Abstract
Phylogenetic studies based on a definite set of marker genes usually reconstruct evolutionary relationships among the prokaryotic species. Based on specific target sequences, such studies represent variations and allow identification of similarities or dissimilarities in organisms. With the advent of completely sequenced genomes and accumulation of information on whole prokaryotic genomes, phylogenetic reconstructions should be considered more reliable if they are ideally based on entire genomes to resolve phylogenetic interest. We applied phylogenomics approaches taking into account completely sequenced cyanobacterial genomes to reconstruct underlying species that represented major taxonomic classes and belonged to distinctly different habitats (freshwater, marine, soils, and rocks). We did not rely on describing phylogeny of all representative class of cyanobacterial species on the basis of only ribosomal gene, 16S rDNA gene. In contrast, we analyzed combined molecular marker and phylogenomics approaches (genome alignment, gene content and gene order, composition vector and protein domain content) for accurately inferring phylogenetic relationship of species. We have shown that this approach reflects the impact of evolution on the organisms and considers connects with the ecological adaptation in cyanobacteria in different habitats. Analysis revealed that the members from marine habitat occupy different profile than those from freshwater. Impact of GC content and genomic repetitiveness over the diversification of cyanobacterial species and their possible role in adaptation was also reflected. Members occupying similar habitats cover more evolutionary distance together and also evolve various strategies for adaptation and survival either through genomic repetitiveness or preferences for genes of particular functions or modified GC content. Genomes undergo different changes for their adaptation in diverse habitats.
Electronic supplementary material
The online version of this article (10.1007/s13205-019-1635-6) contains supplementary material, which is available to authorized users.
Keywords: Cyanobacterial evolution, Phylogeny, Ecological adaptation, Genomic repetitiveness, Functional profile, Phylogenomics, Cyanobacteria
Introduction
The universal ‘tree of life’ constructed on the basis of molecular analysis of ribosomal RNA (rRNA) genes has led to molecular classification of microorganisms (Woese and Fox 1977; Woese 1987). Although the rRNA-based tree of life is the most common and widely accepted approach for microbial identification, it is not quite sufficient to resolve accurate phylogeny of interest (Eisen 2000; Capella-Gutierrez et al. 2014; Adato et al. 2015). The most prominent objection disfavouring this approach was whether a single-gene tree solely represents the evolutionary history of the organisms (Eisen 2000; Sleator 2013). In molecular taxonomy, a single tree of life generally reflects species relatedness through vertical descent. However, not all genes follow similar tendency. Many genes are transferred between lineages horizontally or laterally via ‘Horizontal Gene Transfer’ (HGT), a phenomenon quite frequent in prokaryotic organisms. Such gene transfers within species usually complicate the evolutionary reconstruction based on single gene. They represent that some species are chimeric in nature having diverse histories for different parts of their genome (Eisen 2000; Prasanna and Mehra 2013; Rinke et al. 2013). Multiple approaches were derived to explore phylogeny among organisms on the basis of various structural and functional genes and protein sequences (Sawa et al. 2003; Auch et al. 2010; Lang et al. 2013). Such methods end with troubles in deciphering evolutionary history of organisms due to duplication, deletion or horizontal gene transfer. Another problem in gene(s) based approach lies in the identification of gene to be used for phylogenetic analysis as it should be significantly conserved across the diverse genomes (Sawa et al. 2003). There existed some reports on the phylogenetic analysis of archaea and bacteria using universal genes (Nelson et al. 1999; Makarova et al. 1999). It was, therefore, suggested that the tree generated from universal genes is not necessarily accurate due to factors such as convergence, misidentification of orthologs, gene conversion and ambiguities in sequence alignment (Eisen 2000). The molecular phylogenies that have led to the classification of three primary kingdoms or domains were based on single characteristic gene sequence such as 16S rRNA gene (Weisburg et al. 1991; Li et al 2002; Větrovský and Baldrian 2013). However, using various characteristic gene sequences for phylogeny reconstruction leads to contrast conclusions reflecting problems owing to horizontal gene transfer, unrecognized paralogy and highly variable rates of evolution (Woese 1998; Rudi and Sekelja 2013). Therefore, equating the history of the whole genome with that of a single although highly conserved gene or only a part of the genome or a major region is not conclusive.
Theoretically, the evolutionary history of different genomes can be assessed by comparing phylogenetic trees for each gene in every genome (Eisen 2000). Practically, such approaches have their own limitations because phylogenetic tree reconstruction is cumbersome when every gene in the genome is considered. Ideally, for making the genetic tree reconstruction reliable, the set of genes analyzed should be present in all the species considered, sequence alignments need to be carefully examined and ambiguous or hypervariable alignment regions should be excluded from phylogenetic analysis. Phylogenetic reconstructions based on whole genome sequences are emerging as the most reliable alternatives (Takahashi et al. 2009). It allows comparison of entire gene families or overall gene content, insertions and deletions or HGT events (Sawa et al. 2003). Evaluation of closely related genome sequences can endow with an evidence of macroscopic genome polymorphism which occurs during the phase of recombination processes (Kawai et al. 2006).
Conventional phylogenetics and phylogenomics
Evolutionary biology has witnessed revolution with the disclosure of fact that comparison of gene sequences can reveal the evolutionary history of species (Losos et al. 2013) Ribosomal RNA was analyzed for the interpretation of evolutionary classification of microbes as universal tree of life (Zuckerkandl and Pauling 1965; Woese 1987; Woese and Fox 1977; Eisen 2000). Archaea was recognized as the third domain of life along with well-accepted domains Bacteria and Eukarya on the basis of ssu-rRNA (Rajendhran and Gunasekaran 2011). For evolutionary studies, ssu-rRNA is broadly accepted as ‘phylogenetic marker’ that possesses features such as presence across all three domains of life, highly conserved regions, sequence and structure level conservation and sequence variations to facilitate studies on evolutionary events (Wu et al. 2013). The gene also serves the purpose of ‘ecological marker’ along with ‘phylogenetic marker’ for the analysis of microbes (Wu et al. 2013; Moreno-Hagelsieb et al. 2013). Along with microbes, these gene sequences were also used as phylogenetic marker for other organisms and deciphered valuable information about their phylogenetic relationships. Ribosomal rRNA genes are present in many copies per genome and repeated copies have the similar prototype of concerted evolution which facilitates its analysis either by direct RNA and DNA sequencing or by restriction enzyme methodologies (Hillis and Dixon 1991; Bryant et al. 2013).
Though, there exists many controversies with rRNA gene based tree of life, major concern relates to the use of single gene for defining evolutionary history (Eisen 2000; Daubin and Szöllősi 2016). ssu-rRNA genes also have wide deviation in copy number among different organisms which poses limitation on estimation of their relative abundance across different groups. Primers which are universally used for amplification of ssu-rRNA genes also tend to have slight biasness towards certain taxonomic groups. This gene is reported to influenced by the processes such as HGT, convergent evolution or variations in evolutionary rates in due course of time (Wu et al. 2013). Among phylogeny based on this gene, limited mutable sites and restricted length generates the problem of saturation (Henz et al. 2005). In addition, 16S rRNA gene sequence identity does not correlate with DNA–DNA hybridization (DDH) values which are critical conclusive factor for establishment of new species (Klenk and Göker 2010; Kim et al. 2014). In addition to 16S rRNA genes, 23S rRNA, the β-subunit of F1F0 ATPase, DNA gyrase b gene and elongation factor Tu is also used as phylogenetic marker genes (Ludwig and Schleifer 1999). Though again, the major problem associated with the use of these genes as phylogenetic marker lies in the fact that whether a single gene can resolve evolutionary history of any species. There are experimental facts that rRNA gene is also subjected to horizontal gene transfer (Li et al. 2002; Yabuki et al. 2014) and single-gene-based phylogeny is unable to cope with horizontal gene transfer, unrecognizable paralogy and extremely inconsistent rates of evolution (Wu et al. 2013).
Evolution of gene content in a genome is a complicated and unresolved process (Snel et al. 2002; Comas et al. 2006). Studies revealed that presence or absence of genes between genomes is dependents on the genome size (Snel et al. 1999) and evolutionary distance (Snel et al. 2002). Genome size is ruled by many different factors involving addition/deletion of genetic information, amplifications, horizontal transfer and selection. Prokaryotic genomes tend to have strong deletion biases where DNA lacking adaptive value is rapidly deleted (Mira et al. 2001). This has indicated that the recombination leads to deletions more often than amplifications (Treangen et al. 2009). Larger genomes are reported to preferentially obtain genes involved in regulation, secondary metabolism and energy conversion. Bacteria with larger genomes are ecologically more successful in the environment where resources are varied and poor (Prabha et al. 2016). Complete understanding of these facts will provide detail insight into interaction between ecology and genome evolution (Konstantinidis and Tiedje 2004).
Genomics technologies opened array of scope for microbial biologists to understand factors leading to complex evolutionary patterns of the genomes (Eisen 2000; Wolf et al. 2001; Mirkin et al. 2003; Henz et al. 2005; Zhao et al. 2012). Microbial genomes acquire foreign DNA from surroundings more frequently as compared to higher organisms through HGT events (Capella-Gutierrez et al. 2014). HGT in combination with intra-genomic rearrangements and duplication events leads to bacterial adaptations to different environmental niches and variance in closely related species (Sicheritz-Pontén and Anderssona 2001; Oliveira et al. 2017). Genome size, geometry, GC content and gene number are the important parameters and deviation in any of these parameters reflects typical change in the bacterial genomes (Bentley and Parkhill 2004). Many genomes reflects changes in genome size and gene content as directed by the processes such as deletion, duplication and lateral gene transfer events (Nilsson et al. 2005; Cordero and Hogeweg 2009). Overall, the genome size of bacteria is maintained in an equilibrium between the duplication or HGT and mutations leading to elimination of function followed by deletions (the loss of genes) (Wernegreen et al. 2000). For better understanding of this phenomenon, in-depth phylogenetic analysis is required at genome level (Sicheritz-Pontén and Anderssona 2001). Increasing number of complete genome sequences makes it possible to use wealth of genomic information for phylogenetic reconstruction, focus on entire genome and its genes rather than a single gene or group of genes (Wolf et al. 2001; Mirkin et al. 2003; Henz et al. 2005). Availability of large number of bacterial whole genome sequences facilitated biologists to explore and examine evolutionary hypotheses on a larger scale than ever (Zhao et al. 2012; Land et al. 2015).
Phylogenomics refer to such studies that involve large-scale genomic comparisons for phylogenetic reconstruction. This reflects complex evolutionary pattern for microbes that involve not only vertical descent or lateral gene transfer but also include a mix of recombination, duplication, invention, loss, degradation and convergence of genes during selection processes (Eisen 2000; Meier-Kolthoff et al. 2014). Phylogenomics approaches hold promise for better interpretation of genome organization and function. Recently sequenced genomes provided a clear picture of genome evolution and phylogeny (Medina 2005). Enormous information lead to decipher salient genomic features in terms of gene content and gene order and create reliable phylogenetic reconstructions. Based on the information of gene and gene family content, gene order, protein domain content, protein orthologs and fold information in the genomes, whole genome phylogenetic trees have become more reliable (Klenk and Göker 2010). Though in prokaryotic genomes, gene content and gene order phylogenies are influenced by gene loss and horizontal gene transfer (Klenk and Göker 2010).
Phylogenetic trees are, therefore, valuable means for analyses such as taxonomy assignment, metagenomics studies, inference of co-speciation, identification of ecological trends, epidemiological and biogeographical events, phylogenetic profiling analysis and genomes selection for sequencing (Lang et al. 2013). Currently, reliability in methods, tools, and approaches raises expectations from phylogenomics analysis to infer identification of taxon-specific gene families as a source of explicit physiological features, taxonomic and evolutionary purposes and lacuna in single-gene phylogenies (Klenk and Göker 2010; Lang et al. 2013).
Approaches for phylogenomics analysis
Genome trees were suggested to capture overwhelming information of the phylogeny. Approaches for whole genome-based phylogenies can be broadly classified into three categories as based on (1) sequence alignment, (2) gene content and gene order, and (3) sequence statistics. Certain parameter-free and whole-genome-based composition vector approaches were also developed for phylogenetic reconstruction (Qi et al. 2004; Hao and Qi 2004; Qiang et al. 2010; Bromberg et al. 2016). Thus, phylogenetic trees considering complete genome sequences can be reconstructed into five different ways: (1) alignment-free trees in which statistic properties of genome are considered; (2) gene content trees in which presence or absence of genes are considered; (3) trees based on chromosomal gene order; (4) trees based on average sequence similarity; and (5) phylogenomics based genome trees (Snel et al. 2005). In another study, four major approaches considering (1) gene and/or protein content, (2) gene order, (3) shared gene content, and (4) information theory and genome compression were mentioned for phylogenetic analysis (Khiripet 2005).
Alignment-based methods were in practice from the time of rRNA sequence rooted phylogenetic trees. However, owing to sequence size limitations, alignment-based methods are complicated for application at the entire genome level. Furthermore, it is also not feasible to carry out multiple sequence alignment at whole genome level because of huge sequence size (from millions to billions of base pairs) (Klenk and Göker 2010). Gene duplication affects gene content of genomes and thereby generates inconsistency in phylogeny of closely and distantly related genomes. Gene content tends to have strong phylogenetic signal and assist in removing numerous taxonomic uncertainties (Tekaia et al. 1999; Snel et al. 2005; Anselmetti et al. 2018). It, however, depends up on the availability of complete genomes. In case of incomplete genomes, gene content approach can be used via signature genes, where for each clade, core genes ubiquitous in every genome in a phylogenetically coherent group is identified (Charlebois and Doolittle 2004; Dutilh et al. 2008). Gene order is also used for the assessment of phylogenetic relationship in closely related genomes. However, this parameter lacks resolution as genome rearrangement is not a frequent event in nature (Vishnoi et al. 2010; Gu et al. 2005; Zhou et al. 2017). Alignment-free approaches such as k-string approach or composition vector approach are also available for the construction of genome trees (Xu and Hao 2009). Such approaches are computationally less costly and utilize utmost content of the genomes (Vishnoi et al. 2010).
Cyanobacterial phylogeny
Cyanobacteria are among the most primitive oxygenic photosynthetic organisms. They have been studied extensively for different biological processes including photosynthesis, bioenergetics, nitrogen fixation, environmental stress adaptation and molecular evolution (Koksharova and Wolk 2002; Cassier-Chauvat and Chauvat 2018). Cyanobacterial genomes reveal a complex evolutionary history (Zhaxybayeva et al. 2006; Prabha et al. 2016). These organisms have found their origin in ancient group of photosynthetic prokaryotes. They have shown distinctions in their habitats, cellular differentiation strategies, physiological capacities and metabolic complexity (Beck et al. 2012). Diversity across various cyanobacteria in terms of their size, gene number and GC content is reflected in their whole genome sequences (Larsson et al. 2011; Prabha et al. 2016). This has facilitated studies on the factors governing variations among the organisms and mechanisms responsible for evolutionary diversification. We, therefore, determined phylogenetic relationship within different cyanobacterial species from diverse taxonomic groups by considering their entire genomic sequence and features (genome alignment, gene content and gene order, protein domain content). The study reflected changes in cyanobacterial genomes towards their adaptation in different ecological niches during the evolution. We also compared conventional and phylogenomics approaches reflecting evolutionary history of cyanobacteria and provides light over process of diversification of these organisms.
Materials and methods
Species and genome sequences
Forty-one cyanobacterial species representing five different taxonomic orders (Chroococcales, Prochlorales, Nostacales, Oscillatoriales and Gloeobacterales), for which complete genome sequences were available, were taken into the study. All the genome sequences were downloaded from NCBI Genome database (http://www.ncbi.nlm.nih.gov/genome/). Maximum number of species (22) represented Chroococcales followed by Prochlorales (12). Order Nostacales, Oscillatoriales and Gloeobacterales had 4, 2, and 1 species, respectively (Table 1).
Table 1.
Details of cyanobacterial genomes under consideration [genome size (Mb), GC content (percentage), coding genome (percentage), gene density (genes/Mb), microsatellites (number)]
| S. no. | Taxonomy | Organism name | Abbreviation used | Habitat | Genome Size | GC content | Coding genome | Gene density | Micro-satellites |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Chroococcales | Acaryochloris marina MBIC11017 | Am_MBIC11017 | Marine | 8.36 | 47 | 83.26 | 1025 | 6826 |
| 2 | Cyanothece sp. ATCC 51142 | Cs_ATCC51142 | Marine | 5.46 | 37.9 | 86.8 | 983 | 6540 | |
| 3 | Cyanothece sp. PCC 7424 | Cs_PCC7424 | Fresh water | 6.55 | 38.5 | 81.46 | 907 | 7975 | |
| 4 | Cyanothece sp. PCC 7425 | Cs_PCC7425 | Fresh water | 5.79 | 50.6 | 85.28 | 951 | 7023 | |
| 5 | Cyanothece sp. PCC 7822 | Cs_PCC7822 | Fresh water | 7.84 | 40.1 | 82.83 | 898 | 7512 | |
| 6 | Cyanothece sp. PCC 8801 | Cs_PCC8801 | Fresh water | 4.79 | 39.8 | 84.85 | 964 | 5587 | |
| 7 | Cyanothece sp. PCC 8802 | Cs_PCC8802 | Fresh water | 4.8 | 39.8 | 85.1 | 979 | 5567 | |
| 8 | Microcystis aeruginosa NIES-843 | Ma_NIES_843 | Fresh water | 5.84 | 42.3 | 81.36 | 1090 | 6639 | |
| 9 | Synechococcus sp. CC9311 | Ss_CC9311 | Fresh water | 2.61 | 55.5 | 87.21 | 1128 | 3317 | |
| 10 | Synechococcus sp. CC9605 | Ss_CC9605 | Fresh water | 2.51 | 55.4 | 86.94 | 1098 | 3782 | |
| 11 | Synechococcus sp. CC9902 | Ss_CC9902 | Marine | 2.23 | 52.4 | 90 | 1056 | 2837 | |
| 12 | Synechococcus sp. JA-2-3B’a(2–13) | Ss_JA_2_3Ba | Marine | 3.05 | 59.2 | 85.48 | 965 | 4386 | |
| 13 | Synechococcus sp. JA-3-3Ab | Ss_JA-3-3Ab | Marine | 2.93 | 54.2 | 84.86 | 989 | 4509 | |
| 14 | Synechococcus sp. PCC 7002 | Ss_PCC7002 | Hot spring | 3.41 | 58.5 | 87.64 | 950 | 3426 | |
| 15 | Synechococcus sp. RCC307 | SsRCC307 | Hot spring | 2.22 | 60.2 | 94.16 | 1163 | 3586 | |
| 16 | Synechococcus sp. WH 7803 | Ss_WH7803 | Marine | 2.37 | 49.2 | 93.39 | 1091 | 3425 | |
| 17 | Synechococcus sp. WH 8102 | Ss_WH8102 | Marine | 2.43 | 60.8 | 90.3 | 1062 | 3580 | |
| 18 | Synechococcus elongatus PCC 6301 | Se_PCC6301 | Marine | 2.7 | 60.2 | 88.04 | 956 | 3289 | |
| 19 | Synechococcus elongatus PCC 7942 | Se_PCC7942 | Marine | 2.74 | 59.4 | 89.21 | 991 | 3324 | |
| 20 | Synechocystis sp. PCC 6803 | Sy_PCC6803 | Fresh water | 3.95 | 47.4 | 87.14 | 918 | 4410 | |
| 21 | Thermosynechococcus elongatus BP-1 | Te_BP_1 | Hot spring | 2.59 | 53.9 | 89.99 | 975 | 2956 | |
| 22 | cyanobacterium UCYN-A | C_UCYNA | Marine | 1.44 | 31.1 | 81.41 | 862 | 2349 | |
| 23 | Gloeobacterales | Gloeobacter violaceus PCC 7421 | Gv_PCC7421 | Rock | 4.66 | 62 | 89.36 | 962 | 7078 |
| 24 | Nostacales | Anabaena variabilis ATCC 29413 | Av_ATCC29413 | Multiple | 7.11 | 41.4 | 82.33 | 818 | 7666 |
| 25 | Nostoc sp. PCC 7120 | Ns_PCC7120 | Multiple | 7.21 | 38.3 | 82.5 | 862 | 7627 | |
| 26 | Nostoc punctiforme PCC 73102 | Np_PCC73102 | Fresh water | 9.06 | 41.4 | 77.43 | 791 | 9646 | |
| 27 | ‘Nostoc azollae’ 0708 | Na_0708 | Multiple | 5.49 | 41.3 | 52.13 | 980 | 7175 | |
| 28 | Oscillatoriales | Arthrospira platensis NIES-39 | Ap_NIES-39 | Fresh water | 6.79 | 44.3 | 81.24 | 983 | 7739 |
| 29 | Trichodesmium erythraeum IMS101 | Te_IMS101 | Marine | 7.75 | 34.1 | 60.11 | 661 | 12,530 | |
| 30 | Prochlorales | Prochlorococcus marinus str. AS9601 | Pm_AS9601 | Marine | 1.67 | 31.3 | 91.15 | 1177 | 2967 |
| 31 | Prochlorococcus marinus str. MIT 9211 | Pm_MIT9211 | Marine | 1.69 | 38 | 90.12 | 1124 | 2269 | |
| 32 | Prochlorococcus marinus str. MIT 9215 | Pm_MIT9215 | Marine | 1.74 | 31.1 | 89.62 | 1180 | 3094 | |
| 33 | Prochlorococcus marinus str. MIT 9301 | Pm_MIT9301 | Marine | 1.64 | 31.3 | 91.21 | 1196 | 2798 | |
| 34 | Prochlorococcus marinus str. MIT 9303 | Pm_MIT9303 | Marine | 2.68 | 50 | 84.52 | 1170 | 3650 | |
| 35 | Prochlorococcus marinus str. MIT 9312 | Pm_MIT9312 | Marine | 1.71 | 31.2 | 89.59 | 1085 | 3042 | |
| 36 | Prochlorococcus marinus str. MIT 9313 | Pm_MIT9313 | Marine | 2.41 | 50.7 | 82.23 | 967 | 3234 | |
| 37 | Prochlorococcus marinus str. MIT 9515 | Pm_MIT9515 | Marine | 1.7 | 30.8 | 88.92 | 1155 | 3146 | |
| 38 | Prochlorococcus marinus str. NATL1A | Pm_NATL1A | Marine | 1.86 | 35 | 87.29 | 1210 | 2913 | |
| 39 | Prochlorococcus marinus str. NATL2A | Pm_NATL2A | Marine | 1.84 | 35.1 | 85.62 | 1211 | 2798 | |
| 40 | Prochlorococcus marinus subsp. marinus str. CCMP1375 | Pm_CCMP1375 | Marine | 1.75 | 36.4 | 89.22 | 1103 | 2421 | |
| 41 | Prochlorococcus marinus subsp. pastoris str. CCMP1986 | Pm_CCMP1986 | Marine | 1.66 | 30.8 | 88.42 | 1061 | 3112 |
Genomic features
Genomic features of all of these cyanobacteria were identified.
Genome size, GC content: Information about these parameters was obtained from NCBI Genome database (http://www.ncbi.nlm.nih.gov/genbank/genomes).
%-coding information: Information about how much percentage of genome is coding was calculated as
Gene density: Gene density was calculated as genes/Mb.
Phylogenetic reconstruction on the basis of 16S rRNA gene
For alignment of 16S rRNA gene, MUSCLE (MUltiple Sequence Comparison by Log-Expectation) was used (Edgar 2004). 16S rRNA gene phylogeny was constructed using MEGA 5.0 (Tamura et al. 2011) employing Neighbor-Joining reconstruction with 100 bootstrap iterations. Tree visualization was done via TreeDyn program (Chevenet et al. 2006).
Reconstruction of complete genome-based phylogenies
Distance estimation
On the basis of genome alignment: Alignment of complete genomes was carried out through MUMMER (Delcher et al. 2002) from GGDC web server (Auch et al. 2010) (parameters: coverage algorithm with distance function and 100% identity). DNA–DNA hybridization (DDH) is an extensively used technique for estimation of the overall similarity among the genomes of two organisms (Auch et al. 2010). In fact, the “gold standard” of bacterial species delineation is in general genome similarity identified through DDH, which is a strictly rigorous technique, however, at some instances yields inconsistent results (Colston et al. 2014). In silico approaches for the genome sequence comparison are also available as an alternative of DDH. GGDC web server is based on this only and implies distance methods based as high-scoring segment pairs (HSPs) or maximally unique matches (MUMs) for estimation of similarity among different genomes (Auch et al. 2010).
On the basis of alignment-free composition vector approach: Alignment-free composition vector (CV) approach was used for intergenomic distance estimation from CVTree platform (k-tuple length of 6) (Xu and Hao 2009). Composition vector (CV) approach is an alignment-free method based on K-tuple counting and background subtraction. This approach infers trees from whole-genome data through an alignment-free and parameter-free way (Zuo et al. 2010).
On the basis of overlapping gene content and gene order: For estimation of intergenomic distance on the basis of overlapping genes (OG), OGtree algorithm was used (parameters: 1 for weight of overlapping-gene order and gene content, 1e−9 as threshold of E value and 80% for threshold of alignment coverage in each sequence). OGtree constructs OG distance between the genomes by the analysis of both the OG content and OG order (Jiang et al. 2008). The overlapping genes (OGs) are adjoining genes whose coding sequences are partially or completely overlapped (Jiang et al. 2008). Overlapping genes (OGs) correspond to widely available genomic feature of bacterial genomes and are also used as rare genomic markers for phylogenetic analysis of closely related bacterial species (Zhang and Lin 2015). OGs are potentially involved in different important processes including regulation of gene expression or development of genome compaction (Zhang and Lin 2015). In fact, OGs are much conserved among species rather than non-OGs (Jiang et al. 2008).
On the basis of whole-genome protein domain content: Whole-genome protein domain content approach considers protein domains of the entire genome for the estimation of distances. ProdocTree (http://ibi.cqupt.edu.cn/prodoctree/index.php) was used and it uses the bit score of hmmscan result of each Pfam protein domain for calculation of the coordinate of a dimension of a multi-dimensional space and provides euclidean distances between two points in the space where every point is representative of a species. Protein structural domains represent evolutionary units, the relationships of whom can be assessed along long evolutionary distances (Yang and Bourne 2009). Domain architectures are shown to be conserved at large phylogenetic distances, in prokaryotes and eukaryotes, both (Koehorst et al. 2016).
Tree visualization
Neighbor-Net algorithm (Bryant and Moulton 2004) from SplitsTree software (Huson 1998) was used for phylogenetic network construction for complete genomes.
Functional characterization and COG assignment
Functional characterization of all the cyanobacterial genomes was done using the Clusters of Orthologous Groups (COG) database (Tatusov et al. 2003). For each of the genome, all the genes were subjected to COG assignment using Function Profile tool from IMG database (Markowitz et al. 2012). Function Profile tool assists in identification of the genes associated with a particular function in query genome and thus, genes are expected to share at least the same general functions with their COG matches. Once the genes were assigned COGs, they were clustered in 22 functional categories, which were further grouped in four major classes (Table 2). Self-made Perl scripts were used for the grouping of different COG categories.
Table 2.
Functional distribution of cyanobacterial genomes under consideration (values represents percentage of the particular COG category identified in the cyanobacteria)
COG categories: E amino acid transport and metabolism, G carbohydrate transport and metabolism, F nucleotide transport and metabolism, C energy production and conversion, H coenzyme transport and metabolism, I lipid transport and metabolism, P inorganic ion transport and metabolism, Q secondary metabolites biosynthesis, transport and catabolism, D cell cycle control, cell division, chromosome partitioning, M cell wall/membrane/envelope biogenesis, N cell motility, O posttranslational modification, protein turnover, chaperones, T signal transduction mechanisms, U intracellular trafficking, secretion, and vesicular transport, V defense mechanisms, A RNA processing and modification, B chromatin structure and dynamics, J translation, ribosomal structure and biogenesis, K Transcription, L replication, recombination and repair, S function unknown, R general function prediction only
Repeat identification and analysis
Information about various kinds of microsatellites (Simple Sequence Repeats or SSRs) present in cyanobacterial species was extracted through Imperfect Microsatellite Extractor (IMEx) tool (Mudunuri and Nagarajaram 2007). Parameters taken were, Repeat Type: Perfect; Min. Repeat Number: mono:6, di: 3, tri-hexa:2.
Results and discussion
Habitat and genomic features
Genome size of the cyanobacterial species under study varied from 1.44 Mb (C_UCYN-A) to 9.06 Mb (Np_PCC73102) (Table 1). All the organisms contain single circular chromosome as their major genetic material while Cs_ATCC51142 and Av_ATCC29413 have additional chromosome, i.e., linear chromosome and incision element, respectively. For all cyanobacteria, GC content ranged from 30.8% (Pm_MIT9515) to 62% (Gv_PCC7421). Cyanobacteria from marine habitat has small genome size as compared to the members inhabiting freshwater, soil or multiple habitats (Table 1). Larger genomes showed low gene density as compared to smaller ones (Table 1).
16S Rrna-based phylogenetic analysis
We applied 16S rRNA phylogenetic approach to have an insight on the phylogeny of cyanobacterial species. Phylogenetic tree on the basis of 16S rRNA genes divided all cyanobacteria in two branches (Fig S1). The first branch included four species (all the three thermophilic strains (Ss_JA23Ba, Ss_JA33Ab and Te_BP_1) and the only member of Gloeobacterales (Gv_PCC7421 from rock habitat)), second branch comprised rest of the species. In the second branch, all the marine pico-cyanobacteria of the order Prochlorales were grouped with marine and freshwater species of Synechococcus. All the members of the order Nostacales shared same branch, joint earlier with a branch having two members of Oscillatoriales. Rest members of the order Chroococcales (Cyanothece sps.) clustered on a single branch with members of Nostacales and Oscillatoriales as their nearest neighbor (Fig S1). The tree reflected that cyanobacterial species covering long evolutionary distance together occupy similar habitats and generally possess similar genomic features such as genome size and GC composition as evident from the 16S rRNA-based phylogenetic tree.
Complete genome-based phylogenetic analyses
Genome comparisons suggest that horizontal gene transfer and differential gene loss constitute major evolutionary phenomenon in prokaryotes (Koonin et al. 2001). Whole genome approaches of reconstructing phylogenetic tree have become more apparent due to increasing rate of sequencing projects (Wolf et al. 2002; Delsuc et al. 2005). Considering the entire genome sequences, phylogenetic reconstruction of cyanobacterial species were created using different approaches i.e. genome alignment (Fig S2), alignment-free composition vector approach (Fig S3), overlapping gene content and gene order (Fig S4) and whole-genome protein domain content (Fig S5). Different phylogenomics analyses yielded following results:
Diverse clades for the Order Chroococcales
From the different phylogenomics reconstructions (Fig S1–Fig S5), it is clear that among the order Chroococcales four different clades are identified:
Clade of Cyanothece sps. with members of Nostacales and Oscillatoriales.
Clade of marine Synechococcus sps. with Prochlorococcus sps.
Both the thermophilic Synechococcus sps. occupied same lineage and showed similarity with Gv_PCC7421 (Gloeobacterales).
Clade occupied rest of the Chroococcales species, i.e. Am_MBIC11017, Te_BP_1, C_UCYNA, Se_PCC6301 and Se_PCC7942.
Monophyletic clade for Order Nostacales
In all the phylogenomics reconstructions, members of the Order Nostacales occupied different branches of a single clade along with members of Chroococcales (strains of Cyanothece sps.) and Oscillatoriales as nearest neighbor (Fig S2-S5). In the order Nostacales, Av_ATCC29413 and Ns_PCC7120 cover maximum distance together compared to rest of the two species (Np_PCC73102 and Na_0708) (Fig S2, Fig S4-S5). This is also evident in the phylogenetic reconstruction based on 16S rRNA gene approach (Fig S1). Important to note here is that all the four members of order Nostacales occupied almost similar kind of habitats.
Common clade of marine cyanobacterial species
In all the phylogenetic reconstructions, marine cyanobacterial strains of Synechococcus sps. and Prochlorococcus sps. occupy the same clade, though they represent different taxonomic orders i.e. Chroococcales and Prochlorales, respectively (Fig S1-S5). Both these groups of marine cyanobacteria show similarity in their genomic features and habitat. Furthermore, it was observed that, among the order Prochlorales, all the high light-adapted strains (Pm_CCMP1986, Pm_MIT9515, Pm_MIT9312, Pm_MIT9301, Pm_AS9601, Pm_MIT9215) formed a single clade thereby, supporting their monophyly origin, whereas six low light-adapted strains (Pm_MIT9211, Pm_MIT9303, Pm_MIT9313, Pm_NATL1A, Pm_NATL2A, Pm_CCMP1375) occupied different branches suggesting parallel evolution (Fig S3-S5). It was also observed that Pm_MIT9303 and Pm_MIT9313 shared branching with Synechococcus sps. rather than Prochlorococcus sps. (Fig S5). Marine cyanobacteria C_UCYNA also occupied a branch closer to the Prochlorococcus strains (Fig S3).
Functional genome profile and its role in adaptation and diversification
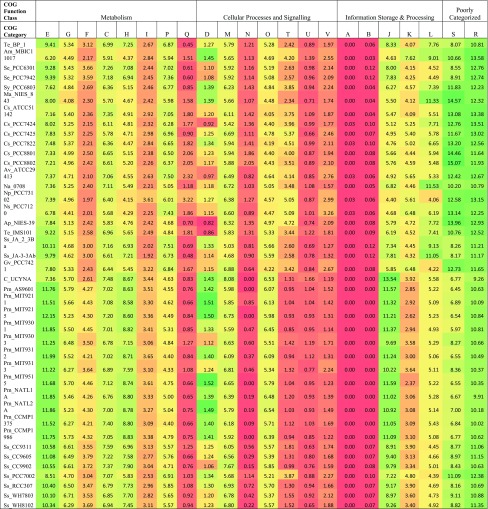
We identified functional profile of all 41 cyanobacterial species which provided insights over the functional composition of each genome. 2406 COGs from 22 different categories of four major functional classes (‘Metabolism’, ‘Cellular processes and signalling’, ‘Information storage & processing’ and ‘Poorly categorized’) were assigned to these species (Table 2). While analyzing distribution of each functional category, it was observed that across all cyanobacteria, genes with metabolic functions gained maximum share (Fig. 1). Next most abundant functional category in most of the cyanobacteria (specifically those inhabiting freshwater and multiple habitat) was that of poorly categorized genes [Function unknown (S) and General function prediction only (R)]. While marine cyanobacteria preferred genes for ‘Information Storage and Processing’ over ‘Cellular Processes and Signalling’, later one is preferred by cyanobacteria from other habitats (Fig. 1). Earlier reports suggested conservation of genes involved in Information processing and signalling in large evolutionary distances (Makarova et al. 1999; Mushegian and Koonin 1996; Azuma and Ota 2009). This may be because they encode for the basic functionalities of the cells (e.g., transcription, translation, repair etc.) and any change in them leads to disruption in normal cellular machinery (Caffrey et al. 2012). In general, habitat seems to influence the functional profile as members from similar habitats possess similar functional profile (Fig. 1). Bacterial genomes contain specific functional gene inventories which are in concurrence with their survival in the particular ecological niche (Allen and Banfield 2005). In the heatmap deduced from the functional profile of cyanobacterial species, cyanobacteria forms two different groups i.e. group I and group II (Fig. 1). Group I majorly includes cyanobacteria from freshwater and other habitats while Group II includes cyanobacteria exclusively from marine habitats. Marine species showed almost different functional genome profile as compared to rest of the cyanobacteria. Most abundant functional category in the cyanobacteria from freshwater and other habitats was Function unknown (S) and General function prediction only (R) (Fig. 1), which possibly reflects that organisms have gained a number of genes, maximum of which remained unknown though they definitely have some important role in their survival.
Fig. 1.
Heatmap based on the percentage distribution of genes in each functional category for all the cyanobacteria under consideration. Color coding varies from black to red, where black represents lowest value and red represents highest. COG categories: E amino acid transport and metabolism, G carbohydrate transport and metabolism, F nucleotide transport and metabolism, C energy production and conversion, H coenzyme transport and metabolism, I lipid transport and metabolism, P inorganic ion transport and metabolism, Q secondary metabolites biosynthesis, transport and catabolism, D cell cycle control, cell division, chromosome partitioning, M cell wall/membrane/ envelope biogenesis, N cell motility, O Posttranslational modification, protein turnover, chaperones, T signal transduction mechanisms, U intracellular trafficking, secretion, and vesicular transport, V defense mechanisms, A RNA processing and modification, B chromatin structure and dynamics, J translation, ribosomal structure and biogenesis, K transcription, L replication, recombination and repair, F function unknown, R general function prediction only
Influence of GC content over adaptation and diversification
GC content is a well-defined compositional feature of organisms and genomic signature considered to be biased across the tree of life (Nakabachi et al. 2006; Nalbantoglu 2011). GC content is the simplest compositional parameter likely to be affected by the environment or lifestyle of any microbial species and is related to phylogenetic variation (Lawrence and Ochman 1997; Nalbantoglu 2011; Dutta and Paul 2012; Bossert et al. 2017). This feature generally remains constant within a microbial species but becomes variable when it comes across the organisms (Dutta and Paul 2012). In the molecular marker based phylogenetic reconstruction, it is evident that members occupying the same clade possess similar genome size, GC composition and also occupy similar kind of habitat (Fig S1). This fact was also identified in the phylogenomics reconstructions (composition vector approach (Fig S3) and gene content and gene order approach (Fig S4)), where it was reflected that members formed clade with other members of same or different taxonomic orders having similar GC content.
Relation between habitat and different phylogenetic analyses
Habitat of cyanobacteria emerge as a major factor behind their grouping in different phylogenetic reconstructions whether it was phylogenetic analysis or phylogenomics approaches. Even though genome alignment-based phylogenomics analysis showed complicated and varied pattern, habitat was a major influencing factor behind the clustering of cyanobacterial species. Cyanobacteria from similar habitats possess similar kind of genomic features such as genome size, GC content (Table 1), genomic repetitiveness and functional profile (Table 2). Among different phylogenomics reconstructions they occupied the same lineages (Fig S1–S5).
Identification of repeats and their role in ecological adaptation and diversification
Microsatellite are widely used for different studies related to strain typing, genetic mapping, population genetics, phylogenetics, and microevolution analysis (Lim et al. 2004). Microsatellite mining has been carried out in bacterial species Escherichia coli (Gur-Arie et al. 2000), Lactobacillus (Basharat and Yasmin 2015), Haemophilus influenzae (Hood et al. 1996) and others (Mrázek et al. 2007) and in fungal species S. cerevisiae (Field and Wills 1998a, b; Kruglyak et al. 2000; Pupko and Graur 1999), Sphaeropsis sapinea (Burgess et al. 2001), Fusarium pseudograminearum (Scott and Chakraborty 2008) and Magnaporthe grisea (Kaye et al. 2003; Lim et al. 2004; Li et al. 2009) etc.
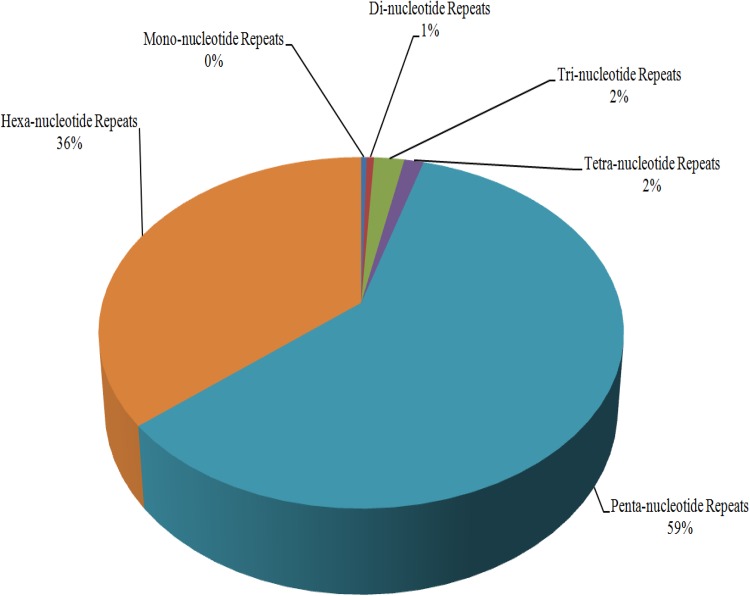
Mono- to hexa-nucleotide repeats were identified for every cyanobacterial species under study. Total 197,750 repeats were identified, ranging from 2269 (Pm_MIT9211) to 12,530 (Te_IMS101) in individual genomes. It was observed that repetitiveness of genome increases with size and smaller genomes tend to have low number of repeats in comparison to larger genome. Furthermore, rather than small-motif repeats (mono- to tetra-nucleotide), penta- and hexa-nucleotide repeats occupied a major proportion of the entire distribution (95%) (Fig. 2). Across all the cyanobacteria, penta-nucleotide repeats occupied 59% of total distribution while hexa-nucleotide repeats represent 36%. Mono- to tetra-nucleotide repeats were present in a very small amount in each genome (Fig. 2). Across all the species, strains of Prochlorococcus marinus (marine pico-cyanobacteria) possesses highest percentage of mono-, tetra- and penta-nucleotide motifs SSRs on an average but when it comes to hexa-nucleotides, they occupy lowest position. Cyanobacteria with large genome size possess larger motif repeats as compared to those with smaller genome size.
Fig. 2.
Distribution of different kind of repeats in cyanobacterial species
Organisms evolve many mechanisms to cope with environmental situations. Presence of repeats is considered as one possible mechanism towards this adaptation (Treangen et al. 2009; Qin et al. 2014). Repeats affect phenotypic variation either through involvement in the gene expression at the transcriptional level (van Ham et al. 1993; Weiser et al. 1989) or by inducing reversible premature ending of translation when present within coding regions (Bayliss et al. 2001; Henderson et al. 1999; Wang et al. 2000). Our analysis has shown that that marine pico-cyanobacteria with small genome size and inhabiting ecological niche where nutrients are available in plenty have repeats of smaller motifs (mono- to tetra-nucleotide). In contrast, cyanobacteria with large genome size and occupying diverse habitats from freshwater to soil or even rock where nutrition resources are scarce and diverse, possess a large repeat numbers and larger motifs (penta- and hexa-nucleotides). Rather than SSRs with larger motifs, shorter motifs SSRs are reported to be less stable, though the phenomenon behind it is still unclear (Mrazek et al. 2007). This could be the consequence of either diverse evolutionary strategies, recombination approaches or both (or none) (Mrazek et al. 2007). One of the major reasons behind the under-representation of SSR of motifs that are not multiples of three nucleotides in coding sequences lies in the fact that recombination may cause frameshifts leading to gene inactivation (Treangen et al. 2009; Field and Wills 1998a, b; Ackermann and Chao 2006; Qi et al. 2015).
Variation of simple sequence repeats within genes should be very significant for regular gene activity. Expansion or contraction of encoding SSR will straightforwardly influence the corresponding gene products and can even lead to phenotypic changes (Li et al. 2004). In short, repeats facilitates invention of novel functions from pre-existing ones through evolutionary tinkering, though they pose problem in chromosome integrity and organization. Owing to these factors, understanding of the role of repeats in any genomes requires in-depth study about their rate of creation and outcomes from a functional and evolutionary perspective (Treangen et al. 2009). Thus, it can be hypothesized that repeats have played an important role in adaptation of cyanobacteria. They help and assist them in survival in diverse ecological conditions while some cyanobacteria have also possibly evolved them for resistance activities.
Conclusion
Genome comparisons suggest that horizontal gene transfer and differential gene loss constitute major evolutionary phenomenon in prokaryotes. The extant of such events makes feasibility of ‘Tree of Life’ reconstruction doubtful as the trees prepared from different genes often state different evolutionary histories. Therefore, alternative approaches to construct tree on the basis of comparisons of complete gene sets or whole genomes can reveal a phylogenetic signal that can support evolutionary history of genomes and suggests the possibility of delineation of undetected clades of organisms. With the increasing rate of sequencing of prokaryotic genomes and the whole genome approaches of reconstructing phylogenetic tree, the concept of universal species tree might be established.
In this study, phylogenetic reconstruction based on the entire genome of different cyanobacteria clearly indicated that clustering of the organisms varied in accordance with their habitats and genome size. Cyanobacteria inhabiting similar habitats tend to have almost similar genome size (and GC-content) and occupied similar lineage during the course of evolution. The study on the evolutionary history of cyanobacterial genomes, even though having several complications, clearly suggested that ecological conditions and the modifications caused within the genomes due to them had great impact on cyanobacterial evolutionary relationships. Habitat also plays an important role in genomic repetitiveness, though, rather than having a direct influence, it majorly affects genome size which eventually is correlated with repeats. Thus, we inferred that maybe large genomes residing in different ecological conditions with scarce and diverse nutritional sources has generated larger repeats (even with larger motifs) which can facilitate development of certain novel function or will play a role in their adaptation. Evolutionary speaking, repeat distribution is a result of selection among different cyanobacterial species and it can be stated that complicated mechanisms are involved in evolution and functioning of repeats. In our study, it was observed that members with different habitats (freshwater, terrestrial or rocks) preferentially accumulate genes for regulation, motility and secondary metabolism in contrast to the genes responsible for informational consequences that are abundant in marine members. A broad metabolic diversity is visible in the large sized cyanobacteria. Furthermore, a large fraction of genes are present in freshwater and terrestrial cyanobacteria, for whom no function is identified till now. The characteristics of gene gain within the genomes can help in understanding the interaction between ecological conditions and genomic evolution. Though, it’s clear that micro-evolutionary processes (functional divergence) couples with macro-evolutionary processes (HGT or genome shrinkage) supports for survival and adaptation of cyanobacterial population to diverse ecological niches.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Authors are grateful to Indian Council of Agricultural Research, New Delhi, India for financial support in the form of “National Agricultural Bioinformatics Grid” (NABG), NAIP. The work is a part of Ph.D. degree program of RP.
Compliance with ethical standards
Conflict of interest
Authors declared no conflict of interest.
References
- Ackermann M, Chao L. DNA sequences shaped by selection for stability. PLoS Genet. 2006;2(2):e22. doi: 10.1371/journal.pgen.0020022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adato O, Ninyo N, Gophna U, Snir S. Detecting horizontal gene transfer between closely related taxa. PLoS Comput Biol. 2015;11(10):e1004408. doi: 10.1371/journal.pcbi.1004408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen EE, Banfield JF. Community genomics in microbial ecology and evolution. Nat Rev Microbiol. 2005;3(6):489–498. doi: 10.1038/nrmicro1157. [DOI] [PubMed] [Google Scholar]
- Anselmetti Y, Duchemin W, Tannier E, Chauve C, Bérard S. Phylogenetic signal from rearrangements in 18 Anopheles species by joint scaffolding extant and ancestral genomes. BMC Genom. 2018;19(Suppl 2):96. doi: 10.1186/s12864-018-4466-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auch AF, von Jan M, Klenk H, Göker M. Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand Genom Sci. 2010;2:117–134. doi: 10.4056/sigs.531120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuma Y, Ota M. An evaluation of minimal cellular functions to sustain a bacterial cell. BMC Syst Biol. 2009;3:111. doi: 10.1186/1752-0509-3-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basharat Z, Yasmin A. Survey of compound microsatellites in multiple Lactobacillus genomes. Can J Microbiol. 2015;61(12):898–902. doi: 10.1139/cjm-2015-0136. [DOI] [PubMed] [Google Scholar]
- Bayliss CD, Field D, Moxon ER. The simple sequence contingency loci of Haemophilus influenzae and Neisseria meningitidis. J Clin Investig. 2001;107:657–666. doi: 10.1172/JCI12557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck C, Knoop H, Axmann IM, Steuer R. The diversity of cyanobacterial metabolism: genome analysis of multiple phototrophic microorganisms. BMC Genom. 2012;13:56. doi: 10.1186/1471-2164-13-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley SD, Parkhill J. Comparative genomic structure of prokaryotes. Annu Rev Genet. 2004;38:771–792. doi: 10.1146/annurev.genet.38.072902.094318. [DOI] [PubMed] [Google Scholar]
- Bossert S, Murray EA, Blaimer BB, Danforth BN. The impact of GC bias on phylogenetic accuracy using targeted enrichment phylogenomic data. Mol Phylogenet Evol. 2017;111:149–157. doi: 10.1016/j.ympev.2017.03.022. [DOI] [PubMed] [Google Scholar]
- Bromberg R, Grishin NV, Otwinowski Z. Phylogeny reconstruction with alignment-free method that corrects for horizontal gene transfer. PLoS Comput Biol. 2016;12(6):e1004985. doi: 10.1371/journal.pcbi.1004985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant D, Moulton V. Neighbor-net: an agglomerative method for the construction of phylogenetic networks. Mol Biol Evol. 2004;21:255–265. doi: 10.1093/molbev/msh018. [DOI] [PubMed] [Google Scholar]
- Bryant WA, Faruqi AA, Pinney JW. Analysis of metabolic evolution in bacteria using whole-genome metabolic models. J Comput Biol. 2013;20:755–764. doi: 10.1089/cmb.2013.0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess T, Wingfield BD, Wingfield MJ. Comparison of genotypic diversity in native and introduced populations of Sphaeropsis sapinea isolated from Pinus radiata. Mycol Res. 2001;105(11):1331–1339. [Google Scholar]
- Caffrey BE, Williams TA, Jiang X, et al. Proteome-wide analysis of functional divergence in bacteria: exploring a host of ecological adaptations. PLoS One. 2012;7(4):e35659. doi: 10.1371/journal.pone.0035659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutierrez S, Kauff F, Gabaldón T. A phylogenomics approach for selecting robust sets of phylogenetic markers. Nucleic Acids Res. 2014;42:e54. doi: 10.1093/nar/gku071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassier-Chauvat C, Chauvat F. Cyanobacteria: wonderful microorganisms for basic and applied research. eLS. 2018 doi: 10.1002/9780470015902.a0027884. [DOI] [Google Scholar]
- Charlebois RL, Doolittle WF. Computing prokaryotic gene ubiquity: rescuing the core from extinction. Genome Res. 2004;14:2469–2477. doi: 10.1101/gr.3024704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevenet F, Brun C, Bañuls AL, Jacq B, Christen R. TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinform. 2006;7:439. doi: 10.1186/1471-2105-7-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colston SM, Fullmer MS, Beka L, Lamy B, Gogarten JP, Graf J. Bioinformatic genome comparisons for taxonomic and phylogenetic assignments using Aeromonas as a test case. MBio. 2014;5(6):e02136. doi: 10.1128/mBio.02136-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comas I, Moya A, Azad RK, Lawrence JG, Gonzalez-Candelas F. The evolutionary origin of xanthomonadales genomes and the nature of the horizontal gene transfer process. Mol Biol Evol. 2006;23(11):2049–2057. doi: 10.1093/molbev/msl075. [DOI] [PubMed] [Google Scholar]
- Cordero OX, Hogeweg P. The impact of long-distance horizontal gene transfer on prokaryotic genome size. Proc Natl Acad Sci USA. 2009;106(51):21748–21753. doi: 10.1073/pnas.0907584106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daubin V, Szöllősi GJ. Horizontal gene transfer and the history of life. Cold Spring Harb Perspect Biol. 2016;8(4):a018036. doi: 10.1101/cshperspect.a018036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcher AL, Phillippy A, Carlton J, Salzberg SL. ‘Fast algorithms for large-scale genome alignment and comparison. Nucleic Acids Res. 2002;30:2478–2483. doi: 10.1093/nar/30.11.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delsuc F, Brinkmann H, Philippe H. Phylogenomics and the reconstruction of the tree of life. Nat Rev Genet. 2005;6(5):361–375. doi: 10.1038/nrg1603. [DOI] [PubMed] [Google Scholar]
- Dutilh BE, Snel B, Ettema TJG, Huynen MA. Signature genes as a phylogenomic tool. Mol Biol Evol. 2008;25:1659–1667. doi: 10.1093/molbev/msn115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta C, Paul S. Microbial lifestyle and genome signatures. Curr Genom. 2012;13(2):153–162. doi: 10.2174/138920212799860698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen JA. Assessing evolutionary relationships among microbes from whole-genome analysis. Curr Opin Microbiol. 2000;3:475–480. doi: 10.1016/s1369-5274(00)00125-9. [DOI] [PubMed] [Google Scholar]
- Field D, Wills C. Abundant microsatellite polymorphism in Saccharomyces cerevisiae, and the different distributions of microsatellites in eight prokaryotes and S. cerevisiae, result from strong mutation pressures and a variety of selective forces. Proc Natl Acad Sci USA. 1998;95(4):1647–1652. doi: 10.1073/pnas.95.4.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field D, Wills C. Abundant microsatellite polymorphism in Saccharomyces cerevisiae and the different distributions of microsatellites in 8 prokaryotes and S. cerevisiae, result from strong mutation pressures and a variety of selective forces. P Natl Acad Sci USA. 1998;95:1647–1652. doi: 10.1073/pnas.95.4.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X, Huang W, Xu D, Zhang H. GeneContent: software for whole-genome phylogenetic analysis. Bioinformatics. 2005;21:1713–1714. doi: 10.1093/bioinformatics/bti208. [DOI] [PubMed] [Google Scholar]
- Gur-Arie R, Cohen CJ, Eitan Y, Shelef L, Hallerman EM, Kashi Y. Simple sequence repeats in Escherichia coli: abundance, distribution, composition, and polymorphism. Genome Res. 2000;10(1):62–71. [PMC free article] [PubMed] [Google Scholar]
- Hao B, Qi J. Prokaryote phylogeny without sequence alignment: from avoidance signature to composition distance. J Bioinform Comput Biol. 2004;2:1–19. doi: 10.1142/s0219720004000442. [DOI] [PubMed] [Google Scholar]
- Henderson IR, Owen P, Nataro JP. Molecular switches—the ON and OFF of bacterial phase variation. Mol Microbiol. 1999;33(9):19–32. doi: 10.1046/j.1365-2958.1999.01555.x. [DOI] [PubMed] [Google Scholar]
- Henz SR, Huson DH, Auch AF, Nieselt-Struwe K, Schuster SC. Whole-genome prokaryotic phylogeny. Bioinformatics. 2005;21:2329–2335. doi: 10.1093/bioinformatics/bth324. [DOI] [PubMed] [Google Scholar]
- Hillis DM, Dixon MT. Ribosomal DNA: molecular evolution and phylogenetic inference. Q Rev Biol. 1991;66:411–453. doi: 10.1086/417338. [DOI] [PubMed] [Google Scholar]
- Hood DW, Deadman ME, Jennings MP, Bisercic M, Fleischmann RD, Venter JC, et al. DNA repeats identify novel virulence genes in Haemophilus influenzae. Proc Natl Acad Sci USA. 1996;93:11121–11125. doi: 10.1073/pnas.93.20.11121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH. SplitsTree: analyzing and visualizing evolutionary data. Bioinformatics. 1998;14:68–73. doi: 10.1093/bioinformatics/14.1.68. [DOI] [PubMed] [Google Scholar]
- Jain R, Rivera MC, Lake JA. Horizontal gene transfer among genomes: the complexity hypothesis. Proc Natl Acad Sci USA. 1999;96:3801–3806. doi: 10.1073/pnas.96.7.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang LW, Lin KL, Lu CL. OGtree: a tool for creating genome trees of prokaryotes based on overlapping genes. Nucleic Acids Res. 2008;36:W475–W480. doi: 10.1093/nar/gkn240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai M, Nakao K, Uchiyama I, Kobayashi I. How genomes rearrange: Genome comparison within bacteria Neisseria suggests roles for mobile elements in formation of complex genome polymorphisms. Gene. 2006;383:52–63. doi: 10.1016/j.gene.2006.07.013. [DOI] [PubMed] [Google Scholar]
- Kaye C, Milazzo J, Rozenfeld S, Lebrun MH, Tharreau D. The development of simple sequence repeat markers for Magnaporthe grisea and their integration into an established genetic linkage map. Fungal Genet Biol. 2003;40:207–214. doi: 10.1016/j.fgb.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Khiripet N (2005) Bacterial whole genome phylogeny using proteome comparison and optimal reversal distance. In: Computational systems bioinformatics conference, 2005, workshops and poster abstracts, IEEE, pp 63–64
- Kim M, Oh HS, Park SC, Chun J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol. 2014;64(Pt 2):346–351. doi: 10.1099/ijs.0.059774-0. [DOI] [PubMed] [Google Scholar]
- Klenk HP, Göker M. En route to a genome-based classification of Archaea and bacteria? Syst Appl Microbiol. 2010;33:175–182. doi: 10.1016/j.syapm.2010.03.003. [DOI] [PubMed] [Google Scholar]
- Koehorst JJ, Saccenti E, Schaap PJ, Martins Dos Santos VAP, Suarez-Diez M. Protein domain architectures provide a fast, efficient and scalable alternative to sequence-based methods for comparative functional genomics. Version 3. F1000Res. 2016;5:1987. doi: 10.12688/f1000research.9416.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koksharova O, Wolk C. Genetic tools for cyanobacteria. Appl Microbiol Biotechnol. 2002;58(2):123–137. doi: 10.1007/s00253-001-0864-9. [DOI] [PubMed] [Google Scholar]
- Konstantinidis KT, Tiedje JM. Trends between gene content and genome size in prokaryotic species with larger genomes. Proc Natl Acad Sci USA. 2004;101(9):3160–3165. doi: 10.1073/pnas.0308653100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Makarova KS, Aravind L. Horizontal gene transfer in prokaryotes: quantification and classification. Annu Rev Microbiol. 2001;55:709–742. doi: 10.1146/annurev.micro.55.1.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruglyak S, Durret R, Shug MC, Aquadro CF. Distribution and abundance of microsatellites in the yeast genome can be explained by a balance slippage between events and point mutations. Mol Biol Evol. 2000;17:1210–1219. doi: 10.1093/oxfordjournals.molbev.a026404. [DOI] [PubMed] [Google Scholar]
- Land M, Hauser L, Jun SR, Nookaew I, Leuze MR, Ahn TH, Karpinets T, Lund O, Kora G, Wassenaar T, Poudel S, Ussery DW. Insights from 20 years of bacterial genome sequencing. Funct Integr Genom. 2015;15(2):141–161. doi: 10.1007/s10142-015-0433-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang JM, Darling AE, Eisen JA. phylogeny of bacterial and archaeal genomes using conserved genes: supertrees and supermatrices. PLoS One. 2013;8:e62510. doi: 10.1371/journal.pone.0062510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson J, Nylander JA, Bergman B. Genome fluctuations in cyanobacteria reflect evolutionary, developmental and adaptive traits. BMC Evol Biol. 2011;11:187. doi: 10.1186/1471-2148-11-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence JG, Ochman H. Amelioration of bacterial genomes: rates of change and exchange. J Mol Evol. 1997;44:383–397. doi: 10.1007/pl00006158. [DOI] [PubMed] [Google Scholar]
- Li W, Fang W, Ling L, Wang J, Xuan Z, Chen R. Phylogeny based on whole genome as inferred from complete information set analysis. J Biol Phys. 2002;28:439–447. doi: 10.1023/A:1020316706928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Korol AB, Fahima T, Nevo E. Microsatellites within genes: structure, function, and evolution. Mol Biol Evol. 2004;21(6):991–1007. doi: 10.1093/molbev/msh073. [DOI] [PubMed] [Google Scholar]
- Li C, Liu L, Yang J, Li J, Su Y, Zhang Y, Wang Y, Zhu Y. Genome wide analysis of microsatellite sequence in seven filamentous fungi. Interdiscip Sci Comput Life Sci. 2009;1:141–150. doi: 10.1007/s12539-009-0014-5. [DOI] [PubMed] [Google Scholar]
- Lim S, Notley-McRobb L, Lim M, Carter DA. A comparison of the nature and abundance of microsatellites in 14 fungal genomes. Fungal Genet Biol. 2004;41:1025–1036. doi: 10.1016/j.fgb.2004.08.004. [DOI] [PubMed] [Google Scholar]
- Losos JB, Arnold SJ, Bejerano G, Brodie ED, III, Hibbett D, Hoekstra HE, Mindell DP, Monteiro A, Moritz C, Orr HA, Petrov DA, Renner SS, Ricklefs RE, Soltis PS, Turner TL. Evolutionary biology for the 21st century. PLoS Biol. 2013;11:e1001466. doi: 10.1371/journal.pbio.1001466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig W, Schleifer KH. Phylogeny of bacteria beyond the 16S rRNA standard. ASM News. 1999;65:752–757. [Google Scholar]
- Makarova KS, Aravind L, Galperin MY, Grishin NV, Tatusov RL, Wolf YI, Koonin EV. Comparative genomics of the Archaea (Euryarchaeota): evolution of conserved protein families, the stable core, and the variable shell. Genome Res. 1999;9:608–628. [PubMed] [Google Scholar]
- Markowitz VM, Chen IM, Palaniappan K, et al. IMG: the integrated microbial genomes database and comparative analysis system. Nucleic Acids Res. 2012;40(Database issue):D115–D122. doi: 10.1093/nar/gkr1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina M. Genomes, phylogeny, and evolutionary systems biology. Proc Natl Acad Sci USA. 2005;102:6630–6635. doi: 10.1073/pnas.0501984102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier-Kolthoff JP, Auch AF, Klenk H, Göker M. Highly parallelized inference of large genome-based phylogenies. Concurr Comput Pract Exp. 2014;26:1715–1729. [Google Scholar]
- Mira A, Ochman H, Moran NA. Deletional bias and the evolution of bacterial genomes. Trends Genet. 2001;17(10):589–596. doi: 10.1016/s0168-9525(01)02447-7. [DOI] [PubMed] [Google Scholar]
- Mirkin BG, Fenner TI, Galperin MY, Koonin EV (2003) Algorithms for computing parsimonious evolutionary scenarios for genome evolution, the last universal common ancestor and dominance of horizontal gene transfer in the evolution of prokaryotes. BMC Evol Biol 3(2) [DOI] [PMC free article] [PubMed]
- Moreno-Hagelsieb G, Wang Z, Walsh S, ElSherbiny A. Phylogenomic clustering for selecting non-redundant genomes for comparative genomics. Bioinformatics. 2013;29:947–949. doi: 10.1093/bioinformatics/btt064. [DOI] [PubMed] [Google Scholar]
- Mrazek J, Guo X, Shah A. Simple sequence repeats in prokaryotic genomes. Proc Natl Acad Sci USA. 2007;104:8472–8477. doi: 10.1073/pnas.0702412104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrázek J, Guo X, Shah A. Simple sequence repeats in prokaryotic genomes. PNAS. 2007;104(20):8472–8477. doi: 10.1073/pnas.0702412104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudunuri SB, Nagarajaram HA. IMEx: imperfect microsatellite extractor. Bioinformatics. 2007;23(10):1181–1187. doi: 10.1093/bioinformatics/btm097. [DOI] [PubMed] [Google Scholar]
- Mushegian AR, Koonin EV. A minimal gene set for cellular life derived by comparison of complete bacterial genomes. Proc Natl Acad Sci USA. 1996;93(19):10268–10273. doi: 10.1073/pnas.93.19.10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabachi A, Yamashita A, Toh H, et al. The 160-kilobase genome of the bacterial endosymbiont Carsonella. Science. 2006;314:267. doi: 10.1126/science.1134196. [DOI] [PubMed] [Google Scholar]
- Nalbantoglu OU (2011) Computational genomic signatures and metagenomics. Electrical engineering. Theses and dissertations, p 19
- Nelson KE, Clayton RA, Gill SR, Gwinn ML, Dodson RJ, Haft DH, Hickey EK, Peterson JD, Nelson WC, Ketchum KA, McDonald L, Utterback TR, Malek JA, Linher KD, Garrett MM, Stewart AM, Cotton MD, Pratt MS, Phillips CA, Richardson D, Heidelberg J, Sutton GG, Fleischmann RD, Eisen JA, White O, Salzberg SL, Smith HO, Venter JC, Fraser CM. Evidence for lateral gene transfer between Archaea and bacteria from genome sequence of Thermotoga maritima. Nature. 1999;399:323–329. doi: 10.1038/20601. [DOI] [PubMed] [Google Scholar]
- Nilsson AI, Koskiniemi S, Eriksson S, et al. Bacterial genome size reduction by experimental evolution. Proc Natl Acad Sci USA. 2005;102(34):12112–12116. doi: 10.1073/pnas.0503654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira PH, Touchon M, Cury J, Rocha EPC. The chromosomal organization of horizontal gene transfer in bacteria. Nat Commun. 2017;8(1):841. doi: 10.1038/s41467-017-00808-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabha R, Singh DP, Somvanshi P, Rai A. Functional profiling of cyanobacterial genomes and its role in ecological adaptations. Genom Data. 2016;9:89–94. doi: 10.1016/j.gdata.2016.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanna AN, Mehra S. Comparative phylogenomics of pathogenic and non-pathogenic Mycobacterium. PLoS One. 2013;8:e71248. doi: 10.1371/journal.pone.0071248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pupko T, Graur D. Evolution of microsatellites in the yeast Saccharomyces cerevisiae: role of length and number of repeated units. J Mol Evol. 1999;48:313–316. doi: 10.1007/pl00006474. [DOI] [PubMed] [Google Scholar]
- Qi J, Luo H, Hao B-L. CVTree: a phylogenetic tree reconstruction tool based on whole genomes. Nucleic Acids Res. 2004;32:W45–W47. doi: 10.1093/nar/gkh362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi W-H, Jiang X-M, Du L-M, Xiao G-S, Hu T-Z, Yue B-S, et al. Genome-wide survey and analysis of microsatellite sequences in bovid species. PLoS One. 2015;10(7):e0133667. doi: 10.1371/journal.pone.0133667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiang, Li, Zhao, Xu, Bailin, Hao Composition vector approach to whole-genome-based prokaryotic phylogeny: success and foundations. J Biotechnol. 2010;149:115–119. doi: 10.1016/j.jbiotec.2009.12.015. [DOI] [PubMed] [Google Scholar]
- Qin L, Zhang Z, Zhao X, Wu X, Chen Y, Tan Z, Li S. Survey and analysis of simple sequence repeats (SSRs) present in the genomes of plant viroids. FEBS Open Bio. 2014;4:185–189. doi: 10.1016/j.fob.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajendhran J, Gunasekaran P. Microbial phylogeny and diversity: small subunit ribosomal RNA sequence analysis and beyond. Microbiol Res. 2011;166(2):99–110. doi: 10.1016/j.micres.2010.02.003. [DOI] [PubMed] [Google Scholar]
- Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng JF, Darling A, Malfatti S, Swan BK, Gies EA, Dodsworth JA, Hedlund BP, Tsiamis G, Sievert SM, Liu WT, Eisen JA, Hallam SJ, Kyrpides NC, Stepanauskas R, Rubin EM, Hugenholtz P, Woyke T. Insights into the phylogeny and coding potential of microbial dark matter. Nature. 2013;499:431–437. doi: 10.1038/nature12352. [DOI] [PubMed] [Google Scholar]
- Rudi K, Sekelja M. High or low correlation between co-occuring gene clusters and 16S rRNA gene phylogeny. FEMS Microbiol Lett. 2013;339:23–29. doi: 10.1111/1574-6968.12042. [DOI] [PubMed] [Google Scholar]
- Sawa G, Dicks J, Roberts IN. Current approaches to whole genome phylogenetic analysis. Brief Bioinform. 2003;4:63–74. doi: 10.1093/bib/4.1.63. [DOI] [PubMed] [Google Scholar]
- Scott JB, Chakraborty S. Identification of 11 polymorphic simple sequence repeat loci in the phytopathogenic fungus Fusarium pseudograminearum as a tool for genetic studies. Mol Ecol Resour. 2008;8(3):628–630. doi: 10.1111/j.1471-8286.2007.02025.x. [DOI] [PubMed] [Google Scholar]
- Sicheritz-Pontén T, Anderssona SGE. A phylogenomic approach to microbial evolution. Nucleic Acids Res. 2001;29:545–552. doi: 10.1093/nar/29.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleator RD. A beginner’s guide to phylogenetics. Microb Ecol. 2013;66:1–4. doi: 10.1007/s00248-013-0236-x. [DOI] [PubMed] [Google Scholar]
- Snel B, Bork P, Huynen MA. Genome phylogeny based on gene content. Nat Genet. 1999;21(1):108–110. doi: 10.1038/5052. [DOI] [PubMed] [Google Scholar]
- Snel B, Bork P, Huynen MA. The identification of functional modules from the genomic association of genes. Proc Natl Acad Sci USA. 2002;99(9):5890–5895. doi: 10.1073/pnas.092632599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snel B, Huynen MA, Dutilh BE. Genome trees and the nature of genome evolution. Ann Rev Microbiol. 2005;59:191–209. doi: 10.1146/annurev.micro.59.030804.121233. [DOI] [PubMed] [Google Scholar]
- Takahashi M, Kryukov K, Saitou N. Estimation of bacterial species phylogeny through oligonucleotide frequency distances. Genomics. 2009;93:525–533. doi: 10.1016/j.ygeno.2009.01.009. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Fedorova ND, Jackson JD, et al. The COG database: an updated version includes eukaryotes. BMC Bioinform. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekaia F, Lazcano A, Dujon B. The genomic tree as revealed from whole proteome comparisons. Genome Res. 1999;9:550–557. [PMC free article] [PubMed] [Google Scholar]
- Treangen TJ, Abraham AL, Touchon M, Rocha EP. Genesis, effects and fates of repeats in prokaryotic genomes. FEMS Microbiol Rev. 2009;33(3):539–571. doi: 10.1111/j.1574-6976.2009.00169.x. [DOI] [PubMed] [Google Scholar]
- van Ham SM, van Alphen L, Mooi FR, van Putten JP. Phase variation of H. influenzae fimbriae: transcriptional control of two divergent genes through a variable combined promoter region. Cell. 1993;73:1187–1196. doi: 10.1016/0092-8674(93)90647-9. [DOI] [PubMed] [Google Scholar]
- Větrovský T, Baldrian P. The Variability of the 16S rRNA gene in bacterial genomes and its consequences for bacterial community analyses. PLoS One. 2013;8:e57923. doi: 10.1371/journal.pone.0057923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishnoi A, Roy R, Prasad HK, Bhattacharya A. Anchor-based whole genome phylogeny (ABWGP): a tool for inferring evolutionary relationship among closely related microorganims. PLoS One. 2010;5:e14159. doi: 10.1371/journal.pone.0014159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Jiang T, Gusfield D. A more efficient approximation scheme for tree alignment. SIAM J Comput. 2000;30(1):283–299. [Google Scholar]
- Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiser JN, Love JM, Moxon ER. The molecular mechanism of phase variation of H. influenzae lipopolysaccharide. Cell. 1989;59:657–665. doi: 10.1016/0092-8674(89)90011-1. [DOI] [PubMed] [Google Scholar]
- Wernegreen JJ, Ochman H, Jones IB, Moran NA. ‘The decoupling of genome size and sequence divergence in a symbiotic bacterium. J Bacteriol. 2000;182:3867–3869. doi: 10.1128/jb.182.13.3867-3869.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. The universal ancestor. Proc Natl Acad Sci USA. 1998;95:6854–6859. doi: 10.1073/pnas.95.12.6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C, Fox G. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc Natl Acad Sci USA. 1977;74:5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf YI, Rogozin IB, Grishin NV, Tatusov RL, Koonin EV. Genome trees constructed using five different approaches suggest new major bacterial clades. BMC Evol Biol. 2001;1:8. doi: 10.1186/1471-2148-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf YI, Rogozin IB, Grishin NV, Koonin EV. Genome trees and the tree of life. Trends Genet. 2002;18:472–479. doi: 10.1016/s0168-9525(02)02744-0. [DOI] [PubMed] [Google Scholar]
- Wu D, Jospin G, Eisen JA. Systematic identification of gene families for use as “markers” for phylogenetic and phylogeny-driven ecological studies of bacteria and archaea and their major subgroups. PLoS One. 2013;8:e77033. doi: 10.1371/journal.pone.0077033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Hao B. CVTree update: a newly designed phylogenetic study platform using composition vectors and whole genomes. Nucleic Acids Res. 2009;37:W174–W178. doi: 10.1093/nar/gkp278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabuki A, Toyofuku T, Takishita K. Lateral transfer of eukaryotic ribosomal RNA genes: an emerging concern for molecular ecology of microbial eukaryotes. ISME J. 2014;8(7):1544–1547. doi: 10.1038/ismej.2013.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Bourne PE. The Evolutionary History of Protein Domains Viewed by Species Phylogeny. PLoS ONE. 2009;4(12):e8378. doi: 10.1371/journal.pone.0008378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YC, Lin K. Phylogeny inference of closely related bacterial genomes: combining the features of both overlapping genes and collinear genomic regions. Evol Bioinform Online. 2015;11(Suppl 2):1–9. doi: 10.4137/EBO.S33491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Wu J, Yang J, et al. PGAP: Pan-genomes analysis pipeline. Bioinformatics. 2012;28:416–418. doi: 10.1093/bioinformatics/btr655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhaxybayeva O, Gogarten JP, Charlebois RL, et al. Phylogenetic analyses of cyanobacterial genomes: quantification of horizontal gene transfer events. Genome Res. 2006;16:1099–1108. doi: 10.1101/gr.5322306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Lin Y, Feng B, Zhao J, Tang J. Phylogeny analysis from gene-order data with massive duplications. BMC Genom. 2017;18(Suppl 7):760. doi: 10.1186/s12864-017-4129-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuckerkandl E, Pauling L. Molecules as documents of evolutionary history. J Theor Biol. 1965;8:357–366. doi: 10.1016/0022-5193(65)90083-4. [DOI] [PubMed] [Google Scholar]
- Zuo G, Xu Z, Yu H, Hao B, Genomics Jackknife and bootstrap tests of the composition vector trees. Proteom Bioinform. 2010;8(4):262–267. doi: 10.1016/S1672-0229(10)60028-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.