Abstract
There is a great deal of interest in ‘personalized,’ ‘individualized,’ or ‘precision’ interventions for disease and health-risk mitigation. This is as true of nutrition-based intervention and prevention strategies as it is for pharmacotherapies and pharmaceutical-oriented prevention strategies. Essentially, technological breakthroughs have enabled researchers to probe an individual’s unique genetic, biochemical, physiological, behavioral and exposure profile, allowing them to identify very specific and often nuanced factors that an individual might possess that may make it more or less likely that he or she will respond favorably to a particular intervention (e.g., nutrient supplementation) or disease prevention strategy (e.g., specific diet). However, as compelling and intuitive as personalized nutrition might be in the current era in which data-intensive biomedical characterization of individuals is possible, appropriately and objectively vetting personalized nutrition strategies is not trivial and will require novel study designs and data analytical methods. These designs and methods must consider a very integrated use of the multiple contemporary biomedical assays and technologies that motivate them, which adds to their complexity. Single subject or ‘N-of-1’ trials can be used to assess the utility of personalized interventions and, in addition, can be crafted in such a way as to accommodate the necessary integrated use of many emerging biomedical technologies and assays. In this review we consider the motivation, design and implementation of N-of-1 trials in translational nutrition research that are meant to assess the utility of personalized nutritional strategies. We provide a number of example studies, discuss appropriate analytical methods given the complex data they will generate and require, and consider how such studies could leverage integration of various biomarker assays and clinical endpoints. Importantly, we also consider the development of strategies and ‘algorithms’ for matching nutritional needs to individual biomedical profiles and the issues surrounding them. Finally, we discuss the limitations of personalized nutrition studies, possible extensions of N-of-1 nutritional intervention studies and areas of future research.
Introduction
The belief that one can tailor interventions, including nutritional interventions, to an individual’s often nuanced and potentially unique genetic, biochemical, behavioral and exposure profile is receiving a great deal of attention. Although some unmitigated success has been observed for specific targeted and ‘individualized’ pharmacotherapies, especially those designed to treat cancers(13, 91), less success has been observed for such ‘personalized,’ ‘individualized,’ or ‘precision’ nutritional interventions. There are at least three interrelated reasons for this lack of success. First, it is likely that not enough time has elapsed since the introduction of high-throughput, data-intensive assays characterizing unique physiologic and exposure profiles (such as DNA sequencing, wireless glucose monitoring, smart-phone application driven diet diaries, etc.) for researchers to have identified definitive connections between the activities or benefits of specific nutrients, diets, and/or nutritional supplements and individual profiles, except in the context of rare, often genetically-mediated overt nutritional deficiencies.(7, 8) Second, identifying and characterizing the molecular and physiologic processes and deficiencies forming the basis for such connections is difficult and may be much more complex than in making connections between, e.g., highly-contrived pharmaceutical products and specific gene products. Third, testing or vetting the utility of a personalized dietary intervention is also non-trivial and will likely require study designs, analytical methods, and overall strategies that differ from those used in the past.
The third reason is actually the focus of this review, although we argue that studies can be designed to simultaneously assess the benefits of ‘personalized’ nutritional interventions for an individual and identify factors that solidify the connection between a specific dietary intervention and an individual’s biochemical, physiological, behavioral and exposure profile. In addition, despite the lack of a large number of success stories proving that personalized nutrition works on a large-scale, there is nothing if not motivation for testing the benefits of personalized nutrition given the availability of high-throughput assays such as DNA sequencing, proteomics, wireless monitoring, etc. and a growing number of insights into how fundamental molecular physiologic processes respond to or require specific nutrients.(55) Thus, questions surrounding how one can best prove that personalized nutritional interventions benefit individuals is of crucial importance. One set of study designs, those falling under the heading of ‘single subject’ or ‘N-of-1’ studies, are highly appropriate in that their focus is on testing whether or not an individual exhibits any evidence, in a well-designed and controlled study, that they responded to a particular intervention, but can be extended and modified in a number of important ways. In this light, we ultimately argue that in an era where ‘personalization’ is emphasized (in, e.g., medicine, nutrition, advertising, general service industries, finance, etc.), nutrition-based clinical trials need to focus on variation in responses exhibited by each participant over the course of the trial as much as variation in responses across participants in the trial. The former may help to identify factors that influence response in a participant-specific way while the latter can shed light on whether or not that factor is shared among others in a way that helps solidify its role in mediating response. Note that we use the term ‘personalized nutrition’ (as opposed to ‘individualized’ or ‘precision’ nutrition) in what follows to refer to attempts to match specific diets, nutrients, or natural-product-based supplements to an individual’s profile except in very specific instances.
The remainder of the review will be broken down into nine broad sections, each with different subsections. The first section provides a general background on the state of clinical trials in nutrition and why different trial designs are needed to advance personalized nutrition. The second considers the biological motivations for personalized nutrition and more appropriate clinical trials designs. The third section discusses basic N-of-1 trial designs and their extensions. The fourth considers how one can aggregate the results of N-of-1 trials to make broader claims about the utility of a nutritional intervention in the population at large. The fifth discusses the problem of determining what to measure in order to assess success when designing a study to test a diet on an individual. The sixth considers monitoring individuals for health status changes either in the wake of providing them an intervention or to determine their general vulnerability to disease. The seventh section considers the increasing interest in vetting or testing ‘matching strategies’ that relate specific diets to individual profiles rather than simply vetting the specific diets themselves. The eighth section focuses on a few of the more intriguing and relevant recently published studies that motivate N-of-1 trials and nutrition and how future studies like them could be modified along the lines discussed in this review. The ninth and last section provides a brief discussion of N-of-1 trials in nutrition and areas of future research.
Personalized Nutrition and Human Clinical Studies
Traditional Population-Based Clinical Trials
Most clinical trials are designed to address questions about the utility or health benefits of a drug or intervention in the population at large, and not necessarily address questions about the unequivocal health benefits for any single individual participating in the trial. In broad terms, population-based trials typically involve providing a particular intervention to a group of individuals while a comparator intervention, often a placebo, is provided to another group of individuals.(31, 36, 41, 60, 77, 90). The average benefit of the intervention across those individuals provided the intervention (e.g., average weight loss; average drop in blood pressure or average cholesterol level; etc.) is compared to the average benefit of those individuals provided a placebo. It is rare in such trials that enough data is collected on any one participant to state unequivocally that the benefit observed for that participant can be attributed to the intervention itself. Although of extreme value in the nutritional sciences (see, e.g., Table 1 for some examples of large-scale nutritional intervention studies), such studies do not often accommodate the quantification of the degree to which the subjects exhibit variation in the response among individuals within the intervention and comparator groups. Population-based clinical trials can actually be designed to explore the benefits of personalized nutritional interventions using, for example, some of the methods discussed in sections III and VI, which are meant to assess the overall benefit of personalized vs. non-personalized interventions. In general, however, traditional designs in population-based trials are not appropriate if the goal is to evaluate the utility of personalization in medicine and nutrition. This is especially important since many of the most often used interventions have been documented not to work in a large fraction of the individuals provided them in population-based studies and the reasons for this are largely unknown, but could be explored with appropriate study designs (see, e.g., the editorial by Schork(86)).
Table 1.
Example Large-Scale Nutritional Intervention Studies.
| Authors (Year) | PMID | Diet and Contrast | N | Study Design | Outcome |
|---|---|---|---|---|---|
| Guasch-Ferre (2013) | 23866098 | Mediterranean Diet (MD) with nuts, olive oil or control group | 7,216 | Randomized Control Trial | Hazard Ratio for death in group eating nuts: 0.61 (0.45 – 0.83). |
| Lippman 2009 SELECT |
19066370 | Selenium and Vitamin E versus placebo | 35,533 | Randomized control trial | No difference in Prostate cancer incidence between intervention and control group. |
| Qiao (2009) (Linxian NIT) |
19318634 | Selenium, beta-carotene, vitamin E | 29,584 | Randomized trial | Decreased overall mortality in group taking vitamins |
| Gaziano (2010) and Sesso (2008) Physician’s Health Study II |
23162860 18997197 |
Vitamin E and C versus placebo | 14,641 | Randomized placebo controlled trial | No decrease in risk of cancers or heart disease. |
| Prentice (2006) Howard (2010) Women’s Health Initiative |
16467232 16467234 |
Intervention directed reduced fat diet versus no intervention | 48,835 | Randomized control trial | No change in the incidence of breast cancer in low-fat diet group. No change in risk of stroke or coronary heart disease. |
| McCullough (2003) American Cancer Society Cancer Prevention Study II – Nutrition Cohort | 12708719 | Calcium, Vitamin D, dairy intake | 127,749 | Longitudinal Cohort Study | Intake of calcium supplements and dietary and supplemental Vitamin D inversely associated with risk for colorectal cancer. |
| Menotti (1999) Seven Countries Study |
10485342 | 18 different diets | 12,763 | Longitudinal Cohort Study | Legumes, fish and alcohol intake had negative correlation with mortality from coronary heart disease. |
| Giovannucii (1998) Nurses’ Health Study |
9758570 | Multivitamins and folate | 88,756 | Longitudinal Cohort Study | 15 years of multivitamin use and dietary folate alone associated with decreased risk of colon cancer. |
Post-Hoc Identification of Responders and Non-Responders
One practice that is pursued often in the context of large-scale population-based clinical trials involves the pursuit of post-hoc analyses that explore the relationships between different factors (i.e., covariates measured on the trial participants) and response. Despite the potential insights that could arise from such analyses they are often frowned upon unless they are pursued as a way of generating hypotheses that could be tested in a more sophisticated way in a future clinical trial.(101) Many researchers have considered using post hoc analyses involving large-scale clinical trial data to identify genetic variants or other biomarkers that may predict response.(38) However, pharmacogenetic analyses of these sorts are complicated by the fact that there are rarely studies that can be used to replicate findings – and replication is considered the sine qua non of genetic association studies.(5, 42, 50, 95) In fact, many studies have been pursued to identify genetic variants that influence response to nutrients, diets or dietary supplements (i.e., ‘nutrigenomics;’ see Table 2). Nutrigenomic findings can motivate focused N-of-1 trials, as discussed later, but also suffer from replication issues.
Table 2.
Example Nutrigenomics Studies Leveraging a Nutritional Intervention.
| Authors (Year) | PMID | Diet and Contrast | N | Study Design | Outcome |
|---|---|---|---|---|---|
| Konstantinidou (2016) | 20179144 | Traditional Mediterranean Diet (TMD) with virgin olive oil versus TMD with washed virgin olive oil versus Habitual Diet | 90 | Randomized Control Trial | TMD with olive oil resulted in down regulation of pro-atherogenic genes INFγ, IL7R, ADRB2, POLK |
| Pu (2015) (COMIT study) |
26806592 | Canola oil High oleic canola oil High oleic canola oil enriched with DHA Flax/safflower oil Corn/safflower oil |
170 | Cross-over randomized control trial | ‘A’ allele carriers at snp rs324420 in FAAH gene had significantly higher DHEA levels than CC genotype carriers, indicating a possible beneficial effect on circulating fatty acid levels. |
| Frankwich (2015) | 25769412 | ‘Nutrigenetic’ guided diet: four types – balanced, low-fat, low-carbohydrate, and Mediterranean Standard versus non-guided balanced diet |
51 | Randomized Control Trial | Participants with low-risk obesity polymorphisms lost significantly more weight. |
| Wojczynski (2015) | 26256467 | High fat diet with 83% fat, 14% carbohydrate and 3% protein | 872 | Genome Wide Association Study | 2 snps identified as significant near the APOA1/C3/A4/A5 gene cluster in lipid metabolism |
| Shab-Bisdar (2015) | 26346470 | Vit D fortified yogurt drink Plain yogurt drink |
60 | Randomized Control Trial | Carriers of the AA genotype of VDR-Cdx-2 had significant decreases in obesity indices compared with carriers of GA and GG genotypes. |
| Goni (2014) | 25870980 | Personalized nutrition diet based on genotype. Diets similar regarding total amount of protein, carbohydrate and vegetables, |
167 | Longitudinal cohort | Carriers of variant alleles in FTO, MC4R and MTNR1B had lower weight loss than non-carriers. Women carriers of variant in MTNR1B, who ate high total protein and high animal protein diets loss less weight than wildtype carriers |
| Renda (2012) | 22170367 | Caffeine consumption | 110 | Observational cohort study | Variants in the ADORA2A and ADRA2B genes associated with increased blood pressure after caffeine consumption. |
One important component of studies designed to determine if an individual has responded to a particular intervention, whether in the context of a pharmacological or nutritional intervention, is the need for internal, individual-specific controls. This can be achieved through the use of cross-over study designs where individuals are provided an intervention and then purposefully provided a comparator intervention (which could be a placebo or sham intervention) to generate an appropriate contrast. Tables 3a and 3b list many studies investigating the benefits of a nutritional intervention, some of which used a crossover design, even though they were designed as traditional population-focused studies. Although there are many issues with the design and conduct of cross-over trials(48, 89), not including a cross-over component in a trial can be highly problematic for making claims about an individual’s unique (if any) response to the intervention of interest. Essentially, in a trial without a cross-over, claims about whether or not a change in the health status of an individual can actually be attributed to the intervention of interest would be based entirely on population-based statistics comparing the health status of that individual to others in the trial. This makes claims about individual rather than group responses to interventions problematic, since any individual may exhibit equivalent responses to other interventions (including a placebo or sham intervention) which undermines confidence that the intervention is working through a unique mechanism and that it is an appropriate intervention for an individual relative to other interventions that could have been chosen.
Table 3a.
Example Nutritional Intervention Studies Focusing on Different Measures and Outcomes.
| Category | Measures | Author (Year) | PMID | N | Nutritional Component | Study Design | Outcome |
|---|---|---|---|---|---|---|---|
| Cognitive, Mental, Behavioral Health | Mood, Quality of Life, Sleep, Sexual Function | Martin (2016) | 27136347 | 218 | Caloric Restriction of 25% versus ad lib diet for 2 years | Randomized Control Trial | Caloric Restriction group had significantly improved mood, reduced tension, sleep duration, sexual drive and general health. |
| Depression Scale | Rahe (2014) | 24468939 | 16 studies | Mediterranean Diet (MD) Western Diet (WD) | Systematic Review | MD - Lower risk of depression WD - Higher risk of depression |
|
| Tiemeier (2002) | 12450964 | 3,884 | Vitamin B12 and Folate deficiency | Cross-sectional survey | Higher risk of depression | ||
| Jacka (2010) | 20048020 | 1,046 | ‘Traditional’ diet with fish, vegetables and fiber | Cross-sectional survey | Lower risk of depression | ||
| Quality of Life, Karnofsky Performance Status, Cognitive Function, Global Health Status | van der Meij (2012) | 22234041 | 40 | Supplements with Polyunsaturated fatty acids | Double-blind Randomized Control Trial | Improvements in all categories for Lung Cancer patients taking supplements versus those not taking supplements. | |
| Cognition scores | Lemaire (2010) | 20712911 | 20 | Frequent small healthy meals versus baseline diet | Cross Over Trial | Score on cognitive testing significantly improved with healthy meals. | |
| Sleep | Afaghi (2008) | 17284739 | 12 | High carbohydrate meals | Cross-over Trial | High carbohydrate meal given 4 hours versus 1 hour before sleep shortened sleep-onset latency | |
| Killer (2015) | 26406911 | 13 | High versus moderate carbohydrate diet | Cross-over trial | Sleep time was significantly higher in moderate carbohydrate group | ||
| Physiologic and Metabolic Function | Body weight, sleep and energy levels | Gill (2015) | 26411343 | 8 | Time-restricted eating | Non-randomized observational cohort study | Decreased body weight, improved sleep and energy levels when eating restricted to 10 hour time periods |
| Insulin resistance, highly sensitive C-reactive Protein (hs-CRP), adiponectin | Jennings (2014) | 24336456 | 1,997 | Flavonoid intake | Cross-sectional survey | High flavone and anthocyanin intake associated with lower peripheral insulin resistance. Higher anthocyanin associated with lower hs-CRP. Higher flavone intake associated with improved adiponectin concentrations. |
|
| Weight loss | Look AHEAD Research Group (2013) | 23796131 | 5145 | Caloric restriction with meal replacement for diabetics versus normal diet. | Randomized Control Trial | Greater weight loss in intervention group. | |
| Brehm (2003) | 12679447 | 53 | Very low carbohydrate versus low fat diet | Randomized Control Trial | Very low carbohydrate group lost more weight. | ||
| Weight loss, Fat Free Mass, Fat Mass | Gomes (2016) | 27885532 | 34 | Whey protein supplementation versus not | Randomized Control Trial | Intervention group lost more weight than control group | |
| Blood Pressure, blood glucose and lipids | Domenech (2014) | 24799608 | 235 | Mediterranean Diet, olive oil or nut supplements | Randomized Control Trial | Reduced blood pressure, blood glucose and lipids in intervention group. | |
| Non-Alcoholic Fatty Liver Disease (NAFLD) | Elias (2010) | 20022466 | 31 | Caloric restriction | Non-randomized observational cohort study | Significant improvements in liver function tests, insulin resistance, visceral fat, liver density and high density lipoprotein levels. | |
| Blood gene expression levels | Leonardson (2010) | 19837700 | 40 | Fasting state versus fed state | Cross-over trial | Significant differences in gene expression between fasted and fed states. |
Table 3b.
Example Nutritional Intervention Studies Focusing on the Gut Microbiome.
| Category | Author (Year) | PMID | N | Nutritional Component | Study Design | Outcome |
|---|---|---|---|---|---|---|
| Microbiome | Dao (2016) | 26100928 | 90 | Energy-restricted high protein diet versus Weight Maintenance diet | Cross-over trial | Participants with higher levels of Akkermansia muciniphila showed greater improvement in metabolic health parameters, e.g., decreased insulin resistance. |
| Brahe (2015) | 26134388 | 58 |
Lactobacillus paracasei versus flax seed mucilage versus placebo |
3-arm single-blind randomized control trial | - Significant alterations in microbiota in flax seed group but not L paracasei arm compared with placebo - Improved insulin sensitivity in flax seed group only |
|
| David (2014) | 24336217 | 11 | Animal-based versus Plant-based | Non-randomized observational cohort study | Significant diet-based alterations in microbial communities. | |
| DeWulf (2013) | 23135760 | 30 | Prebiotics (inulin/oligofructose) versus placebo | Double blind randomized control trial | Prebiotic use lead to: - Increased Bifidobacterium and F. prausnitzii and associated decreased serum lipopolysaccahrides - Decreased Bacteroides associated with slight decrease in fat mass |
|
| Cotillard (2013) | 23985875 | 49 | Energy-restricted high protein diet versus Weight Maintenance diet | Cross-over trial | - Microbiome with greater diversity (high gene count) associated with eating more fruits and vegetables - Individuals with low diversity microbiome changed to high diversity after eating energy-restricted diet. |
|
| Walker (2011) | 24603757 | 14 | Resistant starch, versus non-starch polysaccharides versus low carbohydrate weight loss | Cross-over trial | - Significant alterations in bacterial phylotypes occur rapidly (within 3–4 days) of change in diet. |
N-of-1 Trials and Determining Individual Responses
If the goal of a study is to truly determine whether a particular individual is responding to a specific intervention then, for the reasons discussed, classical population-based clinical trials are not appropriate. Their designs simply do not accommodate the collection of enough information on any one individual over the time the intervention is being administered to lead to unequivocal claims about that individual’s unique response. This, of course, could be changed as emerging and simple, cost-effective and convenient ways of collecting appropriate data through, e.g., wireless devices, could facilitate such studies.(24, 75) However, the actual design of such studies is as crucial as collecting enough data on an individual, since appropriate contrasts that exploit that data must be made in order to draw compelling inferences about the unique response of an individual to an intervention. This suggests that studies focusing on individuals, i.e., ‘N-of-1’ studies, do indeed have their place if emphasis is on assessing those individuals’ unique and nuanced responses to an intervention. We consider specific study designs for N-of-1 studies later, but feel it is important to provide additional historical and biological perspective on the motivation for such designs.
The origins of N-of-1 clinical trials have been discussed by many authors (40, 58, 86) but have been implemented most often in education, behavioral assessment and pain research settings.(58) Of most relevance to this review is the consideration, as noted, of N-of-1 trials in ‘personalized,’ ‘individualized,’ or ‘precision’ medicine including disease prevention and management settings. In this light, a paper by Hogben and Sim (40) published in the early 1950’s described N-of-1 trials as logical extensions of actual clinical practice. The authors argued that physicians often take into consideration the unique and nuanced profile, in terms of medical history, behaviors, and environmental exposures, of patients in making decisions on how to treat them. Essentially, they argued, physicians are accustomed to dealing with patients as individuals in this way, but rarely end up proving to themselves that the nuanced way in which they approach each patient actually worked for that patient, or at least worked better than another approach they could have taken; for example, by treating everyone in exactly the same way. Rather, the information about what whether an intervention worked, or is working, is collected informally in the context of return or follow-up visits, dialog with other hospital staff, mail-in records, etc. – any time at which a new decision about how to treat the patient may arise. The ultimate question Hogben and Sim raised was whether or not this process could be formalized and made more objective. They ultimately argued that one could bring principles of experimental design and data collection into this process in two important ways: 1. By providing the patient with charts they could use to track symptoms over time that may identify important features of their treatment earlier and in a more objective way than standard practice would; and 2. By using control mechanisms and purposeful, possibly pre-specified, data analyses to statistically assess if the patient’s improvement, or lack thereof, could be attributed to the actual intervention in question and not something else (e.g., the placebo effect, a measured or unmeasured covariate, non-compliance, etc.).
The belief that one can make objective claims about an individual response to an intervention using information collected on just that person is backed by the very intuitive notion that it is the number of measures taken on an individual, not the number of individuals being studied, that is important, as well as how, and under what conditions, those measures have been collected to enable statistically and clinically meaningful conclusions to be drawn from them. Consider the fact that many in vitro studies involving, e.g., cellular systems or cell lines, make replicate measures on the cells they are studying under different conditions to draw inferences about the relationships between various factors despite the fact that those cells came from a single individual. Of course, it could be the case that different results would have been observed had a different set of cells been used, perhaps from a different individual, but this possibility actually solidifies the point that there may be individual differences between units of observations (e.g., cells, cell lines, inbred mouse strains, individual humans) that could only be brought to light if those individual units were studied in isolation to identify the phenotypes or outcomes they may not share with others. In other words, one can be just as careful and thought-out in their approach to making objective claims about an individual’s response to an intervention as they could in making claims about the utility of an intervention in the population at large. The actual need for studying individuals in isolation and making claims about their unique and nuanced responses to nutritional interventions is also supported by studies leveraging data-intensive, high-throughput assays, such as DNA sequencing – which clearly show molecular physiologic differences between individuals that likely influence their responses to diets, nutrients and supplements – as well as historical studies documenting the very wide variation individuals exhibit in response to nutritional factors (see, e.g., Table 1 and note the fact that not everyone in those studies seemed to exhibit the same response to the interventions).
Biological Motivation for Individualized Nutrition
The recognition that individuals, whether as patients in a clinical setting or as individuals in the population at large, may exhibit unique responses to nutritional interventions that could only be teased out by studying each of them directly has its roots in a great deal of historical and emerging scientific studies. In fact, many reviews have been written on the biological motivation for personalized nutrition(2, 34, 51, 53) and we therefore provide just a brief overview with a few examples to help put into context the need for N-of-1 studies and study designs. Archibald Garrod is typically attributed with the introduction of the notion of the biochemical individuality of humans.(30) He basically argued that the unique genetic profiles each individual possesses create overt, if not subtle, differences between individuals in the way they respond to the environment, including pharmacological and nutritional interventions. This idea paved the way for the emerging field of pharmacogenetics, whose goal is to identify genetic variants some people possess that influence their unique responses to pharmacologic agents.(76, 85) The insights from pharmacogenetics studies, it is argued, could lead to clinical practices in which pharmacologic interventions for preventing and treating a disease are ‘personalized’ to patients based on their genetic profiles. Pharmacogenetics research has benefitted enormously from the recent and rapid advances of molecular genetic assays, such as DNA sequencing and high-throughput proteomics, and the routine application of those technologies in association studies, especially genome-wide association studies (GWAS) that could lead to connections between genetic variants and phenotypes of all sorts, which may have clinical utility.(12, 37, 83)
The variation individuals exhibit in their responses to pharmacologic agents that may be attributable to genetic or other (e.g., exposure profile) differences between individuals is certainly consistent with the emerging field of nutrigenomics.(46, 47, 52, 64, 70, 102) Many researchers have identified associations between specific genetic variants and response to diets, nutrients, and dietary supplements of all sorts (see Table 2), suggesting that individual responses to nutritional interventions could be as nuanced, if not more so, than responses to pharmacologic agents. Consider, for example, the rare disease Phenylketonuria (PKU) which is caused by genetic mutations in the PAH gene and treatable by manipulating the amount of phenylalanine and protein levels in the diet of an individual with the condition.(8) It is known that there is a complex relationship between mutations in the PAH gene, other genes and genetic variants that could modify the effect of PAH mutations, the severity of the condition, and the response to dietary manipulations to treat the condition.(87) Another very detailed and recent study showed that the indigenous people of Greenland, the Inuit, exhibit evolutionarily-mediated genetic variants at several loci that influence the levels of omega-3 polyunsaturated fatty acids (PUFAs) they possess. Further, these genetic variants were found to be associated with multiple metabolic and anthropometric phenotypes, have large effects on weight and height and modulate fatty acid composition.(29) Complexities in the relationship between various genetic and biochemical factors, as well as behavioral and exposure factors, and responses to nutritional interventions, are also borne out in many studies of different strains of model organisms.(28, 72, 81, 94)
Finally, there is ample evidence for great individual variation in response to nutritional factors that has emerged from highly contrived interventions such as the oral glucose tolerance (OGT) test and related tests.(97) In fact, these tests are designed to determine if, in fact, an individual may possess an inability to process and control products (such as insulin) that are provoked by a specific and highly contrived nutritional challenge.(9, 68, 71)
Basic Study Designs
Objective Data Collection Strategies
The mounting data consistent with the notion that individuals exhibit variation in response to nutritional interventions has broad implications for determining the best way to optimize nutrition and maximize health for an individual. If this variation is attributable to inherent differences between individuals at the genetic and biochemical or behavioral and exposure levels, then obvious strategies for optimizing nutrition can be framed. For example, researchers can identify these factors and test how well they can predict an individual’s response to a nutritional intervention. Clinicians and dieticians can then leverage this information in deciding how best to deal with a particular patient. Unfortunately, there are few instances where a direct, unequivocal relationship between a single (or even set of) identified factors and a nutritional response is known. Therefore, researchers must test an individual’s response to a nutritional intervention empirically and directly using objective and scientifically sound criteria. To do so requires sophisticated N-of-1 study designs.
Block and Period Structure of N-of-1 Trials
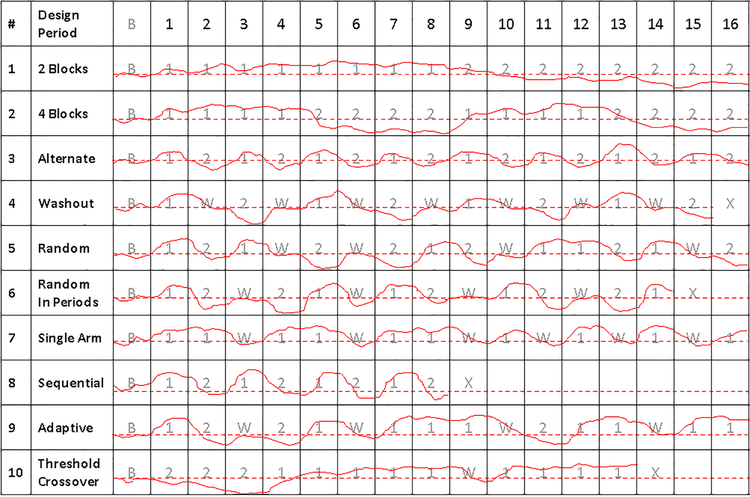
There are many possible N-of-1 clinical trial study designs that can be used to test a nutritional intervention or compare multiple interventions on a single individual. The design of any N-of-1 study, however, should be rooted in the biological issues associated with the primary hypothesis of interest as well the practicalities of the implementation of the study. Figure 1 provides a graphical depiction of 10 different N-of-1 study designs assuming that 2 different interventions, denoted 1 and 2 (e.g., a restricted or supplemented diet vs. an ad libitum or comparator diet), are compared on an individual with respect to a particular quantitative health-related outcome (e.g., lean weight, levels of a particular metabolite, relative mood, etc.). It is further assumed that a crossover design is exploited over a total of 16 periods (e.g., 16 days or weeks) in which observations on the health-related outcome are made on the individual while that individual is either not receiving any intervention or receiving one of the two interventions of interest. Of course the total number of periods, the number and type of measures collected during each period, the length of the periods, the order in which interventions are provided during the various periods, etc. are all important to consider. In addition, the design of an N-of-1 study design must take into consideration practical, scientific and ethical issues as well (e.g., is it practical or ethical to measure something on someone every day for a year? Is the measurement device technically noisy enough to warrant multiple measurements? etc.).
Figure 1.
Hypothetical depiction of 10 different N-of-1 study designs comparing two interventions, denoted 1 and 2. The left most panels name the different designs. The gray numbers and letters in each cell provide which intervention is being administered during 16 different measurement periods in addition to a baseline period, denoted B. The entries in the cells correspond to: 1: intervention 1; 2: intervention 2; W: Washout period; and X: termination of the study prior to completing all 16 periods. The dashed red line corresponds to values of a measure that are not associated with a favorable or unfavorable response to the interventions, but are ambiguous with respect to response. The solid red lines provide the values of hypothetical continuous measures made on an individual, with greater values than the red dashed line indicating a positive response and lesser values indicating a negative response.
The light shaded letters or numbers in each cell in Figure 1 denote which interventions, or lack thereof, the individual is being subjected, where ‘B’ denotes a baseline period in which the individual is not receiving any intervention, ‘1’ denotes periods in which the individual is being subjected to intervention 1, ‘2’ denotes periods in which the individual is being subjected to intervention 2, and ‘W’ denotes a washout period in which the individual is taken off any intervention. ‘X’ denotes termination of the study. The dashed red lines indicate a level of the health outcome that is ‘neutral’ if the measured value stays below that line, that is, it does not indicate a favorable or unfavorable response to an intervention. The solid red lines depict a hypothetical continuous trajectory of a health outcome measures obtained during each period and, for ease of explanation, suggest that intervention 1 is preferable to intervention 2 in each case.
The first two columns of Figure 1 simply index and label the example trial designs. A crucial question for N-of-1 designs concerns how to distribute the interventions across the 16 periods (e.g., provide one intervention for 8 weeks and then the other for 8 weeks?). A related question concerns how often to make measurements within these periods, and this bears on the power of the study, which will be briefly addressed later. With 16 periods, one could, as noted, provide intervention 1 for eight consecutive periods and then intervention 2 for 8 periods. This is the first example study design (study design 1), labeled ‘2 blocks’ since the interventions were provided in 2 blocks of eight periods each. The second design (study design 2; ‘4 blocks’) assumes the interventions are provided in 4 blocks of 4 periods each, such that intervention 1 is provided for 4 consecutive periods, intervention 2 is provided for 4 periods, then intervention 1 is provided again for 4 periods and finally intervention 2 is provided for 4 periods. Study design 3 considers simply alternating the interventions over the 16 periods. This type of design is most often associated with N-of-1 trials, but does suffer from a few issues that will be considered in greater detail later, such as carry-over and order effects.
Randomizing the Order of Interventions and the Use of Washout Periods
Study design 4 in Figure 1 depicts a trial that makes use of washout periods – i.e., periods in which the individual is taken off all interventions to let their body reset or re-acclimate before testing another intervention. The use of washout periods is motivated by many factors (see below) and, although they add to the length and complexity of a study, they help greatly with the interpretation of the study results. Studies 5 and 6 introduce randomization into the designs either with respect to the overall sequence in which the interventions are provided (study design 5) or within specified blocks of periods flanked by washout periods (study design 6). Randomization can be used to avoid a number of thorny issues such as order and carry over effects (see below).
Single Arm, Sequential, Adaptive and Multivariate Designs
There are a number of extensions to the basic N-of-1 designs. For example, interest may be in a single intervention, in which the comparison between the values of the measure taken during the intervention period involves assessing differences with the values taken in the pre-intervention or baseline period, or during washout periods (study design 7). Obviously, using a ‘placebo’ or sham intervention against a single intervention has advantages in reducing biases and confounding that might result if the question is whether or not an intervention has any overall utility or not. Another design involves pursuing the study in a sequential manner, in which stopping boundary rules are set a priori such that if the measure of interest reaches a level outside those bounds, the trial is halted as this would be indicative of overwhelming evidence that one or another of the interventions of interest has a compelling positive, or negative, effect (study design 8). Sequential trials are often pursued with only one intervention. Other studies can be designed and pursued in an adaptive manner, whereby the intervention exhibiting the best evidence for a positive benefit in the trial is applied more often – possibly by changing (e.g., increasing) the probability the individual will receive that intervention as part of a randomization scheme going forward (study design 9). Adaptive study designs of this sort are often referred to as ‘play-the-winner’ designs,(93) and they are thought to be more ethical than many other designs since they minimize the amount of time an individual will spend on what the evidence suggests is an inferior intervention and yet still retain statistical power to make definitive claims about the utility of the interventions relative to one another. Finally, studies can be pursued that combine elements of adaptive and sequential trials, in that they minimize the amount of time an individual spends on what appears to be an inferior treatment and are stopped if the data is overwhelmingly in favor of a better benefit for one or another intervention (study design 10).
Other study designs could involve multivariate outcomes; for example, monitoring weight, mood, microbiome species abundance and blood chemistries simultaneously to assess the more global impact of the intervention, or involve testing the combined effects of interventions to determine their synergy. For example, one might design a study to see if a behavioral intervention, when coupled with a dietary intervention, leads to a better health profile for an individual than either of these interventions alone. Such designs would have to devote certain periods in the study to the individual being assigned each intervention alone to complement the periods when the interventions are combined to assess the non-additive or synergistic effects of the interventions.
Issues Affecting the Power of N-of-1 Designs
There are a number of issues that could affect the yield of an N-of-1 trial. For example, the number of measurements made on a subject will have a profound influence on the power to detect a difference between interventions or a statistically significant change in a measure during an intervention. Obviously, the number of measurements made during any period in an N-of-1 trial will be dictated by the expense of, as well as any logistical issues surrounding, the collection of those measurements. The duration of each period in which an intervention is provided will undoubtedly contribute the total amount of time one has to make relevant measurements. This duration will be dictated, to a large degree, by the half-life, or the amount of time one would likely see an effect, of an intervention. Thus, if it is known that it will take weeks before, e.g., specific diets, will affect the body weight of an individual, then having the periods in which the interventions will be applied only last a few days would not work.
Another crucial factor affecting the power of N-of-1 studies is the serial correlation between the observations. Since the measures will be made on a single individual, they will be correlated over time, especially if the measures are made either continuously or with short intervals between them. This is unlike measures made on a large number of unrelated individuals in free living populations, where the correlations between them will likely be non-existent or negligible. Strong serial correlations between observations can have a profound effect on the power of a study since the lack of independence of the observations reduces the information provided by them.(23, 98) The hypothetical examples in Figure 1 assume that the measurements could be made continuously, which may be possible with wireless devices, such as an actigraph or continuous glucose monitor,(96) but difficult in other settings (e.g., blood draws or whole body imaging to explore body fat distribution).
The likelihood of carryover and order effects are also important to consider in the design of an N-of-1 study. Carry over effects occur when the effect of one intervention lingers over some period of time after that intervention is stopped of changed. This can occur with many pharmaceutical and behavioral interventions as the amount of drug or specific behavior-induced changes, and the effects of that drug or behavior, affect the body going forward. Carryover effects can confound claims about the biological effects of a specific intervention since it becomes difficult to distinguish the effects of the intervention of interest with a previous intervention. Washout periods are often used to avoid carry over effects, but their use adds to the time and complexity of a study. To avoid certain biases even further, one could leverage blinding in a study, such that the individual receiving the interventions would not know to which intervention they were being subjected. This might be very difficult to achieve in practice given, e.g., food tastes, textures, the physiologic effects of certain supplements, etc. since the individual receiving them may recognize which intervention they are being provided based on these features. Blinding could also be applied to the research team and medical overseers by not letting them know which intervention an individual in a trial might be on. Such blinding could avoid conscious or unconscious biases a research group might have about the effects of an intervention.
Order effects are related to carryover effects and occur when one intervention is provided before another systematically. This can create the illusion that one intervention is superior when in fact there could be a learning effect (i.e., an individual recognizes when he or she is on one intervention and that leads to biases in terms of behaviors that might impact interpretation of measures used to assess the differences between two or more interventions), tolerance to one intervention, or a carryover effect that confounds an ability to attribute differences in a measure to that actual intervention and not a bias in the measurements. Order effects can be avoided by randomizing the order in which interventions are provided to an individual.
Finally, two very important considerations in an N-of-1 trial (or any trial, for that matter) involve covariate effects and the statistical analysis methods used. Essentially, if there are factors that could influence a primary measure independently of an intervention, then they must be measured and taken into consideration when analyzing and interpreting the results of the trial. For example, physical activity influences weight and therefore should be considered in a trial investigating the influence of different diets on weight. In this light, it is important to consider whether or not to explicitly control for covariate effects in the design of the trial (e.g., stipulate that activity levels remain constant or are built into the trial as an additional intervention) or simply measure the covariate and accommodate it as such in analyzing the data. In addition, they are a variety of statistical methods that can be leveraged for drawing inferences from N-of-1 trials data, including simple linear models(84) time series analyses, (63) simple comparisons and contrasts using t-tests(35) and bootstrap, permutation and randomization tests.(59) A complete assessment of the statistical methods than can be used is beyond the scope of this review, but an incredibly important component of any N-of-1 trial.(6, 15, 22, 78)
Aggregated N-of-1 Studies
It is possible to aggregate the results of N-of-1 studies and thereby draw more general conclusions about the utility of an intervention in the population at large. Methods for aggregating N-of-1 clinical trial data have been proposed that are based on mixed effects models that can take into consideration population averages as well as individual-specific variations.(79) In this light, aggregated N-of-1 studies may be interpreted in a way that is analogous to population-based trials of the type discussed early, except that they have been designed to ensure that enough data is collected on each individual to make unequivocal claims about their responses (or lack thereof) to the interventions of interest. Such analyses have advantages since they explicitly model and account for regression-to-the mean, measurement error, cryptic or latent variable effects, and other population-level phenomena when drawing inferences about the response of an individual to a specific intervention.
Aggregating N-of-1 trials can facilitate a number of important additional analyses. For example, one could identify, with great precision, individuals that share a response profile and then consider what these individuals might have in common (e.g., a specific set of genetic factors, exposures to certain environmental conditions, etc.). Figure 2 provides a graphical depiction of this concept. This type of analysis suggests that N-of-1 trials can be used to ‘bring out a response phenotype’ in a sophisticated way that could be explored further.
Figure 2.
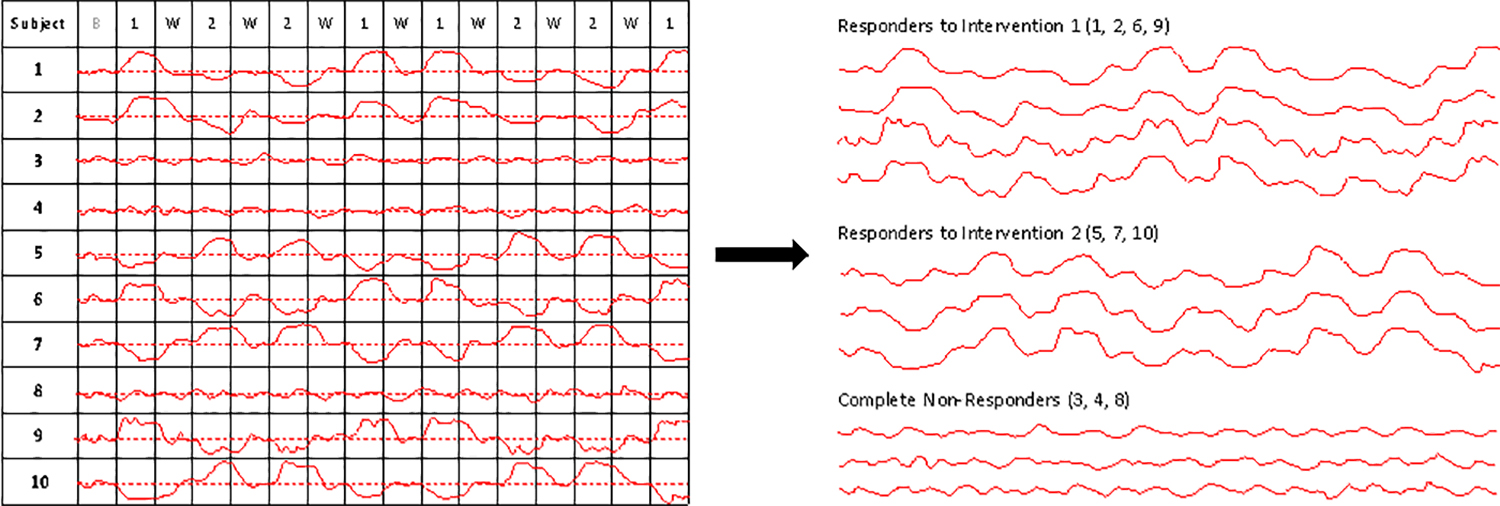
A graphical depiction of the result of aggregating the outcomes of 10 different N-of-1 studies. For the left panel, it is assumed that each individual underwent an N-of-1 trial with a similar design in which interventions were alternated after a baseline and washout periods. As with Figure 1, the dashed red line corresponds to values of a measure that are not associated with a favorable or unfavorable response to the interventions, but are ambiguous with respect to response. The solid red lines provide the values of hypothetical continuous measure made on an individual, with greater values than the red dashed line indicating a positive response and lesser values indicating a negative response. The right panel depicts the results of a clustering of the individual responses, with some individuals exhibiting a greater response to intervention 1 (upper set of curves), some individuals exhibiting a greater response to intervention 2 (middle set of curves) and some individuals exhibiting a lack of response to either intervention.
The ability to aggregate the results of N-of-1 trials ultimately suggests that it is wrong to argue that N-of-1 trials cannot be generalized. In addition, one could always conduct a study to test the hypothesis that conducting N-of-1 trials on individuals leads to better health outcomes for those individuals than not conducting N-of-1 trials on them or providing them interventions based on standard practices. Such a study may simply randomize individuals to a group that will be subjected to an N-of-1 trial for identifying an optimal intervention and a group that will simply be provided interventions based on standard and legacy practices.(88)
What Does It Make Sense to Measure?
There are many different phenotypes or outcomes one could measure in the context of an N-of-1 study. Ultimately, the choice of which phenotype to study would depend on the nature of the condition for which the intervention is being considered (i.e. the reason the individual may be undergoing an N-of-1 study in the first place) as well as the nature of the intervention. Tables 3a and 3b list a number of studies exploring the impact of a nutritional intervention on different phenotypes and outcomes. None of these studies was pursued in the context of an N-of-1 or aggregated N-of-1 study, but clearly could have been pursued as such with appropriate changes in their design and execution. The nature of the nutritional interventions listed in Tables 3a and 3b is also broad and include overall caloric restriction(66), the timing of food consumption on a daily basis(32), food-based diets (Mediterranean, high carbohydrate, etc.) (1, 80), vitamins and supplements (folate, B-12, Omega-3 fatty acids)(43, 99), bioactive compounds (phenols such as resveratrol, phytoestrogens such as isoflavone, carotenoids such as lycopene)(45, 65, 104) and, of increasing interest, probiotics(10).
There are seemingly endless ways in which the impact of a nutritional intervention can be measured, as Tables 3a and 3b make clear. Some of the more obvious measures include body weight and body composition (e.g., fat distribution)(11, 61, 62). Many physiological measures have been studied, for example, blood pressure or heart rate(20). Often considered in nutritional intervention studies are markers obtained from easily accessible tissues such as blood. Blood-based biomarkers that have been considered are gene expression levels(57) and specific factors such as C-reactive protein and adiponectin (45) as well as glucose and insulin(105) and standard clinical chemistries such as cholesterol and triglyceride levels.(69) Many investigations have considered the influence of nutritional interventions on psychological factors such mood (66), cognition scores (56), depression scales(43) as well as sleep(66). As noted in Table 3b, yet another measure that a number of studies have considered involves the microbiome.(18, 19). Given the ease with which fecal samples can be obtained, studies of the gut microbiome are of particular interest given its role in digestion and will therefore likely continue to be pursued.(73)
One additional area where there is growing interest in monitoring post-intervention to assess outcomes and impact involves the use of wireless devices(39, 74). There are many devices that can measure, e.g., activity, mood, sleep quality and length, and related phenotypes, all of which could be used in N-of-1 nutritional intervention studies. In addition, there is no reason one could not consider multiple phenotypes in N-of-1 and aggregated N-of-1 studies (e.g., sleep, blood pressure and heart rate, body composition and mood) as noted previously. The statistical methods for the analysis of the data from such a trial may be more complicated, but they would not be unprecedented. Also, such studies may provide a more complete picture of how an individual is responding to a particular nutritional intervention and hence be more insightful for how to refine or optimize that individual’s nutritional and health profile.
Monitoring and Personal Thresholds
Sequential N-of-1 trials can also be framed as ways of monitoring an individual for a health status change in the wake of taking on an intervention. Determining the levels of a measure collected over time on an individual that would be indicative of a change is not trivial, however. Traditionally, this was done by determining the levels of a measure that are associated with poor outcomes in the population at large – i.e., determining and exploiting ‘population-based thresholds.’ For example, if the cholesterol level of an individual was monitored while on a diet that may increase cholesterol levels, then if the individual exhibited cholesterol readings greater than, say, 200 mg/dL, at a certain point an argument could be made that the individual was exhibiting signs of poor health, given that epidemiologic studies have shown that cholesterol levels > 200 mg/dL are associated with an increased risk of heart disease. However, there are issues with the use of population thresholds for this kind of monitoring that are rooted in the potentially unique physiology each of us possesses. For example, the use of population thresholds ignores the fact that changes in certain measures or biomarkers might reflect physiological disruptions that are simply not consistent with population-based threshold criteria if the change in the biomarker is significant relative to biomarker values collected previously on an individual. Thus, the use of individual-specific or ‘personal thresholds’ for identifying health status changes may be more appropriate.
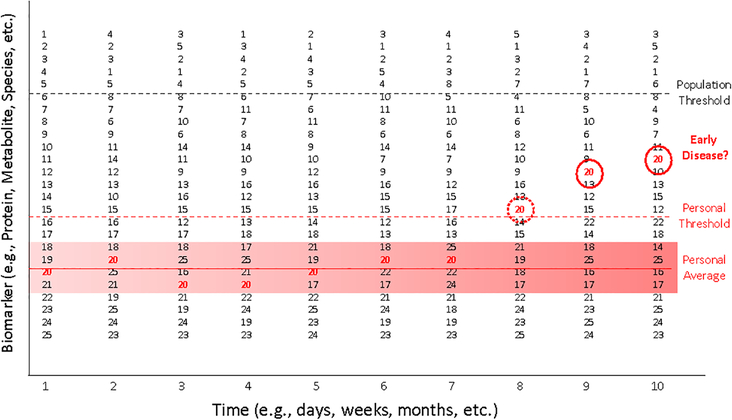
Figure 3 provides a graphical depiction of the concept of personal thresholds. 25 hypothetical individuals have undergone measurements on a phenotype measuring an aspect of health (e.g., cholesterol level or other biomarker). The values of these 25 individuals are ranked and are made at 10 different time points going forward. A ‘population threshold’ (e.g., cholesterol level > 200 mg/dL) is depicted by the dashed black line. The rankings and values of a single individual, individual number 20, is highlighted in red. After enough measures are collected over time, one could calculate a ‘personal average’ for individual 20, which is denoted by the solid red line, as well as error bars representing the bounds of the observed variation in that individual’s values, depicted by the red shading. Based on the errors associated with individual 20’s values, a ‘personal threshold’ can be established for which any value beyond that limit has a low probability of occurring purely by chance (note: these values may be adjusted for certain covariates). This is depicted by the dashed red line. The dashed red circle indicates a value of the biomarker on individual 20 that is outside the personal threshold, and at later time points two additional values, circled in black, get progressively higher. This deviation from Individual 20’s historical or legacy values could be an indication of a health status change, despite being lower than the establish population threshold.
Figure 3.
Graphical depiction of the concept of ‘personalized thresholds’ for making claims about a health status change for an individual. 25 hypothetical individuals have undergone measurements on a phenotype measuring health (e.g., cholesterol level or other biomarker). Their values are ranked and are made at 10 different time points. A ‘population threshold’ (e.g., cholesterol level > 200 units) is depicted by the dashed black line. The rankings and values of a single individual, number 20, is highlighted in red. After enough measures are collected over time, one can calculate a ‘personal average’ for Individual 20, denoted by the solid red line as well as error bars representing variation in that individual’s values, depicted by the red shading. Based on the errors associated with Individual 20, a ‘personal threshold’ can be established for which any value beyond that limit has a low probability of occurring. This is depicted by the dashed red line. The dashed red circle indicates a value outside the personal threshold and at later time points two additional values, circled in black, get progressively higher. This deviation from historical or legacy values on the individual that have a low probability of occurring by chance could be an indication of a health status change despite being lower than the establish population threshold.
The use of personal thresholds to guide inferences about health status changes has been shown to be useful in monitoring CA-125 levels in the blood of individuals at risk for ovarian cancer.(21) It has been argued further that most biomarkers of relevance to health are likely to be better assessed and utilized with personal thresholds rather than population thresholds.(3) Such monitoring could be done with multiple markers simultaneously in a multivariate analysis setting. One important issue with the use of personal thresholds is that if the monitoring is truly done in real time, and not done in a retrospective manner after samples have been collected over time and then processed together to obtain biomarker values for the different time points, then the biomarker assay results must not suffer from assay drift or temporal technical variations (e.g., show great variation depending on the technician performing the biomarker assay). This is to ensure that the values of the measures are comparable. This is not necessarily easy to achieve, as most studies involving longitudinal measurements are done retrospectively, such that the samples collected over time are processed within in a single batch so that the resulting measures avoid having technical variation complicate the interpretation of temporal changes of the measures.
Matching Strategies: Vetting Algorithms vs. Vetting Specific Nutritional Interventions
As an alternative to aggregated N-of-1 trials and combining the results of individual N-of-1 trials for making broad claims about the utility of individualized interventions, one could leverage extensions and offshoots of what have been referred to ‘basket,’ ‘bucket,’ or ‘umbrella’ trial methodology in the cancer clinical trials literature.(82) Essentially, bucket trials assume that there may be many interventions to choose from for an individual based on that individual’s ‘profile,’ whether genetic, microbiome, metabolic, behavioral, some combination of these, etc. If a strategy for matching the individual profiles with the different interventions (e.g., low fat diet for individuals genetically predisposed to heart disease) is set up a priori, then as individuals are enrolled in the trial and their profiles assessed, they are placed into the appropriate intervention ‘buckets’ and provided that intervention. The goal is to then see if the scheme for providing the individuals interventions based on their profiles results in better outcomes compared to a group of individuals that was either not provided any intervention, provided a sham intervention (i.e., a placebo), or provided a single, common ‘one-size-fits-all’ intervention. Obviously, as in any trial, the nature of the outcomes and measures used to assess the success of the interventions is of crucial importance to such trials.
Bucket and related trials conceived in this way suffer from a few major issues. First, they are actually not focusing on the interventions themselves, but rather the strategy or algorithm for matching the interventions to the individual profiles. One could imagine a situation in which an intervention works particularly well, just not for the individuals it was assigned to in the bucket trial because the matching scheme used in the trial was faulty. This could lead to a rejection of a perfectly good intervention. Second, the matching scheme used will only be as good as the biological insights it is based on. Third, and a bit more complicated, is that such trials often ‘lock down’ the strategies for matching the interventions to the patients at the start of the trial to determine how well the strategies in question work. This may compromise one’s ability to incorporate new information about, e.g., the likely effects of a specific nutritional intervention for individuals with a certain profile, since the evidence for this may arise after the initiation of the trial. New insights arising after the initiation of a trial are problematic for any clinical trial, but possibly more so for bucket trials since such trials may be more wide-ranging than a trial focusing on a single, very specific intervention. The issues with how to incorporate the new insights would be varied. For example, statistical issues could arise if the new insight was incorporated into the trial, as it may lead to the creation of a new bucket whose contribution to the overall effect of the set of interventions being tested would have to be considered. In addition, statistical analysis of the trial data may have to accommodate weighting of the individuals, since individuals enrolled later in the trial – if new insights are incorporated into the strategy for assigning individuals to intervention buckets occurs – may benefit from a better intervention strategy than individuals enrolled early in the trial. Finally, ignoring new insights that are truly compelling may create ethical problems if they could really enhance health, since the individuals in the trial could be perceived as being provided likely inferior interventions than what they may have access to.
There are three important extensions of trials seeking to match individuals to nutritional interventions based on their profiles, however defined. First, one may not have to pre-define the intervention ‘buckets’ corresponding to specific features in individual profiles, but rather address or test a much broader question concerning whether or not the profiling itself has any merit for identifying appropriate and effective nutritional interventions. For example, one could, e.g., genetically profile a group of individuals and use that profiling to determine the best nutritional intervention based on, e.g., a panel of experts’ opinions in assessing that profile, as with ‘tumor boards’ in the cancer treatment setting,(54) or an adaptive machine learning strategy whose calculations consider that kind of profiling.(105) The idea would then be to compare how the individuals responded to the interventions provided on the basis of the, e.g., genetically-guided profiling vs. those that may have received expert advice or information resulting from a machine learning strategy that did not consider genetic profiles in their deliberations or as part of the calculations.
Second, one could consider the results of assays and response profiles using biospecimens from the individuals participating in a trial in ex vivo or in vitro settings of particular phenomena to determine what the most appropriate intervention might be for those individuals. For example, establishing cell lines, induced pluripotent cell lines (iPS cells) or organoids from individuals and then exploring how they respond to different nutritional interventions could lead to insights into the best intervention for the individual. Such studies are used routinely to identify treatments for individuals with rare congenital diseases.(100) Obviously, the relevance of the assay system and the measures used to assess the nutritional responses in vitro to the in vivo setting is a crucial concern with such studies. Fenech and colleagues have written extensively on this type of strategy in the context of DNA repair capacity and nutritional schemes to minimize cancer and other disease susceptibility.(26, 27)
Third, an issue related to strategies for assigning or optimizing nutritional interventions to individuals on the basis of ex vivo or in vitro assays that make use of biospecimens obtained from those individuals involves the media in which the, e.g., cellular assays are performed and what, if any, insights might be obtained from the choice of that media. Consider the fact that when cell lines are created and passaged, or when organoids are grown and maintained, they require cell culture media. Often times this media contains a number of factors, such as hormones, growth factors, vitamins, etc.(44, 67) A relevant question in the context of this review is whether or not one could ‘personalize’ the media used to optimally grow cells or create organoids from an individual empirically, by testing different media constituents and comparing the viabilities and functional capabilities of the cells and organoids, and then use the resulting insights to craft better nutritional interventions for the individuals from whom those cells, organoids, etc. were harvested and created.
Recent Studies Motivating N-of-1 Trials in Nutrition
There have been a number of recently published studies investigating the impact of nutritional factors on indicators of health that, although not technically N-of-1 studies, motivate N-of-1 studies because of what they showed and how they were pursued. The study by Alm et al. in which daily microbiome measures were obtained for two individuals over the course of a year is one good example of the potential of N-of-1 studies.(18) Alm et al. also described how the daily eating patterns of the two participants were kept and then correlated with the microbiome. A number of very interesting correlations were found that gave insight into what the two individuals may want to avoid or encourage in the way of food consumption in order to optimize their microbiomes.(18) Although no purposeful nutritional intervention was pursued in the study, it was clear that the results generated some obvious hypotheses about how certain dietary substances may impact the gut microbiome of the two individuals, setting the stage for a bona-fide randomized, blinded (to the degree possible), crossover N-of-1 study of the type emphasized in this review for testing those hypotheses.
Another study which has received considerable attention and is truly reflective of studies exposing nuanced, individual responses to nutritional interventions was the continuous time glucose monitoring study by Segal et al.(105) Essentially, Segal et al. continuously monitored week-long glucose levels in an 800-person cohort, which ultimately considered these individuals’ responses to over 46,000 meals. They found overwhelming evidence for variability in the response to identical meals between individuals, suggesting that individual dietary needs must be based on objective empirical study-based measures of an individual’s response to a change in diet and in this way may be hard to anticipate from any prior information on those individuals. However, the authors did devise a machine-learning algorithm that leveraged blood parameters, dietary habits, anthropometrics, physical activity, and gut microbiota measured on the 800 individuals in the study and showed that it could accurately predict postprandial glycemic response to real-life meals for individuals in the study. The authors went on to validate these predictions in an independent 100-person cohort. To top things off, the authors pursued a blinded, randomized controlled dietary intervention based on the algorithm which resulted in significantly lower postprandial responses and consistent alterations to gut microbiota for the participants who were provided the intervention. The authors concluded that their results suggest that personalized diets may successfully modify elevated postprandial blood glucose and its metabolic consequences. These findings could easily be explored in N-of-1 and aggregated N-of-1 studies of the type envisioned.
Conclusions
The concepts of individualized medicine and individualized nutrition are not likely to be proven useless and disappear soon. Rather, their worth can be evaluated in studies designed to determine the optimal intervention for an individual. The results of these studies can then be aggregated to show that they could not have been anticipated in other ways, or at least that they had to be performed in order to bring out nuanced responses to an intervention whose determinants would have to be explored. N-of-1 trials and aggregated N-of-1 trials of this sort thus have an obvious role to play in the identification of factors that influence response to nutrition that might be shared among a set of individuals. In other words, it could be argued that most variation in response to, e.g., nutritional factors, is likely to be explained by a set of identifiable (but as yet unidentified) genetic, environmental and behavioral factors, with the remaining amount of variation being very individual-specific, the clinical significance of which is in doubt. Since to date it is not clear how much inter-individual variation in nutritional response can be attributed to identifiable, shared factors, more research into individual variation in response to nutritional factors is needed. In addition, since one cannot make confident predictions about an individual’s response to all nutritional interventions because we have yet to identify the factors that might be used for such predictions, an individual’s response must be evaluated empirically to explore variation that could be attributable to those factors, and N-of-1 trials are one vehicle for doing this.
There are a number of trends that could both motivate and enhance N-of-1 trials in nutrition beyond a general interest in personalized health care. First, there is tremendous interest in self-monitoring for health purposes given the changing heath care system, a new focus on disease prevention, and the availability of cheap and relatively sophisticated biomarker and wireless data collection devices. This interest is taken to the extreme by individuals within the ‘quantified self’ movement in which participants knowingly experiment on themselves to determine optimal ways of living.(25, 33, 49, 92, 103) However, most studies pursued by people within the quantified self-movement are anecdotal and lack the scientific rigor of N-of-1 trials, although this could change if the individuals within the movement were exposed to N-of-1 trial methodologies. Second, there is tremendous interest in improving the sophistication of data collection devices for health monitoring purposes, making them more reliable, cost-efficient, and transparent to the user.(14, 106) Such devices can easily enable N-of-1 studies if they collect appropriate information. Third, there is growing interest in ‘Big Data’ and the use of large databases to mine information that might be useful for some purpose or another.(4, 16, 17) One could imagine designing and implementing systems to facilitate the conduct of N-of-1 trials and aggregating their results for pattern discovery and data mining. Ultimately, given these trends, and the biological intuitions behind personalized nutrition and even though they will never be a panacea for all nutritional ills, N-of-1 trials are likely to become very relevant approaches to optimizing individual health and advancing health care generally.
Acknowledgments
The authors would like to acknowledge partial support from the following: National Institutes of Health Grant UL1TR001442 (note that the content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH).
References
- 1.Afaghi A, O’Connor H, Chow CM. 2007. High-glycemic-index carbohydrate meals shorten sleep onset. Am J Clin Nutr 85: 426–30 [DOI] [PubMed] [Google Scholar]
- 2.Allison DB, Bassaganya-Riera J, Burlingame B, Brown AW, le Coutre J, et al. 2015. Goals in Nutrition Science 2015–2020. Front Nutr 2: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson L, Razavi M, Skates S, Anderson NG, Pearson TW. 2016. Squeezing more value from the analytes we have: personal baselines for multiple analytes in serial DBS. Bioanalysis 8: 1539–42 [DOI] [PubMed] [Google Scholar]
- 4.Anoushiravani AA, Patton J, Sayeed Z, El-Othmani MM, Saleh KJ. 2016. Big Data, Big Research: Implementing Population Health-Based Research Models and Integrating Care to Reduce Cost and Improve Outcomes. Orthop Clin North Am 47: 717–24 [DOI] [PubMed] [Google Scholar]
- 5.Aslibekyan S, Claas SA, Arnett DK. 2013. To replicate or not to replicate: the case of pharmacogenetic studies: Establishing validity of pharmacogenomic findings: from replication to triangulation. Circ Cardiovasc Genet 6: 409–12; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bagne CA, Lewis RF. 1992. Evaluating the effects of drugs on behavior and quality of life: an alternative strategy for clinical trials. J Consult Clin Psychol 60: 225–39 [DOI] [PubMed] [Google Scholar]
- 7.Beutler E 1994. G6PD deficiency. Blood 84: 3613–36 [PubMed] [Google Scholar]
- 8.Blau N, van Spronsen FJ, Levy HL. 2010. Phenylketonuria. Lancet 376: 1417–27 [DOI] [PubMed] [Google Scholar]
- 9.Bloomgarden ZT. 2006. Measures of insulin sensitivity. Clin Lab Med 26: 611–33, vi [DOI] [PubMed] [Google Scholar]
- 10.Brahe LK, Le Chatelier E, Prifti E, Pons N, Kennedy S, et al. 2015. Dietary modulation of the gut microbiota--a randomised controlled trial in obese postmenopausal women. Br J Nutr 114: 406–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brehm BJ, Seeley RJ, Daniels SR, D’Alessio DA. 2003. A randomized trial comparing a very low carbohydrate diet and a calorie-restricted low fat diet on body weight and cardiovascular risk factors in healthy women. J Clin Endocrinol Metab 88: 1617–23 [DOI] [PubMed] [Google Scholar]
- 12.Cardon LR, Bell JI. 2001. Association study designs for complex diseases. Nat Rev Genet 2: 91–9 [DOI] [PubMed] [Google Scholar]
- 13.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, et al. 2011. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med 364: 2507–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen LY, Tee BC, Chortos AL, Schwartz G, Tse V, et al. 2014. Continuous wireless pressure monitoring and mapping with ultra-small passive sensors for health monitoring and critical care. Nat Commun 5: 5028. [DOI] [PubMed] [Google Scholar]
- 15.Chen X, Chen P. 2014. A comparison of four methods for the analysis of N-of-1 trials. PLoS One 9: e87752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Y, Elenee Argentinis JD, Weber G. 2016. IBM Watson: How Cognitive Computing Can Be Applied to Big Data Challenges in Life Sciences Research. Clin Ther 38: 688–701 [DOI] [PubMed] [Google Scholar]
- 17.Coveney PV, Dougherty ER, Highfield RR. 2016. Big data need big theory too. Philos Trans A Math Phys Eng Sci 374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.David LA, Materna AC, Friedman J, Campos-Baptista MI, Blackburn MC, et al. 2014. Host lifestyle affects human microbiota on daily timescales. Genome Biol 15: R89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, et al. 2014. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505: 559–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Domenech M, Roman P, Lapetra J, Garcia de la Corte FJ, Sala-Vila A, et al. 2014. Mediterranean diet reduces 24-hour ambulatory blood pressure, blood glucose, and lipids: one-year randomized, clinical trial. Hypertension 64: 69–76 [DOI] [PubMed] [Google Scholar]
- 21.Drescher CW, Shah C, Thorpe J, O’Briant K, Anderson GL, et al. 2013. Longitudinal screening algorithm that incorporates change over time in CA125 levels identifies ovarian cancer earlier than a single-threshold rule. J Clin Oncol 31: 387–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duan N, Kravitz RL, Schmid CH. 2013. Single-patient (n-of-1) trials: a pragmatic clinical decision methodology for patient-centered comparative effectiveness research. J Clin Epidemiol 66: S21–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dugard PF P; Todman J 2011. Single-case and Small-n Experimental Designs: A Practical Guide To Randomization Tests, Second Edition Routledge; 304 pp. [Google Scholar]
- 24.Evenson KR, Goto MM, Furberg RD. 2015. Systematic review of the validity and reliability of consumer-wearable activity trackers. Int J Behav Nutr Phys Act 12: 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fawcett T 2015. Mining the Quantified Self: Personal Knowledge Discovery as a Challenge for Data Science. Big Data 3: 249–66 [DOI] [PubMed] [Google Scholar]
- 26.Fenech M 2001. The role of folic acid and Vitamin B12 in genomic stability of human cells. Mutat Res 475: 57–67 [DOI] [PubMed] [Google Scholar]
- 27.Fenech M, Baghurst P, Luderer W, Turner J, Record S, et al. 2005. Low intake of calcium, folate, nicotinic acid, vitamin E, retinol, beta-carotene and high intake of pantothenic acid, biotin and riboflavin are significantly associated with increased genome instability--results from a dietary intake and micronucleus index survey in South Australia. Carcinogenesis 26: 991–9 [DOI] [PubMed] [Google Scholar]
- 28.Fisher-Wellman KH, Ryan TE, Smith CD, Gilliam LA, Lin CT, et al. 2016. A Direct Comparison of Metabolic Responses to High Fat Diet in C57BL/6J and C57BL/6NJ Mice. Diabetes [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fumagalli M, Moltke I, Grarup N, Racimo F, Bjerregaard P, et al. 2015. Greenlandic Inuit show genetic signatures of diet and climate adaptation. Science 349: 1343–7 [DOI] [PubMed] [Google Scholar]
- 30.Garrod AE. 2002. The incidence of alkaptonuria: a study in chemical individuality. 1902 [classical article]. Yale J Biol Med 75: 221–31 [PMC free article] [PubMed] [Google Scholar]
- 31.Gaziano JM, Sesso HD, Christen WG, Bubes V, Smith JP, et al. 2012. Multivitamins in the prevention of cancer in men: the Physicians’ Health Study II randomized controlled trial. JAMA 308: 1871–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gill S, Panda S. 2015. A Smartphone App Reveals Erratic Diurnal Eating Patterns in Humans that Can Be Modulated for Health Benefits. Cell Metab 22: 789–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gimbert C, Lapointe FJ. 2015. Self-tracking the microbiome: where do we go from here? Microbiome 3: 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gorman U, Mathers JC, Grimaldi KA, Ahlgren J, Nordstrom K. 2013. Do we know enough? A scientific and ethical analysis of the basis for genetic-based personalized nutrition. Genes Nutr 8: 373–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Green AL, Shad A, Watson R, Nandi D, Yianni J, Aziz TZ. 2004. N-of-1 Trials for Assessing the Efficacy of Deep Brain Stimulation in Neuropathic Pain. Neuromodulation 7: 76–81 [DOI] [PubMed] [Google Scholar]
- 36.Guasch-Ferre M, Bullo M, Martinez-Gonzalez MA, Ros E, Corella D, et al. 2013. Frequency of nut consumption and mortality risk in the PREDIMED nutrition intervention trial. BMC Med 11: 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guessous I, Gwinn M, Khoury MJ. 2009. Genome-wide association studies in pharmacogenomics: untapped potential for translation. Genome Med 1: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harper AR, Topol EJ. 2012. Pharmacogenomics in clinical practice and drug development. Nat Biotechnol 30: 1117–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hingle M, Patrick H. 2016. There Are Thousands of Apps for That: Navigating Mobile Technology for Nutrition Education and Behavior. J Nutr Educ Behav 48: 213–8e1 [DOI] [PubMed] [Google Scholar]
- 40.Hogben L, Sim M. 1953. The self-controlled and self-recorded clinical trial for low-grade morbidity. Br J Prev Soc Med 7: 163–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Howard BV, Van Horn L, Hsia J, Manson JE, Stefanick ML, et al. 2006. Low-fat dietary pattern and risk of cardiovascular disease: the Women’s Health Initiative Randomized Controlled Dietary Modification Trial. JAMA 295: 655–66 [DOI] [PubMed] [Google Scholar]
- 42.Ioannidis JP. 2013. To replicate or not to replicate: the case of pharmacogenetic studies: Have pharmacogenomics failed, or do they just need larger-scale evidence and more replication? Circ Cardiovasc Genet 6: 413–8; [DOI] [PubMed] [Google Scholar]
- 43.Jacka FN, Pasco JA, Mykletun A, Williams LJ, Hodge AM, et al. 2010. Association of Western and traditional diets with depression and anxiety in women. Am J Psychiatry 167: 305–11 [DOI] [PubMed] [Google Scholar]
- 44.Jenkins MJ, Farid SS. 2015. Human pluripotent stem cell-derived products: advances towards robust, scalable and cost-effective manufacturing strategies. Biotechnol J 10: 83–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jennings A, Welch AA, Spector T, Macgregor A, Cassidy A. 2014. Intakes of anthocyanins and flavones are associated with biomarkers of insulin resistance and inflammation in women. J Nutr 144: 202–8 [DOI] [PubMed] [Google Scholar]
- 46.Kaput J 2016. Systems-Level Nutrition Approaches to Define Phenotypes Resulting from Complex Gene-Environment Interactions. Nestle Nutr Inst Workshop Ser 84: 1–13 [DOI] [PubMed] [Google Scholar]
- 47.Kaput J, Morine M. 2012. Discovery-based nutritional systems biology: developing N-of-1 nutrigenomic research. Int J Vitam Nutr Res 82: 333–41 [DOI] [PubMed] [Google Scholar]
- 48.Kenward MJ B 2014. Design and Analysis of Cross-Over Trials, Third Edition: Chapman and Hall/CRC [Google Scholar]
- 49.Kim J 2014. Analysis of health consumers’ behavior using self-tracker for activity, sleep, and diet. Telemed J E Health 20: 552–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kraft P, Zeggini E, Ioannidis JP. 2009. Replication in genome-wide association studies. Stat Sci 24: 561–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kussmann M, Affolter M. 2009. Proteomics at the center of nutrigenomics: comprehensive molecular understanding of dietary health effects. Nutrition 25: 1085–93 [DOI] [PubMed] [Google Scholar]
- 52.Kussmann M, Van Bladeren PJ. 2011. The Extended Nutrigenomics - Understanding the Interplay between the Genomes of Food, Gut Microbes, and Human Host. Front Genet 2: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kussmann MF L 2008. Nutrigenomics and personalized nutrition: science and concept. Personalized Medicine 5: 447–55 [DOI] [PubMed] [Google Scholar]
- 54.Lamb BW, Brown KF, Nagpal K, Vincent C, Green JS, Sevdalis N. 2011. Quality of care management decisions by multidisciplinary cancer teams: a systematic review. Ann Surg Oncol 18: 2116–25 [DOI] [PubMed] [Google Scholar]
- 55.Laursen L 2010. Interdisciplinary research: Big science at the table. Nature 468: S2–4 [DOI] [PubMed] [Google Scholar]
- 56.Lemaire JB, Wallace JE, Dinsmore K, Lewin AM, Ghali WA, Roberts D. 2010. Physician nutrition and cognition during work hours: effect of a nutrition based intervention. BMC Health Serv Res 10: 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Leonardson AS, Zhu J, Chen Y, Wang K, Lamb JR, et al. 2010. The effect of food intake on gene expression in human peripheral blood. Hum Mol Genet 19: 159–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lillie EO, Patay B, Diamant J, Issell B, Topol EJ, Schork NJ. 2011. The n-of-1 clinical trial: the ultimate strategy for individualizing medicine? Per Med 8: 161–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lin SX, Morrison L, Smith PW, Hargood C, Weal M, Yardley L. 2016. Properties of bootstrap tests for N-of-1 studies. Br J Math Stat Psychol 69: 276–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lippman SM, Klein EA, Goodman PJ, Lucia MS, Thompson IM, et al. 2009. Effect of selenium and vitamin E on risk of prostate cancer and other cancers: the Selenium and Vitamin E Cancer Prevention Trial (SELECT). JAMA 301: 39–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Look ARG, Wing RR, Bolin P, Brancati FL, Bray GA, et al. 2013. Cardiovascular effects of intensive lifestyle intervention in type 2 diabetes. N Engl J Med 369: 145–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lopes Gomes D, Moehlecke M, Lopes da Silva FB, Dutra ES, D’Agord Schaan B, Baiocchi de Carvalho KM. 2016. Whey Protein Supplementation Enhances Body Fat and Weight Loss in Women Long After Bariatric Surgery: a Randomized Controlled Trial. Obes Surg [DOI] [PubMed] [Google Scholar]
- 63.Macey PM, Schluter PJ, Macey KE, Harper RM. 2016. Detecting variable responses in time-series using repeated measures ANOVA: Application to physiologic challenges. F1000Res 5: 563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Maher M, Pooler AM, Kaput J, Kussmann M. 2016. A systems approach to personalised nutrition: Report on the Keystone Symposium “Human Nutrition, Environment and Health”. Appl Transl Genom 10: 16–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Manach C, Williamson G, Morand C, Scalbert A, Remesy C. 2005. Bioavailability and bioefficacy of polyphenols in humans. I. Review of 97 bioavailability studies. Am J Clin Nutr 81: 230S–42S [DOI] [PubMed] [Google Scholar]
- 66.Martin CK, Bhapkar M, Pittas AG, Pieper CF, Das SK, et al. 2016. Effect of Calorie Restriction on Mood, Quality of Life, Sleep, and Sexual Function in Healthy Nonobese Adults: The CALERIE 2 Randomized Clinical Trial. JAMA Intern Med 176: 743–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Masters JR, Stacey GN. 2007. Changing medium and passaging cell lines. Nat Protoc 2: 2276–84 [DOI] [PubMed] [Google Scholar]
- 68.Matsuda M, DeFronzo RA. 1999. Insulin sensitivity indices obtained from oral glucose tolerance testing: comparison with the euglycemic insulin clamp. Diabetes Care 22: 1462–70 [DOI] [PubMed] [Google Scholar]
- 69.McCann JC, Shigenaga MK, Mietus-Snyder ML, Lal A, Suh JH, et al. 2015. A multicomponent nutrient bar promotes weight loss and improves dyslipidemia and insulin resistance in the overweight/obese: chronic inflammation blunts these improvements. FASEB J 29: 3287–301 [DOI] [PubMed] [Google Scholar]
- 70.Monteiro JP, Kussmann M, Kaput J. 2015. The genomics of micronutrient requirements. Genes Nutr 10: 466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Muniyappa R, Lee S, Chen H, Quon MJ. 2008. Current approaches for assessing insulin sensitivity and resistance in vivo: advantages, limitations, and appropriate usage. Am J Physiol Endocrinol Metab 294: E15–26 [DOI] [PubMed] [Google Scholar]
- 72.Norris KM, Okie W, Kim WK, Adhikari R, Yoo S, et al. 2016. A high-fat diet differentially regulates glutathione phenotypes in the obesity-prone mouse strains DBA/2J, C57BL/6J, and AKR/J. Nutr Res [DOI] [PubMed] [Google Scholar]
- 73.Nunes-Alves C 2016. Microbiome: Microbiota-based nutrition plans. Nat Rev Microbiol 14: 1. [DOI] [PubMed] [Google Scholar]
- 74.Olson CM. 2016. Behavioral Nutrition Interventions Using e- and m-Health Communication Technologies: A Narrative Review. Annu Rev Nutr 36: 647–64 [DOI] [PubMed] [Google Scholar]
- 75.Patrick K, Raab F, Adams MA, Dillon L, Zabinski M, et al. 2009. A text message-based intervention for weight loss: randomized controlled trial. J Med Internet Res 11: e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pirmohamed M 2001. Pharmacogenetics and pharmacogenomics. Br J Clin Pharmacol 52: 345–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Prentice RL, Caan B, Chlebowski RT, Patterson R, Kuller LH, et al. 2006. Low-fat dietary pattern and risk of invasive breast cancer: the Women’s Health Initiative Randomized Controlled Dietary Modification Trial. JAMA 295: 629–42 [DOI] [PubMed] [Google Scholar]
- 78.Punja S, Bukutu C, Shamseer L, Sampson M, Hartling L, et al. 2016. N-of-1 trials are a tapestry of heterogeneity. J Clin Epidemiol 76: 47–56 [DOI] [PubMed] [Google Scholar]
- 79.Punja S, Xu D, Schmid CH, Hartling L, Urichuk L, et al. 2016. N-of-1 trials can be aggregated to generate group mean treatment effects: a systematic review and meta-analysis. J Clin Epidemiol 76: 65–75 [DOI] [PubMed] [Google Scholar]
- 80.Rahe C, Unrath M, Berger K. 2014. Dietary patterns and the risk of depression in adults: a systematic review of observational studies. Eur J Nutr 53: 997–1013 [DOI] [PubMed] [Google Scholar]
- 81.Reed DR. 2008. Animal models of gene-nutrient interactions. Obesity (Silver Spring) 16 Suppl 3: S23–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Renfro LA, Sargent DJ. 2016. Statistical controversies in clinical research: basket trials, umbrella trials, and other master protocols: a review and examples. Ann Oncol [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ritchie MD. 2012. The success of pharmacogenomics in moving genetic association studies from bench to bedside: study design and implementation of precision medicine in the post-GWAS era. Hum Genet 131: 1615–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rochon J 1990. A statistical model for the “N-of-1” study. J Clin Epidemiol 43: 499–508 [DOI] [PubMed] [Google Scholar]
- 85.Roses AD. 2000. Pharmacogenetics and the practice of medicine. Nature 405: 857–65 [DOI] [PubMed] [Google Scholar]
- 86.Schork NJ. 2015. Personalized medicine: Time for one-person trials. Nature 520: 609–11 [DOI] [PubMed] [Google Scholar]
- 87.Scriver CR. 2007. The PAH gene, phenylketonuria, and a paradigm shift. Hum Mutat 28: 831–45 [DOI] [PubMed] [Google Scholar]
- 88.Scuffham PA, Nikles J, Mitchell GK, Yelland MJ, Vine N, et al. 2010. Using N-of-1 trials to improve patient management and save costs. J Gen Intern Med 25: 906–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Senn S 2002. Cross-over Trials in Clinical Research, 2nd edition New York: Wiley; 364 pp. [Google Scholar]
- 90.Sesso HD, Buring JE, Christen WG, Kurth T, Belanger C, et al. 2008. Vitamins E and C in the prevention of cardiovascular disease in men: the Physicians’ Health Study II randomized controlled trial. JAMA 300: 2123–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Seto T, Kato T, Nishio M, Goto K, Atagi S, et al. 2014. Erlotinib alone or with bevacizumab as first-line therapy in patients with advanced non-squamous non-small-cell lung cancer harbouring EGFR mutations (JO25567): an open-label, randomised, multicentre, phase 2 study. Lancet Oncol 15: 1236–44 [DOI] [PubMed] [Google Scholar]
- 92.Shull PB, Jirattigalachote W, Hunt MA, Cutkosky MR, Delp SL. 2014. Quantified self and human movement: a review on the clinical impact of wearable sensing and feedback for gait analysis and intervention. Gait Posture 40: 11–9 [DOI] [PubMed] [Google Scholar]
- 93.Simon R 1977. Adaptive treatment assignment methods and clinical trials. Biometrics 33: 743–9 [PubMed] [Google Scholar]
- 94.Smits SA, Marcobal A, Higginbottom S, Sonnenburg JL, Kashyap PC. 2016. Individualized Responses of Gut Microbiota to Dietary Intervention Modeled in Humanized Mice. mSystems 1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Studies N-NWGoRiA, Chanock SJ, Manolio T, Boehnke M, Boerwinkle E, et al. 2007. Replicating genotype-phenotype associations. Nature 447: 655–60 [DOI] [PubMed] [Google Scholar]
- 96.Stumbo PJ, Weiss R, Newman JW, Pennington JA, Tucker KL, et al. 2010. Web-enabled and improved software tools and data are needed to measure nutrient intakes and physical activity for personalized health research. J Nutr 140: 2104–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Stumvoll M, Mitrakou A, Pimenta W, Jenssen T, Yki-Jarvinen H, et al. 2000. Use of the oral glucose tolerance test to assess insulin release and insulin sensitivity. Diabetes Care 23: 295–301 [DOI] [PubMed] [Google Scholar]
- 98.Tang SMM IB 1993. The Effect of Serial Correlation on Tests for Parameter Change at Unknown Time. The Annals of Statistics 21: 552–75 [Google Scholar]
- 99.Tiemeier H, van Tuijl HR, Hofman A, Meijer J, Kiliaan AJ, Breteler MM. 2002. Vitamin B12, folate, and homocysteine in depression: the Rotterdam Study. Am J Psychiatry 159: 2099–101 [DOI] [PubMed] [Google Scholar]
- 100.Tiscornia G, Vivas EL, Izpisua Belmonte JC. 2011. Diseases in a dish: modeling human genetic disorders using induced pluripotent cells. Nat Med 17: 1570–6 [DOI] [PubMed] [Google Scholar]
- 101.Uryniak TC I; Fedorov V; Jiang Q 2011. Responder Analyses - A PhRMA Position Paper. Statistics in Biopharmaceutical Research 3: 476–87 [Google Scholar]
- 102.van Ommen B, El-Sohemy A, Hesketh J, Kaput J, Fenech M, et al. 2010. The Micronutrient Genomics Project: a community-driven knowledge base for micronutrient research. Genes Nutr 5: 285–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Vesnic-Alujevic L, Breitegger M, Guimaraes Pereira A. 2016. ‘Do-It-Yourself’ Healthcare? Quality of Health and Healthcare Through Wearable Sensors. Sci Eng Ethics [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Williamson G, Manach C. 2005. Bioavailability and bioefficacy of polyphenols in humans. II. Review of 93 intervention studies. Am J Clin Nutr 81: 243S–55S [DOI] [PubMed] [Google Scholar]
- 105.Zeevi D, Korem T, Zmora N, Israeli D, Rothschild D, et al. 2015. Personalized Nutrition by Prediction of Glycemic Responses. Cell 163: 1079–94 [DOI] [PubMed] [Google Scholar]
- 106.Zheng YL, Ding XR, Poon CC, Lo BP, Zhang H, et al. 2014. Unobtrusive sensing and wearable devices for health informatics. IEEE Trans Biomed Eng 61: 1538–54 [DOI] [PMC free article] [PubMed] [Google Scholar]