Abstract
Development, differentiation, and response to environmental stimuli are characterized by sequential changes in cellular state initiated by the de novo binding of regulated transcriptional factors to their cognate genomic sites 1,2,3. The mechanism whereby a given regulatory factor selects a limited number of in vivo targets from myriads of potential genomic binding sites is undetermined. Here we show that up to 95% of induced de novo genomic binding by the glucocorticoid receptor4, a paradigmatic ligand-activated transcription factor, is targeted to pre-existing foci of accessible chromatin. Factor binding invariably potentiates chromatin accessibility. Cell-selective glucocortocoid receptor genomic occupancy patterns appear to be comprehensively pre-determined by cell-specific differences in baseline chromatin accessibility patterns, with secondary contributions from local sequence features. The results define a novel framework for understanding regulatory factor-genome interactions, and provide a molecular basis for the tissue-selectivity of steroid pharmaceuticals and other agents that intersect the living genome.
How regulatory factors interact with the chromatin landscape to effect gene regulation is one of the leading questions in genome biology. Chromatin structure is altered at cis-regulatory regions, resulting in hypersensitivity of the underlying DNA to nuclease attack in vivo 5,6,7. However, how this pre-existing landscape influences de novo binding site selection has not been determined.
Here we address this using a well-controlled model system, the endogenous glucocorticoid hormone response pathway found in most mammalian cells. The cellular actions of glucocorticoids are mediated through the glucocorticoid receptor (GR)4, a hormone-activated transcription factor that rapidly translocates to the nucleus, whereupon its electively engages up to several thousand cognate genomic binding sites9,10. GR signaling thus represents an ideal system for both qualitative and quantitative analysis of de novo transcription factor-genome interactions in a highly controlled fashion.
We first sought to determine the global relationship between the pre-existing chromatin accessibility state of untreated cells and the pattern of GR binding following hormone induction. GR is widely believed to function as a ‘pioneer protein’ that is capable of autonomous binding to genomic DNA target sites resulting in local chromatin remodeling11,12 However, this concept is based largely on qualitative results from a limited set of loci13.
To gain a genome-wide perspective, we used digital DNaseI analysis14,15 and ChIP-seq10,17,18 to map chromatin accessibility and GR occupancy at high resolution both before and after steroid hormone (dexamethasone, Dex) treatment in a well-studied model cell type (mouse 3134 mammary adenocarcinoma cells). Digital DNaseI profiling enables quantitative delineation of chromatin accessibility, including both classical DNaseI hypersensitive sites (DHSs) as well as regions of general chromatin accessibility marked by DNaseI sensitivity16 (Supplementary Figs.1,2).
Genome-wide DNaseI sensitivity and GR occupancy profiles were highly reproducible (Supplementary Fig.3) and revealed a striking correspondence between the locations of GR occupancy post-dexamethasone and the pre-existing pattern of chromatin accessibility in untreated cells (Fig.1 and Supplementary Fig. 3a–c). To quantify this phenomenon, we delineated genomic regions with significantly increased chromatin accessibility over background, and identified 97,717 strongly DNaseI sensitive regions encompassing 2.1% (56.7 Mb) of the genome in untreated cells (Supplementary Tables 1,2 and Supplementary Notes), within which we localized 87,490 DHSs (0.4% of genome at a false discovery rate (FDR) of 1%; Supplementary Tables 1,3).
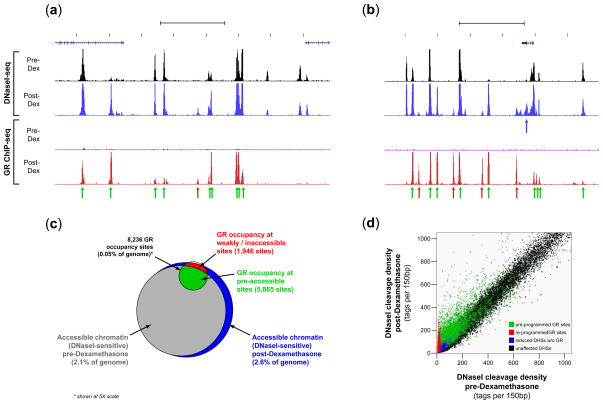
FIGURE 1. Dominant effect of chromatin accessibility on GR occupancy patterns.
(a–b) Examples of DNaseI sensitivity and GR occupancy patterns in relation to dexamethasone exposure (see Supplementary Figure 2a–c for additional examples). Each data track shows tag density (150bp sliding window) from either DNaseI-seq or GR ChIP-seq, normalized to allow comparison across different samples (Online Methods). Green arrows mark sites of post-hormone GR occupancy in pre-existing DNaseI-sensitive chromatin (‘pre-programmed’ sites). Red arrows mark GR occupancy sites in pre-hormone inaccessible chromatin that result in post-hormone chromatin remodeling (‘re-programmed’ sites). Blue arrows mark hormone-induced DHSs not directly associated with GR occupancy (see also Supplementary Fig 4c). (c) Venn diagram summarizing global GR occupancy vs. chromatin accessibility landscape (~25M read depth) in mammary cells (Note: for legibility, GR circle shown at 5X scale). Most GR occupancy occurs within pre-hormone accessible chromatin. A small fraction of generally weak GR peaks (5.2% of total) are not associated with re-programmed or pre-programmed chromatin. (d) DNaseI sensitivity (tag density) pre-hormone (horizontal axis) vs. post-hormone (vertical axis). Colors match those used in panel (c). Black = pre-hormone accessible regions with no post-hormone GR occupancy. Blue = DNaseI-sensitive regions induced post-hormone without GR occupancy (see Supplementary Fig 4c). Green = pre-hormone DNaseI sensitive regions occupied by GR post-hormone (‘pre-programmed’ sites). Red = pre-hormone inaccessible chromatin remodeled by GR occupancy (‘re-programmed’ sites), resulting in marked alteration in DNaseI sensitivity. (see Supplementary Fig 4a–b).
Analysis of GR ChIP-seq data from hormone-treated cells revealed 8,236 sites of GR occupancy (Supplementary Table 4). Performing de novo motif discovery on the top 500 GR occupancy sites recovered a 15bp motif that closely matched the consensus glucocorticoid receptor binding element (GRBE; Figure 2a)19,20. >80% of GR occupancy sites contained some form of this GRBE consensus sequence (at P<10−3), with 50% containing higher stringency matches (P<10−4).
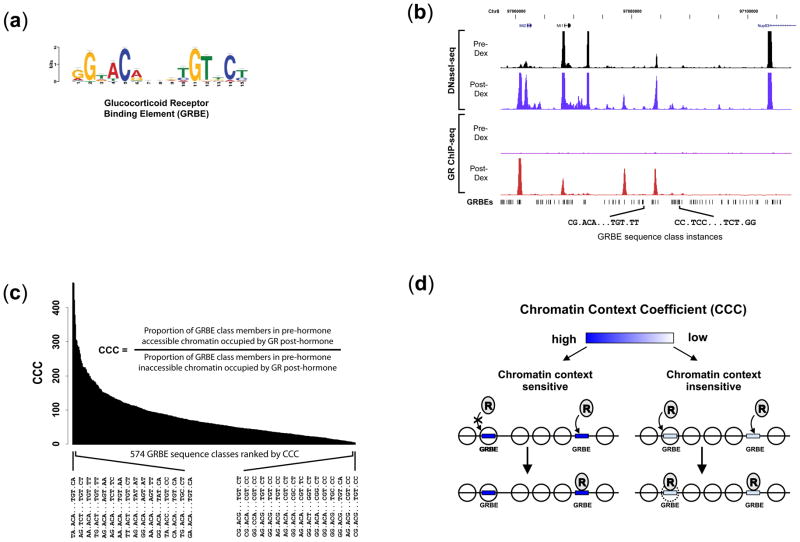
FIGURE 2. Quantitative effect of chromatin context on GR occupancy of GRBEs.
(a) Top scoring motif recovered from de novo motif discovery performed on the top 500 GR occupancy sites by ChIP-seq tag density (MEME E-value: 8.6e−753) closely matches the consensus glucocorticoid receptor binding element (GRBE). (b) 50kb genomic region comparing pre-and post-hormone chromatin accessibility and GR occupancy in relation to GRBE genomic sequence matches (P<10−3). Only a small fraction of the ~2.3×106 GRBE consensus sites are occupied in vivo, and occupied sites differ in their underlying combinations of consensus GRBE motif nucleotides. (c) GRBE sequence classes ranked by Chromatin Context Coefficient (CCC). Genomic GRBE motif matches can be partitioned into discrete sequence classes, each comprising an identical (and distinct) combination of consensus nucleotides. Within each class of identical sequence elements, occurrence of member genomic sequences in a range of pre-hormone DNaseI sensitivity environments (from inaccessible to hyperaccessible) enables quantification of the effect of chromatin context on the probability of post-hormone GR occupancy. Ranking specific GRBE sequence classes by CCC reveals graded sensitivity to chromatin context, from highly context-dependent elements that engender GR occupancy only when situated in accessible chromatin, to relatively context-independent elements associated with sites where GR occupancy induces chromatin remodeling. (d) Model illustrating the contribution of chromatin accessibility to transcription factor binding. CCC encodes the occupancy potential of different GRBE sequence classes relative to accessibility.
The significant majority of GR occupancy sites in 3134 cells (71%, 5,865 sites) were targeted to the 2.1% of the genome defined by pre-existing (i.e., pre-hormone or ‘baseline’) strongly DNaseI sensitive regions (P<10−300). An additional~9% of binding localized to weakly DNaseI sensitive regions, with 80% of GR binding occurring within 4.9% of the genome (Supplementary Fig. 3d). However, this estimate represents a lower limit. For example, increasing the sequencing depth of the pre-hormone DNaseI-seq sample ~8-fold increased the proportion of GR sites falling within pre-hormone accessible chromatin from 71% to 88.3% (P<10−300; Supplementary Notes and Supplementary Fig. 3d). In hormone treated cells,95% of GR occupancy sites (and >99% on deep sequencing) localized to accessible chromatin (P<10−300). Additionally, we observed DHSs unique to hormone-treated cells that were not directly associated with GR binding (Figure 1a, blue arrows; Fig. 1d, blue crescent). Most of these DHSs derived from sites of very weak pre-hormone chromatin accessibility that were potentiated following hormone treatment (Supplementary Fig.4), and may thus represent indirect or ‘network’ effects of GR action.
Taken together, the results indicate that pre-existing patterns of chromatin accessibility exert a dominant, global effect on de novo regulatory factor localization, and that factor occupancy is almost invariably associated with local chromatin remodeling.
In spite of the fact that average pre-hormone chromatin accessibility at promoter regions was high, 93% of GR occupancy sites were observed >2.5kb distal to the nearest transcriptional start site (TSS)(vs. 61% of all DHSs; Supplementary Fig.5). GR sites were also highly clustered along the genome (Supplementary Fig.6). However, we found no clear relationship between GR occupancy patterns and transcriptional activation of nearby genes (Supplementary Table 5 and Supplementary Fig.7), raising the possibility that GR acts through long-range mechanisms or that many GR binding events are opportunistic.
We next asked why, given the dominant influence of chromatin structure, GR occupied only a subset of DNaseI-sensitive regions, and why a small minority of GR binding events could escape the requirement for pre-existing highly accessible chromatin. We first examined the relationship between GRBE motifs and GR occupancy patterns by developing an approach for quantifying the differential sensitivity of different GRBEs to their local chromatin environment. Of 2,296,115 significant GRBE (15bp) matches21 (Fig. 2a) in the non-repetitive mouse genome, only a very small fraction were actually occupied in vivo post-hormone. Standard position weight matrix matching21 to the GRBE consensus was a poor predictor of GR binding, as many GRBEs with a high matching score were not occupied by GR. However, we observed that many occupied GRBEs harbored distinct instantiations of the consensus sequence comprising specific combinations of non-degenerate bases (Fig. 2b).
To quantify the global relationship between these combinations and chromatin reprogramming, we partitioned the ~2.3 million candidate GRBEs into motif sequence classes such that all members of a given class shared identical non-degenerate consensus base sequences. Next, we computed a Chromatin Context Coefficient (CCC) for each GRBE sequence class that quantified its relative dependence on pre-hormone chromatin accessibility as a pre-requisite for post-hormone GR occupancy (Fig. 2c–d and Supplementary Notes). High CCC values denote strong chromatin context-dependence of GR binding, while low values mark classes with potential to override the dominant effect of chromatin structure and initiate local remodeling. Notably, no CCC values <1 were observed, indicating that GR occupancy was universally enhanced by residence of GRBE within pre-hormone accessible chromatin. 526/1,100 statistically well-defined GRBE sequence classes lacked any occupancy at GRBE instances in pre-hormone closed chromatin (i.e., CCC = ∞), indicating an absolute requirement of pre-existing chromatin accessibility for GR occupancy (Supplementary Notes and Supplementary Table 6). Ranking the remaining 574GRBE sequence classes with finite CCC values revealed a hierarchy of chromatin dependence among GRBE elements, with the quantitative effect of pre-existing chromatin accessibility on the probability of GR occupancy ranging from 2-to 473-fold (Fig. 2c and Supplementary Table 6). CCC values and GRBE class size were uncorrelated (R2 = 0.15).
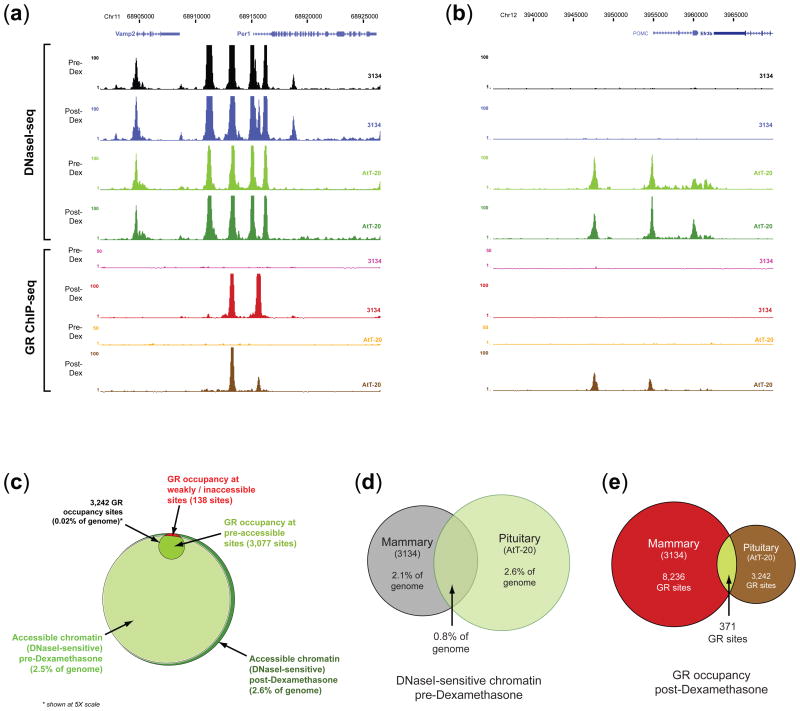
We next profiled both DNaseI sensitivity and GR binding pre-and post-dexamethasone in a highly divergent cell type (mouse pituitary cell line AtT-20), (Fig. 3, Supplementary Fig. 8a–c, and Supplementary Tables 7–10). In pituitary cells, we found an even tighter targeting of de novo GR occupancy to pre-hormone accessible chromatin, with 95% (3,079/3,242) of GR occupancy sites occurring within pre-hormone DNaseI-sensitive regions (Fig. 3c). As in mammary cells, no pre-hormone GR occupancy was observed, and substantially all (99%) post-hormone GR occupancy was accompanied by increased DNaseI sensitivity. Pre-hormone chromatin accessibility patterns in mammary vs. pituitary cells were highly discordant (~30% overlap), consistent with cell type-specific cis-regulatory landscapes (Fig. 3d). The cell-selectivity of GR occupancy was even more pronounced, with only 11.4% (371/3,242) of GR occupancy sites shared between pituitary and mammary cells (Fig. 3e).
FIGURE 3. Cell-specific chromatin landscapes determine cell-selective GR occupancy.
(a–b) Pituitary-specific GR occupancy dictated by pituitary-specific DNaseI sensitivity transitions. Shown are examples of DNaseI sensitivity and GR occupancy patterns in relation to hormone exposure comparing mouse mammary (3134) and pituitary (AtT-20) cells (see Fig.1 legend and Supplementary Fig.8a-c for additional examples). (c) Global GR occupancy vs. chromatin accessibility landscape in pituitary cells. In pituitary cells, virtually all sites of GR occupancy (94.9%, 3,079/3,242 sites) occur within pre-hormone accessible chromatin. The small fraction of re-programmed GR sites (138 GR ChIP peaks, 4.2% of total) is shown in red. As in mammary cells, only a small fraction of pre-hormone accessible chromatin is occupied (note: for legibility, GR circle shown at 5X scale). (d) Significant differences in genomic distribution of pre-hormone DNaseI sensitivity in mammary (grey) vs. pituitary (green) cells; only 0.78% of genome (20.5Mb) is accessible in both cell types. (e) GR occupancy is highly cell-selective. Only 371 GR occupancy sites are shared between mammary and pituitary cells (4.5% of 3134 sites and 11.4% of AtT-20 sites).
83% (473/572) GRBE sequence classes with well-defined CCC values in both 3134 and AtT-20 cells showed statistically significant enhancement of GR binding in both cell types (CCC > 1, Supplementary Fig. 8d). In AtT-20, enhancement of GRBE occupancy by chromatin context ranged from 3-to 596-fold ((Supplementary Table 6). The effects associated with specific GRBE classes were largely stable between cell types (R= 0.48, P<0.01; Supplementary Fig. 8e). Notably, we were unable to identify a unique or specific GRBE sequence class that functioned exclusively to render closed chromatin more accessible.
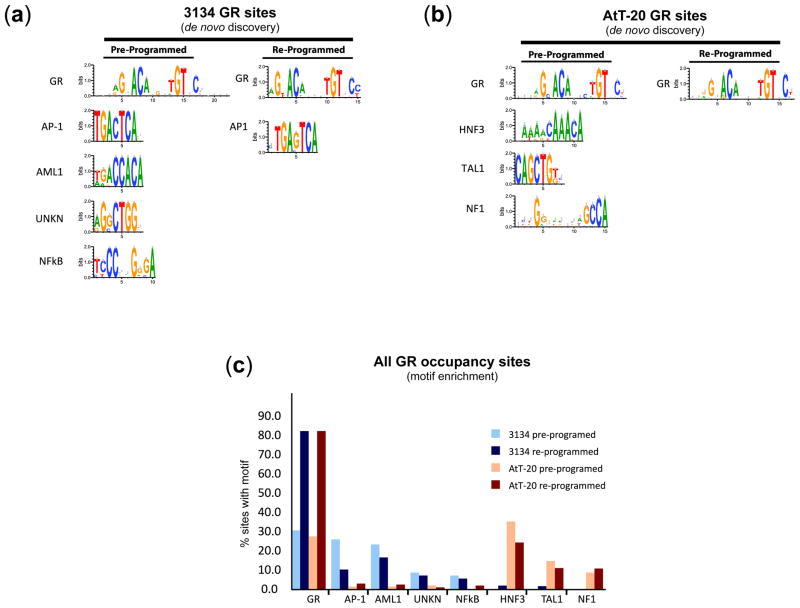
In 3134 cells, ~25% of baseline accessible DHSs contained GRBEs, yet only 23% are occupied by GR, suggesting additional requirements for GR binding. GR has been reported to interact with a number of cell-restricted and ubiquitous transcriptional regulators22. We therefore examined GR sites for evidence of accessory factor motifs by performing de novo motif discovery on pre-programmed vs. re-programmed GR sites from each cell type. This analysis revealed distinct complements of highly significant (e<10−5) motifs enriched in conjunction with classical GRBEs (Fig. 4 and Supplementary Fig.9). In mammary pre-programmed sites, these included AP-1 most prominently, AML1, NF-κB and a novel unassigned motif (Fig. 4a). In pituitary pre-programmed sites, we recovered the canonical GRBE plus consensus motifs for HNF3, TAL1 and NF1 (Fig. 4b). Notably, both HNF3 and NF1 have previously been connected with both nuclear receptor binding generally and with GR interaction specifically 23,24. ChIP analyses confirmed that at least a proportion of the identified sequence motifs were occupied by their cognate factors (Supplementary Fig. 10a–e).
FIGURE 4. Regulatory motifs in GR-occupied regions differ substantially between cell types.
(a–b) Results of de novo motif discovery (see Supplementary Notes) performed on the top 500 GR occupancy sites identified in 3134 (panel a) and AtT-20 (panel b). The GR sites were further separated into pre-programmed (GR occupancy within pre-hormone accessible chromatin) vs. re-programmed (GR occupancy within pre-hormone inaccessible chromatin) sites. Shown are motifs with highly significant enrichment (e<10−5). In all cases, the GRBE is the most highly enriched single motif (8.6e−753). Notably, AP1 and AML1 motifs are enriched in 3134 cells (panel a) while HNF3 and NF1 are correspondingly enriched in AtT-20 (panel b). (c). Motif occurrence patterns across all GR occupancy sites. Bar plots show percentage of all GR occupancy sites (8,236 sites in 3134 cells vs. 3,242 sites in AtT-20) that harbor significant matches to the de novo-identified motifs from panels a–b. Note that canonical GRBEs are highly enriched in re-programmed sites vs. pre-programmed sites (>80% of re-programmed sites vs. <30% of pre-programmed sites, P<10−4).
Analysis of re-programmed GR sites revealed a strikingly different picture. In 3134 cells, we found only the canonical GRBE and AP-1 motifs. GRBEs were found in >80% of re-programmed sites vs. only 29% of pre-programmed sites (P<10−100) (Fig. 4c and Supplementary Figure 9), compatible with direct engagement of DNA following chromatin penetration. By contrast, consensus AP-1 sites were found in~10% of re-programmed sites vs. 26% of pre-programmed sites (P<10−80), and AP-1 and GR motifs were mutually exclusively distributed, such that only 4.8% of pre-programmed sites had both (data not shown). In AtT-20 cells, consensus HNF3 motifs were identified in 34% of pre-programmed vs. 21% in re-programmed GR sites (P<.003) (Fig. 4c and Supplementary Fig.9), with mutual exclusivity between GRBEs and HNF3 in pre-programmed sites (only 5.8% of sites with both, P<10−11), analogous to results with AP-1 in 3134 cells (data not shown). Taken together, these data suggest that in both cell types, common regulatory factors including AP-1 (3134) and HNF3 (AtT-20) – or possibly other factors acting through the same cognate motifs – may be mediating GR occupancy within a subset of pre-hormone accessible chromatin. However, this effect is quantitatively minor compared with that conferred by chromatin accessibility. For example, of the 34,587 positions in the mouse genome where AP-1 motifs and GRBEs co-occur, only 1.8%are occupied by GR post-hormone in 3134 cells, compared with the ~80% of GR binding that occurs with accessible chromatin generically (Supplementary Fig. 10f–g).
In summary, our results reveal the marked dominant effect of pre-existing chromatin structure on de novo regulatory factor binding. This effect may be secondarily modulated by local sequence features such as variations in regulatory factor recognition elements or the presence of accessory sequence motifs for well-known regulators. However, even considered collectively, these additional sequence features likely account for only a minority of the overall effect.
Because of the dramatic dependence of regulatory factor binding on pre-existing chromatin architecture, substantial variations in the baseline pattern of chromatin accessibility between different cell types is expected to expose distinct patterns and genomic locations of regulatory factor recognition sequences. The distribution of such exposed binding elements should, in turn, dictate the genomic distribution of de novo regulatory factor binding.
Corticosteroids are one of the most commonly used pharmaceuticals, and exhibit widely differing effects on different tissues in spite of the fact that most human cell types contain the same glucocorticoid response machinery 4. Our results provide a simple explanation for these effects, namely, that they are a direct consequence of cell type-specific patterns of baseline (i.e., pre-hormone) chromatin accessibility and exposed GR recognition sequences.
A further implication of our results is that sequential factor occupancy during developmental and differentiation may be largely pre-specified by the chromatin landscape as a form of cellular memory. Re-programming of chromatin structure at a limited number of sites may incrementally alter this pattern, and create new potential occupancy sites for subsequently available factors, resulting in a directional process that is difficult to reverse without extraordinary measures such as the simultaneous introduction of multiple potent regulators25.
ONLINE METHODS
Cell lines and culture conditions
The 3134 cell line was derived by transformation of C127, originally isolated from a mammary adenocacinoma tumor of the RIII mouse. The AtT-20 cell line is an anterior pituitary corticotroph of murine origin (ATCC). Both cell lines were maintained in Dulbecco’s Modified Eagle Medium (DMEM) (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (Gemini, Woodland, California), 2 mM L-glutamine, 1 mM sodium pyruvate, 0.1 mM non-essential amino acids, 5 mg/ml penicillin-streptomycin (Invitrogen, Carlsbad, CA) and kept at 37°C incubator with 5% CO2. Cells were transferred to 10% charcoal-dextran-treated, heat-inactivated fetal bovine serum for 48 hrs prior to hormone treatment (1hr with 100 nM dexamethasone)26.
ChIP assays
Chromatin immunoprecipitations were performed as per standard protocols (Upstate)27. Briefly, cells were treated with either vehicle or 100 nM dexamethasone for 1 hr. Cells were cross-linked for 10 min at 37 °C in 1% formaldehyde followed by a quenching step for 10 min with 150 mM glycine. A single chromatin immunoprecipitation contained 400ug of sonicated, soluble chromatin and a cocktail of antibodies to the glucocorticoid receptor (7.5 μg ofPA1-511A antibody, ABR, 15ug of MA1-510 antibody, ABR and 3 ug of sc-1004, Santa Cruz). The ChIP reaction was scaled 5× for ChIP-seq. DNA isolates from immunoprecipitates were used as templates for real-time quantitative PCR amplification or sequenced as described below. All ChIP experiments were performed at least two times.
Digital DNaseI mapping
Digital DNaseI mapping was performed essentially as described in28. Briefly, 3134 and AtT-20 cells were grown as described above. 1×108 cells were pelleted and washed with cold phosphate-buffered saline. We resuspended cell pellets in Buffer A (15 mM Tris-Cl (pH 8.0), 15 mM NaCl, 60 mM KCl, 1 mM EDTA (pH 8.0), 0.5 mM EGTA (pH 8.0), 0.5 mM spermidine, 0.15 mM spermine) to a final concentration of 2×106 cells/ml. Nuclei were obtained by dropwise addition of an equal volume of Buffer A containing .04% NP-40 to the cells, followed by incubation on ice for 10 min. Nuclei were centrifuged at 1,000g for 5 min, and then resuspended and washed with 25 ml of cold Buffer A. Nuclei were resuspended in 2 ml of Buffer A at a final concentration of 1×107 nuclei/ml. We performed DNaseI (Roche, 10–80 U/ml) digests for 3 min at 37 °C in 2 ml volumes of DNase I buffer (60 mM CaCl2, 750 mM NaCl). Reactions were terminated by adding an equal volume (2 ml) of stop buffer (1 M Tris-Cl (pH 8.0), 5 M NaCl, 20% SDS, 0.5 M EDTA (pH 8.0), 10 μg/ml RNase A, Roche) and incubated at 55 °C. After 15 min, we added Proteinase K (25 μg/ml final concentration) to each digest reaction and incubated them overnight at 55 °C. After DNase I treatments, careful phenol-chloroform extractions were performed. Control (untreated) samples were processed as above except for the omission of DNase I. DNaseI double-cut fragments and sequencing libraries constructed as described in 29,30.
High-throughput sequencing data analysis
High-throughput sequencing output is processed similarly for both DNase I and ChIP data. 27bp Illumina sequence reads were mapped to the human genome (UCSC HG18), and only uniquely mapping read positions were considered. For DNaseI sequence tags, 5’ ends represent in vivo cleavage events. Significantly enriched regions were identified in both DNaseI and GR CHiP-seq data sets using a version of the HotSpot algorithm31 (and Thurman et el, in preparation32; see also description below).
Delineation of DNaseI-sensitive regions
DNaseI cleavage sites were represented computationally as the single base pair from the 5’ end of each sequence tag. Enrichment of tags along the genome is gauged in a small window (200–300bp) relative to a local background model based on the binomial distribution, using the observed tags in a 50kb surrounding window. Each mapped tag gets a z-score (explained below) relative to the surrounding small and background windows centered on the tag. A ‘hotspot’ is defined as a succession of neighboring tags within a 250bp window, each of whose z-score is greater than 2. Once a hotspot is identified, the hotspot itself is assigned a z-score relative to the small and background windows centered on the average position of the tags forming the hotspot.
Z-score calculation
Suppose n observed tags are mapped to the small window, and N total tags are mapped to the 50kb surrounding background window (N≥n ). Each tag in the background window is considered an “experiment,” with favorable outcome if it falls in the smaller window. Assuming each base in the 50kb window is equally likely, the probability of success for each tag is therefore p=250/50000 . Not all bases in the 50kb window may be uniquely mappable by 27-mers (the tag length for our data), however, so p is adjusted to account for the number of uniquely mappable bases for that window. Under these assumptions, the binomial distribution applies, and the expected number of tags falling in the smaller window is μ=Np.
The standard deviation of this expected value is
Finally, the z-score for the observed number of tags in the smaller window is z = n−μ/σ.
We also compute the expected number of tags and z-score using the entire genome as background, rather than the 50kb window, and, to be conservative, report the lower of the two z-scores.
Correction for regional DNaseI sensitivity background
In regions of very high enrichment, the resulting hotspots can inflate the background for neighboring regions, and deflate neighboring z-scores. The effect is that regions of otherwise high enrichment can be shadowed by a neighboring extreme hotspot. To address this problem, we implement a two-pass procedure. After the first round of hotspot detection, we delete all tags falling in the first-pass hotspots. We then compute a second round of hotspots with this deleted background. The hotspots from the first and second passes are combined, and all are re-scored using the deleted background: the number of tags in each hotspot is computed using all tags, but 50kb background windows use only the deleted background.
Identification of DNaseI hypersensitive peaks
Hotspots were resolved into discrete 150bp peaks using a peak-finding procedure. First, neighboring hotspots within 150bp of each other are merged. We compute a sliding window tag density (tiled every 20bp in 150bp windows), and then perform peak-finding of the density in each merged hotspot region. Each 150bp peak is assigned the z-score from the unmerged hotspot that contains it. Peak-finding proceeds in two phases, so that each hotspot has at least one peak. Phase-I peaks are local maxima occurring in regions above the 99th percentile of the density and satisfying certain ad-hoc criteria for ensuring a sustained increase to or decrease from the local maxima. For each hotspot that does not contain at least one phase-I peak, a phase-II peak is simply defined as the maximum density value in the hotspot. For details, see the code available from the authors.
False Discovery Rate (FDR) calculations
We assign FDR (false discovery rate) z-score thresholds to a given hotspot set using random data. As a null model, we computationally generate tags uniformly over the uniquely mappable bases of the genome. We use the same number of tags for observed and random data. The random data also coalesce into hotspots, which we identify and score as usual. For a given z-score threshold T, the FDR for the observed hotspots with z-score greater than T is estimated as
Since the numerator, which is calculated on a dataset that is entirely null, likely overestimates the number of false positives in the observed data, this is likely a conservative estimate of the FDR. FDR 0% hotspots are constructed by taking all hotspots with a z-score greater than the maximum z-score attained in the paired random set. We construct FDR-thresholded peak sets by performing peak finding in FDR-thresholded hotspots.
Generation of tables of DNaseI sensitive regions and DHSs for pre- and post-hormone data sets
We observe that Dex- DNase I hotspots (DNase I sensitive regions) that occur outside of Dex+ DNase I hotspots are generally of low intensity and significance. We therefore restrict our published tables of Dex- hotspots and peaks to those that also intersect Dex+ hotspots. For 3134 we pool samples from two replicates for each condition (Dex− and Dex+), whereas for AtT-20, we use a single replicate per condition. See, however, the section on “Replicate concordant sets,” below, which details methods for defining DNase I sets for CCC analysis and aggregate plots.
Analysis of ChIP-seq data
The preceding sections describe procedures for handling DNase I tag data. Modifications are made to this process to account for unique properties of ChIP data. For one, duplicate tags (tags mapped to the same location) are used for DNase I, but unique tags only are retained for ChIP calculations. This is because multiple tags mapping to the same position for DNase I provide biological meaning (the more tags at a given position, the more locally accessible the chromatin is at that location), whereas for ChIP data we expect the relevant information to be only the locations of measured binding. The most important difference between the processing of DNase I and ChIP data is the use of sequence data for the ChIP input experiment, which gives, for each ChIP experiment, a measure of non-binding background signal, which can be significant. We use input tags at the scoring phase for ChIP hotspots. Once two-pass hotspots have been identified as usual, we score each hotspot by first subtracting the number of tags in the paired input experiment from the observed ChIP tags in the hotspot window before applying the binomial model. We normalize the number of input tags subtracted in each window by a factor that brings the total number of input tags to the same number of ChIP tags. We do not subtract input tags from the surrounding 50kb background window, so the scoring should be conservative.
Adjusted scoring for maximum sensitivity analyses using deep sequencing data
When scoring the deeper, 100 million tag datasets, we strive for maximum sensitivity in detecting accessible chromatin, and therefore we make two adjustments in scoring hotspots. First, instead of taking the lower of the two z-scores from using a 50kb local background and the genome-wide background, we use the greater of the two; and second, we lower the initial z-score threshold for hotspot detection from two to one.
For additional Methods see Supplementary Note.
Supplementary Material
Acknowledgments
We would like to thank Tina Miranda, Stephanie Morris, Kip Nalley and Lars Grontved for critical reading of the manuscript. We also thank Molly Weaver, Kristen Lee, Fidencio Neri, Daniel Bates, and Morgan Diegel for technical assistance with DNaseI library preparation and sequencing. This research was supported (in part) by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research and funding from NIH grant 1RC2HG005654 to J.A.S.
Footnotes
AUTHOR CONTRIBUTIONS
SJ, PJS, GLH and JAS designed the experiments. SJ, PJS, SCB and TAJ conducted the DNase-seq, ChIP-seq and expression array experiments. SJ, PJS, RET, MHS, and JAS analyzed the data. SJ, PJS, RET, MHS, GLH and JAS wrote the manuscript. The authors declare no competing financial interests.
DATA AVAILABILITY
All DNaseI and ChIP-seq data are available through the NCBI Sequence Read Archive (SRA), under Study #SRP004871, and the following accession numbers: SRX034804, SRX034802, SRX034811, SRX034818, SRX034860, SRX034861, SRX034862, SRX034863, SRX034864, SRX034865, SRX034837, SRX034838, SRX034867, SRX034868, SRX034869, SRX034870, SRX034871, SRX034872.
References
- 1.Britten RJ, Davidson EH. Gene regulation for higher cells: a theory. Science. 1969;165:349–357. doi: 10.1126/science.165.3891.349. [DOI] [PubMed] [Google Scholar]
- 2.McKenna NJ, O'Malley BW. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell. 2002;108:465–474. doi: 10.1016/s0092-8674(02)00641-4. [DOI] [PubMed] [Google Scholar]
- 3.Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 4.Evans RM. The steroid and thyroid hormone receptor superfamily. Science. 1988;240:889–895. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Felsenfeld G, Groudine M. Controlling the double helix. Nature. 2003;421:448–453. doi: 10.1038/nature01411. [DOI] [PubMed] [Google Scholar]
- 6.Wu C. The 5' ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980;286:854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]
- 7.Gross DS, Garrard WT. Nuclease hypersensitive sites in chromatin. Annu Rev Biochem. 1988;57:159–197. doi: 10.1146/annurev.bi.57.070188.001111. [DOI] [PubMed] [Google Scholar]
- 8.Htun H, Barsony J, Renyi I, Gould DL, Hager GL. Visualization of glucocorticoid receptor translocation and intranuclear organization in living cells with a green fluorescent protein chimera. Proc Natl Acad Sci USA. 1996;93:4845–4850. doi: 10.1073/pnas.93.10.4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.So AYL, Chaivorapol C, Bolton EC, Li H, Yamamoto KR. Determinants of cell-and gene-specific transcriptional regulation by the glucocorticoid receptor. Plos Genetics. 2007;3:e94. doi: 10.1371/journal.pgen.0030094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reddy TE, et al. Genomic determination of the glucocorticoid response reveals unexpected mechanisms of gene regulation. Genome Res. 2009;19:2163–2171. doi: 10.1101/gr.097022.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Richard-Foy H, Hager GL. Sequence-specific positioning of nucleosomes over the steroid-inducible MMTV promoter. EMBO J. 1987;6:2321–2328. doi: 10.1002/j.1460-2075.1987.tb02507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Becker P, Renkawitz R, Schütz G. Tissue-specific DNaseI hypersensitive sites in the 5'-flanking sequences of the tryptophan oxygenase and the tyrosine aminotransferase genes. EMBO J. 1984;3:2015–2020. doi: 10.1002/j.1460-2075.1984.tb02084.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hager GL, et al. Influence of chromatin structure on the binding of transcription factors to DNA. Cold Spring Harbor Symposia on Quantitative Biology. 1993;58:63–71. doi: 10.1101/sqb.1993.058.01.010. [DOI] [PubMed] [Google Scholar]
- 14.Hesselberth JR, et al. Global mapping of protein-DNA interactions in vivo by digital genomic footprinting. Nat Methods. 2009;6:283–289. doi: 10.1038/nmeth.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sekimata M, et al. CCCTC-binding factor and the transcription factor T-bet orchestrate T helper 1 cell-specific structure and function at the interferon-gamma locus. Immunity. 2009;31:551–564. doi: 10.1016/j.immuni.2009.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stalder J, et al. Tissue-specific DNA cleavages in the globin chromatin domain introduced by DNAase I. Cell. 1980;20:451–460. doi: 10.1016/0092-8674(80)90631-5. [DOI] [PubMed] [Google Scholar]
- 17.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 18.Robertson G, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods. 2007;4:651–657. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- 19.von der Ahe D, et al. Glucocorticoid and progesterone receptors bind to the same sites in two hormonally regulated promoters. Nature. 1985;313:706–709. doi: 10.1038/313706a0. [DOI] [PubMed] [Google Scholar]
- 20.Diamond MI, Miner JN, Yoshinaga SK, Yamamoto KR. Transcription factor interactions: selectors of positive or negative regulation from a single DNA element. Science. 1990;249:1266–1272. doi: 10.1126/science.2119054. [DOI] [PubMed] [Google Scholar]
- 21.Bailey TL, Gribskov M. Concerning the accuracy of MAST E-values. Bioinformatics. 2000;16:488–489. doi: 10.1093/bioinformatics/16.5.488. [DOI] [PubMed] [Google Scholar]
- 22.Beck IME, et al. Crosstalk in inflammation: the interplay of glucocorticoid receptor-based mechanisms and kinases and phosphatases. Endocr Rev. 2009;30:830–882. doi: 10.1210/er.2009-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rigaud G, Roux J, Pictet R, Grange T. In vivo footprinting of rat TAT gene: dynamic interplay between the glucocorticoid receptor and a liver-specific factor. Cell. 1991;67:977–986. doi: 10.1016/0092-8674(91)90370-e. [DOI] [PubMed] [Google Scholar]
- 24.Cordingley MG, Hager GL. Binding of multiple factors to the MMTV promoter in crude and fractionated nuclear extracts. Nucleic Acids Research. 1988;16:609–628. doi: 10.1093/nar/16.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 26.John S, et al. Kinetic complexity of the global response to glucocorticoid receptor action. Endocrinology. 2009;150:1766–1774. doi: 10.1210/en.2008-0863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.John S, et al. Interaction of the glucocorticoid receptor with the chromatin landscape. Molecular Cell. 2008;29:611–624. doi: 10.1016/j.molcel.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 28.Sekimata M, et al. CCCTC-binding factor and the transcription factor T-bet orchestrate T helper 1 cell-specific structure and function at the interferon-gamma locus. Immunity. 2009;31:551–564. doi: 10.1016/j.immuni.2009.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sabo PJ, et al. Genome-scale mapping of DNase I sensitivity in vivo using tiling DNA microarrays. Nat Methods. 2006;3:511–518. doi: 10.1038/nmeth890. [DOI] [PubMed] [Google Scholar]
- 30.Hesselberth JR, et al. Global mapping of protein-DNA interactions in vivo by digital genomic footprinting. Nat Methods. 2009;6:283–289. doi: 10.1038/nmeth.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sabo PJ, et al. Discovery of functional noncoding elements by digital analysis of chromatin structure. Proc Natl Acad Sci USA. 2004;101:16837–16842. doi: 10.1073/pnas.0407387101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thurman RE, Stamatoyannopoulos JA. A scan statistic algorithm for identification of genomic regions of chromatin accessibility. (in prep) [Google Scholar]
- 33.Bailey TL, Williams N, Misleh C, Li WW. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Research. 2006;34:W369–373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.