Abstract
The regulation of gene expression occurs through complex relationships between transcription, processing, turnover, and translation, which are only beginning to be elucidated. We know that at least for certain messenger (m) RNAs, processing, modifications, and sequence elements can greatly influence their translational output through recognition by translation and turn-over machinery. Recently, we and others have combined high-throughput sequencing technologies with traditional biochemical methods of studying translation to extend our understanding of these relationships. Additionally, there is growing importance given to how these processes may be regulated across varied cell types as a means to achieve tissue-specific expression of proteins. Here, we provide an in-depth methodology for polysome profiling to dissect the composition of mRNAs and proteins that make up the translatome from both whole tissues and a specific cell type isolated from mammalian tissue. Also, we provide a detailed computational workflow for the analysis of the next-generation sequencing data generated from these experiments.
Keywords: cell-type isolation from tissues, polysome profiling, next-generation sequencing and bioinformatics, ribosomal occupancy shift, translation regulation
1. Introduction
The primary biological function of messenger RNA (mRNA) is to serve as the intermediary molecule between the genome and the functional proteins it encodes. It is often convenient to assume that the abundance of an mRNA is directly related to and tightly correlated with its encoded protein levels, however, this correlation is quite poor under most biological settings [1-5]. In recent years, many rules for the translatability of a mRNA have been determined including its codon optimality, regulatory sites for RNA binding proteins, secondary structures, untranslated regions, the variable inclusion or exclusion of coding and non-coding sequences through alternative splicing as well as covalent modifications to bases and the length of the poly(A) tail [6-14].
Recent technical advances for studying translation have been a boon for studying mRNA regulation. For instance, ribosome profiling uses ribosome footprinting of mRNAs followed by sequencing to measure codon level resolution of ribosome occupancies across transcripts [15, 16]. The limitation is that one cannot obtain data on full-length transcript isoforms, untranslated regions, poly(A) tails, or modifications. While it is possible to extract some splicing data from ribosome profiling reads, it may only be useful for more abundant transcripts [17]. Translating ribosome affinity purification sequencing (TRAP-seq) and RiboTag are powerful techniques that utilize affinity pull-down of tagged-ribosomes to probe the translation status of mRNAs from tissues in a cell-type specific manner [18-20]. The drawback is that it requires genetic modification of the system prior to pull-down, which can be costly and time-consuming, or the use of transfection methods which may have limited efficiency in mammalian systems [21, 22]. Further, there are still improvements to be made either experimentally or computationally to minimize the noise of transcripts from non-targeted cells [23].
Polysome profiling followed by sequencing is a complementary technique that fills the niche of examining translation without the need for exogenous tags. It allows for the investigation of transcripts in their full-length context by utilizing sucrose density gradient fractionation to isolate mRNAs based on ribosome occupancy. When combined with high-throughput sequencing, one can obtain data on full-length transcripts as well as determine the translational status of alternative transcript isoforms [14,24-26]. Further, because the entire context of the transcript is maintained, the influence of post-transcriptional modifications—such as the poly(A) tail length or covalent modifications to bases—on translation can be investigated. However, due to the sensitive nature of using a sucrose gradient, the resolution of profiles can be variable between laboratories or even between experimenters. The impact of this variation may be minimized by pooling peaks during fractionation, as it was shown that transcripts tend to be clustered in either low or high polysome fractions [25]. Moreover, it may not always be possible to deconvolute polysomes from other large RNP complexes in the cytoplasm leading to mischaracterization of ribosome occupancies for transcripts in some instances.
The three methods mentioned above have and will continue to provide valuable insights into the molecular mechanisms of translation, particularly when used in combination. However, a key focus for ongoing research needs to be the physiologic context in which translational regulation occurs. Protein synthesis rates are not static; instead, they vary widely both temporally and between cell types within the same tissue and/or from different tissues [27-30]. Developing methods that optimize the above-mentioned biochemical techniques for use in mammalian systems will allow for the investigation of translation under physiological conditions. Here, we provide a detailed polysome profiling protocol followed by deep sequencing to investigate the translation status of full-length mRNAs isolated from whole tissues and single-cell types (Fig. 1). The mRNAs isolated from this protocol can also be further sub-fractionated prior to sequencing to investigate the roles of a variety of mRNA processing events on translation such poly(A)-tail length and mRNA modifications [31-33]. Further, we designed an easily implemented bioinformatics pipeline for determining relative ribosome occupancy of transcripts within a sample and the differential ribosome occupancy of transcripts across experimental conditions, which we term Shifts in Ribosomal Occupancies (Shirloc).
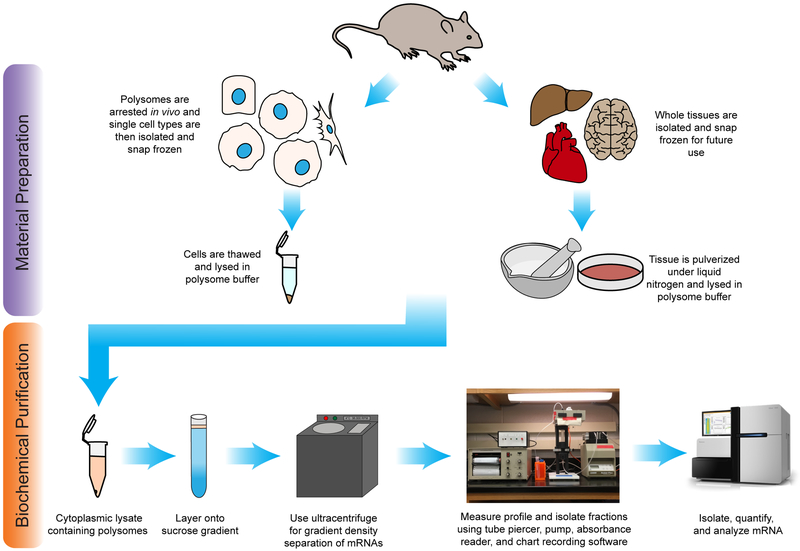
Figure 1. Experimental schematic.
The experimental workflow of polysome profiling from mammalian tissues or isolated cell-types following by next-generation sequencing analysis. Either whole tissue or single cell-types are isolated from the animal. The cells are lysed, nuclei and cell debris are removed, and the cytoplasm is layered onto a sucrose gradient. Following centrifugation, the gradient is flowed through an absorbance detector and the profile is collected. mRNA can then be isolated from fractions for downstream analysis.
2. Materials and Methods
2.1. Buffer stocks and sucrose gradients
The buffers and techniques used in this protocol have been adapted and modified from several other published polysome profiling methods designed for mammalian cell culture [24,25,34,35]. Typical mammalian cell cultures will readily take up cycloheximide and arrest ribosomes during translation elongation, however, adapting that strategy to an entire tissue is not trivial [36,37]. We have developed two separate methods of arresting ribosomes isolated from mammalian tissues both in vivo and ex vivo. Many polysome profiling buffers also contain heparin as a general RNase inhibitor; however, it has also been shown to inhibit reverse transcriptase. Given that the goal of this method is to obtain RNA for sequencing and removal of heparin following RNA isolation greatly reduces yield, we opted to omit it from our preparations and found that using SUPERase•In alone is suitable as an RNase inhibitor as long as other precautions are taken while performing the protocol. Before starting the protocol, clean all workspaces and equipment to maintain an RNase free environment and use the specific materials as noted by an asterisk (Table 1). Prepare all solutions according to the recipes using sterile, RNAse free starting material. The 10X polysome buffer (Table 2), 60% sucrose solution in polysome buffer (Table 3), polysome lysis buffer stock (Table 4), and cycloheximide stock (Table 5) solutions can all be prepared ahead of time and stored accordingly.
Table 1.
Equipment and reagents.
| Material | Manufacturer | Catalog Number |
|---|---|---|
| Tris-HCl | VWR | 97062 |
| KCl | Fisher Scientific | BP366 |
| MgCl2 | Fisher Scientific | M35-500 |
| Sucrose | Fisher Scientific | BP220-1 |
| Sodium deoxycholate | Sigma-Aldrich | D6750 |
| Nonidet-P40‡ | Pharmacia LKB Biotechnology | 1830-202 |
| Cycloheximide | Sigma-Aldrich | C7698 |
| Dithiothreitol (DTT) | Fisher Scientific | FERR0861 |
| Pierce protease inhibitor cocktail (EDTA-Free)* | Thermo Scientific | AM32955 |
| SUPERase•In RNase inhibitor* | Invitrogen | AM2696 |
| Sodium acetate | Sigma-Aldrich | S8750 |
| GlycoBlue Coprecipitant* | Invitrogen | AM9515 |
| Ethanol 200 Proof | Decon Laboratories | 2701 |
| Phosphate buffered saline | Corning | 21040 |
| Dnase I | New England Biolabs | M0303 |
| Acid phenol:chloroform (5:1) | Sigma-Aldrich | P1944 |
| Chloroform:isoamyl alcohol (24:1) | Sigma-Aldrich | 77617 |
| 15 mL conical tubes | Corning | 352096 |
| 50 mL conical tubes | Corning | 352098 |
| 1.5 mL microcentrifuge tubes | Axygen | MCT-150 |
| Thin wall 9/16 × 3 1/2 polypropylene tubes* | Beckman Coulter | 331372 |
| 10 cm cell culture plates | Corning | -- |
| Cell scrapers | Corning | -- |
| Mortar and pestle | VWR | -- |
| SW41Ti swinging bucket rotor* | Beckman Coulter | 331362 |
| Ultracentrifuge | Beckman Coulter | -- |
| Tube piercer w/ density gradient flow cell* | Brandel | BR-184-5 |
| UV Detector with Type 11 Optical Unit and Chart Recorder* | Brandel | UA-6 |
| PeakChart date capture software* | Brandel | 2115-188 |
| Gradient master* | Biocomp | 108 |
| Surgery tool kit | -- | -- |
| Liquid nitrogen | -- | -- |
| Peristatic pump | -- | -- |
Equipment and reagents needed to perform polysome profiling on mammalian tissues or single-cell types. For those materials marked with an asterisk, we highly recommend using the manufacturer and catalog number listed due to the unique nature of the product.
Nonidet-P40 may be no longer commercially available. If a similar substitute is used, the concentrations and lysis times may need to be modified slightly.
Table 2.
10X Polysome buffer stock
| Component | Final Concentration |
|---|---|
| Tris-HCl at pH 7.5 | 100 mM |
| KCl | 1 M |
| MgCl2•6H20 | 50 mM |
| NaOH | pH 7.5 |
| Molecular Grade H20 | -- |
Suggested preparation of 500 mL. Fill a beaker with 450 mL of ddH2O and a stir bar. Add 6.056 g Tris-HCl, 37.275 g KCl, and 1.248 g MgCl2 6H2O solids while stirring. Balance the pH of the solution to 7.5 using 4 N NaOH and then fill remaining volume to 500 mL with ddH2O. Filter through a .22 μm filter and store at room temperature.
Table 3.
60% sucrose in polysome buffer
| Component | Final Concentration |
|---|---|
| Tris-HCl at pH 7.5 | 10 mM |
| KCl | 100 mM |
| MgCl2 | 5 mM |
| Sucrose w/v | 60% w/v |
| Molecular Grade H2O | -- |
Suggested preparation of 500 mL. Add 50 mL of 10X polysome buffer sto ck and 200 mL of ddH2O into a beaker with a hot plate with a stir bar to 80°C. Slowly add 300 g sucrose. Wait until the sucrose has completely dissolved and then let the solution cool with continued stirring. Fill to 500 mL with ddH2O and filter through a .22 μm filter. Store at 4°C for up to a month and check for any microbial growth before using.
Table 4.
Polysome lysis buffer stock
| Component | Final Concentration |
|---|---|
| Tris-HCl at pH 7.5 | 10 mM |
| KCl | 100 mM |
| MgCl2 | 5 mM |
| Sodium deoxycholate | 0.5% w/v |
| Protease inhibitor cocktail | -- |
| Molecular Grade H2O | -- |
Suggested preparation of 10 mL. Combine 1.075 mL of 10X polysome buffer stock with 9 mL of ddH2O. Add 53.75 mg of sodium deoxycholate and 1 protease inhibitor cocktail tablet (EDTA-free). Dissolve completely by vortexing and store as 1 mL aliquots at −20°C for up to a month.
Table 5.
Cycloheximide solution
| Component | Final Concentration |
|---|---|
| Cycloheximide | 150 mg/mL |
| Ethanol | -- |
Suggested preparation of 10 mL. Add 150 mg of cycloheximide to 10 mL of 100% ethanol. Dissolve completely by vortexing and store at −20°C for up to a month.
2.1.1. 1X Polysome lysis buffer prepared fresh
Thaw necessary aliquots of polysome lysis buffer stock and add the components that need to be added fresh (Nonidet-P40, DTT, cycloheximide, and SUPERase•In; Table 6). Keep on ice until you are ready to lyse samples. While optimizing the protocol for use in your laboratory or with a new tissue/cell-type, a negative control should be run in which the lysis buffer and sucrose gradient omit the cycloheximide and instead include 50 mM EDTA.
Table 6.
Polysome lysis buffer prepared fresh
| Component | Final Concentration |
|---|---|
| Tris-HCl at pH 7.5 | 10 mM |
| KCl | 100 mM |
| MgCl2 | 5 mM |
| Sodium deoxycholate | .5% w/ve |
| Nonidet-P40 | 1 % v/v |
| DTT | 10 mM |
| cycloheximide | 150 μg/mL |
| SUPERase In RNase Inhibitor | 1 U/μL |
| Protease inhibitor cocktail | -- |
| Molecular Grade H2O | -- |
This must be prepared the day of the experiment. Each sample will require 1 mL and it is recommended to make an additional milliliter. To prepare 4 mL: thaw 4 aliquots of previously prepared lysis buffer stocks on ice. Combine 3.72 mL into a conical tube. Add 40 μL of Nonidet-P40, 150 μL of 1M DTT, 40 μL of 150 mg/mL cycloheximide in EtOH, and 40 μL of SUPERase•In RNase Inhibitor. Mix well by vortexing and keep on ice until ready for use. ‡Nonidet-P40 is no longer commercially available, and an alternative may need to be used. Final concentrations of detergent for lysis may need to be adjusted to compensate for this or according to your tissue of interest.
2.1.2. Preparation of Sucrose Gradients
Make 15% and 45% sucrose solutions using the 60% stock. To make four gradients, prepare 25 mL of each (15%: 18.75 mL 1X Polysome Buffer and 6.25 mL 60% Sucrose Solution; 45%: 6.25 mL 1X Polysome Buffer and 18.75 mL 60% Sucrose Solution) and supplement with 200 μL of cycloheximide stock and 25 μL of SUPERase•In.
Set up gradient using inch polypropylene ultracentrifugation tubes. Use the marker block provided with the gradient maker to mark the half-full point on each tube for layering.
Using the layering needle and a syringe, add the 15% sucrose solution until it reaches the halfway point (~6.25 mL).
Add the 45% sucrose solution from the bottom until the interface between the two solutions reaches the halfway point (~6 mL).
Using the long rate zonal caps (Biocomp), insert at a shallow angle with the air hole tilted up, allowing the bubble to escape and displace excess sucrose solution.
Remove liquid from the top of the cap and set the gradient master (Biocomp) to: (time:4.06, angle:80, speed:8). Balance tubes in the rotator and press start to form the gradients.
Handle the tubes carefully during the preceding and following steps in the protocol so as not to disrupt the gradient. Special care needs to be taken when making the gradients to ensure accuracy and reproducibility between gradients. Store the gradients in a tube holder at 4°C until lysates are ready to be loaded.
2.2. Collection and preparation of solid mouse tissues for polysome profiling analysis
2.2.1. Collection and storage of tissue
Translation is highly dynamic and quickly responds to both internal and external signals. In order to accurately measure in vivo translation of an entire tissue, we determined that it was imperative to resect and snap freeze the tissue of interest as quickly as possible—on the order of minutes. This minimizes changes in translation due to stress on the tissue and ribosomes falling off the transcripts.
Autoclave necessary surgery tools, prepare an RNase free workspace and fill a dewar with liquid nitrogen.
Quickly sacrifice the animal and remove tissue(s) of interest, following the standard procedures for your institution and laboratory.
Wash the tissue using autoclaved and RNase free PBS, removing any unwanted connective tissue and as much blood as possible.
Flash freeze the tissue in liquid nitrogen.
Store at −80°C for up to three months.
2.2.2. Collection of lysate for polysome analysis of whole tissues
To obtain quality polysomes that will present an accurate representation of ribosome occupancy in downstream applications, great care should be taken during this procedure. Prepare all necessary materials and reagents ahead of time and have them ready in order to minimize the time that samples are left sitting before centrifugation.
Thoroughly wash mortars, pestles, and scoopulas and autoclave them the day before in foil; prepare enough so that you have at least one set for each sample/condition that you will be examining.
Prepare the necessary amount (1 mL per 200-400 mg of tissue) of fresh lysis buffer and make the sucrose gradients as described above.
Turn on and pre-chill the ultracentrifuge.
Set up the workspace with ice, liquid nitrogen, dry ice, and 10 cm cell culture plates.
Fill mortar with liquid nitrogen to pre-chill it and the pestle. Place 10 cm plates on dry ice to pre-chill them.
Pulverize tissue with mortar and pestle under liquid nitrogen until it becomes a fine powder. Take care not to let the liquid nitrogen evaporate during this process. Once complete, let the nitrogen evaporate and scrape the powder into a pre-chilled plate using a chilled scoopula. Keep on dry ice until all samples are pulverized.
Remove plates from dry ice and place onto ice. Add 1 mL of fresh lysis buffer; tilt the plates and gently use a cell scraper to suspend the powder completely. Let it rest on ice for 1-2 minutes and then gently pipet up and down 10 times before transferring it to a clean microcentrifuge tube. Let the cells sit for another 2-3 minutes to ensure complete lysis.
Spin at 2,000×g for 5 min in 4°C to pellet nuclei and tissue debris.
Transfer supernatant to a fresh tube, taking care not to disturb the pellet.
Spin the supernatant at 12,000×g for 5 min at 4°C to pellet any remaining cellular debris.
Transfer the resulting supernatant to a fresh tube.
Measure OD260 and adjust samples accordingly.
Load 400 μL of supernatant onto the gradient (less can be loaded, however, do not load more than 400 μL as that can disrupt the gradient and prevent separation).
Ensure all tubes are balanced.
Centrifuge in a SW-41Ti rotor at 246,606×g (38,000 RPM) for 125 min with low acceleration and the brake off (the centrifuge should reach max speed after ~5 min from starting and then take 45-60 min to stop after the run).
2.3. In vivo-cell type specific polysome profiling
2.3.1. Isolation of single cell type by perfusion
Many protocols exist for purifying single cell types from a tissue for a variety of downstream applications. However, these are often long processes requiring many reagents and steps; ribosome occupancy is highly dynamic in vivo and cells will quickly change their translatome in response to external stresses [38]. We determined that the best way to compensate for this stress and maintain intact polysomes was to pre-treat the cells with cycloheximide in vivo and maintain it in all downstream buffers. Here, we describe polysome profiling of purified hepatocytes from an adult mouse isolated by an adapted perfusion method [30,39]. This protocol can be readily adapted to other perfusion or digestion based methods; however, it would likely not be suitable for longer isolation protocols such as those requiring FACS [40].
Prepare perfusion media fresh according to recipes in Tables 7 and 8.
Prewarm perfusion media in a 39°C water bath.
Prepare clean tubing and cannula. Set-up the surgery tray over a collection bucket as there will be more than 125 mL of liquid that will flow out during the procedure.
Attach tubing to the pump and set at 3.5 mL/min. Flow Solution I through the tubing.
Anesthetize mouse in a chamber (2.5% isoflurane, 1 L/min) and maintain anesthesia during the procedure. Once the mouse is fully anesthetized, secure the animal to the tray. Soak the abdomen with 70% EtOH to aid in the making the incision. Make an incision in the lower abdomen and fold back the skin over the chest to expose the intestines and liver. Move the intestines to the right to expose the portal vein and vena cava.
Insert the cannula into the vena cava. Turn the pump on and let Solution I perfuse the liver. If inserted correctly, the liver should begin to swell—cut the portal vein immediately.
Flow 75 mL of Solution I.
Switch to Solution II and flow 50 mL.
Remove the liver and use two forceps to gently massage the liver in a petridish containing Solution I to release cells.
Pass the cells successively through 70 μm and 40 μm filters to obtain a single-cell suspension. Separate the resulting suspension to two clean 50 mL conical tubes.
Centrifuge suspension at 50 × g for 5 minutes at 4°C to separate live hepatocytes from non-parenchymal cells and dead cells.
Discard the supernatant and resuspend the pellet in Solution I. Repeat this wash 3 times.
During the last wash, aliquot the suspensions into six microcentrifuge tubes. After centrifugation, remove the supernatant. Flash freeze the cell pellets in liquid nitrogen and store them at −80°C.
Table 7.
Hepatocyte perfusion solution A
| Component | Final Concentration |
|---|---|
| HBSS w/ phenol red | 1X |
| Cycloheximide | 300 μg/mL |
Supplement 1X HBSS (w/ phenol red) with the 15 mg/mL cycloheximide stock to a final concentration of 300 μg/mL. Prepare this fresh and warm to 39°C for perfusion.
Table 8.
Hepatocyte perfusion solution B
| HBSS w/ phenol red | 1X |
| Cycloheximide | 300 μg/mL |
| CaCl2 | 5.4 mM |
| Soybean Trypsin Inhibitor | .04 mg/mL |
| Collagenase type I | 90 U/mL |
Supplement 1X HBSS (w/ phenol red) with cycloheximide, CaCl2, soybean trypsin inhibitor, and collagenase type I. Prepare fresh and warm to 39°C for perfusion.
2.3.2. Cell lysis and purification of polysomes by sucrose gradient
Thaw the previously isolated cells on ice in 1 mL of fresh lysis buffer and resuspend the pellet by gently pipetting up and down 10 times. Let the tubes sit on ice 2-5 minutes until cells are lysed.
Spin at 2,000×g for 5 min in 4°C to pellet nuclei and cell debris.
Transfer supernatant to a fresh tube, taking care not to disturb the pellet.
Spin the supernatant at 12,000×g for 5 min at 4°C.
Transfer the resulting supernatant to a fresh tube.
Measure OD260 and adjust samples accordingly.
Load 400 μL of supernatant onto the gradient (less can be loaded, however, do not load more than 400 μL as that can disrupt the gradient and prevent separation).
Ensure all tubes are balanced.
Centrifuge in a SW-41Ti rotor at 246,606×g (38,000 RPM) for 125 min with low acceleration and the brake off (the centrifuge should reach max speed after ~5 min from starting and then take 45-60 min to stop after the run).
2.4. Polysome Gradient Fractionation
Following centrifugation, the sucrose density gradient will have separated cytoplasmic RNA based on the number of ribosomes bound. The RNA/protein complexes can be visualized using a UV absorbance meter. When done correctly, distinct peaks corresponding to the 40S, 60S, 80S monosome, and increasing numbers of polysomes should be distinguishable. The relative intensity of these peaks will vary depending on the tissue being investigated and potentially other experimental conditions (i.e. developmental state, feeding, fasting, activation, knock-down, etc.).
We do not advise attempting to compare absolute absorbance intensities between samples as there is too much potential variability even when loading the same measured absorbance units onto the gradient. However, the relative area under the curve for peaks can be measured, and the ratios of these areas over the total can be used as a comparison between samples to make semi-quantitative observations.
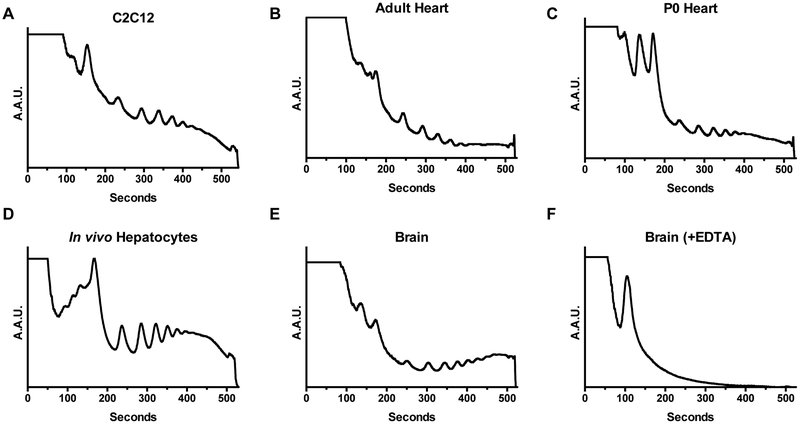
When initially optimizing the protocol for your laboratory or tissue of interest, we recommend measuring the OD260 of the lysate and loading multiple dilutions onto the gradient. This can then be used to inform the amount of starting material needed for subsequent experiments. You may choose to not even measure the OD260 once having optimized the amount of starting material, as the time spent measuring the optical density prior to loading is valuable for maintaining the integrity of the sample. The direct lysate will likely be higher than the accurate detectable range; so, make a series of serial dilutions from roughly 2X to 20X to get an accurate measurement. We found that a range of 1-5 adjusted OD260 provided suitable profiles in the tissues we measured. The optical density will be primarily coming from rRNA, so high values may be an indication of higher ribosome content in a sample. For example, we found that one adult heart or adult brain in 1 mL of lysis buffer yielded an appropriate concentration. Whereas, we would need to pool 2-3 neonatal hearts or brains to achieve similar concentrations. Alternatively, a 200 mg piece of adult liver needed to be diluted. Prior to measuring polysome profiles from mammalian tissues, we recommend performing it on proliferating cells in culture which will have the distinctively high polysome curve. Profiles generated from quiescent and particularly lowly proliferating tissues, will have comparatively less translational activity and may not be suitable for initial troubleshooting. We have provided examples of polysome profile traces for several types of samples as well as what a negative control trace should look like (Fig. 2) [30,31,41].
Figure 2. Polysome profiles from different cells and tissues.
Representative polysome traces measured at 254 nm of (A) undifferentiated C2C12 cells, (B) adult mouse heart, (C) neonatal mouse heart, (D) freshly isolated mouse hepatocytes, and (E) adult mouse brain. (F) A representative negative control trace in which ribosomes are disassociated from RNA by the addition of EDTA.
- Approximately an hour before the centrifugation will finish, begin preparing for fractionation.
- Wash all tubing with RNase free H2O at 3 mL/min by using a clean ultracentrifuge tube.
- Switch on UV lamp to begin warming it up until indicator turns green. Turn on detector, pump, and chart recorder (with computer and accompanying software if desired).
- Flow 60% sucrose through the tubing—including the absorbance reader—and set your baseline.
Carefully remove tubes from the centrifuge and keep them on ice.
Place tube in the piercing apparatus by securing it into the top holder and raising the bottom support. Pierce the tube with the needle by twisting the knob until the second line indicator is fully in the tube.
Set the pump to 1.5 mL/min and start flowing the sucrose; turn on the chart/computer recorder.
The first of the sample will enter the absorbance reader once the chase solution has reached approximately ⅓ of the volume. The first material to come out will be the free RNA and protein, followed by the 40S, 60S, 80S monosome, and increasing polysome peaks.
Collect fractions into microcentrifuge tubes as they exit the reader for use in downstream applications.
Once the last of the sample has been collected, flow several additional milliliters of 60% sucrose through the tubing to clean it out.
Remove tube, taking care to avoid spilling sucrose solution from the hole. This is best done by unscrewing the needle completely (residual pressure from the top of the tube will keep the solution in), placing a beaker underneath the tube, and then gently unscrewing from the top of the tube to let the solution slowly drain out.
Repeat steps 3-8 with remaining sample gradients.
Wash the tubing by flowing several hundred milliliters of RNase free H2O at 3 mL/min as in 2.4.1.a, then turn off all components.
2.5. Downstream analysis of polysome separated RNAs
In addition to the semi-quantitative observations of translation activity that can be made by comparing polysome profiles, there are many quantitative downstream applications to analyze the association of specific proteins and RNA with varying numbers of ribosomes. Here, we will focus on the analysis of RNA. Following extraction from the sucrose gradient fractions, high quality RNA can be obtained for further analysis.
2.5.1. Extraction of RNA from sucrose gradient fractions
Due to the high percentage of sucrose in the solution, a modified protocol must be used to purify the RNA, rather than using standard phenol/chloroform methods[25]. In addition, we recommend two rounds of aqueous phase extractions to remove ribosomes and other proteins that remain bound to the RNA. Once the purified RNA is obtained, downstream applications can be pursued. Because our goal for sequencing was to obtain intact, full-length transcripts we chose to perform poly(A)+ selection using oligo(dT) pull-down. Other groups have had success with rRNA depletion at this step prior to library preparation.
Add a .1 volume of 3 M NaOAc pH 5.2 to the sucrose gradient fraction and mix well.
Add .75 μL of GlycoBlue followed by 2 volumes of 100% EtOH and mix well.
Store at −20°C for at least 2 hours or overnight.
Pellet the precipitated RNA by centrifuging at >13,000 × g in a 4°C microcentrifuge for 30 minutes.
Resuspend pellet in 85 μL of RNase free H2O, 10 μL of 10 X DNase buffer (NEB), 5 μL of DNase I (NEB). Incubate at 37°C for 15 minutes.
Add 200 μL of .3 M NaOAc pH 5.2. Mix well and add 300 μL of Acid phenol:chloroform (5:1). Mix well and then centrifuge at 13,000 × g in a 4°C microcentrifuge for 20 minutes. Carefully remove aqueous layer into a clean tube.
The aqueous layer is then again phase separated using 300 μL Chloroform:Isoamyl Alcohol (24:1). Mix well and then centrifuge at 13,000 × g in a 4°C microcentrifuge for 20 minutes. Remove aqueous layer to a clean tube.
Add 1 μL of GlycoBlue and 1 mL 100% EtOH and mix well. Store at −20°C for at least 2 hours or overnight.
Pellet RNA by centrifuging at >13,000 × g in a 4°C microcentrifuge for 30 minutes.
Remove supernatant. Wash pellet with 1 mL of 75% EtOH. Spin at 13,000 × g for 10 minutes at 4°C. Repeat wash. Remove remaining EtOH using a quick-spin and then leave cap open at room temperature to dry the pellet.
Resuspend pellet in 20-30 μL of RNase free H2O depending on expected yield.
Measure concentration using a Qubit or Nanodrop.
Determine RNA quality by bioanalyzer. RIN numbers above 7.5 are ideal for sequencing.
Proceed with mRNA enrichment for library preparation for deep sequencing using standard methods, reverse transcription for splice assays or qPCR, or further analysis based on modifications.
3. Shirloc workflow for analyzing differential transcript translation
Combining the separation of tissue or cell-type specific RNA ribosome occupancy with the power of deep sequencing has the potential to offer new insights on how translation is regulated at a global scale. Polysome profiling followed by sequencing has the advantage over ribosome profiling that it allows analysis of the entire transcript, rather than only measuring the ribosome footprint [16,24-26]. Here, we detail our bioinformatics pipeline and the generation of figures used to visualize the data obtained from such experiments (Fig. 3). This analysis pipeline is not limited to data gathered from mouse tissue or cells; it can be applied to any polysome profiling experiment performed on a model organism with sufficient annotations and will scale to any number of sequenced fractions. The final output generates a ribosome occupancy score for every transcript isoform, which can then be used to determine shifts in translation between experimental conditions. One can also detect whether alternative transcripts generated from a single gene are moving in similar or opposite directions. The alternative transcripts displaying opposite movement would be interesting, as there might be features on the individual transcript isoforms that are directing their translation differentially and that there might be a biological function for such regulation.
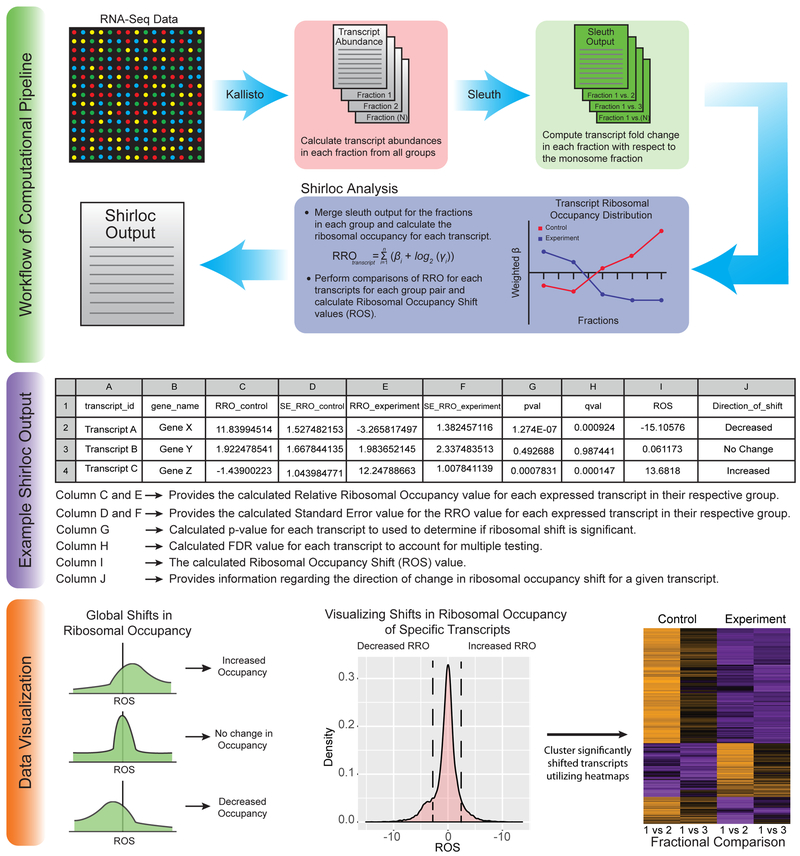
Figure 3. Shirloc bioinformatic analysis pipeline.
Shirloc utilizes Kallisto and Sleuth to determine transcript abundance and calculate fold change of fractions relative to the monosome. A ribosomal occupancy factor is then applied to calculate Ribosomal Occupancy Shift (ROS) values. The final output is given in table form. The data can then be visualized in many different ways, including the examples provided.
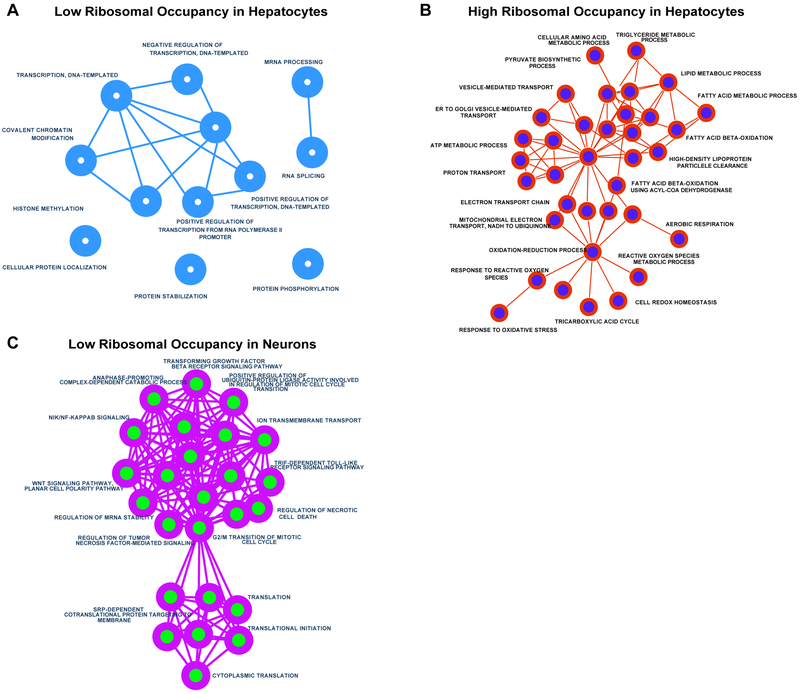
Using the ribosomal occupancy (RO) parameter computed by the Shirloc pipeline, transcripts can be ranked within each sample based on its relative ribosomal occupancy per transcript. This ranking can be used to gain insights into which transcripts have high ribosomal occupancy and vice versa and how these rankings differ between different sample types such as tissues or cells. For example, polysome profiling sequencing dataset from hepatocytes and cultured neurons [26,30] were analyzed using the Shirloc pipeline. Transcripts were then ranked by their RO value and then 1000 transcripts with the lowest and highest RO values were used to generate gene ontology networks for each set (Fig. 4). As expected, the transcripts with the highest occupancy in hepatocytes clustered into processes involved in metabolism and mitochondrial genes. For cultured neurons, transcripts with the least ribosomal occupancy per transcript clustered into processes involved in translation regulation and cell division. The exact scripts used can be found in our package available for download, which is maintained on GitHub (https://github.com/warif2/shirloc).
Figure 4. Gene ontology analysis of cell-type specific ribosome occupancy.
Gene Ontology networks were created using Enrichment Maps plugin for Cytoscape. Networks were generated for 1000 transcripts with the A) lowest relative ribosomal occupancy (RO) in hepatocytes, B) highest RO in hepatocytes and C) lowest RO in cultured neurons. As expected, hepatocytes showed greatest RO for transcripts involved in various metabolic functions. Cultured neurons, on the other hand revealed lowest RO cluster of processes involving translation and cell division. No significant clustering or enrichment was observed for transcripts with high RO scores.
This analysis will require the following interpreters and bioinformatics packages and respective dependencies: Python3, R, Kallisto and Sleuth. These packages were chosen due to their high performance and speed [42,43].
Transcript abundances for each fraction is quantified by processing the FASTQ files generated from each sequenced library using Kallisto.
Using the Kallisto output for each group (e.g., control or experiment), differential transcript analysis is performed between the monosome fraction and each of the higher polysome fractions with the Sleuth program. All replicates should be included into the differential abundance analysis.
For this analysis, the Wald test is performed on the Sleuth model, which computes the beta (β) coefficient for each transcript. This coefficient is a biased estimator of log2 (Fold Change). The comparison is performed between fractional samples within each group.
For each transcript within the group, the Relative Ribosomal Occupancy in regard to the Monosome abundance is calculated using the following equation.
Finally, to calculate the relative Ribosomal Occupancy Shift (ROS) for each transcript between two groups (i.e., control and experiment), the following operation is performed
4. Concluding Remarks
In this article, we present a detailed method for performing cell-type specific and whole tissue polysome profiling. Our analysis pipeline, Shirloc, provides an easy to use tool for analyzing the high-throughput sequencing data generated from polysome profiling sequencing with built in scaling for any number of fractions and determination of ribosome occupancy shifts across experimental conditions. This technique should be valuable for studying translation regulation within a physiologic context and to compare changes in translatome of individual cell-types in response to various environmental and/or developmental signals.
Highlights:
Comparison of different methods to survey translation at a genome-wide scale.
Isolation of tissues and purified cell-types for polysome profiling.
A detailed protocol of polysome fractionation for RNA-sequencing.
Bioinformatics workflow (Shirloc) for determining differential ribosome occupancy of transcripts.
5. Acknowledgements
This work was supported by NIH (R01HL126845) grant to A.K., NIH pre-doctoral NRSA fellowship (F30DK108567) to W.A, and NIH Tissue Microenvironment training program (T32-EB019944) to S.B. J.S. was partly supported by NIH Chemistry-Biology Interface training program (T32-GM070421) and American Heart Association pre-doctoral fellowship (17PRE33670030).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Pascal LE, True LD, Campbell DS, Deutsch EW, Risk M, Coleman IM, et al. , Correlation of mRNA and protein levels: cell type-specific gene expression of cluster designation antigens in the prostate, BMC Genomics. 9 (2008) 246. doi: 10.1186/1471-2164-9-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Maier T, Güell M, Serrano L, Correlation of mRNA and protein in complex biological samples, FEBS Lett 583 (2009) 3966–3973. doi: 10.1016/j.febslet.2009.10.036. [DOI] [PubMed] [Google Scholar]
- [3].Liu Y, Beyer A, Aebersold R, On the Dependency of Cellular Protein Levels on mRNA Abundance, Cell. 165 (2016) 535–550. doi: 10.1016/j.cell.2016.03.014. [DOI] [PubMed] [Google Scholar]
- [4].Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, et al. , Global quantification of mammalian gene expression control, Nature. 473 (2011) 337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- [5].Vogel C, Marcotte EM, Insights into the regulation of protein abundance from proteomic and transcriptomic analyses, Nat. Rev. Genet. 13 (2012) 227–232. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Roy B, Jacobson A, The intimate relationships of mRNA decay and translation, Trends in Genetics. 29 (2013) 691–699. doi: 10.1016/j.tig.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Manning KS, Cooper TA, The roles of RNA processing in translating genotype to phenotype, Nat. Rev. Mol. Cell Biol. 18 (2017) 102–114. doi: 10.1038/nrm.2016.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Zhao BS, Roundtree IA, He C, Post-transcriptional gene regulation by mRNA modifications, Nat. Rev. Mol. Cell Biol. 18 (2017) 31–42. doi: 10.1038/nrm.2016.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Lewis CJT, Pan T, Kalsotra A, RNA modifications and structures cooperate to guide RNA–protein interactions, Nat. Rev. Mol. Cell Biol. 18 (2017) 202–210. doi: 10.1038/nrm.2016.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Hanson G, Coller J, Codon optimality, bias and usage in translation and mRNA decay, Nat. Rev. Mol. Cell Biol. 19 (2017) 20–30. doi: 10.1038/nrm.2017.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Leppek K, Das R, Barna M, Functional 5’ UTR mRNA structures in eukaryotic translation regulation and how to find them, Nat. Rev. Mol. Cell Biol. 19 (2018) 158–174. doi: 10.1038/nrm.2017.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Heck AM, Wilusz J, The Interplay between the RNA Decay and Translation Machinery in Eukaryotes, Cold Spring Harb Perspect Biol. 10 (2018) a032839. doi: 10.1101/cshperspect.a032839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Arthur LL, Djuranovic S, PolyA tracks, polybasic peptides, poly-translational hurdles, Wiley Interdisciplinary Reviews: RNA. 9 (2018) e1486. doi: 10.1002/wrna.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Maslon MM, Heras SR, Bellora N, Eyras E, Caceres JF, The translational landscape of the splicing factor SRSF1 and its role in mitosis, Elife. 3 (2014) e02028. doi: 10.7554/eLife.02028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ingolia NT, Ghaemmaghami S, Newman JRS, Weissman JS, Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling, Science. 324 (2009) 218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ingolia NT, Ribosome profiling: new views of translation, from single codons to genome scale, Nat. Rev. Genet. 15 (2014) 205–213. doi: 10.1038/nrg3645. [DOI] [PubMed] [Google Scholar]
- [17].Weatheritt RJ, Sterne-Weiler T, Blencowe BJ, The ribosome-engaged landscape of alternative splicing, Nat. Struct. Mol. Biol. 23 (2016) 1117–1123. doi: 10.1038/nsmb.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Heiman M, Schaefer A, Gong S, Peterson JD, Day M, Ramsey KE, et al. , A translational profiling approach for the molecular characterization of CNS cell types, Cell. 135 (2008) 738–748. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Heiman M, Kulicke R, Fenster RJ, Greengard P, Heintz N, Cell type-specific mRNA purification by translating ribosome affinity purification (TRAP), Nature Protocols. 9 (2014) 1282–1291. doi: 10.1038/nprot.2014.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Sanz E, Yang L, Su T, Morris DR, McKnight GS, Amieux PS, Cell-type-specific isolation of ribosome-associated mRNA from complex tissues, Proc. Natl. Acad. Sci. U.S.a. 106 (2009) 13939–13944. doi: 10.1073/pnas.0907143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Nectow AR, Moya MV, Ekstrand MI, Mousa A, McGuire KL, Sferrazza CE, et al. , Rapid Molecular Profiling of Defined Cell Types Using Viral TRAP, Cell Rep. 19 (2017) 655–667. doi: 10.1016/j.celrep.2017.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wang AW, Wangensteen KJ, Wang YJ, Zahm AM, Moss NG, Erez N, et al. , TRAP-seq identifies cystine/glutamate antiporter as a driver of recovery from liver injury, J. Clin. Invest. 128 (2018) 2297–2309. doi: 10.1172/JCI95120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Dougherty JD, The Expanding Toolkit of Translating Ribosome Affinity Purification, J. Neurosci. 37 (2017) 12079–12087. doi: 10.1523/JNEUROSCI.1929-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Sterne-Weiler T, Martinez-Nunez RT, Howard JM, Cvitovik I, Katzman S, Tariq MA, et al. , Frac-seq reveals isoform-specific recruitment to polyribosomes, Genome Res. 23 (2013) 1615–1623. doi: 10.1101/gr.148585.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Floor SN, Doudna JA, Tunable protein synthesis by transcript isoforms in human cells, Elife. 5 (2016) 1276. doi: 10.7554/eLife.10921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Blair JD, Hockemeyer D, Doudna JA, Bateup HS, Floor SN, Widespread Translational Remodeling during Human Neuronal Differentiation, Cell Rep. 21 (2017) 2005–2016. doi: 10.1016/j.celrep.2017.10.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lianoglou S, Garg V, Yang JL, Leslie CS, Mayr C, Ubiquitously transcribed genes use alternative polyadenylation to achieve tissue-specific expression, Genes Dev. 27 (2013) 2380–2396. doi: 10.1101/gad.229328.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Buszczak M, Signer RAJ, Morrison SJ, Cellular differences in protein synthesis regulate tissue homeostasis, Cell. 159 (2014) 242–251. doi: 10.1016/j.cell.2014.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Castelo-Szekely V, Arpat AB, Janich P, Gatfield D, Translational contributions to tissue specificity in rhythmic and constitutive gene expression, Genome Biol. 18 (2017) 116. doi: 10.1186/s13059-017-1222-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Bangru S, Arif W, Seimetz J, Bhate A, Chen J, Rashan EH, et al. , Alternative splicing rewires Hippo signaling pathway in hepatocytes to promote liver regeneration, Nat. Struct. Mol. Biol. 25 (2018) 928–939. doi: 10.1038/s41594-018-0129-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Chorghade S, Seimetz J, Emmons R, Yang J, Bresson SM, Lisio MD, et al. , Poly(A) tail length regulates PABPC1 expression to tune translation in the heart, Elife. 6 (2017) 568. doi: 10.7554/eLife.24139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, et al. , N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency, Cell. 161 (2015) 1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, et al. , 5′ UTR m6A Promotes Cap-Independent Translation, Cell. 163 (2015) 999–1010. doi: 10.1016/j.cell.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].He SL, Green R, Polysome analysis of mammalian cells, Meth. Enzymol. 530 (2013) 183–192. doi: 10.1016/B978-0-12-420037-1.00010-5. [DOI] [PubMed] [Google Scholar]
- [35].Gandin V, Sikström K, Alain T, Morita M, McLaughlan S, Larsson O, et al. , Polysome fractionation and analysis of mammalian translatomes on a genome-wide scale, J Vis Exp. (2014). doi: 10.3791/51455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Obrig TG, Culp WJ, McKeehan WL, Hardesty B, The mechanism by which cycloheximide and related glutarimide antibiotics inhibit peptide synthesis on reticulocyte ribosomes, J. Biol. Chem. 246 (1971) 174–181. [PubMed] [Google Scholar]
- [37].Schneider-Poetsch T, Ju J, Eyler DE, Dang Y, Bhat S, Merrick WC, et al. , Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin, Nat. Chem. Biol. 6 (2010) 209–217. doi: 10.1038/nchembio.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Young SK, Willy JA, Wu C, Sachs MS, Wek RC, Ribosome Reinitiation Directs Gene-specific Translation and Regulates the Integrated Stress Response, J. Biol. Chem. 290 (2015) 28257–28271. doi: 10.1074/jbc.M115.693184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Li W-C, Ralphs KL, Tosh D, Isolation and culture of adult mouse hepatocytes, Methods Mol. Biol. 633 (2010) 185–196. doi: 10.1007/978-1-59745-019-5_13. [DOI] [PubMed] [Google Scholar]
- [40].Louch WE, Sheehan KA, Wolska BM, Methods in cardiomyocyte isolation, culture, and gene transfer, J. Mol. Cell. Cardiol. 51 (2011) 288–298. doi: 10.1016/j.yjmcc.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Liu DC, Seimetz J, Lee KY, Kalsotra A, Chung HJ, Lu H, et al. , Mdm2 mediates FMRP- and Gp1 mGluR-dependent Protein Translation and Neural Network Activity, Hum. Mol. Genet. 26 (2017) 3895–3908. doi: 10.1093/hmg/ddx276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Bray NL, Pimentel H, Melsted P, Pachter L, Near-optimal probabilistic RNA-seq quantification, Nature Biotechnology. 34 (2016) 525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- [43].Pimentel H, Bray NL, Puente S, Melsted P, Pachter L, Differential analysis of RNA-seq incorporating quantification uncertainty, Nat. Methods. 14 (2017) 687–690. doi: 10.1038/nmeth.4324. [DOI] [PubMed] [Google Scholar]