Members of the Competibacter lineage are relevant in biotechnology as glycogen-accumulating organisms (GAOs). Here, we document the presence of three Competibacteraceae clades in a full-scale activated sludge wastewater treatment plant and their linkage to specific operational conditions. We find evidence for niche differentiation among the three clades with temporal variability in clade dominance that correlates with operational changes at the treatment plant. Specifically, we observe episodic dominance of a likely motile clade during periods of elevated effluent turbidity, as well as episodic dominance of closely related nonmotile clades that likely enhance floc formation during periods of low effluent turbidity.
KEYWORDS: activated sludge, Competibacteraceae, niche differentiation, wastewater treatment, metagenome, qPCR
ABSTRACT
Multiple clades within a microbial taxon often coexist within natural and engineered environments. Because closely related clades have similar metabolic potential, it is unclear how diversity is sustained and what factors drive niche differentiation. In this study, we retrieved three near-complete Competibacter lineage genomes from activated sludge metagenomes at a full-scale pure oxygen activated sludge wastewater treatment plant. The three genomes represent unique taxa within the Competibacteraceae. A comparison of the genomes revealed differences in capacity for exopolysaccharide (EPS) biosynthesis, glucose fermentation to lactate, and motility. Using quantitative PCR (qPCR), we monitored these clades over a 2-year period. The clade possessing genes for motility and lacking genes for EPS biosynthesis (CPB_P15) was dominant during periods of suspended solids in the effluent. Further analysis of operational parameters indicate that the dominance of the CPB_P15 clade is associated with low-return activated sludge recycle rates and low wasting rates, conditions that maintain relatively high levels of biomass within the system.
IMPORTANCE Members of the Competibacter lineage are relevant in biotechnology as glycogen-accumulating organisms (GAOs). Here, we document the presence of three Competibacteraceae clades in a full-scale activated sludge wastewater treatment plant and their linkage to specific operational conditions. We find evidence for niche differentiation among the three clades with temporal variability in clade dominance that correlates with operational changes at the treatment plant. Specifically, we observe episodic dominance of a likely motile clade during periods of elevated effluent turbidity, as well as episodic dominance of closely related nonmotile clades that likely enhance floc formation during periods of low effluent turbidity.
INTRODUCTION
Activated sludge is the largest application of biotechnology in the world (1). It has achieved this status because under normal operating conditions, it is a robust and flexible method of treating wastewater that can reliably meet effluent regulatory standards. Organic and nitrogenous contaminants are removed from water by establishing aerobic and anoxic regimes within bioreactors that are favorable for the growth of microbial communities that consume waste organic matter and remove nutrients (2). The resulting biomass is settled in clarifiers and recirculated to enable high volumetric rates of contaminant removal. An important operational variable is the biomass wasting rate, which sets a minimum growth rate needed for a species to remain within the system, providing some control over microbial community structure and function. An additional operational control is the ability to change the rates at which settled biomass is recirculated back to the bioreactors. Both contaminant degradation and efficient settling of biomass are needed for consistent and effective process performance.
The Competibacteraceae family is a group of gammaproteobacteria that are favored under conditions of alternating feast and famine (3). Within this family, the Competibacter lineage is a diverse collection of uncultivated organisms which function as glycogen-accumulating organisms (GAOs) that cycle the intracellular storage polymers polyhydroxyalkanoate (PHA) and glycogen under anaerobic-aerobic conditions. This metabolism differs from that of polyphosphate-accumulating organisms (PAOs), which accumulate polyphosphate under aerobic conditions and release it under anaerobic conditions. In fact, GAOs are often viewed as competitors to PAOs in enhanced biological phosphorus removal systems in that they consume carbon substrate without accumulation of phosphorus (3).
To date, 13 clades in the Competibacter lineage have been identified (4). They have been found in lab- and full-scale activated sludge plants, as well as in soil and sediment samples (4). Several clades are observed to coexist within a plant, although the temporal dynamics of each clade may vary substantially (5, 6). Due to the lack of cultured representatives, little is known about factors leading to niche partitioning or temporal dynamics within Competibacter clades. However, recent advances in metagenomic binning and recovery of genomes may enable an assessment of metabolic potential even in the absence of cultivation. The genomes for two members of this lineage, “Candidatus Competibacter denitrificans” and “Candidatus Contendobacter odensis,” obtained from lab-scale reactors in Denmark show significant differences in metabolic potential (7), and entries of additional genomes extracted from metagenomes in public databases will no doubt further expand our understanding of the metabolic diversity of these organisms.
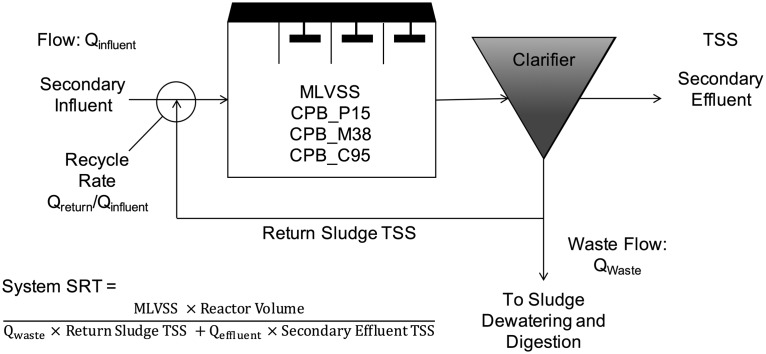
Here, we document the presence of three Competibacteraceae clades at a local wastewater treatment plant in northern California and report on the recovery of three genomes for these organisms. This plant operates a four-stage pure-oxygen activated sludge system, with the first stage acting as an anaerobic selector (Fig. 1). We highlight genetic differences related to floc formation and dispersal and quantitatively monitored these groups over a 2-year time period from January 2014 to December 2015. A correlation with operational parameters suggests that one of these clades may contribute to floc dispersal under conditions of high solids loading in the clarifier. Overall, our results are consistent with previous studies of ecological interactions and possible niche partitioning between Competibacteraceae clades in surveys of full-scale activated sludge plants in Denmark (4–6).
FIG 1.
Schematic of wastewater treatment plant in this study with operational parameters used in statistical analysis. Note that secondary treatment consists of a four-stage process, where the first stage is unaerated and thus operates anaerobically; the remaining three stages are aerated with pure oxygen as an input. TSS, total suspended solids (in milligrams per liter); MLVSS, mixed liquor volatile suspended solids (in milligrams VSS per liter); SRT, solids retention time (in days); Qeffluent = Qinfluent − Qwaste.
RESULTS
Identification of Competibacteraceae phylotypes in secondary treatment.
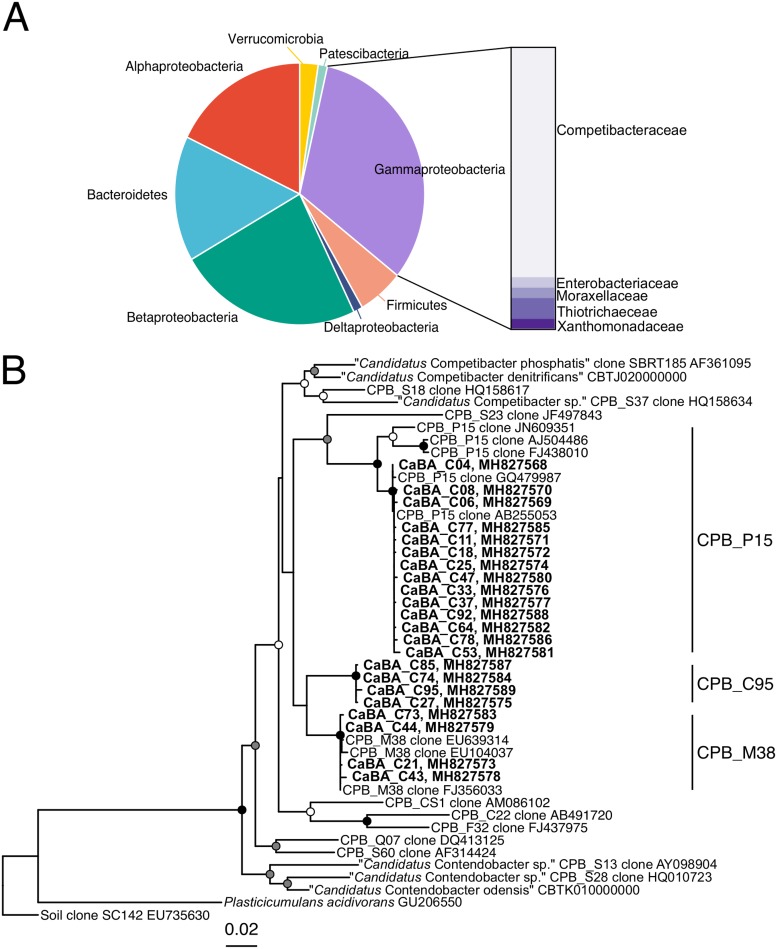
In conducting routine microscopic monitoring of activated sludge, the treatment plant staff observed fluctuations in the abundance of a visually distinct microbe, a nonfilamentous large coccoid cell (3 to 5 μm) present both as part of the floc and as dispersed cells (D. Jenkins, personal communication). Because this cell type occasionally appeared to constitute >50% of the floc by microscopy, we investigated its identity. A 16S rRNA clone library was constructed from a sample taken on 19 September 2013. Partial 16S rRNA sequences were recovered and found to consist of Proteobacteria (Fig. 2A). Over 25% of the sequences recovered were from the gammaproteobacterial family Competibacteraceae, a group that contains cells with a large coccoid morphology previously detected in lab-scale and full-scale activated sludge systems (4, 9). Y. Yang, a researcher at the plant, confirmed the presence of this group using fluorescence in situ hybridization (unpublished data, personal communication).
FIG 2.
16S rRNA clone library results. (A) Phylum-level (or class-level for Proteobacteria) distribution of 84 clones from a sample collected on 19 September 2013. Inset, family-level identification of gammaproteobacteria. (B) Phylogenetic tree of 22 Competibacteraceae sequences. Sequences in bold and listed as “CaBA” are from this study (California Bay Area). White circles at nodes, ≥50% bootstrap support; gray circles, ≥70% bootstrap support; black circles, ≥90% bootstrap support. Clades for the sequences in this study are labeled on the right. Accession numbers are listed for each sequence.
Further examination of the 22 recovered Competibacteraceae sequences revealed the coexistence of three Competibacteraceae clades. The recovered sequences were less than 97% similar, indicating the presence of three distinct species. The addition of these species to the phylogenetic tree proposed by McIlroy et al. (4) indicated that these lineages can be classified as CPB_P15, CPB_M38, and a novel clade within the Competibacter lineage for which we propose the name CPB_C95 (Fig. 2B). Attempts to cultivate these organisms were unsuccessful. Accordingly, we pursued genome recovery and quantitative monitoring of these three groups.
Genome recovery from the metagenome.
Using the differential coverage approach (10), we recovered three Competibacter lineage genomes from four metagenome samples from 2013 to 2015. From two early samples from 25 March 2014 and 19 September 2013, we obtained genome CA14.1. Although two other putative Competibacter lineage bins were observed in the coassembly of these samples (see Fig. S1A in the supplemental material), the low coverage of these groups and the dominance of a closely related species resulted in low-quality draft genomes. Reads mapping to these groups were instead binned in CA23.1 and CA23.3 using the coassembly of samples from 21 July and 27 October 2015 (Fig. S1B). The statistics for the three genomes are given in Table 1. These genomes are >93% complete and contain <5% contamination, as determined by CheckM (11). Average nucleotide identity (ANI) values for pairwise genome comparisons between these three genomes ranged from 76.2 to 77.6% (Table S1 and Fig. S2), which is well below the suggested 95% cutoff value for species delineation (12). Pairwise comparison to all publicly available Competibacteraceae genomes in GenBank also suggested that these genomes are unique species from what has previously been published, with the highest ANI values (77.2% to 79.4%) to the genome of “Candidatus Competibacter” sp. strain UBA3908 (Table 2), which was recovered from a Danish anaerobic digester metagenome sequencing project (13) (BioProject number PRJEB10932).
TABLE 1.
Genome statistics for the Competibacteraceae genomes
| Characteristica | Statistic for genome: |
||
|---|---|---|---|
| CPB_P15 CA14.1 | CPB_M38 CA23.2 | CA23.1 | |
| Genome size (Mbp) | 3.51 | 3.67 | 3.4 |
| No. of contigs | 30 | 143 | 278 |
| Contig N50 (bp) | 259,679 | 44,707 | 25,257 |
| Maximum contig size (bp) | 865,998 | 216,802 | 86,896 |
| Completeness (%) | 96.7 | 97.2 | 93.6 |
| Contamination (%) | 3.2 | 3.7 | 3.8 |
| GC content (%) | 58.0 | 61.5 | 62.3 |
| No. of CDSs | 3,158 | 3,307 | 3,108 |
| Protein-coding density (%) | 90.5 | 90.2 | 90.8 |
| Metagenome coverage by sample date | |||
| 19-09-2013 | 260 | 60 | 90 |
| 25-03-2014 | 3,550 | 7 | 40 |
| 21-07-2015 | 60 | 510 | 580 |
| 27-10-2015 | 20 | 460 | 100 |
Completeness and contamination were estimated using CheckM (11). CDSs, coding sequences.
TABLE 2.
Overview of annotated pathways in Competibacteraceae genomes
| Pathwaya | CPB_P15 CA14.1 | CPB_M38 CA23.2 | CA23.1 | “Candidatus Competibacter denitrificans”b | “Candidatus Contendobacter odensis”b |
|---|---|---|---|---|---|
| Carbohydrate metabolism | |||||
| TCA cycle | + | + | incc | + | + |
| Glyoxylate bypass | + | + | inc | + | + |
| Pentose phosphate (nonoxidative) | + | + | + | + | + |
| Glycolysis EMP pathway | + | + | + | + | + |
| Glycolysis ED pathway | − | − | − | + | − |
| Fermentation (glucose to lactate) | − | + | + | + | − |
| Storage compounds | |||||
| Glycogen synthesis | + | + | + | + | + |
| Trehalose synthesis | + | + | − | − | + |
| PHA synthesis | + | + | + | + | + |
| TAG synthesis | − | − | − | − | + |
| Nitrogen metabolism | |||||
| Nitrogen fixation | + | + | + | − | + |
| Nitrate reduction to nitrite | + | + | + | + | + |
| Nitrite reduction (respiratory) | − | − | − | + | − |
| Nitrate reduction to ammonia (dissimilatory) | + | + | inc | inc | + |
| Motility and dispersion | |||||
| Flagellar motility | + | − | − | + | + |
| Flagellum | + | − | − | + | + |
| EPS cluster | − | − | + | inc | inc |
| Other genes of interest | |||||
| NiFe-hydrogenase | + | − | − | + | + |
EMP, Embden-Meyerhoff-Parnassus; ED,Entner-Doudoroff; TAG, triacylglycerol.
Genomes from McIlroy et al. (7).
inc, pathway is incomplete.
Phylogenetic analysis of the Competibacteraceae genomes.
To assign species taxonomically, we searched for the 16S rRNA gene in genome bins. Only bin CA14.1 contained a full-length 16S rRNA gene sequence. This was associated with the CPB_P15 clade identified with the clone library. A partial 16S rRNA gene was recovered from bin CA23.2 that aligned with the CPB_M38 sequences. Only the CA23.1 bin lacked an associated 16S rRNA gene. However, examination of operational taxonomic unit (OTU) tables constructed from 16S rRNA genes from the metagenome suggested three Competibacteraceae groups within these samples (our unpublished data). Based on the coverage of CA23.1 in the various samples, we suspect that this bin represents the CPB_C95 clade.
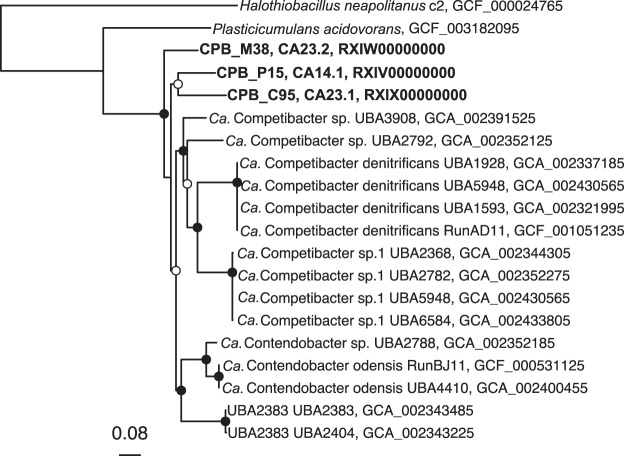
We also examined a phylogenomic tree of concatenated protein sequences to examine where the three California genomes fit in the Competibacteraceae family. According to the Genome Taxonomy Database, there are now 20 genomes for this family (14), of which 15 were available on NCBI. Many of these do not contain 16S rRNA genes. Using either the 120 bacterial marker set from Parks et al. (13) (Fig. 3) or a set of 56 core proteins found in all 18 Competibacter lineage genomes (data not shown), we found that all three genomes are unique and distinctive. The placement of the genomes as closely related groups distinctive from other clades is consistent with the 16S rRNA tree placement of the CPB_P15, CPB_M38, and CPB_C95 groups (Fig. 2B).
FIG 3.
Phylogenomic tree of Competibacteraceae family using the bac120 (13) single-copy genes. Protein-coding sequences were extracted and concatenated in Anvi’o version 4 (56) and were masked with default parameters in Gblocks (58). The tree was made with PhyML (33) using the Le and Gascuel substitution matrix (59), with 100 bootstraps. The genome for Halothiobacillus neapolitanus strain c2 was used as outgroup. White circles at nodes represent ≥50% bootstrap support; black circles, ≥90% bootstrap support. Assembly numbers are provided on the right.
Major metabolic pathways in the California genomes.
McIlroy et al. previously described the metabolism of the Competibacter lineage based on the genomes of “Ca. Competibacter denitrificans” and “Ca. Contendobacter odensis” (7) and documented the capacity for glycogen and PHA accumulation. We examined whether the California genomes contained pathways similar to those identified in the Danish genomes. Not surprisingly, the major metabolic pathways were largely identical, in that genes for glycogen synthesis and PHA synthesis were present in all three genomes, as were genes for the Embden-Meyerhoff-Parnassus glycolysis pathway and the nonoxidative branch of the pentose phosphate pathway (Table 2). Genes were also present for the tricarboxylic acid (TCA) cycle and glyoxylate bypass, although one block for each of these pathways was missing in the CA23.1 bin. We note that this genome is the least complete of the three (Table 1), and these may be among the genes that are missing from the assembly.
In examining the major metabolic pathways, the California genomes appeared to share more pathways in common with “Ca. Contendobacter odensis” than with “Ca. Competibacter denitrificans.” All three California genomes have a complete set of nitrogen fixation genes, but none have the genes required for denitrification from nitrite to nitrogen. The recovered genomes also lack genes for the Entner-Doudoroff pathway for glycolysis.
Genetic differences among the three clades.
Because the three clades occupy a similar niche, we were interested in genetic differences that could enable functional differentiation. The CPB_P15, CPB_M38, and CA23.1 genomes contained 875, 1,030, and 1,017 unique genes relative to the two other genomes, respectively. Two of the three genomes, CPB_M38 and CA23.1, possess the ability to ferment glucose to lactate under anaerobic conditions, a capability shared with the two Danish genomes. CPB_P15 lacked this ability but contains a NiFe-hydrogenase previously annotated in the Danish genomes (7). This hydrogenase is missing in the other two California genomes.
In addition to metabolic properties, properties related to floc formation and settling are also critical for wastewater treatment and could enable niche differentiation among related species in activated sludge systems. In this context, a gene cluster for exopolysaccharide (EPS) genes has recently been identified for the floc-forming organisms Zoogloea resiniphila strain MMB (15) and Aquincola tertiaricarbonis strain RN12 (16). This cluster is present in several gammaproteobacteria, and we detected a cluster of orthologous genes in the CA23.1 bin that was unique to this genome (Table 2). “Ca. Contendobacter odensis” contained a similar cluster of genes, although it is unclear if the genes are colocalized. The full set of related proteins appears to be part of multiple contigs. “Ca. Competibacter denitrificans” contained two partial clusters of genes. The presence of these groups as minor members of activated sludge may be important for the maintenance of desirable flocculation and settling properties.
Further inspection of the genomes revealed that the CPB_P15 genome contained a cluster of taxis-related genes not present in either the CPB_M38 or CA23.1 genome, encoding the assembly and regulation of flagella. Although CPB_M38 and CA23.1 also included some taxis-related genes, including methyl-accepting chemotaxis proteins and the chemotaxis protein CheY, genes for flagellar motility were notably absent. Flagellar motility is not unique to the CPB_P15 genome in the Competibacteraceae family, as genes were present in both the “Ca. Competibacter denitrificans” and “Ca. Contendobacter odensis” genomes (7). Isolates in the Plasticicumulans genus (a second lineage of the Competibacteraceae family) also have flagella (17, 18).
Development of a quantitative assay for Competibacteraceae clades.
To assess how genetic differences among the three Competibacteraceae clades might influence process performance in the activated sludge plant, we developed a monitoring assay for each of the three clades. Primers were developed based on the 22 near-full-length 16S rRNA sequences of Competibacteraceae within the plant for TaqMan quantitative PCR (qPCR) assays targeting each of the three clades (Table 3). An in-house script identified regions of both high and low similarity for these three groups. Primer sets were validated with plasmid DNA containing the 16S rRNA gene sequence using known concentrations. Standard curves produced a linear response from 101 to 107 copies for each of the three clades (Fig. S3), with nontarget Competibacteraceae sequences producing 100-fold lower sequence detection at high concentrations.
TABLE 3.
Parameters for Competibacteraceae qPCR assay
| Assaya | Standard | Primerb | Sequence (5′→3′) | Amplicon size (bp) | R2 | Linear regressionc |
Efficiency (%) | |
|---|---|---|---|---|---|---|---|---|
| m | b | |||||||
| CPB_ P15 | CaBA clone C25 | C25_f1 | GTAGGAATTGGCCCACGAGT | 109 | 0.99 | −3.07 | 35.00 | 112 |
| C25_r1 | CTTGTCCACCAGCGCGA | |||||||
| C25C95_TaqMan | ATGCGGTATTAGCCTGGGTTTCCC | |||||||
| CPB_M38 | CaBA clone C21 | C21_f1 | TAGGAATCTGCCCTGCAGA | 116 | 0.99 | −3.42 | 38.47 | 96 |
| C21_r1 | GACGTAGGCTCCTCCCA | |||||||
| C21_TaqMan | CTCTCTCGAGCGCATGCGGTATT | |||||||
| CPB_C95 | CaBA clone C95 | C95_f1 | CGTAGGAATCTGCCTCGTAGT | 118 | 0.99 | −3.35 | 37.68 | 99 |
| C95_r1 | GACGTAGGCTCATCTCATAGC | |||||||
| C2595_TaqMan | ATGCGGTATTAGCCTGGGTTTCCC | |||||||
Assay targets a subgroup of these clades found at the northern California wastewater treatment plant.
TaqMan probe labeled at 5′ end with 56-FAM, 3′ end with 3IABkFQ, and after position 9 with ZEN.
Values of linear regression for standard curve are given as slope (m) and y-intercept (b).
Comparison of these probes to other clades not found at this plant indicated that although some of the primers may hybridize multiple groups, the combination of primers is specific for a subgroup within CPB_P15, CPB_M38, and CPB_C95.
Temporal dynamics of Competibacteraceae.
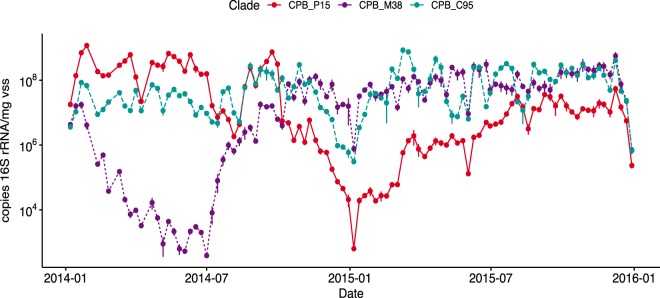
To gain insight into the temporal dynamics of Competibacteraceae populations, we used qPCR to analyze shifts in the abundances of the three clades over a 2-year study period. The results were normalized to milligrams of volatile suspended solids (VSS) and are shown in Fig. 4. Consistent with microscopy results, Competibacteraceae were detected in all samples. Concentrations ranged from 106 to 109 copies of 16S rRNA/mg VSS for the total population and from 102 to 109 copies of 16S rRNA/mg VSS for individual clades. Each clade demonstrated remarkably different dynamics, as follows: the CPB_P15 clade was abundant during most of 2014 but decreased significantly in abundance in the last quarter of that year; subsequently, the other two clades became more prominent; and the CPB_C95 clade varied the least across the 2-year time period (Fig. 4).
FIG 4.
Quantitative PCR results for three Competibacteraceae clades present in the northern California wastewater treatment plant.
Linkage between plant monitoring data and Competibacteraceae community composition.
The only correlation previously detected by other researchers between Competibacteraceae and operational parameters was a linkage to wastewater containing industrial streams (5). This analysis was based on fluorescence in situ hybridization with a general Competibacter lineage probe and was thus unable to identify correlations to individual clades. To determine whether process performance impacts could be attributable to genetic differences among the Competibacteraceae, we correlated Competibacteraceae clade abundance with measured plant operational conditions and environmental parameters across a 2-year study period (see Table S2 for a summary of parameters). Using the Spearman rank correlation coefficient across specific clades and operating parameters, we found that the concentration of the CPB_P15 clade was positively correlated with increased total suspended solids (TSS) and VSS in the secondary system, including TSS in the secondary effluent, but had a slight negative correlation with effluent pH and recycle ratio (Table 4). Influent flow, influent carbonaceous biochemical oxygen demand (cBOD), and solids retention time (SRT) had no significant correlation for CPB_P15. In contrast, the CPB_M38 clade was negatively correlated with the CPB_P15 clade, suggesting a possible competitive interaction. The CPB_M38 clade was negatively correlated with SRT and influent flow, and, in agreement with its negative correlation with CPB_P15, was also negatively correlated to TSS and VSS in the mixed liquor and to TSS in the return sludge, but was positively correlated to effluent pH. The novel CPB_C95 had fewer correlations but was positively correlated with the CPB_M38 clade.
TABLE 4.
Spearman’s rank correlation coefficient between Competibacteraceae clade abundance and environmental variables
| Variable | Spearman's rank correlation coefficient |
||
|---|---|---|---|
| CPB_P15 | CPB_M38 | CPB_C95 | |
| Primary TSS | 0.224a | −0.123 | 0.116 |
| Secondary TSS | 0.534c | −0.252a | 0.044 |
| Mixed liquor TSS | 0.443c | −0.374c | 0.029 |
| Return activated sludge TSS | 0.504c | −0.378c | 0.022 |
| Effluent TSS | 0.510c | −0.216 | 0.102 |
| Flow | −0.012 | −0.384c | −0.318b |
| Recycle rate | −0.245a | 0.111 | 0.005 |
| SRT | −0.017 | −0.314b | −0.164 |
| MLVSS | 0.508c | −0.323b | 0.036 |
| Effluent pH | −0.234a | 0.306b | −0.013 |
| Influent cBOD | 0.089 | 0.154 | 0.118 |
| Effluent cBOD | 0.323b | −0.107 | 0.084 |
| Large coccoid cells | 0.650c | −0.220 | 0.405c |
| CPB_P15 | 1 | −0.402c | 0.162 |
| CPB_M38 | −0.402c | 1 | 0.516c |
| CPB_C95 | 0.162 | 0.516c | 1 |
For the Spearman rank correlation coefficient, significant correlation with a P value of <0.05.
For the Spearman rank correlation coefficient, significant correlation with a P value of <0.01.
For the Spearman rank correlation coefficient, significant correlation with a P value of <0.001.
Although no correlation was observed for CPB_P15 and flow, a significant drop in all three monitored Competibacteraceae clades occurred in December 2014. This coincided with wet-weather events and an increase in flow rates into the plant. It is possible that a hydraulic overload of the secondary system occurred at this time which may have washed out these populations.
Putative linkages to process performance.
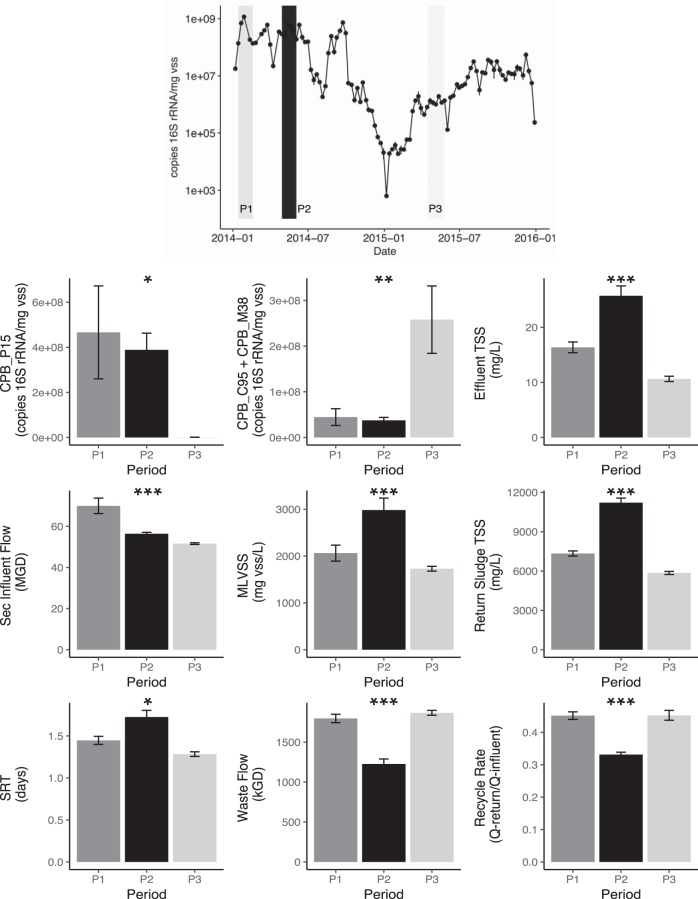
During our study period, process performance was maintained, and the plant remained in compliance with all regulatory limits. However, it is interesting to note that of the three Competibacteraceae clades present in the plant, one clade (CPB_P15) was significantly correlated with secondary effluent TSS (P < 0.001), which fluctuated from 5 to 44 mg/liter, with an average of 14.6 ± 6.7 mg/liter (Table S2). This same clade also demonstrated clear dominance over the other two clades early in 2014 (Fig. 4). Because the Competibacteraceae family was originally identified in the plant due to its observed dispersion from the floc in mixed liquor samples, this group, and specifically, the CPB_P15 clade, could putatively lead to elevated secondary effluent TSS. A comparison of CPB_P15 dynamics and effluent TSS indicated an extended period of time in the first half of 2014 when the population abundance tracked with increases in secondary TSS levels. However, increases in this subgroup did not always coincide with rises in secondary effluent TSS. To determine whether specific parameters might influence the role of CPB_P15 in elevating secondary effluent TSS, we examined which operational parameters (see Fig. 1 for a schematic of sampling sites) significantly differed for the following three time periods: (i) a period when CPB_P15 was dominant and effluent TSS levels were high, (ii) a period when CPB_P15 was dominant but effluent TSS levels remained low, and (iii) a period when both CPB_P15 and TSS levels in the effluent were low (Fig. 5). Examination of the variance in the means of operational parameters during these periods using multivariate analysis of variance (MANOVA) implicated operation at a lower recycle rate and higher concentration of solids in the mixed liquor and return activated sludge in periods with both high Competibacteraceae and high secondary effluent TSS (P < 0.001) (Fig. 5). During the period of high CPB_P15 and high TSS, the secondary system was operating at its peak mixed liquor VSS concentrations for the 2-year period, and the overall TSS concentrations in the mixed liquor were also near their maximum (data not shown).
FIG 5.
Differences in operational and environmental parameters for three time periods representing stable and unstable conditions. Top panel, qPCR results for clade CPB_P15, as shown in Fig. 4. Gray bars represent three periods. Medium gray, period of high CPB_P15 but low-effluent TSS; dark gray/black, unstable period of high CPB_P15 and high-effluent TSS; light gray, period of low CPB_P15 and low-effluent TSS. Bottom, bar charts for various parameters showing the mean and standard error for each time period. Colors are as described above. *, P < 0.05 in MANOVA analysis; **, P < 0.01; ***, P < 0.001. Sec, secondary; kGD, 1,000 gallons per day.
DISCUSSION
Here, we present the recovery of three novel Competibacter lineage genomes from full-scale activated sludge samples and document changes in clade abundances over a 2-year study period. The three genomes include new genomes for clades CPB_P15 and CPB_M38 and one for a novel clade, CPB_C95. We also find evidence for niche differentiation among these three clades with temporal variability that correlates with operational changes at the treatment plant. Specifically, we observe episodic dominance of a likely motile clade, CPB_P15, during periods of elevated effluent turbidity, as well as episodic dominance of closely related nonmotile clades, CPB_M38 and CPB_C95, that likely enhance floc formation during periods of low effluent turbidity.
Although generally associated with enhanced biological phosphorus removal (EBPR), Competibacteraceae are found worldwide in sediments and activated sludge, including non-EBPR treatment plants (4, 5, 7, 9). The particular wastewater treatment plant under study is not operated for phosphorus removal but does include anaerobic-aerobic periods in its treatment train. In particular, this plant utilizes a four-stage secondary system, where the first stage is operated as an anaerobic selector. The influent to the first-stage selector includes primary treated wastewater and anaerobic digester centrate. This is followed by three stages of aeration and recirculation after settling, creating an anaerobic-aerobic cycling regime that is favorable for PHA accumulation (2, 19). Furthermore, because the system is a pure-oxygen system and is covered to prevent loss of oxygen to the atmosphere, carbon dioxide accumulates in the gas phase, thereby maintaining a pH of about 6.8. Lower pH below 7.25 reportedly favors GAOs, such as those of the Competibacteraceae family, over PAOs (3) and may explain selection for this group of organisms in this wastewater treatment plant. Across our 2-year study period, the pH ranged from 6.4 to 7.0 (Table S2), which would maintain selection pressure for GAOs over PAOs.
Within the Competibacteraceae family, three distinct clades were found whose abundances varied dramatically over the study period. Our observations are consistent with other reports on this family in lab- and full-scale activated sludge plants in Denmark, where multiple clades reportedly coexist and vary greatly both spatially and temporally (5, 6, 9). Interestingly, the California plant harbors Competibacter lineage clades that have not been detected at high abundances in other plants. Few 16S rRNA sequences are available for clade CPB_M38 (4), and only one sequence is present in the MiDAS database (20). In a recent survey of EBPR systems in Denmark, the distribution of clade CPB_M38 could not be examined due to the sequence limitation (6). This study provides additional sequences for CPB_M38 and for a proposed novel clade, CPB_C95, and documents their abundances over a 2-year time period.
The third clade identified in the Bay Area plant, CPB_P15, is a clade detected in treatment plants throughout Denmark (6) but not as a dominant clade. In this plant, the CPB_P15 clade was abundant in early 2014, and its increase coincided with elevated TSS in the secondary effluent. The population of CPB_P15 decreased dramatically at the end of 2014, and other clades increased in abundance. Given that the three organisms are closely related and rely on similar metabolisms, we surmise that they may be competing for a similar niche. The significant negative correlation with CPB_M38 supports this hypothesis. The shift in favor of CPB_M38 following a washout may have resulted in the exclusion of CPB_P15, preventing its recovery as dominant clade and suppressing its release into the effluent.
Operational parameters that characterized the period with high CPB_P15 and high effluent TSS include low recycle rates, high concentrations of mixed liquor VSS, and high concentrations of return sludge and low wasting rates. A reduction in recycle rate would imply a smaller fraction of time spent in the aerated portion of the system and the potential for increased accumulation of solids within the clarifier. This accumulation would be magnified by the higher solids loading on the clarifier due to maintenance of high mixed liquor VSS and higher concentration of return sludge TSS. With increased levels of solids in the clarifier, nutrients, such as oxygen, would be depleted more rapidly. We hypothesize that under these operational conditions, genes for flagellar motility may have been expressed, making the CPB_P15 organisms more likely to disperse from the floc and be released into the effluent. Clearly, additional laboratory analyses are needed to confirm this hypothesis. It is likely that there is a confluence of factors occurring during periods of elevated TSS in the clarifier effluent, as genes for flagellar motility are common within activated sludge communities and can contribute to the extracellular matrix, leading to adhesion of cells (21). Flagella are also present in other Competibacter lineage members, but poor settling has not been described. The lack of EPS-producing genes (see below) and hydrogenases may also play a role. Hydrogenases have been implicated in electron balance during anaerobic periods in the PAO “Candidatus Accumulibacter,” and hydrogen production has been observed (22). If CPB_P15 also produces hydrogen, gas production may disrupt integration within the floc, and motility may allow it to access metabolic resources that are unavailable to other floc members.
A factor contributing to the strength of the activated sludge flocs is the abundance of microorganisms that can produce EPS. The biofilm matrix consists of a mixture of polysaccharides, proteins, lipids, extracellular DNA, and other material that connect microbial cells into aggregates (21). In activated sludge, the production of EPS is essential for bioprocess stability, as it leads to formation of well-settling flocs that can be separated from the final effluent and recycled back into the aerated reactor. Recently, a highly conserved EPS biosynthesis gene cluster was identified in Zoogloea resiniphila strain MMB, where mutants in this cluster no longer exhibited a floc-forming phenotype (15). We identified an orthologous gene cluster in the CA23.1 bin, which we believe is the genome for the CPB_C95 clade in the California plant. Partial clusters were also identified in the genomes for “Ca. Competibacter denitrificans” and “Ca. Contendobacter odensis.” Given that Competibacteraceae have been implicated in the synthesis of the polysaccharide “granulan” in aerobic granules (23), further evaluation of these putative EPS gene clusters may provide deeper insight into the nature and stability of this polysaccharide.
In later time periods, CPB_C95 became the dominant Competibacteraceae clade. It is interesting to speculate that CPB_C95 and other EPS-producing organisms exhibited a beneficial role on floc formation, generating EPS that served to retain other organisms in the system. During periods of lower CPB_C95 abundance and higher CPB_P15 abundance, CPB_P15 may have been more likely to disperse. CPB_C95 was not correlated with effluent TSS. Given its ability to produce floc-forming EPS, we might expect a negative correlation. However, additional factors, including the presence of filamentous organisms and chemical and polymer addition, may obscure such a correlation. Continued examination of these genomes, and the retrieval of other genomes from these metagenomic samples, will no doubt provide deeper insight into such interactions, as will future studies examining EPS production and gene expression.
MATERIALS AND METHODS
Site description, sample collection, and operational parameters.
We obtained samples from a wastewater treatment plant in northern California. The plant can treat a maximum of 168 million gallons per day (MGD; 636,000 m3 day−1) with secondary treatment but has an average flow of 63 MGD (240,000 m3 day−1) from both domestic and industrial sources. Dissolved and colloidal organic removal occurs in a four-stage pure-oxygen aeration system, with the first stage operated as an anaerobic selector, prior to removal of microbial biosolids in a clarifier, disinfection of treated wastewater, and discharge into receiving waters. Thickened waste biosolids and high-strength anaerobically digestible material trucked in from offsite undergo anaerobic digestion for energy recovery, and digester centrate is combined with raw wastewater upstream of the primary sedimentation basin. The combined influent to the secondary treatment system therefore consists of raw influent domestic wastewater, anaerobic digester centrate, and filtrate from thickening of biosolids.
Plant staff routinely monitor several operational variables. Flow rates (secondary inflow, secondary outflow, and wasted sludge flow) are recorded with in-line flow meters, and secondary effluent pH is measured in a grab sample. Total suspended solids (TSS) are measured at various locations (influent, secondary influent, secondary effluent, mixed liquor, and waste return activated sludge), using Standard method 2540 D (24). Mixed liquor biosolids are measured as mixed liquor VSS (MLVSS), as per EPA 160.4 (25). Characterization of the dispersed large coccoid cells observed microscopically is per the method of D. Jenkins (26). For this study, because secondary TSS concentrations are reported weekly, but TSS loading in kilopounds (klbs) and secondary flow in million gallons per day are reported daily, TSS concentrations in milligrams per liter were computed from daily loading and flow data and were used for statistical analysis. These estimated values matched with the data reported weekly. Recycle rates were also calculated as the ratio of return flow to influent flow. Return flow was calculated based on influent flow, waste flow, and mass concentrations in the effluent, mixed liquor, and return activated sludge.
From January 2014 to December 2015, weekly grab samples of the activated sludge were collected from the secondary treatment system. Samples (2.0 ml) were centrifuged on-site, the supernatant was removed, and the samples were stored at −20°C until further use.
Clone library DNA extraction, PCR, cloning, and sequencing.
An activated sludge sample was collected on 19 September 2013 for microbial community analysis and identification of major genera. DNA was extracted using the Mo Bio PowerSoil DNA isolation kit (Carlsbad, CA), following the manufacturer’s directions. Near-full-length fragments (1,465 bp) of the 16S rRNA gene were amplified using the primer set 27F.1 (AGAGTTTGATCMTGGCTCAG) and 1492R (GGTTACCTTGTTACGACTT), using a protocol based on a study by Wells et al. (27). Briefly, four replicates of 20-μl PCRs were amplified at different annealing temperatures (48, 51.5, 55, and 58°C) and then pooled prior to cloning. Each PCR contained 300 nM each primer, 1× Fail-Safe PCR buffer F (Epicentre, Madison, WI), 1.25 U AmpliTaq LD Taq polymerase (Applied Biosystems, Foster City, CA), 0.1 μg/μl bovine serum albumin (BSA), and 20 to 50 ng of genomic DNA. The PCR temperature profile was as follows: 95°C for 3 min, and then 15 cycles of 95°C for 30 s, 48 to 55°C for 30 s, and 72°C for 60 s, followed by a final extension at 72°C for 7 min. Amplicons were pooled and gel extracted from a 1.5% agarose gel using the QIAquick gel extraction kit (Qiagen, Valencia, CA) and cloned using the pGEM-T Easy vector system with JM109 competent E. coli cells (Promega, Madison, WI), as per the manufacturer’s instructions. Randomly picked clones were sequenced using T7 and SP6 primers by MCLAB (San Francisco, CA), generating 84 partial 16S rRNA gene sequences. For a subset of these sequences that were identified as Competibacteraceae (see below), clones were sequenced using both the T7 and SP6 primers to yield near-full-length 16S rRNA gene sequences.
Phylogenetic analysis.
The 16S rRNA gene sequences were checked for chimeras using the online Bellerophon tool (28). Suspected chimeras were manually inspected and checked using BLASTN (29) to verify that they matched other sequences available in the database across the whole length of the fragment. Taxonomic identification was then performed concurrently with alignment of the partial 16S rRNA sequences using the online SINA alignment service (30).
Cloned sequences belonging to the Competibacteraceae family and closely related database sequences from McIlroy et al. (4) were aligned with the online SINA alignment service (30) and trimmed, and variable regions were removed using a custom filter from the ARB software (31) (filter by base frequency, 20% to 100%). The program ModelGenerator (32) was used to find an appropriate substitution model to convert the aligned sequences to a tree structure. Based on the Akaike information criterion (AIC), a phylogenetic tree was made using the PhyML (33) plug-in in Geneious version 10.2.2 (34) with the TN93 substitution model (35), with 100 bootstraps. The tree was visualized using the R package ggtree (36).
Metagenome DNA extraction and sequencing.
DNA was extracted from the MLVSS samples for 25 March 2014, 21 July 2015, and 27 October 2015 using the FastDNA Spin kit for soil (MP Biomedicals, Santa Ana, CA), as per the manufacturer’s instructions, with the exception of the initial bead-beating step. Samples were vortexed at maximum speed for 10 min using a Vortex Adapter (Mo Bio Laboratories, Inc., Carlsbad, CA) with the Vortex Genie 2T (Scientific Industries, Inc., Bohemia, NY) to lyse the samples. The extracted DNA was sent to Genewiz (South Plainfield, NJ) for sequencing. Paired-end (PE) (2 × 150 bp) reads were sequenced on the Illumina HiSeq 2500 platform (San Diego, CA) using the TruSeq PCR-free library preparation kit (Illumina). Additionally, the clone library for sample 19 September 2013 was sequenced by MCLAB (Hayward, CA), with 2 × 300-bp PE reads on the Illumina MiSeq platform.
Metagenome assembly and genome binning.
Metagenome assembly and binning followed the protocol for mmgenome (37) based on the differential coverage multimetagenome approach given by Albertsen et al. (10), with modifications for alternate software. First, paired-end reads were imported into the CLC Genomics Workbench version 8.0.3 (CLC bio, Qiagen) and trimmed using a quality limit of 0.04, no ambiguous nucleotides, a minimum read length of 50 bp, and removing adapters if found. Trimmed reads from samples 25 March 2014 and 19 September 2013 were then coassembled with MEGAHIT version 1.1.1 (38), with a k-min of 21, k-max of 100, and a k-step of 10, as used by Vollmers, Wiegand, and Kaster (39). Similarly, trimmed reads from samples 21 July 2015 and 27 October 2015 were coassembled.
The reads for each sample were independently mapped to scaffolds with Bowtie2 version 2.3.2 (40) and SAMtools version 1.3.1 (41) to generate coverage plots using the script calc.coverage.in.bam.depth.pl from the multimetagenome toolkit (10).
Contig information was obtained based on essential genes, 16S rRNA sequences, and scaffold classification. Essential gene prediction followed the pipeline for mmgenome (37) using Prodigal version 2.6.3 (42), HMMER 3.1b2 (43), BLAST (29), MEGAN version 5.11.3 (44), and mmgenome-associated perl scripts. Additionally, 16S rRNA genes were predicted using the workflow described in mmgenome; predicted 16S rRNA sequences were then classified using the online SINA aligner (30). Scaffolds were classified with PhyloPythiaS+ version 1.4 (45). All data were imported into the RStudio integrated development environment (IDE) (46) running R version 3.4.4 (47), and the mmgenome toolkit (37) was used to extract individual genome bins.
Paired-end reads were extracted from the binned scaffolds using extract.fastq.for.reassembly.pl (37) and were used for reassembly using SPAdes version 3.10.1 (48), with k values of 21, 33, 55, and 77, as recommended for PE 2 × 150-bp reads (48). Assembly coverage, GC content, and/or tetranucleotide frequencies were used for an additional round of screening with mmgenome, and assemblies were run through the online web tool for ACDC (49) to remove suspected contaminants. Genome completeness and contamination were estimated with CheckM (11).
Genome annotation and analysis.
Reassembled genomes were annotated with Prokka version 1.12 (50) and annotated with KEGG numbers using the online tool BlastKOALA (51) and the Rapid Annotations using Subsystems Technology (RAST) server (52); 16S rRNAs were predicted using RNAmmer (53).
Genomes for Competibacteraceae were downloaded from GenBank using the accession numbers from the Genome Taxonomy Database (14). Average nucleotide identity (ANI) was calculated with pyani (https://github.com/widdowquinn/pyani) using the BLAST method (29). Orthologous groups were also predicted using Proteinortho version 5.16b (54). The script po2group_stats.py (55) was used to determine the core genome based on the Proteinortho output.
Phylogenomic trees.
Anvi’o version 4 (56) was used to extract and concatenate the bac120 (120 single-copy marker proteins in bacteria) amino acid sequences (13). Amino acids were aligned with MUSCLE (57). The alignment was masked with Gblocks (58), and an appropriate substitution model was chosen using ModelGenerator version 85 (32). A tree was then constructed using the PhyML algorithm (33) using the Le and Gascuel (59) substitution model using SeaView (60), with 100 bootstraps.
Weekly monitoring with DNA extraction.
To select a DNA monitoring tool for routine use by wastewater treatment plant personnel, we compared a heat lysis method to two conventional DNA extraction kits (MP Biomedicals FastDNA Spin kit for soil, Santa Ana, CA; and Mo Bio PowerSoil isolation kit, Carlsbad, CA). No statistically significant difference was observed (data not shown), so heat lysis was adopted for extraction of DNA from activated sludge samples collected over the 2-year time course from 7 January 2014 to 29 December 2015. For the heat lysis method, archived frozen 2.0-ml pellets of biomass samples were resuspended in 1 ml Tris-EDTA (TE) buffer (pH 7.0). Only a small fraction of the pellet was used for DNA extraction, so the sample was first vortexed for 1 min at maximum speed using a Vortex Adapter (Mo Bio Laboratories, Inc., Carlsbad, CA) with the Vortex Genie 2T (Scientific Industries, Inc., Bohemia, NY) to wash and homogenize the sample. Fifty microliters was then aliquoted into a fresh microcentrifuge tube. The 50-μl sample was heated at 95°C for 10 min and then transferred to ice for 5 min. It was then spun for 1 min in a microcentrifuge tube at high speed to pellet any debris. The supernatant containing DNA was transferred to a fresh tube and archived at −20°C until needed.
Quantification of Competibacteraceae 16S rRNA gene abundance.
Quantitative PCR (qPCR) was used to estimate the 16S rRNA gene copy number for three Competibacteraceae clades identified at the Northern California Wastewater Treatment Plant. Based on the 22 sequences from the clone library, regions of the 16S rRNA were identified with either high or low specificity to a subgroup using an in-house script that designed end primers and a corresponding TaqMan probe (Table 2). Plasmid DNAs from the clone library containing representative 16S rRNA segments from the three subgroups were used to test each assay for specificity. These plasmids were also used to generate standard curves and were isolated using the PureLink HiPure plasmid miniprep kit (Thermo Fisher Scientific, Fremont, CA). All standard reactions were carried out in triplicate, and all samples in duplicate, using a StepOnePlus real-time PCR system (Applied Biosystems, Inc., Foster City, CA). A linear response (R2 > 0.98) was observed for plasmids containing specific subgroup 16S rRNA gene between 101 and 107 copies per reaction, with efficiencies from 95% to 112%. Duplicate negative controls containing no-template DNA were included in all experiments to ensure that contamination was not present in the reactions. Each 10-μl reaction mixture contained 1× PrimeTime qPCR assay mix containing two end primers and a fluorescently labeled probe (Integrated DNA Technologies, Coralville, IA), 1× TaqMan universal master mix (Life Technologies, Carlsbad, CA), and 1 μl template. The profiling temperature was as follows: 95°C for 10 min, 40 cycles of 95°C for 15 s and 60°C for 60 s with detection for an additional 10 s, followed by a final extension at 72°C for 10 min.
Attempts were made to normalize the individual Competibacteraceae 16S rRNA qPCR results to total 16S rRNA using the primer set 341f/534r (61). The plasmid sequence for CaBA clone 25 was used as a standard from 103 to 108 copies in a 10-μl reaction mixture containing 0.4 μM each primer, 1× AB Power SYBR master mix (Applied Biosystems, Foster City, CA), and 1 μl template. A linear response was observed for this assay, with an efficiency of 105%. While the signal for each sample was mostly constant for the 2-year time series, values were lower than expected and suggested that stored DNA had degraded over time. Copies of 16S rRNA for each Competibacteraceae clade were therefore normalized to milligrams of VSS as measured by the treatment plant staff to adjust for changes in biomass concentration.
Statistical analysis.
Statistical analyses of plant monitoring data and Competibacteraceae abundance were carried out using the R statistical software version 3.4.4 (47) in RStudio version 1.0.143 (46). For each type of analysis, the Benjamini-Hochberg correction (62) was used to assess significant P values given multiple comparisons.
(i) Linking Competibacteraceae abundance to plant monitoring data.
Correlations of measured plant data to the abundance of Competibacteraceae groups were calculated using Spearman’s rank correlation coefficient using the rcorr function in the Hmisc package in R (63), and the P values were later adjusted with the Benjamini-Hochberg correction (62). Missing values were removed using pairwise comparison.
(ii) MANOVA for Competibacteraceae-associated TSS events.
To assess which variables differed between periods with high and low abundance of clade CPB_P15, a multivariate analysis of variance (MANOVA) was carried out for three time periods, as follows: an initial time period of early 2014, representing a period of high CPB_P15 abundance but low effluent TSS levels (stable conditions); a second period in May to June 2014, representing a period of high CPB_P15 abundance and high effluent TSS levels (unstable conditions); and a final time period in July 2015, representing a period of both low CPB_P15 abundance and low effluent TSS levels (stable conditions). Separate MANOVAs were carried out for microscopy observation data (dispersed large coccoid cells, CPB_P15, and other Competibacter lineage) and operational parameters (TSS levels in the primary and secondary influent, TSS in the effluent, waste return activated sludge, flow rate, mixed liquor VSS, SRT, and recycle rate).
Data availability.
The Competibacteraceae 16S rRNA gene sequences from this study have been deposited in GenBank under the accession numbers MH827568 to MH827589. Short reads from the metagenome and the binned genomes have been uploaded under BioProject number PRJNA509633. The genomes have been deposited at DDBJ/ENA/GenBank under the accession numbers RXIV00000000, RXIW00000000, and RXIX00000000. The versions described in this paper are versions RXIV01000000, RXIW01000000, and RXIX01000000, respectively.
Supplementary Material
ACKNOWLEDGMENTS
We thank the wastewater treatment plant staff for assistance with sampling and for providing additional information on plant operation.
This project was supported by the National Science Foundation (NSF) Engineering Research Center: Reinventing the Nation’s Urban Water Infrastructure (ReNUWIt). V.R.B. was supported in part by an NSF graduate research fellowship and in part by an unrestricted gift from Chevron awarded to C.S.C.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02301-18.
REFERENCES
- 1.Graham DW, Smith VH. 2004. Designed ecosystem services: application of ecological principles in wastewater treatment engineering. Front Ecol Environ 2:199–206. doi: 10.1890/1540-9295(2004)002[0199:DESAOE]2.0.CO;2. [DOI] [Google Scholar]
- 2.Rittmann BE, McCarty PL. 2001. Environmental biotechnology: principles and applications, 1st ed McGraw-Hill Publishers, New York, NY. [Google Scholar]
- 3.Oehmen A, Lemos PC, Carvalho G, Yuan Z, Keller J, Blackall LL, Reis MAM. 2007. Advances in enhanced biological phosphorus removal: from micro to macro scale. Water Res 41:2271–2300. doi: 10.1016/j.watres.2007.02.030. [DOI] [PubMed] [Google Scholar]
- 4.McIlroy SJ, Nittami T, Kanai E, Fukuda J, Saunders AM, Nielsen PH. 2015. Re-appraisal of the phylogeny and fluorescence in situ hybridization probes for the analysis of the Competibacteraceae in wastewater treatment systems. Environ Microbiol Rep 7:166–174. doi: 10.1111/1758-2229.12215. [DOI] [PubMed] [Google Scholar]
- 5.Mielczarek AT, Nguyen HTT, Nielsen JL, Nielsen PH. 2013. Population dynamics of bacteria involved in enhanced biological phosphorus removal in Danish wastewater treatment plants. Water Res 47:1529–1544. doi: 10.1016/j.watres.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 6.Stokholm-Bjerregaard M, McIlroy SJ, Nierychlo M, Karst SM, Albertsen M, Nielsen PH. 2017. A critical assessment of the microorganisms proposed to be important to enhanced biological phosphorus removal in full-scale wastewater treatment systems. Front Microbiol 8:718. doi: 10.3389/fmicb.2017.00718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McIlroy SJ, Albertsen M, Andresen EK, Saunders AM, Kristiansen R, Stokholm-Bjerregaard M, Nielsen KL, Nielsen PH. 2014. “Candidatus Competibacter”-lineage genomes retrieved from metagenomes reveal functional metabolic diversity. ISME J 8:613–624. doi: 10.1038/ismej.2013.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reference deleted.
- 9.Kong Y, Xia Y, Nielsen JL, Nielsen PH. 2006. Ecophysiology of a group of uncultured gammaproteobacterial glycogen-accumulating organisms in full-scale enhanced biological phosphorus removal wastewater treatment plants. Environ Microbiol 8:479–489. doi: 10.1111/j.1462-2920.2005.00914.x. [DOI] [PubMed] [Google Scholar]
- 10.Albertsen M, Hugenholtz P, Skarshewski A, Nielsen KL, Tyson GW, Nielsen PH. 2013. Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat Biotechnol 31:533–538. doi: 10.1038/nbt.2579. [DOI] [PubMed] [Google Scholar]
- 11.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. 2015. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Konstantinidis KT, Tiedje JM. 2005. Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci U S A 102:2567–2572. doi: 10.1073/pnas.0409727102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parks DH, Rinke C, Chuvochina M, Chaumeil P-A, Woodcroft BJ, Evans PN, Hugenholtz P, Tyson GW. 2017. Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life. Nat Microbiol 2:1533–1542. doi: 10.1038/s41564-017-0012-7. [DOI] [PubMed] [Google Scholar]
- 14.Parks DH, Chuvochina M, Waite DW, Rinke C, Skarshewski A, Chaumeil P-A, Hugenholtz P. 2018. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol 36:996–1004. doi: 10.1038/nbt.4229. [DOI] [PubMed] [Google Scholar]
- 15.An W, Guo F, Song Y, Gao N, Bai S, Dai J, Wei H, Zhang L, Yu D, Xia M, Yu Y, Qi M, Tian C, Chen H, Wu Z, Zhang T, Qiu D. 2016. Comparative genomics analyses on EPS biosynthesis genes required for floc formation of Zoogloea resiniphila and other activated sludge bacteria. Water Res 102:494–504. doi: 10.1016/j.watres.2016.06.058. [DOI] [PubMed] [Google Scholar]
- 16.Yu D, Xia M, Zhang L, Song Y, Duan Y, Yuan T, Yao M, Wu L, Tian C, Wu Z, Li X, Zhou J, Qiu D. 2017. RpoN (σ54) is required for floc formation but not for extracellular polysaccharide biosynthesis in a floc-forming Aquincola tertiaricarbonis strain. Appl Environ Microbiol 83:e00709-17. doi: 10.1128/AEM.00709-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang Y, Sorokin DY, Kleerebezem R, Muyzer G, van Loosdrecht M. 2011. Plasticicumulans acidivorans gen. nov., sp. nov., a polyhydroxyalkanoate-accumulating gammaproteobacterium from a sequencing-batch bioreactor. Int J Syst Evol Microbiol 61:2314–2319. doi: 10.1099/ijs.0.021410-0. [DOI] [PubMed] [Google Scholar]
- 18.Jiang Y, Sorokin DY, Junicke H, Kleerebezem R, van Loosdrecht MCM. 2014. Plasticicumulans lactativorans sp. nov., a polyhydroxybutyrate-accumulating gammaproteobacterium from a sequencing-batch bioreactor fed with lactate. Int J Syst Evol Microbiol 64:33–38. doi: 10.1099/ijs.0.051045-0. [DOI] [PubMed] [Google Scholar]
- 19.Tandoi V, Jenkins D, Wanner J. 2015. Activated sludge separation problems: theory, control measures, practical experiences. IWA Publishing, London, United Kingdom. [Google Scholar]
- 20.McIlroy SJ, Kirkegaard RH, McIlroy B, Nierychlo M, Kristensen JM, Karst SM, Albertsen M, Nielsen PH. 2017. MiDAS 2.0: an ecosystem-specific taxonomy and online database for the organisms of wastewater treatment systems expanded for anaerobic digester groups. Database (Oxford) 2017:bax016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flemming H-C, Wingender J. 2010. The biofilm matrix. Nat Rev Microbiol 8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 22.Oyserman BO, Noguera DR, del Rio TG, Tringe SG, McMahon KD. 2016. Metatranscriptomic insights on gene expression and regulatory controls in Candidatus Accumulibacter phosphatis. ISME J 10:810–822. doi: 10.1038/ismej.2015.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seviour TW, Lambert LK, Pijuan M, Yuan Z. 2011. Selectively inducing the synthesis of a key structural exopolysaccharide in aerobic granules by enriching for Candidatus “Competibacter phosphatis. Appl Microbiol Biotechnol 92:1297–1305. doi: 10.1007/s00253-011-3385-1. [DOI] [PubMed] [Google Scholar]
- 24.American Public Health Association, American Water Works Association, Water Environment Federation. 1989. Standard methods for the examination of water and wastewater, 17th ed. American Public Health Association, American Water Works Association, Water Environment Federation, Washington, DC. [Google Scholar]
- 25.United States Environmental Protection Agency. 1971. Method 160.4: residue, volatile (gravimetric, ignition at 550°c) by muffle furnace. United States Environmental Protection Agency, Washington, DC: https://www.epa.gov/sites/production/files/2015-08/documents/method_160-4_1971.pdf. [Google Scholar]
- 26.Richard M, Daigger G, Jenkins D. 2003. Manual on the causes and control of activated sludge bulking, foaming, and other solids separation problems, 3rd ed. CRC Press, Boca Raton, FL. [Google Scholar]
- 27.Wells GF, Park H-D, Eggleston B, Francis CA, Criddle CS. 2011. Fine-scale bacterial community dynamics and the taxa–time relationship within a full-scale activated sludge bioreactor. Water Res 45:5476–5488. doi: 10.1016/j.watres.2011.08.006. [DOI] [PubMed] [Google Scholar]
- 28.Huber T, Faulkner G, Hugenholtz P. 2004. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinforma 20:2317–231910. doi: 10.1093/bioinformatics/bth226. [DOI] [PubMed] [Google Scholar]
- 29.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic Local Alignment Search Tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 30.Pruesse E, Peplies J, Glöckner FO. 2012. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28:1823–182910. doi: 10.1093/bioinformatics/bts252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Westram R, Bader K, Pruesse E, Kumar Y, Meier H, Glockner FO, Ludwig W. 2011. ARB: a software environment for sequence data, p 399–406. In de Bruijn F. (ed), Handbook of molecular microbial ecology I: metagenomics and complementary approaches. John Wiley & Sons, Inc, Hoboken, NJ. [Google Scholar]
- 32.Keane TM, Creevey CJ, Pentony MM, Naughton TJ, Mclnerney JO. 2006. Assessment of methods for amino acid matrix selection and their use on empirical data shows that ad hoc assumptions for choice of matrix are not justified. BMC Evol Biol 6:29. doi: 10.1186/1471-2148-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 34.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tamura K, Nei M. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 36.Yu G, Smith DK, Zhu H, Guan Y, Lam TT-Y. 2017. ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol Evol 8:28–36. doi: 10.1111/2041-210X.12628. [DOI] [Google Scholar]
- 37.Karst SM, Kirkegaard RH, Albertsen M. 2016. mmgenome: a toolbox for reproducible genome extraction from metagenomes. bioRxiv doi: 10.1101/059121. [DOI]
- 38.Li D, Liu C-M, Luo R, Sadakane K, Lam T-W. 2015. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31:1674–1676. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- 39.Vollmers J, Wiegand S, Kaster A-K. 2017. Comparing and evaluating metagenome assembly tools from a microbiologist’s perspective–not only size matters! PLoS One 12:e0169662. doi: 10.1371/journal.pone.0169662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hyatt D, Chen G-L, Locascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eddy SR. 2011. Accelerated profile HMM searches. PLoS Comput Biol 7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huson DH, Auch AF, Qi J, Schuster SC. 2007. MEGAN analysis of metagenomic data. Genome Res 17:377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gregor I, Dröge J, Schirmer M, Quince C, McHardy AC. 2016. PhyloPythiaS+: a self-training method for the rapid reconstruction of low-ranking taxonomic bins from metagenomes. PeerJ 4:e1603. doi: 10.7717/peerj.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.RStudio Team. 2016. RStudio: integrated development for R. 1.0.143. RStudio, Inc, Boston, MA. [Google Scholar]
- 47.R Core Team. 2018. R: a language and environment for statistical computing. 3.4.4. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 48.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lux M, Krüger J, Rinke C, Maus I, Schlüter A, Woyke T, Sczyrba A, Hammer B. 2016. acdc–Automated Contamination Detection and Confidence estimation for single-cell genome data. BMC Bioinformatics 17:543. doi: 10.1186/s12859-016-1397-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 51.Kanehisa M, Sato Y, Morishima K. 2016. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol 428:726–731. doi: 10.1016/j.jmb.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 52.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST server: Rapid Annotations using Subsystems Technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lagesen K, Hallin P, Rødland EA, Stærfeldt H-H, Rognes T, Ussery DW. 2007. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res 35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lechner M, Findeiß S, Steiner L, Marz M, Stadler PF, Prohaska SJ. 2011. Proteinortho: detection of (co-)orthologs in large-scale analysis. BMC Bioinformatics 12:124. doi: 10.1186/1471-2105-12-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Leimbach A. 2016. bac-genomics-scripts: bovine E. coli mastitis comparative genomics edition. https://zenodo.org/record/215824#.XCHtP_lKi1s.
- 56.Eren AM, Esen ÖC, Quince C, Vineis JH, Morrison HG, Sogin ML, Delmont TO. 2015. Anvi’o: an advanced analysis and visualization platform for ’omics data. PeerJ 3:e1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 59.Le SQ, Gascuel O. 2008. An improved general amino acid replacement matrix. Mol Biol Evol 25:1307–1320. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
- 60.Gouy M, Guindon S, Gascuel O. 2010. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol 27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- 61.Yoshida N, Takahashi N, Hiraishi A. 2005. Phylogenetic characterization of a polychlorinated-dioxin- dechlorinating microbial community by use of microcosm studies. Appl Environ Microbiol 71:4325–4334. doi: 10.1128/AEM.71.8.4325-4334.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 57:289–300. [Google Scholar]
- 63.Harrell FE., Jr. 2018. Hmisc: Harrell Miscellaneous. Version 4.1-1. https://cran.r-project.org/web/packages/Hmisc/index.html.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Competibacteraceae 16S rRNA gene sequences from this study have been deposited in GenBank under the accession numbers MH827568 to MH827589. Short reads from the metagenome and the binned genomes have been uploaded under BioProject number PRJNA509633. The genomes have been deposited at DDBJ/ENA/GenBank under the accession numbers RXIV00000000, RXIW00000000, and RXIX00000000. The versions described in this paper are versions RXIV01000000, RXIW01000000, and RXIX01000000, respectively.