Abstract
The role of non-duplex DNA, the guanine-quadruplex structure in particular, is becoming widely appreciated. Increasing evidence in the last decade implicates quadruplexes in important processes such as transcription and replication. Interestingly, more recent work suggests roles for quadruplexes, in association with quadruplex-interacting proteins, in epigenetics through both DNA and histone modifications. Here, we review the effect of the quadruplex structure on post-replication epigenetic memory and quadruplex-induced promoter DNA/histone modifications. Furthermore, we highlight the epigenetic state of the telomerase promoter where quadruplexes could play a key regulatory role. Finally, we discuss the possibility that DNA structures such as quadruplexes, within a largely duplex DNA background, could act as molecular anchors for locally induced epigenetic modifications.
G-Quadruplexes and Their Biological Roles
The most studied form of DNA structure is the double helix. However, it is becoming increasingly evident that non-Watson–Crick base pairing, resulting in alternative DNA secondary structures, is possible while DNA maintains the predominantly duplex state in the genome. These non-canonical structures include Z-DNA (left-handed helix), triple helices, i-motifs, and DNA G-quadruplexes [1–4]. G-quadruplexes, also referred to as the G4 structures (G4 hereafter), comprise G-quartets resulting from self-assembly of Hoogsteen hydrogen-bonded clusters (see Glossary) of four guanines stabilized by monovalent cations (Figure 1) (reviewed in [5,6]). Of limited interest initially, an understanding of G4s was gained from molecular characterization by X-ray fiber diffraction [7,8] and biophysical studies [9,10]. Attention towards G4s surged after they were implicated in fundamental cellular processes such as replication, transcription, translation, and telomere maintenance [11–13] (Box 1).
Glossary.
Bloom’s syndrome: also known as Bloom–Torre–Machacek syndrome, a rare autosomal recessive disorder characterized by short stature; predisposes to the development of cancer and genomic instability.
C9ORF72 locus: gene playing an important role in the regulation of endosomal trafficking implicated in 9p-linked amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD).
CTCF: CCCTC-binding factor that has 11 zinc-finger domains.
DNA methyltransferases: family of enzymes that catalyze the transfer of a methyl group to DNA.
Fragile X mental retardation syndrome: typically due to expansion of the CGG triplet repeats within the fragile X mental retardation 1 (FMR1) gene on the X chromosome.
G4-binding ligands: small molecule ligands that interact with G4s.
Hoogsteen hydrogen-bonded clusters: alternative base pair bonding, N7 position of the purine base (as a hydrogen bond acceptor) and C6 amino group (as a donor) of the pyrimidine base.
Non-metastatic factor NME2: a metastasis suppressor gene, close homolog of NM23-H1.
Nucleosome exclusion: genomic regions outside chromatin packaging DNA–protein complexes; mostly present as open chromatin.
8-Oxoguanine (8oxog): one of the most common DNA lesions resulting from reactive oxygen species; may result in a mismatched pairing with adenine resulting in G-to-T and C-to-A substitutions in the genome; can be caused by ionizing radiation.
REST/co-REST/LSD1 repressor complex: repressor complex primarily discovered for silencing neuronal genes in non-neuronal cells.
Telomere: the (TTAGGG)n repetitive region at each end of a chromosome; it protects chromosomes from end-to-end fusion and prevents loss of essential genetic information during cell division.
Telomere repeat-binding Factor 2 (TRF2): also known as TERF2, a well-known member of the shelterin complex of proteins that prevent telomeres from being detected as DNA double-stranded breaks.
Werner’s syndrome: also known as ‘adult progeria’, a rare, autosomal recessive disorder that is characterized by the appearance of premature aging.
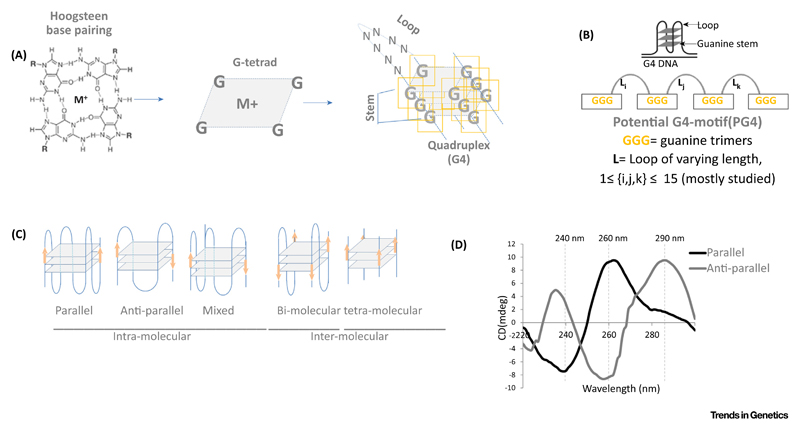
Figure 1. The G4 Topology.
(A) Hoogsteen base pairing gives self-assembly of G-tetrads [assisted by metal (M) ions] in sequences with repetitive G tracts (G-stem). (B) These G-tetrads are separated by stretches of nucleotides called ‘loops’ (L). Two or more G-tetrads form DNA secondary structure G-quadruplex (G4) DNA. (C) Different topologies in which G4s usually exist in intra-molecular and inter-molecular forms. (D) Schematic showing typical circular dichroism (CD) spectral signatures of parallel and anti-parallel G-quadruplexes (G4s).
Box 1. Understanding the G4 Topology.
Observations from gel electrophoresis mobility shift assays indicated formation of intra-molecular and inter-molecular G4s [115]. This was later supported by evidence from X-ray crystallography and NMR spectroscopy [8,9]. In addition, multiple spectroscopic techniques, including UV, fluorescence, and circular dichroism (CD) spectroscopies, as well as surface plasmon resonance are used to characterize G4 structures [115,116]. The folded G4 topology can engage guanine bases from one (unimolecular), two (bimolecular), or as many as four (tetra-molecular) nucleotide chains (see Figure 1 in main text). Canonically, G4 topologies are categorized based on the polarity of one or more of the nucleotide chains involved and the relative location of the loops that link the guanine bases(s). The G-rich strands that remain adjacent to each other in a parallel configuration through connecting loops that join upper and lower G-tetrads give the propeller-type G4 structures (also called ‘parallel G4s’), often identified by CD spectroscopy through the 260 and 240 nm positive and negative peaks, respectively [115]. In cases where at least one of the four strands is oriented opposite in polarity to the others, G4s are designated as ‘anti-parallel’ (identified through positive and negative CD peaks at 290 and 260 nm, respectively) (see Figure 1 in main text).
The prevalence of G4s throughout the genome and their biological relevance has been reviewed extensively [11–15]. However, a discussion about how G4s particularly affect the state of chromatin has received relatively little attention. For several years, the focus on the role of G4s in epigenetics was shaped by how modified DNA bases influenced G4 formation or stability. Recent results, however, implicate the G4 structure itself in regulating DNA (and histone) modifications. This leads to a debate about whether the G4 structure is the cause or consequence of epigenetic modifications. The primary focus of this review article is to discuss these emerging ideas. To underline the important implications, we discuss recent work on telomerase regulation as a case study, emphasizing in particular the epigenetic control of telomerase expression and the role of G4s in the telomerase promoter.
Detection of G4s in the Genome: from Computational Prediction to Intracellular Evidence
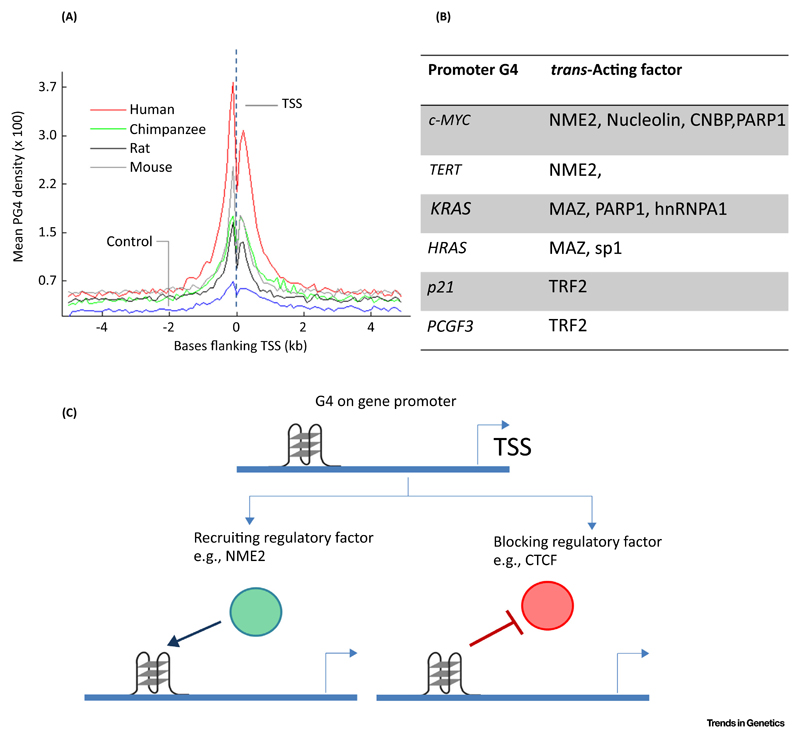
Early work noted the formation of G4s by telomeric DNA and associated the G4 motif to telomeres [16,17]. With advances in sequencing technology stretches of nucleotide sequences that could form a potential G4 (PG4; Box 2) have been found to be spread across genomes of a wide spectrum of organisms such as mammals [18–20], thousands of microbes [21,22], yeasts [23], and plants [24]. The presence of G4s within the promoters of c-MYC and KRAS and evidence supporting their G4-mediated transcription have been reported previously [25,26]. These indications prompted more genome-wide studies. PG4s were found to be non-randomly distributed and enriched at promoters genome-wide in bacteria [21] as well as humans (Figure 2) [27,28]. Furthermore, based on their enrichment within promoters and their conservation across promoters of bacterial species and within the promoters of homologous genes in human, chimpanzee, mouse and rat, a role for G4s in global gene regulation was proposed [20,21,28]. This was also observed experimentally a year later using G4-binding ligands [29]. It is noteworthy that along with enrichment within promoters PG4s were found to be prevalent within the untranslated regions (UTRs) [30] where presence in 5′ UTRs could be related to replication-dependent epigenetic processes.
Box 2. G4 Prediction Using Computational Approaches.
PG4s are typically represented by the sequence Gn-Lm-Gn-Lm-Gn-Lm-Gn, where n represents the number of guanine bases (often called a ‘stem’; n = 2 or more) and m represents the maximum length of the intervening nucleotide sequence (loop) (Figure 1). Computational tools such as Quadparser [18], QGRS mapper [117], Quadfinder [118], QuadBase [20,22], and G4Hunter [119] are used to predict G4s of variable loop and base length of user-provided sequences. Some G4 prediction resources such as QuadBase also provide curated lists of G4s across genomes of many species (both prokaryotes and eukaryotes) [20,22]. G4 prediction tools or resources are also available for RNA sequences [120–122], plant genomes [123,124], and G4-binding ligands [125].
Figure 2. G4 in Gene Regulation.
(A) G-quadruplexes (G4s) are enriched near transcription start sites (TSS) of multiple species. Adapted, with permission, from [28]. (B) Reported trans-acting factors that are recruited to promoter G4s. (C) Schematic showing different mechanisms by which promoter G4s might affect recruitment of regulatory trans-acting factors.
While the availability of sequencing allowed development of computational G4 predictions, G4-specific antibodies presented researchers with the opportunity to detect G4s within cells [17,31–33]. Although antibody cross-reactivity with non-G4 sequences has been reported previously [34], further development in such approaches along with high-resolution confocal microscopy and next-generation sequencing technologies will make more detailed understanding of G4 formation within cells possible [32,33,35,36].
Trans-acting factors that bind to promoter G4s
G4s have been found to be bound by trans-acting factors. One of the first examples of G4-dependent promoter occupancy of a regulatory factor is the non-metastatic factor NME2 (also known as nucleoside diphosphate kinase B or NM23H2). The presence of an intact G4 in the c-MYC promoter was found to be essential for NME2 binding and c-MYC expression [37]. This G4 located within the c-MYC promoter is also recognized by several other trans-acting factors including nucleolin, CNBP, and PARP1 [38–40]. Nucleolin inhibits the expression of c-MYC by binding to the promoter G4. Interestingly, nucleolin also inhibits the Wrn helicase from resolving G4s (Box 3) [41]. In addition to these factors, several other regulatory proteins were found to interact with promoter G4s [42–49]. Moreover, G4 formation in the human telomerase reverse transcriptase (hTERT) promoter prevented CCCTC-binding factor (CTCF) binding, suggesting that G4s might impede function of a regulatory protein [50]. Figure 2 summarizes these studies. It is also noteworthy that regulatory factors such as Telomere repeat-binding Factor 2 (TRF2) and NME2 engage epigenetic modifiers such as the REST-coREST-LSD1 complex [47,49]. Together, therefore, the role of G4s might involve engagement or repulsion of the regulatory factor(s), recruitment of chromatin-modifying enzymes, and/or more indirect impact through nucleosome positioning [51,52] (discussed in subsequent sections).
Box 3. Helicases That Resolve G4 Structures.
Several reports suggest that G4 structures can disrupt the movement of helicases along DNA strands [14,75,78,126–130]. Alternatively, studies also suggest that G4s can promote replication by recruiting specific helicases [130,131]. These include Pif1, Wrn1, FANCJ, BLM, and RTEL1, which have been implicated in resolving G4s to aid replication or suppression of genomic instability at sites containing PG4 motifs [14,75,126–129,132]. G4 helicases play important roles in DNA replication and telomere maintenance [133]. Loss of function mutations in several of the reported G4-resolving helicases results in conditions such as Werner’s syndrome and Bloom’s syndrome [14]. In Bloom’s syndrome, mutations that disrupt the ability of BLM helicase to resolve G4s can lead to alterations in sister chromatid exchange and recombination at sites rich in G4s [134,135]. Defective WRN helicase (and consequent unresolved G4s) has been shown to increase genomic instability as well as affect multiple transcriptional changes that might contribute to Werner’s syndrome pathogenesis [136].
Epigenetic DNA Modifications Affect G4 Stability
A characteristic pattern in the sequence of DNA is required for G4 formation (Box 2). In addition, epigenetic modification(s) (of both DNA bases and histones) that do not alter the genetic sequence of DNA was noted to influence the stability of the folded G4. By contrast, recent studies suggest that the presence of G4s affect epigenetic changes. Both of these aspects, that is, how DNA modifications affect G4 stability and the influence of G4s on epigenetics, are discussed in the following sections.
Early work showed that methylation at the C5 position of cytosine (5mC) residues within G-rich sequences impart higher stability to the folded G4 [53]. In 2017, stabilization of a G4 due to methylation of a specific cytosine within the hTERT gene was implicated in overexpression of hTERT, the ribo-nucleoprotein required for telomere synthesis, commonly observed to be highly expressed in ~90% of human cancers [50,54]. In addition, methylated cytosines within trinucleotide dCGG repeats within G4 regions, and expansion of such repeats, were found to be associated with suppression of the FMR1 gene in the fragile X mental retardation syndrome [55]. On observing the increased stability of the G4s resulting from methylated d(CGG)n oligomers, Fry and Loeb implicated formation of stable methylated G4s in the suppression of FMR1 expression in fragile X syndrome [56].
Two other neurodegenerative diseases, amyotrophic lateral sclerosis (ALS) and fronto-temporal dementia (FTD) are linked to the expansion of the hexanucleotide repeat GGGGCC in the noncoding region of the C9ORF72 locus [57,58] that was found to give folded G4s [59]. RNA transcribed from such regions of expanded G-rich repeats, which could potentially result in RNA G4s, have also been implicated in neurodegenerative conditions [60–62]. In addition, C5 methylation within the repeating units imparted stabilization to the folded structure in vitro, once again suggesting a role of methylation-dependent G4 stabilization in disease [63]. The promoter of the B cell lymphoma-2 (bcl2) gene, the abnormal overexpression of which is associated with a large number of cancers [64], harbors G4s that repress bcl2 transcription and is therefore of therapeutic interest [65,66]. C5 methylation of a bcl2 promoter G4 significantly stabilized the folding of the oligomer; implicating G4 methylation in epigenetic regulation of bcl2 [67].
Apart from this, another DNA modification, a common product of reactive oxygen species-induced base oxidation, 8-oxoguanine (8oxoG), was found within PG4 sequences [68] and also affected G4 stability in solution [69,70]. It was found that the 8oxoG modification on guanines within PG4s increased promoter activity of the vascular endothelial growth factor (VEGF) and endonuclease III-like protein 1 (NTHL1) genes [71,72]. Consistent with this, 8oxoG-dependent promoter G4 stabilization was recently reported to regulate the oncogene KRAS through increased recruitment of the transcription regulatory factors MAZ and hnRNP-A1 [73]. Other than transcription, 8oxoG-induced perturbation of the telomeric DNA G4 increased binding of telomerase to telomeres, resulting in enhanced telomere extension [74].
It must be mentioned that although some studies discuss the extent of in vitro G4 stability after methylation in quantitative terms [53,67], further work is required to understand the impact of DNA methylation-dependent G4 stabilization inside cells within a chromatin setting.
Presence of G4s Can Influence Epigenetic Modifications: The Emerging Contrast
Analyzed together, the observations described above helped build the notion that C5 methylation influences the stability of G4s and that this modification has possible physiological impacts. More recent studies, in contrast, suggest G4s could influence DNA methylation at specific sites. Genetic instability was noted within sequences that adopt G4s, most likely due to a replication-associated error wherein G4s failed to unwind in cells lacking DNA helicases such as BLM, FANCJ, or Pif1 that resolve G4s [75,76] (Box 3). This led to findings implicating G4s in the altered placement of modified histone proteins that package chromatin; another hallmark of epigenetic regulation in addition to modified DNA [77,78].
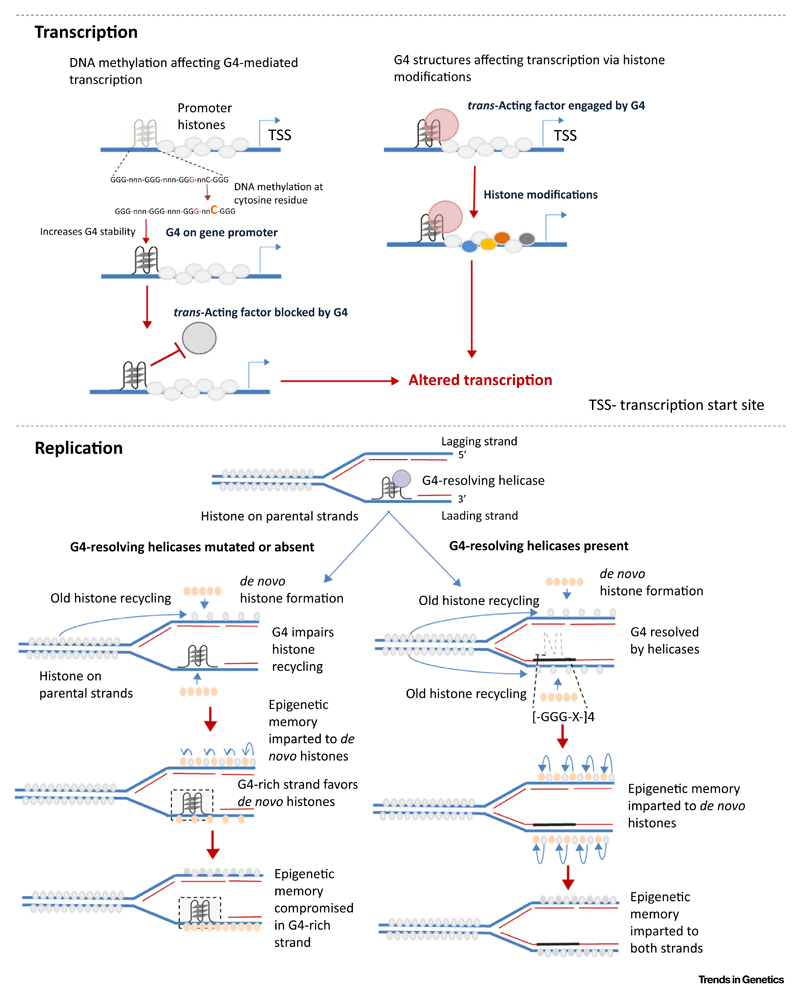
For replication, chromatin needs to be unpacked. Replicative helicases that remove histones are therefore tightly coupled to DNA synthesis. Repackaging of the newly made DNA daughter strand requires precise replacement of parental histone marks. Therefore, fidelity in the chain of events entails coupling of histone eviction by replicative helicases, DNA synthesis, and recycling of histones thereafter for chromatinization. Thus, in cells where G4s remain unresolved due to lack of G4 helicases, there is impediment to the replication process. This was postulated to decouple the critical histone recycling step in the daughter strand, resulting in anomalous or de novo histone modifications [77–79] (Figure 3, Key Figure).
Figure 3. Key Figure: G4s in Epigenetic Modifications.
Schematic of reported mechanisms by which G-quadruplexes (G4s) are linked to epigenetic modifications in transcription and replication.
This was shown in avian DT40 cells devoid of the replicative helicase REV1 [77]. G4s present in the leading-strand template, within the second intron of the β-globin gene, were inefficiently replicated in REV1-deficient cells (rev1). Consequently, the β-globin locus, originally in an epigenetically repressed state, lost dimethylated H3K9 histone-repressive marks resulting in de-repression of β-globin expression. Moreover, an increase in histone H4 N-terminal acetylation, a mark not otherwise associated with β-globin loci, was found in rev1-deficient relative to wild-type cells. This was consistent with the expectation that histone recycling and replication were not closely coordinated (Figure 3). More direct observations came from the introduction of a G4 within the otherwise silent lysozyme C locus lacking endogenous G4s and therefore unaffected by loss of REV-1. Artificial addition of a G4 resulted in activation of lysozyme expression in DT40 rev1 cells [77]. Subsequent studies using the BU-1 locus in rev1-deficient DT40 cells further showed that the presence of a G4 altered H3K4Me3 and H3K9/14ac modifications at the BU-1 promoter [79]. Ligand-induced stabilization of G4s, consistent with the model of G4-dependent replication block, led to the loss of H3K4Me3 and, interestingly, heritable DNA cytosine methylation, at the BU-1 locus in wild-type DT40 cells [80].
Methylation at both histones and DNA cooperate to either recruit or exclude DNA binding proteins [81,82]. Together, these seed the notion of a larger context for G4-dependent histone and/or DNA methylation, suggesting a role of G4s in the assembly of regulatory factors (Figure 3). Two recent observations help make this point. Occupancy of the lysine-specific demethylase LSD1 was G4 dependent in the cyclin-dependent kinase p21 and telomerase (hTERT) promoters [47,49]. This resulted in altered histone modifications at the promoters that, in turn, exerted control on the expression of p21 and hTERT in cancer cells.
G4-Binding Factors Connect G4s to the Epigenetic Machinery
G4-dependent epigenetic modifications raise interesting mechanistic questions about proteins that might associate with G4s and engage modifiers of DNA and/or chromatin (Figure 3). Recent findings begin to address these. For example, the chromatin modifier REST/co-REST/LSD1 repressor complex was engaged at the p21 promoter in a manner that was dependent on the association of the transcription factor TRF2 to the p21 promoter G4 [47]. Recruitment of the repressor complex, in turn, resulted in an increase in repressive histone marks at the p21 promoter. Further support for TRF2-G4 interactions come from an earlier study that found a truncated form of TRF2 to interact with the telomeric G4 in solution [83] and more recently when TRF2 was reported to associate with the PCGF3 promoter G4 [48]. In another study, G4-dependent histone modifications were noted to result from association of NME2 to the hTERT promoter G4 [49] (vide infra). Both NME2 and TRF2 were noted to interact with the REST repressor complex [47,49]. While some components of the complex were common, further work is required to understand the differences/similarities between the interactions and resultant function in TRF2 and NME2 and perhaps other G4-binding proteins.
In addition to these studies, it has been reported that G4s associate avidly with DNA methyltransferases (DNMTs) in vitro [84]. This also finds support from earlier studies. In 1991, recognition of unusual DNA structures by DNMTs was mentioned perhaps for the first time [85]. Almost two decades later, genome-wide correlations were observed: CpG methylation was restricted within G4-forming regions and based on this authors argued about a possible connection between G4s and how DNMTs recognize DNA [86]. Together, these lend support to potential mechanisms of how G4s could impact epigenetic histone and DNA modifications, laying foundations for future work addressing causality.
G4s and Higher-Order Packaging through Nucleosomes
Genome-wide analyses in Saccharomyces cerevisiae [51], Caenorhabditis elegans [52], and human [51,52] introduced a higher level of involvement of G4s in chromatin organization. PG4 motifs were found to be typically present outside nucleosome-occupied regions (reviewed in [87]). This was further substantiated later from G4 ChIP-seq data using G4-specific antibodies [88], genome-wide nuclease footprinting data in mouse B cells [89], and phased presence of G4s and nucleosomes around many human replication origins [90]. Together with new findings showing direct/indirect roles of G4s in epigenetic histone modifications, the involvement of G4s in nucleosomal organization noted in these studies implies a wider framework of G4-mediated chromatin changes. Furthermore, the involvement of G4s in genome instability and possibly related deleterious outcomes such as cancer [91] suggests that modalities of nucleosome exclusion in G4-rich regions be revisited.
The Telomerase Promoter: A Case Study in G4-Related Epigenetic Control
Although tightly restricted in adult somatic cells under unusual and often neoplastic conditions, telomerase is reactivated in most cancers [54,92,93]. Mechanisms involved in the reactivation of hTERT expression remain poorly understood. Contextually, distinct aspects that developed in the past few years, when linked, are notable: (i) clinical findings observing that the hTERT promoter is a hot spot of cancer-specific mutations, (ii) epigenetic changes at the hTERT promoter, and (iii) high G4-forming propensity within the hTERT promoter (Figure 4 and Table 1). These are discussed below as an example of the role of G4s in epigenetic mechanisms and its potential importance.
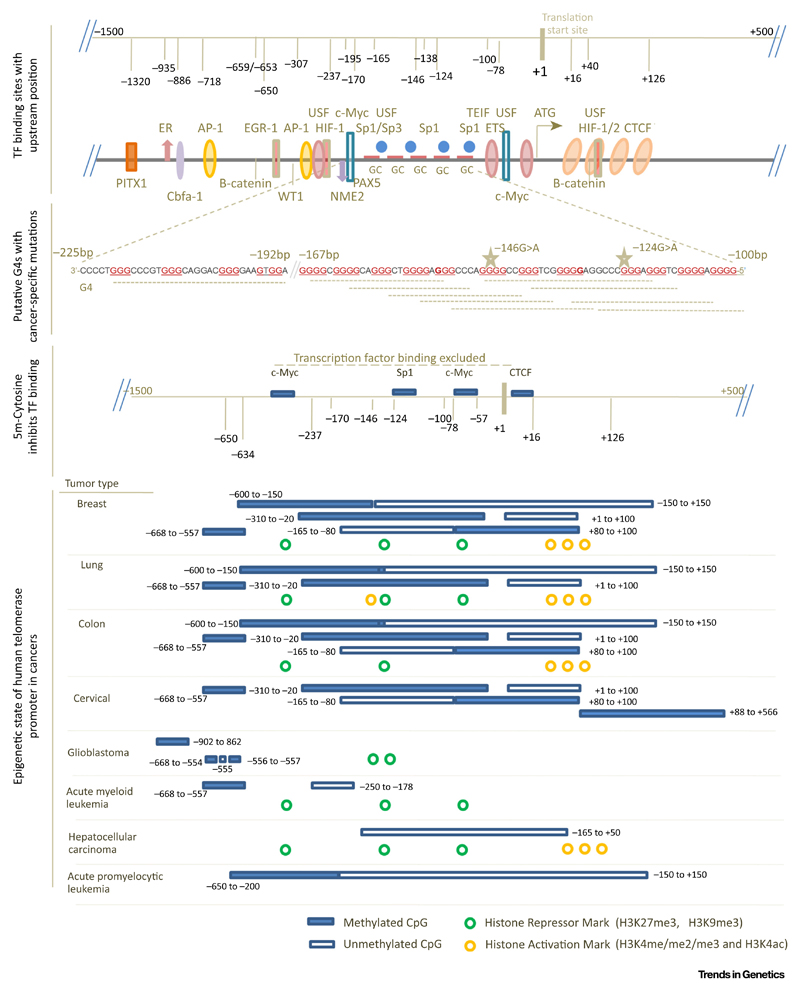
Figure 4. Tandem G4 Structures within Telomerase Promoter Constitute Important Sites of Epigenetic Alterations.
The 2-kb region of hTERT promoter (+500 to −1500 bp) overlaying important transcription factor (TF) binding sites, tandem reported G4s110, specific CpG island methylation resulting in exclusion of transcription factor binding followed by epigenetic changes [103,107,108], that is, histone modifications and DNA methylation observed across cancers. Clinically important cancer-specific mutations −124 G > A and −146G > A are found within G4s [94–98].
Table 1. G4 Motifs (G3L1-7) Mapped on the hTERT Promoter and Positions of Reported Transcription Factors Have Been Marked.
| Upstream distance from translational start site (bp) | Length of G4 motif | G3L1-7 motifs | Transcription factor binding sites |
|---|---|---|---|
| 102 | 18 | GGGGAGGGGCTGGGAGGG | TEIF, Sp1 GABP-α on – 124G > A mutation |
| 18 | GGGGAGGGGCTGGGAGGG | ||
| 27 | GGGGAGGGGCTGGGAGGGCCCGGAGGG | ||
| 27 | GGGGAGGGGCTGGGAGGGCCCGGAGGG | ||
| 28 | GGGGAGGGGCTGGGAGGGCCCGGAGGGG | ||
| 28 | GGGGAGGGGCTGGGAGGGCCCGGAGGGG | ||
| 103 | 17 | GGGAGGGGCTGGGAGGG | |
| 17 | GGGAGGGGCTGGGAGGG | ||
| 26 | GGGAGGGGCTGGGAGGGCCCGGAGGG | ||
| 26 | GGGAGGGGCTGGGAGGGCCCGGAGGG | ||
| 26 | GGGAGGGGCTGGGAGGGCCCGGAGGG | ||
| 27 | GGGAGGGGCTGGGAGGGCCCGGAGGGG | ||
| 27 | GGGAGGGGCTGGGAGGGCCCGGAGGGG | ||
| 27 | GGGAGGGGCTGGGAGGGCCCGGAGGGG | ||
| 107 | 22 | GGGGCTGGGAGGGCCCGGAGGG | |
| 23 | GGGGCTGGGAGGGCCCGGAGGGG | ||
| 29 | GGGGCTGGGAGGGCCCGGAGGGGGCTGGG | ||
| 29 | GGGGCTGGGAGGGCCCGGAGGGGGCTGGG | ||
| 108 | 21 | GGGCTGGGAGGGCCCGGAGGG | |
| 22 | GGGCTGGGAGGGCCCGGAGGGG | ||
| 28 | GGGCTGGGAGGGCCCGGAGGGGGCTGGG | ||
| 28 | GGGCTGGGAGGGCCCGGAGGGGGCTGGG | ||
| 113 | 23 | GGGAGGGCCCGGAGGGGGCTGGG | |
| 23 | GGGAGGGCCCGGAGGGGGCTGGG | ||
| 117 | 24 | GGGCCCGGAGGGGGCTGGGCCGGG | |
| 24 | GGGCCCGGAGGGGGCTGGGCCGGG | ||
| 25 | GGGCCCGGAGGGGGCTGGGCCGGGG | ||
| 25 | GGGCCCGGAGGGGGCTGGGCCGGGG | ||
| 126 | 23 | GGGGGCTGGGCCGGGGACCCGGG | |
| 23 | GGGGGCTGGGCCGGGGACCCGGG | ||
| 127 | 22 | GGGGCTGGGCCGGGGACCCGGG | Sp1 |
| 22 | GGGGCTGGGCCGGGGACCCGGG | ||
| 128 | 21 | GGGCTGGGCCGGGGACCCGGG | |
| 21 | GGGCTGGGCCGGGGACCCGGG | ||
| 25 | GGGCTGGGCCGGGGACCCGGGAGGG | ||
| 26 | GGGCTGGGCCGGGGACCCGGGAGGGG | ||
| 31 | GGGCTGGGCCGGGGACCCGGGAGGGGTCGGG | ||
| 133 | 20 | GGGCCGGGGACCCGGGAGGG | |
| 20 | GGGCCGGGGACCCGGGAGGG | ||
| 21 | GGGCCGGGGACCCGGGAGGGG | ||
| 21 | GGGCCGGGGACCCGGGAGGGG | ||
| 26 | GGGCCGGGGACCCGGGAGGGGTCGGG | ||
| 26 | GGGCCGGGGACCCGGGAGGGGTCGGG | ||
| 138 | 21 | GGGGACCCGGGAGGGGTCGGG | |
| 21 | GGGGACCCGGGAGGGGTCGGG | ||
| 26 | GGGGACCCGGGAGGGGTCGGGACGGG | ||
| 26 | GGGGACCCGGGAGGGGTCGGGACGGG | ||
| 27 | GGGGACCCGGGAGGGGTCGGGACGGGG | ||
| 31 | GGGGACCCGGGAGGGGTCGGGACGGGGCGGG | ||
| 139 | 20 | GGGACCCGGGAGGGGTCGGG | Sp1 GABP-a on -146G > A mutation |
| 20 | GGGACCCGGGAGGGGTCGGG | ||
| 25 | GGGACCCGGGAGGGGTCGGGACGGG | ||
| 25 | GGGACCCGGGAGGGGTCGGGACGGG | ||
| 26 | GGGACCCGGGAGGGGTCGGGACGGGG | ||
| 30 | GGGACCCGGGAGGGGTCGGGACGGGGGGG | ||
| 146 | 18 | GGGAGGGGTCGGGACGGG | |
| 18 | GGGAGGGGTCGGGACGGG | ||
| 19 | GGGAGGGGTCGGGACGGGG | ||
| 19 | GGGAGGGGTCGGGACGGGG | ||
| 23 | GGGAGGGGTCGGGACGGGGCGGG | ||
| 23 | GGGAGGGGTCGGGACGGGGCGGG | ||
| 23 | GGGAGGGGTCGGGACGGGGCGGG | ||
| 23 | GGGAGGGGTCGGGACGGGGCGGG | ||
| 23 | GGGAGGGGTCGGGACGGGGCGGG | ||
| 24 | GGGAGGGGTCGGGACGGGGCGGGG | ||
| 24 | GGGAGGGGTCGGGACGGGGCGGGG | ||
| 24 | GGGAGGGGTCGGGACGGGGCGGGG | ||
| 150 | 19 | GGGGTCGGGACGGGGCGGG | HIF1/2, Sp1, Sp3, E2F1 |
| 19 | GGGGTCGGGACGGGGCGGG | ||
| 20 | GGGGTCGGGACGGGGCGGGG | ||
| 20 | GGGGTCGGGACGGGGCGGGG | ||
| 151 | 18 | GGGTCGGGACGGGGCGGG | |
| 18 | GGGTCGGGACGGGGCGGG | ||
| 19 | GGGTCGGGACGGGGCGGGG | ||
| 19 | GGGTCGGGACGGGGCGGGG |
The Human Telomerase Promoter Is a Hot Spot of Cancer-Specific Mutations
Initially, three papers in 2013 [94–96] and subsequently several reports [97] substantiated the clinical importance of two guanine-to-adenine (G > A) mutations in the hTERT promoter at positions −124 and −146 bp upstream from the translation start site (TSS) (Figure 4). Importantly, these mutations were found to be directly involved in telomerase reactivation across several cancer types [94,96,98]. It has since been shown that the frequency of hTERT promoter mutations is as high as 70–93% in cancer types including skin tumors and glioblastomas and 40–60% in endocrine tumors such as thyroid and renal cancers [97].
Epigenetic Control of Telomerase Reactivation: Histone Modifications and DNA Methylation
The role of histone repressive modifications in restriction of hTERT promoter activity in adult somatic cells has been widely studied [99–105]. Recently, it was observed that the bromodomain-4-mediated histone acetylation of the hTERT promoter resulted in hTERT reactivation in glioblastoma and melanoma cells with hTERT promoter mutations (both, −124G > A and −146G > A) [106]. Importantly, this was not the case in cells of the same lineage but without mutations [106]. These results implicated the role of the promoter mutations in the altered epigenetic status of the hTERT promoter. Furthermore, about a 2-kb region of CpG islands (–1.5 to +0.5 kb of the TSS) is present within the hTERT promoter [103,107]. The actively transcribed hTERT promoter was found to be hypomethylated at the near promoter (–200 to +100 bp of TSS) and hypermethylated further upstream (–650 to −200 bp) [103,107,108] (Figure 4).
Promoter G4s and Epigenetics at the Telomerase Promoter
It was reported in 2009 that the hTERT promoter harbors a tandem array of putative G4s within 330 bp upstream of TSS (Figure 4) [109]. Furthermore, it was shown that G4-binding ligands [110], and base-substitutions that affect G4 stability, impact hTERT expression and promoter activity, suggesting the role of G4s in hTERT regulation [49,111]. The high propensity of G4s in the hTERT promoter, along with literature suggesting the role of G4s in both DNA and histone methylation, prompts questions on whether and how G4s are linked to the epigenetic state of the hTERT promoter.
Recently reported transcriptional repression of hTERT by NME2 in a G4-dependent manner is therefore of interest [49]. It was observed that NME2-G4 interaction induced occupancy of the REST/co-REST/LSD1 chromatin-modifier repressor complex, resulting in repressive histone modifications at the hTERT promoter [49]. We noted with interest another recent report. In this case, it was demonstrated that C5 methylation of a CpG stabilized a G4 within the first exon of hTERT [50]. This excluded binding of the important hTERT repressor factor CTCF, resulting in de-repression of hTERT.
With these in mind, it might be interesting to consider the cancer-specific hTERT promoter mutations −124G > A and −146G > A that occur with high frequency in multiple cancers. First, as depicted in Figure 4, both −124 and −146 Gs constitute G-tetrads required for G4 formation (Table 1). As expected, therefore, G4s formed in vitro by oligonucleotides representing these sequence stretches were disrupted upon including the mutations [111]. Second, both -124 and -146 positions are included within binding motifs of Sp1 (Table 1), which was shown to interact with G4s [112]. Third, these base positions constitute CpG islands within the hTERT promoter where C5 methylations have been reported [113,114]. Fourth, treatment of hTERT promoter constructs and/or cell lines harboring 124G > A or 146G > A mutations with G4-binding ligands resulted in a decrease in hTERT expression [49,111]. This was postulated to be through re-stabilization of the disrupted G4s. Together, therefore, the role of the underlying G4s in the hTERT promoter and its possible impact on the epigenetic state of the hTERT promoter emerge as a potentially important issue.
Concluding Remarks and Future Perspectives
G4s engage regulatory chromatin modification factors. This finds increasing ground from evidence supporting the interaction of G4s with both chromatin modification factors such as REST/LSD1 and DNA modification factors, for example, DNMTs. Although a new concept, this seeds the idea of epigenetic modifications being connected to the underlying DNA sequence, where the sequence folds into DNA structures that bind and engage the chromatin modification machinery. Formation of G4, in a context-specific way, therefore might dictate when and where epigenetic modifications occur. Recent evidence shows that histone and DNA modifications are linked. However, molecular players involved in these possibly concerted epigenetic modifications are not entirely clear. Since results support the role of G4s in engaging factors that are causal for both DNA as well as histone modification(s), the possibility that G4s link both types of epigenetic modifications can be interesting. With further work, it is possible that at a molecular level role of G4s as mediators that may induce or alter site-specific histone and/or DNA modifications can be more clearly understood (see Outstanding Questions).
Outstanding Questions.
Formation, or disruption, of G4s is correlated with promoter cytosine methylation. Questions about causality elucidating the molecular players, for example, whether and how DNA methyltransferases are involved, remain.
DNA methylation and histone modifications are causally associated through factors such as Polycomb repressive complex 2. Therefore, molecular work asking how cytosine methylation-mediated G4 formation/deformation affects histone modification would be important.
Absence of G4 helicases gives de novo histone placements compromising epigenetic memory. This is perceived as a deleterious issue. By contrast, G4-binding factors and/or G4 helicases might regulate where and when de novo histones are placed. This remains to be understood.
G4s engage epigenetic modifiers in promoters. G4 helicases influence chromatin repackaging. Are these linked through common G4-binding proteins?
Two key cancer-specific mutations that occur with high frequency in many cancers disrupt G4 formation in vitro. G4 deformation might result in gain/loss of transcription factor association(s) at the telomerase promoter. Direct evidence of this is yet to be demonstrated.
In parallel, emerging results in the literature show G4s impede replication. G4 helicases, which unwind G4s, are therefore necessary for replication and subsequent replacement of histone marks with fidelity. This helps preserve epigenetic memory. Although speculative at present, the contrasting aspect might be the function of G4s in promoting variation through de novo histone modifications. This can be particularly relevant if this is regulated by G4-binding factors that engage G4 helicases on a case-to-case basis. Furthermore, it also remains to be tested whether G4-mediated modifications during post-replication phases, such as transcription, are linked to the next round of replication thorough common G4-binding factors. Although challenging, this might have important implications for understanding the mechanisms that cause and inherit epigenetic alterations through successive generations.
Note Added in Proof
TRF2 was necessary for error-free replication of peri-centromeres – where TRF2-mediated engagement of the G4-resolving helicase RTEL1 was crucial for replication [137]. Another recent study showed that TRF2 binding at several promoters spread across the genome results in epigenetic changes and altered expression of the target, which interestingly was dependent on the length of telomeres [138]. Genome-wide hypomethylation within CpG islands was reported to be closely associated with presence of G4s [139] – consistent with earlier observations in the same lines [86].
Highlights.
G4s mediate promoter histone modifications. Recent work shows how G4 association of regulatory factors recruits chromatin modifiers, resulting in repressive histone modifications and gene repression.
In the absence of designated helicases, G4s impede DNA replication. This partially delinks DNA replication from repackaging of newly made chromatin. As a result, fidelity in copying parental histone modifications is affected, compromising epigenetic memory.
DNA methylation influences G4 stability, affecting transcription factor binding. At the telomerase promoter, C5 methylation occludes CTCF binding, de-repressing telomerase expression in cancer cells.
Spotlight on tandem array of G4s within the telomerase promoter. These sequence stretches harboring potential G4s are key to the epigenetic status of the tightly repressed telomerase promoter in normal somatic cells and the aberrant high telomerase expression in cancer.
Acknowledgments
A.K.M. and S.S. acknowledge the CSIR for research fellowships. S.C.’s research group is supported by CSIR and partly supported by the Wellcome Trust/DBT India Alliance Fellowship (grant 500127/Z/09/Z) awarded to S.C. from 2011 to 2017. All members of S.C.’s group are acknowledged for engaging discussions and support. The authors acknowledge Munia Ganguli for carefully reading and editing the manuscript.
Footnotes
Supplemental Information
Supplemental information associated with this article can be found, in the online version, at https://doi.org/10.1016/j.tig.2018.11.001.
References
- 1.Dickerson RE, et al. The anatomy of A-, B-, and Z-DNA. Science. 1982;216:475–485. doi: 10.1126/science.7071593. [DOI] [PubMed] [Google Scholar]
- 2.McMurray CT. DNA secondary structure: a common and causative factor for expansion in human disease. Proc Natl Acad Sci U S A. 1999;96:1823–1825. doi: 10.1073/pnas.96.5.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Phan AT, et al. DNA architecture: from G to Z. Curr Opin Struct Biol. 2006;16:288–298. doi: 10.1016/j.sbi.2006.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zeraati M, et al. I-motif DNA structures are formed in the nuclei of human cells. Nat Chem. 2018;10:631–637. doi: 10.1038/s41557-018-0046-3. [DOI] [PubMed] [Google Scholar]
- 5.Huppert JL. Four-stranded nucleic acids: structure, function and targeting of G-quadruplexes. Chem Soc Rev. 2008;37:1375–1384. doi: 10.1039/b702491f. [DOI] [PubMed] [Google Scholar]
- 6.Ghosal G, Muniyappa K. Hoogsteen base-pairing revisited: resolving a role in normal biological processes and human diseases. Biochem Biophys Res Commun. 2006;343:1–7. doi: 10.1016/j.bbrc.2006.02.148. [DOI] [PubMed] [Google Scholar]
- 7.Gellert M, et al. Helix formation by guanylic acid. Proc Natl Acad Sci U S A. 1962;48:2013–2018. doi: 10.1073/pnas.48.12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zimmerman SB, et al. X-ray fiber diffraction and model-building study of polyguanylic acid and polyinosinic acid. J Mol Biol. 1975;92:181–192. doi: 10.1016/0022-2836(75)90222-3. [DOI] [PubMed] [Google Scholar]
- 9.Howard FB, et al. Stable and metastable forms of poly (G) Biopolymers. 1977;16:791–809. doi: 10.1002/bip.1977.360160407. [DOI] [PubMed] [Google Scholar]
- 10.Keniry MA. Quadruplex structures in nucleic acids. Biopolymers. 2000;56:123–146. doi: 10.1002/1097-0282(2000/2001)56:3<123::AID-BIP10010>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 11.Lipps HJ, Rhodes D. G-quadruplex structures: in vivo evidence and function. Trends Cell Biol. 2009;19:414–422. doi: 10.1016/j.tcb.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 12.Murat P, Balasubramanian S. Existence and consequences of G-quadruplex structures in DNA. Curr Opin Genet Dev. 2014;25:22–29. doi: 10.1016/j.gde.2013.10.012. [DOI] [PubMed] [Google Scholar]
- 13.Rhodes D, Lipps HJ. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015;43:8627–8637. doi: 10.1093/nar/gkv862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mendoza O, et al. G-quadruplexes and helicases. Nucleic Acids Res. 2016;44:1989–2006. doi: 10.1093/nar/gkw079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cammas A, Millevoi S. RNA G-quadruplexes: emerging mechanisms in disease. Nucleic Acids Res. 2017;45:1584–1595. doi: 10.1093/nar/gkw1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Balagurumoorthy P, Brahmachari SK. Structure and stability of human telomeric sequence. J Biol Chem. 1994;269:21858–21869. [PubMed] [Google Scholar]
- 17.Paeschke K, et al. Telomere end-binding proteins control the formation of G-quadruplex DNA structures in vivo. Nat Struct Mol Biol. 2005;12:847–854. doi: 10.1038/nsmb982. [DOI] [PubMed] [Google Scholar]
- 18.Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Todd AK, et al. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005;33:2901–2907. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yadav VK, et al. QuadBase: genome-wide database of G4 DNA—occurrence and conservation in human, chimpanzee, mouse and rat promoters and 146 microbes. Nucleic Acids Res. 2008;36:D381–D385. doi: 10.1093/nar/gkm781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rawal P, et al. Genome-wide prediction of G4 DNA as regulatory motifs: role in Escherichia coli global regulation. Genome Res. 2006;16:644–655. doi: 10.1101/gr.4508806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dhapola P, Chowdhury S. QuadBase2: web server for multiplexed guanine quadruplex mining and visualization. Nucleic Acids Res. 2016;44:W277–W283. doi: 10.1093/nar/gkw425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Capra JA, et al. G-quadruplex DNA sequences are evolutionarily conserved and associated with distinct genomic features in Saccharomyces cerevisiae. PLoS Comput Biol. 2010;6:9. doi: 10.1371/journal.pcbi.1000861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Griffin BD, Bass HW. Review: plant G-quadruplex (G4) motifs in DNA and RNA; abundant, intriguing sequences of unknown function. Plant Sci. 2018;269:143–147. doi: 10.1016/j.plantsci.2018.01.011. [DOI] [PubMed] [Google Scholar]
- 25.Siddiqui-Jain A, et al. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc Natl Acad Sci U S A. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cogoi S, Xodo LE. G-quadruplex formation within the promoter of the KRAS proto-oncogene and its effect on transcription. Nucleic Acids Res. 2006;34:2536–2549. doi: 10.1093/nar/gkl286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huppert JL, Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007;35:406–413. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Verma A, et al. Genome-wide computational and expression analyses reveal G-quadruplex DNA motifs as conserved cis-regulatory elements in humanand related species. J Med Chem. 2008;51:5641–5649. doi: 10.1021/jm800448a. [DOI] [PubMed] [Google Scholar]
- 29.Verma A, et al. Evidence of genome-wide G4 DNA-mediated gene expression in human cancer cells. Nucleic Acids Res. 2009;37:4194–4204. doi: 10.1093/nar/gkn1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huppert JL, et al. G-quadruplexes: the beginning and end of UTRs. Nucleic Acids Res. 2008;36:6260–6268. doi: 10.1093/nar/gkn511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schaffitzel C, et al. In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylonychia lemnae macronuclei. Proc Natl Acad Sci U S A. 2001;98:8572–8577. doi: 10.1073/pnas.141229498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Biffi G, et al. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat Chem. 2013;5:182–186. doi: 10.1038/nchem.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Henderson A, et al. Detection of G-quadruplex DNA in mammalian cells. Nucleic Acids Res. 2014;42:860–869. doi: 10.1093/nar/gkt957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kazemier HG, et al. Guanine quadruplex monoclonal antibody 1H6 cross-reacts with restrained thymidine-rich single stranded DNA. Nucleic Acids Res. 2017;45:5913–5919. doi: 10.1093/nar/gkx245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chambers VS, et al. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat Biotechnol. 2015;33:877–881. doi: 10.1038/nbt.3295. [DOI] [PubMed] [Google Scholar]
- 36.Hänsel-Hertsch R, et al. Genome-wide mapping of endogenous G-quadruplex DNA structures by chromatin immunoprecipitation and high-throughput sequencing. Nat Protoc. 2018;13:551–564. doi: 10.1038/nprot.2017.150. [DOI] [PubMed] [Google Scholar]
- 37.Thakur RK, et al. Metastases suppressor NM23-H2 interaction with G-quadruplex DNA within c-MYC promoter nuclease hypersensitive element induces c-MYC expression. Nucleic Acids Res. 2009;37:172–183. doi: 10.1093/nar/gkn919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.González V, et al. Identification and characterization of nucleolin as a c-myc G-quadruplex-binding protein. J Biol Chem. 2009;284:23622–23635. doi: 10.1074/jbc.M109.018028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Borgognone M, et al. Cellular nucleic-acid-binding protein, a transcriptional enhancer of c-Myc, promotes the formation of parallel G-quadruplexes. Biochem J. 2010;428:491–498. doi: 10.1042/BJ20100038. [DOI] [PubMed] [Google Scholar]
- 40.Fekete A, et al. The guanine-quadruplex structure in the human c-myc gene’s promoter is converted into B-DNA form by the human poly(ADP-ribose)polymerase-1. PLoS One. 2012;7:19–22. doi: 10.1371/journal.pone.0042690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Indig FE, et al. Nucleolin inhibits G4 oligonucleotide unwinding by werner helicase. PLoS One. 2012;7:1–12. doi: 10.1371/journal.pone.0035229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cogoi S, et al. Structural polymorphism within a regulatory element of the human KRAS promoter: formation of G4-DNA recognized by nuclear proteins. Nucleic Acids Res. 2008;36:3765–3780. doi: 10.1093/nar/gkn120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paramasivam M, et al. Protein hnRNP A1 and its derivative Up1 unfold quadruplex DNA in the human KRAS promoter: implications for transcription. Nucleic Acids Res. 2009;37:2841–2853. doi: 10.1093/nar/gkp138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cogoi S, et al. The KRAS promoter responds to Myc-associated zinc finger and poly(ADP-ribose) polymerase 1 proteins, which recognize a critical quadruplex-forming GA-element. J Biol Chem. 2010;285:22003–22016. doi: 10.1074/jbc.M110.101923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chu P-C, et al. Regulation of oncogenic KRAS signaling via a novel KRAS-integrin-linked kinase-hnRNPA1 regulatory loop in human pancreatic cancer cells. Oncogene. 2016;35:3897–3908. doi: 10.1038/onc.2015.458. [DOI] [PubMed] [Google Scholar]
- 46.Cogoi S, et al. HRAS is silenced by two neighboring G-quadruplexes and activated by MAZ, a zinc-finger transcription factor with DNA unfolding property. Nucleic Acids Res. 2014;42:8379–8388. doi: 10.1093/nar/gku574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hussain T, et al. Transcription regulation of CDKN1A (p21/CIP1/WAF1) by TRF2 is epigenetically controlled through the REST repressor complex. Sci Rep. 2017;7:11541. doi: 10.1038/s41598-017-11177-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Purohit G, et al. Extratelomeric binding of the telomere binding protein TRF2 at the PCGF3 promoter is G-quadruplex motif-dependent. Biochemistry. 2018;57:2317–2324. doi: 10.1021/acs.biochem.8b00019. [DOI] [PubMed] [Google Scholar]
- 49.Saha D, et al. Epigenetic suppression of human telomerase (hTERT) is mediated by the metastasis suppressor NME2 in a G-quadruplex-dependent fashion. J Biol Chem. 2017;292:15205–15215. doi: 10.1074/jbc.M117.792077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li P-TT, et al. Expression of the human telomerase reverse transcriptase gene is modulated by quadruplex formation in its first exon due to DNA methylation. J Biol Chem. 2017;292:20859–20870. doi: 10.1074/jbc.M117.808022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Halder K, et al. Genome-wide analysis predicts DNA structural motifs as nucleosome exclusion signals. Mol Biosyst. 2009;5:1703–1712. doi: 10.1039/b905132e. [DOI] [PubMed] [Google Scholar]
- 52.Wong HM, Huppert JL. Stable G-quadruplexes are found outside nucleosome-bound regions. Mol Biosyst. 2009;5:1713–1719. doi: 10.1039/b905848f. [DOI] [PubMed] [Google Scholar]
- 53.Hardin CC, et al. Cytosine-cytosine+ base pairing stabilizes DNA quadruplexes and cytosine methylation greatly enhances the effect. Biochemistry. 1993;32:5870–5880. doi: 10.1021/bi00073a021. [DOI] [PubMed] [Google Scholar]
- 54.Kim NW, et al. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 55.Liu XS, et al. Rescue of fragile X syndrome neurons by DNA methylation editing of the FMR1 gene. Cell. 2018;172:979–992.e6. doi: 10.1016/j.cell.2018.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fry M, Loeb LA. The fragile X syndrome d(CGG)n nucleotide repeats form a stable tetrahelical structure. Proc Natl Acad Sci U S A. 1994;91:4950–4954. doi: 10.1073/pnas.91.11.4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Renton AE, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.DeJesus-Hernandez M, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Haeusler AR, et al. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature. 2014;507:195–200. doi: 10.1038/nature13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kumar V, et al. Unraveling the role of RNA mediated toxicity of C9orf72 repeats in C9-FTD/ALS. Front Neurosci. 2017;11:711. doi: 10.3389/fnins.2017.00711. Published online December 15 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Donnelly CJ, et al. RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron. 2013;80:415–428. doi: 10.1016/j.neuron.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schludi MH, Edbauer D. Targeting RNA G-quadruplexes as new treatment strategy for C9orf72 ALS/FTD. EMBO Mol Med. 2018;10:4–6. doi: 10.15252/emmm.201708572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zamiri B, et al. Quadruplex formation by both G-rich and C-rich DNA strands of the C9orf72 (GGGGCC)8•(GGCCCC)8 repeat: effect of CpG methylation. Nucleic Acids Res. 2015;43:10055–10064. doi: 10.1093/nar/gkv1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reed JC. Regulation of apoptosis by bcl-2 family proteins and its role in cancer and chemoresistance. Curr Opin Oncol. 1995;7:541–546. doi: 10.1097/00001622-199511000-00012. [DOI] [PubMed] [Google Scholar]
- 65.Wang XD, et al. Turning off transcription of the bcl-2 gene by stabilizing the bcl-2 promoter quadruplex with quindo-line derivatives. J Med Chem. 2010;53:4390–4398. doi: 10.1021/jm100445e. [DOI] [PubMed] [Google Scholar]
- 66.Dai J, et al. An intramolecular G-quadruplex structure with mixed parallel/antiparallel G-strands formed in the human BCL-2 promoter region in solution. J Am Chem Soc. 2006;128:1096–1098. doi: 10.1021/ja055636a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lin J, et al. Stabilization of G-quadruplex DNA by C-5-methyl-cytosine in bcl-2 promoter: implications for epigenetic regulation. Biochem Biophys Res Commun. 2013;433:368–373. doi: 10.1016/j.bbrc.2012.12.040. [DOI] [PubMed] [Google Scholar]
- 68.Clark DW, et al. Promoter G-quadruplex sequences are targets for base oxidation and strand cleavage during hypoxia-induced transcription. Free Radic Biol Med. 2012;53:51–59. doi: 10.1016/j.freeradbiomed.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Beckett J, et al. Spontaneous DNA lesions modulate DNA structural transitions occurring at nuclease hypersensitive element III(1) of the human c-myc proto-oncogene. Biochemistry. 2012;51:5257–5268. doi: 10.1021/bi300304k. [DOI] [PubMed] [Google Scholar]
- 70.Cheong VV, et al. Xanthine and 8-oxoguanine in G-quadruplexes: formation of a GGXO tetrad. Nucleic Acids Res. 2015;43:10506–10514. doi: 10.1093/nar/gkv826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fleming AM, et al. 8-Oxo-7,8-dihydroguanine in the context of a gene promoter G-quadruplex is an on–off switch for transcription. ACS Chem Biol. 2017;12:2417–2426. doi: 10.1021/acschembio.7b00636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fleming AM, et al. Oxidative DNA damage is epigenetic by regulating gene transcription via base excision repair. Proc Natl Acad Sci U S A. 2017;114:2604–2609. doi: 10.1073/pnas.1619809114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cogoi S, et al. The regulatory G4 motif of the Kirsten ras (KRAS) gene is sensitive to guanine oxidation: implications on transcription. Nucleic Acids Res. 2018;46:661–676. doi: 10.1093/nar/gkx1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee H-T, et al. Molecular mechanisms by which oxidative DNA damage promotes telomerase activity. Nucleic Acids Res. 2017;45:11752–11765. doi: 10.1093/nar/gkx789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wu Y, et al. FANCJ helicase defective in Fanconia anemia and breast cancer unwinds G-quadruplex DNA to defend genomic stability. Mol Cell Biol. 2008;28:4116–4128. doi: 10.1128/MCB.02210-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Maizels N. G4-associated human diseases. EMBO Rep. 2015;16:910–922. doi: 10.15252/embr.201540607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sarkies P, et al. Epigenetic instability due to defective replication of structured DNA. Mol Cell. 2010;40:703–713. doi: 10.1016/j.molcel.2010.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sarkies P, et al. FANCJ coordinates two pathways that maintain epigenetic stability at G-quadruplex DN.A. Nucleic Acids Res. 2012;40:1485–1498. doi: 10.1093/nar/gkr868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schiavone D, et al. Determinants of G quadruplex-induced epigenetic instability in REV1-deficient cells. EMBO J. 2014;33:2507–2520. doi: 10.15252/embj.201488398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Guilbaud G, et al. Local epigenetic reprogramming induced by G-quadruplex ligands. Nat Chem. 2017;9:1110–1117. doi: 10.1038/nchem.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bartke T, et al. Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell. 2010;143:470–484. doi: 10.1016/j.cell.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Borgel J, et al. KDM2A integrates DNA and histone modification signals through a CXXC/PHD module and direct interaction with HP1. Nucleic Acids Res. 2016;45:1114–1129. doi: 10.1093/nar/gkw979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pedroso IM, et al. The effect of the TRF2 N-terminal and TRFH regions on telomeric G-quadruplex structures. Nucleic Acids Res. 2009;37:1541–1554. doi: 10.1093/nar/gkn1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cree SL, et al. DNA G-quadruplexes show strong interaction with DNA methyltransferases in vitro. FEBS Lett. 2016;590:2870–2883. doi: 10.1002/1873-3468.12331. [DOI] [PubMed] [Google Scholar]
- 85.Smith SS, et al. Recognition of unusual DNA structures by human DNA (cytosine-5)methyltransferase. J Mol Biol. 1991;217:39–51. doi: 10.1016/0022-2836(91)90609-a. [DOI] [PubMed] [Google Scholar]
- 86.Halder R, et al. Guanine quadruplex DNA structure restricts methylation of CpG dinucleotides genome-wide. Mol Biosyst. 2010;6:2439–2447. doi: 10.1039/c0mb00009d. [DOI] [PubMed] [Google Scholar]
- 87.Baral A, et al. Emerging trends in G-quadruplex biology–role in epigenetic and evolutionary events. Mol Biosyst. 2013;9:1568–1575. doi: 10.1039/c3mb25492e. [DOI] [PubMed] [Google Scholar]
- 88.Hänsel-Hertsch R, et al. G-quadruplex structures mark human regulatory chromatin. Nat Genet. 2016;48:1267–1272. doi: 10.1038/ng.3662. [DOI] [PubMed] [Google Scholar]
- 89.Kouzine F, et al. Permanganate/S1 nuclease footprinting reveals non-B DNA structures with regulatory potential across a mammalian genome. Cell Syst. 2017;4:344–356.e7. doi: 10.1016/j.cels.2017.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Foulk MS, et al. Characterizing and controlling intrinsic biases of lambda exonuclease in nascent strand sequencing reveals phasing between nucleosomes and G-quadruplex motifs around a subset of human replication origins. Genome Res. 2015;25:725–735. doi: 10.1101/gr.183848.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.De S, Michor F. DNA secondary structures and epigenetic determinants of cancer genome evolution. Nat Struct Mol Biol. 2011;18:950–955. doi: 10.1038/nsmb.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shay JW, Wright WE. Senescence and immortalization: role of telomeres and telomerase. Carcinogenesis. 2005;26:867–874. doi: 10.1093/carcin/bgh296. [DOI] [PubMed] [Google Scholar]
- 93.Greider CW. Molecular biology. Wnt regulates TERT–putting the horse before the cart. Science. 2012;336:1519–1520. doi: 10.1126/science.1223785. [DOI] [PubMed] [Google Scholar]
- 94.Horn S, et al. TERT promoter mutations in familial and sporadic melanoma. Science. 2013;339:959–961. doi: 10.1126/science.1230062. [DOI] [PubMed] [Google Scholar]
- 95.Killela PJ, et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci U S A. 2013;110:6021–6026. doi: 10.1073/pnas.1303607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Huang FW, et al. Highly recurrent TERT promoter mutations in human melanoma. Science. 2013;339:957–959. doi: 10.1126/science.1229259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Liu T, et al. Cancer-specific telomerase reverse transcriptase (TERT) promoter mutations: biological and clinical implications. Genes (Basel) 2016;7:38. doi: 10.3390/genes7070038. Published online July 18, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Vinagre J, et al. Frequency of TERT promoter mutations in human cancers. Nat Commun. 2013;4 doi: 10.1038/ncomms3185. 2185. [DOI] [PubMed] [Google Scholar]
- 99.Daniel M, et al. Regulation of the human catalytic subunit of telomerase (hTERT) Gene. 2012;498:135–146. doi: 10.1016/j.gene.2012.01.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lewis KA, Tollefsbol TO. Regulation of the telomerase reverse transcriptase subunit through epigenetic mechanisms. Front Genet. 2016;7:83. doi: 10.3389/fgene.2016.00083. Publsihed online May 9, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kyo S, et al. Understanding and exploiting hTERT promoter regulation for diagnosis and treatment of human cancers. Cancer Sci. 2008;99:1528–1538. doi: 10.1111/j.1349-7006.2008.00878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ramlee MK, et al. Transcription regulation of the human telomerase reverse transcriptase (hTERT) gene. Genes (Basel) 2016;7:50. doi: 10.3390/genes7080050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Avin BA, et al. Human telomerase reverse transcriptase regulation by DNA methylation, transcription factor binding and alternative splicing (Review) Int J Oncol. 2016;49:2199–2205. doi: 10.3892/ijo.2016.3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Won J, et al. Sp1 and Sp3 recruit histone deacetylase to repress transcription of human telomerase reverse transcriptase (hTERT) promoter in normal human somatic cells. J Biol Chem. 2002;277:38230–38238. doi: 10.1074/jbc.M206064200. [DOI] [PubMed] [Google Scholar]
- 105.Liu C, et al. The telomerase reverse transcriptase (hTERT) gene is a direct target of the histone methyltransferase SMYD3. Cancer Res. 2007;67:2626–2631. doi: 10.1158/0008-5472.CAN-06-4126. [DOI] [PubMed] [Google Scholar]
- 106.Akincilar SC, et al. Long-range chromatin interactions drive mutant TERT promoter activation. Cancer Discov. 2016;6:1276–1291. doi: 10.1158/2159-8290.CD-16-0177. [DOI] [PubMed] [Google Scholar]
- 107.Guilleret I, Benhattar J. Unusual distribution of DNA methylation within the hTERT CpG island in tissues and cell lines. Biochem Biophys Res Commun. 2004;325:1037–1043. doi: 10.1016/j.bbrc.2004.10.137. [DOI] [PubMed] [Google Scholar]
- 108.Stern JL, et al. Allele-specific DNA methylation and its interplay with repressive histone marks at promoter-mutant TERT genes. Cell Rep. 2017;21:3700–3707. doi: 10.1016/j.celrep.2017.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Palumbo SL, et al. Formation of a unique end-to-end stacked pair of G-quadruplexes in the hTERT core promoter with implications for inhibition of telomerase by G-quadruplex-interactive ligands. J Am Chem Soc. 2009;131:10878–10891. doi: 10.1021/ja902281d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Riou J-F, et al. Telomeres and telomerase, new targets for anticancer chemotherapy. Ann Pharm Fr. 2006;64:97–105. doi: 10.1016/s0003-4509(06)75301-9. [DOI] [PubMed] [Google Scholar]
- 111.Kang H-J, et al. A pharmacological chaperone molecule induces cancer cell death by restoring tertiary DNA structures in mutant hTERT promoters. J Am Chem Soc. 2016;138:13673–13692. doi: 10.1021/jacs.6b07598. [DOI] [PubMed] [Google Scholar]
- 112.Raiber E-A, et al. A non-canonical DNA structure is a binding motif for the transcription factor SP1 in vitro. Nucleic Acids Res. 2012;40:1499–1508. doi: 10.1093/nar/gkr882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zinn RL, et al. hTERT is expressed in cancer cell lines despite promoter DNA methylation by preservation of unmethylated DNA and active chromatin around the transcription start site. Cancer Res. 2007;67:194–201. doi: 10.1158/0008-5472.CAN-06-3396. [DOI] [PubMed] [Google Scholar]
- 114.Castelo-Branco P, et al. A cancer specific hypermethylation signature of the TERT promoter predicts biochemical relapse in prostate cancer: a retrospective cohort study. Oncotarget. 2016;7:57726–57736. doi: 10.18632/oncotarget.10639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Burge S, et al. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bochman ML, et al. {DNA} secondary structures: stability and function of G-quadruplex structures. Nat Rev Genet. 2012;13:770–780. doi: 10.1038/nrg3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kikin O, et al. QGRS Mapper: a web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res. 2006;34:676–682. doi: 10.1093/nar/gkl253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Scaria V, et al. Quadfinder: server for identification and analysis of quadruplex-forming motifs in nucleotide sequences. Nucleic Acids Res. 2006;34:W683–W685. doi: 10.1093/nar/gkl299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bedrat A, et al. Re-evaluation of G-quadruplex propensity with G4Hunter. Nucleic Acids Res. 2016;44:1746–1759. doi: 10.1093/nar/gkw006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kikin O, et al. GRSDB2 and GRS_UTRdb: databases of quadruplex forming G-rich sequences in pre-mRNAs and mRNAs. Nucleic Acids Res. 2008;36:141–148. doi: 10.1093/nar/gkm982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Garant JM, et al. G4RNA: an RNA G-quadruplex database. Database. 2015;2015:1–5. doi: 10.1093/database/bav059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kostadinov R. GRSDB: a database of quadruplex forming G-rich sequences in alternatively processed mammalian pre-mRNA sequences. Nucleic Acids Res. 2006;34:D119–D124. doi: 10.1093/nar/gkj073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Garg R, et al. Genome-wide discovery of G-quadruplex forming sequences and their functional relevance in plants. Sci Rep. 2016;6:31–35. doi: 10.1038/srep28211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lexa M, et al. Quadruplex-forming sequences occupy discrete regions inside plant LTR retrotransposons. Nucleic Acids Res. 2014;42:968–978. doi: 10.1093/nar/gkt893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Li Q, et al. G4LDB: a database for discovering and studying G-quadruplex ligands. Nucleic Acids Res. 2013;41:1115–1123. doi: 10.1093/nar/gks1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.London TBC, et al. FANCJ is a structure-specific DNA helicase associated with the maintenance of genomic G/C tracts. J Biol Chem. 2008;283:36132–36139. doi: 10.1074/jbc.M808152200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Paeschke K, et al. DNA replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase. Cell. 2011;145:678–691. doi: 10.1016/j.cell.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Paeschke K, et al. Pif1 family helicases suppress genome instability at G-quadruplex motifs. Nature. 2013;497:458–462. doi: 10.1038/nature12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Chatterjee S, et al. Mechanistic insight into the interaction of BLM helicase with intra-strand G-quadruplex structures. Nat Commun. 2014;5:1–12. doi: 10.1038/ncomms6556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Liu JQ, et al. G-quadruplex hinders translocation of BLM helicase on DNA: a real-time fluorescence spectroscopic unwinding study and comparison with duplex substrates. J Am Chem Soc. 2010;132:10521–10527. doi: 10.1021/ja1038165. [DOI] [PubMed] [Google Scholar]
- 131.Duan XL, et al. G-quadruplexes significantly stimulate Pif1 helicase-catalyzed duplex DNA unwinding. J Biol Chem. 2015;290:7722–7735. doi: 10.1074/jbc.M114.628008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Mendez-Bermudez A, et al. Genome-wide control of heterochromatin replication by the telomere capping protein TRF2. Mol Cell. 2018;70:449–461.e5. doi: 10.1016/j.molcel.2018.03.036. [DOI] [PubMed] [Google Scholar]
- 133.Rhodes D, Lipps HJ. Survey and summary G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015;43:8627–8637. doi: 10.1093/nar/gkv862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.van Wietmarschen N, et al. BLM helicase suppresses recombination at G-quadruplex motifs in transcribed genes. Nat Commun. 2018;9:271. doi: 10.1038/s41467-017-02760-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Sun H, et al. The Bloom’s syndrome helicase unwinds G4 DNA. J Biol Chem. 1998;273:27587–27592. doi: 10.1074/jbc.273.42.27587. [DOI] [PubMed] [Google Scholar]
- 136.Tang W, et al. The Werner syndrome RECQ helicase targets G4 DNA in human cells to modulate transcription. Hum Mol Genet. 2016;25:2060–2069. doi: 10.1093/hmg/ddw079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Mendez-Bermudez A, et al. A genome-wide control of heterochromatin replication by the telomere capping protein TRF2. Mol Cell. 2018;70:449–461.e5. doi: 10.1016/j.molcel.2018.03.036. [DOI] [PubMed] [Google Scholar]
- 138.Mukherjee AK, et al. Telomere length-dependent transcription and epigenetic modifications in promoters remote from telomere ends. PLoS Genet. 2018;14:e1007782. doi: 10.1371/journal.pgen.1007782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Mao SQ, et al. DNA G-quadruplex structures mold the DNA methylome. Nat Struct Mol Biol. 2018;25:951–957. doi: 10.1038/s41594-018-0131-8. [DOI] [PMC free article] [PubMed] [Google Scholar]