Abstract
Reduction of insulin/insulin-like growth factor 1 (IGF1) signaling (IIS) extends the lifespan of various species. So far, several longevity mouse models have been developed containing mutations related to growth signaling deficiency by targeting growth hormone (GH), IGF1, IGF1 receptor, insulin receptor, and insulin receptor substrate. In addition, p70 ribosomal protein S6 kinase 1 (S6K1) knockout leads to lifespan extension. S6K1 encodes an important kinase in the regulation of cell growth. S6K1 is regulated by mechanistic target of rapamycin (mTOR) complex 1. The v-myc myelocytomatosis viral oncogene homolog (MYC)-deficient mice also exhibits a longevity phenotype. The gene expression profiles of these mice models have been measured to identify their longevity mechanisms. Here, we summarize our knowledge of long-lived mouse models related to growth and discuss phenotypic characteristics, including organ-specific gene expression patterns.
Keywords: Gene expression, Growth signaling, Longevity, Mouse model
INTRODUCTION
Insulin/insulin-like growth factor 1 (IGF1) signaling (IIS) is evolutionarily conserved from worms to human (1). Decreased IIS has been related to longevity in an evolutionarily conserved manner (2), showing consistent results in major model organisms, including worms (1), flies (3), and mice (4). Human centenarian studies have revealed that variations are positioned at proteins involved in the IIS system (5). In rodents, IGF1 is mainly produced from the liver by stimulation of growth hormone (GH, also known as somatotropin) secreted from the somatotrophs in the anterior pituitary (6). Circulating IGF1 binds to insulin/IGF1 receptors on the cell membrane of organs and primarily promotes tissue growth at an early developmental stage. Insulin, the other binding substrate of the insulin/IGF1 receptors, is produced by pancreatic β cells in response to postprandial nutrient influx (7). Insulin reduces high blood glucose levels after meals to maintain glucose homeostasis by suppressing glucose production in the liver, and stimulating glucose uptake in muscle and in fat by activation of their carbohydrate metabolism (8). This GH-IIS endocrine cascade axis is an excellent model for aging research to interpret how growth relates to metabolism. Insulin and IGF1 activate the IIS pathway through the insulin receptor (IR), IGF1 receptor (IGF1R), insulin receptor substrate 1 (IRS1), IRS2, phosphoinositide 3-kinase (PI3K), and protein kinase B (AKT) (7). Consequently, the IIS pathway changes gene expression through transcription factors, such as forkhead box O (FOXOs) and the v-myc myelocytomatosis viral oncogene homolog (MYC) (Fig. 1). In this review, we discuss physiological (Table 1) and global gene expression (Table 2) analyses in mice genetically modified to reduce GH-IIS axis in an effort to elucidate the longevity mechanism.
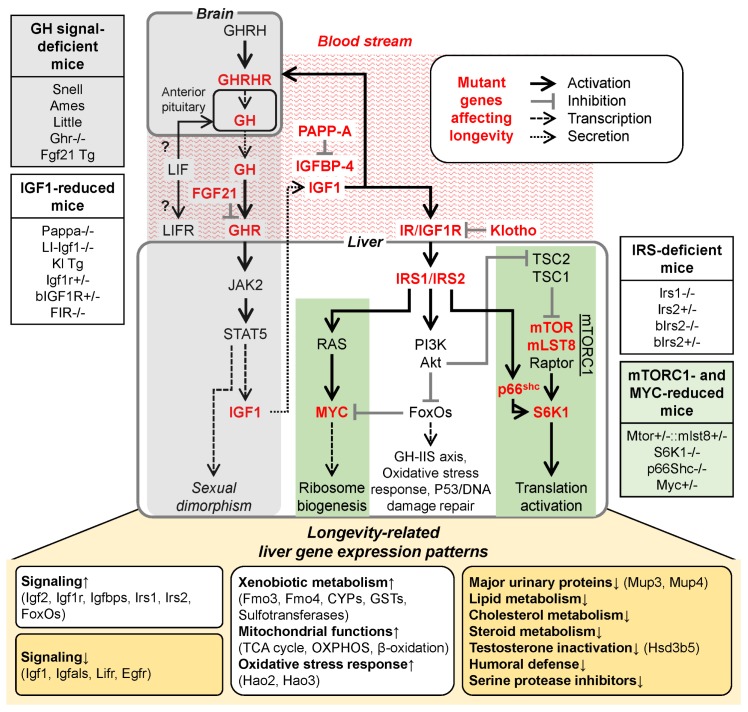
Fig. 1.
GH-IIS axis and hepatic gene expression patterns in long-lived mice. Components of the GH-IIS axis impact on hepatic gene expression and affect longevity. We divided GH-IIS axis genetically-modified mouse models into four groups: 1) GH signal-deficient mice with mutation at genes affecting GH production and GH signaling, 2) IGF1-reduced mice with homozygous and heterozygous mutation at genes involved in IGF1 production, including IGF1R and IR, 3) IRS-deficient mice including homozygous and heterozygous mutations of Irs1 and Irs2, and 4) mTORC1- and MYC-reduced mice with homozygous and heterozygous mutations for reduction of mTORC1 levels, lack of S6K1, p66Shc deficiency, and lower MYC levels. The target genes for long-lived mouse models are written in bolded red. Downstream liver gene expression patterns for longevity are displayed on the bottom panel.
Table 1.
Phenotypic characteristics of mice with mutations affecting the GH-IIS axis and lifespan
| Mice models | Key Physiology | Average Lifespan (%) | Body weight (%) | Body fat | Insulin sensitivity | Tumor incidence | Stress resistance | Plasma GH | Plasma IGF1 | Plasma glucose | References |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Long-lived models | |||||||||||
| Snell (Pou1f1−/−) | GH, PRL and TSH deficiency | F+51 M+29 (11718806) | ↓ (25–33) | ↑ | ↑ | ↓ | ↑ | ↓ | ↓ | ↓ | 16577229 1371619 |
| F and M+38 (15699523) | 11718806 | ||||||||||
| 15699523 | |||||||||||
| Ames (Prop1−/−) | GH, PRL and TSH deficiency | F+68 M+49 (8900272) | ↓ (33) | ↑ | ↑ | ↓ | ↑ | ↓ | ↓ | ↓ | 8900272 11719795 |
| F and M+37 (11719795) | |||||||||||
| Little (Ghrhr−/−) | GH deficiency | F+25 M+23 (11371619) | ↓ (50–67) | ↑ | ↑ | ↓ | ↑ | ↓ | ↓ | ↓ | 1270792 11371619 |
| Ghr−/− | GH resistance | F+38 M+55 (10875265) | ↓ (41–50) | ↑ | ↑ | ↓ | ↑ | ↑ | ↓ | ↓ or ↔ | 10875265 |
| F+25 M+16 (12933651) | 12933651 | ||||||||||
| F+30 M+30 (16682650) | 16682650 | ||||||||||
| Fgf21 Tg* | GH resistance | F+40 M+30 (23066506) | ↓ (60) | ↔ | ↑ | ? | ? | ↑ | ↓ | ↓ | 18585098 |
| 23066506 | |||||||||||
| LI-Igf1−/− | IGF1 reduction 75–85% | F+16 (21799924) | ↓ (75–100) | ↓ | ↓ | ? | ? | ↑ | ↓ | ↔ | 10359843 |
| 11423474 | |||||||||||
| 21799924 | |||||||||||
| Heart Igf1 Tg* | IGF1 excessed | M+23a (17973971) | ↔ (100) | ? | ? | ? | ? | ? | ↑ | ↔ | 17973971 |
| Pappa−/− | IGF1 reduced | F and M+38 (17681037) | ↓ (40) | ? | ↔ | ↓ | ? | ↔ | ↔ | ↔ | 14973274 |
| F+29 M+24 (20351075) | 17681037 | ||||||||||
| 20351075 | |||||||||||
| Klotho Tg | IIS inhibition | F+19 M+31 (16123266) | ↔ (100) | ? | F ↔ | ? | ↑ | ? | ? | ↔ | 16123266 |
| F+19 M+20 (16123266) | M↓ | ||||||||||
| Igf1r+/− | IGF1 resistance | F+33 M NS (12483226) | ↓ (90) | ? | ↓ | ↔ | ↑, F ↑ M | ? | ↔ | ↔ | 12483226 |
| F and M NS (22132081) | ↔ | 22132081 | |||||||||
| Brain IGF1R+/−* | Brain IIS reduction | F+8 M+13b (18959478) | ↓ (90–92) | ↑ | ↓ | ↓ | ↑ | ↓ | ↓ | ↔ | 18959478 |
| FIR−/− | Fat-specific insulin resistance | F and M+11 (12543978) | ↓ (70) | ↓ | ↑ | ? | ? | ? | ? | ↔ | 12110165 |
| 12543978 | |||||||||||
| Irs1−/−* | Insulin resistance or normal | F+16 M+14 (21283571) | ↓ (70) | ↓ | ↓ | ? | ? | ↔ | ↔ | ↔ | 7969452 17928362 |
| 21283571 | |||||||||||
| 29779018 | |||||||||||
| Irs2+/− | IRS2 reduced | F and M+17 (17641201) | ↔ (100) | ? | ↑ or ↔ | ? | ? | ↔ | ↔ | ↓ | 17641201 |
| F and M NS (17928362) | 17928362 | ||||||||||
| Brain Irs2+/− | Brain IRS2 reduced | F and M (+/−)+18 (17641201) | ↔ (100) | ↑ | ↓ | ? | ? | ↔ | ↔ | ? | 17641201 |
| Brain Irs2−/− | F and M (−/−)+14 (17641201) | ||||||||||
| mtor+/−::mlst8+/−* | mTORC1 reduction 50% | F+14 M NS (22461615) | ↔ (100) | ? | ↔ | ? | ? | ? | ? | ↔ | 22461615 |
| S6K1−/−* | S6K1 deficient | F+20 M NS (19797661) | ↓ (48) | ↓ | ↑ | ? | ? | ↔ | ↔ | ↔ | 19797661 |
| p66Shc+/− | IRS inhibition, S6K1 inhibition | F and M (+/−)+7a (10580504) | ↔ (100) | ↓ | ↔ | ? | ↑ | ? | ? | ↔ | 10580504 |
| p66Shc−/− | F and M (−/−)+28a (10580504) | 20624962 | |||||||||
| Myc+/−* | IGF1 reduction and MYC-regulated genes | F+21 M+11a (25619689) | ↓ (81) | ↔ | ? | ↔ | ? | ? | ↓ | ? | 25619689 |
| Normal or short-lived models | |||||||||||
| GHA Tg | GH resistance | NS (12933651) | ↓ (55–70) | ↑ | ↑ | ↓ | ? | ? | ↓ | ↓ or ↔ | 1791834 11487276 |
| 12933651 | |||||||||||
| 15231300 | |||||||||||
| Bovine GH Tg | GH excessed | M-45 (14583653) | ↑ (200) | ↓ | ↓ | ↑ | ? | ↑ | ↑ | ↓ | 1791834 11701438 |
| 14583653 | |||||||||||
| 15231300 | |||||||||||
| 18948397 | |||||||||||
| LID | 75% reduced IGF1 | M-? (22421704) | ↔ (100) | ↑ | ↓ | ↓ | ? | ↑ | ↓ | ↔ | 10377413 |
| 11334415 | |||||||||||
| 22421704 | |||||||||||
| Kl−/−* | IIS activation, growth retardation | 60.7 daysc (9363890) | ↓ (44–47) | ? | ↓ | ? | ? | ? | ? | ? | 9363890 |
| Irs2−/− | Irs2 deletion | F-26 M-84 (9495343) | ↓ (90) | ? | ↓ | ? | ? | ? | ? | ↑ | 9495343 |
New added or updated mice models in this review.
↑ up-regulated; ↓ down-regulated; ↔ no difference; ?, no data available.
Median lifespan.
Significant at censored lifespan data before week 130–135.
No lifespan data for control.
NS, not significant.
Table 2.
Transcriptome studies from long-lived mouse models for the GH-IIS axis
| Genotype | Publication | Design (age: month) | Study Focus | Sex | Back-ground | Tissue | Platform (probesets) | DEG number (statistical criteria) |
|---|---|---|---|---|---|---|---|---|
| GH-deficient mice | ||||||||
| Snell | Dozmorov et al. 2002 11867646 | WT vs Snell (6) | Systemic GH effects | Male | (DW × C3H)F1 | Liver | (2,352) Clontech Mouse Atlas 1.2 array | 17 (t-test FDR < 0.05) |
| Snell | Boylston et al. 2004 15379852 | WT vs Snell (3–5, 24–28) | Systemic GH effects | Male | C3H/HeJ × DW/J F1 or C3DW | Liver | (12,422) Affymetrix MG U74Av2 array | > 50 (t-test, P < 0.05 and FC > 2) |
| Snell | Papaconstantinou et al. 2005 15888324 | Snell ctrl(+/?)vs Snell (3–6, 20–28) | IGF-1 axis protein levels | Male | C3H/HeJ × DW/J F1 or C3DW | Liver | (12,422) Affymetrix MG U74Av2 array | 17 (t-test P < 0.05) |
| Snell | Boylston et al. 2006 19943135 | Snell ctrl(+/?)vs Snell (4–6, 24–26) | Systemic GH effects | Male | C3H/HeJ × DW/J F1 or C3DW | Liver | (12,422) Affymetrix MG U74Av2 array | 205 (t-test FDR < 0.05) |
| Ames | Dozmorov et al. 2001 11213270 | WT vs Ames (5, 13, 22) | Systemic GH effects | ? | ? | Liver | (265) early version of spotting array | 13 (t-test P < 0.01) |
| Ames | Tsuchiya et al. 2004 15039484 | WT vs Ames (2) | Systemic GH effects | Female | ? | Liver | (12,422) Affymetrix MG U74Av2 array | 212 (two-way ANOVA FDR < 0.05) |
| Ames | Amador-Noguez et al. 2004 15569359 | WT littermate vs Ames (3, 6, 12, 24) | Systemic GH effects agaist wt aging patterns | Male | C57Bl/6 | Liver | (> 14,000) Affymetrix MOE340A array | 122 (t-test Bonferroni <0.05 and FC > 2) |
| Ames | Boylston et al. 2006 19943135 | Ames ctrl(+/?)vs Ames (4–6, 12–14, 24–27) | Systemic GH effects | Male | C57Bl/6 | Liver | (> 34,000) Affymetrix MG 430 2.0 arrays | 785 (t-test FDR < 0.05) |
| Little | Amador-Noguez et al. 2004 15569359 | WT vs Little mice (3,6,12,24) | Systemic GH effects agaist wt aging patterns | Male | C57Bl/6 | Liver | (> 14,000) Affymetrix MOE340A array | 104 (t-test Bonferroni <0.05 and FC > 2) |
| bGH Tg | Olsson et al. 2003 12736163 | WT vs bGH-Tg (6) | Systemic GH effects | Male | ? | Liver | (11,000) Affymetrix mouse 11ka and 11kb arrays | 45 (difference call parameter 3–4) |
| Ghr | Miller et al. 2002 12403853 | WT vs Ghr−/− (8–9) | Systemic GH effects | ? | ? | Liver | (2,352) Clontech Mouse Atlas 1.2 array | 10 (t-test FDR < 0.05) |
| Ghr−/− | Rowland et al. 2005 15601831 | WT vs Ghr−/− (1.4) | Systemic GH effects | Male | B6.C3H-6T | Liver | (12,422) Affymetrix MG U74Av2 array | 398 (t-test P < 0.0005 and FC > 1.5) |
| GHR-569, GHR-391 | Rowland et al. 2005 15601831 | WT vs GHR-569, GHR-391 (1.4) | Systemic STAT5 effects | Male | 129/SVJ | Liver | (12,422) Affymetrix MG U74Av2 array | 20 (t-test P < 0.0005 and FC > 1.5) |
| Ghr−/−, GHR-391 | Barclay et al. 2011 21084450 | WT vs Ghr−/−, GHR-391 (4) | STAT5-mediated effects | Male | C57BL/6J | Liver | (46,632) Illumina MouseWG-6 v1.1 array | 55 common genes out of 466 (t-test P < 0.05, FC > 2) |
| Ghr−/− | Stout et al. 2015 26436954 | WT vs Ghr−/− (6) | Systemic GH effects on WAT and BAT | ? | ? | WAT and BAT | (28,853) Affymetrix GeneChip Mouse Gene 1.0 ST Array | 346 in SubQ WAT, 319 in PERI WAT, 192 in EPI WAT, 280 in BAT (t-test FDR < 0.05 and FC >1.5) |
| Ghr−/− | Chang et al. 2016 27064376 | WT vs Ghr−/− (3.7–4.2) | Systemic GH effects on lncRNA and miRNA | Male and Female | C57Bl/b5 | Liver | (25,376 transcript, 31,423 lncRNAs, 3,1000 miRNAs) Agilent Mouse LncRNA Array v2.0 | 3,442 lncRNAs, 1,750 mRNAs, 30 miRNAs in female, 2,642 lncRNAs, 1,489 mRNAs, 57 miRNAs in male (FC≥2) |
| Fgf21-Tg | Zhang et al. 2012 23066506 | WT vs FGF21−/− (3) | Systemic FGF21 effects | Male | C57Bl/6J | Liver, WAT, Muscle | (> 25,600) Illumina mouseRef-8 v2.0 BeadChip | 33 in liver, 8 in muscle, 22 in fat (t-test FDR < 0.1 and FC > 2) |
| Sex-specific liver gene expression | ||||||||
| Ames | Amador-Noguez et al. 2005 15925325 | WT vs Ames (3–6) | Sexual dimorphism of GH effects | Male vs Female | C57Bl/6 | Liver | (> 14,000) Affymetrix MOE430A array | 381 sex-independent, 110 sex-dependent (two-way ANOVA P < 0.001 and FC > 1.5) |
| STAT5b−/− | Clodfelter et al. 2006 16469768 | WT vs STAT5b−/− (1.8–2.1) | Sexual dimorphism of STAT5b effects | Male vs Female | ? | Liver | (23,574) Agilent Sosetta/Merck Mouse TOE 75k array | 85 in female (t-test P <0.05, FC > 1.5) |
| STAT5a−/− | Clodfelter et al. 2007 17536022 | WT vs STAT5a−/− (1.4–2.1) | Sexual dimorphism of STAT5a effects | Male vs Female | 129 x Black/Swiss | Liver | (23,574) Agilent Sosetta/Merck Mouse TOE 75k array | 769 in male, 461 in female (t-test P < 0.05, FC > 1.5) |
| IGF1- and IIS-reduced mice | ||||||||
| FIR−/− | Bluher et al. 2004 15131119 | WT vs WAT (3) | Fat-specific IR & Adipocyte size effect | Male | C57BL/6 x 129/Sv | WAT | (12,488) Affymetric MG-U74A-v2 array | 48 for adipocyte size, 63 for IR signaling (t-test P <0.001 and μ1–μ2 > 2∑SD) |
| FIR−/− | Katic et al. 2007 18001293 | WT vs FIR−/− (6, 18, 30–36) | Fat-specific IR effect | Male | 129sv/C57Bl6/J | WAT | (22,690) Affymetrix mouse 430A array | ? (ANOVA P < 0.01) |
| Irs1−/− | Selman et al. 2008 17928362 | WT vs Irs1−/− (2.6, 15, 22) | Systemic IIS effects | Female | C57BL/6 | Liver | (> 34,000) Affymetrix MG 430 2.0 array | ? (t-test P < 0.05) |
| Irs1−/− | Page et al. 2018 29779018 | WT vs Irs1−/− (15) | Systemic IIS effects | Female | C57BL/6 | Liver, Muscle, Brain, WAT | RNA-sequencing by Thermo Fisher Ion Proton system | 124 (101, specific) in liver, 185 (137) in muscle, 111 (94) in brain, 344 (319) in WAT (t-test FDR < 0.1) |
| mTORC1- and MYC-reduced mice | ||||||||
| S6K1−/− | Selman et al. 2009 19797661 | WT vs S6K1−/− (20) | Systemic S6K1 effect | Female | C57BL/6 | Liver, Muscle, WAT | (> 38,000) Affymetrix MG 430 2.0 array | 1,843 in liver, 2,309 in muscle, 1,970 in WAT (q-value < 0.01) |
| p66Shc−/− | Tomilov et al. 2010 19892704 | WT vs p66Shc−/− (3, 12) | Systemic p66Shc effect | Male and Female | ? | Liver, Spleen, Lungs, WAT, peritoneal macrophage | (> 38,000) Affymetrix MG 430 2.0 array (12,422) Affymetrix MG U74Av2 array |
top 250 genes (t-test P <0.05) |
| Myc+/− | Hofmann et al. 2015 25619689 | WT vs Myc+/− (5, 24) | Systemic MYC effects | Male | C57BL/6NCrl | Liver, Muscle, WAT | (28,853) Affymetrix Mouse 1.0 Gene ST arrays | 307 in liver, 160 in muscle, 412 in WAT (t-test FDR <0.05 and FC > 1.5) |
GH SIGNAL-DEFICIENT MICE
Several dwarf mouse models have been introduced in the aging research field, including Snell (disruption of the POU-domain transcription factor [Pit−/−]), Ames (defect of the homeobox protein prophet of PIT-1 [Prop 1−/−]), and Little (a missense mutation at growth hormone-releasing hormone receptor [Ghrhr−/−]). The Snell and Ames mice are hypopituitary dwarf deficient in GH, prolactin (PRL), and thyroid-stimulating hormone (TSH). These dwarf mice have been shown to live 37–68% longer than their wild-type (WT) littermates (9–11). The Little mice are another GH-deficient model. It was generated by deletion of the Ghrhr gene and the mice have been shown to live 23–25% longer than their WT littermates (9). The Snell, Ames, and Little GH deficient mice commonly have small body sizes and high adiposity, with low IGF1 levels.
The mouse GH receptor (Ghr) gene was disrupted (GHR−/−) to create a mouse model for human Laron syndrome (12). The GHR−/− mouse has been shown to have very low (less than 10% of normal) plasma IGF1 level and severely reduced IGF binding protein 3 (IGFBP3) level (13). A GH receptor antagonist (GHA) transgenic (Tg) mouse was generated by a single amino acid substitution in GH (14). A 50% reduction in plasma IGF1 has been reported in GHA tg mice, compared to their WT littermates. Both GHR−/− and GHA Tg mice have been shown to express obese and slow growth phenotypes (13, 15, 16), as well as small organs, including liver and kidney (17). GHR−/− mice have been reported to live longer (30–55%) than their WT littermates (13), but GHA Tg mice lived as long as their WT littermates (17).
Fibroblast growth factor-21 (FGF21) is an endocrine hormone related to energy metabolism and stress response (18). FGF21 binds to the transmembrane protein βKlotho, which has a protein sequence similar to Klotho (Kl). While studies have shown that Kl Tg mice overexpressing Kl lived longer compared to WT mice, βKlotho−/− mice lived as long as WT mice (19). Fasting has been shown to increase FGF21 levels, causing GH resistance by inactivation of downstream signal transducer and activator of transcription 5 (STAT5), and down-regulation of Igf1 gene (20). Indeed, FGF21-overexpressing Tg mice were shown to have small body sizes (60% of WT), decreased serum insulin and IGF1 levels, and a 36% extended lifespan (21). These results suggest that FGF21 is an upstream regulator of the IIS pathway.
The effect of overexpression of GH in Tg mice has been studied by insertion of the GH gene from other species, including human, rat, bovine, and ovine. Most GH Tg mice showed a high plasma GH level and a giant body size (6, 22). Interestingly, bovine GH (bGH) Tg mice exhibited lean bodies (16) and a 45% shorter lifespan than WT mice (22). In addition, bGH Tg mice had kidney damage, namely nephropathy with glomerular enlargement (23, 24). Human patients having an excess of GH show large hands, feet, face and internal organs, in a condition called acromegaly (25). Acromegaly patients also have lean bodies, heart problems, a high risk of thyroid cancer and colorectal cancer, and often have insulin-resistance or some diabetes (25, 26).
GLOBAL HEPATIC GENE EXPRESSION ALTERATION IN GH-RELATED MODEL MICE
GH stimulates its primary target, the liver, causing it to produce IGF1. A number of microarray analyses have identified genes for longevity mechanism using liver tissues taken from Ames and Snell dwarf mice (27–34). Although early studies identified only a few genes (27, 28), later studies using high-density oligonucleotide microarrays that can detect > 12,000 RNA transcripts identified hundreds of differentially expressed genes (DEGs) as candidates in the aging mechanism. Papaconstantinou et al. reported hepatic DEGs using young (3–6 months of age) Snell male mice. Major down-regulated DEGs included Igf1, Insulin like growth factor binding protein acid labile subunit (Igfals), and Forkhead box C1 (Foxc1). Major up-regulated DEGs included Igf2, Igf1 receptor (Igf1r), Igf binding protein 1 (Igfbp1), Igfbp2, Igfbp3, Irs1, Irs2, and Foxp1 (33).
Tsuchiya et al. identified 212 DEGs (FDR < 0.05) in Ames dwarf mice livers, including two genes encoding Forkhead transcription factors (Foxa3 was up-regulated and Foxa2 was down-regulated), as well as up-regulated xenobiotic metabolism and glutathione synthesis genes (31). Another transcriptome study using Ames dwarf mice livers showed hundreds of genes affected by aging. Amador-Noguez et al. reported that 357 DEGs (P < 0.001) were associated with age progression (3, 6, 12, and 24 months old) in Ames dwarf male mice livers. In particular, they found up-regulated genes for cholesterol and steroid biosynthesis (29). They also found an increase in Igf1 transcript levels during aging, as well as progressive decreases of Igfbp1 and Igfbp2 transcript levels (29). Boylston et al. identified 49 DEGs (FDR < 0.05) in livers from both Snell and Ames dwarf male mice, at both young and old ages. They used 4–6 month old (young age) and 24–26 month old (old age) for Snell mice, and 4–6 month old, 12–14 month old (middle age) and 24–27 month old for Ames mice. Among the DEGs, they found up-regulated genes encoding cytochrome P450 (CYP), and Flavin containing monooxygenase 3 (Fmo3). Fmo3 encodes a flavin adenine dinucleotide (FAD)-dependent monooxygenase that metabolizes various sulfur- and nitrogen-containing metabolites (35). There was a down-regulation of Hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 5 (Hsd3b5). The Hsd3b5 gene plays important roles in the biosynthesis of various steroid hormones (36). The Hydroxyacid oxidase 2 (Hao2) gene, which encodes a flavin mononucleotide (FMN)-dependent peroxisomal 2-hydroxy acid oxidase for long chain 2-hydroxy acid substrates was up-regulated (37).
Using Ames and Little male mice, 547 common DEGs (P < 0.001) were identified in liver during the aging process in 3, 6, 12, and 24 month old mice (29). Gene ontology (GO) analysis showed that the up-regulated DEGs are involved in the oxidative stress response, xenobiotic metabolism, mitochondrial respiration and β-oxidation, and the down-regulated DEGs are involved in humoral defense, including the complement system regulatory genes.
Using GHR−/− male mice (1.5 months old), 330 DEGs were identified in the liver. Up-regulated DEGs were those for β-oxidation, oxidative phosphorylation (OXPHOS), tricarboxylic acid (TCA) cycle, and factors for transcription and translation. Serine protease inhibitors were down-regulated DEGs (38). Using two GHR site-specific truncated mutants, GHR-569 and GHR-391 mouse strains, phosphorylation status of STAT5 was either decreased or removed in the liver by bGH treatment, respectively (38). GHR-STAT5 signaling was severely weakened in these two mutant mice. Using these two mutant mice, 20 common DEGs (P < 0.0005 and FC > 1.5) were identified in the liver (38). Down-regulated DEGs included Igf1, Igfals, Epidermal growth factor receptor (Egfr), and Hsd3b5. Up-regulated DEGs included Sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 2 (Sult2a2), Angiogenin, ribonuclease, RNase A family, 5 (Ang), and Hao2. Sult2a2 and Ang play roles in the sulfation of hydroxysteroid and host defense, respectively. Up-regulated DEGs also included several genes encoding CYPs and glutathione S-transferases (GSTs) (38). Additional studies of GHR-391 and GHR−/− mice livers showed that the loss of GHR-STAT5 signaling caused intracellular lipid accumulation and steatosis by increasing phosphorylation of STAT1 and STAT3 proteins (39). A recent study using young GHR−/− male and female mice (16–18 weeks old) showed a transcript regulatory network for steroid hormone biosynthesis in the liver, composed of several long-noncoding RNA and microRNAs (40).
Using 3 month old young Fgf21 Tg male mice, transcriptome analysis identified 33, 22, and 8 DEGs (FDR < 0.1 and FC > 2) from the liver, epididymal white adipose tissue (WAT), and gastrocnemius, respectively (21). The hepatic DEGs included up-regulation of Fgf21, Fmo3, Cytochrome P450, family 2, subfamily b, polypeptide 9 (Cyp2b9), and Metallothionein 1 (Mt1) genes; and down-regulation of Igfals, Hsd3b5, Major urinary protein 4 (Mup4), Glutathione S-transferase, pi 1 (Gstp1), and Sterol carrier protein 2 (Scp2) genes (21). The authors compared the hepatic DEGs they found to 43 common hepatic DEGs from the four long-lived dwarf strains, including Snell, Ames, Little, and GHR−/− mice (41). They found eight overlapped genes, including down-regulated Igfals, Hairy and enhancer of split 6 (Hes6), Aminolevulinic acid synthase 2, erythroid (Alas2), Cyp4a12b, Mup4, Serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 12 (Serpina12), and Hsd3b5; as well as up-regulated Fmo3 (21). Although only a small number of DEGs were found in the liver, white fat and skeletal muscle, the young long-lived Fgf21 Tg male mice showed dramatic phenotypical and physiological changes, including low body weight and low levels of plasma IGF-1, insulin, and glucose, as well as high levels of plasma ketone bodies (21).
META-ANALYSIS OF TRANSCRIPTOME FROM GH SIGNAL-DEFICIENT MICE
Swindell performed a meta-analysis using microarray data obtained from livers of Snell, Ames, Little, and GHR−/− mice. DEG selection (mean difference significance test P < 0.05) revealed different DEG numbers among these mice models. The GHR−/− mice showed the smallest number of DEGs (46 DEGs), and other mice strains showed hundreds of DEGs: 151–316 DEGs from Snell, 400–987 DEGs from Ames, and 475–650 DEGs from Little (41). The range in number of DEGs was due to different ages of the mice. The results revealed that these mice strains shared strong down-regulation of Igf1 (41). Comparison of gene expression across these four dwarf mice identified 13 common DEGs, including 3 up-regulated genes: Hao3, Sult2a2, and Serine peptidase inhibitor, Kazal type 3 (Spink3). It also identified 10 down-regulated genes: Mup3, Mup4, Igf1, Igfals, Egfr, Socs2, Leukemia inhibitory factor receptor alpha (Lifr), Carboxylesterase 3A (Ces3a: also known as Es31), Hsd3b5, and Kidney expressed gene 1 (Keg1) (41). One down-regulated common DEG, Lifr, has not yet been studied for its role in aging and longevity. The Lifr gene encodes a receptor for the leukemia inhibitory factor (LIF) cytokine that affects various biological functions, including stem cell self-renewal, the reproductive process, and bone modeling (42). Interestingly, LIF can affect the anterior pituitary gland to decrease somatotroph, lactotroph and gonadotroph cells (43).
Malfunction of genes for DNA repair, such as Excision repair cross-complementing rodent repair deficiency, complementation group 1 (Ercc1), Ercc6 (also known as CSB), and Xeroderma pigmentosum, complementation group A (Xpa), caused progeroid syndromes in DNA repair-deficient mouse models. Csbm/m::Xpa−/− and Ercc1−/− mice showed a severe phenotype, Ercc1−/Δ−7 showed intermediate, Csbm/m showed mild, and Xpa−/− showed no phenotype in mouse models (44, 45). Interestingly, Csbm/m::Xpa−/− progeria mice showed reduction of serum IGF1 and glucose levels, and suppression of the GH-IIS axis (45). Schumacher et al. performed a meta-analysis to compare transcriptome data between progeria mice (Csbm/m::Xpa−/−, Ercc1−/−, Ercc1−/Δ−7, Csbm/m and Xpa−/−) and long-lived dwarf mice (Snell, Ames and Little) (46). Strikingly, the hepatic transcriptome data showed a high correlation (r > 0.7) in gene expression patterns. They found a trend for down-regulation of genes involved in the GH-IIS axis, oxidative metabolism, and energy metabolism, and an up-regulation trend in genes involved in the stress response (46).
SEXUAL DIMORPHISM OF TRANSCRIPTOME IN LIVER
At the GH signal-deficient models including Snell, Ames, GHR−/−, and Fgf21 Tg mice, female models showed a greater difference of lifespan than male models (Table 1). GH secretion from the pituitary gland is stimulated by sex-steroids (47), and there is a different pattern of secretion between males and females (48). This indicates sexual dimorphism, including global gene expression in the liver (49–51).
Amador-Noguez et al. studied sexual dimorphism in the liver using 126 DEGs (P < 0.001 and FC > 1.5) between female and male WT mice (32). They found that genes involved in fatty acid biosynthesis and steroid hormone metabolism, especially Hsd3b2, Hsd3b3, Hsd3b5, and Hsd3b6 encoding a hydroxysteroid dehydrogenase, were down-regulated in female WT mice. In contrast, genes for metabolic enzymes including Fmo3, metallothioneins, and peroxisomal acyl-CoA thioesterases were up-regulated in female WT mice. Particularly, xenobiotic metabolizing CYP genes were dominantly up-regulated in female WT mice. Interestingly, sex-specific expression of those 126 DEGs in WT were almost completely lost in Ames dwarf mice (32). The complete loss of sexual dimorphism in the long-lived Ames dwarf mice might reflect a reduction of reproduction that needs a costly physiological investment. This may be a potentially important determinant of the extended longevity of these mice.
JAK2 phosphorylates GHR serving as a docking site for STAT transcription factors in GH signaling in the liver. There are seven mouse STAT proteins, and STAT5a and STAT5b proteins play key roles in sex-specific hepatic gene expression (52, 53). In young (1.4–1.8 month old) mice, STAT5a regulated 23% (89 out of 393 genes) of the sex-specific gene expression, including down-regulation of Y-linked Ddx3y, Eif2s3y, and Jarid1d genes, up-regulation of Sult (sulfotransferase) genes, and both up- and down-regulation of CYP genes in female livers (53). STAT5b regulated 90% (767 out of 950 genes) of down-regulated genes and 61% (461 out of 753) of up-regulated genes in females (52). STAT5b-dependent sex-specific genes were primarily involved in oxidative metabolism, tryptophan and fatty acid metabolism, steroid metabolism, signaling, xenobiotic metabolism, and serine protease inhibition (52).
IGF1 AND IIS-REDUCED MICE
The major proportion of circulating IGF1 is produced by the liver (54). In the liver, by the binding of GH, GHR signaling transmits through JAK-STAT, and consequently activates IGF1 transcription, production, and secretion into the blood stream (6). Circulating IGF1 bioavailability is mediated by IGF binding proteins (IGFBPs). Deletion of the Pappa gene encoding a metalloproteinase, Pappalysin-1 that cleaves IGF1 bound to IGFBP4 decreased bioavailability of IGF1 (55). Indeed, Pappa−/− mice showed 40% smaller body mass, lower cancer incidence, and a 24–38% longer lifespan than WT controls (56–58).
Igf1-deficient (Igf1−/−) mice showed variable neonatal lethality in several different genetic backgrounds such as 129/Sv mice (10% survival), C57BL/6J x 129/Sv F1 hybrids (16% survival), and MF1 x 129/Sv F1 hybrids (68% survival) (59). The surviving Igf1−/− mice showed 24–40% lower body weights and increased maximum lifespan, but no significant change in mean lifespan (60). Although their body mass was lower than WT (WT 39 g vs Igf1−/− 30 g for male; WT 28 g vs Igf1−/− 17 g for female), there was almost no impact on the size of major organs, including liver (Igf1−/− 1.37 g vs WT 1.39 g), kidney (Igf1−/− 0.21 g vs WT 0.21 g), brain (Igf1−/− 0.44 g vs WT 0.47 g), and heart (Igf1−/− 0.16 g vs WT 0.22 g) (60). The IGF1 receptor (IGF1R) deficient (Igf1r−/−) mice show respiratory failure at birth that is 100% lethal (59). Similar to Igf1r−/− mice, homozygous insulin receptor (InsR)-deficient mice suffered from hyperglycemia and died soon after birth (61). Heterozygous InsR knockout (IR+/−) male mice showed heteroinsufficiency, including insulin insensitivity and a 20% extended maximum lifespan, but no significant change in average lifespan (62).
Both inducible liver-specific Igf1-disruption mice (LI-Igf1−/−) (54), and liver-specific Igf1-deficient mice (LID mice) (63) showed a 75% decrease IGF1 levels and high GH levels in serum. Igf1 expression was shut off after ~28 days for LI-Igf1−/− mice (54) and after ~10 days in LID mice (63). LI-Igf1−/− female mice, but not male mice, lived 16% longer than WT mice (64), whereas male LID mice exhibited a shortened lifespan (65).
Heterozygous Igf1r+/− mice exhibited 8% smaller body sizes and ~40% higher serum IGF1 level than those of WT mice (4). Increased serum IGF1 level is a compensatory response to reduced IGF1 receptor levels. Igf1r+/− mice also showed a low cellular IGF1 signaling activity because of glucose tolerance in the liver (4). Igf1r+/− mice lived 26% longer than WT controls, 33% longer for female mice and 16% longer for male mice, though the male difference was not statistically significant (4). Another independent study using Igf1r+/− mice showed negligible changes in lifespan (66). Ladiges et al. discussed that the different results were due to the unusually short lifespan of WT mice in the original study because of sub-optimal husbandry conditions, and may lead to incorrect conclusions about the longevity of Igf1r+/− mice, in comparison (67).
Klotho (Kl) encodes a circulating hormone and functions as an antagonist against IR and IGF1R, increasing insulin resistance (68). Kl−/− mice die early at < 100 days of age and show systemic aging phenotypes: growth retardation (small body size, and atrophy of genital organs and thymus), hypokinesis, arteriosclerosis, ectopic calcification in various organs and arterial walls, osteoporosis, and skin atrophy (69). In contrast, Kl overexpression significantly extended lifespan (19–31%) of Kl Tg mice through GH-independent suppression of the IIS pathway (68). Inhibition of IIS by additional genetic interventions, like Kl−/−::IRS1+/− ameliorated aging-like phenotypes of Kl−/− mice and improved survival (68).
IRS-DEFICIENT MICE
IRS1 and IRS2 are required for the IIS pathway and are evolutionarily-conserved mammalian lifespan regulators (70). IRS1 binds to the IR, activating the PI3K-AKT signaling cascade to regulate glucose metabolism through down-regulation of gluconeogenic enzymes and up-regulation of lipogenic enzymes (71). Irs1−/− mice showed insulin-resistance with defects in insulin signaling (72), and lived 14–16% longer than WT littermates (73). The Irs1−/− mice showed small body size, reduced fat mass, and protection from aging-induced insulin resistance (70). Heterozygous Irs1+/− mice had the same lifespan as WT (70). The lifespan extension seen in the Irs1−/− mice was shorter than that of Snell (29–51%), Ames (37–68%), Little (23–25%), or GHR−/− (16–55%) dwarf mice strains. These lifespan differences suggest that there are IRS1-independent, but GH-dependent mechanism for longevity.
IRS2 is known to play a central role in pancreatic β-cell function (74). Irs2−/− mice die early (maximum 3 months of age) due to their diabetic phenotypes that include defects in hepatic insulin signaling, glucose deregulation, and β-cell failure or apoptosis (70). Several studies have tried to restore the β-cell function of Irs2−/− mice, through mechanisms that included overexpression of transcription factor, Pancreatic and duodenal homeobox 1 (PDX1) to enhance β-cell functions (75), deletion of Protein tyrosine phosphatase, non-receptor type 1 (PTPN1) to reduce insulin sensitivity (76), and reduction of Phosphatase and tensin homolog (PTEN) expression, a potent inhibitor of insulin action (77). These trials achieved some improvements in lifespan, but created additional problems. The lifespan of Irs2−/−::Pdx1 Tg mice increased by 15 months, but they had severe spinal deformities. Irs2−/−::Ptpn1−/− mice had lifespan increases of 8–9 months, but β-cell function deterioration. And Irs2−/−::Pten+/− mice gained 10–12 months of life, but had lymph proliferative disease (78). Heterozygous Irs2+/− mice showed conflicting lifespan results. Taguchi et al. showed 17% longer (79) and Selman et al. showed no significant change in the lifespan of Irs2+/− mice (70). Selman et al. argued that the WT (C57BL/6J) lifespan profile in the study by Taguchi et al. had a significantly different shape compared to other independent WT (C57BL/6J) survival curves (80). KO of liver-specific Irs1 (LIrs1−/−) and Irs2 (LIrs2−/−) in mice exhibited insulin resistance with different meal-timing, after refeeding, and during fasting (81). Interestingly, fasting/fed-responsive genes in the liver were significantly down-regulated in LIrs1−/− mice, but those genes were not changed in LIrs2−/− mice (82). These results suggest that IRS1 is a principal mediator of the regulation of hepatic glucose homeostasis.
Selman et al. measured global hepatic gene expression in female WT and Irs1−/− mice at young (2.6 months old), middle (15 months old), and old (22 months old) ages (70). The results showed up-regulation of several genes in different ages of Irs1−/− mice, including catalase-encoding genes in old age, a glutathione S-transferase gene Gsta4 in young age, an excision repair gene Ercc8 at all ages, and a growth regulator Gadd45b at all ages (70). At both young and middle ages, genes for energy metabolism were up-regulated and they were involved in carbohydrate metabolism, acetyl-CoA metabolism, mitochondrial metabolism, including OXPHOS and TCA cycle; and coenzyme and cofactor catabolism. Down-regulated genes at young and middle ages were involved in immune response and antigen processing (70).
FOXO-MEDIATED LONGEVITY GENES
FOXO transcription factors (FoxOs) are inhibitory downstream targets of IIS through activation of AKT, TSC complex subunit 2 (TSC2) and Glycogen synthase kinase 3 (GSK3) (83). The FoxOs are known as evolutionarily conserved anti-aging effectors (84) with broad roles in glucose homeostasis, tumor suppression, autophagy, and resistance to oxidative stress (84, 85). Among the four mouse FoxO homologs (FoxO1, FoxO3, FoxO4 and FoxO6), FoxO3 was reported to be a longevity factor under dietary restriction condition (86). Lifespan extension by overexpression of FoxOs has not been reported in mice yet. In fresh water polyp Hydra, FoxO achieves immortality through maintaining self-renewal capacity of stem cells (87).
A meta-analysis by Webb et al. using FoxO ChIP-seq data from four mouse cell types (neural progenitor cells, memory CD8+ T-cells, pre-B cells, and T regulatory cells) showed that FoxO commonly binds to the promoter areas (from 5 kb upstream to 1 kb downstream of transcription start site) of both pro-longevity and anti-longevity genes belonging to the GH-IIS axis (e.g., Gh, Igf1r, Akt1, and Irs2), the oxidative stress response (e.g., Cat, Txn1, and Prdx1), and p53/DNA damage repair (e.g., Ercc2, Atr, and Trp53) (88).
TISSUE-SPECIFIC EFFECTS OF GH-IIS AXIS
Recently, Page et al. measured liver, muscle, brain and WAT transcriptomes in middle age (15 months old) female Irs1−/− mice using an RNA-sequencing approach. They identified 124 DEGs (FDR < 0.1) in the liver, 185 DEGs in muscle, 111 DEGs in brain, and 344 DEGs in WAT (89). There was no common DEG across the four different tissues. DEGs from liver, muscle, and brain tissues shared 5 up-regulated genes encoding ribosomal proteins, including Ribosomal protein L37a (Rpl37a), Ribosomal protein S26 (Rps26), and Rpl31; and hemoglobin proteins, including Hemoglobin alpha, adult chain 1 (Hba-a1), and Hemoglobin beta, adult t chain (Hbb-bt). DEGs from muscle and WAT had the largest number of shared genes, including 7 up-regulated genes and 15 down-regulated genes (89). They also found some shared gene ontology terms across tissues. ECM term was shared in down-regulated DEGs across all 4 tissues. Organ development and receptor binding terms were shared from down-regulated DEGs across muscle, brain, and WAT. Ribosomal protein, haptoglobin complex, ECM, and antioxidant activity terms were shared from up-regulated DEGs across liver, muscle, and brain (89). Across both muscle and WAT, DEGs related to fatty acid metabolism were up-regulated, and DEGs related to inflammation were down-regulated. Especially, in liver, up-regulated DEGs were involved in B cell proliferation and biosynthesis of ribosomal small subunits, and included CYP genes for fatty acid metabolism. The down-regulated DEGs were involved in negative regulation of RNA metabolism by transcription factors. In muscle, up-regulated DEGs were involved in mitochondrial functions, including oxidoreductase activity, electron carrier activity, and cytochrome-c oxidase activity. Down-regulated DEGs were involved in inflammatory processes, including leukocyte proliferation, interferon-γ response, and serine-type endopeptidase inhibition. In brain, up-regulated DEGs were involved in regulation of adenylate cyclase and G-protein coupled receptor signaling, and down-regulated DEGs were involved in morphogenesis and angiogenesis. These results indicate that Irs1−/− mice have tissue-specific longevity mechanisms.
Fat-specific reduction of GH-IIS axis
Ames, Snell, and GHR−/− dwarf mice showed an obese phenotype due to their high capacity for lipid storage and activation of pre-adipocyte differentiation (90). Stout et al. measured transcriptomes from brown adipose tissue (BAT) and several WATs at different locations including inguinal, epididymal and perirenal fat from GHR−/− mouse (91). They found opposite gene expression trends between WATs and BAT (91). In BAT, 252 DEGs (FDR < 0.05 and FC > 1.5) were identified. Up-regulated DEGs were involved in several metabolic processes including organic acid metabolism, oxidation-reduction process, cholesterol biosynthesis, and lipid and acetyl-CoA metabolism. Down-regulated DEGs were involved in immune and inflammation, including immune system regulation, innate immune response, inflammation response, and wound healing. In WATs, 346, 319, and 192 DEGs were identified in inguinal, perirenal, and epididymal fat tissues, respectively. Common DEGs of WATs from the three different locations showed up-regulation of dendritic cell-expressed genes; and down-regulation of cellular respiration-related genes, and mitochondrial inner envelope-related genes. The up-regulation of dendritic cell-expressed genes might represent an increase of infiltrated dendritic cells in WATs that correlates with the obese status. Masternak et al. performed surgical removal of visceral fat in both WT and GHR−/− mice (92). Visceral fat removal (VFR) from GHR−/− mice resulted in decreased adiponectin levels, glucose tolerance, and insulin sensitivity, accompanied by increased glucose (92). These results suggest that WAT actually has a beneficial effect on longevity of the GHR−/− mouse. In contrast, WT mouse with VFR showed improved insulin sensitivity, reduced body temperature, and decreased respiration rates; opposite effects, compared to GHR−/− mice with VFR (92).
Fat-specific insulin receptor knockout (FIR−/−) mice showed adipocyte-specific insulin resistance, reduced body fat mass, protection from age- and hyperphagia-associated obesity, and protection from age-related insulin resistance (93). There was an interesting difference in adipocyte size in FIR−/− mouse, compared to WT. Adipocytes isolated from epididymal WAT of FIR−/− mice divided into two sizes, small (< 75 μm) and large (> 100 μm), compared to one medium size (75–100 μm) in WT mice (93). Bluher et al. isolated the small and large adipocytes from 3 month-old young FIR−/− male mice using 75 μm pore size nylon mesh screen and identified 63 DEGs (P < 0.0001) (94). Later, Katic et al. observed that FIR−/− mouse showed an increased metabolic rate and higher oxygen consumption, compared to WT littermates (95). They also measured WAT transcriptome of FIR−/− male mice at young (6 months old), middle (18 months old), and old (30–36 months old) ages. The transcriptome data showed age-associated up-regulation of genes related to mitochondria, including OXPHOS and β-oxidation; and age-associated down-regulation of genes related to defense, external stimuli response, and protein prenylation (95). These results indicate that activated mitochondrial function and its maintenance during the aging process may be key mechanisms for the longevity of FIR−/− mice.
Brain-specific reduction of IIS
As mentioned previously, the Igf1r−/− mice were lethal because of postnatal respiratory failure (59). Kappeler et al. reported that brain (specifically, hypothalamus)-specific Igf1r heterogenic KO (bIGF1R+/−) mice lived 13% longer than WT mice (96). The bIGF1R+/− mouse model showed ~10% growth retardation, and increases in blood lipid, cholesterol, and free fatty acid levels at young ages (10 months old) (96). Similar to the GH-deficient mice (Snell, Ames and Little), bIGF1R+/− mice showed small pituitary glands, low GH levels, and increased body fat mass (96).
Brain-specific Irs2+/− (bIrs2+/−) and bIrs2−/− mice showed lifespan extension by 18% and 14%, respectively (79). During the aging process, these mice showed hyperinsulinemia and insulin resistance, as well as high glucose oxidation, and stable superoxide dismutase 2 (SOD2) levels in the hypothalamus, with stable diurnal rhythm (79). These results indicate that the metabolic rate, but not the obese state or insulin sensitivity might be associated with longevity. Sadagurski et al. reported that genetic intervention to reduce IRS2 signaling decreased incidence of age-associated Huntington disease (HD) (97). These results suggest that because brain-specific IRS2 reduction decreased incidence of neurodegenerative diseases, it could lead to longevity.
Heart-specific Igf1 overexpression
In contrast to the aforementioned studies, Li and Ren argued that aging-associated decreases of circulating IGF1 stimulates cardiac aging. They reported that cardiac-specific Igf1 (cIgf1) Tg male mice showed a 60–80% increase in plasma IGF1 levels and extended their lifespan by 23% (98). cIgf1 Tg mice exhibited improved cardiomyocyte function including increased Ca2+ homeostasis, reduction of protein damage, and decreased apoptosis (98). Indeed, long-lived Ames dwarf mice showed impairment of cardiac excitation-contraction coupling, compared to WT mice (99). Overall, these results suggest that the autocrine/paracrine role of IGF1 is important to cardiac-specific anti-aging effects.
mTORC1- AND MYC-REDUCED MICE
Both IIS and mechanistic target of rapamycin (mTOR) pathways could activate ribosomal protein S6 kinase 1 (S6K1), a regulator of growth and metabolism through activation of protein synthesis (100). Although male S6K1−/− mouse showed a similar lifespan to WT, female S6K1−/− mouse showed a 20% longer lifespan than WT. Other phenotypes of female S6K1−/− mice included lean and small body size, and improved insulin sensitivity (101). Interestingly, their total circulating IGF1, pituitary GH, TSH, and PRL levels were not changed compared to WT (101). Lamming et al. reported that mTOR and mLST8 double heterozygous (mtor+/−::mlst8+/−) female mice showed normal body weight and maintained insulin sensitivity (102). And mtor+/−::mlst8+/− female mice showed decreased mTORC1 activity, but not mTORC2, with a 14% extended lifespan (102). Although both S6K1−/− and mtor+/−::mlst8+/− mice showed female-specific longevity, they showed different phenotypes. These results indicate that the small body size and improved insulin sensitivity in S6K1−/− mouse are independent changes, compared to mTORC1-mediated longevity.
Selman et al. measured transcriptome in liver, skeletal muscle and WAT of young (10 months old) S6K1−/− female mice (101). They identified 1,843 DEGs (q-value < 0.01) in the liver, 2,309 DEGs in muscle, and 1,970 DEGs in WAT. Across these three tissues, DEGs related to adenosine monophosphate-activated protein kinase (AMPK)-mediated signaling were commonly up-regulated.
p66Shc, an isoform of SHC-transforming protein 1, binds to both IRS1 and S6 kinase and can bridge the two signalings (103). p66Shc−/− and p66Shc+/− mice showed 28% and 7% increased median lifespans, respectively (104). The p66Shc−/− mice showed less ROS production, high oxidative stress resistance, and prevention of obesity (104).
Tomilov et al. measured transcriptome in liver, spleen, lung, WATs (retroperitoneal and epididymal), and peritoneal macrophages from young p66Shc−/− mice (3 months old males, and 12 months old males and females) (105). They found that commonly down-regulated DEGs (P < 0.05) were enriched in PI3K signaling, antigen processing, chronic myeloid leukemia, adipokine signaling, anti-apoptosis, and heme transcripts by gene ontology analysis (105). During phagocytosis, phagocytes, including neutrophils, eosinophils and macrophages exhibit an intense consumption of oxygen for NADPH oxidase (PHOX), in a process called a respiratory burst (106). In a comparison of peritoneal macrophages, p66Shc−/− mice showed 31% less PHOX activity and 40% less production of PHOX-dependent superoxide than WT mice (105).
Myc transcription factor, known as a protooncogene regulates various downstream functions, including energy metabolism, ribosome biogenesis, cell cycle, apoptosis, differentiation, and stem cell maintenance (107). Recently, Hofmann et al. reported that heterozygous Myc+/− mice showed a 15% longer lifespan than WT mice, while Myc null mice resulted in an embryonic lethal phenotype (108). Myc+/− mice showed 15–20% less body mass, 56–64% lower serum IGF1 levels, higher metabolic rates, and higher AMP levels in liver, whereas there was no difference in adipose tissue mass, reproductive ability, or incidence of deaths due to cancer, compared to WT mice (108).
Hofmann et al. (2015) measured transcriptome in young (5 months of age) and old (24 months of age) Myc+/− male mice and identified 307, 160 and 412 DEGs (FDR < 0.05 and FC > 1.5) in the liver, skeletal muscle, and gonadal WAT, respectively, compared to those of WT mice (108). The altered DEGs showed tissue-specific functions without a common DEG across the three tissues. In liver, down-regulated DEGs at old age were involved in cholesterol biosynthesis, and Myc+/− mice were protected from age-associated enlargement of hepatic lipid droplets (108). Comparing hepatic DEGs from Myc+/− mice to long-lived Snell, Little, and GHR−/− mice, the DEGs for xenobiotic metabolism enzymes, including CYP genes, Fmo3, Fmo4, Hao2, Mt1, Mt2, and P450 (cytochrome) oxidoreductase (Por) were commonly up-regulated, but the fold changes were orders of magnitudes smaller in Myc+/− mice (108). In addition, Myc+/− mice showed decreases in S6 kinase and AKT activities in both liver and muscle, an increase of AMPK in liver, and decreased serum IGF1 levels, whereas none of the genes related to these pathway components were identified as DEGs (108). Based on these observations, the authors hypothesized that the reduced available energy in the liver of Myc+/− mice activates AMPK, and then reduces mTOR and S6 kinase activities (108), promoting longevity.
CONCLUDING REMARKS
GH signal-deficient mice, including Snell, Ames, Little, GHR−/−, and Fgf21 Tg dwarf mice, showed increased lifespans and smaller body masses than WT mice. Therefore, body size was strongly dependent on GH action. This consistent trend suggests an inverse correlation between size and lifespan (109). However, small size can not be used as a general indicator of longevity, because Kl Tg, Irs2+/−, p66Shc−/−, and mtor+/−::mlst8+/− mice had normal body masses like WT mice, but showed longer lifespans than WT. In addition, GHA Tg mice had normal lifespans like WT mice, and Kl−/− and Irs2−/− mice showing dramatically shorter lifespans also had a dwarfism phenotype.
Snell, Ames, Little, and GHR−/− mice exhibited obesity, caused by loss of lipolytic and anti-lipogenic activities of GH. Especially, long-lived bIGF1R+/−, bIrs2+/−, and bIrs2−/− mice had high body fat. These results indicate brain-mediated endocrinological control of body fat and the IGF1 feedback loop. However, excess weight and obesity are known risk factors of aging and cause critical diseases, including diabetes and heart failure (110). Indeed, LI-Igf1−/−, FIR−/−, Irs1−/−, S6K1−/−, and p66Shc−/− mice showed less body fat than WT animals. In contrast, bGH Tg mice producing excess GH had lean bodies, but shortened lifespans. Surgical removal of visceral fat in GHR−/− mice decreased the fat-induced beneficial effects (92). Therefore, under the different endocrine circumstances, regardless of the body fat mass, adipose tissue itself contributes effects beneficial to longevity.
High insulin sensitivity could be associated with improved health. Long-lived GH signal-deficient mice, as well as FIR−/− and S6K1−/− mice, showed high insulin sensitivity. However, some long-lived mice with a mutation in IIS components, including LI-Igf1−/−, Igf1r+/−, bIGF1R+/−, and Irs1−/−, as well as the Kl Tg mice displayed insulin resistance. In the case of systemic Irs2 deficiency, Irs2−/− mice had diabetes, whereas brain-specific mutant bIrs2−/− mice showed both insulin resistance and longevity. Interestingly, R6/2::Irs2+/−::Irs2βTg mice, genetically modified to be Irs2+/− heterozygous except in β cells, showed a slow Huntington disease (HD) progression (97). In addition, insulin resistant (e.g., IR+/−) male mice showed a 20% extended maximum lifespan, but high insulin sensitivity mice, including PTP-1B−/− and PGC-1α Tg displayed shortened or similar lifespans compared to WT mice, respectively (62). It is clear that manipulation of insulin sensing signaling produces beneficial effects on longevity, however, either impaired or hyper-sensitive insulin signaling can lead to shortened lifespans.
Snell, Ames, Little, GHR−/−, and bIGF1R+/− mice showed both high-stress resistance and less tumor incidence. Pappa−/− mice also showed less tumor incidence, and Kl Tg, Igf1r+/− and p66Shc−/− mice showed high-stress resistance. Hepatic DEGs of Snell, Ames, Little, and GHR−/− mice showed up-regulation of DEGs for xenobiotic metabolism (e.g., Fmo3, CYPs, GSTs, and Sulfotransferases) and oxidative stress response (e.g., Hao2 and Hao3), representing improved oxidative stress response and toxin defense. In addition, similar hepatic DEG patterns were observed in DNA repair-deficient progeroid mice, and these changes might be a compensatory response (46). FoxO genes were up-regulated by inhibition of IIS signaling, and FoxOs regulated gene expression for stress response and DNA damage repair (111) (88). Genetically mTORC1-reduced mice, including mtor+/−;mlst8+/− and S6K1−/− showed extended lifespans with up-regulation of FoxOs (101). And p66Shc−/− mice showed less ROS production and high oxidative stress resistance (104). In addition, hepatic DEGs from Ames, Little, GHR−/−, and Irs1−/− mice commonly showed up-regulation of genes for mitochondrial respiration, including the TCA cycle, OXPHOS, and β-oxidation. Collectively, the gene expression analyses of IIS and mTORC1 reduction commonly represent improved stress response that is a signature of longevity. High resistance to oxidative stress and DNA damage in these mice might be contributing factors to low tumor incidence and increased longevity.
Snell, Ames, Little, GHR−/−, and Fgf21 Tg dwarf mice showed female-biased lifespan extension, and commonly showed STAT5-related gender-different hepatic gene expression. Moreover, female Igf1r+/−, LI-Igf1−/−, Irs1−/−, mtor+/−::mlst8+/−, S6K1−/−, and Myc+/− mice also lived longer than their male mice counterparts. These results suggest that the GH-IIS axis is regulated in a sex-specific manner. Gender is a critical factor in lifespan explained by genes that work differently in males and females in a concept known as sexual antagonistic pleiotropy (SAP) (112). STAT5-mediated liver gene expression analysis showed altered genes in xenobiotic metabolism (e.g., CYPs, GSTs, and sulfotransferases), oxidative stress response, and serine protease inhibitors. They overlap with the aforementioned longevity-associated liver gene expression data. In particular, Hsd3b5 encoding a ketosteroid reductase for steroid hormone biosynthesis, including testosterone was commonly down-regulated in Snell, Ames, Little, GHR−/−, and Fgf21 Tg mice. The expression of Hsd3b5 was associated with activation of GHR-STAT5 signaling, suggesting testosterone inactivation might contribute to the female-biased longevity.
On the other hand, Myc+/− mice showed up-regulated DEGs for xenobiotic metabolism and inhibition of mTOR and S6 kinases, however their tumor incidence was similar to WT mice (108). In addition, Kl Tg, bIGF1R+/−, and Irs1−/− mice showed similar lifespan extension in both males and females. Therefore, more work is needed to elucidate factors contributing to the lifespan of these mice.
ACKNOWLEDGEMENTS
This work was carried out with the support of Bio and Medical Technology Development Program through the National Research Foundation of Korea funded by the Ministry of Education, Science and Technology (NRF-2017R1D1A1A020 19355).
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting interests.
REFERENCES
- 1.Barbieri M, Bonafe M, Franceschi C, Paolisso G. Insulin/IGF-I-signaling pathway: an evolutionarily conserved mechanism of longevity from yeast to humans. Am J Physiol Endocrinol Metab. 2003;285:E1064–1071. doi: 10.1152/ajpendo.00296.2003. [DOI] [PubMed] [Google Scholar]
- 2.Fontana L, Partridge L, Longo VD. Extending healthy life span--from yeast to humans. Science. 2010;328:321–326. doi: 10.1126/science.1172539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hwangbo DS, Gershman B, Tu MP, Palmer M, Tatar M. Drosophila dFOXO controls lifespan and regulates insulin signalling in brain and fat body. Nature. 2004;429:562–566. doi: 10.1038/nature02549. [DOI] [PubMed] [Google Scholar]
- 4.Holzenberger M, Dupont J, Ducos B, et al. IGF-1 receptor regulates lifespan and resistance to oxidative stress in mice. Nature. 2003;421:182–187. doi: 10.1038/nature01298. [DOI] [PubMed] [Google Scholar]
- 5.Bonafe M, Olivieri F. Genetic polymorphism in long-lived people: cues for the presence of an insulin/IGF-pathway-dependent network affecting human longevity. Mol Cell Endocrinol. 2009;299:118–123. doi: 10.1016/j.mce.2008.10.038. [DOI] [PubMed] [Google Scholar]
- 6.Junnila RK, List EO, Berryman DE, Murrey JW, Kopchick JJ. The GH/IGF-1 axis in ageing and longevity. Nat Rev Endocrinol. 2013;9:366–376. doi: 10.1038/nrendo.2013.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang J, Liu F. Tissue-specific insulin signaling in the regulation of metabolism and aging. IUBMB Life. 2014;66:485–495. doi: 10.1002/iub.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rask-Madsen C, Kahn CR. Tissue-specific insulin signaling, metabolic syndrome, and cardiovascular disease. Arterioscler Thromb Vasc Biol. 2012;32:2052–2059. doi: 10.1161/ATVBAHA.111.241919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flurkey K, Papaconstantinou J, Miller RA, Harrison DE. Lifespan extension and delayed immune and collagen aging in mutant mice with defects in growth hormone production. Proc Natl Acad Sci U S A. 2001;98:6736–6741. doi: 10.1073/pnas.111158898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flurkey K, Papaconstantinou J, Harrison DE. The Snell dwarf mutation Pit1(dw) can increase life span in mice. Mech Ageing Dev. 2002;123:121–130. doi: 10.1016/S0047-6374(01)00339-6. [DOI] [PubMed] [Google Scholar]
- 11.Lin L, Hale SP, Schimmel P. Aminoacylation error correction. Nature. 1996;384:33–34. doi: 10.1038/384033b0. [DOI] [PubMed] [Google Scholar]
- 12.Zhou Y, Xu BC, Maheshwari HG, et al. A mammalian model for Laron syndrome produced by targeted disruption of the mouse growth hormone receptor/binding protein gene (the Laron mouse) Proc Natl Acad Sci U S A. 1997;94:13215–13220. doi: 10.1073/pnas.94.24.13215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coschigano KT, Clemmons D, Bellush LL, Kopchick JJ. Assessment of growth parameters and life span of GHR/BP gene-disrupted mice. Endocrinology. 2000;141:2608–2613. doi: 10.1210/endo.141.7.7586. [DOI] [PubMed] [Google Scholar]
- 14.Chen WY, Wight DC, Mehta BV, Wagner TE, Kopchick JJ. Glycine 119 of bovine growth hormone is critical for growth-promoting activity. Mol Endocrinol. 1991;5:1845–1852. doi: 10.1210/mend-5-12-1845. [DOI] [PubMed] [Google Scholar]
- 15.Chen WY, White ME, Wagner TE, Kopchick JJ. Functional antagonism between endogenous mouse growth hormone (GH) and a GH analog results in dwarf transgenic mice. Endocrinology. 1991;129:1402–1408. doi: 10.1210/endo-129-3-1402. [DOI] [PubMed] [Google Scholar]
- 16.Berryman DE, List EO, Coschigano KT, Behar K, Kim JK, Kopchick JJ. Comparing adiposity profiles in three mouse models with altered GH signaling. Growth Horm IGF Res. 2004;14:309–318. doi: 10.1016/j.ghir.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 17.Coschigano KT, Holland AN, Riders ME, List EO, Flyvbjerg A, Kopchick JJ. Deletion, but not antagonism, of the mouse growth hormone receptor results in severely decreased body weights, insulin, and insulin-like growth factor I levels and increased life span. Endocrinology. 2003;144:3799–3810. doi: 10.1210/en.2003-0374. [DOI] [PubMed] [Google Scholar]
- 18.Salminen A, Kaarniranta K, Kauppinen A. Regulation of longevity by FGF21: Interaction between energy metabolism and stress responses. Ageing Res Rev. 2017;37:79–93. doi: 10.1016/j.arr.2017.05.004. [DOI] [PubMed] [Google Scholar]
- 19.Kuro-o M. Klotho and betaKlotho. Adv Exp Med Biol. 2012;728:25–40. doi: 10.1007/978-1-4614-0887-1_2. [DOI] [PubMed] [Google Scholar]
- 20.Inagaki T, Lin VY, Goetz R, Mohammadi M, Mangelsdorf DJ, Kliewer SA. Inhibition of growth hormone signaling by the fasting-induced hormone FGF21. Cell Metab. 2008;8:77–83. doi: 10.1016/j.cmet.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y, Xie Y, Berglund ED, et al. The starvation hormone, fibroblast growth factor-21, extends lifespan in mice. Elife. 2012;1:e00065. doi: 10.7554/eLife.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bartke A. Can growth hormone (GH) accelerate aging? Evidence from GH-transgenic mice. Neuroendocrinology. 2003;78:210–216. doi: 10.1159/000073704. [DOI] [PubMed] [Google Scholar]
- 23.Doi T, Striker LJ, Quaife C, et al. Progressive glomerulosclerosis develops in transgenic mice chronically expressing growth hormone and growth hormone releasing factor but not in those expressing insulinlike growth factor-1. Am J Pathol. 1988;131:398–403. [PMC free article] [PubMed] [Google Scholar]
- 24.Yang CW, Striker LJ, Pesce C, et al. Glomerulosclerosis and body growth are mediated by different portions of bovine growth hormone. Studies in transgenic mice. Lab Invest. 1993;68:62–70. [PubMed] [Google Scholar]
- 25.Chanson P, Salenave S. Acromegaly. Orphanet J Rare Dis. 2008;3:17. doi: 10.1186/1750-1172-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clayton PE, Banerjee I, Murray PG, Renehan AG. Growth hormone, the insulin-like growth factor axis, insulin and cancer risk. Nat Rev Endocrinol. 2011;7:11–24. doi: 10.1038/nrendo.2010.171. [DOI] [PubMed] [Google Scholar]
- 27.Dozmorov I, Bartke A, Miller RA. Array-based expression analysis of mouse liver genes: effect of age and of the longevity mutant Prop1df. J Gerontol A Biol Sci Med Sci. 2001;56:B72–80. doi: 10.1093/gerona/56.2.B72. [DOI] [PubMed] [Google Scholar]
- 28.Dozmorov I, Galecki A, Chang Y, Krzesicki R, Vergara M, Miller RA. Gene expression profile of long-lived snell dwarf mice. J Gerontol A Biol Sci Med Sci. 2002;57:B99–108. doi: 10.1093/gerona/57.3.B99. [DOI] [PubMed] [Google Scholar]
- 29.Amador-Noguez D, Yagi K, Venable S, Darlington G. Gene expression profile of long-lived Ames dwarf mice and Little mice. Aging Cell. 2004;3:423–441. doi: 10.1111/j.1474-9728.2004.00125.x. [DOI] [PubMed] [Google Scholar]
- 30.Boylston WH, Gerstner A, DeFord JH, et al. Altered cholesterologenic and lipogenic transcriptional profile in livers of aging Snell dwarf (Pit1dw/dwJ) mice. Aging Cell. 2004;3:283–296. doi: 10.1111/j.1474-9728.2004.00115.x. [DOI] [PubMed] [Google Scholar]
- 31.Tsuchiya T, Dhahbi JM, Cui X, Mote PL, Bartke A, Spindler SR. Additive regulation of hepatic gene expression by dwarfism and caloric restriction. Physiol Genomics. 2004;17:307–315. doi: 10.1152/physiolgenomics.00039.2004. [DOI] [PubMed] [Google Scholar]
- 32.Amador-Noguez D, Zimmerman J, Venable S, Darlington G. Gender-specific alterations in gene expression and loss of liver sexual dimorphism in the long-lived Ames dwarf mice. Biochem Biophys Res Commun. 2005;332:1086–1100. doi: 10.1016/j.bbrc.2005.05.063. [DOI] [PubMed] [Google Scholar]
- 33.Papaconstantinou J, Deford JH, Gerstner A, et al. Hepatic gene and protein expression of primary components of the IGF-I axis in long lived Snell dwarf mice. Mech Ageing Dev. 2005;126:692–704. doi: 10.1016/j.mad.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 34.Boylston WH, DeFord JH, Papaconstantinou J. Identification of longevity-associated genes in long-lived Snell and Ames dwarf mice. Age (Dordr) 2006;28:125–144. doi: 10.1007/s11357-006-9008-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rossner R, Kaeberlein M, Leiser SF. Flavin-containing monooxygenases in aging and disease: Emerging roles for ancient enzymes. J Biol Chem. 2017;292:11138–11146. doi: 10.1074/jbc.R117.779678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simard J, Ricketts ML, Gingras S, Soucy P, Feltus FA, Melner MH. Molecular biology of the 3beta-hydroxysteroid dehydrogenase/delta5-delta4 isomerase gene family. Endocr Rev. 2005;26:525–582. doi: 10.1210/er.2002-0050. [DOI] [PubMed] [Google Scholar]
- 37.Jones JM, Morrell JC, Gould SJ. Identification and characterization of HAOX1, HAOX2, and HAOX3, three human peroxisomal 2-hydroxy acid oxidases. J Biol Chem. 2000;275:12590–12597. doi: 10.1074/jbc.275.17.12590. [DOI] [PubMed] [Google Scholar]
- 38.Rowland JE, Lichanska AM, Kerr LM, et al. In vivo analysis of growth hormone receptor signaling domains and their associated transcripts. Mol Cell Biol. 2005;25:66–77. doi: 10.1128/MCB.25.1.66-77.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barclay JL, Nelson CN, Ishikawa M, et al. GH-dependent STAT5 signaling plays an important role in hepatic lipid metabolism. Endocrinology. 2011;152:181–192. doi: 10.1210/en.2010-0537. [DOI] [PubMed] [Google Scholar]
- 40.Chang L, Qi H, Xiao Y, et al. Integrated analysis of noncoding RNAs and mRNAs reveals their potential roles in the biological activities of the growth hormone receptor. Growth Horm IGF Res. 2016;29:11–20. doi: 10.1016/j.ghir.2016.03.003. [DOI] [PubMed] [Google Scholar]
- 41.Swindell WR. Gene expression profiling of long-lived dwarf mice: longevity-associated genes and relationships with diet, gender and aging. BMC Genomics. 2007;8:353. doi: 10.1186/1471-2164-8-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nicola NA, Babon JJ. Leukemia inhibitory factor (LIF) Cytokine Growth Factor Rev. 2015;26:533–544. doi: 10.1016/j.cytogfr.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yano H, Readhead C, Nakashima M, Ren SG, Melmed S. Pituitary-directed leukemia inhibitory factor transgene causes Cushing’s syndrome: neuroimmune-endocrine modulation of pituitary development. Mol Endocrinol. 1998;12:1708–1720. doi: 10.1210/mend.12.11.0200. [DOI] [PubMed] [Google Scholar]
- 44.Niedernhofer LJ, Garinis GA, Raams A, et al. A new progeroid syndrome reveals that genotoxic stress suppresses the somatotroph axis. Nature. 2006;444:1038–1043. doi: 10.1038/nature05456. [DOI] [PubMed] [Google Scholar]
- 45.van der Pluijm I, Garinis GA, Brandt RM, et al. Impaired genome maintenance suppresses the growth hormone--insulin-like growth factor 1 axis in mice with Cockayne syndrome. PLoS Biol. 2007;5:e2. doi: 10.1371/journal.pbio.0050002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schumacher B, van der Pluijm I, Moorhouse MJ, et al. Delayed and accelerated aging share common longevity assurance mechanisms. PLoS Genet. 2008;4:e1000161. doi: 10.1371/journal.pgen.1000161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jansson JO, Eden S, Isaksson O. Sexual dimorphism in the control of growth hormone secretion. Endocr Rev. 1985;6:128–150. doi: 10.1210/edrv-6-2-128. [DOI] [PubMed] [Google Scholar]
- 48.MacLeod JN, Pampori NA, Shapiro BH. Sex differences in the ultradian pattern of plasma growth hormone concentrations in mice. J Endocrinol. 1991;131:395–399. doi: 10.1677/joe.0.1310395. [DOI] [PubMed] [Google Scholar]
- 49.Mode A, Gustafsson JA. Sex and the liver - a journey through five decades. Drug Metab Rev. 2006;38:197–207. doi: 10.1080/03602530600570057. [DOI] [PubMed] [Google Scholar]
- 50.Waxman DJ, O’Connor C. Growth hormone regulation of sex-dependent liver gene expression. Mol Endocrinol. 2006;20:2613–2629. doi: 10.1210/me.2006-0007. [DOI] [PubMed] [Google Scholar]
- 51.Yang X, Schadt EE, Wang S, et al. Tissue-specific expression and regulation of sexually dimorphic genes in mice. Genome Res. 2006;16:995–1004. doi: 10.1101/gr.5217506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Clodfelter KH, Holloway MG, Hodor P, Park SH, Ray WJ, Waxman DJ. Sex-dependent liver gene expression is extensive and largely dependent upon signal transducer and activator of transcription 5b (STAT5b): STAT5b-dependent activation of male genes and repression of female genes revealed by microarray analysis. Mol Endocrinol. 2006;20:1333–1351. doi: 10.1210/me.2005-0489. [DOI] [PubMed] [Google Scholar]
- 53.Clodfelter KH, Miles GD, Wauthier V, et al. Role of STAT5a in regulation of sex-specific gene expression in female but not male mouse liver revealed by microarray analysis. Physiol Genomics. 2007;31:63–74. doi: 10.1152/physiolgenomics.00055.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sjogren K, Liu JL, Blad K, et al. Liver-derived insulin-like growth factor I (IGF-I) is the principal source of IGF-I in blood but is not required for postnatal body growth in mice. Proc Natl Acad Sci U S A. 1999;96:7088–7092. doi: 10.1073/pnas.96.12.7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Conover CA. Key questions and answers about pregnancy-associated plasma protein-A. Trends Endocrinol Metab. 2012;23:242–249. doi: 10.1016/j.tem.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Conover CA, Bale LK, Overgaard MT, et al. Metalloproteinase pregnancy-associated plasma protein A is a critical growth regulatory factor during fetal development. Development. 2004;131:1187–1194. doi: 10.1242/dev.00997. [DOI] [PubMed] [Google Scholar]
- 57.Conover CA, Bale LK. Loss of pregnancy-associated plasma protein A extends lifespan in mice. Aging Cell. 2007;6:727–729. doi: 10.1111/j.1474-9726.2007.00328.x. [DOI] [PubMed] [Google Scholar]
- 58.Conover CA, Bale LK, Mader JR, Mason MA, Keenan KP, Marler RJ. Longevity and age-related pathology of mice deficient in pregnancy-associated plasma protein-A. J Gerontol A Biol Sci Med Sci. 2010;65:590–599. doi: 10.1093/gerona/glq032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu JP, Baker J, Perkins AS, Robertson EJ, Efstratiadis A. Mice carrying null mutations of the genes encoding insulin-like growth factor I (Igf-1) and type 1 IGF receptor (Igf1r) Cell. 1993;75:59–72. [PubMed] [Google Scholar]
- 60.Lorenzini A, Salmon AB, Lerner C, et al. Mice producing reduced levels of insulin-like growth factor type 1 display an increase in maximum, but not mean, life span. J Gerontol A Biol Sci Med Sci. 2014;69:410–419. doi: 10.1093/gerona/glt108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Accili D, Drago J, Lee EJ, et al. Early neonatal death in mice homozygous for a null allele of the insulin receptor gene. Nat Genet. 1996;12:106–109. doi: 10.1038/ng0196-106. [DOI] [PubMed] [Google Scholar]
- 62.Nelson JF, Strong R, Bokov A, Diaz V, Ward W. Probing the relationship between insulin sensitivity and longevity using genetically modified mice. J Gerontol A Biol Sci Med Sci. 2012;67:1332–1338. doi: 10.1093/gerona/gls199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yakar S, Liu JL, Stannard B, et al. Normal growth and development in the absence of hepatic insulin-like growth factor I. Proc Natl Acad Sci U S A. 1999;96:7324–7329. doi: 10.1073/pnas.96.13.7324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Svensson J, Sjogren K, Faldt J, et al. Liver-derived IGF-I regulates mean life span in mice. PLoS One. 2011;6:e22640. doi: 10.1371/journal.pone.0022640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Novosyadlyy R, Leroith D. Insulin-like growth factors and insulin: at the crossroad between tumor development and longevity. J Gerontol A Biol Sci Med Sci. 2012;67:640–651. doi: 10.1093/gerona/gls065. [DOI] [PubMed] [Google Scholar]
- 66.Bokov AF, Garg N, Ikeno Y, et al. Does reduced IGF–1R signaling in Igf1r+/− mice alter aging? PLoS One. 2011;6:e26891. doi: 10.1371/journal.pone.0026891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ladiges W, Van Remmen H, Strong R, et al. Lifespan extension in genetically modified mice. Aging Cell. 2009;8:346–352. doi: 10.1111/j.1474-9726.2009.00491.x. [DOI] [PubMed] [Google Scholar]
- 68.Kurosu H, Yamamoto M, Clark JD, et al. Suppression of aging in mice by the hormone Klotho. Science. 2005;309:1829–1833. doi: 10.1126/science.1112766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kuro-o M, Matsumura Y, Aizawa H, et al. Mutation of the mouse klotho gene leads to a syndrome resembling ageing. Nature. 1997;390:45–51. doi: 10.1038/36285. [DOI] [PubMed] [Google Scholar]
- 70.Selman C, Lingard S, Choudhury AI, et al. Evidence for lifespan extension and delayed age-related biomarkers in insulin receptor substrate 1 null mice. FASEB J. 2008;22:807–818. doi: 10.1096/fj.07-9261com. [DOI] [PubMed] [Google Scholar]
- 71.Peng J, He L. IRS posttranslational modifications in regulating insulin signaling. J Mol Endocrinol. 2018;60:R1–R8. doi: 10.1530/JME-17-0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Previs SF, Withers DJ, Ren JM, White MF, Shulman GI. Contrasting effects of IRS-1 versus IRS-2 gene disruption on carbohydrate and lipid metabolism in vivo. J Biol Chem. 2000;275:38990–38994. doi: 10.1074/jbc.M006490200. [DOI] [PubMed] [Google Scholar]
- 73.Selman C, Partridge L, Withers DJ. Replication of extended lifespan phenotype in mice with deletion of insulin receptor substrate 1. PLoS One. 2011;6:e16144. doi: 10.1371/journal.pone.0016144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Withers DJ, Gutierrez JS, Towery H, et al. Disruption of IRS-2 causes type 2 diabetes in mice. Nature. 1998;391:900–904. doi: 10.1038/36116. [DOI] [PubMed] [Google Scholar]
- 75.Kushner JA, Ye J, Schubert M, et al. Pdx1 restores beta cell function in Irs2 knockout mice. J Clin Invest. 2002;109:1193–1201. doi: 10.1172/JCI0214439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kushner JA, Haj FG, Klaman LD, et al. Islet-sparing effects of protein tyrosine phosphatase-1b deficiency delays onset of diabetes in IRS2 knockout mice. Diabetes. 2004;53:61–66. doi: 10.2337/diabetes.53.1.61. [DOI] [PubMed] [Google Scholar]
- 77.Kushner JA, Simpson L, Wartschow LM, et al. Phosphatase and tensin homolog regulation of islet growth and glucose homeostasis. J Biol Chem. 2005;280:39388–39393. doi: 10.1074/jbc.M504155200. [DOI] [PubMed] [Google Scholar]
- 78.Taguchi A, White MF. Insulin-like signaling, nutrient homeostasis, and life span. Annu Rev Physiol. 2008;70:191–212. doi: 10.1146/annurev.physiol.70.113006.100533. [DOI] [PubMed] [Google Scholar]
- 79.Taguchi A, Wartschow LM, White MF. Brain IRS2 signaling coordinates life span and nutrient homeostasis. Science. 2007;317:369–372. doi: 10.1126/science.1142179. [DOI] [PubMed] [Google Scholar]
- 80.Selman C, Lingard S, Gems D, Partridge L, Withers DJ. Comment on “Brain IRS2 signaling coordinates life span and nutrient homeostasis”. Science. 2008;320:1012. doi: 10.1126/science.1152366. author reply 1012. [DOI] [PubMed] [Google Scholar]
- 81.Kubota N, Kubota T, Itoh S, et al. Dynamic functional relay between insulin receptor substrate 1 and 2 in hepatic insulin signaling during fasting and feeding. Cell Metab. 2008;8:49–64. doi: 10.1016/j.cmet.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 82.Guo S, Copps KD, Dong X, et al. The Irs1 branch of the insulin signaling cascade plays a dominant role in hepatic nutrient homeostasis. Mol Cell Biol. 2009;29:5070–5083. doi: 10.1128/MCB.00138-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Brunet A, Bonni A, Zigmond MJ, et al. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–868. doi: 10.1016/S0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 84.Martins R, Lithgow GJ, Link W. Long live FOXO: unraveling the role of FOXO proteins in aging and longevity. Aging Cell. 2016;15:196–207. doi: 10.1111/acel.12427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hannenhalli S, Kaestner KH. The evolution of Fox genes and their role in development and disease. Nat Rev Genet. 2009;10:233–240. doi: 10.1038/nrg2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shimokawa I, Komatsu T, Hayashi N, et al. The life-extending effect of dietary restriction requires Foxo3 in mice. Aging Cell. 2015;14:707–709. doi: 10.1111/acel.12340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Boehm AM, Khalturin K, Anton-Erxleben F, et al. FoxO is a critical regulator of stem cell maintenance in immortal Hydra. Proc Natl Acad Sci U S A. 2012;109:19697–19702. doi: 10.1073/pnas.1209714109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Webb AE, Kundaje A, Brunet A. Characterization of the direct targets of FOXO transcription factors throughout evolution. Aging Cell. 2016;15:673–685. doi: 10.1111/acel.12479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Page MM, Schuster EF, Mudaliar M, Herzyk P, Withers DJ, Selman C. Common and unique transcriptional responses to dietary restriction and loss of insulin receptor substrate 1 (IRS1) in mice. Aging (Albany NY) 2018;10:1027–1052. doi: 10.18632/aging.101446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Stout MB, Tchkonia T, Pirtskhalava T, et al. Growth hormone action predicts age-related white adipose tissue dysfunction and senescent cell burden in mice. Aging (Albany NY) 2014;6:575–586. doi: 10.18632/aging.100681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Stout MB, Swindell WR, Zhi X, et al. Transcriptome profiling reveals divergent expression shifts in brown and white adipose tissue from long-lived GHRKO mice. Oncotarget. 2015;6:26702–26715. doi: 10.18632/oncotarget.5760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Masternak MM, Bartke A, Wang F, et al. Metabolic effects of intra-abdominal fat in GHRKO mice. Aging Cell. 2012;11:73–81. doi: 10.1111/j.1474-9726.2011.00763.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bluher M, Michael MD, Peroni OD, et al. Adipose tissue selective insulin receptor knockout protects against obesity and obesity-related glucose intolerance. Dev Cell. 2002;3:25–38. doi: 10.1016/S1534-5807(02)00199-5. [DOI] [PubMed] [Google Scholar]
- 94.Bluher M, Patti ME, Gesta S, Kahn BB, Kahn CR. Intrinsic heterogeneity in adipose tissue of fat-specific insulin receptor knock-out mice is associated with differences in patterns of gene expression. J Biol Chem. 2004;279:31891–31901. doi: 10.1074/jbc.M404569200. [DOI] [PubMed] [Google Scholar]
- 95.Katic M, Kennedy AR, Leykin I, et al. Mitochondrial gene expression and increased oxidative metabolism: role in increased lifespan of fat-specific insulin receptor knock-out mice. Aging Cell. 2007;6:827–839. doi: 10.1111/j.1474-9726.2007.00346.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kappeler L, De Magalhaes Filho C, Dupont J, et al. Brain IGF-1 receptors control mammalian growth and lifespan through a neuroendocrine mechanism. PLoS Biol. 2008;6:e254. doi: 10.1371/journal.pbio.0060254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sadagurski M, Cheng Z, Rozzo A, et al. IRS2 increases mitochondrial dysfunction and oxidative stress in a mouse model of Huntington disease. J Clin Invest. 2011;121:4070–4081. doi: 10.1172/JCI46305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Li Q, Ren J. Influence of cardiac-specific overexpression of insulin-like growth factor 1 on lifespan and aging-associated changes in cardiac intracellular Ca2+ homeostasis, protein damage and apoptotic protein expression. Aging Cell. 2007;6:799–806. doi: 10.1111/j.1474-9726.2007.00343.x. [DOI] [PubMed] [Google Scholar]
- 99.Ren J, Brown-Borg HM. Impaired cardiac excitation-contraction coupling in ventricular myocytes from Ames dwarf mice with IGF-I deficiency. Growth Horm IGF Res. 2002;12:99–105. doi: 10.1054/ghir.2002.0267. [DOI] [PubMed] [Google Scholar]
- 100.Um SH, D’Alessio D, Thomas G. Nutrient overload, insulin resistance, and ribosomal protein S6 kinase 1, S6K1. Cell Metab. 2006;3:393–402. doi: 10.1016/j.cmet.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 101.Selman C, Tullet JM, Wieser D, et al. Ribosomal protein S6 kinase 1 signaling regulates mammalian life span. Science. 2009;326:140–144. doi: 10.1126/science.1177221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lamming DW, Ye L, Katajisto P, et al. Rapamycin-induced insulin resistance is mediated by mTORC2 loss and uncoupled from longevity. Science. 2012;335:1638–1643. doi: 10.1126/science.1215135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ranieri SC, Fusco S, Panieri E, et al. Mammalian life-span determinant p66shcA mediates obesity-induced insulin resistance. Proc Natl Acad Sci U S A. 2010;107:13420–13425. doi: 10.1073/pnas.1008647107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Migliaccio E, Giorgio M, Mele S, et al. The p66shc adaptor protein controls oxidative stress response and life span in mammals. Nature. 1999;402:309–313. doi: 10.1038/46311. [DOI] [PubMed] [Google Scholar]
- 105.Tomilov AA, Bicocca V, Schoenfeld RA, et al. Decreased superoxide production in macrophages of long-lived p66Shc knock-out mice. J Biol Chem. 2010;285:1153–1165. doi: 10.1074/jbc.M109.017491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wientjes FB, Segal AW. NADPH oxidase and the respiratory burst. Semin Cell Biol. 1995;6:357–365. doi: 10.1016/S1043-4682(05)80006-6. [DOI] [PubMed] [Google Scholar]
- 107.Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hofmann JW, Zhao X, De Cecco M, et al. Reduced expression of MYC increases longevity and enhances healthspan. Cell. 2015;160:477–488. doi: 10.1016/j.cell.2014.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bartke A. Healthy aging: is smaller better? - a mini-review. Gerontology. 2012;58:337–343. doi: 10.1159/000335166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang S, Ren J. Obesity Paradox in Aging: From Prevalence to Pathophysiology. Prog Cardiovasc Dis. 2018;61:182–189. doi: 10.1016/j.pcad.2018.07.011. [DOI] [PubMed] [Google Scholar]
- 111.Murakami S. Stress resistance in long-lived mouse models. Exp Gerontol. 2006;41:1014–1019. doi: 10.1016/j.exger.2006.06.061. [DOI] [PubMed] [Google Scholar]
- 112.Tower J. Sex-Specific Gene Expression and Life Span Regulation. Trends Endocrinol Metab. 2017;28:735–747. doi: 10.1016/j.tem.2017.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]