Chapter Summary
Sortases cleave short peptide motif sequences at the C-terminal end of secreted surface protein precursors and either attach these polypeptides to the peptidoglycan of Gram-positive bacteria or promote their assembly into pilus structures that are also attached to peptidoglycan. Sortase A, the enzyme first identified in the human pathogen Staphylococcus aureus, binds LPXTG motif sorting signals, cleaves between threonine (T) and glycine (G) residues and forms an acyl-enzyme between its active site cysteine thiol and the carboxyl-group of threonine (T). Sortase A acyl enzyme is relieved by the nucleophilic attack of the crossbridge amino group within lipid II, thereby generating surface protein linked to peptidoglycan precursor. Such products are subsequently incorporated into the cell wall envelope by enzymes of the peptidoglycan synthesis pathway. Surface proteins linked to peptidoglycan may be released from the bacterial envelope to diffuse into host tissues and fulfill specific biological functions. S. aureus sortase A is essential for host colonization and for the pathogenesis of invasive diseases. Staphylococcal sortase-anchored surface proteins fulfill key functions during the infectious process and vaccine-induced antibodies targeting surface proteins may provide protection against S. aureus. Alternatively, small molecule inhibitors of sortase may be useful agents for the prevention S. aureus colonization and invasive disease.
Introduction
Prior to bacterial genome sequencing and the genetic analysis of pathogenesis, microbiologists identified molecules on microbial surfaces and studied their role in disease processes (1). Ultimate goal of this research was the identification molecular formulations inciting antibody responses in vaccine recipients that prevented disease yet would otherwise not cause harm (2). Oswald Avery’s discovery of the pneumococcus capsule and the demonstration that capsular polysaccharide vaccine protects against pneumococcal pneumonia, represents an important paradigm (3, 4). Another was Rebecca Lancefield’s characterization of M protein as the determinant of type-specific immunity against Streptococcus pyogenes, the causative agent of streptococcal pharyngitis and rheumatic fever (2). Lancefield and Sjöquist required proteases or peptidoglycan (murein) hydrolases, but not membrane detergents, to solubilize surface proteins of Gram-positive bacteria (2, 5, 6). The underlying reason for this biochemical phenomenon is that surface proteins are covalently linked to peptidoglycan at their C-terminal ends (7, 8).
Whole genome sequencing enabled bioinformatic studies providing rapid answers about the universality of genetic traits among pathogens or about sequence variation in response to host adaptive immune (antibody) responses (9). While bioinformatic analyses have had tremendous impact in supporting or refuting hypotheses about surface proteins in Gram-positive bacteria, experimental work represents the bedrock for hypothesis testing and for the alignment of arguments supporting bacterial vaccine development.
Staphylococcal sortases and their surface protein substrates
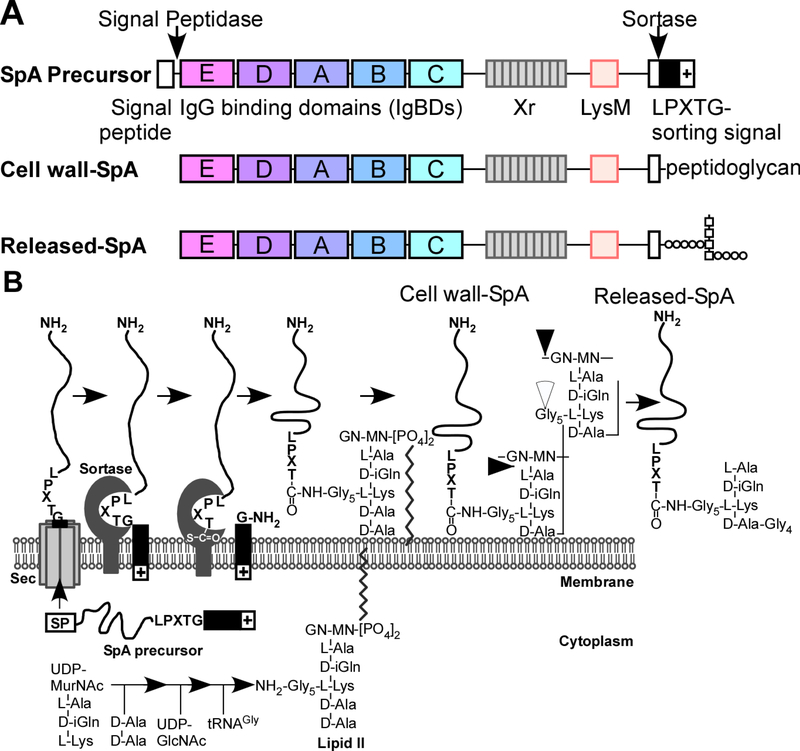
Surface proteins of S. aureus are amide linked to the pentaglycine crossbridge of the bacterial cell wall via their C-terminal threonine residue (8). Precursors of staphylococcal surface proteins are synthesized in the bacterial cytoplasm with N-terminal signal peptides for Sec-mediated secretion and C-terminal LPXTG motif sorting signals that promote cell wall anchoring (FIG. 1A) (10). Sortase A, a type II membrane protein (N-terminal membrane anchor) cleaves the LPXTG motif of the sorting signal between its threonine (T) and glycine (G) residues to form a thioester-linked acyl enzyme intermediate with its active site cysteine thiol (11, 12) (FIG. 1B). The acyl enzyme is relieved by the nucleophilic attack of the amino group of the pentaglycine crossbridge within lipid II, the precursor to peptidoglycan biosynthesis (13, 14) (FIG. 1B). Surface protein-linked to lipid II is subsequently incorporated into the cell wall envelope via the transglycosylation and transpeptidation reactions of bacterial cell wall synthesis (15–18) (FIG. 1B). S. aureus srtA (sortase A) mutants cannot assemble surface proteins into the cell wall envelope (19). The mechanism of action of S. aureus sortase A was validated for Listeria monocytogenes and Bacillus anthracis (20–22) and is considered to be universal in Gram-positive bacteria (23).
FIG. 1.
Sortase-mediated anchoring to the cell wall envelope of Staphylococcus aureus using SpA as a model substrate. (A) Drawing to illustrate the primary structure of the SpA precursor with its N-terminal signal peptide and signal peptidase cleavage site, the five immunoglobulin binding domains (IgBDs), region X (Xr) LysM domain and C-terminal LPXTG motif sorting signal with cleavage site for sortase A. Cell wall-SpA is linked to peptidoglycan via an amide bond between the carboxyl group of the C-terminal threonine and the amino group of the pentaglycine crossbridge. Released-SpA is liberated from the cell wall envelope via the action of several murein hydrolases. (B) Drawing to illustrate S. aureus secretion of SpA precursor, sortase-mediated cleavage of SpA precursor and acyl-enzyme formation, resolution of the acyl-enzyme by lipid II to generate SpA-linked to lipid II, incorporation of SpA into the cell wall via the transpeptidation and transglycosylation reaction, and release of SpA from the cell wall envelope by murein hydrolases. Released SpA bears the overall structure: L-Ala-D-iGln-L-Lys(SpA-LPET-Gly5)-D-Ala-Gly4.
Genome sequences of all clinical S. aureus isolates harbor two sortase genes, srtA and srtB, however the number of surface protein genes is variable (Table 1) (24–26). Sortase A substrates bear the LPXTG motif sorting signal at their C-terminal end (Table 1) (27). Sortase B cleaves the NPQTN sorting signal of IsdC (iron-regulated surface determinant C), a protein that is linked to the cell wall when staphylococci are grown under iron-starvation conditions, as occurs during host invasion (28). Several sortase A substrates have been described as microbial surface components recognizing adherence matrix molecules or MSCRAMMs (29). These include ClfA, ClfB, Cna, FnbpA, FnbpB, and presumably also Pls, SraP, SasG, SrdC and SdrD, albeit that the identify of surface protein ligands in the latter group of proteins remains unclear (Table 1). Each MSCRAMM represents a mosaic of modular domains (30, 31). A surface exposed, N-terminal, A domain is generally endowed with ligand-binding activity. Repeat structural modules allow MSCRAMMs to span the thick peptidoglycan layer of staphylococci (30, 31). ClfA, ClfB, SrdC, SdrD, SdrE, Pls, and SraP each encompass extensively glycosylated serine-aspartate (SD) repeat domains (32–34) (Table 1).
TABLE 1.
Staphylococcus aureus cell wall-anchored surface proteins 1.
| Sortase A anchored protein | Name(s) | Genbank accession number | aa 2 | Ligand(s) 3 | YSIRK Motif 4 | Sorting Motif 5 | Reference |
| Adenosine synthase A | AdsA (SasH) | ABD22278.1 | 772 | Adenosine and dAdo synthesis | No | LPKTG | (106, 108) |
| Clumping Factor A | ClfA | ABD20644.1 | 933 | Fibrinogen (γ chain) Factor I | Yes | LPDTG | (145, 146) |
| Clumping Factor B | ClfB | ABD21326.1 | 899 | Fibrinogen (α chain) Cytokeratin 8 & 10 Loricrin | Yes | LPETG | (97–102) |
| Collagen adhesin | Cna | BAF45800.1 | 1,183 | Collagen C1q | No | LPKTG | (147, 148) |
| Factor affecting methicillin resistance in Triton X-100 B | FmtB (SasB) | ATC68490.1 | 2,478 | Unknown | Yes | LPDTG | (149) |
| Fibronectin binding protein A | FnbpA | ABD21634.1 | 1,018 | Fibronectin Fibrinogen (γ chain) Elastin | Yes | LPETG | (30) |
| Fibronectin binding protein B | FnbpB | ABD22827.1 | 940 | Fibronectin Fibrinogen (α chain) Elastin | Yes | LPETG | (30) |
| Iron-regulated surface determinant A | IsdA (SasE) | ABD21627.1 | 350 | Heme transferred from IsdB/H | No | LPKTG | (35) |
| Iron-regulated surface determinant B | IsdB (SasJ) | ABD21843.1 | 645 | Hemoglobin Heme | Yes | LPQTG | (36–39) |
| Iron-regulated surface determinant H | IsdH (SasI/HarA) | ABD20516.1 | 895 | HaptoglobinHemoglobin Heme | Yes | LPKTG | (36–40) |
| Plasmin sensitive surface protein | Pls | AAD09131.1 | 1,637 | Unknown | Yes | LPDTG | (150, 151) |
| S. aureus surface -protein C | SasC | ABD21355.1 | 2,186 | Promotes intercellular adhesion | Yes | LPNTG | (152) |
| S. aureus surface -protein D | SasD | ABD21427.1 | 241 | Unknown | No | LPAAG | |
| S. aureus surface -protein F | SasF | ABD21199.1 | 635 | Unknown | No | LPKAG | |
| S. aureus surface -protein G | SasG | BAU36055.1 | 1,115 | Unknown | Yes | LPKTG | |
| S. aureus surface -protein K | SasK | ADC38744.1 | 211 | Unknown | No | LPKTG | |
| Serine aspartic repeat -protein C | SdrC | ABD21592.1 | 947 | β-neurexin Homophylic bonds | Yes | LPETG | (153, 154) |
| Serine aspartic repeat -protein D | SdrD | ABD20874.1 | 1,381 | Desmoglein 1 | Yes | LPETG | (155) |
| Serine aspartic repeat -protein E | SdrE | ABD22410.1 | 1,154 | Factor H | Yes | LPETG | (156) |
| S. aureus protein A | SpA | ABD22331.1 | 508 | Immunoglobulin (Fcγ, Fab VH3) | Yes | LPETG | (70, 71, 157, 158) |
| Serine-rich adhesin for platelets | SraP (SasA) | ABD21900.1 | 2,271 | Salivary agglutinin (gp340) | Possibly | LPDTG | (34, 159) |
| Sortase B anchored protein | Name(s) | Genbank accession number | aa1 | Ligand(s)2 | YSIRK motif3 | Sorting motif4 | Reference |
| Iron-regulated surface determinant C | IsdC | ABD20415.1 | 227 | Heme transferred from IsdA | No | NPQTN | (28) |
The number of cell wall-anchored surface proteins varies among strains 272 of S. aureus (26). For example, in strain S. aureus subsp. aureus USA300_FPR3757, genes for Cna, SasK, and Pls are missing; the presence of stop codons results in truncated FmtB (SasB), SasC and SasG products.
aa, protein length in amino acids.
Molecular component(s) recognized and bound by protein, or molecules synthesized in case of AdsA.
Consensus motif found in some signal sequences which presumably accounts for secretion of proteins at the cross walls (62).
Consensus motif recognized by sortases and present in C-terminal cell wall sorting signal.
The srtB and isdC genes are located in the isd locus, which also encodes sortase A-anchored products IsdA and IsdB, the membrane-transporter IsdEF, and the cytoplasmic protein IsdG (35). The structural gene for sortase A anchored IsdH is located outside of the isd locus (36). IsdB and IsdH function as hemophores to remove heme-iron from hemoglobin and haptoglobin when hemoproteins are released from lysed host cells (36–39). IsdH competes with macrophage receptor CD163, the host recycling system for free hemoglobin, for the capture of heme from haptoglobin-hemoglobin (40). Bound heme-iron is transferred from the NEAT (near-iron-transporter) domains of IsdB or IsdH to the NEAT-domain of IsdA for subsequent passage across the cell wall to IsdC and IsdEF-mediated import across the membrane (35). IsdG and its paralogue IsdI cleave the tetrapyrrole ring of heme-iron to liberate iron as a bacterial nutrient and enzyme co-factor (37, 41, 42). The sortase B-IsdC acyl enzyme intermediate is resolved by the nucleophilic attack of assembled peptidoglycan instead of lipid II (43). This mechanism ensures that IsdC is attached to peptidoglycan in the vicinity to the IsdEF membrane transporter, whereas IsdA and IsdB are deposited across the peptidoglycan layer (44).
Sortases and surface protein contributions to S. aureus colonization and disease pathogenesis
S. aureus srtA mutants cannot colonize the nasopharynx and gastrointestinal tract of mice (45, 46). Further, staphylococcal srtA mutants cannot form abscess lesions or survive in mouse tissues (19, 47). Following intravenous S. aureus inoculation to precipitate lethal bacteremia in mice or guinea pigs, srtA mutants are avirulent and cannot cause disease (48, 49). In the mouse skin abscess lesion and pneumonia models, S. aureus srtA mutants display smaller reductions in virulence. We attribute the smaller phenotypic defects to the models’ requirements for large bacterial inocula and α-hemolysin secretion (50–52). S. aureus srtB mutants exhibit small but significant reductions in virulence in the mouse renal abscess, bloodstream and infectious arthritis models; these defects are additive with those of sortase A mutants (53).
Cheng and co-workers isolated S. aureus Newman mutants with insertional lesions any one gene encoding LPXTG motif surface proteins. Unlike srtA variants, all mutants retained the ability to cause renal abscess lesions and lethal bacteremia in mice (47, 48). However, loss of spa (staphylococcal protein A), isdA and isdB resulted in significant reductions in the number of abscess lesions (47). Mutations in the genes for clumping factor A (clfA) or adenosine synthase A (adsA) caused significant delays in time-to-death in the murine model for S. aureus bacteremia (48). When analyzed with human nasal epithelial cells, cotton rats or mice as models for S. aureus colonization, srtA mutants are unable to colonize the nasopharynx and gastrointestinal tract (54–56). In these models, clumping factor B (ClfB) and IsdA, stand out as key contributors to S. aureus colonization (55, 57, 58). Thus, compared to any other virulence gene, srtA mutations exhibit the largest reduction in the ability of S. aureus to colonize and invade its hosts. Further, the sortase substrates AdsA, ClfA, ClfB, IsdA, IsdB, and SpA make important, non-redundant contributions towards colonization, invasion of host tissues or the establishment of abscess lesions.
Staphylococcal protein A (SpA)
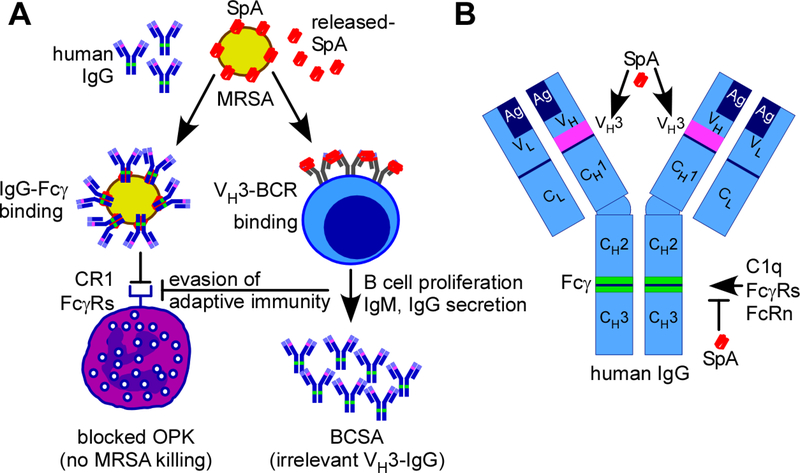
All clinical S. aureus isolates harbor the spa gene, which generates a precursor comprised of an N-terminal YSIRK/GXXS signal peptide, followed by 4–5 immunoglobulin binding domains (IgBDs), the region X repeats (Xr), LysM domain, and LPXTG sorting signal (23, 59, 60) (FIG. 1). SpA precursors enter the secretory pathway at septal membranes via their YSIRK/GXXS signal peptide (61–63). Once SpA is deposited into the cross wall, septal peptidoglycan is split and the cross wall assumes one-half of the spherical surface of S. aureus cells (61, 63). Staphylococci divide perpendicular to previous cell division planes resulting in rapid SpA distribution over the entire bacterial surface (61). During cell division, dedicated murein hydrolases release SpA molecules from the peptidoglycan (64, 65). SpA-linked to cell wall peptide fragments is thereby released into host tissues (66) (FIG. 1). Released SpA activates VH3 idiotype B cell receptors (BCRs) and promotes IgG and IgM secretion in activated plasmablasts (67, 68) (FIG. 2A). When displayed in the bacterial envelope, SpA binds to Fcγ, i.e. the effector domain of IgG, and protects staphylococci from opsonophagocytic killing by immune cells (49, 69) (FIG. 2A). The five IgBDs of SpA each bind to Fcγ of human (IgG1, IgG2 & IgG4) and mouse (IgG1, IgG2a-c & IgG3) IgG (70, 71) (FIG. 2B). Each IgBD also binds VH3 heavy chains of human and mouse immunoglobulin, including IgM (BCRs), IgG, IgE, IgD and IgA (49, 68, 69, 72, 73) (FIG. 2B). Thus, released SpA functions as a B cell superantigen that promotes systemic production of VH3-clonal IgG and IgM antibodies that do not recognize staphylococcal antigens, thereby preventing the development of pathogen-specific antibodies and the establishment of protective immunity (49, 67, 68). In spite of the B cell superantigen activity of SpA, S. aureus colonization and invasive disease in humans is associated with the development of antibody responses against some staphylococcal antigens, predominantly serum IgG4 (74–76). These antibodies are, however, not protective and cannot promote opsonophagocytic killing because they are captured by cell wall anchored SpA (71, 77–80).
FIG. 2.
Biological functions of staphylococcal protein A (SpA). (A) Staphylococcus aureus and its antibiotic-resistant isolates (MRSA) harbor SpA in the cell wall envelope or released into the extracellular milieu (released-SpA). Cell wall-SpA binds Fcγ of human and animal IgG (green segment within blue IgG) and blocks the effector functions of antibodies, thereby preventing opsonophagocytic killing (OPK) of MRSA by immune cells through interference with complement (CR1) and Fcγ receptors (FcγRs). Released-SpA crosslinks VH3-clonal B cell receptors (VH3-BCR on the surface of B cells), triggering B cell proliferation and secretion of VH3-clonal IgM and IgG (pink segments within blue IgG) without antigen-specificity for S. aureus. This B cell superantigen activity (BCSA) of SpA produces irrelevant VH3-clonal IgG and prevents the establishment of protective immunity against S. aureus. (B) Drawing to illustrate the primary structure of human IgG with variable (VL and VH) and conserved (CL and CH1, CH2 & CH3) light (L) and heavy (H) chains, their antigen-binding paratope (Ag), VH3 and Fcγ domains. SpA binding sites at VH3 heavy chains and Fc γ are identified in pink and green color, respectively.
Clumping factors A and B (ClfA and ClfB)
Vascular damage triggers blood coagulation, a process whereby soluble fibrinogen, a 340 kDa dimer of trimers (α-,β-, γ-chains), is converted to insoluble fibrin following cleavage of fibrinopeptides A and B from the α- and γ-chains by thrombin; the prothrombinase complex Va/Xa is responsible for the conversion of prothrombin (PT) to active thrombin (81–83). The hemostatic system also immobilizes microbial invaders for destruction by the immune system (84). However, this does not occur with S. aureus. All clinical S. aureus isolates clot human or animal blood even in the presence of coagulation inhibitors (85). Coagulation is promoted by secreted coagulase (Coa) and von-Willebrand-factor binding protein (vWbp) bound to PT (86). Coa-PT and vWbp-PT complexes cleave the A and B fibrinopeptides of fibrinogen but do not cut any of the other thrombin substrates (FV, FVIII, FXI, FXIII, protein C, antithrombin and plasmin) (87). ClfA triggers S. aureus agglutination by binding to the C-terminal end of the fibrinogen γ-chain (residues 395–411), effectively capping and tethering Coa-PT- and vWbp-PT-polymerized fibrin cables to the staphylococcal surface (48). ClfA, the prototypical MSCRAMM, is comprised of an N-terminal A domain with N1, N2, and N3 subdomains, an EF-hand like calcium binding module and the SD repeat domain with 154 tandem seryl-aspartyl repeats (88). The N2 and N3 domains of ClfA (residues 229–545) assume immunoglobulin-like folds and bind their fibrin/fibrinogen ligand via the “dock, lock, and latch” mechanism (89–93). This interaction prevents further binding between fibrin/fibrinogen and the platelet integrin αIIbβ3 (94, 95). Thus, in addition to binding fibrinogen, ClfA functions as an inhibitor of platelet-fibrin clots. ClfB, which is also conserved among S. aureus isolates, represents a homologue of ClfA. The A domains of the two proteins are 26% identical (96) and both proteins use YSIRK/GXXS signal peptides, glycosylated SD repeats and LPXTG motif sequences as topogenic elements (32, 62). ClfB binds to several host proteins, including the Aα-chain of fibrinogen (97, 98), cytokeratin 8 (99), cytokeratin 10 (100, 101), and loricrin (102) (Table 1). These mammalian proteins harbor a motif sequence, GSSGXG, that represents the binding site for ClfB (103) and and contributes to S. aureus colonization of nasopharynx of mice (102).
Adenosine Synthase A (AdsA)
S. aureus abscess lesions are composed of a bacterial nidus, the staphylococcal abscess community (SAC), encased within a pseudocapsule of fibrin, and surrounded by layers of immune cells (86, 104). In spite of large numbers of infiltrated neutrophils, mice are unable to eliminate staphylococci from abscess lesions and eventually succumb to the persistent infection (47). Although neutrophils use NETosis (extracellular DNA) to entangle staphylococci, NETs are degraded by staphylococcal nuclease (Nuc) and thereby fail to exert bactericidal activities (105). Nuclease digestion of NETs releases 5’ and 3’ monophosphate nucleotides that are converted by S. aureus AdsA into deoxyadenosine (dAdo)(106). AdsA-mediated dAdo production triggers caspase-3 induced apoptosis of mouse and human macrophages and prevents phagocyte entry into the SAC (106). Human equilibrative nucleoside transporter 1 is responsible for the uptake of dAdo in phagocytes (107). Conversion of dAdo to dAMP is catalyzed by deoxycytidine kinase and adenosine kinase, and the subsequent formation of dATP triggers caspase-3 induced cell death (107). AdsA also converts adenosine nucleosides and nucleotides released during host cell lysis into adenosine, which binds adenosine receptors and triggers host immune suppression during bloodstream infection (108, 109).
Using sortases and surface proteins for vaccine development
The contribution of sortases towards S. aureus colonization and invasive disease provoked interest in surface proteins as vaccine antigens. Purified recombinant ClfA (A domain) generates antibodies that neutralize ClfA binding to fibrin/ogen and provide partial protection against lethal bloodstream infection and infectious arthritis in mice (110). Anti-ClfA mouse hybridoma antibody or its cloned humanized variant tefibazumab bind to the ClfA N3 domain, inhibit fibrinogen binding (111, 112) and provide partial protection against lethal bloodstream infection in mice (113). Administration of clinical grade tefibazumab was safe in healthy human volunteers and in patients with methicillin-resistant S. aureus (MRSA) bacteremia but could not improve the clinical outcomes of these patients (114). Using ClfA immunized VelocImmune mice, MEDIMMUNE investigators isolated monoclonal antibody 11H10, with inhibitory activity for ClfA binding to fibrinogen (115). Human 11H10 IgG1 promotes MRSA opsonophagocytic killing with differentiated HL-60 neutrophils (115) and increases the survival of mice with lethal MRSA bloodstream infection (116, 117). MEDIMMUNE seeks to develop 11H10 IgG1 in conjunction with monoclonal antibody against α-hemolysin to improve the outcome of patients with ventilator associated pneumonia and other invasive diseases (115). PFIZER developed SA4Ag, a multicomponent vaccine composed of ClfA, capsular polysaccharide type 5 and 8 conjugates, and manganese transporter C (118). SA4Ag is currently undergoing clinical efficacy evaluation in patients with instrumented posterior spinal fusion to protect against S. aureus surgical site and bloodstream infections (119).
Purified IsdB elicits antibodies that block heme-iron scavenging and provide partial protection against S. aureus bacteremia in preclinical models (120–122). IsdB-specific antibodies may also promote opsonophagocytosis of S. aureus (121, 123). In a phase 3 clinical trial, IsdB (V710) immunization did not protect thoracic surgery patients from S. aureus surgical site infections (124). V710 immunization increased the risk for fatal S. aureus bacteremia five-fold over the control cohort; the molecular basis for this safety concern is not known (124).
Humans and mice cannot generate antibodies against the IgBDs of SpA, however SpA variants, engineered to exhibit reduced immunoglobulin binding, elicit SpA-neutralizing antibody responses (73). Animals with SpA-neutralizing antibodies exhibit dramatic increases in pathogen-specific antibody responses during colonization or invasive disease (46, 49, 69, 73). In fact, the corresponding SpA vaccine can protect against S. aureus colonization, renal abscess formation and lethal bloodstream infection (46, 49, 69, 73). Similarly, SpA-neutralizing monoclonal antibody protects against S. aureus colonization and invasive disease in mice (125, 126). SpA vaccines have not yet been subjected to clinical testing.
Sortase inhibitors
The complete transpeptidation reaction that is carried out by sortases can be recapitulated in vitro (12, 127, 128). However, most screens for sortase inhibitors have been conducted with assays measuring SrtA cleavage of LPXTG peptide (129). These inhibitors are generally not active in vivo, suggesting that in the envelope of S. aureus, sortase A may predominantly exist as an acyl-enzyme (130). Other inhibitors can block sortase A activity in vivo and such compounds abolish surface protein anchoring to the cell wall envelope of S. aureus and protect animals against lethal bloodstream infection (131, 132). Of note, sortase inhibitors may be useful for the prevention of S. aureus disease, as they can be expected to block colonization and invasion. Owing to the fact that the compounds cannot kill S. aureus, sortase inhibitors are unlikely to exhibit a therapeutic effect in individuals with active infectious disease (131).
Sortases in other pathogenic microbes
Gram-positive bacteria often harbor homologs of staphylococcal sortase A or class A sortases; only some microbes express sortase B homologs or class B sortases (133, 134). Based on structural features and substrate specificity, sortase homologs have been classified into six distinct classes A–F (135). Amongst bacterial pathogens, Corynebacterium diphtheriae and Bacillus anthracis harbor class C sortase genes, which are clustered with surface protein genes containing LPXTG- and motif specific sorting signals (136, 137).These genes encode pilus component: adhesin and pilin subunits. Class C sortases link adhesin and pilin subunits together to construct a pilus (136–140). Class C sortases cleave the LPXTG motif of pilins to form acyl-enzyme intermediates that are relieved by the nucleophilic attack of the ε-amino group of a conserved lysine (K) residue within the pilin motif of an incoming subunit (141–143). Pilin protomers are joined progressively to the pilus base; a housekeeping sortase terminates polymerization by transferring the whole structure to the peptidoglycan (142, 144). For additional information on the different classes of sortases and their distribution among various phyla, the reader is referred to a recently published review (135).
In conclusion, sortases are ubiquitous in Gram-positive bacteria, anchoring proteins and pili to peptidoglycan via a conserved transpeptidation mechanism. Sortase-mediated attachment of virulence factors in S. aureus has stimulated searches for sortase inhibitors and protective antigens. These strategies may lead to the development of drugs that can prevent hospital-acquired infections or to protective vaccines that can prevent S. aureus colonization and/or invasive diseases.
Acknowledgements
We thank laboratory members past and present for their contributions to the field of S. aureus sortases and surface proteins. Work on Staphylococcus aureus in the laboratories of the authors is supported by grants AI038897, AI052474, and AI110937 from the National Institute of Allergy and Infectious Diseases. The authors declare conflicts of interest as inventors of patents under commercial license for S. aureus vaccine development. The authors declare no further competing financial interests.
References
- 1.Lancefield RC. 1928. The antigenic complex of Streptococcus hemolyticus. I. Demonstration of a type-specific substance in extracts of Streptococcus hemolyticus. J Exp Med 47:91–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lancefield R 1962. Current knowledge of type-specific M antigens of group A streptococci. J Immunol 89:307–313. [PubMed] [Google Scholar]
- 3.Avery OT. 1915. A further study on the biologic classification of pneumococci. J Exp Med 22:804–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.MacLeod CM, Hodges RG, Heidelberger M, Bernhard WG. 1945. Prevention of pneumococcal pneumonia by immunization with specific capsular polysaccharides. J Exp Med 82:445–465. [PMC free article] [PubMed] [Google Scholar]
- 5.Sjöquist J, Meloun B, Hjelm H. 1972. Protein A isolated from Staphylococcus aureus after digestion with lysostaphin. Eur J Biochem 29:572–578. [DOI] [PubMed] [Google Scholar]
- 6.Fischetti VA. 1989. Streptococcal M protein: molecular design and biological behavior. Clin Microbiol Rev 2:285–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schneewind O, Fowler A, Faull KF. 1995. Structure of the cell wall anchor of surface proteins in Staphylococcus aureus. Science 268:103–106. [DOI] [PubMed] [Google Scholar]
- 8.Marraffini LA, DeDent AC, Schneewind O. 2006. Sortases and the art of anchoring proteins to the envelopes of gram-positive bacteria. Microbiol Mol Biol Rev 70:192–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Musser JM, Shelburne SAr. 2009. A decade of molecular pathogenomic analysis of group A Streptococcus. J Clin Invest 119:2455–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schneewind O, Model P, Fischetti VA. 1992. Sorting of protein A to the staphylococcal cell wall. Cell 70:267–281. [DOI] [PubMed] [Google Scholar]
- 11.Mazmanian SK, Liu G, Ton-That H, Schneewind O. 1999. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science 285:760–763. [DOI] [PubMed] [Google Scholar]
- 12.Ton-That H, Liu G, Mazmanian SK, Faull KF, Schneewind O. 1999. Purification and characterization of sortase, the transpeptidase that cleaves surface proteins of Staphylococcus aureus at the LPXTG motif. Proc Natl Acad Sci USA 96:12424–12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perry AM, Ton-That H, Mazmanian SK, Schneewind O. 2002. Anchoring of surface proteins to the cell wall of Staphylococcus aureus. III. Lipid II is an in vivo peptidoglycan substrate for sortase-catalyzed surface protein anchoring. J Biol Chem 277:16241–16248. [DOI] [PubMed] [Google Scholar]
- 14.Ton-That H, Mazmanian SK, Faull KF, Schneewind O. 2000. Anchoring of surface proteins to the cell wall of Staphylococcus aureus. I. Sortase catalyzed in vitro transpeptidation reaction using LPXTG peptide and NH2-Gly3 substrates. J Biol Chem 275:9876–9881. [DOI] [PubMed] [Google Scholar]
- 15.Ton-That H, Labischinski H, Berger-Bächi B, Schneewind O. 1998. Anchor structure of staphyococcal surface proteins. III. The role of the FemA, FemB, and FemX factors in anchoring surface proteins to the bacterial cell wall. J Biol Chem 273:29143–29149. [DOI] [PubMed] [Google Scholar]
- 16.Ton-That H, Schneewind O. 1999. Anchor structure of staphylococcal surface proteins. IV. Inhibitors of the cell wall sorting reaction. J Biol Chem 274:24316–24320. [DOI] [PubMed] [Google Scholar]
- 17.Ton-That H, Faull KF, Schneewind O. 1997. Anchor structure of staphylococcal surface proteins. I. A branched peptide that links the carboxyl terminus of proteins to the cell wall. J Biol Chem 272:22285–22292. [DOI] [PubMed] [Google Scholar]
- 18.Navarre WW, Ton-That H, Faull KF, Schneewind O. 1998. Anchor structure of staphylococcal surface proteins. II. COOH-terminal structure of muramidase and amidase-solubilized surface protein. J Biol Chem 273:29135–29142. [DOI] [PubMed] [Google Scholar]
- 19.Mazmanian SK, Liu G, Jensen ER, Lenoy E, Schneewind O. 2000. Staphylococcus aureus mutants defective in the display of surface proteins and in the pathogenesis of animal infections. Proc Natl Acad Sci USA 97:5510–5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dhar G, Faull KF, Schneewind O. 2000. Anchor structure of cell wall surface proteins in Listeria monocytogenes. Biochemistry 39:3725–3733. [DOI] [PubMed] [Google Scholar]
- 21.Bierne H, Mazmanian SK, Trost M, Pucciarelli MG, Dehoux P, Jansch L, Portillo FG, G. L, Schneewind O, Cossart P. 2002. Inactivation of the srtA gene in Listeria monocytogenes inhibits anchoring of surface proteins and affects virulence. Mol Microbiol 43:869–881. [DOI] [PubMed] [Google Scholar]
- 22.Gaspar AH, Marraffini LA, Glass EM, DeBord KL, Ton-That H, Schneewind O. 2005. Bacillus anthracis sortase A (SrtA) anchors LPXTG motif-containing surface proteins to the cell wall envelope. J Bacteriol 187:4646–4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mazmanian SK, Ton-That H, Schneewind O. 2001. Sortase-catalyzed anchoring of surface proteins to the cell wall of Staphylococcus aureus. Mol Microbiol 40:1049–1057. [DOI] [PubMed] [Google Scholar]
- 24.Baba T, Bae T, Schneewind O, Takeuchi F, Hiramatsu K. 2007. Genome sequence of Staphylococcus aureus strain Newman and comparative analysis of staphylococcal genomes. J Bacteriol 190:300–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuroda M, Ohta T, Uchiyama I, Baba T, Yuzawa H, Kobayashi I, Cui L, Oguchi A, Aoki K, Nagai Y, Lian J, Ito T, Kanamori M, Matsumaru H, Maruyama A, Murakami H, Hosoyama A, Mitsutani-Ui Y, Kobayashi N, Sawano T, Inoue R, Kaito C, Sekimizu K, Hirakawa H, Kuhara S, Goto S, Yabuzaki J, Kanehisa M, Yamashita A, Oshima K, Furuya K, Yoshino C, Shiba T, Hattori M, Ogasawara N, Hayashi H, Hiramatsu K. 2001. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet 357:1225–1240. [DOI] [PubMed] [Google Scholar]
- 26.McCarthy AJ, Lindsay JA. 2010. Genetic variation in Staphylococcus aureus surface and immune evasion genes is lineage associated: implications for vaccine design and host-pathogen interactions. BMC Microbiol 10:173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schneewind O, Mihaylova-Petkov D, Model P. 1993. Cell wall sorting signals in surface protein of Gram-positive bacteria. EMBO 12:4803–4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mazmanian SK, Ton-That H, Su K, Schneewind O. 2002. An iron-regulated sortase enzyme anchors a class of surface protein during Staphylococcus aureus pathogenesis. Proc Natl Acad Sci USA 99:2293–2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patti JM, Allen BL, McGavin MJ, Hook M. 1994. MSCRAMM-mediated adherence of microorganisms to host tissues. Annu Rev Microbiol 48:585–617. [DOI] [PubMed] [Google Scholar]
- 30.Foster TJ. 2016. The remarkably multifunctional fibronectin binding proteins of Staphylococcus aureus. Eur J Clin Microbiol Infect Dis 35:1923–1931. [DOI] [PubMed] [Google Scholar]
- 31.Foster TJ, Geoghegan JA, Ganesh VK, Hook M. 2014. Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus. Nat Rev Microbiol 12:49–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thomer L, Becker S, Emolo C, Quach A, Kim HK, Rauch S, Faull KF, Schneewind O, Missiakas DM. 2014. N-acetylglucosaminylation of serine-aspartate repeat proteins promotes Staphylococcus aureus blood stream infection. J Biol Chem 289:3478–3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bleiziffer I, Eikmeier J, Pohlentz G, McAulay K, Xia G, Hussain M, Peschel A, Foster S, Peters G, Heilmann C. 2017. The Plasmin-Sensitive Protein Pls in Methicillin-Resistant Staphylococcus aureus (MRSA) Is a Glycoprotein. PLoS Pathog 13:e1006110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Siboo IR, Chambers HF, Sullam PM. 2005. Role of SraP, a Serine-Rich Surface Protein of Staphylococcus aureus, in binding to human platelets. Infect Immun 73:2273–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mazmanian SK, Skaar EP, Gaspar AH, Humayun M, Gornicki P, Jelenska J, Joachmiak A, Missiakas DM, Schneewind O. 2003. Passage of heme-iron across the envelope of Staphylococcus aureus. Science 299:906–909. [DOI] [PubMed] [Google Scholar]
- 36.Dryla A, Gelbmann D, von Gabain A, Nagy E. 2003. Identification of a novel iron regulated staphylococcal surface protein with haptoglobin-haemoglobin binding activity. Mol Microbiol 49:37–53. [DOI] [PubMed] [Google Scholar]
- 37.Skaar EP, Humayun M, Bae T, DeBord KL, Schneewind O. 2004. Iron-source preference of Staphylococcus aureus infections. Science 305:1626–1628. [DOI] [PubMed] [Google Scholar]
- 38.Pishchany G, Sheldon JR, Dickson CF, Alam MT, Read TD, Gell DA, Heinrichs DE, Skaar EP. 2014. IsdB-dependent hemoglobin binding is required for acquisition of heme by Staphylococcus aureus. J Infect Dis 209:1764–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Choby JE, Skaar EP. 2016. Heme Synthesis and Acquisition in Bacterial Pathogens. J Mol Biol 428:3408–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saederup KL, Stodkilde K, Graversen JH, Dickson CF, Etzerodt A, Hansen SW, Fago A, Gell D, Andersen CB, Moestrup SK. 2016. The Staphylococcus aureus Protein IsdH Inhibits Host Hemoglobin Scavenging to Promote Heme Acquisition by the Pathogen. J Biol Chem 291:23989–23998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Skaar EP, Gaspar AH, Schneewind O. 2004. IsdG and IsdI, heme degrading enzymes in the cytoplasm of Staphylococcus aureus. J Biol Chem 279:436–443. [DOI] [PubMed] [Google Scholar]
- 42.Reniere ML, Ukpabi GN, Harry SR, Stec DF, Krull R, Wright DW, Bachmann BO, Murphy ME, Skaar EP. 2010. The IsdG-family of haem oxygenases degrades haem to a novel chromophore. Mol Microbiol 75:1529–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marraffini LA, Schneewind O. 2005. Anchor structure of staphylococcal surface proteins. V. Anchor structure of the sortase B substrate IsdC. J Biol Chem 280:16263–16271. [DOI] [PubMed] [Google Scholar]
- 44.Maresso AW, Schneewind O. 2006. Iron acquisition and transport in Staphylococcus aureus. Biometals 19:193–203. [DOI] [PubMed] [Google Scholar]
- 45.Kiser KB, Cantey-Kiser JM, Lee JC. 1999. Development and characterization of a Staphylococcus aureus nasal colonization model in mice. Infect Immun 67:5001–5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun Y, Emolo CE, Holtfreter S, Wiles S, Kreiswirth B, Missiakas D, Schneewind O. 2018. Staphylococcal protein A is required for persistent colonization of mice with Staphylococcus aureus. J Bacteriol EPub:ahead of press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cheng AG, Kim HK, Burts ML, Krausz T, Schneewind O, Missiakas DM. 2009. Genetic requirements for Staphylococcus aureus abscess formation and persistence in host tissues. FASEB J 23:3393–3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McAdow M, Kim HK, DeDenta AC, Hendrickx APA, Schneewind O, Missiakas DM. 2011. Preventing Staphylococcus aureus sepsis through the inhibition of its agglutination in blood. PLoS Pathog 7:e1002307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim HK, Falugi F, Thomer L, Missiakas DM, Schneewind O. 2015. Protein A suppresses immune responses during Staphylococcus aureus bloodstream infection in guinea pigs. mBio 6:e02369–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bubeck-Wardenburg J, Patel R, Schneewind O. 2007. Surface proteins and exotoxins are required for the pathogenesis of Staphylococcus aureus pneumonia. Infect Immun 74:1040–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bubeck Wardenburg J, Schneewind O. 2008. Vaccine protection against Staphylococcus aureus pneumonia. J Exp Med 205:287–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kennedy AD, Bubeck-Wardenburg J, Gardner DJ, Long D, Whitney AR, Braughton KR, Schneewind O, DeLeo FR. 2010. Targeting of alpha-hemolysin by active or passive immunization decreases severity of USA300 skin infection in a mouse model. J Infect Dis 202:1050–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jonsson IM, Mazmanian SK, Schneewind O, Bremell T, Tarkowski A. 2003. The role of Staphylococcus aureus sortase A and sortase B in murine arthritis. Microb Infect 5:775–780. [DOI] [PubMed] [Google Scholar]
- 54.Corrigan RM, Miajlovic H, Foster TJ. 2009. Surface proteins that promote adherence of Staphylococcus aureus to human desquamated nasal epithelial cells. BMC Microbiol 9:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schaffer AC, Solinga RM, Cocchiaro J, Portoles M, Kiser KB, Risley A, Randall SM, Valtulina V, Speziale P, Walsh E, Foster T, Lee JC. 2006. Immunization with Staphylococcus aureus clumping factor B, a major determinant in nasal carriage, reduces nasal colonization in a murine model. Infect Immun 74:2145–2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Misawa Y, Kelley KA, Wang X, Wang L, Park WB, Birtel J, Saslowsky D, Lee JC. 2015. Staphylococcus aureus colonization of the mouse gastrointestinal tract Is modulated by wall teichoic acid, capsule, and surface proteins. PLoS Pathog 11:e1005061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Clarke SR, Brummell KJ, Horsburgh MJ, McDowell PW, Mohamad SA, Stapleton MR, Acevedo J, Read RC, Day NP, Peacock SJ, Mond JJ, Kokai-Kun JF, Foster SJ. 2006. Identification of in vivo-expressed antigens of Staphylococcus aureus and their use in vaccinations for protection against nasal carriage. J Infect Dis 193:1098–1108. [DOI] [PubMed] [Google Scholar]
- 58.Wertheim HF, Walsh E, Choudhurry R, Melles DC, Boelens HA, Miajlovic H, Verbrugh HA, Foster T, van Belkum A. 2008. Key role for clumping factor B in Staphylococcus aureus nasal colonization of humans. PLoS Med 5:e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Forsgren A 1970. Significance of protein A production by staphylococci. Infect Immun 2:672–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Votintseva AA, Fung R, Miller RR, Knox K, Godwin H, Wyllie DH, Bowden R, Crook DW, Walker AS. 2014. Prevalence of Staphylococcus aureus protein A (spa) mutants in the community and hospitals in Oxfordshire. BMC Microbiol 14:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.DeDent AC, McAdow M, Schneewind O. 2007. Distribution of protein A on the surface of Staphylococcus aureus. J Bacteriol 189:4473–4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.DeDent AC, Missiakas DM, Schneewind O. 2008. Signal peptides direct surface proteins to two distinct envelope locations of Staphylococcus aureus. EMBO J 27:2656–2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yu W, Missiakas D, Schneewind O. 2018. Septal secretion of protein A in Staphylococcus aureus requires SecA and lipoteichoic acid synthesis. Elife 7:e34092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Frankel MB, Hendrickx AP, Missiakas DM, Schneewind O. 2011. LytN, a murein hydrolase in the cross-wall compartment of Staphylococcus aureus, is involved in proper bacterial growth and envelope assembly. J Biol Chem 286:32593–32605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Frankel MB, Schneewind O. 2012. Determinants of murein hydrolase targeting to cross-wall of Staphylococcus aureus peptidoglycan. J Biol Chem 287:10460–10471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Becker S, Frankel MB, Schneewind O, Missiakas DM. 2014. Release of protein A from the cell wall envelope of Staphylococcus aureus. Proc Natl Acad Sci USA 111:1574–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim HK, Falugi F, Missiakas D, Schneewind O. 2016. Peptidoglycan-linked protein A promotes T-cell dependent antibody expansion during Staphylococcus aureus infection. Proc Natl Acad Sci USA 113:5718–5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pauli NT, Kim HK, Falugi F, Huang M, Dulac J, Dunand CH, Zheng NY, Kaur K, Andrews S, Huang Y, Dedent A, Frank K, Charnot-Katsikas A, Schneewind O, Wilson PC. 2014. Staphylococcus aureus infection induces protein A-mediated immune evasion in humans. J Exp Med 211:2331–2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Falugi F, Kim HK, Missiakas DM, Schneewind O. 2013. The role of protein A in the evasion of host adaptive immune responses by Staphylococcus aureus mBio 4:e00575–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Forsgren A, Sjöquist J. 1966. Protein A from S. aureus. I. Pseudo-immune reaction with human gamma-globulin. J Immunol 97:822–827. [PubMed] [Google Scholar]
- 71.Forsgren A 1968. Protein A from Staphylococcus aureus. VI. Reaction with subunits from guinea pig γ1- and γ2-globulin. J Immunol 100:927–930. [PubMed] [Google Scholar]
- 72.Sasso EH, Silverman GJ, Mannik M. 1989. Human IgM molecules that bind staphylococcal protein A contain VHIII H chains. J Immunol 142:2778–2783. [PubMed] [Google Scholar]
- 73.Kim HK, Cheng AG, Kim H-Y, Missiakas DM, Schneewind O. 2010. Non-toxigenic protein A vaccine for methicillin-resistant Staphylococcus aureus infections. J Exp Med 207:1863–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Verkaik NJ, Lebon A, de Vogel CP, Hooijkaas H, Verbrugh HA, Jaddoe VW, Hofman A, Moll HA, van Belkum A, van Wamel WJ. 2010. Induction of antibodies by Staphylococcus aureus nasal colonization in young children. Clin Microbiol Infect 16:1312–1317. [DOI] [PubMed] [Google Scholar]
- 75.Swierstra J, Debets S, de Vogel C, Lemmens-den Toom N, Verkaik N, Ramdani-Bouguessa N, Jonkman MF, van Dijl JM, Fahal A, van Belkum A, van Wamel W. 2015. IgG4 subclass-specific responses to Staphylococcus aureus antigens shed new light on host-pathogen interaction. Infect Immun 83:492–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Holtfreter S, Jursa-Kulesza J, Masiuk H, Verkaik NJ, de Vogel C, Kolata J, Nowosiad M, Steil L, van Wamel W, van Belkum A, Völker U, Giedrys-Kalemba S, Bröker BM. 2011. Antibody responses in furunculosis patients vaccinated with autologous formalin-killed Staphylococcus aureus. Eur J Clin Microbiol Infect Dis 30:707–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kluytmans J, van Belkum A, Verburgh H. 1997. Nasal carriage of Staphylococcus aureus: epidemiology, underlying mechanisms, and associated risks. Clin Microbiol Rev 10:505–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Weinstein HJ. 1959. The relation between nasal-staphylococcal-carrier state and the incidence of postoperative complications. N Engl J Med 260:1303–1308. [DOI] [PubMed] [Google Scholar]
- 79.Wertheim HF, Melles DC, Vos MC, van Leeuwen W, van Belkum A, Verbrugh HA, Nouwen JL. 2005. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect Dis 5:751–62. [DOI] [PubMed] [Google Scholar]
- 80.Forsgren A, Quie PG. 1974. Effects of staphylococcal protein A on heat labile opsonins. J Immunol 112:1177–1180. [PubMed] [Google Scholar]
- 81.Adams RL, Bird RJ. 2009. Review article: Coagulation cascade and therapeutics update: relevance to nephrology. Part 1: Overview of coagulation, thrombophilias and history of anticoagulants. Nephrology 14:462–70. [DOI] [PubMed] [Google Scholar]
- 82.Doolittle RF. 2003. Structural basis of the fibrinogen-fibrin transformation: contributions from X-ray crystallography. Blood Rev 17:33–41. [DOI] [PubMed] [Google Scholar]
- 83.Ware S, Donahue JP, Hawiger J, Anderson WF. 1999. Structure of the fibrinogen gamma-chain integrin binding and factor XIIIa cross-linking sites obtained through carrier protein driven crystallization. Protein Sci 8:2663–2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Levi M, Keller TT, van Gorp E, ten Cate H. 2003. Infection and inflammation and the coagulation system. Cardiovasc Res 60:26–39. [DOI] [PubMed] [Google Scholar]
- 85.Much H 1908. Über eine Vorstufe des Fibrinfermentes in Kulturen von Staphylokokkus aureus. Biochem Z 14:143–155. [Google Scholar]
- 86.Cheng AG, McAdow M, Kim HK, Bae T, Missiakas DM, Schneewind O. 2010. Contribution of coagulases towards Staphylococcus aureus disease and protective immunity. PLoS Pathog 6:e1001036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Thomer L, Schneewind O, Missiakas D. 2016. Pathogenesis of Staphylococcus aureus Bloodstream Infections. Annu Rev Pathol 11:343–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.O’Connell DP, Nanavaty T, McDevitt D, Gurusidappa S, Hook M, Foster TJ. 1998. The fibrinogen-binding MSCRAMM (clumping factor) of Staphylococcus aureus has a Ca2+-dependent inhibitory site. J Biol Chem 273:6821–6829. [DOI] [PubMed] [Google Scholar]
- 89.Strong DD, Laudano AP, Hawiger J, Doolittle RF. 1982. Isolation, characterization and synthesis of peptides from human fibrinogen that block the staphylococcal clumping reaction and construction of a synthetic clumping particle. Biochemistry 21:1414–1420. [DOI] [PubMed] [Google Scholar]
- 90.Ganesh VK, Rivera JJ, Smeds E, Ko Y-P, Bowden MG, Wann ER, Gurusidappa S, Fitzgerald JR, Höök M. 2008. A structural model of the Staphylococcus aureus ClfA-fibrinogen interaction opens new avenues for the design of anti-staphylococcal therapeutics. PLoS Pathog 4:e1000226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ponnuraj K, Bowden MG, Davis S, Gurusiddappa S, Moore D, Choe D, Xu Y, Hook M, Narayana SV. 2003. A “dock, lock, and latch” structural model for a staphylococcal adhesin binding to fibrinogen. Cell 115:217–228. [DOI] [PubMed] [Google Scholar]
- 92.Bowden MG, Heuck AP, Ponnuraj K, Kolosova E, Choe D, Gurusiddappa S, Narayana SV, Johnson AE, Höök M. 2008. Evidence for the “dock, lock, and latch” ligand binding mechanism of the staphylococcal microbial surface component recognizing adhesive matrix molecules (MSCRAMM) SdrG. J Biol Chem 283:638–647. [DOI] [PubMed] [Google Scholar]
- 93.Flick MJ, Du X, Prasad JM, Raghu H, Palumbo JS, Smeds E, Höök M, Degen JL. 2013. Genetic elimination of the binding motif on fibrinogen for the S. aureus virulence factor ClfA improves host survival in septicemia. Blood in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.O’Brien L, Kerrigan SW, Kaw G, Hogan M, Penades J, Litt D, Fitzgerald DJ, Foster TJ, Cox D. 2002. Multiple mechanisms for the activation of human platelet aggregation by Staphylococcus aureus: roles for the clumping factors ClfA and ClfB, the serine-aspartate repeat protein SdrE and protein A. Molecular microbiology 44:1033–44. [DOI] [PubMed] [Google Scholar]
- 95.Loughman A, Fitzgerald JR, Brennan MP, Higgins J, Downer R, Cox D, Foster TJ. 2005. Roles for fibrinogen, immunoglobulin and complement in platelet activation promoted by Staphylococcus aureus clumping factor A. Mol Microbiol 57:804–18. [DOI] [PubMed] [Google Scholar]
- 96.Ní Eidhin D, Perkins S, Francois P, Vaudaux P, Höök M, Foster TJ. 1998. Clumping factor B (ClfB), a new surface-located fibrinogen-binding adhesin of Staphylococcus aureus. Mol Microbiol 30:245–257. [DOI] [PubMed] [Google Scholar]
- 97.Walsh EJ, Miajlovic H, Gorkun OV, Foster TJ. 2008. Identification of the Staphylococcus aureus MSCRAMM clumping factor B (ClfB) binding site in the alphaC-domain of human fibrinogen. Microbiology 154:550–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Perkins S, Walsh EJ, Deivanayagam CC, Narayana SV, Foster TJ, Hook M. 2001. Structural organization of the fibrinogen-binding region of the clumping factor B MSCRAMM of Staphylococcus aureus. J Biol Chem 276:44721–8. [DOI] [PubMed] [Google Scholar]
- 99.Haim M, Trost A, Maier CJ, Achatz G, Feichtner S, Hintner H, Bauer JW, Onder K. 2010. Cytokeratin 8 interacts with clumping factor B: a new possible virulence factor target. Microbiology 156:3710–3721. [DOI] [PubMed] [Google Scholar]
- 100.Walsh EJ, O’Brien LM, Liang X, Hook M, Foster TJ. 2004. Clumping factor B, a fibrinogen-binding MSCRAMM (microbial surface components recognizing adhesive matrix molecules) adhesin of Staphylococcus aureus, also binds to the tail region of type I cytokeratin 10. J Biol Chem 279:50691–9. [DOI] [PubMed] [Google Scholar]
- 101.O’Brien LM, Walsh EJ, Massey RC, Peacock SJ, Foster TJ. 2002. Staphylococcus aureus clumping factor B (ClfB) promotes adherence to human type I cytokeratin 10: implications for nasal colonization. Cell Microbiol 4:759–770. [DOI] [PubMed] [Google Scholar]
- 102.Mulcahy ME, Geoghegan JA, Monk IR, O’Keeffe KM, Walsh EJ, Foster TJ, McLoughlin RM. 2012. Nasal colonisation by Staphylococcus aureus depends upon clumping factor B binding to the squamous epithelial cell envelope protein loricrin. PLoS Pathog 8:e1003092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ganesh VK, Barbu EM, Deivanayagam CC, Le B, Anderson AS, Matsuka YV, Lin SL, Foster TJ, Narayana SV, Höök M. 2011. Structural and biochemical characterization of Staphylococcus aureus clumping factor B/ligand interactions. J Biol Chem 286:25963–25972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cheng AG, DeDent AC, Schneewind O, Missiakas DM. 2011. A play in four acts: Staphylococcus aureus abscess formation. Trends Microbiol 19:225–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Berends ET, Horswill AR, Haste NM, Monestier M, Nizet V, von Köckritz-Blickwede M. 2010. Nuclease expression by Staphylococcus aureus facilitates escape from neutrophil extracellular traps. J Innate Immun 2:576–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Thammavongsa V, Missiakas DM, Schneewind O. 2013. Staphylococcus aureus conversion of neutrophil extracellular traps into deoxyadenosine promotes immune cell death Science 342:863–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Winstel V, Missiakas D, Schneewind O. 2018. Staphylococcus aureus targets the purine salvage pathway to kill phagocytes. Proc Natl Acad Sci USA 115:6846–6851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Thammavongsa V, Kern JW, Missiakas DM, Schneewind O. 2009. Staphylococcus aureus synthesizes adenosine to escape host immune responses. J Exp Med 206:2417–2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Thammavongsa V, Schneewind O, Missiakas DM. 2011. Enzymatic properties of Staphylococcus aureus adenosine synthase (AdsA). BMC Biochem 12:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Josefsson E, Higgins J, Foster TJ, Tarkowski A. 2008. Fibrinogen binding sites P336 and Y338 of clumping factor are crucial for Staphylococcus aureus virulence. PLoS One 3:e2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Domanski PJ, Patel PR, Bayer AS, Zhang L, Hall AE, Syribeys PJ, Gorovits EL, Bryant D, Vernachio JH, Hutchins JT, Patti JM. 2005. Characterization of a humanized monoclonal antibody recognizing clumping factor A expressed by Staphylococcus aureus. Infect Immun 73:5229–5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ganesh VK, Liang X, Geoghegan JA, Cohen AL, Venugopalan N, Foster TJ, Hook M. 2016. Lessons from the crystal structure of the S. aureus surface protein clumping factor A in complex with tefibazumab, an inhibiting monoclonal antibody. EBioMedicine 13:328–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hall AE, Domanski PJ, Patel PR, Vernachio JH, Syribeys PJ, Gorovits EL, Johnson MA, Ross JM, Hutchins JT, Patti JM. 2003. Characterization of a protective monoclonal antibody recognizing Staphylococcus aureus MSCRAMM protein clumping factor A. Infect Immun 71:6864–6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Weems JJ Jr, Steinberg JP, Filler S, Baddley JW, Corey GR, Sampathkumar P, Winston L, John JF, Kubin CJ, Talwani R, Moore T, Patti JM, Hetherington S, Texter M, Wenzel E, Kelley VA, Fowler VG Jr. 2006. Phase II, randomized, double-blind, multicenter study comparing the safety and pharmacokinetics of Tefibazumab to placebo for treatment of Staphylococcus aureus bacteremia. Antimicrob Agents Chemother 50:2751–2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tkaczyk C, Hamilton MM, Sadowska A, Shi Y, Chang CS, Chowdhury P, Buonapane R, Xiao X, Warrener P, Mediavilla J, Kreiswirth B, Suzich J, Stover CK, Sellman BR. 2016. Targeting alpha toxin and ClfA with a multimechanistic monoclonal-antibody-based approach for prophylaxis of serious Staphylococcus aureus disease. MBio 7:e00528–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tkaczyk C, Hua L, Varkey R, Shi Y, Dettinger L, Woods R, Barnes A, MacGill RS, Wilson S, Chowdhury P, Stover CK, Sellman BR. 2012. Identification of anti-alpha toxin monoclonal antibodies that reduce the severity of Staphylococcus aureus dermonecrosis and exhibit a correlation between affinity and potency. Clin Vaccine Immunol 19:377–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tkaczyk C, Kasturirangan S, Minola A, Jones-Nelson O, Gunter V, Shi YY, Rosenthal K, Aleti V, Semenova E, Warrener P, Tabor D, Stover CK, Corti D, Rainey G, Sellman BR. 2017. Multimechanistic Monoclonal Antibodies (MAbs) Targeting Staphylococcus aureus Alpha-Toxin and Clumping Factor A: Activity and Efficacy Comparisons of a MAb Combination and an Engineered Bispecific Antibody Approach. Antimicrob Agents Chemother 61:e00629–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Creech CB, Frenck RWJ, Sheldon EA, Seiden DJ, Kankam MK, Zito ET, Girgenti D, Severs JM, Immermann FW, McNeil LK, Cooper D, Jansen KU, Gruber W, Eiden J, Anderson AS, Baber J. 2017. Safety, tolerability, and immunogenicity of a single dose 4-antigen or 3-antigen Staphylococcus aureus vaccine in healthy older adults: results of a randomised trial. Vaccine 35:385–394. [DOI] [PubMed] [Google Scholar]
- 119.Scully IL, Liberator PA, Jansen KU, Anderson AS. 2014. Covering all the bases: preclinical development of an effective Staphylococcus aureus vaccine. Front Immunol 5:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Stranger-Jones YK, Bae T, Schneewind O. 2006. Vaccine assembly from surface proteins of Staphylococcus aureus. Proc Nat Acad Sci USA 103:16942–16947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kuklin NA, Clark DJ, Secore S, Cook J, Cope LD, McNeely T, Noble L, Brown MJ, Zorman JK, Wang XM, Pancari G, Fan H, Isett K, Burgess B, Bryan J, Brownlow M, George H, Meinz M, Liddell ME, Kelly R, Schultz L, Montgomery D, Onishi J, Losada M, Martin M, Ebert T, Tan CY, Schofield TL, Nagy E, Meineke A, Joyce JG, Kurtz MB, Caulfield MJ, Jansen KU, McClements W, Anderson AS. 2006. A novel Staphylococcus aureus vaccine: iron surface determinant B induces rapid antibody responses in rhesus macaques and specific increased survival in a murine S. aureus sepsis model. Infect Immun 74:2215–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kim HK, DeDent A, Cheng AG, McAdow M, Bagnoli F, Missiakas DM, Schneewind O. 2010. IsdA and IsdB antibodies protect mice against Staphylococcus aureus abscess formation and lethal challenge. Vaccine 28:6382–6392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Brown M, Kowalski R, Zorman J, Wang X-m, Towne V, Zhao Q, Secore S, Finnefrock AC, Ebert T, Pancari G, Isett K, Zhang Y, Anderson AS, Montgomery D, Cope L, McNeely T 2009. Selection and characterization of murine monoclonal antibodies to Staphylococcus aureus iron-regulated surface determinant B with functional activity in vitro and in vivo. Clin Vaccine Immunol 16:1095–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Fowler VG, Allen KB, Moreira ED, Moustafa M, Isgro F, Boucher HW, Corey GR, Carmeli Y, Betts R, Hartzel JS, Chan IS, McNeely TB, Kartsonis NA, Guris D, Onorato MT, Smugar SS, DiNubile MJ, Sobanjo-ter Meulen A. 2013. Effect of an investigational vaccine for preventing Staphylococcus aureus infections after cardiothoracic surgery: a randomized trial. JAMA 309:1368–1378. [DOI] [PubMed] [Google Scholar]
- 125.Kim HK, Emolo C, DeDent AC, Falugi F, Missiakas DM, Schneewind O. 2012. Protein A-specific monoclonal antibodies and the prevention of Staphylococcus aureus disease in mice. Infect Immun 80:3460–3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Thammavongsa V, Rauch S, Kim HK, Missiakas DM, Schneewind O. 2015. Protein A-neutralizing monoclonal antibody protects neonatal mice against Staphylococcus aureus. Vaccine 33:523–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ton-That H, Mazmanian H, Faull KF, Schneewind O. 2000. Anchoring of surface proteins to the cell wall of Staphylococcus aureus. I. Sortase catalyzed in vitro transpeptidation reaction using LPXTG peptide and NH2-Gly3 substrates. J Biol Chem 275:9876–9881. [DOI] [PubMed] [Google Scholar]
- 128.Ton-That H, Mazmanian SK, Alksne L, Schneewind O. 2002. Anchoring of surface proteins to the cell wall of Staphylococcus aureus. II. Cysteine 184 and histidine 120 of sortase A form a thiolate imidazolium ion pair for catalysis. J Biol Chem 277:7447–7452. [DOI] [PubMed] [Google Scholar]
- 129.Maresso AW, Wu R, Kern JW, Zhang R, Janik D, Missiakas DM, Duban ME, Joachimiak A, Schneewind O. 2007. Activation of inhibitors by sortase triggers irreversible modification of the active site. J Biol Chem 282:23129–23139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Maresso AW, Schneewind O. 2008. Sortase as a target of anti-infective therapy. Pharmacol Rev 60:128–141. [DOI] [PubMed] [Google Scholar]
- 131.Zhang J, Liu H, Zhu K, Gong S, Dramsi S, Wang YT, Li J, Chen F, Zhang R, Zhou L, Lan L, Jiang H, Schneewind O, Luo C, Yang CG. 2014. Antiinfective therapy with a small molecule inhibitor of Staphylococcus aureus sortase. Proc Natl Acad Sci USA 111:13517–13522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Oh KB, Nam KW, Ahn H, Shin J, Kim S, Mar W. 2010. Therapeutic effect of (Z)-3-(2,5-dimethoxyphenyl)-2-(4-methoxyphenyl) acrylonitrile (DMMA) against Staphylococcus aureus infection in a murine model. Biochem Biophys Res Commun 396:440–444. [DOI] [PubMed] [Google Scholar]
- 133.Pallen MJ, Lam AC, Antonio M, Dunbar K. 2001. An embarrassment of sortases - a richness of substrates. Trends Microbiol 9:97–101. [DOI] [PubMed] [Google Scholar]
- 134.Hendrickx AP, Budzik JM, Oh SY, Schneewind O. 2011. Architects at the bacterial surface - sortases and the assembly of pili with isopeptide bonds. Nat Rev Microbiol 9:166–176. [DOI] [PubMed] [Google Scholar]
- 135.Jacobitz AW, Kattke MD, Wereszczynski J, Clubb RT. 2017. Sortase Transpeptidases: Structural Biology and Catalytic Mechanism. Adv Protein Chem Struct Biol 109:223–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ton-That H, Schneewind O. 2003. Assembly of pili on the surface of Corynebacterium diphtheriae. Mol Microbiol 50:1429–1438. [DOI] [PubMed] [Google Scholar]
- 137.Mandlik A, Swierczynski A, Das A, Ton-That H. 2007. Corynebacterium diphtheriae employs specific minor pilins to target human pharyngeal epithelial cells. Mol Microbiol 64:111–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Budzik JM, Marraffini LA, Souda P, Whitelegge JP, Faull KF, Schneewind O. 2008. Amide bonds assemble pili on the surface of bacilli. Proc Nat Acad Sci USA 105:10215–10220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Budzik JM, Oh SY, Schneewind O. 2008. Cell wall anchor structure of BcpA pili in Bacillus anthracis. J Biol Chem 283:36676–36686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Budzik JM, Oh SY, Schneewind O. 2009. Sortase D forms the covalent bond that links BcpB to the tip of Bacillus cereus pili. J Biol Chem 284:12989–12997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ton-That H, Marraffini L, Schneewind O. 2004. Sortases and pilin elements involved in pilus assembly of Corynebacterium diphtheriae. Mol Microbiol 53:1147–1156. [DOI] [PubMed] [Google Scholar]
- 142.Swaminathan A, Mandlik A, Swierczynski A, Gaspar A, Das A, Ton-That H. 2007. Housekeeping sortase facilitates the cell wall anchoring of pilus polymers in Corynebacterium diphtheriae. Mol Microbiol 66:961–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Mandlik A, Das A, Ton-That H. 2008. The molecular switch that activates the cell wall anchoring step of pilus assembly in gram-positive bacteria. Proc Natl Acad Sci USA 105:14147–14152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Chang C, Amer BR, Osipiuk J, McConnell SA, Huang IH, Hsieh V, Fu J, Nguyen HH, Muroski J, Flores E, Ogorzalek Loo RR, Loo JA, Putkey JA, Joachimiak A, Das A, Clubb RT, Ton-That H. 2018. In vitro reconstitution of sortase-catalyzed pilus polymerization reveals structural elements involved in pilin cross-linking. Proc Natl Acad Sci USA 115:E5477–E5486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.McDevitt D, Francois P, Vaudaux P, Foster TJ. 1994. Molecular characterization of the clumping factor (fibrinogen receptor) of Staphylococcus aureus. Mol Microbiol 11:237–48. [DOI] [PubMed] [Google Scholar]
- 146.Hair PS, Ward MD, Semmes OJ, Foster TJ, Cunnion KM. 2008. Staphylococcus aureus clumping factor A binds to complement regulator factor I and increases factor I cleavage of C3b. J Infect Dis 198:125–33. [DOI] [PubMed] [Google Scholar]
- 147.Zong Y, Xu Y, Liang X, Keene DR, Hook A, Gurusiddappa S, Hook M, Narayana SV. 2005. A ‘Collagen Hug’ model for Staphylococcus aureus CNA binding to collagen. EMBO J 24:4224–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kang M, Ko YP, Liang X, Ross CL, Liu Q, Murray BE, Hook M. 2013. Collagen-binding microbial surface components recognizing adhesive matrix molecule (MSCRAMM) of Gram-positive bacteria inhibit complement activation via the classical pathway. J Biol Chem 288:20520–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Komatsuzawa H, Sugai M, Ohta K, Fujiwara T, Nakashima S, Suzuki J, Lee CY, Suginaka H. 1997. Cloning and characterization of the fmt gene which affects the methicillin resistance level and autolysis in the presence of triton X-100 in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 41:2355–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kuusela P, Hilden P, Savolainen K, Vuento M, Lyytikainen O, Vuopio-Varkila J. 1994. Rapid detection of methicillin-resistant Staphylococcus aureus strains not identified by slide agglutination tests. J Clin Microbiol 32:143–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kuusela P, Saksela O. 1990. Binding and activation of plasminogen at the surface of Staphylococcus aureus. Increase in affinity after conversion to the Lys form of the ligand. Eur J Biochem 193:759–65. [DOI] [PubMed] [Google Scholar]
- 152.Schroeder K, Jularic M, Horsburgh SM, Hirschhausen N, Neumann C, Bertling A, Schulte A, Foster S, Kehrel BE, Peters G, Heilmann C. 2009. Molecular characterization of a novel Staphylococcus aureus surface protein (SasC) involved in cell aggregation and biofilm accumulation. PLoS One 4:e7567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Barbu EM, Ganesh VK, Gurusiddappa S, Mackenzie RC, Foster TJ, Sudhof TC, Hook M. 2010. beta-Neurexin is a ligand for the Staphylococcus aureus MSCRAMM SdrC. PLoS Pathog 6:e1000726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Feuillie C, Formosa-Dague C, Hays LM, Vervaeck O, Derclaye S, Brennan MP, Foster TJ, Geoghegan JA, Dufrene YF. 2017. Molecular interactions and inhibition of the staphylococcal biofilm-forming protein SdrC. Proc Natl Acad Sci U S A 114:3738–3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Askarian F, Ajayi C, Hanssen AM, van Sorge NM, Pettersen I, Diep DB, Sollid JU, Johannessen M. 2016. The interaction between Staphylococcus aureus SdrD and desmoglein 1 is important for adhesion to host cells. Sci Rep 6:22134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zhang Y, Wu M, Hang T, Wang C, Yang Y, Pan W, Zang J, Zhang M, Zhang X. 2017. Staphylococcus aureus SdrE captures complement factor H’s C-terminus via a novel ‘close, dock, lock and latch’ mechanism for complement evasion. Biochem J 474:1619–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Uhlen M, Guss B, Nilsson B, Gatenbeck S, Philipson L, Lindberg M. 1984. Complete sequence of the staphylococcal gene encoding protein A. A gene evolved through multiple duplications. J Biol Chem 259:1695–702. [PubMed] [Google Scholar]
- 158.Graille M, Stura EA, Corper AL, Sutton BJ, Taussig MJ, Charbonnier JB, Silverman GJ. 2000. Crystal structure of a Staphylococcus aureus protein A domain complexed with the Fab fragment of a human IgM antibody: structural basis for recognition of B-cell receptors and superantigen activity. Proc Natl Acad Sci U S A 97:5399–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kukita K, Kawada-Matsuo M, Oho T, Nagatomo M, Oogai Y, Hashimoto M, Suda Y, Tanaka T, Komatsuzawa H. 2013. Staphylococcus aureus SasA is responsible for binding to the salivary agglutinin gp340, derived from human saliva. Infect Immun 81:1870–9. [DOI] [PMC free article] [PubMed] [Google Scholar]