Abstract
A variety of biomolecules acting on the cell membrane folds into a biologically active structure in the membrane environment, it is, therefore, important to determine the structures and dynamics of such biomolecules in a membrane environment. While several biophysical techniques are used to obtain low-resolution information, solid-state NMR spectroscopy is one of the most powerful means for determining the structure and dynamics of membrane bound biomolecules such as antibacterial biomolecules and amyloidogenic proteins; unlike X-ray crystallography and solution NMR spectroscopy, applications of solid-state NMR spectroscopy are not limited by non-crystalline, non-soluble nature or molecular size of membrane-associated biomolecules. This review article focuses on the applications of solid-state NMR techniques to study a few selected antibacterial and amyloid peptides. Solid-state NMR studies revealing the membrane inserted bent α-helical structure associated with the hemolytic activity of bee venom melittin and the chemical shift oscillation analysis used to determine the transmembrane structure (with α-helix and 310-helix in the N- and C-termini, respectively) of antibiotic peptide alamethicin are discussed in detail. Oligomerization of an amyloidogenic islet amyloid polypeptide (IAPP, or also known as amylin) resulting from its aggregation in a membrane environment, molecular interactions of the antifungal natural product amphotericin B with ergosterol in lipid bilayers, and the mechanism of lipid raft formation by sphingomyelin studied using solid state NMR methods are also discussed in this review article. This article is part of a Special Issue entitled “Biophysical Exploration of Dynamical Ordering of Biomolecular Systems” edited by Dr. Koichi Kato.
Keywords: Antibacterial peptide, Antifungal natural product, Amyloidogenic peptide, IAPP, Membrane environment, Lipid raft, Solid-state NMR
1. Introduction
A variety of biomolecules located and functioning in the membrane that surrounds living cells fold into a biologically active structure in a membrane environment. The cell membrane is in the liquid crystalline state which is an orientationally ordered state and some biomolecules form biologically active structures by interacting with the cell membrane. For example, antimicrobial peptides do not form structures in solution, while they form a structure after they interact with a membrane. Several models (the aggregate, the barrel-stave, the toroidal pore and the carpet models [1]) have been proposed to explain the mechanism by which interaction with a membrane results in peptide ordering. The initial step in all these models of membrane perturbation is the adsorption of the peptide at the ordered membrane surface. In the aggregate model, the peptide acts similar to a detergent and results in the formation of micelles. In the barrel-stave and toroidal pore models, the peptides adopt a transmembrane orientation. In the barrel-stave model the peptides form a structure similar to that of an ion channel and this mechanism of action is favored for long and hydrophobic peptides. For shorter and positively charged peptides, the toroidal pore mechanism is favored since each peptide is surrounded by phospholipids, which limits electrostatic repulsion between charged peptides [2,3]. Finally, the carpet model is similar to the toroidal pore model with the difference that the peptide sits at the surface of the membrane [4]. In both the toroidal and carpet models, the peptides induce a positive curvature of the membrane. In addition, some of the antimicrobial peptides have been shown to translocate across the lipid bi-layers without inducing significant disruption to the cell membrane [5–7].
The structures and orientations of membrane-associated peptides and proteins in membrane environments can be elucidated using structural and orientational constraints derived from magnetically anisotropic interactions as studied by solid-state NMR spectroscopy [8–14]. One such type of interaction is chemical-shift anisotropy (CSA). Usually, this type of interaction appears as a dynamically averaged chemical-shift pattern from which the orientation of peptides and proteins in a lipid-bilayer environment can be derived [15]. Another interaction affording structural constraint is dipolar interaction between coupled nuclei, which can be combined with chemical-shift constraints to provide detailed structural information regarding membrane-associated peptides and proteins [16]. These constraints provide the dynamic structures and orientations of membrane-associated peptides and proteins [17]. Although complete structures have been determined for only a limited number of compounds, these constraints provide detailed local structure and topology as well as orientation.
Solid-state NMR analysis of biomolecules is normally performed using 13C and 15N nuclei, which exhibit spin-1/2. Solid-state NMR spectra yield enormously broadened signals, with line widths on the order of 20 kHz. Magic angle spinning (MAS) experiments combined with cross-polarization (CP) and high-power decoupling (CP-MAS) provide high-resolution 13C and 15N NMR signals for selectively or uniformly labeled membrane-bound peptides and proteins. In solid-state NMR, homomiclear and heteronuclear dipolar interactions can be recoupled using various pulse sequences combined with multidimensional NMR techniques under MAS conditions. These experiments enable the correlation of 13C and 15N signals for assignment to amino acid residues, and chemical shifts and distance constraints. These data can then be used to determine the high-resolution structures of membrane-associated peptides and proteins [18,19].
In this review, we describe solid-state NMR methods used to determine the membrane bound structures, orientations and dynamics of antibacterial peptides, antifungal natural products, and amyloid forming peptides. We have focused on the systems that are investigated in the authors’ laboratories as examples to illustrate the power of various solid-state NMR techniques to study membrane-bound biomolecules.
2. Solid-state NMR spectroscopy for elucidating the structures and orientations of membrane bound biomolecules
2.1. Chemical shift anisotropy
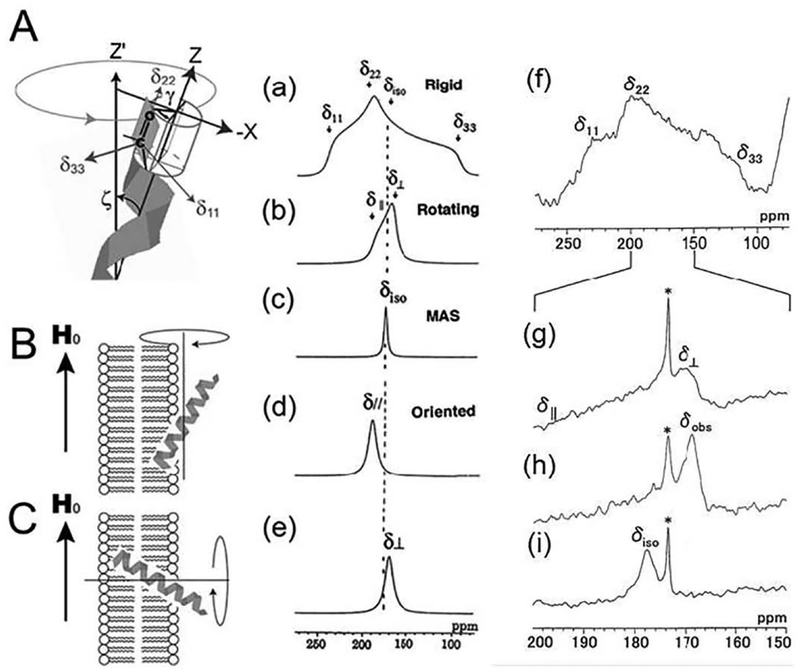
The orientation of peptides bound to a magnetically aligned lipid bilayer can be determined by analyzing the 13C or 15N chemical shift anisotropy (CSA) tensors as reported in the literature. The use of 13C CSA tensors of carbonyl carbons in the peptide chain is illustrated in Fig. 1. When the peptide is completely rigid, 150 ppm of CSA span will be observed in a low-temperature experiment {Fig. 1(a) and (f)). In contrast, when the temperature is increased above the gel-to-liquid crystalline phase transition temperature (Tc), the molecules exhibit a large-amplitude rotational motion about the bilayer normal. In this case, an axially symmetric powder pattern is observed as shown in lecithin-melittin bilayer systems (Fig. 1(b) and (g)) [15,20,21].
Fig. 1.
A: Direction of the principal axis of the 13C chemical shift tensor of the C=0 group. B: The helical axis rotates about the magnetic field (H0). C: the helical axis rotates about the bilayer normal. 13C NMR spectral patterns of the C=0 carbons corresponding to the orientation of the α-helix with respect to the surface of the magnetically oriented lipid bilayers. Simulated spectra were calculated using δ11 = 241, δ22 = 189, and δ33 = 96 ppm for the rigid case (a), rotation about the unique axis without orienting to the magnetic field (slow MAS condition) (b), MAS (c), rotation about the magnetic field (d), and rotation about the bilayer normal (e). 13C NMR of [1-13C]Ile4-melittin-DMPG bilayer system in the rigid state at − 60 °C (f), in the slow MAS condition at 40 °C (g), in the oriented condition at 40 °C (h), and in the fast MAS condition at 40 °C (i) [12].
Dimyristoyl-phosphatidylcholine (DMPC)-melittin lipid bilayers under excess hydration and static conditions have been shown to spontaneously align in the presence of an external magnetic field [12]. The entire melittin molecule forms an α-helix in the membrane-bound state, and the helical axis rapidly rotates about the bilayer normal with a tilt angle ζ and phase angle γ (Fig. 1A). In that case, the 13C NMR signal of the carbonyl carbon in the magnetically oriented lipid bilayer appears at the δ⊥ position of the axially symmetric powder pattern (Fig. 1e). However, it must be stressed that the α-helix does not rotate about the helical axis, but rather about the bilayer normal. In this dynamic state, the CSA, Δδ = δ// − δ⊥, can be expressed as [15]
| (1) |
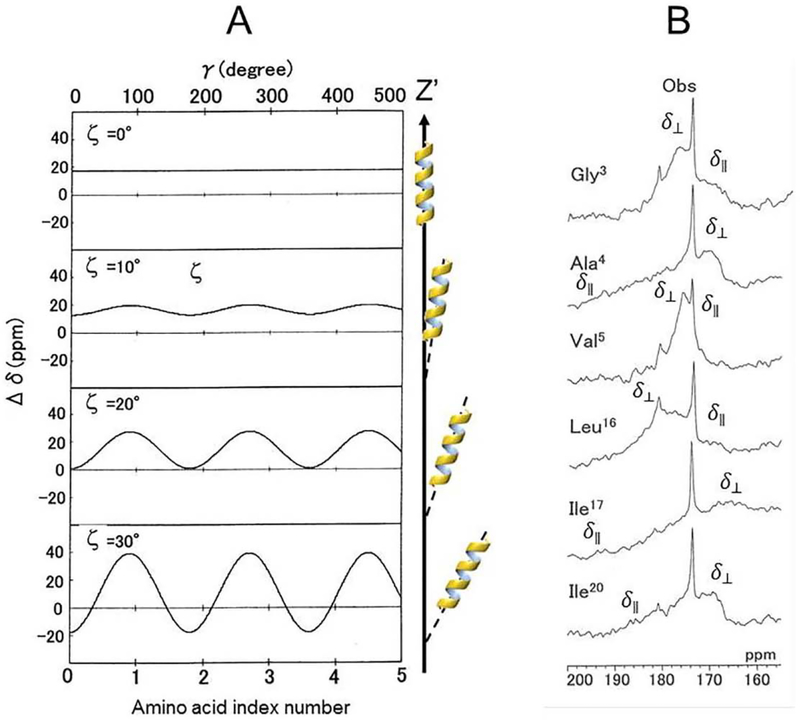
where δ11, δ22, and δ33 are the principal values of the 13C chemical-shift tensor of the carbonyl carbon nucleus in the rigid state. In Eq. (1), Δδ values oscillate as a function of γ, with an oscillation amplitude of (3/2) sin2ζ, referred to as the chemical-shift oscillation [15]. When an α-helical peptide has a large tilt angle ζ, the Δδ value changes significantly, as shown in Fig. 2A. Using this property of Δδ, the tilt angle ζ of the α-helical axis can be determined with respect to the bilayer normal by analyzing the anisotropic 13C chemical-shifts of the carbonyl carbons of the consecutive amino acid residues in the α-helίcal region (Fig. 2). When a peptide forms an ideal α-helical structure, it can be assumed that the inter-peptide plane angle for consecutive peptide planes is 100°. This tilt angle can be accurately obtained by analyzing the root-mean-square deviation (RMSD) for the observed and calculated CSAs using Eq. (2).
| (2) |
where N is the number of 13C labeled residues in the α-helical region.
Fig. 2.
(A): Chemical-shift oscillation curves of chemical-shift anisotropy, Δδ = δ// − δ⊥ against the phase angle, γ, of the peptide plane for the amino acid residues as a function of the tilt angles, ζ of the α-helical axis relative to the bilayer normal (Z’ axis) [12]. (B): 13C chemical shift powder patterns of the [l-13C]Gly3-, [l-13C]Ala4-, [l-13C]Val5, [1-13C] Leu16, [l-13C]Ilel7, and [l-13C]Ile20-melittins-DMPG bilayer systems at 40 °C [21].
In chemical-shift oscillation analyses, lipid bilayers are not necessarily aligned with the magnetic field.
2.2. Dipolar interaction
Similar information can be obtained by observing 15N—lH dipolar interactions in the peptide backbone. Polarization inversion spin exchange at the magic angle (PISEMA), a type of separated-local-field 2D NMR spectroscopy, provides excellent resolution in the dipolar dimension in correlation spectra between 15N chemical-shift values and 15N–lH dipolar interaction [22,23]. PISEMA pulse sequence was developed based on the flip-flop Lee-Goldberg (using phase- and frequency-switched) pulse sequence, which is used to spin-lock 1H spins along the magic angle to suppress homonuclear dipolar interactions among the highly abundant proton spins. Sample heating is a concern in PISEMA experiments, particularly when analyzing hydrated samples containing membrane proteins. This problem can be overcome by drastically reducing the radio frequency (RF) power [24–26] or using a resonance coil designed to minimize sample heating [27]. Offset problems encountered in the PISEMA pulse sequence are also overcome by using the broadband PISEMA technique [28].
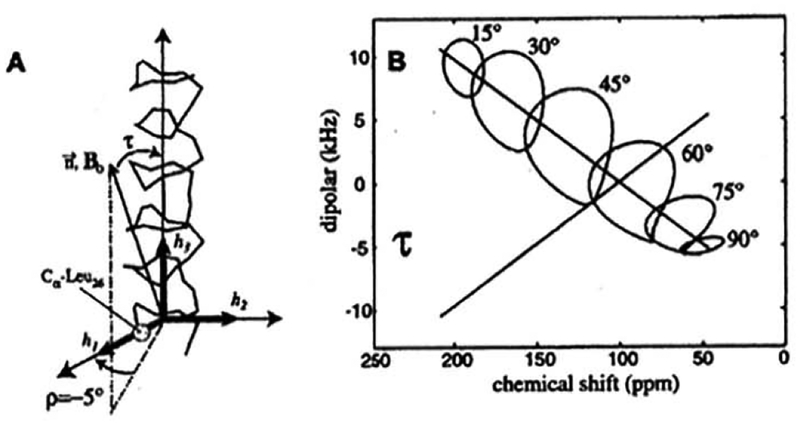
A characteristic circular pattern wheel, known as a polarity index slant angle (PISA), is observed in PISEMA (correlation of 15N chemical shift and 1H–15N dipolar interactions) 2D NMR spectra for a helical structure of a protein embedded in membrane. The shape of the wheel is sensitive to the tilting angle of the α-helix with respect to the lipid bilayer normal [29–31] (Fig. 3). By examining the PISA wheel, one can evaluate the amino acid residues involved in the α-helix, even if the amino acid sequence is unknown. When the helical axis is parallel to the bilayer normal, all the amide sites have a very similar orientation relative to the direction of the applied magnetic field and therefore, all backbone NH resonances overlap with the same 1H-15N dipolar couplings and 15N chemical-shift frequencies. Whereas tilting the helix away from the membrane normal breaks the symmetry about the magnetic field, introducing a variation in the orientations of the amide N–H bond vectors relative to the field direction (Fig. 3B). This is manifested in the observed PISEMA spectra as dispersions of both the 1H–15N dipolar couplings and the 15N chemical-shift frequencies, as shown in Fig. 3B. Thus, the tilt angle of the helical axis can be accurately determined. PISEMA and related multidimensional solid-state NMR techniques have enabled high-resolution structural studies of membrane proteins from fluid lamellar phase lipid bilayers without the need for crystallization or freezing of the sample. Many studies have reported the dynamic structures of a variety of membrane associated peptides and proteins [9,16,23,29–31], Since the interpretation of helical wheels to the structures of transmembrane domains, in particular, is relatively easy, the use of aligned samples in solid-state NMR spectroscopy is attractive and has the potential to be a high throughput approach with the advent of higher magnetic fields and non-uniform sampling approaches.
Fig. 3.
The generation of “PISA wheels”. (A) Definitions of τ and ρ for an α-helix. τ = 0° occurs when the helix axis, h3, is parallel to B0 · ρ = 0° occurs when the projection of B0 onto a plane perpendicular to, h3, makes an angle of 0° with h1, the radial axis of the helix that passes through the Cα carbon of Leu26 of the M2-TMP helix [31]. (B) “Circles” drawn for one of the dipolar transitions rising average values for the 15N chemical shift tensor elements (σ11 = 31.3, σ22 = 55.2, σ33 = 201.8 ppm) and the relative orientations of the 15N–1H dipolar and 15N chemical shift tensor, given by θ = 17°, the angle of the peptide plane between σ22 and υ (parallel to the N–H bond). Note that the center of the PISA wheel falls on a line that passes through the 15N isotropic chemical shift (96 ppm) at 0 kHz on the dipolar scale [31].
2.3. PDSD and DARR 2D MAS NMR spectroscopy
MAS [32] technique seeks to suppress the anisotropic interactions in order to achieve high-resolution “solution-like” spectra by a rapid mechanical rotation of the sample at the magic angle (54.74°). As the anisotropic interactions that give rise to the broadening observation in NMR spectra of solid materials exhibit a (3cos2θ – l)/2 dependence, MAS induces a scaling in the anisotropic interaction. When the sample rotation frequency is slower than the anisotropic interaction, an incomplete averaging occurs, which is manifested in the solid-state NMR spectrum as resonances separated (from the isotropic resonance) by multiples of the rotation speed of the sample, which are known as spinning side bands.
Both 13C and 15N nuclei are frequently observed in membrane-associated peptides and proteins. In this case, CP techniques [33,34] can be used to enhance the intensities of signals from less abundant nuclei by transferring the polarization from abundant protons in the sample. Furthermore, combining CP, high-power proton decoupling, and MAS (CP-MAS) [35] enables the acquisition of high-resolution signals for biological molecules as well demonstrated for numerous systems.
The significant improvements in resolution afforded by MAS comes at the expense of suppressing the valuable anisotropic interactions that contain the rich structural and dynamical information about the system under investigation. To address this, a number of methods have been developed that permit the selective reintroduction of these anisotropic interactions to facilitate analysis. Some of the details about these methods and their applications to biological systems can be found in the literature [14,15].
Single amino acid can be identified by, for example, their characteristic chemical shift and connectivity pattern among carbon resonance. As mentioned above, a number of methods are available for reintroducing homonuclear dipolar 13C–13C couplings. A popular class of experiments relies on the proton-mediated reintroduction of carbon homonuclear dipolar couplings. This approach is usually referred to as longitudinal magnetization transfer by spin diffusion and facilitate zero-quantum flip-flop transition among 13C nuclei. In its basic form, this process is known as proton-driven spin diffusion (PDSD) [36]. In PDSD, the decoupling field on the proton channel is switched off (while 13C magnetization is stored along the z-axis) during the spin diffusion mixing time to reintroduce heteronuclear dipolar couplings, thus increasing the efficiency of flip-flop transitions. This reintroduction occurs, however, only to higher order and scales inversely with the spinning frequency. Other methods, such as dipolar-assisted rotational resonance (DARR) [37,38], the phase-alternated recoupling irradiation scheme [39], or mixed rotational and rotary resonance [40] outperform PDSD with increased spinning speeds of the sample, because the heteronuclear dipolar couplings are reintroduced independent of the MAS frequency. However, because the flip-flop transition appears in the correlation terms of the effective Hamiltonian, the spin diffusion efficiency invariably decreases with increasing spinning frequency, although the transfer can still be efficient at very high MAS frequencies [41].
2.4. REDOR
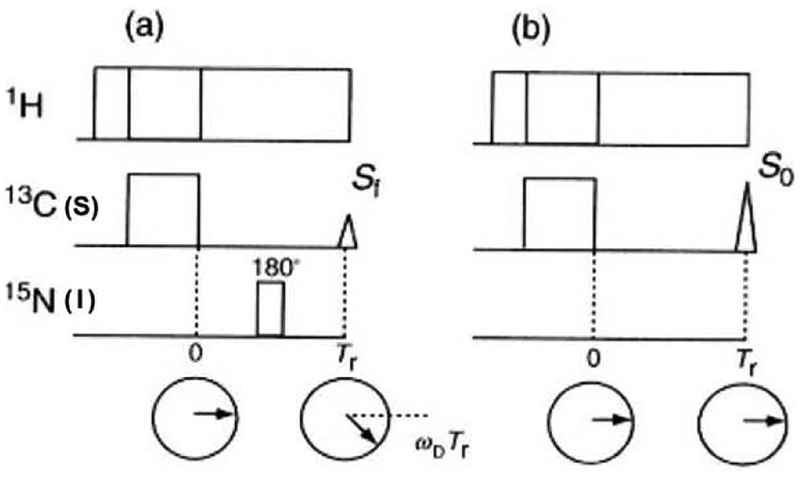
Solid-state NMR allows the determination of inter-nuclear distances quite accurately by measuring recoupled dipolar interactions using re-coupling experiments that are otherwise suppressed by MAS. The experimentally measured interatomic distances can be used to elucidate the three-dimensional (3D) structure of membrane bound biomolecules. The rotational echo double resonance (REDOR) [42] is one such re-coupling MAS technique that has been used to recouple the relatively weak heteronuclear dipolar interactions under MAS by applying a π pulse synchronously with the MAS rotor period. Consequently, the transverse magnetization cannot be refocused completely at the end of the rotor cycle, leading to a reduction in the echo amplitude (Fig. 4a). The extent of the observed reduction in the echo amplitude as a function of the number of rotor periods depends on the strength of the heteronuclear dipolar interaction. This method has extensively been used to determine relatively remote interatomic distances in the range of 2 to 8 Å.
Fig. 4.
Pulse sequence to observe rotational echo. (a) Transverse magnetizations do not refocus at the rotor period Tr in the REDOR pulse sequence, (b) Transverse magnetizations refocus at the rotor period Tr as the rotational echo.
Transverse magnetization which precesses about the static magnetic field due to dipolar interaction under the MAS condition moves back to the same direction at every rotor period because the integration of dipolar frequency, ωD, over a rotor period is zero. Consequently, the rotational echo signals are refocused at the end of each rotor period (Fig. 4b). When a π pulse is applied to the S nucleus, which is coupled with the I nucleus, it inverts the precession direction of the magnetization of the observed I nucleus within a rotor period. Consequently, the magnetization vector of the I nucleus cannot move back to the same direction after one rotor period, therefore, the amplitude of the echo intensity decreases (Fig. 4a). The extent of the reduction of the rotational echo amplitude yields the interatomic distances. To evaluate the REDOR echo amplitude theoretically, one must consider the averaging precession frequency, , in the presence of a π pulse at the center of the rotor period over one rotor cycle as follows:
| (3) |
where α is the azimuthal angle and β is the polar angle defined by the IS intemuclear vector with respect to the rotor axis. Therefore, the phase angle, ΔΦ(α,β), for the NC rotor cycles can be given by
| (4) |
where Tr is one MAS rotor period and NC is the number of rotor cycles. Finally, the echo amplitude can be obtained by averaging over orientations as follows:
| (5) |
Therefore, the normalized echo difference, ΔS/S0 can be given by:
| (6) |
The REDOR echo amplitude can be evaluated more rigorously by using the density matrix approach, and by considering the pulse width in the calculation of the analysis of REDOR [43]. The REDOR based experimental approaches made it possible to elucidate the 3D structure of N-acetyl-Pro-Gly-Phe [43]. It is also important to consider the case where the observed nucleus (I1) can be coupled to two other heteronuclei (S1 and S2) [44]. The experimentally measured ΔS/S0 for such a three-spin system strongly depends not only on I1-S1 and I1-S2 dipolar couplings but also on the S1-I1-S2 angle. REDOR dephasing patterns due to the recoupled dipolar couplings between the selectively labeled carbonyl 13C in bovine lactpferrampin (LFampinB) and 31P in phospholipids were measured by 13C–31P3 multi-spin REDOR analysis, and the results revealed that LFampinB is located in the interfacial region of the membrane [45]. A detailed analysis of the dipolar couplings among multiple spins and Boltzmann statistics analysis of REDOR dephasing curves can be found in the literature [46–48]. Overall, the REDOR based experiments are pretty robust and the results are relatively easy to interpret. Since there are very few steps to be optimized, REDOR is an excellent choice for structural studies on peptides or proteins that are selectively labeled with the required heteronuclear isotopes. The most often used isotopes are a combination of 13C, 15N, 19F and 31P.
3. Dynamic structure of antimicrobial peptides in membrane environments
3.1. Structure determination of membrane-bound antimicrobial peptides
There is considerable interest in understanding the mechanism of action of antimicrobial peptides (AMPs) that function by interacting with the lipid bilayer of the cell membrane of bacteria. The bacterial selectivity and potency of an AMP depends on its structural interactions with the lipid bilayer of the bacteria or mammalian cell. Since the physicochemical properties of the lipid bilayer varies with the lipid composition, and the lipid composition of a bacterial cell membrane is different from that of a mammalian cell membrane, the structural interactions of an AMP can be very different in bacterial vs mammalian membranes. Therefore, while it is important to investigate the high-resolution structures of an AMP in a fluid lipid bilayer that mimics the native membrane, it is highly challenging and such samples are not amenable for most biophysical techniques. Fortunately, solid-state NMR can be used to study the fluid lipid bilayers and the high-resolution structures of AMP.
Structures, topologies, and orientations of a number of membrane-bound peptides in lipid-bilayer environments have been investigated recently using solid-state NMR in order to fully understand their biological functions. Solid-state NMR is particularly well suited for elucidating the dynamics, topologies, orientations, and high resolution structures of peptides in the lipid bilayer environment using membrane mimetics such as lipid vesicles, bicelles or lipid nanodiscs/macrodiscs. While several studies have reported valuable structural and functional insights into a variety of AMPs (including magainins, MSI-78, LL-37, pardaxin, protegrin and other designed peptides), the use of chemical shift oscillation analysis in elucidating the detailed structure and orientation of the membrane bound state of antibacterial peptides such as melittin [15,20,21], bonbolitin II [49], lactoferrampin [45] and alamethicin [50] reported in the literature are highlighted below.
3.1.1. Melittin
Melittin, a hexacosapeptide with the primary structure GIGAVLKVLTTGLPALISTIKRKRQQNH2, is the major component of bee venom [51]. Melittin exhibits potent hemolytic activity [52] in addition to indication of voltage-dependent ion conductance across a planar lipid bilayer at low concentration [53]. Melittin also causes selective micellization of lipid bilayers as well as membrane fusion at high concentration [54]. As the temperature is decreased and approaches the gel phase, the membranes break down into small particles [55]. Upon raising the temperature above the Tc, the small particles reform into unilamellar vesicles and ultimately multilamellar vesicles.
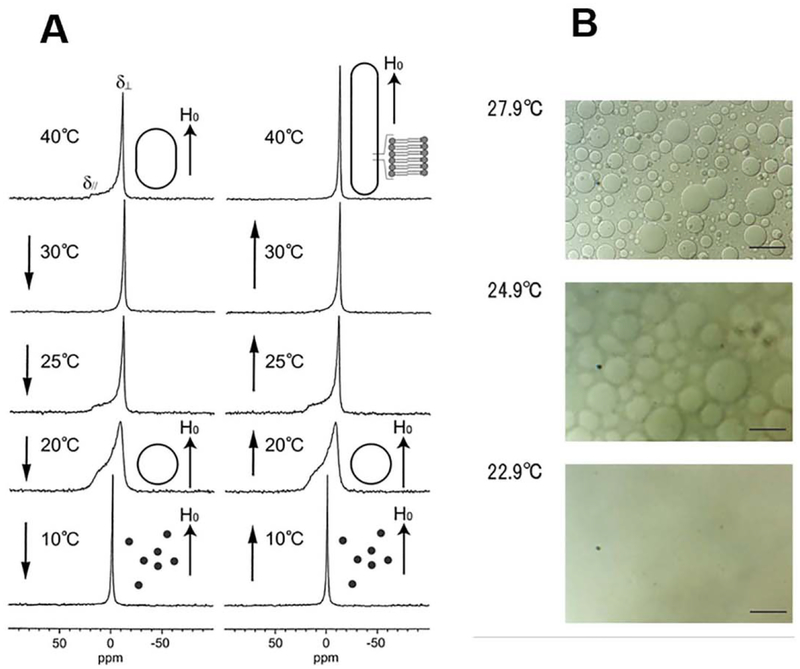
Giant vesicles with diameters of ~ 20 μm were observed upon optical microscopic analysis of melittin-DMPC bilayers at 27.9 °C (Fig. 5B) [20]. When the temperature was lowered to 24.9 °C (Tc = 23 °C for neat DMPC bilayers), the surface of the vesicles became blurred, and dynamic pore formation was visible in microscopic images collected at different exposure times [56]. These vesicles disappeared completely at 22.9 °C. It was thus found that melittin-lecithin bilayers undergo reversible fusion and disruption near the respective Tc.
Fig. 5.
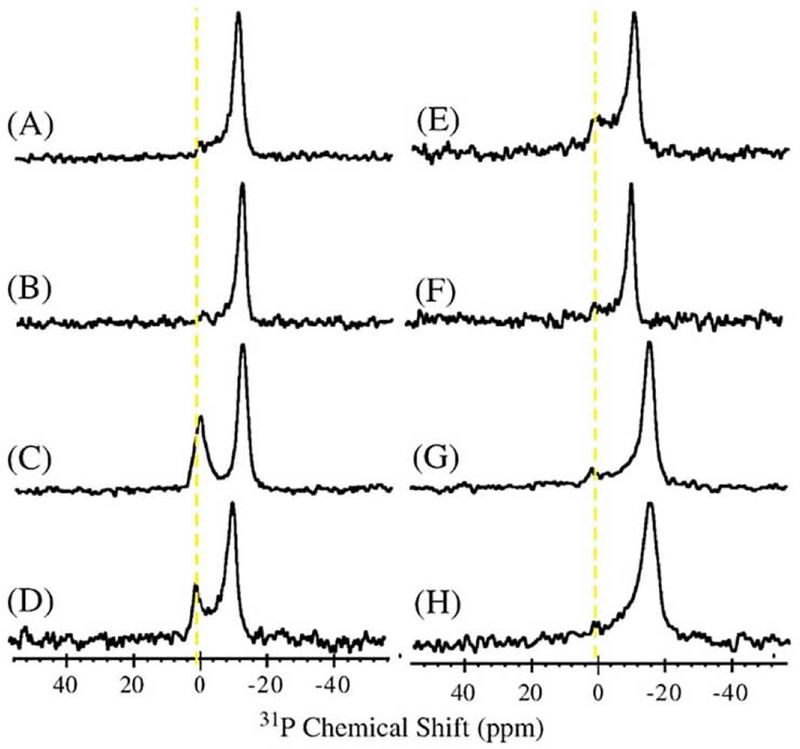
A. Effect of temperature variation on the 31P NMR spectra of melittin-DMPC bilayer systems. Arrows indicate the direction of the temperature variation. H0 indicates the direction of the static magnetic field. The shapes of the vesicles are depicted to the right of the corresponding spectra [56]. B. Optical microscopic pictures of melittin-DMPC vesicle systems at 22.9, 24.9 and 27.9 °C [56].
Static 31P dipolar-decoupled (DD)-MAS spectra of melittin-DMPC bilayers hydrated with Tris buffer were recorded at various temperatures (Fig. 5A) [56]. Immediately after the sample was placed in the magnetic field, the 31P NMR spectrum of an axially symmetric powder pattern characteristic of the liquid-crystalline phase was recorded at 40 °C. The upper field edge (δ⊥) was more intense than the lower field edge (δ//) as compared with a normal axially symmetric powder pattern. This finding indicates that the DMPC bilayer is partially aligned with the applied magnetic field, with the bilayer plane assuming a parallel orientation by forming elongated vesicles, which are referred to as magnetically oriented vesicle systems (MOVS) [56], When the temperature was lowered to 30 °C, the intensity of the upper field edge of the powder pattern increased further, leading to a spectrum exhibiting almost complete alignment with the magnetic field. At 25 °C, the axially symmetric powder pattern appeared again. At 20 °C, this spectrum changed to a broad envelope of the powder pattern, with round edges due to the presence of a large-amplitude motion in addition to a rotational motion about the molecular axis associating with lateral diffusion of the lipid molecules. At 10 °C, the isotropic 31P NMR signal dominated near 0 ppm because of the isotropic rapid tumbling motion of small particles caused by melittin-induced lysis of the larger vesicles. The same axially symmetric powder patterns reappeared as a result of fusion when the temperature was raised from 10 °C to 25 °C, leading to the formation of larger spherical vesicles (Fig. 5A).
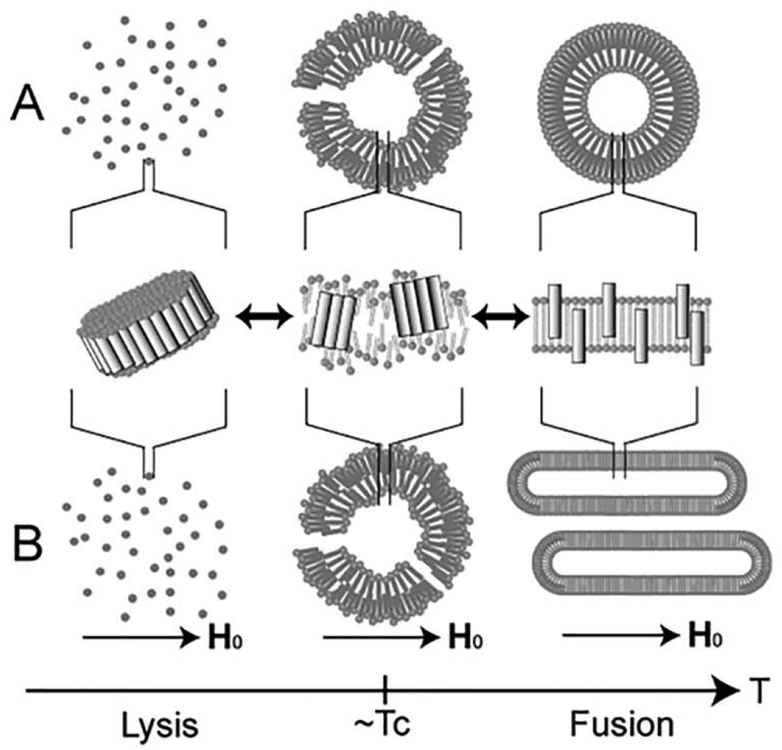
It was shown that melittin forms a pseudo-transmembrane amphophilic α-helix [15]. However, melittin can remain in a homogeneous monomeric form in hydrophobic environments when the lipid bilayer enters the lipid-crystalline phase above the Tc (Fig. 6 right). At temperatures close to the Tc, the hydrophilic sides of melittin molecules associate with one another such that the hydrophobic sides face the lipid molecules, thereby causing greater phase separation and partial disordering of the lipid bilayer (Fig. 6 middle). At temperatures below the Tc, a large number of melittin molecules associate with each other. Consequently, small lipid-bilayer particles became surrounded by a belt of melittin molecules and are released from the vesicle, resulting in membrane disruption. Subsequently, entire vesicles dissolve in the buffer solution (Fig. 6 left).
Fig. 6.
Schematic representation of the process of morphological changes in melittinlecithin bilayer systems in the absence (A) and presence (B) of an applied magnetic field (H0) [56].
The diether lipid, ditetradecylphosphatidylcholine (DTPC) incorporated by 13C labeled melittin, was mechanically aligned between stacked glass plates, then 13C NMR spectra were recorded [57,58] as a function of angle between the bilayer planes and the magnetic field at temperatures above and below Tc. An analysis of the 13C chemical shifts of the carbonyl resonances of each residue, clearly indicated that the peptide was well aligned in the bilayers and reoriented about the bi-layer normal. The reduced CSA and the chemical shift were consistent with melittin adopting a helical conformation with a trans-bilayer orientation in the lipid membranes, with a greater than 120° angle between the helices in the crystal form [59,60].
Fully hydrated melittin-lipid bilayer also spontaneously aligns along a static magnetic field by forming elongated vesicles, with the long axis parallel to the magnetic field. Using this magnetic orientation property of the membrane, the melittin has been determined to be aligned in a transmembrane fashion (Fig. 1C and (h)) [20].
The secondary structure of melittin bound to a DMPC bilayer can be determined based on conformation-dependent 13C chemical shifts referenced to those of model systems [61–63]. The isotropic 13C chemical shifts of the [l-13C]Gly3, [l-13C]Val5, [l-13C]Glyl2, [l-13C]Leul6, and [l-13C]Ile20 residues in melittin were determined to be 172.7, 175.2, 171.6, 175.6, and 174.8 ppm, respectively, indicating that all of these residues are involved in the α-helix [15,20,21].
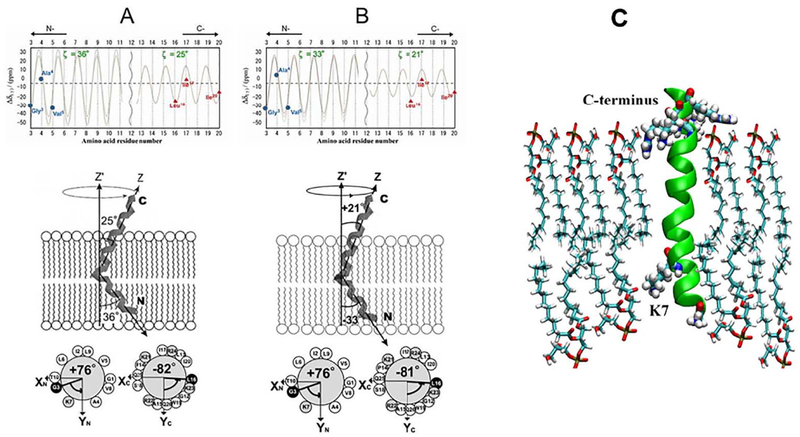
The dynamic structures of melittin bound to dipalmitoylphosphatidylcholine (DPPC), dilauroylphosphatidylcholine (DLPC) and dίmyristoylphosphatίdylgrycerol (DMPG) were investigated by analyzing the 13C anisotropic and isotropic chemical shifts of selectively 13C-labeled carbonyl carbons of melittin under static and MAS conditions [15,21]. Axially symmetric chemical-shift powder patterns were observed for [l-13C]Gly3, [l-13C]Ala4, [l-13C]Val5, [l-13C]Glyl2, [l-13C]Leul6, and [l-13C]Ile20. Analysis of the chemical-shift oscillations (Fig. 7) indicated that melittin molecules form two differently oriented α-helices and diffuse laterally to rotate rapidly around the lipid bilayer normal, with tilt angles of − 33°, − 36° and − 32° for the N-terminal helix, and 21°, 25° and 30° for the C-terminal helix in DLPC, DPPC and DMPG vesicles, respectively. Although the RMSD values could be determined, the tilt angles ζ and − ζ could not be distinguished because of the symmetry relation associated with Eq. (2). As a result, the REDOR method [42] was used to measure the inter-atomic distance between [l-13C]Val8 and [15N]Leul3 to further identify the bending α-helical structure of melittin. The result provided interhelical angles of 126°, 119° and 118° in DLPC, DPPC and DMPG lipid bilayers, respectively [15,21]. The interhelical angle in the range of 139° to 145° was also reported in DTPC lipid bilayers [58]. These analyses led to the conclusion that the α-helices of melittin molecules penetrate the hydrophobic core of the bilayers incompletely as a pseudo-transmembrane structure, which induces fusion and disruption of the vesicles.
Fig. 7.
Schematic representation of the structures and orientations of melittin. (A): Bound to DPPC and (B): DLPC lipid bilayers. (Top): Chemical shift oscillation curves for [1-13C]Gly3, Ala4, Val5, Leu16, Ile17, and Ile20 of melittin in DPPC (A) and DLPC (B) lipid bilayers. (Bottom): The N- and C-terminal helices of melittin bound to DPPC and DLPC lipid bilayers are inserted into the bilayer with tilt angles of 36° and 25° in DPPC and 33° and 21° in DLPC, respectively [15]. (C): Structure of melittin bound to DMPG bilayers obtained from MD simulation [21].
MD simulation for the melittin-DMPG system indicated that the basic residue Lys7 is located slightly within the lipid core region and interacts with one DMPG molecule to disturb the lower surface of the lipid bilayers, and this interaction is associated with melittin’s membrane disruption activity (Fig. 7C) [21].
3.1.2. Alamethicin
Alamethicin is a 20-residue antibiotic peptide from Trichoderma viride [64]. One of the major amino acid sequences is Ac-Aib-Pro-Aib-Ala-Aib-Ala-Gln-Aib-Val-Aib-Gly-Leu-Aib-Pro-Val-Aib-Aib-Gln-Gln-Phol and contains 8 α-amino isobutyric acid (Aib) residues. In addition, the N-terminus is acetylated, and the C-terminus is comprised of an l-phenylalaninol residue [65].
Alamethicin exhibits voltage-dependent ion channel activity in membranes [66]. Alamethicin has a high affinity for lipid bilayers, and binds to the surface of lipid bilayers and inserts into the membrane. Various channel models have been proposed to determine its ion channel activity, such as the barrel stave model [67]. Alamethicin channels are formed by parallel bundles of 3–12 transmembrane helical monomers surrounding a central water-filled pore [68]. The ion channel activity of alamethicin makes it a suitable model for investigating voltage-dependent ion channel proteins [69].
X-ray crystallographic data indicate that alamethicin assumes an overall helical structure with a kink at Prol4, and the N- and C-termini assume α- and 310-helical structures, respectively [67]. In solution, NMR studies have shown that the N-terminus forms an α-helix and that the C-terminus can assume a variety of helical structures depending on the conditions [70,71]. Another study revealed that poly-Aib forms a 310-helix [72].
Two-dimensional separated local-field 15N NMR spectra were obtained using a motion model of alamethicin in which the peptide rotates about the bilayer normal. The spectra revealed an N–H dipolar coupling of 17 kHz, indicating an N–H bond angle of 24° with respect to the magnetic field (Bo). These data are consistent with an α-helical conformation inserted along the bilayer normal [73].
The conformation of alamethicin in mechanically oriented phospholipid bilayers has been studied using 15N solid-state NMR in combination with molecular modeling and MD simulations. Alamethicin variants labeled with 15N at different positions, in combination with substitution of three Aib residues with Ala residues, were examined. Alamethicin with 15N-labeled Ala6, Val9, and Val15 was incorporated into a phospholipid bilayer at a peptide-to-lipid molar ratio of 1:8. Anisotropic 15N chemical shifts and 1H–15N dipolar couplings obtained using this system determined that alamethicin assumes a largely linear α-helical structure that spans the membrane with the molecular axis tilted by 10–20° relative to the bilayer normal. In comparison, a straight α-helix tilted by 17° and a slightly kinked MD structure tilted by 11° relative to the bilayer normal showed a good compatibility with the structure and orientation of membrane bound alamethicin obtained from solid-state NMR [74]. Measurement of the orientation-dependent 1H–15N dipole-dipole coupling, 15N anisotropic chemical shift, and 2H quadrupole coupling parameters for a single residue, combined with analysis of the anisotropic interaction for the Aib8 residue, provided detailed information regarding the helix-tilt angle, wobble, and oscillatory rotation around the helical axis in the membrane-bound state of alamethicin [75].
Proton-decoupled 15N solid-state spectra of a sample of alamethicin uniformly labeled with 15N and reconstituted into oriented palmitoyloleoylphosphatidylcholine (POPC) and DMPC membranes showed that alamethicin adopts a transmembrane orientation [76], Two-dimensional 15N chemical-shift 1H–15N dipolar-coupling solid-state NMR correlation spectroscopy (i.e. PISEMA) analysis suggested that in the transmembrane configuration, alamethicin adapts a mixed α/310-helical structure with a tilt angle of 8.9° with respect to the bilayer normal [77].
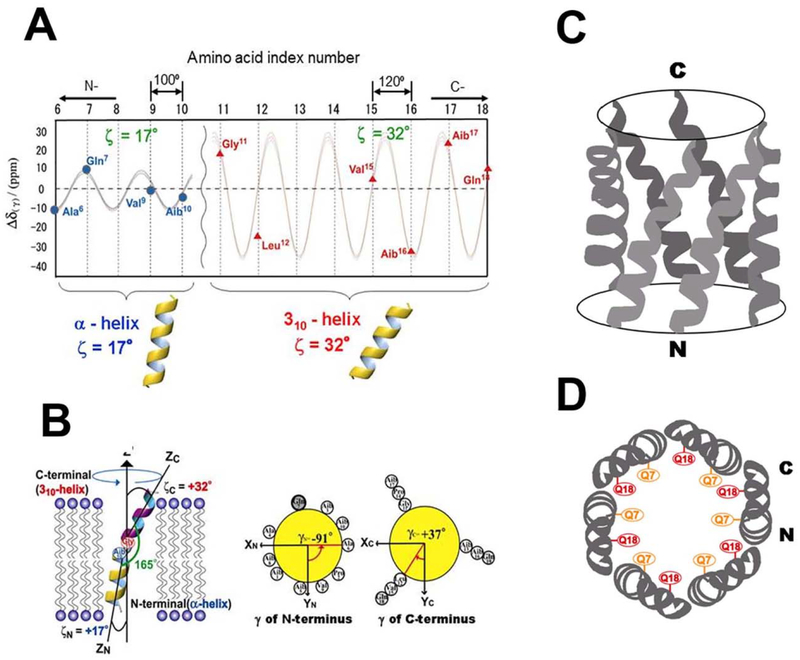
Further detailed structural and orientational analyses of membrane-bound alamethicin have been carried out using solid-state NMR spectroscopy by analyzing the chemical shift oscillation patterns [12,50]. 13C chemical-shift interactions were observed in [1-13C]-labeled alamethicin. The isotropic chemical-shift values indicated that the entire length of alamethicin forms a helical structure. The CSA of the carbonyl carbon of isotopically labeled alamethicin was also analyzed under the assumption that alamethicin molecules rotate rapidly about the bilayer normal. It was concluded that the adjacent peptide plane forms an angle of 100° or 120° upon assumption of an α- or 310-helix, respectively. Anisotropic data were acquired for four and seven different residues at the N- and C-termini, respectively. The observed chemical-shift oscillation patterns for the 13C CSAs of the carbonyl carbons are shown in Fig. 8A. This chemical-shift oscillation pattern clearly indicated that there are two helices with different tilt angles. RMSD analysis indicated that the N- and C-terminal helical axes tilt 17° and 32°, respectively, relative to the lipid bilayer normal (Fig. 8B). The dihedral angles of adjacent peptide planes of the N- and C-termini were found to be 100° and 120°, respectively. It was, therefore, determined that the N- and C-termini form α- and 310-helices, respectively (Fig. 8) [12,50].
Fig. 8.
(A): Chemical shift oscillation patterns of alamethicin. Chemical shift oscillation curves were obtained from the chemical shift anisotropies of the N-terminus (AIa6, Gln7, Val9 and Aib10) and the C-terminus (Val15, Aib16, Aib17, and Gln18). The tilt angles of the N- and C-termini were determined to be 17° and 32°, respectively. The dihedral angles of the peptide planes between the n− and n + 1-residues of the N- and C-termini were determined to be α- and 310-helices, respectively. (B): Structure and topology of alamethicin bound to a DMPC bilayer, as determined from chemical shift oscillation data [50]. Side view (C) and top view (D) of the hexameric oligomer of alamethicin in the membrane environment.
Hexagonal alamethicin ion channel model is illustrated in Fig. 8C and D, based on the previously obtained structure by MD simulation [74]. The membrane bound structure of alamethicin determined by solid-state NMR analysis is used as components of a hexagonal alamethicin oligomer [50]. It is of interest to note that Gln7 and Glnl8 residues face to the inside of the ion channel, and thus the inside of the helical bundle shows a hydrophilic character suggesting the high ion conductivity inside the channel.
4. Protein aggregation and amyloid formation in a membrane environment
4.1. Protein aggregation is implicated in devastating neurodegenerative diseases
Protein aggregation is in general an exciting area of research and has implications in many disciplines including biomedicine and bioengineering. Particularly, misfolding and aggregation of a variety of amyloid proteins are implicated in aging-related diseases such as Alzheimer’s disease, Parkinson’s disease, Huntington’s disease, and type-2 diabetes [78–80], Most of these amyloid proteins are typically unstructured as monomers, and are known to aggregate to form oligomeric intermediates, protoflbers and Anally they end up as amyloid fibers. The process of formation of the species is intriguing and varies with experimental conditions and the nature of the peptide or protein [81]. Physiological studies have shown that the process of aggregation of amyloid-beta (typically containing 39, 40 or 42 residues) causes neuronal cell death and memory loss in Alzheimer’s disease [82], while the aggregation of islet amyloid polypeptide (IAPP, or also known as amylin) kills insulin-producing islet cells only in the case of type-2 diabetics [83]. Therefore, there is considerable interest in understanding the kinetics of aggregation, physicochemical properties and structures of intermediates formed in the aggregation pathway, and polymorphic structures of amyloid fibers. Numerous low-resolution microscopic and diffraction studies and high-resolution solid-state NMR investigations have provided a wealth of information on amyloid fibers [84], Though most of the amyloid fibers reported are found to contain beta-sheet structures, studies have shown polymorphism in the structure of amyloid fibers. Amyloid fibers have also inspired the development of nano-fibril materials for various potential chcmical, biomedical and biotechnological applications. However, for many of these amyloid peptides/proteins, functional studies have demonstrated that oligomeric intermediates are the most toxic species and matured fibers are typically not toxic. To better understand the toxic nature of oligomeric intermediates and to develop chemical compounds to avoid their formation, many research groups are currently investigating the preparation and physicochemical characterization, thermodynamic stability and kinetics of formation, high-resolution structure, and heterogeneity of amyloid oligomers.
4.2. Membrane interaction of an amyloid peptide/protein is similar to that of an antimicrobial peptide
Most importantly, the cellular toxicity induced by the amyloid aggregation is mediated or amplified by the lipid components of the cell membrane [85,86]. In fact, the membrane interaction, oligomerization and cell toxicity of an amyloid protein are somewhat similar to that of the well-studied antimicrobial peptides. In addition to the membrane interaction and aggregation similarities between antimicrobial peptides and amyloid proteins/peptides, studies have reported the amyloid fiber formation by antimicrobial peptides and antibacterial properties of some amyloid proteins. Therefore, the biophysical and biochemical approaches successfully developed to study the mechanism of antimicrobial peptides can be applied to understand the structure and function of amyloid proteins, though amyloid proteins are more challenging.
4.3. Cell membrane catalyzes amyloid aggregation that results in cell toxicity
Because of the link between the cellular toxicity and amyloid aggregation in various aging related amyloid diseases, the role of a lipid membrane on amyloid aggregation must be investigated to fully understand the cellular toxicity of an amyloid peptide or protein; it is remarkable that lipid membrane components dramatically alter most aspects of amyloid aggregation [86–88]. However, these are very difficult tasks to be accomplished, as there are numerous factors that control the formation of oligomers in solution, which include temperature, pH, ionic concentration, metals (such as copper, zinc, aluminum, calcium, and iron), and concentration of protein/peptide [89,90]. In addition to these difficulties, the need for investigating amyloid aggregation in a membrane environment is further complicated by additional variables like the nature of lipids (for example, charge of head group and hydrophobic length of acyl chains), membrane fluidity and curvature, lipid bilayer structure, lipid domains (for example, raft domains), and the presence of other membrane components (for example, cholesterol) [86,87]. Nevertheless, recent studies have made significant contributions in disentangling the many roles of the cell membrane on amyloid aggregation and the formation of toxic species by effectively combining in-vitro and in-vivo studies. The in-vitro studies utilized a combination of various biophysical techniques and membrane mimetics to investigate membrane-protein interactions. Solid-state NMR spectroscopy has elucidated the membrane-induced conformation of Aβ, in which the disordered N-terminal segment is followed by the stable C-terminal β-sheet [91,92]. Membrane binding affinity, low and high resolution structures and topologies of membrane-bound amyloid species, and changes in membrane order/disorder and structure are several important aspects reported to date in the literature [86,87,93–100]. NMR experiments revealed the helical structural domains of Aβ upon interaction with lipid bilayers. Zwitterionic lipid bilayers were used in this study to lower the binding affinity and to slower the aggregation in order to trap the membrane-bound non-fibril structure of Aβ-1–40 [100]. Interestingly, the helical structured membrane-bound region of the peptide is similar to the 310 helical structure determined from a solution sample of Aβ-1–40 [101]. A recent study demonstrated the effect of membrane thinning by probing the interaction of Aβ with lipid bilayers varying in hydrophobic thickness [102]. The thinner membrane enabled easy membrane fragmentation induced by the Aβ peptide; this finding has correlation with the lipid degradation and the formation of fatty acids due to the cell aging of neuronal cells and also due to the physiology of Alzheimer’s disease. We refer the readers to recent review articles reported in the literature that comprehensively cover these aspects [86–88] and to several recent publications that reported novel findings [93–100]. The ability to measure amyloid peptide induced membrane disruption by solid-state NMR experiments is briefly discussed below by using IAPP as an example.
4.4. Solid-state NMR reveals the detergent-type membrane disruption by IAPP
Islet amyloid polypeptide (IAPP, also known as amylin) is a 37-residue peptide hormone whose aggregation to form amyloid fibers is implicated in the loss of insulin producing pancreatic beta-cells [83,103,104]. Thioflavin-based fluorescence experiments, circular dichroism (CD) and NMR experiments have been used to discern the aggregation kinetics, structure and the effects of temperature, pH, zinc and insulin [105–116]. Human-IAPP forms “micelle-like” mesoscopic aggregates whereas the rat-IAPP does not [90]. It is also reported that micelle formation correlated with amyloidogenicity of the peptide: the naturally-occurring C-amidated human-IAPP exhibited about 10 times less CMC than the non-amidated while the human-IAPP-1–19 fragments of both human and rat did not form micelles [109]. Studies have shown that the presence of lipid influences the aggregation of IAPP [86,93–98,117–125]. Both 1–19 and full-length human-IAPP were shown to be toxic to islet cells whereas the 1–19 and full-length rat-IAPP did not exhibit toxicity [116]. A two-step mechanism of membrane disruption has been reported for IAPP: non-fiber structures of IAPP form pores in the first step and fiber-formation dependent “detergent-type” membrane disruption in the second-step [93]. The pore-formation can be suppressed by acidic pH and the fiber formation can be suppressed by insulin, zinc or polyphenolic compounds such as curcumin and EGCG [86,89,90,110]. These studies explain the roles that acidic pH, insulin and zinc play in keeping human-IAPP in a nontoxic mode in non-diabetics. NMR structural studies reported high-resolution three-dimensional structures of membrane-bound IAPP [126–128]. To avoid membrane disruption, detergent micelles were used to trap the helical species of IAPP. To further probe the fiber-formation dependent “detergent-type” membrane disruption by human-IAPP, solid-state NMR experiments were performed on aligned lipid bilayers [128,129]. These NMR studies demonstrated that the C-terminal domain of human-IAPP plays a vital role in fiber formation [108], and thus human-IAPP-20–29 peptide fragment (SNNFGAILSS) which forms fibers [130], was used to demonstrate the membrane disruption process. 31P NMR experiments on anode aluminum oxide (AAO) nanotubes revealed the fragmentation of aligned lipid bilayers in the presence of human IAPP-20–29 (Fig. 9) [129]. The appearance of an isotropic 31P peak illustrates the formation of small “micelle-like” fragments caused by human-IAPP-20–29. At the same time, these results also demonstrated that a non-fiber forming peptide, rat-IAPP-20–29 (SNNLGPVLPP), does not disrupt membrane [131]. These results demonstrate the unique ability of solid-state NMR experiments to probe “detergent-type” membrane disruption by amyloid peptides (Fig. 10). Studies have shown that the presence of anionic lipids enhance the binding affinity [86], negative curvature strain inducing PE lipids enhanced membrane fragmentation [96], and cholesterol to modulate the membrane mediated aggregation of human-IAPP [97]. The presence of calcium ions has been shown to promote human-IAPP aggregation and membrane fragmentation via a carpet-like mechanism [94]. For more details on the biological properties of IAPP, we refer readers to studies reported in the literature [132–138].
Fig. 9.
Phosphorus-31 chemical shift spectra of aligned DMPC bilayers containing various mol% of IAPP in AAO nanotubes: (A) 0%, (B) 0.5% human-IAPP20–29, (C) 0.75% human-IAPP20–29, (D) 1% human-IAPP20–29, (E) 1% human-IAPP20–29 after incubation for 48 h, (F) 1.25% human-IAPP20–29, (G) 0.5% rat-IAPP20–29, (H) 1% rat-IAPP20–29.
Fig. 10.
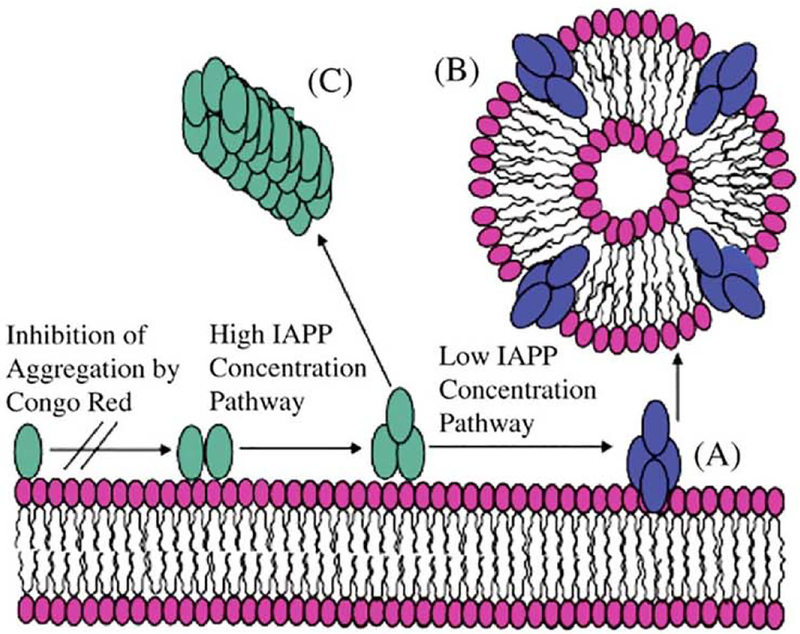
A schematic model for membrane fragmentation by human IAPP20–29. (A) At lower concentrations, the peptide aggregates on the lipid bilayer surface to form an intermediate state that can extract phospholipid molecules from the bilayer, leading to peptide–lipid vesicles (B). At higher concentrations, fiber formation follows an alternate pathway bypassing the formation of this intermediate (C).
5. Dynamics of amphotericin B and sphingomyelin in membrane bilayers
Isotope-labeled peptides and proteins suitable for solid-state NMR measurements are readily available because general methods for isotope labeling are well established. On the other hand, isotope labeling of small molecules such as natural products remains challenging because typically they must be chemically synthesized, and the synthetic route is different for each compound. This makes it more difficult and challenging to apply solid-state NMR to small molecules than to peptides and proteins, and consequently applications of solid-state NMR to membrane-associated natural products remain limited. Thus, studies combining organic synthesis and solid-state NMR are necessary to reveal the dynamic structures of natural products in membrane environments. As examples of such studies, we first present a molecular interaction analysis of amphotericin B with sterol in a hydrated bilaycr, followed by solid-state NMR applications to sphingomyelin, a major component of lipid rafts.
5.1. Amphotericin B
Amphotericin B (AmB) is a clinically important antifungal drug used for over 50 years to treat deep-seated systemic fungal infections [139,140]. Despite its serious side effects, until recently the clinical importance of AmB remained unchanged due to the lack of better alternatives and the rarity of resistant strains. It is generally accepted that AmB forms ion channels with sterols and/or extracts sterols from cell membranes [141,142], leading to cell damage or cell death. Its selective toxicity against fungi over animals is believed to result from its stronger interaction with ergosterol (Erg), an abundant sterol in fungal membranes, than with cholesterol, the major sterol in animal cell membranes [143].
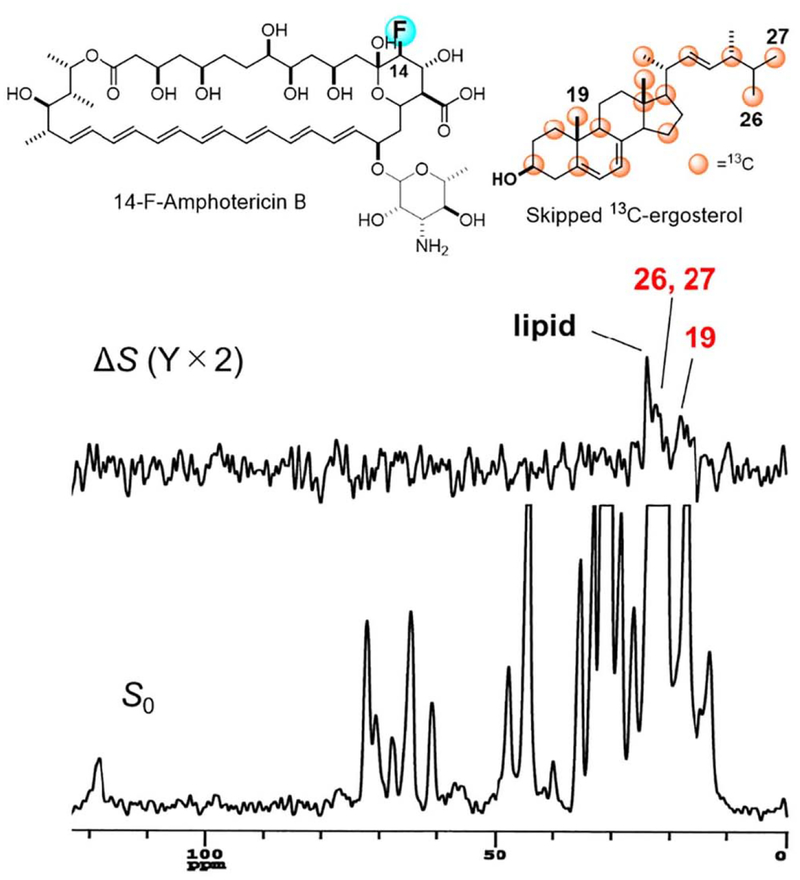
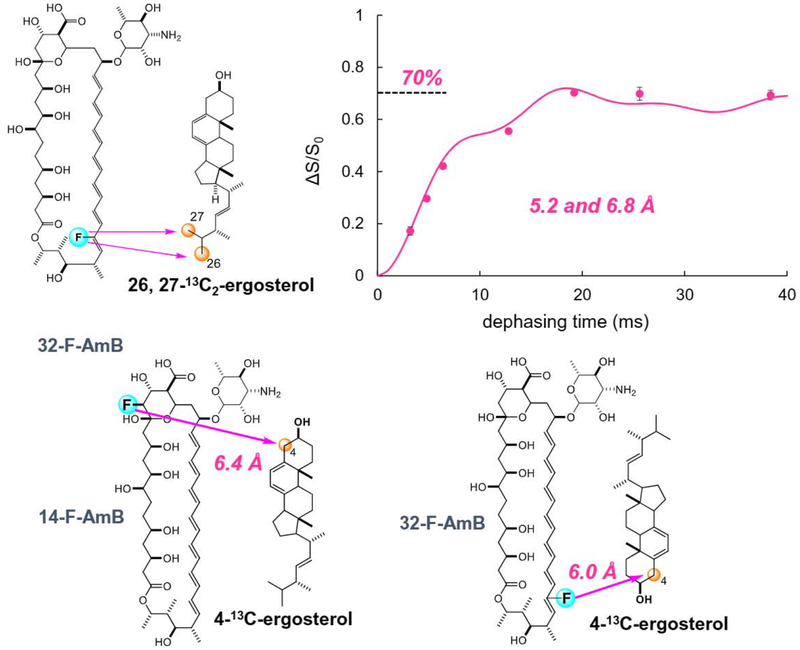
Direct interaction between AmB and sterol, particularly Erg, was verified by 2H NMR measurements of deuterated Erg [144]: the fast axial rotation of deuterated Erg in lipid bilayers was inhibited by the presence of AmB. Insight into the mode of interaction of AmB with Erg were obtained by measuring the intermolecular REDOR between 13C-labeled Erg and F-labeled AmB (Fig. 11) [145]. An F nucleus was chemically introduced at the C-14 position of AmB (14-F-AmB) [146], and Erg was 13C-labeIed by culturing yeast cells in the presence of 2-13C-acetate, furnishing skipped 13C-labeled Erg (Fig. 11). The REDOR experiments unexpectedly suggested that Amp-Erg complexes interacted not only in parallel but also in anti-parallel with each other in lipid bilayers: the REDOR effect from the F nucleus of AmB to the C-19 in Erg indicated a parallel interaction, while that to the C-26/C-27 or C-21 in the Erg side chain suggested an antiparallel alignment in the lipid bilayer (Fig. 11). Previously proposed AmB-Erg assembly models [141,142] suggested that AmB adopts a parallel interaction, but the REDOR results raise the possibility of a different mode of bimolecular recognition, namely, antiparallel interaction, in the AmB-Erg complex.
Fig. 11.
Structures of 14-F AmB and skipped 13C-ergosteroL (top) and intermolecular 13C–19F REDOR spectra of the 1:1 mixture in hydrated bilayers. ΔS is a difference spectrum obtained by subtracting a 19F-irradiated spectrum S from a non-irradiated spectrum, S0 (full echo). Numbers in tile spectra denote the carbon atoms of Erg.
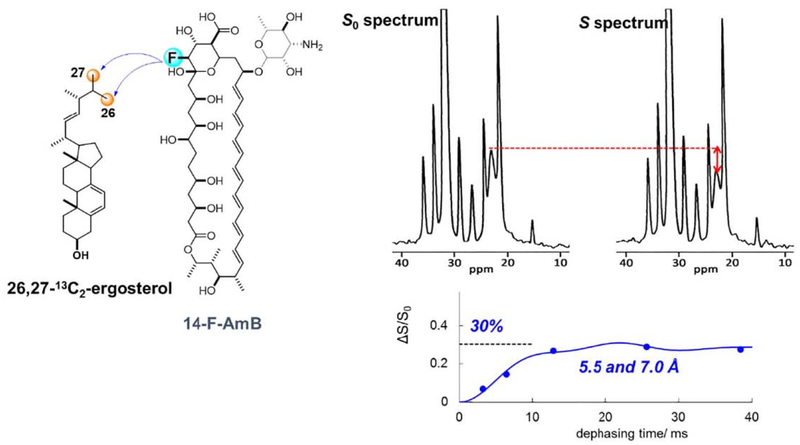
To obtain a clearer evidence for the presence of the antiparallel orientation, we chemically synthesized 26,27-13C2-Erg, in which only the C-26 and C-27 positions were selectively 13C-labeled with 100% enrichment [145]. 13C–19F REDOR experiments with 14-F-AmB and 26,27-13C2-Erg demonstrated that the 13C atoms in the Erg side chain closely approach the 19F atom of the AmB head group (Fig. 12), thus further supporting an antiparallel binary interaction between AmB and Erg in lipid bilayers. The REDOR effect is saturated at ca. 30%, which probably indicates that the antiparallel alignment occurs at ca. 30%. The interatomic distance between the F of AmB and C-26/C-27 of Erg was estimated to be 5.5 and 7.0 Å, where the values are interchangeable, by fitting the REDOR plots to theoretical curves (Fig. 12).
Fig. 12.
Intermolecular 13C–19F REDOR observation between 14-F-AmB and 26,27-13C2-Erg for the antiparallel orientation. The internuclear distances are derived by fitting the REDOR plots to theoretical curves (bottom right).
Next, to obtain distance information for the parallel orientation, 32-F-AmB was synthesized and intermolecular REDOR measurements between 32-F-AmB and 26,27-13C2-Erg were performed [147]. Interestingly, the REDOR effect plateaued at 70% (Fig. 13 top), consistent with the aforementioned 30% saturation of the REDOR effect for the antiparallel complex. The results indicate the coexistence of parallel and antiparallel orientations for AmB and Erg at a ratio of 7:3. Theoretical curve fitting provided estimates of the F-32/C-26 and F-32/C-27 distances of 5.2 and 6.8 Å, or vice versa (Fig. 13 top).
Fig. 13.
Intermolecular 13C–19F REDOR observation between 32-F-AmB and 26,27-13C2-Erg for the parallel orientation (top), and the other two combinations of REDOR measurements using 4-l3C-Erg (bottom).
We further synthesized 4-13C-Erg and conducted intermolecular REDOR measurements of the other two combinations (Fig. 13 bottom) [147,148]: 14-F-AmB and 4-13C-Erg for the parallel orientation, and 32-F-AmB and 4-13C-Erg for the antiparallel orientation. The REDOR data not only supported the coexistence of parallel and antiparallel orientations at a ratio of 7:3, but also provided additional distance information: 6.4 Å for F-14/C-4 and 6.0 Å for F-32/C-4.
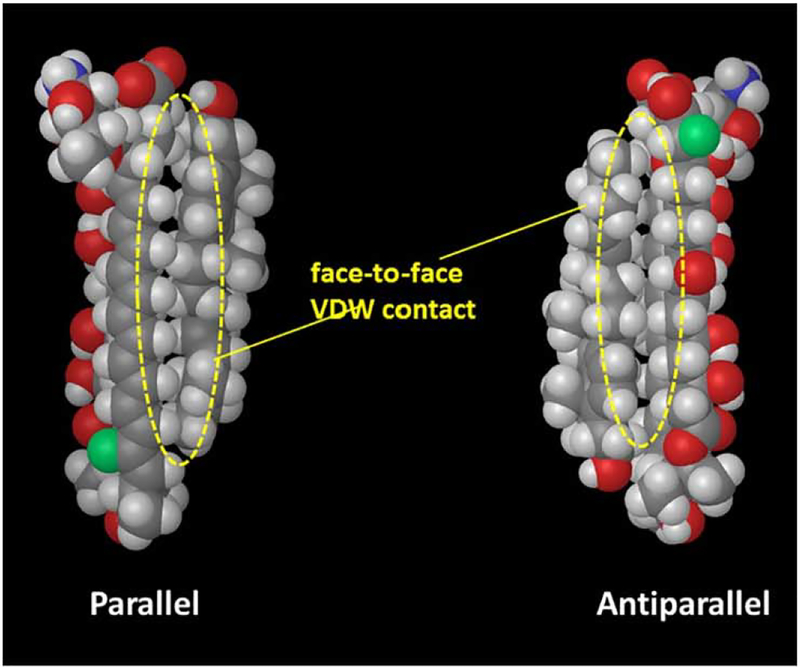
Based on these distance data as well as our previous structure-activity relation study [148], a geometry search for AmB and Erg complexes was performed and the most likely geometries for the AmB-Erg complex were obtained for the parallel and antiparallel orientations (Fig. 14] [147]. The structures show that two molecules are in face-to-face van der Waals (VDW) close contact with each other both in the parallel and antiparallel orientations. The face-to-face contact is less effective with cholesterol, which would explain the weaker activity of AmB in cholesterol-containing animal cell membranes.
Fig. 14.
Structures of the most stable complexes of AmB and Erg for the parallel and antiparallel orientations.
5.2. Sphingomyelin
The lipid rafts hypothesis was proposed in the late 1990s [149], and its implications for membrane processes such as signal transduction, protein sorting, and viral infection have been revealed [150]. In general, cholesterol (Cliol) and sphingolipids such as sphingomyelin (SM), are considered essential for the formation of lipid rafts [151–153]. In addition, SM is known to be the target of some proteinous toxins such as equinatoxins and lysenin [154,155], and it is also reported that the raft formation affects membrane disruptive activities of AMPs [156]. Thus, the dynamics of SM in lipid bilayers is important for understanding not only the mechanism of raft formation but also its influence on the function of membrane-active proteins and peptides including AMPs.
The structure of SM is characterized by the presence of amide and hydroxy groups, which presumably are responsible for the formation of collateral intra- and intermolecular hydrogen bonds in membranes, making SM a preferable raft constituent compared with phosphatidylcholine. In particular, hydrogen bonds between SM and Choi have drawn the most attention as a key factor in raft formation [157–161]. However, the molecular basis of raft formation including inter-molecular hydrogen bonds, has not been fully elucidated, in part, because few NMR experiments have been conducted due to the difficulty of stable isotopical labeling of SM.
We used three approaches to reveal the structure and dynamics of SM molecules in lipid rafts. The first was conformational analysis of SM using bicelles [162]. This study showed that SM forms bicelles in the presence of a detergent, and the conformation of SM in the small bicelles was revealed using liquid state NMR. The second approach was dynamics analysis of the SM alkyl chains using 2H NMR [163–165]. Seventeen different site-selectively deuterated SMs were prepared and site-specific dynamics of SM was revealed in raft and non-raft membranes. In the third approach, the orientation of the SM amide plane in lipid bilayers was analyzed using dipolar couplings and CSAs [166] as described below.
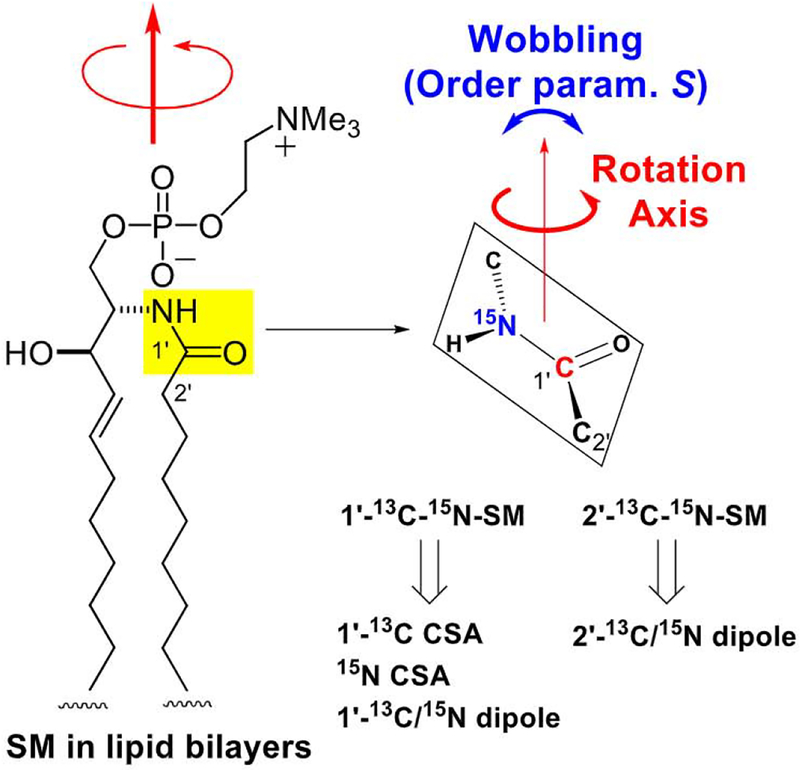
The orientation of the SM amide plane can be defined by the rotation axis direction and the order parameter S (Fig. 15). S represents the degree of wobbling of the rotation axis, and varies from 1 to 0. When S = 1, no molecular wobbling with respect to the rotation axis is present, and when S = 0, every orientation is allowed. To determine the rotation axis direction and order parameter, we prepared the 13C, 15N doubly-labeled SMs, l’−13C-2-15N-SM and 2’−13C-2-15N-SM (Fig. 15). The former sample provided l’−13C and 15N CSAs, and the intramolecular l’−13C-2-15N dipole coupling constant in hydrated bilayers with and without Choi, and the latter gave the intramolecular 2’−13C-2-15N dipole coupling. The CSAs and intramolecular dipole couplings were obtained through slow MAS and REDOR experiments, respectively [164].
Fig. 15.
Schematic summary of orientation analysis of the SM amide plane in lipid bi-layers. The orientation of the SM amide, which is defined by the rotation axis direction and the order parameter S, was analyzed using CSAs and dipole coupling constants, both of which were derived from synthetic 13C and 15N doubly labeled sphingomyelins.
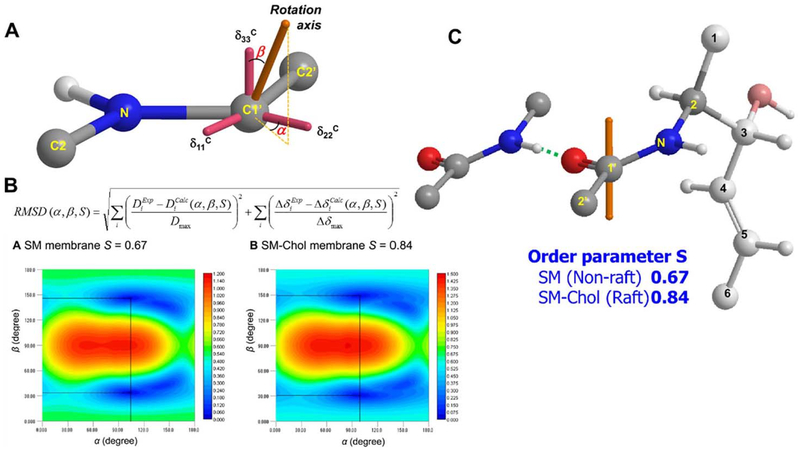
The rotation axis was defined by the polar coordinates α (azimuthal angle) and β (polar angle) with respect to the 13C CSA tensor axes (Fig. 16A). The CSA tensor axis directions were determined by theoretical calculation based on the gauge including atomic orbital (GIAO) method. The CSAs and intramolecular dipole couplings can be calculated using the α, β, and S values, and therefore it is possible to find the combination of α, β, and S that best reproduces the experimental data by minimizing the RMSD. Fig. 16B shows the 2D RMSD plots for SM and SM-Chol membranes at S values of 0.67 and 0.84, respectively, which provide the minimum RMSD values. Using these data, the rotation axis direction and order parameter S of the SM amide in hydrated bilayers were determined, as shown in Fig. 16C. Interestingly, this demonstrated that the SM amide orientation is not affected significantly by the presence or absence of Choi, which suggests that direct hydrogen bonding between the SM amide and Chol may not be as strong as previously proposed. This is also in line with our previous reports that demonstrated that Choi is more deeply positioned in SM than in the PC membrane [163–165], because the formation of a direct hydrogen bond between the SM amide and the OH group of deeply situated Choi should induce some changes in the position and orientation of the amide group. We also noticed that the orientation and conformation around the SM amide is suitable for forming intermolecular hydrogen bonds with neighboring SM amides, as shown in Fig. 16C. Given that the amide group in SM can act as both a hydrogen-bond donor and an acceptor, while the ester groups in phosphatidylcholine function as hydrogen-bond acceptors, it can be assumed that the intermolecular hydrogen-bonds between the SM amide groups, and subsequent formation of a hydrogen-bond network or cluster among SM molecules, is an important molecular basis for raft formation.
Fig. 16.
(A) The rotation-axis direction is defined by the polar coordinates α (azimuthal angle) and β (polar angle) with respect to the 13C CSA tensor axes. The oxygen atom on C-l’ was omitted for clarity. (B) 2D RMSD contour plots for SM at S = 0.67 and SM-Chol at S = 0.84. (C) The rotation-axis direction of SM in hydrated membranes. The orange axes indicate the rotation axes.
What, then, is the role of Chol in raft formation? As demonstrated in this study, Choi significantly enhanced the order parameter S of the SM amide, from 0.67 to 0.84, which means that Chol restricts rotation axis wobbling of the SM amide. On the other hand, as described above, Chol does not change the amide orientation (Fig. 16C), which suggests that Chol is less likely to directly interact with the SM amide group. It was previously demonstrated that Chol is positioned deep in the SM membrane and significantly restricts the motion of the middle region of the SM alkyl chains [163–165]. Therefore, overall, Chol restricts the dynamic motion of the middle region of SM alkyl chains, which indirectly promotes the formation of hydrogen bonds between SM amide groups. These intermolecular hydrogen bonds would restrict the wobbling of the amide group, and lead to the formation of stable lipid rafts.
6. Conclusion
In this review article, the ability of solid-state NMR spectroscopy to provide biologically meaningful information on the structure and function of membrane bound biomolecules arc highlighted. Although there are several reports in the literature on the structure of melittin, the most important dynamic structure of melittin in a membrane environment is obtained using solid-state NMR experiments that reported a pseudo-transmcmbrane bending α-helical structure and the ability of melittin to disrupt the lipid bilayer. The solid-state determined structure of alamethicin was found to be a transmembrane structure with the N- and C-termini forming an α-helix and a 310-helix, respectively, and these helices are associated with the ion channel formation. Solid-state NMR studies demonstrated that the C-terminal domain of human-IAPP plays a vital role in fiber formation, and a human-IAPP-20–29 peptide fragment was used to demonstrate the membrane disruption process, while non-fiber forming rat-IAPP-20–29 does not disrupt the membrane. By using distances measured from solid-state NMR, geometry for the amphotericin B ergosterol complex was found to take both parallel and anti-parallel orientations. The geometry further explained the selective activity of amphotericin B toward an ergosterol-containing fungal membrane over a cholesterol-containing animal cell membrane. The formation of a hydrogen-bond network among sphingomyelin molecules plays an important role in the raft or membrane domain formation. It was also revealed that cholesterol is located deep in the sphingomyelin membrane and significantly restricts the motion of the middle region of sphingomyelin alkyl chains, which lead to the formation of stable lipid rafts. While these are a few selected examples to demonstrate the applications of solid-state NMR techniques, there are many studies that have reported high-resolution structures of membrane proteins that are beyond the scope of this review article. We refer the readers to some of the recent important solid-state NMR approaches for structural and dynamical studies of membrane-associated proteins [167–177]. The advancements in the development of higher field magnets, faster spinning MAS probes, and lipid nanodiscs would significantly overcome the remaining challenges in the membrane arena.
Acknowledgement
Researches in the Naito and Matsumori groups were supported by grants-in-aid for Scientific Research in an Innovative Area 16H00756 (AN) and 16H00773 (NM), and by grants-in-aid for Scientific Research (C) 15K06963 (AN) and (B) 15H03121 (NM) from the Ministry of Education, Culture, Sports, Science and Technology of Japan. Research in the Ramamoorthy laboratory has been supported by funds from the National Institutes of Health: for studies on antimicrobial peptides (AI054515), amyloid proteins (AG048934 and Protein Folding Disease Center at the University of Michigan), and membrane proteins (GM084018). NIH funding supports for the purchase a 600 MHz solid-state NMR spectrometer (to A.R.) and to upgrade a 400 MHz solid-state NMR spectrometer (GM08401816-S1 to A.R.) at the University of Michigan are also acknowledged. A.R. thanks the past and present members of the “Ramyloid” subgroup in the Ramamoorthy laboratory and all the collaborators who have made contributions to overcome the numerous challenges in the exciting amyloid and antimicrobial projects.
Footnotes
Conflict of interest
We do not have any conflict of interest to declare.
This article is part of a Special Issue entitled “Biophysical Exploration of Dynamical Ordering of Biomolecular Systems” edited by Dr. Koichi Kato.
Transparency document
The http://dx.doi.org/10.1016/j.bbagen.2017.06.004 associated with this article can be found, in the online version.
References
- [1].Jenssen H, Hamill P, Hancock BEW, Peptide antimicrobial agents, Clin. Microbiol. Rev 19 (2006) 491–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Matsuzaki K, Murase O, Fujii N, Miyajima K, An antimicrobial peptide, magainin 2, induced rapid flip-flop of phospholipids coupled with pore formation and peptide translocation, Biochemistry 35 (1996) 11361–11368. [DOI] [PubMed] [Google Scholar]
- [3].Henzler Wildman KA, Lee DK, Ramamoorthy A, Mechanism of lipid bilayer disruption by the human antimicrobial peptide, LL-37, Biochemistry 42 (2003) 6545–6558. [DOI] [PubMed] [Google Scholar]
- [4].Pouny Y, Rapaport YD, Mor A, Nicolas P, Shai Y, Interaction of antimicrobial dermaseptin and its fluorescently labeled analogues with phospholipid membrane, Biochemistry 31 (1992) 12416–12423. [DOI] [PubMed] [Google Scholar]
- [5].Sani M-A, Separovic F, How membrane-active peptides get into lipid membranes? Acc. Chem. Ree 49 (2016) 1130–1138. [DOI] [PubMed] [Google Scholar]
- [6].Kichler A, Mason AJ, Bechinger B, Cationic amphipathic histidine-rich peptides for gene delivery, Bίochim. Biophys. Acta 1758 (2006) 301–307. [DOI] [PubMed] [Google Scholar]
- [7].Moulay G, Leborgne C, Mason AJ, Aisenbrey C, Kichler A, Bechinger B, Histidine-rich designer peptides of the LAH4 family promote cell delivery of a multitude of cargo, J. Pept. Sci 23 (4) (April 2017) 320–328. [DOI] [PubMed] [Google Scholar]
- [8].Smith R, Thomas DE, Separovic F, Atkins AR, Cornell BA, Determnation of the structure of a membrane-incorporated ion channel. Solid-state nuclear magnetic resonance of gramicidine A, Biophys. J 56 (1989) 308–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Opella SJ, Marassi FM, Structure determination of membrane proteins by NMR spectroscopy, Chem. Rev 104 (2004) 3587–3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Watts A, Straus SK, Grage SL, Kamihira M, Lam YH, Zhao X, Membrane protein structure determination using solid-state NMR, in; Downing AK (Ed.), Protein NMR, Techniques, Methods in Molecular Biology, Vol. 278 Humana Press, Totowa, New Jersy, 2004, pp. 403–473. [DOI] [PubMed] [Google Scholar]
- [11].Saitô H, Ando L, Naito A, Solid state NMR spectroscopy for biopolymers, Principles and Applications, Springer, Berlin, 2006, pp. 127–149. [Google Scholar]
- [12].Naito A, Structure elucidation of membrane-associated peptides and proteins in oriented bilayers by solid-state NMR spectroscopy, Solid State Nucl. Magn. Reson 36 (2009) 67–76. [DOI] [PubMed] [Google Scholar]
- [13].Opella SJ, Das BB, Determination of the equivalence of solid-state NMR orientational constraints from magnetic and rotational alignment of the coat protein in a filamentous bacteriophage, in: Separopvic F, Naito A (Eds.), Advances in Biological Solid State NMR: Protein and Membrane-active Peptides, Royal Society of Chemistry, Cambridge, UK, 2014, pp. 53–70. [Google Scholar]
- [14].Naito A, Kawamura I, Javkhlantugs N, Recent solid-state NMR studies of membrane-bound peptides and proteins, Annu. Rev. NMR Spectrosc 86 (2015) 333–411. [Google Scholar]
- [15].Toraya S, Nishimura K, Naito A, Dynamic structure of vesicle-bound melittin in a variety of lipid chain lengths by solid-state NMR, Biophys. J 87 (2004) 3323–3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Marassi FM, Ramamoorthy A, Opella SJ, Complete resolution of the solid-state NMR spectrum of a uniformly 15N-labeled membrane protein in phospholipid bilayers, Proc. Natl. Acad. Sci 94 (1997) 8551–8556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Opella SJ, Solid-state NMR and membrane proteins, J. Magn. Reson 253 (2015) 129–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Weingarth M, Buldus M, Introduction to Biological Solid-state NMR, in: Separopvic F, Naito A (Eds.), Advances in Biological Solid State NMR: Protein and Membrane, Royal Society of Chemistry, Cambridge, UK, 2014, pp. 1–17. [Google Scholar]
- [19].Ward ME, Brown LS, Ladizhansky V, Advanced solid-state NMR techniques for characterization of membrane protein structure and dynamics: application to anabaena sensory rhodopsin, J. Magn. Reson 253 (2015) 119–128. [DOI] [PubMed] [Google Scholar]
- [20].Naito A, Nagao T, Norisada K, Mizuno T, Tuzi S, Saito H, Conformation and dynamics of melittin bound to magnetically oriented lipid bilayers by sold-state 31P and 13C NMR spectroscopy, Biophys. J 78 (2000) 2405–2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Norisada K, Javkhlantugs N, Mishima D, Kawamura I, Saito H, Ueda K, Naito A, Dynamic structure and orientation of melittin bound to acidic lipid bilayers, as revealed by solid-state NMR and molecular dynamics simulation, J. Phys. Chem. B 121 (2017) 1802–1811. [DOI] [PubMed] [Google Scholar]
- [22].Wu CH, Ramamoorthy A, Opella SJ, High-resolution heteronuclear dipolar solid-state NMR spectroscopy, J. Magn. Reson A109 (1994) 270–272. [Google Scholar]
- [23].Ramamoorthy A, Wei Y, Lee D-K, PISEMA solid-state NMR spectroscopy, Annu. Rep. NMR Spectrosc 52 (2004) 1–52. [Google Scholar]
- [24].Lee DK, Narasimhaswamy T, Ramamoorthy A, PITAMSEMA, a low-power PISEMA solid-state NMR experiment, Chem. Phys. Lett 399 (2004) 359–362. [Google Scholar]
- [25].Nishimura K, Naito A, Dramatic reduction of the RF power for attenuation of sample heating in 2D-separated local field solid-state NMR spectroscopy, Chem. Phys. Lett 402 (2005) 245–250. [Google Scholar]
- [26].Nishimura K, Naito A, Remarkable reduction of RF power by ATANSEMA and DATANSEMA separated local field solid-state NMR spectroscopy, Chem. Phys. Lett 419 (2006) 120–124. [Google Scholar]
- [27].Gor’kov PL, Chekmenev EY, Li C, Cotton M, Buffy JJ, Trasseth NJ, Vehlia G, Brey WW, Using low-E resonance to reduce RF heating in biological samples for static solid-state NMR up to 900 MHz, J. Magn. Reson 185 (2007) 77–93. [DOI] [PubMed] [Google Scholar]
- [28].Yamamoto K, Lee DK, Ramamoorthy A, Broadband-PISEMA solid-state NMR spectroscopy, Chem. Phys. Lett 144 (2005) 289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Marassi FM, Opella SJ, A solid-state NMR index of helical membrane protein structure and topology, J. Magn. Reson 144 (2000) 150–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Marassi FM, Ma C, Gesell JJ, Opella SJ, Three-dimensional solid-state NMR spectroscopy is essential for resolution of resonances from in-phase residues in uniformly 15N-labeled helical membrane proteins in oriented lipid bilayers, J. Magn. Reson 144 (2000) 156–161. [DOI] [PubMed] [Google Scholar]
- [31].Wang J, Denny J, Tian C, Kim S, Mo Y, Kovacs F, Song Z, Nishimura K, Gan Z, Fu R, Quine JR, Cross TA, Imaging membrane protein helical wheels, J. Magn. Reson 144 (2000) 162–167. [DOI] [PubMed] [Google Scholar]
- [32].Andrew ER, The narrowing of NMR spectra of solids by high-speed specimen rotation and resolution of chemical shift and spin multiplet structure for solids, Prog. Nucl. Magn. Reson. Spectrosc 8 (1971) 1–39. [Google Scholar]
- [33].Hartmann SR, Hahn EL, Nuclear double resonance in the rotating frame, Phys. Rev 128 (1962) 2042–2053. [Google Scholar]
- [34].Pines A, Gibby MG, Waugh JS, Proton-enhanced NMR of dilute spins in solids, J. Chem. Phys 59 (1973) 569–590. [Google Scholar]
- [35].Schaefer J, Stejeskal EO, Carbon-13 nuclear magnetic resonance of polymers spinning at magic angle, J. Am. Chem. Soc 98 (1976) 1031–1032. [Google Scholar]
- [36].Grommek A, Meier BH, Ernst M, Distance information from proton-driven spin diffusion under MAS, Chem. Phys. Lett 427 (2006) 631–637. [Google Scholar]
- [37].Takegoshi K, Nakamura S, Terao T, 13C–1H dipolar-assisted rotational resonance in magic-angle spinning NMR, Chem. Phys. Lett 344 (2001) 631–637. [Google Scholar]
- [38].Takegoshi K, Nakamura S, Terao T, 13C–1H dipolar-driven 13C–13C recoupling without 13C RF irradiation in nuclear magnetic resonance of rotating solids, J. Chem. Phys 118 (2003) 2325. [Google Scholar]
- [39].Weingarth M, Demaco DE, Bodenhausen G, Tekely P, Improved magnetization transfer in solid-state NMR with fast magic angle spinning, Chem. Phys. Lett 469 (2009) 342–348. [Google Scholar]
- [40].Scholz I, Huber M, Manolikas T, Meier BH, Ernst M, MIRROR recoupling and its application to spin diffusion under fast magic-angle spinning, Chem. Phys. Lett 460 (2008) 278–283. [Google Scholar]
- [41].Weingarth M, Masuda Y, Takegoshi K, Bodenhausen G, Tekely P, Sensitive 13C–13C correlation spectra of amyloid fibrils at very high spinning frequencies and magnetic fields, J. Biomol. NMR 50 (2011) 129–136. [DOI] [PubMed] [Google Scholar]
- [42].Gullion T, Schaefer J, Rotational-echo double-resonance, J. Magn. Reson 81 (1989) 196–200. [DOI] [PubMed] [Google Scholar]
- [43].Naito A, Nishimura K, Kimura S, Tuzi S, Aida M, Yasuoka N, Saitô H, Determination of the three-dimensional structure of a new crystalline form of N-acetyl-Pro-Gly-Phe as revealed by 13C REDOR, X-ray diffraction, and molecular dynamics calculation, J. Phys. Chem 100 (1996) 14995–15004. [Google Scholar]
- [44].Naito A, Nishimura K, Tuzi S, Saito H, Inter- and intra-molecular contributions of neighboring dipolar pairs to the precise determination of interatomic distances in a simple [13C, 15N]-peptide by 13C, 15N-REDOR NMR spectroscopy, Chem. Phys. Lett 229 (1994) 506–511. [Google Scholar]
- [45].Tutsumi A, Javkhalantugs N, Kira A, Umeyama M, Kawamura I, Nishimura K, Ueda K, Naito A, Structure and orientation of bovine lactoferranpin in the mimetic bacterial membrane as revealed by solid-state NMR and molecular dynamics simulation, Biophys. J 103 (2012) 1736–1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Nishimura K, Naito A, Tuzi S, Saitô H, Analysis of dipolar dephasing pattern in I-Sn multispin system for obtaining the information of molecular packing and its application to crystalline N-acetyl-Pro-Gly-Phe by REDOR solid state NMR, J. Phys. Chem. B 103 (1999) 8398–8408. [Google Scholar]
- [47].Gehman JD, Separovic F, Lu K, Mehta AK, Boltzmann statistics rotational-echo double-resonance analysis, J. Phys. Chem. B 111 (2007) 7802–7811. [DOI] [PubMed] [Google Scholar]
- [48].Jia L, Liang S, Sackett K, Xie L, Ghosh U, Weliky DP, REDOR solid-state NMR as a probe of the membrane locations of membrane-associated peptides and proteins, J. Magn. Reson 253 (2015) 154–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Toraya S, Javkhlantugs N, Mishima D, Norisada K, Ueda K, Naito A, Dynamic structure of bombolitin II bound to lipid bilayers as revealed by solid-state NMR and molecular-dynamics simulation, Biophys. J 99 (2010) 3282–3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Nagao T, Mishima D, Javkhlantugs N, Wang J, Ishioka D, Yokota K, Norisada K, Kawamura I, Ueda K, Naito A, Structure and orientation of antibiotic peptide alamethicin in phospholipid bilayer as revealed by chemical shift oscillation analysis of solid state nuclear magnetic resonance and molecular dynamics simulation, Biochim. Biophys. Acta 1848 (2015) 2789–2798. [DOI] [PubMed] [Google Scholar]
- [51].Habermann E, Jentsc J, Sequence analysis of melittin from tryptic and peptide degradation products, Hoppe-Seyler’s Z, Physiol. Chem 348 (1967) 37–50. [PubMed] [Google Scholar]
- [52].Sessa G, Free JH, Colacicco G, Weissmann G, Interaction of a lytic polypeptide, melittin, with lipid membrane system, J. Biol. Chem 244 (1969) 3575–3582. [PubMed] [Google Scholar]
- [53].Tosteson MT, Tosteson DC, The sting. Melittin forms channels in lipid bilayers, Biophys. J 36 (1981) 109–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Dempsey CE, The action of melittin on membrane, Biochim. Biophys. Acta 1031 (1990) 143–161. [DOI] [PubMed] [Google Scholar]
- [55].Dufourcq J, Faucon J-F, Fourche G, Dasseux JL, Le Maire M, Gulik-Krywicki T, Morphological change of phosphatidylcholine bilayers induced by melittin vesicularization, fusion, discoidal particles, Biochim. Biophys. Acta 859 (1986) 33–48. [DOI] [PubMed] [Google Scholar]
- [56].Toraya S, Nagao T, Norisada K, Tuzi S, Saito H, Izumi S, Naito A, Morphological behavior of lipid bilayer induced by melittin near the phase transition temperature, Biophys. J 89 (2005) 3214–3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Smith R, Separovic F, Milne TJ, Whittaker A, Bennett FN, Cornell BA, Makriyannis A, Structure and orientation of the pore-forming peptide melittin, in lipid bilayers, J. Mol. Biol 241 (1994) 4566–466. [DOI] [PubMed] [Google Scholar]
- [58].Lam Y-H, Wassail SR, Morton CL, Smith R, Separovic F, Solid-state NMR structure determination of melittin in a lipid environment, Biophys. J 81 (2001) 2752–2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Terwilliger TC, Eisenberg D, The structure of melittin. I. Structure determination and partial refinement, J. Biol. Chem 257 (1982) 6010–6015. [DOI] [PubMed] [Google Scholar]
- [60].Terwiliger TC, Weissman L, Eisenberg D, The structure of melittin in the form I crystals and its implication for melittin’s lytic and surface activities, Biophys. J 37 (1982) 835–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Saitô H, Conformation-dependent 13C chemical shifts: a new means of conformation characterization as obtained by high-resolution solid-state, 13C NMR, Magn. Reson. Chem 24 (1986) 196–200. [Google Scholar]
- [62].Saitô H, Ando I, High-resolution solid-state NMR studies of synthetic and biological macromolecules, Annu. Rep. NMR Spectrosc 21 (1989) 209–290. [Google Scholar]
- [63].Saitô H, Ando I, Ramamoorthy A, Chemical shift tensor — the heart of NMR: insights into biological aspects of proteins, Prog. Nucl. Magn. Reson. Spectrosc 57 (2010) 181–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Meyer CE, Reusser F, A polypeptide antibacterial agent isolated from Trichoderma viride, Experientia 23 (1967) 85–86. [DOI] [PubMed] [Google Scholar]
- [65].Balasubramanîan TM, Kendrick NCE, Taylor M, Marshall GR, Hall JE, Vodyanoy I, Reusser F, Synthesis and characterization of the major component of alamethicin, J. Am. Chem. Soc 103 (1981) 6127–6132. [Google Scholar]
- [66].Muellar P, Rudin DO, Action potentials induced in biomolecular lipid membranes, Nature 217 (1968) 713–719. [DOI] [PubMed] [Google Scholar]
- [67].Fox RO Jr., Richards FM, A voltage-gated ion channel model inferred from the crystal structure of alamethicin at 1.5 Å resolution, Nature 300 (1982) 325–330. [DOI] [PubMed] [Google Scholar]
- [68].Pan J, Tristram-Nagle S, Alamethicin aggregation in lipid membranes, J. Membr. Biol 231 (2009) 11–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Sansom MS, Alamethicin and related peptibols — model ion channels, Eur. Biophys. J 22 (1993) 105–124. [DOI] [PubMed] [Google Scholar]
- [70].Esposito G, Carver JA, Boyd J, Campbell ID, High-resolution lH NMR study of the solution structure of alamethicin, Biochemistry 26 (1987) 1043–1050. [DOI] [PubMed] [Google Scholar]
- [71].Yee AA, O’Neil JDJ, Uniform 15N labeling of a fungal peptide: the structure and dynamics of an alamethicin by 15N and 1H NMR spectroscopy, Biochemistry 31 (1992) 3135–3143. [DOI] [PubMed] [Google Scholar]
- [72].Saito H, Tabeta R, Formaggio F, Crisma M, Toniolo C, High-resolution solid-state 13C-NMR of peptides: a study of chain-length dependence for 3(10)-helix formation, Biochemistry 27 (1988) 1607–1617. [DOI] [PubMed] [Google Scholar]
- [73].North CL, Barranger-Marthys M, Cafiso DS, Membrane orientation of the N-terminal segment of alamethicin detected by solid-state 15N NMR, Biophys. J 69 (1995) 2392–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Bak M, Bywater RP, Hohwy M, K Thomsen J, Adelhorst K, Jakobsen HJ, Sorensen OW, Nielsen NC, Conformation of alamethicin in oriented phospholipid bilayers determined by 15N solid-state nuclear magnetic resonance, Biophys. J 81 (2001) 1684–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Bertelsen K, Paaske B, Thogersen L, Tajkhorshid E, Schiott B, Skrydstrup T, Nielsen NC, Vosegaard T, Residue-specific information about the dynamics of antimicrobial peptides from 1H–15N and 2H solid-state NMR spectroscopy, J. Am. Chem. Soc 131 (2009) 18335–18342. [DOI] [PubMed] [Google Scholar]
- [76].Bechinger B, Skladnev DA, Ogrel A, Li X, Rogozhkina EV, Ovchinnikova TV, O’Neil JDJ, Raap J, 15N and 31P solid-state NMR investigations on the orientation of zervamicin H and alamethicin in phosphatidylcholine membranes, Biochemistry 40 (2001) 9428–9437. [DOI] [PubMed] [Google Scholar]
- [77].Salinlkov ES, Freidrich H, Li X, Bertani P, Reissmann S, Hertweck C, O’Neil JDJ, Raap J, Bechinger B, Structure and alignment of the membrane-associated peptidbols ampullosporin A and alamethicin by oriented 15N and 31P solid-state NMR spectroscopy, Biophys. J 96 (2009) 86–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Selkoe DJ, Hardy J, The amyloid hypothesis of Alzheimer’s disease at 25 years, EMBO Mol. Med 8 (2016) 595–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Selkoe DJ, Weinberger DR, Editorial overview: neurobiology of disease, Curr. Opin. Neurobiol 36 (2016) v–vii, 10.1016/j.conb.2016.01.001. [DOI] [PubMed] [Google Scholar]
- [80].Chiti F, Dobson CM, Protein misfolding, amyloid formation, and human disease: a summary of progress over the last decade, Annu. Rev. Biochem (2017), 10.1146/annurev-biochem-061516-045115. [DOI] [PubMed] [Google Scholar]
- [81].So M, Hall D, Goto Y, Revisiting supersaturation as a factor determining amyloid fibrillation, Curr. Opin. Struct. Biol 36 (2016) 32–39, 10.1016/j.sbi.2015.11.009. [DOI] [PubMed] [Google Scholar]
- [82].Thai DR, Walter J, Saido TC, Fändrich M, Neuropathology and biochemistry of Aβ and its aggregates in Alzheimer’s disease, Acta Neuropathol. 129 (2015) 167–182. [DOI] [PubMed] [Google Scholar]
- [83].Westermark P, Andersson A, Westermark GT, Islet amyloid polypeptide, islet amyloid, and diabetes mellitus, Physiol. Rev 91 (2011) 795–826, http://dx.doi.org/10.1l52/physrev.00042.2009. [DOI] [PubMed] [Google Scholar]
- [84].Knowles TP, Mezzenga R, Amyloid fibrils as building blocks for natural and artificial functional materials, Adv. Mater 28 (2016) 6546–6561, 10.1002/adma.201505961 (Epub 2016 May 11). [DOI] [PubMed] [Google Scholar]
- [85].Evangelisti E, Cascella R, Becatti M, Marrazza G, Dobson CM, Chiti F, Stefani M, Cecchi C, Binding affinity of amyloid oligomers to cellular membranes is a generic indicator of cellular dysfunction in protein misfolding diseases, Sci. Rep 6 (2016) 32721, 10.1038/srep32721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Brender JR, Salamekh S, Ramamoorthy A, Membrane disruption and early events in the aggregation of the diabetes related peptide IAPP from a molecular perspective, Acc. Chem. Res 45 (2012) 454–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Kotier SA, Brender JR, Ramamoorthy A, Differences between amyloid-β aggregation in solution and on the membrane: insights into elucidation of the mechanistic details of Alzheimer’s disease, Chem. Soc. Rev 43 (2014) 6692–6700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Matsuzaki K, How do membranes initiate Alzheimer’s disease? Formation of toxic amyloid fibrils by the amyloid β-protein on ganglioside clusters, Acc. Chem. Res 47 (2014) 2397–2404. [DOI] [PubMed] [Google Scholar]
- [89].Patel HR, Pithadia AS, Brender JR, Fierke CA, Ramamoorthy A, In search of aggregation pathways of IAPP and other amyloidogenic proteins: finding answers through NMR spectroscopy, J. Phys. Chem. Lett 5 (2014) 1864–1870. [DOI] [PubMed] [Google Scholar]
- [90].Pithadia A, Brender JR, Fierke CA, Ramamoorthy A, Inhibition of IAPP aggregation and toxicity by natural products and derivatives, J. Diabetes Res 2016 (2016) 2046327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Yagi-Utsumi M, Kato K, Nishimura K, Membrane-induced dichotomous conformation of amyloid β with the disordered N-terminal segment followed by the stable C-terminal β structure, PLoS One (2016), http://dx.doi.org/10.137l/jounal.pone.0146405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Lau TL, Ambroggio EE, Tew DJ, Cappai R, Masters CL, Fidelio CD, Bamham KJ, Separovic F, Amyloid-beta peptide disruption of lipid membranes and the effect of metal ions, J. Mol. Biol 356 (2006) 759–770. [DOI] [PubMed] [Google Scholar]
- [93].Sciacca MF, Kotier SA, Brender JR, Chen J, Lee DK, Ramamoorthy A, Two-step mechanism of membrane disruption by Aβ through membrane fragmentation and pore formation, Biophys. J 103 (2012) 702–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Sciacca MF, Milard D, Messina GM, Brender JR, Ramamoorthy A, La Rosa C, Cations as switches of amyloid-mediated membrane disruption mechanisms: calcium and IAPP, Biophys. J 104 (2013) 173–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Smith PE, Brender JR, Ramamoorthy A, Induction of negative curvature as a mechanism of cell toxicity by amyloidogenic peptides: the case of islet amyloid polypeptide, J. Am. Chem. Soc 131 (2009) 4470–4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Sciacca MF, Brender JR, Lee DK, Ramamoorthy A, Phosphatidylethanolamine enhances amyloid fiber-dependent membrane fragmentation, Biochemistry 51 (2012) 7676–7684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Sciacca MF, Lolicato F, Di Mauro G, Milardi D, D’Urso L, Satriano C, Ramamoorthy A, La Rosa C, The role of cholesterol in driving IAPP-membrane interactions, Biophys. J 111 (2016) 140–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Zhao J, Hu R, Sciacca MF, Brender JR, Ramamoorthy A, Zheng J, Non-selective ion channel activity of polymorphic human islet amyloid polypeptide (amylin) double channels, Phys. Chem. Chem. Phys 16 (2014) 2368–2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Wakabayashi M, Matsuzaki K, Ganglioside-induced amyloid formation by human islet amyloid polypeptide in lipid rafts, FEBS Lett. 583 (2009) 2854–2858. [DOI] [PubMed] [Google Scholar]
- [100].Korshavn KJ, Bhunia A, Lim MH, Ramamoorthy A, Amyloid-β adopts a conserved, partially folded structure upon binding to zwitterionic lipid bilayers prior to amyloid formation, Chem. Commun. (Camb.) 52 (2016) 882–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Korshavn KJ, Satriano C, Lin Y, Zhang R, Dulchavsky M, Bhunia A, Ivanova MI, Lee YH, La Rosa C, Lim MH, Ramamoorthy A, Reduced lipid bilayer thickness regulates the aggregation and cytotoxicity of amyloid-β, J. Biol. Chem 292 (2017) 4638–4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Vivekanandan S, Brender JR, Lee SY, Ramamoorthy A, A partially folded structure of amyloid-beta(1–40) in an aqueous environment, Biochem. Biophys. Res. Commun 411 (2011) 312–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Rodriguez Camargo DC, Tripsianes K, Buday K, Franko A, Gobi C, Hartlmüller C, Sarkar R, Aichler M, Mettenleiter G, Schulz M, Böddrich A, Erck C, Martens H, Walch AK, Madl T, Wanker EE, Conrad M, de Angelis MH, Reif B, The redox environment triggers conformational changes and aggregation of hIAPP in type II diabetes, Sci. Rep 7 (2017) 44041, 10.1038/srep44041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Zhang X, St Clair JR, London E, Raleigh DH, Islet amyloid polypeptide mem brane interactions: effects of membrane composition, Biochemistry 56 (2017) 376–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Gilead S, Wolfenson H, Gazit E, Molecular mapping of the recognition interface between the islet amyloid polypeptide and insulin, Angew. Chem. Int Ed 45 (2006) 6476–6480. [DOI] [PubMed] [Google Scholar]
- [106].Mazor Y, Gilead S, Benhar I, Gazit E, Identification and characterization of a novel molecular-recognition and self-assembly domain within the islet amyloid polypeptide, J. Mol. Biol 322 (2002) 1013–1024. [DOI] [PubMed] [Google Scholar]
- [107].Bram Y, Frydman-Marom A, Yanai I, Gilead S, Shaltiel-Karyo R, Amdursky N, Gazit E, Apoptosis induced by islet amyloid polypeptide soluble oligomers is neutralized by diabetes-associated specific antibodies, Sci. Rep 4 (2014) 4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Brender JR, Lee EL, Cavitt MA, Gafhi A, Steel DG, Ramamoorthy A, Amyloid fiber formation and membrane disruption are separate processes localized in two distinct regions of IAPP, the tyρe-2-diabetes-related peptide, J. Am. Chem. Soc 130 (2008) 6424–6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Brender JR, Krishnamoorthy J, Sciacca MF, Vivekanandan S, D’Urso L, Chen J, Ra Rosa C, Ramamoorthy A, Probing the sources of the apparent irreproducibility of amyloid formation: drastic changes in kinetics and a switch in mechanism due to miceUelike oligomer formation at critical concentrations of IAPP, J. Phys. Chem. B 119 (2015) 2886–2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Suzuki Y, R Brender J, Hartman PK, Ramamoorthy A, Marsh EN, Alternative pathways of human islet amyloid polypeptide aggregation distinguished by, 9F nuclear magnetic resonance-detected kinetics of monomer consumption, Biochemistry 51 (2012) 8154–8162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Soong R, Brender JR, Macdonald PM, Ramamoorthy A, Association of highly compact type II diabetes related islet amyloid polypeptide intermediate species at physiological temperature revealed by diffusion NMR spectroscopy, J. Am. Chem. Soc 131 (2009) 7079–7085. [DOI] [PubMed] [Google Scholar]
- [112].R Brender J, Hartman K, Nanga RP, Popovych N, de la Salud Bea R, Vivekanandan S, Marsh EN, Ramamoorthy A, Role of zinc in human islet amyloid polypeptide aggregation, J. Am. Chem. Soc 132 (2010) 8973–8983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Salamekh S, Brender JR, Hyimg SJ, Nanga RP, Vivekanandan S, Ruototo BT, Ramamoorthy A, A two-site mechanism for the inhibition of IAPP amyloidogenesis by zinc, J. Mol. Biol 410 (2011) 294–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Brender JR, Krishnamoorthy J, Messina GM, Vivekanandan A, La Rosa C, Penner-Hahn JE, Ramamoorthy A, Zinc stabilization of prefibrillar oligomers of human islet amyloid polypeptide, Chem. Commun. (Camb.) 49 (2013) 3339–3341. [DOI] [PubMed] [Google Scholar]
- [115].Brender JR, Lee EL, Hartmen K, Wong PT, Ramamoorthy A, Steel DG, Gafni A, Biphasic effects of insulin on islet amyloid polypeptide membrane disruption, Biophys. J 100 (2011) 685–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Brender JR, Hartman K, Reid KR, Kennedy RT, Ramamoorthy A, A single mutation in the nonamyloidogenic region of islet amyloid polypeptide greatly reduces toxicity, Biochemistry 47 (2008) 12680–12688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Khemtemourian I, Gazit E, Miranker A, Recent insight in islet amyloid polypeptide morphology, structure, membrane interaction, and toxicity in type 2 diabetes, J. Diabetes Res 2016 (2016) 2535878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Hebda JA, Magzoub M, Miranker AD, Small molecule screening in context: lipid-catalyzed amyloid formation, Protein Sd. 23 (2014) 1341–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Porat Y, Kolusheva S, Jelinek R, Gazit E, The human islet amyloid polypeptide forms transient membrane-active prefibrillar assemblies, Biochemistry 42 (2003) 10971–10977. [DOI] [PubMed] [Google Scholar]
- [120].Hebda JA, Magzoub M, Miranker AD, Islet amyloid polypeptide demonstrates a persistent capacity to disrupt membrane integrity, Proc. Natl. Acad. Sci. U. S. A 108 (2011) 9460–9465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Knight JD, Hebda JA, Miranker AD, Conserved and cooperative assembly of membrane-bound alpha-helical states of islet amyloid polypeptide, Biochemistry 45 (2006) 9496–9508. [DOI] [PubMed] [Google Scholar]
- [122].Gao M, Winter R, The effects of lipid membranes, crowding and osmolytes on the aggregation, and fibrillation propensity of human IAPP, J. Diabetes Res 2015 (2015) 849017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Weise K, Radovan D, Gohlke A, Opitz N, Winter R, Interaction of hIAPP with model raft membranes and pancreatic beta-cells: cytotoxicity of hIAPP oligomers, Chembiochem 11 (2010) 1280–1290. [DOI] [PubMed] [Google Scholar]
- [124].Sellin D, Yan LM, Kapumiotu A, Winter R, Suppression of IAPP fibrillation at anionic lipid membranes via IAPP-derived amyloid inhibitors and insulin, Biophys. Chem 150 (2010) 73–79. [DOI] [PubMed] [Google Scholar]
- [125].Jha S, Sellin D, Seidel R, Winter R, Amyloidogenic propensities and conformational properties of ProIAPP and IAPP in the presence of lipid bilayer membranes, J. Mol. Biol 389 (2009) 907–920. [DOI] [PubMed] [Google Scholar]
- [126].Nanga RP, Brender JR, Xu J, Veglia G, Ramamoorthy A, Structures of rat and human islet amyloid polypeptide IAPP(1–19) in micelles by NMR spectroscopy, Biochemistry 49 (2008) 12689–12697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Nanga RP, Brender JR, Xu J, Hartman K, Subramanian V, Ramamoorthy A, Three-dimensional structure and orientation of rat islet amyloid polypeptide protein in a membrane environment by solution NMR spectroscopy, J. Am. Chem. Soc 131 (2009) 8252–8561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Nanga RP, Brender JR, Vivekanandan S, Ramamoorthy A, Structure and membrane orientation of IAPP in its natively amidated form at physiological pH in a membrane environment, Biochim. Biophys. Acta 1808 (2011) 2337–2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Brender JR, Durr UH, Heyl D, Budarapu MB, Ramamoorthy A, Membrane fragmentation by an amyloidogenic fragment of human Islet Amyloid Polypeptide detected by solid-state NMR spectroscopy of membrane nanotubes, Biochim. Biophys. Acta 1768 (2007) 2026–2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Hajiraissi R, Giner I, Grundmeier G, Keller A, Self-assembly, dynamics, and polymorphism of hIAPP (20–29) aggregates at solid-liquid interfaces, Langmuir 33 (2017) 372–381. [DOI] [PubMed] [Google Scholar]
- [131].R Brender J, Heyl DL, Samisetti S, Kotier SA, Osborne JM, Pesaru RR, Ramamoorthy A, Membrane disordering is not sufficient for membrane permeabilization by islet amyloid polypeptide: studies of IAPP(20–29) fragments, Phys. Chem. Chem. Phys 15 (2013) 8908–8915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Kulak K, Westermark GT, Papac-Milicevic N, Renstrom E, Blom AM, King BC, The human serum protein C4b-binding protein inhibits pancreatic IAPP-induced inflammasome activation, Diabetologia (2017), 10.1007/s00125-017-4286-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Bharadwaj P, Wijesekara N, Liyanapathirana M, Newsholme P, Ittner L, Fraser P, Verdile G, The link between type 2 diabetes and neurodegeneration: roles for amyloid-β, amylin, and tau proteins, J. Alzheimers Dis (2017), 10.3233/JAD-161192. [DOI] [PubMed] [Google Scholar]
- [134].Reiner DJ, Mietlicki-Baase EG, Olivos DR, McGrath LE, Zimmer DJ, Koch Laskowski K, Krawczyk J, Turner CA, Noble EE, Hahn JD, Schmidt HD, Kanoski SE, Hayes MR, Amylin acts in the lateral dorsal tegmental nucleus to regulate energy balance through gamma-aminobutyric acid signaling, Biol. Psychiatry (2017) (pii: S0006-3223(17)30007-0). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Lee SJ, K Kang H, Eum WS, Park J, Choi SY, Kwon HY, Tat-biliverdin reductase A protects INS-1 cells from human islet amyloid polypeptide-induced cytotoxicity by alleviating oxidative stress and ER stress, Cell Biol. Int 41 (2017) 514–524. [DOI] [PubMed] [Google Scholar]
- [136].Wu X, Song Y, Liu W, Wang K, Gao Y, Li S, Duan Z, Shao Z, Yang S, Yang C, IAPP modulates cellular autophagy, apoptosis, and extracellular matrix metabolism in human intervertebral disc cells, Cell Death Dis. 3 (January 30 2017) 16107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Westermark GT, Fändrich M, Lundmark K, Westermark P, Noncerebral amyloidoses: aspects on seeding, cross-seeding, and transmission, Cold Spring Harb. Perspect. Med (2017), 10.1101/cshperspecLa024323 (pii: a024323). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Martel A, Antony L, Gerelli Y, Porcar L, Fluitt A, K Hoffmann I Kiesel, M. Vivaudou, G. Fragneto, J.J. de Pablo, Membrane permeation versus amyloidogenicity: a multitechnique study of islet amyloid polypeptide interaction with model membranes, J. Am. Chem. Soc 139 (1) (January 11 2017) 137–148. [DOI] [PubMed] [Google Scholar]
- [139].Hartsel S, Bolard J, Amphotericin B: new life for an old drug, Trends Pharmacol. Sci 17 (1996) 445–449. [DOI] [PubMed] [Google Scholar]
- [140].Lemke A, Kiderlen AF, Kayser O, Amphotericin B, Appi. Microbiol. Biotechnol 68 (2005) 151–162. [DOI] [PubMed] [Google Scholar]
- [141].De Kruijff B, Demel RA, Polyene antibiotic-sterol interactions in membranes of Acholeplasma laidlawu cells and lecithin liposomes. 3. Molecular structure of the polyene antibiotic-cholesterol complexes, Biochim. Biophys. Acta 339 (1974) 57–70. [DOI] [PubMed] [Google Scholar]
- [142].Anderson TM, Clay MC, Cioffi AG, Diaz KA, Hisao GS, Tuttle MD, Nieuwkoop AJ, Cornelias G, Maryum N, Wang S, Uno BE, Wildeman EL, Gonen T, Rienstra CM, Burke MD, Amphotericin forms an extramembranous and fungicidal sterol sponge, Nat. Chem. Biol 10 (2014) 400–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Bolard J, Legrand P, Heitz F, Cybulska B, One-sided action of amphotericin B on cholesterol-containing membranes is determined by its self-association in the medium, Biochemistry 30 (1991) 5707–5715. [DOI] [PubMed] [Google Scholar]
- [144].Matsumori N, Tahara K, Yamamoto H, Morooka A, Doi M, Oishi T, Murata M, Direct interaction between amphotericin B and ergosterol in lipid bilayers as revealed by 2H NMR spectroscopy, J. Am. Chem. Soc 131 (2009) 11855–11860. [DOI] [PubMed] [Google Scholar]
- [145].Umegawa Y, Nakagawa Y, Tahara K, Tsuchikawa H, Matsumori N, Oishi T, Murata M, Head-to-taίl interaction between amphotericin B and ergosterol occurs in hydrated phospholipid membrane, Biochemistry 51 (2012) 83–89. [DOI] [PubMed] [Google Scholar]
- [146].Matsumori N, Umegawa Y, Oishi T, Murata M, Bioactive fluorinated derivative of amphotericin B, Bioorg. Med. Chem. Lett 15 (2005) 3565–3567. [DOI] [PubMed] [Google Scholar]
- [147].Nakagawa Y, Umegawa Y, Matsushita N, Yamamoto T, Tsuchikawa H, Hanashima S, Oishi T, Matsumori N, Murata M, The structure of the bimolecular complex between amphotericin B and ergosterol in membranes is stabilized by face-to-face van der Waals interaction with their rigid cyclic cores, Biochemistry 55 (2016) 3392–3402. [DOI] [PubMed] [Google Scholar]
- [148].Nakagawa Y, Umegawa Y, Nonomuia K, Matsushita N, Takano T, Tsuchikawa H, Hanashima S, Oishi T, Matsumori N, Murata M, Axial hydrogen at C7 position and bumpy tetracyclic core markedly reduce sterol’s affinity to amphotericin B in membrane, Biochemistry 54 (2015) 303–312. [DOI] [PubMed] [Google Scholar]
- [149].Simons K, Ikonen E, Functional rafts in cell membranes, Nature 387 (1997) 569–572. [DOI] [PubMed] [Google Scholar]
- [150].Simons K, Gerl MJ, Revitalizing membrane rafts: new tools and insights, Nat. Rev. Mol. Cell Biol 11 (2010) 688–699. [DOI] [PubMed] [Google Scholar]
- [151].Ahmed SN, Brown DA, London E, On the origin of sphingolipid/cholesterol-rich detergent-insoluble cell membranes: physiological concentrations of cholesterol and sphingolipid induce formation of a detergent-insoluble, liquid-ordered lipid phase in model membranes, Biochemistry 36 (1997) 10944–10953. [DOI] [PubMed] [Google Scholar]
- [152].Baumgart T, Hess ST, Webb WW, Imaging coexisting fluid domains in bio-membrane models coupling curvature and line tension, Nature 425 (2003) 821–824. [DOI] [PubMed] [Google Scholar]
- [153].Silvius JR, del Giudice D, Lafleur M, Cholesterol at different bilayer concentrations can promote or antagonize lateral segregation of phospholipids of differing acyl chain length, Biochemistry 35 (1996) 15198–15208. [DOI] [PubMed] [Google Scholar]
- [154].Bonev BB, Lam Y-H, Anderluh G, Watts A, Norton RS, Separovic F, Effects of the eukaryotic pore-forming cytolysin equinatoxin II on lipid membranes and the role of sphingomyelin, Biophys. J 84 (2003) 2382–2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Shogomori H, Kobayashi T, Lysenin: a sphingomyelin specific pore-forming toxin, Biochim. Biophys. Acta 1780 (2008) 612–618. [DOI] [PubMed] [Google Scholar]
- [156].McHenry AJ, Sciacca MFM, Brender JR, Ramamoorthy A, Does cholesterol suppress the antimicrobial peptide induced disruption of lipid raft containing membranes? Biochim. Biophys. Acta 1818 (2012) 3019–3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Simons K, Vaz WLC, Model systems, lipid rafts, and cell membranes, Annu. Rev. Biophys. Biomol. Struct 33 (2004) 269–295. [DOI] [PubMed] [Google Scholar]
- [158].Li XM, Momsen MM, Smaby JM, Brockman HL, Brown RE, Cholesterol decreases the interfacial elasticity and detergent solubility of sphingomyelins, Biochemistry 40 (2001) 5954–5963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Bittman R, Kasireddy CR, Mattjus P, Slotte JP, Interaction of cholesterol with sphingomyelin in monolayers and vesicles, Biochemistry 33 (1994) 11776–11781. [DOI] [PubMed] [Google Scholar]
- [160].Sankaram MB, Thompson TE, Interaction of cholesterol with various glycerophospholipids and sphingomyelin, Biochemistry 29 (1990) 10670–10675. [DOI] [PubMed] [Google Scholar]
- [161].Zidar J, Merzel F, Hodošček M, Rebolj K, Sepčić K, Macek P, Janezic D, Liquid ordered phase formation in cholesterol/sphingomyelin bilayers: all-atom molecular dynamics simulations, J. Phys. Chem. B 113 (2009) 15795–15802. [DOI] [PubMed] [Google Scholar]
- [162].Yamaguchi T, Suzuki T, Yasuda T, Oishi T, Matsumori N, Murata M, NMR-based conformational analysis of sphingomyelin in biceiles, Bioorg. Med. Chem 20 (2012) 270–278. [DOI] [PubMed] [Google Scholar]
- [163].Matsumori N, Yasuda T, Okazaki H, Suzuki T, Yamaguchi T, Tsuchikawa H, Doi M, Oishi T, Murata M, Comprehensive molecular motion capture for sphingomyelin by site-specific deuterium labeling, Biochemistry 51 (2012) 8363–8370. [DOI] [PubMed] [Google Scholar]
- [164].Yasuda T, Kinoshita M, Murata M, Matsumori N, Detailed comparison of deuterium quadrupole profiles between sphingomyelin and phosphatidylcholine bi-layers, Biophys. J 106 (2014) 631–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Yasuda T, Tsuchikawa H, Murata M, Matsumori N, Deuterium NMR of raft model membranes reveals domain-specific order profiles and compositional distribution, Biophys. J 108 (2015) 2502–2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Matsumori N, Yamaguchi T, Maeta Y, Murata M, Orientation and order of the amide group of sphingomyelin in bilayers determined by solid-state NMR, Biophys. J 108 (2015) 2816–2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Fu R, Miao Y, Qin H, Cross TA, Probing hydronium ion histidine NH exchange rate constants in the M2 channel via indirect observation of dipolar-dephased 15N signals in magic-angle-spinning NMR, J. Am. Chem. Soc 138 (2016) 15801–15804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Miao Y, Fu R, Zhou HX, Cross TA, Dynamic short hydrogen bonds in histidine tetrad of full-length M2 proton channel reveal tetrameric structural heterogeneity and functional mechanism, Structure 23 (2015) 2300–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Das N, Murray DT, A Cross T, Lipid bilayer preparations of membrane proteins for oriented and magic-angle spinning solid-state NMR samples, Nat. Protoc 8 (2013) 2256–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Murray DT, Das N, A Cross T, Solid state NMR strategy for characterizing native membrane protein structures, Acc. Chem. Res 46 (2013) 2172–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Yao Y, Dutta SK, Park SH, Rai R, Fujimoto LM, Bobkov AA, Opella SJ, Marassi FM, High resolution solid-state NMR spectroscopy of the Yersinia pestis outer membrane protein Ail in lipid membranes, J. Biomol. NMR 67 (2017) 179–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Opella SJ, Relating structure and function of viral membrane-spanning miniproteins, Curr. Opin. Virol 12 (2015) 121–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Gopinath T, Nelson SE, Soller KJ, Veglia G, Probing the conformationally excited states of membrane proteins via 1H-detected MAS solid-state NMR spectroscopy, J. Phys. Chem. B 13 (April 2017), http://dx.doi.org/l0.1021/acs.jpcb.7b03268. [DOI] [PubMed] [Google Scholar]
- [174].Gopinath T, Mote KR, Veglia G, Simultaneous acquisition of 2D and 3D solid-state NMR experiments for sequential assignment of oriented membrane protein samples, J. Biomol. NMR 62 (2015) 53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Wang S, Ing C, Emami S, Jiang Y, Liang H, Pomès R, Brown LS, Ladizhansky V, Structure and dynamics of extracellular loops in human aqua-porin-1 from solid-state NMR and molecular dynamics, J. Phys. Chem. B 120 (2016) 9887–9902. [DOI] [PubMed] [Google Scholar]
- [176].Kimata N, Pope A, Sanchez-Reyes OB, Eilers M, A Opefi C, Ziliox M, Reeves PJ, Smith SO, Free backbone carbonyls mediate rhodopsin activation, Nat Struct. Mol. Biol 23 (2016) 738–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Gayen A, Leninger M, Traaseth NJ, Protonation of a glutamate residue modulates the dynamics of the drug transporter EmrE, Nat Chem. Biol 12 (2016) 141–145. [DOI] [PMC free article] [PubMed] [Google Scholar]