Abstract
Spinal Muscular Atrophy (SMA) is caused by homozygous mutations in the human survival motor neuron 1 (SMN1) gene. SMN protein has a well-characterized role in the biogenesis of small nuclear ribonucleoproteins (snRNPs), core components of the spliceosome. SMN is part of an oligomeric complex with core binding partners, collectively called Gemins. Biochemical and cell biological studies demonstrate that certain Gemins are required for proper snRNP assembly and transport. However, the precise functions of most Gemins are unknown. To gain a deeper understanding of the SMN complex in the context of metazoan evolution, we investigated its composition in Drosophila melanogaster. Using transgenic flies that exclusively express Flag-tagged SMN from its native promoter, we previously found that Gemin2, Gemin3, Gemin5, and all nine classical Sm proteins, including Lsm10 and Lsm11, co-purify with SMN. Here, we show that CG2941 is also highly enriched in the pulldown. Reciprocal co-immunoprecipitation reveals that epitope-tagged CG2941 interacts with endogenous SMN in Schneider2 cells. Bioinformatic comparisons show that CG2941 shares sequence and structural similarity with metazoan Gemin4. Additional analysis shows that three other genes (CG14164, CG31950 and CG2371) are not orthologous to Gemins 6-7-8, respectively, as previously suggested. In D.melanogaster, CG2941 is located within an evolutionarily recent genomic triplication with two other nearly identical paralogous genes (CG32783 and CG32786). RNAi-mediated knockdown of CG2941 and its two close paralogs reveals that Gemin4 is essential for organismal viability.
Keywords: locomotor function, ncRNA, proteomics, RNP assembly, SMN, survival motor neuron, snRNA, snRNP, Spinal Muscular Atrophy, SMA
Spinal muscular atrophy (SMA) is a pediatric neuromuscular disorder caused by mutation or loss of the human survival motor neuron 1 (SMN1) gene (Lefebvre et al. 1995). Approximately 95% of SMA patients have homozygous deletions in SMN1, and the remaining ∼5% are hemizygous for the deletion over a missense mutation in SMN1 (Burghes and Beattie 2009). Despite the great progress that has been made therapeutically, the etiology of SMA remains poorly understood. SMN’s best understood function is in the biogenesis of spliceosomal uridine-rich small nuclear ribonucleoproteins (UsnRNPs), a fundamental process important for all eukaryotic cells (Battle et al. 2006a; Matera et al. 2007; Coady and Lorson 2011; Fischer et al. 2011; Matera and Wang 2014). Additional tissue-specific functions for SMN have also been reported (for reviews, see Fallini et al. 2012; Hamilton and Gillingwater 2013; Shababi et al. 2014; Nash et al. 2016; Chaytow et al. 2018).
To date, no definitive link has been established between a specific function of SMN and SMA pathogenesis. Moving forward, the key question in the field is to identify which of the many potential activities of SMN lie at the root of the neuromuscular dysfunction. Given that SMA is a spectrum disorder with a broad range of phenotypes (Tizzano and Finkel 2017; Groen et al. 2018), it seems likely that the most severe forms of the disease would involve loss of more than one SMN-dependent pathway. Thus, understanding the molecular etiology of the disease is not only important for the basic biology, but also for targeting and refining therapeutic strategies (Groen et al. 2018; Sumner and Crawford 2018).
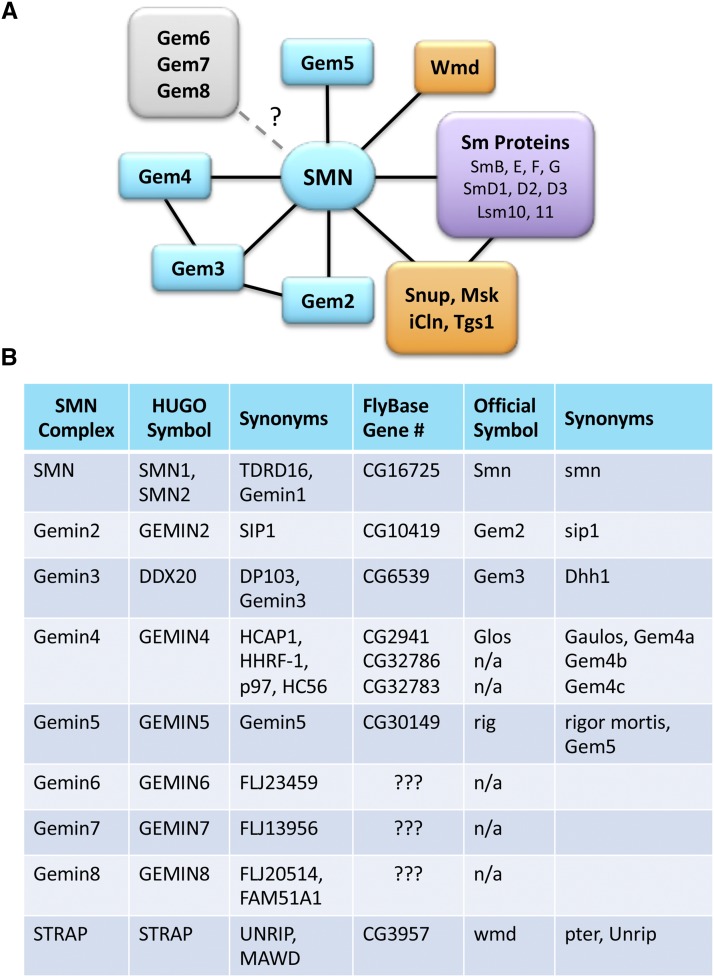
As outlined in Figure 1, SMN works in close partnership with a number of proteins, collectively called Gemins (Paushkin et al. 2002; Shpargel and Matera 2005; Otter et al. 2007; Borg and Cauchi 2013). The N-terminal domain of SMN interacts with a protein called Gemin2 (Liu et al. 1997; Wang and Dreyfuss 2001), whereas the C-terminal region contains a YG-zipper motif (Martin et al. 2012) that drives SMN self-oligomerization, as well as binding to Gemin3 (Lorson et al. 1998; Pellizzoni et al. 1999; Praveen et al. 2014; Gupta et al. 2015). Gemin2 heterodimerizes with SMN and is the only member of the complex that is conserved from budding yeast to humans (Fischer et al. 1997; Kroiss et al. 2008). When provided with in vitro transcribed Sm-class snRNAs, purified recombinant SMN and Gemin2 are sufficient for Sm core assembly activity (Kroiss et al. 2008). In vivo, Gemin5 is thought to play an important role in snRNA substrate recognition (Battle et al. 2006b; Bradrick and Gromeier 2009; Lau et al. 2009; Yong et al. 2010; Jin et al. 2016), but it has also been independently identified as a cellular signaling factor (Gates et al. 2004; Kim et al. 2007) and a translation factor (reviewed in Piñeiro et al. 2015).
Figure 1.
Protein interaction network of the metazoan SMN complex. (A) In addition to the core Gemin family members (shown in teal), SMN is known to form complexes with various Sm protein substrates (purple) and RNP biogenesis factors (orange). Snup, Msk, iCln, Tgs1 and Wmd, correspond to SPN1/Snurportin1, Moleskin/Importin7, CLNS1A/Chloride channel nucleotide-sensitive 1A, TGS1/Trimethylguanosine synthase 1, and STRAP/Unrip, respectively. The presence of three of the Gemins (Gem6, Gem7 and Gem8) within dipteran genomes is uncertain (?). (B) Reference table comparing the names, symbols and synonyms of SMN complex members in humans and flies.
Gemin3/DDX20/Dp103 is a DEAD-box helicase (Charroux et al. 1999; Grundhoff et al. 1999) that interacts with the SMN•Gemin2 hetero-oligomer and is reported to play roles in transcriptional repression (Yan et al. 2003) and microRNA activity (Mourelatos et al. 2002) in addition to its role in Sm-core assembly (Shpargel and Matera 2005), reviewed in (Curmi and Cauchi 2018). Gemin4 is tethered to the SMN complex via direct binding to Gemin3 (Charroux et al. 2000; Meister et al. 2000). Gemin4 has been implicated in nuclear receptor binding (Di et al. 2003; Yang et al. 2015), microRNA biology (Hutvágner and Zamore 2002; Meister et al. 2005) and in nuclear import of the SMN complex (Narayanan et al. 2004; Meier et al. 2018). In human cell lysates, SMN and Gemins2-4 are thought to be essential for proper assembly of the Sm core (Shpargel and Matera 2005). Consistent with this notion, complete loss-of-function mutations in Smn, Gemin2, Gemin3 and Gemin4 in mice result in embryonic lethality (Schrank et al. 1997; Jablonka et al. 2002; Mouillet et al. 2008; Meier et al. 2018). Partial loss-of-function mutations in genes encoding the Gemins have not been reported. Functions of the other members of the SMN complex are largely unknown.
Gemins6-8 (Gem6-7-8) and STRAP (serine-threonine kinase receptor associated protein, a.k.a. UNRIP, UNR-interacting protein; or Wmd/CG3957, wing morphogenesis defect) are peripheral members of the SMN complex and may thus serve in a regulatory capacity. In support of this idea, STRAP/UNRIP is found in a separate complex with a cap-independent translation factor called UNR (Hunt et al. 1999)and has been shown to modulate its function (Carissimi et al. 2005; Grimmler et al. 2005). STRAP binds to the SMN complex via an interaction with Gemin7 (Otter et al. 2007). In Drosophila, mutations in the STRAP ortholog, Wmd/CG3957, cause defects in development of the adult wing (Khokhar et al. 2008), suggesting that this protein may not be essential for basal assembly of spliceosomal snRNPs. Gem6-7-8 forms a subcomplex that is tethered to SMN via interaction with Gemin8 (Carissimi et al. 2006; Otter et al. 2007). Gemin6 and Gemin7 are Sm-like proteins that heterodimerize with one another, but the roles played by these factors in snRNP biogenesis are unknown. Mutations in Gem6-7-8 have yet to be described.
In Drosophila, the SMN complex (as defined by proteins that stably co-purify with SMN) was originally thought to contain only Gemin2 and Gemin3 (Kroiss et al. 2008). Bioinformatic analysis suggested that the gene rigor mortis (Rig) encodes a potential metazoan Gemin5 ortholog, but Gemin5/Rig protein failed to co-purify with SMN in Schneider2 (S2) cells (Kroiss et al. 2008). Notably, transgenic expression of tagged Gemin2 and Gemin5/Rig constructs showed that these two proteins colocalize with endogenous SMN in cytoplasmic structures called U bodies (Cauchi et al. 2010). Consistent with the cytological studies, we found that Gemin5/Rig co-purified with SMN in Drosophila embryos that were engineered to exclusively express Flag-tagged SMN (Gray et al. 2018). However, Drosophila Gem6-7-8 proteins were neither identified bioinformatically nor were they shown to biochemically coprecipitate with SMN in S2 cells (Kroiss et al. 2008).
A recent study presented evidence suggesting the “full conservation” of the SMN complex in fruit flies (Lanfranco et al. 2017), and arguing that Gemin4 and Gemins6-7-8 are present in Diptera. In this report, we re-investigate this issue, showing that among the four novel factors identified by Lanfranco et al. (2017), only Gaulos (CG2941) is orthologous to a metazoan SMN complex protein (Gemin4). Using comparative genomic analysis, we conclusively demonstrate that Hezron (CG14164), Sabbat (CG31950) and Valette (CG2371) are not orthologous to metazoan Gemin6, Gemin7 and Gemin8, respectively. The genes encoding these three Drosophila proteins are actually orthologous to three distinct and highly conserved metazoan genes. The implications of these findings are discussed. Furthermore, we purified SMN complexes from Drosophila embryos and found that endogenous Gaulos co-precipitates with SMN but Valette, Sabbat and Hezron do not. Phylogenetic analysis demonstrates that Gaulos/CG2941 is actually part of a genomic triplication involving two other nearly identical gene copies, CG32783 and CG32786. We interrogate the function of these three factors in vivo using RNA interference analysis, revealing that the function of these redundant genes is essential in flies.
Materials and Methods
Fly stocks
RNAi lines were obtained from the Bloomington TRIP collection. The identifying numbers listed on the chart are stock numbers. Each of the RNAi constructs is expressed from one of five VALIUM vectors and requires Gal4 for expression. All stocks were cultured on molasses and agar at room temperature (25°).
Antibodies and Western blotting
Embryonic lysates were prepared by crushing the animals in lysis buffer (50mM Tris-HCl, pH 7.5, 150 mM NaCl, 1mM EDTA, 1% NP-40) with 1X protease inhibitor cocktail (Invitrogen) and clearing the lysate by centrifugation at 13,000 RPM for 10 min at 4°. S2 cell lysates were prepared by suspending cells in lysis buffer (50mM Tris-HCl, pH 7.5, 150 mM NaCl, 1mM EDTA, 1% NP-40) with 10% glycerol and 1x protease inhibitor cocktail (Invitrogen) and disrupting cell membranes by pulling the suspension through a 25 gauge needle (Becton Dickinson). The lysate was then cleared by centrifugation at 13,000 RPM for 10 min at 4°. Cell fractionation was performed using a standard protocol (West et al. 2008). In brief, following centrifugation, cytoplasmic extracts were taken from the top 0.2mL and the nuclear pellet was resuspended in 0.2mL RIPA buffer. Western blotting on lysates was performed using standard protocols. Rabbit anti-dSMN serum was generated by injecting rabbits with purified, full-length dSMN protein (Pacific Immunology Corp, CA), and was subsequently affinity purified. For Western blotting, dilutions of 1 in 2,500 for the affinity purified anti-dSMN, 1 in 10,000 for monoclonal anti-FLAG (Sigma) were used.
Immunoprecipitation
Lysates were incubated with Anti-FLAG antibody crosslinked to agarose beads (EZview Red Anti-FLAG M2 affinity gel, Sigma) for 2h-ON at 4C with rotation. The beads were washed with RIPA lysis buffer or three times and boiled in SDS gel-loading buffer. Eluted proteins were run on an SDS-PAGE for western blotting.
Drosophila embryo protein lysate and mass spectrometry
0-12h Drosophila embryos were collected from Oregon-R control and Flag-SMN flies, dechorionated, flash frozen, and stored at -80°. Embryos (approx. 1gr) were then homogenized on ice with a Potter tissue grinder in 5 mL of lysis buffer containing 100mM potassium acetate, 30mM HEPES-KOH at pH 7.4, 2mM magnesium acetate, 5mM dithiothreitol (DTT) and protease inhibitor cocktail. Lysates were centrifuged twice at 20,000 rpm for 20min at 4° and dialyzed for 5h at 4° in Buffer D (HEPES 20mM pH 7.9, 100mM KCl, 2.5 mM MgCl2, 20% glycerol, 0.5 mM DTT, PMSF 0.2 mM). Lysates were clarified again by centrifugation at 20000 rpm for 20 min at 4C. Lysates were flash frozen using liquid nitrogen and stored at -80C before use. Lysates were then thawed on ice, centrifuged at 20000 rpm for 20 min at 4C and incubated with rotation with 100 uL of EZview Red Anti-FLAG M2 affinity gel (Sigma) for 2h at 4C. Beads were washed a total of six times using buffer with KCl concentrations ranging from 100mM to 250mM with rotation for 1 min at 4° in between each wash. Finally, Flag proteins were eluted 3 consecutive times with one bed volume of elution buffer (Tris 20mM pH 8, 100 mM KCl, 10% glycerol, 0.5 mM DTT, PMSF 0.2 mM) containing 250ug/mL 3XFLAG peptide (sigma). The eluates were used for mass spectrometric analysis on an Orbitrap Velos instrument, fitted with a Thermo Easy-spray 50cm column.
Viability and Larval Locomotion
Males containing RNAi constructs were crossed to virgin females containing one of the Gal4 constructs balanced by CAG (Tub-Gal4) or TM6BGFP (Da-Gal4). Embryos were collected on molasses agar plates and sorted into vials using lack of GFP fluorescence. A maximum of 50 larvae were sorted into each vial. Viability was assessed based on the number of pupated or eclosed individuals compared to the starting number of larvae in each vial.
To assess larval locomotion, five wandering third instar larvae were set on a large molasses agar plate and placed in a recording chamber. Their crawling movements were recorded for at least 1 min on a digital camera (smartphone) at minimum zoom. Four recording were take for each set of larvae; at least 30 larvae total were recorded for each cross. The videos were transferred to a PC and converted to AVI files using ffmpeg (https://www.ffmpeg.org/). The videos were then opened and converted to binary frames using Fiji/ImageJ. The wrMTrck plugin (http://www.phage.dk/plugins/wrmtrck.html) for ImageJ was used to assess the average speed of each larvae normalized to their body size (body lengths/second or BLPS).
Northern blotting and RT-PCR
Early third instar larvae (73-77 hr post egg-laying) were homogenized in TRIzol reagent (Invitrogen) and RNA was isolated according to the manufacturer’s protocol with the following modifications: a second chloroform extraction was performed, and RNA was precipitated with 0.1 volumes of sodium acetate and 2.5 volumes of ethanol rather than isopropanol. For Northern blotting, 2500 ng of total RNA was separated on Novex 10% TBE-Urea gels (Invitrogen). RNA was transferred to GeneScreen Plus Hybridization Transfer Membrane (PerkinElmer). Blots were dried, UV cross-linked, and pre-hybridized with Rapid-hyb Buffer (GE Healthcare). Probes were prepared by 5′-end labeling oligonucleotides with [γ-32P]ATP (PerkinElmer) using T4 PNK (NEB).
The oligonucleotide probe sequences are as follows:
U1: 5′-GAATAATCGCAGAGGTCAACTCAGCCGAGGT-3′
U2: 5′-TCCGTCTGATTCCAAAAATCAGTTTAACATTTGTTGTCCTCCAAT-3′
U4: 5′-GGGGTATTGGTTAAAGTTTTCAACTAGCAATAATCGCACCTCAGTAG-3′
U5: 5′-GACTCATTAGAGTGTTCCTCTCCACGGAAATCTTTAGTAAAAGGC-3′
U6: 5′-CTTCTCTGTATCGTTCCAATTTTAGTATATGTTCTGCCGAAGCAAGA-3′
U11: 5′-TCGTGATCGGAAACGTGCCAGGACG-3′
U12: 5′-GCCTAGAAGCCAATACTGCCAAGCGATTAGCAAG-3′
U4atac: 5′-AGCAATGTCCTCACTAGACGTTCATTGAACATTTCTGCT-3′
U6atac: 5′-CCTAGCCGACCGTTTATGTGTTCCATCCTTGTCT-3′
Following the PNK reaction, probes were purified using Microspin G-50 Columns (GE Healthcare). For hybridization, the blots were probed with the labeled oligonucleotides at 65°. The blots were then washed twice each in 2X SSC and 0.33X SSC at 60°. Blots were exposed to storage phosphor screens (GE Healthcare) and analyzed with an Amersham Typhoon 5 (GE Healthcare).
To analyze knockdown efficiency, total RNA was treated with TURBO DNase (Invitrogen). Following a phenol/chloroform purification, 350 ng of RNA was converted to cDNA using the SuperScript III First-Strand Synthesis System (Invitrogen). Primers for PCR are as follows:
5S_F: 5′-GCCAACGACCATACCACGCTGAA-3′
5S_R: 5′-AACAACACGCGGTGTTCCCAAGC -3′
CG2941_F: 5′- TGTGGTATTGGCAGGACGGTCT-3′
CG2941_R: 5′- CCTTGTGCTTCAATTTGCTCACTTGGTT -3′
G4a-b-c_F: 5′- CCAGATAGCCTGCATGGAACATCG -3′
G4a-b-c_R: 5′- CTCCCGCTTTAATGGATCATTGAGGG -3′
Data availability
All fly strains, probe sequences and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. Supplemental material available at Figshare: https://doi.org/10.25387/g3.7473359.
Results and Discussion
To identify Drosophila melanogaster proteins that might correspond to those known to be contained within the human SMN complex (Figure 1B), we first carried out in silico bioinformatic analyses. As mentioned above, there have been conflicting reports regarding the conservation (or lack thereof) of certain Gemin proteins in Drosophila (Kroiss et al. 2008; Lanfranco et al. 2017). In particular, Gemin4, Gemin6, Gemin7 and Gemin8 were originally thought to have been lost from Dipteran genomes, although clear orthologs of Gem6-7-8 can been readily identifed in Hymenoptera, and other insects (e.g., search https://www.ncbi.nlm.nih.gov/protein).
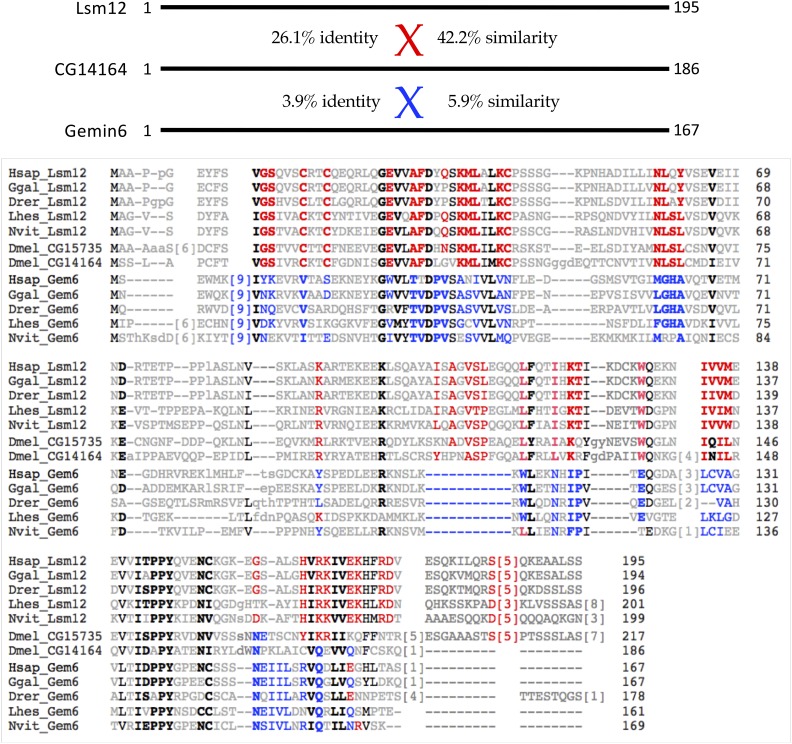
Hezron (CG14164) is orthologous to Lsm12, not Gemin6
Lanfranco et al. (2017) recently suggested that Hezron/CG14164 encodes the Drosophila ortholog of Gemin6. Our bioinformatic analysis suggested that CG14164 is actually more closely related to human Lsm12, an Sm-like protein that contains a C-terminal methyltransferase domain (Albrecht and Lengauer 2004). Because Gemin6 is also an Sm-like protein (Gemin6, Gemin7 and Lsm12 are members of the Sm protein superfamily), we carried out a side-by-side comparison of both Lsm12 and Gemin6 proteins from a variety of vertebrates and invertebrates (Figure 2). That is, we selected Lsm12 and Gemin6 protein pairs from each of five different species (human, chicken, fish, bug and wasp) and aligned them together to identify highly conserved diagnostic amino acid residues in each orthology cluster. Notably, there are two very closely related proteins in D. melanogaster, CG14164 and CG15735, both of which were included in this comparison. As shown in Figure 2, CG14164/Hezron is clearly more closely related to the Lsm12 cluster than it is to that of Gemin6 (compare diagnostic Lsm12 and Gemin6 residues shaded in red and blue, respectively). In nearly every case, CG14164 tracks with the Lsm12 sequences, including the locations of conserved insertions and deletions. Therefore, we conclude that Hezron/CG14164 is orthologous to metazoan Lsm12 and that the ancestral Gemin6 gene has been lost in Drosophila.
Figure 2.
CG14164/Hezron is orthologous to Lsm12, not Gemin6. Top: Schematic of human Lsm12 and Gemin6, with pairwise alignment scores to CG14164 showing %similarity and %identity. Bottom: Amino acid alignment of Lsm12 and Gemin6 (Gem6) protein sequences from a variety of metazoan species, including: Homo sapiens (Hsap), Gallus gallus (Ggal), Danio rerio (Drer), Lygus hesperus (Lhes), and Nasonia vitripennis (Nvit). Two paralogous D. melanogaster proteins CG15735 and CG14164, are shown for comparison. Each of the fruitfly proteins shows a high degree of similarity to Lsm12, as compared to Gemin6. Diagnostic residues shaded in red are highly conserved in Lsm12 orthologs and those shaded in blue are conserved in the Gem6 orthologs. Residues bolded in black are conserved among all of the proteins.
CG15735 was recently shown to function as an Ataxin-2 adaptor (Lee et al. 2017); a genetic analysis of CG14164 has not been reported. We note that CG15735/Lsm12a (217aa) is slightly longer than Hezron/CG14164/Lsm12b (186aa) and that the two proteins begin to diverge only at their respective C-termini (Figure 2). There is the barest hint of similarity between CG14164 and Gemin6 in this region. It is tempting to speculate that an ancestral recombination between Gemin6 and Lsm12 might have created Hezron/CG14164. Additional experiments will be required in order to address these evolutionary relationships, as well as to determine whether or not Hezron/CG14164/Lsm12b protein might have been co-opted into the extant Drosophila SMN complex (see below).
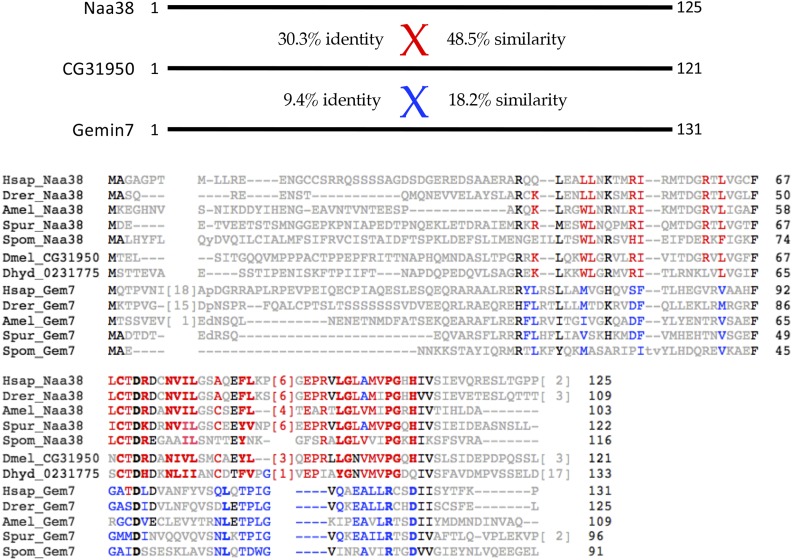
Sabbat (CG31950) is orthologous to Naa38, not Gemin7
CG31950/Sabbat was also identified by Lanfranco et al. (2017) as a potential ortholog to Gemin7 and member of the Drosophila SMN complex. We found that CG31950 was, in fact, more similar to an N-terminal acetyltransferase auxiliary subunit, Naa38 (Varland et al. 2015; Aksnes et al. 2016). An alignment of Naa38 proteins from human, fish, honeybee, sea urchin and fission yeast, along with the Gemin7 orthologs from these same five species reveals that CG31950 is much more similar to the Naa38 orthology cluster than it is to that of Gemin7 (Figure 3). For purposes of comparison, we also include in the alignment the most closely related protein in a second fruitfly genome, D. hydei (XP_023177506.1), along with CG31950 from D. melanogaster (Figure 3). Although the two clusters of proteins share an overall sequence similarity (indeed, human Naa38 is also known to contain an Sm-like fold; see https://www.uniprot.org/uniprot/I3L310) diagnostic residues within the Naa38 orthologs are shared by CG31950. In contrast, the highly conserved regions of Gemin7, including the relative positions of insertions and deletions, do not track with CG31950 (see shaded residues in Figure 3). Hence, we conclude that Sabbat/CG31950 is orthologous to Naa38.
Figure 3.
CG31950/Sabbat is orthologous to Naa38, not Gemin7. Top: Schematic of human Naa38 and Gemin7, with pairwise alignment scores to CG31950 showing %similarity and %identity. Bottom: Amino acid alignment of Gemin7 (Gem7) and Naa38 protein sequences from a variety of metazoan species, including: Homo sapiens (Hsap), Danio rerio (Drer), Apis mellifera (Amel), Strongylocentrotus purpuratus (Spur) and Schizosaccharomyces pombe (Spom). Two orthologous fruitfly proteins from D. melanogaster (Dmel_CG31950) and D. hydei (Dhyd_0231775) are shown for comparison. Diagnostic residues in red are those that are conserved in Naa38 sequences and those in blue are conserved in Gemin7 proteins.
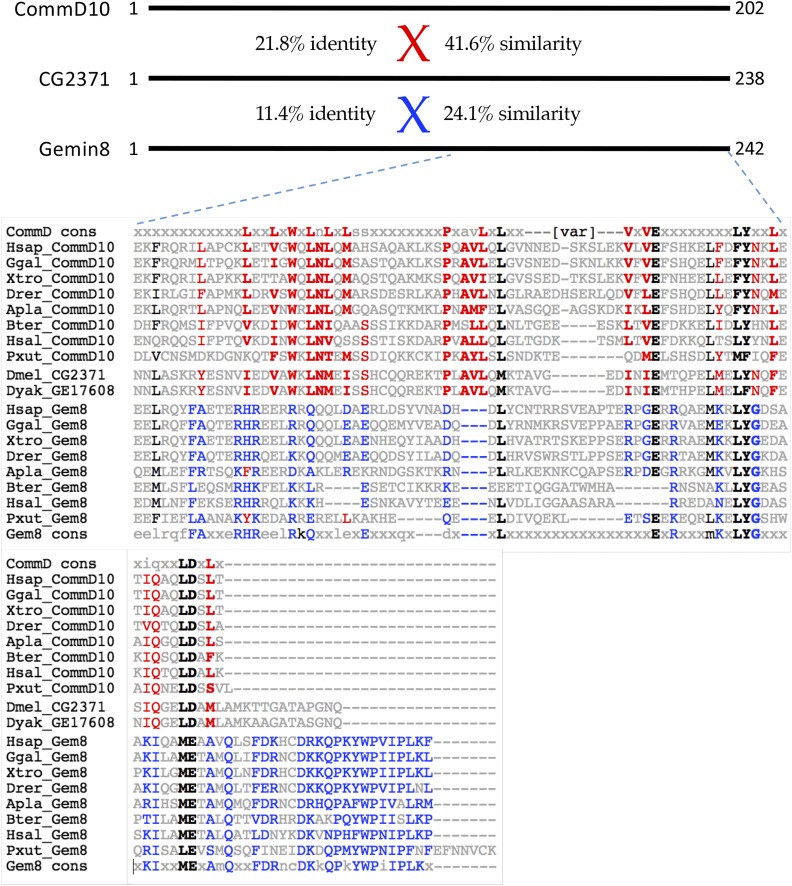
Valette (CG2371) is orthologous to CommD10, not Gemin8
Similar to the situation with Gemin6 and Gemin7, we found that CG2371, identified by Lanfranco et al. (2017) as the potential Gemin8 ortholog, is more closely related to a protein called CommD10 (Figure 4). In humans, there are ten Comm domain paralogs, five of which are conserved in insects (Maine and Burstein 2007). These proteins are characterized by a conserved C-terminal ∼80 aa region called the Comm domain (see discussion below). Gemin8 orthologs are also conserved at their C-termini, but the structure and function of this protein is largely unknown. As shown in Figure 4, D.melanogaster CG2371 and D.yakuba GE17608 proteins are most similar to CommD10 orthologs as compared to the Gemin8 orthologs from a variety of metazoan species. The conservation of diagnostic amino acid residues between CG2371 and CommD10 (Figure 4, shaded residues) leaves little doubt as to the ancestral relationship. Again, the interesting question of whether Valette/CG2371 might have compensated for loss of Gemin8 within the Drosophila lineage is discussed below.
Figure 4.
CG2371/Valette is orthologous to CommD10, not Gemin8. Top: Schematic of human CommD10 and Gemin8, with pairwise alignment scores to CG2371 showing %similarity and %identity. Bottom: Amino acid alignment of the C-terminal domains of CommD10 and Gemin8 (Gem8) from a variety of metazoan species, including: Homo sapiens (Hsap), Gallus gallus (Ggal), Xenopus tropicalis (Xtro), Danio rerio (Drer), Acanthaster planci (Apla), and Bombus terrestris (Bter), H saltator (Hsal), and Papilio xuthus (Xut). Two orthologous fruitfly proteins from D. melanogaster (Dmel_CG2371) and D. yakuba (Dyak_GE17608) are shown for comparison. Diagnostic residues shaded in red text are conserved among CommD10 sequences whereas the blue residues are conserved in Gemin8. For additional comparison, consensus sequences for the C-terminal Comm domain (CommD cons) and C-terminus of Gemin8 (Gem8 cons) are shown at the top and bottom of the alignment, respectively.
Gaulos (CG2941) is orthologous to metazoan Gemin4
In contrast to Gem6-7-8, Gemin4 was originally thought to be lost from insects entirely (Kroiss et al. 2008), however Lanfranco et al. (2017) identified CG2941/Gaulos as a potential Gemin4 ortholog. To investigate this issue, we carried out PSI-BLAST (Position-Specific Iterative Basic Local Alignment Search Tool) analysis (Altschul et al. 1997). Using vertebrate Gemin4 proteins as seed sequences, this procedure readily identified several candidates among the Hymenoptera, but not within the Diptera (or it may take more than six iterations to converge). Anecdotally, we have found that the entire SMN complex is well preserved in many Hymenopteran genomes, and so we used the putative Gemin4 sequence XP_003401506.1 from Bombus terrestris (Buff-tailed bumble bee) as the starting point for PSI-BLAST. We found that this seed sequence readily identified both vertebrate and invertebrate orthologs, along with three nearly identical D. melanogaster proteins, including CG2941, CG32783 and CG32786. An alignment of a subset of these identified proteins is presented in Fig. S1. As shown, the overall conservation of Gemin4 is rather modest. For comparison, the putative Gemin4 ortholog (GH21356) from a distantly related fruitfly, D. grimshawi, is also shown. Notably, an analysis of the predicted secondary structure of Gemin4 orthologs shows a high degree of conservation (see Fig. S1). Thus, despite the fact that the three D. melanogaster proteins (CG2941, CG32786 and CG32783) are most closely related to metazoan Gemin4, it is difficult to assign orthology on the basis of amino acid conservation alone.
Importantly, Lanfranco et al. (2017) did not base their conclusions solely on bioinformatics; these authors also showed that CG2941 interacts with Gemin3 in a targeted genetic modifier screen. Flies expressing low-levels of a dominant-negative Gemin3 construct lacking its N-terminal helicase domain, called Gemin3BART (Borg et al. 2015), were used in combination with a deficiency allele Df(1)ED6716 (Ryder et al. 2007) that spans the 3F2-4B3 interval on the X chromosome that includes CG2941/Gaulos (Lanfranco et al. 2017). Loss of one copy of this region in combination with pan-muscular expression of Gemin3BART led to a marked age-dependent enhancement of the phenotype (Lanfranco et al. 2017). These results were encouraging because Gemin4 was originally identified as a putative Gemin3 cofactor (Charroux et al. 2000), and so a reduction in Gemin4 gene copy-number might reasonably be expected to enhance the phenotype of a Gemin3 hypomorph.
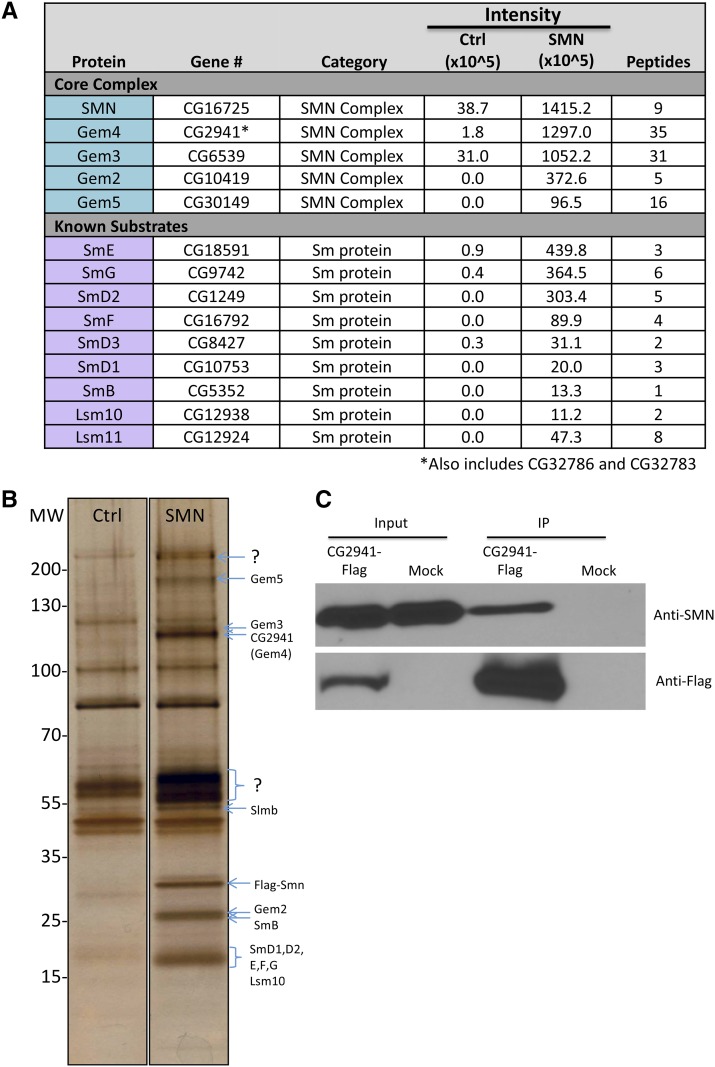
Furthermore, a systematic coaffinity purification analysis of the Drosophila proteome showed that CG2941 is capable of forming a complex in S2 cells transfected with HA-tagged SMN (Guruharsha et al. 2011). Similarly, co-transfection of S2 cells with HA-tagged SMN and GFP-tagged CG2941/Gaulos confirmed this interpretation (Lanfranco et al. 2017). Previously, we carried out proteomic profiling of embryonic lysates from transgenic flies expressing Flag-SMN (Gray et al. 2018). Notably, these animals express SMN protein from the native Smn control regions in an otherwise null background (Praveen et al. 2012; Gray et al. 2018). In order to conclusively determine whether CG2941 forms a complex with SMN under endogenous conditions, we directly analyzed eluates of this purification by label-free mass spectrometry (Figure 5A). We also carried out SDS-PAGE analysis of the purified samples, followed by silver staining (Figure 5B). In addition to identifying all of the known Sm protein substrates of SMN, we also identified Gemin2, Gemin3, Gemin5 and CG2941 as highly-enriched SMN binding partners (Figure 5). Notably, mass spectrometry failed to detect CG14164 (Hezron/Lsm12b), CG31950 (Sabbat/Naa38) or CG2371 (Valette/CommD10) among the co-purified proteins (see below for a discussion).
Figure 5.
Flag-SMN immunopurified lysates contain members of the known SMN complex and known substrates. (A) Flag-purified eluates were analyzed by label-free mass spectrometry. Proteins in the core SMN complex and known substrates were highly enriched in the SMN sample as compared with Ctrl. (B) Lysates from Oregon-R control (Ctrl) Drosophila embryos and those that exclusively express Flag-SMN (SMN) were immunopurified. Protein eluates were separated by SDS-PAGE and silver stained. Band identities were predicted by size. (C) Following transfection of CG2941-Flag in Drosophila S2 cells and immunoprecipitation (IP) of total cell lysates with anti-Flag beads, western analysis was carried out using anti-SMN antibodies. Mock transfected cells were analyzed in parallel. Co-purification of endogenous SMN was detected in the CG2941-Flag IP lane but not in the control lane (Mock).
We also note that CG32786 and CG32783 are so similar to CG2941 that most of their tryptic peptides are indistinguishable from one another. However, we identified 35 peptides corresponding to these three proteins and their overall enrichment in the purified eluates was comparable to that of the other core members of the SMN complex (Figure 5A). To confirm this interaction, we performed a reciprocal coimmunoprecipitation analysis with CG2941-Flag in S2 cells. As shown in Figure 5C, CG2941-Flag also co-purifies with endogenous SMN. On the basis of these findings, we conclude that CG2941/Gaulos is indeed orthologous to human Gemin4.
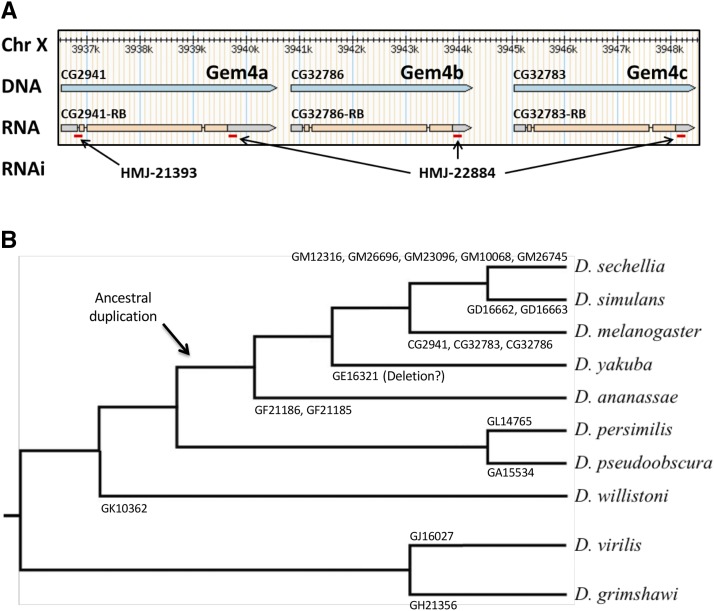
CG2941 is ancestral to CG32786 and CG32783 and is part of a genomic triplication
As shown in Figure 6A, CG2941, CG32786 and CG32783 are tightly linked in the 3F7-3F9 interval on the D. melanogaster X chromosome. A comparison of their DNA sequences reveals that these three genes are extremely similar, with CG32786 and CG32783 being more closely related to each other than they are to CG2941. In contrast, CG2941 shares more sequences with the orthologous sequences in other more distantly related Drosophilids than do CG32786 or CG32783. Thus, we infer that CG2941 is ancestral to the CG32786 and CG32783 gene pair.
Figure 6.
Organization and phylogeny of Gemin4/CG2941-like genes in Drosophilids. (A) Browser shot of D. melanogaster CG2941 paralogs, showing their relative location and exon-intron structure. Red dashes indicate regions targeted by short hairpin (sh)RNAs expressed from Gal4-inducible RNAi constructs, see text for details. (B) Drosophila phylogenetic tree showing gene name and number of respective Gemin4 (Gem4) orthologs within each lineage. Following their divergence from the obscura group (D. pseudoobscura and D. persimilis), potential duplications and deletions within the melanogaster group (D. sechellia, D. melanogaster, D. simulans, D. yakuba, and D. annanasae) are illustrated.
Interestingly, this region of the genome appears to be somewhat fluid, particularly within the melanogaster group, which includes D. sechellia, D. melanogaster, D. simulans, D. yakuba, and D. annanasae. Phylogenetic analysis of the number of CG2941-like genes in various Drosophilid genomes (Figure 6B) suggests that an ancestral duplication of CG2941 occurred sometime between the divergence of the melanogaster group and the obscura group, the latter of which contains D. pseudoobscura and D. persimilis. The hawaiian (represented by D. grimshawi) and virilis (D. virilis) groups each only have one CG2941-like gene and thus serve as outgroups for this analysis. Within the melanogaster group there appears to be ongoing genetic rearrangement of this genomic region, as certain species have up to five different copies of this gene, whereas others have only a single copy (Figure 6B). For ease of future identification, we suggest the following nomenclature for the D. melanogaster genes: Gemin4a (CG2941/Gaulos), Gemin4b (CG32786) and Gemin4c (CG32783).
Gemin4 gene function is essential for viability in Drosophila
The FlyAtlas anatomical and developmental expression database (Chintapalli et al. 2007; Leader et al. 2018) shows that CG2941 is expressed ubiquitously, albeit at relatively low levels. Its highest levels of expression are in the larval central nervous system and the adult ovary. Because the sequences of the three Gemin4 paralogs are so similar, the function and expression levels of the other two genes are unclear. Lanfranco et al. (2017) employed an RNA interference transgene targeting CG2941, but did not mention the existence of the other two paralogous genes in their publication. We note that this transgene (VDRC 52356), obtained from the Vienna Drosophila Resource Center, expresses a 339bp dsRNA that targets all three Gemin4 paralogs (CG2941, CG32786 and CG32783). Furthermore, the deficiency, Df(1)ED6716, used in the original genetic interaction screen also uncovers all three paralogs.
To confirm and extend these studies, we sought to determine whether or not loss of CG2941 might be compensated by the presence of the other paralogs. We therefore carried out RNAi using two different shRNA expressing TRiP lines obtained from the Transgenic RNAi Project (Perkins et al. 2015). As shown in Figure 6A, HMJ-21393 specifically targets a region near the 5′-end of the Gemin4a/CG2941 transcript, whereas the HMJ-22884 construct targets a 3′-UTR sequence that is shared by all three transcripts.
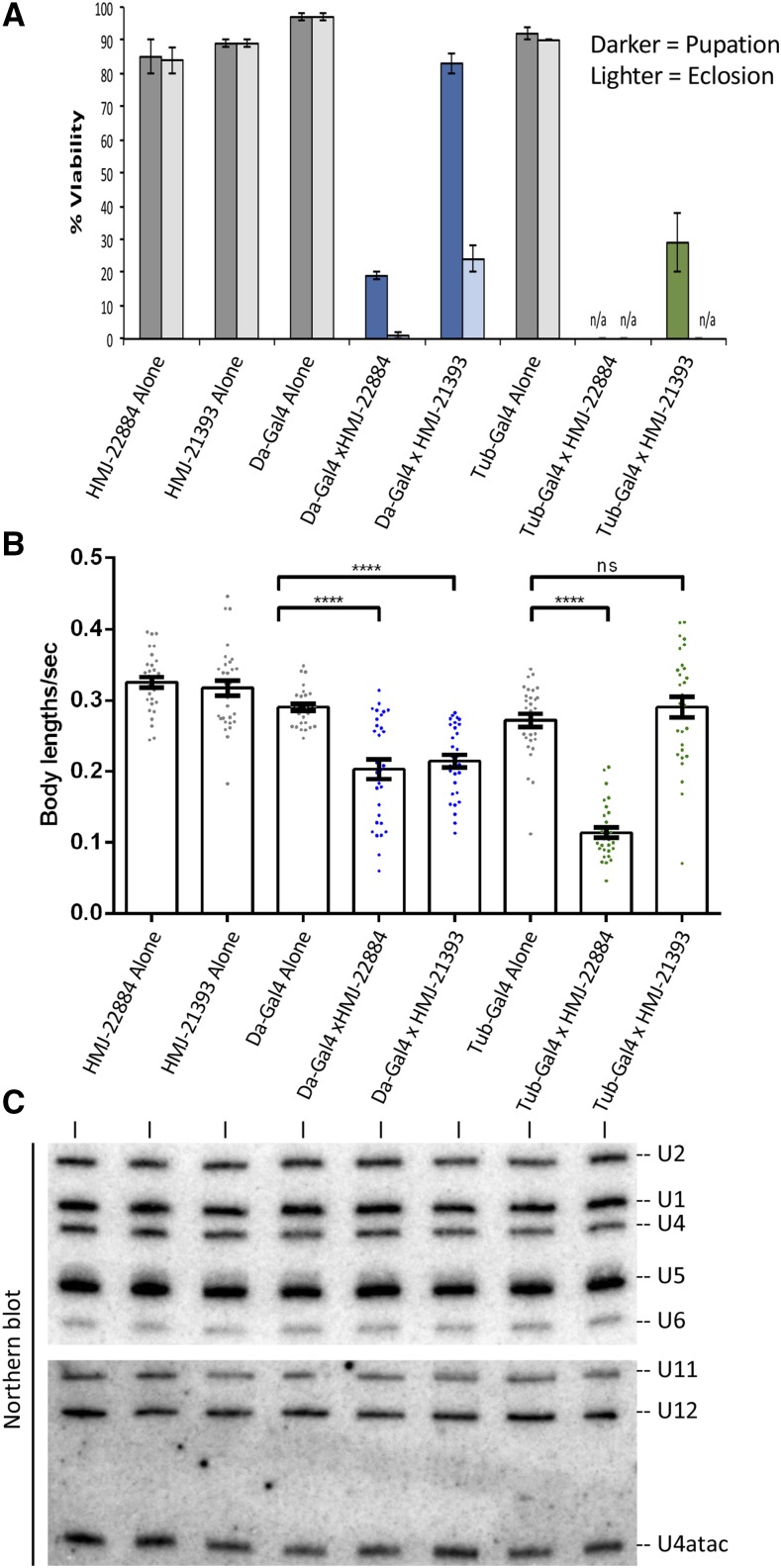
In Figure 7A, we used the Gal4/UAS system to drive these two UAS-shRNA constructs ubiquitously using either daughterless-Gal4 (Da-Gal4) or tubulin-Gal4 (Tub-Gal4). Expression of either drivers or the responders alone had little to no effect on organismal viability (Figure 7A). However, ubiquitous expression of the shRNA that targets all three genes (HMJ-22884) was essentially larval lethal (Figure 7A). The phenotype of animals expressing the HMJ-21933 construct that specifically targets CG2941 was slightly less severe, as most of the animals complete larval development (Da-Gal4 x HMJ-21933), and roughly 20% of them eclose as adults. The phenotype of the Tub-Gal4 x HMJ-21933 cross was even more severe, as only ∼30% of the animals reached pupal stages and none progressed to adulthood (Figure 7A). These findings are consistent with those in mice (Meier et al. 2018), showing that Gemin4 is an essential gene. The data also suggest that Gemin4b/CG32786 and Gemin4c/CG32783 can partially compensate for loss of Gemin4a/CG2941. However, in the absence of specific genetic lesions and complementation analysis, it is difficult to make firm conclusions.
Figure 7.
CG2941 and its paralogs are essential for viability and locomotion independent of snRNP biogenesis. (A) daughterless (Da) or tubulin (Tub) Gal4 drivers were crossed to RNAi lines specifically targeting CG2941 (HMJ-21393) or all three paralogs (HMJ-22884). 100 larvae per cross were selected and viability was measured based on the number of larvae that pupated (darker bars) and that eclosed (lighter bars). “n/a” represents a category in which there were no pupae (pupation) or adults (eclosion) to count. Error bars show standard error of the mean. (B) Wandering third instar larvae from the same crosses in (A) were recorded and locomotor behavior was measured using the wrMTrck plug-in for Fiji/ImageJ. Error bars represent standard error. (C) Early third instar larvae (73-77 hr post-egg laying) from the same crosses in (A) were collected and RNA was extracted to measure snRNA levels via northern blotting. See Fig. S2 for details on the knockdown. Probe sequences are described in Materials and Methods.
To investigate the consequences of Gemin4 loss of function, we carried out larval locomotion assays and northern blotting analysis. Wandering third instar larvae were used to record their movement on a molasses agar plate. The videos were then converted and analyzed using the wrmTrck plug-in of Fiji/ImageJ to generate a measurement of body lengths per second (BLPS), which takes into account the speed and size of each larva. For northern blotting, total RNA was harvested from early third instar (72-76hr) larvae, just prior to the beginning of the lethal phase of the RNAi. As shown in Figure 7C, we did not detect signficant reductions in the levels of either the major (U1, U2, U4 and U5) or the minor (U11, U12 and U4atac) Sm-class snRNAs. Due to the long half-lives of spliceosomal snRNPs in cultured mammalian cells (1-3 days, depending on the snRNA; Sauterer et al. 1988), this finding is perhaps not so surprising. Thus the presumptive loss of Gemin4 function in snRNP biogenesis may not have had time to affect these animals. Given that complete loss of SMN protein only results in a ∼50–60% reduction of U1-U5 snRNAs at this stage of development (Garcia et al. 2016), it is also unsurprising that knockdown of Gemin4 has a less dramatic effect. We conclude that the larval lethality associated with Gemin4 loss of function is not due to a concomitant loss of snRNPs.
Conservation of Gemin4 among Dipteran genomes
In the ten years since Kroiss et al. (2008) first suggested that Gemin4 was missing from the Dipteran SMN complex, there have been numerous hints to the contrary. As early as 2009, raw mass spectrometry data released by the DPiM (Drosophila Protein interaction Map) project showed that CG2941 co-precipitates with ectopically expressed, epitope-tagged SMN in S2 cells (https://interfly.med.harvard.edu). Subsequent quality control steps apparently removed CG2941 from the list of potential SMN interactors despite the fact that it also co-purifies with epitope tagged Gemin2 and Gemin3 (Guruharsha et al. 2011; Guruharsha et al. 2012). On the basis of biochemical purifications from fly embryos and S2 cells, we and others have speculated that CG2941 might well be a bona fide core member of the SMN complex (Sen et al. 2013; Gray et al. 2016). However, in the absence of additional genetic, phylogenetic and biochemical evidence linking endogenous CG2941 to SMN, the conservation of Gemin4 remained an open question.
Three new lines of experimentation demonstrate that CG2941 is indeed Gemin4. First, Lanfranco et al. (2017) found that an N-terminally truncated Gemin3∆N construct interacts genetically with a deficiency that uncovers CG2941/Gemin4a, CG32786/Gemin4b and CG32783/Gemin4c. They also showed that RNAi-mediated knockdown of all three genes enhanced the phenotype of this dominant negative Gemin3∆N transgene. Second, ongoing metazoan genome sequencing efforts allowed us to more confidently predict Gemin4 orthologs on the basis of primary sequence (Fig. S1). Third, we found that endogenous CG2941, CG32786 and CG32783 co-purify with SMN expressed from its native promoter in vivo in fly embryos (Figure 5). The relative enrichment and number of peptides corresponding to CG2941 in the mass spectrometry experiment was similar to that of the other Gemins (Figure 5A). These findings lead us to conclude that Gemin4 has been retained in the genomes of Drosophila and other dipterans.
SMN and the evolution of Gemin subcomplexes
The human SMN complex can be subdivided into several distinct subunits (Battle et al. 2007; Otter et al. 2007). SMN and Gemin2 form an oligomeric heterodimer (SMN•Gemin2)n that makes up the core of the complex (Fischer et al. 1997; Gupta et al. 2015). Gemin3 binds directly and independently to Gemin4, tethering them both to SMN•Gemin2 (Charroux et al. 1999; Charroux et al. 2000). Oligomerization of SMN appears to be required for Gemin3 to enter the complex (Praveen et al. 2014). Gemin5 is a large (175 kD) WD-repeat protein that recruits RNA substrates to the SMN complex (Battle et al. 2006b) via subdomains that bind to the m7G-cap and Sm-site, respectively (Xu et al. 2016). Thus Gemin5 can be viewed as an RNP subunit of the SMN complex. Finally, Gemin6 and Gemin7 heterodimerize (Baccon et al. 2002; Ma et al. 2005) and recruit Gemin8 (Carissimi et al. 2006) to form a Gem6-7-8 subunit, the function of which is unknown. As shown in Figures 2 and 3, these three proteins appear to have been lost from Drosophilids, but retained in the genomes of other insects.
An important question raised by our findings is whether or not the functions normally carried out by Gemin6, Gemin7 and Gemin8 may have been taken over by CG14164 (Hez/Lsm12b), CG31950 (Sbat/Naa38) and CG2371 (Vlet/CommD10). Lanfranco et al. (2017) showed that these three proteins are each capable of forming complexes with exogenously expressed SMN when they are transfected into S2 cells. And given the opportunity to interact in a directed yeast two-hybrid screen, Sbat/Naa38 scored positively for interaction with Gemin3, Hez/Lsm12b and Vlet/CommD10 (Lanfranco et al. 2017). Interestingly, both Hez and Sbat are predicted contain an Sm-fold, also known as a small beta barrel (Youkharibache et al. 2019). This structure is characterized by five short beta strands that form a closed domain wherein the first strand is hydrogen bonded to the last (Arluison et al. 2006).
Small beta barrel containing proteins exhibit a strong tendency to form higher-order structures, as exemplified by the Sm and Lsm proteins, found in all three domains of life (Youkharibache et al. 2019). Thus, despite the fact that Hez, Sbat and Vlet are not orthologous to Gemin6, Gemin7 and Gemin8, respectively, it remains formally possible that they have been evolutionarily co-opted into the SMN complex in flies. Our inability to identify these three proteins as endogenous SMN binding partners by mass spectrometry (Figure 5) argues against this idea. However, stable protein interactions are not required to elicit important biological outcomes, so additional experiments will be needed to conclusively demonstrate a role for Hez/Lsm12b, Sbat/Naa38 and Vlet/CommD10 in SMN biology.
Considerations and Prospects
In the mean time, several interesting possibilities suggest themselves for future investigation. We and others have hypothesized that the SMN complex may function as a hub for various cellular signaling pathways, in addition to its role in chaperoning snRNP biogenesis (Raimer et al. 2017; Gray et al. 2018 and references therein). As shown in Figure 4, the fruitfly CommD10 ortholog is Vlet/CG2371. Intriguingly, human CommD (Copper metabolism Murr1 domain) proteins can form homo- and hetero-dimers (Burstein et al. 2005) and are involved in a variety of cellular pathways including endosomal membrane trafficking and the inhibition of NF-kB signaling (Bartuzi et al. 2013; Mallam and Marcotte 2017). Both SMN (Kim and Choi 2017) and Gemin3 (Shin et al. 2014) have been implicated in NF-kB related pathways. It is tempting to speculate that human Gemin8 might play a role in linking the SMN complex to NF-kB signaling by interacting with, or otherwise functioning as, a CommD-like protein. Given the potential for Sbat/Naa38 to interact with Gemin3 (Lanfranco et al. 2017), perhaps the Gem6-7-8 subcomplex functions as a regulatory subunit that modulates the activity of SMN and/or Gemin3.
Irrespective of any putative role in cellular signaling, the fact that Sbat/Naa38 contains an Sm-fold may help to explain several interesting observations in the literature. Metazoan Naa38 (a.k.a. Lsmd1, Mak31) is an auxiliary subunit of NatC (N-terminal acetyltransferase C; Starheim et al. 2009). N(alpha)-acetyltransferases are enzymes that consist of a catalytic subunit and one or two auxiliary subunits (Aksnes et al. 2016). The auxiliary subunits modulate the activity and substrate specificity of the catalytic subunit. Furthermore, they mediate co-translational binding to the 60S ribosome, in a region that is located near the nascent polypeptide exit tunnel (reviewed in Aksnes et al. 2016). This latter point merits attention for two reasons.
First, Fischer and colleagues recently reported data suggesting that, following their translation, Sm proteins can remain bound to the ribosome near the exit tunnel, dissociating only after binding to the assembly chaperone pICln (Paknia et al. 2016). These authors hypothesize the existence of a quality control hub for chaperone-mediated protein assembly, located on the ribosome. Whether or not Sm protein heterodimers (e.g., Lsm10/11 and SmD1/D2) actually bind to the nascent peptide tunnel region of the ribosome in vivo is unclear. However, the fact that metazoan Naa38 is structurally similar to Sm proteins provides a plausible mechanism for their binding to the ribosome immediately following translation. Given that Gemin6 and Gemin7 are also members of the Sm-like superfamily of proteins (Ma et al. 2005) it is conceivable that Sbat/Naa38 could dimerize with other Sm-like proteins (e.g., Hez/Lsm12b) in Drosophila.
Second, Naa38 might not be a bona fide member of the SMN complex (in flies or any other species), but it could potentially interact with SMN as part of its canonical function in N-terminal protein acetylation. More than 80% of human proteins are co-translationally modified on their N-termini (Arnesen et al. 2009), however the functional impact of this modification is largely unknown. Most proteins do not retain their N-terminal Met residue, and its removal by methionine aminopeptidases frequently leads to acetylation of the resulting N-terminus, particularly if the second residue is Ala, Val, Ser, Thr or Cys (Hwang et al. 2010). Interestingly, the N-terminal Ala2 residue of SMN is known to be acetylated in human cells (Van Damme et al. 2012), and an A2G missense mutation in the SMN1 gene is known to cause a mild form of SMA when SMN2 is present in a single copy (Parsons et al. 1998). This mutation is puzzling because, with the exception of Ala2, the N-terminal 15aa of SMN (i.e., upstream of the Gemin2 binding domain) are very poorly conserved. Moreover, changing Ala2 to Gly is predicted to reduce the probability of N-terminal acetylation and recognition by the N-end-rule proteasomal degradation pathway (Hwang et al. 2010). These findings suggest that the phenotype of the A2G mutation in humans is due to loss of N-terminal acetylation of SMN.
In conclusion, it seems unlikely that three different proteins (CG14164, CG31950, CG2371) derived from three different biological contexts might be co-opted into a novel Gemin subcomplex. A loss of the Gem6-7-8 subunit from the SMN complex in flies would suggest that either this subunit is not essential for basal metazoan viability or that other factors have compensated for deficiency of these proteins in Drosophila. Additional experiments will be needed to rule in, or rule out, any such functional adaptation. In contrast, the identification of Gemin4 via PSI-BLAST in a variety of different insect genomes including the Diptera, Hemiptera, Lepidoptera, and Hymenoptera (this work) indicates that this protein is widely conserved. Moreover, genetic loss of function studies (Figure 7; Lanfranco et al. 2017; Meier et al. 2018) strongly suggest that Gemin4 is essential for metazoan viability.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25387/g3.7473359.
Communicating editor: H. Salz
Literature Cited
- Aksnes H., Drazic A., Marie M., Arnesen T., 2016. First Things First: Vital Protein Marks by N-Terminal Acetyltransferases. Trends Biochem. Sci. 41: 746–760. 10.1016/j.tibs.2016.07.005 [DOI] [PubMed] [Google Scholar]
- Albrecht M., Lengauer T., 2004. Novel Sm-like proteins with long C-terminal tails and associated methyltransferases. FEBS Lett. 569: 18–26. 10.1016/j.febslet.2004.03.126 [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Madden T. L., Schaffer A. A., Zhang J., Zhang Z., et al. , 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arluison V., Mura C., Guzman M. R., Liquier J., Pellegrini O., et al. , 2006. Three-dimensional structures of fibrillar Sm proteins: Hfq and other Sm-like proteins. J. Mol. Biol. 356: 86–96. 10.1016/j.jmb.2005.11.010 [DOI] [PubMed] [Google Scholar]
- Arnesen T., Van Damme P., Polevoda B., Helsens K., Evjenth R., et al. , 2009. Proteomics analyses reveal the evolutionary conservation and divergence of N-terminal acetyltransferases from yeast and humans. Proc. Natl. Acad. Sci. USA 106: 8157–8162. 10.1073/pnas.0901931106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccon J., Pellizzoni L., Rappsilber J., Mann M., Dreyfuss G., 2002. Identification and characterization of Gemin7, a novel component of the survival of motor neuron complex. J. Biol. Chem. 277: 31957–31962. 10.1074/jbc.M203478200 [DOI] [PubMed] [Google Scholar]
- Bartuzi P., Hofker M. H., van de Sluis B., 2013. Tuning NF-kappaB activity: a touch of COMMD proteins. Biochim. Biophys. Acta 1832: 2315–2321. 10.1016/j.bbadis.2013.09.014 [DOI] [PubMed] [Google Scholar]
- Battle D. J., Kasim M., Wang J., Dreyfuss G., 2007. SMN-independent subunits of the SMN complex. Identification of a small nuclear ribonucleoprotein assembly intermediate. J. Biol. Chem. 282: 27953–27959. 10.1074/jbc.M702317200 [DOI] [PubMed] [Google Scholar]
- Battle D. J., Kasim M., Yong J., Lotti F., Lau C. K., et al. , 2006a. The SMN complex: an assembly machine for RNPs. Cold Spring Harb. Symp. Quant. Biol. 71: 313–320. 10.1101/sqb.2006.71.001 [DOI] [PubMed] [Google Scholar]
- Battle D. J., Lau C. K., Wan L., Deng H., Lotti F., et al. , 2006b. The Gemin5 protein of the SMN complex identifies snRNAs. Mol. Cell 23: 273–279. 10.1016/j.molcel.2006.05.036 [DOI] [PubMed] [Google Scholar]
- Borg R., Cauchi R. J., 2013. The Gemin associates of survival motor neuron are required for motor function in Drosophila. PLoS One 8: e83878 10.1371/journal.pone.0083878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borg R. M., Bordonne R., Vassallo N., Cauchi R. J., 2015. Genetic Interactions between the Members of the SMN-Gemins Complex in Drosophila. PLoS One 10: e0130974 (erratum: PLoS ONE 10: e0139649). 10.1371/journal.pone.0130974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradrick S. S., Gromeier M., 2009. Identification of gemin5 as a novel 7-methylguanosine cap-binding protein. PLoS One 4: e7030 10.1371/journal.pone.0007030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burghes A. H., Beattie C. E., 2009. Spinal muscular atrophy: why do low levels of survival motor neuron protein make motor neurons sick? Nat. Rev. Neurosci. 10: 597–609. 10.1038/nrn2670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burstein E., Hoberg J. E., Wilkinson A. S., Rumble J. M., Csomos R. A., et al. , 2005. COMMD proteins, a novel family of structural and functional homologs of MURR1. J. Biol. Chem. 280: 22222–22232. 10.1074/jbc.M501928200 [DOI] [PubMed] [Google Scholar]
- Carissimi C., Baccon J., Straccia M., Chiarella P., Maiolica A., et al. , 2005. Unrip is a component of SMN complexes active in snRNP assembly. FEBS Lett. 579: 2348–2354. 10.1016/j.febslet.2005.03.034 [DOI] [PubMed] [Google Scholar]
- Carissimi C., Saieva L., Baccon J., Chiarella P., Maiolica A., et al. , 2006. Gemin8 is a novel component of the survival motor neuron complex and functions in small nuclear ribonucleoprotein assembly. J. Biol. Chem. 281: 8126–8134. 10.1074/jbc.M512243200 [DOI] [PubMed] [Google Scholar]
- Cauchi R. J., Sanchez-Pulido L., Liu J. L., 2010. Drosophila SMN complex proteins Gemin2, Gemin3, and Gemin5 are components of U bodies. Exp. Cell Res. 316: 2354–2364. 10.1016/j.yexcr.2010.05.001 [DOI] [PubMed] [Google Scholar]
- Charroux B., Pellizzoni L., Perkinson R. A., Shevchenko A., Mann M., et al. , 1999. Gemin3: A novel DEAD box protein that interacts with SMN, the spinal muscular atrophy gene product, and is a component of gems. J. Cell Biol. 147: 1181–1194. 10.1083/jcb.147.6.1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charroux B., Pellizzoni L., Perkinson R. A., Yong J., Shevchenko A., et al. , 2000. Gemin4. A novel component of the SMN complex that is found in both gems and nucleoli. J. Cell Biol. 148: 1177–1186. 10.1083/jcb.148.6.1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaytow H., Huang Y. T., Gillingwater T. H., Faller K. M. E., 2018. The role of survival motor neuron protein (SMN) in protein homeostasis. Cell. Mol. Life Sci. 75: 3877–3894. 10.1007/s00018-018-2849-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintapalli V. R., Wang J., Dow J. A., 2007. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat. Genet. 39: 715–720. 10.1038/ng2049 [DOI] [PubMed] [Google Scholar]
- Coady T. H., Lorson C. L., 2011. SMN in spinal muscular atrophy and snRNP biogenesis. Wiley Interdiscip. Rev. RNA 2: 546–564. 10.1002/wrna.76 [DOI] [PubMed] [Google Scholar]
- Curmi F., Cauchi R. J., 2018. The multiple lives of DEAD-box RNA helicase DP103/DDX20/Gemin3. Biochem. Soc. Trans. 46: 329–341. 10.1042/BST20180016 [DOI] [PubMed] [Google Scholar]
- Di Y., Li J., Zhang Y., He X., Lu H., et al. , 2003. HCC-associated protein HCAP1, a variant of GEMIN4, interacts with zinc-finger proteins. J. Biochem. 133: 713–718. 10.1093/jb/mvg091 [DOI] [PubMed] [Google Scholar]
- Fallini C., Bassell G. J., Rossoll W., 2012. Spinal muscular atrophy: the role of SMN in axonal mRNA regulation. Brain Res. 1462: 81–92. 10.1016/j.brainres.2012.01.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer U., Englbrecht C., Chari A., 2011. Biogenesis of spliceosomal small nuclear ribonucleoproteins. Wiley Interdiscip. Rev. RNA 2: 718–731. 10.1002/wrna.87 [DOI] [PubMed] [Google Scholar]
- Fischer U., Liu Q., Dreyfuss G., 1997. The SMN-SIP1 complex has an essential role in spliceosomal snRNP biogenesis. Cell 90: 1023–1029. 10.1016/S0092-8674(00)80368-2 [DOI] [PubMed] [Google Scholar]
- Garcia E. L., Wen Y., Praveen K., Matera A. G., 2016. Transcriptomic comparison of Drosophila snRNP biogenesis mutants reveals mutant-specific changes in pre-mRNA processing: implications for spinal muscular atrophy. RNA 22: 1215–1227. 10.1261/rna.057208.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gates J., Lam G., Ortiz J. A., Losson R., Thummel C. S., 2004. rigor mortis encodes a novel nuclear receptor interacting protein required for ecdysone signaling during Drosophila larval development. Development 131: 25–36. 10.1242/dev.00920 [DOI] [PubMed] [Google Scholar]
- Gray K. M., Kaifer K. A., Baillat D., Wen Y., Bonacci T. R., et al. , 2018. Self-oligomerization regulates stability of survival motor neuron protein isoforms by sequestering an SCF(Slmb) degron. Mol. Biol. Cell 29: 96–110. 10.1091/mbc.E17-11-0627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray K. M., Wen Y., Matera A. G., 2016. A novel component of the Drosophila SMN complex? CureSMA Proc. 20: 55. [Google Scholar]
- Grimmler M., Otter S., Peter C., Muller F., Chari A., et al. , 2005. Unrip, a factor implicated in cap-independent translation, associates with the cytosolic SMN complex and influences its intracellular localization. Hum. Mol. Genet. 14: 3099–3111. 10.1093/hmg/ddi343 [DOI] [PubMed] [Google Scholar]
- Groen E. J. N., Talbot K., Gillingwater T. H., 2018. Advances in therapy for spinal muscular atrophy: promises and challenges. Nat. Rev. Neurol. 14: 214–224. 10.1038/nrneurol.2018.4 [DOI] [PubMed] [Google Scholar]
- Grundhoff A. T., Kremmer E., Tureci O., Glieden A., Gindorf C., et al. , 1999. Characterization of DP103, a novel DEAD box protein that binds to the Epstein-Barr virus nuclear proteins EBNA2 and EBNA3C. J. Biol. Chem. 274: 19136–19144. 10.1074/jbc.274.27.19136 [DOI] [PubMed] [Google Scholar]
- Gupta K., Martin R., Sharp R., Sarachan K. L., Ninan N. S., et al. , 2015. Oligomeric Properties of Survival Motor Neuron.Gemin2 Complexes. J. Biol. Chem. 290: 20185–20199. 10.1074/jbc.M115.667279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guruharsha K. G., Obar R. A., Mintseris J., Aishwarya K., Krishnan R. T., et al. , 2012. Drosophila protein interaction map (DPiM): a paradigm for metazoan protein complex interactions. Fly (Austin) 6: 246–253. 10.4161/fly.22108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guruharsha K. G., Rual J. F., Zhai B., Mintseris J., Vaidya P., et al. , 2011. A protein complex network of Drosophila melanogaster. Cell 147: 690–703. 10.1016/j.cell.2011.08.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton G., Gillingwater T. H., 2013. Spinal muscular atrophy: going beyond the motor neuron. Trends Mol. Med. 19: 40–50. 10.1016/j.molmed.2012.11.002 [DOI] [PubMed] [Google Scholar]
- Hunt S. L., Hsuan J. J., Totty N., Jackson R. J., 1999. unr, a cellular cytoplasmic RNA-binding protein with five cold-shock domains, is required for internal initiation of translation of human rhinovirus RNA. Genes Dev. 13: 437–448. 10.1101/gad.13.4.437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvágner G., Zamore P. D., 2002. A microRNA in a multiple-turnover RNAi enzyme complex. Science 297: 2056–2060. 10.1126/science.1073827 [DOI] [PubMed] [Google Scholar]
- Hwang C. S., Shemorry A., Varshavsky A., 2010. N-terminal acetylation of cellular proteins creates specific degradation signals. Science 327: 973–977. 10.1126/science.1183147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonka S., Holtmann B., Meister G., Bandilla M., Rossoll W., et al. , 2002. Gene targeting of Gemin2 in mice reveals a correlation between defects in the biogenesis of U snRNPs and motoneuron cell death. Proc. Natl. Acad. Sci. USA 99: 10126–10131. 10.1073/pnas.152318699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W., Wang Y., Liu C. P., Yang N., Jin M., et al. , 2016. Structural basis for snRNA recognition by the double-WD40 repeat domain of Gemin5. Genes Dev. 30: 2391–2403. 10.1101/gad.291377.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khokhar A., Chen N., Yuan J. P., Li Y., Landis G. N., et al. , 2008. Conditional switches for extracellular matrix patterning in Drosophila melanogaster. Genetics 178: 1283–1293. 10.1534/genetics.106.065912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E. K., Choi E. J., 2017. SMN1 functions as a novel inhibitor for TRAF6-mediated NF-kappaB signaling. Biochim Biophys Acta Mol Cell Res 1864: 760–770. 10.1016/j.bbamcr.2017.02.011 [DOI] [PubMed] [Google Scholar]
- Kim E. K., Noh K. T., Yoon J. H., Cho J. H., Yoon K. W., et al. , 2007. Positive regulation of ASK1-mediated c-Jun NH(2)-terminal kinase signaling pathway by the WD-repeat protein Gemin5. Cell Death Differ. 14: 1518–1528. 10.1038/sj.cdd.4402157 [DOI] [PubMed] [Google Scholar]
- Kroiss M., Schultz J., Wiesner J., Chari A., Sickmann A., et al. , 2008. Evolution of an RNP assembly system: a minimal SMN complex facilitates formation of UsnRNPs in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 105: 10045–10050. 10.1073/pnas.0802287105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfranco M., Cacciottolo R., Borg R. M., Vassallo N., Juge F., et al. , 2017. Novel interactors of the Drosophila Survival Motor Neuron (SMN) Complex suggest its full conservation. FEBS Lett. 591: 3600–3614. 10.1002/1873-3468.12853 [DOI] [PubMed] [Google Scholar]
- Lau C. K., Bachorik J. L., Dreyfuss G., 2009. Gemin5-snRNA interaction reveals an RNA binding function for WD repeat domains. Nat. Struct. Mol. Biol. 16: 486–491. 10.1038/nsmb.1584 [DOI] [PubMed] [Google Scholar]
- Leader D. P., Krause S. A., Pandit A., Davies S. A., Dow J. A. T., 2018. FlyAtlas 2: a new version of the Drosophila melanogaster expression atlas with RNA-Seq, miRNA-Seq and sex-specific data. Nucleic Acids Res. 46: D809–D815. 10.1093/nar/gkx976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Yoo E., Lee H., Park K., Hur J. H., et al. , 2017. LSM12 and ME31B/DDX6 Define Distinct Modes of Posttranscriptional Regulation by ATAXIN-2 Protein Complex in Drosophila Circadian Pacemaker Neurons. Mol Cell 66: 129–140 e127. 10.1016/j.molcel.2017.03.004 [DOI] [PubMed] [Google Scholar]
- Lefebvre S., Burglen L., Reboullet S., Clermont O., Burlet P., et al. , 1995. Identification and characterization of a spinal muscular atrophy-determining gene. Cell 80: 155–165. 10.1016/0092-8674(95)90460-3 [DOI] [PubMed] [Google Scholar]
- Liu Q., Fischer U., Wang F., Dreyfuss G., 1997. The spinal muscular atrophy disease gene product, SMN, and its associated protein SIP1 are in a complex with spliceosomal snRNP proteins. Cell 90: 1013–1021. 10.1016/S0092-8674(00)80367-0 [DOI] [PubMed] [Google Scholar]
- Lorson C. L., Strasswimmer J., Yao J. M., Baleja J. D., Hahnen E., et al. , 1998. SMN oligomerization defect correlates with spinal muscular atrophy severity. Nat. Genet. 19: 63–66. 10.1038/ng0598-63 [DOI] [PubMed] [Google Scholar]
- Ma Y., Dostie J., Dreyfuss G., Van Duyne G. D., 2005. The Gemin6-Gemin7 heterodimer from the survival of motor neurons complex has an Sm protein-like structure. Structure 13: 883–892. 10.1016/j.str.2005.03.014 [DOI] [PubMed] [Google Scholar]
- Maine G. N., Burstein E., 2007. COMMD proteins: COMMing to the scene. Cell. Mol. Life Sci. 64: 1997–2005. 10.1007/s00018-007-7078-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallam A. L., Marcotte E. M., 2017. Systems-wide Studies Uncover Commander, a Multiprotein Complex Essential to Human Development. Cell Syst. 4: 483–494. 10.1016/j.cels.2017.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin R., Gupta K., Ninan N. S., Perry K., Van Duyne G. D., 2012. The survival motor neuron protein forms soluble glycine zipper oligomers. Structure 20: 1929–1939. 10.1016/j.str.2012.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera A. G., Terns R. M., Terns M. P., 2007. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell Biol. 8: 209–220. 10.1038/nrm2124 [DOI] [PubMed] [Google Scholar]
- Matera A. G., Wang Z., 2014. A day in the life of the spliceosome. Nat. Rev. Mol. Cell Biol. 15: 108–121 (erratum: Nat. Rev. Mol. Cell Biol. 15: 108–122). 10.1038/nrm3742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier I. D., Walker M. P., Matera A. G., 2018. Gemin4 is an essential gene in mice, and its overexpression in human cells causes relocalization of the SMN complex to the nucleoplasm. Biol. Open 7: bio032409 10.1242/bio.032409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G., Buhler D., Laggerbauer B., Zobawa M., Lottspeich F., et al. , 2000. Characterization of a nuclear 20S complex containing the survival of motor neurons (SMN) protein and a specific subset of spliceosomal Sm proteins. Hum. Mol. Genet. 9: 1977–1986. 10.1093/hmg/9.13.1977 [DOI] [PubMed] [Google Scholar]
- Meister G., Landthaler M., Peters L., Chen P. Y., Urlaub H., et al. , 2005. Identification of novel argonaute-associated proteins. Curr. Biol. 15: 2149–2155. 10.1016/j.cub.2005.10.048 [DOI] [PubMed] [Google Scholar]
- Mouillet J. F., Yan X., Ou Q., Jin L., Muglia L. J., et al. , 2008. DEAD-box protein-103 (DP103, Ddx20) is essential for early embryonic development and modulates ovarian morphology and function. Endocrinology 149: 2168–2175. 10.1210/en.2007-1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourelatos Z., Dostie J., Paushkin S., Sharma A., Charroux B., et al. , 2002. miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev. 16: 720–728. 10.1101/gad.974702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan U., Achsel T., Luhrmann R., Matera A. G., 2004. Coupled in vitro import of U snRNPs and SMN, the spinal muscular atrophy protein. Mol. Cell 16: 223–234. 10.1016/j.molcel.2004.09.024 [DOI] [PubMed] [Google Scholar]
- Nash L. A., Burns J. K., Chardon J. W., Kothary R., Parks R. J., 2016. Spinal Muscular Atrophy: More than a Disease of Motor Neurons? Curr. Mol. Med. 16: 779–792. 10.2174/1566524016666161128113338 [DOI] [PubMed] [Google Scholar]
- Otter S., Grimmler M., Neuenkirchen N., Chari A., Sickmann A., et al. , 2007. A comprehensive interaction map of the human survival of motor neuron (SMN) complex. J. Biol. Chem. 282: 5825–5833. 10.1074/jbc.M608528200 [DOI] [PubMed] [Google Scholar]
- Paknia E., Chari A., Stark H., Fischer U., 2016. The Ribosome Cooperates with the Assembly Chaperone pICln to Initiate Formation of snRNPs. Cell Reports 16: 3103–3112. 10.1016/j.celrep.2016.08.047 [DOI] [PubMed] [Google Scholar]
- Parsons D. W., McAndrew P. E., Iannaccone S. T., Mendell J. R., Burghes A. H., et al. , 1998. Intragenic telSMN mutations: frequency, distribution, evidence of a founder effect, and modification of the spinal muscular atrophy phenotype by cenSMN copy number. Am. J. Hum. Genet. 63: 1712–1723. 10.1086/302160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paushkin S., Gubitz A. K., Massenet S., Dreyfuss G., 2002. The SMN complex, an assemblyosome of ribonucleoproteins. Curr. Opin. Cell Biol. 14: 305–312. 10.1016/S0955-0674(02)00332-0 [DOI] [PubMed] [Google Scholar]
- Pellizzoni L., Charroux B., Dreyfuss G., 1999. SMN mutants of spinal muscular atrophy patients are defective in binding to snRNP proteins. Proc. Natl. Acad. Sci. USA 96: 11167–11172. 10.1073/pnas.96.20.11167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins L. A., Holderbaum L., Tao R., Hu Y., Sopko R., et al. , 2015. The Transgenic RNAi Project at Harvard Medical School: Resources and Validation. Genetics 201: 843–852. 10.1534/genetics.115.180208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piñeiro D., Fernandez-Chamorro J., Francisco-Velilla R., Martinez-Salas E., 2015. Gemin5: A Multitasking RNA-Binding Protein Involved in Translation Control. Biomolecules 5: 528–544. 10.3390/biom5020528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praveen K., Wen Y., Gray K. M., Noto J. J., Patlolla A. R., et al. , 2014. SMA-causing missense mutations in survival motor neuron (Smn) display a wide range of phenotypes when modeled in Drosophila. PLoS Genet. 10: e1004489 10.1371/journal.pgen.1004489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praveen K., Wen Y., Matera A. G., 2012. A Drosophila model of spinal muscular atrophy uncouples snRNP biogenesis functions of survival motor neuron from locomotion and viability defects. Cell Reports 1: 624–631. 10.1016/j.celrep.2012.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raimer A. C., Gray K. M., Matera A. G., 2017. SMN - A chaperone for nuclear RNP social occasions? RNA Biol. 14: 701–711. 10.1080/15476286.2016.1236168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder E., Ashburner M., Bautista-Llacer R., Drummond J., Webster J., et al. , 2007. The DrosDel deletion collection: a Drosophila genomewide chromosomal deficiency resource. Genetics 177: 615–629. 10.1534/genetics.107.076216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauterer R. A., Feeney R. J., Zieve G. W., 1988. Cytoplasmic assembly of snRNP particles from stored proteins and newly transcribed snRNA’s in L929 mouse fibroblasts. Exp. Cell Res. 176: 344–359. 10.1016/0014-4827(88)90336-9 [DOI] [PubMed] [Google Scholar]
- Schrank B., Gotz R., Gunnersen J. M., Ure J. M., Toyka K. V., et al. , 1997. Inactivation of the survival motor neuron gene, a candidate gene for human spinal muscular atrophy, leads to massive cell death in early mouse embryos. Proc. Natl. Acad. Sci. USA 94: 9920–9925. 10.1073/pnas.94.18.9920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen A., Dimlich D. N., Guruharsha K. G., Kankel M. W., Hori K., et al. , 2013. Genetic circuitry of Survival motor neuron, the gene underlying spinal muscular atrophy. Proc. Natl. Acad. Sci. USA 110: E2371–E2380. 10.1073/pnas.1301738110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shababi M., Lorson C. L., Rudnik-Schoneborn S. S., 2014. Spinal muscular atrophy: a motor neuron disorder or a multi-organ disease? J. Anat. 224: 15–28. 10.1111/joa.12083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin E. M., Hay H. S., Lee M. H., Goh J. N., Tan T. Z., et al. , 2014. DEAD-box helicase DP103 defines metastatic potential of human breast cancers. J. Clin. Invest. 124: 3807–3824. 10.1172/JCI73451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpargel K. B., Matera A. G., 2005. Gemin proteins are required for efficient assembly of Sm-class ribonucleoproteins. Proc. Natl. Acad. Sci. USA 102: 17372–17377. 10.1073/pnas.0508947102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starheim K. K., Gromyko D., Evjenth R., Ryningen A., Varhaug J. E., et al. , 2009. Knockdown of human N alpha-terminal acetyltransferase complex C leads to p53-dependent apoptosis and aberrant human Arl8b localization. Mol. Cell. Biol. 29: 3569–3581. 10.1128/MCB.01909-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumner C. J., Crawford T. O., 2018. Two breakthrough gene-targeted treatments for spinal muscular atrophy: challenges remain. J. Clin. Invest. 128: 3219–3227. 10.1172/JCI121658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tizzano E. F., Finkel R. S., 2017. Spinal muscular atrophy: A changing phenotype beyond the clinical trials. Neuromuscul. Disord. 27: 883–889. 10.1016/j.nmd.2017.05.011 [DOI] [PubMed] [Google Scholar]
- Van Damme P., Lasa M., Polevoda B., Gazquez C., Elosegui-Artola A., et al. , 2012. N-terminal acetylome analyses and functional insights of the N-terminal acetyltransferase NatB. Proc. Natl. Acad. Sci. USA 109: 12449–12454. 10.1073/pnas.1210303109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varland S., Osberg C., Arnesen T., 2015. N-terminal modifications of cellular proteins: The enzymes involved, their substrate specificities and biological effects. Proteomics 15: 2385–2401. 10.1002/pmic.201400619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Dreyfuss G., 2001. A cell system with targeted disruption of the SMN gene: functional conservation of the SMN protein and dependence of Gemin2 on SMN. J. Biol. Chem. 276: 9599–9605. 10.1074/jbc.M009162200 [DOI] [PubMed] [Google Scholar]
- West S., Proudfoot N. J., Dye M. J., 2008. Molecular dissection of mammalian RNA polymerase II transcriptional termination. Mol. Cell 29: 600–610. 10.1016/j.molcel.2007.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C., Ishikawa H., Izumikawa K., Li L., He H., et al. , 2016. Structural insights into Gemin5-guided selection of pre-snRNAs for snRNP assembly. Genes Dev. 30: 2376–2390. 10.1101/gad.288340.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X., Mouillet J. F., Ou Q., Sadovsky Y., 2003. A novel domain within the DEAD-box protein DP103 is essential for transcriptional repression and helicase activity. Mol. Cell. Biol. 23: 414–423. 10.1128/MCB.23.1.414-423.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Fuller P. J., Morgan J., Shibata H., Clyne C. D., et al. , 2015. GEMIN4 functions as a coregulator of the mineralocorticoid receptor. J. Mol. Endocrinol. 54: 149–160. 10.1530/JME-14-0078 [DOI] [PubMed] [Google Scholar]
- Yong J., Kasim M., Bachorik J. L., Wan L., Dreyfuss G., 2010. Gemin5 delivers snRNA precursors to the SMN complex for snRNP biogenesis. Mol. Cell 38: 551–562. 10.1016/j.molcel.2010.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youkharibache P., Veretnik S., Li Q., Stanek K. A., Mura C., et al. , 2019. The Small β-barrel Domain: A Survey-based Structural Analysis. Structure 27: (In press). 10.1016/j.str.2018.09.012 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All fly strains, probe sequences and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. Supplemental material available at Figshare: https://doi.org/10.25387/g3.7473359.