65.1. Introduction
Retinitis pigmentosa (RP) is a progressive rod–cone dystrophy that is characterized by night blindness, loss of peripheral vision, which can eventually lead to complete loss of vision (Hartong et al. 2006). RP primarily affects the rod photoreceptors and retinal pigment epithelium (RPE), and is the most common inherited retinal dystrophy, affecting as many as 1:1,000–1:3,500 people worldwide (Bunker et al. 1984; Grondahl 1987; Haim et al. 1992; Xu et al. 2006).
Displaying all modes of Mendelian inheritance, RP is genetically heterogeneous. To date 46 loci have been identified for nonsyndromic RP, leading to the identification of 36 causative genes (RetNet 2009). Genes implicated in RP encode components of the phototransduction cascade, retinal transcription factors, photoreceptor structural proteins, cilia proteins, and ubiquitously expressed components of the spliceosome. Currently, genes encoding five spliceosomal components have been identified in autosomal dominant RP (adRP). These include the pre-mRNA processing factors 3, 8, and 31 (PRPF3, 8, and 31), RP9, and SNRNP200 (Maita et al. 2005; McKie et al. 2001; Vithana et al. 2001; Zhao et al. 2009).
Mutations in splicing factors are of particular interest because these proteins are ubiquitously expressed and required for proper splicing of pre-mRNA in all cell types, yet mutations in PRPF3, 8, and 31, RP9, and SNRNP200 are only known to cause retinal disease. The spliceosome is a dynamic complex consisting of five small nucleolar ribonucleoproteins (snRNPs); U1, U2, U4/U6, and U5 (Grainger and Beggs 2005). The U1 and U2 snRNPs are the first to bind a pre-mRNA by recognizing the 5′ splice site and branch site, respectively. This interaction along with the binding of auxiliary splicing proteins defines the intron/exon boundaries and recruits the U4/U6-U5 tri-snRNP. Following a series of protein and RNA rearrangements, the U4/U6-U5 tri-snRNP becomes the catalytic component that drives splicing (Beggs et al. 1995; Farkas et al. 2010). The five splicing factors implicated in adRP are all components of the U4/U6-U5 tri-snRNP.
65.2. The Retinal Pigment Epithelium Is the Primary Tissue Affected by Mutations in the Pre-mRNA Processing Factors 3, 8, and 31
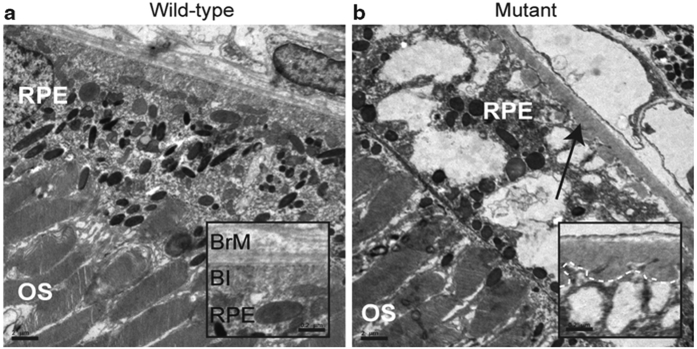
An intriguing aspect of question about RP caused by mutations in spliceosomal components is how the identified mutations result in a tissue-specific disease. Prior to the generation of animal models with single codon mutations in the Prpf3 and 8 genes, mimicking the most common mutations found in patients with this disease and Prpf31-knockout animals that mimic the null alleles found in most PRPF31 patients, it was unclear which retinal cell type was adversely affected (Graziotto et al. 2011). Ultrastructural analyses of the retinas of the gene-targeted mice indicated above showed that the RPE degenerated prior to the photoreceptors. The RPE of wild-type animals appears normal with long apical microvilli interdigitating the photoreceptor outer segments and visible basal infoldings. The RPE of mutant animals, however, shows a loss of basal infoldings, extensive vacuolization, and amorphous deposits between the RPE and Bruch’s membrane (Fig. 65.1).
Fig. 65.1.
Electron microscopy of representative retina and RPE from 2-year-old homozygous Prpf3-T494M, Prpf8-H2309P, and heterozygous 1-year-old Prpf31 knockout mice. (a) Representative wild-type control image for all three mouse models. (b) Representative image for all three mouse models showing vacuoles in the RPE. In the Prpf3-T494M and Prpf8-H2309P mice, loss of basal infoldings and accumulation of amorphous material between Bruch’s membrane and the RPE is evident (arrow). These changes are not evident in the control samples. Scale bars: (a, b) 2 μm; inset scale bars 0.2 μm. OS outer segments; RPE retinal pigment epithelium
Interestingly, the mutations that lead to the RNA splicing factor forms of RP are not unlike disease-causing mutations in other spliceosomal components with regard to the tissue specificity of pathogenesis. A better studied spliceosome-associated disease is spinal muscular atrophy (SMA), which is an autosomal recessive neuro-degenerative disorder characterized by degeneration of α-motor neurons in the spinal cord leading to muscular atrophy, and ultimately paralysis (Zhang et al. 2008). The SMA disease gene produces a ubiquitously expressed SMN1 (survival of motor neurons) protein that is necessary for the biogenesis and assembly of pre-mRNA processing factors, and other ribonucleoprotein complexes, involved in splicing. Mutations in the SMN1 gene lead to this disease resulting in an altered snRNP stoichiometry affecting only motor neurons. It is hypothesized that the altered snRNP composition in turn results in generation of aberrant mRNA transcripts which are responsible for disease (Zhang et al. 2008).
65.3. Identifying Aberrantly Spliced Transcripts Using Next-Generation Sequencing
Similar to mutations in the SMN1 gene, mutations in the pre-mRNA processing factors are believed to cause RP via production of aberrantly spliced transcripts.| Identification of these transcripts is a challenging task given the number of expressed genes and the complexity of the transcriptome with thousands of genes in the mouse, many of which can be alternatively spliced (Bult et al. 2008). Until recently it has been difficult to study splicing-associated diseases in great enough detail to be effective. Exon microar-rays have been useful, but have many disadvantages such as limited probe sets that only cover annotated splice junction boundaries, high levels of background, and low sensitivity (Teng and Xiao 2009). The development of next-generation sequencing (NGS) technologies provided the ability to fully interrogate the transcriptome and has overcome many of the limitations of microarrays (Shendure 2008).
65.3.1. Next-Generation Sequencing Platforms
Multiple platforms exist for NGS-based transcriptome analyses (reviewed in more detail in Simon et al. 2009; Ansorge 2009). Similar to microarrays, NGS analyses provide quantitative expression data. However, NGS analyses also provide data for novel splicing events with greater sensitivity and less background (Shendure 2008). In many cases, the best platform to study splicing is a trade-off between the number of sequencing reads generated and the length of the sequence. Our lab has demonstrated that at least 100 million 108 bp paired-end reads are necessary for full coverage of the mouse neural retina transcriptome (Farkas et al. 2010). This is consistent with data from transcriptome analyses of other tissues and organisms (Blencowe et al. 2009).
65.3.2. An Overview of RNA-Seq
Although the protocols for the generation of cDNA libraries from total RNA are rapidly evolving, the principles underlying the process stay the same. High quality mRNA is isolated and fragmented to a user-defined length (currently 250–350 bp). The fragmented mRNA is then converted to double-stranded cDNA and common adapters are ligated to the ends of the cDNA (Wang et al. 2009). Following sequencing, the data is aligned to the genome or transcriptome and can be analyzed for gene expression, expression of individual exons, alternative/aberrant splicing, insertion/deletions, and polymorphisms.
65.3.3. Bioinformatic Analysis of RNA-Seq Data
The generation of NGS technology applications for RNA sequencing (RNA-seq) applications quickly outpaced the available algorithms for aligning and analyzing the resulting sequence data. Unlike genomic resequencing, RNA-seq requires the ability to align reads that can span multiple exons that can be tens of thousands of bases apart. One option to overcome this obstacle is to align reads to a transcriptome database. While this is a good method for quickly aligning millions of reads that cross annotated splice junctions, it is incapable of identifying reads that cross novel junctions. Identifying novel junctions in normal tissues is an important step toward identifying aberrant transcripts in disease tissues since the two types must be compared to differentiate normal from abnormal.
Algorithms designed to tackle the issues presented by RNA-seq data are continually being developed and adapted (Li and Homer 2010). Most are geared toward aligning data quickly and less compute-intensive. However, this often sacrifices sensitivity and leads to a loss of data. With the advent of Cloud Computing (McPherson 2009; Langmead et al. 2010), there is no longer a need for ultra-efficient algorithms that sacrifice data quality, but rather the ability to develop algorithms that maximize the number reads aligned in a manner that is more sensitive and accurate. We have developed the RNA-seq Ultimate Mapper (RUM) that is based on the alignment algorithms Bowtie to align ungapped reads to the genome and transcriptome followed by a gapped alignment using BLAT (Blast-like Alignment Tool) (Langmead et al. 2009; Kent 2002; Farkas et al. 2010). Using Bowtie, millions of reads that map to both the genome and the annotated transcriptome can quickly be aligned. BLAT is a gapped aligner that is more compute-intensive and slower than Bowtie, but allows for the alignment of reads that span novel junctions. The combination of these two algorithms provides speed, accuracy, and the ability to identify previously unannotated transcripts.
65.4. RNA-Seq Analyses of RNA Splicing Factor Mutant Mice
Preliminary analyses of the neural retinas and RPE of wild-type and splicing factor mutant mice using RUM have revealed that there are thousands of novel/aberrant transcripts being produced in the mutant animals. Since all of the splicing factors are part of the U4/U6-U5 tri-snRNP and they all interact within this complex, we hypothesized that a common splicing mechanism is deficient. Thus, it is logical to compare all data sets for a common set of aberrant transcripts. When this is performed on the preliminary RNA-seq data produced for the Prpf3, 8, and 31 mouse models, as few as 52 transcripts are in common among all three mutant mice. These results suggest that pathogenic splicing errors are not widespread in RP. Two likely outcomes from the generation of aberrant transcripts are production of truncated proteins or the absence of protein production due to nonsense-mediated decay of the aberrant transcript. Given these outcomes, we will test candidate aberrant transcripts for pathogenic effects in the RPE and neural retina using both cell culture and mouse models.
65.5. Future Experimental Approaches
Obvious limitations exist to studying human RNA splicing factor RP disease in gene-targeted mouse models. For example, species-based differences in gene expression and alternative splicing may complicate identification of the cause for RNA splicing factor RP. However, it is not usually possible to obtain high quality ocular tissues from patients affected with RP. A potential solution to this problem is the use of retinal cells differentiated from patient-specific induced pluripotent stem (iPS) cells (Yu et al. 2007, 2009). The generation of iPS-derived RPE and retinal cells has recently been reported (Buchholz et al. 2009; Carr et al. 2009). It is possible that iPS-derived retinal cells from patients with RNA splicing factor RP could provide a source of human cells for transcriptome analyses, although the suitability of these cells for such studies remains to be/will need to be evaluated (Buchholz et al. 2009; Carr et al. 2009; Ozsolak et al. 2010).
RNA-seq is a continuously evolving method for studying molecularly complex diseases such as the RNA splicing factor forms of RP. As the methodology and bioinformatic approaches continue to develop and expand, many questions regarding aberrant splicing and the affect on various tissues may be answered. Furthermore, the characterization of the transcriptomes in normal cells and tissues such as the retina and RPE will help answer other basic biological questions as well. For example, a more thorough annotation of these transcriptomes will aid research into other retinal diseases, as well as provide insight into splice site preference at a single tissue level.
Acknowledgments
This work has been supported by the Ruth-Kirschstein National Research Service Award, Foundation Fighting Blindness, Penn Genome Frontiers Institute, Rosanne Silbermann Foundation, F.M. Kirby Foundation, and Research to Prevent Blindness.
Contributor Information
Michael H. Farkas, Ocular Genomics Institute, Berman Gund Laboratory, Department of Ophthalmology, Massachusetts Eye and Ear Infirmary, 243 Charles St., Boston, MA 02114, USA
Greg R. Grant, Penn Center for Bioinformatics, University of Pennsylvania, Philadelphia, PA 19104, USA
Eric A. Pierce, Ocular Genomics Institute, Berman Gund Laboratory, Department of Ophthalmology, Massachusetts Eye and Ear Infirmary, 243 Charles St., Boston, MA 02114, USA
References
- Ansorge WJ (2009) Next-generation DNA sequencing techniques. New Biotechnol 25:195–203 [DOI] [PubMed] [Google Scholar]
- Beggs JD, Teigelkamp S, Newman AJ (1995) The role of PRP8 protein in nuclear pre-mRNA splicing in yeast. J Cell Sci Suppl 19:101–105 [DOI] [PubMed] [Google Scholar]
- Blencowe BJ, Ahmad S, Lee LJ (2009) Current-generation high-throughput sequencing: deepening insights into mammalian transcriptomes. Genes Dev 23:1379–1386 [DOI] [PubMed] [Google Scholar]
- Buchholz DE, Hikita ST, Rowland TJ et al. (2009) Derivation of functional retinal pigmented epithelium from induced pluripotent stem cells. Stem Cells 27:2427–2434 [DOI] [PubMed] [Google Scholar]
- Bult CJ, Eppig JT, Kadin JA et al. (2008) The Mouse Genome Database (MGD): Mouse biology and model systems. Nuc Acid Res 36:D724–D728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunker CH, Berson EL, Bromley WC et al. (1984) Prevalence of retinitis pigmentosa in Maine. Am J Ophthalmol 97:357–365 [DOI] [PubMed] [Google Scholar]
- Carr AJ, Vugler AA, Hikita ST et al. (2009) Protective effects of human iPS-derived retinal pigment epithelium cell transplantation in the retinal dystrophic rat. PLoS ONE 4:e8152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas MH, Bujakowska K, Krishan A et al. (2010) Characterization of aberrant splicing by next generation high-throughput RNA-seq in mice with targeted mutations in Prpf3, Prpf8, and Prpf31 Invest Ophthalmol Vis Sci 51:ARVO E-Abstract 3667
- Grainger RJ, Beggs JD (2005) Prp8 protein: At the heart of the spliceosome. RNA 11:533–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graziotto JJ, Farkas MH, Bujakowska KM et al. (2011) Three gene targeted mouse models of RNA splicing factor RP show late onset RPE and retinal degeneration. Invest Ophthalmol Vis Sci 52(1):190–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grondahl J (1987) Estimation of prognosis and prevalence of retinitis pigmentosa and Usher syndrome in Norway. Clin Genet 31:255–264 [DOI] [PubMed] [Google Scholar]
- Haim M, Holm NV, Rosenberg T (1992) Prevalence of retinitis pigmentosa and allied disorders in Denmark. I Main results. Acta Ophthalmol (Copenh) 70:178–186 [DOI] [PubMed] [Google Scholar]
- Hartong DT, Berson EL, Dryja TP (2006) Retinitis pigmentosa. Lancet 368:1795–1809 [DOI] [PubMed] [Google Scholar]
- Kent WJ (2002) BLAT – the BLAST-like alignment tool. Genome Res 12:656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Hansen K, Leek J (2010) Cloud-scale RNA-sequencing differential expression analysis with Myrna. Genome Biol 11:R83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M et al. (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Homer N (2010) A survey of sequence alignment algorithms for next-generation sequencing. Brief Bioinform 11:473–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maita H, Kitaura H, Ariga H, Iguchi-Ariga SM (2005) Association of PAP-1 and Prp3p, the products of causative genes of dominant retinitis pigmentosa, in the tri-snRNP complex. Exp Cell Res 302:61–68 [DOI] [PubMed] [Google Scholar]
- McKie AB, McHale JC, Keen TJ et al. (2001) Mutations in the pre-mRNA splicing factor gene PRPC8 in autosomal dominant retinitis pigmentosa (RP13). Hum Mol Genet 10:1555–1562 [DOI] [PubMed] [Google Scholar]
- McPherson JD (2009) Next-generation gap. Nat Meth 6:S2–S5 [DOI] [PubMed] [Google Scholar]
- Ozsolak F, Goren A, Gymrek M et al. (2010) Digital transcriptome profiling from attomole-level RNA samples. Genome Res 20:519–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- RetNet (2009) RetNet Web site address. http://www.sph.uth.tmc.edu/Retnet/
- Shendure J (2008) The beginning of the end for microarrays? Nat Meth 5:585–587 [DOI] [PubMed] [Google Scholar]
- Simon SA, Zhai J, Nandety RS et al. (2009) Short-read sequencing technologies for transcriptional analyses. Annu Rev Plant Biol 60:305–333 [DOI] [PubMed] [Google Scholar]
- Teng X, Xiao H (2009) Perspectives of DNA microarray and next-generation DNA sequencing technologies. Sci C Life Sci 52:7–16 [DOI] [PubMed] [Google Scholar]
- Vithana EN, Abu-Safieh L, Allen MJ et al. (2001) A human homolog of yeast pre-mRNA splicing gene, PRP31, underlies autosomal dominant retinitis pigmentosa on chromosome 19q13.4 (RP11). Mol Cell 8:375–381 [DOI] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M (2009) RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10:57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Hu L, Ma K, et al. (2006) Prevalence of retinitis pigmentosa in urban and rural adult Chinese: The Beijing Eye Study. Eur J Ophthalmol 16:865–866 [DOI] [PubMed] [Google Scholar]
- Yu J, Hu K, Smuga-Otto K et al. (2009) Human induced pluripotent stem cells free of vector and transgene sequences. Science 324:797–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K et al. (2007) Induced pluripotent stem cell lines derived from human somatic cells. Science 318:1917–1920 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Lotti F, Dittmar K et al. (2008) SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell 133:585–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Bellur DL, Lu S et al. (2009) Autosomal-dominant retinitis pigmentosa caused by a mutation in SNRNP200, a gene required for unwinding of U4/U6 snRNAs. Am J Hum Genet 85:617–627 [DOI] [PMC free article] [PubMed] [Google Scholar]