Abstract
Radiation exposure to the gastrointestinal system contributes to the acute radiation syndrome in a dose- and time-dependent manner. Molecular mechanisms that lead to the gastrointestinal acute radiation syndrome remain incompletely understood. Using a murine model of total body irradiation, C57BL/6J male mice were irradiated at 8, 10, 12 and 14 Gy and assayed at day 1, 3, and 6 after exposure and compared to non-irradiated (sham) controls. Tryptic digests of gastrointestinal tissues (upper ileum) were analyzed by liquid chromatography-tandem mass spectrometry on a Waters nanoLC coupled to a Thermo Scientific Q Exactive hybrid quadrupole-orbitrap mass spectrometer. Pathway and gene ontology analysis were performed with Qiagen Ingenuity, Panther GO and DAVID databases. A number of trends were identified in our proteomic data including pronounced protein changes as well as protein changes that were consistently upregulated or downregulated at all time points and dose levels interrogated. Time- and dose-dependent protein changes, canonical pathways affected by irradiation, and changes in proteins that serve as upstream regulators were also identified. Additionally, proteins involved in key processes including inflammation, radiation and retinoic acid signaling were identified. The proteomic profiling conducted here represents an untargeted systems biology approach to identify acute molecular events that will be useful for a greater understanding of animal models and may be potentially useful toward the development of medical countermeasures and/or biomarkers.
Keywords: biological indicators, radiation damage, gastrointestinal tract, whole body irradiation
INTRODUCTION
The gastrointestinal tract is sensitive to injury after exposure to high-dose irradiation in a dose- and time-dependent manner representing a major source of mortality and morbidity (Williams et al. 2010; Booth et al. 2012a; Booth et al. 2012b; MacVittie et al. 2012; Shea-Donohue et al. 2016). Due to its rapidly proliferating cell populations, the gastrointestinal system (GI) contributes significantly to acute radiation syndrome (ARS) (Potten 1990). Characteristics of the injury include damage to the mucosal layer of the small intestine, loss of clonogenic crypt cells, villus blunting, and barrier disruption accompanied by weight loss, diarrhea, dehydration, reduced nutrient absorption and intestinal infection (Booth and Potten 2002). There are no FDA-approved medical countermeasures (MCM) to treat GI-ARS and drug development of MCM for GI-ARS under the FDA Animal Rule requires the use of well-defined animal models given that human studies cannot be ethically conducted (FDA 2015). As such, animal models of GI-ARS have been characterized by the Medical Countermeasures Against Radiological Threats (MCART) Consortium for both mouse and non-human primate (Booth et al. 2012a; Booth et al. 2012b; MacVittie et al. 2012; Jones et al. 2015b).
Proteomic studies of the gastrointestinal system after radiation have been conducted in an intestinal epithelial cell model after 25 Gy and in intestine tissue after 1 Gy or 7 Gy exposure (Bo et al. 2005; Lim et al. 2011; Han et al. 2017). However, a systematic proteomic analysis covering the time- and dose-range of the GI-ARS has not been performed. Proteomic profiling using liquid chromatography-tandem mass spectrometry (LC-MS/MS) yields relative quantitation of protein abundance and allows for systems biology analyses to identify disruption of key signaling pathways and regulatory nodes (Zhang and Chen 2010; Kramer et al. 2014; Karahalil 2016). This type of proteomic approach also provides complementary information from a systems biology perspective to our metabolomic efforts to characterize GI-ARS where changes in metabolites are often the result of alterations in protein function or abundance (Jones et al. 2014). Previously our team has used discovery and targeted metabolomics towards identifying potential biomarkers of GI-ARS, used mass spectrometry imaging to both identify and localize biomarker candidates and mechanistically-relevant molecules in the jejunum of the non-human primate, and correlated biomarker candidates with histological endpoints (Jones JW 2014; Jones et al. 2015a; Jones et al. 2015b; Carter 2018; Jones 2018). Here, an untargeted systems biology approach was employed to identify acute molecular events that could further the mechanistic understanding of the model and potentially serve as biomarkers of GI-ARS.
MATERIALS AND METHODS
Radiation animal model.
Tissues were obtained from Epistem Ltd as part of a previous study (Jones et al. 2015b). Mice (male, strain C57BL/6J) were exposed to a single uniform total-body irradiation (TBI) at doses of 8, 10, 12 and 14 Gy. The established LD50/6 for the C57BL/6J strain was 11.9 Gy (Booth et al. 2012b). Supportive care was not provided in the study design. For each dose, mouse upper ileum samples were taken at day 1, 3 and 6 after exposure (14 Gy at day 5) and compared to non-irradiated controls. Biological triplicates were collected per condition for proteomic analyses.
For the previous study from which these samples were obtained (Jones et al. 2015b), all mouse procedures were certified according to the UK Animal (Scientific Procedures) Act 1986. Male C57BL/6 mice, aged eight to ten weeks were purchased from Harlan UK and allowed to acclimatize for two weeks prior to irradiation. All mice were held in individually ventilated cages (IVCs) in a specific pathogen free (SPF) barrier unit. There were 5 mice per IVC all from the same treatment group. A twelve hour light:dark cycle was maintained with lights being turned on at approximately 07:00 hours and off at approximately 19:00 hours. There was a constant room temperature of 21 ± 2 ºC and a mean relative humidity of 55% ± 10%. The animals received 2918x extruded rodent diet (Harlan) and sterile acidified water (ad libitum; pH 2.0–3.0) from time of arrival and throughout the study. Animals were identified by ear punches in cages labeled with the appropriate information necessary to identify the study, dose, animal number, and treatment groups. Animals were irradiated at 15:00 ± one hour (30 animals per radiation dose). Irradiation was performed using an Xstrahl RS320 X-ray set, operated at 300 kV, 10 mA. The X-ray tube had additional filtration to give a radiation quality of 2.3 mm Copper half-value layer (HVL). Mice were restrained in a compartmentalized plexiglass jig, positioned at a distance of 700 mm below the focus of the X-ray tube. Each rectangular box was divided into twelve ventilated restraints, each holding one animal. For this study, ten mice were irradiated simultaneously. A dosimetry device (ion chamber) was placed within the mouse jig to record the dose received to the animals in each run. All radiation doses delivered were within 2.6% of the intended dose (mean 1.9%). Quality assurance and control procedures were performed prior to and during each irradiation to confirm dose and energy output remained within range. Animals received total-body irradiation (TBI) delivered at a dose rate of 81.2 cGy min-1. All animals were weighed and their well-being inspected daily from the initiation of treatment to the end of the study. Mice were humanely euthanized if the weight loss was sustained at greater than 20% for 24 hours and mice also demonstrated signs of a moribund state (withdrawn behavior, reduced body temperature as judged by feeling cool to touch, lack of grooming and dehydration as judged by a persistent skin tent on pinching). During the peak periods of diarrhea incidence and general decline in well-being, animals were inspected several times during each 24 hour period. On days 1 to 6, five mice per radiation dose were deeply anesthetized with isoflurane and blood collected via cardiac puncture. The mice were then terminated by cervical dislocation. Preparation of mouse intestinal tissue included flushing out contents with PBS. The jejunum and ileum were isolated from the small intestine and snap frozen with liquid nitrogen and stored at −80 °C until analyzed for proteomics.
Sample preparation.
Upper ileum tissues were homogenized in phosphate buffered saline using Precellys CK14 lysing kit (Bertin Corp., Rockville, MD). Proteins were extracted and purified from tissue lysates by trichloroacetic acid precipitation. Protein concentration was measured by bicinchoninic acid assay. 100 ug protein was trypsinolyzed and desalted by C18 tips.
Liquid chromatography-tandem mass spectrometry data acquisition.
Tryptic peptides were separated on a Waters nanoAcquity UPLC system over a 95 minutes linear acetonitrile gradient (3 – 35%) with 0.1 % formic acid, and analyzed on a coupled Thermo Scientific Q Exactive hybrid quadrupole-orbitrap mass spectrometer. Technical triplicates were performed for each biological sample. Full scans were acquired at a resolution of 70,000 with an automatic gain control target value of 5e5 and a maximum injection time of 200 milliseconds, and top 10 most abundant precursors were selected for fragmentation by higher-energy collisional dissociation (normalized collision energy at 35%) and analyzed at a resolution of 17,500 with an automatic gain control target value of 5e4 and a maximum injection time of 50 milliseconds. Interrogated ions were dynamically excluded from re-selection for 30 seconds.
Liquid chromatography-tandem mass spectrometry data analysis.
Tandem mass spectra were searched against a UniProt Mus musculus reference proteome using a SEQUEST HT algorithm described by Eng et al. (Eng et al. 2008) with a maximum precursor mass error tolerance of 20 ppm. Carbamidomethylation of cysteines and deamindations of asparagines and glutamines were treated as static and dynamic modifications, respectively. Resulting hits were validated at a maximum false discovery rate of 0.01 using a semi-supervised machine learning algorithm Percolator developed by Käll et al. (Kall et al. 2007). Abundance ratios were measured by comparing the MS1 peak volumes of peptide ions, whose identities were confirmed by MS2 sequencing as described above. Label-free quantifications were performed using Minora (Thermo Fisher Scientific), an aligned AMRT (Accurate Mass and Retention Time) cluster quantification algorithm.
Bioinformatic analysis.
Pathway and gene ontology analysis were performed with Qiagen Ingenuity, Panther GO and DAVID databases, as described by Krämer et al., Mi et al., and Huang et al., respectively (Huang da et al. 2009; Kramer et al. 2014; Mi et al. 2017). Proteins showing at least a 2-fold change (FC) with a FDR adjusted ANOVA p-value <0.05 were considered significantly changed and used for further analysis. FDR corrected Fisher’s exact p-values of < 0.05 were used in the gene ontology analyses to identify biological processes, molecular functions, and cellular components associated with observed protein changes. Ingenuity Pathway Analysis (IPA) was used to predict canonical pathways and upstream regulators according to the proteins that were significantly different using an absolute activation z-score of > 2 for at least one condition with a Fisher’s exact test p-value < 0.01.
Retinoid analysis.
Sample preparation and analysis methodology are provided for retinoids in the Supporting Information.
RESULTS
Study design.
The study design here assayed the small intestine (ileum) proteome after radiation exposure to identify acute changes that could aid in the further characterization of GI-ARS. Selected total-body irradiation (TBI) doses of 8, 10, 12, and 14 Gy TBI were assayed at 1, 3, and 6 days post-exposure and compared to non-irradiated control. An 8 Gy TBI dose in the mouse model used in these experiments yielded an LD30 at day 8 where the higher TBI doses were an LD50 at day 7 (10 Gy), an LD50 at day 6 (12 Gy), or 100% lethal at day 6 (14 Gy) (Booth et al. 2012b). These time and dose points were selected because they cover a range of lethality relevant to GI-ARS. Previously, our team has interrogated the GI histopathology and its relation to biomarker levels (citrulline) in mice from the same study (Jones et al. 2015b). Metabolomic analyses were also conducted on the small intestine (jejunum) and plasma of the same mice from this study and results are discussed in a manuscript by Jones et al. in this issue (Jones 2018).
Proteins with greatest changes in expression levels after radiation.
Fig. 1 and Supplementary Fig. 1 show heatmaps representing the greatest changes in protein expression after radiation, where an false discovery rate (FDR) of 0.01, a protein expression fold-change (FC) > 10 and the FDR corrected ANOVA p-value cut-off of p < 0.01 were used to identify changes in protein expression. Many proteins showed repeatable patterns, dose-dependency, and/or time-dependency over the times and doses interrogated.
Figure 1. Expression of proteins most changed after radiation.
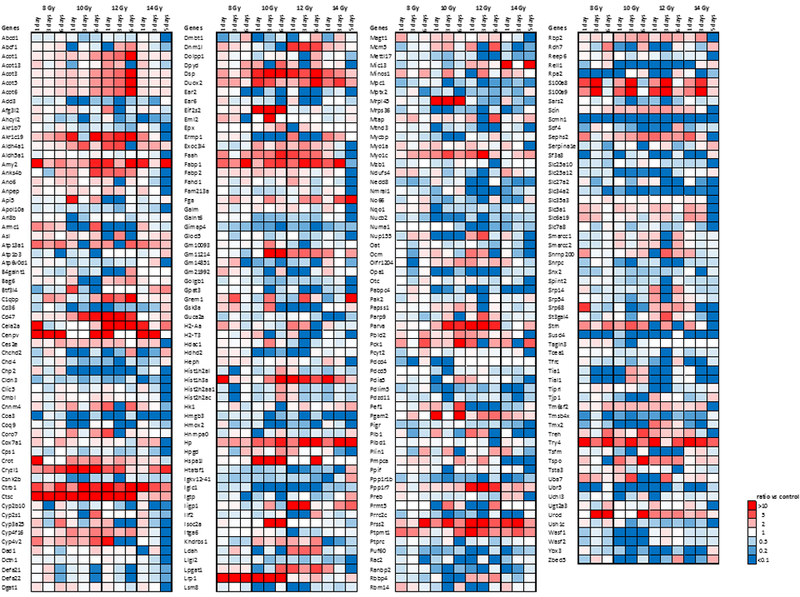
Minimum FC > 10 of expression for at least one condition and FDR adjusted ANOVA p < 0.01. Data shown as a function of time after dose at a given radiation dose. Data shown as a function of dose on a given day after dose are shown in Supplementary Fig. 1.
Canonical pathways and upstream regulators altered by radiation.
Ingenuity Pathway Analysis (IPA) was used to predict canonical pathways and upstream regulators according to the proteins that were significantly different (Kramer et al. 2014). Identification of canonical pathways and upstream regulators provided insight into key events and signaling nodes (Fig. 2, Supplementary Fig. 2 and Fig. 3, Supplementary Fig. 3 respectively). Fig. 2 and Fig. 3 show data as a function of time post-TBI exposure at a given radiation dose. Supplementary Fig. 2 and Supplementary Fig. 3 show data as a function of radiation dose at a given day post-exposure. The calculated z-score is a statistical measure of the match between expected relationship direction from published literature and observed gene expression from the experimental dataset. It was used to infer likely activation states of pathways or upstream regulators based on comparison with a model that assigns random regulation directions. A z-score absolute value of >2 for at least one condition and a p <0.01 was used as criteria for inclusion in Fig. 2 and Fig. 3. Fourteen (14) canonical pathways were altered by radiation according to these criteria (Fig. 2a, Supplementary Fig. 2a). Fig. 2b and Supplementary Fig. 2b show the protein changes associated with the top three pathways altered by radiation using a minimum 2-fold change for at least one condition with a FDR corrected ANOVA p < 0.05 as criteria for inclusion. Fourty-four (44) upstream regulators were identified as either activated or repressed (Fig. 3, Supplementary Fig. 3).
Figure 2. Canonical pathways altered by radiation.
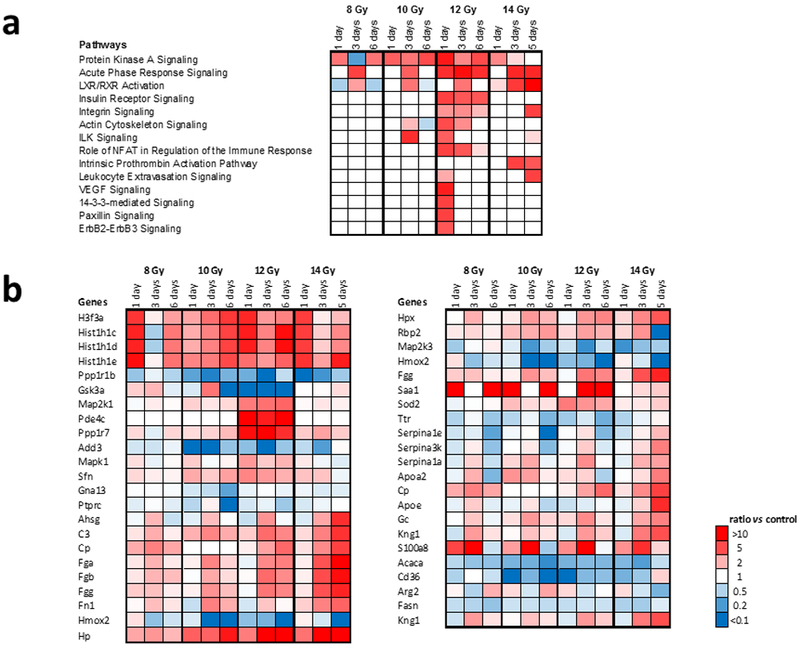
(a.) Canonical pathways altered by radiation where criteria for pathway changes were activation z-score > 2 for at least one condition and Fisher’s exact test p < 0.01. (b.) Protein changes associated with the top three pathways altered by radiation. Minimum FC > 2 for at least one condition with a FDR corrected ANOVA p < 0.05. Data shown as a function of time after dose at a given radiation dose. Data shown as a function of dose on a given day after dose are shown in Supplementary Fig. 2.
Figure 3. Upstream regulators altered by radiation.
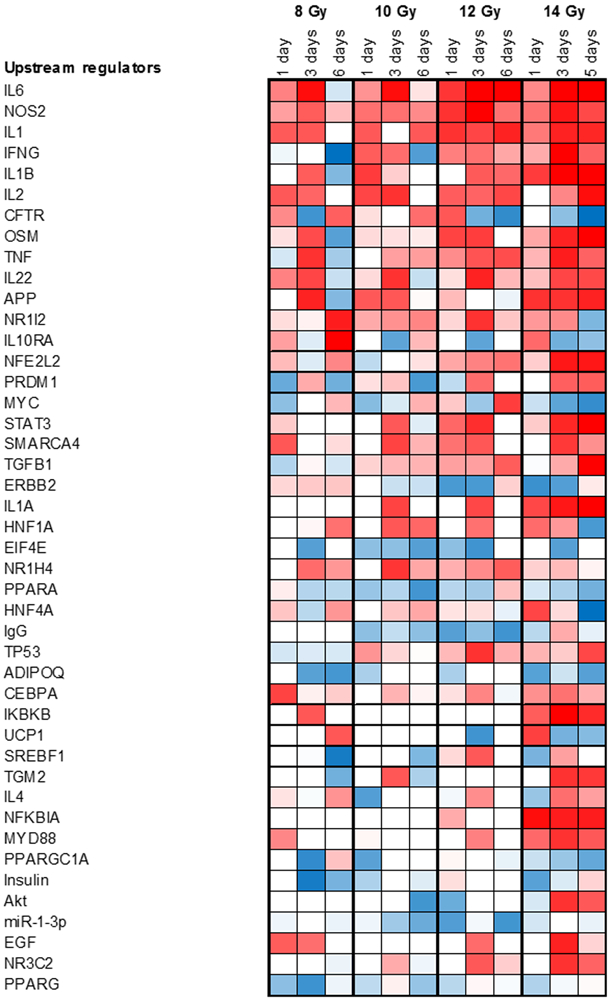
Criteria for transcription regulators were absolute activation z-score > 2 for at least one condition and a Fisher’s exact test p < 0.01. Data shown as a function of time after dose at a given radiation dose. Data shown as a function of dose on a given day after dose are shown in Supplementary Fig. 3.
Proteins exhibiting time-dependent changes after irradiation.
Several proteins that were detected had time-dependent changes in expression over the given dose range (Fig. 4). Among these, some proteins displayed a progressive increase or decrease in abundance over time (highlighted in yellow frames, Fig. 4a). Biological processes associated with progressive changes in proteins over time included chromatin organization, lipid metabolic processes, epigenetic regulation of gene expression, and primary metabolic processes (Fig. 4b). Another population of proteins showed that their protein expression exhibited a transient increase or decrease followed by normalization of protein abundance (highlighted in green frames, Fig. 4a). Protein expression was considered recovered if the difference between day 1 and day 6 was below 30%. Biological processes associated with transient changes in proteins that exhibit normalization/recovery were primarily related to generation of precursor metabolites and energy (Fig. 4c). Venn diagrams describing differentially expressed proteins as a function of time post-exposure at a given radiation dose are shown in Supplementary Fig. 4 – 7.
Figure 4. Proteins exhibiting time-dependent changes after irradiation.
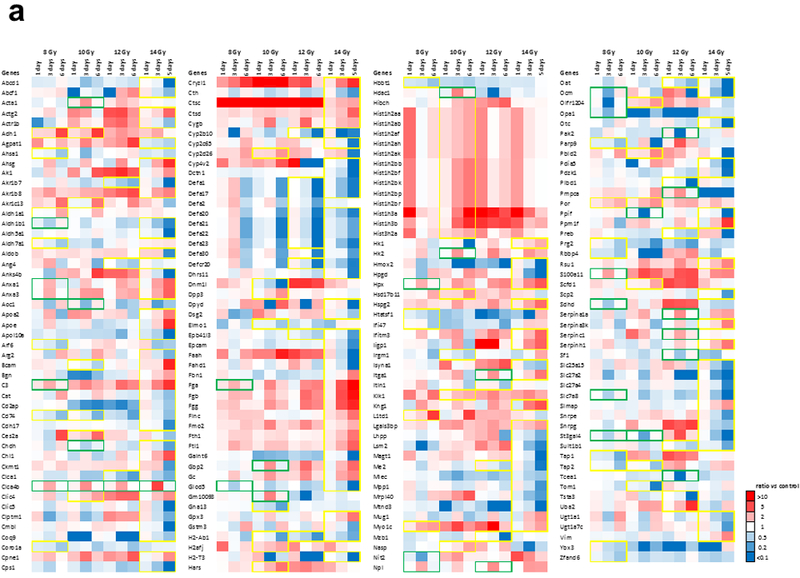
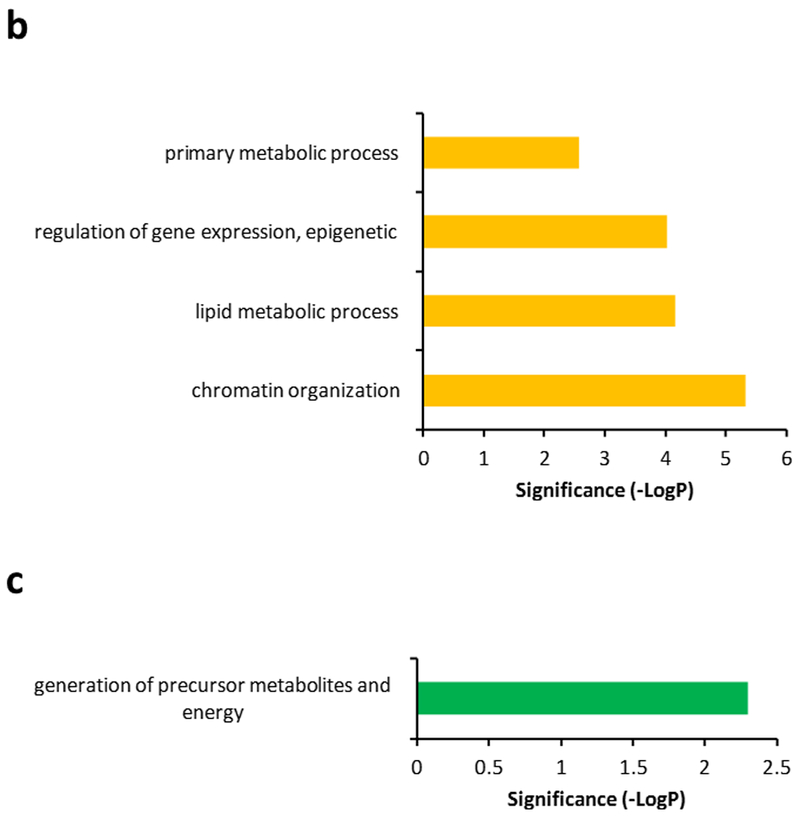
(a.) Yellow framed protein expression exhibits a progressive increase or decrease in abundance over time based on linear regression models with R2 > 0.81. Green framed protein expression exhibits a transient but significant increase or decrease (FC > 2, p < 0.05) followed by normalization of protein abundance. (b.) Biological processes associated with progressive changes in proteins over time. Significance was calculated by FDR corrected Fisher’s exact test (p < 0.05). (c.) Biological processes associated with transient changes in proteins that exhibit normalization/recovery. Significance was calculated by FDR corrected Fisher’s exact test (p < 0.05). Protein expression is considered normalization/recovery if difference between day 1 and day 6 is below 30%.
Proteins exhibiting dose-dependent changes after irradiation.
Several proteins that were detected displayed a dose-dependency (Fig. 5). Those proteins that showed either a progressive increase or decrease in expression on at least one of the days are highlighted in yellow (Fig. 5a). Biological processes associated with dose-dependent changes in abundance included cellular amino acid metabolic processes and generation of precursor metabolites and energy (Fig. 5b). Venn diagrams describing differentially expressed proteins as a function of radiation dose on a given day post-exposure are shown in Supplementary Fig. 8 – 10.
Figure 5. Proteins exhibiting dose-dependent changes after irradiation.
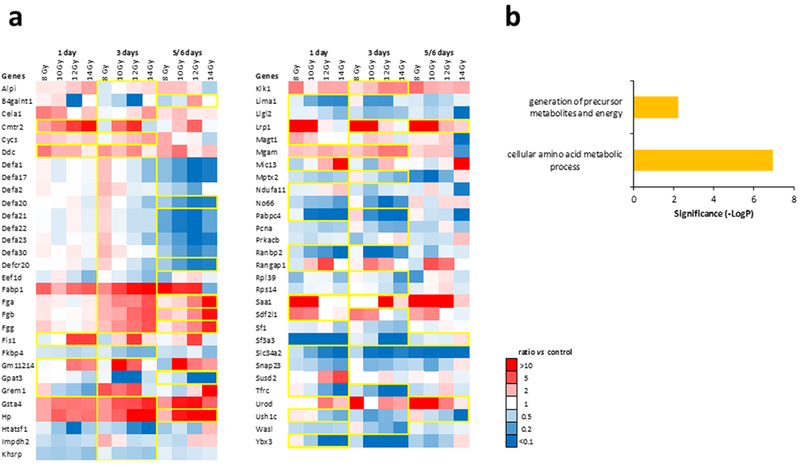
(a.) Yellow framed protein expression exhibits a progressive increase or decrease in abundance according to dose time based on linear regression models with R2 > 0.81. (b.) Biological processes associated with dose-dependent changes in abundance. Significance was calculated by FDR corrected Fisher’s exact test (p < 0.05).
Proteins showing a consistent elevation or depression in expression after radiation.
Fig. 6a and Fig. 7a show heatmaps of proteins with expression that was consistently altered across the radiation doses and days post-exposure studied, i.e., all observed changes were activating or inhibiting. Criteria used to identify these proteins were a FC > 2 and FDR corrected ANOVA p < 0.05 for at least one condition while the rest of the conditions showed changes in the same direction. Forty-six (46) proteins were consistently activated and 57 proteins were consistently inhibited. Further investigation into biological processes that were overrepresented or underrepresented in these proteins that were consistently activated or inhibited during the first 6 days after radiation exposure was carried out (Fig. 6b and Fig. 7b, respectively. Protein FC > 2, FDR corrected ANOVA p < 0.05 for at least one condition. Significant biological process by FDR corrected Fisher’s exact test p < 0.05. Proteolysis nucleosome assembly, and acyl-coA metabolic processes were consistently overrepresented and RNA splicing and mRNA processing were consistently underrepresented. Molecular functions associated with the observed protein changes were also determined (Fig. 6c and Fig. 7c). We found that molecular functions associated with thiolester hydrolase activity, serine-type endopeptidase activity, nucleosomal DNA binding, acyl-CoA hydrolase activity, palmitoyl-CoA hydrolase activity, and carboxylic ester hydrolase activity were significantly elevated and that molecular function associated with poly(A) RNA binding was significantly reduced. Lastly, the association of the observed protein changes with cellular components was investigated (Fig. 6d and Fig. 7d). The extracellular exosome had proteins associated with it that were either significantly activated or significantly decreased. Other cellular components that were significantly associated with activated proteins were extracellular space, nucleosome, and extracellular matrix. Other cellular components that were significantly associated with inhibited proteins after radiation included cytoplasm, spliceosomal complex, nucleus, and intracellular ribonucleoprotein complex.
Figure 6. Proteins showing a consistent elevation in expression after radiation.
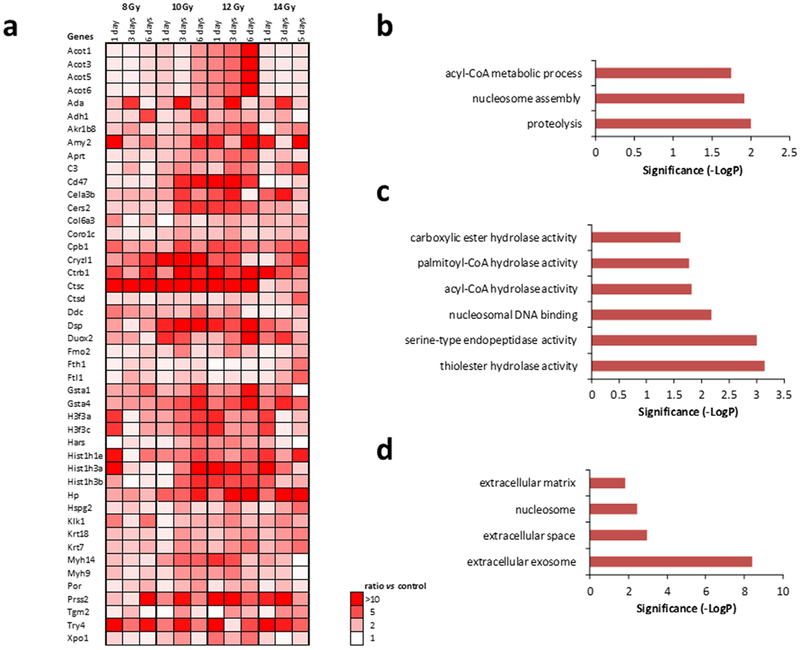
(a.) Protein expression increased after radiation in all doses with FC > 2 for at least one condition and FDR corrected ANOVA p < 0.05. (b.) Biological processes associated with protein increase. Significance was calculated by FDR corrected Fisher’s exact test (p < 0.05). (c.) Molecular function associated with protein increase. Significance was calculated by FDR corrected Fisher’s exact test (p < 0.05). (d.) Cellular component associated with protein increase. Significance was calculated by FDR corrected Fisher’s exact test (p < 0.05).
Figure 7. Proteins showing a consistent depression in expression after radiation.
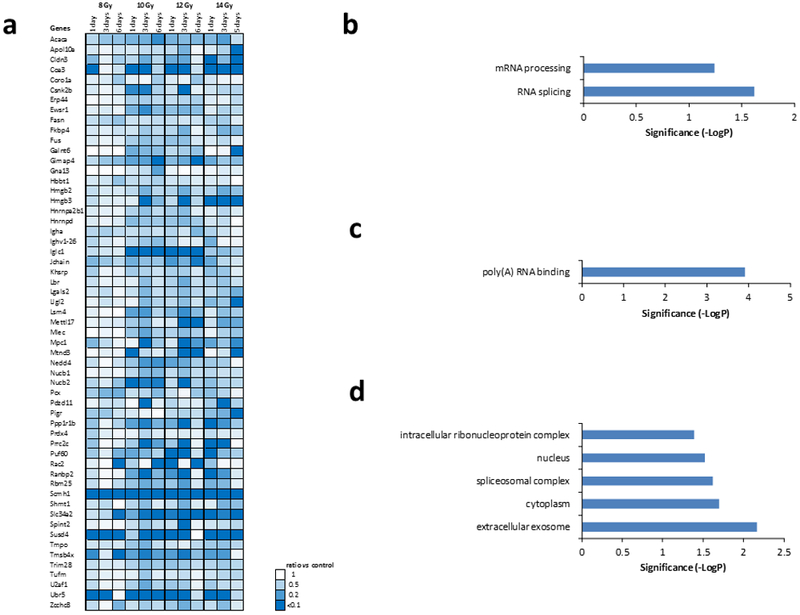
(a.) Protein expression decreased after radiation in all doses with FC > 2 for at least one condition and FDR correct ANOVA p < 0.05. (b.) Biological processes associated with protein increase. (c.) Molecular function associated with protein increase. Significance was calculated by FDR corrected Fisher’s exact test (p < 0.05). (d.) Cellular component associated with protein increase. Significance was calculated by FDR corrected Fisher’s exact test (p < 0.05).
Genes related to retinoic acid, radiation and inflammation.
Retinoic acid, radiation and inflammation related genes were annotated according to their molecular function and biological process involved (Fig. 8). Proteins showing a minimum 2-fold change with a FDR corrected ANOVA p < 0.05 were selected for gene ontology analysis. Retinoic acid is a master regulator of gene expression mainly through ligand-activated control of transcription mediated through retinoic acid receptors (RAR) in the nucleus (Germain et al. 2006). In the small intestine, retinoic acid has important functions regulating differentiation and migration of cell populations required for immune response (Hall et al. 2011; Larange and Cheroutre 2016). As previous reports have shown that retinoic acid is reduced after radiation in lung (Jones et al. 2014), retinoids were quantified in the small intestine (jejunum) as a function of radiation dose and time after irradiation (Supplementary Fig. 11, Supplementary Fig. 12). Small intestine (jejunum) retinoic acid (Supplementary Fig. 11a, Supplementary Fig. 12a) was reduced after radiation across the study dose range with the most pronounced and consistent decreases seen at one and two days after radiation. Sixteen (16) proteins from the proteins exhibiting fold-changes >2 were annotated to be significantly associated with retinoic acid including Aldh1a1, Apoa2, Apoe, Rbp2, Rdh7, Ttr, and a series of Akr proteins. A lesser number (three) of proteins from the proteins exhibiting fold-changes >2 were annotated to be associated with radiation in this type of analysis: Eef1d, Ptprc, and Sod2. Inflammation has been reported to be an important mechanism after radiation insult (Mukherjee et al. 2014). Forty (40) proteins among the proteins exhibiting fold-changes >2 were annotated to be significantly associated with inflammation. One protein associated with inflammation, Apoe (apolipoprotein E), responsible for packaging and transporting lipids especially cholesterol and mediating catabolism of triglyceride-rich lipoproteins, was also associated with retinoic acid.
Figure 8. Genes related to inflammation, retinoic acid, and radiation.
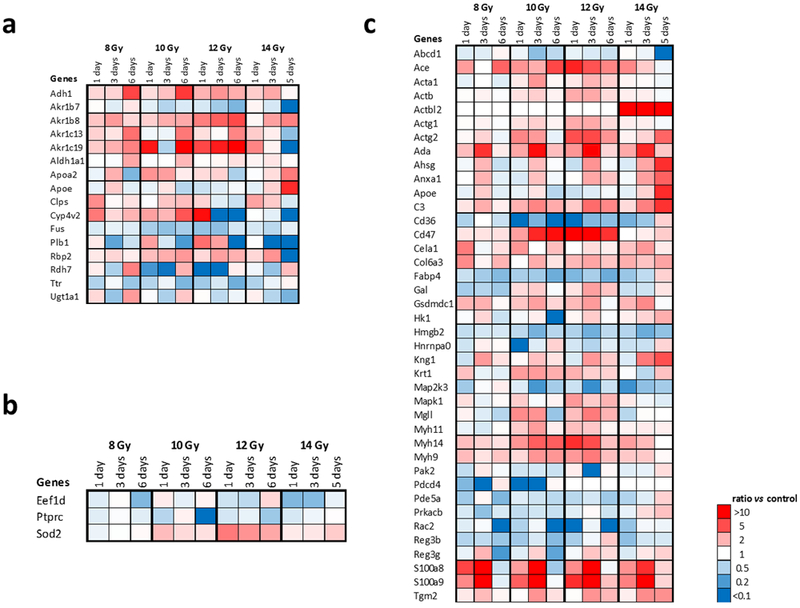
Proteins showing a minimum 2-fold change for at least one condition with a FDR adjusted Fisher’s exact test p < 0.05 were selected for gene ontology clustering. Proteins annotated to have an established relationship are noted: (a) retinoic acid, (b) radiation, (c) inflammation.
DISCUSSION
The murine model of GI-ARS used in these proteomics studies is a well-defined animal model that has been previously characterized (Booth et al. 2012a; Booth et al. 2012b; Jones et al. 2015b). Detailed studies of citrulline and histopathology over the same time and dose course studied here have been conducted over the range of an LD30/8 (8 Gy) to an LD100/6 (14 Gy) (Jones et al. 2015b). For comparison at day 6, the LD30/6 is 11.1 Gy (Booth et al. 2012b). Characterization of this murine model of the GI-ARS includes metabolomic analyses that were also conducted on the small intestine (jejunum) and plasma of the same mice from this study where these results are discussed in a manuscript by Jones et al. in this issue (Jones 2018).
The proteomics analyses conducted here offer the potential for new biomarkers as well as insight into mechanisms of injury. It is important to note that the changes reported here are putative and need to be validated by a secondary method of protein quantification. Time- and dose- dependent changes identified in a number of proteins in Fig. 4 and Fig. 5 that may have potential utility to distinguish radiation dose and/or severity of injury. Proteins that are all up (Fig. 6) or all down (Fig. 7) at the conditions investigated here display a consistent threshold effect with dysregulation after irradiation above 8 Gy and could be useful as markers of high-dose exposure to irradiation. Some of these proteins consistently affected by irradiation above 8 Gy display time- and dose-dependency among the elevated or depressed levels, for example the acyl-coA thioesterases in Fig. 6, Acot1, Acot3, Acot5, and Acot 6. Some proteins displayed similar levels of consistent upregulation or downregulation, such as Scmh1 (scm polycomb group protein homolog 1) and Ctsc (cathepsin C), that indicated high sensitivity to radiation exposure. More pronounced changes were observed in the consistently downregulated proteins between 10 Gy and 14 Gy as compared to 8 Gy (Fig. 7). This observation is consistent with metabolomics trends from the same mouse model of GI-ARS reported by Jones et al. in this issue (Jones 2018).
The top canonical pathways dysregulated by radiation included protein kinase A signaling, acute phase response signaling, and LXR/RXR signaling (Fig. 2). Among the dysregulated proteins from the top three canonical pathways dysregulated by radiation (Fig 2b), there was a number of proteins that were also annotated to be associated with inflammation, radiation and retinoic acid (Fig 8.). Those proteins common to the top canonical pathways that were also annotated to be related to inflammation included Mapk1, Ahsg, C3, Map2k3, Apoe, Kng1, S100a8, and CD36. Those related to top canonical pathways and also annotated to be related to radiation included Ptprc and Sod2 whereas those also annotated to be related to retinoic acid included Rbp2, Ttr, Apa2, and Apoe.
Kinase dysregulation was common to a number of canonical pathways including protein kinase A signaling, integrin signaling, ILK signaling, paxillin signaling, and ErbB2-ErbB3 signaling (Giancotti and Ruoslahti 1999). Protein kinase A regulates metabolism and cell survival and phosphorylates a number of targets including paxillin. Paxillin is involved in epithelial wound repair as it pertains to acute inflammatory conditions in the intestine (Nusrat et al. 1997). Protein kinase A signaling is associated with glucose sensing in the small intestine (Dyer et al. 2003). ILK (integrin-linked kinase) controls the activity of serine/threonine phosphatases and has been associated with cell migration, proliferation, and adhesion (Widmaier et al. 2012). Dysregulation of ErbB2-ErbB3 signaling may also be related to inflammation. ErbB proteins are receptors tyrosine kinases where defects in signaling have been linked to multiple intestinal inflammatory conditions and ErbB ligands have been proposed as therapeutics for inflammatory conditions of the gut (Frey and Brent Polk 2014). Integrin signaling regulates the activities of cytoplasmic kinases, growth factor receptors, and ion channels and controls the organization of the intracellular actin cytoskeleton (Giancotti and Ruoslahti 1999). Related to integrin signaling, actin cytoskeleton signaling was also identified as a canonical pathway that was dysregulated by radiation. Integrins activate various signaling pathways involved in the maintenance of the human intestinal crypt-villus axis (Lussier et al. 2000). Additionally, in the small intestine, mucosal dendritic cells regulate the expression of the gut homing receptors α4β7 integrin and the chemokine receptor CCR9 on activated effector and regulatory lymphocytes in a retinoic acid-dependent manner (Gorfu et al. 2009; Cassani et al. 2012). Therapeutic targeting of α4β7 integrins has been applied to inflammatory bowel disease (IBD) and control of simian immunodeficiency virus (SIV) infection (Gorfu et al. 2009; Villablanca et al. 2011a; Byrareddy et al. 2016).
Acute phase response signaling, a top canonical pathway impacted by radiation, is related to the rapid inflammatory response to infection, tissue injury, trauma, neoplastic growth and immunological disorders. Consistent with acute phase response, which involves an increase in inflammatory cytokines, our upstream regulator analysis identified a number of inflammatory cytokines including IL-1, IL-1A, IL-1B, IL-6, TNF, and INFg. Previous characterization of mechanisms of gastrointestinal damage in the non-human primate identified an upregulation of pro-inflammatory mediators after radiation (Shea-Donohue et al. 2016). LXR/RXR signaling, which was one of the top canonical pathways affected by radiation, is related to regulating metabolism in response to inflammatory cytokine signaling through lipid-sensing nuclear receptors such as LXR and PPAR (Venteclef et al. 2011). This current study identified PPARα and PPARγ as upstream regulators which have been shown to regulate the activation of inflammatory response genes such as IL-2, IL-6, and TNFα (Chinetti et al. 2000). S100a8 (S100 calcium-binding protein A8), which was among the proteins from the top three canonical pathways altered by radiation, has been shown to be affected by acute phase response previously where it was shown to be upregulated during the acute phase of infection in a comparative proteomic analysis of the gut during Cholera infection (Ellis et al. 2015). S100a8 and S100a9 form a heterodimer called calprotectin. Fecal calprotectin has been established as a marker of mucosal inflammation in inflammatory conditions of the small intestine (Kopylov et al. 2016; Koulaouzidis et al. 2016).
Other biological processes that the proteomic analyses indicated are altered by irradiation include many pertaining to metabolism as represented in biological processes, molecular function, and cellular components in Fig. 4 – Fig. 6. Primary biological processes that were identified as altered by radiation include primary metabolic processes, lipid metabolic processes, cellular amino acid metabolic processes, generation of precursor metabolites and energy, and acyl-CoA metabolic process. This is consistent with metabolomics data in this model showing dysregulation of a number of amino acids, biogenic amines, lysophosphatidylcholine (LPC), glycerophosphatidylcholine (PC), and sphingomyelin (SM) species (Jones 2018). After radiation, the dysregulation of cellular amino acid metabolic processes (Fig 5b) is shown in the alteration of the levels of number of amino acids including citrulline (Cit), histidine (His), leucine (Leu), methionine (met), phenylalanine (Phe), serine (Ser), and threonine (Thr) (Jones 2018). We have previously shown a dose-dependent change in citrulline after radiation exposure in a mouse model of GI-ARS (Jones et al. 2015b). In the GI system, amino acids play a critical role in supporting gut barrier integrity and function (Van Der Schoor et al. 2002; Wang et al. 2009). Our mass spectrometry imaging studies of the non-human primate model of GI-ARS has also shown alterations in lipid species consistent with disruption of lipid metabolic processes (Carter 2018). The lipid changes observed in the non-human primate included alterations in phosphatidylinositols, glycosphingolipids, cardiolipins, and phosphatidylglycerols. Changes in these lipids informed on epithelial cell differentiation and maturation, the brush border membrane, mitochondrial integrity and function, and the regulation of immune cell populations, respectively.
In addition to inflammation and energy metabolism, the proteomic analyses of gut tissue reported here revealed several biological processes were noted to be commonly dysregulated amongst multiple tissues after radiation when compared to a similar proteomic analysis of lung (Huang 2018). The biological process of RNA splicing was consistently under-represented in lung as well as gut. Molecular function associated with poly(A) RNA binding was significantly reduced in lung as well as gut. The cellular component of extracellular exosome had elements that were both activated and repressed in both lung and gut. Cellular components of intracellular ribonucleoprotein complex, and cytoplasm were also inhibited in lung and gut.
Previous reports by our team have shown that radiation reduces retinoic acid, an active metabolite of vitamin A, that regulates gene transcription through the ligand-activated nuclear receptors retinoic acid receptor (RAR) and retinoid x receptor (RXR) (Jones et al. 2014; Huang 2018). In the small intestine, retinoic acid is an important regulator of the immune response and essential for differentiation that maintains the crypt-villus axis (Rojanapo et al. 1980; Larange and Cheroutre 2016; Czarnewski et al. 2017). Results here also found that radiation decreased retinoic acid in the small intestine (jejunum) in a murine model of the GI-ARS (Supplementary Fig 11 – 12). Previously studies demonstrated that retinoic acid signaling and MYD88, an upstream regulator identified here, modulate the gastrointestinal activities of dendritic cells (Villablanca et al. 2011b). Interestingly, retinoic acid levels were elevated at higher doses and later time points after dose which could possibly be due to upregulation of ALDH1A1 (an enzyme involved in retinoic acid biosynthesis) observed here, (Fig 4, Fig 8a), or from influx of immune cells in the gut which produce retinoic acid (Iwata and Yokota 2011; Ito et al. 2014; Shea-Donohue et al. 2016). Our recent studies in the non-human primate showed a correlation between retinoic acid concentrations and damage to the intestinal mucosa following infection (Byrareddy et al. 2016). In this regard, models of radiation-induced gut injury and models of immune dysfunction share some similarities in histological damage including loss of epithelial integrity, inflammation, villus blunting, and loss of select cell populations. Other proteins related to the dysregulation in retinoids include Rbp2 and Rdh7. Rbp2 is essential for vitamin A uptake in the intestine and Rdh7 is a retinol dehydrogenase where retinol dehydrogenases are enzymes involved in the biosynthesis of retinoic acid (Napoli 2012; Napoli 2016). Other proteins associated with retinoic acid include Apoa2 and Apoe where Apoa2 was also associated with retinoic acid in the lung (Huang 2018) and Apoe is also associated with inflammation in the gut (Fig. 8).
CONCLUSION
Through an untargeted systems biology approach, proteomic changes that inform on key molecular dysfunction after radiation exposure in a mouse model of GI-ARS have been identified. These data will be useful for a greater understanding of animal models of GI-ARS and may be potentially useful toward the development of medical countermeasures. Identifying molecular mechanisms of injury may also prove useful in the efforts to develop molecular signatures to serve as biomarkers of damage and indicators of recovery and/or therapeutic efficacy.
Supplementary Material
ACKNOWLEDGEMENTS
This project has been funded in whole or in part with Federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under Contract No. HHSN272201000046C and HHSN272201500013I. Additional support was provided by the University of Maryland School of Pharmacy Mass Spectrometry Center (SOP1841-IQB2014). The authors would like to thank the members of the Medical Countermeasures Against Radiological Threats (MCART) consortium for their dedication, support, and guidance. Additionally, we acknowledge Bao Tran for collecting the experimental data and thank the members of the Kane laboratory.
Funding Source:
This project has been funded in whole or in part with Federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under Contract No. HHSN272201000046C and HHSN272201500013I. Additional support was provided by the University of Maryland School of Pharmacy Mass Spectrometry Center (SOP1841-IQB2014).
Footnotes
Conflicts of Interest:
Authors have no conflicts of interest to declare
REFERENCES
- Bo Z, Yongping S, Fengchao W, Guoping A, Yongjiang W. Identification of differentially expressed proteins of gamma-ray irradiated rat intestinal epithelial IEC-6 cells by two-dimensional gel electrophoresis and matrix-assisted laser desorption/ionisation-time of flight mass spectrometry. Proteomics 5: 426–32; 2005. [DOI] [PubMed] [Google Scholar]
- Booth C, Potten CS. The intestine as a model for studying stem-cell behavior. Tumor Models in Cancer Research. Springer; 2002; 337–357. [Google Scholar]
- Booth C, Tudor G, Tonge N, Shea-Donohue T, MacVittie TJ. Evidence of delayed gastrointestinal syndrome in high-dose irradiated mice. Health Phys 103: 400–10; 2012a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth C, Tudor G, Tudor J, Katz BP, MacVittie TJ. Acute gastrointestinal syndrome in high-dose irradiated mice. Health Phys 103: 383–99; 2012b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrareddy SN, Arthos J, Cicala C, Villinger F, Ortiz KT, Little D, Sidell N, Kane MA, Yu J, Jones JW, Santangelo PJ, Zurla C, McKinnon LR, Arnold KB, Woody CE, Walter L, Roos C, Noll A, Van Ryk D, Jelicic K, Cimbro R, Gumber S, Reid MD, Adsay V, Amancha PK, Mayne AE, Parslow TG, Fauci AS, Ansari AA. Sustained virologic control in SIV+ macaques after antiretroviral and alpha4beta7 antibody therapy. Science 354: 197–202; 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter CLH KG; Booth C; Tudor GL; Parker G; Jones JW; Farese AM; MacVittie TJ; Kane MA Characterizing the Natural History of Acute Radiation Syndrome of the Gastrointestinal Tract: Combining high mass and spatial resolution using MALDI-FTICR-MSI. Health Physics; 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassani B, Villablanca EJ, De Calisto J, Wang S, Mora JR. Vitamin A and immune regulation: role of retinoic acid in gut-associated dendritic cell education, immune protection and tolerance. Mol Aspects Med 33: 63–76; 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinetti G, Fruchart JC, Staels B. Peroxisome proliferator-activated receptors (PPARs): nuclear receptors at the crossroads between lipid metabolism and inflammation. Inflamm Res 49: 497–505; 2000. [DOI] [PubMed] [Google Scholar]
- Czarnewski P, Das S, Parigi SM, Villablanca EJ. Retinoic Acid and Its Role in Modulating Intestinal Innate Immunity. Nutrients 9; 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer J, Vayro S, Shirazi-Beechey SP. Mechanism of glucose sensing in the small intestine. Biochem Soc Trans 31: 1140–2; 2003. [DOI] [PubMed] [Google Scholar]
- Ellis CN, LaRocque RC, Uddin T, Krastins B, Mayo-Smith LM, Sarracino D, Karlsson EK, Rahman A, Shirin T, Bhuiyan TR, Chowdhury F, Khan AI, Ryan ET, Calderwood SB, Qadri F, Harris JB. Comparative proteomic analysis reveals activation of mucosal innate immune signaling pathways during cholera. Infect Immun 83: 1089–103; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng JK, Fischer B, Grossmann J, Maccoss MJ. A fast SEQUEST cross correlation algorithm. J Proteome Res 7: 4598–602; 2008. [DOI] [PubMed] [Google Scholar]
- FDA. Product Development Under the Animal Rule: Guidance for Industry. 2015. [Google Scholar]
- Frey MR, Brent Polk D. ErbB receptors and their growth factor ligands in pediatric intestinal inflammation. Pediatr Res 75: 127–32; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain P, Chambon P, Eichele G, Evans RM, Lazar MA, Leid M, De Lera AR, Lotan R, Mangelsdorf DJ, Gronemeyer H. International Union of Pharmacology. LX. Retinoic acid receptors. Pharmacol Rev 58: 712–25; 2006. [DOI] [PubMed] [Google Scholar]
- Giancotti FG, Ruoslahti E. Integrin signaling. Science 285: 1028–32; 1999. [DOI] [PubMed] [Google Scholar]
- Gorfu G, Rivera-Nieves J, Ley K. Role of beta7 integrins in intestinal lymphocyte homing and retention. Curr Mol Med 9: 836–50; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall JA, Grainger JR, Spencer SP, Belkaid Y. The role of retinoic acid in tolerance and immunity. Immunity 35: 13–22; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han YM, Park JM, Choi YS, Jin H, Lee YS, Han NY, Lee H, Hahm KB. The efficacy of human placenta-derived mesenchymal stem cells on radiation enteropathy along with proteomic biomarkers predicting a favorable response. Stem Cell Res Ther 8: 105; 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57; 2009. [DOI] [PubMed] [Google Scholar]
- Huang WY J; Jones JW; Carter CL; Jackson IL; Vujaskovic Z; MacVittie TJ; Kane MA Acute proteomic changes in lung after whole thorax lung irradiation in a mouse model: identification of potential initiating events for delayed effects of acute radiation exposure. Health Physics; 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Zolfaghari R, Hao L, Ross AC. Inflammation rapidly modulates the expression of ALDH1A1 (RALDH1) and vimentin in the liver and hepatic macrophages of rats in vivo. Nutr Metab (Lond) 11: 54; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata M, Yokota A. Retinoic acid production by intestinal dendritic cells. Vitam Horm 86: 127–52; 2011. [DOI] [PubMed] [Google Scholar]
- Jones JW, Bennett A, Carter CL, Tudor G, Hankey KG, Farese AM, Booth C, MacVittie TJ, Kane MA. Citrulline as a Biomarker in the Non-human Primate Total- and Partial-body Irradiation Models: Correlation of Circulating Citrulline to Acute and Prolonged Gastrointestinal Injury. Health Phys 109: 440–51; 2015a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JW SA, Tudor G, Xu PT, Jackson IL, Vujaskovic Z, Booth C, MacVittie TJ, Ernst RK, Kane MA. Identification and quantitation of biomarkers for radiation-induced injury via mass spectrometry. Health Phys 106: 106–19; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JW, Scott AJ, Tudor G, Xu PT, Jackson IL, Vujaskovic Z, Booth C, MacVittie TJ, Ernst RK, Kane MA. Identification and quantitation of biomarkers for radiation-induced injury via mass spectrometry. Health Phys 106: 106–19; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JW, Tudor G, Li F, Tong Y, Katz B, Farese AM, MacVittie TJ, Booth C, Kane MA. Citrulline as a Biomarker in the Murine Total-Body Irradiation Model: Correlation of Circulating and Tissue Citrulline to Small Intestine Epithelial Histopathology. Health Phys 109: 452–65; 2015b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JWC Z; Li F; Tudor G; Farese AM; Booth C; MacVittie TM; Kane MA. Targeted metabolomics reveals metabolomic signatures correlating gastrointestinal tissue to plasma in a mouse total-body irradiation model. Health Physics; 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kall L, Canterbury JD, Weston J, Noble WS, MacCoss MJ. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat Methods 4: 923–5; 2007. [DOI] [PubMed] [Google Scholar]
- Karahalil B Overview of Systems Biology and Omics Technologies. Curr Med Chem 23: 4221–4230; 2016. [DOI] [PubMed] [Google Scholar]
- Kopylov U, Yung DE, Engel T, Avni T, Battat R, Ben-Horin S, Plevris JN, Eliakim R, Koulaouzidis A. Fecal calprotectin for the prediction of small-bowel Crohn’s disease by capsule endoscopy: a systematic review and meta-analysis. Eur J Gastroenterol Hepatol 28: 1137–44; 2016. [DOI] [PubMed] [Google Scholar]
- Koulaouzidis A, Sipponen T, Nemeth A, Makins R, Kopylov U, Nadler M, Giannakou A, Yung DE, Johansson GW, Bartzis L, Thorlacius H, Seidman EG, Eliakim R, Plevris JN, Toth E. Association Between Fecal Calprotectin Levels and Small-bowel Inflammation Score in Capsule Endoscopy: A Multicenter Retrospective Study. Dig Dis Sci 61: 2033–40; 2016. [DOI] [PubMed] [Google Scholar]
- Kramer A, Green J, Pollard J Jr., Tugendreich S Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 30: 523–30; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larange A, Cheroutre H. Retinoic Acid and Retinoic Acid Receptors as Pleiotropic Modulators of the Immune System. Annual review of immunology 34: 369–94; 2016. [DOI] [PubMed] [Google Scholar]
- Lim YB, Pyun BJ, Lee HJ, Jeon SR, Jin YB, Lee YS. Proteomic identification of radiation response markers in mouse intestine and brain. Proteomics 11: 1254–63; 2011. [DOI] [PubMed] [Google Scholar]
- Lussier C, Basora N, Bouatrouss Y, Beaulieu JF. Integrins as mediators of epithelial cell-matrix interactions in the human small intestinal mucosa. Microsc Res Tech 51: 169–78; 2000. [DOI] [PubMed] [Google Scholar]
- MacVittie TJ, Farese AM, Bennett A, Gelfond D, Shea-Donohue T, Tudor G, Booth C, McFarland E, Jackson W 3rd. The acute gastrointestinal subsyndrome of the acute radiation syndrome: a rhesus macaque model. Health Phys 103: 411–26; 2012. [DOI] [PubMed] [Google Scholar]
- Mi H, Huang X, Muruganujan A, Tang H, Mills C, Kang D, Thomas PD. PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res 45: D183–D189; 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee D, Coates PJ, Lorimore SA, Wright EG. Responses to ionizing radiation mediated by inflammatory mechanisms. J Pathol 232: 289–99; 2014. [DOI] [PubMed] [Google Scholar]
- Napoli JL. Physiological insights into all-trans-retinoic acid biosynthesis. Biochim Biophys Acta 1821: 152–67; 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli JL. Functions of Intracellular Retinoid Binding-Proteins. Subcell Biochem 81: 21–76; 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusrat A, Parkos CA, Liang TW, Carnes DK, Madara JL. Neutrophil migration across model intestinal epithelia: monolayer disruption and subsequent events in epithelial repair. Gastroenterology 113: 1489–500; 1997. [DOI] [PubMed] [Google Scholar]
- Potten CS. A comprehensive study of the radiobiological response of the murine (BDF1) small intestine. International journal of radiation biology 58: 925–73; 1990. [DOI] [PubMed] [Google Scholar]
- Rojanapo W, Lamb AJ, Olson JA. The prevalence, metabolism and migration of goblet cells in rat intestine following the induction of rapid, synchronous vitamin A deficiency. J Nutr 110: 178–88; 1980. [DOI] [PubMed] [Google Scholar]
- Shea-Donohue T, Fasano A, Zhao A, Notari L, Yan S, Sun R, Bohl JA, Desai N, Tudor G, Morimoto M, Booth C, Bennett A, Farese AM, MacVittie TJ. Mechanisms Involved in the Development of the Chronic Gastrointestinal Syndrome in Nonhuman Primates after Total-Body Irradiation with Bone Marrow Shielding. Radiat Res 185: 591–603; 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Schoor SR, Reeds PJ, Stoll B, Henry JF, Rosenberger JR, Burrin DG, Van Goudoever JB. The high metabolic cost of a functional gut. Gastroenterology 123: 1931–40; 2002. [DOI] [PubMed] [Google Scholar]
- Venteclef N, Jakobsson T, Steffensen KR, Treuter E. Metabolic nuclear receptor signaling and the inflammatory acute phase response. Trends Endocrinol Metab 22: 333–43; 2011. [DOI] [PubMed] [Google Scholar]
- Villablanca EJ, Cassani B, von Andrian UH, Mora JR. Blocking lymphocyte localization to the gastrointestinal mucosa as a therapeutic strategy for inflammatory bowel diseases. Gastroenterology 140: 1776–84; 2011a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villablanca EJ, Wang S, de Calisto J, Gomes DC, Kane MA, Napoli JL, Blaner WS, Kagechika H, Blomhoff R, Rosemblatt M, Bono MR, von Andrian UH, Mora JR. MyD88 and retinoic acid signaling pathways interact to modulate gastrointestinal activities of dendritic cells. Gastroenterology 141: 176–85; 2011b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WW, Qiao SY, Li DF. Amino acids and gut function. Amino Acids 37: 105–110; 2009. [DOI] [PubMed] [Google Scholar]
- Widmaier M, Rognoni E, Radovanac K, Azimifar SB, Fassler R. Integrin-linked kinase at a glance. J Cell Sci 125: 1839–43; 2012. [DOI] [PubMed] [Google Scholar]
- Williams JP, Brown SL, Georges GE, Hauer-Jensen M, Hill RP, Huser AK, Kirsch DG, Macvittie TJ, Mason KA, Medhora MM, Moulder JE, Okunieff P, Otterson MF, Robbins ME, Smathers JB, McBride WH. Animal models for medical countermeasures to radiation exposure. Radiat Res 173: 557–78; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Chen JY. Discovery of pathway biomarkers from coupled proteomics and systems biology methods. BMC Genomics 11 Suppl 2: S12; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


