shown in italic type. A few residues outside the region shown are also involved in binding: Lys285 of AMP phosphorylase binds to the 5′-phosphate of AMP, and 40-Arg-Gly-41 of adenine phosphoribosyltransferase binds to the diphosphoryl moiety of PRPP or to nucleophilic PPi. (B, left) AMP binding pocket of T. kodakarensis AMP phosphorylase (PDB accession number 4GA6). The helix containing the “aborted” PRPP binding site is shown. (Right) AMP binding site of S. solfataricus adenine phosphoribosyltransferase (PDB accession number 4TS5). Amino acid residues that are labeled in red correspond to those of panel A.
An evolutionary relationship of the two enzymes is presently difficult to establish due to the lack of correct annotation of adenine phosphoribosyltransferases (115).
As mentioned above, AMP phosphorylase is a member of nucleoside phosphorylase family II, which mostly consists of pyrimidine nucleoside phosphorylases. The binding of AMP in AMP phosphorylase resembles the binding of uracil to pyrimidine nucleoside phosphorylase (Fig. 3E). In contrast, AMP phosphorylase differs from the other family II members by cleaving a ribonucleotide rather than a ribonucleoside. The classification is based on comparison of amino acid sequences, three-dimensional structures, and mechanisms of catalysis (118). Thus, the pyrimidine nucleoside phosphorylase family II members have two-domain structures and form active dimers in solution, whereas AMP phosphorylases have a three-domain structure and form multimeric aggregates, as described above (Fig. 3B) (72).
We note that anthranilate phosphoribosyltransferase (type IV phosphoribosyltransferase) appeared in BLAST searches with AMP phosphorylases as the queries. This is consistent with previous studies that revealed conserved sequences among nucleoside phosphorylase family II enzymes and anthranilate phosphoribosyltransferase and suggested an evolutionary relationship between the two enzyme families (118, 119), a relationship that also includes AMP phosphorylase. Figure 7A shows the conservation of amino acid residues that are important for the binding of nucleophilic Pi (AMP phosphorylase) and PPi (anthranilate phosphoribosyltransferase).
Structural and enzymatic similarities between AMP nucleosidase and the hexameric and dimeric members of the nucleoside phosphorylase I family have been demonstrated (120). This is consistent with a lack of conservation of amino acid residues among AMP phosphorylases and AMP nucleosidases. Thus, there appears to be less of a relationship among phosphorolysis and hydrolysis reactions involving AMP.
PRibP versus PRPP
PRibP and PRPP share extended structural resemblance; they are distinguished by only a single phosphoryl group. As described in previous reviews, PRPP is a versatile compound that participates in numerous biological reactions (65, 121). These reactions are of two types: dephosphorylation (PRPP diphosphohydrolase; PRPP → ribose 5-phosphate) (Fig. 2A and C, reaction v) and N-, C-, and O-glycoside bond formation by phosphoribosyltransferases (for example, adenine phosphoribosyltransferase; PRPP + adenine → AMP + PPi) (Fig. 2A, reaction xi, and Fig. 2C, reaction a). N-glycoside bond formation reactions are by far the most abundant among the phosphoribosyltransferase-catalyzed reactions (Fig. 2C, reactions a, xx, and b). Indeed, from this viewpoint, PRibP is even more versatile, as the compound participates in four types of biological reactions: isomerization (PRibP isomerase; PRibP → ribulose 1,5-bisphosphate) (Fig. 2B, reaction xiv), N-glycoside bond formation (AMP phosphorylase; PRibP + adenine → AMP + Pi) (Fig. 2B, reaction vi), phosphoric acid anhydride bond formation (PRibP phosphokinase; PRibP + ATP → PRPP + ADP) (Fig. 2B, reaction xv), and dephosphorylation (PRibP phosphohydrolase; PRibP → ribose 5-phosphate) (Fig. 2B, reaction xvi). In this respect, PRibP contributes to PRPP formation, at least under certain physiological conditions (65, 66).
Contrary to the situation with PRPP, a general binding site for PRibP has not been discovered. Amino acid sequence data are available for three PRibP-utilizing enzymes, PRibP isomerase, AMP phosphorylase, and PRibP phosphokinase. For the former two enzymes, three-dimensional data are also available. However, neither amino acid sequence alignment nor fold alignment has revealed possible similarities of PRibP binding among the three enzymes (data not shown). In the case of PRPP, structural resemblance has been discovered within the various types of phosphoribosyltransferases (type I and PRPP synthase, type II, type III, and type IV) (65, 122).
Both compounds, PRibP and PRPP, are important as effectors in cellular metabolism, as reviewed previously for PRPP (65, 123) and as described below for PRibP.
REGULATORY EFFECTS OF PRibP
As described above, PRibP is an important and versatile intermediate in the catabolism of ribonucleotides, particularly of archaeal species. In addition, PRibP has important properties as an effector molecule in several biochemical pathways, particularly those of mammalian species.
Phosphoribomutase
PRibP was discovered during studies of the mechanism of phosphoribomutase (4). Indeed, the activity of E. coli phosphoribomutase is stimulated 10-fold by the presence of PRibP or 2-deoxyribosyl 1,5-bisphosphate (124). These results are consistent with a reaction mechanism that involves the reversible attachment of a phosphoryl group according to the reaction E-phosphate + ribosyl 1-phosphate → E-PRibP → E-phosphate + ribose 5-phosphate, similar to the reaction mechanism of phosphoglucomutase. Thus, PRibP functions as a coenzyme of the phosphoribomutase-catalyzed reaction (6, 124). A newborn phosphoribomutase presumably is devoid of the enzyme-bound phosphoryl group, and the origin of this modification remains elusive. E. coli phosphoribomutase exists in multiple forms specified by the same gene. None of the forms appeared to be a dephosphorylated form (125).
Effects of PRibP on Metabolism of Brain, Liver, Kidney, and Blood Cells
Activation of brain tissue glycolysis immediately after the introduction of ischemia (i.e., a restriction of the oxygen supply to tissues) is associated not only with the usual “players,” decreases in ATP, citrate, and fructose 6-phosphate concentrations and increases in AMP and fructose 1,6-bisphosphate concentrations, but also with an increase in the concentration of a new effector identified as PRibP. The concentration of PRibP is increased 10-fold approximately 5 s after the onset of ischemia and returns to normal after another 5 s (126). PRibP binds to phosphofructokinase competitively with fructose 2,6-bisphosphate with an activation constant (K0.5) that is 10-fold higher than that of fructose 2,6-bisphosphate (the most potent activator of phosphofructokinase activity) but 6-fold lower than that of fructose 1,6-bisphosphate. Phosphofructokinase from liver and muscle tissue is also stimulated by PRibP (127). Similarly, the level of PRibP increased dramatically in macrophages during hypoxia (i.e., deprivation of oxygen) concomitantly with an increase in anaerobic glycolysis caused by the activation of phosphofructokinase. PRibP, presumably synthesized by the reaction sequence ribose 5-phosphate → PRPP → PRibP, was shown to be the activator that functions after the onset of hypoxia. Furthermore, the stimulatory effect of PRibP was abolished by the presence of calphostin C, a competitor of diacylglycerol binding to protein kinase C, or by the ether lipid 1-O-octadecyl-2-O-methyl-rac-glycero-3-phosphocholine, which inhibits the formation of diacylglycerol from phosphatidyl 4,5-bisphosphate. Thus, an apparent involvement of protein kinase C prompted the authors of that study to suggest that the activities of the three enzymes, (i) catalysis of the conversion of ribose 5-phosphate to PRibP, (ii) PRPP synthase, and (iii) PRPP diphosphohydrolase, are controlled by the phosphatidylinositol-specific phospholipase C signaling pathway (81), which has also been shown to be activated during hypoxia (128).
In vitro analysis of rat liver and kidney and human red cell phosphofructokinase showed that PRibP is an activator of rat kidney and human red cell phosphofructokinase and that PRibP, synergistically with AMP, regulates the activity of phosphofructokinase, because PRibP relieves the inhibition by ATP, increases the affinity for fructose 6-phosphate, and reduces inhibition by citrate. These effects are furthermore enhanced by AMP (129–132). Finally, PRibP inhibits fructose 1,6-bisphosphatase activity of rat kidney, an effect that is also enhanced by AMP (129).
PRibP, Glucose 1,6-Bisphosphate, and Glycerate 2,3-Bisphosphate
Cellular metabolism involves a number of bisphosphate compounds. Some of these, such as glycerate 1,3-bisphosphate and fructose 1,6-bisphosphate, are direct intermediates of biochemical pathways, whereas others, such as glycerate 2,3-bisphosphate, glucose 1,6-bisphosphate, and fructose 2,6-bisphosphate, function as effectors of metabolism. Yet others, such as PRibP, function as both metabolic intermediates and effector molecules although not necessarily within the same organism. Phosphomutases are important in the metabolism of bisphosphate compounds. Crucial to the mechanisms of catalysis of phosphomutases is the formation of phosphoryl-enzyme derivatives. Histidine, threonine, or serine residues have been identified as phosphoryl acceptors and donors in the reversible phosphoryl transfer reactions of phosphomutases.
Human erythrocyte bisphosphoglycerate mutase is responsible for the formation of the allosteric regulator of oxygen binding by hemoglobin, glycerate 2,3-bisphosphate (glycerate 1,3-bisphosphate → glycerate 2,3-bisphosphate [3-phospho-d-glycerate 1,2-phosphomutase]) (Table 2). The active site of bisphosphoglycerate mutase contains His10, which is phosphorylated during catalysis (133). In phosphoribomutase, either a threonine or a serine functions as the phosphorylatable residue. In Bacillus cereus phosphoribomutase, Thr85 is the reactive residue (134), and in human phosphoribomutase 2, the reactive residue is Ser165 (92). Phosphoglucomutase and several other phosphohexomutases contain a conserved serine residue at the active site, Ser117 in rabbit phosphoglucomutase (135).
The catalytic mechanism of phosphomutases is initiated with a phosphorylated enzyme and a phosphorylated compound such as glucose 6-phosphate, E-phosphate + glucose 6-phosphate → E + glucose 1,6-bisphosphate, followed by flipping of the bisphosphate compound within the active site, regeneration of the phosphorylated enzyme, and release of the isomerized product, E + glucose 1,6-phosphate → E-phosphate + glucose 1-phosphate. Both reactions are fully reversible. Although studied in great detail with phosphoglucomutase (135, 136), the reaction is likely to proceed similarly in other phosphomutases as well. It should be noted that some phosphomutases, such as the glycolytic phosphoglycerate mutase (Table 2), are cofactor (i.e., bisphosphate) independent, but their catalytic mechanisms still proceed with the formation of a serine phosphoryl intermediate (E + glycerate 3-phosphate → E-phosphate + glycerate → E + glycerate 2-phosphate) (137).
PHYSIOLOGY OF PENTOSE BISPHOSPHATE METABOLISM
De Novo and Salvage Synthesis of Ribonucleoside 5′-Monophosphates
PRPP is a key intermediate in the biosynthesis of ribonucleotides. PRPP is produced from ribose 5-phosphate, which in turn is produced from ribulose 5-phosphate, the product of the oxidative branch of the pentose phosphate pathway. In general, archaeal species contain an incomplete nonoxidative branch of the pentose phosphate pathway. Specifically, a lack of ribulose phosphate epimerase is widespread. A lack of the oxidative branch is also common among archaeal species. This apparent lack of ribulose 5-phosphate synthesis capability is circumvented by the ribulose monophosphate pathway, which converts fructose 6-phosphate to ribulose 5-phosphate and formaldehyde by the activity of the two enzymes catalyzing the reversible reactions, 6-phospho 3-hexuloisomerase (fructose 6-phosphate → d-arabino 3-hexulose 6-phosphate) and 3-hexulose 6-phosphate synthase (d-arabino 3-hexulose 6-phosphate → ribulose 5-phosphate + formaldehyde) (56) (Fig. 1E). The normal function of this pathway is to scavenge and detoxify formaldehyde by operation in the opposite direction (138). In some organisms, such as T. kodakarensis, the two enzyme activities are carried out by a single polypeptide (TK0475 gene). The pathway appears to function in reverse (i.e., as shown above) in archaeal organisms, and the toxic formaldehyde generated in the process is detoxified in other biochemical processes by conversion to methylene tetrahydromethanopterin or by oxidation to carbon dioxide (138, 139). A knockout of the TK0475 gene in T. kodakarensis results in a ribonucleoside-auxotrophic phenotype. The ribonucleosides adenosine, guanosine, inosine, cytidine, and uridine are all able to promote the growth of the TK0475 knockout strain. The fact that either purine or pyrimidine compounds promote growth suggests a deficiency of an intermediate common to purine and pyrimidine nucleotide biosynthesis. This compound is very likely ribosyl 1-phosphate. The wild-type strain produces ribulose 5-phosphate in processes catalyzed by the TK0475 gene product, and ribulose 5-phosphate is converted to ribose 5-phosphate and then to PRPP, a precursor of purine and pyrimidine nucleotides. The TK0475 knockout strain is unable to produce ribulose 5-phosphate and, hence, ribose 5-phosphate and PRPP. However, when supplied with a ribonucleoside, ribosyl 1-phosphate is produced by the activity of a nucleoside phosphorylase (e.g., adenosine + Pi → adenine + ribosyl 1-phosphate). This ribosyl 1-phosphate can then be used as a ribosyl donor for a second phosphorylase with the formation of a second ribonucleoside (e.g., uracil + ribosyl 1-phosphate → uridine + Pi). As all of the phosphorylases catalyze fully reversible reactions, biosynthetic need determines the direction of flow of the ribose moiety. In addition, the ribosyl 1-phosphate formed by phosphorolysis of ribonucleosides may be converted to ribose 5-phosphate (provided that phosphoribomutase is present) and then to PRPP and finally to ribonucleotides.
Nucleosides may be salvaged by either of two pathways, at least in T. kodakarensis, one pathway involving the activities of nucleoside phosphorylase, ADP-dependent ribosyl 1-phosphate 5-kinase, and AMP phosphorylase and the other involving nucleoside phosphorylase, phosphoribomutase, PRPP synthase, and a phosphoribosyltransferase. Figure 1E shows the two pathways with adenosine as the nucleoside. However, guanosine, inosine, uridine, and cytidine may also be salvaged (139). The PRibP-dependent pathway (adenosine → ribosyl 1-phosphate → PRibP → AMP; the sum of these three reactions is as follows: adenosine + ADP → 2 AMP) is energetically the more favorable than the PRPP-dependent pathway (adenosine → ribosyl 1-phosphate → ribose 5-phosphate → PRPP → AMP; the sum of these four reactions with the inclusion of diphosphatase is as follows: adenosine + ATP → 2 AMP + Pi) due to the consumption of two phosphodiester bonds for the synthesis of PRPP versus one for the synthesis of PRibP. However, this same phenomenon, the hydrolysis of the PPi formed in the PRPP-dependent pathway, contributes to making the pathway essentially irreversible by providing a more negative standard free energy (ΔG0′).
Regulation of Enzyme Activity and Gene Expression
Intuitively, unregulated activity of the pathway leading from AMP to glycerate 3-phosphate may be detrimental to the cell. Indeed, the activity of two of the enzymes of the pathway of T. kodakarensis, AMP phosphorylase and PRibP isomerase, is stimulated by AMP. The activity of AMP phosphorylase exhibits cooperativity in the presence of the substrate AMP, resulting in sigmoidal saturation kinetics in response to AMP, meaning that the activity of the enzyme is stimulated by higher concentrations of AMP. Furthermore, the activity of PRibP isomerase is stimulated by the presence of AMP. In fact, the enzyme is hardly active in the absence of AMP. Altogether, the flux through the pathway responds to the AMP level, which prevents AMP degradation, when the compound is not in surplus. The effects of AMP on AMP phosphorylase and PRibP isomerase are allosteric activation in both cases (52).
Western blot analysis revealed that the synthesis of PRibP isomerase and Rubisco increased in T. kodakarensis following growth in medium with the simultaneous addition of the ribonucleosides adenosine, guanosine, cytidine, and uridine. In contrast, the synthesis of AMP phosphorylase appeared constitutive under these growth conditions (Fig. 1F) (52). Thus, PRibP isomerase and Rubisco function in a pathway for ribonucleoside catabolism. This pathway includes the enzymes ribonucleoside phosphorylase(s) and ADP-dependent ribosyl 1-phosphate 5-kinase as well (Fig. 1A). The regulation of the activity of these enzymes as well as the expression of their genes remains to be established. The fact that AMP stimulates the activity of PRibP isomerase demonstrates that PRibP isomerase and Rubisco (and, of course, AMP phosphorylase) also function in a pathway for AMP catabolism.
EVOLUTION OF PENTOSE BISPHOSPHATE CATABOLISM
Distribution among Species
Metagenomics and single-cell genomic analyses are about to revolutionize the phylogenetic tree of life. Sequencing of genomes of large numbers of uncultivable microbial species, archaea as well as bacteria, reveals the existence of many new phyla unanticipated through studies of organisms cultured in laboratories and therefore designated “candidate” phyla. Examples of these phyla are DPANN and TACK (which are actually superphyla) (140–142), the MSBL1 division of archaea (143), and the candidate phylum radiation of bacteria, a superphylum containing more than 30 phyla (144). The pentose bisphosphate pathway was originally found in class members of the Euryarchaeota phylum (such as the Thermococcus and Pyrococcus genera). Currently, the pentose bisphosphate pathway has been found in representatives of all four main groups of Archaea (Euryarchaeota, TACK, DPANN, and Asgard) (for classification, see references 144 and 145).
Among bacteria, the candidate phylum SR1 consists of cosmopolitan bacteria of diverse origins, marine and terrestrial environments, freshwater lakes, subsurface aquifers, as well as various animal surfaces (146, 147). Analysis of the genomes of SR1 representatives from the human mouth or from a surface aquifer revealed the presence of genes encoding form II/III Rubisco as well as AMP phosphorylase and PRibP isomerase (148, 149). Form II/III Rubisco is an intermediate form that contains important catalytic residues similar to those of form III Rubisco and bears some resemblance to bacterial form II Rubisco (150). Thus, these organisms may contain the pathway AMP → PRibP → ribulose 1,5-bisphosphate → glycerate 3-phosphate. Indeed, a DNA fragment recovered from an isolate of a PER phylum member, cloned and expressed in E. coli, produced an enzyme with bona fide Rubisco activity, providing further evidence for the existence of an AMP catabolic pathway in these uncultivable organisms (151). Similarly, analysis of nucleotide sequences of other candidate phyla (OD1, OP11, PER, and WS6) for the presence of genes encoding form II/III or form III Rubisco also revealed the presence of genes for AMP phosphorylase and PRibP isomerase (151–153). Finally, the genomes of some members of the candidate phyla OP11 and PER also contained the capacity to code for an enzyme homologous to ADP-dependent ribosyl 1-phosphate 5-kinase, and thus, these organisms may contain the pathway ribonucleoside → ribosyl 1-phosphate → PRibP → ribulose 1,5-bisphosphate → glycerate 3-phosphate (151).
Altogether, metagenomic analyses have revealed an expansion of the distribution of the pentose bisphosphate pathway far beyond that originally described among members of the orders Methanococcales, Thermococcales, and Archaeoglobales (61, 154), with the inclusion of members of the archaeal superphyla DPANN and TACK, the division MSBL1, and, perhaps surprisingly, large numbers of bacterial species as well, namely, members of the candidate phylum radiation, such as “Candidatus Parcubacteria.”
Genetic Organization of Pentose Bisphosphate Pathway-Encoding Genes
In many archaeal organisms, the genes encoding AMP phosphorylase, PRibP isomerase, and Rubisco are well separated on the chromosomes. For example, in the T. kodakarensis genome (48), the intragenic distances of the three genes are 169, 139, and 1,777 kbp. Also, in Archaeoglobus species, the three genes are unlinked. Archaeoglobus sulfaticallidus (155), A. fulgidus (156), and several Archaeoglobus species (though not all) contain two AMP phosphorylase-specifying genes located next to one another and arranged with unidirectional transcription. The two A. fulgidus AMP phosphorylase open reading frames (ORFs) (GenBank accession numbers WP_010878838.1 and WP_010878839.1) overlap by 4 bp, and the two A. sulfaticallidus ORFs are separated by 10 bp, whereas other Archaeoglobus sp. AMP phosphorylase ORFs have no intragenic distance or overlap. The two A. fulgidus AMP phosphorylase amino acid sequences are 63% identical (80% similar), whereas they are 45 or 51% identical (63 or 68% similar) to the amino acid sequence of T. kodakarensis AMP phosphorylase.
Contrary to the dispersed location of the three genes, many organisms have the genes encoding AMP phosphorylase, PRibP isomerase, and Rubisco located next to one another in their chromosomes. Figure 8 shows the organization of the genes encoding PRibP isomerase and/or AMP phosphorylase relative to the gene encoding Rubisco (top) or the relative organization of the PRibP isomerase- and AMP phosphorylase-encoding genes (bottom).
FIG 8.
Organization of genes encoding AMP phosphorylase, PRibP isomerase, and Rubisco. Organisms with two or three of the genes grouped together are included. Arrows indicate genes and the direction of transcription. The genes encoding PRibP isomerase are shown in red, those encoding Rubisco are shown in green, and those encoding AMP phosphorylase are shown in yellow. Blue arrows indicate genes encoding PRPP synthase, and gray bars indicate ORFs specifying gene products that appear to be unrelated to the pentose bisphosphate pathway. These ORFs may be transcribed in either direction. In some cases, a gray bar may represent two, three, or four ORFs. The lengths of ORFs and intragenic distances are not drawn to scale. Genes from the following organisms were included in the analysis: “Candidatus Parcubacteria” (“Candidatus Kuenenbacteria”) bacterium (GenBank accession number LCCW01000018.1) (53); “Candidatus Micrarchaeota” archaeon YNP.OP.bin.241 of the “Candidatus Micrarchaeota” archaea (DPANN superphylum) (IMG locus tags Ga0137740_12174 [PRibP isomerase], Ga0137740_12173 [Rubisco], and Ga0137740_12172 [AMP phosphorylase]); members of the Methanomassiliicoccaceae family, including “Candidatus Methanomethylophilus alvus” Mx1201 (GenBank accession number NC_020913.1) (160), Methanomassiliicoccus luminyensis B10 (GenBank accession number CAJE01000001.1) (161), Methanomassiliicoccales archaeon RumEn M1 (GenBank accession number LJKK01000024.1), and “Candidatus Methanomassiliicoccus intestinalis” Issoire-Mx1 (GenBank accession number CP005934.1) (162); “Candidatus Methanoplasma termitum” strain MpT1 (GenBank accession number NZ_CP010070.1); the Thermoplasmatales archaeon BRNA1 (GenBank accession number NC_020892.1); “Candidatus Methanomethylicus mesodigestum” V2 (IMG locus tags Ga0138507_109273 [PRPP synthase], Ga0138507_109274 [PRibP isomerase], Ga0138507_109275 [Rubisco], and Ga0138507_109276 [AMP phosphorylase]) (163); “Candidatus Methanosuratus petracarbonis” V5 (IMG locus tags Ga0138512_10226 [PRPP synthase], Ga0138512_10225 [PRibP isomerase], Ga0138512_10223 and Ga0138512_10224 [incomplete Rubisco-encoding sequence], and Ga0138512_10221 [incomplete AMP phosphorylase-encoding sequence]); “Candidatus Heimdallarchaeota” archaeon LC_2 (GenBank accession number MDVR01000020.1) (164) (the gray bar represents two ORFs); “Candidatus Odinarchaeota” archaeon LCB_4 (GenBank accession number MDVT01000007.1) (164) (the gray bar represents a single ORF that is transcribed toward the Rubisco-encoding gene); the TACK superphylum members “Candidatus Bathyarchaeota” archaeon RBG_13_46_16b (IMG locus tags Ga0154709_15214 [PRibP isomerase], Ga0154709_15213 [Rubisco], and Ga0154709_15211 [AMP phosphorylase], “Candidatus Bathyarchaeota” archaeon JdFR-04 (IMG locus tags Ga0180918_11314 [PRibP isomerase], Ga0180918_11315 [Rubisco], and Ga0180918_11316 [AMP phosphorylase]), and uncultured Crenarchaeota strain C2 (IMG locus tags none_01303 [PRibP isomerase], none_01302 [Rubisco], and none_01301 [AMP phosphorylase]); Halogeometricum species such as H. borinquense PR3 DSM 11551 (GenBank accession number NC_014729.1) (165) and H. rufum strain CGMCC 1.7736 (GenBank accession number NZ_FOYT01000001.1); Halogranum species such as H. salarium strain B-1, (GenBank accession number NZ_ALJD01000003.1) (166) and H. gelatinilyticum strain CGMCC 1.10119 (GenBank accession number NZ_FNHL01000001.1); Haloferax species such as H. volcanii DS2 (GenBank accession number NC_013967.1) (167), H. elongans ATCC BAA-1513 (GenBank accession number NZ_AOLK01000015.1) (168), and H. prahovense TL6 (GenBank accession number NZ_AOLG01000009.1); Halorientalis persicus IBRC-M 10043 (GenBank accession number FOCX01000023.1), Halorientalis regularis IBRC-M 10760 (GenBank accession number NZ_FNBK01000008.1), and Halorientalis sp. strain IM1011 (GenBank accession number NZ_CP019067.1), all of which also contain a gene encoding AMP phosphorylase; Halobellus clavatus CGMCC 1.10118 (GenBank accession number NZ_FNPB01000001.1); Halovenus aranensis IBRC-M10015 (GenBank accession number NZ_FNFC01000006.1); “Candidatus Heimdallarchaeota” archaeon LC_3 (IMG locus tags Ga0197642_10446 [PRibP isomerase] and Ga0197642_10445 [Rubisco] [164], as well as two AMP phosphorylase-encoding genes at separate positions); MSBL1 candidate division members, including the MSBL1 archaeon SCGC-AAA261O19 (IMG locus tags Ga0126901_1651 [PRibP isomerase] and Ga0126901_1652 [Rubisco]) and the MSBL1 archaeon SCGC-AAA261G05 (IMG locus tags Ga0126900_11410 [PRibP isomerase] and Ga0126900_11411 [Rubisco]); members of the order Methanomicrobiales, such as Methanosphaerula palustris E1-9c (GenBank accession number NC_011832.1) (169), Methanospirillum hungatei JF-1 (GenBank accession number NC_007796.1), Methanofollis ethanolicus HASU (GenBank accession number NZ_BCNW01000001.1) (170), and Methanocorpusculum labreanum Z (GenBank accession number NC_008942.1) (171), each of which contains an AMP phosphorylase-encoding gene at separate positions; “Candidatus Lokiarchaeota” archaeon CR_4 (GenBank accession number MBAA01000130.1) (164), with an AMP phosphorylase-encoding gene present at a separate location; Nanoarchaeota archaeon SCGC AAA011-L22 (GenBank accession number AQYW01000059.1) (140), with an AMP phosphorylase-encoding gene present (the gray bar represents three ORFs transcribed toward the Rubisco-encoding gene); Hyperthermus butylicus DSM 5456 (GenBank accession number NC_008818.1) (172), where the gray bar represents three ORFs that are transcribed toward the Rubisco gene and an AMP phosphorylase-encoding gene present at a separate position; the unclassified Crenarchaeota archaeon GMQ bin_5, which contains a cluster of genes for PRPP synthase, PRibP isomerase, and Rubisco (IMG locus tags Ga0180349_10846 [PRPP synthase], Ga0180349_10847 [PRibP isomerase], Ga0180349_10848 [PRibP isomerase], and Ga0180349_10849 [Rubisco]) and an AMP phosphorylase-encoding gene present at a separate position; “Candidatus Bathyarchaeota” archaeon BA1 (a member of the TACK superphylum) (GenBank accession number LIHJ01000032.1) (173), and the archaeon RBG_16_50_20 (GenBank accession number MEMY01000024.1) (174), all of whose genomes also contain AMP phosphorylase-encoding genes; Crenarchaeota group 1 archaeon SG8-32-1 (GenBank accession number LFWU01000010.1) (175), which contains the Rubisco gene next to the AMP phosphorylase gene, with a PRibP isomerase gene present at a separate position; Aciduliprofundum boonei T469 (GenBank accession number NC_013926), which contains PRibP isomerase- and AMP phosphorylase-encoding genes expressed unidirectionally (a second gene encoding PRibP isomerase is located distantly from that close to the AMP phosphorylase-encoding gene, a Rubisco-encoding gene is also present, and a similar gene arrangement is found in Parvarchaeota archaeon JGI [IMG locus tags Ga0098941_1053 {PRibP isomerase} and Ga0098941_1054 {AMP phosphorylase}]; here a Rubisco-encoding gene is not found, but the genome sequence has not been completed); Methanolacinia petrolearia DSM 11571 (GenBank accession number NC_014507.1) (176) and “Candidatus Geothermarchaeota” archaea JdFR-13 (GenBank accession number MTMA01000004.1) and JdFR-14 (GenBank accession number MTMB01000003.1) (IMG locus tags Ga0180860_14459 [PRibP isomerase] and Ga0180860_14458 [AMP phosphorylase]); Methanomicrobium mobile DSM 1539 of the Methanomicrobiales order (GenBank accession number NZ_JOMF01000004.1), which has the AMP phosphorylase and PRibP isomerase genes separated by a single ORF; members of the Desulfurococcales order, such as Thermosphaera aggregans M11TL (GenBank accession number NC_014160.1) (177) and Desulfurococcus kamchatkensis (amylolyticus) 1221n (GenBank accession number NC_011766.1), (178) with two consecutive PRibP isomerase-encoding genes and Rubisco- and AMP phosphorylase-encoding genes at separate positions; and members of the order Thermoproteales, such as the closely related species Thermofilum pendens Hrk 5 (GenBank accession number NC_008698.1) and Thermofilum adornatus 1910b (GenBank accession number NC_022093.1) (179), which contain two consecutive PRibP isomerase-encoding genes with the addition of a PRPP synthase-encoding gene and with Rubisco- and AMP phosphorylase-encoding genes located elsewhere in their genomes. Rare tandem arrangements of AMP phosphorylase-encoding genes are found in species of the Archaeoglobales order, such as A. fulgidus (GenBank accession number NC_000917.1) (156) or A. sulfaticallidus (GenBank accession number NC_021169.1) (155). In contrast, neither Archaeoglobus veneficus (GenBank accession number NC_015320.1) nor Archaeoglobus profundus (GenBank accession number NC_013741.1) has this tandem arrangement. Rather, the former organism has two unlinked AMP phosphorylase-encoding genes, whereas the latter organism has only a single AMP phosphorylase-encoding gene. The GenBank accession numbers for A. fulgidus PRibP isomerase and Rubisco are WP_010879529.1 and WP_010879134.1, respectively (156).
In many organisms with the three genes closely located, some have the three genes transcribed in the same direction, indicating that the three genes may be expressed as operons. This is the case for a bacterium of the “Candidatus Parcubacteria” (“Candidatus Kuenenbacteria”) (OD1) phylum. The distances between each ORF are 174 bp (PRibP isomerase and Rubisco ORFs) and 256 bp (Rubisco and AMP phosphorylase ORFs). This genetic organization is also found in members of the Methanomassiliicoccales order as well as in the Thermoplasmatales archaeon BRNA1. In these organisms, the PRibP isomerase and Rubisco ORFs are separated by 2 to 13 bp, whereas the Rubisco and AMP phosphorylase ORFs share a 4-bp overlap. The genomes of the organisms “Candidatus Methanomethylicus mesodigestum” and “Candidatus Methanosuratus petracarbonis” (both members of the TACK superphylum) contain an arrangement of the PRibP isomerase, Rubisco, and AMP phosphorylase ORFs similar to that of the Thermoplasmata (although without any overlap), with the inclusion of an ORF specifying PRPP synthase (usually designated prs [65, 69]) upstream of the three other ORFs. The “Candidatus Heimdallarchaeota” archaeon LC2 has a different order of the three genes (AMP phosphorylase-PRibP isomerase-Rubisco) and moreover has a three-ORF addition between the PRibP isomerase and Rubisco ORFs. A yet different variant of the order of the three ORFs is found in the “Candidatus Odinarchaeota” archaeon LCB4 (Rubisco-PRibP isomerase-AMP phosphorylase), with the addition of an ORF between those for Rubisco and PRibP isomerase. Some members of the “Candidatus Bathyarchaeota” phylum as well as the uncultured Crenarchaeota archaeon C2 have the gene order PRibP isomerase-Rubisco-AMP phosphorylase but with a reversal of the direction of transcription of the Rubisco ORF relative to that of the “Candidatus Parcubacteria” bacterium described above. In many species of the Halomicrobiales order, the three ORFs for PRibP isomerase, Rubisco, and AMP phosphorylase are located close to one another, with the gene order Rubisco-PRibP isomerase-AMP phosphorylase and with the addition of one, two, or three ORFs between the Rubisco and PRibP isomerase ORFs. This arrangement is particularly widespread among Halogeometricum, Halogranum, and Haloferax species.
In a number of species, only the PRibP isomerase- and Rubisco-encoding genes are linked (Fig. 8). In most cases, an AMP phosphorylase-encoding gene is located elsewhere on the chromosome. In the genomes of a few organisms, an AMP phosphorylase-encoding gene has not been identified. These cases, however, are all permanent draft genome versions, and thus, the apparent lack of an AMP phosphorylase-specifying gene very likely is caused by a lack of sequence information rather than an actual lack of the gene. The members of the Halobacteriales order Halobellus clavatus and Halovenus aranensis are examples of organisms with as-yet-unidentified AMP phosphorylase-encoding genes. The PRibP isomerase- and Rubisco-encoding genes may be organized with convergent transcription, such as in the Halobellus, Halorientalis, and Halovenus species of the Halobacteriales order, or with unidirectional transcription, such as in the “Candidatus Heimdallarchaeota” archaeon LC3, members of the candidate division MBSL1, certain Methanomicrobiales, the “Candidatus Lokiarchaeota” archaeon CR4, a Nanoarchaeota archaeon, a member of the Desulfurococcales order, Hyperthermus butylicus, and Bathyarchaeota archaeon BA1. Occasionally, some other ORFs are found next to or between the Rubisco- and PRibP isomerase-encoding genes, and in some cases, this ORF is a prs gene (“Candidatus Methanosuratus petracarbonis,” the unclassified Crenarchaeota archaeon GMQ, and “Candidatus Bathyarchaeota” archaeon BA1). As is evident from Fig. 8, a number of species contain two PRibP isomerase-specifying genes.
A linkage of the Rubisco- and AMP phosphorylase-encoding genes with a separate location of the PRibP isomerase-encoding gene appears to be rare and represented by only a singe example of a Crenarchaeota group 1 archaeon.
Linkage of the PRibP isomerase- and AMP phosphorylase-encoding genes (without a linked Rubisco-encoding gene) is found in Aciduliprofundum boonei, an obligate thermoacidophilic archaeon of the Euryarchaeota phylum; an archaeon of the Parvarchaeota (a member of the DPANN superphylum), Methanolacinia petrolearia and Methanomicrobium mobile, members of the Methanomicrobiales order, and a “Candidatus Geothermarchaeota” archaeon, a member of the TACK superphylum. Figure 8 shows examples of the linkage of two PRibP isomerase-specifying genes found in several members of the Desulfurococcales and Thermoproteales orders. Species of the latter order often contain a PRPP synthase-encoding gene upstream of the two linked PRibP isomerase-encoding genes. Figure 8 is completed by the Archaeoglobales, some of which contain two consecutively transcribed AMP phosphorylase-specifying genes, as mentioned above.
Evolution and Phylogeny
Much effort has been devoted to analyzing the evolution of Rubisco, primarily to understand the origin of autotrophy. However, there is a great deal of controversy in the field, for example, the question of whether the first cells were autotrophs or, alternatively, whether autotrophs emerged from (chemoorgano)heterotrophs. Form IV Rubisco catalyzing enolization without subsequent carboxylation may represent an ancestral Rubisco to which the carboxylation process was added later in evolution and where forms III, II/III, and I appeared. Additionally, the Calvin-Benson-Bassham cycle may have evolved from ancient nucleotide metabolism (108). In line with this suggestion, Schönheit and colleagues proposed that “a simple interpretation is that the type III Rubisco pathway [i.e., the pentose bisphosphate pathway] is an ancient relic of the first heterotrophic metabolisms in archaea, Rubisco later being transferred to bacteria, where it is less common among obligate heterotrophs, and, where it—in time—ultimately became assimilated into dedicated carbon dioxide fixation in the Calvin[–Benson–Bassham] cycle” and that “ribose was probably the first abundant sugar” (157), and as mentioned above, PRibP may be regarded as an ancient phosphoribosyl donor that became replaced by PRPP in “modern” organisms. A phylogenetic tree of the evolution of Rubisco is shown in Fig. 9A, demonstrating the relationship of the various forms of Rubisco, including forms I and II, participating in photosynthetic carbon dioxide fixation; form III of archaea and various uncultured bacterial species; the intermediate form II/III; and the Rubisco-like proteins, form IV. The data are consistent with those reported previously (151).
FIG 9.
Phylogenetic tree of the evolution of Rubisco and the pentose bisphosphate pathway. Phylogenetic trees were constructed by the neighbor-joining algorithm with MEGA 7 (24). (A) Unrooted phylogenetic tree of Rubisco. The subgroups (IA, IB, IC, and ID) of form I Rubisco are included. Form III consists of two subgroups, Rubisco of Halobacteriales and the rest. (B) Phylogenetic tree of PRibP isomerase, Rubisco, and AMP phosphorylase. The amino acid sequences of the three enzymes PRibP isomerase, Rubisco, and AMP phosphorylase were combined head to tail in the order PRibP isomerase, Rubisco, and AMP phosphorylase for each species or isolate. Alignment was performed with Muscle (22, 23). Bootstrap values were obtained after 1,000 iterations. Values are indicated by purple dots, with the smallest for 70% and the largest for 100%. iToL was used for graphic improvement (25). Leaf labels that are species are color coded to show the orders to which they belong (blue [with decreasing intensity], Methanosarcinales, Methanocellales, Methanomicrobiales, and Methanococcales; red, Halobacteriales; green, Desulfurococcales; magenta, Archaeoglobales; cyan, Methanomassiliicoccales; orange, Thermoproteales; orchid, Thermococcales; olive, “Candidatus Methanomethylicus mesodigestum” and “Candidatus Methanosuratus petracarbonis”). The Parcubacterium phylum and node are shown in lime green for clarity. The outer arc is color coded to show the supergroup to which each species or isolate belongs (blue, TACK; red, Euryarchaeota; plum, DPANN; green-yellow, Asgard; yellow, Eubacteria). Compared to the species shown in Fig. 8, “Candidatus Lokiarchaeota” archaeon CR4 is not included in the phylogenetic tree, as the amino acid sequence is incomplete, and the candidate division MSBL1 archaeon is omitted, as only PRibP isomerase and Rubisco genes could be retrieved from the same isolate. PRibP isomerases from the following organisms were studied: Aciduliprofundum boonei (GenBank accession number WP_008084533.1), A. fulgidus (GenBank accession number WP_048096202.1), “Candidatus Altiarchaeales” archaeon WOR_SM1_79 (GenBank accession number ODS38663.1), “Candidatus Bathyarchaeota” archaeon BA1-A (GenBank accession number KPV64795.1), “Candidatus Bathyarchaeota” archaeon JdFR-04 (IMG locus tag Ga0180918_11314), “Candidatus Bathyarchaeota” archaeon RBG_13_46_16b (GenBank accession number OGD45707.1), “Candidatus Geothermarchaeota” archaeon JdFR-13 (IMG locus tag Ga0180860_14459), “Candidatus Heimdallarchaeota” archaeon LC_2 (GenBank accession number OLS27804.1), “Candidatus Heimdallarchaeota” archaeon LC_3 (GenBank accession number OLS23199.1), “Candidatus Methanomassiliicoccus intestinalis” (GenBank accession number WP_081633149.1), “Candidatus Methanomethylicus mesodigestum” V2 (IMG locus tag Ga0138507_109274), “Candidatus Methanomethylophilus alvus” Mx1201 (GenBank accession number AGI84942.1), “Candidatus Methanoplasma termitum” (GenBank accession number WP_082007275.1), “Candidatus Odinarchaeota” archaeon LCB_4 (GenBank accession number OLS18293.1), “Candidatus Parcubacteria” (“Candidatus Kuenenbacteria”) bacterium GW2011_GWA2_42_15 (GenBank accession number KKS42337.1), Crenarchaeota group 1 archaeon SG8-32-1 (GenBank accession number KON31872.1), Desulfurococcus kamchatkensis (amylolyticus) (GenBank accession number WP_012608127.1), Desulfurococcus mucosus (mobilis) DSM 2161-1 (GenBank accession number WP_013562243.1), Haloarchaeobius iranensis IBRC-M 10013 (GenBank accession number WP_089735480.1), Haloferax elongans (GenBank accession number WP_008323543.1), Haloferax volcanii (GenBank accession number WP_004043979.1), Halogeometricum borinquense (GenBank accession number WP_006056822.1), Halogeometricum rufum (GenBank accession number WP_089805906.1), Halogranum gelatinilyticum (GenBank accession number WP_089695096.1), Halogranum salarium (GenBank accession number WP_009365915.1), Halorientalis regularis (GenBank accession number WP_092692333.1), Haloterrigena limicola JCM 13563 (GenBank accession number WP_008008943.1), Hyperthermus butylicus DSM 5456 (GenBank accession number WP_011821678.1), M. jannaschii DSM 2661 (GenBank accession number Q57586.1), Methanocella paludicola SANAE (GenBank accession number BAI62214.1), Methanococcoides burtonii (GenBank accession number WP_048063643.1), Methanohalophilus mahii (GenBank accession number WP_013036650.1), Methanococcoides vulcani SLH 33 (GenBank accession number WP_091689270.1), Methanoculleus bourgensis (GenBank accession number WP_014867516.1), Methanofollis ethanolicus (GenBank accession number WP_067047982.1), Methanohalophilus euhalobius (GenBank accession number WP_096712556.1), Methanolacinia petrolearia (GenBank accession number WP_013328459.1), Methanolinea tarda (GenBank accession number WP_007314786.1), Methanolobus profundi Mob M (GenBank accession number WP_091936714.1), Methanomassiliicoccus luminyensis (GenBank accession number WP_019176105.1), Methanomicrobium mobile DSM 1539 (GenBank accession number WP_042705231.1), Methanosaeta concilii (GenBank accession number WP_013718165.1), Methanosalsum zhilinae (GenBank accession number WP_013899248.1), Methanosarcina barkeri (GenBank accession number WP_048108174.1), Methanosphaerula palustris (GenBank accession number WP_012616960.1), Methanospirillum hungatei (GenBank accession number WP_011449277.1), Methanotorris igneus Kol5 (GenBank accession number WP_013798611.1), Methanothermococcus thermolithotrophicus (GenBank accession number WP_018153895.1), Methanomassiliicoccales archaeon RumEn M2 (GenBank accession number KQM10928.1), Nanoarchaeota archaeon SCGC AAA011-L22 (IMG locus tag DUSEL4COM1_00971), Natronomonas pharaonis (GenBank accession number WP_011323314.1), Natronorubrum sulfidifaciens (GenBank accession number WP_008160658.1), Palaeococcus pacificus (GenBank accession number WP_048164936.1), Parvarchaeota archaeon JGI (IMG locus tag Ga0098941_1053), Pyrococcus furiosus DSM 3638 (GenBank accession number WP_011011234.1), Pyrodictium occultum (GenBank accession number WP_058371286.1), Staphylothermus marinus (GenBank accession number WP_011839642.1), Theionarchaea archaeon DG-70-1 (GenBank accession number KYK30654.1), T. kodakarensis KOD1 (GenBank accession number WP_011249140.1), Thermofilum pendens (GenBank accession number WP_011752058.1), Thermogladius calderae 1633 (GenBank accession number AFK51564.1), Thermoplasmatales archaeon BRNA1 (GenBank accession number WP_015491730.1), Thermosphaera aggregans (GenBank accession number WP_013129543.1), unclassified “Candidatus Bathyarchaeota” archaeon JZ bin_32 (IMG locus tag Ga0180361_102278), unclassified Crenarchaeota archaeon GMQ bin_5 (IMG locus tag Ga0180349_10848), and uncultured Crenarchaeota archaeon C2 (IMG locus tag none_01303). AMP phosphorylases from the following organisms were studied: Aciduliprofundum boonei T469 (GenBank accession number WP_008084517.1), A. fulgidus (GenBank accession number WP_010878839.1), “Candidatus Altiarchaeales” archaeon WOR_SM1_86-2 (GenBank accession number ODS38082.1), “Candidatus Bathyarchaeota” archaeon BA1 (GenBank accession number KPV62303.1), “Candidatus Bathyarchaeota” archaeon JdFR_04 (IMG locus tag Ga0180918_11316), “Candidatus Bathyarchaeota” archaeon RBG_13_46_16b (GenBank accession number OGD45698.1), “Candidatus Geothermarchaeota” archaeon JdFR-13 (IMG locus tag Ga0180860_14), “Candidatus Heimdallarchaeota” archaeon LC_2 (GenBank accession number OLS27803.1), “Candidatus Heimdallarchaeota” archaeon LC_3 (GenBank accession number OLS18805.1), “Candidatus Methanomethylicus mesodigestum” V2 (IMG locus tag Ga0138507_109276), “Candidatus Methanomethylophilus alvus” Mx1201 (GenBank accession number AGI84944.1), “Candidatus Methanoplasma termitum” (GenBank accession number WP_048113194.1), “Candidatus Odinarchaeota” archaeon LCB_4 (GenBank accession number OLS18294.1), “Candidatus Parcubacteria” (“Candidatus Kuenenbacteria”) bacterium GW2011_GWA2_42_15 (GenBank accession number KKS42335.1), Crenarchaeota group 1 archaeon SG8-32-1 (GenBank accession number KON34322.1), Desulfurococcus kamchatkensis (amylolyticus) (GenBank accession number WP_012607829.1), Desulfurococcus mucosus (mobilis) DSM 2161-1 (GenBank accession number WP_013561832.1), Haloarchaeobius iranensis IBRC-M 10013 (GenBank accession number WP_089735185.1), Haloferax elongans (GenBank accession number WP_008323542.1), Haloferax volcanii (GenBank accession number WP_004043980.1), Halogeometricum borinquense (GenBank accession number WP_006056821.1), Halogeometricum rufum (GenBank accession number WP_089805904.1), Halogranum gelatinilyticum (GenBank accession number WP_089695098.1), Halogranum salarium (GenBank accession number WP_009365914.1), Halorientalis regularis (GenBank accession number WP_092689765.1), Haloterrigena limicola JCM 13563 (GenBank accession number WP_008012656.1), Hyperthermus butylicus DSM 5456 (GenBank accession number WP_011821569.1), M. jannaschii DSM 2661 (GenBank accession number WP_010870172.1), Methanocella paludicola SANAE (GenBank accession number BAI60132.1), Methanococcoides burtonii (GenBank accession number WP_011498424.1), Methanococcoides vulcani SLH 33 (GenBank accession number WP_091690139.1), Methanoculleus bourgensis (GenBank accession number WP_074175757.1), Methanofollis ethanolicus (GenBank accession number WP_067048152.1), Methanohalophilus euhalobius (GenBank accession number WP_096711719.1), Methanohalophilus mahii DSM 5219 (GenBank accession number ADE36782.1), Methanolacinia petrolearia (GenBank accession number WP_013328458.1), Methanolinea tarda (GenBank accession number WP_007314415.1), Methanolobus profundi Mob M (GenBank accession number WP_091932766.1), Methanomassiliicoccales archaeon RumEn M2 (GenBank accession number KQM12620.1), Methanomassiliicoccus luminyensis (GenBank accession number WP_019176103.1), Methanomicrobium mobile DSM 1539 (GenBank accession number WP_042705229.1), Methanosaeta concilii (GenBank accession number WP_013718478.1), Methanosalsum zhilinae (GenBank accession number WP_013897981.1), Methanosarcina barkeri (GenBank accession number WP_048105836.1), Methanosphaerula palustris (GenBank accession number WP_012618742.1), Methanospirillum hungatei (GenBank accession number WP_011449225.1), Methanothermococcus thermolithotrophicus (GenBank accession number WP_018154311.1), Methanotorris igneus Kol5 (GenBank accession number WP_013798979.1), Nanoarchaeota archaeon SCGC AAA011-L22 (IMG locus tag DUSEL4COM1_00910), Natronomonas pharaonis (GenBank accession number WP_011323686.1), Natronorubrum sulfidifaciens (GenBank accession number WP_008163149.1), Palaeococcus pacificus (GenBank accession number WP_048164909.1), Parvarchaeota archaeon JGI (IMG locus tag Ga0098941_1054), Pyrococcus furiosus DSM 3638 (GenBank accession number WP_011012753.1), Pyrodictium occultum (GenBank accession number WP_058370070.1), Staphylothermus marinus (IMG locus tag Smar_1460), T. kodakarensis (GenBank accession number WP_011249307.1), Thermofilum pendens (GenBank accession number WP_011751768.1), Thermogladius calderae 1633 (GenBank accession number WP_014737920.1), Thermoplasmatales archaeon BRNA1 (GenBank accession number WP_015491732.1), Thermosphaera aggregans (GenBank accession number WP_052891646.1), unclassified “Candidatus Bathyarchaeota” archaeon JZbin_32 (IMG locus tag Ga0180361_102215), unclassified Crenarchaeota archaeon GMQ bin_5 (IMG locus tag Ga0180349_10243), and uncultured Crenarchaeota archaeon C2 (IMG locus tag none_01301). Form III Rubiscos from the following organisms were studied: Aciduliprofundum boonei T469 (GenBank accession number WP_008086508.1), A. fulgidus (GenBank accession number WP_010879134.1), “Candidatus Altiarchaeales” archaeon WOR_SM1_86-2 (IMG locus tag Ga0197631_1015), “Candidatus Bathyarchaeota” archaeon BA1 (GenBank accession number KPV64794.1), “Candidatus Bathyarchaeota” archaeon JdFR_4 (IMG locus tag Ga0180918_11315), “Candidatus Bathyarchaeota” archaeon RBG_13_46_16b (IMG locus tag Ga0154709_15213), “Candidatus Geothermarchaeota” archaeon JdFR-13 (IMG locus tag Ga0180860_14283), “Candidatus Heimdallarchaeota” archaeon LC2 (IMG locus tag Ga0197641_102024), “Candidatus Heimdallarchaeota” archaeon LC3 (IMG locus tag Ga0197642_10445), “Candidatus Methanomassiliicoccus intestinalis” (GenBank accession number WP_020449584.1), “Candidatus Methanomethylicus mesodigestum” V2 (IMG locus tag Ga0138507_109275), “Candidatus Methanomethylophilus alvus” (GenBank accession number WP_015504092.1), “Candidatus Methanoplasma termitum” (GenBank accession number WP_048113196.1), “Candidatus Odinarchaeota” archaeon LCB_4 (IMG locus tag Ga0197644_171221), Crenarchaeota group 1 archaeon SG8-32-1 (GenBank accession number KON34323.1), Desulfurococcus kamchatkensis (GenBank accession number WP_048058905.1), Desulfurococcus mobilis (mucosus) (GenBank accession number WP_013562663.1), Haloarchaeobius iranensis IBRC-M 10013 (IMG locus tag Ga0075265_103122), Haloferax elongans (GenBank accession number WP_008323546.1), Haloferax volcanii (GenBank accession number WP_004043974.1), Halogeometricum borinquense (GenBank accession number WP_006056824.1), Halogeometricum rufum (GenBank accession number WP_089805908.1), Halogranum gelatinilyticum (GenBank accession number WP_089695090.1), Halogranum salarium (GenBank accession number WP_009365917.1), Halorientalis regularis (GenBank accession number WP_092692341.1), Haloterrigena limicola JCM 13563 (GenBank accession number ELZ20969.1), Hyperthermus butylicus DSM 5456 (GenBank accession number WP_011821683.1), M. jannaschii DSM 2661 (GenBank accession number WP_010870747.1), Methanocella paludicola SANAE (GenBank accession number BAI61188.1), Methanococcoides burtonii (GenBank accession number WP_011500311.1), Methanococcoides vulcani (GenBank accession number WP_091690387.1), Methanoculleus bourgensis MBBA (IMG locus tag Ga0133540_112304), Methanofollis ethanolicus (GenBank accession number WP_067047979.1), Methanohalophilus euhalobius WG-1MB (GenBank accession number SNY00336.1), Methanohalophilus mahii (GenBank accession number WP_013037391.1), Methanolacinia petrolearia (GenBank accession number WP_013329831.1), Methanolinea tarda (GenBank accession number WP_007314813.1), Methanolobus profundi Mob M (GenBank accession number WP_091933438.1), Methanomassiliicoccales archaeon RumEn M2 (GenBank accession number KQM10927.1), Methanomassiliicoccus luminyensis (GenBank accession number WP_019176104.1), Methanomicrobium mobile (GenBank accession number WP_042706367.1), Methanosaeta concilii (GenBank accession number WP_013720421.1), Methanosalsum zhilinae WeN5 DSM 4017 (GenBank accession number WP_013897574.1), Methanosarcina barkeri 3 (GenBank accession number WP_048108564.1), Methanospirillum hungatei (GenBank accession number WP_011449278.1), Methanosphaerula palustris (GenBank accession number WP_012616961.1), Methanothermococcus thermolithotrophicus DSM 2095 (GenBank accession number WP_018153747.1), Methanotorris igneus Kol5 DSM 5666 (GenBank accession number WP_013799414.1), Nanoarchaeota archaeon SCGC AAA011-L22 (IMG locus tag DUSEL4COM1_00968), Natronomonas pharaonis Gabara DSM 2160 (GenBank accession number WP_011323101.1), Natronorubrum sulfidifaciens JCM 14089 (GenBank accession number WP_008164021.1), Palaeococcus pacificus DY20341 (GenBank accession number WP_048165292.1), “Candidatus Parcubacteria” bacterium (GenBank accession number KKS42336.1), Parvarchaeota archaeon JGI MDM2 ZSSED05-1-D15 (IMG locus tag Ga0191617_11310), Pyrodictium occultum PL-19 1 (IMG locus tag Ga0118740_11925), Pyrococcus furiosus DSM 3638 (GenBank accession number WP_011012296.1), Staphylothermus marinus F1 DSM 3639 (GenBank accession number WP_011839642.1), Theionarchaea archaeon DG-70-1 (IMG locus tag Ga0197638_10453), T. kodakarensis KOD1 (GenBank accession number WP_011251240.1), Thermofilum pendens (GenBank accession number WP_011752890.1), Thermogladius calderae (GenBank accession number WP_014737038.1), Thermoplasmatales archaeon BRNA1 (GenBank accession number WP_015491731.1), Thermosphaera aggregans (GenBank accession number WP_013129771.1), unclassified “Candidatus Bathyarchaeota” archaeon JZ bin_32 (IMG locus tag Ga0180361_101139), unclassified Crenarchaeota archaeon GMQ bin_5 (IMG locus tag Ga0180349_10849), and uncultured Crenarchaeota archaeon C2 (IMG locus tag none_01302). Form I and II Rubiscos from the following species were studied: Spinacia oleracea (GenBank accession number NP_054944.1), Anabaena cylindrica (GenBank accession number WP_015212301.1), Synechococcus lividus (GenBank accession number WP_099799013.1), Chlamydomonas reinhardtii (GenBank accession number ACJ50136.1), Arabidopsis thaliana (GenBank accession number NP_051067.1), Rhodobacter capsulatus (GenBank accession number WP_074553140.1), Thiobacillus denitrificans (GenBank accession number WP_011313136.1), Allochromatium vinosum (GenBank accession number WP_012970574.1), Xanthobacter flavus (GenBank accession number CAA35115.1), Rhodobacter sphaeroides (GenBank accession number WP_011911026.1), Cupriavidus necator (GenBank accession number WP_042878640.1), Galdieria partita (GenBank accession number BAA75796.1), Olisthodiscus luteus (GenBank accession number BAF80663.1), Porphyridium purpureum (GenBank accession number YP_008965646.1), Cyanidium caldarium (GenBank accession number BAA22828.1), Hydrogenovibrio crunogenus (GenBank accession number WP_011369848.1), Hydrogenovibrio marinus (GenBank accession number WP_029912390.1), Rhodoferax ferrireducens (GenBank accession number WP_011463692.1), Riftia pachyptila (endosymbiont) (GenBank accession number AAC38280.1), Gammaproteobacteria bacterium (GenBank accession number PKM43747.1), Rhodopseudomonas palustris (GenBank accession number WP_027276980.1), Magnetospirillum magnetotacticum (GenBank accession number WP_041041009.1), Rhodobacter capsulatus (GenBank accession number WP_023911418.1), and Thiobacillus denitrificans (GenBank accession number WP_011313150.1). Form IV Rubiscos from the following organisms were studied: Chlorobaculum tepidum (GenBank accession number WP_010933432.1) and Bacillus subtilis (GenBank accession number WP_019713909.1).
A phylogenetic analysis of the pentose bisphosphate pathway is presented in Fig. 9B. Representatives of each of the four major groups of Archaea, Asgard, DPANN, Euryarchaeota, and TACK, as well as a “Candidatus Parcubacteria” bacterium are included in this analysis. The phylogenetic tree resembles quite closely that of Archaea in general, i.e., that based on 16S RNA (158), with particular clustering of the pathway of species and isolates of Euryarchaeota and TACK. Members of Asgard and DPANN are also clustered together. The organization of the three genes shown in Fig. 8 is also included in Fig. 9B and shown outside the outer ring. The organization of operon structure is evolutionarily less pronounced, as all of the combinations of organizations or lack thereof appear scattered among the species or isolates.
Formally, the reaction catalyzed by ADP-dependent ribosyl 1-phosphate 5-kinase was included in the pentose bisphosphate pathway, as described above. However, the distribution of this enzyme among species appears to be more restricted. BLAST analysis revealed its presence among species of the Thermococcales order, such as the closely related species T. kodakarensis, Pyrococcus furiosus, and Palaeococcus pacificus (49); the Archaeoglobales order, such as Archaeoglobales archaeon ex4484_92 (GenBank accession number OYT33062.1); as well as the Theionarchaea archaeon DG-70 (GenBank accession number KYK31951.1). In addition, the distribution among organisms of the gene specifying ATP-dependent ribosyl 1-phosphate 5-kinase (Pcal_0041 of P. calidifontis) is restricted to species of the Crenarchaeota phylum (53).
OUTLOOK
As described above, PRibP has important functions in organisms of all three domains of life. Large efforts have been spent on elucidation of the various aspects of PRibP metabolism. However, a number of aspects need clarification. Perhaps most important is the determination of the three-dimensional structure of ADP-dependent ribose 1-phosphate 5-kinase of the pentose bisphosphate pathway as well as other PRibP binding enzymes. Also, determination of the three-dimensional structure of ATP-dependent ribose 1-phosphate 5-kinase is important, with the aim of comparing the mechanisms of the two enzymes. Contrary to the situation with PRPP, apparently there is no well-defined PRibP binding site that is general to all or several PRibP-consuming or -producing enzymes. The elucidation of more three-dimensional structures may change this conclusion in the future. Other important aspects are the molecular mechanism of the regulation of expression of the genes encoding PRibP isomerase, form III Rubisco, and ADP-dependent ribose 1-phosphate 5-kinase. The gene encoding AMP phosphorylase appears to be constitutively expressed.
PRibP furthermore functions as a regulator of several metabolic processes. The dynamics of PRibP synthesis and catabolism of these processes are also important aspects for future studies in order to obtain a full understanding of the physiology of this important compound. Particularly, the possible involvement of PRibP in the phosphatidylinositol-specific phospholipase C signaling pathway deserves further studies.
ACKNOWLEDGMENT
We are grateful to Christian Storm Petersen for invaluable advice on phylogenetic analysis.
Biographies

Bjarne Hove-Jensen began graduate studies at the Enzyme Division, Institute of Biological Chemistry B, University of Copenhagen, in 1976 working on various aspects of purine nucleotide salvage metabolism in E. coli. Serendipitously, he discovered a mutant with an altered phosphoribosyl diphosphate (PRPP) synthase. After receiving his Ph.D. in biochemistry in 1983 at the University of Copenhagen, he continued studies of nucleotide metabolism in various organisms, with emphasis on the physiological role of PRPP synthase. He became Associate Professor of Biochemistry in 1988 at the University of Copenhagen. Fortuitously, around 2000, he discovered that PRPP is also an intermediate in the catabolic pathway by which phosphonic acid-phosphorus is assimilated by the carbon phosphorus-lyase pathway and that this PRPP originated from PRibP. Studies on the origin of PRibP were conducted on sabbaticals at Queen’s University, Kingston, Ontario, Canada, and at Aarhus University. He is now Associate Professor Emeritus at Aarhus University and continues studies of the physiological role of PRibP.

Ditlev E. Brodersen received his Ph.D. in structural biology from Aarhus University, Denmark, in 2000. After postdoctoral studies at the Medical Research Council, Laboratory of Molecular Biology, Cambridge, United Kingdom, working on the structure of the bacterial 30S ribosomal subunits, he returned to the Department of Molecular Biology and Genetics, Aarhus University, Denmark, where he is currently an Associate Professor of Molecular Biology. His main research interests are microbial defense mechanisms, toxin-antitoxin modules, and the catalytic mechanism of phosphonate breakdown.

M. Cemre Manav received her bachelor of science degree in chemistry from Yildiz Technical University, Istanbul, in 2012. In 2013, she received a master of engineering degree in chemistry and biotechnology engineering at Aarhus University School of Engineering, where she worked on the bacterial carbon-phosphorus lyase complex and began investigating bacterial stress response mechanisms. She received her Ph.D. at the Department of Molecular Biology and Genetics, Aarhus University, where she determined the structure of the small alarmone synthetase RelP and the type II bacterial toxin-antitoxin complex by X-ray crystallography. Currently, she is continuing her research at Aarhus University as a Postdoctoral researcher.
REFERENCES
- 1.Abrams A, Klenow H. 1951. On the action of surface forces on phosphoribomutase. Arch Biochem Biophys 34:285–292. [DOI] [PubMed] [Google Scholar]
- 2.Klenow H, Larsen B. 1952. The action of phosphoglucomutase preparations on ribose 1-phosphate. Arch Biochem Biophys 37:488–490. [DOI] [PubMed] [Google Scholar]
- 3.Leloir LF. 1951. Sugar phosphates. Fortschr Chem Org Naturst 8:47–95. [Google Scholar]
- 4.Klenow H. 1953. Some properties of the phosphoribomutase reaction. Arch Biochem Biophys 46:186–200. [DOI] [PubMed] [Google Scholar]
- 5.Klenow H. 1955. On the nature of the salt inhibition of some reactions catalyzed by phosphoglucomutase preparations. Arch Biochem Biophys 58:288–297. [DOI] [PubMed] [Google Scholar]
- 6.Najjar VA, Pullman ME. 1954. The occurrence of a group transfer involving enzyme (phosphoglucomutase) and substrate. Science 119:631–634. [DOI] [PubMed] [Google Scholar]
- 7.Greenberg GR. 1951. De novo synthesis of hypoxanthine via inosine-5-phosphate and inosine. J Biol Chem 190:611–631. [PubMed] [Google Scholar]
- 8.Buchanan JM, Schulman MP. 1953. Biosynthesis of the purines. III. Reactions of formate and inosinic acid and an effect of the citrovorum factor. J Biol Chem 202:241–252. [PubMed] [Google Scholar]
- 9.Saffran M, Scarano E. 1953. Mechanism of the incorporation of adenine into adenosine monophosphate. Nature 172:949–951. [DOI] [PubMed] [Google Scholar]
- 10.Lieberman I, Kornberg A, Simms ES. 1955. Enzymatic synthesis of pyrimidine nucleotides; orotidine-5′-phosphate and uridine-5′-phosphate. J Biol Chem 215:403–451. [PubMed] [Google Scholar]
- 11.Kornberg A, Lieberman I, Simms ES. 1955. Enzymatic synthesis of purine nucleotides. J Biol Chem 215:417–427. [PubMed] [Google Scholar]
- 12.Remy CN, Remy WT, Buchanan JM. 1955. Biosynthesis of the purines. VIII. Enzymatic synthesis and utilization of α-5-phosphoribosylpyrophosphate. J Biol Chem 217:885–895. [PubMed] [Google Scholar]
- 13.Klenow H. 2003. Some selected recollections from a life with biochemistry, p 285–317. In Semenza G, Turner AJ (ed), Comprehensive biochemistry, vol 42 Elsevier Science, Amsterdam, Netherlands. [Google Scholar]
- 14.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 15.Markowitz VM, Chen IM, Chu K, Szeto E, Palaniappan K, Grechkin Y, Ratner A, Jacob B, Pati A, Huntemann M, Liolios K, Pagani I, Anderson I, Mavromatis K, Ivanova NN, Kyrpides NC. 2012. IMG/M: the integrated metagenome data management and comparative analysis system. Nucleic Acids Res 40:D123–D129. doi: 10.1093/nar/gkr975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Markowitz VM, Chen IM, Palaniappan K, Chu K, Szeto E, Grechkin Y, Ratner A, Jacob B, Huang J, Williams P, Huntemann M, Anderson I, Mavromatis K, Ivanova NN, Kyrpides NC. 2012. IMG: the Integrated Microbial Genomes database and comparative analysis system. Nucleic Acids Res 40:D115–D122. doi: 10.1093/nar/gkr1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. 2013. GenBank. Nucleic Acids Res 41:D36–D42. doi: 10.1093/nar/gks1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benson DA, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. 2014. GenBank. Nucleic Acids Res 42:D32–D37. doi: 10.1093/nar/gkt1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marck C. 1988. ‘DNA Strider’: a ‘C’ program for the fast analysis of DNA and protein sequences on the Apple Macintosh family of computers. Nucleic Acids Res 16:1829–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Corpet F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res 16:10881–10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeLano WL. 2002. Unraveling hot spots in binding interfaces: progress and challenges. Curr Opin Struct Biol 12:14–20. [DOI] [PubMed] [Google Scholar]
- 22.Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Letunic I, Bork P. 2011. Interactive Tree of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res 39:W475–W478. doi: 10.1093/nar/gkr201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tener GM, Khorana HG. 1958. Phosphorylated sugars. VI. Syntheses of α-D-ribofuranose 1,5-diphosphate and α-D-ribofuranose 1-pyrophosphate 5-phosphate. J Am Chem Soc 80:1999–2004. doi: 10.1021/ja01541a056. [DOI] [Google Scholar]
- 27.Tarr HLA. 1967. Purine nucleoside phosphorylases: (purine-nucleoside:orthophosphate ribosyltransferase EC 2.3.2.1) purine + R1-P (dR1-P) → ribonucleosides (deoxyribonucleosides) + Pi. Methods Enzymol 12:113–118. [Google Scholar]
- 28.Tarr HLA. 1957. Deoxyribose-1-5-diphosphate. Chem Ind (Lond) 1957(18):562–563. [Google Scholar]
- 29.Hanna R, Mendicino J. 1970. Synthesis of α-D-glucose 1,6-diphosphate, α-D-mannose 1,6-diphosphate, α-D-ribose 1,5 diphosphate, and N-acetyl-α-D-glucosamine 1,6-diphosphate. J Biol Chem 245:4031–4037. [PubMed] [Google Scholar]
- 30.De Chatelet LR, Alpers JB. 1970. Phosphoribokinase from Pseudomonas saccharophila. J Biol Chem 245:3161–3166. [PubMed] [Google Scholar]
- 31.Kalckar HM. 1945. Enzymatic synthesis of a nucleoside. J Biol Chem 158:723–724. [Google Scholar]
- 32.Jensen KF, Houlberg U, Nygaard P. 1979. Thin-layer chromatographic methods to isolate 32P-labeled 5-phosphoribosyl-α-1-pyrophosphate (PRPP): determination of cellular PRPP pools and assay of PRPP synthetase activity. Anal Biochem 98:254–263. [DOI] [PubMed] [Google Scholar]
- 33.Kornberg A, Lieberman I, Simms ES. 1955. Enzymatic synthesis and properties of 5-phosphoribosylpyrophosphate. J Biol Chem 215:389–402. [PubMed] [Google Scholar]
- 34.Sable HZ. 1952. Pentose metabolism in extracts of yeast and mammalian tissues. Biochim Biophys Acta 8:687–697. [DOI] [PubMed] [Google Scholar]
- 35.Khorana HG, Fernandes JF, Kornberg A. 1958. Pyrophosphorylation of ribose 5-phosphate in the enzymatic synthesis of 5-phosphorylribose 1-pyrophosphate. J Biol Chem 230:941–948. [PubMed] [Google Scholar]
- 36.Trembacz H, Jezewska MM. 1990. The route of non-enzymic and enzymic breakdown of 5-phosphoribosyl 1-pyrophosphate to ribose 1-phosphate. Biochem J 271:621–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dennis AL, Puskas M, Stasaitis S, Sandwick RK. 2000. The formation of a 1-5 phosphodiester linkage in the spontaneous breakdown of 5-phosphoribosyl-α-1-pyrophosphate. J Inorg Biochem 81:73–80. [DOI] [PubMed] [Google Scholar]
- 38.Meola M, Yamen B, Weaver K, Sandwick RK. 2003. The catalytic effect of Mg2+ and imidazole on the decomposition of 5-phosphoribosyl-α-1-pyrophosphate in acqueous solution. J Inorg Biochem 93:235–242. [DOI] [PubMed] [Google Scholar]
- 39.Vanderheiden BS. 1961. Ribosediphosphate in the human erythrocyte. Biochem Biophys Res Commun 6:117–123. [DOI] [PubMed] [Google Scholar]
- 40.Vanderheiden BS. 1970. Phosphate esters in human erythrocytes. VII. Further evidence for ribose 1,5-diphosphate as a natural metabolite. Biochim Biophys Acta 215:242–248. [DOI] [PubMed] [Google Scholar]
- 41.Bartlett GR, Bucolo G. 1968. The metabolism of ribonucleoside by the human erythrocyte. Biochim Biophys Acta 156:240–253. [DOI] [PubMed] [Google Scholar]
- 42.Ghodge SV, Cummings JA, Williams HJ, Raushel FM. 2013. Discovery of a cyclic phosphodiesterase that catalyzes the sequential hydrolysis of both ester bonds to phosphorus. J Am Chem Soc 135:16360–16363. doi: 10.1021/ja409376k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kamat SS, Williams HJ, Raushel FM. 2011. Intermediates in the transformation of phosphonates to phosphate by bacteria. Nature 480:570–573. doi: 10.1038/nature10622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hove-Jensen B, McSorley FR, Zechel DL. 2011. Physiological role of phnP-specified phosphoribosyl cyclic phosphodiesterase in catabolism of organophosphonic acids by the carbon-phosphorus lyase pathway. J Am Chem Soc 133:3617–3624. doi: 10.1021/ja1102713. [DOI] [PubMed] [Google Scholar]
- 45.Akouche M, Jaber M, Maurel MC, Lambert JF, Georgelin T. 2017. Phosphoribosyl pyrophosphate: a molecular vestige of the origin of life on minerals. Angew Chem Int Ed Engl 56:7920–7923. doi: 10.1002/anie.201702633. [DOI] [PubMed] [Google Scholar]
- 46.Sutherland JD. 2016. The origin of life—out of the blue. Angew Chem Int Ed Engl 55:104–121. doi: 10.1002/anie.201506585. [DOI] [PubMed] [Google Scholar]
- 47.Atomi H, Fukui T, Kanai T, Morikawa M, Imanaka T. 2004. Description of Thermococcus kodakaraensis sp. nov., a well studied hyperthermophilic archaeon previously reported as Pyrococcus sp. KOD1. Archaea 1:263–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fukui T, Atomi H, Kanai T, Matsumi R, Fujiwara S, Imanaka T. 2005. Complete genome sequence of the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1 and comparison with Pyrococcus genomes. Genome Res 15:352–363. doi: 10.1101/gr.3003105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aono R, Sato T, Imanaka T, Atomi H. 2015. A pentose bisphosphate pathway for nucleoside degradation in Archaea. Nat Chem Biol 11:355–360. doi: 10.1038/nchembio.1786. [DOI] [PubMed] [Google Scholar]
- 50.Sato T, Atomi H, Imanaka T. 2007. Archaeal type III RuBisCOs function in a pathway for AMP metabolism. Science 315:1003–1006. doi: 10.1126/science.1135999. [DOI] [PubMed] [Google Scholar]
- 51.Jensen KF, Dandanell G, Hove-Jensen B, Willemoës M. 18 August 2008, posting date. Nucleotides, nucleosides, and nucleobases. EcoSal Plus 2008 doi: 10.1128/ecosalplus.3.6.2. [DOI] [PubMed] [Google Scholar]
- 52.Aono R, Sato T, Yano A, Yoshida S, Nishitani Y, Miki K, Imanaka T, Atomi H. 2012. Enzymatic characterization of AMP phosphorylase and ribose-1,5-bisphosphate isomerase functioning in an archaeal AMP metabolic pathway. J Bacteriol 194:6847–6855. doi: 10.1128/JB.01335-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aziz I, Bibi T, Rashid N, Aono R, Atomi H, Akhtar M. 25 July 2018. A phosphofructokinase homolog from Pyrobaculum calidifontis displays kinase activity towards pyrimidine nucleosides and ribose 1-phosphate. J Bacteriol doi: 10.1128/JB.00284-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tozzi MG, Camici M, Mascia L, Sgarrella F, Ipata PL. 2006. Pentose phosphates in nucleoside interconversion and catabolism. FEBS J 273:1089–1101. doi: 10.1111/j.1742-4658.2006.05155.x. [DOI] [PubMed] [Google Scholar]
- 55.Soderberg T. 2005. Biosynthesis of ribose-5-phosphate and erythrose-4-phosphate in archaea: a phylogenetic analysis of archaeal genomes. Archaea 1:347–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bräsen C, Esser D, Rauch B, Siebers B. 2014. Carbohydrate metabolism in Archaea: current insights into unusual enzymes and pathways and their regulation. Microbiol Mol Biol Rev 78:89–175. doi: 10.1128/MMBR.00041-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fisher DI, Safrany ST, Strike P, McLennan AG, Cartwright JL. 2002. Nudix hydrolases that degrade dinucleoside and diphosphoinositol polyphosphates also have 5-phosphoribosyl 1-pyrophosphate (PRPP) pyrophosphatase activity that generates the glycolytic activator ribose 1,5-bisphosphate. J Biol Chem 277:47313–47317. doi: 10.1074/jbc.M209795200. [DOI] [PubMed] [Google Scholar]
- 58.Thorsell AG, Persson C, Graslund S, Hammarstrom M, Busam RD, Hallberg BM. 2009. Crystal structure of human diphosphoinositol phosphatase 1. Proteins 77:242–246. doi: 10.1002/prot.22489. [DOI] [PubMed] [Google Scholar]
- 59.Leslie NR, McLennan AG, Safrany ST. 2002. Cloning and characterisation of hAps1 and hAps2, human diadenosine polyphosphate-metabolising Nudix hydrolases. BMC Biochem 3:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bailey S, Sedelnikova SE, Blackburn GM, Abdelghany HM, Baker PJ, McLennan AG, Rafferty JB. 2002. The crystal structure of diadenosine tetraphosphate hydrolase from Caenorhabditis elegans in free and binary complex forms. Structure 10:589–600. [DOI] [PubMed] [Google Scholar]
- 61.Finn MW, Tabita FR. 2004. Modified pathway to synthesize ribulose 1,5-bisphosphate in methanogenic archaea. J Bacteriol 186:6360–6366. doi: 10.1128/JB.186.19.6360-6366.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Eser BE, Zhang X, Chanani PK, Begley TP, Ealick SE. 2016. From suicide enzyme to catalyst: the iron-dependent sulfide transfer in Methanococcus jannaschii thiamin thiazole biosynthesis. J Am Chem Soc 138:3639–3642. doi: 10.1021/jacs.6b00445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang X, Eser BE, Chanani PK, Begley TP, Ealick SE. 2016. Structural basis for iron-mediated sulfur transfer in archael [sic] and yeast thiazole synthases. Biochemistry 55:1826–1838. doi: 10.1021/acs.biochem.6b00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rashid N, Imanaka H, Fukui T, Atomi H, Imanaka T. 2004. Presence of a novel phosphopentomutase and a 2-deoxyribose 5-phosphate aldolase reveals a metabolic link between pentoses and central carbon metabolism in the hyperthermophilic archaeon Thermococcus kodakaraensis. J Bacteriol 186:4185–4191. doi: 10.1128/JB.186.13.4185-4191.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hove-Jensen B, Andersen KR, Kilstrup M, Martinussen J, Switzer RL, Willemoes M. 2017. Phosphoribosyl diphosphate (PRPP): biosynthesis, enzymology, utilization, and metabolic significance. Microbiol Mol Biol Rev 81:e00040-16. doi: 10.1128/MMBR.00040-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hove-Jensen B, Rosenkrantz TJ, Haldimann A, Wanner BL. 2003. Escherichia coli phnN, encoding ribose 1,5-bisphosphokinase activity (phosphoribosyl diphosphate forming): dual role in phosphonate degradation and NAD biosynthesis pathways. J Bacteriol 185:2793–2801. doi: 10.1128/JB.185.9.2793-2801.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hove-Jensen B, Zechel DL, Jochimsen B. 2014. Utilization of glyphosate as phosphate source: biochemistry and genetics of bacterial carbon-phosphorus lyase. Microbiol Mol Biol Rev 78:176–197. doi: 10.1128/MMBR.00040-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Metcalf WW, Wanner BL. 1993. Mutational analysis of an Escherichia coli fourteen-gene operon for phosphonate degradation, using TnphoA′ elements. J Bacteriol 175:3430–3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hove-Jensen B. 1983. Chromosomal location of the gene encoding phosphoribosylpyrophosphate synthetase in Escherichia coli. J Bacteriol 154:177–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hove-Jensen B. 1988. Mutation in the phosphoribosylpyrophosphate synthetase gene (prs) that results in simultaneous requirements for purine and pyrimidine nucleosides, nicotinamide nucleotide, histidine, and tryptophan in Escherichia coli. J Bacteriol 170:1148–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kadziola A, Jepsen CH, Johansson E, McGuire J, Larsen S, Hove-Jensen B. 2005. Novel class III phosphoribosyl diphosphate synthase: structure and properties of the tetrameric, phosphate-activated, non-allosterically inhibited enzyme from Methanocaldococcus jannaschii. J Mol Biol 354:815–828. doi: 10.1016/j.jmb.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 72.Nishitani Y, Aono R, Nakamura A, Sato T, Atomi H, Imanaka T, Miki K. 2013. Structure analysis of archaeal AMP phosphorylase reveals two unique modes of dimerization. J Mol Biol 425:2709–2721. doi: 10.1016/j.jmb.2013.04.026. [DOI] [PubMed] [Google Scholar]
- 73.Xie CH, Yokota A. 2005. Reclassification of Alcaligenes latus strains IAM 12599T and IAM 12664 and Pseudomonas saccharophila as Azohydromonas lata gen. nov., comb. nov., Azohydromonas australica sp. nov. and Pelomonas saccharophila gen. nov., comb. nov., respectively. Int J Syst Evol Microbiol 55:2419–2425. doi: 10.1099/ijs.0.63733-0. [DOI] [PubMed] [Google Scholar]
- 74.Gomila M, Bowien B, Falsen E, Moore ER, Lalucat J. 2007. Description of Pelomonas aquatica sp. nov. and Pelomonas puraquae sp. nov., isolated from industrial and haemodialysis water. Int J Syst Evol Microbiol 57:2629–2635. doi: 10.1099/ijs.0.65149-0. [DOI] [PubMed] [Google Scholar]
- 75.Podzelinska K, He SM, Wathier M, Yakunin A, Proudfoot M, Hove-Jensen B, Zechel DL, Jia Z. 2009. Structure of PhnP, a phosphodiesterase of the carbon-phosphorus lyase pathway for phosphonate degradation. J Biol Chem 284:17216–17226. doi: 10.1074/jbc.M808392200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.He SM, Wathier M, Podzelinska K, Wong M, McSorley FR, Asfaw A, Hove-Jensen B, Jia ZC, Zechel DL. 2011. Structure and mechanism of PhnP, a phosphodiesterase of the carbon-phosphorus lyase pathway. Biochemistry 50:8603–8615. doi: 10.1021/bi2005398. [DOI] [PubMed] [Google Scholar]
- 77.Hove-Jensen B, Rosenkrantz TJ, Zechel DL, Willemoes M. 2010. Accumulation of intermediates of the carbon-phosphorus lyase pathway for phosphonate degradation in phn mutants of Escherichia coli. J Bacteriol 192:370–374. doi: 10.1128/JB.01131-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hove-Jensen B, Nygaard P. 1982. Phosphoribosylpyrophosphate synthetase of Escherichia coli, identification of a mutant enzyme. Eur J Biochem 126:327–332. [DOI] [PubMed] [Google Scholar]
- 79.Horsman GP, Zechel DL. 2017. Phosphonate biochemistry. Chem Rev 117:5704–5783. doi: 10.1021/acs.chemrev.6b00536. [DOI] [PubMed] [Google Scholar]
- 80.Manav MC, Sofos N, Hove-Jensen B, Brodersen DE. 9 September 2018. The Abc of phosphonate breakdown: a mechanism for bacterial survival. Bioessays doi: 10.1002/bies.201800091. [DOI] [PubMed] [Google Scholar]
- 81.Kawaguchi T, Veech RL, Uyeda K. 2001. Regulation of energy metabolism in macrophages during hypoxia. Roles of fructose 2,6-bisphosphate and ribose 1,5-bisphosphate. J Biol Chem 276:28554–28561. doi: 10.1074/jbc.M101396200. [DOI] [PubMed] [Google Scholar]
- 82.Bessman MJ, Frick DN, O’Handley SF. 1996. The MutT proteins or “Nudix” hydrolases, a family of versatile, widely distributed, “housecleaning” enzymes. J Biol Chem 271:25059–25062. [DOI] [PubMed] [Google Scholar]
- 83.Srouji JR, Xu A, Park A, Kirsch JF, Brenner SE. 2017. The evolution of function within the Nudix homology clan. Proteins 85:775–811. doi: 10.1002/prot.25223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sheikh S, O’Handley SF, Dunn CA, Bessman MJ. 1998. Identification and characterization of the Nudix hydrolase from the archaeon, Methanococcus jannaschii, as a highly specific ADP-ribose pyrophosphatase. J Biol Chem 273:20924–20928. [DOI] [PubMed] [Google Scholar]
- 85.Hachisuka SI, Sato T, Atomi H. 5 September 2017. Metabolism dealing with thermal degradation of NAD+ in the hyperthermophilic archaeon Thermococcus kodakarensis. J Bacteriol doi: 10.1128/JB.00162-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.McLennan AG. 2006. The Nudix hydrolase superfamily. Cell Mol Life Sci 63:123–143. doi: 10.1007/s00018-005-5386-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mildvan AS, Xia Z, Azurmendi HF, Saraswat V, Legler PM, Massiah MA, Gabelli SB, Bianchet MA, Kang LW, Amzel LM. 2005. Structures and mechanisms of Nudix hydrolases. Arch Biochem Biophys 433:129–143. doi: 10.1016/j.abb.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 88.Rose IA, Warms JV, Kaklij G. 1975. A specific enzyme for glucose 1,6-bisphosphate synthesis. J Biol Chem 250:3466–3470. [PubMed] [Google Scholar]
- 89.Ueda M, Hirose M, Sasaki R, Chiba H. 1978. Regulation of glucose 1,6-bisphosphate level in liver. III. Purification and properties of bovine glucose 1,6-bisphosphate synthase. J Biochem 83:1721–1730. [DOI] [PubMed] [Google Scholar]
- 90.Guha SK, Rose ZB. 1986. The enzymic synthesis of ribose-1,5-bisphosphate: studies of its role in metabolism. Arch Biochem Biophys 250:513–518. [DOI] [PubMed] [Google Scholar]
- 91.Wong LJ, Rose IA. 1976. Kinetic competence of a phosphoryl enzyme intermediate in the glucose-1,6-P2 synthase-catalyzed reaction. Purification, properties, and kinetic studies. J Biol Chem 251:5431–5439. [PubMed] [Google Scholar]
- 92.Maliekal P, Sokolova T, Vertommen D, Veiga-da-Cunha M, Van Schaftingen E. 2007. Molecular identification of mammalian phosphopentomutase and glucose-1,6-bisphosphate synthase, two members of the α-D-phosphohexomutase family. J Biol Chem 282:31844–31851. doi: 10.1074/jbc.M706818200. [DOI] [PubMed] [Google Scholar]
- 93.Nakamura A, Fujihashi M, Aono R, Sato T, Nishiba Y, Yoshida S, Yano A, Atomi H, Imanaka T, Miki K. 2012. Dynamic, ligand-dependent conformational change triggers reaction of ribose-1,5-bisphosphate isomerase from Thermococcus kodakarensis KOD1. J Biol Chem 287:20784–20796. doi: 10.1074/jbc.M112.349423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gogoi P, Kanaujia SP. 2018. A presumed homologue of the regulatory subunits of eIF2B functions as ribose-1,5-bisphosphate isomerase in Pyrococcus horikoshii OT3. Sci Rep 8:1891. doi: 10.1038/s41598-018-20418-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gogoi P, Srivastava A, Jayaprakash P, Jeyakanthan J, Kanaujia SP. 2016. In silico analysis suggests that PH0702 and PH0208 encode for methylthioribose-1-phosphate isomerase and ribose-1,5-bisphosphate isomerase, respectively, rather than aIF2Bβ and aIF2Bδ. Gene 575:118–126. doi: 10.1016/j.gene.2015.08.048. [DOI] [PubMed] [Google Scholar]
- 96.Kakuta Y, Tahara M, Maetani S, Yao M, Tanaka I, Kimura M. 2004. Crystal structure of the regulatory subunit of archaeal initiation factor 2B (aIF2B) from hyperthermophilic archaeon Pyrococcus horikoshii OT3: a proposed structure of the regulatory subcomplex of eukaryotic IF2B. Biochem Biophys Res Commun 319:725–732. doi: 10.1016/j.bbrc.2004.05.045. [DOI] [PubMed] [Google Scholar]
- 97.Kyrpides NC, Woese CR. 1998. Archaeal translation initiation revisited: the initiation factor 2 and eukaryotic initiation factor 2B α-β-δ subunit families. Proc Natl Acad Sci U S A 95:3726–3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kashiwagi K, Ito T, Yokoyama S. 2017. Crystal structure of eIF2B and insights into eIF2-eIF2B interactions. FEBS J 284:868–874. doi: 10.1111/febs.13896. [DOI] [PubMed] [Google Scholar]
- 99.Schmitt E, Naveau M, Mechulam Y. 2010. Eukaryotic and archaeal translation initiation factor 2: a heterotrimeric tRNA carrier. FEBS Lett 584:405–412. doi: 10.1016/j.febslet.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 100.Sekowska A, Danchin A. 2002. The methionine salvage pathway in Bacillus subtilis. BMC Microbiol 2:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Saito Y, Ashida H, Kojima C, Tamura H, Matsumura H, Kai Y, Yokota A. 2007. Enzymatic characterization of 5-methylthioribose 1-phosphate isomerase from Bacillus subtilis. Biosci Biotechnol Biochem 71:2021–2028. [DOI] [PubMed] [Google Scholar]
- 102.Tamura H, Saito Y, Ashida H, Inoue T, Kai Y, Yokota A, Matsumura H. 2008. Crystal structure of 5-methylthioribose 1-phosphate isomerase product complex from Bacillus subtilis: implications for catalytic mechanism. Protein Sci 17:126–135. doi: 10.1110/ps.073169008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kuhle B, Eulig NK, Ficner R. 2015. Architecture of the eIF2B regulatory subcomplex and its implications for the regulation of guanine nucleotide exchange on eIF2. Nucleic Acids Res 43:9994–10014. doi: 10.1093/nar/gkv930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tabita FR, Hanson TE, Li H, Satagopan S, Singh J, Chan S. 2007. Function, structure, and evolution of the RubisCO-like proteins and their RubisCO homologs. Microbiol Mol Biol Rev 71:576–599. doi: 10.1128/MMBR.00015-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tabita FR, Hanson TE, Satagopan S, Witte BH, Kreel NE. 2008. Phylogenetic and evolutionary relationships of RubisCO and the RubisCO-like proteins and the functional lessons provided by diverse molecular forms. Philos Trans R Soc Lond B Biol Sci 363:2629–2640. doi: 10.1098/rstb.2008.0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Andersson I, Backlund A. 2008. Structure and function of Rubisco. Plant Physiol Biochem 46:275–291. doi: 10.1016/j.plaphy.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 107.Andersson I, Taylor TC. 2003. Structural framework for catalysis and regulation in ribulose-1,5-bisphosphate carboxylase/oxygenase. Arch Biochem Biophys 414:130–140. [DOI] [PubMed] [Google Scholar]
- 108.Erb TJ, Zarzycki J. 2017. A short history of RubisCO: the rise and fall (?) of nature’s predominant CO2 fixing enzyme. Curr Opin Biotechnol 49:100–107. doi: 10.1016/j.copbio.2017.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Watson GM, Tabita FR. 1997. Microbial ribulose 1,5-bisphosphate carboxylase/oxygenase: a molecule for phylogenetic and enzymological investigation. FEMS Microbiol Lett 146:13–22. [DOI] [PubMed] [Google Scholar]
- 110.Ezaki S, Maeda N, Kishimoto T, Atomi H, Imanaka T. 1999. Presence of a structurally novel type ribulose-bisphosphate carboxylase/oxygenase in the hyperthermophilic archaeon, Pyrococcus kodakaraensis KOD1. J Biol Chem 274:5078–5082. [DOI] [PubMed] [Google Scholar]
- 111.Kitano K, Maeda N, Fukui T, Atomi H, Imanaka T, Miki K. 2001. Crystal structure of a novel-type archaeal rubisco with pentagonal symmetry. Structure 9:473–481. [DOI] [PubMed] [Google Scholar]
- 112.Watson GM, Yu JP, Tabita FR. 1999. Unusual ribulose 1,5-bisphosphate carboxylase/oxygenase of anoxic Archaea. J Bacteriol 181:1569–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tabita FR. 1999. Microbial ribulose 1,5-bisphosphate carboxylase/oxygenase: a different perspective. Photosynth Res 60:1–28. doi: 10.1023/A:1006211417981. [DOI] [Google Scholar]
- 114.Mueller-Cajar O, Badger MR. 2007. New roads lead to Rubisco in archaebacteria. Bioessays 29:722–724. doi: 10.1002/bies.20616. [DOI] [PubMed] [Google Scholar]
- 115.Jensen KF, Hansen MR, Jensen KS, Christoffersen S, Poulsen JC, Mølgaard A, Kadziola A. 2015. Adenine phosphoribosyltransferase from Sulfolobus solfataricus is an enzyme with unusual kinetic properties and a crystal structure that suggests it evolved from a 6-oxopurine phosphoribosyltransferase. Biochemistry 54:2323–2334. doi: 10.1021/bi501334m. [DOI] [PubMed] [Google Scholar]
- 116.Hove-Jensen B, Harlow KW, King CJ, Switzer RL. 1986. Phosphoribosylpyrophosphate synthetase of Escherichia coli. Properties of the purified enzyme and primary structure of the prs gene. J Biol Chem 261:6765–6771. [PubMed] [Google Scholar]
- 117.Hershey HV, Taylor MW. 1986. Nucleotide sequence and deduced amino acid sequence of Escherichia coli adenine phosphoribosyltransferase and comparison with other analogous enzymes. Gene 43:287–293. [DOI] [PubMed] [Google Scholar]
- 118.Pugmire MJ, Ealick SE. 2002. Structural analyses reveal two distinct families of nucleoside phosphorylases. Biochem J 361:1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mushegian AR, Koonin EV. 1994. Unexpected sequence similarity between nucleosidases and phosphoribosyltransferases of different specificity. Protein Sci 3:1081–1088. doi: 10.1002/pro.5560030711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhang Y, Cottet SE, Ealick SE. 2004. Structure of Escherichia coli AMP nucleosidase reveals similarity to nucleoside phosphorylases. Structure 12:1383–1394. doi: 10.1016/j.str.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 121.Ashihara H. 2016. Biosynthesis of 5-phosphoribosyl-1-pyrophosphate in plants: a review. Eur Chem Bull 5:314–323. [Google Scholar]
- 122.Schramm VL, Grubmeyer C. 2004. Phosphoribosyltransferase mechanisms and roles in nucleic acid metabolism. Prog Nucleic Acid Res Mol Biol 78:261–304. doi: 10.1016/S0079-6603(04)78007-1. [DOI] [PubMed] [Google Scholar]
- 123.Turnbough CL Jr, Switzer RL. 2008. Regulation of pyrimidine biosynthetic gene expression in bacteria: repression without repressors. Microbiol Mol Biol Rev 72:266–300. doi: 10.1128/MMBR.00001-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hammer-Jespersen K, Munch-Petersen A. 1970. Phosphodeoxyribomutase from Escherichia coli. Purification and some properties. Eur J Biochem 17:397–407. [DOI] [PubMed] [Google Scholar]
- 125.Leer JC, Hammer-Jespersen K. 1975. Multiple forms of phosphodeoxyribomutase from Escherichia coli. Physical and chemical characterization. Biochemistry 14:599–607. [DOI] [PubMed] [Google Scholar]
- 126.Ogushi S, Lawson JW, Dobson GP, Veech RL, Uyeda K. 1990. A new transient activator of phosphofructokinase during initiation of rapid glycolysis in brain. J Biol Chem 265:10943–10949. [PubMed] [Google Scholar]
- 127.Ishikawa E, Ogushi S, Ishikawa T, Uyeda K. 1990. Activation of mammalian phosphofructokinases by ribose 1,5-bisphosphate. J Biol Chem 265:18875–18878. [PubMed] [Google Scholar]
- 128.Derevianko A, Graeber T, D’Amico R, Simms H. 1996. Altered oxygen tension modulates cytokine-induced signal transduction in polymorphonuclear leukocytes: regulation of the GPLC pathway. J Surg Res 62:32–40. doi: 10.1006/jsre.1996.0169. [DOI] [PubMed] [Google Scholar]
- 129.Ozaki I, Mitsui Y, Sugiya H, Furuyama S. 2000. Ribose 1,5-bisphosphate inhibits fructose-1,6-bisphosphatase in rat kidney cortex. Comp Biochem Physiol B Biochem Mol Biol 125:97–102. [DOI] [PubMed] [Google Scholar]
- 130.Ozeki T, Mitsui Y, Sugiya H, Furuyama S. 1999. Ribose 1,5-bisphosphate regulates rat kidney cortex phosphofructokinase. Comp Biochem Physiol B Biochem Mol Biol 124:327–332. [DOI] [PubMed] [Google Scholar]
- 131.Sawada M, Mitsui Y, Sugiya H, Furuyama S. 2000. Ribose 1,5-bisphosphate is a putative regulator of fructose 6-phosphate/fructose 1,6-bisphosphate cycle in liver. Int J Biochem Cell Biol 32:447–454. [DOI] [PubMed] [Google Scholar]
- 132.Rose IA, Warms JV. 1974. Glucose- and mannose-1,6-P2 as activators of phosphofructokinase in red blood cells. Biochem Biophys Res Commun 59:1333–1340. [DOI] [PubMed] [Google Scholar]
- 133.Han CH, Rose ZB. 1979. Active site phosphohistidine peptides from red cell bisphosphoglycerate synthase and yeast phosphoglycerate mutase. J Biol Chem 254:8836–8840. [PubMed] [Google Scholar]
- 134.Panosian TD, Nannemann DP, Watkins GR, Phelan VV, McDonald WH, Wadzinski BE, Bachmann BO, Iverson TM. 2011. Bacillus cereus phosphopentomutase is an alkaline phosphatase family member that exhibits an altered entry point into the catalytic cycle. J Biol Chem 286:8043–8054. doi: 10.1074/jbc.M110.201350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Shackelford GS, Regni CA, Beamer LJ. 2004. Evolutionary trace analysis of the α-D-phosphohexomutase superfamily. Protein Sci 13:2130–2138. doi: 10.1110/ps.04801104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ray WJ Jr, Puvathingal JM, Liu YW. 1991. Formation of substrate and transition-state analogue complexes in crystals of phosphoglucomutase after removing the crystallization salt. Biochemistry 30:6875–6885. [DOI] [PubMed] [Google Scholar]
- 137.Jedrzejas MJ, Chander M, Setlow P, Krishnasamy G. 2000. Mechanism of catalysis of the cofactor-independent phosphoglycerate mutase from Bacillus stearothermophilus. Crystal structure of the complex with 2-phosphoglycerate. J Biol Chem 275:23146–23153. doi: 10.1074/jbc.M002544200. [DOI] [PubMed] [Google Scholar]
- 138.Kato N, Yurimoto H, Thauer RK. 2006. The physiological role of the ribulose monophosphate pathway in bacteria and archaea. Biosci Biotechnol Biochem 70:10–21. doi: 10.1271/bbb.70.10. [DOI] [PubMed] [Google Scholar]
- 139.Orita I, Sato T, Yurimoto H, Kato N, Atomi H, Imanaka T, Sakai Y. 2006. The ribulose monophosphate pathway substitutes for the missing pentose phosphate pathway in the archaeon Thermococcus kodakaraensis. J Bacteriol 188:4698–4704. doi: 10.1128/JB.00492-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng JF, Darling A, Malfatti S, Swan BK, Gies EA, Dodsworth JA, Hedlund BP, Tsiamis G, Sievert SM, Liu WT, Eisen JA, Hallam SJ, Kyrpides NC, Stepanauskas R, Rubin EM, Hugenholtz P, Woyke T. 2013. Insights into the phylogeny and coding potential of microbial dark matter. Nature 499:431–437. doi: 10.1038/nature12352. [DOI] [PubMed] [Google Scholar]
- 141.Castelle CJ, Wrighton KC, Thomas BC, Hug LA, Brown CT, Wilkins MJ, Frischkorn KR, Tringe SG, Singh A, Markillie LM, Taylor RC, Williams KH, Banfield JF. 2015. Genomic expansion of domain Archaea highlights roles for organisms from new phyla in anaerobic carbon cycling. Curr Biol 25:690–701. doi: 10.1016/j.cub.2015.01.014. [DOI] [PubMed] [Google Scholar]
- 142.Solden L, Lloyd K, Wrighton K. 2016. The bright side of microbial dark matter: lessons learned from the uncultivated majority. Curr Opin Microbiol 31:217–226. doi: 10.1016/j.mib.2016.04.020. [DOI] [PubMed] [Google Scholar]
- 143.Mwirichia R, Alam I, Rashid M, Vinu M, Ba-Alawi W, Kamau AA, Kamanda Ngugi D, Goker M, Klenk HP, Bajic V, Stingl U. 2016. Metabolic traits of an uncultured archaeal lineage—MSBL1—from brine pools of the Red Sea. Sci Rep 6:19181. doi: 10.1038/srep19181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hug LA, Baker BJ, Anantharaman K, Brown CT, Probst AJ, Castelle CJ, Butterfield CN, Hernsdorf AW, Amano Y, Ise K, Suzuki Y, Dudek N, Relman DA, Finstad KM, Amundson R, Thomas BC, Banfield JF. 2016. A new view of the tree of life. Nat Microbiol 1:16048. doi: 10.1038/nmicrobiol.2016.48. [DOI] [PubMed] [Google Scholar]
- 145.Wheelis ML, Kandler O, Woese CR. 1992. On the nature of global classification. Proc Natl Acad Sci U S A 89:2930–2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Davis JP, Youssef NH, Elshahed MS. 2009. Assessment of the diversity, abundance, and ecological distribution of members of candidate division SR1 reveals a high level of phylogenetic diversity but limited morphotypic diversity. Appl Environ Microbiol 75:4139–4148. doi: 10.1128/AEM.00137-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Harris JK, Kelley ST, Pace NR. 2004. New perspective on uncultured bacterial phylogenetic division OP11. Appl Environ Microbiol 70:845–849. doi: 10.1128/AEM.70.2.845-849.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Campbell JH, O’Donoghue P, Campbell AG, Schwientek P, Sczyrba A, Woyke T, Söll D, Podar M. 2013. UGA is an additional glycine codon in uncultured SR1 bacteria from the human microbiota. Proc Natl Acad Sci U S A 110:5540–5545. doi: 10.1073/pnas.1303090110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kantor RS, Wrighton KC, Handley KM, Sharon I, Hug LA, Castelle CJ, Thomas BC, Banfield JF. 2013. Small genomes and sparse metabolisms of sediment-associated bacteria from four candidate phyla. mBio 4:e00708-13. doi: 10.1128/mBio.00708-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Alonso H, Blayney MJ, Beck JL, Whitney SM. 2009. Substrate-induced assembly of Methanococcoides burtonii D-ribulose-1,5-bisphosphate carboxylase/oxygenase dimers into decamers. J Biol Chem 284:33876–33882. doi: 10.1074/jbc.M109.050989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wrighton KC, Castelle CJ, Varaljay VA, Satagopan S, Brown CT, Wilkins MJ, Thomas BC, Sharon I, Williams KH, Tabita FR, Banfield JF. 2016. RubisCO of a nucleoside pathway known from Archaea is found in diverse uncultivated phyla in bacteria. ISME J 10:2702–2714. doi: 10.1038/ismej.2016.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wrighton KC, Thomas BC, Sharon I, Miller CS, Castelle CJ, VerBerkmoes NC, Wilkins MJ, Hettich RL, Lipton MS, Williams KH, Long PE, Banfield JF. 2012. Fermentation, hydrogen, and sulfur metabolism in multiple uncultivated bacterial phyla. Science 337:1661–1665. doi: 10.1126/science.1224041. [DOI] [PubMed] [Google Scholar]
- 153.Brown CT, Hug LA, Thomas BC, Sharon I, Castelle CJ, Singh A, Wilkins MJ, Wrighton KC, Williams KH, Banfield JF. 2015. Unusual biology across a group comprising more than 15% of domain Bacteria. Nature 523:208–211. doi: 10.1038/nature14486. [DOI] [PubMed] [Google Scholar]
- 154.Finn MW, Tabita FR. 2003. Synthesis of catalytically active form III ribulose 1,5-bisphosphate carboxylase/oxygenase in archaea. J Bacteriol 185:3049–3059. doi: 10.1128/JB.185.10.3049-3059.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Stokke R, Hocking WP, Steinsbu BO, Steen IH. 2013. Complete genome sequence of the thermophilic and facultatively chemolithoautotrophic sulfate reducer Archaeoglobus sulfaticallidus strain PM70-1T. Genome Announc 1:e00406-13. doi: 10.1128/genomeA.00406-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Klenk HP, Clayton RA, Tomb JF, White O, Nelson KE, Ketchum KA, Dodson RJ, Gwinn M, Hickey EK, Peterson JD, Richardson DL, Kerlavage AR, Graham DE, Kyrpides NC, Fleischmann RD, Quackenbush J, Lee NH, Sutton GG, Gill S, Kirkness EF, Dougherty BA, McKenney K, Adams MD, Loftus B, Peterson S, Reich CI, McNeil LK, Badger JH, Glodek A, Zhou L, Overbeek R, Gocayne JD, Weidman JF, McDonald L, Utterback T, Cotton MD, Spriggs T, Artiach P, Kaine BP, Sykes SM, Sadow PW, D’Andrea KP, Bowman C, Fujii C, Garland SA, Mason TM, Olsen GJ, Fraser CM, et al. 1997. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature 390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 157.Schönheit P, Buckel W, Martin WF. 2016. On the origin of heterotrophy. Trends Microbiol 24:12–25. doi: 10.1016/j.tim.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 158.Eme L, Spang A, Lombard J, Stairs CW, Ettema TJG. 2017. Archaea and the origin of eukaryotes. Nat Rev Microbiol 15:711–723. doi: 10.1038/nrmicro.2017.133. [DOI] [PubMed] [Google Scholar]
- 159.Lee CE, Goodfellow C, Javid-Majd F, Baker EN, Lott JS. 2006. The crystal structure of TrpD, a metabolic enzyme essential for lung colonization by Mycobacterium tuberculosis, in complex with its substrate phosphoribosylpyrophosphate. J Mol Biol 355:784–797. doi: 10.1016/j.jmb.2005.11.016. [DOI] [PubMed] [Google Scholar]
- 160.Borrel G, Harris HM, Tottey W, Mihajlovski A, Parisot N, Peyretaillade E, Peyret P, Gribaldo S, O’Toole PW, Brugere JF. 2012. Genome sequence of “Candidatus Methanomethylophilus alvus” Mx1201, a methanogenic archaeon from the human gut belonging to a seventh order of methanogens. J Bacteriol 194:6944–6945. doi: 10.1128/JB.01867-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Gorlas A, Robert C, Gimenez G, Drancourt M, Raoult D. 2012. Complete genome sequence of Methanomassiliicoccus luminyensis, the largest genome of a human-associated Archaea species. J Bacteriol 194:4745. doi: 10.1128/JB.00956-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Borrel G, Harris HM, Parisot N, Gaci N, Tottey W, Mihajlovski A, Deane J, Gribaldo S, Bardot O, Peyretaillade E, Peyret P, O’Toole PW, Brugere JF. 2013. Genome sequence of “Candidatus Methanomassiliicoccus intestinalis” Issoire-Mx1, a third Thermoplasmatales-related methanogenic archaeon from human feces. Genome Announc 1:e00453-13. doi: 10.1128/genomeA.00453-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Vanwonterghem I, Evans PN, Parks DH, Jensen PD, Woodcroft BJ, Hugenholtz P, Tyson GW. 2016. Methylotrophic methanogenesis discovered in the archaeal phylum Verstraetearchaeota. Nat Microbiol 1:16170. doi: 10.1038/nmicrobiol.2016.170. [DOI] [PubMed] [Google Scholar]
- 164.Zaremba-Niedzwiedzka K, Caceres EF, Saw JH, Backstrom D, Juzokaite L, Vancaester E, Seitz KW, Anantharaman K, Starnawski P, Kjeldsen KU, Stott MB, Nunoura T, Banfield JF, Schramm A, Baker BJ, Spang A, Ettema TJ. 2017. Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature 541:353–358. doi: 10.1038/nature21031. [DOI] [PubMed] [Google Scholar]
- 165.Malfatti S, Tindall BJ, Schneider S, Fahnrich R, Lapidus A, Labuttii K, Copeland A, Glavina Del Rio T, Nolan M, Chen F, Lucas S, Tice H, Cheng JF, Bruce D, Goodwin L, Pitluck S, Anderson I, Pati A, Ivanova N, Mavromatis K, Chen A, Palaniappan K, D’Haeseleer P, Goker M, Bristow J, Eisen JA, Markowitz V, Hugenholtz P, Kyrpides NC, Klenk HP, Chain P. 2009. Complete genome sequence of Halogeometricum borinquense type strain (PR3T). Stand Genomic Sci 1:150–159. doi: 10.4056/sigs.23264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Kim KK, Lee KC, Lee JS. 2012. Draft genome sequence of the extremely halophilic archaeon Halogranum salarium B-1T. J Bacteriol 194:6659. doi: 10.1128/JB.01815-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Hartman AL, Norais C, Badger JH, Delmas S, Haldenby S, Madupu R, Robinson J, Khouri H, Ren Q, Lowe TM, Maupin-Furlow J, Pohlschroder M, Daniels C, Pfeiffer F, Allers T, Eisen JA. 2010. The complete genome sequence of Haloferax volcanii DS2, a model archaeon. PLoS One 5:e9605. doi: 10.1371/journal.pone.0009605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Becker EA, Seitzer PM, Tritt A, Larsen D, Krusor M, Yao AI, Wu D, Madern D, Eisen JA, Darling AE, Facciotti MT. 2014. Phylogenetically driven sequencing of extremely halophilic archaea reveals strategies for static and dynamic osmo-response. PLoS Genet 10:e1004784. doi: 10.1371/journal.pgen.1004784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Cadillo-Quiroz H, Browne P, Kyrpides N, Woyke T, Goodwin L, Detter C, Yavitt JB, Zinder SH. 2015. Complete genome sequence of Methanosphaerula palustris E1-9CT, a hydrogenotrophic methanogen isolated from a minerotrophic fen peatland. Genome Announc 3:e01280-15. doi: 10.1128/genomeA.01280-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Narihiro T, Kusada H, Yoneda Y, Tamaki H. 2016. Draft genome sequences of Methanoculleus horonobensis strain JCM 15517, Methanoculleus thermophilus strain DSM 2373, and Methanofollis ethanolicus strain JCM 15103, hydrogenotrophic methanogens belonging to the family Methanomicrobiaceae. Genome Announc 4:e00199-16. doi: 10.1128/genomeA.00199-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Anderson IJ, Sieprawska-Lupa M, Goltsman E, Lapidus A, Copeland A, Glavina Del Rio T, Tice H, Dalin E, Barry K, Pitluck S, Hauser L, Land M, Lucas S, Richardson P, Whitman WB, Kyrpides NC. 2009. Complete genome sequence of Methanocorpusculum labreanum type strain Z. Stand Genomic Sci 1:197–203. doi: 10.4056/sigs.35575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Brügger K, Chen L, Stark M, Zibat A, Redder P, Ruepp A, Awayez M, She Q, Garrett RA, Klenk HP. 2007. The genome of Hyperthermus butylicus: a sulfur-reducing, peptide fermenting, neutrophilic crenarchaeote growing up to 108 degrees C. Archaea 2:127–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Evans PN, Parks DH, Chadwick GL, Robbins SJ, Orphan VJ, Golding SD, Tyson GW. 2015. Methane metabolism in the archaeal phylum Bathyarchaeota revealed by genome-centric metagenomics. Science 350:434–438. doi: 10.1126/science.aac7745. [DOI] [PubMed] [Google Scholar]
- 174.Anantharaman K, Brown CT, Hug LA, Sharon I, Castelle CJ, Probst AJ, Thomas BC, Singh A, Wilkins MJ, Karaoz U, Brodie EL, Williams KH, Hubbard SS, Banfield JF. 2016. Thousands of microbial genomes shed light on interconnected biogeochemical processes in an aquifer system. Nat Commun 7:13219. doi: 10.1038/ncomms13219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Lazar CS, Baker BJ, Seitz K, Hyde AS, Dick GJ, Hinrichs KU, Teske AP. 2016. Genomic evidence for distinct carbon substrate preferences and ecological niches of Bathyarchaeota in estuarine sediments. Environ Microbiol 18:1200–1211. doi: 10.1111/1462-2920.13142. [DOI] [PubMed] [Google Scholar]
- 176.Brambilla E, Djao OD, Daligault H, Lapidus A, Lucas S, Hammon N, Nolan M, Tice H, Cheng JF, Han C, Tapia R, Goodwin L, Pitluck S, Liolios K, Ivanova N, Mavromatis K, Mikhailova N, Pati A, Chen A, Palaniappan K, Land M, Hauser L, Chang YJ, Jeffries CD, Rohde M, Spring S, Sikorski J, Goker M, Woyke T, Bristow J, Eisen JA, Markowitz V, Hugenholtz P, Kyrpides NC, Klenk HP. 2010. Complete genome sequence of Methanoplanus petrolearius type strain (SEBR 4847T). Stand Genomic Sci 3:203–211. doi: 10.4056/sigs.1183143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Spring S, Rachel R, Lapidus A, Davenport K, Tice H, Copeland A, Cheng JF, Lucas S, Chen F, Nolan M, Bruce D, Goodwin L, Pitluck S, Ivanova N, Mavromatis K, Ovchinnikova G, Pati A, Chen A, Palaniappan K, Land M, Hauser L, Chang YJ, Jeffries CC, Brettin T, Detter JC, Tapia R, Han C, Heimerl T, Weikl F, Brambilla E, Goker M, Bristow J, Eisen JA, Markowitz V, Hugenholtz P, Kyrpides NC, Klenk HP. 2010. Complete genome sequence of Thermosphaera aggregans type strain (M11TLT). Stand Genomic Sci 2:245–259. doi: 10.4056/sigs.821804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Ravin NV, Mardanov AV, Beletsky AV, Kublanov IV, Kolganova TV, Lebedinsky AV, Chernyh NA, Bonch-Osmolovskaya EA, Skryabin KG. 2009. Complete genome sequence of the anaerobic, protein-degrading hyperthermophilic crenarchaeon Desulfurococcus kamchatkensis. J Bacteriol 191:2371–2379. doi: 10.1128/JB.01525-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Dominova IN, Kublanov IV, Podosokorskaya OA, Derbikova KS, Patrushev MV, Toshchakov SV. 2013. Complete genomic sequence of “Thermofilum adornatus” strain 1910bT, a hyperthermophilic anaerobic organotrophic crenarchaeon. Genome Announc 1:e00726-13. doi: 10.1128/genomeA.00726-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Flaks JG, Erwin MJ, Buchanan JM. 1957. Biosynthesis of the purines. XVI. The synthesis of adenosine 5′-phosphate and 5-amino-4-imidazolecarboxamide ribotide by a nucleotide pyrophosphorylase. J Biol Chem 228:201–213. [PubMed] [Google Scholar]
- 181.Hurwitz J, Heppel LA, Horecker BL. 1957. The enzymatic cleavage of adenylic acid to adenine and ribose 5-phosphate. J Biol Chem 226:525–540. [PubMed] [Google Scholar]
- 182.Trackman PC, Abeles RH. 1983. Methionine synthesis from 5′-S-methylthioadenosine. Resolution of enzyme activities and identification of 1-phospho-5-S methylthioribulose. J Biol Chem 258:6717–6720. [PubMed] [Google Scholar]
- 183.Hurwitz J, Weissbach A, Horecker BL, Smyrniotis PZ. 1956. Spinach phosphoribulokinase. J Biol Chem 218:769–783. [PubMed] [Google Scholar]
- 184.Taylor TC, Andersson I. 1997. The structure of the complex between rubisco and its natural substrate ribulose 1,5-bisphosphate. J Mol Biol 265:432–444. doi: 10.1006/jmbi.1996.0738. [DOI] [PubMed] [Google Scholar]
- 185.Lundqvist T, Schneider G. 1991. Crystal structure of activated ribulose-1,5-bisphosphate carboxylase complexed with its substrate, ribulose-1,5-bisphosphate. J Biol Chem 266:12604–12611. [PubMed] [Google Scholar]
- 186.Andersson I, Knight S, Schneider G, Lindqvist Y, Lundqvist T, Bränden CI, Lorimer GH. 1989. Crystal structure of the active site of ribulose-bisphosphate carboxylase. Nature 337:229–234. doi: 10.1038/337229a0. [DOI] [Google Scholar]
- 187.Andersson I. 1996. Large structures at high resolution: the 1.6 A crystal structure of spinach ribulose-1,5-bisphosphate carboxylase/oxygenase complexed with 2-carboxyarabinitol bisphosphate. J Mol Biol 259:160–174. doi: 10.1006/jmbi.1996.0310. [DOI] [PubMed] [Google Scholar]
- 188.Schneider G, Lindqvist Y, Lundqvist T. 1990. Crystallographic refinement and structure of ribulose-1,5-bisphosphate carboxylase from Rhodospirillum rubrum at 1.7 A resolution. J Mol Biol 211:989–1008. doi: 10.1016/0022-2836(90)90088-4. [DOI] [PubMed] [Google Scholar]
- 189.Gunn LH, Valegard K, Andersson I. 2017. A unique structural domain in Methanococcoides burtonii ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) acts as a small subunit mimic. J Biol Chem 292:6838–6850. doi: 10.1074/jbc.M116.767145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Nishitani Y, Yoshida S, Fujihashi M, Kitagawa K, Doi T, Atomi H, Imanaka T, Miki K. 2010. Structure-based catalytic optimization of a type III Rubisco from a hyperthermophile. J Biol Chem 285:39339–39347. doi: 10.1074/jbc.M110.147587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Kreel NE, Tabita FR. 2007. Substitutions at methionine 295 of Archaeoglobus fulgidus ribulose-1,5-bisphosphate carboxylase/oxygenase affect oxygen binding and CO2/O2 specificity. J Biol Chem 282:1341–1351. doi: 10.1074/jbc.M609399200. [DOI] [PubMed] [Google Scholar]